Abstract
Weak association energy can lead to uniform nanostructures: defects can anneal due to subunit lability. What happens when strong association energy leads to particles where defects are trapped? Alphaviruses are enveloped viruses whose icosahedral nucleocapsid core can assemble independently. We used a simplest case system to study Ross River virus (RRV) core-like particle (CLP) self-assembly using purified capsid protein and a short DNA oligomer. We find that capsid protein binds the oligomer with high affinity to form an assembly-competent unit (U). Subsequently, U assembles with concentration dependence into CLPs. We determined that U-U pairwise interactions are very strong (ca. −6 kcal/mol) compared to other virus assembly systems. Nonetheless, assembled RRV CLPs appeared morphologically uniform and cryo-EM image reconstruction with imposed icosahedral symmetry yielded a T=4 structure. However, 2D class averages of the CLPs show that virtually every class had disordered regions. These results suggested that irregular cores may be present in RRV virions. To test this hypothesis, we determined 2D class averages of RRV virions using authentic virions or only the core from intact virions isolated by computational masking. Virion-based class averages were symmetrical, geometric, and corresponded well to projections of image reconstructions. In core-based class averages, cores and envelope proteins in many classes were disordered. These results suggest that partly-disordered components are common even in ostensibly well-ordered viruses, a biological realization of a patchy particle. Biological advantages of partly-disordered complexes may arise from their ease of dissociation and asymmetry.
Keywords: Ross River Virus, capsid, self-assembly, enveloped viruses
About half of known virus families have a spherical (icosahedral) capsid that is composed of tens to thousands of identical subunits. In most of these viruses, subunits are arranged with a quasi-equivalence to make a characteristic pattern of pentamers and hexamers.1
Viruses are an example of an evolved, optimized self-assembling nanoparticle. However, generalizations for nanoparticle assembly should apply equally to viruses, other biological complexes, and abiological nanoparticles. Numerous studies suggest that assembly is based on weak subunit-subunit association energy (Fig 1A).2–4 Weak intersubunit contacts provide the opportunity to thermodynamically correct mistakes to yield a well-ordered product. The resulting complexes, where every multivalent subunit is maximally ligated, have a high barrier to dissociation allowing them to persist even at low concentration. This behavior is ideal for viruses where capsid integrity is critical for infection (e.g. extracellular picornaviruses or intracellular transport of hepadnavirus cores to the nucleus).5 Defects creep into assembly simulations when nucleation is fast compared to elongation or when intersubunit interactions are strong, resulting in trapped intermediates (Fig 1B).4, 6 Formation of trapped incomplete particles can be engineered into particle assembly.7 A third assembly model adds low energy alternative geometries for intersubunit interaction to yield mosaic lattices that do not have quasi-equivalence (Fig 1C).8, 9 Examples of viruses with alternative subunit-subunit interactions include pseudo-T=2 bromoviruses under aggressive assembly conditions.10, 11 Immature human immunodeficiency virus (HIV) provides an example of assembly with numerous trapped defects.12, 13
Figure 1. Models of capsid assembly and trapped assembly.
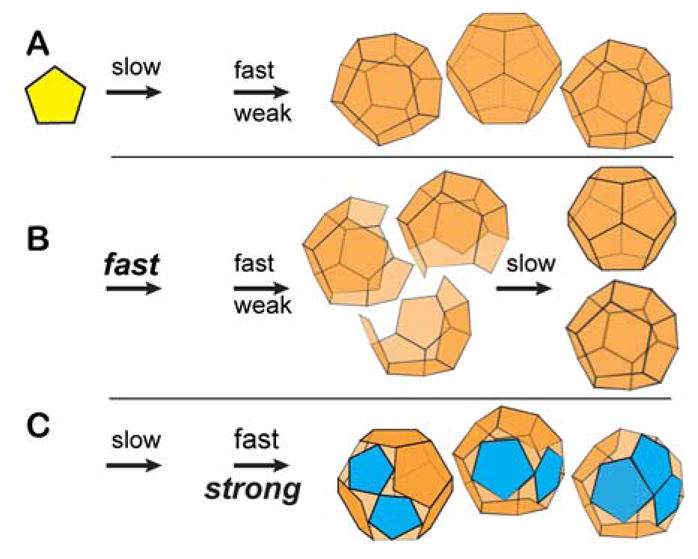
For simplicity, we consider assembly of a dodecahedron; the first arrow indicates the rate of nucleation, the second indicates the rate of elongation (above the arrow) and average association energy (below the arrow).5, 43 Changes in assembly kinetics or thermodynamics are bolded. (A) Assembly is robust when nucleation is relatively slow, elongation fast, and association weak. In this regimen, subunits that add with incorrect geometry readily dissociate to eliminate defects. (B) Incomplete capsids can be trapped when nucleation is relatively fast resulting in so many nuclei that there is insufficient free subunit to complete the nascent capsids. Incomplete capsids can also be trapped with high association energy or high protein concentration. Incomplete capsids can slowly cure by exchange of subunits.6, 7 (C) Capsids can become mosaic when association is fast enough or strong enough to prevent dissociation of subunits with incorrect geometry (blue subunits). Defects will be common when there is a deep local energy minimum for an alternative geometry for subunit-subunit interaction
We chose to evaluate assembly of alphavirus capsid protein (CP) because of the ease of controlling assembly, the apparent lability of particles, and the ease of correlating biophysical and biological observations with this system.14–16 Alphaviruses are enveloped, positive strand RNA viruses that are cyclically transmitted between arthropod vectors and a wide range of vertebrate hosts including humans.17, 18 In both hosts, alphavirus replication and assembly occur in the cytoplasm. Prototype alphaviruses include Sindbis and Ross River virus (RRV). The virion is a ~70 nm diameter particle comprised of a ~40-nm nucleocapsid core, a lipid bilayer, and glycoprotein spikes. The core and glycoprotein layers share T=4 icosahedral symmetry.19–23 The nucleocapsid core consists of 240 copies of capsid protein (CP) and the viral genome. In vivo, one mechanism of alphavirus assembly has independent pathways for core assembly and formation of envelope protein complexes, which merge during budding from the plasma membrane to form mature virion particles.24–26
Alphavirus CPs are highly conserved27 and can be divided into the N- and C-terminal domains. The N-terminus includes ~30 basic residues (27 in RRV)28–30 and an 18-amino-acid region predicted to form a coiled-coil motif.15 The C-terminal domain has a chymotrypsin-like domain and a hydrophobic pocket that interacts with the cytoplasmic domain of the E2 glycoprotein.26, 31–33 In cryo-EM structures of virions, the CP N-terminal residues are disordered while the C-terminal domain can be modeled into density.19–21, 23, 34 In vitro, alphavirus CP requires polyanionic cargo to form core-like particles (CLPs); the protein alone will not self-assemble in response to ionic strength or pH at any concentration yet tested.16, 35–39 In vitro-assembled CLPs are structurally and functionally similar to cores of mature virions14, 16 as they appear to have T=4 symmetry and are able to interact with viral glycoproteins26, 40–42 to form functional virus-like particles.41, 42
Assembly of closed spherical capsids has been characterized by weak subunit-subunit interactions and disassembly under stringent conditions.3, 6, 43 Examples include hepatitis B virus (HBV) and cowpea chlorotic mottle virus. However, in the present study, we found that in vitro alphavirus core assembly follows a different paradigm. We observed strong subunit-subunit interactions but the resulting CLPs readily disassembled. Consistent with these observations, we found that most CLPs had geometric defects. We also found evidence for “imperfect” cores in mature virions. We propose that irregular cores may be common in enveloped viruses where capsid structural integrity is not critical for (and may be detrimental to) the virus lifecycle.
