Abstract
The formation of protein complexes and the co-regulation of the cellular concentrations of proteins are essential mechanisms for cellular signaling and for maintaining homeostasis. Here we use isobaric labeling multiplexed proteomics to analyze protein co-regulation and show that this allows the identification of protein-protein associations with high accuracy. We apply this ‘interactome mapping by high-throughput quantitative proteome analysis’ (IMAHP) method to a panel of 41 breast cancer cell lines and show that deviations of the observed protein co-regulations in specific cell lines from the consensus network impacts to cellular fitness. Furthermore, these aberrant interactions serve as biomarkers predicting drug sensitivity of cell lines in screens across 195 drugs. We expect that IMAHP can be broadly used to gain insight into how changing landscapes of protein-protein associations affect the phenotype of biological systems.
The proteome forms a link between genotype and phenotype, and its exploration provides a wealth of information about the molecular mechanism regulating cellular events1. Mass spectrometry has evolved as the key technology to characterize a broad range of aspects defining the proteome such as protein abundances, post-translational modifications, as well as interaction between proteins. The interaction of a protein reveal its functional network and mapping all protein-protein interactions in a cell – the interactome – and their dynamics will offer unique insights into biological systems and their reaction to perturbations. Major efforts are underway to generate global protein-protein interaction maps by using the yeast-two hybrid (Y2H) assay2 or protein affinity-purification/mass spectrometry (AP-MS)3,4. However, generating a static interaction catalogue of a comprehensive protein-protein interaction network represents a substantial experimental effort, and comprehensively studying network dynamics after perturbation currently seems out of reach. Here, we report the IMAHP technology that uses protein co-regulation analysis to map protein-protein associations and their dysregulation. We further show that interactome dysregulations can allow for the identification of cancer vulnerabilities and sensitivity to drugs.
We used multiplexed quantitative mass spectrometry-based proteomics technology, applying isobaric labeling technology with 10-plex tandem mass tag (TMT) reagents5, to generate quantitative proteome profiles of 41 breast cancer cell lines representing the majority of breast cancer subtypes6 (Supplementary Table 1). A total of 82 proteome samples from two biological replicates were analyzed in 11 experiments, of which each enabled the simultaneous quantification of 10 samples (Fig. 1a). Data were acquired on an Orbitrap Fusion mass spectrometer using the SPS-MS3 method to eliminate ratio distortions known to affect negatively the accuracy and reproducibility of quantitative proteomics data acquired using multiplexed isobaric labeling technology7,8. A total of 10,535 proteins were quantified across all 11 experiments, and on average 9,115 proteins were quantified across the two replicate analyses of each cell line (Fig. 1b and Supplementary Table 2) while requiring less than 10 hours of data acquisition time per cell line.
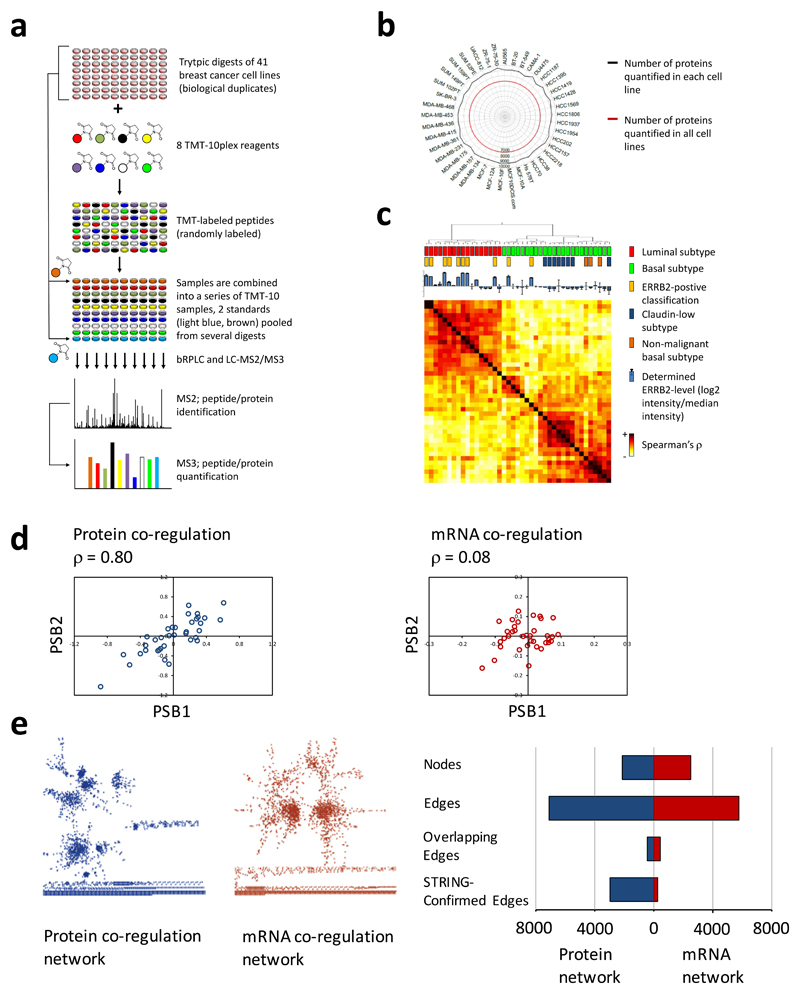
Figure 1. High-throughput multiplexed quantitative proteome mapping of 41 breast cancer cell lines and protein co-regulation analysis to identify protein-protein associations.
(a) Workflow for proteomics analysis. Biological replicates of the proteomes of forty-one cell lines (Supplementary Table 1, only 40 are shown for the sake of simplicity) were quantitatively mapped using mass spectrometry-based proteomics applying TMT-10plex reagents (Supplementary Table 1) and an LC-MS2/MS37,8. (b) Radar chart showing the number of proteins quantified in this study: 10,535 proteins were quantified across all experiments, an average of 9,115 proteins in each of the 41 cell lines, and 6,911 in all cell lines (Supplementary Table 2). (c) Hierarchical clustering of the studied 41 cell lines based on the Spearman’s correlation of proteome patterns measured for each cell line. Clusters of cancer cell line of luminal and basal subtypes are well separated. A prior classification of ERBB2 overexpression was confirmed by the proteomics measurement. (d) Co-regulation analysis was performed on the proteome data from this study and published RNA-seq data11 for 36 cell lines and 6659 gene products for which both datasets were available. Scatterplots showing a high correlation (Spearman’s ρ = 0.80) of protein concentration across the cell lines for two proteasome subunits PSB1 and PSB2 and a much weaker correlation (ρ = 0.08) of the mRNA levels. (e) Co-regulation derived gene association networks of statistically significant associations (FDR 0.05 % by Benjamini-Hochberg) greatly differ when derived from protein (blue) or mRNA (red) profiles and less than 10 % of the associations are found in both networks. Whereas 42 % of associations derived based on the proteome data were confirmed based on high-evidence associations from the STRING database, the portion was only 4 % for associations derived based on the RNA-seq dataset.
