Abstract
Mammalian inner ear comprises of six sensory organs; cochlea, utricle, saccule, and three semicircular canals. The cochlea contains sensory epithelium known as the organ of Corti which senses sound through mechanosensory hair cells. Mammalian inner ear undergoes series of morphogenesis during development beginning thickening of ectoderm nearby hindbrain. These events require tight regulation of multiple signaling cascades including FGF, Wnt, Notch and Bmp signaling. In this review, we will discuss the role of newly emerging signaling, FGF signaling, for its roles required for cochlear development.
Keywords: Cochlea, FGF signaling, Hair cells, Inner ear development, Sensory progenitor cells
INTRODUCTION
Mammalian ear is composed of the outer, middle and inner ear. Sound generated from outside gathers in the outer ear, travels through the middle ear and transfers to the inner ear. The sensory hair cells in the inner ear then convert mechanical sound vibration to electrical signals and transmit those signals to the brain (1). Inner ear development comprises a series of morphogenic events that are orchestrated by a cascade of tightly regulated molecular events (2). During inner ear development, many morphogens play important roles. Fibroblast Growth Factor (FGF) is one of them. FGF signaling plays multiple roles during inner ear development starting as early as embryonic day (E) 8–9 in mouse, when three FGF ligands (FGF3, FGF8 and FGF10) signal to FGFR2 within the otic epithelium for otic placode induction (3). In addition, recent publications indicate the emerging role of FGF signaling during cochlear sensory progenitor proliferation and differentiation (4–7). In this review, we will focus on the various roles of FGF signaling in cochlear development that are identified up-to-date including sensory progenitor proliferation, lateral compartment differentiation, pillar cell differentiation and non-sensory structures development.
FGF SIGNALING
FGF signaling has been implicated in development, metabolism and disease. During vertebrate development, FGFs are widely expressed and regulate multiple diverse processes. FGF family is a group of structurally related polypeptide growth factors. FGFs are typically small ranging about 17–35 kD, secreted, and highly basic proteins (8). In mammalian system, there are 22 members of the FGFs that are classified into 7 subfamilies based on their sequence homology and biochemical properties (9). The canonical FGFs are secreted from the cells and bind to cognate receptors along with heparan sulfate (8, 10). Due to the binding affinity with heparan sulfate, these FGFs function as paracrine or autocrine (11). The hormonal FGFs require novel cofactors, klotho and β-Klotho to bind to their cognate receptors due to lack of binding affinity with heparin sulfate (12, 13). The intracellular FGFs serve as co-factors for voltage gated sodium channels and other molecules (14, 15).
To activate canonical FGF signaling, FGFs bind to their cognate receptors, FGF receptors (FGFR) (8). Binding of FGFs to FGFRs results in receptor dimerization (16, 17). This process is enhanced by heparan sulfate, which forms a tri-molecular complex containing FGF, FGFR and heparin sulfate (8, 18). After dimerization, FGFR auto-phosphorylates to activate itself and phosphorylates intracellular adaptor molecules. FGF/FGFR signaling activates three major downstream signaling pathways: mitogen activated protein kinase (MAPK), phosphatidylinositol-3 kinase (PI3K)/Akt and the phospholipase c-γ (PLC-γ) pathway (19). The most common pathway employed by FGFs is the MAPK pathway. This involves the lipid-anchored docking protein FGFR substrate 2 (FRS2) (20, 21). The two tyrosine auto-phosphorylation sites (Y-653 and Y-654) conduct binding of FRS2 to FGFR. The adaptor protein Grb2 and the protein tyrosine phosphatase Shp2 recognize the FRS2 tyrosine phosphorylation sites and bind (22, 23). Grb2 forms a complex with the guanine nucleotide exchange factor Son of sevenless (SOS) via its SH3 domain (22, 23). Translocation of this complex to the plasma membrane by binding to phosphorylated FRS2 allows SOS to activate Ras by GTP exchange due to its proximity to membrane-bound Ras (21, 24). Once in the active GTP-bound state, Ras interacts with several effector proteins including Raf leading to the activation of the MAPK signaling cascade (21, 24). This cascade leads to phosphorylation of target transcription factors, such as Etv4 and Etv5 (25).
Canonical FGFs are divided by 5 subfamilies and recognize specific cognate receptor isotypes. There are 4 members of FGF receptors in vertebrates and produce many variants using mRNA alternative splicing (26, 27). Among them two major isoforms, the tissue-specific alternative splicing (b and c isoforms) in immunoglobulin (Ig) domain III of FGFRs, have distinct FGFR-binding properties indicating the complexity of FGF signaling (28–33).
The specificity of FGFs for the major splice forms of the FGFRs is critical for both the developmental and pathogenic functions of FGFs. One important observation is that distinct FGFs signal across epithelial-mesenchymal boundaries. For example, in lung development, Fgf9 is expressed in the outermost layer of the lung, the mesothelium, and has been identified as a key factor that signals to mesenchyme to regulate proliferation, differentiation and the expression of other factors that in turn regulate epithelial development (34, 35). Mesenchymal forms of FGFR1 and 2 have been shown to mediate the FGF9 signal (36). In contrast, Fgf10 is expressed in mesenchymal cells surrounding the distal tip of the growing epithelium (37). When FGF10 is locally applied to lung explants on beads, lung epithelium expanded toward the source of FGF10 (37, 38). Fgf10 mutant embryos show lung agenesis due to defects in lung epithelia, indicating that Fgf10 is an essential regulator of branching morphogenesis (39–41). FGF10 functions through epithelium specific FGFR2, which is also critical for lung branching morphogenesis (41, 42).
In this review, we will overview the role of FGFs and FGFRs during inner ear development and focus on recently emerged role of FGF signaling in cochlear sensory progenitor cells. Table 1 summarizes phenotypes of inner ear development from the FGFs and FGFRs knock-out mouse lines.
Table 1.
