Abstract
Recent advances in cryo-electron microscopy (cryo-EM) have opened up the possibility that a large class of biological structures, helical polymers, may now be readily reconstructed at near-atomic resolution. This will have a huge impact, since most of these structures are unlikely to be crystallized. This review focuses on new cryo-EM studies involving three classes of bacterial pili (chaperone-usher, mating, and Type IV) as well as on archaeal flagellar filaments. While it has long been known that one domain within archaeal flagellar filaments is homologous to a domain within bacterial Type IV pilins, the new studies shed light on how homologous and even highly conserved subunits can pack together in different ways with only small changes in sequence.
The current revolution in cryo-electron microscopy (Cryo-EM) [1–3], largely driven by the introduction of direct electron detectors [4], has had a profound impact in our understanding of complexes ranging from integral membrane proteins [5] to the anaphase-promoting complex [6]. Many proteins in bacteria, archaea, eukaryotes and viruses exist as helical polymers, and the impact that these new detectors have had in understanding the structure of such polymers cannot be overestimated. In the case of the anaphase-promoting complex, one might say that the direct electron detectors have allowed people to go from 7–8 Å resolution where secondary structure is visible [7] to 3–4 Å where an atomic model can be reliably built [6]. In the case of many protein and nucleoprotein polymers however, in the absence of a direct electron detector the helical symmetry may not even be determinable, while with the new detectors near-atomic resolution can be readily achieved [8,9]. Further, while structures such as the ribosome or the anaphase-promoting complex can, in practice or in principle, be crystallized, most helical polymers such as pili or flagellar filaments are unlikely to be crystallized due to the fact that they have a symmetry inconsistent with any crystal space group.
The great improvement in throughput provided by automated microscopes such as the Titan Krios allows for massive amounts of data to be collected in an unattended manner, but it has also been shown that one can reach a near-atomic resolution with direct electron detectors installed on more modest microscopes [10,11], albeit with much more effort. Similarly, advances in software for image analysis and reconstruction [12,13] have had an important effect, but people are still using legacy software such as SPIDER [14] to routinely reach near-atomic resolution, as evident from some of the papers that will be discussed.
In this brief review, I will focus on bacterial pili and archaeal flagella, and what structures of these filaments obtained using direct electron detectors have told us about biological function and evolutionary relationships. Bacterial pili are diverse types of thin filaments [15] that protrude from the cell surface, and are involved in a host of activities ranging from adhesion to motility to the transfer of DNA during mating. We have recently learned of an entirely new form of pili [16], suggesting that there are at least six different types of pili that have no common structure or evolutionary origin. It has been known for years that the archaeal flagellar system contains a flagellin with an N-terminal helical domain that is a clear homolog of the N-terminal helical domain in bacterial Type IV pilin [17]. The archaeal flagellar filament has no homology to the bacterial flagellar filament, thus it has been suggested [18] that it be referred to as the “archaellum”. But it also has no homology to the eukaryotic flagellum and should perhaps be given its own designation. However, the bacterial flagellar filament has no homology with the eukaryotic one (discovered many years before the bacterial one). While the bacterial and archaeal flagellar filaments may be built from only one flagellin protein (but some contain several different flagellins), the current best estimate for the number of different proteins in a eukaryotic flagellum is > 400 [19]. If we accept using “archaellum” for the archaeal flagellar filament, then we should also use “bacteriellum” for the bacterial one to distinguish it from the eukaryotic flagellum. We will therefore use flagellum in this review for all three, even though we understand that they have no homology and represent instances of convergent evolution.
