Abstract
Heat shock protein 90 (Hsp90) is differentially expressed in tumor cells including melanoma and involved in proper folding, stabilization and regulation of cellular proteins. We investigated a novobiocin-derived Hsp90 C-terminal inhibitor, KU135, for anti-proliferative effects in melanoma cells. The results indicate that KU135 reduced cell viability and cell proliferation in melanoma cells and IC50 values for A735(DRO), M14(NPA), B16F10 and SKMEL28 cells were 0.82, 0.92, 1.33 and 1.30 M respectively. KU135 induced a more potent anti-proliferative effect in most melanoma cells versus N-terminal Hsp90 inhibitor 17AAG. KU135 induced apoptosis in melanoma cells, as indicated by annexin V/PI staining, reduction in the mitochondrial membrane potential, mitochondrial cytochrome C release and caspase 3 activation. KU135 reduced levels of Hsp90 client proteins Akt, BRAF, RAF-1, cyclin B and cdc25 proteins. Additionally, it reduced Hsp70, Hsp90 paralog, GRP94 and HSF1 levels. KU135 induced strong G2/M cell cycle arrest, associated with decreased expression of cdc25c, cyclin B and increased phosphorylation of cdc25c. These finding show that KU135 reduced cell survival, proliferation, and induces apoptosis in melanoma cells. We suggest that KU135 may be a potential candidate for cancer therapy against melanoma.
Keywords: Hsp90 inhibitor, C-terminal, melanoma
1. Introduction
Melanoma is the most aggressive type of skin cancer and the fifth and sixth most common cancer in men and women in United States, respectively [1]. The life time risk of developing melanoma is estimated to be 1 in 75 [2]. Skin cancer is the third most common human malignancy worldwide and the global incidence of melanoma is rising at an alarming rate [1]. Cancer therapy is most successful when molecules are designed to interfere with the growth and metastasis of cancer cells. The term “oncogenic addiction” is used to explain that cancer cells more heavily rely on key hyperactivated signaling pathway(s) than normal cells do for their abnormal growth. BRAF mutations are present in 50–70% of human skin melanomas [3]. The most common mutation observed in melanoma is BRAFV600E, leading to activation of the RAS-RAF-MEK1/2-ERK1/2 pathway. Although early success was observed with small molecules inhibiting RAF kinase, however, recent findings show that BRAF inhibitors may induce ERK activation in melanoma cells and unsatisfactory results obtained from recent RAF inhibitors phase I clinical trials have pointed to the importance of exploring novel targets for the treatment of melanoma [4,5].
Hsp90 is a chaperone involved in proper folding of several “client” proteins in melanoma cells. BRAF, RAF-1 Akt and MEK which are known to be activated in melanoma are considered Hsp90 client proteins (http://www.picard.ch/downloads/Hsp90interactors.pdf). Furthermore, Hsp90 expression is increased in melanoma cancer when compared to nevi, and this expression has been associated with disease progression [6]. Therefore Hsp90 inhibition may be an effective tool in the treatment of melanoma cancer. Several in vitro studies involving agents that target Hsp90 have been reported [7–9]. Recent human clinical trials involving Hsp90 N-terminal inhibitors revealed that most suffer from limiting side effects [9–11]. Therefore, introduction of novel Hsp90 inhibitors may be an alternative and effective therapy against melanoma. In recent years C-terminal Hsp90 inhibitors have been investigated in several cancer models [12,13]. A novobiocin-derived C-terminal Hsp90 inhibitor, KU135, has recently been investigated for anti-proliferative activity against human leukemic cells [14].
The aim of the present study is to investigate the efficacy of KU135 against melanoma cancer. KU135 induced anti-proliferative activity against several melanoma cells lines. Moreover, KU135 did not change Hsp90, Hsp90, Hsp27 and (TNF) receptor-associated protein 1 (TRAP-1/HSP75) levels. In contrast, KU135 reduced expression of Hsp70 and endoplasmic reticulum Hsp90 paralog glucose-regulated protein 94 (Grp94) in a concentration-dependent manner, which is opposite from the effect of N-terminal Hsp90 inhibitor, 17AAG. KU135 also reduced total and phosphorylated levels of Heat shock factor-1 (HSF1) in melanoma cells. Furthermore, treatment with KU135 was noted to induce apoptosis and G2/M cell cycle arrest in melanoma cells.
2. Materials and methods
2.1. Cell lines and cell culture
A375(DRO) and M14(NPA) cells were provided by Dr. G.J. Juillard (University of California at Los Angeles, Los Angeles CA). DRO and NPA cells were previously thought to be derived from an anaplastic thyroid cancer. However it has been shown that these cell lines are genetically identical to the melanoma A375 and M14 cell lines respectively so they are designated as subtypes of A375, A375(DRO) and M14, M14(NPA) [15]. These cells are known to harbor BRAFV600E genetic alterations. A375(DRO) and M14(NPA) were grown in RPMI 1640 (Sigma-Aldrich, St. Louis, MO), supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO) and 1% penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO). The human melanoma cell line SKMEL28 and murine melanoma cell line B16F10 were purchased from ATCC (Manassas, VA). These cell were grown in DMEM media (Sigma-Aldrich, St. Louis, MO) with 2 mM l-glutamine and 10% (w/v) FBS, supplemented with 1% penicillin/streptomycin. Cells were incubated at 37°C in humidified atmosphere containing 5% CO2. Cells were seeded and incubated overnight to reach 50–60% confluence during each experiment. Cells were treated with KU135 from a 20 mM stock solution (dissolved in DMSO; control cells received DMSO and DMSO levels did not exceed 0.1%).
2.2. Reagents and antibodies
The rabbit polyclonal antibodies against caspase-3 (#9662; 1:1000), Caspase 8 (#9746; 1:1000), Caspase 9 (#9508, 1:1000), poly(ADP-ribose) polymerase (PARP)(#9542; 1:1000), phospho-Akt (Ser473)(#4058, 1:1000), Akt (#9272;1:1000), BRAF (#9434; 1:1000), phospho-ERK1/2(Tyr202/Tyr204) (#4377; 1:1000), cyclin B1 (#4138; 1:1000), phospho-cdc25c(Ser216) (#4901;1:1000), cdc25c (#4688; 1:1000), and cytochrome C (#4280; 1:1000) were purchased from Cell Signaling (Beverly, MA). Rabbit polyclonal antibodies against Hsp27 and mouse monoclonal antibodies against Hsp90 (#SPA840; 1:1000), Hsp90 (#SPA843; 1:1000), Grp94 (#SPA850; 1:1000), HSF-1 (#SPA950; 1:2000) and Hsp70 (#SPA810; 1:1000) were purchased from StressGen (Victoria, BC, Canada). Mouse monoclonal antibody against TRAP1 (#MIA010, 1:1000) was purchased from Affinity Bioreagent – Thermo Scientific, Rockford, IL). Mouse monoclonal antibody against -Actin (#sc1616; 1:1000), polyclonal antibodies against RAF-1 (#sc-227, 1:1000), total ERK2 (#sc-154; 1:5000), and anti-rabbit, and anti-mouse secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Annexin V and propidium iodide were obtained from BD Bioscience (San Jose, CA).