Phenotypes of FGF mutation in mouse
| Gene | Type of mutation | Phenotype | Ref |
|---|---|---|---|
| Fgf3 | Double conventional mutation with Fgf10 | Failure of otic vesicle formation | (72–74) |
| Fgf8 | Conditional mutation with Foxg1Cre | Decrease of pillar cells | (64) |
| Fgf9 | Conventional mutation | Decrease of periotic mesenchyme proliferation | (55) |
| Double conventional mutation with Fgf20 | Decrease of cochlear sensory progenitor proliferation | (5) | |
| Fgf10 | Conventional mutation | Agenesis of posterior vestibular tissue | (67, 75, 76) |
| Double conventional mutation with Fgf3 | Failure of otic vesicle formation | (72–74) | |
| Fgf20 | Conventional mutation | Decrease of cochlear lateral compartment differentiation | (4) |
| Double conventional mutation with Fgf9 | Decrease of cochlear sensory progenitor proliferation | (5) | |
| Fgfr1 | Hypomorph, Conditional mutation with Foxg1Cre, Six1enh21Cre, Emx2Cre and Fgf20Cre | Decrease of cochlear lateral compartment differentiation | (5, 57, 58) |
| Double conditional deletion with Fgfr2 with Twist2Cre | Decrease of sensory progenitor proliferation | (5) | |
| Fgfr2 | IIIC isoform specific mutation | Failure otocyst morphogenesis | (77) |
| Double conditional deletion with Fgfr1 with Twist2Cre | Decrease of sensory progenitor proliferation | (5) | |
| Fgfr3 | Conventional mutation | Loss of pillar cell and increase of outer hair cells | (62, 79) |
INNER EAR DEVELOPMENT
In vertebrates, the inner ear is comprised of two main functional parts: the cochlea that is responsible for sound detection, and the vestibular system that is dedicated to balance. The development of inner ear involves dramatic morphogenic and patterning events that convert simple thickened epithelium to a complex structure connected to the central nervous system. In mice, the inner ear develops from a bilateral thickening (otic placode) within the ectoderm located adjacent to the hindbrain around E8.5 (43). Induction of otic placode from competent pre-placodal ectoderm has been shown to be mediated by FGF signaling where three FGF ligands (FGF3, 8 and 10) signal to FGFR2 within the otic epithelium for otic placode induction (3, 44). One day later, placodal cells invaginate and separate from the surface ectoderm giving rise to the otocyst (43). All cells within the membranous portion of the inner ear are derived from the multipotent progenitor cells initially located within the otocyst (2). Around E10.5, a population of cells delaminates from the ventral region of the otocyst and migrates a short distance ventro-medially. These cells are neuroblasts that will coalesce to form the developing stato-acoustic ganglion (VIII cranial ganglion). Following this event, the spheroidal otocyst undergoes an elaborate series of morphogenic changes resulting in the formation of two main structures: the dorsal vestibular and the ventral cochlear regions (43). During otocyst development, gradients of sonic hedgehog (Shh) and Wnt signaling function to establish positional information across the dorso-ventral axis to confer vestibular and cochlear identities (45, 46).
As the cochlear duct extends and coils, a subset of cells within its ventral aspect begins to develop as the prosensory epithelium (prosensory domain), and these cells become localized to a restricted region of the developing cochlea including a narrow strip that extends along the length of the cochlear duct (43). Some markers of the prosensory domain include the Jag1 (a Notch ligand), and Sox2 transcription factor (47, 48). Cells within the prosensory domain will give rise to both hair cells and supporting cells within the organ of Corti. In mouse, the prosensory domain cells begin to exit the cell cycle starting from the apex around E12, and a wave of cell cycle exit marks with the expression of p27Kip1 then proceeds along the prosensory domain from the apex to the base over the following 48–60 hours (49). Starting at about E13.5, cells in the mid-basal region of the cochlea begin to differentiate to hair cells by expressing the key transcription factor Atoh1, and the region of differentiating cells spreads bidirectionally over the following three days (50). Over the next two weeks, hair cells undergo morphological and biochemical specialization, including the elaboration and polarization of the apical hair bundle stereocilia, the development of mechanosensitivity in hair bundles and the formation of the basal ribbon synapses with neurons of the spiral ganglion (51).
By P0, cochlear sensory epithelium is composed of two types of cells; hair cells and supporting cells. Hair cells are arranged in ordered rows extending the length of the spiral cochlear duct. One row of inner hair cells is located on the medial edge of organ of Corti while three rows of outer hair cells on the lateral edge (43). Supporting cells rest on the basement membrane and send apical projections to the luminal surface. At least five different types of supporting cells are arranged in rows from the outer edge to the inner edge of the organ: Hensen’s cells, Deiters’ cells, pillar cells; inner phalangeal cells; and border cells (2).
COCHLEAR SENSORY PROGENITOR CELL PROLIFERATION
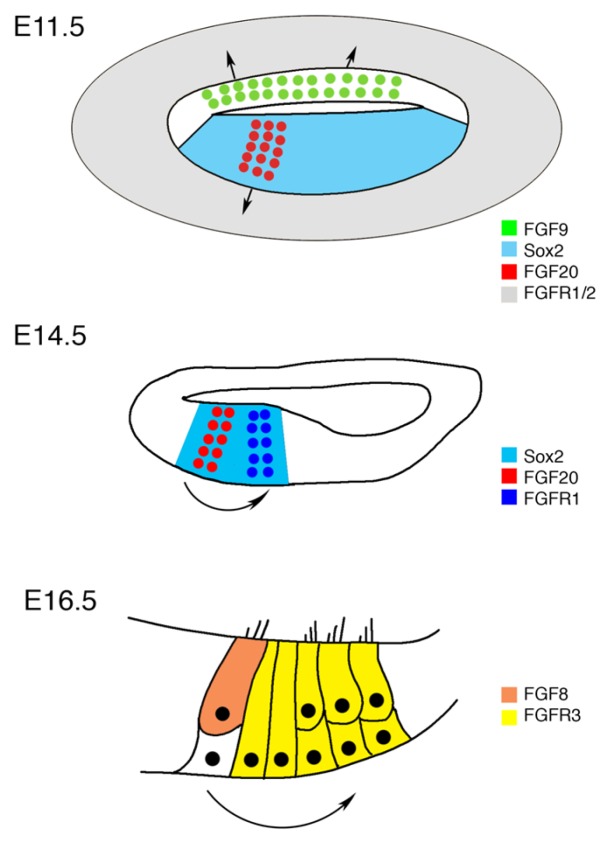
Cochlear sensory progenitor cells start to proliferate beginning E11 (52). Otocysts explanted to ectopic locations in vivo and in vitro do not develop normally unless some periotic mesenchymal tissues are included in the transplant (53, 54). This indicates that factors in mesenchyme are required for cochlear growth. We recently identified that FGF9 and FGF20 are expressed at the non-sensory and sensory epithelium of otic vesicle, respectively and send signal to nearby mesenchymal FGFR1 and FGFR2 to promote cochlear sensory progenitor proliferation and subsequent cochlear growth (Fig. 1) (5). The expression patterns of both Fgf9 and Fgf20 and their potential receptors have been previously studied in the developing cochlea and the surrounding mesenchyme. Fgf9 mRNA is first detected in the otic vesicle epithelium as early as E10.5, specifically in the non-sensory epithelium of the developing cochlear duct in addition to the vestibular components of the inner ear (34). A later study confirmed such expression pattern and showed that Fgf9 continues to be expressed in the ventral wall of the developing cochlear duct that will give rise to the Reissner’s membrane (55). We utilized an Fgf9-βGal reporter allele to monitor Fgf9 expression using βGal activity and antibody analyses showing that there is no overlap between FGF9 and Sox2 expression domains at E11.5 consistent with previously mentioned findings that Fgf9 is expressed in non-sensory epithelium (56).
