Abstract
Cells respond to internal and external cellular stressors by activating stress-response pathways that re-establish homeostasis. If homeostasis is not achieved in a timely manner, stress pathways trigger programmed cell death (apoptosis) to preserve organism integrity. A highly conserved stress pathway is the unfolded protein response (UPR), which senses excessive amounts of unfolded proteins in the ER. While a physiologically beneficial pathway, the UPR requires tight regulation to provide a beneficial outcome and avoid deleterious consequences. Recent work has demonstrated that a conserved and highly selective RNA degradation pathway—nonsense-mediated RNA decay (NMD)—serves as a major regulator of the UPR pathway. NMD degrades mRNAs encoding UPR components to prevent UPR activation in response to innocuous ER stress. In response to strong ER stress, NMD is inhibited by the UPR to allow for a full-magnitude UPR response. Recent studies have indicated that NMD also has other stress-related functions, including promoting the timely termination of the UPR to avoid apoptosis; NMD also regulates responses to non-ER stressors, including hypoxia, amino-acid deprivation, and pathogen infection. NMD regulates stress responses in species across the phylogenetic scale, suggesting that it has conserved roles in shaping stress responses. Stress pathways are frequently constitutively activated or dysregulated in human disease, raising the possibility that “NMD therapy” may provide clinical benefit by downmodulating stress responses.
Keywords: Stress granules, Autophagy, Apoptosis, eIF2α phosphorylation
Introduction
Cellular stress responses are conserved mechanisms that act across the phylogenetic scale to maintain homeostasis in response to changes in the internal or external environment. These stress pathways protect organisms from a wide variety of cellular stressors, including endoplasmic reticulum (ER) stress, hypoxia, osmotic stress, and pathogen-induced stress. If the stress pathway resolves the stress (e.g., rescues homeostasis), then the cell returns to normal operations. However, if the stress cannot be resolved in a timely manner or the stress is overly severe, stress pathways elicit apoptosis. Chronic activation of cellular stress pathways occurs in a variety of circumstances, including neurodegeneration [1], malignancy [2–4], and infections [5, 6]. In these circumstances, cellular stress pathways can contribute to disease. Thus, the magnitude and duration of stress responses must be tightly regulated. In this review, we focus on a RNA regulatory pathway—nonsense-mediated RNA decay (NMD)—which was recently discovered to serve in this manner. This highly conserved and selective RNA degradation mechanism shapes responses to multiple forms of stress, as discussed below.
NMD
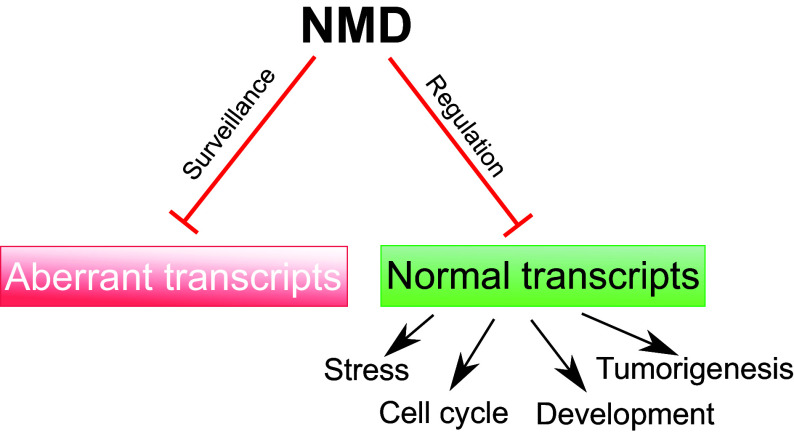
NMD was originally identified as a quality control mechanism that recognizes and degrades aberrant transcripts harboring premature termination codons (PTCs) derived from mutant genes [7–9] (Fig. 1). NMD also degrades PTC-bearing transcripts from normal genes, including alternatively spliced RNAs encoding nonfunctional proteins [10–13]. Later, work revealed that NMD also degrades a subset of mRNAs from normal genes that encode functional proteins (Fig. 1). Specific features in mRNAs elicit their decay by NMD. For example, NMD is triggered when the main open reading frame (ORF) is followed by at least one exon–exon junction [10, 14–17]. Long untranslated regions downstream of the main ORF can also, in some cases, elicit NMD [14, 15, 18–22]. Although the full repertoire of normal mRNAs directly targeted by NMD is not currently known, NMD factor knockout and knockdown experiments suggest that between ~3 and 20% of transcripts in eukaryotes from yeast to man are regulated (directly or indirectly) by NMD [7].
Fig. 1.
NMD degrades a subset of both aberrant and normal RNAs. Its ability to degrade aberrant RNAs with premature termination codons serves as a quality control mechanism. Its ability to degrade normal transcripts allows NMD to influence a variety of events, including those shown
The discovery that NMD regulates batteries of normal transcripts raised the possibility that NMD is not only an RNA surveillance mechanism but also a regulator of normal cellular activities (Fig. 1). In support of this possibility, loss or impairment of NMD causes developmental defects in species spanning the phylogenetic scale. In mice, loss of several NMD factors, including Upf1, Upf2, Upf3a, Smg1, and Smg6, causes early embryonic lethality [11, 23–27], implying that NMD has roles in pre- and/or peri-implantation development. While the underlying mechanism is not known, a recent study showed that NMD strongly influences the differentiation of the primary germ layers from human embryonic stem cells [28]. In addition, studies in Drosophila melanogaster, zebrafish, and mammalian cell lines have shown the importance of NMD in neural development and neural stem cell self-renewal vs. differentiation decision [29–32]. As further evidence for a role of NMD in neural development, mutations in the NMD gene, UPF3B, cause intellectual disability in humans and are associated with neurodevelopmental disorders, including schizophrenia and autism [33, 34]. Furthermore, copy number variations in several NMD genes were recently shown to be associated with human neurodevelopmental disorders and other neural diseases [33, 35]. Other studies suggest that NMD has roles in nonneural lineages, including in hematopoiesis, liver development, and muscle cell development [11, 36, 37].
In order for NMD to serve as a developmental regulator, it stands to reason that its magnitude would have evolved to be altered in a developmentally regulated manner. A shift from a high to a low magnitude of NMD would stabilize its target transcripts, while the opposite shift would elicit NMD target destabilization. Either shift would lead to alterations in the level of batteries of proteins, which would likely have biological effects. Several contexts have been shown to regulate NMD. For example, NMD magnitude varies in different cell types and exhibits tissue- and stage-specific regulation [28, 38–42]. In most known cases, NMD is downregulated as development proceeds [37, 43–45], but notable exceptions are B-cell development and mesoderm differentiation, where NMD is upregulated [28, 38]. Male germ cell development is also likely to be accompanied by shifts in NMD magnitude, but it has not been resolved whether NMD is up- or downregulated [26, 46, 47].
Mechanisms by which NMD is regulated are beginning to be elucidated. For example, neural-specific microRNAs (miRNAs), including miR-9 and -128, have been identified that target NMD factors and thereby repress NMD [29, 44]. These miRNAs operate in a feedback loop with NMD to maintain the neural stem cell state [29]. Differentiation-promoting contexts trigger a switch in this feedback loop to drive differentiation [29]. NMD is also regulated by another RNA decay pathway—Staufen-mediated RNA decay (SMD) [37]. The NMD-specific factor, UPF2, and the SMD-specific factor, STAU1, compete for binding with UPF1, and thus, the NMD and SMD pathways have a competitive relationship. Evidence suggests that this competition influences decisions in both myogenesis and adipogenesis [37, 48]. In addition to developmental decisions, NMD regulation may confer other functions, including in homeostasis [38, 39, 42].
In this review, we focus on the ability of NMD to regulate the sensitivity to stress. In particular, we focus on the role of NMD in the UPR pathway. Not only does NMD regulate the UPR, but UPR represses NMD, which is likely to allow for a more productive response to stress. We also discuss recent evidence that NMD regulates biological responses related to stress, including autophagy, amino-acid transport, and stress granules. The ability of NMD to regulate several stress responses is conserved, underscoring the importance of these regulatory interrelationships.
The UPR
One common form of stress is ER stress [49]. Both intrinsic and extrinsic signals can elicit ER stress. For example, low levels of nutrients elicit ER stress, as do developmental transitions, such as during B-cell differentiation [50, 51]. Some toxicants and low oxygen (hypoxic) conditions can also trigger ER stress [51, 52]. The ER is a major site of protein folding, and thus, a typical cause of ER stress is a high level of unfolded proteins. While misfolded proteins are normally dealt with by constitutive levels of molecular chaperones, protein-coding mutations can create excess unfolded proteins. Other causes of increased ER stress include aberrantly high translation of specific proteins in the ER and defective protein folding capabilities caused by low energy levels or other mechanisms that decrease chaperone protein expression [50–53].
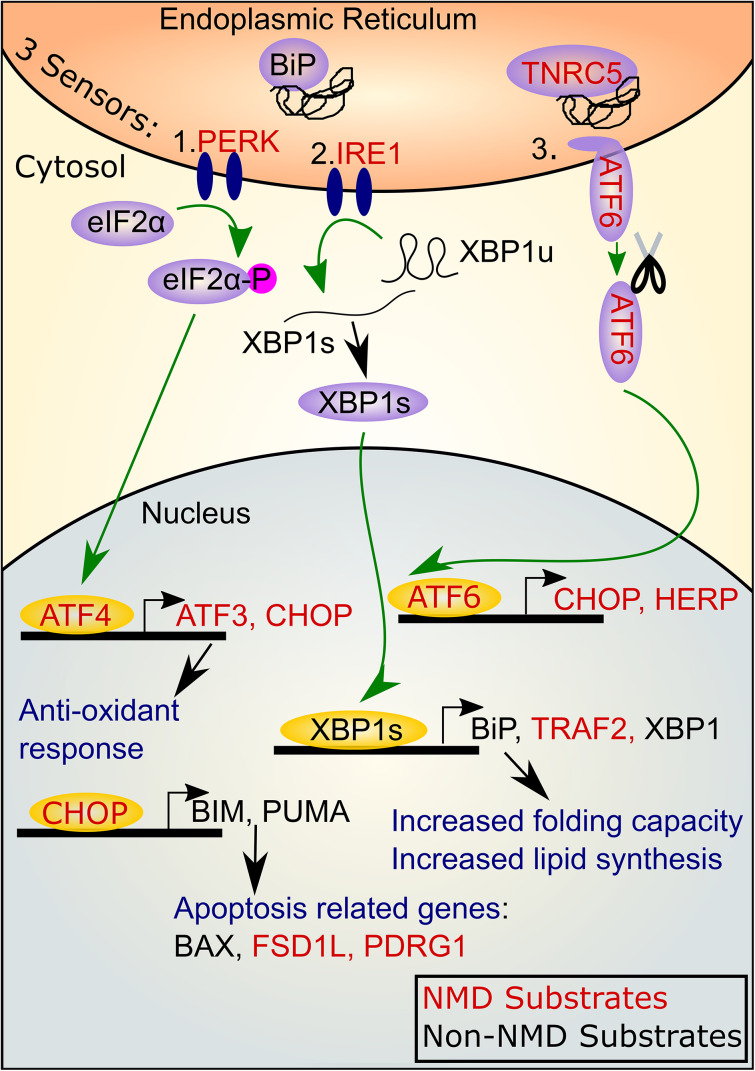
As a result of ER stress, a stress-response pathway termed the unfolded protein response (UPR) is activated. The UPR pathway upregulates factors that allow cells to adapt to the abnormal numbers of unfolded proteins and thereby return to normal homeostasis. Three distinct branches of the UPR pathway have been identified, each of which has a different sensor and activates a different (but sometimes overlapping) array of factors that permit return to normal protein folding capacity (Fig. 2). One UPR sensor is inositol requiring transmembrane kinase/endonuclease1 (IRE1). When it binds to an unfolded protein, IRE1α undergoes trans-autophosphorylation and then drives noncanonical cytoplasmic splicing of the pre-mRNA encoding the X-box-binding protein 1 (XBP1) transcription factor to generate spliced XBP1 mRNA (XBP1s) [54, 55]. XBP1 activates the transcription of genes encoding chaperones, which fold peptide chains into functional proteins. XBP1 also transcriptionally activates genes encoding lipid synthesis proteins, which serve to increase the size of the ER membrane to accommodate increased protein load. XBP1 also induces genes encoding ER-associated degradation (ERAD) proteins, which move unfolded proteins back into the ER [52]. A second UPR sensor is protein kinase RNA (PKR)-like ER Kinase (PERK). When PERK encounters an unfolded protein, it is activated by trans-autophosphorylation, leading this kinase to phosphorylate the α-subunit of eukaryotic translation initiation factor 2 (eIF2α), which, in turn, decreases the rate of protein translation initiation. Decreased translation is an important survival response, as it reduces the translation of unfolded proteins to allow the cell to properly fold the already high load of unfolded proteins. Phosphorylation of eIF2α also increases transcription of genes encoding factors important for responding to ER stress, including ATF4 and its downstream target, C/EBP homologous protein (CHOP) [52]. The third UPR sensor is activating transcription factor 6 (ATF6). Once activated by the exit of the chaperone immunoglobulin-binding protein (BiP; also known as glucose-regulated protein 78) from its binding site, ATF6 is packaged and sent to the Golgi, where it is cleaved and sent to the nucleus to activate the transcription of several genes encoding proteins localized to the ER and important for protein folding, including BiP [51]. The ability of ATF6 to transcriptionally activate the BiP gene, coupled with the fact that BiP suppresses ATF6 activation, creates a negative feedback loop to buffer the UPR pathway. Together, these three sensors allow mammalian cells to react in a comprehensive manner to abnormal levels of unfolded proteins to alleviate ER stress.
Fig. 2.
NMD degrades mRNAs encoding UPR components. A simplified diagram depicting the three UPR pathways, activated by the PERK, IRE1, and ATF6 sensors, respectively. When chaperones, including BiP and TNRC5, leave these sensors to bind unfolded proteins, the sensors activate intracellular signaling, leading to transcriptional activation, as shown. UPR components encoded by high-confidence NMD target RNAs are designated in red font; UPR components encoded by mRNAs that less evidence suggests are NMD target mRNAs are designated in black font. XBP1u XBP1 unspliced, XBP1s XBP1 spliced isoform
While the UPR permits cells exposed to stress to return to normal homeostasis, this pathway is not always able to resolve the stress in a timely manner. If the ensuing chronic stress is sufficiently strong and is sustained (>12 h), cells undergo apoptosis [56]. By analogy, the “guardian of the genome”—p53—elicits cell death if aberrations at the DNA level are not dealt with in a timely manner [57, 58]. The UPR-mediated pro-apoptotic mechanism may serve to preserve resources and prevent deleterious consequences of persistent stress. Each of the three UPR branches described above can trigger apoptosis in the event that there is excessive stress. The most conserved UPR sensor—IRE1α—elicits cell death via activation of the JNK signaling pathway, which leads to caspase-12 activation and subsequent cell death. Caspase-12 activation is specific to UPR induced programmed cell death, as it is not induced by other cellular stressors, including serum deprivation, tumor necrosis factor, or cycloheximide treatment. The PERK and ATF6 UPR sensors induce apoptosis by downregulating the anti-apoptotic protein BCL2 [53]. Given that activation of any of the three sensors can lead to apoptosis, this provides versatility in dealing with chronic stress. However, it comes at a potential cost, as in some circumstances, it is not advantageous to the organism for chronically stressed cells to die, particularly if they are only subject to low-level stress.
