SUMMARY
Ferroptosis is a form of programmed cell death pathogenic to several acute and chronic diseases and executed via oxygenation of polyunsaturated phosphatidylethanolamines (PE) by 15-lipoxygenases (15-LO) that normally use free polyunsaturated fatty acids as substrates. Mechanisms of the altered 15-LO substrate specificity are enigmatic. We sought a common ferroptosis regulator for 15LO. We discovered that PEBP1, a scaffold protein inhibitor of protein kinase cascades, complexes with two 15LO isoforms, 15LO1 and 15LO2, and changes their substrate competence to generate hydroperoxy-PE. Inadequate reduction of hydroperoxy-PE due to insufficiency or dysfunction of a selenoperoxidase, GPX4, leads to ferroptosis. We demonstrated the importance of PEBP1-dependent regulatory mechanisms of ferroptotic death in airway epithelial cells in asthma, kidney epithelial cells in renal failure and cortical and hippocampal neurons in brain trauma. As master regulators of ferroptotic cell death with profound implications for human disease, PEBP1/15LO complexes represent a new target for drug discovery.
Keywords: ferroptosis, phosphatidylethanolamine oxidation, PEBP1/15LO complex, GPX4, asthma, acute kidney injury, brain trauma
In brief
The small scaffolding protein PEBP1 regulates ferroptotic cell death by binding with lipoxygenases and allowing them to generate lipid peroxides
INTRODUCTION
One of the hallmarks of aerobic life is the enzymatic generation of oxygenated polyunsaturated lipids, free polyunsaturated fatty acids (PUFA) and PUFA esterified into phospholipids (PLs) as their diversity made them valuable for signaling (Serhan, 2014). Paradoxically, the usefulness of these oxygenated molecules is also associated with a risk. Indeed, their enzymatic biosynthesis proceeds via the insertion of molecular oxygen to yield the peroxy-derivatives. The specific feature of the peroxy-bond (-O-O-) is the ease with which it decomposes to numerous electrophiles with high reactivity towards nucleophilic protein targets, hence treacherous not only for the cells of their origin but also for the neighbors. Therefore, levels of hydroperoxy (OOH)-lipids are enzymatically controlled by several GSH peroxidases, including a selenoprotein, glutathione peroxidase 4 (GPX4), the only GPX that handles OOH-PLs (Toppo et al., 2009). GPX4 deficiency leads to accumulation of OOH-PEs in kidney epithelial cells leading to acute renal failure (Friedmann Angeli et al., 2014). Consequently, cells with excessive production of these intermediates are subject to elimination via a recently discovered death program, ferroptosis (Dixon et al., 2012).
We recently identified OOH-phosphatidylethanolamines (OOH-PE) as ferroptotic death signals and showed that the balance between the production of OOH-PE metabolites by 15LO and their reduction to hydroxy-(OH)-metabolites by GPX4 serves as a pivot point between cell survival and ferroptosis (Kagan et al., 2017). LOs are effective in oxygenation of free PUFA liberated from PLs by Ca2+-dependent phospholipases A2 feeding the subsequent oxygenation stages. Free PUFAs are minor components of biomembranes - in contrast to a broad range of PUFA-PLs, their most abundant structural components. Thus, oxygenation of an individual PUFA-PL requires selection from diversified PLs present in membranes. The selectivity mechanism remains enigmatic and has emerged as one of important issues of ferroptosis regulation.
A promiscuous small scaffolding protein, phosphatidylethanolamine-binding protein 1 (PEBP1) also known as Raf1 kinase inhibitory protein (RKIP1), is bound to and inhibits the Raf1 kinase cascade under homeostatic conditions. Phosphorylation of PEBP1 liberates it (Yeung et al., 1999) to find new partners, including 15LO. 15LOs catalyze the formation of pro-ferroptotic 15-OOH-eicosatetraenoic (arachidonic, AA) acid (HpETE) esterified into PE (HpETE-PE). Given the preferential accumulation of their reduced derivatives, (15-HETE-PE) (but not free 15-OH-AA (15-HETE)) in human airway epithelial cells (HAECs) stimulated with IL13/IL4 we implicated the involvement of an isoform of 15LO, 15LO1, and 15-HETE-PE in the regulation of Raf1/PEBP1 interactions (Zhao et al., 2011). Assuming the triggering role of 15-HpETE-PE in ferroptosis, we hypothesized that PEBP1/15LO complexes act as master regulators of the oxygenase activity by changing 15LO substrate specificity from free PUFA to PUFA-PE. Here, we deciphered the mechanisms through which PEBP1 acts as a rheostat of the specificity of 15LOs enabling their catalytic competence towards PUFA-PEs and leading to generation of HpETE-PEs. Insufficient reduction of the latter by GPX4 - due to genetic defects, chemical inactivation or GSH depletion – promoted ferroptosis in several types of human and murine cells expressing 15LO1/15LO2, including bronchial and kidney epithelial cells and neurons. We uncovered the relevance of these redox mechanisms of ferroptosis to the pathophysiology of asthma, acute renal failure (AKI) and traumatic brain injury (TBI).
RESULTS
Complexes of 15LOs with PEBP1
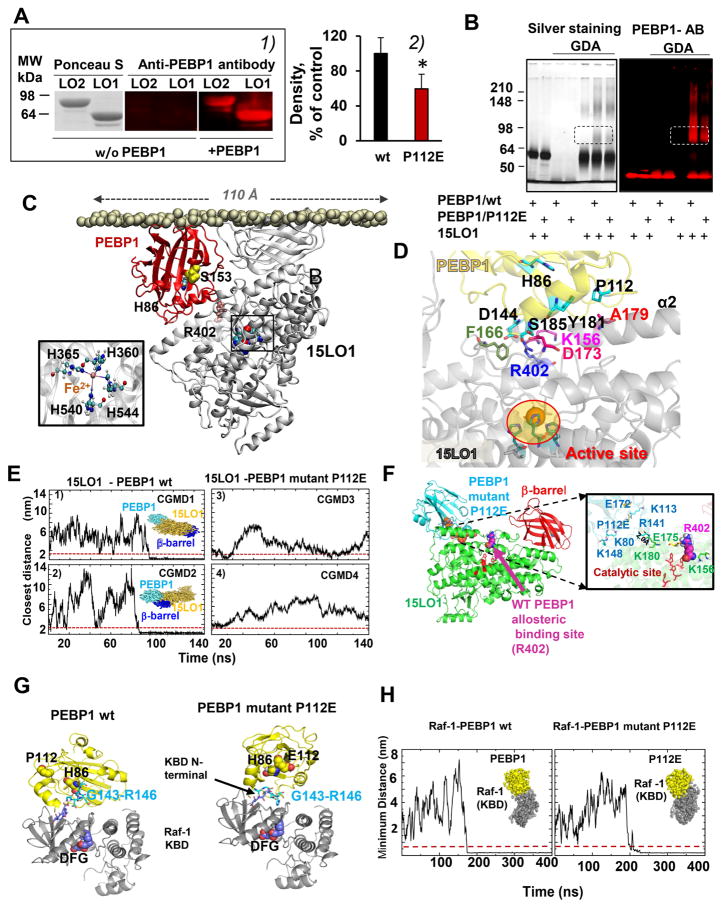
We tested whether PEBP1 forms complexes with 15LO isoforms, 15LO1 and 15LO2, which display differential tissue distribution. 15LO1 is highly expressed in HAECs, (in response to the Type-2 cytokine IL13), and 15LO2 is expressed in renal tubular epithelium and neurons. We used two different protocols to assess PEBP1 binding interactions with 15LO: i) Far-Western blotting (Wu et al., 2007) and ii) crosslinking by an amine-directed reagent, glutaric dialdehyde (GDA). In Far-Western blotting, an antibody-detectable “bait” protein (PEBP1) is used to detect the target “prey” protein (15LO) on the membrane. Using anti-PEBP1 antibody we detected PEBP1 in the bands corresponding to both 15LO1 and 15LO2 (Figure 1A1) indicative of its binding to both isoforms of 15LO1. Formation of PEBP1/15LO1 complexes was supported by cross-linking of the proteins by GDA and the appearance of PEBP1-positive staining in the 15LO1 band and its oligomeric forms (Figure 1B and S1A).
Figure 1. PEBP1 forms complexes with 15LO1 and 15LO2.
(A) 1) Far Western blotting gels showing binding of PEBP1 with 15LO. Ponceau S staining of proteins (left panel). Immuno-detection of PEBP1 bound to 15LO using anti-PEBP1 antibody (middle and right panels). 2) Quantitation of 15LO1 bound to wt PEBP1 and P112E PEBP1 with anti-15LO1 antibody (means±SD, *p < 0.05 vs. wt PEBP1, N=7/group.
(B) SDS-PAGE gels illustrating interactions of 15LO1 with wt PEBP1 and P112E PEBP1 revealed by cross-linking using glutaric dialdehyde (GDA). Proteins were detected by silver-staining or by anti–PEBP1 antibodies.
C) Computational modeling of human PEBP1/15LO1 complex. 15LO1 (gray) and PEBP1 (red) bound to the lipid bilayer are displayed, with the PL head phosphorus atoms in tan spheres, and 15LO1 R402, crucial for PEBP1/15LO1 interactions, in pink spheres. Inset: Coordination of Fe2+ at 15LO1 catalytic byH360, H365, H540 and H544.
(D) Close-up view of PEBP1/15LO1 interface. The interface between PEBP1 (yellow) and 15LO1 (gray) complex closely neighbors the AA binding site located near the catalytic site (highlighted in ellipse). K156, F166, D173, R402 in 15LO1, and P112, D144 and Y181 in PEBP1 make interfacial contacts that stabilize the PEBP1/15LO1 complex.
(E) Coarse-grained molecular dynamics (CGMD) simulations confirm the inability of the P112E to bind 15LO1. Four independent runs are displayed, two for wt PEBP1 (CGMD1 and 2), and two for the mutant (CGMD3 and 4). The ordinates denote the closest distance between the two proteins, originally separated by 20 Å. In CGMD1 and 2, stable bound poses are reached ~80–100 ns; the minimal distance attained and maintained during the remaining 150 ns. For the mutant, there is either no formation of complex (3), or transient binding (4) indicating weaker (if any) binding of P112E to 15LO1.
(F) Binding of P112E to 15LO1 in silico shows the weaker affinity and distinctive binding. Left: the optimal binding pose for mutant P112E is shifted away from the putative allosteric binding site near R402. Right: interfacial interactions. Wt PEBP1 exhibits several close contacts (see also Figure S5), which are broken due to the repulsion between mutated P112 (E112) and E175 on 15LO1. P112E and 15LO1 residues are colored as blue and green, respectively. E112 on PEBP1 and E175 on 15LO1 undergo side chain rotations in the mutant complex that weaken the intermolecular interactions.
(G) Binding of wt PEBP1 and P112E to Raf-1 kinase whose binding domain (KBD) (PDB id: 3c4c) (Tsai et al., 2008) was docked against human PEBP1 (left) and P112E mutant (right). Top docking poses of PEBP1 were in the vicinity of KBD N-terminus. Both wt (left) and P112E (right) utilize the region G143-R146 on PEBP1 for interacting with Raf-1 KBD (Deiss et al., 2012).
(H) CGMD of Raf-1 binding to wt PEBP1 (left) and P112E (right). In both cases, the complex was formed (distance less than 0.5 nm, indicated by dashed redline) at approximately 175ns (left) and 200ns (right).
Computational docking of PEBP1 with 15LO1 (Figure 1C) and 15LO2 (Figure S1B) yielded similar binding poses of PEBP1 onto the two 15LOs, and their similar associations with the membrane lipid bilayer via the β-barrel domain. The interfacial interactions between PEBP1 and the isoforms involved the α2-helix of 15LOs. Closer examination revealed the associations of PEBP1 residues G143-D144-H145 with 15LO1 R402 (highly conserved in both 15LOs) and K156 as well as PEBP1 P112 with 15LO A179, and PEBP1 S185 with 15LO1 D173 (Figure 1D). In the 15LO2/PEBP1 complex, PEBP1 residues D144-H145 and Y185-G186 interacted with L419 and Q425 on 15LO2.
Coarse-grained/molecular dynamics (GGMD) simulations revealed a strong tendency of PEBP1 and 15LO to form complexes (Figure S1C). 15LO1/PEBP1 complexes with both 15LO1 and (top left) and 15LO2 (top right) could rapidly (in <100 ns) bind the lipid bilayer. PEBP1 and 15LOs also displayed the intrinsic ability to form complexes in solution. 15LO1 formed complexes with PEBP1 within 90 ns in two different simulations (Figure 1E). Similarly, two runs showed a stable complex of 15LO2 with PEBP1 for extended durations (Figure S1D).
To assess the specificity of the PEBP1 interactions with 15LO, we generated several PEBP1 mutants in silico, based on the interfacial contacts in the PEBP1/15LO complex and selected the P112E mutation located in a conserved ligand-binding pocket, consisting of 16 residues, at the surface of the protein (Tavel et al., 2012). CGMD simulations revealed the disrupted “landing” of the P112E mutant (P112E) on 15LO1 (Figure 1E). In cases of successful docking, the binding sites did not coincide with the allosteric site (near R402) involved in optimized accommodation of AA-PE within the enzyme’s catalytic site, and the interface exhibited markedly weakened interactions with 15LO1 (Figure 1F) vs. wt PEBP1 (Figure S1E). Experimentally, Far-Western blotting revealed that the complex formation was strongly suppressed when P112E was used in place of wt PEBP1 (Figure 1A2). Similarly, significant cross-linking between wt PEBP1 and 15LO1 was markedly weakened for P112E (Figure 1B and S1A). Comparison of AA-PE oxidation showed a significantly higher potency of 15LO2 in the presence of wt PEBP1 vs P112E (Figure S1F). We also utilized a structurally distinct 15LO from Pseudomonas aeruginosa (LoxA) (Banthiya et al., 2016). Docking (Figure S1G) and CGMD simulations revealed aberrant binding of this bacterial enzyme onto human PEBP1 (Figure S1H). PEBP1-binding regions on P. aeruginosa did not overlap with the interfacial site in the human PEBP1/mammalian 15LO complexes that stimulated AA-PE oxidation.
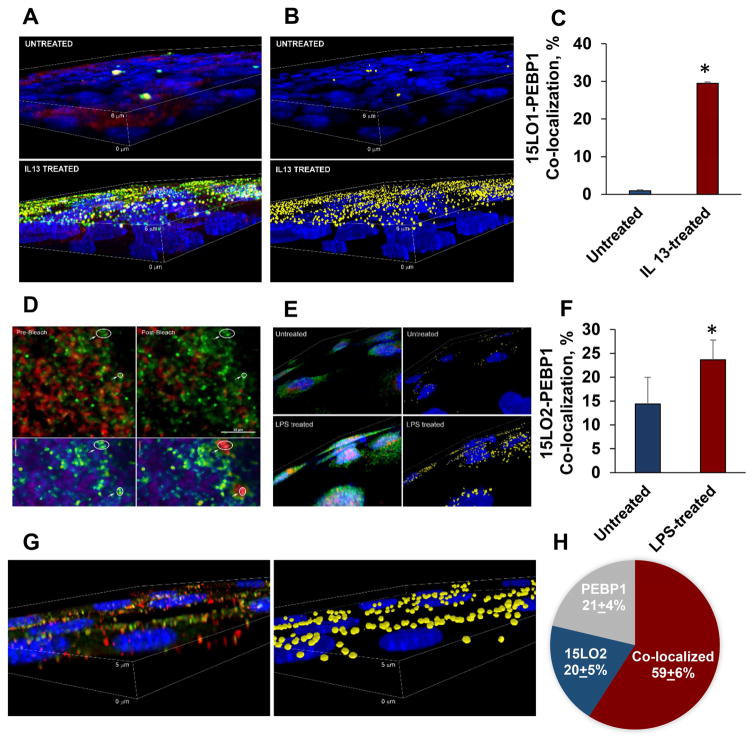
Next, we investigated whether PEBP1 and 15LOs are co-localized in multiple cell types using immunocytochemistry and confocal microscopy, followed by object based co-localization analysis. HAECs express predominantly 15LO1 induced by IL13, while HK2 kidney and HT22 neuronal cells express predominantly 15LO2 (Figure S2A, B). IL13 treatment increased 15LO1 expression (Figure 2A). While untreated HAECs had very few areas where 15LO1 and PEBP1 were co-localized, the number of puncta positive for both 15LO1 and PEBP1 was markedly increased after IL13 stimulation (Figure 2B, 2C and S2C). FRET methods confirmed that 15LO1 and PEBP1 were in very close proximity (within 10Å) in IL13 treated HAECs (Figure 2D). We also found that 15LO2 co-localized with PEBP1 in human kidney epithelial HK2 cells, and that the number of complexes was increased by pretreatment of cells with LPS (Figure 2E, 2F and S2D). Constitutively expressed PEBP1 and 15LO2 displayed co-localization in mouse hippocampal neuronal HT22 cells without additional treatments (Figure 2G and 2H).
Figure 2. PEBP1 co-localizes with15LO1 and 15LO2 in cells.
(A) 3D volume views showing the immuno-localization of PEBP1 (red) with 15LO1 (green) in HAECs (nuclei blue) in untreated (upper panel) and IL13 treated cells (lower panel).
(B) Objects identified as containing both PEBP1 and 15LO1 (yellow) were identified by 3D object-based segmentation and co-localization analysis.
(C) The number of co-localized objects for 15LO1 and PEBP1 expressed as a percentage of the total number of puncta in HAECs following 5 days of culture without or with IL13 (means±SD, *p < 0.05 vs. untreated.
(D) Fluorescence resonance energy transfer (FRET) based analysis showing the close physical proximity (within 10 Á) of 15LO1 with PEBP1 in HAEC (confirmed by proximity ligation assay). Upper panels: the donor (cy3) emissions are colored green and the acceptor (cy5, FRET) colored red. Lower panel: FRET ratio (donor/acceptor) which has been pseudo-colored (range 0–5, blue-red). FRET was confirmed by acceptor photobleaching (right hand panels) with an increase in the ratio following photobleaching of cy5 within the circled regions.
(E) Co-localization of PEBP1 (red) with 15LO2 (green) in HK2 cells exposed to LPS. The yellow shows the spots positive for both PEBP1 and 15LO2. In this example, there were 241 spots positive for both 15LO2 and PEBP1 in untreated cells, and 753 positive for both proteins in LPS treated cells.
(F) Number of co-localized objects positive for both 15LO2 and PEBP1 in untreated and LPS treated HK2 cells, expressed as a percentage of the total number of puncta (means±SD, *p< 0.05 vs. untreated, N=5/group).
(G) Co-localization of 15LO2 and PEBP1 in HT22 cells. Left panel: a 3D volume rendering of immunostaining for 15LO2 (green), and PEBP1 (red) with nuclei in blue.
(H) Percentage of objects identified as positive for 15LO2 alone, PEBP1 alone or both 15LO2 and PEBP1 together (means±SD, N=3/group). See also Figure S2.
Accumulation of Oxygenated PE (PEox) by PEBP1/15LO complexes
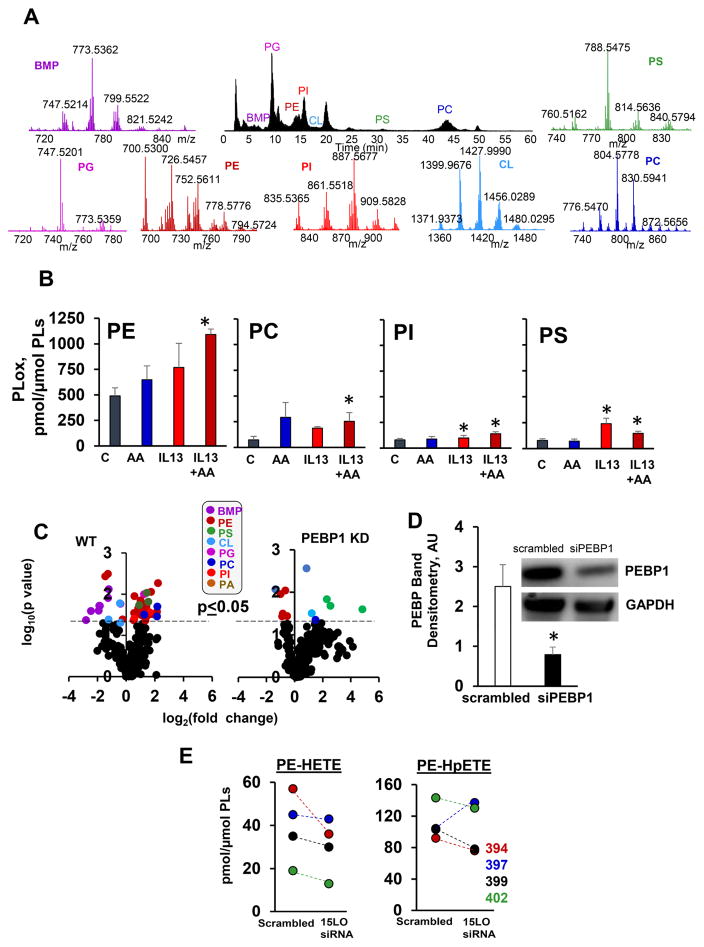
We further employed global redox phospholipidomics (Figure 3A) to explore whether the PEBP1/15LO complexes stimulate PE oxygenation in cells. In HAECs treated with IL13, PEox were the most abundant (both as the number of oxygenated species and their contents) PLs compared to the other classes of PLox (phosphatidylcholine (PC), phosphatidylserine (PS) and phosphatidylinositol (PI)) (Figure 3B). We identified 19 molecular species of PEox vs 3 species of PC, 3 species of PS and 2 species of PI in IL13 stimulated HAECs (Figure 3B, C). PEox species (but not PCox and PSox) were not detected in PEBP1 knockdown (KD) IL13 stimulated HAECs (Figure 3C, D). Similarly, 15LO1 KD in HAECs specifically decreased the content of PEox species in four HAEC cultures (each from a different donor) (Figure 3E). In primary human kidney epithelial cells (PHKC), a GPX4 inhibitor, RSL3, caused robustly increased levels of PEox (Figure S3A). A combination of RSL3 and LPS induced higher levels of PEox (Figure S3B).
Figure 3. 15LO1 catalyzes PEBP1-dependent production of PEox in IL13 stimulated HAECs.
(A) Normal phase LC/MS/MS chromatogram (black) and mass spectra of PLs in HAECs. BMP-bis-monoacylglycero-phosphate; PG-phosphatidylglycerol; PE-phosphatidylethanolamine; PS-phosphatidylserine; CL-cardiolipin; PC-phosphatidylcholine; PI-phosphatidylinositol.
(B) Contents of PLox in HAECs treated with IL13 and AA (means±SD. *p<0.05 vs. control, N=3/group).
(C) Volcano plots of IL13 induced changes of PEox in wt (left plot) and PEBP1 KD HAECs (right plot) (log2 (fold-change) vs. significance (log10 p-value). Cells were exposed to IL13 in the presence of AA.
(D) Quantitation of PEBP1 KD by siRNA in HAECs (means±SD, *p<0.05 scrambled vs. siPEBP1, N=3/group). Insert: A typical Western blot for PEBP1.
(E) 15LO1 KD changes HETE-PE and HpETE-PE in HAECs exposed to IL13 and AA, (N=4, different human donors). See also Figure S3 and S4.