The number of proteins quantified in all cell lines was 6,911, and subsequent analyses were performed on this subset (Supplementary Table 2). When clustered based on the Spearman’s rank correlation coefficient among the proteome profiles, the cell lines were clearly separated into the known breast cancer subtypes: luminal, basal, claudin-low, and nonmalignant subtypes (Fig. 1c and Supplementary Fig. 1). Proteome based clustering was concordant with mRNA-level based clustering9 (Supplementary Fig. 2).
The median Spearman’s rank correlation coefficient between proteome profiles from biological replicates of the same cell line was 0.82, confirming a high reproducibility of the multiplexed proteome quantification technology (Supplementary Figs 3a and 3c). The median correlation coefficient between mRNA and proteome profiles was 0.58 (Supplementary Figs 3b and 3c) and thus slightly higher than reported in other studies10. The analysis was done for 36 cell lines for which published RNA-seq data were available11 (Supplementary Table 3, Supplementary Data 1 and 2).
Next investigated whether mRNA and protein data are different with respect to permitting the identification of functional and physical protein-protein associations through co-regulation analysis across the 36 cell lines. An example of co-regulation for the two proteasome subunits PSB1 and PSB2 is shown in Fig 1d. We observed a high correlation between the protein levels for these known interactors (Spearman’s ρ = 0.80) but very low correlation between mRNA levels (ρ = 0.08). We performed this analysis for each pair of the 6674 gene products for which we had data points in both datasets (Supplementary Table 4). A very strict filter of Benjamini-Hochberg (BH) corrected P value ≤ 5x10-4 was applied, and we considered only positive correlations. Correlation inferred associations are shown in a network form in Fig. 1e with nodes representing proteins or mRNA molecules and edges statistically significant associations. We observed 5748 significant associations among 2494 mRNA molecules and 7086 associations among 2122 proteins (Fig. 1e). Notably, only 431 significant associations between mRNA and proteins encoded by the same gene were found in the overlap of both datasets (< 8 %).
To estimate the accuracy of the mRNA and protein derived networks we used as a benchmark high-confidence associations (score ≥ 0.700) in the STRING database, a compendium of experimentally determined as well as predicted functional protein associations including physical interactions12. We found 2953 (42 %) of the proteome-based associations confirmed by the STRING database but only 250 (4 %) of the associations derived from the mRNA dataset (Fig 1e). An increased relative number of known associations in the proteome derived dataset was confirmed for several precision thresholds (Supplementary Fig. 4 and Supplementary Table 5) indicating that co-regulation analysis applied on proteome profiles has a substantially higher predictive power to identify functional protein-protein associations than co-regulation analysis on transcriptome profiles. These results are supported by a recent report on gene function prediction through co-regulation analysis of proteome and mRNA profiling data on tumors of three cancer types13 and by co-regulation of functionally associated proteins in yeast14 and mouse15.
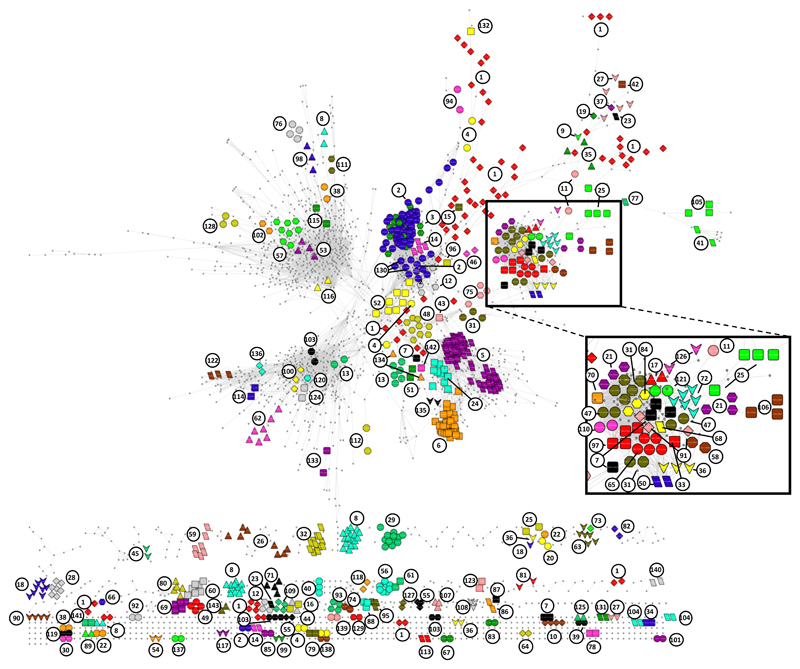
To further explore the proteome inferred network we used co-regulation analysis on the profiles from all 41 cell lines (the 36 lines considered above and an additional 5 for which mRNA data was not available), which revealed 14909 associations among 3024 proteins (BH corrected P ≤ 5x10-4, Supplementary Table 6, Supplementary Data 3). By systematically annotating the correlation data set for known physical protein-protein associations we were able to assign 143 unique complexes from the Comprehensive Resource of Mammalian protein complexes (CORUM) database of high-confidence protein complexes16 (Fig. 2, Supplementary Table 7). The median coverage of CORUM defined components in the complexes with associations observed in our data set was 67 %; for 112 of the complexes, we identified associations between at least 90 % of the components. These data show that protein co-regulation analysis is a useful tool for detection of associations of proteins in multi-protein complexes. Of the 14909 protein-protein associations, 4179 (28 %) were attributed to protein complexes defined in the CORUM database (Supplementary Table 6). The number of observed associations previously defined as high-confidence associations in the STRING database was 5149 (35 %) of which 3032 were overlapping with associations defined by CORUM. High-confidence protein-protein associations from the CORUM and the STRING database confirm 6296 (42 %) of the 14909 associations and 8613 (58 %) associations in this stringently filtered dataset have yet not been reported (Supplementary Table 6). Supporting the validity of these unreported associations, 3636 are linked through an indirect (1 step removed) STRING interaction (Supplementary Fig. 5).
