Abstract
The chemical properties and biological mechanisms of RNAs are determined by their tertiary structures. Exploring the tertiary structure folding processes of RNA enables us to understand and control its biological functions. Here, we report a nanopore snapshot approach combined with coarse-grained molecular dynamics simulation and master equation analysis to elucidate the folding of an RNA pseudoknot structure. In this approach, single RNA molecules captured by the nanopore can freely fold from the unstructured state without constraint and can be programmed to terminate their folding process at different intermediates. By identifying the nanopore signatures and measuring their time-dependent populations, we can “visualize” a series of kinetically important intermediates, track the kinetics of their inter-conversions, and derive the RNA pseudoknot folding pathway. This approach can potentially be developed into a single-molecule toolbox to investigate the biophysical mechanisms of RNA folding and unfolding, its interactions with ligands, and its functions.
While RNA folding is critical for its function, study of this process is challenging. Here, the authors combine nanopore single-molecule manipulation with theoretical analysis to follow the folding of an RNA pseudoknot, monitoring the intermediate states and the kinetics of their interconversion.
Introduction
RNAs play critical roles in the maintenance, transfer, and processing of genetic information and the catalytic control of gene expression1. To perform these biological functions, RNAs must fold into specific tertiary structures2. Thus, knowledge of the RNA folding process is key to understanding the role of RNA structures and manipulating their functionalities3. Various methods have been developed to explore RNA folding mechanisms, including cryo-electron microscopy4, small-angle X-ray scattering5, and NMR6. In addition to these expensive instrument-based methods, chemical probing approaches, such as selective 2ʹ-hydroxyl acylation and primer extension (SHAPE)7 and hydroxyl radical footprinting8, have been widely applied to probe RNA folding structures. As RNA folding is intrinsically an intra-molecular process, various single-molecule methods, such as smFRET9 and optical tweezers10, have been developed to investigate RNA folding. However, due to the diffusive properties of RNA folding and short-lived transition paths, direct experimental observation of the RNA folding process, in particular the capture of intermediate folding states, remains difficult10.
Nanopore is a promising label-free, single-molecule-based, next-generation sequencing technology11–15. By taking advantage of the ability to electrically detect charged biomolecules through a nanometer-wide channel, various nanopore biosensors have been developed, with targets including DNAs16, 17, microRNA biomarkers18–20, tRNA21, peptides22, 23, and proteins24, 25 as well as epigenetic changes such as DNA methylation26–28. In another important application, nanopores have been used as a precise force instrument to explore biomolecular mechanisms such as protein unfolding29, DNA unzipping11, 30, 31, RNA unfolding32, and the binding of nucleic acids to enzymes33. Briefly, single target molecules are driven into the nanopore and produce an ion current “fingerprint” for their dissociation-translocation procedure. The nanopore under this experimental configuration is best suited to the study of unfolding-related biological problems. However, these nanopore methods thus far have not been reported to detect the other half of the biomolecular process—how a biomolecule folds into a functional tertiary structure.
In this report, we propose a generalized nanopore snapshots approach that enables elucidation of the RNA folding mechanism. This approach was established by using the gene 32 messenger RNA of bacteriophage T234 as a model system (Fig. 1a). This 36-nt RNA forms a typical H-type pseudoknot (PK) for regulating gene expression35. Our approach, as shown in Fig. 1b, allows single RNA molecules captured by the nanopore to freely fold outside the pore without constraints, starting from the single-stranded unfolded form. Remarkably, by programming the folding time, we can terminate the folding process at either the PK or any intermediate state and subsequently probe its structure based on the RNA unfolding signature in the nanopore. By characterizing the nanopore snapshots taken at different folding times and combining them with a series of theoretical analyses, we are able to “visualize” various folding intermediates, track their inter-conversions, and derive the folding pathway.
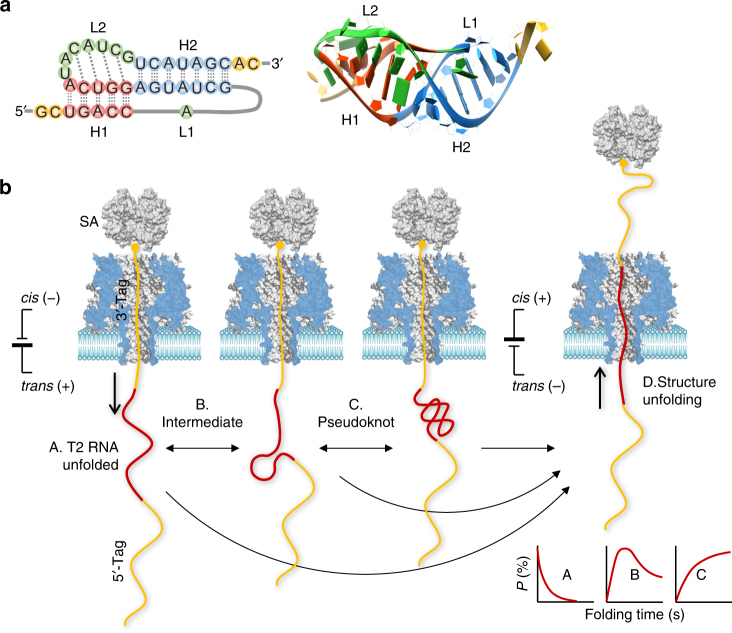
Fig. 1.
Single-molecule detection of RNA folding in a protein nanopore. a 2D (left) and 3D (right) structures of the pseudoknot from the 5ʹ end gene 32 messenger RNA of bacteriophage T2 (PDB id: 2tpk), which contains two helices, H1 (red) and H2 (blue), and two loops (green), L1 and L2. The base triple interactions between loops and helices (identified by RNAview70) and the coaxial stacking between the two helices are illustrated. b Nanopore detection of T2 RNA folding states and the folding pathway. The detection was facilitated by a complex probe containing a chimera of the 5ʹ-DNA tag, T2 RNA, and 3ʹ-DNA tag. The 3ʹ end is attached to streptavidin (SA) to immobilize the probe. Upon immobilization, the T2 RNA starts to fold from the single-stranded form in the trans solution. After a pre-defined folding time, the folding process is terminated at an intermediate state (B) or the pseudoknot structure (C) by using a negative voltage that disrupts the folding structure (D). The unfolding signatures reveal the folding state (B and C) and unfolding state (A). The folding time-dependent population of the identified folding states allows establishment of the pseudoknot folding pathway
Results
Designing a complex probe for RNA folding detection
The T2 RNA PK contains two helices, H1 (5-bp) and H2 (7-bp), that are coaxially stacked on top of one another (Fig. 1a). The loop L2 (7-nt) linking the two helices interacts with H2 by non-canonical base pairing, conferring surprisingly high stability to this long quasi-continuous helix34. To explore the formation of the T2 PK, we designed a multi-functional complex probe, T2 (Table 1 and Fig. 1b). The probe comprises a T2-RNA extended with a poly(CAT)10 DNA tag at both the 5ʹ and 3ʹ ends. The chimera is attached with a streptavidin at the 3ʹ biotinylated end, and the probe is presented in the cis solution of the α-hemolysin protein pore. Under a positive transmembrane voltage applied from the trans side (cis grounded), the chimera is threaded through the nanopore from the cis to trans side. As the attached streptavidin is wider than the pore entrance, it terminates chimera translocation and immobilizes the chain inside the nanopore.
Table 1.
