Abstract
A loop-mediated isothermal amplification (LAMP) technique was employed to develop a simple and rapid method for the detection of tomato leaf curl Bangalore virus (ToLCBaV) in diseased plants of tomato (Solanum lycopersicum). Six sets of primers were designed for LAMP technique targeting the conserved AC1 region and successfully detected ToLCBaV. No reaction was detected in the tissues of healthy plants by either the LAMP or the polymerase chain reaction (PCR). The LAMP products can be visualized by presence or absence of turbidity and staining (0.2 μL for 25 μL LAMP product) directly in the tube with nucleic acid stain dye which allowed easy detection. Sensitivity of LAMP assay is 100 times of conventional PCR technique. Although, both the LAMP and the PCR methods were capable of detecting ToLCBaV in infected tissues of tomato, the LAMP method would be more useful than the PCR method for detection of ToLCBaV infection in tomato plants because it is more rapid, simple and accurate method.
Keywords: LAMP, ToLCBaV, Turbidity, Nucleic acid stain, Sensitivity
Introduction
Tomato is one of the most important widely grown vegetable crops in India and severely infected with leaf curl disease. The members of the genus Begomovirus (family Geminiviridae) cause leaf curling disease in tomato worldwide. However, the virus constantly evolving and threaten horticulture in many of the world’s tropical and subtropical regions. Tomato leaf curl New Delhi virus (ToLCNDV) and tomato leaf curl Bangalore virus (ToLCBaV) are the most damaging in India and they are transmitted by Bemisia tabaci [3]. Epidemics of ToLCV is associated with an upsurge of whiteflies has been frequently reported with up to 100% yield losses [8].
In 1948, Vasudeva and Sam Raj first reported tomato leaf curl disease in northern parts of India [27]. Currently, the disease is a major problem and widespread throughout the country, causing yield loss up to 27–100%, during maturity stage [10]. The symptoms of the disease are leaf curling, vein clearing, reduction in leaf lamina, vein enation and, stunted growth of plants. At least 14 distinct indigenous tomato-infecting begomoviruses that cause major constraints on tomato production in tropical and subtropical regions of the world [6], of which ToLCBaV is widely prevalent in the southern and south-western parts of the country [7, 10].
Several methods have been reported for the detection of the genomic DNA of ToLCV [14, 15, 22, 24]. PCR is the most widely used method for detection of ToLCV DNA from infected plants tissues [14, 15]. However, the PCR method has disadvantages, such as the requirement of thermal cycling and liable to contamination.
Loop mediated isothermal amplification (LAMP) is a novel gene amplification method [11, 12, 18]. The method is functioned by employing a DNA polymerase with strand-displacement activity, along with two inner primers (FIP, BIP) and two outer primers (F3, B3) to form auto-cycling amplification by loop primers. The new assay is quite simple, requiring only a conventional water bath or heat block for incubation under isothermal conditions. In addition, the reaction process is rapid, usually taking 60 min or even less, and low-cost. Generally, four to six primers are designed that recognize several regions of the target sequence, which gives high specificity in LAMP assay. Recently, LAMP assay been applied to the rapid detection of tomato torrado virus in tomato [2]. Several workers found that LAMP shows more sensitive than quantitative real-time PCR assays [19, 21, 25].
However, crop losses can be minimized, and specific treatments can be tailored to combat specific pathogens if plant diseases are correctly diagnosed and identified early. Without proper identification of the disease and the disease-causing agent, disease control measures can be a waste of time and money and can lead to further plant losses. LAMP is a powerful innovative nucleic acid amplification technique emerging as a simple rapid diagnostic tool for advanced detection and identification of virus [20]. While this helps to improve detection research and reduces time effectively for detecting viral pathogens of tomato during field analysis. The objective of this study was to develop a LAMP assay for rapid and efficient detection of ToLCBaV infection in tomato.
Materials and methods
ToLCBaV plants or virus samples
ToLCBaV infected leaf was collected from the vegetable orchard of ICAR—Indian Institute of Horticultural Research (IIHR), Bengaluru. Leaf samples showing foliar curling, shortening of internodes and stunted growth were collected in summer of 2016 and stored under −20 °C. These samples were screened for the presence of ToLCBaV using molecular techniques.