Figure 2. Protein-protein association network revealed by proteomics-based protein co-regulation analysis across 41 breast cancer cell lines.
The network includes 14909 associations across 3024 proteins. Co-regulation was defined using the Spearman’s correlation coefficient and a stringent filter of a Benjamini-Hochberg (BH) corrected P value ≤ 5x10-4 was applied to obtain the network. Only positive correlations are shown. The associations include those of 132 high-confidence known multi-protein complexes from the CORUM database. Proteins of these complexes are shown color and the complexes are labeled with an index number that is referred to in Supplemental Table 12. A dense network region mainly containing DNA associated complexes is enlarged for clarity’s sake.
We have further validated the previously not reported associations by comparing our data with that of large-scale interactome screens using immunaffinty-purification of proteins and identifying associated proteins by mass spectrometry (AP-MS)3,4As shown in Supplementary Fig. 6, we were able to confirm both known and novel associations by this comparison. We found that 20 % of the known and 6 % of the novel associations identified in our study were confirmed in the AP-MS based Bioplex dataset3 when only considering associations where both proteins were identified in both studies. Comparing another large-scale AP-MS dataset4 with the Bioplex data in the same manner showed a similar overlap: 22 % for known and 4 % for novel associations.
To explore how differences in protein network across cell lines can inform the biology of these breast cancer models we sought to identify cell line specific network dysregulation. We used deviations from co-regulation across the 14909 significant correlated protein pairs identified using the 41-cell line dataset. Testing each protein pairs, we performed bivariate outlier testing based on first the Mahalanobis distance followed by the Grubbs test to determine outliers (p≤0.1) corresponding to cell lines with a putative deregulated protein pair.
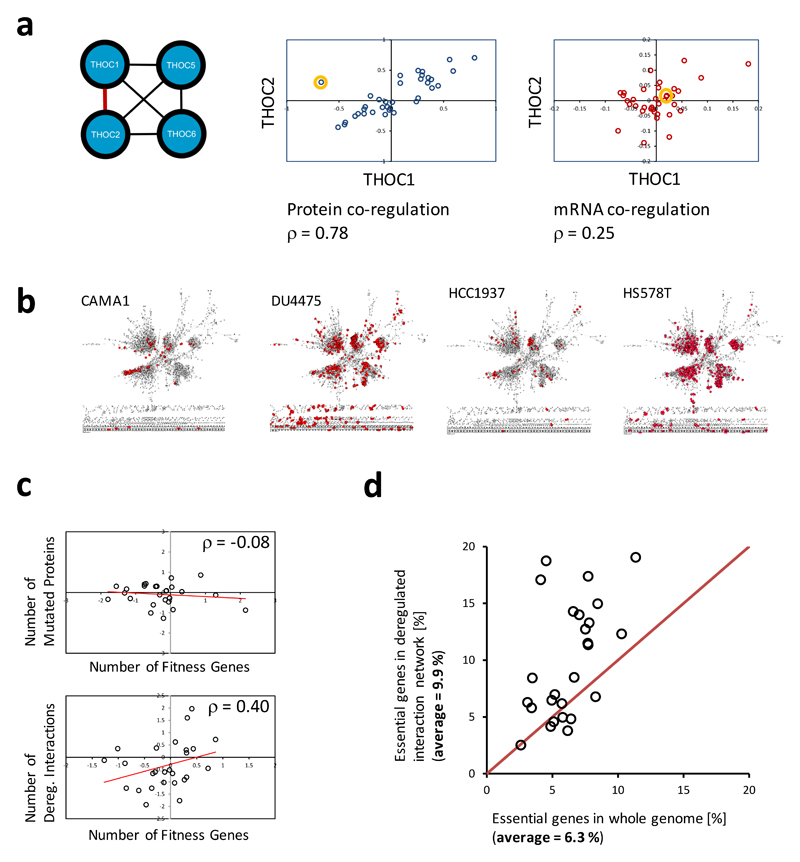
As shown in Fig 3a, we found for example such a deviation in the cell line MDAMB157 for the association between the two proteins THOC1 and THOC2, components of the TREX complex involved in the regulation of transcription, mRNA processing and export17. THOC2 carries a mutation (R1307W) in this cell line, which could underlie this effect by inhibiting the binding between THOC1 and THOC2 leading to degradation of THOC118,19. Consistent with a protein level regulation rather than a change in mRNA levels, THOC1 and THOC2 mRNA levels across the cell lines did not identify MDA-MB-157 as an outlier (Fig 3a).
Figure 3. Deviations of protein co-regulations allow identifying cell-line specific deregulations of protein-protein associations revealing cell-specific vulnerabilities.
(a) Two-dimensional outlier detection based on the Mahalanobis distance was used to define cell-line specific deviations of protein co-regulations for all 14909 identified protein-protein associations in the studied cell lines. Such a deviation (deregulation) was found for the known association between the proteins THOC1 and THOC2 in the MDAMB157 cell line (orange cycle). The deregulation was not identified based on a deviation of mRNA level co-regulation. THOC2 carries a mutation (R1307W) in this cell line19, which may cause the deregulation of the association between the two proteins. (b) Proteins with deregulated associations are highlighted (red) on the background of the determined protein-protein association network (see Fig. 2) for four cell lines (see also Supplementary Table 8) indicating variations in both the number and network areas affected by the deregulations. (c) The number of a cell line’s dysregulated protein-protein associations but not the number of mutated genes is correlating with the number of genes affecting the cell line’s fitness as determined using a genome-wide genetic dropout screen applying pooled shRNA (zGARP ≤ -2)20. Log2 ratios of values/average value are plotted, and correlations are given as Spearman's rank correlation coefficient. The comparison was made for 26 cell lines for which proteome, mutation, and drop-out screen data were available.(d) Proteins with dysregulated associations are enriched for cell line specific proteins when compared to all genes monitored in a genome-wide drop-out screen20 for almost all of the 26 compared cell line. The average enrichment increase is 1.6 fold (9.9 % compared to 6.3 %).
