Abstract
Metabolic reprogramming is a hallmark of cancer. Herein we discovered that the key glycolytic enzyme pyruvate kinase M2 isoform (PKM2), but not the related isoform PKM1, is methylated by co-activator associated arginine methyltransferase 1 (CARM1). PKM2 methylation reversibly shifts the balance of metabolism from oxidative phosphorylation to aerobic glycolysis in breast cancer cells. Oxidative phosphorylation depends on mitochondria calcium concentration, which becomes critical for cancer cell survival when PKM2 methylation is blocked. By interacting with and suppressing the expression of inositol 1, 4, 5-trisphosphate receptors (IP3Rs), methylated PKM2 inhibits the influx of calcium from endoplasmic reticulum (ER) to mitochondria. Inhibiting PKM2 methylation with a competitive peptide delivered by nanoparticle perturbs metabolic energy balance in cancer cells, leading to decrease of cell proliferation, migration, and metastasis. Collectively, the CARM1-PKM2 axis serves as a metabolic reprogramming mechanism in tumorigenesis, and inhibiting PKM2 methylation generates metabolic vulnerability to IP3R-dependent mitochondrial functions.
One hallmark of cancer1, 2 is the “Warburg effect”, where tumor cells rely mainly on aerobic glycolysis for Adenosine-5′-triphosphate (ATP) production, even with sufficient oxygen3. However, metabolic adaptation in tumors extends beyond the “Warburg effect,” including balancing energy needs with equally important needs for macromolecular synthesis and redox homeostasis1, 2, 4. Emerging evidence suggests that mitochondrial respiration is crucial for tumorigenesis and presents a target for cancer therapy5–8.
Pyruvate kinase (PK) catalyzes the final step in glycolysis, converting phosphoenolpyruvate (PEP) to pyruvate while phosphorylating ADP to produce ATP. PK’s M1 and M2 isoforms are produced by mutually exclusive alternative splicing of PKM pre-mRNA9. Although PKM1 and PKM2 differ by only 22 amino acids, PKM1 is not allosterically regulated and exists in tetrameric form with high pyruvate kinase activity. PKM2 shifts between inactive dimeric and active tetrameric forms, modulated by phosphotyrosine signaling10, metabolic intermediates (e.g. FBP, serine and SAICAR) 11, 12 and post-translational modifications13. Switching PKM2 to PKM1 reverses aerobic glycolysis to oxidative phosphorylation and reduces tumor formation in nude mice14, identifying PKM2 as a potential cancer therapy target. However, a recent report challenged PKM2-catalyzed reaction as a rate-limiting step in cancer cell glycolysis15 and a possible protein kinase activity of PKM2 remains controversial16.
Coactivator-associated arginine methyltransferase 1 (CARM1), also known as PRMT4, is a type I protein arginine methyltransferase (PRMT) that asymmetrically dimethylates protein substrates including histones, transcriptional factors and co-regulators, splicing factors and RNA polymerase II17–20. CARM1 is overexpressed in breast cancer to promote cancer growth21, and elevated CARM1 expression correlates with poor prognosis22. Recently, we discovered that chromatin remodeling factor BAF155 methylation by CARM1 promotes breast cancer progression and metastasis23. However, whether CARM1 regulates energy metabolism in cancer cells remains unknown.
Here, we discovered CARM1-PKM2 interaction as a major contributor to metabolic reprogramming in cancer. CARM1 methylates PKM2’s dimeric form at R445/447/455. Methylated PKM2 promotes tumor cell proliferation, migration and lung metastasis by reprogramming oxidative phosphorylation to aerobic glycolysis, and this effect was reversed by a competitive PKM2 peptide delivered using nanoparticles. We showed that methylated PKM2 localized in mitochondria-associated endoplasmic reticulum membrane (MAM), through interaction with inositol 1, 4, 5-trisphosphate receptors (IP3Rs), decreasing mitochondrial membrane potential (ΔΨm) and Ca2+ uptake, which is essential for activating pyruvate dehydrogenase (PDH) to support oxidative phosphorylation24. Blocking PKM2 methylation elevates IP3R expression, increasing mitochondrial Ca2+ uptake, PDH activation and oxidative phosphorylation. Thus, PKM2 methylation represents an important regulator of switching between oxidative phosphorylation to aerobic glycolysis in cancer cells.
RESULTS
CARM1 interacts with and methylates PKM2
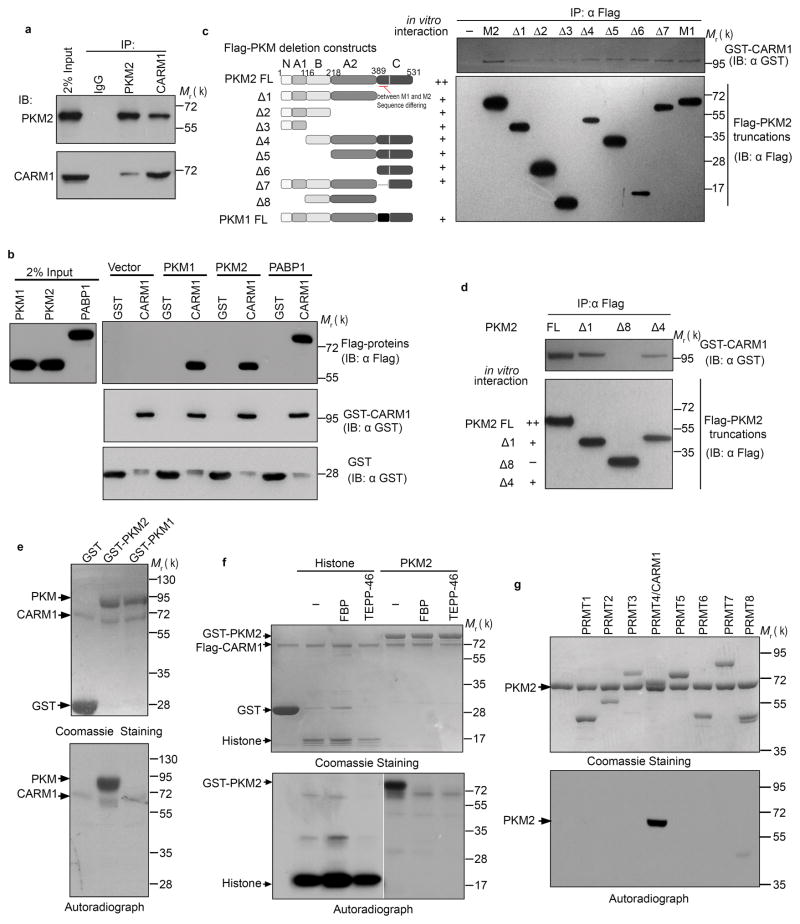
Consistent with CARM1 promotion of tumor development and progression21, 23, knocking out (KO) CARM1 decreased DNA synthesis in MCF7 cells (Supplementary Fig. 1a). CARM1 KO also increased mitochondrial oxygen consumption rate (OCR) but decreased lactate production without affecting glucose uptake (Supplementary Fig. 1b–f). These results prompted us to test whether CARM1 modulates energy metabolism in breast cancer cells. We identified PKM2 as a putative CARM1-interacting protein by mass spectrometry when Halo-tagged CARM1 was overexpressed in HEK293T cells (Supplementary Table 1). Endogenous CARM1-PKM2 interaction was confirmed by reciprocal coimmunoprecipitation in MCF7 cells (Fig. 1a). To determine whether CARM1 directly interacts with PKM2, we performed GST pull-down using recombinant glutathione S-transferase (GST) tagged CARM1, Flag-tagged PKM2 and Flag-PABP1 (positive control)25 and observed that GST-CARM1, but not GST alone, bound PKM2, indicating direct CARM1-PKM2 interaction (Fig. 1b). Interestingly, recombinant PKM1 also associated with CARM1 in vitro (Fig. 1b). To map the PKM2 region that binds CARM1, we expressed truncated Flag-PKM2 using in vitro transcription and translation and performed in vitro interaction assays with GST-CARM1. Deleting PKM2 domain C (Δ1) or N/A1 (Δ4) significantly decreased and truncation of both domains (Δ8) completely abolished PKM2 interaction with CARM1, suggesting that C and N/A1 domains are responsible for the interaction (Fig. 1c, d). Although these domains are identical between PKM1 and PKM2, intriguingly only PKM2, but not PKM1, can be methylated by CARM1 in vitro (Fig. 1e).
Figure 1.
The dimeric form of PKM2 is specially methylated by CARM1. (a) Reciprocal co-immunoprecipitation of PKM2 and CARM1 in MCF7 cells followed by Western blot analysis. (b) Direct interaction of recombinant PKM1, PKM2, and PABP1 (positive control) proteins with CARM1 in GST pull-down assays. GST and GST-CARM1 were detected by anti-GST antibody, Flag-tagged PKM1, PKM2 and PABP1 were detected by anti-Flag antibody, respectively. (c, d) Schematic showing truncations of PKM2 and in vitro interaction assays using full-length PKM and truncated proteins expressed by in vitro transcriptional and translational systems. (e) In vitro methylation assays using recombinant CARM1, GST-PKM1 or PKM2 protein in the presence of 3H-SAM. (f) In vitro methylation assays of PKM2 by CARM1 in the presence of FBP (100μM) or TEPP-46 (1μM). Histone proteins were used as negative controls. (g) In vitro methylation assays of PKM2 by PRMTs. Data represent one of the three independent experiments with similar results. Unprocessed original scans of blots are shown in Supplementary Figure 9.
Since non-methylatable PKM1 only forms tetramers while methylatable PKM2 forms tetramers or dimers, we hypothesized that tetramer formation may prevent CARM1-mediated methylation. TEPP-4626, a PKM2 activator, or FBP stimulated PKM2 tetramer formation (Supplementary Fig. 2a, b), inhibiting PKM2 methylation by CARM1 (Fig. 1f). Neither TEPP-46 nor FBP affected histone H3 methylation by CARM1 (Fig. 1f), excluding the possibility that TEPP-46 and FBP interfere with CARM1 methyltransferase activity. Thus, only PKM2’s dimeric form is methylated by CARM1.
Mammalian genomes encode nine PRMTs that sometimes share the same substrates17. Using purified Halo-tagged23 PRMTs (1–8), we assayed methylation of recombinant PKM2 by PRMTs and showed that PKM2 is uniquely methylated by CARM1 (Fig. 1g).
CARM1 methylates PKM2 at R445/447/455
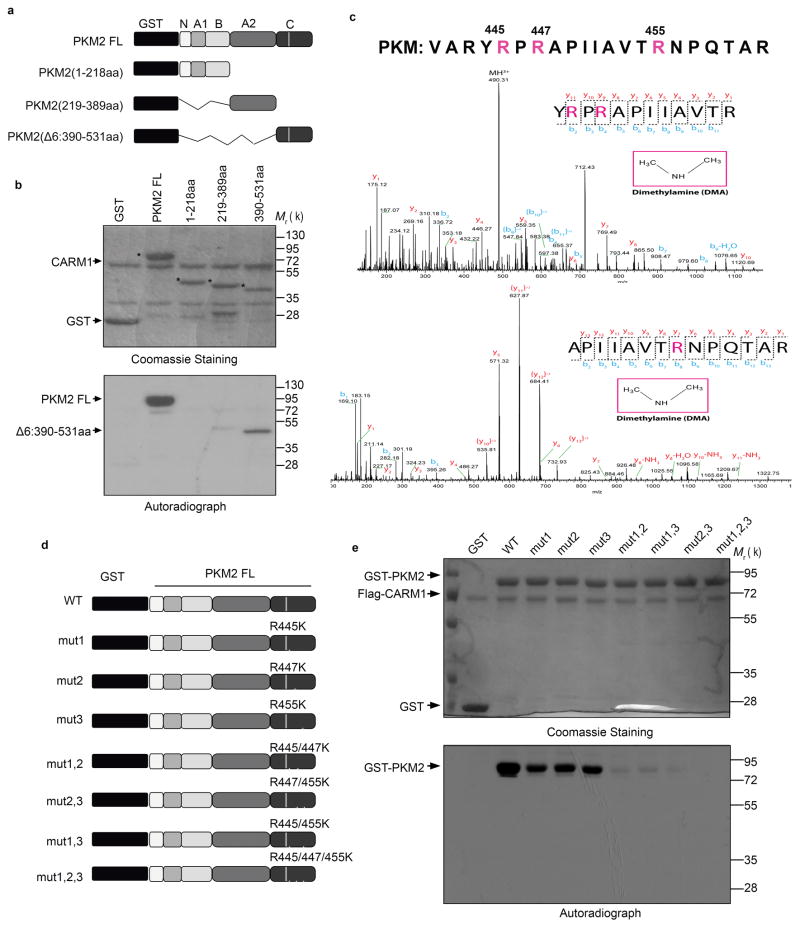
To narrow down the PKM2 methylation sites(s), we assayed methylation in vitro using purified GST tagged full-length (FL) or truncated PKM2 (Fig. 2a). Among the three truncated fragments, only the C-domain was significantly methylated (Fig. 2b), suggesting that it might contain PKM2’s methylation site(s). Three arginine residues (R445, R447 and R455) were identified in the in vitro methylated GST-PKM2 (390-531aa) fragment using liquid chromatography coupled tandem mass spectrometry (LC-MS/MS) (Fig. 2c). To further discern PKM2’s major methylation site(s), we substituted the three methylated arginines to lysine to preserve their positive charge, individually or in combination, in GST-PKM2 (Fig. 2d). While individually mutating each R site slightly affected PKM2 methylation (Fig. 2e), mutating any two sites dramatically decreased methylation, and mutating all three sites abolished methylation (Fig. 2e). While all three R residues reside in the C domain that fosters tetramer formation, none is at the tetrameric interface (Supplementary Fig. 2a). Size exclusion chromatography using purified recombinant His-tagged proteins showed that neither PKM2 (Supplementary Fig. 2b) nor PKM1 (Supplementary Fig. 2c) tetramer formation was affected by mutating R445, R447, and R455 to lysine. Thus, while CARM1 predominantly methylates dimeric PKM2, PKM2 tetramer formation is not affected by PKM2 methylation. PKM1 is not methylated by CARM1 possibly because the corresponding arginines in tetrameric PKM1 are inaccessible to CARM1.