Sequences of various complex probes used in the study
| Probe | Sequence |
|---|---|
| T2a | 5ʹ-(CAT)10-gcugaccagcuaugaggucauacaucgucauagcac-(CAT)10-biotin-3ʹ |
| T2-PolyUb | 5ʹ-(CAT)10-gcugaccagcuaugaggucauuuuuuuucauagcac-(CAT)10-biotin-3ʹ |
| Ref-HPc | 5ʹ-(CAT)10-gcugaccucauagcuuuuuuugcuaugagguca-(CAT)10-biotin-3ʹ |
Unlike previous studies that have used streptavidin-nucleic acid constructs for single-nucleotide discrimination36, 37 and biosensor development38–40, our complex probe was designed to possess multiple functions suited to RNA folding detection (Fig. 1b). (1) The 3ʹ DNA tag (30 nt, ~15 nm) is much longer than the nanopore passage (10 nm)41; thus, when immobilized, it can occupy the entire nanopore. This design guarantees that T2-RNA can be released into the trans solution, enabling RNA to freely fold without constraint from nanopore confinement. (2) Prior to folding in the trans solution, the T2 RNA structure is fully disrupted when passing through the nanopore. Therefore, the folding of all RNA molecules starts from the single-stranded unstructured conformation, regardless of their original folding states in the cis solution. (3) The folding time can be controlled such that the folding process can be terminated at an intermediate state when a negative voltage is applied to reversely pull the probe. After the pulling force unfolds the intermediate state, the generated nanopore signature can take a snapshot (i.e., report the identity) of that state. (4) As any low voltage can be applied, the nanopore can provide a minuscule pulling force (several pN) that disrupts the RNA folding structure. This behavior enables us to discover the sub-populated intermediate structures that otherwise cannot be identified under larger pulling forces. (5) In the nanopore, the folding process of the RNA is conducted at room temperature without a heating–cooling process, allowing the folding mechanism to be studied under near-physiological conditions. (6) The DNA tags of the chimera produce distinct nanopore signatures from RNA. The differences in their unique signals can be used as markers to track the RNA position in the nanopore and determine the intermediate states thereafter32. Note that DNA tags do not affect the RNA structure, as reported previously32. Similar DNA–RNA chimeras have been utilized for RNA folding detection in optical tweezers42. (7) As shown in Fig. 1b, the nanopore facilitated by this probe can simultaneously detect both folding and unfolding of RNA.
Snapshots of RNA folding and intermediate states
The overall strategy for the nanopore folding study is as follows: a positive voltage is first applied to drive a probe into the pore, disrupt its structure, and release the unstructured RNA of the probe into the trans solution. This unstructured molecule is held in the trans solution for a pre-defined duration, i.e., folding time (such as 1 s, 10 s, or 30 s) for re-folding. At the end of the folding time, a negative voltage is applied to pull the folded RNA back into the cis solution. Its folding structure (formed during the folding time) is then inferred from the RNA unfolding signature. Repeating this protocol enables many single-molecule snapshots to be measured, classified, and assigned to specific folding structures with a fractional population. By varying the folding time, time-dependent folding populations can be obtained to elucidate the folding pathways.
Figure 2a shows a representative current signature for the folding–unfolding of a PK in the nanopore. This experiment was performed in 1 M NaCl in the presence of 10 mM Mg2+ (Methods). Figure 2b shows the entire molecular procedure suggested by the multi-level signature, from probe trapping to PK folding and unfolding. The probe was initially trapped in the pore at +120 mV. As the probe passed through the pore, its different domains sequentially occupied the pore lumen, resulting in the stepwise change in the block level (conductance). When the 5ʹ DNA tag was first threaded in the pore, the current was reduced to Level-1 (I/I 0 = 18.6 ± 1.6%, A). Next to the 5ʹ-tag, the T2 RNA was pulled into the pore while disrupting its initial structure, further reducing the current to Level-2 (12.6 ± 1.5%, B). Consistent with the previous finding, the RNA in the nanopore reduced more conductance than DNA in the pore19, 32. As the 3ʹ DNA tag entered and was immobilized (by the attached streptavidin) in the pore in place of T2 RNA, the nanopore current was returned to Level-1 (C). At this moment, the T2 RNA that was released into the trans solution started to re-fold (D). The folding time (t fold) for this illustrated event was 10 s. By the end of t fold, the voltage switched to −60 mV to pull the probe backward (from trans to cis) to unfold the RNA structure. This process produced the characteristic two-level block pattern, a long block at Level-3 (I/I 0 = 9.2 ± 0.5%, E), followed by a short deeper block at Level-4 (I/I 0 = 3.0 ± 0.7%, F). From Level-4, the current returned to the open pore level (G). According to a previous study32, this type of block pattern is the signature for the stepwise unfolding of a PK. The total duration of Level-3 and Level-4 was the unfolding duration (τ) of PK. Here, τ = 3500 ms ± 330 ms at −60 mV (Fig. 3a) and can be dramatically shortened to 39 ms ± 6 ms at −120 mV (Supplementary Fig. 1). To interpret the high stability of the PK, we constructed a reference hairpin ref-HP (Table 1 and Supplementary Fig. 2). Ref-HP utilized all the base pairs of H1 (5 bp) and H2 (7 bp) of the T2 PK to form a long helix (12 bp) but without loop–helix interactions. τ for Ref-HP unfolding was only 18 ms ± 3 ms at −60 mV (Supplementary Fig. 2), consistent with the reported results for hairpin40. Therefore, Ref-HP is 200-fold less stable than PK, verifying the significant contribution of the loop–helix interaction to the stability of the PK32. Overall, through a nanopore folding–unfolding electric signature, we can capture a folding snapshot for the formation of a PK from single-stranded T2 RNA in a given folding time.
Fig. 2.
Nanopore snapshot for T2 RNA pseudoknot formation. a Stepwise multi-level nanopore current signature generated by a complex probe under the +120 mV/t fold = 10 s/−60 mV protocol (the probe was trapped at + 120 mV, the folding time t fold was 10 s, and the folding structure was unfolded at −60 mV). b Model cartoons showing the entire trapping–folding–unfolding procedure revealed by the signature in a: translocation of the 5ʹ DNA tag (A, Level-1), disruption while translocating T2 RNA (B, Level-2), translocation and immobilization of the 3ʹ DNA tag (C, Level-1), folding of the pseudoknot from the single-stranded RNA on the trans side (D) for a pre-defined folding time (t fold = 10 s), unfolding of the folding structure under back-pulling at −60 mV while the 3ʹ DNA tag remains in the pore (E, Level-3), translocation of the partially unfolded T2 RNA through the pore (F, Level-4), and a fully disrupted probe pulled out of the pore and back to the cis solution (G). The time from the beginning of negative voltage (−60 mV) application to the block end is the unfolding duration of an RNA folding state (τ), during which the folding structure was disrupted
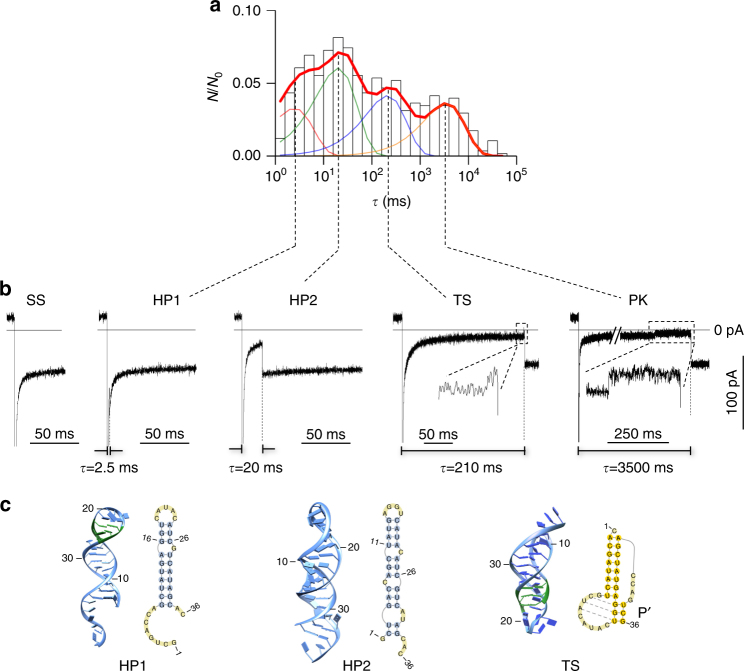
Fig. 3.
