Abstract
Soil salinity is an important stress factor that limits plant growth and productivity. For a given plant species, it is critical to sense and respond to salt stimuli followed by activation of multitude of mechanisms for plants to survive. Halophytes, the wonders of saline soils, have demonstrated ability to withstand and reproduce in at least 200 mM NaCl concentration, which makes them an ideal system to study mechanism of salt adaptation for imparting salt tolerance in glycophytes. Halophytes and salt sensitive glycophytes adapt different defense strategies towards salinity stress. These responses in halophytes are modulated by a well orchestrated network of signaling pathways, including calcium signaling, reactive oxygen species and phytohormones. Moreover, constitutive expression of salt stress response related genes, which is only salt inducible in glycophytes, maintains salt tolerance traits in halophytes. The focus of this review is on the adaptive considerations of halophytes through the genomics approaches from the point of view of sensing and signaling components involved in mediating plant responses to salinity.
Keywords: Halophytes, Salinity, Redox homeostasis, Genomics, miRNAs, Hormonal regulation
1. INTRODUCTION
Soil salinity imposes restrictions on growth and yield of crop plants. Salinity affects nearly 20% of agricultural lands [1]. Plant species are diverse in their ability to tolerate saline soils. While some plants can tolerate high salinity, other plants are sensitive to salinity. Halophytes constituting the native flora of the saline soil and comprising ~1% of total world’s flora [2] are endowed with ability to attain maximum growth and complete their life cycle in 200 mM or more NaCl concentration [3, 4]. Halophytes employ some specific set of traits/strategies over glycophytes which helps them to survive under adverse environmental conditions (Table 1). In general, Effects of salinity on plants include osmotic stress, ion toxicity, nutritional imbalance, oxidative stress, metabolic perturbance, membrane disorganization, genotoxicity, reduction of cell division and expansion [5]. Most of the glycophytes exclude salt ions or prevent uptake of salt ions. In contrast, halophytic plants follow three basic strategies, such as salt exclusion (plant selectively excludes toxic salt ions ex. Rhizopora sp.), salt accumulation (excess salt ions sequestered into vacuoles ex. Sesuvium portulacastrum) and salt excretion (salt is taken up by the roots and excluded from leaves with the help of salt hairs, salt glands or salt bladders ex. Avicennia spp.) [6, 7]. Severe salt stress imposes osmotic stress so that plants unable to take up water and hence loose turgor. Under ionic stress, toxic salt ions are taken up by roots and transported towards aerial parts. These salt ions are accumulated in leaf cells, mostly in mature leaves which prevents ion toxicity to young and expanding leaves. The salt induced effects and plants responses are depicted in Fig. (1).
Table 1. Strategies employed by halophytes and glycophytes [87].
| Strategy | Glycophyte | Halophyte | References |
|---|---|---|---|
| ROS homeostasis | After 72 hrs treatment, no reduction in ROS load in Arabidopsis | Reduced ROS load as early 4 hr of salt treatment in Cakile maritima | [81] |
| Antioxidant enzymes activity | Stress induced | Constitutive expression | [82] |
| Accumulation of reduced ascorbate and glutathione | Low accumulation in Solanum lycopersicum | High accumulation of Lycopersicon pennellii | [83] |
| Osmolytes accumulation | Low basal level and stress inducible in Arabidopsis thaliana | High basal level and high accumulation under stress in T. salsuginea, L. crassifolium, C. maritime | [22, 81, 84, 85] |
| Osmotic adjustment using Sodium ions | ----- | Tecticornia contains 2M intracellular Na+ for osmotic adjustment | [86] |
| Post translational modifications | Not efficient | Highly efficient | [8] |
| Changes in gene sequence | Less complex | Complex with presence of extra transposons and intergenic sequences | [88] |
| Gene duplication | Stress induced expression of NHX8 homologs in Arabidopsis | Constitutive expression of NHX8 homologs in Thellungiella enhances stress tolerance | [88] |
| CBL10 orthologs single copy in Arabidopsis | Three copies of CBL10 orthologs in T. parvula | [56] | |
| Promoter Activities | Low expression of SOS1 and VATD promoters in Arabidopsis | Fivefold and two fold high expression of SOS1 and VATD promoters in T. salsuginea | [89] |
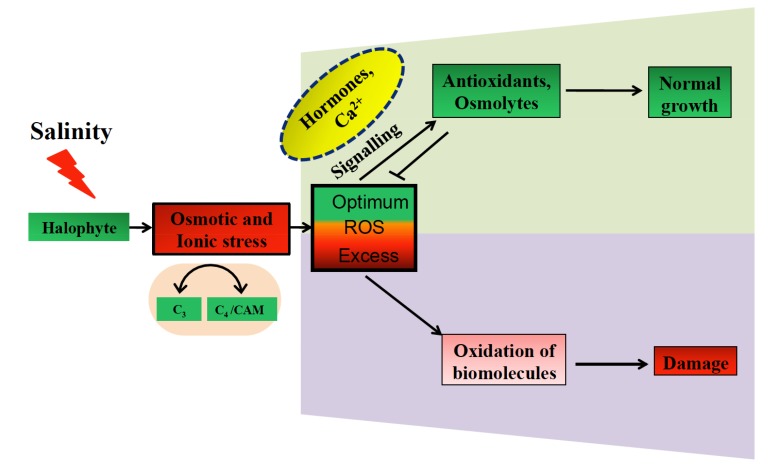
Fig. (1).
General overview of salt induced effects and plats response: Plants face osmotic and ionic stress under salt stress which enhances ROS production and subsequent oxidation of bio molecules, causing irreversible damage to plants. ROS also acts as stress indicator and its enhanced concentration activates downstream processes, like increased antioxidant activity, osmolytes which scavenges ROS and plants show normal growth. In ROS signaling, hormones and calcium play an important role. Some of the halophytes switch their carbon assimilation mode from C3 to C4 or CAM and vice versa.
The maintenance of high K+/Na+ ratio in the cytoplasm and sequestration of Na+ and Cl- in the vacuole is central to tolerance ability in halophytes. In this regard, it is important to consider the role of ion transporters such as vacuolar Na+/H+ exchangers (NHX) or potassium transporters. It is also vital, hence, to prevent potassium loss under saline conditions. Halophytes require some amount of sodium to attain maximum growth, because sodium is one of the cheap sources of osmolytes which help in maintaining osmoticum [8]. This is a low cost strategy of halophytes because it consumes very less energy (only 3.5 ATP) as compared to proline and glycine betaine which costs almost 41 and 50 ATP respectively [9]. At optimum salt concentration, halophytes rely on sodium for osmotic balance and allocate more energy and carbon resources towards normal functioning and growth of halophytes [10, 11]. Zhu [12] opined that halophytes and glycophytes possess almost similar set of strategies for salt tolerance but halophytes respond better to salinity in terms of ability to efficiently utilize energy and carbon resources up to certain level of stress [13]. The current progress on redox controls [14] indicates that, the plasma membrane is central to perception and transduction of environmental change through redox signals; apoplastic redox changes modulate interactions between receptor proteins containing oxidizable thiols in the membrane surface; presence of a steep redox gradient across the plasma membrane, and calcium release and aquaporin or peroxiporin function are triggered through the membrane channel activity [14].
Plant responses to environmental extremities are coordinated via various signaling and stress-response networks, and cross-talk among these pathways. More importantly, the perception of changes in the environment and induction of an adaptive response is critical to halophytes and salt tolerant glycophytes. Thus adaptive responses in halophytes are modulated by well coordinated signaling pathways, including calcium signaling, reactive oxygen species and phytohormones. Genome and transcriptome analyses in halophytes by using advanced genomics technologies like Next Generation Sequencing (NGS) have unraveled several pathways and networks associated with the salt stress responses of extremophiles [15, 16]. Understanding the mechanism of salt tolerance at molecular, physiological or whole plant level and screening of crops which can perform on saline soils is absolutely necessary to realize the promise of crop cultivation in saline soils [17]. This has necessitated studies on the naturally tolerant plants (halophytes) or salt tolerant crop plants [18, 19]. The focus of this review is on the adaptive considerations of halophytes through the genomics approaches from the point of view of sensing and signaling components involved in mediating plant responses to salinity (Fig. 2).