RESULTS AND DISCUSSION
CP and DNA form a complex that assembles into capsids
The goal of the initial studies was to measure CP-CP interaction. We drove assembly with a single-stranded 27mer DNA oligonucleotide. The charge on this short oligo is sufficient to neutralize the N-terminal basic residues of RRV CP, but yet not long enough to crosslink CP subunits or act as a scaffold for binding multiple CP monomers. This system was used to allow assembly to proceed via freely diffusing monomers. Previous studies showed that short oligos supported assembly though not as well as oligonucleotides that could bind several subunits.16
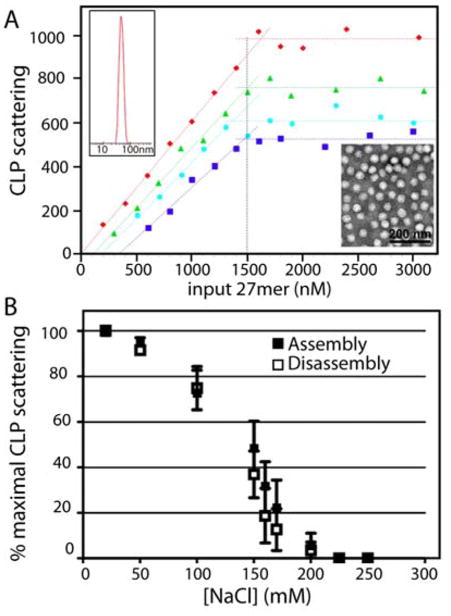
Assembly was measured by static and dynamic light scattering. The intensity of the light scattered by a solute (static light scattering) is proportional to the average molecular weight of the sample. Fluctuations of the light scattering signal (dynamic light scattering or DLS) are proportional to the diffusion coefficient of the solute and thus its hydrodynamic diameter. Solutions of 1.5 μM RRV CP were titrated with the DNA 27mer and monitored by static light scattering and DLS (Fig 2A). Unassembled, monomeric RRV CP molecules had weak static light scattering and a DLS-estimated diameter of ~ 9 nm (Fig S1). The 27mer DNA, at the concentrations used in this work, did not detectably increase the amount of scattered light above background. In titrations of RRV CP by 27mer, light scattering intensity increased and DLS analysis indicated a diameter of 40–50 nm (Figs S2–S5) corresponding to RRV CLPs. In some DLS analyses, large diameter peaks beyond the capsid peak were noted, but these aggregates represented a small amount of the total mass, were irregular in size, were not reproducible, and made little contribution to light scattering intensity. We confirmed the presence of CLPs by TEM (Fig S1). The diameter of RRV CLPs, from negative stain TEM, agrees well with RRV CLPs measured by DLS and nucleocapsid cores from intact virus particles.14, 16, 19, 22, 44
Figure 2. In vitro assembly of RRV CLPs is sensitive to ionic strength.
(A) Static light scattering measurement of titrations of 1.5 μM RRV CP by a 27mer DNA oligo (DLS and TEM in Figs. S1–S5). Samples were at 22°C at various ionic strengths (red=50 mM NaCl, green=110 mM NaCl, cyan=150 mM NaCl, and blue=170 mM NaCl). Data were fit to two straight lines. The pseudo-critical concentration (X-intercept) was determined from this linear fit. In each data set, the RRV CP saturated at about one 27mer DNA oligo per protein. The dotted black vertical line marks where when equimolar amounts of RRV CP and 27mer DNA oligo are mixed. (B) CLPs show no hysteresis. Either CLPs were assembled in increasing concentrations of NaCl (closed squares) or pre-assembled CLPs were disassembled by diluting to lower concentrations of NaCl (open squares). The amount CLP present, measured by light scattering as ionic strength changes, is almost identical for assembly and disassembly.
The dependence of CLP assembly on the concentration of 27mer DNA was determined by static light scattering (Fig S2–S5) at different NaCl concentrations (Fig 2A). We observed that a minimum concentration of DNA was needed to initiate CLP assembly (the X-intercept in Fig 2A, Table 1). Below this pseudo critical concentration there was no measurable CLP (scattering intensities were at noise level and no stable or reproducible DLS peaks were observed) and above it CLP concentration increases linearly with input DNA until it saturates RRV CP at equimolar. With the rising ionic strength, the pseudo critical concentration increases while the maximum CLP assembly decreases, suggesting that consistent with the requirement of anionic cargo for the formation of CLPs because the interactions between CP-DNA complexes have electrostatic contributions.
Table 1.
Association of U to form CLP.
| [NaCl] (mM) | KDapparent (nM) | ΔGcontact intersubunit (kcal/mol)a |
|---|---|---|
| 50 | 0 | tight binding |
| 110 | 108 | −6.25 |
| 150 | 186 | −6.13 |
| 170 | 345 | −5.80 |
From these data, we propose a minimal assembly model where one RRV CP molecule binds to one 27mer oligo to yield an assembly subunit (U), 240 of which can go on to form a CLP. Based on the assembly maximum at an RRV CP:DNA oligo ratio of 1:1 it appears that oligo-binding is essentially quantitative in this concentration range (Fig 2A).
| (1) |
This model (eqn 1) is readily analyzed when 27mer binding is quantitative and the concentration of CLP will vary as a function of the U-U association energy as described by the mass action law for Kcapsid (eqn 2).6, 43 Kcapsid is related to the observed pseudo-critical concentration of assembly, KDapparent. Physically, KDapparent is the equilibrium concentration where the concentrations of free U and U in CLP are equal (KDapparent = [U] = 240[CLP]). Thus at concentrations above KDapparent, addition of oligo results in CLP formation until the protein concentration is saturated.
| (2) |
Kcapsid can be deconstructed to the pairwise association constants between individual subunits (Table 1),6, 43 taking into account that there are 240 subunits in a T=4 capsid each making three intersubunit contacts. Three contacts is the minimum required for forming a 2D lattice and is consistent with the structure of the CLP.14 The resulting association energies between U subunits include contributions from protein-protein and protein-nucleic acid interactions. The nucleic acid may specifically neutralize charge of one CP or may make other contributions to CLP formation.