Detection of intermediate folding states. a Histogram showing the distribution of unfolding durations of various folding states (N 0 = 578). The x-axis is the time with bin width in the log scale. The y-axis is N 0/N, where N 0 is the total number of events in the histogram and N is the number of events within each bin (log time). N/N 0 represents the fractional population for each bin, and the sum of N/N 0 for all bins, i.e., the area of the histogram, is normalized to “1”. After fitting, the area covered by each component is the fractional population of the corresponding structure (Fig. 4). In this histogram, the unfolding duration of each RNA event was determined under the +120 mV/t fold = 10 s/−60 mV protocol. Four components at 2.5 ms, 20 ms, 210 ms, and 3500 ms were identified in the distribution (Methods). Each component corresponds to a folding state. b Representative nanopore current signatures for the four folding states identified from the unfolding duration distribution in panel a. The longest component (3500 ms) is for the pseudoknot (PK). The 2.5 ms, 20 ms, and 210 ms components are intermediate states. The two insets clearly show the two-level conductance patterns, TS and PK, which are distinct from the single-level patterns for the other intermediate states, HP1 and HP2. In addition, the signature of the open pore current (without block) for single-stranded RNA (SS) is shown. c Structures observed in the simulation that correspond to the three intermediate folding states identified in the nanopore experiment, HP1 (2.5 ms), HP2 (20 ms), and TS (210 ms). 2D and 3D structures for the three intermediates were found in the CG MD simulation. Three non-native base pairs in HP1 and the misfolded stem Pʹ in TS are labeled in green
Figure 3a shows the unfolding duration distribution of the folding snapshots with 10 s folding time. We identified the PK state (3500 ms) as well as three less stable components at 2.5 ms ± 2 ms, 20 ms ± 3 ms, and 210 ms ± 25 ms. Each identified component corresponds to a non-PK intermediate state. Figure 3b shows the representative current traces for each type of folding state. Note that the short 2.5 ms and 20 ms states could not be seen at high voltage, such as −120 mV (Supplementary Fig. 1), supporting the notion that the unique small pulling force provided by the nanopore (~5 pN at −60 mV32, 43, 44) is the key to discovering less stable intermediates. In addition to the various folding states identified above, we occasionally observed an open pore current when negative voltage was applied (Fig. 3b). These events were generated by the unfolded T2 RNA in the single-stranded (SS) form that was rapidly pulled back to the cis solution.
To provide insight into the structural details of intermediate folding states, we conducted coarse-grained (CG) molecular dynamics (MD) simulation. For the T2 RNA sequence, the simulation revealed that three intermediate structures (Fig. 3c) emerge before the final formation of the native PK. The initial coil state, SS, undergoes transitions to two possible stem–loop structures, HP1 and HP2. HP1 is a hairpin that contains the native 7-bp helix H2 (the longer stem in the T2 PK), with three additional non-native base pairings (bps) formed in the loop region. HP2 is a fully non-native stem–loop structure consisting of 11 canonical bps along with two or more non-canonical bps. The simulation also revealed a non-native PK-like intermediate, the “TS” state. The TS structure contains the native stem H2 and a misfolded stem Pʹ. A distinct feature of TS is the non-native tertiary interaction between stem Pʹ and the loop. The misfolded HP2 and TS structures are also supported by separate computational models. For example, the Vfold-based free energy analysis for the 2D structures45 and the SimRNA-based 3D structure analysis46 predicted TS and HP2 as misfolded suboptimal states of the sequence.
Overall, the observations revealed by the CG MD simulation are in agreement with the above nanopore experimental results. The nanopore folding snapshots and the CG MD simulation can be combined to construct the structural details for the observed intermediated states. In other words, by considering the features of unfolding nanopore signatures and the stabilities of the intermediated states relative to the ref-HP, we can establish connections between the intermediate states identified by the nanopore folding snapshots and the kinetics intermediates observed in the CG MD simulation. Compared with 12-bp ref-HP (18 ms ± 3 ms at −60 mV), 7-bp HP1 should be less stable and thus is assigned to the 2.5 ms state. The 13-bp HP2 corresponds to the 20 ms state due to it similar stability to ref-HP. In addition, the single-level blocks (−60 mV) for the two states (HP1 and HP2 in Fig. 3b) are consistent with the hairpin structure that is unfolded in one step (Supplementary Fig. 2). TS can be assigned to the 210 ms component because it is much more stable than ref-HP, consistent with the stabilization effect of the loop–helix interactions observed in TS. Moreover, the unfolding of TS produced a two-level block pattern (−60 mV, Fig. 3b), consistent with its PK-like structure obtained in the simulation. In summary, based on the nanopore snapshot and CG MD simulation, we identified a series of T2 RNA folding states and their structures, including single-stranded SS, intermediates HP1, HP2 and TS, and native PK.
RNA folding pathway
After identifying all the folding states, we combined the nanopore snapshot data and theoretical analysis to investigate the time-dependent folding process and the transitions between the different states for the purpose of establishing a PK folding pathway. Experimentally, we investigated how the folding of T2 RNA varied with the folding time. We first obtained a series of nanopore folding snapshots, which are characterized by the unfolding duration distributions, with different folding times ranging from 1 s to 60 s (Supplementary Fig. 3). We then calculated the fractional populations of all the identified states (SS, HP1, HP2, TS, and PK) in the unfolding duration distributions (Supplementary Fig. 3). Finally, we obtained the population kinetics (P–t fold curve) for each state (Fig. 4a and Methods). We found that the population of the SS state sharply decreased with increasing folding time. For HP1, the population increased very quickly in the initial stage, followed by a rapid decrease in the population. For HP2 and TS, the population increased before folding time t fold = 10 s and then slowly decreased. In contrast, the population for the native PK state of T2 RNA continuously increased before finally reaching an equilibrium with t fold = 60 s. All states reached equilibrium around the folding time of 60 s.
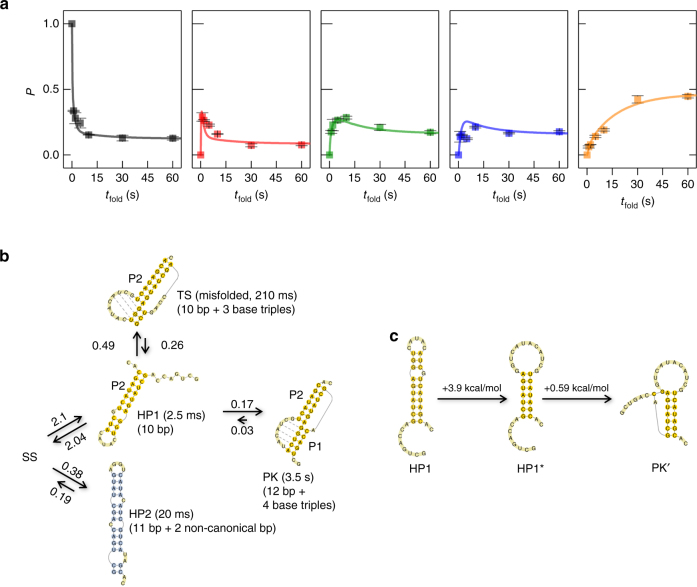
Fig. 4.