Fig. 1.
A schematic model showing diverse roles of FGF signaling in cochlear development. In the E11.5 cochlea, FGF9 expressed in non-sensory epithelium and FGF20 in sensory epithelium signal to FGFR1 and FGFR2 within the surrounding mesenchyme to regulate cochlear progenitor proliferation. Around E14.5–E15.5, FGF20 expressed within the Sox2 prosensory domain signal to FGFR1 expressed in the lateral edge of the prosensory domain to regulate the differentiation of outer hair cells and supporting cells. Around E16.5, FGF8 expressed by inner hair cells signal to FGFR3 in outer hair cells and supporting cells to regulate the pillar cell differentiation.
As for Fgf20, utilizing an Fgf20-βGal reporter allele, βGal activity is detected in the anterio-ventral region of the otic vesicle as early as E10.5. Around E11.5, Fgf20-βGal is expressed within the domain of Sox2-positive sensory progenitor cells (4). In situ hybridization for Fgf20 mRNA showed a wave pattern of expression that spreads from the base (around E13.5) to reach the apex (around E16.5) and then declines in a similar pattern (56). Such expression overlapped with Sox2-expressing sensory epithelium (48). Since both FGF9 and FGF20 belong to the same subfamily which shares the same FGFRs, and the timing of expression within the cochlear epithelium overlaps, they might have functionally redundant roles.
By studying various compound mutants of Fgf9 and Fgf20, we now have better understanding of the redundant role of these FGFs in regulating cochlear progenitor cell proliferation. The cochlear length of the double mutant is reduced more than half compared to double heterozygous controls (5). Furthermore, double mutant cochlea is significantly shorter than that of the single Fgf9 or Fgf20 mutants. Studying the size of the prosensory domain in these mouse models reveals decreased size in double mutant. Quantitation of cell proliferation using phosphor-Histone 3 and EdU labeling of E11.5 embryos shows significant reduction in progenitor cell proliferation (5). Such findings indicate a redundant role of Fgf9 and Fgf20 in regulating cochlear length.
Analysis of Fgf9 mutant mice showed normal length of cochlear duct and normal developing sensory epithelium (55). On the other hand, Fgf20 mutant mice exhibited a cochlear phenotype with a specific deficiency in the formation of outer hair cells and outer supporting cells, which we will discuss later, in addition to a small but significant (10%) decrease in cochlear length when compared to heterozygote controls (4). Deleting either Fgf9 or Fgf20 alone does not cause a proliferation defect, yet lack of both had a significant impact on progenitor proliferation and subsequently cochlear duct length.
For Fgf9 and Fgf20 to function, they require FGFR interaction. Expression of both Fgfr1 and Fgfr2 have been reported in the periotic mesenchyme of the ventral (cochlear) part of the inner ear between E10.5 and E12.5 (55). Mesenchymal FGFR signaling is shown to be both necessary and sufficient for sensory progenitor cell proliferation (5). FGFR1 and FGFR2 shows another evidence of gene redundancy. Deleting either Fgfr1 or Fgfr2 in mesenchymal cells using Twist2Cre results in small but significant decrease of cochlear length compared to control, 7% and 20%, respectively (5). Deletion of both Fgfr1 and Fgfr2 from mesenchymal cells reveals shortening of the cochlea by 55% compared to control at E18.5, which is comparable to the loss of Fgf9 and Fgf20 (5). The size and the proliferation of the Sox2-positive progenitor domain is significantly decreased at E12.5 indicating the necessity of mesenchymal Fgfr1 and Fgfr2 for sensory progenitor proliferation. Gain-of-function experiment expressing a constitutive FGFR1 tyrosine kinase domain in mesenchymal cells results in increased proliferation of sensory progenitors after induction between E10.5 and E12.5 and a 14% increase in cochlear length upon induction from E10.5 to E14.5. Such results indicate the sufficiency of mesenchymal FGFR signaling for progenitor cell proliferation (5). Taken together, FGF9 and FGF20 redundantly signal to mesenchymal FGFR1 and FGFR2 to regulate auditory sensory progenitor cell proliferation in the developing cochlea. To date, the molecular pathways downstream FGF epithelial-mesenchymal interaction for sensory progenitor cell proliferation are not understood.
COCHLEAR LATERAL COMPARTMENT DIFFERENTIATION
Several independent lines of research suggest that FGF20 signals to FGFR1 within the developing sensory epithelium between E13.5 and E14.5 to regulate the differentiation of outer hair cells and supporting cells (Fig. 1) (5, 56–58). As mentioned previously, Fgf20 expression overlaps with Sox2-positive domain within the developing cochlea between E13.5 and 16.5 (56). Fgfr1 mRNA is detected in the ventral wall of the developing cochlear duct which comprises the future prosensory domain at E11.5 (57). This expression level increases over the following few days and becomes concentrated into a cluster of cells at the lateral aspect of the prosensory domain in the region of the developing outer hair cells around E14–15 (59) then starts to decrease concomitant to cellular maturation around E18 (57). Comparing the expression patterns of Fgf20 and Fgfr1, it is obvious that they are close within the prosensory domain around the time of hair cell differentiation.
