Abstract
Introduction
Autism spectrum disorders (ASD) are a group of neurodevelopmental disorders lacking a clinical biomarker for diagnosis. Emerging evidence shows that intestinal microflora from ASD subjects can be distinguished from controls, suggesting metabolite differences due to the action of intestinal microbes may provide a means for identifying potential biomarkers for ASD.
Objectives
The aim of this study was to determine if quantitative differences in levels of stercobilin and stercobilinogen, metabolites produced by biological action of intestinal microflora, exist in the fecal matter between an ASD mouse model population and controls.
Methods
Pairs of fecal samples were collected from two mouse groups, an ASD model group with Timothy syndrome 2 (TS2-NEO) and a gender-matched control group. After centrifugation, supernatant was spiked with an 18O-labeled stercobilin isotopomer and subjected to solid phase extraction for processing. Extracted samples were spotted on a stainless steel plate and subjected to matrix-assisted laser desorption and ionization mass spectrometry using dihydroxybenzoic acid as the matrix (n = 5). Peak areas for bilins and 18O-stercobilin isotopomers were determined in each fecal sample.
Results
A 40–45% depletion in stercobilin in TS2-NEO fecal samples compared with controls was observed with p < 0.05; a less dramatic depletion was observed for stercobilinogen.
Conclusions
The results show that stercobilin depletion in feces is observed for an ASD mouse model vs. controls. This may help to explain recent observations of a less diverse microbiome in humans with ASD and may prove helpful in developing a clinical ASD biomarker.
Keywords: autism spectrum disorders, metabolomics, biomarker, mass spectrometry, MALDI, solid-phase extraction
1 Introduction
Autism spectrum disorders (ASD) (Kanner and Eisenberg 1957) consist of a group of neurobehavioral disorders characterized by impaired social interactions, deficits in communications skills, repetitive behaviors, and other stereotypical behavioral patterns (Rinehart et al. 2002; Lord et al. 2000a; Lord et al. 2000b; Bolte et al. 2002; Eigsti and Shapiro 2003). High rates of diagnosis in the United States (up to 1 in 68 children) (Developmental Disabilities Monitoring Network Surveillance Year Principal et al. 2014) exemplify the seriousness of ASD as a medical concern. Because of the efficacy of early diagnosis and intervention on quality of life for those with ASD (Ospina et al. 2008; Ben-Itzchak and Zachor 2007; Ozonoff et al. 2005), there is great interest in identifying, at a molecular level, biomarkers that could be utilized in ASD diagnosis (Rudolph et al. 2013). Significant effort has been dedicated toward discovery of potential genetic factors in the etiology of ASD. Nevertheless, it is becoming increasingly recognized that non-genetic factors can contribute to ASD etiology (Herbert et al. 2006). The severity of ASD symptoms is also highly heterogeneous (Gotham et al. 2009). Combined, these factors have made determination of a clinical biomarker and a Grand Unified Theory (GUT) for ASD etiology elusive (Rudolph et al. 2013).
While a GUT for ASD etiology may be unrealistic due to the heterogeneity in causation and severity, the search for potential biomarkers with a focus on metabolites has gained considerable momentum in the past decade (Adamo et al. 2014; Altieri et al. 2011; Dager et al. 2015; Dieme et al. 2015; Durieux et al. 2016; Faber et al. 2009; Gabriele et al. 2014; Goldenthal et al. 2015; Gorrindo et al. 2013; Herbert et al. 2006; Jyonouchi et al. 2008; Masi et al. 2017; Ming et al. 2012; Ming et al. 2005; Pastural et al. 2009; Ramsey et al. 2013; Rudolph et al. 2013; Woods et al. 2015; James et al. 2004; Frye et al. 2013; Ngounou Wetie et al. 2015). Expanding the search to identify potential subgroups within ASD has been of interest. One approach toward identifying potential biomarkers has been to consider comorbid conditions (Woods et al. 2015; Adamo et al. 2014; Jyonouchi et al. 2008; Masi et al. 2017). Of particular interest has been gastrointestinal distress, a condition experienced by a high percentage of ASD patients (Bresnahan et al. 2015; Bauman 2010; Kohane et al. 2012). It is known that the microbiome of the gut regulates the homeostasis of the central nervous system (CNS) (Rhee et al. 2009; Sherwin et al. 2016). It is also known that the microbiome has an effect on neuropsychiatric disorders (Rogers et al. 2016; Cryan and Dinan 2012). Therefore the intestinal microbiome and its effects on the gut-brain axis could be a potential source for the discovery and validation of ASD biomarkers (Cryan and Dinan 2012; Krajmalnik-Brown et al. 2015; Sherwin et al. 2016; Mulle et al. 2013).