Total DNA isolation
Approximately 100 mg of diseased leaf tissue was placed into a thick-gauged plastic bag. The tissue was ground using pestle and mortar with 10 volumes (1 mL) of CTAB extraction buffer (2% CTAB, 1.4 M NaCl, 0.05 M EDTA, 0.2 M Tris–HCl with pH 6.5). About 750 μL of the sample was poured into a 1.5 mL eppendorf tube and the samples were heated at 60 °C for 30 min. The samples were mixed with an equal volume (750 μL) of chloroform: isoamyl alcohol (24:1) and centrifuged at 13,000 rpm for 10 min. The top aqueous phase was transferred to a new 1.5 mL eppendorf tube and DNA was precipitated by adding 0.6 volume (300 μL) of cold (−20 °C) isopropanol and incubated at –20 °C for 1 h. The samples were centrifuged at 13,000 rpm at 4 °C for 10 min and the supernatant was discarded. The pellet was washed in 0.5 mL of 70% ethanol by vortexing and then centrifuged for 5 min at 13,000 rpm. The ethanol was removed and the pellet was vacuum dried for 5 min and the dried pellet was suspended in 100 μL 1 × TE buffer and stored at –20 °C. All the DNA extracts were further diluted to ten-fold in single distilled water (SDW) before using for PCR amplification.
PCR amplification and gel electrophoresis
PCR reactions were performed in 25 μL of reaction mix containing 2 μL of template DNA (100 ng), 2 mM dNTPs, 25 mM MgCl2, 3 units of Taq polymerase, 0.4μM of primer and 10X reaction buffer (20 mM (NH4) SO4, 75 mM Tris–HCl (pH 9.0). ToLCBaV primers were used are (5′-TCCCCTTCTTGGCTAACC-3′ Forward and 5′-AGCCAGTTCAAATTAAAGGAG-3′ Reverse). Single distilled water (SDW) control was included in all reactions as the control. The reaction condition consists of for one cycle of 2 min at 94 °C followed by 35 cycles of 1 min at 94 °C, 1 min at 55 °C and 1 min at 72 °C, followed by a final cycle of 10 min at 72 °C in PCR thermal cycler (Eppendorf). A 20 μL of PCR product was mixed with 4 μL of Orange G loading dye [15% (w/v) Ficoll type 400, 0.25% (w/v) Orange-G, 40 mM ethylenediaminete traacetic acid (EDTA), pH 8.0 in 100 mL of SDW] and electrophoresed at 5 V cm−1 through a 0.8% (w/v) agarose gel with ethidium bromide solution (0.5 μg mL−1) in 0.5x TBE buffer (4.5 mM Tris–borate, 0.1 mM EDTA). The DNA size marker 1 kb Ladder (Fermentas) was included to allow the size of the DNA bands to be estimated. Amplified products then visualized and documented by Alpha DigiDoc 1000 system (Alpha Innotech Corporation, USA).
LAMP based detection
Target gene and primer design
Target sequences that are traditionally used to identify ToLCBaV were selected. The LAMP primers were designed to target the AC1 gene from ToLCBaV. Complete set of LAMP primers including loop primers were designed using the software Primer Explorer V4 (http://primerexplorer.jp/elamp4.0.0/index.html). The LAMP primer sequences are listed in Table 1.
Table 1.
Details of LAMP primers designed for the amplification of ToLCBaV
| LAMP assay (gene target) | Primer name | Primer sequence (5′–3′) | Length | Temp and time |
|---|---|---|---|---|
| AC1 gene | F3 | TCCCCTTCTTGGCTAACC | 18 | 60 °C for 60 min |
| B3 | AGCCAGTTCAAATTAAAGGAG | 21 | ||
| FLOOP | AAGAACCATTCTACTCAGGT | 20 | ||
| BLOOP | TCTTTATAGCTGCTGTTAGG | 20 | ||
| FIP | TGCGAGATTCATCACCCTCGTGTGTTGCACTTTGATTGGA | 40 | ||
| BIP | GCTTTAGTGCGTTGTTCTTGTCCTCCCATTATCTTCCTCTGC | 42 |
LAMP reaction and detection
ToLCBaV-LAMP assay were performed in a final reaction volume of 25 μL with 1.4 mM dNTPs (Promega, WI, USA), 8 mM MgSO4, 0.8 M betaine (Sigma-Aldrich, MO, USA) and 8 U of Bsm DNA Polymerase (New England Biolabs, MA, USA). The LAMP reactions also contained 1.6 μM of FIP and BIP primers, 0.2 μM of F3 and B3 primers, 0.4 μM of F Loop and B Loop primers and the corresponding DNA as the template. The templates consisted of 1 μL of high-quality DNA obtained by the rapid DNA extraction procedure as described earlier. The reaction tubes were incubated in equipment [MyCyclerthermocycler (Bio-Rad, CA, USA)]. The LAMP amplicons were visualized by different strategies: (a) 2% agarose gel electrophoresis, staining with 0.5 μg/mL of ethidium bromide and visualized and documented by Alpha DigiDoc 1000 system (Alpha Innotech Corporation, USA); (b) naked-eye inspection by turbidity and nucleic acid stains such as Ethidium bromide, Calcein staining and hydroxynaphthol blue (HNB). For staining, 1 μL of 1/10 nucleic acid stain solution was added directly to each reaction tube after incubation, and the DNA was visualized under naked eye.