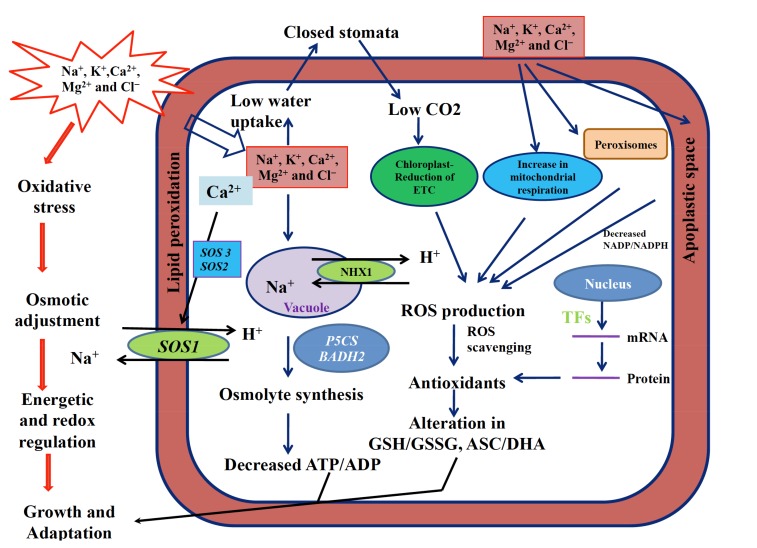
Fig. (2).
Mechanism of salinity tolerance in halophytes: The excess entry of toxic ions disturbs osmotic balance of plants by causing excess ROS production and oxidative damage. Increased concentration of ions in soil imposes water stress which leads to stomatal closure and low CO2 availability to photosynthetic machinery which reduces ETC. This decreases NADP/NADPH ratio and generates ROS in Chloroplast, mitochondria, peroxisomes and apoplastic space. In defense, halophytes increase enzymatic (SOD, CAT, APX, GR, etc.) and non enzymatic (Ascorbate, Glutathione) antioxidants and osmolytes. Also, enhanced expression of salt responsive genes and transcription factors (TFs), is observed. The excessive toxic ions are sequestered in vacuoles. This is an energy dependent process and utilizes energy in the form of ATP which reduces the ATP/ADP ratio. Effective balance between oxidative stress, antioxidant generation and cellular energetics makes halophytes more salt tolerant.
2. GENOMICS LANDSCAPE
Salinity tolerance is a quantitative trait and several genetic circuits are responsible for achieving salt tolerance. Significant progress has been made in the identification of genes and their products, which play an important role in the plant system for overcoming the unfavorable situations of abiotic stresses [20]. A number of studies for the identification and characterization of genes have also been done in halophytes in response to salinity [16]. Comparative genomics has been used to unravel biological significance of genomic regions in halophytes and glycophytes [21] and the information has added to our understanding of Na+ transport and mechanism of salt tolerance. It is now well established that most of the salt tolerance related genes expresses constitutively in halophytes and are stress inducible in glycophytes [22]. Comparative study of DNA sequences with orthologous regions from Arabidopsis and BAC sequences from Thellungiella salsuginea [23] revealed extensive sequence conservation and microcolinearity. The results also showed that T. parvula segments are distinguished from their T. salsuginea counterparts by a pronounced paucity of repeat sequences, resulting in a 30% shorter DNA segment with essentially the same gene content in T. parvula. In T. halophila, superoxide dismutase gene (SOD) is present in much higher levels under non stressed conditions and expressed more under salt stressed condition [22]. This gene encodes superoxide dismutase enzyme which is responsible for dismutation of O2•- into H2O2. SODs are classified according to their metal co factor namely, Fe-SOD, Cu/Zn SOD and Mn SOD. Fe-SOD is generally present in higher amount under non stressed condition [24].
The SOS complex in T. salsuginea is a general feature and the importance of SOS1 in stress tolerance was validated using RNA interference to generate lines with reduced SOS1 transcript and protein abundances [25]. An account of differential gene expression of some of the genes which are stress inducible in glycophytes while constitutively expressed in halophytes in context of oxidative stress has been shown to be the prime mechanism. SOS and NHX genes are Na+/H+ antiporters which maintain low cytoplasmic ion content to minimize salt toxicity. Both the antiporters are membrane bound, NHX present in tonoplast to pump toxic ions into vacuole and SOS is located at plasma membrane to exclude ions into apoplast [26]. Three genes are involved in SOS pathway; SOS3 (Calcium sensor), SOS2 (serine/threonine kinase) and SOS1 (Na+/H+ antiporter). Under stress condition, the increase in Ca2+ content results in the activation of CBL interacting protein kinases. These proteins play a key role in signal transduction to control sodium influx and efflux [27]. Qiu et al. [28] have shown increased activity of Na+/H+ antiporter under salt stress. The SOS4 gene encodes a pyridoxal kinase which is involved in the biosynthesis of pyridoxal-5-phosphate. It is an active form of vitamin B6. This pyridoxal kinase is important for regulation of Na+ and K+ homeostasis in plants [29]. The SOS5 is a putative cell surface adhesion protein which is essential for normal cell expansion. Under salt stress, it helps in the maintenance of cell wall integrity and architecture [30].
3. OSMOTIC ADJUSTMENT
Salinity stress often results in osmotic disturbance which affects cellular, physiological and molecular components. For osmotic adjustment, halophytes synthesize metabolites, such as proline, glycine betaine, pinitol, myo-inositol, mannitol, sorbitol, O-methylmucoinositol, and polyamines [31]. Proline is a scavenger of singlet oxygen, hydrogen peroxide and hydroxyl radicals and protects PSI and PSII. In PSI, it maintains low NADPH to NADP+ ratio, stabilizes mitochondrial respiration and decreases toxic effects of singlet oxygen and hydroxyl ions and prevents programmed cell death during stress. The beneficial effects of proline have been observed in many halophytes like Mesembryanthemum crystallinum, Sesuvium portulacastrum, Thellungiella salsuginea and Lepidium crassifolium [7, 23, 32]. The Δ1-pyrroline-5-carboxylate synthetase (P5CS) and Proline dehydrogenase (PDH) genes are involved in synthesis and degradation of proline. Glycine betaine is a compatible solute that helps plants in osmotic adjustment. During abiotic stress, it prevents dissociation of regulatory extrinsic proteins and there by stabilizes structure and function of PSII (an oxygen evolving complex). Halophytes are hyper accumulators of GB (>90 μmol dry weight) [3]. The Betaine aldehyde dehydrogenase (BADH) and Choline Monooxigenase (CMO) genes are responsible for synthesis of glycine betaine. The transcripts of LcBDH1, LcBDH2 and CMO like were highly up regulated under salt stress in a halophyte Leymus chinensis as compared to that of glycophyte H. vulgare. In Savia fruticosa, S. portulacastrum and many other halophytes it shows constitutive expression [7, 33]. In addition to proline and glycine betaine, it is reported that polyols like sorbitol, mannitol, myoinositol, ononitol and pinitol accumulate under environmental stress. These play significant roles in osmotic adjustment, scavenging of most toxic ROS (•OH-) and also in signaling and protection of cellular structures by interaction with membranes, proteins and enzymes [34]. In M. crystallinum, pinnitol showed two fold scavenging of stable free radical 1,1-diphenyl-2-picrylhydrazl than Lactuca sativa [35]. Myoinositol-1-phosphate synthase gene encodes the L-myo-inositol 1-phosphate synthase (MIPS; EC 5.5.1.4) enzyme. This enzyme catalyzes conversion of glucose -6 phosphate to inositol involving NAD+ and dephosphorylation. It facilitates uptake and long distance transport of sodium in Mesembryanthemum [36]. Myoinositol is most abundant isomer of inositol which plays an important role in protection from oxidative stress and in signaling [37].
