Abstract
A successful HIV cure strategy may require reversing HIV latency to purge hidden viral reservoirs or enhancing HIV latency to permanently silence HIV transcription. Epigenetic modifying agents show promise as antilatency therapeutics in vitro and ex vivo, but also affect other steps in the viral life cycle. In this review, we summarize what we know about cellular DNA and protein methyltransferases (PMTs) as well as demethylases involved in HIV infection. We describe the biology and function of DNA methyltransferases, and their controversial role in HIV infection. We further explain the biology of PMTs and their effects on lysine and arginine methylation of histone and nonhistone proteins. We end with a focus on protein demethylases, their unique modes of action and their emerging influence on HIV infection. An outlook on the use of methylation-modifying agents in investigational HIV cure strategies is provided.
Keywords: : methyltransferase, histone demethylase, methyltransferase inhibitors, demethylase inhibitors, HIV cure, latency reactivation
Introduction
Epigenetic modifications of nucleic acids, histones, and nonhistone proteins are important regulators of gene transcription and expression and are targets in clinical efforts to reverse HIV latency. One such modification is methylation, which involves the transfer of a methyl group with S-adenosyl-L-methionine (SAM) as a methyl donor.1 This modification is a reversible process regulated by methyltransferases (MTs) and demethylases. Both methyltransferases and demethylases are highly specific for the position of their substrate and extent of methylation. Methylation of nucleic acids, histones, and nonhistone proteins regulates gene transcription and expression by altering chromatin packaging, promoting chromatin accessibility, creating DNA–protein interactions, and generating interaction interfaces for the assembly of macromolecular complexes.1,2
Epigenetic regulation of DNA, histone, and nonhistone proteins plays an important role in HIV transcription and is a target in preclinical efforts to reverse HIV latency, a major hurdle to curing HIV infection. HIV latency is established by integrating viral DNA into the host chromatin.3 Once integrated, proviral cDNA is organized into higher-order chromatin and becomes subject to regulation by host chromatin-modifying enzymes, including deacetylases, methyltransferases, and demethylases. Hypoacetylation of histone proteins by histone deacetylases (HDACs) helps to maintain HIV-1 latency by repressing viral transcription.4 Critical cofactors that further contribute to latency include the positive transcription elongation factor b (P-TEFb), NF-κB, and the virally encoded transactivator of transcription (Tat).5–7
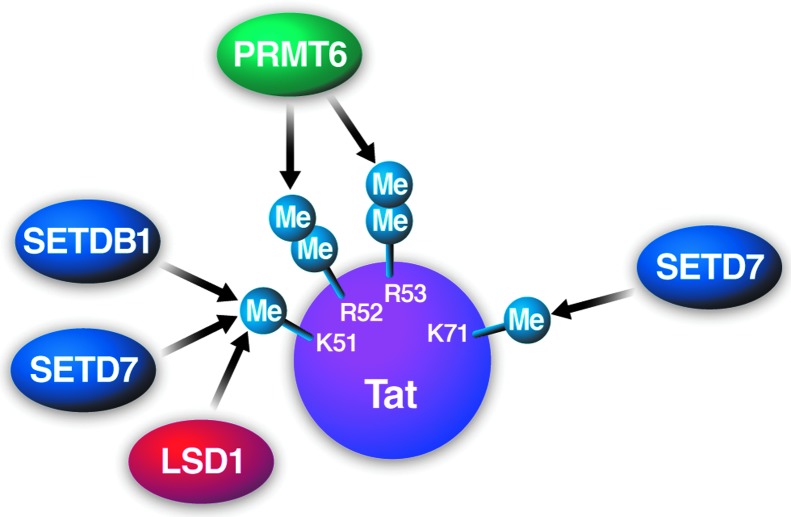
Tat binds to an RNA stem-loop structure called TAR at the 5′ end of all nascent viral transcripts and recruits P-TEFb, which together with other elongation factors, form a “super-elongation complex” at the elongating RNA polymerase II.8–10 Tat itself is subject to reversible methylation by SETD7/SET7/9/KMT7 at lysine 51 in its basic RNA-binding domain11 (Fig. 1). This methylation event positively supports the transcriptional activity of Tat11 and is reversed by the demethylase activity of LSD1/KDM1A.12 Tat is also methylated by the histone lysine methyltransferase SETDB1 at lysine 51,13 by SETD7 at lysine 71,14 and by arginine methyltransferase PRMT6 at R52 and R5315 (Fig. 1).
FIG. 1.
Specificity of protein methyltransferases and demethylase LSD1 that target HIV-1-Tat at K51, R52, R53, and K71.
Latent HIV-1 proviruses are primarily found integrated into actively transcribed genes, but characteristically display heterochromatic features.16,17 The long terminal repeat (LTR) of latent proviruses accumulates HDACs that result in high levels of deacetylated histones.18,19 Besides histone deacetylation, a growing list of MTs and demethylases indicates that methylation of DNA, histones, and nonhistone proteins is essential for HIV-1 transcription. In this review, we summarize the DNA and protein methyltransferases (PMTs) and demethylases involved in HIV infection, and their implications for HIV cure strategies.
Methyltransferases
DNA methyltransferases
DNA methylation is an epigenetic mark that mediates transcriptional activation or repression of genes.20 DNA is methylated within the cytosine–phosphate–guanine (CpG) and non-CpG dinucleotide sites by the addition of a methyl group to the fifth carbon atom of the cytosine residues using SAM as a methyl donor.21 This process is catalyzed by specific DNA methyltransferase (DNMT) enzymes. In mammals, five structurally and functionally distinct DNMTs have been characterized: DNMT1, DNMT2, DNMT3A, DNMT3B, and DNMT3L.21 DNMT3L does not possess catalytic activity, and DNMT2 mainly methylates RNA. DNMT1, DNMT3A, and DNMT3B mediate two different DNA methylation mechanisms: (1) de novo methylation which creates new marks on DNA and (2) maintenance of previously methylated genomic sites during DNA replication.21 At gene promoters, the presence of DNA methylation is linked to chromatin silencing.22,23 The repressive effect of DNA methylation on gene expression is mediated by two mechanisms: (1) the interference of DNA methylation with the recognition of transcription factor-binding sites, which results in impairment of gene activation,24 and (2) the recognition of DNA methylation by specific Methyl-CpG-binding proteins that recruit corepressor protein complexes and thereby mediate silencing.25
The role of DNA methylation in HIV-1 infection is controversial. Kauder et al. identified methyl-CpG-binding domain protein 2 as a regulator of HIV-1 latency and reported the hypermethylation of two CpG islands surrounding the HIV-1 transcriptional start site in latently infected Jurkat cells (J-Lat) and in primary CD4+ T cells.26 Furthermore, methyl-CpG-binding domain protein 2, which is present at one of these CpG islands during latency and absent upon inhibition of cytosine methylation with 5-aza-2′deoxycytidine, possibly recruits transcriptional repressors to methylated DNA, and thereby contributes to HIV-1 latency.26 A separate study showed high levels of CpG methylation of the HIV-1 5′ LTR in an in vitro model of latency and in latently infected T cells isolated from HIV-1-infected patients in a model in which CpG methylation acts as a late event during establishment of HIV-1 latency not required for initial provirus silencing.27
Recently, Maricato et al. reported that HIV-1 infection increases the methylation level of cellular DNA in peripheral blood mononuclear cells (PBMC).28 A separate study found overall low levels of 5′ LTR DNA methylation in resting CD4+ T cells isolated from a group of HIV-infected individuals under antiretroviral treatment for up to 3 years.29 However, in some long-term treated individuals, the authors detected proviral molecules with a high density of 5′ LTR CpG methylation. They proposed that transient stimulation of cells harboring latent proviruses contributes, at least in part, to the methylation of the HIV-1 promoter over time.29 Collectively, these four studies support a model in which DNA methylation contributes to HIV-1 transcriptional gene silencing, likely by inhibiting transcriptional initiation of the integrated provirus and inhibiting viral postintegration reactivation from latency.
In contrast, Blazkova et al. reported very low levels of methylated CpG dinucleotides within the promoter/enhancer region of latent HIV proviruses found in resting CD4+ T cells isolated from aviremic infected individuals.30 The different results can possibly be explained by the use of different primers and the length of antiretroviral treatment of patients.
So far, only DNMT1, DNMT2, and DNMT3B have been implicated in HIV infection.31,32 DNMT1, which is the most abundant DNMT in the cell and transcribed mostly during the S phase of the cell cycle, is primarily responsible for maintaining methylation through transfer of methyl groups to the hemimethylated DNA strands after DNA replication, but also has de novo DNMT activity.33 In addition, DNMT1 influences transcriptional regulation by interacting with HDAC2 and novel corepressors to form a complex at replication foci.34 Furthermore, DNMT1 represses transcription from E2F-responsive promoters by recruitment of transcription factors retinoblastoma protein Rb and E2F1, as well as HDAC1.35 Recently, DNMT2 was shown to relocalize from the nucleus to stress granules and to methylate HIV-1 RNA in a sequence-independent manner, thereby providing posttranscriptional stability to the HIV-1 RNA.32
DNMT3B, which is highly expressed in early embryonic stem (ES) cells, cannot differentiate between unmethylated and hemimethylated CpG sites, and cannot copy or contribute to the maintenance of a specific pattern of methylation.33 However, DNMT3B has an important role in de novo methylation of unmodified cytosine residues.33
Several groups reported that infection of CD4+ T cells with HIV-1 increases expression and activity of DNMTs.31,36–38 One study showed that acute infection with wild-type and integration-defective HIV-1 increases cellular DNMT1 expression and activity, resulting in hypermethylation and reduced expression of the tumor suppressor gene, p16INK4A, in lymphoid cell lines.37 Youngblood and Reich reported that HIV-1 early gene expression caused DNMT1 induction, a process prevented by treatment with the phytoalexin resveratrol thought to interfere with transcription factor AP1 activity.38 The HIV-1 responsive element was mapped to nt–1634 to–1214 in the DNMT1 promoter, which overlaps with AP1 sites.38 An independent study evaluating the effect of HIV-1 infection on cellular vitamin D receptor expression showed that infection enhanced expression of HDAC1, DNMT1, and DNMT3B.31 Recently, Pion et al. reported that HIV-1 infection downregulates expression of master transcription factor Foxp3 in regulatory T cells, which was associated with an increase in the expression of DNMT3B and higher methylation of CpG sites in the FOXP3 locus.39
Others linked the increase in DNMT expression to an overall increase in methylated genomic DNA in HIV-infected cells and the de novo methylation of a single CpG dinucleotide in the gamma interferon (IFN-γ) gene promoter to the downregulation of IFN-γ production in HIV-infected CD4+ T cells.36,37 In an attempt to exploit these findings therapeutically, Martinez-Colom et al. designed a chimeric protein (IN3b) that linked the N-terminal domain of the HIV integrase enzyme with the C-terminal domain of DNMT3B to induce long-term silencing of HIV gene expression in host cells.40 However, reduced DNMT1 and DNMT3A activity was reported in primary oral epithelial cells isolated from aviremic treated HIV+ individuals,41 suggesting that DNA methylation is deregulated, yet not always upregulated, in HIV infection.
