Abstract
The NCI Genomic Data Commons (GDC) was launched in 2016 and makes available over 2 petabytes (PB) of cancer genomic and associated clinical data to the research community. This dataset continues to grow and currently includes over 14,500 patients. The GDC is an example of a biomedical data commons, which collocates biomedical data with storage and computing infrastructure and commonly used web services, software applications, and tools to create a secure, interoperable, and extensible resource for researchers. The GDC is: i) a data repository for downloading data that has been submitted to it, and also a system that: ii) applies a common set of bioinformatics pipelines to submitted data; iii) re-analyzes existing data when new pipelines are developed; and, iv) allows users to build their own applications and systems that interoperate with the GDC using the GDC Application Programming Interface (API). We describe the GDC API and how it has been used both by the GDC itself and by third parties.
Keywords: data commons, genomic data commons, data sharing, cancer genomics
Introduction
The NCI Genomic Data Commons (GDC) was launched in 2016 and makes available over 2 PB of cancer genomic and associated clinical data to the research community (1). GDC data includes raw sequencing data and derived results from a variety of applications including mRNA-Seq, miRNA-Seq, WXS, and WGS sequencing for over 14,500 patients. We expect to add at least another petabyte of data to the GDC by the end of 2017. The GDC is an example of a biomedical data commons, which collocates biomedical data with storage and computing infrastructure and commonly used web services, software applications, and tools to create a secure, interoperable, and extensible resource for the research community (2). The GDC currently has four main functions: i) it is a data repository that allows data to be submitted, processed and downloaded; ii) it is a system that applies a common set of bioinformatics pipelines to submitted data; iii) it re-analyzes the data it contains when new bioinformatics pipelines are developed; and iv) allows users to build their own applications and systems that interoperate with the GDC using the GDC Application Programming Interface (API). All of the data in the GDC is available through the API. In this paper, we describe the API and its use by both the GDC and external groups to harness the extensive data housed at the GDC.
Methods
The GDC API provides programmatic access to GDC functionality, including searching for, accessing, downloading, and submitting data and metadata. Open-access data is available to anyone though the GDC API. In addition, access to controlled-access data is available to anyone who has a NIH eRA Commons account and is authorized by dbGaP to access the data. An eRA Commons account is available to researchers and used to access NIH systems, such as when submitting grants. The database for Genomes and Phenotypes (dbGaP) is a NIH system that manages controlled access genomic and associated phenotype data, including the Data Use Certification Agreements that investigators and their organizations must sign. The GDC interoperates with eRA Commons and dbGaP to support this functionality.
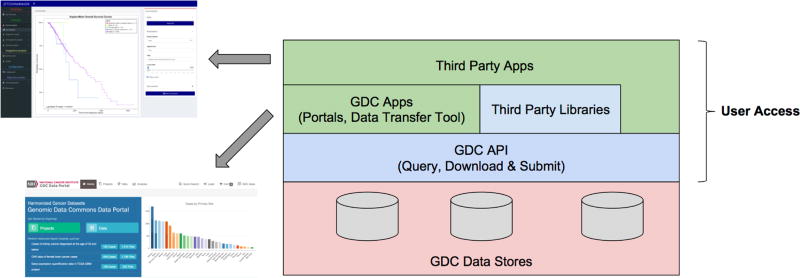
The GDC API uses JSON (3) as its communication format and follows RESTful API conventions (4), including the use of standard HTTP methods like GET, PUT, POST and DELETE. We emphasize that the GDC API is designed to be used by applications, not by researchers writing queries manually, though, of course, this can be done. Figure 1 shows the relationship of the GDC API, GDC Data, internal apps, and external apps.
Figure 1.
GDC API and its relationship to the data managed by the GDC and internal/external applications.
There is extensive online documentation (5) about how to make calls to the API endpoints provided by the GDC. Each GDC API endpoint represents specific API functionality. There are currently 9 endpoints provided by the GDC: status, projects, cases, files, annotations, data, manifest, slicing, and submission. The accompanying video titled “The GDC Application Programming Language (API)” also illustrates how to use the API.
