The field of electron cryo-microscopy (cryoEM) started 40 years ago with the landmark study by Taylor and Glaeser that showed the feasibility of recording data that extended beyond 3 Å resolution from ice-embedded catalase crystals [1]. Improvements in the specimen freezing protocol by Dubochet and colleagues [2] launched an era of structural study of biological single particles embedded in vitreous ice, typically reaching a resolution around 40 Å in the early studies [3]. In these early experiments and most that followed for over a decade, data were recorded on photographic films in a 100 keV electron microscope. Through a series of developments in instrumentation and single particle image processing, the resolution obtained in cryoEM studies was gradually extended from the several nanometers to subnanometer range, at least for highly symmetric particles. In 2008, several near atomic resolution cryoEM structures of large particles with high symmetry were reported [4-7]. However, the acceptance of cryoEM as a high resolution structure tool remained quite limited.
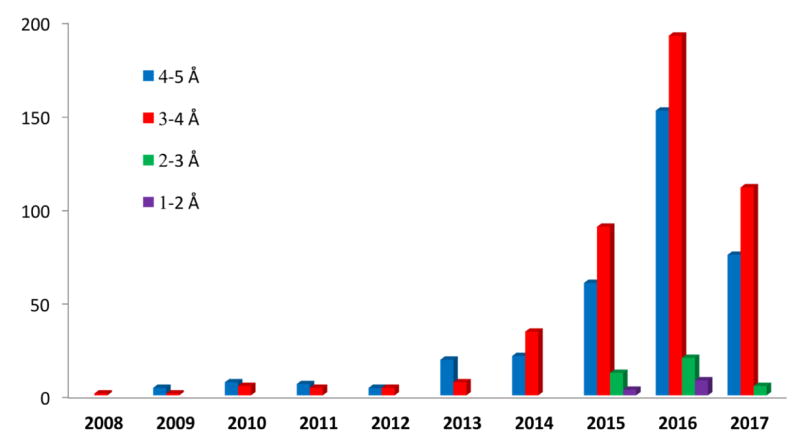
A turning point at which the broad scientific community began to recognize the power of cryoEM came with the development of direct detectors [8-10] and, at about the same time, much improved computational methods for dealing with images of specimens that contain structural (or compositional) heterogeneity [11, 12]. One advantage of the direct detectors is their high frame rate, which allows multiple frames to be collected while imaging a single specimen [13]. These frames can be computationally merged to minimize the beam induced specimen movement effect that had been a bane of working with ice-embedded specimens. The result is that nearly all images are now of a useable quality, where often only a few percent of the images recorded on photographic film or a CCD camera were considered good enough. Another advantage of the direct detectors is their inherently high detective quantum efficiency relative to the previous recording media [14]. This produces images with substantially better signal-to-noise ratio so that particles and other features that were all but invisible previously can now be identified and the particle orientations can be more precisely determined. This has made it possible not only to work with smaller complexes but to resolve small compositional and conformational differences in domains or components within large complexes. Along with the instrumental advances, new software has been developed that is particularly adept at sorting out these differences, so that it is now often possible to obtain a set of “atomic resolution” structures from a complex that represent stages along a biochemical pathway. With the advanced resolution that allows one to see amino acid side chains it is also possible to see drugs and covalent modifications that are on the size of single side chains. The combination of better images, higher throughput in data collection and the ability to deal with heterogeneous samples has led to what has been termed a revolution in cryoEM, with a tremendous expansion of the rate of producing high resolution structures [15, 16]. In the last three years, there has been exponential growth in the number of structures determined by cryoEM that reach 2-5 Å resolution (Figure 1). This review volume highlights a few of the structures of biological machines, many of which were thought to be intractable only a few years ago but that have now been resolved by cryoEM.
Figure 1.
Growth of number of structures from cryoEM. Data from PDB represents the number of atomic models derived from cryoEM maps at near atomic resolutions through the last decade till mid-2017 (Courtesy of Yanyan Zhao at Stanford University).
