Abstract
Despite intense efforts, the cure rates of childhood and adult solid tumors are not satisfactory. Resistance to intensive chemotherapy is common, and targets for molecular therapies are largely undefined. We have now found that the majority of childhood solid tumors, including rhabdoid tumors, neuroblastoma, medulloblastoma and Ewing sarcoma, express an active DNA transposase PGBD5 that can promote site-specific genomic rearrangements in human cells. Using functional genetic approaches, we found that mouse and human cells deficient in non-homologous end joining (NHEJ) DNA repair cannot tolerate the expression of PGBD5. In a chemical screen of DNA damage signaling inhibitors, we identified AZD6738 as a specific sensitizer of PGBD5-dependent DNA damage and apoptosis. We found that expression of PGBD5, but not its nuclease activity-deficient mutant, was sufficient to induce hypersensitivity to AZD6738. Depletion of endogenous PGBD5 conferred resistance to AZD6738 in human tumor cells. PGBD5-expressing tumor cells accumulated unrepaired DNA damage in response to AZD6738 treatment, and underwent apoptosis in both dividing and G1 phase cells in the absence of immediate DNA replication stress. Accordingly, AZD6738 exhibited nanomolar potency against the majority of neuroblastoma, medulloblastoma, Ewing sarcoma and rhabdoid tumor cells tested, while sparing non-transformed human and mouse embryonic fibroblasts in vitro. Finally, treatment with AZD6738 induced apoptosis and regression of human neuroblastoma and medulloblastoma tumors engrafted in immunodeficient mice in vivo. This effect was potentiated by combined treatment with cisplatin, including significant anti-tumor activity against patient-derived primary neuroblastoma xenografts. These findings delineate a therapeutically actionable synthetic dependency induced in PGBD5-expressing solid tumors.
Keywords: PGBD5, AZD6738, ATR, ATM, neuroblastoma, medulloblastoma, Ewing sarcoma, rhabdoid tumors, DNA repair, targeted therapy
Introduction
In spite of the improvements in intensive combination chemotherapy, surgery and radiotherapy, the treatment of the majority of childhood and adult solid tumors remains inadequate. For example, neuroblastomas and medulloblastomas characterized by amplifications of the MYCN and MYC oncogenes, respectively, remain mostly fatal (1–3). Likewise, cancers defined by mutations of the genes encoding the SWI/SNF chromatin remodeling complex, such as rhabdoid tumors, are almost uniformly incurable (4). Finally, the majority of human sarcomas, if they cannot be removed completely by surgery, such as Ewing sarcoma for example, tend to be chemotherapy resistant and lethal (5). The majority of refractory childhood solid tumors are characterized by mutations of factors that regulate gene expression or complex genomic rearrangements, both of which are not generally amenable to current pharmacologic strategies. Thus, new therapeutic approaches are urgently needed to improve the cure rates for these patients.
To enhance the current therapeutic index, synthetic cellular relationships have been leveraged for cancer therapy (6). For example, tumors with inefficient homologous recombination DNA repair due to mutations of BRCA1/2 exhibit synthetic lethality with inhibitors of poly ADP-ribose polymerases (PARPs), enabling significant improvements in the treatment of patients as a result of clinical PARP inhibitors (7, 8). In addition, synthetic dependencies in metabolic function (9), chromatin remodeling (10), and DNA damage signaling (11–13), are beginning to be explored to develop improved targeted therapies. In particular, intrinsic DNA damage due to oncogene or replication stress such as MYC (14), and tumorigenic deficiencies in the DNA damage response due to mutations of TP53, ATM or ATR have been found to confer susceptibility to specific inhibitors of DNA damage repair signaling (15). However, these mutations are generally rare in pediatric cancers, and little is known about therapeutically targetable synthetic dependencies in childhood solid tumors.
Recently, the human piggyBac transposable element derived 5 (PGBD5) was identified as an active DNA transposase that is able to mobilize synthetic DNA transposons in human cells (16). PGBD5-mediated DNA transposition requires the DDD catalytic triad in the PGBD5 transposase domain and specific DNA recognition sequences and target sites (16). In particular, PGBD5 was found to be expressed in the majority of childhood solid tumors, including refractory rhabdoid tumors, where it promotes site-specific genomic rearrangements and mutations of tumor suppressor genes at least in part due to the aberrant targeting of its DNA nuclease activity (17). This tumorigenic nuclease activity of PGBD5 raises the possibility that PGBD5-expressing cells may depend on active DNA damage repair and signaling.
Here, we report that PGBD5 activity confers a functional dependence on the KU complex that binds DNA double-strand breaks (DSBs), and the ATR and ATM kinases that control DNA damage repair signaling in cells. We found that PGBD5 activity is sufficient to confer this synthetic dependence, and endogenous PGBD5 expression is necessary to render childhood solid tumor cells susceptible to inhibitors of DSB repair signaling. As a result, its pharmacologic targeting using selective inhibitors of DNA damage signaling exhibits therapeutic activity in multiple preclinical models of neuroblastoma, medulloblastoma, Ewing sarcoma, and rhabdoid tumors that express PGBD5 in vitro and in vivo. The availability of clinical-grade inhibitors of DNA damage signaling offers immediate potential for translation into clinical trials for patients with refractory childhood solid tumors, the majority of which express PGBD5, as well as distinct subsets of PGBD5-expressing adult solid cancers.
Results
PGBD5-expressing cells do not tolerate deficiency of non-homologous end-joining DNA repair
Eukaryotic DNA transposases rely on cellular DNA repair mechanisms to restore intact target sites upon DNA rearrangements (18). In mammalian cells, this activity is principally carried out by the classic non-homologous end joining (NHEJ) DSB repair apparatus (19). NHEJ repair consists of the heterodimeric KU70/KU80 complex that binds DSB ends, and the DNA damage repair signaling factors including the ataxia telangiectasia mutated (ATM) and ATM- and Rad3-related (ATR) kinases that in concert lead to chromatin reorganization and DNA ligase-mediated DSB repair (20). Depending on the specific molecular features of DNA damage and cell state, assembly of different repair complexes can lead to activation of specific signaling pathways (21–23). This suggests that distinct forms of intrinsic DNA damage in cancer cells can be used selectively for their synthetic lethal targeting.
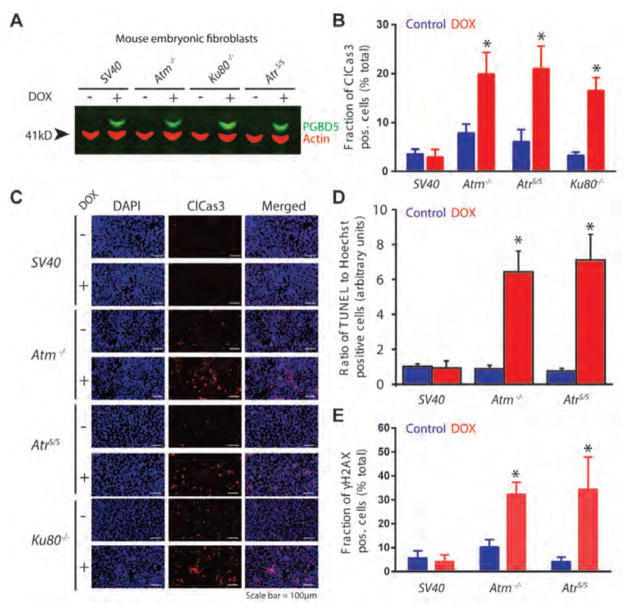
To test the cellular DNA repair requirements of PGBD5, we used mouse embryonic fibroblasts (MEFs) from mice deficient for Ku80−/−, Atm−/−, and hypomorphic Seckel syndrome allele of AtrS/S, and immortalized with the SV40 large T antigen (24–26) (Fig. 1A). Similar experiments using Lig4−/− MEFs could not be performed because of their severe proliferation defect (data not shown) (27). We used a doxycycline-inducible transgene encoding human PGBD5, and confirmed equal PGBD5 protein expression upon doxycycline induction using Western immunoblotting (Fig. 1A). Wild-type SV40 large T antigen-immortalized MEFs exhibited no measureable changes in cell growth upon doxycycline-induced PGBD5 expression (Figs. 1B–E). In contrast, Atm−/−, AtrS/S, and Ku80−/− MEFs underwent cell death, as detected by the significant accumulation of cleaved caspase 3 (p = 1.0e-2, 8.0e-3, and 1.0e-3, respectively, Figs. 1B–C), terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL; p = 1.0e-3 and 2.0e-3, respectively, Fig. 1D), and histone H2AX S139 phosphorylation (γH2AX; p = 3.0e-3 and 2.0e-2, respectively, Fig. 1E). Deficiency of Ku80 that functions in direct DSB binding during NHEJ DNA repair exhibited similar levels of cell death as the respective deficiencies of Atm and Atr that contribute to the activation and propagation of DNA damage signaling (Fig. 1B). Thus, PGBD5 expression requires the cellular NHEJ and DNA damage signaling apparatus.
