Abstract
The association between exposure to smokeless tobacco products (STP) and oral diseases is partially due to the physiological and pathological changes in the composition of the oral microbiome and its metabolic profile. However, it is not clear how STPs affect the physiology and ecology of oral microbiota. A UPLC/QTof-MS-based metabolomics study was employed to analyze metabolic alterations in oral bacterium, Capnocytophaga sputigena as a result of smokeless tobacco exposure and to assess the capability of the bacterium to metabolize nicotine. Pathway analysis of the metabolome profiles indicated that smokeless tobacco extracts caused oxidative stress in the bacterium. The metabolomics data also showed that the argininenitric oxide pathway was perturbed by the smokeless tobacco treatment. Results also showed that LC/MS was useful in identifying STP constituents and additives, including caffeine and many flavoring compounds. No significant changes in levels of nicotine and its major metabolites were found when C. sputigena was cultured in a nutrient rich medium, although hydroxylnicotine and cotinine N-oxide were detected in the bacterial metabolites suggesting that nicotine metabolism might be present as a minor degradation pathway in the bacterium. Study results provide new insights regarding the physiological and toxicological effects of smokeless tobacco on oral bacterium C. sputigena and associated oral health as well as measuring the ability of the oral bacterium to metabolize nicotine.
Keywords: Smokeless tobacco, Toxicology, Oral bacteria, Metabolomics
1. Introduction
Since the 1940s (Pindborg, 1947), it has been recognized that smoking and smokeless tobacco are associated with oral diseases (e.g., periodontal disease) in addition to their well-known impact on other diseases such as cancer and cardiovascular diseases (Wald and Hackshaw, 1996; Siddiqi et al., 2015; Piano et al., 2010). Furthermore, it has been reported that smokeless tobacco users have a high prevalence of gingival recession (25–30% of users) and mucosal lesions (50–60% of users), both of which are localized to the tobacco placement site (Robertson et al., 1990; Greer and Poulson, 1983). The effects of smokeless tobacco products on oral diseases are partly due to the disruption of normal oral microbiota community (Winn, 2001; Meurman and Bascones-Martinez, 2011). Physiological changes in bacteria induced by various stresses can cause loss of cell function and viability of the organisms and result in unbalance of the microbiota and changed in metabolites levels. (Manas and Mackey, 2004; Kapil et al., 2013). For example, extensive studies (Celermajer et al., 1992; Widlansky et al., 2003; Zeiher et al., 1995) in human have pointed that vascular dysfunction induced by smoking is initiated by reduced NO production in oral neutrophils and/or oral bacteria. However, more detailed physiological and toxicological effect of smokeless tobacco on oral microbiota metabolism is still unclear.
Hundreds of trillions of microbes inhabit the human body. On the one hand, these microbes developed a symbiotic relationship with their host that plays an important role in the host’s physiology and pathology (Thompson-Chagoyan et al., 2007; Sokol et al., 2006). These microbial communities can be highly influenced by alterations in the host diet (Turnbaugh et al., 2006; Turnbaugh et al., 2008), antibiotic use, (Swann et al., 2011; Yap et al., 2008; Sun et al., 2013) and other lifestyle factors including travel and tobacco or alcohol use (David et al., 2014). While some researchers have reported no significant microbial species differences between smokers and non-smokers, conflicting results have also been observed (van Winkelhoff et al., 2001; Kamma et al., 1999; Brandsch, 2006). Van Winkelhoff et al. (van Winkelhoff et al., 2001) analyzed subgingival microbial flora profiles and reported that smokers without periodontitis have a higher prevalence of Prevotella intermedia/nigrescens compared with non-smokers with periodontitis; following periodontitis treatment, smokers have a higher prevalence of Bacteroides forsythus, Peptostreptococcus micros, and Campylobacter rectus compared with non-smokers. A separate research group (Kamma et al., 1999) also reported that smokers have different oral microbial profiles and a greater quantity of bacteria compared to non-smokers. While several bacterial species residing in plants and soils can degrade nicotine, the main alkaloid contained in tobacco (Brandsch, 2006), it is not clear whether oral microbiota can metabolize nicotine and its derivatives. For the present study, a liquid chromatography/mass spectrometry (LC/MS)-based metabolomics approach was employed to evaluate the toxicological and physiological effects of smokeless tobacco on one species of oral bacteria metabolism and function as well as to evaluate nicotine metabolism by oral microbiota.
Metabolomic profiling is an emerging powerful technology to measure the metabolic response of living systems to pathophysiological stimuli and genetic modification (Nicholson et al., 1999). Recently (Sun et al., 2013), both LC/MS- and nuclear magnetic resonance (NMR)-based metabolomics were employed to understand host-microbial interactions through evaluating the effects of penicillin on the gut microbiota and the host at metabolite levels. Results indicated that gut microbiota play important roles in the regulation of host metabolism and xenobiotic detoxification mechanism. In a separate study (Wikoff et al., 2009), metabolomics analysis showed that gut microbiota-related metabolites (produced by or derived from the gut microbiota) were observed only in conventional mice but were not present in germ-free mice, suggesting a significant interaction between bacteria and host metabolism. A few NMR-based metabolomics studies (Swann et al., 2011; Yap et al., 2008; Martin et al., 2007) found strong interaction between the gut microbiota and host metabolism. However the interactions at metabolic levels between exposure to STPs and the oral microbiota and its potential impact on host oral health have rarely been assessed.
Capnocytophaga sputigena is an opportunistic pathogen responsible for periodontal infections. It is usually isolated from periodontal pockets, apical and periodontal abscesses where other periodontal bacterial species are found (Murad et al., 2014). In our previous work, we studied the effects of STPs on 38 human oral bacteria in terms of cell growth and viability. Results showed that C. sputigena was one of several oral bacterial species whose growth rates were not significantly affected by STPs (unpublished data). The aim of the present study was to examine the metabolic response of a member of the oral microbiota to smokeless tobacco extract, and to examine the alterations in nicotine and nicotine metabolism by this oral bacterium. The data and method developed from the study could be used as a tool to compare the toxicity between different smokeless tobacco products (i.e., by comparing the difference in metabolome profiles of oral bacteria after being treated with different brands of smokeless tobacco products). An LC/MS-based metabolomics approach was employed to analyze metabolic alterations in cells of C. sputigena and the bacterial culture medium.
2. Materials and methods
2.1. Chemicals
Optima LC/MS grade acetonitrile and water were purchased from Thermo Fisher Scientific (Pittsburgh, PA). Formic acid, leucine-enkephalin, imidazole, pentadecafluorooctanoic acid, L-tryptophan, and all MS standards were obtained from Sigma-Aldrich Corporation (St. Louis, MO). Ten tins of a popular brand of moisture snuff (wintergreen) smokeless tobacco were purchased from local store in Little Rock, Arkansas. This brand of smokeless tobacco is one of the top 10 smokeless tobacco brands (http://www.thetoptens.com/smokeless-tobacco-brands/) and has moderate nicotine content. The samples of the smokeless tobacco were stored at −20 °C. The study was conducted between April and December 2014.
