Abstract
MicroRNA-7 (miR-7) is a non-coding RNA of 23-nucleotides that has been shown to act as a tumor suppressor in various cancers including breast cancer. Although there have been copious studies on the action mechanisms of miR-7, little is known about how the miR is controlled in the mammary cell. In this study, we performed a genome-wide expression analysis in miR-7-transfected MCF-10A breast cell line to explore the upstream regulators of miR-7. Analysis of the dysregulated target gene pool predicted hepatocyte growth factor (HGF) as the most plausible upstream regulator of miR-7. MiR-7 was upregulated in MCF-10A cells by HGF, and subsequently downregulated upon treatment with siRNA against HGF. However, the expression of HGF did not significantly change through either an upregulation or downregulation of miR-7 expression, suggesting that HGF acts upstream of miR-7. In addition, the target genes of miR-7, such as EGFR, KLF4, FAK, PAK1 and SET8, which are all known oncogenes, were downregulated in HGF-treated MCF-10A; in contrast, knocking down HGF recovered their expression. These results indicate that miR-7 mediates the activity of HGF to suppress oncogenic proteins, which inhibits the development of normal cells, at least MCF-10A, into cancerous cells.
Introduction
MicroRNAs (miRs) are small non-coding RNAs that are approximately 20–25 nucleotides long, with over 2500 mature forms identified in the human genome1. MiRs have the capacity to regulate numerous target genes at a post-transcriptional level and sequence-specific manner by binding to the 3′-untranslated regions of target mRNAs2. MiRs are involved in diverse biological process including cell development, proliferation and carcinogenesis, and have thus emerged as potential diagnostic markers and therapeutic targets for various cancers3. MiR-7 is derived from three primary transcripts, namely pri-miR-7-1, pri-miR-7-2 and pri-miR-7-3, which are all encoded from different genomic loci, 9q21, 15q26 and 19q13, respectively4. Downregulation of miR-7 has been revealed in various cancer types including breast, gastric and lung cancer5–7, with lower miR-7 transcript levels associated with poorer prognosis in cancer patients. MiR-7 shows tumor suppressive activity by targeting many oncogenes such as EGFR, FAK, PAK1, KLF4 and SET8 in breast and other cancers8,9. In line with its cancer suppressive activity, the overexpression of miR-7 mimics showed a decreased cell growth and invasion, while those overexpressing miR-7-antisense showed the opposite effects10,11.
Hepatocyte growth factor (HGF) stimulates cell proliferation and differentiation of various cell types12. For example, the differentiation of bone mesenchymal stem cells (BMSCs) can be induced through HGF-activated NF-кB signaling13. In mouse and human mammary glands, HGF acts as a morphogenic factor in mammary epithelial cells, generating tubular branching and well-constructed lumina during differentiation and development14. Upregulation of HGF has been frequently observed in various cancers wherein higher expression of HGF is associated with poorer prognosis in cancer patients15. In contrast, the tumor suppressive functions of HGF in cancer is sparsely known. A recent study has indicated that an HGF-regulated tyrosine kinase substrate harboring tumor suppressive activity is regulated by HGF stimulation16. HGF binds to its only receptor, Met, and thereafter initiates a series of signaling pathways that include the activation of Erk1/2 and Wnt/β-catenin signaling, as well as the AKT pathway17.
A few miRs have been identified to mediate the activity of HGF. One example is miR-124, which is downregulated in HGF-treated mesenchymal stem cells (MSC)18. MiR-124 downregulates Wnt/β-catenin signaling by targeting FZD4 and LRP6, thus suppressing the chemotactic migration of rat MSCs toward HGF. MiR-211 and miR-26b are also regulated by HGF in MSC, but are instead upregulated by HGF to activate PI3K/Akt signaling through targeting PTEN19. In renal cell carcinoma, miR-199a-3p inhibited HGF/c-Met signaling, which included the STAT3, mTOR and ERK1/2 pathways; however, whether miR-199a-3p itself is regulated by HGF has not yet been elucidated20.
Albeit the battery of experimental results that investigate the biological functions of miR-7 and HGF in cancer cells, there are presently no reported association between them. In this study, we explored the target genes of miR-7, which then identified HGF as an upstream regulator of the miR. This observation was then supported by the downregulation of miR-7 after inhibition of HGF using siRNA in MCF-10A mammary cells. In addition, the expression profile of miR-7 target genes was examined after the upregulation or downregulation of HGF.
Results
MiR-7 affects genes involved in the cell cycle, cellular movement, cellular assembly and organization pathways
The tumor suppressive activities of miR-7 act through specific target genes, a few of which have been identified thus far. To comprehensively understand the regulatory mechanism through which miR-7 functions in breast tissues, we performed a genome-wide expression analysis in a mammary epithelial cell line, MCF-10A, after overexpressing miR-7 using a mimic miR. Among the 47000 probes on the Illumina Expression BeadChip, 343 genes satisfied our criteria with P < 0.05 and |fold change| ≥2 compared to the control miR-transfected cells. An Ingenuity Pathway Analysis was conducted on the gene set, which resulted in the “cell cycle, cellular movement, cellular assembly and organization” network with the highest confidence (Fig. 1 and Table 1). Reassuringly, many genes that were previously identified as miR-7 targets appeared in the network, with an expression coincidence as previously claimed. In detail, MUC16, SLC2A5, SMAD6 and NCEH1 were downregulated by miR-7 and are known to possess oncogenic activities in breast cancer21,22, as well as a few other cancers23. Notably, “Mitotic roles of polo-like kinase (PLK)” was identified as the most significant canonical pathway (Fig. 2), which represents a highly conserved family of several serine/threonine kinases that regulate cell division, and are often overexpressed in tumors of various cancers24. In support of this, genes such as PAK1, FBXO5, CDK1 and KIF23, which directly or indirectly interact with PLK to regulate cell division, were observed to be dysregulated in the network (Fig. 1)25. In addition to the IPA, KEGG was also used for the pathway analysis, which identified “cell cycle”-related pathways as the top ones as like as the IPA (Supplementary Fig. S1).