Figure 1. PGBD5-expressing cells do not tolerate deficiency of non-homologous end-joining DNA repair.
(A) Western blot of PGBD5 protein expression after induction with doxycycline (500 ng/ml for 24 hours) of SV40 large T antigen-immortalized mouse embryonic fibroblasts deficient for Atm−/−, AtrS/S or Ku80−/−. Actin serves as loading control. (B) Induction of apoptosis as measured by cleaved caspase-3 staining of mouse embryonic fibroblasts deficient DNA repair factors upon doxycycline-induced (48 hours) expression of PGBD5 (red) as compared to control PBS-treated cells (blue). *p = 0.010, 0.008, and 0.0010 for Atm−/−, AtrS/S, and Ku80−/− of doxycycline vs. control, respectively. (C) Representative photomicrographs of mouse embryonic fibroblasts upon doxycycline-induced PGBD5 expression for 48 hours (+) as compared to PBS-treated controls (−), as stained for DAPI (blue) and cleaved caspase-3 (red). Scale bar = 100 μm. (D–E) Induction of DNA DSB as measured by TUNEL staining (D) and H2AX (E) of mouse embryonic fibroblasts deficient DNA repair factors upon doxycycline-induced (48 hours) expression of PGBD5 (red) as compared to control PBS-treated cells (blue). *p = 0.0010 and 0.0020 for Atm−/− and AtrS/S of doxycycline vs. control, respectively (D). *p = 0.0030 and 0.020 for Atm−/− and AtrS/S of doxycycline vs. control, respectively (E). Error bars represent standard deviations of three biologic replicates.
PGBD5 expression is sufficient to confer susceptibility to inhibition of DNA damage repair signaling
Based on the finding that cells expressing PDBD5 are dependent on active NHEJ DNA damage repair and signaling, we hypothesized that PGBD5 expression might render cells susceptible to pharmacological inhibition of DNA damage signaling. To test this idea, we used primary human retinal pigment epithelial (RPE) cells immortalized with telomerase that allow the investigation of DNA damage response in genomically stable, non-transformed human cells (28). We reasoned that specific PGBD5-dependent DNA damage response requirements may be identified by comparative analysis of growth and survival of RPE cells expressing GFP-PGBD5, as compared to GFP control and its catalytically inactive GFP-PGBD5 D168A/D194A/D386A mutant. This mutant is expressed equally to wild-type PGBD5, as assessed by Western immunoblotting, and physically associates with chromatin, as assessed by chromatin immunoprecipitation and DNA sequencing (ChIP-seq), but does not support DNA transposition in reporter assays (17). To minimize the possible contribution of secondary effects of PGBD5 expression due to its induction of mutagenic DNA rearrangements and cell transformation, we used cells for experiments immediately after transgene transduction (17).
Thus, we screened a panel of commercially available inhibitors of DNA damage signaling in their ability to selectively interfere with the growth and survival of RPE cells expressing GFP-PGBD5 as compared to wild-type or control transgene expressing cells (Fig. 2A & S1, Table S1). In particular, we observed that the ATR- and ATM-selective kinase inhibitors AZD6738 and KU60019 exhibited more than 20-fold and 5-fold enhanced activity, respectively, against RPE cells expressing GFP-PGBD5 as compared GFP control (Fig. 2A). Consistent with the notion that tolerance of PGBD5 DNA nuclease activity requires active DNA repair in cells, mutation of the aspartate catalytic triad (D168A, D194A, D386A) thought to catalyze phosphodiester bond hydrolysis during transposase-induced DNA rearrangements (16), completely abrogated the enhanced susceptibility of RPE cells upon PGBD5 expression (Fig. 2B). We confirmed that the activity of AZD6738 is at least in part due to its selective inhibition of ATR as compared to ATM (29, 30), insofar as Atm−/− and AtrS/S MEFs exhibited respectively increased and diminished susceptibilities to AZD6738, in agreement with the known relationship between ATM and ATR kinase signaling (11–13) (IC50 = 0.023 and 2.0 μM, respectively, as compared to 0.36 μM for wild-type control; Fig. 2C). Thus, PGBD5-dependent effects may be explained by the selective inhibition of DNA damage signaling by ATR- and ATM-selective AZD6738 and KU60019, respectively. Lack of PGBD5-dependent effects of other albeit potent DNA damage signaling inhibitors is presumably related to their respective selectivity profiles, such as for example AZ20 which potently inhibits ATM, ATR and mTOR, and VE-822 (VX-970) which inhibits both ATR and ATM kinases (Table S1).
Figure 2. PGBD5 expression is sufficient to confer sensitivity to DNA damage signaling inhibition.
(A) Ratios of 50% inhibitory concentrations (IC50) upon 120 hours of drug treatment with the DNA damage signaling inhibitors AZD6738, KU60019, AZ20, VE822, and AZD7762 of RPE cells expressing GFP-PGBD5 or GFP control. Ratios of 1 indicate equal susceptibility of GFP-PGBD5 as compared to control GFP-expressing cells. (B) Dose response cell viability curves of RPE cells (blue) expressing GFP-PGBD5 (red) as compared to GFP control (green) or catalytically inactive mutant (D168A/D194A/D386A, black) GFP-PGBD5 treated with AZD6738 for 120 hours. (C) Dose response cell viability curves of wild-type MEFs immortalized with SV40 large T antigen (red), as compared to Atm−/− (brown) and AtrS/S (blue) treated with AZD6738 for 120 hours. (D) Representative photomicrographs of RPE cells upon treatment with 500 nM AZD6738 for 72 hours (+) as compared to DMSO-treated controls (−), and expression of GFP, GFP-PGBD5, or inactive mutant (D168A/D194A/D386A) GFP-PGBD5, as stained for DAPI (blue) and γH2AX (red). Scale bar = 100 μm. (E) Flow cytometric analysis of TUNEL and propidium iodide staining of RPE cells expression GFP-PGBD5 as compared to control GFP or inactive mutant (D168A/D194A/D386A) GFP-PGBD5, and treated with 500 nM AZD6738, as compared to DMSO control for 48 hours. Percentages of TUNEL-positive cells are labeled as indicated. (F–H) Induction of apoptosis and DNA damage, as measured by cleaved caspase 3 (F), γH2AX (G), and TUNEL staining (H) of RPE cells treated with 500 nM AZD6738 for 48 hours (red), as compared to DMSO control (blue). Expression of GFP-PGBD5 as compared to control GFP leads to significant induction of caspase 3 cleavage, γH2AX, and TUNEL *p = 0.00030, 0.040, and 5.6 x 10−6, respectively. Mutation of the catalytic D168A/D194A/D386A (inactive mutant) GFP-PGBD5 rescues this effect **p = 0.030, 0.010, and 1.5 x 10−5, respectively. Expression of GFP-PGBD5 but not its inactive mutant or control GFP causes significant increase in TUNEL staining in the absence of AZD6738 treatment ***p = 1.2 x 10−6. Error bars represent standard deviations of three biologic replicates.
