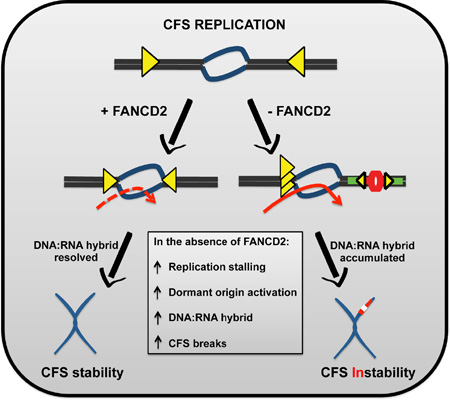
SUMMARY
Common fragile sites (CFSs) are genomic regions that are unstable under conditions of replicative stress. Although the characteristics of CFSs that render them vulnerable to stress are mainly associated with replication, the cellular pathways that protect CFSs during replication remain unclear. Here, we identify and describe a role for FANCD2 as a trans-acting facilitator of CFS replication, in the absence of exogenous replicative stress. In the absence of FANCD2, replication forks stall within the AT-rich fragility core of CFS leading to dormant origin activation. Furthermore, FANCD2 deficiency is associated with DNA:RNA hybrid formation at CFS-FRA16D and inhibition of DNA:RNA hybrid formation suppresses replication perturbation. In addition, we also found that FANCD2 reduces the number of potential sites of replication initiation. Our data demonstrate that FANCD2 protein is required to ensure efficient CFS replication and provide mechanistic insight into how FANCD2 regulates CFS stability.
Graphical Abstract
eTOC Blurb/In Brief
Cells lacking proteins associated with the Fanconi anemia pathway are prone to chromosome breaks at CFSs. Madireddy et al. show that FANCD2 facilitates replication through repeat rich genomic regions such as CFSs by ameliorating DNA:RNA hybrid accumulation and by controlling dormant origin firing, even during unperturbed replication
INTRODUCTION
Nearly two-thirds of the human genome is comprised of repetitive sequences that often challenge DNA replication, which can lead to genomic instability. CFSs are chromosomal regions that are most prone to genomic instability and are implicated in the development and progression of cancer (Arlt et al., 2003; Glover, 2006). Furthermore, CFSs are hotspots for chromosomal structural aberrations such as deletions, duplications, and translocations (Chesi et al., 1998; Finnis et al., 2005; O'Keefe and Richards, 2006). Maintenance of CFS integrity is critical because most of the commonly expressed CFSs contain tumor suppressor genes and proto-oncogenes which when altered are associated with a large spectrum of cancers (Ciullo et al., 2002; Hellman et al., 2002; Siprashvili et al., 1997). The three most prevalent models of CFS instability involve the presence of structure-prone repetitive DNA sequences, the possibility of transcription-associated obstacles and the scarcity of replication initiation events (Le Beau et al., 1998; Lucas et al., 2007). Currently, it is believed that perturbed replication of these regions is at the heart of their fragility. Identifying the factors that alleviate replication perturbation at CFSs is vital to understanding the mechanism(s) leading to CFS instability.
Among the various proteins that have been implicated in CFS breakage are the Fanconi anemia (FA) proteins. FA is a genetic disorder characterized by developmental abnormalities, bone marrow failure, and a high incidence of malignancies. While the repair mediated functions of the FA pathway provide some mechanistic insight, the severe phenotypes observed in some FA patients (Hirsch et al., 2004; Howlett et al., 2002), and FA mouse models (Houghtaling et al., 2003) suggest additional roles for these proteins. Recent reports suggest a role for the FA pathway in DNA replication. The FA pathway is strongly activated in response to replisome stalling that occurs in response to agents that induce replicative stress, such as hydroxyurea (HU) (Petermann et al., 2010; Taniguchi et al., 2002). Furthermore, FANCD2 transiently interacts with the MCM proteins (Lossaint et al., 2013) and stabilizes stalled replication forks (Karanja et al., 2014; Schlacher et al., 2012).
In addition to increased risk of cancer (Kutler et al., 2003; Rosenberg et al., 2003) the absence of FA proteins is associated with elevated chromosomal breaks at CFSs (Howlett et al., 2005), suggesting a link between cancer predisposition and CFS instability in FA patients. The observed but unexplained exacerbation of CFS instability in the absence of FA proteins combined with the inherent replication defects found at CFSs prompted us to determine whether replication associated functions of FA proteins also facilitate CFS replication. If the FA proteins are indeed mediating timely replication of CFSs, then their absence should challenge replication and result in the alteration of the replication program at CFS loci.
Here, we show that the FANCD2 protein is an important trans-acting mediator of CFS replication. By visualizing the in vivo replication dynamics of individual DNA fibers, we found a striking change in the replication program at the CFS loci in FANCD2−/ − patient-derived lymphoblasts. We propose that FANCD2 has a multifaceted role in facilitating replication at difficult to replicate genomic regions such as CFS. It helps the replication machinery navigate past the fragility core of CFS-FRA16D, likely by resolving impediments to replication machinery such as DNA:RNA hydrids. In this manner, FANCD2 appears to maintain CFS stability in the absence of exogenous stress and seems to do so separate from rest of FA core complex proteins and FANCD2 monoubiquitination. Additionally, FANCD2 also ensures optimal firing of dormant rescue origins to facilitate replication completion at CFSs to avoid mitotic instability. These studies provide key mechanistic insight into the role of FANCD2 in maintaining CFS stability and preserving genome integrity.
RESULTS
CFS-FRA16D replication program is altered in the absence of FANCD2
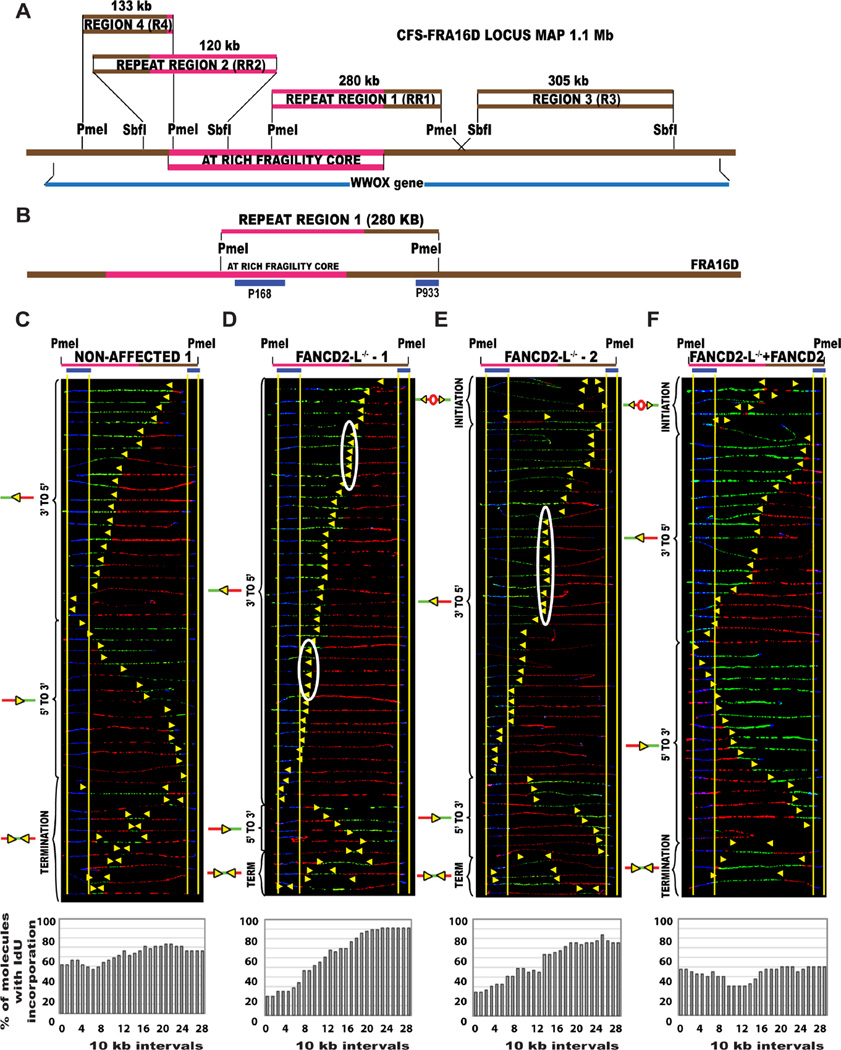
We started our analysis with FANCD2-deficient cell lines since FANCD2 can facilitate DNA replication under conditions of replicative stress (Lossaint et al., 2013). Using single molecule analysis of replicated DNA (SMARD) (Fig. S1a), we examined the endogenous replication program of two CFS loci, FRA16D and FRA6E. Lymphocytes were used because CFSs are usually mapped in lymphocytes (Sutherland and Richards, 1995), and FRA16D and FRA6E are highly expressed in this cell type (Helmrich et al., 2011). To analyze the movement of replication forks through the AT-rich fragility core of CFS-FRA16D, we analyzed a 280 kb PmeI segment of FRA16D that contained a portion of the AT-rich fragility core on the left and flanking DNA sequences on the right; this region will henceforth be referred to as repeat region 1 (RR1) (Fig. 1A–B, Table S1).
Figure 1. The replication profile is altered at the endogenous CFS-FRA16D locus in the absence of FANCD2.
(A) Common fragile site FRA16D locus map Locus map of the CFS-FRA16D (brown line-1.5 Mb) that contains the AT-rich fragility core (pink line–280 kb) and overlaps the WWOX tumor suppressor gene (dark blue line–1.1 Mb). The locus was divided into 4 segments based on restriction enzyme availability. The coordinates of the different regions are summarized in Table S1, providing additional information about fosmids and primers used to identify the regions.
(B) Locus map of RR1-PmeI segment containing a portion of the AT-rich fragility core. The segments are aligned according to the positions of the FISH probes (blue) on the map.
(C–F) Top; Locus map of PmeI digested RR1 segment. Middle; Aligned photomicrograph images of labeled DNA molecules from (C) Non-affected 1 (GM02184), (D) FANCD2−/−-L-1 (PD20), (E) FANCD2−/−-L-2 (2742) and (F) FANCD2−/−-L-1 + FANCD2 (corrected) lymphoblast. The yellow arrows indicate the sites along the molecules where the IdU transitioned to CldU. The molecules are arranged in the following order: molecules with initiation events, molecules with 3’ to 5’ progressing forks, molecules with 5’ to 3’ progressing forks and molecules with termination events. White ovals indicate regions of replication fork pausing and correspond to the pausing peaks listed in Table S2. Bottom; The percentage of molecules incorporating IdU (red) is calculated from the replication program (middle) and is represented as a histogram.