Applying this outlier principle to all globally identified protein-protein association across all cell lines we observed a wide range of dysregulated associations across cell lines ranging from 20 (0.1 %) in ZR751 to 800 dysregulated associations (5.9 %) in Hs578T affecting many different large complexes (see Fig 3b, Supplementary Table 8, Supplementary Figs. 7-9). We term this strategy of interactome mapping by high-throughput quantitative proteome analysis the IMAHP technology.
To test the significance of our findings, we used the data from a genome-wide shRNA-based drop-out screen on breast cancer cell lines20 (Supplementary Table 9) to evaluate the functional consequences of dysregulated protein-protein associations. We analyzed the data of 26 cancer cell lines for which whole genome sequencing data and drop-out screen data were available. We found a significant correlation between the number of dysregulated protein-protein associations in each cell line with the number of proteins whose depletion affects the cell lines fitness (ρ = 0.4, see Methods Section) suggesting that dysregulation of protein associations results in higher level of susceptibilities. Notably, there was no correlation between the number of mutated proteins (Supplementary Table 10) and the number of fitness genes (ρ = -0.08) (Fig. 3c) possibly in part because only a limited number of mutations have functional consequences21. Furthermore, in almost all cell lines fitness gene products were enriched in the group of proteins with dysregulated protein-protein associations compared to all 15309 genes monitored in the dropout screen (Fig 3d)20. The average enrichment in fitness proteins was 64 %. These results show that high-throughput mapping of dysregulated protein-protein associations in cancer cell line with the IMAHP strategy can be used to reveal vulnerabilities of cancer cell lines with high efficiency.
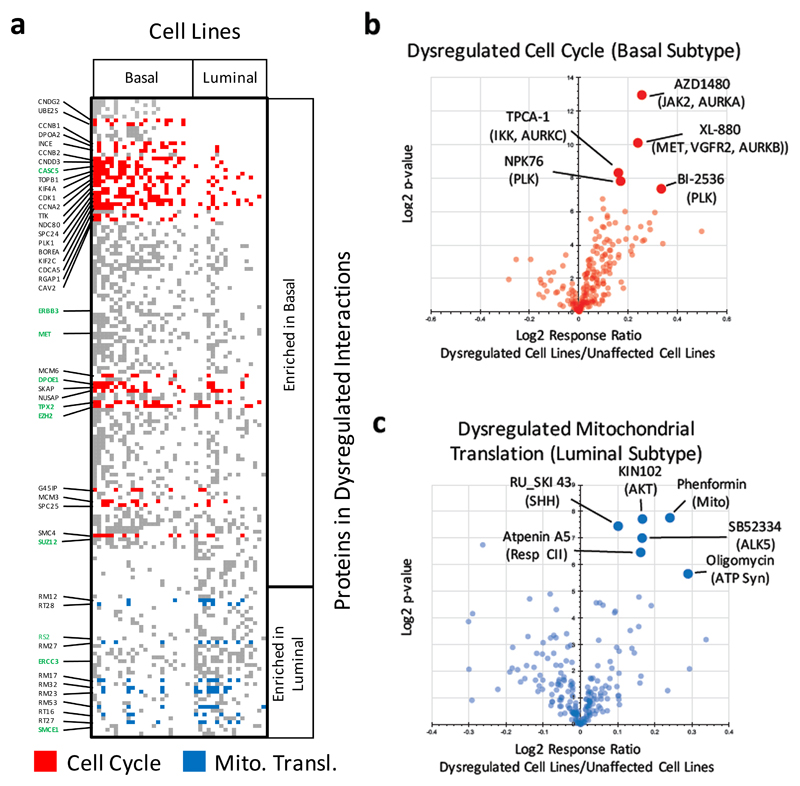
We next mined for differences in functional modules dysregulated in cancer cell lines of the basal and luminal subtypes analyzing data from all 41 studied cell lines. We identified 167 proteins with dysregulated associations in at least 25 % of either luminal (n = 17) or basal (n = 24) cell lines with a significant difference between the two subtypes determined by hypergeometric testing (p ≤ 0.1, Fig. 4a, Supplementary Table 11). Ten of these proteins are encoded by known cancer genes: CASC5, ERBB3, EZH2, DPOE1, MET, TPX2, and SUZ12 with dysregulations enriched in basal subtype cell lines and ERCC3, RS2, and SMCE1 with dysregulations mainly in the luminal cell lines22–24. We used the DAVID Bioinformatics platform25 for Gene Ontology (GO) category analysis of the proteins enriched in the dysregulated protein-protein association network of basal (117 proteins) and luminal cell lines (49 proteins) (Fig 4a). This analysis revealed that 31 cell cycle regulating proteins were affected to diverse extents mainly in the basal cell lines and 9 mitochondrial ribosomal proteins showed dysregulated associations mainly in luminal cell lines (Fig. 3a, Supplementary Table 11).
Figure 4. Breast cancer subtype specific dysregulated functional modules reflect sensitivity to specific therapeutics.
(a) 167 proteins were found in dysregulated associations affecting at least a quarter of either basal (n = 24) or luminal (n = 17) subtype cancer cells and significantly enriched in either subtype (hypergeometric test, p ≤ 0.1). The proteins include ten known cancer genes (bold green) 22–24. Gene Ontology (GO) category analysis revealed cell cycle regulating proteins (red) enriched in the dysregulated network of basal-type cell lines and proteins involved in mitochondrial translation (blue) dysregulated in luminal-type cell lines. Proteins not annotated to either GO category are shown in gray. (b) Monitoring the response to 195 drugs cell lines affected by dysregulation of associations of cell cycle regulation proteins in comparison to unaffected cell lines revealed the most significant response difference for six kinase inhibitors two of which are targeting Polo-like kinases (PLK) known as important cell cycle regulators. Three other inhibitors are targeting the kinases JAK2, IKK, and MET, and have been shown to affect the activity of the cell cycle regulating Aurora kinases. Drug responses were determined based on the area under the dose response curve (see Methods) (c) Cell lines with dysregulated associations of proteins involved in mitochondrial translation showed a significant response to drugs including three therapeutics targeting mitochondrial function: phenformin, oligomycin, and atpenin.