Chaperone-Usher Pathway Pili
This class of pili involves a periplasmic chaperone that is complexed with each pilin subunit prior to assembly, and both the chaperone and the outer membrane usher are essential for having an N-terminal β-strand from one subunit inserted into a β-sheet of a neighboring subunit in the assembled polymer [20]. This class includes the intensively studied P and Type 1 pili of uropathogenic E. coli, responsible for the large majority of urinary tract infections. Without these pili that tenaciously adhere to the lining of the urinary tract, these bacteria would be non-infective and would be flushed from the tract by the large shear forces from urine flow, as are most bacteria. While negative stain and cryo-EM [21] have been used in the past to study these pili, the resolution has been too low to allow for any detailed understanding of the interactions that provide the unusual mechanical properties of these filaments: in the presence of forces, these pili can unwind into a thin strand that is five times the length of the normal coiled filament [22]. Using a direct electron detector, cryo-EM images were obtained which allowed for a reconstruction of the P pilus at 3.8 Å [23] with the legacy SPIDER package [14] and the Iterative Helical Real Space Reconstruction algorithm [24]. The donor strand from one subunit is shown inserted into a β-sheet of a neighboring subunit in Fig. 1. A continuous chain of subunits held together by such a strand-into-sheet arrangement then coils to form a spring-like structure that can be uncoiled by fluid forces. The rather surprising conclusion of this study was that most of the subunit-subunit interactions that hold the “coils in the spring” together are limited and polar, while the interactions that maintain the uncoiled thin filament (involving the β-strand donated by each subunit in the adjacent subunit’s β-sheet) are more extensive and hydrophobic. This allows for rather weak interactions maintaining the coiling, with stronger interactions maintaining the single strand when it is uncoiled.
Figure 1.
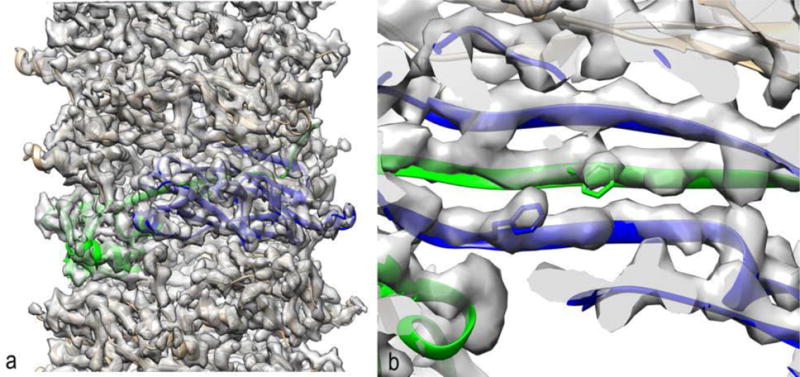
The first near-atomic resolution structure of a chaperone-usher pilus [23]. (a) Two adjacent subunits are colored (green and blue). The contacts with subunits above and below the two colored protomers are much weaker than the contacts between adjacent subunits along the 1-start helix, allowing this structure to uncoil into a thin strand while subunits are still attached to each other. (b) A segment of the P pilus reconstruction and model shows how a β-strand of one subunit (green) is donated to form part of the β-sheet of another subunit (blue). Two aromatic residue are shown (Phe13 in green and Phe35 in blue) to illustrate how threading the sequence through the reconstructed volume was quite unambiguous at this resolution (3.8 Å).
Mating Pili
The pili that play a prominent role in bacterial mating, sometimes called sex pili, have been of interest for many years [25]. These pili are formed from small (~ 60 residue), largely hydrophobic subunits that exist as integral membrane proteins in the bacterial inner membrane prior to assembly. When it was originally observed that these assembled pili were hollow and bore a striking similarity to hollow filamentous bacteriophage [26], it was assumed that DNA was actually transferred through the sex pili between two mating cells. Some subsequent experiments suggested that the main role for these pili was to depolymerize once attached to another cell and bring these two cells into physical juxtaposition before DNA was transferred through a pore. However, a more recent set of experiments [27] has provided support for the original model with DNA actually transferring through the pilus. Prior to direct electron detectors, studies using x-ray fiber diffraction and EM [26,28,29] led to conflicting reports of the helical symmetry of these pili and low resolution models of questionable reliability. With a direct electron detector, cryo-EM images were used [30] to obtain a near-atomic resolution structure of the pED208 sex pilus and a somewhat lower resolution reconstruction of the homologous but more intensively studied F-pilus [31]. The pED208 reconstruction, at 3.6 Å resolution, revealed a very surprising feature: rather than being a simple protein polymer, each subunit was bound stoichiometrically by a single lipid molecule (Fig. 2). Although highly unusual and unexpected, such a stoichiometric lipoprotein complex was also seen in a defensin-lipid oligomeric assembly [32]. While previously it had been assumed that the subunit-subunit interfaces forming this polymer consisted of hydrophobic α-helices from one subunit packed against hydrophobic α-helices from another subunit, it is now clear that these helix-helix interactions are actually mediated in part by the hydrophobic acyl chains of the lipids. More strikingly, the hollow lumen of the pilus is now seen to be lined by the phosphate head groups of the lipids, changing the electrostatic surface of the lumen from positive to negative. It was suggested [30] that the electronegative surface created by the phosphate head groups would repel the DNA from the walls, serving to “lubricate” this channel for the passage of DNA in the same way that repelling a train from tracks by magnetic levitation allows the train to move along the tracks with much less friction.
