Abstract
Introduction:
Drought stress is one of the most important abiotic stresses that negatively influence crop performance and productivity. Plants acclimatize to drought stress conditions through altered molecular, biochemical and physiological responses. Gene and/or protein expression and regulation are thought to be modulated upon stress perception and signal transduction for providing requisite endurance to plants.
Plant growth regulators or phytohormones are important molecules required for various biological processes in plants and are also central to stress signalling pathways. Among various phytohormones, Abscisic Acid (ABA) and Ethylene (ET) are considered to be the most vital growth regulators implicated in drought stress signalling and tolerance. Besides the above two known classical phytohormones, Salicylic Acid (SA) and Jasmonic Acid (JA) have also been found to potentially enhance abiotic stress tolerance particularly that of drought, salinity, and heat stress tolerance in plants. Apart from these several other growth regulators such as Cytokinins (CKs), Auxin (AUX), Gibberellic Acid (GA), Brassinosteroids (BRs) and Strigolactones (SLs) have also been reported to actively participate in abiotic stress responses and tolerance in plants. The abiotic stress signalling in plants regulated by these hormones further depends upon the nature, intensity, and duration of exposure to various environmental stresses. It has been reported that all these phytohormones are also involved in extensive crosstalk and signal transduction among themselves and/or with other factors.
Conclusion:
This review thus summarizes the molecular mechanism of drought signalling and its crosstalk with various phytohormone signalling pathways implicated in abiotic stress response and tolerance.
Keywords: Abiotic stress, Crosstalk, Drought, Functional genomics, Phytohormones, Signal transduction
1. INTRODUCTION
Global climate changes have compounded the multifarious effects of abiotic stresses on crop growth resulting in jeopardized agricultural productivity worldwide. In the present scenario, crops have to endure several abiotic stresses including drought, salinity, heat and chilling stresses in their natural habitat [1, 2]. More than 50% reduction in average yields of major cereal crops has been reported as a consequence of various abiotic stresses [3]. Plants respond to these stresses by activating several complex cellular and molecular responses which help them to adapt under various abiotic and biotic stresses, which ultimately results in better growth and survival.
Drought tolerance, drought avoidance and drought escape are three usual mechanisms adapted by plants exposed to drought stress. Drought escape takes advantage of the short life cycle and/or developmental plasticity of plants, drought avoidance involves increased water uptake and decreased water loss, while drought tolerance applies to exhibition of better osmoprotection, antioxidative capacity and desiccation tolerance [4]. Hence plants must maintain a balance among different drought resistance mechanisms for optimized yield under drought conditions. Drought resistance, therefore, is a complex trait that involves changes manifested at morphological, physiological and molecular levels such as earliness, prolific and deep roots, leaf rolling, reduced leaf area and transpiration, stomatal closure, accumulation of osmolytes and activation of stress responsive genes [5, 6]. Numerous genes have been found to be involved in drought regulatory networks, and are thought to impart stress tolerance by production of vital proteins and metabolites [7]. It is predicted that the genes that are expressed during drought stress help improve cellular tolerance through maintaining osmotic homeostasis or through damage control and repair, and/or by regulating gene expression [8, 9]. Several attempts have been made to identify candidate genes related to drought stress tolerance in a number of plant species [8, 10]. Various functional genomic approaches such as transcriptome analyses and gene expression profiling have proved to be major tools for the identification and validation of stress related genes [7]. Comprehensive transcriptome analyses have led to the identification of two broad categories of genes [11]. The first group includes genes that encode proteins involved in cellular homeostasis and protection from stress such as osmoprotectants, water channels, antioxidative enzymes, metabolic enzymes and lipid-transfer proteins, while the second group largely comprises of kinases and TFs that regulate the stress signal transduction and stress responsive gene expression. The activation and regulation of a large number of genes is also linked to signalling pathway(s) of various phytohormones. Phytohormones or plant growth regulators are signal molecules produced within plants at extremely low concentrations and have the ability to regulate cellular processes both locally as well as distally. Various plant growth regulators such as ABA, ET, AUX, CKs, SA, JA, GA, BRs and SLs have also been found to potentially enhance abiotic stress particularly drought stress tolerance of plants. Thus activation of drought stress responsive genes is thought to be regulated through complex regulatory networks involving Transcription Factors (TFs), kinases and phytohormones. This review thus emphasizes on the role of various components of drought signalling cascade, involvement of phytohormones in gene regulation and their crosstalks.
2. DROUGHT SIGNALLING CASCADE
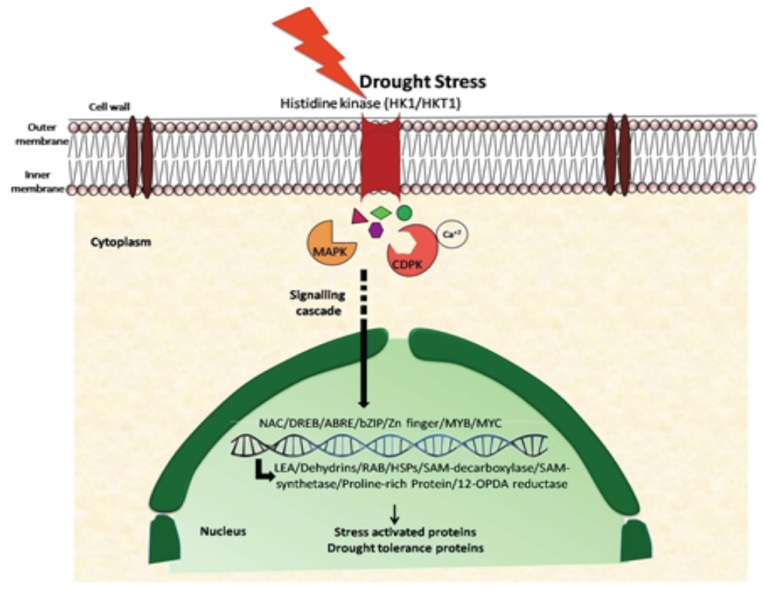
In the last few decades, the research pertaining plant responses to drought stress has rapidly progressed. Several genes that are induced under drought stress have been identified through various functional genomic approaches. However, the functions of several genes are still not known owing to the complexity in plant responses to abiotic stresses particularly drought since it is a quantitative trait. A generalized drought stress signalling pathway initiates with stress perception by plasma membrane receptors as for example transmembrane histidine kinases that bind to signalling molecules or ligands (Fig. 1). In Arabidopsis thaliana, AtHTK1, a histidine kinase domain containing protein has been reported to trigger the downstream signalling cascade resulting in dehydration induced gene expression [12, 13]. The signal transduced downstream thus results in formation of second messengers such as inositol phosphates and reactive oxygen species (ROS) [8]. This leads to perturbation in intracellular calcium levels which are sensed by calcium sensors that are responsible for triggering phosphorylation cascades. These cascades include phosphorylation and dephosphorylation mediated by several protein kinase and phosphatases and finally target several downstream genes that could be either directly involved in protection of cellular machinery or regulation of gene expression. The usual protein kinases that are involved in stress signalling are Ca2+-dependent (CDPK) and mitogen activated (MAPKs) that transduce the water stress signals to the nucleus by implicating various TF regulons such as DREB, NAC, MYB/ MYC, ABRE etc. that further regulate the expression of downstream genes such as LEA, ERD, DHN, RDs (RD19, RD22 and RD29) etc. [9, 11, 14, 15].
Fig. (1).
A schematic representation of drought stress signalling cascade in plants.
Several DREB TFs have been isolated from different plants and have been reported to actively participate in various abiotic stress signal transduction pathways by regulating the stress responsive gene expression [11, 16, 17]. Further genetic manipulation of DREB TFs for generating overexpression transgenic lines in Arabidopsis and rice (Oryza sativa) have shown superior tolerance to drought, salt, and cold stresses [18, 19]. DREB1A/CBF3 when overexpressed using a stress-inducible promoter RD29A in wheat (Triticum aestivum) exhibited improved dehydration tolerance [20]. Transgenic wheat and barley lines expressing DREB2 and DREB3 from wheat under either a constitutive (double 35S) or drought-inducible (Rab17) promoter showed better survival under severe drought stress [21]. Further genome-wide expression profiling of AP2/ERF TFs in foxtail millet indicated stress-specific and temporal responses to various abiotic stresses including drought [22]. These findings thus strengthen the fact that DREBs are crucial to stress endurance of crop plants.
The NAC TFs are also one of the biggest TF families in plants that act in response to various environmental stresses [7]. Several NACs have been reported to be highly activated under drought and osmotic stresses [23]. Further transgenic rice plants overexpressing OsNAC6, OsNAC45 and ONAC022 displayed enhanced drought and salinity tolerance [17, 23, 24]. A Miscanthus NAC TF MINAC9 was also found to increase abiotic stress tolerance in transgenic Arabidopsis suggesting the important role of NAC TFs in physiological adaptation of plants to various abiotic stresses [25].
The MYC/MYB TFs also play important role in abiotic stress signalling as several of these have been found to be induced under dehydration, salinity and osmotic stresses [26]. The transgenic Arabidopsis plants generated by overexpressing various MYB/MYC TFs such as AtMYB2 AtMYB12, AtMYB37, AtMYC2, OsMYB4 etc. exhibit drought, osmotic and chilling tolerance [27-29]. Similarly ABREs and ABFs also actively participate in abiotic stress signalling mostly through ABA-dependent mode [30]. Overexpression of ABRE/ABF genes has shown to considerably improve the drought and salinity tolerance capacity of transgenic rice [6, 17]. Thus these TFs and various other components of stress signalling pathway help plants to cope and adapt for better growth and survival under drought and other abiotic stress. Apart from this, they may also play significant role in the generation and crosstalk of phytohormones like ABA, SA, JA and ET [26, 31]. Several other phytohormones like AUX, CKs, GA, BRs and SLs are also reported to be crucial for abiotic stress signalling. These hormones act as regulatory molecules that interact with various components of drought stress signalling pathway and help in achieving enhanced stress tolerance in plants.
3. PHYTOHORMONES AND REGULATION OF DROUGHT RESPONSIVE GENES
Phytohormones have been time and again implicated in regulation and coordination of plant development as they influence numerous biochemical and physiological processes as well as regulate plant responses to various environmental cues [32, 33]. Phytohormones thus are critical to various abiotic stress responses in plants so that they could escape and/or survive under such conditions. This may however result in reduced growth so that the plants could redirect its resources towards enduring stress [34]. Thus, abiotic stresses may alter production, distribution, or signal transduction of phytohormones leading to activation of precise protective mechanisms in plants. Functional genomic approaches for instance expression profiling, overexpression, knockdown or genomic-assisted breeding offer a powerful platform to identify and characterize genes that play important role in stress response, adaptation and resistance. Thus these approaches could be widely applied to functionally validate various components of drought signalling pathway and their crosstalk with phytohormones. Here we have provided a list of drought stress responsive genes that have been overexpressed in various plants and their response to various plant hormones (Table 1). The role of various phytohormones in drought signalling is discussed below.
Table 1.