The UPR is tightly regulated, so that cells can appropriately respond to stress, but at the same time avoid the deleterious consequences of long-term UPR activation. A large body of research has focused on the role of post-translational pathways (such as phosphorylation events) that regulate the UPR and other stress signaling pathways [51–53]. In this review, we discuss recent work showing that the UPR is regulated by the NMD RNA degradation pathway. As described in detail below, NMD shapes the UPR, as it influences all phases of the UPR, including determining whether it initiates and dictating when it will terminate. NMD also buffers cells from an overactive UPR and thus reduces potentially deleterious consequences of the UPR, including apoptosis. In turn, the UPR regulates NMD to allow for a maximal UPR in response to acute stress.
NMD and stress responses in mammals
NMD suppresses the UPR pathway
The UPR must be regulated to maximize its physiological value. For example, the UPR is inducible to allow it to efficiently respond to high levels of unfolded and misfolded proteins. UPR inducibility is achieved through both signaling and post-translational mechanisms [50, 51, 59]. The UPR is also actively repressed when the stress is no longer encountered. This downregulatory mechanism is critical, as it avoids the potential deleterious effects of this pathway, including apoptosis and decreased translation. The UPR must also be suppressed to avoid deleterious effects when innocuous (e.g., low level) stress is encountered over long periods of time. While transcriptional regulation can, in principle, confer UPR suppression, it suffers from being slow. Furthermore, while transcriptional shut-off of UPR component genes will eliminate the production of new mRNAs encoding these components, it will not eliminate the UPR mRNAs already present. Thus, it is critical that other mechanisms exist to allow complete suppression of the UPR. Below, we discuss the role of mRNA destabilization in UPR regulation. Not only does the regulation of RNA stability permit rapid suppression of the UPR, but it has the potential to drive a more robust and rapid initial UPR by stabilizing mRNAs transcribed from UPR genes induced by stress.
The first hint that the UPR is regulated at the level of RNA stability came from Mendell et al., who found that ATF3 and ATF4 mRNA are stabilized when NMD is suppressed [10] (Fig. 2). This suggested that these two mRNAs, both of which encode transcription factors in the UPR pathway, are direct targets of the NMD pathway. ATF3, which acts in the PERK branch of the UPR and binds cAMP responsive elements, regulates several genes critical for responding to cellular stress. ATF4 acts downstream of PERK and activates the transcription of many genes encoding other UPR pathway factors. Interestingly, one of the targets of ATF4 is the ATF3 gene. Because NMD represses both of these factors, this raises the possibility that NMD could dramatically downregulate the magnitude of the UPR.
The first direct evidence that NMD regulates the magnitude of the UPR came from a study conducted by Gardner [60]. This study utilized the ER stress inducer, tunicamycin, to activate the UPR in U2OS and HeLa cells. To determine the role of NMD, the NMD factor, UPF1, was depleted. This increased the magnitude of the UPR, as measured by the level of several UPR factors, including ATF3, ATF4, CHOP, and GADD34. UPF1 knockdown also increased tunicamycin-induced phosphorylation of eIF2α, a known marker of UPR activation. Conversely, overexpression of UPF1 decreased expression of ATF3/4 and CHOP, indicating that hyperactivation of the NMD pathway suppresses UPR magnitude.
To investigate whether the ability of NMD to suppress the UPR confers a physiological benefit, Karam et al. performed dose–response studies with different levels of ER stress [61]. By investigating the effect of low levels of ER stress, these investigators aimed to determine whether NMD might dampen deleterious responses to innocuous stressors. They found that HeLa cells depleted of the NMD factor, UPF3B, exhibited UPR activation in response to low doses of tunicamycin that triggered a little response or no response in control cells. UPF3B depletion increased the level of XBP1 spliced product, phosphorylation of IRE1α, and ATF6 cleavage. This suggested that the IRE1α and ATF6 branches of the UPR pathway are specifically suppressed by the NMD pathway.
Karam et al. also obtained in vivo evidence that NMD suppresses the UPR. They found that hepatocytes from NMD-deficient mice lacking the NMD factor, UPF3B, had a lower threshold for UPR activation in response to tunicamycin than did control mice. Of note, UPF3B is required for a specific branch of the NMD pathway [62], and thus, this in vivo data, coupled with the in vitro data described above, suggest that this particular arm of NMD is critical for raising the threshold for UPR activation. Unlike the NMD pathway as a whole, the UPF3B-dependent branch of NMD is not required for embryonic development [11, 18, 23–25], but instead has roles in neural differentiation and brain development [30, 34, 63]. This raises the possibility that the UPF3B-dependent branch of NMD regulates the transient UPR activation that occurs during normal neural differentiation [64–66], as well as the chronic UPR activation that accompanies some forms of neural disease [1].
The discovery that NMD suppresses the UPR also raised the possibility that NMD has a role in the termination of the UPR after the stress has been resolved. This is important, because timely termination can prevent apoptosis and other deleterious effects of extended stress responses. Karam et al. tested the role of NMD in UPR termination and found that NMD-deficient HeLa cells exhibited a prolonged stress response when exposed to moderate doses of tunicamycin, as measured by XBP1 s, BIP, and CHOP mRNA levels 15–18 h postexposure [61]. In vivo mouse studies showed that Upf3b-null mouse hepatocytes also exhibited an abnormally long UPR in response to moderate doses of tunicamycin [61]. Together with their other findings, these results from Karam et al. support the notion that NMD protects cells not only from innocuous levels of ER stress, but also prolonged UPR in response to bona fide stress.
UPR transcripts are targeted for decay by the NMD pathway
Recently, progress has been made in determining the molecular mechanism by which NMD suppresses the UPR. As described in the Introduction, NMD specifically recognizes and degrades transcripts that harbor termination codons in specific contexts. The most reliable NMD-inducing feature is an exon–exon junction at least ~50 nt downstream of the stop codon terminating the main ORF. While the underlying molecular basis for this “−50 boundary rule” is not known, the available evidence suggests that it is the consequence of ribosomes displacing a large molecular complex that drives NMD—called the exon junction complex (EJC)—from mRNAs during the first round of translation [67]. The EJC is deposited just upstream of most exon–exon junctions after RNA splicing. According to a consensus model, if all EJCs are displaced during translation, the EJC cannot interact with NMD factors deposited upon translation termination and thus EJC-dependent NMD does not occur. Thus, it is critical that at least one exon–exon junction is downstream of the main ORF to retain at least one EJC. Stop codons closer than ~50 nt do not elicit NMD, probably because they allow the ribosome to displace the EJC, as the latter is typically centered ~24 nt upstream of the exon–exon junction and the footprint of the ribosome is ~20 nt on either side [68, 69]. In addition to exon–exon junctions downstream of the main ORF, open reading frames upstream of the main ORF—called “uORFs”—can, in some circumstances, also elicit NMD [10, 70]. While it is not known how uORFs trigger NMD, it may be through EJCs just downstream of the uORF [10, 16, 70]. A third NMD-inducing feature is long 3′UTRs, which may act by increasing the distance of factors bound at the site of translation termination and those bound at the poly(A) tail [7, 14, 19, 71, 72].
As described above, Mendell et al. were the first to identify UPR component mRNAs that are likely degraded by NMD [10]. They found that both ATF3 and ATF4 mRNA are stabilized by NMD perturbation [10], a finding that was confirmed by Gardner [60]. Gardner went on to demonstrate that uORFs in ATF4 mRNA provide the signal responsible for decay by NMD. He also identified the mRNA encoding the UPR component, CHOP, as another transcript stabilized by NMD suppression, suggesting that CHOP mRNA is another NMD target.
Spurred by these findings, Karam et al. investigated whether other UPR mRNAs might be targeted by NMD [61]. Microarray analysis identified several candidates, which Karam et al. further pursued by investigating their response to depletion of the central NMD factor, UPF1, and the branch-specific factor, UPF3B. Several UPR components were identified that were upregulated by both UPF1 and UPF3B depletion, indicating that they are likely targets of the UPF3B-dependent branch of NMD. This included the previously identified NMD targets—ATF3 and ATF4—as well as new targets—IRE1α, TRAF2, FSD1L, and TNRC5 (Fig. 2). All of these mRNAs were stabilized by UPF1 depletion, providing strong evidence that they are bona fide NMD targets. IRE1α, FSD1L, TRAF2, and TNRC5 all have relatively long 3′UTRs, raising the possibility that they are degraded through this NMD-inducing feature. In addition, TRAF2 has a uORF, and TNRC5 exists as an alternative isoform with a stop codon upstream of an exon–exon junction. Two transcripts—HERP and PERK—were identified that were upregulated and stabilized by UPF1 depletion, not UPF3B depletion (Fig. 2), suggesting that they are degraded by the UPF3B-independent branch of NMD [62]. ATF6, BAX, and PDRG1 mRNA were upregulated and stabilized by UPF3B depletion, but not UPF1 depletion, raising the possibility that these mRNAs are targeted for decay by a UPF3B-dependent mechanism that does not involve NMD. An alternative possibility is that these transcripts are targeted by a putative UPF1-independent branch of NMD. In support, both ATF6 and PDRG1 mRNA have relatively long 3′UTRs and thus may be NMD targets. However, the possibility that they are degraded by a UPF1-independent branch of NMD requires further research, as UPF1 is regarded as a central factor in the NMD pathway [9].
Karam et al. also found that UPR transcripts are regulated by the UPF3B paralog—UPF3A—which was recently shown to act primarily as an NMD repressor [26]. Consistent with this property, UPF3A was found to reverse NMD-driven downregulation of some UPR transcripts—TNRC5, TRAF2, and BAX [61]. In contrast, UPF3A appeared to downregulate some UPR transcripts [61], consistent with the evidence that UPF3A can sometimes serve as an NMD activator [26] and thus act redundantly with UPF3B [62].
The ability of NMD to target UPR component genes was confirmed recently by a proteomic study conducted by Sieber et al. [73]. Through Pulse-SILAC and click-chemistry, these investigators found that several UPR proteins induced by the stressor, Dithiothreitol, were also upregulated in response to UPF1 depletion. Interestingly, only two of the three UPR branches (PERK and IRE1) were impacted, suggesting a selective effect of NMD. This study was also significant, because it showed that NMD-driven shifts in UPR component mRNAs are also manifest at the protein level.
The above studies defined many RNAs encoding UPR components targeted by NMD. Which, if any, of these transcripts must be degraded by NMD to drive UPR suppression? Karam et al. considered IRE1α mRNA as a particularly appealing candidate to be critical for UPR suppression, based on their findings that: (1) IRE1α mRNA was among the most strongly upregulated mRNAs in response to NMD perturbation, (2) IRE1α protein levels were increased and more phosphorylated in NMD-deficient cells, and (3) IRE1α protein was elevated in both NMD-deficient human cells and mice, regardless of which NMD factor was depleted and regardless of whether the UPR was induced [61]. In order for IRE1α mRNA to directly participate in NMD-mediated UPR repression, it is critical that it is directly targeted for decay by NMD. While, as described above, Karam et al. obtained some evidence that this is the case, to definitively evaluate this possibility that they examined whether IRE1α’s long 3′UTR confers NMD-dependent degradation. They inserted its long 3′UTR into a β-globin mini-gene reporter system and showed that this downregulated the reporter mRNA in an UPF1-dependent manner [61]. This confirmed that IRE1α is an NMD direct target and demonstrated that it is mediated by its 3′UTR region. To determine whether downregulation of IRE1α has a role in the ability of NMD to suppress the UPR, Karam et al. performed rescue experiments. In one rescue experiment, they prevented the upregulation of IRE1α that normally occurs in NMD-deficient cells (by depleting IRE1α with RNAi) and found that this increased the UPR activation threshold, as measured by the dosage of tunicamycin required to induce BiP and CHOP expression [61]. They then confirmed this using an IRE1α inhibitor, STF-083010, which inhibited the upregulation of BiP and CHOP in NMD-deficient cells upon exposure to tunicamycin. Together, these data constituted strong evidence that the ability of NMD to degrade IRE1α mRNA is essential for NMD to strongly suppress the UPR. IRE1α mRNA is the first NMD target mRNA shown to have functional consequences in a biological system. Since then, other mRNAs have been defined that must be degraded by NMD to drive biological responses: Smad7 mRNA (its decay maintains the stem cell state in neural precursors) [29], c-myc mRNA (its decay permits mouse embryonic stem cell differentiation) [23], and Gadd45 mRNA (its decay protects from embryonic lethality) [74].
The UPR suppresses the NMD pathway
The finding that NMD suppresses the UPR presents a problem—how can a strong UPR occur in the face of the inhibitory effects of NMD? From a teleological perspective, it would seem counter-productive for NMD to perturb a robust stress response when needed to resolve pervasive ER stress. Indeed, evidence suggests that NMD does not prevent a strong UPR. For example, a deficiency in NMD was found to not significantly impair the UPR of either cultured HeLa cells or mouse hepatocytes in vivo in response to maximal doses of the ER stressor, tunicamycin [61]. A different stressor, thapsigargin, also triggered a normal UPR in NMD-deficient HeLa cells [61]. Together with the data described in the above sections, these results suggest that while NMD raises the threshold of UPR activation and facilitates timely termination of the UPR, it does not inhibit the UPR when cells are challenged with strong ER stress (Fig. 3).
Fig. 3.