Due to early developmental lethality in Fgfr1 null mutants (60, 61), Fgfr1 hypomorph or conditional knockouts were previously utilized as a loss-of-function models. Study of cochleae of hypomorphic Fgfr1 alleles revealed a phenotype ranging from missing third row of outer hair cells throughout the length of cochlear duct to interrupted sensory epithelium with gaps showing no signs of differentiation of hair cells or supporting cells which was more evident towards the apex of the cochlea (57). Conditional deletion of Fgfr1 in the sensory epithelium resulted in a similar yet more severe phenotype compared to hypomorph. Different Cre drivers were used to regionally inactivate Fgfr1 in the epithelium of the developing inner ear at different time points; whole otic epithelium at E9.5 using Foxg1cre (5, 57), or Six1enh21cre (58), sensory progenitor population at E10.5 using Fgf20cre (5) and cochlear duct epithelium around E12.5 using Emx2cre (58). A consistent phenotype observed in all these mouse models is the perturbed organ of Corti with patches of sensory epithelium mainly comprised of inner hair cells and supporting cells separated by gaps of undifferentiated cells. Such phenotype is more severe in the apical half than the basal half of the cochlea. The number of differentiated outer hair cells is significantly reduced compared to control (5, 57, 58). Furthermore, in vitro experiments treating cochlear explant cultures from E14 to E16 with a small molecule inhibitor of FGFRs (SU5402) photocopies the phenotype observed in Fgfr1 conditional knockout experiments with a significant reduction in the number of outer hair cells and supporting cells (56). It is concluded that FGFR1 signaling is required for the differentiation of outer hair cells and supporting cells during cochlear development.
Analyses of the cochlear length, progenitor cell proliferation and prosensory domain size in Fgfr1 conditional knock out mice exhibit variability across studies. Pirvola et al. reported a reduction in the proliferation rate in the ventral wall of the cochlear duct between E12 to E15.5 and a slight decrease in cochlear length at birth in Fgfr1f/f:: Foxg1Cre (57). Ono et al. reported a 41–49% decrease in cochlear length in Fgfr1f/f:: Six1enh21Cre, and Fgfr1f/f::Emx2Cre conditional knockout mice at E18.5 (58). The number of proliferating cells within the developing cochlea was reduced in Fgfr1f/f:: Six1enh21Cre but not in Fgfr1f/f::Emx2Cre at E12.5 (58). We reported a 9% decrease in cochlear length in Fgfr1−/f::Foxg1Cre/+ and a 19% decrease of cochlear length in Fgfr1−/f::Fgf20Cre/+ mice at E18.5 that were not associated with a proliferation defect at E12.5 (5). The size of the Sox2-positive sensory progenitor domain in both mouse models was not affected at E14.5. Such variability across studies could be attributed to mouse models with different backgrounds, different timing of inactivation of Fgfr1 or different methods of analyses.
FGF20 has been suggested by multiple studies to be the ligand for FGFR1 that is responsible for outer hair cell and supporting cell differentiation (4, 56). Both in vivo and in vitro Fgf20 loss of function studies have revealed such role in cochlear development. In vitro studies culturing cochlear explants at E12–13 then treating cultures with antibody against FGF20 using varying concentrations lead to significant dose-dependent reduction of hair cell and supporting cell numbers. Such effect is rescued by the addition of recombinant FGF20 protein (56). Although the effect of Fgf20 loss of function is evident on hair cell development, the study didn’t show whether this effect is specific to outer hair cells or not. A more recent study utilizing an in vivo model shows that at P0, the cochlea of Fgf20 mutants exhibit a similar phenotype as in Fgfr1 deletion (4), where the basal region contains only two rows of outer hair cells, and the middle and apical region showed patches of hair cells. Specifically, outer hair cell number is significantly reduced in Fgf20 mutant cochlea at P4 while inner hair cell number is not affected. Along with the reduction of outer hair cell number, this model also shows a significant reduction in the number of Deiters cells and outer pillar cells (outer supporting cells). In this model, cochleae exhibit normal cell proliferation within the prosensory domain indicating that the reduced number of outer hair cells is due to a differentiation defect rather than a proliferation defect. Treating E13.5–14.5 Fgf20 mutant cochlear explants with exogenous FGF9 (has same biochemical activity as FGF20 in vitro) shows rescue of cochlear phenotype (4). Altogether, there is much evidence that FGF20/FGFR1 signaling is required for lateral compartment differentiation.
PILLAR CELL DIFFERENTIATION
During post-mitotic stages (around E16.5), FGF8 signals from the inner hair cells to FGFR3 in supporting cells to regulate pillar cell differentiation (62–64). The expression of Fgfr3 is detected around E16 in a band of cells extending along the length of the cochlear duct in the region that will develop as the pillar cells, outer hair cells, and Deiters cells. Such expression become restricted to pillar cells by P0 (63, 65). Fgf8 is exclusively expressed in inner hair cells and starts as early as E16 within the basal turn of the cochlea. Such close expression pattern for Fgf8 and Fgfr3 triggered studies to investigate the potential ligand-receptor interaction in pillar cell development (Fig. 1). Deletion of Fgfr3 leads to defects in pillar cell development. In Fgfr3 mutants, two rows of undifferentiated cells are observed in the region between the row of inner hair cells and the first row of outer hair cells that corresponds to the region of pillar cells (62). As in Fgfr3 mutant mice, pillar cell development is disrupted in E13.5–14.5 cochlear explants exposed to the FGFR inhibitor SU5402 for 18 hours in vitro (63). Gain-of-function experiment treating cochlear explants with FGF2 (a ligand for FGFR3) led to a dose-dependent increase in the number of cells that developed as pillar cells (63), suggesting that FGFR3 activation is sufficient to commit progenitor cells to pillar cell fate. Research demonstrates that FGF8 expressed by the inner hair cells signals to the FGFR3 in the sensory epithelium and regulates the development of pillar cells. Loss-of-function experiments either through deletion of Fgf8 or inhibition of binding between Fgf8 and Fgfr3 leads to defects in pillar cell development, whereas overexpression of FGF8 or exogenous FGFR3 activation by adding FGF17 induces ectopic pillar cell formation at the expense of outer hair cell development (64).