2.3. Cell proliferation and viability assays
Cells were plated in 96-well microtiter plates (2×103 cells/well) in growth media. After overnight incubation, cells were treated with varying concentration of KU135 for 72 h. After treatment, the number of viable cells was determined using a colorimetric Cell Proliferation Assay (CellTiter96 Aqueous Nonradioactive Cell Proliferation assay; Promega, Madison WI, USA) according to manufacturer’s instruction. All studies were performed in triplicate. Data was plotted as a function of percent cell viability based upon controls vs. drug concentration (x-axis). The concentration of drug at which cell growth was inhibited by 50% (IC50) was estimated using GraphPad Prism5 software (La Jolla, CA).
2.4. Mitochondrial membrane potential (ΔΨm) measurements
The changes in mitochondrial membrane potential, ΔΨm, were detected by flow cytometry using the fluorescent cationic dye, JC-1 (mitoProbe™, Invitrogen, Carlsbad, CA). In control cells, an intact ΔΨm allows JC-1, bearing a delocalized positive charge, to accumulate and aggregate in the mitochondrial matrix, where it fluoresces red. In cell undergoing apoptosis, the collapse of ΔΨm causes JC-1 to remain in the cytoplasm in a green fluorescent monomeric form. Therefore, mitochondrial depolarization can be detected by a decrease in the red-to-green fluorescence intensity ratio, that is, fluorescence emission shift from red (590 nm) to green (525 nm). Apoptotic cells, as characterized by decreased ΔΨm, exhibited low red-to-fluorescence ratio. A735(DRO) and M14(NPA) cells were treated with 5 and 10 M KU135 for 24 h. As a positive control, untreated control cells were incubated for 5 min, previous to the addition of JC-1, with 5.0 µM uncoupler carbonylcyanide 3-chlorophenylhydrazone (CCCP). Camptothecin was used as a positive drug agent because it is known to reduce mitochondrial membrane potential. Cells were collected and incubated with 5 g/ml of JC-1 at 37°C in a 5% CO2 incubator for 20 min. After washing, cells were analyzed on a FACS Calibur flow cytometer (Becton Dickinson, San Diego, CA).
2.5. Cell Lysis and Western blot analysis of KU135 treated cells
Cells were washed twice with ice-cold 1× PBS and lysed in ice-cold RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% (v/v) Nonidet NP-40, 0.5% (w/v) sodium deoxycholate, 10 mM sodium fluoride, 1 mM sodium orthovanadate, 1 mM PMSF, 10 mM sodium pyrophosphate, 0.1% (w/v), SDS supplemented with 1× protease inhibitor solution (Calbiochem, San Diego, CA)). Protein levels were determined using Protein Assay Reagent (Pierce Rockford, IL). Equal amounts of proteins (15–30 g) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electrotransfered onto a Hybond nitrocellulose membrane (Amersham, Piscataway, NJ). Membranes were blocked with 3% (w/v) non-fat dry milk (Bio-Rad Laboratories, Hercules, CA), and probed with the appropriate dilutions of primary antibodies overnight at 4°C. The blots were incubated with 1:10,000 dilution of secondary antibody and the proteins were visualized by enhanced chemiluminescence West Pico (Pierce Biotechnology, Rockford, IL), then captured on Kodak XAR-5 film (Eastman Kodak, Rochester, NY). Where indicated, the blots were reprobed with antibody against -actin to ensure equal loading and transfer of proteins.
2.6. Mitochondrial fractionation
After treatment, cells were washed with 1× PBS and removed from culture plates by trypsinization. Suspended cells were washed twice with 1× PBS and lysed in buffer A (250 mM sucrose, 20 mM HEPES (pH 7.4), 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT and protease inhibitor (1:500 dilution) and incubated on ice for 20 min. Nuclear faction was removed by centrifugation (750 ×g; 5 min). Supernatant was centrifuged again (10,000 ×g 10 min). The supernatant (cytoplasmic faction) was removed. The pellet was washed once with buffer A and resuspendend in the RIPA buffer.
2.7. Fluorescence-activated cell sorting analysis
Cells were grown overnight, and after KU135 treatment, trypsinized and washed with 0.9% NaCl and fixed with 70% cold ethanol for 30 min at room temperature. Then they were collected by centrifugation (700 ×g, 5 min) and stained with propidium iodide (50 mg/mL in PBS) for 30 min. They were treated with DNAse free RNAse (1 mg/mL) for 30 min and analyzed by flow cytometry. Preliminary studies to evaluate for induction of apoptosis were also performed including annexin V-propidium iodide (PI) co-staining with flow cytometry. An analysis of phosphatidylserine (PS) on the outer leaflet of apoptotic cell membranes was performed using annexin V-FITC and PI to distinguish between apoptotic and necrotic cells. After treatment, cells (1×105 cells/mL) were washed with 1× PBS and trypsinized. Cells were washed and stained with Annexin-V / PI according to the manufacturer’s instruction (BD Pharmingen; San Diego, CA), and the cells were analyzed by flow cytometry (BD LSRII; Becton Dickerson, San Diego, CA).
2.8. Luciferase assays
HSE promoter induction study was carried out using HeLa cells stably expressing the hsp70.1pr-luciferase plasmid (kindly provided by Dr. Sandy Westerheide University of South Florida). The cells were plated at 1.0 × 104 cells/well in 96-well plates overnight before treatment with KU135. For heat treatment, cells were heated at 43°C for 1 h and luciferase activity was measured 24 h after heat treatment. Cells were treated with Bright-Glo reagent (Promega, Madison, WI) and luciferase activity was quantified using Synergy 2 Alpha microplate reader (BioTek, Winooski, VT) according to the manufacturer’s instruction.
2.9. Statistical Analysis
Data were presented as mean values ± standard deviation. Statistical comparisons among groups were performed by Student's t-test. These analyses were run using GraphPad Software (GraphPad Inc., San Diego, CA).