In non-affected lymphoblasts, GM02184 (non-affected 1) and GM03798 (non-affected 2), replication proceeds unperturbed, bidirectionally across the repeats, with equal numbers of 3’ to 5’ and 5’ to 3’ progressing forks replicating the locus (Fig. 1C; Fig. S2B). In contrast, in the FANCD2−/−lymphoblasts (FANCD2−/−-L-1 [PD20], FANCD2−/−-L-2 [2742], and FANCD2−/−-L-3 [2717]), the direction of replication was altered and replication forks progressed predominantly in the 3’ to 5’ direction, into the fragility core, in ~70% of the cells. Very few 5’ to 3’ progressing replication forks managed to reach RR1 at the same time as the 3’ to 5’ progressing forks (Fig. 1D–E; Fig. S2C). Complementation of the FANCD2−/−-L-1 lymphoblasts with wild-type FANCD2 protein (Fig. S3B) restores bidirectional replication program at RR1 (Fig. 1F), similar to the non-affected cells.
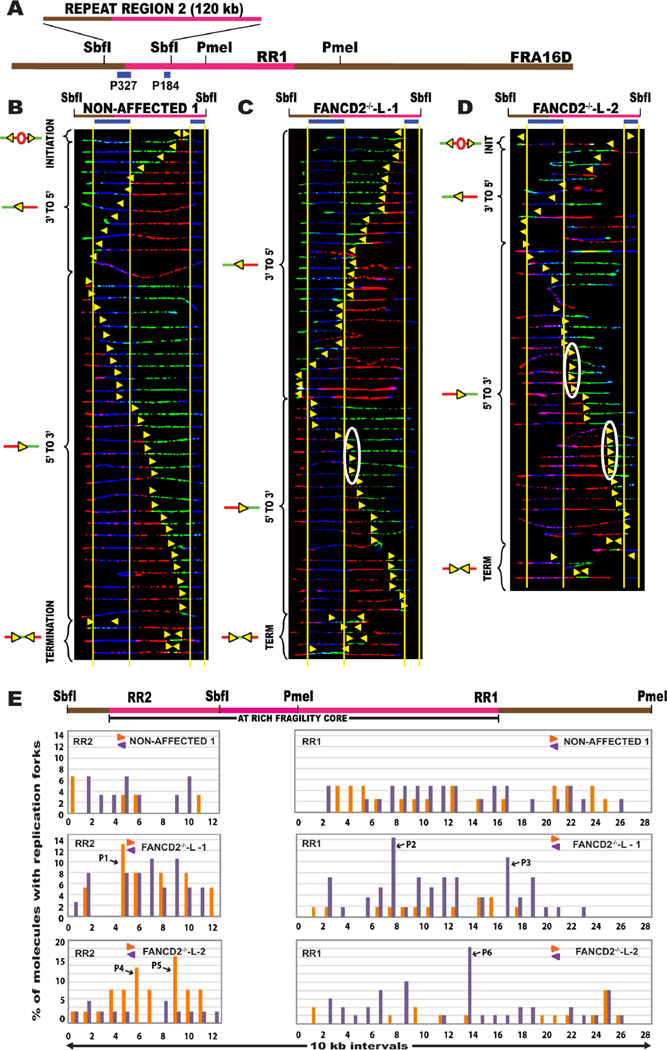
One possible explanation for the inability of the forks progressing in the 5’ to 3’ direction to reach RR1 could be that the forks were stalled at the fragility core (Fig.1A – pink line) of CFS-FRA16D, not included in RR1. To study the progression of the replication forks progressing 5’ to 3’ into RR1, we studied an adjacent segment, to the left of RR1, called the repeat region 2 (RR2) (Fig. 1A; Fig. 2A). Analysis of the replication program of RR2 in non-affected cells revealed that replication proceeds bidirectionally across the repeats in RR2 (Fig. 2B). The FANCD2−/− lymphoblasts also had a bidirectional replication program; however, there appeared to be an accumulation of replication forks at different regions along the 120 kb segment (Fig. 2C–D). Analysis of regions RR1+RR2 collectively revealed that the replication program at the AT-rich fragility core of CFS-FRA16D is altered in the absence of the FANCD2 protein.
Figure 2. DNA replication forks stall within the fragility core of CFS-FRA16D in FANCD2−/− lymphoblasts.
(A) Locus map of RR2-SbfI segment containing a portion of the AT-rich fragility core. The FISH probes that identify the segment are labeled in blue. (See also Figure S2)
(B–D) Top; Locus map of the SbfI digested RR2 segment. Middle; Aligned photomicrograph images of labeled DNA molecules from (B) Non-affected I (GM02184), (C) FANCD2−/−-L-1 (PD20) patient derived lymphoblast, and (D) FANCD2−/−-L-2 (2742) lymphoblasts. White ovals indicate regions of replication fork (yellow arrow) pausing and correspond to the pause peaks listed in Table S2.
(E) Top; Locus map of the RR1 + RR2 regions. The RR1 quantification was included here to enable the visualization of replication pausing along the complete length of the fragility core. Bottom; The percentage of molecules with replication forks at each 10 kb interval of RR2 (left, quantification of molecules shown in Fig. 3B–D) and RR1 (right, quantification of molecules shown in Fig. 2B–D) in the non-affected I (GM02184) line, FANCD2−/−-L-1 (PD20) and the FANCD2−/−-L-2 (2742) lymphoblasts. The replication forks moving in the 3’ to 5’ direction and the forks moving in the 5’ to 3’ direction are denoted by purple < and orange > colors respectively. A high percentage of molecules with replication forks in a particular 10 kb interval is indicative of fork pausing in that interval. Black arrows denote the most prominent pause peaks and correspond to the white ovals in the SMARD profile. Refer Table S2 for the coordinates of the 10 kb region corresponding to the pause peaks.
In the absence of FANCD2, replication forks stall at the fragility core of CFSs
Replication fork pausing can occur at regions of the genome that act as natural impediments to the DNA replication machinery (Mirkin and Mirkin, 2007). To determine whether the altered replication program, at RR1, observed in the absence of FANCD2 is a result of replication pausing, we decided to quantify the replication pause sites at the endogenous fragility core of CFS-FRA16D. To do this, we divided the AT-rich fragility core of CFS-FRA16D (RR1+RR2), collectively spanning a 400 kb region, into 10 kb intervals. We then counted the number of 3’ to 5’ or 5’ to 3’ progressing replication forks that were present at each 10 kb interval at the time of replication. Figure S1B is a schematic representation of how replication pausing is quantified.
In the FANCD2−/− lymphoblasts, the replication forks moving into RR1 (3’ to 5’; purple bars) (Fig. 2E: FANCD2−/−-L-1 and FANCD2−/−-L-2; Fig. S3A: FANCD2−/−-L-3) show increased pausing at the fragility core. In addition, replication forks moving in the 5’ to 3’ direction appear to pause significantly upon entering the AT-rich core, in RR2 (Fig. 2E: FANCD2−/−-L-1 and FANCD2−/−-L-2; orange bars), indicating that FANCD2 is important for replisome movement across the AT-rich regions of CFS-FRA16D.
To further validate the hypothesis that the absence of FANCD2 results in replication fork pausing at CFSs, we quantified replication pausing at the fragility core of a second CFS, FRA6E. We analyzed a 375 kb region that includes the early/late replication transition zone, that corresponds to the fragility core of FRA6E (Palumbo et al., 2010) (Fig. S4A). The SMARD results show increased replication fork pausing, preferentially within the FRA6E fragility core, in FANCD2−/−-L-1 cells (Fig. S4D), indicating FANCD2 is needed for proper replication at CFSs. Together, these results demonstrate that in the absence of FANCD2, the replication machinery finds it difficult to navigate the structure-prone fragility core of CFSs, even in the absence of exogenous replicative stress.
In the absence of FANCD2 dormant replication origins are activated at CFSs
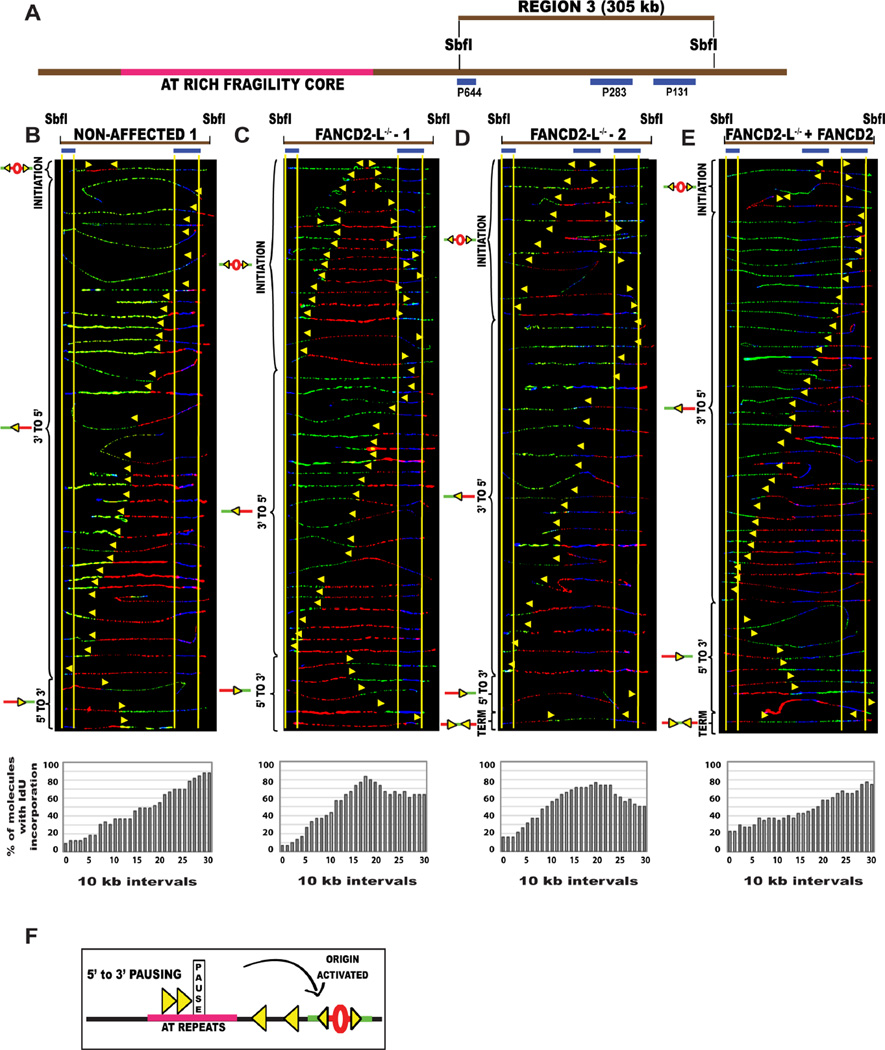
Replication fork stalling or slowing can be accompanied by the activation of dormant origins (Alver et al., 2014; Blow and Ge, 2009). In the absence of FANCD2, increased replication pausing was seen for the forks progressing in the 5’ to 3’ direction into the fragility core of both CFS-FRA16D and CFS-FRA6E (Fig 2E; Fig. S4D). This raises the possibility of identifying dormant initiation events downstream of the pause site, which might have fired to compensate for the replication stalling. Therefore, we examined the replication program of a segment flanking RR1 of FRA16D, to the right, region 3 (R3) (Fig. 1A; Fig. 3A).
