Abstract
The high-throughput extraction of quantitative information from medical images, known as radiomics, has grown in interest due to the current necessity to quantitatively characterize tumour heterogeneity. In this context, texture analysis, consisting of a variety of mathematical techniques that can describe the grey-level patterns of an image, plays an important role in assessing the spatial organization of different tissues and organs. For these reasons, the potentiality of texture analysis in the context of radiotherapy has been widely investigated in several studies, especially for the prediction of the treatment response of tumour and normal tissues. Nonetheless, many different factors can affect the robustness, reproducibility and reliability of textural features, thus limiting the impact of this technique. In this review, an overview of the most recent works that have applied texture analysis in the context of radiotherapy is presented, with particular focus on the assessment of tumour and tissue response to radiations. Preliminary, the main factors that have an influence on features estimation are discussed, highlighting the need of more standardized image acquisition and reconstruction protocols and more accurate methods for region of interest identification. Despite all these limitations, texture analysis is increasingly demonstrating its ability to improve the characterization of intratumour heterogeneity and the prediction of clinical outcome, although prospective studies and clinical trials are required to draw a more complete picture of the full potential of this technique.
INTRODUCTION
In the past decade, there has been an increasing interest in the high-throughput extraction of quantitative information from medical images [in particular, CT, MR and positron emission tomography (PET) images], known as radiomics.1,2
Radiomic features allow the description of structural heterogeneity of tissues, especially referred to tumours, using quantitative indices derived from statistical and mathematical models applied to the images. The interest in this kind of analysis derives from the need to characterize tumour heterogeneity, known to be a relevant factor in tumour prognosis, using non-invasive tools. It has been already reported in several studies that the heterogeneity highlighted by medical images is well correlated to that observed at histopathological, proteomic and genetic levels, and it is linked to intratumoural properties related to cellularity, angiogenesis and the presence of necrosis.2,3 In this sense, the combination of radiomic analysis with genomic data, the so-called radiogenomics, is increasingly growing in interest in the research community. The main purpose of radiogenomics is to exploit the ability of image-based features in characterizing the whole tumour region, overcoming the problem of sampling errors introduced by genomic analysis. In this context, radiomic features can be used for cross-validation of genomic data, helping to understand possible gene expression or mutation status. Moreover, being not completely related to the genomic profile, radiomics can add new independent information about tissue response to radiation treatment. The combination of these two kinds of information can provide new and more robust diagnostic or prognostic models,1 as reported in Aerts et al.4
Texture analysis plays an important role in assessing the spatial organization of different tissues and organs, overcoming the limits of the classical global measures. In fact, global indices such as the mean CT number or the standardized uptake value (SUV) in PET images describe a region of interest (ROI) as a homogeneous structure, thus neglecting its spatial organization.
Therefore, texture analysis has been widely explored in the radiotherapic context, especially for the characterization of tumour in the planning phase and for the prediction of response to treatment.3,5–9 Despite the fact that it seems a promising technique, it has not been completely exploited, since some possible limitations, especially regarding factors that can reduce its repeatability and robustness, need to be explored in more detail.
This review focuses on the use of texture analysis in radiotherapy (RT). Initially, we will discuss textural feature definitions and factors affecting their calculation. Subsequently, we review recent applications of texture analysis within RT.
OVERVIEW ON TEXTURE ANALYSIS APPROACHES
Texture analysis is related to a variety of mathematical techniques that can evaluate the grey-level intensity distribution and spatial organization in an image.3 Textural features can be computed by different methods, which can be divided into three main groups:
Statistical methods: they describe the distribution and relationships of grey-level values in the image, by the computation of histogram and second- and higher-order texture matrices;10–14
Model-based methods: they are based on the most sophisticated and complex mathematical models, including fractals, computed using different algorithms;15–23
Transform-based methods: they analyse texture in a frequency or scale domain, including Fourier transform, wavelet and Gabor filters and Laplacian transform of Gaussian filter.3 This last class allows transformation of the whole image, on which it is possible to extract statistical features on a multiscale approach.
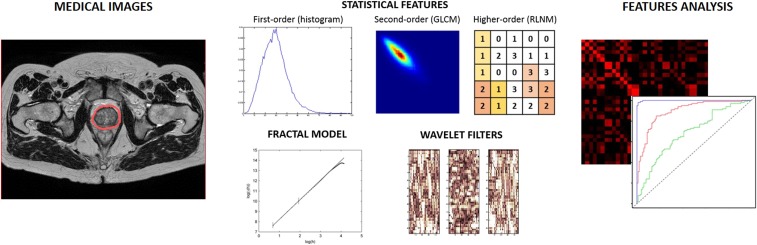
Among these indices, second-order features calculated from the grey-level co-occurrence matrix (GLCM) are the most used due to their simple computation and interpretation. The GLCM represents the joint probability density function of the co-occurrence of pixels with an intensity level i and an intensity level j in a certain direction at a specified distance.10 A typical example of texture analysis workflow is reported in Figure 1, where after the identification of the ROI on the image, features calculated using the three methods (statistical, model based and transform based) are estimated for further statistical analysis to infer physiopathological descriptors. The complete list of the main features and their texture interpretation is reported in Table 1. A more detailed description about their computation can be found in previous reviews.3,5,24
Figure 1.
Texture analysis procedure from tissue identification on medical images to features extraction (here in terms of first-, second- and higher-order statistical features, fractal dimension and wavelet filters) and analysis (such as cross-correlations matrix, receiver operating characteristic curves).
Table 1.
List of the main texture analysis methods
| Class | Type | Method | Texture interpretation | Main estimated features |
|---|---|---|---|---|
| Statistical method | First order | Grey-level histogram | Global distribution of intensity values in terms of spread, symmetry, flatness, uniformity and randomness | Mean |
| Variance | ||||
| Skewness | ||||
| Kurtosis | ||||
| Energy | ||||
| Entropy | ||||
| Second order | GLCM10 | Spatial relationship between pixel in a specific direction, highlighting the properties of uniformity, homogeneity, randomness and linear dependency of the image | Energy | |
| Homogeneity | ||||
| Contrast | ||||
| Entropy | ||||
| Dissimilarity | ||||
| Correlation | ||||
| Higher order | NGTDM11 | Spatial relationship among three or more pixels, closely approaching the human perception of the image | Complexity | |
| Busyness | ||||
| Contrast | ||||
| Coarseness | ||||
| Texture strength | ||||
| Higher order | GLRLM12 | Texture in a specific direction, where fine texture has more short runs whilst coarse texture presents more long runs with different intensity values | Short-run emphasis | |
| Long-run emphasis | ||||
| Grey-level non-uniformity | ||||
| Run length non-uniformity | ||||
| Run percentage | ||||
| Higher order | GLSZM13 | Regional intensity variations or the distribution of homogeneous regions | Zone size emphasis | |
| Grey-level zone emphasis | ||||
| Zone size non-uniformity | ||||
| Grey-level non-uniformity | ||||
| Model-based method | Fractal models | Box counting,15–20 fractional Brownian motion method21,22 and power spectral method21,23 | Complexity of the image. For 2D images, FD ranges from 2 to 3, where more complex pattern presents higher values | FD |
| Fractal abundance | ||||
| Lacunarity | ||||
| Transform-based method | Fourier transform | Analysis of the frequency content
without spatial localization |
||
| Wavelet and Gabor filters | Frequency and spatial
localization |
|||
| LoG | Extraction of areas with increasingly coarse texture patterns | |||
2D, two-dimensional; FD, fractal dimension; GLCM, grey-level co-occurrence matrix; GLRLM, grey-level run-length matrix; GLSZM, grey-level size zone matrix; LoG, Laplacian transform of Gaussian filter; NGTDM, neighbourhood grey-tone difference matrix.
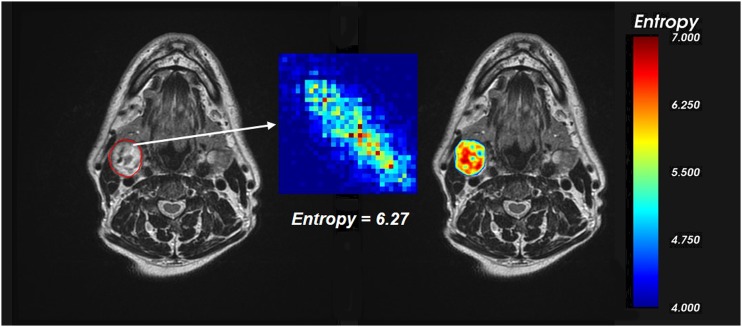
Textural features can be estimated by calculating a single value for the whole ROI or by building a texture map. In the first case, the ROI-based approach provides a unique index for the description of the texture in the whole structure. This is generally performed when there is the need to extract synthetic parameters that can be used for further statistical analysis. In Figure 2 (left), the GLCM is built considering the whole ROI, and it provides a single value for each feature, for example, the entropy, indicating the general organization of the structure. In the second case, the pixel-wise approach allows the estimation of textural features in each pixel, by computing their values considering a square mask (e.g. 5 × 5 pixels) sliding over the image. This is particularly useful in segmentation problems, or for pixel-based statistical analysis, such as correlations with dose maps. Figure 2 (right) shows the map of entropy calculated from a GLCM built on a 5 × 5 mask sliding on the ROI. Here, it is evident that entropy within the region is not uniform and subregions with high heterogeneity can be easily individuated.
