Abstract
We recently reported that CCT chaperonin subunits are upregulated in a cardiac-specific manner under time-restricted feeding (TRF) (Gill et al. Science 2015, 347:1265–9), suggesting that TRiC/CCT has a heart-specific function. To understand the CCT chaperonin function in cardiomyocytes, we performed its cardiac-specific knock-down in the Drosophila melanogaster model. This resulted in disorganization of cardiac actin- and myosin-containing myofibrils and severe physiological dysfunction, including restricted heart diameters, elevated cardiac dysrhythmia and compromised cardiac performance. We also noted that cardiac-specific knock-down of CCT chaperonin significantly shortens lifespans. Additionally, disruption of circadian rhythm yields further deterioration of cardiac function of hypomorphic CCT mutants. Our analysis reveals that both the orchestration of protein folding and circadian rhythms mediated by CCT chaperonin are critical for maintaining heart contractility.
Keywords: TRiC:TCP-1 ring complex chaperonin, Cardiomyopathy, Protein folding and misfolding, Cytoskeletal proteins, Circadian clock, Drosophila genetics
Introduction
The proper folding of proteins is essential to cellular function. Hence, defective protein folding is linked to multiple pathological disorders. Eukaryotic cytoplasmic chaperonins are key elements responsible for this proper folding of proteins, including that of cytoskeletal components. Protein folding is mediated in the eukaryotic cytosol by the TCP-1 ring large multi-subunit complex (TRiC), also called CCT (1–5). The TCP/TRiC/CCT1 chaperonin is a highly conserved, macromolecular enzyme comprised of two hetero-oligomeric stacked rings with eight different subunits each (3–8). Both genetic and biochemical evidence reveals that the TRiC complex is required for folding of many essential proteins, including the cytoskeletal proteins myosin, actin and tubulin (9–15). Additionally, analysis of the transit of newly synthesized proteins through TRiC in intact cells suggests that this chaperonin facilitates folding of a unique subset of cellular proteins (4). The binding site of each subunit has a distinct, evolutionarily conserved pattern of polar and hydrophobic residues specifying the recognition of discrete substrate motifs. Unlike the chaperonin GroEL, TRiC chaperonins display limited substrate specificity (9–15). Interestingly, genetic depletion and inactivation of the TRiC complex prompts human heat shock transcription factor 1 (HSF1) activation, suggesting that TRiC/CCT also regulates HSF1 (16). Based on this study’s results, it may be presumed that protein folding and HSF-1 protect cells from proteotoxicity (16). In addition to HSF1, upstream factors like Hsp70 and Hsp90 work rigorously to transfer their substrates to CCT (17), while the interaction of prefoldin, phosducin-like proteins, and Bag3 protein with CCT is also known to facilitate the protein folding process.
The CCT complex is critical to cytoskeleton protein folding, as C. elegans with reduced CCT function display a defective microtubule cytoskeleton, suggesting these chaperonins are required for folding of microtubule components in metazoan development (15, 18). Human cardiac disease-related mutations in actin interfere with the ability of the TRiC chaperonin to physically interact with actin (19). In addition, the folding of the heavy meromyosin subfragment (HMM) of myosin synthesized in a rabbit reticulocyte lysate was observed to be mediated by TRiC chaperonin (14). The presence of CCT in C2C12 muscle cell cytoplasmic extracts gives rise to more native-like HHM with improved actin binding, indicating CCT facilitates myosin folding (14). Further, mutations of CCT-7 (p. Ser525Leu) encode CCTeta - a mutant TRiC chaperonin associated with increased risk of myocardial infarction (20).
In our Drosophila melanogaster heart RNA-sequencing analysis, we observed consistent cardiac-specific up-regulation of CCT components under time restricted feeding (TRF) (21). Imposing feeding/fasting rhythms with TRF paradigm attenuates age and diet-induced cardiac dysfunction without altering calories intake (21, 22). Under TRF regimen all caloric intakes occur within a consistent ≤12-h every day whereas for the Ad libitum feeding (ALF) group, food is available 24 hour/day (21, 22). The flies are maintained on a 12 h light: 12 h dark cycle at 22°C in humidified incubators. Additionally, our study showed that Drosophila with each of five hypomorphic alleles of CCT components failed to show benefits from TRF during aging, indicating that upregulation of CCT components is important to cardiac improvement derived from TRF (21). However, it is unknown whether TRF-mediated cardiac benefits of TRiC chaperonin are regulated by circadian clock. Further, the pathophysiological roles of the CCT components responsible for maintaining cardiac contractility remain unknown and unexplored. The fact that misregulation and misfolding of cytoskeletal proteins have been associated with cardiac hypertrophy (23) makes the cardiac specific function of TRiC complex members a topic of interest.
The cardiac-specific knock-down (KD) of TRiC chaperonin in the current study resulted in severe physiological and cytological defects including disorganization of cardiac actin- and myosin-containing myofibrils and drastically shortened lifespans. Furthermore, our study demonstrated how the disruption of circadian rhythms further deteriorates cardiac function of TRiC chaperonin mutants. In sum, we show that the orchestration of circadian rhythms and protein folding are essential for maintaining metabolic homeostasis and contractility of the heart.
Experimental Procedures
Drosophila stocks, cardiac specific expression and aging
UAS-RNAi fly lines for the CCT3 (CG8977, VDRC ID, 36071-GD, 106093-KK), CCT4 (CG5525, VDRC ID, 22155-GD, 106099-KK), CCT5 (CG8439 VDRC ID, 24098-GD, 109505-KK), CCT6 (CG8231, VDRC ID, 23751-GD, 109734-KK) and CCT7 (CG8351, VDRC ID, 28895-GD, 108585-KK) genes were obtained from the Vienna Drosophila RNAi Center (VDRC). Two independent lines for each of the five CCTs were tested because all are predicted to have zero off target effects, except CCT5, VDRC ID, 24098-GD. Each RNAi transgene was made with inverted repeats of a respective CCT fragment, driven by the UAS-promoter (24). The cardiac tissue-specific Hand-Gal4 driver was gifted by Eric Olson (25) and the TinCΔ4-Gal4 (TinC) driver was generously donated by Manfred Frasch (26). The GAL4-UAS system (27, 28) was used to knock-down (KD) CCTs in a cardiac specific manner. As previously reported (28), RNAi males or virgin females were crossed to Hand-Gal4 or TinCΔ4-Gal4 flies and incubated at 25°C throughout development. F-1 progeny (both males and females) were separated and allowed to age with food being changed every three days. Control flies included progeny of w1118 flies crossed with Hand-Gal4 or TinCΔ4-Gal4 or w1118 flies crossed with each of the CCTs RNAi transgenic lines. Lifespans were measured for both genders. The number of surviving flies was compared to the original number of flies collected on day 0. A percentage was taken for each day and graphed.