NMD promotes timely termination of the UPR. a The UPR can be thought of as a clock. If the stress is resolved in a timely manner, the result is homeostasis. Alternatively, prolonged stress leads to apoptosis. NMD promotes both maintenance of homeostasis and timely termination of the UPR. b NMD is suppressed by the UPR to allow for maximal activation of the UPR pathway. After the resolution of the stress, the UPR is downregulated, which is thought to rescue normal levels of NMD, thereby leading to further downregulation of the UPR. This regulatory spiral eventually leads to complete shut-off of the UPR pathway. c In chronic stress, the UPR is constitutively activated and NMD is suppressed, which, together, typically leads to apoptosis
How might this be achieved? Several lines of evidence suggest that NMD does not inhibit the UPR in response to a high magnitude of ER stress by virtue of the fact that NMD is repressed under such conditions. Indeed, this has been shown in several cell types. For example, Wang et al. showed that treatment of MEFs with arsenic, a cellular stress inducer, decreased NMD magnitude, as assessed with a NMD β-globin reporter [75]. This reporter system has two forms of the β-globin gene—one with an NMD-inducing feature and the other without—and thus, the ratio of reporter expression from these two genes is a measure of NMD magnitude. Gardner found that hypoxic stress also reduces NMD activity in U2OS cells, as assessed by NMD reporter activity and RNA half-life analysis of known NMD target mRNAs [60]. Another study showed that the ER stress inducer, thapsigargin, reduced NMD activity in C2C12 myoblast cells, as assessed by upregulation of the endogenous NMD substrate Snhg1, reduced Upf1-phosphorylation, and reduced expression of several NMD genes, including Upf1, Eif4A3, and Smg6 [76]. Finally, it was shown that ER stress also suppresses NMD in Neuro-2a neuroblastoma cells; thapsigargin was found to inhibit NMD via the PERK pathway in these neural-lineage cells [77].
How does ER stress elicit NMD suppression? Several lines of evidence suggest that cellular stress achieves this by triggering phosphorylation of the translation initiation factor eIF2α (Fig. 4). One means by which this is accomplished is through the ER stress sensor, PERK, a kinase that directly phosphorylates eIF2α. Extrinsic activation of PERK using the small molecule AP20187—in either murine embryo fibroblasts (MEFs) or Chinese hamster ovary cells (CHOs)—triggered both eIF2α phosphorylation and stabilization of an NMD reporter [75]. In contrast, mutant MEFs with a mutant form of eIF2α that cannot be phosphorylated did not exhibit suppressed NMD in response to PERK activators [75]. As further evidence that eIF2α phosphorylation inhibits NMD, another eIF2α kinase, PKR, also was found to inhibit NMD. Vaccinia virus infection, which activates this kinase, was also found to inhibit NMD, while a mutant vaccinia virus that lacks the ability to phosphorylate eIF2α did not significantly perturb NMD [75] (Fig. 4). As described below, NMD is also inhibited by many other conditions that trigger eIF2α phosphorylation; e.g., hypoxia, amino-acid starvation, double-stranded RNA, and reactive oxygen species. Together, these experiments strongly support the notion that a major means by which stressors repress NMD is through eIF2α phosphorylation.
Fig. 4.
UPR inhibits NMD through eIF2α phosphorylation, a post-translational event that gives rise to stress granules (SGs). Various stressors—including ER stress, hypoxia, chemical agents, and viruses—can trigger eIF2α phosphorylation. This phosphorylation event causes the formation of stress granules, which are enriched in translationally arrested mRNAs. NMD requires translation and thus SGs may be subcellular sites where NMD is repressed. NMD promotes SG formation, suggesting the existence of a negative feedback loop. SGs are also considered to be sites where small RNAs such as miRNAs are functionally silenced
How does eIF2α phosphorylation inhibit NMD? Because NMD depends on in-frame stop codons and thus translation [78, 79], one obvious potential mechanism is the well-known ability of eIF2α phosphorylation to inhibit translation [80]. While an appealing mechanism, eIF2α phosphorylation only decreases translation by 20–45% [75], which casts doubt as to whether this is sufficient to inhibit NMD. Indeed, dose–response studies with several different translation inhibitors (cycloheximide, anisomycin, and emetine) have shown that a well-established NMD substrate (out-of-frame T-cell receptor mRNA) is only upregulated when protein synthesis is inhibited by greater than 70% [78, 81]. Nevertheless, it is possible that the translational repression conferred by eIF2α phosphorylation can influence NMD magnitude in some cell types and/or developmental stages. Thus, the mechanism by which eIF2α phosphorylation inhibits NMD is still under investigation (see also the “Stress Granules” section, below).
Another mechanism by which eIF2α phosphorylation may stabilize mRNAs—particularly stress-response-related mRNAs—is through “local,” rather than “global,” control of NMD. One likely target of this local regulation is ATF4. This UPR transcription factor is encoded by an mRNA with two uORFs, one of which overlaps with the main ORF and thus inhibits ATF4 translation. These uORFs allow ATF4 mRNA to undergo a translational switch in response to different degrees of eIF2α phosphorylation [82]. Under nonstress conditions—when eIF2α phosphorylation is low—the uORFs are translated (by virtue of their being 5′ and thus read first by the scanning 40S subunit), thereby inhibiting translation of the downstream main ORF encoding ATF4 protein. In response to stress, eIF2α becomes hyperphosphorylated, which inhibits translation of the uORFs, allowing for translation of the main ORF and hence ATF4 protein. Not only does eIF2α phosphorylation promote ATF4 translation, but it also likely promotes ATF4 mRNA stability. This follows from the fact that ATF4 mRNA is a direct NMD target that is degraded by NMD through a uORF-dependent mechanism [60]. Thus, translation of the ATF4 uORF under nonstress conditions would be predicted to trigger uORF-dependent NMD, while the switch to main ORF translation in response to stress should allow evasion from NMD, thereby stabilizing the ATF4 mRNA [83]. Another likely example of this type of regulation is conferred on the stress pathway protein GCN4, as it is encoded by an mRNA that has multiple uORFs that inhibit GCN4 translation specifically under nonstress conditions [83]. The potential ability of stress to trigger NMD evasion, coupled with stress-induced increased translation, provides a powerful 2-prong mechanism to increase the expression of these stress pathway proteins.
The NMD and UPR pathways intersect as a means to efficiently respond to ER stress
Together, these findings support a model in which NMD and the UPR regulate each other in a manner that generates an efficient ER stress protective mechanism that exhibits both high specificity and signal-to-noise ratio (Fig. 3). In this model, NMD inhibits the expression of critical UPR components, including IRE1α, to largely prevent inappropriate activation of the UPR pathway in response to innocuous or low levels of ER stress. However, when strong stress is encountered, this leads to inhibited NMD, which relieves its inhibitory effect on the UPR, allowing for maximal UPR pathway stimulation. As the UPR resolves the cellular stress, NMD magnitude is allowed to increase, which facilitates termination of the UPR (Fig. 3).
NMD suppression has differential effects in mammals and C. elegans
The finding by several groups that NMD perturbation upregulates the levels of UPR components in response to ER stress [10, 60, 61, 76] raises the possibility that suppression of NMD alone is sufficient to induce ER stress. Given that NMD degrades large batteries of aberrant mRNAs [9, 14, 27, 72], NMD suppression would allow these aberrant mRNAs to accumulate, which alone could potentially elicit ER stress. To determine whether this was the case, Gardner measured several hallmarks of UPR activation following NMD suppression [60]. To assess PERK activation, eIF2α phosphorylation was measured, and to assess IRE1α activation, XBP1 splicing, and the expression of downstream UPR genes were measured. He found that NMD suppression did not elicit detectable activation of either the PERK or IRE1α branches of the UPR pathway in U2OS and HeLa cells. Another study conducted in HeLa cells came to the same conclusion [73], as did a study performed in C2C12 myoblast cells [76]. In addition, Karam et al. examined whether perturbation of the UPF3B-dependent branch of NMD was sufficient to elicit UPR activation and found no evidence for this either in HeLa cells or mouse hepatocytes in vivo [61]. The collective findings of these many independent studies, conducted in different contexts, provide strong evidence that NMD inhibition is not sufficient to activate the UPR in mammalian cells, i.e., perturbation of the NMD pathway does not cause ER stress. Instead, as described above, many lines of evidence strongly support the notion that NMD perturbation activates the UPR only in the presence of ER stress.
In striking contrast to the above findings in mammalian cells, Sakaki et al. found that—in Caenorhabditis elegans—NMD perturbation is sufficient to elicit ER stress [84]. This study demonstrated that NMD-deficient mutant worms (with mutations in the smg1, smg4, or smg6 gene) exhibited heightened heat shock response activity and increased expression of heat shock response genes in the absence of external stressors. This suggests that NMD suppression itself induces a stress response. The authors speculated that NMD suppression may activate the UPR in C. elegans, because it allows aberrant mRNAs to encode aberrant proteins, including unfolded proteins. Intriguingly, Sakaki et al. also obtained evidence that NMD proteins are, in part, localized to the ER, which places the NMD machinery in the right location to directly respond to ER stress.
Thus, it would appear that C. elegans uses the NMD pathway to actively suppress ER stress. In support of the possibility that this is also the case in mammalian cells, Sakaki et al. found that depletion of the NMD factor, SMG6, was sufficient to induce ER stress in HeLa cells [84]. In particular, they found that SMG6 depletion increased CHOP and GRP78 levels, ATF4 activity, and XBP1 splicing. Conversely, SMG6 overexpression decreased CHOP expression in response to the ER stress inducer tunicamycin [84]. These results suggested that SMG6 suppresses the UPR, which raised the possibility that perturbation of NMD is sufficient to trigger the UPR in mammalian cells, just as it does in C. elegans. This contrasts with the studies described above, which showed that depletion or loss of several other NMD factors from HeLa cells, other mammalian cell lines, and hepatocytes in vivo did not trigger ER stress [60, 61, 76]. One explanation for this discrepancy is that perturbation of a non-NMD SMG6 function is responsible for the ER stress in SMG6-depleted cells. In support of this possibility, SMG6 has been shown to have non-NMD functions, including telomerase binding and chromosome uncapping [85–87]. In further support, Sakaki et al. found that depletion of another NMD factor, SMG1, did not trigger the UPR in HeLa cells as measured by BiP mRNA levels [84]. Thus, the finding that depletion of SMG6 triggered the UPR in mammalian cells may be because of a SMG6 function that is not shared with most other NMD factors.
Thus, the available evidence suggests that the relationship of NMD and the UPR is different in mammals and C. elegans. In mammals, NMD is critical for responding appropriately to exogenous ER stress. NMD shapes the UPR by both increasing the threshold for the initial UPR activation (to avoid inappropriate activation in response to innocuous ER stress) and by triggering the timely termination of the UPR once the stress has been resolved. NMD performs these functions by targeting UPR components for degradation. In contrast, in C. elegans, NMD protects from endogenous ER stress, such as might be triggered by high levels of abnormal proteins translated by C. elegans mRNAs normally degraded by NMD. Thus, NMD serves to actively prevent UPR activation in C. elegans. Further studies may shed light on why there appears to be a fundamental difference in how NMD impacts the UPR pathway in different species.
Stress granules and RNA degradation
Stress granules (SGs) are nonmembrane-bound cytoplasmic bodies that frequently form as a consequence of stress [88]. These dynamic cytoplasmic bodies contain translation elongation factors, other RNA-binding proteins, post-translational modifying enzymes, and signaling components. They form in response to several kinds of mammalian stress, including ER stress, hypoxia, heat shock, DNA damage, oxidative stress, and inflammation [89].
The formation of SGs is triggered by eIF2α phosphorylation [89]. Because this post-translational event also inhibits NMD [75, 90, 91], this raises the hypothesis that cellular stress inhibits NMD through a mechanism involving SGs. An attractive model for how this could occur is supported by three key findings. First, eIF2α phosphorylation is known to lead to inhibited translation of most mRNAs. Indeed, SGs are enriched in mRNAs that are translationally silenced and thus are regarded as sites of translational repression [89, 92, 93] (Fig. 4). Second, NMD requires translation [78]. This follows from the fact that NMD is triggered by stop codons in specific contexts [9]. Third, SGs sequester NMD factors in response to stress. In particular, SG-inducing stressors cause several key NMD factors, including UPF1, SMG1, and UPF2, to become enriched in SGs and thus depleted in the surrounding cytosol [60, 94, 95]. Together, this supports a model in which translationally active mRNAs in the cytosol in stressed cells would not have access to NMD factors and thus would be immune to NMD.
A key feature of this model is that SGs are sites of inactive translation. It remains to be determined how eIF2α phosphorylation leads to this translationally repressed state. One possibility—the “transport model”—is that mRNAs bound by ribosomes associated with phosphorylated eIF2α are marked for transport to SGs [60]. Once in the SG, such mRNAs become translationally repressed and largely immune to NMD. An alternative possibility—the “aggregation model”—posits that eIF2α phosphorylation stalls translation, which, in turn, coaxes messenger ribonucleoproteins (mRNPs) to coalesce into NMD-incompetent SGs. While both models are well supported and thus worthy of further investigation, it is worth noting that there is evidence that eIF2α phosphorylation inhibits NMD by a translation-independent mechanism [75]. Thus, it will also be important to further explore whether this is, indeed, the case and to determine what specific mechanism downstream of eIF2α phosphorylation leads to inhibited NMD.
Not only do SGs appear to influence NMD, but there is evidence that NMD promotes the formation of SGs. Knockdown of SMG1—a key component of the NMD pathway—decreases the number of SGs formed in response to sodium arsenite treatment [94]. Together with the evidence that SG formation inhibits NMD and the finding that NMD factors become concentrated in SGs in response to stress, this supports the existence of a feedback loop that maintains RNA metabolism homeostasis by buffering both RNA decay and translation from stress insults. This is likely to be physiologically important, as selective RNA decay and translational regulatory pathways influence many developmental events and are linked to disease when defective [11, 28–32, 36, 37, 96].
Another RNA degradation pathway—Staufen-mediated mRNA decay (SMD)—has the opposite effect as NMD: it suppresses SG formation. When STAU1 is depleted from stressed cells, the number of SGs is increased [97]. Conversely, STAU1 overexpression reduces the number of SGs induced in response to stress [97]. The finding that SMD has the opposite effect on SG formation as NMD is consistent with the fact that SMD and NMD are competitive pathways that both depend on the RNA helicase UPF1 [37]. In the future, it will be interesting to determine the mechanism by which SMD suppresses SG formation. One possibility is that SMD acts indirectly, e.g., it inhibits SG formation through SMD’s ability to inhibit NMD activity (Fig. 4). A nonmutually exclusive possibility is that SMD or its components inhibit SG formation more directly. In this regard, the protein which is most critical for the SMD pathway—STAU1—is enriched in SGs [97], and thus, it is possible that this RNA-binding protein could directly impact SG formation and/or disassembly.