Figure 1.
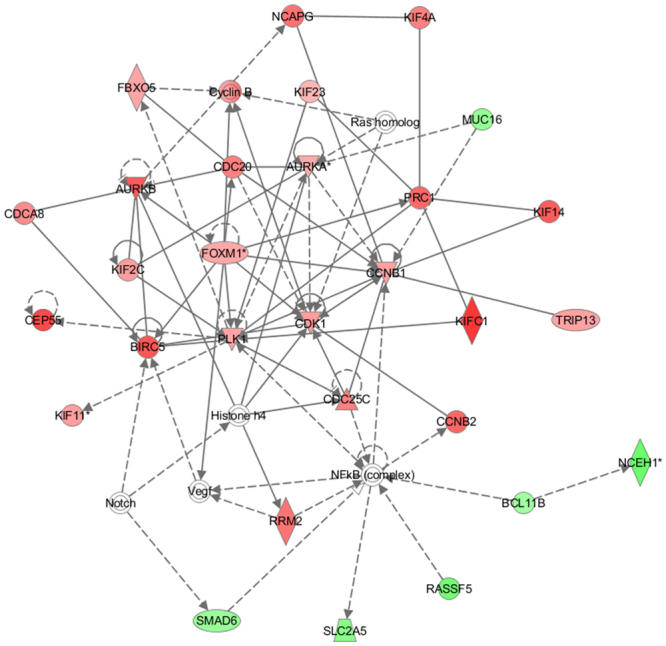
The highest confidence network of genes significantly dysregulated by miR-7. The Ingenuity Pathway Analysis was carried out with the 343 genes that showed significant expression alteration by miR-7 in the MCF-10A cells. The top network identified is “Cell Cycle, Cellular Assembly and Organization, DNA Replication, Recombination, and Repair”. Shapes of red and green color represent upregulated and downregulated genes, respectively. Solid and dashed lines represent direct and indirect interactions, respectively.
Table 1.
Genes in the top IPA network showing altered expression in MCF-10A cells due to miR-7.
| Gene symbol | Accession no. | Description | Expression fold change a |
|---|---|---|---|
| KIFC1 | NM_002263.2 | kinesin family member C1 | 5.478773 |
| CEP55 | NM_018131.3 | centrosomal protein 55 | 4.961733 |
| BIRC5 | NM_001168.2 | baculoviral IAP repeat containing 5 | 4.600387 |
| AURKB | NM_004217.2 | aurora kinase B | 4.471065 |
| KIF14 | NM_014875.1 | kinesin family member 14 | 4.438285 |
| CCNB2 | NM_004701.2 | cyclin B2 | 4.161032 |
| PRC1 | NM_199413.1 | protein regulator of cytokinesis 1 | 4.133515 |
| NCAPG | NM_022346.3 | non-SMC condensin I complex subunit G | 3.857997 |
| RRM2 | NM_001034.1 | ribonucleotide reductase regulatory subunit M2 | 3.80439 |
| KIF4A | NM_012310.3 | kinesin family member 4A | 3.478862 |
| CDC20 | NM_001255.2 | cell division cycle 20 | 3.475324 |
| CDC25C | NM_022809.2 | cell division cycle 25C | 3.46151 |
| CDCA8 | NM_018101.2 | cell division cycle associated 8 | 3.131205 |
| CDK1 | NM_001786.2 | cyclin dependent kinase 1 | 2.689246 |
| KIF2C | NM_006845.2 | kinesin family member 2C | 2.67569 |
| KIF11 | NM_004523.2 | kinesin family member 11 | 2.620285 |
| CCNB1 | NM_031966.2 | cyclin B1 | 2.587337 |
| TRIP13 | NM_004237.2 | thyroid hormone receptor interactor 13 | 2.548576 |
| FBXO5 | NM_012177.2 | F-box protein 5 | 2.448304 |
| PLK1 | NM_005030.3 | polo like kinase 1 | 2.413278 |
| FOXM1 | NM_021953.2 | forkhead box M1 | 2.409883 |
| AURKA | NM_198436.1 | aurora kinase A | 2.226352 |
| KIF23 | NM_004856.4 | kinesin family member 23 | 2.022938 |
| KIFC1 | NM_002263.2 | kinesin family member C1 | 5.478773 |
| CEP55 | NM_018131.3 | centrosomal protein 55 | 4.961733 |
| BIRC5 | NM_001168.2 | baculoviral IAP repeat containing 5 | 4.600387 |
| AURKB | NM_004217.2 | aurora kinase B | 4.471065 |
| KIF14 | NM_014875.1 | kinesin family member 14 | 4.438285 |
| CCNB2 | NM_004701.2 | cyclin B2 | 4.161032 |
| PRC1 | NM_199413.1 | protein regulator of cytokinesis 1 | 4.133515 |
| BCL11B | NM_138576.2 | B cell leukemia/lymphoma 11B | −2.06768 |
| SMAD6 | NM_005585.3 | SMAD family member 6 | −2.25726 |
| MUC16 | NM_024690.2 | mucin 16, cell surface associated | −2.31952 |
| SLC2A5 | NM_003039.1 | solute carrier family 2 member 5 | −2.56213 |
| RASSF5 | NM_182664.2 | Ras association domain family member 5 | −3.02865 |
| NCEH1 | NM_020792.3 | neutral cholesterol ester hydrolase 1 | −3.19179 |
aThe values are obtained by dividing the expression level in miR-7-overexpressed MCF-10A by that in MCF-10A.
Figure 2.
