Summary
Adult stem and progenitor cells are critical to replenishing lost tissue due to injury or normal turnover. How these cells maintain self-renewal and sustain the tissue they populate is an area of active investigation. Here we show that the cohesin complex, which has previously been implicated in regulating chromosome segregation and gene expression, is necessary to promote epidermal stem and progenitor cell self-renewal through cell autonomous mechanisms. Cohesin binds to genomic sites associated with open chromatin including DNase I hypersensitivity sites, RNA polymerase II, and histone marks such as H3K27ac and H3K4me3. Reduced cohesin expression results in spontaneous epidermal differentiation due to loss of open chromatin structure and expression of key self-renewal genes. Our results demonstrate a prominent role for cohesin in modulating chromatin structure to allow for enforcement of a stem and progenitor cell gene expression program.
Keywords: Stem Cell, Differentiation, Epidermis, Skin Differentiation, Cohesin, SMC1A, SMC3, STAG1, STAG2, CTCF, RAD21, Gene Regulation, DNASE, Chromatin Accessibility, Keratinocytes, Self-renewal, Proliferation, NIPBL, ACTL6A, SNAI2, SLUG, SNAIL, SNAI1, UHRF1, DNMT1, KLF4, GRHL3, ZNF750 and Cell Cycle
Graphical abstract
Introduction
Cohesin is a multiprotein complex composed of heterodimers of SMC1A and SMC3 and additional subunits including RAD21 and STAG1/STAG2 which wrap around chromatin (Uhlmann, 2016). This complex forms a ring-like structure necessary for holding together sister chromatids during cell division allowing for proper chromosome segregation and DNA repair (Uhlmann, 2016). More recently, cohesin has been found to regulate gene expression during interphase. This includes regulation of developmental genes such as Runx and Ultrabithorax in zebrafish and Drosophila respectively (Horsfield et al., 2007, Rollins et al., 1999). Its role in regulating gene expression has been attributed to cohesin's ability to promote chromatin looping such as stabilization of enhancer and promoter interactions. Cohesin's role in regulating higher order chromatin has been found to be mediated through interactions with the DNA binding protein CTCF as genome wide mapping has shown high degrees of overlap between their binding sites(Parelho et al., 2008). However, cohesin has also been shown to mediate chromatin looping independent of CTCF(Kagey et al., 2010). Cohesin can also serve as docking sites for transcription factors after cell division to regulate transcription(Yan et al., 2013). Lastly, cohesin may control gene expression by regulating chromatin accessibility. In mammalian cells, a subset of cohesin binding sites overlaps with DNase I hypersensitive sites and global chromatin accessibility is diminished in cohesin mutant cells(Yan et al., 2013, Parelho et al., 2008, Mazumdar et al., 2015).
While the role of cohesin during cell division and regulating gene expression has been well studied, it is still unclear its role in regulating adult mammalian stem cell self-renewal and differentiation. Investigation into this has been hampered by embryonic lethal phenotypes in mouse models where cohesin genes have been knocked out thus limiting its use in deciphering a role in adult tissue maintenance(Remeseiro et al., 2012a). In embryonic stem cells, cohesin is necessary for stem cell self-renewal as loss of complex members results in abolished enhancer-promoter stabilization of key self-renewal genes such as Oct4 and Nanog leading to spontaneous differentiation(Kagey et al., 2010). Recently, through the use of knockdown, haploinsufficient, or mutant cohesin mouse models the importance of the cohesin complex in hematopoiesis was deciphered(Viny et al., 2015, Mullenders et al., 2015, Mazumdar et al., 2015). Insufficient levels of these components resulted in increased self-renewal of hematopoietic stem and progenitor cells due to enhanced site specific chromatin accessibility allowing for transcription factor reinforcement of the stem cell gene expression program(Mazumdar et al., 2015). Whether cohesin acts to promote self-renewal (embryonic stem cells) or inhibit it (hematopoietic stem cells), its mechanism of action in other adult tissues is currently unknown.
The epidermis is an ideal system to study factors that impact self-renewal and differentiation because it is a stratified tissue that is well defined and characterized with the deepest layer containing undifferentiated stem and progenitor cells (basal layer) and the superficial layers containing the most differentiated cells(Watt, 2014). Due to the high turnover rate of the tissue, the stem and progenitor cells must constantly balance self-renewal, proliferation, and differentiation to maintain homeostasis(Donati and Watt, 2015). As a progenitor cell differentiates, it migrates upwards from the basal layer and progresses through the differentiated layers including the suprabasal layer and granular layer. It terminally differentiates upon reaching the stratum corneum where the enucleated corneocytes are shed off the surface of the skin(Sen, 2011, Blanpain and Fuchs, 2009). Epidermal self-renewal is mediated by both transcriptional and post-transcriptional mechanisms(Sen, 2011, Li and Sen, 2016). Transcriptional regulation involves both transcription and epigenetic factors including p63, SNAI2, UHRF1, EZH2, DNMT1, ING5, ACTL6A, SMARCA5, and HDAC1/HDAC2 that promote epidermal stem and progenitor cell self-renewal (Mistry et al., 2014, Mistry et al., 2015, Bao et al., 2013, Mulder et al., 2012, Sen et al., 2010, LeBoeuf et al., 2010, Ezhkova et al., 2009, Yang et al., 1999). Factors such as SNAI2, UHRF1, EZH2, DNMT1, and ACTL6A are necessary for epidermal self-renewal by suppressing the expression of differentiation genes in stem and progenitor cells. While it has been established that these key factors play a critical role in promoting epidermal self-renewal, it is not known what regulates and keeps the expression of these transcriptional regulators high in epidermal stem and progenitor cells.
Here, we determine the role of cohesin in adult human epidermis and found it to be necessary for maintaining epidermal progenitor cell self-renewal while blocking premature differentiation. Cohesin does this by promoting the expression of known epidermal self-renewal genes through maintaining chromatin accessibility at their genomic sites. These findings highlight the importance of the cohesin complex in maintaining adult tissue homeostasis and define mechanisms that promote the expression of self-renewal genes.
