Abstract
Myotonic dystrophy type 1 (DM1) is the most common adult-onset muscular dystrophy, characterized by progressive myopathy, myotonia, and multi-organ involvement. This dystrophy is an inherited autosomal dominant disease caused by a (CTG)n expansion within the 3′ untranslated region of the DMPK gene. Expression of the mutated gene results in production of toxic transcripts that aggregate as nuclear foci and sequester RNA-binding proteins, resulting in mis-splicing of several transcripts, defective translation, and microRNA dysregulation. No effective therapy is yet available for treatment of the disease. In this study, myogenic cell models were generated from myotonic dystrophy patient-derived fibroblasts. These cells exhibit typical disease-associated ribonuclear aggregates, containing CUG repeats and muscleblind-like 1 protein, and alternative splicing alterations. We exploited these cell models to develop new gene therapy strategies aimed at eliminating the toxic mutant repeats. Using the CRISPR/Cas9 gene-editing system, the repeat expansions were removed, therefore preventing nuclear foci formation and splicing alterations. Compared with the previously reported strategies of inhibition/degradation of CUG expanded transcripts by various techniques, the advantage of this approach is that affected cells can be permanently reverted to a normal phenotype.
Keywords: myotonic dystrophy type 1, CRISPR/Cas9, trinucleotide expansion, splicing alterations, phenotypic reversion, DM1 cell models, DMPK, CTG repeats, muscle disease
Introduction
In this work, we describe a genome-editing approach exploited for deletion of trinucleotide expansion in the dystrophia myotonica protein kinase (DMPK) gene involved in the pathogenesis of myotonic dystrophy type 1 (DM1). DM1 is the most common adult-onset muscular dystrophy,1, 2 characterized by progressive skeletal muscle weakness, myotonia, cardiac defects, smooth muscle dysfunction, and neurological abnormalities.1, 2, 3, 4 The disease is caused by the expansion of the (CTG)n triplet repeat in the 3′ UTR of the DMPK gene, which encodes for a myosin kinase. This gene is ubiquitously expressed, but particularly relevant in skeletal and cardiac muscles.2, 5 CTG expansion is characterized by high instability, often resulting in increased repeat size with age and in anticipation of symptoms in successive generations. This tendency of the repeats to further expand is more pronounced in certain tissues compared to others, leading to somatic mosaicism.6 The presence of longer repeats correlates with a more severe pathology.7 The molecular effector of the disease is the DMPK mutant transcript that accumulates into nuclear aggregates (foci) and sequesters RNA-binding proteins, such as muscleblind-like 1 (MBNL1) protein, involved in the regulation of RNA splicing.8, 9, 10 DM1 molecular pathogenesis also involves changes in gene expression and translation efficiency, non-conventional translation, and microRNA deregulation.11, 12, 13 Several mouse models of myotonic dystrophy have been generated, displaying many aspects of human pathology. These models have contributed to clarify the disease mechanisms.14, 15, 16, 17 Nevertheless, cellular models are still needed for evaluation of therapeutic molecules or in vitro strategies and for high-throughput screenings before in vivo validation. DM1 patient-derived cells, both primary cultures and immortalized cell lines, represent valuable models for these studies because the CTG expansions are expressed within their native genomic context and the cells maintain DM1-associated molecular features.18, 19, 20, 21, 22, 23 Understanding of the repeated RNA-induced toxicity in DM1 pathogenesis has led to the rapid development of therapeutic strategies aimed at neutralizing the toxic RNA. It was shown that the major aspects of the DM1 phenotype are potentially reversible by targeting the nuclear CUG repeated mRNA both in cell cultures in vitro and in mouse models in vivo.24, 25, 26 However, these strategies provide only short-term effects unless repeated administration of the inhibitory molecules is applied. To date, there is no effective long-term treatment for DM1.
The recent identification of the clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (Cas9) system has revolutionized the field of genetics and has been proposed as a promising technology to correct the genetic basis of hereditary diseases. CRISPRs were first identified as part of the bacterial immune system, playing a role in viral defense.27 The CRISPR-associated endonuclease Cas9 can be targeted to specific locations in the genome via a RNA-guided system involving single-guide RNAs (sgRNAs) to induce double-strand breaks in regions of interest.28, 29, 30 The double-strand breaks are preferentially repaired by non-homologous end joining (NHEJ). When an additional DNA template is provided, a precise genomic modification can be generated by homology directed repair (HDR). Although HDR is believed to occur infrequently in post-mitotic adult tissues,31 such as heart and skeletal muscle, a recent paper reported HDR-mediated editing in skeletal muscle at a moderate frequency.32 This technology has been recently applied to rescue the reading frame and expression of dystrophin both in cell models derived from Duchenne muscular dystrophy (DMD) patients33, 34, 35 and in mdx mice in vivo. Dystrophin gene editing resulted in restoration of protein production and improvement of muscle function.32, 36, 37, 38 Monogenic diseases resulting from amplification of genomic sequences, such as DM1, can benefit particularly from the multiplexing capability of Cas9. In addition, although NHEJ-mediated repair may lead to imperfect or frameshifted repair junction, the DMPK protein production should not be affected because the repeats are located in noncoding regions of the DMPK gene. Indeed, in a recent paper, published while we were completing our experiments, CRISPR/Cas9 cleavage ability was described to produce large deletions in DMPK repeat regions in vitro.39 Using a similar approach, but different Cas9 nuclease and target sequences, we applied the CRISPR/Cas9 and NHEJ gene-editing system in ad hoc generated cell models from DM1 patients and succeeded in removing pathogenetic CTG expansions permanently, resulting in phenotypic reversion of the edited cells.
Results
Generation and Characterization of Immortalized Human Myogenic Cells Derived from Fibroblasts of DM1 Patients
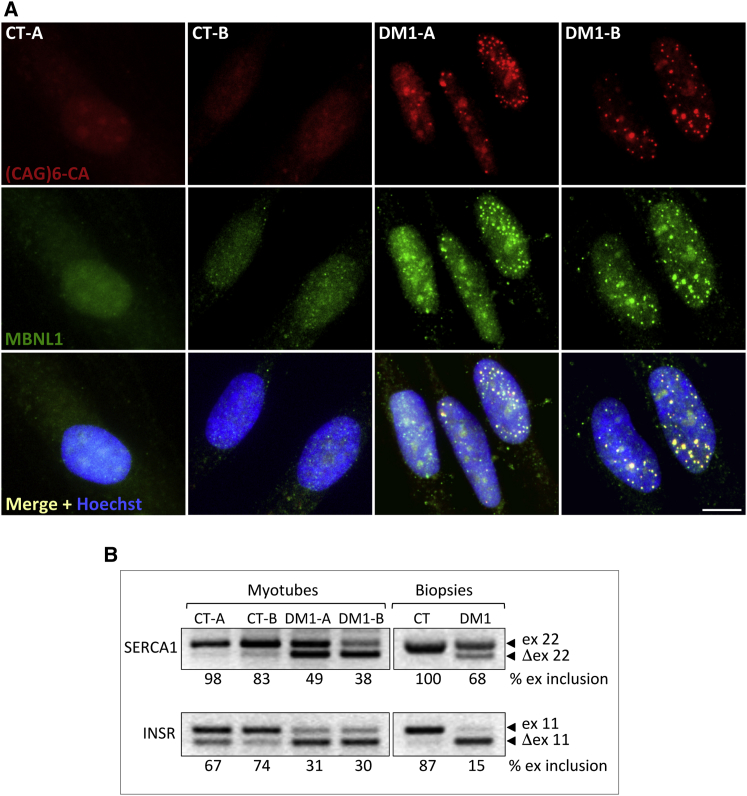
Dermal fibroblasts were derived from 2 healthy individuals (CT-A and CT-B) and 2 DM1 patients diagnosed for displaying abnormal CTG repeats in the 3′ UTR region of the DMPK gene in a single allele (DM1-A and DM1-B). Fibroblasts were immortalized by infection with retroviral vectors carrying the human telomerase (TERT) gene (Figure S1A) and converted to myogenic cells by transduction with retroviruses producing an inducible myogenic differentiation 1 (MYOD1) transcription factor fused to the estrogen receptor (ER) hormone binding domain (MYOD1-ER). Cell immortalization is an essential requirement to allow single cell clonal expansion following CRISPR/Cas9 treatment. The use of TERT to immortalize primary human cells and bypass senescence was demonstrated to be safe because immortalized cells showed a normal karyotype and no evidence of cancer-associated changes.40, 41 After addition of β-estradiol to culture medium, MYOD1-ER translocates to the nucleus and transactivates muscle-specific genes (Figure S1B). We did not observe significant differences in differentiation and fusion among control and DM1 cell lines, as determined by immunofluorescence (Figure S2A) and mRNA/protein expression analyses of muscle-specific transcription factors and structural genes (Figures S2B and S2C). These findings are in agreement with previous reports, in which primary or immortalized myoblasts derived from healthy individuals and DM1 patients were used.19, 21, 22 Differentiated myotubes obtained after MYOD1 induction were analyzed by fluorescent in situ hybridization (FISH) of ribonuclear inclusions containing CUG repeats (nuclear foci), a hallmark of DM1 cell nuclei, through hybridization with a fluorescent (CAG)6CA probe. Staining with antibodies to MBNL1 showed co-localization of the protein in nuclear aggregates exclusively in DM1 cells (Figure 1A). In addition, alternative splicing of insulin receptor (INSR) and sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (SERCA1) transcripts, analyzed by RT-PCR, was found to be defective in DM1 myotubes and muscle biopsies compared to normal controls (Figure 1B), as previously reported in DM1 cells and tissues.42, 43
Figure 1.