4. SIGNALING PATHWAYS
Signaling cascades that regulate different downstream pathways are crucial to plant stress tolerance. ROS plays a contrasting role in living organisms depending on the ability of organisms to tightly regulate their concentration in cell. Besides their damaging effects, ROS acts as signaling molecules which can sense, percept and transduce the stress signal to activate or trigger downstream gene expression and antioxidant machinery (Fig. 3). ROS signaling mechanism deals with Ca2+ and associated proteins (such as calcineurin, calmodulin, GTP binding proteins), phospholipid signaling, MAPKs activation and abscisic acid [38]. Under normal conditions, free calcium levels are extremely low in different organelles and cytoplasm [39] which upon stress sensing, rapidly rise by factor of 10 to 20 within seconds and this elevation is decoded by calcium sensor proteins like CaM, CMLs, CDPKs, CBL/CIPKs which result in stress specific physiological response [40]. Calcineurin B-Like-Interacting Protein Kinase (CIPK) is a kind of plant-specific regulatory protein which interacts with calcineurin B-like (CBL) to form complex and as a signal transducer CIPK interacts with downstream protein, such as SOS1, AKT1 or RBOHF to phosphorylate them to regulate their functions [41]. A novel HbCIPK2-interacting ferredoxin (HbCIPK2) was identified in a halophyte H. brevisubulatum by cDNA-AFLP technique and recently it has been shown that HbCIPK2 could mediate the activities of interacting partners as a signal transducer [42]. Li et al. [43] suggested that HbCIPK2-mediated K+ homeostasis may be different from the SOS pathway, and could be a new mechanism in conferring salt tolerance.
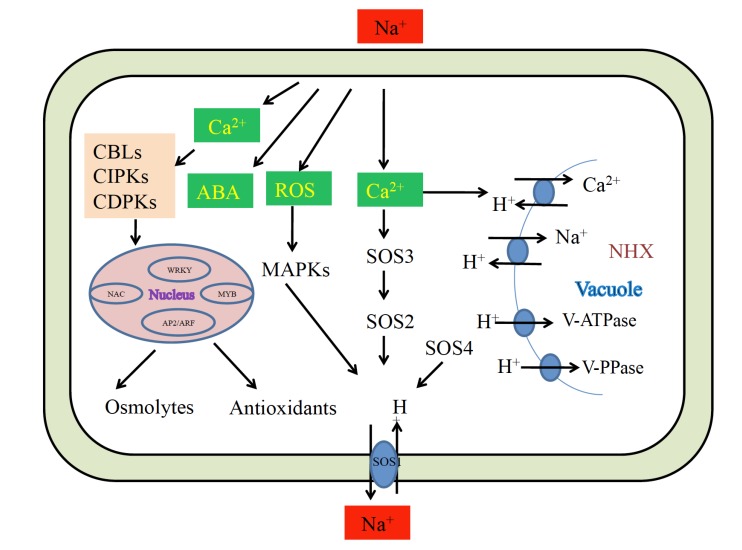
Fig. (3).
Schematic representation of common genetic pathways involved in salt tolerance mechanism of plants. The membrane bound sensors senses increasing salt concentration and activates the signaling molecules like Ca2+, ABA, and ROS. These signaling molecules activate downstream salt responsive genes like CBLs, CIPKs, CDPKs, different transcription factors, membrane bound ATPases, synthesis of osmolytes, antioxidants, activation of SOS and MAPK pathways and finally sequestration of sodium either in to the vacuole or excluded out of the cell.
During salt stress, two salt responsive pathways play important roles in ameliorating salt toxicity in halophytes, namely Salt Overly Sensitive (SOS) Pathway and Mitogen activated Protein Kinase Pathway (MAPK). SOS pathway consists of SOS3, SOS2, SOS1 and SOS4 genes which play a key role in mediating cellular signaling to maintain ion homeostasis under salt stress [44]. This pathway activates with Ca2+ spiking in cytoplasm due to accumulation of excessive Na+ ions in surrounding cells. SOS3 perceives this calcium signal and binds with SOS2. This complex then activates SOS1 which cause extrusion of Na+ from cytoplasm. The SOS1 gene is a typical halophytic trait in T. halophila [24]. The SOS1 gene has been shown to be involved in ion efflux to maintain low Na+ content in root cells [45]. Oh et al. [24] repressed expression of SOS1 gene up to 50% by using RNA interference technique, which resulted in salt toxicity and loss of halophytism in T. halophila. Yadav et al. [46] demonstrated the role of SOS1 gene from S. brachiata in salt tolerance. This gene while encoding a plasma membrane Na+/H+ antiporter also exhibited loading of Na+ to xylem from root and leaf tissues in over expressing tobacco transgenic lines which also showed increased K+ and Ca2+ content in root tissue.
Mitogen activated protein kinase pathway is evolutionarily conserved among eukaryotes and is responsible for alteration in several cellular responses under stress conditions. Three MAPK kinases: MAPKKK, MAPKK and MAPK participate in this pathway. Under salt stress, plasma membrane is stimulated and activates MAPKKK cascade via phosphorylation. MAPKKK are serine threonine kinases which activate MAPKK by phosphorylating its two amino acids S/T-X or -S/T motifs. MAPKK again phosphorylates MAPK in threonine and tyrosine at conserved motif. MAPK kinases have ability to phosphorylate many substrates like other kinases and transcription factors (Fig. 3). SbMAPK gene was isolated from a halophyte S. brachiata, which showed high sequence similarity with MAPK genes from N. benthamiana and L. esculantum. Their role in dehydration, cold and salt tolerance has already been validated [47].
5. TRANSCRIPTOMICS APPLICATIONS
Genome and transcriptome analyses in halophytes by using advanced genomics technologies like Next Generation Sequencing (NGS) have unraveled several pathways and networks associated with the salt stress responses of extremophiles [15]. Initial microarray studies of T. halophila using Arabidopsis cDNA microarray have shown that there is a high level of constitutive gene expression of six candidate genes [22, 48, 49]. In A. marina, using a leaf library of 1,841 ESTs were cloned. Mehta et al. [50] found that 26 genes were up regulated under salt stress and of these, dehydrins were predominant. Similarly, Baisakh et al. [51] found stress responsive 27% ESTs among the 1255 ESTs under salt stress from both leaf and root tissues of S. alterniflora. Novel salt-induced genes were also identified in Bruguiera gymnorhiza using suppression subtractive hybridization [52]. The authors observed up-regulation of genes related to cell communication, signal transduction, lipid metabolic process, photosynthesis, multicellular organismal development, transport and down-regulation of genes for catabolic process. The cDNA subtractive hybridization method was adopted to isolate salt responsive genes in S. brachiata and the results showed that 4.8% ESTs belonged to stress-tolerant gene category [53]. One of the transcription factors, Myb 44 isolated from Salicornia showed binding with different cis-elements of abiotic stress-related promoters and was expressed under abiotic stresses [54]. Although, initially these studies offered good clues about the genes involved in salt adaptation, global transcriptome information remained a challenge which became a reality with the development of NGS based genomics tools.
