Abstract
Background/Aims
Colorectal cancer (CRC) screening using stool DNA was recently found to yield good detection rates. A multi-target stool DNA test (Cologuard®, Exact Sciences), including methylated genes has been recently approved by the U.S. Food and Drug Administration. The aim of this study was to validate these aberrantly methylated genes as stool-based DNA markers for detecting CRC and colorectal advanced adenoma (AA) in the Korean population.
Methods
A single-center study was conducted in 36 patients with AA; 35 patients with CRC; and 40 endoscopically diagnosed healthy controls using CRC screening colonoscopy. The methylation status of the SFRP2, TFPI2, NDRG4, and BMP3 promoters was investigated blindly using bisulfate-modified stool DNA obtained from 111 participants. Methylation status was investigated by methylation-specific polymerase chain reaction.
Results
Methylated SFRP2, TFPI2, NDRG4, and BMP3 promoters were detected in 60.0%, 31.4%, 68.8%, and 40.0% of CRC samples and in 27.8%, 27.8%, 27.8%, and 33.3% of AA samples, respectively. The sensitivities obtained using 4 markers to detect CRC and AA were 94.3% and 72.2%, respectively. The specificity was 55.0%.
Conclusions
Our results demonstrate that the SFRP2, TFPI2, NDRG4, and BMP3 promoter methylation analysis of stool sample DNA showed high sensitivity but low specificity for detecting CRC and AA. Because of the low specificity, 4 methylated markers might not be sufficient for CRC screening in the Korean population. Further large-scale studies are required to validate the methylation of these markers in the Asian population and to find new markers for the Asian population.
Keywords: Colorectal neoplasms, Feces, DNA, Adenoma
INTRODUCTION
Colorectal cancer (CRC) is the 3rd most common cancer worldwide,1 and the incidence and mortality rates of CRC are increasing in Korea.2 The prevention of CRC by screening relies on the effective detection of critical precursor lesions, and thus broadly applied preventive and early detection measures are needed.3,4
In addition to traditional screening methods of CRC, including fecal occult blood test (FOBT) and colonoscopy, stool DNA testing has emerged as a noninvasive, molecular approach for CRC screening.5 Epigenetic alterations in CRC, such as aberrant DNA methylation have been extensively studied; thus, DNA methylation in exfoliated human gastrointestinal cells in stool may serve as a CRC biomarker.6,7,8,9 A multitarget stool DNA test, including methylated genes, has been recently approved by the U.S. Food and Drug Administration (USFDA).10 However, the results of the latest study showed a higher rate of false-positive results,11 and few validation studies have been conducted in Asia, including Korea.12
In the present study, we aimed to investigate stool-based methylated DNA markers for detecting CRC and precancerous lesions in Korean patients, and selected 4 previously identified promoters, including secreted frizzled-related protein 2 (SFRP2), tissue factor pathway inhibitor 2 (TFPI2), N-myc downstream-regulated gene 4 (NDRG4), and bone morphogenetic protein 3 (BMP3).
METHODS
1. Patients
This study included 111 patients who underwent colonoscopy for CRC screening at the Kangbuk Samsung Hospital between August 2012 and March 2014. Of these, 36 and 35 patients were diagnosed with advanced adenoma (AA) and CRC, respectively. A control group consisting of 40 endoscopically diagnosed healthy participants was also included in the study. Exclusion criteria were prior colorectal resection, history of any cancer, clinically apparent polyposis syndrome or hereditary nonpolyposis colon cancer syndrome, incomplete colonoscopic examination, and refusal of consent. All participants provided written informed consent and provided sufficient stool samples for DNA isolation using a self-collection approach. This study was approved by the Institutional Review Board of Kangbuk Samsung Hospital (IRB No. 2013-01-135), South Korea.