Figure 2.
Estimation of entropy from grey-level co-occurrence matrix in a cervical lymph node (in red) using a region of interest-based approach (left) and a pixel-wise approach (right). Colours appear only in the online version.
FACTORS INFLUENCING TEXTURAL FEATURES ESTIMATION
The computation of textural features can be influenced by general variables, such as the ROI identification, or by artefacts specific for the considered imaging modality. A schematic overview of these factors and the works that have studied their impact on features estimation is reported in Table 2.
Table 2.
List of factors influencing textural features estimation
| Factor type | Factor details | References (first author, year) |
|---|---|---|
| General factors | ||
| Features computation | Grey-level discretization | Orlhac et al, 201426; Hatt et al, 201527 |
| Isotropic resampling of the image | Vallières et al, 201528 | |
| Non-standardized nomenclature | Hatt et al, 20169 | |
| Directionality in texture matrices | Hatt et al, 201527 | |
| Specific parameters for texture matrix formulation | Hatt et al, 20169 | |
| Use of already available packages | Nyflot et al, 201534; Hatt et al, 20169 | |
| ROI identification | Manual vs automatic contouring | Orlhac et al, 201426; Parmar et al, 201436 |
| Image registration and contour propagation | Yip and Aerts, 201625; Cunliffe et al, 201238; Cunliffe et al, 201339 | |
| Image-specific factors | ||
| CT images | Presence of metal artefacts | Leijenaar et al, 201541 |
| Noise and blurring | Veenland et al, 199623; Veenland et al, 199843 | |
| Image reconstruction algorithms and contrast injection on CECT | Kim et al, 201644; Yang et al, 201645; He et al, 201646 | |
| PET images | Image acquisition and reconstruction protocols | Galavis et al, 201047; Nyflot et al, 201534; Yan et al, 201548; Lasnon et al, 201649; van Velden et al, 201650 |
| Patient and lesion size | Hatt et al, 201527; Nyflot et al, 201534 | |
| Image smoothing and quantization | Doumou et al, 201551; Lu et al, 201652; Desseroit et al, 201653 | |
| SUV discretization | Leijenaar et al, 201554 | |
| Pre-processing step | Hatt et al, 201355 | |
| Choice of respiration-averaged CT for attenuation correction | Cheng et al, 201656; Grootjans et al, 201657 | |
| MR images | Image acquisition protocols | Mayerhoefer et al, 200959 |
| Robustness of parametric maps | Song et al, 201464 | |
CECT, contrast-enhanced CT; PET, positron emission tomography; ROI, region of interest; SUV, standardized uptake value.
Features computation
There is no acknowledged standardization for textural features computation, which can be affected by different factors:
Grey-level discretization: One of the first choices about features computation is the number of bins used for the discretization of the image. In fact, images are generally acquired with a high number of intensity values (of about 10–12 bits), which should be reduced to a more practical level for computational reasons. For instance in PET images, it is common practice to discretize the intensity range to 2N values, where N ranges from 3 to 8.25 Recently, it has been recommended to have at least 32 discrete values26 but not >64, since it was proven that a higher number of bins do not provide additional prognostic information.27
Isotropic resampling of the image: for the three-dimensional (3D) computation of textural features, in particular, the second- and higher-order features, an isotropic resampling of the image is recommended. This was explained by the fact that the first-order features are computed from the histogram, which counts the number of grey levels in 3D space, and that all higher-order texture measurements explicitly or implicitly involve a distance parameter in the matrix computation.28
Non-standardized nomenclature: there is no standard nomenclature for these indices and thus it is possible that the same parameter in different works referred to different methods. It is the case, for example, for the names “entropy” and “energy” which can refer both to the histogram- or matrix-based parameters. Therefore, Hatt et al9 proposed to indicate the name of the matrix in the feature nomenclature (“entropyHIST” and “entropyGLCM”).
Directionality in texture matrices: regarding GLCM and grey-level run-length matrix (GLRLM) features, which are based on the computation of directional matrices, the choice of directions and whether or not to average them can have an impact on the final result.25,27 For example, some works have treated the GLCMs as separated in each direction,29–31 whereas others have preferred to average the matrices to reduce the number of parameters used in the subsequent statistical analysis.28,32
Specific parameters for texture matrix formulation: the choice of the distance between pixels for the computation of GLCM is another factor that can affect the value and number of the features. However, in this case, it is general practice to consider a distance of 1 pixel,9 even if some works preferred to take into account more than one distance.31,33
Use of already available packages: the availability of different software for the computation of textural features makes texture analysis more appealing and easy to use. However, this leads to doubting if all the considered features were correctly computed. These software, being closed or open source, lack standardization of features selection and computation. Moreover, commercial or freely available tools, such as Mazda (available from: https://www.eletel.p.lodz.pl/mazda/) or the FracLac plugin (Bethesda 2.5, Release 1e; developed by A Karperien, Charles Sturt University, Australia) for ImageJ software (NIH, Bethesda, MD), have the limitation of being “black-box”; thus, it is difficult to know how the features are estimated. This different features calculation can lead to very different results, which can make the comparison of published works unreliable.34 An excellent evaluation and comparison of different available codes for textural features computation can be found in Hatt et al.9
Region of interest identification
One of the main factors influencing textural indices estimation is the identification of the ROI,1,26,35,36 since features depend on the segmented volumes. Moreover, many structures, especially tumours, have indistinct borders and there is no widely and universally acknowledged ground truth. In this context, Parmar et al36 tested the reproducibility of several textural features (shape, histogram, GLCM and GLRLM based) by considering manual and semiautomatic segmentations of the primary tumour in non-small-cell lung cancer (NSCLC) on CT images. They reported a general higher robustness of features extracted from the semi-automatic segmentation, and in fact, it had a higher reproducibility and stability with respect to manual segmentations performed by different observers. In particular, they found that only shape-based features were not significantly different between the two segmentations, whereas histogram and GLCM- and GLRLM-based indices depended on the accuracy of the definition of tumour borders and irregularities, more easily delineated in the automatic way. Similar results were found by Orlhac et al26 in fluorine-18 fludeoxyglucose PET images of patients with colorectal cancer. They compared two different widely clinically accepted tumour segmentation methods and analysed the dependence of textural features from these methods. They found that the more affected indices were histogram-based and GLRLM-based features, whereas the more robust were homogeneity and entropy estimated from GLCM.
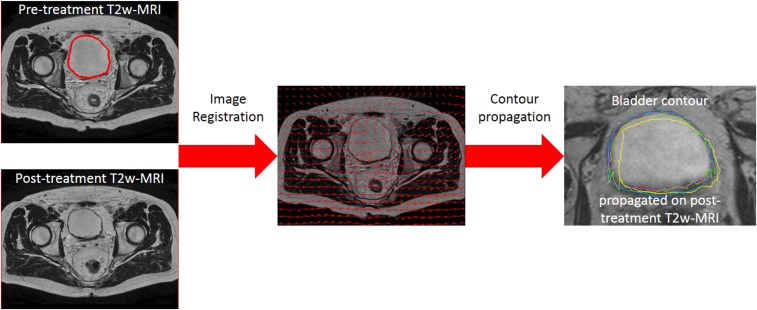
Another relevant role in the ROI identification is played by image registration and contour propagation, as highlighted in Figure 3. In fact, deformable image registration is required to recover growth or shrinkage of structures due to disease progression or radiation effects during treatment, and thus, it is the first step for an accurate contour propagation. If the chosen deformable image registration method is unreliable, texture analysis performed in the automatically propagated volumes can introduce several errors.37 In thoracic CT images, it seems that demons algorithm was more stable with respect to rigid, affine and B-spline deformable registration, leading to more reproducible and robust textural features.38,39 In PET images of patients with oesophageal cancer, Yip et al37 tested a rigid registration and ten different deformable image registration methods for the estimation of textural features in the automatically deformed tumour volumes. They found that the power of features in predicting the final tumour response to treatment was not significantly different for the most part of the deformable image registration methods, except for the fast demons and fast free-form. They concluded that as long as contour propagation accuracy was reasonably good, the predictive power of textural features was robust with respect to image registration.
Figure 3.
Effect of image registration and contour propagation on region of interest identification. Pelvic T2 weighted (T2w)-MR images acquired before and after radiotherapy were co-registered and bladder contour was automatically propagated using four different algorithms, resulting in four different bladder contours, represented by different colours (on the right). Colours appear only in the online version.
Not only the choice of the image registration method has an impact on the contour propagation but also the algorithm used to apply the estimated vector field on the delineated contours. In fact, the deformation field can be applied directly on the binary mask or on a 3D surface generated from the contour. This second choice has the advantage of obtaining a more regular deformed surface due to the intrinsic 3D properties of the mesh. Different algorithms for the 3D surface generation are available, even in commercial and clinical software, giving different performance that can affect the final deformed volumes.40
CT images
Some specific artefacts and source of errors are typical of the imaging modality. In CT images, especially in the head and neck region, textural features can be affected by the presence of metal streaks due to dental fillings.41 There is the possibility to reduce metal artefacts using some promising techniques,42 but as texture analysis relies on extracting meaningful information from medical images, they could introduce artificial texture by modifying image information.
