Abstract
Background
Sensitivity to macrocyclic lactones, which are commonly used in veterinary clinics, was first found in Rough Collies, and was attributed in 2001 to a 4 bp deletion in the MDR1 gene. The list of affected breeds currently includes 13 breeds. Researchers from different countries and continents examined the allelic frequencies of the nt230(del4) MDR1 mutation, emphasizing the clinical importance of this test not only to mutation-prone dogs, but also to their crosses and mongrels, since treatment of a deletion carrier with these compounds may lead to its death.
In this study, the allelic frequencies of nt230(del4) MDR1 mutation in affected breeds, their crosses, unrelated pure breeds and mongrels are reported for the state of Israel (n = 1416 dogs). The Israeli data were compared with reports from the US, Europe, UK, Australia and Japan.
Results
The allelic frequencies of nt230(del4) MDR1 mutation in Israel for Australian, Swiss and German Shepherds (31%, 17% and 2.4%, respectively) are similar to the corresponding frequencies worldwide, much higher for Border Collies (4.8%), twice lower for Rough Collies (28%, compared to 55% or more elsewhere), and ~1% for mongrels. The frequencies for crosses of Australian Shepherd and Border Collies in Israel are 4 and 1.6 times lower, respectively, compared to the frequencies for the respective pure breeds.
Conclusions
This work, that for the first time presents the frequency of nt230(del4) MDR1 mutation in Israel, along with a worldwide survey, has implications for clinicians, owners and breeders of sheepdogs and their crosses and supports the need for extra care in treatment and in future breeding. Of note, the relative proportion of affected breeds, in the overall tested dogs, might be higher than their actual proportion in Israel due to directed samples collection by veterinarians for clinical purposes, as these are mainly limited to certain affected breeds or dogs that resemble them.
Electronic supplementary material
The online version of this article (10.1186/s12917-017-1251-9) contains supplementary material, which is available to authorized users.
Keywords: nt230(del) MDR1 mutation, Macrocyclic lactones, Sheepdogs, Drug sensitivity, Taqman assay
Background
The multi drug resistance 1 (MDR1) gene, a member of the ATP-binding cassette (ABC) transporters superfamily, is a highly conserved ATP-dependent P-glycoprotein (P-gp) membrane transporter [1]. In eukaryotes, the extrusion transporters have a fundamental function as eliminators of several toxic compounds, mainly hydrophilic and amphiphatic ones [2–4]. ABCB1 (ABC subfamily B member 1, also referred to as MDR1 and PGY1) transporters are expressed in several organs and their role in the gut epithelia and the blood-tissue barriers (blood-brain barrier, blood-testis barrier and placenta) of mammals has been studied extensively [5]. While MDR1 activity in the small intestine can limit the bioavailability of various drugs, its inactivity in the luminal membranes of the endothelial cells of the brain can lead to toxic accumulation of xenobiotics in the central nervous system (CNS) and to severe adverse effects, including death [6, 7].
Since the early 1980s, sheepdogs were reported to have sensitivity to ivermectin, a relatively new macrocyclic lactone parasiticide drug [8, 9]. Elevated concentrations of ivermectin were found in the CNS of affected dogs [10]. In 2001, ivermectin sensitivity in Rough Collies was associated with a 4-base pair (bp) deletion mutation in the fourth exon of the canine ABCB1 gene [11]. The deletion, usually annotated ABCB1:c.227_230delATAG or ABCB1–1∆ (MDR1) mutation (hereafter nt230(del4) MDR1 mutation), causes a frameshift that leads to a premature stop codon and a truncated transporter of 91 amino acids as opposed to 1281 amino acids, i.e. only 7.1% of the full-length mature P-gp protein [12]. Later studies showed that nt230(del4) MDR1 mutation heterozygous dogs can be regarded as having an intermediate macrocyclic lactone sensitive phenotype. This is a relevant clinical notion in cases of high-dose protocols [6, 13]. Pertinent macrocyclic lactones include ivermectin, doramectin, moxidectin and milbemycin oxime. Other drugs commonly used in the veterinary clinic, such as P-glycoprotein substrates of the non-macrocyclic lactones type, were also reported as toxic to nt230(del4) MDR1 mutation homozygote dogs (−/−). These include vincristine, digoxin, mexiletine, quinidine, fexofenadine, vinblastine, and loperamide, cyclosporine A, verapamil, paclitexal, doxorubicin, dexamethasone and others [14–19]. Moreover, treating nt230(del4) MDR1 mutation heterozygote dogs (+/−) with reduced doses of P-glycoprotein substrates such as vincristine, vinblastine and doxorubicin (but not a full dosage treatment of a non-P-glycoprotein substrate) may cause delayed drug excretion, and consequently drug toxicosis and myelosuppression [20]. Recently, a collie affected (homozygote) by the Nt230(del4) MDR1 mutation mutation experienced exaggerated CNS depression when treated with apomorphine [21], an opioid which has not been described as a P-glycoprotein substrate. Yet, other opioids, including loperamide and butorphanol, are known substrates for canine P-glycoprotein [22]. A cell line for assessing drugs as canine P-gp substrates was recently developed [23]. Noteworthy, based on its anti-parasitic and anti-inflammatory activities, ivermectin has recently received US Food and Drug Administration (FDA) and EU approval for the treatment of adult human patients for a growing number of indications [24].
