Abstract
Objective
To determine if the molecular profiles of endometriotic lesions contain informative measures of inflammation and immune dysfunction that may contribute to better understanding of the interplay between immune dysfunction and inflammation and their contribution to endometriosis pathogenesis.
Design
Immune and inflammation transcriptomic analysis with the use of the Nanostring nCounter GX Human Immunology V2 platform (579 human immune and inflammation–related genes and 15 housekeeping genes).
Setting
Academic university and teaching hospital.
Intervention(s)
None.
Patient(s)
Stage III–IV endometriosis patients with infertility (n = 8) and fertile disease-free control women undergoing tubal ligation (n = 8). Menstrual stage was matched to secretory phase in all participants.
Main Outcome Measure(s)
Immune and inflammation transcriptomics quantification from ectopic endometriotic lesions and matched eutopic endometrium from patients. Endometria of fertile women served as control subjects.
Result(s)
Our results displayed endometriotic lesions as molecularly distinct entities compared with eutopic endometrium and endometrium of control samples; 396 out of 579 screened immune and inflammation–related genes were significantly different in ectopic tissues compared with control endometrium. Most importantly, eutopic endometrium of the patients displayed a unique molecular profile compared with the control endometrium (91/579 genes were significantly different), particularly of genes involved in regulation of cell apoptosis and decidualization.
Conclusion(s)
We characterize differential expression of immune-inflammation genes in endometriosis patients, and show molecular distinction of eutopic endometrium of patients compared with control fertile women.
Keywords: Angiogenesis, endometriosis, immune genes, infertility, inflammation
Endometriosis is a disease driven by the inflammatory peritoneal environment (1, 2). Characterized by the growth of endometrial-like tissue in ectopic locations, endometriosis affects millions of women worldwide with chronic pelvic pain and subfertility (3). Despite decades of research, pathogenesis of endometriosis is incompletely understood, and multiple theories exist regarding its etiology (4). Since Sampson’s theory of retrograde menstruation, which proposed endometrial fragments as the potential source of endometriosis (5), numerous studies have documented the high prevalence of retrograde menstruation in women with endometriosis (6). However, that theory does not adequately explain disease prevalence, because women with and without endometriosis commonly demonstrate retrograde menstruation (7, 8), but the disease is present in only 2%–10% of women (9). Indeed, dysregulation of the immune response toward endometriotic lesions has been noted in patients, including increased inflammatory cytokines and overreactive macrophages and neutrophils in the peritoneal cavity (10, 11). Additionally, autoimmune diseases are commonly diagnosed in endometriosis patients (12), which further strengthens the notion of dysfunctional immune regulation in women with endometriosis. To provide improved therapeutic options and to increase the diagnostic power of this detrimental disease, it is imperative that we attempt to dissect the molecular profile of endometriosis pathogenesis (13). To this end, we hypothesized that endometriotic lesions have a distinct molecular profile that will provide insights into biologic mechanisms and pathways that govern the immune dysfunction and inflammation in women with endometriosis.
In this study, we profiled the expression of 579 genes involved in immunology and inflammation with the use of the nCounter GX Human Immunology v2 Kit (Nanostring Technologies), from three tissue sources: pelvic endometriotic lesions, matched eutopic endometrium from the patients, and endometrium samples from fertile (disease-free) women as control samples. All of our samples were matched to the secretory phase of the menstrual cycle. To our knowledge, this study is the first to attempt molecular profiling of differential immune and inflammation gene expression between matched ectopic tissues and eutopic endometrium from patients, ectopic and control endometrium, and eutopic and control endometrium that are also matched by menstrual cycle. Our results demonstrate dysregulation of a number of specific pathways involving leukocyte activation and cytokine-cytokine receptor interaction in endometriosis samples. In addition, our data provides insights into immune gene expression profile of eutopic endometrium of women with endometriosis compared with control endometrium samples.
MATERIALS AND METHODS
Ethics Approval
Human eutopic endometrial and ectopic endometriosis tissue samples were collected from endometriosis patients, and endometrial samples were collected from control subjects comprising healthy women after informed consent with the use of a protocol approved by the Institutional Review Committees at Greenville Health Systems, Greenville, South Carolina, and the University of North Carolina, Chapel Hill, North Carolina. Ethics approval for this study was provided by the Health Sciences Research Ethics Board, Queen's University, Kingston, Ontario, Canada.
Sample Collection from Patients and Control Women Undergoing Laparoscopic Surgery
Matched human endometrium (n = 8) and endometriosis samples (n = 8) from patients with stage III–IV endometriosis were provided by Greenville Hospital Systems, Greenville, South Carolina, after informed consent from patients. The mean age and body mass index (BMI) of the patient group were 33.1 ± 7.3 years and 24.2 ± 4.0 kg/m2, respectively. Out of the eight patients, four patients were nulligravida, three were primigravida, and one was multigravida. The stage of endometriosis was determined based on the revised American Society of Reproductive Medicine criteria (14). From each endometriosis patient undergoing laparoscopic removal of the disease, the eutopic endometrium samples were obtained by means of Pipelle sampling. The patient samples used in this study comprised women diagnosed with infertility and/or pelvic pain, and all of the women, including patients and fertile control subjects, were free from hormonal therapy for 3 months before the collection of samples. For control samples, endometrial biopsies (n = 8) were obtained by means of Pipelle sampling from healthy fertile women who underwent tubal ligation at the University of North Carolina. The mean age and BMI of the control group were 26 ± 5.8 years and 23.4 ± 2.6 kg/m2, respectively. All eight healthy control subjects had no history of pregnancy. All samples were snap-frozen with the use of liquid nitrogen and then stored at −80°C until further use. All patients and healthy control subjects were at the secretory phase of the menstrual cycle when samples were obtained.
Gene Expression Profiling with the Use of Nanostring nCounter GX Human Immunology v2 Kit
Total RNA was isolated from all samples with the use of Norgen Biotek Total RNA isolation kit (no. 17200) per the manufacturer’s instructions. In brief, tissues were homogenized in 600 µL lysate buffer by means of an electrical mortar and pestle and then were centrifuged at 15,000 rpm for 1 minute at 20°C. The resulting supernate was collected, mixed with 70% ethanol, and passed through the columns provided with the kit. The concentration and quality of RNA from each sample was assessed with the use of a Nanodrop 2000 Spectrophotometer (Thermo Scientific). All samples were normalized to 25–30 ng/µL with the use of RNAse-free distilled water. Nanostring nCounter analysis system–based gene expression profiling was performed on 100 ng total RNA from each sample as previously reported (15). Briefly. All RNA samples were subjected to analysis by means of nCounter Human Immunology v2 Panel consisting of 579 immune and inflammation–associated genes and 15 housekeeping genes as control samples in a prebuilt panel. The samples were subjected to overnight hybridization reaction at 65°C, where 5 µL of total RNA samples were combined with 20 µL of nCounter Reporter probes in hybridization buffer and 5 µL nCounter capture probes for a total reaction volume of 30 µL. Post-hybridization of probes with targets of interest in the samples, the abundance of target molecules, was quantified with the use of the nCounter Digital Analyzer and assessed with the use of the nSolver platform.