Ribosomes were the main subject of study in the earliest days of the development of single particle processing, and progress in ribosome resolution has marked the progress of the field ever since. While this progress has been fairly steady, the development of direct detectors and more powerful software has led to a sudden advance that has produced a series of atomic models from cryoEM data. This now allows researchers to understand the dynamics and series of conformational states that ribosomes undergo, and even to identify location and pose of small ligands that affect ribosome functions. This already appears to be a boon for antibiotic development and, in addition, the technology has reached the point where small modifications of the ribosome structure itself can be visualized, providing insights on the natural regulation of activity. Loeffelholz et al. give several examples in which ligands bound to ribosomes have been visualized through focused classification, a technique that helps identify local differences among particles in a population, and where modifications such as methylation of the rRNA can be identified. Models directly derived from these efforts are of sufficient quality to guide development of new classes of drugs.
Viruses were the targets in some of the earliest cryoEM applications as well as a driving force in development of much of the technology and software currently in use. As with ribosomes, progress in the field has been marked by improvements in resolution, from the level at which general features of capsid structure could be discerned to the present ability to derive reliable atomic models of a wide range of viruses. Jiang and Tang present a brief review the main developments in the field, pointing out ways in which cryoEM has eclipsed x-ray studies for determining virus structures. Some of the advances have relied on the icosahedral symmetry of the virus capsids, while a few examples demonstrated the feasibility of visualizing the viral genomes without icosahedral symmetry at unprecedented details. These authors discuss ways in which recent specimen preparation developments are already yielding novel information from structures of viruses, as well as expectations for future trends for larger viruses and heterogeneous viral particles.
Vonck and Mills provide an overview of a number of enzymes whose structures have been solved multiple times with gradual improvements from 4.5 to 2 Å resolution over the last few years. Dramatic reduction in data collection time and improvements in structure refinement procedures have led to improved resolution in a number of cases. Examples are given to show the difference in resolvability of side chains, backbone connectivity, ligands and water even within a structure resolution difference of 1 Å. Though only a handful of cryoEM structures of enzymes have been solved so far to between 2-3 Å resolution, there is no reason to doubt that this trend will continue as more samples will be examined by cryoEM.
The tetrameric γ-secretase complex is an example of a specimen that has been studied by several groups that produced reconstructions that seemed incompatible with each other. The limitations in resolution imposed by the technology of even five years ago, as well as possible differences in specimen treatment, led to 3D maps that had little in common. Some of these differences may relate to the distribution of the detergent used in solubilizing and purifying the complex, which can appear like protein density in a map at moderate resolution. The new results described by Yang et al. have resolved these differences by producing atomic models from the cryoEM data in several different states. Mapping the sites of mutations associated with risk of Alzheimer's disease provides better understanding of the functional defects that lead to aberrant cleavage of the amyloid precursor protein. In computing the highest resolution map, the authors used just 10% of the particles identified in the images, suggesting that there is substantial flexibility in the structure. It remains a challenge to take advantage of this resultant image heterogeneity to map out the conformational landscape for this complex, but given the rate of advances in the field one can expect that this will not be far behind.
Baker and co-workers describe the structure of the tetrameric IP3 receptor, which is a 1.2MDa, membrane-bound Ca2+ release channel. The structure was validated with two different image processing protocols and revealed 10 domain folds. Its structure has led to an improved understanding of how the inter- and intra-subunit interactions may affect signal transduction leading to Ca2+ release upon binding of the IP3 ligand. Interestingly, the absence of resolved features in certain parts of the map matches well with the previously known sequence locations of alternative splicing sites and post-translational modifications that contribute to structural heterogeneity. Comparison of the IP3 receptor structure with the larger calcium release channel, ryanodyne receptor, reveals large structural homologies which are not reflected in any substantial sequence homology.