CLP formation shows no detectable hysteresis to dis-assembly
Virus capsids are closed polymers, once complete there is no “edge” to facilitate equilibration between free and bound subunits, which creates an inherent hysteresis to dissociation.45–47 Thus, a simple test for the completeness of a capsid is to compare the ionic strength dependence of assembly and disassembly. If the curves do not overlap it is indicative of hysteresis.
We observed that increasing ionic strength affected CLP stability (Table 1, Fig 2B). To test for hysteresis in RRV CLPs we examined how ionic strength affected stability of pre-assembled particles compared to spontaneous assembly at the same ionic strength (Fig 2B). RRV CP and oligo were assembled in buffers of varying NaCl concentrations and compared to disassembly of pre-formed 1:1 RRV CP:oligo CLPs. Within error, assembly and disassembly showed the same sensitivity to ionic strength for samples measured with one hour of mixing. Between 50 and 200 mM NaCl, CLP stability decreased monotonically. The similarity in ionic strength dependence for both assembly and disassembly was observed regardless of initial RRV CP starting concentration. By comparison, samples of hepatitis B virus capsid protein can persist in the capsid form for months under conditions where they could not assemble.45, 48
In vitro assembled CLPs are heterogeneous
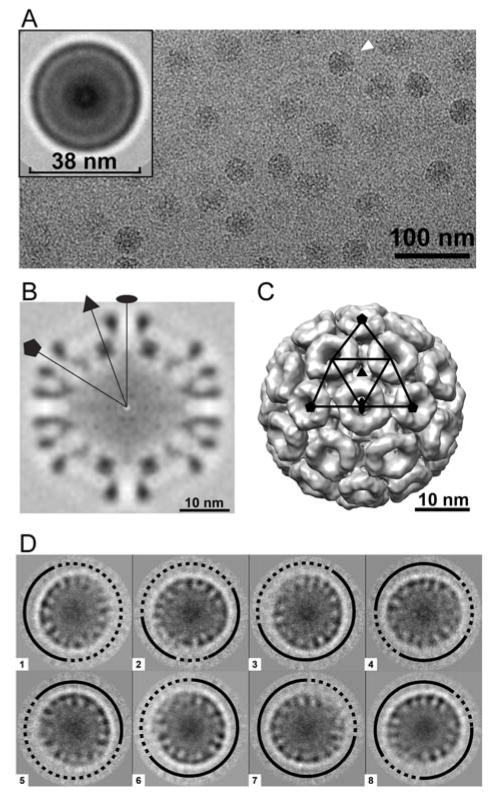
The lack of hysteresis for disassembly suggested that our CLPs had defects.6, 45, 46 A 3D image reconstruction of CLPs had previously been published14 and its low resolution suggested inherent heterogeneity between the particles. Our CLPs in cryo micrographs had spherical morphology with a small number of distorted or smaller particles (Fig 3A). Translationally aligned averaged cryo images show the capsid has an outer diameter of about 38 nm surrounding a weaker internal density layer (Fig 3A, inset), similar to the nucleocapsid in intact Sindbis virus.20, 49 We determined a 3D image reconstruction of our CLPs to 17.9 Å resolution, using the nucleocapsid core from an RRV reconstruction (Fig S6) low-pass filtered to 40 Å to bootstrap particle orientation (Figs 3B, C). This structure shows the well-organized hexamers and pentamers connected to one-another by weaker density as seen in other alphavirus and alphavirus CLP structures.14, 19–23, 44 However, 3D reconstruction imposes symmetry during the alignment process; it shows the presence of a common structural lattice but necessarily obscures irregularities by icosahedral averaging. To test for particle fidelity and uniformity, we examined the CLP images using unbiased 2D classification. Previously, we used 2D classification to identify incomplete kinetically trapped hepatitis B virus capsids, where we found that incomplete particles favored a limited number of discrete sizes.7 We reasoned that reference-free 2D classification would allow CLP defects, not necessarily visible in the raw data, to align.7 Class averaging also allows us to overcome some of the noise inherent in cryo-microscopy at the expense of averaging away some dissimilarities within a class, understating the actual differences. Nonetheless, we observed that a large fraction of particles had partially disordered regions evident in class averages by diffuse density at the capsid surface while the other regions were well demarcated indicating structural order (Fig 3D). This indicates a heterogeneous population of CLPs containing partially organized RRV CP subunits, providing a basis for the lack of hysteresis and fragility observed here and previously.14
Figure 3. The structure of Ross River virus CLPs shows a mixture of complete and partially complete spherical particles.
(A) A cryo-micrograph of RRV CLPs shows many spherical and some distorted particles (white arrow). A translationally aligned averaged image (inset) shows that the RRV CLPs have an average diameter of ~38 nm. (B) The central section of an 17.9 Å resolution 3D reconstruction of an RRV CLP. The oval, triangle, and pentagon indicate locations of twofold, threefold and fivefold axes, respectively. (C) A surface-shaded representation of the RRV CLP rendered at 1σ above the average density, viewed along the icosahedral twofold axis. The CLP shows a T=4 organization. For this reconstruction, icosahedral symmetry was imposed. (D) Reference-free class averages of RRV CLPs. Classes show irregularities (compare solid and dashed lines) on the CLP surface.
Nucleocapsid cores in virions share properties with in vitro assembled CLPs
We hypothesized that high association energy between subunits would lead to a propensity for CLP defects and this was experimentally supported by observations (Fig 2B, 3). Previous work has demonstrated that CLPs interact with glycoproteins and these particles function like mature virions.26, 41, 42 If cores with defects are the standard, then similar defects seen in CLP particles should be observed in virions. Indeed, while the envelope proteins may cause some rearrangement of the CP during budding,26, 50 if the defects seen in the CLPs are observed in virions, then envelope proteins do not completely reorganize the core particle. Because the inner core and outer glycoprotein layers of alphaviruses are aligned,19–21, 23, 34 we reasoned that class averages based on virion cores should yield the same results as class averages based on the whole virus if, and only if, the cores provided sufficient signal and were organized. If the class averages were different between the cores and the entire particle, then the cores are either not intact or have a different organization compared to the glycoprotein spikes. Because of the lack of regularity of anticipated defects, this test will undercount partially disorganized cores.