Immortalized Myogenic Cells Derived from DM1 Patients Exhibit Pathologic Ribonuclear Inclusions and Splicing Alterations
(A) FISH analysis with Texas-Red-labeled (CAG)6CA probe of control and DM1 cell lines following induction to differentiation for 5 days. Cells were stained with anti-MBNL1 antibody and nuclei were counterstained with Hoechst dye. Co-localization of CUG-containing transcripts and MBNL1 protein is visible in nuclear aggregates (merge panels). Scale bar, 10 μm. (B) Splicing analysis of SERCA1 (SERCA1) and INSR (INSR) transcripts in control and DM1-derived myogenic cells (24 hr following induction with β-estradiol) and in muscle biopsies. Percentages of exon inclusion were calculated as the percentage of the total intensity of both isoform signals, taken as 100%.
Design of the CRISPR/Cas9 Constructs to Delete CTG Expansions
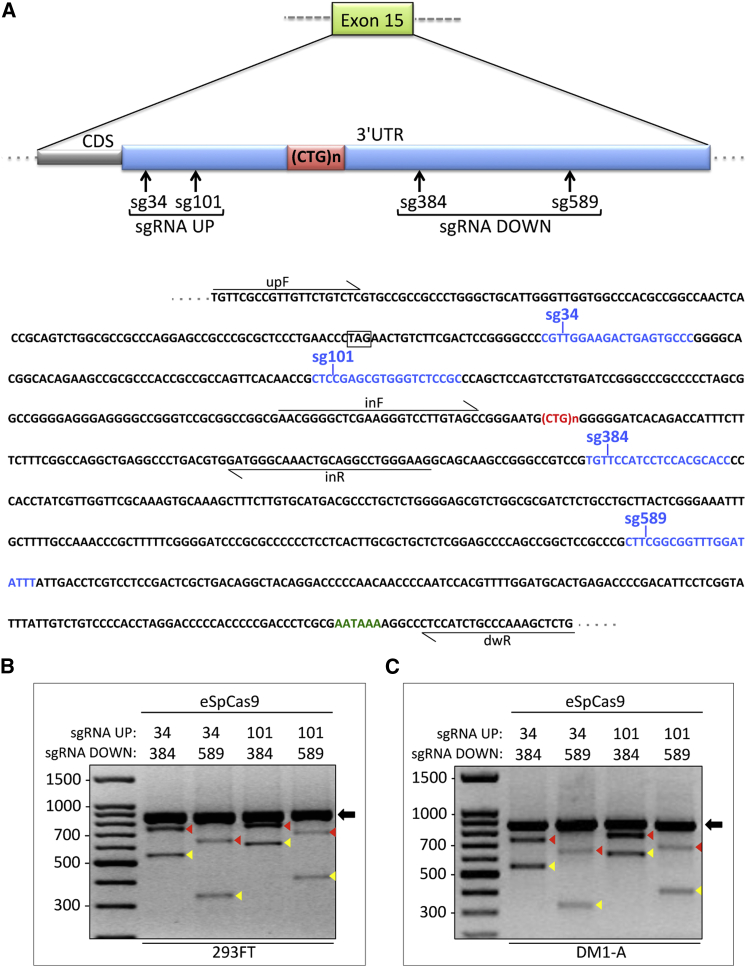
To create genomic deletions of CTG expansions and restore normal DMPK gene expression and function, we chose to apply the CRISPR/Cas9 and NHEJ system to the DM1-A myogenic cell line. In DM1-A and CT-B myogenic cells, PCR amplification of DMPK repeats, followed by Southern blot analysis with a 5′ digoxigenin (DIG)-labeled (CTG)10 probe,44 resulted in small bands around 150 nt, corresponding to fragments containing 5 CTG repeats for the normal allele of DM1-A and 5 and 13 CTG repeats for the two alleles of CT-B (Figure S3A). Higher molecular weight products were amplified from the DM1-A mutated allele (Figure S3A). The number of CTGs in the mutated allele of the DM1-A patient was 290 in peripheral blood cells at diagnosis. However, in primary fibroblasts and engineered DM1-A myogenic cells, the number of repeats was much higher and heterogeneous, ranging from 300 to over one thousand (Figure S3A). This expansion increase was likely due to somatic mosaicism and further amplification during cell culturing. In order to design the sgRNAs for deletion of the CTG repeats in DM1-A cells, first we ruled out the possibility of sequence divergence due to individual variability. We sequenced about 400 bp upstream and downstream of the repeated regions in the DMPK locus, mostly within the 3′ UTR, and found no difference between the wild-type (WT) and mutated allele. In these regions, 4 CRISPR/Cas9 target sequences of 20 nt, 2 upstream and 2 downstream of the CTG repeats, were selected and the corresponding sgRNAs were designed to direct efficient site-specific double-strand breaks (Figure 2A). The sgRNAs were then cloned into the sgRNA expression vector pLB that also expresses GFP. For Cas9 expression, a plasmid encoding the high specificity SpCas9 nuclease45 was used.
Figure 2.
Design and Application of CRISPR/Cas9 Gene Editing to Normal and Mutant Human Cells
(A) Schematic representation of the exon 15 region of the DMPK gene targeted for deletion of CTG repeats by CRISPR/Cas9. sgRNAs specific to different sites upstream (sgRNA UP) and downstream (sgRNA DOWN) of (CTG)n expansion, shown in red, are indicated by arrows (upper panel). Partial sequence of DMPK exon 15 showing cleavage positions and target sequences of the sgRNAs (blue) along the 3′ UTR; TAG stop codon and polyadenylation site are highlighted (black box and green sequence); primer sequences used in PCR analyses are underlined (lower panel). (B and C) PCR analysis of genomic DNA from 293FT cells (B) and DM1-A myogenic cells (C), co-transfected with eSpCas9 and 2 sgRNAs (1 up and 1 down) plasmids in all 4 possible combinations. upF and dwR primer position is indicated in (A). Black arrows indicate amplicons corresponding to the undeleted WT alleles; yellow arrowheads indicate the expected CTG-deleted products; red arrowheads indicate bands likely resulting from heteroduplexes formed by the deletion product strand and unmodified strand. 100-bp ladder molecular weight markers are shown. Note that only WT or edited alleles are visible on gels because large repeats are not amplified efficiently in these PCR conditions.
To determine targeting efficiencies of the different sgRNA pairs, we initially used the human 293FT cells whose DMPK alleles contain five CTG repeats and sequences of the sgRNA target regions identical to those of DM1-A cells. 293FT cells were transfected with the Cas9 plasmid and the 4 possible combinations of sgRNA-encoding plasmid pairs. After 3 days, genomic DNA extracted from each transfected polyclonal population was amplified by PCR using primers designed to reveal all possible editing events. In the 293FT cell line, all sgRNA combinations gave the expected edited products, with comparable efficiencies (Figure 2B).
