Abstract
In the yeast Saccharomyces cerevisiae, the Opi1p repressor controls the expression of INO1 via the Opi1p/Ino2p–Ino4p regulatory circuit. Inositol depletion favors Opi1p interaction with both Scs2p and phosphatidic acid at the endoplasmic reticulum (ER) membrane. Inositol supplementation, however, favors the translocation of Opi1p from the ER into the nucleus, where it interacts with the Ino2p–Ino4p complex, attenuating transcription of INO1. A strain devoid of Scs2p (scs2Δ) and a mutant, OPI1FFAT, lacking the ability to interact with Scs2p were utilized to examine the specific role(s) of the Opi1p–Scs2p interaction in the regulation of INO1 expression and overall lipid metabolism. Loss of the Opi1p–Scs2p interaction reduced INO1 expression and conferred inositol auxotrophy. Moreover, inositol depletion in strains lacking this interaction resulted in Opi1p being localized to sites of lipid droplet formation, coincident with increased synthesis of triacylglycerol. Supplementation of choline to inositol-depleted growth medium led to decreased TAG synthesis in all three strains. However, in strains lacking the Opi1p–Scs2p interaction, Opi1p remained in the nucleus, preventing expression of INO1. These data support the conclusion that a specific pool of phosphatidic acid, associated with lipid droplet formation in the perinuclear ER, is responsible for the initial rapid exit of Opi1p from the nucleus to the ER and is required for INO1 expression in the presence of choline. Moreover, the mitochondria-specific phospholipid, cardiolipin, was significantly reduced in both strains compromised for Opi1p–Scs2p interaction, indicating that this interaction is required for the transfer of phosphatidic acid from the ER to the mitochondria for cardiolipin synthesis.
Keywords: cardiolipin, choline, lipid droplet, phosphatidic acid, triacylglycerol, inositol
Introduction
Many genes encoding enzymes involved in glycerolipid biosynthesis (Fig. 1) in the yeast, Saccharomyces cerevisiae, contain the UASINO promoter element and are, thus, subject to coordinated transcriptional regulation in response to the availability of the phospholipid precursor, inositol, and, to a lesser degree, choline (1–5). UASINO-containing genes are repressed in yeast cells growing in the presence of exogenous inositol and, to a somewhat greater extent, when choline is also present in the medium (6–9). However, inositol and choline enter the pathway for phospholipid biosynthesis by entirely different pathways (Fig. 1) and impact regulation of lipid metabolism in distinctly different ways (2, 3, 6), a major topic addressed further in this paper. INO1, encoding myo-inositol-3-phosphate synthase (10), is the most highly regulated of the UASINO-containing genes (3–5, 8, 11), and its expression is essential for growth in the absence of exogenous inositol (12). The Ino2p–Ino4p activation complex binds to UASINO and is required for activation of INO1 transcription in the absence of exogenous inositol (4, 13). Thus, ino2Δ and ino4Δ mutants are unable to activate INO1 transcription and, consequently, exhibit stringent inositol auxotrophy (Ino− phenotype), comparable with that of the ino1Δ mutant (Fig. 2) (3, 4, 14).
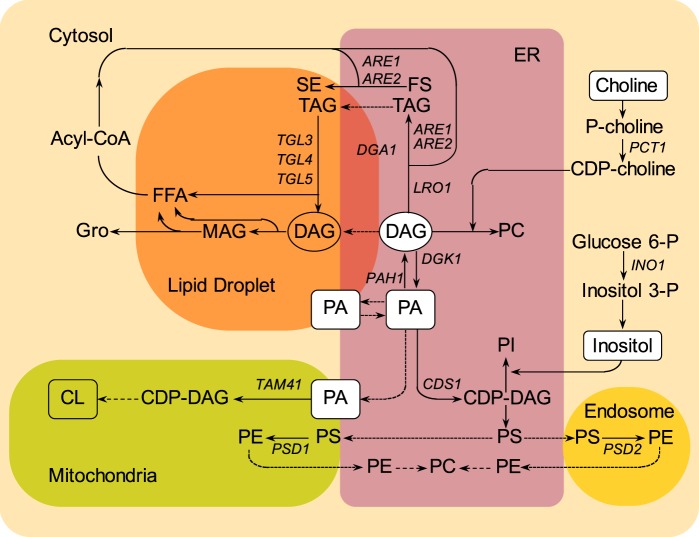
Figure 1.
Localization of key lipids and enzymes, designated by the genes encoding them, in different cellular compartments in S. cerevisiae. PA serves as precursor to both phospholipids and neutral lipids. PA is found in the ER (purple box), in lipid droplets (orange box), and in mitochondria (green box). In the ER, PA serves as immediate precursor of CDP-DAG, precursor to PI and PS. PS is converted into PE by decarboxylation in the mitochondria or in the endosome compartment (accented yellow box). PC is synthesized by methylation of PE or from DAG and CDP-choline. DAG is derived from dephosphorylation of PA or deacylation of TAG. Acyl-CoA is localized in the cytosol (light yellow box) and serves as fatty acid pool for the synthesis of PA, TAG, and SE. PA is also transported from the ER to the mitochondria, where it generates CDP-DAG and CL. The positions of PA within the metabolic network are boxed, the positions of DAG are circled, and the positions at which inositol and choline enter in the metabolic pathway are boxed. The arrows represent routes of metabolic conversion. The names of the structural genes encoding enzymes catalyzing specific metabolic conversions are shown adjacent to the arrows.
Figure 2.
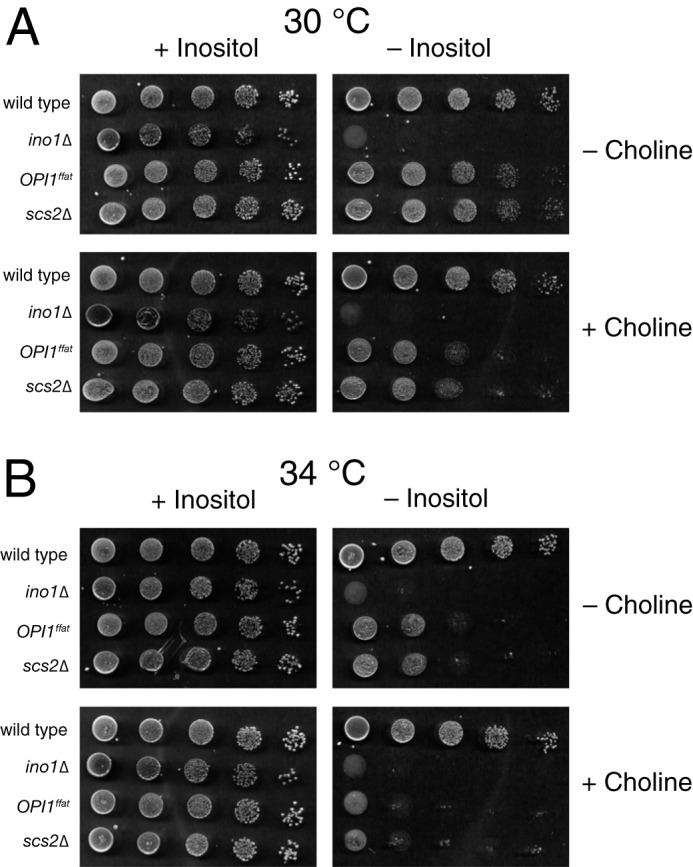
Inositol auxotrophy (Ino− phenotype) of the OPI1ffat and scs2Δ mutants at different temperatures. A suspension of BY4742 (wild type), YCY47 (OPI1ffat), and SJY39 (scs2Δ) cells at a concentration of 1.0 A600 nm/ml and four subsequent 1:10 serial dilutions of strains were spotted on I+C−, I−C−, I+C+, and I−C+ plates and allowed to grow for 2 days at 30 °C (A) or 34 °C (B). SJY425 (ino1Δ) serves as a control for the Ino− phenotype.
Regulated expression of INO1 and other UASINO-containing genes, in response to inositol availability, is controlled by the cellular location of the Opi1p repressor (3, 4, 15). In wild-type cells growing in the absence of exogenous inositol, the Opi1p repressor is retained in the ER,4 in part through its interaction with PA (15), precursor to PI and other phospholipids (Fig. 1). In the absence of exogenous inositol, yeast cells must rely on endogenous synthesis of inositol, catalyzed by inositol 3-phosphate synthase (Ino1p) (Fig. 1). Under these conditions, PI synthesis is reduced, leading to elevated PA levels and retention of Opi1p in the ER, thereby activating transcription of INO1 and other UASINO-containing genes (3, 4, 11). However, when inositol is resupplied to inositol-depleted cultures, PI synthesis rapidly increases, PA levels decline, and Opi1p enters the nucleus (15). Once in the nucleus, Opi1p interacts with the transcriptional activation domain of the Ino2p activator, repressing expression of INO1 (15) and other UASINO-containing genes (3–5, 11, 16).
In addition to elevated levels of PA, retention of Opi1p in the ER also requires its interaction with Scs2p (15). Scs2p, homolog of mammalian synaptobrevin-associated protein (VAMP), is a conserved integral ER protein and a component of a lipid-sensing complex (17–19). Proteins of the VAMP family serve as anchors to the ER for cytoplasmic proteins, including Opi1p, through a conserved motif known as FFAT, two phenylalanines (FF) in an Acidic Tract (19, 20). Many proteins to which VAMP binds are targeted to other intracellular membranes. This implies that VAMP is an important bridge between the ER and a wide variety of organelles at membrane contact sites (21), serving to facilitate lipid transfer and communication between organelles (22–24). For example, in yeast, phosphatidylinositol 4-phosphate levels on the plasma membrane are regulated at membrane contact sites through interaction between Scs2p and Osh3p (25). In addition, deletion of SCS2 leads to a 50% loss in ER-plasma membrane contact sites (26). However, to date, no role for Opi1p in lipid transfer between ER and other membrane compartments has been reported.
Both scs2Δ and opi1Δ mutants exhibit complex, pleiotropic phenotypes, related to lipid metabolism and gene regulation (19, 27–30). The yeast SCS2 gene was originally isolated as a high copy suppressor of choline sensitivity (39), a phenotype associated with an uncharacterized dominant mutation, CSE1 (31). Scs2p was subsequently identified as an integral membrane protein of the ER (17) and homolog of the mammalian VAMP (32). The scs2Δ mutant exhibits a relatively weak Ino− phenotype at 30 °C, which becomes stronger (more visible in plate tests) at growth temperatures of 34 °C or higher (Fig. 2, A and B). The scs2Δ Ino− phenotype also becomes more stringent in the presence of exogenous choline (14) (Fig. 2, A and B) and is suppressed by mutations in the CDP-choline pathway for PC biosynthesis (17, 18). The opi1Δ mutation, in contrast, confers constitutive overexpression of INO1 and other UASINO-containing genes (3–5, 33), resulting in overproduction and excretion of inositol into the growth medium (Opi− phenotype). The opi1Δ mutant also exhibits a Pet− phenotype (i.e. inability to survive complete loss of the mitochondrial genome) and produces very reduced levels of the mitochondrial lipid, CL (34). CL is synthesized in the inner mitochondrial membrane in a multistep pathway (Fig. 1), using PA synthesized in the ER and transferred to mitochondria via the ER–mitochondria encounter structure (ERMES) (35–38).
A major goal of the current study was to determine which of the diverse regulatory and metabolic phenotypes conferred by the scs2Δ and opi1Δ mutations result from the loss of the interaction between Opi1p and Scs2p in the ER. Moreover, given the importance of the FFAT motif in facilitating interorganelle lipid transfer (21), we examined the specific function of the interaction of Opi1p and Scs2p in regulation of lipid metabolism. For this purpose, we used the BY4742 parent strain to mutate the FFAT domain in OPI1, within its genomic locus, to create an OPI1ffat strain. A second goal was to determine the individual and relative roles of inositol and choline on both lipid metabolism and regulation of INO1. In each of three congenic strains, BY4742 (wild type), scs2Δ (SJY39), and OPI1ffat (YCY47), we also tagged OPI1 or OPI1ffat with GFP within the OPI1 genomic locus, to analyze and compare the kinetics of changes in lipid metabolism, Opi1p or Opi1ffatp localization, and INO1 derepression in cells shifted from inositol-containing to inositol-free medium in the presence and/or absence of choline. We report here that the OPI1ffat mutant exhibits relatively weak inositol auxotrophy (Ino− phenotype), which, similar to the phenotype of the scs2Δ mutant (14, 17), becomes stronger at higher growth temperatures and in the presence of choline. Moreover, Opi1p in the scs2Δ strain and Opi1ffatp in the OPI1ffat strains, respectively, did not exit the nucleus following a shift to medium lacking inositol when choline was present, and under these conditions, INO1 failed to derepress. Thus, we conclude that these phenotypes are conferred by the loss of the Opi1p–Scs2p interaction in the ER. Moreover, we observed that loss of the Opi1p–Scs2p interaction in the ER results in reduced content of the mitochondrial lipid CL under all growth conditions tested, indicating that PA transfer from the ER to the mitochondria is impaired in the absence of this interaction. These results suggest a heretofore unrecognized role for Scs2p–Opi1p interaction in lipid transfer from ER to mitochondria.