To evaluate if dysregulated functional modules could predict how affected cells respond to drugs, we determined the response of the 41 cell lines to 195 drugs spanning a wide range of targets (Supplementary Table 12). We identified six therapeutics that produced a significantly higher response in cell lines with a dysregulated cell cycle (≥ 2 cell cycle proteins with disturbed associations) when compared to unaffected cell lines (z-value ≥ 2 considering all p-values of drugs with higher response in affected cell lines) (Fig. 4b). These included two inhibitors – NPK76 (p = 4x10-3) and BI-2536 (p = 6x10-3) – of polo-like kinases known as important cell cycle regulators. Another three drugs have nominal targets that are not directly linked to cell cycle: JAK2 (AZD1480, p = 1x10-4), MET (XL-880, p = 1x10-3), and IKK (TPCA-1, p = 3x10-3). However, all three of them have been shown to potently inhibit Aurora Kinases26–28, and XL-880 (foretinib) also inhibits polo-like Kinase 4 (PLK4)28. Another therapeutic, ponatinib, is targeting ABL1 but is known to inhibit a wide range of kinases29. We also identified dysregulated protein-protein associations encompassing MET enriched in basal cell lines (Supplementary Table 11).
Across the 195 drugs tested, three of the significant six with a stronger response in cell lines with dysregulation of the mitochondrial ribosome protein complex were seen with phenformin (p = 5x10-3) that blocks mitochondrial respiration through inhibition of complex I30, atpenin A5 (p = 1.1x10-2), a mitochondrial complex II inhibitor31, and oligomycin (p = 2x10-2), an inhibitor of ATP synthase32. As a significance cutoff we have defined a z-score of ≥ 2 considering all p-values of drugs with higher response in the affected cell lines. Taken together, these results strongly suggest that predicted dysregulations of functional cellular modules based on deviation from global co-regulation networks is a potentially useful approach to identify drug susceptibilities.
In summary, we have shown that when studying cancer cell lines, protein co-regulation analysis allows for the identification of functional protein-protein associations with an accuracy 10-fold higher than when RNA-seq data are used for co-expression analysis. The high level of correlation that allows for identification of protein complexes using relative expression levels across samples implies a stringent control of protein levels in cells. A likely explanation is that protein degradation leads to appropriate protein concentration in accordance with the functional network. This is concordant with studies showing that proteins from multi-protein complexes are degraded at a higher rate if not embedded into their cognate complex18. In this model differential stability of proteins when they are part of their functional complex or free is linked to appropriate stoichiometry. In keeping with our results, studies on the effects of aneuploidy in yeast and human cell lines have implicated protein degradation in the accurate control of protein levels for complexes between products of genes affected or not by genomic duplication33,34. As shown in Supplementary Fig. 10 for the well-studied 26S proteasome multi-protein complex, mRNA but not protein levels in our dataset correlate well with gene copy number variations (CNV) (Supplementary Table 13). These results are also supported by recent reports comparing CNVs, mRNA and protein levels in colon and breast cancer tumors10,35. The CNV driven anomalies in mRNA levels may also partially explain why mRNA co-regulation analysis is not as predictive of functional relations as proteomic.
We believe that the increased stability of proteins when embedded in complexes compare to their dissociated state is the basis for deviations of the co-regulation of two proteins in individual cell lines: When the interaction of a protein with its partners is perturbed either through a mutation or other dysregulations the protein is subjected to enhanced degradation compared to its regular binding partners. It should be noted that the correlation between mRNA and protein levels was positive for all studied cell lines (median = 0.58, Supplementary Fig. 3c) indicating that, overall, small differences between mRNA and protein levels underlie the diverging results from co-regulation analysis. This is consistent with the similarity in clustering of the cell lines based on their mRNA and protein profiles (Supplementary Fig. 2). Thus, in addition to transcriptional co-regulation a very accurate if often minor posttranscriptional adjustment of protein levels allows to use protein co-regulation analysis to identify interactions between proteins.
Using the IMAHP strategy cell line specific deviations of a co-regulation of protein pairs can be used to identify dysregulations of protein-protein associations and cellular vulnerabilities as revealed here leveraging large RNAi and drug response datasets. The high-throughput capability of the described mapping of protein-protein association dysregulations and its applicability to a wide range of biological samples makes this method a promising tool for a broad number of applications in cell biology and cancer therapeutics studies.
Methods
Cell Culture and Lysis
Breast cancer cell lines were grown to 90 % confluency under indicated culture conditions (Supplementary Table 1). For cell lysis of adherent cells, growth media was removed and cells were rinsed with PBS before being trypsinized to remove from growth plastic. Cells were then counted and 3.0 x 106 cells were transferred to a new tube and pelleted. Media was aspirated and cells were washed with PBS. After re-pelleting cells, the PBS was aspirated and the cells were fast-frozen on dry ice then stored at -80 °C until lysis. For cells growing in suspension, cells were pelleted and re-suspended before counting. Again, 3.0 x 106 cells were pelleted, rinsed and frozen as before.
Cells were lysed with 200 μL of lysis buffer by passing through a 21 gauge needle 20 times. Lysis buffer was composed of 75 mM NaCl, 3 % SDS, 1 mM NaF, 1 mM beta-glycerophosphate, 1 mM sodium orthovanadate, 10 mM sodium pyrophosphate, 1 mM PMSF and 1X Roche Complete Mini EDTA free protease inhibitors in 50 mM HEPES, pH 8.536. Lysates were then sonicated for 5 minutes in a sonicating water bath before cellular debris was pelleted by centrifugation at 14000 rpm for 5 minutes.
Protein Digestion and TMT Labeling
Protein concentration of the cell lysates was determined using a BCA assay (Thermo Scientific). Proteins were then reduced with DTT and alkylated with iodoacetamide as previously described37. Reduced and alkylated proteins were precipitated via methanol-chloroform precipitation38. Precipitated proteins were reconstituted in 300 μL of 1 M urea in 50 mM HEPES, pH 8.5. Vortexing, sonication and manual grinding were used to aid solubility. Solubilized protein was digested in a two-step process starting with overnight digest at room temperature with 3 μg of Lys-C (Wako) followed by six hours of digestion with 3 μg of trypsin (sequencing grade, Promega) at 37 ° C. The digest was acidified with TFA. Digested peptides were desalted with C18 solid-phase extraction (SPE) (Sep-Pak, Waters) as previously described39. The concentration of the desalted peptide solutions was measured with a BCA assay, and peptides were aliquoted into 50 μg portions, which were dried under vacuum and stored at -80 °C until they were labeled with TMT reagents.
