Abstract
Alcohol is a frequently used addictive substance worldwide. Aim of this study is to determine the frequency distribution of SNPs within ADH1B,ADH4,ADH1C,ALDH2, BDNF,OPRM1, and DRD2 genes in a southeastern European Caucasian population from Greece. For this purpose samples of 1276 volunteers were analyzed after deidentification and anonymization. The allele distribution of the examined polymorphisms in the present Greek population cohort was as follows: rs1229984 (ADH1B): GG(wt) = 64.14%, GA = 29.86%, AA = 4.00%; rs1693482 (ADH1C): CC(wt) = 57.45%, CT = 36.76%, TT = 5.80%; rs1799971 (OPRM1): AA(wt) = 72.43%, AG = 28.72%, GG = 1.89%; rs1800497 (DRD2): CC(wt) = 70.84%, CT = 27.18%, TT = 1.98%; rs1800759 (ADH4): CC(wt) = 34.25%, CA = 48.12%, AA = 17.63%; rs6265 (BDNF): GG(wt) = 65.99%, GA = 31.02%, AA = 2.99%; and rs671 (ALDH2): GG(wt) = 99.84% GA = 0.16%, AA = 0.00%. Mutant rs1229984 allele A was ~6.5× more frequent in the Greek than in the European population. Mutant rs1693482 allele T was ~1.7× more frequent in the European than in the Greek population. Mutant alleles for polymorphisms rs1800759 and rs1799971 show similar frequencies in both northern and southern Europeans. One rs671 mutant A allele was detected in the Greek population (0.08%). The mutant rs1800497 allele T was ~1.2× more frequent in the European than in the Greek population and the mutant rs6265 allele A was ~1.1× more frequent in the European than in the Greek population. An alcohol addiction‐specific algorithm was generated (TGS) that may predict alcohol addiction prevalence in a population. According to our findings, the analyzed Southeastern population may differ genetically from north Europeans due to influences from neighboring Asian and African populations and a calculated TGS score >50 indicates individuals with low susceptibility to develop alcohol addiction.
Keywords: Alcohol abuse, alcohol metabolism, dopamine receptor, predisposition, reward pathway, southeastern European population
Abbreviations
- ADH1B
subunit 1B of the ADH gene
- ADH4
alcohol dehydrogenase 4
- ADH
alcohol dehydrogenase
- ALDH
aldehyde dehydrogenase
- ANKK1
ankyrin repeat and kinase‐containing 1
- BDNF
brain‐derived neurotrophic factor
- CNS
central nervous system
- HWE
Hardy–Weinberg equilibrium
- MOR
μ‐opioid receptors
- OPRM1
μ‐opioid receptor gene
- ORs
odd ratios
- PCR
polymerase chain reaction
- SNPs
single‐nucleotide polymorphisms
- TGS
total genotype score
Introduction
Alcohol is one of the most frequently used addictive substances in the world, causing physical and psychological dependence. According to the World Health Organization, alcohol misuse may be the cause of over 3.3 million deaths per year globally. Additionally, excessive use of alcohol may cost more than 249 billion U.S. dollars per year, only in the United States (Sacks et al. 2015). In the European Union the alcohol abuse issue seems to be quite important, as it shows the highest proportion of alcoholics and the highest levels of alcohol consumption per capita in the world and, therefore, the highest level of alcohol‐related health problems.
Alcohols' sedative effects on the central nervous system (CNS) are presented in cerebral cortex and reticular formation through a complex mode of action (Contet et al. 2004; Zaleski et al. 2004). Furthermore, alcohol leads to increased dopamine release in the mesolimbic pathway, through increased release of beta‐endorphin which is binding to μ‐opioid receptors (MOR), inducing activation of the reward pathway, leading to alcohol dependence (Chatterjee et al. 2011; Rahman and Prendergast 2012; Wu et al. 2014; Berrettini 2016).
Alcohol biodegradation is performed mainly in liver through consecutive oxidation steps. It is converted into acetaldehyde by alcohol dehydrogenase (ADH) and then into acetic acid by aldehyde dehydrogenase (ALDH) (Hurley et al. 2002; Zakhari 2006).
Genetic differences have been associated with differences in alcohol response among individuals. The studied single‐nucleotide polymorphisms (SNPs) were rs1229984 (ADH1B), rs1693482 (ADH1C), rs1799971 (OPRM1), rs1800497 (DRD2), rs1800759 (ADH4), rs6265 (BDNF), and rs671 (ALDH2).
The polymorphism rs1229984 (His48Arg) resides at the subunit 1B of the gene encoding ADH (ADH1B). The allele incorporating this SNP is referred as ADH1B*2 and leads to differentiation in enzyme activity. Carriers of G:A or A:A genotype appear to have increased activity of ADH, therefore, metabolizing alcohol faster than carriers of G:G genotype (Lee et al. 2004; Macgregor et al. 2009).
The polymorphism rs1693482 (Arg272Gln) of the ADH1C subunit is located on chromosome 4. Homozygous or heterozygous presence of the mutant ADH1C*2 allele limits the rate of alcohol metabolism and may show increased risk for alcohol dependence due to lower blood acetaldehyde levels (Lee et al. 2004; Macgregor et al. 2009).
Another polymorphism, considered to be participating in alcohol absorption and its biotransformation distribution (usually when alcohol blood levels are elevated) is rs1800759 that represents an SNP located in the promoter region of alcohol dehydrogenase 4 gene (ADH4). This variation causes increased gene transcription and, thus, increased enzyme biosynthesis that leads to heavy drinking. (Luo et al. 2006, 2007; Kimura et al. 2009).