Figure 2.
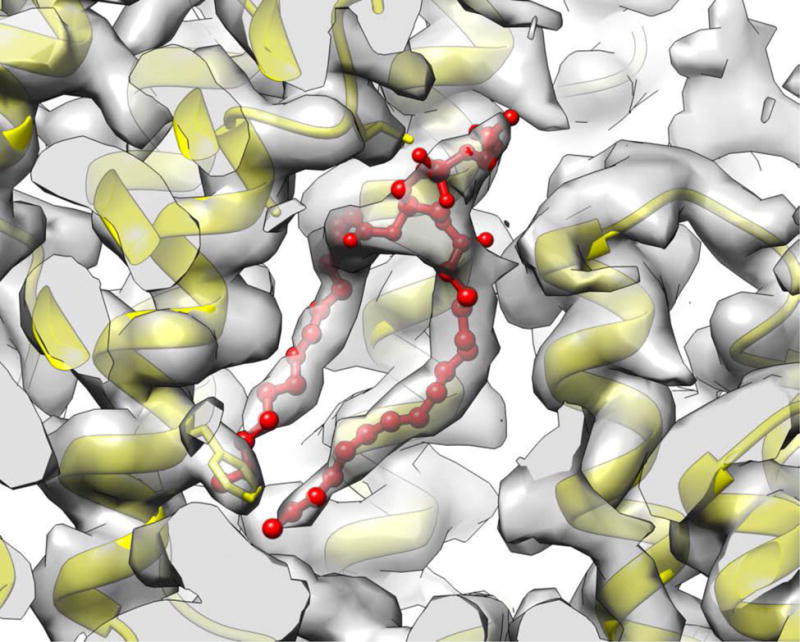
Phospholipid molecules (red) are very clearly resolved in the cryo-EM reconstruction of a pED208 mating pilus [30]. Surprisingly, a substantial part of the interface between the hydrophobic α-helices of the protein subunits (yellow) is mediated by the hydrophobic acyl chains. The polar head groups are facing the lumen of this tubular structure, changing the surface electrostatic potential.
Type IV Pili
Like the subunits that assemble to form the mating pilus, the subunits that form the Type IV pili (T4P) exist as a pool of integral inner membrane proteins both prior to assembly and after depolymerization. Unlike the mating pilin subunits, where most of the residues are inserted into the membrane in two or three helical segments, only the very hydrophobic and largely conserved N-terminal α-helix in each T4P subunit is inserted into the membrane. The larger, and much less conserved, globular domain sits in the periplasmic space before assembly and is on the outside of the filaments after assembly. It is the N-terminal domain that is a homolog of the corresponding N-terminal domain in archaeal flagellar filaments, which also exist as integral membrane proteins prior to assembly [17]. An important advance in understanding T4P structure was made when a crystal structure of a T4P subunit from Neisseria gonorrhoeae was docked as a rigid body into a cryo-EM reconstruction of the N. gonorrhoeae T4P that was at ~ 12 Å resolution [33]. Based upon the tight packing of these N-terminal helices in the core of the filament model that was generated, and the high degree of conservation of this region of the protein, the reasonable prediction was made that this model would be a good model for all T4P, as well as for homologous structures such as the archaeal flagellar filament. This argument might even be strengthened by the subsequent observation that one T4P, from Geobacter sulfurreducens [34], contains almost none of the globular domain found in N. gonorrhoeae, so the filament must be held together mainly by the N-terminal domain.
