Abstract
Transfer of genetic material from cytoplasmic organelles to the nucleus, an ongoing process, has implications in evolution, aging, and human pathologies such as cancer. The transferred mitochondrial DNA (mtDNA) fragments in the nuclear genome are called nuclear mtDNA or NUMTs. We have named the process numtogenesis, defining the term as the transfer of mtDNA into the nuclear genome, or, less specifically, the transfer of mitochondria or mitochondrial components into the nucleus. There is increasing evidence of the involvement of NUMTs in human biology and pathology. Although information pertaining to NUMTs and numtogenesis is sparse, the role of this aspect of mitochondrial biology to human cancers is apparent. In this review, we present available knowledge about the origin and mechanisms of numtogenesis, with special emphasis on the role of NUMTs in human malignancies. We describe studies undertaken in our laboratory and in others and discuss the influence of NUMTs in tumor initiation and progression and in survival of cancer patients. We describe suppressors of numtogenesis and evolutionary conserved mechanisms underlying numtogenesis in cancer. An understanding the emerging field of numtogenesis should allow comprehension of this process in various malignancies and other diseases and, more generally, in human health.
1. Introduction
The eukaryote organelles, mitochondria, and the plant organelles, chloroplasts, have descended, respectively, from proteobacteria and cyanobacteria. As a consequence, in eukaryotic cells, these endosymbionts act as auxiliary DNA-containing compartments. As a result of continual transfer of organelle DNA to the nucleus during the course of evolution, the quantity of DNA in these organelles, however, is less than in their prokaryotic ancestors. This natural transfer of mitochondrial DNA (mtDNA) into the nuclear genomes of eukaryotic cells is an ongoing process; these nuclear copies of mtDNA are called NUMTs (nuclear mtDNA) [57]. Of note, intact mitochondria, containing mtDNA, mitochondrial RNA (mtRNA), and mitochondrial proteins, also localize into the nucleus (Figure 1) [5, 9, 12, 23, 52, 62, 87, 89]. We have named the process numtogenesis, defining the term as the transfer of mtDNA into the nuclear genome, or, less specifically, transfer of mitochondria or mitochondrial components into the nucleus [85]. Like NUMTs, transferred chloroplast DNA fragments are called NUPTs for nuclear chloroplast or plastid DNA [80, 109]. We suggest that the natural transfer of chloroplast DNA or any other chloroplastic components into the nucleus can be described as nuptogenesis.
Figure. 1. Nuclear Mitochondria.
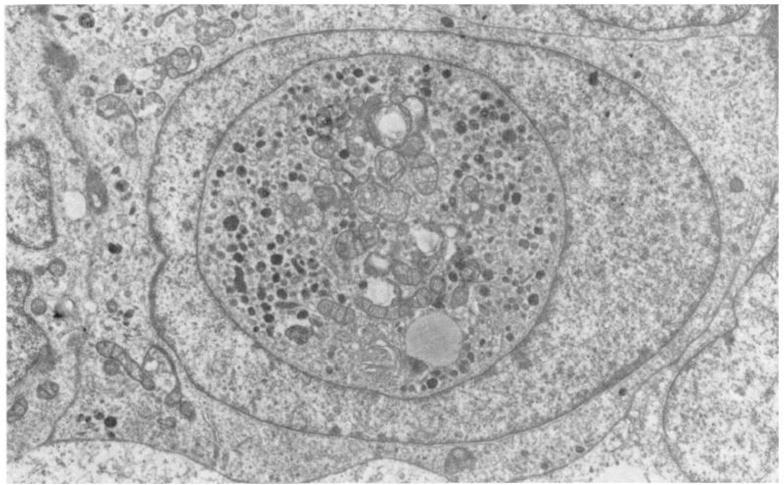
Electron micrograph of a malignant melanoma of the choroid, showing well preserved mitochondria as nuclear inclusions (× 8960). Reproduced from the British Journal of Ophthalmology, 1980, 64: 456. Reprinted with permission from BMJ Publishing Group Ltd.
NUMTs are reported in at least 85 sequenced eukaryotic genomes [39]. In fact, almost all of the eukaryote genomes analysed so far carry NUMTs, which range from a few base pairs in length to entire mitochondrial sequences [39–41]. NUMTs with the longest stretches and highest frequencies are present in genomes of the domestic cat and flowering plants. NUMTs also vary in copy number, from a few copies in Caenorhabditis to a few hundred in flowering plants [70]. Numtogenesis may involve both coding and non-coding regions of mtDNA and insertion of both coding exons and non-coding sequences of the target genome. The significance of NUMTs is manifold: for eukaryotic genomes, insertion of new sequences in nuclear DNA is a mechanism of evolution. Furthermore, in the context of evolutionary biology, NUMTs are “molecular fossils” that have survived due to the lower mutation rate in the nucleus compared to that of mitochondria. As a result, they can be used to trace inter- and intra-species mtDNA ancestry [74]. Although most NUMT insertions occur in the non-coding regions of nuclear genes, some reports suggest their presence in coding sequences as well. This could lead to generation of protein-encoding exons in existing genes; alternatively, insertion of NUMTs in genetic loci can lead to mutations and/or activation of proto-oncogenes. The importance of NUMTs in cancer and other human diseases is underscored by the fact that numtogenesis is an ongoing and frequent biological phenomenon.
2. Historical aspects of numtogenesis
The endosymbiotic theory, which considers mitochondria (and plastids) as former free-living prokaryotes that later invaded the eukaryotic cells, was initially proposed by Margulis in the 1970s [60, 61]. According to this theory, these symbiotic organelles gradually transferred their genes to the eukaryotic genome throughout the course of evolution, resulting in swatches of organelle DNA being integrated into the nuclear genome.
Although mitochondria have retained part of their genome (that controls vital cellular functions), mtDNA is not exclusively localized in the mitochondria. The first evidence of the movement of organelle DNA came from cross-hybridization studies in maize, in which fragments of chloroplast DNA were found in the mitochondrial genome [86]. Soon after this revelation, Ellis et al. described “promiscuous DNA” for the organelle DNA fragments that were present in multiple intra-cellular compartments [24]. In 1983, Farelly and Butow identified, in the yeast nuclear genome, DNA sequences that share extensive homology to mitochondrial sequences [25]. The term NUMT was used by Lopez et al., who made the discovery of the transposition of a 7.9 kb long mtDNA fragment into the nuclear genome of the domestic cat [57]. Since then, whole genomes of many eukaryotes have been sequenced, and NUMTs have been observed in the nuclear genomes of flowering plants, yeast, Caenorhabditis, Drosophila, rat, domestic cat, and primates, including humans [39].
Entire mitochondria are also present in the nucleus. In 1958, Hoffman and Grigg made this discovery in cells of murine lymph nodes [42]. This phenomenon was observed later in 1960 by Mori et al., who detected whole mitochondria in ascitic cancer cells, tongue and pancreas cancer cells, and regenerating liver cells [65]. Thereafter, mitochondria were found in the nuclei of blood lymphocytes [9, 50], Hodgkin’s lymphoma cells [72], leukemic myoblasts [82], and cells of rheumatic carditis [45]. Mitochondrial transfer into the nucleus was analyzed by a Russian group studying the cells of patients with alcohol-induced cardiomyopathies. In the nuclei of cardiomyocytes, they detected an accumulation of mitochondria, along with displacement of chromatin to the periphery of nuclei. Although the mechanism of mitochondrial entry into the nucleus could not be elucidated, the authors found, in electron micrographs of the cells, signs of disrupted nuclear envelopes. They hypothesized that the merging of mitochondria with nuclei is accompanied by the disintegration of the former, which releases mitochondria-encoded apoptotic factors and reactive oxygen species into the cytosol and the nucleoplasm. The resulting degradation of the nuclear proteins may contribute to the cellular pathologies of chronic alcoholism [5].
Regarding the activity of mtDNA proteins in the nucleus, the next relevant discovery was that of humanin, a peptide encoded within the mitochondrial 16S rRNA gene, MT-RNR2. Humanin was found independently by three different groups researching different pathologies. Hashimoto et al. discovered humanin while looking for putative proteins that could protect cells from amyloid beta [38], a protein associated Alzheimer’s disease. Guo et al. found humanin in a screen for proteins that interacted with Bcl-2-associated X protein (Bax), a protein involved in apoptosis [32]. Finally, Ikonen et al. discovered humanin while screening for proteins that interact with IGFBP3 [44]. Apart from the mitochondrial 16s rRNA gene, there is evidence that the humanin peptide is also encoded by NUMTs bearing homology to the humanin open reading frame [88]. Humanin is translated both in the mitochondria and cytosol, although the difference in the translational machinery of the two sites results in peptides of different lengths. Mitochondrially translated humanin has 21 amino acids; the one translated in the cytosol is has 24 amino acids [108]. A recent review by Gong et al. has described the neuro- and cytoprotective roles of humanin in human conditions such as Alzheimer’s disease, Huntington’s disease, prion disease, atherosclerosis, diabetes, and aging [31].