Folding pathway of the T2 RNA pseudoknot. a The fractional population of the different states (from left to right, SS, HP1, HP2, TS, and PK, respectively) as a function of the folding time. The solid squares represent the experimental data. Error bars represent standard deviation of the mean (n ≥ 3). We first obtained the unfolding duration histogram at different folding times (Supplementary Fig. 3). Each histogram was then fitted globally with a sum of four exponentials, and the normalized area of each component is the fractional population of the corresponding state (Methods). The curves in all plots of the panel were theoretically predicted based on the master equation approach. b The predicted pseudoknot folding pathway based on the analysis of the time-dependent population distribution and from the simulation. In the folding pathway of T2 PK, starting from the single-stranded (SS) state, the RNA can form two hairpin structures, HP1 and HP2. From HP2, the RNA can only go back to the SS state. Disruption of the non-native base pairs and loop–helix base triples in the misfolded TS state results in the HP1 structure. Therefore, the TS state can lead to the native PK structure via HP1 as the intermediate. Also shown in the figure are the fitted rate constants (in s−1) for the transitions between the states and the unfolding durations (shown in the parentheses) of each state under the +120 mV/t fold = 10 s/−60 mV protocol. c Two “uphill” steps in the HP1→PK folding process: disruption of the three non-native base pairs in the HP1 hairpin and the loop formation and the subsequent initiation of helix H2 formation in the native PK structure
To understand these time-dependent folding populations, a folding model based on the master equation approach36, 37 was developed (Fig. 4b). Fitting the experimentally determined population kinetics for the five states (Fig. 4a) yielded the estimated rate constants (Fig. 4b and Methods). SS is the initial state of the pathway for all the folding states observed. The RNA chain in the SS state quickly folds into the on-pathway intermediate HP1 and the off-pathway intermediate HP2. The initial folding from SS results in a quick decrease in the SS population and a concurrent initial increase in the HP1 and HP2 populations (Fig. 4a). In this initial process, approximately k SS→HP1/(k SS→HP1 + k SS→HP2) = 84.7% and k SS→HP2/(k SS→HP1 + k SS→HP2) = 15.3% of the population goes from SS to the HP1 and HP2 states, respectively. We note that HP1, TS, and PK share the same long native helix H2 (Fig. 3c), and HP2 is fully non-native and adopts a completely different fold from HP1, TS, and PK. Therefore, apart from going back to the SS state, direct transitions between HP2 and HP1, TS, or PK can be ignored. As a result, the HP2 population experiences a slow decrease and then reaches a plateau (Fig. 4a). From HP1, the RNA can fold directly to either the native PK or the misfolded TS state or back to the initial SS state. After the formation of the HP1 state, 6.3% of the initial HP1 population would fold directly to the native PK, ∼18.1% to the misfolded TS state, and 75.6% back to the SS state. These transitions together cause the decrease of the HP1 population and the increase of the PK and TS populations (Fig. 4a). The TS structure cannot directly fold to PK. Instead, TS returns to HP1 via the disruption of the misfolded base pairs. Therefore, a slow decrease in the TS population occurs as a result of the detrapping transition from TS to HP1, which subsequently folds to the native PK state (Fig. 4b). In addition, based on the Vfold RNA folding model47, 48, the HP1 → PK transition is rate limited by the (mainly enthalpic) barrier (~ 3.8 kcal/mol) to break the three non-native intra-loop base pairs in the HP1 hairpin loop in the HP1 → HP1* transition and the (mainly entropic) barrier (~0.59 kcal/mol) for the formation of the helix stem H1 in the HP1* → PKʹ transition (Fig. 4c). As the former step has a higher barrier, it may be the rate-limiting step for the HP1 → PK folding process.
In summary, the T2 PK folding process follows two possible routes starting from SS (Fig. 4b). In route I, SS first forms a short hairpin HP1. This hairpin contains a 7-bp native helix H2 in the T2 PK and three non-native base pairs inside the loop. The HP1 → PK folding involves two parallel pathways: direct folding to PK and folding through the misfolded TS state. This structure can reversibly return to HP1 and then folds to PK. In Route II, SS folds to a 13-bp hairpin HP2 with three bulges. This structure is stable, and unfolding back to the single-stranded SS state is slow. Route I and Route II together form the folding pathway of T2 RNA.
Regulation of the folding pathway by loop and magnesium ion
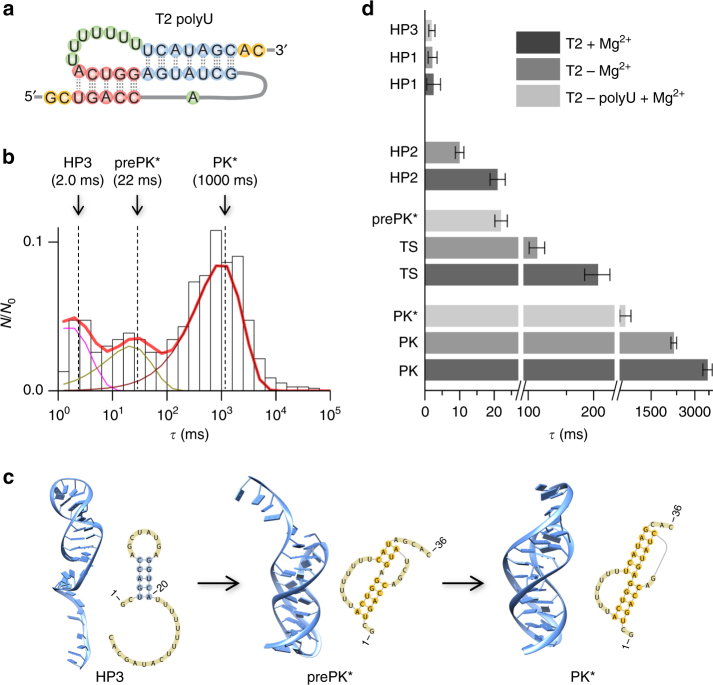
Since the loop–stem interactions in the PK are critical for structural stability3, 49, we investigated how these interactions influence the folding pathway. To weaken the loop–helix tertiary interactions, we changed the sequence of loop L2 into polyU (Fig. 5a, Table 1). Nanopore experimental data showed that this T2-polyU mutant has only three folding states (Fig. 5b). The first state had an unfolding duration of 2.0 ms ± 1.1 ms, which is similar to that of the 7-bp hairpin HP1 (2.5 ms) formed in Route I of the wild-type PK folding. In combination with the CG MD simulation results, we assigned this state to a short hairpin (HP3, Fig. 5b, c) that contains the 5-bp native helix H1 of the native PK. The second intermediate state had an unfolding duration of 22 ms ± 3 ms (Fig. 5b), and the simulation showed that this state may be a PK-like structure, with a native helix H1 and partially folded helix H2 (6–8 bp in total) and without a loop–helix interaction (prePK*, Fig. 5b, c). Compared with the wild-type PK, the helix stem H2 was not fully stabilized because the polyU mutant lacks the loop–helix base triple tertiary interactions in the wild-type PK. Although the polyU loop could significantly disrupt the loop–stem interaction, it may form weak loop–stem interactions to accommodate the folding to the final state of this sequence (PK*), which had an unfolding duration of 1000 ms ± 80 ms in the nanopore (Fig. 5b). This interpretation agrees with our previous finding for the PK unfolding kinetics32. Overall, the loop–stem tertiary interactions are important for the PK and its folding process.
Fig. 5.
