Abstract
Central nervous system injury often leads to functional impairment due, in part, to the formation of an inhibitory glial scar following injury that contributes to poor regeneration. Astrocytes are the major cellular components of the glial scar, which has led to the belief that they are primarily inhibitory following injury. Recent work has challenged this by demonstrating that some astrocytes are required for spinal cord regeneration and astrocytic roles in recovery depend on their phenotype. In this work, two mixed populations containing primarily either fibrous or protoplasmic astrocytes were derived from mouse embryonic stem cells (mESCs). Motoneuron and V2a interneuron growth on live cultures, freeze-lysed cultures, or decellularized extracellular matrix (ECM) from astrocytes were assessed. Both neuronal populations were found to extend significantly longer neurites on protoplasmic-derived substrates than fibrous-derived substrates. Interestingly, neurons extended longer neurites on protoplasmic-derived ECM than fibrous-derived ECM. ECM proteins were compared with in vivo astrocyte expression profiles, and it was found that the ESC-derived ECMs were enriched for astrocyte-specific proteins. Further characterization revealed that protoplasmic ECM had significantly higher levels of axon growth promoting proteins, while fibrous ECM had significantly higher levels of proteins that inhibit axon growth. Supporting this observation, knockdown of spondin-1 improved neurite growth on fibrous ECM, while laminin α5 and γ1 knockdown decreased neurite growth on protoplasmic ECM. These methods allow for scalable production of specific astrocyte subtype-containing populations with different neuronal growth support capacities, and can be used for further studies of the functional importance of astrocyte heterogeneity.
Keywords: : spinal cord injury, interneuron, motor neuron, tissue engineering
Introduction
Central nervous system (CNS) trauma represents a significant healthcare burden in the United States, in part, due to its limited regenerative capacity. Spinal cord injury (SCI) is a good modality for studying CNS regeneration because the cord can be relatively easily accessed, the axonal tracts can be reliably severed by hemisection, and a stereotyped glial scar and lesion cavity form after injury [1]. The lack of regeneration following CNS trauma has historically been partially attributed to the astrocytes present within the glial scar, which represent a physical and chemical barrier to axon growth. This has led to a focus in the field on removing astrocytes to facilitate CNS regeneration.
Astrocyte removal has been achieved in SCI models by using genetic tools to knock out genes required for astrocytes reactivity. In vimentin and glial fibrillary acidic protein (GFAP) double knockout mice, astrocytes do not become appropriately reactive after SCI, leading to impaired glial scar formation and increased bleeding [2]. Similarly, conditional knockout of floxed STAT3, an inducer of astrocyte reactivity, using GFAP-Cre has been found to disrupt the formation of the glial scar, increase macrophage infiltration into the spinal cord, and lead to a larger lesion cavity after SCI [3]. Conditional ablation of reactive astrocytes at the time of injury or 5 weeks after SCI using GFAP-thymidine kinase (TK) + ganciclovir has also been shown to lead to worse functional outcomes due to death of dividing GFAP+ cells. These studies demonstrate that astrocytes are required both acutely to control spread of secondary injury and chronically to facilitate functional recovery [4,5]. Thus, there is a clear role for astrocytes in CNS regeneration and potential for their contribution to regenerative therapies.
Recent work on astrocytes has shown that there are many more distinct astrocyte populations than previously believed [6,7]. Furthermore, the type of insult, inflammatory or ischemic, to the CNS has been shown to affect whether native astrocytes become more proregenerative or more inhibitory, supporting the idea that some astrocytes have proregenerative roles [8].
The physical barrier represented by the glial scar is primarily due to the woven nature of astrocyte processes adjacent to the lesion [9]. Structural studies of astrocytes suggest that not all astrocyte populations form such barriers in response to trauma. In particular, fibrous (white matter) astrocytes have been shown to exhibit process hypertrophy and a significant increase in process overlap following injury in optic nerve [10]; conversely, protoplasmic (gray matter) astrocytes have been demonstrated to exhibit some process hypertrophy after injury, but little to no increase in process overlap [11]. These studies show that fibrous astrocytes adopt a phenotype more consistent with classic glial scar morphology in response to injury, suggesting that these populations may be more involved in glial scar formation. With the lower levels of overlap observed in reactive protoplasmic astrocytes, it is possible that a subset of these astrocytes may be responsible for proregenerative effects of astrocytes following CNS injury.
The concept that astrocyte subtypes have variable roles following injury has been explored using primary glial restricted progenitors (GRPs), isolated from rat, mouse, and human embryonic spinal cords [12]. These cells have been successfully transplanted into rats following a right-sided cervical dorsal column transection SCI, leading to improved recovery [13,14]. Furthermore, GRPs have been predifferentiated into astrocytes using either ciliary neurotrophic factor (CNTF) or bone morphogenic protein 4 (BMP-4) to generate fibrous-like or protoplasmic-like astrocytes [15]. When transplanted following SCI, GRPs differentiated into fibrous astrocytes were found to have a detrimental effect on axon growth into the lesion and functional outcomes, while protoplasmic transplants improved axon growth and functional outcomes [15,16]. While isolation of GRPs from humans has been achieved from a fetal brain and spinal cord, these methodologies are not scalable for clinical use.
The novel component of our work is the derivation of these astrocyte populations from mouse embryonic stem cells (mESCs). This methodology could be applied to human pluripotent cells, and thus represents a scalable cell source for future proregenerative astrocyte-based therapy development. Furthermore, an ESC source of these astrocytes could allow for further elucidation of the mechanisms utilized by proregenerative astrocytes that are absent in astrocyte populations inhibitory to axon growth and vice versa.
Previous protocols to derive astrocytes from ESCs have not focused on deriving specific astrocyte subtypes. In this work, glial cell populations containing primarily fibrous or protoplasmic astrocytes are derived from mESCs. These populations are then demonstrated to have different abilities to support the growth of ESC-derived motoneurons and V2a interneurons, with protoplasmic populations allowing neurons to extend significantly longer neurites than observed on fibrous populations. Finally, these neuronal growth support differences are found to be due, in part, to altered extracellular matrix (ECM) composition with fibrous astrocyte populations depositing more axon growth-inhibitory proteins, while protoplasmic astrocyte populations deposit more axon growth-permissive proteins.
Materials and Methods
mESC culture
RW4 (ATCC, SCRC-1018) mESCs were maintained in complete media (10% fetal bovine serum (Invitrogen), 10% Newborn Calf Serum (Invitrogen), 132 μM beta mercaptoethanol (BME; Sigma, St Louis, MO), and 10,000 U/mL mouse leukemia inhibitory factor (Life Technologies, Carlsbad, CA) and passaged when at 60%–80% confluency. Cells were passaged using 0.25% trypsin-EDTA (Life Technologies) at 37°C for 5 min to dissociate the cells. The trypsin reaction was quenched and cells were seeded into a new T25 flask coated with 0.1% gelatin (Sigma).
Glial population differentiation
1 × 106 RW4 ESCs were cultured in suspension on agar-coated 10 cm dishes in 10 mL DFK5, DMEM/F12 (Life Technologies) plus 5% Knockout Serum Replacement (Life Technologies), 50 μM nonessential amino acids (Life Technologies), 1 × Insulin-Transferrin-Selenium (Life Technologies), 100 μM BME (Sigma), 5 μM thymidine, and 15 μM of the following nucleosides: adenosine, cytosine, guanosine, and uridine (Life Technologies) for 2 days to form embryoid bodies (EBs), followed by 4 days in 10 mL DFK5 plus 2 μM retinoic acid (RA) and 600 nM smoothened agonist (SAG) to confer a spinal identity with a media change after 2 days [17]. On day 6, the EBs were dissociated and 4 × 106 cells were seeded onto a gelatin-coated low adherence 10 cm dish (ThermoFisher) in DFK5 media plus 20 ng/mL epithelial growth factor (Peprotech), 10 ng/mL fibroblast growth factor 1 (FGF-1; Peprotech), and 1 μg/mL laminin for 5 days as described in Benveniste et al. [18]. On day 11, the cultures were switched into lineage-specific media for four additional days on the same plates. Fibrous media: DMEM/F12 plus 1 × G5 supplement (Invitrogen), 10 μg/mL CNTF (Peprotech); Protoplasmic media: DFK5 plus 10 μg/mL FGF-1, 10 μg/mL BMP-4 (Peprotech); and Benveniste astrocyte media: DFK5 plus 10 μg/mL FGF-1, 10 μg/mL platelet-derived growth factor AA (PDGF-AA; Peprotech) [18]. On day 15, the cells were reseeded onto gelatin-coated plates at a density of 20,000 cells/cm2 and maintained in lineage-specific media for six additional days before use (Fig. 1A).
