Abstract
White spot syndrome virus (WSSV) infection is commonly detected by vp28-qPCR assay in wild crayfish, Procambarus clarkii, a widespread crustacean species in the aquatic environment in China. The virions of crayfish WSSV have been isolated and purified. Based on TEM observation, they exhibited morphological structures that are identical to known WSSV. In addition, the WSSV major envelope protein VP28 was observed based on Western blot analysis of the total structural proteins of crayfish WSSV. PCR amplification and sequencing analyses of variable regions of ORF14/15, ORF23/24 and ORF94, along with viral genomic sequencing and phylogenomic analysis, indicated that the crayfish WSSV, named WSSV-CN-Pc, represents a new WSSV genotype. Intramuscular injection bioassay revealed that WSSV-CN-Pc was as virulent as the WSSV Taiwan strain. The WSSV-CN-Pc exhibited characteristics of a dominant genotype, with high infection load (107–108 WSSV/mg) and high prevalence (91.7%, 110 of 120 crayfish samples) observed in the surveyed wild crayfish. WSSV-CN-Pc was also detected, with similar infection pattern as observed in crayfish, in farmed Litopenaeus vannamei shrimp that shared similar ecological niches with the sampled crayfish. Our results indicated that there was horizontal transmission of WSSV-CN-Pc between crayfish and shrimp in nature. Our findings also implicated that crayfish and shrimp farming should be integrated and managed with cautions in order to reduce the risk of spread and circulation of WSSV in the aquatic environment.
Electronic supplementary material
The online version of this article (doi:10.1007/s13337-017-0394-4) contains supplementary material, which is available to authorized users.
Keywords: White spot syndrome virus, Procambarus clarkii, Natural infection, Virulence
Introduction
White spot syndrome virus (WSSV) is an enveloped, double-stranded DNA virus belonging to the genus Whispovirus and family Nimaviridae [17]. Infectious WSSV virions are typically 250–380 nm in length and 120–150 nm in diameter [28]. They usually contain a genome of ~300 kb that encodes ~180 proteins [27]. Since it was first reported in the Fujian province, China in 1991/1992, WSSV has spread rapidly throughout the world, causing heavy economic losses in the aquaculture industry [10].
WSSV infects almost all cultured and wild marine shrimp. Specifically, WSSV is potentially lethal in most of the commercially important species of penaeid shrimp including Litopenaeus vannamei, Penaeus monodon, Fenneropenaeus indicus, Marsupenaeus japonicus, Fe. chinensis, Fe. penicillatus, Farfantepenaeus aztecus, and Fa. merguis [19]. In 1997, Panulirus penicillatus and P. versicolor lobsters were experimentally verified to be infected by WSSV [4], while no lobster has been found to be naturally infected by WSSV. In 1995, a serious disease outbreak was investigated in crayfish, Orconectes punctimanus and Procambarus sp., at the USA National Zoo in Washington D.C. [22]. Histological analysis and transmission electron microscopy (TEM) showed that the pathogen in this outbreak closely resembled WSSV found in shrimp, an observation that was further confirmed by dot blot with specific gene probes, PCR, and bioassays [28]. This is the first report that described natural WSSV infection in crayfish. In addition, seven crayfish species, Procambarus clarkii [4], Astacus leptodactylus, O. limosus [6], Pacifastacus leniusculus [11], Cherax destructor albidus [9], A. astacus [12], and C. quadricarinatus [24] were shown to be infected by WSSV via other analyses.
Among the various crayfish species, P. clarkii is the predominant and most widespread crayfish in aquatic environments, including shrimp farming ponds, in China [26]. Recently, P. clarkii has become one of the most important cultured aquatic species in China. In 2015, the yield of farmed P. clarkii reached 660,000 tons, and the output value was 42 billion dollars [2]. Furthermore, the crayfish farming industry is developing rapidly in China. However, WSSV outbreaks have become more frequent and severe in farmed crayfish, which has become a foremost challenge that the crayfish farming industry has to solve for.
In this study, we identified a high virulent WSSV genotype in naturally infected wild crayfish, P. clarkii, in Shanghai, China. This novel WSSV isolate represents the most dominant and prevalent genotype in the surveyed wild crayfish as well as in farmed L. vannamei shrimp. Our results suggest that the horizontal transmission of WSSV between crayfish and shrimp appears to be plausible.
Materials and methods
Crayfish and shrimp samples
A total of 120 crayfish samples (P. clarkii) (length of 10–13 cm and weight of 25–35 g, 74 males and 46 females) were collected from a neighboring river of shrimp farming ponds in Lingang, Shanghai, China, between February 2015 and August 2015. The samples were subjected to WSSV infection detection. Crayfish were dissected immediately upon arriving at the laboratory. Approximately 30 mg of each of the pleopod and muscle were aseptically removed and stored at −20 °C for subsequent analysis. The remaining tissues were frozen at −80 °C.
Sixty dead shrimp samples (L. vannamei) (length of 11–13 cm and weight of 20–35 g) were collected from a shrimp pond in Lingang, Shanghai, China, between July 2016 and August 2016, and then subjected to WSSV infection detection. Shrimp samples were dissected immediately upon arriving at the laboratory. Approximately 30 mg of muscle was aseptically removed and stored at −20 °C for subsequent analysis. The remaining tissues were frozen at −80 °C.
Bioassay
As for intramuscular injection bioassay, crayfish (P. clarkii) (length of 12–14 cm and weight of 30–40 g, males and females) were purchased from a crayfish farm in Suzhou, Jiangsu province, China, and tested for the presence of WSSV using vp28-qPCR (Table 1) to ensure that they are WSSV-free. WSSV-free crayfish were maintained in tanks (50 × 38 × 25 cm) containing 20 L aerated tap water at 25 °C for 2 weeks and fed a commercial diet at 5% of body weight per day prior to experiments.
Table 1.