A recent review highlights the findings on research detailing connections between the intestinal microbome and ASD and is recommended for a more in-depth account (Ding et al. 2017). Briefly, a number of differences in the microbiota of ASD subjects in comparison to controls have been observed. In a groundbreaking investigation, an order of magnitude higher levels of cultured Clostridium species in fecal samples of ASD subjects relative to healthy controls was observed (Finegold et al. 2002). A subsequent study indicated that clostridial clusters I and XI were elevated in the feces of ASD subjects while cluster XIVab was decreased (Song et al. 2004). Fecal bacterial populations of clostridial clusters I and II (Clostridium histolyticum) were elevated from ASD subjects relative to controls (Parracho et al. 2005). Elevated levels of Sutterella and Ruminococcus torques have been observed from ASD fecal samples versus controls (Wang et al. 2013); moreover, within populations with gastrointestinal disturbances, Sutterella elevation has been detected in gastrointestinal biopsies from ASD subjects. Depleted levels of Prevotella in ASD microbiota have been observed (Kang et al. 2013). One study examining fecal samples of ASD and controls showed significantly higher levels of Bacteroidetes and Desulfovibrio species in ASD specimens, while higher levels of Firmicutes were observed in control specimens (Finegold et al. 2010). One particularly intriguing study showed alterations in the microbiota for the maternal immune activation (MIA) mouse model, which has ASD features; strikingly, administration of Bacteroides fragilis to offspring corrects intestinal permeability, alters the microbiome and metabolomic profiles, and improves behaviors associated with ASD (Hsiao et al. 2013).
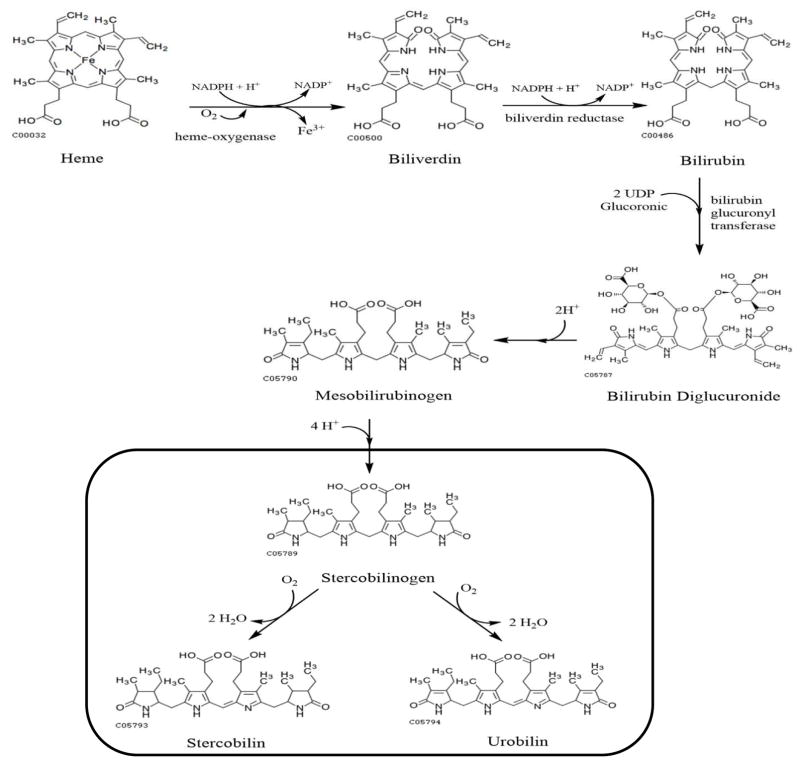
The composition of an organism’s gastrointestinal microbiome is reflected in the bacterial metabolites found in feces. One example is stercobilin, a metabolic product excreted from mammals that is produced from initial catabolism of heme followed by microbial action in the colon on the metabolite urobilinogen (Watson 1936; Watson et al. 1953); the metabolic pathway from urobilinogen to stercobilin by bacterial action is shown in Figure 1. The increasing evidence demonstrating a link between the intestinal microbiome and at least some cases of ASD spurred us to reflect upon an observation made a decade ago from a limited set of ASD and control children, which was that stercobilin was depleted by 67% in urine specimens from ASD subjects relative to controls (Wood et al. 2007).
Figure 1.
Metabolic pathway from heme to stercobilin. The metabolic products within the box are produced by the action of Clostridium perfringens (Vitek et al. 2006).