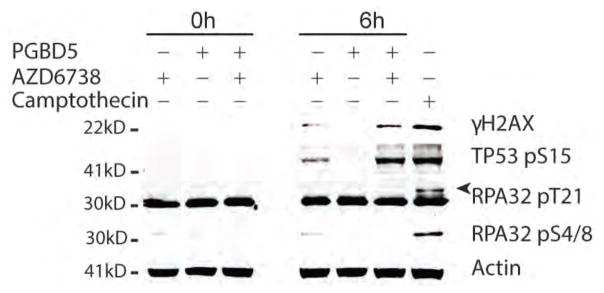
Importantly, as with the functional genetic requirements of DNA repair and DNA damage signaling induced by PGBD5 expression in MEFs (Fig. 1), we found that active DNA damage signaling blocked by AZD6738 is also required for the growth and survival of PGBD5-expressing human RPE cells (Figs. 2D–H). Specifically, we found that RPE cells expressing GFP-PGBD5, but not those expressing GFP or inactive mutant GFP-PGBD5, exhibited significantly increased DNA damage upon treatment with AZD6738, as measured γH2AX staining analysis (p = 0.040, Fig. 2D & G). Similarly, we observed significantly increased TUNEL labeling in RPE cells expressing GFP-PGBD5 upon treatment with AZD6738, whereas cells expressing control GFP or the catalytically inactive GFP-PGBD5 mutant exhibited unperturbed steady-state background TUNEL levels (p = 5.6 x 10−6, Fig. 2E & H). Consistent with the notion that wild-type PGBD5 is actively inducing DSBs in cells, we observed significantly increased TUNEL levels even in the absence of AZD6738 treatment, an effect that was completely abolished by the mutation of its putative nuclease catalytic triad (Figs. 2E & H). Likewise, TUNEL accumulation upon PGBD5 expression and AZD6738 treatment was predominantly observed in G1 phase of the cell cycle (Fig. 2E). In accordance with the accumulation of unrepaired DNA damage upon AZD6738 treatment in PGBD5-expressing cells, we observed significantly increased levels of apoptosis, as measured by caspase 3 cleavage, in cells expressing GFP-PGBD5, as compared to those expressing its inactive mutant or GFP control (p = 3.0e-4, Fig. 2F). Consistent with the lack of apparent effect of AZD6738 on TUNEL levels in S phase cells (Fig. 2E), we confirmed that the PGBD5-specific susceptibility to AZD6738 was not immediately associated with DNA replication stress, as assessed by Western immunoblotting of phosphorylated RPA32 T21 and S4/8, as compared to the DNA topoisomerase I inhibitor camptothecin that predominantly induces DSBs during DNA replication in S phase (31) (Fig. 3). In agreement with this, RPE cells expressing GFP-PGBD5 did not show increased cell cycling as compared to cells expressing GFP control, as measured by 5-ethynyl-2′-deoxyuridine (EdU) incorporation (Fig. S2). Lastly, in agreement with site-specific accumulation of DSBs (32), we observed mostly punctate in contrast to pan-nuclear γH2AX accumulation in PGBD5-expressing cells treated with AZD6738 (Fig. S3). Thus, PGBD5 expression is sufficient to confer specific susceptibility to pharmacologic inhibitors of DNA damage signaling such as AZD6738.
Figure 3. Treatment with AZD6738 induces PGBD5-dependent DNA damage.
Western immunoblot of RPE cells expressing GFP-PGBD5 or GFP and treated with 500 nM AZD6738 for 6 hours, as indicated. PGBD5 expression increases levels of H2AX and TP53 pS15 upon combination with AZD6738 treatment, but not RPA32 pT21 or pS4/8. Actin serves as loading control, and treatment with 1.5 μM camptothecin for 2 hours serves as positive control for replication stress-mediated induction of RPA32 phosphorylation. Arrow head marks the specific RPA32 pT21 band.
Rhabdoid tumor, medulloblastoma, neuroblastoma, and Ewing sarcoma cells that express PGBD5 exhibit enhanced sensitivity to AZD6738
Previously, we observed that PGBD5 is expressed in the majority of childhood solid tumors, including neuroblastoma, medulloblastoma, Ewing sarcoma, and rhabdoid tumors (17). In particular, in rhabdoid tumors, we found that the DNA recombinase activity of PGBD5 was necessary and sufficient to induce genomic rearrangements in both rhabdoid tumor cell lines and patient tumors. We have now found that expression of PGBD5 in non-transformed mouse embryonic fibroblasts and human RPE cells is sufficient to induce DNA damage as measured by TUNEL incorporation in cells, which can be potentiated by DNA damage signaling inhibitor AZD6738. Thus, we reasoned that AZD6738 may exhibit anti-tumor activity against childhood solid tumor cells expressing PGBD5.
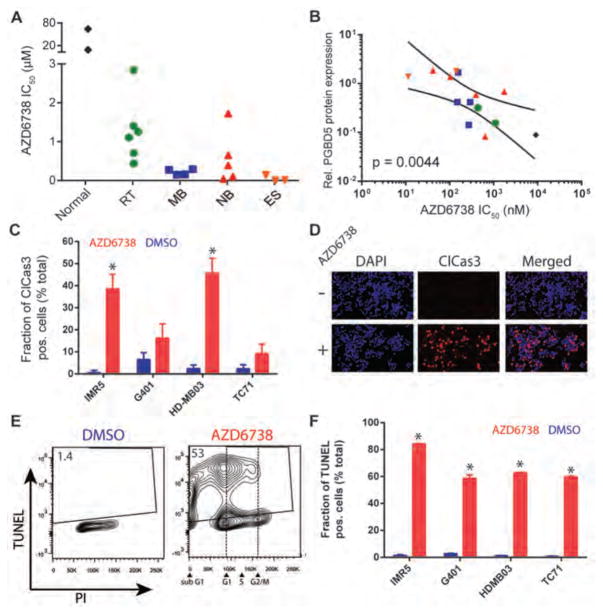
To test this idea, we treated a panel of rhabdoid tumor, neuroblastoma, medulloblastoma and Ewing sarcoma cell lines, as well as non-transformed human RPE and BJ cells with AZD6738. We observed that 19 childhood tumor cell lines tested exhibited enhanced sensitivity to AZD6738, with 50% inhibitory concentrations (IC50) largely in the nanomolar range (Fig. 4A & S4, Table S2). In contrast, non-transformed RPE and BJ cells were relatively resistant to AZD6738, with IC50 values in the high micromolar range (Fig. 4A & S4). Indeed, the susceptibility to AZD6738, as measured by its IC50 values in vitro, exhibited a significant correlation with the level of PGBD5 protein expression, as assessed by quantitative fluorescent Western immunoblotting (p = 4.4e-3, Fig. 4B, S4, S5). This association did not appear to segregate with tumor tissue type (Fig. 4B), or the presence of mutations in genes known to affect DNA damage signaling, such as TP53, ATM, ATR, MYC, MYCN, XRCC3, XRCC5, CHK1, BRCA2, RAS, and ATRX (Fig. S5 and Table S2). In addition, since ATM deficiency can confer increased sensitivity to AZD6738 (12), we confirmed that PGBD5 expression did not affect the expression of ATM itself in RPE cells, and that human tumor cells exhibiting enhanced susceptibility to AZD6738 lacked ATM mutations and retained ATM protein expression (Fig. S6 and Table S2).
Figure 4. PGBD5-expressing rhabdoid tumor, medulloblastoma, neuroblastoma, and Ewing sarcoma cells exhibit enhanced susceptibility to AZD6738.
(A) Susceptibility to AZD6738, as shown by its 50% inhibitory concentration (IC50) upon 120 hour treatment of wild-type MEFs and BJ fibroblasts (Normal), rhabdoid tumor (RT), medulloblastoma (MB), neuroblastoma (NB), and Ewing sarcoma (ES) cell lines. Complete list of cell lines and their dose-response growth curves are shown in Figs. S1 & S3. (B) PGBD5 protein expression as measured by Western immunoblotting (Fig. S4) is significantly associated with the susceptibility to AZD6738 in pediatric tumor cell lines. Lines denote the 95% confidence interval of linear regression (p = 0.0044). Points are labeled according to the color scheme in (A). (C–F) Induction of apoptosis and DNA damage, as measured by caspase 3 cleavage (C–D), and TUNEL (E–F) of IMR5 neuroblastoma, G401 rhabdoid tumor, HD-MB03 medulloblastoma, and TC71 Ewing sarcoma cells with 500 nM AZD6738 for 72 hours (red) as compared to DMSO control (blue). *p = 6.1 x 10−4 and 3.9 x 10−4 for AZD6738 versus DMSO, of IMR5 and HD-MB03, respectively. (D) Representative photomicrographs of IMR5 neuroblastoma cells after treatment with 500 nM AZD6738 (+) or DMSO control (−) for 72 hours, stained for cleaved caspase-3 (red) and DAPI (blue). Scale bar = 100 μm. (E) Representative flow cytometric profile of TUNEL and propidium iodide incorporation into HD-MB03 cells after treatment with 500 nM AZD6738 or DMSO control for 48 hours. (F) Induction of TUNEL upon treatment with 500 nM AZD6738 (red) versus DMSO control (blue) for 48 hours. *p = 0.042, 0.025, 5.17 x 10−9, and 1.98 x 10−5 for IMR5, G401, HDMB03 and TC71, respectively. Error bars represent standard deviations of three biologic replicates.