Finally, the rs671 polymorphism (Glu504Lys), of the gene encoding for ALDH2, has been reported to generate an inactive allele (ALDH2*2). Heterozygous ALDH2*1/*2 (G:A) individuals experience reduced ALDH2 enzyme activity and homozygous polymorphism ALDH2*2/*2 (A:A) carriers are characterized by lack of ALDH2 enzyme activity (Macgregor et al. 2009).
While deficiencies in alcohol metabolizing enzymes may have a protective effect on alcohol dependence, genetic alterations in neurotransmission‐associated genes may affect activation or deactivation of the reward pathway in the CNS, providing a protective alcohol dependence mechanism.
A variation located in the μ‐opioid receptor gene (OPRM1), the rs1799971 (Asn40Asp), is reported to increase the binding affinity of the receptor for beta‐endorphins (Bart et al. 2005; Berrettini and Lerman 2005). This substitution seems to have a significant effect on the mesolimbic pathway in CNS (Berrettini and Lerman 2005; van den Wildenberg et al. 2007; Ehlers et al. 2008).
The TaqIΑ polymorphism (rs1800497) lies more than 10 kb downstream of the DRD2 gene. It is associated with a reduced DRD2 binding and density and its A1 allele is associated with alcoholism (Munafo et al. 2007). Moreover, it is known that this polymorphism is located in the coding region of the ankyrin repeat and kinase‐containing 1 (ANKK1) gene, encoding an enzyme that belongs to the serine/threonine kinases family, and results in a glutamic acid to lysine substitution in residue 713 (Glu713Lys), located at the carboxyl terminal ankyrin repeat domain, in a position associated with interactions between proteins (Huang et al. 2009; Kraschewski et al. 2009).
Lastly, the rs6265 polymorphism (Val66Met), in the gene encoding for Brain‐Derived Neurotrophic Factor (BDNF), is located in the 5′ pro‐BDNF sequence. Although this BDNF polymorphism does not affect mature BDNF protein function, it has been shown to alter the intracellular tracking and packaging of pro‐BDNF and, thus, to affect the regulated secretion of the mature peptide. BDNF may modulate the activity of various neurotransmitters, such as serotonin and dopamine (Gratacos et al. 2007; Wojnar et al. 2009).
This study aims at associating SNPs with alcohol dependence development, by determining their occurrence rates in a southeastern European Caucasian population sample and estimating the total risk of dependence by calculating the total number of alcohol‐associated SNPs for each volunteer.
Subjects and Methods
Samples from 690 women (54.08%) and 586 men (45.92%) (Southeastern European general population of unrelated Caucasians living in Greece, n = 1276) were analyzed. The median age of the participants was 35 years and their ages ranged from 18 to 97 years. Data collected from participants include gender, date of birth, place of residence, and genotype for genes of interest. No additional information on volunteers' health status and dependence on alcohol or any other addictive substances was provided.
The study was approved by the Ethics Committee of the University of Crete (University Hospital of Heraklion Ethics Committee Decision 8195/16) for blood sampling in patients and controls for biochemical and molecular cloning. All participants gave written informed consent to use their data following deidentification and data anonymization.
Epithelial cells were collected from the oral cavity of each volunteer using cotton swabs and DNA extraction was performed using nucleic acid isolation columns. Genotypes were determined by real‐time Polymerase Chain Reaction (PCR), using the LightCycler480 Instrument platform and following LightSnip kit protocols (Roche Diagnostics, Switzerland). Genotypes were classified as homozygous for wild‐type alleles, heterozygous and homozygous for mutant alleles.
Contingency tables 2 × 2 (1 degree of freedom) were designed and odd ratios (ORs) for elevated alcohol risk polymorphisms, as well as the corresponding confidence intervals were calculated. Statistical analysis was performed at a significant level of a = 0.05 and a statistically significance p‐value was calculated by Fisher's exact test. All genotypes were tested for Hardy–Weinberg equilibrium (HWE) using web‐based software (http://www.had2know.com/academics/hardy-weinberg-equilibrium-calculator-2-alleles.html).
For the calculation of dependence susceptibility arising from the combined influence of alcohol‐associated SNPs, the following algorithm was used (Williams and Folland 2008):
TGS = (100/2xN) × (GS1 + GS2 + …. + GSN), where Total Genotype Scores of addiction (TGS) represents the total risk of developing dependence on alcohol. N stands for the number of SNPs associated with the dependence on alcohol and GS (Genotype Score) is the score of each genotype, ranging from 0 to 2, according to the number of susceptible alleles (score 2: two susceptible alleles associated with risk of dependence, score 0: no susceptible allele associated with risk for dependence, and score 1: heterozygous genotype, only one susceptible allele associated with risk for dependence, Table 1).
Table 1.
Total genotype score–grading (TGS‐Grading) for alcohol dependence genes
| SNP | Homozygous wild type | Heterozygous | Homozygous mutated | Risk of alcohol dependence | Reference |
|---|---|---|---|---|---|
| rs1229984 | +2 | +1 | 0 | Decreased | (Chatterjee et al. 2011; Feduccia et al. 2012) |
| rs1693482 | 0 | +1 | +2 | Increased | (Chatterjee et al. 2011; Feduccia et al. 2012) |
| rs1800759 | 0 | +1 | +2 | Increased | (Hendrickson et al. 2009, 2011; Herz 1997) |
| rs671 | +2 | +1 | 0 | Decreased | (Bart et al. 2005; Berrettini and Lerman 2005; Chatterjee et al. 2011; Ehlers et al. 2008) |
| rs1799971 | 0 | +1 | +2 | Increased | (Munafo et al. 2007; Rahman 2013; Rahman and Prendergast 2012; Tizabi et al. 2002; Wu et al. 2014) |
| rs1800497 | 0 | +1 | +2 | Increased | (Hurley et al. 2002; Zakhari 2006) |
| rs6265 | +2 | +1 | 0 | Decreased | (Macgregor et al. 2009; Lee et al. 2004) |
Results
Data collected for polymorphism rs1229984 showed that 64.14% of the 1276 volunteers were homozygous for the wild type (G:G). Polymorphism rs1229984 was in HWE in the volunteer population of this study (x 2 = 0.63) (Table 2).
