Abstract
Background:
Mitochondrial DNA (mtDNA) content measured by different techniques cannot be compared between studies, and age- and tissue-related control values are hardly available. In the present study, we aimed to establish the normal reference range of mtDNA copy number in the Chinese population.
Methods:
Two healthy cohorts of 200 Chinese minors (0.1–18.0 years) and 200 adults (18.0–88.0 years) were recruited. Then, they were further categorized into eight age groups. The absolute mtDNA copy number per cell was measured by a quantitative real-time polymerase chain reaction. We subsequently used this range to evaluate mtDNA content in four patients (0.5–4.0 years) with molecularly proven mitochondrial depletion syndromes (MDSs) and 83 cases of mitochondrial disease patients harboring the m.3243A>G mutation.
Results:
The reference range of mtDNA copy number in peripheral blood was 175–602 copies/cell (mean: 325 copies/cell) in minors and 164–500 copies/cell (mean: 287 copies/cell) in adults. There was a decreasing trend in mtDNA copy number in blood with increasing age, especially in 0–2-year-old and >50-year-old donors. The mean mtDNA copy number level among the mitochondrial disease patients with m.3243A>G mutation was significantly higher than that of healthy controls. The mtDNA content of POLG, DGUOK, TK2, and SUCLA2 genes in blood samples from MDS patients was reduced to 25%, 38%, 32%, and 24%, respectively.
Conclusions:
We primarily establish the reference intervals of mtDNA copy number, which might contribute to the clinical diagnosis and monitoring of mitochondrial disease.
Keywords: Mitochondrial Depletion Syndromes, Mitochondrial Disease, Mitochondrial DNA, Reference Range
INTRODUCTION
The mitochondrion is the cell organelle responsible for energy production. Human mitochondrial DNA (mtDNA) genome is a 16,569 base-pair circular double-stranded molecule. Unlike nuclear DNA, mtDNA is more susceptible to oxidative and other genotoxic damage due to lack of protective histones and efficient repair mechanisms. Each mitochondrion contains 2–10 mtDNA molecules. The content of mtDNA per cell is mainly measured in copy number, which ranges from 102 to 104 depending on the cell type and tissue origin.[1] mtDNA copy number variation in peripheral blood has also been shown to be associated with various disorders. Previous studies indicated that alteration in mtDNA copy number (increased or decreased) might contribute to tumorigenesis.[2,3] Low peripheral blood mtDNA content may be linked to several aging-associated disorders, such as cardiovascular disease,[4] type 2 diabetes,[5] and metabolic syndrome.[6] Moreover, the mtDNA copy number has been shown to be associated with physical and mental health status. It has been reported that increased mtDNA copy number in the blood is associated with higher cognition in elderly women[7] and better health and longevity among the elderly.[8]
It is known that molecular defects in mitochondrial or nuclear genes could lead to mitochondrial genetic disease, resulting in decreased ATP production and increased reactive oxygen species generation in the mitochondria. Altered mtDNA levels may directly contribute to pathogenesis by causing an increased immune response.[9] If mtDNA levels decline or oxidative damage accumulates to a critical point in the cell, mitochondrial dysfunction and defective energy metabolism may appear.[10] It has been shown that an alteration in mtDNA copy number in leukocytes might reflect the same alteration in affected tissues during the progression of mitochondrial diseases. Liu et al.[11] showed that mtDNA copy number in leukocytes of patients with MELAS or MERRF syndrome was significantly higher in young people but lower at an advanced age. Liu et al.[12] found that a low wild-type mtDNA copy number in blood was associated with increased frequencies of three major symptoms, namely, seizures, myopathy, and learning disability, in patients with m.3243 A>G. In conclusion, mtDNA copy number could be a biomarker of mitochondrial dysfunction.[9]
The mitochondrial depletion syndromes (MDSs) are a clinically heterogeneous group of mitochondrial disorders characterized by a severe quantitative reduction of total mtDNA copy number. Several nuclear genes involved in mtDNA biosynthesis and maintenance are associated with MDS. A definitive diagnosis of MDS is difficult due to its genetic and clinical heterogeneity. For clinical purposes, mtDNA depletion has been defined as a residual mtDNA copy number of <30% relative to age-matched controls.[13] However, the mtDNA content data obtained with different methodologies are not comparable between studies, and the age- and tissue-related controls are hardly available.[14,15,16] Thus, the normal reference interval of mtDNA copy number in peripheral blood is of great clinical significance in diagnosis and monitoring of mitochondrial disease and other disorders. We investigated two healthy control cohorts of 200 minors and 200 adults. The aim of this study was to establish the reference range of mtDNA copy number in peripheral blood in healthy Chinese minors and adults. We subsequently used this range to evaluate mtDNA content in patients with molecularly proven MDS and mitochondrial disease patients with the m.3243A>G mutation.