Figure 2.
CARM1 methylates PKM2 at R445, R447 and R455. (a) Schematic diagram of full-length PKM2 and its truncated derivatives. (b) Mapping of the methylation domain of PKM2 by CARM1 in in vitro methylation assays. (c) Identification of di-methylated R445, R447 and R455 of the in vitro methylated GST-PKM2 protein. (d) Schematic showing R→K mutations on GST-PKM2 protein. (e) In vitro methylation assays of PKM2 mutants relative to the wild type PKM2. In b, e, data represent one of the three independent experiments with similar results. Unprocessed original scans of blots are shown in Supplementary Figure 9.
Inhibiting PKM2 methylation decreases breast cancer cell proliferation and migration and tumor growth
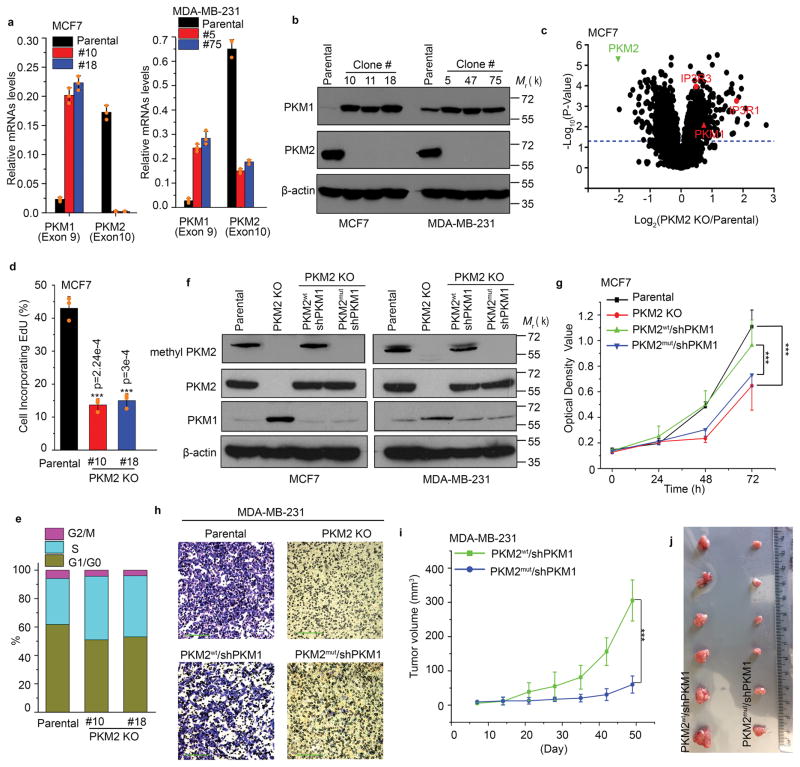
To investigate the function of PKM2 methylation in breast cancer cell lines, we employed CRISPR/Cas9- technology to knock out (KO) endogenous PKM2. Genomic DNA sequencing of two representative PKM2 KO clones of MCF7 or MDA-MB-231 revealed non-homologous end jointing (NHEJ)-induced random insertions or deletions at the targeted site (Supplementary Fig. 3a), causing mRNA degradation and PKM2 protein loss (Fig. 3a, b). Specific PKM2 KO led to compensatory PKM1 induction (Fig. 3a, b), as in primary mouse embryonic fibroblast (MEF) cells27. Whole proteome analysis showed that, among >4000 proteins detected, 158 proteins significantly increased (fold >=1.5, p<0.05), and 261 proteins decreased (fold<=0.7, p<0.05) in PKM2 KO vs. parental cells (Fig. 3c and Supplementary Table 2). In agreement with specific PKM2 KO, PKM2 protein levels decreased by >5-fold and PKM1 increased by >1.6-fold. The total pyruvate kinase (PK) activity in PKM2 KO cells decreased as compared to the parental cells (Supplementary Fig. 3b), possibly because PKM1 restoration in PKM2 KO cells is insufficient to compensate PKM2 loss. No PK activity change was observed in paired parental and CARM1 KO MCF7 and MDA-MB-231 cell lines (Supplementary Fig. 3c), indicating that CARM1-mediated PKM2 methylation does not affect PKM2 PK activity.
Figure 3.
Inhibition of PKM2 methylation decreases cell proliferation and migration. (a) Q-PCR analyses of mRNA levels of PKM1 and PKM2 in parental MCF7 and MDA-MB-231 cells and their respective PKM2 KO clones (n=3 independent experiments). (b) Western blot analyses of PKM1 and PKM2 in parental MCF7 and MDA-MB-231 cells and their respective PKM2 KO clones. (c) Mass spectrometry analysis of global protein changes between parental MCF7 and PKM2 KO cells (n=3 independent experiments). (d) EdU incorporation assays of parental MCF7 and PKM2 KO clones (n=3 independent experiments). (e) Cell cycle analyses of parental MCF7 and MCF7 PKM2 KO clones (n=3 independent experiments). (f) Western blot analysis of PKM2, methyl-PKM2 and PKM1 in parental, PKM2 KO, PKM2 KO restored with PKM2wt and PKM2mut in accompanying with PKM1 knockdown in MCF7 and MDA-MB-231 cells. (g) Measurement of cell proliferation by MTT assays in parental MCF7, PKM2 KO, PKM2wt/shPKM1 and PKM2mut/shPKM1 cells (n=10 independent experiments). (h) Measurement of cell migration by transwell assays in parental MDA-MB-231, PKM2 KO, PKM2wt/shPKM1 and PKM2mut/shPKM1 cells (n=3 independent experiments). (i) The growth curves of MDA-MB-231 PKM2wt/shPKM1 and PKM2mut/shPKM1 xenografts (n=6 biologically independent mice per group). (j) Representative images of the xenograft tumors. Statistical significance was assessed using ANOVA. In a, d, g, i, data are shown as Mean ±SD and statistics source data are available in Supplementary Table 7. Statistical significance was assessed using two-tailed t-test (a and d) and ANOVA (g and i), ***p<0.001. In b, f and h, data represent one of the three independent experiments with similar results. Unprocessed original scans of blots are shown in Supplementary Figure 9.
In MEF cells, deleting PKM2 from one or both allele(s) stimulated PKM1 expression and arrested proliferation, with reduced DNA synthesis due to PKM1 expression, rather than PKM2 loss27. Similarly, EdU incorporation and S-phase accumulation revealed dramatically reduced DNA synthesis when PKM2 was knocked out in MCF7 cells (Fig. 3d, e). However, the mechanism of reduced DNA synthesis in MCF7 cells appears to differ from that of MEF cells. PKM2 KO induced massive reduction of nucleotides in MEF cells27, but not in MCF7 and MDA-MB-231 cells (Supplementary table 3). Thus, reduced DNA synthesis in MCF7 cells was not due to lack of nucleotides, as in MEFs. Also in contrast to MEFs27, PKM1 overexpression in MCF7 cells did not inhibit cell proliferation (Supplementary Fig. 3d) in the presence of PKM2 expression. To delineate the functions of PKM2 methylation on energy metabolism without interference from PKM1, we restored PKM2wt or PKM2mut (where mut = mut1,2,3 from Fig. 2d) in PKM2 KO cell lines, followed by knocking down (KD) PKM1 (Fig. 3f). While knocking down PKM1 60–80% in PKM2 KO cells did not affect cell proliferation and oxidative phosphorylation (Supplementary Fig. 3e–g), cell viability was dramatically reduced when PKM1 knockdown reached nearly 100%, possibly because these cells have insufficient PK to support survival. To ensure that PKM2mut was defective in PKM2 methylation, we generated an antibody against an asymmetrically dimethylated R445 and R447 peptide of PKM2, referred as the methyl-PKM2 antibody. While PKM2wt and PKM2mut were restored to similar levels, methylated PKM2 was only detected in parental and PKM2wt expressing cells, but not in PKM2 KO or PKM2mut-expressing cells (Fig. 3f) or in CARM1 KO cells (Supplementary Fig. 3h), demonstrating the antibody specificity. We used these cell lines to investigate the effects of methyl-PKM2 on cell proliferation and colony formation in MCF7 cells and cell migration in MDA-MB-231. PKM2 methylation defective cells (e.g., PKM2 KO and PKM2mut) elicited reduced cell proliferation and colony formation (Fig. 3g and Supplementary Fig. 3i) compared to parental and PKM2wt MCF7 cells; however, these effects appeared not caused by apoptosis (Supplementary Fig. 3j). PKM2 methylation status also affected cell migration (Fig. 3h) and tumor growth of MDA-MB-231 xenografts (Fig. 3i, j). Therefore, PKM2 methylation is required for promoting cell proliferation, migration and tumor growth in various breast cancer cell models.
Blocking PKM2 methylation results in elevated mitochondrial respiration in breast cancer cells
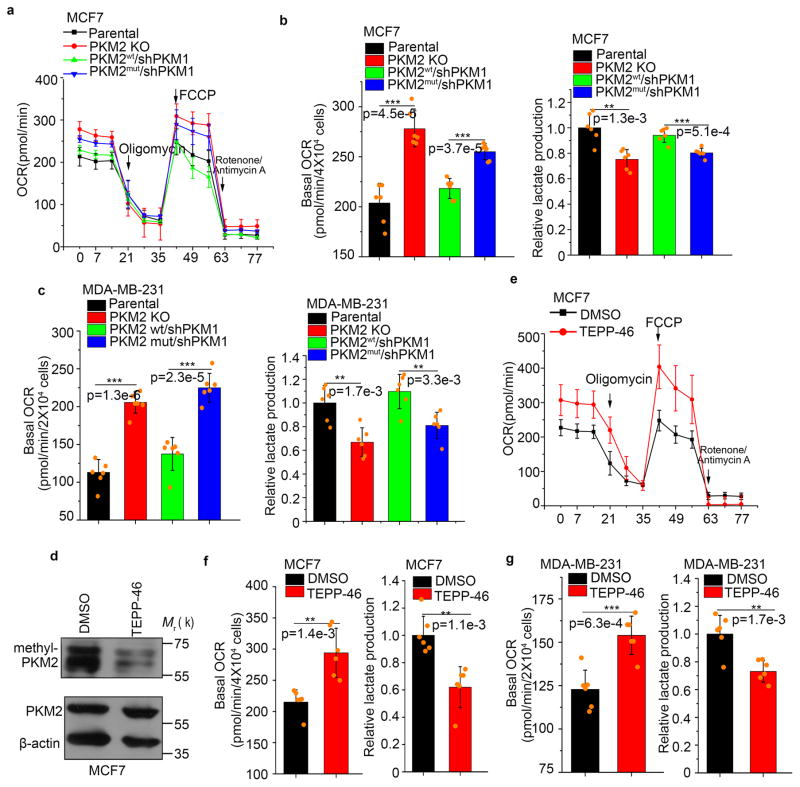
We next examined whether PKM2 methylation regulates metabolic fluxes. The switch from PKM2 to PKM1 in MCF7 and MDA-MB-231 PKM2 KO cells significantly increased OCR and reduced lactate (Fig. 4a–c). Remarkably, the balance of OCR and lactate production could be reversed by restoring PKM2wt, but not PKM2mut (Fig. 4b, c). TEPP-46 treatment, which triggers PKM2 tetramerization, thus blocking its methylation (Fig. 1f and 4d), also significantly increased OCR while decreasing lactate production (Fig. 4e–g). Thus, PKM2 methylation modulates energy metabolism in cancer cells.