3. Numtogenesis in evolution
Although mitochondria retain much of the structure and general composition of their prokaryotic ancestors, their genomes have been downsized to the extent that they code only for a fraction of the organelle proteins; non-coding and non-essential coding sequences have been lost. The genes retained in mitochondria are mainly involved in the electron transport chain and energy transduction, maintenance of redox balance, and controlling the production of reactive oxygen species. All other genes required for the function and integrity of mitochondria reside in the nuclei. Most of the functional mitochondrial genes were transferred to the nucleus early during evolution; this process ceased approximately 1000 million years ago, before the emergence of animals [10]. The transferred genes evolved further to function in the nuclear environment (for instance, developing nuclear-specific regulatory domains) and, in some cases, to perform extra-organelle functions. In addition, many of the parental mitochondrial sequences have been altered due to mutations that occurred during the course of evolution.
On the other hand, the insertion of random and non-coding DNA fragments of mitochondrial origin is thought to be a more recent [8] and an ongoing process [78, 111]. NUMTs usually present themselves in three forms: a) continuous stretches of DNA that align linearly with the original mtDNA sequence, b) scrambled sequences derived from different regions of the mitochondrial genome, and c) re-arranged pieces of mtDNA different mitochondria [55]. Starting with the discovery of NUMTs in yeast [25], scattered reports of mitochondrial fragments in human DNA surfaced [57, 74, 98, 113] until sequencing of the human genome showed hundreds of NUMTs. The first analyses by Mourier et al. [66] and Tourmen et al. [96] revealed between 280 and 296 NUMTs in the human genome, of which 94 were 1000 bp or longer. On the basis of phylogenetic analyses, the conclusions of both studies were essentially the same: that NUMT insertion was a continuous process and fairly rapid. Hazkani-Covo et al. determined the origin of all NUMTs in the human genome, i.e., whether the sequences resulted from (a) the direct insertion from mitochondria to the nuclei or (b) genomic duplication after insertion [41]. By use of pairwise phylogenetic analyses, they concluded that only a third of the total human NUMT repertoire was due to de novo insertions and that the remaining two-thirds arose as a result of the duplications of existing NUMTs. A contradictory study by Bensasson et al., however, indicated that most NUMTs were a result of independent insertion events and in fact originated in a primate ancestor [6]. These scattered studies have not been easily reproducible as the source raw data could not be easily accessed. There was therefore a need to quantify and map all the NUMTs and to build an exhaustive database. Lascaro et al. [54] compared the mtDNA reference sequence [3] and generated the Reference Human NUMTs compilation (RHNUMTs). The initial compilation–RHNUMTs.1–was further improved and validated into the RHNUMTs.2 version by Simone et al. [84]. BLASTING of the human mitochondrial and nuclear genomes revealed 766 high-scoring pairs, meaning mitochondrial sequences that were similar to nuclear sequences. The RHNUMTs.2 compilation was further revised by remapping on the h19 version of the human genome sequence that resulted in identification of 755 NUMTs [14, 84] (Figure 2). At present, the complete length of NUMTs in humans is more than 400 kbp, and they occur at a density of 17 bp per 100 kb of the nuclear genome [40].
Figure 2. NUMT distribution in human chromosomes.
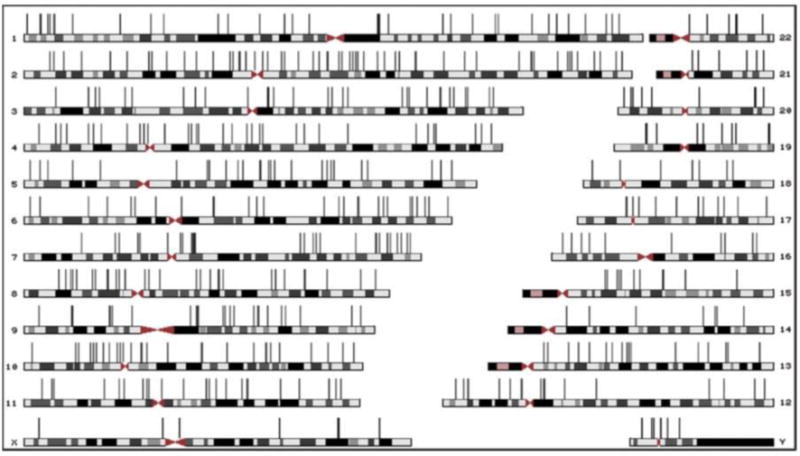
A scale representation of the human chromosomes. The location of human-specific NUMTs is indicated with vertical bars (Adopted from Simone et al., 2011).
Calabrese et al. found a high degree of co-localization of the NUMTs insertions/duplications in the highly repetitive sequences, in humans as well nonhuman primate genomes [14]. This is a difference from yeast, where insertion of NUMTs is largely random in the non-coding host sequences and gives rise to pseudogenes [25]. The preference of human NUMTs for repetitive elements was also shown by other investigators. For example, Mishmar et al. [64] and Qu et al. [75] found Alu sequences (transposable elements) flanking 247 specific human NUMTs. In contrast, other studies found a negative correlation between NUMTs and repetitive sequences in the human genome [28, 46]. A more recent and detailed study of NUMTs of humans, rhesus monkeys, mice, and rats by Tsuji et al. elucidated features of the NUMTs insertion sites [97]. Briefly, they determined that the sequences flanking NUMTs were rich in retrotransposons as well as A-T rich sequences; furthermore, the NUMTs were highly concentrated in the open chromatin areas. Since these sites are closely associated with regulation of gene expression, it is reasonable to expect that insertion of NUMTs has functional consequences, in many cases deleterious. Indeed, the frequency of NUMTs insertion into the human genome is only once in 180,000 years or 5 × 10−6 per germ cell per generation [55]. In contrast, in yeast, NUMTs occur at a higher frequency, 2 × 10−5 per cell per generation. In addition, Tsuji et al. found that NUMTs were under-represented in the chromosomal fragile sites, long genes, and CpG-rich islands [97].
Regarding the source of NUMTs, the studies of Hazkani-Covo [39] and Tourmen [96] pointed to the mtDNA D-loop as the primary contributor to human NUMTs and pseudogenes. The findings of Mournier et al., however, revealed a paucity of the D-loop in the human genome; their study was in fact one of the first to discover large tracts of mtDNA sequences in the nuclear genome [66]. This finding was validated by Tsuji et al., who argued that the faster evolution rate of the D-loop could make detection of D-loop difficult among the human NUMTs [97]. In addition, the regulatory regions of mtDNA, responsible for the replication and gene expression, are also under-represented; one possible explanation relates to the (specific and nonspecific) binding of mitochondrial transcription factor A (TFAM) protein to these regions [35], which protects them from breaking away and forming NUMTs.
The transfer of DNA from the prokaryotic endosymbionts to eukaryotic nuclei brought about changes in the dynamics of the nucleus and the proto-organelles as well as in the overall cellular proteome. Even though most of the functional mitochondrial genes ceased early during evolution, stretches of mtDNA have continuously been transferred into the nucleus. For years, the NUMTs were considered inconsequential in terms of gene evolution and function, as both the NUMTs and the target sequences were largely non-coding [7, 39]. This perception changed when researchers found that the preferred regions of NUMTs insertion in the human genome were relevant to gene regulation (as discussed in the previous sections). In a study by Richetti et al., NUMTs were found to insert in the human genome at the intergenic regions and subsequently to alter the arrangements of exons and introns of certain genes [78]. Thus, NUMTs invasion has three wide-ranging consequences: a) deleterious mutations that can alter or inactivate gene function, b) acquisition of exons that can remodel the existing genes and add new functions to the encoded proteins, and c) no effect on gene sequence or regulation. The association between NUMTs-induced mutations and diseases has been elucidated by recent studies and will be dealt with in the next section.
In regard to the second possibility, there have been reports of NUMTs insertion within genic regions of yeast, Arabidopsis, humans, and several fish species. Antunes et al. discovered numerous mitochondrial pseudogenes in fish genomes and demonstrated that they have a preference for some predicted nuclear genes [4]. The studies of Ricchetti et al. [77, 78] have also shown the preference of both yeast and human NUMTs for insertion into coding regions of genes. Of the 27 NUMTs studied, 23 inserted into the introns of known genes, including the MADH2 tumor suppressor gene. In addition, they also discovered a higher frequency of NUMTs in the genic regions of chromosomes 18 and Y. Noutsos et al. studied the genic insertion pattern of NUMTs in yeast, humans, and Arabidopsis and found several NUMTs in or adjacent to annotated genes [69]. About 45 NUMT insertions were found in the protein-coding exons, which resulted in of functional exons [69].