Other RNA decay pathways also appear to be impacted by SGs. A prime example is the microRNA (miRNA) pathway. AGO2—a key component of the miRNA machinery—is enriched in SGs when cells are incubated with hippuristanol, an eIF4A translation factor inhibitor that induces SG formation [98]. Hippuristanol also causes AGO2 to undergo polyADP-ribosylation, which is likely to cripple AGO2’s ability to interact with target mRNAs [99]. Other stressors that trigger SG formation—including arsenite, heat shock, and glucose deprivation—also trigger AGO2 polyADP-ribosylation [99]. The polymerases responsible for this post-translational modification are also recruited to SGs during stress, so their colocalization in SGs could be a means to post-translationally modify AGO2 and decrease this protein’s ability to support miRNA activity [100]. Another mechanism by which SG formation likely represses miRNAs is by repressing their synthesis. DICER—an enzyme critical for miRNA biogenesis—is decreased in level by stressors that trigger SG formation, such as reactive oxygen species (ROS) [101, 102]. Since a large proportion of miRNAs are encoded by introns, another mechanism by which SGs could regulate miRNAs is through sequestration of RNA splicing proteins. For example, the mRNA splicing factor, T-Cell-Restricted Intracellular Antigen-1 related protein, is abundant in SGs [103], and thus may be sequestered in SGs as a means to promote alternative splicing of miRNA-containing introns and thus alter miRNA levels. Another RNA regulatory mechanism that may be impacted by SGs is the AU-mediated RNA decay pathway, which degrades mRNAs harboring AU-rich elements in their 3′UTR. AUF1, an RNA-binding protein that binds to AU-rich RNA, is recruited to SGs in response to viral infection [104].
In conclusion, the discovery that factors from many different mRNA degradation pathways are enriched in SGs in response to stress raises the possibility that these cytoplasmic bodies are key regulators of RNA stability. This notion is supported by functional studies showing that depletion of SG-inducing proteins decreases mRNA levels globally, consistent with reduced mRNA stability [105, 106]. However, it is important to keep in mind that SGs may, instead, be a consequence of translation repression rather than a cause of translational repression. For example, translationally repressed mRNAs may have intrinsic properties that cause them to aggregate to form SGs, but the SGs do not have a function in RNA decay per se. In support, one study found that inhibition of SG formation had no effect on mRNA degradation rates [107]. One possibility is that eIF2α phosphorylation—the upstream driver of SG formation—is responsible for inhibiting RNA decay, not SGs themselves. Since several steps are required downstream of eIF2α phosphorylation for SG formation [108], it is reasonable that one or more of these steps are involved in regulating RNA decay, not SGs themselves. Thus, further work is required to elucidate whether SGs have an active role in regulating RNA degradation pathways—such as NMD—or they are merely a consequence of such regulation.
Stress and NMD in tumors
NMD magnitude is suppressed in tumors
Tumors typically encounter stressors that come in many forms, including hypoxia and ER stress [2]. As NMD is repressed by some forms of stress, it follows that NMD also might be repressed in tumors. In support, Wang et al. found that subcutaneous injection of PC3 prostate tumor cells into mice caused these cells to exhibit decreased NMD magnitude [75]. Using a B-cell tumor line in which the levels of the oncogene, c-myc, can be manipulated, these investigators found that c-myc has a role in the repression of NMD magnitude [91]. Interestingly, c-myc overexpression induced eIF2α phosphorylation in this B-cell tumor line, suggesting that c-myc suppresses NMD through an eIF2α phosphorylation-dependent mechanism. Wang et al. obtained evidence that c-myc activates phosphorylation of eIF2α through induction of ROS, based on the finding that the ROS scavenger, N-acetylcysteine, reversed c-myc-induced NMD suppression [91].
To further assess whether c-myc inhibits NMD, Wang et al. investigated whether any mRNAs upregulated by c-myc are likely to be NMD targets, based on their being stabilized by both NMD inhibition and translation inhibition. They identified 63 mRNAs that fit these criteria [91]. This degree of overlap was reasonably high, considering that the data sets were from different cell lines (P493 and U2OS cells were used to identify c-myc- and NMD-regulated transcripts, respectively). Together, these data are consistent with the notion that many c-myc "target genes” are actually mRNAs regulated indirectly through the ability of c-myc to suppress NMD.
NMD is a tumor suppressor pathway
The finding that tumors exhibit suppressed NMD raises the possibility that this imparts a selective advantage to tumors, because NMD is a tumor suppressor pathway. In support of this notion, Wang et al. found that overexpression of the NMD factor, UPF1, reduced both the number and size of PC3 prostate tumor cell colonies in soft agar [75]. Clinical evidence for this notion comes from recent studies showing that debilitating mutations in the UPF1 gene are extremely common in pancreatic adenosquamous (ASC) tumors [109] and inflammatory myofibroblastic tumors (IMT) [41]. In both ASC and IMT, the UPF1 mutations are somatic and they are clustered in specific regions of the gene that cause alternative UPF1 splicing, leading to low or undetectable levels of UPF1 protein. Consistent with low UPF1 expression, NMD is greatly suppressed, as shown by the elevated levels of NMD substrate mRNAs in these tumors. In the case of IMT, one elevated NMD substrate is the mRNA encoding mitogen activated protein kinase kinase kinase 14 (MAP3K14/NIK), an enzyme in the NFκb pathway known to trigger inflammatory responses, including chemokine expression. Consistent with this, IMTs with UPF1 mutations were found to have elevated levels of chemokines and infiltrating B cells. Together, these data support a model in which UPF1 mutations downregulate NMD, leading to NIK mRNA upregulation and the consequent immune infiltration characteristic of benign IMTs [41].
In the case of ASC, it is unknown what transcripts dysregulated by NMD contribute to the generation of these malignant tumors. Wang et al. found that the high-confidence NMD target transcripts in another tumor type—osteosarcoma—encode several statistically overrepresented categories of proteins, including those involved in cell cycle, cell growth, growth factor signaling, cell migration, and apoptosis [91]. This raises the possibility that NMD acts as a tumor suppressor pathway by degrading transcripts encoding proteins promoting cell growth and survival. In tumors in which NMD is suppressed, these transcripts would be stabilized, leading to higher expression of growth- and survival-promoting proteins, and thus more favorable conditions for the tumor.
While there is growing evidence that the NMD pathway can suppress tumors, in principle, NMD could also have the opposite effect in specific cell types. In support, Lou et al. showed that overexpression of the NMD factor, UPF1, promoted the growth of P19 embryonal carcinoma cells, while UPF1 depletion had the converse effect [29]. While the underlying mechanism was not determined, UPF1 was found to destabilize transcripts encoding cell cycle inhibitors, consistent with NMD promoting cell growth by decreasing the levels of anti-growth molecules. Another mechanism by which NMD may promote tumors is by inhibiting differentiation. Lou et al. found that UPF1 promoted the decay of many mRNAs encoding pro-neural differentiation proteins, and identified one—Smad7 mRNA—which they found through rescue experiments must be degraded for NMD to maintain cells in the undifferentiated cell state [29]. Given that SMAD7 is a TGFβ signaling inhibitor, this raises the possibility that NMD acts to influence proliferation vs. differentiation decisions by modulating signaling mechanisms. Indeed, a recent study linked the ability of NMD downregulation to trigger endoderm differentiation with its ability to regulate the levels of several RNAs encoding signaling pathway components [28]. Downregulation of NMD appears to be a common theme during development, as it has also been shown to occur during muscle and adipocyte differentiation [37, 110].
Glutathione-mediated defense against ROS stress
A common form of stress elicited under many circumstances, including in the tumor microenvironment, is ROS [111]. Indeed, tumor cells themselves can generate ROS and thereby trigger the death of surrounding cells [111]. ROS-induced damage is counteracted by glutathione (GSH), a well-studied antioxidant that directly neutralizes free radicals and reactive oxygen compounds [111, 112]. GSH is a tripeptide made up of glutamic acid, glycine, and cystine, the latter of which is an oxidized form of cysteine. Cystine is transported to the inside of cells though a cystine-glutamate antiporter called xCT. The critical subunit of this cystine-glutamate transporter is SLC7A11, which is encoded by an RNA that is targeted for decay by NMD [113, 114] (Fig. 5). This has important potential implications, as it predicts that many circumstances that lead to inhibited NMD (as described in the sections above) will raise SLC7A11 level and thus increase the levels of the amino acids needed to generate GSH for protection against ROS-induced damage. Indeed, it has been empirically found that perturbed NMD triggers increased SLC7A11-mediated cystine transport and increased cellular GSH levels [114]. Both hypoxia and the UPR-activating agent, tunicamycin, increased GSH and SLC7A11 mRNA levels, mimicking the effects of NMD inhibition [114]. This effect depends on eIF2α phosphorylation, consistent with hypoxia and tunicamycin acting by inhibiting NMD.
Fig. 5.
Model for how NMD regulates autophagy and amino-acid availability. The PERK arm of the UPR, as well as other cues, including Rapamycin, can trigger autophagy in cells via induction of ATF4, a transcription factor that, in turn, drives the expression of another transcription factor, CHOP. CHOP activates transcription of genes encoding essential autophagy factors, including those shown in the figure. ATF4 and CHOP are both encoded by NMD target mRNAs, providing a molecular basis for how high NMD activity represses autophagy. Since amino acids are recycled by autophagy, one downstream consequence of this regulation is altered amino-acid availability. NMD also impacts amino-acid availability by repressing the expression of the cystine-glutamate transporter xCT. NMD targets SLC7A11 mRNA, which encodes one of the subunits of this amino-acid transporter
These results raise the possibility that NMD is an integral part of an inducible system that provides antioxidant protection. In this system, stress inhibits NMD, which leads to increased cystine transport and consequent increased levels of the antioxidant GSH. If this inducible model is correct, this leads to the prediction that suppressing NMD would enhance the ability of cells to handle H2O2 toxicity. In support, Martin et al. depleted U2OS cells of UPF1 and found that this treatment allowed the cells to survive higher doses of H2O2 [114]. This effect depended on SLC7A11, as depletion of SLC7A11 did not allow NMD-suppressed cells to survive high H2O2 doses [114]. These data indicating that the NMD pathway reduces the ability of cells to survive H2O2 stress contrast with NMD’s ability to increase survival in response to ER and hypoxic stress [60, 61, 76, 84]. The finding that suppressed NMD enhances cell survival to H2O2 stress raises the possibility that combination treatment with both NMD repression therapy and antioxidants is a more efficacious prophylactic treatment for cancer than antioxidants alone.
NMD and autophagy
One important mechanism to adapt to stress is autophagy, a recycling process that replenishes the building blocks required to synthesize proteins and organelles, as well as to generate energy [115]. During autophagy, the cell forms a double-layered membrane around the targeted organelle or targeted protein aggregate; this autophagosome then fuses with lysosomes to degrade the contents for recycling (Fig. 5). Autophagy is a constitutive process but is upregulated under some circumstances, including cellular stress, as a means to replenish/maintain amino-acid levels, maintain protein synthesis, and maintain ATP levels [115]. The well-established finding that the UPR stimulates autophagy [116], coupled with the discovery that NMD suppresses the UPR [60, 61, 76, 84], led Wengrod et al. to hypothesize that NMD represses autophagy. In support of this hypothesis, these investigators inhibited NMD in U2OS cells and observed several hallmarks of increased autophagy [117]. Combination treatment with rapamycin, an autophagy inducer, and NMD inhibition, resulted in more autophagosomes than rapamycin treatment alone, providing evidence that NMD also increases the threshold for induced autophagy [117]. Confidence as to generality was instilled by finding similar results in two independent cell lines and with two in vivo mouse models [117, 118]. Further evidence that NMD suppresses autophagy and increases the threshold for autophagosome formation comes from experiments in which the NMD factor, UPF1, was overexpressed [117]. NMD appears to inhibit autophagy at an early stage (Fig. 5), as a late-stage autophagy inhibitor, chloroquine, did not significantly impair this regulation [117].
Given that autophagy recycles amino acids [115], one potential consequence of the ability of NMD to inhibit autophagy is reduced amino-acid availability (Fig. 5). If correct, this predicts that NMD activation would decrease amino-acid levels. Indeed, Wengrod et al. found that overexpression of the NMD factor, UPF1, led to decreased levels of several amino acids [117]. This is a potentially important consequence of NMD activation that could potentially affect the growth of both normal and malignant cells.
How does NMD suppress autophagy? Wengrod et al. tested whether NMD achieves this by targeting the mRNA encoding the UPR factor, ATF4. ATF4 seemed a particularly good candidate to play this role, as it is a transcription factor that activates the autophagy genes LC3B and ATG5 [115] and is encoded by a well-established direct NMD target [11, 23, 60, 61, 119]. To test its role, Wengrod et al. asked whether ATF4 depletion rescues autophagy inhibition in the face of inhibited NMD. Indeed, they found that depletion of both ATF4 and the NMD factor, UPF2, resulted in less autophagy compared to depletion of UPF2 alone [117]. Together, these data indicate that NMD shapes the autophagy pathway by degrading a specific mRNA.
NMD and apoptosis
NMD protects cells from stress-induced apoptosis
Because NMD is critical for shaping stress responses, it would not be surprising if it also influenced the end stage of prolonged stress—apoptosis. Indeed, several studies have shown that NMD protects cells from stress-induced apoptosis. For example, Sakaki et al. found that SMG6-depleted HeLa cells treated with tunicamycin exhibited ~50% reduced cell survival compared to control cells, while SMG6 overexpression increased HeLa cell viability [84]. The UPF3B-dependent branch of NMD is critical for protection from apoptosis, based on Karam et al.'s finding that UPF3B-depleted HeLa cells exhibited increased apoptosis in response to the ER stressor, tunicamycin, compared to control cells [61]. UPF3B dependence was also observed in vivo, based on the finding that hepatocytes in liver from Upf3b-null mice treated with tunicamycin exhibited increased apoptosis compared to control hepatocytes [61].
Another stress-inducing scenario that has been examined in terms of NMD protection is depressed autophagy, which can lead to increased apoptosis because of reduced amino-acid recycling, increased ROS production, and reduced clearance of dysfunctional organelles or aggregates [120–122]. Evidence that NMD is critical for protection from this form of stress was the finding that NMD factor depletion increased the apoptosis observed following autophagy inhibition [117]. While the underlying mechanism was not investigated, one possibility is that autophagy is important for clearing misfolded truncated proteins translated from mRNAs harboring premature termination codons that are generated at high levels when NMD is inactivated.
Another scenario in which NMD may suppress apoptosis is during the early embryonic development. In flies, zebrafish, and mice, either depletion or loss of any of a number of NMD factors results in massive apoptosis, coupled with early embryonic lethality [11, 23–25, 31]. However, whether NMD is directly responsible for driving cell survival or, instead, it acts indirectly, remains to be determined.
Of note, NMD does not impact the sensitivity to all apoptosis-inducing agents, and thus, there is some selectivity in NMD’s actions. For example, Jia et al. found that depletion of the NMD factors, UPF1 or UPF2, did not significantly influence sensitivity to the apoptosis-inducing agent, staurosporine [123]. One possible explanation for this stems from the fact that staurosporine inhibits NMD [123, 124]. Thus, NMD may be sufficiently compromised by staurosporine action that knockdown of NMD factors has no further impact.