2.2. Bacterial culture and treatment
C. sputigena ATCC 33612 was purchased from the American Type Culture Collection. One single colony of C. sputigena was inoculated into 10 mL brain-heart infusion (BHI) broth (autoclaved for 15 min at 121 °C and 15 psi) and grown at 37 °C overnight in an anaerobic chamber and used as a seed culture. The seed culture was inoculated (10% v/v) into BHI broth that contained 50 mg/mL smokeless tobacco aqueous extracts (STAE, T2) or ddH2O control vehicle (C) and then grown at 37 °C for 48 h in an anaerobic chamber. The number of bacteria in the culture media was counted by flow cytometry. Bacterial culture grown without STAE for 48 h and then treated with STAE for 2 min was used as the T1 sample. Bacterial cells were centrifuged down at 16,060 ×g for 10 min.
2.3. Sample preparation
Cell pellets (~1010 cells) were collected after centrifugation and suspended in 200 μL ice-cold water. A 120 μL aliquot of the cell suspension was transferred into a tube containing 450 μL methanol, vortexed, and then kept at 4 °C for 15 min. The bacterial cells were lysed with 0.1 mm silica spheres by PRECELLYS® 24 homogenizer (Bertin Co., Rockville, MD) at 6800 rpm for 30 s (2×). After centrifugation, the supernatant was then transferred into a clean tube and evaporated to dryness using a SpeedVac concentrator (Thermo Scientific, Waltham, MA). The samples were reconstituted in 200 μL 95:5 water/acetonitrile, vortexed for 2 min, and kept at 4 °C for 20 min. The resulting solution was then centrifuged at 16,060 ×g for 12 min at 4 °C. The supernatant was transferred to autosampler vials for LC/MS analysis.
Media (100 μL aliquot) was mixed with 300 μL methanol, incubated at −20 °C for 20 min, and then centrifuged at 16,060 ×g for 12 min at 4 ° C to precipitate proteins. The supernatant (300 μL) was then transferred to clean tubes and evaporated to dryness using the SpeedVac concentrator. The samples were reconstituted in 200 μL 95:5 water/acetonitrile, vortexed for 2 min, and kept at 4 °C for 20 min. The resulting solution was then centrifuged at 16,060 ×g for 12 min at 4 °C. The supernatant was transferred to autosampler vials for LC/MS analysis.
2.4. Open metabolic profiling
A 3 μL aliquot of cell or media supernatant after methanol precipitation was introduced into a Waters Acquity ultra performance liquid chromatography (UPLC)/QTof system (Waters, Milford, MA) equipped with a Waters bridged ethyl hybrid (BEH) C8 column with a dimension of 2.1 mm × 10 cm and 1.7 μm particle size. The column was held at 40 °C. The UPLC mobile phase consisted of 0.1% formic acid in water (solution A) and 0.1% formic acid in acetonitrile (solution B). While maintaining a constant flow rate of 0.4 mL/min, the metabolites were eluted using gradients of 2–80% solution B from 0 to 15 min, and 80–98% solution B from 15 to 17 min. The final gradient composition was held constant for 2 min, followed by a return to 2% solution B at 19.1 min.
Mass spectrometric data were collected with a Waters QTof Premier mass spectrometer (Waters, Milford, MA) operated in positive and negative ionization electrospray modes (Sun et al., 2010; Sun et al., 2009). Briefly, MSE analysis was performed on a QTof mass spectrometer set up with 5 eV for low collision energy and 20–30 eV for ramp collision energy. Full scan mode from m/z 100 to 900 and from 0 to 22 min was used for data acquisition.
A quality control (QC) sample composed of 40 common chemicals for LC/MS open profiling, was evaluated after every 10 sample runs. UPLC/QTof-MS spectra of pooled bacterial cells or pooled media were acquired from every 10 sample runs for pooled bacterial cells or pooled media to monitor analytical equipment variability.
Raw UPLC/MS data were analyzed using Micromass MarkerLynx XS Application Version 4.1 (Waters, Milford, MA) with extended statistical tools (Sun et al., 2010; Sun et al., 2009). The aligned data from MarkerLynx analysis for QTof-MS data were filtered using the pooled QC samples based on the following criteria: i) ions with a relative standard deviation (RSD) <30% in the pooled QC samples were included; and ii) ions present in ≥70% of QC samples were included. The resulting data set was further normalized based on the cell counts; the normalized data were then analyzed by unsupervised Principal Component Analysis (PCA) and supervised partial least squares discriminant analysis (PLS-DA). The identity of compounds was based on the combined information of accurate mass measurement, fragmentation mass spectra, and compared data from a free online database (www.hmdb.ca). Some compounds were confirmed by authentic standards.
2.5. Statistics
In the metabolome analyses, the values in the treated groups and the respective control group were compared. The data were analyzed by Student’s t-test (MS EXCEL). A value of p < 0.05 was considered statistically significant.
3. Results
The growth of C. sputigena in BHI was not significantly changed in the presence of 50 mg/mL STAE (~2 × 108 cells/mL). Bacterial cultures with no STAE (C) were used as controls, while bacterial cultures mixed with STAE for 2 min followed by immediate cell harvest (T1) were used to account for rapid physical absorption of STAE onto the cell walls or membrane as well as to evaluate the cell response to sudden STAE exposure. T2 are bacterial cultures containing 50 mg/mL smokeless tobacco aqueous extracts grown at 37 °C for 48 h in an anaerobic chamber. No significant changes in cell counts were observed after 48 h incubation with STAE (T2) compared with the corresponding controls. By comparing T1 and T2 samples, the concentration changes in nicotine and its metabolites would be only due to metabolism by oral bacteria.
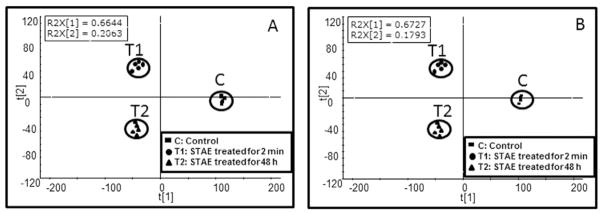
LC/QTof-MS-based metabolomics was employed to analyze the endogenous metabolome in cells and exogenous compounds from STAE treatment in total (i.e., both cells and media), whereas the samples from cells and media were analyzed independently. Fig. 1 shows the scores plot of the PLS-DA model generated from the UPLC/QTof-MS analysis of cell extract in positive (Fig. 1A) and negative (Fig. 1B) ionization modes. For both scores plots, the two tobacco treated groups (T1 and T2) were well separated from the control group (C) along t (Pindborg, 1947), while the T2 group was clearly separated from the T1 group along t (Wald and Hackshaw, 1996). The measured intensity, experimental and theoretical m/z, and formulas of metabolites, which were responsible for group separations, are summarized in Tables 1 & 2. To evaluate bacterial cell function, we compared normalized metabolite levels between STAE treatment and controls in the cells. The detected mass accuracy of all metabolites (Tables 1 & 2) was <6 ppm except for 4-hydroxy-4-(3-pyridyl)-butanoic acid, which was >10 ppm. Metabolites were identified based on mass accuracy and fragment mass spectrum. The identities of some metabolites were further validated with chemical standards. For endogenous metabolites in cells, almost all of the detected metabolites were significantly changed in the T1 (except for adenine, sebacic acid, and pantothenic acid) and T2 groups (except for phenylalanine, tryptophan, tyrosine and leucine) compared with the C group. These findings indicate that the metabolome of the bacteria was changed even for the short (2 min) incubation period with STAE.
