Main Text
CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR associated protein 9) has managed, in just 5 short years, to become the leading and most meaningful world scientific discovery since the implementation of polymerase chain reaction (PCR) as a research tool. The ability of this machinery to interrupt the function of different genes within living organisms has changed the pace of numerous biomedical research branchess especially those concentrated on genetic-driven pathologies.1 We are now holding the ability to potentially cure life-threatening diseases through molecular surgery of the genome. Since the discovery of CRISPR potential in 2012, the number of articles related to this tool has been progressing in a rapid manner, with an extensive number of research papers published in just 5 years. Lately, Chinese scientists have managed to use the technology for the first time in a human patient with lung cancer to cure an advanced form of the disease (ex vivo approach).2 Currently there are several strategies with CRISPR/Cas9 proposed for clinical testing, all of them concentrated on ex vivo approaches. Nevertheless, these forms of last generation therapies are generally based on retrieving immune cells from the patient and subsequent modification of the gene sequences to increase the immune system surveillance against the malignant invasion. The CRISPR/Cas9 genetic-modified and empowered cells are then propagated and characterized before being administrated back into the patient in the direction of malignant impairment. In light of these therapeutic approaches, the US Food and Drug Administration (FDA) recently approved the first CAR-T cell (chimeric antigen receptor T cell) therapy for the treatment of acute lymphoblastic leukemia in children and young adults.3 Despite the fact that this type of strategy uses viral vectors for the modification of immune cells, CRISPR/Cas9 could represent an efficient and flexible alternative for the generation of the personalized empowered T cells.
All these advances in genome editing are writing history at every step, but, even so, the final intention is to deliver the CRISPR system directly into a human body to permanently restore genetic disabilities. But sneaking the editing complex from the immune system and then introducing it into the nucleus of the targeted cells is not an easy task. Despite the immediate availability, flexibility, and multiplex nature of CRISPR/Cas9, it seems that this system will not dodge all the issues associated with previous gene therapy strategies, the constant difficulty remaining the delivery method. In this sense, researchers are trying to find delivery performers able to protect the system from body’s defense mechanisms and conduct the editing tool toward specific sites within the altered genome.4 The fact that we are now literally holding in our hands the machinery to cure numerous pathologies drives scientists to accelerate the process toward safe and efficient carriers for CRISPR/Cas9 within the human organism.
Chira et al.5 comprehensively reviewed the most important approaches in manipulating the effects of the Cas9 enzyme to limit the potential off-target effects. The present manuscript underlines the current strategies for in vivo delivery to offer a complete view of this technology.
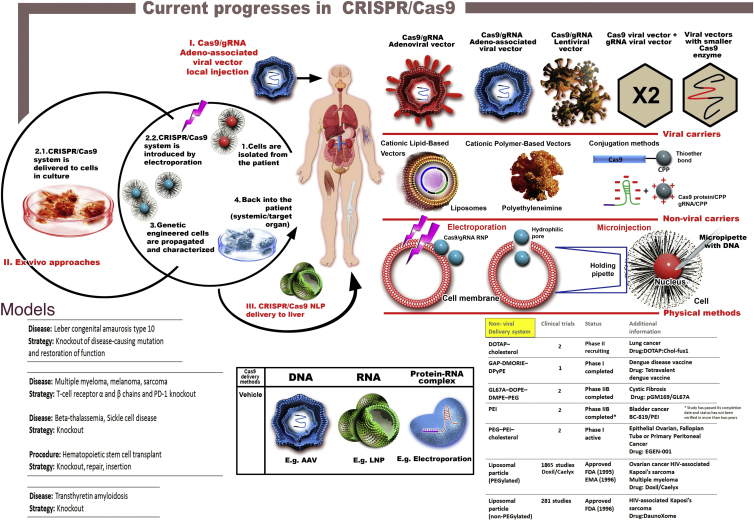
To find a way to inject the CRISPR/Cas9 system into the body, numerous groups are now searching for both viral and non-viral approaches (Figure 1). There are three main ways under which the CRISPR/Cas9 system can be delivered: (1) DNA, where Cas9 enzyme and guideRNA (gRNA) are inserted into a DNA plasmid and usually delivered via an adeno-associated virus (AAV); (2) RNA, where both Cas9 and gRNA are encapsulated as RNA molecules into nanoparticles of diverse origins (e.g., lipid, polymeric origins); and (3) protein-RNA complex, where Cas9 is delivered under the translated form alongside gRNA as a ribonucleoprotein structure.4
Figure 1.