NMD degrades specific RNAs to protect cells from apoptosis
To begin to understand the underlying mechanism by which NMD protects cells from apoptosis, Nelson et al. performed a suppressor screen in D. melanogaster [74]. Their goal was to determine whether mutation of any genes could restore viability to NMD-deficient flies. For their screen, they used Upf2-hypomorphic flies, as ~10% survive to adulthood, thereby providing a baseline for which to compare with [74]. This suppressor screen revealed that mutations in one particular gene—growth arrest and DNA damage inducible 45 (Gadd45)—restored viability to these NMD-deficient mutant flies. Mutations in Gadd45 almost completely restored viability to hypomorphic NMD mutants; and it even improved viability in flies completely lacking the NMD pathway, i.e., in Upf1-null and Upf2-null flies. Nelson et al. also examined eye morphology, as surviving Upf2-hypomorphic flies have smaller clonal patches of eye cells compared to wild-type flies. They found that Gadd45/Upf2 double-mutant flies had significantly larger eye patch size than Upf2-mutant flies, providing further evidence that loss of Gadd45 improves the viability of NMD-deficient cells [74]. These findings, coupled with the fact that GADD45 is a pro-apoptotic molecule [125, 126], and it is encoded by a mRNA that is a direct NMD target transcript [71], led Nelson et al. to propose a model in which NMD normally degrades Gadd45 transcripts to avoid its pro-apoptotic effects. In scenarios in which NMD is suppressed, such as stress, Gadd45 transcripts are stabilized, leading to increased GADD45 protein expression and consequent apoptosis [74].
Gadd45 is known to trigger apoptosis by activating the MTK1 kinase in the MAPK signaling pathway (Fig. 6) [125]. Consistent with this activity, MAPK pathway activation triggers apoptosis in the fly eye, just as Gadd45 overexpression does [126]. This led Nelson et al. to consider the possibility that MAPK pathway activation could also be responsible for the increased embryonic lethality in Upf2-hypomorphic flies. To test this, they investigated lethality of flies null for the Drosophila MTK1 orthologue, Mekk1, crossed with NMD-mutant flies. They found that Mekk1 mutations restored viability to Upf2-hypomorphic flies and even partially suppressed lethality of Upf1 and Upf2-null flies. Together, these data provided strong evidence that Gadd45 promotes apoptosis in flies, and that NMD prevents apoptosis through targeting of the Gadd45 and the MAPK signaling pathway.
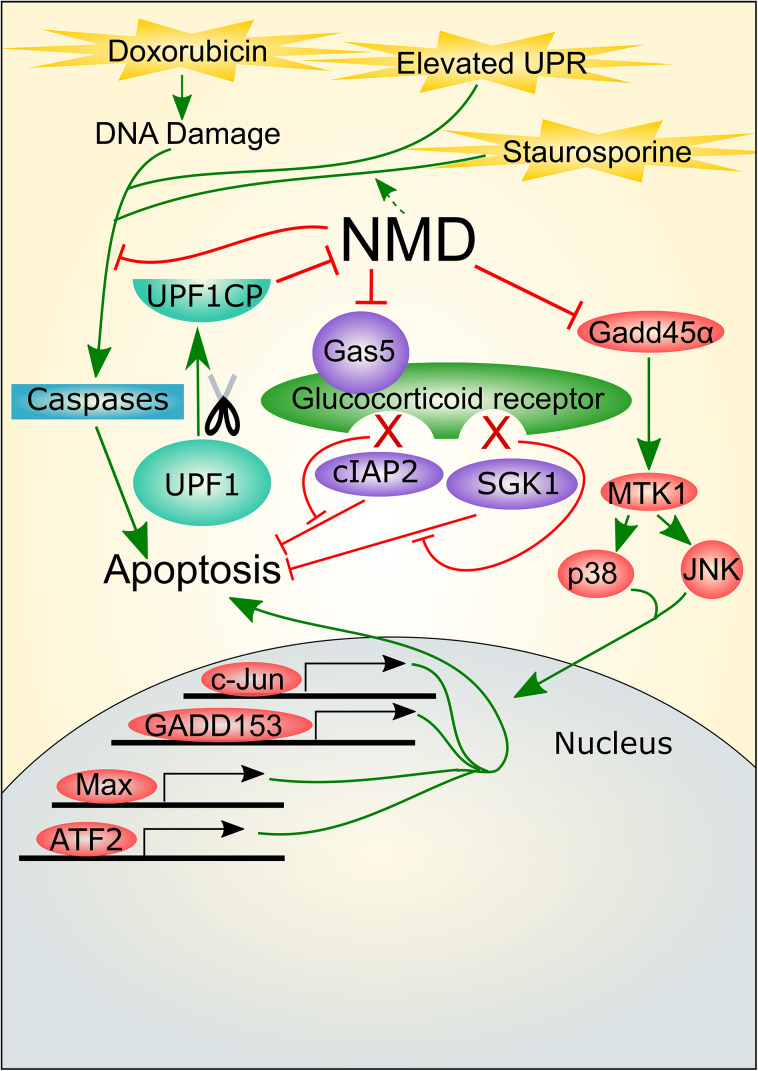
Fig. 6.
NMD and apoptosis have a complex regulatory relationship I. NMD provides protection from apoptosis-inducing agents, including chemotherapeutic drugs. Evidence suggests that NMD achieves this through its targeting of Gas5 and Gadd45 RNAs, both of which promote apoptosis signaling. Not only does NMD regulate apoptosis, but apoptosis-inducing agents can impact NMD. In the example depicted, chemotherapeutic agents trigger cleavage of the NMD factor, UPF1, which downregulates NMD
Given that most NMD target transcripts are not conserved [12, 127], it is notable that GADD45 is one of the few transcripts degraded by NMD in a variety of species [15, 18, 75, 128–130]. This afforded Nelson et al. an opportunity to address whether GADD45 functions in the same manner in mammals as it does in flies. Complicating the situation, however, was the fact that mammals have three GADD45 paralogs (α, β, and γ; or A, B, and G, respectively), whereas Drosophila have only one GADD45 gene. Furthermore, all three mammalian GADD45 gene paralogs express mRNAs that are high-confidence direct NMD targets, based on their being stabilized in response to NMD factor depletion and their harboring NMD-inducing features, such as uORFs and exon–exon junctions downstream of the main ORF stop codon [15, 18, 28, 75, 128–130]. Thus, all 3 GADD45 paralogs have the potential to function in an NMD-based circuit to regulate cell survival. Nelson et al. chose to focus their attention on GADD45B, as it is expressed at a much higher level than the other GADD45 paralogs in HeLa and NIH3T3 cells [131]. They found that GADD45B depletion rescued apoptosis triggered by depletion of the NMD factor, UPF1, in both HeLa and NIH3T3 cells [74]. This supported the notion that GADD45B functions in an NMD-based circuit to control apoptosis in mammalian cells, just as it does in Drosophila. Together, these data suggest that GADD45 and NMD act in a conserved circuit that triggers apoptosis of cells in which conditions have become unfavorable.
Under what conditions might this circuit operate? ER stress is one likely condition, as this triggers NMD downregulation [60, 75–77], and thus, it would likely raise GADD45B/Gadd45 level and elicit cell death if the ER stress is not resolved in a timely manner. Likewise, other conditions that inhibit NMD—including hypoxia, specific NMD modulatory miRNAs, c-myc, and possibly SG formation (see sections above)—would be predicted to induce GADD45B/Gadd45 and thereby trigger apoptosis as a protective response. Nelson et al. speculated that this NMD/Gadd45 circuit could also serve as a defense mechanism to restrict viral growth [74]. Consistent with this possibility, many viruses encode factors that inhibit NMD [132], which could act as a “molecular tripwire” to induce a stress response and cell death through GADD45B/Gadd45 induction.
Because the tumor microenvironment tends to inhibit NMD, this environment could also trigger a pro-apoptotic mechanism. Of note, however, NMD inhibition can also trigger events that favor tumor formation (such as changes in signaling pathways, including the TGFβ/BMP, Wnt, and Notch pathways [28]), and, indeed, evidence suggests that the NMD pathway can act as a tumor suppressor pathway [41, 75, 109]. Thus, whether loss of NMD stimulates or inhibits tumor formation likely depends on a delicate balance of several pro- and anti-tumor mechanisms, with the GADD45B/Gadd45-dependent pro-apoptotic mechanism being just one of many events triggered by loss of NMD.
GADD45B/Gadd45 mRNA is probably not the only pro-apoptotic RNA targeted by NMD to protect cells from death. A long noncoding (lnc) RNA—growth arrest-specific 5 (GAS5)—has been suggested by experiments performed by Tani et al. to act in an NMD-based circuit that is critical for avoiding cell death in response to serum starvation [133]. Tani et al. regarded GAS5 as a good candidate to function in such a circuit, based on four previous observations. First, GAS5 is a well-established NMD target in both human and mouse cell lines [11, 75, 134]. Second, GAS5 is an apoptotic lncRNA that acts by binding to the glucocorticoid receptor (GR) and perturbing this transcription factor from activating its anti-apoptotic program (Fig. 6) [133, 135–137]. Third, GAS5 is induced by serum starvation and, when overexpressed, GAS5 triggers apoptosis and reduced cell cycle progression, suggesting that it acts as a tumor suppressor [137]. Finally, depletion of GAS5 has the opposite effect: it inhibits apoptosis and promotes cell cycle progression [137].
These four qualities of GAS5 led Tani et al. to hypothesize that this lncRNA is a central component in an NMD-regulated circuit that controls apoptosis. They obtained several lines of evidence supporting the existence of such a circuit [133]. First, they used a gold standard method—pulse-chase labeling with BrU—to determine whether, indeed, GAS5 is a direct NMD target. The previous studies had only examined GAS5 steady-state levels [11, 18, 138] or examined GAS5 RNA half-life using transcriptional inhibitors [75, 134], the latter of which is subject to artifacts [134, 139, 140]. Using BrU pulse-chase labeling, they found that GAS5 half-life was significantly longer than previously determined using the conventional methods (i.e., transcriptional inhibitors) in HEK293T cells (6.6 h, instead of 2.6 h [134]). Second, Tani et al. depleted the NMD factor, UPF1, and found that this increased the steady-state level and half-life of GAS5 by ~seven and ~threefold, respectively, confirming that this noncoding RNA is an NMD target. Third, they found that UPF1 knockdown decreased the expression of the two key anti-apoptotic genes in the GR pathway, cIAP2 and SGK1, as predicted given that they are both known to be negatively regulated by GAS5. Fourth, they found that apoptosis itself was also increased by UPF1 knockdown. Finally, Tani et al. examined the effect of serum starvation on GAS5 level, as it was previously shown that serum starvation inhibits NMD through reducing UPF1 phosphorylation [141]. They found that serum starvation increased GAS5 steady-state level and half-life by a magnitude similar to that achieved by UPF1 knockdown. Serum starvation also reduced the expression of cIAP2 and SGK1.
Together, these data supported a model in which GAS5 is constitutively expressed at low level to prevent apoptosis, but in response to stress conditions that inhibit NMD (such as mimicked by serum starvation), GAS5 is dramatically upregulated, leading to activation of the GR pathway and consequent apoptosis. Of note, however, Tani et al. did not perform a rescue experiment to directly test this model. Thus, while their data are consistent with an NMD-GAS5-apoptosis circuit, further work is required to definitively determine its validity.
Pro-apoptotic agents inhibit NMD
Not only does NMD influence apoptosis, but apoptosis activation has been found to affect NMD. Two recent studies—Popp et al. and Jia et al.—showed that several different pro-apoptotic agents—including doxorubicin, etoposide, cycloheximide, and staurosporine—all have the ability to suppress the NMD pathway [123, 124]. NMD magnitude was decreased by two to fourfold. Cell lines from a wide variety of species—human, canine, hamster, bovine, and green monkey—responded to staurosporine and doxorubicin by suppressing NMD, indicating that this NMD suppression response is conserved. Intriguingly, chemotherapeutic agents were found to suppress NMD by triggering cleavage of the NMD factors, UPF1 and UPF2 (Fig. 6). UPF1 is cleaved after the Aspartate at position 37, which is in a well-conserved region just upstream of the UPF2- and eRF3-binding domain—the CH domain [27, 123, 124]. UPF2 is cleaved in the second MIF4G domain, which is thought to be essential for UPF2 activity [142]. Caspase 3 and Caspase 7 were found to be responsible for UPF1 and UPF2 cleavage. Caspase-induced cleavage of UPF1 and UPF2 was relatively specific, as staurosporine treatment did not lead to cleavage of the branch-specific NMD factor, UPF3B, despite its harboring a potential Caspase-cleavage site [124].
How does UPF1 and UPF2 cleavage lead to reduced NMD? One obvious possibility is that the truncated versions of these two UPF proteins are nonfunctional, which both groups found to be the case [123, 124]. For this to lead to reduced NMD magnitude, a significant proportion of full-length functional UPF1 and/or UPF2 would need to be cleaved. Indeed, Jia et al. found the level of both full-length UPF1 and UPF2 was significantly reduced by chemotherapeutic agents. However, this may not be the only mechanism by which NMD magnitude is depressed. Both groups found that the truncated forms of UPF1 and UPF2 generated by Caspase-induced cleavage had dominant-negative activity. Thus, it is likely that chemotherapeutic agents inhibit NMD through a Caspase-mediated mechanism by both reducing the level of full-length UPF proteins and generating an NMD inhibitory cleavage product.
NMD deficiency is known to make cells more likely to undergo apoptosis in response to ER stress and hypoxia [60, 61, 84]. This raised the possibility that inhibited NMD triggered by chemotherapeutic agents leads to a downward spiral—that ensures an apoptotic outcome—by increasing the sensitivity of apoptotic pathways. To assess the validity of this hypothesis, Popp et al. examined whether inhibiting NMD by an independent means rendered cells more sensitive to apoptotic death by chemotherapeutic agents. They found that the NMD inhibitor, NMDI-1—which interferes with the interaction of SMG5 and UPF1—increased the sensitivity to apoptosis triggered by the chemotherapeutic agent doxorubicin [124]. In contrast, when HeLa cells expressed a noncleavable version of UPF1, they were less sensitive to doxorubicin-induced apoptosis and thus had higher viability compared to control cells. These results strongly suggest that UPF1 cleavage induced by chemotherapeutic agents causes increased sensitivity to pro-apoptotic agents.