2. Colonoscopy and Definitions
All polypoid lesions were removed during colonoscopy. The size of each polyp was estimated using open-biopsy forceps that were 7 mm in diameter. All retrieved polyps were sent to the pathology laboratory for histologic evaluation. AA refers to adenomas with diameter ≥10 mm or tubulovillous, villous, or high-grade dysplasia. Patients with intramucosal carcinomas or carcinomas in situ were designated as having high-grade dysplasia. Cancer was defined as the invasion of malignant cells beyond the muscularis mucosa. If one patient had more than 1 lesion, the most advanced lesion was selected for further analysis. The left colon was defined as the rectum, sigmoid, and descending colon, whereas the right colon was defined as the transverse colon, ascending colon, and cecum.
3. Stool DNA Extraction from Stool Samples and Methylation-Specific PCR
Stool samples were collected the day before the colonoscopy (first stool during bowel preparation). Pre-endoscopy stool samples were stored in the patients' household freezers, and the patients were instructed to return the stool samples on the day of endoscopy for long-term storage at −70℃ in our laboratory.
Stool samples were randomly coded before processing to ensure adequate blinding of the clinical information. DNA was purified from feces (250–300 mg) using QIAamp DNA stool mini kit (Qiagen, Hilden, Germany) based on the manufacturer's instructions. This kit is designed for the preferential isolation and purification of DNA from human colonocytes present in the feces. PCR was performed to amplify the human β-actin gene to determine the quality of the isolated DNA. Human β-actin DNA was successfully amplified in all stool samples, confirming that our storage protocol for the samples was appropriate.
Genomic DNA was chemically modified with sodium bisulfite to convert all unmethylated cytosine residues to uracil while leaving the methylated cytosine residues unaltered. Methylation of the SFRP2, TFPI2, NDRG4, and BMP3 promoters in the bisulfite-modified DNA was investigated using methylation-specific PCR (MSP), aimed at either the methylated or the unmethylated alleles. Previously reported primer sequences are listed in Table 1, and commercially available methylated human genomic DNA (CpGenome Universal Methylated DNA; Chemicon International Inc., Temecula, CA, USA) was used as a positive control. The reagents without the isolated DNA served as the negative control. The thermocycler conditions were as follows: 95℃ for 5 minutes, 40 to 45 cycles at 95℃ for 30 seconds at a specific annealing temperature, and final extension at 72℃ for 10 minutes. All MSP assays were repeated at least twice to validate the results. Subsequently, MSP products were separated by horizontal gel electrophoresis in a 1.2% agarose gel, staining with ethidium bromide, and visualization under UV transillumination using the Quantity One image analyzer system (Bio-Rad, Hercules, CA, USA).
Table 1. Primer Sequences, PCR Product Sizes, and Annealing Temperatures Used in MSP Assays.
| PCR product size (bp) | Annealing temperature (℃) | |
|---|---|---|
| SFRP2 | ||
| M S: GGGTCGGAGTTTTTCGGAGTTGCGC | 138 | 62 |
| A: CCGCTCTCTTCGCTAAATACGACTCG | ||
| U S: TTTTGGGTTGGAGTTTTTTGGAGTTGTGT | 145 | 50 |
| A: AACCCACTCTCTTCACTAAATACAACTCA | ||
| BMP3 | ||
| M S: GTTTGGAGTTTAATTTTCGGTTTC | 179 | 54 |
| A: ATAACTTCGATCTCTCTCCCTACG | ||
| U S: GGTTTGGAGTTTAATTTTTGGTTTT | 178 | 54 |
| A: AACTTCAATCTCTCTCCCTACACC | ||
| NDRG4 | ||
| M S: TTTAGGTTCGGTATCGTTTCGC | 110 | 61 |
| A: CGAACTAAAAACGATACGCCG | ||
| U S: GATTAGTTTTAGGTTTGGTATTGTTTTGT | 105 | 61 |
| A: AAAACCAAACTAAAAACAATACACCA | ||
| TFPI2 | ||
| M S: ATTTTTTAGGTTTCGTTTCGGC | 118 | 57 |
| A: GCCTAACGAAAAAAAATACGCG | ||
| U S: TTAGTTATTTTTTAGGTTTTGTTTTGGT | 105 | 57 |
| A: AAAAACACCTAACAAAAAAAAATACACA |
MSP, methylation-specific PCR; SFRP2, secreted frizzled-related protein 2; M, methylated; S, sense; A, antisense; U, unmethylated; BMP3, bone morphogenetic protein 3; NDRG4, N-myc downstream-regulated gene 4; TFPI2, tissue factor pathway inhibitor 2.
