Abstract
Human clear cell renal cell carcinoma (ccRCC) is one of the most common types of malignant adult kidney tumors. We constructed a weighted gene co-expression network to identify gene modules associated with clinical features of ccRCC (n = 97). Six hub genes (CCNB2, CDC20, CEP55, KIF20A, TOP2A and UBE2C) were identified in both co-expression and protein-protein interaction (PPI) networks, which were highly correlated with pathologic stage. The significance of expression of the hub genes in ccRCC was ranked top 4 among all cancers and correlated with poor prognosis. Functional analysis revealed that the hub genes were significantly enriched in cell cycle regulation and cell division. Gene set enrichment analysis suggested that the samples with highly expressed hub gene were correlated with cell cycle and p53 signaling pathway. Taken together, six hub genes were identified to be associated with progression and prognosis of ccRCC, and they might lead to poor prognosis by regulating p53 signaling pathway.
Abbreviations: ccRCC, clear cell renal cell carcinoma; HPA, human protein atlas; GSEA, enrichment analysis and gene set enrichment; WGCNA, weighted gene co-expression network analysis; TCGA, the cancer genome atlas; DEGs, differentially expressed genes; TOM, topological overlap matrix; MEs, module eigengenes; GS, gene significance; MS, module significance; STRING, search tool for the retrieval of interacting genes; PPI, protein-protein interaction; DAVID, Database for Annotation, Visualization and Integrated Discovery; DEG, differentially expressed gene; SAM, significance analysis of microarrays
Keywords: Clear cell renal cell carcinoma (ccRCC), Co-expression network analysis, Hub genes, Progression, Prognosis
1. Introduction
Renal cell carcinoma (RCC) is the most common type of malignant adult kidney tumors, accounting for > 90% of all adult renal tumors. Up to one-third of patients with RCC already suffered with a distant metastasis at the time of diagnosis [1]. Clear cell RCC (ccRCC), taking up about 75%–85% of RCC, is the most common subtype [2]. At present, ccRCC is usually resistant to chemotherapy. Targeted therapies have been exploited for their target specificity and low toxicity so they can be the best choice of non-surgical treatments [3]. Therefore, many biomarkers for clear cell renal cell carcinoma have been discovered including VHL, VEGF, CAIX and HIF1α/2α mutations. Some of which could predict therapeutic effect and clinical prognosis [4]. We know that carcinogenesis is not the result of deregulation of several oncogenes or tumor suppressors; it is the outcome of complex mechanisms, including the high interconnection between genes with similar expression patterns [5]. Thus, it is urgently needed to identify novel molecular biomarkers that can predict disease stage and clinical outcome of ccRCC patients, which could help understand its pathogenesis and provide personalized treatment.
Rapid technological breakthroughs of genome-wide sequencing have shed new light on the research of clinical issues and related pathological mechanisms in various cancers [6]. Nowadays, most studies just concentrated on the screening of differentially expressed genes and not attached enough attention to the high degree of interconnection between genes, where genes with similar expression patterns may be functionally related. The algorithm, weighted gene co-expression network analysis (WGCNA), can construct free-scale gene co-expression networks to explore the relationships between different gene sets or between gene sets and clinical features [7]. WGCNA has been widely applied to finding the hub genes associated with clinical feature in different cancer types [8], [9], [10].
In this study, WGCNA and other analysis methods are adopted to jointly analyze clinical information and microarray data of ccRCC patient samples to identify key genes associated with clinical features (age, gender, and tumor grade). These key genes may have important clinical implications and serve as diagnostic and prognostic biomarkers or therapeutic targets.
2. Materials and methods
2.1. Data collection
Expression profiles of mRNA and related clinical data of clear cell renal carcinoma were downloaded from Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). Dataset GSE40435 performed on Illumina HumanHT-12 V4.0 expression beadchip was used as a training set to construct co-expression networks and identify hub genes in this study. This dataset included 101 pairs of ccRCC tumors and adjacent non-tumor renal tissue from Czech patients (including 22 ccRCC of grade 1, 47 of grade 2, 24 of grade 3, and 8 of grade 4). Another independent dataset of GSE53757 based on the microarray platform of Affymetrix U133 plus 2 was downloaded from GEO and used as a test set to verify our results. This dataset included 72 pairs of clear cell renal carcinoma patients (including ccRCC of grade 1, 2, 3, and 4).
2.2. Data preprocessing
Normalized data were firstly downloaded from GEO database. Microarray quality was assessed by sample clustering according to the distance between different samples in Pearson's correlation matrices and average linkage, and 4 samples (GSM993996, GSM994041, GSM994065, and GSM994069) were removed from subsequent analysis in GSE40435.
2.3. Differentially expressed genes (DEGs) screening
The “limma” (linear models for microarray data) R package was used to screen the DEGs between normal kidney and clear cell renal cell carcinoma. The SAM (significance analysis of microarrays) with FDR (false discovery rate) < 0.05 and | log2 fold change (FC)| > 0.585 were chosen as the cut-off criteria to select genes further considered in the network construction.
2.4. Co-expression network construction
Firstly, expression data profile of DEGs was tested to check if they were the good samples and good genes. Then, we used the “WGCNA” package in R to construct scale-free co-expression network for the DEGs. At first, the Pearson's correlation matrices and average linkage method were both performed for all pair-wise genes. Then, a weighted adjacency matrix was constructed using a power function amn = | cmn |β (cmn = Pearson's correlation between gene m and gene n; amn = adjacency between gene m and gene n). β was a soft-thresholding parameter that could emphasize strong correlations between genes and penalize weak correlations. After choosing the power of β, the adjacency was transformed into a topological overlap matrix (TOM), which could measure the network connectivity of a gene defined as the sum of its adjacency with all other genes for network generation, and the corresponding dissimilarity (1-TOM) was calculated. To classify genes with similar expression profiles into gene modules, average linkage hierarchical clustering was conducted according to the TOM-based dissimilarity measure with a minimum size (gene group) of 50 for the genes dendrogram. To further analyze the module, we calculated the dissimilarity of module eigengenes, chose a cut line for module dendrogram and merged some module.
