Abstract
Background
Cigarette smoking increases risk for multiple diseases. MicroRNAs (miRNAs) regulate gene expression and may play a role in smoking-induced target organ damage. We sought to describe a miRNA signature of cigarette smoking and relate it to smoking-associated clinical phenotypes, gene expression, and lung inflammatory signaling.
Methods and Results
Expression profiling of 283 miRNAs was conducted on whole blood-derived RNA from 5,023 Framingham Heart Study participants (54.0% women, mean age 55±13 years) using TaqMan assays and high-throughput RT-qPCR. Associations of miRNA expression with smoking status and associations of smoking-related miRNAs with inflammatory biomarkers and pulmonary function were tested with linear mixed-effects models. We identified a six-miRNA signature of smoking. Five of the six smoking-related miRNAs were associated with serum levels of C-reactive protein or interleukin-6; miR-1180 was associated with pulmonary function measures at a marginally significant level. Bioinformatic evaluation of smoking-associated genes coexpressed with the miRNA signature of cigarette smoking revealed enrichment for immune-related pathways. Smoking-associated miRNAs altered expression of select inflammatory mediators in cell culture gain-of-function assays.
Conclusions
We characterized a novel miRNA signature of cigarette smoking. The top miRNAs were associated with systemic inflammatory markers and reduced pulmonary function, correlated with expression of genes involved in immune function, and were sufficient to modulate inflammatory signaling. Our results highlight smoking-associated miRNAs and are consistent with the hypothesis that smoking-associated miRNAs serve as mediators of smoking-induced inflammation and target organ damage. These findings call for further mechanistic studies to explore the diagnostic and therapeutic utility of smoking-related miRNAs.
Keywords: smoking, microRNA, inflammation, pulmonary, risk factor
Introduction
Epidemiological studies have established the profound disease burden attributable to cigarette smoking, including coronary heart disease and stroke, chronic obstructive pulmonary disease, and lung cancer. Despite the steady increase in smoking cessation rates in the United States over the past 40 years, the percentage of smokers and number of tobacco-related deaths per year have risen worldwide, and the burden of the tobacco epidemic has shifted toward low- and middle-income countries.1–4 Moreover, the biological mechanisms that mediate the associations between cigarette smoking and a diverse set of morbidities remain unclear.
MicroRNAs (miRNAs) are a class of highly-conserved short non-coding RNAs that function as post-transcriptional regulators of gene expression.5, 6 Over 2,000 miRNA sequences have been detected in the human transcriptome and are predicted to target more than half of all protein-coding genes.7, 8 Because miRNAs exert modest-sized effects on the expression of individual genes and some can target numerous transcripts, miRNAs are purported to coordinate the fine-tuning of gene expression under physiological conditions.9, 10 Supporting this theory, essential roles for miRNAs have been discovered in most cellular processes, including cell proliferation, development, differentiation, and apoptosis.11–14 Moreover, aberrant miRNA expression has emerged as a hallmark of human diseases, starting with cancers but now including cardiovascular disease and inflammatory lung disease.15–18 Most reported miRNA expression patterns in humans come from miRNA detection in cells or tissue from the target organs of disease, including the global downregulation of miRNA expression in alveolar macrophages and bronchial alveolar epithelium from smokers.19, 20
In recent years, miRNAs were discovered to have reproducible signals and to be stable in blood.21,22 These results have motivated efforts to profile circulating miRNAs in disease states, with the goal of harnessing their potential as non-invasive diagnostic biomarkers. Thus far, small cohort studies have profiled miRNA expression in induced sputum and plasma from cigarette smokers.12, 23 We sought to elucidate a comprehensive, whole blood-based miRNA profile for cigarette smoking that may (1) have predictive value for smoking-associated diseases and (2) shed light on the miRNA-mediated alterations in gene expression theorized to link smoking with smoking-related disease phenotypes. We hypothesized that miRNA profiling of whole blood would capture disrupted gene regulatory networks relevant to the inflammatory and immune responses that link the direct effects of cigarette smoking to target organ damage and smoking-related systemic diseases. To this end, we characterized a whole blood-derived miRNA signature of cigarette smoking in 5,023 Framingham Heart Study participants and evaluated the top miRNAs in relation to smoking-associated clinical phenotypes, smoking-associated gene expression, and inflammatory signaling in lung tissue.
Methods
Study Samples
The Framingham Heart Study (FHS), a community-based prospective study of cardiovascular disease and associated risk factors, was initiated in 1948 and has enrolled three generations of participants.24–26 On-site evaluations of the Offspring and Third Generation cohort participants, including anthropometric data and biological sample collections, have been conducted every four to eight years. Samples from Offspring cohort Examination 8 (2005–2008; n=2,571) and Third Generation cohort Examination 2 (2008–2011; n=3,245) with complete smoking status data (n=5,816) were eligible for analysis. Non-fasting blood specimens were collected and stored at −80°C until assayed. Total RNA was isolated from peripheral whole blood samples (mean yield=3.50±1.66 µg) in PAXgene tubes (PreAnalytiX, Hombrechtikon, Switzerland) by Asuragen, Inc. (Austin, TX). RNA quality was assessed by total yield (>2 µg), 260/280 ratio (1.7–2.2) (absorbance reading in spectrophotometer at 260 nm and 280 nm; this ratio gives information about the purity of an RNA sample with 1.6-2.0 ratio showing ideal RNA purity), RNA integrity (RIN 7–10), amplification quality (>5 mcg), and cDNA size (median=400 nucleotides). After excluding 8 samples with poor RNA quality, a total of 5,808 samples were available for further analysis (Supplementary Figure 1A).
Clinical Measures
Participant smoking status was stratified as current, former, or never. Current smokers were defined as those who reported smoking on average at least one cigarette per day in the year prior to the FHS clinic examination, former smokers were individuals who reported smoking on average at least one cigarette per day in the past but had not smoked in the past year, and never smokers reported never smoking. Serum C-reactive protein (CRP) was measured in the Offspring cohort at Examination 8 and Third Generation at Examination 2 using a high sensitivity Dade-Behring BN 100 nephelometer.27 Serum interleukin-6 was quantified in Offspring cohort participants at Examination 8 using the Quantikine HS Interleukin-6 Immunoassay kit (R&D Systems, Bio-Techne, Minneapolis, MN).28 Interleukin-6 was not measured in the Third Generation cohort at Examination 2. Intra-assay coefficients of variation for inflammatory marker measurements were <9.2%.29 Forced expiratory volume at one second (FEV1) and forced vital capacity (FVC) were measured on the Offspring cohort at Examination 8 using a water-filled spirometer (Collins Medical, Inc., Braintree, MA) and personal computer software (S&M Instruments, Doylestown, PA) and on the Third Generation at Examination 2 using a dry rolling-seal spirometer and Collins 2000 Pus/SQL software (Collins Medical, Inc., Braintree, MA). In both examinations the highest value among acceptable efforts was used, as per the American Thoracic Society-European Respiratory Society guidelines.30 In addition, the same staff training and quality control activities were used for both of these examinations. Raw values for FEV1 and FVC were examined as a percentages of the predicted values, with predicted values calculated using standard reference equations derived from healthy never smokers in the NHANES III study.31 The FEV1/FVC ratio was calculated from the highest acceptable individual measures. A dichotomous airflow obstruction variable was defined as the combination of FEV1 <lower limit of normal and the FEV1/FVC ratio <lower limit of normal, with lower limits of normal defined on the basis of published NHANES III data.31
Expression Profiling
miRNA profiling was performed using targeted TaqMan miRNA assays (Applied Biosystems, Foster City, CA). A high-throughput RT-qPCR platform was employed, which uses integrated fluidic circuits to perform miniaturized reactions at high efficiency with a batch size of 96 samples versus 96 miRNA targets (BioMark, Fluidigm Corp, San Francisco, CA). Profiling was completed at Boston University, Boston, MA, and the University of Massachusetts, Worcester, MA (laboratory of Dr. Jane Freedman). Reverse transcription and pre-amplification reactions were conducted in two separate pools (A and B), containing primers for 176 and 157 miRNAs, respectively, and carried out according to standard TaqMan protocols. MiRNAs were assayed in sub-panels (A1–2 and B1–2), each of which contained four “housekeeping” RNAs. Excellent reproducibility was demonstrated in preliminary profiling of 24 samples with 96 miRNA assays (mean CV ~3.5%). The analytic pipeline included an additional pilot study in which 754 miRNAs were measured on 455 Offspring cohort samples (Batch 1). The detectable subset of 345 miRNAs was measured in the remaining Offspring cohort samples (n=2,116) and the Third Generation cohort samples (n=3,245) (Batch 2). Of these, 5,729 samples passed quality control measures for miRNA profiling. Removed samples included those with <100 detectable miRNAs in Batch 1 or <20 detectable miRNAs in Batch 2 as well as nine samples recommended for removal by the laboratory technician during miRNA profiling. Samples with missing cell counts (N=705) and missing smoking status (N=1) were excluded, leaving a final sample size of 5,023 (Supplementary Figure 1A). From the set of detected miRNAs, we filtered out miRNAs (n=56) expressed in <500 participants. Outlier miRNA expression levels [threshold cycle (CT) ≥5 standard deviations from the mean CT] were treated as missing (a total of 32 outliers for six miRNAs were treated as missing). We further excluded six miRNAs that were identified as snoRNAs or tRNA fragments in the latest version of miRBase.7 The final miRNA panel consisted of 283 miRNAs (Supplementary Figure 1B). To normalize miRNA expression data, raw CT values were adjusted for four technical variables: RNA isolation batch, RNA quality, RNA concentration, and 260/280 ratio. This method was chosen for the following reasons. First, reliable reference or housekeeping RNAs have not been well-characterized in whole blood. Second, exploratory models run on pilot study data using (1) adjustment for the small nuclear RNA RNU48 and (2) global mean normalization with mean CT values for ~50 experimental miRNAs both resulted in over-fitting. Third, variance analysis demonstrated that these technical variables accounted for the greatest degree of variation in CT values (40–60%).