Current Progress in CRISPR/Cas9
The main methods for delivering the CRISPR/Cas9 system consist of viral and non-viral carriers or physical approaches. All of them are associated with advantages and limitations. Viral carriers, like adenoviral vectors, are limited by the presence of CAR. AAVs are characterized by limited packaging capacity and lentiviral vectors are associated with increased safety issues due to the possibility of homologous recombination that will generate replication-competent particles. Alternative situations for viral vehicles could be represented by the co-delivery of Cas9 and gRNAs into separate vectors or incorporation of smaller Cas enzymes. However, the co-delivery strategy significantly limits the number of transfected cells, where smaller Cas9 usually requires a larger PAM site. Non-viral delivery through encapsulation of the editing system into nanoparticles of diverse origins, mainly lipid and polymeric, is an alternative for reducing the life of Cas9 inside the targeted cells. However, the potential toxicity of the nanoparticles on the human organism and the affinity for liver and spleen are also limitation factors for systemic delivery of CRISPR/Cas9. The third method, where the CRISPR system is incorporated into cells through electroporation or microinjection is suitable only for ex vivo approaches. There are three main strategies that are currently tested or are emerging in clinical trials comprised of CRISPR/Cas9-mediated gene therapy. (I) Local injection with CRISPR/Cas9 encapsulated in viral vectors (AAVs). (II) Ex vivo approaches where the cells are retrieved from the patient (1), modified through transfection strategies (2.1) or electroporation (2.2), cultured for propagation and characterization (3), and finally administrated back in the patient (4). (III) Systemic delivery of CRISPR/Cas9 inside NLPs (nanolipoprotein particles) for genetic editing of cells within the liver. All of these therapeutic schemes represent significant advances in CRISPR/Cas9 genome surgery, but, even so, they tackle a limited amount of pathologies due to current restrictions in delivery methods.
Each method presents advantages and disadvantages and, for these means, none of them are presently suitable for safe and targeted delivery into human patients.
AAVs have been used for gene delivery because of their reduced immunogenicity and tissue specificity. Problems arise when trying to pack the Cas9 together with gRNA into the virus due to its low packaging capacity. Although this issue has been overcome, where researchers managed to pack the elements together, the similar lengths of the two above-mentioned systems (Cas9 + gRNA ∼4.2 kb and AAV ∼4.5 kb) restrict the possibility of adding supplementary regulatory elements that would make the therapeutic vehicle more precise and efficient.6, 7, 8, 9 Swiech et al.,7 provided an alternative possibility where the functional elements were introduced in separate AAV vectors. Although the method demonstrated a higher packaging efficiency, the necessity of co-delivery of multiple vectors is significantly reducing the number of infected cells and decreases the efficiency of the method because Cas9 cannot function in the absence of gRNA. Smaller Cas9 enzymes could be isolated from Campylobacter jejuni, but these types are more limited in terms of targeted sites due to longer PAM sequences. Other impediments for AAV-mediated delivery are the long persistence of the virus within the organism and inherent constant levels of Cas9, which can determine off-target effects with hazardous effects on the living cells.10
To limit the long-term translation of Cas9 inside the targeted cells, encapsulation of the editing system into nanoparticles of diverse origins, mainly lipid and polymeric, was tested4. Although the life of Cas9 is significantly minimized, the efficiency of the method has emerged as quite restrictive due to the potential toxicity of the nanoparticles on human organism and the liver and spleen affinity of the intravenously injected vehicles9, 11.
The third method consists of direct delivery of the Cas9 protein alongside gRNAs that enables the possibility to precisely control the amount of enzyme that is administrated. This has proven efficient only for ex vivo approaches that rely on electroporation or chemical transfection and also, highly important, lack of immune reaction.12
The immunogenic aspects can be overcome by incorporating the ribonucleoprotein complex into nanoparticles able to fuse with the cell membrane and release the system into the cytoplasm. This situation was evaluated by Mout et al.,11 where they applied the reminded principle and even attached a nuclear recognition complex to the Cas9 protein to direct the editing tool toward the nucleus of the cell. Even if the success rates of the experiments were positive (∼90% delivery efficiency and ∼30% gene editing efficiency), it is important to not overlook the possible toxicity of the administrated particles into human organisms and the predisposition for hepatic localization.
