Abstract
CRISPR–Cas systems defend prokaryotes against bacteriophages and mobile genetic elements and serve as the basis for revolutionary tools for genetic engineering. Class 2 CRISPR–Cas systems use single Cas endonucleases paired with guide RNAs to cleave complementary nucleic acid targets, enabling programmable sequence-specific targeting with minimal machinery. Recent discoveries of previously unidentified CRISPR–Cas systems have uncovered a deep reservoir of potential biotechnological tools beyond the well-characterized Type II Cas9 systems. Here we review the current mechanistic understanding of newly discovered single-protein Cas endonucleases. Comparison of these Cas effectors reveals substantial mechanistic diversity, underscoring the phylogenetic divergence of related CRISPR–Cas systems. This diversity has enabled further expansion of CRISPR–Cas biotechnological toolkits, with wide-ranging applications from genome editing to diagnostic tools based on various Cas endonuclease activities. These advances highlight the exciting prospects for future tools based on the continually expanding set of CRISPR–Cas systems.
Introduction
Bacteria and archaea use CRISPR–Cas (clustered regularly interspaced short palindromic repeats–CRISPR associated) systems to provide immunological memory of prior infections by bacteriophages and mobile genetic elements (Hille and Charpentier, 2016; Marraffini, 2015; Sorek et al., 2013). CRISPR loci comprise repeating sequences that are interrupted by short segments of foreign DNA acquired by the CRISPR–Cas system during infection (Jackson et al., 2017; Sternberg et al., 2016) (Figure 1A). By storing genetic information from viruses and mobile genetic elements in their CRISPR arrays, host cells are tagged for rapid immunological response upon subsequent infections by the invader (Barrangou et al., 2007). Immunity is conferred following transcription and processing of CRISPR RNAs (crRNAs) containing the foreign sequence, which guides Cas effector proteins to the matching region of the invader (Jackson and Wiedenheft, 2015). Following binding, the target is cleaved by the endonuclease activity of the Cas effector, or through the recruitment of trans-acting nucleases (Hille and Charpentier, 2016; Marraffini, 2015; Sorek et al., 2013).
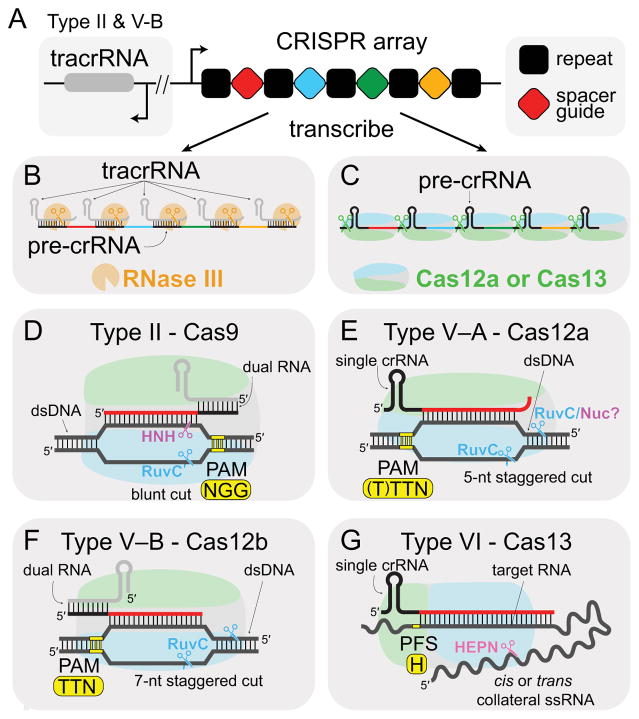
Figure 1. Mechanisms of Class 2 effectors.
(A) Schematic of CRISPR array. Type II and V-B operons also contain tracrRNA loci, which is sometimes expressed from the opposite strand. Types V-A and VI contain only the CRISPR array. (B,C) Processing mechanisms for crRNA. (B) Type II crRNA:tracrRNA duplexes are cleaved by RNase III. It is unknown whether RNase III also processes Type V-B dual-RNA. (C) Cas12a and Cas13 process pre-crRNAs to produce mature guide crRNA. (D–G) Targeting mechanisms of Class 2 effectors. PAM or PFS locations and sequences are shown in yellow. SpCas9 PAM sequence is shown in (D). (D–F) N: A, C, G or T. (G) H: A, C or U.
In response to ever-evolving selective pressure from their viral invaders, CRISPR–Cas systems have diversified into a wide-array of cas genes with varied functionalities (Koonin et al., 2017; Makarova et al., 2015). As a result, Cas effectors are highly diverse, ranging from single-protein endonucleases (Class 2) to large multi-subunit complexes (Class 1). The two classes are divided into a total of six types (I–VI) distinguished by the presence of a unique signature Cas protein, and further subdivided into as many as 32 different sub-types (Koonin et al., 2017). Type II, the first known Class 2 system, contains the signature Cas9 effector protein, which can locate, bind and cleave dsDNA targets complementary to its guide crRNA (Gasiunas et al., 2012; Jinek et al., 2012).
In recent years, Cas9 has become a workhorse for researchers based on its easily programmable DNA binding and cleavage activity (Wang et al., 2016). Cas9 programming is achieved by simply changing the sequence of the guide RNA to match the DNA target of interest. Cas9 has been used extensively for genome editing in a huge variety of organisms (Doudna and Charpentier, 2014; Hsu et al., 2014). In addition, catalytically dead Cas9 (dCas9) can be used as a specific DNA-binding protein for many applications, including transcriptional control and live-cell imaging studies (Wang et al., 2016).
The success of Cas9-based technologies has prompted extensive exploration of bacterial genomes in the hopes of discovering new Class 2 systems for complementary or orthogonal applications (Burstein et al., 2017; Gogleva et al., 2014; Koonin et al., 2017; Shmakov et al., 2017, 2015). In the last two years, the number of Class 2 types and sub-types has exploded, with new systems still likely to be discovered. These include Type V systems containing the signature DNA-targeting Cas12 (formerly Cpf1 or C2c1) effectors and Type VI systems containing the RNA-targeting Cas13 (formerly C2c2) effectors (Shmakov et al., 2015; Zetsche et al., 2015). Together, these Class 2 effectors represent the enormous diversity of CRISPR–Cas systems and immune mechanisms, and offer a formidable toolkit for a large array of technologies. Here we compare the structures, biophysics and mechanisms of effector proteins from each Class 2 sub-type. We also examine the uses and potential applications for each Class 2 effector.
Type II CRISPR-Cas systems
Type II CRISPR–Cas systems have been studied extensively, providing an excellent foundation for comparison with newly discovered Class 2 effectors. Mechanistic differences between the Type II Cas9 and the signature effectors of Type V and VI are highlighted in Figure 1, and underscore the complementary nature of newly characterized Class 2 effectors. The Cas9 mechanism described herein is for the well-characterized ortholog from Streptococcus pyogenes (SpCas9). SpCas9 and other orthologs have also recently been reviewed in detail elsewhere (Jiang and Doudna, 2017).
