Abstract
Introduction:
The high protein value, essential minerals, dietary fibre and notable ability to fix atmospheric nitrogen make chickpea a highly remunerative crop, particularly in low-input food production systems. Of the variety of constraints challenging chickpea productivity worldwide, salinity remains of prime concern owing to the intrinsic sensitivity of the crop. In view of the projected expansion of chickpea into arable and salt-stressed land by 2050, increasing attention is being placed on improving the salt tolerance of this crop. Considerable effort is currently underway to address salinity stress and substantial breeding progress is being made despite the seemingly highly-complex and environment-dependent nature of the tolerance trait.
Conclusion:
This review aims to provide a holistic view of recent advances in breeding chickpea for salt tolerance. Initially, we focus on the identification of novel genetic resources for salt tolerance via extensive germplasm screening. We then expand on the use of genome-wide and cost-effective techniques to gain new insights into the genetic control of salt tolerance, including the responsive genes/QTL(s), gene(s) networks/cross talk and intricate signalling cascades.
Keywords: Chickpea, DNA markers, Genomics, Molecular breeding, RNA-Seq, Salinity, Tolerance, Transcript, QTL
1. INtroduction
1.1. Chickpea as a Global Food Staple
Chickpea (Cicer arietinum L.) is the second most important grain legume and earliest known cultivated species [1]. Among grain legume crops, chickpea is highly valued for its dietary proteins, low fat, vitamins and essential minerals [2, 3]. Chickpea accessions exhibit large variance in genetic and phenotypic characteristics across the Mediterranean region, western Asia, central Asia and India, and recent breeding advances has led to its adoption to temperate regions [1, 4, 5]. The leading chickpea producing countries include India, Australia, Pakistan, Myanmar and Ethiopia, which collectively contribute more than 85 per cent of the global production. Over the past 50 years, annual chickpea production has increased from 6.4 to 14.2 million tonnes [6]. From years 2009 to 2013, the global area under chickpea increased 25 per cent from 11.5 to 13.5 million hectares [6]. The long tap root system and ability to establish symbiotic relationships with Rhizobia enables chickpea to fix atmospheric nitrogen and improve soil health. Given its high nutritional content, market value, adaptability and nitrogen fixation ability, chickpea is being increasingly recognised as a staple food crop of the future.
1.1.1. The Increasing Importance of Salinity Tolerance in Chickpea
Chickpea has an indeterminate growth habit, i.e. it continues to grow vegetatively even after flower initiation [7]. This renders it sensitive to a number of environmental factors such as salinity, drought, heat and cold [8, 9]. Soil salinity is a major constraint that limits crop productivity and almost 80 million ha of the worlds’ arable land is prone to this stress [10]. Globally, 20 per cent (45 million ha) of irrigated and 2 per cent (32 million ha) of dry land are constrained by salinity [11]. This is predicted to expand to 50 per cent worldwide by the second half of the 21st century [12]. In conjunction with the predicted marked expansion of salinity-affected area, an additional two billion people are anticipated to inhabit the planet by 2050. Therefore, soil salinity is a major stumbling block to meeting the predicted global food demand by 2050. Abiotic stresses account for about 6.4 million tonnes in crop yield losses every year, where soil salinity is a major environmental stress [13]. The enormity of the current challenge of sustaining or increasing productivity to meet yield demands in the face of increasing salinity has been well highlighted [11, 14-16]. This translates into an urgent requirement for improved crop production by almost 70 per cent [6, 17]. However, salinity limits the plant growth and severely affects the reproductive processes, resulting in lowered crop yields [18] and chickpea is intrinsically salt sensitive unlike cereals [10]. The imperative is therefore to elucidate the genetic architecture of salinity tolerance and in particular the molecular mechanisms underpinning the tolerance responses.
The phenotypic responses to a stress are due to a plant’s genetic constitution and genotype interaction with the environmental variables [19, 20]. Existence of large genetic variation within a gene pool would enable genotypes conferring desirable traits to be identified, including those that are able to withstand adverse saline conditions [21, 22]. Salinity tolerance is conferred by several physiological factors which have been recorded in response to salt stress to identify tolerant/sensitive genotypes [23, 24]. To enable more strategic and precise selection of tolerant genotypes, frontier genomics technologies, such as gene expression profiling, have the potential to identify robust transcripts/candidate gene(s) and their alleles that condition tolerance [25-28]. Recent advances have substantially illuminated the mechanisms of salinity tolerance in chickpea, thereby paving the way towards the strategic incorporation of tolerance-imparting component traits into elite genetic backgrounds. Here, we offer a critical overview of the different genomics approaches that have been used to address salinity in chickpea. Later follows a brief discussion on targeted breeding aided by genomics tools, concluding with the future research needs for developing salinity tolerant chickpea.
2. QUANTIFYING GENETIC VARIATION FOR SALINITY STRESS TOLERANCE
Chickpea is a salt sensitive crop [10], and broad germplasm has been assessed for tolerance and subsequent deployment as parental genotypes in breeding programs [3, 21, 24, 29, 30]. Salinity tolerance has been studied at different developmental stages such as germination [31], vegetative growth [29, 32] and reproduction. Large genotypic variation was observed among chickpea landraces [29], and core collection [33], based on shoot biomass in soil treated with NaCl at vegetative and maturity stage in glasshouse experiments [21, 24]. The chickpea plant was reported to suffer most severely under salinity stress during the reproductive stage [7, 18, 19, 21, 34]. A greater number of flowers are considered to be a more important measure of tolerance during stress rather than acquiring greater root or shoot biomass [24, 35]. For example, Vadez [18], found that tolerant lines produced 70 per cent and 30 per cent more flowers under salt stress at sowing and flowering, respectively [35, 36]. Physiological studies have shown that salinity delays flowering time and severely affected during the pod filling stage. Despite pollen viability, sensitive genotypes show higher occurrence of empty pods and seed abortions [24, 36]. This observation suggests failure in ovule fertilisation as the main reason for pod abortion or empty pods, despite the viable pollen and pollen tube growth.
Further, differences in tissue ion-regulation in root, shoot and floral parts are reported among genotypes in studies that investigated the tissue/stage specific effect of salinity by measuring the Na+/Cl-/K+ ion concentrations [7, 18, 20, 24, 35, 37]. Therefore chickpea has been shown to exhibit variable responses to high ion levels that in turn condition their ability to be tolerant or sensitive when the ionic concentration reaches a physiological threshold.
Plants employ different mechanisms such as ion-exclusion and tissue tolerance to overcome ionic stress [11, 37-39]. Reducing the (Na+, K+ and Cl-) ion accumulation in shoots by manipulating the root ion transport processes was used to explain the ion-exclusion mechanism in plants [11, 40]. However, this may not be the case with chickpea where tolerant and sensitive genotypes have equal ionic concentrations in shoots [36, 37]. Also, fully expanded leaves maintained high Na+ and Cl- ion concentrations compared to reproductive organs during the pod filling stage and eventually restricted ions from accumulating in flowers and developing ovules [11, 41]. This suggests that the salinity effect in chickpea is minimised by compartmentalisation of toxic ions in leaf vacuoles, a process that has not yet been studied [42-45]. There is a need to investigate these cellular events in detail, to help unravel the salt tolerance mechanism in chickpea [37]. Indeed, during the pod filling stage, there is no accumulation of Na+, Cl- ions in reproductive organs such as petals, stamens or ovules in contrast to that detected in fully expanded leaves [24, 36]. It is possible that high ion-accumulation in leaves resulted in decreased photosynthesis efficiency and therefore tolerant genotypes (such as cv. Genesis836, JG11), differ from sensitive genotypes (such as cv. Rupali) in terms of chlorophyll content when subjected to saline conditions [46, 47]. Importantly, as mentioned shoot ion accumulation does not translate into reduced shoot biomass and seed yield [18], therefore tissue ion regulation in leaves during the reproductive stage is likely responsible for imparting a major part of the tolerance. Apart from identifying genotypic variation in already existing germplasm, there is a need to identify the molecular mechanisms pertaining to ion regulation.
3. APPROACHES TO ELUCIDATE SALINITY-ASSOCIATED CANDIDATE GENES/DNA MARKERS IN CHICKPEA FOR BREEDING APPLICATIONS
3.1. Functional Genomics
Cultivated chickpea has a narrow genetic base [1], and possesses phenotypic plasticity [48], which makes it difficult to identify the salt stress responsive and potentially tolerance gene(s) especially when plants adopt cross-talk to respond to various simultaneous stresses [49]. Therefore, there it is important to identify the transcript variants specific to salt tolerance by analysing variation in the spatio-temporal gene expression of tolerant/sensitive genotypes within a controlled environment setting. Accordingly, a large set of 20,162 Expressed Sequence Tags (ESTs) was identified from NaCl-treated roots of salt tolerant (JG11) and sensitive (ICCV2) genotypes using Sanger sequencing [50]. The functional annotation of the ESTs was achieved by similarity searches against model legume datasets (Medicago, Glycine, Lotus, and Arachis) and model plants (Oryza, Arabidopsis). Differentially expressed genes were reported to be associated with “cellular processes”, “cell transport” and “osmotic adjustments”. Interestingly, cDNA libraries of the sensitive genotype ICCV2 presented a higher number of up-regulated gene(s) having putative functions like heat shock proteins, metallothionein and abscisic acid production [3, 28, 50]. This suggested that chickpea plants adopt an ion-transport mechanism for regulating cellular homoeostasis through trans-membrane protein conformational changes [51], and detoxify the metal ions accumulating beyond a threshold level. However, ESTs only represent low transcript abundances [52], and do not provide tissue or developmental stage differential gene expression within the genotypes, which are thought important to understand salinity tolerance. Serial analysis of gene(s) expression (SAGE) represents another approach to quantify transcripts exhibiting differential responses to salt stress in root and nodule tissues. SuperSAGE and DeepSAGE were used in chickpea to identify over 3,000 stress-responsive transcripts, where several SOS gene candidates were expressed spatially in roots [38, 53]. Subsequent microarray-based gene expression profiling of chickpea genotypes revealed a set of differentially expressed genes [54], at different time-points and in different tissue-types [38]. Earlier, Mantri et al. [55], reported the tolerant genotype (CPI 060546) had a higher number of repressed gene(s) than the sensitive one (CPI 60527) at different time-points (Table 1), potentially indicating the slowing down or turning off of other non-essential metabolic processes to redirect resources towards the tolerance mechanisms.