Cardiac physiological analysis
Semi-intact hearts were prepared as previously described (21, 28–31). Direct immersion optics were used in conjunction with a digital high-speed camera (up to 200 frame/sec, Hamamatsu EM-CCD) to record 30 s movies of beating hearts; images were captured using HC Image (Hamamatsu Corp.). Cardiac function was analyzed from the high speed movies using a semi-automatic optical heartbeat analysis software that quantifies heart period, diastolic and systolic diameters, diastolic and systolic intervals, cardiac rhythmicity, fractional shortening and produces M-mode records (21, 28–31).
Cardiac cytological analysis
As reported previously, (28, 31, 32) dissected hearts (from 1 and 3 week old flies) were briefly exposed to 10 mM EGTA for 1 min prior to being fixed with 4% paraformaldehyde in PBS for 25 min. Fixed hearts were probed with muscle myosin antibody followed by goat anti mouse Dylight 647 (Thermofisher), and Alexa555-phalloidin (Invitrogen, Carlsbad, CA) to stain F-actin. Confocal images were taken using a Zeiss 710 microscope.
Disruption of circadian clocks
Disruption of circadian rhythms was carried out as previously reported (33). Briefly, as previously described (21) flies with hypomorphic alleles of CCT components (Cct3 BL# 17293, Cct5 BL# 10393, Cct6 BL # 11475 and Cct7 BL# 13446) were obtained from Bloomington Stock Center. TRiC/CCT mutant stocks (carrying markers and/or balancers) were crossed to w1118 wildtype flies and F1 male progeny that were heterozygous for the TRiC/CCT mutation and devoid of balancers and markers were used for ALF and TRF experiments. Control and heterozygous CCT mutant flies were housed beginning at the third day after eclosion under constant light (LL) and 25°C to disrupt their circadian rhythms until they reached 3-weeks old. Age-matched flies were kept in 12hL: 12hD light-dark (LD) cycles during that time period at 25°C. Cardiac physiological parameters of 3-week old wild-type and mutant CCT flies under LL and LD were collected as described above.
Statistical analysis
For all quantitation except lifespan analysis, statistical significance was determined by one-way analysis of variance (ANOVA) followed by Dunnett’s post-hoc test to determine significance between groups using Prism 6.0 (Graph Pad) software (31). Significant differences were assumed for p<0.05. For lifespan studies, data were analyzed using the Gehan-Breslow-Wicoxon test followed by multiple comparisons between control and experimental groups (31). Significance was taken at p values less than the Bonferroni-corrected threshold of p<0.0125.
Results
TRiC/CCT chaperonin complex structural subunits, cardiac specific upregulation and conservation with human TRiC/CCT
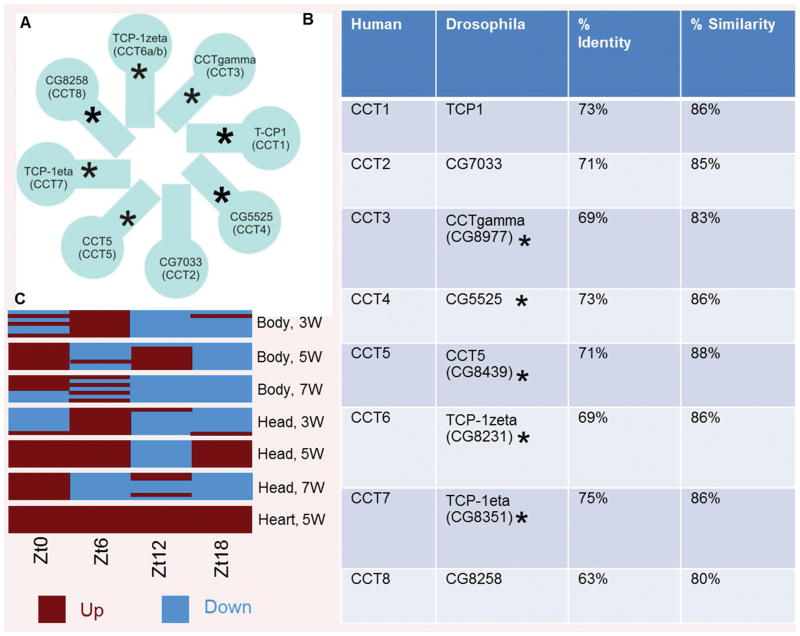
The eukaryotic cytoplasmic chaperonin TCP/TRiC/CCT forms a barrel-like structure, which is composed of 8 related subunits that are each present twice (Fig. 1A). TRiC chaperonins play a crucial role in the proper folding of several proteins relevant to cardiac health, such as cytoskeletal components (9–15). As shown in Fig. 1B, TRiC chaperonin subunits are highly conserved between Drosophila and humans. However, their cardiac-specific function remains unknown and is the subject of this investigation. As shown in Fig. 1C, time restricted feeding (TRF) conditions (21) produced transcripts of TRiC chaperonin that increased in a cardiac-specific manner (relative expression levels of seven subunits tested at four different time points, i.e., Zt0: 12 PM, Zt6: 6 PM, Zt12: 12 AM and Zt18: 6 AM, at 6 h intervals over 24 h are shown) in 5-week old fly hearts. In contrast, the average expression of TRiC chaperonin components at all of the time points tested was not upregulated in the head and body of the 3, 5 and 7 week-old flies under TRF (Fig. 1C). Furthermore, we have shown that TRiC mutants did not display TRF-mediated cardiac benefits, possibly due to the lack of organization of actin-containing myofibrils in the TRF hearts (21).
Figure 1. TRiC/CCT structure illustration, cardiac-specific upregulation under TRF and structural homology of TRiC/CCT between Drosophila and human.
A) TRiC/CCT chaperonin is composed of eight homologous but distinct subunits (CCT 1–8). Asterisks are used to represent the seven out of eight TRiC subunits upregulated in a cardiac-specific manner under TRF. B) Representation of % identity and similarity between Drosophila and human TRiC chaperonin subunits, which demonstrates a high level of structural conservation. Asterisks are used to represent the five RNAi subunits tested in the present study. C) Relative expression levels of TRiC chaperonin subunits represent transcripts from 5-week old fly hearts that are only showing increased expression (red) under TRF compared to age-matched hearts under Ad libitum feeding (ALF). However, expression levels were alternatively up-(red) and down-(blue) regulated in the head and body in 3, 5 and 7 week-old TRF flies compared to age matched ALF flies. Relative transcript levels are shown at different Zeitgeber times (ZT) in fly hearts, heads and bodies collected at 6 h intervals (i.e., Zt0: 12 PM, Zt6: 6 PM, Zt12: 12 AM and Zt18: 6 AM) over 24 h at the indicated age.