Results
Detection of ToLCBaV from infected tomato samples by polymerase chain reaction (PCR)
Tomato samples showing viral infections were collected from ICAR-IIHR, Bengaluru, India and screened for the presence of ToLCBaV DNA through PCR technique. The most consistent amplification of 0.7 kb DNA fragment was obtained with the ToLCBaV specific primer with samples from ToLCBaV infected tomato plants. The result revealed that there is the presence of ToLCBaV DNA with this disease and the virus could be detected by using specific primers (Fig. 1a).
Fig. 1.
a PCR detection of ToLCBaV specific 700 bp in virus infected tomato samples, electrophoresis on a 0.8% agarose gel and staining by ethidium bromide. Lane M Marker (1 kb DNA ladder, Fermentas), Lane 1 & 2 positive sample for ToLCBaV, 3–6 virus infected tomato samples, 7 Healthy tomato leaf sample. b LAMP-PCR detection of ToLCBaV in virus infected tomato samples, electrophoresis on a 2.0% agarose gel and staining by ethidium bromide. Lane M Marker (1 kb DNA ladder, Fermentas), Lane 1 positive sample for ToLCBaV, 2–4 virus infected tomato samples, 5 Healthy tomato leaf sample, 6 Control (Water). c Sensitivity of LAMP assay for detecting ToLCBaV in tomato sample. Dilutions of target viral nucleic acids, standards ranging from 100 to 10−4. Lane M 100 bp DNA ladder, (Lane 1–5) amplification products with ten-fold serially diluted total DNA of tomato from 100 to 10−4
Visual examination of LAMP products
Turbidity
Tomato samples showing viral infection were collected from ICAR-IIHR, Bengaluru and screened for the presence of ToLCBaV DNA using LAMP technique. LAMP reaction was carried out using Bsm DNA polymerase and incubated for 60 min under isothermal condition @ 60 °C. Positive samples are visualized by the presence of turbidity caused by increasing the quantity of magnesium pyrophosphate in reaction tubes.
Nucleic acid stains
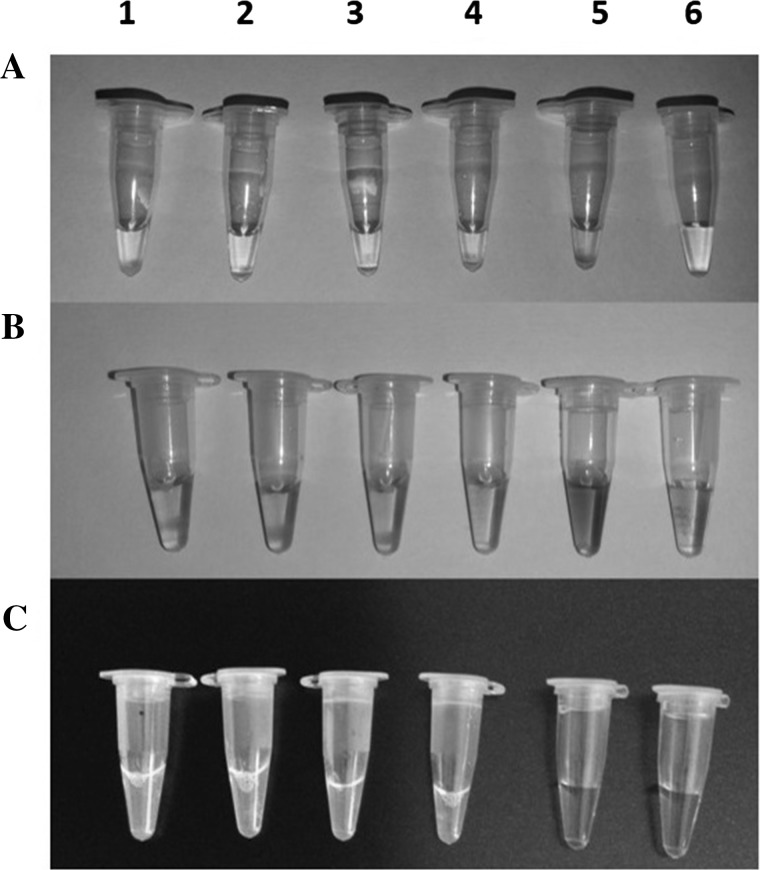
Viral nucleic acids were subjected to isothermal amplification to evaluate the assay’s detection ability utilizing virus-specific LAMP primers-sets using nucleic acid stain. The reaction tubes having LAMP products were mixed with 1 μL of 1/10 nucleic acid stain solution like Ethidium bromide, HNB and Calcein and visualize the colour change under the naked eye in normal light (Fig. 2). The result revealed an intense colour change in tubes with amplified product of ToLCBaV but no colour change in the control tube.
