Abstract
Automatic segmentation of brain tissues and white matter hyperintensities of presumed vascular origin (WMH) in MRI of older patients is widely described in the literature. Although brain abnormalities and motion artefacts are common in this age group, most segmentation methods are not evaluated in a setting that includes these items. In the present study, our tissue segmentation method for brain MRI was extended and evaluated for additional WMH segmentation. Furthermore, our method was evaluated in two large cohorts with a realistic variation in brain abnormalities and motion artefacts.
The method uses a multi-scale convolutional neural network with a T1-weighted image, a T2-weighted fluid attenuated inversion recovery (FLAIR) image and a T1-weighted inversion recovery (IR) image as input. The method automatically segments white matter (WM), cortical grey matter (cGM), basal ganglia and thalami (BGT), cerebellum (CB), brain stem (BS), lateral ventricular cerebrospinal fluid (lvCSF), peripheral cerebrospinal fluid (pCSF), and WMH.
Our method was evaluated quantitatively with images publicly available from the MRBrainS13 challenge (n = 20), quantitatively and qualitatively in relatively healthy older subjects (n = 96), and qualitatively in patients from a memory clinic (n = 110). The method can accurately segment WMH (Overall Dice coefficient in the MRBrainS13 data of 0.67) without compromising performance for tissue segmentations (Overall Dice coefficients in the MRBrainS13 data of 0.87 for WM, 0.85 for cGM, 0.82 for BGT, 0.93 for CB, 0.92 for BS, 0.93 for lvCSF, 0.76 for pCSF). Furthermore, the automatic WMH volumes showed a high correlation with manual WMH volumes (Spearman's ρ = 0.83 for relatively healthy older subjects). In both cohorts, our method produced reliable segmentations (as determined by a human observer) in most images (relatively healthy/memory clinic: tissues 88%/77% reliable, WMH 85%/84% reliable) despite various degrees of brain abnormalities and motion artefacts.
In conclusion, this study shows that a convolutional neural network-based segmentation method can accurately segment brain tissues and WMH in MR images of older patients with varying degrees of brain abnormalities and motion artefacts.
Keywords: Brain MRI, Segmentation, White matter hyperintensities, Deep learning, Convolutional neural networks, Motion artefacts, Brain atrophy
Highlights
-
•
A method for brain tissue segmentation was extended for additional WMH segmentation.
-
•
Additional WMH segmentation does not compromise tissue segmentation performance.
-
•
High overlap and volume correlation between automatic and manual segmentations.
-
•
Evaluation in two cohorts with a varying degree of abnormalities and artefacts.
-
•
Accurate segmentations despite brain abnormalities and motion artefacts.
1. Introduction
Segmentation of brain tissues and white matter hyperintensities of presumed vascular origin (WMH) is widely being performed in MR images of older patients and is especially relevant in the context of neurovascular and neurodegenerative diseases De Groot et al., 2000, Ikram et al., 2008, De Bresser et al., 2010a, De Bresser et al., 2010b, Driscoll et al., 2009, Giorgio and De Stefano, 2013, Wardlaw et al., 2013.
Numerous automatic brain tissue segmentation methods already exist, with varying performance De Boer et al., 2010, De Bresser et al., 2011, Mendrik et al., 2015. The segmentation performance depends for example on image acquisition factors, such as MR field strength (Heinen et al., 2016) and MRI motion artefacts, and patient specific factors, such as brain abnormalities. Although brain abnormalities (e.g. WMH) and MRI motion artefacts are common in older patients, brain segmentation methods are not commonly evaluated in a setting that includes these items.
Previous work on automatic WMH segmentation (Caligiuri et al., 2015) consists of methods that use supervised classification Anbeek et al., 2004, Klöppel et al., 2011, Steenwijk et al., 2013, Ghafoorian et al., 2016 or detection of WMH as outliers of tissue segmentation Van Leemput et al., 2001, De Boer et al., 2009, Schmidt et al., 2012, Sudre et al., 2014, Sudre et al., 2015, Roura et al., 2015, Jain et al., 2015, Kuijf et al., 2016. Segmentation of WMH often has a lower performance than segmentation of brain tissues, because of the larger heterogeneity of WMH. Furthermore, compared with brain tissue segmentations, WMH segmentations are likely more susceptible to the presence of motion artefacts and other brain abnormalities, such as brain infarcts.
Convolutional neural networks have gained a lot of attention in the recent years because of their effectiveness in learning layers of convolution kernels directly from training images, instead of relying on explicitly defined features. In the field of MR brain image segmentation, convolutional neural networks have been used for brain tissue segmentation Zhang et al., 2015, Moeskops et al., 2016a, Moeskops et al., 2016b and various brain abnormality segmentation tasks Pereira et al., 2016, Brosch et al., 2016, Havaei et al., 2017, Havaei et al., 2016, Kamnitsas et al., 2017, Ghafoorian et al., 2017a, Ghafoorian et al., 2017b, Valverde et al., 2017.
We have previously developed a segmentation method that uses a convolutional neural network for brain tissue segmentation in neonatal and adult brain MRI (Moeskops et al., 2016a). In the present study, this brain tissue segmentation method was extended and evaluated in data from the MRBrainS13 challenge (Mendrik et al., 2015) to additionally include WMH segmentation. Furthermore, our method was evaluated in two large cohorts (relatively healthy older subjects and patients from a memory clinic), with a realistic variation in brain abnormalities and motion artefacts. The cohorts are therefore representative data sets for clinical application in a wide range of elderly patients.
2. Methods
2.1. Data
The method was evaluated with images from three different data sets. For all three data sets, MR images were acquired with a Philips Achieva 3T scanner using the same acquisition protocol: a 3D T1-weighted image (TR: 7.9 ms, TE: 4.5 ms), a T1-weighted inversion recovery (IR) image (TR: 4416 ms, TE: 15 ms, TI: 400 ms), and a T2-weighted fluid attenuated inversion recovery (FLAIR) image (TR: 11,000 ms, TE: 125 ms, TI: 2800 ms) (Mendrik et al., 2015). The 3D T1-weighted image and the T1-weighted IR image were registered to the T2-weighted FLAIR image with elastix (Klein et al., 2010). After registration, all images had a voxel size of 0.96 × 0.96 × 3.0 mm3. The images were corrected for MR field bias and brain masks were generated with SPM12 (Ashburner and Friston, 2005).
2.1.1. MRBrainS13
The MRBrainS131 (Mendrik et al., 2015) framework consists of MR images and manual segmentations from 20 patients (age, mean ± standard deviation: 71 ± 4 years; 10 male, 10 female). The MR images were manually segmented in eight classes: white matter (WM), cortical grey matter (cGM), basal ganglia and thalami (BGT), cerebellum (CB), brain stem (BS), lateral ventricular cerebrospinal fluid (lvCSF), peripheral cerebrospinal fluid (pCSF), and WMH. Note that the MRBrainS13 challenge only includes evaluation of three combined tissue classes: white matter (including WMH), grey matter (including BGT) and CSF (pCSF and lvCSF) instead of all eight classes.
2.1.2. Relatively healthy older subjects
Patients with type 2 diabetes mellitus and healthy controls were included from the Utrecht Diabetic Encephalopathy Study part 2 (UDES2) (Reijmer et al., 2013). The images used in MRBrainS13 were selected from the UDES2 cohort. From the UDES2 cohort we analysed images from 96 additional patients (age, mean ± standard deviation: 71 ± 5 years; 58 male, 38 female; 51 with type 2 diabetes mellitus and 45 healthy controls). Reference segmentations of WMH were performed by manual outlining on the FLAIR images using relatively strict criteria (Brundel et al., 2014).
2.1.3. Patients from a memory clinic
Patients with cognitive impairment from a memory clinic were included from the Dutch Parelsnoer Study (Aalten et al., 2014). From the Parelsnoer cohort we analysed 110 patients (age, mean ± standard deviation: 76 ± 8 years; 56 male, 54 female) that were included at the University Medical Center Utrecht. No manual reference segmentations were performed for these images.
2.2. Automatic segmentation method
Our previously described automatic segmentation method (Moeskops et al., 2016a) was extended to include WMH as an additional segmentation class, resulting in 9 output nodes (WM, cGM, BGT, CB, BS, lvCSF, pCSF, WMH and background). In contrast to the previously described approach that used a single input image, the current method uses three input images: a T1-weighted image, a T2-weighted FLAIR image and a T1-weighted IR image.
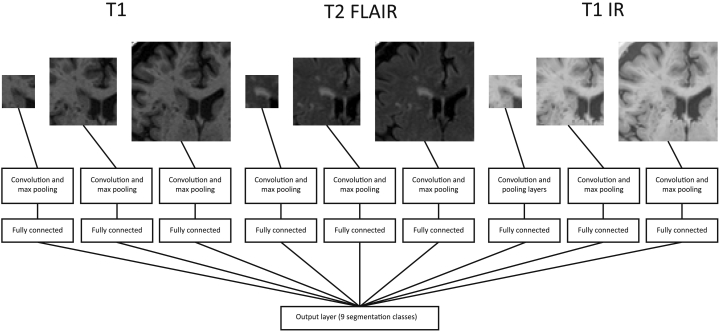
From each of these three images, 2D patches of three different sizes (25 × 25, 51 × 51 and 75 × 75 voxels) are extracted centred around each voxel, therefore resulting in 9 inputs. A CNN architecture with 9 branches (one for each of these 9 inputs) is used, corresponding to the architecture used in the previous work for orthogonal inputs. In total, this network architecture has 2,267,721 trainable parameters. A schematic of the network is shown in Fig. 1.