Statistical Analysis, Marker Selection Analysis, and Hierarchic Clustering with the Use of GENE-E Software
The nCounter human Immunology v2 panel included 15 housekeeping genes, eight negative and six positive control samples, that were used for background subtraction and normalization of the raw mRNA transcript counts of all samples. With the use of nSolver data analysis software provided by Nanostring technologies, the raw data were normalized with the use of the geomean of all six positive control samples, in addition to the geomean of the housekeeping genes that displayed percentage coefficient of variation (%CV) of ≤55. The list of ten housekeeping genes used in the normalization process is shown in Supplemental Table 1 (Supplemental Tables 1–6 and Supplemental Figs. 1–7 are available online at www.fertstert.org). After normalization and background noise subtraction with the use of in-house negative control samples, the data were transferred into Graphpad Prism to perform multiple t tests with correction for multiple comparisons by means of the Holm-Sidak method. For multiple t tests, alpha was set to 0.05. We further selected the 299 genes that were significantly differentially expressed between the comparisons of ectopic versus control and ectopic versus eutopic and attributed them to be ectopic-specific genes. Additionally, the 299 genes were ranked by signal-to-noise ratio by means of marker selection analysis provided by GENE-E (www.broadinstitute.org/cancer/software/GENE-E/). Heat maps for visualization of differential gene expression patterns for the comparisons of ectopic versus control, ectopic versus eutopic, and eutopic versus control were also generated for all 579 genes (Supplemental Fig. 1). We used DAVID bioinformatics resources 6.7 (https://david.ncifcrf.gov/) for KEGG pathway analysis and Gene Ontology (GO) terms. Series of two-tailed paired (ectopic vs. eutopic) or nonpaired (ectopic vs. control, eutopic vs. control) t tests were also performed on genes that showed statistical significance from multiple t tests. Outlier identifying test was conducted on comparisons of ectopic versus control, ectopic versus eutopic, and eutopic versus control, which was followed by nonparametric t test (Mann-Whitney U test) after removal of the identified outliers with the use of Graphpad Prism. Nonparametric Wilcoxon signed rank test was performed on comparisons of ectopic versus eutopic. A two-tailed P value of ≤.05 was considered to be statistically significant on the t tests. Fold difference of >1.5 was considered to have biologic significance and was taken into account when analyzing the data.
RESULTS
By means of the relatively novel method of quantifying transcriptomics with minimal sample manipulation, our study set out to investigate the differential gene transcript counts from matched ectopic endometriotic tissue samples and eutopic endometrium, as well as endometrium from healthy women, focusing on genes involved in inflammation and cellular immunity. After normalization of the raw counts based on the ten housekeeping genes and six positive control samples, distinct gene expression between comparisons were visualized with the use of heat maps for the following groups: ectopic tissues (ectopic) compared with control endometrium (control), eutopic endometrium (eutopic) compared with ectopic, and eutopic compared with control. Marker selection analysis with the use of GENE-E was then performed to rank all 579 genes from most to least differentially expressed between the comparisons, ranking them based on signal-to-noise ratio. The list of genes significantly differentially expressed in each comparisons were used to generate KEGG pathways with the use of DAVID Bioinformatics resources 6.7 (https://david.ncifcrf.gov/home.jsp). This additionally provided GO terms, which were useful in gaining insights into biologic processes of the gene clusters that showed statistical significance between comparisons.
Heat Map Analysis Shows that Ectopic Tissue is Uniquely Different from Matched Eutopic Endometrium and from Control Endometrium
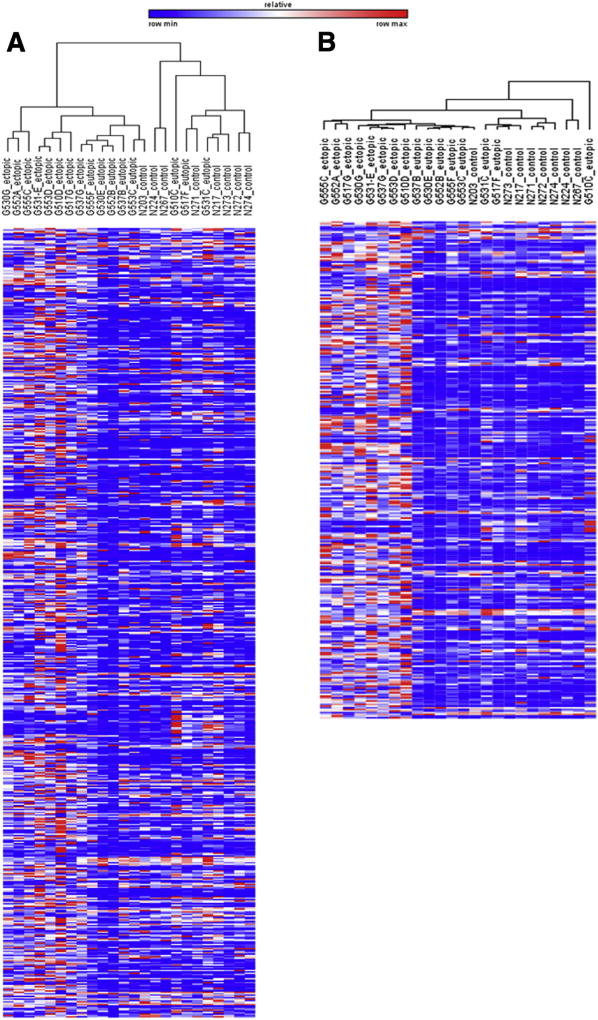
We first conducted unsupervised hierarchic clustering of 24 samples (eight ectopic, eight eutopic, and eight control) on all 579 screened genes (Fig. 1A) to cluster our samples by differential expression patterns. As seen by the dendogram, ectopic endometriotic tissues completely segregated from both eutopic endometrium and control endometrium tissues, suggesting molecular abnormality of endometriotic tissues, even from the molecular profile of a patient’s own eutopic endometrium. Surprisingly, the eutopic endometrium from patients did not cluster separately from the endometrium of control women. This may be due to the menstrual cycle being controlled to the secretory phase. To visualize differentially expressed genes further, heat maps were generated by conducting marker selection analysis with the use of GENE-E for the following comparisons: ectopic versus control, ectopic versus eutopic, and eutopic versus control (Supplemental Fig. 1). This analysis calculated the differences in gene expression with the use of test statistics and then estimated the significance of the test score. The analysis also corrected for multiple hypotheses testing by computing both the false discovery rate (FDR; Q < 0.01) and the familywise error rate (FWER). The output of the analysis ranked 579 genes from the most to least differentially expressed in each comparison.
FIGURE 1.