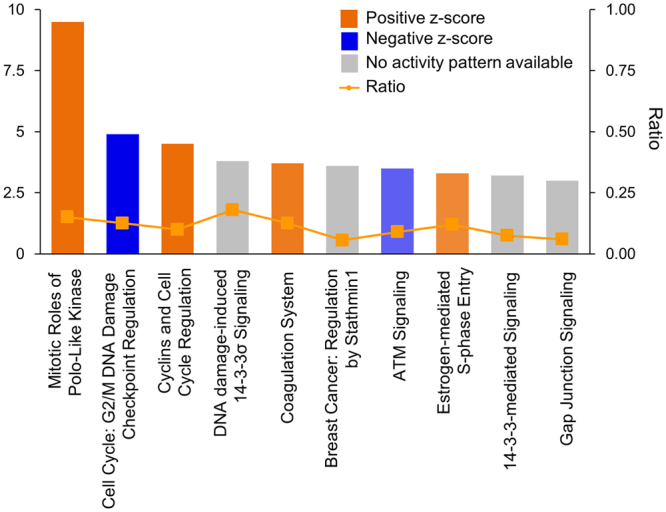
Top 10 canonical pathways of the genes significantly dysregulated by miR-7. The most significant canonical pathway is “Mitotic Roles of Polo-Like Kinase”. Pathways with positive and negative z-scores indicate that the pathways are activated and inhibited, respectively. Ratio is calculated as the number of genes that overlap with the corresponding pathway.
HGF acts as an upstream regulator of miR-7
In order to investigate how miR-7 is modulated, we identified upstream regulators of the miR by performing an IPA’s upstream regulator/mediator analysis of the 343 deregulated genes in the pool. Among 20 putative regulators, HGF had the highest activation z-score (Table 2), with 32 genes affected when HGF was activated or inactivated (Supplementary Table S1). For example, SERPINE1, which was downregulated by miR-7 (2.5-fold decrease), is known to suppress HGF by activating the cleavage of HGF26. For the upregulated genes, TPX2 (2.8-fold increase) plays a critical role in the chromosome segregation machinery during mitosis and suppresses tumor cell growth27.
Table 2.
Potential upstream regulators or mediators of miR-7 predicted by target gene analysis.
| Upstream regulator | Molecular type | Predicted activation state a | Activation z-score | P-value of overlap |
|---|---|---|---|---|
| HGF | growth factor | Activated | 4.953 | 2.25E-16 |
| RABL6 | other | Activated | 4.796 | 4.91E-25 |
| CSF2 | cytokine | Activated | 4.7 | 9.42E-16 |
| PTGER2 | g-protein coupled receptor | Activated | 4.459 | 1.52E-28 |
| Vegf | group | Activated | 4.31 | 1.96E-14 |
| RARA | ligand-dependent nuclear receptor | Activated | 4.123 | 5.77E-08 |
| ESR1 | ligand-dependent nuclear receptor | Activated | 3.963 | 8.66E-10 |
| FOXM1 | transcription regulator | Activated | 3.696 | 4.82E-19 |
| E2f | group | Activated | 3.687 | 1.86E-16 |
| MITF | transcription regulator | Activated | 3.483 | 4.47E-13 |
| BNIP3L | other | Inhibited | −3.317 | 8.12E-10 |
| CDKN1A | kinase | Inhibited | −3.519 | 2.07E-33 |
| Irgm1 | other | Inhibited | −3.592 | 1.24E-14 |
| KDM5B | transcription regulator | Inhibited | −3.601 | 3.74E-08 |
| let-7 | microrna | Inhibited | −3.734 | 4.12E-15 |
| CDKN2A | transcription regulator | Inhibited | −3.764 | 3.21E-11 |
| phorbol myristate acetate | chemical drug | Inhibited | −4.448 | 2.89E-07 |
| TP53 | transcription regulator | Inhibited | −4.529 | 5.91E-26 |
| NUPR1 | transcription regulator | Inhibited | −5.488 | 6.02E-13 |
| calcitriol | chemical drug | Inhibited | −5.68 | 1.86E-25 |
aThe status of the regulator that results in the same expression profile of the target genes of miR-7 in the miR-7-overexpressing MCF-10A.
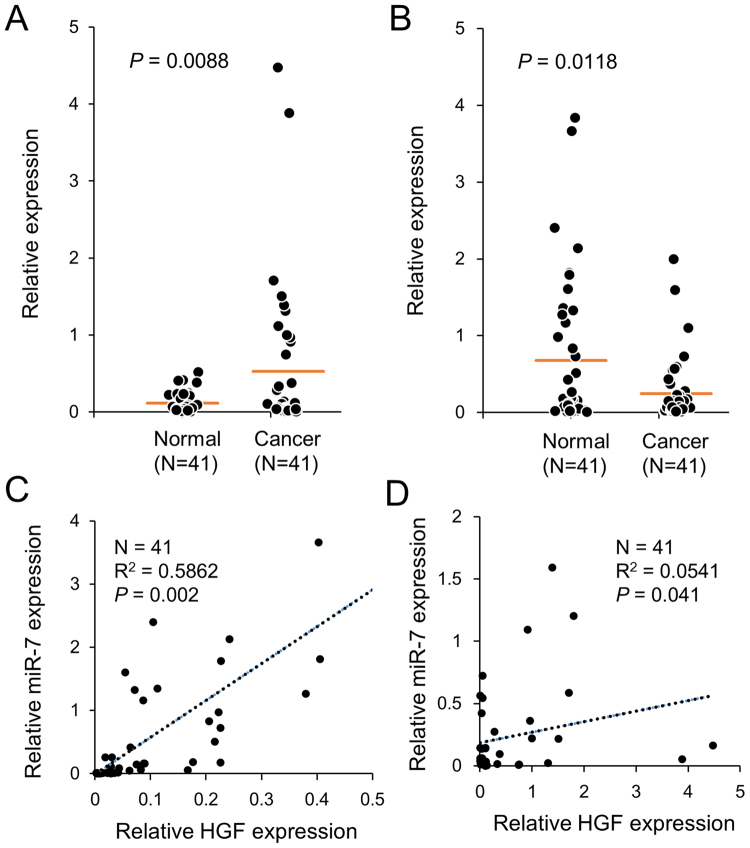
The regulatory relationship between miR-7 and HGF was also investigated by examining the expression of HGF and miR-7 in 41 pairs of breast cancer and normal tissues. HGF showed an increased expression in cancer tissues compared to normal tissues, while miR-7 showed the opposite pattern, confirming the observations in previous studies (Fig. 3A,B). Notably, comparing the expression between the two genes revealed a high positive association in the normal tissues (R 2 = 0.58, P < 0.01) but not in the cancer tissues (R 2 = 0.05, P < 0.05) (Fig. 3C,D). When examined in a few other tissues including liver, head and neck, colon, and bladder, no remarkable association was found regardless normal or caner tissue (Supplementary Fig. S2). These results imply that the association of HGF and miR-7 expression is stronger in normal tissues than in cancer tissues for breast and further that it is not a general phenomenon through body tissues.