Fig. 1.
Overview of the network with nine branches, using three different input patch sizes from three different images. Details can be found in the paper by Moeskops et al. (2016a).
Similar to the previously described method, the network is trained in 10 epochs, where in each epoch 50,000 randomly selected samples are extracted from every class in each of the training images. If fewer than 50,000 samples are available for a certain class in a certain image, all available samples are included in the training set. In our training set, the number of WMH samples ranged from 0 to 12,880 per patient, resulting in inclusion of all WMH samples. The weights in the network are optimised with RMSprop (Tieleman and Hinton, 2012) using categorical cross-entropy as loss function. Dropout (Srivastava et al., 2014) is used on the fully connected layers to decrease overfitting.
2.3. Experiments
The method was trained using images from MRBrainS13 (n = 20). First, it was trained using the 5 images as training data and evaluated on the 15 test images, corresponding to the training and test sets in the MRBrainS13 challenge. Second, the method was trained in leave-one-subject-out over all 20 images to allow comparison with previous work on WMH segmentation using the same data Kuijf et al., 2014, Raidou et al., 2016. Third, the method was trained using all 20 images in MRBrainS13 and evaluated on a set of relatively healthy older subjects (n = 96) and patients from a memory clinic (n = 110), to evaluate the method in the presence of motion artefacts and brain abnormalities.
2.4. Evaluation of the automatic segmentation method
To evaluate the performance of the method, the brain tissue and WMH segmentations were quantitatively evaluated with the images from MRBrainS13 using:
-
•
Dice coefficients computed for each segmentation class in each image and subsequently averaged over all test images, to evaluate the overlap between the automatic and reference segmentations. This evaluation allows comparison with the MRBrainS13 challenge and our previous work on tissue segmentation (Moeskops et al., 2016a).
-
•
Dice coefficients computed for each segmentation class over all test images combined. This evaluation allows direct comparison with previous work on WMH segmentation using the same data Kuijf et al., 2014, Raidou et al., 2016.
-
•
Mean surface distances (MSD) computed for each segmentation class in each image and subsequently averaged over all test images, to evaluate the surface distance between the automatic and reference segmentations.
Because not all three input images may be acquired in every study, we also evaluate the performance using less than these three input images. To this end, we evaluate the method using only the T1-weighted and T2-weighted FLAIR images as input (i.e. leaving out the T1-weighted IR images) and using only the T2-weighted FLAIR images as input (i.e. leaving out both the T1-weighted and the T1-weighted IR images). In these cases, the images are, respectively, input for a network with six branches (three patch sizes from two images) and three branches (three patch sizes from one image) instead of nine branches. Furthermore, to assess if the method is not overfitting as a result of the large number of parameters in the network with nine branches, we also evaluate the method when the three images were used as three-channel input for three branches instead of nine separate branches.
2.5. Evaluation of the segmentation method in the presence of brain abnormalities and motion artefacts
To evaluate the performance of the method in the presence of motion artefacts and brain abnormalities, quantitative and qualitative evaluation of the MR images from the two large patient cohorts was performed.
2.5.1. Quantitative evaluation of WMH segmentation in relatively healthy older subjects
Quantitatively, the WMH segmentations in the MR images from relatively healthy older subjects (n = 96) were evaluated with:
-
•
Correlation analysis between the automatic and manual reference WMH volumes, to evaluate the level of correspondence between the automatic and reference volumes. Spearman's rank correlation was used instead of Pearson's correlation because the WMH volumes are not normally distributed. Instead, the WMH volumes follow a distribution skewed to the large volumes, i.e. most patients have a small lesion volume and only a few patients have a large lesion volume.
-
•
Sensitivity on the level of WMH lesion detection, i.e. quantifying the number of lesions that were detected, not taking into account if the volume or shape matched the reference segmentation. Therefore, a detected lesion (i.e. a 3D connected component) was considered a true positive when it (partially) overlapped with the manual reference segmentation.
-
•
Free-response receiver operating characteristic (FROC) curves on the level of WMH lesions, showing sensitivity versus number of false positive WMH detections per patient. Because of the class imbalance, which results in a high specificity, an FROC curve on the level of lesion detection provides a more insightful evaluation metric than a standard, voxel-based, ROC curve. Because of this lesion-based analysis, small false positive detections could have a large influence on the performance. To assess this influence, the FROC curves were computed with and without a greyscale opening operation on the probabilistic output. Greyscale opening was performed using a spherical structuring element with a radius of 1 mm. In the evaluated images, this resulted in only in-plane horizontal and vertical neighbours, i.e. 4-connectivity.
-
•
Overall detection error rate (DER) and overall outline error rate (OER) Wack et al., 2012, Steenwijk et al., 2013, to evaluate the error caused by missed lesions (DER) or different outlining of lesions (OER), where .
To allow comparison with other WMH segmentation methods, two publicly available methods were evaluated. First, we have evaluated the lesion prediction algorithm (LPA) as implemented in the LST toolbox version 2.0.15 for SPM (Schmidt et al., 2012). Second, we have evaluated the cascaded CNN as proposed by Valverde et al. (2017). We have trained this cascaded CNN using the same training set as we have used for our method, i.e. all 20 patients in MRBrainS13. Because the method balances positive and negative samples per image, no samples were selected from the image that did not have WMH, resulting in training samples from 19 patients.
2.5.2. Qualitative evaluation in relatively healthy older subjects and patients from a memory clinic
Qualitatively, the MR images from the relatively healthy older subjects (n = 96) and the MR images from the patients from a memory clinic (n = 110) were visually scored by an observer with 10 years of experience in neuroimaging and brain segmentation (JdB) using:
-
•
Conventional visual scoring methods, including the Fazekas scale for deep WMH (Fazekas et al., 1987) and the global cortical atrophy (GCA) scale (Pasquier et al., 1996). For both measures, scoring is performed in four classes (0–3).
-
•
A custom visual scoring method for motion artefacts, where all three images per patient were separately scored in four classes: no motion (0), low motion (1), medium motion (2), high motion (3) (see Fig. 2).
-
•
Visual scoring of the segmentation quality for tissue segmentation and WMH segmentation, as being reliable (1) or not reliable (0). Segmentation errors were considered relative to the volume of the affected brain tissue. Relatively small segmentation errors were considered reliable segmentations. Relatively medium to large segmentation errors were considered not reliable segmentations.
Fig. 2.
Different classes of motion artefacts in T1-weighted (top row), T1-weighted IR (middle row) and T2-weighted FLAIR images (bottom row).
For each of the qualitative scores, the intra-rater variability is assessed by performing the score twice for 20 randomly selected patients (10 relatively healthy older subjects and 10 patients from a memory clinic). The agreement is computed with Cohen's linearly weighted κ coefficients.
3. Results
3.1. Evaluation of the automatic segmentation method
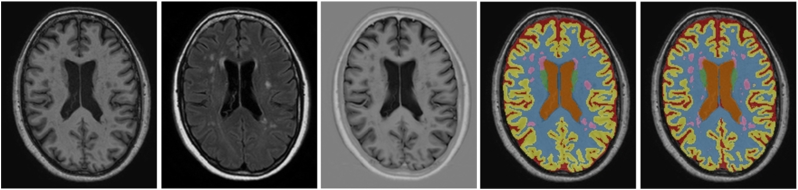
Quantitative evaluation was performed using the MR images from MRBrainS13 (n = 20) with segmentations of eight classes (WM, cGM, BGT, CB, BS, lvCSF, pCSF and WMH). An example segmentation result is shown in Fig. 3. The results when trained with 5 images and evaluated with 15 test images are shown in Table 1, top left, the results when trained in leave-one-subject-out cross-validation are shown in Table 2.
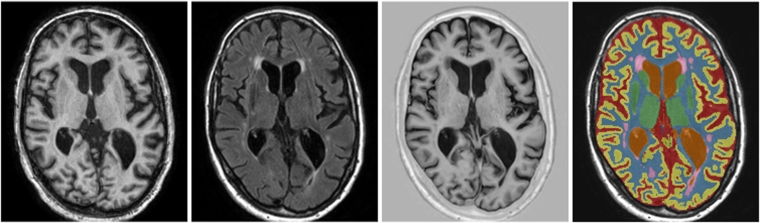
Fig. 3.
Example segmentation for one of the test images from the MRBrainS13 challenge, trained using the 5 training images available within MRBrainS13. From left to right: T1-weighted image, T2-weighted FLAIR image, T1-weighted IR image, reference segmentation and automatic segmentation.
Table 1.