Earl and co-workers review three advances in high resolution cryoEM methodology: 3D classification, membrane protein preparation and cryo-specimen preparation. When these are all optimized, it is now possible for the resolution with cryoEM to exceed that of x-ray crystallography. The dramatic advance in resolution is illustrated by the 1.8 Å structure of glutamate dehydrogenase achieved by cryoEM, compared to the best x-ray structure at 2.7 Å resolution. With this complex it was also possible to identify conformations never seen by crystallography. Similar results in separating conformers in other complexes are highlighted, including the finding that the AAA+ ATPase P97 coexists in three biochemical states, and visualization of the IRES RNA moiety as it progresses through a series of binding sites in the ribosome. There are still challenges in working with membrane proteins, and some new approaches for preservation, such as with nanodiscs and amphipols, are discussed. Work on developing methods to obtain a suitable orientational distribution of particles in just the right thickness of ice is also reviewed.
The spliceosome is such a dynamic complex, with so many subunits that recruit and displace each other through its functional cycle, that electron microscopy seemed to be a far better approach to understanding its structure than crystallography. Some early successes gave limited insights on the structure, but the advent of the new cryoEM technology has produced a shower of results that now reveal much about the mechanisms involved in RNA splicing. Scheres and Nagai review the multiple near-atomic resolution cryoEM structures of spliceosomes that have been obtained in several key chemical states. These results now allow one to probe the mechanism of removing introns from eukaryotic RNA precursors by two trans-esterification reactions. There is certainly still much to learn about this process, but this paper presents an impressive story on the mechanisms as they are being revealed by cryoEM.
Microtubules have been the subject of study by cryoEM since at least 1990. Since around 2000 several dozen structures of microtubules, some in complex with various motor molecules and other microtubule-associated proteins, have been published, until recently reaching a maximum resolution around 8 Å. Nogales and Kellogg review more recent work that has broken this barrier and produced maps with substantially better quality than those used to derive the tubulin structure by electron crystallography. The progressive improvements have led to visualizing MAP complexes at 3.4 Å. Among the surprising discoveries was the fact that in the apo state the microtubule wall is not cylindrical, an effect that was masked by the usual imposition of symmetry and which limited resolution to some degree. In the most recent work it has been possible to visualize several of the microtubule-stabilizing drugs. This opens up structure based development with microtubules, although the resolution still requires a bit more improvement.
Many filamentous structures are much thinner than microtubules and barely visible in the electron microscope. Impressive results have been obtained with filaments of moderate size, such as bacterial flagella, using the technology that pre-dates the direct detectors. But results with thinner structures such as bacterial pili and archaeal flagella have been at substantially lower resolution, in those cases where the symmetry could be identified so that image processing could even begin. Egelman reviews some results with the current generation of detectors, which have produced such a substantial improvement in signal-to-noise ratio that atomic models have been generated with systems previously considered impossible to deal with. These results highlight the ways in which sequence and structural homologies inform our understanding of the relationships among various systems such as bacterial pili and archaeal flagella. In this paper filamentous structures from three kingdoms are reviewed. The near atomic resolution structures led to unexpected hypotheses about how these flagella and pili function differently in different cells. Multiple new structures across species show nicely how much structural similarity and difference can exist independent of similarities in the protein sequences. The visualization of lipid phosphate head groups lining the hollow lumen of the bacterial mating pili led to an intriguing channel lubricating mechanism for the DNA transfer from one cell to another.
Development of phase plates that allow in-focus imaging with good contrast has been hailed as one of the major recent advance for cryoEM. Danev and Baumeister demonstrate the power of increasing image contrast using a phase plate to retrieve near atomic resolution structure for proteins as small as 64 kDa as well as for the much larger structures generally dealt with in electron tomography. Challenges remain, including that the phase shift is not constant and that one needs to determine and compensate for the effective defocus in order to obtain a correct structure. Still, the current results represent a notable step towards reaching the theoretical limit of the lowest molecular weight complex for which the atomic structure can be determined by cryoEM.