The presence of noise and blurring due to acquisition parameters can also affect texture analysis.23,43 As expected, the more noisy and blurred the image is, the less robust are the features. Therefore, a homogeneous image acquisition protocol is suggested in order to reduce possible errors due to these limitations.
The impact of reconstruction algorithms in contrast-enhanced CT (CECT) was assessed by Kim et al44 in 42 patients with lung tumours. Shape, histogram and GLCM-based features were calculated, and they found that the most robust indices were shape-based metrics and homogeneity and entropy from GLCM, being not significantly different and showing low variability between the two image-reconstruction algorithms.
Regarding CECT images, the dependency on the time after contrast injection and the feature reproducibility between scans were assessed again on quantitative imaging features extracted from patients with lung cancer, finding little relationship between these variables.45 However, the authors claimed that their study was limited in the number of estimated features and in not considering the amount of contrast and therefore their results can be considered only representative. A larger data set (240 subjects) of patients with solitary pulmonary nodule was assessed by He et al46 for the evaluation of the diagnostic performance of textural features in differentiating benign and malignant solitary pulmonary nodule. About 150 features (from histogram and GLCM) were calculated at different scales, using a Laplacian of Gaussian spatial band-pass filter. The effects of contrast enhancement, reconstruction slice thickness and convolution kernel were evaluated, and it was found that non-contrast, thin-slice and standard convolution kernel-based CT images were more informative.
Positron emission tomography images
Texture analysis in PET images has reached great interest in the past few years. However, these images present different factors that can strongly affect textural features estimation, more than other imaging modalities. The impact of these factors, especially image acquisition and reconstruction, has been addressed in several recent studies.
One of the first studies that assessed the effect of PET image acquisition and reconstruction on textural features found that among 50 different features, only entropy (estimated from histogram), energy and maximal correlation coefficient (estimated from GLCM), and low grey-level run emphasis (estimated from GLRLM) presented variations <5%.47 Next, Nyflot et al found that skewness of the intensity histogram, autocorrelation and cluster prominence of the GLCM, contrast, complexity, strength of the neighbourhood grey-tone difference matrix (NGTDM), and the grey-zone emphasis subset of the GLRLM showed large variation despite no change in the underlying ground truth image.34 Yan et al48 analysed the effect of reconstruction settings, especially relating to time-of-flight and point-spread function modelling, finding that the iteration number and full width at half maximum of the Gaussian filter have a similar impact on the image features, whereas grid size has a larger impact. The features that exhibited large variations such as skewness, cluster shade and zone percentage should be used with caution, whereas the most robust features were similar to those found in other works.47,49 Finally, a very recent work of van Velden et al50 investigated the impact of reconstruction and delineation on features reproducibility in 11 NSCLC fluorine-18 fludeoxyglucose PET/CT studies. 105 radiomic features were calculated on 19 ROIs, delineated twice—once on CT and once on PET images—and considering two different image reconstruction methods. 63 features showed high values of reproducibility. Moreover, it seems that changes in delineation had a higher impact than changes in reconstruction.
Other factors were also assessed, such as image-acquisition parameters,34,47 patient and lesion size,27,34 image smoothing and quantization,51–53 SUV discretization,54 pre-processing steps55 and the choice of respiration-averaged CT for attenuation correction.56,57 The impact of tumour delineation on textural features computation was also assessed in PET images.50–52,55 Finally, an attempt of interpreting PET textural indices was carried out in a very recent work, where heterogeneity quantified by texture analysis was investigated on simulated patient data.58 Here, the authors found that homogeneity, entropy, short-run emphasis and long-run emphasis were sensitive to the presence of uptake heterogeneity, whereas high grey-level zone emphasis and low grey-level zone emphasis were mostly sensitive to the average uptake. All these indices were sensitive to the pixel size and edge effects.
MR images
Texture analysis on MR images has not been completely exploited,25 and in fact, the repeatability of MR-based textural features has been investigated in very few studies. The effect of image acquisition parameters was assessed by Mayerhoefer et al59 on phantom images considering different textural features, and their results showed that all features were increasingly sensitive to acquisition parameter variations with increasing spatial resolution. Nevertheless, as long as the spatial resolution was sufficiently high, these variations had little effect on pattern discrimination. In addition, GLCM-based features were demonstrated to be superior to other indices.
The limitation of the non-quantitative value of pixel intensities in MR images can be reduced by the application of some pre-processing steps, as for example, the correction of magnetic field inhomogeneities60,61 and the intensity normalization across intersubject or intrasubject acquisitions.62
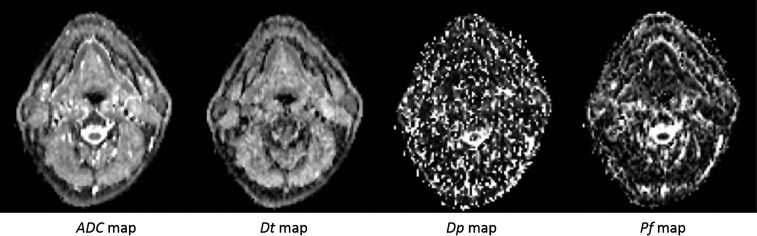
MR images can be used for the estimation of quantitative parameters that describe functional properties. In particular from diffusion-weighted MRI (DW-MRI) and dynamic contrast-enhanced MRI (DCE-MRI), parametric maps can be derived by the estimation of these indices pixel-by-pixel, using model-fitting algorithms. Texture analysis of these maps has been introduced in some recent works to assess tissue heterogeneity from the diffusion and perfusion point of view.63–65 However, the reliability of textural features calculated on these maps depends on the robustness of fitting. This is a research area which is just starting to be explored; only few works assessed the relationship between the reproducibility of parametric maps estimated from DW-MRI and DCE-MRI and the reliability of textural indices. In particular, Song et al64 studied the reproducibility of histogram and texture parameters derived from intravoxel incoherent motion66 DW-MRI. In this model, from DW-MR images acquired at different b-values, it is possible to derive maps of apparent diffusion coefficient (ADC), true diffusion coefficients (Dt), pseudodiffusion coefficient (Dp) and perfusion fraction (Pf). It has been reported that ADC and Dt maps are more robust than Dp and Pf maps67 (Figure 4), and consequently, histogram and texture parameters derived from ADC and Dt maps were confirmed to be more reproducible than those from Dp and Pf maps. Understanding the stability of textural features derived from parametric maps deserves more attention and further investigations.
Figure 4.
Perfusion maps derived from intravoxel incoherent motion images. From left to right: apparent diffusion coefficient (ADC), true diffusion coefficient (Dt), pseudodiffusion coefficient (Dp) and perfusion fraction (Pf). The ADC and Dt maps are more robust and less noisy than Dp and Pf maps.
From all these studies, it can be highlighted that there is a strong heterogeneity in the computation of textural features and that the results are mostly dependent on different factors. In some works, some features resulted to be more stable and robust with respect to others, but the same features can show an opposite behaviour in other studies. Therefore, it is difficult to obtain a clear message due to the difference in parameter settings and in the metrics used for variability estimation. Nevertheless, it is important to pursue this trend and this kind of analysis to better understand the reliability of texture analysis, which is becoming more and more appealing in this context.
APPLICATION OF TEXTURE ANALYSIS IN RADIOTHERAPY
In the context of RT, one of the first problems addressed by texture analysis was the characterization and identification of tumoural lesions. This issue, which was extensively explored within the oncological context,30,68–71 had the primary aim to improve radiation targeting in the planning phase. The automatic organs outline for RT planning of prostate cancer using texture analysis on CT images was primarily faced by Nailon et al.29 Next, Yu et al72,73 combined the analysis of PET and CT images in patients with head and neck cancer for the estimation of textural features, demonstrating its potential in improving the accuracy of gross tumour volume identification. CT coarseness, CT business and PET coarseness calculated from NGTDM have shown better results in the discrimination of normal and abnormal tissues compared with those of an expert observation and with mean CT intensity and mean SUV. Moreover, using the maps generated with these textural features, an automatic classifier performed a pixelwise radiation targeting, with an accuracy similar to manual contours.
Currently, the main application of texture analysis is focused on the assessment of the response of tumour and organs at risk to treatment to identify possible biomarkers, which could predict how the tissues will respond to radiations. This was highlighted in some recent reviews,5,7,74,75 and some of the latest results in the field are schematically summarized in Table 3 and discussed in the following sections.
Table 3.