Together, these studies suggest that there is an intimate interplay between apoptosis and NMD. Conditions that favor apoptosis suppress NMD, while NMD protects cells from apoptosis (Fig. 7). What might be the physiological significance of this? As indicated above, we suggest that suppression of NMD by conditions favoring apoptosis would further sensitize these cells towards apoptosis engagement, thereby creating a positive feedback loop to ensure that the apoptosis mechanism is triggered. This would also potentially tend to drive more rapid apoptosis induction, allowing for faster recycling of cellular components for neighboring healthy cells. By promoting rapid and efficient death of diseased and stressed cells, NMD would allow an organism to survive.
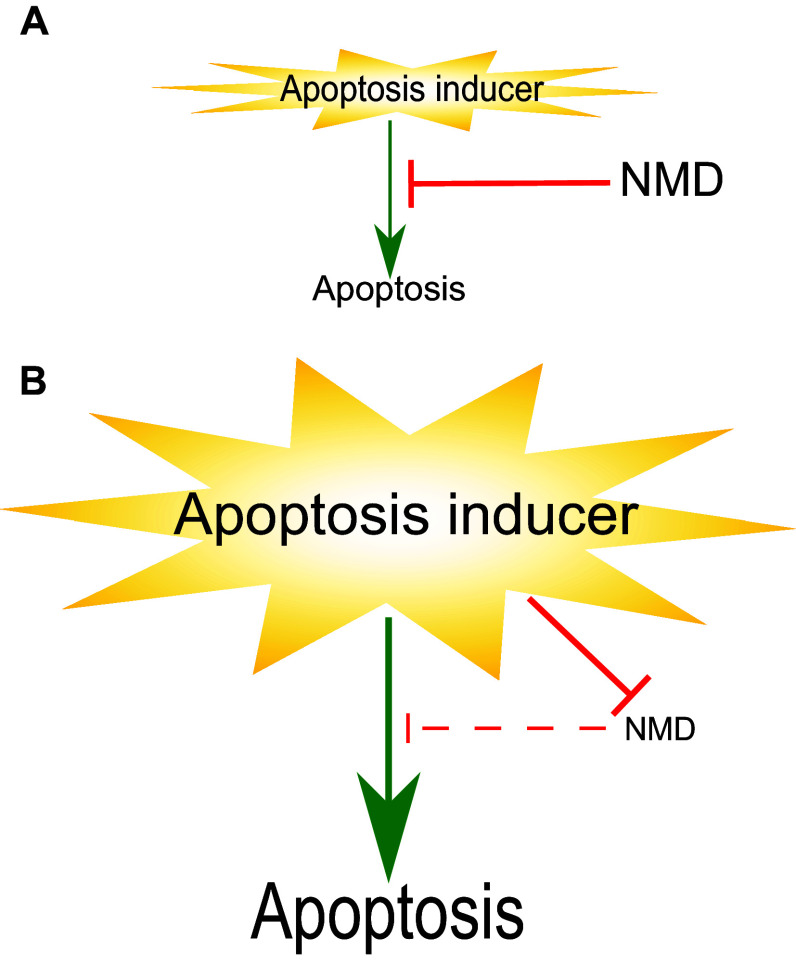
Fig. 7.
NMD and apoptosis have a complex regulatory relationship II. a NMD inhibits cell death triggered by apoptosis-inducing agent. b Sustained and/or strong stress leads to suppression of NMD, thereby triggering apoptosis
NMD in stress responses in plants
It has recently become apparent that NMD not only modulates stress responses in animals, but also in plants. NMD in plants has been best studied in Arabidopsis, which has homologues of several mammalian NMD genes, including UPF1, UPF2, UPF3, and SMG7 [143]. As expected, Arabidopsis harboring mutations in these NMD genes are NMD-deficient, but they are viable and exhibit few defects under normal light and soil conditions. However, these NMD-deficient plants exhibit dysregulated expression of many genes related to the host-pathogen response and have elevated resistance to various pathogens, including the tomato virus Pseudomonas syringae [143]. Such NMD-deficient plants express elevated levels of the stress response chemical, salicylic acid, and have fewer Pseudomonas colonies forming on their leaves than wild-type plants challenged with the same virus [143]. While it is counter intuitive why the NMD pathway would reduce viral resistance, one possible explanation is that this conserves energy under nonstress conditions.
Evidence also suggests that Arabidopsis harboring mutations in NMD genes are more sensitive to “solar stress” (long periods of daylight) than their wild-type counterparts [144]. Shi et al. found that under long-day conditions (16 h of light per day), NMD-deficient Arabidopsis had more narrow, epinastic leaves and had smaller rosette sizes and stunted growth compared to controls [144]. In contrast, under short-day conditions (10 h of light per day), NMD-deficient plants and control plants were indistinguishable in phenotype. To determine whether solar stress also makes plants more susceptible to other forms of stress such as pathogen infection, NMD-deficient plants were grown under long-day conditions and inoculated with tomato virus or received physical stress in their leaves. Under long-day conditions combined with virus inoculation or physical stress, these plants expressed more of the stress response chemicals, salicylic acid and jasmonic acid, as compared to wild-type plants responding to the same sources of stress. This suggests that NMD is critical for defending plants against solar stress, and that when under solar stress, NMD-deficient plants are also more sensitive to other stressors.
NMD recognizes similar features in RNAs in plants as in mammals. Premature termination codons [145–147], long 3′UTRs [148, 149], downstream exon junctions [149, 150], and uORFs [151, 152] have all been shown to lead to mRNA downregulation through the NMD pathway in Arabidopsis. Interestingly, a recent study showed that putative NMD target transcripts encoding plant pathogen response factors contained conserved uORFs [143]. This raises the possibility that these ORFs not only trigger NMD but encode functional peptides. It is also possible that specific sequences in these conserved uORF are critical for triggering NMD. In mammals, only a small fraction of mRNAs with uORFs are downregulated by NMD [16, 153, 154], and thus, a future area of investigation is to determine what distinguishes NMD-inducing uORFs from non-NMD-inducing uORFs.
Perspectives
In this review, we discussed the interplay of stress responses with post-transcriptional pathways. Our focus was on how the highly conserved and selective NMD RNA degradation pathway impacts stress responses; and how, in turn, stress-response pathways regulate NMD. Much has been learned about the interplay between stress and NMD, but there are also many future questions, some of which we address below:
While it is clear that NMD has a profound impact on many cellular adaptations to stress—including the UPR, the hypoxic stress response, and autophagy- and apoptosis-associated stress responses—it is not known whether NMD’s reach stretches beyond this. For example, NMD may also shape stress responses to heat, pH alterations, high concentrations of heavy metals, and amino-acid under-availability. In this review, we focused on mammalian stress responses. Does NMD also influence adaptations to stress in non-mammalian species? NMD influences the ability of mammalian cells to survive oxidative stress by targeting the mRNA encoding a glutathione factor transporter, SLC7A11, for degradation [114]. The yeast, Schizosaccharomyces pombe, has also evolved an NMD-regulated oxidative stress response pathway. NMD-deficient (Upf1-mutant) S. pombe are more vulnerable than controls to oxidative stress in response to hydrogen peroxide-induced oxidative stress [155]. This protection conferred by NMD from oxidative stress is specific, as it does not extend to protection from osmotic shock or UV irradiation. It remains for the future to determine whether oxidative stress protection in S. pombe and mammalian cells is mediated by a conserved NMD-dependent mechanism. In other cases, it is very likely that species-specific NMD-dependent strategies have evolved to shape stress responses. In part, this follows from the fact that species have unique needs, and therefore, they need to adapt to different forms of stress. For example, plants and yeast may be more sensitive than mammals to changes in temperature, pH alterations, heavy metals, changes in sunlight availability, and amino-acid availability, because they have fewer defenses from the external environment than mammals.
It is important to note that some factors critical for the NMD RNA degradation pathway have other functions, including in other RNA decay pathways, telomere maintenance, and genome surveillance [86, 156, 157]. Thus, the effects of depleting NMD factors on stress responses could result from perturbation of one or more of these other mechanisms, rather than NMD. Mitigating this possibility, it has been shown that the UPR is impacted by depletion of any of several different NMD factors [60, 61, 84]. This supports the notion that NMD—not other pathways—is most critical for UPR regulation. It is worth noting, however, that the ability of NMD factors to promote RNA decay may not be their primary role. It has been suggested that the primary function of NMD factors is to promote ribosome recycling and regulate ribosome density [158]. Thus, the ability of NMD factors to enhance RNA decay may be merely a secondary consequence of these effects on translation. Future experiments will be required to unravel precisely why NMD factors have critical roles in stress responses, as well as associated processes, including autophagy and apoptosis.
Not only do NMD factors impact stress responses, but stress responses impact NMD. We discussed one possible mechanism by which stress inhibits NMD—through eIF2α phosphorylation and SG formation. However, it will be important to determine whether other mechanisms might also be involved. For example, the discovery that exogenous pro-apoptotic agents inhibit NMD by triggering caspase-dependent UPF1 and UPF2 cleavage [123, 124] (Fig. 6) raises the possibility that this mechanism might also contribute to NMD inhibition in response to endogenous, naturally occurring signals. For example, might excess ER stress inhibit NMD through not only induction of eIF2α phosphorylation [60, 75], but through UPF cleavage? In addition, UPF cleavage-mediated NMD inhibition could provide a mechanism by which stressors promote timely apoptosis during the early embryonic development in the nervous system and immune system, and in wound healing in the skin [159–161]. Indeed, even circumstances that require NMD downregulation for normal biological events not necessarily involving apoptosis—such as neural and endoderm differentiation [28, 29, 44]—could involve UPF cleavage.
Another avenue for future research is to investigate why NMD tends to be inhibited, rather than activated, by stress in mammals. In this regard, it is worth noting that—in Saccharomyces cerevisiae—evidence suggests that osmotic stress increases NMD magnitude [162, 163]. This may be a mechanism to degrade ribosomal protein mRNAs by NMD and thereby redirect the translational apparatus to stress-response mRNAs, many of which are immune to NMD.
The ability of NMD to both regulate and shape stress responses raises the possibility that NMD modulatory therapy could be clinically beneficial in some disease settings. One example is cancer. NMD activator therapy would be predicted to be efficacious for the treatment of tumors, as increasing evidence suggests that NMD is a tumor suppressor pathway [41, 91, 109] and tumors are known to have suppressed NMD as a result of hypoxia and ER stress [60, 75]. NMD activator therapy could also potentially protect normal cells from chemotherapy-induced death. This follows from the findings that (1) chemotherapeutic agents reduce NMD factor expression and NMD activity in mice in vivo [123] and (2) NMD protects normal cells from apoptosis [74, 124, 133]. If, indeed, future studies show that NMD activator therapy increases normal cell survival, this may allow higher doses of chemotherapeutic agents to be administered to cancer patients, and it could facilitate recovery after chemotherapy treatment.
NMD activator therapy could also potentially be used to treat some forms of neurodegenerative disease. This follows from the fact that the hallmark of several neurodegenerative diseases—including Alzheimer’s and Parkinson’s disease—is chronic UPR activity [1]. Because the UPR pathway inhibits NMD [60, 61, 75], it is likely (though not yet directly tested) that NMD is suppressed in neural cells from such patients. Suppressed NMD is known to cause apoptosis [74, 133] and, indeed, neurodegenerative disease is often accompanied by neuron loss [164]. By restoring high levels of NMD, NMD activator therapy has the potential to promote neuron survival. Neurodegenerative disease is also commonly accompanied by autophagy dysregulation [164], which may have a role in the formation of potentially toxic aggregates—such as amyloid plaques, tau tangles, and Lewy bodies—which are often present in diseased neurons [164]. One approach to deplete such bodies is NMD inhibitory therapy, as this treatment would be predicted to stimulate autophagy [117]. Conversely, NMD activator therapy could provide clinical benefit for neural diseases in which autophagy is abnormally elevated [164]. Signaling pathways are also known to be dysregulated in several neurodegenerative diseases [165–167], which is interesting in light of the recent work demonstrating that the NMD pathway regulates TGF-β signaling and possibly Wnt and Notch signaling [28, 29]. This raises the possibility that NMD activation or inhibition therapy that reverses dysregulated signaling in neurodegenerative disease could provide clinical benefit. Future studies will reveal the full range of disease states in which NMD modulatory therapy reverses stress-related defects for therapeutic purposes.
Acknowledgements
We thank the NIH (RO1 GM111838) for financial support. The first author was also supported by the NIH P42 Superfund Training grant (ES010337).
Footnotes
An erratum to this article is available at https://doi.org/10.1007/s00018-017-2642-6.
Change history
9/13/2017
The original version of this article unfortunately contained errors in the section entitled “NMD in stress responses in plants”.