The nt230(del4) MDR1 mutation has to date been detected in diverse dog breeds, including Rough Collies and other related sheepdogs, such as Australian Shepherds, miniature Australian Shepherds, Border Collies, Shetland Sheepdogs, Old English Sheepdogs, English Shepherd, German Shepherd, White Swiss Shepherd, McNab as well as Wäller and two sighthounds, Longhaired Whippet and Silken Windhound. From a genealogy perspective, all of the above breeds share a common ancestor that lived in Great Britain before the breeds were classified and registered in 1873 [15, 25]. This notion is consistent with genomic data that cluster these breeds in common clusters or sometimes even fail to differentiate between close varieties that may have undergone recent selection based on size, such as in the case of Rough Collies and Shetland Sheepdogs [26, 27].
A fast and simple detection method using allele-specific loop-mediated isothermal amplification (AS-LAMP) [28] as well as PCR-based diagnostic tests [29, 30] and the TaqMan allelic discrimination assay [15] were designed as a result of the high clinical importance of nt230(del4) MDR1 mutation detection. Efforts to map allele frequencies in affected pure breed dogs, some of their crosses, and unrelated purebred dogs have been made in different parts of the world [19, 25, 31–40]. These studies supply clinicians with regional information on the mutation distribution in their countries and highlight the need for a genetic test prior to any treatment involving common parasiticides in affected breeds and their crosses.
In Israel, spirocercosis is a widespread disease and dogs are routinely treated with macrocyclic lactones parasiticide. On average, more than 20 dogs are diagnosed annually, with the highest infection rates in the center of the country [41, 42]. The aim of this study was to determine the frequency of the mutant allele in Israel in the affected breeds, their crosses and undefined dogs, and to compare the data with recent worldwide studies. To the best of our knowledge, this is the first report on nt230(del4) MDR1 mutation frequency in Israel and it may provide a comparative perspective. It should be noted that the evaluation of nt230(del4) MDR1 mutation genetic disposition in diverse canine breeds was performed in order to inform medical and clinical professionals, rather than to illuminate the relationship of different genotypes with the clinical signs in each breed. The latter is not within the scope of this study.
Methods
Animals
Biological samples from 1416 dogs (blood, blood on paper and hair roots) were voluntarily and non-selectively obtained from veterinarians, breeders and owners from all regions of the country, and analyzed for the nt230(del4) MDR1 mutation as part of the diagnostic research service at our institute. A total of 481 samples from 7 purebred breeds known to bear the nt230(del4) MDR1 mutation allele (including 5 crosses of two of these breeds) and 280 crosses of these same breeds (dogs for which one parent was known to be a purebred dog, often either Australian Shepherd or Border Collie), 61 unrelated purebred dogs and 594 unclassified crosses (mongrels, dogs for which either both parents were known to be non-purebred dogs or the parentage was unclassified) were obtained. Breed status was reported by the veterinarian or owner, and was not confirmed by other inspection.
DNA extraction
DNA from blood and blood on paper samples was extracted with the Chelex method using Bio-Rad, ‘InstaGeneTM Matrix’. Briefly, 5 μL of whole blood (or 0.25 cm2 paper with blood) were incubated at room temperature for 15–30 min and then centrifuged at 12,000 rpm for 3 min. The supernatant was carefully withdrawn and 200 μL of InstaGene matrix were added to the pellet and incubated at 56 °C for 15–30 min, vortexed and incubated in a 100 °C heat block for 8 min. After incubation, spin down was performed at 12,000 rpm for 3 min. The resulting supernatant contained the DNA. DNA from hair roots was extracted with the nexttec ‘Special Protocol DNA Isolation Hair by nexttec™ 1-Step’. Briefly, three to five hair roots were dissolved with Proteinase K and DTT. Samples were then incubated in a shaker (at 56 °C, 200 rpm for 2 h). Then, 100 μL of the lysates were incubated for 3 min at room temperature and later centrifuged and eluted at 700×g for 1 min. All procedures were performed according to these kits’ extraction manuals and according to the facility guidelines.