Fig. 1.
The scores plot from PLS-DA analysis of LC/MS data in positive (A) and negative (B) ionization modes from cell extracts after treatment with water (control) and 50 mg/mL STAE, incubated for 2 min or 48 h. Four and five replicates were included in controls and two STAE treated groups, respectively. Notes: C: control; T1: STAE treated for 2 min; T2: STAE treated for 48 h.
Table 1.
Total intensity (Mean ± STD) and percentage of nicotine and its eight metabolites as well as minor tobacco alkaloids present in cells and media.
| Mean ± STD | Percentage (%) | ||||
|---|---|---|---|---|---|
| Nicotine metabolites | C | T1 | T2 | T1 | T2 |
| 2′-Hydroxynicotine | 0.00 ± 0.00 | 33.01 ± 23.19 | 63.77 ± 7.08a | 0.05 | 0.10 |
| Cotinine | 6.14 + 5.01 | 2660.30 + 195.70 | 2511.26 ± 395.94 | 4.34 | 3.85 |
| Nicotine | 106.31 + 68.59 | 43,490.38 + 2060.07 | 45,962.58 + 3319.22 | 70.97 | 70.45 |
| Nicotine-1′-N-oxide | 5.88 + 4.21 | 3653.93 + 794.39 | 4030.06 ± 1058.47 | 5.96 | 6.18 |
| Nornicotine | 70.76 + 6.61 | 681.86 + 28.57 | 598.00 ± 28.02a | 1.11 | 0.92 |
| 4-(3-Pyridyl)-3-butenoic acid | 81.87 + 11.12 | 191.73 + 29.32 | 186.18 ± 15.17 | 0.31 | 0.29 |
| 4-Hydroxy-4-(3-pyridyl)-butanoic acid | 0.00 + 0.00 | 501.66 + 41.90 | 501.21 ± 25.30 | 0.82 | 0.77 |
| Cotinine N-oxide | 47.60 + 15.26 | 6641.62 + 1103.40 | 7977.95 ± 316.14a | 10.84 | 12.23 |
| Hydroxycotinine | 65.58 + 4.63 | 3427.32 + 469.96 | 3407.78 ± 520.05 | 5.59 | 5.22 |
| Anabasine | 34.55 ± 16.15 | 1421.67 ± 376.24 | 1033.02 ± 390.58 | – | – |
| Anatabine | 22.08 ± 5.70 | 1022.95 ± 36.10 | 929.47 ± 80.65a | – | – |
Indicates statistically significant differences at p < 0.05 in T2 compared to T1; all of the nicotine metabolites were significantly changed in groups of T1 and T2 compared to controls.
Table 2.
Intensity (Mean ± STD) of the metabolites in cells detected by LC/QTof-MS.
| Metabolites | Ionization | Experimental | Theoretical | Formula | C | T1 | T2 |
|---|---|---|---|---|---|---|---|
| Tobacco compounds | |||||||
| Anabasine | POS | 163.1238 | 163.1235 | C10H14N2 | 0 | 84.83 ± 19.32 Δ | 88.66 ± 34.54* |
| Anatabine | POS | 161.1084 | 161.1079 | C10H12N2 | 22.08 ± 5.70 | 253.89 ± 10.92 Δ | 254.14 ± 29.52* |
| 2-(1,2,3,4-Tetrahydroxybutyl)-6-(2,3,4-trihydroxybutyl)pyrazine | POS | 305.1348 | 305.1349 | C12H20N2O7 | 10.70 ± 3.76 | 115.65 ± 8.58 Δ | 139.79 ± 16.09*¥ |
| Caffeine | POS | 195.0583 | 195.0882 | C8H10N4O2 | 0 | 6.37 ± 3.89 Δ | 3.84 ± 3.53* |
| Dimethylpyrazine | POS | 109.0765 | 109.0766 | C6H8N2 | 0 | 7.89 ± 1.88 Δ | 17.24 ± 4.32*¥ |
| Periandrin I (1) | POS | 453.3380 | 453.3369 | C30H44O3 | 7.18 ± 1.03 | 2206.21 ± 217.30 Δ | 1957.24 ± 288.99* |
| Periandrin I (2) | POS | 453.3379 | 453.3369 | C30H44O3 | 3.23 ± 0.30 | 845.07 ± 102.00 Δ | 864.87 ± 97.37* |
| Periandrin I (3) | POS | 453.3366 | 453.3369 | C30H44O3 | 0 | 174.25 ± 18.39 Δ | 124.96 ± 13.28*¥ |
| 3-Indoleacetonitrile | POS | 157.0766 | 157.0766 | C10H8N2 | 0 | 173.44 ± 10.43 Δ | 237.30 ± 30.75*¥ |
| Caffeic acid | Neg | 179.0345 | 179.0344 | C9H8O4 | N/A | N/A | N/A |
| Guaiacyl acetate | Neg | 165.0550 | 165.0552 | C9H10O3 | 0 | 45.14 ± 4.12 Δ | 89.67 ± 15.62*¥ |
| Glycerol tributanoate | Neg | 301.1649 | 301.1651 | C15H26O6 | 0 | 23.07 ± 5.33 Δ | 55.88 ± 8.96*¥ |
| Monomenthyl succinate | Neg | 255.1594 | 255.1596 | C14H24O4 | 0 | 17.97 ± 3.97 Δ | 37.33 ± 7.86*¥ |
| 21-Hydroxyisoglabrolide | Neg | 483.3095 | 483.311 | C30H44O5 | 0.63 ± 1.26 | 55.45 ± 3.30 Δ | 33.33 ± 9.39*¥ |
| Vitamins | |||||||
| Riboflavin | POS | 377.1465 | 377.1461 | C17H20N4O6 | 0 | 7.16 ± 1.63 Δ | 6.35 ± 3.65* |
| Ascorbic acid | Neg | 175.0303 | 175.0243 | C6H8O6 | 0 | 6.54 ± 1.94 Δ | 13.49 ± 1.88*¥ |
| Pantothenic acid | Neg | 218.1031 | 218.1028 | C9H17NO5 | 0.66 ± 1.32 | 2.25 ± 1.30 | 7.51 ± 4.66*¥ |
| Bacterial metabolites | |||||||
| Heptaprenyl diphosphate | Neg | 653.3744 | 653.3736 | C35H60O7P2 | 470.67 ± 66.92 | 27.60 ± 7.22 Δ | 0.32 ± 0.71*¥ |
| Hydroxyphenylpyruvic acid sulfate | Neg | 259.0121 | 258.9912 | C9H8SO7 | 26.17 ± 10.14 | 5.87 ± 3.31 Δ | 1.61 ± 3.59* |
| Leucinic acid | Neg | 131.0702 | 131.0708 | C6H12O3 | 405.13 ± 43.16 | 175.73 ± 15.61 Δ | 165.98 ± 78.39* |