4. Statistical Analysis
Continuous variables were expressed as mean±SD, and comparisons between groups were performed using one-way ANOVA. Variables not normally distributed were compared using Kruskal-Wallis test. Categorical variables were expressed as percentages, and comparisons between groups were performed using chi-square test. The sensitivity and specificity (including 95% CI) of the stool DNA assay were calculated using a manual method. The sensitivity and specificity were reported for each marker and combination of the 4 markers, which was defined as at least 1 methylation among the SFRP2, TFPI2, NDRG4, and BMP3 promoters. P-values <0.05 were considered significant. All data analyses were performed using SPSS version 18.0 (SPSS Inc., Chicago, IL, USA).
RESULTS
1. Patients
The stool samples were obtained from 40 endoscopically diagnosed healthy controls, 36 patients with AA, and 35 patients with CRC. The mean age was 55.7 years in normal patients, 63.2 years in patients with AA, and 60.6 years in patients with CRC, showing a statistically significant difference (P=0.03). No significant differences were found between sexes (Table 2).
Table 2. Characteristics of the Patients Enrolled in the Study.
| Colorectal cancer | Advanced adenoma | Normal | P-value | |
|---|---|---|---|---|
| No. of patients | 35 | 36 | 40 | - |
| Male sex | 25 (62.5) | 223 (65.7) | 25 (69.4) | 0.81 |
| Age (yr) | 60.60±13.04 | 63.20±10.16 | 55.70±14.64 | 0.03 |
Values are presented as number (%) or mean±SD.
2. Hypermethylation of SFRP2, TFPI2, NDRG4, and BMP3 Promoters in Patients with CRC
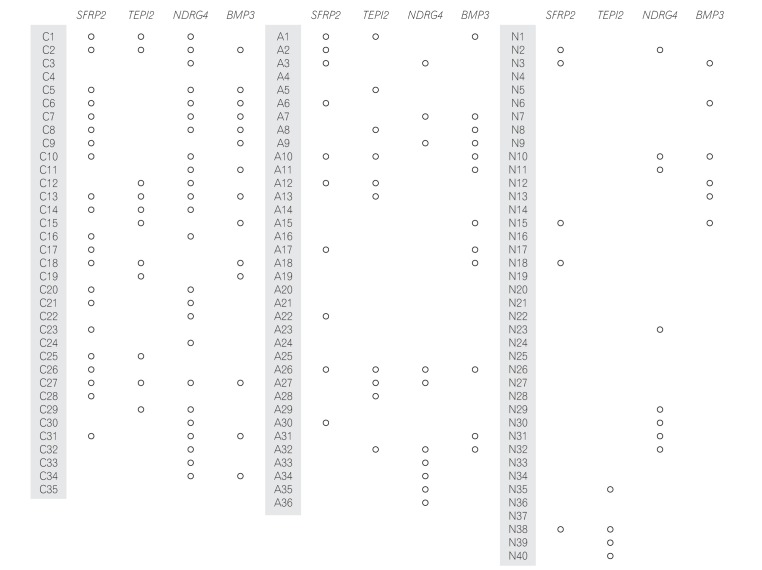
MSP was performed in 35 patients with CRC (Fig. 1). Methylation of the SFRP2, TFPI2, NDRG4, and BMP3 promoters was detected in 21 (60.0%), 11 (31.4%), 24 (68.8%), and 14 (40.0%) CRC samples, respectively. The sensitivity of combination of the 4 markers for detecting CRC was 94.3% (95% CI, 86.2–100.0) (Table 3).