References
- 1.Hoozemans JJM, Scheper W. Endoplasmic reticulum: the unfolded protein response is tangled in neurodegeneration. Int J Biochem Cell Biol. 2012;44:1295–1298. doi: 10.1016/j.biocel.2012.04.023. [DOI] [PubMed] [Google Scholar]
- 2.Giampietri C, Petrungaro S, Conti S, et al. Cancer microenvironment and endoplasmic reticulum stress response. Mediat Inflamm. 2015;2015:417281. doi: 10.1155/2015/417281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodvold JJ, Mahadevan NR, Zanetti M. Immune modulation by ER stress and inflammation in the tumor microenvironment. Cancer Lett. 2015 doi: 10.1016/j.canlet.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Hasmim M, Messai Y, Ziani L, et al. Critical role of tumor microenvironment in shaping NK cell functions: implication of hypoxic stress. Front Immunol. 2015;6:482. doi: 10.3389/fimmu.2015.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan S-W. The unfolded protein response in virus infections. Front Microbiol. 2014;5:518. doi: 10.3389/fmicb.2014.00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duwi Fanata WI, Lee SY, Lee KO. The unfolded protein response in plants: a fundamental adaptive cellular response to internal and external stresses. J Proteom. 2013;93:356–368. doi: 10.1016/j.jprot.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 7.Peccarelli M, Kebaara BW. Regulation of natural mRNAs by the nonsense-mediated mRNA decay pathway. Eukaryot Cell. 2014;13:1126–1135. doi: 10.1128/EC.00090-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang Y-F, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 9.Fatscher T, Boehm V, Gehring NH. Mechanism, factors, and physiological role of nonsense-mediated mRNA decay. Cell Mol Life Sci. 2015 doi: 10.1007/s00018-015-2017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendell JT, Sharifi NA, Meyers JL, et al. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 11.Weischenfeldt J, Damgaard I, Bryder D, et al. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 2008;22:1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lareau LF, Inada M, Green RE, et al. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 13.Ni JZ, Grate L, Donohue JP, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurt JA, Robertson AD, Burge CB. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res. 2013;23:1636–1650. doi: 10.1101/gr.157354.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yepiskoposyan H, Aeschimann F, Nilsson D, et al. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA. 2011;17:2108–2118. doi: 10.1261/rna.030247.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Somers J, Pöyry T, Willis AE. A perspective on mammalian upstream open reading frame function. Int J Biochem Cell Biol. 2013;45:1690–1700. doi: 10.1016/j.biocel.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramani AK, Nelson AC, Kapranov P, et al. High resolution transcriptome maps for wild-type and nonsense-mediated decay-defective Caenorhabditis elegans . Genome Biol. 2009;10:R101. doi: 10.1186/gb-2009-10-9-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang L, Lou C-H, Chan W, et al. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol Cell. 2011;43:950–961. doi: 10.1016/j.molcel.2011.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hogg JR, Goff SP. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell. 2010;143:379–389. doi: 10.1016/j.cell.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eberle AB, Stalder L, Mathys H, et al. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol. 2008;6:e92. doi: 10.1371/journal.pbio.0060092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh G, Rebbapragada I, Lykke-Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 2008;6:e111. doi: 10.1371/journal.pbio.0060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buhler M, Steiner S, Mohn F, et al. EJC-independent degradation of nonsense immunoglobulin-μ mRNA depends on 3′UTR length. Nat Struct Mol Biol. 2006;13:462–464. doi: 10.1038/nsmb1081. [DOI] [PubMed] [Google Scholar]
- 23.Li T, Shi Y, Wang P, et al. Smg6/Est1 licenses embryonic stem cell differentiation via nonsense-mediated mRNA decay. EMBO J. 2015;34:1630–1647. doi: 10.15252/embj.201489947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McIlwain DR, Pan Q, Reilly PT, et al. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc Natl Acad Sci USA. 2010;107:12186–12191. doi: 10.1073/pnas.1007336107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medghalchi SM, Frischmeyer PA, Mendell JT, et al. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- 26.Shum EY, Jones SH, Shao A, et al. The antagonistic gene paralogs Upf3a and Upf3b govern nonsense-mediated RNA decay. Cell. 2016;165:382–395. doi: 10.1016/j.cell.2016.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang J, Maquat LE. Nonsense-mediated mRNA decay (NMD) in animal embryogenesis: to die or not to die, that is the question. Curr Opin Genet Dev. 2011;21:422–430. doi: 10.1016/j.gde.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lou C, Dumdie J, Goetz A, et al. Nonsense-mediated RNA decay influences human embryonic stem cell fate. Stem Cell Reports. 2016;6:844–857. doi: 10.1016/j.stemcr.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lou CH, Shao A, Shum EY, et al. Posttranscriptional control of the stem cell and neurogenic programs by the nonsense-mediated rna decay pathway. Cell Rep. 2014;6:748–764. doi: 10.1016/j.celrep.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jolly LA, Homan CC, Jacob R, et al. The UPF3B gene, implicated in intellectual disability, autism, ADHD and childhood onset schizophrenia regulates neural progenitor cell behaviour and neuronal outgrowth. Hum Mol Genet. 2013;22:4673–4687. doi: 10.1093/hmg/ddt315. [DOI] [PubMed] [Google Scholar]
- 31.Metzstein MM, Krasnow MA. Functions of the nonsense-mediated mRNA decay pathway in Drosophila development. PLoS Genet. 2006;2:e180. doi: 10.1371/journal.pgen.0020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wittkopp N, Huntzinger E, Weiler C, et al. Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol Cell Biol. 2009;29:3517–3528. doi: 10.1128/MCB.00177-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen LS, Wilkinson MF, Gecz J. Nonsense-mediated mRNA decay: inter-individual variability and human disease. Neurosci Biobehav Rev. 2014;46(Pt 2):175–186. doi: 10.1016/j.neubiorev.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tarpey PS, Raymond FL, Nguyen LS, et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39:1127–1133. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen LS, Kim H-G, Rosenfeld JA, et al. Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neuro-developmental disorders. Hum Mol Genet. 2013;22:1816–1825. doi: 10.1093/hmg/ddt035. [DOI] [PubMed] [Google Scholar]
- 36.Thoren LA, Nørgaard GA, Weischenfeldt J, et al. UPF2 is a critical regulator of liver development, function and regeneration. PLoS One. 2010;5:e11650. doi: 10.1371/journal.pone.0011650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong C, Kim YK, Woeller CF, et al. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev. 2009;23:54–66. doi: 10.1101/gad.1717309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang L, Wilkinson MF. Regulation of nonsense-mediated mRNA decay. Wiley Interdiscip Rev RNA. 2012;3:807–828. doi: 10.1002/wrna.1137. [DOI] [PubMed] [Google Scholar]
- 39.Karam R, Wengrod J, Gardner LB, Wilkinson MF. Regulation of nonsense-mediated mRNA decay: implications for physiology and disease. Biochim Biophys Acta Gene Regul Mech. 2013;1829:624–633. doi: 10.1016/j.bbagrm.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zetoune AB, Fontanière S, Magnin D, et al. Comparison of nonsense-mediated mRNA decay efficiency in various murine tissues. BMC Genet. 2008;9:83. doi: 10.1186/1471-2156-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu J, Plank T-D, Su F, et al. The nonsense-mediated RNA decay pathway is disrupted in inflammatory myofibroblastic tumors. J Clin Invest. 2016;126:3058–3062. doi: 10.1172/JCI86508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hug N, Longman D, Caceres JF. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 2016;44:1483–1495. doi: 10.1093/nar/gkw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alonso CR, Akam M. A Hox gene mutation that triggers nonsense-mediated RNA decay and affects alternative splicing during Drosophila development. Nucleic Acids Res. 2003;31:3873–3880. doi: 10.1093/nar/gkg482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bruno IG, Karam R, Huang L, et al. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell. 2011;42:500–510. doi: 10.1016/j.molcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barberan-soler S, Lambert NJ, Zahler AM. Global analysis of alternative splicing uncovers developmental regulation of nonsense-mediated decay in C. elegans . RNA. 2009;15:1652–1660. doi: 10.1261/rna.1711109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bao J, Vitting-seerup K, Waage J, Tang C. UPF2-dependent nonsense-mediated mRNA decay pathway is essential for spermatogenesis by selectively eliminating longer 3′UTR transcripts. PLoS Genet. 2016;12:e1005863. doi: 10.1371/journal.pgen.1005863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fanourgakis G, Lesche M, Akpinar M, Dahl A. Chromatoid body protein TDRD6 supports long 3′UTR triggered nonsense mediated mRNA decay. PLoS Genet. 2016;12:e1005857. doi: 10.1371/journal.pgen.1005857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cho H, Kim KM, Han S, et al. Staufen1-mediated mRNA decay functions in adipogenesis. Mol Cell. 2012;46:495–506. doi: 10.1016/j.molcel.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 49.Hetz C, Martinon F, Rodriguez D, Glimcher LH. The unfolded protein response: integrating stress signals through the stress sensor IRE1α. Physiol Rev. 2011;91:1219–1243. doi: 10.1152/physrev.00001.2011. [DOI] [PubMed] [Google Scholar]
- 50.Moore KA, Hollien J. The unfolded protein response in secretory cell function. Annu Rev Genet. 2011;46:120830114430006. doi: 10.1146/annurev-genet-110711-155644. [DOI] [PubMed] [Google Scholar]
- 51.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 52.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 53.Ma Y, Hendershot LM. The role of the unfolded protein response in tumour development: friend or foe? Nat Rev Cancer. 2004;4:966–977. doi: 10.1038/nrc1505. [DOI] [PubMed] [Google Scholar]
- 54.Yoshida H, Matsui T, Yamamoto A, et al. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/S0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 55.Calfon M, Zeng H, Urano F, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 56.Oslowski CM, Urano F. Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol. 2011;490:71–92. doi: 10.1016/B978-0-12-385114-7.00004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshida K, Miki Y. The cell death machinery governed by the p53 tumor suppressor in response to DNA damage. Cancer Sci. 2010;101:831–835. doi: 10.1111/j.1349-7006.2009.01488.x. [DOI] [PubMed] [Google Scholar]
- 58.Sun T, Cui J. Dynamics of P53 in response to DNA damage: mathematical modeling and perspective. Prog Biophys Mol Biol. 2015;119:175–182. doi: 10.1016/j.pbiomolbio.2015.08.017. [DOI] [PubMed] [Google Scholar]
- 59.Brewer JW. Regulatory crosstalk within the mammalian unfolded protein response. Cell Mol Life Sci. 2014;71:1067–1079. doi: 10.1007/s00018-013-1490-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gardner LB. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol Cell Biol. 2008;28:3729–3741. doi: 10.1128/MCB.02284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karam R, Lou C-H, Kroeger H, et al. The unfolded protein response is shaped by the NMD pathway. EMBO Rep. 2015;16:599–609. doi: 10.15252/embr.201439696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan W-K, Huang L, Gudikote JP, et al. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 2007;26:1820–1830. doi: 10.1038/sj.emboj.7601628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nguyen LS, Wilkinson MF, Gecz J. Nonsense-mediated mRNA decay: inter-individual variability and human disease. Neurosci Biobehav Rev. 2013;46:175–186. doi: 10.1016/j.neubiorev.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weng WC, Lee WT, Hsu WM, et al. Role of glucose-regulated protein 78 in embryonic development and neurological disorders. J Formos Med Assoc. 2011;110:428–437. doi: 10.1016/S0929-6646(11)60064-8. [DOI] [PubMed] [Google Scholar]
- 65.Zhao L, Longo-Guess C, Harris BS, et al. Protein accumulation and neurodegeneration in the woozy mutant mouse is caused by disruption of SIL1, a cochaperone of BiP. Nat Genet. 2005;37:974–979. doi: 10.1038/ng1620. [DOI] [PubMed] [Google Scholar]
- 66.Luo S, Mao C, Lee B, Lee AS. GRP78/BiP is required for cell proliferation and protecting the inner cell mass from apoptosis during early mouse embryonic development. Mol Cell Biol. 2006;26:5688–5697. doi: 10.1128/MCB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dostie J, Dreyfuss G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr Biol. 2002;12:1060–1067. doi: 10.1016/S0960-9822(02)00902-8. [DOI] [PubMed] [Google Scholar]
- 68.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kataoka N, Yong J, Kim VN, et al. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell. 2000;6:673–682. doi: 10.1016/S1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 70.He F, Li X, Spatrick P, et al. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell. 2003;12:1439–1452. doi: 10.1016/S1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- 71.Chapin A, Hu H, Rynearson SG, et al. In vivo determination of direct targets of the nonsense-mediated decay pathway in Drosophila . G3 (Bethesda) 2014;4:485–496. doi: 10.1534/g3.113.009357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Popp MW-L, Maquat LE. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet. 2013;47:139–165. doi: 10.1146/annurev-genet-111212-133424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sieber J, Hauer C, Bhuvanagiri M, et al. Proteomic analysis reveals branch-specific regulation of the unfolded protein response by nonsense-mediated mRNA decay. Mol Cell Proteom. 2016 doi: 10.1074/mcp.M115.054056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nelson JO, Moore KA, Chapin A, et al. Degradation of Gadd45 mRNA by nonsense-mediated decay is essential for viability. Elife. 2016 doi: 10.7554/eLife.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang D, Zavadil J, Martin L, et al. Inhibition of nonsense-mediated RNA decay by the tumor microenvironment promotes tumorigenesis. Mol Cell Biol. 2011;31:3670–3680. doi: 10.1128/MCB.05704-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Usuki F, Fujimura M, Yamashita A. Endoplasmic reticulum stress preconditioning attenuates methylmercury-induced cellular damage by inducing favorable stress responses. Sci Rep. 2013;3:2346. doi: 10.1038/srep02346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Z, Vuong JK, Zhang M, et al. Inhibition of nonsense-mediated RNA decay by ER stress. RNA. 2016 doi: 10.1261/rna.058040.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carter MS, Doskow J, Morris P, et al. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- 79.Li S, Leonard D, Wilkinson MF. T cell receptor (TCR) mini-gene mRNA expression regulated by nonsense codons: a nuclear-associated translation-like mechanism. J Exp Med. 1997;185:985–992. doi: 10.1084/jem.185.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wek RC, Jiang H-Y, Anthony TG, et al. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST0340007. [DOI] [PubMed] [Google Scholar]
- 81.Qian L, Vu MN, Carter MS, et al. T cell receptor-beta mRNA splicing during thymic maturation in vivo and in an inducible T cell clone in vitro. J Immunol. 1993;151:6801–6814. [PubMed] [Google Scholar]
- 82.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 84.Sakaki K, Yoshina S, Shen X, et al. RNA surveillance is required for endoplasmic reticulum homeostasis. Proc Natl Acad Sci. 2012;109:8079–8084. doi: 10.1073/pnas.1110589109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mascarenhas R, Dougherty JA, Schoenberg DR. SMG6 cleavage generates metastable decay intermediates from nonsense-containing β-globin mRNA. PLoS One. 2013;8:e74791. doi: 10.1371/journal.pone.0074791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Frizzell KA, Rynearson SG, Metzstein MM. Drosophila mutants show NMD pathway activity is reduced, but not eliminated, in the absence of Smg6. RNA. 2012;18:1475–1486. doi: 10.1261/rna.032821.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Reichenbach P, Höss M, Azzalin CM, et al. A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr Biol. 2003;13:568–574. doi: 10.1016/S0960-9822(03)00173-8. [DOI] [PubMed] [Google Scholar]