Studies who have applied texture analysis in the context of radiotherapy
| Radiotherapic aim | District | Imaging modalities | Treatment type | References (first author, year) |
|---|---|---|---|---|
| Radiation targeting in RT planning | Head and neck | PET/CT | IMRT | Yu et al, 200972,73 |
| Prostate | CT | IMRT | Nailon et al, 200829 | |
| Tumour response to treatment | Lung | PET | SABR | Pyka et al, 201582 |
| PET | CRT | Cook et al, 201383 | ||
| CT | SABR | Huynh et al, 201685; Mattonen et al, 201479; Mattonen et al, 201580; Mattonen et al, 201681; | ||
| CT | CRT | Coroller et al, 201684 | ||
| Oesophagus | PET | CRT | Tixier et al, 201186; Nakajo et al, 201687; Yip, et al 201637 | |
| CT | CRT | Yip et al, 201488 | ||
| Head and neck | PET | CRT | El Naqa et al, 200989 | |
| mp-MRI | CRT | Liu et al, 201690; Scalco et al, 201691 | ||
| DCE-MRI | IMRT | Jansen et al, 201663 | ||
| Prostate | T2w-MRI | EBRT | Gnep et al, 201694 | |
| Rectum | PET | CRT | Bundschuh et al, 201495 | |
| T2w-MRI | CRT | De Cecco et al, 201596 | ||
| mp-MRI | CRT | Nie et al, 201697 | ||
| Brain | MRI | SRT | Nardone et al, 201699 | |
| Soft-tissue sarcoma | CT | CRT | Tian et al, 201598 | |
| Radiation-induced effects on normal tissue | Lung | CT | SBRT | Mattonen et al, 201479; Mattonen et al, 201580; |
| CT | Oesophageal RT | Cunliffe et al, 2015100 | ||
| Parotid glands | Ultrasound | Head–neck RT | Yang et al, 2012101 | |
| CT | IMRT | Scalco et al, 2013102; Scalco et al, 2015104; Pota et al, 2015103 |
CRT, chemoradiotherapy; DCE-MRI, dynamic contrast-enhanced MRI; EBRT, external beam radiotherapy; IMRT, intensity-modulated radiotherapy; mp-MRI, multiparametric MRI; PET, positron emission tomography; RT, radiotherapy; SABR, stereotactic ablative radiation therapy; SRT, stereotactic radiotherapy; T2w, T2 weighted.
Tumour response to treatment
Effects of radiations in lung region in NSCLC is one of the most studied subject in this context, as reported in a very recent review.75 In fact, the evaluation of treatment response is complicated by the possible presence of radiation-induced lung injuries (RILIs), such as radiation pneumonitis and fibrosis, which appear as an increase in lung density on CT, similar to tumour recurrence.6 Although most parts of the works in this context faced the detection of RILI by the evaluation of simple CT density,76–78 Mattonen et al79–81 proposed texture analysis for an automatic classification of tumour recurrence and lung injuries. They found a radiomic signature consisting of five textural features (minimum grey level, grey-level uniformity, GLCM homogeneity, GLCM correlation and GLCM energy) after stereotactic ablative radiation therapy (SABR) in consolidative and periconsolidative regions. With this combination of features, they could predict recurrence within 6 months post-SABR with an error of 24%, a false-positive rate of 24.0% and a false-negative rate of 23.1% on a data set of 45 patients. They compared these results with performance of three radiation oncologists and three radiologists (mean error of 35%, false-positive rate of 1% and false-negative rate of 99%), suggesting that texture analysis can detect early changes associated with local recurrence that are not typically considered by physicians.81 Other works focused on prediction of lung recurrence. For example, from PET images, it was found that heterogeneity measures, such as entropy, can predict disease-specific survival,82,83 whereas CT images were used for the assessment of pathologic response,84 overall survival and distant metastases,85 finding that texture analysis can outperform conventional indices (as tumour volume and diameter).
The response of oesophageal cancer to chemo-RT (CRT), classified as complete responder, partial responder and non-responder, was assessed on PET images using textural features from histogram, GLCM, NGTDM and GLRLM,86 improving the stratification performance obtained by simple SUV measures. Other more recent works explored the potential of texture analysis on PET images for the prediction of tumour response to CRT on patients with oesophageal cancer. Nakajo et al87 found that intensity variability and size-zone variability were higher in patients who were non-responders than responders. Yip et al37 studied the impact of different image registration methods for the definition of tumour volume after CRT on the estimation of several textural features. The variation of these features after the treatment was then used to predict tumour response. They found that variations in short run emphasis remained predictive of pathologic response for all image registration algorithms used for contour propagation. In a previous study,88 the same group considered a combination of textural and morphological properties, finding that lower post-treatment intratumoural heterogeneity on CECT images was associated with improved overall survival time in patients who completed definitive CRT for primary oesophageal cancer. Moreover, they observed that the combination of pre-treatment entropy, uniformity and proportional difference in entropy with maximal wall thickness changes performed better than morphological indices alone in predicting overall survival.
For the prediction of response of head and neck tumours, histogram-based features, and entropy and homogeneity from GLCM estimated on PET images were able to capture tissue heterogeneity and to predict the overall survival. However, the necessity for a standardized image acquisition and reconstruction protocol and accurate target delineation was pointed out.89 Focusing on multiparametric models, two recent works tried to predict tumour and lymph node response of nasopharyngeal cancer to CRT. In particular, Liu et al proposed a model, composed of second- and higher-order features and Gabor filters extracted from T1 weighted MRI, T2 weighted (T2w) MRI and DW-MRI, which could reach an accuracy >90%.90 Scalco et al91 evaluated the combination of first- and second-order features and fractal dimension extracted from T2w-MRI with ADC estimated from DW-MRI, for the characterization of cervical lymph nodes. In this case, pre-treatment ADC combined with pre-treatment fractal dimension classified responder and non-responder lymph nodes with an accuracy of 82%, highlighting higher values of ADC and lower values of fractal dimension in non-responders. Quantitative maps derived from dynamic contrast-enhanced images and acquired during treatment were instead considered for the calculation of heterogeneity features, which demonstrated their ability in predicting the response of head and neck squamous-cell carcinoma, highlighting a reduction in heterogeneity during CRT.63 In this work, they did not find any correlation between pre-treatment features and the final outcome, contrary to the results reported by other groups about the use of textural features from parametric perfusion maps. In fact, in these studies, it was found that heterogeneity measures at baseline, such as coherence and fractal dimension, can be predictive of the final response for limb92 and colorectal cancer.93
The application of texture analysis on MRI images was also proposed in the pelvic region for the prediction of prostate recurrence after external beam RT, based on T2w-MRI and ADC maps.94 74 patients were analysed and textural features, shape and volume indices were estimated within the prostate tumour. They found significant correlations between tumour recurrences and some GLCM parameters (as contrast and difference variance) calculated on T2w-MRI, whereas ADC features were poorly associated with biochemical recurrence. This was explained by the different spatial resolution of the two images: T2w-MRI offers a higher resolution, and thus, it is richer in texture information.
Regarding rectal cancer, tumour response to CRT was firstly evaluated using PET/CT images acquired before, during and after the treatment in 27 patients. It was reported that tumour heterogeneity assessed by the coefficient of variation was superior to the conventional parameters and was an important predictive factor of the pathologic response.95 Two more recent studies evaluated the performance of texture analysis on MR images; in particular, De Cecco et al96 considered textural features estimated on T2w-MRI acquired before and at mid-treatment, whereas Nie et al97 evaluated multiparametric MRI (T1 weighted-MRI, T2w-MRI, DW-MRI and DCE-MRI) acquired before CRT. They both concluded that radiomic analysis performed on MRI images may improve the prediction of the pathologic response of rectal cancer to treatment. The pathologic response prediction, assessed on CECT images using texture analysis, was also improved in patients affected by soft-tissue sarcoma, with respect to classical indices, such as tumour size, density and perfusion.98
Finally, the assessment of prognostic value of MRI texture analysis in brain NSCLC oligometastases undergoing stereotactic irradiation was explored by Nardone et al99 to help drive the best treatment in these patients.
Radiation-induced effects on normal tissues
A less explored application of texture analysis concerns the prediction of radiation-induced effects on organs at risk. In this context, two organs were mostly considered: lungs in the treatment of oesophageal cancer and NSCLC and parotid glands in the treatment of head and neck squamous-cell carcinoma.
Radiation pneumonitis is one of the drawbacks of lung irradiation, and it was investigated by Cunliffe et al100 on patients affected by oesophageal cancer, considering CT images acquired before and after RT. A relationship between dose and texture variations was observed, and in particular, these changes were significantly related to radiation pneumonitis development in 12 features, consisting of the first and second order, fractal and Law's filter parameters. As previously described, Mattonen et al79,80 developed a method based on texture analysis on the consolidative and ground-glass opacity regions to automatically differentiate tumour recurrence and RILI. At 2–5 months after SABR, they found an increase in energyGLCM and a decrease in entropyGLCM in lung injuries with respect to local recurrence.
A side effect of the irradiation of head and neck tumours is represented by the shrinkage of parotid glands and the development of toxicity, such as xerostomia. The early prediction of these events can be of interest both to better plan radiation treatment, by considering anatomical variations of the glands and to identify those patients who can benefit from personalized care. In this case, echographic101 and CT102−104 images of the parotid tissue were examined and GLCM features and fractal dimension were calculated. On echographic images, an increased entropy was observed with respect to normal glands, whereas on CT images, a decrease in mean CT density, entropy and fractal dimension was found in the first half of RT. These results, apparently discordant, were concordant in highlighting a less-complex tissue organization of the glands due to the loss of acinar cells after irradiation. Moreover, the addition of textural features to the prediction models has improved the accuracy with respect to the simple volume measure.
LIMITATIONS OF TEXTURE ANALYSIS
Although texture analysis is increasingly adopted in the context of RT, it remains quite a novel technique, not completely explored and still having a lot of open challenges. One of the main limitations is associated with the study design. In fact, texture analysis has been used in most part in retrospective studies, needed in this first phase to provide proof of concept for further investigations.25 Retrospective analyses lack complete control on data acquisition and management, which can negatively affect reproducibility and robustness of results. A rigorous study design should foresee a prospective analysis in which an optimal protocol can be planned in terms of data acquisition and management and a further model validation can be provided.