Cas9 is a large, multi-domain RNA-dependent endonuclease that targets dsDNA, cleaving both strands through the activity of separate HNH and RuvC nuclease domains (Gasiunas et al., 2012; Jinek et al., 2012). Like all Cas effectors, Cas9 relies on a crRNA for guidance to the complementary strand of the dsDNA target. However, Cas9 is one of a small subset of Cas effectors that requires a second, trans-acting crRNA (tracrRNA) (Deltcheva et al., 2011; Jinek et al., 2012). During crRNA biogenesis, the CRISPR locus is transcribed as a long precursor crRNA (pre-crRNA), and the repetitive regions of the transcript base pairs with the tracrRNA, leaving the guide spacer region free (Figure 1A–B). The resulting RNA duplex is cleaved by the host RNase III to form a mature dual-RNA that activates Cas9 (Deltcheva et al., 2011) (Figure 1B). The two RNAs required for Cas9 activation can also be combined into a single-guide RNA (sgRNA) rendering a two-component system suitable for biotechnological applications (Jinek et al., 2012).
Upon dual-RNA activation, Cas9 searches for targets by locating the protospacer adjacent motif (PAM), a short, conserved sequence that is just downstream of the non-complementary strand of the target dsDNA (Mojica et al., 2009; Sternberg et al., 2014) (Figure 1D). Recognition of the PAM initiates dsDNA unwinding, enabling crRNA strand invasion and base pairing with the target (Sternberg et al., 2014). The subsequent formation of an R-loop proceeds directionally away from the PAM (Szczelkun et al., 2014), and the first 8–10 base pairs that form between the crRNA and target DNA are disproportionately important for target recognition and binding (Jinek et al., 2012). Mismatches in this region significantly reduce Cas9-target binding affinity, while mismatches in the PAM-distal region of the target are generally tolerated for binding (Kuscu et al., 2014; Singh et al., 2016; Wu et al., 2014). However, cleavage is inhibited for targets with multiple PAM-distal mismatches based on a conformational switch that regulates the fidelity of Cas9 (Dagdas et al., 2017; Sternberg et al., 2015). Structures of Cas9 have revealed that the HNH domain is mobile (Anders et al., 2014; Jiang et al., 2016, 2015; Jinek et al., 2014; Nishimasu et al., 2014), and the catalytic conformation is preferentially adopted when Cas9 is bound to a target with perfect complementarity (Dagdas et al., 2017; Sternberg et al., 2015). Movement of the HNH domain also allosterically regulates cleavage by the RuvC domain, ensuring high fidelity cleavage for both strands (Sternberg et al., 2015).
Type V CRISPR-Cas systems
Five additional RNA-guided DNA-targeting Class 2 effectors have been recently discovered and assigned to the newly designated Type V–A-E (Koonin et al., 2017) (Table 1). Based on the similarity of domain organization, effectors associated with these five sub-types have been classified as variants of the Cas12 family. Only Cas12a (formerly Cpf1) and Cas12b (formerly C2c1) have been extensively characterized both structurally and functionally (Dong et al., 2016; Gao et al., 2016; Liu et al., 2017a; Shmakov et al., 2015; Stella et al., 2017; Swarts et al., 2017b; Wu et al., 2017; Yamano et al., 2016; Yang et al., 2016; Zetsche et al., 2015). Two additional Type V effectors, Cas12d (formerly CasY) and the compact 980 amino acid Cas12e (formerly CasX), have demonstrated RNA-dependent DNA interference activity in in vivo studies (Burstein et al., 2017), while activity of Cas12c (formerly C2c3) has not been established (Shmakov et al., 2015). Intriguingly, several small, uncharacterized effectors have also been identified (Type V–U1-5), with some as small as ~400 amino acids (Types V–U3-4) (Shmakov et al., 2017). While the activity of these putative effector proteins has yet to be determined, their diminutive size make them highly attractive for potential biotechnology applications.
Table 1.
Current characterization of Type V and VI effectors.
| Effector/Subtypea | Other names | Target | Nuclease domainb | Pre-crRNA processing | tracrRNA | Cut nature | PAM/PFSc |
|---|---|---|---|---|---|---|---|
| Cas9/II-A | Csn1 | dsDNA | RuvC-NTS, HNH-TS | RNase III | Yes | Blunt | 3′ G-rich PAMd |
| Cas12a/V-A | Cpf1 | dsDNA | RuvC-NTS,TS Nuc(?) | Self-WED III | No | Staggered, 5 nt overhangs | 5′ T-rich PAM |
| Cas12b/V-B | C2c1 | dsDNA | RuvC-NTS,TS | RNase III? | Yes | Staggered, 7 nt overhangs | 5′ T-rich PAM |
| Cas12d/V-D | CasY | dsDNA | RuvC | ? | No | ? | 5′ Y-rich PAM |
| Cas12e/V-E | CasX | dsDNA | RuvC | ? | Yes | ? | 5′ TA PAM |
| Cas13a/VI-A | C2c2 | ssRNA | 2 HEPN | Self-Helical 1? | No | Collateral activity | 3′ PFS: H |
| Cas13b/VI-B | C2c6 | ssRNA | 2 HEPN | Self-Helical 1? | No | Collateral activity | 5′ PFS: D 3′ PFS: NAN or NNA |
Uncharacterized types including V-C, V-U1-U5 and VI-C are omitted (Koonin et al., 2017)
TS, NTS indicate domain that cleaves target strand (TS) or non-target strand (NTS).
N: A, C, G or T (or U), Y: C or T, D: A, G, or U, H: A, C or U
SpCas9 PAM
Type V domain organization and architecture
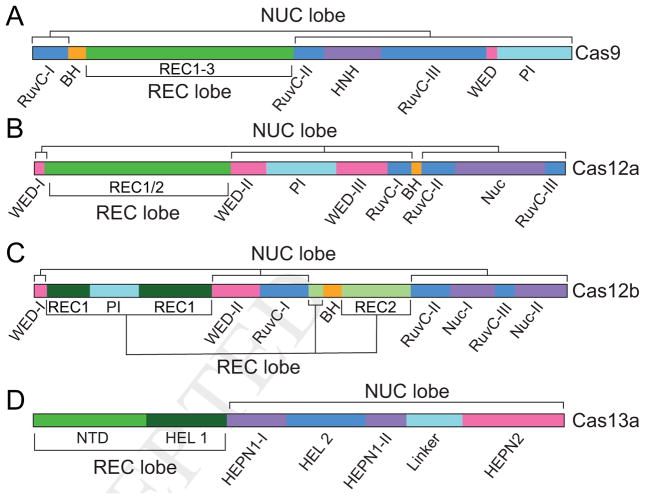
A common feature of Type II and V effectors is the RuvC domain, composed of three motifs that form a single catalytic domain in the tertiary structure (Figure 2A–C). However, unlike Type II effectors, Type V effectors lack HNH domains, substituting it with a previously uncharacterized nuclease (Nuc) domain (Yamano et al., 2016). Despite the substantial differences in domain composition and their placement in the primary structure, the overall structures of Cas9 and Cas12 contain several similar features (Figure 3A–F). Both adopt a bi-lobed architecture comprising two major lobes: a recognition (REC) lobe containing alpha-helical REC motifs; and a nuclease (NUC) lobe containing the RuvC and HNH/Nuc domains (Figure 3A, C, E). A positively charged channel formed between the lobes binds the crRNA-DNA hybrid. The bridge helix (BH), a characteristic feature of Type II and V effector proteins, connects the REC and NUC lobes in the tertiary structure. Two additional domains, the PAM interacting (PI) and wedge (WED) domains, are involved in PAM recognition, although their structures and sequences vary substantially between different Class 2 effectors (Yamano et al., 2016).