3. Results
3.1. KU135 reduces cell viability and cell proliferation in melanoma cells
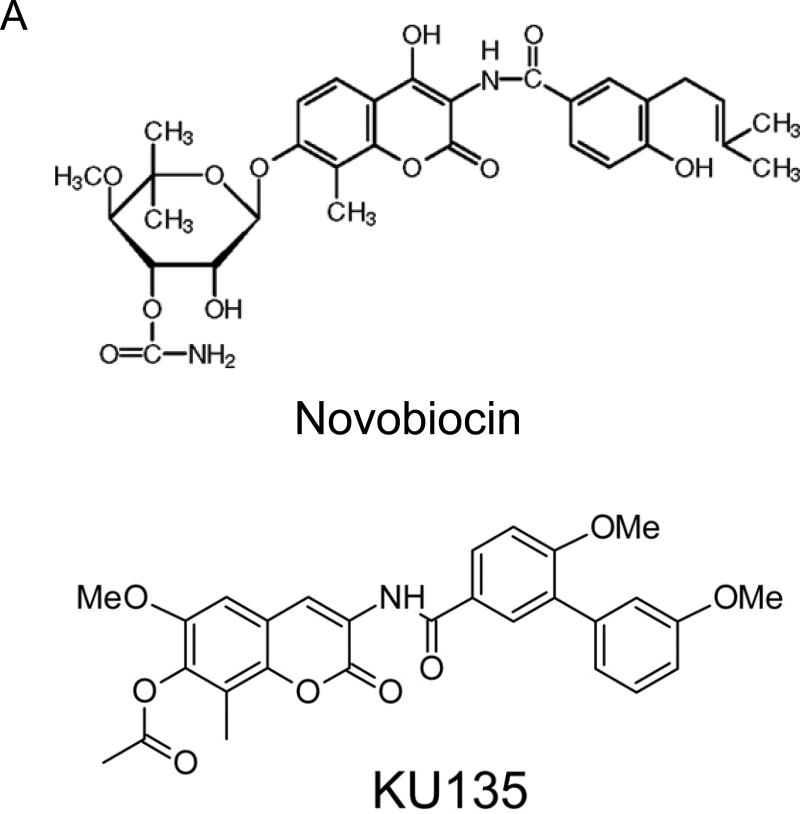
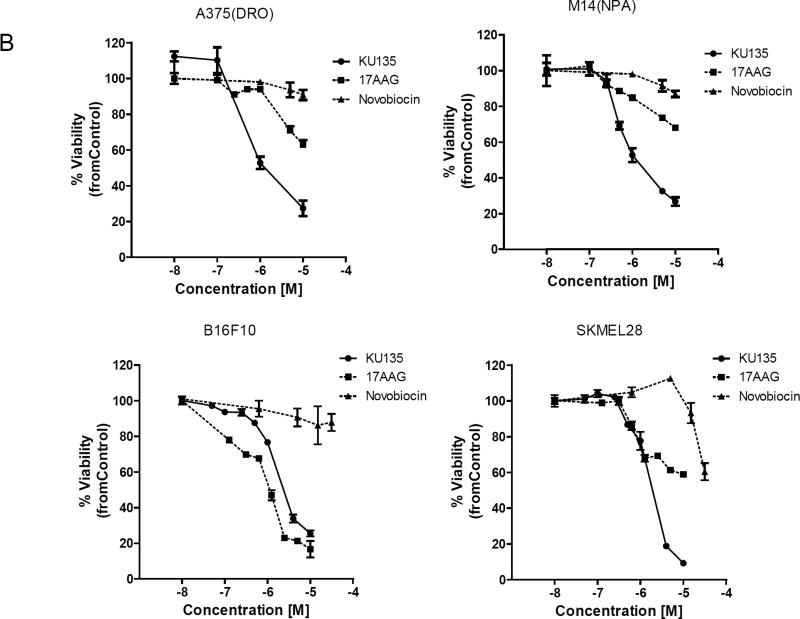
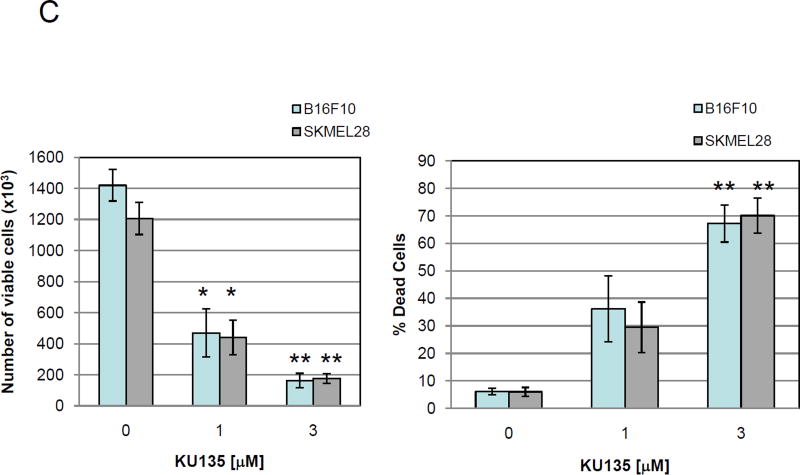
KU135 is a small molecule C-terminal Hsp90 inhibitor, derived from the antibiotic novobiocin, which has been structurally optimized to convert KU135 into a potent Hsp90 inhibitor [14] (Fig. 1A, structure of novobiocin and KU135). To investigate the biological effect of Hsp90 inhibition by KU135 in melanoma cells, we incubated melanoma cells with increasing concentrations of KU135 and DMSO (0.1% final concentration), and viability was determined using the MTS assay. KU135 inhibited cell viability in a dose-dependent manner in A735(DRO), M14(NPA), B16F10 and SKMEL28 cells (Fig. 1B). The IC50 of KU135 was determined to be 0.82 M, 0.93 M, 1.33 M and 1.30 M against A735(DRO), M14(NPA), B16F10 and SKMEL28 cells, respectively after 72 h of treatment. KU135 was shown to be a more potent drug to reduce cell viability in A375(DRO), M14(NPA) and SKMEL28 cells compared to Hsp90 N-terminal inhibitor 17AAG and novobiocin. However, 17AAG was more potent reducing cell viability in B16F10 murine melanoma cells compared to KU135. Trypan blue staining was also used to investigate cell proliferation and cell death in cell. B16F10 and SKMEL28 cells were treated with different concentrations of KU135. KU135 reduced cell proliferation and induced cell death in B16F10 and SKMEL28 cells in a concentration-dependent manner (Fig. 1C). Treatment of B16F10 and SKMEL28 cells with KU135 reduced cell proliferation by 3- and 6-fold at 1 M and 3 M, respectively. In addition, the percentage of trypan blue positive cells increased from 4–5% in untreated cells to approximately 30% and 65% in melanoma cells treated with 1 M and 3 M KU135, respectively. Similar results were obtained in A735(DRO) and M14(NPA) cells (data not shown).
Fig. 1.