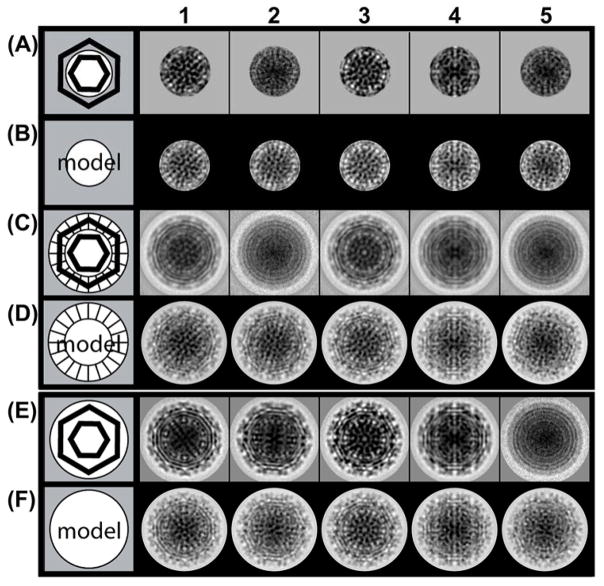
To focus on the core and minimize contribution of the glycoprotein envelope, reference free class averages were determined by limiting the data used in classification to a radius of ≤22 nm (Fig. 4A). 6560 cryo-EM images of virions were separated into twelve classes. Class averages of the core region compared to varying degrees with projections of core density (from a 15 Å resolution RRV image reconstruction (Fig. S6B)) were aligned to achieve the best match (Figs. 4B and S7). Most of the class averages (Fig 4A) and projections (Fig 4B) showed matching geometrical features but a substantial fraction did not.
Figure 4. Selected virion class averages show evidence of core disorder.
A group of 6560 images of freshly prepared RRV virions were grouped into 12 classes (see Fig S7 for the complete set) and compared with a 3D model. (A) Experimental core-based class averages used only the core for the class averaging. The schematic of a virion (left most column) shows the glycoprotein masked out (grey area). (B) The nucleocapsid density from a 3D reconstruction of RRV was isolated computationally (white area of cartoon) and projections were calculated to best match the class averages in row A. (C) Experimental class averages of RRV virions, classified and aligned based on their cores from row A, but including the glycoprotein layer. The hashed outer ring in the cartoon indicates that the glycoprotein layer is shown in the images to the right but not used for classification. In several classes the envelope is regular and geometric (e.g. column 1). In others, it is misaligned resulting in concentric circles (e.g. column 2). (D) To mimic the images in row C, projections were calculated using the entire RRV image reconstruction and the orientations from row B. In these projections, the glycoprotein envelopes are distinctly geometric, even in cases where row C showed an irregular core. (E and F) In control experiments, classification of RRV based on the entire virion (E) generally shows a well-organized periphery (i.e. the glycoprotein layer) and matches well with corresponding projections of the RRV image reconstruction (F). The core region of E generally appears to be well-organized. However, here are some classes that appear poorly organized (e.g. column 5).
To examine how well the glycoprotein envelopes were aligned with cores, images in the core-based class averages (Fig. 4A) and model projections (Fig. 4B) were expanded to include the glycoprotein layers (Fig. 4C, D). In the projections, the highly geometric features of the glycoprotein layer were unambiguous (Fig. 4D). The similarities and differences between the class averages and the projections were striking (Compare Fig. 4C and 4D). For example, in the class shown in column 1, the correspondence between the project and class average were good for both the core (rows A and B) and the whole virus (rows C and D). On the other hand, column 2 tells a different story: the class average and the core projection appear consistent (compare rows A and B), but the glycoprotein layer, averaged over the class, is so misaligned that it is reduced to featureless concentric circles. We see evidence of this disconnect in 40% of the particles, based on class averages (classes 2, 4, 5, 8, 11, and 12 in Fig S7). To examine the organization of the whole virion for comparison to the core-based averages, the same images were sorted into classes using a cutoff radius of 37 nm to include the envelope with minimal background (Fig. 4E). Most, but not all, of these averages (accounting for 88% of the images) show detailed geometrical features at high radius consistent with a well-ordered glycoprotein layer. The geometrical averages correspond extremely well with projections (Fig. 4F).
Strikingly, in some cases, the core in the core-based class average (column 4, rows A–D) matches very well with the core in the glycoprotein-based class (column 4, rows E–F). However, the glycoprotein layer for the core-based averages is poorly ordered (row C).
Taken together, it appears that virion-based class averages, dominated by the glycoprotein layer, generally yield ordered results. A small fraction of viruses (12%) were in virion-based class averages that had featureless glycoprotein layers, suggestive of irregular particles. A larger fraction of core-based class averages showed weak alignment of the glycoprotein layers. To the best of our knowledge, this is the first example where masked 2D class averages have been used to look for defective virus components. This result suggests that cores in particular, as well as some viruses, have a heterogeneity of organization inconsistent with icosahedral symmetry. It is not clear how symmetry correlates with infectivity. For RRV from BHK cells, the ratio of particles to plaque forming units is anywhere from 100 to 500. We cannot distinguish whether infectivity is attributable to a particular subset of asymmetric particles, the rare symmetrical particles, or particles that have some specific feature not readily obvious from this analysis.
Context for a high-error assembly model
In summary, we quantitatively described alphavirus CLP assembly and disassembly with a minimal system composed of RRV CP and a short DNA oligomer, where the DNA had sufficient charge to neutralize the RRV CP RNA-binding domain;51–53 in addition to nucleotide binding, RRV CLP assembly appears to include attractive electrostatic interactions (Table 1). We determined that RRV CP-DNA subunits had a pairwise association energy of approximately −6 kcal/mol per pairwise contact (Table 1), much stronger than the typical value of −3 to −4 kcal/mol observed for other viruses.5, 6, 46 We observed no evidence of hysteresis between assembly and disassembly (Fig 2B). While assembled CLPs had T=4 organization (Fig 3), we discovered many had defects or disordered regions. We found evidence that a fraction of RRV virions also have irregular cores, correlating our biophysical study with biology (Fig 4).
All of our results are consistent with a CLP that has flaws in its geometry. We suggest that these arise from subunits assembling incorrectly due to strong association energy between subunits. In simulations, when individual interactions between capsid subunits are weak, “incorrect” subunits can dissociate and re-associate correctly (thermodynamic editing); conversely, when association energy is strong, defects in the packing of subunits are anticipated because subunits become trapped and the growing complex cannot self-repair (Fig. 1C).6, 46, 54 Conceptually, this can create a mosaic capsid, a capsid with defects, regions of order and disorder, and even alternative lattices. We observe evidence that many RRV CLPs and cores do not form an ideal shell (Figs 3D, 4A, and C). The icosahedral averaging used to determine structures can disguise these geometric flaws, giving an impression of perfect symmetry.