Genotyping by Taqman assay
nt230(del4) MDR1 mutation was screened using the Fluidigm BioMark apparatus (Fluidigm Corporation US) with an Assay-specific TaqMan fluorescence probe mix. The probe sequences, which were designed following NC_006596.3, are as follows: VIC Probe Sequence was ATGACAGATAGCTTTGCAA (wt), FAM Probe Sequence AACATGACAGCTTTGCAAA (mutant); the forward and reverse primers are CCATCATCCATGGAGCTGC and CACAAATAATACTTACTTTCATTAATTATAACTGG, respectively, amplicon size 133 bp. Assay along with PCR master mix were run in duplicate by loading 5 μl into each well of the primed 96.96 Fluidigm Chip. The chip was then placed in the integrated fluidic circuit controller and loaded before analysis with the BioMark reader. The following thermal cycling protocol was used: 50°C (2 min), 70°C (30 min), 25°C (10 min), 50°C (2 min), and 95°C (4 min). This was followed by 40 cycles of 95°C (10 s) and 61°C (30 s). The initial cycle [50°C (2 min), 70°C (30 min), 25°C (10 min)]. Data were analyzed and cycle threshold (CT) values were determined using BioMark real-time PCR analysis software (Fluidigm Corp.), and automated mutation calling was carried out using an algorithm based on the change in CT (DCT) values between the wild-type and the mutant.
Validation by PCR and Sanger sequencing
Each nt230(del4) MDR1 mutated allele was validated by PCR and Sanger sequencing. The nt230(del4) MDR1 mutation region was amplified using a touchdown reaction (4 cycles: 94°C (30 s), 58°C (30 s), 72°C (30 s); 30 cycles: 94°C (30 s), 56°C (30 s), 72°C (30 s)) with the following forward and reverse primers: TTTTTAGTTTCGCTATTCAAATTGGC and CAAACTTATTACCAATATTAACTGTAGCTC, respectively. Sequencing reactions for both DNA strands were performed with BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA) and analyzed with an automatic sequencer (Applied Biosystems).
Data analysis
Breeds were classified into two main groups according to nt230(del4) MDR1 mutated allele breed genealogy: Related (as detailed in the introduction) and non-related (all others). Within each group, breeds were subsequently separated into pure breeds and mix breeds. Worldwide mapping of the allelic frequency of nt230(del4) MDR1 mutation only includes breeds for which at least 50 dogs were genotyped.
Results and discussion
nt230(del4) MDR1 mutation allelic frequencies in Israel
Raw data is provided in Additional file 1 (for each sample ID, the breed and MDR1 genotype status are indicated). Table 1 summarizes the allelic frequencies of the canine nt230(del4) MDR1 mutation in Israel in all mentioned subgroups. The nt230(del4) MDR1 mutation frequency varied markedly by breed. Among the relatively abundant breeds (sample size greater than 10 dogs, not including 37.5% for Old English Sheepdog where the sample size was only 4 dogs), it was highest for Australian Shepherds (31.4%, where 9.0% of 145 dogs were homozygotes for the mutation), Rough Collies (27.5%, n = 20 and 5.0%, respectively), and Shetland Sheepdog (26.9%, n = 13 and 0%, respectively), followed by White Swiss Shepherd (16.7%, n = 12 and 8.3%, respectively), Border Collie (4.8%, n = 269 and 2.3%, respectively), and German Shepherd (2.4%, n = 21 and 0%, respectively). As expected, an intermediate frequency (10.0%) was observed in crosses of pure Australian Shepherds and pure Border Collies (n = 5). The respective allelic frequencies among crosses of these breeds were much lower compared to their purebred counterpart, as expected: 4 and 1.6 times lower in Australian Shepherd (7.8%, n = 32) and Border Collie (3.0%, n = 183), respectively. The nt230(del4) MDR1 mutation allele was not detected in any of the unrelated purebred dogs (n = 61), as expected [43]. Almost 2% (n = 594) of the mongrels are affected (heterozygotes for the mutation, allelic frequency of almost 0.9%), a finding that justifies a more cautious approach in the clinic in case of unspecified dogs. This value is at the lower end of the previously reported 2–7% [32, 37].
Table 1.