Regarding data acquisition, image acquisition and reconstruction protocols should be clearly defined and standardized along the study, as previously discussed. Moreover, in this phase, the sample size should be defined by considering the high number of image-based features that can be estimated from the acquired images. In general, this issue has not been correctly faced, since the number of features is often greater than the number of considered patients, and consequently, a high risk of false-positive discovery rate can be easily found, as recently highlighted by Yip and Aerts25 and deeply discussed in Chalkidou et al.105
Regarding the statistical data analysis, it is evident that texture analysis can provide a very complex and large set of data, which can present high correlations among them. It is thus necessary to reduce the number of features to avoid the risk of overfitting analysis and to build a classifier or prediction model. There is no consensus about the unsupervised approach to obtain the best results.1 In a recent work of Parmar et al,106 a large panel of machine-learning approaches for radiomics-based survival prediction was investigated, considering both features selection and classification methods. Their variability analysis pointed out that the choice of the classification method has a higher impact on performance variation in predicting the overall survival (>30% of the total variation) with respect to the choice of features selection method (only 6% of variation). Another factor that can impact on the classification performance is the choice of the optimal cut-off to stratify patients into a binary model.105 This choice is highly dependent on the studied data set, and thus it is difficult to be applied in external populations.25
Another issue that should be faced during the study design phase is how to generalize and validate the radiomic signature found in the assessed patients population. Validation on an external cohort of patients should be considered as the gold standard, as recently reported in some works. In fact, these studies reported high values of accuracy, considering the radiomic signature estimated in a selected population and applied on an independent one from another institute, or from another body region, reporting high values of accuracy.4,41,107 If this kind of validation is not feasible, some other evaluations should be performed, for example, using test and training sets, which should be taken from a separated cohort or at least considering leave-one-out cross-validation.9
Even if texture analysis was carried out in the most rigorous way and if significant models that could predict tissue response to radiation or provide a characterization of tumour heterogeneity were found, the biological interpretation of these parameters remains one of the major questions about texture analysis. In fact, up to now, the biological origin of the features is still not completely understood, as reported in the review of Alobaidli et al.5 In this context, some studies try to assess possible correlations between texture analysis and genetic or proteomic data4,108,109 or between texture and histological image analysis.30 In particular, it was found that pathophysiological properties determined by genomic analysis were significantly related to texture analysis on CT,4 PET109 and DCE-MRI images,108 suggesting that heterogeneity features were strongly correlated with cell cycling pathways.
CONCLUSION
As highlighted in this review, despite its drawbacks and limitations, there is growing interest in texture analysis, even in the context of RT. What is emerging from the high number of studies in this field is that the major limitation of this approach resides in the quite discordant results, the non-standardization of their protocols and the lack of prospective studies. In this sense, future developments should be addressed towards a general uniformity in the image-processing workflow, with particular attention to image acquisition and reconstruction protocols and textural features computation. The use of different software (commercial, open-source or “in-house”) also requires careful attention as they may vary in the manner in which the features are calculated, and thus they could lead to different results.
Future directions about texture analysis should be addressed towards prospective studies and clinical trials to validate the radiomic signature found in previous works and to prove the true potential of texture analysis in clinical RT practice. At the same time, the knowledge of texture interpretation would be essential to improve the quality of this kind of analysis. In this way, texture analysis can better demonstrate its ability to improve the characterization of intratumoural heterogeneity and the prediction of clinical outcome to obtain robust and reliable image-based biomarkers for a real patient-specific treatment care.
Acknowledgments
ACKNOWLEDGMENTS
The authors thank the Radiology and Radiotherapy Department of Regina Elena National Cancer Institute, Rome, Italy; the Prostate Cancer Program of Fondazione IRCCS Istituto Nazionale dei Tumori, Milan, Italy; and the Department of Radiology of ASST degli Spedali Civili di Brescia, Brescia, Italy, for providing MRI images.
Contributor Information
Elisa Scalco, Email: elisa.scalco@ibfm.cnr.it.
Giovanna Rizzo, Email: giovanna.rizzo@ibfm.cnr.it.
FUNDING
The work was partially funded by “Ministero degli Affari Esteri e della Cooperazione Internazionale, Direzione Generale per la Promozione del Sistema Paese (MAECI–DGSP)”.
REFERENCES
- 1.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016; 278: 578–84. doi: https://doi.org/10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012; 48: 441–6. doi: https://doi.org/10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davnall F, Yip CS, Ljungqvist G, Selmi M, Ng F, Sanghera B, et al. Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging 2012; 3: 573–89. doi: https://doi.org/10.1007/s13244-012-0196-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Cavalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014; 5: 4006. doi: https://doi.org/10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alobaidli S, McQuaid S, South C, Prakash V, Evans P, Nisbet A. The role of texture analysis in imaging as an outcome predictor and potential tool in radiotherapy treatment planning. Br J Radiol 2014; 87: 5–14. doi: https://doi.org/10.1259/bjr.20140369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mattonen SA, Ward AD, Palma DA. Pulmonary imaging after stereotactic radiotherapy—does RECIST still apply? Br J Radiol 2016; 89: 20160113. doi: https://doi.org/10.1259/bjr.20160113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu W, Wang J, Zhang HH. Computerized PET/CT image analysis in the evaluation of tumour response to therapy. Br J Radiol 2015; 88: 20140625. doi: https://doi.org/10.1259/bjr.20140625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chicklore S, Goh V, Siddique M, Roy A, Marsden PK, Cook GJ. Quantifying tumour heterogeneity in 18F-FDG PET/CT imaging by texture analysis. Eur J Nucl Med Mol Imaging 2013; 40: 133–40. doi: https://doi.org/10.1007/s00259-012-2247-0 [DOI] [PubMed] [Google Scholar]
- 9.Hatt M, Tixier F, Pierce L, Kinahan PE, Le Rest CC, Visvikis D. Characterization of PET/CT images using texture analysis: the past, the present… any future? Eur J Nucl Med Mol Imaging 2017; 44: 151–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybernet 1973; 3: 610–21. doi: https://doi.org/10.1109/tsmc.1973.4309314 [Google Scholar]
- 11.Haralick RM. Statistical and structural approach to texture. Proc IEEE 1979; 67: 786–804. doi: https://doi.org/10.1109/PROC.1979.11328 [Google Scholar]
- 12.Galloway MM. Texture analysis using gray level run lengths. Comput Graph Image Process 1975; 4: 172–9. doi: https://doi.org/10.1016/s0146-664x(75)80008-6 [Google Scholar]
- 13.Tang X. Texture information in run-length matrices. IEEE Trans Image Process 1998; 7: 1602–9. doi: https://doi.org/10.1109/83.725367 [DOI] [PubMed] [Google Scholar]
- 14.Sun C, Wee WG. Neighboring gray level dependence matrix for texture classification. Cambridge, MA: Academic Press; 1983. [Google Scholar]
- 15.Russell DA, Hanson JD, Ott E. Dimension of strange attractors. Phys Rev Lett 1980; 45: 1175–8. doi: https://doi.org/10.1103/physrevlett.45.1175 [Google Scholar]
- 16.Buczkowski S, Kyriacos S, Nekka F, Lo C. The modified box-counting method: analysis of some characteristic parameters. Pattern Recognit 1998; 31: 411–18. doi: https://doi.org/10.1016/S0031-3203(97)00054-X [Google Scholar]
- 17.Chen WS, Yuan SY, Hsieh CM. Two algorithms to estimate fractal dimension of gray-level images. Opt Eng 2003; 42: 2452. doi: https://doi.org/10.1117/1.1585061 [Google Scholar]