Unsupervised hierarchic clustering of 24 samples with the use of GENE-E software for 579 inflammation and immune-related genes. Ectopic endometrium (n = 8), eutopic endometrium (n = 8), and control endometrium samples (n = 8) were subjected to unsupervised hierarchical clustering analysis for (A) all 579 analyzed genes followed by (B) the same analysis using 299 selected genes to assess differential expression of transcriptomics in each group. The dendogram on the right-hand side shows complete segregation of the ectopic endometriotic samples (ectopic 1–8) from the eutopic endometrium (eutopic 1–8) and control endometrium tissue samples (control 1–8). The list of significantly differentially expressed genes for ectopic versus control (396 genes significantly differentially expressed) and ectopic versus eutopic (322 genes significantly differentially expressed) were further compared to identify transcriptomes that were differentially measured in ectopic endometriotic tissues only. This led to the identification of 299 genes that were commonly differentially expressed in transcript counts in the ectopic endometriotic tissue sample (B). The color scale bar denotes maximum counts in red and minimal counts in blue.
Genes Involved in Cytokine–Cytokine Receptor Interaction, Cellular Adhesion, Immune Cell Recruitment, and Apoptosis are Significantly Increased in Endometriotic Lesions
First, to profile the molecular differences in the immune and inflammation composition of the ectopic endometriotic tissues, we compared the molecular gene profile associated with ectopic tissues with that of matched eutopic endometrium and control endometrium samples. Compared with the control endometrium samples, the ectopic tissues significantly differentially expressed 396 immune and inflammation– related genes out of 579 genes screened (Supplemental Table 2). We expected the ectopic lesions to be molecularly different, but this number was surprising. Such a large amount of differentially expressed genes poses challenges to dissecting out potential biomarkers that could be used as a therapeutic strategy. To discern the list of genes that were specific to the ectopic samples, we compared the list of significantly differentially expressed genes in ectopic samples to both eutopic endometrium and control endometrium (Supplemental Fig. 2). This process produced 299 genes that we presumed to be specific to the ectopic samples and as such could be discerned as perhaps “signature genes” for ectopic samples (Fig. 1B). Inputting 299 genes into the DAVID functional annotation tool generated GO terms including immune response, positive regulation of immune system process, defense response, and inflammatory response (Supplemental Table 3). Furthermore, 299 genes clustered into cellular processes relating to increased lymphocyte and immune system process activation. These genes were also involved in KEGG pathways including cytokine-cytokine receptor interaction, primary immunodeficiency, cell adhesion molecules, and allograft rejection (Supplemental Table 4).
In particular, genes involved in cytokine–cytokine receptor signaling, including IL18, IL18R1, IL18RAP, CCL5, CCR2, and S100A8 (Fig. 2A–2F, respectively), and the expression of tumor necrosis factor (TNF) superfamily genes, including FAS, TNFRSF1B, TNFSF13B, TNFSF8, TNFSF4, TNFSF12, and CD27 (Fig. 2G–2M) were significantly increased in ectopic tissues compared with both matched eutopic endometrium from the same patients and control endometrium samples. Furthermore, genes associated with cell surface markers of immune cells, including CD4, CD45R0, CD8A, CD40, and CD48, (Fig. 2N–2R), were significantly increased in ectopic tissues compared with both eutopic endometrium and control endometrium. Additionally, expression of genes for cell adhesion molecules such as ICAM-1 and VCAM-1 were significantly increased in ectopic tissues compared with both eutopic endometrium and control endometrial samples (Fig. 2S and T). Finally, HLA genes, including HLA-DPA1, HLA-DPB1, HLA-DQA1, HLA-DRA, and HLA-DMA were also significantly increased in expression in ectopic tissues (Fig. 2U–Y).
FIGURE 2.
Representative immune genes were significantly increased in ectopic endometriotic lesions. The expression of genes encoding for (A–F) inflammatory cytokines and receptors, (G) receptor for FAS, (H–L) TNF superfamily, (M–R) cell surface markers for T-cell activation and antigen-presenting cell activation, (P and Q) chemoattractants for endothelial progenitor cell recruitment, (S and T) cell adhesion molecules, and (U–Y) HLA molecules were significantly increased in the ectopic tissues compared with the control. The graphs represent the scatter plots of all samples. The middle bar denotes the mean of the samples, and the error bars represent the standard deviation. *P≤.05 between ectopic versus eutopic and between ectopic versus control.
It is well established that endometriotic lesions recruit endothelial progenitor cells to the site to establish its own network of vasculature. Indeed, the expression of CXCL12 and PECAM-1, two genes involved in the recruitment of the endothelial progenitor cells and other immune cells to the site of damage to promote angiogenesis, were significantly increased in ectopic tissues compared with control endometrium (Supplemental Fig. 3A and 3B). Additionally, prostaglandins are associated with the promotion of pelvic pain in women with endometriosis. The gene encoding one of the receptors for prostaglandin E2, PTGER4, is significantly increased in the ectopic tissue compared to control endometrium (Supplemental Fig. 3C). Furthermore, endometriosis is often referred to as sterile inflammation, and recent research has shown involvement of inflammasome complex in the pathogenesis of endometriosis (13). Indeed, we were intrigued to find that the genes involved in the formation of inflammasome complex, NLRP3 and CASP1, were increased in expression in ectopic tissues compared with control endometrium (Supplemental Fig. 3D and 3E).
Genes Involved in Natural Killer and T-Cell Cytotoxicity, Cell Signaling, and Regulation of Inflammatory Responses are Decreased in Expression in Endometriotic Lesions
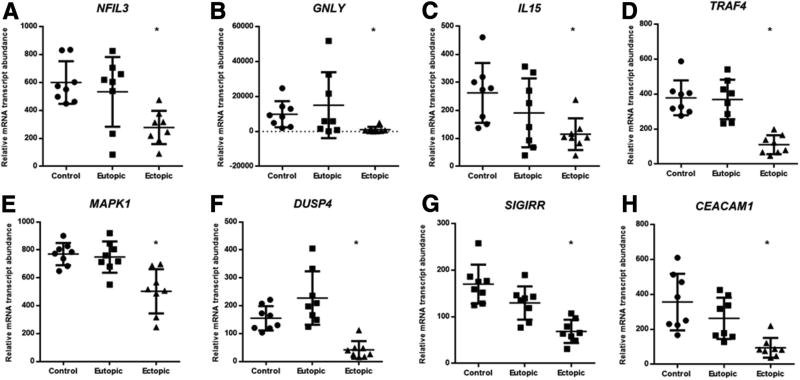
Dysfunctional cytotoxicity of natural killer (NK) cells and cytotoxic T cells toward endometrial fragments are well documented. Indeed, genes involved in NK cell differentiation, cytotoxicity, and recruitment, including NFIL3, GNLY, and IL15, were significantly decreased in expression in ectopic tissues compared with control endometrium (Fig. 3A–3C). Genes involved in TNF signaling, and mitogen-activated protein kinase signaling pathway, including TRAF4, MAPK1, and DUSP4, were significantly decreased in expression in ectopic tissues compared to control endometrium only (Fig. 3D–F). Furthermore, two genes involved in the regulation of inflammatory response in immune cells, SIGIRR and CEA-CAM-1 were also significantly decreased in ectopic tissues compared with both eutopic endometrium and control endometrium.