Figure 3. In the absence of FANCD2, cells activate dormant origins associated with replication stalling at the AT-rich fragility core of CFS. (See also Figure S2, S4).
(A) Locus map of R3-SbfI segment that lies outside the AT-rich fragility core to the right. The FISH probes that identify the segment are labeled in blue. Combinations of two-three probes were used to identify the R3 segment.
(B–E) Top; Locus map of the SbfI digested Region 3 (R3) segment. Middle; Aligned photomicrograph images of labeled DNA molecules from (B) non-affected I (GM02184), (C) FANCD2−/−-L-1 (PD20), (D) FANCD2−/−-L-2 (2742), (E) FANCD2−/−-L-1 + FANCD2 (corrected) patient derived lymphoblast. The molecules are arranged as in Fig. 2. Bottom; The percentage of molecules incorporating IdU (red) is calculated from the replication program (middle) and is represented as a histogram.
(F) Schematic representation of dormant origin activation when replication forks pause in the 5’ to 3’ direction
Analysis of the non-affected cells revealed that only 2 of the 73 molecules displayed initiation events in the 305 kb R3 segment (Fig. 3B, Fig. S2D). In comparison, all three FANCD2−/− lymphoblasts activated a prominent dormant origin in more than a third of the R3 molecules (Fig. 3C–D; Fig. S2E). Moreover, when FANCD2 protein expression was restored in the FANCD2−/−-1 patient cells (Fig. S3B), the dormant origin activation response was suppressed in the R3 segment (Fig. 3E). Importantly, the appearance of this origin in the FANCD2-deficient cells supports the idea that pausing of forks progressing 5’ to 3’ through RR1 is accompanied by the activation of dormant origins downstream of the pause site (Fig. 3F). To further test this prediction, we analyzed the replication program of CFS-FRA6E for dormant origin activity. In non-affected lymphoblasts, we found very few initiation events occurring along the 375 kb region of CFS-FRA6E (1 out of 40 molecules in Fig. S4B, S4E; grey bar). In contrast, the FANCD2−/−-1 lymphoblasts had a prominent dormant origin activated in ~12% of the molecules (Fig. S4C, S4E; red bar), downstream of the replication pause site. The results were similar to those obtained for FRA16D, which strengthens the mechanism we propose.
Next, we wanted to determine whether the pausing observed in the 3’ to 5’ progressing forks in FRA16D resulted in the activation of dormant origins upstream of the fragility core. Thus, we analyzed region 4, adjacent to RR2 (left of RR2 in Fig. 1A), in FANCD2−/− lymphoblasts. However, we did not find any detectable dormant origin activation upstream of the repeats. Interestingly, we found some distinct replication pausing in the 5’ to 3’ direction, within 20–40 kb of the repeats (Fig. S3C–D). These results collectively indicate that in the absence of FANCD2, the activation of the origins in region 3 appears to generate the replication forks (3’ to 5’) required to complete replication of RR1 in order to compensate for the 5’ to 3’ replication stalling observed in the fragility core of CFS-FRA16D.
Replication perturbation at CFS-FRA16D is specific to FANCD2-deficient lymphoblasts, not fibroblasts
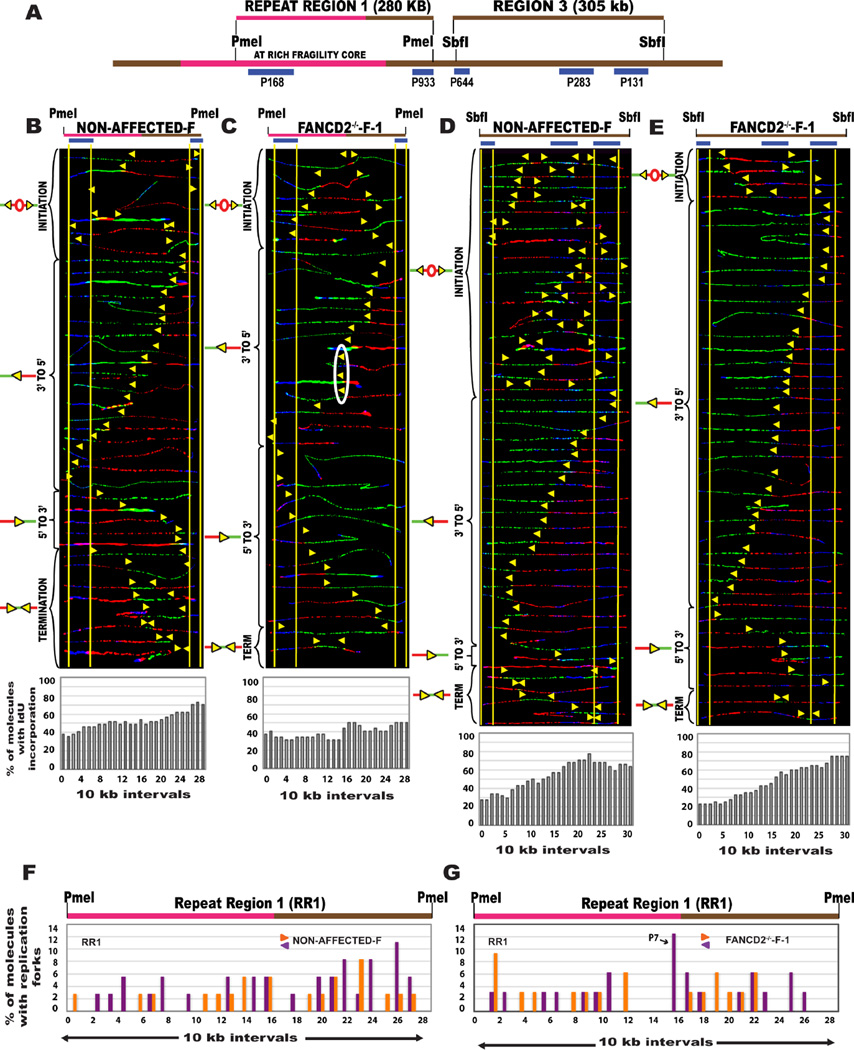
CFS instability arises as a consequence of incomplete replication that could result from the lack of replication initiation events at the locus (Letessier et al., 2011). Accordingly, CFSs are known to be less fragile/unstable in fibroblasts that have an abundance of initiation events (Durkin and Glover, 2007; Le Tallec et al., 2011). So we next asked whether the absence of FANCD2 perturbs replication at FRA16D in fibroblasts. Non-affected fibroblasts had an abundance of replication origins in both RR1 and R3 regions of FRA16D (Fig. 4A–B, 4D), in contrast to the paucity of initiation events observed in the non-affected lymphoblasts. This delineates the inherent differences in CFS-FRA16D replication between lymphocytes and fibroblasts, under unperturbed conditions. Similarly, the replication of FRA16D in FANCD2-deficient fibroblasts (FANCD2−/−-F-1) was distinctly different from that of FANCD2-deficient lymphoblasts (Fig. 4C). Despite having a bidirectional replication program, similar to non-affected fibroblasts (Fig. 4B), the FANCD2-deficient fibroblasts had some replication pausing, likely due to the repetitive DNA sequences at FRA16D (Fig. 4G).
Figure 4. The absence of FANCD2 protein affects the replication program of CFS-FRA16D in lymphoblasts but not in fibroblasts.
(A) Locus map of the RR1-PmeI and R3-SbfI segments. The FISH probes that identify the segment are labeled in blue.
(B–C) Top; Locus map of PmeI digested RR1 segment. Middle; Aligned photomicrograph images of labeled DNA molecules from (B) Non-affected-F (IMR90 fibroblast), (C) FANCD2−/−-F-1 (PD20) patient fibroblasts. The yellow arrows indicate the sites along the molecules where the IdU transitioned to CldU. The molecules are arranged as in Fig. 2. Bottom; The percentage of molecules incorporating IdU (red) is represented as a histogram.
(D–E) Top; Locus map of SbfI digested R3 segment. Middle; Aligned photomicrograph images of labeled DNA molecules from (D) Non-affected-F (IMR90 fibroblast), (E) FANCD2−/−-F-1 (PD20) patient fibroblasts.
(F–G) Top; Locus map of the RR1 region. Bottom; The percentage of molecules with replication forks at each 10 kb interval of RR1 (quantification of molecules shown in Fig. 4B–C) in (F) Non-affected-F (IMR90 fibroblast), (G) FANCD2−/−-F-1 (PD20) patient fibroblasts. Black arrows denote the most prominent pause peaks and correspond to the white ovals in the SMARD profile.
In R3 of FANCD2-deficient fibroblasts, there was a decrease in replication origins (Fig. 4E) as compared to non-affected fibroblasts (Fig. 4D). Importantly, FANCD2−/− fibroblasts did not activate the strong dormant origin observed in FANCD2−/− lymphoblasts (Fig. 3C). Complementation of FANCD2−/− fibroblasts with wild-type protein (Fig. S6B) led to an increase in replication initiation events (9 in 40 examined) at CFS-FRA16D (Fig. S6A), indicating that the ectopic expression of FANCD2 rescues the FANCD2-associated initiation defect. These results clearly demonstrate that FANCD2 deficiency prominently alters CFS replication only in cell types that express CFS (lymphoblasts) and have a paucity of replication initiation events at CFS.
Replication pausing at CFS-FRA16D, in the absence of FANCD2 is attributed to DNA:RNA hybrids
Fragility of CFS (and the cell type specific nature of these breaks) has also been attributed to differential expression of genes underlying CFS loci (Helmrich et al., 2011). In agreement with this hypothesis, we observed higher WWOX expression in lymphoblasts compared to fibroblasts (Figure S6C–D). Collision of transcription and replication machinery and DNA:RNA hybrid formation have been implicated in instability at common fragile site loci, which harbor long transcribed genes (Garcia-Muse and Aguilera, 2016; Helmrich et al., 2013). Recent reports suggest that the FA pathway plays a role in protecting cells from the deleterious effects of DNA:RNA hybrids (Garcia-Rubio et al., 2015; Schwab et al., 2015).