| Phenyllactic acid | Neg | 165.0548 | 165.0552 | C9H10O3 | 91.25 ± 24.57 | 48.15 ± 6.26 Δ | 43.27 ± 21.36* |
| Endogenous metabolites | |||||||
| Oxidative stress | |||||||
| Glutamate | Neg | 128.0338 | 128.0348 | C5H7NO3 | 455.80 ± 15.98 | 168.63 ± 12.95 Δ | 85.71 ± 83.16* |
| Glutathione | POS | 308.1641 | 308.0916 | C10H17N3O6S | 47.80 ± 5.02 | 13.01 ± 2.73 Δ | 11.42 ± 3.95* |
| Methionine | POS | 150.0600 | 150.0589 | C5H11NO2S | 71.78 ± 8.20 | 49.52 ± 10.70 Δ | 32.78 ± 6.27*¥ |
| S-Adenosylmethionine | POS | 399.1335 | 399.1451 | C15H22N6O5S | 34.93 ± 5.99 | 0.86 ± 1.93 Δ | 0* |
| Oxidized Glutathione | Neg | 611.1387 | 611.1441 | C20H32N6O12S2 | 42.73 ± 5.87 | 7.14 ± 2.47 Δ | 2.21 ± 1.40*¥ |
| Pyroglutamic acid | Neg | 128.0344 | 128.0348 | C5H7NO3 | 273.04 ± 36.89 | 153.27 ± 14.19 Δ | 321.44 ± 25.92*¥ |
| Taurine | Neg | 124.0071 | 124.0068 | C2H7NO3S | 82.05 ± 14.44 | 25.89 ± 3.83 Δ | 29.42 ± 8.42* |
| Arginine pathway | |||||||
| Arginine | POS | 175.1131 | 175.1195 | C6H14N4O2 | 33.43 ± 8.08 | 14.49 ± 2.30 Δ | 10.47 ± 3.36* |
| Citrulline | POS | 176.1045 | 176.1035 | C6H13N3O3 | 61.43 ± 8.48 | 35.81 ± 7.84 Δ | 27.17 ± 22.50* |
| Creatinine | POS | 114.0663 | 114.0667 | C4H7N3O | 98.87 ± 7.99 | 30.65 ± 7.25 Δ | 19.62 ± 15.52* |
| Phenylalanine | POS | 120.0816 | 120.0813 | C8H9N | 1396.42 ± 236.33 | 744.99 ± 222.41 Δ | 1356.23 ± 156.93¥ |
| Proline | POS | 116.0715 | 116.0712 | C5H9NO2 | 382.18 ± 19.78 | 258.07 ± 75.78 Δ | 116.73 ± 6.40*¥ |
| Aspartic acid | Neg | 132.0292 | 132.0297 | C4H7NO4 | 1195.03 ± 73.40 | 696.20 ± 71.92 Δ | 302.94 ± 283.39*¥ |
| Glutamate | Neg | 128.0338 | 128.0348 | C5H7NO3 | 455.80 ± 15.98 | 168.63 ± 12.95 Δ | 85.71 ± 83.16* |
| Others | |||||||
| Tryptophan | POS | 188.0714 | 188.0712 | C11H9NO2 | 1092.41 ± 50.32 | 619.00 ± 97.68 Δ | 1147.30 ± 76.73¥ |
| Tyrosine | POS | 136.0765 | 136.0762 | C8H9NO | 92.09 ± 11.96 | 59.98 ± 9.47 Δ | 79.59 ± 5.10¥ |
| Leucine | Neg | 130.0864 | 130.0868 | C6H13NO2 | 76.71 ± 13.97 | 18.94 ± 12.84 Δ | 57.84 ± 7.21¥ |
| 5′-Methylthioadenosine | POS | 136.0629 | 136.0623 | C5H5N5 | 179.00 ± 17.11 | 31.78 ± 21.38 Δ | 3.06 ± 4.19*¥ |
| Adenine | POS | 136.0646 | 136.0623 | C5H5N5 | 1044.92 ± 48.06 | 691.30 ± 405.55 | 269.30 ± 42.96*¥ |
| Inosine | Neg | 267.0725 | 267.0729 | C10H12N4O5 | 35.85 ± 3.42 | 5.14 ± 3.08 Δ | 10.91 ± 6.88* |
| Asparaginyl-Glutamine | POS | 261.1242 | 261.1199 | C9H16N4O5 | 184.21 ± 13.98 | 102.65 ± 19.25 Δ | 144.18 ± 4.20*¥ |
| Carnitine | POS | 162.1125 | 162.113 | C7H15NO3 | 110.64 ± 6.57 | 40.66 ± 4.83 Δ | 35.28 ± 5.84* |
| Azelaic acid | Neg | 187.0967 | 187.097 | C9H16O4 | 31.64 ± 1.51 | 82.36 ± 13.22 Δ | 142.34 ± 18.50*¥ |
| Cytidine monophosphate | Neg | 322.0478 | 322.024 | C9H14N3O8P | 134.98 ± 15.85 | 9.58 ± 5.04 Δ | 5.34 ± 3.41* |
| Gluconic acid | Neg | 195.0502 | 195.0505 | C6H12O7 | 77.76 ± 21.18 | 373.26 ± 22.85 Δ | 314.57 ± 137.85* |
| Glucose or other sugar | Neg | 179.0551 | 179.0556 | C6H12O6 | 435.53 ± 55.94 | 180.56 ± 42.81 Δ | 163.71 ± 38.26* |
| Myristic acid | Neg | 227.2014 | 227.2011 | C14H28O2 | 162.41 ± 23.06 | 44.69 ± 11.32 Δ | 16.68 ± 6.60*¥ |
| Sebacic acid | Neg | 201.1111 | 201.1127 | C10H18O4 | 19.48 ± 1.64 | 19.46 ± 3.67 | 32.10 ± 3.69*¥ |
| Undecanedionic acid | Neg | 215.1282 | 215.1283 | C11H20O4 | 11.01 ± 4.29 | 33.06 ± 8.17 Δ | 75.84 ± 14.48*¥ |
| Methylmalonic acid | Neg | 117.0159 | 117.0188 | C4H6O4 | 0 | 42.74 ± 3.57 Δ | 86.09 ± 19.69*¥ |
| Cytochalasin Npho | Neg | 492.2721 | 492.275 | C30H39NO5 | 180.10 ± 17.42 | 109.97 ± 12.66 Δ | 35.28 ± 14.17*¥ |
| LysoPC(14:1) | POS | 466.2933 | 466.2934 | C22H44NO7P | 242.67 ± 10.36 | 99.88 ± 1.49 Δ | 5.57 ± 0.49*¥ |
| peptide containing Asparaginyl-Proline | POS | 668.3610 | Unknown | 129.69 ± 2.81 | 66.58 ± 1.50 Δ | 171.24 ± 3.02*¥ | |
| peptide containing Asparaginyl-Proline (2) | POS | 230.1146 | Unknown | 87.08 ± 0.88 | 34.89 ± 1.03 Δ | 90.10 ± 1.98¥ | |
| peptide containing Leucyl-Proline | POS | 229.1552 | Unknown | 392.82 ± 6.56 | 110.27 ± 3.25 Δ | 385.23 ± 20.18¥ | |
Asterisks indicate statistically significant differences at p < 0.05 *: T2 vs C and ¥: T2 vs T1, while Δ: T1 vs C. Measures for which differences between controls and T1 or T2 are not significant are highlighted.