As with many good ideas, this prediction has been refuted by a number of observations, such as that the PulG pseudo-pilus is packed quite differently [35] and that the core of the archaeal flagellar filaments containing the homologous N-terminal domain are also packed differently [36]. A similar situation exists in bacterial flagellar filaments, where the highly conserved N- and C-terminal coiled-coils led to the reasonable prediction that the structural model from Salmonella [37] would be a useful model for all flagellar filaments [38]. This has turned out not be the case [39]. The new direct electron detectors have enabled us to image at high resolution an in vitro system that shows this incredible lability of quaternary structure. Using a 29-residue peptide that self assembles into helical nanotubes, a single semi-conservative mutation in one residue of these 29 (Arg to Lys) dramatically changes the quaternary structure, including doubling the diameter of the filaments and changing the asymmetric unit from one peptide to two [40]. In the absence of a direct electron detector the symmetry of these tubes could not even be determined, while with the new detectors a near-atomic level of resolution was reached for one of the two states.
Extending the work done in 2006 on the N. gonorrhoeae filaments [33] to N. meningitidis T4P using a direct electron detector [41], it has become apparent that rigid-body docking of crystallographic subunits has its limitations. A continuous N-terminal α-helix is seen in crystal structures (prepared with detergents), and this α-helix was also assumed to be continuous when inserted into the bacterial inner membrane, but actually contains a melted region of nine residues (15–23) when assembled into the pilus (Fig. 3). This melting has a number of consequences. First, the residues that are involved in the subunit-subunit interface will be quite different from what was assumed by rigid-body docking [33]. Second, this structure now provides insights into the unusual mechanical properties of T4P. It was observed that under force these pili can extend to ~ 300% of their normal length, and relax back to their normal length when this force is removed [42]. The new cryo-EM based model provides a simple explanation for these observations. As the pili are stretched, the N-terminal region containing residues 1–14 maintains a rather fixed contact with the globular domain of another subunit (Fig. 3b), but the force leads to a full extension of residues 15–23 and a further melting of a few additional residues beyond 23. This extends the pili (changing the axial rise per subunit from ~ 10 Å to ~ 30 Å), and when the force is removed the residues recoil into the native state.
Figure 3.
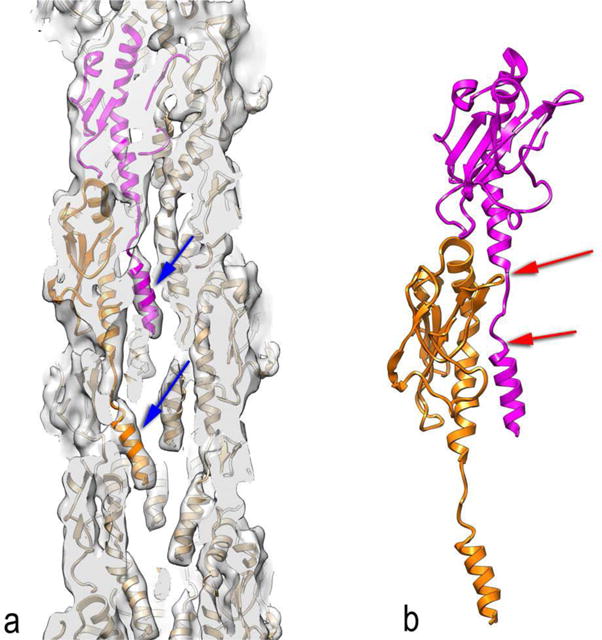
A bacterial Type IV pilus. (a) Although the resolution of the cryo-EM reconstruction (~ 6 Å) was modest [41], it is clear that the very N-terminal portion of the subunits (blue arrows) are not part of a single long continuous α-helix as seen in several crystal structures. Rather, this region is separated from the remainder of the α-helix by a melted region seen above the blue arrows. (b) Two subunits from the model in (a) are shown, where the melted region is indicated by the two red arrows. This melting allows the very N-terminal portion of the magenta subunit to maintain contacts with the globular region of the orange subunit, while the melted region can be stretched. In addition, further residues may also become part of this melted region.
Archaeal Flagellar Filaments
The bacterial flagellar system has been the icon of creationists in the United States, based upon an argument of “irreducible complexity” [43]. The argument is that the Salmonella flagellar system is built from ~ 40 different proteins, and if only one of these proteins were to be removed, the system would be non-functional. Therefore, the system could not have had a simpler precursor and thus could not have evolved but must have instead been created (by an intelligent “creator”). There are many problems with this argument, starting with the fact that many bacterial flagellar systems contain fewer proteins [44], a diversity which is typical of evolutionarily related systems. The implicit argument is also that a simpler system, containing fewer different proteins, could never accomplish the function of the bacterial flagellar system. The archaeal flagellar system is therefore quite interesting, because although it is not homologous to the bacterial one it can contain as few as 10 different proteins [18].