Figure 3.
Expression of HGF and miR-7 is strongly associated in normal breast tissue. Expression of HGF (A) and miR-7 (B) was examined by qPCR in 41 pairs of breast cancer tissues and nearby normal tissues and represented by dot plots. Association between HGF and miR-7 was determined in breast normal (C) and cancer tissues (D) by linear regression, and represented by the coefficient of determination (R 2).
HGF controls expression of MiR-7 and its target genes
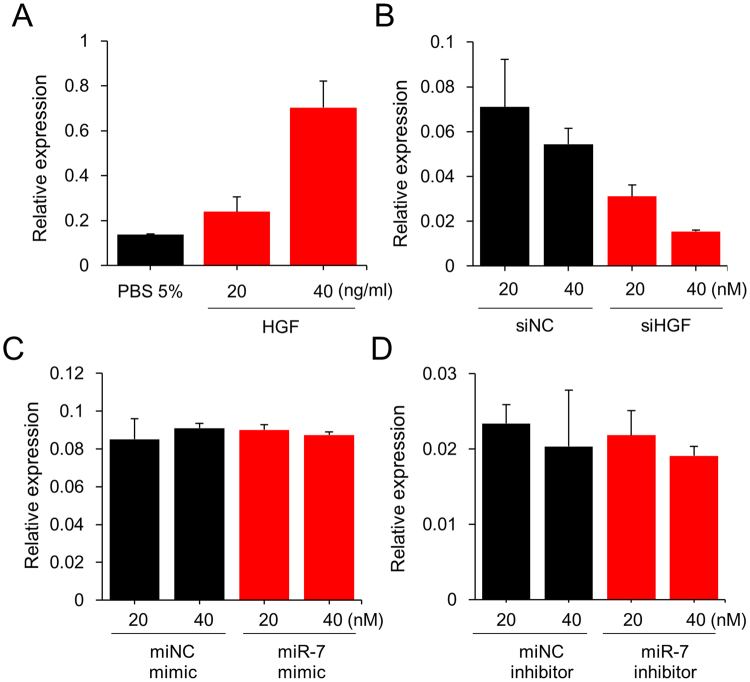
Based on the results that HGF and miR-7 share many target genes in common and their expression is strongly associated in normal breast tissue, we examined the effects of HGF in a normal breast cell line, MCF-10A. First, the effect of HGF and miR-7 on cell proliferation was examined. As results, HGF alone increased the growth rate of the cell, while miR-7 mimic alone decreased the growth rate. When miR-7 mimic was co-treated with HGF, it deteriorated the growth-stimulation effect of HGF (Supplementary Fig. S3). Next, we investigated the influence of HGF on the expression of miR-7 as well as its target genes such as FAK, PAK1, EGFR, KLF4 and SET8, which were all validated in our microarray assay, as well as in previous studies5,28–31. When MCF-10A cells were treated with 20 or 40 ng/ml HGF, a resulting upregulation of miR-7 was observed (Fig. 4A); on the other hand, a downregulation of miR-7 was also observed when cells were treated with siRNA against HGF (Fig. 4B and Supplementary Fig. S4A). Both occasions showed a dose-dependent response.
Figure 4.
HGF upregulates miR-7 in MCF-10A cells. (A) Cells were treated with HGF for 24 hours, after which the expression level of miR-7 was measured by qPCR. (B) HGF expression was downregulated with siRNA (siHGF) and the expression level of miR-7 was measured by qPCR 24 hours after transfection. siNC, control siRNA. (C and D) miR-7 was overexpressed using mimic miR or downregulated using an inhibitor miR. Twenty-four hours after treatment, the expression level of HGF was measured by qPCR. The expression levels of HGF and miR were normalized to GAPDH and U6 RNA, respectively. All experiments were performed in triplicate and indicated as mean ± SE. siNC, siRNA negative control; miNC, microRNA negative control.
To investigate whether the regulation of miR-7 by HGF functions in a feedback loop, we performed a pulse-chase experiment to check for HGF expression after miR-7 was either upregulated through mimic miR or downregulated through an inhibitor miR (Supplementary Fig. S4B,C). In both cases, no significant expression changes in HGF was observed, implying that there is no regulation of HGF by miR-7 (Fig. 4C,D).
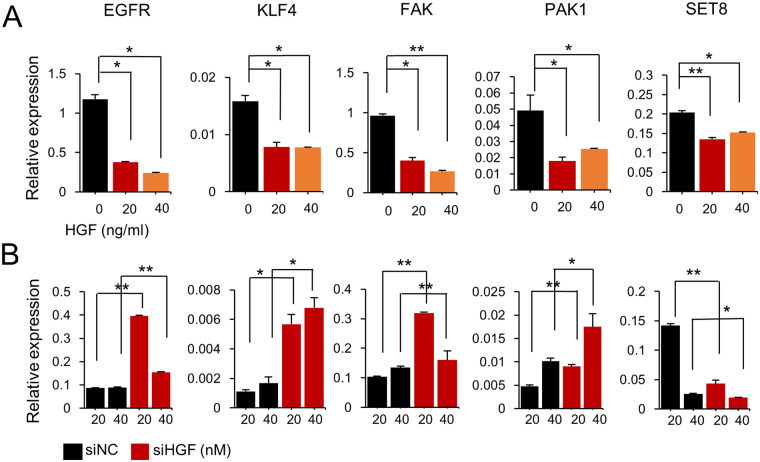
Five genes that are known to be direct targets of miR-7 were then selected, and the effect of HGF on their expression was monitored by qPCR. Treatment of HGF to MCF-10A cells resulted in a downregulation in all genes (Fig. 5A). On the other hand, when HGF was inhibited by siRNA, an upregulation was observed in the expression of all genes except for SET8 (Fig. 5B). Taken together, these results suggest that HGF downregulates cell proliferation-related genes by upregulating miR-7, which acts to suppress the cancer progression of normal breast cells.