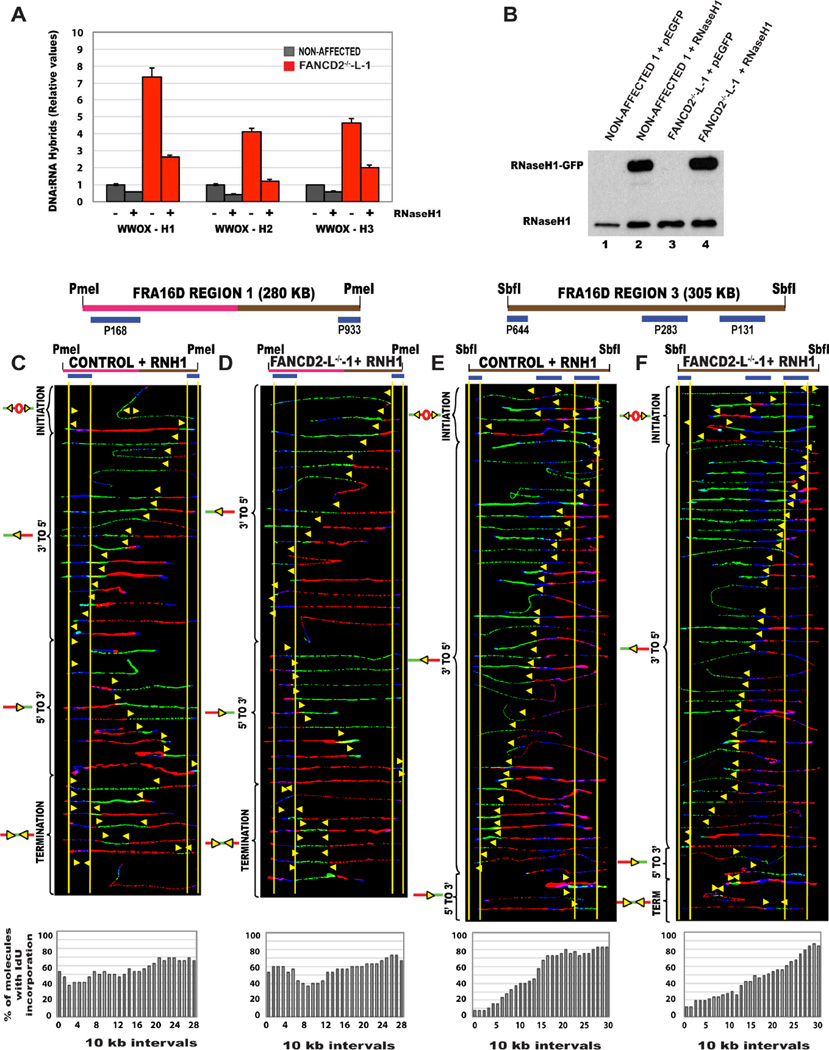
In order to determine whether the replication pausing observed in the absence of FANCD2 is due to DNA:RNA hybrid formation, we performed a DNA:RNA hybrid immunoprecipitation (DRIP) analysis at three sites (chosen based on RDIP-seq databases from (Nadel et al., 2015)) of CFS-FRA16D (Table S3). At all three sites analyzed, there was an accumulation of DNA:RNA hybrids, preferentially in the absence of FANCD2 (Fig. 5A; red bars) Treatment with RNaseH1, which cleave the RNA component of DNA:RNA hybrids, resulted in a marked reduction in the DNA:RNA hybrid signal obtained in FANCD2-deficient cells (Fig. 5A). These results demonstrate that DNA:RNA hybrids do indeed accumulate at CFS loci, in the absence of FANCD2.
Figure 5. Replication pausing at CFS-FRA16D, observed in the absence of FANCD2 is associated with the accumulation of DNA:RNA hydrids.
(A) DNA:RNA hybrid accumulation in FANCD2-deficient lymphoblasts. DRIP-qPCR using the anti-DNA:RNA hybrid S9.6 monoclonal antibody, in non-affected I (GM02184-grey bar), FANCD2−/−-L (red bars) lymphoblasts. The samples obtained by immunoprecipitation with either treated (+) or not treated (−), with RNase H1, as indicated. Signal values of DNA:DNA hybrids, immunoprecipitated in each region are normalized to input values. Data represent mean ± SEM from three independent experiments.
(B) Immunoblot analysis to detect the relative levels of the RNaseH1 protein expression in the non-affected 1 (GM02184) cells transfected with the control GFP vector (Lane 1-CONTROL + GFP), non-affected 1 (GM02184) cells transfected with the RNaseH1 overexpression vector (Lane 2-CONTROL+RNH1), FANCD2−/−-L lymphoblast cells transfected with the control GFP vector (Lane 3-FANCD2−/−-L-1+GFP) and FANCD2−/−-L lymphoblast cells transfected with the RNaseH1 overexpression vector (Lane 4-FANCD2−/−-L-1+RNH1). Proteins from whole cell extracts were separated, immunoblotted and detected with RNaseH1 antibody.
(C–D) Top; Locus map of PmeI digested RR1 segment. Middle; Aligned photomicrograph images of labeled DNA molecules from (C) non-affected I or control + RNH1, (D) FANCD2−/−-L-1+RNH1 lymphoblast. The molecules are arranged as in Fig. 2. Bottom; The percentage of molecules incorporating IdU (red) is represented as a histogram.
(E–F) Top; Locus map of the SbfI digested Region 3 (R3) segment. Middle; Aligned photomicrograph images of labeled DNA molecules from (E) Non-affected I or Control + RNH1, (F) FANCD2−/−-L-1+RNH1 lymphoblast. (See also Figure S7)
If DNA:RNA hybrids are indeed responsible for replication perturbation at CFS-FRA16D, then overexpressing RNaseH1 should eliminate the source of stalling. To test this, we generated non-affected/control and FANCD2−/−-L-1 lymphoblasts expressing either the control eGFP vector (Fig. 5B; Lanes 1,3), or the eGFP-tagged RNaseH1 vector (Lanes 2,4). The RR1 segment, in the presence of RNaseH1 over expression, was replicated by equal numbers of 5’ to 3’ and 3’ to 5’ progressing replication forks, in FANCD2−/− lymphoblasts, with no significant replication pausing (Fig. 5D). Importantly, the replication program of the RNaseH1 overexpressing FANCD2−/− lymphoblasts closely resembled the replication program of both non-affected lymphoblasts (Fig. 1C; S2B) and also the non-affected lymphoblasts overexpressing RNaseH1 (Fig. 5C). This clearly shows that eliminating DNA:RNA hybrids alleviates pausing and restores bidirectional replication fork movement across the fragility core of FRA16D.
If the overexpression of RNaseH1 truly alleviates replication pausing, it should suppress dormant origins that fire to rescue replication. Accordingly, FANCD2−/− lymphoblasts overexpressing RNaseH1 do not activate the strong dormant origin that was observed in region 3 of FRA16D in FANCD2-deficient cells and very closely resembled the replication program of non-affected cells (Fig. 5F). The few initiation events observed in both the non-affected and FANCD2-deficient lymphoblasts (Fig. 5E–F) over expressing RNaseH1 could be attributed to the presence of increased amounts of RNaseH1 protein. Furthermore, no change in the replication program was observed at the R3 region in non-affected and FANCD2−/− lymphoblasts expressing GFP control (Fig. S7A–B). Elimination of the source of replication perturbation (DNA:RNA hybrids), appears to have suppressed the need for rescue (dormant origins). These results clearly demonstrate that replication pausing at CFS-FRA16D in the absence of FANCD2 is associated with DNA:RNA hybrids.
Disruption of the FA pathway results in replication pausing at CFS-FRA16D
FANCD2's role in CFS-FRA16D replication could stem from its involvement in the FA/BRCA pathway. If this were the case, deficiency in other FA proteins would be predicted to have a similar effect on CFS-FRA16D replication as seen in the absence of FANCD2. Alternately, FANCD2 could be functioning independently of the FA/BRCA pathway to facilitate CFS-FRA16D replication. To discriminate between these possibilities, we wanted to analyze the replication program of CFS-FRA16D in other FA patient-derived lymphoblastoid lines and in the absence of FANCD2/FANCI monoubiquitination. First, we measured the relative levels of FANCD2 protein expression and found that FANCD2 protein was not expressed in all three FANCD2−/− cell lines (Fig. S5A). FANCD2 monoubiquitination was absent in cells expressing FANCI-K523R (FANCImonoub−/−) and FANCD2-K561R (FANCD2monoub−/−) (Fig. S3B; S5A).
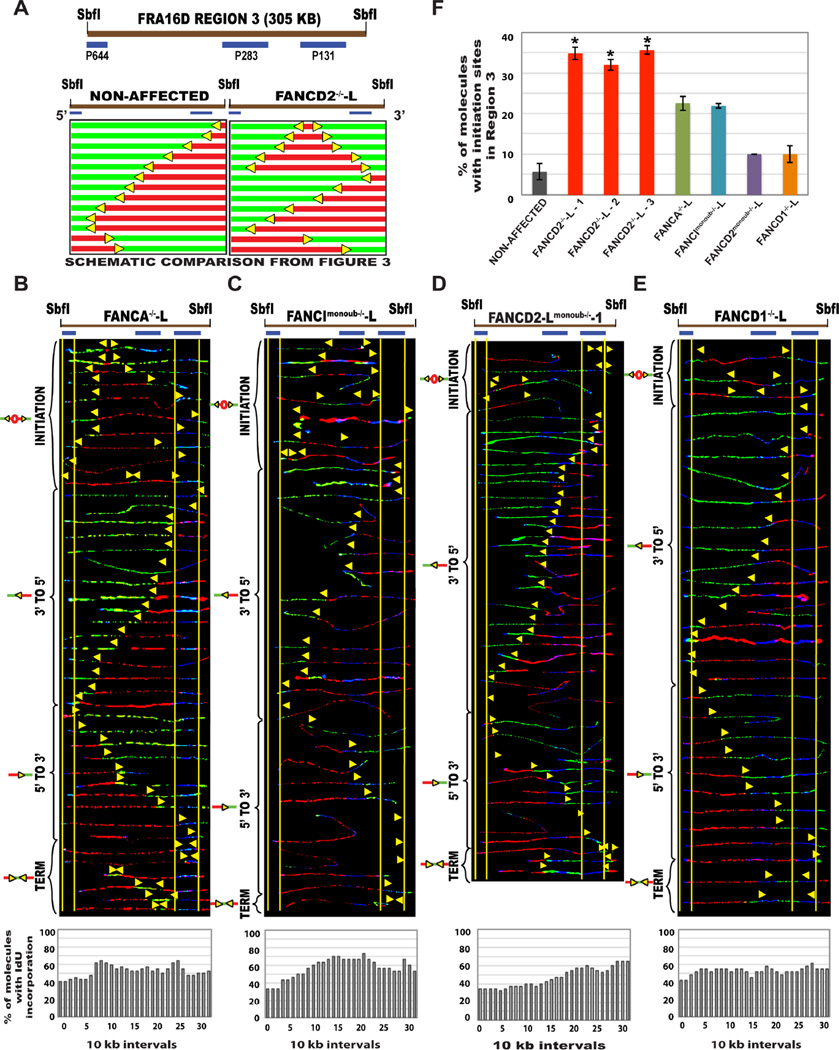
In the absence of the FA core complex protein FANCA, equal numbers of 5’ to 3’ and 3’ to 5’ progressing replication forks replicated the RR1 region of CFS-FRA16D (Fig. S5C). This was associated with a ~10% decrease in dormant initiation events (Fig. 6F, green bar), as compared to FANCD2-deficient lymphoblasts (red bars). However, in the absence of the downstream FA protein BRCA2/FANCD1, forks progressing predominantly in the 3’ to 5’ direction replicated RR1 (Fig. S5E), similar to FANCD2-deficient cells. However, both FANCA−/− and FANCD1−/− lymphoblasts had some replication fork pausing at RR1 (Fig. S5F, S5G).