Results
The scs2Δ and OPI1ffat mutations confer inositol auxotrophy that is strengthened in the presence of choline and at higher growth temperatures
In this study, we sought to investigate the root cause of the choline-sensitive inositol auxotrophy phenotype of scs2Δ. The scs2Δ mutant exhibits reduced growth in the absence of inositol (Ino− phenotype), which is most evident at higher growth temperatures (Fig. 2, A and B) (18, 19, 40) and in the presence of exogenous choline (14) (Fig. 3). The scs2Δ Ino− phenotype is also suppressed by mutations in the CDP-choline pathway for PC biosynthesis (18), indicating that incorporation of choline into phospholipids via this pathway is involved in the Ino− phenotype of scs2Δ (Fig. 3).
Figure 3.
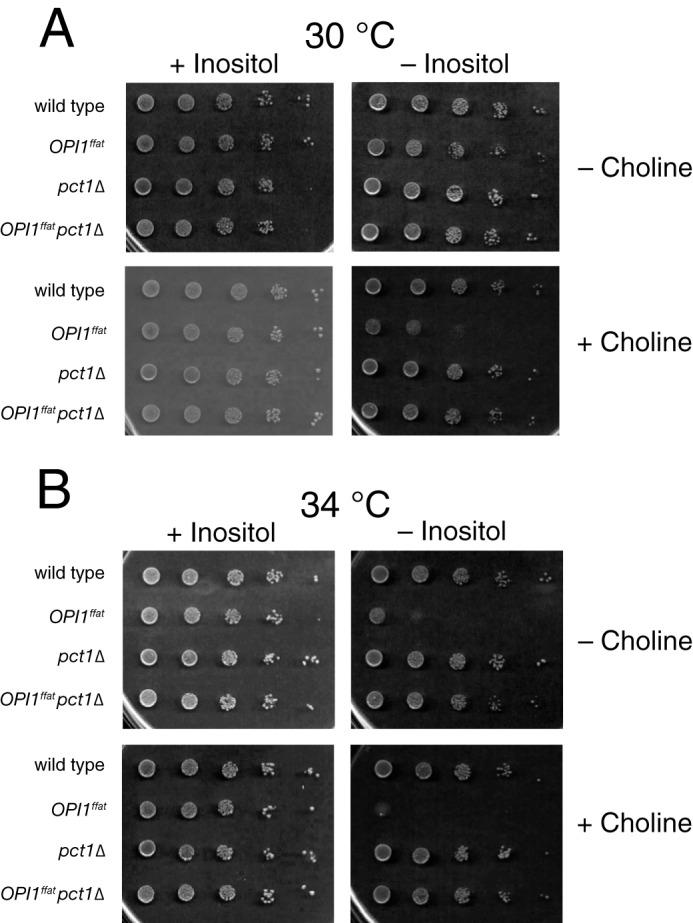
The pct1Δ mutation suppresses the Ino− phenotype of the OPI1ffat strain at all temperatures and in the presence of choline. Suspension of BY4742 (wild type), YCY47 (OPI1ffat), LGY169 (pct1Δ), and LGY541 (OPI1ffat pct1Δ) cells at a concentration of 1.0 A600 nm/ml and four subsequent 1:10 serial dilutions of strains were spotted on I+C−, I−C−, I+C+, and I−C+ plates and allowed to grow for 2 days at 30 °C (A) or 34 °C (B).
Because a functional FFAT domain is necessary for interaction of Opi1p with Scs2p in the ER, we asked whether an OPI1ffat strain exhibits phenotypes similar to those observed in the scs2Δ strain. We reasoned that shared phenotypes in these two strains indicate a functional role for this important interaction. To test this hypothesis, we used the BY4742 parent strain to mutate the FFAT domain in OPI1, within its genomic locus, thereby creating the OPI1ffat strain. The OPI1ffat (YCY47) and scs2Δ (SJY39) strains were then tested and compared in standard plate assays for their ability to grow in the absence of inositol at 30 and 34 °C in the presence and/or absence of choline. The OPI1ffat and scs2Δ strains both exhibit similar (weak) inositol auxotrophy at 30 °C. (Compare the stringent Ino− phenotype of ino1Δ with the weak Ino− phenotypes of scs2Δ and OPI1ffat at 30 °C; Fig. 2A). The Ino− phenotype of both scs2Δ and OPI1ffat is strengthened at 34 °C and further enhanced when choline is present at 34 °C (Fig. 2B). Because scs2Δ and OPI1ffat SCS2 share only one defect in common, namely the lack of a functional Scs2p–Opi1p interaction in the ER, we conclude that loss of this interaction is responsible for these growth phenotypes. Thus, interaction of Opi1p with Scs2p in the ER is required, in addition to its interaction with PA, for optimal growth in the absence of inositol, especially at higher growth temperatures and in the presence of choline.
We also deleted PCT1, encoding choline-phosphate cytidylyltransferase (41), in the OPI1ffat and scs2Δ strains, thereby blocking incorporation of choline via the CDP-choline (Kennedy) pathway for PC synthesis (42) (Fig. 1). The pct1Δ mutation suppressed choline-sensitive inositol auxotrophy phenotype in both OPI1ffat and scs2Δ strains (Fig. 3). Thus, choline has to enter the CDP-choline pathway for PC synthesis to influence the Ino− phenotype of the scs2Δ and OPI1ffat strains. PC synthesized in the ER via CDP-choline pathway utilizes diacylglycerol (DAG), which also serves as the immediate precursor to triacylglycerol (TAG), a major constituent of lipid droplets (Fig. 1). In addition, we constructed and tested a diploid strain, OPI1ffat/OPI1, to determine whether the OPI1ffat mutation was dominant or recessive in terms of its growth in the presence and absence of inositol and choline at 30 and 34 °C. The diploid strain, OPI1ffat/OPI1, exhibited a slight growth reduction on I−C− and I−C+ medium (data not shown), a phenotype intermediate between that observed in the wild-type and OPI1ffat strains, indicating that the OPI1ffat mutation is semidominant with respect to its sensitivity to choline in the absence of inositol. This result implies that both gene products are expressed from the native promoter of OPI1 and contribute proportionately to the phenotype of the diploid.
The effects of the OPI1ffat and scs2Δ mutations on INO1 expression and Opi1p localization following a shift to inositol-free medium in the absence of choline
Manipulation of PA levels following withdrawal of inositol and its subsequent readdition in the presence or absence of choline provides a powerful method for analyzing the relative effects of PA on Opi1p function (15, 43). To this end, we analyzed and compared the relative timing of changes in lipid metabolism, Opi1p localization, and INO1 expression in the wild-type (YCY3), OPI1ffat (YCY5), and scs2Δ (YCY7) strains under these growth conditions.
As expected, before the shift to inositol-free medium in the absence of choline (i.e. from I+C− to I−C− medium) at 30 °C, INO1 was fully repressed in the wild-type strain (repressed level set arbitrarily at 1 unit of expression) (Fig. 4A). Under these conditions, Opi1p-GFP was localized exclusively to the nucleus (Fig. 4B, 0 h, left panel). At 1 h following the shift to I−C− medium, Opi1p-GFP, in the wild-type strain, had largely exited the nucleus and translocated, primarily to the perinuclear ER region, with a slight residual pool of fluorescence remaining visible within the nucleus (Fig. 4B, 1 h, left panel). During this interval, consistent with the partial exit of Opi1p-GFP from the nucleus (Fig. 4B, 1 h, left panel), INO1 expression increased about 50-fold (Fig. 4A). Within the interval from 1 to 2 h following the shift to I−C− medium, expression of INO1 increased from 50-fold to about 375-fold in the wild-type strain (Fig. 4A). After 2 h in I−C− medium, the rate of increase in INO1 expression leveled off, rising slightly above 410-fold (Fig. 4A). At this time point, a significant pool of Opi1p-GFP was still visible in the perinuclear ER region (Fig. 4B, 2 h, left panel).
Figure 4.
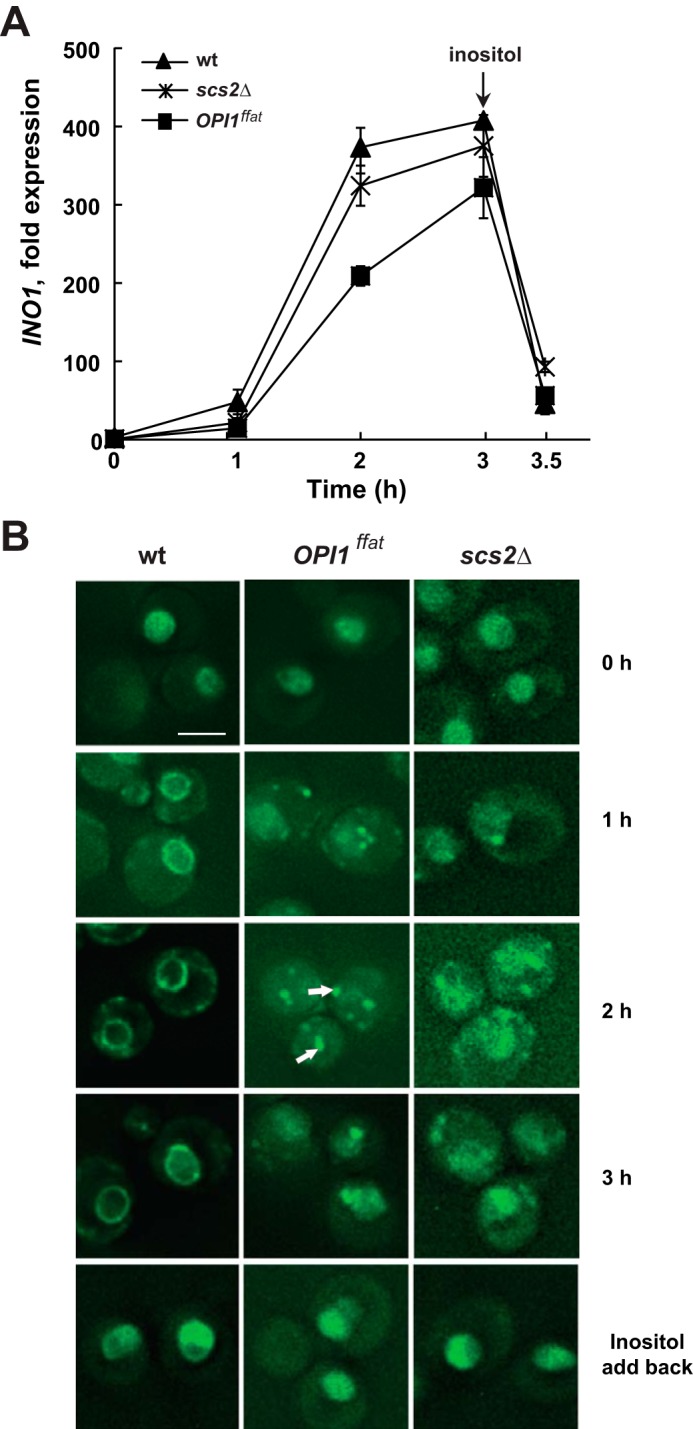
Derepression of the INO1 gene and localization of Opi1p-GFP in wild type, OPI1ffat, and scs2Δ strains following a shift to medium lacking inositol and choline. A, overnight cultures of YCY3 (wild type), YCY5 (OPI1ffat), and YCY7 (scs2Δ) expressing genomic Opi1p-GFP were diluted to A600 nm = 0.2 in I+C− medium and allowed to grow to mid-logarithmic phase at 30 °C. Cells were harvested by centrifugation and washed and resuspended in I− medium, followed by incubation for 3 h at the same temperature. Inositol was added back after 3 h of inositol starvation. Samples were taken at 0, 1, 2, and 3 h of inositol starvation and 30 min after adding back inositol. Total RNA was isolated and analyzed by RT-PCR as described under “Experimental procedures.” Solid triangles, wild type; solid squares, OPI1ffat; solid crosses, scs2Δ. B, Opi1p-GFP localization over the same time course. Cells were imaged by fluorescence microscopy. A representative z-section is chosen for each image. Scale bar, 5 μm. White arrows, Opi1p-GFP associated with distinctive puncta.