Table 2.
Comparative frequency and distribution rates of alleles and genotypes of the rs1229984 (ADH1B), rs1693482 (ADH1C), rs1800759 (ADH4), rs671 (ALDH2), rs1799971 (OPRM1), rs1800497 (DRD2), and rs6265 (BDNF) polymorphisms in a global, a European, and a Greek population (http://browser.1000genomes.org)
| Population | % Allele | % Genotype | ||||
|---|---|---|---|---|---|---|
| rs1229984 | G | A | G:G | G:A | A:A | |
| Global | 84.15 | 15.85 | 78.24 | 11.82 | 9.94 | |
| European | 97.12 | 2.88 | 94.43 | 5.37 | 0.20 | |
| Greek | 81.07 | 18.93 | 66.14 | 29.86 | 4.00 | |
| rs1693482 | C | T | C:C | C:T | T:T | |
| Global | 78.57 | 21.43 | 63.78 | 29.59 | 6.63 | |
| European | 59.54 | 40.46 | 37.57 | 43.94 | 18.49 | |
| Greek | 75.82 | 24.18 | 57.44 | 36.76 | 5.80 | |
| rs1800759 | C | A | C:C | C:A | A:A | |
| Global | 56.21 | 43.79 | 38.90 | 34.62 | 26.48 | |
| European | 60.44 | 39.56 | 35.79 | 49.30 | 14.91 | |
| Greek | 41.69 | 58.31 | 17.63 | 48.12 | 34.25 | |
| rs671 | G | A | G:G | G:A | A:A | |
| Global | 96.43 | 3.57 | 93.65 | 5.55 | 0.80 | |
| European | 100 | 0 | 100 | 0 | 0 | |
| Greek | 99.92 | 0.08 | 99.84 | 0.16 | 0 | |
| rs1799971 | G | A | G:G | G:A | A:A | |
| Global | 22.34 | 77.66 | 7.15 | 30.39 | 62.46 | |
| European | 16.20 | 83.80 | 2.58 | 27.24 | 70.18 | |
| Greek | 14.73 | 85.27 | 1.88 | 25.69 | 72.43 | |
| rs1800497 | C | T | C:C | C:T | T:T | |
| Global | 67.43 | 32.57 | 46.57 | 41.73 | 11.70 | |
| European | 81.21 | 18.79 | 67.00 | 28.43 | 4.57 | |
| Greek | 84.43 | 15.57 | 70.84 | 27.18 | 1.98 | |
| rs6265 | G | A | G:G | G:A | A:A | |
| Global | 79.87 | 20.13 | 66.65 | 26.44 | 6.91 | |
| European | 80.32 | 19.68 | 64.21 | 32.21 | 3.58 | |
| Greek | 81.50 | 18.50 | 65.99 | 31.03 | 2.98 | |
For polymorphism rs1693482, 94.21% of the volunteers were either homozygous for the wild‐type genotype (C:C) or heterozygous (C:T). HWE was calculated for this polymorphism in this study (x 2 = 0.63) (Table 2).
Data collected for polymorphism rs1800759 showed that 17.63% were homozygous for the mutant allele (A:A). HWE was detected for polymorphism rs1800759 as well (x 2 = 0.1352) (Table 2).
According to the data collected for the polymorphism rs671, the vast majority (99.84%) of the volunteers were homozygous for the wild‐type genotype (G:G). HWE could not be accurately determined for polymorphism rs671 due to small population sample used (Table 2).
For the polymorphism rs1799971 data were obtained from 1,273 volunteers. Of these, 27.58% were homozygous for the mutant genotype (G:G) or heterozygous (G:A). HWE was calculated for polymorphism rs1799971 (x 2 = 0.6521) (Table 2).
Data were obtained only from 1262 volunteers for polymorphism rs1800497. Of these, 29.16% were heterozygous (C:T) or homozygous for the mutant allele (T:T). Polymorphism rs1800497 was in HWE in this study (x 2 = 1.4371) (Table 2).
Finally, for polymorphism rs6265 data were obtained from 1273 volunteers. Of those, 65.69% were homozygous for the wild‐type genotype (G:G). HWE was detected for polymorphism rs6265 in the volunteer Greek population of our study (x 2 = 1.0715) (Table 2).
No association between the polymorphisms rs1229984, rs1693482, rs1800759, rs671, rs1799971, rs1800497, and rs6265 and gender could be established.
TGS of addiction was calculated according to Table 1. The population of this study is normally distributed (Fig. 1). The population sample that had a TGS equal to or bigger than 50 was 92.09%, with only a few being in the very high‐risk group of TGS of 71‐100. Most volunteers participating in this study achieved TGSs between 20 and 70. Examining separately the SNPs for alcohol metabolism and the CNS, a right shift of the alcohol metabolism normal distribution curve and a left shift of the CNS normal distribution curve are observed. (Fig. 1G, H and I).
Figure 1.