Two new archaeal cryo-EM structures, one of a bona fide flagellar filament [45] and the other of a flagellar-like filament [46], provide new perspectives on the evolution of these polymers. Previous studies of the Ignicoccus hospitalis Iho670 flagellar-like filament [36,47] referred to it as an “adhesion filament” since I. hospitalis is not motile, and the Iho670 filament subunit, apart from the N-terminal helix that shows homology to the corresponding N-terminal helical domain in T4P, displayed no homology with any other proteins, including archaeal flagellins. The cryo-EM reconstruction of this “adhesion filament” at ~ 4 Å resolution (Fig. 4), and the atomic model that was built from this [46], showed that the large globular domain has an Ig-like fold. The only protein found by the Dali server [48] as having structural homology was FlaF [49], which is an archaeal flagellar protein. While FlaF is not the archaeal flagellin, which is FlaB, FlaF displays clear sequence homology with FlaB. So if Iho670 is homologous with FlaF, and FlaF is homologous with FlaB, then Iho670 must be homologous with FlaB. This has been confirmed by the cryo-EM reconstruction of a true archaeal flagellar filament [45], which shows that a true flagellin, FlaB, has the same overall fold as Iho670. In addition, the Iho670 filament and the FlaB true flagellar filament have very similar helical parameters, with both having a twist per subunit of 107–108° and a rise of 5.3–5.5 Å. We can now see that the archaeal flagellin arose from a gene fusion between a bacterial T4P N-terminal domain and an ancestral Ig-like domain. However, in contrast to the N. meningitidis bacterial pili where a portion of this N-terminal domain is melted, it is all α-helical in the archaeal flagellar and flagellar-like filaments, showing that homology dictates neither the packing nor even the secondary structure.
Figure 4.
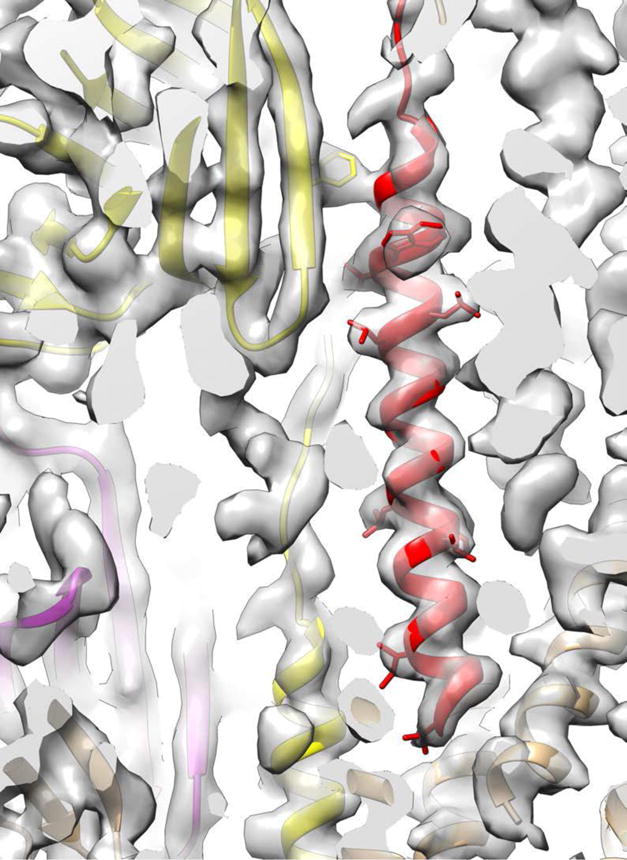
A small portion of the Iho670 flagellar-like filament reconstruction and atomic model [46], with a few sidechains shown. The red N-terminal helix is homologous with the N-terminal helix present in bacterial Type IV pili, but the packing of helices in these filaments is very different, and this helix is continuous in the flagellar-like filament but not in at least some bacterial pili.