To determine the timeline for NUMT insertions in humans, alignments of genomes of humans and non-human primates have been performed. This has revealed NUMTs that a) exist in both species and were acquired prior to the divergence of humans and chimpanzees and b) are unique to humans and acquired mostly from insertions after the two species diverged. There is also evidence of creation of NUMTs by tandem duplication of pre-existing NUMTs. This kind of phylogenetic analysis is necessary in order to distinguish older or germ-line NUMTs from the more recently acquired or somatic NUMTs [40, 41, 53, 66]. Although the germline NUMTs have been evolutionarily conserved for tens of millions of years, the somatic NUMTs have been acquired in more recent history [6, 28, 53, 97, 112]. According to a report by Perna and Kocher, at a rough estimate, 5.7 NUMTs have been inserted every 1 million years and have been retained in the human genome [74]. Since the nucleus is less susceptible to mutations relative to mitochondria, the NUMTs are molecular fossils of mtDNA. They can therefore be used as markers in phylogenetic studies and to trace mtDNA inheritance in the human population [74].
4. Numtogenesis in health, diversity, disease and cancer
4.1 Numtogenesis in health and diversity
An important aspect of numtogenesis is the degree of polymorphism in the NUMTs. Genetic allele variants of NUMTs may provide markers for human phylogenetic analyses and contribute susceptibility to cancer/genetic disorders. The first study to address NUMT polymorphism was conducted by Ricchetti et al., who investigated 41 NUMTs and found that 6 of them showed insertion polymorphisms [78]. One or more of these polymorphic NUMTs was present in every ethnic group, indicating that these NUMTs were probably inserted soon after modern humans evolved. Even when a particular NUMT was shared between more than one ethnic group, the allelic frequency was not equal among the various ethnic groups. They also commented on polymorphism in the insertion sites (the micro-homology sites) of the nuclear genome by comparing the alleles with a NUMT insertion with those that lacked an insertion. A seminal study by Lang et al. involved investigation of 14 polymorphic NUMTs whose individual variants existed in various native populations [53]. The allele frequencies were calculated using the Human Genome Project data for 13 different populations. They found correlations between the genetic diversity of the NUMTs and their geographical distribution. A more recent large-scale analysis of 23 polymorphic NUMTs was performed by Dayama et al., wherein they established that most of the polymorphic NUMTs were integrated within the past million years [22]. More than 50% of those NUMTs were present in low frequencies (>0.1%), indicating that their insertions were even more recent. Another finding was the higher percentage of GC content in the polymorphic NUMTs compared to the parental mitochondrial sequence. In contrast to the findings of Tsuji et al. [97], this study found D-loop sequences in several polymorphic NUMTs and could not detect any preference of the NUMTs for AT-rich or free-chromatin regions of the genome. Natural polymorphic variants of NUMTs have been associated with diseases, including cancer.
4.2 Numtogenesis in genetic disorders
The role of de novo numtogenesis in certain human genetic disorders has been established (table 1). For instance, Turner et al. presented a case study of the Pallister-Hall syndrome in which they identified a 72-bp NUMT insertion into exon 14 of the GLI3 gene as the causative mutation [99]; the NUMT insertion created a premature stop codon that resulted in a truncated protein product. A 251-bp NUMT insertion in the plasma factor VII gene results in a splice site mutation that causes severe plasma factor VII deficiency and bleeding disease [11]. In a case of mucolipidosis IV, a 93-bp NUMT fragment was found to be inserted into exon 2 of the MCOLN1 gene, preventing the gene’s proper splicing [30]. Further, Usher syndrome type IC has been linked to a 36-bp insertion in exon 9 of the USH1C gene [2]. Interestingly, the mitochondrial genome was reported to be intact in theses afflicted individuals [39]. A study by Muradian et al. observed, for several mammalian species, a strong correlation between the number of NUMT insertions and longevity, although the authors did not provide a convincing mechanistic explanation [67].
Table 1.
NUMTs that cause diseases
4.3 Numtogenesis in cancer
In many cancers, somatic changes in regard to mtDNA copy number and D-loop and mitochondrial gene(s) alterations have been implicated as drivers in tumorigenesis [114]. For cancer cells, the somatic insertion of NUMTs into the nuclear genome has been implicated in genetic instability. mtDNA fragments that escape from the mitochondria and invade the nucleus can bring about changes in the nuclear genome through insertional mutagenesis. In the context of tumorigenesis, these changes can manifest as any of the following: a) inactivation of a tumor suppressor gene, b) activation of a proto-oncogene, c) epigenetic dysregulation of either type of gene, and d) introduction of a constitutively active oncogene. A slow and steady accumulation of mtDNA in the nuclear genome could lead to progressive changes in the overall cellular proteome and cause cancer [87].
An early indication of the role of mitochondrial insertions in tumorigenesis came from the experiments by Kamimura et al. on HeLa cells [48]. They hybridized the entire HeLa genomic library to the total human mtDNA and focused on three nuclear fragments that showed homology to parts of mitochondrial 12s rRNA, cytochrome oxidase I, and NADH dehydrogenase. These pseudogenes were found in other human cancer cell lines and in the human placenta; this was the first report correlating acquisition of mtDNA with malignant transformation. In a later study by the same group [83], a fragment of the mitochondrial cytochrome-c oxidase subunit III (coxIII) was found to be inserted in exons 2 and 3 of the c-myc gene. Nielsen et al. followed up the study with the finding that, upon transfection into HeLa cells, the NUMT insertion had autonomous replication capacity [68]. With rodent models, Hadler et al. provided evidence that NUMTs were present at a higher frequency in the tumor tissues relative to normal tissues [33, 34]. Xianglong et al. also detected mtDNA fragments in cancerous gastric mucosal cells but did not establish a causal mechanism [105]. Chen et al. could not detect any somatic NUMTs in normal cervical epithelial cells; however, the frequency of NUMTs in malignant cells was 27% [19]. In addition, the frequency of c-myc alterations was directly proportional to the concentration of NUMTs. They hypothesized that insertion of mtDNA fragments in the nuclei of cervical epithelial cells acted as the driver of tumorigenesis by activating the c-myc gene, although they did not show direct physical proximity of the NUMTs with c-myc.
Our study [85] demonstrated increased numtogenesis in colorectal cancers (CRCs). Since there is a causal relationship between mtDNA copy number [95] and germline variants [90] with CRC susceptibility, we studied the role of NUMTs and numtogenesis in CRCs. We performed a comprehensive quantitative analysis on the abundance of somatic NUMTs in the genomes of CRC cells relative to that in normal cells. On a genome-wide scale, the frequency of NUMTs in the tumor cells was 4.42 times higher than in normal cells. This abundance of NUMTs in the cancerous state also correlated with a poorer prognosis and survival of the patients, more so for women. In addition, we identified NUMT insertion hotspots in the tumor genomes. When tallied with the Candidate Cancer Gene Database, some of the sites corresponded to known cancer-associated genes. Furthermore, the source of the NUMTs was comprised of certain fragile sites in the mitochondrial genome which housed the ND1, CoxI, and CoxIII genes, which are linked with CRC risk and progression. We also observed a high rate of the mutated YME1L1 gene, which produces an ATP-dependent metalloprotease that normally functions in the maintenance of mitochondrial morphology, in those CRC cases that also had a high frequency of NUMTs. These results were validated by analysis of the YME1L1 gene in the TCGA database of CRC genes. This was noteworthy, since inactivation of this gene in yeast was implicated in increased mtDNA escape and numtogenesis. We showed that, upon knockout of the human YME1L1 gene, the amount of mtDNA fragments increased rapidly in the nuclear genome. In other words, our study was the first to identify a bona fide suppressor of NUMTs, the inactivation of which leads to increased numtogenesis. Furthermore, in Yme1-1 yeast cells (Yme1 inactivated strain), we partially blocked the escape of mtDNA by transforming the latter with the human YME1L1 gene, thus strengthening our hypothesis.
Ju et al. reported that de novo somatic NUMTs were often present along with other somatic rearrangements of the nuclear genome [47]. These co-localizations were strongest in breast cancer tissues with NUMTs insertions in the fifth intron of the potassium channel KCNMA1 gene, which is frequently amplified in breast and prostate cancers [70]. Also, triple-negative breast cancer tissues displayed more NUMTs compared to the estrogen receptor-positive tissues, pointing to a positive correlation between chromosomal rearrangements and numtogenesis.