Peptides were labeled with 10-plex tandem mass tag (TMT) reagents (Thermo Scientific)8,40 in principle as previously described7. TMT reagents were suspended in dry acetonitrile (ACN) at a concentration of 20 μg/μL. Dried peptides (50 μg) were re-suspended in 30 % ACN in 200 mM HEPES, pH 8.5 and 5 μL of the appropriate TMT reagent was added to the sample. TMT reagents 126 and 131 were reserved for “bridge” samples (see below), the remaining TMT reagents (127c, 127n, 128c, 128n, 129c, 129n, 130c, 130n) were used to label the digests from the individual cell lines in a random order. Peptides were incubated for with the reagents for 1 hour at room temperature. The labeling reaction was quenched by adding 6 μL of 5 % hydroxylamine. Labeled samples were then acidified by adding 50 μL of 1 % TFA and the peptide mixtures were pooled into 11 10-plex TMT samples (Supplementary Table 1), with the bridge samples carrying 126 and 131 labels. The pooled samples were desalted via C18 SPE on Sep-Pak cartridges as described above.
Basic pH Reversed-Phase Liquid Chromatography (bRPLC) Sample Fractionation
Sample fractionation was performed by basic pH reversed-phase liquid chromatography (bRPLC)41 with concatenated fraction combining as previously described39. Briefly, samples were re-suspended in 5 % formic acid/5 % ACN and separated over a 4.6 mm x 250 mm ZORBAX Extend C18 column (5 μm, 80 Å, Agilent Technologies) on an Agilent 1260 HPLC system outfitted with a fraction collector, degasser and variable wavelength detector. The separation was performed applying a gradient build from 22 to 35 % ACN in 10 mM ammonium bicarbonate in 60 minutes at a flowrate of 0.5 mL/minute. A total of 96 fractions, which were combined as previously described39. The combined fractions were dried under vacuum, re-constituted with 5 % formic acid/5 % ACN, and then analyzed by LC-MS2/MS3 for identification and quantification.
Liquid Chromatography Mass Spectrometry
All LC-MS2/MS3 experiments were conducted on an Orbitrap Fusion (Thermo Fisher Scientific) coupled to an Easy-nLC 1000 (Thermo Fisher Scientific) with chilled autosampler. Peptides were separated on an in-house pulled, in-house packed microcapillary column (inner diameter, 100 μm; outer diameter, 360 μm). Columns were packed first with approximately 0.5 cm of Magic C4 resin (5 μm, 100 Å, Michrom Bioresources) followed by approximately 0.5 cm of Maccel C18 AQ resin (3 μm, 200 Å, Nest Group) and then to a final length of 30 cm with GP-C18 (1.8 μm, 120 Å, Sepax Technologies). Peptides were eluted with a linear gradient from 11 to 30 % ACN in 0.125 % formic acid over 165 minutes at a flow rate of 300 nL/minute while the column was heated to 60 ° C. Electrospray ionization was achieved by applying 1800 V through a PEEK T-junction at the inlet of the microcapillary column.
The Orbitrap Fusion was operated in data-dependent mode, with a survey scan performed over an m/z range of 500-1,200 at a resolution of 6x104 in the Orbitrap. For the MS1 survey scan, automatic gain control (AGC) was set to 5 x 105 and a maximum injection time of 100 ms. The S-lens was set to an RF of 60 and data was centroided. The most abundant ions detected in the survey scan were subjected to MS2 and MS3 experiments using the Top Speed setting that enables a maximum number of spectra to be acquired in a 5 seconds experimental cycle before the next cycle is initiated with another survey full-MS scan.
For MS2 analysis, the decision tree option was enabled, whereby precursors were selected based on charge state and m/z range. Doubly charged ions were selected from an m/z range of 600-1200, for triply and quadruply charged ions had to be detected in an m/z range of 500-1200. The ion intensity threshold was set to 5x105. When acquiring MS2 spectra ions were isolated applying a 0.5 m/z window using the quadrupole and fragmented using CID at a normalized collision energy of 30 %. Fragment ions were detected in the ion trap at a rapid scan rate. The AGC target was set to 1 x 104 and the maximum ion injection time was 35 ms. Centroided data was collected.
MS3 analysis was performed using synchronous precursor selection (MultiNotch MS3) enabled to maximize sensitivity for quantification of TMT reporter ions8. Up to 10 MS2 precursors were simultaneously isolated and fragmented for MS3 analysis. The isolation window was set to 2.5 m/z and fragmentation was carried out by HCD at a normalized collision energy of 50 %. Fragment ions in the MS3 spectra were detected in the Orbitrap at a resolution of 60,000 at an m/z of ≥ 110. The AGC target was set to 5 x 104 ions and the maximum ion injection time to 250 ms. Centroided data were collected. Fragment ions in the MS2 spectra with an m/z of 40 m/z below and 15 m/z above the precursor m/z were excluded from being selected for MS3 analysis.
Data Processing and Analysis
Data were processed using an in-house developed software suite42. RAW files were converted into the mzXML format using a modified version of ReAdW.exe (http://www.ionsource.com/functional_reviews/readw/t2x_update_readw.htm). Spectral assignments of MS2 data were made using the Sequest algorithm43 to search the Uniprot database (02/04/2014) of human protein sequences including known contaminants such as trypsin. The database was appended to include a decoy database consisting of all protein sequences in reverse order44–46. Searches were performed with a 50 ppm precursor mass tolerance. Static modifications included ten-plex TMT tags on lysine residues and peptide n-termini (+229.162932 Da) and carbamidomethylation of cysteines (+57.02146 Da). Oxidation of methionine (+15.99492 Da) was included as a variable modification. Data were filtered to a peptide and protein false discovery rate of less than 1 % using the target-decoy search strategy46. This was achieved by first applying a linear discriminator analysis to filter peptide annotations (peptide-spectral matches) using a combined score from the following peptide and spectral properties: XCorr, ΔCn, missed tryptic cleavages, peptide mass accuracy, and peptide length42. The probability of a peptide-spectral match to be correct was calculated using a posterior error histogram and the probabilities of all peptides assigned to one specific protein were combined through multiplication and the dataset was re-filtered to a protein assignment FDR of less than 1 % 42 for the entire dataset of all proteins identified across all analyzed samples. Peptides that matched to more than one protein were assigned to that protein containing the largest number of matched redundant peptide sequences following the law of parsimony42.