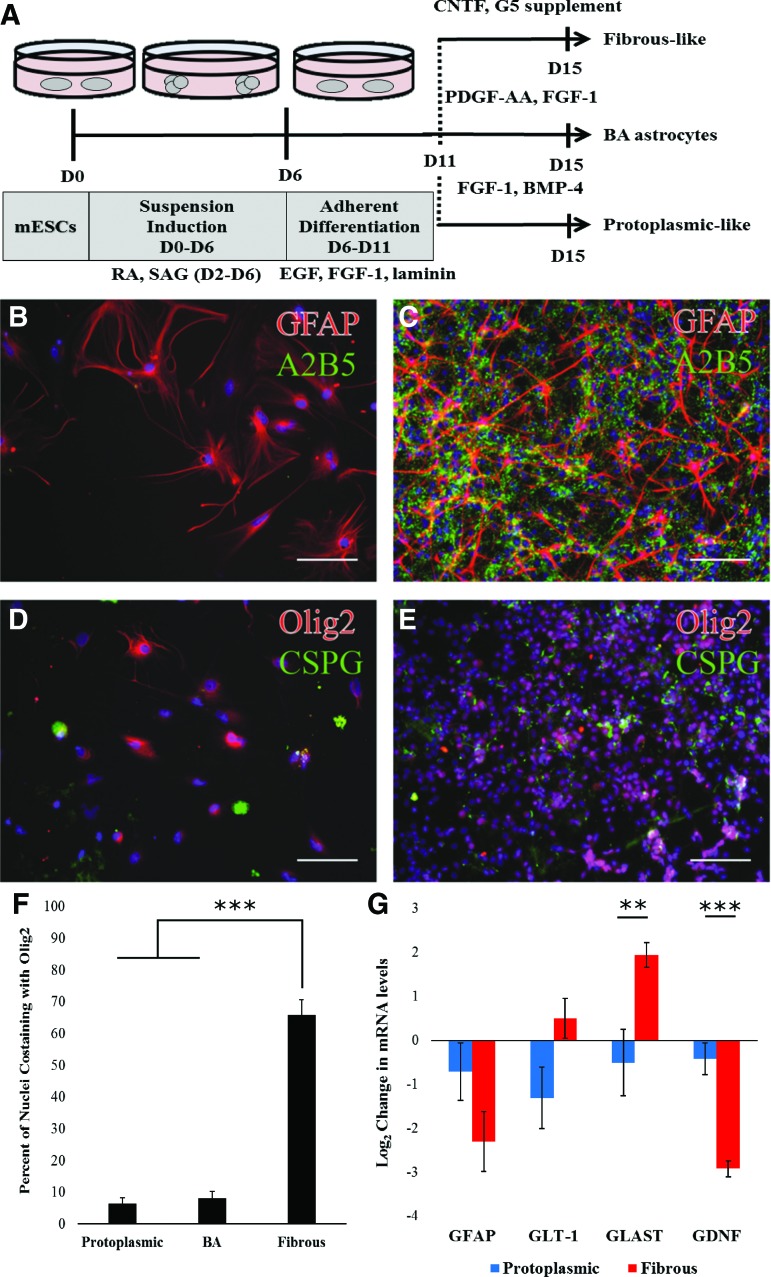
FIG. 1.
mESCs can be selectively differentiated into mixed populations containing either protoplasmic or fibrous astrocytes. (A) Schematic representation of the astrocyte differentiation protocols used in this work. All astrocytes are subjected to the same initial 11 days after which media factors are changed so that fibrous, protoplasmic, or the BA (Benveniste protocol [18]) population develops. (B, C) GFAP (red) and A2B5 (green) staining in protoplasmic (B) or fibrous (C) cultures at the end of differentiation. (D, E) Olig2 (red) and CSPG (green) staining in protoplasmic (D) or fibrous cultures (E) at end of differentiation. Nuclei stained with Hoechst (blue). Scale bar: 100 μm. (F) Quantification showing the percent of nuclei that colocalized with Olig2 staining in different cultures. Error bars: standard error, n = 9–12. (G) Log2 expression difference in mRNA levels for fibrous and protoplasmic astrocyte markers compared to BA control population. Error bars represent standard error, n = 6. **P < 0.01, ***P < 0.001. BMP-4, bone morphogenetic protein 4; CNTF, ciliary neurotrophic factor; CSPG, chondroitin sulfate proteoglycan; EGF, epithelial growth factor; FGF-1, fibroblast growth factor 1; GFAP, glial fibrillary acidic protein; mESC, mouse embryonic stem cells; PDGF-AA, platelet-derived growth factor AA; RA, retinoic acid; SAG, smoothened agonist. Color images available online at www.liebertpub.com/scd
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde (Sigma) for 20 min and then permeabilized in 0.1% Triton-X (Sigma) for 15 min. Cells were then blocked with 5% of an appropriate serum [Goat (Sigma) or Donkey (Sigma)] in phosphate-buffered saline (PBS) for 1 h. Primary antibodies were used at the following dilutions: GFAP 1:100 (Immunostar), A2B5 1:25 (DSHB), aquaporin 4 (Aqp-4) 1:100 (Santa Cruz Biotechnology), Olig2 1:1,000 (Santa Cruz Biotech), CS56 1:250 (Sigma), Spondin-1 1:100 (Abcam), Collagen XIIα1 1:100 (Santa Cruz Biotechnology), Sox2 1:100 (Santa Cruz Biotechnology), HSPG2 1:100 (Fisher), O4 1:100 (Millipore), S100 1:100 (Dako, Santa Clara, CA), SSEA-1 1:25 (DSHB), Oct 3/4 1:200 (Santa Cruz Biotech), β-tubulin 1:1,000 (Biolegend), Neurofilament 1:10 (DSHB), and Nestin 1:10 (DSHB). Primary antibody incubation was carried out overnight in 2% of appropriate serum in PBS. Secondary antibodies (Life Technologies) were all used at a 1:1,000 dilution and incubated in 2% of appropriate serum in PBS for 1 h at room temperature. 1:1,000 Hoechst (Invitrogen) in PBS was incubated with the cells for 15 min before imaging. Nuclear colocalization was determined using Cell Profiler software (Broad Institute).
Flow cytometry
Fibrous and protoplasmic populations were removed from the plate surface with 0.25% trypsin-EDTA and fixed using 1% paraformaldehyde for 15 min followed by blocking in 5% goat serum in PBS for 20 min. Primary antibody incubation was performed for 45 min using the same dilutions as immunocytochemistry (ICC). Cells were washed once with PBS and secondary incubation was performed for 45 min before washing thrice with PBS. Cells were then analyzed using a Guava EasyCyte (Millipore), and data were quantified using FlowJo. For analysis, a cell gate was drawn based on forward scatter and side scatter. Staining graphs were restricted to events within this cell gate and stain gates were drawn based on secondary only controls to exclude 99% of control events from the gate to minimize the false positive rate. Quadrant gates for double-stained samples were drawn based on secondary controls as well as single-stained populations.
Quantitative reverse transcriptase–polymerase chain reaction
Day 21 glial populations were removed from the culture plate by 0.25% trypsin treatment followed by quenching with conditioned media (CM). The resulting cell suspension was then spun down at 300 rcf at 4°C for 5 min, and the RNA was extracted using a Qiagen RNA extraction kit following manufacturer's instructions. RNA concentration was determined using an Implen Nanophotomer and 500 ng of RNA from each sample was converted to cDNA using a High-Capacity RNA to cDNA kit (Applied Biosystems). Finally, the mRNA levels of the genes of interest were determined using specific TaqMan assays following manufacturer's instructions (Life Tech, Table 1). β-Actin was used as the normalization control in all samples.
Table 1.
Quantitative Reverse Transcriptase–Polymerase Chain Reaction Primers
| Target | Primer ID (ThermoFischer) |
|---|---|
| β-Actin | Mm02619580 |
| GFAP | Mm01253033 |
| GLT-1 | Mm01275814 |
| GLAST | Mm00600697 |
| GDNF | Mm00599849 |
GDNF, glial-derived neurotrophic factor; GFAP, glial fibrillary acidic protein; GLAST, glutamate and aspartate transporter 1; GLT-1, glutamate transporter 1.
Viral knockdown
On day 15 of differentiation, cells were seeded at 10,000 cells/cm2 in appropriate media. Cells were then infected with mission shRNA lentiviral particles (Sigma) (Table 2) at a multiplicity of infection of 2.5 on day 16. Infection was allowed to proceed for 1 day and media were then replaced and the cells cultured for an additional day. To ensure only infected cells were present, cultures were selected with 2 μg/mL puromycin for either 1 day for the fibrous cultures or 2 days for the protoplasmic cultures. On day 21, all cells were removed from the plate using trypsin and reseeded onto gelatin-coated 48-well plates. Cells were then cultured for 6 days with media changes every other day to allow for ECM deposition. After ECM deposition, cells were removed by decellularization as described in substrate preparation or stained for ICC.
Table 2.
shRNAs used for ECM Component Knockdown Experiments
| Target | TRC number |
|---|---|
| Spondin-1 | 0000090520 |
| Laminin γ1 | 0000055421 |
| Laminin α5 | 0000252850 |
| Perlecan | 0000256980 |
| Collagen XIIα1 1 | 0000091115 |
| Collagen XIIα1 2 | 0000091116 |
| Collagen XIIα1 3 | 0000335258 |
| Collagen XIIα1 4 | 0000335319 |
| Collagen XIIα1 5 | 0000335320 |
| Collagen VIα3 1 | 0000091854 |
| Collagen VIα3 2 | 0000091855 |
| Collagen VIα3 3 | 0000091856 |
| Collagen VIα3 4 | 0000091857 |
| Laminin β1 1 | 0000094314 |
| Laminin β1 2 | 0000094315 |
| Laminin β1 3 | 0000094316 |
| Laminin β1 4 | 0000094317 |
| Laminin β1 5 | 0000094318 |
Substrate preparation
Day 21 glial cells were seeded onto gelatin-coated 48-well plates at a density of 20,000 cells/cm2 and cultured for 6 days in appropriate media to allow for matrix deposition. Following the 6-day culture period, cells were removed by decellularization (using a modified Hudson protocol [19]), lysed by freezing, or left alive. For frozen cells, the plates were sealed with Parafilm (Bemis, Neenah, WI) and placed in the freezer overnight. All plates were washed once with DMEM containing 25 mM HEPES (Life Technologies) before neuron seeding.
Motoneuron culture
Motoneurons were derived from Hb9-Puro CAG-TdTomato mESCs that constitutively express TdTomato under the control of the synthetic CAG promoter, as previously described [20]. To obtain pure motoneuron cultures, EBs were selected with 4 μg/mL puromycin from day 5 to 6 of differentiation before dissociation seeding onto the desired substrate at a density of 20,000 cells/cm2. Motoneurons were cultured in half DFK5 and half Neurobasal media (Life Tech) plus 1 × B27. Cultures were then imaged at regular time points over 2 days.