METHODS
Ethical approval
This study was approved by the Medical Ethics Committee of Peking University First Hospital. Informed written consent was obtained from all healthy controls and patients or their respective legal guardians.
Controls and patients cohorts
This study was conducted during 2015 to 2016. Four hundred blood samples (anticoagulated with EDTA-K2) from healthy Chinese donors were collected from Peking University First Hospital and Beijing Children's Hospital during a medical examination. Mitochondrial or other genetic diseases, as well as neurological diseases, hypertension, and diabetes mellitus were excluded. The age range was from 0.1 to 88.0 years. Donor samples were divided into two groups: the minors and adults groups. The minors group included 200 cases with a male:female ratio of 1:1 and an average age of 6.2 years (0.1–17.9 years). The mean age of the adults group was 45.2 years (18.0–88.0 years) and the male: female ratio was 1.06:1. To study the correlation between mtDNA copy number and age, the samples were further categorized into eight groups, namely, 0–2.0, 2.1–6.0, 6.1–10.0, 10.1–17.9, 18.0–30.0, 30.1–40.0, 40.1–50.0, and >50.0 years.
A total of 83 patients diagnosed with mitochondrial disease carrying the m.3243A>G mutation were included in this study. Their average age was 11.5 years (0.2–48.0 years). The whole blood samples of these patients were collected from 2009 to 2013. The total DNA was extracted and kept at −80°C until measurements. We also analyzed four minor patients (0.5–4.0 years) with MDS carrying two mutations in one of four nuclear genes that cause mtDNA depletion (POLG, DGUOK, TK2, and SUCLA2).
Mitochondrial DNA copy number quantification by real-time quantitative polymerase chain reaction
For all the whole blood samples, total DNA (including nuclear and mitochondrial genomic gene) was extracted by TIANamp Blood DNA Kit (TIANGEN Biotech, Beijing, China). The absolute mtDNA copy number was measured by a quantitative real-time polymerase chain reaction (PCR)-based method as previously described.[12,17] mtDNA was quantified as the ratio of a mitochondrial gene copy number (ND1) to a single-copy nuclear gene (human β-globin gene, HBB). mtDNA copy number per cell was calculated by the formula 2×ND1/HBB. The specific primers and probes used were as follows: ND1: F 5’-ATTCGATGTTGAAGCCTGAGACT-3’, R 5’-TGACCCTTGGCCATAATATGATT-3’, probe 5’-FAM-TTCGGACTCCCCTTCG GCAAGG-TAMRA-3’; HBB: F 5’-ACCTCAAGGGCACCTTTGC-3’, R 5’-AAAACATCAAGCGTCCCATAGAC-3’, probe 5’-FAM-CACTGTGACAAGCTG CACGTGGATCC-TAMRA-3’. The 20-μl PCR reaction solution contained 2× Taqman Universal PCR Master Mix, 500 nmol/L of each primer, 200 nmol/L Taqman Probe, and 20-100 ng of total DNA. PCR conditions were 2 min at 50°C, 10 min at 95°C, followed by 40 cycles of 15 s denaturation at 95°C and 60 s annealing/extension at 60°C. Real-time quantitative analysis was performed on Roche LightCycler 480. Two recombinant plasmids containing ND1 and HBB genes were used as standards for quantitative PCR (qPCR). The concentration of the purified plasmid DNA was determined by NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The plasmid copy number in solution was derived from its molar concentration and the Avogadro constant. The standard curves were constructed using a serial dilution of 108, 107, 106, 105, 104, and 103 copy number standards for ND1 and HBB genes.
Each DNA sample was quantified in triplicates using either ND1 or HBB primers in parallel reactions. The acceptable coefficient of variance of the triplicate threshold cycle (Ct) values was set at 5%. If a result was out of the acceptable range, the run was repeated for that particular sample. The result of mtDNA copy number per cell is shown in a logarithmic scale (lg).