Table 1. List of recently developed functional genomic resources in chickpea relevant to major abiotic stresses.
| Stress | Tissue | Method | Gene/ESTs/transcripts | References |
|---|---|---|---|---|
| Salinity | Root | RNA-Sequencing | 5545 | [28] |
| Seedlings | Subtractive cDNA libraries/ Yeast One-hybrid assay / Northern and western blot analyses | CaZF gene (C2H2-zinc finger family protein) | [56] | |
| Seedlings | Semi-quantitative RT-PCR | CapLEA-1 CapLEA-2, CarLEA-4 (late embryogenesis abundant protein) | [57] | |
| Hooks, epicotyls, mesocotyls from seedlings. Stems Leaves, pods, flowers and roots | Northern/Southern blot analyses | CapLTP (lipid transfer proteins), CapLEA-1 CapLEA-2, CarLEA-4 (late embryogenesis abundant protein) | [58] | |
| Seedlings | cDNA | SOD (cytosolic superoxide dismutase) | [58] | |
| Seedlings | RNA-Blot Hybridization and RT-PCR/ Site-directed mutagenesis/ Yeast One-hybrid assay | CAP2 gene (an APETALA2-family transcription factor) | [59] | |
| Root, shoot, leaves, stem, flowers, young pod and seedlings | RNA-Sequencing | 1163 genes | [26] | |
| Root and nodules | deepsuperSAGE | 21401 transcripts | [38] | |
| Salinity and Drought | Leaves, apical meristem, shoots, roots, buds, flowers, pods, embryo | Tentative unique sequences (TUSs) using Roche454 and Sanger ESTs/ Illumina/Solexa sequencing | 103 215 ESTs | [25] |
| Leaves, roots, flowers | ‘Pulse Chip’ microarray/RT-PCR | 266 (salt responsive transcripts) | [54] | |
| Root | ESTs | 20162 cDNAs | [50] | |
| SuperSAGE | 3000 transcripts | [60] | ||
| Seedlings | subcellular localization/qRT-PCR | CarF-box1(CarF-box1 protein) | [61] | |
| Salinity, Drought, Cold | Leaves, roots, flowers from tolerant | ‘Pulse Chip’ microarray/RT-PCR | 386 (salt responsive transcripts) | [55] |
| Salinity, Heat, Environmental stress | seedlings | Quantitative real-time PCR/Northern Analysis | CaMIPS1, CaMIPS2 (L- myo-inositol 1-phosphate synthase) | [62] |
Importantly, genotypes differ in temporal gene regulation and a greater number of genes were down-regulated in the tolerant genotype in all of the tissue-types analysed in response to salt stress. This provided great insight into the differential transcriptional response programming among tissue types that is activated on perceiving the salt stress [28, 54]. For example, trans-membrane channels such as aquaporin genes, which transport water, were repressed much earlier in tolerant roots (24 hpt) than sensitive roots (48 hpt) to restrict the salt uptake along with water on exposure to salt treatment [55]. Importantly, both physiological and genomic screening demonstrated that a large genetic variation for salinity tolerance exists among and within the tolerant and sensitive genotypes. As an example, in microarray gene profiling, aquaporin genes were induced in tolerant-1 and repressed in tolerant-2, simultaneously [36, 55]. Other candidates such as heat shock proteins, proline-osmolytes, senescence-associated genes and ripening-related genes were repressed in tolerant roots/shoots, while the same genes were induced in sensitive roots/shoots. The identification of tissue- specific differentially expressed transcripts suggests major transcriptional reprogramming at the cellular level, which in turn confers the genetic variation by altering the plant’s physiological responses to other processes such as photosynthesis [37] and senescence [24], to impart tolerance. Although these techniques provided remarkable information to identify the candidates for salt tolerance, they were restricted by the lack of a chickpea reference genome at the time of their application [63] and hence were limited to assessing known transcripts such as those represented on microarrays. They were also relatively low-throughput and failed to detect low expressed or rare transcripts, splicing events and gene isoforms which are thought to be master switches in regulating stress responses.
In recent years, advances in Next Generation Sequencing (NGS) have enabled easy access to identify thousands of gene(s) that are regulated in response to abiotic stresses in plants [28, 64-66]. A global view of the salt-stressed transcriptome using RNA-Seq would enable the measuring of gene expression responses at the whole genome level [26, 28], and provide an in-depth understanding of molecular mechanisms and pathways conferring salt tolerance. In this approach, total RNA from control and stressed root tissues are fragmented and cDNAs are sequenced with enough depth (~40 million reads per sample) to generate short (~100 bp -150 bp length) sequence reads. The generated reads are then mapped to the now publically available chickpea reference genome (http://dx.doi.org/10.5524/100076) to obtain the “gene count values” in order to measure the differential gene expressions. Along the same lines of technology, RNA-Seq is an innovative approach which allows identification of novel transcribed loci, exon and intron boundaries and splice isoforms through reference-guided assembly. Unlike ESTs and microarrays, RNA-Seq presents a robust in-depth list of candidate genes along with rare and low expressed transcripts that facilitates understanding of the tissue/stage-specific transcriptional reprogramming in response to salt stress [52]. In a recent RNA-Seq study, an additional 15 per cent of novel transcribed loci were identified than previously annotated in the chickpea (CDC Frontier) reference genome [28]. Until now, important biological events such as alternative splicing which control the transcriptional regulation of gene isoforms during the stress were not researched [67, 68]. A transcribed locus on an average can encode for more than two exons, i.e. one gene can encode more than one protein and several biological processes like water transport, protein modification and defence response are reported to be alternatively spliced at different developmental stages during salt stress [28, 69, 70].
Deep-sequencing technologies generate large genic datasets to unravel the transcriptome response and understand its diversity based on developmental stage, tissue type, genotype and salt treatment [28, 66]. Several studies have demonstrated the use of RNA-Seq to study the transcriptome response of a specific cell-type such as radial patterning of root growth, anther maturity and pollen tube growth during abiotic stress responses in plants [71]. Large set of genes (5,523) were reported to be differentially expressed in chickpea in response to salinity, mostly at the late reproductive stage in root tissues [28]. This is in accordance with phenological studies where salinity is most disastrous at reproductive stages and therefore genes differentially expressed amongst sensitive and tolerant genotypes at this particular stage provide an important basis to uncover the underlying molecular mechanisms [72].
It is important to draw useful biological meaning from huge dataset generated through RNA-Seq and therefore co-expression of these genes are further analysed using computational methods to assign Gene Ontology (GO) terms to categories such as cell wall biogenesis, oxidative stress, protein folding, redox-signalling and transport [28, 63]. A set of genes enriched in particular ontology categories will enable the identification of the master regulators of complex pathways regulating salt stress responses [73, 74]. Further, given the sensitivity of the RNA-Seq approach, it is now trivial to identify the exon-junctions and intergenic non-coding regions to further study the role of small RNA molecules such as miRNA, siRNA and lincRNA, which are thought to manipulate gene functions during stress responses [75, 76]. This gene information can be used to understand the dynamics of gene networks in response to salt stress. Also, these genes can be used for developing DNA marker resources [25, 50]
or act as candidates for genomics-assisted breeding and prediction of phenotype from genotype [77]. The RNA-Sequencing technique is continuously improving to overcome issues like sequence coverage or 3’ fragmentation bias through longer read length, paired-end sequencing, cation-heat fragmentation, stranded library and random hexamer priming during cDNA synthesis [52, 66].
3.2. Mapping of Genomic Regions Relevant to Salinity Tolerance
Salinity tolerance is a physiological and biochemical trait controlled by many genes/quantitative trait loci (QTLs) exerting variable contributions to the tolerance phenomenon [3, 18, 19]. Limitations of conventional genetic analyses in precisely delineating causative gene(s)/genomic segments conditioning tolerance to salinity are largely due to the complex interactive mechanisms at play. The lack of selectable phenotypic variation for salinity tolerance in the cultigen has greatly impeded successful breeding of tolerant cultivars [78, 79]. Alternatively, selection of the genetic components underpinning the tolerance traits would potentially speed up the accuracy and timing of tolerance breeding. Several methods including QTL discovery, using family- or population- based mapping, were applied to identify the genomic position of the genetic determinants governing the various aspects of the salinity tolerance trait. For example, Samineni et al. [80] identified a set of QTLs/genes related to yield, seed size and shoot biomass under salt-stress using a RIL population (ICC6263 × ICC1431). Although 20 candidate genomic segments were implicated, these accounted for just 9 per cent of the total trait variation, highlighting the complex and multigenic nature of the salinity tolerance trait. Similarly, Vadez et al. [18], identified multiple QTLs for salinity tolerance using another RIL population (JG62 × ICCV2) on linkage groups (LGs) 3 and 6, including a major QTL on LG 6 that accounted for 37 per cent of the variation in seed number.
Most recently, genomic regions associated with salinity tolerance were identified on LGs 5 and 7 using the RIL population (ICCV2 × JG11) and these segments corresponded to 48 putative candidate tolerance genes [3]. These QTL regions explained higher reproductive success under salinity and can be potentially used in marker-assisted breeding. By using the chickpea reference genome sequence information, the putative full-length candidate gene(s) underlying these QTL regions could be identified and annotated with respect to their structural and functional features. The syntenic regions of tightly linked markers to QTL were BLAST against whole genome sequence to find candidate genes [3], which have putative role in phytohormone signaling pathways and Na+/K+ antiporter ion channels such as AKT1 [3, 81, 82].