In agreement with observations of MEFs and RPE cells expressing PGBD5, human tumor cell lines expressing endogenous PGBD5 and treated with AZD6738 underwent apoptosis and accumulated unrepaired DNA damage, as measured by caspase 3 cleavage and TUNEL staining (Figs. 4C–F). Most of the TUNEL incorporation induced by AZD6738 in PGBD5-expressing tumor cells was observed in G1 cells (Figs. 4E & S7). Thus, AZD6738 exhibits anti-tumor efficacy against PGBD5-expressing childhood solid tumors in vitro.
Endogenous PGBD5 is necessary to confer enhanced susceptibility of tumor cells to inhibitors of DNA damage signaling
Since ectopic PGBD5 expression may induce DNA damage and signaling dependencies not present in human tumors with endogenous PGBD5, we sought to determine if endogenous PGBD5 expression is required for the enhanced susceptibility to inhibitors of DNA damage signaling. To achieve this, we identified two independent lentiviral RNA interference vectors expressing short hairpin RNA (shRNA) against human PGBD5 that substantially depleted endogenous PGBD5 both at the mRNA and protein levels, as compared to control vectors targeting GFP (Fig. 5A) (17). The degree of PGBD5 depletion appeared to depend on tumor type, with rhabdoid G401 cells exhibiting more than 15-fold reduction in mean PGBD5 levels, whereas neuroblastoma IMR5 and medulloblastoma HD-MB03 cells exhibited reduced effects (Fig. 5A). We found that depletion of PGBD5 induced significant resistance to AZD6738 as compared to wild-type cells or cells transduced with control vector targeting GFP (shGFP), as evidenced by the relative increase in the IC50 values of rhabdoid tumor G401, neuroblastoma IMR5, and medulloblastoma HD-MB03 cells (Fig. 5B–D, Table S3). Likewise, depletion of endogenous PGBD5 also conferred relative resistance to the ATM-selective inhibitor KU60019, supporting the notion that PGBD5 induces intrinsic DNA damage and requires ongoing DNA damage signaling in tumor cells (Figure S8). Notably, PGBD5-dependent resistance to inhibition of DNA damage signaling appeared to depend on tumor type, consistent with the notion that intrinsic differences in DNA repair and DNA damage signaling may impact therapeutic effects of targeting PGBD5-induced DNA repair dependencies. Thus, PGBD5 expression is necessary to confer susceptibility to inhibitors of DNA damage signaling such as AZD6738 in diverse childhood solid tumors.
Figure 5. PGBD5 expression is necessary for tumor cell susceptibility to AZD6738.
(A) PGBD5 mRNA expression in cells transduced with shRNA against PGBD5, as compared to shGFP and untransduced control cells. (B–D) Dose-response of childhood tumor cell lines HDMB-03 (B), IMR5 (C) and G401 (D) treated with varying concentrations of AZD6738 for 120 hours upon PGBD5 depletion. Error bars represent standard deviations of three replicates.
AZD6738 induces DNA damage, apoptosis, and exhibits anti-tumor efficacy in xenograft models of high-risk human neuroblastoma and medulloblastoma in vivo
Compelled by the finding that AZD6738 induced PGBD5-dependent DNA damage and apoptosis in pediatric solid tumor cell lines in vitro, we set out to test whether single-agent AZD6738 treatment has anti-tumor activity in preclinical models of pediatric solid tumors in vivo. First, we chose to investigate its activity against mouse xenografts of high-risk MYCN-amplified neuroblastoma (IMR5), high-risk MYC group 3 medulloblastoma (HD-MB03), refractory rhabdoid tumor (G401) and Ewing sarcoma (TC-71) cells, as they represent the most common refractory childhood solid tumors (33, 34). Thus, we transplanted IMR5, HD-MB03, G401, TC-71 cells subcutaneously in athymic nude Foxn1nu immunodeficient mice, and monitored tumor growth upon oral treatment of mice with 50 mg/kg/day of AZD6738 (Fig. 6). We found that AZD6738 significantly impaired the growth of both neuroblastoma IMR5 and medulloblastoma HD-MB03 tumors, as compared to vehicle control treated mice in vivo (p = 4.9e-3 and 5.5e-6 at day 28 respectively, Fig. 6A & B). We did not observe significant toxicity of this treatment, as evidenced by the unchanged animal body weights (Fig. S9). The magnitude of this effect appeared as substantial as compared to the previously reported effects of AZD6738 against tumors with genetic deficiencies of ATM, XRCC1 or ERCC1 (35–38). Similar effects were observed using Kaplan-Meier survival analysis (log-rank p = 1.0e-3 and 1.0e-4, respectively, Figure S11). Single-agent anti-tumor activity of AZD6738 against G401 and TC-71 cells were less pronounced in vivo, suggesting that tumor-specific differences in DNA repair and DNA damage signaling may impact therapeutic targeting of PGBD5-induced DNA repair dependencies (Fig. S6). Residual tumor cells isolated from mice upon the completion of 20 days of AZD6738 treatment exhibited significantly reduced proliferation, as measured by Ki67 staining (p = 3.1e-5 and 1.0e-3 for IMR5 and HD-MB03 respectively, Fig. 6C & D), and increased DNA damage and apoptosis, as measured by γH2AX and cleaved caspase 3 staining, respectively (p = 1.4e-3, 4.3e-4, 1.0e-3 and 4.9e-4, respectively, Figs. 6C, E, F). In addition, we also assessed the activity of VE-822, an ATR/ATM inhibitor that is currently in clinical trials (39), against neuroblastoma IMR5 and rhabdoid G401 cell line xenografts. In contrast to the ATR-selective AZD6738, single-agent treatment with VE-822 had no significant effects on tumor growth in vivo (Fig. S11), supporting the notion that pharmacologic and selectivity properties of inhibitors of DNA damage signaling may also affect the efficacy of PGBD5-dependent therapeutic targeting.
Figure 6. AZD6738 induces DNA damage, apoptosis, and exhibits anti-tumor efficacy in xenograft models of high-risk medulloblastoma and neuroblastoma in vivo.
(A–B) Tumor volumes over time of nude mice harboring IMR5 (A) and HD-MB03 (B) subcutaneously xenografted tumors treated with AZD6738 by oral gavage at 50 mg/kg body weight/day (red) or as compared to vehicle control (blue). Asterisks denote p < 0.05. Error bars represent standard deviations of 10 individual xenograft mice per group. Arrows denote start of treatment. (C) Representative photomicrographs of sections from IMR5 (top) or HD-MB03 (bottom) tumors upon completion of treatment with AZD6738 (50 mg/kg/day) or vehicle control in vivo, and stained for hematoxylin and eosin H&E, Ki67, cleaved caspase-3, and H2AX, as indicated. Scale bar = 100 μm. (D–F) Quantification of the number of cells positively stained for Ki67 (D), cleaved caspase-3 (E) and H2AX (F) in IMR5 or HD-MB03 xenograft tumors upon completion of treatment with AZD6738 (50 mg/kg/day, red) or vehicle control (blue) in vivo. *p = 3.1 x 10−5 and 0.001 for Ki67 in AZD6738 versus vehicle-treated IMR5 and HD-MB03, respectively. *p = 0.001 and 4.9 x 10−4 for cleaved caspase-3 in AZD6738 versus vehicle-treated IMR5 and HD-MB03, respectively. *p = 0.014 and 4.3 x 10−4 for H2AX in AZD6738 versus vehicle-treated IMR5 and HD-MB03, respectively. Error bars represent standard deviations of 3 independent fields analyzed.