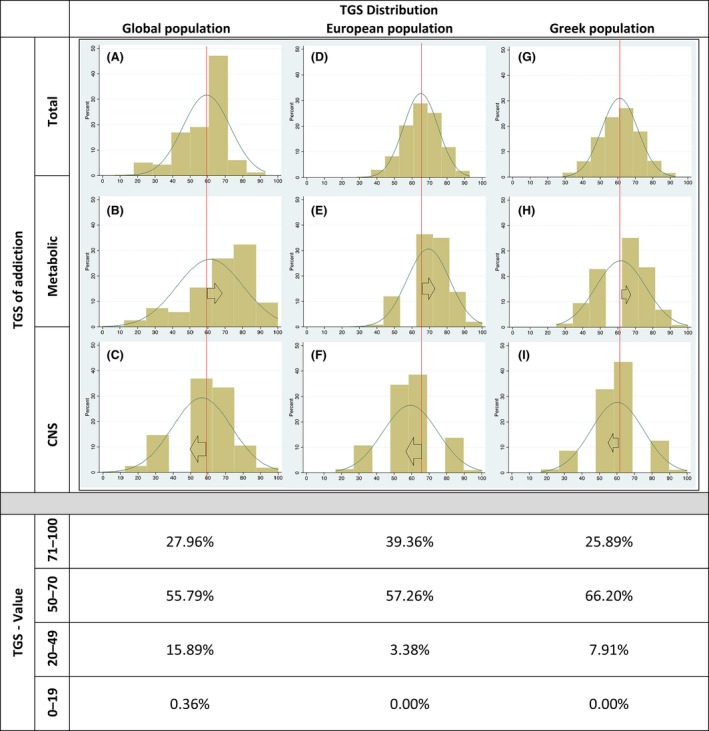
Comparative distribution of the Total Genotype Score of addiction in the global, the European, and the Greek populations of this study (A, D, and G). Distribution of the TGS of addiction for the SNPs associated with alcohol metabolism (B/E/H). A right shift of the distribution can be seen. Distribution of the TGS of addiction for the SNPs associated with Central Nervous System (C/F/I). A left shift of the distribution can be seen. TGS values for global, European, and Greek populations of this study.
Discussion
Alcohol abuse has a great social, as well as economic impact, as it may be responsible for inaptitude behaviors, such as reduced reflexes, which often lead to misjudgment of certain situations (https://www.cdc.gov/alcohol/fact-sheets/alcohol-use.htm). Alcohol effects depend on many factors, one of the more important being the amount of alcohol consumed and how fast it is metabolized in the human body. Even though there are studies suggesting a beneficial effect of mild alcohol consumption (Hvidtfeldt et al. 2010; Brien et al. 2011; Ronksley et al. 2011), if this happens on a more regular basis, it will most likely lead to addiction (https://www.drugabuse.gov/drugs-abuse/alcohol). The possibility of developing alcohol addiction could be predicted by genetic predisposition analysis.
According to the literature, some of the studied genes are associated with increased risk of dependence, while others seem to be protective against alcohol dependence (Luu et al. 1995; Shen et al. 1997; Wall et al. 1997; Lee et al. 2004; Luo et al. 2006, 2007; Gratacos et al. 2007; Kimura et al. 2009; Macgregor et al. 2009; Wojnar et al. 2009). In this study, the correlation between seven SNPs in genes, associated with the development of dependence on alcohol and their incidence was investigated. Then it was compared with published data for global and North (http://browser.1000genomes.org/).
The rs1229984 (ADH1B) polymorphism leads to a decreased risk for alcohol addiction (Lee et al. 2004). The mutant allele (A) occurs in 15.85% of the global population and 2.88% of the European population. This study revealed an A allele of the population of this study, similar to that in the global population and much higher than that in the European population (Table 3).
Table 3.
Correlation of alcohol dependence, allele, and genotype distributions between Greek and global populations and between Greek and European populations for the polymorphisms rs1229984, rs1693482, rs1800759, rs671, rs1799971, rs1800497, and rs6265
| Gene | Polymorphism | Odds ratio | 95% confidence interval | P | |
|---|---|---|---|---|---|
| Allele distribution | |||||
| Greek and global populations | ADH1B | rs1229984 | 0.80 | 0.71–0.91 | 0.0007 |
| ADH1C | rs1693482 | 1.16 | 1.04–1.39 | 0.006 | |
| ADH4 | rs1800759 | 0.91 | 0.83–1.01 | n.s | |
| ADLH2 | rs671 | 47.26 | 11.70–190 | <0.0001 | |
| OPRM1 | rs1799971 | 0.60 | 0.52–0.68 | <0.0001 | |
| DRD2 | rs1800497 | 0.38 | 0.33–0.43 | <0.0001 | |
| BDNF | rs6265 | 1.11 | 0.98–1.25 | n.s | |
| Greek and European populations | ADH1B | rs1229984 | 0.12 | 0.08–0.18 | <0.0001 |
| ADH1C | rs1693482 | 0.46 | 0.40–0.54 | <0.0001 | |
| ADH4 | rs1800759 | 1.09 | 0.93–1.27 | n.s. | |
| ADLH2 | rs671 | n.a. | n.a. | n.a. | |
| OPRM1 | rs1799971 | 0.89 | 0.72–1.09 | n.s. | |
| DRD2 | rs1800497 | 0.79 | 0.65–0.96 | 0.02 | |
| BDNF | r s6265 | 1.07 | 0.89–1.30 | n.s. | |
| Genotype distribution | |||||
| Greek and global populations | ADH1B | rs1229984 | 0.54 | 0.46–0.63 | <0.0001 |
| ADH1C | rs1693482 | 1.30 | 1.13–1.49 | 0.0001 | |
| ADH4 | rs1800759 | 0.59 | 0.49–0.70 | <0.0001 | |
| ADLH2 | rs671 | 44.14 | 11.90–368.02 | <0.0001 | |
| OPRM1 | rs1799971 | 0.62 | 0.53–0.72 | <0.0001 | |
| DRD2 | rs1800497 | 0.35 | 0.30–0.41 | <0.0001 | |
| BDNF | rs6265 | 0.95 | 0.83–1.10 | n.s. | |
| Greek and European populations | ADH1B | rs1229984 | 0.11 | 0.07–0.17 | <0.0001 |
| ADH1C | rs1693482 | 0.44 | 0.35–0.55 | <0.0001 | |
| ADH4 | rs1800759 | 1.21 | 0.90–1.63 | n.s. | |
| ADLH2 | rs671 | n.a. | n.a. | n.a. | |
| OPRM1 | rs1799971 | 0.87 | 0.69–1.11 | n.s. | |
| DRD2 | rs1800497 | 0.83 | 0.66–1.04 | n.s. | |
| BDNF | rs6265 | 7 | 5.89–8.31 | <0.0001 | |
The genetic predisposition to developing alcohol addiction is reported to be lower for individuals carrying at least one mutant allele (G:A or A:A genotype) (Lee et al. 2004). Those genotypes appear in 21.76% of the global population and in 5.57% of the European population. In the population of this study, the results revealed an increased frequency of combined genotypes G:A and A:A, as compared to both global and European populations. This observation may indicate a lower predisposition to developing alcohol addiction for Caucasians of southeast Europe (Table 3).