Observed frequencies of nt230(del4) MDR1 mutation in related (affected known breeds, pure and crossed ones), and non-related (un-affected known breeds and mongrels) breeds in Israel
| nt230(del4) MDR1 mutation allele breed genealogy | Breed | Frequency of nt230(del4) MDR1 mutation allele status [%] | |||||
|---|---|---|---|---|---|---|---|
| Name | Status | Sample size | Homozygote (−/−) | Heterozygote (+/−) | Normal (+/+) | Frequency of nt230(del4) MDR1 mutation allele | |
| Related | Australian Shepherd | Pure | 145 | 9.0% | 44.8% | 46.2% | 31.4% |
| Australian Shepherd | Crosses | 32 | 0.0% | 15.6% | 84.4% | 7.8% | |
| Border Collie | Pure | 261 | 2.3% | 5.0% | 92.7% | 4.8% | |
| Border Collie | Crosses | 183 | 0.0% | 6.0% | 94.0% | 3.0% | |
| Border Collie X Australian Shepherd | Pure | 5 | 0.0% | 20.0% | 80.0% | 10.0% | |
| Rough Collie | Pure | 20 | 5.0% | 45.0% | 50.0% | 27.5% | |
| Rough Collie | Crosses | 49 | 2.0% | 4.1% | 93.9% | 4.1% | |
| German Shepherd | Pure | 21 | 0.0% | 4.8% | 95.2% | 2.4% | |
| German Shepherd | Crosses | 11 | 0.0% | 0.0% | 90.9% | 0.0% | |
| White Swiss Shepherd | Pure | 12 | 8.3% | 16.7% | 75.0% | 16.7% | |
| White Swiss Shepherd | Crosses | 4 | 0.0% | 0.0% | 75.0% | 0.0% | |
| Shetland Sheepdog | Pure | 13 | 0.0% | 53.8% | 46.2% | 26.9% | |
| Shetland Sheepdog | Crosses | 1 | 0.0% | 0.0% | 100.0% | 0.0% | |
| Old English Sheepdog | Pure | 4 | 0.0% | 75.0% | 25.0% | 37.5% | |
| Old English Sheepdog | Crosses | 0 | 0% | 0.0% | 0.0% | 0% | |
| Non-Related | Others | Pure | 61 | 0.0% | 0.0% | 100.0% | 0.0% |
| Others (mongrels) | Crosses | 594 | 0.0% | 1.9% | 98.1% | 0.9% | |
‘+’ stands for WT allele; ‘-’ stands for nt230(del4) MDR1 mutation allele
A worldwide view – Comparative perspective
Figure 1 presents a comparative view of the Israeli and the worldwide nt230(del4) MDR1 mutation allelic frequencies. In most cases, the frequencies in Israel seem to be similar to those in other countries, whereas in other cases the range of frequencies is wide [also see 6]. These values might also be biased by the sample size, which varied greatly in different studies, and/or by the collection method. The frequencies observed for Australian Shepherd dogs living in Israel, US, Europe and Japan are similar, ranging from 25% to 33%, while higher frequencies were observed in the UK and Australia (~45%). Allelic frequencies for Border Collie are below 1% across the world, except 2.3% in the UK, compared to a much higher frequency in Israel (4.8%). This might be an indication for over inbreeding of carrier dogs in Israel. In contradistinction, the worldwide frequencies for Rough Collies are very high (55% or higher) compared to an almost 2-fold lower rate in Israel (28%, however the sample size was small). This study provides evidence for the existence of the mutation in the popular German Shepherd, for the first time outside the US, with frequencies ranging from ~2.5% to ~4%. Similarly the nt230(del4) MDR1 mutation allele is reported in White Swiss Shepherd outside Europe, for the first time, with a similar frequency in Israel (~15%). These two latter pure breeds, the German Shepherd and the White Swiss Shepherd, arise from the same lineage, as older breed standards of the German Shepherd allowed all color varieties, including white [38]. The differences in allelic frequencies between these breeds might indicate over inbreeding in the case of the White Swiss Shepherd or for a founder effect in the breeds founding stock, back in the 1960s and 1970s [38]. The allelic frequency observed for Shetland Sheepdogs in Israel is very similar to that in Europe (27% and 30%, respectively), close to that in Australia (21%) and the UK (36%), while the frequencies in the US (7.1%) and Japan (1.2%) are much lower.
Fig. 1.
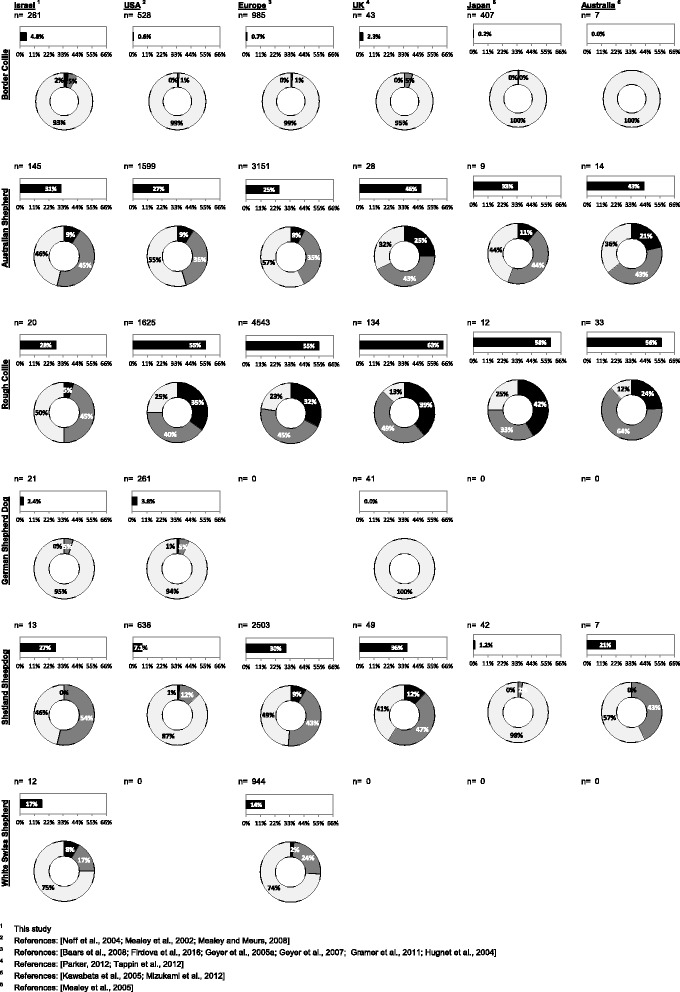
Allelic frequencies of nt230(del4) MDR1 mutation in six related breeds (from top to bottom: Border Collie, Australian Shepherd, Collie, German Shepherd Dog, Shetland Sheepdog and White Swiss Shepherd) in Israel, USA, Europe, UK, Japan and Australia (footnotes 1 to 6 respectively with the relevant literature in the bottom of the panel). Dogs number is given on top of each panel as n. Bar graph indicate allele frequency of each breed, pie graph indicate allelic frequencies of nt230(del4) MDR1 mutation as follows: pale gray for normal (+/+), gray for heterozygote (+/−) and black for homozygote (−/−)
It is pertinent to state that the relative proportion of affected (predominantly pure) breeds, in the overall tested dogs in this study, might be higher than their actual proportion in Israel. The bias is related to samples collection by professional veterinarians for clinical purposes, as these are mainly limited to certain affected breeds, or dogs that resemble these breeds, from specific areas of the country where spirocercosis is more widespread [41]. Nevertheless, the allelic frequency of nt230(del4) MDR1 mutation within each affected breed is probably not significantly (if at all) affected by this bias, while it can be affected in the non-related breeds.