In humans, a gain-of-function mutation in FGFR3 causes Muenke syndrome which comprises hearing loss. The mutation is within the extracellular domain of FGFR3 and increases its binding affinity for certain FGFs. Mouse model of the Muenke syndrome exhibits transformation of two rows of Deiters cells to two rows of pillar cells yielding to four rows of pillar cells and one row of Deiters cells (66). A recent study investigating the underlying molecular mechanisms of this transformation found that such switch occurs sequentially between E17.5 and P3. Surprisingly, supporting cell fate transformation was not rescued by reducing Fgf8. Instead, Fgf10 inhibition was sufficient for rescue of cochlear structure and function. It is concluded that the Muenke syndrome mutation changes the specificity of FGFR3b and FGFR3c such that both acquire responsiveness to FGF10 that normally binds to FGFR1b and FGFR2b (7).
NON-SENSORY DOMAIN DEVELOPMENT
Reciprocal epithelium-mesenchyme signaling is a fundamental process for the morphogenesis of multiple organs. In inner ear, FGF9 expressed in non-sensory domain of otic epithelium signals to the FGFR1 and FGFR2 (IIIc spliceform) in the surrounding mesenchyme between E11.5 and E13.5 to regulate mesenchymal cell proliferation and subsequent condensation (55). Inner ears of Fgf9 null mice show hypoplastic vestibular component of the otic capsule and absent semicircular ducts mainly due to reduced proliferation of the prechondrogenic mesenchyme causing capsular hypoplasticity (55). Although the length of the cochlear duct and the structure of cochlear sensory epithelium was not affected in Fgf9 null mutants at birth, cochlear mesenchyme shows failure in the development of scala vestibule (55). It is concluded that epithelial FGF9 signaling is required for stimulating growth and remodeling of the otic mesenchyme.
A recent study suggests that FGF10 expressed in otic epithelium signals to FGFR2b expressed in adjacent epithelial regions to induce Reissner’s membrane and outer sulcus development between E12.5–E15.5 (67). Fgf10 is expressed in a sensory-competent domain early in cochlear development, eventually becoming restricted to Kolliker’s organ (44), while Fgfr2b is expressed in the entire non-sensory domain of the otic epithelium, including the prospective Reissner’s membrane, stria vascularis and outer sulcus (67). Examining the cochlea of Fgf10 mutants at E18.5, cochlear ducts appeared to lack Reissner’s membrane and most of the outer sulcus. Immunostaining for specific markers for these two regions confirmed the phenotype. Since FGFR2b receptors are expressed in these two regions, it is suggested that FGF10 signal to FGFR2b receptors to induce these two non-sensory domains, Reissner’s membrane and the outer sulcus.
In human, heterozygous mutations in FGF10 cause LADD (lacrimo-auriculo-dento-digital) syndrome, and 55% of LADD syndrome subjects has hearing loss (68). LADD syndrome is caused by heterozygous mutations in FGFR2 and FGFR3 that reduce FGF signaling activity.
CONCLUSION AND FUTURE PERSPECTIVE
It is quite evident that FGF signaling plays diverse roles during cochlear development in a context-dependent manner. Different members of the FGF family function either individually or redundantly to regulate progenitor cell number, mediate sensory epithelial patterning, and induce specification and differentiation of different cell populations within the developing cochlea. Developmental defects result from aberrant activity of FGF signaling pathway confirm the importance of such pathway during different stages of cochlear development. However, there is a few discrepancies among published data that need to be addressed.
The phenotype severity of loss of Fgf20 in vivo is less than that observed upon treating cochlear explants with FGF20 antibody that exhibit more hair cell loss (4, 56). Possible explanations would be 1) different genetic background, 2) artifact of the explant, or 3) existence of other FGF functioning with FGF20 to promote lateral compartment differentiation. Our recent experiments show that treatment of Fgf20 null cochlear explant with FGFR inhibitor decreases number of hair cells compared to Fgf20 null untreated explant (unpublished). This suggests that there might be other FGF functioning with FGF20 to promote cochlear lateral compartment differentiation.
Another unsolved question is the splice variant of FGFR1 to which FGF20 binds for regulating lateral compartment differentiation. FGF20 belongs to FGF9 subfamily that comprises FGF9, FGF16 and FGF20. They bind to c splice variants of FGFR1, FGFR2, and both b and c splice variant of FGFR3 (30). The phenotype of Fgf20 mutant mice recapitulates epithelial Fgfr1 deletion mutants (57), yet Fgfr1b mutant mice are viable and do not have gross defects (69). In addition, number of hair cells from Fgfr1b mutant cochlea is comparable to control (unpublished). This raises possibility that FGFR1C might be expressed in the cochlear sensory epithelium and receives signal from FGF20 to regulate lateral compartment differentiation. Indeed, FGFR2C is expressed in developing lung epithelium and receive signals from FGF9 (70). Therefore, further study is required to better understand the mechanism by which FGF20 regulates cochlear sensory differentiation.
Given the different roles FGF signaling plays during cochlear sensory epithelial development, it is a potential pathway that can be manipulated for hair cell regeneration. Evidence of FGF signaling role in regeneration comes from chicken and zebrafish models that are capable of spontaneously regenerating their sensory epithelium. Transcriptional profiling of regenerating chicken cochlear sensory epithelia after ototoxic injury revealed that Fgf20 expression is low initially during the proliferative phase of regenerating supporting cells, then increase later when proliferation stops (71). Such finding indicates a role of FGF20 in the differentiation of proliferating supporting cells during hair cell regeneration, which is comparable to the role of FGF20 in lateral compartment differentiation during normal development. For mammalian hair cell regeneration, a possible way to utilize FGF20 might be first inducing proliferation of supporting cells using Wnt signaling then applying FGF20 to guide hair cell differentiation in cochlear explants with hair cell insult.
Since the mammalian cochlea lacks the capability to regenerate hair cells after damage, the ultimate goal for understanding the role of FGF signaling as well as other signaling pathways is to manipulate these pathways to induce hair cell regeneration. A learned lesson from the diverse roles of FGF signaling is that the context is the main determinant of FGF signaling function. Such concept must be taken into consideration for utilizing this signaling pathway to induce hair cell regeneration.