PCR primers used in this study
| Primer pair | Sequence (5′-3′) | Viral sequence coordinate | Size of PCR product (bp) and reference |
|---|---|---|---|
| WSSV PCR-757 | |||
| FP | TTGCCAATTGTCCTGTTACGTACTCG | 244,110–244,136 | 757 [30] |
| RP | ACGATTTATTTACTCGGTCTCAGTGC | 244,866–244,840 | |
| (WSSV-TW) | |||
| VP28 | |||
| VP28–140F | AGGTGTGGAACAACACATCAAG | 244,610–244,631 | 140 [18] |
| VP28–140R | TGCCAACTTCATCCTCATCA | 244,750–244,731 | |
| (WSSV-TW) | |||
| ORF14/15 | |||
| F (14/15) | CCGCCAGGAGAGATGCTAAG | 22,416–22,436 | 2151 (this study) |
| R (14/15) | TGGACCAAATGACCTGGACG | 24,567–24,547 | |
| (WSSV-TH) | |||
| ORF23/24 | |||
| F (23/24) | GGTAGGAGAAGGTACGCACG | 29,999–30,019 | 4025 (this study) |
| R (23/24) | GCCCAGATTGGTCATGTCCA | 34,024–34,004 | |
| (WSSV-TH) | |||
| ORF 94 | |||
| F (ORF94) | GGGCAAACGTACACCTGAGA | 141,982–142,002 | 1770 (this study) |
| R (ORF94) | ATCTCTGTGGCTTGCTTTGC | 143,752–143,732 | |
| (WSSV-TH) | |||
| WSSV-CN-Pc-2203 | |||
| F-PC | GGGACGTTTCCATCTAGGGT | 279,871–279,891 | 2203 (this study) |
| R-PC | GAGGCGAGACTTGCAGAACT | 282,074–282,054 | |
| (WSSV-CN-Pc) | |||
| WSSV-CN-Pc-166 | |||
| F (PC) | AGGCATATGGATAGTACAAATTGTCT | 281,081–281,107 | 166 (this study) |
| R (PC) | TCACCTTTTATTGTACTGCATGGT | 281,247–281,223 | |
| (WSSV-CN-Pc) | |||
Primer WSSV PCR-757 was used for the PCR-cloning of VP28
Primer VP28 was used in qPCR assay of all WSSV
Primer WSSV-CN-Pc-2203 was used for the PCR-cloning of WSSV-CN-Pc specific sequence
Primer WSSV-CN-Pc-166 was used in qPCR assay of WSSV-CN-Pc
DNA extraction
TIANamp Marine Animals DNA Kit (TIANGEN) was used to extract total genomic DNA from crayfish pleopod and muscle, and shrimp muscle. Genomic DNA of purified WSSV virions was extracted with the TIANamp Virus DNA/RNA Kit (TIANGEN). All DNA samples were frozen at −20 °C before use.
Preparation of recombination plasmid standard
The vp28 plasmid standard for quantitative detection of all WSSV strains was constructed as previously described [30]. The plasmid standard for detection of WSSV-CN-Pc was prepared following the procedures previously described [30]. In brief, a 2203 bp fragment specific for WSSV-CN-Pc was amplified and cloned. The 50 μl PCR reaction contained 4 μl WSSV-CN-Pc DNA (44 ng), 1.0 μl each of the 10 mM primers WSSV-CN-Pc-2203 (Table 1), 25 μl of 2 × PCR mix (TaKaRa), and 19 μl double distilled water. The thermal cycle program was as follows: 94 °C for 4 min, followed by 30 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 60 s and a final extension step of 72 °C for 10 min. Amplification products were verified by 1.5% agarose gel electrophoresis. The PCR products containing specific target bands were purified using Universal DNA Purification Kit (TIANGEN) according to the manufacturer’s instruction. The purified amplicons were ligated into the pGM-T vector using the pGM-T Cloning Kit (TIANGEN). The recombinant plasmid was transformed into competent Escherichia coli TOP10 cells (TIANGEN) following the manufacturer’s instruction. Recombinant plasmids were identified by colony PCR. The correct insert was verified by sequencing and quantified using the Quant-iTPicoGreen dsDNA Reagent and Kits (Invitrogen). The concentration of the recombinant plasmid was converted to copy number based on the following equation: Number of copies = (M × 6.02 × 1023 × 10−9)/(n × 660). M is the amount of DNA in nanogram, n is the number of recombinant plasmid in base pair, and the average weight of one base pair is assumed to be 660 Da. The prepared plasmid standard was aliquoted and stored at −80 °C before use.
PCR and qPCR
Primers ORF 14/15 and ORF 23/24 (Table 1) were used to characterize variable regions of WSSV ORF14/15 and ORF23/24, respectively. VNTR in WSSV ORF94 was analyzed with primer ORF94 (Table 1). The 25 μl PCR reaction contained 12.5 μl of 2 × PCR mix (TaKaRa), 0.5 μl of each of the 10 mM primers, 9.5 μl double distilled water and 2 μl DNA template (approximately 200 ng). The thermal cycle program was as follows: 94 °C for 4 min, followed by 35 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 90 s and a final extension step of 72 °C for 10 min. Amplification products were verified by 1.5% agarose gel electrophoresis.
The crayfish samples and shrimp samples were investigated for WSSV infection by qPCR using the primer VP28 for all WSSV strains and WSSV-CN-Pc-166 for WSSV-CN-Pc (Table 1). The recombinant plasmid standard was serial-diluted to a range of 10 to 108 copies per microliter. Two microliters of each dilution were used in qPCR assay on an ABI 7500 Fast qPCR system with 2.0.1 version software (Applied Biosystems). The reaction mix (20 μl) contained 2 μl DNA, 0.6 μl of each of the 10 mM primers (Table 1), 10 μl 2 × SYBR Green qPCR Mix (Roche) and 6.8 μl double distilled water. The thermal cycle program was described as follows: 95 °C for 10 min, followed by 40 cycles of 94 °C for 15 s, 60 °C for 60 s.
The prevalence of WSSV was investigated by analyzing boxplot that was generated using the package of R language’s ggplot 2.
Purification of WSSV-CN-Pc virions
WSSV-CN-Pc virions were purified from crayfish that were infected with a single WSSV-CN-Pc isolate as previously described [31]. The viral concentration was quantified by qPCR using primer VP28 (Table 1) and then stored at −80 °C.
Transmission electron microscopy observation
TEM sample preparation and observation were performed as previously described [25].
Western blot analysis
Twenty microliters of purified WSSV-CN-Pc virions were added to 5 μl SDS-PAGE sample loading buffer (5×) (Sangon Biotech) and then were boiled at 100 °C for 15 min. The viral proteins were separated on 12% SDS-PAGE and then transferred to PVDF membrane (Immobilon-PSQ, Millipore). The membrane was blocked with skim milk at 4 °C for 12 h. Anti-VP28 antibody (rabbit) was added, and the membrane was incubated at RT for 1 h. Subsequently, secondary antibody IgG-AP (goat) (Thermo Fisher Scientific) was added, and the membrane was incubated at RT for 1 h. After washing three times with TBST, positive signals were developed by adding BCIP/NBT solution (Sangon Biotech).
Sequencing and analysis of the whole genome of WSSV-CN-Pc
Twenty micrograms of genomic DNA of WSSV-CN-Pc were sequenced on an Illumina MiSeq sequencer (Majorbio Bio-Pharm Technology Co, Ltd, Shanghai, China). Sequences were first subjected to quality control using NGSQC toolkit (v 2.3.3) with default parameters. SOAP denovo v2.04 (http://soap.genomics.org.cn/) was used to perform de novo assembly. The obtained scaffolds were mapped to WSSV-TH (GenBank accession no. AF369029) using of Geneious Pro (version 6.1.2).