Here, we test if stercobilin is depleted in fecal matter from a mouse model of ASD. To test this hypothesis, a Timothy syndrome (TS) mouse model for ASD (Bader et al. 2011) was employed. TS is a rare disorder caused by a mutation in the L-type voltage-gated calcium channel (Cav1.2). ASD occurs in about 75% of all individuals with TS arrhythmia disorder (Splawski et al. 2005). TS mice have demonstrated a triad of ASD behaviors: repetitive/perseverative behavior, impaired social behavior, and impaired communication (Bader et al. 2011). No difference was observed in gross brain anatomy of TS mice and littermate controls (Bett et al. 2012) but changes in dendritic retraction were observed (Krey et al. 2013). Thus, this study examines whether fecal stercobilin levels can be distinguished between TS mouse (ASD model) and control populations.
2 Experimental
2.1 Materials
The following materials were utilized throughout: stercobilin hydrochloride (Frontier Scientific, Logan, UT, USA), isotopically labeled water (18O, 97%) (Cambridge Isotope Laboratories, Inc., Tewksbury, MA, USA), methanol (HPLC grade ≥99.9%) (Sigma-Aldrich, St. Louis, MO, USA), acetonitrile (Sigma-Aldrich), ammonium hydroxide (J.T. Baker, Central Valley, PA, USA), trifluoroacetic acid (TFA) (EMD Millipore, Darmstadt, Germany), formic acid (FA) (99%) (Sigma-Aldrich), LCMS-grade water (HPLC Grade submicron filtered) (Fisher Scientific, Fair Lawn, NJ, USA), 2,5-dihydroxybenzoic acid (DHB) (Sigma-Aldrich), and Oasis© MAX extraction cartridges (1cc/30mg) (Waters, Milford, MA, USA).
2.2 Instrumentation
All samples were analyzed using a Bruker Daltonics (Billerica, MA) 12T Solarix Fourier transform-ion cyclotron resonance (FT-ICR) mass spectrometer equipped with a SmartBeam II frequency-tripled (355 nm) Nd:YAG matrix-assisted laser desorption and ionization (MALDI) source. The number of laser shots per pixel was set to 1000 at the same location with a laser power of 55% and a small focus. A saturated solution of DHB in 20:80 (v/v) ACN:H2O was utilized as the MALDI matrix. All samples were analyzed in positive ion mode using 1M word time-domain datasets for high resolution. Samples of 2.5 μL were spotted on a stainless steel MALDI plate (5 replicates for each sample) unless otherwise noted.
2.3 Animals
All procedures performed in the collection of fecal material of mice were in accordance with the ethical standards of the institution’s Division of Comparative Medicine and Laboratory Animal Facilities. In this study, we used a mouse population of TS, which exhibits ASD behavioral traits (Bader et al. 2011), and compared to a control population to examine relative levels of stercobilin in the fecal material of the mice. In this case, TS2 mice, a more severe form of TS caused by a missense mutation, were utilized. Nine pairs of mice that were age-matched were selected for this study. Details of the two populations are provided in Table 1.
Table 1.
Characteristics of Mice Used.
| Controls | TS2-NEO | |
|---|---|---|
| Total (n) | 9 | 9 |
| Female | 3 | 3 |
| Male | 6 | 6 |
| Mean Age ± (SD), Days | 232 ± 109 | 206 ± 54 |
In TS2 mice, a missense mutation occurs in either GR406R or G406S in exon 8. To determine if the mice have TS2, mice are genotyped to detect any mutations at the time in which they are tagged for identification (Bader et al. 2011). In the set of parents, the strain of TS can come from either the sire or the dam but is determined by which of the mice in the pair has been previously identified to have TS. Because only heterozygous mice that were allowed to keep an inverted neomycin cassette in exon 8A (TS2-NEO) survived through adulthood (Bader et al. 2011), it was actually the TS2-NEO population used here.
2.4 Methods
2.4.1 Synthesis of Isotopomer for Quantitation
A scaled up version of a previously described procedure for synthesis of stercobilin isotopomers (Rudolph et al. 2016) was employed here; this method is based upon an approach described by Bergmann et al. to create an isotopic standard for fumonisin B1 (Bergmann et al. 2013). For this experiment, ca. 5 × 10−6 mol of stercobilin was mixed with 10 μL 5% (v/v) TFA and 95 μL of H218O in an Agilent autosampler vial with screw cap lid. In this scale up procedure, 10,000x greater stercobilin, 10x greater TFA, and 19x greater H218O was used than previously reported (Rudolph et al. 2016). The vial was placed in an incubator at 70°C for 8 hours. Following the reaction, the sample was dried down under air and reconstituted in 100 mL of 20:80 (v/v) ACN/H2O to produce a stock solution.