- 18.Sarkar N, Chaudhuri BB. An efficient differential box-counting approach to compute fractal dimension of image. IEEE Trans Syst Man Cybernet 1994; 24: 115–20. doi: https://doi.org/10.1109/21.259692 [Google Scholar]
- 19.Li J, Du Q, Sun C. An improved box-counting method for image fractal dimension estimation. Pattern Recognit 2009; 42: 2460–9. doi: https://doi.org/10.1016/j.patcog.2009.03.001 [Google Scholar]
- 20.Chaudhuri BB, Sarkar N. Texture segmentation using fractal dimension. IEEE Trans Pattern Anal Mach Intell 1995; 17: 72–7. doi: https://doi.org/10.1109/34.368149 [Google Scholar]
- 21.Pentland AP. Fractal-based description of natural scenes. IEEE Trans Pattern Anal Mach Intell 1984; 6: 661–74. doi: https://doi.org/10.1109/tpami.1984.4767591 [DOI] [PubMed] [Google Scholar]
- 22.Soille P, Rivest JF. On the validity of fractal dimension measurements in image analysis. J Vis Commun Image Represent 1996; 7: 217–29. doi: https://doi.org/10.1006/jvci.1996.0020 [Google Scholar]
- 23.Veenland JF, Grashius JL, van der Meer F, Beckers AL, Gelsema ES. Estimation of fractal dimension in radiographs. Med Phys 1996; 23: 585–94. doi: https://doi.org/10.1118/1.597816 [DOI] [PubMed] [Google Scholar]
- 24.Depeursinge A, Foncubierta-Rodriguez A, van de Ville D, Müller H. Three-dimensional solid texture analysis in biomedical imaging: review and opportunities. Med Image Anal 2014; 18: 176–96. doi: https://doi.org/10.1016/j.media.2013.10.005 [DOI] [PubMed] [Google Scholar]
- 25.Yip SS, Aerts HJ. Applications and limitations of radiomics. Phys Med Biol 2016; 61: R150–66. doi: https://doi.org/10.1088/0031-9155/61/13/R150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orlhac F, Soussan M, Maisonobe JA, Garcia CA, Vanderlinden B, Buvat I. Tumor texture analysis in 18F-FDG PET: relationships between texture parameters, histogram indices, standardized uptake values, metabolic volumes, and total lesion glycolysis. J Nucl Med 2014; 55: 414–22. doi: https://doi.org/10.2967/jnumed.113.129858 [DOI] [PubMed] [Google Scholar]
- 27.Hatt M, Majdoub M, Vallières M, Tixier F, Le Rest CC, Groheux D, et al. 18F-FDG PET uptake characterization through texture analysis: investigating the complementary nature of heterogeneity and functional tumor volume in a multi-cancer site patient cohort. J Nucl Med 2015; 56: 38–44. doi: https://doi.org/10.2967/jnumed.114.144055 [DOI] [PubMed] [Google Scholar]
- 28.Vallières M, Freeman CR, Skamene SR, El Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol 2015; 60: 5471–96. [DOI] [PubMed] [Google Scholar]
- 29.Nailon WH, Redpath AT, McLaren DB. Characterisation of radiotherapy planning volumes using textural analysis. Acta Oncol 2008; 47: 1303–8. doi: https://doi.org/10.1080/02841860802256467 [DOI] [PubMed] [Google Scholar]
- 30.Khalvati F, Wong A, Haider MA. Automated prostate cancer detection via comprehensive multi-parametric magnetic resonance imaging texture feature models. BMC Med Imaging 2015; 15: 27. doi: https://doi.org/10.1186/s12880-015-0069-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown AM, Nagala S, Mclean MA, Lu Y, Scoffings D, Apte A, et al. Multi-institutional validation of a novel textural analysis tool for preoperative stratification of suspected thyroid tumors on diffusion-weighted MRI. Magn Reson Med 2016; 75: 1708–16. doi: https://doi.org/10.1002/mrm.25743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vignati A, Mazzetti S, Giannini V, Russo F, Bollito E, Porpiglia F, et al. Texture features on T2-weighted magnetic resonance imaging: new potential biomarkers for prostate cancer aggressiveness. Phys Med Biol 2015; 60: 2685–701. [DOI] [PubMed] [Google Scholar]
- 33.Park SY, Park JM, Sung W, Kim IH, Ye SJ. Texture analysis on the edge-enhanced fluence of VMAT. Radiat Oncol 2015; 10: 74. doi: https://doi.org/10.1186/s13014-015-0382-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nyflot MJ, Yang F, Byrd D, Bowen SR, Sandison GA, Kinahan PE. Quantitative radiomics: impact of stochastic effects on textural feature analysis implies the need for standards. J Med Imaging (Bellingham) 2015; 2: 041002. doi: https://doi.org/10.1117/1.JMI.2.4.041002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balagurunathan Y, Gu Y, Wang H, Kumar V, Grove O, Hawkins S, et al. Reproducibility and prognosis of quantitative features extracted from CT images. Transl Oncol 2014; 7: 72–87. doi: https://doi.org/10.1593/tlo.13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parmar C, Velazquez ER, Leijenaar R, Jermoumi M, Carvalho S, Mak RH, et al. Robust radiomics feature quantification using semiautomatic volumetric segmentation. PLoS One 2014; 9: 1–8. doi: https://doi.org/10.1371/journal.pone.0102107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yip SS, Coroller TP, Sanford NN, Huynh E, Mamon H, Aerts HJ, et al. Use of registration-based contour propagation in texture analysis for esophageal cancer pathologic response prediction. Phys Med Biol 2016; 61: 906–22. doi: https://doi.org/10.1088/0031-9155/61/2/906 [DOI] [PubMed] [Google Scholar]
- 38.Cunliffe AR, Al-Hallaq HA, Labby ZE, Pelizzari CA, Straus C, Sensakovic WF, et al. Lung texture in serial thoracic CT scans: assessment of change introduced by image registration. Med Phys 2012; 39: 4679. doi: https://doi.org/10.1118/1.4730505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cunliffe AR, Armato SG, Fei XM, Tuohy RE, Al-Hallaq HA. Lung texture in serial thoracic CT scans: registration-based methods to compare anatomically matched regions. Med Phys 2013; 40: 61906. doi: https://doi.org/10.1118/1.4805110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moriconi S, Scalco E, Rancati T, Messina A, Giandini T, Valdagni R, et al. Application and evaluation of wavelet-based surface reconstruction for contour propagation in radiotherapy. In: Proceedings of MICCAI workshop on imaging and computer assistance in radiation therapy (ICART). Munich, Germany; 2015. pp. 58–65. [Google Scholar]
- 41.Leijenaar RT, Carvalho S, Hoebers FJ, Aerts HJ, van Elmpt WJ, Huang SH, et al. External validation of a prognostic CT-based radiomic signature in oropharyngeal squamous cell carcinoma. Acta Oncol 2015; 54: 1423–9. doi: https://doi.org/10.3109/0284186X.2015.1061214 [DOI] [PubMed] [Google Scholar]
- 42.Meyer E, Raupach R, Lell MM, Schmidt B, Kachelrieb M. Normalized metal artifact reduction (NMAR) in computed tomography. Med Phys 2010; 37: 5482–93. doi: https://doi.org/10.1118/1.3484090 [DOI] [PubMed] [Google Scholar]
- 43.Veenland JF, Grashuis JL, Gelsema ES. Texture analysis in radiographs: the influence of modulation transfer function and noise on the discriminative ability of texture features. Med Phys 1998; 25: 922–36. doi: https://doi.org/10.1118/1.598271 [DOI] [PubMed] [Google Scholar]
- 44.Kim H, Park CM, Lee M, Park SJ, Song YS, Lee JH, et al. Impact of reconstruction algorithms on CT radiomic features of pulmonary tumors: analysis of intra- and inter-reader variability and inter-reconstruction algorithm variability. PLoS One 2016; 11: e0164924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J, Zhang L, Fave XJ, Fried DV, Stingo FC, Ng CS, et al. Uncertainty analysis of quantitative imaging features extracted from contrast-enhanced CT in lung tumors. Comput Med Imaging Graph 2016; 48: 1–8. doi: https://doi.org/10.1016/j.compmedimag.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He L, Huang Y, Ma Z, Liang C, Liang C, Liu Z. Effects of contrast-enhancement, reconstruction slice thickness and convolution kernel on the diagnostic performance of radiomics signature in solitary pulmonary nodule. Sci Rep 2016; 6: 34921. doi: https://doi.org/10.1038/srep34921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galavis PE, Hollensen C, Jallow N, Paliwal B, Jeraj R. Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta Oncol 2010; 49: 1012–16. doi: https://doi.org/10.3109/0284186X.2010.498437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan J, Chu-Shern JL, Loi HY, Khor LK, Sinha AK, Quek ST, et al. Impact of image reconstruction settings on texture features in 18F-FDG PET. J Nucl Med 2015; 56: 1667–73. doi: https://doi.org/10.2967/jnumed.115.156927 [DOI] [PubMed] [Google Scholar]
- 49.Lasnon C, Majdoub M, Lavigne B, Do P. 18F-FDG PET/CT heterogeneity quantification through textural features in the era of harmonisation programs: a focus on lung cancer. Eur J Nucl Med Mol Imaging 2016; 43: 2324–35. [DOI] [PubMed] [Google Scholar]
- 50.van Velden FH, Kramer GM, Frings V, Nissen IA, Mulder ER, de Langen AJ, et al. Repeatability of radiomic features in non-small-cell lung cancer [(18)F]FDG-PET/CT studies: impact of reconstruction and delineation. Mol Imaging Biol 2016; 18: 788–95. doi: https://doi.org/10.1007/s11307-016-0940-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doumou G, Siddique M, Tsoumpas C, Goh V, Cook GJ. The precision of textural analysis in 18F-FDG-PET scans of oesophageal cancer. Eur Radiol 2015; 25: 2805–12. doi: https://doi.org/10.1007/s00330-015-3681-8 [DOI] [PubMed] [Google Scholar]