PEBP1/15LO complexes contribute to ferroptosis
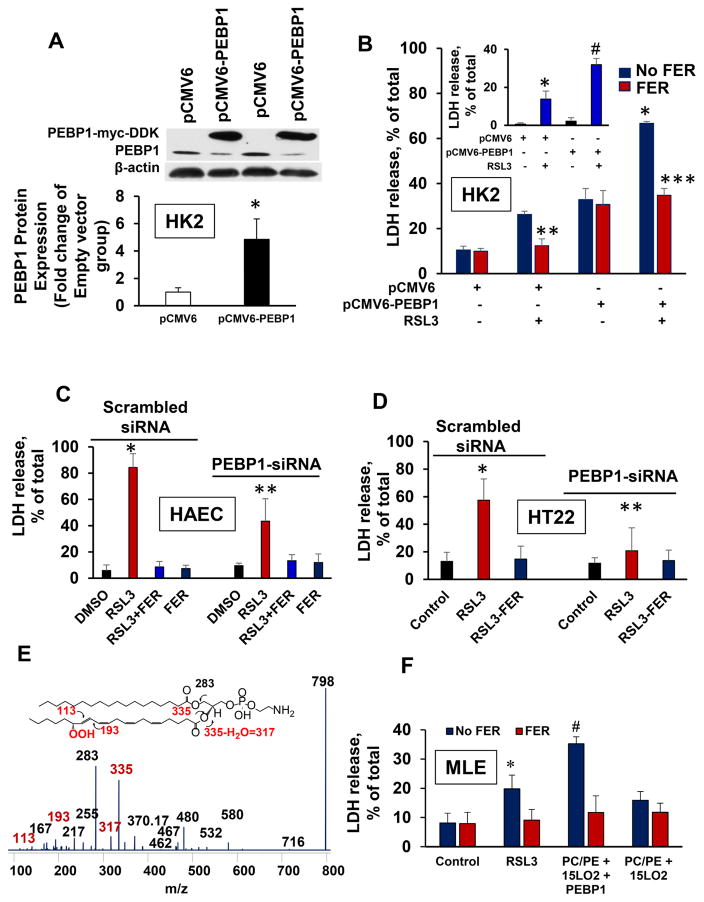
HAEC cells stimulated with IL13 (Figure S3C) as well as neuronal and kidney cells (HT22 (Figure S3D), PHKC (Figure S3E) and HK2 (Figure S3F)) express high levels of GPX4 that will reduce 15-HpETE-PEs generated by PEBP1/15LO complexes, hence prevent ferroptotic cell death. Therefore, we employed a GPX4 inhibitor, RSL3 (which chemically inactivates the enzyme and suppresses its expression), to block the enzymatic reduction of 15-HpETE-PE (Kagan et al., 2017; Skouta et al., 2014). RSL3 induced death of HAEC, PHKC, HK2 and HT22 cells (Figure S3) that was prevented by several prototypical inhibitors of ferroptosis, including ferrostatin-1 (Figure S3C–F). Similar results were obtained by assaying cell death by PI staining or MTT assay of cell survival (Figure S4). Inactivation of GPX4 by RSL3 in HAECs stimulated with IL13 (Figure S3G) or PHKCs (Figure S3A) caused a robust accumulation of different oxygenated PEs, including the pro-ferroptotic 15-HpETE-PEs (Figure S3H). In PHKCs, the effect of RSL3 was completely inhibited by ferrostatin-1 (Figure S3A). To further scrutinize PEBP1’s role in ferroptosis, we overexpressed PEBP1 (in HK2 cells, Figure 4A) or KD PEBP1 (in HAEC cells, Figure 3D and HT22cells, Figure S5A). Elevated levels of PEBP1 resulted in increased sensitivity of HK2 cells to RSL3 (Figure 4B) whereas lowered contents of PEBP1 in HAEC and HT22 cells were associated with decreased sensitivity to ferroptosis (Figure 4C, D).
Figure 4. RSL3-induced ferroptosis depends on endogenous or exogenously added PEBP1.
(A) Quantitation of PEBP1 (normalized to actin) in overexpressing (pCMV6-PEBP1) and empty vector (pCMV6) cells. Note, PEBP1-myc-FLAG and constitutive PEBP1 migrated differently due to the differences in molecular masses. Insert: Western blots show overexpressed PEBP1 (upper band, PEBP1-myc-FLAG) in transfcted HK2 cells.
(B) HK2 cells with elevated PEBP1 are more sensitive to ferroptosis. Insert: Ferrostatin-1-inhibitable cell death (means±SD, *p<0.05 vs. pCMV6, **p< 0.05 vs. pCMV6/RSL3, ***p< 0.05 vs pCMV6-PEBP1/RSL3, #p<0.05 vs. pCMV6/RSL3, N=3/group). Insert: quantitation of ferroptosis as differences in LDH in ±FER-treated cells.
(C) PEBP1 KD suppresses RSL3-induced ferroptosis in HAEC cells (means±SD, *p<0.05 RSL3 (wt) vs. DMSO (wt) **p<0.05 PEBP1 KD vs. RSL3 (wt), N=3/group).
(D) PEBP1 KD suppresses RSL3-induced ferroptosis in HT22 cells (means±SD, *p<0.05 vs. scrambled siRNA control, **p<0.05 vs. scrambled siRNA treated cells+RSL3, N=3/group).
(E) Composite MS3 spectrum of HpETE-PE with m/z 798.47 from supernatants after incubations of MLE cells exposed to AA-PE/15LO2/PEBP1. Fragments of HpETE-PE are shown in red. Insert: structural formula of OOH-AA-PE and fragments formed during MS2/MS3 analysis (red).
(F) PEBP1 enhances RSL3-induced ferroptosis in MLE cells incubated with exogenously added AA-PE/15LO2 (means±SD, *p<0.05 vs. control, #p< 0.05 vs. RSL3. N=3/group). See also Figure S5.
PEBP1 is required for ferroptosis induced by exogenous 15-LO2
Multiplicity of different regulatory pathways in which PEBP1 is engaged obscures its specific effects on 15LO dependent ferroptosis. To circumvent this, we employed exogenously added 15LO2 to generate the proximate ferroptotic signals, 15-HpETE-PE, from the precursor AA-PE in the presence and absence of PEBP1 and assessed ferroptosis in target mouse lung epithelial (MLE) cells pretreated with RSL3. LC/MS analysis of the supernatants demonstrated that 15-HpETE-PE was the most abundant AA-PE oxidation product (Figure 4E) whose formation was enhanced by PEBP1 from 15.6±0.3 to 19.0±0.6 pmol/nmol of cell PLs (p<0.05 vs untreated cell samples, N=3/group). Approximately 5% of thus generated 15-HpETE-PE was detected in cells. Ferroptosis (inhibitable by ferrostatin-1) was strongly enhanced by PEBP1 (Figure 4F and S5B) and was not observed in the absence of exogenous 15LO2. The pro-ferroptotic effect was specific to AA-PE (but not free AA) oxidation (data not shown). As an additional control for specificity we tested the effects of LoxA from P. aeruginosa that has a weak catalytic activity towards oxygenation of AA-PE and was not affected by human PEBP1 (Table S1). Human PEBP1 did not enhance ferroptotic response of MLE cells to P. aeruginosa LoxA, in stark contrast to its strong effect in combination with mammalian 15LO2 (Figure 4F and S5B). P112E that maintained its binding capacity towards Raf1 kinase but displayed markedly weakened interactions with 15LO, did not cause death of MLE cells in the presence of 15LO2, and AA-PE, in contrast to wt PEBP1 (Figure S5C). Docking of the PEBP1 with Raf1 kinase demonstrated that P112E mutation did not affect the interaction with the kinase binding domain at the N-terminal region near D448-D449 whereby G143-R146 residues of the scaffold protein were engaged in the interactions (Figure 1G) (Deiss et al., 2012). In the CG-MD simulations (Figure 1H), the complexes of Raf1 kinase were formed with both the wt and P112E and stably remained in the complex. In separate experiments, MLE cells transfected with P112E displayed a markedly decreased sensitivity to RSL3 pro-ferroptotic stimulation than cells transfected with wt PEBP1 (Figure S5D).
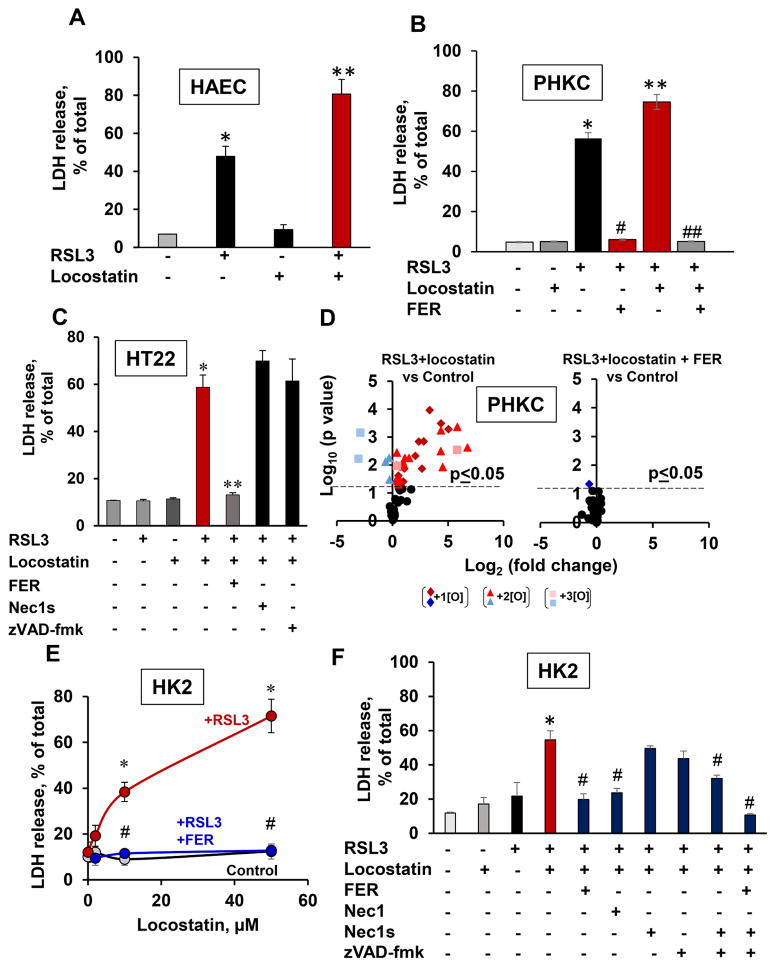
Synergistic stimulation of ferroptosis by locostatin
Disruption of PEBP1 interactions with Raf1 kinase would liberate the protein and make it available for binding 15LO. A small molecule, locostatin, covalently interacts with a highly conserved His86 in the PEBP1 ligand binding pocket and dissociates the scaffold protein from Raf1 kinase (Rudnitskaya et al., 2012), hence makes PEBP1 available for pro-ferroptotic interactions with 15LO. We selected low concentrations of RSL3 inducing mild ferroptosis and explored the combined effects of RSL3 plus locostatin (Figure 5). In HAECs (predominantly 15LO1), PHKC, HK2 and HT22 cells (predominantly 15LO2), locostatin induced synergistic cell death preventable by ferrostatin-1 (Figure 5). Accumulation of PEox species was also blocked by ferrostatin-1 (Figure 5D). Specific inhibitors of necroptosis (necrostatin-1s, but not necrostatin-1) and apoptosis (zVAD-fmk) were ineffective in preventing ferroptosis in HK2 and HT22 cells (Figure 5C, F).
Figure 5. Locostatin enhances RSL3-induced ferroptosis.
(A) HAECs (means±SD, *p<0.05 vs. no RSL3/no locostatin, **p<0.05 vs. locostatin, N=3/group)
(B) PHKCs (means±SD, *p<0.05 vs. no RSL3/no locostatin/no FER, **p<0.05 vs. RSL3, #p<0.05 vs. RSL3, ##p<0.05 vs. RSL3/locostatin, N=3/group).
(C) Ferrostatin-1 (but not zVAD-fmk or Necrostatin-1s) inhibits RSL3+locostatin induced ferroptosis in HT22 cells (means±SD, *p<0.05 vs. no RSL3/no locostatin/no FER, **p<0.05 vs. RSL3/locostatin, N=3/group).
(D) Ferrostatin-1 (FER) suppresses RSL3-induced PEox in PHKCs in the presence of locostatin. Volcano plot of PEox changes (log2 (fold-change) vs. significance (−log10 (p-value)), N=3/group.
(E) Locostatin enhances RSL3-induced ferroptosis in HK2 cells (means±SD, *p<0.05 vs. control (no RSL3, no FER, no locostatin), #p<0.05 vs. RSL3, N=3/group).
(F) Necrostatin-1s and zVAD-fmk do not suppress RSL3/locostatin induced death in HK2 cells (means±SD, *p<0.05 vs RSL3 only; #p<0.05 vs. RSL3 plus locostatin, N=3/group).
Decoding the mechanisms of PEBP1 regulation of 15LO
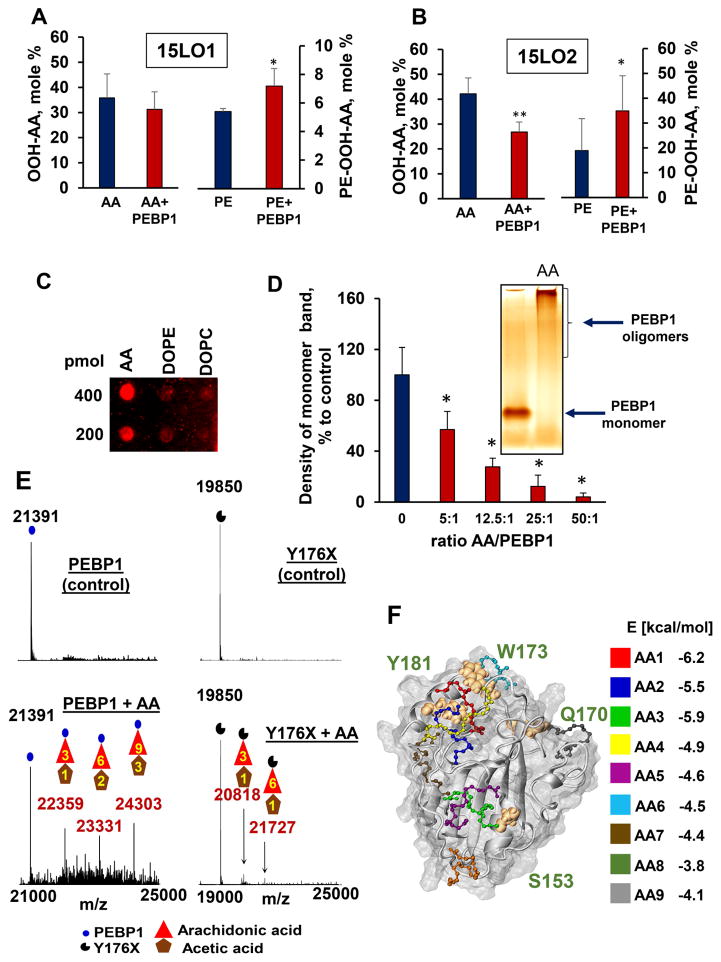
Further, we studied effects of PEBP1 on enzymatic oxidation of free AA vs AA-PE using porcine 15LO1, which has a high homology (86%) to human 15LO1, and recombinant human 15LO2. In the absence of PEBP1, 15LO1 exerted high activity towards free AA and very low or no detectable activity towards esterified AA-PE (Figure 6A). Upon addition of PEBP1, oxidation of AA-PE was markedly increased. Similarly, 15LO2 alone used AA almost exclusively as its substrate (Figure 6B). When a combination of 15LO2 plus PEBP1 was tested, significant oxidation of AA-PE occurred along with the oxidation of free AA. Measurements of kinetic characteristics of AA-PE oxidation vs free AA oxidation by 15LO1 and 15LO2 in the presence and absence of human PEBP1 for both enzymes, showed that the action of PEBP1 was unidirectional although the effects were more robust for 15LO2 than 15LO1 (Table S1). Vmax, Km, Kcat and Kcat/Km for free AA as a substrate in the absence of PEBP1 were in reasonable agreement with the previously published data for 15LO2 (Green et al., 2016). When PEBP1 was added to 15LO2, we found no significant changes in Vmax, a 1.7-fold increase in Km and a slight decrease of Kcat/Km for free AA. In contrast, when AA-PE was used as a substrate, a higher Vmax (~2.3-fold), a lower Km, and a 3.5 fold higher Kcat/Km was observed. For 15LO1 in the absence of PEBP1, Vmax was lower and Km higher for free AA and AA-PE compared to those for 15LO2. This may reflect the known higher substrate specificity of 15LO1 towards C18:2 PUFA vs C20:4 PUFA (Gata et al., 1996). In combination with PEBP1, 15LO1 had a 1.5-fold increased Vmax towards AA-PE without significant changes in Km and an increased Kcat/Km. Assessments of the specificity of these effects using LoxA from P. aeruginosa showed that human PEBP1 did not increase catalytic potency of AA or AA-PE oxygenation.
Figure 6. Binding of free AA by PEBP1 facilitates 15LO-catalyzed oxygenation of AA-PE.
PEBP1 stimulates accumulation of OOH-AA-PE (but not free AA-OOH) catalyzed by: (A) porcine 15LO1 and (B) human recombinant 15LO2 (means±SD, *p<0.05 vs. PE, ** p<0.05 vs. AA, N=4/group).
(C) A typical dot-blot illustrates binding of PEBP1 with free AA (3 separate experiments).
(D) AA causes concentration-dependent decrease of monomeric PEBP1 and increase of its oligomers (means±SD, *p<0.05 vs. PEBP1 without AA, N=4/group). Insert: a typical Blue native PAGE gel (silver-staining),
(E) Electrospray ionization mass spectrometry (ESI-MS) demonstrates AA binding by PEBP1. ESI-MS spectra before and after incubation with AA (left panels). PEBP1 mass was 21391 Da. AA + PEBP1 yielded 3 additional species with masses 22359, 23331 and 24303 Da corresponding to PEBP1 plus 3, 6 and 9 AA molecules (with 1, 2 and 3 acetic acid molecules), respectively. AA binding to the PEBP1 mutant Y176X (right panels). MS spectra of Y176X before and after incubation with AA. Mutant mass was 19850 Da. AA + Y176X mutant yielded 2 additional species, albeit at much lower levels, with masses 20818 and 21727 corresponding to Y176X plus 3 AA and 6AA ligands, respectively (plus 1 acetic acid molecule). Shown are typical spectra from at least four independent experiments.
(F) Computational docking shows multiple binding sites for AA on PEBP1. Up to nine AA molecules bind to PEBP1 with energies ranging from −6.2 to −4.0 kcal/mol. See also Figure S6 and Tables S1, S2 and S3.
PEBP1 binds free PUFA
In our assessments of the substrate specificity of 15LO1 towards free AA vs AA-PE, we noticed that PEBP1 binds free (but not esterified) AA. Assuming that this may lead to elimination of free AA as a substrate for 15LO, we determined binding of recombinant purified PEBP1 with phospholipids and AA using a dot-blot assay and found that only AA demonstrated marked binding (Figure 6C). Furthermore, on native PAGE gels, free AA interacted with PEBP1, caused its oligomerization and decreased the amounts of its monomeric forms (Figure 6D).
We further used mass spectrometry (MS) to assess whether complexes of PEBP1 with AA are formed and can be stable enough to be detectable in the gas phase (Hopper and Robinson, 2014). We detected complexes of PEBP1 with three, six and nine bound AA molecules (Figure 6E). Docking studies established that PEBP1 has multiple sites for binding free fatty acids (Figure 6F and S6A). Up to nine AA binding sites (labeled AA1–AA9) with relatively strong binding energies (from −6.2 to −3.8 kcal/mol) were identified (Figure 6F). MD simulations (two independent sets for each of the 9 bound poses) confirmed that at least seven of these sites stably retained the bound AA for over 200 ns (Figure S6B). A number of highly stable AA-binding poses (e.g. AA1 and AA6) involve the coordination of the AA by PEBP1 residues (G110–G116, Y181, Q183, D173) that participate in the interface with 15LO1 (Figure S6C). To evaluate the specificity of PEBP1 interactions with free AA, we computationally generated virtual PEBP1 mutants, tested their capacities of AA binding (Table S2) and selected a mutant with truncated C-terminal α-helix starting from Y176 (designated as Y176X) for further testing. Computationally, we showed that the mutant was able to bind up to 6 AA molecules (vs 9 in wt protein, Table S2, and Figure S6A) with markedly increased total binding energy vs. wt PEBP1. Binding of AA to Y176X was less favorable than to wt PEBP1 by a free energy difference (ΔΔG) of about 10 kcal/mol (Table S3). Direct MS evaluations demonstrated that deletion of the C-terminal fragment starting from Y176 greatly reduced AA binding (Figure 6E). Using AA-amide as a negative control, we found computationally that its binding to wt PEBP1 was considerably weaker than that of AA (data not shown). Accordingly, MS testing revealed the complete lack of AA-amide binding capacity by wt PEBP1.
To directly assess the ability of PEBP1 to bind AA in cells, we transfected MLE cells and HT22 cells with FLAG-tagged PEBP1 and confirmed its increased levels by Western blotting using anti-PEBP1 antibody (Figure S5E). Then, we immuno-precipitated the Flag-tagged PEBP1 using FLAG-tagged M2 agarose beads (Figure S5F). The presence of PEBP1 in the pulldown eluates was confirmed by Western blotting using anti-FLAG antibody. LC-MS analysis of the eluates revealed the presence of AA (Figure S5G). Quantitatively, the contents of AA in the immuno-precipitated complexes from transfected MLE and HT22 cells were 324±14 and 292±83 fmol/mg protein, respectively.
We then asked whether binding of free AA by PEBP1 and “depletion” of endogenous AA might act as a mechanism controlling the switch from AA to AA-PE oxidation for 15LOs. We measured the endogenous levels of free AA and AA-PE species as oxidation substrates by PEBP1/15LO complexes in HAEC cells with manipulated expression of PEBP1 (siRNA). We found that the cells with decreased levels of PEBP1 had higher contents of free AA (Figure S3I). In contrast, the esterified AA-PE levels were not affected by KD of PEBP1 (Figure S3I). These data are compatible with a possible role of PEBP1 binding in regulating the endogenous levels of free AA. In addition, LC/MS analysis, along with the ELISA estimates of PEBP1, showed that the ratio of free AA to PEBP1 was 4:1 (mol/mol) in HAEC cells. Based on our estimated stoichiometry of PEBP1/AA binding of 9:1 (Figure 6F), PEBP1 can deplete free AA as oxidation substrate. In contrast, the ratio of AA-PE to PEBP1 was 5,600:1 (mol/mol). The estimated amounts of both substrates in HAEC cells showed that the content of free AA constituted ~0.01–0.2 mol% of the levels of oxidizable AA-PE species as potential substrates for 15LO oxidation. Given that PEBP1 does not bind PE at physiological pH (Shemon et al., 2010), this suggests that AA-PE is the major oxidation substrate for PEBP1/15LO complexes in HAECs. In line with this, the levels of 15LO1-induced 15-HpETE-PE in wt HAEC cells were markedly higher than those of free 15-HpETE: 61.4 ± 4.3 and 1.6 ± 0.9 pmol/mg protein, respectively. Notably, PEBP1 KD, (to 68.7 ± 18.2%, p<0.04 vs. scrambled control, N=5), was accompanied by lowered levels of 15-HpETE-PE (down to 33.2 ± 8.8% of its content in wt cells). Based on the established kinetic parameters for 15LO1 (Table S1), we calculated that generation of the detected amounts of 15-HpETE-PE by 15LO1 in IL-13 stimulated HAECs will occur within ~190 min.
Computational modeling reveals PEBP1 induced cooperative motion allosterically favoring AA-PE binding by 15LO
Comparison of the structural dynamics of 15LO1 alone and in complex with PEBP1 (Figure S7) revealed a new mode of motion cooperatively gained upon complexation. In this energetically favorable mode predicted by the anisotropic network model (ANM) (Eyal et al., 2015) (Figure S6D), the complex undergoes a large bending movement between PEBP1 and 15LO1 affecting the surface properties of 15LO1 near the catalytic site and exposing new epitopes that can bind AA-PE molecules and promote the production of AA-PEox species. Analysis using the Gaussian network model (Figure S6E–G) showed that this movement divides the structure into two anticorrelated substructures (red and blue in Figure S6F). The anticorrelated substructures move together, but in opposite directions (Figure S6E). The first (blue) includes all 15LO1, except for its β-barrel domain and helix α2 which are integrated with PEBP1 to take part in the substructure 2 (red). 15LO1 helix α2 thus plays a pivotal role, not only by coupling the two proteins, but also lying at their interface, thus forming a hinge-bending site (Movies S1, S2).