Evaluation of the MRBrainS13 images in terms of Dice coefficients (mean ± standard deviation), overall Dice coefficient and MSD [mm] (mean ± standard deviation). Four different experiments are shown, each using 5 training subjects and 15 test subjects. Top left: the network using all 3 input images in 9 network branches. Top right: the network using 2 input images (leaving out the T1-weighted IR image) in 6 network branches. Bottom left: the network using 1 input image (only the T2-weighted FLAIR image) in 3 network branches. Bottom right: the network using all 3 input images in 3 network branches with 3-channel input. For each experiment, from top to bottom: white matter (WM), cortical grey matter (cGM), basal ganglia and thalami (BGT), cerebellum (CB), brain stem (BS), lateral ventricular cerebrospinal fluid (lvCSF), peripheral cerebrospinal fluid (pCSF) and white matter hyperintensities (WMH). *Because no WMH were found in 1 of the 20 subjects, the average Dice and average MSD for WMH were computed over 14 of the 15 test subjects. The overall Dice for WMH was computed over all test subjects.
| 3 inputs, 9 branches |
2 inputs, 6 branches (no IR) |
|||||
|---|---|---|---|---|---|---|
| Dice | Overall Dice | MSD [mm] | Dice | Overall Dice | MSD [mm] | |
| WM | 0.87 ± 0.02 | 0.87 | 0.31 ± 0.05 | 0.88 ± 0.02 | 0.88 | 0.27 ± 0.03 |
| cGM | 0.84 ± 0.01 | 0.84 | 0.24 ± 0.04 | 0.85 ± 0.01 | 0.85 | 0.21 ± 0.02 |
| BGT | 0.80 ± 0.03 | 0.79 | 0.64 ± 0.13 | 0.82 ± 0.02 | 0.82 | 0.63 ± 0.11 |
| CB | 0.91 ± 0.02 | 0.91 | 0.66 ± 0.27 | 0.91 ± 0.02 | 0.91 | 0.97 ± 0.66 |
| BS | 0.89 ± 0.02 | 0.89 | 0.67 ± 0.23 | 0.90 ± 0.02 | 0.90 | 0.62 ± 0.21 |
| lvCSF | 0.91 ± 0.03 | 0.90 | 0.30 ± 0.19 | 0.92 ± 0.03 | 0.93 | 0.30 ± 0.09 |
| pCSF | 0.74 ± 0.03 | 0.74 | 0.53 ± 0.10 | 0.71 ± 0.03 | 0.72 | 0.57 ± 0.11 |
| WMH | 0.54 ± 0.13* | 0.63 | 3.15 ± 1.82* | 0.53 ± 0.14* | 0.64 | 2.04 ± 1.13* |
| 1 input, 3 branches (only FLAIR) |
3 inputs, 3 branches with 3 channels |
|||||
|---|---|---|---|---|---|---|
| Dice | Overall Dice | MSD [mm] | Dice | Overall Dice | MSD [mm] | |
| WM | 0.80 ± 0.03 | 0.81 | 0.57 ± 0.09 | 0.88 ± 0.02 | 0.88 | 0.28 ± 0.04 |
| cGM | 0.76 ± 0.01 | 0.76 | 0.39 ± 0.04 | 0.84 ± 0.01 | 0.84 | 0.22 ± 0.02 |
| BGT | 0.77 ± 0.02 | 0.77 | 0.87 ± 0.13 | 0.81 ± 0.02 | 0.81 | 0.70 ± 0.12 |
| CB | 0.88 ± 0.03 | 0.88 | 1.64 ± 1.07 | 0.90 ± 0.03 | 0.90 | 1.64 ± 0.96 |
| BS | 0.85 ± 0.03 | 0.85 | 0.92 ± 0.28 | 0.90 ± 0.02 | 0.90 | 0.68 ± 0.51 |
| lvCSF | 0.87 ± 0.05 | 0.89 | 0.65 ± 0.37 | 0.91 ± 0.04 | 0.92 | 0.29 ± 0.09 |
| pCSF | 0.67 ± 0.04 | 0.68 | 0.71 ± 0.11 | 0.73 ± 0.04 | 0.73 | 0.54 ± 0.12 |
| WMH | 0.51 ± 0.14* | 0.62 | 2.11 ± 1.01* | 0.43 ± 0.14* | 0.54 | 3.15 ± 1.64* |
Table 2.
Evaluation of the MRBrainS13 images in terms of Dice coefficients (mean ± standard deviation), overall Dice coefficient and MSD [mm] (mean ± standard deviation). Leave-one-subject-out (LOSO) cross-validation over all 20 subjects for the network using all 3 input images in 9 network branches. From top to bottom: white matter (WM), cortical grey matter (cGM), basal ganglia and thalami (BGT), cerebellum (CB), brain stem (BS), lateral ventricular cerebrospinal fluid (lvCSF), peripheral cerebrospinal fluid (pCSF) and white matter hyperintensities (WMH). *Because no WMH were found in 1 of the 20 subjects, the average Dice and average MSD for WMH were computed over 19 of the 20 test subjects. The overall Dice for WMH was computed over all test subjects.
| 3 inputs, 9 branches (LOSO) |
|||
|---|---|---|---|
| Dice | Overall Dice | MSD [mm] | |
| WM | 0.87 ± 0.02 | 0.87 | 0.27 ± 0.04 |
| cGM | 0.85 ± 0.02 | 0.85 | 0.20 ± 0.02 |
| BGT | 0.82 ± 0.03 | 0.82 | 0.63 ± 0.17 |
| CB | 0.93 ± 0.02 | 0.93 | 0.65 ± 0.22 |
| BS | 0.92 ± 0.03 | 0.92 | 0.45 ± 0.22 |
| lvCSF | 0.93 ± 0.03 | 0.93 | 0.22 ± 0.07 |
| pCSF | 0.76 ± 0.04 | 0.76 | 0.46 ± 0.12 |
| WMH | 0.59 ± 0.19* | 0.67 | 4.14 ± 12.07* |
The results using only the T1-weighted and T2-weighted FLAIR images as input, i.e. leaving out the T1-weighted IR images, are listed in Table 1, top right. The performance was similar to the results with all three images, in some cases even slightly better. However, the average Dice coefficient for pCSF decreased from 0.74 ± 0.03 to 0.71 ± 0.03. This could be explained by the outer border of pCSF that has a large intensity differene with the bone on the T1-weighted IR images. The results using only the T2-weighted FLAIR image as input are listed in Table 1, bottom left. The performance was, however, poorer than when three or two input images were used.
The results of the experiment where the three images were used as input for three branches with a three-channel input instead of nine separate branches are shown in Table 1, bottom right. The performance was similar for most segmentation classes, with WMH as a clear exception. The average Dice coefficient for WMH decreased from 0.54 ± 0.13 to 0.43 ± 0.14. When using a 3-channel input instead of separate branches, the information of all three images is combined in a single set of features after the first convolution layer, instead of allowing each of the branches to focus on extracting the relevant information from each of the images.
Compared with our previous work on brain tissue segmentation (Moeskops et al., 2016a) using the same data and evaluation (MRBrainS13 with 5 training images and 15 test images), the extension of the method for additional segmentation of WMH resulted in a similar performance for the brain tissue segmentations (Average Dice coefficients of (with WMH vs. without WMH) 0.87 vs. 0.88 for WM, 0.84 vs. 0.84 for cGM, 0.80 vs. 0.81 for BGT, 0.91 vs. 0.90 for CB, 0.89 vs. 0.90 for BS, 0.91 vs. 0.92 for lvCSF, and 0.74 vs. 0.76 for pCSF).
Compared with previous work on WMH segmentation Kuijf et al., 2014, Raidou et al., 2016 using the same data and evaluation (MRBrainS13 with leave-one-subject-out cross-validation over 20 images), the overall Dice coefficients for WMH achieved by our method were substantially higher (0.67 vs. 0.57 (Kuijf et al., 2014) and 0.58 (Raidou et al., 2016)), even though the previous work also included additional features based on diffusion-weighted MR images.
The MRBrainS13 challenge provides a framework to evaluate three combined segmentation classes (WM, GM and CSF) instead of the eight segmentation classes that were evaluated in our work. When we combined the eight segmentation classes of our method to WM, GM and CSF, the Dice coefficients in the MRBrainS13 framework were 0.88 for WM, 0.84 for GM and 0.77 for CSF. The most recent submissions to MRBrainS13 are reported on the website2.
3.2. Evaluation of the segmentation method in the presence of brain abnormalities and motion artefacts
Data from two large cohort studies is evaluated: relatively healthy older subjects and patients from a memory clinic.
3.2.1. Quantitative evaluation of WMH segmentation in relatively healthy older subjects
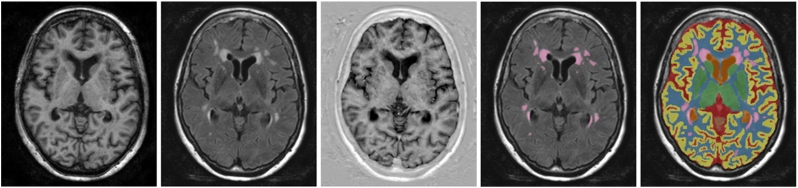
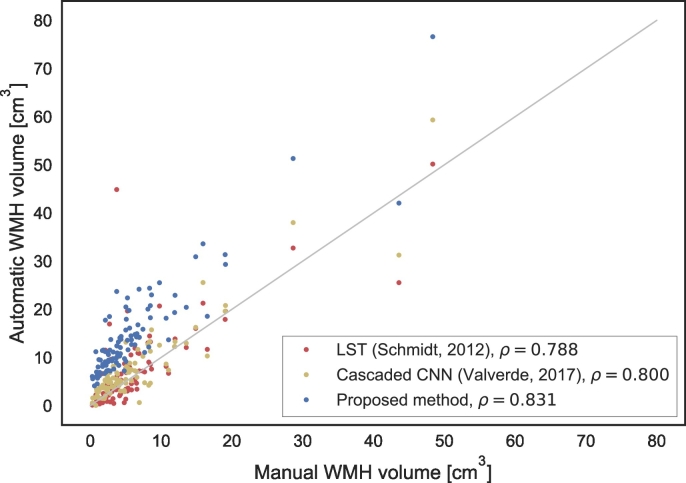
An example of the automatic segmentation compared with the reference WMH segmentation for one of the relatively older subjects with motion artefacts in the MR images is shown in Fig. 4. Despite these motion artefacts, the segmentation is visually of good quality. Furthermore, from this example it can be observed that the automatic segmentation generally produces somewhat larger segmentations of WMH than the reference segmentation. This originates from the MRBrainS13 data, which was used to train the method and where a larger definition of WMH for the manual segmentations is used. The same effect can be seen in Fig. 5, where the automatic and reference volumes are compared. Although a consistently larger WMH segmentation volume was produced by the automatic method, a high correlation (ρ = 0.83) was obtained (Fig. 5).