Figure 5.
HGF regulates target genes of miR-7. The expression of selected target genes for miR-7 was examined after treatment of the MCF-10A cells with HGF protein (A) and siRNA (B) to induce upregulation or downregulation of HGF, respectively. Twenty-four hours after treatment, the expression level of target genes was measured by qPCR. All the target genes were downregulated by HGF but upregulated by siRNA against HGF, except for SET8. All experiments were performed in triplicates and indicated as mean ± SE. *P < 0.05; **P < 0.001.
Discussion
This study aimed to explore upstream regulators of miR-7, a microRNA that has shown tumor suppressor functions in many cancers including breast cancer. To accomplish this, we adopted a strategy wherein potential upstream regulators or mediators of miR-7 were identified by screening for commonly affected genes by miR-7 and its regulators. The potential regulators appeared in diverse subcellular locations, i.e., ligands such as HGF and VEGF, receptors on the cell membrane such as PTGER2, signaling kinases such as CDKN1A and CDKN2A, and transcription factors such as TP53 and MITF.
One remarkable characteristic of the identified potential regulators is sharing of the PI3K/AKT, MAPK, or JNK in common en route the signaling pathway. Especially, 13 genes including VEGF32, RARA33 and ESR134 activate the signaling pathway by regulating MAPK. A previous study also identified TGF-β signaling to be responsible for miR-7 inhibition in the MCF-7 breast cancer cell line35. The regulatory pathways from ligands and/or receptors therefore become complicated, and the detailed pathway from HGF to miR-7 should be identified in a future study. A few of the potential mediators of miR-7 have already been known to affect or be affected by miR-7. MiR-7 was shown to be regulated by estrogen and to target signaling intermediates such as EGFR, IGF1R and IRS-236. KLF4, a direct target of miR-7, acts on the VEGF promoter to induce its mRNA and protein levels. This miR-7-KLF4-VEGF signaling axis contributes to the regulation of angiogenesis in human umbilical vein endothelial cells37. Upregulation of miR-7 targeted genes implicated in the TP53 pathway, such as Ak1 and p21, also led to the controlled growth of cortical neural progenitors38.
HGF triggers multiple signaling pathways, including the conventional PI3K/AKT and MAPK pathways, coupled with the Hic-5-reactive oxygen species (ROS)-c-jun-N-terminal kinase (JNK) cascade39. These signaling events eventually increase the expression of a group of genes such as N-cadherin, vimentin and Zeb1, which trigger metastatic changes including epithelial mesenchymal transition (EMT), enhancement of motility and the invasiveness of tumor cells40. The majority of target genes activated by HGF have pro-proliferation activities, while those suppressed by HGF have anti-proliferation activities. HGF, therefore, has been generally considered as an oncogenic growth factor. HGF also has specific roles associated with cell proliferation in normal cells, even though these are less well-known than its oncogenic roles. Previous studies indicate that HGF increased mammary epithelial cell proliferation by acting through the PI3K-including mitogenic pathway, which thus induced a tubulo-ductal morphological response41.
In contrast to previous studies, the four selected oncogenes, EGFR, KLF4, FAK and PAK142–45 were downregulated by HGF in MCF-10A cells. Because our result suggest that the downregulation of the genes are mediated via miR-7, we speculate that HGF acts as a double-edged sword depending on cellular status or cell type. This idea is supported by the upregulation of miR-7 in the HGF-treated MCF-10A cells, and by the strong association between miR-7 and HGF expression in normal mammary tissues. There have also been previous studies indicating the tumor-suppressive activity of HGF by abrogating the oncogenic effects of c-Myc during early stages of liver carcinogenesis46, as well as enhancing the differentiation activity in mammary glands14 and hepatocytes13. Therefore, during tumorigenesis, upregulated HGF would drive the expression of oncogenes, while in normal cells, miR-7 would mediate HGF to downregulate the same oncogenes. Recently, effective computational models such as PBMDA47, HGIMDA48, and RKNNMDA49 have been constructed to identify disease-related miRNA biomarkers. These bioinformatics-based approaches should help to gain further insight into the molecular mechanisms of HGF and miR-7. In addition, dynamic feedback modeling50 and Cancer Hallmark Network Framework51 could give us an insight to better understand the feedback loop of miR-7 in breast cancer.
Our genome-wide analysis revealed that a set of oncogenes was downregulated by miR-7, while many tumor suppressors were upregulated, supporting the tumor suppressive activity of miR-7. In addition, a group of genes that was not previously identified as targets of miR-7 was discovered to rank at the top of the network. For example, FAM83A (2.4-fold decrease) is an oncogene that activates CRAF/MAPK signaling and drives epithelial transformation52, while MUC16 (2.3-fold decrease) is a tumor marker that induces breast cancer cell proliferation by interacting with JAK221. Whether these genes are directly regulated by miR-7, however, should be elucidated in a further study.
SET8, being different from other target genes of miR-7, was downregulated regardless of HGF overexpression or inhibition. This observation suggests the existence of other regulatory pathways in between HGF and SET8, which are independent of miR-7. SET8 is the sole protein lysine methyltransferase to monomethylate histone 4 lysine 20 (H4K20) and its function has been implicated in normal cell cycle progression and cancer metastasis53. Recently, a study revealed that miR-502 directly targets SET8 to suppress cell proliferation and cell cycle54.
In conclusion, HGF was identified as an upstream regulator of miR-7 due to their sharing of a group of target genes that showed similar gene expression changes. These genes include oncogenes such as EGRF, KLF4, FAK and PAK1, which were downregulated by either HGF or miR-7. In addition, there seems to be no feedback regulation of HGF by miR-7. Because miR-7 acts as a tumor suppressor, the HGF/miR-7 pathway could potentially explain the tumor-suppressive effects of HGF in normal cells.