Messenger RNA (mRNA) expression profiling of whole blood-derived RNA using the Affymetrix Human Exon Array ST 1.0 was performed as described previously.32, 33 Briefly, amplified cDNA was generated using the WT-Ovation Pico RNA Amplification System and processed with the WT-Ovation Exon Module (NuGEN, San Carlos, CA) in an automated genechip array station. Affymetrix Human Exon 1.0 ST microarrays were hybridized, washed, and scanned (Affymetrix, Santa Clara, CA). Microarray data for 17,873 genes were collected using the robust multi-chip average (RMA) method, and 5,626 samples passed mRNA quality controls (Supplementary Figure 1A). Data were normalized with RMA and further adjusted for technical covariates, as described previously.32,33
Cell Culture, miRNA Transfection, & Cytokine Analysis
To evaluate the impact of smoking-associated miRNAs on the expression of inflammatory mediators in the lung, human lung epithelial A549 (ATCC CCL-185) cells were maintained in Ham’s F12 media (Life Technologies, Thermo Fischer Scientific Corp., Carlsbad, CA) supplemented with 10% FBS (Life Technologies) and 1x penicillin/streptomycin at 37°C and 5% CO2. Cells grown in six-well tissue culture dishes were transfected with human miRNA mimetics (Dharmacon, Inc., Lafayette, CO) of non-targeting (NT) sequence (CN-001000-01-05) or one of three smoking-associated miRNAs—miR-744-3p (MIMAT0004946), miR-1180-3p (MIMAT0005825), or miR-1285-3p (MIMAT0005876)—using DharmaFECT1 to achieve a final concentration of 50 nM. Smoking- associated miRNAs were selected for this functional experiment based on the strength of observed associations with smoking status, inflammatory biomarkers, and/or lung function and the putative involvement of these miRNAs in inflammatory and immune pathways based on gene ontology results from coexpression and target prediction analyses. After 24 hours of transfection, cells were stimulated with 10 ng/mL recombinant human TNF-α (R&D Systems, Bio-Techne, Minneapolis, MN) or vehicle for six hours. A panel of cytokines was measured in the collected cell supernatants using a multiplex bead array (R&D Systems). Cytokine expression was analyzed using a Liquichip100 (Qiagen, Hilden, Germany).
Statistical Methods
Statistical analyses were performed using SAS software version 9.2 (SAS Institute Inc., Cary, NC, USA) or R software version 3.1.1 (R Foundation for Statistical Computing, Vienna, Austria).34 Linear mixed effects models with a random effect term to account for covariance due to familial relationships (the “lmekin” function in R) were used to test the association of miRNA expression with cigarette smoking as a three-level categorical variable (current/former/never). Expression of each miRNA was the dependent variable and smoking status was the exposure. Covariates included age, sex, cohort, four technical covariates (RNA isolation batch [50 levels], RNA quality, RNA concentration, and 260/280 ratio), and complete blood cell (CBC) count variables. CBC counts were imputed from mRNA expression values via Partial Least Square (PLS) prediction, with cross-validated prediction accuracy estimates ranging from 0.25–0.89.
False discovery rate (FDR) and q-values were estimated using the Benjamini-Hochberg method.35 An FDR threshold of 0.10 was applied to identify significant miRNAs in the primary analysis. We then performed pairwise comparisons in (1) current vs. never smokers, (2) current vs. former smokers, and (3) former vs. never smokers. MiRNAs that passed the screening threshold were considered significant in pairwise comparisons at FDR<0.10. To demonstrate the reproducibility of observed associations, we conducted internal validation by splitting our sample into two groups of equal size by family into discovery and validation sets, with ~2,500 individuals in each, and analyzing miRNA associations in each set independently.
We used linear mixed effects models with a random effect term to account for covariance due to familial relationships (the “lmekin” function in R) to assess the association of smoking-related miRNAs with inflammatory markers and pulmonary function measures. MiRNA expression level was treated as the exposure variable. For inflammatory biomarker analyses, the loge-transformed serum concentration of CRP or interleukin-6 was treated as the outcome. For pulmonary function analyses, the outcome variables were FEV1, FVC, the ratio of FEV1/FVC, and airflow obstruction, with raw values of FEV1 and FVC examined as a percentages of the predicted values as described above.31 All models were adjusted for age, sex, cohort, smoking status, imputed CBC counts, and the four technical covariates described above. For associations with clinical phenotypes, the number of tests was modest, so the Bonferroni method was used to correct for multiple testing. The p-value threshold (p<1.4×10−3) was calculated by dividing 0.05 by the number of significant smoking-associated miRNAs (6) times the six clinical phenotypes tested [0.05/(6*6)].
For miRNA-mRNA coexpression analysis, pairwise coexpression of all profiled miRNAs and mRNAs was conducted using linear mixed effects models with fix effect terms including age, sex, and technical covariates, and with a random effect term to account for covariance due to genetic relationships in extended pedigrees (the “lmekin” function in R). MiRNA expression was the dependent variable, mRNA expression was the independent variable, and models were adjusted for age, sex, cohort, technical covariates, and imputed CBC counts. Split sample analysis was conducted, with significant coexpressed pairs in both discovery and validation sets (~2,800 samples each) identified from the large number of coexpressed pairs at the conservative threshold of FDR<0.01. The coexpression results were then overlapped with the set of significant miRNAs (FDR<0.10) and significant mRNAs (FDR<0.05) to identify an mRNA-miRNA coexpression signature for cigarette smoking. The more stringent FDR threshold for significant mRNAs than for significant miRNAs reflects the far greater number of mRNAs associated with smoking. To further interrogate the identified mRNA-miRNA correlations, we cross-referenced these pairs with three miRNA target databases: miRDB and miRNAorg (predicted targets) and miRTarBase (experimentally validated targets).
Separate gene ontology (GO) enrichment analyses were conducted on the sets of smoking-associated mRNAs coexpressed with each of our smoking-associated miRNAs based on functional classifications using all biological process terms in the GO Consortium database.36 The enrichment p-value threshold for significance (p<6.1×10−5) was determined by the Bonferroni method, adjusting for 825 biological process terms in the GO database. GO analysis was also performed on coexpressed genes in which the mRNA was a predicted or validated target of its coexpressed miRNA. Due to the much smaller sample size, the analysis was performed on the aggregate results (rather than separately for each individual miRNA) and used a more liberal threshold (FDR<0.05) to define significant pathways. Network figures were generated using the JAVA-based visual analytic software tool ProteoLens.37
For determining the potential effects of miRNA mimetic transfection on cytokine expression in lung epithelial cells, data were pooled from independent miRNA transfection experiments (n=3), which were analyzed and presented together. Analyses used loge-transformed data in a two-way ANOVA with random block design across both factors (transfection and stimulation) to account for experiment-to-experiment variability. Dunnett’s test was applied post hoc to compare cytokine concentrations from cells transfected with miRNA mimetics to similarly stimulated cells that were instead transfected with NT control. Statistical analyses were performed using Prism 6.0 (GraphPad Software, Inc., San Diego, CA). A conservative p-value threshold was set at p<0.01.
Study Approval
All participants gave informed consent for participation in this study and collection of biosamples for genetic/genomic analysis. The study protocol was approved by the Boston University Medical Center Institutional Review Board.
Results
Study Sample Characteristics
Out of 5,023 participants (54.0% women, mean age 55±13 years) with data for miRNA profiling, 10% were current cigarette smokers (n=524), 41% were former smokers (n=2,079), and 48% were never smokers (n=2,420) (Table 1). Former smokers (mean age=60 years) were older than current smokers (51 years) or never smokers (52 years). Imputed WBC count was higher in current smokers (mean WBC=7.2) versus former smokers (mean WBC=6.1) and never smokers (mean WBC=5.9). Levels of inflammatory markers were higher and pulmonary function measures were lower in current versus former versus never smokers. Airflow obstruction was highest in current smokers (14.9%), lower in former smokers (6.2%), and lowest in never smokers (2.8%).