Figure 2. Domain arrangement of Class 2 effectors.
Domains comprising recognition (REC) lobe and nuclease (NUC) lobes are labeled. PI: PAM interacting, WED: wedge domain, BH: bridge helix, Nuc: nuclease domain, HEPN: Higher Eukaryotes and Prokaryotes Nucleotide-binding, HEL: Helical.
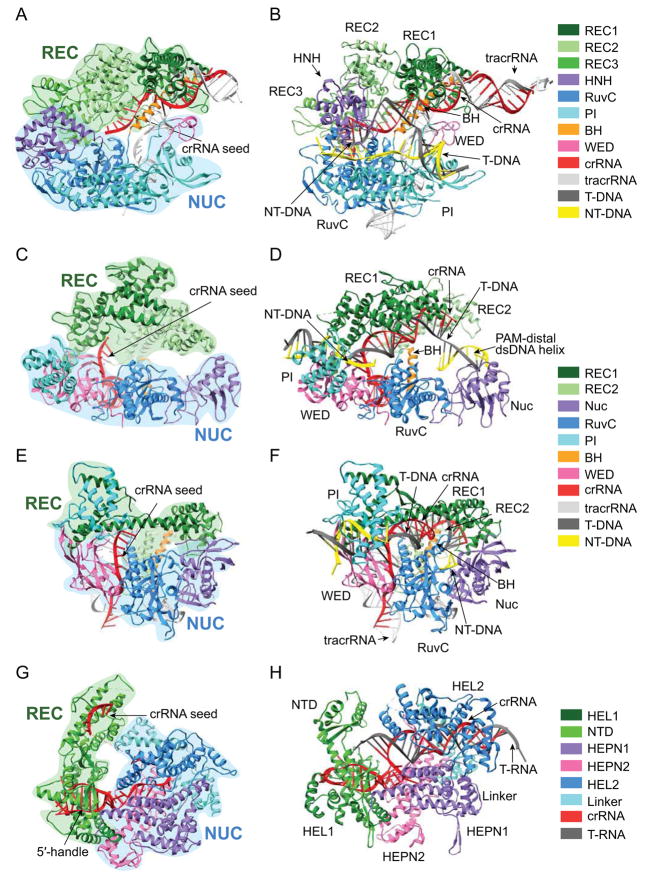
Figure 3. Structures of Class 2 effector binary and ternary complexes.
(A) SpCas9 with sgRNA (PDB 4ZT0) and (B) SpCas9-sgRNA bound to dsDNA (PDB 5F9R). (C) Francisella novicida Cas12a (FnCas12a) bound to crRNA (PDB 5NG6) and (D) FnCas12a-pre-crRNA bound to dsDNA (PDB 5NFV). (E) Alicyclobacillus acidoterrestris Cas12b (AacCas12b) bound to sgRNA (PDB 5U34) and (F) AacCas12b-sgRNA bound to dsDNA with extended non-target strand (PDB 5U33). (G) Leptotrichia shahii Cas13a (LshCas13a) bound to crRNA (PDB 5WTK) and (H) Leptotrichia buccalis Cas13a (LbuCas13a) and crRNA bound to target ssRNA (PDB 5XWP). T-DNA: Target DNA strand, NT-DNA: Non-target DNA strand, T-RNA: Target RNA strand.
Divergent RNA requirements of Type V systems
Initial mechanistic studies of Type V systems have revealed the RNA requirements of several Cas12 effectors (Figure 1B–F, Table 1). Like Cas9, Cas12b and Cas12e require both crRNA and tracrRNA for activation, although it is unknown whether endogenous RNase III is responsible for their processing as in the Type II systems (Burstein et al., 2017; Shmakov et al., 2015). Cas12a has no tracrRNA requirement, utilizing only a single ~44-nucleotide (nt) crRNA for activation (Zetsche et al., 2015). It is also likely that Cas12c and Cas12d only require a single crRNA, as no tracrRNA loci have been identified for Type V-C and V-D CRISPR–Cas systems (Burstein et al., 2017; Shmakov et al., 2015).
The lack of tracrRNA in some Type V systems indicates that pre-crRNA processing occurs through a different mechanism than in Type II systems, and indeed crRNA maturation has been demonstrated to be dependent on cleavage by Cas12a in Type V-A (Fonfara et al., 2016) (Figure 1B). The pre-crRNA is cleaved within the repeat region, which adopts a pseudoknot structure (Swarts et al., 2017; Yamano et al., 2016). In the structure of Cas12a bound to a pre-crRNA, the scissile phosphate is directly positioned between the sidechains of highly-conserved histidine and lysine residues within the WED domain (Swarts et al., 2017). Although early studies suggested that pre-crRNA cleavage requires divalent metal ions (Fonfara et al., 2016), subsequent studies showed that Mg2+ ions help stabilize Cas12a binding to the RNA but are not required for catalysis (Swarts et al., 2017). Cleavage proceeds through a divalent-metal-ion independent general acid-base catalytic mechanism based on nucleophilic attack of the adjacent 2′-OH at the scissile phosphate, leading to formation of a 2′, 3′–cyclic phosphate. This mechanism is reminiscent of metal-independent Cas5 and Cas6 endoribonucleases that are responsible for pre-crRNA processing in Class 1 systems (Hochstrasser and Doudna, 2015), an example of the functional convergence that is commonly observed across CRISPR–Cas subtypes.
A common strategy of RNA-guided protein complexes is the presentation of the guide RNA in a pre-ordered conformation poised for pairing with the nucleic acid target (Gorski et al., 2017). As in Type I CRISPR–Cascade and both prokaryotic and eukaryotic Argonaute complexes, the Type II and V effectors pre-order the PAM-proximal “seed” region of the guide in an A-form conformation to facilitate target search (Jackson et al., 2014; Jiang et al., 2015; Swarts et al., 2017a, 2014; Yang et al., 2016; Zhao et al., 2014) (Figure 3A, C, E). The binary complexes of Cas9 and Cas12b with sgRNA have pre-organized DNA binding pockets that can accommodate the RNA:DNA duplex without altering the conformation of the binding channel (Jiang et al., 2016, 2015; Yang et al., 2016). In contrast, the Cas12a-crRNA complex must undergo extensive rearrangements to accommodate the DNA, with concerted conformational changes occurring within the two REC domains upon PAM recognition (Yamano et al., 2016). These differences likely reflect the scaffolding nature of the tracrRNA in Cas9 and Cas12b, in which interactions with the RNA secondary structure position the REC domains prior to DNA binding (Figure 3A, E). The shorter crRNA of Cas12a lacks interactions with the REC domains, necessitating the rearrangement of the REC lobe to accommodate the DNA (Figure 3C).
Target binding by Type V effectors
One of the most remarkable features of Cas effectors is their ability to efficiently locate, unwind and bind dsDNA targets. This process is facilitated through a mechanism based on PAM-searching and local unwinding, which is common to sub-types in both Class 1 and 2 CRISPR–Cas systems (Redding et al., 2015; Sternberg et al., 2014). By coupling PAM recognition with DNA unwinding, Cas effectors only locally unwind DNA at possible target sites, which significantly increases the efficiency of the search process.