Fig. 4.
Example segmentation for one of the relatively healthy older subjects with motion artefacts in the MR images, trained using all 20 patients of MRBrainS13. From left to right: T1-weighted image, T2-weighted FLAIR image, T1-weighted IR image, reference segmentation and automatic segmentation.
Fig. 5.
Correlation between automatic and manual WMH volumes for the relatively healthy older subjects (n = 96) in terms of Spearman's ρ. The method was trained using all 20 patients of MRBrainS13. The method is compared with the lesion prediction algorithm of LST (Schmidt et al., 2012) and a cascaded CNN (Valverde et al., 2017).
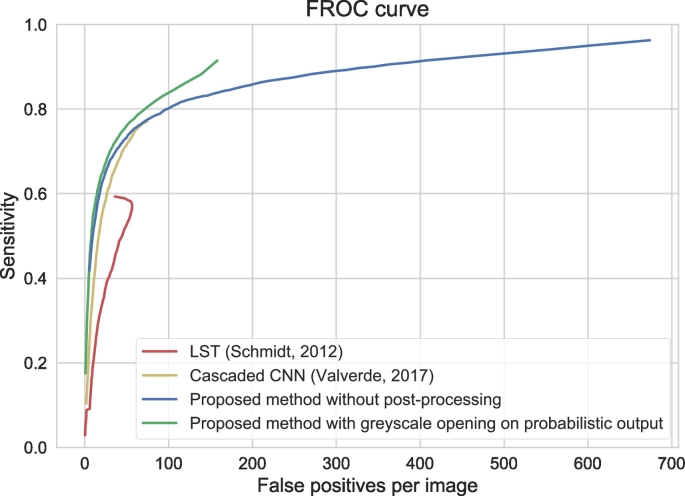
FROC analysis for lesion detection (i.e. if a lesion is detected or not) is shown in Fig. 6, showing that a high sensitivity is achieved at the cost of a number of false positive detections by the automatic method, and that a simple greyscale opening operation on the probabilistic output can decrease the number of small clusters of false positive voxels.
Fig. 6.
Free-response ROC curve for detection of individual WMH lesions for the relatively healthy older subjects (n = 96), showing sensitivity versus false positive detections. The method was trained using all 20 patients of MRBrainS13. The results are shown with (green) and without (blue) a greyscale opening operation that uses 4-connectity in the imaging plane as structuring element. The results are further compared with the lesion prediction algorithm of LST (Schmidt et al., 2012) (red) and a cascaded CNN (Valverde et al., 2017) (yellow). For LST, the number of false positives decreases again at about 60 false positives per image, because lesions start merging, which decreases the number of false positive detections but increases the sensitivity.
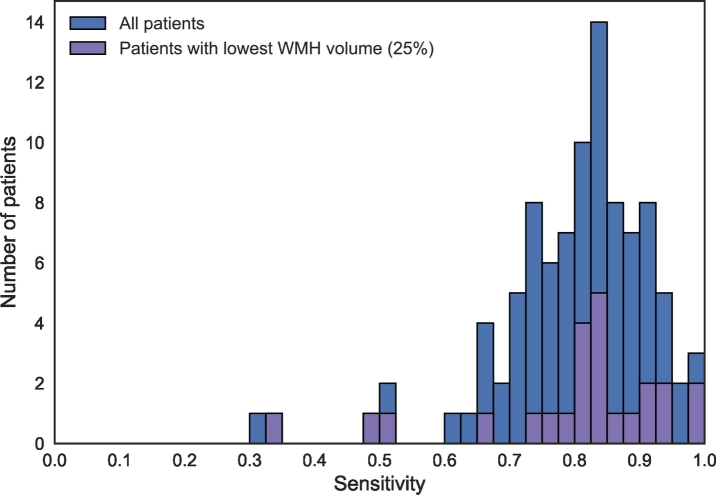
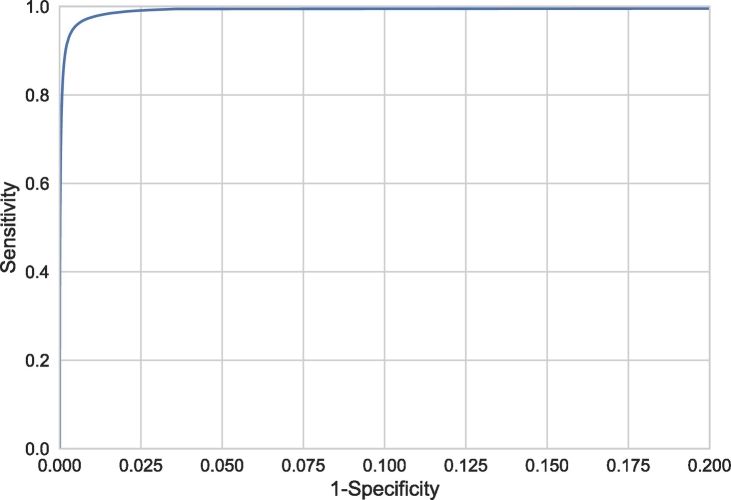
The sensitivity in WMH lesion detection is shown as a histogram in Fig. 7. The results in this figure were obtained by assigning each voxel to the class with the highest probability, without post-processing. A median sensitivity of 0.82 was obtained for WMH lesion detection. This demonstrates that even though the automatically estimated volume might be different than the manually determined volume, most of the lesions in the reference segmentations were detected by the automatic method. Fig. 7 further separately shows the 25% of patients with the lowest reference WMH volume. This results in 24 patients with a reference WMH volume <2.3 cm3. This subgroup of patients shows a similar sensitivity distribution as the whole cohort, which indicates a similar performance for patients with a low WMH volume. Fig. 8 shows the standard voxel-based ROC curve.
Fig. 7.
Histogram of the sensitivity for detection of individual WMH lesions for the relatively healthy older subjects (n = 96). This figure shows the number of patients where the automatic detection obtained a particular sensitivity level. The results are shown for all patients (blue) as well as for the 25% with the lowest reference WMH volume (purple). The method was trained using all 20 patients of MRBrainS13.
Fig. 8.
Voxel-based ROC curve, showing the sensitivity and specificity for detection of WMH voxels instead of WMH lesions. Note that the range of the x-axis is from 0 to 0.2 to better visualise the relevant part of the curve.
The same effects can be seen from the overall detection error rate (DER = 0.16) and the overall outline error rate (OER = 0.83) Wack et al., 2012, Steenwijk et al., 2013. DER quantifies the number of false positive and false negative voxels because of lesions that were missed completely. OER quantifies the number of false positive and false negative voxels because of lesions that were detected but outlined differently. The DER is small while the OER is larger, indicating that most lesions were detected and that the error mostly originates from different outlining of the lesions.
The results for LST (Schmidt et al., 2012) and the cascaded CNN (Valverde et al., 2017) are shown in terms of volume correlations (Fig. 5) and FROC curves (Fig. 6). Our method performs better in terms of Spearman's rank correlation: 0.83 for our method, 0.80 for the cascaded CNN and 0.79 for LST. Moreover, the FROC curve is higher than the FROC curves of the two evaluated methods. At the point of about 50 false positives per image, the performance of the cascaded CNN is similar to our method without post-processing. In line with the observation that the automatic volumes are consistently higher than the manual reference volumes (Fig. 5), the voxel-based Dice coefficients averaged over patients are relatively low for these data: proposed method without post-processing: 0.43 ± 0.15, proposed method with post-processing: 0.47 ± 0.15, LST: 0.51 ± 0.16, cascaded CNN: 0.52 ± 0.16.
