Abstract
Purpose
Sarcomatoid renal cell carcinoma (SRCC) ranks among the most aggressive clinicopathologic phenotypes of RCC. However, the paucity of high-quality, genome-wide molecular examinations of SRCC has hindered our understanding of this entity.
Experimental Design
We interrogated the mutational, copy number, and transcriptional characteristics of SRCC and compared these data with those of non-sarcomatoid RCC (RCC). We evaluated whole exome sequencing, single nucleotide polymorphism, and RNA sequencing data from patients with SRCC (n=65) and RCC (n=598) across different parent RCC subtypes, including clear cell RCC, papillary RCC, and chromophobe RCC subtypes.
Results
SRCC was molecularly discrete from RCC and clustered according to its parent RCC subtype, though with upregulation of TGFβ signaling across all subtypes. The epithelioid (E-) and spindled (S-) histologic components of SRCC did not show differences in mutational load among cancer related genes, despite a higher mutational burden in S-. Notably, sarcomatoid clear cell RCC (SccRCC) showed significantly fewer deletions at 3p21–25, a lower rate of two-hit loss for VHL and PBRM1, and more mutations in PTEN, TP53, and RELN compared to clear cell RCC (ccRCC). A two-hit loss involving VHL predicted for ccRCC and a better prognosis whereas mutations in PTEN, TP53, or RELN predicted for SccRCC and worse prognosis.
Conclusions
Sarcomatoid RCC segregates by parent subtype and SccRCC has a fundamentally different early molecular pathogenesis, usually lacking the classic 3p21–25 deletion and showing distinctive mutational and transcriptional profiles. These features prompt a more precise molecular classification of RCC with diagnostic, prognostic, and therapeutic implications.
Keywords: sarcomatoid, renal, carcinoma, sequencing
INTRODUCTION
Renal cell carcinoma (RCC) is an important contributor to cancer-specific mortality (1). Sarcomatoid RCC (SRCC), which is characterized by a biphasic epithelioid carcinomatous (E-) component and a spindle cell sarcomatous (S-) component, represents an important and understudied subset of RCC. SRCC comprises up to 20% of stage IV RCC cases and shows aggressive behavior, irrespective of the parent subtype with which it is shares histological characteristics (2–4). There are no established therapies that are effective against SRCC (5). Current clinical thinking conceptualizes SRCC as a single entity (6); however, a thorough molecular understanding of the disease has been limited by the dearth of genome-wide examinations, both within specific RCC subtypes and across different parent RCC subtypes.
Large-scale multi-institutional, multiplatform genomic studies of RCC have been reported, notably The Cancer Genome Atlas (TCGA) data on clear cell RCC (ccRCC), papillary RCC (PapRCC), and chromophobe RCC (ChRCC) (7–10). Genome wide molecular characterization of SRCC has heretofore been based on formalin-fixed, paraffin-embedded (FFPE) tissues and largely confined to the sarcomatoid ccRCC subtype (SccRCC) (11–14).
The aim of this study was to comprehensively examine the mutational, copy number, and transcriptional characteristics of SRCC and to compare these data with those of non-sarcomatoid RCC (RCC). We found that SRCC is not a single entity but is rather highly influenced by the parent RCC subtype with which it shares histological characteristics. Moreover, we show that SccRCC exhibits a different early molecular pathogenesis, driver mutation, and transcriptional profile than ccRCC. These findings will improve the molecular classification of RCC and provide a means for predicting aggressive sarcomatoid tumors preoperatively as well as suggesting therapeutic targets, thus aiding in the management of this disease.
MATERIALS AND METHODS
Patient and tumor characteristics
For this retrospective study, we obtained 40 frozen SRCC samples and non-neoplastic kidney control samples from the tissue bank of The University of Texas MD Anderson Cancer Center (Houston, TX) after informed consent and using an institutional review board-approved protocol (IRB# LAB 08-670). The study was conducted in accordance with the Declaration of Helsinki. Lesional foci representing the E- or S- component of SRCC as well as non-neoplastic kidney controls were marked on H&E-stained slides, with all cases reviewed by at least two genitourinary pathologists (Figure S1). These foci were macrodissected from the frozen samples prior to the extraction of tissue analytes. We identified a second cohort of SRCC cases (n=32) by examining surgical pathology reports and digital pathologic images from TCGA. The clinicopathologic features of the SRCCs derived from the MD Anderson and TCGA cohorts are summarized in Table S1.
Whole exome sequencing and analysis
Genomic DNA was extracted using the AllPrep DNA/RNA Mini kit according to the manufacturer's instructions (Qiagen). DNA concentration was quantified using a NanoDrop 1000 spectrophotometer (NanoDrop Technologies) and the Quant-iT Picogreen kit (Life Technologies) individually. Paired-end whole exome sequencing (WEX) was performed on frozen sarcomatoid renal tumors and corresponding normal controls using the HiSeq2000 platform. DNA mutations and copy numbers were assessed from WEX data using in-house pipelines. Data were analyzed within and across sample subtypes with details provided in Supplemental Methods 1.
RNA-seq expression profiling and analysis
Total RNA was isolated using the miRNeasy Mini kit according to the manufacturer’s instructions (Qiagen). RNA was quantitatively assessed using the NanoDrop 1000 spectrophotometer (Nanodrop Technologies) and qualitatively assessed using the Agilent 2100 bioanalyzer (Agilent Technologies).
RNA seq was performed on frozen and FFPE tissues with paired-end sequencing using the HiSeq2000 platform. Gene expression was assessed using an in-house pipeline that uses tophat (15) and htseq (16), among other tools. The details are provided in Supplemental Methods 2.
DNA methylation profiling and analysis
DNA methylation profiling was performed from genomic DNA extracted from frozen sarcomatoid renal tumors and corresponding normal kidney controls using the Illumina Infinium Methyl EPIC Beadchip platform. The details are provided in Supplemental Methods 3. Assessment of gene silencing by DNA methylation was carried out as previously described (17).
Fluorescence In Situ Hybridization
Fluorescence in situ hybridization (FISH) using a commercial probe set (VHL/CEN3q - Abnova, Catalog #: FG0029) was performed on FFPE derived tissue microarray sections from two patient cohorts. The first cohort comprised patients with sarcomatoid ccRCC: n=88 patients, median age at surgery was 59 with 83% dead of disease and median follow-up of 11 months. The second cohort comprised patients with ccRCC lacking sarcomatoid features: n=66 patients (Fuhrman grade 3, n=57; Fuhrman grade 2, n=9), median age at surgery was 58 with 23% dead of disease and median follow-up of 70 months. The detailed FISH methodology is provided in Supplemental Methods 4.
In silico analysis of The Cancer Genome Atlas data
The TCGA exome sequencing and RNA seq data derived from SRCC and non-sarcomatoid RCC cases was evaluated using the Broad Institute fire-hose pipeline. TCGA data downloaded from the TCGA data coordinating center (DCC) site were also used to compare SccRCC with non-sarcomatoid ccRCC cases in terms of gene-level copy number, arm level chromosomal copy number, exome-based somatic mutations, and VHL methylation status.