2.5. Identification of clinically significant modules
Two approaches were used to identify modules related to clinical traits of ccRCC. First, gene significance (GS) was defined as the log10 transformation of the P value (GS = lgP) in the linear regression between gene expression and the clinical traits. In addition, module significance (MS) was defined as the average GS for all the genes in a module. In general, the module with the absolute MS ranked first or second among all the selected modules was considered as the one related with clinical trait. Module eigengenes (MEs) were considered as the major component in the principal component analysis for each gene module, and the expression patterns of all genes could be summarized into a single characteristic expression profile within a given module. In addition, we calculated the correlation between MEs and clinical traits to identify the relevant module. The module with the maximal absolute MS among all the selected modules was usually considered as the one related with clinical trait. Finally, the module highly correlated with certain clinical trait was selected for further analysis.
2.6. Identification of hub genes
Hub genes, highly interconnected with nodes in a module, have been shown to be functionally significant. In this study, hub genes were defined by module connectivity, measured by absolute value of the Pearson's correlation (cor.geneModuleMembership > 0.8) and clinical trait relationship, measured by absolute value of the Pearson's correlation (cor.geneTraitSignificance > 0.2). We identified hub genes in module which were highly correlated with certain clinical trait. Furthermore, we uploaded all genes in the hub module to the STRING database, choosing confidence > 0.4 to construct protein-protein interaction (PPI). In the PPI network, genes with a connectivity degree of ≥ 8 were also defined hub genes. The common hub genes both in co-expression network and PPI network were regarded as “real” hub genes for further analysis.
2.7. Hub genes validation
In the test set of GSE53757, linear regression analyses were performed to validate the role of hub genes in the progression of ccRCC as well as the transcriptional levels in normal kidney and ccRCC samples. Moreover, we used 2 other databases: Gene Expression Profiling Interactive Analysis (http://gepia.cancer-pku.cn) and Oncolnc (http://www.oncolnc.org) to perform validation of cancer specific expression and prognosis of the candidate hub genes [11], [12].
2.8. Functional and pathway enrichment analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/) is an online program providing a comprehensive set of functional annotation tools for investigators to understand biological meaning behind large list of genes [13]. We uploaded DEGs in hub module to perform gene ontology and KEGG pathway enrichment analysis. Those terms including hub genes were selected as the key biological process and pathways. P < 0.05 was set as the cut-off criterion.
2.9. Gene set enrichment analysis (GSEA)
In the test set, ccRCC we firstly seek out the median expression of each hub gene and according to the median, 72 ccRCC samples were divided into two groups. To identify potential function of the hub gene, GSEA (http://software.broadinstitute.org/gsea/index.jsp) was conducted to detect whether a series of a priori defined biological processes were enriched in the gene rank derived from DEGs between the two groups [14], [15]. For use with GSEA software, the collection of annotated gene sets of c2.cp.kegg.v6.0.symbols.gmt in Molecular Signatures Database (MSigDB, http://software.broadinstitute.org/gsea/msigdb/index.jsp) was chosen as the reference gene sets. FDR < 0.05 was chosen as the cut-off criterion.