In contrast to the relatively rapid initial increase in INO1 expression in the wild-type strain, INO1 expression in the OPI1ffat and scs2Δ strains increased by only about 15-fold during the first hour following the shift to I−C− medium (Fig. 4A). After the shift to inositol-free medium in the absence of choline (Fig. 4), the nuclear pool of Opi1ffatp-GFP had become less intense by 2 h but was still visible in most cells (Fig. 4B, 2 h, middle panel). After 2 h in I−C− medium, Opi1pffat-GFP in the OPI1ffat strain was associated with distinctive “puncta” (about 3–6/cell), and the nuclear pool of Opi1ffatp-GFP had become less intense but was still visible in most cells (Fig. 4B, 2 h, middle panel). At this time, INO1 expression in the OPI1ffat strain had increased by 210-fold, still significantly lagging the 375-fold increase seen in wild type (Fig. 4A). However, within 3 h following the shift to I−C− medium, INO1 expression in OPI1ffat had increased 320-fold, and signal from Opi1ffatp-GFP had reappeared to some extent in the nucleus, with fewer distinctive puncta (1–2/cell) remaining visible (Fig. 4B, 3 h, middle panel). In the scs2Δ strain, Opi1p-GFP fluorescence was still visible in the nucleus at 1 h following the shift to inositol-free medium (Fig. 4B, 1 h, right panel). At this time, one or two puncta, similar in appearance to those associated with Opi1ffatp-GFP in the OPI1ffat strain, were visible adjacent to the nucleus or plasma membrane in the scs2Δ strain (Fig. 4B). By 2 h following the shift to inositol-free medium, Opi1p-GFP in the scs2Δ strain had translocated more prominently to puncta adjacent to nucleus and plasma membrane. However, unlike either wild type or OPI1ffat (Fig. 4B, 2 h, left and middle panels), a diffuse pattern of Opi1p-GFP also persisted throughout the cell in the scs2Δ strain, possibly reflecting a reduction in cortical ER associated with the scs2Δ mutation, as reported by Loewen et al. (29). At 3 h, a diffuse pattern of Opi1p-GFP localization also remained visible in the nuclear region in the scs2Δ strain.
However, despite differences between the scs2Δ and wild-type strains in the pattern of Opi1p-GFP localization (Fig. 4B) following the shift to I−C− medium, overall expression of INO1 in the scs2Δ strain reached levels only slightly lower than those seen in wild type (Fig. 4A). By 2 h following the shift to I−C− medium, INO1 expression had increased by 325-fold in the scs2Δ strain, a level close to the 375-fold increase observed in the wild-type control during the same interval. Within 3 h following the shift to I−C− medium, INO1 expression had increased by 375-fold in the scs2Δ strain, a level that is not significantly different in comparison with the 410-fold increase observed in the wild-type strain (Fig. 4A). In contrast, INO1 expression in the OPI1ffat strain remained somewhat lower in comparison with the other two strains at each time point following the shift to inositol-free medium (Fig. 4A). After 3 h following the shift to I−C− medium, inositol was added back to each of the cultures. INO1 expression in all three strains decreased within about 30 min to values similar to those observed in these same strains between 0 and 1 h following the shift from I+C− to I−C− medium (Fig. 4A). Translocation of Opi1-GFP or Opi1ffatp-GFP back into the nuclei of the respective strains was observed within this same 30-min interval (Fig. 4B).
The puncta associated with Opi1ffatp-GFP, following a shift to medium lacking both inositol and choline, are ER-bound lipid droplets
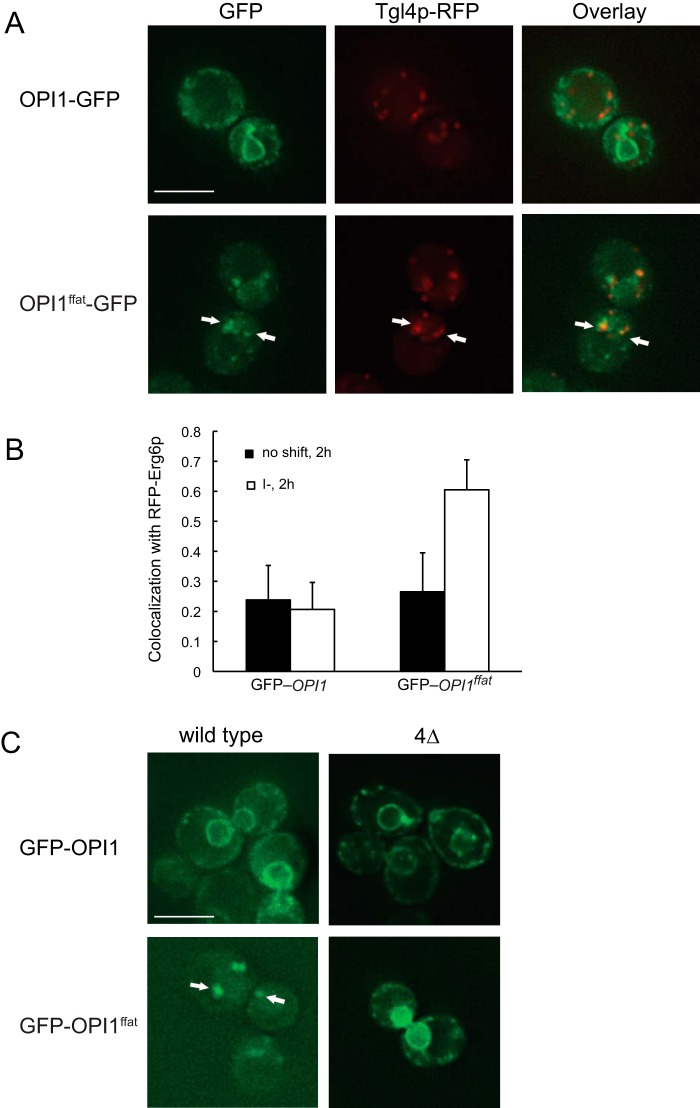
As described above, within 2 h following a shift of the OPI1ffat strain to I−C− medium, a fraction of the fluorescence of Opi1ffatp-GFP was associated with distinctive puncta, adjacent to the perinuclear ER (Fig. 4B, 2 h, middle panel). In addition to its role as precursor to PI and other membrane forming phospholipids, PA also serves as precursor, via DAG, in the synthesis of TAG (Fig. 1), a major constituent of lipid droplets. The appearance of Opi1ffatp-GFP–associated puncta in cells deprived of inositol is consistent with the report of Han et al. (44) showing that Opi1ffatp associates with areas of nascent lipid droplet formation. Therefore, given the morphology of the Opi1ffatp-GFP–labeled puncta and their proximity to the perinuclear ER, we suspected that these puncta represent a pool of PA associated with nascent lipid droplets. To test this hypothesis, we created cells co-expressing Tgl4p-RFP, a lipid droplet marker (45). We observed a significant overlap of Tgl4p-RFP– and Opi1ffatp-GFP–associated puncta following a 2-h shift to I−C− medium (Fig. 5A, right), supporting the identification of the puncta as lipid droplets. After 3 h of inositol starvation, Opi1ffatp-GFP had translocated in part back to the nucleoplasm. Following inositol addition, Opi1ffatp-GFP relocated completely to the nucleus within 30 min (Fig. 4B). To further verify and quantify Opi1ffatp co-localization with lipid droplets, we examined the localization of GFP-Opi1ffatp, expressed from a centromeric plasmid, in cells also expressing Erg6p-RFP, a protein that is abundant in yeast lipid droplets (46, 47). Co-localization of Erg6p and Opi1p was quantified using Pearson's correlation coefficient (PCC) (48) in 30 cells that exhibited clear expression of the plasmid versions of GFP-Opi1p or GFP-Opi1ffatp. The co-localization of GFP-Opi1p and RFP-Egr6p, both before and 2 h after the shift from I+C− to I−C− medium, was not significant (PCC = 0.24 and 0.21, respectively) (Fig. 5B). In contrast, in the strain co-expressing GFP-Opi1ffatp and RFP-Erg6p, PCC was 0.27 before the shift and 0.61 by 2 h following the shift to I−C− medium (Fig. 5B), indicating a significant increase in Opi1ffatp co-localization with lipid droplets at 2 h following the shift to I−C− medium. These observations are consistent with previous studies indicating that a pool of PA in the ER and/or lipid droplets (49), used for synthesis of DAG, precursor to TAG (Fig. 1), competes with the pool of PA used in the synthesis of CDP-DAG, precursor to PI and other membrane phospholipids. Thus, in cells grown in the presence of inositol, increased PI synthesis is correlated with decreased TAG accumulation (4, 43, 50, 51).
Figure 5.
Opi1pffat-GFP transiently colocalizes with lipid droplets following a shift to medium lacking inositol and choline. Overnight cultures grown in I+C− medium at 30 °C were diluted to A600 nm = 0.2 and allowed to grow to mid-logarithmic phase in I+C− medium at 30 °C. Cells were harvested by centrifugation, washed and resuspended in I−C− medium, incubated for 2 h, and analyzed by fluorescence microscopy. A, Tgl4-RFP, carried on a plasmid, was co-expressed with genomic Opi1p-GFP in wild type (top), or with genomic Opi1pffat-GFP in OPI1ffat (bottom). B, the CMY564 strain expressing genomic RFP-Erg6p was transformed with plasmids expressing either GFP-Opi1p or GFP-Opi1pffat. Colocalization of Tgl4-RFP with genomic Opi1p-GFP or Opi1pffat-GFP, respectively, was assessed using PCC (n = 30) between wavelength 528 nm (GFP) and 617 nm (RFP). PCC > 0.5 indicates significant correlation. C, the wild-type and dga1Δlro1Δare1Δare2Δ quadruple mutant (4Δ) strains were transformed with plasmids expressing either GFP-Opi1p (top) or GFP-Opi1pffat (bottom). Cells were grown and collected as indicated above and analyzed by fluorescence microscopy. A representative z-section was chosen for each image. Scale bars, 5 μm. Error bars, S.D.
To further test this hypothesis, translocation of Opi1p-GFP and Opi1ffatp-GFP was also examined in a dga1Δ lro1Δ are1Δ are2Δ quadruple mutant (Table 1), which is unable to make lipid droplets (52). A plasmid version of GFP-Opi1p or GFP-Opi1ffatp (as described under “Experimental procedures”; see Table 2) was transformed into the wild-type strain and the quadruple mutant. In both strains, GFP-Opi1p translocated to ER membranes within 2 h following transfer to I−C− medium (Fig. 5C, top), following a pattern and kinetics similar to that observed using the genomic Opi1p-GFP construct (Fig. 4B, 2 h, left panel). Following the shift of the OPI1ffat strain expressing the plasmid-borne GFP-Opi1ffatp to I−C− medium, fluorescence was also associated with puncta. However, in the dga1Δlro1Δare1Δare2Δ strain, Opi1ffatp was mainly localized in the nucleus and the perinuclear region, and no puncta were observed by 2 h following the shift to I−C− medium (Fig. 5C, bottom right), indicating that formation of the puncta is most likely related to lipid droplet formation.
Table 1.
Yeast strains used
| Strain | Genotype/description | Source/reference |
|---|---|---|
| BY4742 (parent strain) | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 (S288C) | Ref. 67 |
| YCY47 | BY4742 OPI1ffat | This study |
| SJY39 | BY4742 scs2Δ::KanMX6 | This study |
| SJY425 | BY4742 ino1Δ::HIS3 | This study |
| YCY3 | BY4742 OPI1 GFP::KanMX6 | This study |
| YCY5 | BY4742 OPI1ffat GFP::KanMX6 | This study |
| YCY7 | BY4742 OPI1 GFP::KanMX6 scs2Δ::URA3 | This study |
| LGY169 | BY4742 pct1Δ::LEU2 | This study |
| LGY541 | BY4742 OPI1ffat pct1Δ::LEU2 | This study |
| YCY45 | BY4742 with pBP73-C[GFP-OPI1] | This study |
| YCY46 | BY4742 with pBP73-C[GFP-OPI1ffat] | This study |
| YJP1078 | BY4742 dga1Δ::KanMX4 lro1Δ::KanMX4 are1Δ::KanMX4 are2Δ::KanMX4 | Ref. 71 |
| YCY36 | YJP1078 with pBP73-C[GFP-OPI1] | This study |
| YCY37 | YJP1078 with pBP73-C[GFP-OPI1ffat] | This study |
| CMY564 | BY4742 RFP-ERG6::KanMX | Ref. 47 |
| YCY96 | CMY564 with pBP73-C[GFP-OPI1] | This study |
| YCY97 | CMY564 with pBP73-C[GFP-OPI1ffat] | This study |
Table 2.
Plasmids used
| Plasmid | Genotype/description | Source/reference |
|---|---|---|
| pFA6a-GFP(S65T)-kanMX | Kanamycin resistance cassette for genomic N terminus tagging of GFP | Ref. 69 |
| pCRCU[TGL4-RFP] | URA3 cassette with TGL4 cloned at NotI-ClaI | Gift of S. Kohlwein |
| pBP73-C | pRS416 with the CPY1 promotor cloned at SacI-XbaI and GFP at Xba-BamHI | Ref. 72 |
| pBP73-C[GFP-OPI1] | pBP73-C with OPI1 cloned at BamHI-HindIII | This study |
| pBP73-C[GFP-OPI1ffat] | pBP73-C with OPI1ffat cloned at BamHI-HindIII | This study |
Following a shift to inositol-free medium in the presence of choline, INO1 fails to derepress in the OPI1ffat and scs2Δ strains
In wild-type cells grown in medium containing both inositol and choline and shifted to medium lacking inositol but containing choline (i.e. a shift from I+C+ to I−C+ medium; Fig. 6), derepression of INO1 was delayed by about 30–60 min in comparison with the same cells shifted to inositol-free medium in the absence of choline (i.e. from I+C− to I−C− medium; Fig. 4A). Within 2 h following the shift from I+C+ to I−C+ medium, Opi1p-GFP in the wild-type strain had exited the nucleus and was distinctly observed in the perinuclear ER (Fig. 6B). At this time point, INO1 expression had increased by about 100-fold (Fig. 4A), and by 3 h INO1 expression had increased by about 400-fold (Fig. 6A), an increase comparable with that seen in this same strain after the shift from I+C− to I−C− for 3 h (Fig. 4A).