Conclusions
Here, we report for the first time on the allelic frequencies of the nt230(del4) MDR1 mutation in affected breeds, as well as in their crosses and even in mongrels, in Israel. The Israeli data was compared to a worldwide literature survey which summarizes the corresponding allele status in the USA, Europe, UK, Australia and Japan. The main findings are:
nt230(del4) MDR1 mutation allele frequencies in Israel resemble, per breed, the status in the world. Exceptions to this finding are the Israeli Border Collies and Rough Collies which have the highest (4.8%) and lowest (28%) frequency rates in the world.
The first report on the existence of the nt230(del4) MDR1 mutated allele outside the USA and Europe on German Shepherd Dogs and White Swiss Shepherds, respectively.
High prevalence rate of the nt230(del4) MDR1 mutated allele in mixes of prone breeds.
The nt230(del4) MDR1 mutation frequency in dogs which were described as unrelated mongrels is 1%.
Noteworthy, due to inherent bias in the sample collection by veterinarians for clinical purposes, the relative proportion of affected breeds, in the overall tested dogs, might be higher than their actual proportion in Israel. Nevertheless, the allelic frequency of nt230(del4) MDR1 mutation within each affected breed is probably not significantly affected by this bias, while it can be affected in the non-related breeds.
The allelic frequencies of the nt230(del4) MDR1 mutation in affected breeds seems to vary slightly across the world. This notion is in agreement with the strict registry breeding procedures in breeding clubs that do not permit “new genes” to enter the breeding stock. The breeding aims at meeting the breed standard, which is a combination of phenotypic and characteristic traits with less emphasis on genetic predisposition for ailments.
This work, that for the first time presents the nt230(del4) MDR1 mutation frequency status in Israel, along with a worldwide survey, has implications for clinicians, owners and breeders of sheepdogs and their crosses and supports the need for extra care in treatment and in future breeding.
Acknowledgements
The authors thank Mrs. Esther Furman for linguistic editing and editorial assistance.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Availability of data and materials
All data analyzed during this study are included in this published article. For Intellectual Property reasons, details of dogs and owners cannot be shared.
Abbreviations
- ABC
ATP-Binding Cassette transporters superfamily;
- ABCB1
ABC subfamily B member 1
- AS-LAMP
Allele-Specific Loop-Mediated isothermal amplification
- CNS
Central Nervous System
- FDA
Food and Drug Administration
- MDR1
Multi Drug Resistance 1
- P-gp
P-glycoprotein
Additional file
List of all tested dogs, stratified by breed. For each sample ID, the breed and MDR1 genotyping status is indicated. (XLSX 60 kb)
Authors’ contributions
We hereby declare that all authors made substantial contributions to conception and design / acquisition of data / analysis and interpretation of data; were involved in drafting the article / revising it critically for important intellectual content; gave final approval of the version to be published. Therefore, each of the authors takes public responsibility for the content, and agrees to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. More specifically, these authors led the following tasks: Conceptualization: YD DB(2). Formal analysis and interpretation of data: YM YD. Investigation: AS DB(1) AA EK ONP YD DB(2) YP. Project administration: YD DB(2) YP. Resources: GE YD YP. Supervision: YD DB(2) IBS YP. Validation: YD YM. Visualization: YM. Writing – original draft: YD YM. Writing – review and editing: YD YM DB(2) ISB YP EK AS AA DB(1) ONP GE. All authors read and approved the final manuscript.