Table 1.
Clinical Characteristics
| Clinical Characteristic* | Available Sample† |
Current Smokers |
Former Smokers |
Never Smokers |
P-value | Total Sample |
|---|---|---|---|---|---|---|
| Sample size (#) | 5023 | 524 (10%) | 2079 (41%) | 2420 (48%) | 5023 | |
| Age (years) | 5023 | 51±11 | 60±13 | 52±13 | <0.0001 | 55±13 |
| Sex (% women) | 5023 | 52% | 55% | 54% | 0.52 | 54% |
| BMI (kg/m2) | 5017 | 27.8±5.9 | 28.5±5.5 | 28.0±5.7 | 0.002 | 28.2±5.6 |
| SBP (mm Hg) | 5023 | 120±16 | 124±17 | 120±17 | <0.0001 | 122±17 |
| DBP (mm Hg) | 5021 | 74±10 | 74±10 | 75±10 | 0.002 | 74±10 |
| Total cholesterol (mg/dL) | 5023 | 190±37 | 186±36 | 187±36 | 0.02 | 187±36 |
| HDL-cholesterol (mg/dL) | 5018 | 57±17 | 59±18 | 59.2±18 | 0.01 | 59±18 |
| Glucose (mg/dL) | 5023 | 101±21 | 103±22 | 99±21 | <0.0001 | 101±21 |
| Diabetes (% yes) | 5018 | 6% | 12% | 7% | <0.0001 | 9% |
| BP Med. (% yes) | 5023 | 26% | 43% | 29% | <0.0001 | 34% |
| Lipid Med. (% yes) | 5023 | 24% | 36% | 22% | <0.0001 | 28% |
| Diabetes Med. (% yes) | 5023 | 4% | 8% | 5% | <0.0001 | 6% |
| WBC (billion cells/L) | 5023 | 7.2±1.4 | 6.1±1.3 | 5.9±1.3 | <0.0001 | 6.1±1.4 |
| CRP (mg/L) | 5014 | 3.8±6.4 | 3.0±5.7 | 2.7±4.9 | <0.0001 | 2.9±5.5 |
| IL-6 (mg/L) | 2,123 | 3.1±2.8 | 2.8±3.3 | 2.2±2.3 | 0.0001 | 2.6±3.0 |
| FEV1 (pp‡) | 4,648 | 0.92±0.16 | 0.97±0.16 | 1.00±0.14 | <0.0001 | 0.98±0.15 |
| FVC (pp‡) | 4,648 | 1.00±0.13 | 1.02±0.14 | 1.03±0.13 | 0.0001 | 1.02±0.14 |
| FEV1/FVC (pp†) | 4,648 | 0.91±0.10 | 0.95±0.09 | 0.97±0.08 | <0.0001 | 0.95±0.09 |
| Airflow Obstruction (% pos) | 4,648 | 15% | 6% | 3% | <0.0001 | 5% |
Clinical characteristics are described as mean ± standard deviation except sample size, cohort, sex, diabetes, and medication use.
The maximum available sample size (N=5,023) reflects the number of subjects with miRNA data (N=5,729), excluding those with missing cell counts (N=705) and missing smoking status (N=1). Smaller available samples reflect missing data for the variable of interest.
FEV1, FVC, and FEV1/FVC were examined as percentages of predicted values (pp), based on standard reference equations derived from healthy never smokers in the NHANES III study.
MiRNA Associations with Smoking Status
Six miRNAs were associated with three-level smoking status (Table 2). Each was detectable in over 4,400 participant samples with the exception of miR-342-5p, which was measurable in 1,115 samples. Five of these miRNAs were associated with current vs. never smokers, with all but one (miR-342-5p) underexpressed in current smokers. MiR-423-5p was the only marker differentially expressed in former versus never smokers, with decreased expression in former smokers. The distribution of miRNA CT values by smoking status is shown in Supplementary Figure 2. Split sample analyses demonstrated a consistent pattern for t-statistics of miRNAs considered separately in the discovery and validation sets. For each significant miRNA, the directionality and scale of effect sizes in discovery and validation sets were consistent with the combined results (Supplementary Table 1).
Table 2.
MiRNA Associations with Smoking Status
| Three-level comparison |
Pairwise comparison | |||||||
|---|---|---|---|---|---|---|---|---|
| miRNA | Sample size | P value | q-value* | Groups | Estimated β† | SE | P-value | Fold Change |
| C v N | 0.31 | 0.06 | 1.5×10−6 | 0.81 | ||||
| miR-1180 | 4905 | 8.1×10−6 | 0.003 | C v F | 0.24 | 0.07 | 2.7×10−4 | 0.85 |
| F v N | 0.07 | 0.04 | 7.2×10−2 | 0.95 | ||||
| miR-181a-2-3p | 4892 | 5.4×10−5 | 0.009 | C v N | 0.27 | 0.06 | 1.6×10−5 | 0.83 |
| C v F | 0.19 | 0.06 | 3.3×10−3 | 0.88 | ||||
| F v N | 0.08 | 0.04 | 3.1×10−2 | 0.95 | ||||
| miR-423-5p | 4418 | 1.3×10−4 | 0.014 | C v N | 0.16 | 0.1 | 1.0×10−1 | 0.90 |
| C v F | −0.09 | 0.1 | 3.3×10−1 | 1.06 | ||||
| F v N | 0.25 | 0.06 | 2.7×10−5 | 0.84 | ||||
| miR-25-5p | 4853 | 1.6×10−4 | 0.014 | C v N | 0.36 | 0.09 | 4.9×10−5 | 0.78 |
| C v F | 0.25 | 0.09 | 5.8×10−3 | 0.84 | ||||
| F v N | 0.11 | 0.05 | 4.2×10−2 | 0.93 | ||||
| miR-1285-3p | 4636 | 8.6×10−4 | 0.057 | C v N | 0.32 | 0.08 | 1.8×10−4 | 0.80 |
| C v F | 0.25 | 0.09 | 3.9×10−3 | 0.84 | ||||
| F v N | 0.07 | 0.05 | 1.9×10−1 | 0.95 | ||||
| miR-342-5p | 1180 | 1.4 ×10−3 | 0.075 | C v N | −0.64 | 0.18 | 4.6×10−4 | 1.56 |
| C v F | −0.41 | 0.19 | 2.8×10−2 | 1.33 | ||||
| F v N | −0.23 | 0.12 | 5.5×10−2 | 1.17 | ||||
We used the Benjamini-Hochberg method to calculate q-values (SAS Proc MULTTEST) and we applied a threshold of FDR<0.10 for the three-level analysis. MiRNAs are sorted by ascending q-value.
Estimated β coefficients represent the mean differences in miRNA CT values between specific smoking groups, where higher CT values reflect lower miRNA expression.
Abbreviations: C=current; F=former; N=never
MiRNA Associations with Smoking-related Clinical Phenotypes
Four smoking-associated miRNAs showed negative associations with loge-transformed serum concentration of CRP: miR-1180 (p=2.0×10−19), miR-181a-2-3p (p=6.7 ×10−6), miR-423-5p (p=2.0×10−8), and miR-25-5p (p=1.3×10−7) (Table 3). One miRNA in our smoking signature, miR-1285-3p, was positively associated with the pro-inflammatory biomarker interleukin-6 (p=7.9×10−6). Analysis of significant miRNAs in relation to pulmonary function measures revealed that miR-1180 was negatively associated with prevalence of airflow obstruction (p=1.8×10−3) and positively associated with FEV1 (p=9.5×10−3) at a marginally significant level (Table 4).
Table 3.
MiRNA Associations with Inflammatory Markers
| loge CRP | loge Interleukin-6 | |||||||
|---|---|---|---|---|---|---|---|---|
| miRNA | Samples | Estimated β* | SE | P-value† | Samples | Estimated β* | SE | P-value† |
|
| ||||||||
| miR-1180 | 5496 | 0.07 | 0.01 | 2.0×10−9 | 2434 | −0.02 | 0.01 | 0.18 |
|
| ||||||||
| miR-181a-2-3p | 5484 | 0.05 | 0.01 | 6.7×10−6 | 2389 | −0.02 | 0.01 | 0.21 |
|
| ||||||||
| miR-423-5p | 4975 | 0.05 | 0.01 | 2.0×10−8 | 1838 | 4.3×10−4 | 0.01 | 0.97 |
|
| ||||||||
| miR-25-5p | 5435 | 0.04 | 0.01 | 1.3×10−7 | 2383 | −0.01 | 0.01 | 0.23 |
|
| ||||||||
| miR-1285-3p | 5145 | 0.01 | 0.01 | 0.27 | 2244 | −0.05 | 0.01 | 7.9×10−6 |
|
| ||||||||
| miR-342-5p | 1325 | −0.01 | 0.02 | 0.40 | 593 | 0.04 | 0.02 | 0.02 |
Estimated β coefficients represent the mean differences in log concentration of the stated inflammatory marker per one standard deviation difference in miRNA CT value, where higher CT values reflect lower miRNA expression.