Statistical analysis
For RNA-seq data differentially expressed gene analysis, we used ‘edgeR’ package (18) in R to calculate the fold change and to adjust the p-value. We analyzed canonical pathways and functional networks with Ingenuity Pathways Analysis (Ingenuity Systems, Mountain View, CA). Kaplan-Meier survival analysis was used with the log-rank test to assess the statistical significance of the differences between stratified survival groups using the ‘Survival’ package in R. Other standard statistical tests were used to analyze the clinical and genomics data, including the Chi-square test, Fisher’s exact test, Mann-Whitney-Wilcoxon test and Cox proportional hazard analysis. Significance was defined as P < 0.05. Analyses were primarily performed using R. Heat maps were plotted based on the pheatmap function in pheatmap package.
RESULTS
Sarcomatoid RCC segregates molecularly according to underlying parent subtype
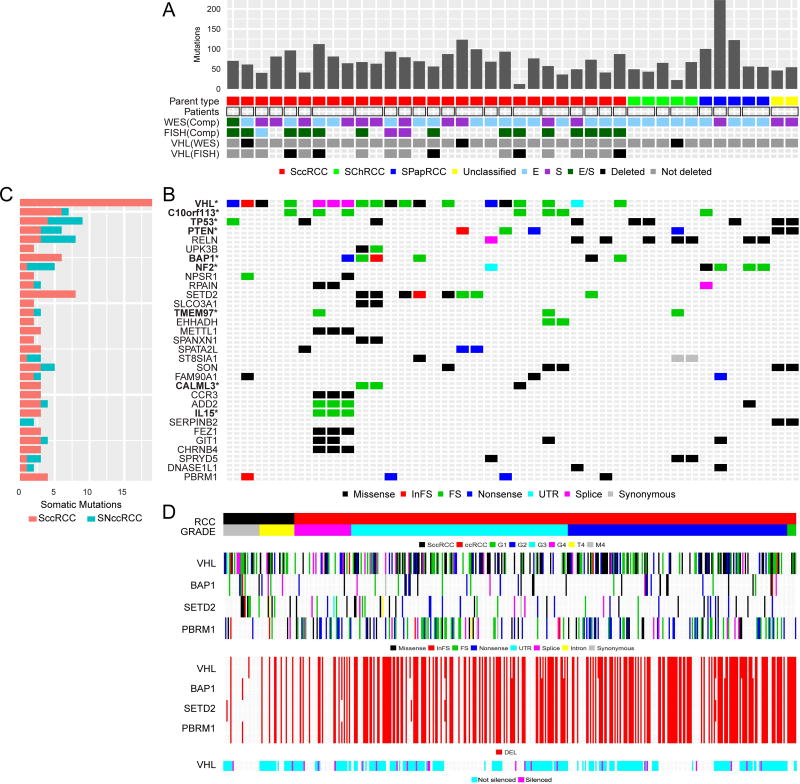
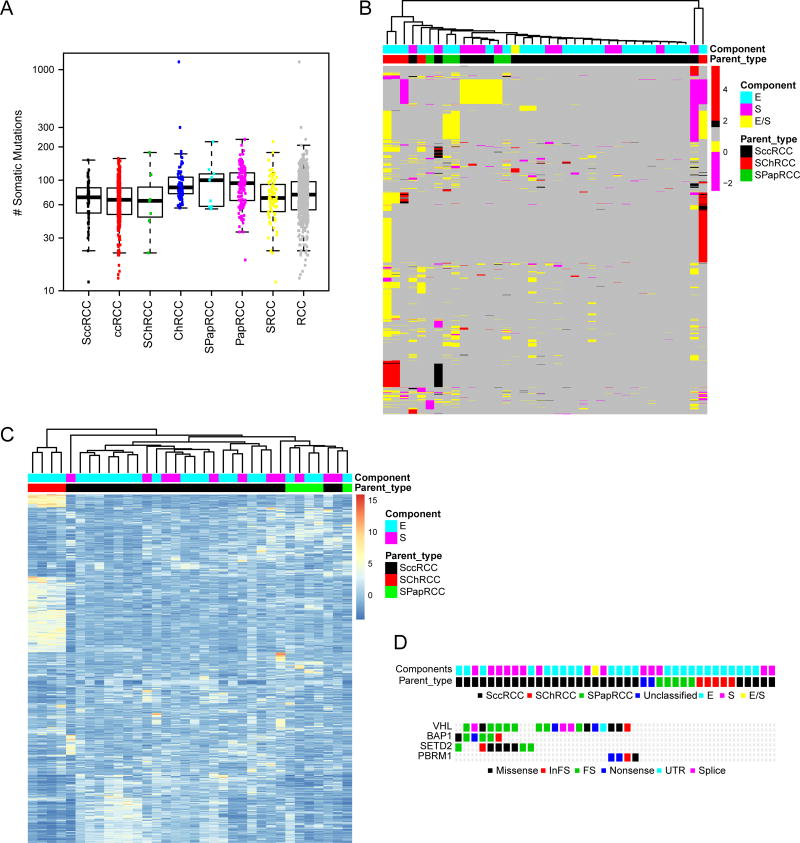
We examined SRCC derived from three different parent subtypes (sarcomatoid clear cell RCC or SccRCC; sarcomatoid chromophobe RCC or SChRCC; and sarcomatoid papillary RCC or SPapRCC) and their biphasic E- and S- morphological components. The overall mutational landscape of SRCC found in our cohort is schematically represented in Figure 1a. Table S2 summarizes the mutations found in our cohort; detailed sequencing data are in the process of being deposited in database of Genotypes and Phenotypes (dbGaP, accession numbers pending)..” Using both local and TCGA data, we found sarcomatoid RCC of different subtypes to show a mutational burden similar to that of RCC (7–9) (Figure 2a). Unsupervised clustering of copy number and transcriptional data showed that SRCC segregated according to the parent subtype rather than to the E- or S- morphologic components, as illustrated in Figures 2b–c. Potentially actionable mutations, involving pan-cancer T200 panel genes (19), were distributed across all SRCC subtypes (Table S2). However, mutations in 3p21–25 genes (VHL, PBRM1, SETD2, and BAP1) were only seen in SccRCC (Figure 2d).
Figure 1. Mutational, copy number and methylation landscape of sarcomatoid RCC.
(A) Mutation frequency sorted by parent RCC subtype (SccRCC, SPapRCC, SChRCC, and Unclassified); unique patient samples indicated by square borders with paired patient samples indicated by rectangular borders; E- and/or S- components from each patient assayed by whole exome sequencing (WES) or Fluorescence in situ hybridization (FISH) with VHL deletions assessed by WES and FISH. (B) Types of somatic mutations involving specific genes, including significantly mutated genes by mutation frequency and found by MutSig*. (C) Bar plot shows mutations grouped by sarcomatoid clear cell (SccRCC) or sarcomatoid non-clear cell RCC (SNccRCC) subtype. (D) Sarcomatoid clear cell RCC derived from MD Anderson (M4) and TCGA (T4) cohorts and clear cell RCC (Grades 1–4) from TCGA in terms of the “hits” on 3p genes (VHL, PBRM1, SETD2, BAP1) consisting of mutations, deletions (DEL) and epigenetic silencing by methylation.