Cardiac-specific knock-down of TRiC/CCT chaperonins results in cardiac dysfunction and shortening of lifespan
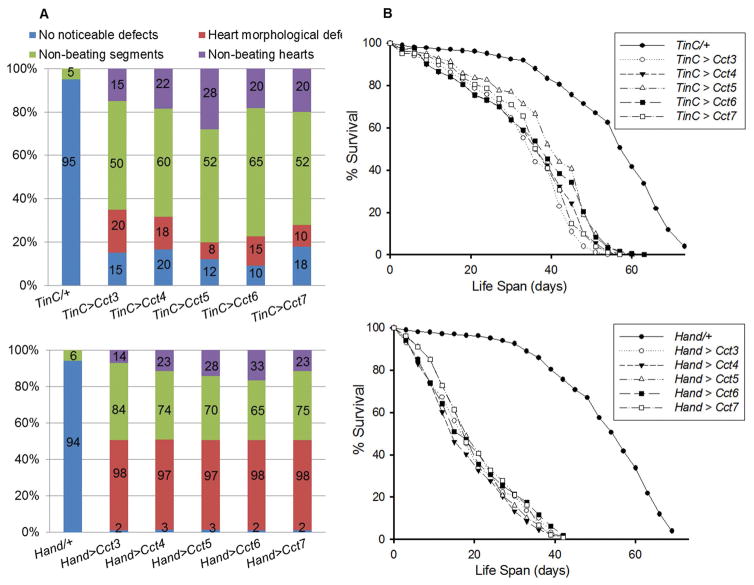
To determine the mechanistic basis of cardiomyopathy, cardiac-specific KD of Cct3, Cct4, Cct5, Cct6 and Cct7 was carried out using two different cardiac drivers and UAS-RNAi constructs. We observed severe cardiac defects upon the cardiac-specific KD of TRiC chaperonin with TinCΔ4-Gal4 (expressed in myocardium) and Hand-Gal4 driver (expressed in both pericardium and myocardium) in one-week old hearts, whereas presence of each driver in the absence of UAS-RNAi constructs (progeny after crossing with w1118 strain, referred to as TinC/+ or Hand/+ hereafter) had no obvious effects (Fig. 2A). KD of Cct3, Cct4, Cct5, Cct6 or Cct7 with the TinCΔ4 driver resulted in cardiac morphological defects in one or more non-beating regions of the heart and completely non-beating hearts (Fig. 2A). However, these defects were not observed with TinC/+. Moreover, defects were even more severe with KD of Cct3, Cct4, Cct5, Cct6 or Cct7 with the Hand-Gal4 driver, whereas Hand/+ hearts did not show these defects (Fig. 2A). For example, as shown in Fig 2A (bottom panel), almost all hearts showed severe cardiac morphological defects. Among these defects, the majority of the hearts showed significant cardiac contractility defects in more than one heart segment (65–84% of hearts). Additionally 14–23% of the hearts completely lacked a beating pattern upon KD of the Cct component with the Hand-Gal4 driver. In addition to cardiac defects, cardiac-specific KD of Cct3, Cct4, Cct5, Cct6 or Cct7 had a drastic impact on the lifespan of the flies (female and male combined) compared to TinCΔ4/+ and Hand/+ flies (Fig. 2B). Extremely severe cardiac dysfunction observed with Hand/+ upon cardiac-specific KD of Cct3, Cct4, Cct5, Cct6 or Cct7, correlated with further shortening of lifespan compared to TinCΔ4/+ (Fig. 2B). Ultimately, our results demonstrate the necessity of CCT chaperonin for preserving both cardiac integrity and lifespans.
Figure 2. TRiC chaperonin is required for cardiac remodeling, maintaining cardiac contractility and normal life span.
A) Summary of the qualitative cardiac defects from 1-week old driver control (TinCΔ4/+) and KD of TRiC chaperonin (Cct3, Cct4, Cct5, Cct6 and Cct7) male and female flies showing the percent of hearts exhibiting morphological defects, one or more non-contractile regions and non-beating hearts. A greater severity of cardiac morphological defects in one or more non-contractile regions and non-beating hearts was observed upon KD of TRiC chaperonin with the Hand-Gal4 driver (bottom). Representation of n = 100 (male and female combined) for each group is shown and percentage of the hearts indicated under each category (i.e., percentage of the hearts with no noticeable defects, heart morphological defects, non-beating heart segments and non-beating hearts). B) Cardiac-specific KD of the Cct3, Cct4, Cct5, Cct6 and Cct7 resulted in a significant reduction (p<0.001) in lifespan, compared to driver control flies TinCΔ4/+ or Hand/+ (male and female combined). Graph plots % survival of flies (n = 200 for each group) vs. time post-eclosion.
Cardiac-specific knock-down of TRiC/CCT chaperonins results in progressive cardiac physiological dysfunction
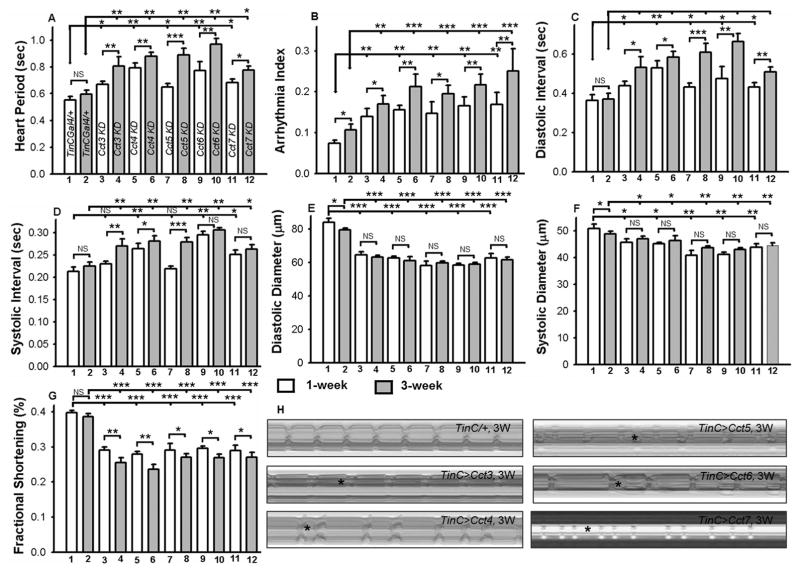
Quantitative analysis of additional cardiac physiological parameters (Fig. 3) confirmed progressive and severe cardiac defects upon cardiac-specific KD of Cct3, Cct4, Cct5, Cct6 or Cct7 with the TinCΔ4-Gal4 driver. For example, compared to driver control (TinCΔ4/+), the heart period (Fig. 3A), arrhythmia index (Fig. 3B), diastolic (Fig. 3C) and systolic intervals (Fig. 3D) of 1 and 3-week old female fly hearts were significantly elevated in the age-matched KD carried out with TinCΔ4-Gal4 driver with RNAi for Cct3, Cct4, Cct5, Cct6 or Cct7. Furthermore, these parameters in the TRiC chaperonin KD hearts significantly deteriorated in 3-week old flies compared to 1-week old flies, meaning that the phenotype is progressive in nature (Fig. 3A–D). Additionally, the cardiac restriction caused by KD of Cct3, Cct4, Cct5, Cct6 or Cct7, as measured by both diastolic (Fig. 3E) and systolic diameters (Fig. 3F), was present at both ages (1 and 3 weeks-old) compared to TinCΔ4/+ controls. Upon KD of Cct components, both diastolic and systolic diameters were reduced compared to age-matched control (TinC/+), however, diastolic diameters were reduced more drastically compared to systolic diameters (Fig. 3E–F). These results imply that KD of these TRiC components results in contraction defects at both ages. The KD of Cct3, Cct4, Cct5, Cct6 or Cct7 resulted in major reductions in heart contractility as evidenced by decreased fractional shortening (Fig. 3G). A similar effect was also seen in the male flies (not shown).