The p-value threshold for significance (p<1.4×10−3) was determined by the Bonferroni method [0.05/(6 smoking-associated miRNAs*6 clinical phenotypes tested)]. Significant p-values are shown in bold.
Table 4.
MiRNA Associations with Pulmonary Function Measures
| FEV1 | FVC | |||||||
|---|---|---|---|---|---|---|---|---|
| miRNA | Samples | Estimated β* | SE | P-value† | Samples | Estimated β* | SE | P-value† |
|
| ||||||||
| miR-1180 | 4539 | −15.1 | 5.8 | 9.5×10−3 | 4539 | −17.9 | 7.2 | 0.01 |
|
| ||||||||
| miR-181a-2-3p | 4530 | −1.1 | 6.1 | 0.86 | 4530 | −2.3 | 7.5 | 0.76 |
|
| ||||||||
| miR-423-5p | 4092 | −6.4 | 4.4 | 0.14 | 4092 | −8.7 | 5.4 | 0.11 |
|
| ||||||||
| miR-25-5p | 4492 | −2.6 | 4.2 | 0.53 | 4492 | −8.6 | 5.3 | 0.10 |
|
| ||||||||
| miR-1285-3p | 4299 | 4.7 | 4.6 | 0.30 | 4299 | 1.6 | 5.7 | 0.78 |
|
| ||||||||
| miR-342-5p | 1100 | −1.3 | 9.0 | 0.89 | 1100 | 5.4 | 11.1 | 0.63 |
|
| ||||||||
| FEV1/FVC | Airflow Obstruction | |||||||
| miRNA | Samples | Estimated β* | SE | P-value† | Samples | Estimated β* | SE | P-value† |
|
| ||||||||
| miR-1180 | 4539 | −2.8×10−4 | 8.0×10−4 | 0.72 | 4539 | 0.01 | 2.7×10−3 | 1.8×10−3 |
|
| ||||||||
| miR-181a-2-3p | 4530 | 7.2×10−6 | 8.3×10−4 | 0.99 | 4530 | 1.9×10−3 | 2.8×10−3 | 0.50 |
|
| ||||||||
| miR-423-5p | 4092 | 1.6×10−4 | 5.9×10−4 | 0.78 | 4092 | 4.2×10−3 | 2.0×10−3 | 0.03 |
|
| ||||||||
| miR-25-5p | 4492 | 8.3×10−4 | 5.7×10−4 | 0.15 | 4492 | −1.3×10−3 | 1.9×10−3 | 0.51 |
|
| ||||||||
| miR-1285-3p | 4299 | 5.9×10−4 | 6.3×10−4 | 0.35 | 4299 | 1.1×10−3 | 2.1×10−3 | 0.60 |
|
| ||||||||
| miR-342-5p | 1100 | −1.8×10−3 | 1.1×10−3 | 0.13 | 1100 | 0.01 | 4.1×10−3 | 0.10 |
Estimated β coefficients represent the mean differences in the stated pulmonary function measure per one standard deviation difference in miRNA CT value, where higher CT values reflect lower miRNA expression.
The p-value threshold for significance (p<1.4×10−3) was determined by the Bonferroni method [0.05/(6 smoking-associated miRNAs*6 clinical phenotypes tested)]. Marginally significant p-values are shown in bold.
MiRNA Correlations with Smoking-Related Genes
Expression levels of approximately 8,200 genes (nearly 50% of those measured) were associated with cigarette smoking at FDR<0.05. The majority of highly differentially expressed genes were upregulated in the context of cigarette smoking. Coexpression analysis identified 4,137 smoking-associated gene transcripts that were coexpressed with at least one of the 345 miRNAs measured and 2,116 smoking-associated gene transcripts that were coexpressed with one or more of the six smoking-associated miRNAs (at FDR<0.01 in the discovery and validation sets). There were 1,666 genes coexpressed with miR-25-5p, 524 with miR-1285-3p, 491 with miR-1180, 446 with miR-181a-2-3p, 275 with miR-423-5p, and none with miR-342-5p. We further identified 116 coexpressed pairs in which the mRNA was a predicted (n=111) or validated (n=5) target of the correlated miRNA. There were 17 predicted targets for miR-1180, 13 predicted for miR-181a-2-3p, 32 predicted for miR-1285-3p, five predicted for miR-25-5p, and five validated and 44 predicted for miR-423-5p (Supplementary Table 2).
GO Analysis
Independent GO enrichment analyses were conducted on the sets of smoking-associated mRNA transcripts that were coexpressed with each of our top miRNAs. Figure 1 shows the network of GO categories for the coexpressed mRNAs. Supplementary Tables 3 and 4 provide detailed information on the GO analysis and the overlapping genes in each category. The expression of miR-1180, our top smoking-associated miRNA, was negatively correlated with the expression of genes involved in monocyte differentiation (enrichment p=3.1×10−5). MiR-181a-2-3p expression was positively correlated with expression of genes that function in immune cell mediated cytotoxicity (enrichment p=5.7×10−5). MiR-181a-2-3p was negatively correlated with genes involved in viral response (enrichment p=4.7×10−6). Coexpressed mRNAs for the remaining miRNAs were enriched for a variety of common pathways, e.g., regulation of transcription and protein processing, which are known to be preferentially targeted by randomly selected sets of miRNAs.38
Figure 1.
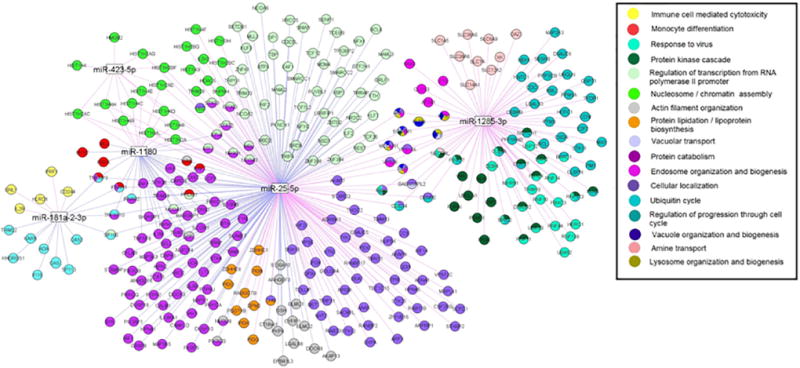
Network of Gene Ontology Categories for Smoking-Associated mRNAs Coexpressed with Smoking-Associated miRNAs. Smoking-associated gene transcripts that are coexpressed with one or more smoking-associated miRNAs and represented by one or more GO categories. Only gene transcripts belonging to GO categories meeting the Bonferroni-corrected enrichment p-value threshold of p<10−5, are included (see Supplementary Tables 3 and 4 for GO results). Blue lines denote negative correlations, while purple lines indicate positive correlations between miRNA and mRNA expression. This figure was drawn by ProteoLens.37
We also performed GO enrichment analysis on the set of miRNA-mRNA coexpressed pairs in which the mRNA was a predicted (n=112) or validated (n=5) target of the correlated miRNA. In the aggregate results, the GO term with the greatest fold enrichment was T cell homeostasis (enrichment p=4.2×10−4 ; q-value=0.038) (Supplementary Table 5). Two of the four genes in the T cell homeostasis pathway were predicted targets of miR-1180: BAX (coexpression q-value=7.2×10−8) and P2RX7 (coexpression q-value=6.1×10−8); expression of miR-1180 was negatively correlated with expression of BAX and positively correlated with expression of P2RX7 (Supplementary Table 2). Other enriched GO terms, such as regulation of gene expression, represent common functional pathways, as described above.
miRNA Effects on Inflammatory Mediators
To determine whether individual miRNAs that are associated with cigarette smoking status and smoking-induced inflammation could be sufficient to modulate the expression of inflammatory mediators, we measured cytokine elaboration by human lung epithelial cells separately transfected with mimetics for miR-1180 and miR-1285-3p A non-targeting (NT) miRNA mimetic served as a negative control. Expression levels of eight cytokines were quantified in cell supernatants: interleukin-6, interleukin-8, granulocyte macrophage colony-stimulating factor (GM-CSF), granulocyte colony-stimulating factor (G-CSF), chemokine C-C motif ligand 2 (CCL2), chemokine C-C motif ligand 20 (CCL20), chemokine C-X-C motif ligand 5 (CXCL5), and chemokine C-X-C motif ligand 6 (CXCL6). Transfection of miR-1180 reduced production of GM-CSF (p=0.01) in stimulated cells (Figure 2). Basal levels of CXCL5 and CXCL6 were increased in response to transfection of both miRNAs, and miR-1285-3p transfection also increased production of CXCL5 (p=0.003) in stimulated cells. Other cytokines tested, including interleukin-6, interleukin-8, G-CSF, CCL20, and CCL2, were not significantly affected by the miRNAs examined.
Figure 2.