Conclusion
The incredible improvements in gene sequencing throughput have led to an exponential increase in the amount of sequence data that we have available, and these sequences have provided many new insights into protein evolution. However, there are limits to how different two sequences can be and still be detected as homologs when “intermediate” sequences are lacking. In contrast, structural studies continue to show that proteins that display no detectable sequence homology can share the same fold and are clearly homologous. The revolution in cryo-EM means that we will see an exponential increase in the number of large structures that can now be solved with atomic models built de novo, providing many new insights into the evolution of systems such as pili and flagella.
Highlights.
Direct electron detectors allow for near-atomic resolution for many polymers.
Three types of bacterial pili discussed: chaperone-usher, mating, and Type IV.
Homology of an archaeal “adhesion filament” to a flagellar filament has been shown.
Insights into homology of N-terminal domains in Type IV pili and archaeal flagella
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
I have no conflicts of interest.
References
- 1.Egelman EH. The current revolution in cryo-em. Biophys J. 2016;110(5):1008–1012. doi: 10.1016/j.bpj.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Subramaniam S, Earl LA, Falconieri V, Milne JL, Egelman EH. Resolution advances in cryo-em enable application to drug discovery. Curr Opin Struct Biol. 2016:41, 194–202. doi: 10.1016/j.sbi.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai XC, McMullan G, Scheres SH. How cryo-em is revolutionizing structural biology. Trends Biochem Sci. 2015;40(1):49–57. doi: 10.1016/j.tibs.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-em. Nature methods. 2013;10(6):584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liao M, Cao E, Julius D, Cheng Y. Structure of the trpv1 ion channel determined by electron cryo-microscopy. Nature. 2013;504(7478):107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Atomic structure of the apc/c and its mechanism of protein ubiquitination. Nature. 2015;522(7557):450–454. doi: 10.1038/nature14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Molecular architecture and mechanism of the anaphase-promoting complex. Nature. 2014;513(7518):388–393. doi: 10.1038/nature13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiMaio F, Yu X, Rensen E, Krupovic M, Prangishvili D, Egelman EH. A virus that infects a hyperthermophile encapsidates a-form DNA. Science. 2015:348, 914–917. doi: 10.1126/science.aaa4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schroder GF, Fitzgerald KA, Wu H, Egelman EH. Unified polymerization mechanism for the assembly of asc-dependent inflammasomes. Cell. 2014;156(6):1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu B, Peisley A, Tetrault D, Li Z, Egelman EH, Magor KE, Walz T, Penczek PA, Hur S. Molecular imprinting as a signal-activation mechanism of the viral rna sensor rig-i. Mol Cell. 2014;55(4):511–523. doi: 10.1016/j.molcel.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell MG, Kearney BM, Cheng A, Potter CS, Johnson JE, Carragher B, Veesler D. Near-atomic resolution reconstructions using a mid-range electron microscope operated at 200 kv. J Struct Biol. 2014;188(2):183–187. doi: 10.1016/j.jsb.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheres SH. Processing of structurally heterogeneous cryo-em data in relion. Methods Enzymol. 2016:579, 125–157. doi: 10.1016/bs.mie.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 13.Scheres SH. Relion: Implementation of a bayesian approach to cryo-em structure determination. J Struct Biol. 2012;180(3):519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. Spider and web: Processing and visualization of images in 3d electron microscopy and related fields. Journal of Structural Biology. 1996;116(1):190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 15.Fronzes R, Remaut H, Waksman G. Architectures and biogenesis of non-flagellar protein appendages in gram-negative bacteria. EMBO J. 2008;27(17):2271–2280. doi: 10.1038/emboj.2008.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16**.Xu Q, Shoji M, Shibata S, Naito M, Sato K, Elsliger MA, Grant JC, Axelrod HL, Chiu HJ, Farr CL, Jaroszewski L, et al. A distinct type of pilus from the human microbiome. Cell. 2016;165(3):690–703. doi: 10.1016/j.cell.2016.03.016. This paper describes crystallographic studies of a newly discovered type of pilin isolated from the human microbiome. It highlights the fact that we do not need to collect samples from exotic locations to still learn very new things in biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bayley DP, Jarrell KF. Further evidence to suggest that archaeal flagella are related to bacterial type iv pili. JMolEvol. 1998;46(3):370–373. [PubMed] [Google Scholar]