Figure 2. Sarcomatoid RCC segregates according to parent subtype.
Sarcomatoid RCC (SRCC) of different parent subtypes (SccRCC, SPapRCC, and SChRCC) demonstrates (A) similar mutational burdens compared to non-sarcomatoid RCC (RCC) of different parent subtypes (ccRCC, PapRCC, and ChRCC). SRCCs segregate according to parent subtype, as shown by an unsupervised clustering analysis of (B) copy number, 1500 top genes; and (C) mRNA expression, 1000 genes. (D) Mutations of 3p21–25 tumor suppressor genes are confined to SccRCC.
Subsets of genes are mutated across different parent subtypes of SRCC
Despite the large number of non-synonymous mutations that were found in SRCC, relatively few genes were recurrently mutated. Indeed, among the top 30 recurrently mutated genes, only nine were significant in SRCC according to the MutSig algorithm (q< 0.1, Figure 1b and Table S3a). Some gene mutations were shared across different subtypes of SRCC, most notably TP53, PTEN, NF2, and RELN (Figure 1c).
We next assessed whether mutations in the aforementioned genes were significantly enriched in sarcomatoid tumors compared to non-sarcomatoid tumors within the same parent RCC subtype using institutional and TCGA derived data (Table S3b). PTEN was more frequently mutated in SccRCC versus ccRCC; TP53 mutations were associated with SccRCC and SPapRCC; and NF2 mutations were associated with SPapRCC only. RELN mutations were significantly higher in SRCC across all subtypes.
Sarcomatoid clear cell RCC shows a different molecular pathogenesis compared to lower grade clear cell RCC
Biallelic loss of tumor suppressor genes at 3p21–25 is fundamental to the pathogenesis of ccRCC (20,21). Loss of VHL is thought to be the initiating event, followed by inactivation of other 3p21–25 genes, such as PBRM1 and BAP1 (22). Numerous additional chromosomal copy number alterations have been proposed as contributing to the progression of ccRCC (23). In terms of arm-spanning copy number changes, fewer chromosomal losses at 3p were associated with high-grade and SccRCC (Table S4). Differences in gene level copy number were more dramatic, with significantly less segmental 3p21–25 deletion seen in high-grade and SccRCC spanning the VHL, PBRM1, BAP1, and SETD2 genes (Table S5).
This finding of markedly less 3p21–25 deletions in SccRCC was maintained across a broad range of bioinformatics thresholds used for calling copy loss. We next considered whether a greater burden of contaminating normal diploid cells may have confounded our results. However, the distribution of tumor purities did not differ significantly between high grade (SccRCC + G4) versus lower grades (G1–3) of ccRCC (P=0.28). We then used the Fluorescence in situ hybridization (FISH) platform to assay sarcomatoid ccRCC and ccRCC for VHL copy number. We found that sarcomatoid features correlated with significantly higher VHL/3q ratios as shown in Figure S2. Using FISH as the basis for copy number calls, SccRCC showed fewer VHL deletions compared to ccRCC (P= 0.001) and high grade ccRCC showed fewer VHL deletions compared to lower grade ccRCC tumors (P=0.000049).
Finally, we considered whether copy neutral loss of heterozygosity (LOH) contributed to LOH at 3p in those cases that did not show simple 3p21–25 loss. Our analysis of SNP data from TCGA showed copy neutral LOH to be an exceedingly rare event in ccRCC (~1%) and to involve only 1 case of SccRCC (Supplemental Methods 5 and Figures S3–S7). We assessed independent ccRCC samples assayed by SNP array (GSE12808, GSE14670, GSE11447 and GSE9469) and also found a low incidence of copy neutral LOH (~2%) that was not correlated to tumor grade.
SccRCC exhibited less concurrent mutation and copy loss (i.e., two-hit loss) of the VHL and PBRM1 genes than did ccRCC across different histological grades (Table 1). Gene silencing by methylation is another mechanism of gene inactivation. Epigenetic silencing has been reported to be mutually exclusive with mutation for VHL in ccRCC (7,24), consistent with our data where VHL mutations and methylation did not overlap (Figure 1d). We only found methylation of the VHL gene and not for the PBRM1, SETD2, or BAP1 genes (Figure S8). Notably, after taking methylation status into account as well as mutation and loss of heterozygosity, SccRCC still showed fewer two-hit losses of VHL compared to ccRCC (P<0.001; OR: 0.22).
Table 1.
Fewer concurrent loss of heterozygosity and mutations of the VHL and PBRM1 genes in sarcomatoid ccRCC than in ccRCC, by P value (odds ratio)
| 3p 21–25 gene | ||||
|---|---|---|---|---|
|
|
||||
| Test | VHL | BAP1 | SETD2 | PBRM1 |
| SccRCC vs grade 1 ccRCC | 3.97E-02 (0.13) | 1 (1) | 1 (1) | 1.93E-03 (0.04) |
| SccRCC vs grade 2 ccRCC | 2.7E-06 (0.14) | 1 (1.03) | 2.11E-01 (0.43) | 1.53E-05 (0.11) |
| SccRCC vs grade 3 ccRCC | 1.49E-06 (0.13) | 1 (1.01) | 5.72E-01 (0.63) | 1.66E-04 (0.14) |
| SccRCC vs grade 4 ccRCC | 5.25E-02 (0.3) | 6.97E-01 (0.58) | 1 (1.21) | 7.99E-03 (0.17) |
| SccRCC vs all ccRCC | 1.79E-06 (0.15) | 1 (0.96) | 4.52E-01 (0.56) | 1.79E-05 (0.13) |
Mutational profile of clear cell RCC is predictive of sarcomatoid change and poor prognosis
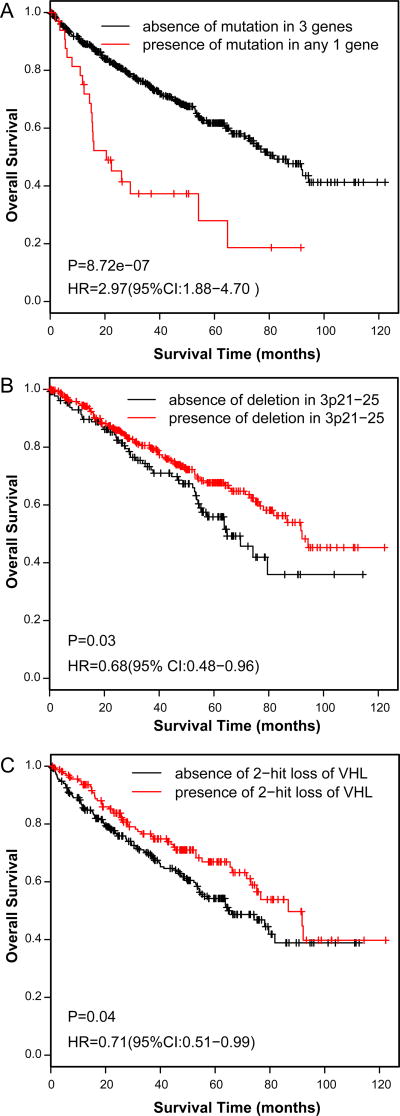
With respect to ccRCC, we found that mutations in TP53, PTEN, RELN, BAP1 and SETD2 were individually associated with or anti-correlated (PBRM1) with sarcomatoid change (Table 2). A mutation in TP53, PTEN, or RELN was correlated with sarcomatoid change in ccRCC (P=1.64E-06, OR=6.53), including grade 4 (P=4.68E-02, OR=3.2) (Table S6). Further, mutant allelic frequencies in TP53, PTEN, and RELN were lower than in VHL or BAP1 (P=0.035), suggesting they occurred later in the evolution of the tumor. When comparing the mutational landscape of SccRCC versus ccRCC, profiling of only the E- component of SccRCC revealed similar results compared to profiling of both E- and S- components (Table 2 and Table S6). A survival analysis showed that a mutation in TP53, PTEN, or RELN was associated with reduced overall survival in ccRCC (Figure 3a, P=8.72E-07). By contrast, deletion of 3p21–25 genes or a two-hit loss of VHL was associated with increased overall survival (Figures 3b and 3c).