3.2.2. Qualitative evaluation in relatively healthy older subjects and patients from a memory clinic
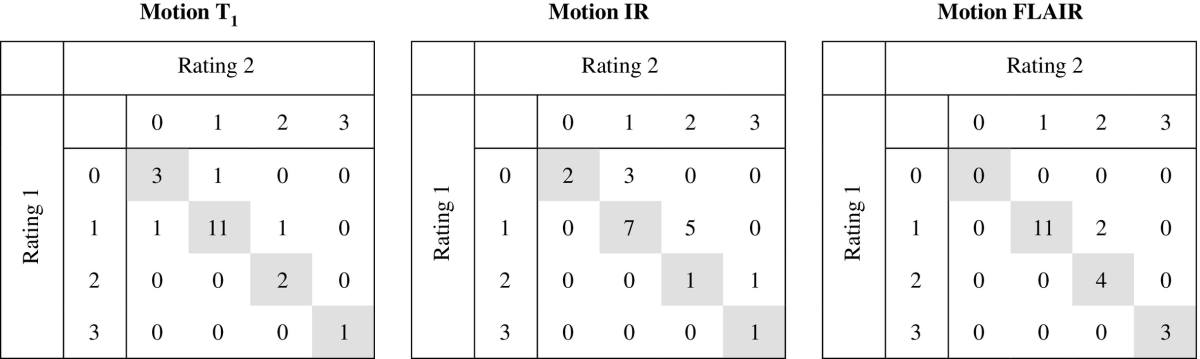
To assess the intra-rater agreement of the visual scoring used for the qualitative evaluation, 20 subjects were rated twice (by the same rater, in a different session) for each of the scores, including: segmentation reliability (Table 3), brain abnormalities (WMH load (Fazekas scale) and brain atrophy severity (GCA scale), Table 4) and motion artefacts (Table 5). In most of the cases, the exact same classification was given in the second rating and it never differed more than one class from the first rating. The Cohen's linearly weighted κ coefficients, showing the agreement between the two ratings, were 0.57 for WMH segmentation reliability, 0.73 for brain tissue segmentation reliability, 0.86 for Fazekas, 0.58 for GCA, 0.78 for motion in the T1-weighted images, 0.58 for motion in the T1-weighted IR images and 0.86 for motion in the T2-weighted FLAIR images.
Table 3.
Intra-rater confusion tables for the WMH and brain tissue segmentation quality scoring in 20 subjects. The exact same classification was given in 17 of 20 patients for the WMH segmentation scoring (κ = 0.57) and in 18 of 20 patients for the tissue segmentation scoring (κ = 0.73).
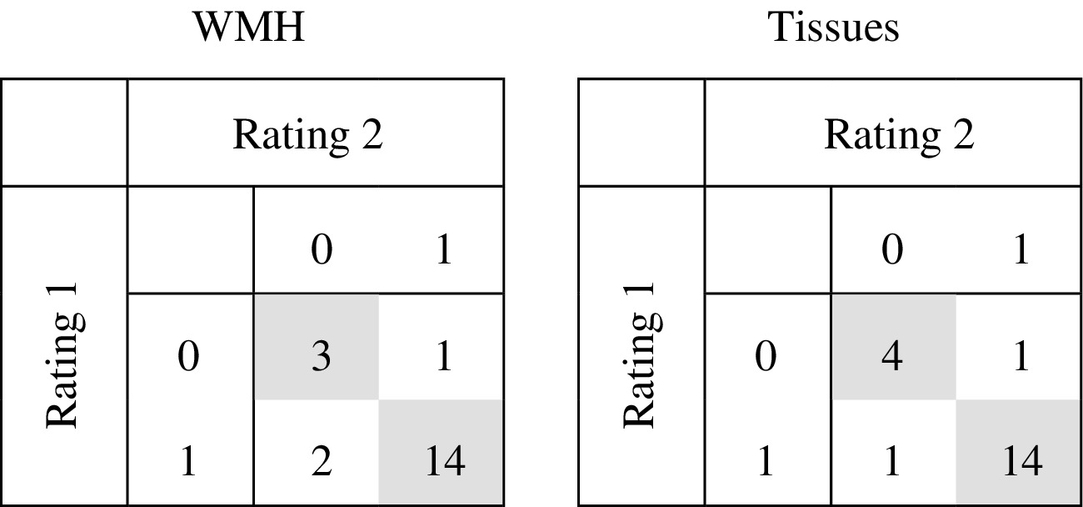
Table 4.
Intra-rater confusion tables for the Fazekas and GCA scales in 20 subjects. The exact same classification was given in 18 of 20 patients for the Fazekas scale (linearly weighted κ = 0.86) and in 15 of 20 patients for the GCA scale (linearly weighted κ = 0.58). In the other cases, the classifications differed by only one class.
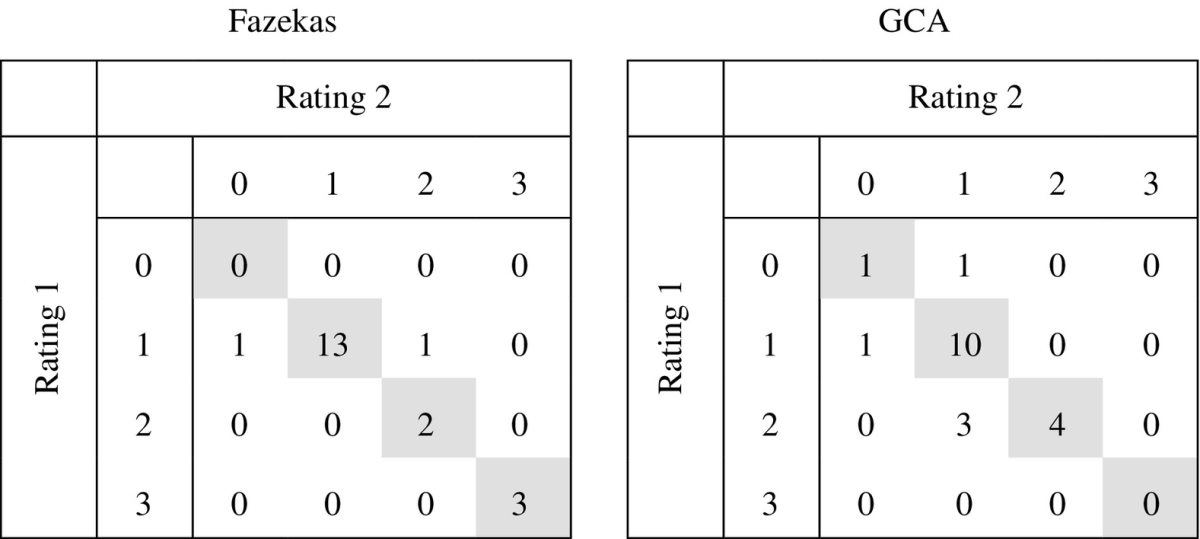
Table 5.
Intra-rater confusion tables for the rating of motion in 20 subjects. The exact same classification was given in 17 of 20 T1-weighted images (linearly weighted κ = 0.78), 11 of 20 T1-weighted IR images (linearly weighted κ = 0.48) and 18 of 20 T2-weighted FLAIR images (linearly weighted κ = 0.86). In the other cases, the classifications differed by only one class.
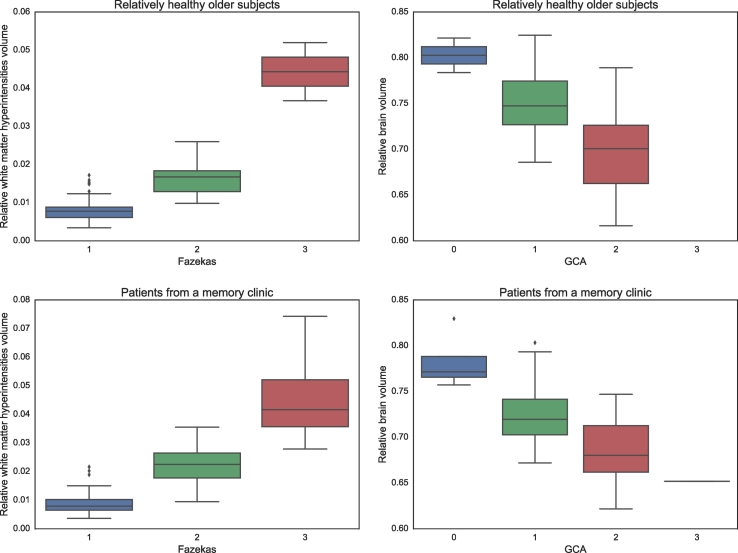
As expected, the patients from a memory clinic overall have a larger WMH volume relative to the intracranial volume (median: 1.58%, range: 0.37–7.43%) than the relatively healthy older subjects (median: 0.83%, range: 0.34–5.20%). The same can be seen from the Fazekas scales, where more patients are in the highest scales for the cohort of patients from a memory clinic (Fazekas 0: 0%, 1: 48%, 2: 35%, 3: 17%) than for the cohort of relatively healthy older subjects (Fazekas 0: 0%, 1: 78%, 2: 20%, 3: 2%). In both cohorts, an association between the automatically obtained WMH volumes and the Fazekas scoring can be observed (Fig. 9, left).
Fig. 9.
WMH volume relative to the intracranial volume for the different Fazekas scales (left column) and total brain volume relative to the intracranial volume for the different GCA scales (right column) for relatively healthy older subjects (top row) and the patients from a memory clinic (bottom row).
Furthermore, the patients from a memory clinic overall have a smaller brain volume relative to the intracranial volume (mean ± standard deviation: 70.7 ± 3.9%) than the relatively healthy older subjects (mean ± standard deviation: 74.0 ± 4.0%), indicating more brain atrophy. The same can be seen from the GCA scales, where more patients are in the highest scales for the cohort of patients from a memory clinic (GCA 0: 4%, 1: 50%, 2: 45%, 3: 1%) than for the cohort of relatively healthy older subjects (GCA 0: 2%, 1: 77%, 2: 21%, 3: 0%). In both cohorts, an association between the automatically obtained brain volumes and the visual atrophy scoring can be observed (Fig. 9, right).
Reliable segmentations, as determined by a human observer, for the relatively healthy older subjects were obtained in 84/96 patients (88%) for brain tissues and in 82/96 patients (85%) for WMH. For the patients from a memory clinic, reliable segmentations were obtained in 85/110 patients (77%) for brain tissues and in 92/110 patients (84%) for WMH. An example segmentation result for a patient from a memory clinic with motion artefacts in the MR images is shown in Fig. 10.