Ethics approval
This research does not involve human subjects, human material, or human data. It is not an experimental research on vertebrates or any regulated invertebrates. It only involved analysis of genetic data, which was not generated within the framework of this research. Yet, we provide all relevant information on the process of gathering the biological samples and generating the genetic data. These have been done in compliance with relevant guidelines. In addition, this research was carried out in strict accordance with the recommendations in applicable national and international guidelines. Biological samples were given from professional veterinarians from January 2014 to December 2016 for clinical purposes. No animal procedures or additional genetics tests were performed on these samples by the researchers. The whole study protocol, of obtaining samples and analysing the finding of the genetic tests for research purposes, was approved by the Israel Nature and Parks authority (Permit Number: 41,584) during January 2017, prior to the beginning of the research.
Consent for publication
Not applicable.
Competing interests
The author declares that he/she has no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12917-017-1251-9) contains supplementary material, which is available to authorized users.
Contributor Information
Yaron Dekel, Phone: +972 4 6123901, Email: Yarond@gri.org.il.
Yossy Machluf, Email: yossy.machluf@gmail.com.
Aviad Stoler, Email: aviad@gga.org.il.
Arava Aderet, Email: arava@gga.org.il.
Daniel Baumel, Email: daniel@gga.org.il.
Efrat Kellerman, Email: efratk@gga.org.il.
Yoram Plotsky, Email: yoram@gga.org.il.
Oshrat Noked Partouche, Email: oshratyotam@gmail.com.
Gal Elhalal, Email: gal.elhalal@vetmarket.co.il.
Izhar Ben-Shlomo, Email: izhar.benshlomo@gmail.com.
Dani Bercovich, Email: dannyb@telhai.ac.il.
References
- 1.Saier MH, Jr, Paulsen IT, Sliwinski MK, Pao SS, Skurray RA, Nikaido H. Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. 1998;12(3):265–274. doi: 10.1096/fasebj.12.3.265. [DOI] [PubMed] [Google Scholar]
- 2.Annilo T, Chen ZQ, Shulenin S, Costantino J, Thomas L, Lou H, Stefanov S, Dean M. Evolution of the vertebrate ABC gene family: analysis of gene birth and death. Genomics. 2006;88(1):1–11. doi: 10.1016/j.ygeno.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Dean M, Annilo T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu Rev Genomics Hum Genet. 2005;6:123–142. doi: 10.1146/annurev.genom.6.080604.162122. [DOI] [PubMed] [Google Scholar]
- 4.Fromm MF. P-glycoprotein: a defense mechanism limiting oral bioavailability and CNS accumulation of drugs. Int J Clin Pharmacol Ther. 2000;38(2):69–74. doi: 10.5414/CPP38069. [DOI] [PubMed] [Google Scholar]
- 5.Fromm MF. Importance of P-glycoprotein at blood-tissue barriers. Trends Pharmacol Sci. 2004;25(8):423–429. doi: 10.1016/j.tips.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Geyer J, Janko C. Treatment of MDR1 mutant dogs with macrocyclic lactones. Curr Pharm Biotechnol. 2012;13(6):969–986. doi: 10.2174/138920112800399301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thuerauf N, Fromm MF. The role of the transporter P-glycoprotein for disposition and effects of centrally acting drugs and for the pathogenesis of CNS diseases. Eur Arch Psychiatry Clin Neurosci. 2006;256(5):281–286. doi: 10.1007/s00406-006-0662-6. [DOI] [PubMed] [Google Scholar]
- 8.Preston JM. Adverse reactions to unapproved applications. The Veterinary record. 1983;112(12):286. doi: 10.1136/vr.112.12.286. [DOI] [PubMed] [Google Scholar]
- 9.Seward RL. Reactions in dogs given ivermectin. J Am Vet Med Assoc. 1983;183(5):493. [PubMed] [Google Scholar]
- 10.Pulliam JD, Seward RL, Henry RT, Steinberg SA. Investigating ivermectin toxicity in collies. Veterinary Medicine. 1985;80(7):33–40. [Google Scholar]
- 11.Mealey KL, Bentjen SA, Gay JM, Cantor GH. Ivermectin sensitivity in collies is associated with a deletion mutation of the mdr1 gene. Pharmacogenetics. 2001;11(8):727–733. doi: 10.1097/00008571-200111000-00012. [DOI] [PubMed] [Google Scholar]
- 12.Roulet A, Puel O, Gesta S, Lepage JF, Drag M, Soll M, Alvinerie M, Pineau T. MDR1-deficient genotype in collie dogs hypersensitive to the P-glycoprotein substrate ivermectin. Eur J Pharmacol. 2003;460(2–3):85–91. doi: 10.1016/S0014-2999(02)02955-2. [DOI] [PubMed] [Google Scholar]