To explore whether 15LO1 cooperative changes exposed new epitopes for binding AA-PE, we performed docking simulations. Simulations with PEBP1/15LO1 complex as rigid target (Figure S6H) indicated three hot spots (black circles/ellipses) where AA-PEs preferentially bound, none of which was sufficiently close to the catalytic site. Simulations with an ensemble of conformers along the newly gained movement (flexible docking) on the other hand yielded two additional sites, including one closely neighboring the catalytic site. Thus complexation with PEBP1 resulted in a gain of a new catalytic competence towards oxygenating PUFA-PE.
GPX4 regulates ferroptosis by PEox
Reduction of ferroptotic 15-HpETE-PEs by GPX4 can occur in the vicinity of the PEBP1/15LO complexes or at remote locations via the diffusion-controlled reduction. We used confocal microscopy and object based co-localization analysis and identified co-localization of all three proteins: PEBP1, 15LO1 and GPX4 in HAEC stimulated with IL13 (which was not observed in the absence of IL13 pre-treatment) (Figure S2E, F). These triple protein complexes were less abundant (Figure S2E) than the double combinations of PEBP1/15LO (Figure 2B). Similar to 15LO1, IL13 also induced GPX4 (Figure S2E, F). PEBP1 KD and, to a lesser degree, 15LO1 KD also reduced GPX4 levels, supporting an interdependency (Figure S2G). GPX4 KD decreased both 15LO1 and PEBP1 but possibly due to the expected increased cell death (Figure S2G).
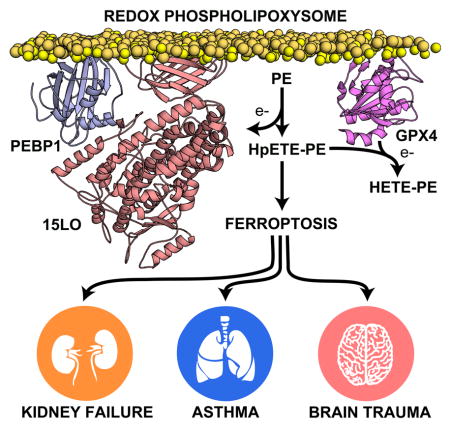
Overall, our data can be rationalized if we assume the existence of functional and structural arrangements – redox phospholipoxysomes – of five major components: three redox/regulatory proteins (15LO1-15LO2/PEBP1/GPX4) and two PL constituents (oxygenated PUFA-PEs and non-oxygenated PUFA-PEs). Notably, matrix-assisted laser desorption/ionization (MALDI-MS) imaging revealed that pulmonary airway epithelium, kidney tubular epithelium and rat brain cortex and hippocampus are enriched in AA-PE species (Figure S7) – substrates of enzymatic oxygenation reactions. These oxidizable PE species are not co-localized with oxidizable polyunsaturated PC species or non-oxidizable PE species. Similarly, LC/MS studies demonstrated high abundance of AA-PE species in neuronal cells (Figure S7).
Role of PEBP1/15LO Complexes in Disease
We further explored the presence and dysregulation of redox phospholipoxysomes with potential clinical manifestations of ferroptosis in: i) asthma, ii) AKI, and iii) traumatic injury of the brain.
Asthma
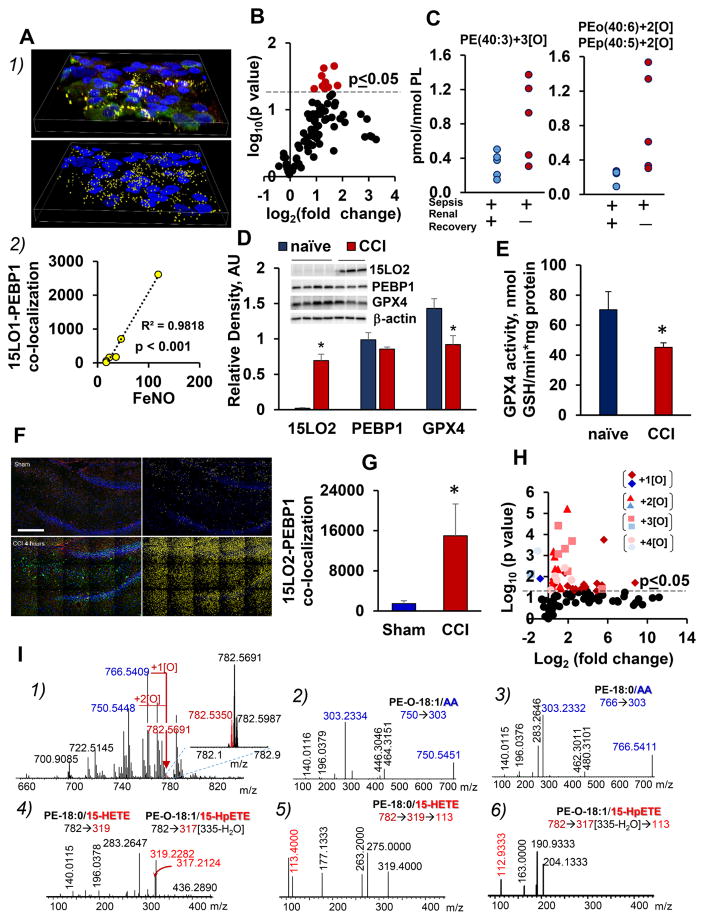
A type-2 (IL13) signature is known to exist in about 50% of asthma patients (Wenzel, 2012). Evaluating fresh HAECs obtained by bronchoscopic brushing of patients with controlled, non-exacerbating asthma, we revealed co-localization of PEBP1 with 15LO1 (Figure 7A1). We found a very strong correlation (r2=0.98) between the number of co-localized PEBP1/15LO1 puncta in the freshly brushed airway epithelial cells and the fractional exhaled nitric oxide (FeNO) (Dweik et al., 2011), in 5 patients and one healthy control case (Figure 7A2). A significant correlation of total 15LO1 (or PEBP1) with FeNO was not detected (p>0.15). Thus PEBP1/15LO1-driven AA-PE oxidation is important in the T2 airway inflammation associated with asthma.
Figure 7. PEBP1/15LO complexes and PEox are detectable in vivo in pro-inflammatory disease conditions.
(A) Increased co-localization of PEBP1/15LO1 correlates with asthmatic T2 inflammation in freshly brushed HAECs. 1) Fresh HAEC cytospin from a patient with asthma. Top panel: a volume view of the immunolocalization of 15LO1 (green), PEBP1 (red), and nuclei (blue). Lower panel: the complexes containing both PEBP1 and 15LO1 (yellow). 2) The number of objects identified as positive for both 15LO1 and PEBP1 correlates strongly with fraction of exhaled nitric oxide (FeNO) in human subjects (N=6, p<0.001)
(B) Increased contents of PEox in urine cell pellets from patients with acute kidney injury (AKI). Volcano plot of changes in PEox (log2 (fold-change) vs. significance (−log10 (p-value)) in urine pellet samples from patients with AKI (N=5/group).
(C) Scatter plots of PEox species from the plot in B (as indicated) in urine cell pellets from patients with AKI.
(D) Changes in ferroptotic protein expression in rat brain cortex after CCI. Note increased 15LO2 levels (*p<0.05; N=3–4/group), and decreased GPX4 levels (*p<0.05; N =3–4/group) at 4 h after CCI in ipsilateral cortex of injured vs. naïve rats. The amount of PEBP1 remained unchanged.
(E) Changes in GPX4 activity in rat brain cortex at 4 h after CCI (means±SD, *p<0.05 vs. naïve rats, N=3/group).
(F) Co-localization of PEBP1 and 15LO2 in brain tissue. Stitched image showing high resolution large area confocal scanning of 3x5 image fields. Left panels: the overlayed emissions for the immunolocalization of PEBP1 (red), 15LO2 (green) and nuclei (blue). Right panels: co-localization analysis for 15LO2 and PEBP1, with the number of spots having both proteins appearing yellow. Scale bar is 200 microns.
(G) Number of co-localized 15LO2 and PEBP1 in brain tissue 4 h after CCI (means±SD, *p<0.001 vs. sham, N=5/group).
(H) Volcano plot demonstrates changes in the content of PEox at 1h post CCI (N=4/group).
(I) Identification of pro-ferroptotic PEox in rat brain cortex after CCI using high resolution Orbitrap Fusion™ Lumos™ Tribrid™ Mass Spectrometer.
1) Full mass spectrum of PE (rat brain after CCI). Insert: Spectrum in the range of m/z from 782.1 to 782.9. Molecular ion with m/z 782.5350 (PEox) is shown in red. 2)-3) MS2 spectra of precursor ions with m/z 750.5451 and 766.5411 containing AA and corresponding to PE-O-18:1/20:4 and PE-18:0/20:4, respectively. 4) MS2 spectra of PEox with m/z 782.5350. Note 2 species with 1 and 2 oxygens formed after oxidation of PE(18:1/20:4) and PE(18:0/20:4) respectively. 5) and 6) Fragmentation patterns of ions with m/z 319 and m/z 317 (335-H2O) generated by MS3 analysis of PEox with m/z 782.5350. The fragment with m/z 113 is diagnostic of the OH- and OOH-groups at 15th carbon of AA. See also Figure S7 and S8.
Acute Kidney Injury
As ferroptosis of kidney epithelial cells may contribute to the severity of AKI and reduce chances for recovery, we characterized PL oxidation products in cell pellets from urine samples obtained from patients enrolled in the Biologic Markers of Recovery for the Kidney (BioMaRK) study (Network et al., 2008). Using patients (5/group) from four groups (AKI ± recovery ± sepsis), we found that PEox characteristic of ferroptosis were detectable in samples from patients obtained at the time of initiation of dialysis (Figure 7B). PEox were increased in AKI patients who did not recover renal function (defined as persistent dialysis requirement) vs. patients who recovered renal function regardless of the etiology of AKI (sepsis or not) (Figure 7C).
We further employed folic acid (FA) induced kidney injury model leading to ferroptosis preventable by ferrostatin-1 (Martin-Sanchez et al., 2017). TUNEL staining (Figure S8A) showed that FA induced cell death was suppressed by treatment with an anti-ferroptotic agent, ferrostatin-1 (Figure S8A, B). LC/MS revealed higher levels of doubly-oxygenated AA-PE species in kidneys of FA-exposed mice (Figure S8C, D). In ferrostatin-1 treated animals, the contents of these products were not different from the control mice.
Traumatic brain injury
We utilized a pediatric rat controlled cortical impact (CCI) model to detect changes in co-localization of pro-ferroptotic proteins and oxygenated AA-PEs in sham vs. injured animals. At early time points after CCI (1–4 h), ipsilateral cortical expression of 15LO2 was elevated vs naïve animals (Figure 7D) whereas GPX4 levels and the enzymatic activity (Figure 7E and S8E) were decreased. Similar protein profiles were observed in ipsilateral hippocampus after CCI (Figure S8F). High-resolution large area confocal immuno-fluorescence microscopy assessments of PEBP1/15LO2 co-localizations (Figure 7F, G) demonstrated a remarkably higher abundance of co-localized puncta in the injured brain compared to the brain from sham animals. We detected a significant increase in PEox – including 15-HETE-PE and 15-HpETE-PE - in contusional cortex as early as 1h after CCI vs. sham-operated controls (Figure 7H, I).
Discussion
Harmonized multicellular life relies on sophisticated signaling by free oxygenated PUFA (Dennis and Norris, 2015). While historically these free oxygenated PUFA were associated with many signaling functions, lately it has become obvious that a more diversified group of PLox represent a much richer signaling language albeit with a poorly understood role. It has been shown that AA-PEox are required for normal functions of many types of leukocytes (Aldrovandi et al., 2017; Morgan et al., 2009), are meaningful signals in phagocytosis of apoptotic cells (Uderhardt et al., 2012) and represent crucial death signals in ferroptosis (Kagan et al., 2017).
As PUFA-PLs make up the most abundant structural membrane components, oxygenation of AA-PE requires a highly selective mechanism. The reverse hexagonal phase arrangements of PEs – less ordered than the bilayer membrane regions – are likely the preferred areas of 15LO-driven oxygenation reactions (Kagan et al., 2017). Here we discovered that a redox phospholipoxysome, comprising PUFA-PE and PUFA-PEox as the substrates and products of oxygenation reactions as well as catalytic/regulatory proteins, 15LO, PEBP1 and GPX4, constitutes a redox assembly regulating ferroptosis. This regulatory mechanism operates with both isoforms, 15LO1 and 15LO2, that may respond differently to alternate stimulatory and suppressive signals. The complex generates 15-HpETE-PEs, which – if not controlled by GPX4- – act as proximal ferroptosis signals.
The role of ferroptosis as an important pathogenic factor has emerged in several experimental models of disease. While necroptosis has been considered as the common pathway in ischemia-reperfusion injury (Linkermann et al., 2013), recent studies discovered that ferroptotic death may represent the major mechanism of injury in several tissues (eg, kidney (Friedmann Angeli et al., 2014; Linkermann et al., 2014; Martin-Sanchez et al., 2017) and liver (Friedmann Angeli et al., 2014)). However, the contribution of ferroptosis to human disease, although actively discussed, is less clear. Our new mechanistic understanding of the essential interactions between the components the redox phospholipoxysome may provide a valuable connection with the pathogenesis of human disease.
Recent targeted biologic interventions confirm the importance of T2 inflammation with IL4/IL13-driven increased 15LO1 expression and PEBP1 binding in vivo to asthma exacerbations (Zhao et al., 2011). Thus, asthmatic airway epithelial cells generating high 15HpETE-PE levels are poised to undergo cell death. However, reflecting the in vitro relationships in IL13-cultured HAECs, freshly brushed HAECs from stable, non-exacerbating asthmatic patients show upregulation of both 15LO1 and a high degree of co-localization of 15LO1 with PEBP1. This co-localization in asthma is strongly correlated with the fraction of exhaled NO (FeNO), a marker of T2 inflammation (Wenzel, 2012), suggesting that in the “steady state” of T2-High asthma, high 15-HETE-PE generation is mitigated by the peroxidase activity of GPX4. In this setting any reduction in GPX4 activity through increased oxidative stress, as seen in asthma exacerbations, cigarette smoking or infection, could substantially dysregulate the structural and functional balance of the redox phospholipoxysome and contribute to epithelial dysfunction, death and loss of asthma control.
In severe AKI, ferroptotic PEox were increased in the urine pellets from patients who did not recover renal function compared to those who did. These results were consistent regardless of whether the inciting event for AKI was sepsis or some other insult. Nearly 75% of patients developing AKI in the setting of septic shock already have evidence of AKI at presentation and non-recovery from this AKI was associated with a 3-fold increase in the odds of death by 1-year (Kellum et al., 2017). Ferroptosis may therefore characterize the phenotype of non-recovery after AKI. This phenotype may not be fully established for several days following injury — late recovery and stuttering course are common in AKI patients (Kellum et al., 2017), suggesting that there is potential window of several days where strategies to limit ferroptosis can be deployed.
Sufficient GSH and high levels of GPX4 are essential for the maintenance of redox neuro-homeostasis (Bayir et al., 2002; Ji et al., 2012) and control of 15-HpETE-PE. GSH depletion caused by exposure of neurons to high concentrations of glutamate generates cytotoxic responses that are similar to ferroptotic death. The role of ferroptosis in acute brain injury in vivo has not been investigated so far. Our demonstration of the high levels of 15-HpETE-PE in the brain cortex and hippocampus after TBI along with increased expression of 15LO2 and decreased levels of GPX4 strongly suggest the possibility of ferroptotic death.
The structure and function of the redox phospholipoxysome, coupled with the emerging data on its importance to disease states, indeed opens up multiple potential opportunities to inhibit ferroptosis. Drugs, which enhance GPX4, given perhaps at the onset of brain or kidney injury, or at the initiation of an asthma exacerbation could prevent ferroptosis. As these studies suggest, inhibiting the ability of PEBP1/15LO complexes to form 15-HpETE-PE could lead to novel anti-ferroptotic approaches.
STAR Methods text
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed and will be fulfilled by the Lead Contact (kagan@pitt.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Subjects
Acute Kidney Injury Urinary Cell Pellets
The urine pellets were obtained from 20 subjects enrolled into an IRB approved study- BioMaRK. The median age of the subjects was 55 years (IQR of 45-75), 4 of 20 were women. All samples were de-identified prior analysis. The BioMaRK study was a multicenter, prospective, nested, observational cohort study conducted as an ancillary study to a large multicenter, randomized clinical trial ((ATN study (NCT00076219), n=1124) comparing intensive and less-intensive dialysis strategies in critically ill patients. Details of the trial are described in detail elsewhere (Network et al., 2008). Patients with chronic kidney disease (defined as premorbid serum creatinine >2 mg/dL in men and >1.5 mg/dL in women) or prior kidney transplantation were excluded. We obtained approval from the institutional review boards of the University of Pittsburgh and all other participating sites for sample and data collection. Fresh urine samples were obtained on day 1 of dialysis. A well-mixed, 30 ml sample was obtained from the Foley catheter. A protease inhibitor cocktail tablet (Roche Diagnostics Corporation, IN) was added to the urine specimen immediately on collection. Urine samples were then poured into 50 ml conical centrifuge tubes, centrifuged for 5 minutes at 1000 g at 4°C, and the supernatants were separated from the pellets and frozen separately at −80 °C until shipped to the central laboratory. Samples were then stored at −80 °C until analyzed.
Bronchoscopically obtained airway epithelial brushings
All human samples were obtained from human volunteers who provided written informed consent for participation in studies of inflammatory processes in asthma. All studies were approved by the Institutional Review Board (IRB) of the University of Pittsburgh. Epithelial brushings were obtained via bronchoscopy from 7 subjects (six asthmatics, one healthy control). The median age for the asthmatics was 33 years (IQR 26-45), 5 of 6 were women. The healthy control subject was a 20 year old woman. Cells were dispersed in saline prior to confocal immunofluorescent microscopy studies of 15LO1 and PEBP1 as outlined below.
Distribution and availability of patient-derived primary cells and samples
The urinary pellets and airway epithelial cells used for this work are depleted. Primary human kidney proximal tubule epithelial cells (see below) used for this work are a limited resource. Investigators interested in their use should contact the Lead Contact and a Material Transfer Agreement (MTA) put in place as per University of Pittsburgh policies.
Animals
Controlled cortical impact model
All experiments were approved by the Institutional Animal Care and Use Committee at the University of Pittsburgh. Male postnatal day (PND) 10 Sprague–Dawley rats were purchased with lactating mothers from Envigo (Indianapolis, IN). The rats were housed for 1 week before the initiation of the experiments. The rats were fed standard rat chow and provided water ad libitum. Ambient temperature was controlled at 20–22°C, and lighting was on a 12-h/12-h cycle. The rats were randomly assigned to an established controlled cortical impact (CCI) model of TBI (Ji et al., 2012)or sham. Anesthesia was induced with 3.5 % isoflurane and maintained with 2 % isoflurane with N2O/O2 (2:1) via nose cone. Rats were placed in a stereotaxic frame and temperature maintained at 37 °C. The bone overlying the left parietal cortex was removed using a high-speed dental drill. A vertically-directed CCI was delivered using a flat 6 mm pneumatically-driven impactor tip (4.0 ± 0.2 m/s, 50 ms dwell time, 2.5 mm depth). After injury, the bone flap was replaced and sealed with dental cement, and the scalp incision was closed. Anesthesia was discontinued and rats were monitored with supplemental O2 for 1 h. Sham animals underwent identical anesthesia and preparation without injury. Rats were sacrificed at 1 or 4 h post-injury, brains were perfused with heparinized saline, the contusional cortex and hippocampus were dissected, and samples were snap-frozen in liquid nitrogen for western blotting or lipid extraction. For immunohistochemistry experiments, animals were sacrificed at 24 h post-injury. Brains were perfused with heparinized saline followed by perfusion with 2 % paraformaldehyde, immersion-fixed for 2 h, cryoprotected in 30 % sucrose, then frozen in dry ice-cooled methylbutane. Coronal brain sections (10 μm) through dorsal hippocampus were cut on a freezing sliding microtome.
Folic Acid AKI model
All experiments were approved by the Institutional Animal Care and Use Committee at the University of Pittsburgh. Male 20 weeks old CD-1 mice were obtained from Charles River Laboratories. The mice were housed in a temperature (20–22°C) and light (on a 12-h/12-h cycle) controlled environment for 1 week before the commencement of the experiments. The mice were given standard rodent chow and had access to water ad libitum. The mice were randomly assigned (using random number generator in Excel) to receive 5 mg/kg Fer-1 (Abcam, cat #ab146169) or 1.5% DMSO (vehicle) 60 minutes before injection of folic acid (Sigma-Aldrich) of 250 mg/kg in 0.3 mol/L sodium bicarbonate intraperitoneally (Wen et al., 2012). Mice were euthanized 48 h later. The doses and 48 h time point were on the basis of prior report, which showed prevention of renal injury by Fer-1 in biochemical and histological assessments (Martin-Sanchez et al., 2017). Kidneys were perfused in situ with cold saline before removal. One kidney was snap frozen in liquid nitrogen for redox lipidomics studies, and the other was fixed and paraffin embedded. The kidneys were cut into 3 micron parallel sections perpendicular to the longitudinal axis for detection of Terminal deoxynucleotidyl transferase (TdT)-mediated dUTP nick end labeling (TUNEL) positive cells using the ApopTag Peroxidase In Situ Apoptosis Detection Kit according to the manufacturer’s instructions (S7100; MerckMillipore).
Tissue for MALDI-MS Imaging
All procedures were pre-approved and performed according to the protocols approved by the Institutional Animal Care and Use Committee of the University of Pittsburgh. Lungs (left lobe) from adult male C57B6 mice, and brains and kidneys from male PND 17 Sprague–Dawley rats (Envigo, Indianapolis, IN) were harvested and prepared for MALDI-MS imaging analysis. Most conventional histological methods for tissue preparation are not compatible with lipid MALDI-MS of tissue sections, therefore modified techniques were used. Gelatin (porcine skin - approximately 300 bloom, Sigma) was prepared as an 8 % solution in Hank’s buffered saline solution (HBSS) without calcium or magnesium (ThermoFisher Scientific). Mouse lungs were inflated with the gelatin solution, harvested, frozen in liquid N2, placed in cryostat molds containing more gelatin solution, cooled on ice to solidify and the molds were frozen at −20 °C for sectioning. Rat kidneys were harvested, frozen in liquid N2 and embedded in gelatin as above. Rat brains were harvested and frozen in liquid N2 without embedding. The frozen tissue and embedded blocks were attached to cryotome stages with minimal Tissue-Tek OCT (Sakura FineTek USA). The blade and working surfaces of the cryotome were cleaned with methanol immediately prior to all cutting, and at no time did the blade come into contact with the OCT. Sections from lung (−19 °C, 7 μm), kidney and brain (both −20 °C, 10 μm) were applied to special glass slides coated with Indium-Tin Oxide (ITO, Bruker Daltonics). Semi-serial sections were applied to plain glass slides for conventional Hematoxylin and Eosin (H&E) Histology (Harris Hematoxylin, Eosin Y, and Bluing Reagent all from ThermoFisher Scientific) to determine cutting depth. H&E-stained sections were scanned and montaged using a Nikon 90i upright microscope using Nikon Elements software with 4X and 10X dry plan apochromat objectives and a motorized stage.
Primary Cell Cultures
Primary Human Airway Epithelial Cell Culture in Air–Liquid Interface
HAECs were obtained by bronchoscopic brushing of 27 asthmatic and healthy control airways. The median age of the participants was 39 years (IQR of 20-49). Seventy-five percent of participants were women. HAECs were cultured in air–liquid interface (ALI) under serum-free condition as previously described (Zhao et al., 2011). Briefly, fresh bronchoscopic brushing primary HAECs were cultured under immersed condition for proliferation. When 80–90% confluent, cells are trypsinized and plated on Transwell plate for submerged stage culture with 200 μl culture medium in upper insert and 1000 μl culture medium in lower chamber. When cells reached 100% confluence, cells went into ALI culture by reducing the upper volume to 50 μl. Cells were stimulated with IL13 (10 ng/ml) under ALI culture for 7 days supplemented with 7.2 μM AA unless specified otherwise.