Salinity tolerance is also influenced by epistatic and environmental (G×E) interactions offering an additional set of challenges to breeding efforts [20, 33, 83]. Future functional validation of these candidate sequences will determine the magnitude of their functional relevance to salinity tolerance across a broad set of genotypes and treatment environments, thus substantiating their suitability as selection tools within breeding programs.
3.3. Molecular Mechanisms Underpinning Salinity Tolerance in Chickpea
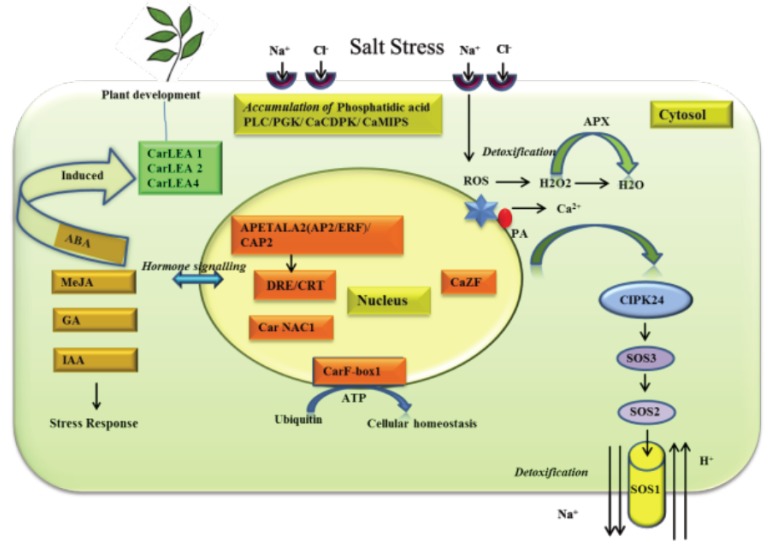
Plants have developed different sensory mechanisms to cope up with salt stress. The signalling mechanism is regulated first at the plant hormonal level [84], second by activation of transcription factors for gene expression and third by activation of metabolic pathways (Fig. 1).
Fig. (1).
Overview of the proposed salinity tolerance mechanism in Cicer arietinum L. Upon salt stress, Ca2+, ROS and hormone signalling are activated. AP2/ERF, CAP2, CarNAC, CarF box-1 type transcription factors have been reported to overexpress at the stress reception. The Salt Overly Sensitive (SOS) pathway regulates the Na+/ H+ antiporters. Ubiquitin, Ionositol, ABA, MeJA and salicylic acid pathways are induced by gene(s) such as CarLEA (Cicer arietinum late embryogenesis protein).
The plasma membrane is the first line of defence which perceives the stress through trans-membrane protein sensors [39]. The phospholipids like Phosphatidylcholine (PC) and Phosphatic Acid (PA) receive the signal when high extracellular NaCl concentration occurs at ionic receptors of the root cell [85]. These phospholipids have specific roles in regulating activation of calcium-dependent protein kinase CaCDPK1 genes, which are involved in the release of signalling messengers such as calcium ions (Ca2+) [86, 87] through the control of transcription factors during the saline stress responses in chickpea [88]. Transcription factors bind to the promoter regions of the genes to facilitate the RNA polymerase to start the transcription and subsequent translation of the gene products [89]. They are important regulators of stress response and have been widely found to show differential expressions in salt-challenged tissues [25, 26, 90]. Crops like Arabidopsis, Oryza and Glycine present a robust suite of functionally annotated candidate gene(s) and have been extensively studied to understand molecular mechanisms involved in abiotic response through gene manipulations. A number of transcription factors in these crops have been identified to be associated in the activation of genes responsible for osmotic adjustments [59, 91, 92]. Transcription factors such as CAP2/AP2, CarNAC1, CaZF and CarF that are known to up-regulate the CaCDPK1 genes have been identified in chickpea [26, 61].
Recently, the chickpea F-box gene CarF-box1, isolated from a cDNA library of polyethylene glycol (PEG)-treated chickpea seedling leaves, was significantly induced in roots following drought and salinity stresses [61]. Several transcription factors such as CAP2/AP2, CarNAC1, CaZF and CarF were identified from stressed tissues through generation of cDNA [61, 93] and genomic libraries [26]. These TFs were reported to regulate gene expression mediating hormonal biosynthesis and subsequent plant growth under stressed conditions [94, 95]. Overexpression of the CAP2 gene resulted in an increased tolerance to dehydration and salt stress [59, 93]. Another chickpea abiotic stress transcription factor NAC gene, CarNAC1 and CaZF, which imparted high salinity tolerance when expressed in tobacco plants, was isolated from a cDNA library constructed with PEG-treated seedlings [56, 91, 96]. Concordant with the earlier reports, chickpea is known to accumulate increased inositol during dehydration stress [97, 98], and these TFs regulate expression of MIPS (Myo-inositol-1-phosphate synthase) genes, aiding the cell to maintain the osmotic environment [62]. Previously, CaMIPS2 was reported to be present as an ABA-inducible early dehydration-responsive gene in chickpea and therefore, an important component of hormonal signalling [49].
Further, during the response to salinity stress in chickpea, Lipid Transfer Proteins (LTPs) form an impermeable layer, which obstructs water loss and retains cell turgidity [57]. The CapLTP gene in chickpea was expressed in young tissues and during early developmental stages in response to water stress, suggesting implications for protecting cellular functions from damage caused by high ion concentration.
The activation of TFs and gene transcription is highly dependent on up-regulated plant hormones such as ABA, Indole Acetic Acid (IAA), gibberellic acid (GA) and methyl jasmonate (MeJA) [49, 99], which induce expression of CarLEA genes. Accordingly, CarLEA genes (CarLEA1, CarLEA2 and CarLEA4) isolated from chickpea cDNA libraries, were found to impart desiccation tolerance during seed development, thereby protecting plants against a variety of stresses, including drought, salinity and freezing [57, 58].
Plant hormones such as ABA, SA, JA induce signalling cascades and protect cells from osmotic imbalance and dehydration. Several ABA-responsive transcripts were identified in relation to various abiotic stresses in chickpea [88], however the role of this hormone in the signalling for salt stress tolerance remains unclear. Likewise, Salicylic Acid (SA) is recognized as an endogenous signalling molecule and major elicitor of ROS, Phosphatidic Acid (PA) and cellular proteins like annexin [84]. SA evokes environmental stress responses by regulating nutrient uptake, photosynthesis, osmotic balance and seed germination. Several ion-channel and membrane transporters are located in the plasma membrane, and elucidation of this pathway triggered by PA production as a result of SA accumulation could be very important in the salt stress response.
As indicated in the preceding sections, there is substantial evidence that ions accumulate in different tissues and cell organelles under salinity stress [45, 100, 101]. Plants deploy various pathways to regulate the ionic detoxification process by importing proton (H+) and exporting Na+ out of the cell [102]. One major ion-exchange protein (NHX) has been located within the plasma membrane and proposed to control the detoxification process by the efflux of excess Na+ ions and influx of H+ ion in Arabidopsis [103-105]. However, not enough is known about the molecular mechanism and pathways that are involved in these detoxification processes, the sequestration of ions into vacuoles through compartmentalisation or ion-exclusion in chickpea.
To date, few studies have uncovered candidate genes in chickpea such as SOS1/Na+/H+ antiporter, SOS2/CIPK24, SOS3/CBL, associated with the SOS pathway. These have a putative role in excretion of Na+ ion suggesting ion-exclusion mechanisms occur under high salinity concentrations in chickpea [38]. However, more comprehensive studies are required to identify the members of SOS signalling pathways that invoke the salt stress response in chickpea. There is a great need to employ technologies like RNA sequencing to elucidate in more depth the signalling cascades and gene-networks that are crucial for salt tolerance.
4. POTENTIAL STRATEGIES TO BREED SALT STRESS TOLERANT CHICKPEA
An in-depth understanding about the genetic determinants of salt tolerance is an essential prerequisite for selective biotechnological manipulation or molecular breeding towards developing salt tolerant cultivars. In important crops like barley, strategies have focused on targeted modification of osmotic tolerance and ion exclusion mechanisms, and these have resulted in high tolerance to tissue ion concentrations [104, 106]. Given that high tissue ion tolerance was recently identified as a key mechanism for salt tolerance in chickpea, it is logical to propose that a strategy to improve the ion channel regulation mechanisms would be most relevant for improving salt tolerance in this species.
Molecular breeding methods are now available that facilitate effective translation of the genomic knowledge for the development of tolerant varieties. Screening the available germplasm to identify tolerant accessions is the foremost approach. As previously discussed, chickpea germplasm collections were examined by many researchers with the aim to discover salt-tolerant genotypes [21]. This has resulted in the identification of a set of salt tolerant genotypes, which could be used as a genetic reservoir to mine salt responsive and potentially tolerance-related gene(s)/QTLs, and also, to facilitate transfer of the corresponding alleles into elite yet vulnerable elite genetic backgrounds [13]. A set of specific physiological indicators may be selected while assessing salt tolerance among chickpea genotypes. With the advent of next generation phenomics platforms such as robotic field sensors high resolution multi-spectral mapping using UAVs and laser light back scattering technology, it has become easier to study developmental stages of plant more precisely and analyse the multi-dimensional large volume of bio-imaging data [107-109]. The important parameters involve germination, biomass, leaf necrosis, nodulation and nitrogen fixation, death and senescence, ion concentrations, osmoregulation, plant growth and yield. Each has been used previously with varying effectiveness for the selection of salinity tolerant plants [36]. The factors that challenge accurate evaluation of genotypic tolerance include: 1) the complex genetic control of salinity tolerance 2) time consuming screening protocols and 3) substantial G × E effects.