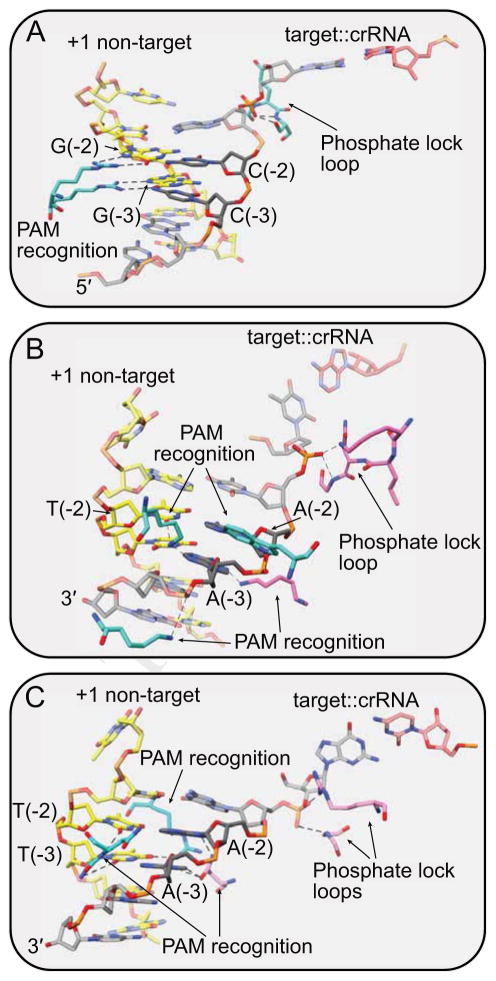
Type V systems also require PAM sequences (Table 1), and Cas12 likely employs a similar mechanism for target searching. Both Type V–A and V–B effectors recognize T-rich PAM sequences, and notably these motifs are located just upstream of the non-complementary strand of the target, the opposite end in comparison to Type II (Shmakov et al., 2015; Zetsche et al., 2015) (Figure 1D–F). Cas9 reads the PAM through sequence-specific major groove interactions of two arginine residues within the dedicated PI domain (Anders et al., 2014) (Figure 4A). For Cas12, the T-rich PAM helix causes the minor groove to narrow, enabling PAM readout through sequence- and shape-dependent interactions with both the major and minor grooves (Yamano et al., 2016; Yang et al., 2016) (Figure 4B–C).
Figure 4. PAM recognition and phosphate locking mechanisms in Type II and V effectors.
Close-ups of PAM regions for (A) SpCas9 (PDB 5F9R) (B) FnCas12a (PDB 5NFV) (C) AacCas12b (PDB 5U33). (A) SpCas9 reads the G-rich PAM sequence through major groove interactions. (B,C) FnCas12a and AacCas12b read T-rich PAM sequences through sequence and shape-specific interactions in major and minor groove. All three effectors stabilize the rotated phosphate of the PAM-proximal deoxyribonucleotide using a phosphate lock loop. Residues from the effectors are colored by domain as in Figure 3.
Upon PAM recognition, local destabilization of the DNA duplex is facilitated through a “phosphate locking” mechanism that is conserved in all Cas effectors that bind dsDNA (Anders et al., 2014; Hayes et al., 2016; Yamano et al., 2016; Yang et al., 2016). Residues within phosphate lock loops stabilize the phosphate group of the PAM-proximal deoxyribonucleotide in a rotated conformation that distorts the target strand and allows for base pairing with the complementary crRNA ribonucleotide (Figure 4). Formation of the RNA:DNA heteroduplex is stabilized through backbone interactions within the cleft formed between the REC and NUC lobes. Interestingly, although the Cas12a crRNA guide region is 24 nt, 4 nt longer than Cas12b, RNA:DNA heteroduplex formation is interrupted after 20 base pairs by the insertion of an aromatic sidechain, resulting in a duplex that is the same length as in Cas12b (Swarts et al., 2017b; Yamano et al., 2016) (Figure 1F–G, 3D, F).
R-loop formation is also dependent on stabilization of the separated non-target strand to prevent reannealing with the target strand. In Cas9, the PAM-proximal region of the non-target strand is bound within a tight channel through the NUC lobe, which places the strand within the RuvC active site and positions the scissile phosphate for cleavage (Jiang et al., 2016) (Figure 3B). Notably, in Cas12a and Cas12b crystal structures, much of the non-target strand is disordered beyond the first few PAM-proximal nucleotides, suggesting that the central region of the strand does not interact stably with the protein (Swarts et al., 2017b; Yang et al., 2016) (Figure 3D, F). Instead, the non-target strand may be stabilized through interactions on the PAM-distal end, near the cleavage site. Consistent with this model, a short section of the PAM-distal region has also been observed in the Cas12b RuvC active site (Yang et al., 2016) (Figure 3F, 5B). The relative lack of stabilizing interactions with the single-stranded non-target region suggests that Cas12 R-loop formation may be less energetically favorable than for Cas9. Intriguingly, genome editing studies have indicated that Cas12a is more specific than Cas9 (Kim et al., 2016; Kleinstiver et al., 2016b), which could be a consequence of lower Cas12a off-target binding affinity. Future studies of R-loop formation energetics will be vital for understanding the relative specificities of Cas9 and Cas12 effectors.
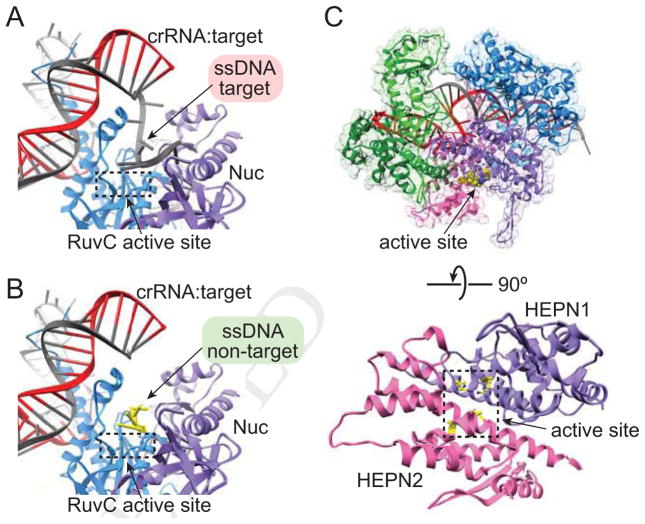
Figure 5. Class 2 effector active sites.
(A–B) Cas12b active sites with (A) target strand (PDB 5U30) or (B) non-target strand (PDB 5U33) accommodated in RuvC active site. (C) Solvent exposed Cas13a HEPN1/2 domain active site (PDB 5XWP).
Target cleavage by Type V effectors
A consequence of the divergent domain organization and PAM locations is the substantial differences in cleavage sites by Cas9 and Cas12 endonucleases (Figure 1D–F). While Cas9 produces blunt cuts proximal to the PAM (Gasiunas et al., 2012; Jinek et al., 2012) (Figure 1D), both Cas12a and Cas12b generate staggered cuts at the PAM distal end of the target DNA (Yang et al., 2016; Zetsche et al., 2015) (Figure 1E–F). Cas9 cleavage of the target and non-target strands is mediated by the metal-ion dependent HNH and RuvC endonuclease domains, respectively (Gasiunas et al., 2012; Jinek et al., 2012). Similarly, the RuvC domain in Cas12 is required for cleavage, although the lack of HNH domain has obscured the exact catalytic mechanism of dsDNA cleavage for these effectors. The Nuc domain has been hypothesized to be the second endonuclease site, and early studies of Cas12a implicated this domain in target strand cleavage (Yamano et al., 2016). However, mutations in the RuvC active site block cleavage of both dsDNA strands (Swarts et al., 2017b; Yamano et al., 2016; Zetsche et al., 2015), and structures of Cas12b reveal that both strands can be individually accommodated in the RuvC active site (Yang et al., 2016) (Figure 5A–B). These data suggest that the RuvC domain may be the sole endonuclease in Cas12 effectors, while the Nuc domain may be critical for positioning the scissile phosphates for cleavage but may not contribute directly to catalysis (Swarts et al., 2017b).