Another polymorphism of the ADH1 gene, rs1693482, increases alcohol addiction susceptibility (Lee et al. 2004; Macgregor et al. 2009). The mutant allele (T) occurs worldwide at a rate of 21.43% and at a rate of 40.46% in Europe. The frequency of mutant T allele in the study population was similar to that in the global population, but much lower than that in the European population (Table 3).
Thus, genetic predisposition to developing alcohol addiction is lower for those carrying the homozygous wild‐type C:C genotype (Lee et al. 2004; Macgregor et al. 2009). This genotype is found in 63.78% of the global population and in 37.57% of the European population. In the population, of this study the results revealed a similar frequency compared to the global population, indicating a better alcohol addiction protection than that of other Caucasians living in the northern regions of Europe (Table 3).
The rs1800759 (ADH4) polymorphism enhances the tendency to develop alcohol addiction (Luo et al. 2006, 2007; Kimura et al. 2009). The A allele appears in 43.79% of the global population, while in the European population, it is found at a rate of 39.56%. The frequency rate in this study population is higher than both populations (Table 3).
The percentage of homozygous rs1800759 A:A carriers in the global population is 26.48 and in the European population 14.91, both lower than that of this study population. These carriers may be predisposed to consuming more alcohol and thus may have a higher risk of developing alcohol addiction (Table 3) (Luo et al. 2006, 2007; Kimura et al. 2009).
The rs671 (ALDH2) polymorphism leads to a decreased possibility to develop alcohol addiction (Macgregor et al. 2009). The mutant allele (A) occurs in 3.57% in the global population and seems to be completely absent in the European population. This study revealed the existence of A allele in the study population, in a rate close to that in the European population but much lower than that in the global population. The genetic predisposition to developing alcohol addiction may be lower for individuals carrying at least one mutant allele (G:A or A:A genotype) (Macgregor et al. 2009). Those genotypes appear in 6.35% of the global population, while they do not appear in the European population. In the population of this study, only one heterozygous carrier was detected (Table 3).
The rs1799971 (OPRM1) polymorphism leads to an increased probability of developing alcohol addiction (Berrettini and Lerman 2005; van den Wildenberg et al. 2007; Ehlers et al. 2008). The mutant allele (A) occurs in 77.66% of the global population and in 83.80% of the European population. This study revealed an A allele frequency which is slightly higher when compared to both global and European populations (Table 3).
Individuals carrying at least one OPRM1 gene mutant allele (A) may enhance genetic predisposition to develop alcohol addiction (Berrettini and Lerman 2005; van den Wildenberg et al. 2007; Ehlers et al. 2008). Such genotypes appear in 92.85% of the global population and in 97.42% of the European population. In the population of this study, the results revealed increased prevalence of the heterozygous (G:A) and homozygous mutant (A:A) genotypes (Table 3).
The rs1800497 polymorphism is associated with increased susceptibility to develop alcohol addiction (Munafo et al. 2007). The mutant allele (T) occurs in 32.57% of the global population and in 18.79% of the European population. The frequency of the T allele in this study population was similar to that of the European population (Table 2).
Developing alcohol addiction seems to be lower for individuals carrying the wild‐type C:C genotype. The existence of even one mutant allele seems to play a decisive role in the CNS reward pathway because dopaminergic neurotransmission is known to be essential for the alcohol mechanism of action (Munafo et al. 2007). The C:C genotype is found in 46.57% of the global population and in 67.00% of the European population. In this study, the results revealed an increased frequency of C:C genotype compared to both global and European populations. According to the participants of this study, it is obvious that the T allele is present twice as much in the global population, while homozygous mutated individuals (T:T) appear 2.5 times more frequent in the European than in the Greek population and more than 5 time more frequent in the global population (Table 3).
The rs6265 (BDNF) polymorphism has been associated with decreased risk of alcohol dependence (Gratacos et al. 2007; Wojnar et al. 2009). The mutant allele (A) occurs in 20.13% of the global population and in 19.68% of the European population. This study revealed an A allele frequency similar to both populations (Table 3).
The susceptibility to developing alcohol addiction is higher for individuals carrying the homozygous wild‐type G:G genotype of the rs6265 polymorphism (Gratacos et al. 2007; Wojnar et al. 2009). A percentage of 66.65 of the global population and 64.21 of the European population are homozygous for wild‐type G:G genotype. In the population of this study, the results revealed an increased prevalence of the homozygous wild‐type genotype G:G (Table 3).