Results
The Cohesin Complex Promotes Self-renewal of Epidermal Progenitor Cells
To determine potential roles for the cohesin complex in epidermal self-renewal and differentiation, the core ring components of the complex (SMC1A and SMC3) were knocked down. To avoid being misled by off-target RNAi effects, two distinct shRNAs targeting different regions of SMC1A and SMC3 were generated and delivered via retroviruses to primary human keratinocytes (SMC1Ai, SMC1A-Bi, SMC3i, and SMC3-Bi) (Figures 1A and S1A). Notably, knockdown of one component did not impact the mRNA levels of the other (Figures 1A and S1A). Knockdown of SMC1A or SMC3 resulted in the spontaneous differentiation of primary human epidermal progenitor cells with increased expression of differentiation protein transglutaminase I and differentiation genes (SBSN, TGM1, FLG, IVL, S100P, LCE3D, GRHL3, and SCEL) including those that have been implicated in human skin diseases (Figure 1A-C and Figure S1A-B)(Schmuth et al., 2007). Loss of cohesin also impaired epidermal cell proliferation compared to control (CTL) shRNA transduced cells (Figures 1D and S1C). While not statistically significant, cohesin knockdown increased the percentage of apoptotic cells (Figure 1E). In clonogenic assays, SMC1Ai and SMC3i cells failed to proliferate into large colonies and produced only small colonies (Figure 1F-G). Regenerated organotypic human skin with cohesin knockdown also showed a hypoplastic epidermis compared with controls (Figure S1D-E). Importantly, the basal layer was much smaller in the SMC1A and SMC3 knockdown tissue with around one cell layer whereas in control tissue there were several layers of undifferentiated basal layer cells (Figure S1D).
Figure 1. Cohesin is necessary for epidermal progenitor cell function.
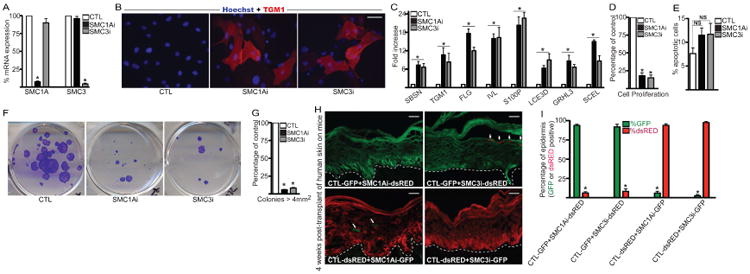
(A) Epidermal progenitor cells were knocked down for control (CTL), SMC1A shRNAs (SMC1Ai), or SMC3 shRNAs (SMC3i) through retroviral transduction. RT-QPCR was used to determine the extent of SMC1A and SMC3 knockdown. QPCR results were normalized to GAPDH levels. Mean ± SEM, n=3. (B) Staining for differentiation protein transglutaminase I (TGMI) is shown in red and hoechst staining in blue marks the nuclei. Scale bar= 15mm. (C) RT-QPCR for differentiation gene expression in CTL, SMC1Ai, and SMC3i cells. QPCR results were normalized to GAPDH levels. Mean ± SEM; n=3. (D) Control and knockdown cells were seeded at 100,000 cells and counted 7 days later. Cell number for each gene knockdown is represented as a percentage of control cell number. Mean ± SEM, n=3. (E) Annexin V staining of CTL, SMC1Ai, and SMC3i cells. The percent of Annexin V positive cells (apoptotic cells) was determined by flow cytometry. Mean ± SEM, n=3. N.S.=Not significant, p>0.05 (t-test) comparing CTL with SMC1Ai or SMC3i samples. (F) Clonogenic assay of CTL, SMC1Ai, and SMC3i keratinocytes plated at limiting dilution (1,000 cells per plate) on mitomycin treated fibroblasts. Cells were stained with crystal violet 4 weeks after plating. (G) Quantification of colonies > 4 mm2 for samples shown in (F) (n=3 per group), Mean ± SEM. (H) In-vivo human epidermal progenitor cell competition assay. GFP expressing CTL keratinocytes were mixed at a 1:1 ratio with dsRed expressing SMC1Ai or SMC3i cells. The reverse experiment was also performed by taking dsRed-CTL cells and mixing at a 1:1 ratio with GFP expressing SMC1Ai or SMC3i cells. The mixed cells were used to regenerate human epidermis on immune deficient mice. GFP expressing cells are shown in green while dsRed cells are shown in red. Tissue was harvested 4 weeks post-grafting on mice. Scale bar=20mm; n=3 grafted mice for each of the 4 groups. The dashed white lines denote the basement membrane zone. White arrowheads mark remaining SMC1Ai or SMC3i cells in the epidermis. (I) Quantitation of GFP and dsRED cells in the epidermis. 12 independent sections were quantitated using Image J per group to determine percent of GFP and dsRED contribution to the epidermis. *=p<0.05 (t-test) comparing CTL with SMC1Ai or SMC3i samples.
To determine whether cohesin is necessary for progenitor cell function in-vivo as well as whether cohesin is acting through cell or non-cell autonomous mechanisms we used the in-vivo progenitor cell competition assay that we previously developed (Sen et al., 2010, Mistry et al., 2012, Wang et al., 2015). Epidermal cells were first infected with retroviruses expressing GFP or dsRED. dsRED expressing cells were then knocked down for SMC1A or SMC3 and mixed with CTL GFP cells at a 1:1 ratio. Conversely to rule out impacts from the fluorescent proteins, GFP expressing SMC1Ai or SMC3i cells were also mixed with CTL dsRED cells. These cell mixtures were then used to regenerate human epidermis on immune compromised mice. Initially, regenerated epidermis from CTL-GFP cells mixed with either SMC3i-dsRED or SMC1Ai-dsRED cells were present in the epidermis at similar percentages (Figure S1F-G). Similar results were also obtained when the epidermis was regenerated with CTL-dsRED cells mixed with SMC1Ai-GFP or SMC3i-GFP cells (Figure S1F-G). However by 4 weeks post-grafting, epidermis regenerated from CTL-GFP cells mixed with SMC1Ai-dsRED or SMC3i-dsRED cells showed that the dsRED cells were depleted from the epidermis (Figure 1H:top panel and Figure 1I). Similarly when CTL-dsRED cells were mixed with SMC1Ai-GFP or SMC3i-GFP cells, the GFP cells were depleted after 4 weeks in-vivo (Figure 1H:bottom panel and Figure 1I). The SMC1Ai or SMC3i expressing cells that remained in the epidermis (white arrowheads) were all found in the upper more differentiated layers suggesting that cohesin is necessary to prevent premature differentiation as well as promote self-renewal of progenitor cells. Cohesin did this through cell autonomous mechanisms since the CTL cells could not rescue the loss of SMC1Ai or SMC3i cells.