Structural studies of Cas12b provide important clues as to how these effectors may use a single active site to cleave each strand (Yang et al., 2016). When the target strand is elongated past the end of the 20-bp RNA:DNA duplex, the single-stranded PAM-distal end adopts a kinked conformation that places the scissile phosphate into the active site (Figure 5A). However, the target strand cleavage site is past the end of the R-loop and likely double-stranded (Figure 1F), and it remains unclear if and how the PAM distal dsDNA helix is unwound to expose the target strand for cleavage. For the non-target strand, a short segment of the PAM-distal end also travels through the RuvC domain (Figure 3F, 5B), although the exact mechanism dictating cleavage site selection has not been determined. While Cas9 guides the non-target strand to the RuvC active site through extensive interactions within the NUC lobe (Figure 3B), the apparent lack of interactions between Cas12 and the central region of the non-target strand obscure the mechanism of strand placement in the active site. It is possible that non-target cleavage site selection may be guided by interactions with the dsDNA helix flanking the PAM-distal end of the R-loop. Indeed, such interactions have been observed for Cas12a, where the flanking dsDNA is bound at the end of the REC/NUC channel (Swarts et al., 2017b) (Figure 3D).
It remains unclear how Cas12a cleaves both strands using a single active site. The crystal structures of both the pre-cleavage and post-cleavage forms show that the scissile phosphates of both strands are almost 30 Å away from the RuvC catalytic site (Stella et al., 2017; Swarts et al., 2017b; Yamano et al., 2016) (Figure 3D). It is therefore likely that further conformational changes are essential for the RuvC domain to cleave both strands, and that Cas12a may use a similar strategy to Cas12b to accommodate each strand in the active site.
In addition to questions regarding the conformational rearrangements required for cleavage, another important question is whether target and non-target cleavage by the RuvC domain occurs sequentially or in a random order. These two questions will be critical for understanding whether the putative conformational changes that dictate cleavage of each strand help to regulate cleavage fidelity, as has been observed for the Cas9 HNH domain (Dagdas et al., 2017; Sternberg et al., 2015). It is possible that the single-active-site mechanism may provide more stringent control of Cas12a cleavage fidelity, resulting in the higher specificity observed in genome editing experiments (Kim et al., 2016; Kleinstiver et al., 2016b). Future studies of this mechanism will help to illuminate these disparate specificities, and could facilitate the design of Type V effectors with higher specificity, as has been achieved for Cas9 (Chen et al., 2017; Kleinstiver et al., 2016a; Slaymaker et al., 2016).
Type VI CRISPR–Cas systems
Of the recently discovered CRISPR–Cas systems, Type VI contains the most highly divergent effectors both in sequence and in function (Abudayyeh et al., 2016; Shmakov et al., 2015) (Figure 1G, 2D). Cas13, the signature effectors of Type VI systems, targets and cleaves RNA rather than DNA, the only CRISPR–Cas systems to target RNA outside of the Class 1 Type III systems (Abudayyeh et al., 2016; Tamulaitis et al., 2017). To date, three Type VI sub-types (VIA-C) have been discovered (Koonin et al., 2017) (Table I), with the founding type VI–A and corresponding Cas13a effector (formerly C2c2) the best characterized to date.
Type VI domain organization and architecture
Similar to the other Class 2 effectors, Cas13a adopts a bilobed structure consisting of the crRNA recognition (REC) and nuclease (NUC) lobes (Liu et al., 2017b, 2017c) (Figure 2D, 3G–H). However, consistent with its divergent functional role, Cas13 has no sequence or structural similarity with other Class 2 effector proteins, lacking the signature RuvC domain common to both Cas9 and Cas12 effectors. Instead, Cas13 endoribonucleases contain two Higher Eukaryotes and Prokaryotes Nucleotide-binding (HEPN) RNase domains that are also a characteristic feature in Csm6 proteins of the Class 1, type III–A system (Abudayyeh et al., 2016; Niewoehner and Jinek, 2016) (Figure 2D).
Activation of Cas13a by crRNA
Cas13a is activated by a single crRNA and, similar to Cas12a, is required for pre-crRNA processing (East-Seletsky et al., 2016). The location of the pre-crRNA processing active site varies between different Cas13a orthologs that belong to two separate clades, and is found in either the helical 1 domain or in a region of the HEPN2 domain that is distinct from the target-cleavage active site (East-Seletsky et al., 2017; Liu et al., 2017b, 2017c). The different sites of cleavage may be related to the length of the repeat-derived 5′ handle. Nevertheless, the active sites are similar in both domains, comprising arginine and lysine residues that surround the scissile phosphate (Liu et al., 2017b, 2017c). Cas13a cleaves pre-crRNA in a metal-ion-independent fashion, likely proceeding through a mechanism similar to Type I Cas6 and Cas12a enzymes (Hochstrasser and Doudna, 2015, Swarts et al., 2017).
Upon cleavage, the crRNA remains bound to Cas13a and forms multiple contacts with the protein (Liu et al., 2017b, 2017c). The 5′-end of the guide spacer is buried within the NUC lobe, while the central region forms extensive backbone contacts with the protein but is otherwise solvent exposed. In this configuration, the central region of the spacer is presented for nucleation of target binding (Figure 3G). Consistent with these structural observations, functional studies have suggested that Cas13a has a central seed region, as mutations in the middle of the target is more deleterious than mutations at either end (Abudayyeh et al., 2016). This mechanism stands in strong contrast to Cas9 and Cas12 effectors that present terminal PAM-proximal regions of the spacer for RNA:DNA heteroduplex nucleation (Jiang et al., 2015; Swarts et al., 2017b; Yang et al., 2016) (Figure 3A, C, E).
Target binding by Type VI effectors
The central seed region reflects the unique target-binding properties of Cas13 effectors that stem from targeting single-stranded rather than double-stranded nucleic acids. Cas13 has no requirement for a double-stranded unwinding mechanism. Consistently, Cas13 targets do not contain PAM sequences (Abudayyeh et al., 2016), which are associated with dsDNA unwinding in Type I, II and V systems. Similarly, the RNA-targeting Type III effectors are the only other characterized CRISPR–Cas systems to use a PAM-independent mechanism for target recognition (Tamulaitis et al., 2017).
Instead, Cas13 effectors display differential targeting based on protospacer flanking sites (PFS) (Abudayyeh et al., 2016; Smargon et al., 2017) (Table 1). For the well-characterized Leptotrichia buccalis Cas13a, targets flanked by G on the 3′ end are cleaved with lower efficiency than targets with A, C or U at that position (Figure 1G). Notably, the last nucleotide of the 5′ repeat-derived handle within the crRNA (−1 position) is a C, suggesting that PFS non-complementarity is important for targeting. The ternary structure of Cas13a in complex with crRNA and target ssRNA reveals that the 3′ PFS nucleotide in the target RNA is bound within a cleft between the NTD and helical 1 domain, preventing base pairing between the non-guide region of the crRNA and target RNA (Liu et al., 2017b). A potential base pair between the PFS and 5′-handle could disrupt crRNA conformation or target binding leading to the observed defects in target cleavage.