2.4.2 Determination of Response Factor
A calibration curve was constructed to cover the range of 5.05 × 10−11 mol to 6.14 × 10−8 mol of stercobilin (five replicates for each level). A 90 μL spike of 2.77 × 10−6 M labeled stercobilin was added to 6 mL of LCMS-grade H2O, 10 μL of 5% (v/v) NH4OH, and an appropriate amount of 4.99 × 10−6 M stercobilin in 20:80 (v/v) ACN/H2O for values within our range. The solution is processed by solid phase extraction (SPE) analysis using a Waters Oasis MAX cartridge that is first conditioned with 3 mL of methanol, then equilibrated with 3 mL LCMS-grade H2O. Sample solutions are then loaded (~ 0.5 mL/min) and the sample is eluted with 1.2 mL 5% (v/v) FA in methanol. Samples are then dried down with air and reconstituted in 1 mL of 20:80 (v/v) ACN/H2O. A 5 μL aliquot of the reconstituted sample was mixed with 20 μL of saturated DHB matrix solution. Finally, 2.5 μL of the sample and matrix solution were spotted onto a ground steel matrix plate and analyzed on a 12 tesla SolariX Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer (Bruker Daltonics, Billerica, MA) using matrix-assisted laser desorption/ionization (MALDI) with the SmartBeam II frequency-tripled Nd:YAG laser.
2.4.3 Limits of Detection and Quantitation
An aliquot of our labeled stercobilin was diluted in 20:80 (v/v) ACN/H2O to the same concentration as would be observed in our samples. Subsequently 5 μL of the diluted labeled stercobilin in 20:80 (v/v) ACN/H2O was mixed with 20 μL of saturated DHB matrix. Next, 2.5 μL of the labeled sample and matrix solution were spotted onto a ground steel matrix plate seven times and analyzed by MALDI FT-ICR mass spectrometry.
2.4.4 Fecal Material Preparation
Fecal samples were obtained from a mouse colony housing TS2NEO and wild-type (control) populations and were stored in labeled centrifuge tubes at -4°C until use, within seven days. Each sample was weighed (typically 12–14 mg droppings, but up to 54 mg) and placed into a 1.5 mL microcentrifuge tube with approximately 1 mL of LCMS-grade H2O. Samples were then vortexed for 1 minute at 1800 rpm followed by centrifugation for 5 minutes at 6000 RPM. After centrifugation, the supernatant is removed and the process is repeated two more times to obtain 3 mL of supernatant per fecal sample. The 3 mL of supernatant were diluted with 3 mL of LCMS-grade H2O. A 90 μL spike of 2.77 × 10−6 M 18O-labeled stercobilin is added to the diluted sample along with 10 μL of 5% (v/v) NH4OH. The solution is then processed by SPE analysis and spotted on the MALDI matrix plate using the same method mentioned in the calibration curve section with FT-ICR mass spectrometry.
2.4.5 Data Analysis
All data analysis was performed using Bruker Data Analysis software version 4.2 for extracting mass spectra and peak areas for the labeled and unlabeled stercobilin species. To correct for the presence of the small but detectable levels of unlabeled stercobilin in the 18O-labeled standard (6% of total, Table 2), the peak area ratio of m/z 595 due to this unlabeled material was subtracted from the overall m/z 595 peak area in the fecal samples based on upon the ratio of m/z 595:601 and m/z 595:603 in the labeled material.
Table 2.
Oxygen-18 Labeled Isotope peaks for stercobilin detected with the percent labeled as compared to all peak areas.
| m/z Peak (18On) | % Labelling |
|---|---|
| 595 (18O0) | 6 ± 2 |
| 597 (18O1) | 13 ± 4 |
| 599 (18O2) | 26 ± 7 |
| 601 (18O3) | 27 ± 14 |
| 603 (18O4) | 28 ± 1 |
Statistical analysis was performed using SigmaPlot© 13 software. Data graphing was performed in Excel 2016.