Fig. 2.
Visualization of LAMP product using nucleic acid stains a Ethidium Bromide b HNB c Calcein. Tube 1 positive sample for ToLCBaV, 2–4 virus infected tomato sample, 5 Healthy tomato sample, 6 control (water). The reaction products were visualized under naked eye and revealed colour change in tubes with amplified products (colour figure online)
Gel electrophoresis
ToLCBaV DNA was amplified from total genomic DNA extracts of the individual ToLCBaV-infected tomato plant by LAMP. The result revealed electrophoretic analysis of reaction products amplification of the targeted viral pathogen by their specific LAMP primer set and produced characteristic ladder-like banding patterns for ToLCBaV infected sample in the 2% agarose gel electrophoresis (Fig. 1b).
LAMP sensitivity assay
To determine the limit of the LAMP assay, total nucleic acids were extracted from both ToLCBaV- infected and healthy tomato cultivars and diluted from 100 to 10−4 and used in amplification reaction to investigate the sensitivity of both PCR and LAMP assay. Results of electrophoretic gel analysis revealed detection limit of ToLCBaV is 100 to 10−2 and sensitivity of the LAMP assay is 100 times that of PCR (Fig. 1c).
Discussion
Several detection and diagnosis methods have been operated for the amplification of ToLCV [14, 16, 22, 23]. Among these techniques, LAMP method has the advantage of rapid amplification rate and high sensitivity. The LAMP method is functioned by four different sets of primer to recognize the six distinct regions of the target pathogen. In that F3 and B3 act as an outer primer for the strand displacing activity and FIP & BIP act as an inner primer which helps to form stem loop structure or dumbbell structures at the ends. These stem loop structure pay way to auto cyclic amplification by involving loop primers which shorten time and increase sensitivity. Bst and Bsm DNA polymerase is the enzyme play key role in LAMP amplification process [1]. The LAMP reaction process proceeds at isothermal conditions vary from 60 to 65 °C for 30–60 min [9, 12, 18]. LAMP reaction can be supervised by observing the white precipitation of magnesium pyrophosphate produced as outcome of combination with magnesium ion in the reaction solution [9, 26]. The turbidity in the reaction tubes shows the formation of magnesium pyrophosphate precipitate which indicates the presence of ToLCBaV DNA in the sample and switches the visual examination of the LAMP reaction product to replace the gel electrophoresis.
In this experiment, we have collected fifteen tomato leaf curl disease infected samples from ICAR-IIHR, Bengaluru, India for the amplification of genomic DNA of ToLCBaV by the use of LAMP method. Six sets of primers were designed for targeting the conserved AC1 region of ToLCBaV by using the primer explorer V4 software. Appropriate design of the primers is an important key in gene amplification using the LAMP method [26]. LAMP reaction was carried out using Bsm DNA polymerase and incubated @ 60 °C for 60 min. No reaction was detected in the healthy sample either by LAMP technique or PCR. The positive reaction tubes show the presence of turbidity in the LAMP product. Colorimetric detection of LAMP using the metal ion-binding indicator dye HNB, which is known to bind the Mg2+ ion and changes its colour depending upon the pH and Mg2+ concentration present in the reaction master mix [4].
The visual examination was carried out using nucleic acid stains like Ethidium bromide, HNB and Calcein and colour differentiation was observed between healthy and ToLCBaV—infected LAMP products. A higher sensitivity of detection of LAMP products by the colour change method using HNB than by turbidity measurement has also been reported by others [4]. Previous studies revealed that 0.8 M betaine resulted in elevated sensitivity and increased effectiveness of the LAMP assay [13, 17]. To determine the limit of LAMP assay, total nucleic acid were extracted from both ToLCBaV—infected and healthy tomato cultivar and diluted from 10 to 10−6. The sensitivity of the LAMP assay is ten-fold that of conventional PCR technique [5, 24, 28]. This result shows that LAMP assay for ToLCBaV will straighten the ability to diagnose the viral infection in tomato cultivar under various circumstances.