Upon target binding, the Cas13a crRNA forms an extended A-form helix with the target RNA that is bound in a positively charged channel within the NUC lobe (Liu et al., 2017b) (Figure 3H). To accommodate the duplex RNA, the NUC domain undergoes significant structural rearrangements that bring the catalytic residues of the two HEPN domains into close proximity. The two HEPN domains form a single active site that cooperate in cleaving the target ssRNA (Figure 5C), as mutation of either domain is sufficient to block cleavage activity (Abudayyeh et al., 2016). In the binary Cas13a-crRNA complex, the active site residues of the HEPN domains are >5 Å further apart, preventing promiscuous RNA cleavage (Liu et al., 2017b, 2017c). In this way, target binding activates Cas13a for ssRNA cleavage by correctly positioning the HEPN domains to create a functional active site.
Collateral ssRNA cleavage by Type VI Cas13 effectors
Interestingly, unlike Cas9 and Cas12, the Cas13a ternary complex does not cleave the target at a specific site, instead cleaving non-specifically at U-rich regions (Abudayyeh et al., 2016; East-Seletsky et al., 2016). This suggests that the target may be cleaved in trans by neighboring Cas13a enzymes, a model supported by recent structural studies (Liu et al., 2017b). In addition to target-RNA cleavage, activated Cas13 can degrade non-target RNA through a promiscuous, “collateral” cleavage activity (Abudayyeh et al., 2016). The active site of the activated dual-HEPN domains is solvent-exposed (Figure 5C), allowing easy access for collateral RNA cleavage (Liu et al., 2017b). Cas13a has been shown to cleave non-specific RNAs once activated by binding the target. This non-specific activity can occur in cis at other regions of the targeted RNA, as well as on non-target RNAs provided in trans (Abudayyeh et al., 2016; East-Seletsky et al., 2016) (Figure 1G).
It is unclear whether Cas13 has a mechanism for distinguishing host- and phage-derived transcripts during collateral RNA cleavage. Promiscuous RNA cleavage may therefore damage host cells, leading to cellular dormancy or programmed cell death to slow the spread of infection amongst the population (Abudayyeh et al., 2016). Type VI systems also contain mechanisms for regulating Cas13 cleavage that could provide further control to reduce damage against host cells or induce programmed cell death. In Type VI–B systems, the host cas operon encodes one of two accessory protein, Csx27 or Csx28. Intriguingly, these proteins regulate Cas13b function, as Csx27 inhibits target RNA cleavage while Csx28 enhances collateral RNA cleavage (Smargon et al., 2017). The inhibitory activity of Csx27 is intriguingly similar to phage-encoded anti-CRISPR proteins that turn off Cas effector activity, providing phage an advantage in the host-pathogen interaction (Borges et al., 2017). The presence of a regulatory inhibitor in Type VI systems suggests that host expression of anti-CRISPR-like proteins may be wider-spread mechanisms for regulation of CRISPR–Cas activity.
Recent studies have revealed an immune mechanism in the Class 1 Type III–A system that parallels the target-activated collateral cleavage by Cas13 (Kazlauskiene et al., 2017; Niewoehner et al., 2017). In this system, the multi-subunit Csm effector complex binds RNA transcripts that match the guide crRNA. Upon target binding, the Csm complex Cas10 subunit converts ATP into a cyclic oligoadenylate. This signaling molecule then binds and activates the non-specific CRISPR-associated ribonuclease Csm6, which cleaves phage-derived transcripts leading to enhanced immunity. Thus, in both the Type III and VI systems, potentially promiscuous RNases are activated only in the presence of invader target RNA.
Applications based on Type V and VI systems
The fundamental mechanistic understanding of Type V and VI systems has enabled rapid development of biotechnology tools based on Cas12a and Cas13a (Gootenberg et al., 2017; Hur et al., 2016; Tang et al., 2017; Zetsche et al., 2017). The distinct mechanisms of Cas9 and Cas12a increases the functionality of CRISPR-Cas genome editing, while the orthogonal RNA-targeting activity of Cas13a allows for tool development in creative new directions.
Type V applications
Differences at every level of Cas9 and Cas12 mechanism contribute to the complementary nature of the two tools, and provide several potential advantages in the case of Cas12a (Figure 1D–E). While Cas9 sgRNAs are large (~100 nt), the single, short Cas12a crRNA provides a significant advantage, especially for the production of synthetic guide RNAs. The shorter size of both Cas12a and the crRNA are also attractive for viral delivery systems. In addition, the unique mechanism for Type V–A crRNA processing provides an additional tool when using Cas12a for biotechnology. Because Cas12a processes its own crRNAs, it can be used to process multiple crRNAs from a single transcript for multiplex gene editing (Zetsche et al., 2016; Zhong et al., 2017). The difference in PAM requirements also substantially increases the range of genome editing targets. Cas12a is especially useful for editing AT rich genomes and target genes due to its T-rich PAM sequence.
The differences in active sites and cleavage site selection between Cas9 and Cas12a are also significant for genome editing applications. Following DNA cutting by the Cas effector, endogenous DNA repair mechanisms introduce mutations, either through the error-prone non-homologous end joining (NHEJ) mechanism or through specific homology directed repair (HDR) pathways if a template is provided (Doudna and Charpentier, 2014). The staggered cuts produced by Cas12a may additionally allow for directional insertion of gene fragments with complementary “sticky ends”, similar to traditional restriction enzyme cloning methods (Zetsche et al., 2015). It has also been proposed that Cas12a may help to promote HDR, which occurs less frequently than NHEJ (Zetsche et al., 2015). Because Cas12a cleaves at the PAM-distal end, indels formed through NHEJ will be less disruptive for further Cas12a targeting, enabling multiple cutting events at the same site that could improve the chances of repair through HDR.
Specificity is a critical consideration as genome editing technologies mature to the stage where they may be used in human therapeutics. Off-target studies of Cas effectors have revealed that Cas9 can produce significant off-target edits at sites with high homology to the target (Tsai and Joung, 2016), while Cas12a has very low or non-detectable off-target editing activity (Kim et al., 2016; Kleinstiver et al., 2016b). Although several high-fidelity variants of Cas9 have been engineered (Chen et al., 2017; Kleinstiver et al., 2016a; Slaymaker et al., 2016), the intrinsic higher specificity of Cas12a is an attractive quality for genome editing applications. Conversely, it is likely that Cas9 may provide broader defense in endogenous CRISPR–Cas immunity, as the relatively relaxed specificity may enable continued targeting of rapidly evolving phage.
Apart from genome editing, catalytically dead Cas9 (dCas9) has been widely used as a programmable DNA-binding protein, enabling in situ imagining of genomic loci (Shao et al., 2016) and precise control of transcription through repression or activation (Wang et al., 2016). Recent studies demonstrated that DNase-dead Cas12a can also be used for gene regulation in bacteria and plants (Tang et al., 2017; Zhang et al., 2017), suggesting that a wider range of applications will also be enabled by dCas12a.