Primer sets were designed based on the assembled contigs of WSSV-CN-Pc. PCR and Sanger sequencing (Sangon Biotech, Technology Co, Ltd, Shanghai, China) were performed to fill in the gaps and to confirm overlaps between scaffolds. The prediction and annotation of open reading frames (ORFs) in WSSV-CN-Pc were conducted according to the procedures described previously [13]. Each predicted ORF encompassed a start codon ATG, exhibited a minimum size of 120 bp of standard genetic code, and contained a stop codon. BlastP program (http://blast.ncbi.nlm.nih.gov/) was used for sequence similarity comparisons of all predicted ORFs to NCBI non-redundant protein sequence (nr) database.
Intramuscular injection bioassay
Sixty WSSV-free crayfish were divided into 3 groups (20 per group) and injected with WSSV-CN-Pc, WSSV-TW and TM buffer (negative control), respectively. The total amount of virus administered per crayfish (108 copies in 200 μl) remained constant in all groups. Mortality was recorded twice daily for 7 days, and dead crayfish were tested for the presence of WSSV by qPCR using primer VP28 for WSSV-TW and WSSV-CN-Pc-166 for WSSV-CN-Pc, respectively (Table 1). The experiment was repeated twice.
Genome sequence comparison analysis of different WSSV genotypes
Variable region sequences of WSSV-CN-Pc were compared with those of the four main WSSV genotypes using Geneious Pro (version 6.1.2). The four main WSSV genomes were downloaded from the NCBI database (http://www.ncbi.nlm.nih.gov/). The GenBank accession numbers of these four WSSV genotypes were as follows: WSSV-TW (AF440570), WSSV-CN (AF332093), WSSV-Korea (JX515788) and WSSV-TH (AF369029).
Sequence identities of the WSSV genomes were compared using Mauve alignment (Geneious Pro). Plots were generated using BRIG [1] with 70 and 50% as the upper and lower identity threshold, respectively.
Phylogenomic analysis
All nine WSSV genomic sequences available in GenBank (Table 2) were aligned with WSSV-CN-Pc using Geneious Pro. Then, phylogenetic analysis was conducted using MEGA 6 with maximum-likelihood and neighbor-joining algorithms, respectively.
Table 2.
Genomic sequence identity based on Mauve alignment of WSSV-CN-Pc with other WSSV genomes
| Isolate | Length (bp) | Accession no. | Identity (%) |
|---|---|---|---|
| WSSV-TW | 307,287 | AF440570 | 98.9 |
| WSSV-CN | 305,119 | AF332093 | 98.8 |
| WSSV-EG3 | 305,119 | KR083866 | 98.8 |
| WSSV-CN01 | 309,286 | KT995472 | 98.5 |
| WSSV-Korea | 295,884 | JX515788 | 98.1 |
| WSSV-CN02 | 294,261 | KT995470 | 97.1 |
| WSSV-TH | 292,967 | AF369029 | 96.5 |
| WSSV-MEX2008 | 293,183 | KU216744 | 96.3 |
| WSSV-CN03 | 284,148 | KT995471 | 93.3 |
Results
Detection of WSSV in wild crayfish
VP28-PCR
A total of 120 wild crayfish were subjected to PCR and sequence analysis. The results showed that 119 of them were positive for WSSV infection. The PCR amplicons were sequenced, and found to be 140 bp in length and share 100% identity with WSSV vp28 sequences in GenBank (Fig. 1).
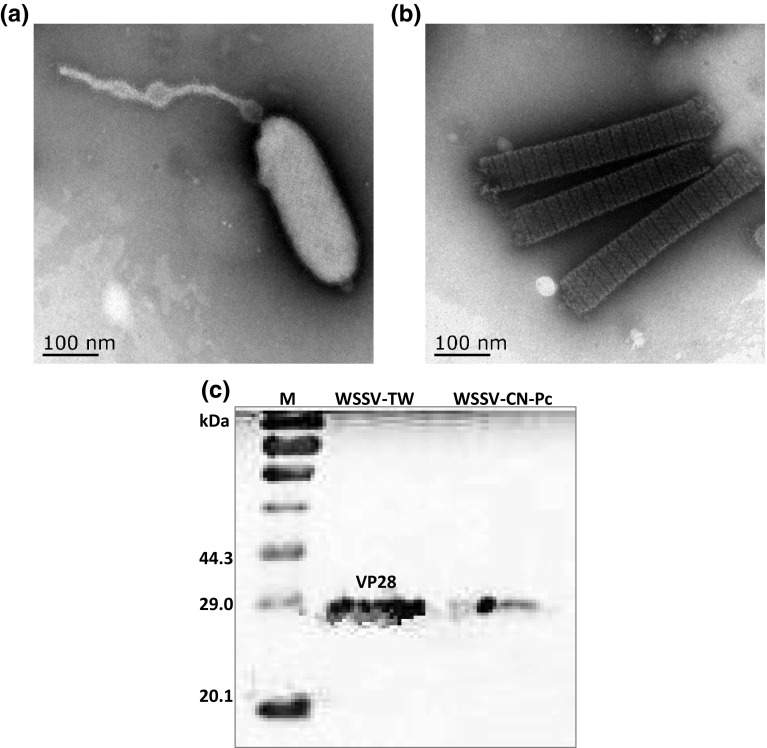
Fig. 1.
Electron micrograph and Western blot of WSSV-CN-Pc purified from crayfish samples. a Enveloped virion, scale bar 100 nm; b nucleocapsid (naked) virus (the viral envelope was peeled off due to ultracentrifugation), scale bar 100 nm; c Western blot analysis of WSSV structural protein VP28. WSSV-TW was used as positive control. M protein marker
Variable region ORF14/15
Primers were designed for PCR amplification of variable region ORF14/15 in WSSV-CN-Pc using WSSV-TH as the reference sequence (Table 1). An amplicon of 950 bp was obtained and sequenced. Our analysis revealed that the variable region ORF14/15 contained a deletion of 541, 706, 1591 and 698 bp when compared to WSSV-TH, WSSV-CN, WSSV-Korea and WSSV-TW, respectively (Fig. 2a).
Fig. 2.