Genome Editing Application to Human DM1 Cells for Generation of Clonal Cell Lines with Deleted CTG Expansions
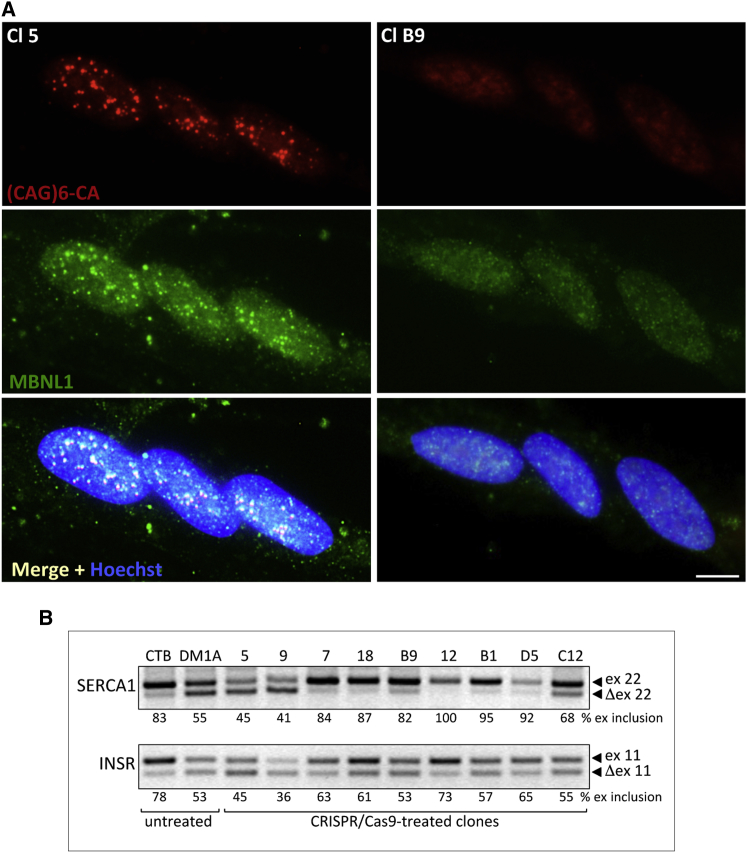
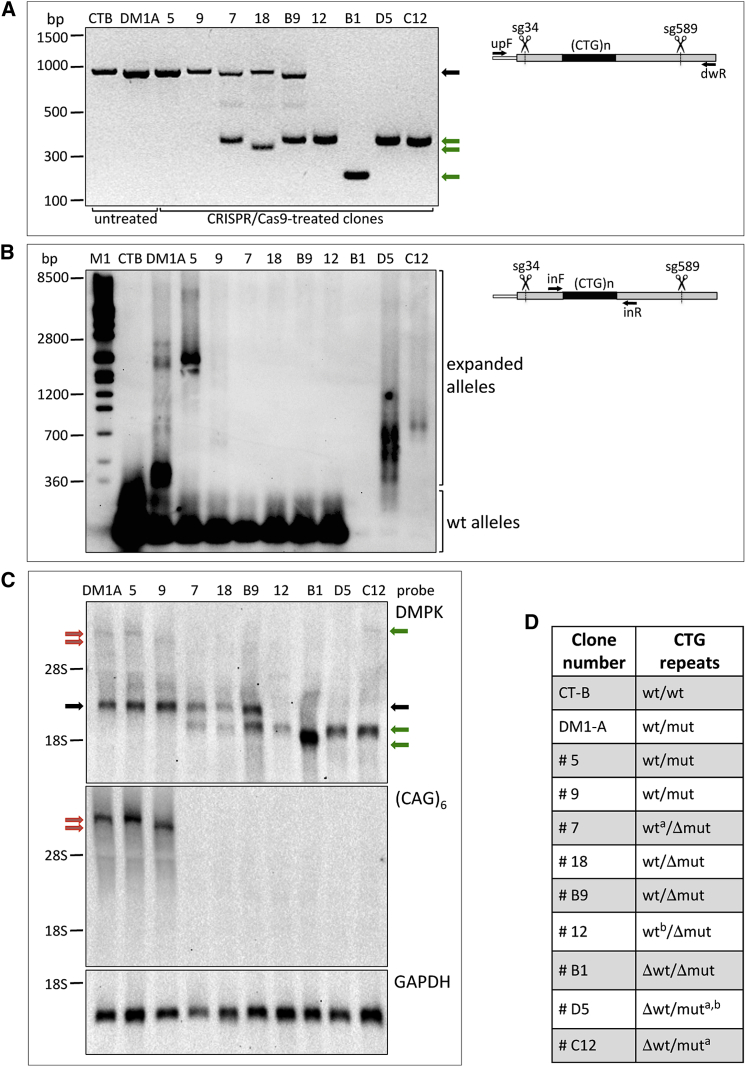
We next tested the sgRNAs for deletion of the CTG expansion in the DM1-A cell line. Because transfection efficiency was very low in these cells, we electroporated the cells with the 4 Cas9/sgRNA plasmid combinations and, 3 days later, selected GFP-positive cells by flow cytometry. Sorted cells were either grown as polyclonal populations or plated as single cells into 96-well plates for clonal expansion. PCR analysis of genomic DNA from the polyclonal cell populations showed a similar editing efficiency for the 4 sgRNA combinations (Figure 2C). Note that only amplicons derived from WT or edited alleles could be obtained with the adopted PCR conditions, whereas large repeat-containing regions were not amplified at detectable levels. The sgRNA combination sg34-sg589 targeting more distal sequences relative to the CTG expansions was selected for further analysis, assuming that CRISPR/Cas9 activity in regions more distant from the CTG repeats should be less influenced by DNA hairpin structures formed by the repeated sequences. A total of 85 clones were analyzed by PCR, and 12 of them showed bands compatible with a deletion in a single allele or both (14% editing efficiency). The 12 clones were subjected to FISH analysis and MBNL1 staining to assess the presence of nuclear foci (representative clones are shown in Figure 3A). Seven clones (7, 18, B9, 12, B1, D5, and C12) were negative for foci and were further analyzed by PCR and sequencing to verify the occurrence of gene editing, along with two undeleted clones (5 and 9); untreated parental cell lines (CT-B and DM1-A) were used as controls (Figure 4A). To confirm deletion of CTG expansions in the selected clones, genomic DNA was analyzed by long-range PCR, followed by Southern blot analysis using probes to detect CTGs. PCR reactions and Southern hybridizations were performed using two different protocols and both produced very similar results, although with different PCR efficiencies. For clarity, some samples are shown in duplicate (Figures 4B and S3B). The amplified repeat regions of the mutated alleles containing long repeats of heterogeneous size (higher smeared bands) migrate slower and can be distinguished from the short repeats of the WT alleles (lower bands). Minor bands and smears detectable in Southern blots likely arise from inappropriate reannealing during PCR reactions. Deletion of CTG repeats resulted in the lack of amplified bands (Figures 4B and S3B). The combined results of PCR, Southern hybridization, and sequencing analyses of CRISPR/Cas9-treated clones are summarized in Table 1 and Figure 4D. In 5 out of 7 clones negative for nuclear foci in FISH analysis, the mutated alleles were completely deleted (clones 7, 18, B9, 12, and B1). Clone 7, 18, B9, and 12 maintained WT repeats. However, in clone 7, an inversion of the WT repeated region occurred. In clone 12, only one PCR product was detectable when distal primers were used, suggesting deletions in both alleles (Figure 4A). However, PCR with primers adjacent to the repeats, followed by Southern hybridization, identified a band corresponding to the WT repeat size (Figures 4B and S3B). Further PCR and sequencing analysis demonstrated that the WT allele contained a big insertion in the sg34 target site, explaining why it could be not amplified by the PCR conditions previously used (Figure 4A). In clone B1, both the mutated and WT alleles were deleted (Figures 4A and 4B). Clone D5 showed repeat deletion in the WT allele and inversion of partial CTG expansion with additional insertions in the mutated allele. Clone C12 showed repeat deletion in the WT allele and inversion of the entire repeat expansion of the mutated allele. Interestingly, small indels in one or both sgRNA target sites were revealed by sequence analysis also in undeleted regions. CRISPR-Cas9 editing events leading to CTG-repeat deletions resulted in re-joining of the double-strand breaks at cleavage sites either without indels or with indels, ranging from 1 to 151 nt (Table 1).
Figure 3.
CRISPR/Cas9-Mediated Deletion of CTG Expansions in DM1 Myogenic Clones Results in Phenotypic Reversion
(A) FISH analysis of CRISPR/Cas9-treated clones 5 and B9 following induction to differentiation for 2 days. Cells were stained with anti-MBNL1 antibody and nuclei counterstained with Hoechst dye. Co-localization of CUG-containing transcripts and MBNL1 protein is visible in nuclear aggregates (merge panels). Scale bar, 10 μm. (B) Splicing analysis of SERCA1 (SERCA1) and INSR (INSR) transcripts in CT-B, DM1-A parental cells, and CRISPR/Cas9-treated clones. Percentages of exon inclusion, calculated as in Figure 1, are shown.
Figure 4.