Cohesin Regulates a Gene Expression Program that Blocks Premature Differentiation while Promoting Proliferation
To assess the impacts of cohesin on epidermal growth and differentiation, global gene expression profiling was performed on control (CTL) and cohesin (SMC1Ai and SMC3i) knockdown cells. 1,798 genes significantly changed in SMC1Ai cells with 780 genes upregulated and 1,018 genes downregulated (Table S1). Comparison with our previously generated dataset (Sen et al., 2010) of genes that changed during calcium-induced differentiation (CTL DM: Differentiation medium) revealed a significant overlap of 1,075 genes (Figure 2A, Left Panel). The vast majority (91%: 983/1,075) of the genes in the overlapped SMC1Ai gene signature (SMC1Ai) were regulated in the same direction as cells undergoing calcium-induced differentiation suggesting that loss of SMC1A triggers the epidermal differentiation program (Figure 2A, Right Panel). Upregulated genes were enriched for gene ontology (GO) terms such as epidermal cell differentiation, cornified envelope, and keratinization while downregulated genes were involved in cell cycle and mitosis (Figure 2B-C). Knockdown of SMC3 had similar effects as SMC1A with 1,305 genes changing and 840 genes overlapping with the differentiation signature (Figure 2D and Table S2). Again the vast majority of the genes (89%: 744/840) in the overlapped SMC3i gene signature changed in the same direction as the differentiation signature (Figure 2D, Right Panel). Upregulated genes were enriched in GO terms associated with epidermal differentiation while downregulated genes were associated with proliferation (Figure 2E-F). Overlap of the SMC1Ai and SMC3i gene expression signatures demonstrated that a preponderance of the genes are co-regulated (88%: 1,147/1,305) (Figure 2G). These results suggest that the cohesin complex is necessary to sustain epidermal proliferation while inhibiting premature differentiation.
Figure 2. Cohesin promotes epidermal growth and suppresses differentiation.
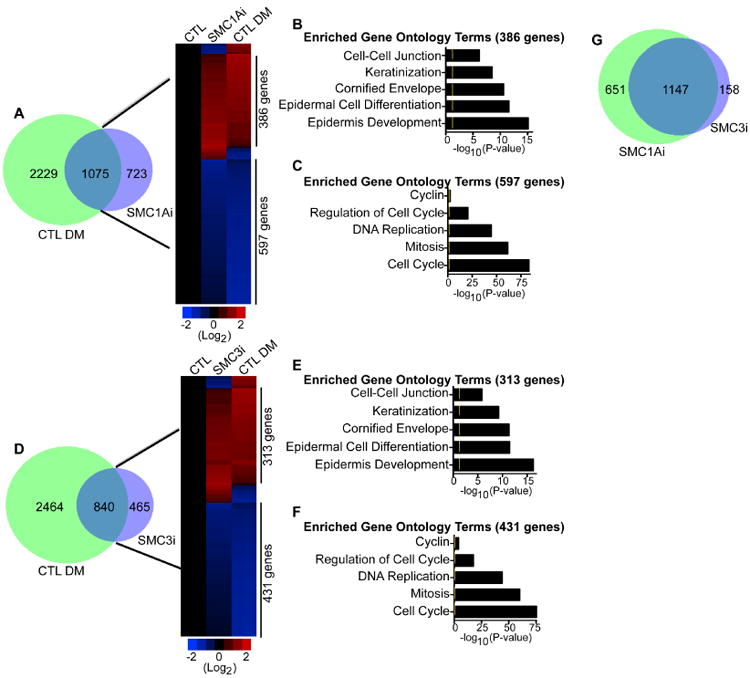
(A) Overlap (left panel) of the differentiation gene signature (CTL DM: 3,304 genes) with the genes that change upon knockdown of SMC1A in cells cultured in growth medium (SMC1Ai: 1,798 genes). The differentiation gene signature (CTL DM) is the differentially expressed genes when epidermal cells are induced to differentiate in high calcium for 3 days. Heat map (right panel) of the 1,075 genes that overlap. Differentiated control cells (CTL DM) were compared to control (CTL) and SMC1A knockdown (SMC1Ai) cells cultured in growth conditions. Heat map is shown in red (induced genes) and blue (repressed genes) on a log 2-based scale. (B) Gene ontology analysis of genes with increased expression that are co-regulated by SMC1Ai and CTL DM samples. Yellow mark in bar graphs demark p-value= 0.05. (C) Gene ontology analysis of coregulated genes with decreased expression. (D) Overlap (left panel) of the SMC3i (cells cultured in growth medium) gene signature with the differentiation signature (CTL DM). Heat map (right panel) of the 840 genes that overlap which shows CTL and SMC3 knockdown (SMC3i) cells cultured in growth medium compared to differentiated cells (CTL DM). (E) Gene ontology analysis of genes with increased expression that are co-regulated by SMC3i and CTL DM samples. (F) Gene ontology analysis of co-regulated genes with decreased expression. (G) Overlap between the SMC1A and SMC3 knockdown gene expression signatures.