Figure 3. Cardiac specific knock-down of TRiC chaperonin leads to severe and progressive cardiac physiological dysfunction.
Quantification of heart period (A), arrhythmia index (B), diastolic and systolic intervals (C and D), diastolic and systolic parameters (E and F) and cardiac performance represented as fractional shortening (G), from 1 and 3-week-old female flies (n=50 for each genotype) of driver control (TinCΔ4/+) and KD of TRiC chaperonin (Cct3, Cct4, Cct5, Cct6 and Cct7). There were only minor differences between 1 and 3 week old cardiac parameters of the TinCΔ4/+ fly hearts, however, this difference was more significant in Cct subunits (A–G). Additionally, all the listed cardiac parameters (A–G) of the Cct3, Cct4, Cct5, Cct6 and Cct7 KD hearts were significantly altered in 1 and 3-week flies compared to age-matched TinCΔ4/+ flies. Names in panel A correspond to the numbers 1–12 in panels B to G. H) M-mode records of dissected hearts for 5 sec time periods from 3-week-old female TinCΔ4/+ and from flies with cardiac-specific KD of TRiC chaperonin. As apparent in M-mode analysis, KD of TRiC chaperonin hearts resulted in restricted heart morphology and cardiac arrhythmias. All the data are shown as average ± SEM; statistical significance was determined using one-way ANOVA and Tukey’s post hoc test and compared with age-match control (TinC/+). For all parameters, statistical significance is denoted as: * = p < 0.05; ** = p < 0.01; *** = p < 0.001; NS = not significant.
As shown with M-mode analysis of the hearts from 3-week-old female flies upon KD of Cct3, Cct4, Cct5, Cct6 or Cct7, cardiac restriction and dysrhythmic beating patterns were prevalent compared to the age-matched hearts expressing TinCΔ4/+ (Fig. 3H). Additionally, the heart phenotypes obtained with KD of Cct3, Cct4, Cct5, Cct6 or Cct7 with Hand-Gal4 driver were extremely compromised, rendering them impractical for software analysis. These CCT subunits are structurally conserved and possibly interchangeable in function. Therefore, their cardiac-specific KD resulted in similar phenotypes (Figs. 1–3). Thus, our results indicate that cardiac-specific KD of TRiC chaperonin causes severe and progressive contractility-related cardiac phenotypes.
TRiC/CCT chaperonins are essential for actin and myosin containing myofibril integrity
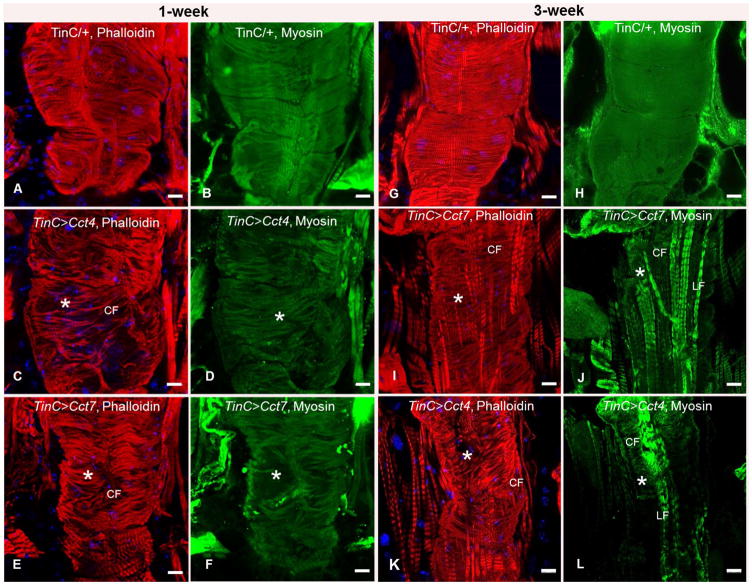
To examine the impact of cardiac-specific KD of TRiC chaperonin components on cardiac cytology, dissected hearts from 1 and 3 weeks old flies were fixed under relaxed conditions, probed with specific antibodies and/or fluorescent dyes, and analyzed by confocal microscopy. Interestingly, cardiac-specific KD of Cct4 and Cct7 (These were chosen for analysis since they display the highest percent identify to human forms) resulted in severe disorganization of actin-containing myofibrils (Fig. 4C, E) in 1-week old hearts. However, TinCΔ4/+ (driver control) hearts resulted in organized actin-containing myofibrils in age-matched hearts (Fig. 4A). Our results also demonstrated that cardiac-specific KD of Cct4 and Cct7 in 1-week old hearts caused disorganization of myosin-containing myofibrils (Fig. 4D, F), whereas TinCΔ4/+ hearts provided more organized myosin-containing myofibrils (Fig. 4B). Even greater disorganization of actin-containing myofibrils was observed with cardiac specific KD of Cct4 and Cct7 in 3-week old fly hearts (Fig. 4I, K) compared to age-matched TinCΔ4/+ (Fig. 4G). Furthermore, compared to TinCΔ4/+ hearts (4H), cardiac-specific KD of Cct4 and Cct7 in 3-week old fly hearts when probed with muscle myosin antibody resulted in near-complete loss of the contractile circumferential (CF) myosin-containing myofibrils; however, non-contractile longitudinal (LF) myosin-containing myofibrils were still present (Fig. 4J, L). These findings indicate that CCT subunits are vital for initiating/maintaining normal actin and myosin-containing myofibrils.
Figure 4. Cardiac-specific knock-down of TRiC chaperonin subunits causes disorganization of actin- and myosin-containing myofibrils.