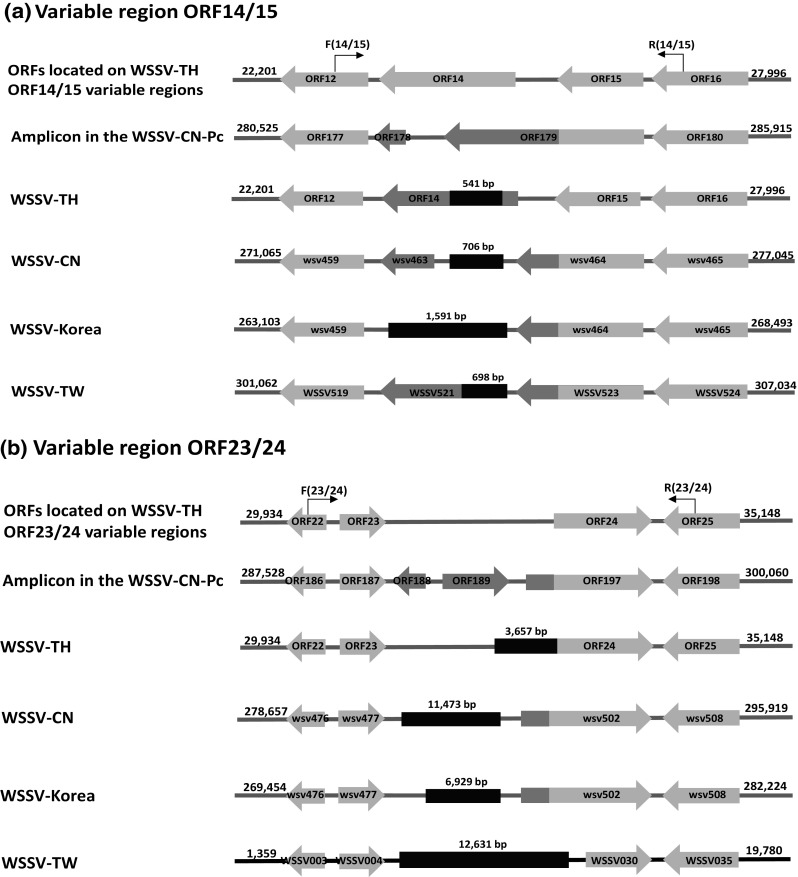
Schematic representation of the variable regions in the WSSV genomes. The deep red indicates the corresponding deletion in WSSV-CN-Pc compared with a particular genotype, and the length of the deletion is indicated above the box. The ORFs are shown in arrows. The ORFs in orange or light orange represent homologous regions and their numbering refers to the relevant published papers (color figure online)
Variable region ORF23/24
PCR amplification with the ORF23/24 primers designed using WSSV-TH as a reference sequence (Table 1) yielded an amplicon of 673 bp for WSSV-CN-Pc. Our analysis revealed that the variable region ORF23/24 contained a deletion of 3657, 11,473, 6929 and 12,631 bp when compared to WSSV-TH, WSSV-CN, WSSV-Korea and WSSV-TW, respectively (Fig. 2b).
VNTR loci ORF94
PCR amplification with the primer ORF94 (Table 1) and sequence analysis showed that WSSV-CN-Pc shared the highest sequence identity (99.3%) with WSSV-Korea, followed by WSSV-TH and WSSV-TW (82.8% sequence identity). Sequence identity shared with the other genotypes ranged from 32.2 to 69.3%. In addition, 54 bp of repeat unit (RU) was not identified in WSSV-CN-Pc.
Isolation and characterization of WSSV-CN-Pc
Viral particles
WSSV-CN-Pc exhibited a rod-shape morphology typical of WSSV, with a tail-like appendage at one end of the virion (Fig. 1). Both enveloped virion (Fig. 1a) and naked nucleocapsid (Fig. 1b) were observed in the purified virions. In addition, Western blot assay targeting WSSV envelop structural protein VP28 further confirmed that the virus isolated from P. clarkii crayfish belonged to Whispovirus at the viral protein level (Fig. 1c).
Whole genome sequencing
The complete sequence of WSSV-CN-Pc (accession no. KX686117) was obtained using MiSeq and Sanger sequencing. It is 300,223 bp in size with 199 predicted open reading frames (ORFs). Among the 199 ORFs, no BLASTp hit in GenBank nr database was obtained on 4 small ORFs of 159–180 bp in size. All the other ORF were homologous to WSSV genes, including 187 ORFs with 97–100% of sequence identity and 8 ORFs with 56–91% of sequence identity to WSSV genes (Fig. 3; Table S1).
Fig. 3.
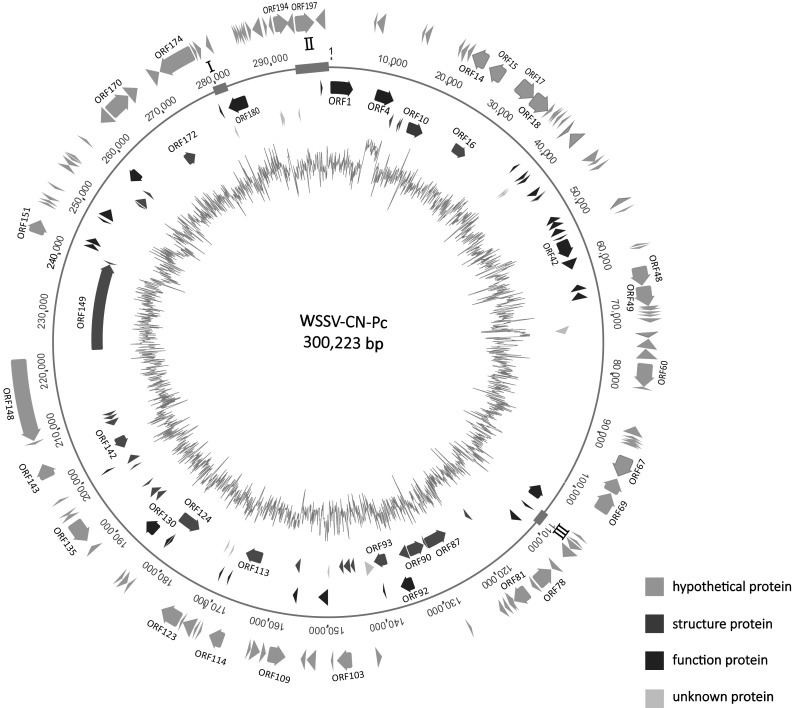
Sequence map of WSSV-CN-Pc. All predicted open reading frames (ORFs) are shown as arrows in different colors. Direction of arrowhead represents the transcriptional orientation. From inner to outer: GC plot, structure proteins, function proteins, and hypothetical proteins. The unknown proteins are shown in green. I variable region ORF14/15, II variable region ORF23/24, III variable region ORF94 (color figure online)
Genomic alignment analysis revealed 12 distinct regions in WSSV-CN-Pc and 9 other WSSV genomic sequences available in GenBank. These included 4 homologous repeat regions and 3 functional regions (collagen-like protein, immediate-early protein, E3 ligase-immediate-early protein). Although WSSV-CN-Pc shared 93.3–98.9% of sequence identity with other WSSV genomes (Fig. 4; Table 2), it possessed significant sequence differences in the variable regions of ORF14/15, ORF23/24 and ORF94 (Fig. 2), compared with other WSSV genomic sequences.
Fig. 4.