Fig. 10.
Example segmentation for one of the patients from the memory clinic with motion artefacts in the MR images. The method was trained using all 20 patients of MRBrainS13. From left to right: T1-weighted image, T2-weighted FLAIR image, T1-weighted IR image and automatic segmentation.
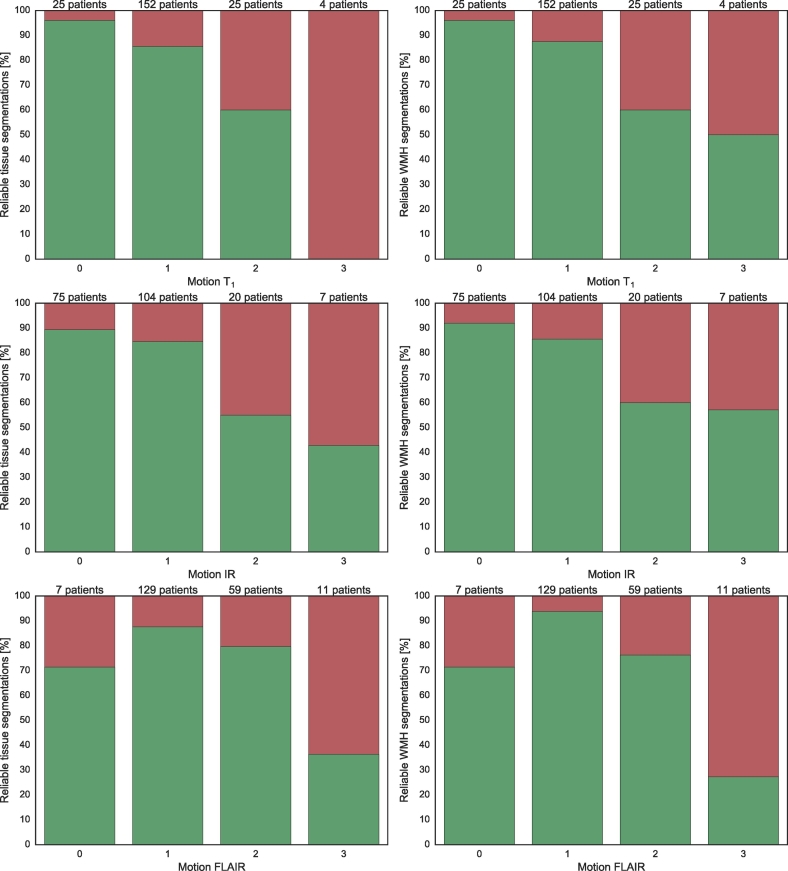
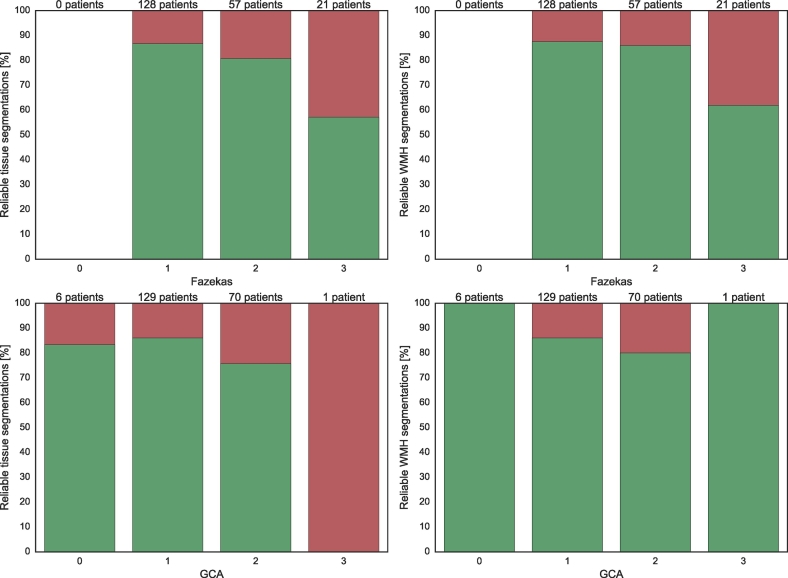
Fig. 11, Fig. 12 show the number of reliable brain tissue (left panels) and WMH (right panels) segmentations for different degrees of motion artefacts and brain abnormalities, respectively. The patients from both cohorts were combined (n = 206) in these figures. It can be observed that the reliability generally decreased with an increasing severity of motion artefacts or brain abnormalities. Other reasons for unreliable segmentations included (lacunar) infarctions (n = 14, 6.8%) and arachnoid cysts (n = 1, 0.5%).
Fig. 11.
Brain tissue (left column) and WMH (right column) segmentation reliability for different severities of motion artefacts: no motion (0), low motion (1), medium motion (2) and high motion (3) for the relatively healthy older subjects and the patients from a memory clinic combined (n = 206). From top to bottom: motion in the T1-weighted image, motion in the T1-weighted IR image and motion in the T2-weighted FLAIR image. Green indicates the percentage of reliable segmentations and red indicates the percentage of unreliable segmentations.
Fig. 12.
Brain tissue (left column) and WMH (right column) segmentation reliability for different classes of the Fazekas scales (top) and GCA scales (bottom) for the relatively healthy older subjects and the patients from a memory clinic combined (n = 206). Green indicates the percentage of reliable segmentations and red indicates the percentage of unreliable segmentations.
4. Discussion
This paper has presented the evaluation of an automatic segmentation method for brain tissues and WMH in MRI using a convolutional neural network. We have shown that our brain tissue segmentation approach can be extended to include WMH as an additional segmentation class, therefore performing segmentation of brain tissues and WMH at the same time. The evaluation performed on MR images from relatively healthy older subjects (n = 96) and MR images from patients from a memory clinic (n = 110) showed that the method can perform accurate segmentation of brain tissues and WMH in MR images with varying degrees of brain abnormalities and motion artefacts.
4.1. Evaluation of the segmentation method
Unlike other methods that perform WMH segmentation, our method performs WMH segmentation as well as tissue segmentation. The inclusion of WMH as an additional segmentation class did not result in a decreased performance for tissue segmentation. We further show that the method is not limited to three input images (T1-weighted, T2-weighted FLAIR and T1 IR), but achieved similar performance when two input images were used (T1-weighted and T2-weighted FLAIR). The method could therefore also be applied in studies where no T1-weighted IR images are acquired. Furthermore, reasonable results were even obtained when the network was trained using only the T2-weighted FLAIR images as input.
Recently, several CNN-based methods for WMH segmentation were proposed in the literature. Most of the papers were evaluated in images of patients with multiple sclerosis. Brosch et al. (2016) presented a method using a convolutional and a deconvolutional pathway with skip-connections to allow multi-scale feature integration. Havaei et al. (2016) presented a method that allows an arbitrary number of input images by computing the mean and variance over the feature maps of the available images. Valverde et al. (2017) presented a method that uses a sequence of two CNNs where the first network is used for an initial segmentation and the second network is used to finetune the segmentations. Ghafoorian et al. (2017b) presented a multi-scale network similar to our network and evaluate different methods of fusing the branches with different inputs. All papers report accurate results. An advantage of our method over these papers is that we show that the same method can be used to perform tissue segmentation as well as WMH segmentation at the same time. If there is only an interest in WMH segmentation for a new study, our method could also be trained to only perform WMH segmentation.
4.2. Evaluation of the segmentation method in the presence of brain abnormalities and motion artefacts
In contrast to most previous studies (see e.g. the review by Caligiuri et al. (2015)), we have evaluated the performance of our method in MR images from two large cohorts with a realistic varying degree of brain abnormalities and motion artefacts. Brain abnormalities were assessed with conventional visual scoring methods (Fazekas and GCA) and motion artefacts were assessed with a custom visual scoring method that was validated for intra-observer variability.
The automatically obtained WMH volumes for the relatively healthy older subjects showed a high correlation with the manually obtained volumes (Fig. 5). However, because of the more strict definition of WMH in the MR images of relatively healthy older subjects compared with the MRBrainS13 training data that was used, the automatically obtained WMH volumes were consistently overestimated compared with the reference volumes. The definition of the lesion boundaries, and therefore the WMH volume, is highly influenced by inter-observer variability. However, a high rank correlation (Spearman's ρ = 0.83) between the manual and automatic volumes shows that the patients can be accurately ranked amongst each other and could in this way for example be classified in risk categories.
Furthermore, in terms of lesion detection it can be seen that, even though the volume might be different, most of the lesions were detected by the automatic method. This sensitivity for lesion detection did however result in a number of false positive detections. In some cases, this also included false positive detections in the automatic segmentation, which were in fact small lesions that were below the strict definition of WMH that was used by the observers. In clinical research, an interactive system where the user quickly goes through all possible WMH lesions identified by the automatic method and labels them as being correct or not could be beneficial to increase both sensitivity and specificity of WMH lesion detection (Wolterink et al., 2015). In addition, the data generated by such an approach could be used as additional training data to improve the automatic method. A simple greyscale opening operation reduced a number of very small false positive detections. With this approach, small isolated detections were suppressed and such voxels could be relabelled to the class with the new highest probability.