Statistical analysis
Statistical analyses were performed with SPSS version 16.0 (Chicago, IL, USA). The mtDNA copy numbers were log-transformed into a normal distribution. Normality was tested using the Kolmogorov-Smirnov method. Independent t-test was used to compare mtDNA copy numbers between males and females. A linear regression analysis was used to investigate the effect of age on mtDNA copy number.[18] One-way analysis of variance (ANOVA), followed by the least significant difference (LSD) test, was used to compare the mean copy number among the eight age groups. A P < 0.05 was considered statistically significant.
RESULTS
Accuracy of the real-time quantitative polymerase chain reaction method for measurement of mitochondrial DNA copy number
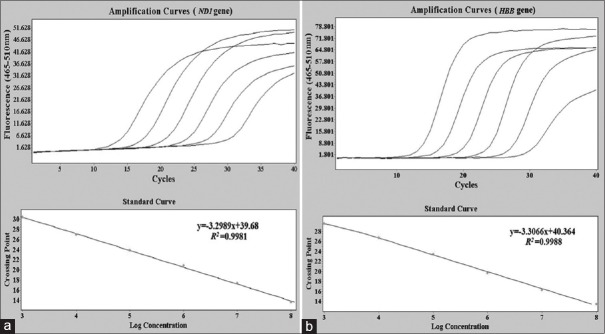
The amplification and standard curves of ND1 nuclear [Figure 1a] and HBB mitochondrial genes [Figure 1b] in plasmids are shown in Figure 1. For each standard curve, a straight linear correlation was observed with R2 > 0.998. Three samples were assayed to assess the reproducibility of qPCR. To assess intra-assay variation, we assayed each of the three DNA samples in six replicates in one run. Furthermore, we assayed the same three DNA samples in triplicates in six separate runs to evaluate inter-assay variation. Good reproducibility was observed in both intra- and inter-assays. The intra-assay coefficients of variation of Ct values ranged 1.6–2.0% and 1.4–2.1% for ND1 and HBB genes, respectively. The inter-assay coefficients of variation of Ct values ranged 2.9–3.3% and 2.7–3.4% for ND1 and HBB genes, respectively.
Figure 1.
Amplification and standard curves for nuclear and mitochondrial genes in plasmids. The amplification and standard curves were obtained by a dilution of standards ranging 103−108 for ND1 nuclear gene (a) and HBB mitochondrial gene (b) separately.
Reference range of mitochondrial DNA copy number in peripheral blood
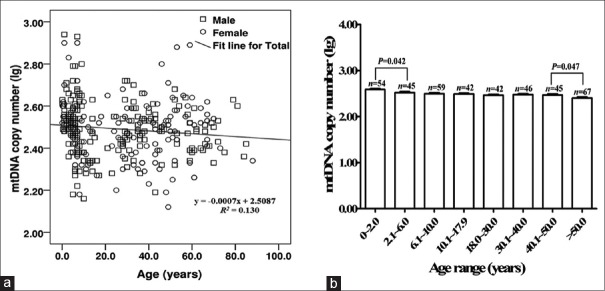
The mtDNA copy number range in both the minor and adult control groups followed a normal distribution when transformed to lg values [Figure 2a]. The results of normality test were as follows: minor group P = 0.062 and adult group P = 0.200. In blood, mtDNA copy number decreased with age in healthy population (R2 = 0.130, P = 0.043). The mtDNA copy number (lg) in the minor group was significantly higher than that in adults (t = 2.086, P = 0.038). The mean and standard deviation (SD) from both groups were expressed as 2.51 ± 0.14 and 2.46 ± 0.12, respectively. There were no differences in copy numbers between males and females in both two groups (P > 0.05). The reference range of mtDNA copy number in peripheral blood was calculated as mean ± 1.96 SD, which ranged 175–602 copies/cell (mean: 325 copies/cell) in minors and 164–500 copies/cell (mean: 287 copies/cell) in adults.
Figure 2.
Mitochondrial DNA copy number in blood from 400 healthy Chinese individuals. (a) Linear regression analysis of mitochondrial DNA copy number with age and sex (n=400). (b) Mitochondrial DNA copy number in blood of various age groups.
The total control samples were further categorized into eight groups according to age, with nearly 50 cases in each group [Figure 2b]. The mean mtDNA copy number (lg) in each group was 2.58, 2.52, 2.49, 2.49, 2.46, 2.48, 2.47, and 2.41, respectively. The ANOVA result showed a significant difference in mean copy numbers among groups (F = 4.470, P < 0.05). Furthermore, the LSD t-test results showed that the mtDNA copy number significantly decreases since the first 2 years of life (t = 2.040, P = 0.042) and remained almost unaltered before 50 years old. Individuals after 50 years old showed the lowest level of mtDNA content (t = 2.230, P = 0.047).