A recent study aimed to investigate the influence of antiretroviral therapy on methylation markers found in PBMCs isolated from blood from a group of HIV-infected, ART-treated patients that the percentage of 5-methylcytosine was inversely correlated with proviral DNA and active replication, whereas DNMT1 and DNMT3A were independently correlated with active viral replication.42 Differential DNA methylation associated with HIV infection was also reported in a recent large-scale epigenome-wide study, where HIV-infected and uninfected patients from the Veteran Aging Cohort Study (VACS) were profiled for CpG sites in DNA extracted from the blood.43 The authors identified 20 epigenome-wide significant CpGs for HIV-1 infection, including two CpGs in the promoter of the NLR family, CARD domain containing gene 5 (NLRC5), a key regulator of major histocompatibility complex class I gene expression, which showed significantly lower methylation in HIV-infected subjects than in uninfected subjects and which was negatively correlated with viral load in the HIV-infected samples.43
Protein methyltransferases
PMTs are enzymes that covalently transfer methyl groups from the cofactor SAM to specific amino acid residues within nuclear and cytoplasmic proteins. Based on their structure, SAM-dependent MTs are divided into three classes: PMTs with seven-strand twisted β-sheet structure,44 SET (SuVar3-9, enhancer of Zeste, Trithorax) domain lysine MTs,45 and membrane-associated MTs.46 These enzymes mediate protein methylation through O-methylation or N-methylation. Methylations of glutamic acid and aspartic acid are a type of O-methylation, and N-methylation involves methylation of lysine, arginine, histidine, alanine, proline, glutamine, phenylalanine, asparagine, and methionine. Among these, methylation of lysines and arginines is most common.47 Lysines can be mono-, di-, or trimethylated, but arginines can be monomethylated or symmetrically or asymmetrically dimethylated. Protein lysine and arginine methylation results either in gene activation or repression, depending on the amino acid residue that becomes methylated and the state of methylation.
Lysine methyltransferases (KMT)
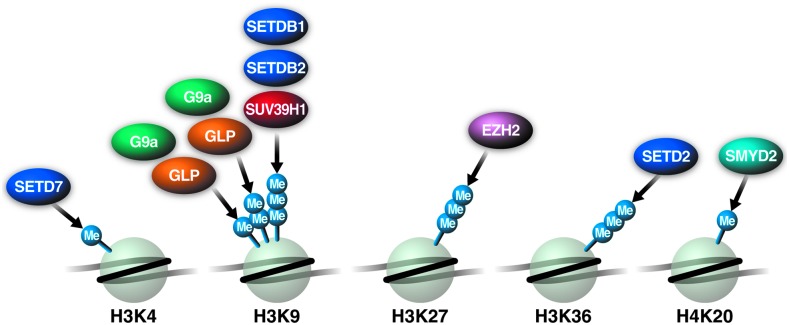
More than 50 human KMTs have been found that methylate histones and nonhistone substrates. Based on their catalytic domain, KMTs are grouped into two different families: (1) DOT1-like (DOT1L) that does not possess a SET domain, and (2) the SET domain-containing MTs (reviewed in Ref.48). SET domain-containing MTs can be further divided into four families: SET1, SET2, SUV39, and RIZ.48 Except for family members EZH1 and EZH2, the SET1 family is characterized by the SET domain, followed by a post-SET domain. SET2 class members are NSD1-3, SETD2, and the SMYD family, all of which have a SET domain positioned between a post-SET and an AWS domain. The SUV39 family members SUV39H1, SUV39H2, G9a, GLP, SETDB1, and SETDB2 all contain a pre-SET domain. Finally, the RIZ family members BLIMP1, PFM1, and RIZ1 are characterized by a SET domain at the amino terminus. In addition, several SET domain-containing methyltransferases include SET7/9, SET8, SUV4-20H1, and SUV4-20H2, which do not belong to any of these families. Figure 2 shows the KMT that have been implicated in HIV infection (Fig. 2).
FIG. 2.
Histone lysine methyltransferases (KMT) and histone methyl marks implicated in HIV infection, showing residue-specific KMTs for H3K4/9/27/36 and H4K20. The majority of KMTs are highly specific for a single histone residue, whereas a few enzymes target multiple residues, as indicated.
Enhancer of zeste homolog 2 (EZH2)
EZH2 belongs to the SET1 family of KMTs and trimethylates histone H3 lysine 27 (H3K27me3).49 This subunit of the polycomb repressor complex 2 (PRC2) is an important regulator of chromatin state involved in the maintenance of transcriptional silencing.50 EZH2 lacks enzymatic activity as an isolated protein and only methylates lysine residues when in complex with EED and SUZ12.51 EZH2 is modified through O-GlcNAcylation by O-linked N-acetylglucosamine transferase, resulting in increased stability of EZH2 and higher cellular histone H3 lysine 27 trimethylation levels.52
EZH2 associates with the promoter/enhancer region of HIV-1 proviruses in latently infected Jurkat T cell lines and is found with the corresponding H3K27me3 mark.53 Knockdown of EZH2 with shRNA or treatment with the chemical inhibitor 3-Deazaneplanocin A efficiently reactivates a significant portion of silenced proviruses. Furthermore, knockdown of EZH2 sensitizes latent proviruses to external stimuli, such as T cell receptor stimulation, and interferes with the reversion of reactivated proviruses back to latency, underscoring its restrictive role in HIV-1 latency.53 In addition, Nguyen et al. showed that treatment with newer inhibitors of EZH2 (GSK-343, EPZ-6438) was sufficient to induce the reactivation of latent proviruses in a primary T cell model using primarily latently infected Th17-polarized cells or in resting memory T cells isolated from aviremic HIV-1-infected patients.54 In contrast, Tripathy et al. showed that GSK343 reduced trimethylation of HIV provirus-associated histone H3K27 without increasing proviral expression in latently infected resting CD4+ T cells.55 However, after reduction of H3K27 methylation at the HIV-1 LTR, subsequent exposure to the HDAC inhibitors, suberoylanilide hydroxamic acid (SAHA) or vorinostat resulted in increases in viral gag RNA and p24 antigen production, pointing to the combination of HMT and HDAC inhibitors as a promising reversal strategy in HIV latency.55
Euchromatin histone methyltransferases EHMT1 (GLP) and EHMT2 (G9a)
Euchromatin histone methyltransferases (EHMTs) are SUV39 family members. They are evolutionarily conserved proteins that mono- and dimethylate histone 3 at lysine 9 (H3K9me1 and H3K9me2) in euchromatic regions of the genome.56 In mammals, there are two EHMT proteins: G9a-like protein GLP is encoded by EHMT1, and G9a is encoded by EHMT2.56 EHMTs are characterized by an N-terminal SET and Pre-SET domain and a series of ankyrin repeats that are required for protein–protein interactions, particularly with H3K9me1 and H3K9me2, the marks that EHMT proteins generate.57 G9a methylates itself and mediates the interaction with the epigenetic regulator heterochromatin protein 1 (HP1).58 G9a heterodimerizes with GLP through its carboxyl terminal SET domain to catalyze methylation.59,60
In addition to H3K9, both proteins, independently and in the complex, mono- and dimethylate histone H3 lysine 27 (H3K27)61 and histone H1 isotype 4 (H1.4).62,63 Additionally, during the G1 phase G9a monomethylates histone H3 lysine 56 (H3K56me1) to regulate DNA replication.64 Furthermore, G9a and GLP methylate a number of nonhistone substrates, including CEBPβ, DNMT1, HDAC1, KLF12,65 and lysine 373 of p53.66
In HIV-1 latency, both G9a and GLP are responsible for transcriptional repression of the HIV-LTR by promoting repressive dimethylation at H3K9.67 Imai et al. reported that wild-type, but not mutant, G9a lacking the SET domain significantly inhibited basal and tumor necrosis factor alpha (TNFα) or Tat-induced HIV-1 gene expression in ACH-2 and OM10.1 T cell lines. Treatment with BIX01294, a chemical inhibitor of G9a, reactivated HIV-1 in latently infected cells. When G9a expression was knocked down by small interfering RNAs, HIV-1 replication was augmented from cells transiently transfected with a full-length HIV-1 clone. The authors also used chromatin immunoprecipitation assays to show G9a and H3K9me2 on histones in the vicinity of the HIV-1 LTR.67 H3K9 dimethylation was also decreased after treatment with 5′-deoxy-5′-methylthioadenosine, a broad-spectrum histone methyltransferase inhibitor, causing reactivation of latent HIV-1 in C11 cells.67
In addition, chemical inhibition of G9a by the compound UNC-0638 reactivated latent proviruses in the Th17 primary T cell model of HIV latency, as well as in resting memory T cells isolated from aviremic HIV-1-infected individuals.54 As Ding et al. showed that GLP knockdown also induces HIV-1 LTR expression,68 the two EHMT enzymes may be significant in maintaining HIV-1 latency by catalyzing dimethylation of H3K9.68
SETD2
The histone-lysine N-methyltransferase SETD2, also referred to as HYPB, trimethylates lysine 36 on histone 3 (H3K36me3), using dimethylated H3K36me2 as substrate.69 H3K36me3 is a signature chromatin mark associated with active transcription implicated in coupling transcription with mRNA splicing.70 Furthermore, SETD2 has been linked for maintenance of genomic integrity through coordination of homologous recombination repair after double strand breaks, suggesting a role as tumor suppressor.71–73
In HIV infection, SETD2 is recruited to the RNA polymerase II (RNAPII) elongation complex in HLM107 cells, a HeLa-derived cell line that contains a single integrated, Rev-defective, HIV-1 provirus.74 Previously, Yoh et al. reported that Spt6, a transcription elongation factor and histone H3 chaperone, binds serine 2 (Ser2P) of the C-terminal domain of RNAPII and recruits the “interacts-with-Spt6” (Iws1) transcription factor and the REF1/Aly mRNA export adaptor to facilitate mRNA export.75 Subsequently, the same authors showed that Iws1 recruits SETD2 to the RNAPII elongation complex, which is required for H3K36 trimethylation across the transcribed region of the HIV-1 gene in HLM107 cells.74
Knockdown of Iws1 disrupts binding of SETD2, but not Spt6, to the coding region of the integrated HIV-1 provirus, whereas depletion of SETD2 did not affect binding of these factors. These data suggest a mechanism by which Iws1 connects SETD2-mediated H3K36me3 with Spt6-driven nucleosome reassembly. Both are thought to depend upon the histone H3 chaperone activity of Spt6, thereby affecting mRNA export and the histone modification state of actively transcribed HIV-1.74
SET domain containing lysine methyltransferase SETD7 (SET7/9, KMT7)
SETD7 was first identified to specifically monomethylate histone 3 lysine 4 (H3K4me1), a marker for transcriptional activation.76,77 In addition, SETD7 methylates several nonhistone proteins, including p53,78 retinoblastoma (RB),79 NF-κB subunit p65,80 TAF10,81 ARTD1,82 estrogen receptor (ER),83 HIF-1α,84 STAT3,85 DNMT1,86 SOX2,87 SUV39H1,88 PGC-1α,89 β-catenin,90 IFITM3,91 FOXO3,92 and YY1.93 Despite the large number of substrates, SETD7's exact in vivo function remains largely undetermined. SETD7 knockout mouse models have no obvious developmental defects and show no apparent deficiencies in DNA damage and p53 responses.94,95
SETD7 functions as a coactivator of HIV transcription.11 The enzyme associates in vivo with the HIV promoter in reactivated J-Lat T cell models of HIV latency and monomethylates lysines 51 and 71, two highly conserved residues in Tat.11,14 Knockdown of SETD7 suppresses Tat transactivation of the HIV promoter, but does not affect the transcriptional activity of methylation-deficient Tat (K51A or K71R).11,14 SETD7 itself binds TAR RNA and forms a complex with Tat and P-TEFb, suggesting a positive role of SETD7-mediated Tat methylation in early steps of the Tat transactivation cycle.11
SET domain, bifurcated 1 (SETDB1)
SETDB1 regulates the trimethylation of histone H3 on lysine 9 (H3K9me3), a specific mark for gene silencing and transcriptional repression.77–79 In mouse ES cells, SETDB1 recruited to histone 3 by the Krüppel-associated box-associated protein 1 (KAP1/TRIM28/TIF1B), results in H3K9 trimethylation and subsequent silencing of endogenous and introduced retroviruses.80 The human SETDB1 protein is organized into six domains: the tandem Tudor domains in the N-terminal region,96 the methyl-CpG-binding domain in the middle region,78 and the pre-SET, SET, and post-SET domains in the C-terminus.77,78 Interestingly, to function as an H3K9 MT, SETDB1 needs to undergo posttranslational modifications.78 The C-terminal region of SETDB1 is ubiquitinated at lysine 867, which is necessary for full H3K9 MT activity in mammalian cells.97
SETDB1 associates with the HIV-Tat protein.13 SiRNA knockdown of SETDB1 in cell systems with both transient and integrated LTR reporter genes results in increased transcription of the HIV-LTR in the presence of suboptimal levels of Tat, indicating a repressive role for SETDB1 in HIV transcription.13 In vitro methylation assays with Tat peptides and SETDB1 show increased incorporation of methyl groups on lysine 51; however, lysine 50 was also susceptible for methylation.13 The association of Tat with histone methyltransferases and the ability for Tat to be differentially methylated at K51 suggest a very sophisticated mechanism of transcriptional regulation of the HIV-1 promoter as monomethylation by SETD7 acts as an activator and trimethylation by SETDB1 presumably as an inhibitory mark in Tat.