FIGURE 3.
Representative immune genes were significantly decreased in ectopic endometriotic lesions compared with matched eutopic endometrium. Genes involved in (A–C) the regulation of natural killer and cytotoxic T-cell activity and (D–F) the cell signaling mechanism for proliferation and apoptosis were significantly decreased in expression in ectopic tissues compared with control. The expression of genes involved in the regulation of T-cell activity and inflammation were significantly decreased in ectopic tissues compared with control (G and H). The graphs represent scatter plots of all samples. *P≤.05 between ectopic versus eutopic and between ectopic versus control.
Genes Involved in Both Classic and Alternate Complement Pathways are Aberrantly Expressed in Ectopic Tissues Compared with Control Endometrium Samples
Genes involved in complement pathways were aberrantly expressed in ectopic tissues compared with control endometrium. Genes encoding for proteins of classic and alternate complement pathways, including C1QA, C1QB, C1R, C1S, C2, C3, C4A/B, C5, C6, C7, C8A, CFB, CFH, and CFI (Supplemental Fig. 4A–4N) were significantly increased in expression, whereas C4BPA (Supplemental Fig. 4O) was significantly decreased in ectopic tissues compared with control samples.
Eutopic Endometrium of Patients Aberrantly Express Genes Involved in Regulation of Decidualization, Cellular Adhesion, Cytokine–Cytokine Receptor Interaction, and Apoptosis
To gain better understanding of the immune microenvironment in eutopic endometrium obtained from endometriosis patients, we also compared the immune transcriptome of eutopic endometrium with that of control endometrium from healthy women. Using the same approach as above, we found 91 genes out of 579 to be differentially expressed in the eutopic endometrium compared with the endometrium of the control subjects (Supplemental Table 5). Although distinct differences in the level of expression can be visualized in the heat maps comparing ectopic versus eutopic and ectopic versus control, the molecular distinction of the eutopic endometrium of patients compared with that of control endometrium was not clearly apparent (Supplemental Fig. 1). Nevertheless, a number of genes were differentially expressed in the eutopic endometrium. These included genes encoding for proinflammatory cytokines, chemokines, and receptors, such as CXCL1, CX3CL1, CXCL9, CXCL10, IL32, CXCR2, and IL7R (Supplemental Fig. 5A–5G), as well as genes encoding for cell adhesion molecules, such as ICAM3 and SELL (Supplemental Fig. 5H and 5I), which were all significantly increased in expression in eutopic endometrium of patients compared with control subjects. Furthermore, genes related to allograft rejection, including HLA-DPA1, HLA-DPB1, and HLA-DRA, were also significantly increased in expression in eutopic endometrium of patients (Supplemental Fig. 5J–5L). Additionally, 31 genes out of 91 were significantly decreased in expression in eutopic endometrium compared with control endometrium. We inputted these genes into DAVID bioinformatics resources to assess GO terms. Interestingly, the top five GO terms were related to the regulation of apoptosis, programmed cell death, regulation of cell death, regulation of cytokine production, and response to wound healing (Supplemental Table 6). The genes found in the top five GO terms were BCL6, CD44, SMAD3, CARD9, IKBKG, IL6R, PML, and RELA (Fig. 4A–4H), which were significantly decreased in expression in eutopic endometrium compared with control endometrium. Additionally, BAX, which encodes for the protein that functions with Bcl-2 as a promoter of antiapoptosis, was significantly increased in expression in eutopic endometrium compared with control endometrium. Most importantly, eutopic endometrium of patients had significant decrease in NOTCH1 and NOTCH2, genes involved in the process of decidualization, compared with control endometrium (Fig. 4I–4K).
FIGURE 4.
Genes involved in the cell signaling pathway for apoptosis and cell death were significantly decreased in the eutopic endometrium. Genes involved in (A–H) the apoptosis and cell death signaling pathway and (J and K) decidualization were significantly decreased in eutopic endometrium compared with control. (I) BAX, a gene encoding for a protein involved in apoptosis, was significantly increased in eutopic endometrium compared with control. *P≤.05.
Eutopic Endometrium Shows Dysregulated Immune Activation Compared with Control Endometrium
Genes involved in the complement pathways, including C4A/B, CFB, CFH, and CFI were aberrantly expressed in eutopic endometrium of patients compared with control endometrium (Supplemental Fig. 6A–6D). In addition to increased expression of genes encoding for inflammatory cytokines and chemokines, eutopic endometrium of women with endometriosis showed molecular expression characteristics suggesting dysregulation of immune cell regulation and cell signaling, especially B-cell signaling. We found that genes including NFKBIA, CD19, CD28, CD160, SYK, SLAMF1, SH2D1A, and STAT1 (Supplemental Fig. 6E–6L) were significantly increased, whereas genes including IKBKAP, STAT6, and STAT5B (Supplemental Fig. 6M–6O) were significantly decreased in eutopic endometrium compared with control endometrium. The gene expressions for immune cell surface markers CD40 and CD48 (Supplemental Figs. 5P and 5Q) were increased in the endometrium of patients compared with the control endometrium.
DISCUSSION
Peritoneal inflammation and immune system dysregulation are two key components that drive the pathogenesis of endometriosis (9, 16). Earlier studies have reported on the contribution of increased concentration of proinflammatory, growth-promoting, and angiogenic factors to the pathogenesis of endometriosis (17). Furthermore, aberrant activities of the immune system are emphasized in studies that show increased activation of macrophages that drive the inflammatory network (18) and suppression of NK cell cytotoxicity and differentiation (19, 20) in women with endometriosis. Studies of genome-wide association (21, 22) and microarrays have been conducted in the hopes of identifying nonhormonal therapeutic regimens and identifying biologic pathways relevant to endometriosis pathogenesis (23). Those studies, however, focused on illuminating genetic susceptibility to endometriosis development or on generalized pathways particularly targeting genes regulated by estrogen and progesterone. We conducted our study to fill in the gap of knowledge by focusing on the role of inflammation and the immune system in the pathogenesis of endometriosis. Specifically, we wanted to dissect the immune microenvironment contributing to inflammatory processes in patients with endometriosis to aid in finding the answers to pertinent questions regarding disease etiology and pathogenesis.