2.10. Analysis of DEGs screening between grade I-III and grade IV
One hundred one samples were divided into 2 groups (grade I-III and grade IV). The FDR < 0.05 and | log2 fold change (FC)| > 0.585 were chosen as DEGs between low grade ccRCC and high grade ccRCC. Then DEGs were uploaded to DAVID database to perform functional enrichment analysis.
3. Results
3.1. DEGs screening
After data preprocessing and quality assessment, the expression matrices were obtained from the 202 samples in training set GSE40435. Under the threshold of FDR < 0.05 and | log2FC | > 0.585, a total of 2756 DEGs (1314 up-regulated and 1442 down-regulated) were selected for subsequent analysis.
3.2. Weighted co-expression network construction and key modules identification
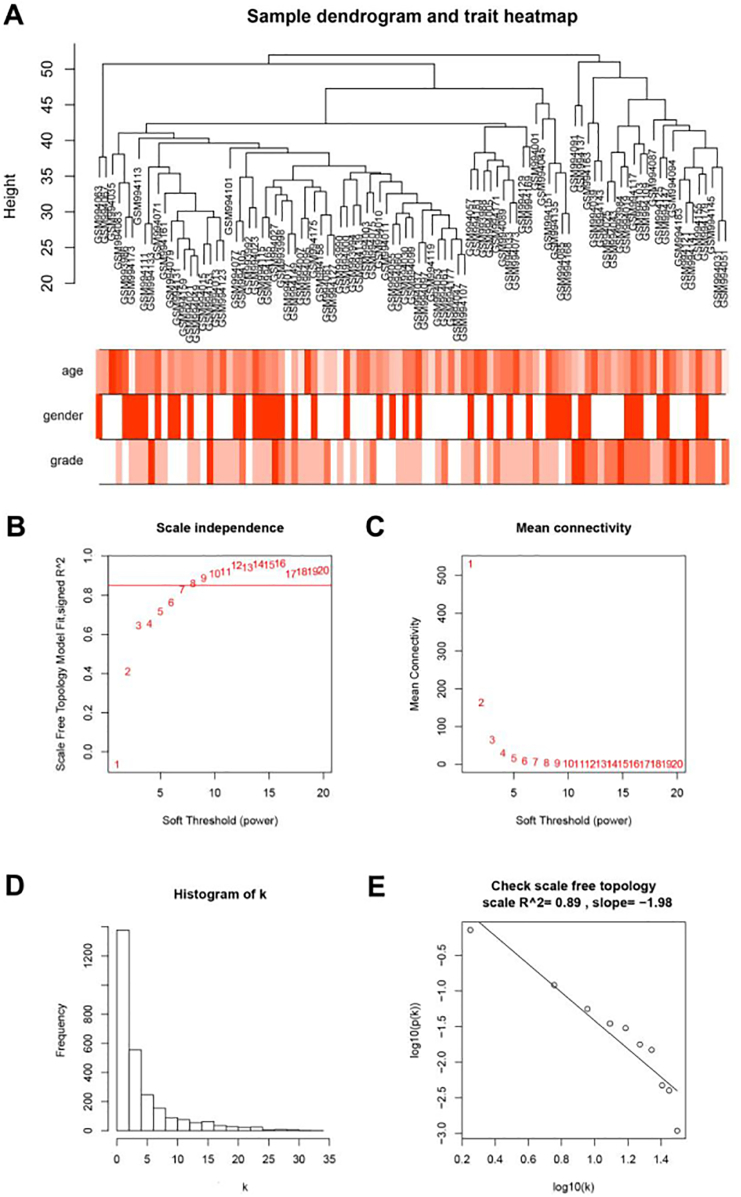
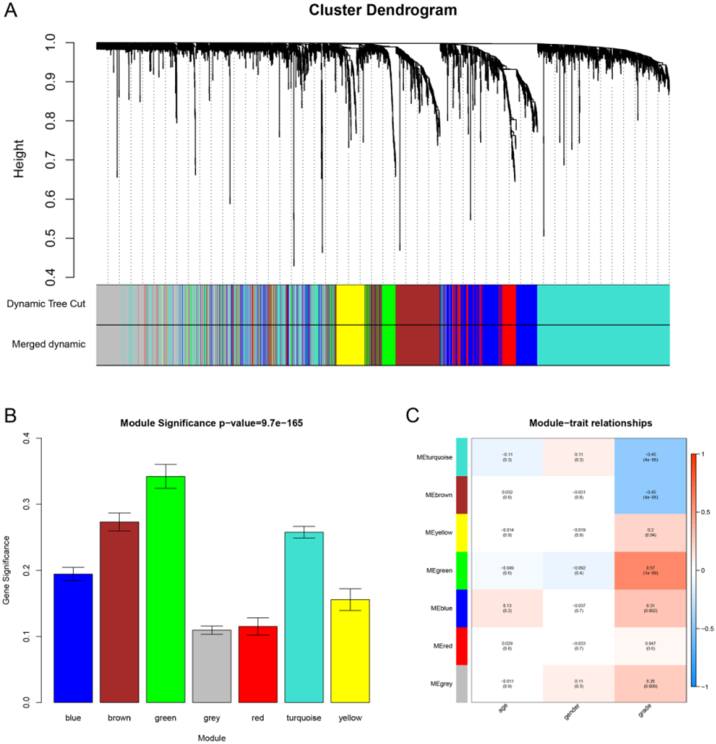
Ninety-four over one hundred one samples with clinical data were included in co-expression analysis (Fig. 1A). Using “WGCNA” package in R, the DEGs with similar expression patterns were grouped into modules via the average linkage hierarchical clustering. In this study, the power of β = 8 (scale free R2 = 0.89) was selected as the soft-thresholding to ensure a scale-free network (Fig. 1B-E). A total of 7 modules were identified (Fig. 2A). Two methods were used to test the relevance between each module and the ccRCC progression. Firstly, modules with greater MS were considered to have more connection with the disease progression. However, most of the correlations were low to moderate (R2 < 0.5), and we found that the MS of green module (P = 1 × 10− 9, R2 = 0.57) was higher than those of any other MS (Fig. 2B). Afterwards, the ME in the green module showed a higher correlation with disease progression than other modules (Fig. 2C). Based on the two methods, the green module with tumor progression was identified as the clinical significant module, which was extracted for further analysis.
Fig. 1.
Clustering dendrogram of 97 tumor samples and the clinical traits. (A) The clustering was based on the expression data of differentially expressed genes between tumor samples and non-tumor samples in ccRCC. The color intensity was proportional to older age and higher Furhman grade. (B-D) Determination of soft-thresholding power in the weighted gene co-expression network analysis (WGCNA). (B) Analysis of the scale-free fit index for various soft-thresholding powers (β). (C) Analysis of the mean connectivity for various soft-thresholding powers. (D) Histogram of connectivity distribution when β = 8. (E) Checking the scale free topology when β = 8. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 2.
Identification of modules associated with clinical information. (A) Dendrogram of all differentially expressed genes clustered based on a dissimilarity measure (1-TOM). (B) Distribution of average gene significance and errors in the modules associated with the Furhman grade of ccRCC. (C) Heatmap of the correlation between module eigengenes and different clinical information of ccRCC (age, gender, and Furhman grade).