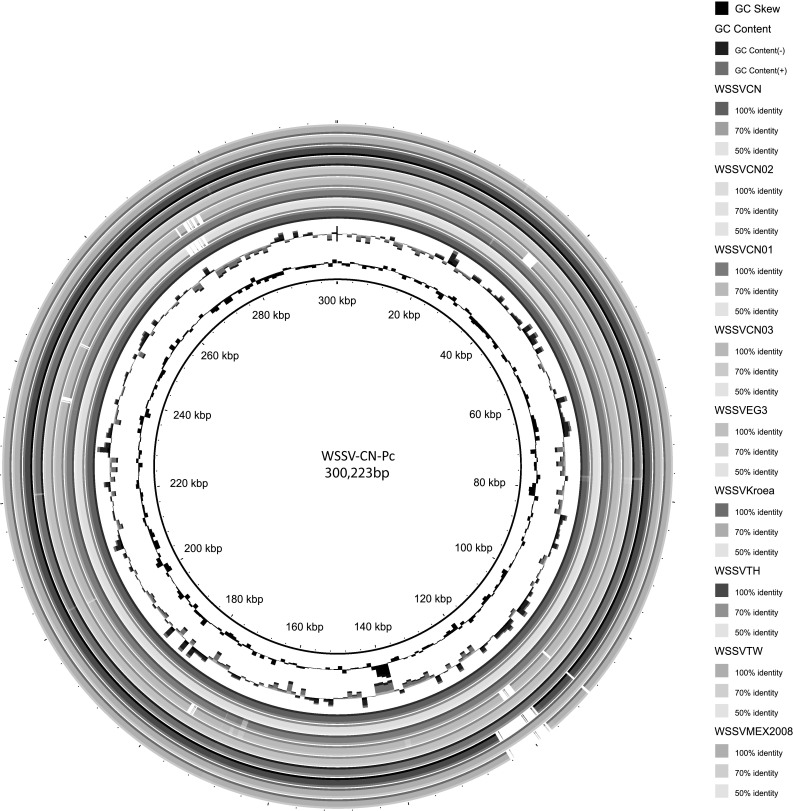
Whole genomic sequence similarity comparison between WSSV-CN-Pc and other WSSV strains. Sequence identity plots of other WSSV strains are generated based on comparison to WSSV-CN-Pc
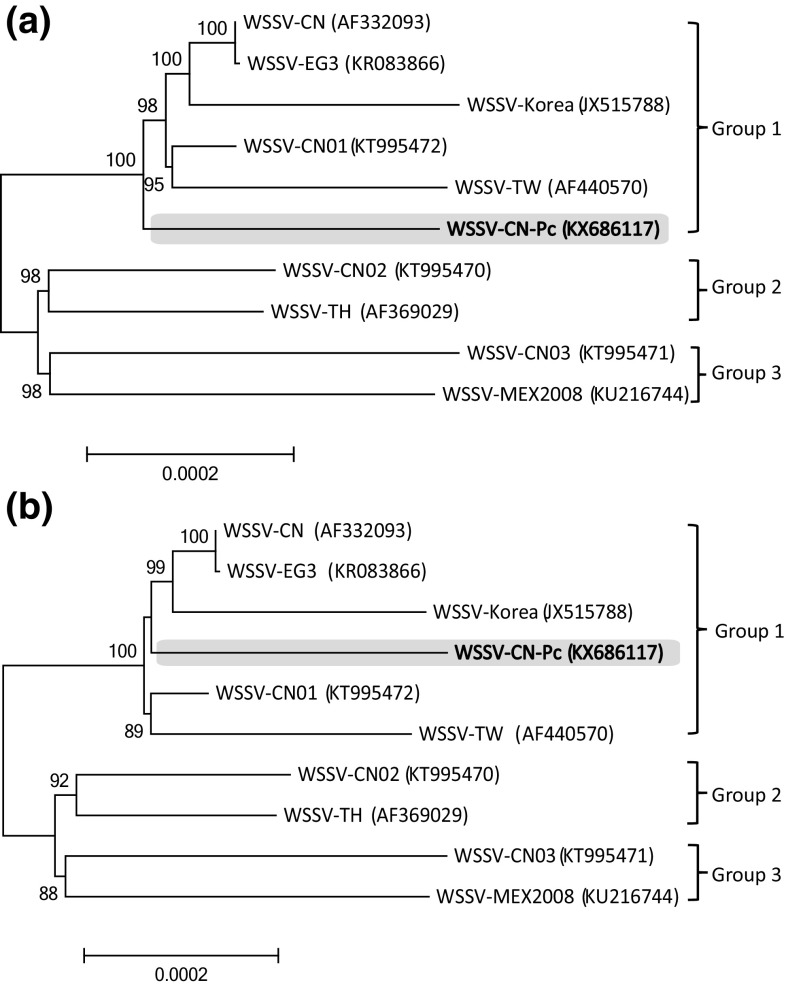
Phylogenomic analysis
On both the neighbor-joining and maximum-likelihood trees, 3 groups of WSSV were observed with significant bootstrap value support (Fig. 5). The WSSV-CN-Pc belonged to Group 1 and represented a novel genotype in comparison to the other strains within Group 1 (Fig. 5).
Fig. 5.
Phylogenomic analyses of the evolutionary relationship of WSSV strains. Both a neighbor-joining tree and b maximum-likelihood tree were reconstructed based on 272,682 informative positions. The Chinese strains are shown in bold face. The WSSV-CN-Pc are in grey background. The GenBank accession numbers of the WSSV genomes are shown in the parentheses
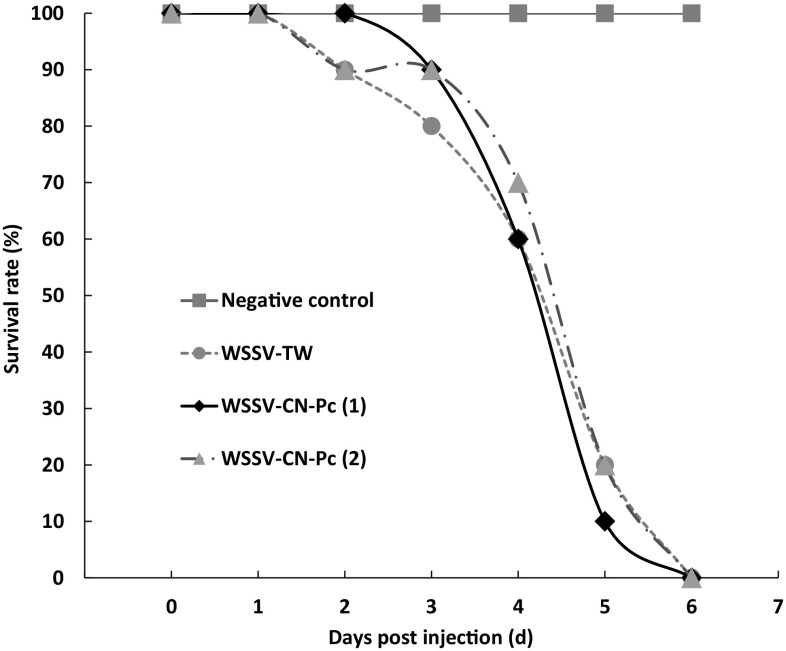
Virulence
Intramuscular injection bioassay was carried out to investigate the virulence of WSSV-CN-Pc. As shown in Fig. 5, the curves of cumulative survival rate of crayfish were quite similar for both WSSV-CN-Pc and WSSV-TW infection. On day 6 of post-injection, no live crayfish were observed in both the WSSV-TW and WSSV-CN-Pc injection groups. In the negative control group, crayfish that were injected with TM buffer all survived (Fig. 6). The LT50 [16] was about 4.2 days for WSSV-TW and 4.1–4.4 days for WSSV-CN-Pc (Fig. 6).