Type VI applications
The orthogonal activity of Type VI Cas13 provides exciting new opportunities for applications based on programmable RNA targeting. Cas9 has been adapted for RNA binding and cleavage, but requires an additional short DNA complementary to the PAM region for activity (O’Connell et al., 2014). In contrast, Cas13a requires only the crRNA to target an RNA of interest, providing a simpler system with an array of potential applications based on site-specific RNA binding or cleavage. Catalytically dead Cas13a could be used to bind mRNAs to modulate processing or translation, or to visualize RNA movement and localization.
The unique collateral cleavage activity of Cas13a provides an additional functionality that has been exploited for nucleic acid detection based on a signal released upon collateral cleavage of an RNA reporter. Early studies of Cas13a utilized a quenched fluorescent RNA reporter to release a signal upon detection of a target RNA within a complex mixture of RNA sequences. Upon binding to the target, Cas13a collaterally cleaves the reporter, producing a fluorescent signal only upon detection of specific RNA molecules (East-Seletsky et al., 2016). These early studies revealed the potential for highly sensitive RNA detection, enabling precise detection of picomolar quantities of RNA. More recently, Cas13a has been further developed into SHERLOCK (Specific High-Sensitivity Enzymatic Reporter UnLOCKing), a diagnostic tool that enables precise, rapid and sensitive detection of nucleic acid in environmental or clinical samples (Gootenberg et al., 2017). In this system, nucleic acid samples are first amplified then transcribed to produce target RNA for detection by Cas13a. In initial studies, SHERLOCK could be used to discriminate between closely related Zika and Dengue viruses, detect pathogenic bacteria, and identify mutations in cell-free tumor DNA with attomolar sensitivity (Gootenberg et al., 2017). Although Cas13a tool development is still in its infancy, the results are already remarkable and indicate the potential for another powerful CRISPR-based tool.
Future prospects
CRISPR-Cas systems have already provided an impressive set of applications, and the future discovery and characterization of divergent systems will undoubtedly lead to further expansion of CRISPR-based tools. While Cas12a and Cas13a have had an immediate impact on technology development, other newly discovered Class2 effectors, including Cas13b, are still in the initial stages of characterization. It remains to be seen whether they will be useful for biotechnological applications. The continued combination of detailed molecular studies and tool development will be vital for defining the mechanistic differences that could further enhance the CRISPR-Cas toolkit.
Acknowledgments
We thank members of the Sashital and Rajan labs for helpful discussions and Yanli Wang for sharing the Cas13a ternary complex manuscript and structure prior to their publication. D.G.S. acknowledges support of NIH R01 GM115874, NSF CAREER 1652661, and USDA NIFA 2016-06247. R.R acknowledges support from the Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103640.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DBT, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016:353. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Borges AL, Davidson AR, Bondy-Denomy J. The Discovery, Mechanisms, and Evolutionary Impact of Anti-CRISPRs. Annu Rev Virol. 2017 doi: 10.1146/annurev-virology-101416-041616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein D, Harrington LB, Strutt SC, Probst AJ, Anantharaman K, Thomas BC, Doudna JA, Banfield JF. New CRISPR-Cas systems from uncultivated microbes. Nature. 2017;542:237–241. doi: 10.1038/nature21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Harrington LB, Sternberg SH, Joung JK, Yildiz A, Doudna JA. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. 2017 doi: 10.1038/nature24268. bioRxiv 160036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagdas YS, Chen JS, Sternberg SH, Doudna JA, Yildiz A. A Conformational Checkpoint Between DNA Binding And Cleavage By CRISPR-Cas9. Sci Adv. 2017:3. doi: 10.1126/sciadv.aao0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong D, Ren K, Qiu X, Zheng J, Guo M, Guan X, Liu H, Li N, Zhang B, Yang D, Ma C, Wang S, Wu D, Ma Y, Fan S, Wang J, Gao N, Huang Z. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature. 2016;532:522–526. doi: 10.1038/nature17944. [DOI] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- East-Seletsky A, O’Connell MR, Burstein D, Knott GJ, Doudna JA. RNA Targeting by Functionally Orthogonal Type VI-A CRISPR-Cas Enzymes. Mol Cell. 2017;66:373–383e3. doi: 10.1016/j.molcel.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East-Seletsky A, O’Connell MR, Knight SC, Burstein D, Cate JHD, Tjian R, Doudna JA. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538:270–273. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, Richter H, Bratovic M, Le Rhun A, Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532:517–521. doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- Gao P, Yang H, Rajashankar KR, Huang Z, Patel DJ. Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition. Cell Res. 2016;26:901–913. doi: 10.1038/cr.2016.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogleva AA, Gelfand MS, Artamonova II. Comparative analysis of CRISPR cassettes from the human gut metagenomic contigs. BMC Genomics. 2014;15:202. doi: 10.1186/1471-2164-15-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, Myhrvold C, Bhattacharyya RP, Livny J, Regev A, Koonin EV, Hung DT, Sabeti PC, Collins JJ, Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski SA, Vogel J, Doudna JA. RNA-based recognition and targeting: sowing the seeds of specificity. Nat Rev Mol Cell Biol. 2017;18:215–228. doi: 10.1038/nrm.2016.174. [DOI] [PubMed] [Google Scholar]
- Hayes RP, Xiao Y, Ding F, van Erp PBG, Rajashankar K, Bailey S, Wiedenheft B, Ke A. Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli. Nature. 2016;530:499–503. doi: 10.1038/nature16995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille F, Charpentier E. CRISPR-Cas: biology, mechanisms and relevance. Philos Trans R Soc Lond B Biol Sci. 2016:371. doi: 10.1098/rstb.2015.0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser ML, Doudna JA. Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem Sci. 2015;40:58–66. doi: 10.1016/j.tibs.2014.10.007. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur JK, Kim K, Been KW, Baek G, Ye S, Hur JW, Ryu S-M, Lee YS, Kim J-S. Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleoproteins. Nat Biotechnol. 2016:1–2. doi: 10.1038/nbt.3596. [DOI] [PubMed] [Google Scholar]
- Jackson RN, Golden SM, van Erp PB, Carter J, Westra ER, Brouns SJ, van der Oost J, Terwilliger TC, Read RJ, Wiedenheft B. Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. 2014;345:1473–1479. doi: 10.1126/science.1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, Wiedenheft B. A Conserved Structural Chassis for Mounting Versatile CRISPR RNA-Guided Immune Responses. Mol Cell. 2015;58:722–728. doi: 10.1016/j.molcel.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SA, McKenzie RE, Fagerlund RD, Kieper SN, Fineran PC, Brouns SJJ. CRISPR-Cas: Adapting to change. Science. 2017:356. doi: 10.1126/science.aal5056. [DOI] [PubMed] [Google Scholar]
- Jiang F, Doudna JA. CRISPR-Cas9 Structures and Mechanisms. Annu Rev Biophys. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ, Nogales E, Doudna JA. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 2016;351:867–871. doi: 10.1126/science.aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. STRUCTURAL BIOLOGY. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015;348:1477–1481. doi: 10.1126/science.aab1452. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, Kaplan M, Iavarone AT, Charpentier E, Nogales E, Doudna JA. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskiene M, Kostiuk G, Venclovas C, Tamulaitis G, Siksnys V. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science. 2017 doi: 10.1126/science.aao0100. [DOI] [PubMed] [Google Scholar]