Details of overexpressing stress responsive genes in plants under drought stress and their response to various phytohormones in last five years.
| S. No. | Gene | Source | Transgenic | Stress Tolerance |
Phytohormone
Responsiveness |
Ref. | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | APX6 | T. salsuginea | A. thaliana | Drought, Salt | ABA | [35] | |||||
| 2 | MYB12 | A. thaliana | A. thaliana | Drought, Salt | ABA | [27] | |||||
| 3 | EXPA2 | T. aestivum | N. tabacum | Drought | ABA, JA, SA, GA | [36] | |||||
| 4 | NAC2 | G. herbaceum | A. thaliana | Drought | ABA, JA, ET | [37] | |||||
| 5 | CKX1 | A. thaliana | H. vulgare | Drought | CK | [38] | |||||
| 6 | HB12 | C. arabica. | A. thaliana | Drought | ABA | [39] | |||||
| 7 | NINV | P. trifoliata | N. tabacum | Drought, Salt, Cold | ABA | [40] | |||||
| 8 | SIK1 | M. sativa | A. thaliana | Drought | ABA, GA | [41] | |||||
| 9 | VIN | N. tabacum | A. thaliana | Drought | ABA | [42] | |||||
| 10 | BCG25 | A. thaliana | A. thaliana | Drought | ABA | [43] | |||||
| 11 | HDG11 | A. thaliana | T. aestivum | Drought | ABA | [44] | |||||
| 12 | HD2D | A. thaliana | A. thaliana | Drought, Salt, Cold | ABA | [45] | |||||
| 13 | EBP1 | A. canescens | A. thaliana | Drought, Cold | ABA | [46] | |||||
| 14 | EPF2 | P. deltoids | A. thaliana | Drought | ABA | [47] | |||||
| 15 | LEA4-1 | A. thaliana | B. juncea | Drought, Salt | ABA, GA, JA, IAA | [48] | |||||
| 16 | WRKY1 | T. aestivum | N. tabacum | Drought | ABA | [49] | |||||
| 17 | bZIP23 | O. sativa | O. sativa | Drought | ABA | [50] | |||||
| 18 | DHN | V. vinifera | N. tabacum | Drought, Salt | GA, CK, ABA | [51] | |||||
| 19 | MYB37 | A. thaliana | A. thaliana | Drought | ABA | [28] | |||||
| 20 | FIN1 | M. sativa | A. hypogaea | Drought | ABA | [52] | |||||
| 21 | DH45 | P. sativum | A. hypogaea | Drought | ABA | [52] | |||||
| 22 | HSF4 | P. glaucum | A. hypogaea | Drought | ABA | [52] | |||||
| 23 | JAZ14 | Zea mays | A. thaliana | Drought | ABA, JA, GA | [53] | |||||
| 24 | DREB2 | Glycine max | N. tabacum | Drought | ABA | [54] | |||||
| 25 | CBF3 | G. hirsutum | A. thaliana | Drought, Salt | ABA | [55] | |||||
| 26 | MLP43 | A. thaliana | A. thaliana | Drought | ABA | [56] | |||||
| 27 | NF-YA1 | S. italica | N. tabacum | Drought, Salt | ABA | [57] | |||||
| 28 | ERD4 | B. juncea | A. thaliana | Drought | ABA, SA | [58] | |||||
| 29 | ARGOS1 | Zea mays | A. thaliana | Drought | ET | [59] | |||||
| 30 | YUCCA6 | A. thaliana | Poplar | Drought | AUX | [60] | |||||
| 31 | LOX1 | C. annumm | A. thaliana | Drought, Salt | ABA | [61] | |||||
| 32 | DREB2A | V. radiata | A. thaliana | Drought, Salt | ABA | [18] | |||||
| 33 | NAC6 | O. sativa | O. sativa | Drought, Salt | ABA | [17] | |||||
| 34 | ETOL1 | O. sativa | O. sativa | Drought, Submergence | ET, GA | [62] | |||||
| 35 | EXPA4 | R. hybrida | A. thaliana | Drought, Salt | ABA | [63] | |||||
| S. No. | Gene | Source | Transgenic | Stress Tolerance |
Phytohormone Responsiveness |
Ref. | |||||
| 36 | EDT1/HDG11 | A. thaliana | O. sativa | Drought | ABA | [64] | |||||
| 37 | EXPB23 | T. aestivum | N. tabacum | Drought, Salt | ABA, JA, GA, ET, AUX | [65] | |||||
| 38 | PIN3t | O. sativa | O. sativa | Drought | AUX | [66] | |||||
| 39 | Sce9 | S. alterniflora | A. thaliana | Drought, Salt, Cold | ABA | [67] | |||||
| 40 | ERF5 | S. lycopersicum | S. lycopersicum | Drought, Salt | ET | [68] | |||||
| 41 | bHLH148 | O. sativa | O. sativa | Drought | JA | [69] | |||||
| 42 | MYB1R-1 | S. tuberosum | S. tuberosum | Drought | ABA, JA | [70] | |||||
| 43 | PARG1 | A. thaliana | A. thaliana | Drought, Osmotic | ABA | [71] | |||||
| 44 | FTL1/DDF1 | A. thaliana | A. thaliana | Drought, Cold, Heat | GA | [72] | |||||
| 45 | RDCP1 | O. sativa | O. sativa | Drought | JA | [73] | |||||
| 46 | MKK4 | Zea mays | N. tabacum | Drought | ABA | [74] | |||||
| 47 | DREB2A | O. sativa | O. sativa | Drought | ABA | [19] | |||||
| 48 | NAC10 | O. sativa | O. sativa | Drought | ABA | [75] | |||||
| 49 | DREB2C | A. thaliana | A. thaliana | Drought | ABA | [76] | |||||
| 50 | VAMP71 | A. thaliana | A. thaliana | Drought | ABA | [77] | |||||
3.1. Abscisic Acid
ABA has been proposed to play the role of a central regulator in developmental processes as well as stress response and tolerance in plants [7, 11, 78]. ABA has been reported to control the expression of several drought responsive genes [8]. Most of the transcripts for genes encoding ABA biosynthesis enzymes displayed enhanced expression under drought, oxidative, and temperature stresses [79, 80]. However, a large number of genes respond to various abiotic stresses including drought but not to ABA suggesting the existence of both ABA-dependent and ABA-independent mode of gene expression [8, 11]. In rice all OsLEA genes that are regulated by ABA contain ABRE motifs while drought-inducible OsDHODH1 gene has both ABRE and DRE/CRT motifs [18, 81]. Similarly a PEG and ABA-inducible ABRE binding TF, OsAREB1 could regulate downstream genes in an ABA-dependent manner [5]. Various other drought related TF regulons for example DREB, NAC and MYB that recognize and bind to DRE/CRT, NACRS and MYBRS motifs, respectively, but not ABRE may modulate downstream gene expression through ABA-independent pathway [78]. Transgenic Arabidopsis expressing wheat TaRK2.8 showed enhanced tolerance to drought, salt and cold stress by activating both ABA-dependent e.g. ABA1, ABA2, RD20A and RD29B, and ABA-independent e.g. CBF1, CBF2 and CBF3 genes [82, 83]. It was reported that EmBP-1, a bZIP family TF when bound to ABRE motif can lead to activation of ABA-induced genes under drought stress [1]. A novel NAC TF gene, TaNAC29 isolated from bread wheat when overexpressed in Arabidopsis showed better drought and salt stress tolerance, and also exhibited an ABA-hypersensitive response [84]. In rice overexpression of DSM2, a drought inducible gene encoding a putative β-carotene hydroxylase led to considerably augmented tolerance to drought and oxidative stresses most likely by increasing the levels of zeaxanthin, a carotenoid precursor of ABA [80]. The transgenic rice also displayed increased ABA levels under the stress conditions. Further ABA triggers stomatal closure and also leads to accumulation of numerous proteins, such as LEAs that help in osmotic adjustment, and as a result improve tolerance to drought [85].
3.2. Salicylic Acid
SA is essentially a phenolic compound produced by plants that functions as a growth regulator, and plays a crucial role in regulating plant growth, development, and biotic interactions [86, 87]. The majority of genes that respond positively to severe SA treatment have been found to be related to stress and signal transduction pathways such as those encoding antioxidants, chaperones, heat shock proteins, and genes for secondary metabolites biosynthesis [88]. Earlier the key role of SA in plants was considered to be the regulation of biotic stress responses only; however, now it is well accepted that SA is also implicated in a number of abiotic stress responses like drought and salinity [9, 87, 89]. Elevated expression of an SA-responsive gene PR-1 in chickpea has been reported under drought stress confirms that pathogenesis related genes (PR) are not only induced under both biotic but also under abiotic stresses [9]. Several WRKY TFs such as WRKY70 and WRKY54 are found to be governed primarily by SA [90]. WRKY70 and WRKY54 as key components in biotic stress response, also co-operatively act as negative regulators of osmotic stress tolerance and stomatal closure in Arabidopsis suggesting their important role in abiotic stress signalling.SA-related Arabidopsis mutant has also been used to explain the role of SA under drought stress [91]. In tomato and bean plants, lower concentrations of SA when applied exogenously seemed to be useful in improving drought tolerance [87]. Further there have been reports that correlated SA levels and/or SA signalling with positive regulation of plant responses to drought stress and SA-induced stomatal closure was proposed as the most possible mechanism coupled with the maintenance of water content in leaves [89]. Further various gene expression profiling studies have shown that several TFs such as AP2/ERFs, leucine zipper, Zn fingers etc. responded to SA as well as drought and salt stress suggesting the important role of this phytohormone in stress response [22, 92].
3.3. Jasmonates
JA is an important plant hormone and growth regulator involved in seed germination, callus and primary root growth, flowering, fertilization, and senescence [93]. JA is also found to be involved in plant responses to wounding caused by insects, pathogen infections, and several environmental stresses [94]. There have been many reports on the involvement of JA in drought stress [9, 85]. A significant activation of a jasmonate signalling pathway gene MYC2 in a tolerant chickpea cultivar under drought stress ascertained the role of jasmonates in early drought stress signalling and tolerance [9, 95]. Drought-induced regulation of a rice JA receptor protein, CORONATINE INSENSITIVE 1, OsCOI1a and a jasmonic acid ZIM-domain protein, OsJAZ which are key regulators of JA signalling indicate significant amalgamation of JA metabolism and signalling in abiotic stress responses of plants [96, 97]. Further it has been reported that the induction of MYC2 relies on a JA-Ile receptor COI1 [98]. In rice, OsbHLH148 interacts with OsJAZs in response to drought, and its constitutive expression improves drought tolerance by inducing OsDREB1 expression [69, 99]. In another study, JAR1 expression in wild type Arabidopsis plants significantly increased throughout in response to water stress, but in jar1-1 mutant seedling its expression remained constant [100]. Similarly overexpression of maize gene ZmJAZ14 in Arabidopsis enhanced tolerance to PEG- induced drought stress [53]. In rice leaves and roots, the JA levels were found to be increased under both drought and high salinity, leading to the activation of stress-related PR and JA-biosynthetic genes [93]. These data showed that JA is not only involved in plant defense during wounding and pathogen attack but also during drought.