We have compared the method to two publicly available methods, LST (Schmidt et al., 2012) and a cascaded CNN (Valverde et al., 2017). Our method outperformed both methods on our data set, in terms of volume correlation (Fig. 5) and FROC analysis (Fig. 6). Similar to our method, the cascaded CNN overestimates the lesion volume compared with the reference volumes. LST provides volumes that are more similar to the reference volumes, but achieves a lower volume correlation than both CNN-based methods. We have only performed the quantitative evaluation for the other methods and not the qualitative evaluation. Qualitatively comparing different methods could however be an interesting future study.
The cascaded CNN uses 3D convolutions, which could be advantageous, especially for the isotropic images that were used in their study. The images in our paper are however anisotropic (0.96 mm in-plane voxel size vs. 3.0 mm slice thickness), which could explain the lower performance on our data set. An advantage of LST over our method and the cascaded CNN is that it is unsupervised. Applying supervised methods to a new data set might, depending on the difference between the training set and the new data, require retraining on representative data or the use of a transfer learning approach.
Qualitative evaluation showed that the reliability of the automatic segmentations decreased with an increasing degree of motion artefacts and with an increasing degree of brain abnormalities, but that in most cases the method obtained accurate segmentations despite the artefacts or abnormalities being visible in the images. Motion artefacts are common in most patient cohorts and it is therefore advantageous when the influence on the segmentation performance is limited. Moreover, the ability to perform accurate segmentations in patients with brain abnormalities is especially important in an ageing population as this facilitates the use of brain tissue and WMH volumes as markers for treatment effect and disease progression in future studies.
5. Conclusion
This paper showed that a convolutional neural network-based segmentation method can accurately segment brain tissues and WMH in MR images of older patients with varying degrees of brain abnormalities and motion artefacts.
Acknowledgements
The manual WMH segmentations that were part of the evaluation in this study were performed by W.H. Bouvy and M. Brundel.
The MR images were collected by the Utrecht Vascular Cognitive Impairment Study Group. Members of the group involved in the patient recruitment and MRI acquisition include (in alphabetical order by department): University Medical Center Utrecht, the Netherlands, Department of Neurology: E. van den Berg, G.J. Biessels, M. Brundel, W.H. Bouvy, S.M. Heringa, L.J. Kappelle, Y.D. Reijmer, L.E.M. Wisse; Department of Radiology/Image Sciences Institute: J. de Bresser, H.J. Kuijf, A. Leemans, P.R. Luijten, W.P.Th.M. Mali, M.A. Viergever, K.L. Vincken, J.J.M. Zwanenburg; Department of Geriatrics: H.L. Koek, J.E. de Wit; Hospital Diakonessenhuis Zeist, the Netherlands: M. Hamaker, R. Faaij, M. Pleizier, E. Vriens; and Julius Center for Health Sciences and Primary Care: A. Algra, M.I. Geerlings, G.E.H.M. Rutten.
The authors gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan X Pascal GPU that is used in this research.
This work was financially supported by the project Brainbox (Quantitative analysis of MR brain images for cerebrovascular disease management), funded by the Netherlands Organisation for Health Research and Development (ZonMw) in the framework of the research programme IMDI (Innovative Medical Devices Initiative); project 104002002.
Footnotes
References
- Aalten P., Ramakers I.H., Biessels G.J., de Deyn P.P., Koek H.L., OldeRikkert M.G., Oleksik A.M., Richard E., Smits L.L., van Swieten J.C. The Dutch Parelsnoer Institute–Neurodegenerative diseases; methods, design and baseline results. BMC Neurol. 2014;14:254. doi: 10.1186/s12883-014-0254-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbeek P., Vincken K.L., van Osch M.J.P., Bisschops R.H.C., van der Grond J. Automatic segmentation of different-sized white matter lesions by voxel probability estimation. Med. Image Anal. 2004;8:205–215. doi: 10.1016/j.media.2004.06.019. [DOI] [PubMed] [Google Scholar]
- Ashburner J., Friston K.J. Unified segmentation. NeuroImage. 2005;26(3):839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Brosch T., Tang L.Y., Yoo Y., Li D.K., Traboulsee A., Tam R. Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. IEEE Trans. Med. Imaging. 2016;35(5):1229–1239. doi: 10.1109/TMI.2016.2528821. [DOI] [PubMed] [Google Scholar]
- Brundel M., Reijmer Y.D., van Veluw S.J., Kuijf H.J., Luijten P.R., Kappelle L.J., Biessels G.J. Cerebral microvascular lesions on high-resolution 7-Tesla MRI in patients with type 2 diabetes. Diabetes. 2014;63(10):3523–3529. doi: 10.2337/db14-0122. [DOI] [PubMed] [Google Scholar]
- Caligiuri M.E., Perrotta P., Augimeri A., Rocca F., Quattrone A., Cherubini A. Automatic detection of white matter hyperintensities in healthy aging and pathology using magnetic resonance imaging: a review. Neuroinformatics. 2015;13(3):261. doi: 10.1007/s12021-015-9260-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer R., Vrooman H.A., Ikram M.A., Vernooij M.W., Breteler M.M., van der Lugt A., Niessen W.J. Accuracy and reproducibility study of automatic MRI brain tissue segmentation methods. NeuroImage. 2010;51(3):1047–1056. doi: 10.1016/j.neuroimage.2010.03.012. [DOI] [PubMed] [Google Scholar]
- De Boer R., Vrooman H.A., Van der Lijn F., Vernooij M.W., Ikram M.A., Van der Lugt A., Breteler M., Niessen W.J. White matter lesion extension to automatic brain tissue segmentation on MRI. NeuroImage. 2009;45(4):1151–1161. doi: 10.1016/j.neuroimage.2009.01.011. [DOI] [PubMed] [Google Scholar]
- De Bresser J., Portegies M.P., Leemans A., Biessels G.J., Kappelle L.J., Viergever M.A. A comparison of MR based segmentation methods for measuring brain atrophy progression. NeuroImage. 2011;54(2):760–768. doi: 10.1016/j.neuroimage.2010.09.060. [DOI] [PubMed] [Google Scholar]
- De Bresser J., Reijmer Y.D., Van Den Berg E., Breedijk M.A., Kappelle L.J., Viergever M.A., Biessels G.J., Utrecht Diabetic Encephalopathy Study Group Microvascular determinants of cognitive decline and brain volume change in elderly patients with type 2 diabetes. Dement. Geriatr. Cogn. Disord. 2010;30(5):381–386. doi: 10.1159/000321354. [DOI] [PubMed] [Google Scholar]
- De Bresser J., Tiehuis A.M., Van Den Berg E., Reijmer Y.D., Jongen C., Kappelle L.J., Mali W.P., Viergever M.A., Biessels G.J., Utrecht Diabetic Encephalopathy Study Group Progression of cerebral atrophy and white matter hyperintensities in patients with type 2 diabetes. Diabetes Care. 2010;33(6):1309–1314. doi: 10.2337/dc09-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groot J.C., Oudkerk M., Gijn J. v., Hofman A., Jolles J., Breteler M. Cerebral white matter lesions and cognitive function: the Rotterdam Scan Study. Ann. Neurol. 2000;47(2):145–151. doi: 10.1002/1531-8249(200002)47:2<145::aid-ana3>3.3.co;2-g. [DOI] [PubMed] [Google Scholar]
- Driscoll I., Davatzikos C., An Y., Wu X., Shen D., Kraut M., Resnick S. Longitudinal pattern of regional brain volume change differentiates normal aging from MCI. Neurology. 2009;72(22):1906–1913. doi: 10.1212/WNL.0b013e3181a82634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazekas F., Chawluk J.B., Alavi A., Hurtig H.I., Zimmerman R.A. MR signal abnormalities at 1.5 T in Alzheimer's dementia and normal aging. Am. J. Neuroradiol. 1987;8(3):421–426. doi: 10.2214/ajr.149.2.351. [DOI] [PubMed] [Google Scholar]
- Ghafoorian M., Karssemeijer N., Heskes T., Bergkamp M., Wissink J., Obels J., Keizer K., de Leeuw F.-E., van Ginneken B., Marchiori E., Platel B. Deep multi-scale location-aware 3D convolutional neural networks for automated detection of lacunes of presumed vascular origin. NeuroImage: Clin. 2017;14:391–399. doi: 10.1016/j.nicl.2017.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghafoorian M., Karssemeijer N., Heskes T., van Uden I.W., Sanchez C.I., Litjens G., de Leeuw F.-E., van Ginneken B., Marchiori E., Platel B. Location sensitive deep convolutional neural networks for segmentation of white matter hyperintensities. Sci. Rep. 2017;7:5110. doi: 10.1038/s41598-017-05300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghafoorian M., Karssemeijer N., van Uden I.W., de Leeuw F.-E., Heskes T., Marchiori E., Platel B. Automated detection of white matter hyperintensities of all sizes in cerebral small vessel disease. Med. Phys. 2016;43(12):6246–6258. doi: 10.1118/1.4966029. [DOI] [PubMed] [Google Scholar]