KU135 reduces cell viability and proliferation in melanoma cells. (A) Chemical structure of novobiocin and active analogue KU135. (B) Cytotoxic effect of KU135 on melanoma cells. Melanoma cells (2×103) were seeded in 96 well plates and incubated for 24 h. After treatment for 72 h, cell viability was determined by MTS viability assay. Results are expressed as percentage of cell viability compared to control. (C), Cell proliferation was carried out using trypan blue staining. Cells (3×104) were incubated overnight in 60 mm culture plates and treated with KU135 for 24 h. Cells were stained with trypan blue and counted using hemocytometer. All experiments were carried out in triplicate and performed three times. KU135 reduces cell proliferation and induces cell death. Data are presented as mean±SD of viable cells and % of dead cells (trypan blue stained). * p<0.05 and **p<0.001
3.2. KU135 induces mitochondrial membrane potential change and cytochrome C release
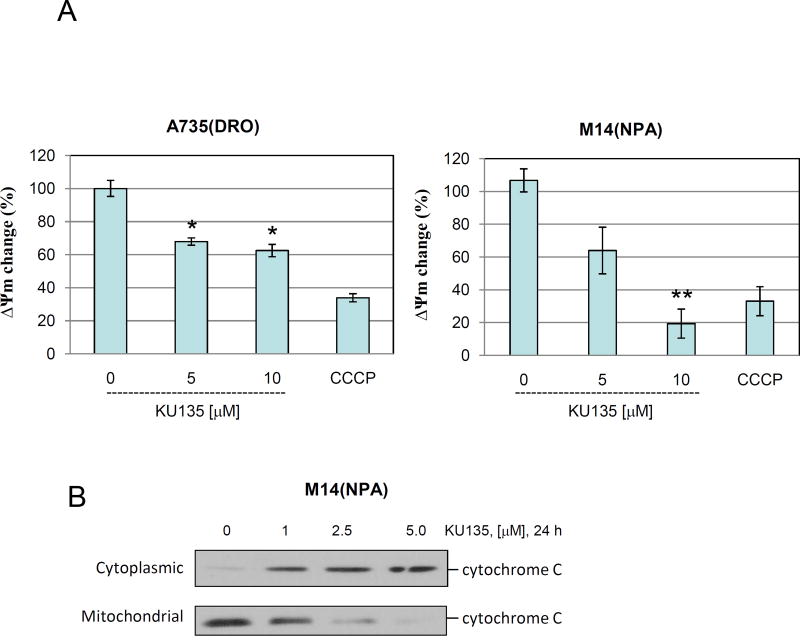
In order to confirm the cell viability and proliferation results, we performed mitochondrial membrane potential and cytochrome C assay. KU135 reduced mitochondrial membrane potential in a concentration-dependent manner (Fig. 2A). These results indicate that M14(NPA) cells were more responsive to mitochondrial membrane changes compared to A735(DRO) cells upon KU135 treatment. Furthermore, treatment of melanoma cells with KU135 resulted in mitochondrial cytochrome C release in a concentration-dependent manner (Fig. 2B). These results showed that mitochondrial cytochrome C release occurred at lower KU135 concentration (1.0 M) than mitochondrial membrane potential change.
Fig. 2.
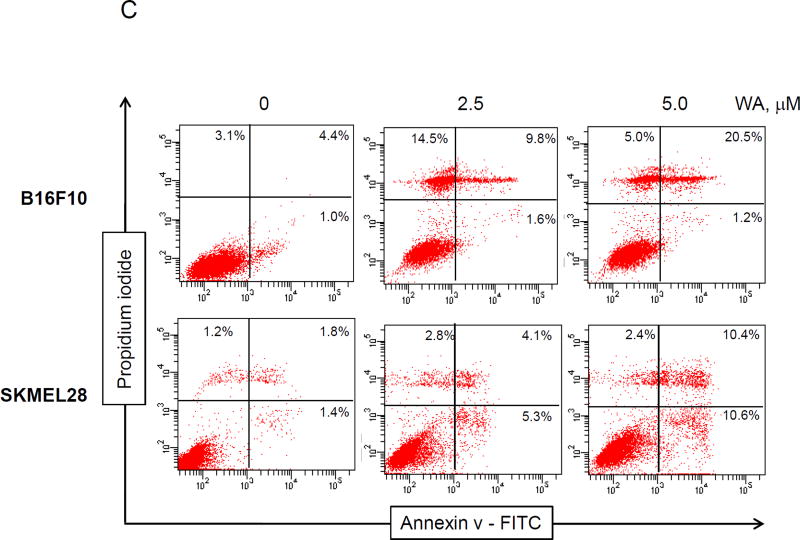
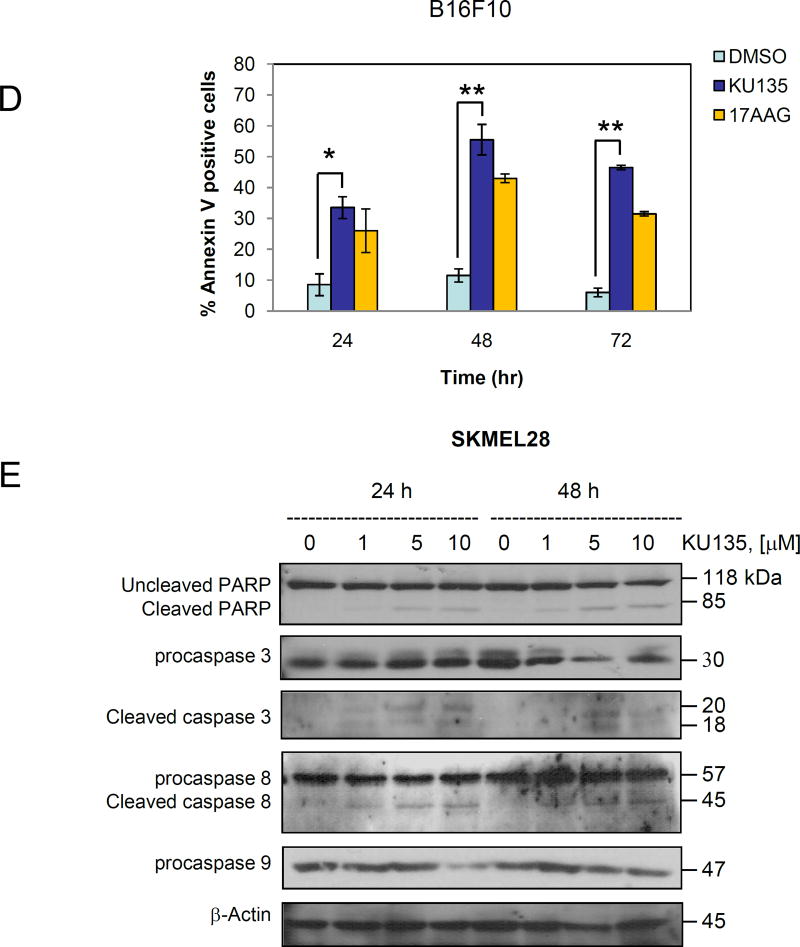
KU135 induces apoptosis in melanoma cells. (A) A735(DRO) and M14(NPA) cells (1×105) were treated with KU135 and then treated with JC-1 according to the manufacturer’s instruction (see Methods and material section). Stained cells were analyzed by flow cytometry. All experiments were carried out in triplicates and presented as mean±SD. KU135 reduced mitochondrial membrane potential in a concentration-dependent manner. (B) M14(NPA) cells were treated with increasing concentration of KU135 for 24 h. Treated cells were removed from plates by trypsinization and cytoplasmic and mitochondrial fractions were separated. Western blot analysis was carried out to examine cytoplasmic and mitochondrial cytochrome c levels. Additionally, KU135 induced cytoplasmic release of cytochrome C. (C) KU135 increased annexin V/PI staining in melanoma cells in a concentration-dependent manner. Melanoma cells (SKMEL28 and B16F10) were incubated overnight and treated with different concentrations of KU135. Cells (1×105) were washed and stained with annexin V/PI. Apoptotic cells were counted by flow cytometry. All experiments were carried out in triplicate. (D) KU135 a stronger inducer of apoptosis measured by annexin V/PI staining compared to the N-terminal Hsp90 inhibitor 17AAG in a time-dependent manner. B16F10 cells were treated 5.0 M KU135 and 5.0 M 17AAG for 24, 48 and 72 h. Each experiment was carried in triplicate. (E) SKMEL28 cell were treated with increasing concentration of KU135 for 24 and 48 h. Cell lysates were analyzed by Western blotting and probed for caspase 3, PARP caspase 8 and caspase 9. -Actin was used as a loading control. KU135 increase activation of caspase 8, 9. Furthermore KU135 induced caspase 3 activation which is signified by reduction of uncleaved caspase 3 and cleavage of PARP in melanoma cells. * p<0.05 and **p<0.001
3.3. KU135 induces melanoma cell death