As mentioned in the preceding section, comparative genomics studies have shed light on differential gene expression profiles in halophytes [55]. It is surmised that halophytes while possessing the same genetic machinery for ion transport and other mechanisms of salt adaptation, may have better and highly expressive gene expression modules. Comparison of gene expression profiles of salt tolerance genes in the highly tolerant salt accumulating halophyte S. dolichostachya and the taxonomically related glycophytic Spinacia oleracea, found that SOS1 was highly expressed constitutively, with no detectable HKT1; 1 expression, suggesting that the constitutive high level of shoot salt accumulation in S. dolichostachya is accomplished through enhancement of SOS1-mediated Na(+) xylem loading, in combination with complete suppression of HKT1; 1-mediated Na(+) retrieval from the xylem. Using a genome-wide comparison of A. thaliana with T. parvula, Dassanayake et al. [56] suggested that halophytic behavior had a possible basis of the tandem duplications of genes. In another halophyte, P. coarctata, transcriptome analysis based on NGS led to revelation that functionally for its adaptation under high salinity and submergence conditions, the species has genes involved in diverse cellular processes including amino acid biosynthesis, hormone biosynthesis, secondary metabolite biosynthesis, carbohydrate metabolism and cell wall structures [57]. In a dicot halophyte, R. trigyna, genes for ion transport and reactive oxygen species scavenging system were shown to be highly expressed under salinity [58]. Extensive sequencing and gene-annotation analysis of S. europaea L. shoots under NaCl Treatment provided insights that in this halophyte, genes involved in ion homeostasis and osmotic adjustment, including cation transporters and proteins for the synthesis of low-molecular weight compounds play important role in halophilic mechanisms [59].
6. ROLE OF MicroRNA
MicroRNA plays a dual role in growth and development as well as plant stress tolerance. They regulate gene expression at both transcriptional as well as post-transcriptional levels [60]. In halophytic plant species, there have been few studies on miRNA expression based on Deep sRNA sequencing and high-throughput sRNA sequencing data in A. marina [61], S. europaea [62], S. brachiata [63], S. maritima [64], H. caspica [65], O. coarctata [66] and T. halophila [67]. These few halophytes and several glycophytes have elucidated the role of conserved miRNAs and novel miRNAs under salt stress and their regulatory roles in the modulation of several signaling and metabolic pathways. The role of miRNAs in signaling under salt stress and their putative targets are given in Table 2 Feng et al. [62] performed high throughput sequencing to identify salt responsive miRNAs in S. europaea. They reported 43 conserved and 13 novel differentially expressed miRNA under salt stress. Out of all the differentially expressed miRNAs Seu-miR160 and Seu-miR5targeted two and one ARF genes, respectively and Seu-miR164 targeted NAC TFs. These miRNAs were down regulated under salt treatment and therefore releases miRNA mediated repression of ARF genes and triggers auxin signaling to activate downstream components of various salt tolerance related pathways [62].
Table 2. Role of miRNAs, their targets in stress response and signaling under salt stress in halophytes.
| Halophyte | miRNA | Target Gene | Role | References |
|---|---|---|---|---|
| Avicennia marina | Am-miR159 and Am-mir319 | MYBs | Potassium starvation signaling | [61] |
| Am-miR160 and Am-miR167 | auxin response factor | Sulphur metabolism | ||
| Am-miR164 | NAC domain protein |
Abiotic stress and hormone signaling | ||
| Am-miR165 and Am-miR166 | HD-ZIPs transcription factor | Lipid biosynthesis and transport | ||
| Am-miR169 | Nuclear transcription factor Y |
Stress responsive | ||
| Am-miR395 | ATP sulfurylase 3 | Auxin signaling | ||
| Am-miR398 | Copper/Zinc Superoxide Dismutase 1 |
Antixidant | ||
|
Halostachys caspica |
Hca-miR2619b-5p | PAK, ANT | Calcium signaling | [65] |
| Hca-miR5077 | Phospho lipase C | Calcium signaling | ||
| Hca-miR167d-5p | Auxin Responsive factor F | Hormonal signaling | ||
| Hca-miR393b-5p | TIR1 | Hormonal signaling | ||
| Hca-miR902c-3p | MAPK | MAPK signaling | ||
| Hca-miR159a | Heat shock proteins 72 | MAPK signaling | ||
| Hca-miR2867-3p | PP5 | MAPK signaling | ||
| Hca-miR393b-5p | TIR1 and AFB2 receptor TIR 1 and AFB 2 receptors |
Auxin signaling and redox regulation | ||
| Oryza corctata | Oca-miR1432-5p | Calmodulin binding protein | Calcium signaling | [66] |
| Oco-miR164d | Calmodulin (CAM) | Calcium signaling | ||
| Oco-miR528-3p | Serine/threinone kinase | MAPK signaling | ||
| Oco-miR159a | Heat shock proteins | MAPK signaling | ||
| Oco-miR079-3p | Cation transporter | Ion homeostasis | ||
| Oco-miR087-5p | Peroxidase (POX) | Antioxidant | ||
| Saeuda maritima | Sma-miR2 | 4- coumarate-CoA ligase | Environmental interaction | [64] |
| Sma-miR7 | Auxin responsive factor | Auxin signaling | ||
|
Salicornia brachiata |
Sb-miRNA10 | NF-YA transcription factor | Stress responsive | [63] |
| Sb-miRNA6 and Sb-miRNA7 | Cytochrome P450-like TATA box binding protein | ROS signaling | ||
| Sb-miRNA9 | serine/threonine protein phosphatase | ABA signal transduction | ||
|
Salicornia europaea |
Sea-miR1 | NBS-LRR | Stress response, signaling | [62] |
| Sea-miR8 | Heat shock proteins | Stress response, signaling | ||
| Sea-miR2 | Kinases | Stress response, signaling | ||
| Sea-miR160/miR5 | Auxin responsive factor | Auxin signaling | ||
| Sea-miR164 | NAC | Auxin signaling |
Another species of Salicornia also studied for identification of salt responsive miRNAs in S. brachiata [63]. They found out Sb-miR10 targeted NF-YA transcription factor which is a salt stress responsive and SbmiRNA7 targeted cytochrome P450-like TATA box binding protein which is involved in ROS signaling. This miRNA-Target relationship indicates effective stress perception and signaling playing an important role in salt adaptation mechanism of this halophyte [63]. In A. marina miRNAs and their putative targets were identified using high throughput sequencing but the library preparation was done from different plant parts and not under different salt treatments [61] and in T. halophila the miRNAs and their respective targets were identified using computational tools [67]. But these miRNAs were not validated under different salt treatments. In H. caspica, five miRNAs HcmiR160a, HcmiR167d-5p, HcmiR393b-5p, HcmiR529a, HcmiR169b were found to be induced by salt stress and negatively regulated their respective targets such as ARF18, ARF8, TIR1 (transport inhibition factor1), SPL (Squmosa promoter binding protein like) and NF-YA (Nuclear factor Y subunit A), respectively. Many of the miRNAs were involved in stress induced pathways, such as auxin signal pathway, MAPK signaling pathway, plant hormone signal transduction, flavonoid biosynthesis, ubiquitin-mediated proteolysis, apoptosis, ABC transporter [65]. In S. maritima Gharat and Shaw [64] identified two miRNAs, sma-miR2 and sma-miR5 which showed salt specific expression. These miRNAs showed expression only in salt treatment which might be due to their metabolic regulatory role in saline environments. Their behavior may be mediated by altered expression of some genes, protein modification and production of secondary metabolites which is revealed by miRNA target prediction [64]. In O. coarctata [66] seven miRNAs namely oco-miR166e-3p, oco-miR169g, oco-miR169o, oco-miR393a, oco-mi396c, oco-miR020-3p and oco-miR014-3p showed negative correlation with their respective targets and possible salt responsive miRNAs of this species [66]. The miRNAs oco-miR393a, oco-miR396c, oco-miR014-3p targets oxidoreductase, cullin family domain containing protein, F-box domain containing protein respectively and down regulated under salt stress. The oco-miR169g, oco-miR020-3p, oco-miR160b targets nuclear transcription factor Y subunit, histone-lysine N- methyltransferase, lysine-9 specific SUVH1 and auxin response factor and were down regulated [66]. These studies established the important role of miRNAs and will provide a new dimension for unraveling salt tolerance mechanism in halophytes.
