Abstract
Plants, being sessile organisms, constantly withstand environmental fluctuations, including low-temperature, also referred as cold stress. Whereas cold poses serious challenges at both physiological and developmental levels to plants growing in tropical or sub-tropical regions, plants from temperate climatic regions can withstand chilling or freezing temperatures. Several cold inducible genes have already been isolated and used in transgenic approach to generate cold tolerant plants. The conventional breeding methods and marker assisted selection have helped in developing plant with improved cold tolerance, however, the development of freezing tolerant plants through cold acclimation remains an unaccomplished task. Therefore, it is essential to have a clear understanding of how low temperature sensing strategies and corresponding signal transduction act during cold acclimation process. Herein, we synthesize the available information on the molecular mechanisms underlying cold sensing and signaling with an aim that the summarized literature will help develop efficient strategies to obtain cold tolerant plants.
Keywords: Cold, Calcium, Protein kinase, CBF, Signaling, Phosphorylation
1. INTRODUCTION
In contrast to animals which preferentially opt for the ‘avoid’ response, plants being sessile usually adopt the ‘overcome’ strategy to counter any extreme environmental fluctuation. Temperature stress is one such factor which plants experience from their surrounding environment. Plant grow best at their optimum temperature range and any extreme fluctuation from this optima affects their growth and final yield [1]. Due to its wide-spread occurrence, low temperature or cold stress affects several facets of plant’s life and causes extreme economic losses in agriculture. Plant endurance to low temperatures can be grouped into two types i.e. chilling tolerance (above 0°C) and freezing tolerance (below 0°C) [2]. Among various abiotic stresses, chilling stress influences the production and quality of economically important crops the most, especially in the tropical and subtropical climatic zones [3, 4]. In contrast, plants originating from temperate climatic regions are considered chilling tolerant because they can increase their freezing tolerance by cold acclimation process. Plants of tropical and sub-tropical origins lack this mechanism and are more prone to the adverse effect of chilling stress [5-8]. Depending on their response to low temperature stress, plants have been categorized into chilling sensitive, chilling tolerant, and freezing tolerant. Chilling sensitive plants show metabolic complications on the exposure to the temperatures below their optima.
Important crops, such as rice, maize, soybean, cotton and tomato are chilling sensitive. On the contrary, chilling tolerant plants are able to survive the lower range of temperatures but not freezing temperature. The freezing tolerant plants can survive even in the freezing conditions [9].
2. PHYSIOLOGICAL CONSEQUENCES OF COLD STRESS ON PLANTS AND COLD TOLERANCE MECHANISM
Exposure of plants to chilling results in a number of transient biochemical perturbations, including thermodynamic slowdown of the kinetics of metabolic reactions. Further, it modulates the thermodynamic equilibrium of the cell which might cause the non-polar side chains of proteins to reorient towards the aqueous medium. Such reorientation impacts on the solubility and stability of globular proteins [10]. Low temperatures also cause rigidification of membrane, resulting in the disturbance of all membrane-related processes, and induction of cold-associated genes [11]. Chilling also leads to higher reactive oxygen species (ROS) accumulation as the ROS scavenging system does not function properly; due to reduced enzymes activity. In turn, ROS over accumulation has harmful effects on membranes and leads to ion leakage. Altogether, low temperature directly affects DNA secondary structure, lowers enzymatic activities involved in fundamental processes such as transcription and translation, induces membrane rigidification, destabilizes protein complexes, stabilizes RNA secondary structure, impairs photosynthesis, and leads to ion leakage across membranes [7, 12-16].
Below sub-zero temperature, ice formation first takes place in the intercellular spaces of plant tissues and finally culminates in the intracellular symplastic freezing. The extracellular ice formation results in a drop in water potential and causes intracellular water to move out of the cell. It is followed by dehydration and shrinkage of cells causing freezing injuries [8, 17, 18]. The level of such dehydration is determined by the severity of temperature drop which increases with the decreasing temperatures. Finally, ice can penetrate the symplast and spoil the intracellular structures resulting in death of the tissue [19, 20]. Cold-induced dehydration results in various physiological effects, for example, precipitation of molecules, protein denaturation, membrane damaging effects and cell lysis [8, 21]. The signs and symptoms associated with chilling-induced stress injuries in chilling sensitive plants are varied and generally express within 48 to 72 h of exposure to the stress. The phenotypic signs of chilling stress consist of reduced leaf expansion, chlorosis, wilting, and necrosis [22]. Chilling also causes defect in plant reproductive development. For example, rice plants displayed sterility upon exposure to chilling temperatures during anthesis [23]. The severity of plant damage depends on multiple factors such as the developmental stage of plant, duration of the frost, rates of cooling and rewarming, and the extent of ice formation [24]. As a consequence, plants adopt two strategies to resist the frost-related injuries: first, prevention of ice formation in tissues; and second, tolerance to apoplastic extracellular ice. Further, an individual plant may utilize both types of mechanisms for frost resistance in different tissues [25].
3. COLD ACCLIMATION
Cold acclimation is a process where temperate plants acquire freezing tolerance upon previous exposure to low but non-freezing temperatures [26-28]. This ability is not found in plants from the tropical and sub-tropical regions. As a consequence, these plants cannot tolerate ice formation in their tissues. This process is considered as the first line of defence and has an important role in stabilizing plasma membranes against cold-induced injury [7, 29-31]. Several mechanisms, including processes like change in lipid composition [30] and accumulation of simple sugars [32] contribute to this stabilization. Additionally, LEA (late embryogenic abundant) and hydrophilic proteins help stabilize membranes against cold-induced damage. Several chilling-related injuries can also be associated with ROS, especially in chilling sensitive plants [7, 33-35]. As the counteractive mechanisms, plants develop effective oxygen-scavenging systems which consist of several antioxidant enzymes such as ascorbate peroxidase, Superoxide Dismutase (SOD), Glutathione Reductase (GR) and catalase. Moreover, non-enzymatic antioxidants, such as ascorbic acid and reduced glutathione also help in minimizing the negative effects attributed by high ROS levels [36-38].
3.1. Perception of Cold
Stress perception determines the specificity of the signal transduction network and adaptation of plant’s physiology in a particular environment [21]. Plants exhibit a variety of responses to the environmental temperature in a time based fashion. While a few of these responses are short term, other responses require long exposure to low temperature; sometimes for several days or weeks as in the case of vernalization process [39]. Evidence suggests that plant cell preferentially senses the rate of temperature change (dT/dt) over the absolute temperature as fast cooling of temperature between 22°C and 16°C have been found to generate strong depolarization of membrane than the slower cooling in cucumber [40]. Although the mechanism of sensing low temperature in plants remains unclear, the temperature dependent modifications of membrane fluidity were initially considered as the primary temperature sensor in cyanobacteria and yeast [11, 41].
Plasma membrane, being the interface between internal and external environment of the cell, is considered as a site for the perception of temperature change [7, 42-44]. In plants, the potential sensors of cold include Ca+2 influx channels, two-component histidine kinases and receptors associated with G-proteins [45]. The initial evidences established that plant response to low temperature involved an influx of Ca+2 from apoplast into the cytosol and a positive correlation exists between Ca+2 influx and the rate of temperature drop [7, 46-49]. Cellular Ca+2 dynamics are detected in response to cold within 40 s through a novel aquaporin-based Ca+2 signaling mechanism in Arabidopsis [50].
It has been suggested that the cold induced calcium transients may occur downstream to membrane rigidification and cytoskeletal reorganization in signal transduction pathway [7, 21, 51-53]. Further, stabilization and destabilization of cytoskeletal components such as actin microfilament at different temperatures, 4°C and 25°C respectively, can control cold stress responses by preventing both expression of COR genes and Ca2+ influx [51]. It is known that drugs that strengthen microfilaments decrease cold sensitivity. In contrast, the drugs that destabilize microfilaments can stimulate cold-dependent downstream processes even in the absence of cold, suggesting that cytoskeletal reorganization possibly participates in the sensor mechanisms [51]. Nonetheless, cytoskeletal reorganization was found not to be an absolute requirement for cold-induced responses and therefore it is not considered as the primary cold sensor [28].
Additionally, expression of the two-component response regulator-like proteins has been implicated in low temperature responses in Arabidopsis [54, 55] and Synechocystis [56-58]. Low temperature induced receptor-like protein kinases have also been suggested as possible cold sensors [7, 59, 60]. The extracellular domains of these proteins undergo a temperature-induced conformational modification and leads to induction of their kinase activity in cytoplasm. Analysis of cold-induced gene expression in transgenic or mutant plants with changes in membrane lipid saturation or sterol content revealed that membrane fluidity is a part of cold sensing mechanisms in higher plants. Besides, recent evidence also suggests an important role of chromatin remodelling in low temperature sensing [7, 61, 62]. The histone remodelling proteins such as HOS15 (high expression of osmotically responsive genes), and AGC1 (aspartate/glutamate carrier) or histone subunits may play an equivalent role in cold sensing at lower temperatures. Taken together, it can be concluded that more than one thermo sensors are involved during cold sensing in plant cells [28].
3.2. Signal Transduction Mechanism
In general, the signal produced by physical factors such as cold converted to a genetic response which initiates changes in gene expression leading to physiological and metabolic changes in the cell and culminates in a response. Upon experiencing low temperature, plants identify the induced signal and transduce it to the nucleus.
3.2.1. Role of Calcium
Calcium (Ca+2) is the most ubiquitous secondary messenger in eukaryotes [63, 64]. Within seconds of low temperature exposure, free cytosolic Ca+2 level is elevated by its influx from apoplast or vacuole [46, 49]. Such cytosolic Ca+2 oscillations can be recognized within a short time span, just few seconds or minutes after transferring the plant to low temperature. The extent of these Ca+2 oscillations also depends on the previous exposure of temperature stress to plants as recurring experience of low temperature leads to reduced Ca+2 oscillations, suggesting that plants have a Ca+2 signature memory associated with earlier temperature experiences [65]. The prompt Ca+2 influx is induced by activation of ionophores or Ca+2 channels agonists which in turn leads to activation of cold-acclimation-specific genes [7, 66, 67]. The cytosolic influx of Ca+2 during stress is needed for the expression of some cold-induced genes like COR6 and KIN1 (Knotted1) n Arabidopsis thaliana [47-49]. The Ca+2 released from internal cellular reserves, mediated by inositol triphosphate, is upstream to the expression of C-repeat Binding Factors (CBFs) and Cold Responsive (COR) genes in the cold-signaling pathway [53, 68, 69].
3.2.1.1. Ca+2 Regulated Proteins (Decoders of the Calcium Signature)
The signal-specific Ca+2 signatures (cytosolic Ca+2 changes) are decoded by a large number of Ca+2 binding proteins in plants [70]. These proteins change their phosphorylation status on the elevation of intracellular Ca+2 level and function as Ca+2 sensors [7, 48, 71, 72]. The majority of Ca+2 sensors possess high affinity Ca+2 binding helix-turn-helix structures; also known as EF-hands [71, 73]. Ca+2 sensors have been broadly divided into two classes: sensor relays and sensor responders [74, 75]. Bonafide sensor relay proteins do not possess any known enzymatic or functional domains. Instead, upon binding to Ca+2, these interact with another group of proteins and regulate their activities in downstream signaling. Some of the major sensors included in this group are calmodulin (CaM), CaM-like (CMLs) and calcineurin B-like proteins (CBLs) [76-79].
The sensor responders are protein kinases which consist of one or more EF-hand motif and whose activity is controlled by binding of Ca+2 to EF hand motifs. In this group, sensing via EF-motif and responses via protein kinase function are combined within a single protein. It includes Ca+2-Dependent Protein Kinases (CDPKs), [80, 81], Ca+2 and Ca+2/CaM-dependent protein kinase (CCaMK) [82-84], cytosolic phospholipase A2 (cPLA2), phospholipase C (PLC) and some lipid (AtCLB, Caleiosins, PLD, Annexins) or DNA binding proteins (SUB1, Calreticulin) [71, 85-88].
Calmodulin (CaM) is one of the most conserved and best characterized small acidic Ca+2 binding proteins found in eukaryotes [89]. Its binding to Ca+2 induces a structural change and relay the signal to downstream components. CaM activity is essential for the expression of cold inducible genes in many systems, including Arabidopsis and Alfalfa [90]. CaMs are constituted by a small gene family in plants [91-94]. Over-expression of Arabidopsis CaM3 hinders cold induction of some of key cold responsive genes, including RD29A, KIN1(Knotted1 Induced1) and KIN2 (Knotted2 Induced2) [95]. In plants, CaM also presents an example of indirect regulation of gene expression by mediating through a CaM-binding protein kinase and a CaM-binding protein phosphatase [96].