Fig. 1. Results of the stool DNA methylation-specific PCR assay performed with samples obtained from patients with colorectal cancer (C) and advanced adenoma (A) and healthy controls (N). SFRP2, secreted frizzled-related protein 2; TFPI2, tissue factor pathway inhibitor 2; NDRG4, N-myc downstream-regulated gene 4; BMP3, bone morphogenetic protein 3.
Table 3. Sensitivity and Specificity of Methylated Stool DNA among Subgroups.
| SFRP2 | TFPI2 | NDRG4 | BMP3 | Combinationa | |
|---|---|---|---|---|---|
| Colorectal cancer (n=35) | |||||
| No. of positive results | 21 | 11 | 24 | 14 | 33 |
| Sensitivity (%) | 60.0 | 31.4 | 68.8 | 40.0 | 94.3 |
| 95% CI | 43.0–77.0 | 15.0–48.0 | 52.0–85.0 | 23.0–57.0 | 86.2–100.0 |
| Advanced adenoma (n=36) | |||||
| No. of positive results | 10 | 10 | 10 | 12 | 26 |
| Sensitivity (%) | 27.8 | 27.8 | 27.8 | 33.3 | 72.2 |
| 95% CI | 12.4–43.2 | 12.4–43.2 | 12.4–43.2 | 17.2–49.5 | 43.0–77.0 |
| Normal (n=40) | |||||
| No. of positive results | 5 | 4 | 8 | 6 | 18 |
| Specificity (%) | 87.5 | 90.0 | 80.0 | 85.0 | 55.0 |
| 95% CI | 76.8–98.2 | 80.3–99.7 | 67.0–92.9 | 73.4–96.5 | 38.9–71.1 |
aHypermethylation of 1 of the 4 promoters (SFRP2, TFPI2, NDRG4, and BMP3).
SFRP2, secreted frizzled-related protein 2; TFPI2, tissue factor pathway inhibitor 2; NDRG4, N-myc downstream-regulated gene 4; BMP3, bone morphogenetic protein 3.
In patients with CRC, the sensitivity using a combination of the 4 markers in left-sided CRC was 95.7%, and this difference was not significant compared with the sensitivity in right-sided CRC. The sensitivity of combination of the 4 markers in patients with stage III/IV CRC was 100%, but the sensitivity did not vary significantly based on CRC stage (Table 4).
Table 4. Correlation between Clinicopathologic Findings and Sensitivity of Methylated Stool DNA.
| No. | MSPa | Sensitivity (%)b | P-value | |
|---|---|---|---|---|
| Colorectal cancer | 35 | |||
| Location | 0.99 | |||
| Right | 12 | 11 | 91.7 | |
| Left | 23 | 22 | 95.7 | |
| Stage | 0.23 | |||
| I/II | 17 | 15 | 88.2 | |
| III/IV | 18 | 18 | 100.0 | |
| Advanced adenoma | 36 | |||
| Size (mm) | 0.28 | |||
| >10 | 35 | 26 | 74.3 | |
| ≤10 | 1 | 0 | 0 | |
| Histology | 0.68 | |||
| Villous | 27 | 20 | 74.1 | |
| Tubular | 9 | 6 | 66.7 | |
| Dysplasia | 0.37 | |||
| High grade | 7 | 4 | 57.1 | |
| Low grade | 29 | 22 | 75.9 |
aHypermethylation of 1 of the 4 promoters (SFRP2, TFPI2, NDRG4, and BMP3).
bCombination of 4 promoters (SFRP2, TFPI2, NDRG4, and BMP3).
MSP, methylation-specific PCR; SFRP2, secreted frizzled-related protein 2; TFPI2, tissue factor pathway inhibitor 2; NDRG4, N-myc downstream-regulated gene 4; BMP3, bone morphogenetic protein 3.