3.1. Changes in nicotine and nicotine-related compounds
Exogenous nicotine and its metabolites were analyzed for total changes (i.e., sum of the intensity of a compound in cells and media) to examine the nicotine metabolism changes by C. sputigena. Total intensity levels (mean ± STD) of nicotine and its eight major metabolites and their percentages are listed in Table 1. The percentage of individual metabolite was calculated based on the metabolite intensity divided by the sum of intensities of all nine metabolites (i.e., percentage (%) of nicotine = Intensity of nicotine/Σ[Intensity of all nine metabolites] × 100). This percentage only provides a semi-quantitative estimate of nicotine metabolism changes based on the assumption that all of the nicotine metabolites have the same ionization efficiency (which is frequently not the case). However, a semi-quantitative analysis of nicotine metabolism changes can be used to estimate the role of bacterial cells in nicotine metabolism. Since the half-life in humans is about 2 h for nicotine and about 12 h for cotinine, it is reasonable to use nicotine and its eight metabolites in T1 at 2 min as a baseline to account for rapid physical absorption of STAE onto the cell walls or membranes. The total levels of all detected nicotine metabolites were significantly different between treatment and control groups; 2′-hydroxynicotine and cotinine N-oxide significantly increased by 0.05% and 0.22%, respectively, and nornicotine decreased in the T2 group by 0.37% compared with T1 at 2 min. Table 1 also shows that anatabine, a minor alkaloid, was significantly decreased in the T2 group compared to the T1 group. The findings indicate that the bacterial cells might not influence the major nicotine metabolism pathway but might be involved in the minor nicotine metabolism pathway. Fig. 2 depicts the major and minor nicotine metabolism pathways in humans. Fig. 2 shows intensity and percentage changes in nicotine and its eight metabolites in control, T1 and T2 groups. It must be noted that N-nitrosonornicotine (NNN), a tobacco specific nitrosamine, was also detected; however, the detection signal is below the lower limit of quantitation (LLOQ).
Fig. 2.
Observed total intensity and percentage changes in nicotine and its major metabolites involved in nicotine metabolism. Whereas, the major nicotine metabolism pathways in humans are shown in bold black arrow, while the minor pathways are in grey arrows. Changes in STAE treatment for 48 h are compared with STAE treatment for 2 min to account for physical absorption on the membrane and/or bacterial wall. *: p < 0.05, T2 vs C; ¥: p < 0.05, T1 vs T2. No significant changes were observed in total nicotine levels after 48 h incubation with the oral bacteria compared with 2 min incubation.
Other tobacco-containing compounds were also detected in STAE-treated samples but absent in the control samples (Table 2). These compounds include additives like caffeine and flavor compounds such as dimethylpyrazine, periandrin I, guaiacyl acetate, glycerol tributanoate, and monomenthyl succinate. Furthermore, three vitamins (ascorbic acid, riboflavin, and pantothenic acid) were only observed in the cell extracts after STAE treatment. However, it is not certain that these vitamins originated from STAE, since they were also detected in the media from the control groups. Nonetheless, the levels of the three detected vitamins in the treatment groups were all significantly higher than those in the controls for both cell and media extracts.
3.2. Changes in bacteria-related metabolites
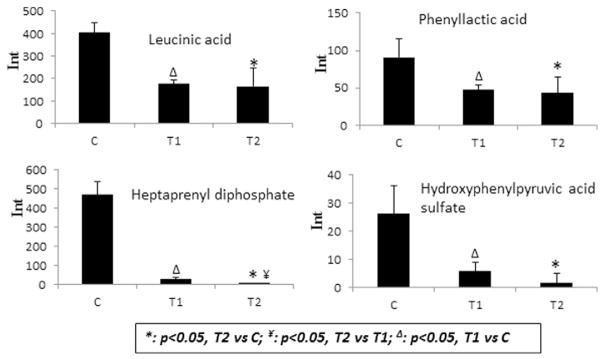
The production of bacteria-related metabolites is mediated by bacterial cells and dependent on the presence of bacteria. Therefore, the changes in the bacteria-related metabolites can be indicators for the disturbance of bacterial function due to smokeless tobacco exposure. Levels of four bacteria-related metabolites – leucinic acid, phenyllactic acid, heptaprenyl diphosphate and hydroxyphenylpyruvic acid sulfate (www.hmdb.ca) – were significantly decreased in cells after STAE treatment at 2 min and 48 h incubation (Fig. 3). Furthermore, levels of heptaprenyl diphosphate decreased significantly after 48 h incubation compared with the 2 min treatment.
Fig. 3.
Observed intensity changes in bacteria-related metabolites from cell extracts. Exampled metabolites include leucinic acid, phenyllactic acid, heptaprenyl diphosphate and hydroxyphenylpyruvic acid sulfate. *: p < 0.05, T2 vs C; ¥: p < 0.05, T2 vs T1; Δ: p < 0.05, T1 vs C. Significant changes were observed in all four metabolites in the treated groups compared with controls.
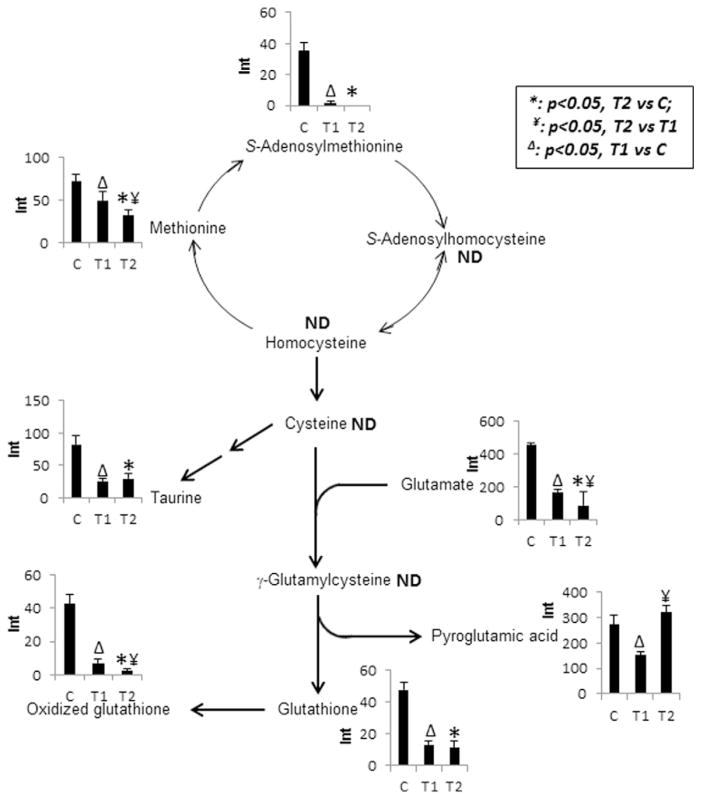
3.3. Changes in oxidative stress-related metabolites
Fig. 4 shows the changes in metabolites involved in the oxidative stress pathway. These metabolites included glutathione (GSH), methionine, S-adenosylmethionine, oxidized glutathione, taurine, glutamate, and pyroglutamic acid. The first five metabolites (sulfur-containing compounds) and glutamate were all significantly decreased in the STAE-treated samples compared to controls, while glutamate was significantly decreased in the T2 group compared to the T1 group at 2 min. Pyroglutamic acid, a product of GSH consumption, was significantly increased in T2 vs T1. These findings indicate that STAE treatment might cause oxidative stress on bacterial cells.