Figure 6. The absence of other key FA proteins or the monoubiquitination of FANCI or FANCD2 only moderately alters CFS-FRA16D replication. (See also Figure S5).
(A) Schematic representation of results from Fig. 4 (SbfI segment), for comparison of replication program at the R3 segment of CFS-FRA16D, Top; the locus map, Bottom Left; replication profile of the non-affected-L cell line Bottom Right; replication profile of the FANCD2−/−-L lymphoblast.
(B–E) Top; Locus maps of SbfI digested R3 segment. Middle; Aligned photomicrograph images of labeled DNA molecules from (B) FANCA−/−-L, (C) FANCImonoub−/−-L, (D) FANCD2monoub−/−-L and (E) FANCD1−/−-L patient derived lymphoblast. The molecules are arranged as in Fig. 2. Bottom; The percentage of molecules incorporating IdU (red) is represented as a histogram.
(F) Percentage of molecules with initiation sites in Region 3 of non-affected I (GM02184 - grey bar), FANCD2−/−-L (red bars), FANCA−/−-L (green bar), FANCImonoub−/−-L (blue bar), FANCD2monoub−/−-L (purple bar) and FANCD1−/−-L (orange bar) patient derived lymphoblast. Error bars represent mean ± s.d. from two independent experiments (*P<0.05).
Analysis of the FANCImonoub−/− and FANCD2monoub−/− cells revealed that the RR1 region was replicated by forks progressing in both 3’ to 5’ and the 5’ to 3’ directions, in FANCImonoub−/− lymphoblasts (Fig. S5D), in contrast to FANCD2-deficient cells. Furthermore, both FANCImonoub−/− and FANCD2monoub−/− cell lines activated fewer numbers of dormant origins at R3 of CFS-FRA16D (Fig. 6C–D, 6F). In contrast to FANCD2-deficient cells, only 10% of the FANCD2monoub−/− cells activated origins in R3 of FRA16D (Fig. 6F – purple bar). Furthermore, the replication program of the FANCD2monoub−/− lymphoblasts at R3 (Fig. 6D) is similar to non-affected lymphoblasts. This indicates that the monoubiquitination of FANCD2 is not essential to facilitate CFS replication under unperturbed conditions.
Despite the replication pausing observed, the bidirectional replication fork movement at RR1 in FANCA−/− and FANCImonoub−/− lymphoblasts indicates that replication fork movement is only partly hindered in the absence of the FA core complex proteins and perhaps FANCI/FANCD2 monoubiquitination, under unperturbed conditions. The replication program in R3 further supports this idea. Interestingly, the severity of the absence of downstream FA proteins (BRCA2/FANCD1) closely resembles FANCD2 deficiency. This indeed fits nicely with the observation that FANCD2 and the downstream FA protein BRCA2/FANCD1 are involved in replication fork restart independent of the FA pathway (Raghunandan et al., 2015). In summary, these results show that while other FA proteins play a role in replication at the FRA16D locus, this role is not exactly the same as FANCD2.
Replication initiation events are strongly associated with CFS fragility
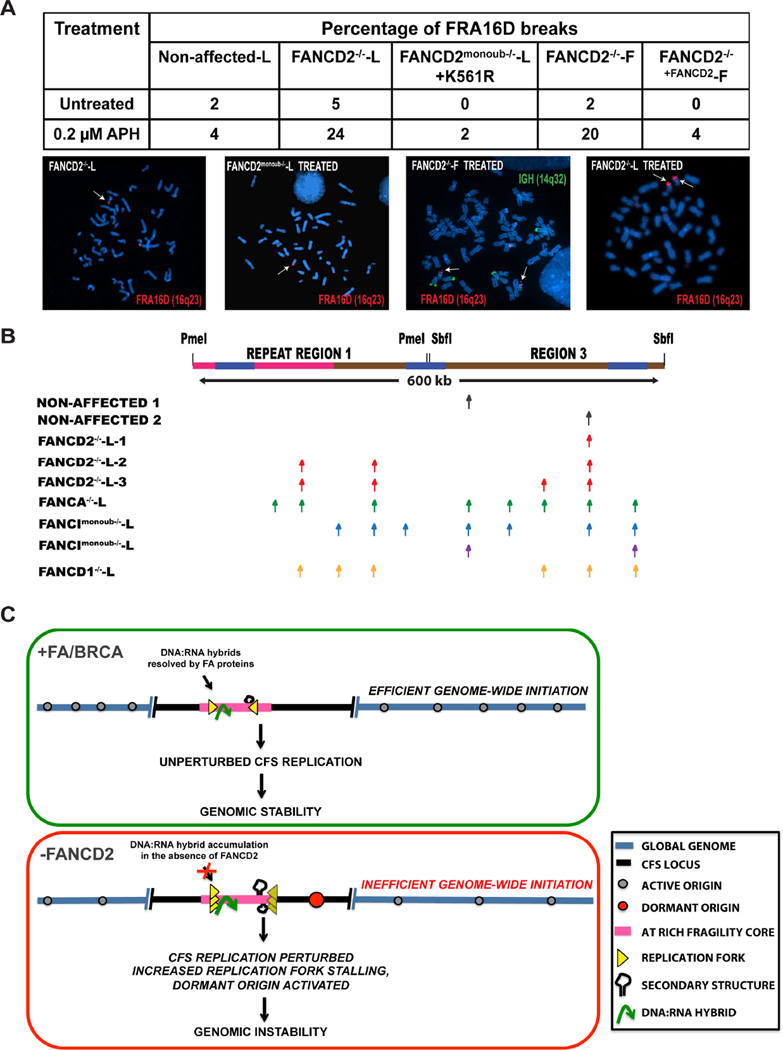
To understand the implications of FANCD2-associated CFS replication perturbation on CFS fragility, we compared CFS-FRA16D fragility in FANCD2-deficient lymphoblasts and fibroblasts. Results indicated that spontaneous FRA16D breaks accumulate in FANCD2−/− cells, even in the absence of exogenous replicative stress. Treatment with mild doses (0.2 µM) of the replication inhibitor aphidicolin (APH) resulted in a ~five-fold increase in FRA16D breaks in FANCD2-deficient lymphoblasts (Fig. 7A). In comparison, FANCD2-deficient fibroblasts (FANCD2−/−-F-1) did not display spontaneous breaks at FRA16D (Fig. 7A). However, treatment with APH resulted in a significant increase (20%) in FRA16D breaks in FANCD2−/−-F-1 cells (Fig. 7A). This is possibly due to the initiation defect observed in the R3 segment of the FANCD2−/−-F-1 cells since there is a strong correlation between the abundance of initiation events at CFS loci and fragility (Letessier et al., 2011). Furthermore, the secondary structure prone sequences at FRA16D still obstruct replication forks and lead to pausing (Fig. 4G), likely contributing to the fragility still observed in FANCD2-deficient fibroblasts (Fig. 7A). Complementation of FANCD2-deficient fibroblasts with wild-type FANCD2 suppressed FRA16D breaks.
Figure 7. FANCD2 deficiency is associated with altered replication initiation and instability at CFS loci.
(A) Cytogenetic FISH analysis using the IGH/MAF probe set, to detect and compare breaks at CFS-FRA16D in lymphoblasts and fibroblasts, in the presence and absence of Aphidicolin. The MAF probe set consist of two probes that flank the FRA16D locus such that a split signal (two red dots within a chromatid) represents a FRA16D break (modified from (Bergsagel and Kuehl, 2001)). Top; Table representing the percentage of FRA16D breaks in the presence and absence of 0.2 µM aphidicolin in Non-affected-L, FANCD2−/−-L (PD20), FANCD2−/−+FANCD2-L-1, FANCD2monoub−/−-L-1, FANCD2−/−-F-1 (PD20), FANCD2−/−+FANCD2-F-1. Bottom; Representative images of FRA16D breaks. White arrows indicate FRA16D breaks.
(B) Top; Locus map of CFS-FRA16D spanning the ~600 kb, RR1+R3 segments. Bottom; Observed sites of initiation in non-affected 1 (GM02184 – grey arrow), non-affected 2 (GM03798 - grey arrow), FANCD2−/−-L-1 (PD20) (red arrow), FANCD2−/−-L-2 (2742) lymphoblasts (red arrows), FANCD2−/−-L-3 (2717) (red arrow), FANCA−/−-L (green arrows), FANCImonoub−/−-L (blue arrows), FANCD2monoub−/−-L (purple arrows) and FANCD1−/−-L (orange arrows). (See also Figure S6)
(C) Model depicting the consequence of FANCD2 deficiency on CFS replication. Under unperturbed replicative conditions (+FA/BRCA), replication forks progressing from distantly fired origins replicate the CFS locus (black line). Upon reaching the AT-rich fragility core (pink line), replication forks efficiently replicate through WWOX transcription associated DNA:RNA hybrids (green) and AT associated secondary structures (black hairpins) likely aided by the FANCD2 protein, in association with the FA/BRCA pathway. This ensures CFS replication completion genomic stability.