3.3. Identification of hub genes for tumor progression in the green module
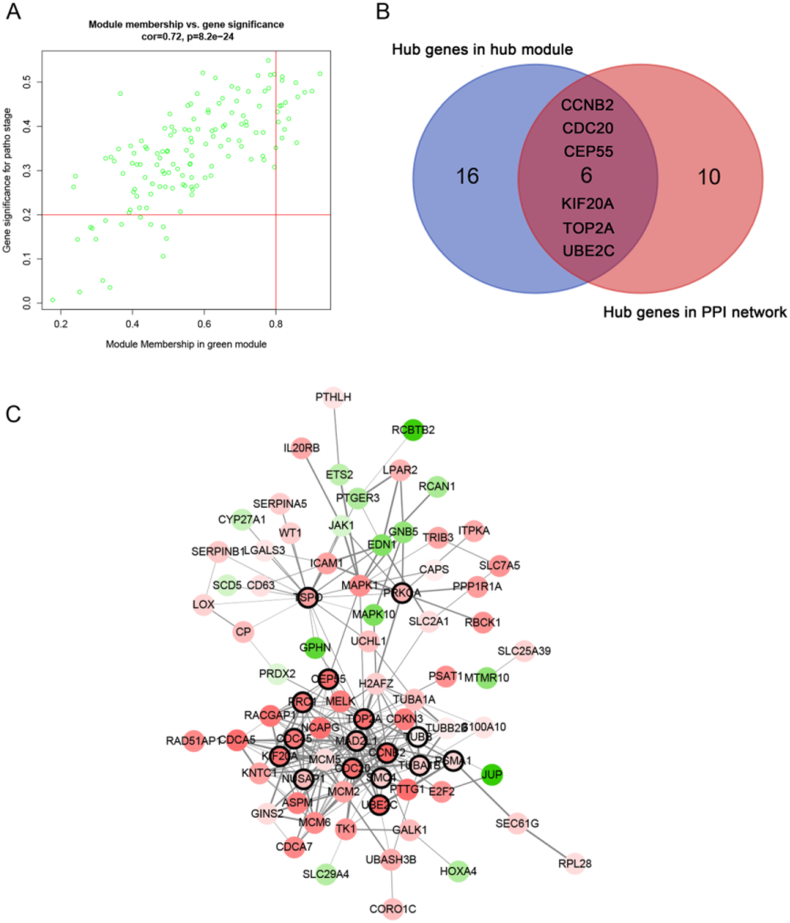
Highly connected hub genes in a module play important roles in the biological processes. Defined by module connectivity, measured by absolute value of the Pearson's correlation (cor.geneModuleMembership > 0.8) and clinical trait relationship, measured by absolute value of the Pearson's correlation (cor.geneTraitSignificance > 0.2), 22 genes with the high connectivity in green module were taken as hub genes (TOP2A, UHRF1, RAD51AP1, NR3C2, MCM6, PTTG1, NCAPG, CDCA5, UBE2C, PDGFD, RGS5, CEP55, CCNB2, CDKN3, CCDC109B, PTTG3P, RACGAP1, KIF20A, ALDH3A2, CDC20, EDNRB, and EMCN) (Fig. 3A). As to the PPI network, under the cutoff of confidence > 0.4 and connectivity degree of ≥ 8, 16 genes were identified as hub genes (CCNB2, CDC20, CDC45, CEP55, KIF20A, MAD2L1, NUSAP1, PRC1, PRKCA, PSMA1, SMC4, TOP2A, TSPO, TUBA1B, TUBB, and UBE2C) (Fig. 3C). CCNB2, CDC20, CEP55, KIF20A, TOP2A, and UBE2C were identified both in PPI network and co-expression network (Fig. 3B). Eventually, these six genes were regarded as “real” hub genes for tumor progression, which were chosen for further analysis.
Fig. 3.
Hub genes detection and protein-protein network (PPI). (A) Scatter plot of module eigengenes in green module. (B) The Venn diagram of co-expression hub genes and PPI network hub genes. (C) Protein–protein interaction network of genes in the green module. The color intensity in each node was proportional to fold change of expression in comparison to normal kidney samples (up-regulation in red and down-regulation in green). The nodes with bold circle represented network hub genes identified by WGCNA. The edge width was proportional to the score of protein-protein interaction based on the STRING database. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Hub gene validation
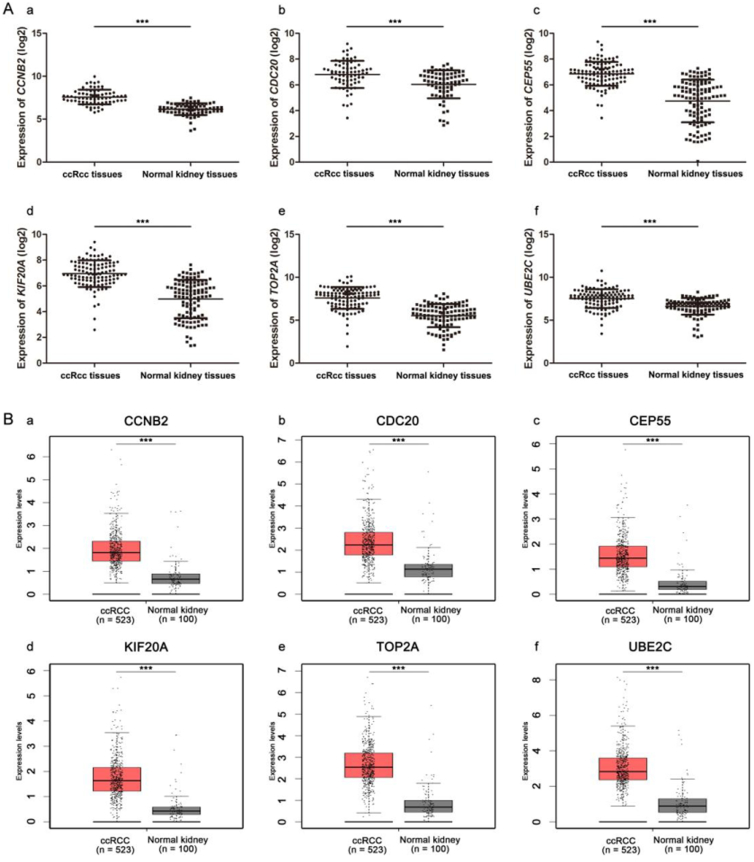
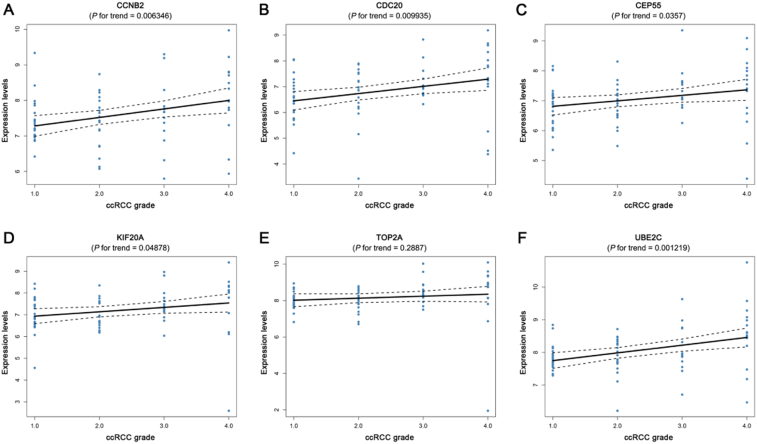
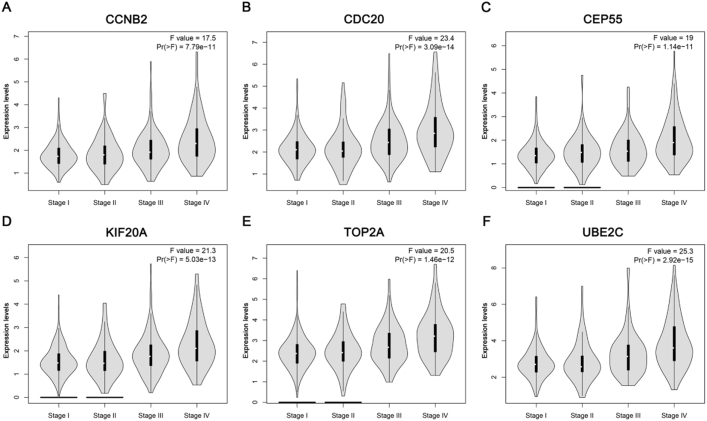
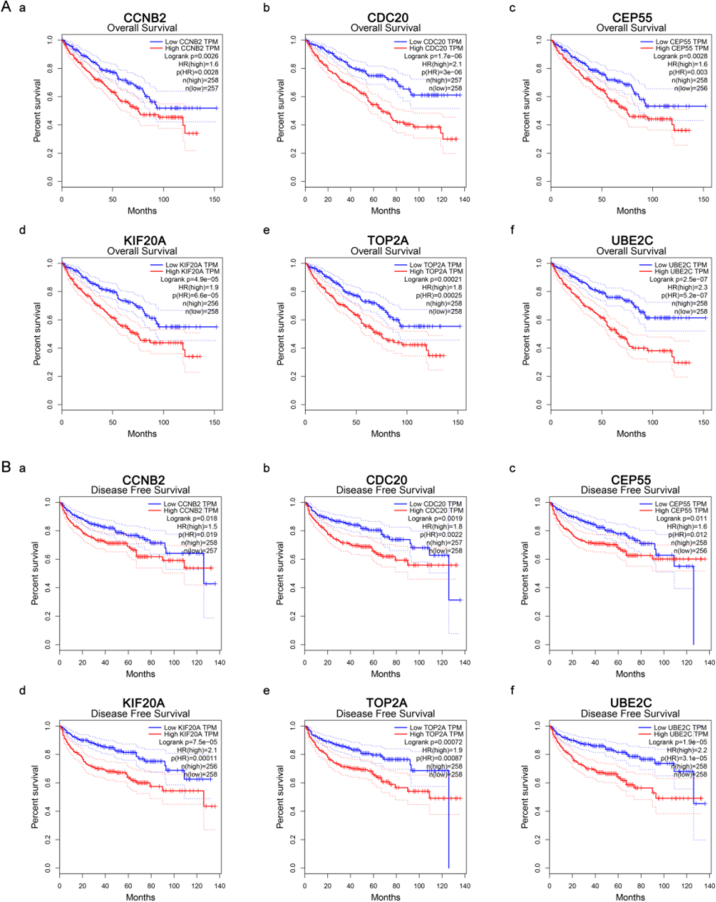