Despite reports on the role of NUMTs in tumorigenesis, some caution is needed to interpret the results. Yao et al. illustrated this concern as the contribution of the NUMTs can often be ignored when some mtDNA mutations are encountered in cancer [106]. Second, the effects of NUMTs and NUMT polymorphisms are often downplayed in studies of mitochondrial heteroplasmy [22, 107], defined as cellular differences in individual mitochondrial genomes due to high rates of mutations [63]. Since mitochondrial heteroplasmy has been implicated in aging and cancer [26, 81], it is the effect of NUMTs is appropriate to minimized on these processes. Dayama et al. identified several sites in the mitochondrial genome that have hitherto been mistaken as mutated when the mutations were actually present in NUMTs [22]. In addition, there are reports of different frequencies of NUMTs. This is mainly because a) any particular NUMT can undergo post-insertional modifications such as deletions and substitutions and may escape the detection primers used in that specific study and b) there is, at the moment, no unified criteria and standardized primers for the detection and study of NUMTs. The latter issue can be solved by creating a NUMT database [76] and regularly updating it parallel to the human genome and mtDNA databases. Taken together, the data available so far on the role of NUMTs in cancer suggest that increased numtogenesis is a factor in determining cancer risk and prognosis. Further studies are required to refine the detection of NUMTs, to elucidate the mechanisms of numtogenesis, and to establish causal relationships between NUMT insertions and tumorigenesis.
5. Environmental numtogenesis
Environmental factors influence the movement of genetic material from the mitochondria (or chloroplasts) into the nucleus. Such physical/chemical/biological stresses can be described as “numtogens.” In principle, agents that increase the mitotoxicity or mitostress that causes mitochondrial disintegration are potential numtogens, since mitochondrial damage is the first step in the process of numtogenesis. Gaziev and Shaikhaev showed that radiation functions as a numtogen, since mtDNA fragments escaped to the cytoplasm at a higher rate in irradiated cells [27]. A possible mechanism involves inducing double-strand breaks in the mtDNA due to action of the ionizing radiation, followed by the integration of these fragments into the nuclear genome. In a later study by the same group [1], a greater frequency of numtogenesis was noted in the Gallus (chicken) egg nuclear genome exposed to X-rays. The NUMTs in control and irradiated cells were detected using primers specific for mtDNA. Although the nDNA in the control group did not show the presence of NUMTs, the irradiated egg cells showed NUMT insertions in 3–4 different loci, depending on the embryos. Chan et al. showed that ionizing radiation increased the frequency of non-homologous end joining (NHEJ), with the irradiated yeast cells showing a frequency of NUMTs twice as high relative to non-irradiated cells [18].
Caro et al. detected CoxIII and 16s rRNA sequences from mtDNA in the brain and liver nuclei of young and old rats; in both tissues, the density of mtDNA sequences in nuclear DNA increased with age [17]. Using Saccharomyces cerevisiae as a model, Cheng and Ivessa also showed that the rate of mtDNA transfer into the nucleus increased with age of the yeast cells [20]. In both studies, the driving numtogens were free radicals (Figure 3), which increase with age and can cause mitochondrial instability leading to cancer. Heat stress has been identified as a numtogen for tobacco, as temperatures greater than 55°C increase the double-strand breaks in the cytoplasmic organelle DNA and result in its transfer and integration into the nuclear genome [102].
Figure 3. Probable mechanism(s) underlying numtogenesis.
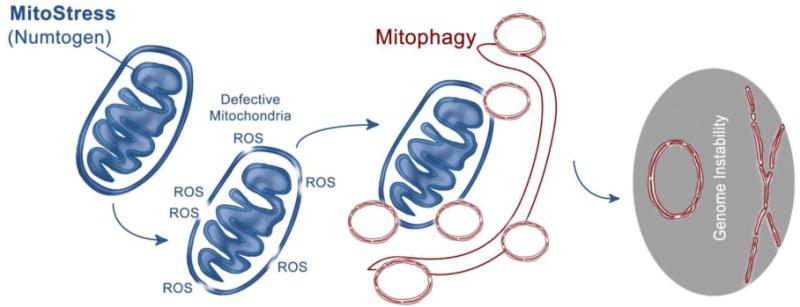
Mitostress by an intracellular or extracellular agent (numtogen) may lead to dysfunctional mitochondria, which, if not removed by mitophagy, may result in transfer of mitochondria and/or mtDNA-induced numtogenesis and tumor development.
6. Molecular mechanisms underlying numtogenesis
6.1 The types of NUMTs
Mitochondrial genes integrated into the nuclear genome differ from the parental genes in that: a) the nuclear copies often lack introns typical of the mitochondrial versions, b) they are flanked by regulatory regions that respond to nucleus-specific DNA binding proteins, and c) depending on the (non-mitochondrial) site of function, they carry specific targeting sequences. Nugent et al. established that the nuclear CoxII gene of flowering plants resembles the edited mitochondrial transcripts more than mitochondrial CoxII [70]. Since then, it has been a common observation in flowering plants that the nuclear copies show greater similarity to the edited mitochondrial mRNA than to the corresponding DNA [13, 21, 51]. This has led scientists to consider that mitochondrial genes (at least in flowering plants) may be transferred to the nucleus in the form of cDNA via an intermediate step of reverse transcription of edited mitochondrial mRNA. Indeed, there is evidence of RNA editing in plant mitochondria, usually involving the conversion of uracil residues in the primary mRNA into cytosines in the edited mRNA. This process has been studied most extensively in the mitochondrial genome of Arabidopsis [29], which contains about 441 RNA-editing sites. However, the presence of these sites do not assure an mRNA-mediated transfer of mitochondrial genes. First, for the mitochondrial gene to be decoded to the correct protein, the unedited version of the gene must be transferred. Second, cDNA reverse transcribed from the edited mitochondrial mRNA can recombine with mtDNA and consequently delete the editing sites in the mitochondrial genomes. Similarly, the absence of introns in the nuclear copies of mitochondrial genes does not necessarily point to an mRNA/cDNA intermediate; it could simply mean that the particular gene was transferred before it acquired any introns. Furthermore, introns can be removed after the gene has been transferred. e.g., the RPS10 intron of the 26s rRNA of Marchantia [59, 103].
Apart from conjectures and hypotheses, there is evidence that transfer of mitochondrial DNA to the nucleus occurs without any cDNA intermediate, that is, in the form of chunks of the mitochondrial genome seen in nuclear genomes of eukaryotes such as yeast and Arabidopsis. In Arabidopsis, for instance, a large portion of the mitochondrial genome (>75%) is nestled in chromosome 2 as a complete, continuous piece, including the introns, tRNAs, and non-coding regulatory regions. In addition, Thorsness and Fox found that, in yeast, mtDNA moves directly into the nucleus [91]. Finally, analysis of human NUMTs has not uncovered any evidence of pre-insertion splicing and editing [104], indicating that direct DNA transfer is the predominant mechanism in human numtogenesis.
6.2 mtDNA egress from mitochondria
There are three main ways for mitochondrial genetic material to escape from mitochondria and enter the nucleus: a) mitochondrial disintegration followed by DNA uptake by the nuclear import apparatus, b) fusion between the mitochondria and nucleus, and c) engulfment of mitochondria by the nucleus. Events causing disintegration of mitochondrial membranes, such as excessive production of reactive oxygen species, cytochrome c release, and mitophagy facilitate the escape of mtDNA from mitochondria (Figure 3)). Although the first method is, more or less, a random occurrence, the latter two are seen during gametogenesis and are, therefore, largely programmed [103, 110].
The mechanisms for export of mtDNA to the nucleus have been studied most extensively in yeast. With Saccharomyces cerevisiae, Thorsness and Fox measured the rate of transfer of mtDNA from the mitochondria to the nucleus [91]. A ura3−/− yeast strain deficient in uracil biosynthesis was transfected with an ura3+/+ plasmid that was maintained in the mitochondrion. When this yeast strain was propagated in a uracil-deficient medium, the plasmid DNA moved from the mitochondria to the nucleus and restored uracil biosynthesis. The rate of DNA transfer from the mitochondria was measured as a function of cell growth. In a screen for mutations that increased mtDNA migrations in yeast, the nuclear gene YME1 was identified along with five others [92, 93]. The Yme1 protein, a member of a family of ATPases, is homologous to the E. coli FtsH protein, which is involved in septum formation during cell division. A deletion in the YME1 gene increased the rate of mtDNA escape. This points to a role for YME1 in maintaining the integrity of the mitochondrial membrane. A biochemical analysis of the YME1 protein revealed its association with the matrix side of the mitochondrial inner membrane [94]. These investigators also reported stabilization of the unassembled subunit II of cytochrome oxidase in the yme1 mutant strain, indicating that coxII is a likely substrate of yme1p. By use of complementation assays, the second gene affecting mtDNA escape, YME2, was discovered. Inactivation of the YME2 gene led to an increased rate of DNA transfer from mitochondria to the nucleus [94]. YME2 encodes an inner mitochondrial membrane protein whose larger C-terminal domain faces the intermembrane space. A particular allele of the YME2 gene, yme2-4, decreases the formation of mtDNA nucleoids as well as the sensitivity of mtDNA to digestion by DNA exonuclease [73].