Similar to CaMs, Calcineurin B like (CBL) proteins (also called SOS3-like Ca+2 binding proteins, ScaBLs) form another group of Ca+2 sensors. These proteins (AtCBLs/ SCaBPs) were first identified in Arabidopsis and lack any enzymatic activity [97-101]. Overexpression of AtCBL1 has been found to confer increased stress tolerance in transgenic Arabidopsis plants. These plants showed decreased rate of water loss with upregulated CBF/DREB transcription factors and other related genes in non-stressed plants [102]. Whereas mutation of this gene affected the transcript levels of cold regulated genes, its overexpression induced the expression of early stress responsive transcription factors [102]. However, CBLs alone cannot function and require a group of serine/threonine protein kinases, named CBL-interacting protein kinases (CIPKs), to impart their roles under cold stress conditions [103]. In total, 10 CBLs and 26 CIPKs genes have been identified in Arabidopsis [104]. Likewise, 10 CBLs and 33 CIPKs are present in rice [101, 105]. Interactions CBLs and CIPKs have been found to be Ca+2 dependent in cold stress responses. [99, 106, 107]. For example, CIPK3 has been suggested to act downstream of the Ca+2 signal [108]. Further, CIPK7 is induced by cold and interacts with CBL1 both in vitro and in vivo conditions [103]. Moreover, overexpression of OsCIPK3 in transgenic lines of rice, showed an improved cold tolerance and found better survival rate [109].
Calcium-dependent protein kinases (CDPKs) are other important sensors which are involved in response to abiotic stresses, including cold [81, 110-113]. These genes are also multigene family members. For example, it has been reported that this gene family is comprised of 34 CDPK members in Arabidopsis [81], 20 members in wheat [114], 29 members in tomato [115] and 31 members in rice [116]. CDPKs often have a conserved structure with an N-terminal variable domain, which mediate isoform specificity and localization [117, 118], a middle catalytic protein kinase domain, which is linked to a junction sequence, and C- terminally located CaM-like domain, which canonically harbours four EF-hands. A junction sequence acts like an auto-inhibitory region and keeps the kinase inactive using pseudosubstrate-binding mechanism [119-121]. Under low cytoplasmic calcium concentration, CDPK remains inactive due to the blocking of catalytic site by auto-inhibitory region. Upon stress perception and calcium influx into the cell, calcium binding to the EF-hands triggers the intramolecular interaction between CaM-like domain and the auto-inhibitory domain causing the conformational change that leads to activation of the enzyme [120, 122, 123]. Subsequently, downstream responses such as phosphorylation and activation of many regulatory proteins including transcription factors, changes in ion fluxes across membranes, accumulation of stress-related metabolites and developmental growth processes are induced [124-127]. Transient transactivation assays of stress-responsive CDPKs-reporter gene constructs in transformed maize (Zea mays) protoplasts provided the first evidence of their involvement in specific signal/response pathways [128]. In rice, a membrane associated CDPK is activated after exposure to cold [129]. Similarly, OsCPK7/OsCDPK13 or OsCPK13/OsCDPK7 is activated by a 3 h cold treatment [7]. Further, overexpression of OsCPK7/OsCDPK13 OsCPK13/OsCDPK7 confers cold tolerance in transgenic rice [7, 130]. These studies suggested the possibility of involvement of CDPKs in Ca+2 mediated signaling during acquisition of cold tolerance. Genetic analysis also proved that CDPKs act as positive regulators [130] whereas CaM3 acts as a negative regulator of gene expression during cold tolerance [95]. Similar to CIPKs, CDPKs perform their functions by binding to their targets and a few potential targets of CPK3 such as RARE COLD INDUCIBLE 1A (RCI1A) and ALCOHOL DEHYDROGENASE 1 (ADH1) have already been discovered in Arabidopsis. Further, cold inducible CPK4 targets a b-ZIP transcription factor ABF1, encoded by an ABA- and cold-inducible gene, for phosphorylation. It shows that ABF1 might participate in ABA-mediated cold acclimation in plants [131].
3.2.2. MAPK Cascade
MAPK family includes a large family of serine/threonine protein kinases in plants. A typical MAPK cascade is composed of three protein kinases. Inactive Mitogen Activated Protein Kinase Kinase Kinases (MAPKKKs) are activated by a stress signal messenger. Three kinds of MAPKKKs, including CTR1, ANP1-3 and MEKK exist in Arabidopsis thaliana. Among these, MEKK is expressed in response to various abiotic stresses, including cold. Upon activation, they activate MAPKKs by phosphorylation at conserved serine/threonine residue. Activated MAPKKs in turn activate MAPKs by phosphorylating MAPK at both threonine and tyrosine residues in the TXY motif; which leads to phosphorylation of various effector proteins like enzymes or transcription factors [132]. MAPK cascade is conserved among eukaryotes and transduces extracellular stimuli for cellular responses [133]. MAPK pathways are also triggered by various abiotic stresses [134]. The role of MAPKs in cold acclimation was demonstrated in Arabidopsis by a MAPK pathway mediated by Ca2+/CaM-CRLK1-MEKK1-MKK2-MPK4/6 under cold acclimation. A positive regulator of the cold tolerance, Ca2+/Calmodulin-Regulated Receptor-Like Kinase (CRLK1) has been reported in plants [135]. CRLK1 has been found to interact and phosphorylate MEKK1 [136, 137]. In turn, MEKK1, which is induced by cold, phosphorylates MKK2 during cold treatment [136]. It has been suggested that Ca2+ signaling occurs upstream of the MEKK1–MKK2 pathway [137]. Further, enzymatic activity of MEKK1 is increased kinase in the presence of MKK2 after cold treatment. MKK2 also interacts with MPK4/MPK6 during cold signaling [138]. Such interaction was also validated genetically as mkk2 mutant showed freezing sensitive phenotype, as no interaction of MKK2 with MPK4 nor MPK6, suggesting that MKK2 is present upstream to these two proteins during cold signaling [138]. Evidence suggests that MAP kinase pathways may also act independently of Ca+2. More specifically, MPK4 and MPK6 were found to operate independently of CPKs [139, 140]. However, it is not clear how Ca+2-dependent and Ca+2-independent (MAP kinase) pathways leading from cold perception affect post-translational modifications of TFs responsible for the regulation of cold gene expression [69] and the interplay between calcium and MAPK signaling pathways warrants future investigation [141].
3.2.3. Transcription Factor CBF/DREB Regulon
Changes in gene expression profiles upon exposure to low temperatures are well established and many genes which are either up- or down-regulated have been identified [142, 143]. A multidisciplinary approach in tomato suggested the role of transcriptome reprogramming in cold acclimation. Early response (after a few hours of suboptimal growth temperature exposure), resulted in changes in the expression levels of stress-related proteins including those belonging to transcription factors, hormone biosynthesis and signaling. In contrast, a late response (after 24 h of exposure) induced stable changes in the gene expression resulted in extensive adjustment of metabolism, photosystems, transcription and translation machineries by stable changes in gene expression [144]. Homologs of DREB1/CBFs have been identified in many agronomic crops, such as rice, maize, soybean (Glycine max) and wheat (Triticum aestivum) [145]. DREB1/ CBFs acts upstream to the cold-inducible genes, including cold-regulated genes (COR) and regulate their expression [142, 146]. Initially, promoter analysis of a cold inducible gene, COR15A, identified a region that conferred cold, abscisic acid (ABA) and drought responsive expression [147, 148]. Promoters of many other ABA-independent, cold- and drought-induced proteins contain one or more copies of Dehydration-Responsive/C-Repeat Element/Low Temperature Responsive Element (CRT or DRE or LTRE) cis-acting element. This element is attributed by the presence of CCGAC as its core sequence [147, 149, 150]. Additionally, many COR genes have ABA-responsive elements (ABREs) in their promoter, however, expression of COR genes is not strictly correlated by its presence as their transcript levels are regulated by both ABA-independent and ABA-dependent pathways [151, 152].
The CRT/DRE/LTRE element is recognized by a group of three similar cold induced transcription factors, known as either C-repeat binding factors, CBF1-3 or dehydration-responsive element binding factors DREB1A-C. These TFs control ABA-independent expression of COR genes in response to cold stress [150, 153-156]. The CBF pathway of Arabidopsis remains the best-understood regulatory pathway with its role in freezing tolerance [28, 157]. As reported, CBF1(DREB1b), CBF2 (DREB1C) and CBF3(DREB1A) are strongly and rapidly upregulated at the transcriptional level by low temperature and possess overlapping effects on COR gene regulation [158]. The CRT/DRE is also recognized by a group of drought-inducible transcriptional activators such as DREB2A-B, which are structurally not related to the CBF/DREB1 group. Overexpression of the CBF/DREB genes has resulted in enhancement of cold tolerance in Arabidopsis, tobacco and other agricultural important species such as rice and wheat [153, 159]. Overexpression of OsDREB1 in rice plants showed higher survival rate of transgenic plants in chilling (2°C) stress condition in comparison to the wild type [160]. Similarly, overexpression of a sweet pepper (Capsicum annuum) CBF3 gene in tobacco plants showed enhanced chilling (4°C) tolerance through higher accumulations of proline, soluble sugars, unsaturated fatty acids, and lower accumulations of ROS [161]. Arabidopsis plant overexpressing HbCBF1 gene of Hevea brasiliensis showed chilling resistance and activated expression of CBF pathway downstream target genes, such as AtCOR15a and AtRD29a [162]. AtCBF3 overexpressing Arabidopsis transgenic plants also demonstrated enhanced tolerance to freezing stress [163, 164]. It was recently reported that freezing tolerance of Muscadinia rotundifolia CBF2-overexpressing transgenic Arabidopsis lines was enhanced along with an increased expression of the cold regulated genes AtCOR47, AtCOR15A, AtRD29A, AtKIN1 and AtSuSy (Arabidopsis sucrose synthase 2) [165]. However, role of constitutive overexpression of either AtCBF1 or AtCBF3 genes in improving cold tolerance is not universal as their overexpression do not improve freezing tolerance in freezing-sensitive tomato plants. Further, studies in CBF2-deficient mutant have revealed that CBF2, which negatively regulate CBF1 and CBF3, may have distinct function in freezing tolerance from the other CBFs [166]. However, it can also not be ruled out that coordinated action of CBFs are required for cold acclimation [167]. While CBF1 and CBF3 have a concerted additive effect in the induction of whole CBF regulon, they both are simultaneously required for the induction of CBF target genes [167, 168].
Transcription of CBF genes is subjected to both positive and negative regulation. Several regulators have already been identified as inducer of CBF expression 1 (ICE 1) proteins [169], inducer of CBF expression 2 (ICE2) proteins [170], MYB15 [171], calmodulin binding transcription activator 3 (CAMTA3) [172], ZAT12 [168] and Ethylene Insensitive 3 (EIN3) [173]. The promoter regions of the CBF/DREB1 TFs lack DRE element, suggesting that they do not regulate their own expression. Further, mutational screens have determined the regulatory aspect of CBFs and identified additional components involved in the regulation of the cold-induced expression of CBFs [174]. ICE1 and ICE2 encode a MYC-like basic helix-loop-helix (bHLH) transcriptional activator and positively regulate CBF1, CBF2 and CBF3 by binding to the MYC recognition sequences present in their promoters. These TFs are known to act upstream to the most of other factors involved in cold stress responses. ICE1 and ICE2 are the master regulators of cold responses and control COR genes and CBF/DREB1 [170, 175]. In response to low temperature, ICE1 is modified (stabilized and activated) by sumoylation involving the SIZ1 SUMO E3 ligase. This promotes ICE1 binding to the CBF3 promoter and increases CBF3 expression [176]. Further, over-expression of ICE1 enhanced the expression of CBF regulon under cold stress and improved the cold tolerance of transgenic Arabidopsis plants [169]. In contrast, ice1 mutant showed inhibition of cold-induced transcription of a CBF3 gene. Further, a dominant negative mutation of ICE1 eliminates the cold-induced CBF3 expression. These mutant plants also showed loss of freezing tolerance [169, 175]. However, the limited impact on the accumulation of CBF2 transcripts in ice1 indicated occurrence of diverse activation mechanisms within the CBF/DREB1 family [169]. Similarly, over-expression of SlICE1 led to improved chilling tolerance by inducing the expression of dehydrin Ci7 homolog (SlDRCi7), SlCBF1 and Δ1-pyrroline-5-carboxylase synthase (SlP5CS) genes in tomato [177]. Recently, it has been reported that a central component of ABA signaling pathway, OPEN STOMATA 1 (OST1), plays a crucial role in cold response. OST1 is induced by cold. OST1 is known to contribute to the increased plant tolerance to freezing by phosphorylating ICE1; a biochemical event which enhances its stability [178]. Another important gene, SCRM, can function as an inducer of CBF expression1 (ICE 1). This gene along with its homolog SCRM2 is essential for the functions of SPEECHLESS (SPCH), MUTE, and FAMA during stomatal development. Both ICE1/SCRM and SCRM2 are redundant proteins and have overlapping functions. The evidences point towards a possible link between the transcriptional regulation of environmental adaptation and stomatal development in plants [179].