SET domain, bifurcated 2 (SETDB2)
SETDB2, also referred to as CLLD8/KMT1F, belongs to the SUV39 subfamily of histone 3 lysine 9 MTs. SETDB2 specifically trimethylates H3K9me3. The H3K9 histone methylation mark is associated with gene silencing, and recent work has linked SETDB2 to antiviral and anti-inflammatory responses through negative regulation of lipopolysaccharide and IFNβ-induced genes in macrophages.98,99 Furthermore, SETDB2 has been associated with embryonic development and cell division.100–102
SETDB2 coimmunoprecipitates with the HIV-Tat protein, but the function of this in vitro association remains to be determined.13 A separate study evaluating epigenetic modifications in primary blood cells after HIV-1 infection found SETDB2 transcripts highly upregulated in activated as well as resting cells, indicating that HIV-1 infection could be linked to the expression of this gene.28
SET and MYND domain-containing protein 2 (SMYD2)
SMYD2 is a member of the SMYD family of five methyltransferases. SMYD1–5 contain a catalytic SET domain that is split by a zinc finger that contains the myeloid translocation protein-8, Nervy, and DEAF-1 (MYND) motif, followed by a cysteine-rich post-SET domain.103 SMYD2 regulates transcription by methylating H3K36 and H3K4, functioning as a repressor or activator, respectively, depending on the presence of heat shock protein 90 (HSP90).103,104 SMYD2 itself methylates HSP90 and, in muscle cells, controls protein complex formation in the cytoplasm.105 Furthermore, SMYD2 inhibits p53 function by methylating lysine 370 (K370)106 as well as K810 and K860 within the RB tumor suppressor (citation). Other nonhistone substrates of SMYD2 include ERα, poly-(ADP-ribose) polymerase 1 (PARP1), BTF3, PDAP1, AHNAK, and AHNAK2.107
In latently infected T cell lines and in primary CD4+ T cells knockdown of SMYD2 or its pharmacological inhibition reactivates latent HIV-1, identifying SMYD2 as a repressor of HIV transcription.108 This repressive function is associated with the enrichment of monomethylated lysine 20 at histone H4 (H4K20me1) at the latent promoter, a modification catalyzed by SMYD2 in vitro and in cells. The H4K20me1 “reader protein” lethal 3 malignant brain tumor 1 (L3MBTL1), which has chromatin-compacting properties, is recruited to the latent HIV-1 promoter in a SMYD2-dependent manner, supporting the model that the SMYD2-H4K20me1-L3MBTL1 axis promotes latency in HIV infection.108
Suppressor of variegation 3–9 homolog 1 (SUV39H1)
SUV39H1, also known as KMT1A, was the first identified human lysine MT.109 It specifically catalyzes trimethylation on histone H3 lysine 9 (H3K9me3) using monomethylated H3K9 as substrate.109 H3K9me3 is a hallmark of facultative and constitutive heterochromatin and is enriched in silenced genes.110 SUV39H1 exerts its function in complexes with other H3K9 MTs, such as G9a, GLP, and SETDB1, by creating a binding site for HP1α, which binds H3K9me2/3 through its chromodomain.111 HP1α further recruits SUV4–20H enzymes to heterochromatic regions, which generate H4K20me3, another heterochromatic histone modification.112 SUV39H1 consists of a catalytic SET domain and a chromodomain, a reader domain for trimethylated H3K9.113–115 The catalytic activity of SUV39H1 is reduced by trimethylated H3K4 confirming an observation in Drosophila that H3K9 methylation-dependent heterochromatin formation is initiated through active removal of H3K4 methylation.116,117 So far, only histones have been identified as substrates for SUV39H1.
Several studies link SUV39H1 to HIV-1 latency. Marban et al. reported that, in microglial cells, DNA-bound CTIP2 associates with SUV39H1, which increases local H3K9 methylation.118 This results in the recruitment of HP1 proteins to the viral promoter and formation of local heterochromatin, leading to HIV-1 silencing.118 Similarly, du Chene et al. showed that SUV39H1, HP1γ, and H3K9me3 are associated with chromatin-mediated repression of integrated HIV-1 gene expression in several cell systems, including HeLa cells containing a single integrated copy of an HIV-1 LTR reporter gene and peripheral blood mononucleated cells (PBMCs) isolated from infected individuals.119 The importance of H3K9 trimethylation has also been shown in recent work using chaetocin, an SUV39H1/G9a histone MT inhibitor (HMTI).120
Chaetocin treatment caused a 25-fold induction of latent HIV-1 expression in Jurkat cells containing a pTY-LAI-luciferase reporter virus, while exhibiting minimal toxicity and T cell activation. Induction of HIV-1 gene expression is associated with loss of H3K9 trimethylation at the viral LTR, and a corresponding increase in H3K9 acetylation, a marker for gene activation.1,120 A separate study evaluating the therapeutic potential of MT inhibitors in HIV infection found that chaetocin induced HIV-1 transcription in 50% of CD8+-depleted PBMCs and in 86% of resting CD4+ T cell cultures isolated from aviremic HIV-1-infected individuals.121 Besides chaetocin, multiple short-chain fatty acids from periodontal pathogens and also therapeutic doses of irradiation reactivate HIV-1 and reduce the presence of SUV39H1 at the HIV-1 promoter.122,123
B Lymphocyte-induced maturation protein-1 (Blimp-1)
The RIZ family member BLIMP-1, also referred to as PR domain zinc finger protein 1 (PRDM1), is a transcription factor that contains five Krüppel-like zinc fingers that mediate DNA binding, nuclear import, and recruitment of histone-modifying enzymes, as well as a PR/SET domain at the amino terminus (reviewed in Ref.124). BLIMP-1 is critical for the differentiation of mature B cells into immunoglobulin-secreting cells and is also expressed in dendritic cells, macrophages, keratinocytes, and T cells.125–129 In T cells, BLIMP-1 regulates the activation and generation of CD4+ and CD8+ T cell effector populations.129–131 BLIMP-1 represses the transcription of several regulatory factors, including Bcl-6, T-bet, IL-2, IFN-γ, and IFN-β, while enhancing the transcription of IL-10.132–135
Several studies address the role of BLIMP-1 in HIV-1 infection in different cell populations.136–139 De Masson et al. showed that BLIMP-1 overexpression is associated with low viral transcription levels in central memory CD4+ T cells from HIV-1 elite controllers, a rare group of HIV-positive individuals who maintain undetectable viral loads in the absence of any treatment, suggesting that induction of BLIMP-1 may reduce the size of the HIV-1 reservoirs.139 Other studies showed that BLIMP-1 expression is increased in chronically infected HIV-1 patients and correlates with enhanced expression of negative regulators of T cell activation, including PD-1, LAG3, and CTLA-4, as well as with T cell exhaustion and apoptosis.136,137
Seddiki et al. reported higher BLIMP-1 levels in total CD4+ T cells from HIV-1 progressors than long-term nonprogressors or healthy controls, implying a positive role in HIV infection.138 In support of this, the HIV-1 transactivator Tat was shown to induce BLIMP-1 expression in activated, but not in resting, CD8+ T cells.140 Furthermore, Tat upregulates, in both activated CD4+ and CD8+ T cells, the release of IL-2,140 which is an inducer of BLIMP-1,141 supporting a model that in activated T cells BLIMP1 could support HIV infection.
At the molecular level, the HIV-1 LTR includes binding sites for BLIMP-1, suggesting that this factor directly binds proviral DNA.142 BLIMP-1 was found highly expressed in memory, as compared with naive CD4+ T cells, where it represses basal and Tat-mediated HIV-1 transcription.143 To do this, BLIMP-1 binds an interferon-stimulated response element within the HIV-1 provirus under latent conditions and is displaced after T cell activation. Reduction of BLIMP-1 in infected primary T cells increases RNA polymerase II processivity, histone acetylation, and basal HIV-1 transcription.143 It remains unknown whether and how the SET domain of BLIMP1 is involved. However, it is intriguing that BLIMP1 has these strong repressive activities and we speculate that high BLIMP-1 levels in memory T cells contribute to the establishment and maintenance of latent HIV-1.
Protein arginine N-methyltransferases
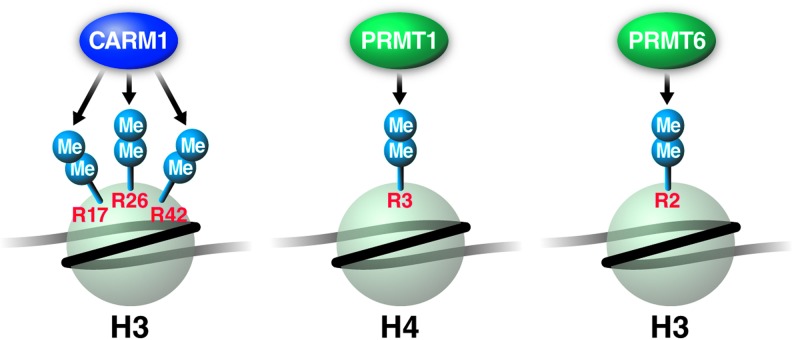
In mammals, nine protein arginine methyltransferases (PRMT1-9) transfer methyl groups from SAM to the terminal guanidino nitrogens of arginine residues in target proteins44 (Fig. 3). PRMT family members are characterized by four different motifs (I, post-I, II, and III) and a conserved threonine-histidine-tryptophan-loop important for the formation of the AdoMet-binding pocket.144 Type I PRMTs form asymmetric (ω-NG,ω-NG) dimethyl-arginine, whereas type II enzymes give rise to symmetric (ω-NG,ω-N‘G) dimethyl-arginine.145 Type III PRMTs generate monomethyl arginine at terminal nitrogen atoms (ω-NG-methylarginine), which is an intermediate of PRMT type I and II reactions.146 Methylation of an arginine does not alter the positive charge of the guanidinium side chain, but changes its structure and the affinity between the substrate and its binding partners, resulting in promotion or inhibition of interactions.147 So far, CARM1, PRMT1, and PRMT6 have been implicated in HIV infection (Fig. 3).