Similar to other cultivated legume species, chickpea has a narrow genetic base, which has been severely impacted through domestication and subsequent selection events [1]. Alternatively, mutation breeding is an attractive approach to broaden the available genetic diversity, and to create novel variability. This approach has already been employed to develop several chickpea varieties [110], and the modern variants of mutation-detection systems like TILLING-by-sequencing [111], create novel opportunities to tap variations related to abiotic stress tolerances. Other potential approaches enabling “siphoning” of exotic/wild alleles include the use of crossable wild relatives. Wild chickpeas do possess tolerance to a number of abiotic stresses. However, potential of these wild relatives has to be realized concerning salt tolerance using conventional breeding protocols. Toward this end, recent genomic techniques like advanced backcross (AB)-QTL and introgression libraries may be particularly relevant for capturing the beneficial yet previously unnoticed exotic alleles [112].
Advancements in the field of crop genomics, with copious sequence data now available, help scientists to identify, isolate and deploy the genes associated with the tolerance traits [77, 113]. Once candidate sequences have been identified and functionally validated, salt tolerant chickpea cultivars may be routinely developed using modern breeding techniques such as Marker-Assisted Selection (MAS), marker-assisted backcrossing (MABC) and marker-assisted recurrent selection (MARS) [114]. Also, with the availability of high-density marker genotyping assays and high-throughput phenotyping platforms, opportunities are created to employ Genome Wide Association Studies (GWAS) and Genomic Selection (GS) to counter the issue of QTL with relatively smaller effects for salt tolerance [115, 116]. The latest applications of NGS technology have rendered identification and mapping of DNA markers a rapid and cost-effective procedure [117]. The high density genotyping is a common occurrence now with the availability of diverse protocols such as Reduced Representation Libraries, Restriction site Associated DNA sequencing (RAD-seq), Genotyping by sequencing (GBS), low-coverage Whole Genome Re-sequencing (WGRS) or genome skimming [118], and a more recent, Single Locus Amplified Fragment Sequencing (SLAF-Seq) [119]. Also, development of genome-scale catalogues of genetic variants such as SNP chips including SoySNP50K iSelect BeadChip [120], SoySNP6K Infinium BeadChip [121], Axiom SoyaSNP array for nearly 180,000 SNPs greatly assists genetic analyses [122]. The genome-wide SNPs in combination with high-quality phenotyping records permit genetic resolution of trait mapping at an unprecedented scale. For instance, WGRS recombination bin map based QTL analysis in chickpea allowed splitting of the single 3-Mb QTL hotspot region into two precisely delineated genomic regions (139.22 kb and 153.36 kb). Concerning salinity tolerance, a refined trait dissection facilitated by high-density SNP data is evident in a recent GBS assay of RILs, which elucidated salinity tolerance in rice to be controlled not only by additive but also epistatic interactions [123]. The average QTL interval size was 132kb. In another study using BSA with 50K SNP array, authors discovered known as well as novel QTLs for salinity tolerance in rice, with an average QTL region of 2.3 cM [124]. In soybean, the WGRS of RI panel (W05 × C08) at 1x revealed a 978-kb QTL region for salt tolerance, which was further narrowed down to 388-kb. The corresponding 388-kb of W05 with William 82 led authors to propose a major dominant gene Glyma03g32900 (GmCHX1) as the causal one [125]. This locus was also later detected by Patil et al. [126] through conducting GWAS on publically available WGRS data of 106 diverse soybean lines, leading to design of KASPar assay to support breeding for salt tolerance. In view of the availability of the reference and re-sequenced genomes in chickpea, such approaches could be extended to comprehend the genetic makeup of salinity tolerance in chickpea followed by fine-mapping, prioritization of the candidate genes and pyramiding of candidate genes.
In view of the deluge of genome-scale sequencing data as described above, now is an opportune time to characterise the ion-transport channel genes such as Hydrogen/potassium Exchanger (HKT), Cation/proton Exchanger (CHX), sodium/hydrogen exchanger (NHX), as has been done in major crops like wheat [127], and soybean [81]. Experimental evidences collected so far support the crucial role of these genes in conferring salinity tolerance and improved yield [128]. In parallel, different metalloenzymes such as Superoxide Dismutase (SOD), Ascorbate Peroxidase (APX), peroxidase (POX) that is elicited during stress could be targeted [117-119, 129-131]. Quantifying expression levels through RNA sequencing or Digital Expression Analysis (DGA) can generate unprecedented insights into the molecular cascades of mechanisms that lead to ion-exclusion inside cells.
Developments in different next generation omics platforms have generated huge information which is useful to understand the complex genetic and physiological nature of abiotic stress tolerance. An efficient integration of genomics, proteomics, metabolomics, ionomics and phenomics will enrich our biological understanding of the salt tolerance response however it still remains a challenge in legume crops [132]. Recently, an integrated transcriptome and metabolome study in Dendrobium officinale provided deeper understanding of gene to metabolite network regulating energy metabolism through oxidation of carbohydrates during the cold stress [133]. Computational biology has emerged as a powerful approach to make the different omics data accessible to the community through creation of public databases. Establishment of such databases allow easy retrieval of information to examine to study the correlation of a gene to its functional protein, syntenic regions to a chromosome of model plants, end product metabolites or ionic regulation in order to predict and shape an improved phenotype for stress tolerance. Hence, there is a great need to develop databases containing information on ionomics, metabolomics and phenomics databases especially in legume crops like soybean and chickpea which have been greatly benefitted with current genomics and transcriptomics advances.
In the near future, high-resolution and annotated transcriptome/genome sequence data will lead to the development of large-scale selective breeding tools for accurate and fast salt tolerance selection. The genomic toolkit that underpins breeding for salinity-tolerant chickpea will encompass a suite of robust molecular resources that have been validated through multiple treatments and environments as well as across diverse genotypes. The selective breeding for improved production will be strengthened and help assure that chickpea remains a major and secure food source in the face of increasing salinity stress worldwide.
ACKNOWLEDGEMENTS
The authors are thankful to Australia-India Strategic Research Fund (AISRF) for supporting this research project.
LIST OF ABBREVIATIONS
- AGP
Arabinogalactan class proteins
- CAP2
Adenylyl Cyclase-Associated Protein
- CaZF
Cicer arietinum zinc finger
- CDPK1
Calcium-dependent protein kinase
- CIPK
Calcineurin B-like interacting protein kinases
- DRE/CRT
Dehydration responsive element/C-repeat element
- EST
Expressed sequence tag
- LEA
Late-embryogenesis abundant
- MeJA
Methyl jasmonate
- MIPS1
Myo-inositol-1-phosphate synthase
- P5CS
Pyrroline-5-carboxylate synthetase gene
- QTL
Quantitative trait loci
- RGA
Resistance gene analogues
- RIL
Recombinant inbred line
- ROS
Reactive oxygen species
- SAGE
Serial Analysis of Gene Expression
- SNP
Single nucleotide polymorphism
- SOS
Salt overly sensitive
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Varshney R.K., Song C., Saxena R.K., Azam S., Yu S., Sharpe A.G., Cannon S., Baek J., Rosen B.D., Tar’an B., Millan T., Zhang X., Ramsay L.D., Iwata A., Wang Y., Nelson W., Farmer A.D., Gaur P.M., Soderlund C., Penmetsa R.V., Xu C., Bharti A.K., He W., Winter P., Zhao S., Hane J.K., Carrasquilla-Garcia N., Condie J.A., Upadhyaya H.D., Luo M.C., Thudi M., Gowda C.L., Singh N.P., Lichtenzveig J., Gali K.K., Rubio J., Nadarajan N., Dolezel J., Bansal K.C., Xu X., Edwards D., Zhang G., Kahl G., Gil J., Singh K.B., Datta S.K., Jackson S.A., Wang J., Cook D.R. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 2013;31(3):240–246. doi: 10.1038/nbt.2491. [DOI] [PubMed] [Google Scholar]
- 2.Alarcon-Valdez C., Milan-Carrillo J., Cardenas-Valenzuela O.G., Mora-Escobedo R., Bello-Perez L.A., Reyes-Moreno C. Infant food from quality protein maize and chickpea: optimization for preparing and nutritional properties. Int. J. Food Sci. Nutr. 2005;56(4):273–285. doi: 10.1080/09637480500146804. [DOI] [PubMed] [Google Scholar]
- 3.Pushpavalli R., Krishnamurthy L., Thudi M., Gaur P.M., Rao M.V., Siddique K.H., Colmer T.D., Turner N.C., Varshney R.K., Vadez V. Two key genomic regions harbour QTLs for salinity tolerance in ICCV 2 x JG 11 derived chickpea (Cicer arietinum L.) recombinant inbred lines. BMC Plant Biol. 2015;15:124. doi: 10.1186/s12870-015-0491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abbo S., Saranga Y., Peleg Z., Kerem Z., Lev-Yadun S., Gopher A. Reconsidering domestication of legumes versus cereals in the ancient near east. Q. Rev. Biol. 2009;84(1):29–50. doi: 10.1086/596462. [DOI] [PubMed] [Google Scholar]
- 5.Milan-Carrillo J., Valdez-Alarcon C., Gutierrez-Dorado R., Cardenas-Valenzuela O.G., Mora-Escobedo R., Garzon-Tiznado J.A., Reyes-Moreno C. Nutritional properties of quality protein maize and chickpea extruded based weaning food. Plant Foods Hum. Nutr. 2007;62(1):31–37. doi: 10.1007/s11130-006-0039-z. [DOI] [PubMed] [Google Scholar]
- 6.2014 http://faostat.fao.org/ Faostat, Food and Agriculture Organization of the United Nations: USA, 2014.