Feedback repression of transcription factors that regulates cold-responsive gene expression also seems to be an important mechanism for sustaining perfect cold-induced transcriptome (Fig. 1). For example, high expression of osmotically sensitive (HOS1) gene acts as a negative regulator of ICE1. HOS1 encodes a RING E3 ligase and targets ICE1 and ICE2 for their proteasome-mediated degradation [170, 180]. One of the member of Zn finger transcription factor family, ZAT12, also acts as a negative regulator of DREB1/CBFs, though it is generally induced in this timeframe in Arabidopsis [168, 181]. CBFs are also negatively regulated by MYB15 transcription factor. Knockout mutants of myb15 showed enhanced CBF expression and freezing tolerance after cold acclimation, whereas overexpression showed the opposite effect [171]. CAMTA3 (Calmodulin-binding transcription activator 3) was shown to be a positive regulator of Arabidopsis DREB1C/CBF2, through its binding to the CM2 cis-motif present in the promoter of that gene [172]. A quantitative trait locus COLD1 also contribute in the regulation of signaling pathway of cold tolerance. It encodes a regulator of G-protein signaling and localizes on plasma membrane and endoplasmic reticulum (ER). Its interaction with the α-subunit of G-protein activates Ca2+ channel for sensing decreased temperature and to accelerate G-protein GTPase activity [182].
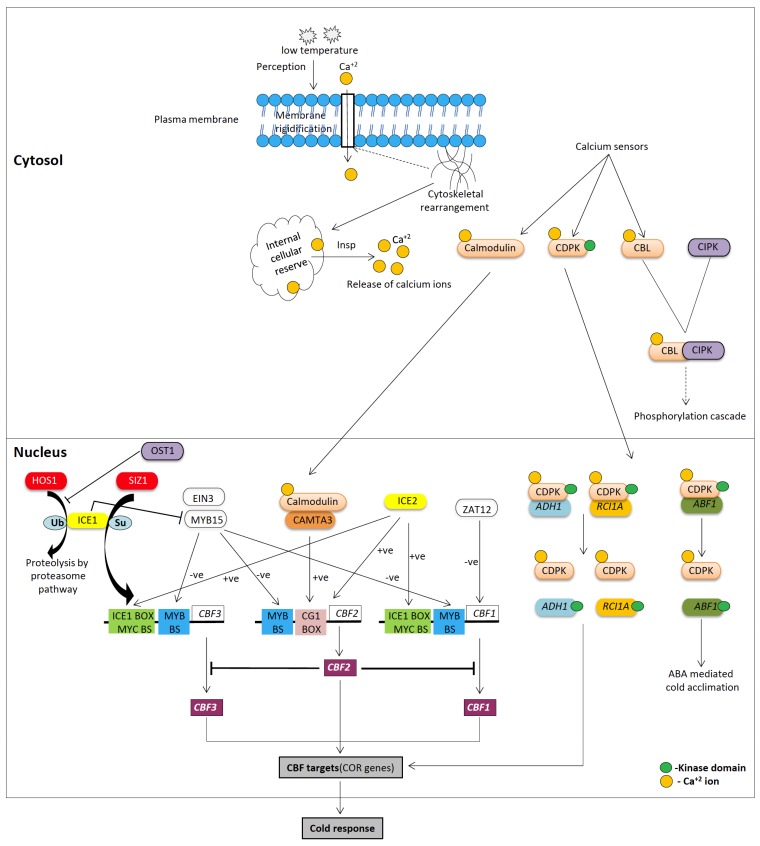
Fig. (1).
Schematic representation of signaling network in response to low temperature stress. It shows the early events of cold perception, leading expression of COR genes regulated by CBF transcription factors. Characterized Ca+2 sensor proteins and their target proteins are also shown. Refer to text for detailed descriptions. CBF/DREB, C-repeat binding factors/DRE-binding proteins; CBL/CIPK, calcineurin B-like protein/CBL-interacting protein kinase; CAMTA, Calmodulin-binding transcription activator; COR, cold-regulated; HOS, high expression of osmotically responsive gene; ICE1, inducer of CBF expression 1; RCI1A, Rare cold inducible 1A and ADH1, Alcohol dehydrogenase 1; ABF1, ABRE binding factor 1; EIN3, Ethylene Insensitive; HOS1, high expression of osmotically responsive genes; SIZ1, E3 SUMO ligase.
Interactions between low temperature and other abiotic stresses are also known to have an effect on cold-regulated gene expression. For example, pre-exposure of plants to NaCl has been found to delay the low-temperature induced expression of cold-inductive genes [183]. Some of these interactions may be mediated by the cold-inducible and drought-inducible transcription factors via interacting with DRE elements. However, the role of other nodes within the signal transduction network has also been suggested and their validation warrants further investigations [65]. In such a similar study, transcriptome profiling of the model legume, Lotus japonicus, under cold stress condition identified different types of cold-inducible transcription factors such as AP2/ERF, NAC, MYB, and WRKY families and other putative novel transcription factors. The findings of this study can serve as a template for future research [184]. Likewise, comparative transcriptome profiling of three maize inbred lines indicated that cold acclimation process in plants involves modifications in the photosynthetic apparatus, cell wall properties and developmental processes [185]. Comparative transcriptomic and proteomic analyses have identified specific regulatory targets during CA (cold acclimation) and DA (de acclimation) processes in Arabidopsis. Based on the accumulated evidences, it is expected that identification of several putative targets of translational regulation under cold stress will help understand the mechanism of RNA regulation during CA and DA in other plants [186]. High-throughput transcriptome analysis of rice germinating seeds of two indica genotypes further identified novel genes related to cold tolerance [187]. Altogether, understanding the cold signaling regulon will help in the transcriptome engineering of crop plants for enhanced tolerance to multiple abiotic stresses.
3.2.4. ABA
ABA plays a central role in abiotic stress tolerance as it is involved in the integration of various stress signals [22, 188, 189]. Cold stress is known to mildly enhance endogenous ABA levels in plants. Further evidence suggests that exogenous application of ABA induces cold tolerance in herbaceous plants [16, 188, 190]. Global transcriptional response to cold stress in chilling tolerant Japonica rice suggested a role of ABA signaling in chilling tolerance [191]. Gene expression analyses have further identified a common set of stress-responsive genes induced under both cold and ABA [45, 192]. These observations suggested that ABA accumulates in response to low temperature and it is important for providing improved freezing tolerance. Further, promoter analysis of cold-inducible genes of Arabidopsis plants revealed the abundance of ABRE in the promoters [149, 193]. Reduced cold acclimation was also reported in ABA-deficient aba1 and ABA-insensitive abi1-1 mutants of Arabidopsis and stress was found to induce lethality in these mutants [152].
Similarly, ABA application was found to induce expression of temperature responsive genes (ZmCOI6.1, ZmACA1, ZmDREB2A and ZmERF3) in Zea mays [194]. Two major cis-acting elements, ABRE and CRT/DRE (C-repeat/DREs), which function in ABA-dependent and ABA-independent manner, respectively, contribute independently or in concert with the ABA-induced gene expression [195]. ABA-dependent gene expression is usually controlled by transcription factors that are part of bZIP (ABRE-binding factors or AREB’s), MYC and MYB families [69]. A global promoter analysis indicated that both ABRE and CRT/DRE are conserved in cold-inducible promoters of soybean and Arabidopsis. Though, ABRE is also conserved in rice, CRT/DREs show variation in cold inducible promoters [196]. ABA can also enhance the expression level of CBF1, CBF2, CBF3, and ICE1 genes, but such induction is considerably lower than that caused by cold [197]. ICE1 also participate in ABA-dependent pathways (glucose and ABA signaling), suggesting that ICE1 might play a new role in cross-talk between ABA-independent and ABA-dependent pathways [198]. Many experts have reported that both ABA-independent and ABA-dependent pathways regulate cold-responsive genes [69, 199].
3.2.5. Cytokinins
Cytokinin signal transduction pathway plays a significant role in cold signaling [200, 201]. The multistep phosphorelay cytokinin signaling pathway is composed of sensor histidine kinases (AHK2, AHK3, AHK4), histidine phosphotransfer proteins (AHPs), and downstream response regulators (ARRs). Transcription of Cytokinin Response Factors (CRFs), the downstream component of this pathway, is induced after exposure to cold (4 °C). Likewise, in comparison to the CRF4 overexpressing plants, the crf4 mutant (lacking the expression of CRF4) plants showed more sensitivity to freezing temperatures [202]. A number of temperature responsive proteins such as LL-diaminopimelate aminotransferase and peroxisomal malate dehydrogenase are involved in early response to cytokinin. Role of calcium has been implicated in the cytokinin-mediated responses under cold stress and a molecular link between cytokinin and calcium signaling has been established. It was observed that inhibition of calcium signaling affected the cytokinin-mediated regulation of several phosphoproteins [203]. Another hint that cytokinin is involved in the regulation of cold tolerance is provided by the fact that both cytokinin- and temperature shocks-altered proteomes share a high proportion of co-regulated proteins [204]. Numerous studies have also established the role of cytokinins in cold mediated adaptive mechanisms in response to the increased concentration of cytokinin [200, 201, 205, 206].
3.2.6. H2O2
Over the years, H2O2 has become an established signaling molecule. Its small size mobile, long half-life (1 ms) and high permeability across membrane allow H2O2 to traverse through cellular membranes and migrate to different compartments to mediate different biological outcomes, including the one that leads to its own synthesis [207-213]. H2O2 performs a vital role in induction of physiological, biochemical and molecular responses under stress conditions in plants [214].
At low concentration, it functions generally as a mediator of signaling pathways and results in stress acclimation; however, at higher concentration it causes cellular damage and cell death. The multi-functionality of H2O2 actions such as stress alleviation on one side and the risks at higher concentrations requires a very strict control of H2O2 concentration in plant cells. H2O2 has been found to accumulate in response to various biotic and abiotic stresses, including cold [33, 215]. Likewise, exogenous application of H2O2 induces low temperature stress tolerance in maize. During cold acclimation H2O2 act as a signal to induce synthesis of ROS-scavenging enzymes [216, 217]. H2O2 enhance the antioxidant capacity of cells by alleviating the activities of antioxidant enzymes, such as Ascorbate Peroxidase (APX), Catalase (CAT), and Superoxide Dismutase (SOD) [218]. It has been
observed that exogenous application of H2O2 resulted in increased activities of APX, CAT, GPX, and GST in manila grass and APX, POD and GR activities in mascarene which protect plants against damage by chilling [219]. Additional studies revealed that H2O2 functions as a mediator in stress responses, via interacting with many other important signal molecules (Ca+2, SA, ABA, JA, ethylene, NO) [220-224]. Functional characterization of a cold induced MfSAMS1 (Medicago sativa subsp. falcata S-adenosyl methionine synthetase 1) in tobacco showed that H2O2, ABA and NO interactions mediated its cold-induced expression and cold acclimation in falcate. Overexpression of MfSAMS1 favoured polyamine synthesis and oxidation. As a consequence, it improved H2O2-induced antioxidant protection and led to enhanced cold tolerance in transgenic plants [225]. H2O2 has also been observed to communicate with ethylene in response to cold stress [226]. Notably, H2O2 regulates the activities of many signaling components such as protein kinases, protein phosphatases and various transcription factors (TFs) [69, 227].
3.2.7. Cytoskeleton Rearrangement
Plant cytoskeleton maintains proximity with the plasma membrane that provides an important platform for signal perception and transduction [228, 229]. The bond between plasma membrane and cytoskeleton arises through a hydrophobic domain, which is present either on the tubulin molecule or it is facilitated indirectly through interaction with an integral membrane protein [230]. For example, Phospholipase D (PLD) has been confirmed for having the ability to make structural and signaling connections between cortical microtubules and the plasma membrane [231-234]. Role of cytoskeletal reorganization and PLD activation in cold acclimation has also been reported [235, 236]. In this mechanism, activation of PLD leads to cytoskeletal reorganization by releasing the cortical array of microtubules from plasma membrane [232]. Further, the enhanced production of PLD has been found to confer improved frost tolerance after cold acclimation [237].
The cytoskeleton plays a key role as low temperature sensor in plants during cold stress signaling and acclimation process [238, 239]. The activities of various ion channels in plant cells have been analysed to show the importance of cytoskeleton reorganization [239-242]. Based on the Ca+2 channel activity under cold, it was found that specific type of cytoskeletal components (microtubules and actin filaments) are involved in cold sensing by regulating the activity of these ion channels. The subsequent membrane rigidification further assists this hypothesis [51, 243, 244]. A synergistic increase in Ca+2 influx in the cold shocked tobacco plants treated with oryzalin and cytochalasin (destabilizers of microtubules and microfilaments, respectively) further established the link between cytoskeleton and Ca+2 ions [245]. Exposure of plants to low temperatures has been found to result in destabilization and depolymerisation of microtubules. Upon continuous exposure to low temperature, cold labile microtubules are swapped with cold stable microtubules. In cold stress-treated root tip cells of cucumber (Cucumis sativus L.), stable cortical microtubules were found to be located both under the plasma membrane as well as in the cytoplasm. It was suggested that these additional microtubules might be associated with organelles [246]. In wheat (T. aestivum L.), three members of α-tubulin gene family were induced during cold acclimation. A fourth member showed increased mRNA level for up to 14 days during cold acclimation and had decreased levels after 36 days of cold treatment [247]. In tobacco, a mutational screen identified both aryl carbamate (a blocker of microtubule assembly) and chilling tolerant mutants. The carbamate tolerant mutants were also resistant to chilling stress. It was observed that the stability of microtubules in cold treatment can be improved by Microtubule Associated Proteins (MAPs). In case of Arabidopsis thaliana, nine such genes constitute the evolutionarily conserved MAP65 family and the presence of AtMAP65-1 provides more resistance to microtubules in cold stress [248]. An Actin Depolymerizing Factor (ADF) has also been characterized for its role in cold acclimation in wheat. The accumulation of ADF was higher in freezing tolerant wheat cultivars compared to less tolerant cultivar. Thus, cytoskeletal rearrangements and again remodelling of the actin cytoskeleton was proposed to be very important for the improvement of frost tolerance in plants [249].