Figure 4.
Inhibiting PKM2 methylation increased mitochondrial oxidative phosphorylation. (a) The OCR curves in parental MCF7, PKM2 KO, PKM2wt/shPKM1 and PKM2mut/shPKM1 cells treated with oligomycin, FCCP, and rotenone/antimycin A (n=6 independent experiments). (b) Basal OCR and lactate production normalized to the cell numbers in parental MCF7, PKM2 KO, PKM2wt/shPKM1 and PKM2mut/shPKM1 cells (n=6 independent experiments). (c) Basal OCR and lactate production normalized to cell numbers in parental MDA-MB-231, PKM2 KO, PKM2wt/shPKM1 and PKM2mut/shPKM1 cells (n=6 independent experiments). (d) Western blot analysis of methyl-PKM2 in MCF7 cells treated with DMSO or TEPP-46. (e) The OCR curves in parental MCF7 cells treated with DMSO or TEPP-46 (n=6 independent experiments). (f, g) Basal OCR and lactate production normalized to cell numbers in MCF7 cells (f) or MDA-MB-231 (g) treated with DMSO or TEPP-46 (n=6 independent experiments). In a, b, c, e–g, data are shown as Mean ±SD and statistics source data are available in Supplementary Table 7. Statistical significance was assessed using two-tailed t-test (f and g) and ANOVA (b and c), *p<0.05, **p<0.01, ***p<0.001. In d, data represent one of the three independent experiments with similar results. Unprocessed original scans of blots are shown in Supplementary Figure 9.
Reactive oxygen species (ROS), an inevitable byproduct of mitochondrial oxidative phosphorylation, are often scavenged by nicotinamide adenine dinucleotide phosphate (NADPH) and glutathione (GSH)1, 28. Because blocking PKM2 methylation increases oxidative phosphorylation, it may elevate ROS and deplete NADPH and GSH. Indeed, ROS levels were elevated by knocking out PKM2 (Supplementary Fig. 4a), mutating PKM2 methylation sites (Supplementary Fig. 4b) or knocking out CARM1 (Supplementary Fig. 4c) in MCF7 cells relative to corresponding controls, and increased ROS was accompanied by decreased NADPH/NADP+ ratio (Supplementary Fig. 4d, e) and GSH concentration (Supplementary Fig. 4f, g). Similarly, NADPH/NADP+ ratio (Supplementary Fig. 4h) and GSH concentration (Supplementary Fig. 4i) were higher in parental and PKM2wt expressing cells than PKM2 KO and PKM2mut expressing MDA-MB-231 cells. However, mitoTEMPO, a specific scavenger of mitochondrial superoxide, and glutathione did not alter cell proliferation and migration in ROS highly producing cells (i.e., PKM2 KO expressing PKM2mut and CARM1 KO cells) (Supplementary Fig. 4j–o). Thus, glycolytic metabolism and growth effects regulated by PKM2 methylation are largely independent of ROS.
Inhibiting PKM2 methylation increases Ca2+ uptake and mitochondrial membrane potential
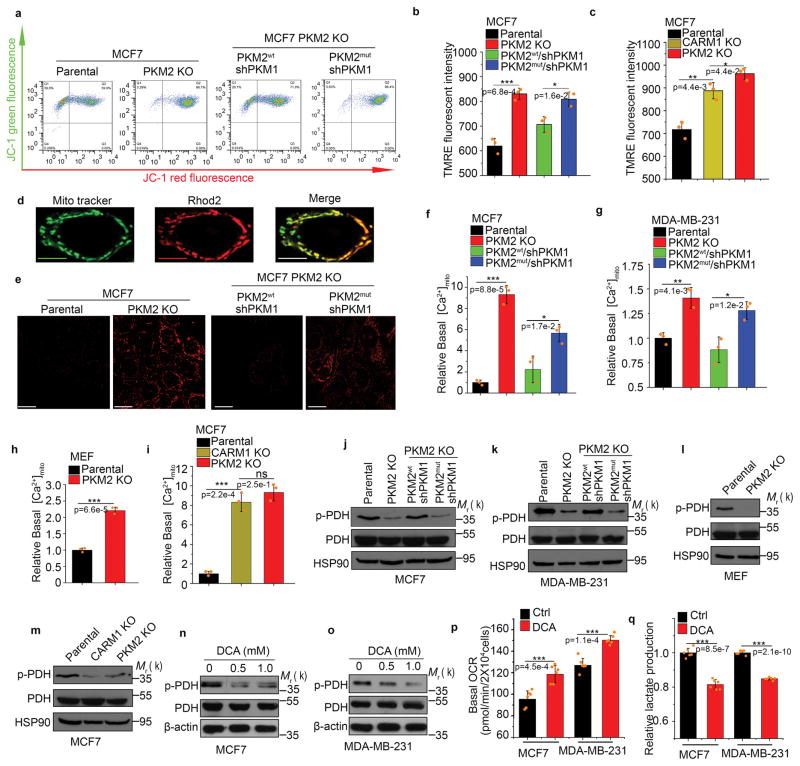
To investigate the mechanism by which mitochondrial respiration was elevated in PKM2 methylation-defective cells relative to PKM2 methylation-competent cells, we evaluated the effects of PKM2 methylation on mitochondrial membrane potential (ΔΨm), an indicator of oxidative energy metabolism. Incorporation of mitochondrial-specific JC-1 dye followed by flow cytometry showed that ΔΨm increased upon knocking out PKM2 in MCF7 cells, and that restoring PKM2wt, but not PKM2mut, in PKM2 KO cells abrogated increased ΔΨm (Fig. 5a). To validate this, we used tetramethylrhodamine ethyl ester (TMRE) to measure ΔΨm. Similar results were obtained in both MCF7 (Fig. 5b) and MDA-MB-231 cells (Supplementary Fig. 5a). In accordance with the PKM2 methylation-dependent ΔΨm change, CARM1 KO also increased ΔΨm in MCF7 cells (Fig. 5c). Increased mitochondrial DNA copy number is another indicator of increased mitochondrial activity. Ablating PKM2 expression or PKM2 methylation resulted in increased mitochondrial DNA content in MCF7 cells (Supplementary Fig. 5b). Therefore, PKM2 methylation suppresses mitochondrial function and loss of PKM2 methylation releases this suppression and elevates ΔΨm.
Figure 5.
Inhibiting PKM2 methylation increases mitochondrial membrane potential and [Ca2+]mito. (a–c) Measurement of mitochondrial membrane potential (ΔΨ) by incorporation of JC-1 dye (a) or TMRE dye (b, c) followed by flow cytometry. The ΔΨ was measured by incorporation of JC-1 (a) or TMRE dyes (b) in parental MCF7, PKM2 KO, PKM2wt/shPKM1 and PKM2mut/shPKM1 cells (n=3 independent experiments). Alternatively, the ΔΨ was measured by TMRE dye incorporation in parental MCF7, PKM2 KO and CARM1 KO cells (c) (n=3 independent experiments). (d) Representative images of co-localized mitochondrial tracker and Rhod-2 in MCF7 PKM2 KO cells. Scale bars, 5μm. (e) Representative images of Rhod-2-labeled mitochondria in parental MCF7, PKM2 KO, PKM2wt/shPKM1 and PKM2mut/shPKM1 cells. Scale bars, 10μm. (f–i) Relative basal [Ca2+]mito in Rhod-2-labeled parental, PKM2 KO, PKM2wt/shPKM1 and PKM2mut/shPKM1 MCF7 (f) (n=3 independent experiments) or corresponding MDA-MB-231 cells (g) (n=3 independent experiments); or parental MEF and PKM2 KO cells (h) (n=3 independent experiments); or parental MCF7, CARM1 KO and PKM2 KO cells (i) (n=3 independent experiments). (j–m) Western blotting of phosphorylated PDH and total PDH in indicated MCF7 (j) or corresponding MDA-MB-231 cells (k); or parental MEF and PKM2 KO (l); or parental MCF7, CARM1 KO and PKM2 KO cells (m). (n, o) Western blot analysis of phosphorylated PDH and total PDH in MCF7 (n) or MDA-MB-231 (o) cells treated with DCA. (p, q) Basal OCR (p) and lactate production (q) normalized to the cell numbers in MCF7 or MDA-MB-231 cells treated with DCA (n=6 independent experiments). In b, c, f–h and p–q, data are shown as Mean ±SD and statistics source data are available in Supplementary Table 7. Statistical significance was assessed using two-tailed t-test (h, p and q) and ANOVA (b, c f, g and i), *p<0.05, **p<0.01, ***p<0.001. In d and e, data represent one of the two independent experiments with similar results. In a and j–o, data represent one of the three independent experiments with similar results. Unprocessed original scans of blots are shown in Supplementary Figure 9.
Mitochondrial Ca2+, primarily driven by ΔΨm, stimulates oxidative phosphorylation to maintain cellular energy homeostasis24, 29. ΔΨm sensitivity to PKM2 methylation prompted us to assess basal mitochondrial Ca2+ concentration ([Ca2+]mito) using a Ca2+-sensitive Rhod-2 AM dye and confocal imaging (Fig. 5d). PKM2 KO profoundly increased basal [Ca2+]mito in MCF7 (Fig. 5e, f) and MDA-MB-231 (Fig. 5g) cells, The effect is not cancer-cell specific as [Ca2+]mito also increased in PKM2 KO MEF cells (PKM2fl/fl, Cre-estrogen receptor)27 (Fig. 5h). Restoring PKM2wt, but not PKM2mut, abrogated the elevated basal [Ca2+]mito caused by PKM2 KO in MCF7 and MDA-MB-231 cells (Fig. 5f, g). Basal [Ca2+]mito was also elevated in CARM1 KO MCF7 cells (Fig. 5i). Thus, methylated PKM2 suppresses mitochondrial Ca2+ uptake. Mitochondrial matrix calcium regulates oxidative phosphorylation through activating several dehydrogenases, including pyruvate dehydrogenase (PDH) that couples glycolysis to the tricarboxylic acid (TCA) cycle by pyruvate decarboxylation24. PDH activity is suppressed by phosphorylation by PDH kinase and enhanced by dephosphorylation by Ca2+-dependent pyruvate phosphatases24, 30. To investigate if altered [Ca2+]mito levels changes PDH activity, we measured phosphorylated PDH levels in PKM2 KO, CARM1 KO and PKM2wt- or PKM2mut-restored cell lines by Western blot. PKM2 KO dramatically decreased PDH phosphorylation in MCF7 (Fig. 5j), MDA-MB-231 (Fig. 5k) and MEFs (Fig. 5l) cells, implying increased PDH activity by [Ca2+]mito influx. Restoring PKM2wt, but not PKM2mut, increased phosphorylated PDH in MCF7 and MDA-MB-231 cells (Fig. 5j, k), implying attenuated PDH activity. CARM1 KO similarly decreased PDH phosphorylation in MCF7 (Fig. 5m). As expected, treating cells with dichloroacetate (DCA), a PDH kinase inhibitor, also decreased PDH phosphorylation and lactate production while increasing oxidative phosphorylation (Fig. 5n–q). Thus, PKM2 methylation is critical for restraining mitochondrial oxidative phosphorylation via decreasing mitochondrial ΔΨm and Ca2+ uptake, and increasing PDH phosphorylation.