V2a interneuron culture
V2a interneurons were generated from Chx10-PAC bact-TdTomato mESCs as previously described [21]. After induction, EBs were dissociated with 0.25% trypsin and 2.5 × 107 cells were seeded onto a poly-l-ornithine-/laminin-coated T25 flask. Chx10+ cells were then selected in half neural basal-half DFK5 media with 1 × GlutaMAX (Life Tech), 1 × B27, 2 μg/mL puromycin, and 10 ng/mL of the following growth factors for 24 h: glial-derived neurotrophic factor (GDNF; Peprotech), neurotrophin-3 (NT-3; Peprotech), and brain-derived neurotrophic factor (BDNF; Peprotech). After selection, neurons were lifted from the flasks using Accutase® (Sigma) treatment for 30 min and then reseeded at 50,000 cells/cm2 onto the desired substrate in half neuro basal-half DFK5 media plus 1 × GlutaMAX, 1 × B27, and 10 ng/mL of the following growth factors: BDNF, GDNF, and NT-3. Neurite extension for both neuronal types was determined using Cell Profiler.
Conditioned media
After day 15 of differentiation, each population was seeded in its appropriate media for 2 days to condition the media. CM was subsequently harvested and spun down at 3,000 rcf for 25 min. Motoneurons were seeded at 84,000 cells/cm2 onto poly-L-ornithine-coated six-well polystyrene dishes in one of three media conditions: 50% unconditioned protoplasmic media and 50% unconditioned fibrous media, 50% protoplasmic CM and 50% unconditioned fibrous media, or 50% unconditioned protoplasmic media and 50% fibrous CM. This 50:50 mixing strategy was used so CM effects could be directly compared without having the growth factors in the astrocyte media affect neuronal growth outcomes. Following the addition of one of the three media solutions to motoneuron cultures, five images of each condition were acquired in a random spatial orientation every 24 h for 3 days, and the neurite area per nucleus was quantified with Cell Profiler.
Proteomics and western blotting
Day 21 cells were seeded at 20,000 cells/cm2 onto gelatin-coated plates and allowed to grow for 6 days before decellularization. After decellularization, the residual proteins were scraped off the plate into Milli-Q water and lyophilized overnight. The resulting powder was dissolved in 4% sodium dodecyl sulfate (SDS; Sigma) and 100 mM Tris-HCl, pH 7.6 (ThermoFisher). Protein concentration of this solution was determined using a BCA Assay Kit (ThermoFisher) and then dithiothreitol (DTT; ThermoFisher) was added to the samples to a final concentration of 100 mM. Fifteen micrograms of ECM protein was used for proteomics analysis. For western analysis, the samples were run on 4%–15% mini-PROTEAN TGX gradient gels (Bio-Rad) at 130 V for 1.5 h. Western tank transfer was performed at 30 V overnight using 10% methanol (Sigma), 25 mM Tris Base (ThermoFisher), and 192 mM glycine (Sigma) transfer buffer to a polyvinylidene difluoride membrane (Millipore). The membrane was then probed with the following antibody dilutions overnight: Spondin-1 1:150, Collagen XIIα1 1:200, and HSGP2 1:100. Licor goat anti-rabbit and goat anti-rat secondary antibodies were used at a dilution of 1:15,000 in PBS and were incubated for 20 min before imaging.
Preparation of peptides for liquid chromatography-mass spectrometry
Fifteen micrograms of ECM proteins solubilized in 30 μL SDT buffer (4%SDS, 100 mM Tris-HCl, pH 7.6, and 100 mM DTT) was diluted with 200 μL of 100 mM Tris-HCL buffer, pH 8.5 containing 8 M urea. Detergent was removed by buffer exchange in filter unit with a 30K MWCO (Part No. MRCF0R030; Millipore). Iterative centrifugations were performed at 14,000 rcf for 15 min with the addition of 200 μL of 100 mM Tris-HCL buffer, pH 8.5 containing 8 M urea, to the top filter unit. The proteins were alkylated with 100 μL of 50 mM iodoacetamide directly to the top of the filter, mixing at 25 rcf, and incubating at room temperature for 20 min in the dark. The filter was spun at 14,000 rcf for 10 min and the flow through discarded. Unreacted iodoacetamide was washed through the filter with application of 2 × 200 μL of 100 mM Tris-HCL buffer, pH 8.5 containing 8 M urea, with centrifugation for 10 min after each addition. The urea buffer was exchanged into 100 mM ammonium bicarbonate buffer, pH 8, with two additions of 200 μL each and spinning after each addition. The filters were transferred to a new collection tube and 100 μL of 0.05 μg/μL trypsin, dissolved in 100 mM ammonium bicarbonate buffer, was added to each filter. The samples were digested overnight at 37°C in a humidity chamber. An additional aliquot of trypsin (1 μg) was added and digestion was continued for 4 h. The filter units were spun for 15 min and the digest was collected in the lower unit. The filter was washed with 50 μL 0.5 M sodium chloride and the wash was collected with the peptides. Residual detergent was removed by ethyl acetate transfer, followed by acidification to 5% formic acid final concentration in preparation for desalting. The peptides were desalted using micro-tips (C4, BIOMEKNT3C04, and porous graphite carbon, BIOMETNT3CAR) (Glygen) on a Beckman robot (Biomek NX) as previously described [22], for analysis using liquid chromatography-mass spectrometry (LC-MS).
LC/MS analysis
LC-ESI/MS/MS analysis was performed using a Q-Exactive™ Plus Hybrid Quadrupole-Orbitrap™ Plus mass spectrometer (ThermoFisher) coupled to an EASY-nanoLC 1000 system (ThermoFisher). The samples were loaded (2 μL) onto a 75 μm i.d. × 25 cm Acclaim® PepMap 100 RP column (ThermoFisher). The peptides were eluted at a flow rate of 300 nL/min with an acetonitrile gradient in aqueous formic acid (0.1%) as mobile phase A. Peptide elution occurred in the following sequence: 0%–4% B (buffer B) for 1 min, 4%–12% B over 127 min, 12%–22% B over 112 min, 22%–30% B over 40 min, 30%–70% B over 6 min, hold at 70% B for 6 min, followed by increase in B to 95% B over 1 min, and an isocratic wash at 95% B for 6 min. Full-scan mass spectra were acquired using the Orbitrap™ mass analyzer in the mass-to-charge ratio (m/z) of 375–1,500 and with a mass resolving power set to 70,000. Ten data-dependent high-energy collisional dissociations were performed with a mass resolving power set to 35,000, a fixed first m/z 100, an isolation width of 2.0 m/z, and the normalized collision energy setting of 27.
The maximum injection time was 120 ms for parent ion analysis and 120 ms for product ion analysis. Target ions already selected for MS/MS were dynamically excluded for 30 s. An automatic gain control target value of 3 × 106 ions was used for full MS scans and 5 × 105 ions for MS/MS scans. Peptide ions with charge states of one or greater than seven were excluded from MS/MS acquisition. The tandem mass spectra were processed using Matrix Science Distiller version 2.5 without charge state deconvolution and deisotoping. The processed files were used for protein database searches using Mascot (version 2.5.1; Matrix Science, London, United Kingdom). The UniProt Mouse Reference database (downloaded May 3, 2014, 69,021 entries) was used. A parent ion tolerance and MS2 fragment tolerance were set to 10 ppm and 0.05 Da, respectively. Carbamidomethyl of cysteine was specified as a fixed modification and oxidation of methionine was set as a variable modification.
Protein identifications were performed using Scaffold, version 4.4.8 (Proteome Software, Inc., Portland, OR), implementing the Protein and Peptide Prophet algorithms [23,24]. Peptide identifications were accepted with >90.0% probability. Protein identifications were accepted if they could be established at >95.0% probability and contained at least two peptides with unique sequences. Protein probabilities were assigned using the Protein Prophet algorithm. Proteins that contained similar peptides, but could not be differentiated based on identification of unique peptide sequences, were grouped to satisfy the principles of parsimony.
Translating ribosome affinity purification-Seq analysis
Translating ribosome affinity purification (TRAP) libraries were sequenced on an Illumina HiSeq 2500 and reads were analyzed as previously described [25]. Differential expression analysis of cortex TRAP versus Pre-IP was performed using the edgeR package. Raw and analyzed RNA-sequencing data are available at GEO: GSE74456. For this study, only cortex Pre-IP and TRAP samples were used (GSM1920988-1920993). Candidate ECM components were mapped to Ensembl gene IDs using the biomaRt package, based on gene symbol. Out of the 638 ECM components with ≥99% protein identification probability, 559 were robustly expressed in the TRAP-seq samples and thus were used for comparative analysis.
Statistics
Statistical analysis was performed using Minitab software for neurite length measurements [one-way analysis of variance (ANOVA) with Bonferroni correction]. Excel was used for Quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR; pairwise t-test) and colocalization data (Student's t-test). Proteomics data were analyzed using Scaffold 4, significance determined with Fisher's exact test using the Benjamini–Hochberg false positive correction method.
Results
Generation of fibrous astrocyte or protoplasmic astrocyte containing glial populations from mESCs
To determine the effect of fibrous and protoplasmic populations on neuron growth, methods were developed to obtain these astrocyte subtypes from mESCs. In particular, this protocol was based on prior ESC-derived astrocyte protocols described in Benveniste et al. [18] and Roybon et al. [17], and the GRP-derived astrocyte protocol described in Davies et al. [14]. This protocol was designed to generate spinal populations by first caudalizing mESCs with RA treatment [26] followed by BMP-4 treatment to obtain cultures containing protoplasmic astrocytes and CNTF treatment to obtain cultures containing fibrous astrocytes (Fig. 1A). To allow for direct comparison of protein expression between the fibrous and protoplasmic populations, a second population containing protoplasmic-like astrocytes was generated using the protocol described in Benveniste et al. [18]. This protocol was not used for future studies because the continued use of PDGF-AA led to significant oligodendrocyte presence within the cultures.
To determine the dominant phenotype of the astrocytes within these cultures, the expression of known protoplasmic or fibrous markers was assessed using ICC and qRT-PCR. In particular, A2B5 [27] and nuclear Olig2 [15] are expected to be elevated in fibrous astrocytes and oligodendrocyte precursors. ICC demonstrated that cells exposed to CNTF and G5 containing media expressed both A2B5 and nuclear Olig2, as expected for fibrous astrocytes (Fig. 1B–E). Quantification of Olig2+ nuclei in both fibrous and protoplasmic cultures revealed that a significantly higher percentage of nuclei were Olig2+ in fibrous cultures than protoplasmic cultures (Fig. 1F).