CONCLUSION AND FUTURE PROSPECTS
Research on cold tolerance mechanism has made our understanding better on adaptation of plants under cold stress but there is more to be discovered in this field. The current review covers the involvement and acting mechanisms of different players in cold signal transduction by which plants develop cold tolerance. Briefly, sensing of low temperature is initiated by plasma membrane rigidification or by histidine two component system or calcium channels which lead to influx of calcium ions into cytosol. The signal is then transferred to the nucleus by decoders of calcium for transcriptional regulation through CBF-dependent or CBF-independent mechanisms. Though, the role of master regulator ICE1 in cold signaling is largely known, more research about the mechanism of modification and regulation of ICE1 is necessary. Moreover, better understanding of the molecular mechanisms underlying the cross talk among different signaling pathways at various points is necessary and remains an area of intense research in the near future.
ACKNOWLEDGEMENTS
Research in the author’s laboratory is supported by the grants from Department of Biotechnology, Govt. of India and Department of Science and Technology, Govt. of India (IFA-LSPA-15). RK thanks Prof. R. P. Sharma, Department of Plant Sciences, University of Hyderabad, for his support in carrying out the research work.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Rathore A.C., Raizada A., Prakash J.J., Sharda V.N. Impact of chilling injury on common fruit plants in the Doon Valley. Curr. Sci. 2012;102(8):1107–1111. [Google Scholar]
- 2.Miura K., Furumoto T. Cold signaling and cold response in plants. Int. J. Mol. Sci. 2013;14(3):5312–5337. doi: 10.3390/ijms14035312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solanke A.U., Sharma A.K. Signal transduction during cold stress in plants. Physiol. Mol. Biol. Plants. 2008;14(1-2):69–79. doi: 10.1007/s12298-008-0006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu Z., Zhang R., Wang D., Qiu M., Feng H., Zhang N., Shen Q. Enhanced control of cucumber wilt disease by Bacillus amyloliquefaciens SQR9 by altering the regulation of Its DegU phosphorylation. Appl. Environ. Microbiol. 2014;80(9):2941–2950. doi: 10.1128/AEM.03943-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prasad T.K., Anderson M.D., Martin B.A., Stewart C.R. Evidence for chilling-induced oxidative stress in maize seedlings and a regulatory role for hydrogen-peroxide. Plant Cell. 1994;6(1):65–74. doi: 10.1105/tpc.6.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foyer C.H. LopezDelgado, H.; Dat, J.F.; Scott, I.M. Hydrogen peroxide- and glutathione-associated mechanisms of acclimatory stress tolerance and signalling. Physiol. Plant. 1997;100(2):241–254. [Google Scholar]
- 7.Abbasi F., Onodera H., Toki S., Tanaka H., Komatsu S. OsCDPKI3, a calcium-dependent protein kinase gene from rice, is induced by cold and gibberellin in rice leaf sheath. Plant Mol. Biol. 2004;55(4):541–552. doi: 10.1007/s11103-004-1178-y. [DOI] [PubMed] [Google Scholar]
- 8.Thomashow M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- 9.Sanghera G.S., Wani S.H., Hussain W., Singh N.B. Engineering cold stress tolerance in crop plants. Curr. Genomics. 2011;12(1):30–43. doi: 10.2174/138920211794520178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siddiqui K.S., Cavicchioli R. Cold-adapted enzymes. Annu. Rev. Biochem. 2006;75:403–433. doi: 10.1146/annurev.biochem.75.103004.142723. [DOI] [PubMed] [Google Scholar]
- 11.Murata N., Los D.A. Membrane fluidity and temperature perception. Plant Physiol. 1997;115(3):875–879. doi: 10.1104/pp.115.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nayyar H., Chander K., Kumar S., Bains T. Glycine betaine mitigates cold stress damage in Chickpea. Agron. Sustain. Dev. 2005;25(3):381–388. [Google Scholar]
- 13.Nayyar H., Bains T., Kumar S. Low temperature induced floral abortion in chickpea: relationship to abscisic acid and cryoprotectants in reproductive organs. Environ. Exp. Bot. 2005;53(1):39–47. [Google Scholar]
- 14.Nayyar H., Bains T.S., Kumar S. Chilling stressed chickpea seedlings: effect of cold acclimation, calcium and abscisic acid on cryoprotective solutes and oxidative damage. Environ. Exp. Bot. 2005;54(3):275–285. [Google Scholar]
- 15.Nayyar H., Bains T.S., Kumar S., Kaur G. Chilling effects during seed filling on accumulation of seed reserves and yield of chickpea. J. Sci. Food Agric. 2005;85(11):1925–1930. [Google Scholar]
- 16.Nayyar H., Chander S. Protective effects of polyamines against oxidative stress induced by water and cold stress in chickpea. J. Agron. Crop Sci. 2004;190(5):355–365. [Google Scholar]
- 17.Dowgert M.F., Steponkus P.L. Behavior of the plasma membrane of isolated protoplasts during a freeze-thaw cycle. Plant Physiol. 1984;75(4):1139–1151. doi: 10.1104/pp.75.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olien C.R., Smith M.N. Ice adhesions in relation to freeze stress. Plant Physiol. 1977;60(4):499–503. doi: 10.1104/pp.60.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gusta L.V., Wisniewski M., Nesbitt N.T., Gusta M.L. The effect of water, sugars, and proteins on the pattern of ice nucleation and propagation in acclimated and nonacclimated canola leaves. Plant Physiol. 2004;135(3):1642–1653. doi: 10.1104/pp.103.028308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruelland E., Vaultier M.N., Zachowski A., Hurry V. Cold signalling and cold acclimation in plants. Adv. Bot. Res. 2009;49:35–150. [Google Scholar]
- 21.Smallwood M., Bowles D.J. Plants in a cold climate. Philosophical Transactions of the Royal Society of London Series B-Biol. Sci. 2002;357(1423):831–846. doi: 10.1098/rstb.2002.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahajan S., Tuteja N. Cold, salinity and drought stresses: An overview. Arch. Biochem. Biophys. 2005;444(2):139–158. doi: 10.1016/j.abb.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 23.Wen J.Q., Oono K., Imai R. Two novel mitogen-activated protein signaling components, OsMEK1 and OsMAP1, are involved in a moderate low-temperature signaling pathway in rice. Plant Physiol. 2002;129(4):1880–1891. doi: 10.1104/pp.006072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beck E.H., Heim R., Hansen J. Plant resistance to cold stress: Mechanisms and environmental signals triggering frost hardening and dehardening. J. Biosci. 2004;29(4):449–459. doi: 10.1007/BF02712118. [DOI] [PubMed] [Google Scholar]
- 25.Sarkar A.K. Ice formation and propagation in plants of cold climate region. Int. J. Rec. Sci. Res. 2015;6(10):7098–7102. [Google Scholar]
- 26.Hughes M.A., Dunn M.A. The molecular biology of plant acclimation to low temperature. J. Exp. Bot. 1996;47(296):291–305. [Google Scholar]
- 27.Mohapatra S.S., Poole R.J., Dhindsa R.S. Abscisic Acid-regulated gene expression in relation to freezing tolerance in alfalfa. Plant Physiol. 1988;87(2):468–473. doi: 10.1104/pp.87.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knight M.R., Knight H. Low-temperature perception leading to gene expression and cold tolerance in higher plants. New Phytol. 2012;195(4):737–751. doi: 10.1111/j.1469-8137.2012.04239.x. [DOI] [PubMed] [Google Scholar]
- 29.Webb M.S., Uemura M., Steponkus P.L. A comparison of freezing-injury in oat and rye: two cereals at the extremes of freezing tolerance. Plant Physiol. 1994;104(2):467–478. doi: 10.1104/pp.104.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uemura M., Steponkus P.L. Effect of cold acclimation on the lipid composition of the inner and outer membrane of the chloroplast envelope isolated from rye leaves. Plant Physiol. 1997;114(4):1493–1500. doi: 10.1104/pp.114.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao C., Zhang Z., Xie S., Si T., Li Y., Zhu J.K. Mutational evidence for the critical role of CBF genes in cold acclimation in Arabidopsis. Plant Physiol. 2016;171:2744–2759. doi: 10.1104/pp.16.00533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strauss G., Hauser H. Stabilization of lipid bilayer vesicles by sucrose during freezing. Proc. Natl. Acad. Sci. USA. 1986;83(8):2422–2426. doi: 10.1073/pnas.83.8.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.OKane D.; Gill, V.; Boyd, P.; Burdon, B. Chilling, oxidative stress and antioxidant responses in Arabidopsis thaliana callus. Planta. 1996;198(3):371–377. doi: 10.1007/BF00620053. [DOI] [PubMed] [Google Scholar]
- 34.Guo Z., Ou W., Lu S., Zhong Q. Differential responses of antioxidative system to chilling and drought in four rice cultivars differing in sensitivity. Plant Physiol. Biochem. 2006;44(11-12):828–836. doi: 10.1016/j.plaphy.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 35.Guo Z., Ou W., Lu S., Zhong Q. Differential responses of antioxidative system to chilling and drought in four rice cultivars differing in sensitivity. Plant Physiol. Biochem. 2006;44(11-12):828–836. doi: 10.1016/j.plaphy.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 36.Anderson M.D., Prasad T.K., Martin B.A., Stewart C.R. Differential gene-expression in chilling-acclimated maize seedlings and evidence for the involvement of abscisic-acid in chilling tolerance. Plant Physiol. 1994;105(1):331–339. doi: 10.1104/pp.105.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo Z.F., Tan H.Q., Zhu Z.H., Lu S.Y., Zhou B.Y. Effect of intermediates on ascorbic acid and oxalate biosynthesis of rice and in relation to its stress resistance. Plant Physiol. Biochem. 2005;43(10-11):955–962. doi: 10.1016/j.plaphy.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki N., Koussevitzky S., Mittler R., Miller G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 2012;35(2):259–270. doi: 10.1111/j.1365-3040.2011.02336.x. [DOI] [PubMed] [Google Scholar]
- 39.Medina J., Catala R., Salinas J. The CBFs: Three Arabidopsis transcription factors to cold acclimate. Plant Sci. 2011;180(1):3–11. doi: 10.1016/j.plantsci.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 40.Minorsky P.V. Temperature sensing by plants - a review and hypothesis. Plant Cell Environ. 1989;12(2):119–135. [Google Scholar]
- 41.Vigh L., Los D.A., Horvath I., Murata N. The primary signal in the biological perception of temperature: Pd-catalyzed hydrogenation of membrane lipids stimulated the expression of the desA gene in Synechocystis PCC6803. Proc. Natl. Acad. Sci. USA. 1993;90(19):9090–9094. doi: 10.1073/pnas.90.19.9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uemura M., Tominaga Y., Nakagawara C., Shigematsu S., Minami A., Kawamura Y. Responses of the plasma membrane to low temperatures. Physiol. Plant. 2006;126(1):81–89. [Google Scholar]
- 43.Vaultier M.N., Cantrel C., Vergnolle C., Justin A.M., Demandre C., Benhassaine-Kesri G., Cicek D., Zachowski A., Ruelland E. Desaturase mutants reveal that membrane rigidification acts as a cold perception mechanism upstream of the diacylglycerol kinase pathway in Arabidopsis cells. FEBS Lett. 2006;580(17):4218–4223. doi: 10.1016/j.febslet.2006.06.083. [DOI] [PubMed] [Google Scholar]
- 44.Wang X.M., Li W.Q., Li M.Y., Welti R. Profiling lipid changes in plant response to low temperatures. Physiol. Plant. 2006;126(1):90–96. [Google Scholar]
- 45.Xiong L.M., Schumaker K.S., Zhu J.K. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002;14:S165–S183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knight M.R., Campbell A.K., Smith S.M., Trewavas A.J. Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature. 1991;352(6335):524–526. doi: 10.1038/352524a0. [DOI] [PubMed] [Google Scholar]