Synergistic targeting of PGBD5-induced DNA repair dependency in patient-derived primary neuroblastoma xenografts in vivo
Considering that AZD6738 exhibited potent single-agent activity against high-risk neuroblastoma and medulloblastoma cell line xenografts, with reduced apparent activity against rhabdoid and Ewing sarcoma xenografts, we reasoned that specific agents that induce DNA damage may be used to selectively potentiate the anti-tumor effects of AZD6738. To test this hypothesis, we analyzed the combination of AZD6738 with cisplatin, a chemotherapeutic drug that crosslinks DNA and is frequently used to treat childhood solid tumors (40, 41) (Fig. 7). We found significant synergy between cisplatin and AZD6738 at all drug concentrations tested, as indicated by their low combination indices for the neuroblastoma, medulloblastoma, rhabdoid tumor, and Ewing sarcoma cell lines (42) (Fig. 7A).
Figure 7. Synergistic targeting of PGBD5-induced DNA repair dependency in primary patient derived high-risk neuroblastoma xenografts in vivo.
(A) Combination indices (CI) for IMR5, HDMB-03 and TC71 cells treated with cisplatin and AZD6738. (B) PGBD5 mRNA expression in two patient derived high-risk neuroblastoma xenografts; complete demographic and molecular features are described in Table S4 (C) Tumor volumes over time of mice engrafted with PDX1 and treated with 50 mg/kg AZD6738 by daily oral gavage (red), as compared to vehicle control (blue), 2 mg/kg cisplatin by weekly IP (black), or combination of AZD6738 and cisplatin (violet). (D) Tumor volumes over time of mice engrafted with PDX2 and treated with 50 mg/kg AZD6738 by daily oral gavage (red), as compared to vehicle control (blue), 2 mg/kg cisplatin by weekly IP (black), or combination of AZD6738 and cisplatin (violet). Asterisks denote p < 0.05. Error bars represent standard deviations of 4 individual xenograft mice per group.
To investigate the potential therapeutic benefit of combining AZD6738 with cisplatin therapy as a prelude to its clinical testing in patients, we established two patient-derived primary xenografts PDX1 and PDX2 from non-MYCN amplified, stage IV metastatic neuroblastoma tumors obtained at diagnosis prior to therapy (Figure 7B, Table S4). In agreement with results in human tumor cell lines (Fig. 4), we observed varying levels of PGBD5 expression in the two neuroblastoma xenografts (Fig. 7B). Subsequently, we transplanted these tumor specimens subcutaneously into NOD.Cg-Prkdcscid Il2rgtm1Sug/JicTac (NOG) immunodeficient mice, and upon tumor engraftment as evidenced by tumor volumetric measurements, randomized recipient mice to be treated with single-agent AZD6738 (50 mg/kg daily PO for 14 days), single-agent cisplatin (2 mg/kg IP every 7 days), combination of AZD6738 and cisplatin, or vehicle control (Fig. 7C & D). Consistent with the relatively low level of PGBD5 expression in PDX1 (Fig. 7B), we found that single-agent treatment with AZD6738 or cisplatin had limited effects, whereas combination of AZD6738 with cisplatin exhibited significant reduction in tumor growth, as compared to single-agent or vehicle control treatments (p = 1.9e-3, Fig. 7C). Likewise, for PDX2 that had relatively high PGBD5 expression (Fig. 7B), we observed significant reduction in tumor growth upon single-agent AZD6738 and cisplatin treatment, as compared to vehicle control (p = 0.032 and 0.079, respectively, Fig. 7D). Altogether, these results indicate that AZD6738 exhibits significant single-agent and cisplatin-combination efficacy against PGBD5-expressing solid tumors.
Discussion
We have now found that the PGBD5 DNA transposase expressed in the majority of childhood solid tumors confers a synthetic dependency on DNA damage repair and signaling. Consistent with the genomic rearrangements promoted by PGBD5 in rhabdoid tumors (17), expression of PGBD5 induces DNA damage, which requires both DNA damage repair and DNA damage signaling, resulting in apoptosis if impaired by their selective inhibitors (Fig. 8). Indeed, both primary mouse and human cells engineered to express PGBD5, as well as PGBD5-expressing childhood solid tumors, accumulated unrepaired DNA damage and underwent apoptosis upon treatment with selective inhibitors of DNA damage signaling. This effect was due to the specific nuclease activity of PGBD5, insofar as mutation of its putative catalytic nuclease residues completely abrogated the dependence on DNA damage signaling. In turn, single-agent treatment with the DNA damage signaling inhibitor AZD6738 exhibited potent anti-tumor activity against high-risk neuroblastomas and medulloblastomas that express high levels of PGBD5 in preclinical mouse models in vivo.
Figure 8. Model of therapeutic targeting of PGBD5-induced DNA damage signaling synthetic dependency.

Tumors with increasing levels of PGBD5 expression and DNA recombinase activity accumulate DNA damage, in concert with other intrinsic sources of cellular DNA damage such as replication stress. PGBD5-dependent DNA damage leads to the generation of various DNA damage structure and DNA damage signals, which in turn activate distinct arms of the DNA damage signaling. Consequently, PGBD5-dependent DNA damage signaling can be inhibited using selective pharmacologic inhibitors, inducing accumulation of DNA damage, impairing DNA repair, and leading to cell death.
Human cancers harbor numerous mechanisms of endogenous DNA damage as a source of genetic mutations and requirements for active DNA damage repair (43). As a result, selective inhibitors of DNA damage signaling exhibit anti-tumor activities in various cancers (44–47). In particular, selective inhibitors of ATR and ATM kinases have been used to target tumors with intrinsic deficiencies in DNA repair (43), as well as DNA damage susceptibility, such as that induced by oncogene and replication stress (47). Our current work now revealed a specific synthetic dependency conferred by the endogenous DNA transposase PGBD5 in the majority of childhood solid tumors. Genetic experiments showed equivalent functional requirements for the scaffolding KU complex that directly binds DSBs, and the ATR and ATM kinases that mediate DNA damage signaling. However, chemical DNA damage signaling kinase inhibitors exhibited a specific profile with the ATR- and ATM-selective AZD6738 and KU60019 inhibitors exhibiting enhanced PGBD5-dependent activity. Given their varied potency and selectivity, it is possible that other selective DNA damage signaling inhibitors can also be used to effectively target PGBD5-induced tumor synthetic dependencies. Since ATR and ATM are activated by specific DNA structures, including single-stranded DNA bound by RPA (48), modified bases (49), and chromatin assemblies (50), the apparent activity of selective inhibitors of DNA damage signaling may be due to the PGBD5-induced generation of as of yet unrecognized DNA or chromatin structures that recruit specific components of the DNA damage response (51). While PGBD5-dependent DNA damage and apoptosis induced by AZD6738 appeared to occur in G1 phase without measureable replication stress or ATM depletion, it is also possible that inhibition of ATR or additional kinases by AZD6738 in S phase leads to apoptosis in the subsequent G1 phase of the cell cycle. Lastly, the susceptibility of PGBD5-expressing tumors to selective inhibitors of DNA damage signaling is expected to cooperate with other sources of DNA damage in tumors, such as oncogene or replication stress.
We anticipate that susceptibility to AZD6738 and other therapeutic strategies to target PGBD5-dependent DNA repair requirements may depend on the tumor-specific mechanisms of DNA repair, including variability in the expression and activity of non-homologous end-joining and homologous recombination, as determined by variation in tumorigenic mechanisms and cells of origin (15). For example, neuroblastoma cells can have varying activity of NHEJ repair as partly affected by neurotrophin receptor signaling (52, 53), which may impact the therapeutic efficacy of targeting PGBD5-induced DNA repair dependencies. Such a model, in which intrinsic differences in DNA repair and DNA damage signaling impact therapeutic targeting of PGBD5-induced DNA repair dependencies, is supported both by the apparent variation in tumor response to AZD6738 as well as its synergistic potentiation by cisplatin therapy (Fig. 4 & 8).