DMPK Gene Editing and mRNA Expression in CRISPR/Cas9-Treated Clones
(A) PCR analysis of CTG expansions in genomic DNA from untreated CT-B and DM1-A parental cells and CRISPR/Cas9 selected clones. Positions of the primers used for amplification are shown in the scheme. Black arrow indicates amplicons corresponding to the untreated/undeleted WT alleles; green arrows indicate the amplification products obtained following CTG deletion. The faint bands above the 500-nt marker are likely due to heteroduplexes formed by the deletion product strand and the unmodified strand. (B) Southern blot hybridization of PCR products amplified with the primers shown in the scheme from genomic DNA of CRISPR/Cas9-treated clones and untreated CT-B and DM1-A cells using a DIG-labeled (CAG)6 probe. Bands corresponding to WT alleles and alleles with expansions of multiple sizes can be visualized. Molecular weight markers are indicated: 700 bp correspond to about 190 triplets, 1,200 bp correspond to about 340 triplets, and 2,800 bp correspond to about 890 triplets. Sequences of the primers upF and dwR used in (A) and inF and inR used in (B) are shown underlined in Figure 2A. (C) Northern blot hybridization of polyadenylated RNA from differentiated myoblasts of CRISPR/Cas9-treated clones and untreated DM1-A cells using a probe for the DMPK coding region (DMPK), a probe for the CTG expansions ((CAG)6), and a probe for GAPDH (GAPDH) as a loading control. Black arrows indicate undeleted DMPK WT transcripts; red arrows indicate undeleted expanded transcripts; green arrows indicate transcripts derived from edited WT or expanded alleles. (D) Summary of the state of the CTG repeat regions in DMPK WT and mutated alleles of CRISPR/Cas9-treated clones and untreated CT-B and DM1-A, resulting from the experiments showed in (A)–(C) and sequence analyses. Δ, deletion; a, inversion; b, insertion.
Table 1.
Description of CRISPR/Cas9-Editing Events in WT and Mutated Alleles
| Clone Number | CTG Repeats | Indel Types |
|---|---|---|
| # 5 | not altered in WT and mutated alleles (WT/mut) | WT, sg34 site: +2 nt |
| mut, sg34 site: +1 nt | ||
| # 9 | not altered in WT and mutated alleles (WT/mut) | WT, sg34 site: −1 nt |
| mut, sg34 site: +1 nt | ||
| # 7 | inverted in WT allele and deleted in mutated allele (WTa/Δmut) | WT, sg34 site: none; sg589 site: none |
| mut, sg34 site: none; sg589 site: none | ||
| # 18 | not altered in WT and deleted in mutated allele (WT/Δmut) | WT, sg34 site: +1 nt |
| mut, sg34 site: −20 nt; sg589 site: −15 nt | ||
| # B9 | not altered in WT and deleted in mutated allele (WT/Δmut) | WT, sg34 site: +1 nt; sg589 site: −26 nt |
| mut, sg34 site: +1 nt; sg589 site: none | ||
| # 12 | not altered in WT allele with insertion in sg34 site and deleted in mutated allele (WTb/Δmut) | WT, sg34 site: insertion; sg589 site: −1 nt |
| mut, sg34 site: none; sg589 site: none | ||
| # B1 | deleted in WT and mutated alleles (ΔWT/Δmut) | WT/mut,c sg34 site: −3 nt; sg589 site: −151 nt |
| # D5 | deleted in WT allele and rearranged with inversions/insertions in mutated allele (ΔWT/muta,b) | WT, sg34 site: +1 nt; sg589 site: none |
| mut, sg34 site: +1 nt; sg589 site: none | ||
| # C12 | deleted in WT allele and inverted in mutated allele (ΔWT/muta) | WT, sg34 site: none; sg589 site: none |
| mut, sg34 site: none; sg589 site: none |
Only indels derived from detectable re-joining events are indicated. Δ, deletion.
Inversion
Insertion
PCR performed using primers located about 700 nt upstream and downstream the sgRNA target sites did not produce further amplicons, suggesting that cleavage is identical in both alleles.
To test whether gene editing altered the expression of the DMPK gene, Northern blot analysis of untreated and CRISPR/Cas9-treated clones was performed. Hybridization with a probe to the DMPK coding region allowed detection of the expected transcripts (WT, expanded, deleted, and inverted), with the exception of clone 12 and D5, where only the deleted mutated and WT transcripts, respectively, were visible (Figure 4C, top panel). The other transcripts of clones 12 and D5 could not be detected, probably due to low expression levels. Only transcripts containing large expansions in DM1-A, clone 5 and 9, could be detected using the (CAG)6 probe (Figure 4C, middle panel). Clone C12 produced a transcript from the mutated allele, unaltered in size and visible with the DMPK probe, but undetectable with the (CAG)6 probe due to the repeat inversion. A GAPDH probe was used as loading control (Figure 4C, bottom panel).
Although infrequent in primary human cells,33, 34 CRISPR/Cas9-induced off-target mutations have been reported.46, 47, 48 To verify these effects in our cell context, analysis of possible off-target induced mutations was performed by PCR and sequencing of the top seven homologous sites for each sgRNA determined by COSMID and Cas-OFFinder software (Table S1) in parental DM1-A cells and in some CRISPR/Cas9-treated clones (clone 9, clone B9, clone B1, and clone 12). This analysis revealed no off-target mutations in edited cells (Table S1).
Splicing and Differentiation Analysis in CRISPR/Cas9 Edited Clones
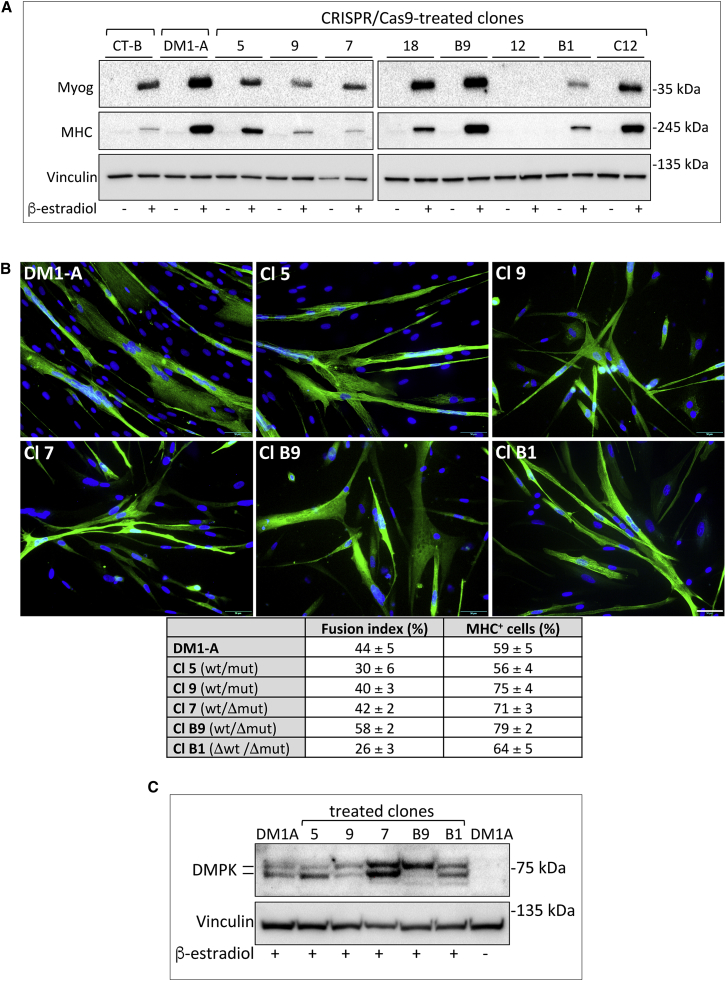
Clones with or without repeats were further analyzed for re-establishment of normal splicing function by RT-PCR amplification of alternatively spliced SERCA1 and INSR transcripts. As expected, clones negative for nuclear foci and deleted for CTG repeats exhibited normal splicing of SERCA1 and INSR transcripts compared to clones positive for foci and retaining full expansions (Figure 3B). Clone C12 showed a partial phenotype. We then determined whether the clones with deleted repeats maintained myogenic differentiation capacity upon MYOD1-ER induction and expressed DMPK protein. Most clones without repeats differentiated and fused normally, as compared to parental DM1-A and control CT-B cells and clones with intact repeats (Figures 5A and 5B), and maintained DMPK protein expression (Figure 5C), indicating that repeat deletion neither affected myogenic differentiation capacity nor production of DMPK protein. Note that DMPK protein is barely expressed in the absence of β-estradiol treatment, in agreement with a previous report showing detectable DMPK protein accumulation only in skeletal and heart tissues and differentiated myoblasts in culture.49 Only one clone failed to accumulate muscle-specific proteins (clone 12, Figure 5A); this clone expressed a hormone-insensitive MYOD1-ER protein that translocated poorly to the nucleus failing to induce detectable myosin and DMPK gene expression (Figure S4 and not shown). Clone D5 could not be analyzed for myogenic differentiation because cells underwent senescence upon prolonged passaging in culture. Characteristics of all the CRISPR/Cas9-treated clones are summarized in Table 2.