- 7.Samineni S., Siddique K.H., Gaur P.M., Colmer T.D. Salt sensitivity of the vegetative and reproductive stages in chickpea (Cicer arietinum L.): Podding is a particularly sensitive stage. Environ. Exp. Bot. 2011;71(2):260–268. [Google Scholar]
- 8.Turner N.C., Abbo S., Berger J.D., Chaturvedi S.K., French R.J., Ludwig C., Mannur D.M., Singh S.J., Yadava H.S. Osmotic adjustment in chickpea (Cicer arietinum L.) results in no yield benefit under terminal drought. J. Exp. Bot. 2007;58(2):187–194. doi: 10.1093/jxb/erl192. [DOI] [PubMed] [Google Scholar]
- 9.Devasirvatham V., Tan D.K., Gaur P.M., Raju T.N., Trethowan R.M. High temperature tolerance in chickpea and its implications for plant improvement. Crop Pasture Sci. 2012;63:419–428. [Google Scholar]
- 10.Flowers T.J., Gaur P.M., Gowda C.L., Krishnamurthy L., Samineni S., Siddique K.H., Turner N.C., Vadez V., Varshney R.K., Colmer T.D. Salt sensitivity in chickpea. Plant Cell Environ. 2010;33(4):490–509. doi: 10.1111/j.1365-3040.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- 11.Munns R., Tester M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 12.Ladeiro B. Saline agriculture in the 21st century: Using salt contaminated resources to cope food requirements. J. Bot. 2012;•••:7. [Google Scholar]
- 13.Jha U.C., Chaturvedi S.K., Bohra A., Basu P.S., Khan M.S., Barh D. Abiotic stresses, constraints and improvement strategies in chickpea. Plant Breed. 2014;133(2):163–178. [Google Scholar]
- 14.Teakle N.L., Tyerman S.D. Mechanisms of Cl(-) transport contributing to salt tolerance. Plant Cell Environ. 2010;33(4):566–589. doi: 10.1111/j.1365-3040.2009.02060.x. [DOI] [PubMed] [Google Scholar]
- 15.Amin U.S., Biswas S., Elias S.M., Razzaque S., Haque T., Malo R., Seraj Z.I. Enhanced salt tolerance conferred by the complete 2.3 kb cDNA of the rice vacuolar Na(+)/H(+) antiporter gene compared to 1.9 kb coding region with 5′ UTR in transgenic lines of rice. Front. Plant Sci. 2016;7:14. doi: 10.3389/fpls.2016.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong Y., Zhang H., Huang L., Li D., Song F. Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front. Plant Sci. 2016;7:4. doi: 10.3389/fpls.2016.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tester M., Langridge P. Breeding technologies to increase crop production in a changing world. Science. 2010;327(5967):818–822. doi: 10.1126/science.1183700. [DOI] [PubMed] [Google Scholar]
- 18.Vadez V., Krishnamurthy L., Thudi M., Anuradha C., Colmer T., Turner N., Siddique K., Gaur P.M., Varshney R.K. Assessment of ICCV 2 × JG 62 chickpea progenies shows sensitivity of reproduction to salt stress and reveals QTL for seed yield and yield components. Mol. Breed. 2012;30:9–21. [Google Scholar]
- 19.Krishnamurthy L., Turner N.C., Gaur P.M., Upadhyaya H.D., Varshney R.K., Siddique K.H., Vadez V. Consistent variation across soil types in salinity resistance of a diverse range of chickpea (Cicer arietinum L.) genotypes. J. Agron. Crop Sci. 2011;197(3):214–227. [Google Scholar]
- 20.Samineni S., Gaur P.M., Colmer T.D., Krishnamurthy L., Vadez V., Siddique K.H. Estimation of genetic components of variation for salt tolerance in chickpea using the generation mean analysis. Euphytica. 2011;182(1):73–86. [Google Scholar]
- 21.Vadez V., Krishnamurthy L., Serraj R., Gaur P.M., Upadhyaya H.D., Hoisington D.A., Varshney R.K., Turner N.C., Siddique K.H. Large variation in salinity tolerance in chickpea is explained by differences in sensitivity at the reproductive stage. Field Crops Res. 2007;104(1-3):123–129. [Google Scholar]
- 22.Sohrabi Y., Heidari G., Esmailpoor B. Effect of salinity on growth and yield of Desi and Kabuli chickpea cultivars. Pak. J. Biol. Sci: PJBS. 2008;11(4):664–667. doi: 10.3923/pjbs.2008.664.667. [DOI] [PubMed] [Google Scholar]
- 23.Moses F.A., Maliro D.M., Bob R., Kollmorgen J.F., Chris P. Sampling strategies and screening of chickpea (Cicer arietinum L.) germplasm for salt tolerance. Genet. Resour. Crop Evol. 2008;55(1):53–63. [Google Scholar]
- 24.Turner N.C., Colmer T.D., Quealy J., Pushpavalli R., Krishnamurthy L., Kaur J., Singh G., Siddique K.H., Vadez V. Salinity tolerance and ion accumulation in chickpea (Cicer arietinum L.) subjected to salt stress. Plant Soil. 2013;365(1):347–361. [Google Scholar]
- 25.Hiremath P.J., Farmer A., Cannon S.B., Woodward J., Kudapa H., Tuteja R., Kumar A., Bhanuprakash A., Mulaosmanovic B., Gujaria N., Krishnamurthy L., Gaur P.M., Kavikishor P.B., Shah T., Srinivasan R., Lohse M., Xiao Y., Town C.D., Cook D.R., May G.D., Varshney R.K. Large-scale transcriptome analysis in chickpea (Cicer arietinum L.), an orphan legume crop of the semi-arid tropics of Asia and Africa. Plant Biotechnol. J. 2011;9(8):922–931. doi: 10.1111/j.1467-7652.2011.00625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain M., Misra G., Patel R.K., Priya P., Jhanwar S., Khan A.W., Shah N., Singh V.K., Garg R., Jeena G., Yadav M., Kant C., Sharma P., Yadav G., Bhatia S., Tyagi A.K., Chattopadhyay D. A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant J. 2013;74(5):715–729. doi: 10.1111/tpj.12173. [DOI] [PubMed] [Google Scholar]
- 27.Kudapa H., Azam S., Sharpe A.G., Taran B., Li R., Deonovic B., Cameron C., Farmer A.D., Cannon S.B., Varshney R.K. Comprehensive transcriptome assembly of Chickpea (Cicer arietinum L.) using sanger and next generation sequencing platforms: development and applications. PLoS One. 2014;9(1):e86039. doi: 10.1371/journal.pone.0086039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garg R., Shankar R., Thakkar B., Kudapa H., Krishnamurthy L., Mantri N., Varshney R.K., Bhatia S., Jain M. Transcriptome analyses reveal genotype- and developmental stage-specific molecular responses to drought and salinity stresses in chickpea. Sci. Rep. 2016;6:19228. doi: 10.1038/srep19228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maliro M.F., McNeil D., Kollmorgen J., Pittock C., Redden B. Proceedings of the 4th International Crop Science Congress, New directions for a diverse planet; Brisbane. September 26; Brisbane, Australia. 2004. [Google Scholar]
- 30.Roy S.J., Tucker E.J., Tester M. Genetic analysis of abiotic stress tolerance in crops. Curr. Opin. Plant Biol. 2011;14(3):232–239. doi: 10.1016/j.pbi.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Kaya M., Kaya G., Kaya M.D., Atak M., Saglam S., Khawar K.M., Ciftci C.Y. Interaction between seed size and NaCl on germination and early seedling growth of some Turkish cultivars of chickpea (Cicer arietinum L.). J. Zhejiang Univ. Sci. B. 2008;9(5):371–377. doi: 10.1631/jzus.B0720268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karajeh F., Hamdy A., Bruggeman A., Touchan H., Oweis T. Regional Action Programme (RAP): Water Resources Management And Water Saving in Irrigated Agriculture (WASIA PROJECT); Vol. 44. CIHEAMIAMB; Italy: 2003. pp. 163–169. [Google Scholar]
- 33.Upadhyaya H.D., Kashiwagi J., Varshney R.K., Gaur P.M., Saxena K.B., Krishnamurthy L., Gowda C.L., Pundir R.P., Chaturvedi S.K., Basu P.S., Singh I.P. Phenotyping chickpeas and pigeonpeas for adaptation to drought. Front. Physiol. 2012;3:179. doi: 10.3389/fphys.2012.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katerji N., van Hoorn J.W., Hamdy A., Mastrorilli M., Nachit M.M., Oweis T. Salt tolerance analysis of chickpea, faba bean and durum wheat varieties. II.Durum wheat. Agric. Water Manage. 2005;72:195–207. [Google Scholar]
- 35.Vadez V., Rashmi M., Sindhu K., Muralidharan M., Pushpavalli R., Turner N.C., Krishnamurthy L., Gaur P.M., Colmer T.D. Large number of flowers and tertiary branches, and higher reproductive success increase yields under salt stress in chickpea. Eur. J. Agron. 2012;41:42–51. [Google Scholar]
- 36.Kotula L., Khan H.A., Quealy J., Turner N.C., Vadez V., Siddique K.H., Clode P.L., Colmer T.D. Salt sensitivity in chickpea (Cicer arietinum L.): ions in reproductive tissues and yield components in contrasting genotypes. Plant Cell Environ. 2015;38(8):1565–1577. doi: 10.1111/pce.12506. [DOI] [PubMed] [Google Scholar]
- 37.Khan H.A., Siddique K.H., Munir R., Colmer T.D. Salt sensitivity in chickpea: Growth, photosynthesis, seed yield components and tissue ion regulation in contrasting genotypes. J. Plant Physiol. 2015;182:1–12. doi: 10.1016/j.jplph.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 38.Molina C., Zaman-Allah M., Khan F., Fatnassi N., Horres R., Rotter B., Steinhauer D., Amenc L., Drevon J.J., Winter P., Kahl G. The salt-responsive transcriptome of chickpea roots and nodules via deepSuperSAGE. BMC Plant Biol. 2011;11:31. doi: 10.1186/1471-2229-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deinlein U., Stephan A.B., Horie T., Luo W., Xu G., Schroeder J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014;19(6):371–379. doi: 10.1016/j.tplants.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mudgal V., Madaan N. Mudgal, Anurag.; Mishra, S.; Singh, A.; Singh P.K. Changes in growth and metabolic profile of Chickpea under salt stress. J. Appl. Biosci. 2009;23:1436–1446. [Google Scholar]
