Abstract
A wide variety of experimental techniques can be used for understanding the precise molecular mechanisms underlying the activities of cellular assemblies. The inherent limitations of a single experimental technique often requires integration of data from complementary approaches, to gain sufficient insights into the assembly structure and function. Here, we review popular computational approaches for integrative modelling of cellular assemblies, including protein complexes and genomic assemblies. We provide recent examples of integrative models generated for such assemblies by different experimental techniques, especially including 3D electron microscopy (3D-EM) and HiC data, respectively. We highlight general concepts in integrative modelling and discuss the need for careful formulation and merging of different types of information.
Keywords: integrative modelling, macromolecular assemblies, genome organization, hybrid methods, structure modelling, cellular assemblies
Introduction
Understanding the structure and dynamics of the interacting biomolecules can give great insights into the cell’s function and integrity. These macromolecules are organized at different levels, from small assemblies (involving 2–3 proteins) to very large ones (tens to hundreds of proteins), some of which are organised further, at the supramolecular level: for example, the assembly of ribosomes into polysomes, the assembly of viral particles, and on an even larger scale the organization of chromosome structures and entire organelles such as the nucleome.
Unfortunately, structure determination of assemblies, in particular large ones, at resolutions that enable meaningful function inference is not always viable using a single biophysical technique. This problem can be overcome if the information from several techniques (e.g., assembly subunit composition, topology and overall architecture and dynamics) is combined efficiently using a variety of computational methods. Integrative modelling based on complementary experimental data is increasingly being used as the strategy to compute structural models for large cellular assemblies. Multiple sources of structural information can reduce the ambiguity and noise associated with low-resolution and/or heterogeneous data. In this review, we focus on popular experimental techniques and computational methods used in integrative modelling of cellular assemblies.
Sources of spatial information
High-resolution structures of cellular assemblies have been traditionally characterized in vitro by X-ray crystallography, the success of which is often limited by the ability to form crystals (Fig. 1). Cryo electron microscopy (cryo EM) techniques have been generally used to determine the structures of large assemblies (above ~150–200kD) that could not be crystallised (with the advantage of being carried out at near-native conditions) though at substantially lower resolutions. However, recent developments (including direct electron detectors and advanced image processing methods) have brought a substantial increase in subnanometer and near-atomic resolution structures even of smaller size [1].
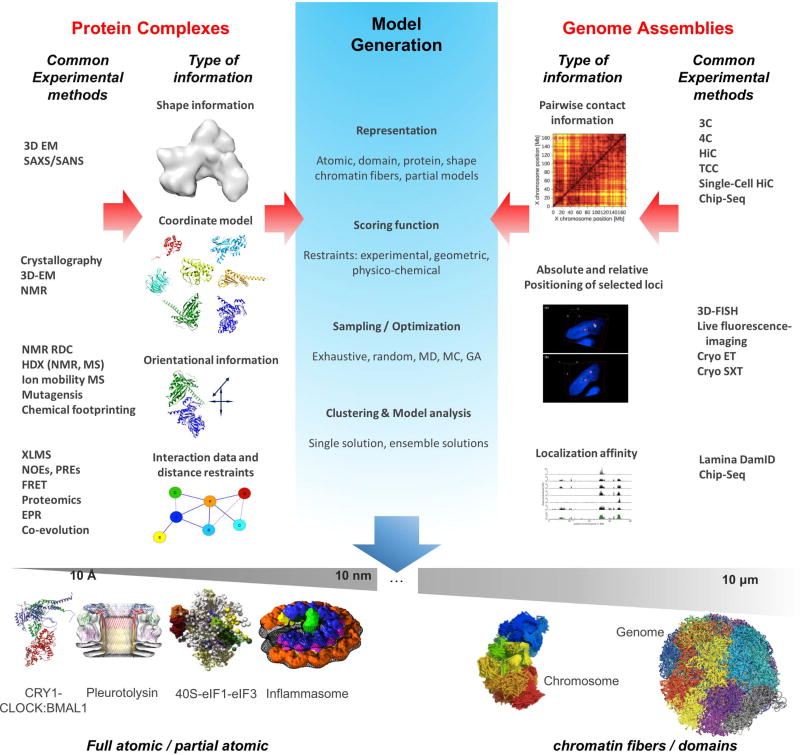
Figure 1.
Top: Examples of different types of information from various experimental sources that are integrated for structure determination of protein complexes and genome assemblies. The important steps involved in integrative modeling process are highlighted in the central panel (model generation).
Bottom: Examples of recently-determined integrative models of cellular assemblies across resolution and size scales (from sub-nanometer to ~10 micron range). From left to right: transcription repressive complex CLOCK:BMAL1 (brain and muscle Arnt-like protein 1) bound to CRY1 (cryptochrome-1) (adapted from [78]), atomic model built based on data from SAXS, NMR, Size Exclusion Chromatography and X-ray crystallography; structure of the fungal toxin Pleurotolysin (adapted from [38]), model built based on 11 Å resolution cryo EM map and X-ray crystallography; model of 40S-eIF1-eIF3 complex built using data from, XL-MS and X-ray crystallography (~25 Å negative stain EM map used for validation) (adapted from [9]); model of flagellin-induced NAIP5/NLRC4 inflammasome built based on data from 4nm resolution subtomogram average and X-ray crystallography (adapted from [79]); Ensemble of mouse X chromosome conformations from single cell HiC at 500kb resolution (adapted from [74]); example of a genome model for haploid mouse embryonic stem cells from HiC experiments using chain particles representing 100kb DNA (adapted from [75]).
