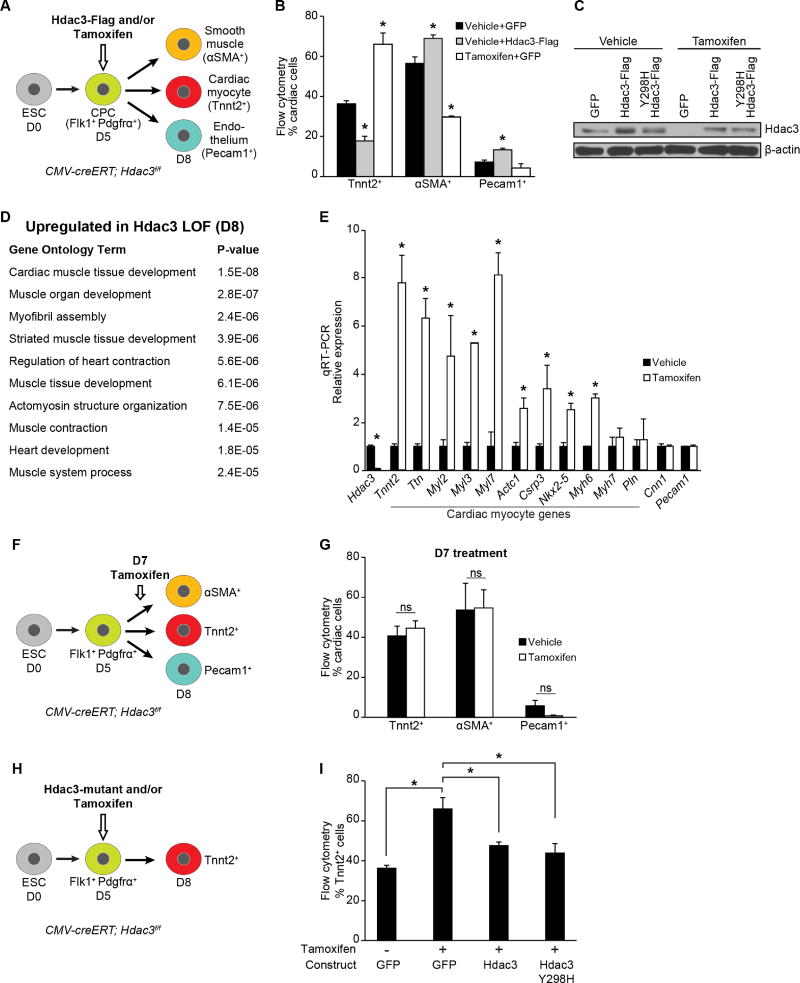
Figure 1. Hdac3 represses differentiation of cardiac progenitor into cardiomyocytes.
(A) Schema of cardiac differentiation of mouse ESCs into multipotent progenitors that give rise to indicated differentiated cell types. (B) Percentage of each cell type measured by flow cytometry at day 8. Cells overexpressing Hdac3 (vehicle+Hdac3-Flag) at day 5 of differentiation show a decrease in Tnnt2+ CMs; tamoxifen-mediated Hdac3 deletion (Tamoxifen+GFP) increases the number of Tnnt2+ CMs. (C) Immunoblot of total Hdac3 and loading control (β-actin) following transduction of CMV-creERT; Hdac3f/f ESCs with GFP control or indicated Hdac3 construct with vehicle or tamoxifen treatment. (D) GO analysis of gene expression data from day 8 tamoxifen-mediated Hdac3 deletion compared to control cultures identifies cardiac-specific GO terms associated with Hdac3 loss-of-function. (E) Gene expression (qRT-PCR) of candidate genes chosen from GO terms in (D) relative to Gapdh. (F) Schema of tamoxifen-mediated Hdac3 deletion at day 7 of differentiation. (G) Tamoxifen-mediated Hdac3 deletion starting at day 7 of cardiac differentiation shows no difference in percentage of each cell type measured by flow cytometry at day 8 compared to vehicle. (H) Schema of tamoxifen-mediated Hdac3 deletion and transduction of wildtype or mutant constructs at day 5 of differentiation. (I) Percentage of Tnnt2+ CMs measured by flow cytometry at day 8 (cells treated at day 5 with tamoxifen and transduced with wild-type Hdac3 or Hdac3Y298H mutant). Data represent mean ± SEM, n≥3 replicates for (B, E, G, I). (B, I) analyzed by one-way ANOVA, with a Tukey post-hoc test. (E, G) analyzed by two-tailed Student’s t-test; statistical comparisons are to vehicle. * p< 0.05. See Fig. S1, Table S1.