- 47.Knight H., Trewavas A.J., Knight M.R. Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell. 1996;8(3):489–503. doi: 10.1105/tpc.8.3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Monroy A.F., Sarhan F., Dhindsa R.S. Cold-induced changes in freezing tolerance, protein-phosphorylation, and gene-expression - evidence for a role of calcium. Plant Physiol. 1993;102(4):1227–1235. doi: 10.1104/pp.102.4.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monroy A.F., Dhindsa R.S. Low-temperature signal-transduction - induction of cold acclimation-specific genes of alfalfa by calcium at 25-degrees-C. Plant Cell. 1995;7(3):321–331. doi: 10.1105/tpc.7.3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu X.H., Feng Y., Liang G.M., Liu N., Zhu J.K. Aequorin-based luminescence imaging reveals stimulus- and tissue-specific Ca2+ dynamics in Arabidopsis plants. Mol. Plant. 2013;6(2):444–455. doi: 10.1093/mp/sst013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orvar B.L., Sangwan V., Omann F., Dhindsa R.S. Early steps in cold sensing by plant cells: the role of actin cytoskeleton and membrane fluidity. Plant J. 2000;23(6):785–794. doi: 10.1046/j.1365-313x.2000.00845.x. [DOI] [PubMed] [Google Scholar]
- 52.Sangwan V., Foulds I., Singh J., Dhindsa R.S. Cold-activation of Brassica napus BN115 promoter is mediated by structural changes in membranes and cytoskeleton, and requires Ca2+ influx. Plant J. 2001;27(1):1–12. doi: 10.1046/j.1365-313x.2001.01052.x. [DOI] [PubMed] [Google Scholar]
- 53.Chinnusamy V., Zhu J.K., Sunkar R. Gene regulation during cold stress acclimation in plants. Methods Mol. Biol. 2010;639:39–55. doi: 10.1007/978-1-60761-702-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Urao T., Yakubov B., Yamaguchi-Shinozaki K., Shinozaki K. Stress-responsive expression of genes for two-component response regulator-like proteins in Arabidopsis thaliana. FEBS Lett. 1998;427(2):175–178. doi: 10.1016/s0014-5793(98)00418-9. [DOI] [PubMed] [Google Scholar]
- 55.Wulfetange K., Lomin S.N., Romanov G.A., Stolz A., Heyl A., Schmulling T. The cytokinin receptors of Arabidopsis are located mainly to the endoplasmic reticulum. Plant Physiol. 2011;156(4):1808–1818. doi: 10.1104/pp.111.180539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suzuki I., Los D.A., Kanesaki Y., Mikami K., Murata N. The pathway for perception and transduction of low-temperature signals in Synechocystis. EMBO J. 2000;19(6):1327–1334. doi: 10.1093/emboj/19.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki I., Kanesaki Y., Mikami K., Kanehisa M., Murata N. Cold-regulated genes under control of the cold sensor Hik33 in Synechocystis. Mol. Microbiol. 2001;40(1):235–244. doi: 10.1046/j.1365-2958.2001.02379.x. [DOI] [PubMed] [Google Scholar]
- 58.Cybulski L.E., de Mendoza D. Bilayer hydrophobic thickness and integral membrane protein function. Curr. Protein Pept. Sci. 2011;12(8):760–766. doi: 10.2174/138920311798841681. [DOI] [PubMed] [Google Scholar]
- 59.Hong S.W., Jon J.H., Kwak J.M., Nam H.G. Identification of a receptor-like protein kinase gene rapidly induced by abscisic acid, dehydration, high salt, and cold treatments in Arabidopsis thaliana. Plant Physiol. 1997;113(4):1203–1212. doi: 10.1104/pp.113.4.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kreps J.A., Wu Y.J., Chang H.S., Zhu T., Wang X., Harper J.F. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130(4):2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar S.V., Wigge P.A. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140(1):136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 62.Kumar S.V., Lucyshyn D., Jaeger K.E., Alos E., Alvey E., Harberd N.P., Wigge P.A. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature. 2012;484(7393):242–U127. doi: 10.1038/nature10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Galon Y., Finkler A., Fromm H. Calcium-regulated transcription in plants. Mol. Plant. 2010;3(4):653–669. doi: 10.1093/mp/ssq019. [DOI] [PubMed] [Google Scholar]
- 64.Ranty B., Aldon D., Cotelle V., Galaud J.P., Thuleau P., Mazars C. Calcium sensors as key hubs in plant responses to biotic and abiotic stresses. Front. Plant Sci. 2016;7:327. doi: 10.3389/fpls.2016.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Knight H., Knight M.R. Abiotic stress signalling pathways: specificity and cross-talk. Trends Plant Sci. 2001;6(6):262–267. doi: 10.1016/s1360-1385(01)01946-x. [DOI] [PubMed] [Google Scholar]
- 66.Jenkins G.I. Signal transduction networks and the integration of responses to environmental stimuli. Adv. Bot. Res. Adv. Plant Patho. 1999;29:53–73. [Google Scholar]
- 67.Malho R., Moutinho A., van der Luit A., Trewavas A.J. Spatial characteristics of calcium signalling: the calcium wave as a basic unit in plant cell calcium signalling. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1998;353(1374):1463–1473. [Google Scholar]
- 68.Chinnusamy V., Zhu J., Zhu J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007;12(10):444–451. doi: 10.1016/j.tplants.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 69.Thakur P., Nayyar H. Facing the Cold Stress by Plants in the Changing Environment: Sensing, Signaling, and Defending Mechanisms. In: Tuteja N., Singh Gill S., editors. Plant Acclimation to Environmental Stress. New York, NY: Springer New York; 2013. pp. 29–69. [Google Scholar]
- 70.Poovaiah B.W., Du L., Wang H., Yang T. Recent advances in calcium/calmodulin-mediated signaling with an emphasis on plant-microbe interactions. Plant Physiol. 2013;163(2):531–542. doi: 10.1104/pp.113.220780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Day I.S., Reddy V.S., Ali G.S., Reddy A.S.N. 2002.
- 72.Boonburapong B., Buaboocha T. Genome-wide identification and analyses of the rice calmodulin and related potential calcium sensor proteins. BMC Plant Biol. 2007;7:4. doi: 10.1186/1471-2229-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reddy A.S., Ali G.S., Celesnik H., Day I.S. Coping with stresses: Roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell. 2011;23(6):2010–2032. doi: 10.1105/tpc.111.084988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sanders D., Pelloux J., Brownlee C., Harper J.F. Calcium at the crossroads of signaling. Plant Cell. 2002;14:S401–S417. doi: 10.1105/tpc.002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kudla J., Batistic O., Hashimoto K. Calcium signals: The lead currency of plant information processing. Plant Cell. 2010;22(3):541–563. doi: 10.1105/tpc.109.072686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reddy V.S., Day I.S., Thomas T., Reddy A.S. KIC, a novel Ca2+ binding protein with one EF-hand motif, interacts with a microtubule motor protein and regulates trichome morphogenesis. Plant Cell. 2004;16(1):185–200. doi: 10.1105/tpc.016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McCormack E., Tsai Y.C., Braam J. Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 2005;10(8):383–389. doi: 10.1016/j.tplants.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 78.Luan S. The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 2009;14(1):37–42. doi: 10.1016/j.tplants.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 79.DeFalco T.A., Bender K.W., Snedden W.A. Breaking the code: Ca2+ sensors in plant signalling. Biochem. J. 2010;425:27–40. doi: 10.1042/BJ20091147. [DOI] [PubMed] [Google Scholar]
- 80.Roberts D.M., Harmon A.C. Calcium-modulated proteins - targets of intracellular calcium signals in higher-plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1992;43:375–414. [Google Scholar]
- 81.Cheng S.H., Willmann M.R., Chen H.C., Sheen J. Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 2002;129(2):469–485. doi: 10.1104/pp.005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shimazaki K., Kinoshita T., Nishimura M. Involvement of calmodulin and calmodulin-dependent myosin light chain kinase in blue light-dependent H+ pumping by guard-cell protoplasts from vicia-faba L. Plant Physiol. 1992;99(4):1416–1421. doi: 10.1104/pp.99.4.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pandey S., Tiwari S.B., Tyagi W., Reddy M.K., Upadhyaya K.C., Sopory S.K. Ca2+/CaM-dependent kinase from pea is stress regulated and in vitro phosphorylates a protein that binds to AtCaM5 promoter. Eur. J. Biochem. 2002;269(13):3193–3204. doi: 10.1046/j.1432-1033.2002.02994.x. [DOI] [PubMed] [Google Scholar]
- 84.Wang J.P., Munyampundu J.P., Xu Y.P., Cai X.Z. Phylogeny of plant calcium and calmodulin-dependent protein kinases(CCaMKs) and functional analyses of tomato CCaMK in disease resistance. Front. Plant Sci. 2015;6:1075. doi: 10.3389/fpls.2015.01075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang T.B., Poovaiah B.W. Calcium/calmodulin-mediated signal network in plants. Trends Plant Sci. 2003;8(10):505–512. doi: 10.1016/j.tplants.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 86.Harper J.F., Harmon A. Plants, symbiosis and parasites: A calcium signalling connection. Nat. Rev. Mol. Cell Biol. 2005;6(7):555–566. doi: 10.1038/nrm1679. [DOI] [PubMed] [Google Scholar]
- 87.de Silva K., Laska B., Brown C., Sederoff H.W., Khodakovskaya M. Arabidopsis thaliana calcium-dependent lipid-binding protein(AtCLB): a novel repressor of abiotic stress response. J. Exp. Bot. 2011;62(8):2679–2689. doi: 10.1093/jxb/erq468. [DOI] [PubMed] [Google Scholar]
- 88.Tuteja N. In: Integrated Calcium Signaling in Plants. Signaling in Plants, Mancuso, S. Baluka F.E., editor. Berlin, Heidelberg: Springer Berlin Heidelberg; 2009. pp. 29–49. [Google Scholar]
- 89.Kim M.C., Chung W.S., Yun D.J., Cho M.J. Calcium and calmodulin-mediated regulation of gene expression in plants. Mol. Plant. 2009;2(1):13–21. doi: 10.1093/mp/ssn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tahtiharju S., Sangwan V., Monroy A.F., Dhindsa R.S., Borg M. The induction of kin genes in cold-acclimating Arabidopsis thaliana. Evidence of a role for calcium. Planta. 1997;203(4):442–447. doi: 10.1007/s004250050212. [DOI] [PubMed] [Google Scholar]
- 91.Bender K.W., Snedden W.A. Calmodulin-related proteins step out from the shadow of their namesake. Plant Physiol. 2013;163(2):486–495. doi: 10.1104/pp.113.221069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhu X., Dunand C., Snedden W., Galaud J.P. CaM and CML emergence in the green lineage. Trends Plant Sci. 2015;20(8):483–489. doi: 10.1016/j.tplants.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 93.Boonburapong B., Buaboocha T. Genome-wide identification and analyses of the rice calmodulin and related potential calcium sensor proteins. BMC Plant Biol. 2007;7:4. doi: 10.1186/1471-2229-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao Y., Liu W., Xu Y.P., Cao J.Y., Braam J., Cai X.Z. Genome-wide identification and functional analyses of calmodulin genes in Solanaceous species. BMC Plant Biol. 2013;13:70. doi: 10.1186/1471-2229-13-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Townley H.E., Knight M.R. Calmodulin as a potential negative regulator of Arabidopsis COR gene expression. Plant Physiol. 2002;128(4):1169–1172. doi: 10.1104/pp.010814. [DOI] [PubMed] [Google Scholar]
- 96.Liu H.T., Li G.L., Chang H., Sun D.Y., Zhou R.G., Li B. Calmodulin-binding protein phosphatase PP7 is involved in thermotolerance in Arabidopsis. Plant Cell Environ. 2007;30(2):156–164. doi: 10.1111/j.1365-3040.2006.01613.x. [DOI] [PubMed] [Google Scholar]
- 97.Liu J.P., Zhu J.K. A calcium sensor homolog required for plant salt tolerance. Science. 1998;280(5371):1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- 98.Kudla J., Xu Q., Harter K., Gruissem W., Luan S. Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc. Natl. Acad. Sci. USA. 1999;96(8):4718–4723. doi: 10.1073/pnas.96.8.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gong D.M., Guo Y., Schumaker K.S., Zhu J.K. The SOS3 family of calcium sensors and SOS2 family of protein kinases in Arabidopsis. Plant Physiol. 2004;134(3):919–926. doi: 10.1104/pp.103.037440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Luan S., Kudla J., Rodriguez-Concepcion M., Yalovsky S., Gruissem W. Calmodulins and calcineurin B-like proteins: Calcium sensors for specific signal response coupling in plants. Plant Cell. 2002;14:S389–S400. doi: 10.1105/tpc.001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kolukisaoglu U., Weinl S., Blazevic D., Batistic O., Kudla J. Calcium sensors and their interacting protein kinases: Genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 2004;134(1):43–58. doi: 10.1104/pp.103.033068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Albrecht V., Weinl S., Blazevic D., D’Angelo C., Batistic O., Kolukisaoglu U., Bock R., Schulz B., Harter K., Kudla J. The calcium sensor CBL1 integrates plant responses to abiotic stresses. Plant J. 2003;36(4):457–470. doi: 10.1046/j.1365-313x.2003.01892.x. [DOI] [PubMed] [Google Scholar]