Primarily, ectopic tissues were significantly different in immune gene signatures compared with both matched eutopic endometrium and control endometrium. Genes encoding for proinflammatory cytokines and receptors were significantly increased in ectopic tissues compared with normal endometrial samples. Indeed, increased mRNA expression of IL18 and its receptors IL18RAP and IL18R1 was reported in adenomyosis patients (24), which may suggest an adenomyosis-like invasive phenotype of endometriosis lesions. Additionally, the expression of TNF superfamily genes was significantly increased in ectopic tissues compared with both matched eutopic endometrium and control endometrium samples. TNF receptor superfamily signaling is involved not only in apoptosis and inflammation, but also in pathways associated with proliferation, survival, and differentiation (25, 26). Furthermore, increased expression of genes associated with leukocytes, including CD4, CD45R0, CD8A, CD3D, CD48, and costimulatory molecules such as CD40, in ectopic tissues suggest that the lesions were potentially infiltrated with T cells and antigen-presenting cells. Increased expression of HLA genes in ectopic tissues compared with matched eutopic endometrium and control endometrium also suggests increased accumulation of antigen-presenting cells in ectopic tissues, which express HLA class II molecules on the surface. In addition, expression of genes for cell adhesion molecules, such as ICAM-1, VCAM-1, and SELL, were also increased, potentially aiding adhesion or invasion of ectopic tissues onto peritoneum or other surfaces. Interestingly, in concordance with the recent report by Suryawanshi et al., genes encoding for complementary proteins were significantly increased in the ectopic tissues compared with the control tissues (27). The contribution of complement proteins toward the pathogenesis of endometriosis is unknown; however, we can speculate, from its known functions, that they participate not only as a part of immune surveillance system in identifying diseased tissues, but also participate in the processes of angiogenesis, hematopoietic stem cell mobilization, and regeneration of damaged tissues (28). Not surprisingly, we also observed significant increase in PECAM1, CXCL10, and CXCL12 (genes known to play role in angiogenesis) in ectopic tissues compared with eutopic and control endometrium. Indeed, we previously reported increased expression of CXCL12 protein in human endometriotic lesions compared with matched eutopic endometrium samples and demonstrated that blocking of CXCL12 in a xenograft mouse model of endometriosis affects neovascularization and survival of endometriotic lesions (29). To this end, these immune gene profiles further support that ectopic endometriotic lesion is viewed as foreign, elicits local inflammation, establishes vasculatures, and participates in the generation of pain for some patients, as suggested by increased PTGER4 expression in ectopic tissues.
We also observed that the genes involved in regulation of inflammation, NK and cytotoxic T-cell activity, and cellular apoptosis are aberrantly expressed in the ectopic tissues. This is not surprising as endometriosis is associated with peritoneal inflammation. Currently, it is unknown whether women that develop endometriosis inherently harbor a proinflammatory peritoneal microenvironment or it is the presence of the disease that leads to the inflammatory milieu. What was interesting was to find that two novel genes previously unrelated to endometriosis, SIGIRR and CEACAM-1, were significantly decreased in expression in ectopic tissues compared with control tissues. Mice deficient in SIGIRR gene showed enhanced inflammatory response following interleukin (IL) 1 injection and showed increased response to certain Toll ligands (30). Furthermore, a recent study demonstrated the role of CEACAM-1 protein in protumor activity and that T cells deficient in CEACAM-1 gene are hyperinflammatory in nature, leading to severe experimental colitis in mice (31). Consequently, decreased expression of both SIGIRR and CEACAM-1 genes in ectopic tissues may explain the dysregulated inflammatory cytokine elevation and T-cell activation observed in endometriosis patients. Furthermore, decreased expression of NFIL3 (32), IL15 (33), and GNLY (34) in ectopic lesions support diminished cytotoxicity of NK and CD8+ T cells in women with endometriosis toward autologous endometrial fragments (35, 36). The expression of genes involved in signaling pathways, including TRAF4, MAPK1, and DUSP4, were significantly decreased in expression in ectopic tissues compared with control endometrium. These genes may be involved in the regulation of cellular proliferation and apoptosis, and may aid in the survival of the endometriotic fragments in ectopic locations.
Interestingly, gene expression of FAS, which encodes for FAS, or death, receptor, was significantly increased in ectopic tissues compared with disease-free endometrium. The significant elevation of FAS in ectopic tissues contradicts a recent study that showed, by means of immunohistochemistry, decreased FAS expression in the epithelial cells of ectopic tissues (37). As such, we attribute increased FAS expression to the resident immune cells, which are likely subjected to apoptosis by increased FasL in the peritoneal fluid of endometriosis patients (38). Overall, our data characterize endometriosis as a disease where many biologic and immune-related pathways are dysregulated.
We previously reported the potential involvement of IL-17A in the pathogenesis of endometriosis (39) and speculated that TH17 cells, or other cells of immune system with the capacity to produce IL-17A, must be contributing to the recruitment of proinflammatory immune cells into the peritoneal cavity. Interestingly, in the present study, we observed that the gene expression of RORC, which encodes for the transcription factor RORγt that determines TH17 lineage from naïve CD4+ T cells, was significantly decreased in ectopic tissues compared with disease-free endometrium (Supplemental Fig. 7A). RORC expression was also decreased in the matched eutopic endometrium of patients compared with the disease-free endometrium (Supplemental Fig. 7B). However, expression of IL-17A, which encodes for the main effector cytokine IL-17A, was significantly increased in the ectopic tissues and in the matched eutopic endometrium of patients compared with the disease-free endometrium (Supplemental Fig. 7C and 7D). These data suggest the involvement of other immune cell subtypes that express IL-17A and potentially participate in the promotion of inflammation.
To further contribute to the growing evidence showing molecular differences inherent in endometrium of women with endometriosis (40), we compared the gene expression of eutopic endometrium of patients with the expression profile obtained from the endometrium of healthy control subjects. As previously reported (41), genes encoding for cell adhesion molecules, including ICAM3 and SELL, were significantly increased in expression in the eutopic endometrium of patients compared with control subjects, which supports their enhanced capacity to adhere to the mesothelium. We also observed increased gene expression of innate immune cell chemotactic factors such as CXCL1, CXCL9, and CXCL10. However, it was interesting to find increased gene expression of CD40 and CD48, which are costimulatory molecules used by antigen-presenting cells to activate T and B cells (42). Additionally, we found increased B-cell activation–related genes, including SYK, SH2D1A, SLAMF1, and CD19, in eutopic endometrium compared with control. Increased expression of SYK, SH2D1A, and SLAMF1 is especially intriguing, because SYK plays a role in autoimmune disease development, and SH2D1A and SLAMF1 are implicated in T-cell differentiation and function and regulation of T-cell–dependent B-cell activation, respectively (43, 44). In particular, the proteins encoded by SLAMF1 and SH2D1A, CD150 and SAP, interact to promote differentiation of TH17 and TC17 cells and have been shown to contribute to the pathogenesis of autoimmunity in experimental autoimmune encephalomyelitis (45). Furthermore, interesting B-cell signaling is shown in eutopic endometrium of patients by increased expression of CD40 and SYK and decreased expression of BCL6. Down-regulation of BCL6 expression by CD40 signaling has been documented in B-cell lymphoma (46). Abnormal B-cell activity in eutopic endometrium of patients, as suggested by our data, further substantiates endometriosis as an autoimmune disease.