Accessing data from the GDC
To get basic information about a particular file or entity in the GDC a user may query the associated UUID (Universally Unique IDentifier). UUIDs are designed to be globally unique. The GDC assigns UUIDs to all files as well as other entities such as samples and cases (i.e. patients). An example query for a particular variant calling format (VCF) file is listed below. A VCF is the main file output by GDC somatic variant callers; it lists the location of each identified mutation in an individual’s analyzed tumor sample.
https://api.gdc.cancer.gov/files/ee3f77ff-7347-49b4-8729-29a0d5fd029f
Request parameters can be also supplied to further customize the API request and response. These additional parameters include: filters, format, fields, pretty, size, from, sort, expand, and facets. The expand parameter returns additional metadata associated with related entities in the GDC Data Model (6). The following request asks for information regarding the case’s diagnosis and the sample from which the DNA and downstream analysis files were derived.
Using filters to query certain types of data from the GDC
Filters allow the user to limit the results returned by an API POST or GET request. Filters are supplied in a JSON-formatted payload. In the following example, a GET request is made to return only files derived from male cases.
Filters can be combined and nested to create more complex queries. Options for filtering can be easily displayed using the mapping endpoint (e.g. https://api.gdc.cancer.gov/files/_mapping). The following request asks to return RNA-Seq files from cases in which patients were 40 or older at the time of diagnosis.
Controlled-access files can be downloaded via the API if a user has the necessary permissions and a token is supplied as part of a request header. An example using curl is shown below.
curl -H "X-Auth-Token:$TOKEN" http://api.gdc.cancer.gov/data/003cb96e-d759-4304-8d07-17e859f5d9f1
Submitting data to the GDC
The API can also be used to submit data to the GDC. Once a project has been created, the API can be used to upload metadata, register files containing molecular data for uploading, and upload the registered files. In the example below, a file, TCGA_BRCA_1.BAM, is being uploaded after it was first registered in the system.
curl -H "X-Auth-Token: $TOKEN" -- data-binary@TCGA_BRCA_1.BAM https://api.gdc.cancer.gov/v0/submission/TCGA/BRCA/files/c414a205-376e-4993-af48-2a4689eb433e
The GDC API currently responds to approximately 100,000 requests per day and from nearly 100 countries every month. These may be in the form of direct queries, GDC developed application queries, or third party application queries.
Results
The GDC API is designed to be powerful, versatile, and easy to use. It has been used by both the internal GDC Team as well as external collaborators and third party app developers. Some examples are described below.
Internal GDC Applications
The GDC team has created multiple applications and tools using our own internal API. These include the GDC Data Portal, GDC Submission Portal, GDC Legacy Archive, and the GDC Data Transfer Tool, all available via https://gdc.cancer.gov. Each of these systems uses the same API commands and endpoints that are available to external users. In addition to the online user guide, users may easily learn about the functionality of the API by using standard web browser developer tools (4) while exploring any of the GDC applications.
High volume data submitters
Those groups currently submitting large volumes of data to the GDC, including genome sequencing centers, biospecimen core repositories, etc, also use the GDC API.
NCI Cloud Pilots
The NCI Cloud Pilots program was created to allow users to run their own computational analyses with their own data alongside data from the The Cancer Genome Atlas (TCGA) project and newly harmonized data stored in the GDC, avoiding large data transfer costs and the need for in-house high performance computing architecture. There are three cloud pilots: FireCloud developed by the Broad Institute, the Cancer Genomics Cloud developed by Seven Bridges Genomics, and Cancer Genomics Cloud developed by Institute for Systems Biology (ISB). These organizations have all made extensive use of the GDC API to query and download the data from the GDC. While the cloud pilots have much of the original TCGA data and metadata in their own infrastructure, they pass some queries to the GDC in real time, which enable access to updated and GDC-harmonized data they do not house themselves. FireCloud, for example, will allow “just-in-time” download of BAM sequence alignment data files from GDC storage rather than storing all of these files on their own servers (7).