Methylated PKM2 decreases IP3Rs expression
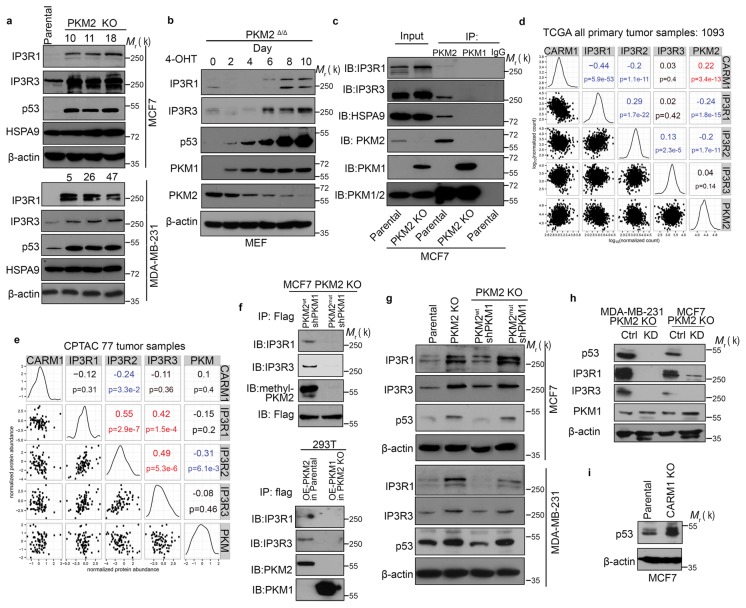
A previous study reported PKM2 in mitochondria31. We confirmed PKM2 in the mitochondrial fraction by subcellular fractionation (Supplementary Fig. 6a) and observed a portion of PKM2 co-localizing with HSPA9 in mitochondrial outer membrane (Supplementary Fig. 6b). However, PKM2’s mitochondrial localization appeared not affected by its methylation (Supplementary Fig. 6c). To elucidate how methylated PKM2 modulates mitochondrial oxidative phosphorylation, we overexpressed Flag-tagged PKM2wt or PKM2mut in HEK293T cells, and performed MS analyses on Flag-PKM2 co-immunoprecipitated proteins to identify differentially interacting proteins. Many interacting proteins were identical between PKM2wt and PKM2mut (Supplementary Table 4, 5); however, ER calcium-releasing proteins IP3R1, 2, 3 showed the most notable difference between PKM2wt and PKM2mut (Supplementary Fig. 6d). Interestingly, IP3R1 and IP3R3 are not only putative PKM2-interacting proteins but their levels also increased in PKM2 KO MCF7 cells (Fig. 3c and Supplementary Fig. 6e). To confirm the proteomics results, we examined IP3R1 and IP3R3 expression levels by western blotting in different PKM2 KO clones from MCF7 and MDA-MB-231 cells. Both IP3R1 and IP3R3 were significantly elevated in three different PKM2 KO clones of two cell lines (Fig. 6a). As a negative control, levels of HSPA9, another PKM2-interacting protein, were insensitive to PKM2 KO. To test if increased IP3Rs resulted from increased PKM1 in PKM2 KO cells, we measured the kinetics of protein changes in immortalized, tamoxifen-inducible PKM2 KO (PKM2fl/fl, Cre-estrogen receptor) MEFs. As reported27, PKM1 expression was elevated after a two-day 4-hydroxytamoxifen (4-OHT) treatment and plateaued after four-day treatment (Fig. 6b). However, increased IP3R1 and IP3R3 levels were only detected later when PKM2 was substantially lower (Fig. 6b). Moreover, overexpressing PKM1 failed to increase IP3Rs expression (Supplementary Fig. 6f), reinforcing that IP3Rs are regulated by PKM2 not PKM1. Co-immunoprecipitation showed that, PKM2, but not PKM1, interacted with the endogenous IP3R1 and IP3R3 in breast cancer cells (Fig. 6c and Supplementary Fig. 6g). Thus, elevated IP3Rs in PKM2 KO cells likely results from PKM2 loss rather than PKM1 gain. Inverse expression changes of IP3Rs and PKM2 were also observed in The Cancer Genome Atlas (TCGA) breast tumor specimens32. Across 1093 primary breast tumors in TCGA, CARM1 and PKM2 mRNA levels were positively correlated but negatively correlated with IP3R1 and IP3R2 expression (Fig. 6d). Similarly, in the CPTAC 77 breast tumor proteogenomics database 33, IP3R2 protein level was negatively correlated with CARM1 or PKM (Fig. 6e).
Figure 6.
Methylated PKM2 decreases IP3Rs expression. (a) Western blotting of IP3R1, IP3R3, p53 and HSPA9 in parental and PKM2 KO MCF7 or MDA-MB-231 cells. (b) Western blotting of IP3R1, IP3R3, p53, PKM1 and PKM2 in MEF (PKM2fl/fl, Cre-ER) cells treated with 4-OHT for the indicated time. (c) Co-immunoprecipitation of IP3Rs and HSPA9 with PKM2 but not PKM1. Flag-tagged PKM1 or PKM2 are immunoprecipitated from cell lysates derived from parental MCF7 or PKM2 KO cells using α-Flag antibody followed by detection of PKM1, PKM2, IP3R1, IP3R3 and HSPA9 by Western blot. (d) mRNA correlation of CARM1, IP3R1, IP3R2, IP3R3 and PKM2 in 1093 primary breast tumors of TCGA (n=1093 biologically independent patient samples). (e) Protein correlation of CARM1, IP3R1, IP3R2, IP3R3 and PKM in 77 breast tumors of CPTAC collection33 (n=77 biologically independent patient samples). (f) Co-immunoprecipitation of IP3R1 and IP3R3 with PKM2wt but not PKM2mut in MCF7 (left panel) and overexpressed PKM1 in HEK293T PKM2 KO cells (right panel). IP3R1, IP3R3, PKM1, PKM2 and methyl-PKM2 are detected by Western blot in Flag-PKM1/2 immunoprecipitates using corresponding antibodies. (g) Western blotting of IP3Rs and p53 in parental MCF7 (or MDA-MB-231), PKM2 KO, PKM2wt/shPKM1 and PKM2mut/shPKM1 cells. (h) Western blotting of p53, IP3R1, IP3R3 and PKM1 in MCF7 PKM2 KO and MDA-MB-231 PKM2 KO cells expressing control shRNA and p53 shRNA. (i) Western blotting of p53 in parental MCF7 and CARM1 KO cells. In a–c and f–i, data represent one of the three independent experiments with similar results. Unprocessed original scans of blots are shown in Supplementary Figure 9.
To determine if PKM2-IP3Rs interaction is affected by PKM2 methylation, we precipitated PKM2 from MCF7 cells stably expressing Flag-tagged PKM2wt or PKM2mut using anti-Flag M2 resin. PKM2-IP3Rs interaction was detected in PKM2wt- but not PKM2mut-expressing cells (Fig. 6f, upper panel). Even when Flag-PKM1 was overexpressed in HEK293T-PKM2 KO cells, no PKM1-IP3Rs interaction was detected (Fig. 6f, lower panel). Interestingly, IP3Rs expression appears sensitive to PKM2 methylation since restoring PKM2wt, but not PKM2mut, abrogated elevated IP3Rs expression in PKM2 KO cells (Fig. 6g). Furthermore, blocking PKM2 methylation by knocking out CARM1 or TEPP-46 treatment elevated IP3R3 expression (Supplementary Fig. 6h). Thus, IP3Rs expression is inversely correlated with methylated PKM2. ChIP-seq34 revealed p53 binding to IP3Rs promoters, indicating that IP3Rs could be direct p53 target genes. IP3Rs mRNA levels were increased by PKM2 KO, particularly in MDA-MB-231 cells (Supplementary Fig. 6i), consistent with increased p53 protein levels in PKM2 KO clones of MCF7, MDA-MB-231 and MEF cells (Fig. 6a, b). Knocking down p53 using shRNAs significantly decreased IP3Rs protein levels (Fig. 6h). p53 protein levels were sensitive to PKM2 methylation as restoring PKM2wt but not PKM2mut reduced p53 expression (Fig. 6g). Conversely, CARM1 knockout induced p53 expression (Fig. 6i). Accordingly, methylated PKM2, via down-regulating p53, is one means to control IP3Rs levels.
Methylated PKM2 restrains mitochondrial addiction to Ca2+ through IP3Rs
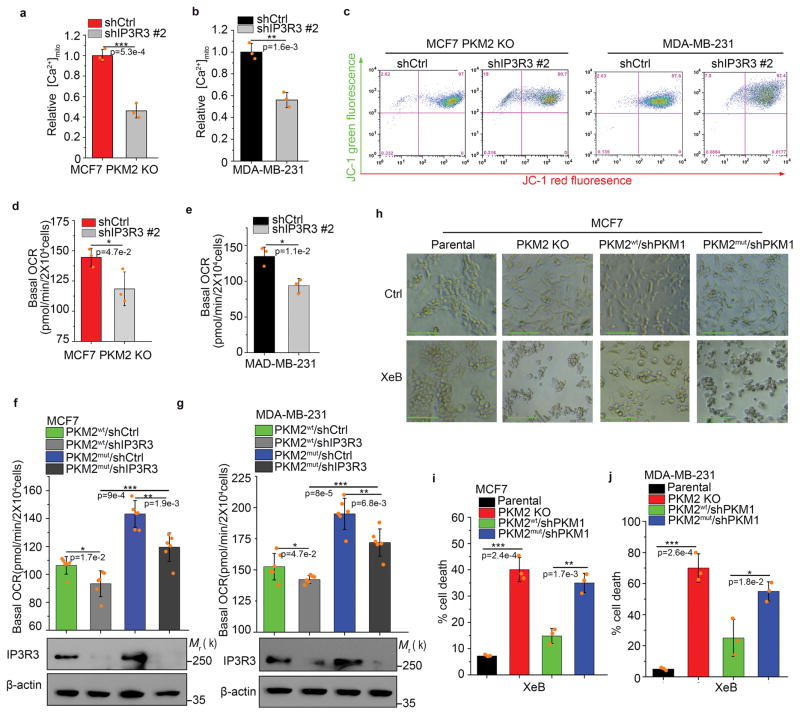
To test if methylated PKM2 regulates mitochondrial functions through modulating IP3Rs, which sustain mitochondrial functions, we stably knocked down IP3R3 in MCF7 PKM2 KO and MDA-MB-231 cell lines, which highly express IP3R3 (Supplementary Fig. 7a, b). IP3R3 KD reduced basal [Ca2+]mito level (Fig. 7a, b) and ΔΨm (Fig. 7c and Supplementary Fig. 7c). Similarly, OCR was decreased by knocking down IP3R3 in MCF7 PKM2 KO (Fig. 7d) and MDA-MB-231 cells (Fig. 7e). To delineate IP3Rs roles in PKM2-modulated mitochondrial activity, we knocked down IP3R3 in PKM2wt or PKM2mut-expressing cells. IP3R3 KD significantly reduced OCR in PKM2wt or PKM2mut cells. Basal OCR in PKM2mut /shIP3R3 cells was higher than in PKM2wt/shIP3R3 cells, possibly due to other IP3Rs (IP3R1 and 2) that remain abundant in PKM2mut cells (Fig. 7f, g). Calcium transport between ER and mitochondria is essential for Ca2+ homeostasis and cell survival29. To assess the importance of calcium hemostasis and oxidative phosphorylation to cell survival in PKM2 KO or methylation defective cells, we treated cells with Xestospongin B (XeB), a specific IP3Rs inhibitor, to inhibit IP3Rs-mediated ER Ca2+ release. PKM2 KO or PKM2mut cells were more vulnerable to XeB than parental and PKM2wt cells (Fig. 7h–j and Supplementary Fig. 7d), indicating that addiction to oxidative phosphorylation upon loss of PKM2 methylation plays essential roles in cell survival. Thus, methylated PKM2 repressed mitochondrial addiction to Ca2+ via interacting with and suppressing the expression of IP3Rs.
Figure 7.
Methylated PKM2 restrains mitochondrial addiction to Ca2+ through IP3Rs. (a, b) Relative basal [Ca2+]mito levels in MCF7 PKM2 KO and MDA-MB-231 cells with ctrl shRNA or IP3R3 shRNA knockdown (n=3 independent experiments). (c) ΔΨ measurement in MCF7 PKM2 KO and MDA-MB-231 cells with ctrl shRNA or IP3R3 shRNA knockdown. (d–g) Normalized basal OCR values in MCF7 PKM2 KO and MDA-MB-231 cells (d, e) and MCF7 (or MDA-MB-231) PKM2wt/shPKM1 and PKM2mut/shPKM1 cells (f, g) with ctrl shRNA or IP3R3 shRNA knockdown (n=6 independent experiments). (h) Representative images of parental MCF7, PKM2 KO, PKM2wt/shPKM1 and PKM2mut/shPKM1 cells after 3.5 μM IP3Rs inhibitor XeB treatment for 24 hours. Scale bars, 50μm. (i, j) Cell death measured by PI staining in parental MCF7, PKM2 KO, PKM2wt/shPKM1 and PKM2mut/shPKM1 cells after treatment with 3.5 μM XeB for 24 hours (i) or in parental MDA-MB-231, PKM2 KO, PKM2wt/shPKM1 and PKM2mut/shPKM1 cells after treatment with 5 μM XeB for 24 hours (n=3 independent experiments). In a, b, d–g and i–j, data are shown as Mean ±SD and statistics source data are available in Supplementary Table 7. Statistical significance was assessed using two-tailed t-test (a, b, d and e) and ANOVA (f, g, i and j), *p<0.05, **p<0.01. In f (lower panel) and g (lower panel), data represent one of the three independent experiments with similar results. Unprocessed original scans of blots are shown in Supplementary Figure 9.