Figure 6.
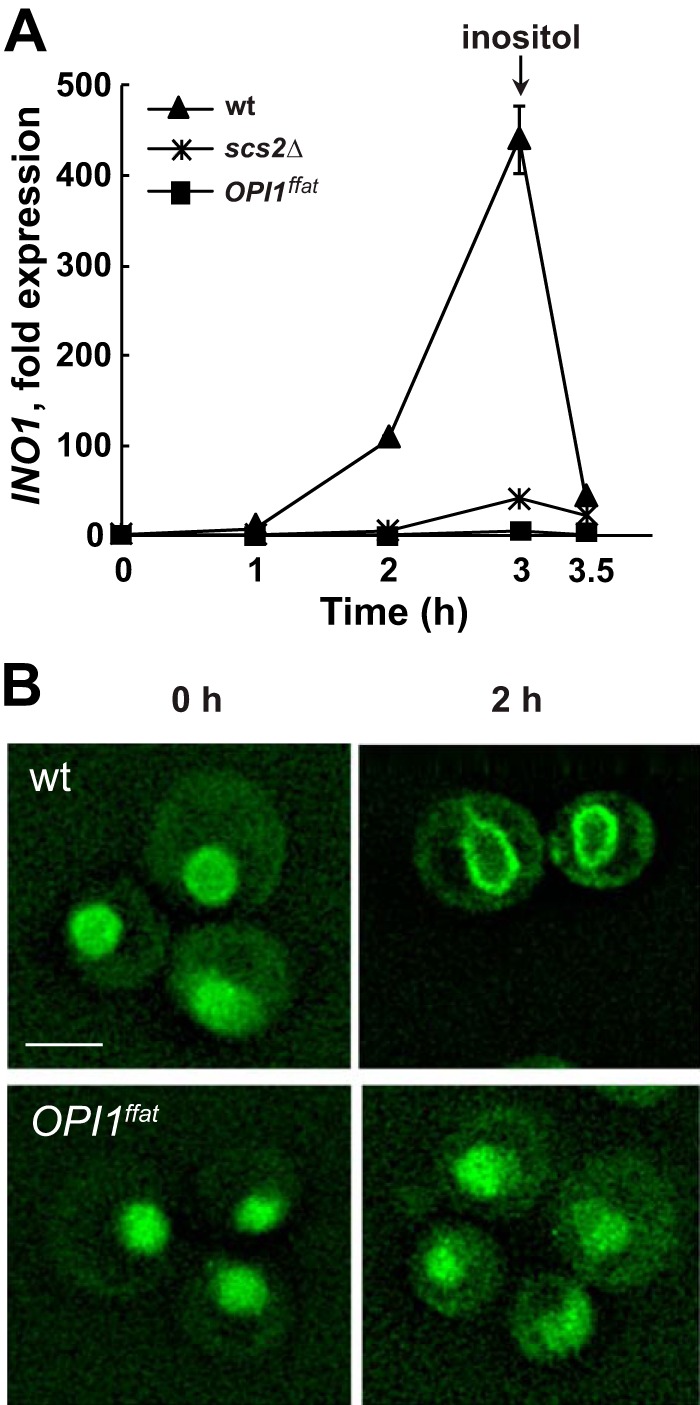
INO1 expression in wild-type, OPI1ffat, and scs2Δ strains and localization of Opi1p-GFP in wild-type or Opi1ffatp-GFP in scs2Δ and OPI1ffat strains, respectively, after a shift to I−C+ medium. A, overnight cultures of YCY3 (wild type), YCY5 (OPI1ffat), and YCY7 (scs2Δ) expressing genomic Opi1p-GFP or Opi1pffat-GFP, respectively, were diluted to A600 = 0.2 in I+C+ medium and allowed to grow to mid-logarithmic phase at 30 °C. Cells were harvested by centrifugation, washed and resuspended in I−C+ medium, and incubated for 3 h. Inositol was added back after 3 h of inositol starvation. Samples were taken at 0, 1, 2, and 3 h of inositol starvation and 30 min after adding back inositol. Total RNA was isolated and analyzed by RT-PCR as described under “Experimental procedures.” Solid triangles, wild type; solid squares, OPI1ffat; solid crosses, scs2Δ. B, Opi1p-GFP localization in wild type and Opi1pffat-GFP in OPI1ffat at 0 and 2 h after a shift to I−C+ medium. Cells were imaged by fluorescence microscopy. A representative z-section was chosen for each image. Scale bar, 5 μm.
In marked contrast to the behavior of Opi1p-GFP in the wild-type strain, Opi1ffatp-GFP in the OPI1ffat strain completely failed to exit the nucleus following a shift to inositol-free medium in the presence of choline (i.e. from I+C+ to I−C+ medium; Fig. 6B). Significantly, the Opi1ffatp-GFP–labeled puncta, observed in association with lipid droplets following the shift of OPI1ffat from I+C− to I−C− medium (Fig. 4B, 2 h, middle panel), were not observed after the shift from I+C+ to I−C+ medium (Fig. 6B). Similar to Opi1ffatp-GFP in the OPI1ffat strain, Opi1p-GFP in scs2Δ also failed to exit the nucleus following the shift from I+C+ to I−C+ medium. Moreover, the Opi1p-GFP–associated puncta, seen in this strain after the shift from I+C− to I−C− medium, were also not observed (data not shown). Furthermore, INO1 failed to derepress in both the OPI1ffat and scs2Δ strains following the shift to I−C+ medium (Fig. 6A). These observations suggest that a pool of PA, associated with the synthesis of DAG, precursor to TAG in nascent lipid droplets (44), is sufficient to attract Opi1ffatp or Opi1p from the nuclei of the OPI1ffat and scs2Δ strains, respectively, in the absence of inositol, but only when choline is also absent (compare Fig. 4B with Fig. 6B). This result is consistent with the hypothesis that synthesis of PC via the CDP-choline pathway (Fig. 1) competes for a pool of DAG derived from PA in the ER, a pool that, in the absence of exogenous choline, is available for increased production of TAG (Fig. 1) and lipid droplet formation (Fig. 4B).
Overall cellular PA levels increase in all three strains following a shift from I+ medium to I− medium, whether choline is present or not, but do not correlate with INO1 expression in the OPI1ffat and scs2Δ strains when choline is present
To determine whether the lack of the FFAT domain in Opi1p affects PA content, we performed lipid analysis under all growth conditions described above. PA levels in the wild-type strain increased in a comparable fashion following a shift from I+ to I− medium, whether choline was present or not (Fig. 7, compare A with B and C with D). There was also no significant difference in the levels of PA in the wild-type strain growing in I+C− medium versus I+C+ medium (Fig. 7, compare A with C). After the shift to inositol-free medium in the absence or presence of choline (i.e. from I+C− to I−C− medium or from I+C+ to I−C+ medium), PA levels rose significantly in the wild-type strain. However, the kinetics of the increase in PA levels were somewhat affected by the presence of choline, initially spiking higher at 2 h following the shift to I−C+ (Fig. 7D) and then dropping down by 3 h (Fig. 7E) to a level comparable with that seen at 2 h after the shift to I−C− medium (Fig. 7B). At 2 h after the shift to I−C+ medium, INO1 expression in wild type was significantly lower (Fig. 6A) than the level seen by 2 h following the shift to I−C− medium (Fig. 4A). However, by 3 h following the shift to I−C+ medium (Fig. 6A), INO1 expression in wild type had reached levels as high as or higher than those observed in this strain after the shift to I−C− medium (Fig. 4). This transient contrast between the kinetics of rising PA levels and INO1 derepression, in the presence versus the absence of choline, suggests that the rising pool of PA, accompanying the removal of inositol, is not as rapidly available for interaction with Opi1p as it is when choline is absent, an issue to be taken up under “Discussion.”
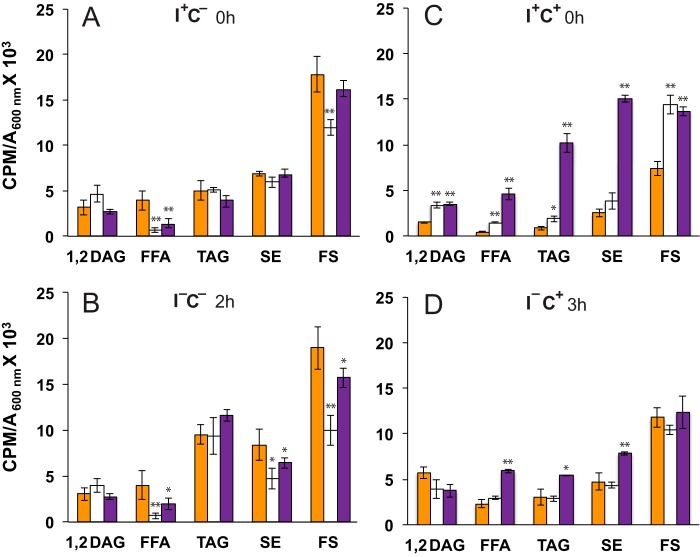
Figure 7.
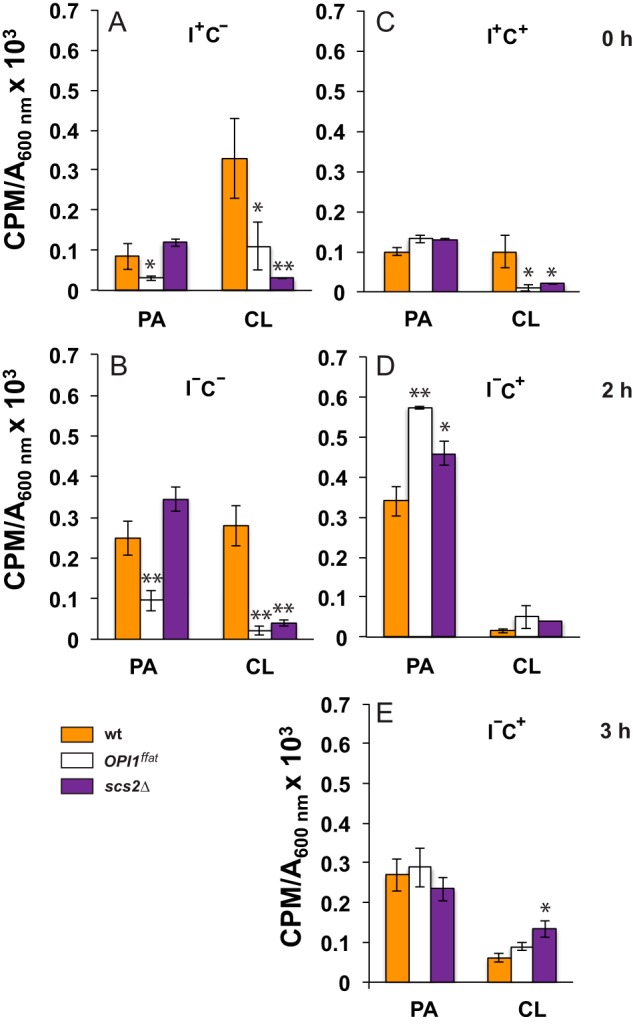
PA and CL levels in wild-type and mutant strains following a shift to medium lacking inositol. Overnight cultures grown in I+C− or I+C+ medium at 30 °C in the presence of 32P as described under “Experimental procedures” were diluted to A600 nm = 0.2 in I+C− or I+C+ medium at 30 °C maintaining label constant. Cells were allowed to grow to mid-logarithmic phase, and samples (time 0) were collected for lipid analysis (A and C). The remaining cultures were filtered and resuspended in I−C− medium or I−C+ medium maintaining label constant. Cultures growing in I−C− were allowed to continue growing for an additional 2 h (B), whereas I−C+ cultures were allowed to grow for an additional 2 and 3 h (D and E, respectively). At these time points, cells were collected for lipid analysis. Data are expressed as cpm of radiolabel 32P incorporated into total phospholipids per OD unit in the cell culture. Data are expressed as mean ± S.D. (error bars) (n = 3). **, p < 0.005; *, p < 0.05.