Mapping of SMC1A Binding Sites Across the Genome
To gain an understanding of the direct targets of cohesin, chromatin immunoprecipitations (ChIP) followed by deep sequencing (ChIP-Seq) was performed using a SMC1A antibody on epidermal progenitor cells. 23,097 peaks were enriched with SMC1A binding with 33.59%, 23.42%, and 21.42% of the peaks binding within intronic, intergenic, and promoter regions respectively (Figure 3A and Table S3). Lower percentages of binding were found in the 5′UTR (9.76%), CDS (5.62%), and 3′UTR (2.25%). The peaks mapped back to 7,517 genes which were enriched for GO terms such as alternative splicing, cell adhesion, and regulation of epidermal proliferation and differentiation (Figure 3B and Table S3). Mapping of SMC1A bound peaks from -5kb to +5kb from the transcriptional start site (TSS) of genes showed that SMC1A binding is localized primarily to the TSS (Figure 3C-D). Prior studies have suggested that cohesin regulates chromatin accessibility and occupies a subset of DNase I hypersensitive sites to control gene expression (Parelho et al., 2008, Mazumdar et al., 2015, Yan et al., 2013). To determine if this type of regulation is true in keratinocytes, the DNase I hypersensitive genome wide data set for primary human keratinocytes from the ENCODE consortium was compared to our SMC1A ChIP-Seq data (Gerstein et al., 2012). The SMC1A binding sites overlapped with the DNase I hypersensitive sites at the majority of the sites and co-localized at or near the TSS (Figure 3C-D). Comparison of the SMC1A binding sites with the ENCODE data set for histone marks of active transcription and open chromatin such as H3K27ac and H3K4me3 (Kimura, 2013) also showed substantial overlap (Figure 3D). A majority of the SMC1A sites across the TSS of genes also showed overlap with RNA Polymerase II binding suggesting that SMC1A correlates with open chromatin and active transcription (Figure 3D). Consistent with this, histone marks for closed chromatin/heterochromatin such as H3K9me3 and H3K27me3 (Kimura, 2013) were significantly depleted from SMC1A bound sites in primary human keratinocytes (Figure 3D). Since SMC1A regulation of chromatin accessibility near the TSS may potentially impact epidermal growth and differentiation gene expression, the genes bound by SMC1A were overlapped with the genes that are differentially expressed upon SMC1A knockdown. 692 genes were found in the overlap which were enriched in GO terms such as cell cycle, epithelial cell differentiation, and epidermis development (Figure 3E-F, Table S1 and S3).
Figure 3. ChIP-Seq analysis of SMC1A binding sites.
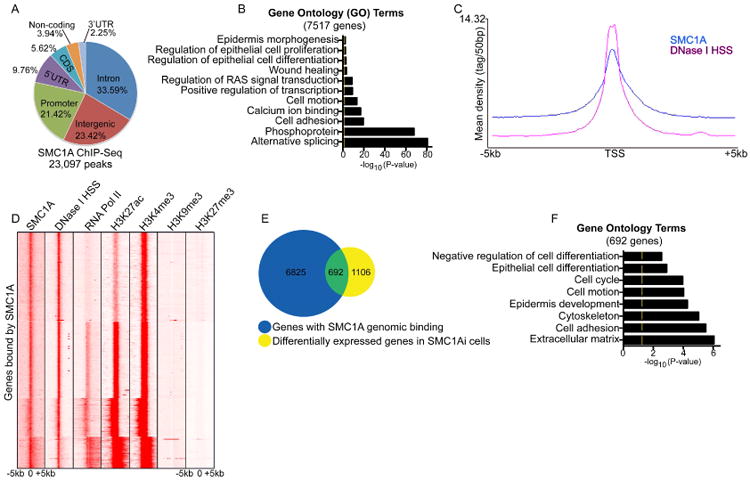
(A) Distribution of SMC1A binding sites in human epidermal progenitor cells. Chromatin immunoprecipitation followed by deep sequencing (ChIP-Seq) was performed using an SMC1A antibody. The total number of SMC1A bound peaks was identified and the percentage of peaks found within various regions of the genome calculated. (B) The 23,097 SMC1A peaks were mapped back to there nearest genes (7,517 genes) and gene ontology analysis performed to determine gene enrichment. Yellow mark in bar graphs demark p-value= 0.05. (C) The distribution of SMC1A binding and DNase I hypersensitivity sites (DNase I HSS) from +/-5kb to the transcriptional start site (TSS) of genes is shown. The x-axis denotes position from TSS and y-axis shows signal strength (mean density of the reads). (D) Heat map of genes bound by SMC1A. Each row shows the +/- 5kb regions centered around the TSS binding for SMC1A, H3K27ac, H3K4me3, RNA polymerase II (RNA Pol II), H3K9me3, H3K27me3 and DNase I HSS. (E) Overlap between the SMC1A bound genes (blue circle: 7,517 genes) with the SMC1A knockdown gene expression signature (yellow circle: 1,798 genes). (F) Gene ontology analysis of the 692 gene found in the overlap in (E).
Several of the genes in the overlap have previously been demonstrated by our group and others to be necessary for epidermal self-renewal including ACTL6A, UHRF1, and SNAI2 whose loss in expression may contribute to the SMC1Ai premature differentiation phenotype (Mistry et al., 2014, Mistry et al., 2015, Bao et al., 2013, Mulder et al., 2012, Sen et al., 2010). Analysis of SMC1A binding to these genes revealed enrichment at gene promoters, introns, and at the TSS (Figure S2A-B). Knockdown of SMC1A decreased binding to those regions demonstrating the specificity of SMC1A binding to the genomic regions of ACTL6A, UHRF1, and SNAI2 (Figure S2B). Furthermore, loss of SMC1A binding to those genes also resulted in their decreased mRNA expression (Figure S2C, Table S1 and S3). Comparison of the SMC1A knockdown gene expression signature with the previously published knockdown signatures of ACTL6A, UHRF1, and SNAI2 showed significant overlap between SMC1A and each of the three self-renewal genes suggesting that SMC1A promotes self-renewal of the epidermis through maintaining the expression of these genes (Mistry et al., 2014, Mistry et al., 2015, Bao et al., 2013, Mulder et al., 2012, Sen et al., 2010) (Figure S2D).