Figure 5.
CRISPR/Cas9 Edited Clones Maintain Differentiation and Fusion Capacity upon MYOD1-ER Induction
(A) Western blot analysis of muscle-specific myogenin (Myog), myosin (MHC), and constitutively expressed vinculin (Vinculin) in CT-B and DM1-A parental cells and CRISPR/Cas9-edited clones in the absence or presence of β-estradiol to induce differentiation for 5 days. (B) Immunofluorescence analysis of parental DM1-A myogenic cells and representative CRISPR/Cas9-treated clones, with (Cl 5 and Cl 9) or without (Cl 7, Cl B9, and Cl B1) CTG expansions, allowed to differentiate for 5 days and stained with anti-MHC antibody and Hoechst dye (scale bar, 50 μM). The fusion index and percentage of MHC-positive cells is shown in the table below (average ± SE, n ≥ 4). The fusion index is calculated as the percentage of nuclei in MHC-positive myotubes (containing ≥2 nuclei) over the total number of cells. At least 300 cells were counted for each cell type in each experiment. (C) Western blot analysis of DMPK and vinculin in DM1-A parental cells and representative CRISPR/Cas9-treated clones, treated as above.
Table 2.
Characteristics of CRISPR/Cas9-Edited Clones and Parental Cells
| Parental Cells/ Clone Number | CTG Repeats | Nuclear Foci | Myogenic Differentiation | DMPK WT/mut mRNA | DMPK Protein | Normal Splicing SERCA1 | Normal Splicing INSR |
|---|---|---|---|---|---|---|---|
| CT-B | WT/WT | − | ++ | ND | + | + | + |
| DM1-A | WT/mut | + | +++ | +/+ | + | − | − |
| # 5 | WT/mut | + | ++ | +/+ | + | − | − |
| # 9 | WT/mut | + | ++ | +/+ | + | − | − |
| # 7 | WTa/Δmut | − | ++ | +/Δ+ | + | + | + |
| # 18 | WT/Δmut | − | +++ | +/Δ+ | ++ | + | + |
| # B9 | WT/Δmut | − | +++ | +/Δ+ | ++ | + | +/− |
| # 12 | WTb/Δmut | − | − | −/Δ+ | −/+ | + | + |
| # B1 | ΔWT/Δmut | − | ++ | Δ+/Δ+ | + | + | +/− |
| # D5 | ΔWT/muta,b | − | ND | Δ+/− | ND | + | + |
| # C12 | ΔWT/muta | − | ++ | Δ+/+ | + | + | +/− |
Δ, deletion; ND, not determined.
Inversion
Insertion
Discussion
Among genome-editing technologies, the CRISPR/Cas9 gene-editing system can be easily used to target virtually any genomic location. The multiplex capability of CRISPR/Cas9 gene editing allows large deletion of unwanted genomic sequences by eliciting two simultaneous double-strand breaks. This capacity has been recently exploited in myogenic cell models and in mdx mouse models of DMD by different research groups.32, 36, 37, 38 In all cases, CRISPR/Cas9-mediated deletion resulted in exon skipping and production of a shorter but functional dystrophin protein in vitro and in rescuing of muscular strength in diseased animals. In the present study, we tested whether CRISPR/Cas9 and the NHEJ system can be effectively used to excise CTG repeats in DM1 patient-derived cells, leading to normalization of DMPK gene expression and abrogation of RNA toxicity in edited cells. To this aim, we generated immortalized myogenic cells from primary fibroblasts derived from DM1 patients, which can synchronously differentiate and fuse into multinucleated myotubes upon activation of an ectopically expressed hormone-inducible MYOD1. These cells present typical DM1-associated alterations, such as ribonuclear aggregates containing CUG repeats and MBNL1 protein, and alternative splicing abnormalities. Compared to primary myoblast cultures, our cell models offer a number of advantages. Fibroblasts can be easily obtained from skin biopsies, can be grown indefinitely by immortalization with TERT, and can be induced to differentiate synchronously and efficiently by conditional MYOD1 activation. Importantly, these cells maintain their myogenic potential in the absence of induction, unlike primary myoblasts that tend to decrease their differentiation efficiency during propagation in culture because of spontaneous differentiation. Similar cell models of muscular dystrophies have been generated by other groups for studies on disease mechanisms and for testing therapeutic strategies.20, 22, 23 Here, we show that CRISPR/Cas9 and the NHEJ gene-editing strategy can be successfully applied to our DM1 cell models and leads to restoration of a normal phenotype. While we were completing our experiments, similar results were reported by van Agtmaal et al.,39 with some differences. Unlike these authors, we used the high specificity SpCas9 nuclease and designed different sgRNAs pairs to target sequences spanning about 200 nt upstream and 320 nt downstream the expansion flanking regions. In addition, the cell models were generated from different donor DM1 patients, from a different tissue and using a different protocol. Nonetheless, in both studies, editing of the expanded repeats occurred efficiently, further supporting the robustness and versatility of the CRISPR/Cas9 and NHEJ gene-editing system. In CRISPR/Cas9-treated clones, deletions, inversions of the excised repeat regions, and insertions were found. Sequencing analysis of the edited clones revealed small indels at sgRNA target sites, also in the absence of deletion of the repeat regions. When single cleavage events occurred in the mutated allele, we did not observe deletion of the repeat regions, as described by van Agtmaal et al.39 This may be due to the different positions of the sgRNA target sites, with respect to the CTG repeats in each case.
Because Cas9 shows occasional off-target activity, we analyzed off-target editing on selected genomic sites for both sgRNAs in our CRISPR/Cas9-treated clones and found no effects, confirming previous reports describing a low tendency of Cas9 to cut on off-target genomic regions in human patient-derived cells.33, 34 However, considering the number of clones analyzed in our and others’ experiments, inversion, insertions, or other uncontrolled events around sgRNA on-target sites appear to be rather frequent, raising an important concern on the future application of this technology in gene therapy. With regard to detrimental Cas9 editing activity, a more stringent analysis of Cas9 off-target effects using, for example, whole genome sequencing may be necessary to identify off-target sites that cannot be predicted by sequence comparison. Improvement of the targeting specificity and adjustable expression of Cas9 nuclease, both tissue specific and time controlled, will also be important for therapeutic use because genome editing leads to permanent modifications.
Recently, a genome therapy approach for DM1 has been applied to patient-derived induced pluripotent stem cells (iPSCs) using the site-specific transcription activator-like effector nuclease (TALEN) to insert a polyA signal upstream of DMPK CTG repeats, producing a shorter transcript that lacks the repeated region. Treated iPSCs maintained pluripotency and exhibited a reversed phenotype.50 However, expanded CTG repeats in the DMPK gene, which have been shown to affect transcription of adjacent genomic regions,51 were not removed and, due to sequence constraint for TALEN activity, editing resulted in the production of a DMPK-truncated protein.
The great advantage and novelty of the CRISPR/Cas9 genome-editing system is that it makes possible deletions up to several kilobases of repeated DNA. In addition, it leads to permanent rescue of normal cell function, in contrast to transient effects observed when using antisense approaches to repeated transcripts.24, 25, 26 Importantly for future application, we have shown that this technology has no apparent negative effects in cell cultures because differentiation and fusion capability were not affected. Likewise, DMPK transcript and protein production in edited cells were similar to untreated cells. We did not observe differences in myotube formation among control, DM1-derived myoblasts, and clones with deleted repeats, in contrast to van Agtmaal et al.,39 who described increased fusion in their CRISPR/Cas9-edited myoblasts.39 Our results support previous work showing that differentiation and fusion are not significantly different in primary and immortalized myoblasts derived from DM1 patients or unaffected individuals.19, 21 Modest reduction of fusion capacity in DM1-derived myoblasts has been recently reported,23 and increased apoptosis and autophagy have been observed in DM1 myotubes after prolonged time in culture.19
Once it is determined that CRISPR/Cas9 can achieve therapeutic reversal of DM1-related alterations in cell cultures, the next step will be to test this approach in preclinical models, with the future goal to apply it in DM patient tissues for somatic gene therapy. The use of CRISPR/Cas9 gene-editing strategies in animal models will require the use of improved Cas9 nuclease and sgRNAs, limiting possible off-target effects. In addition, Cas9 size should be reduced to fit into the genome of recombinant vectors, such as adeno associated viral (AAV) vector for in vivo transduction experiments. AAVs have been successfully engineered as gene-delivery vectors for efficient transduction of post-mitotic cells, including skeletal muscles.52 An interesting AAV vector carrying a short Cas9 (saCas9) under a muscle-specific promoter and 2 sgRNA expression cassettes in a single vector has been recently generated and used for recovering dystrophin expression in heart and skeletal muscle of mdx mice.32 In the same paper, CRISPR/Cas9 combined with the more specific HDR repair system has also been exploited and proved functional in muscle cells, although at a lower efficiency compared to NHEJ.32 Overall, the results obtained by us and others clearly highlight the potential of the CRISPR/Cas9 gene-editing strategy as a powerful tool for future gene therapy application in DM1 pathology.