In contrast to both the wild-type and scs2Δ strains, PA levels in the OPI1ffat strain were significantly lower when growing in medium lacking choline, whether inositol was present or not (compare PA levels in OPI1ffat in comparison with wild type and scs2Δ; Fig. 7, A and B). This observation is consistent with the somewhat reduced level of INO1 expression observed in this strain in comparison with wild type after the shift to I−C− medium (Fig. 4A). Nevertheless, in each of the three strains, PA levels increased about 2-fold following the shift from I+C− medium to I−C− medium (Fig. 7). Furthermore, in each of the three strains, in the absence of choline, the increase in PA that followed the shift from I+C− to I−C− medium was correlated with the exit of Opi1p-GFP or Opi1pffat-GFP, respectively, from the nucleus (Fig. 4B) and with derepression of INO1 (Fig. 4A). Strikingly, however, after a shift to inositol-free medium in the presence of choline, Opi1p-GFP and Opi1pffat-GFP in the OPI1ffat and scs2Δ strains, respectively, failed to exit the nucleus, and INO1 failed to derepress (Fig. 6). Significantly, the failure of INO1 derepression in these two strains under these conditions occurred despite rising PA levels, comparable with or higher than those observed in the wild-type strain after the shift to I−C+ medium (compare PA levels in all three strains; Fig. 7, B, D, and E). Thus, when choline is present, direct interaction of Opi1p and Scs2p in the ER, in addition to rising cellular PA levels, is essential for INO1 expression.
CL levels are significantly reduced in the OPI1ffat and scs2Δ strains
In yeast, PA serves as precursor to two separate pools of CDP-DAG. In the ER, Cds1p catalyzes the production of CDP-DAG used in the synthesis of PI and PS (Fig. 1). PA used in the synthesis of CDP-DAG in the inner mitochondrial membrane is synthesized in the ER and must be transferred to the outer mitochondrial membrane (53–55) (Fig. 1), primarily via the ERMES (35–38). From the outer mitochondrial membrane, PA must be transferred to the inner mitochondrial membrane by a process requiring the Ups1p–Mdm35 complex (36, 53, 56). Once transferred to the inner mitochondrial membrane, PA serves as precursor to CDP-DAG, catalyzed by the mitochondrial CDP-DAG synthase, Tam41p (Fig. 1) (54). CDP-DAG is then converted to phosphatidylglycerol phosphate in the mitochondria. Phosphatidylglycerol phosphate is then dephosphorylated to form phosphatidylglycerol, the immediate precursor of CL (54).
Despite the significant increase in the wild-type strain in the level of PA, CL content was not greatly affected by the shift from I+C− to I−C− medium (in Fig. 7, compare relative changes in PA and CL levels in A with those in B). Moreover, in comparison with wild type, PA levels in the OPI1ffat strain were somewhat reduced, whereas those in scs2Δ were elevated, both before and after the shift to I−C− medium (Fig. 7, A and B). However, CL levels were greatly reduced in both OPI1ffat and scs2Δ, in comparison with wild type, both before and after the shift from I+C− to I−C− medium (Fig. 7, compare data in A with data in B). Because low CL content was reported in the opi1Δ strain in combination with a Pet− phenotype (34), we assessed growth of the scs2Δ and OPI1ffat strains, in comparison with wild type, on YPD plates supplemented with ethidium bromide, for the Pet− phenotype. Growth of all three strains was comparable under these conditions (data not shown). Thus, basic mitochondrial function was not compromised by the changes in lipid metabolism observed in scs2Δ and OPI1ffat strains. Consistent with these observations, the scs2Δ mutant was not identified among the mutations conferring the Pet− phenotype in a genome-wide screen conducted by Dunn et al. (57).
Whereas the shift of the wild-type strain from I+C− to I−C− medium had little impact on CL levels (Fig. 7, compare A with B), the presence of choline together with inositol (i.e. I+C+ versus I+C− medium) resulted in a significant decrease in CL in comparison with inositol alone (Fig. 7, compare CL levels in wild type in A with those in C). However, CL levels in the OPI1ffat and scs2Δ strains were significantly lower than those seen in the wild-type strain under these same conditions (i.e. in I+C+ medium; Fig. 7C). Following the shift from I+C+ to I−C+ medium, CL level in wild type decreased by an additional 3-fold in the first 2 h (Fig. 7, compare C with D) but recovered somewhat during the interval from 2 to 3 h (Fig. 7E). As described above, in contrast, the level of PA, precursor to CL, increased significantly in the wild-type strain after the shift from I+C+ to I−C+ medium (Fig. 7, compare C with D and E). Thus, the continuing low level of CL after the shift to I−C+ medium suggests that a specific pool of PA, accessible for transport from the ER to mitochondria in wild-type cells, is specifically impacted in the presence of exogenous choline.
TAG levels increased in all three strains after a shift to inositol-free medium in the absence of choline; however, when inositol and choline were both present, neutral lipid composition was affected in distinctly different ways in each of the three strains
TAG levels rose significantly in all three strains after the shift to inositol-free medium in the absence of choline (i.e. I+C− medium to I−C− medium; Fig. 8, compare A with B). However, DAG, the immediate precursor of TAG (Fig. 1), showed relatively little change in any of the three strains following a shift from I+C− medium to I−C− medium (Fig. 8, compare DAG levels in A with those in B). Indeed, in the wild-type strain, shifting from I+C− medium to I−C− medium had relatively little effect on any single neutral lipid category other than TAG (Fig. 8, A and B), as reported previously (43, 50). In contrast, in the OPI1ffat and scs2Δ strains, free fatty acids (FFA) were reduced, in comparison with wild type, both before and after the shift from I+C− medium to I−C− medium, and steryl ester (SE) levels in these two strains were reduced only after the shift to I−C− medium. Free sterols were also significantly reduced in the OPI1ffat SCS2 strain in comparison with OPI1 SCS2 both before and after the shift from I+C− medium to I−C− medium (Fig. 8, compare A with B).
Figure 8.
Neutral lipid levels in wild-type and mutant strains following a shift to medium lacking inositol. A–D, analysis of neutral lipid classes of YCY3 (wild type), YCY5 (OPI1ffat), and YCY7 (scs2Δ) strains grown in the presence of [1-14C]acetate in the same conditions described in the legend to Fig. 7. Data are expressed as cpm of radiolabel 1-14C incorporated into neutral lipids per OD unit in the cell culture. Data are expressed as mean ± S.D. (error bars) (n = 3). **, p < 0.005; *, p < 0.05.
However, in the presence of both choline and inositol (I+C+ medium; Fig. 8C), as compared with inositol alone (I+C− medium; Fig. 8A), the wild-type strain exhibited reductions of 50% or more in essentially all neutral lipids. In stark contrast to wild type, in the scs2Δ strain, every category of neutral lipid, except free sterols, was elevated in I+C+ medium (Fig. 8C), as compared with I+C− medium (Fig. 8A). In the OPI1ffat strain, the levels of essentially all of the individual neutral lipids growing in I+C+ medium were intermediate between the other two strains. However, neutral lipid levels in the OPI1ffat strain were more similar to those seen in wild type than in scs2Δ. We conclude that the higher levels of FFA, TAG, and SE observed in the scs2Δ strain growing in I+C+ medium are attributable to functions of Scs2p beyond its interaction with Opi1p in the ER.
After the shift to inositol-free medium in the presence of choline (i.e. from I+C+ to I−C+ medium), essentially all categories of neutral lipids in the wild-type strain increased (Fig. 8, compare C with D) while remaining generally lower, especially with respect to TAG, SE, and FFA, than the levels seen in this same strain after the shift from I+C− medium to I−C− medium (Fig. 8, compare data in B with data in D). However, DAG, immediate precursor to both PC via the CDP-choline pathway and TAG, a major constituent of lipid droplets (Fig. 1), is an exception. DAG levels were significantly lower in the wild-type strain growing in I+C+ versus I+C− medium (Fig. 8, compare A with C) but increased about 2-fold after the shift from I+C+ to I−C+ medium (compare data in Fig. 8, C and D). In contrast, no significant change in DAG levels occurred in the wild-type strain following the shift to inositol-free medium in the absence of choline (i.e. from I+C− to I−C− medium; Fig. 8, compare data in A with data in B). DAG levels in the OPI1ffat and scs2Δ strains, which were higher in I+C+ medium than those seen in wild type, also did not change significantly after the shift from to I−C+ medium.
However, following the shift from I+C+ to I−C+ medium, both TAG and SE levels underwent significant reductions in the scs2Δ strain, changes that were far more dramatic than those observed in OPI1ffat or wild type (Fig. 8, compare data in C with data in D). The level of FFA was also significantly elevated in scs2Δ, in comparison with the other two strains, both before and after the shift from I+C+ to I−C+ medium (Fig. 8, compare data in C with data in D). Again, these changes are presumably attributable to functions of Scs2p beyond those controlled by its interaction with Opi1p in the ER.
Effects of inositol and choline on phospholipid composition
Under steady-state growth conditions in the presence of inositol and absence of choline (I+C− medium), all three strains exhibited comparable levels of CDP-DAG, PS, and PE (Fig. 9A). PI and PC levels were also comparable in the wild-type and OPI1ffat strains in I+C− medium. However, in the scs2Δ strain, PI and PC levels were about 20 and 30% lower, respectively, in comparison with the levels observed in the other two strains (Fig. 9A). Following the shift to inositol-free medium in the absence of choline (I+C− to I−C− medium), levels of PI in all three strains declined significantly, as reported previously for wild-type strains (50), reaching levels equivalent to about 20% of the PI level seen in the wild-type strain before the shift to I−C− medium (Fig. 9B). The level of PS, precursor to PE (Fig. 1), was comparable in all three strains in I+C− medium and increased slightly in all three strains after the shift to I−C− medium (Fig. 9, compare data in A with data in B). However, a significant reduction in PE content was observed in all three strains when growing in the presence of choline, independent of inositol supplementation (compare PE levels in cells grown in the absence of choline (Fig. 9, A and B) with levels in cells grown in its presence (Fig. 9, C–E)) PE is synthesized from PS via two distinct pathways, localized to different cellular compartments, catalyzed either by Psd2p in the endosome or by Psd1p in the mitochondria (58) (Fig. 1). The Psd1p protein precursor is synthesized on cytoplasmic ribosomes and is processed in the mitochondria (59). Transcription of PSD1 is regulated by the Ino4p, Ino2p, and Opi1p transcription factors in response to inositol availability, whereas PSD2 is not (7).
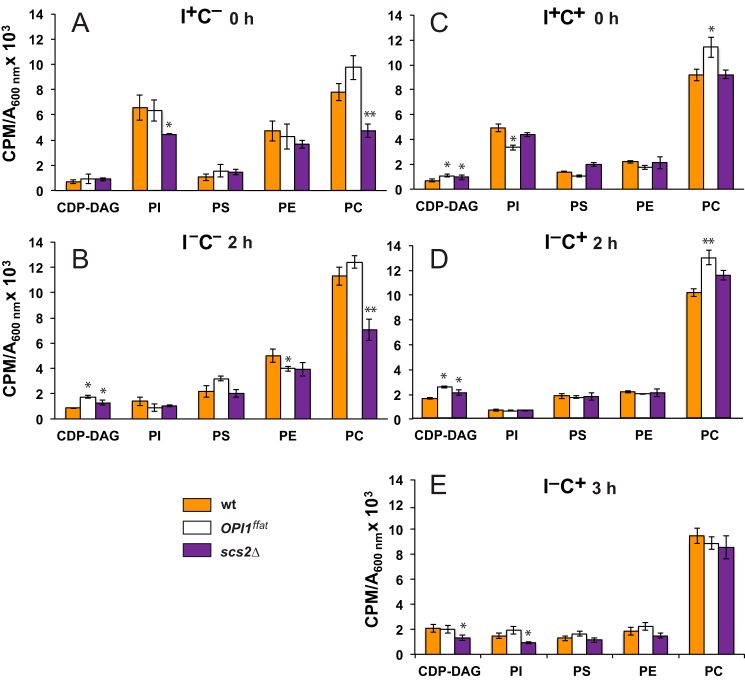
Figure 9.
CDP-DAG, PI, PS, PE, and PC levels in wild-type and mutant strains following a shift to medium lacking inositol. YCY3 (wild type), YCY5 (OPI1ffat), and YCY7 (scs2Δ) strains were grown in the presence of 32P in the same conditions described in the legend to Fig. 7. CDP-DAG, PI, PS, PE, and PC levels are expressed as cpm of radiolabel 32P incorporated into total phospholipids per OD unit in the cell culture. The legend for strains is the same as in Figs. 7 and 8. Data are expressed as mean ± S.D. (error bars) (n = 3). **, p < 0.005; *, p < 0.05.
The level of PC increased significantly in the wild-type strain (Fig. 9, A and B) when shifted to inositol-free medium in the absence of choline (i.e. from I+C− to I−C− medium), consistent with previous studies (50). However, when shifted to inositol-free medium in the presence of choline (i.e. from I+C+ to I−C+ medium; Fig. 9, compare data in C with data in D and E), PC levels in wild type changed minimally if at all. PC levels were significantly lower in the scs2Δ strain than in the other two strains, both before and after the shift to I−C− medium (Fig. 9, A and B). However, PC in the scs2Δ strain, when growing in I+C+ medium, was also significantly higher than in I+C− medium and was comparable with the level observed in the wild-type strain growing in I+C+ medium (Fig. 9A).