3.4. Ethylene
Ethylene is a gaseous plant hormone known for inducing responses to various stresses and leaf abscission. Its role in different metabolic pathways under abiotic stresses has been elucidated in several plants [101]. ET is produced from methionine via AdoMet (S-adenosyl- L-methionine) and ACC, a cyclic non-protein amino acid precursor, 1-aminocyclo- propane-1-carboxylate. The conversion of AdoMet to ethylene is carried out by enzymes ACC synthase (ACS) and ACC oxidase (ACO) [102]. In soybean, the expression of MAT (Methionine Adenosyl Transferase) gene, the first enzyme in the biosynthesis pathway responsible for production of AdoMet, was enhanced under water stress [102]. It was found that the expression of ACO and ACS were also enhanced under drought stress in chickpea [9] and soybean [102]. There was a reduction in the level of expression of ETR (ET receptors) and CTR genes under drought stress in soybean [102, 103]. CTR is a key negative regulator of the ethylene signal transduction pathway, keeping the downstream signalling components EIN2 inactive through CTR1-kinase-dependent phosphorylation under drought stress [103]. The Arabidopsis ethylene insensitive mutants, ein2-5 and ein3-1, were found to be more susceptible to drought stress as compared to wild type Col-0 plants [104]. In another study, SlERF5, AtERF5 and AtERF6 were established as master regulators after sudden exposure to salt and drought stress [68, 105]. Gene expression of these TFs was induced very rapidly in actively growing leaves. AtERF6 also induces the expression of various osmotic stress-responsive genes, including the STZ, MYB51, and WRKY33. The enhanced transcript accumulation for various ERF genes have also been reported under drought, salt and cold stresses [79]. Further several genome-wide investigations on TFs and transcriptome analyses of crop plants subjected to drought stress have shown significant up-regulation of ethylene responsive genes [22, 106, 107]. The five ET receptor genes in Arabidopsis namely, ETR1, ERS1, ETR2, EIN4, and ERS2 were able to bind to ET [102]. Interestingly ETR1 has also a His kinase activity. Thus it can be concluded that ethylene signalling is very important for regulating plant growth and stress responses and ET functions via its receptors.
3.5. Indole-3-acetic Acid
IAA is considered to be the first plant hormone to be identified and it plays an important role in plant growth regulation via cell elongation, cell and vascular tissue differentiation, axial elongation and apical dominance [96]. However an increase in IAA level has apparently been linked to diminished growth, suggesting altered hormonal balance to be the cause of growth reduction under stress conditions [1]. Several auxin-responsive genes have been isolated and characterized from various plant species such as rice, Arabidopsis, soybean etc. [108]. An auxin-inducible GST, PjGSTU1 from Prosopis juliflora was found to confer drought tolerance in transgenic tobacco [109]. The activation of YUCCA6, a flavin monooxygenase encoding gene that functions in the tryptophan-dependent auxin biosynthetic pathway led to enhanced endogenous levels of auxin and improved drought tolerance in Arabidopsis [60, 110]. Similarly overexpression of YUCCA7 resulted in better drought tolerance in Arabidopsis [111]. Conversly induction of OsGH3-2 gene that encodes an enzyme for IAA inactivation exhibited reduced free IAA content as well as diverse alterations in drought and cold resistance pattern along with hypersensitive response in transgenic rice [112, 113]. Two rice OsPIN genes, namely OsPIN2 and OsPIN5b were found to be up-regulated by drought and heat stress [96]. Overexpression of an IAA-amido synthetase encoding gene, TLD1/OsGH3.13 enhanced the expression of LEA genes, leading to increased drought tolerance of rice seedlings [114, 115]. This shows that transcription of majority of genes of different metabolic pathways are interconnected and stimulated by auxins.
3.6. Cytokinins
CKs are crucial for several plant growth and developmental processes including cell division, leaf senescence, nutrient mobilization, vascular and shoot differentiation, anthocyanin biosynthesis as well as photomorphogenic development [116]. CKs are also found to regulate responses to various environmental stimuli such as drought and salt stress [6, 117]. CKs have both positive and negative effects on drought tolerance depending on stress duration or intensity. Isopentenyl transferase (IPTs) catalyse rate-limiting step of CKs biosynthesis. In transgenic cotton enhanced IPT expression led to higher endogenous level of CKs which is responsible for delay senescence in plant and showed improved drought tolerance [118]. CKs are thought to be a negative regulator of root growth and branching, and thus a root-specific degradation of CKs may also augment primary root growth and branching under drought stress [119]. On exposure to drought, induced expression of AtMYB2 in Arabidopsis downregulates IPTs gene expression and as consequence endogenous level of CKs. Arabidopsis plants exhibiting enhanced expression of cytokinin oxidase/dehydrogenase1 (CKX1) genes, established that an increase in CKs degradation in the roots boost both primary root length as well as lateral root formation during drought stress [119]. Further partial cytokinin insensitivity induced in transgenic barley lines overexpressing cytokinin dehydrogenase was found to enhance drought tolerance [38]. The transgenic barley roots also showed a higher auxin turnover. Functional analyses of CKs receptor mutants revealed that all three Arabidopsis CKs receptors namely, AHK2, AHK3, and CRE1/AHK4 function as negative regulators of osmotic stress [12, 120]. Furthermore CKs inducible typeA ARR4 and ARR5 were found to be activated by dehydration, cold and high salinity [6].
3.7. Brassinosteroids
BRs constitute a group of novel steroidal phytohormones that regulate plant growth and development by manifesting an array of physiological and morphological changes [1, 121]. They are also identified in alleviating various abiotic stresses including drought, salinity, and heat stresses, and also play an important role in biotic stress response [122]. Microarray examination of BR-deficient Arabidopsis plants revealed strong expression of thioredoxin and monodehydroascorbate reductase and lower expression of COR78 while BR-treated plant showed strong induction of COR47 indicating that BR-regulating genes are involved in regulation of cold and drought tolerance [123]. Application of BRs also led to increased seedling growth sorghum cultivars subjected to osmotic stress, which was manifested in terms of seedling length and fresh and dry weights [124]. A knockout mutant of rice OsGSK1, a BIN2 ortholog, improved tolerance to different abiotic stresses including drought [125]. The role of BR in drought stress has further been validated in Arabidopsis and B. napus by using BR-deficient mutants, det2-1 and dwf4 where treatment of epibrrasinolide improved drought tolerance than untreated mutants [126]. Further the drought responsive gene regulatory network in Brassica napus suggests the involvement of its various components in biosynthesis and signalling of various phytohormones including ABA, auxins and BRs indicating their important role in stress response and tolerance [127].
3.8. Gibberellic Acid
GAs are a group of tetracyclic diterpene phytohormones that are crucial for plant development such as germination, cell elongation, leaf expansion, and flower and fruit development as well as responses to several environmental stresses [128, 129]. They can also influence several photosynthetic enzymes, and thereby improving the light interception, photosynthetic efficiency, leaf area index, and nutrient use efficiency of plants [130]. It has been reported that GA could invert the morphological and stress protective consequences of triazoles signifying a close link between GA and stress protection in plants [131, 132]. GA has also been reported to alleviate the adverse effects of environmental stresses on plant water relations [128]. As for example, in potato a GA stimulated transcripts in Arabidopsis 6 gene encoding GASA6 has been related to drought and water stress stimulus response [133]. DELLA proteins, the major regulators of GA responses and also act as suppressors, have been shown to contribute towards osmotic stress tolerance in GA deficient Arabidopsis mutants [131, 134]. A GA-GID1-DELLA mechanism provides an explanation for both plant growth and stress responses. In Arabidopsis GA signalling is mediated by binding of GA to GA INSENSITIVE DWARF1 or GID1a/b/c, the receptor ortholog of which in rice is called OsGID1 [17, 155]. DELLA proteins when interact with GID1 stimulates degradation of DELLA proteins and activates the regulatory roles of GA [17, 130]. However a GRAS/SCL TF, PeSCL7 from poplar was highly induced by drought and salt, but was repressed by the application of GA in leaves [135-137]. Further SPINDLY (SPY) gene that acts as a negative regulator of GA signalling in plants was found to negatively regulate abiotic stress responses; most probably by integrating environmental stress signals through GA and CKs crosstalk [138]. Thus different approaches to modulate GA levels in plants can be integrated to form the basis for novel crop improvement strategies under stress conditions.
3.9. Strigolactones
Recently a new class of phytohormones have been identified which are carotenoid derived compounds and are referred to as strigolactones (SLs) [139]. They are well-known for their role in plant-microbe interactions. SLs maintain root and shoot architecture under various environmental stimulus. They act as a positive regulator under both drought and salt stress [139, 140]. More axillary growth (MAX) genes namely MAX1, MAX3 and MAX4 encode enzymes that play role in SLs biosynthesis [140]. Evidence supported that SLs plays role in suppression of shoot branching. In Arabidopsis exogenous application of GR24 (synthetic SL analogue) rescued drought sensitive effect of the SL-deficient mutant (max3 and max4) but not of the SL-response mutant (max2) and in wild type SL treatments enhanced drought tolerance of plants [140]. Genome-wide expression analysis of carotenoid oxygenase genes in response to salt, drought, cold, ABA, and SL treatments in Brassica rapa and Brassica oleracea led to the identification of various genes related to carotenoid metabolism such as carotenoid cleavage dioxygenase (CCD)1, CCD4, CCC7, CCD8, NCED (9-cis-epoxycarotenoid dioxygenase) 2, NCED3, NCED5, NCED 6, and NCED9 [141]. Further NCED2, NCED 6 and NCED9 were found to be responsive to both ABA and SL treatments indicating their important role in abiotic stress signalling. In Lotus japonicus SL-depleted plants showed increased stomatal conductance under both normal and osmotic stress conditions [142]. These evidences showed that SLs are major signalling molecules which play role in plant development and adaptation during environmental challenges. However, detailed study of SLs role in abiotic stress tolerance need to be investigated.