Mitochondrial DNA copy number in mitochondrial disease patients
Mitochondrial disease patients with the m.3243A>G mutation were divided into two groups, namely, 70 minors and 13 adult patients. The mtDNA copy number in m.3243A>G patients was significantly higher than that in the control group [Figure 3]. The minor patients had a 32% increase in mtDNA copy number than the corresponding controls (2.63 ± 0.21). Adult patients had a 38% increase over the corresponding controls (2.60 ± 0.16).
Figure 3.
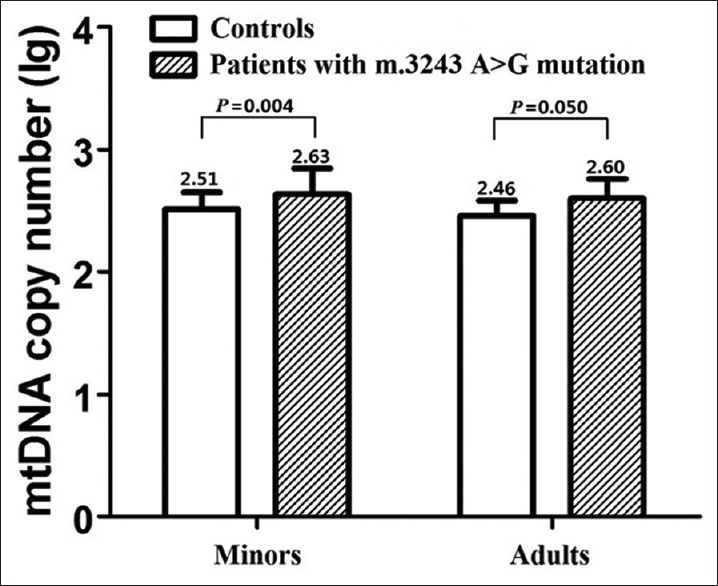
Mitochondrial DNA copy number in blood of healthy controls and mitochondrial disease patients with the m.3243A>G mutation. P value was determined by independent samples t-test.
Minor patients with two mutations in POLG, DGUOK, TK2, and SUCLA2 genes showed a wide phenotypic spectrum, including Alpers’ syndrome, early-onset hepatocerebral disease, myopathy, and ophthalmoparesis. Their mtDNA copy number was 81, 123, 104, and 79 copies/cell, respectively. Compared with the mean mtDNA copy number (325 copies/cell) in healthy minors, the mtDNA content in MDS patients was reduced to 25%, 38%, 32%, and 24%, respectively.
DISCUSSION
Several studies have reported changes in mtDNA copy number in blood for various diseases.[2,3] However, normal range comparable data between studies are not available as a result of different quantitative methods and lack of appropriate healthy controls. Aiming to establish clinical reference ranges of mtDNA copy number in blood to be used in clinical diagnosis and monitoring, we studied 400 healthy Chinese controls, between 0.1 and 88.0 years old. To study the correlation between mtDNA copy number and age, we divided the normal population into several age groups, as in previous studies.[11,16] We found that there is a decreasing trend in the mtDNA copy number in blood with increasing age, particularly in 0–2-year-old and >50-year-old individuals. The mtDNA copy number negatively correlated with age in healthy populations, in accordance with previous studies.[19,20] Mengel-From et al.[8] found that, in a Danish population, an age-related decline in mtDNA copy number was observed from the age of 48 years. However, the population of this study consisted of individuals between 18 and 93 years of age, excluding the minor population. Another study showed no association between mtDNA copy number and age in cases and controls, probably due to the narrow age span of the subjects (3–6 years old).[21] The precise mechanisms of mtDNA copy number decrease in blood with aging are not clear. A previous study confirmed that maintenance of mtDNA copy number is essential for the preservation of mitochondrial function and cell growth.[22] It was speculated that low mtDNA copy number among the elderly could indicate a low ATP production and mitochondrial gene expression.[8] A slight decrease in mtDNA copy number could result in a slowdown of the mitochondrial metabolism. Reduced mitochondrial activity leads to decreased reactive oxygen species production, in turn, protecting mitochondria against aging.[23]
We used real-time qPCR with fluorescent probes which provided greater specificity than SYBR green to quantify mtDNA copy number. We performed an absolute quantification using a standard curve, which was more accurate than the relative determination calculated by the difference in Ct values between nuclear and mtDNA genes.[24] In this study, the mean mtDNA copy number in peripheral blood samples was 325 copies per cell in children and 287 copies per cell in adults. The control samples’ absolute value was similar to previous reports. In previous study, the mean mtDNA content was 330 copies per cell in blood samples.[15] Liou et al.[25] reported about 350 mtDNA copies per cell in leukocytes.