7. HORMONAL REGULATION
Plant hormones such as abscisic acid, jasmonic acid, ethylene and salicylic acid play a significant role in abiotic stress tolerance mechanism of plants. In halophytes, there is paucity of information about the role of hormones in salt adaptation.
ABA plays an important role in stress perception and during osmotic phase of salt stress. It is accumulated in plant cells and controls stomatal closure and activates the transcription of salt responsive genes [68]. The Regulatory components of ABA-receptor/Pyrabactin resistant Protein/PYR-like (PYR/ RCAR/PYL proteins) family proteins acts as ABA receptors; they recognize ABA and bind to group A PP2C (type 2C protein phosphatase) molecules. These molecules then release SNF1 related protein kinase 2 by negative regulation to phosphorylate ABA- responsive element- binding transcription factors (ABRE- ABFs). The OPEN STOMATA1 (OST1) gene in the presence of ABA gets activated and phosphorylates SLOW ANION CHANNEL-ASSOCIATED1 anion channel which closes stomata by releasing stomata from guard cells (Reviewed by [69]. This OST1 gene also interacts with NADPH oxidase which catalyzes ROS production, such as superoxides and hydrogen peroxides. This ROS burst again activates OST1 gene by inactivating PP2C in a positive feedback loop. The ROS burst and ABA function is interlinked as ABA accumulation decreases with decrease in GSH level in guard cells [70]. Similarly, in a halophyte, K. caspica, differential transcriptomics showed up regulation of orthologs of PYL, PP2C and SnRK2 genes under salt treatment indicating role of ABA signaling in salt tolerance mechanism of halophytes [42].
Jasmonic acid (JA) is one of the oxidation products of linolenic acid [71]. JA is also involved in stress induced stomatal closure by stimulating Nitrous Oxide (NO) production [72]. It increases cytosolic pH of guard cells which favors ROS production and thus plays role in co-ordination with ABA and ROS signaling to regulate stress induced stomatal movement [73]. In model plant, Arabidopsis, ROS level and JA synthesis does not show direct relationship while in the halophytes, C. maritima and T. salsuginea increased levels of JA are seen. This change may be a result of efficient channeling of linolenic acid oxidation towards synthesis of JA. This may improve oxidative stress response in halophytes.
Ethylene is a gaseous hormone which plays an important role in leaf senescence, fruit ripening, biotic and abiotic stress tolerance. The ethylene responsive transcription factor 1(ERF1) inhibits superoxide dismutase and peroxidase enzymes and increases ROS production, while ERF6 negatively regulates ROS accumulation and its signaling occurs through MAPK cascade [74]. In two halophytes C. maritima and T. salsuginea significant accumulation of ACC (precursor molecule of ethylene) was observed under salt stress. In halophytes, the direct link between ROS production and plant hormone is not yet established. But since these may act as signaling agents in stress condition, there must be direct link.
The Salicylic Acid (SA) is a phenolic hormone, involved in various physiological processes during growth, development and stress management [69]. Under stress regime, SA alters cellular redox state by stimulating ROS production and shifts redox towards oxidative stress. It also mediates ROS detoxification via antioxidant action and subsequent restoration of reductive phase [75]. Ca2+ signaling indirectly mediate ROS mediated regulation of SA signaling. The induction of ICS 1 gene (codes for SA producing enzyme, isochorismate synthase) is associated with calcium dependant kinases and TFs like CBP60, SARD1 and WRKY/8/28/48 which are modulated by calmodulin [76-78]. The SA plays both roles as pro-oxidant as well as antioxidant role in concert with GSH in response to salinity in plants [76]. Accumulation of SA increases GSH redox activity, while its inhibition decreases GSH/GSSG ratio under stress condition [79]. Role of SA in salt tolerance mechanism of halophytes is not very well understood. In the halophyte, Prosopis strombulifera, salicylic acid accumulation is correlated with the damaging effect of sulfate anion and low pH on plant growth under salinity stress [80].
CONCLUSION
The inherent salt tolerance ability of halophytes makes them a model for understanding the mechanism of their adaptation to high salinity. The ability to sense and respond to salt stimuli followed by signaling and activation of multitude of stress-protective mechanisms, are critical to halophyte survival in saline environment. The stress defense includes ROS-scavenging machinery, osmolytes and other protective gear. High salinity stress also induces phytohormone synthesis, particularly ABA and regulation of specific gene expression and metabolite synthesis. Besides being generated under stress, ROS also take part in cellular signaling involving Ca2+ and associated proteins (such as calcinurin, calmodulin, GTP binding proteins), phospholipid signaling and MAPKs activation and abscisic acid. Continued research efforts are required to completely understand these salt loving or preferring plant species. Molecular approaches including genomics and proteomics have provided clues to unravel the genome-wide gene expression, miRNAs and novel, essential proteins under salt stress in halophytes. There has been some success in the use of genes from halophytes to engineer salt tolerance in glycophytes especially crop plants, however, intensive efforts will have to be made to transfer useful genes and evaluate their utility under field conditions. Advances in genomics, whole genome sequencing and comparative genomics will pave the way for the integration of results on genome organization, gene and transcript structures, non coding regulatory RNAs, regulatory mechanisms, biochemical complexity, and hormone- or metabolite-based signaling networks. Such information resource should facilitate the understanding of the genetic and evolutionary processes that enabled halophytic adaptations in saline environment.
ACKNOWLEDGEMENTS
GN is thankful to Bhabha Atomic Research Centre and Savitribai Phule Pune University collaborative Ph.D programme and Department of Atomic Energy for providing fellowship.