To further confirm the presence of either fibrous or protoplasmic astrocytes within these mixed cultures, quantitative polymerase chain reaction was used to determine mRNA levels of genes known to be differentially expressed between these astrocyte populations, specifically GFAP (general reactive astrocyte marker), glutamate transporter 1 (GLT-1), glutamate and aspartate transporter 1 (GLAST), and GDNF. Based on primary cell characterizations and analysis of GRPs in culture, GLT-1 is slightly elevated in fibrous astrocytes [28], GLAST is significantly elevated in fibrous astrocytes [28,29], and GDNF is elevated in protoplasmic astrocytes [16]. Consistent with these observations, the ESC-derived populations showed significant upregulation of GLAST and slight upregulation of GLT-1 in the fibrous population, and significant upregulation of GDNF in the protoplasmic population, suggesting that astrocytes within these populations are either protoplasmic or fibrous depending on media exposure (Fig. 1G).
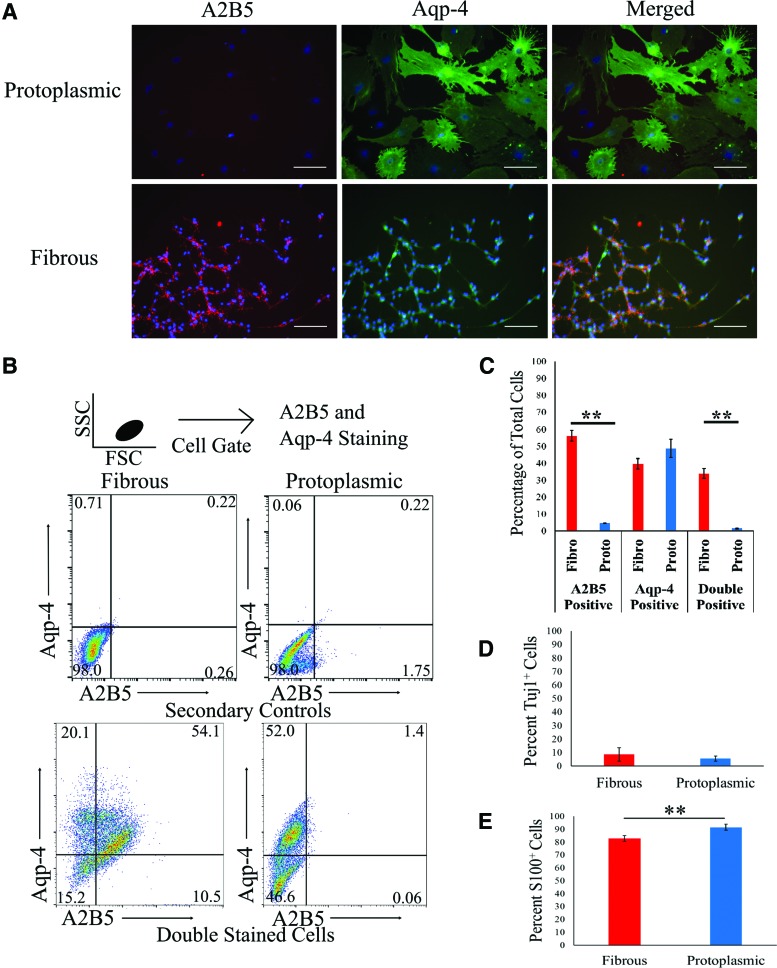
To determine the percentage of cells in these cultures expressing mature astrocyte markers, flow cytometry was performed for Aqp-4 and A2B5 expression [6] (Fig. 2B). Aqp-4 is known to be specifically expressed on the cell membranes of both mature fibrous and mature protoplasmic astrocytes, but not radial glia radial glia in vivo [30]. This makes it an appealing marker to determine the percentage of mature astrocytes in these cultures by flow cytometry. A notable drawback to the use of Aqp-4 is that Aqp-4 is known to have variable expression levels both in cultured astrocytes and in vivo. Thus, Aqp-4 quantification may result in an underestimation of the percent of astrocytes present in the culture (Fig. 2A) [31]. Quantification of flow cytometry data demonstrated 48.8% ± 0.8% of cells staining Aqp-4+ in protoplasmic cultures and 39.7% ± 0.5% of cells staining Aqp-4+ in fibrous cultures, with 34.0% ± 0.4% of cells in fibrous cultures staining both Aqp-4+ and A2B5+ (Fig. 2B, C). Overall, fibrous cultures had a significantly higher percentage of A2B5+ cells than protoplasmic cultures and a significantly higher percentage of cells, both Aqp-4+ and A2B5+, consistent with the expected fibrous astrocyte phenotype (Fig. 2).
FIG. 2.
Quantification of astrocytes present in mESC cultures. (A) Representative images showing A2B5, Aqp-4, and merged staining in protoplasmic and fibrous cultures 6 days after differentiation. Blue represents Hoechst nuclear stain. Scale bar: 100 μm. (B) Representation flow cytometry graphs of secondary controls and samples stained for Aqp-4 and A2B5 for both fibrous and protoplasmic cultures. Each graph represents 10,000 cells based on an FSC and SSC gate. Quadrant gates drawn based on single stained samples and secondary controls. Numbers represent the percent of cell events within each quadrant (C) average percentage of cells staining for A2B5, Aqp-4, and both markers in fibrous and protoplasmic cultures. n = 5, error bars: standard error, **P < 0.01. (D, E) Average percent of total cells staining β-tubulin+ (neurons) (D) or S100+ (E). N = 4, error bars: 95% confidence interval. **P < 0.01. Aqp-4, aquaporin 4; FSC, forward scatter; SSC, side scatter. Color images available online at www.liebertpub.com/scd
Since flow cytometry quantification indicated that other cell types may be present, ICC staining was used to determine what nonastrocyte populations were present and to cast a larger net for astrocytes. Cultures were stained for S100, which has been shown to be upregulated in astrocytes as they mature [32], β-tubulin (neurons), O4 (oligodendrocytes), SSEA-1 (mESCs), and Oct 3/4 (mESCs). It is important to note that S100 is also present in some radial glial populations and early oligodendrocyte lineages based on lineage tracing experiments [33], although it is generally used as an astrocyte marker. We found that fibrous populations contained 8.6% ± 4.9% neurons based on β-tubulin staining and 82.8% ± 2.2% of the cells were S100+. Protoplasmic populations were found to have 5.6% ± 1.9% β-tubulin+ cells and 91.5% ± 2.3% S100+ cells (Fig. 2D, E). There was no observable staining of nuclear S100 in either culture, which has been linked to oligodendrocyte differentiation [34], indicating that, combined with a few to no cells staining O4+, both cultures do not contain significant oligodendrocyte presence [35]. There were also few to no cells staining SSEA-1+ or Oct 3/4+, demonstrating limited presence of undifferentiated stem cells. Taken together, these data suggest that these glial cultures are primarily composed of cells within the astrocyte lineage with some presence of gliogenic radial glia and neurons.
Astrocyte-derived substrates modify neuronal growth
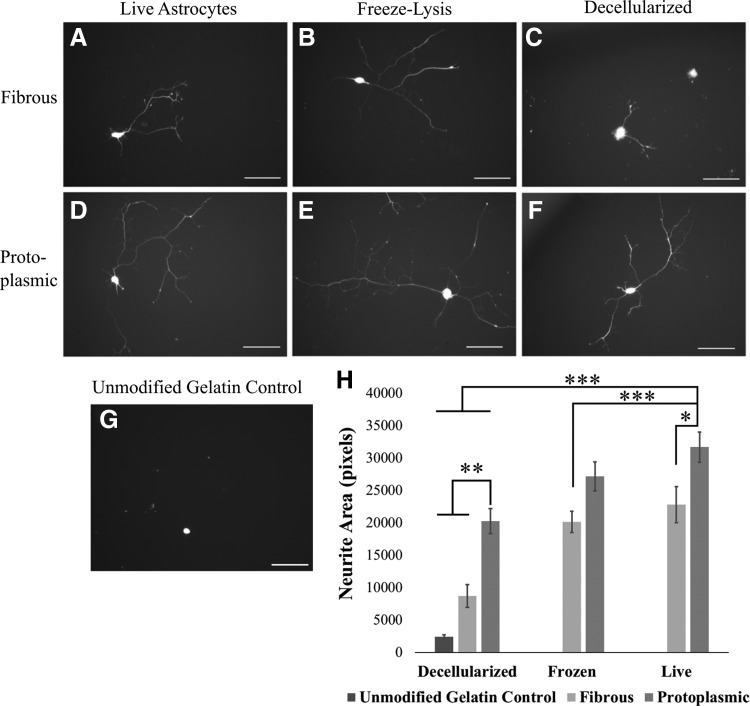
Next, the ability of these two populations to serve as supportive substrates for neuronal growth was assessed and it was observed that live astrocyte substrates exhibiting either phenotype were able support motoneuron neurite outgrowth (Fig. 3A, D). To determine what aspects of these substrates contributed to neurite outgrowth, modified substrates were produced using decellularization or freeze lysis. Decellularization leaves only ECM on the plates, while freeze lysis leaves behind ECM and cell membranes. The neurite outgrowth area per nucleus from motoneurons was measured on live, decellularized, and frozen substrates to determine the relative contribution of ECM, membrane, and secreted factors. Motoneurons could extend neurites on both live (Fig. 3A, D) and frozen (Fig. 3B, E) substrates; however, the motoneurons exhibited only limited neurite extension on decellularized fibrous ECM (Fig. 3C), while showing robust growth on protoplasmic ECM (Fig. 3F). Neurite area was also found to be significantly greater on live protoplasmic substrates than decellularized protoplasmic ECM or any fibrous substrate after 2 days of culture (Fig. 3H). Furthermore, decellularized protoplasmic ECM exhibited significantly greater growth than either unmodified gelatin or decellularized fibrous ECM (Fig. 3H).