Discussion
In the present study, we compared changes in lipid metabolism and gene regulation in the wild-type, scs2Δ, and OPI1ffat strains to determine the individual and combined effects of exogenous inositol and choline on lipid metabolism, Opi1p localization, and INO1 expression. Fig. 10 provides a visual summary of the major findings and the significance of this work. One striking outcome of this study was the discovery that the mitochondrial lipid, CL, is significantly reduced in the OPI1ffat and scs2Δ strains in comparison with the wild-type strain (Fig. 7). These observations, coupled with the earlier report by Luévano-Martinez et al. (34) that the opi1Δ mutant exhibits similarly low CL levels, indicate that the reductions in CL content in these strains are most likely due to the loss of the Opi1p–Scs2p interaction in the ER, the one interaction these two proteins share in common. Moreover, we report that the scs2Δ and OPI1ffat mutations both confer weak Ino− phenotypes at 30 °C, phenotypes that are strengthened at higher growth temperatures and in the presence of choline (Fig. 2). In contrast, the opi1Δ mutant exhibits unregulated high-level constitutive expression of INO1 and other UASINO-containing genes and excretes excess inositol into the growth medium (4, 33, 60). Thus, the opi1Δ mutant shares no reported metabolic or regulatory phenotype with scs2Δ other than low CL levels. To support CL biosynthesis, PA synthesized in the ER must be transferred to the outer mitochondrial membrane and transferred to the inner mitochondrial membrane to be converted to CDP-DAG by Tam41p, the mitochondrial CDP-DAG synthase (35, 53, 54) (Fig. 1). Optimal synthesis of CL requires transfer of PA from the ER to the outer mitochondrial membrane via the ERMES complex (38). The mitochondrial pathway for PE synthesis, similarly, requires the transit of PS from the ER to the mitochondria (55), and deletion of several genes encoding ERMES subunits results in reduced levels of both CL and PE (38). For example, deletion of the MMM1 gene, encoding an ERMES subunit, containing a conserved “synaptotagmin-like mitochondrial lipid-binding” (SMP) domain (61), is associated with significantly reduced levels of both CL and PE in the mitochondria (38). However, deletion of MDM34, encoding an outer mitochondrial membrane protein, is associated with reduced levels of CL but does not affect PE levels (38).
Figure 10.
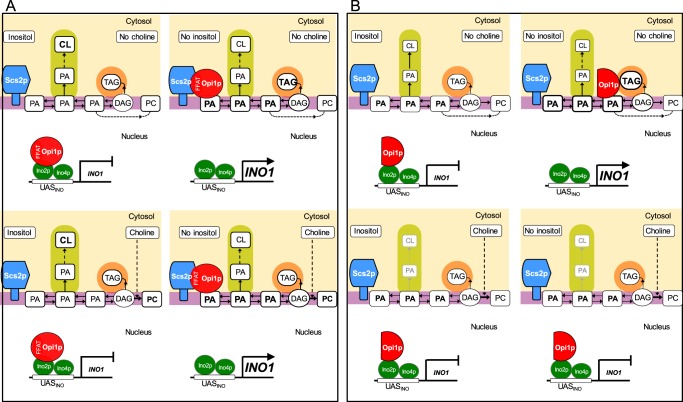
Effect of the Opi1p–Scs2p interaction on lipid metabolism and the regulation of phospholipid biosynthetic genes. A, wild-type cells exhibit normal repression and derepression of INO1 expression in response to changing levels of exogenous inositol. CL content is not affected by inositol in the wild type. Following a shift to medium lacking inositol with or without choline, Opi1p (depicted as a red circle with a FFAT domain) exits the nucleus and interacts with both Scs2p and PA in the ER. Moreover, TAG levels increase after the shift to medium lacking both inositol and choline. However, in the presence of choline, DAG consumption increases to synthesize PC, resulting in diminished TAG content. The presence of choline also results in a reduction in CL. B, when the OPI1ffat strain is shifted to medium lacking inositol, Opi1ffatp (depicted in a red semicircle, lacking a FFAT domain) leaves the nucleus and localizes to sites of lipid droplet formation, in contrast to the perinuclear ER, as is the case for Opi1p (as shown in A). In addition, in the presence of choline, CL content is also dramatically decreased. However, the presence of choline results in both reduced TAG levels and reduced lipid droplet formation. Under these conditions, the OPI1ffat strain is unable to interact with sites of lipid droplet formation in the perinuclear ER and remains in the nucleus, preventing INO1 expression. Purple box, ER; orange box, lipid droplet; green box, mitochondria; light yellow box, cytosol.
The reductions in CL content that we observed in scs2Δ and OPI1ffat strains relative to wild type (Fig. 7) are proportionately comparable with the reductions in CL reported in the mitochondrial lipids of mutants carrying deletions of ERMES subunits (38). Thus, the reductions in CL synthesis that we observed in the scs2Δ and OPI1ffat strains are consistent with the hypothesis that the interaction of Opi1p with Scs2p facilitates transfer of PA from the ER to the mitochondria. Moreover, simultaneous disruption of both ERMES and vCLAMP (vacuole and mitochondrial patch) resulted in a higher reduction of CL synthesis than disruption of ERMES alone (62). Thus, the residual synthesis of CL, which we observed in both the scs2Δ and OPI1ffat strains, compared with wild type (Fig. 7), could be the result of compensatory vCLAMP facilitation of PA transfer to mitochondria from the vacuole. Regardless, the low CL levels detected in the scs2Δ and OPI1ffat strains strongly support the hypothesis that the interaction of Opi1p and Scs2p facilitates optimal transfer of PA from the ER to mitochondria. However, when growing in the presence of choline, CL levels were reduced in all strains, and the relative reduction in CL levels in the presence of choline was proportionally much greater in the wild-type strain than in the OPI1ffat and scs2Δ strains, in which CL levels were already greatly reduced due to the loss of the Opi1p–Scs2p interaction (Fig. 7). As we discuss below, the presence of exogenous choline exerts distinctly different effects on specific pools of PA in the ER.
The presence of choline alone has a significant impact on TAG content. The substantial reduction in the level of TAG observed in the wild-type strain growing in the presence of choline is consistent with the hypothesis that PC synthesis via the CDP-choline pathway competes directly for a pool of DAG that serves as precursor to TAG synthesis. Also, in contrast to the dramatic increase in PI levels observed in the presence of inositol (Fig. 9), choline is associated with only a modest increase in PC levels in the wild-type strain, regardless of inositol supplementation. These relatively small changes in PC levels in the wild-type strain growing in the presence of choline are consistent with previous reports that choline induces turnover of PC by deacylation, via the Nte1p phospholipase B (63–66). However, in both the wild-type and OPI1ffat strains, supplementation with both choline and inositol was associated with a reduction in most neutral lipids, especially TAG. In contrast, most neutral lipids were markedly elevated in the scs2Δ strain in I+C+ medium, as compared with the other two strains as well as with its own neutral lipid composition in I+C− medium. These data are indicative of significant additional perturbations of neutral lipid metabolism related to the total loss of Scs2p function.
A major issue that remains to be discussed is the root cause of the choline-sensitive inositol auxotrophy of the OPI1ffat and scs2Δ strains. Both strains exhibit Ino− phenotypes that are more evident both in the presence of choline and at the higher growth temperature of 34 °C (Fig. 2). This phenotype is essentially identical to the phenotype of “choline-sensitive inositol auxotrophy” as originally described in association with the CSE mutant used in the isolation of the SCS2 gene as a high copy suppressor (31). Retention of the Opi1p repressor in the ER requires its interaction with PA in the ER, and this interaction is essential both for expression of INO1 and growth of wild-type cells in the absence of inositol (15). Opi1ffatp-GFP in the OPI1ffat strain and Opi1p-GFP in the scs2Δ strains, respectively, both retain the ability to interact with PA in the ER. However, neither the OPI1ffat nor the scs2Δ strain has the capacity for direct interaction between Opi1p and Scs2p in the ER. Importantly, after the shift to inositol-free medium in the absence of choline, Opi1ffatp-GFP in the OPI1ffat strain exited the nucleus and localized to distinctive puncta associated with synthesis of TAG in lipid droplet formation. Moreover, the level of INO1 derepression in the OPI1ffat strain following the shift from I+C− to I−C− medium was only slightly lower than that supported by Opi1p in the wild-type strain under the same conditions. Thus, we conclude that the pool of PA associated with lipid droplet formation in the ER, which is created by diversion of PA from PI to increasing TAG synthesis after the shift from I+C− to I−C− medium, is sufficiently robust to serve as a signal for the exit of Opi1ffatp from the nucleus. However, when choline is present in the absence of inositol, increased PC synthesis competes directly for the pool of DAG derived from PA, a pool of DAG in the ER that also serves as the immediate precursor for TAG. Thus, the presence of choline results in both reduced TAG levels and reduced lipid droplet formation. Under these conditions, the Opi1ffatp in the OPI1ffat strain is unable to interact with sites of lipid droplet formation in the perinuclear ER and remains in the nucleus, preventing INO1 expression. This, indeed, is the root cause of the choline-sensitive inositol auxotrophy shown in Fig. 2.
Experimental procedures
Strains
Yeast strains used are listed in Table 1. The parent strain BY4742 (MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0) derived from S288C (67) and mutants derived from BY4742 were used. All strains were maintained on YPD plates (1% yeast extract, 2% bactopeptone, 2% glucose, and 2% agar). Mutations in the FFAT motif (OPI1ffat) (residues 200–203 mutated from EFFD to ALLA) within the genomic OPI1 locus were created using the delitto perfetto method (68). The mutated sites are as described by Loewen et al. (15). Deletion mutant strains for PCT1 were generated in wild-type, scs2Δ, and OPI1ffat strains by PCR-mediated gene replacement, as described previously (69). The plasmid pRS315 was used as a template to generate a PCR fragment for the PCT1 gene disruption. The entire open reading frame of the PCT1 gene was replaced with the LEU2 marker gene. Leucine prototrophs were screened by colony PCR to verify integration at the correct genetic locus.
Construction of strains expressing Opi1p-GFP and Opi1pffat-GFP fusion proteins tagged at the C terminus of the OPI1 genomic locus
Fusions of GFP to the C termini of wild-type, scs2Δ, and OPI1ffat strains were constructed by PCR-mediated gene integration at the genomic OPI1 locus using the template plasmid pFA6a-GFP (S65T)-kanMX6 (a gift from M. Longtine) (69). The insertion of GFP was confirmed by PCR, and expression of genomic Opi1p-GFP and its variants was confirmed by Western blotting.
Analysis of Ino− phenotypes on solid media
The presence of choline has been shown to increase the severity of Ino− phenotypes (i.e. reduce residual growth in the absence of inositol) in a number of mutant strains in plating assays, including scs2Δ (14). Chromatographic assessment of residual choline, performed in our laboratory on samples of agar sourced from several vendors, indicated that all of these agar samples contained varying trace levels of choline (data not shown). However, in our analysis, agar sourced from Sigma contained the lowest residual trace of choline and was therefore used exclusively in all plate assays for assessing Ino− phenotypes in this study. Yeast strains were grown to mid-logarithmic phase in synthetic complete medium containing 75 μm inositol (I+ medium), harvested, washed with sterile distilled water, and resuspended in sterile distilled water at a concentration of 1.0 A600 nm/ml. 10-Fold serial dilutions were then spotted onto plates containing 0 μm inositol (I−) or 75 μm inositol (I+) with or without 1 mm choline (C+) and incubated at the indicated temperature of 30 or 34 °C for 3 days.
Protocol for assessing the Pet− phenotype
The opi1Δ mutation was shown to confer the Pet− phenotype (34), namely the inability to tolerate the loss of the mitochondrial genome (57). Accordingly, we tested the OPI1ffat strain for ethidium bromide sensitivity, a phenotype that is associated with complete loss of the mitochondrial genome, following the procedure used by Luévano-Martinez et al. (34). In brief, strains were cultured in YPD supplemented with 25 μg/ml filtered sterilized ethidium bromide at 30 °C to an A600 nm = 0.5. At this optical density, samples were subjected to serial dilution in sterile distilled water and spotted onto YPD and YPD plus 25 μg/ml ethidium bromide agar plates. Strains were grown for 2 days on the plates, whereupon the plates were examined for growth conditions under which the Pet− strains fail to grow.
Protocol for growth in liquid medium and shifting cells from medium containing inositol to medium lacking inositol
All studies on cells grown in liquid medium were conducted at 30 °C. As described by Gaspar et al. (43), cells were pregrown overnight in medium containing 75 μm inositol with or without 1 mm choline (i.e. I+C+ or I+C− medium). The following day, cultures were diluted back to A600 = 0.2 in the same medium at the same temperature and allowed to grow to mid-logarithmic growth phase, A600 = 0.5–0.6, in I+C− or I+C+ medium, respectively. Cells were then collected by filtration, washed with I−C− or I−C+ medium prewarmed to 30 °C, and resuspended in I−C− or I−C+ medium at the same temperature. Samples were harvested by filtration or centrifugation immediately after 0, 1, 2, and 3 h of growth. Inositol was then reintroduced to the cell cultures at a concentration of 75 μm, and cells were harvested 30 min after inositol readdition. Harvested cells were flash frozen on dry ice after harvesting and stored at −80 °C for RNA extraction. Changes in lipid metabolism and gene expression, as described below, were also measured over the same interval in a strain carrying Opi1-GFP (wild type; YCY3) and compared with its parent, BY4742 (Table 1), expressing untagged Opi1p. The level of expression of INO1 at each time point was not statistically different in the BY4742 strain, in comparison with the YCY3 strain at any time point before or after a shift from I+C− to I−C− medium or from I+C+ to I−C+ medium or following the readdition of inositol (data not shown). On this basis, the YCY3 strain was used as the “wild-type” control for analysis of gene expression, Opi1p localization, and lipid metabolism in response to inositol and choline supplementation.