Although at much lower resolutions, cryo electron tomography (cryo ET) allows in-situ visualization of cellular architectures as well as assembly structures they contain if sub-tomogram averaging is possible [2]. Using fluorescently labelled biomolecules, dynamic interactions of assemblies can also be studied in living cells by super-resolution microscopy, at resolutions in the order of 10nm.
Small-angle X-ray and Neutron scattering (SAXS and SANS) have proven useful in the structural characterization of assemblies under near-native solution conditions, providing information on pairwise electron (or nuclear) distances, which can be used for computing 3D shapes and related features [3]. Nuclear Magnetic Resonance (NMR) is typically limited by sample size, but for large assemblies it can provide information such as direct interactions between monomers and relative orientations [4]. Electron Paramagnetic Resonance (EPR) allows determination of distance distributions (typically 1–10nm) that are generally more precise compared to fluorescence based methods [5].
Mass Spectrometry (MS) approaches have been used to obtain spatial information, stoichiometry and connectivity on entire assemblies in the gas phase (assuming they maintain their interactions) with the advantage of dealing with limited sample amounts. Advancements in protein and peptide separation techniques in MS make the method tolerant to a high degree of sample heterogeneity [6] and samples involving small protein mixtures and even large viral assemblies can be characterized. Ion mobility MS (IM-MS) allows separation of coexisting forms of the same complex. The time taken by an ion to traverse the ion mobility cell is related to its mass, charge, and their rotationally averaged collision cross-section (which gives a measure of the overall shape) [7]. Chemical cross-linking coupled to MS (XL-MS) is very popular as it can reveal interactions within or between biomolecules by providing an upper-limit distance between specific residues [8]. A number of methods have been used to calculate the optimal cross-link distances on a given atomistic model and score these against XL-MS data [9–12]. Hydrogen-Deuterium Exchange (HDX) experiments combined with either NMR or MS provide information on biomolecular interaction surfaces and conformational changes [4,13].
Protein interaction data can also be inferred using evolutionary approaches. Recent advances in the detection of correlated mutations in sequences across species enable prediction of residue pairs that interact not only within but also between proteins [14]. Experimental interaction data can be combined with evolutionary information, by mapping known interactions [15–17] and the 3D structure of the related complex if available [18,19].
Information about the nucleome organisation comes from recent technologies. For example, Chromosome Conformation Capture (3C) and related technologies (4C, 5C, HiC, in situ HiC, TCC) detect the genome-wide frequencies of spatial co-location between non-consecutive genomic regions in a population of cells [20]. Mapping of chromatin interactions is also possible in single cells, although at relatively lower coverage in comparison to ensemble HiC [21]. Lamina-DamID (DNA adenine methyltransferase identification) experiments provide probabilities with which chromatin regions are exposed to the lamina at the nuclear envelope.
Multi-color fluorescence in situ hybridization (FISH) uses fluorescent probes to detect the positions of targeted genomic loci in single cells; this decades old technique has lately witnessed impressive improvements in the number of simultaneous target loci, making it possible to trace entire chromosomes [22]. Finally, imaging technologies such as soft X-ray cryo tomography (cryo SXT) provide insights into spatial distributions of chromatin in different functional states [23], while cryo ET can reveal folding patterns of individual nucleosomal chromatin fibers.
Approaches to integrative modelling
It remains a major challenge to develop computational methods that can generate reliable models of cellular assemblies, ranging from small protein complexes to structural maps of entire organelles such as the nucleome. Below we describe modelling approaches driven by integration of different types of experimental data (Figure 1).
Protein complexes
Proteins and their assemblies are usually characterised by one or a few stable observable conformations under specific experimental conditions (except loops and disordered regions). A typical strategy for integrative modelling of protein assemblies (including domain-domain, protein-protein, protein-RNA or protein-DNA interactions) relies on minimizing a scoring function, which measures the agreement between the model and the experimental data (as well as penalizing violations from known standard geometries). The model can be represented as full-atomic, coarse-grained (e.g. secondary structure elements, domains, whole protein), or partial (ie. some parts of the model are missing). Experimental information is typically represented in the form of restraints that help in reducing the sampling space. The reliability of individual experimental restraints can be considered, for example, by adding weights in the scoring function or by selectively using subsets of them at different stages of the optimisation [24,25]. Alternatively, several initial models may be generated by using only a fraction of restraints (random subset) each time, the size of the fraction being a crude indicator of the reliability of restraints [26]. Integrative approaches yield a model or an ensemble of models that will have minimal restraints violations. The generated models can be clustered and ranked based on a score or a consensus among multiple scores and/or size of the clusters [27]. For cross validation, part of the experimental data can be excluded during the modelling process.