In summary, LAMP-PCR based detection of plant virus uses 4–6 primers recognizing 6–8 distinct regions of target DNA. A strand-displacing DNA polymerase initiates synthesis by (F3 & B3) outer primer and (FIP & BIP) inner primers to form loop structures which facilitate auto cyclic amplification. LAMP-PCR is simple, rapid, sensitive, and amplification is so extensive that the magnesium pyrophosphate produced during the reaction can be seen by naked eye, making LAMP well-suited for field diagnostics.
Acknowledgement
Funding was provided by Indian Council of Agricultural Research (Grant Number ICAR- Consortia Research Platform (CRP) on Vaccines and diagnostics. F. No. 16-18/PP/ICAR CRP/16-17/18).
References
- 1.Abdullahi UF, Naim R, Taib WR, Saleh A, Muazu A, Aliyu S, Baig AA. Loop-mediated isothermal amplification (LAMP), an innovation in gene amplification: bridging the gap in molecular diagnostics; a review. Indian J Sci Technol. 2015;8(17):55767. doi: 10.17485/ijst/2015/v8i17/55767. [DOI] [Google Scholar]
- 2.Budziszewska M, Wieczorek P, Obrępalska Stęplowska A. One-step reverse transcription loop-mediated isothermal amplification (RT-LAMP) for detection of tomato torrado virus. Arch Virol. 2016;161(5):1359–1364. doi: 10.1007/s00705-016-2774-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chowda Reddy RV, Colvin J, Muniyappa V, Seal S. Diversity and distribution of Begomoviruses infecting tomato in India. Arch Virol. 2004;150:845–867. doi: 10.1007/s00705-004-0486-5. [DOI] [PubMed] [Google Scholar]
- 4.Goto M, Honda E, Ogura A, Nomoto A, Hanaki K. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxynaphthol blue. Biotechniques. 2009;46:167–172. doi: 10.2144/000113072. [DOI] [PubMed] [Google Scholar]
- 5.Imai M, Ninomiya A, Minekawa H, Notomi T, Ishizaki T, Tashiro M, Odagiri T. Development of H5-RT-LAMP (loop-mediated isothermal amplification) system for rapid diagnosis of H5 avian influenza virus infection. Vaccine. 2006;24:6679–6682. doi: 10.1016/j.vaccine.2006.05.046. [DOI] [PubMed] [Google Scholar]
- 6.Inoue-Nagata AK, Lima MF, Gilbertson RL. A review of geminivirus diseases in vegetables and other crops in Brazil: current status and approaches for management. Hortic Bras. 2016;34:1. doi: 10.1590/S0102-053620160000100002. [DOI] [Google Scholar]
- 7.Kirthi N, Maiya SP, Murthy MR, Savithri HS. Evidence for recombination among the tomato leaf curl virus strains/species from Bangalore, India. Arch Virol. 2002;147:255–272. doi: 10.1007/s705-002-8318-8. [DOI] [PubMed] [Google Scholar]
- 8.Maruthi MN, Rekha AR, Cork A, Colvin J, Alam SN, Kader KA. First report of tomato leaf curl New Delhi virus infecting tomato in Bangladesh. Plant Dis. 2005;89(9):1011. doi: 10.1094/PD-89-1011C. [DOI] [PubMed] [Google Scholar]
- 9.Mori Y, Nagamine K, Tomita N. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 10.Muniyappa V, Venkatesh HM, Ramappa HK, Kulkarni RS, Zeiden M, Tarba C-Y, Ghanim M, Czosnek H. Tomato leaf curl virus from Bangalore (ToLCV-Ban4): sequence comparison with Indian ToLCV isolates, detection in plants and insects, and vector relationships. Arch Virol. 2000;145:1583–1598. doi: 10.1007/s007050070078. [DOI] [PubMed] [Google Scholar]
- 11.Nagamine K, Hase T, Notomi T. Loop-mediated isothermal amplification reaction using a nondenatured template. Clin Chem. 2001;47:1742–1743. [PubMed] [Google Scholar]
- 12.Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- 13.Nagdev KJ, Kashyap RS, Parida MM, Kapgate RC, Purohit HJ, Taori GM. Loop-mediated isothermal amplification for rapid and reliable diagnosis of tuberculous meningitis. J Clin Microbiol. 2011;49(5):1861–1865. doi: 10.1128/JCM.00824-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Navas-Castillo J, Dıaz JA, Sánchez-Campos S, Moriones E. Improvement of the print-capture polymerase chain reaction procedure for efficient amplification of DNA virus genomes from plants and insect vectors. J Virol Methods. 1998;75:195–198. doi: 10.1016/S0166-0934(98)00110-4. [DOI] [PubMed] [Google Scholar]