In the absence of FANCD2, replication forks pause (overlapping yellow arrows) within the fragility core and this is accompanied by the activation of a dormant origin (red circle). Replication pausing likely persists due to the absence of a functional FA/BRCA pathway to resolve DNA:RNA hybrids (green arrow) and other AT associated secondary structures (black hairpins). Furthermore, the absence of FANCD2 further affects CFS replication by restraining the number of sites at which dormant origins fire, collectively leading to genomic instability.
In summary, perturbed replication at CFS in the absence of FANCD2 is associated with increased CFS fragility. The relative abundance of origins in fibroblasts (Fig. 4) is potentially one of the reasons why CFS-FRA16D is less fragile in fibroblasts, as compared to lymphoblasts. These results highlight the importance of FANCD2 to CFS-FRA16D replication, specifically in lymphoblasts, where these sites are fragile even in the absence of exogenous stress.
FANCD2 deficiency is associated with a decrease in replication initiation sites
Replication initiation events are strongly associated with CFS fragility. Despite the strong dormant origin activated to rescue replication, the FRA16D locus persistently breaks in lymphoblasts deficient for FANCD2. Furthermore, although FANCD2-deficient fibroblasts display a relatively unperturbed replication program at FRA16D, they too are fragile under stress. To understand this better, we studied the effect of FANCD2 deficiency on replication initiation by enumerating the number of sites at which initiation events occur at FRA16D. Figures 7B and S6E illustrate the locus map of the RR1+R3 segments of CFS-FRA16D and summarize the observed locations of initiation events in all eight lymphoblastoid cell lines (Fig. 7B) and fibroblast lines (Fig. S6E).
FANCD2deficiency is associated with initiation events at fewer sites of FRA16D (red arrows-Figure 7B, S6E). This alteration in replication initiation sites is consistent between lymphoblasts and fibroblasts, indicating that this FANCD2-associated replication defect likely occurs genomic wide. These results suggest that while the FA pathway proteins share some common functions in alleviating replication pausing at CFS-FRA16D, FANCD2 appears to have an additional role in replication initiation, leading to the pronounced alterations in replication program at CFSs, in FANCD2-deficient lymphoblasts.
DISCUSSION
While it has been clearly established that stress-induced replication intermediates occur at CFS (Chan et al., 2009), the replicative difficulties that lead to incomplete replication and the mechanisms that promote replication completion have been elusive. The present study provides mechanistic insight into the multifaceted role of FANCD2 in enabling efficient replication of structure-prone CFS loci, by alleviating transcription:replication-associated conflicts and by possibly ensuring efficient replication initiation.
Our results suggest a model in which FANCD2 and the other FA proteins act as facilitators of CFS replication, even under unperturbed conditions (Fig 7C). In the presence of a functional FA/BRCA pathway, forks that appear to have originated from initiation events outside the CFS loci mediate replication. Upon reaching the AT-rich fragility core, replication forks manage to efficiently replicate the region and ensure replication completion. In the absence of FANCD2, replication is perturbed at CFS even in the absence of exogenous replicative stress. This manifests as replication fork pausing at the fragility core of FRA16D, preferentially at sites of DNA:RNA hybrid accumulation, accompanied by dormant origin activation. This defect is further exacerbated by the observed reduction in the potential sites of replication initiation in the FANCD2-deficient lymphoblasts. Based on these observations we propose that FANCD2 has two unique roles in facilitating CFS replication to prevent genomic instability: (i) facilitating the movement of replication forks across secondary structures such as DNA:RNA hybrids and (ii) efficient replication initiation.
In this study, we provide in vivo evidence of replication pausing at endogenous CFS loci, in human FA patient lymphoblasts. In vitro studies suggest that [AT]n or [AT/TA]nflexible sequences found at CFSs (Glover, 2006; Zhang and Freudenreich, 2007), pose a challenge to the replicative DNA polymerase δ (Shah et al., 2010) and leads to polymerase pausing at CFS repeat sequences (Walsh et al., 2013). This implies that additional polymerases and/or accessory proteins are required for proper replication of CFS sequences. Strong candidates for this role are the FA/BRCA proteins (Howlett et al., 2005), helicases (Chaudhury et al., 2013; Kamath-Loeb et al., 2000; Pellicioli and Muzi-Falconi, 2013), translesion polymerases (Bergoglio et al., 2013; Rey et al., 2009) and nucleases (Ying et al., 2013). It has been shown that FANCD2, in association with endonucleases and Bloom syndrome helicase, resolves intermediates resulting from incomplete CFS replication in G2/M (Naim et al., 2013). However, it is possible that FANCD2’s role in preserving CFS stability begins earlier in the cell cycle, during CFS replication.
During repair, FANCD2 can recruit additional polymerases to sites of damage (Fu et al., 2013). The numerous replication pause sites observed in the absence of FANCD2 implies that it may be involved in recruiting proteins that assist in replicating CFS regions by a similar mechanism. Bidirectional replication fork movement observed in FANCD2monoub−/− and FANCImonoub−/− lymphoblasts indicates that the monoubiquitination of FANCD2 is perhaps not essential for its role in facilitating CFS replication under unperturbed conditions. However, under conditions of severe replication stress, monoubiquitination of FANCD2 may be necessary to recruit endonucleases, e.g. FAN1 (Lachaud et al., 2016), to CFS loci since the monoubiquitination of FANCD2 is critical for FA/BRCA pathway activation (Garcia-Higuera et al., 2001; Rajendra et al., 2014).
In addition to AT-associated secondary structures, transcription-associated obstacles at CFSs are a major cause of instability. DNA:RNA hybrids can lead to genomic instability by obstructing the progression of replication machinery or by making the cell more susceptible to genotoxic stress (Aguilera and Garcia-Muse, 2012). Our results not only demonstrate that DNA:RNA hybrids form at CFSs, but also show that replication forks tend to stall due to DNA:RNA hybrid accumulation. Collectively, our data suggest that transcription-associated conflicts are a major source of replication perturbation at CFSs and suggest that FANCD2 is key to alleviating these conflicts at CFSs. Interestingly, BRCA2, also prevents DNA:RNA induced genetic instability (Bhatia et al., 2014).
FANCD2 influences the efficiency of replication initiation
Under conditions of replicative stress, FANCD2 plays an important role in suppressing dormant origin firing and this function is independent of the monoubiquitination of FANCD2 (Chen et al., 2015). It is possible that this role of FANCD2 is contributing to the increased dormant origin activation (Fig. 6F - ~40%) observed in the absence of FANCD2. However, FANCD2-deficient fibroblasts from the same patient show decreased origin firing at the same genomic locus (Fig. 4E). Furthermore, similar to recent reports, the most prominent effect of an inability to monoubiquitinate FANCD2 was a decrease in dormant origin firing (Fig. 6D, (Panneerselvam et al., 2014). These results imply that in the absence of FANCD2 monoubiquitination, the activation of the dormant origin in R3 is perhaps not required to rescue replication, under unperturbed conditions.
Irrespective of cell type, all FANCD2-deficient cell lines appeared to activate origins at fewer regions at the CFS-FRA16D locus (Fig. 7B; S6E). These results delineate a role for FANCD2 in efficient replication origin firing. Since changes in origin usage can be attributed to changes in chromatin looping (Buongiorno-Nardelli et al., 1982; Courbet et al., 2008), this role of FANCD2 is possibly associated with changes to chromatin looping and/or with the histone chaperone activity of FANCD2 (Sato et al., 2012).
Fanconi anemia, CFS instability and cancer
The results from this study demonstrate that replication perturbation due to DNA:RNA hybrids and defective replication initiation, collectively contribute to CFS instability. Importantly, we propose that FANCD2 is a central regulator that overcomes these threats to CFS replication. The replication-associated functions of FANCD2 are particularly important at CFS loci, which are hypersensitive to replicative stress (Yunis et al., 1987). Through our results we propose that FANCD2 and other FA proteins protect CFS from endogenous sources of replicative stress and ensure efficient replication completion at CFS, to preserve genome integrity. However, FANCD2 and the downstream FA proteins have a more prominent role in replication at CFSs as compared to the FA core complex proteins. Given the implicated role of CFS instability in oncogenesis, our results provide vital mechanistic insights into the increased cancer risk of FA patients.
EXPERIMENTAL PROCEDURE
Cell Culture
GM02184 (non-affected 1), GM03798 (non-affected 2), GM16756 (PD20-FANCD2−/−-L-1), GM13022 (FANCA−/−-L), and GM13023 (FANCD1−/−-L) Epstein–Barr virus-transformed lymphoblasts were obtained from Coriell Cell Repositories and were grown in RPMI 1640 medium supplemented with 15% FBS. The FANCA−/−-L, FANCImonoub−/−-L, FANCD1−/−-L, 2741-FANCD2−/−-L-2, 2717-FANCD2−/−-L-3, FANCD2−/−-L-1 lymphoblasts and the complemented cell line, FANCD2−/−+FANCD2-L-2 lymphoblasts were grown in RPMI 1640 medium supplemented with 15% FBS v/v and 1 µg/ml puromycin. AG03204 (IMR90) SV40-transformed fibroblasts (Coriell Cell Repositories), PD20-F (FANCD2−/− fibroblasts) and the completed cell line, FANCD2−/−+FANCD2 fibroblasts were maintained in DMEM supplemented with 15% FBS.