3 Results
3.1 Synthesis of Isotopomer
In the approach to synthesize 18O-labeled stercobilin for quantification, the low yield was insufficient to run many trials (Rudolph et al. 2016). Therefore, it was a primary concern to create greater quantities of 18O-labeled stercobilin. To improve 18O-stercobilin isotopomer yield, a higher temperature than that used by Bergmann et al. was used, which produces a higher yield of 18O-labeled stercobilin while maintaining the concentrations of materials constant with those of Bergmann et al. (2013). With these considerations in mind, the labeling efficiency of stercobilin was found to be 64.8% with percent labeled for each 18O isotope shown in Table 2. One matter of concern was the potential for labeled stercobilin to revert back to its original form due to the labile sites of the oxygen. To overcome this, the label is stored at -4°C, which has been shown to preserve isotopic labeling over a period of 20 days (Rudolph et al. 2016).
3.2 Response Factor Calculations
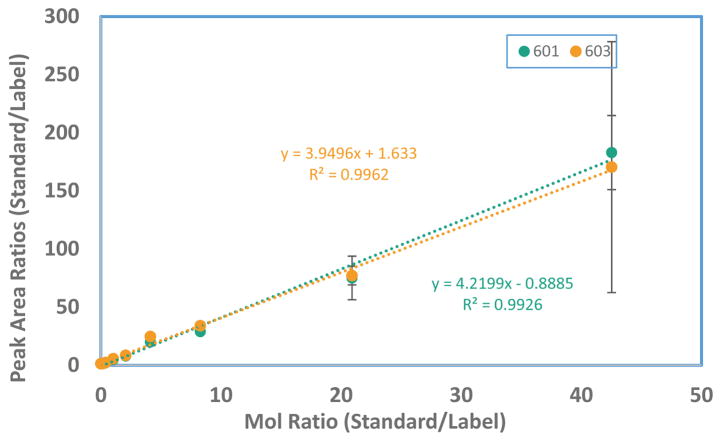
To determine the unknown levels of stercobilin or stercobilinogen in the fecal material of TS2-NEO mice, a calibration curve of varying amounts of stercobilin with a constant spike of labeled stercobilin was created (Figure 2). A range of 5.06 × 10−11mol to 6.14 × 10−8 mol was utilized. By comparison of peak areas to concentrations, a response factor can be determined. As shown in Figure 2, the slopes were slightly different but comparable whether the m/z 601 (18O3 isotopomer) or m/z 603 (18O4 isotopomer) was used to determine the response factor. The large error bars for m/z 603 may be the result of detector saturation and hence non-linear response.
Figure 2.
Response factor calculations based on the peak areas of m/z 601 and 603. Error bars represent one standard deviation from the mean of the stercobilin/labeled stercobilin ratio.
3.3 Limits of Detection and Quantitation
To calculate the limits of detection and quantitation, we followed calculations as detailed by Mermet for finding the method detection limit of our instrument (Mermet 2008); the limit of detection (LOD) was 1.37 × 10−11 mole for quantitation using m/z 601 and 1.38 × 10−11 mole for quantitation using m/z 603. The limit of quantitation (LOQ) was 4.37 × 10−11 mole using either m/z 601 or m/z 603.
3.4 Fecal Material Analysis
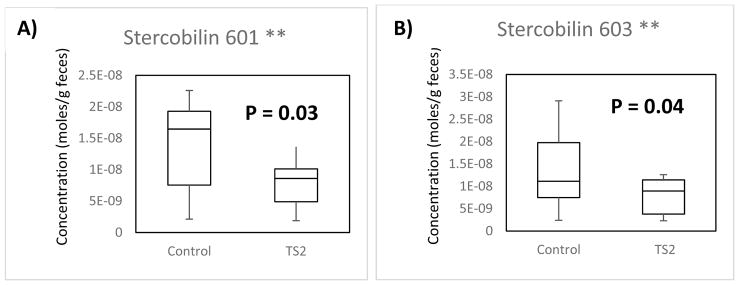
Utilizing the calculated response factors, the stercobilin levels in nine gender-matched pairs of TS2-NEO:wild-type mice were calculated based on both the m/z 601 and m/z 603 labeled peaks as shown in the box-whisker plots in Figure 3. All concentrations were normalized per gram of fecal material to account for the differences in mass of the individual samples. The unpaired t-test calculation was performed to compute p values and assess whether the two populations have different means.
Fig. 3.
(A) Comparison of the normalized concentration of moles of stercobilin of WT mice (left) to TS2-NEO mice (right) using the m/z 601 labeled stercobilin. (B) Comparison of the normalized concentration of moles of stercobilin of WT mice (left) to TS2-NEO mice (right) using the m/z 603 labeled stercobilin. The line represents the median in each sample population.