Immunofluorescence images of 1-week (A–F) and 3-week (G–L) old hearts probed with phalloidin (F-actin, red) and DAPI (blue, DNA). Cardiac specific knock-down of TRiC chaperonin (Cct4 or Cct7) resulted in severe disorganization of actin-containing myofibrils (C, E, asterisks) in 1-week old fly hearts. Severe myofibril disorganization was also observed upon cardiac-specific knock-down of TRiC chaperonin (Cct4 or Cct7) when probed with muscle myosin antibody (green, D, F, asterisks). More severe myofibril defects (actin-containing myofibril disorganization is represented with asterisks) were observed in 3-week old fly hearts (I, K). Cardiac specific knock-down of TRiC chaperonin (Cct4 or Cct7) when probed with muscle myosin antibody resulted in near complete loss of contractile circumferential myofibrils (CF) in 3-week old hearts, whereas non-contractile longitudinal myofibrils (LF) were still present (J, L). Scale bars are 20 μm.
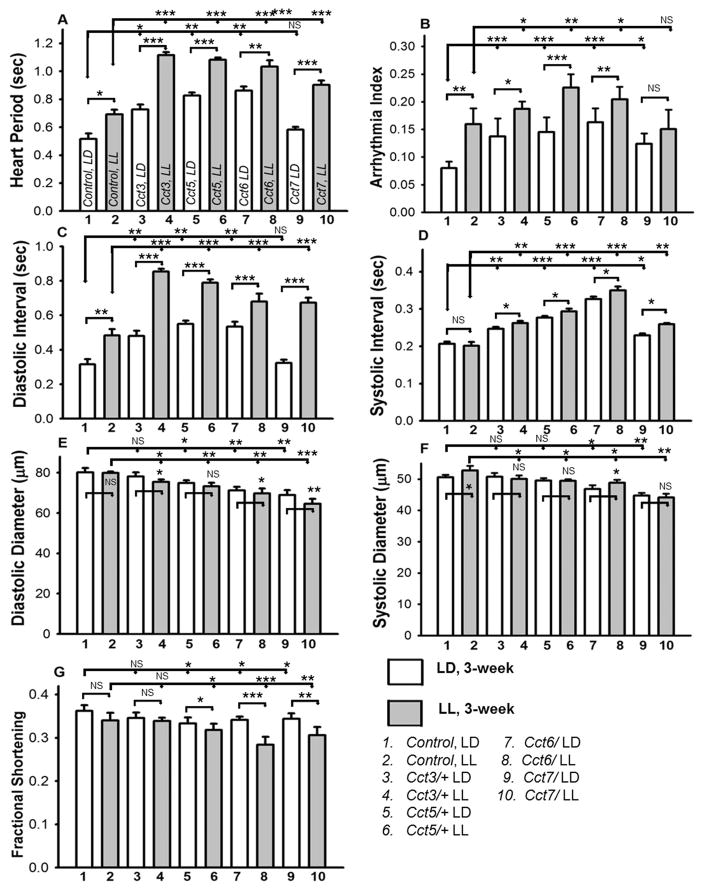
Disruption of circadian rhythm further deteriorates mutants’ TRiC/CCT-induced cardiac physiological dysfunction
Previous work using hypomorphic P element insertion mutants showed that TRiC/CCT chaperonin reduction yields subtle cardiac dysfunction and that these mutants lost the TRF-mediated cardiac benefit during aging (21). Furthermore, the mRNAs encoding all 8 subunits of the TRiC chaperonin exhibited synchronous circadian oscillation in the suprachiasmatic nucleus (SCN) (34) and liver of mice (35), which is mediated by direct binding of clock (Clk) to the promoter of TRiC genes. This appears to be conserved between flies and mice (36). Based on these observations, we explored whether disruption of circadian rhythms further exacerbates TRiC-induced cardiac dysfunction of hypomorphic mutants. As described in the methods section, circadian rhythms were disrupted by housing flies in constant light (LL) from the third day after eclosion until they reached 3-weeks old. Age-matched flies were kept in a 12hL: 12hD (LD) cycle during that time period. Cardiac physiology measurements of 3-week old flies were carried out. As shown in Fig. 5, compared to LD flies, age-matched LL flies showed more severe cardiac dysfunction and reduced cardiac contractility in TRiC hypomorphic mutants (Cct3, Cct5, Cct6 and Cct7). Furthermore, cardiac function of 3-week old w1118 (wild-type) flies under LL was also compromised to some extent compared to age-matched LD flies. However, the extent of deterioration was less in wild type than it was for mutant TRiC subunits (Fig. 5). For example, the already-compromised heart period (Fig. 5A), cardiac dysrhythmia (Fig. 5B) and diastolic intervals (Fig. 5C) of TRiC chaperonin mutants (Cct3, Cct5, Cct6 and Cct7) worsened even further under LL compared to age-matched hearts under LD for TCP mutants and wild-type. Other cardiac parameters such as systolic intervals (Fig. 5D) and diastolic diameters (Fig. 5E) were not changed in the wild-type flies under LL compared to LD. Nonetheless, significant alterations were observed in the TRiC mutants under LL when compared to their age-matched LD counterparts. Systolic diameters (Fig. 5F) of the wild-type or TRiC mutants changed only slightly in LL relative to that of LD. Furthermore, the cardiac contractility (fractional shortening) of TRiC mutants was significantly reduced under LL compared to age-matched LD while the cardiac contractility of the wild-type remained unchanged under LL compared to LD condition (Fig. 5G). Our results suggest that the disruption of circadian clocks compromises cardiac health, i.e., more severe phenotype under LL compared to LD. Additionally, this alteration is more significant in the TRiC mutants compared to w1118 (wild-type), which implies that the synchronous circadian oscillation of the TRiC complex and the presence of all subunits at normal levels is important to cardiac health.
Figure 5. Disruption of circadian rhythm further deteriorates reduced TRiC/CCT-induced cardiac dysfunction.
Cardiac parameters of 3-week old females w1118 (wild-type) and TRiC chaperonin mutants (hypomorphic P element insertion mutants) housed since 3 days after eclosion under 12hL: 12hD (LD) or constant light (LL), to disrupt circadian rhythms. Quantification of heart period (A), arrhythmia index (B), diastolic and systolic intervals (C and D), diastolic and systolic parameters (E and F) and cardiac performance represented as fractional shortening (G) was carried out under LL and LD conditions. Relative to age-matched wild-type flies under LD conditions, LL flies displayed diminished cardiac parameters. However, more severe deterioration of cardiac parameters was observed under LL compared to age-matched LD in mutant TRiC chaperonin flies. Additionally, all the listed cardiac parameters (A–G) of the Cct3, Cct5, Cct6 and Cct7 mutants were significantly (at least p < 0.05) altered under LL and LD conditions in 3 week old flies compared to wild-type flies under LL and LD respectively. All the data are shown as average ± SEM; statistical significance was determined using one-way ANOVA and Tukey’s post hoc test. For all parameters, statistical significance is denoted as: * = p < 0.05; ** = p < 0.01; *** = p < 0.001; NS = not significant.