The mean mtDNA copy number level in mitochondrial disease patients harboring the m.3243A>G mutation was significantly higher than that of minor and adult controls. The m.3243A>G mutation is the most frequent mitochondrial point mutation, which is often associated with multiorgan dysfunction. This mutation might result in the reduction of mtDNA-encoded proteins and oxidative phosphorylation activity in mitochondrial translation. The dysfunctional mitochondria fail to generate sufficient ATP to meet the energy needs of various organs resulting in the multiorgan disease. Energy deficiency may stimulate mitochondrial proliferation leading to an increase in mtDNA copy number as a compensatory response.[26] However, altered mtDNA levels may contribute to enhanced oxidative stress and inflammation possibly playing a pathogenic role in mitochondrial dysfunction and disease.[9] Thus, the severity of the mitochondrial disease is related to mtDNA copy number in leukocytes.[11,16] Therefore, mtDNA copy number could be a predictive biomarker as well as a monitoring biomarker of mitochondrial dysfunction.
MDSs are a group of clinically heterogeneous autosomal recessive disorders that are characterized by a severe reduction in mtDNA copy number in the affected tissues and organs.[27] MDS has been associated with mtDNA maintenance caused by mutations in nuclear genes that function in either mitochondrial nucleotide synthesis or mtDNA replication. Several mutations in 12 nuclear genes have been found as responsible for encoding vital proteins to mtDNA maintenance.[28,29] Among these genes, TK2, DGUOK, and SUCLA2 genes encode proteins that maintain the mitochondrial dNTP pool, and POLG gene is essential for mtDNA replication. Mutations in TK2 are associated with mitochondrial myopathy, mutations in SUCLA2 are associated with mitochondrial encephalopathy, and mutations in POLG and DGUOK are associated with mitochondrial hepatoencephalopathy.[29] In the study, four MDS patients harboring two mutations in one of these genes, including POLG, DGUOK, TK2, and SUCLA2, showed a decrease to 25%, 38%, 32%, and 24% of mtDNA content in blood samples compared to the mean value in the reference population, respectively. Real-time qPCR could be an effective method for detecting mtDNA depletion. With a 50% mtDNA content cutoff in healthy controls’ liver tissue, mtDNA depletion was detected with a sensitivity of 100% and a specificity of 97%.[16] Despite the lack of sensitivity in blood samples, its specificity for the detection of MDS patients was also demonstrated in their study.[16] We suggest that mtDNA copy number in blood can be used to assist MDS diagnosis. However, the cutoff value in blood should be evaluated with a larger sample size.
Although mtDNA copy number can be measured in various tissues and body fluids, blood is a stable and easy-to-collect sample. As blood cells are in contact with the whole body and organs, they could reflect changes in mitochondrial function. The establishment of mtDNA copy number reference intervals in blood contributes to its application in clinical diagnosis and monitoring of mitochondrial disease as well as other diseases. mtDNA copy number in infants of 0–2 years and adults aged >50 years was significantly different from the other age groups. It is suggested to establish reference ranges for these two population groups independently. However, given our limited sample size for these two groups, we can only establish the reference intervals in minors and adults for clinical purposes.
Financial support and sponsorship
This study was supported by grants from the National Natural Science Foundation of China (No. 81271256 and No. 81471153) and Beijing Municipal Science and Technology Commission (No. Z131107002213062).
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
We thank Dr. Wei-Xing Feng and Dr. Fang Fang from the Department of Neurology, Beijing Children's Hospital, for the collection of data assistance in this study.