Inhibiting PKM2 methylation with a nanoparticle-delivered competitive peptide blocks cancer cell metastasis in vivo
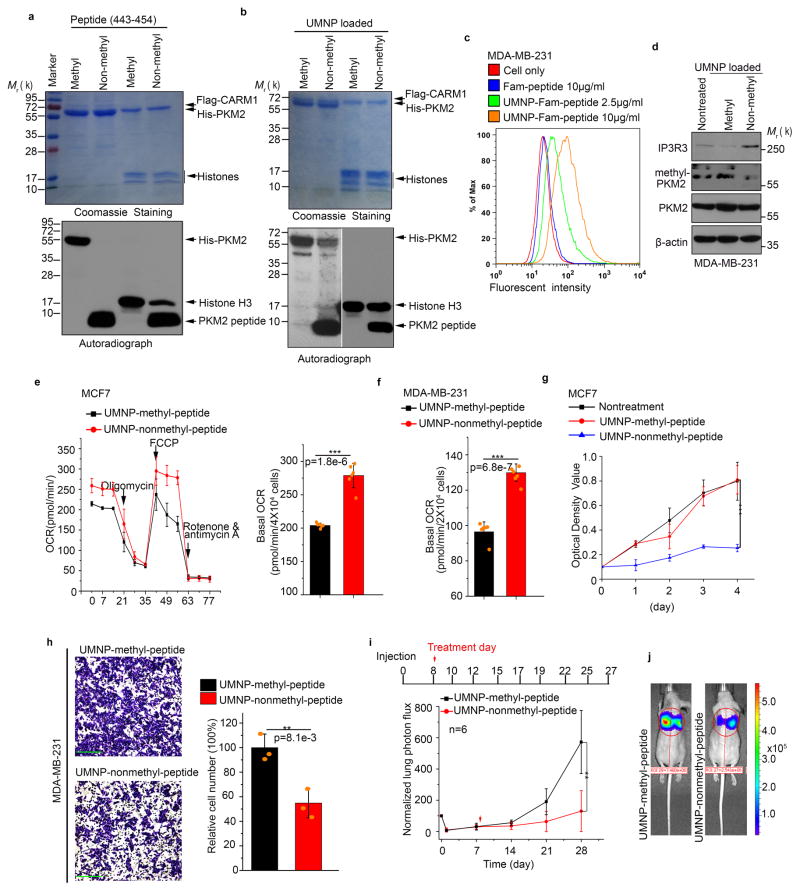
Having established that PKM2 methylation controls ER-mitochondrial Ca2+ signaling and promotes breast cancer cell proliferation and migration, we investigated if PKM2 methylation can be therapeutically targeted. To assess PKM2 methylation dynamics, we estimated the extent of endogenous PKM2 methylation in cancer cells. We used excess methyl-PKM2 antibody for immunoprecipitation and measured the proportion of PKM2 in the supernatant and pellets (i.e., in methylated form) in MCF7 cells. Approximately 40% of endogenous PKM2 was methylated (Supplementary Fig. 8a). This partial PKM2 methylation in cancer cells implies that PKM2 methylation is dynamic and regulatable. Peptide drugs have made huge impacts on cancer treatment35. We evaluated if a competitive, non-methylated PKM2 peptide encompassing the methylation sites could inhibit endogenous PKM2 methylation and reversing aerobic glycolysis to oxidative phosphorylation. As a negative control, we also synthesized a corresponding peptide with R445 and R447 asymmetrically di-methylated. In vitro, the non-methyl-peptide, but not the control methyl-peptide, abrogated CARM1-mediated methylation of PKM2 (Fig. 8a). In contrast, the peptides only partially inhibited methylation of histone H3, a control CARM1 substrate, suggesting that PKM2 is the primary target of inhibition by this peptide.
Figure 8.
Inhibiting PKM2 methylation using a competitive PKM2 peptide reduces proliferation, migration, and lung metastasis of cancer cells due to increased oxidative phosphorylation. (a) In vitro methylation assays showing the inhibitory effects of the methyl- or non-methyl PKM2 peptides on the methylation of PKM2 or a control histone H3 protein. (b) In vitro methylation assays showing the inhibitory effects of the methyl- or non-methyl PKM2 peptides encapsulated in the UMNPs on the methylation of PKM2 or a control histone H3 protein. (c) Flow cytometric measurement of FAM-labelled peptide uptake delivered by UMNP in MDA-MB-231 cells. (d) Western blotting of endogenous PKM2 methylation and the IP3R3 protein levels upon cellular uptake of UMNP-methyl peptide or UMNP-non-methyl peptide. (e, f) Normalized basal OCR values in MCF7 (e) or MDA-MB-231 (f) cells treated with UMNP-methyl-peptide or UMNP-non-methyl peptide (n=6 independent experiments). (g) MTT assays of MCF7 cells under indicated treatment conditions (n=10 independent experiments). (h) The relative migratory cell numbers of MDA-MB-231 cells treated with UMNP-methyl-peptide or UMNP-non-methyl-peptide. Scale bars, 50μm. (n=3 independent experiments). (i) Bioluminescence in lungs of mice treated with UMNP-methyl-peptide or UMNP-non-methyl-PKM2 peptide (n=6 biologically independent mice per group). (j) Representative bioluminescence images of mice after 28 days treatment. The color scale depicts the photon flux (photons per second) emitted from the lung. In e–I, data are shown as Mean ±SD and statistics source data are available in Supplementary Table 7. Significance was assessed using two-tailed t-test (e, f and h) and ANOVA (g and i), **p<0.01, ***p<0.001. In a, b, and d, data represent one of the three independent experiments with similar results. Unprocessed original scans of blots are shown in Supplementary Figure 9.
We employed unimolecular nanoparticle (UMNP) to deliver PKM2 methylation inhibitory peptide in vivo (Supplementary Fig. 8b–f). To ensure that UMNP encapsulation did not alter the PKM2 peptide’s inhibitory activity, we performed PKM2 in vitro methylation assay in the presence of non-methyl-peptide loaded UMNP or methyl-peptide loaded UMNP. The UMNP-non-methyl-peptide, but not UMNP-methyl-peptide, inhibited CARM1-mediated PKM2 methylation (Fig. 8b). Under this condition, histone H3 methylation was not inhibited. The results imply that UMNP-non-methyl-peptide, similar to expressing PKM2mut and TEPP-46, selectively inhibited PKM2 methylation. MDA-MB-231 cells efficiently uptook the 6-carboxyfluorescein-conjugated peptide (FAM-peptide) loaded UMNPs in a dose-dependent manner (Fig. 8c). Additionally, only UMNP-non-methyl peptide inhibited endogenous PKM2 methylation, increasing IP3R3 protein levels (Fig. 8d). Treatment with UMNP-non-methyl-peptide, significantly increased OCR in MCF7 and MDA-MB-231 cells relative to UMNP-methyl-peptide control (Fig. 8e, f). Just as CARM1 KO did not affect PK activity (Fig. S3c), inhibiting PKM2 methylation by UMNP-non-methyl-peptide did not alter PKM2 PK activity (Supplementary Fig. 8g). Moreover, the non-methyl-peptide, but not methyl-peptide, inhibited MCF7 cell proliferation (Fig. 8g) and MDA-MB-231 cell migration (Fig. 8h). To test whether the competitive PKM2 peptide inhibits breast cancer lung metastasis in vivo, LM2 cells36, a metastatic MDA-MB-231 derivative clone, were tail-vein injected into nude mice. While the majority of cells did not survive one day after injection, the remaining cells colonized in the lungs and reached 30–40% of the initial cell numbers by day 7. The mice were treated with UMNPs loaded with methyl-peptide or non-methyl-peptide on day 8 and treatment continued for 3-weeks. LM2 colonization and outgrowth in the lungs of the two cohorts were monitored by bioluminescence imaging23. Bioluminescence intensities in the UMNP-non-methyl peptide-treated group were significantly decreased compared to those in the UMNP-methyl-peptide-treated group, indicating that non-methyl peptide inhibited LM2 lung colonization (Fig. 8i, j). Thus, targeting PKM2 methylation is a feasible therapeutic strategy to reverse oncogenic processes.
DISCUSSION
We show here that reversible PKM2 methylation reprograms cancer metabolism from oxidative phosphorylation to aerobic glycolysis. PKM2 methylation by CARM1 inhibits Ca2+ influx from ER to mitochondria. In breast cancer cells, mitochondrial oxidative phosphorylation dramatically increased upon loss of CARM1, PKM2, or PKM2 methylation, increasing basal mitochondrial [Ca2+] and ΔΨm (Supplementary Fig. 8h). These findings provide mechanistic insights into the metabolic reprogramming controlled by the CARM1-PKM2 axis in breast cancer cells and show that inhibiting PKM2 methylation has therapeutic applications.
PKM2 plays an important role in aerobic glycolysis by distributing glycolytic intermediates for anabolic and catabolic purposes in cancer cells11. Several post-translational modifications of PKM237,38 have been reported to modulate PKM2 function by inhibiting PK activity. However, a recent study challenged PKM2-catalyzed reaction as a rate-limiting step in cancer cell glycolysis15. Consistent with this, PKM2 PK activity was not affected by KO CARM1 (Supplementary Fig. 3c), or by inhibiting PKM2 methylation using non-methyl PKM2 peptide (Supplementary Fig. 8g), suggesting that PKM2 methylation has little effect on its PK activity. Mitochondrial oxidative phosphorylation dramatically increased in CARM1 KO, PKM2 KO or PKM2mut expressing breast cancer cells, suggesting that non-glycolytic function of PKM2 regulates aerobic glycolysis rather than PK activity. Indeed, we found that PKM2 methylation elicits profound effects on energy production by altering mitochondrial oxidative phosphorylation. Notably, mitochondria have well-recognized roles in producing ATP and intermediates for macromolecule biosynthesis in normal and cancerous cells, and are promising chemotherapeutic targets8. In breast cancer cells, knockdown of mitochondrial p32, a critical regulator of tumor metabolism via maintenance of oxidative phosphorylation, shifted metabolism from oxidative phosphorylation to glycolysis, yet tumorigenesis was impaired7, suggesting that high levels of glycolysis without adequate oxidative phosphorylation does not always benefit tumor growth. Thus, our results support the notion that the balance between aerobic glycolysis and mitochondrial respiration is essential for tumor progression.
Cancer cells rely on mitochondria for TCA cycle intermediates to fuel lipid, nucleic acid, and protein biosynthesis essential for growth8. The TCA cycle is regulated by mitochondrial Ca2+, which activates matrix dehydrogenases including pyruvate-, α-ketoglutarate- and isocitrate-dehydrogenases29, to promote oxidative phosphorylation and ATP production39. Mitochondrial Ca2+ is primarily taken from ER at mitochondria-associated ER membrane (MAM) contacts. A minor fraction is from cytosol through low-affinity mitochondrial calcium uniporters (MCUs). Both processes are tightly controlled by IP3Rs, the ubiquitous family of ER Ca2+ release channels40. Interestingly, we found that IP3Rs expression levels are inversely associated with PKM2 expression, and are sensitive to PKM2 methylation, i.e., high IP3Rs in PKM2 methylation defective cells. Accordingly, mitochondrial Ca2+ uptake increases in PKM2-methylation defective cells, activating PDH and increasing oxidative phosphorylation. PKM2 methylation, on the contrary, decreases IP3Rs expression and [Ca2+]mito, increasing PDH phosphorylation and inactivation, decreasing ΔΨm, and switching energy homeostasis from mitochondrial respiration to aerobic glycolysis. We found that methylated PKM2 suppresses the expression of IP3Rs via negatively regulating p53, transcription factor regulating IP3R expression. In addition to controlling IP3R expression, methylated PKM2 co-precipitates with IP3R1 and IP3R3 (Fig. 6f). Thus, methylated PKM2, through regulating IP3Rs expression and interaction, delicately controls Ca2+ uptake by mitochondria.
Mitochondrial Ca2+ addiction was recently identified as a feature of cancer cells41. While inhibiting ER-to-mitochondria Ca2+ transfer creates a bioenergetic crisis in normal and tumor cells, normal cells trigger autophagy to sustain survival, whereas the same autophagic response in tumor cells is insufficient for survival. Tumor cell survival uniquely depends on IP3R-regulated, constitutive ER-to-mitochondria Ca2+ transfer, since inhibiting IP3R activity reduces cancer cell line proliferative potential in vitro and impairs tumor growth in vivo41. Accordingly, increased IP3R expression and/or activity are associated with cancer cell proliferation, survival and invasiveness. All three IP3R subtypes are expressed in breast cancer cells at various levels to regulate intracellular Ca2+ release, which is essential for growth control of these cells41. Although inhibiting PKM2 methylation appears to reduce tumor cell growth, migration and metastasis in various breast cancer cell lines, it is insufficient to alter cell survival (Supplementary Fig. 3j), whereas inhibiting both PKM2 methylation and IP3Rs activity are lethal to cancer cells (Fig. 7h–j). The results underscore gain-of-dependence of cancer cells to IP3R-mediated Ca2+ transport for maintaining cell viability. The acquired mitochondrial addiction to Ca2+ renders cancer cells susceptible to therapies based on inhibiting IP3R (e.g. XeB). Thus, combinatory inhibition of IP3R activity and PKM2 methylation may elicit synergistic therapeutic effects. Targeting cancer-specific metabolism pathways (i.e., aerobic glycolysis and ER-to-mitochondria Ca2+ transfer) should provide new therapeutic avenues for cancer treatment, as exemplified by the UMNP peptide delivery system here.