When calculating the average moles of stercobilin for m/z 601 of WT and TS2-NEO mice, levels of 1.52 × 10−8 ± 6.2 × 10−9 mole/g and 9.11 × 10−9 ± 4.6 × 10−9 mole/g were observed for WT and TS2-NEO populations, respectively. These values show a depletion of stercobilin of 40% in TS2-NEO mice (p = 0.03). With m/z 603, levels of stercobilin of 1.52 × 10−8 ± 8.4 × 10−9 mole/g and 8.34 × 10−9 ± 3.9 × 10−9 mole/g are found for WT and TS2-NEO, respectively. These values show a depletion of stercobilin of 45% in TS2-NEO mice (p = 0.04).
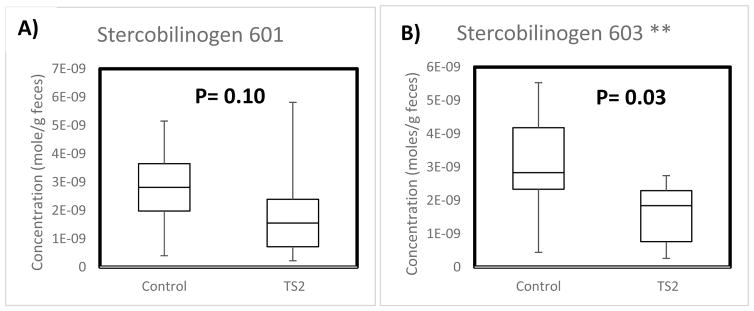
Stercobilin (C33H46N4O6, PubChem CID 5280818) is an oxidative product of stercobilinogen (C33H48N4O6, PubChem CID 9548718) by bacterial action in the colon (Vitek et al. 2006). Although these two compounds differ in mass by two H atoms, the high mass accuracy of FT-ICR makes possible distinguishing isotope peaks of stercobilin from stercobilinogen. The identity of stercobilin was further verified by tandem mass spectrometry using collision induced dissociation (Quinn et al. 2012). Thus, the levels of stercobilinogen in the fecal material of TS2-NEO and control mice populations were also quantified. The results are displayed in the box-whisker plots in Figure 4.
Fig. 4.
(A) Comparison of the normalized concentration of moles of stercobilinogen of WT mice (left) to TS2 mice (right) using the m/z 601 labeled stercobilin. (B) Comparison of the normalized concentration of moles of stercobilinogen of WT mice (left) to TS2 mice (right) using the m/z 603 labeled stercobilin. The line represents the median in each sample population.
The results indicate that stercobilinogen, the precursor to stercobilin, is depleted in TS2-NEO mice by 40% utilizing both m/z 601 and m/z 603 stercobilin isotopomer peaks. However, it is not clear if how meaningful the observed depletion is based on m/z 601 (p = 0.10); quantification by m/z 603 was more significant (p = 0.03) but needs additional testing from a larger population. Other bilins such as urobilin and urobilinogen were not consistently detected across all samples.
4 Discussion
The 40–45% depletion of stercobilin in the fecal material of ASD-model mice relative to controls at a greater than 95% confidence level suggests that depletion fecal stercobilin may serve as a potential ASD biomarker in humans. These results are consistent with the observation of stercobilin depletion in the urine samples of ASD subjects relative to controls (Wood et al. 2007). It is also noteworthy that depletion was observed in seven of the nine pairs of mice; it should be noted that there is not 100% concordance of TS2 and ASD in humans, but it is about 75% (Splawski et al. 2005).
Thus, it is possible that there are TS2-NEO mice amongst the cohort that might not fit the diagnostic criteria for ASD, even though as a population, TS2-NEO mice do exhibit behavioral features of ASD (Bader et al. 2011). There is the possibility that the level of stercobilin depletion may correlate to the severity of ASD. Unfortunately, with the current animal model it is not possible to quantify the severity of ASD symptoms. While less statistically significant, it appears that stercobilinogen, the metabolic precursor to stercobilin, is also depleted in TS2-NEO mouse fecal material relative to controls. The stronger correlation of stercobilin depletion relative to stercobilinogen depletion is an indicator that the check point in the metabolic process is the transformation from stercobilinogen to stercobilin. The concordance of the depletion of both stercobilin and stercobilinogen implies the importance of metabolism differences between TS2-NEO and WT mice. Because of the high number of differences observed in the gut microbiome of ASD and control subjects (Ding et al. 2017), it is tantalizing to speculate that bacterial population variations observed could lead to differences in bilin production. Feces can be collected non-invasively. The results here suggest that microbiome analysis coupled to molecular analysis of bilins from fecal material is warranted, and may provide a vehicle for a combined biochemical and molecular biological approach to ASD diagnosis.