4. CROSSTALK: DROUGHT SIGNALLING CASCADE AND PHYTOHORMONES
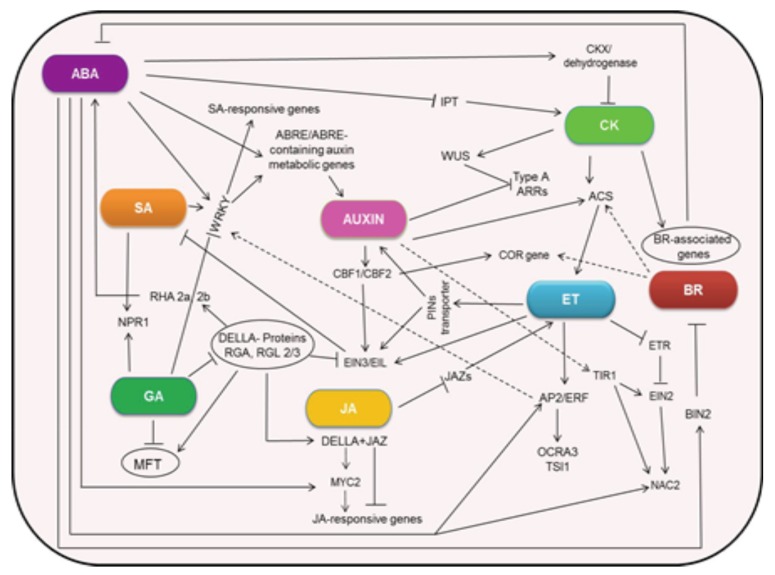
Evidences supporting phytohormone crosstalk during plant response and acclimatization to abiotic stresses comes mainly from their synergistic and/or antagonistic action and the coordinated regulation of plant hormone biosynthetic pathways. ABA, SA, JA and ET are central to abiotic stress responses, with ABA play major role in regulating osmotic stresses [11]. CKs, IAA, GA, BRs and SLs also interact with other phytohormones and stress-related genes to maintain balanced growth under abiotic stresses. Functional genomic approaches have led to the identification of several phytohormone-mediated signalling pathways in Arabidopsis and in numerous other plants under abiotic stresses, suggesting interconnection between phytohormones at various levels for stress tolerance by the plants [143] (Fig. 2). An increase in endogenous ABA level in plants under drought stress was correlated to the activation of ABA biosynthetic genes [80, 144], triggering stomatal closure and accumulation of osmoprotectants, thereby improving drought tolerance [85]. JA and SA have also been implicated as a key player in drought stress signalling based on its accumulation during drought and its positive regulatory role in stomatal closure [9, 145, 146]. In Arabidopsis, JAZ and AtMYC2 together regulate the expression of JA-responsive genes. Interaction between DELLA/RGL3 and JAZ repressors prevents JA-mediated degradation of JAZ [147, 148], and triggers the induction of MYC2 [149]. AtMYC2 has been found to repress JA/ET-responsive genes under pathogen stress but in case of oxidative stress the JA-responsive genes are positively regulated [150, 151]. These studies suggested that JA and GA interact in an ABA-dependent response to drought. GA also interacts with SA during stress response. The exogenous application of GA3 induced expression levels of NPR1, SA biosynthetic genes and genes involved in SA action, [152]. Transgenic Arabidopsis plants constitutively expressing a Fagus sylvatica GA responsive gene FsGASA4, a member of the GA3 gene family, showed improved oxidative stress
Fig. (2).
Phytohormone crosstalk related to drought stress response in plants. Arrows and dashed lines indicate positive regulation and blocked arrows indicate negative regulation. The phytohormones and gene interactions shown here is only a fraction of stress signal crosstalk occurring in plants when exposed to drought stress and different other crosstalk points are yet to be discovered.
tolerance which was linked to enhanced endogenous levels of SA [152]. ABA could also regulate BR-mediated signalling through BIN2 or its upstream components via protein phosphatase 2C (PP2C) family of genes [153]. ABA was also found to restrain BR-induced responses in plants exposed to drought stress [90]. CKs have also been reported to regulate the responses to various abiotic stresses such as drought and salt stress [117]. Drought stress has generally been found to decrease the production and transport of CKs [117]. Arabidopsis plants when subjected to drought stress showed an increased ABA content in roots that were thought to inhibit ethylene production and stimulate CKs degradation maintaining primary root growth [154]. Of late CKs has also been shown to be a positive regulator of auxin biosynthesis, and it was proposed that both CKs and IAA signalling are involved in a homeostatic feedback regulatory loop that acts to retain proper CKs and IAA concentrations in developing roots and shoots [155]. On the other hand auxin was found to influence ethylene biosynthesis as for example, several members of the ACS gene family that encode rate-limiting enzymes in ethylene biosynthetic pathway, were regulated by application of auxin [102, 114]. Together with auxins, BRs and MeJA could also activate ACO enzymes, increasing ethylene production in maize and olive plants [102]. Mutations in AHK2 and AHK3 that encode CKs receptors induced both drought and ABA-responsive genes in Arabidopsis [12]. AHK5 was found to counteract ET and ABA regulated growth in stomatal guard cells [156, 157]. The expression of Arabidopsis CKs inducible typeA ARR4 and ARR5 were induced by dehydration, high salinity and cold [38, 158]. Furthermore, the expression of the typeA OsRR6 was found to be up-regulated by ABA, drought, salt and cold stresses [159]. In Arabidopsis, SLs alter abiotic stress response through both ABA-dependent and ABA-independent signalling pathways [140]. SLs along with ABA plays regulatory role in stomatal closure, senescence of leaves and thus stress response, whereas CKs delays the senescence. Impaired SLs signalling in Arabidopsis also led to down regulation of CKX gene which is responsible for CKs degradation [140]. Collectively these results indicated co-ordination between phytohormones that led to better adaption in plants during environmental stresses. However still a lot of investigations need to be carried out to discover and understand various crosstalk points among phytohormones and stress signalling cascades so that they could be effectively utilized for developing varieties with improved stress tolerance.
CONCLUSION
Drought is one of the major threats for agriculture globally as it can directly impact quality of crops production and yield. Global climate change has further compounded the adverse effects of drought stress. However plants have developed mechanisms to counter the effects of drought which are manifested at morphological, physiological, cellular and molecular levels. Understanding the molecular mechanism of drought stress responses is very important as it helps in manipulating plants to enhance stress tolerance and yield potential. Several transcriptomic and functional genomic approaches have revealed that plants respond, adapt and resist to drought by activating numerous genes including receptor kinases, TFs and stress responsive genes. In addition the expression of these genes may be directly or indirectly influenced by several phytohormones. Several components of abiotic stress signalling especially drought signalling cascade have been found to be regulated through phytohormones. Key plant hormones such as ABA, SA, JA and ET are well known for their regulatory response in drought stress. However recently IAA, CKs, GA, BRs and SLs have also reported to affect the drought signalling cascade and could be integrated with other hormones for better plant survival. It has also been anticipated that phytohormone synthesis and signalling play central role in response and adaptation to adverse environmental conditions.
FUTURE PERSPECTIVES
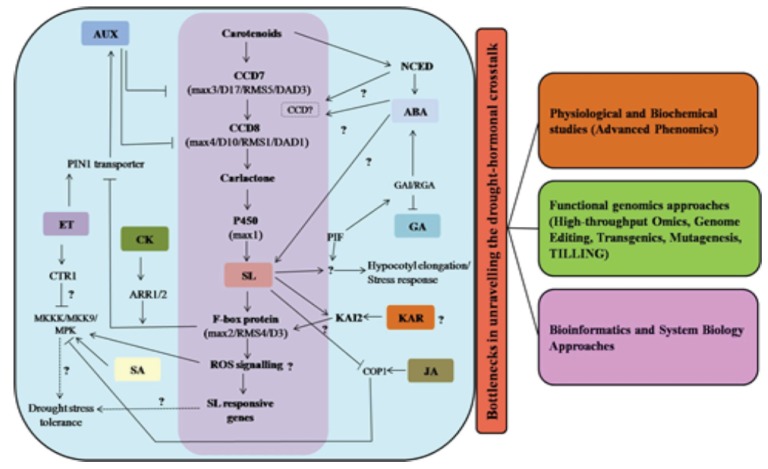
Despite several findings till date, various steps of gene regulatory networks via phytohormones and their crosstalk in drought stress signal transduction are still unknown, and more investigations need to be done for unravelling the crosstalk among these components in different crops. Recent studies suggested positive role of SLs in plant adaptation under abiotic stresses [139] wherein MAX (MAX2, MAX3, MAX4) genes involved in SLs signalling were found to play major role during drought and salt stress response. However, still more SLs responsive genes and their role in stress signalling need to be elucidated. Karrikins are the newest member in the growing list of phytohormones. These are found to be involved in promotion of seed germination, cotyledon expansion and hypocotyls elongation [160]. Recent studies showed that MAX2 gene is also involved in karrikins signalling suggesting an investigation regarding the role of karrikins in plant adaptation under abiotic stresses. Some known common and specific genes (e.g., AtD14, KAI2, D53, SMAX1, and SMXLs) help us to better understand the role of SLs and karrikins and their interaction during abiotic stresses. Other plant hormones like SA, JA and BRs signalling pathways are unclear at one or more points till date. After perception of stress, phytohormone signalling and activation of different ROS species and their downstream signalling pathways remain an enigma. Various functional genomic approaches could be helpful in showing inter-relatedness and crosstalk among phytohormones and their regulation under stress (Fig. 3). Genome editing tools provide opportunities for modification(s) in targeted genome(s) and would be helpful in understanding the complexity of regulatory networks in plants under various stresses. Further in future, the use of phytohormones may be envisaged as a management tool for agricultural crops for mitigating environmental constraints and improving production and productivity. However still a lot of work need to be done to better understand the phytohormone biosynthesis, mode of action, and their role in regulation of various abiotic stress responsive genes. The discovery of various other crosstalk points between stress-related genes and phytohormones in future can thus offer effective strategies for genetic improvement of crop plants.
Fig. (3).
A schematic representation of significant bottlenecks in unravelling the drought-hormonal crosstalk, and the strategies to address them.
Consent for Publication
Not applicable.
ACKNOWLEDGeMENTS
CL acknowledges Department of Science &Technology, Govt. of India for the INSPIRE Faculty Award [Grant No.: IFA-11LSPA-01]. The study was also partly supported by New Initiative (as a Cross Flow Technology project) “Root Biology and Its Correlation to Sustainable Plant Development and Soil Fertility” (RootSF; BSC0204) from the Council of Scientific and Industrial Research (CSIR), New Delhi, India. MP acknowledges core grant from National Institute of Plant Genome Research, New Delhi for the support and work in this area.