Methods
Meterials
The antibodies, reagents, shRNAs and primers are listed in the supplementary table 6.
Cell culture and generation of PKM2 knockout cells
MCF7, MDA-MB-231, HEK293T cell lines were purchased from ATCC, LM2 was provided by Dr. Joan Massagué, and immortalized MEF (PKM2fl/fl, Cre-ER) were kindly provided by Dr. Matthew Heiden and were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) purchased from Gibco (Gaithersburg, MD). None of the cell lines used in this study was found in the database of commonly misidentified cell lines that are maintained by ICLAC and NCBI Biosample. Cell lines were authenticated by short tandem repeat profiling and were routinely tested for mycoplasma contamination. PKM2 knockout cell lines were generated using a PKM2 specific CRISPR/Cas9/eGFP plasmid. For PKM2 KO in MEF cells, MEF cells were treated with 1μM 4-hydroxytamoxifen (4-OHT) at least for 8–10 days to allow complete knockout of PKM2.
Virus packaging and stable cell line generation
For lentivirus packaging, three plasmids (PAX2, VSVG and pLKO-shRNA expressing plasmid) were transfected into HEK293T cells. For retrovirus packaging, the three plasmids PHIT60, VSVG and pLNCX-PKM2 expressing plasmid were employed. Supernatant containing the virus was harvested for cell line infection after 48 hours. To generate PKM2 wild type or methylation-defective mutant stably expressing cell lines accompanying with PKM1 knockdown, 2 x 105 PKM2 KO cells were seeded into 6-well plate. For next day’s infection, 1 ml retrovirus was mixed with 1 ml fresh cell culture medium; polybrene was added at a final concentration of 5 μg/ml to increase the infection efficiency. On the second day, cells were cultured with fresh medium containing 400 μg/ml G418 (or 200 μg/ml G418 for MDA-MB-231) for at least 4 weeks to obtain the PKM2 wild type or methylation-defective mutant stably expressing cell lines. To knockdown PKM1, the above cell lines were infected with pLKO-PKM1 shRNA lentivirus and selected with 2 μg/ml puromycin for at least 2 weeks. The expression of PKM2 in stable cell lines were individually examined by western blotting.
Co-immunoprecipitation
Co-immunoprecipitation was performed as previously described42.
In vitro methylation assay
In vitro methylation assay was performed as previously described42.
Quantitative Real-time PCR
q-RT PCR was performed as described42.
Gel Filtration
Recombinant His-tagged PKM2 protein was incubated with TEPP-46 (10 μM) for 1 hour on ice, then separated in the Superdex 200 Increase 10/300 GL column (GE Healthcare) in 0.01 M phosphate buffer and 0.14 M NaCl at pH 7.4. The speed rate of flow is 0.5 ml/min. 0.3 ml fractions were collected and analyzed by UV absorbance or SDS-PAGE and Western blot.
Pyruvate Kinase and Lactate Dehydrogenase Assays
Pyruvate kinase activity was measured as described43. Briefly, 2 μg whole cell lysate was incubated in 1 ml buffer (Tris pH 7.5 (50 mM), KCl (100 mM), MgCl2 (5 mM), ADP (0.6 mM), PEP (0.5 mM), NADH (180 μM), and LDH (8 units). The change in absorbance at 340 nm owing to the oxidation of NADH was measured using a Nano drop ND-2000 1-position spectrophotometer (Thermo). LDH activity was determined by measuring the decreased fluorescence intensity at 340nm from the NADH oxidation in buffer (Tris pH 7.5 (50 mM), KCl (100 mM), MgCl2 (5 mM), pyruvate (20 mM), NADH (180 μM).
Subcellular Fractionation
The isolation of cytosol and mitochondria was conducted as described44. Briefly, the cell pellets were resuspended in 11 mL ice-cold RSB hypo buffer (10 mM NaCl 1.5 mM MgCl2 10 mM Tris-HCl (pH 7.5)) and allow the cells to swell for 5–10min, then the swollen cells were broken open with several strokes in the presence of 8mL of 2.5× MS homogenization buffer (525 mM mannitol, 175 mM sucrose, 12.5 mM Tris-HCl, 2.5 mM EDTA, pH 7.5) to give a final concentration of 1× MS homogenization buffer. The homogenate was centrifuged at 1300g for 5 min and repeated for several times. After centrifugation at 7,000g for 15 min, the supernatant (cytosolic fraction) and the crude mitochondria fraction were separated. The pellet was re-suspended with 1× MS homogenization buffer followed by the 7,000g sedimentation several times.
Confocal Imaging
MAD-MB-231 cells were fixed with 4% paraformaldehyde in culture media for 15 min at 37°C and permeabilized with 0.2% Triton X-100 for 10 min at room temperature. The nonspecific binding was blocked by incubation with 4% BSA in PBS for 60 min, and cells were subsequently stained with primary PKM2 and HSPA9 antibodies overnight at 4 °C. The slides were washed in PBS three times (5min/each time) and were incubated for 1 hour with the following secondary antibodies: FITC-conjugated goat anti-mouse IgG and Dylight®594 conjugated goat anti-rabbit IgG. After being washed three time in PBS and air-dried, the coverslips were mounted in ProLong Gold anti-fade reagent with DAPI (Invitrogen). Fluorescence was examined using a Leica SP8 3X STED Super-resolution microscope (Buffalo Grove, IL) equipped with a 63x objective lens with laser excitation at 405nm, 488nm or 592nm. For z-stack analysis, optical sections were obtained along the z-axis at 0.5-μm intervals. Images were analyzed with the ImageJ software.
Expression and purification of recombinant proteins
Human PRMT1-8 cDNAs were cloned into pFN21K HaloTag CMV Flexi Vector (Promega) and the corresponding proteins were purified as described42.
GST Pull-down Assays
GST and GST-CARM1 proteins were expressed in E. coli BL-21 competent cells and purified by glutathione sepharose 4B resin (GE Healthcare Life Sciences). Recombinant, Flag-tagged PKM2 proteins were purified from HEK 293T cells. GST pull down experiment was conducted as described42.
In vitro Protein-Protein Interaction Assay
Flag-tagged full length PKM2 and its truncations constructs were translated by T7 Quick Coupled Translation / Transcription system (Promega). Interaction with GST-CARM1 fusion protein (1μg/mL) was conducted as described42.
Cell Proliferation and DNA synthesis assays
MTT assay was conducted as described42. For 5-Ethynyl-2′-deoxyuridine (EdU) incorporation assay, 3x105 cells were seeded into 6-well plate and incubated with 10μM EdU for 1 hour, followed by procedures described in the Click-iT® EdU cytometry assay kit on a BD LSRII. For clonogenicity assay, 1000 viable transfected cells were cultured in six-well plates for 2 weeks. Colonies were washed with PBS and fixed with 3.7% formaldehyde at room temperature for 15 min, then stained for 20 min with 0.05 % crystal violet.
Transwell Cell Migration Assays
Transwell cell migration assay was performed as described42. Briefly, 1x105 cells in 100μL serum free media were added into the upper chamber, 500μL culture media with 20% FBS was in the lower well. After 12-hour culture, cells on the upper surface of the membrane were removed and migrant cells on the lower surface were fixed with 3.7% formaldehyde in culture media at 37°C for 15 min, and then stained with 0.05 % crystal violet for 20 min.
Generation of methylated PKM2 (methyl-PKM2) specific antibody
Methyl-PKM2 specific anti-peptide antibody was generated by Genemed Synthesis Inc., TX. The KLH-conjugated PKM2 peptide RYR(as)PR(as)APIIAVTC, with R445 and R447 asymmetrically dimethylated was synthesized. This peptide corresponding to human PKM2 (aa 443-454) was used to immunize rabbits. Purification of antibody was conducted as described42.
Measurement of Oxygen Consumption Rate
The OCR was measured in an XF96 extracellular analyzer (Seahorse Bioscience). 2x 104 cells per well were seeded into the 96-well plate and incubated in DMEM media with 10% FBS at 37°C. On the next day, the media was changed to analysis media containing 10mM glucose. The cells were incubated in a CO2-free incubator at 37°C for 1 hour. Cells were sequentially exposed to oligomycin (1 μM), FCCP (1 μM) and rotenone (0.5μM). Each point in traces represent the average of measurement from six different wells.
Measurement of Glucose Uptake
Cells were washed and subsequently studied in a modified balanced salt solution (MBSS) containing (in mM): 140 NaCl, 5.4 KCl, 0.5 MgCl2, 0.4 MgSO4, 3.3 NaHCO3, 2.0 CaCl2, 10 HEPES, 5.5 glucose, pH 7.4, 2-NBDG (0.1–0.3 mM) was added to the bathing media, after 20min incubation and several washes, uptake of 2-NBDG was measured by fluorescence spectrometry.
Mass Spectrometry Analysis of Arginine Methylation
In gel digestions
Experiments were performed with previously described protocol with modifications1. Briefly, the gel was destained twice with 100 mM (NH4)HCO3/50% Methanol, followed by dehydration with 25 mM (NH4)HCO3 / 50% Acetonitrile and 100% Acetonitrile. The gel particles were rehydrated with freshly prepared 25 mM dithiothreitol at 56°C. The proteins were then alkylated with 55 mM iodoacetamide in the dark. The gel pieces were incubated with 50 ng of trypsin in 25 mM (NH4)HCO3 / 0.01% ProteasMAXTM (Promega) at 42°C. The tryptic peptides were extracted by adding 2.5% TFA solution.
Samples were analyzed on a Waters nanoAcquity UPLC system coupled to a Q-Exactive quadrupole orbitrap mass spectrometer (Thermo). Tryptic peptides were loaded onto the Waters Symmetry C18 trap column (180 μm x 20 mm, 5 μm) and separated over a 70min gradient on the fabricated column (150 mm x 75 μm, 1.7 μm). The data was acquired under data dependent acquisition mode (DDA, top 20). Mass spectrometric conditions were as follows: normalized high-energy collision dissociation (HCD) 30%, 70K and 17.5K resolution for full scan and MS/MS scan, automatic gain control (AGC) of 2e5, maximum ion injection time of 100 ms, isolation window of 2 m/z, and fixed first mass of 100 m/z.
Mass spectral data were searched in PEAKS Studio (Bioinformatics) against a home-made FASTA database containing the PKM2 protein sequence. Trypsin was selected and two missed cleavage sites were allowed. PEAKS searches were performed with a precursor mass tolerance of 20 ppm and a fragment mass tolerance of 0.05 Da. The fixed modifications consisted of carbamidomethylation (57.0215 Da) of cysteine residues. The variable modifications consisted of dimethylation (28.0313 Da) of arginine residues and oxidation (15.9949 Da) of methionine. Peptide spectral matches were validated based on p-value of 1% false discovery rate.
Quantitative proteomics analysis
Cells were lysed in lysis buffer containing 8 M urea, 50 mM Tris-HCl, 30 mM NaCl, and protease inhibitor tablet. The lysates were homogenized and processed by centrifugation. Protein content were determined by bicinchoninic acid (BCA) assay (Thermo). Protein extract was diluted to a urea concentration of 0.9 M and subjected to digestion with trypsin. The samples were quenched with 10% TFA and desalted with Sep-Pak C18 cartridge (Waters).
DiLeu labeling was performed according to previous studies2. Briefly, each DiLeu tag was dissolved in anhydrous DMF and activated with DMTMM and NMM. The supernatant was used immediately to label the peptides and reaction was quenched with hydroxylamine. Samples were pooled at 1:1 ratio across all the channels and dried in vacuo.
Strong cation exchange (SCX) fractionation was performed on a Waters Alliance e2695 HPLC (Milford, MA) with a flow rate of 0.2 ml/ min. Tryptic peptides were loaded onto a polySULFOETHYL A (200 mm × 2.1 mm, 5 μm) column. The samples were offline separated over 90min gradient, collected every 1.5 min and concatenated into 10 fractions.