We also observed significant increase in HLA genes in the eutopic endometrium compared with the control endometrium, which may confer the ability to escape immune surveillance to the exuded endometrial fragments in the peritoneal cavity (47). Specifically, HLA-DP proteins are perhaps of importance because they are expressed on activated antigen-presenting cells and function as peptide-antigen presentation elements to CD4+ T cells (48). Increased expression of certain HLA genes may also contribute to the theory of endometriosis as autoimmune disease (12), and potentially explain the mechanism behind how fragments may escape the immune surveillance.
Furthermore, the eutopic endometrium of patients showed diminished spontaneous apoptosis compared with the endometrium from women without disease (49), and increased expression of antiapoptotic genes in the eutopic endometrium from patients has been previously reported (50). Here, we also observed that BAX, a gene encoding for the protein that functions with Bcl-2 as a promoter of antiapoptosis, was increased in the eutopic endometrium compared with control. Such gene expression profile presents eutopic endometrium of patients as a curious entity infiltrated with activated T, B, and dendritic cells with increased capacity for cellular adhesion and diminished potential for apoptosis with the ability to evade immune surveillance. Our study is unique regarding the nature of the patient cohort, consisting of those diagnosed with infertility, which may explain the decreased gene expression of notch signaling molecules, NOTCH1 and NOTCH2, in endometrium of patients compared with disease-free endometrium of control subjects. Our observation supports previous findings (51) that linked diminished notch signaling to defective endometrium decidualization in endometriosis patients.
Recent research into the infertility associated with endometriosis patients revealed that eutopic endometrium of patients is molecularly different compared with the endometrium of healthy control subjects. Indeed, women with endometriosis exhibit menstrual cycle irregularities and their endometria have diminished ability of decidualization (40). Our data suggests that once an endometrial fragment is exteriorized into the peritoneal cavity, it becomes pathogenic by undergoing molecular changes. It is still unknown whether the peritoneal inflammatory milieu is an inherent feature of women with endometriosis, or the presence of endometrial fragments initiates sterile inflammation that triggers endometriosis development. Clearly, aberrant molecular features of the eutopic endometrium may play a mechanistic role toward the formation of endometriotic lesions in the peritoneal cavity. By means of cDNA microarray technology and microdissection with the use of 9,600 genes/expressed sequence tags, Wu et al. (52) profiled gene expression differences between ovarian endometrioma and eutopic endometria from 12 women adjusted for menstrual phase. Despite the belief that ectopic lesions arise from the patient’s own eutopic endometrium, the researchers were surprised to find significant transcriptional differences between what used to be the same tissue in different locations. We also found in our study that out of 579 genes, 322 genes were significantly differentially expressed when eutopic endometrium was compared with ectopic tissues. Undoubtedly, the endometrium of infertile women with endometriosis, which is molecularly different from that of healthy control subjects, undergoes further changes in the peritoneal cavity that transform it to an endometriotic lesion.
CONCLUSION
One of the important limitations of this study is the use of whole-tissue lysates to discern immune dysfunction and inflammation in women with endometriosis. An endometriotic tissue contains myriads of both innate and adoptive immune cells that are actively participating in peritoneal inflammation and lesion establishment. Besides, an endometriotic lesion is composed of complex epithelial, stromal, and endothelial cells known to respond distinctly to hormonal stimuli. To truly investigate the immune aspect of pathophysiology, we need to isolate the infiltrating immune cells and compare the differences in their activation status between the disease and control cohorts. This task is challenging and to our knowledge has not been attempted as of yet. We partially addressed this aspect by consulting a paper on immune response in silico (53) that elegantly compiled a list of markers specific to immune cells, which can be used to study the immune component from nonimmune tissues. That article demonstrated that the means of identifying immune-specific markers from nonimmune tissues, such as endometriotic lesions or endometrium, are not impossible if we use the markers that are specific to immunity.
Another important issue that needs to be considered is the complexity of endometriosis in terms of stage and type of the disease, pain, age of the individual, and the impact of hormonal interventions on the disease state. The disease can definitely be diagnosed and staged with the use of surgery, adding further complexity to establishing true control samples. For these reasons, the pathophysiology of endometriosis remains elusive. Furthermore, our study specifically focused on the secretory phase of endometrium. To fully map out the course of endometriosis development throughout the menstrual cycle, we need to analyze tissue samples from both proliferative and secretary phases of the menstrual cycle to determine whether the differences in immune gene profiles are influenced by the menstrual stage/hormones. Finally, low sample numbers used in the study did not permit us to differentiate between the specific disease phenotype (ovarian endometriosis and deep infiltrating endometriosis). Therefore the conclusions derived from the findings of this study are restricted to endometriosis patients with infertility related problems.
To this end, the present study, for the first time to our knowledge, provides a molecular profile of ectopic endometriotic lesions compared with the matched eutopic tissues and control endometrium focused on genes involved in inflammation and the immune system. We show aberrant transcription of genes involved in decidualization of endometrium, which may explain why our endometriosis patients were referred for infertility-related problems. Fundamentally, our data provide critical insights into why endometrial fragments of women with endometriosis are able to implant, proliferate, establish localized blood supply, and elicit inflammatory stimulus. Our data also provide explanation, evident from molecular profiling, of why endometrial fragments of women with endometriosis are able to implant, proliferate, establish localized blood supply, and elicit inflammatory stimulus. Indeed, our data validate the presence of immune dysregulation often observed in the peritoneal fluid of women with endometriosis, and further emphasize the importance of using immune gene expression profiles as a guide to better define all the facets that contribute to the pathogenesis of endometriosis.
Supplementary Material
Acknowledgments
Supported with funds from Queen's University Principals Development Fund (grant 366705; to M.K.), Canadian Institutes of Health Research (CIHR 394924; C.T.), and National Institutes of Health (NIH R01-067721; S.L.Y. and B.A.L.).
Footnotes
S.H.A. has nothing to disclose. K.K. has nothing to disclose. S.L.Y. has nothing to disclose. B.A.L. has nothing to disclose. M.K. has nothing to disclose. C.T. has nothing to disclose.