Regulation of the pseudoknot folding pathway by RNA sequence and magnesium ions. a 2D structure of T2 RNA with a polyU loop. b Histogram of the unfolding duration for T2 polyU mutant folding states after the folding time of t fold = 10 s (N 0 = 232). Three components at 2.0 ms for HP3, 22 ms for prePK*, and 1000 ms for PK* were identified from the histogram. c 2D and 3D structures for the intermediate states HP3 and prePK* and the final state PK* obtained from the simulation study. d Comparison of unfolding durations of various states in wild-type T2 with magnesium ions, wild-type T2 without magnesium and T2 with polyU loop with magnesium ions. Error bars represent standard deviation of the mean duration (n ≥ 3). Magnesium ions are at the concentration of 10 mM, and the unfolding voltage in trans is −60 mV. All the data were collected from at least three different nanopores, and the errors are given as standard deviations
Magnesium ions (Mg2+) can regulate RNA structure formation and stabilize the pocket motifs in RNA or DNA tertiary structures50–52. To explore the effect of Mg2+ on the PK folding process, we repeated the nanopore snapshots in 1 M NaCl in the absence of Mg2+ (Methods). With 10 s folding time, T2 RNA without Mg2+ folded to four states, similar to T2 with Mg2+ (Fig. 5d). The unfolding durations of the HP1 and HP2 states (at −60 mV) were similar to those with Mg2+ ions, indicating that the two hairpin-based states are less influenced by Mg2+. In contrast, without Mg2+, the two PK-like states, TS and PK, were less stable, with the unfolding durations shortened from 3500 ms ± 330 ms to 1900 ms ± 170 ms for PK and from 210 ms ± 25 ms to 80 ms ± 9 ms for TS, respectively (Fig. 5d). This change might have been caused by a reduction in ion binding and charge neutralization as well as the loss of interactions between Mg2+ and the PK loops. The Mg2+–loop interaction stabilizes the PK structure but not the first two less stable states (hairpins HP1 and HP2), which lack loop–helix interactions and have much weaker stabilizing interactions with Mg2+.
Discussion
The nanopore snapshot-based kinetic detection, combined with CG molecular dynamics-based structural modeling, master equation-based rate constant estimation, and population kinetics analysis, provides a tool to investigate the RNA folding process. Through a programmable RNA disruption–refolding–disruption procedure in the nanopore, a series of intermediates that may be inaccessible in equilibrium experiments can be captured based on their unfolding signatures and the population kinetics for each state can be measured. The nanopore is able to capture a wide range of intermediates, with the unfolding duration ranging from milliseconds to seconds and minutes. The CG MD simulations can identify the potential intermediate structures and kinetic pathways, which are supported by the nanopore signatures. Based on the identified structures, the master equation approach can provide the transition rates between different states from the population kinetics. The combination of the above three methods leads to a reliable construction of the folding kinetics. The reliability of the results is supported by the theory–experiment consistency for the different measured properties. The integrated approach above can potentially be adapted for the study of RNAs with unknown structures. For RNAs with unknown structures, in addition to the structure information provided from the computational studies, nanopore data for various designed mutants would be highly useful for the probing and confirmation of the structures, stabilities, and folding pathways. The theory–experiment comparisons for the designed mutants may lead to reliable identification of the key factors that determine the kinetics.
Our study suggests five-state folding kinetics for the T2 RNA PK. Similar to the unfolding kinetics for the same molecule revealed in a previous study53, the folding kinetics are multi-state and non-cooperative and involve off-pathway misfolded intermediates and the kinetic pathways are strongly influenced by the helix–loop tertiary interactions. In addition, the comparative studies for the wild-type T2 PK and the polyU mutant indicate that the sequence forming the same PK may fold through very different pathways54. Among the three folding intermediates identified, two are hairpins, and the third is a PK-like structure that adopts a short misfolded stem for one of two helices in the native structure. The rate-limiting step of the whole folding pathway is the formation of the native PK from the hairpin structure that contains a native helix and three extra base pairs in the hairpin loop region. The folding equilibrium within an RNA PK is highly sequence dependent (mutation of one loop with polyU can change the stability of the PK) and can be regulated by magnesium ions. These results have important implications for the interplay between RNA structures and their functions in cells, since the regulatory functions of RNA molecules are often related to conformational transitions55.
To identify more intermediate states (such as the pre-PK structure with two native helices formed but without loop–helix tertiary interactions53, 54) and to develop further applications, the resolution and accuracy of the nanopore system need improvement. First, due to the resistor–capacitor (RC) time of the setup, the voltage change will simultaneously cause a curved charging current (Figs. 2a and 3b). Rapidly unfolding events occurring in this sharply changing current region may be unidentifiable. To improve the temporal resolution for the detection of fast events, the RC time of the setup requires optimization. Meanwhile, the application of low voltage can significantly prolong the unfolding duration for identifying fast, partially folded structures. Second, it could be difficult to discriminate different folding structures that are similar in unfolding duration (Fig. 3a) due to their large population overlap in the duration histogram. One solution is to use the blocking level as an “identifier” to discriminate different folding structures. As verified in Supplementary Fig. 4, different lengths and positions of RNA in a DNA–RNA chimera can be clearly discriminated based on their characteristic blocking levels in the nanopore, providing the potential to use the blocking level for precisely identifying RNA folding states. Third, how to apply this approach for long RNA investigations remains an issue. We anticipate that combining it with alternative methods, such as nanopore sequencing14, may help to precisely read RNA positions and report various folding states. Notably, synthetic nanopores with tunable dimensions have been developed recently to characterize drug-induced RNA conformational changes56. Combined with machine learning, synthetic nanopores can discriminate different tRNAs57.
From the theoretical aspect, a more physically reliable force field in the MD simulation is helpful to provide structural details for various potential folding intermediate states and to understand the nanopore unfolding current signature. The key points include non-canonical base pair interaction, multiple base–base interaction, nucleotide-ion interaction, freedom from native bias, the ability to account for the whole folding landscape, and more efficient conformational space sampling. Furthermore, a direct simulation of the nanopore unfolding and translocation experiment using methods such as steered molecular dynamics could be helpful to understand the unfolding electric signatures of various structures and to establish a direct connection between the nanopore signature and RNA structure. These efforts would significantly improve the resolution of the nanopore technique, making it possible to more accurately reveal RNA intermediate states and folding pathways.
The improved system has the potential to study a variety of disease-relevant RNA and DNA tertiary structures, from hairpins58 and PKs59 to kissing-loops60 and G-quadruplexes61, and RNA-based interactions, from microRNA–target RNA interactions to ligand–RNA interactions, such as RNA repeats62 and riboswitches63. In addition to mutant construction, modified nucleotides can be introduced to detect chemical-specific folding procedures in the nanopore. Therefore, the nanopore can be potentially combined with chemical approaches, such as selective 2ʹ-hydroxyl acylation analyzed by primer extension (SHAPE)64, for joint structure exploration. Most notably, our nanopore system can provide very small forces (by applying low voltage) to detect weak interactions in small RNA structures. As demonstrated by this and previous studies32, this system is also able to characterize non-canonical base pairs involved in tertiary structure formation and dissociation. Finally, this system may be generalized to investigate the folding of other biomolecules, including polymers, peptides, and even proteins. Overall, this method can find applications in biomolecular folding investigations and related areas such as pharmaceutical kinetic investigations and drug development.