Materials and Methods
Cell culture and transfection
The normal epithelial breast cell line MCF-10A was purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA) and cultured in MEBM basal medium (Lonza, Basel, Switzerland) supplemented with the MEGM Single Quot Kit (Lonza) and cholera toxin (List Biological Labs, Campbell, CA) under a humid environment with 5% CO2 at 37 °C. MiR-7-5p mimic, control miR (miNC), miR-7-5p inhibitor, control inhibitor (miNC inhibitor), siHGF and siRNA control (siNC) were synthesized by Bioneer (Korea). All miRs, inhibitors and siRNAs were diluted in OptiMEM I Medium (Gibco, Los Angeles, CA, USA) and transiently transfected into cells at a final concentrations of 20 and 40 nM using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA).
Study subjects
Forty-one breast cancer tissues were obtained from patients who underwent surgery between 2013 and 2014 at the National Cancer Center (NCC) in Korea. All patients provided written informed consent to donate removed tissue to NCC, and samples were obtained according to protocols approved by the Research Ethics Board of NCC.
RNA extraction and real-time qPCR
Total RNA was harvested from miR- or siRNA-transfected cells and breast tissues using the ZR-Duet DNA/RNA MiniPrep kit (Zymo research, Irvine, CA, USA) according to the manufacturer’s recommendations. In order to quantify the levels of mature miR-7, the extracted RNA was reverse transcribed to cDNA using the miScript II RT Kit (Qiagen, Valencia, CA, USA). Afterwards, quantitative RT-PCR (qPCR) was performed with the miScript SYBR Green PCR Kit (Qiagen) and miScript Primer Assays as the primers. For the quantification of protein coding gene’s expression level, reverse transcription was carried out with ReverTra Ace qPCR RT Master Mix with gDNA remover (Toyobo, Japan) and PCR was performed with Kapa SYBR Fast qPCR Kit Master Mix ABI Prism (Kapa Biosystems, Inc., Wilmington, MA, USA). The reactions were assayed in triplicate on an ABI 7300 instrument (Applied Biosystems, Foster City, CA, USA). The expression of miR-7 and protein coding genes was normalized using endogenous U6 and GAPDH with the 2−ΔΔCt calculation, respectively. The primers used for amplification of miRs and coding genes are listed in Supplementary Table S2.
HGF treatment
3 × 103 MCF-10A cells in culture media were seeded in each well of a 96-well plate, and were treated with HGF dissolved in PBS to a final concentrations of 20 and 40 ng/ml. As a control, PBS was used to a final concentration of 5%. The cells were cultured for 24 hours and total RNA was isolated for the expression analysis of miR-7 and coding genes. To verify the cell activation by HGF, the cell growth rate was measured using Cell Counting Kit-8 (Dojindo, Japan) at 450 and 600 nm 2 h after incubation with 10 μl of CCK-8 reagent to a well. The absorption value at 450 nm was subtracted from the value at 600 nm for turbidity removal.
Expression microarrays and pathway analysis
One microgram of total RNA from miR-7- or miNC-transfected MCF-10A cells was used for the expression microarray of Illumina Human HT-12 v4 Expression BeadChip (Illumina, San Diego, CA). Among 47000 probes on the chip, probes with detection P-value < 0.05 and |fold change| ≥ 2 were screened as significantly deregulated genes. Relevant networks and canonical pathways were generated using the Ingenuity Pathway Analysis (IPA) (Qiagen). The KEGG pathway enrichment analysis was performed by the KEGG Orthology Based Annotation System (KOBAS)55. The expression microarray data were deposited into the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) with the series accession number GSE102758. The mining of upstream regulators of miR-7 was carried out on the IPA platform which analyzed commonly regulated genes by HGF and miR-7.
Statistical analysis
Gene expression data were represented as the mean ± standard error of three independent experiments and analyzed by Student’s t-test using SPSS for Windows, version 17.0 (SPSS, Chicago, IL, USA). Differences were considered statistically significant when the P-value is lower than 0.05. Linear regression was conducted to calculate the coefficient of determination (R 2) and the statistical significance of the correlation between miR-7 and HGF expression.
Electronic supplementary material
Acknowledgements
This study was supported by the Basic Science Research Program (2016R1D1A1B01009235) through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science, and Technology. Dr. H. S. Kang was supported by a grant provided by the National Cancer Center, Korea.