Fig. 6.
Intramuscular injection bioassay of WSSV-CN-Pc. WSSV-TW strain was used as positive control. WSSV-CN-Pc (1) and (2) stand for two times of repeat
All experimental crayfish were subjected to WSSV detection by vp-28 qPCR. Our results showed that 1.1 × 107–1.7 × 108 WSSV/mg of crayfish tissue were detected in each crayfish from the infected groups (n = 60), indicating that the crayfish indeed died from WSSV infection. No WSSV was detected in the crayfish in the negative control group (n = 20).
Prevalence of WSSV-CN-Pc
WSSV-CN-Pc in wild crayfish
Using vp28-qPCR assay for total WSSV, and WSSV-CN-Pc-166-qPCR specific for WSSV-CN-Pc (Table 1), WSSV-CN-Pc and total WSSVs were quantified in the wild crayfish samples. Results showed that 119 of the 120 crayfish samples (99.2%) were WSSV positive, and 80 of these 119 positive samples (67.8%) contained more than 1 × 107 WSSV/mg of tissue. In addition, only 1 sample was WSSV free. As for WSSV-CN-Pc, 110 of 120 (91.7%) were WSSV-CN-Pc positive, and 75 of these 110 positive samples (68.2%) had more than 1 × 107 WSSV-CN-Pc/mg of tissue. WSSV-CN-Pc was absent in 10 samples (Fig. 7a).
Fig. 7.
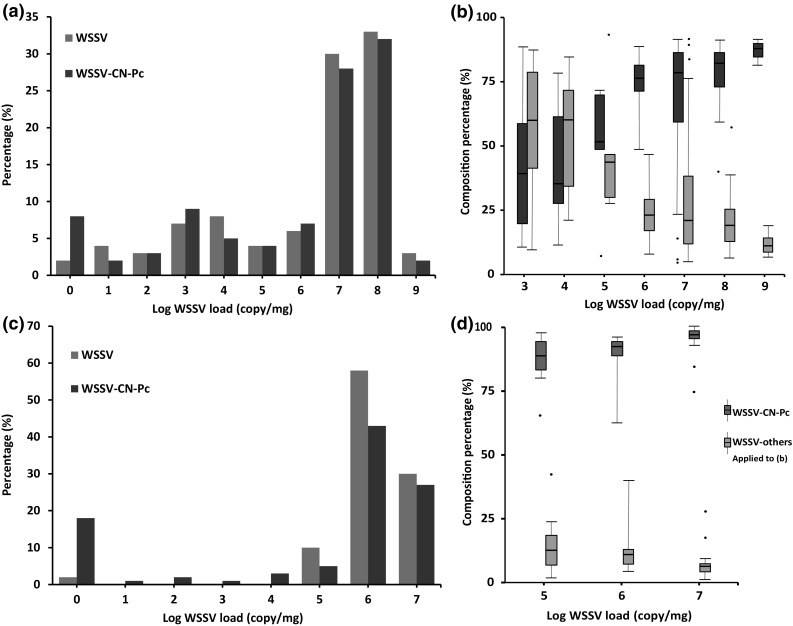
a Percentages of all WSSV stains and WSSV-CN-Pc in various WSSV loads in crayfish samples; b composition percentage distribution of other WSSV strains and WSSV-CN-Pc in various WSSV loads in crayfish samples. c Percentage of all WSSV stains and WSSV-CN-Pc in various WSSV loads in shrimp samples; d composition percentage distribution of other WSSV strains and WSSV-CN-Pc in various WSSV loads in shrimp samples
The composition percentage, the average percentage and the floating range of WSSV-CN-Pc and total of the other WSSVs were calculated for different viral infection load groups. Results showed that the composition percentages of total other WSSVs were high, but the floating range was also high at concentration of 1 × 103–1 × 105 WSSV/mg of tissue. Notably, the composition percentage of total other WSSVs gradually declined as the viral load reached over 1 × 106 WSSV/mg of tissue. In contrast, the composition percentage of WSSV-CN-Pc obviously increased as the viral load increased. In most samples, the composition percentage of WSSV-CN-Pc exceeded 90%, along with low floating ranges, especially at 1 × 109 WSSV/mg (Fig. 7b).
WSSV-CN-Pc in farmed penaeid shrimp
Similar to that in the crayfish samples, the prevalence of total WSSVs and WSSV-CN-Pc were analyzed in the penaeid shrimp samples. We found that 59 of 60 shrimp samples (98.3%) were WSSV positive, and 55 of these 59 positive samples (93.2%) contained more than 1 × 106 WSSV/mg of tissue. In addition, only one sample was WSSV free. Furthermore, 49 of 60 shrimp samples (81.7%) were WSSV-CN-Pc positive, and 42 of these 49 positive samples (85.7%) contained more than 1 × 106 WSSV-CN-Pc/mg of tissue. No WSSV-CN-Pc was detected in the 11 WSSV-CN-Pc negative samples (Fig. 7c).
The composition percentage, average of percentage and floating range of both total other WSSVs and WSSV-CN-Pc were calculated for the different levels of viral load in shrimp. The composition percentage of WSSV-CN-Pc obviously increased in response to a higher load of WSSV. In most samples, the composition percentage of WSSV-CN-Pc was more than 90%, especially at viral load of 1 × 107 WSSV/mg of tissue, along with low floating ranges. In contrast, the total other WSSVs exhibited a low (about 10%) composition percentage, and only 3–7% of other WSSVs-infected shrimp contained viral load of 1 × 107 WSSV/mg (Fig. 7d).