In yeast, proteins in the Yme family are suppressed by various proteins involved in mtDNA transfer. One YME suppressor is the YNT1 protein, a homologue of a 26S protease subunit [15]. The metabolic and morphological alterations of the yme1 mutant yeast cells can be compensated for by mutant ynt1. The protease activity of YNT1, which is necessary for progression through the cell cycle, has been implicated in the regulation of transcription. The YNT20 protein, a 3′-5′ exonuclease localized in yeast mitochondria, is also a suppressor of YME; in the yme1 and yme2 mutant yeast strains, inactivation of the YNT20 gene leads to reductions in mtDNA [37]. mtDNA is inherited as a protein-DNA complex (the nucleoid). The yeast enzyme ILV5 associates with the nucleoid. Deletions of ILV5 lead to destabilization of mtDNA, and overproduction stabilizes mtDNA. Binding of Ilv5p to mtDNA is apparently necessary for the maintenance and stability of mtDNA [58]. Park et al. showed that a missense and a null allele of the ILV5 gene acted as suppressors of the YME2-4 allele and prevented the escape of mtDNA to the nucleus [73]. By examining electron micrographs, Campbell and Thorsness showed that the mitochondria of the YME1 mutant yeast strain were degraded through vacuolar engulfment, which then increased the rate of mtDNA passage from the mitochondria to the nucleus [16]. The vacuole-dependent mitochondrial turnover is regulated by vacuolar alkaline phosphatase and the vacuolar protease PEP4. By blocking the vacuolar function, mutations in the PEP4 gene lead to a decrease in mtDNA escape. TIM17, a potential suppressor of NUMTs, is present in human cells, and its overexpression increases mtDNA stability and prevents mtDNA loss [43].
6.3 Ingress and integration of mtDNA into the nuclear genome
Experiments with yeast revealed micro-homologies (2–5 bp) adjacent to the NUMT insertion sites, suggesting that mtDNA integrates into the nuclear genome by NHEJ repair of double-strand breaks (DSBs) [77]. To repair DSBs, sequence homology between the respective termini is not a requirement; even non-complementary ends can be combined [100]. These terminal micro-homologies in the NUMTs insertion sites are evident in humans [78] and Arabidopsis [49], indicating that, in eukaryotes, NHEJ may be the most common mechanism for NUMT integration. As determined by Hazkani-Covo [39], NUMTs apparently prevented chromosomal deletions during primate evolution through NHEJ. According to Ju et al. [47], blunt end repair and micro-homology-mediated break induced replication (MMBIR) [56] are implicated as other mechanisms for joining NUMTs. It is likely that environmental stimuli that induce DSBs induce numtogenesis (Figure 3).
7. Conclusion and future prospective
To date, only one case of de novo NUMT insertion, which led to a sporadic case of Pallister-Hall syndrome, has been reported [99]. As shown in this review, our investigations [85] as well as investigations by others provide evidence for paradigm-shifting roles for numtogenesis in cancer. It is unclear how numtogenesis alters functions of the nuclear genome. Nevertheless, insertion of NUMTs into the nuclear genome of somatic cells can disrupt the function of tumor suppressor genes and pathways that contribute to tumorigenesis. NUMTs can also activate oncogenes or induce chimeric gene fusion and thereby drive tumor development. Identification of genes disrupted by NUMTs and establishment of the role of these genes in tumor initiation and progression and metastasis will help advance the cancer field. It will also pave the way for development of biomarkers and will facilitate diagnosis, prognosis, and treatment of cancer [115]. The new field of numtogenesis should allow comprehension of the function of this process in malignancies and, generally, in human health and disease. An understanding of the process for insertion of mtDNA into nuclear genomes of other organisms may elucidate mechanisms of numtogenesis that relate to other diseases, and studies with model organisms such as yeast may provide mechanistic insights underlying numtogenesis in humans. In conclusion, future efforts will likely establish a new scientific paradigm relating to the role of numtogenesis in general and, more specifically, to cancer and a variety of other human diseases.
Acknowledgments
This study was supported by grants NIH R01 CA204430, Veterans Administration 1I01BX001716, and a UAB NCTN LAPS pilot project to KKS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abdullaev SA, Fomenko LA, Kuznetsova EA, Gaziev AI. Experimental detection of integration of mTDNA in the nuclear genome induced by ionizing radiation. Radiats Biol Radioecol. 2013;53:380–8. [PubMed] [Google Scholar]
- 2.Ahmed ZM, Smith TN, Riazuddin S, Makishima T, Ghosh M, et al. Nonsyndromic recessive deafness DFNB18 and Usher syndrome type IC are allelic mutations of USHIC. Hum Genet. 2002;110:527–531. doi: 10.1007/s00439-002-0732-4. [DOI] [PubMed] [Google Scholar]
- 3.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 4.Antunes A, Ramos MJ. Discovery of a large number of previously unrecognized mitochondrial pseudogenes in fish genomes. Genomics. 2005;86:708–17. doi: 10.1016/j.ygeno.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Bakeeva LE, Skulachev VP, Sudarikova YV, Tsyplenkova VG. Mitochondria enter the nucleus (one further problem in chronic alcoholism) Biochemistry (Mosc) 2001;66:1335–41. doi: 10.1023/a:1013374410540. [DOI] [PubMed] [Google Scholar]
- 6.Bensasson D, Feldman MW, Petrov DA. Rates of DNA duplication and mitochondrial DNA insertion in the human genome. J Mol Evol. 2003;57:343–354. doi: 10.1007/s00239-003-2485-7. [DOI] [PubMed] [Google Scholar]
- 7.Bensasson D, Zhang D, Hartl DL, Hewitt GM. Mitochondrial pseudogenes: evolution’s misplaced witnesses. Trends Ecol Evol. 2001;16:314–21. doi: 10.1016/s0169-5347(01)02151-6. [DOI] [PubMed] [Google Scholar]
- 8.Blanchard JL, Schmidt GW. Mitochondrial DNA migration events in yeast and humans: integration by a common end-joining mechanism and alternative perspectives on nucleotide substitution patterns. Mol Biol Evol. 1996;13:537–48. doi: 10.1093/oxfordjournals.molbev.a025614. [DOI] [PubMed] [Google Scholar]
- 9.Bloom GD. A nucleus with cytoplasmic features. J Cell Biol. 1967;35:266–8. doi: 10.1083/jcb.35.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–80. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borensztajn K, Chafa O, Alhenc-Gelas M, Salha S, Reghis A, et al. Characterization of two novel splice site mutations in human factor VII gene causing severe plasma factor VII deficiency and bleeding diathesis. Br J Haematol. 2002;117:168–71. doi: 10.1046/j.1365-2141.2002.03397.x. [DOI] [PubMed] [Google Scholar]
- 12.Brandes D, Schofield BH, Anton E. Nuclear mitochondria? Science. 1965;149:1373–4. doi: 10.1126/science.149.3690.1373. [DOI] [PubMed] [Google Scholar]
- 13.Brennicke A, Grohmann L, Hiesel R, Knoop V, Schuster W. The mitochondrial genome on its way to the nucleus: different stages of gene transfer in higher plants. FEBS Lett. 1993;325:140–5. doi: 10.1016/0014-5793(93)81430-8. [DOI] [PubMed] [Google Scholar]
- 14.Calabrese FM, Simone D, Attimonelli M. Primates and mouse NumtS in the UCSC Genome Browser. BMC Bioinformatics. 2012;13(Suppl 4):S15. doi: 10.1186/1471-2105-13-S4-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell CL, Tanaka N, White KH, Thorsness PE. The mitochondrial genome on its way to the nucleus: different stages of gene transfer in higher plants. Mol Biol Cell. 1994;5:899–905. doi: 10.1091/mbc.5.8.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell CL, Thorsness PE. Escape of mitochondrial DNA to the nucleus in yme1 yeast is mediated by vacuolar-dependent turnover of abnormal mitochondrial compartments. J Cell Sci. 1998;111:2455–64. doi: 10.1242/jcs.111.16.2455. [DOI] [PubMed] [Google Scholar]