- 103.Huang C.L., Ding S., Zhang H., Du H., An L.Z. CIPK7 is involved in cold response by interacting with CBL1 in Arabidopsis thaliana. Plant Sci. 2011;181(1):57–64. doi: 10.1016/j.plantsci.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 104.Lyzenga W.J., Liu H., Schofield A., Muise-Hennessey A., Stone S.L. Arabidopsis CIPK26 interacts with KEG, components of the ABA signalling network and is degraded by the ubiquitin-proteasome system. J. Exp. Bot. 2013;64(10):2779–2791. doi: 10.1093/jxb/ert123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Meena M.K., Ghawana S., Dwivedi V., Roy A., Chattopadhyay D. Expression of chickpea CIPK25 enhances root growth and tolerance to dehydration and salt stress in transgenic tobacco. Front. Plant Sci. 2015;6:683. doi: 10.3389/fpls.2015.00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shin H.S., Brown R.M. GTPase activity and biochemical characterization of a recombinant cotton fiber annexin. Plant Physiol. 1999;119(3):925–934. doi: 10.1104/pp.119.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim M.C., Panstruga R., Elliott C., Muller J., Devoto A., Yoon H.W., Park H.C., Cho M.J., Schulze-Lefert P. Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature. 2002;416(6879):447–450. doi: 10.1038/416447a. [DOI] [PubMed] [Google Scholar]
- 108.Kim K.N., Cheong Y.H., Grant J.J., Pandey G.K., Luan S. CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell. 2003;15(2):411–423. doi: 10.1105/tpc.006858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xiang Y., Huang Y., Xiong L. Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 2007;144(3):1416–1428. doi: 10.1104/pp.107.101295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Harmon A.C., Gribskov M., Harper J.F. CDPKs - a kinase for every Ca2+ signal? Trends Plant Sci. 2000;5(4):154–159. doi: 10.1016/s1360-1385(00)01577-6. [DOI] [PubMed] [Google Scholar]
- 111.Ludwig A.A., Romeis T., Jones J.D. CDPK-mediated signalling pathways: Specificity and cross-talk. J. Exp. Bot. 2004;55(395):181–188. doi: 10.1093/jxb/erh008. [DOI] [PubMed] [Google Scholar]
- 112.Chinnusamy V., Schumaker K., Zhu J.K. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J. Exp. Bot. 2004;55(395):225–236. doi: 10.1093/jxb/erh005. [DOI] [PubMed] [Google Scholar]
- 113.Klimecka M., Muszynska G. Structure and functions of plant calcium-dependent protein kinases. Acta Biochim. Pol. 2007;54(2):219–233. [PubMed] [Google Scholar]
- 114.Li A.L., Zhu Y.F., Tan X.M., Wang X., Wei B., Guo H.Z., Zhang Z.L., Chen X.B., Zhao G.Y., Kong X.Y., Jia J.Z., Mao L. Evolutionary and functional study of the CDPK gene family in wheat (Triticum aestivum L.). Plant Mol. Biol. 2008;66(4):429–443. doi: 10.1007/s11103-007-9281-5. [DOI] [PubMed] [Google Scholar]
- 115.Hu Z., Lv X., Xia X., Zhou J., Shi K., Yu J., Zhou Y. Genome-wide identification and expression analysis of calcium-dependent protein kinase in tomato. Front. Plant Sci. 2016;7:469. doi: 10.3389/fpls.2016.00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ray S., Agarwal P., Arora R., Kapoor S., Tyagi A.K. Expression analysis of calcium-dependent protein kinase gene family during reproductive development and abiotic stress conditions in rice (Oryza sativa L. ssp indica). Mol. Genet. Genomics. 2007;278(5):493–505. doi: 10.1007/s00438-007-0267-4. [DOI] [PubMed] [Google Scholar]
- 117.Harmon A.C., Gribskov M., Harper J.F. CDPKs - a kinase for every Ca2+ signal? Trends Plant Sci. 2000;5(4):154–159. doi: 10.1016/s1360-1385(00)01577-6. [DOI] [PubMed] [Google Scholar]
- 118.Ito T., Nakata M., Fukazawa J., Ishida S., Takahashi Y. Alteration of substrate specificity: the variable N-terminal domain of tobacco Ca2+-dependent protein kinase is important for substrate recognition. Plant Cell. 2010;22(5):1592–1604. doi: 10.1105/tpc.109.073577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Christodoulou J., Malmendal A., Harper J.F., Chazin W.J. Evidence for differing roles for each lobe of the calmodulin-like domain in a calcium-dependent protein kinase. J. Biol. Chem. 2004;279(28):29092–29100. doi: 10.1074/jbc.M401297200. [DOI] [PubMed] [Google Scholar]
- 120.Wernimont A.K., Amani M., Qiu W., Pizarro J.C., Artz J.D., Lin Y.H., Lew J., Hutchinson A., Hui R. Structures of parasitic CDPK domains point to a common mechanism of activation. Proteins-Struct. Funct. Bioinfo. 2011;79(3):803–820. doi: 10.1002/prot.22919. [DOI] [PubMed] [Google Scholar]
- 121.Liese A., Romeis T. Biochemical regulation of in vivo function of plant calcium-dependent protein kinases(CDPK). B. B. A. Mol. Cell Res. 2013;1833(7):1582–1589. doi: 10.1016/j.bbamcr.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 122.Harper J.E., Breton G., Harmon A. Decoding Ca2+ signals through plant protein kinases. Annu. Rev. Plant Biol. 2004;55:263–288. doi: 10.1146/annurev.arplant.55.031903.141627. [DOI] [PubMed] [Google Scholar]
- 123.Gao X., Cox K.L. Jr.; He, P. Functions of calcium-dependent protein kinases in plant innate immunity. Plants(Basel) 2014;3(1):160–176. doi: 10.3390/plants3010160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Weckwerth P., Ehlert B., Romeis T. ZmCPK1, a calcium-independent kinase member of the Zea mays CDPK gene family, functions as a negative regulator in cold stress signalling. Plant Cell Environ. 2015;38(3):544–558. doi: 10.1111/pce.12414. [DOI] [PubMed] [Google Scholar]
- 125.Tuteja N., Mahajan S. Calcium signaling network in plants: an overview. Plant Signal. Behav. 2007;2(2):79–85. doi: 10.4161/psb.2.2.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mori I.C., Murata Y., Yang Y.Z., Munemasa S., Wang Y.F., Andreoli S., Tiriac H., Alonso J.M., Harper J.F., Ecker J.R., Kwak J.M., Schroeder J.I. CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca2+-permeable channels and stomatal closure. PLoS Biol. 2006;4(10):1749–1762. doi: 10.1371/journal.pbio.0040327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dodd A.N., Kudla J., Sanders D. The language of calcium signaling. Annu. Rev. Plant Biol. 2010;61:593–620. doi: 10.1146/annurev-arplant-070109-104628. [DOI] [PubMed] [Google Scholar]
- 128.Sheen J. Ca2+-dependent protein kinases and stress signal transduction in plants. Science. 1996;274(5294):1900–1902. doi: 10.1126/science.274.5294.1900. [DOI] [PubMed] [Google Scholar]
- 129.Martin M.L., Busconi L. A rice membrane-bound calcium-dependent protein kinase is activated in response to low temperature. Plant Physiol. 2001;125(3):1442–1449. doi: 10.1104/pp.125.3.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Saijo Y., Hata S., Kyozuka J., Shimamoto K., Izui K. Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 2000;23(3):319–327. doi: 10.1046/j.1365-313x.2000.00787.x. [DOI] [PubMed] [Google Scholar]
- 131.Barrero-Gil J., Salinas J. Post-translational regulation of cold acclimation response. Plant Sci. 2013;205-206:48–54. doi: 10.1016/j.plantsci.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 132.Rodriguez M.C., Petersen M., Mundy J. Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 2010;61:621–649. doi: 10.1146/annurev-arplant-042809-112252. [DOI] [PubMed] [Google Scholar]
- 133.Nakagami H., Pitzschke A., Hirt H. Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci. 2005;10(7):339–346. doi: 10.1016/j.tplants.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 134.Ligterink W., Hirt H. Mitogen-activated protein(MAP) kinase pathways in plants: Versatile signaling tools. Int. Rev. Cytol. 2001;201:209–275. doi: 10.1016/s0074-7696(01)01004-x. [DOI] [PubMed] [Google Scholar]
- 135.Yang T.B., Chaudhuri S., Yang L.H., Du L.Q., Poovaiah B.W. A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. J. Biol. Chem. 2010;285(10):7119–7126. doi: 10.1074/jbc.M109.035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Furuya T., Matsuoka D., Nanmori T. Phosphorylation of Arabidopsis thaliana MEKK1 via Ca2+ signaling as a part of the cold stress response. J. Plant Res. 2013;126(6):833–840. doi: 10.1007/s10265-013-0576-0. [DOI] [PubMed] [Google Scholar]
- 137.Furuya T., Matsuoka D., Nanmori T. Membrane rigidification functions upstream of the MEKK1-MKK2-MPK4 cascade during cold acclimation in Arabidopsis thaliana. FEBS Lett. 2014;588(11):2025–2030. doi: 10.1016/j.febslet.2014.04.032. [DOI] [PubMed] [Google Scholar]
- 138.Teige M., Scheikl E., Eulgem T., Doczi F., Ichimura K., Shinozaki K., Dangl J.L., Hirt H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell. 2004;15(1):141–152. doi: 10.1016/j.molcel.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 139.Mehlmer N., Wurzinger B., Stael S., Hofmann-Rodrigues D., Csaszar E., Pfister B., Bayer R., Teige M. The Ca2+-dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. Plant J. 2010;63(3):484–498. doi: 10.1111/j.1365-313X.2010.04257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wurzinger B., Mair A., Pfister B., Teige M. Cross-talk of calcium-dependent protein kinase and MAP kinase signaling. Plant Signal. Behav. 2011;6(1):8–12. doi: 10.4161/psb.6.1.14012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Saidi Y., Finka A., Goloubinoff P. Heat perception and signalling in plants: a tortuous path to thermotolerance. New Phytol. 2011;190(3):556–565. doi: 10.1111/j.1469-8137.2010.03571.x. [DOI] [PubMed] [Google Scholar]
- 142.Fowler S., Thomashow M.F. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 2002;14(8):1675–1690. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Seki M., Narusaka M., Abe H., Kasuga M., Yamaguchi-Shinozaki K., Carninci P., Hayashizaki Y., Shinozaki K. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell. 2001;13(1):61–72. doi: 10.1105/tpc.13.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Barrero-Gil J., Huertas R., Rambla J.L., Granell A., Salinas J. Tomato plants increase their tolerance to low temperature in a chilling acclimation process entailing comprehensive transcriptional and metabolic adjustments. Plant Cell Environ. 2016;39(10):2303–2318. doi: 10.1111/pce.12799. [DOI] [PubMed] [Google Scholar]
- 145.Zhuang L., Yuan X., Chen Y., Xu B., Yang Z., Huang B. PpCBF3 from cold-tolerant kentucky bluegrass involved in freezing tolerance associated with up-regulation of cold-related genes in transgenic Arabidopsis thaliana. PLoS One. 2015;10(7):e0132928. doi: 10.1371/journal.pone.0132928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Maruyama K., Sakuma Y., Kasuga M., Ito Y., Seki M., Goda H., Shimada Y., Yoshida S., Shinozaki K., Yamaguchi-Shinozaki K. Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 2004;38(6):982–993. doi: 10.1111/j.1365-313X.2004.02100.x. [DOI] [PubMed] [Google Scholar]
- 147.Baker S.S., Wilhelm K.S., Thomashow M.F. The 5′-Region of Arabidopsis-Thaliana Cor15a has Cis-Acting elements that confer cold-regulated, drought-regulated and ABA-regulated gene-expression. Plant Mol. Biol. 1994;24(5):701–713. doi: 10.1007/BF00029852. [DOI] [PubMed] [Google Scholar]
- 148.Novillo F., Alonso J.M., Ecker J.R., Salinas J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2004;101(11):3985–3990. doi: 10.1073/pnas.0303029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yamaguchishinozaki K., Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6(2):251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Maruyama K., Todaka D., Mizoi J., Yoshida T., Kidokoro S., Matsukura S., Takasaki H., Sakurai T., Yamamoto Y.Y., Yoshiwara K., Kojima M., Sakakibara H., Shinozaki K., Yamaguchi-Shinozaki K. Identification of cis-acting promoter elements in cold- and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 2012;19(1):37–49. doi: 10.1093/dnares/dsr040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ishitani M., Liu J.P., Halfter U., Kim C.S., Shi W.M., Zhu J.K. SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell. 2000;12(9):1667–1677. doi: 10.1105/tpc.12.9.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Shinozaki K., Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 2000;3(3):217–223. [PubMed] [Google Scholar]