A classical example of integrative modeling is to obtain an atomic model from cryo EM data by fitting individual assembly components into the 3D map. This usually involves a global search to get the best fit between the model/s and the map, for which many programs have been developed [7,28], such as the Situs package (which can also fit models to SAXS data) [29]. Geometric features from the 3D-EM map can also be used as restraints, instead of directly fitting the subunit models into the EM density [30,31]. Interactive fitting of atomic models is popular, and is enabled by visualization tools like Coot [32] and Chimera [33]. Chimera provides methods for fitting atomic models in 3D shapes from cryo EM/ET through both global and local searches in a user-friendly and interactive manner.
TEMPy is a python package for integrative modeling primarily based on EM density [27] (but also crosslinking data). Simultaneous fitting of multiple components is carried out by a genetic algorithm, which uses the mutual information to account for the goodness-of-fit and penalises for steric clashes [34]. This is followed by a real-space refinement in the density using MODELLER/Flex-EM [35,36]. TEMPy integrates various scoring functions, including correlation-based scores, surface based-scores and statistical scores. In a recent application, low-resolution density from sub-tomogram averaging was combined with chemical cross-linking to identify the oligomerization state of the HSV-1 glycoprotein B assembly in vesicle membranes. Multiple methods of hierarchical constrained density-fitting were employed for building atomic models for the pre- and post-fusion conformations [37]. Models were generated and scored by TEMPy using an ensemble of alternate conformations and validated using insertion mutational data, revealing a surprising pre-fusion conformation. Integrative modelling using a combination of density fitting with other experimental data has also been useful in the intermediate resolution range [38,39].
Another related Monte-Carlo based method – PyRy3D [40] – uses a scoring function based on a combination of restraints, fitness to the shape data (EM, SAXS) and penalties for steric clashes. This method was recently used to model the structure of CCR4-NOT complex, based on EM data and component interaction restraints from X-ray and biochemical experiments [41]. POWER employs particle swarm optimization to predict the component arrangement in a symmetric assembly, based on a few geometric or distance restraints [42]. The program uses ensembles of subunits from molecular dynamics simulations or experiments to explore conformational changes of subunits during assembly optimization. Using this method, intrinsic flexibility derived from X-ray structures of subunits was integrated with cryo EM data of the assembly, to build near-atomistic models of aerolysin at different stages of pore formation [43].
A few integrative methods are also developed to work primarily on SAXS data [44]. For example, the ATSAS suite provides a collection of methods for processing and analysis of both SAXS and SANS data, including tools for integrative modelling and flexible optimization [45]. SAXS, like EM, is often used in combination with other techniques for integrative modelling. Recently, SAXS, HDX and RNA SHAPE (selective 2’-hydroxyl acylation analysed by primer extension) data based restraints were used to provide a dynamic model of a machinery involved in HIV-1 pro-viral transcription [46]. The SHAPE data highlights RNA segments that are potentially involved in interaction based on changes in reactivity of nucleotides upon assembly.
The Rosetta software suite also uses a Monte-Carlo approach for conformational sampling, which recombines frequently occurring protein backbone structural features with different types of restraints, such as sparse NMR data based on NOE, chemical shifts, and Residual dipolar coupling (RDCs, as orientation restraint) as well as XL-MS and cryo EM [25]. The reliability of NMR restraints is considered by their relative contributions in the scoring function. Distance constraints can be enforced (e.g., [47]) as part of the scoring function involving statistical interaction potentials and evaluation of standard geometry (atoms and secondary structures) and clashes. In a recent structure of S. flexneri type III secretion system (T3SS) needle several solid-state NMR (ssNMR) distance restraints were used to build models with a 7.7 Å cryo EM density map [48]. Intra and inter-subunit distance constraints from ssNMR experiment were integrated in iterative modeling and the final models were identified using a weighted sum of the Rosetta scoring function, number of constraint violations and correlation with EM density.
In Haddock, sparse experimental data can be added as ambiguous interaction restraints and random sets of these are used to generate initial configurations. Rigid-body protein-protein docking and energy minimization is followed by the Haddock flexible refinement, with an additional density cross-correlation score added to non-bonded interaction terms, for integrating EM data [49]. For example, chemical shift perturbations were used as restraints to map the potential active site and model the interaction between bacterial outer membrane receptor FusA and host cell ferredoxin using a comparative model of FusA and crystal structures of ferredoxin [50].