- 17.Caro P, Gómez J, Arduini A, González-Sánchez M, González-García M, Borrás C, Viña J, Puertas MJ, Sastre J, Barja G. Mitochondrial DNA sequences are present inside nuclear DNA in rat tissues and increase with age. Mitochondrion. 2010;10:479–86. doi: 10.1016/j.mito.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Chan CY, Kiechle M, Manivasakam P, Schiestl RH. Ionizing radiation and restriction enzymes induce microhomology-mediated illegitimate recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:5051–59. doi: 10.1093/nar/gkm442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen D, Xue W, Xiang J. The intra-nucleus integration of mitochondrial DNA (mtDNA) in cervical mucosa cells and its relation with c-myc expression. Journal Exp Clin Cancer Res. 2008;27:36–41. doi: 10.1186/1756-9966-27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng X, Ivessa AS. The migration of mitochondrial DNA fragments to the nucleus affects the chronological aging process of Saccharomyces cerevisiae. Aging Cell. 2010;9:919–23. doi: 10.1111/j.1474-9726.2010.00607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Covello PS, Gray MW. Silent mitochondrial and active nuclear genes for subunit 2 of cytochrome c oxidase (cox2) in soybean: evidence for RNA-mediated gene transfer. EMBO J. 1992;11:3815–20. doi: 10.1002/j.1460-2075.1992.tb05473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dayama G, Emery SB, Kidd JM, Mills RE. The genomic landscape of polymorphic human nuclear mitochondrial insertions. Nucleic Acids Res. 2014;42:12640–12649. doi: 10.1093/nar/gku1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Vos WH, Houben F, Kamps M, Malhas A, Verheyen F, Cox J, et al. Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum Mol Genet. 2011;20:4175–86. doi: 10.1093/hmg/ddr344. [DOI] [PubMed] [Google Scholar]
- 24.Ellis J. Promiscuous DNA-chloroplast genes inside plant mitochondria. Nature. 1982;299:678–79. doi: 10.1038/299678a0. [DOI] [PubMed] [Google Scholar]
- 25.Farrelly F, Butow RA. Rearranged mitochondrial genes in the yeast nuclear genome. Nature. 1983;301:296–301. doi: 10.1038/301296a0. [DOI] [PubMed] [Google Scholar]
- 26.Gasparre G, Porcelli AM, Lenaz G, Romeo G. Relevance of mitochondrial genetics and metabolism in cancer development. Cold Spring Harbor perspectives in biology. 2013:5. doi: 10.1101/cshperspect.a011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaziev AI, Shaĭkhaev GO. Ionizing radiation can activate the insertion of mitochondrial DNA fragments in the nuclear genome. Radiats Biol Radioecol. 2007;47:673–83. [PubMed] [Google Scholar]
- 28.Gherman A, Chen PE, Teslovich TM, Stankiewicz P, Withers M, Kashuk CS, Chakravarti A, Lupski JR, Cutler DJ, Katsanis N. Population bottlenecks as a potential major shaping force of human genome architecture. PLoS Genet. 2007;3:e119. doi: 10.1371/journal.pgen.0030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giegé P, Brennicke A. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc Natl Acad Sci U S A. 1999;96:15324–9. doi: 10.1073/pnas.96.26.15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldin E, Stahl S, Cooney AM, Kaneski CR, Gupta S, Brady RO, Ellis JR, Schiffmann R. Transfer of a mitochondrial DNA fragment to MCOLN1 causes an inherited case of mucolipidosis IV. Hum Mutat. 2004;24:460–5. doi: 10.1002/humu.20094. [DOI] [PubMed] [Google Scholar]
- 31.Gong Z, Tas E, Muzumdar R. Humanin and age-related diseases: a new link? Front Endocrinol (Lausanne) 2014;5:210. doi: 10.3389/fendo.2014.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo B, Zhai D, Cabezas E, Welsh K, Nouraini S, Satterthwait AC, Reed JC. Humanin peptide suppresses apoptosis by interfering with Bax activation. Nature. 2003;423:456–61. doi: 10.1038/nature01627. [DOI] [PubMed] [Google Scholar]
- 33.Hadler HI, Daniel BG, Pratt RD. The induction of ATP energized mitochondrial volume changes by carcinogenic N-hydroxy-N-acetyl-aminofluorenes when combined with showdomycin. A unitary hypothesis for carcinogenesis. J Antibiot (Tokyo) 1971;24:405–17. doi: 10.7164/antibiotics.24.405. [DOI] [PubMed] [Google Scholar]
- 34.Hadler HI, Devadas K, Mahalingam R. Selected nuclear Line elements with mitochondrial-DNA-like inserts are more plentiful and mobile in tumor than in normal tissye of mouse and rat. J Cell Biochem. 1998;68:100–109. doi: 10.1002/(sici)1097-4644(19980101)68:1<100::aid-jcb10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 35.Hallberg BM, Larsson NG. TFAM forces mtDNA to make a U-turn. Nat Struct Mol Biol. 2011;18:1179–81. doi: 10.1038/nsmb.2167. [DOI] [PubMed] [Google Scholar]
- 36.Hanekamp T, Thorsness PE. Inactivation of YME2/RNA12, which encodes an integral inner mitochondrial membrane protein, causes increased escape of DNA from mitochondria to the nucleus in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2764–71. doi: 10.1128/mcb.16.6.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanekamp T, Thorsness PE. YNT20, a bypass suppressor of yme1 yme2, encodes a putative 3′–5′ exonuclease localized in mitochondria of Saccharomyces cerevisiae. Curr Genet. 1999;34:438–48. doi: 10.1007/s002940050418. [DOI] [PubMed] [Google Scholar]
- 38.Hashimoto Y, Ito Y, Niikura T, Shao Z, Hata M, Oyama F, et al. Mechanisms of neuroprotection by a novel rescue factor humanin from Swedish mutant amyloid precurso rprotein. Biochem Biophys Res Commun. 2001;283:460–8. doi: 10.1006/bbrc.2001.4765. [DOI] [PubMed] [Google Scholar]
- 39.Hazkani-Covo E, Covo S. Numt-mediated double-strand break repair mitigates deletions during primate genome evolution. PLoS Genet. 2008;4:e1000237. doi: 10.1371/journal.pgen.1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hazkani-Covo E, Graur D. A comparative analysis of numt evolution in human and chimpanzee. Mol Biol Evol. 2007;24:13–8. doi: 10.1093/molbev/msl149. [DOI] [PubMed] [Google Scholar]
- 41.Hazkani-Covo E, Sorek R, Graur D. Evolutionary dynamics of large numts in the human genome: rarity of independent insertions and abundance of post-insertion duplications. J Mol Evol. 2003;56:169–74. doi: 10.1007/s00239-002-2390-5. [DOI] [PubMed] [Google Scholar]
- 42.Hoffman H, Grigg GW. An electron microscopic study of mitochondria formation. Exp Cell Res. 1958;15:118–31. doi: 10.1016/0014-4827(58)90068-5. [DOI] [PubMed] [Google Scholar]
- 43.Iacovino M, Granycome C, Sembongi H, Bokori-Brown M, Butow RA, Holt IJ, Bateman JM. The conserved translocase Tim17 prevents mitochondrial DNA loss. Hum Mol Genet. 2009;18:65–74. doi: 10.1093/hmg/ddn313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ikonen M, Liu B, Hashimoto Y, Ma L, Lee KW, Niikura T, et al. Interaction between the Alzheimer’s survival peptide humanin and insulin-like growth factor-binding protein3 regulates cell survival and apoptosis. Proc Natl Acad Sci USA. 2003;100:13042–7. doi: 10.1073/pnas.2135111100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jensen H, Engedal H, Saetersdal TS. Ultrastructure of mitochondria-containining nuclei in human myocardial cells. Virchows Arch B Cell Pathol. 1976;1:1–12. doi: 10.1007/BF02899139. [DOI] [PubMed] [Google Scholar]