R packages
Several R packages have also been developed that leverage the GDC API to provide convenient access to TCGA data (8, 9, 10). Querying data in R allows users to interact more directly with the data in a comfortable environment and move seamlessly into their favorite bioinformatics tools that are available via CRAN or Bioconductor. An example from the TCGAbiolinks package (8) is shown below, where the user queries and downloads gene expression data for two particular aliquots from the TCGA Glioblastoma project.
query <- GDCquery(project = "TCGA-GBM",
data.category = "Transcriptome Profiling",
data.type = "Gene Expression Quantification",
workflow.type = "HTSeq - Counts",
barcode = c("TCGA-14-0736-02A-01R-2005-01",
"TCGA-06-0211-02A-02R-2005-01"))
GDCdownload(query, method = "client")
Discussion
The NCI Genomic Data Commons (GDC) houses and distributes over 2 PB of cancer genomic data. The GDC is not just a data repository, as it also provides the results of many standard cancer bioinformatics analyses including somatic variant calling, copy number variation, mRNA-Seq and miRNA-Seq expression, and methylation array analysis. To maximize the usefulness of this resource, the GDC has created an API that allows users to have the same programmatic access to the data as internal developers at the GDC. Future plans include allowing more access to the underlying data via the API such as filtering for specific mutations or genes. External groups have made steady use of the API since its inception, building interesting applications and resources on top of it. As more data is deposited and harmonized at the GDC in the coming years, the GDC API will open up this data to applications from the cancer genomics research community.
Supplementary Material
Acknowledgments
We would like to thank the TCGAbiolinks team. We would also like to commemorate Sergey V. Marechek’s contribution to the GDC.
Financial support: This project has been funded in whole or in part with Federal funds from the NCI, NIH, under Contract No. HHSN261200800001E. The content above doesn’t necessarily reflect views of policies of the DHHS, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Conflict of interest statement: The authors declare no potential conflicts of interest.
References
- 1.Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, Staudt LM. Toward a shared vision for cancer genomic data. N Engl J Med. 2016;375:1109–1112. doi: 10.1056/NEJMp1607591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grossman RL, Heath AP, Murphy M, Patterson M, Wells W. A case for data commons: Toward data science as a service. Comput Sci Eng. 2016;18(5):10–20. doi: 10.1109/MCSE.2016.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The javascript object notation (json) data interchange format [Internet] Bray T, editor. [cited 2017 Jan 10];Internet Engineering Task Force. c2014 Available from: https://tools.ietf.org/html/rfc7159.
- 4.Richardson L, Amundsen M, Ruby S. RESTful Web APIs. Newton, MA: O'Reilly Media; 2013. [Google Scholar]
- 5.The GDC Application Programming Interface (API): An Overview [Internet] [retrieved on January 10, 2017];Genomic Data Commons. https://docs.gdc.cancer.gov/API/Users_Guide/Getting_Started/
- 6.The GDC Data Model [Internet] [retrieved on January 10, 2017];Genomic Data Commons Project Team. https://docs.gdc.cancer.gov/Data/Data_Model/GDC_Data_Model/
- 7.Firebrowse.org [Internet] Cambridge, MA: Broad Institute of MIT & Harvard; c2016. [cited 2017 Jan 10]. Available from: http://firebrowse.org/ [Google Scholar]
- 8.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016 May 5;44(8):e71. doi: 10.1093/nar/gkv1507. Epub 2015 Dec 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. [cited 2017 Jan 10];Bioconductor's Genomic Data Commons Package. Davis S and Morgan M. c2017 Available from: https://github.com/Bioconductor/GenomicDataCommons.
- 10.Zhu, Qiu, Ji Nature Methods. 2014;11(6):599–600. doi: 10.1038/nmeth.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.