In IMP, assembly components can be represented as sets of particles of different types, depending on the available information and structural resolution. Such representations can be useful when some high-resolution information is missing (e.g., there are no atomic models for some of the proteins/domains in the complex). These components can be assembled using different optimization schemes (such as Monte Carlo, Conjugate Gradients, Molecular dynamics) and many different types of spatial restraints [24], such as connectivity and interaction restraints from various MS techniques [51]. A notable example of an IMP application is a model of the Yeast Mediator complex (YMC), which performs an essential regulatory role in eukaryotic transcription initiation [52]. Regions of known atomic structure or comparative models were modelled as rigid bodies, whereas unknown regions were represented as a flexible string of beads. Positions and orientations of rigid and polymer beads were iteratively sampled to minimize a scoring function which accounted for excluded volume, sequence connectivity, cryo EM, and crosslinking restraints. A Bayesian framework was used to assign confidence to subsets of cross-linking data based on relative consistency [9]. Finally, a single cluster with the best average score was chosen. The model was successfully validated with independent experimental data on protein–protein interactions. XL-MOD [53] also uses a Bayesian framework to reweight conflicting or ambiguous distance restraints from XL-MS, represented as log-harmonic potentials. An elastic network model based on a low-resolution representation of the subunit structures is used to sample conformational changes upon assembly and the component positions and orientations are sampled by a Monte-Carlo optimization scheme followed by relaxation using molecular dynamics.
Genome assemblies
A common challenge in resolving structures of genomes is the wide range of spatial and time scales involved compared to protein assemblies: the human genome counts ~6 billion basepairs in a diploid cell and its structure arguably never reaches thermodynamic equilibrium [54]. These complexities demand a level of coarse graining, ranging from models of the 10nm chromatin fiber to chromatin domains. Chromatin can be segmented into self-interacting chromatin domains (e.g. topologically associated domains (TAD) [55] or contact domains [56]), often bordered by chromatin loops. Chromatin domains with similar functional properties form sub-compartments and chromosomes organize in territories with stochastic preferences in their nuclear locations.
Recent physics-based polymer simulations of chromosomal regions revealed potential mechanisms into chromatin loop and TAD domain formation [57–60]. However, our focus will be based on data-driven (or restrained-based) approaches. Those methods use experimental data (mostly from HiC experiments) as input information for generating genome assembly structures. The interpretation of the data and its use differs widely depending on the chosen approach.
Some schemes generate a single representative structure (a consensus model) from ensemble HiC data. Contact frequencies are usually mapped to spatial distances, used to generate a 3D structure by optimizing a scoring function [61], Bayesian inference (BACH) [62] or multidimensional scaling (ShRec3D) [63]. These consensus models represent data averaged over millions of cells and cannot describe the actual physical nature of individual genomes, including the considerable structural variability between cells observed in 3D FISH and single-cell HiC experiments [21].
In contrast, resampling approaches perform many independent optimizations of a single scoring function from random starting configurations to resample an ensemble of structures. In TADBit, models are calculated by IMP, followed by a cluster analysis of the ensemble [64]. It has been used to investigate structures of TADs and bacterial genomes [65]. Resampling is also applied in several other examples (reviewed in [66]). Chrom3D relies on contact restraints for the most significant HiC contacts and lamina DamID data [67].
Population-based modelling (PM) techniques differ conceptually from resampling ones, in that they attempt to de-convolve ensemble HiC data into a population of individual structures. These methods explicitly address the variability of genome structures by creating a large population of models so that the cumulated chromatin contacts of all the structures reconstitute the ensemble HiC data rather than each structure individually. Such approaches avoid unphysical structures from simultaneous enforcement of conflicting restraints. One of the first HiC-based PM methods for modelling complete diploid genomes was introduced by Kalhor et al [68] and refined in a recent publication [69] (PGS software: https://www.github.com/alberlab/PGS). PGS has been applied to complete diploid genomes of human, mouse and Drosophila melanogastor cells [69,70].
Other PM approaches use a maximum entropy method combined with molecular dynamics sampling to construct effective energy landscapes that reproduced experimental HiC maps of individual chromosomes [71]. In another example, chromatin was represented by a few functional states and chromosome models were generated using chromatin state binding affinities as parameters [72]. In an earlier method a polymer model combined with Monte Carlo sampling was used to study chromatin conformations within TADs from ensemble 5C data [73].
Recently, the development of single cell HiC assays enabled the modelling of genomes of individual cells [21,74,75]. In Stevens et al. 10 individual genome structures of haploid mouse embryonic stem cells were modelled from single cell HiC data at 100kb resolution and validated by fluorescence imaging [75].
A challenge remains in the analysis of structure populations, either generated from deconvolution or single cell modeling. Dai et al. addressed this challenge by identifying frequently occurring chromatin clusters [76], which are often enriched in binding of specific regulatory factors.
Concluding paragraph
With the developments in biophysical techniques for structure determination of cellular assemblies, the need to integrate these data for solving a biological problem is becoming increasingly evident. Many of the approaches discussed in this review reflect recent advancements in this fields of integrative modelling of protein and genome assemblies. Although providing models of the two types of assemblies often requires different representations and different experimental information overall, the methodology used for data integration has many overlaps. The data used in integrative modelling is often sparse, noisy and ambiguous. Therefore, it is important to choose an adequate representation and formulation of the restraints (and their relative weights) to represent the data as accurately as possible. Thorough sampling is another important requirement to ascertain that optimal or near-optimal models are generated. The final model(s) should also reflect uncertainty and completeness of input information, apart from providing relevant structural details of the assembly. Cross-validation against an independent set of data is recommended for model assessment and the detection of over-fitting. Finally, large conformational variability in cellular assemblies often affect the consistency between different experimental data and this requires generation of an ensemble of models to more accurately represent the data. New community effort aims at addressing issues related to standardization of representation, validation and archiving of integrative models [77].