The prevalence of polymorphisms favoring alcohol dependence (rs1693482, rs1800759, rs1799971, and rs1800497), which may eventually lead to alcohol addiction in the South Eastern European Greek population, seems to be lower than the respective one in the Northern European populations (Lee et al. 2004; Berrettini and Lerman 2005; Luo et al. 2006, 2007; Munafo et al. 2007; van den Wildenberg et al. 2007; Ehlers et al. 2008; Kimura et al. 2009; Macgregor et al. 2009). According to these results, the Greek population may be more protected against addiction development.
According to the HWE law, genotype frequencies are functions of allele frequencies and a large random mating population is at equilibrium given that there is no migration, natural selection, or genetic drift (Namipashaki et al. 2015). In this study, HWE was detected for all polymorphisms except rs671. This could be interpreted as a stability regarding the mechanisms of evolution mentioned above, which means that evolution did not occur and, theoretically, the gene pool frequencies remained unaltered. However, it cannot safely be assumed that the next generations will also follow the HWE because evolution is an inevitable result. Moreover, it is important to note that for the rs671 polymorphism the chi square calculated was zero. This could possibly mean that the population sample used was too small to detect a Hardy–Weinberg disequilibrium for very rare polymorphisms.
In order to determine the existence of a significant difference between frequency distribution rates of the alleles and genotypes of Greek, global and European populations chi‐square test was conducted between those three populations, separately for the alleles and the genotypes that favor the development of alcohol addiction (Table 3).
The allele frequencies of this study population were significantly different (P < 0.05) when compared to the global population for polymorphisms rs1229984 (ADH1B), rs1799971 (OPRM1), rs1800497 (DRD2), rs1693482 (ADH1C), and rs671 (ALDH2). This study population seems to be better protected against alcohol addiction for polymorphisms rs1229984, rs1799971, and rs1800497, but seems to be more exposed to alcohol addiction compared to the global population due to their rs1693482 and rs671 genotype profile. (Table 3).
Compared to the European population, the Greek population of this study is significantly different in polymorphisms rs1229984, rs1693482, and rs1800497. Thus, individuals of this study population seem to be more protected than their northern European counterparts (Table 3).
Frequency distribution of rs1229984, rs1693482, rs1800759, rs671, rs1799971, rs1800497, and rs6265 genotypes in this study population was compared to the respective frequency distribution of the northern European and the global population. The population of this study seems to be significantly more protected against alcohol addiction for polymorphisms rs1229984, rs1800759, rs1799971, rs1800497, but significantly more exposed to alcohol addiction due to their genotypic profile of polymorphisms rs1693482 and rs671, when compared to the global population (Table 3).
Compared to the European population, the Greek population appeared to be more protected for polymorphisms rs1229984 and rs1693482, but seems to be more exposed to alcohol addiction due to their rs6265 genotype profile (Table 3).
Individuals generating TGS scores above 50 seem to have higher predisposition to alcohol addiction. Almost all volunteers of this study had TGS higher than 50, but only a small portion of the investigated population was in the very high‐risk group of TGS score between 71 and 100. The majority was concentrated in the medium‐risk region of 50–70 (Fig. 1).
Total genotype score for global and Northern European populations has been calculated. The results revealed that in both populations more than 80% have a TGS score higher than 50 (Fig. 1). Of the European population, 39.36% is in the very high‐risk group of TGS score between 71 and 100 and 57.26% is in the medium‐risk region of 50–70 (Fig. 1). In the global population, 27.96% are in the very high‐risk group of TGS score between 71 and 100 and 55.79% in the medium‐risk region of 50–70 (Fig. 1). These findings indicate that volunteers of this study population may be more protected against developing alcohol dependence, compared to the north European population, which are consistent with a recent study (Labhart et al. 2017).
Alcohol‐associated metabolic‐ and CNS‐polymorphism analysis in the global (Fig. 1A, B and C) and the European populations (Fig. 1D, E and F) revealed a right shift of the alcohol metabolism normal distribution curve and a left shift of the CNS normal distribution curve, similar – but less distinguishable – to the Greek sample population of this study (Fig. 1G, H and I). This finding indicates a greater impact of alcohol metabolism polymorphisms than CNS polymorphisms on TGS.
Conclusions
The investigated Southeastern population of this study may differ when compared to global and other European populations. In some of the studied SNPs there is a greater similarity to the global population [rs1229984 (ADH1B), rs1693482 (ADH1C), and rs1800759 (ADH4)], whereas in other polymorphisms, there is more similarity to the European population [rs671 (ALDH2), rs1799971 (OPRM1), rs1800497 (DRD2), and rs6265 (BDNF)]. The geographical position of Greece, on the crossroad between three continents (Europe, Asia, and Africa), could provide a plausible explanation for genetic influences from Asian and African populations on the Greek population.
The extremely low incidence (0.08%) of rs671 polymorphism in ALDH2 gene in the population of this study seems to be rather important. Polymorphism rs671 has been associated with reduced desire for alcohol consumption and protective role against alcohol dependence (Takeuchi et al. 2011). Another noteworthy finding is the incidence of rs1800759 polymorphism of ADH4 gene, related to increased alcohol consumption, is rather high in comparison with other SNPs.
The investigation of polymorphisms associated with dependence could be useful in the prevention of alcohol dependence development. TGS calculation may predict individual response to alcohol, as it enables to identify potential alteration in the alcohol mechanism of action and biotransformation, providing additional tools in dependence prevention.
Analysis of alcohol‐associated genetic polymorphisms may also be valuable for the treatment of alcohol withdrawal symptoms, enabling the prediction of the most effective therapeutic method or agent for each individual. In addition, SNPs may be useful tools to predict the potential posttreatment relapse risk.