LIST OF ABBREVIATIONS
- ABRE/ ABF
ABA-Responsive Elements
- AP2/ERF
APETALA2/Ethylene Responsive Factor
- ARR
Arabidopsis Response Regulator
- COR
Cold-Regulated
- DHN
Dehydrin
- DHODH1
Dihydroorotate Dehydrogenase 1
- DRE/CRT
Dehydration-Responsive Element/ C-Repeat
- DREB
Dehydration Responsive Element Binding
- EIN
Ethylene-Insensitive
- ERD
Early Responsive to Dehydration
- ERS
Ethylene Response Sensor
- GID
Gibberellin Insensitive Dwarf
- KAI
Karrikin-Insensitive
- LEA
Late Embryogenesis Abundant
- MYB/MYC
Myeloblastosis/Myelocytomatosis
- NAC
NAM, ATAF and CUC 1
- NPR
Non-expressor of Pathogenesis-Related
- RD
Dehydration Responsive
- Rab
Responsive to ABA
- STZ
Salt Tolerance Zinc Finger
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Fahad S., Hussain S., Bano A., Saud S., Hassan S., Shan D., Khan F.A., Khan F., Chen Y., Wu C. Potential role of phytohormones and plant growth-promoting rhizobacteria in abiotic stresses, consequences for changing environment. Environ. Sci. Pollut. Res. Int. 2015;22:4907–4921. doi: 10.1007/s11356-014-3754-2. [DOI] [PubMed] [Google Scholar]
- 2.Tardieu F., Tuberosa R. Dissection and modelling of abiotic stress tolerance in plants. Curr. Opin. Plant Biol. 2010;13:206–212. doi: 10.1016/j.pbi.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 3.Rao N.K., Laxman R.H., Shivashankara K.S. Abiotic Stress Physiology of Horticultural Crops. India: Springer; 2016. Physiological and morphological responses of horticultural crops to abiotic stresses. pp. 3–17. [Google Scholar]
- 4.Fang Y., Xiong L. General mechanisms of drought response and their application in drought resistance improvement in plants. Cell. Mol. Life Sci. 2015;72(4):673–689. doi: 10.1007/s00018-014-1767-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin X.F., Xiong A.S., Peng R.H., Liu J.G., Gao F., Chen J.M., Yao Q.H. OsAREB1, an ABRE-binding protein responding to ABA and glucose; has multiple functions in Arabidopsis. BMB Rep. 2010;43:34–39. doi: 10.5483/bmbrep.2010.43.1.034. [DOI] [PubMed] [Google Scholar]
- 6.Hadiarto T., Tran L.S. Progress studies of drought-responsive genes in rice. Plant Cell Rep. 2011;30:297–310. doi: 10.1007/s00299-010-0956-z. [DOI] [PubMed] [Google Scholar]
- 7.Lata C., Yadav A., Prasad M. 2011. Role of plant transcription factors in abiotic stress tolerance. [Google Scholar]
- 8.Lata C., Muthamilarasan M., Prasad M. 2015. Drought stress responses and signal transduction in plants. [Google Scholar]
- 9.Tiwari S., Lata C., Chauhan P.S., Nautiyal C.S. Pseudomonas putida attunes morphophysiological; biochemical and molecular responses in Cicer arietinum L. during drought stress and recovery. Plant Physiol. Biochem. 2016;99:108–117. doi: 10.1016/j.plaphy.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Lata C., Sahu P.P., Prasad M. Comparative transcriptome analysis of differentially expressed genes in foxtail millet (Setaria italica L.) during dehydration stress. Biochem. Biophys. Res. Commun. 2010;393:720–727. doi: 10.1016/j.bbrc.2010.02.068. [DOI] [PubMed] [Google Scholar]
- 11.Lata C., Prasad M. Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 2011;62:4731–4748. doi: 10.1093/jxb/err210. [DOI] [PubMed] [Google Scholar]
- 12.Osakabe Y., Yamaguchi-Shinozaki K., Shinozaki K., Tran L.S. Sensing the environment: key roles of membrane-localized kinases in plant perception and response to abiotic stress. J. Exp. Bot. 2013;64(2):445–458. doi: 10.1093/jxb/ers354. [DOI] [PubMed] [Google Scholar]
- 13.Kuromori T., Mizoi J., Umezawa T., Yamaguchi-Shinozaki K., Shinozaki K. Molecular Biology. New York: Springer; 2014. Drought stress signaling network. pp. 383–409. [Google Scholar]
- 14.Zhang S., Zhang L., Zhao Z., Li Y., Zhou K., Su L., Zhou Q. Root transcriptome sequencing and differentially expressed drought-responsive genes in the Platycladus orientalis (L.). Tree Genet. Genomes. 2016;12(4):79. [Google Scholar]
- 15.Golldack D., Li C., Mohan H., Probst N. Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front. Plant Sci. 2014;5:151. doi: 10.3389/fpls.2014.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lata C., Bhutty S., Bahadur R.P., Majee M., Prasad M. Association of an SNP in a novel DREB2-like gene SiDREB2 with stress tolerance in foxtail millet. J. Exp. Bot. 2011;62:3387–3401. doi: 10.1093/jxb/err016. [Setaria italica (L.)]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakashima K., Jan A., Todaka D., Maruyama K., Goto S., Shinozaki K., Yamaguchi-Shinozaki K. Comparative functional analysis of six drought-responsive promoters in transgenic rice. Planta. 2014;239:47–60. doi: 10.1007/s00425-013-1960-7. [DOI] [PubMed] [Google Scholar]
- 18.Chen H., Liu L., Wang L., Wang S., Cheng X. VrDREB2A, a DREB-binding transcription factor from Vigna radiata, increased drought and high-salt tolerance in transgenic Arabidopsis thaliana. J. Plant Res. 2015;•••:1–11. doi: 10.1007/s10265-015-0773-0. [DOI] [PubMed] [Google Scholar]
- 19.Cui M., Zhang W., Zhang Q., Xu Z., Zhu Z., Duan F., Wu R. Induced over-expression of the transcription factor OsDREB2A improves drought tolerance in rice. Plant Physiol. Biochem. 2011;49:1384–1391. doi: 10.1016/j.plaphy.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 20.Pellegrineschi A., Reynolds M., Pacheco M., Brito R.M., Almeraya R., Yamaguchi-Shinozaki K., Hoisington D. Stress-induced expression in wheat of the Arabidopsis thaliana DREB1A gene delays water stress symptoms under greenhouse conditions. Genome. 2004;47:493–500. doi: 10.1139/g03-140. [DOI] [PubMed] [Google Scholar]
- 21.Morran S., Eini O., Pyvovarenko T., Parent B., Singh R., Ismagul A., Eliby S., Shirley N., Langridge P., Lopato S. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J. 2011;9(2):230–249. doi: 10.1111/j.1467-7652.2010.00547.x. [DOI] [PubMed] [Google Scholar]
- 22.Lata C., Mishra A.K., Muthamilarasan M., Bonthala V.S., Khan Y., Prasad M. Genome-wide investigation and expression profiling of AP2/ERF transcription factor superfamily in foxtail millet (Setaria italica L.). PLoS One. 2014;9:e113092. doi: 10.1371/journal.pone.0113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakshi S., Dewan D. Status of transgenic cereal crops: A review. Clon. Transgen. 2013;3:119. [Google Scholar]
- 24.Hong Y., Zhang H., Huang L., Li D., Song F. Overexpression of a stress responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front. Plant Sci. 2016;7:4. doi: 10.3389/fpls.2016.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao X., Yang X., Pei S., He G., Wang X., Tang Q., Jia C., Lu Y., Hu R., Zhou G. The Miscanthus NAC transcription factor MlNAC9 enhances abiotic stress tolerance in transgenic Arabidopsis. Gene. 2016;586:158–169. doi: 10.1016/j.gene.2016.04.028. [DOI] [PubMed] [Google Scholar]
- 26.Lata C. Advances in omics for enhancing abiotic stress tolerance in millets. Proc. Indian Natn. Sci. Acad. 2015;81:397–417. [Google Scholar]
- 27.Wang F., Kong W., Wong G., Fu L., Peng R., Li Z., Yao Q. AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana. Mol. Genet. Genomics. 2016;•••:1–15. doi: 10.1007/s00438-016-1203-2. [DOI] [PubMed] [Google Scholar]
- 28.Yu Y.T., Wu Z., Lu K., Bi C., Liang S., Wang X.F., Zhang D.P. Overexpression of the MYB37 transcription factor enhances abscisic acid sensitivity, and improves both drought tolerance and seed productivity in Arabidopsis thaliana. Plant Mol. Biol. 2016;90:267–279. doi: 10.1007/s11103-015-0411-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh D., Laxmi A. Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front. Plant Sci. 2015;6:895. doi: 10.3389/fpls.2015.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monika D., Chinnusamy V. Elucidation of abiotic stress signaling in plants. New York: Springer; 2015. ABA receptors: prospects for enhancing biotic and abiotic stress tolerance of crops. pp. 271–298. [Google Scholar]
- 31.Roy S. Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal. Behav. 2016;11(1):e1117723. doi: 10.1080/15592324.2015.1117723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fahad S., Hussain S., Matloob A., Khan F.A., Khaliq A., Saud S., Hassan S., Shan D., Khan F., Ullah N. Phytohormones and plant responses to salinity stress, a review. Plant Growth Regul. 2015;75:391–404. [Google Scholar]
- 33.Großkinsky D.K., van der Graaff E., Roitsch T. Plant Pathogen Resistance Biotechnology. New York: Wiley & Sons; 2016. Regulation of abiotic and biotic stress responses by plant hormones. pp. 131–134. [Google Scholar]
- 34.Skirycz A., Inzé D. More from less, plant growth under limited water. Curr. Opin. Biotechnol. 2010;21:197–203. doi: 10.1016/j.copbio.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 35.Li Z., Zhang J., Li J., Li H., Zhang G. The Functional and regulatory mechanisms of the Thellungiela salsuginea ascorbate peroxidase 6 (TsAPX6) in response to salinity and water deficit stresses. PLoS One. 2016;11:e0154042. doi: 10.1371/journal.pone.0154042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y., Han Y., Zhang M., Zhou S., Kong X., Wang W. Overexpression of the wheat expansin gene TaEXPA2 improved seed production and drought tolerance in transgenic tobacco plants. PLoS One. 2016;11:e0153494. doi: 10.1371/journal.pone.0153494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gunapati S., Naresh R., Ranjan S., Nigam D., Hans A., Verma P.C., Gadre R., Pathre U.V., Sane A.P., Sane V.A. Expression of GhNAC2 from G. herbaceum, improves root growth and imparts tolerance to drought in transgenic cotton and Arabidopsis. Sci. Rep. 2016;6:24978. doi: 10.1038/srep24978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vojta P., Kokáš F., Husičková A., Grúz J., Bergougnoux V., Marchetti C.F., Jiskrová E., Ježilová E., Mik V., Ikeda Y. Whole transcriptome analysis of transgenic barley with altered cytokinin homeostasis and increased tolerance to drought stress. N. Biotechnol. 2016;33:676–691. doi: 10.1016/j.nbt.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 39.Nobres P., Patreze C.M., Waltenberg F.P., Correa M.F., Tavano E.C., Mendes B.M., Alves-Ferreira M. Characterization of the Promoter of the Homeobox Gene CaHB12 in Coffea arabica. Trop. Plant Biol. 2016;9:50–62. [Google Scholar]
- 40.Dahro B., Wang F., Peng T., Liu J.H. PtrA/NINV, an alkaline/neutral invertase gene of Poncirus trifoliata, confers enhanced tolerance to multiple abiotic stresses by modulating ROS levels and maintaining photosynthetic efficiency. BMC Plant Biol. 2016;16:1. doi: 10.1186/s12870-016-0761-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo P., Wei H., Zhang W., Yang B., Bao Y. The dehydration-induced ERECTA gene, MsSIK1, from alfalfa improved water use efficiency in transgenic Arabidopsis. Acta Physiol. Plant. 2016;38:1–12. [Google Scholar]
- 42.Chen S.F., Liang K., Yin D.M., Ni D.A., Zhang Z.G., Ruan Y.L. Ectopic expression of a tobacco vacuolar invertase inhibitor in guard cells confers drought tolerance in Arabidopsis. J. Enzyme Inhib. Med. Chem. 2016;•••:1–5. doi: 10.3109/14756366.2016.1142981. [DOI] [PubMed] [Google Scholar]
- 43.Kuromori T., Fujita M., Urano K., Tanabata T., Sugimoto E., Shinozaki K. Overexpression of AtABCG25 enhances the abscisic acid signal in guard cells and improves plant water use efficiency. Plant Sci. 2016;251:75–81. doi: 10.1016/j.plantsci.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 44.Li L., Zheng M., Deng G., Liang J., Zhang H., Pan Z., Long H., Yu M. Overexpression of AtHDG11 enhanced drought tolerance in wheat (Triticum aestivum L.). Mol. Breed. 2016;36:1–10. [Google Scholar]
- 45.Han Z., Yu H., Zhao Z., Hunter D., Luo X., Duan J., Tian L. AtHD2D gene plays a role in plant growth, development, and response to abiotic stresses in Arabidopsis thaliana. Front. Plant Sci. 2016;7:310. doi: 10.3389/fpls.2016.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J., Yu G., Sun X., Zhang X., Liu J., Pan H. AcEBP1, an ErbB3-Binding Protein (EBP1) from halophyte Atriplex canescens, negatively regulates cell growth and stress responses in Arabidopsis. Plant Sci. 2016;248:64–74. doi: 10.1016/j.plantsci.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 47.Liu S., Wang C., Jia F., An Y., Liu C., Xia X., Yin W. Secretory peptide PdEPF2 enhances drought tolerance by modulating stomatal density and regulates ABA response in transgenic Arabidopsis thaliana. Plant Cell Tissue Organ Cult. 2016;125(3):419–431. [Google Scholar]
- 48.Saha B., Mishra S., Awasthi J.P., Sahoo L., Panda S.K. Enhanced drought and salinity tolerance in transgenic mustard [Brassica juncea (L.) Czern & Coss.] overexpressing Arabidopsis group 4 late embryogenesis abundant gene (AtLEA4-1). Environ. Exp. Bot. 2016;128:99–111. [Google Scholar]
- 49.Ding W., Fang W., Shi S., Zhao Y., Li X., Xiao K. 2016.