An intact core is not actually necessary for the construction of an alphavirus-like particle. This was demonstrated with a Semliki Forest virus mutant that carried a capsid protein with defects in protein-protein interaction.55 The mutant virus formed an enveloped nucleoprotein complex in which the glycoprotein envelope was nearly identical to that of wild type virion and the core was largely disordered although the virus-like particles had the correct stoichiometry of components. Similar organized glycoprotein layers and disorganized cores are seen in flaviviruses (e.g. West Nile and Dengue).56, 57 High-resolution icosahedrally averaged structures are available for two alphaviruses, Chikungunya virus23 and Venezuelan Equine Encephalitis virus.19 As with lower resolution virion structures, intra-capsomer contacts (fivefold and quasi-sixfold complexes) were more apparent than inter-capsomer contacts. Well-defined interactions were also found between the core and the cytoplasmic domain of glycoprotein E2, suggesting an ordering influence.26, 32, 33, 40 In the Chikungunya virus structure the core density is weaker than that of the glycoprotein and the first ~115 residues are disordered. Indeed, they have been suggested to form a non-icosahedral internal scaffold.23
Closed, spherical capsids may be advantageous when the capsid is exposed to destabilizing environments, e.g. during transmission (Fig. 1A). We suggest that capsid integrity correlates with particle persistence. For viruses that rely on their envelope for stability and envelope proteins for the transmission, a different assembly mechanism (Fig. 1C) may be advantageous. We speculate that there is a biological advantage to RRV capsids with structural defects. In the virion, the nucleocapsid core appears to be a well-ordered, closed shell. Indeed, interaction with well-ordered glycoprotein may correct the misassembled capsids.55 However, once the nucleocapsid core is released into the cytoplasm a flawed capsid can readily disassemble. A virus would not survive if being “sloppy” did not provide an evolutionary advantage.
Assembly of virus capsids is not restricted to weak interactions. However, when viruses use strong interactions we find the irregularity predicted from models.3–6, 8, 46, 47, 58, 59 Irregularity is not necessarily a bad thing. Models based on cones, irregular structures, and spheres show that novel interactions will be accommodated.8, 9, 58 Biology clearly accommodates defects.10–13 Advantages of a mosaic particle include responsiveness to a destabilizing environment and also provide a basis for asymmetry from a symmetric lattice.
METHODS
Expression and purification of Ross River capsid protein (RRV CP)
Ross River virus capsid protein (RRV CP) was cloned into a pET29b, expressed in Rosetta 2 cells (Novagen, EMD Chemical Inc. Gobbstown, NJ) and purified as described previously (Fig. S8).35, 42 For the protein samples used in the dissociation experiments the lysis, loading, and elution buffer at pH 6.8 rather than pH 7.5 and subsequently concentrated and exchanged into 20 mM HEPES, pH=7.4, 150 mM NaCl, and 0.1 mM EDTA using Vivaspin 2 filters with a 10kDa cut of filter (Vivaproducts, Inc., Littleton, MA). The protein concentration was determined by absorbance using an extinction coefficient of 39,670 M-1cm-1 at 280 nm. Concentrated protein was aliquotted and stored at 4°C or −20°C; frozen aliquots were not thawed more than twice.
Core-like particle (CLP) formation
For assembly titrations, CP at a final concentration of 1.5μM was mixed with 27mer single-stranded DNA oligo (5′-TAC CCA CGC TCT CGC AGT CAT AAT TCG). For these reactions, the NaCl concentration was varied as described in text. For comparisons of assembly and disassembly, CLPs were assembled by adding 1–3 μM of RRV CP to DNA oligo in the appropriate ionic strength. In disassembly reactions, CLPs were assembled at 0 mM NaCl and aliquots diluted into buffer containing NaCl. The extent of assembly was determined by static light scattering, dynamic light scattering, and transmission electron microscopy.
Light scattering and dynamic light scattering (DLS)
Light scattering and DLS experiments were performed with Malvern Zetasizer Nano ZS ZEN3600 at 22 ˚C using the manufacturer’s quartz cuvette, ZEN2112. Light scattering is reported as “raw” counts from this instrument. Particle size information was determined using the manufacturer’s software. Each measurement consisted of at least 10 measurements and triplicate samples. Each experiment was reproduced using at least two different protein preparations.
Virus purification
RRV was generated by electroporating BHK cells with T48 strain of Ross River virus as described previously.60 Twenty-four hours post electroporation the media was collected and cell debris was removed by centrifugation. The media was spun at 5000 × g overnight at 4°C. The media was removed and the pellet was resuspended in 20 mM HEPES, pH 7.4, 150 mM NaCl, and 0,1 mM EDTA. When necessary, virions were concentrated using Vivaspin 6 100kDa filters at 5000 × g at 4°C.
Transmission electron microscopy (TEM)
For negative stained EM, sample was applied to a hydrophilic glow-discharged carbon-coated 300-mesh copper grid and stained with 2% uranyl acetate. Images were acquired on an 80-kV JEOL-1010 transmission electron microscopy equipped with a Gatan 4k × 4 k CCD camera. For cryo-EM, sample was applied to glow-discharged 300-mesh copper grids and plunge-frozen using an FEI Vitrobot. Cryo-images were collected using a 300-kV JEOL-3200FS electron microscope with an in-column energy filter at a nominal magnification of 60,000× (equal to 1.84 Å per pixel at the specimen space). Exposure was less than 25 e−/Å2 to avoid beam-induced damage. Images with minimal specimen drift and astigmatism were selected for further analysis. Reconstructions were calculated using EMAN2 and AUTO3DEM software as previously described.61–63 For RRV virion, 6829 particles were initially selected and the initial model was built de novo. The final 3D reconstruction, computed from 6146 particles, was estimated to 15 Å in resolution using a Fourier Shell Correlation (FSC) of 0.5. For the RRV CLP (Fig 3B, C), the initial model was based on the nucleocapsid layer (radii of 12 – 22 nm) of the cryo-EM density map of the RRV virus low pass filtered to 40Å. In total, 4124 particles were selected and 3713 particles were used in the final 3D reconstruction. The resolution was estimated to be 17.9 Å based on a FSC of 0.5. The 3D reconstructions were rendered and visualized using RobEM64 and UCSF Chimera.65 The 2D reference-free classification was computed using XMIPP.66 The glycoprotein layer of the 3D model of RRV virus was masked by using PCUT program to remove the density beyond radius of 22 nm. To mask the glycoprotein layer of the 2D images of RRV virus, the particle images were first centered using Cenalignint67 and then masked by xmipp_transform_mask to remove the density beyond 22 nm in radius. The correlation coefficient between model projections and class averages were calculated using Parallel Polar Fourier Transform (PPFT) program, part of the AUTO3DEM software.