Single molecule analysis of replicated DNA (SMARD)
SMARD analysis was carried our using a procedure described previously (Madireddy et al., 2016; Norio and Schildkraut, 2001)(Gerhardt et al., 2014). Briefly, exponentially growing cells were cultured in media containing 30 µM 5-iodo-2′-deoxyuridine (IdU) at 37°C for 4 h (Sigma-Aldrich, St. Louis, MO). After 4 h, the cells were centrifuged at 800 rpm for 5 min and the media containing IdU was removed. The cells were then cultured in fresh RPMI medium containing 30 µM 5-chloro-2′-deoxyuridine (CIdU) (Sigma-Aldrich, St. Louis, MO) and the cells were incubated for an additional 4 h. After 4 h, the cells were then collected by centrifugation, and they were resuspended at 3 × 107 cells per ml in PBS. The cells were then resuspended in an equal volume of molten 1% InCert agarose (Lonza Rockland, Inc., Rockland, ME) in PBS. DNA gel plugs were made by pipetting the cell-agarose mixture into a chilled plastic mold with 0.5- by 0.2-cm wells with a depth of 0.9 cm. The gel plugs were allowed to solidify on ice for 30 min. The cells in the plugs were lysed in buffer containing 1% n-lauroylsarcosine (Sigma-Aldrich), 0.5 M EDTA, and 20 mg/ml proteinase K. The gel plugs were incubated at 50°C for 3 days and were treated with fresh proteinase K at 20 mg/ml concentration (Roche Diagnostics), every 24 h. The plugs were then rinsed in Tris-EDTA (TE) and subjected to phenylmethanesulfonyl fluoride (PMSF) (Sigma-Aldrich) treatment. To prepare the cells for restriction enzyme digestion, the plugs were washed with 10 mM MgCl2 and 10 mM Tris-HCl (pH 8.0) and the genomic DNA in the gel plugs was digested with 80 units of PmeI (New England BioLabs Inc.) at 37°C overnight. The d igested gel plugs were rinsed with TE and cast into a 0.7% SeaPlaque GTG agarose gel (Lonza Rockland, Inc.) for size separation of DNA by pulse field gel electrophoresis. Gel slices from the appropriate positions in the pulsed-field electrophoresis gel were melted at 72°C for 20 min. The melted agarose was digested with GELase enzyme (Epicentre Biotechnologies 1 unit per 50 µl of agarose suspension) by incubating the GELase-DNA-agarose mixture at 45°C for 4 h. The resulting DNA was pipetted along one side of a coverslip that had been placed on top of a 3-aminopropyltriethoxysilane (Sigma-Aldrich)-coated glass slide and allowed to enter by capillary action. The DNA was denatured with sodium hydroxide in ethanol and then fixed with glutaraldehyde.
The slides containing the DNA were hybridized overnight with biotinylated probes (represented as blue bars on the CFS-FRA16D locus map). The next day, the slides were rinsed in 2 × SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) 1% SDS and washed in 40% formamide solution containing 2 × SSC at 45°C for 5 min and rinsed in 2 × SSC-0.1% IGEPAL CA-630. Following several detergent rinses (4 times in 4× SSC-0.1% IGEPAL CA-630), the slides were blocked with 1% BSA for at least 20 min and treated with Avidin Alexa Fluor 350 (Invitrogen Molecular Probes) for 20 min.
The slides were rinsed with PBS containing 0.03% IGEPAL CA-630, treated with biotinylated anti-avidin D (Vector Laboratories) for 20 min, and rinsed again. The slides were then treated with Avidin Alexa Fluor 350 for 20 min and rinsed again, as in the previous step. The slides were incubated with the IdU antibody, a mouse anti-bromodeoxyuridine (Becton Dickinson Immunocytometry Systems), the antibody specific for CldU, a monoclonal rat anti-bromodeoxyuridine (anti-BrdU) (Accurate Chemical and Scientific Corporation) and biotinylated anti-avidin D for 1 h. This was followed by incubation with Avidin Alexa Fluor 350 and secondary antibodies, Alexa Fluor 568 goat anti-mouse IgG (H+L) (Invitrogen Molecular Probes), and Alexa Fluor 488 goat anti-rat IgG (H+L) (Invitrogen Molecular Probes) for 1 h. The coverslips were mounted with ProLong gold antifade reagent (Invitrogen) after a final PBS/CA630 rinse. Fluorescence microscopy was carried out using a Zeiss fluorescence microscope to monitor the IdU/CIdU nucleoside incorporation.
DNA-RNA immunoprecipitation (DRIP)
DRIP was perfomed mainly as described (Herrera-Moyano et al., 2014) with few differences. 5×106 cells were collected, washed with PBS, resuspended in 1.6 ml of TE and treated overnight with 41.5 ml of 20 %SDS and 5 ml of proteinase K (Roche). DNA was extracted with phenol-chloroform. Precipitated DNA was spooled, washed with 70% EtOH, resuspended gently in TE and digested overnight with 50 U of HindIII, EcoRI, BsrGI, XbaI and SspI, and bovine serum albumin (BSA). For the negative control, half of the DNA was treated with 4 µl RNAse H1 (Ginno et al., 2012) (New England BioLabs) overnight. 5mg of the digested DNA was bound to 10 µl of S9.6 antibody (1mg/ml) in 500 µl binding buffer (10mM NaPO4, 140 mM NaCl, 0.05% triton X-100) overnight at 4°C. DNA-antibody complexes were immunoprecipitated using Dynabeads Protein A (invitrogen) for 2 h at 4°C and washed 3 times with binding buffer. DNA was eluted with 50mM Tris pH 8.0, 10mM EDTA, 0.5% SDS, treated for 45 min with 7 µl proteinase K at 55°C and cleaned with NucleoSpin Gel and PCR Clean-up (Macherey-Nagel). The enrichment for each qPCR of interest was normalized with respect to the corresponding ratios of the input.
Supplementary Material
Highlights.
FANCD2 regulates CFS replication even in the absence of replicative stress
Replication forks stall at endogenous CFS loci in FANCD2 patient-derived cells
DNA:RNA hybrid removal restores normal replication at CFS-FRA16D
FANCD2-deficiency is associated with altered replication initiation
Acknowledgments
We thank Paul R. Andreassen at Cincinnati Children’s Hospital Medical Center for PD20 lymphoblasts stably complemented with wild-type FANCD2 or monoubiquitination-defective FANCD2-K561R. We thank Dr. William C. Drosopoulos for editing the manuscript and Grace Wang for helping with data collection. This work was supported by NIH/NIGMS 5R01-GM045751 and Empire State Stem Cell Fund through New York State contract C024348 and the Tri-Institutional Stem Cell Initiative funded by the Starr Foundation to C.L.S., NIH/NHLBI R01HL101977 to N.G.H and Spanish Ministry of Economy and Competitiveness BFU2013-15687 and European ERC-2014-ADG 669898 TARLOOP grants to A.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
A.M and C.L.S are co-corresponding authors of this manuscript. A.M. conceived, designed, performed the experiments and analyzed the results. S.T.K. helped perform multiple SMARD experiments. J.G. and Z.Y. helped perform the FANCA SMARD experiments. M.L.G-R. performed the DRIP experiments. E.H-M. generated and pulsed lymphoblasts overexpressing RNase H1 for SMARD. E.A.V and R.A.B performed the immunoblotting experiments. A.M. and C.L.S wrote the manuscript. C.L.S., A.A, N.H, and J.G. discussed the work and edited the manuscript.
The authors have NO commercial affiliations or conflicts of interest.
REFERENCES
- Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Molecular cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Alver RC, Chadha GS, Blow JJ. The contribution of dormant origins to genome stability: from cell biology to human genetics. DNA Repair (Amst) 2014;19:182–189. doi: 10.1016/j.dnarep.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt MF, Casper AM, Glover TW. Common fragile sites. Cytogenetic and genome research. 2003;100:92–100. doi: 10.1159/000072843. [DOI] [PubMed] [Google Scholar]
- Bergoglio V, Boyer AS, Walsh E, Naim V, Legube G, Lee MY, Rey L, Rosselli F, Cazaux C, Eckert KA, et al. DNA synthesis by Pol eta promotes fragile site stability by preventing under-replicated DNA in mitosis. The Journal of cell biology. 2013;201:395–408. doi: 10.1083/jcb.201207066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsagel PL, Kuehl WM. Chromosome translocations in multiple myeloma. Oncogene. 2001;20:5611–5622. doi: 10.1038/sj.onc.1204641. [DOI] [PubMed] [Google Scholar]
- Bhatia V, Barroso SI, Garcia-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014;511:362–365. doi: 10.1038/nature13374. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Ge XQ. A model for DNA replication showing how dormant origins safeguard against replication fork failure. EMBO reports. 2009;10:406–412. doi: 10.1038/embor.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buongiorno-Nardelli M, Micheli G, Carri MT, Marilley M. A relationship between replicon size and supercoiled loop domains in the eukaryotic genome. Nature. 1982;298:100–102. doi: 10.1038/298100a0. [DOI] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nature cell biology. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- Chaudhury I, Sareen A, Raghunandan M, Sobeck A. FANCD2 regulates BLM complex functions independently of FANCI to promote replication fork recovery. Nucleic acids research. 2013;41:6444–6459. doi: 10.1093/nar/gkt348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Jones MJ, Yin Y, Crist SB, Colnaghi L, Sims RJ, 3rd, Rothenberg E, Jallepalli PV, Huang TT. ATR-mediated phosphorylation of FANCI regulates dormant origin firing in response to replication stress. Molecular cell. 2015;58:323–338. doi: 10.1016/j.molcel.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesi M, Bergsagel PL, Shonukan OO, Martelli ML, Brents LA, Chen T, Schrock E, Ried T, Kuehl WM. Frequent dysregulation of the c-maf proto-oncogene at 16q23 by translocation to an Ig locus in multiple myeloma. Blood. 1998;91:4457–4463. [PubMed] [Google Scholar]
- Ciullo M, Debily MA, Rozier L, Autiero M, Billault A, Mayau V, El Marhomy S, Guardiola J, Bernheim A, Coullin P, et al. Initiation of the breakage-fusion-bridge mechanism through common fragile site activation in human breast cancer cells: the model of PIP gene duplication from a break at FRA7I. Human molecular genetics. 2002;11:2887–2894. doi: 10.1093/hmg/11.23.2887. [DOI] [PubMed] [Google Scholar]
- Courbet S, Gay S, Arnoult N, Wronka G, Anglana M, Brison O, Debatisse M. Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature. 2008;455:557–560. doi: 10.1038/nature07233. [DOI] [PubMed] [Google Scholar]
- Durkin SG, Glover TW. Chromosome fragile sites. Annual review of genetics. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- Finnis M, Dayan S, Hobson L, Chenevix-Trench G, Friend K, Ried K, Venter D, Woollatt E, Baker E, Richards RI. Common chromosomal fragile site FRA16D mutation in cancer cells. Human molecular genetics. 2005;14:1341–1349. doi: 10.1093/hmg/ddi144. [DOI] [PubMed] [Google Scholar]
- Fu D, Dudimah FD, Zhang J, Pickering A, Paneerselvam J, Palrasu M, Wang H, Fei P. Recruitment of DNA polymerase eta by FANCD2 in the early response to DNA damage. Cell Cycle. 2013;12:803–809. doi: 10.4161/cc.23755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Molecular cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- Garcia-Muse T, Aguilera A. Transcription-replication conflicts: how they occur and how they are resolved. Nat Rev Mol Cell Biol. 2016 doi: 10.1038/nrm.2016.88. [DOI] [PubMed] [Google Scholar]