- 50.Dey A., Samanta M.K., Gayen S., Sen S.K., Maiti M.K. Enhanced gene expression rather than natural polymorphism in coding sequence of the OsbZIP23 determines drought tolerance and yield improvement in rice genotypes. PLoS One. 2016;11:e0150763. doi: 10.1371/journal.pone.0150763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jardak-Jamoussi R., Zarrouk O., Salem A.B., Zoghlami N., Mejri S., Gandoura S., Khiari B., Mliki A., Chaves M., Ghorbel A. Overexpressing Vitis vinifera YSK 2 dehydrin in tobacco improves plant performance. Agric. Water Manage. 2016;164:176–189. [Google Scholar]
- 52.Ramu V.S., Swetha T.N., Sheela S.H., Babitha C.K., Rohini S., Reddy M.K., Tuteja N., Reddy C.P., Prasad T.G., Udayakumar M. Simultaneous expression of regulatory genes associated with specific drought‐adaptive traits improves drought adaptation in peanut. Plant Biotechnol. J. 2016;14:1008–1020. doi: 10.1111/pbi.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou X., Yan S., Sun C., Li S., Li J., Xu M., Liu X., Zhang S., Zhao Q., Li Y. A maize jasmonate Zim-domain protein, ZmJAZ14, associates with the JA, ABA, and GA signaling pathways in transgenic Arabidopsis. PLoS One. 2015;10:e0121824. doi: 10.1371/journal.pone.0121824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan D.X., Tuong H.M., Thuy V.T., Son L.V., Mau C.H. Cloning and overexpression of GmDREB2 gene from a Vietnamese drought-resistant soybean variety. Braz. Arch. Biol. Technol. 2015;58:651–657. [Google Scholar]
- 55.Ma L.F., Li Y., Chen Y., Li X.B. Improved drought and salt tolerance of Arabidopsis thaliana by ectopic expression of a cotton (Gossypium hirsutum) CBF gene. Plant Cell Tissue Organ Cult. 2015;•••:1–16. [Google Scholar]
- 56.Wang Y., Yang L., Chen X., Ye T., Zhong B., Liu R., Wu Y., Chan Z. Major latex protein-like protein 43 (MLP43) functions as a positive regulator during abscisic acid responses and confers drought tolerance in Arabidopsis thaliana. J. Exp. Bot. 2015;•••:erv477. doi: 10.1093/jxb/erv477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng Z.J., He G.H., Zheng W.J., Lu P.P., Chen M., Gong Y.M., Ma Y.Z., Xu Z.S. Foxtail millet NF-Y families: genome-wide survey and evolution analyses identified two functional genes important in abiotic stresses. Front. Plant Sci. 2015;6:1142. doi: 10.3389/fpls.2015.01142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rai A.N., Tamirisa S., Rao K., Kumar V., Suprasanna P. Brassica RNA binding protein ERD4 is involved in conferring salt, drought tolerance and enhancing plant growth in Arabidopsis. Plant Mol. Biol. 2015;•••:1–13. doi: 10.1007/s11103-015-0423-x. [DOI] [PubMed] [Google Scholar]
- 59.Shi J., Habben J.E., Archibald R.L., Drummond B.J., Chamberlin M.A., Williams R.W., Lafitte H.R., Weers B.P. Overexpression of ARGOS genes modifies plant sensitivity to ethylene, leading to improved drought tolerance in both Arabidopsis and maize. Plant Physiol. 2015;169:266–282. doi: 10.1104/pp.15.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ke Q., Wang Z., Ji C.Y., Jeong J.C., Lee H.S., Li H., Xu B., Deng X., Kwak S.S. Transgenic poplar expressing Arabidopsis YUCCA6 exhibits auxin-overproduction phenotypes and increased tolerance to abiotic stress. Plant Physiol. Biochem. 2015;94:19–27. doi: 10.1016/j.plaphy.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 61.Lim C.W., Han S.W., Hwang I.S., Kim D.S., Hwang B.K., Lee S.C. The pepper lipoxygenase CaLOX1 plays a role in osmotic, drought, and high salinity. Plant Cell Physiol. 2015;•••:pcv020. doi: 10.1093/pcp/pcv020. [DOI] [PubMed] [Google Scholar]
- 62.Du H., Wu N., Cui F., You L., Li X., Xiong L. A homolog of Ethylene Overproducer, Osetol1, differentially modulates drought and submergence tolerance in rice. Plant J. 2014;78:834–849. doi: 10.1111/tpj.12508. [DOI] [PubMed] [Google Scholar]
- 63.Lü P., Kang M., Jiang X., Dai F., Gao J., Zhang C. RhEXPA4, a rose expansin gene, modulates leaf growth and confers drought and salt tolerance to Arabidopsis. Planta. 2013;237:1547–1559. doi: 10.1007/s00425-013-1867-3. [DOI] [PubMed] [Google Scholar]
- 64.Yu L., Chen X., Wang Z., Wang S., Wang Y., Zhu Q., Li S., Xiang C. Arabidopsis enhanced drought tolerance1 /Homeodomain Glabrous11 confers drought tolerance in transgenic rice without yield penalty. Plant Physiol. 2013;162:1378–1391. doi: 10.1104/pp.113.217596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han Y.Y., Li A.X., Li F., Zhao M.R., Wang W. Characterization of a wheat (Triticum aestivum L.) expansin gene, TaEXPB23, involved in the abiotic stress response and phytohormone regulation. Plant Physiol. Biochem. 2012;54:49–58. doi: 10.1016/j.plaphy.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Q., Li J., Zhang W., Yan S., Wang R., Zhao J., Li Y., Qi Z., Sun Z., Zhu Z. The putative auxin efflux carrier OsPIN3t is involved in the drought stress response and drought tolerance. Plant J. 2012;72:805–816. doi: 10.1111/j.1365-313X.2012.05121.x. [DOI] [PubMed] [Google Scholar]
- 67.Karan R., Subudhi P.K. A stress inducible SUMO conjugating enzyme gene (SaSce9) from a grass halophyte Spartina alterniflora enhances salinity and drought stress tolerance in Arabidopsis. BMC Plant Biol. 2012;12:187. doi: 10.1186/1471-2229-12-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pan Y., Seymour G.B., Lu C., Hu Z., Chen X., Chen G. An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rep. 2012;31:349–360. doi: 10.1007/s00299-011-1170-3. [DOI] [PubMed] [Google Scholar]
- 69.Seo J.S., Joo J., Kim M.J., Kim Y.K., Nahm B.H., Song S. Cheong, J.J.; Lee, J.S.; Kim, J.K.; Choi, Y.D. OsbHLH148, a basic helix‐loop‐helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011;65:907–921. doi: 10.1111/j.1365-313X.2010.04477.x. [DOI] [PubMed] [Google Scholar]
- 70.Shin D., Moon S.J., Han S., Kim B.G., Park S.R., Lee S.K., Yoon H.J., Lee H.E., Kwon H.B., Baek D. Expression of StMYB1R-1, a novel potato single MYB-like domain transcription factor, increases drought tolerance. Plant Physiol. 2011;155:421–432. doi: 10.1104/pp.110.163634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li G., Nasar V., Yang Y., Li W., Liu B., Sun L., Li D., Song F. Arabidopsis poly(ADP-ribose) glycohydrolase 1 is required for drought, osmotic and oxidative stress responses. Plant Sci. 2011;180:283–291. doi: 10.1016/j.plantsci.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 72.Kang H.G., Kim J., Kim B., Jeong H., Choi S.H., Kim E.K., Lee H.Y., Lim P.O. Overexpression of FTL1/DDF1, an AP2 transcription factor, enhances tolerance to cold, drought, and heat stresses in Arabidopsis thaliana. Plant Sci. 2011;180:634–641. doi: 10.1016/j.plantsci.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 73.Bae H., Kim S.K., Cho S.K., Kang B.G., Kim W.T. Overexpression of OsRDCP1, a rice RING domain-containing E3 ubiquitin ligase, increased tolerance to drought stress in rice (Oryza sativa L.). Plant Sci. 2011;180:775–782. doi: 10.1016/j.plantsci.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 74.Kong X., Pan J., Zhang M., Xing X., Zhou Y., Liu Y., Li D., Li D. ZmMKK4, a novel group C mitogen‐activated protein kinase kinase in maize (Zea mays), confers salt and cold tolerance in transgenic Arabidopsis. Plant Cell Environ. 2011;34:1291–1303. doi: 10.1111/j.1365-3040.2011.02329.x. [DOI] [PubMed] [Google Scholar]
- 75.Jeong J.S., Kim Y.S., Baek K.H., Jung H., Ha S.H., Do Choi Y., Kim M., Reuzeau C., Kim J.K. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010;153:185–197. doi: 10.1104/pp.110.154773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee S.J., Kang J.Y., Park H.J., Kim M.D., Bae M.S., Choi H.I., Kim S.Y. DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol. 2010;153:716–727. doi: 10.1104/pp.110.154617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leshem Y., Golani Y., Kaye Y., Levine A. Reduced expression of the v-SNAREs AtVAMP71/AtVAMP7C gene family in Arabidopsis reduces drought tolerance by suppression of abscisic acid-dependent stomatal closure. J. Exp. Bot. 2010;•••:erq099. doi: 10.1093/jxb/erq099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim J.S., Mizoi J., Yoshida T., Fujita Y., Nakajima J., Ohori T., Todaka D., Nakashima K., Hirayama T., Shinozaki K., Yamaguchi-Shinozaki K. An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol. 2011;52(12):2136–2146. doi: 10.1093/pcp/pcr143. [DOI] [PubMed] [Google Scholar]
- 79.Miao Z., Xu W., Li D., Hu X., Liu J., Zhang R., Tong Z., Dong J., Su Z., Zhang L. De novo transcriptome analysis of Medicago falcata reveals novel insights about the mechanisms underlying abiotic stress-responsive pathway. BMC Genomics. 2015;16:818. doi: 10.1186/s12864-015-2019-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Du H., Wang N., Cui F., Li X., Xiao J., Xiong L. Characterization of the β-carotene hydroxylase gene DSM2 conferring drought and oxidative stress resistance by increasing xanthophylls and abscisic acid synthesis in rice. Plant Physiol. 2010;154:1304–1318. doi: 10.1104/pp.110.163741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ryu H., Cho Y.G. Plant hormones in salt stress tolerance. J. Plant Biol. 2015;58(3):147–155. [Google Scholar]
- 82.Padmalatha K.V., Dhandapani G., Kanakachari M., Kumar S., Dass A., Patil D.P., Rajamani V., Kumar K., Pathak R., Rawat B. Genome-wide transcriptomic analysis of cotton under drought stress reveal significant down-regulation of genes and pathways involved in fibre elongation and up-regulation of defense responsive genes. Plant Mol. Biol. 2012;78:223–246. doi: 10.1007/s11103-011-9857-y. [DOI] [PubMed] [Google Scholar]