Methods
Materials and probe formation
All chemicals, including NaCl, MgCl2, 3-(N-morpholino) propanesulfonic acid (MOPS), and diethylpyrocarbonate (DEPC), were purchased from Sigma-Aldrich (St. Louis, MO, USA) and used as received. Lipid 1,2-diphytanoyl-sn-glycero-3-phosphocholine for bilayer formation was purchased from Avanti Polar Lipids (Alabaster, AL, USA) and used without further purification. All biotinylated RNA–DNA chimeras (Table 1) were synthesized and purified by Integrated DNA Technologies Inc. (Coralville, IA, USA), and dissolved in Millipore water to a stock concentration of 100 μM. Streptavidin was purchased from ProSpec-Tany Technogene Ltd. (East Brunswick, NJ, USA). The recording solution contained 1 M NaCl, 25 mM MOPS, and 10 mM MgCl2 (pH 7.4). In the magnesium effect experiment, different concentrations of MgCl2 from 0 to 10 mM were used in the recording solution. In total, 5 μL of the chimera stock solution was directly released to the cis solution to reach a final concentration of 250 nM. Then, 25 μL of 100 μM streptavidin solution (in 100 mM NaCl, 25 mM MOPS, pH 7.4) was added to the cis compartment. The mixture was incubated for 15 min in order for streptavidin to bind the chimera and form the complex.
Nanopore formation and measurement
The nanopore recording chamber was assembled by two Teflon compartments that were separated by a thin Teflon partition film (Goodfellow Corp., Coraopolis, PA). Each compartment was filled with the recording solution to support the lipid bilayer formation and facilitate the ionic current flow. A lipid bilayer membrane was formed spanning a 150 μm orifice fabricated in the center of the partition. About 1 μL of the α-hemolysin protein solution was released into the cis compartment. The protein can be spontaneously inserted into the lipid membrane to assemble a single nanopore. The ionic current through the nanopore at various transmembrane voltages was monitored by an Axopatch 200B amplifier (Molecular Devices Inc., Sunnyvale, CA, USA) and acquired by a DigiData 1440A A/D converter (Molecular Devices), filtered with a built-in 4-pole low-pass Bessel filter at 5 kHz with a sampling rate of 20 kHz. A Clampex software (Molecular Devices) was used for the data recording and acquisition, and a Clampfit software (Molecular Devices) was used for analyzing the nanopore current traces, including event duration histogram analysis and amplitude histogram analysis.
Extraction of folding state lifetimes
The folding of RNA to a specific tertiary state in the pathway is a time-dependent procedure. Tracking the time-dependent formation of each state in the nanopore allows the establishment of the RNA folding pathway. This process is realized by stopping the folding procedure after a given folding time (t fold), and each folding state can occur with a specific probability (P). The folding procedure can be stopped at an intermediate structure or the native PK structure with a probability (P) by pulling the probe reversely in the trans-to-cis direction with a negative voltage (–60 mV). Under this pulling force, the formed folding structure can be disrupted. The duration from the beginning of voltage application to the block end for structure disruption is measured as the lifetime of the structure (τ). Each folding structure has a specific stability and can be identified from its lifetime. The lifetime of a folding structure follows the exponential distribution. According to previous studies65, 66, if the bin is log(t), the distribution can be expressed as
| M1 |
where the amplitude c is equivalent to the area covered by the distribution. For multiple states, the lifetime distribution can be expressed as
| M2 |
The fractional population of the state i, P i, can be obtained from
| M3 |
where P 0 is the population of the unfolded single-stranded state (SS), which is the beginning state of all RNA molecules measured, and 1–P 0 is the total fraction of RNA molecules that form various folding structures (including intermediate and PK structures). P 0 is obtained by measuring the number of SS events (N 0, events without blocks) divided by the total number of events collected.
We first constructed the lifetime distributions (histograms) for RNAs experiencing a folding time of 1 s, 2 s, 5 s, 10 s, 30 s, and 60 s and then fit each histogram with four components (HP1, HP2, TS, and KP) by using Eq. (M2). Finally, the fractional populations were obtained using Eq. (M3). For τ and fractional population, the mean and standard deviation (SD) were obtained from at least three experiments (n ≥ 3) with independent nanopores. All nanopore pulling experiments were conducted at room temperature (22 ± 1 °C).
Molecular dynamics simulation method
Based on a CG RNA model, where the pyrimidines/purines are represented by 4/5 CG beads and a knowledge-based force field, we simulated the folding of the wild-type T2 and T2-polyU mutation RNAs using Langevin dynamics with the modified LAMMPS packages67. To enhance the conformational sampling, we used Replica-Exchange MD (REMD) with 10 replicas for temperatures ranging from 175 K to 400 K. In the simulation, the total simulation time per replica was set to t = 1 μs, with an integration time-step Δt = 0.5 fs. To monitor the folding process, the conformational snapshots were collected every 50 ps. For every 25 ns simulation time interval, the collected snapshots (5000 structures in total) were submitted to the clustering procedure. Based on the pairwise root mean square deviation (RMSD) for all the CG beads within a cutoff of 5.0 Å, the top 50% low-energy structures (2500 structures for every simulation interval) were clustered to identify the typical intermediate states. The sequence of centroid structures of the clusters gave the folding pathway.
Master equation method
Based on the initial unfolded state, the native PK state, and the intermediates states, we constructed the master equation68,69 to estimate the transition rates between the different states. For a system of ω states, the master equation method considers the rate of population changes () for the ith state as the difference between the rates of entering and leaving the state:
| M4 |
where is the population of the ith state and and are the rate constants for the transitions from states j to i and from i to j, respectively. With a column vector =col (, …, ) to represent the population of each state, the master equation can be transformed into a matrix form . In that formula, is a rate matrix. Off-diagonal elements inside the matrix are defined as , and the diagonal elements are .
To obtain the time-dependent population of each state , we found the eigenvalues ( = 1,2, …, ω) and eigenvectors ( = 1,2, …, ω) of rate matrix . The time-dependent population of the different states can be calculated as , where the coefficients are determined from the initial population of each state.
For the T2 PK folding, our simulations suggested three intermediate states in addition to the initial unfolded and final folding states (see main text for details). For such a five-state system, the master equation involves a 5×5 rate matrix. By solving the master equation, we estimated the rate constants from the best fitted, experiment-determined population kinetics.
Parameter optimization
The fitness function F is defined as the RMSD between the experimentally determined population and the theoretically predicted population for each state:
| M5 |
where and are the experimentally measured and the theoretically predicted populations for state i at time t, respectively, and N=30 is the total count of experiment data (i.e., the number of time points sampled in the sum over time t in the fitness function).
One of the states in the master equation calculation is the unfolded state, denoted as SS (single-stranded state). In the experiment, the SS state may be a fraction of the total ensemble of “unfolded” conformations identified from the electric current. In the experiment, the population of the SS state is estimated from the events where the electric current is not blocked. Limited by the experimental conditions, such as the purity of the RNA sequence and the effect of DNA extension, it is possible that not all the SS RNA chains can be involved in the folding process. There exists a buffer state in the experiment, SS0, which should be deducted from the apparent total SS population in order to calculate the effective population of SS. Therefore, in the theory–experiment fitting process, the effective SS population is equal to the apparent (experimental data-derived) total SS population minus the population of the buffer state SS0. Our estimation based on the experimental data for the populations of the different states (see main text for the details) suggested that [SS0] is ∼4.2% of the total initial SS population. To avoid the possible trapping in local minima in the optimization procedure for the fitness function F, the parameter search process was repeated with different initial values. Additionally, to further confirm the fitted rate constants, we used alternative algorithms, such as the genetic algorithm, for the optimization. Comparisons between the different methods also led to the same consistent results.
Data availability
All data related to this manuscript are included in the main text and supplementary information and will also be available from the corresponding authors upon reasonable request.
Electronic supplementary material
Acknowledgements
We are grateful to the Coulter Translation Program at the University of Missouri and the National Institutes of Health for support of this work through HG009338 (L.-Q.G.), GM114204 (L.-Q.G.) and GM063732 (S.-J.C.).