Primary human kidney proximal tubule epithelial cells
All reagents for cell culture were from Life Technologies unless mentioned otherwise. Proximal tubule cells were procured, isolated, and characterized from cortices of whole adult human kidneys obtained from the Center for Organ Recovery and Education (CORE, Pittsburgh, PA, USA) through a University of Pittsburgh approved protocol as previously described (Emlet et al., 2017). Renal cortices from four male donors with a median age of 54 years (IQR of 44-64) were subjected to enzymatic and mechanical dissociation by razor blade mincing and digestion with 200 u/ml Collagenase IV, and 100 u/ml DNAse in HBSS for 1 h at 37 °C, followed by sieving with a 250 μm sieve (Gilson, Lewis Center, OH, USA) to remove undigested material. The resultant slurry was cultured in DMEM/F12 with the addition of 5 % Fetal Bovine Serum, Insulin, Transferrin, Selenium, Glutamax, and Penicillin-Streptomycin in 150 cm2 flasks coated with 5 μg/cm2 Rat Tail collagen-1 at 37 °C, 5 % CO2, to generate heterogeneous cell stocks. Primary proximal tubule epithelial cells were isolated from the heterogeneous cell pools using immunoaffinity isolation via the Dynal Pan-Mouse IgG magnetic bead system (Thermo-Fisher Scientific) per the manufacturer’s instructions, using an antibody directed against Aminopeptidase N (CD13, Cat. #555691, BD Biosciences). Isolated cells were characterized as proximal tubule cells using a panel of antibodies directed against three proximal tubule markers (Gamma Glutamyl-transpeptidase, 1:20000, Santa Cruz Biotechnology (SCB), Dallas, TX, USA; Aquaporin-1 (1:250, SCB); Sodium-Hydrogen exchanger 3 (1:500, Thermo Scientific, Rockford, IL, USA), one distal tubule marker (E-Cadherin (1:20000, BD Biosciences), and two general tubule markers to identify that the cells were functional (Na+-K+-ATPase, 1:1000, Alpha 1 subunit, Abcam; Vacuolar H+-ATPase, E subunit, 1:500, Sigma-Aldrich, Saint Louis, MO, USA). For all experimentation, immunoaffinity isolated cells from passages 2–6 were used, and each passage was characterized for consistency.
Cell Lines
HT22 cells
Ht22 cells were a generous gift from Dr. David Schubert (The Salk Institute, La Jolla, CA). These cells were maintained and differentiated in Dulbecco’s modified Eagle’s medium (DMEM) supplied with 10% FBS.
HK2 cells
All reagents for cell culture were from Life Technologies unless mentioned otherwise. HK2 (from American Type Culture Collection (ATCC)) and isolated primary cells were propagated in DMEM/F12 with the addition of 5 % Fetal Bovine Serum, 10 μg/ml Insulin, 6.7 ng/ml Selenium, 5.5 μg/ml Transferrin, 2 mM Glutamax, and 10,000 u/ml Pennicillin-Streptomycin at 37 °C and 5 % CO2. During experimentation, cells were cultured in the similar media as described for propagation except with the addition of 2 % Fetal Bovine Serum, 10 ng/ml EGF, 40 ng/ml Hydrocortisone (Sigma-Aldrich), and 4 pg/ml Triodothyronine (Sigma).
MLE-12 cells
The murine lung epithelial cell line MLE-12 purchased from ATCC, was cultured at 37 °C and 5 % CO 2 in DMEM/F-12 (1:1) medium supplemented with 0.005 mg/ml insulin, 0.01 mg/ml transferrin, 30 nM sodium selenite (Life Technologies, Grand Island, NY, USA), 10 nM hydrocortisone, 10 nM beta-estradiol (Sigma-Aldrich, Saint Louis, MO), 2 mM L-glutamine, 10 mM HEPES, and 10% FBS (Life Technologies, Grand Island, NY).
METHOD DETAILS
Protein Expression and Purification
PEBP1 expression and purification
Full-length human PEBP1 and related mutants were cloned into a pET21-derived (EMD Millipore, Billerica, MA) bacterial expression plasmid modified to express PEBP1 with N-terminal His10- and mRuby2 tags (Lam et al., 2012). All PEBP1 constructs were cloned into the modified pET21-mRuby2 vector by Gibson Assembly (New England Biolabs, Billerica, MA) using primers with homology at the upstream (sense) NdeI site (5′-GGTCTGAGGGGATACACTCATATG-3′) and downstream (antisense) EcoRI site (5′-GCTTGTCGACGGAGCTCGAATTC-3′) of the vector. Mutations were prepared using the primers and complimentary internal primers (sense and antisense) specific to each mutation as listed in Key Resource Table. All primers were ordered from Life Technologies (Carlsbad, CA). Clones were evaluated by diagnostic restriction digest and sequencing (Genewiz, South Plainfield, NJ). Protein expression was performed in the Escherichia coli strain BL21 (DE3) Codon+ (Agilent, Santa Clara, CA). Following initial growth to OD600 of 0.6, cells were induced using 0.2 mM isopropylthio-beta-galactosidase (IPTG) and cultured overnight at room temperature. Cells were harvested by centrifugation and lysed in buffer containing 20 mM Tris (pH 8.0), 500 mM NaCl, 5 % glycerol, 5 mM imidazole, 1 mM β-mercaptoethanol, and protease inhibitors. Lysates were cleared by centrifugation at 29,000 G. Protein was purified by nickel affinity chromatography (Qiagen, Hilden, Germany) and the N-terminal His10-mRuby2 tag was subsequently removed by overnight digestion with TEV protease. After digestion, PEBP1 was isolated via a second round of nickel affinity chromatography. The protein was then dialyzed overnight at 4 °C into buffer containing 20mM sodium acetate (pH 5.5) before cation exchange chromatography was performed in sodium acetate (pH 5.5) using a HiTrap SP column (GE Healthcare, Little Chalfont, UK). The resulting fractions were further refined by size exclusion chromatography in 20 mM HEPES (pH 8.0), 500 mM NaCl buffer using a Sephacryl S-200 column (GE Healthcare). Before performing experiments, PEBP1 was desalted into 5 mM Bis-Tris (pH 6.5), 25 mM NaCl as described previously.
KEY RESOURCE TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Horseradish peroxidase (HRP)-conjugated anti-rabbit IgG H&L | ThermoFisher Scientific | Cat#65-6120 |
| Anti-Rabbit IgG (whole molecule)–Peroxidase antibody produced in goat | Sigma-Aldrich | Cat#A0545 |
| Monoclonal Anti-β-Actin antibody produced in mouse AC-74, monoclonal, from purified immunoglobulin | Sigma-Aldrich | Cat#A2228 |
| Monoclonal Anti-β-Actin Peroxidase antibody produced in mouse AC-15, monoclonal, from purified immunoglobulin | ThermoFisher Scientific | Cat#A3854 |
| Anti-Mouse IgG (whole molecule)–Peroxidase antibody produced in rabbit | Sigma-Aldrich | Cat#A9044 |
| Anti-Glutathione Peroxidase 4 antibody [EPNCIR144] | Abcam | Cat#ab125066 |
| RKIP (PEBP1) Antibody (FL-187), rabbit polyclonal IgG | Santa Cruz Biotechnology | Cat#sc-28837 |
| 15-LO2 Antibody (M-50), rabbit polyclonal IgG | Santa Cruz Biotechnology | Cat#sc-67143 |
| Rabbit Polyclonal to Human ALOX15 (aa539-588) | Life Span Biosciences | Cat#LS-111783 |
| PEBP1 (RKIP) antibody, mouse monoclonal IgG1 | Santa Cruz Biotechnology | Cat#sc-376925 |
| GPx4 antibody, goat polyclonal | Abcam | Cat#51944 |
| 15-LO1 antibody | Dr. Doug Conrad, University of California, San Diego | n/a |
| Aminopeptidase N (CD13) | BD Biosciences | Cat#555691 |
| Aquaporin-1 | Santa Cruz Biotechnology | Cat# sc-25287 |
| E-Cadherin | BD Biosciences | Cat# 610181 |
| Gamma Glutamyl-transpeptidase | Santa Cruz Biotechnology | Cat# sc-166908 |
| Na+-K+-ATPase Alpha 1 subunit | Abcam | Cat# ab7671 |
| Sodium-Hydrogen exchanger 3 | Thermo-Fisher Scientific | Cat# MA1-46355 |
| Vacuolar H+-ATPase, E subunit | Sigma-Aldrich | Cat# GW22284F |
| Alexa Fluor 488 donkey anti-goat IgG (H+L) | Thermo-Fisher Scietific | Cat#A-11055 |
| Cy3 AffiniPure donkey anti-mouse IgG (H+L) | Jackson Immuno | Cat#715-165-151 |
| Cy5 AffiniPure donkey anti-rabbit IgG (H+L) | Jackson Immuno | Cat#711-175-152 |
| Monoclonal Anti-Flag M2 Antibody (Mouse) | Sigma-Aldrich | Cat#F-3165 |
| HRP conjugated donkey anti-mouse IgG (H+L) | Jackson Immuno | Cat# 715-035-151 |
| Peroxidase AffiniPure Donkey Anti-Chicken IgY (IgG) (H+L) | Jackson Immuno | Cat# 715-035-155 |
| Peroxidase AffiniPure Donkey Anti-rabbit IgG (IgG) (H+L) | Jackson Immuno | Cat# 715-035-152 |
| Bacterial and Virus Strains | ||
| Escherichia coli strain BL21 (DE3) Codon+ | Aligent | argU (AGA, AGG), ileY (AUA), leuW (CUA) |
| Biological Samples | ||
| Kidney cortex from adult humans | Center for Organ Recovery and Education | n/a |
| Airway epithelial cells from adult humans | University of Pittsburgh Asthma Institute | n/a |
| Epithelial brushings obtained by bronchoscopy from adult humans | University of Pittsburgh Asthma Institute | n/a |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 1-(10Z-heptadecenoyl)-sn-glycero-3-phospho-(1′-myo-inositol) (ammonium | Avanti Polar Lipids | Cat#110718 |
| 1-(10Z-heptadecenoyl)-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) | Avanti Polar Lipids | Cat#110712 |
| 1-(10Z-heptadecenoyl)-sn-glycero-3-phosphocholine | Avanti Polar Lipids | Cat#110905 |
| 1-(10Z-heptadecenoyl)-sn-glycero-3-phosphoethanolamine | Avanti Polar Lipids | Cat#110699 |
| 1-(10Z-heptadecenoyl)-sn-glycero-3-phospho-L-serine (sodium salt) | Avanti Polar Lipids | Cat#110724 |
| 1,2-dioleoyl-sn-glycero-3-phosphocholine | Avanti Polar Lipids | Cat#850375 |
| 10x Tris Buffered Saline (TBS) | Bio-Rad | Cat#1706435 |
| 10x Tris/Glycine Buffer for Western Blots and Native Gels | Bio-Rad | Cat#1610771 |
| 10x Tris/Glycine/SDS | Bio-Rad | Cat#1610772 |
| 15(S)-HpETE | Cayman Chemical | Cat#44720 |
| 15-Lipoxygenase-2 (human recombinant) | Cayman Chemical | Cat#10011263 |
| 1-heptadecanoyl-sn-glycero-3-phosphate (sodium salt) | Avanti Polar Lipids | Cat#110679 |
| 1-hexadecanoyl(d31)-2-(9Z-octadecenoyl)-sn-glycero-3-[phospho-L-serine] (sodium salt) | Avanti Polar Lipids | Cat#110922 |
| 1-hexadecanoyl(d31)-2-(9Z-octadecenoyl)-sn-glycero-3-[phospho-rac-(1-glycerol)] (sodium salt) | Avanti Polar Lipids | Cat#110919 |
| 1-hexadecanoyl(d31)-2-(9Z-octadecenoyl)-sn-glycero-3-phosphocholine | Avanti Polar Lipids | Cat#110918 |
| 1-hexadecanoyl(d31)-2-(9Z-octadecenoyl)-sn-glycero-3-phosphoethanolamine | Avanti Polar Lipids | Cat#110921 |
| 1-hexadecanoyl(d31)-2-(9Z-octadecenoyl)-sn-glycero-3-phospho-myo-inositol (ammonium salt) | Avanti Polar Lipids | Cat#110923 |
| 1-stearoyl-2-arachidonoyl-sn-glycero-3-phosphocholine | Avanti Polar Lipids | Cat#850469 |
| 1-stearoyl-2-arachidonoyl-sn-glycero-3-phosphoethanolamine | Avanti Polar Lipids | Cat#850804 |
| 2,5-dihydroxybenzoic acid (DHB) | Sigma-Aldrich | Cat#149357 |
| 2-Mercaptoethanol | Sigma-Aldrich | Cat#M6250 |
| 2-Methylbutane (Certified ACS), Fisher Chemical | Fisher Scientific | Cat#O3551-4 |
| 2-Propanol | Fisher Chemical | Cat#A461-4 |
| 4x Laemmli Sample Buffer | Bio-Rad | Cat#161-0747 |
| 8–16% Mini-PROTEAN® TGX™Precast Protein Gels, 12-well, 20 μl | Bio-Rad | Cat#4561105 |
| Acetonitrile | Fisher Chemical | Cat#A955-4 |
| alpha-tochopherol | Sigma-Aldrich | Cat#T3251 |
| Ammomium acetate | Sigma-Aldrich | Cat#238074-500G |
| Ammonium hydroxide | Fluka | Cat#44273-100ml-F |
| Arachidonic acid | Cayman Chemical | Cat#90010 |
| Blotting Grade Blocker Non Fat Dry Milk | Bio-Rad | Cat#1706404XTU |
| Bluing Reagent | ThermoFisher Scientific | Cat#6769001 |
| Bovine Serum Albumin | Sigma-Aldrich | Cat#A4503-50G |
| Chloroform | Fisher Chemical | Cat#C606-4 |
| Chloroform, HPLC grade | Sigma-Aldrich | Cat#270636 |
| Deferoxamine | Sigma-Aldrich | Cat#D9533 |
| Diethylenetriamine pentaacetic acid | Sigma-Aldrich | Cat#D6518-10G |
| DMEM/F12 | Life Technologies | Cat#11330-032 |
| EGF | ThermoFisher Scientific | Cat#PHG0311 |
| Eosin Y | ThermoFisher Scientific | Cat#6766010 |
| Ferrostatin-1 | Sigma-Aldrich | Cat#SML0583 |
| Ferrostatin-1 | Abcam | Cat#ab146169 |
| Fetal Bovine Serum | Life Technologies | Cat#10437-028 |
| Folic acid | Sigma-Aldrich | Cat#F8758 |
| Glutamax | Life Technologies | Cat#35050-061 |
| Guanidine hydrochloride | ThermoFisher Scientific | Cat#BP 178 |
| Halt™ Protease and Phosphatase Inhibitor Cocktail (100X) | ThermoFisher Scientific | Cat#78440 |
| Hank’s buffered saline solution (HBSS) without calcium or magnesium | ThermoFisher Scientific | Cat#14175079 |
| Harris Hematoxylin | ThermoFisher Scientific | Cat#6765003 |
| Hepadecanoic acid | Fluka | Cat#51610 |
| HEPES | Life Technologies | Cat#15630-080 |
| HEPES | Sigma-Aldrich | Cat#H3375-250G |
| Hexanes | Fisher Chemical | Cat#H303-4 |
| Hydrocortizone | Sigma-Aldrich | Cat#H0396 |
| Insulin Selenium Transferrin | Life Technologies | Cat#41400-045 |
| L-Glutathione reduced | Sigma-Aldrich | Cat#G4251 |
| Lipofectamine transfection reagent | Invitrogen | Cat#18324012 |
| Locostatin | Sigma-Aldrich | Cat#L5670 |
| Methanol | Fisher Chemical | Cat#A456-4 |
| Methanol (Certified ACS), Fisher Chemical | Fisher Scientific | Cat#A412-4 |
| ML351 | Cayman Chemical | Cat#847163-28-4 |
| MTT (Thiazolyl blue tetrazolium bromide) | Sigma-Aldrich | Cat#M5655 |
| Necrostatin-1 | Sigma-Aldrich | Cat#N9037 |
| Necrostatin-1s | BioVision | Cat#2263 |
| Paraformaldehyde, reagent grade, crystalline | Sigma-Aldrich | Cat#P6148 |
| Paraformaldehyde | Fisher Scientific | Cat#O4042-500 |
| PD146176 | Cayman Chemical | Cat#4079-26-9 |
| Penicillin-Streptomycin | Life Technologies | Cat#15140122 |
| Porcine skin gelatin -approximately 300 bloom | Sigma | Cat#G1890 |
| Precision Plus Protein Kaleidoscope Prestained Protein Standards | Bio-Rad | Cat#1610375 |
| Propidium Iodide (PI) | Life Technologies | Cat#P3566 |
| Protease inhibitor cocktail tablet | Roche Diagnostics | Cat#11 836 153 001 |
| RSL3 | Selleck chemicals | Cat#S8155 |
| Sodium selenite | Life Technologies | Cat#S5261 |
| SuperSignal™ West Pico Chemiluminescent Substrate | ThermoFisher Scientific | Cat#34087 |
| Thermo Scientific™ Pierce™ GelCode SilverSNAP | ThermoFisher Scientific | Cat#P24612 |
| Thermo Scientific™ SuperSignal™ West Femto Chemiluminescent Substrate | ThermoFisher Scientific | Cat#PI34095 |
| Thermo Scientific™ SuperSignal™ West Pico Chemiluminescent Substrate | ThermoFisher Scientific | Cat#PI34077 |
| Tissue-Tek® O.C.T. Compound, Sakura® Finetek | VWR | Cat#25608-930 |
| Triiodothyronine | Sigma-Aldrich | Cat#T5516 |
| Triton X-100 | Fisher Scientific | Cat#BP151-500 |
| zVAD-fmk | Enzo Life Sciences | Cat#ALX-260-020-M001 |
| β-Estradiol | Sigma-Aldrich | Cat#E2758 |
| β-Nicotinamide adenine dinucleotide 2′-phosphate reduced tetrasodium salt hydrate | Sigma-Aldrich | Cat#N7505 |
| Sodium Chloride | Sigma-Aldrich | Cat#S9888 |
| Glycine | Sigma-Aldrich | Cat#G8898 |
| EDTA | Boston Bioproducts | Cat#BM-150 |
| Glycerol | Acros | Cat#56-81-5 |
| Collagenase IV | Gibco | Cat# 17104-019 |
| DMEM | Thermo-Fisher Scientific | Cat#11995-065 |
| DNAse | Thermo Fisher Scientific | Cat# EN0523 |
| Dynal Pan-Mouse IgG magnetic bead system | Invitrogen | Cat# 11531D |
| Glutamine | Life Technologies | Cat#25030081 |
| Hoescht | Sigma-Aldrich | Cat#B2883 |
| Hydrocortisone | Sigma-Aldrich | Cat#H0888 |
| Interleukin 13 | R&D Systems | Cat#213-ILB-005 |
| Nitrocellulose Membrane | Bio-Rad | Cat#1620112 |
| Rat Tail collagen-1 | Gibco | Cat# A10483-01 |
| Platinum PFX DNA polymerase | Invitrogen | Cat # 11708013 |
| pet151-d-TOPO | Invitrogen | Cat# K15101 |
| Critical Commercial Assays | ||
| CytoTox-ONE™ Homogeneous Membrane Integrity Assay(LDH) | Promega Corporation | Cat#G7890 |
| Human 15-LO1 (Human 15-Lipoxygenase) ELISA Kit | MyBioSource | Cat#MBS263359 |
| Human PEBP1 (Phosphatidylethanolamine Binding Protein 1) ELISA Kit | MyBioSource | Cat#MBS452262 |
| Amersham Plus Western Blotting Detection Reagents | GE Health Care | Cat#PRPN2132 |
| Anti-Flag-M2-Agarose beads | Sigma-Aldrich | Cat#A1205 |
| Deposited Data | ||
| MALDI-MS imaging raw data | This paper; Mendeley Data | http://dx.doi.org/10.17632/trpc5kjpn5.2 |
| Experimental Models: Cell Lines | ||
| Immortalized mouse hippocampal cell line (HT22) | Dr. David Schubert, The Salk Institute | HT22 |
| MLE-12 | ATCC | Cat#CRL-2110 |
| HK2 cells | ATCC | Cat#CRL-2190 |
| Experimental Models: Organisms/Strains | ||
| Hsd:Sprague Dawley® SD® outbred Rat | Envigo | Order#002 |
| CD-1 mice | Charles River Laboratories | Crl:CD1(ICR) |
| Oligonucleotides | ||
| 5′-UCAUUCUGUUGUUGAUAUAAAAAA-3′ and 3′-UCAGUAAGACAACAACUAUAUUUUUUU-5′ | Integrated DNA-Technologies (IDT) | mm.Ri.pebp1.13.1 |
| 5-AGUCAUUCUGUUGUUGAUAUAAAAA-3′ and 3-CCUCAGUAAGACAACAACUAUAUUUUU-5′ | Integrated DNA-Technologies (IDT) | mm.Ri.pebp1.13.2 |
| P112E (5′-GTGGGCTCGGGGCCTGAAAAGGGCACAGGCCTCC-3′ and 5′-GGAGGCCTGTGCCCTTTTCAGGCCCCGAGCCCAC-3′), | Life Technologies | n/a |
| Y176X (5′-GGCCGAGTGGGATGACTAGGTGCCCAAACTGTACGAGCA G-3′ and 5′-CTGCTCGTACAGTTTGGGCACCTAGTCATCCCACTCGGCC-3′) | Life Technologies | n/a |
| pLO151f cacc gaa ttc atg AAT GAC TCG ATA TTC TTT TCA CCC | Invitrogen | n/a |
| pLO151r, gcg ctc gag aag ctt tta tca GAT ATT GGT GCT CGC CGG GAT C. | Invitrogen | n/a |
| Recombinant DNA | ||
| pCMV6-PEBP1-Flag | OriGene | Cat#RC206355 |
| pCMV6-PEPB1P112E -Flag | This Study | n/a |
| Software and Algorithms | ||
| FlexImaging 4.0 | Bruker Daltonics | www.bruker.com |
| MSiReader 0.06 | Robichaud et al., JASMS 2013 | www.msireader.com |
| Compound discoverer 2.0 | ThermoFisher Scientific | mycompounddiscoverer.com |
| CHARMM-GUI | Im research lab, Lehigh University | www.charmm-gui.org |
| AUTODOCK-VINA | Molecular Graphics Lab at Scripps Research Institute | vina.scripps.edu |
| ANM | Bahar research lab, University of Pittsburgh | anm.csb.pitt.edu |
| GROMACS | Science for Life Lab, Stockholm University | www.gromacs.org |
| GRAMMX | Vasker lab, University of Kansas | vakser.compbio.ku.edu/resources/gramm/grammx/ |
| OPM | Lomize lab, University of Michigan | opm.phar.umich.edu/server.php |
| GNM | Bahar research lab, University of Pittsburgh; Li et al., 2016 | Gnmdb.csb.pitt.edu |
| PRODY API | Bahar research lab, University of Pittsburgh | Prody.csb.pitt.edu |
| NAMD | NIH Center for Macromolecular Modeling and Bioinformatics, University of Illinois at Urbana-Champaign | www.ks.uiuc.edu/Research/namd |
| GraphPad Prism | GraphPad Software | www.graphpad.com/prism |
| SPSS 18.0 | IBM Corporation | www.ibm.com/us-en/marketplace/spss-statistics |
| ImageJ | NIH | imagej.nih.gov/ij/ |
| Other | ||
| Leica Cryocut 1800 Research Cryostat Microtome CM1800 | Leica Biosystems | CM1800 |
| Molecular Imager ChemiDoc XRS System | Bio-Rad | 170-8071 |
| Indium-Tin Oxide (ITO) coated glass slides for MALDI-MS imaging | Bruker Daltonics | Cat#237001 |
| Immobilon®-P PVDF Membrane | EMD Millipore | Cat#IPVH00005 |
15LO1 expression and purification
Plasmids for expressing the catalytic domain (residues 112-663) of porcine 15LO1 was generously provided to us by Max Funk. Purification was carried out as previously described (Xu et al., 2012), with minor modifications. Protein expression was carried out in the Escherichia coli strain BL21 (DE3) Codon+ (Agilent, Santa Clara, CA), and cultures were grown in standard lysogeny broth (LB) supplemented with 50 μM iron (III) chloride. Following initial growth to OD600 of 1.0, cells were induced with 0.2 mM isopropylthio-beta-galactosidase (IPTG) and cultured overnight at room temperature. Cells were harvested by centrifugation and lysed in buffer containing 20 mM Tris (pH 7.5), 500 mM NaCl, 5 % glycerol, 5 mM imidazole, 1 mM Tris (2-carboxyethyl) phosphine (TCEP), and protease inhibitors. Lysates were cleared by centrifugation at 29,000 g. Protein was purified by nickel affinity chromatography (Qiagen, Hilden, Germany). Eluates from the column containing 15LO1 were dialyzed overnight against buffer containing 10 mM Tris (pH 7.5) and 1mM TCEP before anion exchange chromatography using a HiTrap Q column (GE Healthcare, Little Chalfont, UK). The resulting fractions were further refined by size exclusion chromatography in 10mM Tris (pH 7.5), 500 mM NaCl, and 1 mM TCEP using a Sephacryl S-200 column (GE Healthcare).