FIG. 3.
Protoplasmic astrocyte-derived substrates are permissive to motoneuron growth and neurite extension. (A–G) Representative images of motoneurons on different astrocyte-derived substrates after 48 h in culture. (A–C) Fibrous substrates, (D–F) protoplasmic substrates. (A, D) Live astrocyte substrate, (B, E) freeze-lysed astrocyte substrate, (C, F) decellularized astrocyte substrate. (G) Gelatin control shows no clear neurites. Scale bar: 100 μm (H) quantification of neurite extension from motoneurons cultured for 48 h on different astrocyte substrates. Error bars: standard error, n = 19–52. *P < 0.05, **P < 0.01, ***P < 0.001. Dark bars = protoplasmic substrate, light bars = fibrous substrate.
The effect of fibrous and protoplasmic CM on neurite extension from motoneurons was also tested to determine if there were any growth benefits of the factors secreted by the astrocyte populations. To allow for direct comparison between fibrous and protoplasmic CMs and control for the effect of the astrocyte growth factors on the neurons, motoneurons were grown in ½ fibrous media and ½ protoplasmic media. These studies found that protoplasmic CM slightly, but significantly, improved neurite extension from motoneurons compared to unconditioned media after 72 h (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/scd).
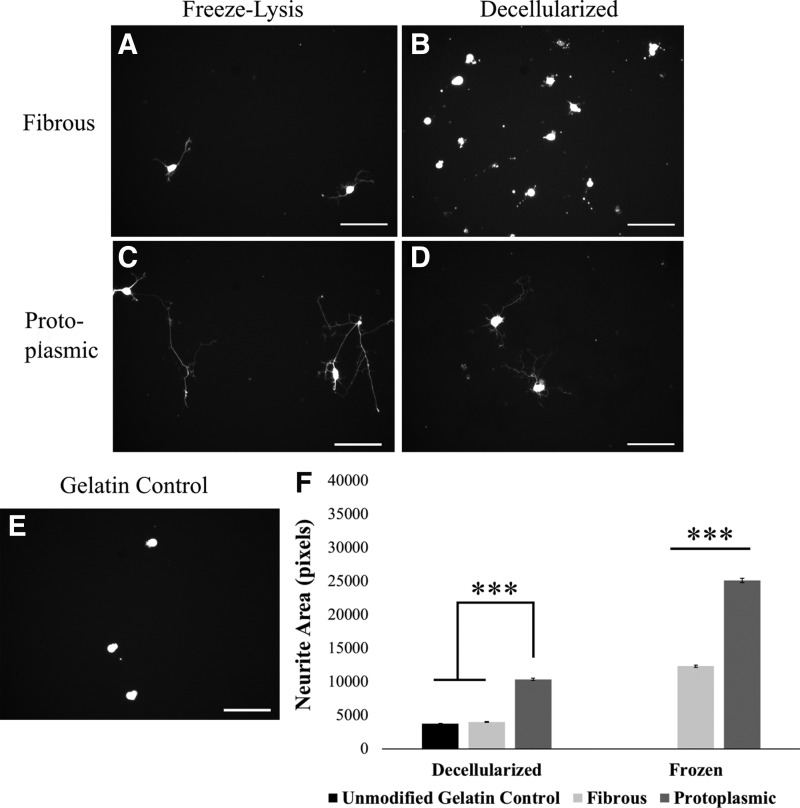
To confirm that these growth differences were not specific to motoneurons, the effect of frozen and decellularized astrocyte substrates on mESC-derived V2a interneuron neurite extension was tested. V2a interneurons were derived from mESCs using a Chx10-PAC cell line with constitutively active TdTomato expression [21]. It was found that the interneurons, similar to motoneurons, exhibited significantly longer neurite outgrowth on protoplasmic (Fig. 4 C, D) versus fibrous substrates (Fig. 4 A, B); in addition, the magnitude of the ECM effect was found to be greater on the interneuron cultures than the motoneuron cultures (Fig. 4F).
FIG. 4.
Protoplasmic astrocyte-derived substrates are more permissive to V2a interneuron growth than fibrous astrocyte-derived substrates. (A–E) Representative images of V2a interneurons after 72 h in culture on different astrocyte-derived substrates. (A, B) Fibrous substrates, (D, E) Protoplasmic substrates, (A, D) freeze-lysed astrocyte substrate, (B, E) decellularized astrocyte substrate. (C) Unmodified gelatin control. Scale bar: 100 μm. (F) Quantification of neurite extension from V2a interneurons cultured for 3 days on different astrocyte substrates. Error bars: Standard error, n = 40–56, ***P < 0.001. Dark bars = protoplasmic substrate, light bars = fibrous substrate.
Fibrous and protoplasmic astrocytes deposit distinct ECMs
Since the ECM deposited by these predominantly glial cultures was sufficient to support neuron growth, label-free LC/MS proteomics was performed to determine what proteins are present in ECMs harvested from both protoplasmic and fibrous populations. Identification of these proteins will allow for better understanding of the reasons for the observed neuronal growth differences and potentially allow for customized substrate design. ECMs harvested from mESC-derived populations were found to be largely similar with 530 of the 638 proteins identified not being expressed at different levels; however, there were significant expression differences of several key axon growth-related proteins (Fig. 5A, Table 3, Supplementary Table S1).
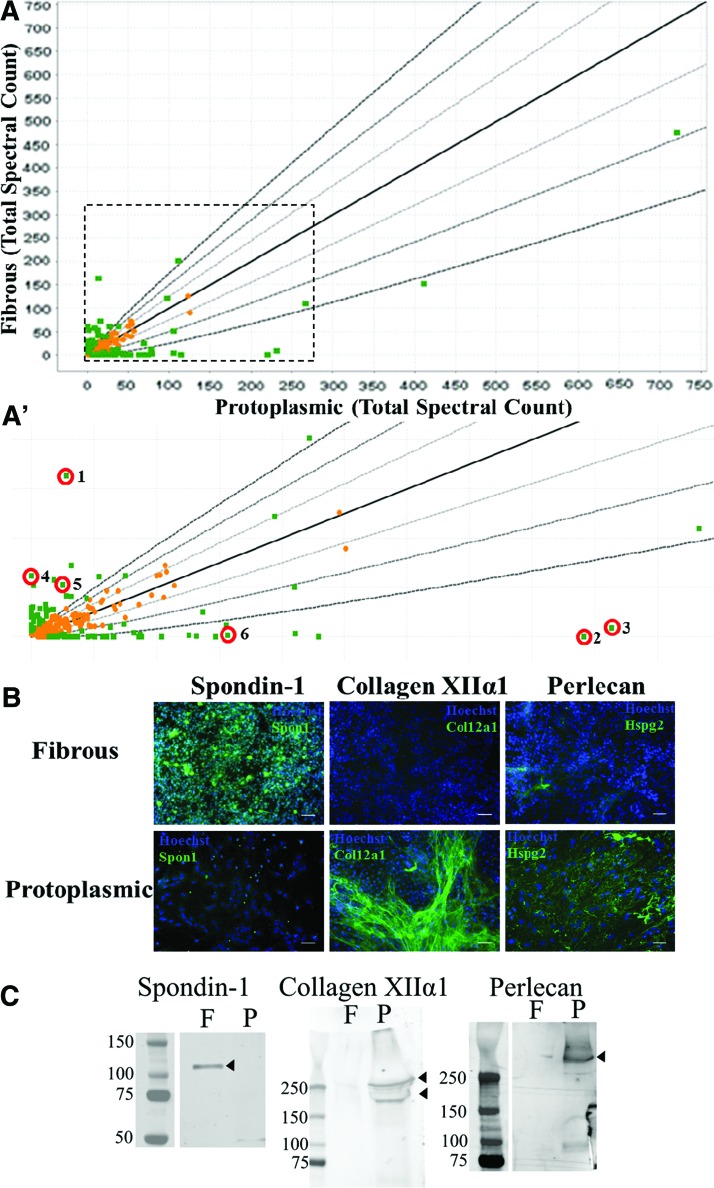
FIG. 5.
Protoplasmic and fibrous astrocytes have distinct ECM composition. (A) Graphical representation of proteomics results. Each point represents the average spectral count of the identified protein from two independent runs. Lines denote one, two, or three standard deviations away from equal spectral count of the specific protein detected. Proteins found to be significantly different between protoplasmic and fibrous ECMs are denoted by green squares, Fisher's exact test with Benjamini–Hochberg correction, P < 0.001, n = 2. Orange squares denote proteins that did not differ significantly between astrocyte ECMs. (A′) Blowup of indicated area in (A). Red circles mark proteins of interest: 1: spondin-1, 2: collagen XIIα1, 3: perlecan, 4: neurocan, 5: versican, 6: laminin α5. (B) Confirmation of proteomics findings for spondin-1, collagen XIIα1, and perlecan with immunohistochemistry (target protein in green and nuclei in blue) showing an expression pattern consistent with the proteomics data. Scale bar = 100 μm. (C) Western blots of ECMs harvested from decellularized plates also show the expected pattern spondin-1, collagen XIIα1, and perlecan. F: Fibrous ECM, P: protoplasmic ECM, Arrowhead denotes protein band. ECM, extracellular matrix. Color images available online at www.liebertpub.com/scd
Table 3.
Known Axon Growth Modulatory Proteins Identified in Astrocyte Extracellular Matrix Proteomics Data
| Protein | Significantly upregulated in | Axon growth role |
|---|---|---|
| Neurocan | Fibrous | Inhibitory |
| Versican | Fibrous | Inhibitory |
| Netrin-1 | Fibrous | Inhibitory |
| Semaphorin-3A | Fibrous | Inhibitory |
| Tenascin C | Fibrous | Permissive |
| Laminin α1 | Protoplasmic | Permissive |
| Laminin α5 | Protoplasmic | Permissive |
| Laminin β1 | Protoplasmic | Permissive |
| Laminin γ1 | Protoplasmic | Permissive |
| Collagen 4α1 | Protoplasmic | Permissive |
| Fibronectin | Protoplasmic | Permissive |
| Perlecan | Protoplasmic | Permissive |
| Matrillin-2 | N.S. | Permissive |
N.S., not significant.