FIG. 3.
Histone arginine methyltransferases (PRMT) and histone methyl marks implicated in HIV infection, showing residue-specific PRMTs for H3 and H4.
Coactivator-associated arginine methyltransferase 1 (CARM1)
CARM1, also called PRMT4, produces mainly asymmetric dimethylated arginines (Rme2a) and methylates histone H3 primarily on two sites, R17 and R26, with an additional methylation site R42 recently identified.148–151 CARM1 is recruited to promoters upon gene activation and methylates multiple nonhistone proteins involved in gene transcription, including transcriptional coactivators CBP/p300 and SRCs; mediator Med12; RNA pol II; RNA-binding proteins PABP1, HuR, and HuD, as well as the splicing factors CA150, SAP49, and SmB.152–159
A recent study, investigating how H3K27 acetylation (H3K27ac) regulates HIV-1 transcription, found that acetylation at H3K27 (H3K27ac) increases H3R26 methylation (H3K26me2a) catalyzed by CARM1.160 H3K27ac is a key epigenetic mark that correlates with gene transcriptional activation.161,162 It recruits the super elongation complex (SEC), which is essential for HIV-1 LTR-mediated transcription.160 As H3K27ac stimulates H3R26 methylation, this methylation mark interferes with SEC recruitment, initiating a negative regulatory feedback loop.160 In line with CARM1's negative effect on SEC recruitment, its inhibition resulted in reactivation of HIV-1 transcription in several HIV latency cell models, including in primary resting CD4+ T cells, and acted synergistically with other latency reversing agents, such as the BET inhibitor JQ1 and HDAC inhibitor SAHA.160
PRMT1
PRMT1 is a type I protein arginine methyltransferase that is primarily responsible for asymmetric dimethylation of H4R3me2a, a mark of transcriptional activation.163 In addition, PRMT1 modifies a large number of other proteins, including histone H2, H3, hnRNPs, RNA helicase A, ERα, PIAS1, CITED2, FOXO1, and TAF15 (reviewed in Ref.44).
In HIV-1 transcription, PRMT1 participates in NF-κB-dependent gene expression.164 Hassa et al. showed that, under TNFα stimulation, PRMT1 synergistically coactivates the HIV-1 LTR together with p300/CREB-binding protein, coactivator-associated arginine methyltransferase 1 (CARM1), and poly (ADP-ribose) polymerase 1(PARP1). PRMT1 forms a nuclear aggregate with p65 and PARP1 and is recruited to p65-containing complexes at the HIV promoter.164 However, the exact mechanism of crosstalk between these coactivators is still unclear. PRMT1 might directly methylate NF-κB, p300, CARM1, or PARP1 or exert its function through dimethylation of H4R3me2a, which has been shown to be essential to maintain “active” chromatin.163
PRMT6
The type I protein arginine methyltransferase PRMT6 catalyzes asymmetric dimethylation of H3R2me2a, using monomethylated arginine as a substrate.165 Dimethylation of H3R2 results in transcriptional repression and is also a negative regulator of H3K4 trimethylation (H3K4me3), as it inhibits the activity of the H3K4 methyltransferase MLL1 and blocks recruitment of the MLL-complex subunit WDR5 to histone H3.165–167 In addition, PRMT6 dimethylates H3R42me2a151 and H2AR292a,168 leading to either transcriptional repression or activation. Similar to other PRMTs, PRMT6 typically targets glycine–arginine-rich (GAR) motifs in substrates.169,170 However, PRMT6 methylates also non-GAR motifs, such as in the HIV-1 Tat protein leading to restriction of viral transcription and replication.171
Interestingly, PRMT6 was also reported to automethylate itself in a non-GAR motif, which results in increased protein stability and activity.172 In agreement with its primary nuclear localization, PRMT6 methylates HMGA1a, a high-mobility group protein involved in transcriptional regulation of genes.169,173 In addition, PRMT6 controls gene expression by directly interacting with transcription factors, including NF-κB and G-protein pathway suppressor 2.174,175 Because those two molecules are directly involved in inflammatory responses, it is possible that PRMT6 also plays a role in inflammation responses.
PRMT6 restricts HIV infection by methylating and impairing the function of various HIV-1 proteins, such as Tat, Rev, and nucleocapsid protein p7 (NC). In the context of HIV-1, PRMT6 was first identified to associate and methylate Tat within cells.171 Overexpression of wild-type PRMT6 decreases Tat transactivation of an HIV-1 LTR reporter plasmid in a dose-dependent manner, and knockdown of PRMT6 increases HIV-1 production in HEK293T cells, indicating that PRMT6 acts as a restriction factor for HIV replication.171 Subsequently, Xie et al. reported that PRMT6 targets R52 and R53 in Tat, which leads to decreased interaction with the Tat transactivation region (TAR) in viral RNAs and also negatively affects complex formation of Tat-TAR with the critical cofactor cyclin T1.15
Interestingly, PRMT6 increases Tat half-life, which could play a critical role in Tat persisting within the infected cell and the extracellular environment.176 Furthermore, overexpression of PRMT6 leads to the exclusion of R52/53-methylated Tat from the nucleolus.177 Experiments using fluorescence recovery after photobleaching indicate that Tat's nucleolar accumulation is largely mediated through binding to nucleolar components, which is prevented by methylation of Tat by PRMT6.177 PRMT6 also dimethylates Rev at a single arginine in the N-terminal portion of its arginine-rich motif and associates with Rev in vivo.178 This study also showed that PRMT6 significantly decreases Rev-mediated viral RNA export from the nucleus to the cytoplasm in a dose-dependent manner.178
Beyond that, PRMT6 dimethylates HIV NC in each of its two basic regions at positions R10 and R32, leading to decreased RNA annealing and diminished initiation of reverse transcription.179 Thus, PRMT6 is an HIV restriction factor that acts at multiple steps of the viral lifecycle. Recently, inhibitors of PRMT6 have been developed for cancer therapy and could also have potential for treatment of HIV infection.180,181
Other methyltransferases
Protein-L-isoaspartate (D-aspartate) O-methyltransferase (PCMT1)
PCMT1, also referred to as PIMT1, is an enzyme that catalyzes methyl esterification of L-isoaspartyl (L-isoAsp) and D-isoaspartyl (D-isoAsp) residues to L-aspartic acid (L-Asp).182 During stress-related conditions, oxidative environmental conditions, and with age, L-Asp and L-Asn residues are nonenzymatically modified through dehydration (L-Asp) or deamidation (L-Asn), resulting in formation and accumulation of L-isoAsp, D-isoAsp, and D-aspartic acid (D-Asp) residues.182 These changes result in structurally nonfunctional proteins.183 In this context, PCMT1 functions as a chaperone or repair enzyme of aged or damaged proteins and facilitates the restoration to aspartate residues.184
PCMT1 is ubiquitously expressed in all living organisms. Since its expression decreases with age, it has been associated with age-related diseases such as Alzheimer's dementia.185 Furthermore, PCMT1-deficient mice show acceleration of aging characterized by induced accumulation of L-isoAsp, D-isoAsp, and D-Asp residues in proteins and dysfunction of proteins.186
PCMT1 methylates histone H4 at aspartic acid 24 (H4D24me).187 This histone mark is recognized by VprBP (HIV-1 viral protein R [Vpr]-binding protein), also known as DCAF1 (DDB1- and CUL4-associated factor 1), which is a chromodomain-containing protein.187 In HIV infection, VprBP is recruited by HIV-1 Vpr to hijack the CUL4A-RBX1-DDB1-DCAF1/VprBP complex leading to cell-cycle arrest in G2 phase, and also to protect the viral protein from proteasomal degradation.188
Trimethylguanosine synthase
Human trimethylguanosine synthase (TGS1) was originally identified as an interaction partner of PRIP (PPAR-interacting protein) and, therefore, named PIMT (PRIP-interacting protein with methyltransferase domain).189 TGS1 is specific for guanine and catalyzes two successive methyl-transfer reactions from AdoMet to the N2 position of 7-methylguanosine.190,191 The enzyme adds two methyl groups to RNAPII-transcribed small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), and telomerase RNA, for the conversion of 7-monomethylguanosine caps to 2,2,7-trimethylguanosine caps.190,191 The 5′-mRNA capping affects pre-mRNA synthesis and splicing, RNA transport to the cytoplasm, and mRNA translation and turnover.
Yedavalli and Jeang showed that, like snRNAs and snoRNAs, some Rev/RRE-dependent HIV-1 RNAs are TMG-capped by TGS1 and proposed a new regulatory mechanism for selective expression.192 The study showed that TGS1 enhances HIV-1 gene expression and that intracellular HIV-1 RNAs are 7-methylguanosine and trimethylguanosine capped. Furthermore, TGS1 selectively modulates the expression p55 and p24 HIV-1 proteins encoded by Rev/RRE-dependent RNAs. Activation of PBMCs or purified CD4+ T cells significantly increased the expression of TGS1, indicating that TGS1 might be a limiting factor in quiescent cells.192
Demethylases
Up to date, two classes of lysine-specific KDMs are known: (1) the amine-oxidase type lysine-specific demethylases 1 and 2 (LSD1 and 2; also known as KDM1A and B, respectively), which are both dependent on flavin adenine dinucleotide (FAD) as a cofactor; and (2) the JumonjiC (JMJC) domain-containing histone demethylase, in which the demethylase activity is dependent on Fe(II) and α-ketoglutarate (2-oxoglutarate) (reviewed in Ref.48). The latter consists of a group with over 30 members and can be divided, based on the JMJC-domain homology, into seven subfamilies (KDM2–8).48,193–195
The LSD family members generate an imine intermediate that is hydrolyzed to the demethylated lysine and formaldehyde.193 Upon recycling of the cofactor FAD, hydrogen peroxide is formed as a byproduct of demethylation. As these enzymes require a free electron pair on the lysine ɛ-nitrogen atom to initiate demethylation, LSD1 and 2 demethylate only mono- and dimethylated, but not trimethylated, lysines.193 In contrast, the JMJC domain-containing demethylases are able to remove methyl groups from all three methyl lysine states, with concomitant production of succinate, carbon dioxide, the demethylated lysine and formaldehyde.196,197 The target specificity of KDMs is regulated by their participation in different complexes.48
So far, only LSD1 and the JMJC protein UTX have been implicated in HIV infection (Fig. 4). No reports link HIV to ten-eleven translocation (TET) enzymes involved in active DNA demethylation.198 A subset of JMJC proteins also act as arginine demethylases.199
FIG. 4.
Histone demethylases and histone methyl marks implicated in HIV infection, showing residue-specific KMTs for H3K4, H3K9, and H3K27.