Finally, our findings may explain the apparent activity of other kinase inhibitors, such as for example the observed anti-tumor activity of dactolisib (NVP-BEZ235) in neuroblastoma, medulloblastoma and Ewing sarcoma (54–56), given its potent kinase inhibition of ATR (12). Inhibitors of DNA damage signaling are currently being investigated in clinical trials, including AZD6738 (NCT02264678, NCT02223923, NCT02630199). Our findings warrant their immediate investigation in clinical trials for children with solid tumors, the majority of which express PGBD5 and should be susceptible to targeted therapy of this synthetic DNA damage signaling dependency. We anticipate that improved understanding of the molecular synthetic dependencies and their targeting by emerging selective pharmacologic inhibitors should lead to rational therapeutic strategies for refractory solid tumors.
Material and Methods
Reagents
All reagents were obtained from Sigma Aldrich if not otherwise specified. Synthetic oligonucleotides were obtained from Eurofins (Eurofins MWG Operon, Huntsville, AL, USA) and purified by HPLC. All small molecule inhibitors used were dissolved in dimethyl sulfoxide at 10 mM (Fisher Scientific, Watham, MA, USA) and stored at −20 °C until further use: AZD6738 (Astra Zeneca, London, UK), KU60019 (Selleck Chemicals LLC, Munich, Germany), AZ20 (Tocris, Bristol, UK), VE822 (Selleck Chemicals LLC), AZD7762 (Selleck Chemicals LLC).
Plasmid constructs
Human PGBD5 cDNA (Refseq ID: NM_001258311.1) was cloned into the lentiviral vector in frame with N-terminal GFP to generate pRecLV103-GFP-PGBD5 (GeneCopoeia, Rockville, MD, USA). pReceiver-Lv103 encoding GFP was used as a negative control in all experiments. Missense GFP-PGBD5 mutants were generated using site-directed mutagenesis according to the manufacturer’s instructions (QuikChange Lightning, Agilent, Santa Clara, CA, USA) as described (16). Doxycycline-inducible pINDUCER21 vector was obtained from Thomas Westbrook (57), and used to generate pINDUCER21-PGBD5 using Gateway cloning, according to the manufacturer’s instructions (Fisher Scientific, Watham, MA, USA). pLKO.1 shRNA vectors targeting PGBD5 (TRCN0000138412, TRCN0000135121) and control shGFP were obtained from the RNAi Consortium (Broad Institute, Cambridge, MA).
Lentivirus production and cell transduction
Lentivirus production was carried out as previously described (58). Briefly, HEK293T cells were transfected using TransIT-LT1 with 2:1:1 ratio of the lentiviral vector, and psPAX2 and pMD2.G packaging plasmids, according to manufacturer’s instructions (Mirus, Madison, WI, USA). Virus supernatant was collected at 48 and 72 hours post-transfection, pooled, filtered and stored at −80 °C. RPE and BJ cells were transduced with pRecLV103 virus particles at a multiplicity of infection (MOI) of 5 in the presence of 8 μg/ml hexadimethrine bromide. Transduced cells were selected for 2 days with puromycin hydrochloride (RPE cells at 10 μg/ml, BJ cells at 2 μg/ml) or G418 sulfate (2 mg/ml), depending on the vector-mediated resistance. For pINDUCER21 viruses, cells were transduced at a MOI of 1, and isolated using fluorescence-activated cell sorting (FACSAria III, BD Bioscience, San Jose, CA, USA). For lentiviral transduction of shRNAs, cells were selected for 2 days with puromycin hydrochloride at 2 μg/ml.
Cell culture
All cell lines were obtained from the American Type Culture Collection if not otherwise specified (ATCC, Manassas, Virginia, USA). Ku80−/−, AtrS/S, and Atm−/− mouse embryonic fibroblasts were generated by transduction with SV40 large T antigen (24–26). All neuroblastoma cell lines were provided by Johannes H. Schulte. HD-MB03 cells were generated by Milde et al. (59). Rhabdoid tumor cell lines KP-MRT-NS, KP-MRT-YM, KP-MRT-RY and MP-MRT-AN were generated by Yasumichi Kuwahara and Hajime Hosoi. The identity of all cell lines was verified by STR analysis (Genetica DNA Laboratories, Burlington, NC, USA) and absence of Mycoplasma sp. contamination was determined using Lonza MycoAlert (Lonza Walkersville, Inc., Walkersville, MD, USA). Cell lines were cultured in 5% CO2 in a humidified atmosphere at 37 °C in Dulbecco’s Modified Eagle medium with high glucose (DMEM-HG) supplemented with 10 % fetal bovine serum (FBS) and antibiotics (100 U / ml penicillin and 100 μg / ml streptomycin). Cell viability was assessed using CellTiter-Glo according to the manufacture’s protocol (Promega, Madison, WI, USA).
Quantitative RT-PCR
RNA was isolated using RNeasy Mini, according to manufacturer’s instructions (Qiagen, Venlo, Netherlands). cDNA was synthesized using the SuperScript III First-Strand Synthesis System, according to the manufacturer’s instructions (Invitrogen, Waltham, MA, USA). Quantitative real-time PCR was performed using the KAPA SYBR FAST PCR polymerase with 20 ng template and 200 nM primers, according to the manufacturer’s instructions (Kapa Biosystems, Wilmington, MA, USA). PCR primers are listed in Table S5. Ct values were calculated using ROX normalization using the ViiA 7 software (Applied Biosystems).
Western blotting
To analyze protein expression by Western immunoblotting, 1 million cells were suspended in 80 μl of lysis buffer (4% sodium dodecyl sulfate, 7% glycerol, 1.25% beta-mercaptoethanol, 0.2 mg/ml Bromophenol Blue, 30 mM Tris-HCl, pH 6.8) and incubated at 95 °C for 10 minutes. Cell suspensions were lysed using Covaris S220 adaptive focused sonicator, according to the manufacturer’s instructions (Covaris, Woburn, CA). Lysates were cleared by centrifugation at 16,000 g for 10 minutes at 4 °C. Clarified lysates (30 μl) were resolved using sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and electroeluted using the Immobilon FL PVDF membranes (Millipore, Billerica, MA, USA). Membranes were blocked using the Odyssey Blocking buffer (Li-Cor, Lincoln, Nebraska, USA), and blotted using antibodies listed in Table S6. Blotted membranes were visualized using goat secondary antibodies (Table S6) conjugated to IRDye 800CW or IRDye 680RD and the Odyssey CLx fluorescence scanner, according to manufacturer’s instructions (Li-Cor, Lincoln, Nebraska, USA). Image analysis was done using the Li-Cor Image Studio software (version 4). Relative protein expression was calculated as fluorescence intensity of the protein of interest relative to the fluorescence intensity of the loading control protein. At least 3 blots were analyzed for each condition.
Flow cytometry
Cells were fixed using neutral-buffered formalin for 10 min on ice, washed with PBS, resuspended in 0.1% Triton-X100 in PBS, and incubated for 15 min at room temperature. Permeabilized cells were washed twice with PBS, and resuspended in 100 μl of Hank’s balanced salt solution (HBSS) with 0.1% bovine serum albumin and 2 μl of Alexa Fluor 647-conjugated antibody against cleaved caspase-3 (Table S6). Cells were incubated for 30 min at room temperature in the dark washed twice with PBS and stained with 1 μg/ml DAPI. Cells were analyzed on the Fortessa LSR as described before (BD Bioscience) (60, 61). TUNEL staining was done using the APO-BrdU TUNEL Assay Kit, according to the manufacturer’s protocol (Fisher Scientific). EdU labeling was done using the EdU Click-iT kit according to the manufacture’s protocol (Fisher Sceintific).
Histological staining
Histologic processing and staining was done as described previously (62, 63). In short, cells were plated on 8-well glass Millicell EZ chamber slides at 5000 cells/well, grown for 24 hours, and fixed using 4% paraformaldehyde for 10 min at room temperature (Millipore). Tumor xenograft tissue was fixed using 4% paraformaldehyde for 24 hours at room temperature. Tissues were embedded in paraffin using the ASP6025 tissue processor (Leica, Wetzlar, Germany), sectioned at 5 μm using the RM2265 microtome (Leica), and collected on SuperfrostPlus slides (Fisher Scientific). Tissue sections were deparaffinized with EZPrep buffer (Ventana Medical Systems). Antigen retrieval was performed with Cell Conditioning 1 buffer (Ventana Medical Systems), and sections were blocked for 30 minutes with Background Buster solution (Innovex, Norwood, MA, USA). Primary antibodies were applied for 5 hours at 1 μg/ml (Table S6). Secondary antibodies were applied for 60 minutes (Table S6).