Fig. 4.
Observed intensity changes in metabolites mapped onto the glutathione pathway. *: p < 0.05, T2 vs C; ¥: p < 0.05, T2 vs T1; Δ: p < 0.05, T1 vs C. Significant decreases were observed in all sulfur-containing metabolites in the treated groups compared with controls.
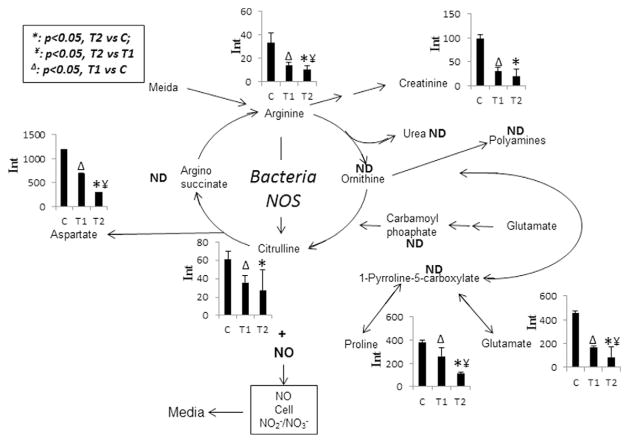
3.4. Changes in arginine pathway and other metabolites
Fig. 5 shows the changes in metabolites involved in the arginine pathway. Citrulline and NO are co-produced from arginine catalyzed by bacterial nitric oxide synthases (NOSs). Citrulline level was significantly decreased in the STAE treated samples at 2 min and 48 h, which indicated that NO production was likely reduced by STAE treatment. Levels of other amino acids including arginine, creatinine, aspartate, proline, and glutamate were all significantly decreased in both the T1 and T2 groups compared to the control; levels of arginine, aspartate, proline, and glutamate were also significantly decreased in the T2 group compared to the T1 group.
Fig. 5.
Observed intensity changes in metabolites mapped onto the arginine metabolism pathway. *: p < 0.05, T2 vs C; ¥: p < 0.05, T2 vs T1; Δ: p < 0.05, T1 vs C. Significant decreases were observed in all detected metabolites in the treated groups compared with controls.
Changes in other endogenous metabolites – including three amino acids, three nucleic acids, two glucose-related metabolites, five short-to medium-chain fatty acids, one lysophosphatidylcholine, and three unknown compounds – were also detected (Table 2). Changes in glucose and fatty acid levels indicated that the energy pathway was disturbed by STAE treatment.
4. Discussion
Use of tobacco products, including smokeless tobacco, causes a broad spectrum of pathologies such as cancer, cardiovascular disease, and respiratory problems, and has been linked to oral disease (Wald and Hackshaw, 1996). Users place smokeless tobacco in contact with mucous membranes, where gingival recession and mucosal lesions often occur. Furthermore, these oral diseases have been attributed to alterations in the oral microbiota as a result of smokeless tobacco exposure, as well as viability and composition changes in the oral microbiota population (Kamma et al., 1999).
Nicotine (the principal tobacco alkaloid) is extensively metabolized by the liver cytochrome P450 system in mammalian species (Benowitz and Jacob, 1994). In humans (Benowitz et al., 1994; Byrd et al., 1992), about 70–80% of nicotine is converted to cotinine and about 10–15% of nicotine excretes as cotinine; the remainder excretes as nicotine N-oxide (NNO, 4–7%), nicotine glucuronide (3–5%), trans-3-hydroxycotinine (33–40%), cotinine glucuronide (12–17%, and trans-3-hydrocycotinine glucuronide (7–9%). Not surprisingly, the current study showed that the pathway of nicotine degradation is different in C. sputigena compared to mammalian species. Glucuronide conjugates of nicotine were not observed in this study. In the present study, nicotine and its major metabolites, including cotinine, NNO, hydroxycotinine, 4-(3-pyridyl)-3-butenoic acid, and 4-hydroxy-4-(3-pyridyl)-butanoic acid, remained almost unchanged after STAE treatment for 48 h (Fig. 2). However, the significant increases in hydroxynicotine and cotinine N-oxide (CNO) indicated that the bacterial cells might have degraded nicotine to a small extent. Robert and Cole (Roberts and Cole, 1979) reported that the growth of Haemophilus influenzae, a Gram-negative bacterium, was stimulated by tobacco or nicotine in a phosphate-buffered saline agar with a nutrient-scarce condition from visual observation; this finding suggests that bacteria might use nicotine as a nutrient source when other nutrients are not sufficient. Our observation of unchanged levels of nicotine and its major metabolites supports that hypothesis that bacteria incubated in the rich culture media used in the present study did not utilize nicotine as a major energy source. Furthermore, significant decreases in nornicotine and anatabine and decreases in anabasine were observed after 48 h exposure to 50 mg STAE/mL. Besides, nicotine, these three compounds are the most abundant minor alkaloids present in tobacco. Consistent with our result, it has been reported that bacteria can degrade the minor alkaloids by oxidation during tobacco processing (Leete, 1983). Based on the above-mentioned discussion, C. sputigena did not metabolize nicotine except to a minor extent of degradation (0.05% for hydroxynicotine and 1.39% for CNO) under the current cultivation condition. However, this bacterium can degrade minor alkaloids including nornicotine, anatabine and anabasine to some extent (<30% after 48 h incubation with the bacterial cells).
To date, no other studies have evaluated the effects of smokeless tobacco on the oral microbiota at the metabolite level. Although the viability of C. sputigena was not changed by the STAE treatments, whether the health status of the bacterium was changed by the STAE treatments is not clear. In the present study, decreases in the bacteria-related metabolites, including levels of leucinic acid, phenylacetic acid, heptaprenyl diphosphate, and hydroxyphenylpyruvic acid sulfate, were observed after STAE treatments (Fig. 3). Wikoff et al. (Wikoff et al., 2009) reported that several organic acids containing phenyl groups (e.g. phenylacetic acid) and other bacteria-related compounds were observed only in conventional mice but not in germ-free animals. In our previous penicillin study (Sun et al., 2013), time- and dose-dependent decreases in bacteria-related metabolites (including indole-containing metabolites, organic acids containing phenyl groups, sulfate, and glucuronide conjugates) were observed in the penicillin-dosed rats, which indicated that the gut microbiota population was suppressed. Therefore, bacteria-related metabolite decreases in the present study indicated that bacterial functions might be significantly compromised by the STAE treatments even after only 2 min exposure.