Fibronectin-1 was found to be the most abundant protein in both ECMs with significantly more spectral counts detected in the protoplasmic samples (Fig. 5A). Fibronectin has been described as being expressed by primary astrocytes in culture [36], is transiently expressed away from the blood vessels during CNS development [37], and has been linked to improved neuron growth in adult white matter [38]. The proteins with the greatest differential expression between the protoplasmic and fibrous populations, were spondin-1 (spon1), Collagen XIIα1 (col12a1), and perlecan (hspg2). Spondin-1, also known as F-spondin, was expressed at significantly greater levels in fibrous ECM and is known to be involved in axonal pathfinding and turning during development [39] (Fig. 5A′, #1). Col12a1 and perlecan were both found at significantly greater levels in protoplasmic ECM (Fig. 5A′, #2, #3). Perlecan-coated plates have been demonstrated to promote neurite extension in vitro [40].
Also of note is that the chondroitin sulfate proteoglycans (neurocan and versican), which are known inhibitory molecules [41], were found to be significantly more prevalent in fibrous ECM (Fig. 5A′, #4, #5), while laminins, known to be axon growth promoting, were more prevalent within protoplasmic ECM (Fig. 5A′, #6). To confirm the validity of our proteomics results, ICC was performed on both protoplasmic and fibrous cultures for collagen XIIα1, perlecan, and spondin-1 (Fig. 5B). To further validate the proteomics, western blotting was performed on ECMs harvested from decellularized astrocyte plates (Fig. 5C). The expression pattern of these proteins matched the profiles found in the LC/MS data demonstrating the validity of the proteomic results (Fig. 5B, C).
ECM produced by ESC-derived cultures is consistent with in vivo astrocyte protein expression
The cultures used for this work are not purely astrocytic as there is a clear presence of both neurons and glial progenitors. Thus, it was important to determine if the harvested ECMs were consistent with in vivo astrocyte expression profiles to demonstrate if the harvested ECMs are similar to native astrocyte ECM production. This was achieved by comparing the proteins found in astrocyte ECMs to a set of genes identified as expressed in astrocytes in vivo. In vivo astrocyte expression profiles were determined using a previously described TRAP mouse line that targets all in vivo astrocyte populations using ALDH1 L1 [42], coupled with RNAseq. This BAC-TRAP methodology compares the prevalence of mRNA transcripts in the total brain isolate to the prevalence of the same mRNA transcripts when only the mRNAs attached to astrocyte ribosomes are present. This approach allows for any transcript that is enriched in the astrocyte (TRAP) fraction to be considered astrocyte specific or, at least, more highly expressed by astrocytes than other CNS populations. Genes that are relatively depleted in the TRAP fraction (enriched in the pre-IP fraction) can be considered more highly expressed by a different population within the CNS or not expressed by astrocytes. The distinction between enrichment and population-specific expression was made based on the fold change in expression level of the particular gene between the TRAP and pre-IP fractions.
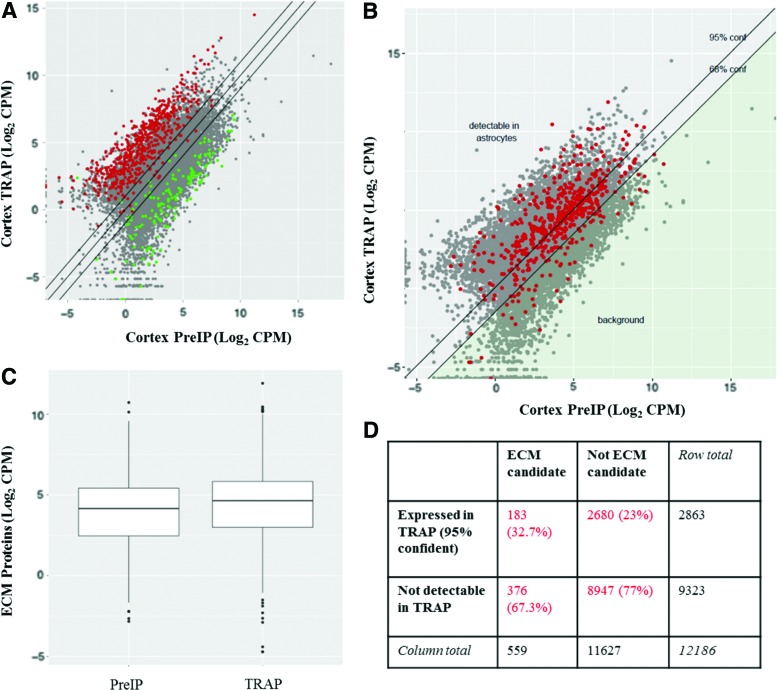
The validity of the TRAP-Seq method was confirmed by locating all genes previously identified as astrocyte or neuron specific within the data set [43]. It was found that 84% of genes previously identified as astrocyte specific were enriched in the astrocyte fraction, and 81% of those previously identified as neuron specific were depleted in the astrocyte fraction, confirming a successful TRAP (Fig. 6A). Based on the mean fold change of the neuronal markers between the astrocyte fraction and whole brain isolate, a threshold was calculated to allow for genes to be considered astrocyte specific with 95% confidence. With a threshold established, the mRNAs that encode for proteins found within the mESC-derived ECMs were located within the BAC-TRAP RNA-seq samples. Based on this methodology, 183 (33%) of the ECM proteins identified can be considered to be astrocyte specific [44] and 159 of these were at least two-fold enriched in the TRAP fraction (Fig. 6B). The majority of these astrocyte-specific ECM proteins were found in both protoplasmic and fibrous ECMs.
FIG. 6.
ECM components generated by ESC-derived astrocytes are found in cortical astrocytes based on BAC-TRAP. (A) Astrocyte-specific genes (red) are enriched in the TRAP fraction, while neuronal-specific genes (green) are enriched in the Pre-IP samples. n = 3. (B) ECM candidate genes are enriched within the TRAP fraction compared to Pre-IP sample. Lines are 95% and 68% confidence of astrocyte-specific expression. Gray points are all genes detected in the BAC-TRAP data, red points denote genes that code for proteins detected in ECM proteomics data. (C) ECM proteins are significantly enriched in TRAP fraction compared to the Pre-IP fraction. Paired t-test, P = 5.66 × 10−9. (D) Comparison of identified proteins with the proteomics and BAC-TRAP data. Proteins were considered astrocyte expressed if there was 95% confidence that the mRNA was enriched within the TRAP fraction compared to the Pre-IP fraction. The ECM proteins identified in proteomics were enriched for protein expression in astrocytes based on BAC-TRAP at a significantly higher level than expected by chance. Fisher's exact test, P = 2. × 10−7. TRAP, translating ribosome affinity purification. Color images available online at www.liebertpub.com/scd
In addition to containing astrocyte-specific proteins, ECM components were also found to be more highly expressed in the TRAP than in the Pre-IP fraction (Fig. 6C, paired t-test, P < 0.0001). Among those genes detectable in RNA-seq samples, ECM components were more likely than chance to be identified as expressed by astrocytes in the BAC-TRAP data (Fig. 6B, D, Fisher's exact test, P < 0.001). Furthermore, ECM components were more likely than chance to be identified as having two-fold higher expression in astrocytes in vivo (Fisher's exact test, P < 0.05). These data show that the proteins found within the ESC-derived ECMs are consistent with in vivo astrocyte expression profiles and suggest that the mESC-derived ECMs from both fibrous and protoplasmic populations are consistent with in vivo astrocyte ECMs.
Motoneuron growth depends on the presence and absence of specific ECM proteins
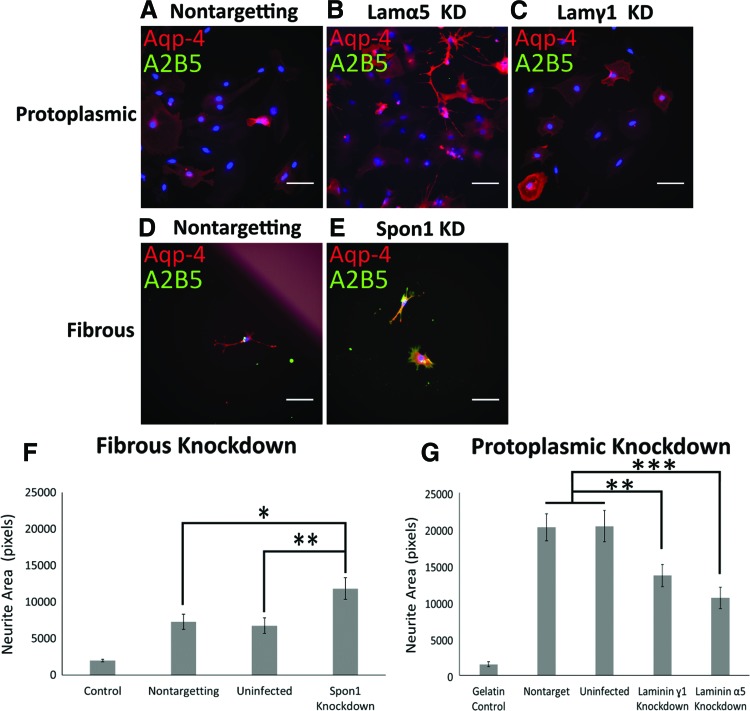
To test which proteins within the mESC-derived ECMs are important for motoneuron growth, proteins with known roles in neurite growth found to be highly expressed in one ECM, but not the other, were targeted with shRNA knockdown. Targets were preferentially chosen from protoplasmic ECM to determine growth promoting factors for potential future work using a small number of specific proteins. In particular, collagen XIIα1, collagen VIα3, laminin β1, perlecan, laminin α5, and laminin γ1 were targeted in protoplasmic ECM, while spondin-1 was targeted in fibrous ECM. Since matrix deposition takes multiple days, shRNAs were stably expressed using lentiviral transduction. Staining of astrocytes following infection showed that both fibrous and protoplasmic astrocytes maintained the expected morphology and phenotype, although cell density was significantly lower than previously observed (Fig. 7A–E).