ACKNOWLEDGEMENTS
This work was supported by Mary & Dick Holland Regenerative Medicine Program, NIH grants DC012825 and GM110768, and Edna Ittner Pediatric Research Support.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Lopez-Poveda EAPARMR. The neurophysiological bases of auditory perception. Springer; New York: 2010. [DOI] [Google Scholar]
- 2.Driver EC, Kelley MW. Specification of cell fate in the mammalian cochlea. Birth Defects Res C Embryo Today. 2009;87:212–221. doi: 10.1002/bdrc.20154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schimmang T. Expression and functions of FGF ligands during early otic development. Int J Dev Biol. 2007;51:473–481. doi: 10.1387/ijdb.072334ts. [DOI] [PubMed] [Google Scholar]
- 4.Huh SH, Jones J, Warchol ME, Ornitz DM. Differentiation of the lateral compartment of the cochlea requires a temporally restricted FGF20 signal. PLoS Biol. 2012;10:e1001231. doi: 10.1371/journal.pbio.1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huh SH, Warchol ME, Ornitz DM. Cochlear progenitor number is controlled through mesenchymal FGF receptor signaling. eLife. 2015;4:e05921. doi: 10.7554/eLife.05921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ono K, Kita T, Sato S, et al. FGFR1-Frs2/3 signalling maintains sensory progenitors during inner ear hair cell formation. PLoS Genet. 2014;10:e1004118. doi: 10.1371/journal.pgen.1004118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mansour SL, Li C, Urness LD. Genetic rescue of Muenke syndrome model hearing loss reveals prolonged FGF-dependent plasticity in cochlear supporting cell fates. Genes Dev. 2013;27:2320–2331. doi: 10.1101/gad.228957.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ornitz DM. FGFs, heparan sulfate and FGFRs: complex interactions essential for development. Bioessays. 2000;22:108–112. doi: 10.1002/(SICI)1521-1878(200002)22:2<108::AID-BIES2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 9.Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2:reviews3005.3001–reviews3005.3012. doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rapraeger AC, Krufka A, Olwin BB. Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science (New York, N.Y.) 1991;252:1705–1708. doi: 10.1126/science.1646484. [DOI] [PubMed] [Google Scholar]
- 11.Ornitz DM, Itoh N. The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip Rev Dev Biol. 2015;4:215–266. doi: 10.1002/wdev.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potthoff MJ, Kliewer SA, Mangelsdorf DJ. Endocrine fibroblast growth factors 15/19 and 21: from feast to famine. Genes Dev. 2012;26:312–324. doi: 10.1101/gad.184788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith ER, McMahon LP, Holt SG. Fibroblast growth factor 23. Ann Clin Biochem. 2014;51:203–227. doi: 10.1177/0004563213510708. [DOI] [PubMed] [Google Scholar]
- 14.Goldfarb M. Fibroblast growth factor homologous factors: evolution, structure, and function. Cytokine Growth Factor Rev. 2005;16:215–220. doi: 10.1016/j.cytogfr.2005.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoorlemmer J, Goldfarb M. Fibroblast growth factor homologous factors and the islet brain-2 scaffold protein regulate activation of a stress-activated protein kinase. J Biol Chem. 2002;277:49111–49119. doi: 10.1074/jbc.M205520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herr AB, Ornitz DM, Sasisekharan R, Venkataraman G, Waksman G. Heparin-induced self-association of fibroblast growth factor-2. Evidence for two oligomerization processes. J Biol Chem. 1997;272:16382–16389. doi: 10.1074/jbc.272.26.16382. [DOI] [PubMed] [Google Scholar]
- 17.Mach H, Volkin DB, Burke CJ, et al. Nature of the interaction of heparin with acidic fibroblast growth factor. Biochemistry. 1993;32:5480–5489. doi: 10.1021/bi00071a026. [DOI] [PubMed] [Google Scholar]
- 18.McKeehan WL, Wang F, Kan M. The heparan sulfate-fibroblast growth factor family: diversity of structure and function. Prog Nucleic Acid Res Mol Biol. 1998;59:135–176. doi: 10.1016/S0079-6603(08)61031-4. [DOI] [PubMed] [Google Scholar]
- 19.Bottcher RT, Niehrs C. Fibroblast growth factor signaling during early vertebrate development. Endocr Rev. 2005;26:63–77. doi: 10.1210/er.2003-0040. [DOI] [PubMed] [Google Scholar]
- 20.Wang JK, Xu H, Li HC, Goldfarb M. Broadly Expressed Snt-Like Proteins Link Fgf Receptor Stimulation to Activators Of Ras. Oncogene. 1996;13:721–729. [PubMed] [Google Scholar]
- 21.Ong SH, Guy GR, Hadari YR, et al. FRS2 proteins recruit intracellular signaling pathways by binding to diverse targets on fibroblast growth factor and nerve growth factor receptors. Mol Cell Biol. 2000;20:979–989. doi: 10.1128/MCB.20.3.979-989.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kouhara H, Hadari YR, Spivakkroizman T, et al. A Lipid-Anchored Grb2-Binding Protein That Links Fgf-Receptor Activation to the Ras/Mapk Signaling Pathway. Cell. 1997;89:693–702. doi: 10.1016/S0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 23.Hadari YR, Gotoh N, Kouhara H, Lax I, Schlessinger J. Critical role for the docking-protein FRS2 alpha in FGF receptor-mediated signal transduction pathways. Proc Natl Acad Sci U S A. 2001;98:8578–8583. doi: 10.1073/pnas.161259898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouhara H, Kasayama S, Saito H, Matsumoto K, Sato B. Expression cDNA cloning of fibroblast growth factor (FGF) receptor in mouse breast cancer cells: A variant form in FGF-responsive transformed cells. Biochem Biophys Res Comm. 1991;176:31–37. doi: 10.1016/0006-291X(91)90885-B. [DOI] [PubMed] [Google Scholar]
- 25.Janknecht R, Monte D, Baert JL, de Launoit Y. The ETS-related transcription factor ERM is a nuclear target of signaling cascades involving MAPK and PKA. Oncogene. 1996;13:1745–1754. [PubMed] [Google Scholar]
