Abstract
Purpose
The activity of epithelial lactase (LCT) is associated with a polymorphism 13910 bp upstream in the lactase encoding gene. Because the association between the LCT-13910 polymorphism and the risk for colorectal cancer is not clear, we investigated the role of the LCT-13910 polymorphism as a potential risk factor for colorectal cancer and colorectal polyps in the Turkish population.
Methods
One hundred sixty-six subjects (74 with polyps, 44 with colorectal cancer, 48 controls), who had undergone a total colonoscopy between January 2012 and November 2012 in our endoscopy unit were genotyped for the LCT-13910 polymorphism by using the polymerase chain reaction and minisequencing.
Results
The CC genotype in the lactose gene 13910 locus, which is accepted as the genetic indicator of lactase deficiency, was determined as 83.7%. The CC genotype rate was determined as 89.1% in patients who had a history of lactose intolerance and 81.5% in those without a history of lactose intolerance (P = 0.236). No difference was detected between the patients who had colorectal polyp(s) and/or cancer and the controls with regard to the LCT-13910 polymorphism. No differences were determined between groups when they were compared with regard to the C or the T allele.
Conclusion
No differences were detected between the patients who had colorectal polyp(s) and/or cancer and those with normal colonoscopy findings with regard to lactase gene polymorphisms. No differences were determined between the groups when they were compared with regard to the C or the T allele.
Keywords: Adenocarcinoma, Colorectal neoplasms, LCT gene, Lactose intolerance
INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer in the world. The effect of milk and dairy products on CRC is controversial [1]. Several cohort studies have shown that consumption of milk reduces the risk of CRC. This positive effect is known to be due to the components of milk, including calcium, vitamin D, lactoferrin, and conjugated linoleic acid. In the epithelial cells of the intestine, calcium and vitamin D regulate cell growth and promote cell differentiation by stimulating calcium-sensing receptors. Also, in the intestinal lumen, calcium may bind complex fatty acids and secondary bile acids, thereby reducing their cytotoxic effects [2,3,4].
Lactose is the main sugar of milk, and lactose intolerance (LI) is common. Symptoms of LI include diarrhea, abdominal pain, and flatulence after drinking or eating milk or products that contains milk. These symptoms are caused by low levels of small intestinal lactase due to mucosal injury or, more often, due to reduced genetic expression of the enzyme lactase-phlorizin hydrolase. Lactase deficiency is defined as an intestinal brush border lactase enzyme activity that is lower than that of normal infants. Lactose malabsorption means the failure of the small bowel to absorb a sizable fraction of ingested lactose. LI is a clinical syndrome that includes many symptoms (e.g., abdominal pain, bloating, flatulence, and diarrhea) due to lactose malabsorption [5]. Lactose malabsorbers may consume small amounts of milk and dairy products, which may result in low calcium intake, a risk factor for the development of CRC.
The lactase gene codes an enzyme called lactase-phlorizin hydrolase that helps the digestion of milk. The level of lactase enzyme remains high until weaning, after which it falls to lower levels in the adult population where it is called adult type hypolactasia. In some populations like northern Europe, high levels of lactase enzyme are maintained in adulthood, which is called lactase persistence (LP) [6]. Multiple single nucleotide polymorphisms (SNPs) in the DNA sequence of the coding region and the regulatory region of the lactase gene on 2q21 have been identified. The SNP LCT-13910C > T (rs4988235), located in the minichromosome maintenance-6 gene upstream from the LCT-encoding gene, was found to be associated with adult-type hypolactasia. Genetic analysis has established that LP in adulthood is inherited as an autosomal dominant trait. The presence of the T allele of the SNP located at -13.9-kb upstream of the lactase gene has been strongly correlated with LP. The CC genotype defines lactose malabsorbers; CT and TT genotypes define lactose absorbers [7,8,9,10,11,12]. Recent studies showed a significant association between the CC genotype and an elevated CRC risk among members of the Finnish population, but it was not confirmed in British, Spanish, Hungarian, or Italian patients [13,14,15]. For those reasons, in this study, we aimed to investigate the role of LCT-13910 polymorphisms as a potential risk factor for CRC and colorectal polyps in the Turkish population.