Limitations of the study
Further larger‐scale investigations may be needed in order to precisely determine alcohol‐associated polymorphisms incidence rates and to correlate them with alcohol dependence. Furthermore, in order to adjust the TGS algorithm, larger population studies for each alcohol addiction relevant gene may be necessary to better scale alcohol dependence susceptibility by TGS. This would enable accurate prediction of alcohol addiction developing prevalence to serve as individualized prevention tool.
This study was conducted in a general population. Further studies should be initialized to investigate the frequency distribution of these polymorphisms in alcohol addicted individuals. TGS was calculated assuming equal impact of each polymorphism to the score. In following studies, each polymorphism score should be weighted according to its contribution to alcohol dependence development. This would enhance the results and provide a more accurate tool for the physicians.
Disclosure
There are no competing interests.
Katsarou M.‐S., Karakonstantis K., Demertzis N., Vourakis E., Skarpathioti A., Nosyrev A. E., Tsatsakis A., Kalogridis T., Drakoulis N.. Effect of single‐nucleotide polymorphisms in ADH1B, ADH4, ADH1C, OPRM1, DRD2, BDNF, and ALDH2 genes on alcohol dependence in a Caucasian population. Pharma Res Per, 5(4), 2017, e00326, https://doi.org/10.1002/prp2.326
References
- Bart G, Kreek MJ, Ott J, LaForge KS, Proudnikov D, Pollak L, et al. (2005). Increased attributable risk related to a functional mu‐opioid receptor gene polymorphism in association with alcohol dependence in central Sweden. Neuropsychopharmacology 30: 417–422. [DOI] [PubMed] [Google Scholar]
- Berrettini W (2016). Opioid neuroscience for addiction medicine: from animal models to FDA approval for alcohol addiction. Prog Brain Res 223: 253–267. [DOI] [PubMed] [Google Scholar]
- Berrettini WH, Lerman CE (2005). Pharmacotherapy and pharmacogenetics of nicotine dependence. Am J Psychiatry 162: 1441–1451. [DOI] [PubMed] [Google Scholar]
- Brien SE, Ronksley PE, Turner BJ, Mukamal KJ, Ghali WA (2011). Effect of alcohol consumption on biological markers associated with risk of coronary heart disease: systematic review and meta‐analysis of interventional studies. BMJ 342: d636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Steensland P, Simms JA, Holgate J, Coe JW, Hurst RS, et al. (2011). Partial agonists of the alpha3beta4* neuronal nicotinic acetylcholine receptor reduce ethanol consumption and seeking in rats. Neuropsychopharmacology 36: 603–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contet C, Kieffer BL, Befort K (2004). Mu opioid receptor: a gateway to drug addiction. Curr Opin Neurobiol 14: 370–378. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Lind PA, Wilhelmsen KC (2008). Association between single nucleotide polymorphisms in the mu opioid receptor gene (OPRM1) and self‐reported responses to alcohol in American Indians. BMC Med Genet 9: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feduccia AA, Chatterjee S, Bartlett SE (2012). Neuronal nicotinic acetylcholine receptors: neuroplastic changes underlying alcohol and nicotine addictions. Front Mol Neurosci 5: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratacos M, Gonzalez JR, Mercader JM, de Cid R, Urretavizcaya M, Estivill X (2007). Brain‐derived neurotrophic factor Val66Met and psychiatric disorders: meta‐analysis of case‐control studies confirm association to substance‐related disorders, eating disorders, and schizophrenia. Biol Psychiatry 61: 911–922. [DOI] [PubMed] [Google Scholar]
- Hendrickson LM, Zhao‐Shea R, Tapper AR (2009). Modulation of ethanol drinking‐in‐the‐dark by mecamylamine and nicotinic acetylcholine receptor agonists in C57BL/6J mice. Psychopharmacology 204: 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson LM, Guildford MJ, Tapper AR (2013). Neuronal nicotinic acetylcholine receptors: common molecular substrates of nicotine and alcohol dependence. Front Psychiatry 4: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz A (1997). Endogenous opioid systems and alcohol addiction. Psychopharmacology 129: 99–111. [DOI] [PubMed] [Google Scholar]
- Huang W, Payne TJ, Ma JZ, Beuten J, Dupont RT, Inohara N, et al. (2009). Significant association of ANKK1 and detection of a functional polymorphism with nicotine dependence in an African‐American sample. Neuropsychopharmacology 34: 319–330. [DOI] [PubMed] [Google Scholar]
- Hurley TD, Edenberg HJ, Li T‐K (2002). Pharmacogenomics of alcoholism, in pharmacogenomics: the search for individualized therapies. Wiley‐VCH Verlag GmbH & Co KGaA, Weinheim, FRG. [Google Scholar]
- Hvidtfeldt UA, Tolstrup JS, Jakobsen MU, Heitmann BL, Gronbaek M, O'Reilly E, et al. (2010). Alcohol intake and risk of coronary heart disease in younger, middle‐aged, and older adults. Circulation 121: 1589–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Nishimura FT, Abe S, Fukunaga T, Tanii H, Saijoh K (2009). Polymorphisms in the promoter region of the human class II alcohol dehydrogenase (ADH4) gene affect both transcriptional activity and ethanol metabolism in Japanese subjects. J Toxicol Sci 34: 89–97. [DOI] [PubMed] [Google Scholar]