Discussion
A novel WSSV genotype, WSSV-CN-Pc, was identified in naturally infected wild crayfish (P. clarkii) sampled in Shanghai, China. WSSV-CN-Pc contains a genome of 300,223 bp, which is of a medium size relative to the other WSSV genomes that range from approximately 312 to 284 kbp. Analysis of the WSSV-CN-Pc genome reveals clear differences regarding the variable regions (WSSV-TH ORF14/15 and ORF23/24) and the variable numbers of tandem repeats (VNTR) (WSSV-TH ORF94) [7, 15, 21]. To decipher the potential evolutionary relationship of the various WSSV strains, phylogenomic analysis was performed in this study. Our results clearly indicate that the WSSV genotypes/strains discovered thus far are affiliated with three geno-groups (Fig. 4). WSSV-CN-Pc belongs to Group 1, which contains the most numbers of WSSV strains with fully sequenced genomes. With the exception of crayfish WSSV-CN-Pc, all the other WSSVs were isolated from infected penaeid shrimp. Interestingly, phylogenomic analysis also suggests that WSSV may have originated from China and subsequently spread to other regions and countries worldwide. Sequence analysis of additional WSSV strains from different infected animals and outbreak geographical locations would further shed light on the mechanism of WSSV transmission among diverse hosts and across the globe.
Variable region ORF14/15 is located within the WSSV-TH segment coding frames for ORF14 and ORF15, and has been confirmed to be prone to recombination [7, 15, 21, 29]. Variable region ORF23/24, a hotspot deletion region, is within the ORF23 and ORF24 coding frames of WSSV-TH genome [7, 15]. Large deletions were found in ORF14/15 and ORF23/24 of WSSV-CN-Pc when compared with four other WSSV genotypes (Fig. 2). For example, compared with WSSV-TW, an approximately 12,631 bp deletion was observed in the ORF23/24 region of WSSV-CN-Pc. This deletion region includes the VP35 gene that encodes nucleocapsid protein VP35 [5]. The absence of the VP35 gene was also observed in other WSSV strains. For example, relative to WSSV-TW and WSSV-TH-96-II, a 10,970 bp deletion in the ORF 23/24 region was found in the Indian strains [21]. Interestingly, it has been demonstrated that VP35 does not contribute to intracellular protective immunity in P. monodon shrimp [23]. Taken together, it is reasonable to speculate that the structural protein VP35 is not an essential protein involved in WSSV infection and intracellular propagation in both crayfish and penaeid shrimp. Notably, the source of VP35 gene, viral versus cellular, remains unknown. In addition, few homologous genes are shared between WSSV and shrimp, which lower the likelihood of shrimp acting as the natural hosts of WSSV.
A wide host range, especially in regards to crustacean hosts, has been reported in WSSV, with the exceptions of shrimp, crayfish and lobster. For example, WSSV was detected in wild crabs, Calappa lophos, Portunus sanguinolentus [3], and Helice tridens [14]. In 2015, WSSV was detected in naturally infected farmed mitten crabs, Eriocheir sinensis in China [8]. Meanwhile, WSSV was also detected in crayfish that cohabit with the crab in the same pond [8]. Moreover, WSSV was found in the infection of 34 species of crab in another study [20]. Interestingly, WSSV was also detected in copepod [14], insects [14] and rotifera [32] in nature. It is unclear why WSSV is able to infect and transmit among such diverse hosts given its relatively conserved genome. One explanation might be that WSSV might have emerged before or in parallel to the divergence of arthropods, and might have packaged the appropriate genetic contents for interacting with their broad range of modern day hosts. This speculation is supported by the evidence that few homologous genes are shared between WSSV and other viruses as well as animals, and the orphan nature of WSSV based on viral classification.
In our study, WSSV-CN-Pc was detected, with high prevalence and high viral load, in penaeid shrimp samples collected from farming ponds adjacent to the crayfish sampling locations. These suggest that horizontal transmission of WSSV-CN-Pc had likely occurred between crayfish and shrimp, although the probability of a third party involvement in the spread of WSSV-CN-Pc could not be completely ruled out. Therefore, it remains unclear what the source of WSSV-CN-Pc was and how such interspecies transmission occurred. Future comprehensive investigation is required to better understand WSSV transmission and its source in the aquatic environment.
In conclusion, the high prevalence of the novel WSSV-CN-Pc genotype in crayfish and its presence in farmed penaeid shrimp indicate the broad circulation and persistence of WSSV in the aquatic environment. Crayfish may serve as an important reservoir in the spread of WSSV in the nature, especially under the large-scaled and intensive cultivation environment for both crayfish and shrimp currently established in China. Therefore, extra cautions should be exercised to prevent possible horizontal transmission of WSSV between crayfish and shrimp in order to avoid the risk of cross-species and multi-WSSV strain infection, as well as the spread of communicable diseases.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Prof. Chu-Fang Lo of National Cheng Kung University, Tainan, Taiwan for providing VP28 antibody and WSSV-TW strain. This work was supported partially by the National Natural Science Foundation of China (41376135, 31570112, 31601570), Doctoral Fund of Ministry of Education of China (20133104110006), Innovation Program of Shanghai Municipal Education Commission (14ZZ144), China.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s13337-017-0394-4) contains supplementary material, which is available to authorized users.
References
- 1.Alikhan NF, Petty NK, Zakour NLB, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genom. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CFSY. China fishery statistics yearbook 2015. China Agriculture Press; 2015.