Materials and Methods
Antibodies
Mouse monoclonal antibody (mAb) to human MBNL1 (3A4), mouse mAb to human DMPK (9-RY26), and rabbit polyclonal Ab to mouse MYOD1 (C-20) were from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse mAb to vinculin was purchased from Sigma-Aldrich (St. Louis, MO). Mouse mAb to fast myosin heavy chain (MF20) was obtained from D. Fischman, and mouse mAb to myogenin (F5D) was a gift from G. Cossu. TRITC-conjugated goat anti-rabbit antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA). Alexa Fluor 488 goat anti-mouse antibody was from Thermo Fisher Scientific (Waltham, MA). Horseradish-peroxidase-conjugated goat anti-mouse and anti-rabbit antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA).
Design and Construction of CRISPR/Cas9 Components
For Cas9 expression, we selected a plasmid expressing the Streptococcus pyogenes (Sp) high specificity nuclease (eSpCas9(1.1),45 Addgene #71814, Cambridge, MA). Target sites for CRISPR/Cas9 across the trinucleotide repeats in exon 15 of DMPK (NCBI, Gene Database, GeneID: 1760, nucleotides 45,770,345–45,770,148) were selected using the CRISPR sgRNA design web tool at: https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design. Different target sites were scored based on the number of predicted off-targets and whether off-targets were perfect hits or contained mismatches. PCRs for amplification and sequencing of the flanking regions upstream and downstream the CTG repeats were performed using upF and upR and dwF and dwR primers (Table S2). Complementary DNA oligonucleotides specifying the 20-nt guide RNA sequence and an optimized Cas9 nuclease-recruiting sequence53 (Table S2) were cloned into the sgRNA expression vector pLB also expressing GFP (Addgene #11619) (Cambridge, MA). Proper insertion of the oligonucleotides into the vector was verified by sequencing.
Cell Culture, Viral Infections, and DNA Transfection
Immortalized human myoblasts CT-A and CT-B were derived from primary dermal fibroblasts of control individuals unaffected with DM1. Immortalized human DM1 myogenic cells DM1-A and DM1-B were derived from primary dermal fibroblasts of two DM1 patients, with one DMPK normal allele and one mutated allele containing 290 and 520 CTG amplifications at diagnosis, respectively. All human specimens were obtained after receiving written informed consent. This study was authorized by the Institutional Ethics Committee (ASL MI2-Melegnano via VIII Giugno, Milan) and was conducted in accordance with the principles expressed in the Declaration of Helsinki, the institutional regulation, and Italian laws and guidelines. The skin biopsies were cut into small pieces and placed into an empty 10-cm Petri dish for approximately 10–15 min to promote attachment. Then sufficient growth medium (DMEM supplemented with 10% FBS) was gently added, and the cells were incubated in a humidified incubator at 37°C and 5% CO2 atmosphere. The medium was changed every 2 to 3 days to remove cell debris and maintain a physiological pH. Skin explants were kept in culture for a few weeks and transferred to new dishes every week to allow complete fibroblast outgrowth. Cells were grown to confluence and then removed by trypsinization, counted, and cryopreserved. Fibroblasts were sequentially infected with retroviral vectors carrying TERT and Hygromicin selection54 (Addgene #1773) (Cambridge, MA), and with retroviruses carrying estrogen-inducible mouse Myod1 and puromicin selection55 (Addgene #13494) (Cambridge, MA). TERT- and MYOD1-expressing cells were propagated in DMEM without phenol red (Gibco, Thermo Fisher Scientific, Waltham, MA) supplemented with 15% FBS (Gibco, Thermo Fisher Scientific, Waltham, MA). Differentiation to myotubes was induced by growing cells to confluency on dishes coated with 0.5% gelatin (Sigma-Aldrich, St. Louis, MO) and replacing the proliferation medium with differentiation medium consisting of DMEM without phenol red supplemented with 10 μg/mL insulin (Sigma-Aldrich, St. Louis, MO), 100 μg/mL transferrin (Gibco, Thermo Fisher Scientific, Waltham, MA), and 10−7 M β-estradiol (Sigma-Aldrich, St. Louis, MO). Human 293FT cells were propagated in DMEM (Gibco, Thermo Fisher Scientific, Waltham, MA) supplemented with 10% FBS. All cells were incubated under a 5% CO2 atmosphere at 37°C. Cell trasfection of 293FT cells was performed with Lipofectamin 2000 (Invitrogen, Thermo Fisher Scientific, Carlsbad, CA), whereas myoblasts were electroporated according to the manufacturers’ instructions (Amaxa Basic Nucleofector Kit VP1-1002, Lonza, Basel, Switzerland). Electroporated cells co-expressing GFP were sorted by flow cytometry at the Flow Cytometry Facility at EMBL, Monterotondo, Italy.
Northern Blot Analysis
Polyadenylated mRNA was isolated from differentiated myoblasts using the GeneElute Direct mRNA Miniprep kit (Sigma-Aldrich, St. Louis, MO), following the manufacturer’s instructions. Northern blotting was performed according to standard procedures. 1 to 2 μg of polyadenylated RNA was subjected to electrophoresis in a 1.2% agarose gel under denaturing conditions. RNA was transferred to positively charged nylon membranes (Roche, Sigma-Aldrich, St. Louis, MO) by capillary transfer in 20 × SSC and hybridized with 5′ DIG-labeled (CAG)6 LNA-oligonucleotide (Eurogentec, Liège, Belgium) and with DIG-labeled probes to DMPK (NM_001081563.2, nt 1145-1526) and GAPDH (NM_001289745, nt 133-617), generated with DIG-High Prime (Roche, Sigma-Aldrich, St. Louis, MO) according to the random primed labeling technique. The probe-target hybrids were visualized by chemiluminescent assay using the CDP-Star substrate (Roche, Sigma-Aldrich, St. Louis, MO) and the ChemiDoc Imaging System (Bio-Rad, Hercules, CA).
Long PCR and Southern Blot Hybridization
First protocol: long PCR was performed as previously described44 using the Myotonic Dystrophy SB kit (Experteam, Venice, Italy) with 100 ng genomic DNA for each cell line. PCR conditions were one cycle of 1 min at 94°C; 28 cycles of 20 s at 94°C and 7 min at 62°C; and finally 10 min at 72°C. For Southern blot analysis, PCR products were separated by electrophoresis on 1% agarose gels, along with DIG-labeled DNA molecular weight markers VII and VIII (Roche, Sigma-Aldrich, St. Louis, MO), transferred to nylon membranes (Roche, Sigma-Aldrich, St. Louis, MO), and hybridized overnight with a nonradioactive Digoxigenin-based probe, 5′ DIG-labeled (CTG)10 according to manufacturer instructions (Roche, Sigma-Aldrich, St. Louis, MO). Second protocol: genomic DNA was digested with BamHI restriction enzyme and 15 ng were amplified using primers inF and inR (Table S2). PCR conditions were one cycle of 10 min at 95°C; 28 cycles of 45 s at 95°C, 45 s at 65°C, and 3 min at 72°C; and finally 45 s at 65°C and 10 min at 72°C. PCR products were separated on 1.5% agarose gels, along with DIG-labeled markers VII. Hybridization was performed as previously reported56 using a 5′ DIG-labeled (CAG)6 LNA probe. The chemiluminescence signal was visualized on the ChemiDoc Instrument (Bio-Rad, Hercules, CA).
PCR Amplification Analysis of On-Target and Off-Target Genome-Editing Events
Amplifications of the on-target regions of the edited polyclonal population and individual clones were performed from genomic DNA using the KAPA2G Fast HS Genotyping Mix (2X) (Roche, Sigma-Aldrich, St. Louis, MO), with specifically designed primers (upF and dwR; Table S2). The putative off-target sites corresponding to different genomic locations for sg34 and sg589 were determined by COSMID public software at https://CRISPR.bme.gatech.edu/,48 adopting the following criteria: NGG PAM, 3 mismatches with no indels and 2 mismatches with 1-base deletions or insertions. Genomic DNA extracted from the parental and four different edited clonal cell lines was amplified using the KAPA2G Fast HS Genotyping Mix (2X) (Roche, Sigma-Aldrich, St. Louis, MO), with primers specific for 7 top potential off-target sites (Table S2). The selected sites were also identified by the Cas-OFFinder software (http://www.rgenome.net/cas-offinder). Primer synthesis and sequencing were performed by Eurofins Scientific (Luxembourg, Luxembourg). DNA sequences were aligned using Clustal Omega-Multiple Sequence Alignment public software (EMBL-EBI, Hinxton, UK).