While electron diffraction analysis of sub-micron sized crystals is not an inherently single particle cryoEM technique, the technique known as “MicroED” has been applied so far just to protein and peptide crystals. The method involves collecting full diffraction data sets from sub-micron sized crystals in the microscope, with the most recent implementation collecting data while the sample is tilting to cover a finite angular range within each diffraction pattern. Because of the high electron scattering power of biological material, it is possible to collect this data from crystals many orders of magnitude smaller than with x-rays, which provides the capability to solve structures of some samples for which macroscopic crystallization has failed. The paper by Rodriguez and Gonen highlights results with MicroED since its first demonstration in 2013. Over this time the data collection and processing techniques have improved with resultant improvements in resolution. It is now fairly routine to achieve resolution in the 1-Å range with proteins of a wide range of sizes, from a few hundred Daltons to at least several hundred thousand Daltons. It is already apparent that the usual notion that the sample should be thin enough to behave as a weak phase object may be overly conservative, but the effects of crystal thickness are still to be explored.
A recurring theme of the structures in this review is the construction of atomic models of the highly purified macromolecular assemblies. It has suddenly become almost routine to reach a resolution at which, for example, secondary structure is well resolved, and where enough of the bulky side chains can be identified to allow threading a peptide sequence onto the precisely positioned backbone. It has long been a distinct advantage of cryoEM that interpretable results can be obtained at any resolution, but the field has rapidly moved on from the days of “blobology”, where understanding the arrangement of domains within a complex was the usual target, to being able to understand the structure and interactions of the domains and components at the atomic level. This level of interpretability is reached with resolution in the 3-4 Å range, which is now in reach for a wide variety of samples as shown in this review. As in x-ray crystallography, though, in this range there are few constraints on details such as side chain rotamers, so what we call “atomic” models are rather imprecise. As the resolution reaches 2.5 Å or better it becomes possible to place atoms with sufficient precision that the chemistry of their interactions is revealed. This resolution has been achieved with several specimens by single-particle cryoEM, which opens the door to using this technique in drug characterization and rational design. There is still often variability of resolvability in different regions of the structures, so the atomic models generally need to be interpreted with caution. In cases where the structures of components of a complex are known through crystallography, model building can proceed more quickly and from a lower resolution map. Crystallography often provides a static view that may not be most relevant to the context of the biological entity but is still an excellent starting point for fitting to a cryoEM density map. It is a great advantage of cryoEM that an ensemble of structures can now be extracted from a single preparation, providing insight on the mechanical pathways that a complex undergoes as it performs its function. As shown in the articles presented in this issue, the results give a solid basis for merging mechanistic hypotheses, which can settle long standing puzzles or wrong conceptual conjectures that had developed prior to the cryoEM structures.
Even with the impressive progress in cryo-EM made in many laboratories around the globe as reflected in this issue, technical challenges remain in cryoEM. Among these would be better macromolecule purification and cryo-specimen preservation with optimal ice thickness and random particle orientation, improvements in phase plate design, enhancement of data recording with phase plates, extending routine resolution beyond 3 Å for reliable interpretation of functional mechanisms at the atom level, and further development of software to handle complex structural/compositional heterogeneity. Interestingly, some specimens are being pursued in more than one lab, and it is not uncommon to observe differences in structures or structure interpretation from the different groups. These differences can sometimes be attributed to specimen conformation variation under different chemical and preparation conditions, but it can also be that different models for structural details arise from differences in data quality, image processing, or interpretation of the map. This leads to a final point, that it is utterly important to have a more robust protocol to validate cryoEM structures. A community effort is being made to derive the best practice to represent the atomic models properly with a set of reliability indices through an open challenge for validating cryoEM maps and models (challenges.emdatabank.org).
The recent developments in cryoEM have ushered in a new era in structural biology, as illustrated by the papers in this volume. These results represent the first wave of what we predict will be a veritable flood. We can expect that, over the next few years as the field continues to develop and expand, cryoEM will produce ever greater insights on mechanistic biology.
Acknowledgments
WC is supported by NIH grant P41GM103832; KHD is supported by NIH grant P01GM051487.