Lysine demethylases
Lysine-specific histone demethylase 1A (KDM1A, LSD1)
LSD1 is the first discovered histone demethylase and belongs to the family of FAD-dependent amine oxidases.193 LSD1 contains an N-terminal SWIRM domain, a tower domain and an amine oxidase domain at the C-terminus.200–202 In the presence of the RCOR1/CoREST complex, LSD1 catalyzes demethylation of mono- and dimethylated H3K4.193 In addition, LSD1 interacts with JMJD2C, a histone tridemethylase, and cooperatively removes methyl groups from mono-, di- and trimethylated H3K9.203,204 LSD1 also demethylates several nonhistone proteins, including K370 in p53, K185 of E2F1, and K1096, in the DNA methylase DNMT1.205–207
LSD1 is an HIV-Tat K51-specific demethylase, which is required for the activation of HIV-1 transcription in latently infected CD4+ T cells.12 LSD1 and its cofactor CoREST associate with the HIV promoter in vivo and activate Tat transcriptional activity in a K51-dependent manner in T cells.12 In addition, small hairpin RNAs directed against LSD1 or inhibition of its activity with the monoamine oxidase inhibitor phenelzine suppresses the activation of HIV transcription in latently infected T cells, indicating that the LSD1/CoREST complex, normally known as a transcriptional repressor, acts as a coactivator of HIV transcription in infected T cells by demethylating K51 in Tat.12
In contrast, in microglial cells, an important HIV-1 target in the central nervous system, LSD1 represses HIV-1 transcription and viral expression in a synergistic manner with the COUP-TF interacting protein 2 (CTIP2).208 CTIP2 forces heterochromatin formation and HIV-1 gene silencing by recruiting HDAC and HMT activities at the integrated viral promoter.118 Le Douce et al. further showed that recruitment of LSD1 at the HIV-1 proximal promoter is associated with H3K4me3 and H3K9me3 epigenetic marks and that LSD1-induced H3K4 trimethylation is linked to SETD1A recruitment at the integrated provirus.208 The cell-type-specific role of LSD1 in HIV-1 infection remains an interesting subject of investigation.
However, in latently infected T cells LSD1 inhibitors function to suppress reactivation of HIV transcription, in accordance with a proposed coactivator role of LSD1 in this cell type. Interestingly, they also suppress reactivation of latent infections of α-herpesvirus,209 suggesting a broader role of LSD1 in activating viral gene expression.
Lysine-specific demethylase 6A (KDM6A, UTX)
KDM6A, also referred to as UTX, specifically demethylates lysine 27 of histone 3 (H3K27).210,211 UTX demethylates di- and trimethylated, but not monomethylated, H3K27.212 Since H3K27me2 and H3K27me3 are highly correlated with genomic silencing and repression of transcription, removal of these marks by UTX results in gene activation.213 In addition, UTX interacts with protein complexes that are associated with H3K4 methylation, a mark of active transcription.214,215 Furthermore, UTX binds the SWI/SNF chromatin-remodeling complex by engaging the BRG1 catalytic subdomain, resulting in nucleosome remodeling and gene activation of T-box factors.216
At the HIV-1 LTR, the removal of the H3K27me3 mark by UTX is important for the robust induction of many specific genes during Tat-mediated HIV-1 transactivation.217 Zhang et al. reported that Tat upregulates the expression of UTX and downregulates the H3K27me3 mark in TZM-bl cells.217 Using chromatin immunoprecipitation assays, the authors found that UTX associates with nucleosomes at the 0 and +1 position relative to the start of transcription and downregulates the H3K27me2 and H3K27me3 states.217 Also, UTX promoted HIV-1 gene expression by enhancing NF-κB p65 nuclear translocation, suggesting a novel function of UTX in the timely transition from poised to active chromatin in HIV-1 infection.217
Concluding remarks
Current antiretroviral therapy suppresses HIV viral replication, requiring life-long adherence to continuously limit viral loads, but does not typically eradicate virus from the host. Successful sterilizing cure of HIV will require elimination of persistent viral reservoirs. One approach is to reactivate proviral genomes in latently infected cells to “purge” viral reservoirs.218 Another is to find ways to permanently silence HIV in latent reservoirs so that likelihood of reactivation is diminished. Several epigenetic modifying agents, including inhibitors of HDAC, HMT, and DNMT, show great promise as antilatency therapeutics in vitro and ex vivo, and some, including valproic acid and SAHA/vorinostat, are currently being examined in clinical trials. Although the role of DNA methylation in HIV-1 infection is controversial, DNMT inhibitors are considered as reactivating agents. For example, the DNMT inhibitors Decitabine (5-aza-2′ deoxycytidine, aza-CdR) and its analog azacitidine (5-azacytidine, Vidaza®), which are approved by the FDA for the treatment of myelodysplastic syndrome, are inducers of latent HIV-1.219 In addition, decitabine synergizes with TNF-α and prostratin to significantly increase viral gene expression in several J-Lat cell lines.26
Similarly, PMTs and demethylases are regulators with emerging impacts on HIV infection. Both groups harbor repressors (EZH2,53 G9a,67 SETDB1,13 SMYD2,108 SUV39H1,118,119 CARM1,160 and PRMT615,171) and coactivators (SETD7,11,14 LSD1,12,208 UTX217) for HIV infection through histone and nonhistone protein modification, most importantly Tat and NF-κB. Several inhibitors, such as BIX01294,67 Chaetocin,120 UNC-0638,54 3-deazaneplanocin,53 AZ391,108 GSK-343,54,55 and EPZ-6438,54 have been tested in preclinical model cell lines for latency reactivation as well as in cells derived from patients.67 Interestingly, some HMT inhibitors, as discussed above, can enhance proviral reactivation in combination with bromodomain inhibitors, such as JQ1,108 or PKC agonists (prostratin), or HDAC inhibitors (such as vorinostat/SAHA),55 making this class of compounds possible candidates for combinatorial treatments.
Although research into HMT inhibitors and HIV latency reversal is less advanced than HDAC inhibitors, some methyltransferase inhibitors are now in clinical trials for the treatment of malignancies, making future clinical use in HIV infection more feasible. For example, the EZH2 inhibitors GSK126, tazemetostat, and CPI-1205, are currently in phase I clinical trials for the treatment of B cell lymphomas bearing EZH2-activating mutations (reviewed in Ref.48). Similarly, LSD1 inhibitors TCP, ORY-1001, GSK2879552, and 4SC-202 are in current trials for the treatment of acute myeloid leukemia (AML), acute leukemia (phase I/IIA), for AML and small cell lung cancer (phase I), and hematological malignancies (phase I), respectively (reviewed in Ref.48).
Acknowledgments
The authors thank the members of the Ott laboratory for helpful discussions. They thank Gary Howard and Lisa Loeb Stanga for editorial, John Carrol for graphics, and Veronica Fonseca for administrative assistance. This publication was made possible with the help from the amfAR Institute for HIV Cure Research, with funding from amfAR grant number 109301. They gratefully acknowledge support from the California HIV/AIDS Research Program (award no.: F13-GI-316) to D.B., as well as grant support from the CARE Collaboratory (U19 AI096113) and the NIH (RO1 AI083139 and RO1 DA043142) to M.O.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Kouzarides T: Chromatin modifications and their function. Cell 2007;128:693–705 [DOI] [PubMed] [Google Scholar]
- 2.Allis CD, Jenuwein T: The molecular hallmarks of epigenetic control. Nat Rev Genet 2016;17:487–500 [DOI] [PubMed] [Google Scholar]
- 3.Mbonye U, Karn J: Transcriptional control of HIV latency: Cellular signaling pathways, epigenetics, happenstance and the hope for a cure. Virology 2014;454–455:328–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Lint C, Emiliani S, Ott M, Verdin E: Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J 1996;15:1112–1120 [PMC free article] [PubMed] [Google Scholar]
- 5.Budhiraja S, Famiglietti M, Bosque A, Planelles V, Rice AP: Cyclin T1 and CDK9 T-loop phosphorylation are downregulated during establishment of HIV-1 latency in primary resting memory CD4+ T cells. J Virol 2013;87:1211–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duverger A, Jones J, May J, et al. : Determinants of the establishment of human immunodeficiency virus type 1 latency. J Virol 2009;83:3078–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tyagi M, Pearson RJ, Karn J: Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J Virol 2010;84:6425–6437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sobhian B, Laguette N, Yatim A, et al. : HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell 2010;38:439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marzio G, Tyagi M, Gutierrez MI, Giacca M: HIV-1 tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc Natl Acad Sci U S A 1998;95:13519–13524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lusic M, Marcello A, Cereseto A, Giacca M: Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J 2003;22:6550–6561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pagans S, Kauder SE, Kaehlcke K, et al. : The Cellular lysine methyltransferase Set7/9-KMT7 binds HIV-1 TAR RNA, monomethylates the viral transactivator Tat, and enhances HIV transcription. Cell Host Microbe 2010;7:234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakane N, Kwon HS, Pagans S, et al. : Activation of HIV transcription by the viral Tat protein requires a demethylation step mediated by lysine-specific demethylase 1 (LSD1/KDM1). PLoS Pathog 2011;7:e1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Duyne R, Easley R, Wu W, et al. : Lysine methylation of HIV-1 Tat regulates transcriptional activity of the viral LTR. Retrovirology 2008;5:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ali I, Ramage H, Boehm D, et al. : The HIV-1 Tat protein is monomethylated at lysine 71 by the lysine methyltransferase KMT7. J Biol Chem 2016;291:16240–16248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie B, Invernizzi CF, Richard S, Wainberg MA: Arginine methylation of the human immunodeficiency virus type 1 Tat protein by PRMT6 negatively affects Tat Interactions with both cyclin T1 and the Tat transactivation region. J Virol Apr 2007;81:4226–4234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han Y, Lassen K, Monie D, et al. : Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J Virol 2004;78:6122–6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewinski MK, Yamashita M, Emerman M, et al. : Retroviral DNA integration: Viral and cellular determinants of target-site selection. PLoS Pathog 2006;2:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ylisastigui L, Archin NM, Lehrman G, Bosch RJ, Margolis DM: Coaxing HIV-1 from resting CD4 T cells: Histone deacetylase inhibition allows latent viral expression. AIDS 2004;18:1101–1108 [DOI] [PubMed] [Google Scholar]
- 19.Williams SA, Chen LF, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC: NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J 2006;25:139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones PA, Liang G: Rethinking how DNA methylation patterns are maintained. Nat Rev Genet 2009;10:805–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen T, Li E: Structure and function of eukaryotic DNA methyltransferases. Curr Top Dev Biol 2004;60:55–89 [DOI] [PubMed] [Google Scholar]
- 22.Hsieh CL: Dependence of transcriptional repression on CpG methylation density. Mol Cell Biol 1994;14:5487–5494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siegfried Z, Eden S, Mendelsohn M, Feng X, Tsuberi BZ, Cedar H: DNA methylation represses transcription in vivo. Nat Genet 1999;22:203–206 [DOI] [PubMed] [Google Scholar]
- 24.Domcke S, Bardet AF, Adrian Ginno P, Hartl D, Burger L, Schubeler D: Competition between DNA methylation and transcription factors determines binding of NRF1. Nature 2015;528:575–579 [DOI] [PubMed] [Google Scholar]
- 25.Nan X, Ng HH, Johnson CA, et al. : Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 1998;393:386–389 [DOI] [PubMed] [Google Scholar]