The masked particles allowed us to make predictions for different scenarios. If the particles were well-ordered, we would see well-defined arrangement of density. If the particles had defects with a specific geometric relationship to the underlying icosahedral symmetry, we would expect to see partially organized and partially smeared density. If the particles had random defects we would expect classification to fail completely resulting in a smeared ring. We have used this method to identify incomplete particles in kinetically trapped HBV capsid assembly reactions. For ordered virions consistent with symmetrized reconstructions, we expected both glycan and core layers to show order in common orientations. Whereas cores with defects would have a tendency to allow alternative orientations for the glycan layer, resulting in disordered core or glycan layers.
The cryo-EM density maps reported in this paper have been deposited in the EMDataBank database. The accession number for RRV CLP is EMD-2964 and for RRV virion is EMD-2965.
Acknowledgments
This research was supported by the NSF through Award MCB 1157716 to SM and the NIH through R01-AI077688 to AZ. We thank the Electron Microscopy Center at Indiana University. We also recognize the Indiana University cyberinfrastructure, which was supported in part by Lilly Endowment, Inc., through its support for the Indiana University Pervasive Technology Institute, and in part by the Indiana METACyt Initiative. The Indiana METACyt Initiative at IU is also supported in part by Lilly Endowment, Inc.
Footnotes
Supporting information
DLS and EM data for a typical sample of CLPs (S1). DLS size data for titrations at various NaCl concentrations (S2–S5). A reconstruction of RRV showing a surface shaded view, a central section, and resolution data (S6). A full set of class averages (some of which are displayed in figure 4) for CLPs and virions (S7). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Caspar DL, Klug A. Physical Principles in the Construction of Regular Viruses. Cold Spring Harbor Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 2.Katen SP, Chirapu SR, Finn MG, Zlotnick A. Trapping of Hepatitis B Virus Capsid Assembly Intermediates by Phenylpropenamide Assembly Accelerators. ACS Chem Biol. 2010;5:1125–1136. doi: 10.1021/cb100275b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perlmutter JD, Hagan MF. Mechanisms of Virus Assembly. 2014:1407. doi: 10.1146/annurev-physchem-040214-121637. arXiv. preprint arXiv:1407.3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitesides GM, Boncheva M. Beyond Molecules: Self-Assembly of Mesoscopic and Macroscopic Components. Proc Natl Acad Sci U S A. 2002;99:4769–4774. doi: 10.1073/pnas.082065899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zlotnick A, Mukhopadhyay S. Virus Assembly, Allostery and Antivirals. Trends Microbiol. 2011;19:14–23. doi: 10.1016/j.tim.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zlotnick A. Are Weak Protein-Protein Interactions the General Rule in Capsid Assembly? Virology. 2003;315:269–274. doi: 10.1016/s0042-6822(03)00586-5. [DOI] [PubMed] [Google Scholar]
- 7.Pierson EE, Keifer DZ, Selzer L, Lee LS, Contino NC, Wang JC, Zlotnick A, Jarrold MF. Detection of Late Intermediates in Virus Capsid Assembly by Charge Detection Mass Spectrometry. J Am Chem Soc. 2014;136:3536–3541. doi: 10.1021/ja411460w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zandi R, Reguera D, Bruinsma RF, Gelbart WM, Rudnick J. Origin of Icosahedral Symmetry in Viruses. Proc Natl Acad Sci U S A. 2004;101:15556–15560. doi: 10.1073/pnas.0405844101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen T, Zhang Z, Glotzer SC. A Precise Packing Sequence for Self-Assembled Convex Structures. Proc Natl Acad Sci U S A. 2007;104:717–722. doi: 10.1073/pnas.0604239104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krol MA, Olson NH, Tate J, Johnson JE, Baker TS, Ahlquist P. Rna-Controlled Polymorphism in the in Vivo Assembly of 180-Subunit and 120-Subunit Virions from a Single Capsid Protein. Proc Natl Acad Sci U S A. 1999;96:13650–13655. doi: 10.1073/pnas.96.24.13650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson JM, Tang J, Nyame Y, Willits D, Young MJ, Zlotnick A. Regulating Self-Assembly of Spherical Oligomers. Nano Lett. 2005;5:765–770. doi: 10.1021/nl050274q. [DOI] [PubMed] [Google Scholar]
- 12.Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Krausslich HG. Structure and Assembly of Immature Hiv. Proc Natl Acad Sci U S A. 2009;106:11090–11095. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu Z, Dobro MJ, Woodward CL, Levandovsky A, Danielson CM, Sandrin V, Shi J, Aiken C, Zandi R, Hope TJ, et al. Unclosed Hiv-1 Capsids Suggest a Curled Sheet Model of Assembly. J Mol Biol. 2013;425:112–123. doi: 10.1016/j.jmb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukhopadhyay S, Chipman PR, Hong EM, Kuhn RJ, Rossmann MG. In Vitro-Assembled Alphavirus Core-Like Particles Maintain a Structure Similar to That of Nucleocapsid Cores in Mature Virus. J Virol. 2002;76:11128–11132. doi: 10.1128/JVI.76.21.11128-11132.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perera R, Owen KE, Tellinghuisen TL, Gorbalenya AE, Kuhn RJ. Alphavirus Nucleocapsid Protein Contains a Putative Coiled Coil Alpha-Helix Important for Core Assembly. J Virol. 2001;75:1–10. doi: 10.1128/JVI.75.1.1-10.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tellinghuisen TL, Hamburger AE, Fisher BR, Ostendorp R, Kuhn RJ. In Vitro Assembly of Alphavirus Cores by Using Nucleocapsid Protein Expressed in Escherichia Coli. J Virol. 1999;73:5309–5319. doi: 10.1128/jvi.73.7.5309-5319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strauss JH, Strauss EG. The Alphaviruses: Gene Expression, Replication, and Evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhn RJ. Togaviradae: The Viruses and Their Replication. In: Knipe DM, Howley PM, editors. Fields’ Virology. Lippincott Williams; Philadelphia: 2007. pp. 1001–1022. [Google Scholar]