METHODS
We performed a total colonoscopy on all patients between January 2012 and November 2012 in our endoscopy unit and screened all of them for LCT-13910 polymorphisms. Inclusion criteria were Turkish ethnicity, age >25 years, and informed consent to be interviewed. A partial colonoscopy, inflammatory bowel disease, familial polyposis, non–CRC-related previous bowel surgery, history of a neoplastic disease other than CRC or adenoma served as exclusion criteria. Seventy-seven of 92 patients with adenomatous polyps and 45 of 52 patients with colon cancer gave informed consent. Fifty patients with normal colonoscopy whose ages were similar to the ages of the colon carcinoma patients were selected as controls. All patients filled out a questionnaire about their symptoms of LI, such as abdominal pain, bloating, flatulence, and diarrhea. Following colonoscopy, anticoagulated blood was drawn from the patients into ethylenediaminetetraacetic acid tubes for genetic analyses. Genetic analysis failed in 3 patients with adenomatous polyps, 1 with a colon carcinoma, and in 2 controls; then, 74, 44, and 48 patients were analyzed in the polyp, cancer, and control groups, respectively. Approval for this research was obtained from the local Ethics Committee.
DNA was extracted from mononuclear cells of the peripheral blood. We used Invitrogen Blood DNA Isolation Kits (Lot#: 1388070, Purelink Genomic DNA Kit, Life Technologies, Camarillo, CA, USA) for extraction of DNA. Under the conditions recommended by the manufacturer (95℃ for 10 minutes to activate the AmpliTaq Gold enzyme, followed by 40 cycles of 15 seconds at 95℃ and 60 seconds at 60℃), the 25-µL polymerase chain reaction, with 2x TaqMan Genotyping Master Mix (Applied Biosystems, Waltham, MA, USA), 20x probe and primers assay mix, and 5 ng of genomic DNA, was performed in 96-well plates in a real time-PCR System (7900 HT, Applied Biosystems). Both primers and probes were obtained from Applied Biosystems.
The data were collected and analyzed using the SPSS ver. 15.0 (SPSS Inc., Chicago, IL, USA). The associations of age, sex, job, smoking, alcohol use, history of LI, presence of diabetes, marital status, the LCT-13910 genotype (CC, CT, TT), and allele frequency with colon cancer, colon polyps or normal colonoscopy were investigated. The categorical variables of colon cancer were compared using the chi-square test, and the continuous variables were analyzed using the analysis of variance test or the t-test. The patients with colon cancer and colon polyp were gathered in a group as colon neoplasms, and the associations were investigated.
RESULTS
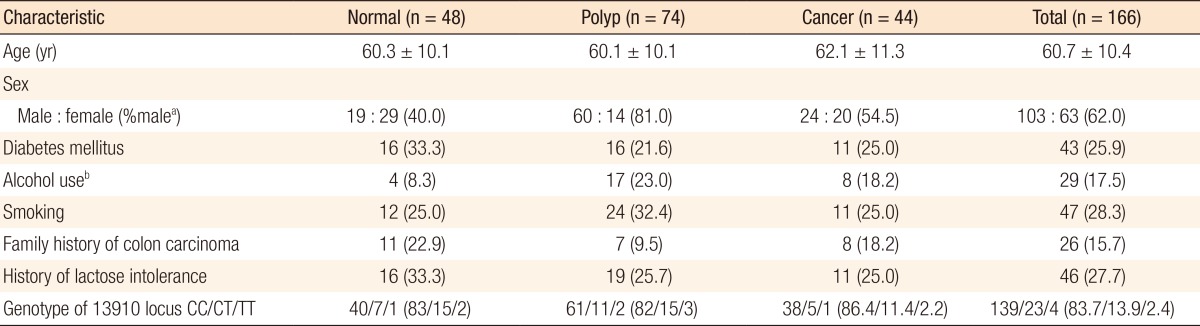
One hundred sixty-six patients were included in the study (male, 62%; mean age, 60.7 ± 10.4 years). No differences with regard to age, presence of diabetes, alcohol use, smoking, and history of LI were found between the patients who had an adenomatous polyp or cancer and those whose colon was normal (Table 1). The percent of the male sex was lower in the controls. When patients with polyps and with cancer were approached as a single group and compared with the controls, the percents of the male sex (71.2%, P < 0.0001) and of alcohol use (21.2%, P = 0.048) were found to be higher in the neoplasia group.