Footnotes
Edited by: Qiang Shi
REFERENCES
- 1.Veltri KL, Espiritu M, Singh G. Distinct genomic copy number in mitochondria of different mammalian organs. J Cell Physiol. 1990;143:160–4. doi: 10.1002/jcp.1041430122. doi: 10.1002/jcp.1041430122. [DOI] [PubMed] [Google Scholar]
- 2.Shen J, Platek M, Mahasneh A, Ambrosone CB, Zhao H. Mitochondrial copy number and risk of breast cancer: A pilot study. Mitochondrion. 2010;10:62–8. doi: 10.1016/j.mito.2009.09.004. doi: 10.1016/j.mito.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egan K, Kusao I, Troelstrup D, Agsalda M, Shiramizu B. Mitochondrial DNA in residual leukemia cells in cerebrospinal fluid in children with acute lymphoblastic leukemia. J Clin Med Res. 2010;2:225–9. doi: 10.4021/jocmr443w. doi: 10.4021/jocmr443w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carreira RS, Lee P, Gottlieb RA. Mitochondrial therapeutics for cardioprotection. Curr Pharm Des. 2011;17:2017–35. doi: 10.2174/138161211796904777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee HK, Song JH, Shin CS, Park DJ, Park KS, Lee KU, et al. Decreased mitochondrial DNA content in peripheral blood precedes the development of non-insulin-dependent diabetes mellitus. Diabetes Res Clin Pract. 1998;42:161–7. doi: 10.1016/s0168-8227(98)00110-7. doi: 10.1016/S0168-8227(98) 00110-7. [DOI] [PubMed] [Google Scholar]
- 6.Huang CH, Su SL, Hsieh MC, Cheng WL, Chang CC, Wu HL, et al. Depleted leukocyte mitochondrial DNA copy number in metabolic syndrome. J Atheroscler Thromb. 2011;18:867–73. doi: 10.5551/jat.8698. doi: 10.5551/jat.8698. [DOI] [PubMed] [Google Scholar]
- 7.Lee JW, Park KD, Im JA, Kim MY, Lee DC. Mitochondrial DNA copy number in peripheral blood is associated with cognitive function in apparently healthy elderly women. Clin Chim Acta. 2010;411:592–6. doi: 10.1016/j.cca.2010.01.024. doi: 10.1016/j.cca.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 8.Mengel-From J, Thinggaard M, Dalgård C, Kyvik KO, Christensen K, Christiansen L. Mitochondrial DNA copy number in peripheral blood cells declines with age and is associated with general health among elderly. Hum Genet. 2014;133:1149–59. doi: 10.1007/s00439-014-1458-9. doi: 10.1007/s00439-014-1458-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malik AN, Czajka A. Is mitochondrial DNA content a potential biomarker of mitochondrial dysfunction? Mitochondrion. 2013;13:481–92. doi: 10.1016/j.mito.2012.10.011. doi: 10.1016/j.mito.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 10.Lee HC, Wei YH. Mitochondrial alterations, cellular response to oxidative stress and defective degradation of proteins in aging. Biogerontology. 2001;2:231–44. doi: 10.1023/a:1013270512172. [DOI] [PubMed] [Google Scholar]
- 11.Liu CS, Cheng WL, Lee CF, Ma YS, Lin CY, Huang CC, et al. Alteration in the copy number of mitochondrial DNA in leukocytes of patients with mitochondrial encephalomyopathies. Acta Neurol Scand. 2006;113:334–41. doi: 10.1111/j.1600-0404.2006.00586.x. doi: 10.1111/j.1600-0404.2006.00586.x. [DOI] [PubMed] [Google Scholar]
- 12.Liu H, Ma Y, Fang F, Zhang Y, Zou L, Yang Y, et al. Wild-type mitochondrial DNA copy number in urinary cells as a useful marker for diagnosing severity of the mitochondrial diseases. PLoS One. 2013;8:e67146. doi: 10.1371/journal.pone.0067146. doi: 10.1371/journal.pone.0067146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahman S, Poulton J. Diagnosis of mitochondrial DNA depletion syndromes. Arch Dis Child. 2009;94:3–5. doi: 10.1136/adc.2008.147983. doi: 10.1136/adc.2008.147983. [DOI] [PubMed] [Google Scholar]
- 14.Bai RK, Perng CL, Hsu CH, Wong LJ. Quantitative PCR analysis of mitochondrial DNA content in patients with mitochondrial disease. Ann N Y Acad Sci. 2004;1011:304–9. doi: 10.1007/978-3-662-41088-2_29. doi: 10.1196/annals.1293.029. [DOI] [PubMed] [Google Scholar]