- 19.Zhang R, Hryc CF, Cong Y, Liu X, Jakana J, Gorchakov R, Baker ML, Weaver SC, Chiu W. 4.4 a Cryo-Em Structure of an Enveloped Alphavirus Venezuelan Equine Encephalitis Virus. Embo J. 2011 doi: 10.1038/emboj.2011.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang J, Jose J, Chipman P, Zhang W, Kuhn RJ, Baker TS. Molecular Links between the E2 Envelope Glycoprotein and Nucleocapsid Core in Sindbis Virus. J Mol Biol. 2011;414:442–459. doi: 10.1016/j.jmb.2011.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kostyuchenko VA, Jakana J, Liu X, Haddow AD, Aung M, Weaver SC, Chiu W, Lok SM. The Structure of Barmah Forest Virus as Revealed by Cryo-Electron Microscopy at a 6-Angstrom Resolution Has Detailed Transmembrane Protein Architecture and Interactions. J Virol. 2011;85:9327–9333. doi: 10.1128/JVI.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng RH, Kuhn RJ, Olson NH, Rossmann MG, Choi HK, Smith TJ, Baker TS. Nucleocapsid and Glycoprotein Organization in an Enveloped Virus. Cell. 1995;80:621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun S, Xiang Y, Akahata W, Holdaway H, Pal P, Zhang X, Diamond MS, Nabel GJ, Rossmann MG. Structural Analyses at Pseudo Atomic Resolution of Chikungunya Virus and Antibodies Show Mechanisms of Neutralization. Elife. 2013;2:e00435. doi: 10.7554/eLife.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suomalainen M, Garoff H. Alphavirus Spike-Nucleocapsid Interaction and Network Antibodies. J Virol. 1992;66:5106–5109. doi: 10.1128/jvi.66.8.5106-5109.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang J, Jose J, Chipman P, Zhang W, Kuhn RJ, Baker TS. Molecular Links between the E2 Envelope Glycoprotein and Nucleocapsid Core in Sindbis Virus. J Mol Biol. 2011;414:442–459. doi: 10.1016/j.jmb.2011.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jose J, Przybyla L, Edwards TJ, Perera R, Burgner JW, 2nd, Kuhn RJ. Interactions of the Cytoplasmic Domain of Sindbis Virus E2 with Nucleocapsid Cores Promote Alphavirus Budding. J Virol. 2012;86:2585–2599. doi: 10.1128/JVI.05860-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X Version 2. 0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 28.Garoff H, Frischauf AM, Simons K, Lehrach H, Delius H. The Capsid Protein of Semliki Forest Virus Has Clusters of Basic Amino Acids and Prolines in Its Amino-Terminal Region. Proc Natl Acad Sci U S A. 1980;77:6376–6380. doi: 10.1073/pnas.77.11.6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bell JR, Strauss EG, Strauss JH. Purification and Amino Acid Compositions of the Structural Proteins of Sindbis Virus. Virology. 1979;97:287–294. doi: 10.1016/0042-6822(79)90340-4. [DOI] [PubMed] [Google Scholar]
- 30.Bell JR, Bond MW, Hunkapiller MW, Strauss EG, Strauss JH, Yamamoto K, Simizu B. Structural Proteins of Western Equine Encephalitis Virus: Amino Acid Compositions and N-Terminal Sequences. J Virol. 1983;45:708–714. doi: 10.1128/jvi.45.2.708-714.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi HK, Tong L, Minor W, Dumas P, Boege U, Rossmann MG, Wengler G. Structure of Sindbis Virus Core Protein Reveals a Chymotrypsin-Like Serine Proteinase and the Organization of the Virion. Nature. 1991;354:37–43. doi: 10.1038/354037a0. [DOI] [PubMed] [Google Scholar]
- 32.Lee S, Owen KE, Choi HK, Lee H, Lu G, Wengler G, Brown DT, Rossmann MG, Kuhn RJ. Identification of a Protein Binding Site on the Surface of the Alphavirus Nucleocapsid and Its Implication in Virus Assembly. Structure. 1996;4:531–541. doi: 10.1016/s0969-2126(96)00059-7. [DOI] [PubMed] [Google Scholar]
- 33.Skoging U, Vihinen M, Nilsson L, Liljestrom P. Aromatic Interactions Define the Binding of the Alphavirus Spike to Its Nucleocapsid. Structure. 1996;4:519–529. doi: 10.1016/s0969-2126(96)00058-5. [DOI] [PubMed] [Google Scholar]
- 34.Mukhopadhyay S, Zhang W, Gabler S, Chipman PR, Strauss EG, Strauss JH, Baker TS, Kuhn RJ, Rossmann MG. Mapping the Structure and Function of the E1 and E2 Glycoproteins in Alphaviruses. Structure. 2006;14:63–73. doi: 10.1016/j.str.2005.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng F, Tsvetkova IB, Khuong YL, Moore AW, Arnold RJ, Goicochea NL, Dragnea B, Mukhopadhyay S. The Packaging of Different Cargo into Enveloped Viral Nanoparticles. Mol Pharmaceutics. 2013;10:51–58. doi: 10.1021/mp3002667. [DOI] [PubMed] [Google Scholar]
- 36.Goicochea NL, De M, Rotello VM, Mukhopadhyay S, Dragnea B. Core-Like Particles of an Enveloped Animal Virus Can Self-Assemble Efficiently on Artificial Templates. Nano Lett. 2007;7:2281–2290. doi: 10.1021/nl070860e. [DOI] [PubMed] [Google Scholar]
- 37.Wengler G, Boege U, Wahn K. Establishment and Analysis of a System Which Allows Assembly and Disassembly of Alphavirus Core-Like Particles under Physiological Conditions in Vitro. Virology. 1984;132:401–412. doi: 10.1016/0042-6822(84)90045-x. [DOI] [PubMed] [Google Scholar]
- 38.Wengler G, Wengler G. In Vitro Analysis of Factors Involved in the Disassembly of Sindbis Virus Cores by 60s Ribosomal Subunits Identifies a Possible Role of Low Ph. J Gen Virol. 2002;83:2417–2426. doi: 10.1099/0022-1317-83-10-2417. [DOI] [PubMed] [Google Scholar]
- 39.Wengler G. The Regulation of Disassembly of Alphavirus Cores. Arch Virol. 2009;154:381–390. doi: 10.1007/s00705-009-0333-9. [DOI] [PubMed] [Google Scholar]
- 40.Wilkinson TA, Tellinghuisen TL, Kuhn RJ, Post CB. Association of Sindbis Virus Capsid Protein with Phospholipid Membranes and the E2 Glycoprotein: Implications for Alphavirus Assembly. Biochemistry. 2005;44:2800–2810. doi: 10.1021/bi0479961. [DOI] [PubMed] [Google Scholar]