Table 2.
Mutations in sarcomatoid ccRCC versus ccRCC across different Fuhrman grades, by P value (Odds Ratio)
| Both S- and E-components of sarcomatoid ccRCC vs. ccRCC | ||||||
|
| ||||||
| Gene | Grade 1 | Grade 2 | Grade 3 | Grade 4 | All ccRCC | Grades 1–3* |
|
| ||||||
| VHL | 0.691(1.72) | 0.757(1.12) | 0.877(1.07) | 0.316(1.55) | 0.667(1.15) | 0.732(0.91) |
| BAP1 | 0.586(3.46) | 0.033(2.93) | 0.082(2.27) | 0.464(0.67) | 0.066(2.05) | 0.0004(3.17) |
| SETD2 | 0.328(4.76) | 0.085(2.08) | 0.044(2.43) | 0.305(1.96) | 0.032(2.25) | 0.068(1.76) |
| PBRM1 | 0.022(0.13) | 0.004(0.32) | 0.01(0.35) | 0.005(0.25) | 0.002(0.32) | 0.146(0.69) |
| PTEN | 1(1.97) | 0.015(5.08) | 0.115(2.45) | 0.727(1.67) | 0.034(3.05) | 0.03(2.78) |
| NF2 | 1(0.41) | 0.244(9.39) | 1(1.02) | 1(0.8) | 0.485(1.78) | 0.31(2.35) |
| RELN | 0.462(0.47) | 0.003(29.8) | 0.014(13.1) | 0.378(3.37) | 0.006(10.1) | 0.007(9.12) |
| TP53 | 1(2.32) | 0.002(8.12) | 0.001(12.1) | 0.291(3.06) | 0.001(7.96) | 0.001(6.8) |
| UPK3B | 0.306(0.23) | 0.059(15.9) | 0.151(6.3) | 0.501(4.16) | 0.077(7.3) | 0.216(3.54) |
| HIF1A | 1(0.7) | 0.149(6.38) | 0.06(15.75) | 1(0.79) | 0.118(4.86) | 0.01(14.48) |
| PTPRD | 1(0.41) | 0.571(1.56) | 0.247(9.28) | 1(2.45) | 0.328(3.58) | 0.532(1.75) |
|
| ||||||
| Only E-component of sarcomatoid ccRCC vs. ccRCC | ||||||
|
| ||||||
| Gene | Grade 1 | Grade 2 | Grade 3 | Grade 4 | All ccRCC | Grades 1–3* |
|
| ||||||
| VHL | 1(1.4) | 0.865(0.91) | 0.733(0.87) | 0.67(1.26) | 0.873(0.93) | 0.4(0.8) |
| BAP1 | 0.573(3.08) | 0.072(2.56) | 0.168(1.98) | 0.429(0.58) | 0.189(1.8) | 0.001(3.07) |
| SETD2 | 0.58(3.59) | 0.334(1.54) | 0.207(1.79) | 0.572(1.45) | 0.229(1.66) | 0.285(1.42) |
| PBRM1 | 0.048(0.17) | 0.045(0.43) | 0.092(0.47) | 0.034(0.33) | 0.04(0.43) | 0.525(0.82) |
| PTEN | 1(2.14) | 0.018(5.46) | 0.146(2.63) | 0.484(1.8) | 0.039(3.28) | 0.04(2.81) |
| NF2 | 1(0.53) | 0.202(12.0) | 1(1.31) | 1(1.02) | 0.41(2.29) | 0.265(2.68) |
| RELN | 0.464(0.45) | 0.008(29.5) | 0.027(12.5) | 0.36(3.23) | 0.015(9.65) | 0.017(8.27) |
| TP53 | 1(1.3) | 0.098(4.17) | 0.059(6.22) | 0.676(1.57) | 0.067(4.09) | 0.032(4.15) |
| UPK3B | 0.14(0.05) | 1(3.92) | 1(1.28) | 1(1.02) | 1(1.78) | 1(0.79) |
| HIF1A | 1(0.9) | 0.104(8.24) | 0.041(20.3) | 1(1.02) | 0.08(6.28) | 0.007(16.6) |
| PTPRD | 1(0.53) | 0.493(2) | 0.204(11.9) | 0.494(3.14) | 0.27(4.61) | 0.492(1.99) |
Sarcomatoid and grade 4 ccRCC vs grade 1–3 ccRCC.
Figure 3. Mutational and copy number profiles associated with sarcomatoid ccRCC are prognostic.
A mutation in PTEN, RELN, or TP53 is associated with significantly lower overall survival (A), whereas deletion of 3p21–25 (B) or a two-hit loss of VHL (C) was associated with increased overall survival.