For immunohistochemistry staining, diaminobenzidine (DAB) detection was performed with the DAB detection kit according to manufacturer instruction (Ventana Medical Systems). Slides were counterstained with hematoxylin and a cover slip was mounted with Permount (Fisher Scientific).
For immunofluorescence staining, the detection was performed with Streptavidin-HRP D (Ventana Medical Systems), followed by incubation with Tyramide Alexa Fluor 647 prepared according to manufacturer instruction (Invitrogen). Slides were then counterstained with 5 μg/ml DAPI for 10 min and a cover slip was mounted with Mowiol (Sigma Aldrich). TUNEL and Hoechst staining was done using Click-iT TUNEL Alexa Fluor Imaging Assay according to the manufacture’s protocol.
Image acquisition and analysis
Bright-field images were acquired on an Axio Observer microscope (Carl Zeiss Microimaging, Oberkochen, Germany). Epifluorescence images were acquired using the EVOS FL microscope (Thermo Fisher). Slides were scanned using the Pannoramic 250 slide scanner and images analyzed using the Pannoramic Viewer (3DHistech, Budapest, Hungary). At least 100 stained cells were counted in at least 3 independent fields of view.
Xenografts
All mouse experiments were carried out in accordance with institutional animal protocols. Five million cells were suspended in 200 μl Matrigel (BD Bioscience, Heidelberg, Germany) and injected subcutaneously into the left flank of 6-week-old female athymic nude Foxn1nu mice (The Jackson Laboratory, Bar Harbor, Maine, USA). Patient-derived xenografts were performed using NOD.Cg-Prkdcscid Il2rgtm1Sug/JicTac (NOG) mice (Taconic). Patient tumors were serially transplanted in mice three times before experiments. Tumor growth was monitored using caliper measurements, and tumor volume was calculated using the formula 3.14159 x length x width 2 / 6000. Mice were sacrificed by CO2 asphyxiation when tumor size exceeded 2,000 mm3. Mice were treated with AZD6738 at 50 mg/kg body weight once a day per oral gavage. For in vivo treatment, AZD6738 was dissolved in DMSO at 62.5 mg/ml and mixed 1:10 in 40% propylene glycol and 50% sterile water resulting in a final AZD6738 concentration of 6.25 mg/ml. VE-822 was dissolved in 10% Vitamin E d-alpha tocopheryl polyethylene glycol 1000 succinate and administered by oral gavage. Cisplatin was dissolved in phosphate buffered saline and administered intraperitoneally at 2 mg/kg body weight once every 7 days.
Statistical analysis
For comparisons between two sample sets, statistical analysis of means was performed using 2-tailed, unpaired Student’s t-tests. Survival analysis was done using the Kaplan-Meier method, as assessed using a log-rank test.
Supplementary Material
Acknowledgments
We thank Alejandro Gutierrez, Marc Mansour, Michael Kharas, and John Poirier for critical discussions, and Wong Wai for technical assistance. We thank the Experimental Pharmacology & Oncology Berlin-Buch GmbH for assistance with patient-derived xenografts.
Funding: This work was supported by the NIH K08 CA160660, P30 CA008748, Cycle for Survival, Geoffrey Beene Cancer Research Center, Starr Cancer Consortium, Burroughs Wellcome Fund, Sarcoma Foundation of America, Matthew Larson Foundation, Josie Robertson Investigator Program, Rita Allen Foundation, and the European ERA-Net ERACoSysMed project OPTIMIZE-NB (031L0087B). A.G.H. is supported by the Berliner Krebsgesellschaft e.V. and the Berlin Institute of Health. A.K. is the Damon Runyon-Richard Lumsden Foundation Clinical Investigator.
Footnotes
Author Contributions: AGH: study design, collection and interpretation of the data; CR, HDG, IM, JVS, PH, EJ, JK, WW, EDS, YK, HH, NG, AK, JP, JS: data collection; AK: study design and data analysis. AGH and AK wrote the manuscript with contributions from all authors.
Competing Interests: There are no competing interests of any of the authors.
Data and materials availability: Data are openly available at the Zenodo digital repository (https://zenodo.org/).
References
- 1.Matthay KK, et al. Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. Children’s Cancer Group. The New England journal of medicine. 1999 Oct 14;341:1165. doi: 10.1056/NEJM199910143411601. [DOI] [PubMed] [Google Scholar]
- 2.Morgenstern DA, Baruchel S, Irwin MS. Current and future strategies for relapsed neuroblastoma: challenges on the road to precision therapy. Journal of pediatric hematology/oncology. 2013 Jul;35:337. doi: 10.1097/MPH.0b013e318299d637. [DOI] [PubMed] [Google Scholar]
- 3.Robinson G, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012 Aug 2;488:43. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee RS, Roberts CW. Rhabdoid tumors: an initial clue to the role of chromatin remodeling in cancer. Brain Pathol. 2013 Mar;23:200. doi: 10.1111/bpa.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Granowetter L, et al. Dose-intensified compared with standard chemotherapy for nonmetastatic Ewing sarcoma family of tumors: a Children’s Oncology Group Study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009 May 20;27:2536. doi: 10.1200/JCO.2008.19.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartwell LH, Szankasi P, Roberts CJ, Murray AW, Friend SH. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997 Nov 07;278:1064. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- 7.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005 Apr 14;434:917. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 8.Fong PC, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. The New England journal of medicine. 2009 Jul 9;361:123. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 9.Muller FL, et al. Passenger deletions generate therapeutic vulnerabilities in cancer. Nature. 2012 Aug 16;488:337. doi: 10.1038/nature11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao D, et al. Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer. Nature. 2017 Feb 06; doi: 10.1038/nature21357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwok M, et al. Synthetic lethality in chronic lymphocytic leukaemia with DNA damage response defects by targeting the ATR pathway. Lancet. 2015 Feb 26;385(Suppl 1):S58. doi: 10.1016/S0140-6736(15)60373-7. [DOI] [PubMed] [Google Scholar]
- 12.Toledo LI, et al. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol. 2011 Jun;18:721. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reaper PM, et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol. 2011 Jul;7:428. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 14.Colicchia V, et al. PARP inhibitors enhance replication stress and cause mitotic catastrophe in MYCN-dependent neuroblastoma. Oncogene. 2017 Apr 10; doi: 10.1038/onc.2017.40. [DOI] [PubMed] [Google Scholar]
- 15.Middleton FK, et al. Common cancer-associated imbalances in the DNA damage response confer sensitivity to single agent ATR inhibition. Oncotarget. 2015 Oct 20;6:32396. doi: 10.18632/oncotarget.6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henssen AG, et al. Genomic DNA transposition induced by human PGBD5. eLife. 2015;4 doi: 10.7554/eLife.10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henssen AG, et al. PGBD5 promotes site-specific oncogenic mutations in human tumors. Nature genetics. 2017 Jul;49:1005. doi: 10.1038/ng.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marechal A, Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harbor perspectives in biology. 2013 Sep;5 doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nature reviews Molecular cell biology. 2008 Aug;9:616. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gamper AM, et al. ATR kinase activation in G1 phase facilitates the repair of ionizing radiation-induced DNA damage. Nucleic acids research. 2013 Dec;41:10334. doi: 10.1093/nar/gkt833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beucher A, et al. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. The EMBO journal. 2009 Nov 04;28:3413. doi: 10.1038/emboj.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Difilippantonio MJ, et al. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000 Mar 30;404:510. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murga M, et al. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nature genetics. 2009 Aug;41:891. doi: 10.1038/ng.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zha S, Sekiguchi J, Brush JW, Bassing CH, Alt FW. Complementary functions of ATM and H2AX in development and suppression of genomic instability. Proceedings of the National Academy of Sciences of the United States of America. 2008 Jul 08;105:9302. doi: 10.1073/pnas.0803520105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferguson DO, et al. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proceedings of the National Academy of Sciences of the United States of America. 2000 Jun 06;97:6630. doi: 10.1073/pnas.110152897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hahn WC, et al. Creation of human tumour cells with defined genetic elements. Nature. 1999 Jul 29;400:464. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 29.Foote KM, Lau A, Nissink JW. Drugging ATR: progress in the development of specific inhibitors for the treatment of cancer. Future Med Chem. 2015;7:873. doi: 10.4155/fmc.15.33. [DOI] [PubMed] [Google Scholar]