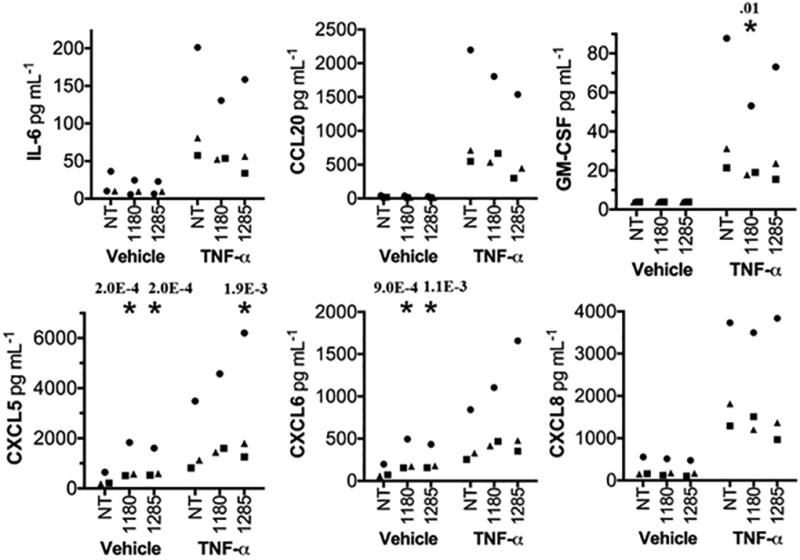
Smoking-Related miRNAs Alter Cytokine Expression in Vitro. Multiplex cytokine bead array analysis of A549 cell culture supernatants after transfection with non-targeting (NT), miR-1180, or miR-1285-3p mimetics. Cells were stimulated 24 hours post-transfection with vehicle or 10 ng/mL rhTNF-α for 6 hours. Squares, triangles, and circles represent data collected from three independent experiments, normalized to the cytokine concentrations in the negative control group (NT vehicle). The * indicates p<0.01 (significant p-values shown) when compared to NT control within a given stimulation group as determined by a two-way ANOVA followed by Dunnett’s test, using raw rather than normalized values.
Discussion
Cigarette smoking acutely promotes oxidative stress, as evidenced by increased plasma peroxides and decreased antioxidants, and chronic smoking leads to systemic inflammation characterized by the upregulation of circulating inflammatory cells (primarily neutrophils, lymphocytes, and platelets), inflammatory mediators, and acute-phase proteins.39, 40 To test our hypothesis that miRNAs may have predictive value for smoking-associated diseases, we characterized a comprehensive miRNA signature of cigarette smoking using high-throughput RT-qPCR profiling of whole blood RNA from a large, population-based sample.
We identified six miRNAs that were characteristically dysregulated in the context of smoking, five of which were downregulated in smokers versus non-smokers (Table 2). These results are supported by the widespread downregulation of miRNAs found in cigarette smoke-exposed lung tissue from mouse models and human samples.12, 20, 41–43 Our whole blood-derived miRNA signature of cigarette smoking is novel, and the results show little overlap with previous studies. This lack of concordance is common across miRNA-smoking studies, and can be explained by diverse tissue sources, non-uniform tobacco exposures, and different miRNA platforms. Two of our top hits —miR-1180 and miR-1285-3p— are recently identified miRNAs44, 45 that have been linked to cancer-specific gene regulatory networks; miR-1180 has been shown to inhibit cell apoptosis in human hepatocellular carcinoma both in vivo and in vitro, while miR-1285-3p inhibits the expression of tumor suppressor p53.46, 47 MiR-181a-2-3p is involved in the immune response as a positive regulator of B-cell development and T-cell sensitivity.48, 49 The remaining three miRNAs have been associated with multiple human cancers, and miR-423-5p and miR-25-5p have been linked to heart disease.50, 51 Dysregulation of circulating miR-342-5p has been found in autoimmune conditions; miR-342-5p has been shown to promote inflammatory activation of macrophages in atherosclerotic lesions, consistent with our finding that current cigarette smokers exhibited higher miR-342-5p expression compared to never smokers.52–54
Whole blood is readily accessible and can be stored for later RNA extraction. This stable form of blood-derived RNA can be isolated with relative ease and high purity, making miRNA results in whole blood particularly amenable to clinical translation. miRNAs detected in whole blood may be present in leukocytes, platelets, erythrocytes, and/or cell-free plasma.55 In previous work using the same miRNA detection platform and a small sample of healthy volunteers, several of our coauthors defined the distributions of over 600 circulating miRNAs in whole blood, peripheral blood mononuclear cells (PBMCs), platelets, plasma, and serum.56 Of our significant miRNAs, three (miR-181a-2-3p, miR-25-5p, and miR-423-5p) were detectable in whole blood and PBMCs only, while the remaining three (miR-1180, miR-1285-3p, and miR-342-5p) were additionally detectable in platelets.56 These data suggest that our miRNA expression patterns are largely derived from leukocytes, and secondarily from platelets. We may also have captured signals from small amounts of cell-free miRNAs, which can be released from or targeted to smoking-relevant tissues. Because miRNA profiling in whole blood cannot distinguish cell type-specific effects, further studies are needed to localize the observed miRNA patterns to specific blood components.
To explore the functional significance of the cigarette smoking-related miRNAs, we evaluated their associations with smoking-related clinical phenotypes: two biomarkers of systemic inflammation and four measures of pulmonary function. Population-based studies have demonstrated that the systemic inflammatory state induced by cigarette smoking is associated with increased levels of acute-phase proteins (CRP, fibrinogen, etc.) and pro-inflammatory cytokines (interleukin-6, TNF-α, etc.) in the peripheral circulation.57–59 In the Framingham Heart Study, serum CRP and interleukin-6 levels were significantly higher in current cigarette smokers than never smokers.60 We investigated the relations of smoking-associated miRNAs to systemic inflammation by relating our significant miRNAs to serum concentrations of CRP and interleukin-6. Four miRNAs were negatively associated with CRP: miR-1180, miR-181a-2-3p, miR-423-5p, and miR-25-5p (Table 3). In support of these findings, mir-181a-2-3p has been shown to downregulate inflammatory factors in monocytes and macrophages.61 MiR-25-5p is downregulated in human airway smooth muscle cells in response to pro-inflammatory stimuli and is proposed to regulate expression of multiple inflammatory mediators in these cells.62 Elevated levels of miR-423-5p were found in surgical lung biopsies of idiopathic pulmonary fibrosis, in which inflammation may play a critical role.63, 64 One miRNA, miR-1285-3p, was positively associated with interleukin-6 (Table 3). Additional studies may shed light on the discrepancy between miRNA associations with CRP and with interleukin-6 observed in our cross-sectional analysis.
Cigarette smoking is an established cause of impaired pulmonary function.65 Smoking is the major risk factor for chronic obstructive pulmonary disease (COPD), which is defined by irreversible airflow obstruction and is the fourth leading cause of death worldwide.66 Smoking-induced COPD is driven by inflammation, as the activation of neutrophils, macrophages, and lymphocytes and associated secretion of inflammatory mediators trigger a chronic inflammatory response in the lung.67 Premature senescence of epithelial and immune cells, the latter of which heightens inflammation, has also been implicated in the pathogenesis of smoking-induced COPD.68 Nevertheless, the causal mechanisms and molecular players underlying COPD progression in the lungs of smokers have yet to be elucidated. Analysis of our significant miRNAs in relation to pulmonary function measures revealed that miR-1180 was negatively associated with airflow obstruction and positively associated with FEV1 at a marginally significant level (Table 4). Although our cross-sectional results suggest that miRNAs may serve as biomarkers for smoking-related inflammation and target organ damage, we cannot deduce with certainty the directionality of observed associations.
To better understand the mechanistic connections between dysregulated miRNA expression, systemic inflammation, and lung dysfunction in the context of cigarette smoking, we identified over 1,600 smoking-associated genes that were coexpressed with one or more of our top miRNAs in whole blood. GO analysis of the mRNAs correlated with each miRNA showed enrichment for immune-related pathways among genes coexpressed with miR-1180 and miR-181a-2-3p (Figure 1 and Supplementary Table 3). MiR-1180 expression was negatively correlated with expression of genes involved in differentiation of several immune cell types. MiR-181a-2-3p level was positively correlated with genes for immune cell-mediated cytotoxicity and negatively correlated with viral response genes (Figure 1 and Supplementary Table 3). We further interrogated the regulatory networks that these miRNAs may be involved in by cross-referencing our coexpressed pairs with miRNA target databases. We identified 116 coexpressed pairs in which the mRNA was a predicted or validated target of its correlated miRNA. GO analysis of the predicted targets highlighted T-cell homeostasis, and two of four overlapping genes in this pathway were predicted targets of miR-1180 (Supplementary Tables 2 and 5).