- 46.Jensen-Seaman MI, Wildschutte JH, Soto-Calderon ID, Anthony NM. A comparative approach shows differences in patterns of numt insertion during hominoid evolution. J Mol Evol. 2009;68:688–99. doi: 10.1007/s00239-009-9243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ju YS, Tubio JMC, Stratton MR, et al. Frequent somatic transfer of mitochondrial DNA into the nuclear genome of human cancer cells. Genome Res. 2015;25(6):814–24. doi: 10.1101/gr.190470.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kamimura N, Ishii S, Ma LD, Shay JW. Three separate mitochondrial DNA sequences are contiguous in human genome cDNA. J Mol Biol. 1989;210:703–707. doi: 10.1016/0022-2836(89)90103-4. [DOI] [PubMed] [Google Scholar]
- 49.Kirik A, Salomon S, Puchta H. Species-specific double-strand break repair and genome evolution in plants. EMBO J. 2000;19:5562–5566. doi: 10.1093/emboj/19.20.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klug H. On the occurrence of mitochondria in the cell nucleus. Naturwissenschaften. 1966;53:339. doi: 10.1007/BF00631212. [DOI] [PubMed] [Google Scholar]
- 51.Knoop V, Ehrhardt T, Lattig K, Brennicke A. The gene for ribosomal protein S10 is present in mitochondria of pea and potato but absent from those of Arabidopsis and Oenothera. Curr Genet. 1995;27:559–564. doi: 10.1007/BF00314448. [DOI] [PubMed] [Google Scholar]
- 52.Landerer E, Villegas J, Burzio VA, Oliveira L, Villota C, Lopez C, et al. Nuclear localization of the mitochondrial ncRNAs in normal and cancer cells. Cell Oncol (Dordr) 2011;34:297–305. doi: 10.1007/s13402-011-0018-8. [DOI] [PubMed] [Google Scholar]
- 53.Lang M, Sazzini M, Calabrese FM, Gasparre G. Polymorphic NumtS trace human population relationships. Hum Genet. 2012;131:757–771. doi: 10.1007/s00439-011-1125-3. 2012. [DOI] [PubMed] [Google Scholar]
- 54.Lascaro D, Castellana S, Gasparre G, Romeo G, Saccone C, Attimonelli M. The RHNumtS compilation: features and bioinformatics approaches to locate and quantify Human NumtS. BMC Genomics. 2008;9:267. doi: 10.1186/1471-2164-9-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leister D. Origin, evolution and genetic effects of nuclear insertions of organelle DNA. Trends Genet. 2005;21:655–63. doi: 10.1016/j.tig.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 56.Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lopez JV, Yuhki N, Masuda R, Modi W, O’Brien SJ. Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. J Mol Evol. 1994;39:174–90. doi: 10.1007/BF00163806. [DOI] [PubMed] [Google Scholar]
- 58.Macierzanka M, Plotka M, Pryputniewicz-Drobinska D, Lewandowska A, Lightowlers R, Marszalek J. Maintenance and stabilization of mtDNA can be facilitated by the DNA-binding activity of Ilv5p. Biochim Biophys Acta. 2008;1783:107–17. doi: 10.1016/j.bbamcr.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 59.Malek O, Brennicke A, Knoop V. Evolution of trans-splicing plant mitochondrial introns in pre-Permian times. Proc Natl Acad Sci U S A. 1997;94:553–8. doi: 10.1073/pnas.94.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Margulis L. Symbiosis and evolution. Sci Am. 1971;225:48–57. doi: 10.1038/scientificamerican0871-48. [DOI] [PubMed] [Google Scholar]
- 61.Margulis L. Symbiotic theory of the origin of eukaryotic organelles; criteria for proof. Symp Soc Exp Biol. 1975;29:21–38. [PubMed] [Google Scholar]
- 62.Matsuyama M, Suzuki H. Seizing mechanism and fate of intranuclear mitochondria. Experientia. 1972;28:1347–8. doi: 10.1007/BF01965337. [DOI] [PubMed] [Google Scholar]
- 63.Meyer M, Fu Q, Aximu-Petri A, Glocke I, Nickel B, Arsuaga JL, Martinez I, Gracia A, de Castro JM, Carbonell E, et al. A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature. 2014;505:403–06. doi: 10.1038/nature12788. [DOI] [PubMed] [Google Scholar]
- 64.Mishmar D, Ruiz-Pesini E, Brandon M, Wallace DC. Mitochondrial DNA-like sequences in the nucleus (NUMTs): insights into our African origins and the mechanism of foreign DNA integration. Hum Mutat. 2004;23:125–33. doi: 10.1002/humu.10304. [DOI] [PubMed] [Google Scholar]
- 65.Mori H. Fukushima. J Med Sci. 1960;7:21–32. [Google Scholar]
- 66.Mourier T, Hansen AJ, Willerslev E, Arctander P. The Human Genome Project reveals a continuous transfer of large mitochondrial fragments to the nucleus. Mol Biol Evol. 2001;18:1833–37. doi: 10.1093/oxfordjournals.molbev.a003971. [DOI] [PubMed] [Google Scholar]
- 67.Muradian KK, Lehmann G, Fraifeld VE. NUMT (“new mighty”) hypothesis of longevity. Rejuvenation Res. 2010;13:152–5. doi: 10.1089/rej.2009.0974. [DOI] [PubMed] [Google Scholar]
- 68.Nielsen T, Piatyszek M, Shay J, Pearson C, Zannishadjopoulos M, Price G. Autonomous replication activity of a human mitochondrial-DNA sequence inserted into genomic DNA. Int J Oncol. 1994;5:1003–8. doi: 10.3892/ijo.5.5.1003. [DOI] [PubMed] [Google Scholar]
- 69.Noutsos C, Kleine T, Armbruster U, DalCorso G, Leister D. Nuclear insertions of organellar DNA can create novel patches of functional exon sequences. Trends Genet. 2007;23:597–601. doi: 10.1016/j.tig.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 70.Nugent JM, Palmer JD. RNA-mediated transfer of the gene coxII from the mitochondrion to the nucleus during flowering plant evolution. Cell. 1991;66:473–81. doi: 10.1016/0092-8674(81)90011-8. [DOI] [PubMed] [Google Scholar]
- 71.Oeggerli M, Tian Y, Ruiz C, Wijker B, Sauter G, Obermann E, Guth U, Zlobec I, Sausbier M, Kunzelmann K, et al. Role of KCNMA1 in breast cancer. PLoS One. 2012;7:e41664. doi: 10.1371/journal.pone.0041664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oliva H, Valle A, Flores LD, Rivas MC. Intranuclear mitochondriae in Hodgkin’s disease. Virchows Arch B Cell Pathol. 1973;12:189–94. doi: 10.1007/BF02893997. [DOI] [PubMed] [Google Scholar]
- 73.Park S, Hanekamp T, Thorsness MK, Thorsness PE. Yme2p is a mediator of nucleoid structure and number in mitochondria of the yeast Saccharomyces cerevisiae. Curr Genet. 2006;50:173–182. doi: 10.1007/s00294-006-0087-9. [DOI] [PubMed] [Google Scholar]
- 74.Perna NT, Kocher TD. Mitochondrial DNA: molecular fossils in the nucleus. Curr Biol. 1996;6:128–29. doi: 10.1016/s0960-9822(02)00441-4. [DOI] [PubMed] [Google Scholar]
- 75.Qu H, Ma F, Li Q. Comparative analysis of mitochondrial fragments transferred to the nucleus in vertebrate. J Genet Genomics. 2008;35:485–490. doi: 10.1016/S1673-8527(08)60066-1. [DOI] [PubMed] [Google Scholar]
- 76.Ramos A, Barbena E, Santos C, et al. Nuclear insertions of mitochondrial origin: Database updating and usefulness in cancer studies. Mitochondrion. 2011;11:946–953. doi: 10.1016/j.mito.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 77.Ricchetti M, Fairhead C, Dujon B. Mitochondrial DNA repairs double-strand breaks in yeast chromosomes. Nature. 1999;402:96–100. doi: 10.1038/47076. [DOI] [PubMed] [Google Scholar]
- 78.Ricchetti M, Tekaia F, Dujon B. Continued colonization of the human genome by mitochondrial DNA. PLoS Biol. 2004;2:E273. doi: 10.1371/journal.pbio.0020273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Richter C. Do mitochondrial DNA fragments promote cancer and aging? FEBS Lett. 1988;241:1–5. doi: 10.1016/0014-5793(88)81018-4. [DOI] [PubMed] [Google Scholar]
- 80.Roark LM, Hui AY, Donnelly L, Birchler JA, Newton KJ. Recent and frequent insertions of chloroplast DNA into maize nuclear chromosomes. Cytogenet Genome Res. 2010 Jul;129(1–3):17–23. doi: 10.1159/000312724. [DOI] [PubMed] [Google Scholar]
- 81.Ross JM, Stewart JB, Hagstrom E, Brene S, Mourier A, Coppotelli G, Freyer C, Lagouge M, Hoffer BJ, Olson L, et al. Germline mitochondrial DNA mutations aggravate ageing and can impair brain development. Nature. 2013;501:412–15. doi: 10.1038/nature12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schumacher HR, Szekely IE, Patel SB, Fisher DR. Leukemic mitochondria. I. Acute myeloblastic leukemia. Am J Pathol. 1974;74:71–82. [PMC free article] [PubMed] [Google Scholar]