- 15.Morten KJ, Ashley N, Wijburg F, Hadzic N, Parr J, Jayawant S, et al. Liver mtDNA content increases during development: A comparison of methods and the importance of age- and tissue-specific controls for the diagnosis of mtDNA depletion. Mitochondrion. 2007;7:386–95. doi: 10.1016/j.mito.2007.09.001. doi: 10.1016/j.mito.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Dimmock D, Tang LY, Schmitt ES, Wong LJ. Quantitative evaluation of the mitochondrial DNA depletion syndrome. Clin Chem. 2010;56:1119–27. doi: 10.1373/clinchem.2009.141549. doi: 10.1373/clinchem.2009.141549. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Ma Y, Bu D, Liu H, Xia C, Zhang Y, et al. Deletion of a 4977-bp fragment in the mitochondrial genome is associated with mitochondrial disease severity. PLoS One. 2015;10:e0128624. doi: 10.1371/journal.pone.0128624. doi: 10.1371/journal.pone.0128624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
- 19.van Leeuwen N, Beekman M, Deelen J, van den Akker EB, de Craen AJ, Slagboom PE, et al. Low mitochondrial DNA content associates with familial longevity: The Leiden Longevity Study. Age. 2014;36:1463–70. doi: 10.1007/s11357-014-9629-0. doi: 10.1007/s11357-014-9629-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding J, Sidore C, Butler TJ, Wing MK, Qian Y, Meirelles O, et al. Assessing mitochondrial DNA variation and copy number in lymphocytes of ~2,000 Sardinians using tailored sequencing analysis tools. PLoS genetics. 2015;11:e1005306. doi: 10.1371/journal.pgen.1005306. doi: 10.1371/journal.pgen.1005306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen S, Li Z, He Y, Zhang F, Li H7, Liao Y, et al. Elevated mitochondrial DNA copy number in peripheral blood cells is associated with childhood autism. BMC Psychiatry. 2015;15:50. doi: 10.1186/s12888-015-0432-y. doi: 10.1186/s12888-015-0432-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeng JY, Yeh TS, Lee JW, Lin SH, Fong TH, Hsieh RH. Maintenance of mitochondrial DNA copy number and expression are essential for preservation of mitochondrial function and cell growth. J Cell Biochem. 2008;103:347–57. doi: 10.1002/jcb.21625. doi: 10.1002/jcb.21625. [DOI] [PubMed] [Google Scholar]
- 23.Conley KE, Amara CE, Jubrias SA, Marcinek DJ. Mitochondrial function, fibre types and ageing: New insights from human muscle in vivo. Exp Physiol. 2007;92:333–9. doi: 10.1113/expphysiol.2006.034330. doi: 10.1113/expphysiol.2006.034330. [DOI] [PubMed] [Google Scholar]
- 24.Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: Real-time monitoring of DNA amplification reactions. Biotechnology (N Y) 1993;11:1026–30. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 25.Liou CW, Lin TK, Chen JB, Tiao MM, Weng SW, Chen SD, et al. Association between a common mitochondrial DNA D-loop polycytosine variant and alteration of mitochondrial copy number in human peripheral blood cells. J Med Genet. 2010;47:723–8. doi: 10.1136/jmg.2010.077552. doi: 10.1136/jmg.2010.077552. [DOI] [PubMed] [Google Scholar]
- 26.James AM, Murphy MP. How mitochondrial damage affects cell function. J Biomed Sci. 2002;9(6 Pt 1):475–87. doi: 10.1159/000064721. doi: 10.1007/BF02254975. [DOI] [PubMed] [Google Scholar]
- 27.Alberio S, Mineri R, Tiranti V, Zeviani M. Depletion of mtDNA: Syndromes and genes. Mitochondrion. 2007;7:6–12. doi: 10.1016/j.mito.2006.11.010. doi: 10.1016/j.mito.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 28.Suomalainen A, Isohanni P. Mitochondrial DNA depletion syndromes – Many genes, common mechanisms. Neuromuscul Disord. 2010;20:429–37. doi: 10.1016/j.nmd.2010.03.017. doi: 10.1016/j.nmd.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 29.El-Hattab AW, Scaglia F. Mitochondrial DNA depletion syndromes: Review and updates of genetic basis, manifestations, and therapeutic options. Neurotherapeutics. 2013;10:186–98. doi: 10.1007/s13311-013-0177-6. doi: 10.1007/s13311-013-0177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]