All samples were reconstituted in 0.1% FA, 3% ACN and loaded onto the fabricated column (150 mm x 75 μm, 1.7 μm). Peptides were online separated with Dionex UltiMate 3000 LC system before entering the Orbitrap Fusion Lumos tribrid mass spectrometer (Thermo). Survey scan (300–1500 m/z) were acquired at resolution of 60K, followed by top 15 HCD fragmentation (normalized collision energy 30, isolation window 1 m/z). 2×105 and 5×105 were selected as the AGC target for MS and MS/MS scans, respectively. The maximum injection time was set to 100 ms.
The OMSSA proteomic Analysis Software Suite (COMPASS) was used for peptide identification. Raw files were searched against the Homo sapiens Uniprot database (December 2015). Trypsin was selected as the enzyme and maximum of two missed cleavages were allowed. Precursor and fragment ion tolerance was set to 25 ppm and 0.02 Da. DiLeu labeling on peptide N-termini and lysine residue (+145.1267748 Da), and carbamidomethylation of cysteine residues (+57.02146 Da) were chosen as static modifications. Methionine oxidation (+15.99492 Da) and DiLeu labeling on tyrosine residue (+145.1267748) were selected as variable modifications. Search results were filtered to 1% false discovery rate (FDR) at both peptide and protein levels. Quantification was performed using an in-house software called DiLeu tool. Reporter ion abundances are corrected for isotope impurities with python script.
Cellular metabolite extraction and analysis
Global metabolomic analysis of nucleotides was performed by Metabolon Inc. (Durham, NC, USA) as described45.
Measurement of Mitochondrial Membrane Potential (ΔΨ) and DNA content
Cells was loaded with JC-1 (10μg/mL) for 10 min or Tetramethylrhodamine, Ethyl Ester (TMRE) for 20 min in phenol red free media at 37°C followed by three washes with PBS. Trypsinized cells were subjected to flow cytometry analysis. The relative quantification of mitochondrial DNA levels were determined by the ratio of mitochondrial tRNA Leu to the nuclear-encoded B2-microglobulin 46.
Measurement of Calcium Flux
Cell were loaded with Rhod2 dye (MCF7 (1μM), MDA-MB-231 and MEFs (0.45μM)) and 0.002% Pluronic F-127 and incubated for 10 minutes at room temperature, then changed the buffer with Rhod2 free buffer and further incubated 30 minutes. Calcium imaging was carried out by confocal microscope at 561 nm excitation using a ×63 oil objective. Images were analyzed and quantified using ImageJ (NIH).
Cell apoptosis and death analyses
Cell apoptosis and death assays were conducted as described47. Cell apoptosis was determined by annexin V/APC and propidium iodide (PI) staining. Cell death was determined by PI staining using flow cytometry.
Unimolecular nanoparticle (UMNP) synthesis
β-benzyl L-aspartate N-carboxyanhydride (BLA-NCA) monomer, poly (β-benzyl L-aspartate)-poly(ethylene glycol) (PBLA-PEG) block copolymers (i.e., PBLA-mPEG and PBLA-PEG-Maleimide (Mal)), as well as poly(amidoamine)-poly(β-benzyl L-aspartate)-poly(ethylene glycol)-OCH3/Mal (PAMAM-PBLA-PEG-Mal) were all synthesized following the methods as previously reported 48, 49. 1H NMR (400 MHz, CDCl3). BLA-NCA: 7.42–7.28 (5H, m, Ar-H), 6.30 (1H, s, NH), 5.20 (2H, s, CH2-Ar), 4.6 (1H, t, CH), and 2.9 (2H, t, COCHCH2) ppm. 1H NMR (400 MHz, DMSO-D6). PBLA-mPEG: 7.26–7.38 (102H, m, Ar-H), 5.00–5.10 (40H, s, CH2-Ar), 4.55–4.68 (20H, m, COCHCH2), 3.35–3.53 (450H, m, CH2CH2O from PEG), and 2.48–2.90 (41H, m, COCHCH2) ppm. PBLA-PEG-Mal: 7.26–7.38 (102H, m, Ar-H), 6.95 (2H, s, Mal), 5.00–5.10 (40H, s, CH2-Ar), 4.55–4.68 (20H, m, COCHCH2), 3.35–3.53 (450H, m, CH2CH2O from PEG), and 2.48–2.90 (41H, m, COCHCH2) ppm. PAMAM-PBLA-PEG-Mal: 7.30–7.10 (105H, m, Ar-H), 6.90 (0.5H, s, Mal), 4.90–5.00 (40H, s, CH2-Ar), 4.60–4.50 (20H, m, COCHCH2), 3.22–3.50 (450H, m, CH2CH2O from PEG), and 2.50–2.80 (41H, m, COCHCH2) ppm.
To synthesize poly(amidoamine)-poly(aspartate diethyltriamine)-poly(ethylene glycol)-OCH3/Mal (PAMAM-PAsp(DET)-PEG-Mal), PAMAM-PBLA-PEG-Mal (20 mg) was dissolved in 5 ml DMF. Diethyltriamine (224 μl) was added to the solution dropwise at 4 °C and the resulting mixture was stirred at room temperature for 4 h. Thereafter, it was added in 10 mL DI water, neutralized by 1 M HCl, and dialyzed against DI water (MWCO =15 kDa) for 48 h. 1H NMR (400 MHz, D2O): 6.90 (0.5H, s, Mal), 4.80–4.50 (20H, s, COCHCH2), 4.00–3.60 (450H, m, CH2CH2O from PEG), 3.30 (40H, s, CONHCH2), 3.10–2.98 (81H, m, CH2NHCH2), 2.75 (41H, CH2NH2), and 2.40-2.25 (m, 84H, COCHCH2) ppm.
To synthesize poly(amidoamine)-poly(aspartate diethyltriamine-r-imidazole)-poly(ethylene glycol)-OCH3/Mal (PAMAM-PAsp(DET-r-Im)-PEG-Mal), PAMAM-PAsp(DET)-PEG-Mal (20 mg), 4-imidazolecarboxylic acid (1.13 mg), 1,3-dicyclohexyl-carbodiimide (2.06 mg), and N-hydroxysuccinimide (1.15 mg) were dissolved in 5 ml DMF and stirred at room temperature for 24 h at pH 6.5 followed by dialysis against DI water (MWCO 15 kDa) for 48 h. 1H NMR (400 MHz, D2O): 8.24-8.22 (5H, s, Im), 7.53–7.52 (5H, d, Im), 6.90 (0.5 H, s, Mal), 4.80–4.50 (20H, s, COCHCH2), 4.00–3.50 (450H, m, CH2CH2O from PEG), 3.30 (40H, s, CONHCH2), 3.10–2.98 (80H, m, CH2NHCH2), 2.75 (40H, CH2NH2), and 2.70-2.50 (m, 81H, COCHCH2) ppm.
Poly(amidoamine)-poly(aspartate diethyltriamine-aconitic acid-r-imidazole)-poly(ethylene glycol)-OCH3/Mal (PAMAM-PAsp(DET-Aco-r-Im)-PEG-Mal) was synthesized using PAMAM-PAsp(DET-r-Im)-PEG/Mal following a previously reported method.50 1H NMR (400 MHz, D2O): 8.24-8.22 (5H, s, Im), 7.53–7.52 (5H, s, Im), 6.90 (0.5H, s, Mal), 5.82-5.72 (15H, s, COCHC(COOH)CH2COOH), 4.80–4.50 (20H, s, COCHCH2), 4.00–3.50 (450H, m, CH2CH2O from PEG), 3.30 (40H, s, CONHCH2), 3.10–2.98 (82H, m, CH2NHCH2), 2.75 (41H, CH2NH2), 2.70-2.50 (m, 81H, COCHCH2), 1.80 (30H, s, COCHC(COOH)CH2COOH) ppm.
To synthesize PAMAM-PAsp(DET-Aco-r-Im)-PEG-TAT, PAMAM-PAsp(DET-Aco-r-Im)-PEG (20 mg), TAT (sequence CYGRKKRRQRRR, 0.3 mg) and tris(2-carboxyethyl)phosphine hydrochloride (0.57 mg) were dissolved in PBS buffer (pH 7.4, 5 ml). After 24 h stirring, the solution was purified by dialysis against DI water (MWCO 15 kDa). 1H NMR (400 MHz, D2O). 8.24-8.22 (5H, s, Im), 7.55–7.30 (15H, m, Im and TAT), 5.82-5.72 (15H, s, COCHC(COOH)CH2COOH), 4.80–4.50 (20H, s, COCHCH2), 4.00–3.50 (450H, m, CH2CH2O from PEG), 3.30 (40H, s, CONHCH2), 3.10–2.98 (82H, m, CH2NHCH2), 2.75 (41H, CH2NH2), 2.70-2.50 (m, 81H, COCHCH2), 1.80 (30H, s, COCHC(COOH)CH2COOH) ppm.
To prepare PKM2 peptide-loaded UMNPs, 6-carboxyfluorescein (FAM)-conjugated PKM2 peptide (FAM-PKM2 peptide, 0.5 mg) was dissolved in 0.5 ml DI water under stirring, while the PAMAM-PAsp(DET-Aco-r-Im)-PEG-TAT polymer (UMNP, 2 mg) was dissolved in 1 ml DI water at pH 7. The UMNP solution was slowly added to the peptide solution under stirring at room temperature for 4 h and dialyzed against DI water (MWCO = 100 kDa). The peptide loading level was determined by a UV-Vis spectrometer (Cary 5000 UV-Vis-NIR, Agilent Technologies) with the absorbance of FAM at 495 nm. UMNP-non-methyl peptide and UMNP-methyl peptide without FAM conjugation were prepared using the same method.
Characterization: The 1 H NMR spectra were collected on a Bruker Advance 400 NMR spectrometer. The hydrodynamic size distribution and zeta-potential of the UMNPs were characterized using a dynamic light scattering (DLS) spectrometer (Malvern Zetasizer Nano ZS) at a polymer concentration of 0.1 mg/ml.
Principle of PKM2 peptide delivery using UMNP: Positively charged PKM2 peptides were loaded onto the charge conversional polyanionic (PAsp(DET-Aco-r-Im)) segments through electrostatic interactions 51. Under neutral pH (e.g., the blood stream), the PAsp (DET-Aco-r-Im) segments carried negative charges, allowing for the complexation of the PKM2 peptides. Once the UMNPs were endocytosed by target cancer cells the polyanionic PAsp(DET-Aco-r-Im) segments were converted to polycationic PAsp(DET-r-Im) segments in the acidic endocytic compartments due to the acid-induced cleavage of the aconitic acid side groups, thereby facilitating the release of PKM2 peptides. The imidazole functional groups were incorporated into the PAsp(DET-Aco-r-Im) segments to enhance the endosomal escape capability, thereby preventing potential damage of the peptides in the acidic endosomes/lysosomes52. A cell penetrating peptide TAT was conjugated onto the surfaces of the nanocarriers to enhance their cellular uptake53.
Animal Experiments
All animal work was performed in accordance with protocols approved by Research Animal Resource Center of UW-Madison and that the study was compliant with ethical regulations regarding animal research. Balb/c nude female mice at 4–6 week old were used for all xenograft experiments (Harlan, Madison, WI). For xenograft tumors assays, 1x106 cells (MDA-MB-231 PKM2wt/shPKM1 or PKM2mut/shPKM1 cells) were injected into the inguinal mammary fat pads of nude mice (n=6 per group). Tumor size was determined using caliper measurement and the tumor volume was calculated using the formula 1/2 × L × W2. For lung metastasis assays, 1×105 LM2, a metastatic MDA-MB-231 derivative clone, was resuspended in 0.1 ml PBS and tail-vein injected into mice. Mice were imaged for luciferase activity immediately after injection (day 0) to exclude any mouse that were not successfully xenografted. Luciferase-based noninvasive bioluminescent imaging and analysis were performed as described54 using an IVIS Imaging System (Caliper Life Sciences, Hopkinton, MA). Briefly, mice were anaesthetized and injected intraperitoneally with 2 mg D-luciferin (20 mg/ml in PBS) (Gold Biotechnology). Imaging was completed between 5 to 15 min after injection. For bioluminescence plots, total photon flux was calculated for each mouse in the gated areas. Then, the mice were retro-orbitally injected with UMNP-methyl-peptide or UMNP-non-methyl-peptide (100μl, 1g/L) at the indicated time interval. Imaging was taken every week and endpoint assays were conducted 4 weeks after injection.
Statistics and reproducibility
Statistical testing was performed using the unpaired two-tailed Student’s t-test and/or ANOVA analysis. All experiments were repeated at least three times unless otherwise indicated. N numbers are indicated in the figure legends. p value < 0.05 (*) was considered as statistically significant.