- 3.Chakraborty A, Otta SK, Joseph B, Kumar S, Hossain MS, Karunasagar I. Prevalence of white spot syndrome virus in wild crustaceans along the coast of India. Curr Sci. 2002;82:1392–1397. [Google Scholar]
- 4.Chang P, Chen H, Wang Y. Detection of white spot syndrome associated baculovirus in experimentally infected wild shrimp, crab and lobsters by in situ hybridization. Aquaculture. 1998;164:233–242. doi: 10.1016/S0044-8486(98)00189-6. [DOI] [Google Scholar]
- 5.Chen LL, Leu JH, Huang CJ, Chou CM, Chen SM, Wang CH, et al. Identification of a nucleocapsid protein (VP35) gene of shrimp white spot syndrome virus and characterization of the motif important for targeting VP35 to the nuclei of transfected insect cells. Virology. 2002;293(1):44–53. doi: 10.1006/viro.2001.1273. [DOI] [PubMed] [Google Scholar]
- 6.Corbel V, Zuprizal Z, Huang C, Arcier JMS, Bonami JM. Experimental infection of European crustaceans with white spot syndrome virus (WSSV) J Fish Dis. 2001;24:377–382. doi: 10.1046/j.1365-2761.2001.00302.x. [DOI] [Google Scholar]
- 7.Dieu BT, Marks H, Siebenga JJ, Goldbach RW, Zuidema D, Duong TP, et al. Molecular epidemiology of white spot syndrome virus within Vietnam. J Gen Virol. 2004;85(Pt 12):3607–3618. doi: 10.1099/vir.0.80344-0. [DOI] [PubMed] [Google Scholar]
- 8.Ding Z, Yao Y, Zhang F, Wan J, Sun M, Liu H, et al. The first detection of white spot syndrome virus in naturally infected cultured Chinese mitten crabs, Eriocheir sinensis in China. J Virol Methods. 2015;220:49–54. doi: 10.1016/j.jviromet.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 9.Edgerton BF. Susceptibility of the Australian freshwater crayfish Cherax destructor albidus to white spot syndrome virus (WSSV) Dis Aquat Org. 2004;59:87–93. doi: 10.3354/dao059187. [DOI] [PubMed] [Google Scholar]
- 10.Flegel TW. Special topic review: major viral diseases of the black tiger prawn (Penaeus monodon) in Thailand. World J Microbiol Biotechnol. 1997;13:433–442. doi: 10.1023/A:1018580301578. [DOI] [Google Scholar]
- 11.Jiravanichpaisal P, Bangyeekhun E, Soderhall K, Soderhall I. Experimental infection of white spot syndrome virus in freshwater crayfish Pacifastacus leniusculus. Dis Aquat Org. 2001;47:15–19. doi: 10.3354/dao047151. [DOI] [PubMed] [Google Scholar]
- 12.Jiravanichpaisal P, Soderhall K, Soderhall I. Characterization of white spot syndrome virus replication in in vitro-cultured haematopoietic stem cells of freshwater crayfish, Pacifastacus leniusculus. J Gen Virol. 2006;87(Pt 4):847–854. doi: 10.1099/vir.0.81758-0. [DOI] [PubMed] [Google Scholar]
- 13.Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20(2):265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo CF, Leu JH, Ho CH, Chen CH, Peng SE, Chen YT, et al. Detection of baculovirus associated with white spot syndrome (WSBV) in penaeid shrimps using polymerase chain reaction. Dis Aquat Org. 1996;25:33–41. doi: 10.3354/dao025133. [DOI] [Google Scholar]
- 15.Marks H, Goldbach RW, Vlak JM, van Hulten MC. Genetic variation among isolates of white spot syndrome virus. Arch Virol. 2004;149(4):673–697. doi: 10.1007/s00705-003-0248-9. [DOI] [PubMed] [Google Scholar]
- 16.Marks H, van Duijse JJ, Zuidema D, van Hulten MC, Vlak JM. Fitness and virulence of an ancestral white spot syndrome virus isolate from shrimp. Virus Res. 2005;110(1–2):9–20. doi: 10.1016/j.virusres.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Mayo MA. A summary of taxonomic changes recently approved by ICTV. Arch Virol. 2002;147:1655–1656. doi: 10.1007/s007050200039. [DOI] [PubMed] [Google Scholar]
- 18.Moser JR, Álvarez DAG, Cano FM, Garcia TE, Molina DEC, Clark GP, et al. Water temperature influences viral load and detection of white spot syndrome virus (WSSV) in Litopenaeus vannamei and wild crustaceans. Aquaculture. 2012;326–329:9–14. doi: 10.1016/j.aquaculture.2011.10.033. [DOI] [Google Scholar]
- 19.OIE . Diagnostic manual for aquatic animal diseases. Paris: Office International des Epizooties (OIE); 2016. [PubMed] [Google Scholar]
- 20.Pradeep B, Rai P, Mohan SA, Shekhar MS, Karunasagar I. Biology, host range, pathogenesis and diagnosis of white spot syndrome virus. Indian J Virol. 2012;23(2):161–174. doi: 10.1007/s13337-012-0079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pradeep B, Shekar M, Karunasagar I, Karunasagar I. Characterization of variable genomic regions of Indian white spot syndrome virus. Virology. 2008;376(1):24–30. doi: 10.1016/j.virol.2008.02.037. [DOI] [PubMed] [Google Scholar]
- 22.Richman LK, Montali RJ, Nichols DK, Lightner DV. A newly recognized fatal baculovirus infection in freshwater crayfish. In: Baer CK, editor. Proceedings American Association of zoo veterinarians, Oct 26–30, Houston, vol 2. 1997. p. 262–264.
- 23.Rout N, Kumar S, Jaganmohan S, Murugan V. DNA vaccines encoding viral envelope proteins confer protective immunity against WSSV in black tiger shrimp. Vaccine. 2007;25(15):2778–2786. doi: 10.1016/j.vaccine.2006.12.056. [DOI] [PubMed] [Google Scholar]
- 24.Shekhar MS, Gopikrishna G. Development of immunodot blot assay for the detection of white spot syndrome virus infection in shrimps (Penaeus monodon) Aquac Res. 2010;41(11):1683–1690. doi: 10.1111/j.1365-2109.2010.02553.x. [DOI] [Google Scholar]
- 25.Sun G, Xiao J, Wang H, Gong C, Pan Y, Yan S, et al. Efficient purification and concentration of viruses from a large body of high turbidity seawater. MethodsX. 2014;1:197–206. doi: 10.1016/j.mex.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan SH, Deng XY, Jiang WM. Effects of high level chromium on antioxidant enzyme system in gill and hepatopancreas of Procambarus clarkii. J Agro-Environ Sci. 2007;23:24–34. [Google Scholar]
- 27.van Hulten MC, Witteveldt J, Peters S, Kloosterboer N, Tarchini R, Fiers M, et al. The white spot syndrome virus DNA genome sequence. Virology. 2001;286(1):7–22. doi: 10.1006/viro.2001.1002. [DOI] [PubMed] [Google Scholar]
- 28.Wang C, Yang H, Yang C, Lu C, Kou G, Lo C. Ultrastructure of white spot syndrome virus development in primary lymphoid organ cell cultures. Dis Aquat Org. 2000;41:91–104. doi: 10.3354/dao041091. [DOI] [PubMed] [Google Scholar]
- 29.Wongteerasupaya C, Pungchai P, Withyachumnarnkul B, Boonsaeng V, Panyim S, Flegel TW, et al. High variation in repetitive DNA fragment length for white spot syndrome virus (WSSV) isolates in Thailand. Dis Aquat Org. 2003;54:253–257. doi: 10.3354/dao054253. [DOI] [PubMed] [Google Scholar]
- 30.Xia X, Yu Y, Weidmann M, Pan Y, Yan S, Wang Y. Rapid detection of shrimp white spot syndrome virus by real time, isothermal recombinase polymerase amplification assay. PLoS ONE. 2014;9:e104667. doi:10.1371/journal.pone.0104667.t001; doi:10.1371/journal.pone.0104667.t002. [DOI] [PMC free article] [PubMed]
- 31.Xie X, Li H, Xu L, Yang F. A simple and efficient method for purification of intact white spot syndrome virus (WSSV) viral particles. Virus Res. 2005;108(1–2):63–67. doi: 10.1016/j.virusres.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Yan D, Dong S, Huang J, Yu X, Feng M, Liu X. White spot syndrome virus (WSSV) detected by PCR in rotifers and rotifer resting eggs from shrimp pond sediments. Dis Aquat Org. 2004;59:69–73. doi: 10.3354/dao059069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.