- 30.Vendetti FP, et al. The orally active and bioavailable ATR kinase inhibitor AZD6738 potentiates the anti-tumor effects of cisplatin to resolve ATM-deficient non-small cell lung cancer in vivo. Oncotarget. 2015 Dec 29;6:44289. doi: 10.18632/oncotarget.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Connell BC, et al. A genome-wide camptothecin sensitivity screen identifies a mammalian MMS22L-NFKBIL2 complex required for genomic stability. Molecular cell. 2010 Nov 24;40:645. doi: 10.1016/j.molcel.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Feraudy S, Revet I, Bezrookove V, Feeney L, Cleaver JE. A minority of foci or pan-nuclear apoptotic staining of gammaH2AX in the S phase after UV damage contain DNA double-strand breaks. Proceedings of the National Academy of Sciences of the United States of America. 2010 Apr 13;107:6870. doi: 10.1073/pnas.1002175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheung NK, Dyer MA. Neuroblastoma: developmental biology, cancer genomics and immunotherapy. Nature reviews Cancer. 2013 Jun;13:397. doi: 10.1038/nrc3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pui CH, Gajjar AJ, Kane JR, Qaddoumi IA, Pappo AS. Challenging issues in pediatric oncology. Nature reviews Clinical oncology. 2011 Sep;8:540. doi: 10.1038/nrclinonc.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Min A, et al. AZD6738, A Novel Oral Inhibitor of ATR, Induces Synthetic Lethality with ATM Deficiency in Gastric Cancer Cells. Molecular cancer therapeutics. 2017 Apr;16:566. doi: 10.1158/1535-7163.MCT-16-0378. [DOI] [PubMed] [Google Scholar]
- 36.Kwok M, et al. ATR inhibition induces synthetic lethality and overcomes chemoresistance in TP53- or ATM-defective chronic lymphocytic leukemia cells. Blood. 2016 Feb 04;127:582. doi: 10.1182/blood-2015-05-644872. [DOI] [PubMed] [Google Scholar]
- 37.Mohni KN, et al. A Synthetic Lethal Screen Identifies DNA Repair Pathways that Sensitize Cancer Cells to Combined ATR Inhibition and Cisplatin Treatments. PloS one. 2015;10:e0125482. doi: 10.1371/journal.pone.0125482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohni KN, Kavanaugh GM, Cortez D. ATR pathway inhibition is synthetically lethal in cancer cells with ERCC1 deficiency. Cancer research. 2014 May 15;74:2835. doi: 10.1158/0008-5472.CAN-13-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Charrier JD, et al. Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. Journal of medicinal chemistry. 2011 Apr 14;54:2320. doi: 10.1021/jm101488z. [DOI] [PubMed] [Google Scholar]
- 40.Dasari S, Tchounwou PB. Cisplatin in cancer therapy: molecular mechanisms of action. European journal of pharmacology. 2014 Oct 05;740:364. doi: 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruggiero A, et al. Platinum compounds in children with cancer: toxicity and clinical management. Anti-cancer drugs. 2013 Nov;24:1007. doi: 10.1097/CAD.0b013e3283650bda. [DOI] [PubMed] [Google Scholar]
- 42.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacological reviews. 2006 Sep;58:621. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 43.Tubbs A, Nussenzweig A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell. 2017 Feb 09;168:644. doi: 10.1016/j.cell.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karnitz LM, Zou L. Molecular Pathways: Targeting ATR in Cancer Therapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015 Nov 1;21:4780. doi: 10.1158/1078-0432.CCR-15-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nieto-Soler M, et al. Efficacy of ATR inhibitors as single agents in Ewing sarcoma. Oncotarget. 2016 Aug 26; doi: 10.18632/oncotarget.11643. [DOI] [PMC free article] [PubMed]
- 46.David L, et al. CHK1 as a therapeutic target to bypass chemoresistance in AML. Science signaling. 2016;9:ra90. doi: 10.1126/scisignal.aac9704. [DOI] [PubMed] [Google Scholar]
- 47.Morgado-Palacin I, et al. Targeting the kinase activities of ATR and ATM exhibits antitumoral activity in mouse models of MLL-rearranged AML. Science signaling. 2016;9:ra91. doi: 10.1126/scisignal.aad8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003 Jun 06;300:1542. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 49.Yoshioka K, Yoshioka Y, Hsieh P. ATR kinase activation mediated by MutSalpha and MutLalpha in response to cytotoxic O6-methylguanine adducts. Molecular cell. 2006 May 19;22:501. doi: 10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hermand D, Nurse P. Cdc18 enforces long-term maintenance of the S phase checkpoint by anchoring the Rad3-Rad26 complex to chromatin. Molecular cell. 2007 May 25;26:553. doi: 10.1016/j.molcel.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 51.Kemp MG. DNA damage-induced ATR kinase activation in non-replicating cells is regulated by the XPB subunit of transcription factor II-H (TFIIH) J Biol Chem. 2017 Jun 07; doi: 10.1074/jbc.M117.788406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schulte JH, et al. Expression of the TrkA or TrkB receptor tyrosine kinase alters the double-strand break (DSB) repair capacity of SY5Y neuroblastoma cells. DNA repair. 2008 Oct 01;7:1757. doi: 10.1016/j.dnarep.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 53.Newman EA, et al. Alternative NHEJ Pathway Components Are Therapeutic Targets in High-Risk Neuroblastoma. Mol Cancer Res. 2015 Mar;13:470. doi: 10.1158/1541-7786.MCR-14-0337. [DOI] [PubMed] [Google Scholar]
- 54.Pei Y, et al. An animal model of MYC-driven medulloblastoma. Cancer cell. 2012 Feb 14;21:155. doi: 10.1016/j.ccr.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chanthery YH, et al. Paracrine signaling through MYCN enhances tumor-vascular interactions in neuroblastoma. Sci Transl Med. 2012 Jan 04;4:115ra3. doi: 10.1126/scitranslmed.3002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giorgi C, et al. PI3K/AKT signaling modulates transcriptional expression of EWS/FLI1 through specificity protein 1. Oncotarget. 2015 Oct 06;6:28895. doi: 10.18632/oncotarget.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Westbrook TF, Stegmeier F, Elledge SJ. Dissecting cancer pathways and vulnerabilities with RNAi. Cold Spring Harb Symp Quant Biol. 2005;70:435. doi: 10.1101/sqb.2005.70.031. [DOI] [PubMed] [Google Scholar]
- 58.Kentsis A, et al. Autocrine activation of the MET receptor tyrosine kinase in acute myeloid leukemia. Nat Med. 2012 Jul;18:1118. doi: 10.1038/nm.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Milde T, et al. HD-MB03 is a novel Group 3 medulloblastoma model demonstrating sensitivity to histone deacetylase inhibitor treatment. J Neurooncol. 2012 Dec;110:335. doi: 10.1007/s11060-012-0978-1. [DOI] [PubMed] [Google Scholar]
- 60.Fox R, Aubert M. Flow cytometric detection of activated caspase-3. Methods in molecular biology. 2008;414:47. doi: 10.1007/978-1-59745-339-4_5. [DOI] [PubMed] [Google Scholar]
- 61.Sordet O, et al. Specific involvement of caspases in the differentiation of monocytes into macrophages. Blood. 2002 Dec 15;100:4446. doi: 10.1182/blood-2002-06-1778. [DOI] [PubMed] [Google Scholar]
- 62.Yarilin D, et al. Machine-based method for multiplex in situ molecular characterization of tissues by immunofluorescence detection. Scientific reports. 2015;5:9534. doi: 10.1038/srep09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fujisawa S, Turkekul M, Barlas A, Fan N, Manova K. Double in situ detection of sonic hedgehog mRNA and pMAPK protein in examining the cell proliferation signaling pathway in mouse embryo. Methods in molecular biology. 2011;717:257. doi: 10.1007/978-1-61779-024-9_15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.