Table 1. Characteristics of the patients and results of the genetic analyses.
aHigher proportion of males in the polyp group (P < 0.05). b>40 g/day alcohol use.
Although a history consistent with LI was seen at a rate of 27.7%, the CC genotype in the lactose gene 13910 locus, which was accepted as the genetic indicator of lactase deficiency, was determined as 83.7%. The CC genotype rates were determined as 89.1% in patients who had a history of LI and 81.5% in those without a history of LI (P = 0.236). No differences with regard to lactase gene polymorphisms were detected between the patients who had a colorectal polyp or cancer and those with normal colonoscopy findings. No differences were found between the groups when they were compared with regard to the C or the T allele.
DISCUSSION
The main mechanism of lactose malabsorption in healthy individuals is a genetically regulated reduction in the lactase enzyme activity, which is caused by racial or ethnic factors. Most of the world's populations have low intestinal lactase levels during midchildhood. This characteristic is most common in the Asian and the African populations; in contrast, most Caucasians, particularly of northern European background, have elevated lactase activity into adulthood. LP has also been observed in populations that historically domesticated cows or other milk providers and consumed products of milk into adulthood. Evidence exists for a convergent evolution of LP in some of these groups. LP in adulthood is inherited as an autosomal dominant trait, but the molecular basis is not sufficiently clear [16,17,18].
Genetic polymorphisms were found to be related with lactase deficiency and are being widely used to determine that condition. In this study, a CT polymorphism located 13.9-kb upstream of exon 1 of the lactase gene, which is the most commonly polymorphism used worldwide and shows about 90% concordance with lactase deficiency in the white population, was used. This polymorphism is closely related with LP in the Finnish population, where it was first determined, and in the German, English, Italian, and even Indian populations [19,20,21]. However, no relationship could be found between lactase deficiency and the 13910 CT polymorphism in non-White populations such as Africans, the Chinese, and the Japanese [21,22,23,24,25]. The relationship between lactase deficiency and the 13910 CT polymorphism in the Turkish population is not known. In 2 studies in the Turkish population, the rates of lactase deficiency diagnosed with the H2 breath test, the lactose tolerance test, and the urine galactose/creatinine ratio were found to fall in the range from 71% to 85% [26,27]. LP seems to be 6% according to the Global Lactase persistence Association Database. Namely, lactase deficiency is seen in 94% of the adult population [28].
We investigated if differences with regard to lactose gene polymorphism could be found between the 44 patients with CRC, the 74 patients with an adenomatous polyp of the colon, and the 48 patients with normal colonoscopy findings. The 13910 CC genotype, which was found to be related with lactose nonpersistence, was identified in more than 80% of the patients in each subgroup, and no differences were found between the groups. No differences were found between the groups when they were compared with regard to the C or the T allele. The CC genotype in the lactose gene 13910 locus, which was accepted as the genetic indicator of lactase deficiency, was determined as 83.7%. The CC genotype rates were determined as 89.1% in patients who had a history of LI and 81.5% in those without a history of LI (P = 0.236).
A literature search about ‘LCT-13910 polymorphisms and colorectal cancer,’ revealed a few studies. Firstly, Rasinperä et al. [13] found that the CC genotype occurred more frequently in the patient group in the Finnish population (CRC patients, 23.5%; controls, 18%), but in the same study, these findings were not supported in the British (CRC patients, 9.2%; controls, 8.8%) and the Spanish (CRC patients, 31.9%; controls, 36.7%) populations. Among Hungarians, the prevalence of the CC genotype was studied by Bácsi et al. [14]; they found the frequency of the CC genotype to be higher in the Hungarian population (patients, 41%; controls, 36%) than in the Finnish population, but they found no statistically significant association between CRC and the CC genotype. In the Italian population, Tarabra et al. [15] reported that the LCT-13910C > T polymorphism was not associated with an increased risk for CRC or polyps (CC genotype: CRC patients, 62.09%; patients with polyp, 64.77%; controls, 63.02%). Our results were compatible with those found in studies done on the Italian and the Hungarian populations (CC genotype: CRC patients, 86.4%; patients with polyp, 82%; controls, 83%).