LIST OF ABBREVIATIONS
- ABF
Ascaris suum antibacterial factor
- ACC
1-aminocyclopropane-1-carboxylic acid
- ADP
Adenosine diphosphate
- AKT
RAC-alpha serine/threonine-protein kinase
- ANT
Adenosine nucleoside transporter
- AOX
Alternative oxidase
- ATAF
Arabidopsis transcription activation factor
- CaM
Calmodulin
- CAM
Crassulacian Acid Metabolism
- CBL
Cancinurin B like
- CBP
Calmodulin binding protein
- CDPK
Calcium dependant protein kinase
- CIPK
CBL-interacting protein kinase
- CML
Calmodulin-like proteins
- CSD
Superoxide dismutase [Cu-Zn]
- CUC
Cup shaped cotyledon
- ERF
Ethylene responsive factor
- H2O2
Hydrogen peroxide
- ICS
Isochorismate synthase
- MAPK
Mitogen activated protein kinase
- Myb
Myeloblastosis transcription factor
- NHX
Na+/H+ exchangers
- PAK
P21-Activated kinase 1
- PP5
Protein phosphatase 5
- RBOH
Respiratory Burst Oxidase Homologs
- ROS
Reactive oxygen species
- SARD
Systemic acquired resistance deficient
- SNF
Sugar non fermenting
- SOS
Salt overly sensitive
- TIR
Toll-interleukin 1 repeat protein
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Food and Agriculture Organization 2008 http://www.fao.org/ ag.agl/agll/spush
- 2.Rozema J., Flowers T. Ecology. Crops for a salinized world. Science. 2008;5(5907):1478–1480. doi: 10.1126/science.1168572. [DOI] [PubMed] [Google Scholar]
- 3.Flowers T.J., Colmer T.D. Salinity tolerance in halophytes. New Phytol. 2008;179:945–963. doi: 10.1111/j.1469-8137.2008.02531.x. [DOI] [PubMed] [Google Scholar]
- 4.Flowers T.J., Troke P.F., Yeo A.R. The mechanism of salt tolerance in halophytes. Ann. Rev. Plant. Phy. 1977;28:89–121. [Google Scholar]
- 5.Zhu J.K. Plant salt stress: John wiley and sons, Ltd. A. K. Parida, A.B. Das. Salt tolerance and salinity effects on plants: A review. Eco. Env. Safety. 2007;60(3):324–349. doi: 10.1016/j.ecoenv.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 6.Yensen N.P. Halophyte uses for the twenty first century. In: Khan M.A., Weber D.J., editors. Ecophysiology of high salinity tolerant plants. Dordrecht, The Netherlands: Springer; 2008. [Google Scholar]
- 7.Lokhande V.H., Gor B.K., Desai N.S., Nikam T.D., Suprasanna P. Sesuvium portulacastrum, a plant for drought, salt stress, and fixation, food and phytoremediation. A review. Agron. Sustain. Dev. 2013;33:329–348. [Google Scholar]
- 8.Bose J., Rodrigo-Moreno A., Lai D., Xie Y., Shen W., Shabala S. Rapid regulation of the plasma membrane H+-ATPase activity is essential to salinity tolerance in two halophyte species, Atriplex lentiformis and Chenopodium quinoa. Ann. Bot. 2015;115:481–494. doi: 10.1093/aob/mcu219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raven J.A. Regulation of pH and generation of osmolarity in vascular land plants: costs and benefits in relation to efficiency of use of water, energy and nitrogen. New Phytol. 1985;101:25–77. doi: 10.1111/j.1469-8137.1985.tb02816.x. [DOI] [PubMed] [Google Scholar]
- 10.Lokhande V.H., Srivastava A.K., Srivastava S., Nikam T.D., Suprasanna P. Regulated alterations in redox and energetic status are the key mediators of salinity tolerance in the halophyte Sesuvium portulacastrum (L.) L. Plant Growth Regul. 2011;65(2):287–298. [Google Scholar]
- 11.Naidoo G., Rughunanan R. Salt tolerance in the succulent, coastal halophyte, Salicornia natalensis. J. Exp. Bot. 1990;41:497–502. [Google Scholar]
- 12.Zhu J.K. Plant salt tolerance. Trends Plant Sci. 2001;6:66–71. doi: 10.1016/s1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
- 13.Munns R., Tester M. Mechanism of salinity tolerance. Annu. Rev. Plant Biol. 2008;59:651–581. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 14.Foyer C.H., Noctor G. Redox sensing and signaling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol. Plant. 2003;119:355–364. [Google Scholar]
- 15.Bressan R.A., Park H.C., Orsini F., Oh D., Dassanayake M., Inan G., Yun D.J., Bohnert H.J., Maggio A. Biotechnology for mechanisms that counteract salt stress in extremophile species: a genome-based view. Plant Biotechnol. Rep. 2013;7:27–37. [Google Scholar]
- 16.Joshi R., Mangu V.R., Bedre R., Sanchez L., Pilcher W., Zandkarimi H., Baisakh N. In: Elucidation of Abiotic Stress Signaling in Plants, Functional genomics perspective. Pandey G.K., editor. Springer; 2015. [Google Scholar]
- 17.Cheeseman J.M. The evolution of halophytes, glycophytes and crops, and its implications for food security under saline conditions. New Phytol. 2014;206:557–570. doi: 10.1111/nph.13217. [DOI] [PubMed] [Google Scholar]
- 18.Amtmann A. Learning from evolution: Thellungiella generates new knowledge on essential and critical components of abiotic stress tolerance in plants. Mol. Plant. 2009;2(1):3–12. doi: 10.1093/mp/ssn094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flowers T.J., Colmer T.D. Plant salt tolerance: adaptations in halophytes. Ann. Bot. 2015;115:327–331. doi: 10.1093/aob/mcu267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bohnert H., Cushman J.J. The ice plant cometh: lessons in Abiotic stress tolerence. Plant Growth Regul. 2000;19:334. [Google Scholar]
- 21.Nah G., Pagliarulo C.L., Mohr P.G., Luo M., Sisneros N., Yu Y., Collura K., Currie J., Goicoechea J.L., Wing R.A., Schumaker K.S. Comparative sequence analysis of the SALT OVERLY SENSITIVE1 orthologous region in Thellungiella halophila and Arabidopsis thaliana. Genomics. 2009;94(3):196–203. doi: 10.1016/j.ygeno.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 22.Taji T., Seki M., Satou M., Sakurai T., Kobayashi M., Ishiyama K., Narusaka Y., Narusaka M., Zhu K.K., Shinozaki K. Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis related halophyte salt cress using Arabidopsis microarray. Plant Physiol. 2004;135:1697–1709. doi: 10.1104/pp.104.039909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh D.H., Dassanayake M., Haas J.S., Kropornika A., Wright C., d’Urzo M.P., Hong H., Ali S. Hernandez, Hernandez, A.; Lambert, G.M.; Inan, G.; Galbraith, D.W.; Bressan, R.A.; Yun, D.J.; Zhu, JK.; Cheeseman, J.M.; Bohnert, H.J. Genome structures and halophyte specific gene expression of the extremophile Thellungiella parvula in comparison with Thellungiella salsuginea (Thellungiella halophila) and Arabidopsis. Plant Physiol. 2010;154(3):1040–1052. doi: 10.1104/pp.110.163923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holmberg N., Bulow L. Improving stress tolerance in plants by gene transfer. Trends Plant Sci. 1998;3:61–66. [Google Scholar]
- 25.Oh D.H., Leidi E., Zhang Q., Hwang S.M., Li Y., Quintero F.J., Jiang X., d’Urzo M.P., Lee S.Y., Zhao Y., Bahk J.D., Bressan R.A., Yun D.J., Pardo J.M., Bohnert H.J. Loss of halophytism by interference with SOS1 expression. Plant Physiol. 2009;151:210–222. doi: 10.1104/pp.109.137802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blumwald E., Aharon G.S., Apse M.P. Sodium transport in plant cells. Biochim. Biophys. Acta. 2000;1465:140–151. doi: 10.1016/s0005-2736(00)00135-8. [DOI] [PubMed] [Google Scholar]
- 27.Pardo J.M., Reddy M.P., Yang S., Maggio A., Huh G.H., Matsumoto T., Coca M.A., Paino-D’Urzo M., Koiwa H., Yun D.J., Watad A.A., Bressan R.A., Hasegawa P.M. Stress signaling through Ca2+ / calmodulin-dependant protein phosphatase calcinurin mediates salt adaptation in plants. Proc. Natl. Acad. Sci. USA. 1998;95:9681–9686. doi: 10.1073/pnas.95.16.9681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu N., Chen M., Guo J., Bao H., Ma X., Wang B. Coordinate up-regulation of V-H+-ATPase and vacuolar Na+/H+ antiporter as a response to NaCl treatment in a C3 halophyte Suaeda salsa. Plant Sci. 2007;172:1218–1225. [Google Scholar]