- 18.Jarrell KF, Albers SV. The archaellum: An old motility structure with a new name. Trends in microbiology. 2012;20(7):307–312. doi: 10.1016/j.tim.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 19.Pazour GJ, Agrin N, Leszyk J, Witman GB. Proteomic analysis of a eukaryotic cilium. The Journal of cell biology. 2005;170(1):103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waksman G, Hultgren SJ. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nature reviews Microbiology. 2009;7(11):765–774. doi: 10.1038/nrmicro2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mu XQ, Bullitt E. Structure and assembly of p-pili: A protruding hinge region used for assembly of a bacterial adhesion filament. ProcNatlAcadSciUSA. 2006;103(26):9861–9866. doi: 10.1073/pnas.0509620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bullitt E, Makowski L. Structural polymorphism of bacterial adhesion pili. Nature. 1995;373(6510):164–167. doi: 10.1038/373164a0. [DOI] [PubMed] [Google Scholar]
- 23**.Hospenthal MK, Redzej A, Dodson K, Ukleja M, Frenz B, Rodrigues C, Hultgren SJ, DiMaio F, Egelman EH, Waksman G. Structure of a chaperone-usher pilus reveals the molecular basis of rod uncoiling. Cell. 2016;164(1–2):269–278. doi: 10.1016/j.cell.2015.11.049. Cryo-EM has been used to reach a near-atomic level of resolution for the P pilus from uropathogenic bacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000:85, 225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 25.Silverman PM. Towards a structural biology of bacterial conjugation. MolMicrobiol. 1997;23(3):423–429. doi: 10.1046/j.1365-2958.1997.2411604.x. [DOI] [PubMed] [Google Scholar]
- 26.Folkhard W, Leonard KR, Malsey S, Marvin DA, Dubochet J, Engel A, Achtman M, Helmuth R. X-ray diffraction and electron microscope studies on the structure of bacterial f pili. Journal of Molecular Biology. 1979;130(2):145–160. doi: 10.1016/0022-2836(79)90423-6. [DOI] [PubMed] [Google Scholar]
- 27.Babic A, Lindner AB, Vulic M, Stewart EJ, Radman M. Direct visualization of horizontal gene transfer. Science. 2008;319(5869):1533–1536. doi: 10.1126/science.1153498. [DOI] [PubMed] [Google Scholar]
- 28.Marvin DA, Folkhard W. Structure of f-pili: Reassessment of the symmetry. Journal of Molecular Biology. 1986;191(2):299–300. doi: 10.1016/0022-2836(86)90267-6. [DOI] [PubMed] [Google Scholar]
- 29.Wang YA, Yu X, Silverman PM, Harris RL, Egelman EH. The structure of f-pili. J Mol Biol. 2009;385(1):22–29. doi: 10.1016/j.jmb.2008.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30**.Costa TR, Ilangovan A, Ukleja M, Redzej A, Santini JM, Smith TK, Egelman EH, Waksman G. Structure of the bacterial sex f pilus reveals an assembly of a stoichiometric protein-phospholipid complex. Cell. 2016;166(6):1436–1444. e1410. doi: 10.1016/j.cell.2016.08.025. The cryo-EM structures of the F-pilus, and at higher resolution, the pED208 pilus, have been determined. The reconstructions allow for a de novo atomic model of the subunits and filaments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arutyunov D, Frost LS. F conjugation: Back to the beginning. Plasmid. 2013;70(1):18–32. doi: 10.1016/j.plasmid.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 32**.Kvansakul M, Lay FT, Adda CG, Veneer PK, Baxter AA, Phan TK, Poon IK, Hulett MD. Binding of phosphatidic acid by nsd7 mediates the formation of helical defensin-lipid oligomeric assemblies and membrane permeabilization. Proc Natl Acad Sci U S A. 2016;113(40):11202–11207. doi: 10.1073/pnas.1607855113. Crystal structures show how a single lipid molecule is bound stoichiometrically to each defensin protein subunit in a helical assembly, similar to what has been seen for the sex pili. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. Type iv pilus structure by cryo-electron microscopy and crystallography: Implications for pilus assembly and functions. Molecular Cell. 2006;23(5):651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Reardon PN, Mueller KT. Structure of the type iva major pilin from the electrically conductive bacterial nanowires of geobacter sulfurreducens. J Biol Chem. 2013;288(41):29260–29266. doi: 10.1074/jbc.M113.498527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nivaskumar M, Bouvier G, Campos M, Nadeau N, Yu X, Egelman EH, Nilges M, Francetic O. Distinct docking and stabilization steps of the pseudopilus conformational transition path suggest rotational assembly of type iv pilus-like fibers. Structure. 2014;22(5):685–696. doi: 10.1016/j.str.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu X, Goforth C, Meyer C, Rachel R, Wirth R, Schroder GF, Egelman EH. Filaments from ignicoccus hospitalis show diversity of packing in proteins containing n-terminal type iv pilin helices. Journal of Molecular Biology. 2012:422, 274–281. doi: 10.1016/j.jmb.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424(6949):643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- 38.Beatson SA, Minamino T, Pallen MJ. Variation in bacterial flagellins: From sequence to structure. Trends in microbiology. 2006;14(4):151–155. doi: 10.1016/j.tim.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 39.Galkin VE, Yu X, Bielnicki J, Heuser J, Ewing CP, Guerry P, Egelman EH. Divergence of quaternary structures among bacterial flagellar filaments. Science. 2008;320(5874):382–385. doi: 10.1126/science.1155307. [DOI] [PubMed] [Google Scholar]
- 40.Egelman EH, Xu C, DiMaio F, Magnotti E, Modlin C, Yu X, Wright E, Baker D, Conticello VP. Structural plasticity of helical nanotubes based on coiled-coil assemblies. Structure. 2015;23(2):280–289. doi: 10.1016/j.str.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41**.Kolappan S, Coureuil M, Yu X, Nassif X, Egelman EH, Craig L. Structure of the Neisseria meningitidis type IV pilus. Nature communications. 2016;7:13015. doi: 10.1038/ncomms13015. A cryo-EM reconstruction of this Type IV pilus reveals new details about the packing of the highly conserved N-terminal α-helix and reveals that a portion of it is melted in the filament. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biais N, Higashi DL, Brujic J, So M, Sheetz MP. Force-dependent polymorphism in type iv pili reveals hidden epitopes. Proc Natl Acad Sci U S A. 2010;107(25):11358–11363. doi: 10.1073/pnas.0911328107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller KR. Deconstructing design: A strategy for defending science. Cold Spring HarbSympQuantBiol. 2009 doi: 10.1101/sqb.2009.74.012. [DOI] [PubMed] [Google Scholar]
- 44.Pallen MJ, Gophna U. Bacterial flagella and type iii secretion: Case studies in the evolution of complexity. Genome Dyn. 2007:3, 30–47. doi: 10.1159/000107602. [DOI] [PubMed] [Google Scholar]
- 45**.Poweleit N, Ge P, Nguyen HH, Loo RR, Gunsalus RP, Zhou ZH. Cryo-EM structure of the Methanospirillum hungatei archaellum reveals structural features distinct from the bacterial flagellum and type iv pili. Nat Microbiol. 2016;2:16222. doi: 10.1038/nmicrobiol.2016.222. The cryo-EM structure of an archaeal flagellum is quite similar to that of the Iho670 “adhesion filament” [46] which displays no sequence homology outside of the N-terminal Type IV pilin-like domain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46**.Braun T, Vos MR, Kalisman N, Sherman NE, Rachel R, Wirth R, Schroder GF, Egelman EH. Archaeal flagellin combines a bacterial type IV pilin domain with an ig-like domain. Proc Natl Acad Sci U S A. 2016;113(37):10352–10357. doi: 10.1073/pnas.1607756113. Although these I. hospitalis filaments displayed no sequence homology to archaeal flagellins and the archaea are non-motile, a cryo-EM reconstruction and atomic model revealed that they are flagellar-like. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Müller DW, Meyer C, Gürster S, Küper U, Huber H, Rachel R, Wanner G, Wirth R, Bellack A. The iho670 fibers of ignicoccus hospitalis: A new type of archaeal cell surface appendage. Journal of Bacteriology. 2009;191(20):6465–6468. doi: 10.1128/JB.00858-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3d. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banerjee A, Tsai CL, Chaudhury P, Tripp P, Arvai AS, Ishida JP, Tainer JA, Albers SV. Flaf is a beta-sandwich protein that anchors the archaellum in the archaeal cell envelope by binding the s-layer protein. Structure. 2015;23(5):863–872. doi: 10.1016/j.str.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]