Our study does have some limitations. Firstly, the small number of the subjects in this study may have hindered the demonstration of the relationship between the LCT-13910 polymorphism and an increased risk for colorectal polyps and cancer. We did not use a method such as lactase expression in biopsy to show directly lactase deficiency; this could have masked a real relationship. According to the results of this study, the LCT-13910 polymorphisms (CC/CT/TT) are not associated with an increased risk for development of CRC in the study population. Further studies including larger populations are needed to confirm our findings.
Footnotes
CONFLICT OF INTEREST: No potential conflict of interest relevant to this article was reported.
References
- 1.Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132:1133–1145. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- 2.Holt PR. New insights into calcium, dairy and colon cancer. World J Gastroenterol. 2008;14:4429–4433. doi: 10.3748/wjg.14.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van der Meer R, Lapré JA, Govers MJ, Kleibeuker JH. Mechanisms of the intestinal effects of dietary fats and milk products on colon carcinogenesis. Cancer Lett. 1997;114:75–83. doi: 10.1016/s0304-3835(97)04629-6. [DOI] [PubMed] [Google Scholar]
- 4.Govers MJ, Termont DS, Lapré JA, Kleibeuker JH, Vonk RJ, Van der Meer R. Calcium in milk products precipitates intestinal fatty acids and secondary bile acids and thus inhibits colonic cytotoxicity in humans. Cancer Res. 1996;56:3270–3275. [PubMed] [Google Scholar]
- 5.Suchy FJ, Brannon PM, Carpenter TO, Fernandez JR, Gilsanz V, Gould JB, et al. National Institutes of Health Consensus Development Conference: lactose intolerance and health. Ann Intern Med. 2010;152:792–796. doi: 10.7326/0003-4819-152-12-201006150-00248. [DOI] [PubMed] [Google Scholar]
- 6.Itan Y, Jones BL, Ingram CJ, Swallow DM, Thomas MG. A worldwide correlation of lactase persistence phenotype and genotypes. BMC Evol Biol. 2010;10:36. doi: 10.1186/1471-2148-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harvey CB, Hollox EJ, Poulter M, Wang Y, Rossi M, Auricchio S, et al. Lactase haplotype frequencies in Caucasians: association with the lactase persistence/non-persistence polymorphism. Ann Hum Genet. 1998;62(Pt 3):215–223. doi: 10.1046/j.1469-1809.1998.6230215.x. [DOI] [PubMed] [Google Scholar]
- 8.Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Järvelä I. Identification of a variant associated with adult-type hypolactasia. Nat Genet. 2002;30:233–237. doi: 10.1038/ng826. [DOI] [PubMed] [Google Scholar]
- 9.Kuokkanen M, Enattah NS, Oksanen A, Savilahti E, Orpana A, Järvelä I. Transcriptional regulation of the lactase-phlorizin hydrolase gene by polymorphisms associated with adult-type hypolactasia. Gut. 2003;52:647–652. doi: 10.1136/gut.52.5.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olds LC, Sibley E. Lactase persistence DNA variant enhances lactase promoter activity in vitro: functional role as a cis regulatory element. Hum Mol Genet. 2003;12:2333–2340. doi: 10.1093/hmg/ddg244. [DOI] [PubMed] [Google Scholar]
- 11.Troelsen JT, Olsen J, Moller J, Sjostrom H. An upstream polymorphism associated with lactase persistence has increased enhancer activity. Gastroenterology. 2003;125:1686–1694. doi: 10.1053/j.gastro.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 12.Lewinsky RH, Jensen TG, Møller J, Stensballe A, Olsen J, Troelsen JT. T-13910 DNA variant associated with lactase persistence interacts with Oct-1 and stimulates lactase promoter activity in vitro. Hum Mol Genet. 2005;14:3945–3953. doi: 10.1093/hmg/ddi418. [DOI] [PubMed] [Google Scholar]