Highlights.
Approaches to integrative modeling of protein complexes and genomic assemblies.
Advances in experimental and computational methods help integrative structure determination of cellular assemblies
Multiple models provide insights into dynamics and structural variations of cellular assemblies.
Recent examples of integrative modelling.
Acknowledgments
We thank the support from the Medical Research Council project grant MR/M019292/1 (MT) and the Arnold and Mabel Beckman foundation (BYI program), NIH U54DK107981, R01GM096089, NSF CAREER (1150287) (FA).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflict of interest to declare.
References
- 1.Kuhlbrandt W. Biochemistry. The resolution revolution. Science. 2014;343:1443–1444. doi: 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- 2.Beck M, Baumeister W. Cryo-Electron Tomography: Can it Reveal the Molecular Sociology of Cells in Atomic Detail? Trends Cell Biol. 2016;26:825–837. doi: 10.1016/j.tcb.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Trewhella J. Small-angle scattering and 3D structure interpretation. Curr Opin Struct Biol. 2016;40:1–7. doi: 10.1016/j.sbi.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 4.van Ingen H, Bonvin AM. Information-driven modeling of large macromolecular assemblies using NMR data. J Magn Reson. 2014;241:103–114. doi: 10.1016/j.jmr.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 5.Sahu ID, McCarrick RM, Lorigan GA. Use of electron paramagnetic resonance to solve biochemical problems. Biochemistry. 2013;52:5967–5984. doi: 10.1021/bi400834a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lossl P, van de Waterbeemd M, Heck AJ. The diverse and expanding role of mass spectrometry in structural and molecular biology. EMBO J. 2016;35:2634–2657. doi: 10.15252/embj.201694818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thalassinos K, Pandurangan AP, Xu M, Alber F, Topf M. Conformational States of macromolecular assemblies explored by integrative structure calculation. Structure. 2013;21:1500–1508. doi: 10.1016/j.str.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faini M, Stengel F, Aebersold R. The Evolving Contribution of Mass Spectrometry to Integrative Structural Biology. J Am Soc Mass Spectrom. 2016;27:966–974. doi: 10.1007/s13361-016-1382-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erzberger JP, Stengel F, Pellarin R, Zhang S, Schaefer T, Aylett CH, Cimermancic P, Boehringer D, Sali A, Aebersold R, et al. Molecular architecture of the 40SeIF1eIF3 translation initiation complex. Cell. 2014;158:1123–1135. doi: 10.1016/j.cell.2014.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kahraman A, Herzog F, Leitner A, Rosenberger G, Aebersold R, Malmstrom L. Cross-link guided molecular modeling with ROSETTA. PLoS One. 2013;8:e73411. doi: 10.1371/journal.pone.0073411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bullock JMA, Schwab J, Thalassinos K, Topf M. The importance of non-accessible crosslinks and solvent accessible surface distance in modelling proteins with restraints from crosslinking mass spectrometry. Molecular and Cellular Proteomics. 2016;9:2491–2500. doi: 10.1074/mcp.M116.058560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kahraman A, Malmstrom L, Aebersold R. Xwalk: computing and visualizing distances in cross-linking experiments. Bioinformatics. 2011;27:2163–2164. doi: 10.1093/bioinformatics/btr348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrison RA, Engen JR. Conformational insight into multi-protein signaling assemblies by hydrogen-deuterium exchange mass spectrometry. Curr Opin Struct Biol. 2016;41:187–193. doi: 10.1016/j.sbi.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hopf TA, Scharfe CP, Rodrigues JP, Green AG, Kohlbacher O, Sander C, Bonvin AM, Marks DS. Sequence co-evolution gives 3D contacts and structures of protein complexes. Elife. 2014;3 doi: 10.7554/eLife.03430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashford P, Hernandez A, Greco TM, Buch A, Sodeik B, Cristea IM, Grunewald K, Shepherd A, Topf M. HVint: A Strategy for Identifying Novel Protein-Protein Interactions in Herpes Simplex Virus Type 1. Mol Cell Proteomics. 2016;15:2939–2953. doi: 10.1074/mcp.M116.058552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segura J, Sanchez-Garcia R, Tabas-Madrid D, Cuenca-Alba J, Sorzano CO, Carazo JM. 3DIANA:3D Domain Interaction Analysis: A Toolbox for Quaternary Structure Modeling. Biophys J. 2016;110:766–775. doi: 10.1016/j.bpj.2015.11.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsuji T, Yoda T, Shirai T. Deciphering Supramolecular Structures with Protein-Protein Interaction Network Modeling. Sci Rep. 2015;5:16341. doi: 10.1038/srep16341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosca R, Ceol A, Aloy P. Interactome3D: adding structural details to protein networks. Nat Methods. 2013;10:47–53. doi: 10.1038/nmeth.2289. [DOI] [PubMed] [Google Scholar]