- 52.Lu L, Lv W, Jiang J, Ma J, Feng Q, Rahmim A, et al. Robustness of radiomic features in (11C)Choline and (18F)FDG PET/CT imaging of nasopharyngeal carcinoma: impact of segmentation and discretization. Mol Imaging Biol 2016; 18: 935–45. [DOI] [PubMed] [Google Scholar]
- 53.Desseroit MC, Tixier F, Weber W, Siegel BA, Le Rest CC, Visvikis D, et al. Reliability of PET/CT shape and heterogeneity features in functional and morphological components of non-small cell lung cancer tumors: a repeatability analysis in a prospective multi-center cohort. J Nucl Med 2016; jnumed.116.180919 [Epub ahead of print]. doi: https://doi.org/10.2967/jnumed.116.180919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leijenaar RT, Nalbantov G, Carvalho S, van Elmpt WJ, Troost EG, Boellaard R, et al. The effect of SUV discretization in quantitative FDG-PET radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep 2015; 5: 11075. doi: https://doi.org/10.1038/srep11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hatt M, Tixier F, Le Rest CC, Pradier O, Visvikis D. Robustness of intratumour 18F-FDG PET uptake heterogeneity quantification for therapy response prediction in oesophageal carcinoma. Eur J Nucl Med Mol Imaging 2013; 40: 1662–71. doi: https://doi.org/10.1007/s00259-013-2486-8 [DOI] [PubMed] [Google Scholar]
- 56.Cheng NM, Fang YH, Tsan DL, Hsu CH, Yen TC. Respiration-averaged CT for attenuation correction of PET images—impact on PET texture features in non-small cell lung cancer patients. PLoS One 2016; 11: e0150509. doi: https://doi.org/10.1371/journal.pone.0150509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grootjans W, Tixier F, van der Vos CS, Vriens D, Le Rest CC, Bussink J, et al. The impact of optimal respiratory gating and image noise on evaluation of intra-tumor heterogeneity in 18F-FDG positron emission tomography imaging of lung cancer. J Nucl Med 2016; 57: 1692–8. doi: https://doi.org/10.2967/jnumed.116.173112 [DOI] [PubMed] [Google Scholar]
- 58.Orlhac F, Nioche C, Soussan M, Buvat I. Understanding changes in tumor textural indices in PET: a comparison between visual assessment and index values in simulated and patient data. J Nucl Med 2016; jnumed.116.181859 [Epub ahead of print]. doi: https://doi.org/10.2967/jnumed.116.181859 [DOI] [PubMed] [Google Scholar]
- 59.Mayerhoefer ME, Szomolanyi P, Jirak D, Materka A, Trattnig S. Effects of MRI acquisition parameter variations and protocol heterogeneity on the results of texture analysis and pattern discrimination: an application-oriented study. Med Phys 2009; 36: 1236. doi: https://doi.org/10.1118/1.3081408 [DOI] [PubMed] [Google Scholar]
- 60.Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, et al. N4ITK: Improved N3 Bias Correction. IEEE Trans Med Imaging 2010 29: 1310–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giannini V, Mazzetti S, Vignati A, Russo F, Bollito E, Porpiglia F, et al. A fully automatic computer aided diagnosis system for peripheral zone prostate cancer detection using multi-parametric magnetic resonance imaging. Comput Med Imaging Graph 2015; 46: 219–26. doi: https://doi.org/10.1016/j.compmedimag.2015.09.001 [DOI] [PubMed] [Google Scholar]
- 62.Lemaitre G, Martì R, Freixenet J, Vilanova JC, Walker PM, Meriaudeau F. Computer-aided detection and diagnosis for prostate cancer based on mono and multi-parametric MRI: a review. Comput Biol Med 2015; 60: 8–31. doi: https://doi.org/10.1016/j.compbiomed.2015.02.009 [DOI] [PubMed] [Google Scholar]
- 63.Jansen JF, Lu Y, Gupta G, Lee NY, Stambuk HE, Mazaheri Y, et al. Texture analysis on parametric maps derived from dynamic contrast-enhanced magnetic resonance imaging in head and neck cancer. World J Radiol 2016; 8: 90–7. doi: https://doi.org/10.4329/wjr.v8.i1.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Song YS, Park CM, Lee SM, Park SJ, Cho HR, Choi SH, et al. Reproducibility of histogram and texture parameters derived from intravoxel incoherent motion diffusion-weighted MRI of FN13762 rat breast carcinomas. Anticancer Res 2014; 34: 2135–44. [PubMed] [Google Scholar]
- 65.Brynolfsson P, Nilsson D, Henriksson R, Hauksson J, Karlsson M, Garpebring A, et al. ADC texture-an imaging biomarker for high-grade glioma? Med Phys 2014; 41: 101903. doi: https://doi.org/10.1118/1.4894812 [DOI] [PubMed] [Google Scholar]
- 66.Le Bihan D, Breton E, Lallemand D, Grenier P, Cabanis E, Laval-Jeantet M. MR imaging of intravoxel incoherent motions: application to diffusion and perfusion in neurologic disorders. Radiology 1986; 161: 401–7. doi: https://doi.org/10.1148/radiology.161.2.3763909 [DOI] [PubMed] [Google Scholar]
- 67.Andreou A, Koh DM, Collins DJ, Blackledge M, Wallace T, Leach MO, et al. Measurement reproducibility of perfusion fraction and pseudodiffusion coefficient derived by intravoxel incoherent motion diffusion-weighted MR imaging in normal liver and metastases. Eur Radiol 2013; 23: 428–34. doi: https://doi.org/10.1007/s00330-012-2604-1 [DOI] [PubMed] [Google Scholar]
- 68.Raja JV, Khan M, Ramachandra VK, Al-Kadi O. Texture analysis of CT images in the characterization of oral cancers involving buccal mucosa. Dentomaxillofac Radiol 2012; 41: 475–80. doi: https://doi.org/10.1259/dmfr/83345935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alvarenga AV, Infantosi AF, Pereira WC, Azevedo CM. Assessing the combined performance of texture and morphological parameters in distinguishing breast tumors in ultrasound images. Med Phys 2012; 39: 7350–8. doi: https://doi.org/10.1118/1.4766268 [DOI] [PubMed] [Google Scholar]
- 70.Fehr D, Veeraraghavan H, Wibmer A, Gondo T, Matsumoto K, Vargas HA, et al. Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proc Natl Acad Sci USA 2015; 112: E6265–73. doi: https://doi.org/10.1073/pnas.1505935112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Al-Kadi OS, Watson D. Texture analysis of aggressive and nonaggressive lung tumor CE CT images. IEEE Trans Biomed Eng 2008; 55: 1822–30. doi: https://doi.org/10.1109/TBME.2008.919735 [DOI] [PubMed] [Google Scholar]
- 72.Yu H, Caldwell C, Mah K, Mozeg D. Coregistered FDG PET/CT-based textural characterization of head and neck cancer for radiation treatment planning. IEEE Trans Med Imaging 2009; 28: 374–83. doi: https://doi.org/10.1109/TMI.2008.2004425 [DOI] [PubMed] [Google Scholar]
- 73.Yu H, Caldwell C, Mah K, Poon I, Balogh J, MacKenzie R, et al. Automated radiation targeting in head-and-neck cancer using region-based texture analysis of PET and CT images. Int J Radiat Oncol Biol Phys 2009; 75: 618–25. doi: https://doi.org/10.1016/j.ijrobp.2009.04.043 [DOI] [PubMed] [Google Scholar]
- 74.Wong AJ, Kanwar A, Mohamed AS, Fuller CD. Radiomics in head and neck cancer: from exploration to application. Transl Cancer Res 2016; 5: 371–82. doi: https://doi.org/10.21037/tcr.2016.07.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scrivener M, De Jong EE, van Timmeren JE, Pieters T, Geets X. Radiomics applied to lung cancer: a review. Transl Cancer Res 2016; 5: 398–409. doi: https://doi.org/10.21037/tcr.2016.06.18 [Google Scholar]
- 76.De Ruysscher D, Sharifi H, Defraene G, Kerns SL, Christiaens M, De Ruyck K, et al. Quantification of radiation-induced lung damage with CT scans: the possible benefit for radiogenomics. Acta Oncol 2013; 52: 1405–10. doi: https://doi.org/10.3109/0284186X.2013.813074 [DOI] [PubMed] [Google Scholar]
- 77.Palma DA, van Sörnsen de Koste J, Verbakel WF, Vincent A, Senan S. Lung density changes after stereotactic radiotherapy: a quantitative analysis in 50 patients. Int J Radiat Oncol 2011; 81: 974–8. doi: https://doi.org/10.1016/j.ijrobp.2010.07.025 [DOI] [PubMed] [Google Scholar]
- 78.Defraene G, van Elmpt W, Crijns W, Slagmolen P, De Ruysscher D. CT characteristics allow identification of patient-specific susceptibility for radiation-induced lung damage. Radiother Oncol 2015; 117: 29–35. doi: https://doi.org/10.1016/j.radonc.2015.07.033 [DOI] [PubMed] [Google Scholar]
- 79.Mattonen SA, Palma DA, Haasbeek CJ, Senan S, Ward AD. Early prediction of tumor recurrence based on CT texture changes after stereotactic ablative radiotherapy (SABR) for lung cancer. Med Phys 2014; 41: 33502. doi: https://doi.org/10.1118/1.4866219 [DOI] [PubMed] [Google Scholar]