References
- 1.Podgaec S, Abrao MS, Dias JA, Jr, Rizzo LV, de Oliveira RM, Baracat EC. Endometriosis: an inflammatory disease with a TH2 immune response component. Hum Reprod. 2007;22:1373–9. doi: 10.1093/humrep/del516. [DOI] [PubMed] [Google Scholar]
- 2.Lousse JC, van Langendonckt A, Defrere S, Ramos RG, Colette S, Donnez J. Peritoneal endometriosis is an inflammatory disease. Front Biosci (Elite Ed) 2012;4:23–40. doi: 10.2741/e358. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy S, Berqqvist A, Chapron C, d'Hooghe T, Dunselman G, Greb R, et al. ESHRE Special Interest Group for Endometriosis and Endometrium Guideline Development Group. ESHRE guideline for the diagnosis and treatment of endometriosis. Hum Reprod. 2005;20:2698–704. doi: 10.1093/humrep/dei135. [DOI] [PubMed] [Google Scholar]
- 4.Vercellini P, Vigano P, Somigliana E, Fedele L. Endometriosis: pathogenesis and treatment. Nat Rev Endocrinol. 2014;10:261–75. doi: 10.1038/nrendo.2013.255. [DOI] [PubMed] [Google Scholar]
- 5.Sampson AJ. The development of the implantation theory for the origin of peritoneal endometriosis. Am J Obstet Gynecol. 1940;40:549–57. [Google Scholar]
- 6.d'Hooghe T, Debrock S. Endometriosis, retrograde menstruation and peritoneal inflammation in women and in baboons. Hum Reprod Update. 2002;8:84–8. doi: 10.1093/humupd/8.1.84. [DOI] [PubMed] [Google Scholar]
- 7.Halme J, Hammond MG, Hulka JF, Raj SG, Talbert LM. Retrograde menstruation in healthy women and inpatients with endometriosis. Obstet Gynecol. 1984;64:151–4. [PubMed] [Google Scholar]
- 8.Bartosik D, Jacobs SL, Kelly LJ. Endometrial tissue in peritoneal fluid. Fertil Steril. 1986;46:796–800. doi: 10.1016/s0015-0282(16)49813-4. [DOI] [PubMed] [Google Scholar]
- 9.Giudice LC, Kao LC. Endometriosis. Lancet. 2004;364:1789–99. doi: 10.1016/S0140-6736(04)17403-5. [DOI] [PubMed] [Google Scholar]
- 10.Kyama CM, Debrock S, Mwenda JM, d'Hooghe TM. Potential involvement of the immune system in the development of endometriosis. Reprod Biol Endocrinol. 2003;1:123. doi: 10.1186/1477-7827-1-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrington JL, Bruner-Tran KL, Lucas JA, Osteen KG. Immune interactions in endometriosis. Expert Rev Clin Immunol. 2011;7:611–26. doi: 10.1586/eci.11.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenberg VH, Zolti M, Soriano D. Is there an association between autoimmunity and endometriosis? Autoimmun Rev. 2012;11:806–14. doi: 10.1016/j.autrev.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Han SJ, Jung SY, Wu S-P, Hawkins SM, Park MJ, Kyo S, et al. Estrogen receptor β modulates apoptosis complexes and the inflammasome to drive the pathogenesis of endometriosis. Cell. 2015;163:960–74. doi: 10.1016/j.cell.2015.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.American Society for Reproductive Medicine. Revised American Society for Reproductive Medicine classification of endometriosis: 1996. Fertil Steril. 1997;67:817–21. doi: 10.1016/s0015-0282(97)81391-x. [DOI] [PubMed] [Google Scholar]
- 15.Koti M, Siu A, Clement I, Bidarimath M, Turashvili G, Edwards A, et al. A distinct pre-existing inflammatory tumour microenvironment is associated with chemotherapy resistance in high-grade serous epithelial ovarian cancer. Br J Cancer. 2015;112:1215–22. doi: 10.1038/bjc.2015.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKinnon BD, Bertschi D, Bersinger NA, Mueller MD. Inflammation and nerve fiber interaction in endometriotic pain. Trends Endocrinol Metab. 2015;26:1–10. doi: 10.1016/j.tem.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Lebovic DI, Mueller MD, Taylor RN. Immunobiology of endometriosis. Fertil Steril. 2001;75:1–10. doi: 10.1016/s0015-0282(00)01630-7. [DOI] [PubMed] [Google Scholar]
- 18.Beste MT, Pfaffle-Doyle N, Prentice EA, Morris SN, Lauffenburger DA, Isaacson KB, et al. Molecular network analysis of endometriosis reveals a role for c-Jun-regulated macrophage activation. Sci Transl Med. 2014;222:1–13. doi: 10.1126/scitranslmed.3007988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu MY, Yang JH, Chao KH, Hwang JL, Yang YS, Ho HN. Increase in the expression of killer cell inhibitory receptors on peritoneal natural killer cells in women with endometriosis. Fertil Steril. 2000;74:1187–91. doi: 10.1016/s0015-0282(00)01592-2. [DOI] [PubMed] [Google Scholar]
- 20.Kang YJ, Jeung IC, Park A, Park YJ, Jung H, Kim TD, et al. An increased level of IL-6 suppresses NK cell activity in peritoneal fluid of patients with endometriosis via regulation of SHP-2 expression. Hum Reprod. 2014;29:2176–89. doi: 10.1093/humrep/deu172. [DOI] [PubMed] [Google Scholar]
- 21.Nyholt DR, Low SK, Anderson CA, Painter JN, Uno S, Morris AP, et al. Genome-wide association meta-analysis identifies new endometriosis risk loci. Nat Genet. 2012;44:1355–9. doi: 10.1038/ng.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rahmioglu N, Nyholt DR, Morris AP, Missmer SA, Montgomery GW, Zondervan KT. Genetic variants underlying risk of endometriosis: insights from meta-analysis of eight genome-wide association and replication datasets. Hum Reprod Update. 2014;20:702–16. doi: 10.1093/humupd/dmu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fung JN, Rogers PA, Montgomery GW. Identifying the biological basis of GWAS hits for endometriosis. Biol Reprod. 2015;92:1–12. doi: 10.1095/biolreprod.114.126458. [DOI] [PubMed] [Google Scholar]
- 24.Huang H-Y, Yu H-T, Chan S-H, Lee C-L, Wang H-S, Soong Y-K. Eutopic endometrial interleukin-18 system mRNA and protein expression at the level of endometrial-myometrial interface in adenomyosis patients. Fertil Steril. 2010;94:33–9. doi: 10.1016/j.fertnstert.2009.01.132. [DOI] [PubMed] [Google Scholar]
- 25.Wajant H, Pfizenmaier K, Scheurich P. Tumor necrosis factor signaling. Cell Death Differ. 2003;10:45–65. doi: 10.1038/sj.cdd.4401189. [DOI] [PubMed] [Google Scholar]
- 26.Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol. 2003;3:745–56. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- 27.Suryawanshi S, Huang X, Elishaev E, Budiu RA, Zhang L, Kim S, et al. Complement pathway is frequently altered in endometriosis and endometriosis-associated ovarian cancer. Clin Cancer Res. 2014;20:6163–74. doi: 10.1158/1078-0432.CCR-14-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–97. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Virani A, Edwards SK, Thomas R, Childs T, Tayade C. Blocking of stromal cell–derived factor-1 reduces neoangiogenesis in human endometriosis lesions in a mouse model. Am J Reprod Immunol. 2013;70:386–97. doi: 10.1111/aji.12134. [DOI] [PubMed] [Google Scholar]