FIG. 7.
Viral knockdown modifies motoneuron growth on astrocyte ECMs. (A–E) Aqp-4 (red) and A2B5 (green) staining of infected astrocytes. (A–C) Protoplasmic astrocytes infected with nontargeting (A), laminin α5 targeting (B), or laminin γ1 targeting (C) virus. (D, E) Fibrous astrocytes infected with nontargeting (D), or spondin-1 targeting (E) virus. Scale bar: 100 μm. (F) Spondin-1 knockdown in fibrous ECM leads to a significant increase in motoneuron growth. n = 39–58. (G) Laminin α5 or laminin γ1 knockdown decreases motoneuron growth on protoplasmic ECM. n = 44–104. Error bars: standard error; *P < 0.05; **P < 0.01; ***P < 0.001. Color images available online at www.liebertpub.com/scd
Quantification of motoneuron growth on knockdown ECMs found that knockdown of spondin-1 in fibrous cultures increased neurite extension (Fig. 7F), while laminin α5 or laminin γ1 knockdown in protoplasmic astrocytes decreased neurite extension (Fig. 7G). There were no significant effects observed for any of the shRNA constructs targeting collagen XIIα1, collagen VIα3, laminin β1, or perlecan; however, knockdown was not verified in these cultures due to the lack of effect (Supplementary Fig. S2). Using western blotting and qPCR, spon1 shRNA treatment was found to decrease expression of spondin-1 to 53% of the uninfected control and decreased mRNA levels to 44% of the nontargeted control (Supplementary Fig. S3). The laminin α5 and γ1 shRNAs used have been previously validated by Sigma to reduce expression of the target protein by 67% and 95%, respectively; so no further validation was performed on these constructs. Interestingly, the decreases observed in the knockdown ECMs do not fully account for the growth differences observed between protoplasmic and fibrous ECMs, suggesting that the neuronal growth effects are multifactorial and due to many different proteins within the ECMs.
Discussion
CNS injury leads to a significant economic and psychological burden for patients due, in part, to the lack of spontaneous regeneration within the CNS [45]. Classically, this lack of recovery has been thought to be partially caused by the formation of a glial scar that contains woven astrocyte processes and a large number of proteins that inhibit axon growth [1]. Recently, it has been observed that astrocytes are also involved in spontaneous recovery in mice and zebrafish [46–48] and astrocyte transplants can improve recovery following SCI, if they have a proregenerative phenotype [15,49]. Since astrocytes have been successfully used for SCI treatment, a scalable methodology for the derivation of proregenerative astrocytes could have a significant clinical impact.
This work demonstrates a scalable method to produce either fibrous or protoplasmic astrocytes from mESCs (Figs. 1 and 2), with around 40% astrocytes in fibrous cultures and 49% astrocytes in protoplasmic cultures based on Aqp-4 staining (Fig. 2). This is likely a conservative estimate, as Aqp-4 staining is known to vary across astrocyte cultures, which is clear in our staining as well (Fig. 2). Quantification of S100 staining suggests that fibrous cultures contain around 83% astrocytic-lineage cells and protoplasmic cultures contain around 92% astrocytic-lineage cells, even if not all the astrocytes express sufficient Aqp-4 for detection with flow cytometry (Fig. 2). The significant differences in GLAST, GDNF, and A2B5 expression, coupled with the cell morphology differences, indicate that these methods generate astrocytes that are consistent with in vivo fibrous and protoplasmic astrocyte phenotypes, and fibrous populations contain mostly white matter astrocytes, while protoplasmic populations contain mostly gray matter astrocytes (Figs. 1 and 2). Astrocyte presence within these cultures was further confirmed with BAC-TRAP demonstrating that ECMs derived from fibrous and protoplasmic cultures were consistent with in vivo astrocyte expression profiles during development (Fig. 6).
Functionally these predominantly astrocytic cultures were found to have differing abilities to support neuronal growth with protoplasmic-derived substrates tending to be more supportive of neuron growth than fibrous substrates. This was especially apparent in the case of decellularized ECMs (Figs. 3 and 4). It is important to note that transplanted immature, but not mature, astrocytes have been found to improve outcomes following SCI, while demonstrating further migration from the transplant area and reduction of the glial scar [50,51]. Due to these observations, and the relatively immature nature of these cultures, it is possible that the functional differences observed in the mESC cultures are applicable to embryonic, but not adult, astrocyte populations. However, this protocol does provide a readily scalable method to produce populations that can be used to support neuronal growth.
It would be potentially interesting to explore the effects of inflammatory stimulus on the observed neuronal growth support properties of these astrocytes. Recent work has demonstrated that astrocytes exhibit a continuum of reactive phenotypes following injury, with some exhibiting proregenerative phenotypes and others exhibiting inhibitory phenotypes [52]. It has been observed that proinflammatory and anti-inflammatory stimuli push reactive astrocyte phenotypes one way or the other on this continuum. In particular, inflammatory injury caused by lipopolysaccharide causes astrocytes to adopt a more inhibitory phenotype [8]. In addition, interleukin-6 (IL-6), a proinflammatory cytokine, leads to an increase in astrocyte proliferation and scar formation in vivo [53], while, IL-10, an anti-inflammatory cytokine, has been to shown improve functional recovery following CNS injury when delivered intrathecally or intramuscularly [54]. Future experiments could explore the effects of these inflammatory and anti-inflammatory stimuli on the neuronal growth support capacities of these mESC-derived fibrous and protoplasmic populations.
Recent work by Anderson et al. showed that astrocytes are required for recovery following SCI in mice [4]. As part of this work, they compared axon growth permissive and inhibitory proteins produced by astrocytes and nonastrocytes following injury. The proteomics data set collected in this study was searched for these proteins, and it was found that, if present within the ECMs, permissive proteins tended to be significantly upregulated in protoplasmic ECM, while inhibitory proteins were upregulated in fibrous ECM (Table 3). This suggests that the neuron growth differences observed on the ECMs derived in this study are likely due to both the more inhibitory nature of the fibrous ECM and the more permissive nature of the protoplasmic ECM. This concept is supported by the observed decrease in motoneuron growth on protoplasmic ECM following knockdown of laminin α5 and laminin γ1 and the increase in motoneuron growth on fibrous ECM following spondin-1 knockdown (Fig. 7). Future work on which proteins are the most important for neuron growth in these ECMs could allow for a small subset of ECM components to be mass produced and delivered to improve recovery following CNS injury.
Supplementary Material
Acknowledgments
The technical expertise of Petra Erdmann-Gilmore is gratefully acknowledged. This work was supported by NINDS R01 NS090617-02, MH099798-01, the Washington University Medical Scientist Training Program (NIH T32 GM07200) and graduate program (T32 GM008151), and the Children's Discovery Institute CDI (MD-II-2013-269). The LC-MS analysis and protein identifications were performed at the Washington University Proteomics Shared Resource (WU-PSR). The WU-PSR is supported by the WU Institute of Clinical and Translational Sciences (NCATS UL1 TR000448), the WU Mass Spectrometry Research Resource (NIGMS P41 GM103422), and the Siteman Comprehensive Cancer Center (NCI P30 CA091842). Sequencing of Illumina libraries was performed by the Washington University Genome Technology Access Center (supported by NIH grants P30 CA91842 and UL1TR000448).
Author Disclosure Statement
J.D.D. has received royalties related to TRAP in the past. The remaining authors declare no competing financial interests.