For quantitative analysis TMT reporter ion intensities were extracted from the MS3 spectra selecting the most intense ion within a 0.003 m/z window centered at the predicted m/z value for each reporter ion and a signal-to-noise (S/N) values were extracted from the RAW files. Spectra were used for quantification if the sum of the S/N values of all reporter ions was ≥ 386 and the isolation specificity for the precursor ion was ≥ 0.757. Protein intensities were calculated by summing the TMT reporter ions for all peptides assigned to a protein. Normalization of the quantitative data followed a multi-step process. Intensities were first normalized using the intensity measured for the bridge sample labeled with the 126 TMT-reagent and then independently normalized using the intensity measured for the bridge sample labeled with the 131 TMT-reagent. The median bridge channel intensity measured across all 11 TMT experiments was used for the normalization. An average value was calculated for the protein intensity by averaging the two intensities from the independent bridge-sample normalizations. Taking account of slightly different protein amounts analyzed in each TMT channels we then added an additional normalization step by normalizing the protein intensities measured for each sample by the median of the median protein intensities measured in these samples. The proteome profiles from the analyses of two biological replicates were combined by calculating the average intensity if the protein was quantified in both replicates but also including intensities of proteins that were only quantified for one replicate. For further data analysis the normalized intensities were converted into log2 ratios of the intensities over the median intensity measured for each protein across all cell lines. This conversion was also performed for the transcriptome (Supplementary Table 3)11 and gene copy number variation (CNV) data (Supplementary Table 11) (cansar.icr.ac.uk).
Spearman’s Correlation Based Clustering
Spearman’s correlations of proteome or transcript or CNV profiles were calculated in the R environment using the cor.prob function47. Unsupervised clustering of profiles was done using the statistical software JMP (version Pro 11) using the Ward method without standardizing the data.
Protein-Protein Association Network Construction
For protein and RNA based networks, abundance profiles were correlated using Spearman’s rank correlation. Correlation coefficients, p-values and BH adjusted p-values were calculated in the R environment employing the cor.prob function. Resultant correlation tables were filtered for positive correlations and BH adjusted p-values ≤ 5x10-4.
Evaluation of Protein-Protein Associations
Correlation pairs were ordered and redundancies removed before comparing to a non-redundant version of the STRING database (downloaded on 08/07/2014) to find STRING annotated interactions. Only STRING interactions of high confidence (score ≥ 0.700) were considered.
For comparison with entries in the CORUM database (downloaded on 11/18/2014), all theoretical connections within a complex were determined in the R environment for each complex containing two or more unique constituents. These interactions were then ordered and compared to the generated theoretical interactions based on Spearman’s correlations. Redundant subunits of complexes were assigned to the largest complex in which they were contained (Fig. 3). Predicted CORUM interactions were also compared with STRING associations to define an overlap between CORUM and STRING.
Constructed networks were visualized in Cytoscape48 (available at www.cytoscape.org). Cys files for constructed networks are available as Supplementary Data Files 1-3, which can be opened directly in Cytoscape for visualization.
Protein-protein Association Dysregulation Screening
Cell line specific deviations from co-regulation for each protein pair identified to be associated through co-regulation analysis we first calculated the Mahalanobis distance for each cell line in a scatterplot of the protein concentration of each protein pair using excel. We then used the Grubb’s test (p ≤ 0.1) to identify cell lines as outliers inferring that the monitored protein-protein association was dysregulated in these cell lines.
Genome-Wide Pooled shRNA Dropout Screen Data
The data were published in Marcotte et al., 201620, and were provided by Dr. Benjamin Neel in a format that allowed to annotate the screening data to each cell line. The used dropout scores were zGARP scores as described in Marcotte et al., 201249. Genes defined by Marcotte et al.20 as “general essential” (essential in all cell lines) were not considered when comparing proteins in dysregulated associations with protein dropout scores. Genes with a zGARP score smaller than -2 were defined as cell line specific fitness genes.
Drug screen across large cell line collection
High-throughput drug screening was performed essentially as described previously50. Cells were grown in the media specified in Supplementary Table 1. Briefly, cells were seeded in 384 well plates at variable density to insure optimal proliferation during the assay. Drugs were added to the cells the day after seeding for adherent cell lines and the day of seeding for suspension cell lines. A series of nine doses was used with 2-fold dilution steps for a total concentration range of 256 fold. For each drug the maximum concentration was chosen based on prior knowledge of activity on target and in cells or previous data acquired in the Benes laboratory. Viability was determined using CellTiter-Glo (Promega) after 3 days of drug exposure. All plates were submitted to stringent quality control with the coefficient of variation of replicate control wells (cells with no drug) within a plate <20% and a signal (control wells) / noise (blank wells; no cells) ratio > 50. At least 2 biological replicates (different plating days) were acquired for each drug. Drugs were sourced from reputable commercial vendors with accompanying quality control documentation or were generously given by the laboratory of Dr. Nathanael Gray (Harvard Medical School). An estimator of the response to drugs (AUC: Area Under the dose response Curve) was obtained using drexplorer 1.1.0 in R (R3.1.0)51.
Gene Ontology (GO) category analysis of proteins with dysregulated protein-protein associations enriched in cells of either basal or luminal subtype were done separately for each subtype using the DAVID platform52 only considering “biological process” categories enriched with a Benjamini-Hochberg corrected p-value of ≤ 0.1 and using all proteins in the network of associated proteins determined in this study (Supplementary Table 8) as background. An unpaired, unequal variance, two tailed t-test was used to identified drugs significantly differentially affecting cell lines with defects in either mitochondrial biology (corresponding to the GO term Mitochondrial Translation) or the cell cycle (GO term Cell Cycle). For these tests cell lines were designated as normal or dysregulated using a threshold of number of dysregulated proteins leading to balanced number of cell lines in the two groups compared. The thresholds were more than one cell cycle protein and at least one mitochondrial translation protein per with dysregulated protein-protein associations per cell line. The drug response estimated by average of the AUC values of biological replicates were used for these tests. Drugs decreasing the viability of cell lines (positive log2 response ratio) associated with dysregulated cell cycle or mitochondrial function were sorted based on their Z-value transformed log2 t-test p-values. Drugs with a Z-value greater than 2 were selected as strongly affecting the survival of dysregulated cell lines and are discussed further in the text.