Cohesin Promotes Progenitor Function by Maintaining Open Chromatin to Allow for Expression of Self-renewal Genes Such as SNAI2
Since the SNAI2 knockdown gene expression signature had the highest number of genes shared with the SMC1Ai signature, the regulation of SNAI2 expression by SMC1A was explored. SMC1A bound to three distinct regions within the SNAI2 genomic locus at -2.1kb (peak C), -351bp (peak B), and around the TSS (peak A) which correlated with marks of open chromatin and gene transcription (Figure 4A). To test if SMC1A is necessary to sustain the open chromatin structure, DNase I was used to digest DNA isolated from CTL and SMC1Ai cells. Loss of SMC1A resulted in a 3-5 fold (Sites SNAI2 A-C) increase in DNase I resistance/chromatin compaction of the SNAI2 locus with no changes in the control B-actin (ACTB) gene locus (Figure 4B). SMC1A knockdown also decreased RNA Pol II loading on to the promoter and TSS of SNAI2 as well as decreased H3K27Ac which resulted in decreased protein expression of SNAI2 (Figure 4C-E). To determine if SMC1A blocks premature differentiation of progenitor cells through the expression of SNAI2, a retrovirus was used to express exogenous SNAI2 in SMC1Ai cells. Restoration of SNAI2 expression in SMC1Ai cells (SMC1Ai + SNAI2) blocked the expression of differentiation genes that is seen with SMC1Ai cells (SMC1Ai + LACZ) and partially rescued the premature differentiation phenotype similar to control levels (CTL +LACZ) (Figure 4F). Interestingly, expression of SNAI2 didn't restore the proliferative capacity of the SMC1Ai cells suggesting that SMC1A regulation of other self-renewal genes such as ACTL6A or UHRF1 may potentially mediate its impacts on proliferation (Figure 4G, 4H, S2). It is also unlikely that the full SMC1A knockdown phenotype can be fully rescued by any one factor due to the vast number of genes it regulates.
Figure 4. SMC1A promotes the expression of SNAI2 by maintaining open chromatin to prevent premature epidermal differentiation.
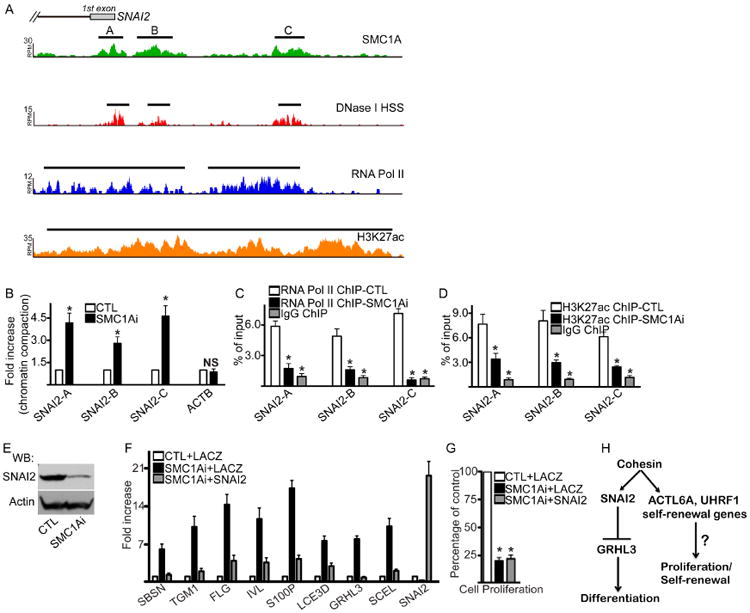
(A) Gene tracks showing SMC1A occupancy at the promoter and TSS of SNAI2. Binding of RNA Pol II, H3K27ac, and DNase I HSS along the same region is also shown. The x-axis denotes genomic position and y-axis shows signal strength (RPM: reads per million). Black bars over peaks indicate significantly bound peaks with FDR < 0.05. (B) DNase I was used to treat DNA harvested from control (CTL) and SMC1A knockdown (SMC1Ai) cells. QPCR was used to amplify genomic regions SNAI2 A-C with their locations shown in (A). The promoter region of B-actin (ACTB) was used as a control. Increases in chromatin compaction were calculated as the fold increase of resistance to DNase I digestion in SMC1Ai cells compared to controls. Mean ± SEM, n=3. (C) ChIP was performed on CTL (white bar) and SMC1Ai (black bar) cells using an RNA Pol II antibody or IgG (grey bar). RNA Pol II binding to respective SNAI2 genomic regions were calculated as a percent of input. Mean ± SEM, n=3. (D) ChIP on CTL (white bar) and SMC1Ai (black bar) cells using an H3K27ac antibody. (E) Western blot on SNAI2 expression in CTL and SMC1A knockdown cells. Actin was used as a loading control. n=3. (F) LACZ or SNAI2 overexpressing cells were knocked down for CTL or SMC1A. Groups are defined as LACZ expressing cells with control knockdown (CTL+LACZ), LACZ expressing cells with SMC1A knockdown (SMC1Ai+LACZ), and SNAI2 expressing cells with SMC1A knockdown (SMC1Ai +SNAI2). RT-QPCR was used to assay gene expression. Results were normalized to GAPDH. Mean ± SEM, n=3. (G) Samples in (F) were seeded at 100,000 cells and counted 7 days later. Cell number for each gene knockdown is represented as a percentage of control (CTL+LACZ). Mean ± SEM, n=3. (H) Model of cohesin mediated epidermal self-renewal. *=p<0.05 (t-test) comparing CTL with SMC1Ai samples.