Author Contributions
Conceived and designed the experiments: S.J.K. Performed the experiments: D.J., J.H., S.P., S.L. and H.L. Analyzed the data: H.S.K. and S.J.K. Wrote the paper: D.J., S.L. and S.J.K.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-15846-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fabbri M, Croce CM, Calin GA. MicroRNAs. Cancer J. 2008;14:1–6. doi: 10.1097/PPO.0b013e318164145e. [DOI] [PubMed] [Google Scholar]
- 3.Bushati N, Cohen S. M. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 4.Kalinowski FC, et al. microRNA-7: a tumor suppressor miRNA with therapeutic potential. Int J Biochem Cell Biol. 2014;54:312–317. doi: 10.1016/j.biocel.2014.05.040. [DOI] [PubMed] [Google Scholar]
- 5.Kong X, et al. MicroRNA-7 inhibits epithelial-to-mesenchymal transition and metastasis of breast cancer cells via targeting FAK expression. PLoS One. 2012;7:e41523. doi: 10.1371/journal.pone.0041523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie J, et al. miR-7 inhibits the invasion and metastasis of gastric cancer cells by suppressing epidermal growth factor receptor expression. Oncol Rep. 2014;31:1715–1722. doi: 10.3892/or.2014.3052. [DOI] [PubMed] [Google Scholar]
- 7.Zhao J, et al. Promoter mutation of tumor suppressor microRNA-7 is associated with poor prognosis of lung cancer. Mol Clin Oncol. 2015;3:1329–1336. doi: 10.3892/mco.2015.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J, et al. MicroRNA-7: a promising new target in cancer therapy. Cancer Cell Int. 2015;15:103. doi: 10.1186/s12935-015-0259-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giles KM, et al. microRNA-7-5p inhibits melanoma cell proliferation and metastasis by suppressing RelA/NF-kappaB. Oncotarget. 2016;7:31663–31680. doi: 10.18632/oncotarget.9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu W, Liu S, Liang Y, Zhou Z, Liu X. MiR-7 inhibits progression of hepatocarcinoma by targeting KLF-4 and promises a novel diagnostic biomarker. Cancer Cell Int. 2017;17:31. doi: 10.1186/s12935-017-0386-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hua K, et al. MicroRNA-7 inhibits proliferation, migration and invasion of thyroid papillary cancer cells via targeting CKS2. Int J Oncol. 2016;49:1531–1540. doi: 10.3892/ijo.2016.3660. [DOI] [PubMed] [Google Scholar]
- 12.Kermorgant S, Aparicio T, Dessirier V, Lewin MJ, Lehy T. Hepatocyte growth factor induces colonic cancer cell invasiveness via enhanced motility and protease overproduction. Evidence for PI3 kinase and PKC involvement. Carcinogenesis. 2001;22:1035–1042. doi: 10.1093/carcin/22.7.1035. [DOI] [PubMed] [Google Scholar]
- 13.Yang T, Wang Y, Jiang S, Liu X, Yu Z. Hepatocyte growth factor-induced differentiation of bone mesenchymal stem cells toward hepatocyte-like cells occurs through nuclear factor-kappa B signaling in vitro. Cell Biol Int. 2016;40:1017–1023. doi: 10.1002/cbin.10630. [DOI] [PubMed] [Google Scholar]
- 14.Niranjan B, et al. HGF/SF: a potent cytokine for mammary growth, morphogenesis and development. Development. 1995;121:2897–2908. doi: 10.1242/dev.121.9.2897. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Li Q, Zhu L. Expression of the hepatocyte growth factor and c-Met in colon cancer: correlation with clinicopathological features and overall survival. Tumori. 2012;98:105–112. doi: 10.1177/030089161209800115. [DOI] [PubMed] [Google Scholar]
- 16.Row PE, Clague MJ, Urbe S. Growth factors induce differential phosphorylation profiles of the Hrs-STAM complex: a common node in signalling networks with signal-specific properties. Biochem J. 2005;389:629–636. doi: 10.1042/BJ20050067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakai K, Aoki S, Matsumoto K. Hepatocyte growth factor and Met in drug discovery. J Biochem. 2015;157:271–284. doi: 10.1093/jb/mvv027. [DOI] [PubMed] [Google Scholar]
- 18.Yue Q, et al. MiR-124 suppresses the chemotactic migration of rat mesenchymal stem cells toward HGF by downregulating Wnt/beta-catenin signaling. Eur J Cell Biol. 2016;95:342–353. doi: 10.1016/j.ejcb.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 19.Zhu A, et al. MiR-221 and miR-26b Regulate Chemotactic Migration of MSCs Toward HGF Through Activation of Akt and FAK. J Cell Biochem. 2016;117:1370–1383. doi: 10.1002/jcb.25428. [DOI] [PubMed] [Google Scholar]
- 20.Huang J, et al. miR-199a-3p inhibits hepatocyte growth factor/c-Met signaling in renal cancer carcinoma. Tumour Biol. 2014;35:5833–5843. doi: 10.1007/s13277-014-1774-7. [DOI] [PubMed] [Google Scholar]
- 21.Lakshmanan I, et al. MUC16 induced rapid G2/M transition via interactions with JAK2 for increased proliferation and anti-apoptosis in breast cancer cells. Oncogene. 2012;31:805–817. doi: 10.1038/onc.2011.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan KK, Chan JY, Chung KK, Fung KP. Inhibition of cell proliferation in human breast tumor cells by antisense oligonucleotides against facilitative glucose transporter 5. J Cell Biochem. 2004;93:1134–1142. doi: 10.1002/jcb.20270. [DOI] [PubMed] [Google Scholar]
- 23.Jeon HS, et al. SMAD6 contributes to patient survival in non-small cell lung cancer and its knockdown reestablishes TGF-beta homeostasis in lung cancer cells. Cancer Res. 2008;68:9686–9692. doi: 10.1158/0008-5472.CAN-08-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juntermanns B, et al. Polo-like kinase 3 is associated with improved overall survival in cholangiocarcinoma. Liver Int. 2015;35:2448–2457. doi: 10.1111/liv.12839. [DOI] [PubMed] [Google Scholar]
- 25.Donaldson MM, Tavares AA, Hagan IM, Nigg EA, Glover DM. The mitotic roles of Polo-like kinase. J Cell Sci. 2001;114:2357–2358. doi: 10.1242/jcs.114.13.2357. [DOI] [PubMed] [Google Scholar]