- 13.Bissonnette S, Paradis M, Daneau I, Silversides DW. The ABCB1-1Delta mutation is not responsible for subchronic neurotoxicity seen in dogs of non-collie breeds following macrocyclic lactone treatment for generalized demodicosis. Vet Dermatol. 2009;20(1):60–66. doi: 10.1111/j.1365-3164.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- 14.Kitamura Y, Koto H, Matsuura S, Kawabata T, Tsuchiya H, Kusuhara H, Tsujimoto H, Sugiyama Y. Modest effect of impaired P-glycoprotein on the plasma concentrations of fexofenadine, quinidine, and loperamide following oral administration in collies. Drug metabolism and disposition: the biological fate of chemicals. 2008;36(5):807–810. doi: 10.1124/dmd.107.017624. [DOI] [PubMed] [Google Scholar]
- 15.Klintzsch S, Meerkamp K, Doring B, Geyer J. Detection of the nt230[del4] MDR1 mutation in dogs by a fluorogenic 5′ nuclease TaqMan allelic discrimination method. Veterinary journal (London, England : 1997) 2010;185(3):272–277. doi: 10.1016/j.tvjl.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 16.Krugman L, Bryan JN, Mealey KL, Chen A. Vincristine-induced central neurotoxicity in a collie homozygous for the ABCB1Delta mutation. J Small Anim Pract. 2012;53(3):185–187. doi: 10.1111/j.1748-5827.2011.01155.x. [DOI] [PubMed] [Google Scholar]
- 17.Henik RA, Kellum HB, Bentjen SA, Mealey KL. Digoxin and mexiletine sensitivity in a collie with the MDR1 mutation. Journal of veterinary internal medicine / American College of Veterinary Internal Medicine. 2006;20(2):415–417. doi: 10.1111/j.1939-1676.2006.tb02878.x. [DOI] [PubMed] [Google Scholar]
- 18.Sartor LL, Bentjen SA, Trepanier L, Mealey KL. Loperamide toxicity in a collie with the MDR1 mutation associated with ivermectin sensitivity. Journal of veterinary internal medicine / American College of Veterinary Internal Medicine. 2004;18(1):117–118. doi: 10.1111/j.1939-1676.2004.tb00145.x. [DOI] [PubMed] [Google Scholar]
- 19.Geyer J, Doring B, Godoy JR, Leidolf R, Moritz A, Petzinger E. Frequency of the nt230 (del4) MDR1 mutation in collies and related dog breeds in Germany. J Vet Pharmacol Ther. 2005;28(6):545–551. doi: 10.1111/j.1365-2885.2005.00692.x. [DOI] [PubMed] [Google Scholar]
- 20.Mealey KL, Northrup NC, Bentjen SA. Increased toxicity of P-glycoprotein-substrate chemotherapeutic agents in a dog with the MDR1 deletion mutation associated with ivermectin sensitivity. J Am Vet Med Assoc. 2003;223(10):1453. doi: 10.2460/javma.2003.223.1453. [DOI] [PubMed] [Google Scholar]
- 21.Campbell O, de Lorimier LP, Mealey KL. Adverse reaction to apomorphine in a collie homozygous for the ABCB1-1 (MDR1) mutation. J Small Anim Pract. 2017;58(2):119. doi: 10.1111/jsap.12618. [DOI] [PubMed] [Google Scholar]
- 22.Mealey KL, Fidel J. P-glycoprotein mediated drug interactions in animals and humans with cancer. Journal of veterinary internal medicine / American College of Veterinary Internal Medicine. 2015;29(1):1–6. doi: 10.1111/jvim.12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mealey KL, Dassanayake S, Burke NS. Establishment of a cell line for assessing drugs as canine P-glycoprotein substrates: proof of principle. J Vet Pharmacol Ther. 2017;40(5):545–51. [DOI] [PubMed]
- 24.Kircik LH, Del Rosso JQ, Layton AM, Schauber J. Over 25 years of clinical experience with Ivermectin: an overview of safety for an increasing number of indications. Journal of drugs in dermatology. 2016;15(3):325–332. [PubMed] [Google Scholar]
- 25.Neff MW, Robertson KR, Wong AK, Safra N, Broman KW, Slatkin M, Mealey KL, Pedersen NC. Breed distribution and history of canine mdr1-1Delta, a pharmacogenetic mutation that marks the emergence of breeds from the collie lineage. Proc Natl Acad Sci U S A. 2004;101(32):11725–11730. doi: 10.1073/pnas.0402374101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parker HG, Kim LV, Sutter NB, Carlson S, Lorentzen TD, Malek TB, Johnson GS, DeFrance HB, Ostrander EA, Kruglyak L. Genetic structure of the purebred domestic dog. Science (New York, NY) 2004;304(5674):1160–1164. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- 27.Parker HG. Genomic analyses of modern dog breeds. Mamm Genome. 2012;23(1–2):19–27. doi: 10.1007/s00335-011-9387-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stiedl CP, Weber K. Fast and simple detection methods for the 4-base pair deletion of canine MDR1/ ABCB1 gene by PCR and isothermal amplification. J Vet Diagn Investig. 2017;29(2):176–180. doi: 10.1177/1040638716683213. [DOI] [PubMed] [Google Scholar]