- 83.Zhang H., Mao X., Wang C., Jing R. Overexpression of a common wheat gene TaSnRK2.8 enhances tolerance to drought, salt and low temperature in Arabidopsis. PLoS One. 2010;5:e16041. doi: 10.1371/journal.pone.0016041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang Q., Wang Y., Li B., Chang J., Chen M., Li K., Yang G., He G. TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 2015;15:268. doi: 10.1186/s12870-015-0644-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.de Ollas C., Dodd I.C. Physiological impacts of ABA-JA interactions under water-limitation. Plant Mol. Biol. 2016;91(6):641–650. doi: 10.1007/s11103-016-0503-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hayat Q., Hayat S., Irfan M., Ahmad A. Effect of exogenous salicylic acid under changing environment, a review. Environ. Exp. Bot. 2010;68:14–25. [Google Scholar]
- 87.Singh P.K., Gautam S. Role of salicylic acid on physiological and biochemical mechanism of salinity stress tolerance in plants. Acta Physiol. Plant. 2013;35(8):2345–2353. [Google Scholar]
- 88.Jumali S.S., Said I.M., Ismail I., Zainal Z. Genes induced by high concentration of salicylic acid in Mitragyna speciosa. Aust. J. Crop Sci. 2011;5:296. [Google Scholar]
- 89.Miura K., Tada Y. Regulation of water, salinity, and cold stress responses by salicylic acid. Front. Plant Sci. 2014;5:4. doi: 10.3389/fpls.2014.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Divi U.K., Rahman T., Krishna P. Brassinosteroid-mediated stress tolerance in Arabidopsis shows interactions with abscisic acid; ethylene and salicylic acid pathways. BMC Plant Biol. 2010;10:1. doi: 10.1186/1471-2229-10-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.He Q., Zhao S., Ma Q., Zhang Y., Huang L., Li G., Hao L. Endogenous salicylic acid levels and signaling positively regulate Arabidopsis response to polyethylene glycol-simulated drought stress. J. Plant Growth Regul. 2014;33:871–880. [Google Scholar]
- 92.Gao Y., Gao S., Xiong C., Yu G., Chang J., Ye Z., Yang C. Comprehensive analysis and expression profile of the homeodomain leucine zipper IV transcription factor family in tomato. Plant Physiol. Biochem. 2015;96:141–153. doi: 10.1016/j.plaphy.2015.07.025. [DOI] [PubMed] [Google Scholar]
- 93.Tani T., Sobajima H., Okada K., Chujo T., Arimura S.i., Tsutsumi N., Nishimura M., Seto H., Nojiri H., Yamane H. Identification of the OsOPR7 gene encoding 12-oxophytodienoate reductase involved in the biosynthesis of jasmonic acid in rice. Planta. 2008;227:517–526. doi: 10.1007/s00425-007-0635-7. [DOI] [PubMed] [Google Scholar]
- 94.Yang F., Zhang Y., Huang Q., Yin G., Pennerman K.K., Yu J., Liu Z., Li D., Guo A. Analysis of key genes of jasmonic acid mediated signal pathway for defense against insect damages by comparative transcriptome sequencing. Sci. Rep. 2015;5:16500. doi: 10.1038/srep16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.De Domenico S., Bonsegna S., Horres R., Pastor V., Taurino M., Poltronieri P., Imtiaz M., Kahl G., Flors V., Winter P. Transcriptomic analysis of oxylipin biosynthesis genes and chemical profiling reveal an early induction of jasmonates in chickpea roots under drought stress. Plant Physiol. Biochem. 2012;61:115–122. doi: 10.1016/j.plaphy.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 96.Du H., Liu H., Xiong L. Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 2013;4:397. doi: 10.3389/fpls.2013.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee H.Y., Seo J.S., Cho J.H., Jung H., Kim J.K., Lee J.S., Rhee S., Do Choi Y. Oryza sativa COI homologues restore jasmonate signal transduction in Arabidopsis coi1-1 mutants. PLoS One. 2013;8:52802. doi: 10.1371/journal.pone.0052802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fernández-Arbaizar A., Regalado J.J., Lorenzo O. Isolation and characterization of novel mutant loci suppressing the ABA hypersensitivity of the Arabidopsis coronatine insensitive 1-16 (coi1-16) mutant during germination and seedling growth. Plant Cell Physiol. 2012;53:53–63. doi: 10.1093/pcp/pcr174. [DOI] [PubMed] [Google Scholar]
- 99.de Ollas C., Hernando B., Arbona V., Gómez‐Cadenas A. Jasmonic acid transient accumulation is needed for abscisic acid increase in citrus roots under drought stress conditions. Physiol. Plant. 2013;147:296–306. doi: 10.1111/j.1399-3054.2012.01659.x. [DOI] [PubMed] [Google Scholar]
- 100.de Ollas C., Arbona V. GóMez‐Cadenas, A. Jasmonoyl isoleucine accumulation is needed for abscisic acid build‐up in roots of Arabidopsis under water stress conditions. Plant Cell Environ. 2015;38:2157–2170. doi: 10.1111/pce.12536. [DOI] [PubMed] [Google Scholar]
- 101.Gamalero E., Glick B.R. Ethylene and abiotic stress tolerance in plants. In: Ahmad P., Prasad M.N., editors. Environmental adaptations and stress tolerance of plants in the era of climate change. New York: Springer; 2012. pp. 395–412. [Google Scholar]
- 102.Arraes F.B., Beneventi M.A., de Sa M.E., Paixao J.F., Albuquerque E.V., Marin S.R., Purgatto E., Nepomuceno A.L., Grossi-de-Sa M.F. Implications of ethylene biosynthesis and signaling in soybean drought stress tolerance. BMC Plant Biol. 2015;15:1. doi: 10.1186/s12870-015-0597-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Eppel A., Rachmilevitch S. Photosynthesis and photoprotection under drought in the annual desert plant Anastatica hierochuntica. Photosynthetica. 2016;54(1):143–147. [Google Scholar]
- 104.Cui M., Lin Y., Zu Y., Efferth T., Li D., Tang Z. Ethylene increases accumulation of compatible solutes and decreases oxidative stress to improve plant tolerance to water stress in Arabidopsis. J. Plant Biol. 2015;58:193–201. [Google Scholar]
- 105.Dubois M., Skirycz A., Claeys H., Maleux K., Dhondt S., De Bodt S., Bossche R.V., De Milde L., Yoshizumi T., Matsui M. Ethylene response factor6 acts as a central regulator of leaf growth under water-limiting conditions in Arabidopsis. Plant Physiol. 2013;162:319–332. doi: 10.1104/pp.113.216341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hopper D.W., Ghan R., Schlauch K.A., Cramer G.R. Transcriptomic network analyses of leaf dehydration responses identify highly connected ABA and ethylene signaling hubs in three grapevine species differing in drought tolerance. BMC Plant Biol. 2016;16:118. doi: 10.1186/s12870-016-0804-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu S.C., Jin J.Q., Ma J.Q., Yao M.Z., Ma C.L., Li C.F., Ding Z.T., Chen L. Transcriptomic analysis of tea plant responding to drought stress and recovery. PLoS One. 2016;11:e0147306. doi: 10.1371/journal.pone.0147306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hagen G. Auxin signal transduction. Essays Biochem. 2015;58:1–12. doi: 10.1042/bse0580001. [DOI] [PubMed] [Google Scholar]
- 109.George S., Venkataraman G., Parida A. A chloroplast-localized and auxin-induced glutathione S-transferase from phreatophyte Prosopis juliflora confer drought tolerance on tobacco. J. Plant Physiol. 2010;167:311–318. doi: 10.1016/j.jplph.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 110.Kim J.I., Baek D., Park H.C., Chun H.J., Oh D.H., Lee M.K., Cha J.Y., Kim W.Y., Kim M.C., Chung W.S. Overexpression of Arabidopsis YUCCA6 in potato results in high auxin developmental phenotypes and enhanced resistance to water deficit. Mol. Plant. 2013;6:337–349. doi: 10.1093/mp/sss100. [DOI] [PubMed] [Google Scholar]
- 111.Lee M., Jung J.H., Han D.Y., Seo P.J., Park W.J., Park C.M. Activation of a flavin monooxygenase gene YUCCA7 enhances drought resistance in Arabidopsis. Planta. 2012;235:923–938. doi: 10.1007/s00425-011-1552-3. [DOI] [PubMed] [Google Scholar]
- 112.Du H., Wu N., Fu J., Wang S., Li X., Xiao J., Xiong L.A. GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J. Exp. Bot. 2012;63:6467–6480. doi: 10.1093/jxb/ers300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yang C., Liu J., Dong X., Cai Z., Tian W., Wang X. Short-term and continuing stresses differentially interplay with multiple hormones to regulate plant survival and growth. Mol. Plant. 2014;7:841–855. doi: 10.1093/mp/ssu013. [DOI] [PubMed] [Google Scholar]
- 114.Peleg Z., Blumwald E. Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Biol. 2011;14:290–295. doi: 10.1016/j.pbi.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 115.Zhang S.W., Li C.H., Cao J., Zhang Y.C., Zhang S.Q., Xia Y.F., Sun D.Y., Sun Y. Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3. 13 activation. Plant Physiol. 2009;151:1889–1901. doi: 10.1104/pp.109.146803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Davies P.J. The plant hormones: their nature, occurrence, and functions. In: Davies P.J., editor. Plant hormones: biosynthesis, signal transduction, action! Boston: Kluwer Academic Publishers M.A; 2004. pp. 1–15. [Google Scholar]
- 117.Tran L.S., Shinozaki K., Yamaguchi-Shinozaki K. Role of cytokinin responsive two component system in ABA and osmotic stress signalings. Plant Signal. Behav. 2010;5:148–150. doi: 10.4161/psb.5.2.10411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kuppu S., Mishra N., Hu R.B., Sun L., Zhu X.L., Shen G.X., Blumwald E., Payton P., Zhang H. Water-deficit inducible expression of a cytokinin biosynthetic gene IPT improves drought tolerance in cotton. PLoS One. 2013;8(5):e64190. doi: 10.1371/journal.pone.0064190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Werner T., Nehnevajova E., Köllmer I., Novák O., Strnad M., Krämer U., Schmülling T. Root-specific reduction of cytokinin causes enhanced root growth; drought tolerance; and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell. 2010;22:3905–3920. doi: 10.1105/tpc.109.072694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Harrison M.A. Crosstalk between phytohormone signaling pathways under both optimal and stressful environmental conditions. In: Khan N.A., Nazar R., Iqbal N., Anjum N.A., editors. Phytohormones and Abiotic Stress Tolerance in Plants. Springer Berlin Heidelberg; 2012. pp. 49–76. [Google Scholar]
- 121.Khripach V., Zhabinskii V., de Groot A. Twenty years of brassinosteroids, steroidal plant hormones warrant better crops for the XXI century. Ann. Bot. 2000;86:441–447. [Google Scholar]