Cohesin Prevents Premature Differentiation Through SNAI2 Mediated Repression of GRHL3 Expression
SNAI2 is a transcriptional repressor that is necessary to prevent premature epidermal differentiation in progenitor cells by binding to the TSS of GRHL3 and suppressing its expression (Figure S3A)(Mistry et al., 2014). GRHL3 is a Grainyhead-like transcription factor that is critical for turning on the differentiation signature with knockout mice lacking the skin barrier (Ting et al., 2005, Hopkin et al., 2012). Since SNAI2 expression is reduced in SMC1A knockdown cells, it may impact SNAI2's binding at the GRHL3 locus (Figure 4E). ChIP was performed using a SNAI2 antibody on control and SMC1A knockdown cells. Loss of SMC1A resulted in a 3-fold decrease in SNAI2 binding to the TSS of GRHL3 which led to a dramatic increase in RNA Pol II binding (Figure S3B-C). This loss of SNAI2 binding and increased RNA POL II led to increased GRHL3 mRNA and protein levels in SMC1Ai cells (Figure S3D-E). To determine if the premature differentiation seen in SMC1A knockdown cells is due to the increased expression of GRHL3 levels, double knockdown experiments were performed. Double knockdown of GRHL3 and SMC1A rescued the increased differentiation phenotype seen in SMC1Ai cells to levels similar to control (Figure S3F). These results suggest that SMC1A inhibits differentiation gene expression in part through SNAI2 blockade of GRHL3 expression (Figure 4H).
Discussion
Our results show that cohesin is necessary for epidermal self-renewal by maintaining the expression of self-renewal genes such as SNAI2. We found that loss of SMC1A resulted in a reduction of open chromatin at the SNAI2 genomic locus. This increased chromatin compaction led to decreased RNA polymerase II binding to the SNAI2 locus and reduced gene expression. This is similar to other studies where knockdown of cohesin leads to global loss of open chromatin (Mazumdar et al., 2015, Yan et al., 2013). While there is genome-wide loss of DNA accessibility in cohesin mutant and knockdown hematopoietic cells, the stem cell program is enforced due to the actions of key hematopoietic transcription factors ERG, GATA2, and RUNX1 (Mazumdar et al., 2015, Wang et al., 2007, Lichtinger et al., 2012, Chen et al., 2013). Mazumdar et al. proposed that these pioneer factors are able to bind DNA and drive the stem cell gene expression program even with global loss of open chromatin(Mazumdar et al., 2015). This was evidenced by the increased accessibility of chromatin with site-specific DNA binding motifs for ERG, GATA2, and RUNX1 in mutant cohesin cells that had global chromatin compaction (Mazumdar et al., 2015). In contrast to hematopoietic cells, epidermal cells have no defined pioneer transcription factors to reinforce the stem cell gene expression program due to cohesin loss. This makes epidermal self-renewal gene exquisitely sensitive to closed chromatin and without cohesin to maintain the open chromatin structure, genes such as SNAI2, UHRF1, and ACTL6A are downregulated leading to loss of epidermal self-renewal.
Interestingly, cohesin mutations have also been linked to human disease such as Cornelia de Lange syndrome which is characterized by cognitive defects, reduced body size, limb defects, cleft palate, hearing loss, and organ abnormalities(Krantz et al., 2004, Tonkin et al., 2004). Specifically, heterozygous mutations in the cohesin loader, NIPBL has been linked to this disorder as well as mutations in SMC1A and SMC3(Krantz et al., 2004, Tonkin et al., 2004, Deardorff et al., 2007). While no epidermal phenotypes have been reported in humans with cohesin mutations, this is likely due to the mutations still producing functional complexes (SMC1A/SMC3) that may alter their DNA binding sites or haplosufficient (NIPBL) function (Krantz et al., 2004, Tonkin et al., 2004, Deardorff et al., 2007). The altered chromatin binding sites in the cohesin mutants may still allow enough binding and regulation of epidermal self-renewal genes to allow for skin homeostasis. In support of a role for the cohesin complex in skin development and maintenance, STAG1 null mouse embryos have extremely thin skin which could potentially be due to a depletion of skin stem cells(Remeseiro et al., 2012b, Remeseiro et al., 2012a). It will be interesting to determine the long-term physiologic impacts of loss of function cohesin in adult mouse skin using conditional knockouts or through the inducible knockdown of grafted human skin. The grafted human skin does have the limitation of not accounting for the immune system since immune deficient mice must be used for the xenografts.
Here, we have addressed two major unresolved issues including determining the role of cohesin in adult tissue homeostasis as well as defining cohesin as a master regulator of epidermal self-renewal gene expression. These results point to the prominent role of cohesin in maintaining adult stem and progenitor cell function.
Experimental Procedures
The Supplemental Experimental Procedures section which includes a detailed description of the protocols as well as a list of retroviral constructs, siRNAs, antibodies, and primer sequences is available online.
Tissue culture
Primary human epidermal keratinocytes were derived from neonatal foreskin and cultured in growth medium (Life Technologies, KSFM) as previously described (Mistry et al., 2012, Wang et al., 2015). Phoenix cells were cultured in DMEM with 10% fetal bovine serum.
Retroviral constructs
shRNA retroviral constructs to knockdown genes were generated by cloning oligos into the pSuper retroviral vector as previously described (Mistry et al., 2012, Wang et al., 2015). The full-length open reading frame of SNAI2 was cloned into the LZRS retroviral vector as previously described (Mistry et al., 2014).
Microarray analysis
Cells knocked down for SMC1A, SMC3, or control were harvested 10 days after puromycin selection (selects for cells integrated with the pSuper retroviral construct). Microarray analysis using Affymetrix HG-U133 2.0 plus arrays was performed on duplicate samples. Significantly changed genes were identified as previously described (Sen et al., 2010, Mistry et al., 2012, Mistry et al., 2015).
Statistical Analysis
Graph data are represented as mean ± SEM. Statistical analysis were performed using GraphPad Prism. Student's t-test were used comparing one parameter and significant changes were defined as p<0.05. Number of biological experiments was performed as indicated by n in the figure legends.
Supplementary Material
Table S1. Global Gene Expression Profiling Comparing Control and SMC1Ai Cells. Related to Figure 2. All values are shown in Log2 scale except for the last column which is shown as a fold change between control and SMC1Ai samples. Samples were performed in biological duplicates. The average value for each sample is shown in Log2 scale. Significant changes between control (CTL) and SMC1Ai cells were identified by Significance Analysis of Microarrays with a FDR of less than or equal to 5% and an average fold change of greater than or equal to 2 (which is a change of equal to or greater than 1 in log2 scale).