- 41.Flowers T.J., Munns R., Colmer T.D. Sodium chloride toxicity and the cellular basis of salt tolerance in halophytes. Ann. Bot. 2015;115(3):419–431. doi: 10.1093/aob/mcu217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mullan D.J., Colmer T.D., Francki M.G. Arabidopsis-rice-wheat gene orthologues for Na+ transport and transcript analysis in wheat-L. elongatum aneuploids under salt stress. Mol. Genet. Genomics: MGG. 2007;277(2):199–212. doi: 10.1007/s00438-006-0184-y. [DOI] [PubMed] [Google Scholar]
- 43.Chen S., Polle A. Salinity tolerance of Populus. Plant Biol. 2010;12(2):317–333. doi: 10.1111/j.1438-8677.2009.00301.x. [DOI] [PubMed] [Google Scholar]
- 44.Battelli R., Lombardi L., Picciarelli P., Lorenzi R., Frigerio L., Rogers H.J. Expression and localisation of a senescence-associated KDEL-cysteine protease from Lilium longiflorum tepals. Plant Sci. 2014;214:38–46. doi: 10.1016/j.plantsci.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 45.Reginato M., Sosa L., Llanes A., Hampp E., Vettorazzi N., Reinoso H., Luna V. Growth responses and ion accumulation in the halophytic legume Prosopis strombulifera are determined by Na2SO4 and NaCl. Plant Biol. 2014;16(1):97–106. doi: 10.1111/plb.12001. [DOI] [PubMed] [Google Scholar]
- 46.Najafi F., Khavari-Nejad R.A., Rastgar-Jazii F., Sticklen M. Growth and some physiological attributes of pea (Pisum sativum L.) as affected by salinity. Pak. J. Biol. Sci.: PJBS. 2007;10(16):2752–2755. doi: 10.3923/pjbs.2007.2752.2755. [DOI] [PubMed] [Google Scholar]
- 47.Teakle N.L., Colmer T.D., Pedersen O. Leaf gas films delay salt entry and enhance underwater photosynthesis and internal aeration of Melilotus siculus submerged in saline water. Plant Cell Environ. 2014;37(10):2339–2349. doi: 10.1111/pce.12269. [DOI] [PubMed] [Google Scholar]
- 48.Berger J.D., Ali M., Basu P.S., Chaudhary B.D., Chaturvedi S.K., Deshmukh P.S., Dharmaraj P.S., Dwivedi S.K., Gangadhar G.C., Gaur P.M., Kumar J., Pannu R.K., Siddique K.H., Singh D.N., Singh D.P., Singh S.J., Turner N.C., Yadav H.S., Yadav S.S. Genotype by environment studies demonstrate the critical role of phenology in adaptation of chickpea (Cicer arietinum L.) to high and low yielding environments of India. Field Crops Res. 2006;98(2-3):230–244. [Google Scholar]
- 49.Tuteja N. Mechanisms of high salinity tolerance in plants. Methods Enzymol. 2007;428:419–438. doi: 10.1016/S0076-6879(07)28024-3. [DOI] [PubMed] [Google Scholar]
- 50.Varshney R.K., Hiremath P.J., Lekha P., Kashiwagi J., Balaji J., Deokar A.A., Vadez V., Xiao Y., Srinivasan R., Gaur P.M., Siddique K.H., Town C.D., Hoisington D.A. A comprehensive resource of drought- and salinity- responsive ESTs for gene discovery and marker development in chickpea (Cicer arietinum L.). BMC Genomics. 2009;10:523. doi: 10.1186/1471-2164-10-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao J., Zhu Y., Zhou W., Molinier J., Dong A., Shen W.H. NAP1 family histone chaperones are required for somatic homologous recombination in Arabidopsis. Plant Cell. 2012;24(4):1437–1447. doi: 10.1105/tpc.112.096792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laetitia B.B., Martin Z.F., Giovannoni J.J., Rose J.K. Catalyzing plant science research with RNA-Seq. Front. Plant Sci. 2013;4:66. doi: 10.3389/fpls.2013.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Molina C., Rotter B., Horres R., Udupa S.M., Besser B., Bellarmino L., Baum M., Matsumura H., Terauchi R., Kahl G., Winter P. SuperSAGE: the drought stress-responsive transcriptome of chickpea roots. BMC Genomics. 2008;9:553. doi: 10.1186/1471-2164-9-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mantri N.L., Ford R., Coram T.E., Pang E.C. Evidence of unique and shared responses to major biotic and abiotic stresses in chickpea. Environ. Exp. Bot. 2010;69(3):286–292. [Google Scholar]
- 55.Mantri N.L., Ford R., Coram T.E., Pang E.C. Transcriptional profiling of chickpea genes differentially regulated in response to high-salinity, cold and drought. BMC Genomics. 2007;8:303. doi: 10.1186/1471-2164-8-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jain D., Roy N., Chattopadhyay D. CaZF, a plant transcription factor functions through and parallel to HOG and calcineurin pathways in Saccharomyces cerevisiae to provide osmotolerance. PLoS One. 2009;4(4):e5154. doi: 10.1371/journal.pone.0005154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gu H., Jia Y., Wang X., Chen Q., Shi S., Ma L., Zhang J., Zhang H., Ma H. Identification and characterization of a LEA family gene CarLEA4 from chickpea (Cicer arietinum L.). Mol. Biol. Rep. 2012;39(4):3565–3572. doi: 10.1007/s11033-011-1130-6. [DOI] [PubMed] [Google Scholar]
- 58.Romo S., Labrador E., Dopico B. Water stress-regulated gene expression in Cicer arietinum seedlings and plants. Plant Physiol. Biochem. 2001;39:1017–1026. [Google Scholar]
- 59.Shukla R.K., Raha S., Tripathi V., Chattopadhyay D. Expression of CAP2, an APETALA2-family transcription factor from chickpea, enhances growth and tolerance to dehydration and salt stress in transgenic tobacco. Plant Physiol. 2006;142(1):113–123. doi: 10.1104/pp.106.081752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kahl G., Molina C., Udupa S.M. Plant and animal genome XV conference. San Diego, CA, USA; 2007. Super SAGE, exploring the stress transcriptome in chickpea. [Google Scholar]
- 61.Jia Y., Gu H., Wang X., Chen Q., Shi S., Zhang J., Ma L., Zhang H., Ma H. Molecular cloning and characterization of an F-box family gene CarF-box1 from chickpea (Cicer arietinum L.). Mol. Biol. Rep. 2012;39(3):2337–2345. doi: 10.1007/s11033-011-0984-y. [DOI] [PubMed] [Google Scholar]
- 62.Kaur H., Shukla R.K., Yadav G., Chattopadhyay D., Majee M. Two divergent genes encoding L-myo-inositol 1-phosphate synthase1 (CaMIPS1) and 2 (CaMIPS2) are differentially expressed in chickpea. Plant Cell Environ. 2008;31(11):1701–1716. doi: 10.1111/j.1365-3040.2008.01877.x. [DOI] [PubMed] [Google Scholar]
- 63.Garg R., Jain M. Pyrosequencing data reveals tissue-specific expression of lineage-specific transcripts in chickpea. Plant Signal. Behav. 2011;6(11):1868–1870. doi: 10.4161/psb.6.11.17879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barrera-Figueroa B.E., Gao L., Wu Z., Zhou X., Zhu J., Jin H., Liu R., Zhu J.K. High throughput sequencing reveals novel and abiotic stress-regulated microRNAs in the inflorescences of rice. BMC Plant Biol. 2012;12:132. doi: 10.1186/1471-2229-12-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fan X., Guo Q., Xu P., Gong Y., Shu H., Yang Y., Ni W., Zhang X., Shen X. Transcriptome-wide identification of salt-responsive members of the WRKY gene family in Gossypium aridum. PLoS One. 2015;10(5):e0126148. doi: 10.1371/journal.pone.0126148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Filichkin S.A., Cumbie J.S., Dharmawadhana J.P., Jaiswal P., Chang J.H., Palusa S.G., Reddy A.S., Megraw M., Mockler T.C. Environmental stresses modulate abundance and timing of alternatively spliced circadian transcripts in Arabidopsis. Mol. Plant. 2014;8(2):207–227. doi: 10.1016/j.molp.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 67.Carvalho R.F., Feijao C.V., Duque P. On the physiological significance of alternative splicing events in higher plants. Protoplasma. 2013;250(3):639–650. doi: 10.1007/s00709-012-0448-9. [DOI] [PubMed] [Google Scholar]
- 68.Ding F., Cui P., Wang Z., Zhang S., Ali S., Xiong L. Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genomics. 2014;15:431. doi: 10.1186/1471-2164-15-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.E, Z.; Wang, L.; Zhou, J. Splicing and alternative splicing in rice and humans. BMB Rep. 2013;46(9):439–447. doi: 10.5483/BMBRep.2013.46.9.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shen Y., Wu X., Liu D., Song S., Liu D., Wang H. Cold-dependent alternative splicing of a Jumonji C domain-containing gene MtJMJC5 in Medicago truncatula. Biochem. Biophys. Res. Commun. 2016;474(2):271–276. doi: 10.1016/j.bbrc.2016.04.062. [DOI] [PubMed] [Google Scholar]
- 71.Chen C., Farmer A.D., Langley R.J., Mudge J., Crow J.A., May G.D., et al. Meiosis-specific gene discovery in plants: RNA-Seq applied to isolated Arabidopsis male meiocytes. Plant Biol. 2010;10:280. doi: 10.1186/1471-2229-10-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Filichkin S.A., Priest H.D., Givan S.A., Shen R., Bryant D.W., Fox S.E., Wong W.K., Mockler T.C. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20(1):45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu R., Wang Y., Zheng H., Lu W., Wu C., Huang J., Yan K., Yang G., Zheng C. Salt-induced transcription factor MYB74 is regulated by the RNA-directed DNA methylation pathway in Arabidopsis. J. Exp. Bot. 2015;66(19):5997–6008. doi: 10.1093/jxb/erv312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matsui A., Mizunashi K., Tanaka M., Kaminuma E., Nguyen A.H., Nakajima M., Kim J.M., Nguyen D.V., Toyoda T., Seki M. 2014. [DOI] [PMC free article] [PubMed]