The types of proposed delivery methods for CRISPR/Cas9 are numerous and also increasing at a rapid pace.13, 14 The constant issue is represented by the ability to safely, efficiently, and specifically deliver the tool inside the altered cells within the human body without disrupting healthy cells or causing side effects. For the moment, there is no treatment scheme comprising the systemic administration of CRISPR/Cas9 into clinical patients approved by the FDA. Even if ex vivo approaches are making their way into clinical trials, these strategies are effective to a certain limit, where numerous pathologies are not suitable for this approach manner. Therefore, to make the most of this new and innovative machinery, it is imperative to find a way to release the tool directly into the human body and inside altered cells.
The most promising approaches are those that combine the available techniques to limit the disadvantages of both methods and take advantage of the positive aspects. One example is the research conducted by Yin et al.,9 where they used a two-step program it to cure Tyrosinemia type I in mice. First, they injected into the mouse a viral vector compromising the replacement gene and the gRNAs and waited 7 days to permit the organism to multiply the received sequences. By eliminating the administration of Cas9 from the first steps, the DNA template and gRNAs become ineffective and unable to produce any modification within the genome of the cell. Also, the multiplicative ability of the viral vectors and implicit high production of inserted molecules is canceled by the harmless properties of the administrated sequences in the absence of Cas9 enzyme. After this initial step, Cas9 mRNA was administered inside lipid nanoparticles, activating the ribonucleoprotein complex. The downside of this method consists of the affinity of the nanovehicles for hepatic sites and their potential toxicity to the human body.
Significant advancements are also made with bacteriophages, bacteria infecting specific viruses, which can be engineered to target mammalian cells and are safe for systemic administration (previously used for antibiotic therapy in children through systemic administration). Even so, the lack of eukaryotic optimization in terms of gene delivery and expression (due to bacteria specificity) has redirected the attention toward other types of viral vectors for experimental gene therapy15. However, Hajitou and colleagues16, 17, 18 are making promising progress with tumor-targeted AAVP (AAV/phage), a hybrid system between AAV and the filamentous M13 bacteriophage that is able to specifically and efficiently infect cancer cells by combining the advantages from both “worlds” in one single platform.
Other genome editing tools, like transcription activator-like effector nucleases (TALENs) and zinc-finger nucleases (ZFNs) have also made advancements toward the clinical area. SB-FIX, a ZFN-related therapeutic tool administrated in patients with severe hemophilia B to obtain long term expression of Factor IX (ClinicalTrials.gov: NCT02695160), is the first strategy that has entered clinical trials. At the present time, the conductors are evaluating the safety and tolerability of the therapeutic formulation concomitant with effects of ascending doses of SB-FIX. In this case, the small size of the genome editing tool permits the packaging into AAVs, and the targeted locus for introduction of the donor gene, albumin gene locus, allows the accelerated transcription based on the increased activity of the original gene in hepatocytes.19, 20 Two other clinical trials based on the same in vivo administration platform are now recruiting patients: SB-318 for the treatment of Mucopolysaccharidosis I through insertion of a functional IDUA (α-L-iduronidase) gene (ClinicalTrials.gov: NCT02702115) and SB-913 for the treatment of Mucopolysaccharidosis II through insertion of a functional IDS (iduronate 2-sulfatase) gene (ClinicalTrials.gov: NCT03041324); both strategies are using rAAV2/6 for the delivery of ZFN. Even so, compared with these last two genome editing tools, CRISPR/Cas9 is still a new technique, where ZFNs and TALENs had the time to “maturate” toward safe and efficient in vivo administration options.
The future requires that we take advantages of the already tested concepts (for ZFNs and TALENs) and implement a combination of them that works in favor of the new technology. Most likely, time does not allow the implementation of a completely new delivery particle, considering the extensive period necessary for the evaluation of a new vehicle. The rapid development of CRISPR/Cas9, a machinery that was presented to the public as a genome editing tool only in 2012, makes us believe that these strategies are closer than we think. Once this step is achieved, the possibilities are unlimited and pathologies that were previously thought incurable will be treated through molecular surgery techniques.
Author Contributions
I.B.N. together with D.G. designed the concept of the present article. Also, D.G. wrote the manuscript, while I.B.N. critically revised and improved the final version before publication.
Conflicts of Interest
The authors have no conflicting financial interests.
Acknowledgments
This work was supported by POC grant 35/01.09.2016, ID 37_796, entitled “Clinical and economical impact of personalized targeted anti-microRNA therapies in reconverting lung cancer chemoresistance” – CANTEMIR and Project No. 164/2017, entitled “Addressing the complex exposome profile in hormone-dependent cancers of the breast and prostate and its influence on tumoral genome.”