Author contributions
X.Z., K.T. and L.-Q.G. designed the experiments. X.Z., K.T., R.S., X.D., A.J.B. and J.W. performed the experiments and analyzed the data. D.Z., C.Z. and S.-J.C. designed the simulations. D.Z. and C.Z. performed all the simulations. S.-J.C. and L.-Q.G. built the pathway model. X.Z., D.Z., S.-J.C. and L.-Q.G. wrote the paper. All authors contributed to the manuscript revision.
Competing interests
The authors declare no competing financial interests.
Footnotes
Xinyue Zhang and Dong Zhang contributed equally to this work.
Electronic supplementary material
Supplementary Information accompanies this paper at doi:10.1038/s41467-017-01588-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shi-Jie Chen, Email: chenshi@missouri.edu.
Li-Qun Gu, Email: gul@missouri.edu.
References
- 1.Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Mortimer SA, Kidwell MA, Doudna JA. Insights into RNA structure and function from genome-wide studies. Nat. Rev. Genet. 2014;15:469–479. doi: 10.1038/nrg3681. [DOI] [PubMed] [Google Scholar]
- 3.Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 4.Baird NJ, et al. Discrete structure of an RNA folding intermediate revealed by cryo-electron microscopy. J. Am. Chem. Soc. 2010;132:16352–16353. doi: 10.1021/ja107492b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russell R, Millett IS, Doniach S, Herschlag D. Small angle X-ray scattering reveals a compact intermediate in RNA folding. Nat. Struct. Biol. 2000;7:367–370. doi: 10.1038/80691. [DOI] [PubMed] [Google Scholar]
- 6.Xue Y, Gracia B, Herschlag D, Al-Hashimi HM, Russell R. Visualizing the formation of an RNA folding intermediate through a fast highly modular secondary structure switch. Nat. Commun. 2000;7:11768. doi: 10.1038/ncomms11768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haller A, Soulière MF, Micura R. The dynamic nature of RNA as key to understanding riboswitch mechanisms. Acc. Chem. Res. 2011;44:1339–1348. doi: 10.1021/ar200035g. [DOI] [PubMed] [Google Scholar]
- 8.Woodson SA. RNA folding pathways and the self-assembly of ribosomes. Acc. Chem. Res. 2011;44:1312–1319. doi: 10.1021/ar2000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keller BG, Kobitski A, Jäschke A, Nienhaus GU, Noé F. Complex RNA folding kinetics revealed by single-molecule FRET and hidden markov models. J. Am. Chem. Soc. 2014;136:4534–4543. doi: 10.1021/ja4098719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neupane K, et al. Direct observation of transition paths during the folding of proteins and nucleic acids. Science. 2016;352:239–242. doi: 10.1126/science.aad0637. [DOI] [PubMed] [Google Scholar]
- 11.Cherf GM, et al. Automated forward and reverse ratcheting of DNA in a nanopore at 5-A precision. Nat. Biotechnol. 2012;30:344–348. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke J, et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 2009;4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 13.Manrao EA, et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat. Biotechnol. 2012;30:349–353. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laszlo AH, et al. Decoding long nanopore sequencing reads of natural DNA. Nat. Biotechnol. 2014;32:829–833. doi: 10.1038/nbt.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuller CW, et al. Real-time single-molecule electronic DNA sequencing by synthesis using polymer-tagged nucleotides on a nanopore array. Proc. Natl Acad. Sci. USA. 2016;113:5233–5238. doi: 10.1073/pnas.1601782113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao C, et al. Discrimination of oligonucleotides of different lengths with a wild-type aerolysin nanopore. Nat. Nanotechnol. 2016;11:713–718. doi: 10.1038/nnano.2016.66. [DOI] [PubMed] [Google Scholar]
- 17.Liu L, Wu H-C. DNA-based nanopore sensing. Angew. Chem. Int. Ed. 2016;55:15216–15222. doi: 10.1002/anie.201604405. [DOI] [PubMed] [Google Scholar]
- 18.Wanunu M, et al. Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors. Nat. Nanotechnol. 2010;5:807–814. doi: 10.1038/nnano.2010.202. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Zheng D, Tan Q, Wang MX, Gu L-Q. Nanopore-based detection of circulating microRNAs in lung cancer patients. Nat. Nanotechnol. 2011;6:668–674. doi: 10.1038/nnano.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Wang Y, Fricke BL, Gu L-Q. Programming nanopore ion flow for encoded multiplex microRNA detection. ACS Nano. 2014;8:3444–3450. doi: 10.1021/nn406339n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith AM, Abu-Shumays R, Akeson M, Bernick DL. Capture, unfolding, and detection of individual tRNA molecules using a nanopore device. Front. Bioeng. Biotechnol. 2015;3:91. doi: 10.3389/fbioe.2015.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Q, de Zoysa RSS, Wang D, Jayawardhana DA, Guan X. Real-time monitoring of peptide cleavage using a nanopore probe. J. Am. Chem. Soc. 2009;131:6324–6325. doi: 10.1021/ja9004893. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, et al. ACS Appl. Mater. Interfaces. 2015. Nanopore sensing of botulinum toxin type b by discriminating an enzymatically cleaved peptide from a synaptic protein synaptobrevin 2 derivative; pp. 184–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bell NAW, Keyser UF. Digitally encoded DNA nanostructures for multiplexed, single-molecule protein sensing with nanopores. Nat. Nanotechnol. 2016;11:645–651. doi: 10.1038/nnano.2016.50. [DOI] [PubMed] [Google Scholar]
- 25.Soskine M, Biesemans A, Maglia G. Single-molecule analyte recognition with ClyA nanopores equipped with internal protein adaptors. J. Am. Chem. Soc. 2015;137:5793–5797. doi: 10.1021/jacs.5b01520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shim J, et al. Detection and quantification of methylation in DNA using solid-state nanopores. Sci. Rep. 2013;3:1389. doi: 10.1038/srep01389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laszlo AH, et al. Detection and mapping of 5-methylcytosine and 5-hydroxymethylcytosine with nanopore MspA. Proc. Natl Acad. Sci. USA. 2013;110:18904–18909. doi: 10.1073/pnas.1310240110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, et al. Single molecule investigation of Ag+ interactions with single cytosine-, methylcytosine- and hydroxymethylcytosine-cytosine mismatches in a nanopore. Sci. Rep. 2014;4:5883. doi: 10.1038/srep05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez-Larrea D, Bayley H. Protein co-translocational unfolding depends on the direction of pulling. Nat. Commun. 2014;5:4841. doi: 10.1038/ncomms5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin Q, Fleming AM, Burrows CJ, White HS. Unzipping kinetics of duplex DNA containing oxidized lesions in an α-hemolysin nanopore. J. Am. Chem. Soc. 2012;134:11006–11011. doi: 10.1021/ja304169n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Tian K, Hunter LL, Ritzo B, Gu L-Q. Probing molecular pathways for DNA orientational trapping, unzipping and translocation in nanopores by using a tunable overhang sensor. Nanoscale. 2014;6:11372–11379. doi: 10.1039/C4NR03195D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, et al. Mimicking ribosomal unfolding of RNA Pseudoknot in a protein channel. J. Am. Chem. Soc. 2015;137:15742–15752. doi: 10.1021/jacs.5b07910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hornblower B, et al. Single-molecule analysis of DNA-protein complexes using nanopores. Nat. Methods. 2007;4:315–317. doi: 10.1038/nmeth1021. [DOI] [PubMed] [Google Scholar]