- 30.Wald D, Qin J, Zhao Z, Qian Y, Naramura M, Tian L, et al. SIGIRR, a negative regulator of Toll-like receptor-interleukin 1 receptor signaling. Nat Immunol. 2003;4:920–7. doi: 10.1038/ni968. [DOI] [PubMed] [Google Scholar]
- 31.Huang Y-H, Zhu C, Kondo Y, Anderson AC, Gandhi A, Russell A, et al. CEA-CAM1 regulates TIM-3–mediated tolerance and exhaustion. Nature. 2015;517:386–90. doi: 10.1038/nature13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seillet C, Huntington ND, Gangatirkar P, Axelsson E, Minnich M, Brady HJ, et al. Differential requirement of Nfil3 during NK cell development. J Immunol. 2014;192:2667–76. doi: 10.4049/jimmunol.1302605. [DOI] [PubMed] [Google Scholar]
- 33.Allavena P, Giardina G, Bianchi G, Mantovani A. IL-15 is chemotactic for natural killer cells and stimulates their adhesion to vascular endothelium. J Leukoc Biol. 1997;61:729–35. doi: 10.1002/jlb.61.6.729. [DOI] [PubMed] [Google Scholar]
- 34.Deng A, Chen S, Li Q, Lyu SC, Clayberger C, Krensky AM. Granulysin, a cytolytic molecule, is also a chemoattractant and proinflammatory activator. J Immunol. 2005;174:5243–8. doi: 10.4049/jimmunol.174.9.5243. [DOI] [PubMed] [Google Scholar]
- 35.Thiruchelvam U, Wingfield M, O'Farrelly C. Natural killer cells: key players in endometriosis. Am J Reprod Immunol. 2015;74:291–301. doi: 10.1111/aji.12408. [DOI] [PubMed] [Google Scholar]
- 36.Slabe N, Meden-Vrtovec H, Verdenik I, Kosir-Pogacnik R, Ihan A. Cytotoxic T cells in peripheral blood in women with endometriosis. Geburtshilfe Frauenheilkd. 2013;73:1042–8. doi: 10.1055/s-0033-1350702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sbracia M, Valeri C, Antonini G, Biagiotti G, Pacchiarotti A, Pacchiarotti A. Fas and Fas-ligand in eutopic and ectopic endometrium of women with endometriosis: the possible immune privilege of ectopic endometrium. Reprod Sci. 2016;23:81–6. doi: 10.1177/1933719115594019. [DOI] [PubMed] [Google Scholar]
- 38.Selam B, Kayisli UA, Akbas GE, Basar M, Arici A. Regulation of FAS ligand expression by chemokine ligand 2 in human endometrial cells. Biol Reprod. 2006;75:203–9. doi: 10.1095/biolreprod.105.045716. [DOI] [PubMed] [Google Scholar]
- 39.Ahn SH, Edwards AK, Singh SS, Young SL, Lessey BA, Tayade C. IL-17A contributes to the pathogenesis of endometriosis by triggering proinflammatory cytokines and angiogenic growth factors. J Immunol. 2015;195:2591–600. doi: 10.4049/jimmunol.1501138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brosens I, Brosens JJ, Benagiano G. The eutopic endometrium in endometriosis: are the changes of clinical significance? Reprod Biomed Online. 2012;24:496–502. doi: 10.1016/j.rbmo.2012.01.022. [DOI] [PubMed] [Google Scholar]
- 41.Vigano P, Gaffuri B, Somigliana E, Busacca M, Blasio AMD, Vignali M. Expression of intercellular adhesion molecule (ICAM)-1 mRNA and protein is enhanced in endometriosis versus endometrial stromal cells in culture. Mol Hum Reprod. 1998;4:1150–6. doi: 10.1093/molehr/4.12.1150. [DOI] [PubMed] [Google Scholar]
- 42.Klyushnenkova EN, Li L, Armitage RJ, Choi YS. CD48 delivers an accessory signal for CD40-mediated activation of human B cells. Cell Immunol. 1996;174:90–8. doi: 10.1006/cimm.1996.0297. [DOI] [PubMed] [Google Scholar]
- 43.Colonna L, Catalano G, Chew C, d'Agati V, Thomas JW, Wong FS, et al. Therapeutic targeting of Syk in autoimmune diabetes. J Immunol. 2010;185:1532–43. doi: 10.4049/jimmunol.1000983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hron JD, Caplan L, Gerth AJ, Schwartzberg PL, Peng SL. SH2D1A regulates T-dependent humoral autoimmunity. J Exp Med. 2004;200:261–6. doi: 10.1084/jem.20040526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang YH, Tsai K, Ma C, Vallance BA, Priatel JJ, Tan R. SLAM-SAP signaling promotes differentiation of IL-17 producing T cells and progression of experimental autoimmune encephalomyelitis. J Immunol. 2014;193:5841–53. doi: 10.4049/jimmunol.1301435. [DOI] [PubMed] [Google Scholar]
- 46.Saito M, Gao J, Basso K, Kitagawa Y, Smith PM, Bhagat G, et al. A signaling pathwaymediatingdownregulationofBCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell. 2007;12:280–92. doi: 10.1016/j.ccr.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 47.Liu H, Lang JH. Is abnormal eutopic endometrium the cause of endometriosis? The role of eutopic endometrium in pathogenesis of endometriosis. Med Sci Monit. 2011;17:RA92–9. doi: 10.12659/MSM.881707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fleischhauer K. Immunogenetics of HLA-DP—A new view of permissible mismatches. N Engl J Med. 2015;373:669–72. doi: 10.1056/NEJMe1505539. [DOI] [PubMed] [Google Scholar]
- 49.Meresman FG, Vighi S, Buquet RA, Contreras-Ortiz O, Tesone M, Rumi LS. Apoptosis and expression of Bcl-2 and Bax in eutopic endometrium from women with endometriosis. Fertil Steril. 2000;74:760–6. doi: 10.1016/s0015-0282(00)01522-3. [DOI] [PubMed] [Google Scholar]
- 50.Burney RO, Talbi S, Hamilton AE, Vo KC, Nyegaard M, Nezat CR, et al. Gene expression analysis of endometrium reveals progesterone resistance and candidate susceptibility genes in women with endometriosis. Endocrinology. 2007;148:3814–26. doi: 10.1210/en.2006-1692. [DOI] [PubMed] [Google Scholar]
- 51.Su R-W, Strug MR, Joshi NR, Jeong J-W, Miele L, Lessey BA, et al. Decreased notch pathway signaling in the endometrium of women with endometriosis impairs decidualization. J Clin Endocrinol Metab. 2015;100:E433–42. doi: 10.1210/jc.2014-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu Y, Kajdacsy-Balla A, Strawn E, Basir Z, Halverson G, Jailwala P, et al. Transcriptional characterizations of differences between eutopic and ectopic endometrium. Endocrinology. 2006;147:232–46. doi: 10.1210/en.2005-0426. [DOI] [PubMed] [Google Scholar]
- 53.Abbas D, Baldwin AR, Ma Y, Ouyang W, Gurney A, Martin F, et al. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 2005;6:319–31. doi: 10.1038/sj.gene.6364173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.