RNA isolation and RT-PCR analysis
Strains were pregrown as described above in I+C− or I+C+ liquid medium at 30 °C and then shifted to I−C− or I−C+ medium, respectively, maintaining constant growth temperature. Total RNA was isolated using the RNeasy® minikit, including a DNA digestion with an RNase-free DNase set (both from Qiagen). 1 μg of RNA was transcribed into cDNA using oligo(dT)12–18 primer (0.5 μg), PCR grade dNTP mix (0.5 μm), First Strand Buffer (1×), DTT (10 mm), and 100 units of MaximaTM reverse transcriptase (Fermentas). Real-time PCR was performed on a StepOnePlusTM real-time PCR system (Applied Biosystems) using Maxima® probe/ROX qPCR Master Mix, No AmpErase® UNG (Applied Biosystems), and the following TaqMan® probes and primers: INO1, TaqMan® probe, 5′-FAM-CTGTTG CCC ATG GTT AGC CCA AAC G-TAMRA-3′; forward primer, 5′-GGA ATG ACG TTT ATG CTC CTT TTA A-3′; reverse primer, 5′-GTC CCA ACC AGA GAC GAC AAA-3′; ACT1, TaqMan® probe, 5′-FAM-TGC AAA CCG CTG CTC AAT CTT CTT CAAT-TAMRA-3′; forward primer, 5′-CGC CTT GGA CTT CGA ACA AG-3′; reverse primer, 5′-GAC CAT CTG GAA GTT CGT AGG ATT-3′. The ACT1 gene served as an internal standard for normalization.
In brief, the reaction mix in a volume of 25 μl consisted of 0.5 μm primers, 0.2 μm TaqMan® probe, 1× Master Mix, and 10 ng of cDNA. All reactions were performed in technical duplicate. Non-template control (10 ng of RNA) and non-reaction control (RNase-free water) were routinely performed. The thermal program for the PCR included stage 1 (95 °C for 10 min), stage 2 (95 °C for 0.5 min and 60 °C for 1 min for a total of 40 cycles), and stage 3 (hold at 4 °C). Relative quantitation was done using the ΔΔCt method (see the StepOnePlusTM user manual, Applied Biosystems). The ΔΔCt represents the change in mRNA expression after ACT1 normalization relative to the wild-type control calculated as follows: 2−(Gene Ctx − ACT1 Ctx) − (Gene Ctcr − ACT1 Ctcr), where Gene represents the mRNA under study (INO1), x refers to the strain from which the mRNA to be tested was derived (i.e. wild type or mutant), and cr refers to the control mRNA, the value of which was derived from the level of mRNA in the BY4742 parent strain, pregrown as described above in I+ medium at 30 °C, shifted to I− medium at the same temperature at time 0 h. The Ct (cycle threshold) is defined as the number of cycles required for the fluorescent signal to cross the threshold (i.e. to exceed background level). Each RT-PCR experiment was performed in triplicate.
Fluorescence microscopy
Wild type, scs2Δ, and OPI1ffat cells, expressing genomic or plasmid versions of GFP-tagged OPI1 or OPI1ffat, were grown overnight, as described above, at 30 °C in I+C− or I+C+ medium. Before microscopy, overnight cultures were diluted to A600 = 0.2 and allowed to continue to grow to mid-logarithmic phase in I+C− or I+C+ medium at 30 °C. After reaching A600 = 0.5–0.6, cultures were shifted, as described above, to I−C− or I−C+ medium, respectively, and allowed to grow for 2 h (A600 = 0.8–1.0) for single time point observations or to 4–5 h (A600 = 1.5–2) for time course observations. Cells were then concentrated to OD = 12.5 by centrifugation, and 3.5-μl samples of concentrated cultures were subjected to deconvolution fluorescence microscopy using a Deltaversion RT microscopy system (Applied Precision, LLC). Cells were viewed using a X71 Olympus microscope equipped with a PlanApo 100× objective (1.35 numeric aperture, Olympus), FITC, and rhodamine filters and a Cool Snap HQ digital camera (Photometrics). GFP images were acquired with the FITC filter set, and RFP/mCherry images were acquired with the RD-TR-PE filter set. Five or six z-sections from each strain were inspected. The acquired images were deconvolved using soft-WoRX version 3.5.0 software (Applied Precision, LLC). The colocalization of GFP-Opi1p or GFP-Opi1pffat with the lipid droplet marker RFP-Erg6p (Table 1) was analyzed with PCC (48) in soft-WoRX version 3.5.0. The number of prominent punctate structures associated with Opi1pffat-GFP was manually counted in 50 cells by scanning through the z-sections.
Phospholipid composition assessed by [32P]orthophosphate steady-state labeling
Changes in the composition of cellular phospholipids were determined over a time course of 3 h following a shift of actively growing cells from medium containing inositol to medium lacking inositol, followed by subsequent reintroduction of inositol into the medium. For this purpose, cells were grown overnight, as described above, in I+C− or I+C+ medium at 30 °C in the presence of 10 μCi/ml [32P]orthophosphate. The following day, cultures were diluted to A600 nm = 0.1 in I+C− or I+C+ medium at 30 °C maintaining label at 10 μCi/ml [32P]orthophosphate and allowed to grow to mid-logarithmic phase (A600 = 0.5–0.6). At this cell density, each culture was divided in half. One-half of each culture was filtered, washed with prewarmed medium containing inositol, and resuspended in I+C− or I+C+ medium at 30 °C, maintaining label at 10 μCi/ml [32P]orthophosphate. The other half was also filtered and then washed with prewarmed medium lacking inositol and resuspended in I−C− or I−C+ medium at 30 °C maintaining label at 10 μCi/ml [32P]orthophosphate. Samples from each culture were taken at 2 or 3 h following the shift. Labeled lipids were extracted as described by Gaspar et al. (50). The individual phospholipid species were resolved by two-dimensional thin layer chromatography (50).
For the assessment of CL content, lipids were labeled with [32P]orthophosphate and extracted, as described above, and the lipid extract was analyzed by one-dimensional thin layer chromatography according to the method developed by Vaden et al. (70). Briefly, Sigma-Aldrich Silicagel on TLC plates (layer thickness 250 μm) were dipped in 1.8% boric acid prepared in 100% ethanol, dried for 5 min, and baked for 15 min at 100 °C. Phospholipids were separated using the solvent system chloroform/ethanol/water/triethylamine (30/35/7/35, v/v/v/v) for at least 2 h. Phospholipid identity was based on the mobility of known standards and quantified on a STORM 860 PhosphorImager (Amersham Biosciences).
Neutral lipid composition assessed by steady-state labeling with [14C]acetate
Cells were grown as described above for 32P steady-state labeling, except that they were labeled in the presence of 2 μCi/ml [1-14C]acetate (specific activity, 57 mCi/mmol) to steady state in I+C− or I+C+ medium at 30 °C and then shifted to I−C− or I−C+ at 30 °C medium respectively, maintaining label, as described above, at 2 μCi/ml [1-14C]acetate. Changes in neutral lipid composition were monitored over a set time course of 2 or 3 h after the shift to medium lacking inositol, followed by inositol readdition. Samples were taken at 0 and 2 h following the shift to I−C− or I−C+ medium. 5-ml samples were mixed with 0.5 ml of 50% trichloroacetic acid and allowed to stand on ice for 20 min. Lipids were extracted and analyzed as described by Gaspar et al. (50). Labeled lipids on the chromatograms were quantified on a STORM 860 PhosphorImager (Amersham Biosciences), and metabolites were identified as described previously (50).
Author contributions
M. L. G., Y.-F. C., and S. A. J. performed the experiments and prepared the manuscript. M. A. performed experiments. S. A. H. directed the research and contributed to the preparation of the manuscript.
This work was supported by National Institutes of Health Grant GM-19629 (to S. A. H.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
- ER
- endoplasmic reticulum
- TAG
- triacylglycerols
- PI
- phosphatidylinositol
- PA
- phosphatidic acid
- DAG
- diacylglycerol
- PS
- phosphatidylserine
- PE
- phosphatidylethanolamine
- PC
- phosphatidylcholine
- FFA
- free fatty acids
- SE
- steryl ester
- CL
- cardiolipin
- I
- inositol
- C
- choline
- ERMES
- ER–mitochondria encounter structure
- PCC
- Pearson's correlation coefficient
- FAM
- 6-carboxyfluorescein
- TAMRA
- tetramethylrhodamine
- PCC
- Pearson correlation coefficient.