Discussion
Molecular chaperones are machines vital to the proper folding and function of numerous proteins. TRiC chaperonin is an essential 1 MDa eukaryotic chaperonin composed of eight homologous, yet distinct subunits (CCT 1–8, Fig. 1A). CCTs are required for folding of cytoskeletal proteins in vitro and also essential for maintaining microtubule cytoskeleton in C. elegans (15, 18). However, the pathophysiological function (including cardiac-specific roles) of these folding machines is yet to be explored. Our results have shown that cardiac-specific KD of TRiC-chaperonin leads to severe and progressive cardiac phenotypes as well as significant disorganization of actin and myosin-containing myofibrils (Figs. 2–4). Additionally, our study has proven that cardiac-specific KD of these TRiC-chaperonin shortens lifespans (Fig. 2), while the disruption of circadian rhythms further deteriorates cardiac function of these chaperonins (Fig. 5). Thus, our results elucidate the requirement of TRiC-chaperonin for cardiac contractions, myofibrillar integrity and viability. Moreover, we have established a link of TRiC-chaperonin with circadian clocks in maintaining cardiac function.
Using an in vivo cardiac model, we are the first to establish that KD of TRiC chaperonin likely leads to improper contractile protein folding and results in myofibril disorganization and failure to maintain cardiac contractility (Figs. 2–4). It is well established that the integrity of contractile proteins is essential to sustaining mitochondrial organization and cardiomyocyte viability (31). An actin mutation associated with human cardiac disease appeared to yield inability to bind with TRiC chaperonin possibly due to defective folding (19). Moreover, potentially due to the inability to fold client substrates, mutation of CCT7 (p. Ser525Leu) has been associated with increased risks of myocardial infarction (20). Previously, we have reported that the cardiac-specific KD of UNC-45, a myosin specific chaperone, like the TRiC KD, causes severe disorganization of myosin-actin containing myofibrils and sarcomeres (28). We have also established that the cardiac-specific expression of mutant Huntingtin (Htt), interferes with folding in the cardiomyocytes and results in the disorganization of actin and myosin-containing containing myofibrils consequent of contractile defects observed with KD of TRiC chaperonin (31). Overall our results along with previous findings reveal that the folding of contractile proteins is dependent on chaperones and suggest that the compromised folding results in myofibril disorganization and defective cardiac function.
Although hypomorphic mutants of TRiC chaperonin show compromised cardiac function during aging and failure to attain TRF-mediated cardiac benefits (21), it is unclear whether the effect is cell autonomous or not. Thus, we have generated flies with heart-specific knockdown of TRiC components with two cardiac specific drivers to test for cell autonomous cardiac function. We observed more severe cardiac phenotypes and significantly reduced lifespans when using the Hand-driver (expressed in both myocardium and pericardium), compared to the TinCΔ4-driver (expressed in only in myocardium) (Fig. 2). KD with the Hand-driver may result in more reduction of TRiC chaperonin in myocardial cells compared to that with TinCΔ4-driver. It is also possible that expression of the Hand-driver in pericardial cells led to cell non-autonomous effects on the myocardium, which may account for the greater severity. However, we have also seen more drastic cardiac morphological defects with KD of TCPs with the Hand-driver, which potential indicates that the effect is cell autonomous. Previous studies have shown that the Drosophila heart undergoes remodeling during metamorphosis with significant morphological and structural alterations (37–39). Like we have shown for UNC-45 KD, these TRiC chaperonin subunits may be responsible for cardiac remodeling (28). More severe cardiac remodeling and contractile defects of TRiC chaperonin KD with the Hand-driver could have resulted in more significantly reduced lifespans.
Our RNA Seq. data show that that the increase in TRiC component mRNA in TRF flies was not accompanied by any significant increase in mRNAs encoding cytoskeletal proteins (21). Rather, the TRF heart showed 20–40% reduction in mRNAs encoding Myosin heavy chain, Myosin Light chain 2, Myosin Alkali light chain 1, Tropomyosin 1, Tropomyosin 2, Actin 57B, and Actin 87E (21). The parallel increase in CCT chaperonins and reduction in cytoskeletal components is likely necessary for the optimum cytoskeletal function in cardiac tissues (21). In addition to protein misfolding, reactive oxygen species (ROS) are known to cause cytoskeletal disorganization, as heart diseases are often associated with compensatory increases in cytoskeletal gene expression (40). Conversely, pharmacological stabilization of microtubules improves cardiac functional recovery during post-ischemic reperfusion in rodents (41). TRF reduces mRNA levels of several cytoskeletal genes and increases expression of TRiC chaperonin mRNA, which promotes better organized actin-containing and myosin containing myofibrils (21).
We have pioneered the connection between circadian clocks with TRiC chaperonin and have now tested the relevance of TRiC components for cytoskeletal integrity and function in cardiomyocytes. Our experimental evidence reveals that hypomorphic TRiC mutant-induced cardiac dysfunction further diminishes upon disruption of circadian rhythm (Fig. 5). Additionally, mRNAs encoding data for the all 8 subunits of the TRiC chaperonin exhibit synchronous circadian oscillation in the suprachiasmatic nucleus (SCN) in the mouse liver (34). Another study revealed direct binding of Clock to the promoter of TRiC chaperonin genes (35), as well as the fact that this phenomenon is conserved between flies and mice (36). The lack of significant change in expression of circadian clock components from analysis of heart transcripts from TRiC mutants suggests their function is downstream of the clock (21). Overall, these data support a model in which the circadian clock operates upstream of metabolic homeostasis, mitochondrial function, and TRiC function.
We have now shown the cardiac-specific significance of TRiC chaperonin in sustaining cardiac contractility and lifespan. Myosin, actin and other cytoskeletal proteins essential for maintaining cardiac contractility and their deficiency/disorganization may lead to cardiac remodeling defects. It has been previously reported that Drosophila heart undergoes remodeling during metamorphosis (37–39), and we have shown the necessity of Unc-45 cardiac for the accumulation of and folding to de novo myosin (28). Additionally, folding and accumulation of these contractile proteins in the cardiomyocytes is dependent on TRiC and other chaperones. TRiC/CCT also regulates HSF1, which protects cells from proteotoxicity (16), while upstream factors such as Hsp70 and Hsp90 facilitate CCT’s transfer of the substrate. Thus, depletion of CCT causes mis-coordination of the entire chaperonin cycle, and ultimately, severe cardiac dysfunction (17). Our findings also revealed the importance of circadian clocks for TRiC chaperonin function; however, it is still unclear whether this is through the central clock or the cardiac specific peripheral clock. Future genetic based studies will be essential to dissecting this relationship among components of TRiC and circadian clocks. Overall, our findings showed that the cardiac specific function of highly conserved TCP is critical for sustaining cardiomyocyte contractility.