Sarcomatoid RCC shows a distinct transcriptional program
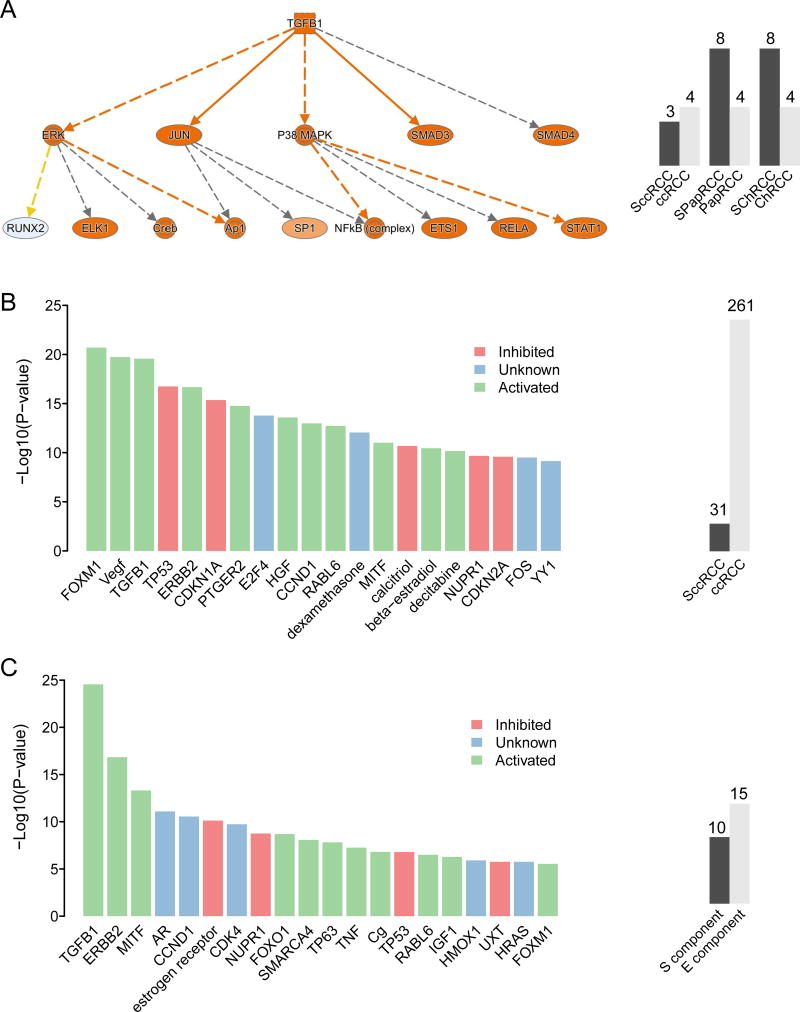
Since SRCC practically always presents as advanced-stage disease, we compared the transcriptional profile of sarcomatoid tumors with that of advanced-stage non-sarcomatoid tumors across all three RCC subtypes. SRCC showed enrichment of several pathways, including TGFβ1 signaling (activated, P=5.67E-25) (Figure 4a and Table S7a).
Figure 4. Sarcomatoid RCC shows a distinct transcriptional program.
A. Sarcomatoid RCC shows enrichment of TGFβ1 signaling across different parent subtypes. B. Sarcomatoid ccRCC shows differential regulation of various signaling pathways compared to advanced-stage (stages III and IV) non-sarcomatoid ccRCC. C. Expression differences between the E- and S- components of sarcomatoid ccRCC translate into discrete pathway alterations. Barplots indicate number of cases analyzed across each contrast.
We next assessed only SccRCC versus advanced-stage ccRCC: using TCGA data, we found a large number of differentially expressed genes (n=2362 genes, FDR<0.1) that translated into differential regulation of numerous signaling cascades, as illustrated in Figure 4b and Table S7b. Notably, alteration of the VEGF (activated, P=1.8E-20), TGFβ1 (activated, P=2.7E-20), and TP53 (inhibited, P=1.8E-17) pathways was seen in SccRCC. At the level of cellular function, SccRCC showed enrichment of cell growth, movement, and proliferation processes. We performed the same analysis using expression profiling data from local samples (GSE59266) derived from FFPE tissues and found approximately 600 genes that overlapped between the two independent datasets. A pathway analysis based on the overlapping genes revealed enrichment of similar cellular functions and pathways (Figure S9).
Biphasic E- and S- components of sarcomatoid RCC show regional genetic differences
Multiple regions of a given tumor were sequenced in a subset of SRCC cases encompassing different RCC subtypes and different histologic components (E- or S-), as shown in Figure 1a and Table S8. The S- component showed a significantly higher overall mutational load than did the E- component across all RCC subtypes (P=0.0215) and in the ccRCC subtype (P=0.0199) (Figure S10a and S10b). The mutational load among cancer related genes (25), however, did not show significant differences between E-and S-. Moreover, despite the biphasic components harboring private mutations that were unique to that region, no single gene mutation was significantly enriched in E- or S- regions. Interestingly, mutations in cancer related genes were more commonly shared among the E- and S- components as compared to all gene mutations: eg. 66% shared cancer related mutations in SRCC vs 19% shared total mutations in RCC, P=1.15 E-09; and 62% shared cancer related mutations in SccRCC vs 19% shared total mutations in ccRCC, P= 3.57 E-05. An Ingenuity pathway analysis of the mRNA expression profile of paired E- and S- components of SccRCC revealed activation of TGFβ1 signaling (P=2.85E-25) as the top upstream regulator (Figure 4c and Table S7c). Network functions that were altered included those involved in cellular assembly and organization as well as cell movement. Together these pathways provide potential targets for future clinical trials.
DISCUSSION
SRCC is often considered and managed as a single clinical entity, regardless of the underlying parent subtype with which it is associated. For instance, the National Comprehensive Cancer Network guidelines do not distinguish between sarcomatoid tumors of clear cell or non-clear cell origin in terms of recommendations for management (6). Molecular characterization of SRCC has generally been limited to the clear cell subtype using targeted single platform studies (13,14,26,27). Our multiplatform, genome wide analysis shows subtype specific differences and that SRCC clusters according to parent subtype. In support of our observations, Tickoo et al. demonstrated upregulation of hypoxia inducible factor (HIF) pathway markers in only the clear cell subtype of SRCC (28). These results suggest that SRCC is not a single disease entity but rather that its molecular biology reflects its parent subtype, which should be taken into account when managing this disease.
The signal molecular alteration in ccRCC is loss of VHL on 3p (through deletion, mutation, or methylation), leading to activation of HIF pathway molecules such as VEGF and CAIX (22). This has led to the implementation of targeted anti-angiogenic agents that abrogate HIF signaling with improvements in progression free survival (29). Other 3p genes involved in chromatin remodeling (PBRM1, SETD2, and BAP1) were subsequently described as important tumor suppressors that are lost during the pathogenesis of ccRCC (22). The relationship between VHL’s mutational status and tumor progression is controversial (30–33) and we did not find a mutation in VHL to correlate with sarcomatoid features or with aggressive disease (Table 2). However, sarcomatoid ccRCC differs from non-sarcomatoid ccRCC molecularly in that there is significantly less 3p21–25 deletion spanning the VHL, PBRM1, BAP1, and SETD2 genes and less two-copy loss of VHL and PBRM1. This is consistent with historical cytogenetic studies that have shown the presence of 3p deletions to be associated with lower grade and lower stage disease, absence of sarcomatoid features (34) and with improved survival outcomes in ccRCC (34–36). More recent molecular evidence also supports our findings in that loss of heterozygosity at the VHL locus (37) and two-hit loss of the VHL gene in ccRCC (24) were associated with lower stage, lower grade disease and with absence of sarcomatoid features. Regulation of PBRM1 is postulated to be influenced by tumor differentiation, with fewer mutations in higher grade and more aggressive tumors (12,38,39). By contrast, BAP1 and SETD2 have been reported to show more mutations with disease progression (7,12,40). Taken together, it appears that high-grade ccRCC tumors—exemplified by SccRCC—are prone to not show loss of heterozygosity at 3p21–25 and, notably, to retain a copy of the wild-type VHL and PBRM1 genes. Our analyses challenge the dogma that all ccRCC tumors show a 3p LOH/deletion event (22,41). Rather, it appears that SccRCC tumor clones show a distinct early molecular pathogenesis, as illustrated in Figure 5. This suggests that while SccRCC and ccRCC may originate from a common precursor, the molecular events are different, and SccRCC does not evolve linearly from low grade ccRCC. Consequently, diagnostic molecular assays that fail to demonstrate a 3p or VHL deletion in a suspected ccRCC case do not necessarily rule out a clear cell subtype of RCC, particularly when associated with higher grade or sarcomatoid features.