- 26.Johnson DE, Williams LT. Structural and functional diversity in the FGF receptor multigene family. Adv Cancer Res. 1993;60:1–41. doi: 10.1016/s0065-230x(08)60821-0. [DOI] [PubMed] [Google Scholar]
- 27.Groth C, Lardelli M. The structure and function of vertebrate fibroblast growth factor receptor 1. Int J Dev Biol. 2002;46:393–400. [PubMed] [Google Scholar]
- 28.Ornitz DM, Xu J, Colvin JS, et al. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 29.Xu J, Liu Z, Ornitz DM. Temporal and spatial gradients of Fgf8 and Fgf17 regulate proliferation and differentiation of midline cerebellar structures. Development. 2000;127:1833–1843. doi: 10.1242/dev.127.9.1833. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Avivi A, Yayon A, Givol D. A novel form of FGF receptor-3 using an alternative exon in the immunoglobulin domain III. FEBS Lett. 1993;330:249–252. doi: 10.1016/0014-5793(93)80882-U. [DOI] [PubMed] [Google Scholar]
- 32.Gilbert E, Del Gatto F, Champion-Arnaud P, Gesnel MC, Breathnach R. Control of BEK and K-SAM splice sites in alternative splicing of the fibroblast growth factor receptor 2 pre-mRNA. Mol Cell Biol. 1993;13:5461–5468. doi: 10.1128/MCB.13.9.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA. sprouty Encodes a Novel Antagonist of FGF Signaling that Patterns Apical Branching of the Drosophila Airways. Cell. 1998;92:253–263. doi: 10.1016/S0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- 34.Colvin JS, Feldman B, Nadeau JH, Goldfarb M, Ornitz DM. Genomic organization and embryonic expression of the mouse fibroblast growth factor 9 gene. Dev Dyn. 1999;216:72–88. doi: 10.1002/(SICI)1097-0177(199909)216:1<72::AID-DVDY9>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 35.Colvin JS, White A, Pratt SJ, Ornitz DM. Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development. 2001;128:2095–2106. doi: 10.1242/dev.128.11.2095. [DOI] [PubMed] [Google Scholar]
- 36.White AC, Xu J, Yin Y, Smith C, Schmid G, Ornitz DM. FGF9 and SHH signaling coordinate lung growth and development through regulation of distinct mesenchymal domains. Development. 2006;133:1507–1517. doi: 10.1242/dev.02313. [DOI] [PubMed] [Google Scholar]
- 37.Bellusci S, Grindley J, Emoto H, Itoh N, Hogan BL. Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development. 1997;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- 38.Park WY, Miranda B, Lebeche D, Hashimoto G, Cardoso WV. FGF-10 is a chemotactic factor for distal epithelial buds during lung development. Dev Biol. 1998;201:125–134. doi: 10.1006/dbio.1998.8994. [DOI] [PubMed] [Google Scholar]
- 39.Min H, Danilenko DM, Scully SA, et al. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sekine K, Ohuchi H, Fujiwara M, et al. Fgf10 is essential for limb and lung formation. Nat Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- 41.Ohuchi H, Hori Y, Yamasaki M, et al. FGF10 acts as a major ligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem Biophys Res Commun. 2000;277:643–649. doi: 10.1006/bbrc.2000.3721. [DOI] [PubMed] [Google Scholar]
- 42.Arman E, Haffner-Krausz R, Gorivodsky M, Lonai P. Fgfr2 is required for limb outgrowth and lung-branching morphogenesis. Proc Natl Acad Sci U S A. 1999;96:11895–11899. doi: 10.1073/pnas.96.21.11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morsli H, Choo D, Ryan A, Johnson R, Wu DK. Development of the mouse inner ear and origin of its sensory organs. J Neurosci. 1998;18:3327–3335. doi: 10.1523/JNEUROSCI.18-09-03327.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohyama T, Basch ML, Mishina Y, Lyons KM, Segil N, Groves AK. BMP signaling is necessary for patterning the sensory and nonsensory regions of the developing mammalian cochlea. J Neurosci. 2010;30:15044–15051. doi: 10.1523/JNEUROSCI.3547-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riccomagno MM, Martinu L, Mulheisen M, Wu DK, Epstein DJ. Specification of the mammalian cochlea is dependent on Sonic hedgehog. Genes Dev. 2002;16:2365–2378. doi: 10.1101/gad.1013302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riccomagno MM, Takada S, Epstein DJ. Wnt-dependent regulation of inner ear morphogenesis is balanced by the opposing and supporting roles of Shh. Genes Dev. 2005;19:1612–1623. doi: 10.1101/gad.1303905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiernan AE, Xu J, Gridley T. The Notch ligand JAG1 is required for sensory progenitor development in the mammalian inner ear. PLoS Genet. 2006;2:e4. doi: 10.1371/journal.pgen.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kiernan AE, Pelling AL, Leung KK, et al. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005;434:1031–1035. doi: 10.1038/nature03487. [DOI] [PubMed] [Google Scholar]
- 49.Lee YS, Liu F, Segil N. A morphogenetic wave of p27Kip1 transcription directs cell cycle exit during organ of Corti development. Development. 2006;133:2817–2826. doi: 10.1242/dev.02453. [DOI] [PubMed] [Google Scholar]
- 50.Chen P, Johnson JE, Zoghbi HY, Segil N. The role of Math1 in inner ear development: Uncoupling the establishment of the sensory primordium from hair cell fate determination. Development. 2002;129:2495–2505. doi: 10.1242/dev.129.10.2495. [DOI] [PubMed] [Google Scholar]
- 51.Kelley MW. Regulation of cell fate in the sensory epithelia of the inner ear. Nat Rev Neurosci. 2006;7:837–849. doi: 10.1038/nrn1987. [DOI] [PubMed] [Google Scholar]
- 52.Zhai S, Shi L, Wang BE, et al. Isolation and culture of hair cell progenitors from postnatal rat cochleae. J Neurobiol. 2005;65:282–293. doi: 10.1002/neu.20190. [DOI] [PubMed] [Google Scholar]