Wah Chiu received his BA and PHD at the University of California, Berkeley. He recently transitioned his professorship from Baylor College of Medicine to Stanford University. He is the founder of the National Center for Macromolecular Imaging supported by NIH, and has been a leader in the development and application of cryoEM techniques that allow visualization of high resolution structures of large molecular complexes such as viruses, membrane proteins and chaperonins. He is a member of the National Academy of Sciences and a recipient of the Distinguished Scientist Award of the Microscopy Society of America.
Kenneth Downing received his BS and PhD from Cornell University. After a time at the Institute for Cell Biology at the ETH, Zurich, he moved to the Lawrence Berkeley National Laboratory, where he is a now senior scientist. He has contributed to the development of techniques for electron microscopy study of soft materials. He was involved with developing some of the first atomic model of proteins by electron crystallography and is now extending these methods to studies of polymer systems. He is a Fellow of the Microscopy Society of America and recipient of their Distinguished Scientist Award, as well as the Berkeley Lab Prize.
Contributor Information
Wah Chiu, Departments of Bioengineering and of Microbiology and Immunology, Stanford University, Stanford, California 94305 and SLAC National Accelerator Laboratory, Stanford University, Menlo Park, California 94025 USA.
Kenneth H. Downing, Molecular Biophysics and Integrated Bioimaging Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720 USA
References
- 1.Taylor KA, Glaeser RM. Electron diffraction of frozen, hydrated protein crystals. Science. 1974;186(4168):1036–1037. doi: 10.1126/science.186.4168.1036. [DOI] [PubMed] [Google Scholar]
- 2.Dubochet J, Lepault J, Freeman R, Berriman JA, Homo JC. Electron microscopy of frozen water and aqueous solutions. J Microscopy. 1982;128:219–237. [Google Scholar]
- 3.Adrian M, Dubochet J, Lepault J, McDowall AW. Cryo-electron microscopy of viruses. Nature. 1984;308(5954):32–36. doi: 10.1038/308032a0. [DOI] [PubMed] [Google Scholar]
- 4.Yu X, Jin L, Zhou ZH. 3.88 Å structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453(7193):415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ludtke SJ, Baker ML, Chen DH, Song JL, Chuang DT, Chiu W. De Novo backbone trace of GroEL from single particle electron cryomicroscopy. Structure. 2008;16(3):441–448. doi: 10.1016/j.str.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Jiang W, Baker ML, Jakana J, Weigele PR, King J, Chiu W. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451(7182):1130–1134. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, Harrison SC, Grigorieff N. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci U S A. 2008;105(6):1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milazzo AC, Cheng A, Moeller A, Lyumkis D, Jacovetty E, Polukas J, Ellisman MH, Xuong NH, Carragher B, Potter CS. Initial evaluation of a direct detection device detector for single particle cryo-electron microscopy. J Struct Biol. 2011;176(3):404–408. doi: 10.1016/j.jsb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bammes BE, Rochat RH, Jakana J, Chen DH, Chiu W. Direct electron detection yields cryo-EM reconstructions at resolutions beyond 3/4 Nyquist frequency. J Struct Biol. 2012;177(3):589–601. doi: 10.1016/j.jsb.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10(6):584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ludtke SJ. Single-Particle Refinement and Variability Analysis in EMAN2.1. Methods Enzymol. 2016;579:159–189. doi: 10.1016/bs.mie.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheres SH. Processing of Structurally Heterogeneous Cryo-EM Data in RELION. Methods Enzymol. 2016;579:125–157. doi: 10.1016/bs.mie.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 13.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504(7478):107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McMullan G, Faruqi AR, Clare D, Henderson R. Comparison of optimal performance at 300keV of three direct electron detectors for use in low dose electron microscopy. Ultramicroscopy. 2014;147:156–163. doi: 10.1016/j.ultramic.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhlbrandt W. Biochemistry. The resolution revolution. Science. 2014;343(6178):1443–1444. doi: 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- 16.Nogales E, Scheres SH. Cryo-EM: A Unique Tool for the Visualization of Macromolecular Complexity. Mol Cell. 2015;58(4):677–689. doi: 10.1016/j.molcel.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]