Data availability
Mass spectrometry data have been deposited in ProteomeXchange with the primary accession code PXD007671 [http://www.ebi.ac.uk/pride/archive/]55. Global metabolomic nucleotides data have been deposited in MetaboLights with the primary accession code MTBLS533 [http://www.ebi.ac.uk/metabolights/index]56. The RNA-seq data of 1,093 human breast tumor samples were derived from the TCGA Research Network: http://cancergenome.nih.gov/ and it is available in [Fire Browse (http://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/BRCA/20160128/gdac.broadinstitute.org_BRCA.Merge_rnaseqv2__illuminahiseq_rnaseqv2__unc_edu__Level_3__RSEM_genes_normalized__data.Level_3.2016012800.0.0.tar.gz)]. The proteomics data of 77 human breast tumour samples were derived from the CPTAC Research Network (https://proteomics.cancer.gov/programs/cptac) and it is available in [http://prot-shiny-vm.broadinstitute.org:3838/BC2016/]. Data related to CARM1 or PKM2 interacting proteins was provided in Supplementary Table 1, 4 and 5. Source data for Figs 3a, d, g, i, 4a–c, e–g, 5b, c, f–i, p, q, 7a, b, d–g, i, j, 8e–i and Supplementary Fig. 1a–f, 2b–d, f, g, 4a–l, 5a, b, 6i and 8g have been provided in Supplementary Table 7. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments
We thank Dr. Matthew G. Vander Heiden for kindly providing the immortalized MEF (PKM2fl/fl, Cre-estrogen receptor) cells, Dr. Joan Massagué for kindly providing the LM2 cells, Dr. Nathan Sherer for fluorescence microscopy, Drs. Carlos Coriano and Jing Fan for comments, Dr. Paul Ahlquist for editing. This project is supported by NCI RO1 CA213293 to W.X. and R21 CA196653 to W.X and S.Q.G and supported in part by NIH/NCI P30CA014520 -UW Carbone Cancer Center Grant, and NIH R01 DK071801 to L.L., S10RR029531, and P41GM108538.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests
AUTHOR CONTRIBUTIONS
W.X. and F.B.L. conceived the project, designed the experiments, analyzed the data and wrote the manuscript. F.B.L. performed the experiments with assistance from H.Z., B.B.L., J.Y.J. and Y.D.W. F.F.M., H.L and C.X.J. performed the mass spectrometry experiments; Y.Y.W., G.J.C. and S.Q.G. designed the nanoparticles. P.L. and I.M.O. performed bioinformatics analyses; W.X., L.J.L. and S.Q.G. directed and supervised all aspects of the study, all authors discussed the results and commented on the manuscript.
References
- 1.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 2.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 4.Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 5.Guo JY, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinberg F, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fogal V, et al. Mitochondrial p32 protein is a critical regulator of tumor metabolism via maintenance of oxidative phosphorylation. Mol Cell Biol. 2010;30:1303–1318. doi: 10.1128/MCB.01101-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinberg SE, Chandel NS. Targeting mitochondria metabolism for cancer therapy. Nat Chem Biol. 2015;11:9–15. doi: 10.1038/nchembio.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noguchi T, Inoue H, Tanaka T. The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J Biol Chem. 1986;261:13807–13812. [PubMed] [Google Scholar]
- 10.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 11.Tamada M, Suematsu M, Saya H. Pyruvate kinase M2: multiple faces for conferring benefits on cancer cells. Clin Cancer Res. 2012;18:5554–5561. doi: 10.1158/1078-0432.CCR-12-0859. [DOI] [PubMed] [Google Scholar]
- 12.Keller KE, Tan IS, Lee YS. SAICAR stimulates pyruvate kinase isoform M2 and promotes cancer cell survival in glucose-limited conditions. Science. 2012;338:1069–1072. doi: 10.1126/science.1224409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z, Yang P, Li Z. The multifaceted regulation and functions of PKM2 in tumor progression. Biochim Biophys Acta. 2014;1846:285–296. doi: 10.1016/j.bbcan.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 15.Xie J, Dai C, Hu X. Evidence That Does Not Support Pyruvate Kinase M2 (PKM2)-catalyzed Reaction as a Rate-limiting Step in Cancer Cell Glycolysis. J Biol Chem. 2016;291:8987–8999. doi: 10.1074/jbc.M115.704825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosios AM, Fiske BP, Gui DY, Vander Heiden MG. Lack of Evidence for PKM2 Protein Kinase Activity. Mol Cell. 2015;59:850–857. doi: 10.1016/j.molcel.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 18.Kawabe Y, Wang YX, McKinnell IW, Bedford MT, Rudnicki MA. Carm1 regulates Pax7 transcriptional activity through MLL1/2 recruitment during asymmetric satellite stem cell divisions. Cell Stem Cell. 2012;11:333–345. doi: 10.1016/j.stem.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hein K, et al. Site-specific methylation of Notch1 controls the amplitude and duration of the Notch1 response. Sci Signal. 2015;8:ra30. doi: 10.1126/scisignal.2005892. [DOI] [PubMed] [Google Scholar]
- 20.Shishkova E, et al. Global mapping of CARM1 substrates defines enzyme specificity and substrate recognition. Nat Commun. 2017;8:15571. doi: 10.1038/ncomms15571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frietze S, Lupien M, Silver PA, Brown M. CARM1 regulates estrogen-stimulated breast cancer growth through up-regulation of E2F1. Cancer Res. 2008;68:301–306. doi: 10.1158/0008-5472.CAN-07-1983. [DOI] [PubMed] [Google Scholar]
- 22.Cheng H, et al. Overexpression of CARM1 in breast cancer is correlated with poorly characterized clinicopathologic parameters and molecular subtypes. Diagn Pathol. 2013;8:129. doi: 10.1186/1746-1596-8-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, et al. CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis. Cancer Cell. 2014;25:21–36. doi: 10.1016/j.ccr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cardenas C, et al. Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell. 2010;142:270–283. doi: 10.1016/j.cell.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J, Bedford MT. PABP1 identified as an arginine methyltransferase substrate using high-density protein arrays. EMBO Rep. 2002;3:268–273. doi: 10.1093/embo-reports/kvf052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anastasiou D, et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012;8:839–847. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lunt SY, et al. Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Mol Cell. 2015;57:95–107. doi: 10.1016/j.molcel.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patra KC, Hay N. The pentose phosphate pathway and cancer. Trends Biochem Sci. 2014;39:347–354. doi: 10.1016/j.tibs.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rizzuto R, De Stefani D, Raffaello A, Mammucari C. Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol. 2012;13:566–578. doi: 10.1038/nrm3412. [DOI] [PubMed] [Google Scholar]
- 30.Patel MS, Korotchkina LG. Regulation of the pyruvate dehydrogenase complex. Biochem Soc Trans. 2006;34:217–222. doi: 10.1042/BST20060217. [DOI] [PubMed] [Google Scholar]
- 31.Qattan AT, Radulovic M, Crawford M, Godovac-Zimmermann J. Spatial distribution of cellular function: the partitioning of proteins between mitochondria and the nucleus in MCF7 breast cancer cells. J Proteome Res. 2012;11:6080–6101. doi: 10.1021/pr300736v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koboldt DC, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mertins P, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez Y, et al. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat Commun. 2014;5:5812. doi: 10.1038/ncomms6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thundimadathil J. Cancer treatment using peptides: current therapies and future prospects. J Amino Acids. 2012;2012:967347. doi: 10.1155/2012/967347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hitosugi T, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anastasiou D, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Territo PR, Mootha VK, French SA, Balaban RS. Ca(2+) activation of heart mitochondrial oxidative phosphorylation: role of the F(0)/F(1)-ATPase. Am J Physiol Cell Physiol. 2000;278:C423–435. doi: 10.1152/ajpcell.2000.278.2.C423. [DOI] [PubMed] [Google Scholar]
- 40.Foskett JK, White C, Cheung KH, Mak DO. Inositol trisphosphate receptor Ca2+ release channels. Physiol Rev. 2007;87:593–658. doi: 10.1152/physrev.00035.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cardenas C, et al. Selective Vulnerability of Cancer Cells by Inhibition of Ca(2+) Transfer from Endoplasmic Reticulum to Mitochondria. Cell Rep. 2016;14:2313–2324. doi: 10.1016/j.celrep.2016.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L, et al. CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis. Cancer Cell. 2014;25:21–36. doi: 10.1016/j.ccr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 44.Clayton DA, Shadel GS. Isolation of mitochondria from tissue culture cells. Cold Spring Harb Protoc. 2014;2014 doi: 10.1101/pdb.prot080002. pdb prot080002. [DOI] [PubMed] [Google Scholar]
- 45.Kanaan YM, et al. Metabolic Profile of Triple-negative Breast Cancer in African-American Women Reveals Potential Biomarkers of Aggressive Disease. Cancer Genom Proteom. 2014;11:279–294. [PubMed] [Google Scholar]
- 46.Bai RK, Wong LJ. Simultaneous detection and quantification of mitochondrial DNA deletion(s), depletion, and over-replication in patients with mitochondrial disease. J Mol Diagn. 2005;7:613–622. doi: 10.1016/S1525-1578(10)60595-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cardenas C, et al. Selective Vulnerability of Cancer Cells by Inhibition of Ca(2+) Transfer from Endoplasmic Reticulum to Mitochondria. Cell Rep. 2016;14:2313–2324. doi: 10.1016/j.celrep.2016.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prabaharan M, Grailer JJ, Pilla S, Steeber DA, Gong S. Amphiphilic multi-arm-block copolymer conjugated with doxorubicin via pH-sensitive hydrazone bond for tumor-targeted drug delivery. Biomaterials. 2009;30:5757–5766. doi: 10.1016/j.biomaterials.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 49.Chen G, Wang Y, Xie R, Gong S. Tumor-targeted pH/redox dual-sensitive unimolecular nanoparticles for efficient siRNA delivery. Journal of Controlled Release. 2017 doi: 10.1016/j.jconrel.2017.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee Y, et al. Charge-Conversion Ternary Polyplex with Endosome Disruption Moiety: A Technique for Efficient and Safe Gene Delivery. Angewandte Chemie. 2008;120:5241–5244. doi: 10.1002/anie.200800963. [DOI] [PubMed] [Google Scholar]
- 51.Pittella F, et al. Enhanced endosomal escape of siRNA-incorporating hybrid nanoparticles from calcium phosphate and PEG-block charge-conversional polymer for efficient gene knockdown with negligible cytotoxicity. Biomaterials. 2011;32:3106–3114. doi: 10.1016/j.biomaterials.2010.12.057. [DOI] [PubMed] [Google Scholar]
- 52.Chang KL, Higuchi Y, Kawakami S, Yamashita F, Hashida M. Efficient Gene Transfection by Histidine-Modified Chitosan through Enhancement of Endosomal Escape. Bioconjugate Chemistry. 2010;21:1087–1095. doi: 10.1021/bc1000609. [DOI] [PubMed] [Google Scholar]
- 53.Ziegler A, Nervi P, Durrenberger M, Seelig J. The cationic cell-penetrating peptide CPP(TAT) derived from the HIV-1 protein TAT is rapidly transported into living fibroblasts: optical, biophysical, and metabolic evidence. Biochemistry. 2005;44:138–148. doi: 10.1021/bi0491604. [DOI] [PubMed] [Google Scholar]
- 54.Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vizcaino JA, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haug K, et al. MetaboLights--an open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Res. 2013;41:D781–786. doi: 10.1093/nar/gks1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry data have been deposited in ProteomeXchange with the primary accession code PXD007671 [http://www.ebi.ac.uk/pride/archive/]55. Global metabolomic nucleotides data have been deposited in MetaboLights with the primary accession code MTBLS533 [http://www.ebi.ac.uk/metabolights/index]56. The RNA-seq data of 1,093 human breast tumor samples were derived from the TCGA Research Network: http://cancergenome.nih.gov/ and it is available in [Fire Browse (http://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/BRCA/20160128/gdac.broadinstitute.org_BRCA.Merge_rnaseqv2__illuminahiseq_rnaseqv2__unc_edu__Level_3__RSEM_genes_normalized__data.Level_3.2016012800.0.0.tar.gz)]. The proteomics data of 77 human breast tumour samples were derived from the CPTAC Research Network (https://proteomics.cancer.gov/programs/cptac) and it is available in [http://prot-shiny-vm.broadinstitute.org:3838/BC2016/]. Data related to CARM1 or PKM2 interacting proteins was provided in Supplementary Table 1, 4 and 5. Source data for Figs 3a, d, g, i, 4a–c, e–g, 5b, c, f–i, p, q, 7a, b, d–g, i, j, 8e–i and Supplementary Fig. 1a–f, 2b–d, f, g, 4a–l, 5a, b, 6i and 8g have been provided in Supplementary Table 7. All other data supporting the findings of this study are available from the corresponding author on reasonable request.