3. Hypermethylation of SFRP2, TFPI2, NDRG4, and BMP3 Promoters in Patients with AA
MSP was performed in 36 patients with AA. Methylation of the SFRP2, TFPI2, NDRG4, and BMP3 promoters was detected in 10 (27.8%), 10 (27.8%), 10 (27.8%), and 12 (33.3%) of the AA samples, respectively. The sensitivity of combination of the 4 markers for detecting AA was 72.2% (95% CI, 43.0–77.0) (Table 3).
In patients with AA, the sensitivities of combination of the 4 markers for detecting adenoma with >10 mm, villous histology, and high-grade dysplasia were 74.3%, 74.1%, and 57.1%, respectively. No statistically significant association was found between stool DNA hypermethylation and size, histology, or dysplasia (Table 4).
4. Hypermethylation of SFRP2, TFPI2, NDRG4, and BMP3 Promoters in Normal Patients
MSP was performed in 40 normal patients (Fig. 1). Methylation of the SFRP2, TFPI2, NDRG4, and BMP3 promoters was detected in 5 (12.5%), 4 (10%), 8 (20%), and 6 (15%) normal patients. Thus, their specificities were 87.5%, 90.0%, 80.0%, and 85.0%, respectively. The specificity of combination of the 4 markers for detecting CRC or AA was 55.0% (Table 3). When we compared the clinical characteristics in patients with true negative (n=22) and false negative (n=10) using combination of the 4 markers, no difference was found in the proportion of male patients (60.0% vs. 72.7%, P=0.47) and the mean age (57.1±13.7 years vs. 53.9±16.6 years, P=0.67).
DISCUSSION
In this study, we investigated the feasibility of detecting methylated stool DNA as a screening tool for CRC and precancerous lesions (AA). Using 4 methylation markers (SFRP2, TFPI2, NDRG4, and BMP3), 94.3% of patients with CRC and 72.2% of patients with AA showed methylated DNA in their stool samples. However, the specificity of our testing method was only 55.0%.
In analyzing different screening tests for CRC, the sensitivity, specificity, and particularly compliance must be considered. A screening test must not only be accurate, but also be widely accessible to provide effective lesion detection at a population level. To screen for CRC using stool samples, FOBT is widely performed and is the only screening method shown to reduce the mortality rate of CRC.13 However, FOBT has limitations, such as interference by dietary components, low sensitivity, and the requirement of multiple samplings.14 Assaying stool DNA for CRC screening has the advantages of continuous marker release and production from the neoplasm with better sensitivity than FOBT.15 Furthermore, the detection accuracy of stool DNA testing is not affected by the anatomic site of the target lesions16 and is not affected by diet components.17 Therefore, the acceptance and compliance for stool DNA testing are likely to be superior to those of FOBT.
The feasibility of testing stool DNA to screen for CRC was initially examined using mutation-based DNA, such as mutant KRAS.18 Methylation-based testing, involving the use of specific markers that are representative of an epigenetic signature of neoplastic alterations has been recently examined.14 A Cologuard® (Exact Sciences, Madison, WI, USA), a multitarget stool DNA test, including molecular assays for aberrantly methylated BMP3 and NDRG4 gene promoter regions, has been recently approved by the USFDA.10
In our study, we selected 4 methylation markers, SFRP2, TFPI2, NDRG4, and BMP3, which are known to be involved in CRC and were included in a recent multitarget stool DNA test.3,11 SFRP2 is a tumor suppressor gene, and its protein product contains domains similar to those in Wnt-receptor frizzled proteins. Promoter hypermethylation induces epigenetic inactivation of SFRP2 by constitutive Wnt signaling, which is observed in approximately 90% of CRC cases.19,20 In previous studies, the sensitivity and specificity of a single marker for SFRP2 in CRC patient stool samples were 63% to 94% and 91% to 96%, respectively.14 TFPI2 is a tumor suppressor gene that may be predisposed to aberrant DNA methylation in CRC carcinogenesis.21 In a previous study, TFPI2 