- Garcia-Rubio ML, Perez-Calero C, Barroso SI, Tumini E, Herrera-Moyano E, Rosado IV, Aguilera A. The Fanconi Anemia Pathway Protects Genome Integrity from R-loops. PLoS genetics. 2015;11:e1005674. doi: 10.1371/journal.pgen.1005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt J, Zaninovic N, Zhan Q, Madireddy A, Nolin SL, Ersalesi N, Yan Z, Rosenwaks Z, Schildkraut CL. Cis-acting DNA sequence at a replication origin promotes repeat expansion to fragile X full mutation. The Journal of cell biology. 2014;206:599–607. doi: 10.1083/jcb.201404157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Molecular cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover TW. Common fragile sites. Cancer letters. 2006;232:4–12. doi: 10.1016/j.canlet.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Hellman A, Zlotorynski E, Scherer SW, Cheung J, Vincent JB, Smith DI, Trakhtenbrot L, Kerem B. A role for common fragile site induction in amplification of human oncogenes. Cancer cell. 2002;1:89–97. doi: 10.1016/s1535-6108(02)00017-x. [DOI] [PubMed] [Google Scholar]
- Helmrich A, Ballarino M, Nudler E, Tora L. Transcription-replication encounters, consequences and genomic instability. Nat Struct Mol Biol. 2013;20:412–418. doi: 10.1038/nsmb.2543. [DOI] [PubMed] [Google Scholar]
- Helmrich A, Ballarino M, Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Molecular cell. 2011;44:966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Herrera-Moyano E, Mergui X, Garcia-Rubio ML, Barroso S, Aguilera A. The yeast and human FACT chromatin-reorganizing complexes solve R-loop-mediated transcription-replication conflicts. Genes & development. 2014;28:735–748. doi: 10.1101/gad.234070.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch B, Shimamura A, Moreau L, Baldinger S, Hag-alshiekh M, Bostrom B, Sencer S, D'Andrea AD. Association of biallelic BRCA2/FANCD1 mutations with spontaneous chromosomal instability and solid tumors of childhood. Blood. 2004;103:2554–2559. doi: 10.1182/blood-2003-06-1970. [DOI] [PubMed] [Google Scholar]
- Houghtaling S, Timmers C, Noll M, Finegold MJ, Jones SN, Meyn MS, Grompe M. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes & development. 2003;17:2021–2035. doi: 10.1101/gad.1103403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Durkin SG, D'Andrea AD, Glover TW. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Human molecular genetics. 2005;14:693–701. doi: 10.1093/hmg/ddi065. [DOI] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- Kamath-Loeb AS, Johansson E, Burgers PM, Loeb LA. Functional interaction between the Werner Syndrome protein and DNA polymerase delta. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:4603–4608. doi: 10.1073/pnas.97.9.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanja KK, Lee EH, Hendrickson EA, Campbell JL. Preventing over-resection by DNA2 helicase/nuclease suppresses repair defects in Fanconi anemia cells. Cell Cycle. 2014;13:1540–1550. doi: 10.4161/cc.28476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutler DI, Auerbach AD, Satagopan J, Giampietro PF, Batish SD, Huvos AG, Goberdhan A, Shah JP, Singh B. High incidence of head and neck squamous cell carcinoma in patients with Fanconi anemia. Archives of otolaryngology--head & neck surgery. 2003;129:106–112. doi: 10.1001/archotol.129.1.106. [DOI] [PubMed] [Google Scholar]
- Lachaud C, Moreno A, Marchesi F, Toth R, Blow JJ, Rouse J. Ubiquitinated Fancd2 recruits Fan1 to stalled replication forks to prevent genome instability. Science. 2016;351:846–849. doi: 10.1126/science.aad5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Beau MM, Rassool FV, Neilly ME, Espinosa R, 3rd, Glover TW, Smith DI, McKeithan TW. Replication of a common fragile site, FRA3B, occurs late in S phase and is delayed further upon induction: implications for the mechanism of fragile site induction. Human molecular genetics. 1998;7:755–761. doi: 10.1093/hmg/7.4.755. [DOI] [PubMed] [Google Scholar]
- Le Tallec B, Dutrillaux B, Lachages AM, Millot GA, Brison O, Debatisse M. Molecular profiling of common fragile sites in human fibroblasts. Nat Struct Mol Biol. 2011;18:1421–1423. doi: 10.1038/nsmb.2155. [DOI] [PubMed] [Google Scholar]
- Letessier A, Millot GA, Koundrioukoff S, Lachages AM, Vogt N, Hansen RS, Malfoy B, Brison O, Debatisse M. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470:120–123. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- Lossaint G, Larroque M, Ribeyre C, Bec N, Larroque C, Decaillet C, Gari K, Constantinou A. FANCD2 binds MCM proteins and controls replisome function upon activation of s phase checkpoint signaling. Molecular cell. 2013;51:678–690. doi: 10.1016/j.molcel.2013.07.023. [DOI] [PubMed] [Google Scholar]
- Lucas I, Palakodeti A, Jiang Y, Young DJ, Jiang N, Fernald AA, Le Beau MM. High-throughput mapping of origins of replication in human cells. EMBO reports. 2007;8:770–777. doi: 10.1038/sj.embor.7401026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madireddy A, Purushothaman P, Loosbroock CP, Robertson ES, Schildkraut CL, Verma SC. G-quadruplex-interacting compounds alter latent DNA replication and episomal persistence of KSHV. Nucleic acids research. 2016 doi: 10.1093/nar/gkw038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiology and molecular biology reviews : MMBR. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadel J, Athanasiadou R, Lemetre C, Wijetunga NA, P OB, Sato H, Zhang Z, Jeddeloh J, Montagna C, Golden A, et al. RNA:DNA hybrids in the human genome have distinctive nucleotide characteristics, chromatin composition, and transcriptional relationships. Epigenetics & chromatin. 2015;8:46. doi: 10.1186/s13072-015-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim V, Wilhelm T, Debatisse M, Rosselli F. ERCC1 and MUS81-EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nature cell biology. 2013;15:1008–1015. doi: 10.1038/ncb2793. [DOI] [PubMed] [Google Scholar]
- Norio P, Schildkraut CL. Visualization of DNA replication on individual Epstein-Barr virus episomes. Science. 2001;294:2361–2364. doi: 10.1126/science.1064603. [DOI] [PubMed] [Google Scholar]
- O'Keefe LV, Richards RI. Common chromosomal fragile sites and cancer: focus on FRA16D. Cancer letters. 2006;232:37–47. doi: 10.1016/j.canlet.2005.07.041. [DOI] [PubMed] [Google Scholar]
- Palumbo E, Matricardi L, Tosoni E, Bensimon A, Russo A. Replication dynamics at common fragile site FRA6E. Chromosoma. 2010;119:575–587. doi: 10.1007/s00412-010-0279-4. [DOI] [PubMed] [Google Scholar]
- Panneerselvam J, Pickering A, Han B, Li L, Zheng J, Zhang J, Zhang Y, Fei P. Basal level of FANCD2 monoubiquitination is required for the maintenance of a sufficient number of licensed-replication origins to fire at a normal rate. Oncotarget. 2014;5:1326–1337. doi: 10.18632/oncotarget.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A, Muzi-Falconi M. A blooming resolvase at chromosomal fragile sites. Nature cell biology. 2013;15:883–885. doi: 10.1038/ncb2812. [DOI] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Molecular cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunandan M, Chaudhury I, Kelich SL, Hanenberg H, Sobeck A. FANCD2, FANCJ and BRCA2 cooperate to promote replication fork recovery independently of the Fanconi Anemia core complex. Cell Cycle. 2015;14:342–353. doi: 10.4161/15384101.2014.987614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendra E, Oestergaard VH, Langevin F, Wang M, Dornan GL, Patel KJ, Passmore LA. The genetic and biochemical basis of FANCD2 monoubiquitination. Molecular cell. 2014;54:858–869. doi: 10.1016/j.molcel.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey L, Sidorova JM, Puget N, Boudsocq F, Biard DS, Monnat RJ, Jr, Cazaux C, Hoffmann JS. Human DNA polymerase eta is required for common fragile site stability during unperturbed DNA replication. Molecular and cellular biology. 2009;29:3344–3354. doi: 10.1128/MCB.00115-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg PS, Greene MH, Alter BP. Cancer incidence in persons with Fanconi anemia. Blood. 2003;101:822–826. doi: 10.1182/blood-2002-05-1498. [DOI] [PubMed] [Google Scholar]
- Sato K, Ishiai M, Toda K, Furukoshi S, Osakabe A, Tachiwana H, Takizawa Y, Kagawa W, Kitao H, Dohmae N, et al. Histone chaperone activity of Fanconi anemia proteins, FANCD2 and FANCI, is required for DNA crosslink repair. The EMBO journal. 2012;31:3524–3536. doi: 10.1038/emboj.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab RA, Nieminuszczy J, Shah F, Langton J, Lopez Martinez D, Liang CC, Cohn MA, Gibbons RJ, Deans AJ, Niedzwiedz W. The Fanconi Anemia Pathway Maintains Genome Stability by Coordinating Replication and Transcription. Molecular cell. 2015;60:351–361. doi: 10.1016/j.molcel.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SN, Opresko PL, Meng X, Lee MY, Eckert KA. DNA structure and the Werner protein modulate human DNA polymerase delta-dependent replication dynamics within the common fragile site FRA16D. Nucleic acids research. 2010;38:1149–1162. doi: 10.1093/nar/gkp1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siprashvili Z, Sozzi G, Barnes LD, McCue P, Robinson AK, Eryomin V, Sard L, Tagliabue E, Greco A, Fusetti L, et al. Replacement of Fhit in cancer cells suppresses tumorigenicity. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:13771–13776. doi: 10.1073/pnas.94.25.13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland GR, Richards RI. The molecular basis of fragile sites in human chromosomes. Current opinion in genetics & development. 1995;5:323–327. doi: 10.1016/0959-437x(95)80046-8. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D'Andrea AD. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- Walsh E, Wang X, Lee MY, Eckert KA. Mechanism of replicative DNA polymerase delta pausing and a potential role for DNA polymerase kappa in common fragile site replication. Journal of molecular biology. 2013;425:232–243. doi: 10.1016/j.jmb.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying S, Minocherhomji S, Chan KL, Palmai-Pallag T, Chu WK, Wass T, Mankouri HW, Liu Y, Hickson ID. MUS81 promotes common fragile site expression. Nature cell biology. 2013;15:1001–1007. doi: 10.1038/ncb2773. [DOI] [PubMed] [Google Scholar]
- Yunis JJ, Soreng AL, Bowe AE. Fragile sites are targets of diverse mutagens and carcinogens. Oncogene. 1987;1:59–69. [PubMed] [Google Scholar]
- Zhang H, Freudenreich CH. An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in. S. cerevisiae. Molecular cell. 2007;27:367–379. doi: 10.1016/j.molcel.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.