- 34.Holland JA, Hansen MR, Du Z, Hoffman DW. An examination of coaxial stacking of helical stems in a pseudoknot motif: the gene 32 messenger RNA pseudoknot of bacteriophage T2. RNA. 1999;5:257–271. doi: 10.1017/S1355838299981360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nixon PL, Giedroc DP. Equilibrium unfolding (folding) pathway of a model H-type Pseudoknotted RNA: the role of magnesium ions in stability. Biochemistry. 1998;37:16116–16129. doi: 10.1021/bi981726z. [DOI] [PubMed] [Google Scholar]
- 36.Stoddart D, Heron AJ, Mikhailova E, Maglia G, Bayley H. Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proc. Natl Acad. Sci. USA. 2009;106:7702–7707. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakane J, Wiggin M, Marziali A. A Nanosensor for transmembrane capture and identification of single nucleic acid molecules. Biophys. J. 2004;87:615–621. doi: 10.1529/biophysj.104.040212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schibel AEP, et al. Nanopore Detection of 8-oxo-7,8-dihydro-2ʹdeoxyguanosine in immobilized single-stranded DNA via adduct formation to the DNA damage site. J. Am. Chem. Soc. 2010;132:17992–17995. doi: 10.1021/ja109501x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henrickson SE, DiMarzio EA, Wang Q, Stanford VM, Kasianowicz JJ. Probing single nanometer-scale pores with polymeric molecular rulers. J. Chem. Phys. 2010;132:135101. doi: 10.1063/1.3328875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schink S, et al. Quantitative analysis of the nanopore translocation dynamics of simple structured polynucleotides. Biophys. J. 2012;102:85–95. doi: 10.1016/j.bpj.2011.11.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song L, et al. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1865. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 42.Frieda KL, Block SM. Direct observation of cotranscriptional folding in an adenine riboswitch. Science. 2012;338:397–400. doi: 10.1126/science.1225722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding Y, Fleming AM, He L, Burrows CJ. Unfolding kinetics of the human telomere i-Motif under a 10 pN force imposed by the α-hemolysin nanopore identify transient folded-state lifetimes at physiological pH. J. Am. Chem. Soc. 2015;137:9053–9060. doi: 10.1021/jacs.5b03912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Dorp S, Keyser UF, Dekker NH, Dekker C, Lemay SG. Origin of the electrophoretic force on DNA in solid-state nanopores. Nat. Phys. 2009;5:347–351. doi: 10.1038/nphys1230. [DOI] [Google Scholar]
- 45.Xu X, Zhao P, Chen S-J. Vfold: a web server for RNA structure and folding thermodynamics prediction. PLoS ONE. 2014;9:e107504. doi: 10.1371/journal.pone.0107504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boniecki MJ, et al. SimRNA: a coarse-grained method for RNA folding simulations and 3D structure prediction. Nucleic Acids Res. 2016;44:e63. doi: 10.1093/nar/gkv1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao S, Chen S-J. Predicting RNA folding thermodynamics with a reduced chain representation model. RNA. 2005;11:1884–1897. doi: 10.1261/rna.2109105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao S, Chen S-J. Predicting RNA pseudoknot folding thermodynamics. Nucleic Acids Res. 2006;34:2634–2652. doi: 10.1093/nar/gkl346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cornish PV, Hennig M, Giedroc DP. A loop 2 cytidine-stem 1 minor groove interaction as a positive determinant for pseudoknot-stimulated –1 ribosomal frameshifting. Proc. Natl Acad. Sci. USA. 2005;102:12694–12699. doi: 10.1073/pnas.0506166102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y, Wilson TJ, Lilley DMJ. The structure of a nucleolytic ribozyme that employs a catalytic metal ion. Nat. Chem. Biol. 2017;13:508–513. doi: 10.1038/nchembio.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu Z, et al. Directional mechanical stability of Bacteriophage φ29 motor’s 3WJ-pRNA: Extraordinary robustness along portal axis. Sci. Adv. 2017;3:e1601684. doi: 10.1126/sciadv.1601684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Batra VK, et al. Magnesium-induced assembly of a complete DNA polymerase catalytic complex. Structure. 2006;14:757–766. doi: 10.1016/j.str.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, Zhang J, Wang W. Atomistic analysis of pseudoknotted RNA unfolding. J. Am. Chem. Soc. 2011;133:6882–6885. doi: 10.1021/ja1109425. [DOI] [PubMed] [Google Scholar]
- 54.Cho SS, Pincus DL, Thirumalai D. Assembly mechanisms of RNA pseudoknots are determined by the stabilities of constituent secondary structures. Proc. Natl Acad. Sci. USA. 2009;106:17349–17354. doi: 10.1073/pnas.0906625106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. Functional complexity and regulation through RNA dynamics. Nature. 2012;482:322–330. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shasha C, et al. Nanopore-based conformational analysis of a viral RNA drug target. ACS Nano. 2014;8:6425–6430. doi: 10.1021/nn501969r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gareau D, Desrosiers A, Vallée-Bélisle A. Programmable quantitative DNA nanothermometers. Nano Lett. 2016;16:3976–3981. doi: 10.1021/acs.nanolett.6b00156. [DOI] [PubMed] [Google Scholar]
- 58.Marquis Gacy A, Goellner G, Juranić N, Macura S, McMurray CT. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 59.Vulliamy T, Marrone A, Dokal I, Mason PJ. Association between aplastic anaemia and mutations in telomerase RNA. Lancet. 2002;359:2168–2170. doi: 10.1016/S0140-6736(02)09087-6. [DOI] [PubMed] [Google Scholar]
- 60.Tsetsarkin KA, Liu G, Shen K, Pletnev AG. Kissing-loop interaction between 5′ and 3′ ends of tick-borne Langat virus genome “bridges the gap” between mosquito- and tick-borne flaviviruses in mechanisms of viral RNA cyclization: applications for virus attenuation and vaccine development. Nucleic Acids Res. 2016;44:3330–3350. doi: 10.1093/nar/gkw061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cammas A, Millevoi S. RNA G-quadruplexes: emerging mechanisms in disease. Nucleic Acids Res. 2017;45:1584–1595. doi: 10.1093/nar/gkw1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Błaszczyk L, Rypniewski W, Kiliszek A. Structures of RNA repeats associated with neurological diseases. Wiley Interdiscip. Rev. RNA. 2017;8:e1412. doi: 10.1002/wrna.1412. [DOI] [PubMed] [Google Scholar]
- 63.Garst AD, Edwards AL, Batey RT. Riboswitches: structures and mechanisms. Cold Spring Harb. Perspect. Biol. 2011;3:a003533. doi: 10.1101/cshperspect.a003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 65.Blatz AL, Magleby KL. Quantitative description of three modes of activity of fast chloride channels from rat skeletal muscle. J. Physiol. 1986;378:141–174. doi: 10.1113/jphysiol.1986.sp016212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sigworth FJ, Sine SM. Data transformations for improved display and fitting of single-channel dwell time histograms. Biophys. J. 1987;52:1047–1054. doi: 10.1016/S0006-3495(87)83298-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Plimpton S. Fast parallel algorithms for short-range molecular dynamics. J. Comput. Phys. 1995;117:1–19. doi: 10.1006/jcph.1995.1039. [DOI] [Google Scholar]
- 68.Zhang W, Chen S-J. RNA hairpin-folding kinetics. Proc. Natl Acad. Sci. USA. 2002;99:1931–1936. doi: 10.1073/pnas.032443099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cieplak M, Henkel M, Karbowski J, Banavar JR. Master equation approach to protein folding and kinetic traps. Phys. Rev. Lett. 1998;80:3654–3657. doi: 10.1103/PhysRevLett.80.3654. [DOI] [Google Scholar]
- 70.Yang H, et al. Tools for the automatic identification and classification of RNA base pairs. Nucleic Acids Res. 2003;31:3450–3460. doi: 10.1093/nar/gkg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data related to this manuscript are included in the main text and supplementary information and will also be available from the corresponding authors upon reasonable request.