Cloning, expression and pruirification of PA-LoxA
LoxA with a His6 tag was amplified by PCR with Platinum PFX DNA polymerase (Invitrogen) using the following primer set: pLO151f, cacc gaa ttc atg AAT GAC TCG ATA TTC TTT TCA CCC; pLO151r, gcg ctc gag aag ctt tta tca GAT ATT GGT GCT CGC CGG GAT C. The PCR amplified fragment was cloned into pet151-d-TOPO to generate pet151/His6-LoxA. The construct was checked by restriction digestion and confirmed by DNA sequencing facility of University of California (Berkeley, CA). LoxA was purified by standard NTA-Ni affinity chromatography. Briefly, pet151/His6-LoxA plasmid was expressed in E. coli BL21(DE3). The culture was grown to an OD of 0.6 at 37 °C and expression was induced by incubating the culture at low temperature 20 °C for overnight (16 h). Culture was harvested at 5,000g and the pellet snap-frozen in liquid nitrogen. For lysis cell pellet was resuspended in buffer A [25 mM HEPES (pH 7.5) containing 150 mM NaCl] and lysed using sonication. The cell lysate was cleared by centrifugation at 40,000g for 25 min and the supernatant was loaded onto an NTA-Ni affinity column. After washing with lysis buffer, protein was eluted with a gradient of 0 to 500 mM imidazole in buffer A. LoxA fractions were collected and pooled together with a purity of >90%. Protein was preserved in 10% (v/v) glycerol and frozen for further use.
Biochemical Assays
Oxidation of AA and AA-PE induced by 15LO1 or 15LO2 in a model system
Lipoxygenase activity was assessed by formation of primary products of AA oxidation: 15-HpETE or PE (C18:0/15-HpETE) that were detected by reverse phase LC/MS. To prevent conversion of 15-HpETE to secondary products during incubation, the HEPES buffer included 100 μM DTPA (for transition metals chelation) that has been saturated with oxygen (before addition of 15LO1 or 15LO2). The enzyme concentrations were 400 nM 15LO1 (porcine) or 80 and 400 nM 15LO2 (human recombinant, Cayman Chemicals, Ann Arbor, MI) for AA and AA-PE, respectively, at four different substrate concentrations (10, 20, 50 and 100 μM). Basic kinetic parameters for AA and AA-PE oxygenation were estimated by Graphpad Prism software. 15LO activity was estimated in settings that reflect physiological conditions. Aliquots of AA in organic solvent were mixed with DOPC in a ratio 1:10 (or 1:1 when AA-PE was mixed with PC). Organic solvent was dried, vortexed and sonicated in the buffer to form liposomes. For AA-PE ratio with PC was 1:1. Then AA or PE (SA) were incubated with 15LO1 or 15LO2 in the presence of 0.5 μM 15-HpETE, 100 μM DTPA in 20 mM HEPES pH 7.4 at 37 °C during 10 min in the absence or in the presence of PEBP1 (ratio PEBP1/15LO was 1:1). At the end of incubation AA and PE as well as their oxygenated products were extracted by the Folch procedure and analyzed by LC/MS.
Far-western blotting
The interaction between PEBP and 15LO in vitro was examined with far-western blotting. Human 15LO2 (18 μl of 3 μM solution) (Cayman Chemicals, Cat. # 10011263) and porcine 15LO1 (18 μl of 3 μM solution) (expressed, and purified as described above) were separated on 10 % Tris-glycine SDS-PAGE and electrically transferred to a nitrocellulose membrane. Proteins were renatured by incubation of the membrane in denaturing and renaturing buffer (100 mM NaCl, 20 mM Tris (pH 7.6), 0.5 mM EDTA, 10 % glycerol, 0.1 % Tween-20, 2 % skim milk powder and 1 mM DTT) containing decreasing concentration of guanidine HCl (6 M, 3 M, 1 M, 0.1 M, 0 M) and the membrane was blocked with 5 % skim milk. Then the membrane was incubated in 0.5 ml of protein-binding buffer (100 mM NaCl, 20 mM Tris (pH 7.6), 0.5 mM EDTA, 10 % glycerol, 0.1 % Tween-20, 2 % skim milk powder and 1 mM DTT) containing 5 μM of recombinant PEBP1 (expressed, and purified as described below) and DOPE/DOPC liposomes (1:1) (25:1 ratio of DOPE to PEBP) at 4 °C overnight. Excess PEBP1 was removed and bound PEBP was immunodetected with PEBP-specific antibodies (Santa Cruz, #SC-28837, Rabbit, 1:1000) after 1.5 h incubation at room temperature. Thereafter, HRP-conjugated goat anti-rabbit IgG H&L (Sigma, #A0545, 1:1000) was applied as the secondary antibody, and positive bands were detected using SuperSignal West Femto Maximum Sensitivity Substrate (ThermoFisher Scientific).
To compare the interaction of wild type and mutated PEBP1 with 15LO1, both wt PEBP1 and P112E mutant PEBP1 (4 μM) were separated on 10 % Tris-glycine SDS-PAGE and electrically transferred to a nitrocellulose membrane. The membrane-bound proteins were denatured by guanidine HCl with subsequent renaturation and incubated in protein-binding buffer containing 3 μM porcine 15LO1 and DOPE/DOPC liposomes (1:1) (ratio DOPA to 15 LO1 25:1) at 4 °C overnight. Excess 15LO1 was removed and bound 15LO1 was immunodetected with 15LO-specific antibodies (Life Span Biosciences Inc, #LS-111783, 1:2000) after 1.5 h incubation at room temperature. HRP-conjugated goat anti-rabbit IgG H&L (Sigma, #A0545,1:1000), was applied as the secondary antibody, and bands were detected using SuperSignal West Femto Maximum Sensitivity Substrate (ThermoFisher Scientific).
Cross-linking
Recombinant human PEBP1 (2 μM) and porcine 15 LO1 (2 μM) cross-linking was performed by incubation with 0.1 % glutaraldehyde in 20 mM HEPES (pH 7.4) for 15 min. The reaction was stopped by addition of tris (pH 7.5) (final concentration 200 mM) and samples were incubated for 15 min at room temperature. SDS PAGE of samples was performed in 7.5 % running gel and proteins were stained by GelCode SilverSNAP kit (ThermoFisher Scientific). In some cases proteins were electro-transferred to nitrocellulose membrane, blocked by 5 % skim milk and PEBP was immunodetected using PEBP-specific antibodies (Santa Cruz, # SC-28837, 1:1000), HRP-conjugated goat anti-rabbit IgG H&L (Sigma, #A0545, 1:1000) as secondary antibodies and SuperSignal West Pico Substrate (ThermoFisher Scientific). The densities of protein bands were assessed by an open source image processing program: Image J.
Native blue gel shift assay
Recombinant human PEBP1 (4 μM) was incubated in 25 mM HEPES buffer (pH 7.4) with different ratios of AA for 10 min at room temperature, followed by Native Blue electrophoresis in 4–16 % bis-tris gel, and silver staining using the GelCode SilverSNAP kit (ThermoFisher Scientific). The density of the monomeric band was assessed by an open source image processing program: Image J.
Assessment of binding of PEBP1 with AA and phospholipids using dot-blot assay
Lipids (200 pmol and 400 pmol per dot) were loaded onto nitrocellulose membranes in 50 mM tris HCl (pH 7.4) using the Bio-Dot Microfiltration Apparatus (Bio Rad, Hercules, CA). The membranes were blocked by 3 % BSA in tris-saline buffer, 0.1 % tween-20 (TBS-T), and then incubated with recombinant PEBP1 (3 μM) in TBS-T containing 3 % BSA for 3 h at room temperature. Excess PEBP1 was removed and bound PEBP1 was detected using anti-PEBP specific antibodies (Santa Cruz, #SC-28837,1:1000) and HRP-conjugated goat anti-rabbit IgG H&L (Sigma, #A0545,1:1000) as the secondary antibody. Dots were detected using SuperSignal West Femto Maximum Sensitivity Substrate (ThermoFisher Scientific).
Measurement of GPX4 activity in brain cortex homogenates was performed in 0.1 M Tris –HCl (pH 8.0), containing 0.5 mM EDTA and 1.25 % Triton X-100, 0.12 mM NADPH, 3 mM glutathione, glutathione reductase (1 U/ml), and 0.12 mg of protein. Oxidized tetralinoleoyl cardiolipin (final concentration 50 μM) was used as a substrate. The amount of consumed NADPH was assessed by fluorescence (λexc 340 nm, λem 460 nm). The amount of oxidized GSH was calculated based on the amount of NADPH consumed during GSSG reduction.
Liposome preparation
Liposomes of DOPC/SAPE (1:1) were prepared under sterile conditions using the sonication method. Briefly, 1,2-dioleoyl-PC (DOPC) and 1-stearoyl-2-arachidonyl-PE (SAPE) (Avanti Polar Lipids Inc., Alabaster, AL), lipids were dried with a stream of nitrogen gas and resuspended in PBS (pH 7.4) prepared fresh under oxygen flow and filtered before adding to a dried film of lipids. Lipids were mixed well and then sonicated using a Torbeo ultra cell disruptor (36180-series) with maximum power for 10 minutes on ice.
PEBP1 stimulated ferroptosis of MLE-cells by exogenous 15LO2
The freshly prepared liposomes were added to MLE-12 cells (10 μM each) in a serum free media, along with recombinant human proteins 15LO2 (Cayman Chemicals, Ann Arbor, MI) and wt PEBP1 or PEBP1 P112E mutant (0.5 μM each) and RSL3 (Selleck chemicals, Houston, TX). The reaction was initiated by adding 0.5 μM of 15(S)-HpETE (Cayman Chemicals, Ann Arbor, MI). After 2 h of incubation in serum free media, serum was added back to cells at 10 % final concentration and incubated further for 20 h before determination of cell death by LDH release assay.
Cell Death assays
Cell death was determined by measuring released lactate dehydrogenase (LDH) activity. LDH activity was quantified using the Cytotoxicity Detection Kit (LDH) according to the manufacturer’s instructions (Promega Corporation, Madison, WI). Cell death was also monitored by propidium iodide (PI) (Life Technologies, Grand Island, NY) staining using flow-cytometry as described previously (Kagan et al., 2017) and by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) (Sigma-Aldrich, Saint Louis, MO) cell viability assay. Cell death was calculated as per instructions of the manufacturer.
In studies evaluating cell death in Figure 4, HK2 cells were transfected with plasmids containing either pCMV6 or pCMV6-PEBP1 were exposed to RSL3 (200 nM, 18 h) in the absence or presence of a ferroptosis inhibitor, ferrostatin-1 (FER, 0.4 μM). MLE cells were incubated with RSL3 (200 nM) alone or in combination with liposomes (DOPC/SAPE; 1:1) (10 μM), 15LO2 (0.5 μM) and PEBP1 (0.5 μM) or with only liposomes (DOPC/SAPE) and 15LO2 with or without ferrostatin-1 (FER, 0.4 μM) for 20 h, 5 % CO2 at 37 °C.
In studies evaluating cell death in Figure 5, HAECs were stimulated with IL13 for 72 h and then exposed to RSL3 (10 μM) in the absence or in the presence of locostatin (10 μM) for 24 h. PHKCs were treated with RSL3 (750 nM) for 5 h in the absence or in the presence of locostatin (10 μM); ferrostatin-1 (FER, 0.4 μM) completely abolished the effects of RSL3 and RSL3+locostatin. HT22 cells were exposed to RSL3 (25 nM) for 24 h in the absence or in the presence of locostatin (12.5 μM); ferrostatin-1 (FER, 0.4 μM) completely abolished the effects of RSL3 and RSL3+locostatin. Necrostatin-1s (Nec1s, 20μM) and zVAD-fmk (50μM) were ineffective. PHKCs were treated with RSL3 (750 nM, 5 h) in the presence of locostatin (750 nM, 5 h) and FER (0.4 μM, 5 h). HK2 cells were treated with RSL3 (100 nM, 20 h) in the presence of locostatin; ferrostatin (FER, 0.2 μM, 20 h) completely abolished the effects of RSL3+locostatin. Incubation conditions: RSL3 (100 nM, 20 h) in the presence of locostatin (50 μM). FER (0.4 μM, ferrostatin-1), Nec1 (50 μM, necrostatin-1, RIP kinase 1 inhibitor), Nec1s (50 μM, necrostatin-1s) and zVAD-fmk (50 μM, caspase inhibitor). In the combination groups, the concentrations of Nec1s, zVAD-fmk, and FER were 25 μM, 25 μM, and 0.4 μM, respectively.
Knock down experiments
1) Dicer-substrate short interfering RNAs (DsiRNAs) transfection of HAECs was performed using Lipofectamine transfection reagent. Briefly, DsiRNA was pre-mixed with Lipofectamine transfection reagent (ThermoFisher Scientific) for 20 min at room temperature before pooled together with HAECs suspension and seeded onto transwells for incubations. After 24 h, the transfection mixture was removed and cells were switched to ALI culture for 7 days. 2) siRNA transfection of HT22 cells was performed as follows: Chemically synthesized, double-stranded siRNAs were purchased from integrated DNA technologies (IDT, Coralville, IA). Two sequences corresponding to mouse PEBP1 were used as targets for siRNAs: 5′-UCAUUCUGUUGUUGAUAUAAAAAA-3′ and 3′-UCAGUAAGACAACAACUAUAUUUUUUU-5′ (mm. Ri. pebp1.13.1), and 5′-AGUCAUUCUGUUGUUGAUAUAAAAA-3′ and 3′-CCUCAGUAAGACAACAACUAUAUUUUU-5′ (mm. Ri.pebp1.13.2), One unrelated sequence that shows no significant homology to any mouse known gene was used as a control (scrambled –siRNA) and purchased from IDT. siRNAs were transfected into cells using Lipofectamine transfection reagent according to the manufacturers’ instructions (Cat. #18324012, Invitrogen, Carlsbad, CA). Briefly, cells were 70–80% confluent at the time of transfection, siRNA was pre-mixed with Lipofectamine transfection reagent for 45 min at room temperature before pooled together with HT22 cell suspension and seeded onto 6 well-plates at 1–2x105 of cells/1.5ml/well for incubations up 7 h, then the transfection mixture was removed and cells were switched to DMEM supplied with 10 % FBS for 24 h. Knock down efficiency was detected at 72 h by western blot analysis: anti-PEBP1 (Santa Cruz, Cat.#SC-28837,1:500) primary antibody, horseradish peroxidase (HRP)-conjugated anti-rabbit IgG H&L (Sigma_Aldrich, Cat. #A0545, 1:1000) secondary antibody and anti-β-Actin-Peroxidase antibody (ThermoFisher Scientific, Cat. #A3854,1:1000).
Transient wt PEBP1 and PEBP1 P112E mutant cDNAs Transfection
HK2, MLE and HT22 cells were transfected with either wt PEBP1 cDNA (1 or 2.5 μg, 24 h; OriGene, Rockville, MD) or PEBP1 P112E mutant cDNA (2.5 μg, 24 h) using Lipofectamine™ (ThermoFisher Scientific, Waltham, MA) transfection reagent. PEBP1 cDNA was incubated with Lipofectamine™ and P3000™ transfection reagents for 15 minutes at room temperature before the complex was added to the adherent cells. For cell death experiments, media was replaced with complete media and cells were reseeded and attached before treatment was given. For immunoprecipitation and expression, cells were collected after 48 h of transfection and processed for further experiments.
Immunoprecipitation of Flag-tagged PEBP1
Cells (MLE-12 and HT22) were transfected with PEBP1 DNA (2.5 ug) and after 48 h of transfection cell pellets were resuspended in extraction buffer (300 mM NaCl, 50 mM Tris, 1 mM EDTA and 10 % glycerol) containing protease inhibitors (1X). Cells were lysed by freeze-thaw method (3 times) in liquid nitrogen and further incubated on ice for 30 min and centrifuged at high speed (14000 rpm, 10 min) to prepare the lysates. To set-up the pull-down, the lysates were diluted (1:1) with no salt buffer (50 mM Tris, 1 mM EDTA and 10 % glycerol) to make final salt concentration of 150 mM NaCl, and mixed with Anti-Flag M2-agarose beads (25 μl/per reaction) equilibrated with low-salt buffer (150 mM NaCl, 50 mM Tris, 1 mM EDTA and 10 % glycerol). Pull-down samples were incubated overnight at 4 °C, washed 4 times with buffer (150 mM NaCl, 50 mM Tris, 1 mM EDTA and 10 % glycerol) and eluted with 0.1 M glycine (pH 2.3). Eluted samples were used for determination of AA by MS analysis.
Detection of 15LO1 and PEBP1 by ELISA
ELISA kits specific for human 15LO1 (MyBioSource, San Diego, CA, Cat. # MBS263359), 15LO2 (Cat. #MBS914061) and PEBP1 (Cat. #MBS452262) were used for in vitro quantitative measurement of 15LO1 and PEBP1 in HAECs cells or 15LO2 and PEBP1 in HK2 cells. All the measurements were performed as per manufacturer’s instructions.
Western blot analysis
Proteins were separated by gradient 8–16 % Tris-Glycine Gels (Precise™ 8–16 % Tris-Glycine Gels, 25 μl/well, 15 well, Life Technologies) and electrically transferred to a polyvinylidene difluoride or a nitrocellulose membrane (Bio-Rad, Hercules, CA). After the membrane was blocked with 5 % skim milk, target proteins were immunodetected using GPX4 (1:500, ab125066, Abcam), 15LO2 (1:1000, sc-67143, Santa Cruz), PEBP1 (1:1000, SC-28837, Santa Cruz), and β-actin (1:1000, A2228, Sigma) antibodies following overnight incubation at 4 °C. Thereafter, the horseradish peroxidase (HRP)-conjugated anti-rabbit IgG H&L at 1: 2000 (Abcam) or was applied as the secondary antibody, and the positive bands were detected using Amersham ECL plus Western blotting detection reagents (GE Health Care, Piscataway, NJ).
Fluorescent Microscopy
Immunostaining and Confocal Microscopy
Cells were fixed in 2 % paraformaldehyde at 4 °C for 15 min, permeabilized with 0.1 % Triton X-100 in PBS+ 0.5 % BSA (PBB) for 15 min. Tissue sections and cells were blocked with 5 % donkey serum for 45 min and incubated for 2 h at room temperature with the primary antibodies (GPx4: Abcam, ab51944 (Goat, 1:100); PEBP1: Santa Cruz, SC-376925 (Mouse,1:200) 15LO1: 15LO2: Santa Cruz, SC-67143 (Rabbit, 1:100). Alexa 488, cy3 and cy5 conjugated secondary antibodies were from Thermo-Fisher Scientific and Jackson Immuno, respectively. 15LO1 antibody was a gift from Dr. Doug Conrad, University of California, San Diego (Rabbit 1:200). Cells were counterstained with hoescht (Sigma B2883) 1mg/100ml dH20, mounted using Gelvatol, and imaged in 200 nm z-sections collected using a Nikon A1R equipped with GAsP detectors and a 60X (1.4NA) objective.
Object Based Co-localization Analyses
Pixel (intensity) based approaches for assessing co-localization, such as Pearson’s correlations are only appropriate when dealing with proteins that have similar levels of expression (Dunn et al., 2011), whereas object-based methods take into account the spatial and intensity distribution of the fluorescent signal. 3D confocal (Nikon A1, 60X, 1.4NA) stacks (200 nm optical sections) were processed using blind deconvolution (10 iterations, NIS Elements, Nikon Inc., Melville NY) in order to maximize the information in each image and improve the accuracy of the subsequent object based analysis (Biggs, 2010). Puncta (objects) were then segmented based on size and intensity using the 3D spot detection tool in NIS Elements, which generated a binary layer containing the spatial positioning of each object in the x-, y- and z- axis for each individual protein label (i.e. 15LO1/15LO2, PEBP1 and GPX4). Co-localization was tested using a Boolean “and” operation within NIS Elements. This logical argument tests the intersection of selected binary layers, and will identify objects that have both protein A “and” protein B (e.g. 15LO1 and PEBP1).
Fluorescence Resonance Energy Transfer
FRET based analyses can resolve the proximity of interacting proteins within 1–10 nm. The FRET phenomenon is the non-radiative (dipole-dipole) energy transfer from a fluorescent donor to an acceptor fluorophore. Because the efficiency of FRET decreases as the sixth power of the distance between the donor and acceptor, when the proteins are separated by >10 nm, no FRET occurs. 15LO1 and PEBP1 were labeled with cy3 (donor) and cy5 (acceptor) and imaged at 10X zoom using a Leica SP8 spectral confocal microscope equipped with a 60X (1.4NA) optic. FRET was confirmed by acceptor (cy5) photo-bleaching. With this method, if true FRET is present, the donor will show an increased intensity in the bleached region.
Mass Spectrometry
MS analysis of PEBP1 and PEBP1 mutants
PEBP1 (42 μM) or Y176X (14.5 μM) was incubated with or without equimolar amounts of arachidonic acid (AA) in 10 mM ammonium acetate, pH 6.9. The arachidonic acid stock solution was prepared in DMSO such that the final concentration of DMSO was less than 1 % when AA was incubated with PEBP-1. For PEBP1/AA-amide interactions, a concentration of 9 μM was used. The mixtures were incubated for 30 min at room temperature. Acetic acid was then added (1 % final concentration) for charging purposes. The resulting solution was loaded into a metal-coated PicoTip nanospray emitter (New Objective, Inc. Woburn, MA). The nanospray emitter was fitted with a conductive elastomer sleeve and mounted on the nanospray probe of a Quattro II triple quadrupole mass spectrometer (Waters, Inc. (formerly Micromass, Inc.). The probe depth was adjusted such that the distance from the probe stop to the nanovial tip was 170 mm. The source temperature was set to 25 °C and the analyzer pressure was 1.8 e-5 mbar. Capillary voltage was adjusted to maintain a stable ion current (1.5–2.0 kV) with a cone voltage of 20 V in the positive ion mode. The data was processed using a Maximum Entropy software program (Waters, Inc.).
LC/MS analysis of phospholipids
Lipids were extracted by using the Folch procedure. Lipid phosphorus was determined by a micro-method. MS analysis of PLs was performed on a Q-Exactive hybrid-quadrupole-orbitrap mass spectrometer (ThermoFisher Scientific). Phospholipids were separated on a normal phase column (Luna 3 μm Silica (2) 100 Å, 150 x 2.0 mm, (Phenomenex)) at a flow rate of 0.2 mL/min on a Dionex Ultimate 3000 HPLC system. The column was maintained at 35 °C. The analysis was performed using gradient solvents (A and B) containing 10 mM ammonium acetate. Solvent A contained propanol:hexane:water (285:215:5, v/v/v) and solvent B contained propanol:hexane:water (285:215:40, v/v/v). All solvents were LC/MS grade. The column was eluted for 0–23 min with a linear gradient from 10 % to 32 % B; 23–32 min using a linear gradient of 32–65 % B; 32–35 min with a linear gradient of 65–100 % B; 35–62 min held at 100 % B; 62–64 min with a linear gradient from 100 % to 10 % B followed by an equilibration from 64 to 80 min at 10 % B. Analysis was performed in negative ion mode at a resolution of 140,000 for the full MS scan in a data-dependent mode. The scan range for MS analysis was m/z 400–1800 with a maximum injection time of 128 ms using 1 microscan. An isolation window of 1.0 Da was set for the MS and MS2 scans. Capillary spray voltage was set at 3.5 kV, and capillary temperature was 320 °C. The S-lens Rf level was set to 60. Analysis of LC/MS data was performed using software package Compound DiscovererTM (ThermoFisher Scientific, San Jose, CA) with an in-house generated analysis workflow and oxidized phospholipid database. Briefly, peaks with S/N ratio of more than 3 were identified and searched against oxidized phospholipid database. Lipids were further filtered by retention time and confirmed by a fragmentation mass spectrum. The species assignments were done based on three criteria: retention time, exact mass and fragmentation pattern. The retention time of each lipid class was determined based on the retention time of the exogenously added internal standard (± 2 min). The list of internal standards used in the analysis is presented in the method section. Values for m/z were matched within 5 ppm to identify the lipid species. The structure of identified lipids was confirmed by fragmentation analysis.