The effects of KU135 on induction of apoptosis were examined using the annexin V/PI assay and Western blot analysis of caspase activation. These data showed that KU135 (24 h treatment) increased cell death at 2.5 M in B16F10 cells, at which 11.4% of cells were stained with annexin V and 14.5% of cells were stained with PI, characteristic of necrosis only (Fig. 2C). Treatment of B16F10 cells with higher concentrations of KU135 increased the number of cells at late stages of apoptosis leading to annexin V staining of 22.7%. Treatment of SKMEL28 human melanoma cells with KU135 increased apoptosis (Fig. 2C) from basal level of 2.2% for untreated cells to 9.4% for cells treated with 2.5 M and 21% for cells treated with 5.0 M of KU135. The efficacy of KU135-mediated induction of apoptosis was compared to treatment with 17AAG, in a time-dependent manner. KU135 (5.0 M) exhibited a more potent cytotoxic effect when compared to 17AAG (5.0 M) at 24, 48 and 72 h time points (Fig. 2D) against melanoma cells.
We next examined the activation of caspase 3 upon treatment of melanoma cells with KU135. Melanoma cells were treated with increasing concentrations of KU135 and the activation of caspase was assessed by examining increasing levels of cleaved endogenous caspase 3 substrate PARP and reduction in the level of uncleaved caspase 3 (Fig. 2E). KU135-mediated PARP cleavage was detected at 5 and 10 M after 24 h of melanoma treatment while PARP cleavage was detected at 1, 5 and 10 M of KU135 after 48 h treatment. Caspase 3 activation pattern indicated that KU135 may not induce apoptosis at concentrations close to its IC50 when treated for a short period of time, however longer treatment induced apoptosis in melanoma cells. These results show that caspase 3 activation was detectable after 24 h of treatment, whereas long-term treatment with KU135 increased caspase 3 activation. Furthermore, when comparing the time required for mitochondrial membrane potential change and cytochrome C release with that for caspase 3 activation, KU135 induced mitochondrial membrane potential change and cytochrome C release at a faster rate (after 24 h treatment). These results indicate that the C-terminal Hsp90 inhibitor, KU135, reduced cell viability and cell proliferation in melanoma cells, and these events were accompanied with mitochondrial membrane changes, cytochrome C release, and caspase 3 activation. Furthermore, Caspase 3 upstream effectors caspase 8 and caspase 9 were activated in KU135-treated cells. KU135 induced activation of caspase 8 which indicates that KU135 activates extrinsic apoptotic pathway (Fig. 2E). Caspase 8 cleavage was observed at low KU135 (1.0 M) concentration after 24 h and 48 h treatment. Furthermore, KU135 induced cleavage of caspase 9 in a concentration- and time-dependent manner (Fig. 2E). These studies indicated that KU135 induced apoptosis in melanoma cells by inducing mitochondrial membrane potential change, release of mitochondrial cytochrome C, activation of caspase 8, caspase 9 and caspase 3.
3.4. KU135 inhibits Akt and ERK1/2 MAP kinase activation and reduces Hsp90 client proteins
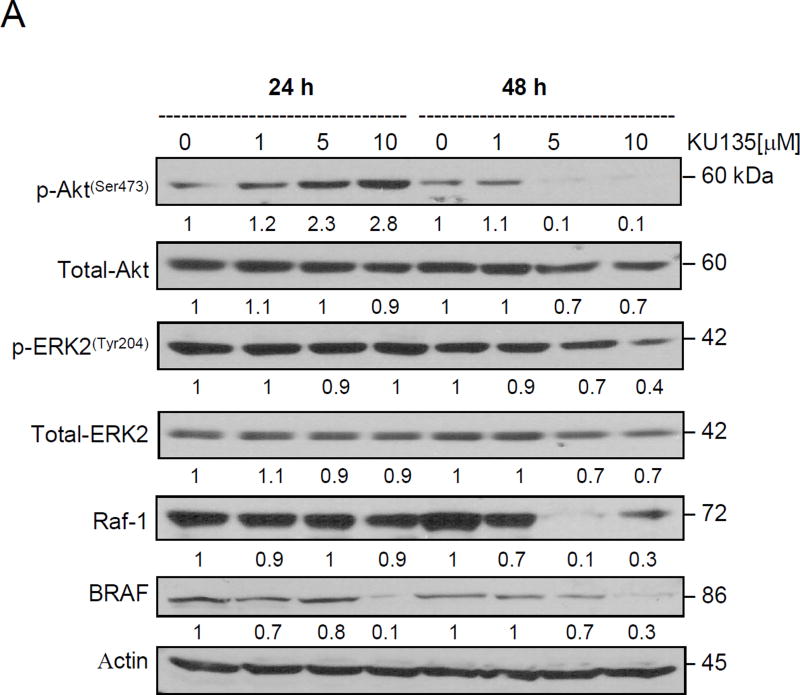
We next examined the effect of KU135 on proliferative pathways PI3Kinase/Akt and ERK1/2 MAP kinase signaling. Melanoma cells carry the BRAF(V600E) mutation, [3,15] resulting in activation of the RAF/MEK/ERK signaling pathway. KU135 reduced Akt phosphorylation (Ser473) and total Akt levels in melanoma cells. Short KU135 treatment increased Akt phosphorylation while long treatment time (48 h) reduced Akt phosphorylation (Fig. 3A). KU135 did not significantly alter phosphorylation of ERK1/2 after 24 h treatment, whereas it reduced ERK activation after 48 h. Total ERK levels were unchanged at either time points. Moreover, treatment of melanoma cells with KU135 reduced BRAF and Raf-1 levels. BRAF and Raf-1 are Hsp90 client proteins. KU135 reduced BRAF and Raf-1 levels in a concentration and time-dependent manner in melanoma cells (Fig. 3A).
Fig. 3.