- 26.Campbell DB, Li C, Sutcliffe JS, Persico AM, Levitt P. Genetic evidence implicating multiple genes in the MET receptor tyrosine kinase pathway in autism spectrum disorder. Autism Res. 2008;1:159–168. doi: 10.1002/aur.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan HW, Su HH, Hsu CW, Huang GJ, Wu TT. Targeted TPX2 increases chromosome missegregation and suppresses tumor cell growth in human prostate cancer. Onco Targets Ther. 2017;10:3531–3543. doi: 10.2147/OTT.S136491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reddy SD, Ohshiro K, Rayala SK, Kumar R. MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res. 2008;68:8195–8200. doi: 10.1158/0008-5472.CAN-08-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Webster RJ, et al. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem. 2009;284:5731–5741. doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- 30.Okuda H, et al. miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res. 2013;73:1434–1444. doi: 10.1158/0008-5472.CAN-12-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Q, Zhu F, Chen P. miR-7 and miR-218 epigenetically control tumor suppressor genes RASSF1A and Claudin-6 by targeting HoxB3 in breast cancer. Biochem Biophys Res Commun. 2012;424:28–33. doi: 10.1016/j.bbrc.2012.06.028. [DOI] [PubMed] [Google Scholar]
- 32.Wang W, et al. MicroRNA-497 suppresses angiogenesis by targeting vascular endothelial growth factor A through the PI3K/AKT and MAPK/ERK pathways in ovarian cancer. Oncol Rep. 2014;32:2127–2133. doi: 10.3892/or.2014.3439. [DOI] [PubMed] [Google Scholar]
- 33.Xiao C, et al. NLS-RARalpha Inhibits the Effects of All-trans Retinoic Acid on NB4 Cells by Interacting with P38alpha MAPK. Int J Med Sci. 2016;13:611–619. doi: 10.7150/ijms.15374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lucas TF, et al. 17beta-estradiol induces the translocation of the estrogen receptors ESR1 and ESR2 to the cell membrane, MAPK3/1 phosphorylation and proliferation of cultured immature rat Sertoli cells. Biol Reprod. 2008;78:101–114. doi: 10.1095/biolreprod.107.063909. [DOI] [PubMed] [Google Scholar]
- 35.Akalay I, et al. Targeting WNT1-inducible signaling pathway protein 2 alters human breast cancer cell susceptibility to specific lysis through regulation of KLF-4 and miR-7 expression. Oncogene. 2015;34:2261–2271. doi: 10.1038/onc.2014.151. [DOI] [PubMed] [Google Scholar]
- 36.Cochrane DR, et al. MicroRNAs link estrogen receptor alpha status and Dicer levels in breast cancer. Horm Cancer. 2010;1:306–319. doi: 10.1007/s12672-010-0043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li YZ, et al. Inhibition of miR-7 promotes angiogenesis in human umbilical vein endothelial cells by upregulating VEGF via KLF4. Oncol Rep. 2016;36:1569–1575. doi: 10.3892/or.2016.4912. [DOI] [PubMed] [Google Scholar]
- 38.Pollock A, Bian S, Zhang C, Chen Z, Sun T. Growth of the developing cerebral cortex is controlled by microRNA-7 through the p53 pathway. Cell Rep. 2014;7:1184–1196. doi: 10.1016/j.celrep.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozaki M, Haga S, Zhang HQ, Irani K, Suzuki S. Inhibition of hypoxia/reoxygenation-induced oxidative stress in HGF-stimulated antiapoptotic signaling: role of PI3-K and Akt kinase upon rac1. Cell Death Differ. 2003;10:508–515. doi: 10.1038/sj.cdd.4401172. [DOI] [PubMed] [Google Scholar]
- 40.Matsumoto R, et al. Adaptor protein CRK induces epithelial-mesenchymal transition and metastasis of bladder cancer cells through HGF/c-Met feedback loop. Cancer Sci. 2015;106:709–717. doi: 10.1111/cas.12662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson M, Kochhar K, Nakamura T, Iyer A. Hepatocyte growth factor-induced signal transduction in two normal mouse epithelial cell lines. Biochem Mol Biol Int. 1995;36:465–474. [PubMed] [Google Scholar]
- 42.Cataisson C, et al. MET signaling in keratinocytes activates EGFR and initiates squamous carcinogenesis. Sci Signal. 2016;9:ra62. doi: 10.1126/scisignal.aaf5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lai JK, et al. Kruppel-like factor 4 is involved in cell scattering induced by hepatocyte growth factor. J Cell Sci. 2012;125:4853–4864. doi: 10.1242/jcs.108910. [DOI] [PubMed] [Google Scholar]
- 44.Beviglia L, Kramer RH. HGF induces FAK activation and integrin-mediated adhesion in MTLn3 breast carcinoma cells. Int J Cancer. 1999;83:640–649. doi: 10.1002/(SICI)1097-0215(19991126)83:5<640::AID-IJC13>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 45.Ro TB, et al. HGF and IGF-1 synergize with SDF-1alpha in promoting migration of myeloma cells by cooperative activation of p21-activated kinase. Exp Hematol. 2013;41:646–655. doi: 10.1016/j.exphem.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Santoni-Rugiu E, et al. Inhibition of neoplastic development in the liver by hepatocyte growth factor in a transgenic mouse model. Proc Natl Acad Sci USA. 1996;93:9577–9582. doi: 10.1073/pnas.93.18.9577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.You ZH, et al. PBMDA: A novel and effective path-based computational model for miRNA-disease association prediction. PLoS Comput Biol. 2017;13:e1005455. doi: 10.1371/journal.pcbi.1005455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen X, et al. HGIMDA: Heterogeneous graph inference for miRNA-disease association prediction. Oncotarget. 2016;7:65257–65269. doi: 10.18632/oncotarget.11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen X, Wu QF, Yan GY. RKNNMDA: Ranking-based KNN for MiRNA-Disease Association prediction. RNA Biol. 2017;14:952–962. doi: 10.1080/15476286.2017.1312226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cloutier M, Wang E. Dynamic modeling and analysis of cancer cellular network motifs. Integr Biol (Camb) 2011;3:724–732. doi: 10.1039/c0ib00145g. [DOI] [PubMed] [Google Scholar]
- 51.McGee SR, Tibiche C, Trifiro M, Wang E. Network Analysis Reveals A Signaling Regulatory Loop in the PIK3CA-mutated Breast Cancer Predicting Survival Outcome. Genomics Proteomics Bioinformatics. 2017;15:121–129. doi: 10.1016/j.gpb.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cipriano R, et al. Conserved oncogenic behavior of the FAM83 family regulates MAPK signaling in human cancer. Mol Cancer Res. 2014;12:1156–1165. doi: 10.1158/1541-7786.MCR-13-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Houston SI, et al. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J Biol Chem. 2008;283:19478–19488. doi: 10.1074/jbc.M710579200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu B, et al. MiR-502/SET8 regulatory circuit in pathobiology of breast cancer. Cancer Lett. 2016;376:259–267. doi: 10.1016/j.canlet.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 55.Wu J, Mao X, Cai T, Luo J, Wei L. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006;34:W720–724. doi: 10.1093/nar/gkl167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.