- 26.Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E: Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog 2009;5:e1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blazkova J, Trejbalova K, Gondois-Rey F, et al. : CpG methylation controls reactivation of HIV from latency. PLoS Pathog 2009;5:e1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maricato JT, Furtado MN, Takenaka MC, et al. : Epigenetic modulations in activated cells early after HIV-1 infection and their possible functional consequences. PLoS One 2015;10:e0119234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trejbalova K, Kovarova D, Blazkova J, et al. : Development of 5′ LTR DNA methylation of latent HIV-1 provirus in cell line models and in long-term-infected individuals. Clin Epigenetics 2016;8:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blazkova J, Murray D, Justement JS, et al. : Paucity of HIV DNA methylation in latently infected, resting CD4+ T cells from infected individuals receiving antiretroviral therapy. J Virol 2012;86:5390–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandel N, Ayasolla KS, Lan X, et al. : Epigenetic modulation of human podocyte vitamin D receptor in HIV Milieu. J Mol Biol 2015;427:3201–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dev RR, Ganji R, Singh SP, Mahalingam S, Banerjee S, Khosla S: Cytosine methylation by DNMT2 facilitates stability and survival of HIV-1 RNA in the host cell during infection. Biochem J 2017;474:2009–2026 [DOI] [PubMed] [Google Scholar]
- 33.Subramaniam D, Thombre R, Dhar A, Anant S: DNA methyltransferases: A novel target for prevention and therapy. Front Oncol 2014;4:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rountree MR, Bachman KE, Baylin SB: DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet 2000;25:269–277 [DOI] [PubMed] [Google Scholar]
- 35.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP: DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet 2000;25:338–342 [DOI] [PubMed] [Google Scholar]
- 36.Mikovits JA, Young HA, Vertino P, et al. : Infection with human immunodeficiency virus type 1 upregulates DNA methyltransferase, resulting in de novo methylation of the gamma interferon (IFN-gamma) promoter and subsequent downregulation of IFN-gamma production. Mol Cell Biol 1998;18:5166–5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fang JY, Mikovits JA, Bagni R, Petrow-Sadowski CL, Ruscetti FW: Infection of lymphoid cells by integration-defective human immunodeficiency virus type 1 increases de novo methylation. J Virol 2001;75:9753–9761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Youngblood B, Reich NO: The early expressed HIV-1 genes regulate DNMT1 expression. Epigenetics 2008;3:149–156 [DOI] [PubMed] [Google Scholar]
- 39.Pion M, Jaramillo-Ruiz D, Martinez A, Munoz-Fernandez MA, Correa-Rocha R: HIV infection of human regulatory T cells downregulates Foxp3 expression by increasing DNMT3b levels and DNA methylation in the FOXP3 gene. AIDS 2013;27:2019–2029 [DOI] [PubMed] [Google Scholar]
- 40.Martinez-Colom A, Lasarte S, Fernandez-Pineda A, Relloso M, Munoz-Fernandez MA: A new chimeric protein represses HIV-1 LTR-mediated expression by DNA methylase. Antiviral Res 2013;98:394–400 [DOI] [PubMed] [Google Scholar]
- 41.Ghosh SK, McCormick TS, Eapen BL, Yohannes E, Chance MR, Weinberg A: Comparison of epigenetic profiles of human oral epithelial cells from HIV-positive (on HAART) and HIV-negative subjects. Epigenetics 2013;8:703–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosca A, Anton G, Ene L, et al. : Immunoassay and molecular methods to investigate DNA methylation changes in peripheral blood mononuclear cells in HIV infected patients on cART. J Immunoassay Immunochem 2017;38:299–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, Justice AC, Hu Y, et al. : Epigenome-wide differential DNA methylation between HIV-infected and uninfected individuals. Epigenetics 2016;1–11:750–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim JH, Yoo BC, Yang WS, Kim E, Hong S, Cho JY: The role of protein arginine methyltransferases in inflammatory responses. Mediators Inflamm 2016;2016:4028353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeates TO: Structures of SET domain proteins: Protein lysine methyltransferases make their mark. Cell Oct 4 2002;111:5–7 [DOI] [PubMed] [Google Scholar]
- 46.Romano JD, Michaelis S: Topological and mutational analysis of Saccharomyces cerevisiae Ste14p, founding member of the isoprenylcysteine carboxyl methyltransferase family. Mol Biol Cell 2001;12:1957–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E: PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res 2015;43(Database issue):D512–D520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morera L, Lubbert M, Jung M: Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin Epigenetics 2016;8:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao R, Zhang Y: The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev 2004;14:155–164 [DOI] [PubMed] [Google Scholar]
- 50.Gall Troselj K, Novak Kujundzic R, Ugarkovic D: Polycomb repressive complex's evolutionary conserved function: The role of EZH2 status and cellular background. Clin Epigenetics 2016;8:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao R, Zhang Y: SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell 2004;15:57–67 [DOI] [PubMed] [Google Scholar]
- 52.Chu CS, Lo PW, Yeh YH, et al. : O-GlcNAcylation regulates EZH2 protein stability and function. Proc Natl Acad Sci U S A 2014;111:1355–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Friedman J, Cho WK, Chu CK, et al. : Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of Zeste 2. J Virol 2011;85:9078–9089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen K, Das B, Dobrowolski C, Karn J: Multiple histone lysine methyltransferases are required for the establishment and maintenance of HIV-1 latency. MBio 2017;8:e00133–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tripathy MK, McManamy ME, Burch BD, Archin NM, Margolis DM: H3K27 demethylation at the proviral promoter sensitizes latent HIV to the effects of vorinostat in ex vivo cultures of resting CD4+ T Cells. J Virol 2015;89:8392–8405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kramer JM: Regulation of cell differentiation and function by the euchromatin histone methyltranserfases G9a and GLP. Biochem Cell Biol 2016;94:26–32 [DOI] [PubMed] [Google Scholar]
- 57.Collins RE, Northrop JP, Horton JR, et al. : The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat Struct Mol Biol 2008;15:245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sampath SC, Marazzi I, Yap KL, et al. : Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol Cell 2007;27:596–608 [DOI] [PubMed] [Google Scholar]
- 59.Chang Y, Zhang X, Horton JR, et al. : Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nat Struct Mol Biol 2009;16:312–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tachibana M, Ueda J, Fukuda M, et al. : Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3–K9. Genes Dev 2005;19:815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y: Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem 2001;276:25309–25317 [DOI] [PubMed] [Google Scholar]
- 62.Trojer P, Zhang J, Yonezawa M, et al. : Dynamic Histone H1 isotype 4 methylation and demethylation by histone lysine methyltransferase G9a/KMT1C and the jumonji domain-containing JMJD2/KDM4 Proteins. J Biol Chem 2009;284:8395–8405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weiss T, Hergeth S, Zeissler U, et al. : Histone H1 variant-specific lysine methylation by G9a/KMT1C and Glp1/KMT1D. Epigenetics Chromatin 2010;3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu Y, Song C, Zhang Q, et al. : Histone H3 lysine 56 methylation regulates DNA replication through its interaction with PCNA. Mol Cell 2012;46:7–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shankar SR, Bahirvani AG, Rao VK, Bharathy N, Ow JR, Taneja R: G9a, a multipotent regulator of gene expression. Epigenetics 2013;8:16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang J, Dorsey J, Chuikov S, et al. : G9a and Glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem 2010;285:9636–9641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Imai K, Togami H, Okamoto T: Involvement of histone H3 lysine 9 (H3K9) methyltransferase G9a in the maintenance of HIV-1 latency and its reactivation by BIX01294. J Biol Chem 2010;285:16538–16545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ding D, Qu X, Li L, et al. : Involvement of histone methyltransferase GLP in HIV-1 latency through catalysis of H3K9 dimethylation. Virology 2013;440:182–189 [DOI] [PubMed] [Google Scholar]
- 69.Edmunds JW, Mahadevan LC, Clayton AL: Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J 2008;27:406–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pokholok DK, Harbison CT, Levine S, et al. : Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 2005;122:517–527 [DOI] [PubMed] [Google Scholar]
- 71.Pfister SX, Ahrabi S, Zalmas LP, et al. : SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep 2014;7:2006–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carvalho S, Vitor AC, Sridhara SC, et al. : SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. Elife 2014;3:e02482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Duns G, van den Berg E, van Duivenbode I, et al. : Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res 2010;70:4287–4291 [DOI] [PubMed] [Google Scholar]
- 74.Yoh SM, Lucas JS, Jones KA: The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev 2008;22:3422–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoh SM, Cho H, Pickle L, Evans RM, Jones KA: The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev 2007;21:160–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang H, Cao R, Xia L, et al. : Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell 2001;8:1207–1217 [DOI] [PubMed] [Google Scholar]
- 77.Xiao B, Jing C, Wilson JR, et al. : Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature 2003;421:652–656 [DOI] [PubMed] [Google Scholar]
- 78.Chuikov S, Kurash JK, Wilson JR, et al. : Regulation of p53 activity through lysine methylation. Nature 2004;432:353–360 [DOI] [PubMed] [Google Scholar]
- 79.Munro S, Khaire N, Inche A, Carr S, La Thangue NB: Lysine methylation regulates the pRb tumour suppressor protein. Oncogene 2010;29:2357–2367 [DOI] [PubMed] [Google Scholar]
- 80.Yang XD, Huang B, Li M, Lamb A, Kelleher NL, Chen LF: Negative regulation of NF-kappaB action by Set9-mediated lysine methylation of the RelA subunit. EMBO J 2009;28:1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kouskouti A, Scheer E, Staub A, Tora L, Talianidis I: Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol Cell 2004;14:175–182 [DOI] [PubMed] [Google Scholar]
- 82.Kassner I, Andersson A, Fey M, Tomas M, Ferrando-May E, Hottiger MO: SET7/9-dependent methylation of ARTD1 at K508 stimulates poly-ADP-ribose formation after oxidative stress. Open Biol 2013;3:120173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Subramanian K, Jia D, Kapoor-Vazirani P, et al. : Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol Cell 2008;30:336–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim Y, Nam HJ, Lee J, et al. : Methylation-dependent regulation of HIF-1alpha stability restricts retinal and tumour angiogenesis. Nat Commun 2016;7:10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang J, Huang J, Dasgupta M, et al. : Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci U S A 2010;107:21499–21504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Esteve PO, Chin HG, Benner J, et al. : Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc Natl Acad Sci U S A 2009;106:5076–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fang L, Zhang L, Wei W, et al. : A methylation-phosphorylation switch determines Sox2 stability and function in ESC maintenance or differentiation. Mol Cell 2014;55:537–551 [DOI] [PubMed] [Google Scholar]
- 88.Wang D, Zhou J, Liu X, et al. : Methylation of SUV39H1 by SET7/9 results in heterochromatin relaxation and genome instability. Proc Natl Acad Sci U S A 2013;110:5516–5521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Aguilo F, Li S, Balasubramaniyan N, et al. : Deposition of 5-methylcytosine on enhancer RNAs enables the coactivator function of PGC-1alpha. Cell Rep 2016;14:479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shen C, Wang D, Liu X, et al. : SET7/9 regulates cancer cell proliferation by influencing beta-catenin stability. FASEB J 2015;29:4313–4323 [DOI] [PubMed] [Google Scholar]