Many compounds present in smokeless tobacco are oxidative stress-inducing compounds (e.g., oxygen, nitrogen radicals or non-radicals species, reactive carbonyl compounds) that can cause damage by binding biological macromolecules, resulting in collapsing cell structure or cell dysfunction (Pryor and Stone, 1993; Hoffmann et al., 2001; Colombo et al., 2012). In the present study, the observation of decreases in glutamate and sulfur-containing metabolites as well as increases in pyroglutamate indicated that STAE caused oxidative stress in C. sputigena (Fig. 4). GSH is an important antioxidant endogenous metabolite in bacteria, preventing impairment to critical cellular components by scavenging reactive oxygen species. The data from this in vitro study are consistent with a previous report (Bizon and Milnerowicz, 2012) that blood GSH levels were significantly decreased in smokers using ≥20 cigarettes per day. Reduced levels of glutathione and ascorbic acid and increased lipid peroxide were observed in adult male smokers (Banerjee et al., 1998). STAE treatments enhanced oxidative stress in the oral bacterial cells, which might further induce more physical damage (e.g., periodontitis).
Disruption in the arginine-NO pathway was also observed in the bacterial cells after STAE exposure (Fig. 5). NO, a powerful vasodilator with a half-life of a few seconds is biosynthesized endogenously from arginine by various NOSs in the bacterial cells. In mammalian species, NO is used by endothelium of blood vessels to relax the surrounding smooth muscles, resulting in increasing blood flow. In the present study, all of the detected metabolites involved in the arginine pathway showed a time-dependent decrease in T1 and T2 groups. Most importantly, citrulline, a co-product with NO from arginine, was significantly decreased in STAE-treated cells. This result suggests that NO production might be reduced by STAE treatments. Extensive studies (Celermajer et al., 1992; Widlansky et al., 2003; Zeiher et al., 1995) in humans have consistently reported that smoking-induced vascular dysfunction is initiated by reduced NO production followed by cascade progression. In addition to the initiation of cardiovascular disease, NO is a potent bacterial compound. Its generation in oral neutrophils and/or oral bacteria is important for protecting hosts against invading microbes and tissue inflammation (Sato et al., 2008).
5. Conclusion
An LC/MS-based metabolomic approach was utilized to evaluate the dynamic relationship between smokeless tobacco exposure and the alteration of metabolism in the oral bacterium, C. sputigena. The absence of significant changes in the levels of nicotine and its major metabolites in the presence of this bacterium suggest that it does not degrade nicotine to a significant extent when cultured in nutrient-rich medium. Furthermore, LC/MS based metabolome profiling was proven to be a powerful tool to identify metabolomics changes in bacteria and presence of the additives and constituents in smokeless tobacco products. The detected additives included caffeine and multiple flavoring compounds. Finally, the physiology and toxicology effects of the smokeless tobacco on the oral bacterium were revealed by pathway analysis. Results showed that smokeless tobacco extracts caused oxidative stress in the bacterium. In addition, perturbation in the arginine-NO pathway indicated that smokeless tobacco could cause NO reduction. Further studies to screen more oral bacterial species and test various brands of smokeless tobacco products on the oral bacterial physiology and metabolism are warranted. Data provided here could be useful in assessing the relative toxicity of various smokeless tobacco products to the oral microbiome.
Acknowledgments
We thank Drs. Steven L. Foley, John B. Sutherland and Hans Rosenfeldt for their critical review of this manuscript. We also thank CTP writer Deborah Neveleff for proof reading the manuscript. This study was funded by the Center for Tobacco Products, U.S. Food and Drug Administration (E0747201). Dr. Jinshan Jin was supported through the Oak Ridge Institute for Science and Education. The findings and conclusions in this publication are those of the authors and do not represent FDA positions or policies.
Footnotes
Transparency document
The Transparency document associated with this article can be found, in online version.
References
- Banerjee KK, Marimuthu P, Sarkar A, Chaudhuri RN. Influence of cigarette smoking on vitamin C, glutathione and lipid peroxidation status. Indian J Public Health. 1998;42:20–23. [PubMed] [Google Scholar]
- Benowitz NL, Jacob P., 3rd Metabolism of nicotine to cotinine studied by a dual stable isotope method. Clin Pharmacol Ther. 1994;56:483–493. doi: 10.1038/clpt.1994.169. [DOI] [PubMed] [Google Scholar]
- Benowitz NL, Jacob P, 3rd, Fong I, Gupta S. Nicotine metabolic profile in man: comparison of cigarette smoking and transdermal nicotine. J Pharmacol Exp Ther. 1994;268:296–303. [PubMed] [Google Scholar]
- Bizon A, Milnerowicz H. Effect of tobacco smoking on glutathione concentration in the blood. Przegl Lek. 2012;69:809–811. [PubMed] [Google Scholar]
- Brandsch R. Microbiology and biochemistry of nicotine degradation. Appl Microbiol Biotechnol. 2006;69:493–498. doi: 10.1007/s00253-005-0226-0. [DOI] [PubMed] [Google Scholar]
- Byrd GD, Chang KM, Greene JM, deBethizy JD. Evidence for urinary excretion of glucuronide conjugates of nicotine, cotinine, and trans-3′-hydroxycotinine in smokers. Drug Metab Dispos. 1992;20:192–197. [PubMed] [Google Scholar]
- Celermajer DS, Sorensen KE, Gooch VM, Spiegelhalter DJ, Miller OI, Sullivan ID, Lloyd JK, Deanfield JE. Non-invasive detection of endothelial dysfunction in children and adults at risk of atherosclerosis. Lancet. 1992;340:1111–1115. doi: 10.1016/0140-6736(92)93147-f. [DOI] [PubMed] [Google Scholar]
- Colombo G, Clerici M, Giustarini D, Rossi R, Milzani A, Dalle-Donne I. Redox albuminomics: oxidized albumin in human diseases. Antioxid Redox Signal. 2012;17:1515–1527. doi: 10.1089/ars.2012.4702. [DOI] [PubMed] [Google Scholar]
- David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, Erdman SE, Alm EJ. Host lifestyle affects human microbiota on daily time-scales. Genome Biol. 2014;15:R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer RO, Jr, Poulson TC. Oral tissue alterations associated with the use of smokeless tobacco by teen-agers. Part I Clinical findings. Oral Surg Oral Med Oral Pathol. 1983;56:275–284. doi: 10.1016/0030-4220(83)90009-9. [DOI] [PubMed] [Google Scholar]