- 29.Geyer J, Doring B, Godoy JR, Moritz A, Petzinger E. Development of a PCR-based diagnostic test detecting a nt230(del4) MDR1 mutation in dogs: verification in a moxidectin-sensitive Australian shepherd. J Vet Pharmacol Ther. 2005;28(1):95–99. doi: 10.1111/j.1365-2885.2004.00625.x. [DOI] [PubMed] [Google Scholar]
- 30.Baars C, Leeb T, von Klopmann T, Tipold A, Potschka H. Allele-specific polymerase chain reaction diagnostic test for the functional MDR1 polymorphism in dogs. Veterinary journal (London, England : 1997) 2008;177(3):394–397. doi: 10.1016/j.tvjl.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 31.Firdova Z, Turnova E, Bielikova M, Turna J, Dudas A. The prevalence of ABCB1:c.227_230delATAG mutation in affected dog breeds from European countries. Res Vet Sci. 2016;106:89–92. doi: 10.1016/j.rvsc.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 32.Gramer I, Leidolf R, Doring B, Klintzsch S, Kramer EM, Yalcin E, Petzinger E, Geyer J. Breed distribution of the nt230(del4) MDR1 mutation in dogs. Veterinary journal (London, England : 1997) 2011;189(1):67–71. doi: 10.1016/j.tvjl.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 33.Hugnet C, Bentjen SA, Mealey KL. Frequency of the mutant MDR1 allele associated with multidrug sensitivity in a sample of collies from France. J Vet Pharmacol Ther. 2004;27(4):227–229. doi: 10.1111/j.1365-2885.2004.00585.x. [DOI] [PubMed] [Google Scholar]
- 34.Mealey KL, Bentjen SA, Waiting DK. Frequency of the mutant MDR1 allele associated with ivermectin sensitivity in a sample population of collies from the northwestern United States. Am J Vet Res. 2002;63(4):479–481. doi: 10.2460/ajvr.2002.63.479. [DOI] [PubMed] [Google Scholar]
- 35.Mizukami K, Chang HS, Yabuki A, Kawamichi T, Hossain MA, Rahman MM, Uddin MM, Yamato O. Rapid genotyping assays for the 4-base pair deletion of canine MDR1/ABCB1 gene and low frequency of the mutant allele in border collie dogs. J Vet Diagn Investig. 2012;24(1):127–134. doi: 10.1177/1040638711425591. [DOI] [PubMed] [Google Scholar]
- 36.Tappin SW, Goodfellow MR, Peters IR, Day MJ, Hall EJ, Mealey KL. Frequency of the mutant MDR1 allele in dogs in the UK. The Veterinary record. 2012;171(3):72. doi: 10.1136/vr.100633. [DOI] [PubMed] [Google Scholar]
- 37.Mealey KL, Meurs KM. Breed distribution of the ABCB1-1Delta (multidrug sensitivity) polymorphism among dogs undergoing ABCB1 genotyping. J Am Vet Med Assoc. 2008;233(6):921–924. doi: 10.2460/javma.233.6.921. [DOI] [PubMed] [Google Scholar]
- 38.Geyer J, Klintzsch S, Meerkamp K, Wohlke A, Distl O, Moritz A, Petzinger E. Detection of the nt230(del4) MDR1 mutation in white Swiss shepherd dogs: case reports of doramectin toxicosis, breed predisposition, and microsatellite analysis. J Vet Pharmacol Ther. 2007;30(5):482–485. doi: 10.1111/j.1365-2885.2007.00885.x. [DOI] [PubMed] [Google Scholar]
- 39.Kawabata A, Momoi Y, Inoue-Murayama M, Iwasaki T. Canine mdr1 gene mutation in Japan. The Journal of veterinary medical science / the Japanese Society of Veterinary Science. 2005;67(11):1103–1107. doi: 10.1292/jvms.67.1103. [DOI] [PubMed] [Google Scholar]
- 40.Mealey KL, Munyard KA, Bentjen SA. Frequency of the mutant MDR1 allele associated with multidrug sensitivity in a sample of herding breed dogs living in Australia. Vet Parasitol. 2005;131(3–4):193–196. doi: 10.1016/j.vetpar.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 41.Aroch I, Markovics A, Mazaki-Tovi M, Kuzi S, Harrus S, Yas E, Baneth G, Bar-El M, Bdolah-Abram T, Segev G, et al. Spirocercosis in dogs in Israel: a retrospective case-control study (2004-2009) Vet Parasitol. 2015;211(3–4):234–240. doi: 10.1016/j.vetpar.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 42.Gottlieb Y, Lavy E, Kaufman M, Markovics A, Ghanim M, Aroch I. A novel bacterial symbiont in the nematode Spirocerca lupi. BMC Microbiol. 2012;12:133. doi: 10.1186/1471-2180-12-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merola VM, Eubig PA. Toxicology of avermectins and milbemycins (macrocylic lactones) and the role of P-glycoprotein in dogs and cats. Vet Clin North Am Small Anim Pract. 2012;42(2):313–333. doi: 10.1016/j.cvsm.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data analyzed during this study are included in this published article. For Intellectual Property reasons, details of dogs and owners cannot be shared.