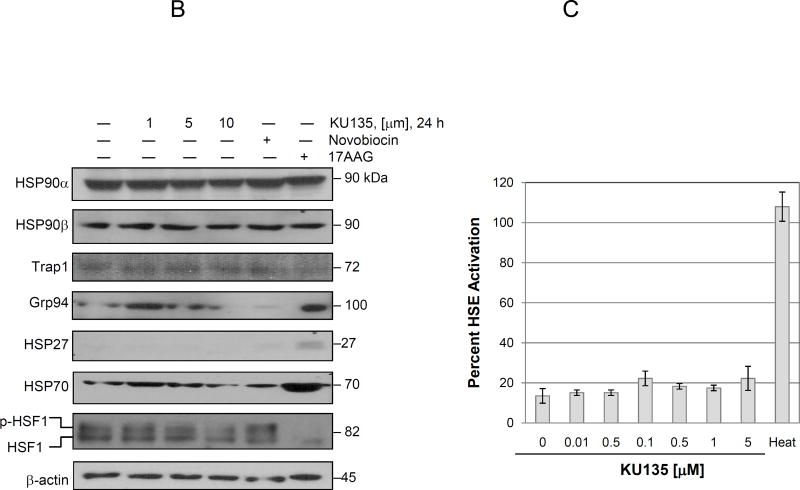
KU135 modulates proliferative signaling and heat shock proteins. (A) SKMEL28 melanoma cells were treated with increasing concentration of KU135 for 24 and 48 h. Cell lysates were prepared and probed for phospho-Akt (Ser473), total Akt, phospho-ERK2(Tyr 204), total ERK2, RAF-1 and BRAF. (B) KU135-mediated modulation of heat shock proteins were examined by Western blotting. (C) KU135-mediated HSE promoter activity was analysed using Hela cells stably expressing hsp70.1pr-luciferase plasmid. Hela cells were plated at 1.0×104 cells/well in 96-well plates overnight before treatment with KU135. Cell were treated for 24 h with increased concentration of KU135 or heat-treated at 43°C for 1 h and luciferase activity was measured 24 h after heat treatment. Luciferase activity was measured by Bright-Glo reagent. KU135-mediated luciferase activity was compared to heat- induced luciferase activity (100%). All experiments were carried in triplicate.
3.5. KU135 reduces levels of heat shock proteins
We next examined KU135-mediated changes in the levels of heat shock proteins. KU135 did not alter Hsp90 and Hsp90 levels in melanoma cells (Fig. 3B). Furthermore, levels of TRAP1, the mitochondrial Prolag of Hsp90 did not significantly change upon KU135 treatment. However, KU135 reduced the endoplasmic reticulum Hsp90 Prolag, Grp94, levels in melanoma cells, which is the opposite of the effect of treatment with 17AAG. Hsp27 levels remained unchanged upon KU135 treatment, which is in contrast to treatment with 17AAG which significantly induced Hsp27 expression in melanoma cells (Fig. 3B). Hsp90 inhibition is often compensated for by increased expression of Hsp90 and Hsp70 as well as other heat shock proteins. KU135 slightly increased Hsp70 expression at 1 M whereas, at higher concentrations, Hsp70 expression was reduced (Fig. 3B). In contrast, 17AAG treatment of melanoma cells substantially induced Hsp70 expression in melanoma cells. Expression of heat shock proteins is primarily controlled by transcription factor heat shock factor 1 (HSF1). Upon stress signaling, HSF1 is released from the Hsp90-Hsp70 multiprotein complex in the cytoplasm, whereupon it then trimmerizes, translocates into the nucleus. It is phosphorylated and binds to cognate heat shock response element (HSRE) on the promoter region of HSF1 regulated proteins. Similar to N-terminal Hsp90 inhibitors, KU135 decreased the level of HSF1 phosphorylation in a concentration-dependent manner, whereas, less potent C-terminal Hsp90 inhibitor, novobiocin, did not significantly change HSF1 phosphorylation (Fig. 3B).
To confirm that KU135 did not significantly induced Hsp70 levels in treated cells, HeLa cells stably expressing hsp70.1pr-luciferase plasmid were employed. This experiment is an accurate representation of HSF1 transcriptional activity in KU135-treated cells since it measures binding of HSF1 and induction of RNA polymerase activity in the cell. As a positive control, cells were heat shocked for 1 hr to induce HSF1 activation. Results showed that KU135 did not induced luciferase activity and, therefore did not affect induction of Hsp70 expression (Fig. 3C). Overall these results show that KU135 decreased Hsp70, Grp94 and HSF1 protein levels, while expression of other heat shock proteins did not significantly change. Furthermore, KU135 did not affect Hsp70 promoter activation.
3.6. KU135 induces G2/M cell cycle arrest in melanoma cells
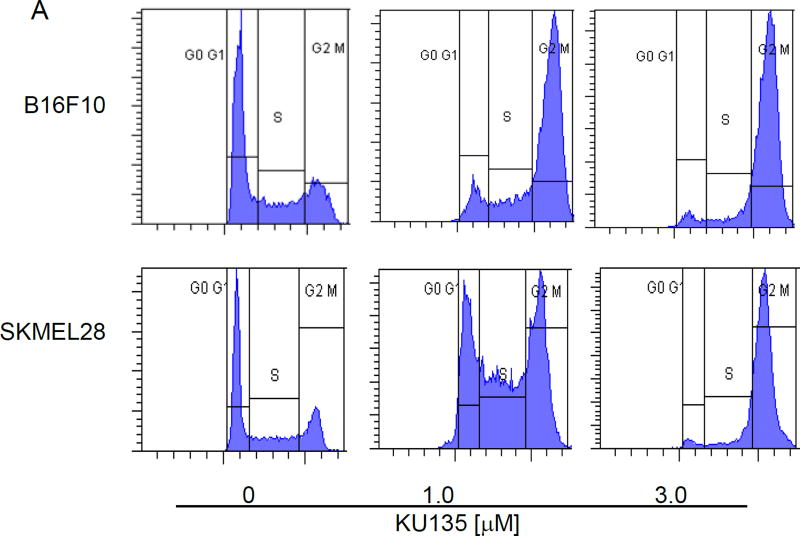
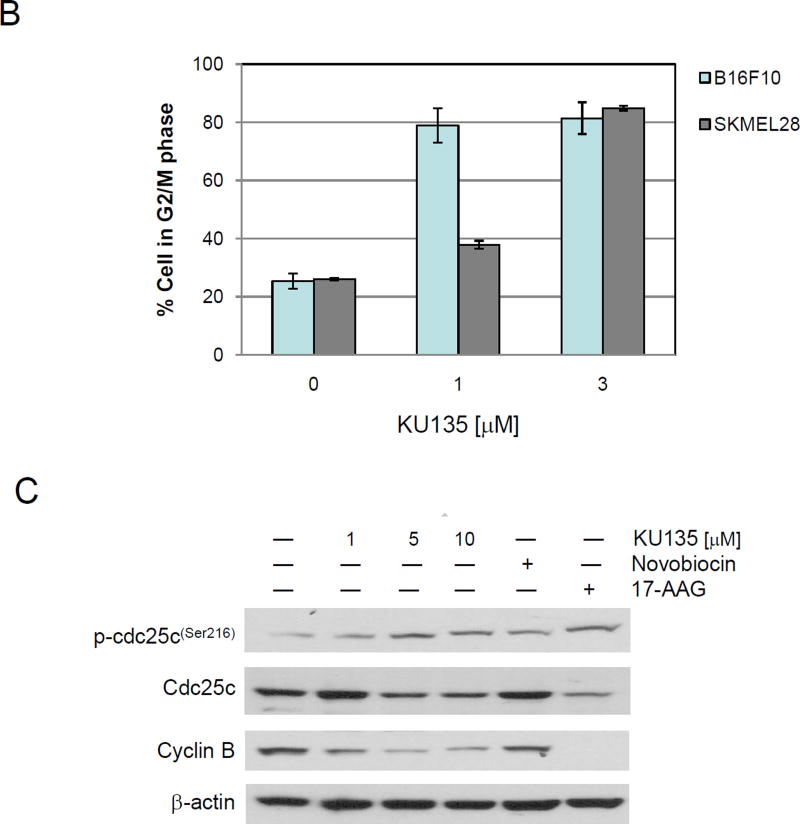
Melanoma cells were treated with increasing concentration of KU135 for 24 h and the distribution of cell cycle was detected by flow cytometry. KU135 shifted G2/M cell cycle from 25.4% and 26.1% in untreated cells to 78.9% and 37.8% at 1.0 M and 81.4% and 84.9% at 3.0 M of treatment with KU135 for B16F10 and SKMEL28 cells, respectively (Fig. 4A). Figure 4B illustrates the strong shift from G0/G1 to G2/M cell cycle shift upon KU135 treatment. These data show that KU135 induced G2/M cell cycle arrest at concentrations below the IC50 for KU135 (Fig. 4B). To further investigate molecular mechanism involved in KU135-mediated cell cycle regulation, melanoma cells were treated with increasing concentrations of KU135 as well as novobiocin and 17AAG. We first examined the activation levels of cdc25C phosphatase in KU135-treated cells. KU135 treatment increased phosphorylation of cdc25c phosphatase, decreased cellular cdc25c levels and reduced cyclin B levels in a concentration dependent manner (Fig. 4C). These results indicate that KU135 promoted strong G2/M cell cycle arrest in melanoma cells by increased phosphorylation of cdc25c, reduction of cdc25c and cyclin B levels.