- 91.Shan Z, Han Q, Nie J, et al. : Negative regulation of interferon-induced transmembrane protein 3 by SET7-mediated lysine monomethylation. J Biol Chem 2013;288:35093–35103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xie Q, Hao Y, Tao L, et al. : Lysine methylation of FOXO3 regulates oxidative stress-induced neuronal cell death. EMBO Rep 2012;13:371–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang WJ, Wu XN, Shi TT, et al. : Regulation of transcription factor Yin Yang 1 by SET7/9-mediated lysine methylation. Sci Rep 2016;6:21718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lehnertz B, Rogalski JC, Schulze FM, et al. : p53-dependent transcription and tumor suppression are not affected in Set7/9-deficient mice. Mol Cell 2011;43:673–680 [DOI] [PubMed] [Google Scholar]
- 95.Campaner S, Spreafico F, Burgold T, et al. : The methyltransferase Set7/9 (Setd7) is dispensable for the p53-mediated DNA damage response in vivo. Mol Cell 2011;43:681–688 [DOI] [PubMed] [Google Scholar]
- 96.Pek JW, Anand A, Kai T: Tudor domain proteins in development. Development 2012;139:2255–2266 [DOI] [PubMed] [Google Scholar]
- 97.Ishimoto K, Kawamata N, Uchihara Y, et al. : Ubiquitination of lysine 867 of the human SETDB1 protein upregulates its histone H3 lysine 9 (H3K9) methyltransferase activity. PLoS One 2016;11:e0165766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kroetz DN, Allen RM, Schaller MA, Cavallaro C, Ito T, Kunkel SL: Type I interferon induced epigenetic regulation of macrophages suppresses innate and adaptive immunity in acute respiratory viral infection. PLoS Pathog 2015;11:e1005338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schliehe C, Flynn EK, Vilagos B, et al. : The methyltransferase Setdb2 mediates virus-induced susceptibility to bacterial superinfection. Nat Immunol 2015;16:67–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xu PF, Zhu KY, Jin Y, et al. : Setdb2 restricts dorsal organizer territory and regulates left-right asymmetry through suppressing fgf8 activity. Proc Natl Acad Sci U S A 2010;107:2521–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Falandry C, Fourel G, Galy V, et al. : CLLD8/KMT1F is a lysine methyltransferase that is important for chromosome segregation. J Biol Chem 2010;285:20234–20241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hogarth CA, Mitchell D, Evanoff R, Small C, Griswold M: Identification and expression of potential regulators of the mammalian mitotic-to-meiotic transition. Biol Reprod 2011;84:34–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brown MA, Sims RJ, 3rd, Gottlieb PD, Tucker PW: Identification and characterization of Smyd2: A split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol Cancer 2006;5:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Abu-Farha M, Lambert JP, Al-Madhoun AS, Elisma F, Skerjanc IS, Figeys D: The tale of two domains: Proteomics and genomics analysis of SMYD2, a new histone methyltransferase. Mol Cell Proteomics 2008;7:560–572 [DOI] [PubMed] [Google Scholar]
- 105.Donlin LT, Andresen C, Just S, et al. : Smyd2 controls cytoplasmic lysine methylation of Hsp90 and myofilament organization. Genes Dev 2012;26:114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huang J, Perez-Burgos L, Placek BJ, et al. : Repression of p53 activity by Smyd2-mediated methylation. Nature 2006;444:629–632 [DOI] [PubMed] [Google Scholar]
- 107.Olsen JB, Cao XJ, Han B, et al. : Quantitative profiling of the activity of protein lysine methyltransferase SMYD2 using SILAC-based proteomics. Mol Cell Proteomics 2016;15:892–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Boehm D, Jeng M, Camus G, et al. : SMYD2-mediated histone methylation contributes to HIV-1 latency. Cell Host Microbe 2017;21:569–579 e566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rea S, Eisenhaber F, O'Carroll D, et al. : Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000;406:593–599 [DOI] [PubMed] [Google Scholar]
- 110.Peters AH, O'Carroll D, Scherthan H, et al. : Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 2001;107:323–337 [DOI] [PubMed] [Google Scholar]
- 111.Fritsch L, Robin P, Mathieu JR, et al. : A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell 2010;37:46–56 [DOI] [PubMed] [Google Scholar]
- 112.Schotta G, Lachner M, Sarma K, et al. : A silencing pathway to induce H3–K9 and H4–K20 trimethylation at constitutive heterochromatin. Genes Dev 2004;18:1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Krouwels IM, Wiesmeijer K, Abraham TE, et al. : A glue for heterochromatin maintenance: Stable SUV39H1 binding to heterochromatin is reinforced by the SET domain. J Cell Biol 2005;170:537–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chin HG, Patnaik D, Esteve PO, Jacobsen SE, Pradhan S: Catalytic properties and kinetic mechanism of human recombinant Lys-9 histone H3 methyltransferase SUV39H1: Participation of the chromodomain in enzymatic catalysis. Biochemistry 2006;45:3272–3284 [DOI] [PubMed] [Google Scholar]
- 115.Wang T, Xu C, Liu Y, et al. : Crystal structure of the human SUV39H1 chromodomain and its recognition of histone H3K9me2/3. PLoS One 2012;7:e52977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Binda O, LeRoy G, Bua DJ, Garcia BA, Gozani O, Richard S: Trimethylation of histone H3 lysine 4 impairs methylation of histone H3 lysine 9: Regulation of lysine methyltransferases by physical interaction with their substrates. Epigenetics 2010;5:767–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rudolph T, Yonezawa M, Lein S, et al. : Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3–3. Mol Cell 2007;26:103–115 [DOI] [PubMed] [Google Scholar]
- 118.Marban C, Suzanne S, Dequiedt F, et al. : Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J 2007;26:412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.du Chene I, Basyuk E, Lin YL, et al. : Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J 2007;26:424–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bernhard W, Barreto K, Saunders A, Dahabieh MS, Johnson P, Sadowski I: The Suv39H1 methyltransferase inhibitor chaetocin causes induction of integrated HIV-1 without producing a T cell response. FEBS Lett 2011;585:3549–3554 [DOI] [PubMed] [Google Scholar]
- 121.Bouchat S, Gatot JS, Kabeya K, et al. : Histone methyltransferase inhibitors induce HIV-1 recovery in resting CD4(+) T cells from HIV-1-infected HAART-treated patients. AIDS 2012;26:1473–1482 [DOI] [PubMed] [Google Scholar]
- 122.Das B, Dobrowolski C, Shahir AM, et al. : Short chain fatty acids potently induce latent HIV-1 in T-cells by activating P-TEFb and multiple histone modifications. Virology 2015;474:65–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Iordanskiy S, Van Duyne R, Sampey GC, et al. : Therapeutic doses of irradiation activate viral transcription and induce apoptosis in HIV-1 infected cells. Virology 2015;485:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Boi M, Zucca E, Inghirami G, Bertoni F: PRDM1/BLIMP1: A tumor suppressor gene in B and T cell lymphomas. Leuk Lymphoma 2015;56:1223–1228 [DOI] [PubMed] [Google Scholar]
- 125.Turner CA, Jr, Mack DH, Davis MM: Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell 1994;77:297–306 [DOI] [PubMed] [Google Scholar]
- 126.Chan YH, Chiang MF, Tsai YC, et al. : Absence of the transcriptional repressor Blimp-1 in hematopoietic lineages reveals its role in dendritic cell homeostatic development and function. J Immunol 2009;183:7039–7046 [DOI] [PubMed] [Google Scholar]
- 127.Chang DH, Angelin-Duclos C, Calame K: BLIMP-1: Trigger for differentiation of myeloid lineage. Nat Immunol 2000;1:169–176 [DOI] [PubMed] [Google Scholar]
- 128.Magnusdottir E, Kalachikov S, Mizukoshi K, et al. : Epidermal terminal differentiation depends on B lymphocyte-induced maturation protein-1. Proc Natl Acad Sci U S A 2007;104:14988–14993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Martins GA, Cimmino L, Shapiro-Shelef M, et al. : Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat Immunol 2006;7:457–465 [DOI] [PubMed] [Google Scholar]
- 130.Kallies A, Xin A, Belz GT, Nutt SL: Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity 2009;31:283–295 [DOI] [PubMed] [Google Scholar]
- 131.Hua L, Yao S, Pham D, et al. : Cytokine-dependent induction of CD4+ T cells with cytotoxic potential during influenza virus infection. J Virol 2013;87:11884–11893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Keller AD, Maniatis T: Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev 1991;5:868–879 [DOI] [PubMed] [Google Scholar]
- 133.Martins GA, Cimmino L, Liao J, Magnusdottir E, Calame K: Blimp-1 directly represses Il2 and the Il2 activator Fos, attenuating T cell proliferation and survival. J Exp Med 2008;205:1959–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cimmino L, Martins GA, Liao J, et al. : Blimp-1 attenuates Th1 differentiation by repression of ifng, tbx21, and bcl6 gene expression. J Immunol 2008;181:2338–2347 [DOI] [PubMed] [Google Scholar]
- 135.Iwasaki Y, Fujio K, Okamura T, et al. : Egr-2 transcription factor is required for Blimp-1-mediated IL-10 production in IL-27-stimulated CD4+ T cells. Eur J Immunol 2013;43:1063–1073 [DOI] [PubMed] [Google Scholar]
- 136.Che KF, Shankar EM, Muthu S, et al. : p38 Mitogen-activated protein kinase/signal transducer and activator of transcription-3 pathway signaling regulates expression of inhibitory molecules in T cells activated by HIV-1-exposed dendritic cells. Mol Med 2012;18:1169–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shankar EM, Che KF, Messmer D, Lifson JD, Larsson M: Expression of a broad array of negative costimulatory molecules and Blimp-1 in T cells following priming by HIV-1 pulsed dendritic cells. Mol Med 2011;17:229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Seddiki N, Phetsouphanh C, Swaminathan S, et al. : The microRNA-9/B-lymphocyte-induced maturation protein-1/IL-2 axis is differentially regulated in progressive HIV infection. Eur J Immunol 2013;43:510–520 [DOI] [PubMed] [Google Scholar]
- 139.de Masson A, Kirilovsky A, Zoorob R, et al. : Blimp-1 overexpression is associated with low HIV-1 reservoir and transcription levels in central memory CD4+ T cells from elite controllers. AIDS 2014;28:1567–1577 [DOI] [PubMed] [Google Scholar]
- 140.Sforza F, Nicoli F, Gallerani E, et al. : HIV-1 Tat affects the programming and functionality of human CD8(+) T cells by modulating the expression of T-box transcription factors. AIDS 2014;28:1729–1738 [DOI] [PubMed] [Google Scholar]
- 141.Crotty S, Johnston RJ, Schoenberger SP: Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat Immunol 2010;11:114–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kaczmarek K, Morales A, Henderson AJ: T cell transcription factors and their impact on HIV expression. Virology (Auckl) 2013;2013:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kaczmarek Michaels K, Natarajan M, Euler Z, Alter G, Viglianti G, Henderson AJ: Blimp-1, an intrinsic factor that represses HIV-1 proviral transcription in memory CD4+ T cells. J Immunol 2015;194:3267–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Katz JE, Dlakic M, Clarke S: Automated identification of putative methyltransferases from genomic open reading frames. Mol Cell Proteomics 2003;2:525–540 [DOI] [PubMed] [Google Scholar]
- 145.Di Lorenzo A, Bedford MT: Histone arginine methylation. FEBS Lett 2011;585:2024–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zurita-Lopez CI, Sandberg T, Kelly R, Clarke SG: Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming omega-NG-monomethylated arginine residues. J Biol Chem 2012;287:7859–7870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gayatri S, Bedford MT: Readers of histone methylarginine marks. Biochim Biophys Acta 2014;1839:702–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chen D, Huang SM, Stallcup MR: Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J Biol Chem 2000;275:40810–40816 [DOI] [PubMed] [Google Scholar]
- 149.Ma H, Baumann CT, Li H, et al. : Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr Biol 2001;11:1981–1985 [DOI] [PubMed] [Google Scholar]
- 150.Schurter BT, Koh SS, Chen D, et al. : Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry 2001;40:5747–5756 [DOI] [PubMed] [Google Scholar]
- 151.Casadio F, Lu X, Pollock SB, et al. : H3R42me2a is a histone modification with positive transcriptional effects. Proc Natl Acad Sci U S A 2013;110:14894–14899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cheng D, Cote J, Shaaban S, Bedford MT: The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell 2007;25:71–83 [DOI] [PubMed] [Google Scholar]