- 53.Montcouquiol M, Kelley MW. Planar and vertical signals control cellular differentiation and patterning in the mammalian cochlea. J Neurobiol. 2003;23:9469–9478. doi: 10.1523/JNEUROSCI.23-28-09469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swanson GJ, Howard M, Lewis J. Epithelial autonomy in the development of the inner ear of a bird embryo. Dev Biol. 1990;137:243–257. doi: 10.1016/0012-1606(90)90251-D. [DOI] [PubMed] [Google Scholar]
- 55.Pirvola U, Zhang X, Mantela J, Ornitz DM, Ylikoski J. Fgf9 signaling regulates inner ear morphogenesis through epithelial-mesenchymal interactions. Dev Biol. 2004;273:350–360. doi: 10.1016/j.ydbio.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 56.Hayashi T, Ray CA, Bermingham-McDonogh O. Fgf20 is required for sensory epithelial specification in the developing cochlea. J Neurosci. 2008;28:5991–5999. doi: 10.1523/JNEUROSCI.1690-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pirvola U, Ylikoski J, Trokovic R, Hebert JM, McConnell SK, Partanen J. FGFR1 is required for the development of the auditory sensory epithelium. Neuron. 2002;35:671–680. doi: 10.1016/S0896-6273(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 58.Ono K, Kita T, Sato S, et al. FGFR1-Frs2/3 Signalling Maintains Sensory Progenitors during Inner Ear Hair Cell Formation. PLoS Genet. 2014;10:e1004118. doi: 10.1371/journal.pgen.1004118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hayashi T, Ray CA, Younkins C, Bermingham-McDonogh O. Expression patterns of FGF receptors in the developing mammalian cochlea. Dev Dyn. 2010;239:1019–1026. doi: 10.1002/dvdy.22236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deng CX, Wynshaw-Boris A, Shen MM, Daugherty C, Ornitz DM, Leder P. Murine FGFR-1 is required for early postimplantation growth and axial organization. Genes Dev. 1994;8:3045–3057. doi: 10.1101/gad.8.24.3045. [DOI] [PubMed] [Google Scholar]
- 61.Yamaguchi TP, Harpal K, Henkemeyer M, Rossant J. Fgfr-1 is Required for Embryonic Growth and Mesodermal Patterning during Mouse Gastrulation. Genes Dev. 1994;8:3032–3044. doi: 10.1101/gad.8.24.3032. [DOI] [PubMed] [Google Scholar]
- 62.Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- 63.Mueller KL, Jacques BE, Kelley MW. Fibroblast growth factor signaling regulates pillar cell development in the organ of corti. J Neurosci. 2002;22:9368–9377. doi: 10.1523/JNEUROSCI.22-21-09368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jacques BE, Montcouquiol ME, Layman EM, Lewandoski M, Kelley MW. Fgf8 induces pillar cell fate and regulates cellular patterning in the mammalian cochlea. Development. 2007;134:3021–3029. doi: 10.1242/dev.02874. [DOI] [PubMed] [Google Scholar]
- 65.Peters K, Ornitz D, Werner S, Williams L. Unique expression pattern of the FGF receptor 3 gene during mouse organogenesis. Dev Biol. 1993;155:423–430. doi: 10.1006/dbio.1993.1040. [DOI] [PubMed] [Google Scholar]
- 66.Mansour SL, Twigg SR, Freeland RM, Wall SA, Li C, Wilkie AO. Hearing loss in a mouse model of Muenke syndrome. Hum Mol Genet. 2009;18:43–50. doi: 10.1093/hmg/ddn311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Urness LD, Wang X, Shibata S, Ohyama T, Mansour SL. Fgf10 is required for specification of non-sensory regions of the cochlear epithelium. Dev Biol. 2015;400:59–71. doi: 10.1016/j.ydbio.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shams I, Rohmann E, Eswarakumar VP, et al. Lacrimo-auriculo-dento-digital syndrome is caused by reduced activity of the fibroblast growth factor 10 (FGF10)-FGF receptor 2 signaling pathway. Mol Cell Biol. 2007;27:6903–6912. doi: 10.1128/MCB.00544-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang H, Dessimoz J, Beyer TA, et al. Fibroblast growth factor receptor 1-IIIb is dispensable for skin morphogenesis and wound healing. Eur J Cell Biol. 2004;83:3–11. doi: 10.1078/0171-9335-00355. [DOI] [PubMed] [Google Scholar]
- 70.del Moral PM, De Langhe SP, Sala FG, et al. Differential role of FGF9 on epithelium and mesenchyme in mouse embryonic lung. Dev Biol. 2006;293:77–89. doi: 10.1016/j.ydbio.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 71.Ku YC, Renaud NA, Veile RA, et al. The transcriptome of utricle hair cell regeneration in the avian inner ear. J Neurosci. 2014;34:3523–3535. doi: 10.1523/JNEUROSCI.2606-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alvarez Y, Alonso MT, Vendrell V, et al. Requirements for FGF3 and FGF10 during inner ear formation. Development. 2003;130:6329–6338. doi: 10.1242/dev.00881. [DOI] [PubMed] [Google Scholar]
- 73.Wright TJ, Mansour SL. Fgf3 and Fgf10 are required for mouse otic placode induction. Development. 2003;130:3379–3390. doi: 10.1242/dev.00555. [DOI] [PubMed] [Google Scholar]
- 74.Urness LD, Paxton CN, Wang X, Schoenwolf GC, Mansour SL. FGF signaling regulates otic placode induction and refinement by controlling both ectodermal target genes and hindbrain Wnt8a. Dev Biol. 2010;340:595–604. doi: 10.1016/j.ydbio.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ohuchi H, Yasue A, Ono K, et al. Identification of cis-element regulating expression of the mouse Fgf10 gene during inner ear development. Dev Dyn. 2005;233:177–187. doi: 10.1002/dvdy.20319. [DOI] [PubMed] [Google Scholar]
- 76.Pauley S, Wright TJ, Pirvola U, Ornitz D, Beisel K, Fritzsch B. Expression and function of FGF10 in mammalian inner ear development. Dev Dyn. 2003;227:203–215. doi: 10.1002/dvdy.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pirvola U, Spencer-Dene B, Xing-Qun L, et al. FGF/FGFR-2(IIIb) signaling is essential for inner ear morphogenesis. J Neurosci. 2000;20:6125–6134. doi: 10.1523/JNEUROSCI.20-16-06125.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hayashi T, Cunningham D, Bermingham-McDonogh O. Loss of Fgfr3 leads to excess hair cell development in the mouse organ of Corti. Dev Dyn. 2007;236:525–533. doi: 10.1002/dvdy.21026. [DOI] [PubMed] [Google Scholar]