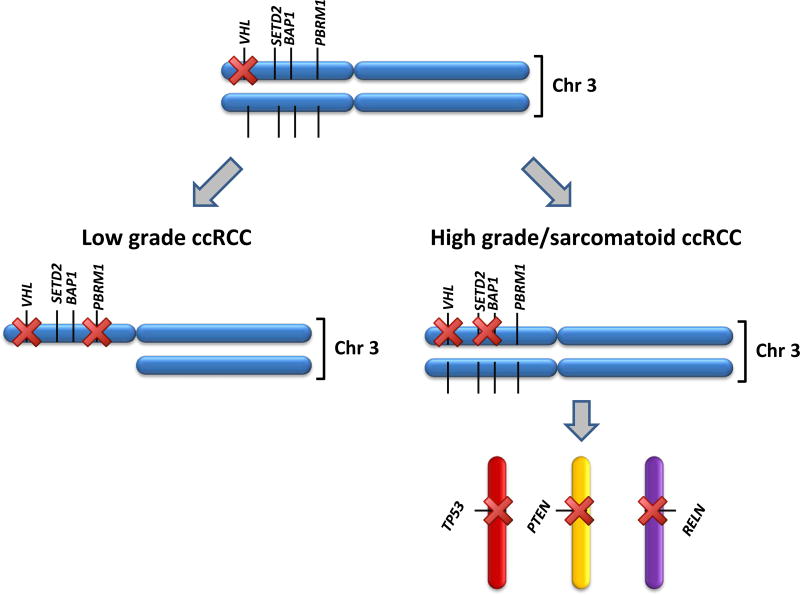
Figure 5. High-grade SccRCC and low-grade ccRCC show a distinct molecular pathogenesis.
Schematic illustration of the proposed sequence of events in low-grade ccRCC versus high-grade SccRCC.
Whereas mutations in 3p genes were confined to the ccRCC subtype, a handful of other recurrent mutations were shared by SRCC of different subtypes, namely, PTEN, TP53, NF2, and RELN. Although mutations in these genes occurred in all SRCC subtypes, their specific association with sarcomatoid change within a given subtype varied according to the gene. Among ccRCC, a mutation in PTEN, TP53, or RELN was predictive of sarcomatoid histological type, whereas a two-hit inactivation of VHL or PBRM1 was predictive of non-sarcomatoid ccRCC histologic type (Figure 5). Knowing whether a patient with advanced disease has sarcomatoid features is relevant because of the frequent explosive growth of sarcomatoid tumors that dissuades clinicians from offering cytoreductive nephrectomy prior to initiating systemic therapy. The ability to predict SccRCC is not feasible at present owing to the lack of radiological criteria or tissue biomarkers. Moreover, biopsy prediction of RCC tumor grade is notoriously unreliable, including predicting for sarcomatoid change (42).
Assaying for a handful of mutations is feasible using a clinical platform in a CLIA-certified environment and may be an informative test, with prognostic and management implications. However, the intratumoral mutational heterogeneity of ccRCC, resulting from a putative pattern of branched evolution (41), limits the utility of any mutational screen based on single biopsies because the majority of mutations are subclonal and therefore would not be detected by sampling a single region. Nevertheless, VHL is considered as a clonal mutation, and PBRM1 is sometimes clonal and often shared among different regions (41,43); thus, assays for these mutations or deletions are less likely to be impacted by intratumoral heterogeneity than are assays for the other genes that we have proposed. Sampling strategies have not yet been devised to capture subclonal mutations, although early work is promising (43). In this context, our finding, supported by another independent cohort (14), that the S- component showed a higher mutational burden suggests that the higher grade component of a tumor should be assayed as part of a multiregional sampling approach.
The mutational pattern in SccRCC is likely a key driver of its response to existing therapies and may provide insight into new treatment approaches. TP53 mutations have been associated with decreased therapeutic response to antiangiogenic therapies and to PI3K pathway targeting agents (44,45). Our recent publication testing the efficacy of PI3K pathway targeted agents, including MK-2206 and everolimus, demonstrated that mutations in TP53 were associated with reduced response to either agent (45). Duration of response to antiangiogenic agents also appears to be shorter in SccRCC than in RCC (46). Interestingly, sarcomatoid ccRCC is associated with increased tissue expression of PD-L1 (47), and emerging data suggest SccRCC may be associated with heightened response to checkpoint antibodies (unpublished observations). The mechanistic link between the unique genomic features of SccRCC and response to checkpoint antibodies is being actively investigated. For PTEN-deficient tumors, several drugs are currently under clinical development in this setting including PI3K inhibitors, AKT inhibitors and PARP inhibitors (48). Circulating biomarkers for SccRCC have not yet been formally developed, but a recent study by Pal et al. detected a high rate of TP53 mutations (64%) in the circulating tumor DNA of patients with clear-cell RCC treated with later lines of vascular endothelial growth factor inhibitors versus first-line treatment (31%) (49). Whether this finding is related to sarcomatoid dedifferentiation remains to be determined.
Transcriptional differences between SccRCC and ccRCC may also be exploited to develop biomarkers of sarcomatoid change, particularly as current technologies enable high-throughput mRNA assays using FFPE material. The major advantage of a transcriptional analysis would appear to be the relatively stable gene expression signature among different regions (12,13,41). Our transcriptional analyses suggested that different pathways are enriched in SRCC, particularly TGFβ signaling. These may provide opportunities for therapeutic targeting in addition to biomarkers of tissue of origin.
In summary, we have shown that SRCC is not a single entity but rather multiple diseases that segregate according to the underlying parent subtype. With respect to the most common ccRCC subtype, our findings challenge the dogma that loss of heterozygosity at 3p21–25 is an event common to all ccRCC (20,22,41). Rather it appears that clinically aggressive, sarcomatoid ccRCC has a distinct tumorigenesis, generally lacking copy losses at 3p21–25. Together with its distinct mutational and transcriptomic landscape, this line of investigation will herald a more precise molecular classification of RCC with diagnostic, prognostic and therapeutic implications.
Supplementary Material
Translational Relevance.