- 29.Shi H., Xiong L., Stevenson B., Lu T., Zhu J.K. The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance. Plant Cell. 2002;14:575–588. doi: 10.1105/tpc.010417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi H., Kim Y., Guo Y., Stevenson B., Zhu J.K. The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell. 2003;15:19–32. doi: 10.1105/tpc.007872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumari A., Das P., Parida A.K., Agarwal P.K. Proteomics, metabolomics, and ionomics perspectives of salinity tolerance in halophytes. Front. Plant Sci. 2015;6:537. doi: 10.3389/fpls.2015.00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanada Y., Ueda H., Kuribayashi K., Andoh T., Hayashi F., Tamai N., Wada K. Novel light–dark change of proline levels in halophyte (Mesembryanthemum crystallinum L.) and glycophyte (Hordeum vulgare L. and Triticum aestivum L.) leaves and roots under salt stress. Plant Cell Physiol. 1995;36:965–970. [Google Scholar]
- 33.Khan M.J., Ungar I.A., Showalter A.M., Dewald H.D. NaCl induced accumulation of glycine betaine in four subtropical halophytes from Pakistan. Physiol. Plant. 1998;102:487–492. [Google Scholar]
- 34.Valluru R., Van den Ende W. Myo-inositol and beyond emerging networks under stress. Plant Sci. 2011;181:387–400. doi: 10.1016/j.plantsci.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 35.Agarie S., Kawaguchi A., Kodera A., Sunagawa H., Kojima H., Nose A., Nakahara T. Potential of the common ice plant, Mesembryanthemum crystallinum as a new high-functional food as evaluated by polyol accumulation. Plant Prod. Sci. 2009;12:37–46. [Google Scholar]
- 36.Nelson D.E., Koukoumanos M., Bohnert H.J. Myo-inositol-dependent sodium uptake in ice plant. Plant Physiol. 1999;119(1):165–172. doi: 10.1104/pp.119.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sureshan K.M., Riley A.M., Rossi A.M., Tovey S.C., Skarlatos G.D., Taylor C.W., Barry V.L. Activation of IP3 receptors by synthetic bisphosphate ligands. Chem. Commun. 2009;14(10):1204–1206. doi: 10.1039/b819328b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mittler R. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;9(10):490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 39.Medvedev S.S. Calcium signaling system in plants. Russ. J. Plant Physiol. 2005;52:249–270. [Google Scholar]
- 40.DeFalco T.A., Bender K.W., Snedden W.A. Breaking the code: Ca2+ sensors in plant signaling. Biochem. J. 2010;425:27–40. doi: 10.1042/BJ20091147. [DOI] [PubMed] [Google Scholar]
- 41.Hashimoto K., Eckert C., Anschutz U., Scholz M., Held K., Waadt R., Reyer A., Hippler M., Becker D., Kudla J. Phosphorylation of calcineurin B-like (CBL) calcium sensor proteins by their CBL-interacting protein kinases (CIPKs) is required for full activity of CBL-CIPK complexes toward their target proteins. J. Biol. Chem. 2012;287:7956–7968. doi: 10.1074/jbc.M111.279331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang C., Ge R., Zhang J., Chen Y., Wang H., Wei J., Li R. Identification and expression analysis of a novel HbCIPK2-interacting ferredoxin from halophyte H. brevisubulatum. PLoS One. 2015;10(12):e0144132. doi: 10.1371/journal.pone.0144132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li R., Zhang J., Wu G., Wang H., Chen Y., Wei J. HbCIPK2, a novel CBL-interacting protein kinase from halophyte Hordeum brevisubulatum, confers salt and osmotic stress tolerance. Plant Cell Environ. 2012;35(9):1582–1600. doi: 10.1111/j.1365-3040.2012.02511.x. [DOI] [PubMed] [Google Scholar]
- 44.Ji H. Jose, Pardo, J.M.; Batellic, G.; Van Oostend, M.J.; Bressane, R.A.; Lia, X. The salt overly sensitive (SOS) pathway: established and emerging roles. Mol. Plant. 2013;6(2):275–286. doi: 10.1093/mp/sst017. [DOI] [PubMed] [Google Scholar]
- 45.Shi H., Quintero F.J., Pardo J.M., Zhu J.K. the putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell. 2002;14(2):465–477. doi: 10.1105/tpc.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yadav N.S., Shukla P.S., Jha A., Agarwal P.K., Jha B. The SbSOS1 gene from the extreme halophyte Salicornia brachiata enhances Na(+) loading in xylem and confers salt tolerance in transgenic tobacco. BMC Plant Biol. 2012;12:188. doi: 10.1186/1471-2229-12-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agarwal P.K., Gupta K., Jha B. Molecular characterization of the Salicornia brachiata SbMAPKK gene and its expression by abiotic stress. Mol. Biol. Rep. 2010;37(2):981–986. doi: 10.1007/s11033-009-9774-1. [DOI] [PubMed] [Google Scholar]
- 48.Gong Q., Li P., Ma S., Rupassara S.I., Bohnert H.J. Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J. 2005;44:826–839. doi: 10.1111/j.1365-313X.2005.02587.x. [DOI] [PubMed] [Google Scholar]
- 49.Du J., Huang Y.P., Xi J., Cao M.J., Ni W.S., Chen X., Zhu J.K., Oliver D.J., Xiang C.B. Functional gene-mining for salt-tolerance genes with the power of Arabidopsis. Plant J. 2008;56:653–664. doi: 10.1111/j.1365-313X.2008.03602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mehta P.A., Sivaprakash K., Parani M., Venkataraman G., Parida A.K. Generation and analysis of expressed sequence tags from the salt tolerant mangrove species Avicennia marina (Forsk) Vierh. Theor. Appl. Genet. 2005;110:416–424. doi: 10.1007/s00122-004-1801-y. [DOI] [PubMed] [Google Scholar]
- 51.Baisakh N., Subudhi P.K., Varadwaj P. Primary responses to salt stress in a halophyte, smooth cordgrass (Spartina alterniflora Loisel.). Funct. Integr. Genomics. 2008;8:287–300. doi: 10.1007/s10142-008-0075-x. [DOI] [PubMed] [Google Scholar]
- 52.Miyama M., Shimizu H., Sugiyama M., Hanagata N. Sequencing and analysis of 14,842 expressed sequence tags of burma mangrove, Bruguiera gymnorrhiza. Plant Sci. 2006;171:241–324. [Google Scholar]
- 53.Jha B., Agarwal P.K., Reddy P.S., Lal S., Sopory S.K., Reddy M.K. Identification of salt-induced genes from Salicornia brachiata, an extreme halophyte through expressed sequence tags analysis. Genes Genet. Syst. 2009;84(2):111–120. doi: 10.1266/ggs.84.111. [DOI] [PubMed] [Google Scholar]
- 54.Shukla P.S., Agarwal P., Guptaa K., Agarwal P.K. Molecular characterisation of a MYB transcription factor from a succulent halophyte involved in stress tolerance. AoB Plants. 2015;7:plv054. doi: 10.1093/aobpla/plv054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katschnig D., Bliek T., Rozema J., Schat H. Constitutive high-level SOS1 expression and absence of HKT1;1 expression in the salt-accumulating halophyte Salicornia dolichostachya. Plant Sci. 2015;234:144–154. doi: 10.1016/j.plantsci.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 56.Dassanayake M., Oh D.H., Hong H., Bohnert H.J., Cheeseman J.M. Transcription strength and halophytic lifestyle. Trends Plant Sci. 2011;16:1–3. doi: 10.1016/j.tplants.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 57.Garg R., Verma M., Agrawal S., Shankar R., Majee M., Jain M. Deep transcriptome sequencing of wild halophyte rice, Porteresia coarctata, provides novel insights into the salinity and submergence tolerance factors. DNA Res. 2013;•••:1–16. doi: 10.1093/dnares/dst042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dang Z.H., Zheng L., Wang J., Gao Z., Wu S., Qi Z., Wang Y.C. Transcriptomic profiling of the salt-stress response in the wild recretohalophyte Reaumuria trigyna. BMC Genomics. 2013;14:29. doi: 10.1186/1471-2164-14-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma J., Zhang M., Xiao X., You J., Wang J., Wang T., Yao Y., Tian C. Global transcriptome profiling of Salicornia europaea L. shoots under NaCl treatment. PLoS One. 2013;8(6):e65877. doi: 10.1371/journal.pone.0065877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 61.Khraiwesh B., Pugalenthi G., Fedoroff N.V. Identification and analysis of red sea mangrove (Avicennia marina) microRNAs by high-throughput sequencing and their association with stress responses. PLoS One. 2013;8:e60774. doi: 10.1371/journal.pone.0060774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feng J., Wang J., Fan P., Jia W., Nie L., Jiang P., Chen X., Lv S., Wan L., Chang S., Li S., Li Y. High throughput deep sequencing reveals that microRNAs play important roles in salt tolerance of euhalophyte Salicornia europaea. BMC Plant Biol. 2015;15:63. doi: 10.1186/s12870-015-0451-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singh D., Jha B. The isolation and identification of salt-responsive novel microRNAs from Salicornia brachiata, an extreme halophyte. Plant Biotechnol. Rep. 2014;8:325–336. [Google Scholar]