- Kraschewski A, Reese J, Anghelescu I, Winterer G, Schmidt LG, Gallinat J, et al. (2009). Association of the dopamine D2 receptor gene with alcohol dependence: haplotypes and subgroups of alcoholics as key factors for understanding receptor function. Pharmacogenet Genomics 19: 513–527. [DOI] [PubMed] [Google Scholar]
- Labhart F, Ferris J, Winstock A, Kuntsche E (2017). The country‐level effects of drinking, heavy drinking and drink prices on pre‐drinking: an international comparison of 25 countries. Drug Alcohol Rev. https://doi.org/10.1111/dar.12525. [DOI] [PubMed] [Google Scholar]
- Lee SL, Hoog JO, Yin SJ (2004). Functionality of allelic variations in human alcohol dehydrogenase gene family: assessment of a functional window for protection against alcoholism. Pharmacogenetics 14: 725–732. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Lappalainen J, Yang BZ, Gelernter J (2006). ADH4 gene variation is associated with alcohol dependence and drug dependence in European Americans: results from HWD tests and case‐control association studies. Neuropsychopharmacology 31: 1085–1095. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Wang S, Gelernter J (2007). Personality traits of agreeableness and extraversion are associated with ADH4 variation. Biol Psychiatry 61: 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu SU, Wang MF, Lin DL, Kao MH, Chen ML, Chiang CH, et al. (1995). Ethanol and acetaldehyde metabolism in chinese with different aldehyde dehydrogenase‐2 genotypes. Proc Natl Sci Counc Repub China B 19: 129–136. [PubMed] [Google Scholar]
- Macgregor S, Lind PA, Bucholz KK, Hansell NK, Madden PA, Richter MM, et al. (2009). Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption and dependence: an integrated analysis. Hum Mol Genet 18: 580–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafo MR, Matheson IJ, Flint J (2007). Association of the DRD2 gene Taq1A polymorphism and alcoholism: a meta‐analysis of case‐control studies and evidence of publication bias. Mol Psychiatry 12: 454–461. [DOI] [PubMed] [Google Scholar]
- Namipashaki A, Razaghi‐Moghadam Z, Ansari‐Pour N (2015). The Essentiality of Reporting Hardy‐Weinberg Equilibrium Calculations in Population‐Based Genetic Association Studies. Cell J 17: 187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Prendergast MA (2012). Cholinergic receptor system as a target for treating alcohol abuse and dependence. Recent Pat CNS Drug Discov 7: 145–150. [DOI] [PubMed] [Google Scholar]
- Rahman S (2013). Nicotinic receptors as therapeutic targets for drug addictive disorders. CNS Neurol Disord Drug Targets 12: 633–640. [DOI] [PubMed] [Google Scholar]
- Ronksley PE, Brien SE, Turner BJ, Mukamal KJ, Ghali WA (2011). Association of alcohol consumption with selected cardiovascular disease outcomes: a systematic review and meta‐analysis. BMJ 342: d671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks JJ, Gonzales KR, Bouchery EE, Tomedi LE, Brewer RD (2015). 2010 National and State Costs of Excessive Alcohol Consumption. Am J Prev Med 49: e73–e79. [DOI] [PubMed] [Google Scholar]
- Shen YC, Fan JH, Edenberg HJ, Li TK, Cui YH, Wang YF, et al. (1997). Polymorphism of ADH and ALDH genes among four ethnic groups in China and effects upon the risk for alcoholism. Alcohol Clin Exp Res 21: 1272–1277. [PubMed] [Google Scholar]
- Takeuchi F, Isono M, Nabika T, Katsuya T, Sugiyama T, Yamaguchi S, et al. (2011). Confirmation of ALDH2 as a Major locus of drinking behavior and of its variants regulating multiple metabolic phenotypes in a Japanese population. Circ J 75: 911–918. [DOI] [PubMed] [Google Scholar]
- Tizabi Y, Copeland RL Jr, Louis VA, Taylor RE (2002). Effects of combined systemic alcohol and central nicotine administration into ventral tegmental area on dopamine release in the nucleus accumbens. Alcohol Clin Exp Res 26: 394–399. [PubMed] [Google Scholar]
- Wall TL, Peterson CM, Peterson KP, Johnson ML, Thomasson HR, Cole M, et al. (1997). Alcohol metabolism in Asian‐American men with genetic polymorphisms of aldehyde dehydrogenase. Ann Intern Med 127: 376–379. [DOI] [PubMed] [Google Scholar]
- van den Wildenberg E, Wiers RW, Dessers J, Janssen RG, Lambrichs EH, Smeets HJ, et al. (2007). A functional polymorphism of the mu‐opioid receptor gene (OPRM1) influences cue‐induced craving for alcohol in male heavy drinkers. Alcohol Clin Exp Res 31: 1–10. [DOI] [PubMed] [Google Scholar]
- Williams AG, Folland JP (2008). Similarity of polygenic profiles limits the potential for elite human physical performance. J Physiol 586: 113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojnar M, Brower KJ, Strobbe S, Ilgen M, Matsumoto H, Nowosad I, et al. (2009). Association between Val66Met brain‐derived neurotrophic factor (BDNF) gene polymorphism and post‐treatment relapse in alcohol dependence. Alcohol Clin Exp Res 33: 693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Gao M, Taylor DH (2014). Neuronal nicotinic acetylcholine receptors are important targets for alcohol reward and dependence. Acta Pharmacol Sin 35: 311–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakhari S (2006). Overview: how is alcohol metabolized by the body? Alcohol Res Health 29: 245–254. [PMC free article] [PubMed] [Google Scholar]
- Zaleski M, Morato GS, Silva VA, Lemos T (2004). Neuropharmacological aspects of chronic alcohol use and withdrawal syndrome. Rev Bras Psiquiatr 26(Suppl 1): S40–S42. [DOI] [PubMed] [Google Scholar]