methylation was detected in stool DNA from CRC patients with sensitivity and specificity of 76% and 79%, respectively.21 NDRG4 is a tumor suppressor gene in CRC whose expression is frequently inactivated by promoter methylation. In stool DNA, methylated NDRG4 showed a sensitivity of 61% for detecting CRC, with a corresponding specificity of 93%.22 BMP3 is a transforming growth factor-β of cytokine, and aberrant hypermethylation of this gene has been reported to downregulate the expression of BMP3 in CRC.23 Although no studies have investigated methylated BMP3 in stool as a single marker for CRC detection, multi-target stool DNA assay to measure β-actin, mutant KRAS, methylated BMP3, and NDRG4 showed 90% specificity, with 98% sensitivity in patients with CRC.16
In the present study, although the specificity for each marker was as high as 80% to 90%, the specificity for the combined 4 markers was as low as 55%. If we only included 2 markers (BMP3 and NDRG4) similar to the previous study,11 specificity was increased to 70%. The reason for the high false-positive rate might be related with field effect, as we previously reported.24 However, although recently developed DNA panels include not only methylated promoters, but also mutant KRAS and an immunochemical assay for human hemoglobin,11 their specificity is lower than that for fecal immunochemical test (FIT), resulting in more false-positive results, more diagnostic colonoscopies, and more associated adverse events per screening test.25
This study had several strengths. We investigated methylated markers that were included in a multitarget stool DNA test, which has been recently approved by the USFDA, in the Asian population. In addition, the use of stool collected from well-characterized patients undergoing colonoscopy for average risk CRC screening in a blinded design, and specimens were uniformly collected and analyzed.
The limitations of this study include a relatively small number of specimens and age differences between groups. Because a previous study demonstrated the effect of age on methylation markers,7 a large study should be performed to examine the role of stool methylated DNA for CRC screening related with age. Second, we did not perform FOBT or FIT simultaneously. Third, because we collected the first stool during bowel preparation, the fecal specimen might have been contaminated with the bowel preparation solution, and it might have affected the sensitivity of a single target. Since a few stool DNA marker studies have been conducted in Asia, large studies investigating stool DNA markers and comparing sensitivity and specificity with FIT or FOBT in Asian populations are needed.
In conclusion, the combination of 4 methylated markers might not be sufficient for CRC screening in Korea because of the low specificity. Further studies are needed to validate the methylation of these markers in the Asian population.
Footnotes
Financial support: This work was supported by a Ferring grant of the Korean Association for the Study of Intestinal Diseases for 2008, and Medical Research Funds from Kangbuk Samsung Hospital and Hyoseok Medical Research Fund.
Conflict of interest: None.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Jung KW, Park S, Kong HJ, et al. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2009. Cancer Res Treat. 2012;44:11–24. doi: 10.4143/crt.2012.44.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahlquist DA, Zou H, Domanico M, et al. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology. 2012;142:248–256. doi: 10.1053/j.gastro.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME, Colorectal Cancer Study Group Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351:2704–2714. doi: 10.1056/NEJMoa033403. [DOI] [PubMed] [Google Scholar]