- 41.Snyder JE, Azizgolshani O, Wu B, He Y, Lee AC, Jose J, Suter DM, Knobler CM, Gelbart WM, Kuhn RJ. Rescue of Infectious Particles from Preassembled Alphavirus Nucleocapsid Cores. J Virol. 2011;85:5773–5781. doi: 10.1128/JVI.00039-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng F, Mukhopadhyay S. Generating Enveloped Virus-Like Particles with in Vitro Assembled Cores. Virology. 2011;413:153–160. doi: 10.1016/j.virol.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 43.Katen SP, Zlotnick A. Thermodynamics of Virus Capsid Assembly. Methods Enzymol. 2009;455:395–417. doi: 10.1016/S0076-6879(08)04214-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paredes A, Alwell-Warda K, Weaver SC, Chiu W, Watowich SJ. Structure of Isolated Nucleocapsids from Venezuelan Equine Encephalitis Virus and Implications for Assembly and Disassembly of Enveloped Virus. J Virol. 2003;77:659–664. doi: 10.1128/JVI.77.1.659-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh S, Zlotnick A. Observed Hysteresis of Virus Capsid Disassembly Is Implicit in Kinetic Models of Assembly. J Biol Chem. 2003;278:18249–18255. doi: 10.1074/jbc.M211408200. [DOI] [PubMed] [Google Scholar]
- 46.Hagan MF, Chandler D. Dynamic Pathways for Viral Capsid Assembly. Biophys J. 2006;91:42–54. doi: 10.1529/biophysj.105.076851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Schoot P, Zandi R. Kinetic Theory of Virus Capsid Assembly. Phys Biol. 2007;4:296–304. doi: 10.1088/1478-3975/4/4/006. [DOI] [PubMed] [Google Scholar]
- 48.Uetrecht C, Watts NR, Stahl SJ, Wingfield PT, Steven AC, Heck AJ. Subunit Exchange Rates in Hepatitis B Virus Capsids Are Geometry- and Temperature-Dependent. Phys Chem Chem Phys. 2010;12:13368–13371. doi: 10.1039/c0cp00692k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang W, Mukhopadhyay S, Pletnev SV, Baker TS, Kuhn RJ, Rossmann MG. Placement of the Structural Proteins in Sindbis Virus. J Virol. 2002;76:11645–11658. doi: 10.1128/JVI.76.22.11645-11658.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coombs K, Brown B, Brown DT. Evidence for a Change in Capsid Morphology During Sindbis Virus Envelopment. Virus Res. 1984;1:297–302. doi: 10.1016/0168-1702(84)90018-2. [DOI] [PubMed] [Google Scholar]
- 51.Geigenmuller-Gnirke U, Nitschko H, Schlesinger S. Deletion Analysis of the Capsid Protein of Sindbis Virus: Identification of the Rna Binding Region. J Virol. 1993;67:1620–1626. doi: 10.1128/jvi.67.3.1620-1626.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Owen KE, Kuhn RJ. Identification of a Region in the Sindbis Virus Nucleocapsid Protein That Is Involved in Specificity of Rna Encapsidation. J Virol. 1996;70:2757–2763. doi: 10.1128/jvi.70.5.2757-2763.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Warrier R, Linger BR, Golden BL, Kuhn RJ. Role of Sindbis Virus Capsid Protein Region Ii in Nucleocapsid Core Assembly and Encapsidation of Genomic Rna. J Virol. 2008;82:4461–4470. doi: 10.1128/JVI.01936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zandi R, Reguera D. Mechanical Properties of Viral Capsids. Phys Rev E: Stat Nonlinear, Soft Matter Phys. 2005;72:021917. doi: 10.1103/PhysRevE.72.021917. [DOI] [PubMed] [Google Scholar]
- 55.Forsell K, Xing L, Kozlovska T, Cheng RH, Garoff H. Membrane Proteins Organize a Symmetrical Virus. EMBO J. 2000;19:5081–5091. doi: 10.1093/emboj/19.19.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mukhopadhyay S, Kim BS, Chipman PR, Rossmann MG, Kuhn RJ. Structure of West Nile Virus. Science. 2003;302:248. doi: 10.1126/science.1089316. [DOI] [PubMed] [Google Scholar]
- 57.Pokidysheva E, Zhang Y, Battisti AJ, Bator-Kelly CM, Chipman PR, Xiao C, Gregorio GG, Hendrickson WA, Kuhn RJ, Rossmann MG. Cryo-Em Reconstruction of Dengue Virus in Complex with the Carbohydrate Recognition Domain of Dc-Sign. Cell. 2006;124:485–493. doi: 10.1016/j.cell.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 58.Glotzer SC, Solomon MJ. Anisotropy of Building Blocks and Their Assembly into Complex Structures. Nat Mater. 2007;6:557–562. doi: 10.1038/nmat1949. [DOI] [PubMed] [Google Scholar]
- 59.Luque A, Reguera D, Morozov A, Rudnick J, Bruinsma R. Physics of Shell Assembly: Line Tension, Hole Implosion, and Closure Catastrophe. J Chem Phys. 2012;136:184507. doi: 10.1063/1.4712304. [DOI] [PubMed] [Google Scholar]
- 60.Snyder AJ, Mukhopadhyay S. The Alphavirus E3 Glycoprotein Functions in a Clade-Specific Manner. J Virol. 2012;86:13609–13620. doi: 10.1128/JVI.01805-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang JC, Dhason MS, Zlotnick A. Structural Organization of Pregenomic Rna and the Carboxy-Terminal Domain of the Capsid Protein of Hepatitis B Virus. PLoS Pathog. 2012;8:e1002919. doi: 10.1371/journal.ppat.1002919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yan X, Sinkovits RS, Baker TS. Auto3dem - an Automated and High Throughput Program for Image Reconstruction of Icosahedral Particles. J Struct Biol. 2007;157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. Eman2: An Extensible Image Processing Suite for Electron Microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 64.Yan X, Sinkovits RS, Baker TS. Auto3dem--an Automated and High Throughput Program for Image Reconstruction of Icosahedral Particles. J Struct Biol. 2007;157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. Ucsf Chimera--a Visualization System for Exploratory Research and Analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 66.Sorzano CO, Marabini R, Velazquez-Muriel J, Bilbao-Castro JR, Scheres SH, Carazo JM, Pascual-Montano A. Xmipp: A New Generation of an Open-Source Image Processing Package for Electron Microscopy. J Struct Biol. 2004;148:194–204. doi: 10.1016/j.jsb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 67.Ludtke SJ, Baldwin PR, Chiu W. Eman: Semiautomated Software for High-Resolution Single-Particle Reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]