- 19.Kuzu G, Keskin O, Nussinov R, Gursoy A. PRISM-EM: template interface-based modelling of multiprotein complexes guided by cryo-electron microscopy density maps. Acta Crystallogr D Struct Biol. 2016;72:1137–1148. doi: 10.1107/S2059798316013541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denker A, De Laat W. The second decade of 3C technologies: Detailed insights into nuclear organization. Vol. 30. Cold Spring Harbor Laboratory Press; 2016. pp. 1357–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang S, Su JH, Beliveau BJ, Bintu B, Moffitt JR, Wu CT, Zhuang X. Spatial organization of chromatin domains and compartments in single chromosomes. Science. 2016;353:598–602. doi: 10.1126/science.aaf8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Gros MA, Clowney EJ, Magklara A, Yen A, Markenscoff-Papadimitriou E, Colquitt B, Myllys M, Kellis M, Lomvardas S, Larabell CA. Soft X-Ray Tomography Reveals Gradual Chromatin Compaction and Reorganization during Neurogenesis In Vivo. Cell Rep. 2016;17:2125–2136. doi: 10.1016/j.celrep.2016.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russel D, Lasker K, Webb B, Velazquez-Muriel J, Tjioe E, Schneidman-Duhovny D, Peterson B, Sali A. Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol. 2012;10:e1001244. doi: 10.1371/journal.pbio.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Zundert GC, Rodrigues JP, Trellet M, Schmitz C, Kastritis PL, Karaca E, Melquiond AS, van Dijk M, de Vries SJ, Bonvin AM. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J Mol Biol. 2016;428:720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 27.Farabella I, Vasishtan D, Joseph AP, Pandurangan AP, Sahota H, Topf M. TEMPy: a Python library for assessment of three-dimensional electron microscopy density fits. J Appl Crystallogr. 2015;48:1314–1323. doi: 10.1107/S1600576715010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villa E, Lasker K. Finding the right fit: chiseling structures out of cryo-electron microscopy maps. Curr Opin Struct Biol. 2014;25:118–125. doi: 10.1016/j.sbi.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Wriggers W. Conventions and workflows for using Situs. Acta Crystallogr D Biol Crystallogr. 2012;68:344–351. doi: 10.1107/S0907444911049791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campos M, Francetic O, Nilges M. Modeling pilus structures from sparse data. J Struct Biol. 2011;173:436–444. doi: 10.1016/j.jsb.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 31.Fleishman SJ, Harrington S, Friesner RA, Honig B, Ben-Tal N. An automatic method for predicting transmembrane protein structures using cryo-EM and evolutionary data. Biophys J. 2004;87:3448–3459. doi: 10.1529/biophysj.104.046417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Pandurangan AP, Vasishtan D, Alber F, Topf M. gamma-TEMPy: Simultaneous Fitting of Components in 3D-EM Maps of Their Assembly Using a Genetic Algorithm. Structure. 2015;23:2365–2376. doi: 10.1016/j.str.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joseph AP, Malhotra S, Burnley T, Wood C, Clare DK, Winn M, Topf M. Refinement of atomic models in high resolution EM reconstructions using Flex-EM and local assessment. Methods. 2016;100:42–49. doi: 10.1016/j.ymeth.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Topf M, Lasker K, Webb B, Wolfson H, Chiu W, Sali A. Protein structure fitting and refinement guided by cryo-EM density. Structure. 2008;16:295–307. doi: 10.1016/j.str.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeev-Ben-Mordehai T, Vasishtan D, Hernandez Duran A, Vollmer B, White P, Prasad Pandurangan A, Siebert CA, Topf M, Grunewald K. Two distinct trimeric conformations of natively membrane-anchored full-length herpes simplex virus 1 glycoprotein B. Proc Natl Acad Sci U S A. 2016;113:4176–4181. doi: 10.1073/pnas.1523234113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *38.Lukoyanova N, Kondos SC, Farabella I, Law RH, Reboul CF, Caradoc-Davies TT, Spicer BA, Kleifeld O, Traore DA, Ekkel SM, et al. Conformational changes during pore formation by the perforin-related protein pleurotolysin. PLoS Biol. 2015;13:e1002049. doi: 10.1371/journal.pbio.1002049. The mechanism of action of a pore-forming protein from the MACPF superfamily is shown for the first time based on data from X-ray crystallography, cryo electron microscopy, fluorescence spectroscopy and cross-linking experiments combined with molecular modelling and flexible fitting with TEMPy and MODELLER/Flex-EM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *39.Plaschka C, Lariviere L, Wenzeck L, Seizl M, Hemann M, Tegunov D, Petrotchenko EV, Borchers CH, Baumeister W, Herzog F, et al. Architecture of the RNA polymerase II-Mediator core initiation complex. Nature. 2015;518:376–380. doi: 10.1038/nature14229. Using cryo EM maps at intermediate resolutions and distance restraints from XL-MS data, the authors determine the structure of RNA polymerase II-Mediator core initiation complex. [DOI] [PubMed] [Google Scholar]
- 40.Kasprzak JM, Dobrychłop M, Bujnicki J. PyRy3D. http://genesilico.pl/pyry3d.