- 88.Protter DSW, Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;26:668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 90.Gardner LB. Nonsense-mediated RNA decay regulation by cellular stress: implications for tumorigenesis. Mol Cancer Res. 2010;8:295–308. doi: 10.1158/1541-7786.MCR-09-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang D, Wengrod J, Gardner LB. Overexpression of the c-myc oncogene inhibits nonsense-mediated RNA decay in B lymphocytes. J Biol Chem. 2011;286:40038–40043. doi: 10.1074/jbc.M111.266361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brown JAL, Roberts TL, Richards R, et al. A novel role for hSMG-1 in stress granule formation. Mol Cell Biol. 2011;31:4417–4429. doi: 10.1128/MCB.05987-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Abrahamyan LG, Chatel-Chaix L, Ajamian L, et al. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J Cell Sci. 2010;123:369–383. doi: 10.1242/jcs.055897. [DOI] [PubMed] [Google Scholar]
- 96.Louros SR, Osterweil EK. Perturbed proteostasis in autism spectrum disorders. J Neurochem. 2016 doi: 10.1111/jnc.13723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Thomas MG, Martinez Tosar LJ, Desbats MA, et al. Mammalian Staufen 1 is recruited to stress granules and impairs their assembly. J Cell Sci. 2009;122:563–573. doi: 10.1242/jcs.038208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Leung AKL, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci USA. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Leung AKL, Vyas S, Rood JE, et al. Poly(ADP-Ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell. 2011;42:489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Leung AKL. The whereabouts of microRNA actions: cytoplasm and beyond. Trends Cell Biol. 2015;25:601–610. doi: 10.1016/j.tcb.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wiesen JL, Tomasi TB. Dicer is regulated by cellular stresses and interferons. Mol Immunol. 2009;46:1222–1228. doi: 10.1016/j.molimm.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mori MA, Raghavan P, Thomou T, et al. Role of microRNA processing in adipose tissue in stress defense and longevity. Cell Metab. 2012;16:336–347. doi: 10.1016/j.cmet.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Anderson P, Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones. 2002;7:213–221. doi: 10.1379/1466-1268(2002)007<0213:VSTROE>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wu S, Lin L, Zhao W, et al. AUF1 is recruited to the stress granules induced by coxsackievirus B3. Virus Res. 2014;192:52–61. doi: 10.1016/j.virusres.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 105.Fred RG, Mehrabi S, Adams CM, Welsh N. PTB and TIAR binding to insulin mRNA 3′- and 5′UTRs; implications for insulin biosynthesis and messenger stability. Heliyon. 2016 doi: 10.1016/j.heliyon.2016.e00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Aulas A, Caron G, Gkogkas CG, et al. G3BP1 promotes stress-induced RNA granule interactions to preserve polyadenylated mRNA. J Cell Biol. 2015;209:73–84. doi: 10.1083/jcb.201408092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bley N, Lederer M, Pfalz B, et al. Stress granules are dispensable for mRNA stabilization during cellular stress. Nucleic Acids Res. 2015;43:e26. doi: 10.1093/nar/gku1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kedersha N, Panas MD, Achom C, et al. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol. 2016;212:845–860. doi: 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu C, Karam R, Zhou Y, et al. The UPF1 RNA surveillance gene is commonly mutated in pancreatic adenosquamous carcinoma. Nat Med. 2014;20:596–598. doi: 10.1038/nm.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cho H, Han S, Park OH, Kim YK. SMG1 regulates adipogenesis via targeting of staufen1-mediated mRNA decay. Biochim Biophys Acta. 2013;1829:1276–1287. doi: 10.1016/j.bbagrm.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 111.Estrela JM, Ortega A, Obrador E. Glutathione in cancer biology and therapy. Crit Rev Clin Lab Sci. 2006;43:143–181. doi: 10.1080/10408360500523878. [DOI] [PubMed] [Google Scholar]
- 112.Valko M, Leibfritz D, Moncol J, et al. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 113.Lewerenz J, Hewett SJ, Huang Y, et al. The cystine/glutamate antiporter system x(c)(-) in health and disease: from molecular mechanisms to novel therapeutic opportunities. Antioxid Redox Signal. 2013;18:522–555. doi: 10.1089/ars.2011.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Martin L, Gardner LB. Stress-induced inhibition of nonsense-mediated RNA decay regulates intracellular cystine transport and intracellular glutathione through regulation of the cystine/glutamate exchanger SLC7A11. Oncogene. 2015;34:4211–4218. doi: 10.1038/onc.2014.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Garcia-Huerta P, Troncoso-Escudero P, Jerez C, et al. The intersection between growth factors, autophagy and ER stress: a new target to treat neurodegenerative diseases? Brain Res. 2016 doi: 10.1016/j.brainres.2016.02.052. [DOI] [PubMed] [Google Scholar]
- 116.Kania E, Pająk B, Orzechowski A. Calcium homeostasis and ER stress in control of autophagy in cancer cells. Biomed Res Int. 2015;2015:352794. doi: 10.1155/2015/352794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wengrod J, Martin L, Wang D, et al. Inhibition of nonsense-mediated RNA decay activates autophagy. Mol Cell Biol. 2013;33:2128–2135. doi: 10.1128/MCB.00174-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sun X, Perlick HA, Dietz HC, Maquat LE. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc Natl Acad Sci USA. 1998;95:10009–10014. doi: 10.1073/pnas.95.17.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Oren YS, McClure ML, Rowe SM, et al. The unfolded protein response affects readthrough of premature termination codons. EMBO Mol Med. 2014;6:685–701. doi: 10.1002/emmm.201303347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rouschop KMA, Ramaekers CHMA, Schaaf MBE, et al. Autophagy is required during cycling hypoxia to lower production of reactive oxygen species. Radiother Oncol. 2009;92:411–416. doi: 10.1016/j.radonc.2009.06.029. [DOI] [PubMed] [Google Scholar]
- 121.Ondrej M, Cechakova L, Durisova K, et al. To live or let die: unclear task of autophagy in the radiosensitization battle. Radiother Oncol. 2016;119:265–275. doi: 10.1016/j.radonc.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 122.Xie W-Y, Zhou X-D, Yang J, et al. Inhibition of autophagy enhances heat-induced apoptosis in human non-small cell lung cancer cells through ER stress pathways. Arch Biochem Biophys. 2016;607:55–66. doi: 10.1016/j.abb.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 123.Jia J, Furlan A, Gonzalez-Hilarion S, et al. Caspases shutdown nonsense-mediated mRNA decay during apoptosis. Cell Death Differ. 2015;22:1754–1763. doi: 10.1038/cdd.2015.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Popp MW, Maquat LE. Attenuation of nonsense-mediated mRNA decay facilitates the response to chemotherapeutics. Nat Commun. 2015;6:6632. doi: 10.1038/ncomms7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Takekawa M, Saito H. A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell. 1998;95:521–530. doi: 10.1016/S0092-8674(00)81619-0. [DOI] [PubMed] [Google Scholar]
- 126.Peretz G, Bakhrat A, Abdu U. Expression of the Drosophila melanogaster GADD45 homolog (CG11086) affects egg asymmetric development that is mediated by the c-Jun N-terminal kinase pathway. Genetics. 2007;177:1691–1702. doi: 10.1534/genetics.107.079517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rehwinkel J, Letunic I, Raes J, et al. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tani H, Imamachi N, Salam KA, et al. Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biol. 2012;9:1370–1379. doi: 10.4161/rna.22360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kurosaki T, Li W, Hoque M, et al. A post-translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev. 2014;28:1900–1916. doi: 10.1101/gad.245506.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Viegas MH, Gehring NH, Breit S, et al. The abundance of RNPS1, a protein component of the exon junction complex, can determine the variability in efficiency of the nonsense mediated decay pathway. Nucleic Acids Res. 2007;35:4542–4551. doi: 10.1093/nar/gkm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yue F, Cheng Y, Breschi A, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mocquet V, Neusiedler J, Rende F, et al. The human T-lymphotropic virus type 1 tax protein inhibits nonsense-mediated mRNA decay by interacting with INT6/EIF3E and UPF1. J Virol. 2012;86:7530–7543. doi: 10.1128/JVI.07021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tani H, Torimura M, Akimitsu N. The RNA degradation pathway regulates the function of GAS5 a non-coding RNA in mammalian cells. PLoS One. 2013;8:e55684. doi: 10.1371/journal.pone.0055684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tani H, Mizutani R, Salam KA, et al. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012;22:947–956. doi: 10.1101/gr.130559.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Deveraux QL, Reed JC. IAP family proteins—suppressors of apoptosis. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 136.Mikosz CA, Brickley DR, Sharkey MS, et al. Glucocorticoid receptor-mediated protection from apoptosis is associated with induction of the serine/threonine survival kinase gene, sgk-1. J Biol Chem. 2001;276:16649–16654. doi: 10.1074/jbc.M010842200. [DOI] [PubMed] [Google Scholar]
- 137.Kino T, Hurt DE, Ichijo T, et al. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ideue T, Sasaki YTF, Hagiwara M, Hirose T. Introns play an essential role in splicing-dependent formation of the exon junction complex. Genes Dev. 2007;21:1993–1998. doi: 10.1101/gad.1557907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Blattner C, Kannouche P, Litfin M, et al. UV-Induced stabilization of c-fos and other short-lived mRNAs. Mol Cell Biol. 2000;20:3616–3625. doi: 10.1128/MCB.20.10.3616-3625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Friedel CC, Dölken L, Ruzsics Z, et al. Conserved principles of mammalian transcriptional regulation revealed by RNA half-life. Nucleic Acids Res. 2009;37:e115. doi: 10.1093/nar/gkp542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pal M, Ishigaki Y, Nagy E, Maquat LE. Evidence that phosphorylation of human Upfl protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA. 2001;7:5–15. doi: 10.1017/S1355838201000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Clerici M, Deniaud A, Boehm V, et al. Structural and functional analysis of the three MIF4G domains of nonsense-mediated decay factor UPF2. Nucleic Acids Res. 2014;42:2673–2686. doi: 10.1093/nar/gkt1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rayson S, Arciga-Reyes L, Wootton L, et al. A role for nonsense-mediated mRNA decay in plants: pathogen responses are induced in Arabidopsis thaliana NMD mutants. PLoS One. 2012;7:e31917. doi: 10.1371/journal.pone.0031917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shi C, Baldwin IT, Wu J. Arabidopsis plants having defects in nonsense-mediated mRNA decay factors UPF1, UPF2, and UPF3 show photoperiod-dependent phenotypes in development and stress responses. J Integr Plant Biol. 2012;54:99–114. doi: 10.1111/j.1744-7909.2012.01093.x. [DOI] [PubMed] [Google Scholar]
- 145.Yoine M, Ohto M, Onai K, et al. The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis . Plant J. 2006;47:49–62. doi: 10.1111/j.1365-313X.2006.02771.x. [DOI] [PubMed] [Google Scholar]
- 146.Arciga-Reyes L, Wootton L, Kieffer M, Davies B. UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis . Plant J. 2006;47:480–489. doi: 10.1111/j.1365-313X.2006.02802.x. [DOI] [PubMed] [Google Scholar]
- 147.Hori K, Watanabe Y. UPF3 suppresses aberrant spliced mRNA in Arabidopsis . Plant J. 2005;43:530–540. doi: 10.1111/j.1365-313X.2005.02473.x. [DOI] [PubMed] [Google Scholar]
- 148.Kalyna M, Simpson CG, Syed NH, et al. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis . Nucleic Acids Res. 2012;40:2454–2469. doi: 10.1093/nar/gkr932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kertész S, Kerényi Z, Mérai Z, et al. Both introns and long 3′-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res. 2006;34:6147–6157. doi: 10.1093/nar/gkl737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nyikó T, Kerényi F, Szabadkai L, et al. Plant nonsense-mediated mRNA decay is controlled by different autoregulatory circuits and can be induced by an EJC-like complex. Nucleic Acids Res. 2013;41:6715–6728. doi: 10.1093/nar/gkt366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nyikó T, Sonkoly B, Mérai Z, et al. Plant upstream ORFs can trigger nonsense-mediated mRNA decay in a size-dependent manner. Plant Mol Biol. 2009;71:367–378. doi: 10.1007/s11103-009-9528-4. [DOI] [PubMed] [Google Scholar]
- 152.Saul H, Elharrar E, Gaash R, et al. The upstream open reading frame of the Arabidopsis AtMHX gene has a strong impact on transcript accumulation through the nonsense-mediated mRNA decay pathway. Plant J. 2009;60:1031–1042. doi: 10.1111/j.1365-313X.2009.04021.x. [DOI] [PubMed] [Google Scholar]
- 153.Toma KG, Rebbapragada I, Durand S, Lykke-Andersen J. Identification of elements in human long 3′UTRs that inhibit nonsense-mediated decay. RNA. 2015;21:887–897. doi: 10.1261/rna.048637.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mocquet V, Durand S, Jalinot P. How retroviruses escape the nonsense-mediated mRNA decay. AIDS Res Hum Retrovir. 2015;31:948–958. doi: 10.1089/aid.2014.0326. [DOI] [PubMed] [Google Scholar]
- 155.Rodríguez-Gabriel MA, Watt S, Bähler J, Russell P. Upf1, an RNA helicase required for nonsense-mediated mRNA decay, modulates the transcriptional response to oxidative stress in fission yeast. Mol Cell Biol. 2006;26:6347–6356. doi: 10.1128/MCB.00286-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat Rev Genet. 2008;9:699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Azzalin CM, Lingner J. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol. 2006;16:433–439. doi: 10.1016/j.cub.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 158.Brogna S, McLeod T, Petric M. The meaning of NMD: translate or perish. Trends Genet. 2016;32:395–407. doi: 10.1016/j.tig.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 159.Nijhawan D, Honarpour N, Wang X. Apoptosis in neural development and disease. Annu Rev Neurosci. 2000;23:73–87. doi: 10.1146/annurev.neuro.23.1.73. [DOI] [PubMed] [Google Scholar]
- 160.Opferman JT, Korsmeyer SJ. Apoptosis in the development and maintenance of the immune system. Nat Immunol. 2003;4:410–415. doi: 10.1038/ni0503-410. [DOI] [PubMed] [Google Scholar]
- 161.Greenhalgh DG. The role of apoptosis in wound healing. Int J Biochem Cell Biol. 1998;30:1019–1030. doi: 10.1016/S1357-2725(98)00058-2. [DOI] [PubMed] [Google Scholar]
- 162.Kawashima T, Douglass S, Gabunilas J, et al. Widespread use of non-productive alternative splice sites in Saccharomyces cerevisiae . PLoS Genet. 2014;10:e1004249. doi: 10.1371/journal.pgen.1004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Garre E, Romero-Santacreu L, Barneo-Muñoz M, et al. Nonsense-mediated mRNA decay controls the changes in yeast ribosomal protein pre-mRNAs levels upon osmotic stress. PLoS One. 2013;8:e61240. doi: 10.1371/journal.pone.0061240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lim Y, Cho H, Kim E-K. Brain metabolism as a modulator of autophagy in neurodegeneration. Brain Res. 2016;1649:158–165. doi: 10.1016/j.brainres.2016.02.049. [DOI] [PubMed] [Google Scholar]
- 165.Atwood CS, Bowen RL. A unified hypothesis of early- and late-onset Alzheimer’s disease pathogenesis. J Alzheimers Dis. 2015;47:33–47. doi: 10.3233/JAD-143210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Singh S, Mishra A, Srivastava N, Shukla S. MK-801 (Dizocilpine) regulates multiple steps of adult hippocampal neurogenesis and alters psychological symptoms via Wnt/β-catenin signaling in parkinsonian rats. ACS Chem Neurosci. 2016 doi: 10.1021/acschemneuro.6b00354. [DOI] [PubMed] [Google Scholar]
- 167.Eixarch H, Calvo-Barreiro L, Montalban X, Espejo C. Bone morphogenetic proteins in multiple sclerosis: role in neuroinflammation. Brain Behav Immun. 2017 doi: 10.1016/j.bbi.2017.02.019. [DOI] [PubMed] [Google Scholar]