Fig. 4.
KU135 strongly induces G2/M cell cycle arrest in melanoma cells. (A) Cells were treated with increasing concentration of KU135 and stained with propidium iodide. (B) The level of G2/M cell cycle arrest was shown when SKMEL28 and B16F10 cells were treated with 1.0 and 3.0 M KU135. (C) KU135-mediated modulation of G2/M proteins phospho-cdc25c, cdc25c and cyclin B were probed using Western blotting.
4. Discussion
Small molecule kinase-targeting drugs have emerged as a promising class of cancer therapeutics. However, cancer cells invariably display drug resistance to these single-targeted therapies and therefore it is important to identify novel targets. Targets that regulate multiple proteins and signal transduction pathways are especially promising in the pursuit of anti-cancer agents. Hsp90 is a molecular chaperone that promotes proper folding and stability of several classes of proteins. Hsp90 expression is increased in several cancer types including melanoma [6,16]. N-terminal Hsp90 inhibitors have been used to target melanoma cells [17,18]. However, clinical trials with N-terminal Hsp90 inhibitors have been associated with severe side effects highlighted the need for novel Hsp90 inhibitors cancer therapy [5,12]. The present study investigated the effect of a novel novobiocin-derived C-terminal Hsp90 inhibitor in melanoma cells. KU135 offers a distinct mechanism of Hsp90 inhibition [14]. KU135 binds to Hsp90 C-terminus and inhibits Hsp90 chaperone activity. KU135 reduced cell viability and proliferation in melanoma cells, and reduced mitochondrial membrane potential as well as release of mitochondrial cytochrome C. KU135 induced apoptosis through caspase 8, 9 and 3 activation. Additionally, KU135 exerts its anti-proliferative activity by reducing Hsp70 expression as well as reduction of total and phosphorylated HSF1. KU135 failed to activate Hsp70 promoter activity. Furthermore, heat shock proteins Hsp90 Hsp90 and TRAP1 expression were unaffected upon KU135 treatment.
Additionally, KU135 shifted melanoma cells into the G2/M phase. Several proteins are involved in regulation of the cell cycle. Recent investigation has highlighted an important role for cdc25 phosphatase in G2/M cell progression [19,20]. Cdc25c phosphatase is required for the assembly of Ckd1/cyclin B and the transition out of the G2/M phase. In eukaryotic cells, progression through different stage of cell cycle is regulated by cyclin-dependent kinases (Cdks). Activation of cdk1 is required for the progression of cells through mitosis [21]. Cdk1 is activated by cdc25c phosphatase activity. Phosphorylation of cdc25c (Ser216) during the interphase and G2/M cell cycle arrest increases cytoplasmic localization and inactivation of cdc25c phosphatase activity. We have shown that KU135 increased Ser216 cdc25c phosphorylation in a concentration-dependent manner, while reducing total cdc25c and cyclin B protein levels. Previous reports showed that Hsp90 is involved in proper localization of cyclin B during mitosis [22]. However, the mechanism of KU135-mediated phosphorylation and cdc25c inactivation is not known at this point and requires further investigation. Additionally a recent report indicates that N-terminal Hsp90 inhibitors reduce cdc25 levels in cancer cells [23,24], therefore KU135 may destabilized cdc25c through Hsp90 inhibition.
KU135, a novel C-terminal Hsp90 inhibitor [14] exhibited potent anticancer activity against melanoma cells. These results indicate that KU135 is a chemotherapeutic alternative to N-terminal Hsp90 inhibitors. These data indicated that KU135 modulates cell proliferation and cell cycle progression. Ongoing studies seek to characterize KU135-induced cell cycle progression and the mechanism by which it activates caspase 3.
Highlights.
-
>
Novel c-terminal Hsp90 inhibitor, KU135 is an effective drug against melanoma.
-
>
It inhibited proliferation and induced G2/M cell cycle arrest, and apoptosis.
-
>
KU135 reduced Hsp90 client proteins Akt, BRAF, RAF-1, cyclin B and cdc25 proteins.