Table S2. Global Gene Expression Profiling Comparing Control and SMC3i Cells. Related to Figure 2. All values are shown in Log2 scale except for the last column which is shown as a fold change between control and SMC3i samples. Samples were performed in biological duplicates. The average value for each sample is shown in Log2 scale. Significant changes between control (CTL) and SMC3i cells were identified by Significance Analysis of Microarrays with a FDR of less than or equal to 5% and an average fold change of greater than or equal to 2 (which is a change of equal to or greater than 1 in log2 scale).
Table S3. SMC1A binding across the genome. Related to Figure 3. ChIP-Seq of SMC1A in primary human keratinocytes grown in proliferation conditions. The start and end of each SMC1A peak mapped back to its nearest gene is shown. SMC1A binding intensity is shown as reads per million (RPM).
Acknowledgments
This work was supported by grants from the National Institutes of Health (NIH R01AR066530-01A1 and 1R01EY025090-01A1) to G.L. Sen and the UCSD Dermatologist Investigator Training Program (1T32-AR062497-01) to M. Noutsou.
Footnotes
Accession Numbers: Microarray data was deposited in GEO with accession number: GSE83870. SMC1A ChIP-seq data was deposited in GEO with accession number: GSE85526. The SNAI2 ChIP-seq data was previously published by our group (Mistry et al., 2014).
Author Contributions: M.N., J.Li, J.Ling, J.J., Y.C., and Y.W. conducted the experiments. M.N. and G.L.S designed the experiments. G.L.S. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bao X, Tang J, Lopez-Pajares V, Tao S, Qu K, Crabtree GR, Khavari PA. ACTL6a enforces the epidermal progenitor state by suppressing SWI/SNF-dependent induction of KLF4. Cell Stem Cell. 2013;12:193–203. doi: 10.1016/j.stem.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat Rev Mol Cell Biol. 2009;10:207–17. doi: 10.1038/nrm2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chi P, Rockowitz S, Iaquinta PJ, Shamu T, Shukla S, Gao D, Sirota I, Carver BS, Wongvipat J, Scher HI, Zheng D, Sawyers CL. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat Med. 2013;19:1023–9. doi: 10.1038/nm.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff MA, Kaur M, Yaeger D, Rampuria A, Korolev S, Pie J, Gil-Rodriguez C, Arnedo M, Loeys B, Kline AD, Wilson M, Lillquist K, Siu V, Ramos FJ, Musio A, Jackson LS, Dorsett D, Krantz ID. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet. 2007;80:485–94. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati G, Watt FM. Stem cell heterogeneity and plasticity in epithelia. Cell Stem Cell. 2015;16:465–76. doi: 10.1016/j.stem.2015.04.014. [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–35. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, Min R, Alves P, Abyzov A, Addleman N, Bhardwaj N, Boyle AP, Cayting P, Charos A, Chen DZ, Cheng Y, Clarke D, Eastman C, Euskirchen G, Frietze S, Fu Y, Gertz J, Grubert F, Harmanci A, Jain P, Kasowski M, Lacroute P, Leng J, Lian J, Monahan H, O'geen H, Ouyang Z, Partridge EC, Patacsil D, Pauli F, Raha D, Ramirez L, Reddy TE, Reed B, Shi M, Slifer T, Wang J, Wu L, Yang X, Yip KY, Zilberman-Schapira G, Batzoglou S, Sidow A, Farnham PJ, MyerS RM, Weissman SM, Snyder M. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkin AS, Gordon W, Klein RH, Espitia F, Daily K, Zeller M, Baldi P, Andersen B. GRHL3/GET1 and trithorax group members collaborate to activate the epidermal progenitor differentiation program. PLoS Genet. 2012;8:e1002829. doi: 10.1371/journal.pgen.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsfield JA, Anagnostou SH, Hu JK, Cho KH, Geisler R, Lieschke G, Crosier KE, Crosier PS. Cohesin-dependent regulation of Runx genes. Development. 2007;134:2639–49. doi: 10.1242/dev.002485. [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, Van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–5. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H. Histone modifications for human epigenome analysis. J Hum Genet. 2013;58:439–45. doi: 10.1038/jhg.2013.66. [DOI] [PubMed] [Google Scholar]
- Krantz ID, Mccallum J, Descipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, Nowaczyk MJ, Toriello H, Bamshad MJ, Carey JC, Rappaport E, Kawauchi S, Lander AD, Calof AL, Li HH, Devoto M, Jackson LG. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet. 2004;36:631–5. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leboeuf M, Terrell A, Trivedi S, Sinha S, Epstein JA, Olson EN, Morrisey EE, Millar SE. Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells. Dev Cell. 2010;19:807–18. doi: 10.1016/j.devcel.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Sen GL. Post-Transcriptional Mechanisms Regulating Epidermal Stem and Progenitor Cell Self-Renewal and Differentiation. J Invest Dermatol. 2016;136:746–52. doi: 10.1016/j.jid.2015.12.030. [DOI] [PubMed] [Google Scholar]
- Lichtinger M, Ingram R, Hannah R, Muller D, Clarke D, Assi SA, Lie ALM, Noailles L, Vijayabaskar MS, Wu M, Tenen DG, Westhead DR, Kouskoff V, Lacaud G, Gottgens B, Bonifer C. RUNX1 reshapes the epigenetic landscape at the onset of haematopoiesis. EMBO J. 2012;31:4318–33. doi: 10.1038/emboj.2012.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar C, Shen Y, Xavy S, Zhao F, Reinisch A, Li R, Corces MR, Flynn RA, Buenrostro JD, Chan SM, Thomas D, Koenig JL, Hong WJ, Chang HY, Majeti R. Leukemia-Associated Cohesin Mutants Dominantly Enforce Stem Cell Programs and Impair Human Hematopoietic Progenitor Differentiation. Cell Stem Cell. 2015;17:675–88. doi: 10.1016/j.stem.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry DS, Chen Y, Sen GL. Progenitor function in self-renewing human epidermis is maintained by the exosome. Cell Stem Cell. 2012;11:127–35. doi: 10.1016/j.stem.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry DS, Chen Y, Wang Y, Sen GL. Transcriptional profiling of SNAI2 regulated genes in primary human keratinocytes. Genom Data. 2015;4:43–46. doi: 10.1016/j.gdata.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry DS, Chen Y, Wang Y, Zhang K, Sen GL. SNAI2 controls the undifferentiated state of human epidermal progenitor cells. Stem Cells. 2014;32:3209–18. doi: 10.1002/stem.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder KW, Wang X, Escriu C, Ito Y, Schwarz RF, Gillis J, Sirokmany G, Donati G, Uribe-Lewis S, Pavlidis P, Murrell A, Markowetz F, Watt FM. Diverse epigenetic strategies interact to control epidermal differentiation. Nat Cell Biol. 2012;14:753–63. doi: 10.1038/ncb2520. [DOI] [PubMed] [Google Scholar]