- 80.Mattonen SA, Palma DA, Haasbeek CJ, Senan S, Ward AD. Texture analysis of automatic graph cuts segmentations for detection of lung cancer recurrence after stereotactic radiotherapy. SPIE Med Imaging 2015; 94: 941719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mattonen SA, Palma DA, Johnson C, Louie AV, Landis M, Rodrigues G, et al. Detection of local cancer recurrence after stereotactic ablative radiation therapy for lung cancer: physician performance versus radiomic assessment. Int J Radiat Oncol 2016; 94: 1121–8. doi: https://doi.org/10.1016/j.ijrobp.2015.12.369 [DOI] [PubMed] [Google Scholar]
- 82.Pyka T, Bundschuh RA, Andratschke N, Mayer B, Specht HM, Papp L, et al. Textural features in pre-treatment (F18)-FDG-PET/CT are correlated with risk of local recurrence and disease-specific survival in early stage NSCLC patients receiving primary stereotactic radiation therapy. Radiat Oncol 2015; 10: 1–9. doi: https://doi.org/10.1186/s13014-015-0407-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cook GJ, Yip C, Siddique M, Goh V, Chicklore S, Roy A, et al. Are pretreatment 18F-FDG PET tumor textural features in non-small cell lung cancer associated with response and survival after chemoradiotherapy? J Nucl Med 2013; 54: 19–26. doi: https://doi.org/10.2967/jnumed.112.107375 [DOI] [PubMed] [Google Scholar]
- 84.Coroller TP, Agrawal V, Narayan V, Hou Y, Grossmann P, Lee SW, et al. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother Oncol 2016; 119: 480–6. doi: https://doi.org/10.1016/j.radonc.2016.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huynh E, Coroller TP, Narayan V, Agrawal V, Hou Y, Romano J, et al. CT-based radiomic analysis of stereotactic body radiation therapy patients with lung cancer. Radiother Oncol 2016; 120: 258–66. doi: https://doi.org/10.1016/j.radonc.2016.05.024 [DOI] [PubMed] [Google Scholar]
- 86.Tixier F, Le Rest CC, Hatt M, Albarghach N, Pradier O, Metges JP, et al. Intratumor heterogeneity characterized by textural features on baseline 18F-FDG PET images predicts response to concomitant radiochemotherapy in esophageal cancer. J Nucl Med 2011; 52: 369–78. doi: https://doi.org/10.2967/jnumed.110.082404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nakajo M, Jinguji M, Nakabeppu Y, Nakajo M, Higashi R, Fukukura Y, et al. Texture analysis of 18F-FDG PET/CT to predict tumour response and prognosis of patients with esophageal cancer treated by chemoradiotherapy. Eur J Nucl Med Mol Imaging 2016; [Epub ahead of print]. doi: https://doi.org/10.1007/s00259-016-3506-2 [DOI] [PubMed] [Google Scholar]
- 88.Yip C, Landau D, Kozarski R, Ganeshan B, Thomas R, Michaelidou A, et al. Primary esophageal cancer: heterogeneity as potential prognostic biomarker in patients treated with definitive chemotherapy and radiation therapy. Radiology 2014; 270: 141–8. doi: https://doi.org/10.1148/radiol.13122869 [DOI] [PubMed] [Google Scholar]
- 89.El Naqa I, Grigsby P, Apte A, Kidd E, Donnelly E, Khullar D, et al. Exploring feature-based approaches in PET images for predicting cancer treatment outcomes. Pattern Recognit 2009; 42: 1162–71. doi: https://doi.org/10.1016/j.patcog.2008.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu J, Mao Y, Li Z, Zhang D, Zhang Z, Hao S, et al. Use of texture analysis based on contrast-enhanced MRI to predict treatment response to chemoradiotherapy in nasopharyngeal carcinoma. J Magn Reson Imaging 2016; 44: 445–55. doi: https://doi.org/10.1002/jmri.25156 [DOI] [PubMed] [Google Scholar]
- 91.Scalco E, Marzi S, Sanguineti G, Vidiri A, Rizzo G. Characterization of cervical lymph-nodes using a multi-parametric and multi-modal approach for an early prediction of tumor response to chemo-radiotherapy. Phys Med 2016; [Epub ahead of print]. doi: https://doi.org/10.1016/j.ejmp.2016.09.003 [DOI] [PubMed] [Google Scholar]
- 92.Alic L, van Vliet M, van Dijke CF, Eggermont MM, Veenland JF, Niessen WJ. Heterogeneity in DCE-MRI parametric maps: a biomarker for treatment response? Phys Med Biol 2011; 56: 1601–16. doi: https://doi.org/10.1088/0031-9155/56/6/006 [DOI] [PubMed] [Google Scholar]
- 93.O'Connor JP, Rose CJ, Jackson A, Watson Y, Cheung S, Maders F, et al. DCE-MRI biomarkers of tumour heterogeneity predict CRC liver metastasis shrinkage following bevacizumab and FOLFOX-6. Br J Cancer 2011; 105: 139–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gnep K, Fargeas A, Gutiérrez-Carvajal RE, Commandeur F, Mathieu R, Ospina JD, et al. Haralick textural features on T2-weighted MRI are associated with biochemical recurrence following radiotherapy for peripheral zone prostate cancer. J Magn Reson Imaging 2016; [Epub ahead of print]. doi: https://doi.org/10.1002/jmri.25335 [DOI] [PubMed] [Google Scholar]
- 95.Bundschuh RA, Dinges J, Neumann L, Seyfried M, Zsótér N, Papp L, et al. Textural parameters of tumor heterogeneity in 18F-FDG PET/CT for therapy response assessment and prognosis in patients with locally advanced rectal cancer. J Nucl Med 2014; 55: 891–7. doi: https://doi.org/10.2967/jnumed.113.127340 [DOI] [PubMed] [Google Scholar]
- 96.De Cecco CN, Ganeshan B, Ciolina M, Rengo M, Meinel FG, Musio D, et al. Texture analysis as imaging biomarker of tumoral response to neoadjuvant chemoradiotherapy in rectal cancer patients studied with 3-T magnetic resonance. Invest Radiol 2015; 50: 239–45. doi: https://doi.org/10.1097/rli.0000000000000116 [DOI] [PubMed] [Google Scholar]
- 97.Nie K, Shi L, Chen Q, Hu X, Jabbour S, Yue N, et al. Rectal cancer: assessment of neoadjuvant chemo-radiation outcome based on radiomics of multi-parametric MRI. Clin Cancer Res 2016; 22: 5256–64. doi: https://doi.org/10.1158/1078-0432.CCR-15-2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tian F, Hayano K, Kambadakone AR, Sahani DV. Response assessment to neoadjuvant therapy in soft tissue sarcomas: using CT texture analysis in comparison to tumor size, density, and perfusion. Abdom Imaging 2015; 40: 1705–12. doi: https://doi.org/10.1007/s00261-014-0318-3 [DOI] [PubMed] [Google Scholar]
- 99.Nardone V, Tini P, Biondi M, Sebaste L, Vanzi E, De Otto G, et al. Prognostic value of MR imaging texture analysis in brain non-small cell lung cancer oligo-metastases undergoing stereotactic irradiation. Cureus 2016; 8: e584. doi: https://doi.org/10.7759/cureus.584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cunliffe A, Armato SG, Castillo R, Pham N, Guerrero T, Al-Hallaq HA. Lung texture in serial thoracic computed tomography scans: correlation of radiomics-based features with radiation therapy dose and radiation pneumonitis development. Int J Radiat Oncol 2015; 91: 1048–56. doi: https://doi.org/10.1016/j.ijrobp.2014.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang X, Tridandapani S, Beitler JJ, Yu DS, Yoshida EJ, Curran WJ, et al. Ultrasound GLCM texture analysis of radiation-induced parotid-gland injury in head-and-neck cancer radiotherapy: an in vivo study of late toxicity. Med Phys 2012; 39: 5732. doi: https://doi.org/10.1118/1.4747526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Scalco E, Fiorino C, Cattaneo GM, Sanguineti G, Rizzo G. Texture analysis for the assessment of structural changes in parotid glands induced by radiotherapy. Radiother Oncol 2013; 109: 384–7. doi: https://doi.org/10.1016/j.radonc.2013.09.019 [DOI] [PubMed] [Google Scholar]
- 103.Pota M, Scalco E, Sanguineti G, Cattaneo GM, Esposito M, Rizzo G. Early classification of parotid glands shrinkage in radiotherapy patients: a comparative study. Biosyst Eng 2015; 138: 77–89. doi: https://doi.org/10.1016/j.biosystemseng.2015.06.007 [Google Scholar]
- 104.Scalco E, Moriconi S, Rizzo G. Texture analysis to assess structural modifications induced by radiotherapy. In: Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS 2015: 5219–22. [DOI] [PubMed]
- 105.Chalkidou A, O'Doherty MJ, Marsden PK. False discovery rates in PET and CT studies with texture features: a systematic review. PLoS One 2015; 10: 1–18. doi: https://doi.org/10.1371/journal.pone.0124165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJ, Doroshow J, et al. Machine learning methods for quantitative radiomic biomarkers. Sci Rep 2015; 5: 649. doi: https://doi.org/10.1038/srep13087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Huang YQ, Liang CH, He L, Tian J, Liang CS, Chen X, et al. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol 2016; 34: 2157–64. doi: https://doi.org/10.1200/JCO.2015.65.9128 [DOI] [PubMed] [Google Scholar]
- 108.Wan T, Bloch BN, Plecha D, Thompson CL, Gilmore H, Jaffe C, et al. A radio-genomics approach for identifying high risk estrogen receptor-positive breast cancers on DCE-MRI: preliminary results in predicting OncotypeDX risk scores. Sci Rep 2016; 6: 21394. doi: https://doi.org/10.1038/srep21394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tixier F, Hatt M, Le Rest CC, Simon B, Key S, Corcos L, et al. Signaling pathways alteration involved in head and neck cancer can be identified through textural features analysis in 18F-FDG PET images: a prospective study. J Nucl Med 2015; 56: 449. [Google Scholar]