- 83.Shay JW, Werbin H. New evidence for the insertion of mitochondrial DNA into the human genome: significance for cancerand aging. Mutat Res. 1992;2752:227–35. doi: 10.1016/0921-8734(92)90026-l. [DOI] [PubMed] [Google Scholar]
- 84.Simone D, Calabrese FM, Lang M, Gasparre G, Attimonelli M. The reference human nuclear mitochondrial sequences compilation validated and implemented on the UCSC genome browser. BMC Genomics. 2011;12:517. doi: 10.1186/1471-2164-12-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Srinivasainagendra V, Sandel MW, Singh B, Sundaresan A, Mooga VP, Bajpai P, Tiwari HK, Singh KK. Migration of mitochondrial DNA in the nuclear genome of colorectal adenocarcinoma. Genome Med. 2017;9:31. doi: 10.1186/s13073-017-0420-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stern DB, Lonsdale DM. Mitochondrial and chloroplast genomes of maize have a 12-kb DNA sequence in common. Nature. 1982;299:698–702. doi: 10.1038/299698a0. [DOI] [PubMed] [Google Scholar]
- 86.Sunba MS, Rahi AH, Morgan G. Tumors of the anterior uvea. II. Intranuclear cytoplasmic inclusions in malignant melanoma of the iris. Br J Ophthalmol. 1980;64:453–6. doi: 10.1136/bjo.64.6.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tajima H, Niikura T, Hashimoto Y, Ito Y, Kita Y, Terashita K, et al. Evidence for in vivo production of humanin peptide, a neuroprotective factor against Alzheimer’s disease-related insults. Neurosci Lett. 2002;324:227–31. doi: 10.1016/s0304-3940(02)00199-4. [DOI] [PubMed] [Google Scholar]
- 88.Takemura G, Takatsu Y, Sakaguchi H, Fujiwara H. Intranuclear mitochondria in human myocardial cells. Pathol Res Pract. 1997;193:305–11. doi: 10.1016/s0344-0338(97)80008-8. [DOI] [PubMed] [Google Scholar]
- 89.Theodoratou E, Campbell H, Farrington SM, et al. A large-scale meta-analysis to refine colorectal cancer risk estimates associated with MUTYH variants. Br J Cancer. 2010;103:1875–84. doi: 10.1038/sj.bjc.6605966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thorsness PE, Fox TD. Escape of DNA from mitochondria to the nucleus in Saccharomyces cerevisiae. Nature. 1990;346:376–79. doi: 10.1038/346376a0. [DOI] [PubMed] [Google Scholar]
- 91.Thorsness PE, Fox TD. Nuclear mutations in Saccharomyces cerevisiae that affect the escape of DNA from mitochondria to the nucleus. Genetics. 1993;134:21–28. doi: 10.1093/genetics/134.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thorsness PE, White KH, Fox TD. Inactivation of YME1, a member of the ftsH-SEC18-PAS1-CDC48 family of putative ATPase-encoding genes, causes increased escape of DNA from mitochondria in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:5418–26. doi: 10.1128/mcb.13.9.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thorsness PE, Weber ER. Escape and migration of nucleic acids between chloroplasts, mitochondria and the nucleus. Int Rev Cytol. 1996;165:207–34. doi: 10.1016/s0074-7696(08)62223-8. [DOI] [PubMed] [Google Scholar]
- 94.Thyagarajan B, Wang R, Barcelo H, Koh WP, Yuan JM. Mitochondrial copy number is associated with colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2012;21:1574–81. doi: 10.1158/1055-9965.EPI-12-0138-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tourmen Y, Baris O, Dessen P, Jacques C, Malthiery Y, Reynier P. Structure and chromosomal distribution of human mitochondrial pseudogenes. Genomics. 2002;80:71–77. doi: 10.1006/geno.2002.6798. [DOI] [PubMed] [Google Scholar]
- 96.Tsuji J, Frith MC, Tomii K, Horton P. Mammalian NUMT insertion is nonrandom. Nucleic Acids Res. 2012;40:9073–88. doi: 10.1093/nar/gks424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tsuzuki T, Nomiyama H, Setoyama C, Maeda S, Shimada K. Presence of mitochondrial-DNA-like sequences in the human nuclear DNA. Gene. 1983;25:223–229. doi: 10.1016/0378-1119(83)90226-3. [DOI] [PubMed] [Google Scholar]
- 98.Turner C, Killoran C, Thomas NS, Rosenberg M, Chuzhanova NA, Johnston J, Kemel Y, Cooper DN, Biesecker LG. Human genetic disease caused by de novo mitochondrial-nuclear DNA transfer. Hum Genet. 2003;112:303–309. doi: 10.1007/s00439-002-0892-2. [DOI] [PubMed] [Google Scholar]
- 99.van Gent DC, Hoeijmakers JH, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet. 2001;2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- 100.Wallace DC. Mitochondria and cancer: Warburg addressed. Cold Spring Harb Symp Quant Biol. 2005;70:363–74. doi: 10.1101/sqb.2005.70.035. [DOI] [PubMed] [Google Scholar]
- 101.Wang D, Lloyd AH, Timmis JN. Environmental stress increases the entry of cytoplasmic organellar DNA into the nucleus in plants. Proc Natl Acad Sci U S A. 2012;109:2444–8. doi: 10.1073/pnas.1117890109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wischmann C, Schuster W. Transfer of rps10 from the mitochondrion to the nucleus in Arabidopsis thaliana: evidence for RNA-mediated transfer and exon shuffling at the integration site. FEBS Lett. 1995;374:152–6. doi: 10.1016/0014-5793(95)01100-s. [DOI] [PubMed] [Google Scholar]
- 103.Woischnik M, Moraes CT. Pattern of organization of human mitochondrial pseudogenes in the nuclear genome. Genome Res. 2002;12:885–93. doi: 10.1101/gr.227202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xianlong L, Dianchun F, Xiaodong Z. Relationship between integration of mtDNA fragments in the nuclei of gastric mucosal cells and Helicobacterpyiori infection. Acta Academiae Medicinae Militaris Tertiae. 2001;23:1043–46. [Google Scholar]
- 105.Yao YG, Bravi CM, Bandelt HJ. A call for mtDNA data quality control in forensic science. Forensic Sci Int. 2004;141:1–6. doi: 10.1016/j.forsciint.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 106.Yao YG, Kong QP, Salas A, Bandelt HJ. Pseudomitochondrial genome haunts disease studies. J Med Genet. 2008;45:769–72. doi: 10.1136/jmg.2008.059782. [DOI] [PubMed] [Google Scholar]
- 107.Yen K, Lee C, Mehta H, Cohen P. The emerging role of the mitochondrial-derived peptide humanin in stress resistance. J Mol Endocrinol. 2013;50:11–9. doi: 10.1530/JME-12-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yoshida T, Furihata HY, Kawabe A. Patterns of genomic integration of nuclear chloroplast DNA fragments in plant species. DNA Res. 2014;21(2):127–40. doi: 10.1093/dnares/dst045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yu HS, Russell SD. Occurrence of mitochondria in the nuclei of tobacco sperm cells. Plant Cell. 1994;6:1477–84. doi: 10.1105/tpc.6.10.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yu X, Gabriel A. Patching broken chromosomes with extranuclear cellular DNA. Mol Cell. 1999;4:873–81. doi: 10.1016/s1097-2765(00)80397-4. [DOI] [PubMed] [Google Scholar]
- 111.Zhang Q, et al. Examination of the cytoplasmic DNA in male reproductive cells to determine the potential for cytoplasmic inheritance in 295 angiosperm species. Plant Cell Physiol. 2003;44:941–951. doi: 10.1093/pcp/pcg121. [DOI] [PubMed] [Google Scholar]
- 112.Zischler H, Hoss M, Handt O, von Haeseler A, van der Kuyl AC, et al. Detecting dinosaur DNA. Science. 1995;268:1192–1193. doi: 10.1126/science.7605504. [DOI] [PubMed] [Google Scholar]
- 115.Zong WX, Rabinowitz JD, White E. Mitochondria and Cancer. Mol Cell. 2016;61:667–76. doi: 10.1016/j.molcel.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; Koo DH, Singh B, Jiang J, Friebe B, Gill BS, Chastain PD, Manne U, Tiwari HK, Singh KK. Single molecule mtDNA fiber FISH for analyzing numtogenesis. Anal Biochem. 2017 doi: 10.1016/j.ab.2017.03.015. pii: S0003-2697(17)30131-8. [DOI] [PMC free article] [PubMed] [Google Scholar]