Acknowledgments
We gratefully acknowledge the financial support of the National Institutes of Health through the National Center for Research Resources (Grant #S10-RR029517-01) for providing funding used to obtain the instrumentation used, the Mark Diamond Research Fund (Grant # FA-14-17), and the University at Buffalo for financial support for this research.
Funding: This study was funded by the National Institutes of Health through the National Center for Research Resources (Grant #S10-RR029517-01), the Mark Diamond Research Fund (Grant # FA-14-17).
Footnotes
Compliance with Ethical Standards:
Conflict of Interest: The authors declare that they have no conflict of interest.
Ethical approval: All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All procedures performed in studies involving animals were in accordance with the ethical standards of the institution or practice at which the studies were conducted.
This article does not contain any studies with human participants performed by any of the authors.
References
- Adamo N, Huo L, Adelsberg S, Petkova E, Castellanos FX, Di Martino A. Response time intra-subject variability: commonalities between children with autism spectrum disorders and children with ADHD. Eur Child Adolesc Psychiatry. 2014;23(2):69–79. doi: 10.1007/s00787-013-0428-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altieri L, Neri C, Sacco R, Curatolo P, Benvenuto A, Muratori F, et al. Urinary p-cresol is elevated in small children with severe autism spectrum disorder. Biomarkers. 2011;16(3):252–260. doi: 10.3109/1354750X.2010.548010. [DOI] [PubMed] [Google Scholar]
- Bader PL, Faizi M, Kim LH, Owen SF, Tadross MR, Alfa RW, et al. Mouse model of Timothy syndrome recapitulates triad of autistic traits. Proc Natl Acad Sci U S A. 2011;108(37):15432–15437. doi: 10.1073/pnas.1112667108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman ML. Medical comorbidities in autism: challenges to diagnosis and treatment. Neurotherapeutics. 2010;7(3):320–327. doi: 10.1016/j.nurt.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Itzchak E, Zachor DA. The effects of intellectual functioning and autism severity on outcome of early behavioral intervention for children with autism. Res Dev Disabil. 2007;28(3):287–303. doi: 10.1016/j.ridd.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Bergmann D, Hubner F, Humpf HU. Stable Isotope Dilution Analysis of Small Molecules with Carboxylic Acid Functions Using O-18 Labeling for HPLC-ESI-MS/MS: Analysis of Fumonisin B-1. Journal of Agricultural and Food Chemistry. 2013;61(33):7904–7908. doi: 10.1021/jf4022702. [DOI] [PubMed] [Google Scholar]
- Bett GC, Lis A, Wersinger SR, Baizer JS, Duffey ME, Rasmusson RL. A Mouse Model of Timothy Syndrome: a Complex Autistic Disorder Resulting from a Point Mutation in Cav1.2. N Am J Med Sci (Boston) 2012;5(3):135–140. doi: 10.7156/najms.2012.053135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Rudolf L, Poustka F. The cognitive structure of higher functioning autism and schizophrenia: a comparative study. Compr Psychiatry. 2002;43(4):325–330. doi: 10.1053/comp.2002.33490. [DOI] [PubMed] [Google Scholar]
- Bresnahan M, Hornig M, Schultz AF, Gunnes N, Hirtz D, Lie KK, et al. Association of maternal report of infant and toddler gastrointestinal symptoms with autism: evidence from a prospective birth cohort. JAMA Psychiatry. 2015;72(5):466–474. doi: 10.1001/jamapsychiatry.2014.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13(10):701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- Dager SR, Corrigan NM, Shaw DW. Brain lactate as a potential biomarker for comorbid anxiety disorder in autism spectrum disorder. JAMA Psychiatry. 2015;72(2):190. doi: 10.1001/jamapsychiatry.2014.2419. [DOI] [PubMed] [Google Scholar]
- Developmental Disabilities Monitoring Network Surveillance Year Principal I, Centers for Disease C & Prevention. Prevalence of autism spectrum disorder among children aged 8 years - autism and developmental disabilities monitoring network, 11 sites, United States, 2010. MMWR Surveill Summ. 2014;63(2):1–21. [PubMed] [Google Scholar]