Acknowledgments
We would like to thank Dr. Satchidananda Panda, Salk Institute for Biological Studies, for RNA Seq. and providing critiques in the manuscript. We also like to thank Luis Pablos for his help with genetic crosses and maintaining stocks. This work was supported by National Institutes of Health (NIH) grant AG049494 to GCM and by NIH grant AR055958 to SIB. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Abbreviations
- TRiC
TCP-1 ring complex chaperonin
- ALF
Ad libitum feeding: time-restricted feeding
- M-mode
mechanical mode
- RNA-Seq
RNA sequencing
- LD
Light-Dark cycle
- LL
Light-Light cycle
References
- 1.Dunn AY, Melville MW, Frydman J. Review: cellular substrates of the eukaryotic chaperonin TRiC/CCT. Journal of structural biology. 2001;135(2):176–84. doi: 10.1006/jsbi.2001.4380. Epub 2001/10/03. [DOI] [PubMed] [Google Scholar]
- 2.Yam AY, Xia Y, Lin HT, Burlingame A, Gerstein M, Frydman J. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nature structural & molecular biology. 2008;15(12):1255–62. doi: 10.1038/nsmb.1515. Epub 2008/11/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russmann F, Stemp MJ, Monkemeyer L, Etchells SA, Bracher A, Hartl FU. Folding of large multidomain proteins by partial encapsulation in the chaperonin TRiC/CCT. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(52):21208–15. doi: 10.1073/pnas.1218836109. Epub 2012/12/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joachimiak LA, Walzthoeni T, Liu CW, Aebersold R, Frydman J. The structural basis of substrate recognition by the eukaryotic chaperonin TRiC/CCT. Cell. 2014;159(5):1042–55. doi: 10.1016/j.cell.2014.10.042. Epub 2014/11/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nadler-Holly M, Breker M, Gruber R, Azia A, Gymrek M, Eisenstein M, et al. Interactions of subunit CCT3 in the yeast chaperonin CCT/TRiC with Q/N-rich proteins revealed by high-throughput microscopy analysis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(46):18833–8. doi: 10.1073/pnas.1209277109. Epub 2012/11/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sevigny G, Lemieux N, Steyaert A, Bibor-Hardy V. Structure of the gene coding for the mouse TRiC-P5 subunit of the cytosolic chaperonin TRiC. Genomics. 1996;31(1):107–10. doi: 10.1006/geno.1996.0015. Epub 1996/01/01. [DOI] [PubMed] [Google Scholar]
- 7.Leroux MR, Hartl FU. Protein folding: versatility of the cytosolic chaperonin TRiC/CCT. Current biology : CB. 2000;10(7):R260–4. doi: 10.1016/s0960-9822(00)00432-2. Epub 2001/02/07. [DOI] [PubMed] [Google Scholar]
- 8.Leitner A, Joachimiak LA, Bracher A, Monkemeyer L, Walzthoeni T, Chen B, et al. The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure. 2012;20(5):814–25. doi: 10.1016/j.str.2012.03.007. Epub 2012/04/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sternlicht H, Farr GW, Sternlicht ML, Driscoll JK, Willison K, Yaffe MB. The t-complex polypeptide 1 complex is a chaperonin for tubulin and actin in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(20):9422–6. doi: 10.1073/pnas.90.20.9422. Epub 1993/10/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yaffe MB, Farr GW, Miklos D, Horwich AL, Sternlicht ML, Sternlicht H. TCP1 complex is a molecular chaperone in tubulin biogenesis. Nature. 1992;358(6383):245–8. doi: 10.1038/358245a0. Epub 1992/07/16. [DOI] [PubMed] [Google Scholar]
- 11.Gao Y, Thomas JO, Chow RL, Lee GH, Cowan NJ. A cytoplasmic chaperonin that catalyzes beta-actin folding. Cell. 1992;69(6):1043–50. doi: 10.1016/0092-8674(92)90622-j. Epub 1992/06/12. [DOI] [PubMed] [Google Scholar]
- 12.Liang P, MacRae TH. Molecular chaperones and the cytoskeleton. Journal of cell science. 1997;110( Pt 13):1431–40. doi: 10.1242/jcs.110.13.1431. Epub 1997/07/01. [DOI] [PubMed] [Google Scholar]
- 13.Kubota H, Hynes G, Willison K. The chaperonin containing t-complex polypeptide 1 (TCP-1). Multisubunit machinery assisting in protein folding and assembly in the eukaryotic cytosol. European journal of biochemistry / FEBS. 1995;230(1):3–16. doi: 10.1111/j.1432-1033.1995.tb20527.x. Epub 1995/05/15. [DOI] [PubMed] [Google Scholar]
- 14.Srikakulam R, Winkelmann DA. Myosin II folding is mediated by a molecular chaperonin. The Journal of biological chemistry. 1999;274(38):27265–73. doi: 10.1074/jbc.274.38.27265. Epub 1999/09/10. [DOI] [PubMed] [Google Scholar]
- 15.Lundin VF, Leroux MR, Stirling PC. Quality control of cytoskeletal proteins and human disease. Trends in biochemical sciences. 2010;35(5):288–97. doi: 10.1016/j.tibs.2009.12.007. Epub 2010/02/02. [DOI] [PubMed] [Google Scholar]
- 16.Neef DW, Jaeger AM, Gomez-Pastor R, Willmund F, Frydman J, Thiele DJ. A direct regulatory interaction between chaperonin TRiC and stress-responsive transcription factor HSF1. Cell reports. 2014;9(3):955–66. doi: 10.1016/j.celrep.2014.09.056. Epub 2014/12/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kabir MA, Uddin W, Narayanan A, Reddy PK, Jairajpuri MA, Sherman F, et al. Functional Subunits of Eukaryotic Chaperonin CCT/TRiC in Protein Folding. Journal of amino acids. 2011;2011:843206. doi: 10.4061/2011/843206. Epub 2012/02/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lundin VF, Srayko M, Hyman AA, Leroux MR. Efficient chaperone-mediated tubulin biogenesis is essential for cell division and cell migration in C. elegans. Developmental biology. 2008;313(1):320–34. doi: 10.1016/j.ydbio.2007.10.022. Epub 2007/12/08. [DOI] [PubMed] [Google Scholar]
- 19.Vang S, Corydon TJ, Borglum AD, Scott MD, Frydman J, Mogensen J, et al. Actin mutations in hypertrophic and dilated cardiomyopathy cause inefficient protein folding and perturbed filament formation. The FEBS journal. 2005;272(8):2037–49. doi: 10.1111/j.1742-4658.2005.04630.x. Epub 2005/04/12. [DOI] [PubMed] [Google Scholar]