- 13.Rasinpera H, Forsblom C, Enattah NS, Halonen P, Salo K, Victorzon M, et al. The C/C-13910 genotype of adult-type hypolactasia is associated with an increased risk of colorectal cancer in the Finnish population. Gut. 2005;54:643–647. doi: 10.1136/gut.2004.055939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bacsi K, Hitre E, Kosa JP, Horvath H, Lazary A, Lakatos PL, et al. Effects of the lactase 13910 C/T and calcium-sensor receptor A986S G/T gene polymorphisms on the incidence and recurrence of colorectal cancer in Hungarian population. BMC Cancer. 2008;8:317. doi: 10.1186/1471-2407-8-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarabra E, Pazienza P, Borghesio E, Actis GC, Tappero G, Framarin L, et al. LCT-13910C>T polymorphism-associated lactose malabsorption and risk for colorectal cancer in Italy. Dig Liver Dis. 2010;42:741–743. doi: 10.1016/j.dld.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 16.Tishkoff SA, Reed FA, Ranciaro A, Voight BF, Babbitt CC, Silverman JS, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scrimshaw NS, Murray EB. The acceptability of milk and milk products in populations with a high prevalence of lactose intolerance. Am J Clin Nutr. 1988;48(4 Suppl):1079–1159. doi: 10.1093/ajcn/48.4.1142. [DOI] [PubMed] [Google Scholar]
- 18.Sahi T, Isokoski M, Jussila J, Launiala K, Pyorala K. Recessive inheritance of adult-type lactose malabsorption. Lancet. 1973;2:823–826. doi: 10.1016/s0140-6736(73)90862-3. [DOI] [PubMed] [Google Scholar]
- 19.Szilagyi A, Malolepszy P, Hamard E, Xue X, Hilzenrat N, Ponniah M, et al. Comparison of a real-time polymerase chain reaction assay for lactase genetic polymorphism with standard indirect tests for lactose maldigestion. Clin Gastroenterol Hepatol. 2007;5:192–196. doi: 10.1016/j.cgh.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 20.Schirru E, Corona V, Usai-Satta P, Scarpa M, Oppia F, Loriga F, et al. Genetic testing improves the diagnosis of adult type hypolactasia in the Mediterranean population of Sardinia. Eur J Clin Nutr. 2007;61:1220–1225. doi: 10.1038/sj.ejcn.1602638. [DOI] [PubMed] [Google Scholar]
- 21.Babu J, Kumar S, Babu P, Prasad JH, Ghoshal UC. Frequency of lactose malabsorption among healthy southern and northern Indian populations by genetic analysis and lactose hydrogen breath and tolerance tests. Am J Clin Nutr. 2010;91:140–146. doi: 10.3945/ajcn.2009.27946. [DOI] [PubMed] [Google Scholar]
- 22.Mattar R, Monteiro Mdo S, Silva JM, Carrilho FJ. LCT-22018G>A single nucleotide polymorphism is a better predictor of adult-type hypolactasia/lactase persistence in Japanese-Brazilians than LCT-13910C>T. Clinics (Sao Paulo) 2010;65:1399. doi: 10.1590/S1807-59322010001200030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mulcare CA, Weale ME, Jones AL, Connell B, Zeitlyn D, Tarekegn A, et al. The T allele of a single-nucleotide polymorphism 13.9 kb upstream of the lactase gene (LCT) (C-13.9kbT) does not predict or cause the lactase-persistence phenotype in Africans. Am J Hum Genet. 2004;74:1102–1110. doi: 10.1086/421050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun HM, Qiao YD, Chen F, Xu LD, Bai J, Fu SB. The lactase gene -13910T allele can not predict the lactase-persistence phenotype in north China. Asia Pac J Clin Nutr. 2007;16:598–601. [PubMed] [Google Scholar]
- 25.Mendoza Torres E, Varela Prieto LL, Villarreal Camacho JL, Villanueva Torregroza DA. Diagnosis of adult-type hypolactasia/lactase persistence: genotyping of single nucleotide polymorphism (SNP C/T-13910) is not consistent with breath test in Colombian Caribbean population. Arq Gastroenterol. 2012;49:5–8. doi: 10.1590/s0004-28032012000100002. [DOI] [PubMed] [Google Scholar]
- 26.Flatz G, Henze HJ, Palabiyikoglu E, Dagalp K, Turkkan T. Distribution of the adult lactase phenotypes in Turkey. Trop Geogr Med. 1986;38:255–258. [PubMed] [Google Scholar]
- 27.Kurt I, AbouGhoush MW, Hasimi A, Serdar MA, Kutluay T. Comparison of indirect methods for lactose malabsorption. Turk J Med Sci. 2003;33:103–110. [Google Scholar]
- 28.Global Lactase persistance Association Database [Internet] London: University College London; c1999-2017. [cited 2013 Jul 12]. Available from: http://www.ucl.ac.uk/mace-lab/resources/glad. [Google Scholar]