- Mullenders J, Aranda-Orgilles B, Lhoumaud P, Keller M, Pae J, Wang K, Kayembe C, Rocha PP, Raviram R, Gong Y, Premsrirut PK, Tsirigos A, Bonneau R, Skok JA, Cimmino L, Hoehn D, Aifantis I. Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J Exp Med. 2015;212:1833–50. doi: 10.1084/jem.20151323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–33. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Remeseiro S, Cuadrado A, Carretero M, Martinez P, Drosopoulos WC, Canamero M, Schildkraut CL, Blasco MA, Losada A. Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 2012a;31:2076–89. doi: 10.1038/emboj.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remeseiro S, Cuadrado A, Gomez-Lopez G, Pisano DG, Losada A. A unique role of cohesin-SA1 in gene regulation and development. EMBO J. 2012b;31:2090–102. doi: 10.1038/emboj.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152:577–93. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmuth M, Gruber R, Elias PM, Williams ML. Ichthyosis update: towards a function-driven model of pathogenesis of the disorders of cornification and the role of corneocyte proteins in these disorders. Adv Dermatol. 2007;23:231–56. doi: 10.1016/j.yadr.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL. Remembering one's identity: the epigenetic basis of stem cell fate decisions. FASEB J. 2011;25:2123–8. doi: 10.1096/fj.11-182774. [DOI] [PubMed] [Google Scholar]
- Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463:563–7. doi: 10.1038/nature08683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting SB, Caddy J, Hislop N, Wilanowski T, Auden A, Zhao LL, Ellis S, Kaur P, Uchida Y, Holleran WM, Elias PM, Cunningham JM, Jane SM. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science. 2005;308:411–3. doi: 10.1126/science.1107511. [DOI] [PubMed] [Google Scholar]
- Tonkin ET, Smith M, Eichhorn P, Jones S, Imamwerdi B, Lindsay S, Jackson M, Wang TJ, Ireland M, Burn J, Krantz ID, Carr P, Strachan T. A giant novel gene undergoing extensive alternative splicing is severed by a Cornelia de Lange-associated translocation breakpoint at 3q26.3. Hum Genet. 2004;115:139–48. doi: 10.1007/s00439-004-1134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F. SMC complexes: from DNA to chromosomes. Nat Rev Mol Cell Biol. 2016 doi: 10.1038/nrm.2016.30. [DOI] [PubMed] [Google Scholar]
- Viny AD, Ott CJ, Spitzer B, Rivas M, Meydan C, Papalexi E, Yelin D, Shank K, Reyes J, Chiu A, Romin Y, Boyko V, Thota S, Maciejewski JP, Melnick A, Bradner JE, Levine RL. Dose-dependent role of the cohesin complex in normal and malignant hematopoiesis. J Exp Med. 2015;212:1819–32. doi: 10.1084/jem.20151317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li W, Liu XS, Carroll JS, Janne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27:380–92. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Arribas-Layton M, Chen Y, Lykke-Andersen J, Sen GL. DDX6 Orchestrates Mammalian Progenitor Function through the mRNA Degradation and Translation Pathways. Mol Cell. 2015;60:118–30. doi: 10.1016/j.molcel.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt FM. Mammalian skin cell biology: at the interface between laboratory and clinic. Science. 2014;346:937–40. doi: 10.1126/science.1253734. [DOI] [PubMed] [Google Scholar]
- Yan J, Enge M, Whitington T, Dave K, Liu J, Sur I, Schmierer B, Jolma A, Kivioja T, Taipale M, Taipale J. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell. 2013;154:801–13. doi: 10.1016/j.cell.2013.07.034. [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, Mckeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–8. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Global Gene Expression Profiling Comparing Control and SMC1Ai Cells. Related to Figure 2. All values are shown in Log2 scale except for the last column which is shown as a fold change between control and SMC1Ai samples. Samples were performed in biological duplicates. The average value for each sample is shown in Log2 scale. Significant changes between control (CTL) and SMC1Ai cells were identified by Significance Analysis of Microarrays with a FDR of less than or equal to 5% and an average fold change of greater than or equal to 2 (which is a change of equal to or greater than 1 in log2 scale).
Table S2. Global Gene Expression Profiling Comparing Control and SMC3i Cells. Related to Figure 2. All values are shown in Log2 scale except for the last column which is shown as a fold change between control and SMC3i samples. Samples were performed in biological duplicates. The average value for each sample is shown in Log2 scale. Significant changes between control (CTL) and SMC3i cells were identified by Significance Analysis of Microarrays with a FDR of less than or equal to 5% and an average fold change of greater than or equal to 2 (which is a change of equal to or greater than 1 in log2 scale).
Table S3. SMC1A binding across the genome. Related to Figure 3. ChIP-Seq of SMC1A in primary human keratinocytes grown in proliferation conditions. The start and end of each SMC1A peak mapped back to its nearest gene is shown. SMC1A binding intensity is shown as reads per million (RPM).