Identification of PE oxygenated products
Oxygenated PE products were identified by MS/MS analysis using an Orbitrap Fusion Lumos mass spectrometer (ThermoFisher Scientific). The instrument was operated with an electrospray ionization probe in negative polarity mode. Ion source conditions were set as follows: Spray voltage = 4 kV, Sheath gas = 20 (arbitrary unit), Auxiliary gas = 4 (arb. unit), Sweep gas = 0 (arb. unit), Transfer tube temperature = 300 °C, RF-Lens level = 50 %. Data were acquired in data-dependant-MS2 targeted-MS3 mode with a Cycle Time setting of 3 s. For MS scan events, the parameters were set as follow: Ion detection = Orbitrap, Mass resolution = 120,000, Scan range = m/z 400 – 1800, AGC target = 1e5. The most intense ion was selected for the data-dependent MS2 scan. The dynamic exclusion was set to 9 s. The exclusion mass list for MS2 was created from solvent blank injection data. For MS2 scan event(s), the parameters were set as follows: Quadrupole isolation = 1 Da, First Mass = m/z 87, Activation type = HCD, Collision energy = 24 %, Ion detection = Orbitrap, Mass resolution = 15,000, Max Injection time = 250 ms, AGC target = 2e4. Product ions from a targeted mass list were selected for MS3 scan. The target mass list for MS3 included 74 ions for a variety of potential oxidized fatty acyl products. For MS3 scan event(s), the parameters were set as follows: Isolation window = 2 Da, Activation type = CID, Collision energy = 35 %, Ion detection = Ion Trap, Ion trap Scan rate = Rapid, Max Injection time = 500 ms, AGC target = 5e4.
LC/MS analysis of AA and its oxidation products
AA and AAox were analyzed by LC/MS using a Dionex Ultimate™ 3000 HPLC system coupled on-line to a Q-Exactive hybrid quadrupole-orbitrap mass spectrometer (ThermoFisher Scientific, San Jose, CA) using a C18 column (Accliam PepMap RSLC, 300 μm 15 cm, Thermo Scientific). Gradient solvents (A: Methanol (20 %)/Water (80 %) (v/v) and B: Methanol (90 %)/Water (10 %) (v/v) both containing 5 mM ammonium acetate was used. The column was eluted at a flow rate of 12 μL/min using a linear gradient from 30 % solvent B to 95 % solvent B over 70 min, held at 95 % B from 70 to 80 min followed by a return to initial conditions by 83 min and re-equilibration for an additional 7 min. Spectra were acquired in negative ion mode. The scan range for MS analysis was m/z 150–600 with a maximum injection time of 100 ms using 1 microscan and a resolution of 140,000. An isolation window of 1.0 Da was set for the MS and MS2 scans with an inclusion list of 102 potential oxidized and non-oxidized fatty acyl products. Capillary spray voltage was set at 2.6 kV, and capillary temperature was 250 °C. The S-lens Rf level was set to 60. Analytical data were acquired and analyzed using Xcalibur software.
MALDI-MS imaging
DHB (2,5-dihydroxybenzoic acid, Sigma-Aldrich) was used as a matrix. Kidney sections for high spatial resolution analysis were coated with 40 mg/ml DHB in 50 % methanol using a TM Imaging Sprayer (HTX Technologies). Brain and lung sections were coated with DHB as a 0.5 M solution in 2:1 chloroform (HPLC grade, Sigma-Aldrich) to methanol (LC/MS grade, ThermoFisher Scientific) according to our previously published protocol (Sparvero et al., 2016). In brief, the matrix solution was loaded into a capillary-sprayer aerosol device. This device was constructed in-house and placed in a PCR chamber (Coy Laboratories, Grass Lake, MI) inside a fume hood in order to minimize variability from airflow cross-currents. The capillary-sprayer used an electrospray source from a Thermo-Finnigan TSQ 7000 mass spectrometer. The existing capillary was replaced with 75 μm i.d. polyimide coated capillary tubing (Scientific Instrument Services, Ringoes, NJ). Sheath gas (grade 5.0 N2) was regulated to 80 psi. The nozzle height was 120 mm with a spray time of 10 min without interruption at a flow rate of 2400 μL/h.
Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) imaging analyses were all performed in reflector mode and m/z 600–1000. Lung sections were analyzed using an Ultraflex II mass spectrometer (Bruker Daltonics), kidney with a rapifleXtm MALDI Tissuetypertm (Bruker Daltonics) and brain with an ABI 4800 MALDI (AB Sciex). Lung images were acquired at 50 μm lateral resolution in both positive and negative polarity, brain in positive polarity at 200 μm, and kidney in positive polarity at 5 μm. MALDI-MS images from Bruker instruments were produced using FlexImaging 4.0 software (Bruker Daltonics), and those from the AB Sciex with MSI Reader v0.06 software (www.msireader.com) using our previously published protocol (Sparvero et al., 2016). Heat maps for a given mass/charge (m/z) value ± 0.25 Da were generated either as a rainbow display (for single species) or red/green (for co-localization of two different species). Rainbow heat maps range from black-blue (low intensity) to red-white (high intensity). Lipid identification was based on theoretical monoisotopic mass values: [M+K]+ for positive polarity and [M−H]− for negative polarity.
Molecular Docking and Molecular Dynamics Simulations
Protein-protein docking of 15LO and PEBP1
Simulations of 15LO/PEBP1 complex predictions have been performed using the Gramm-X package respectively with PDB structures: 1lox (15LO1), 4nre (15LO2) and 1beh (PEBP1). The 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE)/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) membrane bilayer has been added to the complex with the CHARMM-GUI package with a priori prediction of 15LO1/PEBP1 orientation in the membrane obtained from OPM server.
AA-PE docking to PEBP1/15LO1 complex
Molecular docking has been performed with the SMINA package which is a fork of AutoDock Vina; customized to better support scoring function development and high-performance energy minimization. For dynamic complexes, we prepared 10 structures (frames) along ANM mode 1 (see Movie S1). For each structure 10 independent runs were performed.
Preparation of PEBP1 mutants
The1beh structure and the NAMD package were used to prepare mutated models of PEBP1. Mutation of selected residues was performed using the NAMD psfgen package and further minimized with 5000 steps.
PEBP1-multiple AA docking
A “Multiple molecular docking” approach was used to identify the binding sites of nine molecules of arachidonic acid (AA) on PEBP1 (1beh (Banfield et al., 1998)) structure. PEBP1-AA docking has been performed using the SMINA package. For each system 5 independent runs were performed and the AA binding pose with the highest binding affinity was chosen for the next calculation. Analysis and visualization has been made using VMD (Humphrey et al., 1996), ProDy (Bakan et al., 2011).
All-atom Molecular Dynamics Simulations
All-atom MD simulations were performed for human PEBP1 (1beh) in complex with nine molecules of (AA) using the NAMD MD simulation software with CHARMM27 force fields. The PEBP1-AA interaction has been identified using the molecular docking approach described above. In preparatory simulations with explicit water models (TIP3P), we adopted the following protocol: 0.2 ns of water equilibration, 10,000 steps of minimization, 0.35 ns of heating from 0 to 300 K, and 0.15 ns equilibration of the whole system before initiating production MD run. Three independent MD trajectories were generated, each 200 ns duration with 2 fs time step. A cutoff of 12 Å for non-bonded interactions was applied. Langevin dynamics and the Langevin piston algorithm were used to maintain the temperature at 300 K and the pressure at 1 atm. VMD were used for visualizing and GROMACS (Lindahl et al., 2001) for analyzing trajectories.
Coarse-Grained Molecular Dynamics Simulations
The MARTINI force field version 2.2 (Marrink et al., 2007) was used to simulate protein-protein interactions. The crystal structures of 15LO2 (4nre) and PEBP1 (1beh) were used to build the coarse-grained models. For LoxA, the crystal structure 5lc8 (Banthiya et al., 2016; Kalms et al., 2017) was used. Two simulations were set up, one in which the PEBP1 was place 2.5 nm above the helical bundle, of the 15LO2 (or LoxA) protein and another in which it was place below the alpha-helical bundle, away from the N-terminal beta-barrel (or N-terminal helix in LoxA). The CHARMM-GUI interface was used to set up the simulation models. GROMACS version 5.1.4 was used to perform the molecular dynamics simulations with the standard parameter settings for the MARTINI force field. The system was simulated using the isothermal-isobaric (NPT) ensemble.
Prior to the production run of 150 ns, the simulation system was energy minimized and a series of equilibration simulations were performed to relax the protein and the solvent system. During the equilibration phase the force-constant of 4000 kJ mol−1 nm−2 is used and a standard value of 0.9 nm is the cut-off. The simulation temperature was 303.15 K. The v-rescale thermostat was used to control the temperature with a coupling constant τp = 1.0 ps. The pressure was isotropically coupled to an external bath of τp = 5.0 ps and compressibility of 4.5 x 10−5 bar−1 using the Berendsen barostat. The electrostatic interactions were calculated using a shifted potential with a cut-off of 1.2 nm while for the van der Waals interactions a shifted potential was used with a cutoff of 1.2 nm and switch at 0.9 nm. The interaction between the proteins is calculated as a minimum distance between any atom pair on the respective proteins.
Analysis of collective modes using the Elastic Network Models - ANM and GNM
ANM (Anisotropic Network Model) (Eyal et al., 2015) and GNM (Gaussian Network Model) (Li et al., 2016) analyses were performed using ProDy API. We used GNM to identify the relative motions of different regions of 15LO1 after complexation with PEBP1 along the principal mode coordinate. Positive (red) and negative (blue) regions delineate the structural regions subject to concerted anticorrelated (coupled but opposite direction) motions. We employed the anisotropic network model (ANM) (Eyal et al., 2015), developed for predicting the collective dynamics of known structures, and determining the most cooperative rearrangements of subunits/domains which usually enable functional mechanisms. The correlation-cosine map between ANM modes (1 to 10 modes) of 15LO1 only and the PEBP1-15LO1 complex have been calculated and visualized using ProDy API. ANM and GNM theory can be found in (Eyal et al., 2015).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical Analysis
The data in figure legends are presented as means±SD values. In general the exact value of sample size (n) is presented in figure legends and reflects either the number of experimental replications with cells or biochemical model systems or the number of animals used in in vivo experiments. For measurements on human samples n represents the number of patients. Statistical analyses were performed by either Student’s t-test or one-way ANOVA for normally distributed data and by Kruskal-Wallis one-way ANOVA on ranks for data that were not normally distributed. When the overall ANOVA revealed a significant effect, the data were further analyzed with the Tukey/Dunn post-hoc test to determine specific group differences. The statistical significance of differences was set at p < 0.05. Phospholipids were quantified from the full scan LC/MS spectra with ratiometric comparison to the pre-selected internal standard using corresponding standard curve for each phospholipid class. Differences between the groups were analyzed by one-way ANOVA with Tukey post-hoc analyses by SPSS 18.0 software (SPSS Inc).
DATA AND SOFTWARE AVAILABILITY
Raw data from the MALDI-MS imaging is available online (http://dx.doi.org/10.17632/trpc5kjpn5.2).
Supplementary Material
An overall bending motion enabled by the interfacial hinge region is observed, shown from the front view. PEBP1 and 15LO1 are colored as yellow and grey transparent spheres, respectively. Ribbon diagram colors indicate the change in position driven by ANM mode 1 (coded from blue (most rigid) to red (most flexible)). The movement allows PEBP1 to come into close proximity with the catalytic region of 15LO1.
(A) Cross-linking by glutaric dialdehyde (GDA) confirms the interaction of 15LO1 with wt PEBP1 and suppressed interaction with P112E mutant PEBP1. Samples containing 15LO1 and wt PEBP1 or P112E mutant PEBP1 were treated with 0.1% glutaraldehyde for 20 min at room temperature and the reaction was stopped by the addition 200 mM of Tris–HCl (pH 7.5). Samples were run in SDS-PAGE. Proteins were detected by staining with silver or by incubation with anti–PEBP1 antibodies after electro-transfer (see Figure 1B). Cross-linked bands appeared after GDA treatment of PEBP1/15LO1 mixture (see Figure 1B), but not after treatment of PEBP1 or 15LO1 alone. The cross-linking was suppressed for P112E mutant PEBP1 vs wt PEBP1. Data are mean ± SD, *p < 0.05 vs. wt PEBP1, N=5/group.
(B) Computational modeling of PEBP1-15LO2 interactions. Human PEBP1 (red)/15LO2 (gray) complex near a POPE/POPC lipid membrane. The hydrophobic heads of the lipid molecules are displayed as green spheres (lower leaflet only). The model contains over 170,000 atoms including water, lipids, and ions. Water molecules and the remaining portions of the lipid bilayer have been deleted for clarity. The catalytic site residues on 15LO2 (H373, H378, H553) are highlighted in blue and enclosed in a black square. The conserved active site residues on PEBP1, H86 and S153 are highlighted in pink and yellow, respectively, and labeled.
(C) Coarse-grained Molecular Dynamics (CGMD) simulations of PEBP1-15LO binding in the presence of a phospholipid bilayer. Two independent runs were performed for each pair of proteins, 15LO1/PEBP1 (left; top and bottom) and 15LO2/PEBP1 (right top and bottom), using the MARTINI force field. In the first run, the complex was placed at 2.5 nm (top) separation from the lipid bilayer, and in the second, the complex was placed at 0.5 nm (bottom) away from the bilayer. The dotted line represents the minimum distance required for an interaction to take place. The distance between the PEBP1-15LO1/LO2 complex and the lipid bilayer show large fluctuations in the top two panels, coming into close proximity multiple times during the total duration (> 500 ns) of the simulations. The bilayer consisted of 50% POPC and 50% POPE with a total of 127 lipids in each leaflet. The inset shows the snapshots of the complex and the bilayer at the indicated times. The red spheres represent PEBP1, and the yellow and blue (b-barrel) spheres represent 15LO1/15LO2. Water molecules (included in simulations) are not shown for clarity.
(D) Coarse-grained molecular dynamics simulations of PEBP1/15LO2 binding in solution. Results from docking simulations performed for the complexation of PEBP1 with 15LO2. The simulations were performed using the MARTINI force field. PEBP1 was placed at −2.5 nm (top) and +2.5 nm (bottom) from 15LO2, in the absence of lipid bilayer. PEBP1 moves towards 15LO2 and forms a tight complex within 60–80 nm, and the complex remains stable during the rest of simulations (up to 150 ns).
(E) Comparison of the binding of wt PEBP1 to 15LO1 in silico shows the weaker affinity and distinctive binding pose of the P112E mutant. Left panel displays the optimal binding poses for wt PEBP1. The right panel shows the interface in greater detail, where wt PEBP1 exhibits several close contacts (atom-atom contact distances given). PEBP1 and 15LO1 residue labels are colored blue and green, respectively.
(F) Accumulation of PE-OOH species in PC/PE liposomes catalyzed by 15LO2 in the absence and in the presence of either wt PEBP1 or P112E mutant PEBP1. Data are mean ± SD, *p < 0.05 vs. control (no PEBP1), **p < 0.05 vs. wt PEBP1, N=5/group (for control and PEBP1), N=3/group (for P122E mutant).
(G and H) Results from coarse-grained MD simulations confirm the inability of human wt PEBP1 to stably bind Pseudomonas aeruginosa 15LOXA at the allosteric site. Results from docking simulations (G) and two independent coarse-grained MD runs CGMD1 and CGMD2 (H) are presented. In panel A, the two proteins are represented using ribbon diagrams and the N-terminal helix of P. aeruginosa 15LOXA and C-terminal helix of wt PEBP1 are labeled and colored red and blue, respectively. Comparison to Figure 1E shows the distinctive binding pose of PEBP1, as indicated by the position of the wt PEBP1 C-terminal helix. In panel B, the ordinate denotes the closest distance of approach between the two proteins, which originally were separated by 20 Å. During the CGMD simulations, although the two proteins find a stably bound pose at approximately 50 or 160 ns as evidenced by the fixed (minimal) distance, the binding poses (space-filling representations in insets) are different from those observed in wt PEBP1 and 15LO1.
Table S1. Basic kinetic parameters for AA and AA-PE oxygenation by 15LO1, 15LO2 and LoxA in the absence and in the presence of PEBP1, Related to Figure 6.
Data are mean ± SD
Table S2. “Multiple docking” calculations of arachidonic acid (AA) to PEBP1 mutants, Related to Figure 6.
(a) DDG = change in binding free energy with respect to that of wt PEBP1 (i.e. DG (mutant) – DG (wt). A positive value indicates an increase in binding energy, which is unfavorable.
* Number of residues located with 3.5 Å from all nine AA ligands and present in a wt PEBP1.
↑ Excluding AAs (energies colored in red) which are interacting only with AA (lack of residues within 3 Å)
Table S3. Results on AA-binding (to PEBP1 (case 1) or Y176X (case 2)) energetics(a), Related to Figure 6.
(a) The entries in the table show the interaction energy between wt PEBP1 or Y176X mutant PEBP1 and AA (or AA-amide) molecules, upon binding one small molecule at a time, up to 9 molecules binding. Red entries refer to interactions of AA (or AA-amide) molecules with previously bound AA (or AA-amide) molecules; gray entries are the same, except for the involvement of 1 PEBP1 residues. (b) Total DG is obtained by summing up all the black entries in the corresponding run. (c) Average change in free energy, relative to that obtained for the wt PEBP1 + 9 AA molecules
Movie represents the ANM mode 1 shown in Movie S1, displayed from a different perspective
(A) Western blots of 15LO2 in HT22 cells. Human recombinant 15LO2 protein (Cayman Chemicals) is presented as a positive control. (B) Western blots of 15LO2 in HK2 cells. Human recombinant 15LO2 protein (Cayman Chemicals) is presented as a positive control.
(C) Pie charts showing % of 15LO1, PEBP1, and co-localized puncta normalized to the total number of puncta in HEAC cells with or without IL13 treatment.
(D) Pie charts showing % of 15LO2, PEBP1, and co-localized puncta normalized to the total number of puncta in HK2 cells with or without LPS treatment.
(E) Co-localization of PEBP1, 15LO1 and GPX4 in HAECs treated with IL13. The upper panel shows a 3D volume view of 15LO1 (green), PEBP1 (red), GPX4 (pink) and nuclei (blue) in IL13-treated cells. The lower panel shows the co-localized puncta (objects, white) identified by object based segmentation and co-localization analysis. (F) The number of objects identified as positive for 15LO1, PEBP1, GPX4 or complexes that were positive for all three proteins 15LO1, PEBP1 and GPX4 (co-localized) in HAECs treated with IL13. Media control=4±3, IL13 823±693, p≤0.05 vs. untreated, N=3/group.
(G) Structural and functional interdependency of 15LO1, PEBP1 and GPX4. Upper panel: IL13 (10 ng/ml) for 72 h induced both 15LO1 and GPX4 in parallel in HAECs, with KD of PEABP1 and to a lesser degree 15LO1 reducing GPX4 levels, further supporting their interdependency. GPX4 KD decreased both 15LO1 and PEBP. Lower panel: GPX4 KD in HAECs led to the expected increase in cell death.
(A) Effect of RSL3 (750 nM, 5 h) in the absence or in the presence of ferrostatin-1 (FER, 0.4 μM) on the accumulation of PE oxygenated species in PHKCs. Scatter plot of changes in the levels of oxygenated PE species showing log2(fold-change) vs significance (−log10 (p value)), N=3/group.
(B) Effect of LPS (50 μg/ml, 24 h) in the absence or in the presence of RSL3 (750 nM, 5 h) and ferrostatin-1 (FER, 0.4 μM) on the accumulation of PE oxygenated species in PHKCs. Scatter plot of changes in the levels of oxygenated PE species showing log2(fold-change) vs significance (−log10 (p value)), N=3/group
(C) Effect of a ferroptosis inhibitor, ferrostatin (FER, 1 μM), on RSL3 (10 μM) induced cell death in HAECs. Data are mean ± SD, *p < 0.05 vs control; **p < 0.05 vs. RSL3, N=3/group. (Insert) Western blot analysis shows the increased expression of GPX4 following IL13 (10 ng/ml) in fresh bronchial epithelial cells.
(D) Effect of FER (0.4 μM) on RSL3 (50 nM, 24 h) induced death of HT22 cells. Data are mean ± SD, *p < 0.05 vs. control; **p < 0.05 vs. RSL3, N=3/group. (Insert) Western blot analysis demonstrates high expression of GPX4 in HT22 cells, M: molecular weight marker.
(E) Effect of different ferroptosis inhibitors on RSL3 (200 nM, 24 h)-induced death in PHKCs. Conditions: ferroptosis inhibitors: FER (0.2 μM); deferoxamine (DFO, 25μM); LO15 inhibitors: ML351 (0.5 μM); PD146176 (0.5 μM). Data are mean ± SD, *p < 0.05 vs. control; #p < 0.05 vs. RSL3, N=3/group. (Insert) Western blot analysis demonstrates high expression of GPX4 in PHKC cells. PHKCs were isolated from proximal tubule epithelial cells by immuno-affinity method.
(F) Effect of different ferroptosis inhibitors on RSL3 induced death in HK2 cells. Cells were exposed to RSL3 (200 nM, 24 h) in the absence or in the presence of the ferroptosis inhibitors ferrostatin (FER, 0.2 μM) and deferoxamine (DFO, 25 μM) and vitamin E (vit E, 25 μM), as well as a 15LO inhibitor, PD146176 (0.5 μM). Data are mean ± SD. *p < 0.05 vs. control; #p < 0.05 vs. RSL3. N=4/group. (Insert) Western blot analysis demonstrates high expression of GPX4 in HK2 cells and effects of RSL3 (200 nM, 24 h) on the expression of GPX4.
(G). IL13-dependent ferroptosis in RSL3- and AA-treated HAECs. Human airway epithelial cells (HAECs) with or without IL13 (10ng/ml) were incubated with AA (10 μM) and then treated with RSL3 (10 μM) in the presence or absence ferrostatin-1 (1 μM). Cell death was assessed 20–24 h after treatment with RSL3 by the amount of LDH released. Data are mean ± SD, *p < 0.05 vs. control; #p < 0.05 vs. RSL3 + AA, No IL13, N=3/group.
(H) Effects of IL13 and RSL3 on the accumulation of PEox in HAECs. Incubation (72 h) conditions: IL13 (10 ng/ml), RSL3 (10 μM) or their combination. Scatter plot of changes in the levels of oxygenated PE species (log2 (fold-change) vs. significance (−log10 (p-value), (N=3/group or 4/group).
(I) HAECs with KD PEBP1 contain higher levels of free AA but not AA esterified into PE. Closed bars indicate the number of samples with increased or decreased AA (blue) or AA-PE (red) after PEBP1 KD (primary y-axis). Open bars depict the average changes relative to control ± SEM (secondary y-axis). The number of samples with increased AA was significantly different from the number of samples with increased AA-PE (p=0.029, z-test): 40±16% increase in AA while the changes in AA-PE were −12±13% after PEBP1 KD.
Mouse lung epithelial (MLE-12) or HT22 cells were treated with RSL3 (250 nM or 50 nM, respectively) in the presence or absence of a ferroptosis inhibitor, ferrostatin 1 (0.4 μM), for 20 h. Effect of RSL3 was monitored by LDH release (A and D), Propidium iodide (PI) staining (B and E) using flow cytometry, or MTT assay (C and F). Data are mean ± SD, *p < 0.05 vs. control, N=3/group.