- 20.Erdmann J, Stark K, Esslinger UB, Rumpf PM, Koesling D, de Wit C, et al. Dysfunctional nitric oxide signalling increases risk of myocardial infarction. Nature. 2013;504(7480):432–6. doi: 10.1038/nature12722. Epub 2013/11/12. [DOI] [PubMed] [Google Scholar]
- 21.Gill S, Le HD, Melkani GC, Panda S. Time-restricted feeding attenuates age-related cardiac decline in Drosophila. Science. 2015;347(6227):1265–9. doi: 10.1126/science.1256682. Epub 2015/03/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melkani GC, Panda S. Time-restricted feeding for prevention and treatment of cardiometabolic disorders. The Journal of physiology. 2017;595(12):3691–700. doi: 10.1113/JP273094. Epub 2017/03/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willis MS, Patterson C. Proteotoxicity and cardiac dysfunction--Alzheimer’s disease of the heart? The New England journal of medicine. 2013;368(5):455–64. doi: 10.1056/NEJMra1106180. Epub 2013/02/01. [DOI] [PubMed] [Google Scholar]
- 24.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448(7150):151–6. doi: 10.1038/nature05954. Epub 2007/07/13. [DOI] [PubMed] [Google Scholar]
- 25.Han Z, Olson EN. Hand is a direct target of Tinman and GATA factors during Drosophila cardiogenesis and hematopoiesis. Development. 2005;132(15):3525–36. doi: 10.1242/dev.01899. Epub 2005/06/25. [DOI] [PubMed] [Google Scholar]
- 26.Lo PC, Frasch M. A role for the COUP-TF-related gene seven-up in the diversification of cardioblast identities in the dorsal vessel of Drosophila. Mechanisms of development. 2001;104(1–2):49–60. doi: 10.1016/s0925-4773(01)00361-6. Epub 2001/06/19. [DOI] [PubMed] [Google Scholar]
- 27.Duffy JB. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis. 2002;34(1–2):1–15. doi: 10.1002/gene.10150. Epub 2002/09/27. [DOI] [PubMed] [Google Scholar]
- 28.Melkani GC, Bodmer R, Ocorr K, Bernstein SI. The UNC-45 chaperone is critical for establishing myosin-based myofibrillar organization and cardiac contractility in the Drosophila heart model. PloS one. 2011;6(7):e22579. doi: 10.1371/journal.pone.0022579. Epub 2011/07/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fink M, Callol-Massot C, Chu A, Ruiz-Lozano P, Izpisua Belmonte JC, Giles W, et al. A new method for detection and quantification of heartbeat parameters in Drosophila, zebrafish, and embryonic mouse hearts. BioTechniques. 2009;46(2):101–13. doi: 10.2144/000113078. Epub 2009/03/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ocorr K, Reeves NL, Wessells RJ, Fink M, Chen HS, Akasaka T, et al. KCNQ potassium channel mutations cause cardiac arrhythmias in Drosophila that mimic the effects of aging. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(10):3943–8. doi: 10.1073/pnas.0609278104. Epub 2007/03/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melkani GC, Trujillo AS, Ramos R, Bodmer R, Bernstein SI, Ocorr K. Huntington’s Disease Induced Cardiac Amyloidosis Is Reversed by Modulating Protein Folding and Oxidative Stress Pathways in the Drosophila Heart. PLoS Genet. 2013;9(12):e1004024. doi: 10.1371/journal.pgen.1004024. Epub 2013/12/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alayari NN, Vogler G, Taghli-Lamallem O, Ocorr K, Bodmer R, Cammarato A. Fluorescent labeling of Drosophila heart structures. Journal of visualized experiments : JoVE. 2009;(32) doi: 10.3791/1423. Epub 2009/10/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katewa SD, Akagi K, Bose N, Rakshit K, Camarella T, Zheng X, et al. Peripheral Circadian Clocks Mediate Dietary Restriction-Dependent Changes in Lifespan and Fat Metabolism in Drosophila. Cell metabolism. 2016;23(1):143–54. doi: 10.1016/j.cmet.2015.10.014. Epub 2015/12/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hatori M, Gill S, Mure LS, Goulding M, O’Leary DD, Panda S. Lhx1 maintains synchrony among circadian oscillator neurons of the SCN. eLife. 2014;3:e03357. doi: 10.7554/eLife.03357. Epub 2014/07/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, et al. Harmonics of circadian gene transcription in mammals. PLoS genetics. 2009;5(4):e1000442. doi: 10.1371/journal.pgen.1000442. Epub 2009/04/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–54. doi: 10.1126/science.1226339. Epub 2012/09/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Curtis NJ, Ringo JM, Dowse HB. Morphology of the pupal heart, adult heart, and associated tissues in the fruit fly, Drosophila melanogaster. Journal of morphology. 1999;240(3):225–35. doi: 10.1002/(SICI)1097-4687(199906)240:3<225::AID-JMOR2>3.0.CO;2-V. Epub 1999/06/15. [DOI] [PubMed] [Google Scholar]
- 38.Molina MR, Cripps RM. Ostia, the inflow tracts of the Drosophila heart, develop from a genetically distinct subset of cardial cells. Mechanisms of development. 2001;109(1):51–9. doi: 10.1016/s0925-4773(01)00509-3. Epub 2001/10/26. [DOI] [PubMed] [Google Scholar]
- 39.Zeitouni B, Senatore S, Severac D, Aknin C, Semeriva M, Perrin L. Signalling pathways involved in adult heart formation revealed by gene expression profiling in Drosophila. PLoS genetics. 2007;3(10):1907–21. doi: 10.1371/journal.pgen.0030174. Epub 2007/10/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heusch P, Aker S, Boengler K, Deindl E, van de Sand A, Klein K, et al. Increased inducible nitric oxide synthase and arginase II expression in heart failure: no net nitrite/nitrate production and protein S-nitrosylation. American journal of physiology Heart and circulatory physiology. 2010;299(2):H446–53. doi: 10.1152/ajpheart.01034.2009. Epub 2010/06/01. [DOI] [PubMed] [Google Scholar]
- 41.Xiao J, Liang D, Liu Y, Zhang H, Liu Y, Zhao H, et al. Taxol, a microtubule stabilizer, improves cardiac functional recovery during postischemic reperfusion in rat in vitro. Cardiovascular therapeutics. 2012;30(1):12–30. doi: 10.1111/j.1755-5922.2010.00163.x. Epub 2010/06/18. [DOI] [PubMed] [Google Scholar]