The presence of sarcomatoid histology is associated with a poor prognosis in cancers arising from various organs and sarcomatoid renal cell carcinoma (SRCC) represents the most aggressive, treatment-resistant group of RCC. The molecular features that characterize and potentially drive the aggressive behavior of SRCC are poorly understood. Our study of SRCC suggests that it is not a single disease entity but rather multiple diseases that segregate according to the underlying parent RCC subtype. Notably, sarcomatoid clear cell RCC (SccRCC) has a distinct tumorigenesis, often lacking copy losses at 3p21–25, and showing distinct mutational and transcriptomic features. The molecular landscape of SccRCC is likely related to its traditionally poor response to current therapies and may inform future treatments. Our findings improve the molecular classification of RCC, propose biomarkers that may aid in predicting aggressive sarcomatoid tumors preoperatively and provide opportunities for therapeutic targeting of SRCC.
Acknowledgments
We would like to acknowledge Kim-Anh Vu, Camille Sanchez and Ann Sutton for technical and secretarial assistance.
Funding
K. Sircar is grateful for funding from the The University of Texas MD Anderson Cancer Center (Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation and CCSG grant CA016672, the Khalifa Bin Zayed Al Nahyan Foundation), and the Kidney Cancer Research Program (Monteleone Foundation). C. J. Creighton is supported by NIH grant CA125123. P.Trevisan is supported partially from CAPES Foundation and M. Varella-Garcia has support from NCI- P30CA046934 to the Cancer Center Molecular Pathology Shared Resource. Zixing Wang and Ken Chen are partially supported by NIH R01CA172652 (P.I. Ken Chen).
Footnotes
Authors’ Disclosure Of Potential Conflicts Of Interest
FM is an employee and stock holder at Castle Biosciences, a commercial laboratory that develops molecular diagnostics tests for cancer. The other authors indicate no potential conflicts of interest.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Shuch B, Said J, La Rochelle JC, Zhou Y, Li G, Klatte T, et al. Cytoreductive nephrectomy for kidney cancer with sarcomatoid histology--is up-front resection indicated and, if not, is it avoidable? The Journal of urology. 2009;182(5):2164–71. doi: 10.1016/j.juro.2009.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shuch B, Bratslavsky G, Shih J, Vourganti S, Finley D, Castor B, et al. Impact of pathological tumour characteristics in patients with sarcomatoid renal cell carcinoma. BJU international. 2012;109(11):1600–6. doi: 10.1111/j.1464-410X.2011.10785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheville JC, Lohse CM, Zincke H, Weaver AL, Leibovich BC, Frank I, et al. Sarcomatoid renal cell carcinoma: an examination of underlying histologic subtype and an analysis of associations with patient outcome. Am J Surg Pathol. 2004;28(4):435–41. doi: 10.1097/00000478-200404000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Shuch B, Bratslavsky G, Linehan WM, Srinivasan R. Sarcomatoid renal cell carcinoma: a comprehensive review of the biology and current treatment strategies. Oncologist. 2012;17(1):46–54. doi: 10.1634/theoncologist.2011-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Motzer RJ, Jonasch E, Agarwal N, Beard C, Bhayani S, Bolger GB, et al. Kidney cancer, version 3.2015. Journal of the National Comprehensive Cancer Network : JNCCN. 2015;13(2):151–9. doi: 10.6004/jnccn.2015.0022. [DOI] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–9. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linehan WM, Spellman PT, Ricketts CJ, Creighton CJ, Fei SS, Davis C, et al. Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma. The New England journal of medicine. 2015 doi: 10.1056/NEJMoa1505917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis CF, Ricketts CJ, Wang M, Yang L, Cherniack AD, Shen H, et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer cell. 2014;26(3):319–30. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durinck S, Stawiski EW, Pavia-Jimenez A, Modrusan Z, Kapur P, Jaiswal BS, et al. Spectrum of diverse genomic alterations define non-clear cell renal carcinoma subtypes. Nature genetics. 2015;47(1):13–21. doi: 10.1038/ng.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pal SK, He ML, Tong T, Wu HQ, Liu XL, Lau C, et al. RNA-seq Reveals Aurora Kinase-Driven mTOR Pathway Activation in Patients with Sarcomatoid Metastatic Renal Cell Carcinoma. Mol Cancer Res. 2015;13(1):130–37. doi: 10.1158/1541-7786.MCR-14-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh RR, Murugan P, Patel LR, Voicu H, Yoo SY, Majewski T, et al. Intratumoral morphologic and molecular heterogeneity of rhabdoid renal cell carcinoma: challenges for personalized therapy. Modern Pathol. 2015;28(9):1225–35. doi: 10.1038/modpathol.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sircar K, Yoo S-Y, Majewski T, Wani K, Patel LR, Voicu H, et al. Biphasic Components of Sarcomatoid Clear Cell Renal Cell Carcinomas Are Molecularly Similar but Distinct from Non-Sarcomatoid Renal Carcinomas. The Journal of Pathology: Clinical Research. 2015;1:212–24. doi: 10.1002/cjp2.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bi M, Zhao S, Said JW, Merino MJ, Adeniran AJ, Xie Z, et al. Genomic characterization of sarcomatoid transformation in clear cell renal cell carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(8):2170–5. doi: 10.1073/pnas.1525735113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England) 2015;31(2):166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen F, Zhang Y, Senbabaoglu Y, Ciriello G, Yang L, Reznik E, et al. Multilevel Genomics-Based Taxonomy of Renal Cell Carcinoma. Cell reports. 2016;14(10):2476–89. doi: 10.1016/j.celrep.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen K, Meric-Bernstam F, Zhao H, Zhang Q, Ezzeddine N, Tang LY, et al. Clinical actionability enhanced through deep targeted sequencing of solid tumors. Clinical chemistry. 2015;61(3):544–53. doi: 10.1373/clinchem.2014.231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nature genetics. 2013;45(8):860–7. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 21.Nickerson ML, Jaeger E, Shi Y, Durocher JA, Mahurkar S, Zaridze D, et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin Cancer Res. 2008;14(15):4726–34. doi: 10.1158/1078-0432.CCR-07-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brugarolas J. Molecular genetics of clear-cell renal cell carcinoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32(18):1968–76. doi: 10.1200/JCO.2012.45.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagenkord JM, Gatalica Z, Jonasch E, Monzon FA. Clinical genomics of renal epithelial tumors. Cancer Genet. 2011;204(6):285–97. doi: 10.1016/j.cancergen.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Dagher J, Kammerer-Jacquet SF, Brunot A, Pladys A, Patard J, Bensalah K, et al. Wild-Type VHL Clear Cell Renal Carcinomas Are a Distinct Clinical and Histologic Entity: A 10 Year Follow-Up. European urology focus. 2016;1:284–90. doi: 10.1016/j.euf.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 25.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–9. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malouf GG, Ali SM, Wang K, Balasubramanian S, Ross JS, Miller VA, et al. Genomic Characterization of Renal Cell Carcinoma with Sarcomatoid Dedifferentiation Pinpoints Recurrent Genomic Alterations. Eur Urol. 2016 doi: 10.1016/j.eururo.2016.01.051. [DOI] [PubMed] [Google Scholar]