We hypothesize that our miRNA-mRNA coexpressed pairs, especially those involving predicted targets, describe gene regulatory networks primarily localized to leukocytes and platelets. Loss-of-function experiments have demonstrated the broad importance of miRNAs in development, including the developmental programs of B cells, T cells, and other leukocytes.69 Moreover, specific “immuno-miRNAs,” including miR-181a-2-3p, have been shown to regulate lymphocyte differentiation and function.70 Although platelets are anuclear, they contain cytoplasmic mRNA, carry out protein translation, and express over 280 miRNAs, and platelet miRNA-mRNA coexpression profiles have been correlated with platelet reactivity.71, 72 Taken together, these bioinformatic results suggest that smoking-induced dysregulation of the aforementioned miRNAs may play a mediating role in inflammatory responses by modulating miRNA-specific gene regulatory networks in leukocytes and, to a lesser extent, platelets.
Our study has several strengths, including the large community-based cohort of young, middle-aged, and older adults and the ascertainment of miRNAs, inflammatory markers, and pulmonary function measures adhering to rigorous quality control protocols. Our cross-sectional results highlight smoking-associated miRNAs that have the potential to serve as non-invasive biomarkers of smoking exposure or smoking-related diseases. Moreover, our functional evidence suggesting that these miRNAs can mediate lung inflammation combined with our bioinformatic identification of coexpressed predicted gene targets is a first step toward determining disease-relevant miRNA targets that may be funneled into emerging clinical pipelines for RNA interference-based therapeutics.73 Our results highlight select smoking-associated miRNAs and are consistent with the hypothesis that smoking-associated miRNAs function as mediators of cigarette smoking-induced inflammation and downstream target organ damage (Figure 3). Additional functional studies are needed to assess whether these miRNAs are causally associated with smoking-related phenotypes.
Figure 3.
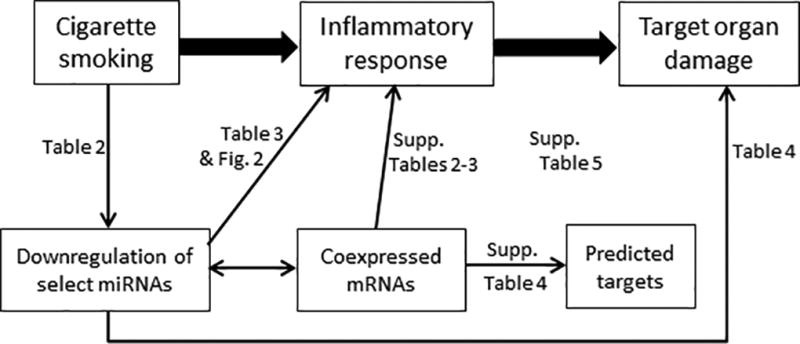
Proposed Roles for Smoking-Associated miRNAs in Mediating Target Organ Damage. Thick arrows indicate established relationships, while thin arrows indicate observed associations or correlations based on epidemiological analyses (Tables 2–4), bioinformatics approaches (Supplementary Tables 2–5), and functional experiment (Figure 2).
We also acknowledge several limitations. Our study participants were largely of European ancestry, and the generalizability of our findings to other races/ethnicities is uncertain. Because we analyzed results across a large number of miRNAs, mRNAs, and miRNA-mRNA coexpressed pairs, our study is susceptible to both false positive and false negative findings despite adjustment for multiple testing. MiRNA profiling was conducted on whole blood and as such may not reflect tissue-specific patterns of expression – even though we adjusted for differential blood cell composition imputed from mRNA expression values. Although we present evidence of robust validation of our cross-sectional results based on analysis of split samples for discovery and validation, our study lacks external replication. We cannot exclude residual confounding or establish causal associations in the cross-sectional results. Our functional study also has limitations: small sample size (n=3 independent experiments); single time-point collection; restriction to one cell type; and reliance on miRNA overexpression, which could target genes that would not be affected under physiological conditions.
The proposed identification of gene regulatory networks involving coexpressed miRNA-mRNA pairs assumes temporal sequence and spatial co-localization, neither of which can be proven by cross-sectional correlations in whole blood. It is possible that different cell types or blood components account for changes in miRNA and mRNA expression and that tissue specificity precludes their joint participation in intracellular regulatory networks. Observed correlations may also indicate common consequences of smoking-induced damage rather than mutual involvement in mediating pathways. We addressed these issues in part by identifying coexpressed pairs in which the mRNA was a predicted target of the correlated miRNA. However, we were underpowered to study miRNA-specific pathway enrichment, and the functional relevance of computational target prediction remains unclear. Because gene repression by miRNAs is generally modest and tunable, whether a target is actually affected depends on the local concentrations of both miRNA and mRNA, such that predicted relationships may be irrelevant in a given tissue.74 Furthermore, predicted target analysis only addresses direct targeting relationships outside the context of intact cells, but miRNAs act within complex regulatory networks that cannot be fully captured by first-order correlations.75
Because our bioinformatics analysis linked the smoking-associated miRNAs to inflammation, and because miRNAs can target transcripts for cytokines and for signaling intermediates upstream of cytokine expression,76–80 we tested whether these miRNAs were capable of modulating inflammatory cytokines relevant to cigarette smoking. To our knowledge, none of these miRNAs has previously been connected to pulmonary inflammation. We observed miRNA-specific effects on cytokine elaboration by lung cells for the two miRNAs tested, with miR-1180 reducing production of GM-CSF, and miR-1180 and miR-1285-3p increasing levels of CXCL5 and CXCL6 (Figure 2). Although miR-1285-3p was positively associated with interleukin-6 in the cross-sectional analysis, overexpression of miR-1285-3p in lung cells did not significantly affect interleukin-6 levels. GM-CSF drives alveolar macrophage maturation and innate immune activities in the lung.81 CXCL5 and CXCL6 are lung epithelium-derived ligands for the CXCR2 receptor on neutrophils; these chemokines recruit their cognate leukocytes to the lungs during pulmonary inflammation.82,83 Each of these cytokine pathways influences cigarette smoking-induced inflammation and disease,84–88 so their regulation by miRNA changes could impact the health of cigarette smokers. The cytokine changes suggest that decreased levels of these miRNAs could skew smoking-induced inflammation towards mononuclear cell inflammation and away from neutrophilic infiltration. Future studies are needed to determine the mechanisms responsible for regulating miRNA levels affected by cigarette smoking, to identify the transcripts targeted by these miRNAs that modulate cytokine expression, and to elucidate the net effects of these multiple miRNA changes on integrated inflammatory responses to cigarette smoke. Our results provide evidence in support of the hypothesis that the miRNAs associated with smoking, inflammation, and pulmonary function in an observational study are functionally capable of dictating expression of the cytokines governing inflammation and disease in cigarette smokers.
Supplementary Material
Clinical Perspective.
Cigarette smoking increases risk for cancer, lung disease, and cardiovascular disease. MicroRNAs (miRNAs) regulate gene expression and may play a role in smoking-induced target organ damage. We sought to identify a miRNA signature of cigarette smoking and relate it to smoking-associated clinical phenotypes, gene expression, and lung inflammatory signaling. By studying miRNA expression levels in approximately 5000 people with cigarette smoking histories, we identified a six-miRNA signature of smoking. The top miRNAs were associated with systemic inflammatory markers and reduced pulmonary function, correlated with expression of genes involved in immune function, and were sufficient to modulate inflammatory signaling. Our results highlight smoking-associated miRNAs and are consistent with the hypothesis that smoking-associated miRNAs serve as mediators of smoking-induced inflammation and target organ damage. These findings call for further mechanistic studies to explore the diagnostic and therapeutic utility of smoking-related miRNAs.
Acknowledgments
Funding Source: The Framingham Heart Study is funded by National Institutes of Health contract HHSN268201500001I; N01-HC-25195. Laboratory work for this investigation was funded by the Division of Intramural Research, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD. The analytical component of this project was funded by the Division of Intramural Research, National Heart, Lung, and Blood Institute, and the Center for Information Technology, National Institutes of Health, Bethesda, MD. The inflammatory biomarkers measured at Examination 8 were funded by R01 AG028321. Drs. Freedman and Tanriverdi are supported in part by the NIH Common Fund 1 UH2TR000921-02 and 1 U01 OD019771. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov). Dr. Benjamin is supported by 1P50HL120163.