- 122.Sirhindi G., Kumar M., Kumar S., Bhardwaj R. 2016. [Google Scholar]
- 123.Müssig C., Fischer S., Altmann T. Brassinosteroid-regulated gene expression. Plant Physiol. 2002;129:1241–1251. doi: 10.1104/pp.011003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vardhini B.V., Rao S.S. Amelioration of osmotic stress by brassinosteroids on seed germination and seedling growth of three varieties of sorghum. Plant Growth Regul. 2003;41:25–31. [Google Scholar]
- 125.Koh S., Lee S.C., Kim M.K., Koh J.H., Lee S., An G., Choe S., Kim S.R. T-DNA tagged knockout mutation of rice OsGSK1, an orthologue of Arabidopsis BIN2, with enhanced tolerance to various abiotic stresses. Plant Mol. Biol. 2007;65:453–466. doi: 10.1007/s11103-007-9213-4. [DOI] [PubMed] [Google Scholar]
- 126.Kagale S., Divi U.K., Krochko J.E., Keller W.A., Krishna P. Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta. 2007;225:353–364. doi: 10.1007/s00425-006-0361-6. [DOI] [PubMed] [Google Scholar]
- 127.Shamloo-Dashtpagerdi R., Razi H., Ebrahimie E. Mining expressed sequence tags of rapeseed (Brassica napus L.) to predict the drought responsive regulatory network. Physiol. Mol. Biol. Plants. 2015;21:329–340. doi: 10.1007/s12298-015-0311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yamaguchi S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- 129.Iqbal N., Nazar R., Khan M.I., Masood A., Khan N.A. Role of gibberellins in regulation of source-sink relations under optimal and limiting environmental conditions. Curr. Sci. 2011;100:998–1007. [Google Scholar]
- 130.Hedden P., Thomas S.G. Annual Plant Reviews, The Gibberellins. Vol. 49 John Wiley & Sons; 2016. [Google Scholar]
- 131.Plaza-Wüthrich S., Bloesch R., Rindisbacher A., Cannarozzi G., Tadele Z. Gibberellin deficiency confers both lodging and drought tolerance in small cereals. Front. Plant Sci. 2016;7:643. doi: 10.3389/fpls.2016.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lü P., Zhang C., Liu J., Liu X., Jiang G., Jiang X., Khan M.A., Wang L., Hong B., Gao J. RhHB1 mediates the antagonism of gibberellins to ABA and ethylene during rose (Rosa hybrida) petal senescence. Plant J. 2014;78:578–590. doi: 10.1111/tpj.12494. [DOI] [PubMed] [Google Scholar]
- 133.Gong L., Zhang H., Gan X., Zhang L., Chen Y., Nie F., Shi L., Li M., Guo Z., Zhang G. Transcriptome profiling of the potato (Solanum tuberosum L.) plant under drought stress and water stimulus conditions. PLoS One. 2015;10:e0128041. doi: 10.1371/journal.pone.0128041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., Van Der Straeten D., Peng J., Harberd N.P. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311:91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- 135.Feng S., Martinez C., Gusmaroli G., Wang Y., Zhou J., Wang F., Chen L., Yu L., Iglesias-Pedraz J.M., Kircher S. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Xu K., Chen S., Li T., Ma X., Liang X., Ding X., Liu H., Luo L. OsGRAS23, a rice GRAS transcription factor gene, is involved in drought stress response through regulating expression of stress-responsive genes. BMC Plant Biol. 2015;15:141. doi: 10.1186/s12870-015-0532-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ma H.S., Liang D., Shuai P., Xia X.L., Yin W.L. The salt- and drought-inducible poplar GRAS protein SCL7 confers salt and drought tolerance in Arabidopsis thaliana. J. Exp. Bot. 2010;61:4011–4019. doi: 10.1093/jxb/erq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Qin F., Kodaira K.S., Maruyama K., Mizoi J., Tran L.S., Fujita Y., Morimoto K., Shinozaki K., Yamaguchi-Shinozaki K. SPINDLY, a negative regulator of gibberellic acid signaling, is involved in the plant abiotic stress response. Plant Physiol. 2011;157:1900–1913. doi: 10.1104/pp.111.187302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pandey A., Sharma M., Pandey G.K. Emerging roles of strigolactones in plant responses to stress and development. Front. Plant Sci. 2016;7:434. doi: 10.3389/fpls.2016.00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Van Ha C., Leyva-González M.A., Osakabe Y., Tran U.T., Nishiyama R., Watanabe Y., Tanaka M., Seki M., Yamaguchi S., Van Dong N., Yamaguchi-Shinozaki K. Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc. Natl. Acad. Sci. USA. 2014;111(2):851–856. doi: 10.1073/pnas.1322135111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kim Y., Hwang I., Jung H.J., Park J.I., Kang J.G., Nou I.S. Genome-wide classification and abiotic stress-responsive expression profiling of carotenoid oxygenase genes in Brassica rapa and Brassica oleracea. J. Plant Growth Regul. 2016;35(1):202–214. [Google Scholar]
- 142.Liu J., He H., Vitali M., Visentin I., Charnikhova T., Haider I., Schubert A., Ruyter-spira C., Cardinale F. Osmotic stress represses strigolactone biosynthesis in Lotus japonicus roots: exploring the interaction between strigolactones and ABA under abiotic stress. Planta. 2015;241:1435–1451. doi: 10.1007/s00425-015-2266-8. [DOI] [PubMed] [Google Scholar]
- 143.Kohli A., Sreenivasulu N., Lakshmanan P., Kumar P.P. The phytohormone crosstalk paradigm takes center stage in understanding how plants respond to abiotic stress. Plant Cell Rep. 2013;32:945–957. doi: 10.1007/s00299-013-1461-y. [DOI] [PubMed] [Google Scholar]
- 144.Trivedi D.K., Gill S.S., Tuteja N. 2016. [Google Scholar]
- 145.Savchenko T., Kolla V.A., Wang C.Q., Nasafi Z., Hicks D.R., Phadungchob B., Chehab W.E., Brandizzi F., Froehlich J., Dehesh K. Functional convergence of oxylipin and abscisic acid pathways controls stomatal closure in response to drought. Plant Physiol. 2014;164:1151–1160. doi: 10.1104/pp.113.234310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Miura K., Okamoto H., Okuma E., Shiba H., Kamada H., Hasegawa P.M., Murata Y. SIZ1 deficiency causes reduced stomatal aperture andenhanced drought tolerance via controlling salicylic acid-induced accumulation of reactive oxygen species in Arabidopsis. Plant J. 2013;73:91–104. doi: 10.1111/tpj.12014. [DOI] [PubMed] [Google Scholar]
- 147.Shyu C., Figueroa P., Depew C.L., Cooke T.F., Sheard L.B., Moreno J.E., Katsir L., Zheng N., Browse J., Howe G.A. JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell. 2012;24:536–550. doi: 10.1105/tpc.111.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wild M., Davière J.M., Cheminant S., Regnault T., Baumberger N., Heintz D., Baltz R., Genschik P., Achard P. The Arabidopsis Della Rga-Like3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell. 2012;24:3307–3319. doi: 10.1105/tpc.112.101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Colebrook E.H., Thomas S.G., Phillips A.L., Hedden P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014;217:67–75. doi: 10.1242/jeb.089938. [DOI] [PubMed] [Google Scholar]
- 150.Cheng M.C., Liao P.M., Kuo W.W., Lin T.P. The Arabidopsis Ethylene response factor1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 2013;162:1566–1582. doi: 10.1104/pp.113.221911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Niu Y., Figueroa P., Browse J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 2011;62:2143–2154. doi: 10.1093/jxb/erq408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Alonso-Ramírez A., Rodríguez D., Reyes D., Jiménez J.A., Nicolás G., López-Climent M., Gómez-Cadenas A., Nicolás C. Evidence for a role of gibberellins in salicylic acid-modulated early plant responses to abiotic stress in Arabidopsis seeds. Plant Physiol. 2009;150:1335–1344. doi: 10.1104/pp.109.139352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhang S., Cai Z., Wang X. The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc. Natl. Acad. Sci. USA. 2009;106:4543–4548. doi: 10.1073/pnas.0900349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tanaka Y., Sano T., Tamaoki M., Nakajima N., Kondo N., Hasezawa S. Cytokinin and auxin inhibit abscisic acid-induced stomatal closure by enhancing ethylene production in Arabidopsis. J. Exp. Bot. 2006;57:2259–2266. doi: 10.1093/jxb/erj193. [DOI] [PubMed] [Google Scholar]
- 155.Jones B., Gunnerås S.A., Petersson S.V., Tarkowski P., Graham N., May S., Dolezal K., Sandberg G., Ljung K. Cytokinin regulation of auxin synthesis in Arabidopsis involves a homeostatic feedback loop regulated via auxin and cytokinin signal transduction. Plant Cell. 2010;22:2956–2969. doi: 10.1105/tpc.110.074856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Desikan R., Horák J., Chaban C., Mira-Rodado V., Witthöft J., Elgass K., Grefen C., Cheung M.K., Meixner A.J., Hooley R. The histidine kinase AHK5 integrates endogenous and environmental signals in Arabidopsis guard cells. PLoS One. 2008;3:2491. doi: 10.1371/journal.pone.0002491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kang N.Y., Cho C., Kim N.Y., Kim J. Cytokinin receptor-dependent and receptor-independent pathways in the dehydration response of Arabidopsis thaliana. J. Plant Physiol. 2012;169:1382–1391. doi: 10.1016/j.jplph.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 158.Nguyen K.H., Van Ha C., Nishiyama R., Watanabe Y., Leyva-González M.A., Fujita Y., Tran U.T., Li W., Tanaka M., Seki M., Schaller G.E. Arabidopsis type B cytokinin response regulators ARR1, ARR10, and ARR12 negatively regulate plant responses to drought. Proc. Natl. Acad. Sci. USA. 2016;113:3090–3095. doi: 10.1073/pnas.1600399113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jain M., Tyagi A.K., Khurana J.P. Molecular characterization and differential expression of cytokinin-responsive type-A response regulators in rice (Oryza sativa). BMC Plant Biol. 2006;6:1. doi: 10.1186/1471-2229-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Smith S.M., Li J. Signalling and responses to strigolactones and karrikins. Curr. Opin. Plant Biol. 2014;21:23–29. doi: 10.1016/j.pbi.2014.06.003. [DOI] [PubMed] [Google Scholar]