- 75.Arikit S., Zhai J., Meyers B.C. Biogenesis and function of rice small RNAs from non-coding RNA precursors. Curr. Opin. Plant Biol. 2013;16(2):170–179. doi: 10.1016/j.pbi.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 76.Wang T.Z., Liu M., Zhao M.G., Chen R., Zhang W.H. Identification and characterization of long non-coding RNAs involved in osmotic and salt stress in Medicago truncatula using genome-wide high-throughput sequencing. BMC Plant Biol. 2015;15:131. doi: 10.1186/s12870-015-0530-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Varshney R.K., Mohan S.M., Gaur P.M., Gangarao N.V., Pandey M.K., Bohra A., Sawargaonkar S.L., Chitikineni A., Kimurto P.K., Janila P., Saxena K.B., Fikre A., Sharma M., Rathore A., Pratap A., Tripathi S., Datta S., Chaturvedi S.K., Mallikarjuna N., Anuradha G., Babbar A., Choudhary A.K., Mhase M.B., Bharadwaj C., Mannur D.M., Harer P.N., Guo B., Liang X., Nadarajan N., Gowda C.L. Achievements and prospects of genomics-assisted breeding in three legume crops of the semi-arid tropics. Biotechnol. Adv. 2013;31(8):1120–1134. doi: 10.1016/j.biotechadv.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 78.Hamwieh A., Imtiaz M., Malhotra R.S. Multi-environment QTL analyses for drought-related traits in a recombinant inbred population of chickpea (Cicer arietinum L.). Theor. Appl. Genet. 2013;126(4):1025–1038. doi: 10.1007/s00122-012-2034-0. [DOI] [PubMed] [Google Scholar]
- 79.Varshney R.K., Thudi M., Nayak S.N., Gaur P.M., Kashiwagi J., Krishnamurthy L., Jaganathan D., Koppolu J., Bohra A., Tripathi S., Rathore A., Jukanti A.K., Jayalakshmi V., Vemula A., Singh S.J., Yasin M., Sheshshayee M.S., Viswanatha K.P. Genetic dissection of drought tolerance in chickpea (Cicer arietinum L.). TAG. Theor. Appl. Genet. 2014;127(2):445–462. doi: 10.1007/s00122-013-2230-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Samineni S. Physiology, genetics and molecular mapping of salt tolerance in chickpea. The University of Western Australia Australia; 2010. [Google Scholar]
- 81.Chen H.T., Chen X., Wu B.Y., Yuan X.X., Zhang H.M., Cui X.Y. Whole-genome identification and expression analysis of K+ efflux antiporter (KEA) and Na+/H+ antiporter (NHX) families under abiotic stress in soybean. J. Integr. Agric. 2015;14(6):1171–1183. [Google Scholar]
- 82.Zhou H., Zhou J., Yang Y., Chen C., Liu Y., Jin X., Chen L., Li X., Deng X.W., Schumaker K.S., Guo Y. Ubiquitin-specific protease16 modulates salt tolerance in Arabidopsis by regulating Na(+)/H(+) antiport activity and serine hydroxymethyltransferase stability. Plant Cell. 2012;24(12):5106–5122. doi: 10.1105/tpc.112.106393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang W.J., Niu Y., Bu S.H., Li M., Feng J.Y., Zhang J., Yang S.X., Odinga M.M., Wei S.P., Liu X.F., Zhang Y.M. Epistatic association mapping for alkaline and salinity tolerance traits in the soybean germination stage. PLoS One. 2014;9(1):e84750. doi: 10.1371/journal.pone.0084750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.War A.R., Paulraj M.G., War M.Y., Ignacimuthu S. Role of salicylic acid in induction of plant defense system in chickpea (Cicer arietinum L.). Plant Signal. Behav. 2011;6(11):1787–1792. doi: 10.4161/psb.6.11.17685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zepeda-Jazo I., Velarde-Buendia A.M., Enriquez-Figueroa R., Bose J., Shabala S., Muniz-Murguia J., Pottosin I.I. Polyamines interact with hydroxyl radicals in activating Ca(2+) and K(+) transport across the root epidermal plasma membranes. Plant Physiol. 2011;157(4):2167–2180. doi: 10.1104/pp.111.179671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Syam Prakash S.R., Jayabaskaran C. Heterologous expression and biochemical characterization of two calcium-dependent protein kinase isoforms CaCPK1 and CaCPK2 from chickpea. J. Plant Physiol. 2006;163(11):1083–1093. doi: 10.1016/j.jplph.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 87.Geiger D., Scherzer S., Mumm P., Marten I., Ache P., Matschi S., Liese A., Wellmann C., Al-Rasheid K.A., Grill E., Romeis T., Hedrich R. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. USA. 2010;107(17):8023–8028. doi: 10.1073/pnas.0912030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dixit A.K., Jayabaskaran C. Phospholipid mediated activation of calcium dependent protein kinase 1 (CaCDPK1) from chickpea: a new paradigm of regulation. PLoS One. 2012;7(12):e51591. doi: 10.1371/journal.pone.0051591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Franco-Zorrillaa J.M., Lopez V.I., Carrasco J.L., Godoy M., Vera P., Solano R. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA. 2014;111(6):2367–2372. doi: 10.1073/pnas.1316278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kang H.G., Kim J., Kim B., Jeong H., Choi S.H., Kim E.K., Lee H.Y., Lim P.O. Overexpression of FTL1/DDF1, an AP2 transcription factor, enhances tolerance to cold, drought, and heat stresses in Arabidopsis thaliana. Plant Sci. 2011;180(4):634–641. doi: 10.1016/j.plantsci.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 91.Peng H., Cheng H.Y., Chen C., Yu X.W., Yang J.N., Gao W.R., Shi Q.H., Zhang H., Li J.G., Ma H. A NAC transcription factor gene of Chickpea (Cicer arietinum), CarNAC3, is involved in drought stress response and various developmental processes. J. Plant Physiol. 2009;166(17):1934–1945. doi: 10.1016/j.jplph.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 92.Guo D., Qin G. EXB1/WRKY71 transcription factor regulates both shoot branching and responses to abiotic stresses. Plant Signal. Behav. 2016;11(3):e1150404. doi: 10.1080/15592324.2016.1150404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shukla R.K., Tripathi V., Jain D., Yadav R.K., Chattopadhyay D. CAP2 enhances germination of transgenic tobacco seeds at high temperature and promotes heat stress tolerance in yeast. FEBS J. 2009;276(18):5252–5262. doi: 10.1111/j.1742-4658.2009.07219.x. [DOI] [PubMed] [Google Scholar]
- 94.Jin H., Xu G., Meng Q., Huang F., Yu D. GmNAC5, a NAC transcription factor, is a transient response regulator induced by abiotic stress in soybean. 2013. [DOI] [PMC free article] [PubMed]
- 95.de Zelicourt A., Diet A., Marion J., Laffont C., Ariel F., Moison M., Zahaf O., Crespi M., Gruber V., Frugier F. Dual involvement of a Medicago truncatula NAC transcription factor in root abiotic stress response and symbiotic nodule senescence. Plant J. 2012;70(2):220–230. doi: 10.1111/j.1365-313X.2011.04859.x. [DOI] [PubMed] [Google Scholar]
- 96.Peng H., Yu X., Cheng H., Shi Q., Zhang H., Li J., Ma H. Cloning and characterization of a novel NAC family gene CarNAC1 from chickpea (Cicer arietinum L.). Mol. Biotechnol. 2010;44(1):30–40. doi: 10.1007/s12033-009-9202-8. [DOI] [PubMed] [Google Scholar]
- 97.Boominathan P., Shukla R., Kumar A., Manna D., Negi D., Verma P.K., Chattopadhyay D. Long term transcript accumulation during the development of dehydration adaptation in Cicer arietinum. Plant Physiol. 2004;135(3):1608–1620. doi: 10.1104/pp.104.043141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kaur H., Verma P., Petla B.P., Rao V., Saxena S.C., Majee M. Ectopic expression of the ABA-inducible dehydration-responsive chickpea L-myo-inositol 1-phosphate synthase 2 (CaMIPS2) in Arabidopsis enhances tolerance to salinity and dehydration stress. Planta. 2013;237(1):321–335. doi: 10.1007/s00425-012-1781-0. [DOI] [PubMed] [Google Scholar]
- 99.Vincente M.R., Plasencia J. Salicylic acid beyond defence: its role in plant growth and development. J. Exp. Bot. 2011;62(10):3321–3338. doi: 10.1093/jxb/err031. [DOI] [PubMed] [Google Scholar]
- 100.Ren S., Weeda S., Li H., Whitehead B., Guo Y., Atalay A., Parry J. Salt tolerance in soybean WF-7 is partially regulated by ABA and ROS signaling and involves withholding toxic Cl- ions from aerial tissues. Plant Cell Rep. 2012;31(8):1527–1533. doi: 10.1007/s00299-012-1268-2. [DOI] [PubMed] [Google Scholar]
- 101.Tavakkoli E., Rengasamy P., McDonald G.K. High concentrations of Na+ and Cl- ions in soil solution have simultaneous detrimental effects on growth of faba bean under salinity stress. J. Exp. Bot. 2010;61(15):4449–4459. doi: 10.1093/jxb/erq251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Qiu Q.S., Barkla B.J., Vera-Estrella R., Zhu J.K., Schumaker K.S. Na+/H+ exchange activity in the plasma membrane of Arabidopsis. Plant Physiol. 2003;132(2):1041–1052. doi: 10.1104/pp.102.010421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wu X., Ebine K., Ueda T., Qiu Q.S. AtNHX5 and AtNHX6 are required for the subcellular localization of the SNARE complex that mediates the trafficking of seed storage proteins in Arabidopsis. PLoS One. 2016;11(3):e0151658. doi: 10.1371/journal.pone.0151658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang L., Wu X., Liu Y., Qiu Q.S. AtNHX5 and AtNHX6 control cellular K+ and pH homeostasis in Arabidopsis: three conserved acidic residues are essential for K+ transport. PLoS One. 2015;10(12):e0144716. doi: 10.1371/journal.pone.0144716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Qiu Q.S. Plant and yeast NHX antiporters: roles in membrane trafficking. J. Integr. Plant Biol. 2012;54(2):66–72. doi: 10.1111/j.1744-7909.2012.01097.x. [DOI] [PubMed] [Google Scholar]