RNA Analysis by RT-PCR
RNAs from polyclonal and clonal cell lines were extracted with TRIzol reagent (Invitrogen, Thermo Scientific, Carlsbad, CA) and retro-transcribed with the SuperScript IV First-Strand Synthesis System (Invitrogen) using oligo (dT) and random primers. All RT-PCR analyses were performed using GoTaq Flexi DNA Polymerase (Promega, Fitchburg, WI) for 35 cycles using the specific primers listed in Table S2. Quantitation of amplified bands on gel images was performed using ImageJ software. Exon inclusion was quantified as the percentage of the total intensity of isoform signals.
RNA FISH and Immunofluorescence Staining
Cells were fixed with 2% formaldehyde and subjected to FISH using a (CAG)6 probe labeled with Texas Red at the 5′ end (IDT, Coralville, IA) in combination with immunofluorescence staining, as described previously.56 To verify the co-localization of MBNL1 in ribonuclear inclusions, following the last post-hybridization wash, cells were incubated in PBS containing 3% BSA for 15 min and stained sequentially with antibodies to MBNL1, diluted 1:250 in PBS containing 3% BSA, and with goat anti-mouse antibody conjugated Alexa Fluor 488. For labeling with antibodies to MHC and MYOD1, cultures were fixed with 80% acetone and washed thoroughly before incubation with primary and secondary antibody. Nuclei were visualized with Hoechst 33258 dye. The samples were examined with an Olympus AX70 immunofluorescence microscope. Images were recorded on an Olympus XM10 camera and processed using the Olympus CellSens Standard 1.8.1 software. Cell scoring was carried out using ImageJ software.
Whole-Cell Extracts and Western Blot Analysis
Cells were lysed in RIPA buffer (140 mM NaCl, 3 mM MgCl2, 1 mM EDTA, and 15 mM HEPES, pH 7.2, also containing 0.5% sodium deoxycholate, 1% NP-40, and 0.1% SDS) supplemented with a cocktail of protease inhibitors (Roche, Sigma-Aldrich, St. Louis, MO). Western blots were carried out using horseradish-peroxidase-conjugated goat anti-rabbit and anti-mouse antibodies and revealed with a chemiluminescence detection system by Cyanagen (Bologna, Italy). Imaging and quantitation of the bands were carried out by the ChemiDoc XRS Western Blot Imaging System using the ImageLab 4.0 software (Bio-Rad, Hercules, CA).
Author Contributions
G.F. conceived the investigation; G.F., B.C., C.P., and M.C. designed the experiments and analyzed the results; C.P., M.C., B.C., and R.V. performed the experiments; R.C. and G.M. provided patient-derived fibroblasts and muscle biopsies; G.F., C.P., B.C., and F.M. discussed the results; G.F. and B.C. wrote the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
We thank Cora Chadick for cell sorting at the EMBL flow cytometry facility in Monterotondo, Italy, and James Sawitzke, EMBL Monterotondo, Italy, for proofreading the manuscript. We gratefully acknowledge the financial support of Telethon-Italy (grant no. GGP14092 to F.M. and G.F.), AFM-Telethon (grant no. 18477 to F.M. and G.F.), and Ministero della Salute (to F.M.). M.C. is a recipient of a fellowship from Telethon-Italy.
Footnotes
Supplemental Information includes four figures and two tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2017.10.006.
Contributor Information
Beatrice Cardinali, Email: beatrice.cardinali@cnr.it.
Germana Falcone, Email: germana.falcone@cnr.it.
Supplemental Information
References
- 1.Turner C., Hilton-Jones D. Myotonic dystrophy: diagnosis, management and new therapies. Curr. Opin. Neurol. 2014;27:599–606. doi: 10.1097/WCO.0000000000000128. [DOI] [PubMed] [Google Scholar]
- 2.Meola G., Cardani R. Myotonic dystrophies: an update on clinical aspects, genetic, pathology, and molecular pathomechanisms. Biochim. Biophys. Acta. 2015;1852:594–606. doi: 10.1016/j.bbadis.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 3.Harper P.S. Myotonic dystrophy. In: Karpati G., Hilton-Jones D., Griggs R.C., editors. Disorders of Voluntary Muscle. Cambridge University Press; 2001. pp. 541–559. [Google Scholar]
- 4.Wahbi K., Babuty D., Probst V., Wissocque L., Labombarda F., Porcher R., Bécane H.M., Lazarus A., Bécane H.M., Lazarus A. Incidence and predictors of sudden death, major conduction defects and sustained ventricular tachyarrhythmias in 1388 patients with myotonic dystrophy type 1. Eur. Heart J. 2017;38:751–758. doi: 10.1093/eurheartj/ehw569. [DOI] [PubMed] [Google Scholar]
- 5.Mahadevan M., Tsilfidis C., Sabourin L., Shutler G., Amemiya C., Jansen G., Neville C., Narang M., Barceló J., O’Hoy K. Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- 6.Lavedan C., Hofmann-Radvanyi H., Shelbourne P., Rabes J.P., Duros C., Savoy D., Dehaupas I., Luce S., Johnson K., Junien C. Myotonic dystrophy: size- and sex-dependent dynamics of CTG meiotic instability, and somatic mosaicism. Am. J. Hum. Genet. 1993;52:875–883. [PMC free article] [PubMed] [Google Scholar]
- 7.Groh W.J., Groh M.R., Shen C., Monckton D.G., Bodkin C.L., Pascuzzi R.M. Survival and CTG repeat expansion in adults with myotonic dystrophy type 1. Muscle Nerve. 2011;43:648–651. doi: 10.1002/mus.21934. [DOI] [PubMed] [Google Scholar]
- 8.Nakamori M., Sobczak K., Puwanant A., Welle S., Eichinger K., Pandya S., Dekdebrun J., Heatwole C.R., McDermott M.P., Chen T. Splicing biomarkers of disease severity in myotonic dystrophy. Ann. Neurol. 2013;74:862–872. doi: 10.1002/ana.23992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang H., Mankodi A., Swanson M.S., Moxley R.T., Thornton C.A. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum. Mol. Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- 10.Konieczny P., Stepniak-Konieczna E., Sobczak K. MBNL proteins and their target RNAs, interaction and splicing regulation. Nucleic Acids Res. 2014;42:10873–10887. doi: 10.1093/nar/gku767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zu T., Gibbens B., Doty N.S., Gomes-Pereira M., Huguet A., Stone M.D., Margolis J., Peterson M., Markowski T.W., Ingram M.A. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. USA. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perbellini R., Greco S., Sarra-Ferraris G., Cardani R., Capogrossi M.C., Meola G., Martelli F. Dysregulation and cellular mislocalization of specific miRNAs in myotonic dystrophy type 1. Neuromuscul. Disord. 2011;21:81–88. doi: 10.1016/j.nmd.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 13.Falcone G., Perfetti A., Cardinali B., Martelli F. Noncoding RNAs: emerging players in muscular dystrophies. Biomed. Res. Int. 2014;2014:503634. doi: 10.1155/2014/503634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seznec H., Lia-Baldini A.S., Duros C., Fouquet C., Lacroix C., Hofmann-Radvanyi H., Junien C., Gourdon G. Transgenic mice carrying large human genomic sequences with expanded CTG repeat mimic closely the DM CTG repeat intergenerational and somatic instability. Hum. Mol. Genet. 2000;9:1185–1194. doi: 10.1093/hmg/9.8.1185. [DOI] [PubMed] [Google Scholar]
- 15.Mankodi A., Logigian E., Callahan L., McClain C., White R., Henderson D., Krym M., Thornton C.A. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 16.Wang G.S., Kearney D.L., De Biasi M., Taffet G., Cooper T.A. Elevation of RNA-binding protein CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy. J. Clin. Invest. 2007;117:2802–2811. doi: 10.1172/JCI32308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomes-Pereira M., Cooper T.A., Gourdon G. Myotonic dystrophy mouse models: towards rational therapy development. Trends Mol. Med. 2011;17:506–517. doi: 10.1016/j.molmed.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holt I., Mittal S., Furling D., Butler-Browne G.S., Brook J.D., Morris G.E. Defective mRNA in myotonic dystrophy accumulates at the periphery of nuclear splicing speckles. Genes Cells. 2007;12:1035–1048. doi: 10.1111/j.1365-2443.2007.01112.x. [DOI] [PubMed] [Google Scholar]
- 19.Loro E., Rinaldi F., Malena A., Masiero E., Novelli G., Angelini C., Romeo V., Sandri M., Botta A., Vergani L. Normal myogenesis and increased apoptosis in myotonic dystrophy type-1 muscle cells. Cell Death Differ. 2010;17:1315–1324. doi: 10.1038/cdd.2010.33. [DOI] [PubMed] [Google Scholar]