- 27.Pal SK, He M, Tong T, Wu H, Liu X, Lau C, et al. RNA-seq reveals aurora kinase-driven mTOR pathway activation in patients with sarcomatoid metastatic renal cell carcinoma. Mol Cancer Res. 2015;13(1):130–7. doi: 10.1158/1541-7786.MCR-14-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tickoo SK, Alden D, Olgac S, Fine SW, Russo P, Kondagunta GV, et al. Immunohistochemical expression of hypoxia inducible factor-1alpha and its downstream molecules in sarcomatoid renal cell carcinoma. The Journal of urology. 2007;177(4):1258–63. doi: 10.1016/j.juro.2006.11.100. [DOI] [PubMed] [Google Scholar]
- 29.Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Rixe O, et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. The New England journal of medicine. 2007;356(2):115–24. doi: 10.1056/NEJMoa065044. [DOI] [PubMed] [Google Scholar]
- 30.Patard JJ, Fergelot P, Karakiewicz PI, Klatte T, Trinh QD, Rioux-Leclercq N, et al. Low CAIX expression and absence of VHL gene mutation are associated with tumor aggressiveness and poor survival of clear cell renal cell carcinoma. International journal of cancer Journal international du cancer. 2008;123(2):395–400. doi: 10.1002/ijc.23496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao M, Yoshida M, Kishida T, Nakaigawa N, Baba M, Kobayashi K, et al. VHL tumor suppressor gene alterations associated with good prognosis in sporadic clear-cell renal carcinoma. Journal of the National Cancer Institute. 2002;94(20):1569–75. doi: 10.1093/jnci/94.20.1569. [DOI] [PubMed] [Google Scholar]
- 32.Brauch H, Weirich G, Brieger J, Glavac D, Rodl H, Eichinger M, et al. VHL alterations in human clear cell renal cell carcinoma: association with advanced tumor stage and a novel hot spot mutation. Cancer Res. 2000;60(7):1942–8. [PubMed] [Google Scholar]
- 33.Kim JH, Jung CW, Cho YH, Lee J, Lee SH, Kim HY, et al. Somatic VHL alteration and its impact on prognosis in patients with clear cell renal cell carcinoma. Oncology reports. 2005;13(5):859–64. [PubMed] [Google Scholar]
- 34.Klatte T, Rao PN, de Martino M, LaRochelle J, Shuch B, Zomorodian N, et al. Cytogenetic profile predicts prognosis of patients with clear cell renal cell carcinoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27(5):746–53. doi: 10.1200/JCO.2007.15.8345. [DOI] [PubMed] [Google Scholar]
- 35.Kroeger N, Klatte T, Chamie K, Rao PN, Birkhauser FD, Sonn GA, et al. Deletions of chromosomes 3p and 14q molecularly subclassify clear cell renal cell carcinoma. Cancer. 2013;119(8):1547–54. doi: 10.1002/cncr.27947. [DOI] [PubMed] [Google Scholar]
- 36.Dagher J, Dugay F, Verhoest G, Cabillic F, Jaillard S, Henry C, et al. Histologic prognostic factors associated with chromosomal imbalances in a contemporary series of 89 clear cell renal cell carcinomas. Human pathology. 2013;44(10):2106–15. doi: 10.1016/j.humpath.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 37.Young AC, Craven RA, Cohen D, Taylor C, Booth C, Harnden P, et al. Analysis of VHL Gene Alterations and their Relationship to Clinical Parameters in Sporadic Conventional Renal Cell Carcinoma. Clin Cancer Res. 2009;15(24):7582–92. doi: 10.1158/1078-0432.CCR-09-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kapur P, Pena-Llopis S, Christie A, Zhrebker L, Pavia-Jimenez A, Rathmell WK, et al. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal-cell carcinoma: a retrospective analysis with independent validation. The Lancet Oncology. 2013;14(2):159–67. doi: 10.1016/S1470-2045(12)70584-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, Leng N, Pavia-Jimenez A, Wang S, et al. BAP1 loss defines a new class of renal cell carcinoma. Nature genetics. 2012;44(7):751–9. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hakimi AA, Ostrovnaya I, Reva B, Schultz N, Chen YB, Gonen M, et al. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: a report by MSKCC and the KIRC TCGA research network. Clin Cancer Res. 2013;19(12):3259–67. doi: 10.1158/1078-0432.CCR-12-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nature genetics. 2014;46(3):225–33. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abel EJ, Culp SH, Matin SF, Tamboli P, Wallace MJ, Jonasch E, et al. Percutaneous biopsy of primary tumor in metastatic renal cell carcinoma to predict high risk pathological features: comparison with nephrectomy assessment. The Journal of urology. 2010;184(5):1877–81. doi: 10.1016/j.juro.2010.06.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sankin A, Hakimi AA, Mikkilineni N, Ostrovnaya I, Silk MT, Liang Y, et al. The impact of genetic heterogeneity on biomarker development in kidney cancer assessed by multiregional sampling. Cancer medicine. 2014;3(6):1485–92. doi: 10.1002/cam4.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu JL, Rak JW, Coomber BL, Hicklin DJ, Kerbel RS. Effect of p53 status on tumor response to antiangiogenic therapy. Science. 2002;295(5559):1526–8. doi: 10.1126/science.1068327. [DOI] [PubMed] [Google Scholar]
- 45.Jonasch E, Hasanov E, Corn PG, Moss T, Shaw KR, Stovall S, et al. A randomized phase 2 study of MK-2206 versus everolimus in refractory renal cell carcinoma. Ann Oncol. 2017;28(4):804–08. doi: 10.1093/annonc/mdw676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jonasch E, Lal LS, Atkinson BJ, Byfield SD, Miller LA, Pagliaro LC, et al. Treatment of metastatic renal carcinoma patients with the combination of gemcitabine, capecitabine and bevacizumab at a tertiary cancer centre. BJU international. 2011;107(5):741–7. doi: 10.1111/j.1464-410X.2010.09626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joseph RW, Millis SZ, Carballido EM, Bryant D, Gatalica Z, Reddy S, et al. PD-1 and PD-L1 Expression in Renal Cell Carcinoma with Sarcomatoid Differentiation. Cancer immunology research. 2015;3(12):1303–7. doi: 10.1158/2326-6066.CIR-15-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dillon LM, Miller TW. Therapeutic targeting of cancers with loss of PTEN function. Current drug targets. 2014;15(1):65–79. doi: 10.2174/1389450114666140106100909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pal SK, Sonpavde G, Agarwal N, Vogelzang NJ, Srinivas S, Haas NB, et al. Evolution of Circulating Tumor DNA Profile from First-line to Subsequent Therapy in Metastatic Renal Cell Carcinoma. Eur Urol. 2017 doi: 10.1016/j.eururo.2017.03.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.