Gene expression validations were performed, and all of six hub genes were also oncogenes in the test set and TCGA data (Fig. 4). Then, linear regression analyses were conducted to validate hub genes in the test set GSE53757, most of which except TOP2A showed positive correlation with ccRCC progression (Furhman grade) (P for trend < 0.05) (Fig. 5). Moreover, based on the TCGA data, we investigated that all of six genes were highly-correlated with pathologic stage (Fig. 6). As the tumor progression always affected the tumor prognosis, we also validated the six hub genes by investigating their roles in ccRCC prognosis including overall survival time and disease free survival time. We found a shorter overall survival time (Fig. 7A) and disease-free survival time in patients with higher expression levels of CCNB2, CDC20, CEP55, KIF20A, TOP2A, and UBE2C (Fig. 7B). In addition, based on the Oncolnc database, we found a cancer-specific expression of the six hub genes, which demonstrated that the significance of expression of CCNB2, CDC20, CEP55, KIF20A, TOP2A, and UBE2C was top 4 among all cancers (Suplplementary Fig. S1).
Fig. 4.
Validation of the gene expression levels of CCNB2, CDC20, CEP55, KIF20A, TOP2A and UBE2C between normal kidney and ccRCC samples. (A) Validation based on microarray data of GSE53757. (a) CCNB2, (b) CDC20, (c) CEP55, (d) KIF20A, (e) TOP2A, and (f) UBE2C. (B) Validation based on TCGA data in GEPIA. (a) CCNB2, (b) CDC20, (c) CEP55, (d) KIF20A, (e) TOP2A, and (f) UBE2C.
Fig. 5.
Validation of the correlation between the expression levels of CCNB2, CDC20, CEP55, KIF20A, TOP2A, and UBE2C and the Furhman grade of ccRCC (based on microarray data of GSE53757). (A) CCNB2, (B) CDC20, (C) CEP55, (D) KIF20A, (E) TOP2A, and (F) UBE2C.
Fig. 6.
Validation of the correlation between the expression levels of CCNB2, CDC20, CEP55, KIF20A, TOP2A and UBE2C and the pathologic stage of ccRCC (based on TCGA data in GEPIA). (A) CCNB2, (B) CDC20, (C) CEP55, (D) KIF20A, (E) TOP2A, and (F) UBE2C.
Fig. 7.