Acknowledgments
Authors gratefully acknowledge support of this project by the NIH/NCI (CA120458) to BSB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
None declared
Role of funding sources
None declared
References
- 1.Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- 2.Ibrahim N, Haluska FG. Molecular pathogenesis of cutaneous melanocytic neoplasms. Annu Rev Pathol. 2009;4:551–579. doi: 10.1146/annurev.pathol.3.121806.151541. [DOI] [PubMed] [Google Scholar]
- 3.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 4.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer CJ, Pritchard C, Marais R. Kinase-Dead BRAF and Oncogenic RAS Cooperate to Drive Tumor Progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCarthy MM, Pick E, Kluger Y, Gould-Rothberg B, Lazova R, Camp RL, Rimm DL, Kluger HM. HSP90 as a marker of progression in melanoma. Ann Oncol. 2008;19:590–594. doi: 10.1093/annonc/mdm545. [DOI] [PubMed] [Google Scholar]
- 7.Smith V, Sausville EA, Camalier RF, Fiebig HH, Burger AM. Comparison of 17-dimethyl-aminoethylamino-17-demethoxy-geldanamycin (17DMAG) and 17-allylamino-17-demethoxygeldanamycin (17AAG) in vitro: effects on Hsp90 and client proteins in melanoma models. Cancer Chemother Pharmacol. 2005;56:126–137. doi: 10.1007/s00280-004-0947-2. [DOI] [PubMed] [Google Scholar]
- 8.Faingold D, Marshall JC, Antecka E, Di Cesare S, Odashiro AN, Bakalian S, Fernandes BF, Burnier MN., Jr Immune expression and inhibition of heat shock protein 90 in uveal melanoma. Clin Cancer Res. 2008;14:847–855. doi: 10.1158/1078-0432.CCR-07-0926. [DOI] [PubMed] [Google Scholar]
- 9.Solit DB, Osman I, Polsky D, Panageas KS, Daud A, Goydos JS, Teitcher J, Wolchok JD, Germino FJ, Krown SE, Coit D, Rosen N, Chapman PB. Phase II trial of 17-allylamino-17-demethoxygeldanamycin in patients with metastatic melanoma. Clin Cancer Res. 2008;14:8302–8307. doi: 10.1158/1078-0432.CCR-08-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pacey S, Gore M, Chao D, Banerji U, Larkin J, Sarker S, Owen K, Asad Y, Raynaud F, Walton M, Judson I, Workman P, Eisen T. A Phase II trial of 17-allylamino, 17-demethoxygeldanamycin (17-AAG, tanespimycin) in patients with metastatic melanoma. Invest New Drugs. 2010 doi: 10.1007/s10637-010-9493-4. [DOI] [PubMed] [Google Scholar]
- 11.Banerji U, Affolter A, Judson I, Marais R, Workman P. BRAF and NRAS mutations in melanoma: potential relationships to clinical response to HSP90 inhibitors. Mol Cancer Ther. 2008;7:737–739. doi: 10.1158/1535-7163.MCT-08-0145. [DOI] [PubMed] [Google Scholar]
- 12.Kimura H, Yukitake H, Tajima Y, Suzuki H, Chikatsu T, Morimoto S, Funabashi Y, Omae H, Ito T, Yoneda Y, Takizawa M. ITZ-1, a client-selective Hsp90 inhibitor, efficiently induces heat shock factor 1 activation. Chem Biol. 2010;17:18–27. doi: 10.1016/j.chembiol.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 13.Zhang T, Li Y, Yu Y, Zou P, Jiang Y, Sun D. Characterization of celastrol to inhibit hsp90 and cdc37 interaction. J Biol Chem. 2009;284:35381–35389. doi: 10.1074/jbc.M109.051532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shelton SN, Shawgo ME, Matthews SB, Lu Y, Donnelly AC, Szabla K, Tanol M, Vielhauer GA, Rajewski RA, Matts RL, Blagg BS, Robertson JD. KU135, a novel novobiocin-derived C-terminal inhibitor of the 90-kDa heat shock protein, exerts potent antiproliferative effects in human leukemic cells. Mol Pharmacol. 2009;76:1314–1322. doi: 10.1124/mol.109.058545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schweppe RE, Klopper JP, Korch C, Pugazhenthi U, Benezra M, Knauf JA, Fagin JA, Marlow LA, Copland JA, Smallridge RC, Haugen BR. Deoxyribonucleic acid profiling analysis of 40 human thyroid cancer cell lines reveals cross-contamination resulting in cell line redundancy and misidentification. J Clin Endocrinol Metab. 2008;93:4331–4341. doi: 10.1210/jc.2008-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lebret T, Watson RW, Molinie V, O'Neill A, Gabriel C, Fitzpatrick JM, Botto H. Heat shock proteins HSP27, HSP60, HSP70, and HSP90: expression in bladder carcinoma. Cancer. 2003;98:970–977. doi: 10.1002/cncr.11594. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Neely L, Lundgren K, Yang YC, Lough R, Timple N, Burrows F. BIIB021, a synthetic Hsp90 inhibitor, has broad application against tumors with acquired multidrug resistance. Int J Cancer. 2010;126:1226–1234. doi: 10.1002/ijc.24825. [DOI] [PubMed] [Google Scholar]
- 18.Sauvageot CM, Weatherbee JL, Kesari S, Winters SE, Barnes J, Dellagatta J, Ramakrishna NR, Stiles CD, Kung AL, Kieran MW, Wen PY. Efficacy of the HSP90 inhibitor 17-AAG in human glioma cell lines and tumorigenic glioma stem cells. Neuro Oncol. 2009;11:109–121. doi: 10.1215/15228517-2008-060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galaktionov K, Lee AK, Eckstein J, Draetta G, Meckler J, Loda M, Beach D. CDC25 phosphatases as potential human oncogenes. Science. 1995;269:1575–1577. doi: 10.1126/science.7667636. [DOI] [PubMed] [Google Scholar]
- 20.Forester CM, Maddox J, Louis JV, Goris J, Virshup DM. Control of mitotic exit by PP2A regulation of Cdc25C and Cdk1. Proc Natl Acad Sci U S A. 2007;104:19867–19872. doi: 10.1073/pnas.0709879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Timofeev O, Cizmecioglu O, Settele F, Kempf T, Hoffmann I. Cdc25 phosphatases are required for timely assembly of CDK1-cyclin B at the G2/M transition. J Biol Chem. 2010;285:16978–16990. doi: 10.1074/jbc.M109.096552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basto R, Gergely F, Draviam VM, Ohkura H, Liley K, Raff JW. Hsp90 is required to localise cyclin B and Msps/ch-TOG to the mitotic spindle in Drosophila and humans. J Cell Sci. 2007;120:1278–1287. doi: 10.1242/jcs.000604. [DOI] [PubMed] [Google Scholar]
- 23.Aressy B, Ducommun B. Cell cycle control by the CDC25 phosphatases. Anticancer Agents Med Chem. 2008;8:818–824. doi: 10.2174/187152008786847756. [DOI] [PubMed] [Google Scholar]
- 24.Senju M, Sueoka N, Sato A, Iwanaga K, Sakao Y, Tomimitsu S, Tominaga M, Irie K, Hayashi S, Sueoka E. Hsp90 inhibitors cause G2/M arrest associated with the reduction of Cdc25C and Cdc2 in lung cancer cell lines. J Cancer Res Clin Oncol. 2006;132:150–158. doi: 10.1007/s00432-005-0047-7. [DOI] [PubMed] [Google Scholar]