- 153.Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10(8):1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Shinwari Z.K., Nakashima K., Miura S., Kasuga M., Seki M., Yamaguchi-Shinozaki K., Shinozaki K. An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem. Biophys. Res. Commun. 1998;250(1):161–170. doi: 10.1006/bbrc.1998.9267. [DOI] [PubMed] [Google Scholar]
- 155.Gilmour S.J., Zarka D.G., Stockinger E.J., Salazar M.P., Houghton J.M., Thomashow M.F. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998;16(4):433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- 156.Stockinger E.J., Gilmour S.J., Thomashow M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA. 1997;94(3):1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Thomashow M.F. Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol. 2010;154(2):571–577. doi: 10.1104/pp.110.161794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gilmour S.J., Fowler S.G., Thomashow M.F. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 2004;54(5):767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4. [DOI] [PubMed] [Google Scholar]
- 159.Jaglo-Ottosen K.R., Gilmour S.J., Zarka D.G., Schabenberger O., Thomashow M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280(5360):104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- 160.Ito Y., Katsura K., Maruyama K., Taji T., Kobayashi M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 2006;47(1):141–153. doi: 10.1093/pcp/pci230. [DOI] [PubMed] [Google Scholar]
- 161.Yang S., Tang X.F., Ma N.N., Wang L.Y., Meng Q.W. Heterology expression of the sweet pepper CBF3 gene confers elevated tolerance to chilling stress in transgenic tobacco. J. Plant Physiol. 2011;168(15):1804–1812. doi: 10.1016/j.jplph.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 162.Cheng H., Cai H., Fu H., An Z., Fang J., Hu Y., Guo D., Huang H. Functional characterization of hevea brasiliensis CRT/DRE binding factor 1 gene revealed regulation potential in the CBF pathway of tropical perennial tree. PLoS One. 2015;10(9):e0137634. doi: 10.1371/journal.pone.0137634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jaglo-Ottosen K.R., Gilmour S.J., Zarka D.G., Schabenberger O., Thomashow M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280(5360):104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- 164.Kasuga M., Liu Q., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999;17(3):287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- 165.Wu J., Folta K.M., Xie Y., Jiang W., Lu J., Zhang Y. Overexpression of Muscadinia rotundifolia CBF2 gene enhances biotic and abiotic stress tolerance in Arabidopsis. Protoplasma. 2016;254(1):239–251. doi: 10.1007/s00709-015-0939-6. [DOI] [PubMed] [Google Scholar]
- 166.Novillo F., Alonso J.M., Ecker J.R., Salinas J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in, stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2004;101(11):3985–3990. doi: 10.1073/pnas.0303029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Novillo F., Medina J., Salinas J. Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl. Acad. Sci. USA. 2007;104(52):21002–21007. doi: 10.1073/pnas.0705639105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Maruyama K., Takeda M., Kidokoro S., Yamada K., Sakuma Y., Urano K., Fujita M., Yoshiwara K., Matsukura S., Morishita Y., Sasaki R., Suzuki H., Saito K., Shibata D., Shinozaki K., Yamaguchi-Shinozaki K. Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 2009;150(4):1972–1980. doi: 10.1104/pp.109.135327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Chinnusamy V., Ohta M., Kanrar S., Lee B.H., Hong X.H., Agarwal M., Zhu J.K. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17(8):1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kim Y.S., Lee M., Lee J.H., Lee H.J., Park C.M. The unified ICE-CBF pathway provides a transcriptional feedback control of freezing tolerance during cold acclimation in Arabidopsis. Plant Mol. Biol. 2015;89(1-2):187–201. doi: 10.1007/s11103-015-0365-3. [DOI] [PubMed] [Google Scholar]
- 171.Agarwal M., Hao Y., Kapoor A., Dong C.H., Fujii H., Zheng X., Zhu J.K. R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006;281(49):37636–37645. doi: 10.1074/jbc.M605895200. [DOI] [PubMed] [Google Scholar]
- 172.Doherty C.J., Van Buskirk H.A., Myers S.J., Thomashow M.F. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell. 2009;21(3):972–984. doi: 10.1105/tpc.108.063958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shi Y., Tian S., Hou L., Huang X., Zhang X., Guo H., Yang S. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell. 2012;24(6):2578–2595. doi: 10.1105/tpc.112.098640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ishitani M., Xiong L., Stevenson B., Zhu J.K. Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell. 1997;9(11):1935–1949. doi: 10.1105/tpc.9.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lee B.H., Henderson D.A., Zhu J.K. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell. 2005;17(11):3155–3175. doi: 10.1105/tpc.105.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Miura K., Jin J.B., Lee J., Yoo C.Y., Stirm V., Miura T., Ashworth E.N., Bressan R.A., Yun D.J., Hasegawa P.M. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell. 2007;19(4):1403–1414. doi: 10.1105/tpc.106.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Miura K., Shiba H., Ohta M., Kang S.W., Sato A., Yuasa T., Iwaya-Inoue M., Kamada H., Ezura H. SlICE1 encoding a MYC-type transcription factor controls cold tolerance in tomato. Plant Biotechnol. J. 2012;29(3):253–260. [Google Scholar]
- 178.Ding Y., Li H., Zhang X., Xie Q., Gong Z., Yang S. OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell. 2015;32(3):278–289. doi: 10.1016/j.devcel.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 179.Kanaoka M.M., Pillitteri L.J., Fujii H., Yoshida Y., Bogenschutz N.L., Takabayashi J., Zhu J.K., Torii K.U. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell. 2008;20(7):1775–1785. doi: 10.1105/tpc.108.060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Dong C.H., Agarwal M., Zhang Y.Y., Xie Q., Zhu J.K. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA. 2006;103(21):8281–8286. doi: 10.1073/pnas.0602874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Figueiredo D.D., Barros P.M., Cordeiro A.M., Serra T.S., Lourenco T., Chander S., Oliveira M.M., Saibo N.J. Seven zinc-finger transcription factors are novel regulators of the stress responsive gene OsDREB1B. J. Exp. Bot. 2012;63(10):3643–3656. doi: 10.1093/jxb/ers035. [DOI] [PubMed] [Google Scholar]
- 182.Ma Y., Dai X.Y., Xu Y.Y., Luo W., Zheng X.M., Zeng D.L., Pan Y.J., Lin X.L., Liu H.H., Zhang D.J., Xiao J., Guo X.Y., Xu S.J., Niu Y.D., Jin J.B., Zhang H., Xu X., Li L.G., Wang W., Qian Q., Ge S., Chong K. COLD1 confers chilling tolerance in rice. Cell. 2015;162(1):222–222. doi: 10.1016/j.cell.2015.01.046. [DOI] [PubMed] [Google Scholar]
- 183.Xiong L.M., Ishitani M., Zhu J.K. Interaction of osmotic stress, temperature, and abscisic acid in the regulation of gene expression in Arabidopsis. Plant Physiol. 1999;119(1):205–211. doi: 10.1104/pp.119.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Calzadilla P.I., Maiale S.J., Ruiz O.A., Escaray F.J. Transcriptome response mediated by cold stress in Lotus japonicus. Front. Plant Sci. 2016;7:374. doi: 10.3389/fpls.2016.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sobkowiak A., Jonczyk M., Adamczyk J., Szczepanik J., Solecka D., Kuciara I., Hetmanczyk K., Trzcinska-Danielewicz J., Grzybowski M., Skoneczny M., Fronk J., Sowinski P. Molecular foundations of chilling-tolerance of modern maize. BMC Genomics. 2016;17:125. doi: 10.1186/s12864-016-2453-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Nakaminami K., Matsui A., Nakagami H., Minami A., Nomura Y., Tanaka M., Morosawa T., Ishida J., Takahashi S., Uemura M., Shirasu K., Seki M. Analysis of differential expression patterns of mRNA and protein during cold-acclimation and de-acclimation in Arabidopsis. Mol. Cell. Proteomics. 2014;13(12):3602–3611. doi: 10.1074/mcp.M114.039081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Dametto A., Sperotto R.A., Adamski J.M., Blasi E.A., Cargnelutti D., de Oliveira L.F., Ricachenevsky F.K., Fregonezi J.N., Mariath J.E., da Cruz R.P., Margis R., Fett J.P. Cold tolerance in rice germinating seeds revealed by deep RNAseq analysis of contrasting indica genotypes. Plant Sci. 2015;238:1–12. doi: 10.1016/j.plantsci.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 188.Swamy P.M., Smith B.N. Role of abscisic acid in plant stress tolerance. Curr. Sci. 1999;76(9):1220–1227. [Google Scholar]
- 189.Sah S.K., Reddy K.R., Li J. Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 2016;7:571. doi: 10.3389/fpls.2016.00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Xing W.B., Rajashekar C.B. Glycine betaine involvement in freezing tolerance and water stress in Arabidopsis thaliana. Environ. Exp. Bot. 2001;46(1):21–28. doi: 10.1016/s0098-8472(01)00078-8. [DOI] [PubMed] [Google Scholar]
- 191.Chawade A., Lindlof A., Olsson B., Olsson O. Global expression profiling of low temperature induced genes in the chilling tolerant japonica rice Jumli Marshi. PLoS One. 2013;8(12):e81729. doi: 10.1371/journal.pone.0081729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Nakamura T., Yazaki J., Kishimoto N., Kikuchi S., Robertson A.J., Gusta L.V., Ishikawa M. Comparison of long-term up-regulated genes during induction of freezing tolerance by cold and ABA in bromegrass cell cultures revealed by microarray analyses. J. Plant Growth Regul. 2013;71(2):113–136. [Google Scholar]
- 193.Seki M., Narusaka M., Ishida J., Kamiya A., Nakajima M., Enju A., Oono Y., Satou M., Akiyama K., Sakurai T., Fujita M., Nanjo T., Yamaguchi-Shinozaki K., Carninci P., Kawai J., Hayashizaki Y., Shinozaki K. Monitoring the expression pattern of Arabidopsis genes using full-length CDNA microarray. Plant Cell Physiol. 2002;43:S1–S1. [Google Scholar]
- 194.Nguyen H.T., Leipner J., Stamp P., Guerra-Peraza O. Low temperature stress in maize (Zea mays L.) induces genes involved in photosynthesis and signal transduction as studied by suppression subtractive hybridization. Plant Physiol. Biochem. 2009;47(2):116–122. doi: 10.1016/j.plaphy.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 195.Narusaka Y., Nakashima K., Shinwari Z.K., Sakuma Y., Furihata T., Abe H., Narusaka M., Shinozaki K., Yamaguchi-Shinozaki K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003;34(2):137–148. doi: 10.1046/j.1365-313x.2003.01708.x. [DOI] [PubMed] [Google Scholar]
- 196.Maruyama K., Todaka D., Mizoi J., Yoshida T., Kidokoro S., Matsukura S., Takasaki H., Sakurai T., Yamamoto Y.Y., Yoshiwara K., Kojima M., Sakakibara H., Shinozaki K., Yamaguchi-Shinozaki K. Identification of cis-acting promoter elements in cold and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 2012;19(1):37–49. doi: 10.1093/dnares/dsr040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Theocharis A., Clement C., Barka E.A. Physiological and molecular changes in plants grown at low temperatures. Planta. 2012;235(6):1091–1105. doi: 10.1007/s00425-012-1641-y. [DOI] [PubMed] [Google Scholar]
- 198.Liang C.H., Yang C.C. Identification of ICE1 as a negative regulator of ABA-dependent pathways in seeds and seedlings of Arabidopsis. Plant Mol. Biol. 2015;88(4-5):459–470. doi: 10.1007/s11103-015-0335-9. [DOI] [PubMed] [Google Scholar]
- 199.Xiong L., Ishitani M., Zhu J.K. Interaction of osmotic stress, temperature, and abscisic acid in the regulation of gene expression in Arabidopsis. Plant Physiol. 1999;119(1):205–212. doi: 10.1104/pp.119.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Jeon J., Kim N.Y., Kim S., Kang N.Y., Novak O., Ku S.J., Cho C., Lee D.J., Lee E.J., Strnad M., Kim J. A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J. Biol. Chem. 2010;285(30):23371–23386. doi: 10.1074/jbc.M109.096644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Jeon J., Kim J. Arabidopsis response Regulator1 and Arabidopsis histidine phosphotransfer Protein2(AHP2), AHP3, and AHP5 function in cold signaling. Plant Physiol. 2013;161(1):408–424. doi: 10.1104/pp.112.207621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Zwack P.J., Compton M.A., Adams C.I., Rashotte A.M. Cytokinin response factor 4(CRF4) is induced by cold and involved in freezing tolerance. Plant Cell Rep. 2016;35(3):573–584. doi: 10.1007/s00299-015-1904-8. [DOI] [PubMed] [Google Scholar]