References
- 1.Cregg JM, DePaul MA, Filous AR, Lang BT, Tran A. and Silver J. (2014). Functional regeneration beyond the glial scar. Exp Neurol 253:197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pekny M, Johansson CB, Eliasson C, Stakeberg J, Wallén A, Perlmann T, Lendahl U, Betsholtz C, Berthold CH. and Frisén J. (1999). Abnormal reaction to central nervous system injury in mice lacking glial fibrillary acidic protein and vime. J Cell Biol 145:503–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herrmann JE, Imura T, Song B, Qi J, Ao Y, Nguyen TK, Korsak RA, Takeda K, Akira S. and Sofroniew MV. (2008). STAT3 is a critical regulator of astrogliosis and scar formation after spinal cord injury. J Neurosci 28:7231–7243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson MA, Burda JE, Ren Y, Ao Y, O'Shea TM, Kawaguchi R, Coppola G, Khakh BS, Deming TJ. and Sofroniew MV. (2016). Astrocyte scar formation aids central nervous system axon regeneration. Nature 532:195–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faulkner JR, Herrmann JE, Woo MJ, Tansey KE, Doan NB. and Sofroniew MV. (2004). Reactive astrocytes protect tissue and preserve function after spinal cord injury. J Neurosci 24:2143–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ. and Barres BA. (2008). A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci 28:264–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oberheim NA, Goldman SA. and Nedergaard M. (2012). Heterogeneity of astrocytic form and function. Methods Mol Biol 814:23–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zamanian JL, Xu L, Foo LC, Nouri N, Zhou L, Giffard RG. and Barres BA. (2012). Genomic analysis of reactive astrogliosis. J Neurosci 32:6391–6410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun D. and Jakobs TC. (2012). Structural remodeling of astrocytes in the injured CNS. Neuroscientist 18:567–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun D, Lye-Barthel M, Masland RH. and Jakobs TC. (2010). Structural remodeling of fibrous astrocytes after axonal injury. J Neurosci 30:14008–14019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilhelmsson U, Bushong EA, Price DL, Smarr BL, Phung V, Terada M, Ellisman MH. and Pekny M. (2006). Redefining the concept of reactive astrocytes as cells that remain within their unique domains upon reaction to injury. Proc Natl Acad Sci U S A 103:17513–17518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rao MS, Noble M. and Mayer-Pröschel M. (1998). A tripotential glial precursor cell is present in the developing spinal cord. Proc Natl Acad Sci U S A 95:3996–4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin Y, Bouyer J, Shumsky JS, Haas C. and Fischer I. (2016). Transplantation of neural progenitor cells in chronic spinal cord injury. Neuroscience 320:69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies JE, Huang C, Proschel C, Noble M, Mayer-Proschel M. and Davies SJA. (2006). Astrocytes derived from glial-restricted precursors promote spinal cord repair. J Biol 5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies JE, Pröschel C, Zhang N, Noble M, Mayer-Pröschel M. and Davies SJA. (2008). Transplanted astrocytes derived from BMP- or CNTF-treated glial-restricted precursors have opposite effects on recovery and allodynia after spinal cord injury. J Biol 7:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies SJA, Shih C-H, Noble M, Mayer-Proschel M, Davies JE. and Proschel C. (2011). Transplantation of specific human astrocytes promotes functional recovery after spinal cord injury. PLoS One 6:e17328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roybon L, Lamas N, Garcia-Diaz A, Yang E, Sattler R, Jackson-Lewis V, Kim Y, Kachel CA, Rothstein J, et al. (2013). Human stem cell-derived spinal cord astrocytes with defined mature or reactive phenotypes. Cell Rep 4:1035–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benveniste RJ, Keller G. and Germano I. (2005). Embryonic stem cell-derived astrocytes expressing drug-inducible transgenes: differentiation and transplantion into the mouse brain. J Neurosurg 103:115–123 [DOI] [PubMed] [Google Scholar]
- 19.Hudson TW, Liu SY. and Schmidt CE. (2004). Engineering an improved acellular nerve graft via optimized chemical processing. Tissue Eng 10:1346–1358 [DOI] [PubMed] [Google Scholar]
- 20.McCreedy DA, Wilems TS, Xu H, Butts JC, Brown CR, Smith AW. and Sakiyama-Elbert SE. (2014). Survival, differentiation, and migration of high-purity mouse embryonic stem cell-derived progenitor motor neurons in fibrin scaffolds after sub-acute spinal cord injury. Biomater Sci 2:1672–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iyer NR, Huettner JE, Butts JC, Brown CR. and Sakiyama-Elbert SE. (2016). Generation of highly enriched V2a interneurons from mouse embryonic stem cells. Exp Neurol 277:305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Z-W, Fuchs K, Sieghart W, Townsend RR. and Evers AS. (2012). Deep amino acid sequencing of native brain GABAA receptors using high-resolution mass spectrometry. Mol Cell Proteomics 11:M111.011445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keller A, Nesvizhskii AI, Kolker E. and Aebersold R. (2002). Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74:5383–5392 [DOI] [PubMed] [Google Scholar]
- 24.Nesvizhskii AI, Keller A, Kolker E. and Aebersold R. (2003). A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75:4646–4658 [DOI] [PubMed] [Google Scholar]
- 25.Reddy AS, Brien DO', Pisat N, Weichselbaum CT, Sakers K, Lisci M, Dalal JS. and Dougherty JD. (2016). A comprehensive analysis of cell type–specific nuclear RNA from neurons and glia of the brain. Biol Psychiatry 81:252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wichterle H, Wichterle H, Lieberam I, Lieberam I, Porter JA, and Jessell TM. (2002). Directed differentiation of embryonic stem cells into motor neurons. Cell 110:385–397 [DOI] [PubMed] [Google Scholar]
- 27.Holland EC. (2001). Gliomagenesis: genetic alterations and mouse models. Nat Rev Genet 2:120–129 [DOI] [PubMed] [Google Scholar]
- 28.Goursaud S, Kozlova EN, Maloteaux J-M. and Hermans E. (2009). Cultured astrocytes derived from corpus callosum or cortical grey matter show distinct glutamate handling properties. J Neurochem 108:1442–1452 [DOI] [PubMed] [Google Scholar]
- 29.Vanhoutte N, de Hemptinne I, Vermeiren C, Maloteaux J-M. and Hermans E. (2004). In vitro differentiated neural stem cells express functional glial glutamate transporters. Neurosci Lett 370:230–235 [DOI] [PubMed] [Google Scholar]
- 30.Nagelhus EA, Veruki ML, Torp R, Haug FM, Laake JH, Nielsen S, Agre P. and Ottersen OP. (1998). Aquaporin-4 water channel protein in the rat retina and optic nerve: polarized expression in Müller cells and fibrous astrocytes. J Neurosci 18:2506–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kleiderman S, Sá JV, Teixeira AP, Brito C, Gutbier S, Evje LG, Hadera MG, Glaab E, Henry M, et al. (2016). Functional and phenotypic differences of pure populations of stem cell-derived astrocytes and neuronal precursor cells. Glia 64:695–715 [DOI] [PubMed] [Google Scholar]
- 32.Raponi E, Agenes F, Delphin C, Assard N, Baudier J, Legraverend C. and Deloulme J-C. (2007). S100B expression defines a state in which GFAP-expressing cells lose their neural stem cell potential and acquire a more mature developmental stage. Glia 55:165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hachem S, Aguirre A, Vives V, Marks A, Gallo V. and Legraverend C. (2005). Spatial and temporal expression of S100B in cells of oligodendrocyte lineage. Glia 51:81–97 [DOI] [PubMed] [Google Scholar]
- 34.Deloulme JC, Raponi E, Gentil BJ, Bertacchi N, Marks A, Labourdette G. and Baudier J. (2004). Nuclear expression of S100B in oligodendrocyte progenitor cells correlates with differentiation toward the oligodendroglial lineage and modulates oligodendrocytes maturation. Mol Cell Neurosci 27:453–465 [DOI] [PubMed] [Google Scholar]
- 35.Brunne B, Zhao S, Derouiche A, Herz J, May P, Frotscher M. and Bock HH. (2010). Origin, maturation, and astroglial transformation of secondary radial glial cells in the developing dentate gyrus. Glia 58:1553–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liesi P, Kirkwood T. and Vaheri A. (1986). Fibronectin is expressed by astrocytes cultured from embryonic and early postnatal rat brain. Exp Cell Res 163:175–185 [DOI] [PubMed] [Google Scholar]
- 37.Stewart GR. and Pearlman AL. (1987). Fibronectin-like immunoreactivity in the developing cerebral cortex. J Neurosci 7:3325–3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tom VJ, Doller CM, Malouf AT. and Silver J. (2004). Astrocyte-associated fibronectin is critical for axonal regeneration in adult white matter. J Neurosci 24:9282–9290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burstyn-Cohen T, Tzarfaty V, Frumkin A, Feinstein Y, Stoeckli E. and Klar A. (1999). F-spondin is required for accurate pathfinding of commissural axons at the floor plate. Neuron 23:233–246 [DOI] [PubMed] [Google Scholar]
- 40.Nakamura R, Nakamura F. and Fukunaga S. (2015). Diverse functions of perlecan in central nervous system cells in vitro. Anim Sci J 86:904–911 [DOI] [PubMed] [Google Scholar]
- 41.Oohira A, Matsui F. and Katoh-Semba R. (1991). Inhibitory effects of brain chondroitin sulfate proteoglycans on neurite outgrowth from PC12D cells. J Neurosci 11:822–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, et al. (2008). Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135:749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tien A-C, Tsai H-H, Molofsky AV, McMahon M, Foo LC, Kaul A, Dougherty JD, Heintz N, Gutmann DH, Barres BA. and Rowitch DH. (2012). Regulated temporal-spatial astrocyte precursor cell proliferation involves BRAF signalling in mammalian spinal cord. Development 139:2477–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dougherty JD, Schmidt EF, Nakajima M. and Heintz N. (2010). Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res 38:4218–4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boakye M, Leigh BC. and Skelly AC. (2012). Quality of life in persons with spinal cord injury: comparisons with other populations. J Neurosurg Spine 17:29–37 [DOI] [PubMed] [Google Scholar]
- 46.Goldshmit Y, Frisca F, Pinto AR, Pébay A, Tang J-KKY, Siegel AL, Kaslin J. and Currie PD. (2014). Fgf2 improves functional recovery-decreasing gliosis and increasing radial glia and neural progenitor cells after spinal cord injury. Brain Behav 4:187–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zukor K, Belin S, Wang C, Keelan N, Wang X. and He Z. (2013). Short hairpin RNA against PTEN enhances regenerative growth of corticospinal tract axons after spinal cord injury. J Neurosci 33:15350–15361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haas C, Neuhuber B, Yamagami T, Rao M. and Fischer I. (2012). Phenotypic analysis of astrocytes derived from glial restricted precursors and their impact on axon regeneration. Exp Neurol 233:717–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chu T, Zhou H, Li F, Wang T, Lu L. and Feng S. (2014). Astrocyte transplantation for spinal cord injury: current status and perspective. Brain Res Bull 107:18–30 [DOI] [PubMed] [Google Scholar]
- 50.Smith GM. and Silver J. (1988). Transplantation of immature and mature astrocytes and their effect on scar formation in the lesioned central nervous system. Prog Brain Res 78:353–361 [DOI] [PubMed] [Google Scholar]
- 51.Smith GM. and Miller RH. (1991). Immature type-1 astrocytes suppress glial scar formation, are motile and interact with blood vessels. Brain Res 543:111–122 [DOI] [PubMed] [Google Scholar]
- 52.Liddelow SA. and Barres BA. (2017). Reactive astrocytes: production, function, and therapeutic potential. Immunity 46:957–967 [DOI] [PubMed] [Google Scholar]
- 53.Nakamura M, Okada S, Toyama Y. and Okano H. (2005). Role of IL-6 in spinal cord injury in a mouse model. Clin Rev Allergy Immunol 28:197–204 [DOI] [PubMed] [Google Scholar]
- 54.Jackson CA, Messinger J, Peduzzi JD, Ansardi DC. and Morrow CD. (2005). Enhanced functional recovery from spinal cord injury following intrathecal or intramuscular administration of poliovirus replicons encoding IL-10. Virology 336:173–183 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.