- Giorgio A., De Stefano N. Clinical use of brain volumetry. J. Magn. Reson. Imaging. 2013;37(1):1–14. doi: 10.1002/jmri.23671. [DOI] [PubMed] [Google Scholar]
- Havaei M., Davy A., Warde-Farley D., Biard A., Courville A., Bengio Y., Pal C., Jodoin P.-M., Larochelle H. Brain tumor segmentation with deep neural networks. Med. Image Anal. 2017;35:18–31. doi: 10.1016/j.media.2016.05.004. [DOI] [PubMed] [Google Scholar]
- Havaei M., Guizard N., Chapados N., Bengio Y. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2016. HeMIS: Hetero-modal image segmentation; pp. 469–477. [Google Scholar]
- Heinen R., Bouvy W.H., Mendrik A.M., Viergever M.A., Biessels G.J., de Bresser J. Robustness of automated methods for brain volume measurements across different MRI field strengths. PLOS ONE. 2016;11(10):e0165719. doi: 10.1371/journal.pone.0165719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikram M.A., Vrooman H.A., Vernooij M.W., van der Lijn F., Hofman A., van der Lugt A., Niessen W.J., Breteler M.M. Brain tissue volumes in the general elderly population: the Rotterdam Scan Study. Neurobiol. Aging. 2008;29(6):882–890. doi: 10.1016/j.neurobiolaging.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Jain S., Sima D.M., Ribbens A., Cambron M., Maertens A., Van Hecke W., De Mey J., Barkhof F., Steenwijk M.D., Daams M. Automatic segmentation and volumetry of multiple sclerosis brain lesions from MR images. NeuroImage: Clin. 2015;8:367–375. doi: 10.1016/j.nicl.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamnitsas K., Ledig C., Newcombe V.F., Simpson J.P., Kane A.D., Menon D.K., Rueckert D., Glocker B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 2017;36:61–78. doi: 10.1016/j.media.2016.10.004. [DOI] [PubMed] [Google Scholar]
- Klein S., Staring M., Murphy K., Viergever M.A., Pluim J.P.W. elastix: a toolbox for intensity-based medical image registration. IEEE Trans. Med. Imaging. 2010;29(1):196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- Klöppel S., Abdulkadir A., Hadjidemetriou S., Issleib S., Frings L., Thanh T.N., Mader I., Teipel S.J., Hüll M., Ronneberger O. A comparison of different automated methods for the detection of white matter lesions in MRI data. NeuroImage. 2011;57(2):416–422. doi: 10.1016/j.neuroimage.2011.04.053. [DOI] [PubMed] [Google Scholar]
- Kuijf H.J., Moeskops P., de Vos B.D., Bouvy W.H., de Bresser J., Biessels G.J., Viergever M.A., Vincken K.L. SPIE Medical Imaging. International Society for Optics and Photonics; 2016. Supervised novelty detection in brain tissue classification with an application to white matter hyperintensities; p. 978421. [Google Scholar]
- Kuijf H.J., Tax C.M.W., Zaanen L.K., Bouvy W.H., Bresser J., Leemans A., Viergever M.A., Biessels G.J., Vincken K.L. MICCAI workshop on Computational Diffusion MRI. Springer; 2014. The added value of diffusion tensor imaging for automated white matter hyperintensity segmentation; pp. 45–53. [Google Scholar]
- Mendrik A.M., Vincken K.L., Kuijf H.J., Breeuwer M., Bouvy W.H., de Bresser J., Alansary A., de Bruijne M., Carass A., El-Baz A., Jog A., Katyal R., Khan A.R., van der Lijn F., Mahmood Q., Mukherjee R., van Opbroek A., Paneri S., Pereira S., Persson M., Rajchl M., Sarikaya D., Smedby Örjan, Silva C.A., Vrooman H.A., Vyas S., Wang C., Zhao L., Biessels G.J., Viergever M.A. MRBrainS challenge: online evaluation framework for brain image segmentation in 3T MRI scans. Comput. Intell. Neurosci. 2015:813696. doi: 10.1155/2015/813696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeskops P., Viergever M.A., Mendrik A.M., de Vries L.S., Benders M.J., Išgum I. Automatic segmentation of MR brain images with a convolutional neural network. IEEE Trans. Med. Imaging. 2016;35(5):1252–1261. doi: 10.1109/TMI.2016.2548501. [DOI] [PubMed] [Google Scholar]
- Moeskops P., Wolterink J.M., van der Velden B.H., Gilhuijs K.G., Leiner T., Viergever M.A., Išgum I. MICCAI. Springer; 2016. Deep learning for multi-task medical image segmentation in multiple modalities; pp. 478–486. [Google Scholar]
- Pasquier F., Leys D., Weerts J.G., Mounier-Vehier F., Barkhof F., Scheltens P. Inter-and intraobserver reproducibility of cerebral atrophy assessment on MRI scans with hemispheric infarcts. Eur. Neurol. 1996;36(5):268–272. doi: 10.1159/000117270. [DOI] [PubMed] [Google Scholar]
- Pereira S., Pinto A., Alves V., Silva C.A. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans. Med. Imaging. 2016;35(5):1240–1251. doi: 10.1109/TMI.2016.2538465. [DOI] [PubMed] [Google Scholar]
- Raidou R.G., Kuijf H.J., Sepasian N., Pezzotti N., Bouvy W.H., Breeuwer M., Vilanova A. Medical Image Computing and Computer-Assisted Intervention. Springer; 2016. Employing visual analytics to aid the design of white matter hyperintensity classifiers; pp. 97–105. [Google Scholar]
- Reijmer Y.D., Brundel M., De Bresser J., Kappelle L.J., Leemans A., Biessels G.J., Utrecht Vascular Cognitive Impairment Study Group Microstructural white matter abnormalities and cognitive functioning in type 2 diabetes: a diffusion tensor imaging study. Diabetes Care. 2013;36(1):137–144. doi: 10.2337/dc12-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roura E., Oliver A., Cabezas M., Valverde S., Pareto D., Vilanova J.C., Ramió-Torrentà L., Rovira À., Lladó X. A toolbox for multiple sclerosis lesion segmentation. Neuroradiology. 2015;57(10):1031–1043. doi: 10.1007/s00234-015-1552-2. [DOI] [PubMed] [Google Scholar]
- Schmidt P., Gaser C., Arsic M., Buck D., Förschler A., Berthele A., Hoshi M., Ilg R., Schmid V.J., Zimmer C. An automated tool for detection of FLAIR-hyperintense white-matter lesions in multiple sclerosis. NeuroImage. 2012;59(4):3774–3783. doi: 10.1016/j.neuroimage.2011.11.032. [DOI] [PubMed] [Google Scholar]
- Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014;15(1):1929–1958. [Google Scholar]
- Steenwijk M.D., Pouwels P.J., Daams M., van Dalen J.W., Caan M.W., Richard E., Barkhof F., Vrenken H. Accurate white matter lesion segmentation by k nearest neighbor classification with tissue type priors (kNN-TTPs) NeuroImage: Clin. 2013;3:462–469. doi: 10.1016/j.nicl.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudre C.H., Cardoso M.J., Bouvy W., Biessels G.J., Barnes J., Ourselin S. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2014. Bayesian model selection for pathological data; pp. 323–330. [DOI] [PubMed] [Google Scholar]
- Sudre C.H., Cardoso M.J., Bouvy W.H., Biessels G.J., Barnes J., Ourselin S. Bayesian model selection for pathological neuroimaging data applied to white matter lesion segmentation. IEEE Trans. Med. Imaging. 2015;34(10):2079–2102. doi: 10.1109/TMI.2015.2419072. [DOI] [PubMed] [Google Scholar]
- Tieleman T., Hinton G. COURSERA: Neural Networks for Machine Learning. vol. 4. 2012. Lecture 6.5-rmsprop: divide the gradient by a running average of its recent magnitude. [Google Scholar]
- Valverde S., Cabezas M., Roura E., González-Villà S., Pareto D., Vilanova J.C., Ramió-Torrentà L., Rovira À., Oliver A., Lladó X. Improving automated multiple sclerosis lesion segmentation with a cascaded 3D convolutional neural network approach. NeuroImage. 2017;155:159–168. doi: 10.1016/j.neuroimage.2017.04.034. [DOI] [PubMed] [Google Scholar]
- Van Leemput K., Maes F., Vandermeulen D., Colchester A., Suetens P. Automated segmentation of multiple sclerosis lesions by model outlier detection. IEEE Trans. Med. Imaging. 2001;20(8):677–688. doi: 10.1109/42.938237. [DOI] [PubMed] [Google Scholar]
- Wack D.S., Dwyer M.G., Bergsland N., Di Perri C., Ranza L., Hussein S., Ramasamy D., Poloni G., Zivadinov R. Improved assessment of multiple sclerosis lesion segmentation agreement via detection and outline error estimates. BMC Med. Imaging. 2012;12(1):17. doi: 10.1186/1471-2342-12-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardlaw J.M., Smith E.E., Biessels G.J., Cordonnier C., Fazekas F., Frayne R., Lindley R.I., T O’Brien J., Barkhof F., Benavente O.R. Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol. 2013;12(8):822–838. doi: 10.1016/S1474-4422(13)70124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolterink J.M., Leiner T., Takx R.A.P., Viergever M.A., Išgum I. Automatic coronary calcium scoring in non-contrast-enhanced ECG-triggered cardiac CT with ambiguity detection. IEEE Trans. Med. Imaging. 2015;34(9):1867–1878. doi: 10.1109/TMI.2015.2412651. [DOI] [PubMed] [Google Scholar]
- Zhang W., Li R., Deng H., Wang L., Lin W., Ji S., Shen D. Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. NeuroImage. 2015;108:214–224. doi: 10.1016/j.neuroimage.2014.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]