Survival analysis of six hub genes in ccRCC (based on TCGA data in GEPIA). (A) Overall survival analysis. (a) CCNB2, (b) CDC20, (c) CEP55, (d) KIF20A, (e) TOP2A, and (f) UBE2C. (B) Disease free survival analysis. (a) CCNB2, (b) CDC20, (c) CEP55, (d) KIF20A, (e) TOP2A, and (f) UBE2C. Red line represented the samples with gene highly expressed and blue line was for the samples with gene lowly expressed. HR: hazard ratio. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5. Functional and pathway enrichment analysis
To obtain further insight into the function of DEGs in hub module, they were uploaded to the DAVID database. GO analysis results showed that hub genes were enriched in the top 10 biological process (BP), including proteasome-mediated ubiquitin-dependent protein catabolic process, negative regulation of blood coagulation, anaphase-promoting complex-dependent catabolic process, DNA unwinding involved in DNA replication, microtubule-based process, DNA replication initiation, mitotic cytokinesis, G1/S transition of mitotic cell cycle, mitotic chromosome condensation, and cell division. Moreover, hub genes were also overrepresented in these top 10 KEGG pathways, including phagosome, progesterone-mediated oocyte maturation, DNA replication, Hepatitis B, pancreatic cancer, pathways in cancer, HTLV-1 infection, pathogenic Escherichia coli infection, gap junction, and cell cycle (Supplementary Fig. S2). Among the functional and pathway enrichment analysis, cell division and cell cycle were most significantly enriched.
3.6. Gene set enrichment analysis
To identify the potential function of those hub genes in ccRCC, GSEA was conducted to search biological processes enriched in any hub gene highly-expressed samples. Six gene sets were enriched in the samples with all hub genes highly expressed, including “cell cycle”, “p53 signaling pathway”, “DNA replication”, “primary immunodeficiency”, “homologous recombination”, and “base excision repair” (Supplementary Fig. S3).
3.7. Grade-associated DEGs analysis
Firstly, sample cluster was performed (Supplementary Fig. S4A).Under the threshold of FDR < 0.05 and | log2FC | > 0.585, a total of 48 DEGs (35 up-regulated and 13 down-regulated) were identified (Supplementary Fig. S4B and Supplementary Table 1). And they were significantly enriched in following terms listed in Supplementary Table 2.
4. Discussion
The progression and prognosis of clear cell renal cell carcinoma were quite variable. Though a number of prognostic models had been proposed, most of them were based on clinical parameters and lacked accuracy. In the era of precise medicine, better biomarkers for cancer specific prognosis and progression were urgently needed and provided more accurate clinical information that could significantly enhance decision making for patient management. Here, we used an integrated analysis to screen progression and prognosis related biomarkers.
WGCNA was performed to identify gene co-expression modules related with the progression of ccRCC. The green module was identified, and 22 hub genes were derived from the module. Furthermore, relating the results of PPI network, 16 hub nodes were identified. Therefore, CCNB2, CDC20, CEP55, KIF20A, TOP2A, and UBE2C in common networks, were real hub genes, indicating that they had high connection with clinical trait as well as vital biological processes. Few of them were identified in clear cell renal cell carcinoma.
Cyclin B2 (CCNB2) is a member of the cyclin family, specially the B-type cyclins. Takashima S et al. reported that stronger expression of cyclin B2 mRNA in tumor cells was an independent predictor of a poor prognosis in patients with adenocarcinoma of the lung [16]. Furthermore, Shubbar E et al. also found that CCNB2 might function as an oncogene and could serve as a potential biomarker of unfavorable prognosis over short-term follow-up in breast cancer [17]. Cell Division Cycle 20 (CDC20), acted as a regulatory protein interacting with several other proteins at multiple points in the cell cycle. Many studies demonstrated that CDC20 might be a potential biomarker for both therapeutic target and prognosis in a number of cancers [18], [19], [20], [21]. Centrosomal protein 55 (CEP55), played a role in mitotic exit and cytokinesis, which was important in carcinogenesis as well. Tao J. et al. suggested that CEP55 could suppress cellular proliferation by regulating cell cycle arrest at G2/M phase in human gastric cancer [22]. CEP55 overexpression has been found to significantly correlate with tumor stage, aggressiveness, metastasis, and poor prognosis across multiple tumor types and therefore has been included as part of several prognostic ‘gene signatures’ for cancer [23]. Kinesin Family Member 20A (KIF20A) might act as a motor required for the retrograde RAB6 regulated transport of Golgi membranes and associated vesicles along microtubules. Shi C. et al. found that Gli2-KIF20A axis was essential for the proliferation and growth of human hepatocellular carcinoma cells as well as a potential target for future therapeutic intervention and as an independent prognostic biomarker for hepatocellular carcinoma [24]. KIF20A was also reported that its over-expression correlates with HPV infection, clinical stage, tumor recurrence, lymphovascular space involvement, pelvic lymph node metastasis, and poor outcome in early-stage cervical squamous cell carcinoma patients [25]. Topoisomerase (DNA) II Alpha (TOP2A) encoded a DNA topoisomerase, an enzyme that controlled and altered the topologic states of DNA during transcription. Study had demonstrated that TOP2A identified and provided epigenetic rationale for novel combination therapeutic strategies for aggressive prostate cancer [26]. Moreover, we could find the prognostic value of TOP2A in nasopharyngeal carcinoma and breast cancer [27], [28]. Ubiquitin-conjugating enzyme E2 C (UBE2C), a member of the E2 ubiquitin-conjugating enzyme family, was required for the destruction of mitotic cyclins and for cell cycle progression and may be involved in cancer progression. It was reported to be a promising biomarker in tumor progression and prognosis [29], [30], [31], [32].
Based on many studies about six hub genes, we could find the promising values of tumor progression and prognosis. According to the tumor specific expression (Supplementary Fig. S1), we found that the p value of each hub gene in ccRCC ranked top4 in all kinds of cancers based on the Oncolnc database, which demonstrated that those hub genes were strongly correlated with ccRCC as well as with great diagnostic value for ccRCC.