- Kim D, Kim J, Hur JK, Been KW, Yoon S-H, Kim J-S. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol. 2016 doi: 10.1038/nbt.3609. [DOI] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016a;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Tsai SQ, Prew MS, Nguyen NT, Welch MM, Lopez JM, McCaw ZR, Aryee MJ, Joung JK. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol. 2016b doi: 10.1038/nbt.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32:677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- Liu L, Chen P, Wang M, Li X, Wang J, Yin M, Wang Y. C2c1-sgRNA Complex Structure Reveals RNA-Guided DNA Cleavage Mechanism. Mol Cell. 2017a;65:310–322. doi: 10.1016/j.molcel.2016.11.040. [DOI] [PubMed] [Google Scholar]
- Liu L, Li X, Ma J, Li Z, You L, Wang J, Wang M, Zhang X, Wang Y. The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a. Cell. 2017b;170:1–13. doi: 10.1016/j.cell.2017.06.050. [DOI] [PubMed] [Google Scholar]
- Liu L, Li X, Wang J, Wang M, Chen P, Yin M, Li J, Sheng G, Wang Y. Two Distant Catalytic Sites Are Responsible for C2c2 RNase Activities. Cell. 2017c;168:121–134. e12. doi: 10.1016/j.cell.2016.12.031. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJM, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- Mojica FJM, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- Niewoehner O, Garcia-Doval C, Rostol JT, Berk C, Schwede F, Bigler L, Hall J, Marraffini LA, Jinek M. Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature. 2017 doi: 10.1038/nature23467. [DOI] [PubMed] [Google Scholar]
- Niewoehner O, Jinek M. Structural basis for the endoribonuclease activity of the type III-A CRISPR-associated protein Csm6. RNA. 2016;22:318–329. doi: 10.1261/rna.054098.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516:263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding S, Sternberg SH, Marshall M, Gibb B, Bhat P, Guegler CK, Wiedenheft B, Doudna JA, Greene EC. Surveillance and Processing of Foreign DNA by the Escherichia coli CRISPR-Cas System. Cell. 2015 doi: 10.1016/j.cell.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, Zhang W, Hu H, Xue B, Qin J, Sun C, Sun Y, Wei W, Sun Y. Long-term dual-color tracking of genomic loci by modified sgRNAs of the CRISPR/Cas9 system. 2016:44. doi: 10.1093/nar/gkw066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, Zhang F, Koonin EV. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol Cell. 2015 doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, Severinov K, Zhang F, Koonin EV. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017;15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D, Sternberg SH, Fei J, Ha T, Doudna JA. Real-time observation of DNA recognition and rejection by the RNA-guided endonuclease Cas9. Nat Commun. 2016;7:1–8. doi: 10.1101/048371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smargon AA, Cox DBT, Pyzocha NK, Zheng K, Slaymaker IM, Gootenberg JS, Abudayyeh OA, Essletzbichler P, Shmakov S, Makarova KS, Koonin EV, Zhang F. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol Cell. 2017;65:618–630. e7. doi: 10.1016/j.molcel.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–66. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- Stella S, Alcon P, Montoya G. Structure of the Cpf1 endonuclease R-loop complex after target DNA cleavage. Nature. 2017;546:559–563. doi: 10.1038/nature22398. [DOI] [PubMed] [Google Scholar]
- Sternberg SH, LaFrance B, Kaplan M, Doudna JA. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature. 2015;527:110–113. doi: 10.1038/nature15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–7. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Richter H, Charpentier E, Qimron U. Adaptation in CRISPR-Cas Systems. Mol Cell. 2016;61:797–808. doi: 10.1016/j.molcel.2016.01.030. [DOI] [PubMed] [Google Scholar]
- Swarts DC, Makarova K, Wang Y, Nakanishi K, Ketting RF, Koonin EV, Patel DJ, van der Oost J. The evolutionary journey of Argonaute proteins. Nat Struct Mol Biol. 2014;21:743–753. doi: 10.1038/nsmb.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Szczepaniak M, Sheng G, Chandradoss SD, Zhu Y, Timmers EM, Zhang Y, Zhao H, Lou J, Wang Y, Joo C, van der Oost J. Autonomous generation and loading of DNA guides by bacterial Argonaute. Mol Cell. 2017a doi: 10.1016/j.molcel.2017.01.033. This issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, van der Oost J, Jinek M. Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a. Mol Cell. 2017b;66:221–233. e4. doi: 10.1016/j.molcel.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczelkun MD, Tikhomirova MS, Sinkunas T, Gasiunas G, Karvelis T, Pschera P, Siksnys V, Seidel R. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci. 2014;111:9798–803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamulaitis G, Venclovas C, Siksnys V. Type III CRISPR-Cas Immunity: Major Differences Brushed Aside. Trends Microbiol. 2017;25:49–61. doi: 10.1016/j.tim.2016.09.012. [DOI] [PubMed] [Google Scholar]
- Tang X, Lowder LG, Zhang T, Malzahn AA, Zheng X, Voytas DF, Zhong Z, Chen Y, Ren Q, Li Q, Kirkland ER, Zhang Y, Qi Y. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat plants. 2017 doi: 10.1038/nplants.2017.103. [DOI] [PubMed] [Google Scholar]
- Tsai SQ, Joung JK. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat Rev Genet. 2016;17:300–312. doi: 10.1038/nrg.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, La Russa M, Qi LS. CRISPR/Cas9 in Genome Editing and Beyond. Annu Rev Biochem. 2016;85:227–264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- Wu D, Guan X, Zhu Y, Ren K, Huang Z. Structural basis of stringent PAM recognition by CRISPR-C2c1 in complex with sgRNA. Cell Res. 2017;27:705–708. doi: 10.1038/cr.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen S, Jaenisch R, Zhang F, Sharp PA. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T, Nishimasu H, Zetsche B, Hirano H, Slaymaker IM, Li Y, Fedorova I, Nakane T, Makarova KS, Koonin EV, Ishitani R, Zhang F, Nureki O. Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell. 2016;165:949–962. doi: 10.1016/j.cell.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Gao P, Rajashankar KR, Patel DJ. PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease. Cell. 2016;167:1814–1828. e12. doi: 10.1016/j.cell.2016.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Heidenreich M, Mohanraju P, Fedorova I, Kneppers J, Degennaro EM, Winblad N, Choudhury SR, Abudayyeh OO, Gootenberg JS, Wu WY, Scott DA, Severinov K, Oost J, Van Der Zhang F. Multiplex gene editing by CRISPR – Cpf1 using a single crRNA array. 2016 doi: 10.1038/nbt.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Heidenreich M, Mohanraju P, Fedorova I, Kneppers J, DeGennaro EM, Winblad N, Choudhury SR, Abudayyeh OO, Gootenberg JS, Wu WY, Scott DA, Severinov K, van der Oost J, Zhang F. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol. 2017;35:31–34. doi: 10.1038/nbt.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang J, Cheng Q, Zheng X, Zhao G, Wang J. Multiplex gene regulation by CRISPR-ddCpf1. Nat Publ Gr. 2017:1–9. doi: 10.1038/celldisc.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sheng G, Wang J, Wang M, Bunkoczi G, Gong W, Wei Z, Wang Y. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature. 2014;515:147–150. doi: 10.1038/nature13733. [DOI] [PubMed] [Google Scholar]
- Zhong G, Wang H, Li Y, Tran MH, Farzan M. Cpf1 proteins excise CRISPR RNAs from mRNA transcripts in mammalian cells. Nat Chem Biol. 2017 doi: 10.1038/nchembio.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]