References
- 1. Bachhawat N., Ouyang Q., and Henry S. A. (1995) Functional characterization of an inositol-sensitive upstream activation sequence in yeast: a cis-regulatory element responsible for inositol-choline mediated regulation of phospholipid biosynthesis. J. Biol. Chem. 270, 25087–25095 [DOI] [PubMed] [Google Scholar]
- 2. Carman G. M., and Henry S. A. (1999) Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog. Lipid Res. 38, 361–399 [DOI] [PubMed] [Google Scholar]
- 3. Henry S. A., Gaspar M. L., and Jesch S. A. (2014) The response to inositol: regulation of glycerolipid metabolism and stress response signaling in yeast. Chem. Phys. Lipids 180, 23–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Henry S. A., Kohlwein S. D., and Carman G. M. (2012) Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 190, 317–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jesch S. A., Zhao X., Wells M. T., and Henry S. A. (2005) Genome-wide analysis reveals inositol, not choline, as the major effector of Ino2p-Ino4p and unfolded protein response target gene expression in yeast. J. Biol. Chem. 280, 9106–9118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hirsch J. P., and Henry S. A. (1986) Expression of the Saccharomyces cerevisiae inositol-1-phosphate synthase (INO1) gene is regulated by factors that affect phospholipid synthesis. Mol. Cell. Biol. 6, 3320–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jesch SAa H., SA. (2005) Yeast inositol phospholipids: synthesis, regulation and involvement in membrane trafficking and lipid signaling. in Cell Biology and Dynamics of Yeast Lipids (Daum G., ed.) pp. 105–131, Research Signpost, Kerala, India [Google Scholar]
- 8. Santiago T. C., and Mamoun C. B. (2003) Genome expression analysis in yeast reveals novel transcriptional regulation by inositol and choline and new regulatory functions for Opi1p, Ino2p, and Ino4p. J. Biol. Chem. 278, 38723–38730 [DOI] [PubMed] [Google Scholar]
- 9. Schüller H. J., Hahn A., Tröster F., Schütz A., and Schweizer E. (1992) Coordinate genetic control of yeast fatty acid synthase genes FAS1 and FAS2 by an upstream activation site common to genes involved in membrane lipid biosynthesis. EMBO J. 11, 107–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Donahue T. F., and Henry S. A. (1981) myo-Inositol-1-phosphate synthase: characteristics of the enzyme and identification of its structural gene in yeast. J. Biol. Chem. 256, 7077–7085 [PubMed] [Google Scholar]
- 11. Jesch S. A., Liu P., Zhao X., Wells M. T., and Henry S. A. (2006) Multiple endoplasmic reticulum-to-nucleus signaling pathways coordinate phospholipid metabolism with gene expression by distinct mechanisms. J. Biol. Chem. 281, 24070–24083 [DOI] [PubMed] [Google Scholar]
- 12. Culbertson M. R., and Henry S. A. (1975) Inositol-requiring mutants of Saccharomyces cerevisiae. Genetics 80, 23–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ambroziak J., and Henry S. A. (1994) INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J. Biol. Chem. 269, 15344–15349 [PubMed] [Google Scholar]
- 14. Villa-García M. J., Choi M. S., Hinz F. I., Gaspar M. L., Jesch S. A., and Henry S. A. (2011) Genome-wide screen for inositol auxotrophy in Saccharomyces cerevisiae implicates lipid metabolism in stress response signaling. Mol. Genet. Genomics 285, 125–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Loewen C. J., Gaspar M. L., Jesch S. A., Delon C., Ktistakis N. T., Henry S. A., and Levine T. P. (2004) Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science 304, 1644–1647 [DOI] [PubMed] [Google Scholar]
- 16. Wagner C., Dietz M., Wittmann J., Albrecht A., and Schüller H. J. (2001) The negative regulator Opi1 of phospholipid biosynthesis in yeast contacts the pleiotropic repressor Sin3 and the transcriptional activator Ino2. Mol. Microbiol. 41, 155–166 [DOI] [PubMed] [Google Scholar]
- 17. Kagiwada S., Hosaka K., Murata M., Nikawa J., and Takatsuki A. (1998) The Saccharomyces cerevisiae SCS2 gene product, a homolog of a synaptobrevin-associated protein, is an integral membrane protein of the endoplasmic reticulum and is required for inositol metabolism. J. Bacteriol. 180, 1700–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kagiwada S., and Zen R. (2003) Role of the yeast VAP homolog, Scs2p, in INO1 expression and phospholipid metabolism. J. Biochem. 133, 515–522 [DOI] [PubMed] [Google Scholar]
- 19. Loewen C. J., Roy A., and Levine T. P. (2003) A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 22, 2025–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Loewen C. J., and Levine T. P. (2005) A highly conserved binding site in vesicle-associated membrane protein-associated protein (VAP) for the FFAT motif of lipid-binding proteins. J. Biol. Chem. 280, 14097–14104 [DOI] [PubMed] [Google Scholar]
- 21. Murphy S. E., and Levine T. P. (2016) VAP, a versatile access point for the endoplasmic reticulum: review and analysis of FFAT-like motifs in the VAPome. Biochim. Biophys. Acta 1861, 952–961 [DOI] [PubMed] [Google Scholar]
- 22. Holthuis J. C., and Menon A. K. (2014) Lipid landscapes and pipelines in membrane homeostasis. Nature 510, 48–57 [DOI] [PubMed] [Google Scholar]
- 23. Levine T. (2004) Short-range intracellular trafficking of small molecules across endoplasmic reticulum junctions. Trends Cell Biol. 14, 483–490 [DOI] [PubMed] [Google Scholar]
- 24. Prinz W. A. (2014) Bridging the gap: membrane contact sites in signaling, metabolism, and organelle dynamics. J. Cell Biol. 205, 759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stefan C. J., Manford A. G., Baird D., Yamada-Hanff J., Mao Y., and Emr S. D. (2011) Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell 144, 389–401 [DOI] [PubMed] [Google Scholar]
- 26. Manford A. G., Stefan C. J., Yuan H. L., Macgurn J. A., and Emr S. D. (2012) ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell 23, 1129–1140 [DOI] [PubMed] [Google Scholar]
- 27. Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 28. Levine T. P., and Munro S. (2001) Dual targeting of Osh1p, a yeast homologue of oxysterol-binding protein, to both the Golgi and the nucleus-vacuole junction. Mol. Biol. Cell 12, 1633–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Loewen C. J., Young B. P., Tavassoli S., and Levine T. P. (2007) Inheritance of cortical ER in yeast is required for normal septin organization. J. Cell Biol. 179, 467–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wilson J. D., Thompson S. L., and Barlowe C. (2011) Yet1p-Yet3p interacts with Scs2p-Opi1p to regulate ER localization of the Opi1p repressor. Mol. Biol. Cell 22, 1430–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hosaka K., Nikawa J., Kodaki T., and Yamashita S. (1992) A dominant mutation that alters the regulation of INO1 expression in Saccharomyces cerevisiae. J. Biochem. 111, 352–358 [DOI] [PubMed] [Google Scholar]
- 32. Skehel P. A., Martin K. C., Kandel E. R., and Bartsch D. (1995) A VAMP-binding protein from Aplysia required for neurotransmitter release. Science 269, 1580–1583 [DOI] [PubMed] [Google Scholar]
- 33. Greenberg M. L., Goldwasser P., and Henry S. A. (1982) Characterization of a yeast regulatory mutant constitutive for synthesis of inositol-1-phosphate synthase. Mol. Gen. Genet. 186, 157–163 [DOI] [PubMed] [Google Scholar]
- 34. Luévano-Martinez L. A., Appolinario P., Miyamoto S., Uribe-Carvajal S., and Kowaltowski A. J. (2013) Deletion of the transcriptional regulator opi1p decreases cardiolipin content and disrupts mitochondrial metabolism in Saccharomyces cerevisiae. Fungal Genet. Biol. 60, 150–158 [DOI] [PubMed] [Google Scholar]
- 35. Baile M. G., Lu Y. W., and Claypool S. M. (2014) The topology and regulation of cardiolipin biosynthesis and remodeling in yeast. Chem. Phys. Lipids 179, 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Connerth M., Tatsuta T., Haag M., Klecker T., Westermann B., and Langer T. (2012) Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science 338, 815–818 [DOI] [PubMed] [Google Scholar]
- 37. Kornmann B., Currie E., Collins S. R., Schuldiner M., Nunnari J., Weissman J. S., and Walter P. (2009) An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325, 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Osman C., Haag M., Potting C., Rodenfels J., Dip P. V., Wieland F. T., Brügger B., Westermann B., and Langer T. (2009) The genetic interactome of prohibitins: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J. Cell Biol. 184, 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nikawa J., Murakami A., Esumi E., and Hosaka K. (1995) Cloning and sequence of the SCS2 gene, which can suppress the defect of INO1 expression in an inositol auxotrophic mutant of Saccharomyces cerevisiae. J. Biochem. 118, 39–45 [DOI] [PubMed] [Google Scholar]
- 40. Chao J. T., Wong A. K., Tavassoli S., Young B. P., Chruscicki A., Fang N. N., Howe L. J., Mayor T., Foster L. J., and Loewen C. J. (2014) Polarization of the endoplasmic reticulum by ER-septin tethering. Cell 158, 620–632 [DOI] [PubMed] [Google Scholar]
- 41. Tsukagoshi Y., Nikawa J., and Yamashita S. (1987) Molecular cloning and characterization of the gene encoding cholinephosphate cytidylyltransferase in Saccharomyces cerevisiae. Eur. J. Biochem. 169, 477–486 [DOI] [PubMed] [Google Scholar]
- 42. Weiss S. B., Smith S. W., and Kennedy E. P. (1958) The enzymatic formation of lecithin from cytidine diphosphate choline and d-1,2-diglyceride. J. Biol. Chem. 231, 53–64 [PubMed] [Google Scholar]
- 43. Gaspar M. L., Hofbauer H. F., Kohlwein S. D., and Henry S. A. (2011) Coordination of storage lipid synthesis and membrane biogenesis: evidence for cross-talk between triacylglycerol metabolism and phosphatidylinositol synthesis. J. Biol. Chem. 286, 1696–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Han S., Binns D. D., Chang Y. F., and Goodman J. M. (2015) Dissecting seipin function: the localized accumulation of phosphatidic acid at ER/LD junctions in the absence of seipin is suppressed by Sei1p(DeltaNterm) only in combination with Ldb16p. BMC Cell Biol. 16, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kurat C. F., Wolinski H., Petschnigg J., Kaluarachchi S., Andrews B., Natter K., and Kohlwein S. D. (2009) Cdk1/Cdc28-dependent activation of the major triacylglycerol lipase Tgl4 in yeast links lipolysis to cell-cycle progression. Mol. Cell 33, 53–63 [DOI] [PubMed] [Google Scholar]
- 46. Athenstaedt K., Zweytick D., Jandrositz A., Kohlwein S. D., and Daum G. (1999) Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J. Bacteriol. 181, 6441–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Curwin A. J., Fairn G. D., and McMaster C. R. (2009) Phospholipid transfer protein Sec14 is required for trafficking from endosomes and regulates distinct trans-Golgi export pathways. J. Biol. Chem. 284, 7364–7375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Manders E. M., Stap J., Brakenhoff G. J., van Driel R., and Aten J. A. (1992) Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J. Cell Sci. 103, 857–862 [DOI] [PubMed] [Google Scholar]
- 49. Grillitsch K., Connerth M., Köfeler H., Arrey T. N., Rietschel B., Wagner B., Karas M., and Daum G. (2011) Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited: lipidome meets proteome. Biochim. Biophys. Acta 1811, 1165–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gaspar M. L., Aregullin M. A., Jesch S. A., and Henry S. A. (2006) Inositol induces a profound alteration in the pattern and rate of synthesis and turnover of membrane lipids in Saccharomyces cerevisiae. J. Biol. Chem. 281, 22773–22785 [DOI] [PubMed] [Google Scholar]
- 51. Gaspar M. L., Jesch S. A., Viswanatha R., Antosh A. L., Brown W. J., Kohlwein S. D., and Henry S. A. (2008) A block in endoplasmic reticulum-to-Golgi trafficking inhibits phospholipid synthesis and induces neutral lipid accumulation. J. Biol. Chem. 283, 25735–25751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sandager L., Gustavsson M. H., Ståhl U., Dahlqvist A., Wiberg E., Banas A., Lenman M., Ronne H., and Stymne S. (2002) Storage lipid synthesis is non-essential in yeast. J. Biol. Chem. 277, 6478–6482 [DOI] [PubMed] [Google Scholar]
- 53. Tatsuta T., Scharwey M., and Langer T. (2014) Mitochondrial lipid trafficking. Trends Cell Biol. 24, 44–52 [DOI] [PubMed] [Google Scholar]
- 54. Tamura Y., Harada Y., Nishikawa S., Yamano K., Kamiya M., Shiota T., Kuroda T., Kuge O., Sesaki H., Imai K., Tomii K., and Endo T. (2013) Tam41 is a CDP-diacylglycerol synthase required for cardiolipin biosynthesis in mitochondria. Cell Metab. 17, 709–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tamura Y., Sesaki H., and Endo T. (2014) Phospholipid transport via mitochondria. Traffic 15, 933–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Watanabe Y., Tamura Y., Kawano S., and Endo T. (2015) Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria. Nat. Commun. 6, 7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dunn C. D., Lee M. S., Spencer F. A., and Jensen R. E. (2006) A genomewide screen for petite-negative yeast strains yields a new subunit of the i-AAA protease complex. Mol. Biol. Cell 17, 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kannan M., Riekhof W. R., and Voelker D. R. (2015) Transport of phosphatidylserine from the endoplasmic reticulum to the site of phosphatidylserine decarboxylase2 in yeast. Traffic 16, 123–134 [DOI] [PubMed] [Google Scholar]
- 59. Di Bartolomeo F., Wagner A., and Daum G. (2017) Cell biology, physiology and enzymology of phosphatidylserine decarboxylase. Biochim. Biophys. Acta 1862, 25–38 [DOI] [PubMed] [Google Scholar]
- 60. Greenberg M. L., Reiner B., and Henry S. A. (1982) Regulatory mutations of inositol biosynthesis in yeast: isolation of inositol-excreting mutants. Genetics 100, 19–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. AhYoung A. P., Jiang J., Zhang J., Khoi Dang X., Loo J. A., Zhou Z. H., and Egea P. F. (2015) Conserved SMP domains of the ERMES complex bind phospholipids and mediate tether assembly. Proc. Natl. Acad. Sci. U.S.A. 112, E3179–E3188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Elbaz-Alon Y., Rosenfeld-Gur E., Shinder V., Futerman A. H., Geiger T., and Schuldiner M. (2014) A dynamic interface between vacuoles and mitochondria in yeast. Dev. Cell 30, 95–102 [DOI] [PubMed] [Google Scholar]
- 63. Dowd S. R., Bier M. E., and Patton-Vogt J. L. (2001) Turnover of phosphatidylcholine in Saccharomyces cerevisiae: the role of the CDP-choline pathway. J. Biol. Chem. 276, 3756–3763 [DOI] [PubMed] [Google Scholar]
- 64. Fernández-Murray J. P., and McMaster C. R. (2005) Glycerophosphocholine catabolism as a new route for choline formation for phosphatidylcholine synthesis by the Kennedy pathway. J. Biol. Chem. 280, 38290–38296 [DOI] [PubMed] [Google Scholar]
- 65. Fernández-Murray J. P., and McMaster C. R. (2007) Phosphatidylcholine synthesis and its catabolism by yeast neuropathy target esterase 1. Biochim. Biophys. Acta 1771, 331–336 [DOI] [PubMed] [Google Scholar]
- 66. Zaccheo O., Dinsdale D., Meacock P. A., and Glynn P. (2004) Neuropathy target esterase and its yeast homologue degrade phosphatidylcholine to glycerophosphocholine in living cells. J. Biol. Chem. 279, 24024–24033 [DOI] [PubMed] [Google Scholar]
- 67. Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., and Boeke J. D. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 [DOI] [PubMed] [Google Scholar]
- 68. Storici F., Lewis L. K., and Resnick M. A. (2001) In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 19, 773–776 [DOI] [PubMed] [Google Scholar]
- 69. Longtine M. S., McKenzie A. 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., and Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 70. Vaden D. L., Gohil V. M., Gu Z., and Greenberg M. L. (2005) Separation of yeast phospholipids using one-dimensional thin-layer chromatography. Anal. Biochem. 338, 162–164 [DOI] [PubMed] [Google Scholar]
- 71. Petschnigg J., Wolinski H., Kolb D., Zellnig G., Kurat C. F., Natter K., and Kohlwein S. D. (2009) Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast. J. Biol. Chem. 284, 30981–30993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rue S. M., Mattei S., Saksena S., and Emr S. D. (2008) Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol. Biol. Cell 19, 475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]