Footnotes
Disclosures: None
References
- 1.US Department of Health and Human Services. The health consequences of smoking: A report of the surgeon general. Washington: US Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health; 2004. [Google Scholar]
- 2.Bjartveit K, Tverdal A. Health consequences of smoking 1–4 cigarettes per day. Tobacco Control. 2005;14:315–320. doi: 10.1136/tc.2005.011932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mackay J. The global epidemiology of tobacco and related chronic diseases. Public Health. 2012;126:199–201. doi: 10.1016/j.puhe.2012.01.027. [DOI] [PubMed] [Google Scholar]
- 4.Chapman S. Public health advocacy and tobacco control: Making smoking history. Wiley.com. 2008 [PubMed] [Google Scholar]
- 5.Wahid F, Shehzad A, Khan T, Kim YY. MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim Biophys Acta. 2010;1803:1231–43. doi: 10.1016/j.bbamcr.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 6.Sand M. The pathway of miRNA maturation. Methods Mol Biol. 2014;1095:3–10. doi: 10.1007/978-1-62703-703-7_1. [DOI] [PubMed] [Google Scholar]
- 7.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–87. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 11.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi K, Yokota S, Tatsumi N, Fukami T, Yokoi T, Nakajima M. Cigarette smoking substantially alters plasma microRNA profiles in healthy subjects. Toxicol Appl Pharmacol. 2013;272:154–60. doi: 10.1016/j.taap.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 13.Meltzer PS. Cancer genomics: small RNAs with big impacts. Nature. 2005;435:745–6. doi: 10.1038/435745a. [DOI] [PubMed] [Google Scholar]
- 14.Bhaskaran M, Mohan M. MicroRNAs: history, biogenesis, and their evolving role in animal development and disease. Vet Pathol. 2014;51:759–74. doi: 10.1177/0300985813502820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, et al. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 17.Small EM, Frost RJ, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 2010;121:1022–32. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oglesby IK, Mcelvaney NG, Greene CM. MicroRNAs in inflammatory lung disease--master regulators or target practice? Respir Res. 2010;11:148. doi: 10.1186/1465-9921-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graff JW, Powers LS, Dickson AM, Kim J, Reisetter AC, Hassan IH, et al. Cigarette smoking decreases global microRNA expression in human alveolar macrophages. PLoS ONE. 2012;7:e44066. doi: 10.1371/journal.pone.0044066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schembri F, Sridhar S, Perdomo C, Gustafson AM, Zhang X, Ergun A, et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc Natl Acad Sci USA. 2009;106:2319–24. doi: 10.1073/pnas.0806383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van pottelberge GR, Mestdagh P, Bracke KR, Thas O, van Durme YM, Joos GF, et al. MicroRNA expression in induced sputum of smokers and patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2011;183(7):898–906. doi: 10.1164/rccm.201002-0304OC. [DOI] [PubMed] [Google Scholar]
- 24.Dawber TR, Meadors GF, Moore FE. Epidemiological approaches to heart disease: the Framingham Study. Am J Public Health Nations Health. 1951;41:279–81. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feinleib M, Kannel WB, Garrison RJ, Mcnamara PM, Castelli WP. The Framingham Offspring Study. Design and preliminary data. Prev Med. 1975;4:518–25. doi: 10.1016/0091-7435(75)90037-7. [DOI] [PubMed] [Google Scholar]
- 26.Splansky GL, Corey D, Yang Q, Atwood LD, Cupples LA, Benjamin EJ, et al. The Third Generation Cohort of the National Heart, Lung, and Blood Institute’s Framingham Heart Study: design, recruitment, and initial examination. Am J Epidemiol. 2007;165:1328–35. doi: 10.1093/aje/kwm021. [DOI] [PubMed] [Google Scholar]
- 27.Wang TJ, Nam BH, Wilson PW, Wolf PA, Levy D, Polak JF, et al. Association of C-reactive protein with carotid atherosclerosis in men and women: the Framingham Heart Study. Arterioscler Thromb Vasc Biol. 2002;22:1662–7. doi: 10.1161/01.atv.0000034543.78801.69. [DOI] [PubMed] [Google Scholar]
- 28.Schnabel R, Larson MG, Dupuis J, Lunetta KL, Lipinska I, Meigs JB, et al. Relations of inflammatory biomarkers and common genetic variants with arterial stiffness and wave reflection. Hypertension. 2008;51:1651–7. doi: 10.1161/HYPERTENSIONAHA.107.105668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McManus DD, Beaulieu LM, Mick E, Tanriverdi K, Larson MG, Keaney JF, Jr, et al. Relationship among circulating inflammatory proteins, platelet gene expression, and cardiovascular risk. Arterioscler Thromb Vasc Biol. 2013;33:2666–73. doi: 10.1161/ATVBAHA.112.301112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller MR, Hankinson J, Brusasco V, Burgos F, Casaburi R, Coates A, et al. Standardisation of spirometry. Eur Respir J. 2005;26:319–38. doi: 10.1183/09031936.05.00034805. [DOI] [PubMed] [Google Scholar]
- 31.Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med. 1999;159:179–87. doi: 10.1164/ajrccm.159.1.9712108. [DOI] [PubMed] [Google Scholar]
- 32.Joehanes R, Ying S, Huan T, Johnson AD, Raghavachari N, Wang R, et al. Gene expression signatures of coronary heart disease. Arterioscler Thromb Vasc Biol. 2013;33:1418–26. doi: 10.1161/ATVBAHA.112.301169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huan T, Zhang B, Wang Z, Joehanes R, Zhu J, Johnson AD, et al. A systems biology framework identifies molecular underpinnings of coronary heart disease. Arterioscler Thromb Vasc Biol. 2013;33:1427–34. doi: 10.1161/ATVBAHA.112.300112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing. 2012 [Google Scholar]
- 35.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;B:289–300. [Google Scholar]
- 36.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huan T, Sivachenko AY, Harrison SH, Chen JY. ProteoLens: a visual analytic tool for multi-scale database-driven biological network data mining. BMC Bioinformatics. 2008;9:S5. doi: 10.1186/1471-2105-9-S9-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bleazard T, Lamb JA, Griffiths-Jones S. Bias in microRNA functional enrichment analysis. Bioinformatics. 2015;31:1592–8. doi: 10.1093/bioinformatics/btv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yanbaeva DG, Dentener MA, Creutzberg EC, Wesseling G, Wouters EF. Systemic effects of smoking. Chest. 2007;131:1557–66. doi: 10.1378/chest.06-2179. [DOI] [PubMed] [Google Scholar]
- 40.Jensen EJ, Pedersen B, Frederiksen R, Dahl R. Prospective study on the effect of smoking and nicotine substitution on leucocyte blood counts and relation between blood leucocytes and lung function. Thorax. 1998;53:784–9. doi: 10.1136/thx.53.9.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2009;23:806–12. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christenson S, Schembri F, Sridhar S, Campbell J, Zhang X, Lenburg M, et al. Impact of smoking on miRNA expression in buccal and nasal epithelium. American Journal of Respiratory and Critical Care Medicine. 2009:179. [Google Scholar]
- 43.Russ R, Slack FJ. Cigarette-Smoke-Induced Dysregulation of MicroRNA Expression and Its Role in Lung Carcinogenesis. Pulm Med. 2012;2012:791234. doi: 10.1155/2012/791234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morin RD, O’Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18(4):610–21. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bar M, Wyman SK, Fritz BR, Qi J, Garg KS, Parkin RK, et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells. 2008;26(10):2496–505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan G, Wu L, Tan J, Zhang B, Tai WC, Xiong S, et al. MiR-1180 promotes apoptotic resistance to human hepatocellular carcinoma via activation of NF-κB signaling pathway. Sci Rep. 2016;6:22328. doi: 10.1038/srep22328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian S, Huang S, Wu S, Guo W, Li J, He X. MicroRNA-1285 inhibits the expression of p53 by directly targeting its 3’ untranslated region. Biochem Biophys Res Commun. 2010;396(2):435–9. doi: 10.1016/j.bbrc.2010.04.112. [DOI] [PubMed] [Google Scholar]
- 48.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–6. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 49.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–61. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 50.Wahlquist C, Jeong D, Rojas-Muñoz A, Kho C, Lee A, Mitsuyama S, et al. Inhibition of miR-25 improves cardiac contractility in the failing heart. Nature. 2014;508:531–5. doi: 10.1038/nature13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nabiałek E, Wańha W, Kula D, Jadczyk T, Krajewska M, Kowalówka A, et al. Circulating microRNAs (miR-423-5p, miR-208a and miR-1) in acute myocardial infarction and stable coronary heart disease. Minerva Cardioangiol. 2013;61:627–37. [PubMed] [Google Scholar]