Supplementary Material
Acknowledgements
We thank Steven Gygi, Harvard Medical School, for access to computational software and facilities to process the proteomics data. We are grateful to Jessica Boisvert for her help with culturing the cell lines and we also acknowledge all members of the Haas and Benes laboratories for valuable discussions. C.H.B. is supported by grant 102696 from the Wellcome Trust. Cell lines were purchased with funds from the NIH LINCS phase 1 grant, HG006097. The data reported in this manuscript are tabulated in the Supplementary Materials. The mass spectrometry
Footnotes
Author Contributions
C.H.B. and W.H. conceived and designed the study. J.D.L., C.H.B., and W.H. wrote the manuscript. J.D.L. and W.H. performed the proteomics experiments. P.G. and C.H.B. performed the drug screen and analyzed the drug screen data. J.D.L, R.M., A.A., I.P-M., C.H.B, and W.H. performed the analysis of the proteomics data.
Author Information
The mass spectrometry proteomics data have been deposited in the MassIVE proteomics data repository under the accession number MSV000081383.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 2.Rolland T, et al. A proteome-scale map of the human interactome network. Cell. 2014;159:1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huttlin EL, et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hein MY, et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 5.McAlister GC, et al. Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal Chem. 2012;84:7469–7478. doi: 10.1021/ac301572t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heiser LM, et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci U S A. 2012;109:2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ting L, Rad R, Gygi SP, Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Methods. 2011;8:937–940. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McAlister GC, et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal Chem. 2014;86:7150–7158. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang B, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382–387. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klijn C, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol. 2015;33:306–312. doi: 10.1038/nbt.3080. [DOI] [PubMed] [Google Scholar]
- 12.Szklarczyk D, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, et al. Proteome profiling outperforms transcriptome profiling for co-expression based gene function prediction. Mol Cell Proteomics. 2016 doi: 10.1074/mcp.M116.060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stefely JA, et al. Mitochondrial protein functions elucidated by multi-omic mass spectrometry profiling. Nat Biotechnol. 2016;34:1191–1197. doi: 10.1038/nbt.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chick JM, et al. Defining the consequences of genetic variation on a proteome-wide scale. Nature. 2016;534:500–505. doi: 10.1038/nature18270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruepp A, et al. CORUM: the comprehensive resource of mammalian protein complexes--2009. Nucleic Acids Res. 2010;38:D497–501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heath CG, Viphakone N, Wilson SA. The role of TREX in gene expression and disease. Biochem J. 2016;473:2911–2935. doi: 10.1042/BCJ20160010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldberg AL, Dice JF. Intracellular protein degradation in mammalian and bacterial cells. Annu Rev Biochem. 1974;43:835–869. doi: 10.1146/annurev.bi.43.070174.004155. [DOI] [PubMed] [Google Scholar]
- 19.cansar.icr.ac.uk.
- 20.Marcotte R, et al. Functional Genomic Landscape of Human Breast Cancer Drivers, Vulnerabilities, and Resistance. Cell. 2016;164:293–309. doi: 10.1016/j.cell.2015.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berger AH, et al. High-throughput Phenotyping of Lung Cancer Somatic Mutations. Cancer Cell. 2016;30:214–228. doi: 10.1016/j.ccell.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.cancer.sanger.ac.uk/census/.
- 23.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Ioannidis S, et al. Discovery of 5-chloro-N2-[(1S)-1-(5-fluoropyrimidin-2-yl)ethyl]-N4-(5-methyl-1H-pyrazol-3-yl)p yrimidine-2,4-diamine (AZD1480) as a novel inhibitor of the Jak/Stat pathway. J Med Chem. 2011;54:262–276. doi: 10.1021/jm1011319. [DOI] [PubMed] [Google Scholar]
- 27.Jester BW, Gaj A, Shomin CD, Cox KJ, Ghosh I. Testing the promiscuity of commercial kinase inhibitors against the AGC kinase group using a split-luciferase screen. J Med Chem. 2012;55:1526–1537. doi: 10.1021/jm201265f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis MI, et al. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 29.O'Hare T, et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009;16:401–412. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El-Mir MY, et al. Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J Biol Chem. 2000;275:223–228. doi: 10.1074/jbc.275.1.223. [DOI] [PubMed] [Google Scholar]
- 31.Miyadera H, et al. Atpenins, potent and specific inhibitors of mitochondrial complex II (succinate-ubiquinone oxidoreductase) Proc Natl Acad Sci U S A. 2003;100:473–477. doi: 10.1073/pnas.0237315100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kagawa Y, Racker E. Partial resolution of the enzymes catalyzing oxidative phosphorylation. 8. Properties of a factor conferring oligomycin sensitivity on mitochondrial adenosine triphosphatase. J Biol Chem. 1966;241:2461–2466. [PubMed] [Google Scholar]
- 33.Dephoure N, et al. Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. Elife. 2014;3:e03023. doi: 10.7554/eLife.03023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stingele S, et al. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol. 2012;8:608. doi: 10.1038/msb.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mertins P, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villen J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc. 2008;3:1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haas W, et al. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol Cell Proteomics. 2006;5:1326–1337. doi: 10.1074/mcp.M500339-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 39.Edwards A, Haas W. Multiplexed Quantitative Proteomics for High-Throughput Comprehensive Proteome Comparisons of Human Cell Lines. Methods Mol Biol. 2016;1394:1–13. doi: 10.1007/978-1-4939-3341-9_1. [DOI] [PubMed] [Google Scholar]
- 40.Thompson A, et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, et al. Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics. 2011;11:2019–2026. doi: 10.1002/pmic.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huttlin EL, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eng JK, McCormack AL, Yates JRI. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 44.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 45.Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods. 2005;2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 46.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 47.R_Developmental_Core_Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- 48.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marcotte R, et al. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov. 2012;2:172–189. doi: 10.1158/2159-8290.CD-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garnett MJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tong P, et al. drexplorer: A tool to explore dose-response relationships and drug-drug interactions. Bioinformatics. 2015;31:1692–1694. doi: 10.1093/bioinformatics/btv028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang DW, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.