References
- 1.Carter J., Hoffman C., Wiedenheft B. The Interfaces of Genetic Conflict Are Hot Spots for Innovation. Cell. 2017;168:9–11. doi: 10.1016/j.cell.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cyranoski D. CRISPR gene-editing tested in a person for the first time. Nature. 2016;539:479. doi: 10.1038/nature.2016.20988. [DOI] [PubMed] [Google Scholar]
- 3.NCI. CAR T-Cell Therapy Approved for Some Children and Young Adults with Leukemia. https://www.cancer.gov/news-events/cancer-currents-blog/2017/tisagenlecleucel-fda-childhood-leukemia.
- 4.Cross R. CRISPR’s breakthrough problem. Chem. Eng. News. 2017;95:28–33. [Google Scholar]
- 5.Chira S., Gulei D., Hajitou A., Zimta A.A., Cordelier P., Berindan-Neagoe I. CRISPR/Cas9: Transcending the Reality of Genome Editing. Mol. Ther. Nucleic Acids. 2017;7:211–222. doi: 10.1016/j.omtn.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swiech L., Heidenreich M., Banerjee A., Habib N., Li Y., Trombetta J., Sur M., Zhang F. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 2015;33:102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Senís E., Fatouros C., Große S., Wiedtke E., Niopek D., Mueller A.K., Börner K., Grimm D. CRISPR/Cas9-mediated genome engineering: an adeno-associated viral (AAV) vector toolbox. Biotechnol. J. 2014;9:1402–1412. doi: 10.1002/biot.201400046. [DOI] [PubMed] [Google Scholar]
- 9.Yin H., Song C.Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang C., Mei M., Li B., Zhu X., Zu W., Tian Y., Wang Q., Guo Y., Dong Y., Tan X. A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and pcsk9 in vivo. Cell Res. 2017;27:440–443. doi: 10.1038/cr.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mout R., Ray M., Yesilbag Tonga G., Lee Y.W., Tay T., Sasaki K., Rotello V.M. Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing. ACS Nano. 2017;11:2452–2458. doi: 10.1021/acsnano.6b07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang W., Kutny P.M., Byers S.L., Longstaff C.J., DaCosta M.J., Pang C., Zhang Y., Taft R.A., Buaas F.W., Wang H. Delivery of Cas9 Protein into Mouse Zygotes through a Series of Electroporation Dramatically Increases the Efficiency of Model Creation. J. Genet. Genomics. 2016;43:319–327. doi: 10.1016/j.jgg.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komor A.C., Badran A.H., Liu D.R. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pranjol M.Z., Hajitou A. Bacteriophage-derived vectors for targeted cancer gene therapy. Viruses. 2015;7:268–284. doi: 10.3390/v7010268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hajitou A., Trepel M., Lilley C.E., Soghomonyan S., Alauddin M.M., Marini F.C., 3rd, Restel B.H., Ozawa M.G., Moya C.A., Rangel R. A hybrid vector for ligand-directed tumor targeting and molecular imaging. Cell. 2006;125:385–398. doi: 10.1016/j.cell.2006.02.042. [DOI] [PubMed] [Google Scholar]
- 17.Paoloni M.C., Tandle A., Mazcko C., Hanna E., Kachala S., Leblanc A., Newman S., Vail D., Henry C., Thamm D. Launching a novel preclinical infrastructure: comparative oncology trials consortium directed therapeutic targeting of TNFalpha to cancer vasculature. PLoS ONE. 2009;4:e4972. doi: 10.1371/journal.pone.0004972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yata T., Lee E.L., Suwan K., Syed N., Asavarut P., Hajitou A. Modulation of extracellular matrix in cancer is associated with enhanced tumor cell targeting by bacteriophage vectors. Mol. Cancer. 2015;14:110. doi: 10.1186/s12943-015-0383-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anguela X.M., Sharma R., Doyon Y., Miller J.C., Li H., Haurigot V., Rohde M.E., Wong S.Y., Davidson R.J., Zhou S. Robust ZFN-mediated genome editing in adult hemophilic mice. Blood. 2013;122:3283–3287. doi: 10.1182/blood-2013-04-497354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma R., Anguela X.M., Doyon Y., Wechsler T., DeKelver R.C., Sproul S., Paschon D.E., Miller J.C., Davidson R.J., Shivak D. In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood. 2015;126:1777–1784. doi: 10.1182/blood-2014-12-615492. [DOI] [PMC free article] [PubMed] [Google Scholar]