- Hoffmann D, Hoffmann I, El-Bayoumy K. The less harmful cigarette: a controversial issue. A tribute to Ernst L Wynder. Chem Res Toxicol. 2001;14:767–790. doi: 10.1021/tx000260u. [DOI] [PubMed] [Google Scholar]
- Kamma JJ, Nakou M, Baehni PC. Clinical and microbiological characteristics of smokers with early onset periodontitis. J Periodontal Res. 1999;34:25–33. doi: 10.1111/j.1600-0765.1999.tb02218.x. [DOI] [PubMed] [Google Scholar]
- Kapil V, Haydar SM, Pearl V, Lundberg JO, Weitzberg E, Ahluwalia A. Physiological role for nitrate-reducing oral bacteria in blood pressure control. Free Radic Biol Med. 2013;55:93–100. doi: 10.1016/j.freeradbiomed.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leete E. Biosynthesis and metabolism of the tobacco alkaloids, in Alkaloids. In: SWP, editor. Chemical and Biological Perspectives. John Wiley & Sons; New York: 1983. pp. 85–152. [Google Scholar]
- Manas P, Mackey BM. Morphological and physiological changes induced by high hydrostatic pressure in exponential- and stationary-phase cells of Escherichia coli: relationship with cell death. Appl Environ Microbiol. 2004;70:1545–1554. doi: 10.1128/AEM.70.3.1545-1554.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FP, Wang Y, Sprenger N, Holmes E, Lindon JC, Kochhar S, Nicholson JK. Effects of probiotic Lactobacillus paracasei treatment on the host gut tissue metabolic profiles probed via magic-angle-spinning NMR spectroscopy. J Proteome Res. 2007;6:1471–1481. doi: 10.1021/pr060596a. [DOI] [PubMed] [Google Scholar]
- Meurman JH, Bascones-Martinez A. Are oral and dental diseases linked to cancer? Oral Dis. 2011;17:779–784. doi: 10.1111/j.1601-0825.2011.01837.x. [DOI] [PubMed] [Google Scholar]
- Murad CF, Sassone LM, Faveri M, Hirata R, Jr, Figueiredo L, Feres M. Microbial diversity in persistent root canal infections investigated by checkerboard DNA-DNA hybridization. J Endod. 2014;40:899–906. doi: 10.1016/j.joen.2014.02.010. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- Piano MR, Benowitz NL, Fitzgerald GA, Corbridge S, Heath J, Hahn E, Pechacek TF, Howard G. American Heart Association Council on Cardiovascular N: impact of smokeless tobacco products on cardiovascular disease: implications for policy, prevention, and treatment: a policy statement from the American Heart Association. Circulation. 2010;122:1520–1544. doi: 10.1161/CIR.0b013e3181f432c3. [DOI] [PubMed] [Google Scholar]
- Pindborg JJ. Tobacco and gingivitis: statistical examination of the significance of tobacco in the development of ulceromembranous gingivitis and in the formation of calculus. J Dent Res. 1947;26:261–264. doi: 10.1177/00220345470260030901. [DOI] [PubMed] [Google Scholar]
- Pryor WA, Stone K. Oxidants in cigarette smoke. Radicals, hydrogen peroxide, peroxynitrate, and peroxynitrite. Ann N Y Acad Sci. 1993;686:12–27. doi: 10.1111/j.1749-6632.1993.tb39148.x. discussion 27–18. [DOI] [PubMed] [Google Scholar]
- Roberts D, Cole P. Effect of tobacco and nicotine on growth of Haemophilus influenzae in vitro. J Clin Pathol. 1979;32:728–731. doi: 10.1136/jcp.32.7.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson PB, Walsh M, Greene J, Ernster V, Grady D, Hauck W. Periodontal effects associated with the use of smokeless tobacco. J Periodontol. 1990;61:438–443. doi: 10.1902/jop.1990.61.7.438. [DOI] [PubMed] [Google Scholar]
- Sato EF, Choudhury T, Nishikawa T, Inoue M. Dynamic aspect of reactive oxygen and nitric oxide in oral cavity. J Clin Biochem Nutr. 2008;42:8–13. doi: 10.3164/jcbn.2008002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi K, Shah S, Abbas SM, Vidyasagaran A, Jawad M, Dogar O, Sheikh A. Global burden of disease due to smokeless tobacco consumption in adults: analysis of data from 113 countries. BMC Med. 2015;13:194. doi: 10.1186/s12916-015-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Seksik P, Rigottier-Gois L, Lay C, Lepage P, Podglajen I, Marteau P, Dore J. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm Bowel Dis. 2006;12:106–111. doi: 10.1097/01.MIB.0000200323.38139.c6. [DOI] [PubMed] [Google Scholar]
- Sun J, Von Tungeln LS, Hines W, Beger RD. Identification of metabolite profiles of the catechol-O-methyl transferase inhibitor tolcapone in rat urine using LC/MS-based metabonomics analysis. J Chromatogr B Anal Technol Biomed Life Sci. 2009;877:2557–2565. doi: 10.1016/j.jchromb.2009.06.033. [DOI] [PubMed] [Google Scholar]
- Sun J, Schnackenberg LK, Hansen DK, Beger RD. Study of valproic acid-induced endogenous and exogenous metabolite alterations using LC-MS-based metabolomics. Bioanalysis. 2010;2:207–216. doi: 10.4155/bio.09.173. [DOI] [PubMed] [Google Scholar]
- Sun J, Schnackenberg LK, Khare S, Yang X, Greenhaw J, Salminen W, Mendrick DL, Beger RD. Evaluating effects of penicillin treatment on the metabolome of rats. J Chromatogr B Anal Technol Biomed Life Sci. 2013;932:134–143. doi: 10.1016/j.jchromb.2013.05.030. [DOI] [PubMed] [Google Scholar]
- Swann JR, Tuohy KM, Lindfors P, Brown DT, Gibson GR, Wilson ID, Sidaway J, Nicholson JK, Holmes E. Variation in antibiotic-induced microbial recolonization impacts on the host metabolic phenotypes of rats. J Proteome Res. 2011;10:3590–3603. doi: 10.1021/pr200243t. [DOI] [PubMed] [Google Scholar]
- Thompson-Chagoyan OC, Maldonado J, Gil A. Colonization and impact of disease and other factors on intestinal microbiota. Dig Dis Sci. 2007;52:2069–2077. doi: 10.1007/s10620-006-9285-z. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Winkelhoff AJ, Bosch-Tijhof CJ, Winkel EG, van der Reijden WA. Smoking affects the subgingival microflora in periodontitis. J Periodontol. 2001;72:666–671. doi: 10.1902/jop.2001.72.5.666. [DOI] [PubMed] [Google Scholar]
- Wald NJ, Hackshaw AK. Cigarette smoking: an epidemiological overview. Br Med Bull. 1996;52:3–11. doi: 10.1093/oxfordjournals.bmb.a011530. [DOI] [PubMed] [Google Scholar]
- Widlansky ME, Gokce N, Keaney JF, Jr, Vita JA. The clinical implications of en-dothelial dysfunction. J Am Coll Cardiol. 2003;42:1149–1160. doi: 10.1016/s0735-1097(03)00994-x. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn DM. Tobacco use and oral disease. J Dent Educ. 2001;65:306–312. [PubMed] [Google Scholar]
- Yap IK, Li JV, Saric J, Martin FP, Davies H, Wang Y, Wilson ID, Nicholson JK, Utzinger J, Marchesi JR, Holmes E. Metabonomic and microbiological analysis of the dynamic effect of vancomycin-induced gut microbiota modification in the mouse. J Proteome Res. 2008;7:3718–3728. doi: 10.1021/pr700864x. [DOI] [PubMed] [Google Scholar]
- Zeiher AM, Schachinger V, Minners J. Long-term cigarette smoking impairs endothelium-dependent coronary arterial vasodilator function. Circulation. 1995;92:1094–1100. doi: 10.1161/01.cir.92.5.1094. [DOI] [PubMed] [Google Scholar]