(A) PEBP1 KD in HT22 cells using siRNA. Bar graphs show quantitative assessments of PEBP expression. Data are mean ± SD, *p < 0.05 vs. control or scrambled siRNA, N=3/group. (Insert) Representative western blot in control, PEBP1-siRNA treated cells, and scrambled siRNA treated cells.
(B) Ferroptotic death of RSL3-treated MLE cells induced by exogenously added 15LO2, AA-PE and PEBP1. Cell death was determined as the difference between the total levels of death and ferrostatin-1 eliminatable cell death (see Figure 4F). Data are mean ± SD, *p < 0.05 vs. RSL3, N=3/group.
(C) Ferroptotic death of RSL3-treated MLE cells induced by exogenously added 15LO2, AA-PE and PEBP1 P112E mutant. Cell death was determined by LDH release. Data are means ± SD, *p < 0.05 vs. RSL3, N=3/group.
(D) RSL3-induced ferroptosis in MLE cells transfected with wt PEBP1 or PEBP1 P112E mutant. Cells were treated with RSL3 (200nM, 24h) in the presence or absence of FER (ferrostatin, 0.4 μM). Cell death was determined by LDH release. Data are means ± SD, *p<0.05 vs control; #p<0.05 vs RSL3 treated cells transfected with wt PEBP1. N=3/group.
(E) Western blots documenting expression of FLAG-tagged PEBP1 in MLE cells and HT22 cells using anti-PEBP1 anti-body. pCMV6 – empty vector; pCMV6-PEBP1- plasmid containing PEBP1.
(F) Westerns blots revealing the presence of Flag-tagged PEBP1 using anti-FLAG anti-body in eluates of MLE and HT22 cells after immuno-precipitation using anti-FLAG M2 agarose beads.
(G) Typical base profile of LC-MS signals corresponding to AA in eluates after immune-precipitation of PEBP1 from MLE cells and HT22 cells. Four independent experiments with each type of cells have been performed.
(A) The detailed interactions of each of the nine AA with PEBP1 in the PEBP1-AA9 complex. Each panel shows the interaction between respective AA (cyan) and PEBP1 (gray). In each panel, PEBP1 residues that are in close proximity (within 5 Å) of the respective AA (indicated by red arrow) are labeled in blue and shown as stick representations. In some of the panels, other AA which occupy the same binding site as the cyan one are indicated in different colors (e.g. red, blue or green in AA2, AA4, AA5 and AA6).
(B) Examination of the stability of PEBP1-bound states of AA in two sets of independent runs, MD1 (left 3x3 panels) and MD2 (right 3x3 panels). In each case the time evolution of the instantaneous position of the mass center is shown for the labeled AA over simulations of 200 ns. The original binding energy in kcal/mol is shown in parentheses. The majority of the binding poses are highly stable: even if there are occasional rearrangements, the poses are quickly restored. AA8 and AA5 are relatively unstable.
(C) Decreased AA-binding affinity of PEBP1 with the truncated C-terminal helix (Y176X mutant). Results from five independent runs are displayed in each case, each run consisted of successive bindings of up to nine AAs. Left panel displays the binding sites (or corresponding clusters of bound AAs) observed in those multiple runs (see color code in the inset). Right panel displays the same results for the Y176X mutant PEBP1. Two of the clusters (C2 and C3) observed to bind AAs in the wt PEBP1 disappeared in Y176X, due to the elimination of the C-terminal helix that otherwise showed high AA-binding propensity. Clusters C1 and C2 are maintained in both cases, although C1 appears to be more broadly distributed in the mutant. Cluster C5 shows a reduced AA-binding probability in the mutant. PEBP1 residues that exhibited high AA-binding propensities are shown in space-filling (cyan) representations. Simulations showed a decrease from nine to six AAs bound to PEBP1 in the Y176X mutant protein.
(D) Correlation cosines between the ANM modes of motions accessible to 15LO1 in isolation (ordinate) and those accessible in its complex with PEBP1 (abscissa). The subset of the 10 softest (most cooperative) modes of motions are shown in each case. The correlations are color coded, from red (strongest) to blue (weakest). Note that the soft modes (e.g. modes 1–6) accessible to the uncomplexed 15LO1 are usually maintained in the complex with slight perturbations (cumulative correlation > 0.7, in general). However, the softest mode accessible in the complex (indicated by the arrow) is unique to the complexed form. The shape of this mode is illustrated in Fig 6. See also the Supplementary Movie S1 for the cooperative movements of 15LO1 and PEBP1 driven by this mode.
(E–I) A cooperative mode of motion induced by PEBP1/15LO1 complexation favors AA-PE binding near the catalytic site of the enzyme.
(E) Mode profile calculated with GNM. The curve shows the relative motions of different regions of 15LO1 in a complex with PEBP1 along the principal mode of motion gained in the complex. Positive (red) and negative (blue) regions indicate the structural regions subject to anticorrelated (coupled but opposite direction) motions. The zero-crossing points serve as hinges (white) between anti-correlated regions. These include 15LO1 K156, D173 and R402, interacting with PEBP1 P112, D144, Y181.
(F) Ribbon diagram of 15LO1/PEBP1 complex, color-coded by substructures that undergo anticorrelated movements in the GNM principal mode.
(G) Cross-correlation map describing the pairs of structural elements undergoing correlated (dark red) and anticorrelated (dark blue) movements in the cooperative motion acquired in the complex.
(H and I) Comparison of binding poses of AA-PE onto the PEBP1/15LO1 complex, obtained for the rigid/static complex (H) and dynamic/flexible complex (I) The results from 10 runs for each frame (total of 10 frames along ANM mode 1) are superimposed in panel E. The optimal (highest-scoring or lowest energy) binding pauses of AA-PE molecules are displayed with licorice representation (carbons in cyan, oxygens in red and nitrogens in blue). Three binding sites (hot spots) enclosed in ellipses/circles are observed for the rigid docking case (H). Notably, two additional hot spots, enclosed in red ellipses/circles, are observed when the complex is allowed to reconfigure along mode 1: one at the interface between PEBP1 and 15LO1, and the second in close proximity of the active site (H). The active site is indicated by the arrow (Fe2+ (pink sphere) coordinated by 4 histidines (green sticks)). Docking simulations were performed using Smina, visualization using VMD, and clustering using ProDy.
(A) AA-containing PE-species are abundant in airway epithelium. Mouse lungs (left lobe) were inflated and embedded with 8% gelatin in order to be MALDI-compatible and sectioned on special conductive ITO (indium tin oxide) slides for MALDI-MS imaging with DHB (2,5-dihydroxybenzoic acid) matrix. Semi-serial H&E stained section (left) shows that morphology is preserved. MALDI positive ion mode (center) shows highly saturated species (such as PC(32:0)) localizing broadly, while species containing 20:4 or 22:4 (such as PE(38:4) and PE(40:5)) localizing to the epithelium. MALDI negative ion mode (right) detects 20:4-containing PI(38:4) that also localizes to the epithelium. Scale bar = 500 μm. Ions are shown as a rainbow heat map of relative intensities ranging from black-blue (low intensity) to red-white (high intensity). MALDI-MS imaging was performed on a Bruker Ultraflex II instrument at a spatial resolution of 50 μm per pixel. Ions were detected as [M+K]+ in positive mode and [M−H]− in negative mode.
(B) AA-containing PE species are abundant in kidney tubular epithelium. High resolution MALDI-MS images of rat kidney lipids were generated using a Bruker Daltonics rapifleXtm MALDI Tissuetypertm mass spectrometer. Fresh-frozen kidneys from 17-day old male rats were sectioned coronally and placed onto special conductive ITO (indium tin oxide) slides for MALDI-MS imaging with DHB (2,5-dihydroxybenzoic acid) matrix. Note the differences in localizations of pro-ferroptotic precursors PE species vs non-pro-ferroptotic PEs as well as non-pro-ferroptotic PC species. Precursors of pro-ferroptotic signals: arachidonoyl-PE species PE(38:4); PE(40:6); PE(40:4); non-pro-ferroptotic PE species: PE(34:0); PE(36:2); non- pro-ferroptic PC species; PC(32:0); PC(34:1). The top left panel shows an overlay of single-color ion intensity images of pro-ferrototic PE(38:4) (red) and non-pro-ferroptotic PC(34:1) (green) which confirms separate localizations. Scale bar = 500 μm. Ion intensities are shown as a rainbow heat map of absolute intensities ranging from black-blue (low intensity) to red-white (high intensity). Analysis was performed in positive ion mode with a spatial resolution of 5 microns per pixel. Ions were detected as [M+K]+.
(C) AA-containing PE species are abundant in rat brain. MALDI-MS imaging of rat brain PE. Left. Optical image of an H&E stained semi-serial tissue section to confirm cutting depth; Right. MALDI-MS image of 20:4-containing PE(38:4) localization, displayed as a relative intensity (0 % to 100 %) rainbow heat map. Scale bar = 1000 μm.
(D) Representative spectrum of PEs in HT22 cells from LC-MS/MS analysis. Ions containing at least one AA species are labeled in red. Inset: Pie chart showing the fraction of total PE in which at least one of the species contains AA.
(E–H) Fragmentation spectra of PE species such as PEp(36:5) (E), PEp(38:4) (F), PE(38:4) (G), PE(40:6) (H) confirming the integrated AA-residue. The 303.233 (marked in red) corresponds to the AA ion.
(I) Table showing the prominent AA-containing PE species in HT22 cells (more than 1% of total PEs)
(A) Ferrostatin-1 (FER) attenuated cell death in folic acid-induced acute kidney injury (AKI). CD-1 mice were pretreated with ferrostatin-1 (FER) 60 minutes before AKI induction by an overdose of folic acid, and euthanized at 48 h. (A) Assessment of cell death by TUNEL.
(B) Quantification of TUNEL positivity. Mean ± SD. *p <0.05 vs. control and AKI+FER, one-way ANOVA on ranks, since data were not normally distributed. N=6/group.
(C) Folic acid induced changes (left panel) in PE species containing oxygenated arachidonic acid.
(D) Folic acid induced accumulation of PE oxygenated molecular species in mouse kidney. Data are mean ± SD, *p < 0.05 vs. control, **p < 0.05 vs Folic acid plus FER, N=6/group.
(E) Assessment of GPX4 activity in brain cortex show decreased activity at 4 h after CCI. The activity measurements in brain cortex homogenates were performed in 0.1 M Tris–HCl (pH 8.0), containing 0.5 mM EDTA and 1.25 % Triton X-100, 0.12 mM NADPH, 3 mM glutathione, glutathione reductase (1 U/ml), and 0.12 mg of protein. Hydroperoxy-tetralinoleoyl-cardiolipin (final concentration 50 μM) was used as a substrate. Amount of consumed NADPH was assessed by fluorescence (λex 340 nm, λem 460 nm). The amount of oxidized GSH was calculated based on the amount of NADPH consumed during GSSG reduction.
(F). Changes of ferroptotic protein expression in the hippocampus after CCI. 15LO2 expression was significantly elevated 4 h after CCI (*p < 0.05 vs. naïve, N=3–4/group). PEBP1 and GPX4 were not significantly changed (p < 0.05; N=3–4/group) in the ipsilateral hippocampi of injured vs. naïve rats. Data are mean ± SD. Inset shows representative western blot images of each protein.
Highlights.
A new pro-ferroptotic role for an old actor, PEBP1 - catalytic reshuffle of 15LO
Epithelial redox phospholipoxysome oxidizes unsaturated phosphatidylethanolamines
PEBP1 and GPX4 are master regulators of ferroptosis in diverse epithelial cells
PEBP1/15LO-driven ferroptosis occurs in asthma, kidney injury and brain trauma
Acknowledgments
Supported by NIH (HL114453, U19AI068021, NS076511, NS061817, ES020693, GM097204, AI106684, HL098177, HL109152, DK070910, DK083961, HG008540, P30 DA035778, S10OD021540, and Human Frontier Science Program). We thank John C. Freedman (UCSC) for help in expression and purification of P. LoxA
Footnotes
Author Contributions: SEW, HB and VEK designed/planned the study and wrote the paper; JZ, GM, EK, QY, HD, DE, YM performed experiments with five types of cells, analyzed data; YYT, TSA, FQ, AAM, LJS performed different MS measurements and analyzed data; YYT participated in working on paper; CSC and SW performed imaging analysis; VAT, AAK, JCR and APVD performed in vitro biochemical experiments; TRH isolated and purified P. aeruginosa LoxA and participated in discussion of related experiments; KMR, HIS performed computational modeling; IB supervised computational studies and participated in paper writing; JAK, HB and XW performed and analyzed clinical and experimental AKI data, EK, HB, CSC, and TSA performed and analyzed TBI data.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aldrovandi M, Hinz C, Lauder SN, Podmore H, Hornshaw M, Slatter DA, Tyrrell VJ, Clark SR, Marnett LJ, Collins PW, et al. DioxolaneA3-phosphatidylethanolamines are generated by human platelets and stimulate neutrophil integrin expression. Redox Biol. 2017;11:663–672. doi: 10.1016/j.redox.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakan A, Meireles LM, Bahar I. ProDy: protein dynamics inferred from theory and experiments. Bioinformatics. 2011;27:1575–1577. doi: 10.1093/bioinformatics/btr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfield MJ, Barker JJ, Perry AC, Brady RL. Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction. Structure. 1998;6:1245–1254. doi: 10.1016/s0969-2126(98)00125-7. [DOI] [PubMed] [Google Scholar]
- Banthiya S, Kalms J, Galemou Yoga E, Ivanov I, Carpena X, Hamberg M, Kuhn H, Scheerer P. Structural and functional basis of phospholipid oxygenase activity of bacterial lipoxygenase from Pseudomonas aeruginosa. Biochim Biophys Acta. 2016;1861:1681–1692. doi: 10.1016/j.bbalip.2016.08.002. [DOI] [PubMed] [Google Scholar]
- Bayir H, Kagan VE, Tyurina YY, Tyurin V, Ruppel RA, Adelson PD, Graham SH, Janesko K, Clark RS, Kochanek PM. Assessment of antioxidant reserves and oxidative stress in cerebrospinal fluid after severe traumatic brain injury in infants and children. Pediatr Res. 2002;51:571–578. doi: 10.1203/00006450-200205000-00005. [DOI] [PubMed] [Google Scholar]
- Biggs DS. 3D deconvolution microscopy. Curr Protoc Cytom. 2010;Chapter 12(Unit 12 19):11–20. doi: 10.1002/0471142956.cy1219s52. [DOI] [PubMed] [Google Scholar]
- Deiss K, Kisker C, Lohse MJ, Lorenz K. Raf Kinase Inhibitor Protein (RKIP) Dimer Formation Controls Its Target Switch from Raf1 to G Protein-coupled Receptor Kinase (GRK) 2. Journal of Biological Chemistry. 2012;287:23407–23417. doi: 10.1074/jbc.M112.363812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis EA, Norris PC. Eicosanoid storm in infection and inflammation. Nat Rev Immunol. 2015;15:511–523. doi: 10.1038/nri3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn KW, Kamocka MM, McDonald JH. A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol. 2011;300:C723–742. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dweik RA, Boggs PB, Erzurum SC, Irvin CG, Leigh MW, Lundberg JO, Olin AC, Plummer AL, Taylor DR American Thoracic Society Committee on Interpretation of Exhaled Nitric Oxide Levels for Clinical, A. An official ATS clinical practice guideline: interpretation of exhaled nitric oxide levels (FENO) for clinical applications. Am J Respir Crit Care Med. 2011;184:602–615. doi: 10.1164/rccm.9120-11ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emlet DR, Pastor-Soler N, Marciszyn A, Wen X, Gomez H, Humphries WHt, Morrisroe S, Volpe JK, Kellum JA. Insulin-like growth factor binding protein 7 and tissue inhibitor of metalloproteinases-2: differential expression and secretion in human kidney tubule cells. American journal of physiology Renal physiology. 2017;312:F284–F296. doi: 10.1152/ajprenal.00271.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyal E, Lum G, Bahar I. The anisotropic network model web server at 2015 (ANM 2.0) Bioinformatics. 2015;31:1487–1489. doi: 10.1093/bioinformatics/btu847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann Angeli JP, Schneider M, Proneth B, Tyurina YY, Tyurin VA, Hammond VJ, Herbach N, Aichler M, Walch A, Eggenhofer E, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nature cell biology. 2014;16:1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gata JL, Pinto MC, Macias P. Lipoxygenase activity in pig muscle: Purification and partial characterization. J Agr Food Chem. 1996;44:2573–2577. [Google Scholar]
- Green AR, Barbour S, Horn T, Carlos J, Raskatov JA, Holman TR. Strict Regiospecificity of Human Epithelial 15-Lipoxygenase-2 Delineates Its Transcellular Synthesis Potential. Biochemistry. 2016;55:2832–2840. doi: 10.1021/acs.biochem.5b01339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper JT, Robinson CV. Mass spectrometry quantifies protein interactions--from molecular chaperones to membrane porins. Angew Chem Int Ed Engl. 2014;53:14002–14015. doi: 10.1002/anie.201403741. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Ji J, Kline AE, Amoscato A, Samhan-Arias AK, Sparvero LJ, Tyurin VA, Tyurina YY, Fink B, Manole MD, Puccio AM, et al. Lipidomics identifies cardiolipin oxidation as a mitochondrial target for redox therapy of brain injury. Nature neuroscience. 2012;15:1407–1413. doi: 10.1038/nn.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan VE, Mao G, Qu F, Angeli JP, Doll S, Croix CS, Dar HH, Liu B, Tyurin VA, Ritov VB, et al. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat Chem Biol. 2017;13:81–90. doi: 10.1038/nchembio.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalms J, Banthiya S, Galemou Yoga E, Hamberg M, Holzhutter HG, Kuhn H, Scheerer P. The crystal structure of Pseudomonas aeruginosa lipoxygenase Ala420Gly mutant explains the improved oxygen affinity and the altered reaction specificity. Biochim Biophys Acta. 2017;1862:463–473. doi: 10.1016/j.bbalip.2017.01.003. [DOI] [PubMed] [Google Scholar]
- Kellum JA, Sileanu FE, Bihorac A, Hoste EA, Chawla LS. Recovery after Acute Kidney Injury. Am J Respir Crit Care Med. 2017;195:784–791. doi: 10.1164/rccm.201604-0799OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam AJ, St-Pierre F, Gong Y, Marshall JD, Cranfill PJ, Baird MA, McKeown MR, Wiedenmann J, Davidson MW, Schnitzer MJ, et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Chang YY, Yang LW, Bahar I. iGNM 2.0: the Gaussian network model database for biomolecular structural dynamics. Nucleic Acids Res. 2016;44:D415–422. doi: 10.1093/nar/gkv1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl E, Hess B, van der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J Mol Model. 2001;7:306–317. [Google Scholar]
- Linkermann A, Brasen JH, Darding M, Jin MK, Sanz AB, Heller JO, De Zen F, Weinlich R, Ortiz A, Walczak H, et al. Two independent pathways of regulated necrosis mediate ischemia-reperfusion injury. Proc Natl Acad Sci U S A. 2013;110:12024–12029. doi: 10.1073/pnas.1305538110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkermann A, Skouta R, Himmerkus N, Mulay SR, Dewitz C, De Zen F, Prokai A, Zuchtriegel G, Krombach F, Welz PS, et al. Synchronized renal tubular cell death involves ferroptosis. Proc Natl Acad Sci U S A. 2014;111:16836–16841. doi: 10.1073/pnas.1415518111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The MARTINI force field: coarse grained model for biomolecular simulations. J Phys Chem B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- Martin-Sanchez D, Ruiz-Andres O, Poveda J, Carrasco S, Cannata-Ortiz P, Sanchez-Nino MD, Ruiz Ortega M, Egido J, Linkermann A, Ortiz A, et al. Ferroptosis, but Not Necroptosis, Is Important in Nephrotoxic Folic Acid-Induced AKI. Journal of the American Society of Nephrology: JASN. 2017;28:218–229. doi: 10.1681/ASN.2015121376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan AH, Dioszeghy V, Maskrey BH, Thomas CP, Clark SR, Mathie SA, Lloyd CM, Kuhn H, Topley N, Coles BC, et al. Phosphatidylethanolamine-esterified eicosanoids in the mouse: tissue localization and inflammation-dependent formation in Th-2 disease. J Biol Chem. 2009;284:21185–21191. doi: 10.1074/jbc.M109.021634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network VNARFT, Palevsky PM, Zhang JH, O’Connor TZ, Chertow GM, Crowley ST, Choudhury D, Finkel K, Kellum JA, Paganini E, et al. Intensity of renal support in critically ill patients with acute kidney injury. N Engl J Med. 2008;359:7–20. doi: 10.1056/NEJMoa0802639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnitskaya AN, Eddy NA, Fenteany G, Gascon JA. Recognition and reactivity in the binding between Raf kinase inhibitor protein and its small-molecule inhibitor locostatin. J Phys Chem B. 2012;116:10176–10181. doi: 10.1021/jp303140j. [DOI] [PubMed] [Google Scholar]
- Serhan CN. Pro-resolving lipid mediators are leads for resolution physiology. Nature. 2014;510:92–101. doi: 10.1038/nature13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemon AN, Heil GL, Granovsky AE, Clark MM, McElheny D, Chimon A, Rosner MR, Koide S. Characterization of the Raf kinase inhibitory protein (RKIP) binding pocket: NMR-based screening identifies small-molecule ligands. PLoS One. 2010;5:e10479. doi: 10.1371/journal.pone.0010479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skouta R, Dixon SJ, Wang J, Dunn DE, Orman M, Shimada K, Rosenberg PA, Lo DC, Weinberg JM, Linkermann A, et al. Ferrostatins inhibit oxidative lipid damage and cell death in diverse disease models. J Am Chem Soc. 2014;136:4551–4556. doi: 10.1021/ja411006a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparvero LJ, Amoscato AA, Fink AB, Anthonymuthu T, New LA, Kochanek PM, Watkins S, Kagan VE, Bayir H. Imaging mass spectrometry reveals loss of polyunsaturated cardiolipins in the cortical contusion, hippocampus, and thalamus after traumatic brain injury. J Neurochem. 2016;139:659–675. doi: 10.1111/jnc.13840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavel L, Jaquillard L, Karsisiotis AI, Saab F, Jouvensal L, Brans A, Delmas AF, Schoentgen F, Cadene M, Damblon C. Ligand binding study of human PEBP1/RKIP: interaction with nucleotides and Raf-1 peptides evidenced by NMR and mass spectrometry. PLoS One. 2012;7:e36187. doi: 10.1371/journal.pone.0036187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toppo S, Flohe L, Ursini F, Vanin S, Maiorino M. Catalytic mechanisms and specificities of glutathione peroxidases: variations of a basic scheme. Biochim Biophys Acta. 2009;1790:1486–1500. doi: 10.1016/j.bbagen.2009.04.007. [DOI] [PubMed] [Google Scholar]
- Uderhardt S, Herrmann M, Oskolkova OV, Aschermann S, Bicker W, Ipseiz N, Sarter K, Frey B, Rothe T, Voll R, et al. 12/15-lipoxygenase orchestrates the clearance of apoptotic cells and maintains immunologic tolerance. Immunity. 2012;36:834–846. doi: 10.1016/j.immuni.2012.03.010. [DOI] [PubMed] [Google Scholar]
- Wen X, Peng Z, Li Y, Wang H, Bishop JV, Chedwick LR, Singbartl K, Kellum JA. One dose of cyclosporine A is protective at initiation of folic acid-induced acute kidney injury in mice. Nephrol Dial Transplant. 2012;27:3100–3109. doi: 10.1093/ndt/gfr766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012;18:716–725. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- Wu Y, Li Q, Chen XZ. Detecting protein-protein interactions by Far western blotting. Nat Protoc. 2007;2:3278–3284. doi: 10.1038/nprot.2007.459. [DOI] [PubMed] [Google Scholar]
- Xu S, Mueser TC, Marnett LJ, Funk MO., Jr Crystal structure of 12-lipoxygenase catalytic-domain-inhibitor complex identifies a substrate-binding channel for catalysis. Structure. 2012;20:1490–1497. doi: 10.1016/j.str.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung K, Seitz T, Li S, Janosch P, McFerran B, Kaiser C, Fee F, Katsanakis KD, Rose DW, Mischak H, et al. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature. 1999;401:173–177. doi: 10.1038/43686. [DOI] [PubMed] [Google Scholar]
- Zhao J, O’Donnell VB, Balzar S, St Croix CM, Trudeau JB, Wenzel SE. 15-Lipoxygenase 1 interacts with phosphatidylethanolamine-binding protein to regulate MAPK signaling in human airway epithelial cells. Proc Natl Acad Sci U S A. 2011;108:14246–14251. doi: 10.1073/pnas.1018075108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
An overall bending motion enabled by the interfacial hinge region is observed, shown from the front view. PEBP1 and 15LO1 are colored as yellow and grey transparent spheres, respectively. Ribbon diagram colors indicate the change in position driven by ANM mode 1 (coded from blue (most rigid) to red (most flexible)). The movement allows PEBP1 to come into close proximity with the catalytic region of 15LO1.