- 52.Carlsen AL, Schetter AJ, Nielsen CT, Lood C, Knudsen S, Voss A, et al. Circulating microRNA expression profiles associated with systemic lupus erythematosus. Arthritis Rheum. 2013;65:1324–34. doi: 10.1002/art.37890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi YL, Weiland M, Lim HW, Mi QS, Zhou L. Serum miRNA expression profiles change in autoimmune vitiligo in mice. Exp Dermatol. 2014;23:140–2. doi: 10.1111/exd.12319. [DOI] [PubMed] [Google Scholar]
- 54.Wei Y, Nazari-jahantigh M, Chan L, Zhu M, Heyll K, Corbalán-Campos J, et al. The microRNA-342-5p fosters inflammatory macrophage activation through an Akt1- and microRNA-155-dependent pathway during atherosclerosis. Circulation. 2013;127:1609–19. doi: 10.1161/CIRCULATIONAHA.112.000736. [DOI] [PubMed] [Google Scholar]
- 55.Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110:483–95. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 56.Freedman JE, Ercan B, Morin KM, Liu CT, Tamer L, Ayaz L, et al. The distribution of circulating microRNA and their relation to coronary disease. F1000Res. 2012;1:50. doi: 10.12688/f1000research.1-50.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fröhlich M, Sund M, Löwel H, Imhof A, Hoffmeister A, Koenig W. Independent association of various smoking characteristics with markers of systemic inflammation in men. Results from a representative sample of the general population (MONICA Augsburg Survey 1994/95) Eur Heart J. 2003;24:1365–72. doi: 10.1016/s0195-668x(03)00260-4. [DOI] [PubMed] [Google Scholar]
- 58.Wannamethee SG, Lowe GD, Shaper AG, Rumley A, Lennon L, Whincup PH. Associations between cigarette smoking, pipe/cigar smoking, and smoking cessation, and haemostatic and inflammatory markers for cardiovascular disease. Eur Heart J. 2005;26:1765–73. doi: 10.1093/eurheartj/ehi183. [DOI] [PubMed] [Google Scholar]
- 59.Bermudez EA, Rifai N, Buring JE, Manson JE, Ridker PM. Relation between markers of systemic vascular inflammation and smoking in women. Am J Cardiol. 2002;89:1117–9. doi: 10.1016/s0002-9149(02)02284-1. [DOI] [PubMed] [Google Scholar]
- 60.Levitzky YS, Guo CY, Rong J, Larson MG, Walter RE, Keaney JF, Jr, et al. Relation of smoking status to a panel of inflammatory markers: the Framingham offspring. Atherosclerosis. 2008;201:217–24. doi: 10.1016/j.atherosclerosis.2007.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie W, Li M, Xu N, Lv Q, Huang N, He J, et al. MiR-181a regulates inflammation responses in monocytes and macrophages. PLoS ONE. 2013;8:e58639. doi: 10.1371/journal.pone.0058639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuhn AR, Schlauch K, Lao R, Halayko AJ, Gerthoffer WT, Singer CA. MicroRNA expression in human airway smooth muscle cells: role of miR-25 in regulation of airway smooth muscle phenotype. Am J Respir Cell Mol Biol. 2010;42:506–13. doi: 10.1165/rcmb.2009-0123OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oak SR, Murray L, Herath A, Sleeman M, Anderson I, Joshi AD, et al. A micro RNA processing defect in rapidly progressing idiopathic pulmonary fibrosis. PLoS ONE. 2011;6:e21253. doi: 10.1371/journal.pone.0021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bringardner BD, Baran CP, Eubank TD, Marsh CB. The role of inflammation in the pathogenesis of idiopathic pulmonary fibrosis. Antioxid Redox Signal. 2008;10:287–301. doi: 10.1089/ars.2007.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anthonisen NR, Connett JE, Murray RP. Smoking and lung function of Lung Health Study participants after 11 years. Am J Respir Crit Care Med. 2002;166:675–9. doi: 10.1164/rccm.2112096. [DOI] [PubMed] [Google Scholar]
- 66.Pauwels RA, Buist AS, Calverley PM, Jenkins CR, Hurd SS. Global strategy for the diagnosis, management, prevention of chronic obstructive pulmonary disease. NHLBI/WHO Global Initiative for Chronic Obstructive Lung Disease (GOLD) Workshop summary. Am J Respir Crit Care Med. 2001;163:1256–76. doi: 10.1164/ajrccm.163.5.2101039. [DOI] [PubMed] [Google Scholar]
- 67.Gonçalves RB, Coletta RD, Silvério KG, Benevides L, Casati MZ, da Silva JS, et al. Impact of smoking on inflammation: overview of molecular mechanisms. Inflamm Res. 2011;60:409–24. doi: 10.1007/s00011-011-0308-7. [DOI] [PubMed] [Google Scholar]
- 68.John-schuster G, Günter S, Hager K, Conlon TM, Eickelberg O, Yildirim AÖ. Inflammaging increases susceptibility to cigarette smoke-induced COPD. Oncotarget. 2015 doi: 10.18632/oncotarget.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luck ME, Muljo SA, Collins CB. Prospects for Therapeutic Targeting of MicroRNAs in Human Immunological Diseases. J Immunol. 2015;194:5047–52. doi: 10.4049/jimmunol.1403146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kroesen BJ, Teteloshvili N, Smigielska-czepiel K, Brouwer E, Boots AM, van den Berg A, et al. Immuno-miRs: critical regulators of T-cell development, function and ageing. Immunology. 2015;144:1–10. doi: 10.1111/imm.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stakos DA, Gatsiou A, Stamatelopoulos K, Tselepis AD, Stellos K. Platelet microRNAs: From platelet biology to possible disease biomarkers and therapeutic targets. Platelets. 2013;24:579–89. doi: 10.3109/09537104.2012.724483. [DOI] [PubMed] [Google Scholar]
- 72.Nagalla S, Shaw C, Kong X, Kondkar AA, Edelstein LC, Ma L, et al. Platelet microRNA-mRNA coexpression profiles correlate with platelet reactivity. Blood. 2011;117:5189–97. doi: 10.1182/blood-2010-09-299719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haussecker D, Kay MA. RNAinterference. Drugging RNAi. Science. 2015;347:1069–70. doi: 10.1126/science.1252967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–24. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peláez N, Carthew RW. Biological robustness and the role of microRNAs: a network perspective. Curr Top Dev Biol. 2012;99:237–55. doi: 10.1016/B978-0-12-387038-4.00009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jones MR, Quinton LJ, Blahna MT, Neilson JR, Fu S, Ivanov AR, et al. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat Cell Biol. 2009;11:1157–63. doi: 10.1038/ncb1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aguado LC, Schmid S, Sachs D, Shim JV, Lim JK, Tenoever BR. microRNA Function Is Limited to Cytokine Control in the Acute Response to Virus Infection. Cell Host Microbe. 2015;18:714–22. doi: 10.1016/j.chom.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Escobar TM, Kanellopoulou C, Kugler DG, Kilaru G, Nguyen CK, Nagarajan V, et al. miR-155 activates cytokine gene expression in Th17 cells by regulating the DNA-binding protein Jarid2 to relieve polycomb-mediated repression. Immunity. 2014;40:865–79. doi: 10.1016/j.immuni.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schulte LN, Eulalio A, Mollenkopf HJ, Reinhardt R, Vogel J. Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J. 2011;30:1977–89. doi: 10.1038/emboj.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shibata Y, Berclaz PY, Chroneos ZC, Yoshida M, Whitsett JA, Trapnell BC. GM-CSF regulates alveolar macrophage differentiation and innate immunity in the lung through PU.1. Immunity. 2001;15:557–67. doi: 10.1016/s1074-7613(01)00218-7. [DOI] [PubMed] [Google Scholar]
- 82.Mei J, Liu Y, Dai N, Favara M, Greene T, Jeyaseelan S, et al. CXCL5 regulates chemokine scavenging and pulmonary host defense to bacterial infection. Immunity. 2010;33:106–17. doi: 10.1016/j.immuni.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Besnard AG, Struyf S, Guabiraba R, Fauconnier L, Rouxel N, Proost P, et al. CXCL6 antibody neutralization prevents lung inflammation and fibrosis in mice in the bleomycin model. J Leukoc Biol. 2013;94:1317–23. doi: 10.1189/jlb.0313140. [DOI] [PubMed] [Google Scholar]
- 84.Crotty alexander LE, Shin S, Hwang JH. Inflammatory Diseases of the Lung Induced by Conventional Cigarette Smoke: A Review. Chest. 2015;148:1307–22. doi: 10.1378/chest.15-0409. [DOI] [PubMed] [Google Scholar]
- 85.Balamayooran G, Batra S, Cai S, Mei J, Worthen GS, Penn AL, et al. Role of CXCL5 in leukocyte recruitment to the lungs during secondhand smoke exposure. Am J Respir Cell Mol Biol. 2012;47:104–11. doi: 10.1165/rcmb.2011-0260OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bracke KR, D’hulst AI, Maes T, Moerloose KB, Demedts IK, Lebecque S, et al. Cigarette smoke-induced pulmonary inflammation and emphysema are attenuated in CCR6-deficient mice. J Immunol. 2006;177:4350–9. doi: 10.4049/jimmunol.177.7.4350. [DOI] [PubMed] [Google Scholar]
- 87.Thatcher TH, Mchugh NA, Egan RW, Chapman RW, Hey JA, Turner CK, et al. Role of CXCR2 in cigarette smoke-induced lung inflammation. Am J Physiol Lung Cell Mol Physiol. 2005;289:L322–8. doi: 10.1152/ajplung.00039.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vlahos R, Bozinovski S, Chan SP, Ivanov S, Lindén A, Hamilton JA, et al. Neutralizing granulocyte/macrophage colony-stimulating factor inhibits cigarette smoke-induced lung inflammation. Am J Respir Crit Care Med. 2010;182:34–40. doi: 10.1164/rccm.200912-1794OC. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