- 15.Navot N, Pichersky E, Zeidan M, Zamir D, Czosnek H. Tomato yellow leaf curl virus: a whitefly transmitted geminivirus with a single genomic component. Virology. 1991;185:151–161. doi: 10.1016/0042-6822(91)90763-2. [DOI] [PubMed] [Google Scholar]
- 16.Navot N, Zeiden M, Pichersky E, Zaimer D, Czosnek H. Use of the polymerase chain reaction to amplify tomato yellow leaf curl virus DNA from infected plants and viruliferous whiteflies. Phytopathology. 1992;82:1199–1202. doi: 10.1094/Phyto-82-1199. [DOI] [Google Scholar]
- 17.Njiru ZK. Rapid and sensitive detection of human African trypanosomiasis by loop-mediated isothermal amplification combined with a lateral-flow dipstick. Diagn Microbiol Infect Dis. 2011;69:205–209. doi: 10.1016/j.diagmicrobio.2010.08.026. [DOI] [PubMed] [Google Scholar]
- 18.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parida M, Posadas G, Inoue S, Hasebe F, Morita K. Real-time reverse transcription loop mediated isothermal amplification for rapid detection of West Nile virus. J Clin Microbiol. 2004;42(1):257–263. doi: 10.1128/JCM.42.1.257-263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parida MM, Horioke K, Ishida H, Dash PK, Saxena P, Jana AM, Islam MA, Inoue S, Hosaka N, Morita K. Rapid detection and differentiation of dengue virus serotypes by a real-time reverse transcription-loop-mediated isothermal amplification assay. J Clin Microbiol. 2005;43:2895–2903. doi: 10.1128/JCM.43.6.2895-2903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parida M, Sannarangaiah S, Kumar Dash P, Rao PVL, Morita K. Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev Med Virol. 2008;18:407–421. doi: 10.1002/rmv.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rojas MR, Gilbertson RL, Russell DR, Maxwell DP. Use of degenerate primers in the polymerase chain reaction to detect whitefly-transmitted geminiviruses. Plant Dis. 1993;77:340–347. doi: 10.1094/PD-77-0340. [DOI] [Google Scholar]
- 23.Salati R, Nahkla MK, Rojas MR, Guzman P, Jaquez J, Maxwell DP, Gilbertson RL. Tomato yellow leaf curl virus in the Dominican Republic: characterization of an infectious clone, virus monitoring in whiteflies, and identification of reservoir hosts. Phytopathology. 2002;92:487–496. doi: 10.1094/PHYTO.2002.92.5.487. [DOI] [PubMed] [Google Scholar]
- 24.Salih DA, Liu Z, Bakheit MA, Ali AM, El Hussein AM, Unger H, Viljoen G, Seitzer U, Ahmed JS. Development and evaluation of a loop-mediated isothermal amplification method for diagnosis of tropical theileriosis. Transbound Emerg Dis. 2008;55:238–243. doi: 10.1111/j.1865-1682.2008.01033.x. [DOI] [PubMed] [Google Scholar]
- 25.Thai HTC, Le MQ, Vuong CD, Parida M, Minekawa H, Notomi T, Hasebe F, Morita K. Development and evaluation of a novel loop mediated isothermal amplification (LAMP) method for rapid detection of SARS Corona virus. J Clin Microbiol. 2004;42(5):1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomita N, Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of product. Nat Protoc. 2008;3(5):877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 27.Vasudeva RS, Sam Raj J. A leaf curl disease of tomato. Phytopathology. 1948;38:364–369. [Google Scholar]
- 28.Zhou D, Guo J, Xu L, Gao S, Lin Q, Wu Q, Wu L, Que Y. Establishment and application of a loop-mediated isothermal amplification (LAMP) system for detection of cry1Ac transgenic sugarcane. Sci Rep. 2014;4:4912. doi: 10.1038/srep04912. [DOI] [PMC free article] [PubMed] [Google Scholar]