To further study their mechanism of regulating tumorigenesis, we perform the gene ontology and KEGG pathway analysis as well as GSEA analysis. In GO analysis, we found that hub genes were significantly enriched in terms of regarding cell cycle and cell cycle regulation; meanwhile, the KEGG analysis showed that cell cycle was the most significant pathway. Interestingly, through GSEA analysis, we found that hub genes were commonly enriched in cell cycle and p53 signaling pathway. Several researchers had demonstrated that the presence of p53 was necessary for DNA-damaged cells to arrest, repair the damage, and reenter the cell cycle [33]. Thus, we might suppose that those six hub genes played certain role in the progression of ccRCC and influenced the prognosis probably by regulating p53 signaling pathway, which contributed to the poor prognosis of ccRCC.
5. Conclusions
Our study used weighted gene co-expression analysis to construct a gene co-expression network, identify and validate network hub genes associated with the progression and poor prognosis of ccRCC. Eventually, six hub genes including CCNB2, CDC20, CEP55, KIF20A, TOP2A, and UBE2C were identified and validated in association with the progression and poor prognosis of ccRCC probably by regulating p53 signaling pathway.
The following are the supplementary data related to this article.
Grade associated DEGs.
Enrichment analysis for grade associated DEGs.
Supplementary figures
Acknowledgments
Acknowledgments
The excellent technical assistance of Shanshan Zhang and Danni Shan is gratefully acknowledged. We also would like to acknowledge the KEGG database developed by Kanehisa Laboratories. This study was supported in part by grants from the Hubei Province Health and Family Planning Scientific Research Project (grant number WJ2017H0002 to Yu Xiao), Wuhan Clinical Cancer Research Center of Urology and Male Reproduction (grant number 303-230100055 to Xinghuan Wang) and Hubei Province Urological Clinical Research Center of Laparoscopy (grant number ZN-31 to Xinghuan Wang). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Declaration of conflicts of interest
The authors declare no conflicts of interest.
Contributor Information
Xinghuan Wang, Email: wangxinghuan@whu.edu.cn.
Yu Xiao, Email: yu.xiao@whu.edu.cn.
References
- 1.Gupta K., Miller J.D., Li J.Z., Russell M.W., Charbonneau C. Epidemiologic and socioeconomic burden of metastatic renal cell carcinoma (mRCC): a literature review. Cancer Treat. Rev. 2008;34:193–205. doi: 10.1016/j.ctrv.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Baldewijns M.M., van Vlodrop I.J., Schouten L.J., Soetekouw P.M., de Bruine A.P., van Engeland M. Genetics and epigenetics of renal cell cancer. Biochim. Biophys. Acta. 2008;1785:133–155. doi: 10.1016/j.bbcan.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Vera-Badillo F.E., Templeton A.J., Duran I., Ocana A., de Gouveia P., Aneja P., Knox J.J., Tannock I.F., Escudier B., Amir E. Systemic therapy for non-clear cell renal cell carcinomas: a systematic review and meta-analysis. Eur. Urol. 2015;67:740–749. doi: 10.1016/j.eururo.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 4.Chan J.Y., Choudhury Y., Tan M.H. Predictive molecular biomarkers to guide clinical decision making in kidney cancer: current progress and future challenges. Expert. Rev. Mol. Diagn. 2015;15:631–646. doi: 10.1586/14737159.2015.1032261. [DOI] [PubMed] [Google Scholar]
- 5.Tavazoie S., Hughes J.D., Campbell M.J., Cho R.J., Church G.M. Systematic determination of genetic network architecture. Nat. Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- 6.Stratton M.R., Campbell P.J., Futreal P.A. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi K., Bing Z.T., Cao G.Q., Guo L., Cao Y.N., Jiang H.O., Zhang M.X. Identify the signature genes for diagnose of uveal melanoma by weight gene co-expression network analysis. Int. J. Ophthalmol. 2015;8:269–274. doi: 10.3980/j.issn.2222-3959.2015.02.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu X.L., Ai Z.H., Wang J., YL Xu, Teng Y.C. Weighted gene co-expression network analysis in identification of endometrial cancer prognosis markers. Asian Pac. J. Cancer Prev. 2012;13:4607–4611. doi: 10.7314/apjcp.2012.13.9.4607. [DOI] [PubMed] [Google Scholar]
- 10.Wang Q.L., Chen X., Zhang M.H., Shen Q.H., Qin Z.M. Identification of hub genes and pathways associated with retinoblastoma based on co-expression network analysis. Genet. Mol. Res. 2015;14:16151–16161. doi: 10.4238/2015.December.8.4. [DOI] [PubMed] [Google Scholar]
- 11.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx247. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anaya J. Oncolnc: linking tcga survival data to mrnas, mirnas, and lncrnas. PeerJ Computer. Sci. 2016;2 [Google Scholar]
- 13.Dennis G., Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 14.Croken M.M., Qiu W., White M.W., Kim K. Gene set enrichment analysis (GSEA) of toxoplasma gondii expression datasets links cell cycle progression and the bradyzoite developmental program. BMC Genomics. 2014;15:515. doi: 10.1186/1471-2164-15-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takashima S., Saito H., Takahashi N., Imai K., Kudo S., Atari M., Saito Y., Motoyama S., Minamiya Y. Strong expression of cyclin B2 mRNA correlates with a poor prognosis in patients with non-small cell lung cancer. Tumour Biol. 2014;35:4257–4265. doi: 10.1007/s13277-013-1556-7. [DOI] [PubMed] [Google Scholar]