- 6.Lansdorp-Vogelaar I, Kuntz KM, Knudsen AB, Wilschut JA, Zauber AG, van Ballegooijen M. Stool DNA testing to screen for colorectal cancer in the Medicare population: a cost-effectiveness analysis. Ann Intern Med. 2010;153:368–377. doi: 10.1059/0003-4819-153-6-201009210-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim ER, Kim YH. Clinical application of genetics in management of colorectal cancer. Intest Res. 2014;12:184–193. doi: 10.5217/ir.2014.12.3.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagasaka T, Tanaka N, Cullings HM, et al. Analysis of fecal DNA methylation to detect gastrointestinal neoplasia. J Natl Cancer Inst. 2009;101:1244–1258. doi: 10.1093/jnci/djp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Maio G, Rengucci C, Zoli W, Calistri D. Circulating and stool nucleic acid analysis for colorectal cancer diagnosis. World J Gastroenterol. 2014;20:957–967. doi: 10.3748/wjg.v20.i4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.A stool DNA test (Cologuard) for colorectal cancer screening. JAMA. 2014;312:2566. doi: 10.1001/jama.2014.15746. [DOI] [PubMed] [Google Scholar]
- 11.Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 12.Chang E, Park DI, Kim YJ, et al. Detection of colorectal neoplasm using promoter methylation of ITGA4, SFRP2, and p16 in stool samples: a preliminary report in Korean patients. Hepatogastroenterology. 2010;57:720–727. [PubMed] [Google Scholar]
- 13.Mandel JS, Bond JH, Church TR, et al. Reducing mortality from colorectal cancer by screening for fecal occult blood: Minnesota Colon Cancer Control study. N Engl J Med. 1993;328:1365–1371. doi: 10.1056/NEJM199305133281901. [DOI] [PubMed] [Google Scholar]
- 14.Ahlquist DA. Molecular detection of colorectal neoplasia. Gastroenterology. 2010;138:2127–2139. doi: 10.1053/j.gastro.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 15.Osborn NK, Ahlquist DA. Stool screening for colorectal cancer: molecular approaches. Gastroenterology. 2005;128:192–206. doi: 10.1053/j.gastro.2004.10.041. [DOI] [PubMed] [Google Scholar]
- 16.Lidgard GP, Domanico MJ, Bruinsma JJ, et al. Clinical performance of an automated stool DNA assay for detection of colorectal neoplasia. Clin Gastroenterol Hepatol. 2013;11:1313–1318. doi: 10.1016/j.cgh.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 17.Itzkowitz S, Brand R, Jandorf L, et al. A simplified, noninvasive stool DNA test for colorectal cancer detection. Am J Gastroenterol. 2008;103:2862–2870. doi: 10.1111/j.1572-0241.2008.02088.x. [DOI] [PubMed] [Google Scholar]
- 18.Sidransky D, Tokino T, Hamilton SR, et al. Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors. Science. 1992;256:102–105. doi: 10.1126/science.1566048. [DOI] [PubMed] [Google Scholar]
- 19.Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat Rev Cancer. 2001;1:55–67. doi: 10.1038/35094067. [DOI] [PubMed] [Google Scholar]
- 20.Xu Q, D'Amore PA, Sokol SY. Functional and biochemical interactions of Wnts with FrzA, a secreted Wnt antagonist. Development. 1998;125:4767–4776. doi: 10.1242/dev.125.23.4767. [DOI] [PubMed] [Google Scholar]
- 21.Glöckner SC, Dhir M, Yi JM, et al. Methylation of TFPI2 in stool DNA: a potential novel biomarker for the detection of colorectal cancer. Cancer Res. 2009;69:4691–4699. doi: 10.1158/0008-5472.CAN-08-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melotte V, Lentjes MH, van den Bosch SM, et al. N-myc downstream-regulated gene 4 (NDRG4): a candidate tumor suppressor gene and potential biomarker for colorectal cancer. J Natl Cancer Inst. 2009;101:916–927. doi: 10.1093/jnci/djp131. [DOI] [PubMed] [Google Scholar]
- 23.Loh K, Chia JA, Greco S, et al. Bone morphogenic protein 3 inactivation is an early and frequent event in colorectal cancer development. Genes Chromosomes Cancer. 2008;47:449–460. doi: 10.1002/gcc.20552. [DOI] [PubMed] [Google Scholar]
- 24.Park SK, Song CS, Yang HJ, et al. Field cancerization in sporadic colon cancer. Gut Liver. 2016;10:773–780. doi: 10.5009/gnl15334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.US Preventive Services Task Force. Bibbins-Domingo K, Grossman DC, et al. Screening for colorectal cancer: US Preventive Services Task Force recommendation statement. JAMA. 2016;315:2564–2575. doi: 10.1001/jama.2016.5989. [DOI] [PubMed] [Google Scholar]