- 106.Moller I.S., Gilliham M., Jha D., Mayo G.M., Roy S.J., Coates J.C., Haseloff J., Tester M. Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell. 2009;21(7):2163–2178. doi: 10.1105/tpc.108.064568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mielewczik M., Friedli M., Kirchgessner N., Walter A. Diel leaf growth of soybean: a novel method to analyze two-dimensional leaf expansion in high temporal resolution based on a marker tracking approach (Martrack Leaf). Plant Methods. 2013;9:30. doi: 10.1186/1746-4811-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Harfouche A., Meilan R., Altman A. Molecular and physiological responses to abiotic stress in forest trees and their relevance to tree improvement. Tree Physiol. 2014;34(11):1181–1198. doi: 10.1093/treephys/tpu012. [DOI] [PubMed] [Google Scholar]
- 109.Cai J., Okamoto M., Atieno J., Sutton T., Li Y., Miklavcic S.J. Quantifying the onset and progression of plant senescence by color image analysis for high throughput applications. PLoS One. 2016;11(6):e0157102. doi: 10.1371/journal.pone.0157102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gaur P.M., Gowda C.L., Knights E.J., Warkentin T., Acikgoz N., Yadav S.S., Kumar J. Chickpea and breeding management. Wallingford, UK: CABI; 2007. [Google Scholar]
- 111.Guo Y., Abernathy B., Zeng Y., Ozias-Akins P. TILLING by sequencing to identify induced mutations in stress resistance genes of peanut (Arachis hypogaea). BMC Genomics. 2015;16:157. doi: 10.1186/s12864-015-1348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bohra A., Pandey M.K., Jha U.C., Singh B., Singh I.P., Datta D., Chaturvedi S.K., Nadarajan N., Varshney R.K. Genomics-assisted breeding in four major pulse crops of developing countries: present status and prospects. Theor. Appl. Genet. 2014;127(6):1263–1291. doi: 10.1007/s00122-014-2301-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Smykal P., Coyne C.J., Ambrose M.J., Maxted N., Schaefer H., Blair M.W., Berger J., Greene S.L., Nelson M.N., Besharat N., Vymyslický T., Toker C., Saxena R.K., Roorkiwal M., Pandey M.K., Hu J., Li Y.H., Wang L.X., Guo Y., Qiu L.J., Redden R.J., Varshney R.K. Legume crops phylogeny and genetic diversity for science and breeding. Crit. Rev. Plant Sci. 2015;34(1-3):43–104. [Google Scholar]
- 114.Bohra A., Jha U.C., Kavi Kishor P.B., Pandey S., Singh N.P. Genomics and molecular breeding in lesser explored pulse crops, Current trends and future opportunities. Biotechnol. Adv. 2014;32:1410–1428. doi: 10.1016/j.biotechadv.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 115.Upadhyaya H.D., Thudi M., Dronavallia N., Gujaria N., Singh S., Sharma S., Varshney R.K. Genomic tools and germplasm diversity for chickpea improvement. Plant Genet. Resour. 2011;9(1):45–58. [Google Scholar]
- 116.Deokar A.A., Ramsay L., Sharpe A.G., Diapari M., Sindhu A., Bett K., Warkentin T.D., Taran B. Genome wide SNP identification in chickpea for use in development of a high density genetic map and improvement of chickpea reference genome assembly. BMC Genomics. 2014;15:708. doi: 10.1186/1471-2164-15-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Davey J.W., Hohenlohe P.A., Etter P.D., Boone J.Q., Catchen J.M., Blaxter M.L. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 2011;12(7):499–510. doi: 10.1038/nrg3012. [DOI] [PubMed] [Google Scholar]
- 118.Straub S.C., Parks M., Weitemier K., Fishbein M., Cronn R.C., Liston A. Navigating the tip of the genomic iceberg: Next-generation sequencing for plant systematics. Am. J. Bot. 2012;99(2):349–364. doi: 10.3732/ajb.1100335. [DOI] [PubMed] [Google Scholar]
- 119.Sun X., Liu D., Zhang X., Li W., Liu H., Hong W., Jiang C., Guan N., Ma C., Zeng H., Xu C., Song J., Huang L., Wang C., Shi J., Wang R., Zheng X., Lu C., Wang X., Zheng H. SLAF-seq: An efficient method of large-scale De Novo SNP discovery and genotyping using high-throughput sequencing. PLoS One. 2013;8(3):e58700. doi: 10.1371/journal.pone.0058700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Song Q., Hyten D.L., Jia G., Quigley C.V., Fickus E.W., Nelson R.L., Cregan P.B. Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PLoS One. 2013;8(1):e54985. doi: 10.1371/journal.pone.0054985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Akond M., Schoener L., Kantartzi S., Meksem K., Song Q., Wang D., Liu S., Anderson J.A., Kantartzi S.K., Wen Z., Lightfoot D.A., Kassem A. A SNP-based genetic linkage map of soybean using the SoySNP6K Illumina Infinium BeadChip genotyping array. J. Plant Genome Sci. 2013;1:80–89. [Google Scholar]
- 122.Lee Y.G., Jeong N., Kim J.H., Lee K., Kim K.H., Pirani A., Ha B.K., Kang S.T., Park B.S., Moon J.K., Kim N., Jeong S.C. Development, validation and genetic analysis of a large soybean SNP genotyping array. Plant J. 2015;81:625–636. doi: 10.1111/tpj.12755. [DOI] [PubMed] [Google Scholar]
- 123.De Leon T.B., Linscombe S., Subudhi P.K. Molecular dissection of seedling salinity tolerance in rice (Oryza sativa L.) using a High-density GBS-based SNP linkage map. Rice (N. Y.) 2016;9:52. doi: 10.1186/s12284-016-0125-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tiwari S., Krishnamurthy S.L., Kumar V., Singh B., Rao A., Amitha Mithra S.V., Rai V., Singh A.K., Singh N.K. Mapping QTLs for salt tolerance in rice (Oryza sativa L.) by bulked segregant analysis of recombinant inbred lines using 50K SNP chip. PLoS One. 2016;11(4):e0153610. doi: 10.1371/journal.pone.0153610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Qi X., Li M.W., Xie M., Liu X., Ni M., Shao G., Song C., Kay Yuen Yim A., Tao Y., Wong F.L., Isobe S., Wong C.F., Wong K.S., Xu C., Li C., Wang Y., Guan R., Sun F., Fan G., Xiao Z., Zhou F., Phang T.H., Liu X., Tong S.W., Chan T.F., Yiu S.M., Tabata S., Wang J., Xu X., Lam H.M. Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat. Commun. 2014;5:4340. doi: 10.1038/ncomms5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Patil G., Do T., Vuong T.D., Valliyodan B., Lee J.D., Chaudhary J., Shannon J.G., Nguyen H.T. Genomic-assisted haplotype analysis and the development of high-throughput SNP markers for salinity tolerance in soybean. Sci. Rep. 2016;6:19199. doi: 10.1038/srep19199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Munns R., James R.A., Xu B., Athman A., Conn S.J., Jordans C., Byrt C.S., Hare R.A., Tyerman S.D., Tester M., Plett D., Gilliham M. Wheat grain yield on saline soils is improved by an ancestral Na(+) transporter gene. Nat. Biotechnol. 2012;30(4):360–364. doi: 10.1038/nbt.2120. [DOI] [PubMed] [Google Scholar]
- 128.Qi X., Li M.W., Xie M., Liu X., Ni M., Shao G., Song C., Kay-Yuen Yim A., Tao Y., Wong F.L., Isobe S., Wong C.F., Wong K.S., Xu C., Li C., Wang Y., Guan R., Sun F., Fan G., Xiao Z., Zhou F., Phang T.H., Liu X., Tong S.W., Chan T.F., Yiu S.M., Tabata S., Wang J., Xu X., Lam H.M. Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat. Commun. 2014;5:4340. doi: 10.1038/ncomms5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dauch A.L., Jabaji-Hare S.H. Metallothionein and bZIP transcription factor genes from velvetleaf and their differential expression following colletotrichum coccodes infection. Phytopathology. 2006;96(10):1116–1123. doi: 10.1094/PHYTO-96-1116. [DOI] [PubMed] [Google Scholar]
- 130.Kumari A., Kumar A., Wany A., Prajapati G.K., Pandey D.M. Identification and annotation of abiotic stress responsive candidate genes in peanut ESTs. Bioinformation. 2012;8(24):1211–1219. doi: 10.6026/97320630081211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pagani M.A., Tomas M., Carrillo J., Bofill R., Capdevila M., Atrian S., Andreo C.S. The response of the different soybean metallothionein isoforms to cadmium intoxication. J. Inorg. Biochem. 2012;117:306–315. doi: 10.1016/j.jinorgbio.2012.08.020. [DOI] [PubMed] [Google Scholar]
- 132.Deshmukh R., Sonah H., Patil G., Chen W., Prince S., Mutava R., Vuong T., Valliyodan B., Nguyen H.T. Integrating omic approaches for abiotic stress tolerance in soybean. Front. Plant Sci. 2014;5:244. doi: 10.3389/fpls.2014.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wu Z.G., Wu J., Chen S.L., Mantri N., Tao Z.M., Jiang C.X. Insights from the cold transcriptome and metabolome of dendrobium officinale: global reprogramming of metabolic and gene regulation networks during cold acclimation. Front. Plant Sci. 2016;7:1653. doi: 10.3389/fpls.2016.01653. [DOI] [PMC free article] [PubMed] [Google Scholar]