- 20.Larsen J., Pettersson O.J., Jakobsen M., Thomsen R., Pedersen C.B., Hertz J.M., Gregersen N., Corydon T.J., Jensen T.G. Myoblasts generated by lentiviral mediated MyoD transduction of myotonic dystrophy type 1 (DM1) fibroblasts can be used for assays of therapeutic molecules. BMC Res. Notes. 2011;4:490. doi: 10.1186/1756-0500-4-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Botta A., Malena A., Loro E., Del Moro G., Suman M., Pantic B., Szabadkai G., Vergani L. Altered Ca2+ homeostasis and endoplasmic reticulum stress in myotonic dystrophy type 1 muscle cells. Genes (Basel) 2013;4:275–292. doi: 10.3390/genes4020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pantic B., Borgia D., Giunco S., Malena A., Kiyono T., Salvatori S., De Rossi A., Giardina E., Sangiuolo F., Pegoraro E. Reliable and versatile immortal muscle cell models from healthy and myotonic dystrophy type 1 primary human myoblasts. Exp. Cell Res. 2016;342:39–51. doi: 10.1016/j.yexcr.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Arandel L., Polay Espinoza M., Matloka M., Bazinet A., De Dea Diniz D., Naouar N., Rau F., Jollet A., Edom-Vovard F., Mamchaoui K. Immortalized human myotonic dystrophy muscle cell lines to assess therapeutic compounds. Dis. Model. Mech. 2017;10:487–497. doi: 10.1242/dmm.027367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wheeler T.M., Leger A.J., Pandey S.K., MacLeod A.R., Nakamori M., Cheng S.H., Wentworth B.M., Bennett C.F., Thornton C.A. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wojtkowiak-Szlachcic A., Taylor K., Stepniak-Konieczna E., Sznajder L.J., Mykowska A., Sroka J., Thornton C.A., Sobczak K. Short antisense-locked nucleic acids (all-LNAs) correct alternative splicing abnormalities in myotonic dystrophy. Nucleic Acids Res. 2015;43:3318–3331. doi: 10.1093/nar/gkv163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bisset D.R., Stepniak-Konieczna E.A., Zavaljevski M., Wei J., Carter G.T., Weiss M.D., Chamberlain J.R. Therapeutic impact of systemic AAV-mediated RNA interference in a mouse model of myotonic dystrophy. Hum. Mol. Genet. 2015;24:4971–4983. doi: 10.1093/hmg/ddv219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 28.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bengtsson N.E., Hall J.K., Odom G.L., Phelps M.P., Andrus C.R., Hawkins R.D., Hauschka S.D., Chamberlain J.R., Chamberlain J.S. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat. Commun. 2017;8:14454. doi: 10.1038/ncomms14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ousterout D.G., Kabadi A.M., Thakore P.I., Majoros W.H., Reddy T.E., Gersbach C.A. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat. Commun. 2015;6:6244. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young C.S., Hicks M.R., Ermolova N.V., Nakano H., Jan M., Younesi S., Karumbayaram S., Kumagai-Cresse C., Wang D., Zack J.A. A single CRISPR-Cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hiPSC-derived muscle cells. Cell Stem Cell. 2016;18:533–540. doi: 10.1016/j.stem.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maggio I., Stefanucci L., Janssen J.M., Liu J., Chen X., Mouly V., Gonçalves M.A. Selection-free gene repair after adenoviral vector transduction of designer nucleases: rescue of dystrophin synthesis in DMD muscle cell populations. Nucleic Acids Res. 2016;44:1449–1470. doi: 10.1093/nar/gkv1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Long C., Amoasii L., Mireault A.A., McAnally J.R., Li H., Sanchez-Ortiz E., Bhattacharyya S., Shelton J.M., Bassel-Duby R., Olson E.N. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Castellanos Rivera R.M., Madhavan S., Pan X., Ran F.A., Yan W.X. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tabebordbar M., Zhu K., Cheng J.K.W., Chew W.L., Widrick J.J., Yan W.X., Maesner C., Wu E.Y., Xiao R., Ran F.A. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Agtmaal E.L., André L.M., Willemse M., Cumming S.A., van Kessel I.D.G., van den Broek W.J.A.A., Gourdon G., Furling D., Mouly V., Monckton D.G. CRISPR/Cas9-induced (CTG⋅CAG)n repeat instability in the myotonic dystrophy type 1 locus: implications for therapeutic genome editing. Mol. Ther. 2017;25:24–43. doi: 10.1016/j.ymthe.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morales C.P., Holt S.E., Ouellette M., Kaur K.J., Yan Y., Wilson K.S., White M.A., Wright W.E., Shay J.W. Absence of cancer-associated changes in human fibroblasts immortalized with telomerase. Nat. Genet. 1999;21:115–118. doi: 10.1038/5063. [DOI] [PubMed] [Google Scholar]
- 41.Falcone G., Mazzola A., Michelini F., Bossi G., Censi F., Biferi M.G., Minghetti L., Floridia G., Federico M., Musio A. Cytogenetic analysis of human cells reveals specific patterns of DNA damage in replicative and oncogene-induced senescence. Aging Cell. 2013;12:312–315. doi: 10.1111/acel.12034. [DOI] [PubMed] [Google Scholar]
- 42.Savkur R.S., Philips A.V., Cooper T.A. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 43.Santoro M., Piacentini R., Masciullo M., Bianchi M.L., Modoni A., Podda M.V., Ricci E., Silvestri G., Grassi C. Alternative splicing alterations of Ca2+ handling genes are associated with Ca2+ signal dysregulation in myotonic dystrophy type 1 (DM1) and type 2 (DM2) myotubes. Neuropathol. Appl. Neurobiol. 2014;40:464–476. doi: 10.1111/nan.12076. [DOI] [PubMed] [Google Scholar]
- 44.Valaperta R., Sansone V., Lombardi F., Verdelli C., Colombo A., Valisi M., Brigonzi E., Costa E., Meola G. Identification and characterization of DM1 patients by a new diagnostic certified assay: neuromuscular and cardiac assessments. Biomed. Res. Int. 2013;2013:958510. doi: 10.1155/2013/958510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cradick T.J., Fine E.J., Antico C.J., Bao G. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41:9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lam L.T., Pham Y.C., Nguyen T.M., Morris G.E. Characterization of a monoclonal antibody panel shows that the myotonic dystrophy protein kinase, DMPK, is expressed almost exclusively in muscle and heart. Hum. Mol. Genet. 2000;9:2167–2173. doi: 10.1093/hmg/9.14.2167. [DOI] [PubMed] [Google Scholar]
- 50.Gao Y., Guo X., Santostefano K., Wang Y., Reid T., Zeng D., Terada N., Ashizawa T., Xia G. Genome therapy of myotonic dystrophy type 1 iPS cells for development of autologous stem cell therapy. Mol. Ther. 2016;24:1378–1387. doi: 10.1038/mt.2016.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brouwer J.R., Huguet A., Nicole A., Munnich A., Gourdon G. Transcriptionally repressive chromatin remodelling and CpG methylation in the presence of expanded CTG-repeats at the DM1 locus. J. Nucleic Acids. 2013;2013:567435. doi: 10.1155/2013/567435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kotterman M.A., Schaffer D.V. Engineering adeno-associated viruses for clinical gene therapy. Nat. Rev. Genet. 2014;15:445–451. doi: 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dang Y., Jia G., Choi J., Ma H., Anaya E., Ye C., Shankar P., Wu H. Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol. 2015;16:280. doi: 10.1186/s13059-015-0846-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Counter C.M., Meyerson M., Eaton E.N., Ellisen L.W., Caddle S.D., Haber D.A., Weinberg R.A. Telomerase activity is restored in human cells by ectopic expression of hTERT (hEST2), the catalytic subunit of telomerase. Oncogene. 1998;16:1217–1222. doi: 10.1038/sj.onc.1201882. [DOI] [PubMed] [Google Scholar]
- 55.Hollenberg S.M., Cheng P.F., Weintraub H. Use of a conditional MyoD transcription factor in studies of MyoD trans-activation and muscle determination. Proc. Natl. Acad. Sci. USA. 1993;90:8028–8032. doi: 10.1073/pnas.90.17.8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cardani R., Mancinelli E., Sansone V., Rotondo G., Meola G. Biomolecular identification of (CCTG)n mutation in myotonic dystrophy type 2 (DM2) by FISH on muscle biopsy. Eur. J. Histochem. 2004;48:437–442. doi: 10.4081/918. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.