- 17.Shubbar E., Kovacs A., Hajizadeh S., Parris T.Z., Nemes S., Gunnarsdottir K., Einbeigi Z., Karlsson P., Helou K. Elevated cyclin B2 expression in invasive breast carcinoma is associated with unfavorable clinical outcome. BMC Cancer. 2013;13:1. doi: 10.1186/1471-2407-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karra H., Repo H., Ahonen I., Loyttyniemi E., Pitkanen R., Lintunen M., Kuopio T., Soderstrom M., Kronqvist P. Cdc20 and securin overexpression predict short-term breast cancer survival. Br. J. Cancer. 2014;110:2905–2913. doi: 10.1038/bjc.2014.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding Z.Y., HR Wu, Zhang J.M., Huang G.R., Ji D.D. Expression characteristics of CDC20 in gastric cancer and its correlation with poor prognosis. Int. J. Clin. Exp. Pathol. 2014;7:722–727. [PMC free article] [PubMed] [Google Scholar]
- 20.WJ Wu, KS Hu, Wang D.S., Zeng Z.L., Zhang D.S., Chen D.L., Bai L., Xu R.H. CDC20 overexpression predicts a poor prognosis for patients with colorectal cancer. J. Transl. Med. 2013;11:142. doi: 10.1186/1479-5876-11-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z., Wan L., Zhong J., Inuzuka H., Liu P., Sarkar F.H., Wei W. Cdc20: a potential novel therapeutic target for cancer treatment. Curr. Pharm. Des. 2013;19:3210–3214. doi: 10.2174/1381612811319180005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tao J., Zhi X., Tian Y., Li Z., Zhu Y., Wang W., Xie K., Tang J., Zhang X., Wang L., Xu Z. CEP55 contributes to human gastric carcinoma by regulating cell proliferation. Tumour Biol. 2014;35:4389–4399. doi: 10.1007/s13277-013-1578-1. [DOI] [PubMed] [Google Scholar]
- 23.Jeffery J., Sinha D., Srihari S., Kalimutho M., Khanna K.K. Beyond cytokinesis: the emerging roles of CEP55 in tumorigenesis. Oncogene. 2016;35:683–690. doi: 10.1038/onc.2015.128. [DOI] [PubMed] [Google Scholar]
- 24.Shi C., Huang D., Lu N., Chen D., Zhang M., Yan Y., Deng L., Lu Q., Lu H., Luo S. Aberrantly activated Gli2-KIF20A axis is crucial for growth of hepatocellular carcinoma and predicts poor prognosis. Oncotarget. 2016;7:26206–26219. doi: 10.18632/oncotarget.8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang W., He W., Shi Y., Gu H., Li M., Liu Z., Feng Y., Zheng N., Xie C., Zhang Y. High expression of KIF20A is associated with poor overall survival and tumor progression in early-stage cervical squamous cell carcinoma. PLoS One. 2016;11 doi: 10.1371/journal.pone.0167449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirk J.S., Schaarschuch K., Dalimov Z., Lasorsa E., Ku S., Ramakrishnan S., Hu Q., Azabdaftari G., Wang J., Pili R., Ellis L. Top2a identifies and provides epigenetic rationale for novel combination therapeutic strategies for aggressive prostate cancer. Oncotarget. 2015;6:3136–3146. doi: 10.18632/oncotarget.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lan J., Huang H.Y., Lee S.W., Chen T.J., Tai H.C., Hsu H.P., Chang K.Y., Li C.F. TOP2A overexpression as a poor prognostic factor in patients with nasopharyngeal carcinoma. Tumour Biol. 2014;35:179–187. doi: 10.1007/s13277-013-1022-6. [DOI] [PubMed] [Google Scholar]
- 28.Xu Y.C., Zhang F.C., Li J.J., Dai J.Q., Liu Q., Tang L., Ma Y., Xu Q., Lin X.L., Fan H.B., Wang H.X. RRM1, TUBB3, TOP2A, CYP19A1, CYP2D6: difference between mRNA and protein expression in predicting prognosis of breast cancer patients. Oncol. Rep. 2015;34:1883–1894. doi: 10.3892/or.2015.4183. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z., Liu P., Wang J., Gong T., Zhang F., Ma J., Han N. Ubiquitin-conjugating enzyme E2C regulates apoptosis-dependent tumor progression of non-small cell lung cancer via ERK pathway. Med. Oncol. 2015;32:149. doi: 10.1007/s12032-015-0609-8. [DOI] [PubMed] [Google Scholar]
- 30.Morikawa T., Kawai T., Abe H., Kume H., Homma Y., Fukayama M. UBE2C is a marker of unfavorable prognosis in bladder cancer after radical cystectomy. Int. J. Clin. Exp. Pathol. 2013;6:1367–1374. [PMC free article] [PubMed] [Google Scholar]
- 31.Psyrri A., Kalogeras K.T., Kronenwett R., Wirtz R.M., Batistatou A., Bournakis E., Timotheadou E., Gogas H., Aravantinos G., Christodoulou C., Makatsoris T., Linardou H., Pectasides D., Pavlidis N., Economopoulos T., Fountzilas G. Prognostic significance of UBE2C mRNA expression in high-risk early breast cancer. A Hellenic Cooperative Oncology Group (HeCOG) study. Ann. Oncol. 2012;23:1422–1427. doi: 10.1093/annonc/mdr527. [DOI] [PubMed] [Google Scholar]
- 32.Loussouarn D., Campion L., Leclair F., Campone M., Charbonnel C., Ricolleau G., Gouraud W., Bataille R., Jezequel P. Validation of UBE2C protein as a prognostic marker in node-positive breast cancer. Br. J. Cancer. 2009;101:166–173. doi: 10.1038/sj.bjc.6605122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukin D.J., Carvajal L.A., Liu W.J., Resnick-Silverman L., Manfredi J.J. p53 Promotes cell survival due to the reversibility of its cell-cycle checkpoints. Mol. Cancer Res. 2015;13:16–28. doi: 10.1158/1541-7786.MCR-14-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Grade associated DEGs.
Enrichment analysis for grade associated DEGs.
Supplementary figures