- 41.Ukleja M, Cuellar J, Siwaszek A, Kasprzak JM, Czarnocki-Cieciura M, Bujnicki JM, Dziembowski A, Valpuesta JM. The architecture of the Schizosaccharomyces pombe CCR4-NOT complex. Nat Commun. 2016;7:10433. doi: 10.1038/ncomms10433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Degiacomi MT, Dal Peraro M. Macromolecular symmetric assembly prediction using swarm intelligence dynamic modeling. Structure. 2013;21:1097–1106. doi: 10.1016/j.str.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 43.Degiacomi MT, Iacovache I, Pernot L, Chami M, Kudryashev M, Stahlberg H, van der Goot FG, Dal Peraro M. Molecular assembly of the aerolysin pore reveals a swirling membrane-insertion mechanism. Nat Chem Biol. 2013;9:623–629. doi: 10.1038/nchembio.1312. [DOI] [PubMed] [Google Scholar]
- 44.Graewert MA, Svergun DI. Impact and progress in small and wide angle X-ray scattering (SAXS and WAXS) Curr Opin Struct Biol. 2013;23:748–754. doi: 10.1016/j.sbi.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 45.Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HD, Konarev PV, Svergun DI. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schulze-Gahmen U, Echeverria I, Stjepanovic G, Bai Y, Lu H, Schneidman-Duhovny D, Doudna JA, Zhou Q, Sali A, Hurley JH. Insights into HIV-1 proviral transcription from integrative structure and dynamics of the Tat:AFF4:P-TEFb:TAR complex. Elife. 2016;5 doi: 10.7554/eLife.15910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lossl P, Kolbel K, Tanzler D, Nannemann D, Ihling CH, Keller MV, Schneider M, Zaucke F, Meiler J, Sinz A. Analysis of nidogen-1/laminin gamma1 interaction by cross-linking, mass spectrometry, and computational modeling reveals multiple binding modes. PLoS One. 2014;9:e112886. doi: 10.1371/journal.pone.0112886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **48.Demers JP, Habenstein B, Loquet A, Kumar Vasa S, Giller K, Becker S, Baker D, Lange A, Sgourakis NG. High-resolution structure of the Shigella type-III secretion needle by solid-state NMR and cryo-electron microscopy. Nat Commun. 2014;5:4976. doi: 10.1038/ncomms5976. Using cryo-EM and ssNMR restraints, the authors determine a high-resolution structure of the Shigella type-III secretion needle with Rosetta structure calculations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Zundert GCP, Bonvin AMJJ. Fast and sensitive rigid-body fitting into cryo-EM density maps with PowerFit. AIMS Biophysics. 2015;2:73–87. [Google Scholar]
- 50.Grinter R, Josts I, Mosbahi K, Roszak AW, Cogdell RJ, Bonvin AM, Milner JJ, Kelly SM, Byron O, Smith BO, et al. Structure of the bacterial plant-ferredoxin receptor FusA. Nat Commun. 2016;7:13308. doi: 10.1038/ncomms13308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Politis A, Schmidt C, Tjioe E, Sandercock AM, Lasker K, Gordiyenko Y, Russel D, Sali A, Robinson CV. Topological models of heteromeric protein assemblies from mass spectrometry: application to the yeast eIF3:eIF5 complex. Chem Biol. 2015;22:117–128. doi: 10.1016/j.chembiol.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **52.Robinson PJ, Trnka MJ, Pellarin R, Greenberg CH, Bushnell DA, Davis R, Burlingame AL, Sali A, Kornberg RD. Molecular architecture of the yeast Mediator complex. Elife. 2015;4 doi: 10.7554/eLife.08719. An excellent example of integrative modeling, where a variety of experimental information (including cryo electron microscopy and crosslinking data) were used to characterize the structure of 21-subunit Mediator complex Yeast Mediator complex using IMP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *53.Ferber M, Kosinski J, Ori A, Rashid UJ, Moreno-Morcillo M, Simon B, Bouvier G, Batista PR, Muller CW, Beck M, et al. Automated structure modeling of large protein assemblies using crosslinks as distance restraints. Nat Methods. 2016;13:515–520. doi: 10.1038/nmeth.3838. The authors developed XL-MOD, an automated method for modeling large assemblies by weighting and optimizing spatial restraints from XL-MS data, and allowing flexibility of subunits. [DOI] [PubMed] [Google Scholar]
- 54.Rosa A, Everaers R. Structure and dynamics of interphase chromosomes. PLoS Computational Biology. 2008;4:e1000153–e1000153. doi: 10.1371/journal.pcbi.1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barbieri M, Chotalia M, Fraser J, Lavitas LM, Dostie J, Pombo A, Nicodemi M. Complexity of chromatin folding is captured by the strings and binders switch model. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:16173–16178. doi: 10.1073/pnas.1204799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brackley CA, Johnson J, Kelly S, Cook PR, Marenduzzo D. Simulated binding of transcription factors to active and inactive regions folds human chromosomes into loops, rosettes and topological domains. Nucleic Acids Research. 2016;44:3503–3512. doi: 10.1093/nar/gkw135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of Chromosomal Domains by Loop Extrusion. Cell Reports. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanborn AL, Rao SSP, Huang S-C, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proceedings of the National Academy of Sciences. 2015;112:201518552–201518552. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS. A three-dimensional model of the yeast genome. Nature. 2010;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu M, Deng K, Qin Z, Dixon J, Selvaraj S, Fang J, Ren B, Liu JS. Bayesian Inference of Spatial Organizations of Chromosomes. PLoS Computational Biology. 2013;9:e1002893–e1002893. doi: 10.1371/journal.pcbi.1002893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lesne A, Riposo J, Roger P, Cournac A, Mozziconacci J. 3D genome reconstruction from chromosomal contacts. Nat Methods. 2014;11:1141–1143. doi: 10.1038/nmeth.3104. [DOI] [PubMed] [Google Scholar]
- 64.Baù D, Marti-Renom MA. Genome structure determination via 3C-based data integration by the Integrative Modeling Platform. Methods. 2012;58:300–306. doi: 10.1016/j.ymeth.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 65.Trussart M, Yus E, Martinez S, Bau D, Tahara YO, Pengo T, Widjaja M, Kretschmer S, Swoger J, Djordjevic S, et al. Defined chromosome structure in the genome-reduced bacterium Mycoplasma pneumoniae. Nat Commun. 2017;8:14665. doi: 10.1038/ncomms14665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Serra F, Di Stefano M, Spill YG, Cuartero Y, Goodstadt M, Baù D, Marti-Renom MA. Restraint-based three-dimensional modeling of genomes and genomic domains. FEBS Letters. 2015;589:2987–2995. doi: 10.1016/j.febslet.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 67.Paulsen J, Sekelja M, Oldenburg AR, Barateau A, Briand N, Delbarre E, Shah A, Sørensen AL, Vigouroux C, Buendia B, et al. Chrom3D: three-dimensional genome modeling from Hi-C and nuclear lamin-genome contacts. Genome Biology. 2017;18:21–21. doi: 10.1186/s13059-016-1146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol. 2012;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tjong H, Li W, Kalhor R, Dai C, Hao S, Gong K, Zhou Y, Li H, Zhou XJ, Le Gros MA, et al. Population-based 3D genome structure analysis reveals driving forces in spatial genome organization. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E1663–1672. doi: 10.1073/pnas.1512577113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu Y, Gong K, Denholtz M, Chandra V, Kamps MP, Alber F, Murre C. Comprehensive characterization of neutrophil genome topology. Genes Dev. 2017;31:141–153. doi: 10.1101/gad.293910.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang B, Wolynes PG. Topology, structures, and energy landscapes of human chromosomes. Proceedings of the National Academy of Sciences. 2015;112:6062–6067. doi: 10.1073/pnas.1506257112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Di Pierro M, Zhang B, Aiden EL, Wolynes PG, Onuchic JN. Transferable model for chromosome architecture. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:12168–12173. doi: 10.1073/pnas.1613607113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Giorgetti L, Galupa R, Nora EP, Piolot T, Lam F, Dekker J, Tiana G, Heard E. Predictive Polymer Modeling Reveals Coupled Fluctuations in Chromosome Conformation and Transcription. Cell. 2014;157:950–963. doi: 10.1016/j.cell.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carstens S, Nilges M, Habeck M. Inferential Structure Determination of Chromosomes from Single-Cell Hi-C Data. PLOS Computational Biology. 2016;12:e1005292–e1005292. doi: 10.1371/journal.pcbi.1005292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stevens TJ, Lando D, Basu S, Atkinson LP, Cao Y, Lee SF, Leeb M, Wohlfahrt KJ, Boucher W, O'Shaughnessy-Kirwan A, et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature. 2017 doi: 10.1038/nature21429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dai C, Li W, Tjong H, Hao S, Zhou Y, Li Q, Chen L, Zhu B, Alber F, Jasmine Zhou X. Mining 3D genome structure populations identifies major factors governing the stability of regulatory communities. Nature Communications. 2016;7:11549–11549. doi: 10.1038/ncomms11549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sali A, Berman HM, Schwede T, Trewhella J, Kleywegt G, Burley SK, Markley J, Nakamura H, Adams P, Bonvin AM, et al. Outcome of the First wwPDB Hybrid/Integrative Methods Task Force Workshop. Structure. 2015;23:1156–1167. doi: 10.1016/j.str.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Michael AK, Fribourgh JL, Chelliah Y, Sandate CR, Hura GL, Schneidman-Duhovny D, Tripathi SM, Takahashi JS, Partch CL. Formation of a repressive complex in the mammalian circadian clock is mediated by the secondary pocket of CRY1. Proc Natl Acad Sci U S A. 2017;114:1560–1565. doi: 10.1073/pnas.1615310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Diebolder CA, Halff EF, Koster AJ, Huizinga EG, Koning RI. Cryoelectron Tomography of the NAIP5/NLRC4 Inflammasome: Implications for NLR Activation. Structure. 2015;23:2349–2357. doi: 10.1016/j.str.2015.10.001. [DOI] [PubMed] [Google Scholar]