- 203.Cerny M., Dycka F., Bobal’ova J., Brzobohaty B. Early cytokinin response proteins and phosphoproteins of Arabidopsis thaliana identified by proteome and phosphoproteome profiling. J. Exp. Bot. 2011;62(3):921–937. doi: 10.1093/jxb/erq322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Cerny M., Jedelsky P.L., Novak J., Schlosser A., Brzobohaty B. Cytokinin modulates proteomic, transcriptomic and growth responses to temperature shocks in Arabidopsis. Plant Cell Environ. 2014;37(7):1641–1655. doi: 10.1111/pce.12270. [DOI] [PubMed] [Google Scholar]
- 205.Kosova K., Prasil I.T., Vitamvas P., Dobrev P., Motyka V., Flokova K., Novak O., Tureckova V., Rolcik J., Pesek B., Travnickova A., Gaudinova A., Galiba G., Janda T., Vlasakova E., Prasilova P., Vankova R. Complex phytohormone responses during the cold acclimation of two wheat cultivars differing in cold tolerance, winter Samanta and spring Sandra. J. Plant Physiol. 2012;169(6):567–576. doi: 10.1016/j.jplph.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 206.Belintani N.G., Guerzoni J.T., Moreira R.M., Vieira L.G. Improving low-temperature tolerance in sugarcane by expressing the ipt gene under a cold inducible promoter. Biol. Plant. 2012;56(1):71–77. [Google Scholar]
- 207.Bienert G.P., Schjoerring J.K., Jahn T.P. Membrane transport of hydrogen peroxide. Biochim. Biophys. Acta Biomembr. 2006;1758(8):994–1003. doi: 10.1016/j.bbamem.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 208.Neill S., Desikan R., Hancock J. Hydrogen peroxide signalling. Curr. Opin. Plant Biol. 2002;5(5):388–395. doi: 10.1016/s1369-5266(02)00282-0. [DOI] [PubMed] [Google Scholar]
- 209.Huang C.S., Li J.X., Ke Q.D., Leonard S.S., Jiang B.H., Zhong X.S., Costa M., Castranova V., Shi X.L. Ultraviolet-induced phosphorylation of p70(S6K) at Thr(389) and Thr(421)/Ser(424) involves hydrogen peroxide and mammalian target of rapamycin but not Akt and atypical protein kinase C. Cancer Res. 2002;62(20):5689–5697. [PubMed] [Google Scholar]
- 210.Yang T., Poovaiah B.W. Hydrogen peroxide homeostasis: Activation of plant catalase by calcium/calmodulin. Proc. Natl. Acad. Sci. USA. 2002;99(6):4097–4102. doi: 10.1073/pnas.052564899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002;7(9):405–410. doi: 10.1016/s1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- 212.de Pinto M.C., Paradiso A., Leonetti P., De Gara L. Hydrogen peroxide, nitric oxide and cytosolic ascorbate peroxidase at the crossroad between defence and cell death. Plant J. 2006;48(5):784–795. doi: 10.1111/j.1365-313X.2006.02919.x. [DOI] [PubMed] [Google Scholar]
- 213.Van Breusegem F., Dat J.F. Reactive oxygen species in plant cell death. Plant Physiol. 2006;141(2):384–390. doi: 10.1104/pp.106.078295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Dat J., Vandenabeele S., Vranova E., Van Montagu M., Inze D., Van Breusegem F. Dual action of the active oxygen species during plant stress responses. Cell. Mol. Life Sci. 2000;57(5):779–795. doi: 10.1007/s000180050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Laloi C., Apel K., Danon A. Reactive oxygen signalling: the latest news. Curr. Opin. Plant Biol. 2004;7(3):323–328. doi: 10.1016/j.pbi.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 216.Hossain M.A., Bhattacharjee S., Armin S.M., Qian P., Xin W., Li H.Y., Burritt D.J., Fujita M., Tran L.S. Hydrogen peroxide priming modulates abiotic oxidative stress tolerance: insights from ROS detoxification and scavenging. Front. Plant Sci. 2015;6:420. doi: 10.3389/fpls.2015.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Prasad T.K., Anderson M.D., Martin B.A., Stewart C.R. Evidence for chilling-induced oxidative stress in maize seedlings and a regulatory role for hydrogen peroxide. Plant Cell. 1994;6(1):65–74. doi: 10.1105/tpc.6.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Xia X.J., Wang Y.J., Zhou Y.H., Tao Y., Mao W.H., Shi K., Asami T., Chen Z.X., Yu J.Q. Reactive oxygen species are involved in brassinosteroid- induced stress tolerance in cucumber. Plant Physiol. 2009;150(2):801–814. doi: 10.1104/pp.109.138230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Wang Y., Li J., Wang J., Li Z. Exogenous H2O2 improves the chilling tolerance of manilagrass and mascarene grass by activating the antioxidative system. J. Plant Growth Regul. 2010;61(2):195–204. [Google Scholar]
- 220.Gundlach H., Muller M.J., Kutchan T.M., Zenk M.H. Jasmonic acid is a signal transducer in elicitor-induced plant-cell cultures. Proc. Natl. Acad. Sci. USA. 1992;89(6):2389–2393. doi: 10.1073/pnas.89.6.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Dempsey D.A., Klessig D.F. Signals in Plant-Disease Resistance. Bull. Inst. Pasteur. 1995;93(3):167–186. [Google Scholar]
- 222.Desikan R., Cheung M.K., Clarke A., Golding S., Sagi M., Fluhr R., Rock C., Hancock J., Neill S. Hydrogen peroxide is a common signal for darkness- and ABA-induced stomatal closure in Pisum sativum. Funct. Plant Biol. 2004;31(9):913–920. doi: 10.1071/FP04035. [DOI] [PubMed] [Google Scholar]
- 223.Wendehenne D., Durner J., Klessig D.F. Nitric oxide: a new player in plant signalling and defence responses. Curr. Opin. Plant Biol. 2004;7(4):449–455. doi: 10.1016/j.pbi.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 224.Liu Q., Zhou G.Y., Wen C.K. Ethylene signal transduction in Arabidopsis. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao. 2004;30(3):241–250. [PubMed] [Google Scholar]
- 225.Guo Z.F., Tan J.L., Zhuo C.L., Wang C.Y., Xiang X., Wang Z.Y. Abscisic acid, H2O2 and nitric oxide interactions mediated cold-induced S-adenosylmethionine synthetase in Medicago sativa subsp falcata that confers cold tolerance through up-regulating polyamine oxidation. Plant Biotechnol. J. 2014;12(5):601–612. doi: 10.1111/pbi.12166. [DOI] [PubMed] [Google Scholar]
- 226.Neill S.J., Desikan R., Clarke A., Hancock J.T. Nitric oxide is a novel component of abscisic acid signaling in stomatal guard cells. Plant Physiol. 2002;128(1):13–16. [PMC free article] [PubMed] [Google Scholar]
- 227.Cheng Y.L., Song C.P. Hydrogen peroxide homeostasis and signaling in plant cells. Sci. China C Life Sci. 2006;49(1):1–11. doi: 10.1007/s11427-005-0071-5. [DOI] [PubMed] [Google Scholar]
- 228.Gilroy S., Trewavas A. Signal processing and transduction in plant cells: the end of the beginning? Nat. Rev. Mol. Cell Biol. 2001;2(4):307–314. doi: 10.1038/35067109. [DOI] [PubMed] [Google Scholar]
- 229.Wasteneys G.O., Galway M.E. Remodelling the cytoskeleton for growth and form: An overview with some new views. Annu. Rev. Plant Biol. 2003;54:691–722. doi: 10.1146/annurev.arplant.54.031902.134818. [DOI] [PubMed] [Google Scholar]
- 230.Sonesson A., Berglund M., Staxen I., Widell S. The characterization of plasma membrane-bound tubulin of cauliflower using triton X-114 fractionation. Plant Physiol. 1997;115(3):1001–1007. doi: 10.1104/pp.115.3.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Gardiner J.C., Harper J.D., Weerakoon N.D., Collings D.A., Ritchie S., Gilroy S., Cyr R.J., Marc J.A. 90-kD phospholipase D from tobacco binds to microtubules and the plasma membrane. Plant Cell. 2001;13(9):2143–2158. doi: 10.1105/TPC.010114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Dhonukshe P., Laxalt A.M., Goedhart J., Gadella T.W., Munnik T. Phospholipase D activation correlates with microtubule reorganization in living plant cells. Plant Cell. 2003;15(11):2666–2679. doi: 10.1105/tpc.014977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Drobak B.K., Franklin-Tong V.E., Staiger C.J. The role of the actin cytoskeleton in plant cell signaling. New Phytol. 2004;163(1):13–30. doi: 10.1111/j.1469-8137.2004.01076.x. [DOI] [PubMed] [Google Scholar]
- 234.Hong Y.Y., Zheng S.Q., Wang X.M. Dual functions of phospholipase D alpha 1 in plant response to drought. Mol. Plant. 2008;1(2):262–269. doi: 10.1093/mp/ssm025. [DOI] [PubMed] [Google Scholar]
- 235.Ruelland E., Cantrel C., Gawer M., Kader J.C., Zachowski A. Activation of phospholipases C and D is an early response to a cold exposure in Arabidopsis suspension cells. Plant Physiol. 2002;130(2):999–1007. doi: 10.1104/pp.006080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Welti R., Li W.Q., Li M.Y., Sang Y.M., Biesiada H., Zhou H.E., Rajashekar C.B., Williams T.D., Wang X.M. Profiling membrane lipids in plant stress responses - Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 2002;277(35):31994–32002. doi: 10.1074/jbc.M205375200. [DOI] [PubMed] [Google Scholar]
- 237.Li W.Q., Li M.Y., Zhang W.H., Welti R., Wang X.M. The plasma membrane-bound phospholipase D delta enhances freezing tolerance in Arabidopsis thaliana. Nat. Biotechnol. 2004;22(4):427–433. doi: 10.1038/nbt949. [DOI] [PubMed] [Google Scholar]
- 238.Dodd A.N., Jakobsen M.K., Baker A.J., Telzerow A., Hou S.W., Laplaze L., Barrot L., Poethig R.S., Haseloff J., Webb A.A. Time of day modulates low-temperature Ca2+ signals in Arabidopsis. Plant J. 2006;48(6):962–973. doi: 10.1111/j.1365-313X.2006.02933.x. [DOI] [PubMed] [Google Scholar]
- 239.Thion L., Mazars C., Thuleau P., Graziana A., Rossignol M., Moreau M., Ranjeva R. Activation of plasma membrane voltage-dependent calcium-permeable channels by disruption of microtubules in carrot cells. FEBS Lett. 1996;393(1):13–18. doi: 10.1016/0014-5793(96)00844-7. [DOI] [PubMed] [Google Scholar]
- 240.Thion L., Mazars C., Nacry P., Bouchez D., Moreau M., Ranjeva R., Thuleau P. Plasma membrane depolarization-activated calcium channels, stimulated by microtubule-depolymerizing drugs in wild-type Arabidopsis thaliana protoplasts, display constitutively large activities and a longer half-life in ton 2 mutant cells affected in the organization of cortical microtubules. Plant J. 1998;13(5):603–610. doi: 10.1046/j.1365-313x.1998.00062.x. [DOI] [PubMed] [Google Scholar]
- 241.Thuleau P., Schroeder J.I., Ranjeva R. Recent advances in the regulation of plant calcium channels: evidence for regulation by G-proteins, the cytoskeleton and second messengers. Curr. Opin. Plant Biol. 1998;1(5):424–427. doi: 10.1016/s1369-5266(98)80267-7. [DOI] [PubMed] [Google Scholar]
- 242.Zimmermann S., Ehrhardt T., Plesch G., Muller-Rober B. Ion channels in plant signaling. Cell. Mol. Life Sci. 1999;55(2):183–203. doi: 10.1007/s000180050284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Abdrakhamanova A., Wang Q.Y., Khokhlova L., Nick P. Is microtubule disassembly a trigger for cold acclimation? Plant Cell Physiol. 2003;44(7):676–686. doi: 10.1093/pcp/pcg097. [DOI] [PubMed] [Google Scholar]
- 244.Nick P., Heuing A., Ehmann B. Plant chaperonins: a role in microtubule-dependent wall formation? Protoplasma. 2000;211(3-4):234–244. [Google Scholar]
- 245.Mazars C., Thion L., Thuleau P., Graziana A., Knight M.R., Moreau M., Ranjeva R. Organization of cytoskeleton controls the changes in cytosolic calcium of cold-shocked Nicotiana plumbaginifolia protoplasts. Cell Calcium. 1997;22(5):413–420. doi: 10.1016/s0143-4160(97)90025-7. [DOI] [PubMed] [Google Scholar]
- 246.Zhao Y.D., Dai X.H., Blackwell H.E., Schreiber S.L., Chory J. SIR1, an upstream component in auxin signaling identified by chemical genetics. Science. 2003;301(5636):1107–1110. doi: 10.1126/science.1084161. [DOI] [PubMed] [Google Scholar]
- 247.Farajalla M.R., Gulick P.J. The alpha-tubulin gene family in wheat (Triticum aestivum L.) and differential gene expression during cold acclimation. Genome. 2007;50(5):502–510. doi: 10.1139/g07-027. [DOI] [PubMed] [Google Scholar]
- 248.Mao G.J., Chan J., Calder G., Doonan J.H., Lloyd C.W. Modulated targeting of GFP-AtMAP65-1 to central spindle microtubules during division. Plant J. 2005;43(4):469–478. doi: 10.1111/j.1365-313X.2005.02464.x. [DOI] [PubMed] [Google Scholar]
- 249.Ouellet F., Carpentier E., Cope M.J., Monroy A.F., Sarhan F. Regulation of a wheat actin-depolymerizing factor during cold acclimation. Plant Physiol. 2001;125(1):360–368. doi: 10.1104/pp.125.1.360. [DOI] [PMC free article] [PubMed] [Google Scholar]