- 64.Gharat S.A., Shaw B.P. Novel and conserved miRNAs in the halophyte Suaeda maritima identified by deep sequencing and computational predictions using the ESTs of two mangrove plants. BMC Plant Biol. 2015;15:301. doi: 10.1186/s12870-015-0682-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang R., Zeng Y., Yi X., Zhao L., Zhang Y. Small RNA deep sequencing reveals the important role of microRNAs in the halophyte Halostachys caspica. Plant Biotechnol. 2015;13:395–408. doi: 10.1111/pbi.12337. [DOI] [PubMed] [Google Scholar]
- 66.Mondal T.K., Ganie S.A., Debnath A.B. Identification of novel and conserved miRNAs from extreme halophyte, Oryza coarctata, a wild relative of Rice. PLoS One. 2015;10(10):e0140675. doi: 10.1371/journal.pone.0140675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Panahi B., Mogammadi S.A., Abrahimie E. Identification of miRNAs and their potential targets in halophyte plant Thellungiella halophila. BioTechnologia. 2013;94(3):285–290. [Google Scholar]
- 68.Wilkinson S., Davies W.J. ABA-based chemical signaling: the co-ordination of responses to stress in plants. Plant Cell Environ. 2002;25:195–210. doi: 10.1046/j.0016-8025.2001.00824.x. [DOI] [PubMed] [Google Scholar]
- 69.Srivastava A.K., Redij T., Sharma B., Suprasanna P. 2015. In press. [Google Scholar]
- 70.Okuma E., Jahan M.S., Munemasa S., Hossain M.A., Muroyama D., Islam M.M., Ogawa K., Watanabe S.M., Nakamura Y., Shimoishi Y., Mori I.C., Murata Y. Negative regulation of abscisic acid-induced stomatal closure by glutathione in Arabidopsis. J. Plant Physiol. 2011;168:2048–2055. doi: 10.1016/j.jplph.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 71.Wasternack C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 2007;100(4):681–697. doi: 10.1093/aob/mcm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saito N., Nakamura Y., Mori I.C., Murata Y. Nitric oxide functions in both methyl jasmonate signaling and abscisic acid signaling in Arabidopsis guard cells. Plant Signal. Behav. 2009;4(2):119–120. doi: 10.4161/psb.4.2.7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gonugunta V.K., Srivastava N., Raghavendra A.S. Cytosolic alkalinization is a common and early messenger preceding the production of ROS and NO during stomatal closure by variable signals, including abscisic acid, methyl jasmonate and chitosan. Plant Signal. Behav. 2009;4:561–564. doi: 10.4161/psb.4.6.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang F., Cui X., Sun Y., Dong C.H. Ethylene signaling and regulation in plant growth and stress responses. Plant Cell Rep. 2013;32:1099–1109. doi: 10.1007/s00299-013-1421-6. [DOI] [PubMed] [Google Scholar]
- 75.Herrera-Vasquez A., Salinas P., Holuigue L. Salicylic acid and reactive oxygen species interplay in the transcriptional control of defense genes expression. Front. Plant Sci. 2015;6:171. doi: 10.3389/fpls.2015.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Du L., Ali G.S., Simons K.A., Hou J., Yang T., Reddy A.S., Poovaiah B.W. Ca(2+)/calmodulin regulates salicylic-acid-mediated plant immunity. Nature. 2009;457:1154–1158. doi: 10.1038/nature07612. [DOI] [PubMed] [Google Scholar]
- 77.Gao X., Chen X., Lin W., Chen S., Lu D., Niu Y., Li L., Cheng C., McCormack M., Sheen J., Shan L., He P. Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog. 2013;9:e1003127. doi: 10.1371/journal.ppat.1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Truman W., Sreekanta S., Lu Y., Bethke G., Tsuda K., Katagiri F., Glazebrook J. The calmodulin-binding protein60 family includes both negative and positive regulators of plant immunity. Plant Physiol. 2013;163:1741–1751. doi: 10.1104/pp.113.227108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Noctor G., Mhamdi A., Foyer C.H. The roles of reactive oxygen metabolism in drought: not so cut and dried. Plant Physiol. 2014;164(4):1636–1648. doi: 10.1104/pp.113.233478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Devinar G., Llanes A., Masciarelli O., Luna V. Different relative humidity conditions combined with chloride and sulfate salinity treatments modify abscisic acid and salicylic acid levels in the halophyte Prosopis strombulifera. Plant Growth Regul. 2013;70(3):247–256. [Google Scholar]
- 81.Ellouzi H., Ben Hamed K., Cela J., Munne-Bosch S., Abdelly C. Early effects of salt stress on the physiological and oxidative status of Cakile maritima (halophyte) and Arabidopsis thaliana (glycophyte). Physiol. Plant. 2011;142:128–143. doi: 10.1111/j.1399-3054.2011.01450.x. [DOI] [PubMed] [Google Scholar]
- 82.Bose J., Rodrigo-Moreno A., Shabala S. ROS homeostasis in halophytes in the context of salinity stress tolerance. J. Exp. Bot. 2014;65(5):1241–1257. doi: 10.1093/jxb/ert430. [DOI] [PubMed] [Google Scholar]
- 83.Shalata A., Mittova V., Volokita M., Guy M., Tal M. Response of the cultivated tomato and its wild salt-tolerant relative Lycopersicon pennellii to salt-dependent oxidative stress: the root antioxidative system. Physiol. Plant. 2001;112:487–494. doi: 10.1034/j.1399-3054.2001.1120405.x. [DOI] [PubMed] [Google Scholar]
- 84.Murakeözy É.P., Nagy Z., Duhazé C., Bouchereau A., Tuba Z. Seasonal changes in the levels of compatible osmolytes in three halophytic species of inland saline vegetation in Hungary. J. Plant Physiol. 2003;160:395–401. doi: 10.1078/0176-1617-00790. [DOI] [PubMed] [Google Scholar]
- 85.Ghars M.A., Richard L., Lefebvre-De V.D., Leprince A.S., Parre E., Bordenave M., Abdelly C., Savoure A. Phospholipases C and D modulate proline accumulation in Thellungiella halophila/salsuginea differently according to the severity of salt or hyperosmotic stress. Plant Cell Physiol. 2012;53:183–192. doi: 10.1093/pcp/pcr164. [DOI] [PubMed] [Google Scholar]
- 86.English J.P., Colmer T.D. Tolerance of extreme salinity in two stem succulent halophytes (Tecticornia species). Funct. Plant Biol. 2013;40(9):897–912. doi: 10.1071/FP12304. [DOI] [PubMed] [Google Scholar]
- 87.Himabindu Y., Chakradhar T., Reddy M.C., Kanygin A., Redding K.E., Chandrasekhar T. Salt-tolerant genes from halophytes are potential key players of salt tolerance in glycophytes. Environ. Exp. Bot. 2016;124:39–63. [Google Scholar]
- 88.An R., Chen Q.J., Chai M.F., Lu P.L., Su Z., Qin Z.X., Chen J., Wang X.C. AtNHX8, a member of the monovalent cation: proton antiporter-1 family in Arabidopsis thaliana, encodes a putative Li+/H+ antiporter. Plant J. 2007;49:718–728. doi: 10.1111/j.1365-313X.2006.02990.x. [DOI] [PubMed] [Google Scholar]
- 89.Nawaz I., Iqbal M., Hakvoort H.W., Bliek M., de Boer B., Schat H. Expression levels and promoter activities of candidate salt tolerance genes in halophytic and glycophytic Brassicaceae. Environ. Exp. Bot. 2007;99:59–66. [Google Scholar]