(A) Cross-linking by glutaric dialdehyde (GDA) confirms the interaction of 15LO1 with wt PEBP1 and suppressed interaction with P112E mutant PEBP1. Samples containing 15LO1 and wt PEBP1 or P112E mutant PEBP1 were treated with 0.1% glutaraldehyde for 20 min at room temperature and the reaction was stopped by the addition 200 mM of Tris–HCl (pH 7.5). Samples were run in SDS-PAGE. Proteins were detected by staining with silver or by incubation with anti–PEBP1 antibodies after electro-transfer (see Figure 1B). Cross-linked bands appeared after GDA treatment of PEBP1/15LO1 mixture (see Figure 1B), but not after treatment of PEBP1 or 15LO1 alone. The cross-linking was suppressed for P112E mutant PEBP1 vs wt PEBP1. Data are mean ± SD, *p < 0.05 vs. wt PEBP1, N=5/group.
(B) Computational modeling of PEBP1-15LO2 interactions. Human PEBP1 (red)/15LO2 (gray) complex near a POPE/POPC lipid membrane. The hydrophobic heads of the lipid molecules are displayed as green spheres (lower leaflet only). The model contains over 170,000 atoms including water, lipids, and ions. Water molecules and the remaining portions of the lipid bilayer have been deleted for clarity. The catalytic site residues on 15LO2 (H373, H378, H553) are highlighted in blue and enclosed in a black square. The conserved active site residues on PEBP1, H86 and S153 are highlighted in pink and yellow, respectively, and labeled.
(C) Coarse-grained Molecular Dynamics (CGMD) simulations of PEBP1-15LO binding in the presence of a phospholipid bilayer. Two independent runs were performed for each pair of proteins, 15LO1/PEBP1 (left; top and bottom) and 15LO2/PEBP1 (right top and bottom), using the MARTINI force field. In the first run, the complex was placed at 2.5 nm (top) separation from the lipid bilayer, and in the second, the complex was placed at 0.5 nm (bottom) away from the bilayer. The dotted line represents the minimum distance required for an interaction to take place. The distance between the PEBP1-15LO1/LO2 complex and the lipid bilayer show large fluctuations in the top two panels, coming into close proximity multiple times during the total duration (> 500 ns) of the simulations. The bilayer consisted of 50% POPC and 50% POPE with a total of 127 lipids in each leaflet. The inset shows the snapshots of the complex and the bilayer at the indicated times. The red spheres represent PEBP1, and the yellow and blue (b-barrel) spheres represent 15LO1/15LO2. Water molecules (included in simulations) are not shown for clarity.
(D) Coarse-grained molecular dynamics simulations of PEBP1/15LO2 binding in solution. Results from docking simulations performed for the complexation of PEBP1 with 15LO2. The simulations were performed using the MARTINI force field. PEBP1 was placed at −2.5 nm (top) and +2.5 nm (bottom) from 15LO2, in the absence of lipid bilayer. PEBP1 moves towards 15LO2 and forms a tight complex within 60–80 nm, and the complex remains stable during the rest of simulations (up to 150 ns).
(E) Comparison of the binding of wt PEBP1 to 15LO1 in silico shows the weaker affinity and distinctive binding pose of the P112E mutant. Left panel displays the optimal binding poses for wt PEBP1. The right panel shows the interface in greater detail, where wt PEBP1 exhibits several close contacts (atom-atom contact distances given). PEBP1 and 15LO1 residue labels are colored blue and green, respectively.
(F) Accumulation of PE-OOH species in PC/PE liposomes catalyzed by 15LO2 in the absence and in the presence of either wt PEBP1 or P112E mutant PEBP1. Data are mean ± SD, *p < 0.05 vs. control (no PEBP1), **p < 0.05 vs. wt PEBP1, N=5/group (for control and PEBP1), N=3/group (for P122E mutant).
(G and H) Results from coarse-grained MD simulations confirm the inability of human wt PEBP1 to stably bind Pseudomonas aeruginosa 15LOXA at the allosteric site. Results from docking simulations (G) and two independent coarse-grained MD runs CGMD1 and CGMD2 (H) are presented. In panel A, the two proteins are represented using ribbon diagrams and the N-terminal helix of P. aeruginosa 15LOXA and C-terminal helix of wt PEBP1 are labeled and colored red and blue, respectively. Comparison to Figure 1E shows the distinctive binding pose of PEBP1, as indicated by the position of the wt PEBP1 C-terminal helix. In panel B, the ordinate denotes the closest distance of approach between the two proteins, which originally were separated by 20 Å. During the CGMD simulations, although the two proteins find a stably bound pose at approximately 50 or 160 ns as evidenced by the fixed (minimal) distance, the binding poses (space-filling representations in insets) are different from those observed in wt PEBP1 and 15LO1.
Table S1. Basic kinetic parameters for AA and AA-PE oxygenation by 15LO1, 15LO2 and LoxA in the absence and in the presence of PEBP1, Related to Figure 6.
Data are mean ± SD
Table S2. “Multiple docking” calculations of arachidonic acid (AA) to PEBP1 mutants, Related to Figure 6.
(a) DDG = change in binding free energy with respect to that of wt PEBP1 (i.e. DG (mutant) – DG (wt). A positive value indicates an increase in binding energy, which is unfavorable.
* Number of residues located with 3.5 Å from all nine AA ligands and present in a wt PEBP1.
↑ Excluding AAs (energies colored in red) which are interacting only with AA (lack of residues within 3 Å)
Table S3. Results on AA-binding (to PEBP1 (case 1) or Y176X (case 2)) energetics(a), Related to Figure 6.
(a) The entries in the table show the interaction energy between wt PEBP1 or Y176X mutant PEBP1 and AA (or AA-amide) molecules, upon binding one small molecule at a time, up to 9 molecules binding. Red entries refer to interactions of AA (or AA-amide) molecules with previously bound AA (or AA-amide) molecules; gray entries are the same, except for the involvement of 1 PEBP1 residues. (b) Total DG is obtained by summing up all the black entries in the corresponding run. (c) Average change in free energy, relative to that obtained for the wt PEBP1 + 9 AA molecules
Movie represents the ANM mode 1 shown in Movie S1, displayed from a different perspective
(A) Western blots of 15LO2 in HT22 cells. Human recombinant 15LO2 protein (Cayman Chemicals) is presented as a positive control. (B) Western blots of 15LO2 in HK2 cells. Human recombinant 15LO2 protein (Cayman Chemicals) is presented as a positive control.
(C) Pie charts showing % of 15LO1, PEBP1, and co-localized puncta normalized to the total number of puncta in HEAC cells with or without IL13 treatment.
(D) Pie charts showing % of 15LO2, PEBP1, and co-localized puncta normalized to the total number of puncta in HK2 cells with or without LPS treatment.
(E) Co-localization of PEBP1, 15LO1 and GPX4 in HAECs treated with IL13. The upper panel shows a 3D volume view of 15LO1 (green), PEBP1 (red), GPX4 (pink) and nuclei (blue) in IL13-treated cells. The lower panel shows the co-localized puncta (objects, white) identified by object based segmentation and co-localization analysis. (F) The number of objects identified as positive for 15LO1, PEBP1, GPX4 or complexes that were positive for all three proteins 15LO1, PEBP1 and GPX4 (co-localized) in HAECs treated with IL13. Media control=4±3, IL13 823±693, p≤0.05 vs. untreated, N=3/group.
(G) Structural and functional interdependency of 15LO1, PEBP1 and GPX4. Upper panel: IL13 (10 ng/ml) for 72 h induced both 15LO1 and GPX4 in parallel in HAECs, with KD of PEABP1 and to a lesser degree 15LO1 reducing GPX4 levels, further supporting their interdependency. GPX4 KD decreased both 15LO1 and PEBP. Lower panel: GPX4 KD in HAECs led to the expected increase in cell death.
(A) Effect of RSL3 (750 nM, 5 h) in the absence or in the presence of ferrostatin-1 (FER, 0.4 μM) on the accumulation of PE oxygenated species in PHKCs. Scatter plot of changes in the levels of oxygenated PE species showing log2(fold-change) vs significance (−log10 (p value)), N=3/group.
(B) Effect of LPS (50 μg/ml, 24 h) in the absence or in the presence of RSL3 (750 nM, 5 h) and ferrostatin-1 (FER, 0.4 μM) on the accumulation of PE oxygenated species in PHKCs. Scatter plot of changes in the levels of oxygenated PE species showing log2(fold-change) vs significance (−log10 (p value)), N=3/group
(C) Effect of a ferroptosis inhibitor, ferrostatin (FER, 1 μM), on RSL3 (10 μM) induced cell death in HAECs. Data are mean ± SD, *p < 0.05 vs control; **p < 0.05 vs. RSL3, N=3/group. (Insert) Western blot analysis shows the increased expression of GPX4 following IL13 (10 ng/ml) in fresh bronchial epithelial cells.
(D) Effect of FER (0.4 μM) on RSL3 (50 nM, 24 h) induced death of HT22 cells. Data are mean ± SD, *p < 0.05 vs. control; **p < 0.05 vs. RSL3, N=3/group. (Insert) Western blot analysis demonstrates high expression of GPX4 in HT22 cells, M: molecular weight marker.
(E) Effect of different ferroptosis inhibitors on RSL3 (200 nM, 24 h)-induced death in PHKCs. Conditions: ferroptosis inhibitors: FER (0.2 μM); deferoxamine (DFO, 25μM); LO15 inhibitors: ML351 (0.5 μM); PD146176 (0.5 μM). Data are mean ± SD, *p < 0.05 vs. control; #p < 0.05 vs. RSL3, N=3/group. (Insert) Western blot analysis demonstrates high expression of GPX4 in PHKC cells. PHKCs were isolated from proximal tubule epithelial cells by immuno-affinity method.
(F) Effect of different ferroptosis inhibitors on RSL3 induced death in HK2 cells. Cells were exposed to RSL3 (200 nM, 24 h) in the absence or in the presence of the ferroptosis inhibitors ferrostatin (FER, 0.2 μM) and deferoxamine (DFO, 25 μM) and vitamin E (vit E, 25 μM), as well as a 15LO inhibitor, PD146176 (0.5 μM). Data are mean ± SD. *p < 0.05 vs. control; #p < 0.05 vs. RSL3. N=4/group. (Insert) Western blot analysis demonstrates high expression of GPX4 in HK2 cells and effects of RSL3 (200 nM, 24 h) on the expression of GPX4.
(G). IL13-dependent ferroptosis in RSL3- and AA-treated HAECs. Human airway epithelial cells (HAECs) with or without IL13 (10ng/ml) were incubated with AA (10 μM) and then treated with RSL3 (10 μM) in the presence or absence ferrostatin-1 (1 μM). Cell death was assessed 20–24 h after treatment with RSL3 by the amount of LDH released. Data are mean ± SD, *p < 0.05 vs. control; #p < 0.05 vs. RSL3 + AA, No IL13, N=3/group.
(H) Effects of IL13 and RSL3 on the accumulation of PEox in HAECs. Incubation (72 h) conditions: IL13 (10 ng/ml), RSL3 (10 μM) or their combination. Scatter plot of changes in the levels of oxygenated PE species (log2 (fold-change) vs. significance (−log10 (p-value), (N=3/group or 4/group).
(I) HAECs with KD PEBP1 contain higher levels of free AA but not AA esterified into PE. Closed bars indicate the number of samples with increased or decreased AA (blue) or AA-PE (red) after PEBP1 KD (primary y-axis). Open bars depict the average changes relative to control ± SEM (secondary y-axis). The number of samples with increased AA was significantly different from the number of samples with increased AA-PE (p=0.029, z-test): 40±16% increase in AA while the changes in AA-PE were −12±13% after PEBP1 KD.
Mouse lung epithelial (MLE-12) or HT22 cells were treated with RSL3 (250 nM or 50 nM, respectively) in the presence or absence of a ferroptosis inhibitor, ferrostatin 1 (0.4 μM), for 20 h. Effect of RSL3 was monitored by LDH release (A and D), Propidium iodide (PI) staining (B and E) using flow cytometry, or MTT assay (C and F). Data are mean ± SD, *p < 0.05 vs. control, N=3/group.
(A) PEBP1 KD in HT22 cells using siRNA. Bar graphs show quantitative assessments of PEBP expression. Data are mean ± SD, *p < 0.05 vs. control or scrambled siRNA, N=3/group. (Insert) Representative western blot in control, PEBP1-siRNA treated cells, and scrambled siRNA treated cells.
(B) Ferroptotic death of RSL3-treated MLE cells induced by exogenously added 15LO2, AA-PE and PEBP1. Cell death was determined as the difference between the total levels of death and ferrostatin-1 eliminatable cell death (see Figure 4F). Data are mean ± SD, *p < 0.05 vs. RSL3, N=3/group.
(C) Ferroptotic death of RSL3-treated MLE cells induced by exogenously added 15LO2, AA-PE and PEBP1 P112E mutant. Cell death was determined by LDH release. Data are means ± SD, *p < 0.05 vs. RSL3, N=3/group.
(D) RSL3-induced ferroptosis in MLE cells transfected with wt PEBP1 or PEBP1 P112E mutant. Cells were treated with RSL3 (200nM, 24h) in the presence or absence of FER (ferrostatin, 0.4 μM). Cell death was determined by LDH release. Data are means ± SD, *p<0.05 vs control; #p<0.05 vs RSL3 treated cells transfected with wt PEBP1. N=3/group.
(E) Western blots documenting expression of FLAG-tagged PEBP1 in MLE cells and HT22 cells using anti-PEBP1 anti-body. pCMV6 – empty vector; pCMV6-PEBP1- plasmid containing PEBP1.
(F) Westerns blots revealing the presence of Flag-tagged PEBP1 using anti-FLAG anti-body in eluates of MLE and HT22 cells after immuno-precipitation using anti-FLAG M2 agarose beads.
(G) Typical base profile of LC-MS signals corresponding to AA in eluates after immune-precipitation of PEBP1 from MLE cells and HT22 cells. Four independent experiments with each type of cells have been performed.
(A) The detailed interactions of each of the nine AA with PEBP1 in the PEBP1-AA9 complex. Each panel shows the interaction between respective AA (cyan) and PEBP1 (gray). In each panel, PEBP1 residues that are in close proximity (within 5 Å) of the respective AA (indicated by red arrow) are labeled in blue and shown as stick representations. In some of the panels, other AA which occupy the same binding site as the cyan one are indicated in different colors (e.g. red, blue or green in AA2, AA4, AA5 and AA6).
(B) Examination of the stability of PEBP1-bound states of AA in two sets of independent runs, MD1 (left 3x3 panels) and MD2 (right 3x3 panels). In each case the time evolution of the instantaneous position of the mass center is shown for the labeled AA over simulations of 200 ns. The original binding energy in kcal/mol is shown in parentheses. The majority of the binding poses are highly stable: even if there are occasional rearrangements, the poses are quickly restored. AA8 and AA5 are relatively unstable.
(C) Decreased AA-binding affinity of PEBP1 with the truncated C-terminal helix (Y176X mutant). Results from five independent runs are displayed in each case, each run consisted of successive bindings of up to nine AAs. Left panel displays the binding sites (or corresponding clusters of bound AAs) observed in those multiple runs (see color code in the inset). Right panel displays the same results for the Y176X mutant PEBP1. Two of the clusters (C2 and C3) observed to bind AAs in the wt PEBP1 disappeared in Y176X, due to the elimination of the C-terminal helix that otherwise showed high AA-binding propensity. Clusters C1 and C2 are maintained in both cases, although C1 appears to be more broadly distributed in the mutant. Cluster C5 shows a reduced AA-binding probability in the mutant. PEBP1 residues that exhibited high AA-binding propensities are shown in space-filling (cyan) representations. Simulations showed a decrease from nine to six AAs bound to PEBP1 in the Y176X mutant protein.
(D) Correlation cosines between the ANM modes of motions accessible to 15LO1 in isolation (ordinate) and those accessible in its complex with PEBP1 (abscissa). The subset of the 10 softest (most cooperative) modes of motions are shown in each case. The correlations are color coded, from red (strongest) to blue (weakest). Note that the soft modes (e.g. modes 1–6) accessible to the uncomplexed 15LO1 are usually maintained in the complex with slight perturbations (cumulative correlation > 0.7, in general). However, the softest mode accessible in the complex (indicated by the arrow) is unique to the complexed form. The shape of this mode is illustrated in Fig 6. See also the Supplementary Movie S1 for the cooperative movements of 15LO1 and PEBP1 driven by this mode.
(E–I) A cooperative mode of motion induced by PEBP1/15LO1 complexation favors AA-PE binding near the catalytic site of the enzyme.
(E) Mode profile calculated with GNM. The curve shows the relative motions of different regions of 15LO1 in a complex with PEBP1 along the principal mode of motion gained in the complex. Positive (red) and negative (blue) regions indicate the structural regions subject to anticorrelated (coupled but opposite direction) motions. The zero-crossing points serve as hinges (white) between anti-correlated regions. These include 15LO1 K156, D173 and R402, interacting with PEBP1 P112, D144, Y181.
(F) Ribbon diagram of 15LO1/PEBP1 complex, color-coded by substructures that undergo anticorrelated movements in the GNM principal mode.
(G) Cross-correlation map describing the pairs of structural elements undergoing correlated (dark red) and anticorrelated (dark blue) movements in the cooperative motion acquired in the complex.
(H and I) Comparison of binding poses of AA-PE onto the PEBP1/15LO1 complex, obtained for the rigid/static complex (H) and dynamic/flexible complex (I) The results from 10 runs for each frame (total of 10 frames along ANM mode 1) are superimposed in panel E. The optimal (highest-scoring or lowest energy) binding pauses of AA-PE molecules are displayed with licorice representation (carbons in cyan, oxygens in red and nitrogens in blue). Three binding sites (hot spots) enclosed in ellipses/circles are observed for the rigid docking case (H). Notably, two additional hot spots, enclosed in red ellipses/circles, are observed when the complex is allowed to reconfigure along mode 1: one at the interface between PEBP1 and 15LO1, and the second in close proximity of the active site (H). The active site is indicated by the arrow (Fe2+ (pink sphere) coordinated by 4 histidines (green sticks)). Docking simulations were performed using Smina, visualization using VMD, and clustering using ProDy.
(A) AA-containing PE-species are abundant in airway epithelium. Mouse lungs (left lobe) were inflated and embedded with 8% gelatin in order to be MALDI-compatible and sectioned on special conductive ITO (indium tin oxide) slides for MALDI-MS imaging with DHB (2,5-dihydroxybenzoic acid) matrix. Semi-serial H&E stained section (left) shows that morphology is preserved. MALDI positive ion mode (center) shows highly saturated species (such as PC(32:0)) localizing broadly, while species containing 20:4 or 22:4 (such as PE(38:4) and PE(40:5)) localizing to the epithelium. MALDI negative ion mode (right) detects 20:4-containing PI(38:4) that also localizes to the epithelium. Scale bar = 500 μm. Ions are shown as a rainbow heat map of relative intensities ranging from black-blue (low intensity) to red-white (high intensity). MALDI-MS imaging was performed on a Bruker Ultraflex II instrument at a spatial resolution of 50 μm per pixel. Ions were detected as [M+K]+ in positive mode and [M−H]− in negative mode.
(B) AA-containing PE species are abundant in kidney tubular epithelium. High resolution MALDI-MS images of rat kidney lipids were generated using a Bruker Daltonics rapifleXtm MALDI Tissuetypertm mass spectrometer. Fresh-frozen kidneys from 17-day old male rats were sectioned coronally and placed onto special conductive ITO (indium tin oxide) slides for MALDI-MS imaging with DHB (2,5-dihydroxybenzoic acid) matrix. Note the differences in localizations of pro-ferroptotic precursors PE species vs non-pro-ferroptotic PEs as well as non-pro-ferroptotic PC species. Precursors of pro-ferroptotic signals: arachidonoyl-PE species PE(38:4); PE(40:6); PE(40:4); non-pro-ferroptotic PE species: PE(34:0); PE(36:2); non- pro-ferroptic PC species; PC(32:0); PC(34:1). The top left panel shows an overlay of single-color ion intensity images of pro-ferrototic PE(38:4) (red) and non-pro-ferroptotic PC(34:1) (green) which confirms separate localizations. Scale bar = 500 μm. Ion intensities are shown as a rainbow heat map of absolute intensities ranging from black-blue (low intensity) to red-white (high intensity). Analysis was performed in positive ion mode with a spatial resolution of 5 microns per pixel. Ions were detected as [M+K]+.
(C) AA-containing PE species are abundant in rat brain. MALDI-MS imaging of rat brain PE. Left. Optical image of an H&E stained semi-serial tissue section to confirm cutting depth; Right. MALDI-MS image of 20:4-containing PE(38:4) localization, displayed as a relative intensity (0 % to 100 %) rainbow heat map. Scale bar = 1000 μm.
(D) Representative spectrum of PEs in HT22 cells from LC-MS/MS analysis. Ions containing at least one AA species are labeled in red. Inset: Pie chart showing the fraction of total PE in which at least one of the species contains AA.
(E–H) Fragmentation spectra of PE species such as PEp(36:5) (E), PEp(38:4) (F), PE(38:4) (G), PE(40:6) (H) confirming the integrated AA-residue. The 303.233 (marked in red) corresponds to the AA ion.
(I) Table showing the prominent AA-containing PE species in HT22 cells (more than 1% of total PEs)
(A) Ferrostatin-1 (FER) attenuated cell death in folic acid-induced acute kidney injury (AKI). CD-1 mice were pretreated with ferrostatin-1 (FER) 60 minutes before AKI induction by an overdose of folic acid, and euthanized at 48 h. (A) Assessment of cell death by TUNEL.
(B) Quantification of TUNEL positivity. Mean ± SD. *p <0.05 vs. control and AKI+FER, one-way ANOVA on ranks, since data were not normally distributed. N=6/group.
(C) Folic acid induced changes (left panel) in PE species containing oxygenated arachidonic acid.
(D) Folic acid induced accumulation of PE oxygenated molecular species in mouse kidney. Data are mean ± SD, *p < 0.05 vs. control, **p < 0.05 vs Folic acid plus FER, N=6/group.
(E) Assessment of GPX4 activity in brain cortex show decreased activity at 4 h after CCI. The activity measurements in brain cortex homogenates were performed in 0.1 M Tris–HCl (pH 8.0), containing 0.5 mM EDTA and 1.25 % Triton X-100, 0.12 mM NADPH, 3 mM glutathione, glutathione reductase (1 U/ml), and 0.12 mg of protein. Hydroperoxy-tetralinoleoyl-cardiolipin (final concentration 50 μM) was used as a substrate. Amount of consumed NADPH was assessed by fluorescence (λex 340 nm, λem 460 nm). The amount of oxidized GSH was calculated based on the amount of NADPH consumed during GSSG reduction.
(F). Changes of ferroptotic protein expression in the hippocampus after CCI. 15LO2 expression was significantly elevated 4 h after CCI (*p < 0.05 vs. naïve, N=3–4/group). PEBP1 and GPX4 were not significantly changed (p < 0.05; N=3–4/group) in the ipsilateral hippocampi of injured vs. naïve rats. Data are mean ± SD. Inset shows representative western blot images of each protein.