- 153.Daujat S, Bauer UM, Shah V, Turner B, Berger S, Kouzarides T: Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr Biol 2002;12:2090–2097 [DOI] [PubMed] [Google Scholar]
- 154.Feng Q, Yi P, Wong J, O'Malley BW: Signaling within a coactivator complex: Methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol Cell Biol 2006;26:7846–7857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fujiwara T, Mori Y, Chu DL, et al. : CARM1 regulates proliferation of PC12 cells by methylating HuD. Mol Cell Biol 2006;26:2273–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Naeem H, Cheng D, Zhao Q, et al. : The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol Cell Biol 2007;27:120–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sims RJ, 3rd, Rojas LA, Beck D, et al. : The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science 2011;332:99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wei Y, Horng JC, Vendel AC, Raleigh DP, Lumb KJ: Contribution to stability and folding of a buried polar residue at the CARM1 methylation site of the KIX domain of CBP. Biochemistry 2003;42:7044–7049 [DOI] [PubMed] [Google Scholar]
- 159.Wang L, Zeng H, Wang Q, et al. : MED12 methylation by CARM1 sensitizes human breast cancer cells to chemotherapy drugs. Sci Adv 2015;1:e1500463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang Z, Nikolai BC, Gates LA, et al. : Crosstalk between histone modifications indicates that inhibition of arginine methyltransferase CARM1 activity reverses HIV latency. Nucleic Acids Res 2017;45:9348–9360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kimura H: Histone modifications for human epigenome analysis. J Hum Genet 2013;58:439–445 [DOI] [PubMed] [Google Scholar]
- 162.Wang Z, Zang C, Rosenfeld JA, et al. : Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 2008;40:897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang H, Huang ZQ, Xia L, et al. : Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 2001;293:853–857 [DOI] [PubMed] [Google Scholar]
- 164.Hassa PO, Covic M, Bedford MT, Hottiger MO: Protein arginine methyltransferase 1 coactivates NF-kappaB-dependent gene expression synergistically with CARM1 and PARP1. J Mol Biol 2008;377:668–678 [DOI] [PubMed] [Google Scholar]
- 165.Hyllus D, Stein C, Schnabel K, et al. : PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev 2007;21:3369–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Guccione E, Bassi C, Casadio F, et al. : Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 2007;449:933–937 [DOI] [PubMed] [Google Scholar]
- 167.Iberg AN, Espejo A, Cheng D, et al. : Arginine methylation of the histone H3 tail impedes effector binding. J Biol Chem 2008;283:3006–3010 [DOI] [PubMed] [Google Scholar]
- 168.Waldmann T, Izzo A, Kamieniarz K, et al. : Methylation of H2AR29 is a novel repressive PRMT6 target. Epigenetics Chromatin 2011;4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Frankel A, Yadav N, Lee J, Branscombe TL, Clarke S, Bedford MT: The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J Biol Chem 2002;277:3537–3543 [DOI] [PubMed] [Google Scholar]
- 170.Thandapani P, O'Connor TR, Bailey TL, Richard S: Defining the RGG/RG motif. Mol Cell 2013;50:613–623 [DOI] [PubMed] [Google Scholar]
- 171.Boulanger MC, Liang C, Russell RS, et al. : Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression. J Virol 2005;79:124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Singhroy DN, Mesplede T, Sabbah A, Quashie PK, Falgueyret JP, Wainberg MA: Automethylation of protein arginine methyltransferase 6 (PRMT6) regulates its stability and its anti-HIV-1 activity. Retrovirology 2013;10:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Miranda TB, Webb KJ, Edberg DD, Reeves R, Clarke S: Protein arginine methyltransferase 6 specifically methylates the nonhistone chromatin protein HMGA1a. Biochem Biophys Res Commun 2005;336:831–835 [DOI] [PubMed] [Google Scholar]
- 174.Di Lorenzo A, Yang Y, Macaluso M, Bedford MT: A gain-of-function mouse model identifies PRMT6 as a NF-kappaB coactivator. Nucleic Acids Res 2014;42:8297–8309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Huang J, Cardamone MD, Johnson HE, et al. : Exchange factor TBL1 and arginine methyltransferase PRMT6 cooperate in protecting G Protein Pathway Suppressor 2 (GPS2) from proteasomal degradation. J Biol Chem 2015;290:19044–19054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sivakumaran H, van der Horst A, Fulcher AJ, et al. : Arginine methylation increases the stability of human immunodeficiency virus type 1 Tat. J Virol 2009;83:11694–11703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fulcher AJ, Sivakumaran H, Jin H, Rawle DJ, Harrich D, Jans DA: The protein arginine methyltransferase PRMT6 inhibits HIV-1 Tat nucleolar retention. Biochim Biophys Acta 2016;1863:254–262 [DOI] [PubMed] [Google Scholar]
- 178.Invernizzi CF, Xie B, Richard S, Wainberg MA: PRMT6 diminishes HIV-1 Rev binding to and export of viral RNA. Retrovirology 2006;3:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Invernizzi CF, Xie B, Frankel FA, et al. : Arginine methylation of the HIV-1 nucleocapsid protein results in its diminished function. AIDS 2007;21:795–805 [DOI] [PubMed] [Google Scholar]
- 180.Mitchell LH, Drew AE, Ribich SA, et al. : Aryl Pyrazoles as potent inhibitors of arginine methyltransferases: Identification of the first PRMT6 tool compound. ACS Med Chem Lett 2015;6:655–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Shen Y, Szewczyk MM, Eram MS, et al. : Discovery of a potent, selective, and cell-active dual inhibitor of protein arginine methyltransferase 4 and protein arginine methyltransferase 6. J Med Chem 2016;59:9124–9139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Shimizu T, Matsuoka Y, Shirasawa T: Biological significance of isoaspartate and its repair system. Biol Pharm Bull 2005;28:1590–1596 [DOI] [PubMed] [Google Scholar]
- 183.Weintraub SJ, Deverman BE: Chronoregulation by asparagine deamidation. Sci STKE 2007;2007:re7. [DOI] [PubMed] [Google Scholar]
- 184.McFadden PN, Clarke S: Methylation at D-aspartyl residues in erythrocytes: Possible step in the repair of aged membrane proteins. Proc Natl Acad Sci U S A 1982;79:2460–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Shimizu T, Watanabe A, Ogawara M, Mori H, Shirasawa T: Isoaspartate formation and neurodegeneration in Alzheimer's disease. Arch Biochem Biophys 2000;381:225–234 [DOI] [PubMed] [Google Scholar]
- 186.Mamula MJ, Gee RJ, Elliott JI, et al. : Isoaspartyl post-translational modification triggers autoimmune responses to self-proteins. J Biol Chem 1999;274:22321–22327 [DOI] [PubMed] [Google Scholar]
- 187.Biterge B, Richter F, Mittler G, Schneider R: Methylation of histone H4 at aspartate 24 by protein L-isoaspartate O-methyltransferase (PCMT1) links histone modifications with protein homeostasis. Sci Rep 2014;4:6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Le Rouzic E, Belaidouni N, Estrabaud E, et al. : HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle 2007;6:182–188 [DOI] [PubMed] [Google Scholar]
- 189.Boon KL, Pearson MD, Kos M: Self-association of Trimethylguanosine Synthase Tgs1 is required for efficient snRNA/snoRNA trimethylation and pre-rRNA processing. Sci Rep 2015;5:11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Mouaikel J, Verheggen C, Bertrand E, Tazi J, Bordonne R: Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol Cell 2002;9:891–901 [DOI] [PubMed] [Google Scholar]
- 191.Monecke T, Dickmanns A, Ficner R: Structural basis for m7G-cap hypermethylation of small nuclear, small nucleolar and telomerase RNA by the dimethyltransferase TGS1. Nucleic Acids Res 2009;37:3865–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yedavalli VS, Jeang KT: Trimethylguanosine capping selectively promotes expression of Rev-dependent HIV-1 RNAs. Proc Natl Acad Sci U S A 2010;107:14787–14792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Shi Y, Lan F, Matson C, et al. : Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004;119:941–953 [DOI] [PubMed] [Google Scholar]
- 194.Karytinos A, Forneris F, Profumo A, et al. : A novel mammalian flavin-dependent histone demethylase. J Biol Chem 2009;284:17775–17782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Tsukada Y, Fang J, Erdjument-Bromage H, et al. : Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006;439:811–816 [DOI] [PubMed] [Google Scholar]
- 196.Tian X, Fang J: Current perspectives on histone demethylases. Acta Biochim Biophys Sin (Shanghai) 2007;39:81–88 [DOI] [PubMed] [Google Scholar]
- 197.Dong C, Zhang H, Xu C, Arrowsmith CH, Min J: Structure and function of dioxygenases in histone demethylation and DNA/RNA demethylation. IUCrJ 2014;1(Pt 6):540–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Pastor WA, Aravind L, Rao A: TETonic shift: Biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol 2013;14:341–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Walport LJ, Hopkinson RJ, Chowdhury R, et al. : Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat Commun 2016;7:11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Anand R, Marmorstein R: Structure and mechanism of lysine-specific demethylase enzymes. J Biol Chem 2007;282:35425–35429 [DOI] [PubMed] [Google Scholar]
- 201.Kong X, Ouyang S, Liang Z, et al. : Catalytic mechanism investigation of lysine-specific demethylase 1 (LSD1): A computational study. PLoS One 2011;6:e25444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Stavropoulos P, Blobel G, Hoelz A: Crystal structure and mechanism of human lysine-specific demethylase-1. Nat Struct Mol Biol 2006;13:626–632 [DOI] [PubMed] [Google Scholar]
- 203.Metzger E, Wissmann M, Yin N, et al. : LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 2005;437:436–439 [DOI] [PubMed] [Google Scholar]
- 204.Wissmann M, Yin N, Muller JM, et al. : Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol 2007;9:347–353 [DOI] [PubMed] [Google Scholar]
- 205.Huang J, Sengupta R, Espejo AB, et al. : p53 is regulated by the lysine demethylase LSD1. Nature 2007;449:105–108 [DOI] [PubMed] [Google Scholar]
- 206.Xie Q, Bai Y, Wu J, et al. : Methylation-mediated regulation of E2F1 in DNA damage-induced cell death. J Recept Signal Transduct Res 2011;31:139–146 [DOI] [PubMed] [Google Scholar]
- 207.Wang J, Hevi S, Kurash JK, et al. : The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet 2009;41:125–129 [DOI] [PubMed] [Google Scholar]
- 208.Le Douce V, Colin L, Redel L, et al. : LSD1 cooperates with CTIP2 to promote HIV-1 transcriptional silencing. Nucleic Acids Res 2012;40:1904–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM: Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat Med 2009;15:1312–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Agger K, Cloos PA, Christensen J, et al. : UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 2007;449:731–734 [DOI] [PubMed] [Google Scholar]
- 211.Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K: Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A 2007;104:18439–18444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lan F, Bayliss PE, Rinn JL, et al. : A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 2007;449:689–694 [DOI] [PubMed] [Google Scholar]
- 213.Faralli H, Wang C, Nakka K, et al. : UTX demethylase activity is required for satellite cell-mediated muscle regeneration. J Clin Invest 2016;126:1555–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lee MG, Villa R, Trojer P, et al. : Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 2007;318:447–450 [DOI] [PubMed] [Google Scholar]
- 215.Cho YW, Hong T, Hong S, et al. : PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem 2007;282:20395–20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Miller SA, Mohn SE, Weinmann AS: Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell 2010;40:594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zhang HS, Du GY, Liu Y, et al. : UTX-1 regulates Tat-induced HIV-1 transactivation via changing the methylated status of histone H3. Int J Biochem Cell Biol 2016;80:51–56 [DOI] [PubMed] [Google Scholar]
- 218.Deeks SG: HIV: Shock and kill. Nature 2012;487:439–440 [DOI] [PubMed] [Google Scholar]
- 219.Fenaux P: Inhibitors of DNA methylation: Beyond myelodysplastic syndromes. Nat Clin Pract Oncol 2005;2 Suppl 1:S36–S44 [DOI] [PubMed] [Google Scholar]