- Dieme B, Mavel S, Blasco H, Tripi G, Bonnet-Brilhault F, Malvy J, et al. Metabolomics Study of Urine in Autism Spectrum Disorders Using a Multiplatform Analytical Methodology. J Proteome Res. 2015;14(12):5273–5282. doi: 10.1021/acs.jproteome.5b00699. [DOI] [PubMed] [Google Scholar]
- Ding HT, Taur Y, Walkup JT. Gut Microbiota and Autism: Key Concepts and Findings. J Autism Dev Disord. 2017;47(2):480–489. doi: 10.1007/s10803-016-2960-9. [DOI] [PubMed] [Google Scholar]
- Durieux AM, Horder J, Mendez MA, Egerton A, Williams SC, Wilson CE, et al. Cortical and subcortical glutathione levels in adults with autism spectrum disorder. Autism Res. 2016;9(4):429–435. doi: 10.1002/aur.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigsti IM, Shapiro T. A systems neuroscience approach to autism: biological, cognitive, and clinical perspectives. Ment Retard Dev Disabil Res Rev. 2003;9(3):205–215. doi: 10.1002/mrdd.10081. [DOI] [PubMed] [Google Scholar]
- Faber S, Zinn GM, Kern JC, 2nd, Kingston HM. The plasma zinc/serum copper ratio as a biomarker in children with autism spectrum disorders. Biomarkers. 2009;14(3):171–180. doi: 10.1080/13547500902783747. [DOI] [PubMed] [Google Scholar]
- Finegold SM, Dowd SE, Gontcharova V, Liu CX, Henley KE, Wolcott RD, et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe. 2010;16(4):444–453. doi: 10.1016/j.anaerobe.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Finegold SM, Molitoris D, Song Y, Liu C, Vaisanen ML, Bolte E, et al. Gastrointestinal microflora studies in late-onset autism. Clin Infect Dis. 2002;35(Suppl 1):S6–S16. doi: 10.1086/341914. [DOI] [PubMed] [Google Scholar]
- Frye RE, Delatorre R, Taylor H, Slattery J, Melnyk S, Chowdhury N, et al. Redox metabolism abnormalities in autistic children associated with mitochondrial disease. Transl Psychiatry. 2013;3:e273. doi: 10.1038/tp.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriele S, Sacco R, Cerullo S, Neri C, Urbani A, Tripi G, et al. Urinary p-cresol is elevated in young French children with autism spectrum disorder: a replication study. Biomarkers. 2014;19(6):463–470. doi: 10.3109/1354750X.2014.936911. [DOI] [PubMed] [Google Scholar]
- Goldenthal MJ, Damle S, Sheth S, Shah N, Melvin J, Jethva R, et al. Mitochondrial enzyme dysfunction in autism spectrum disorders; a novel biomarker revealed from buccal swab analysis. Biomark Med. 2015;9(10):957–965. doi: 10.2217/bmm.15.72. [DOI] [PubMed] [Google Scholar]
- Gorrindo P, Lane CJ, Lee EB, McLaughlin B, Levitt P. Enrichment of elevated plasma F2t-isoprostane levels in individuals with autism who are stratified by presence of gastrointestinal dysfunction. PLoS One. 2013;8(7):e68444. doi: 10.1371/journal.pone.0068444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotham K, Pickles A, Lord C. Standardizing ADOS scores for a measure of severity in autism spectrum disorders. J Autism Dev Disord. 2009;39(5):693–705. doi: 10.1007/s10803-008-0674-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert MR, Russo JP, Yang S, Roohi J, Blaxill M, Kahler SG, et al. Autism and environmental genomics. Neurotoxicology. 2006;27(5):671–684. doi: 10.1016/j.neuro.2006.03.017. [DOI] [PubMed] [Google Scholar]
- Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155(7):1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James SJ, Cutler P, Melnyk S, Jernigan S, Janak L, Gaylor DW, et al. Metabolic biomarkers of increased oxidative stress and impaired methylation capacity in children with autism. Am J Clin Nutr. 2004;80(6):1611–1617. doi: 10.1093/ajcn/80.6.1611. [DOI] [PubMed] [Google Scholar]
- Jyonouchi H, Geng L, Cushing-Ruby A, Quraishi H. Impact of innate immunity in a subset of children with autism spectrum disorders: a case control study. J Neuroinflammation. 2008;5:52. doi: 10.1186/1742-2094-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DW, Park JG, Ilhan ZE, Wallstrom G, Labaer J, Adams JB, et al. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS One. 2013;8(7):e68322. doi: 10.1371/journal.pone.0068322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanner L, Eisenberg L. Early infantile autism, 1943–1955. Psychiatr Res Rep Am Psychiatr Assoc. 1957;7:55–65. doi: 10.4159/harvard.9780674367012.c2. [DOI] [PubMed] [Google Scholar]
- Kohane IS, McMurry A, Weber G, MacFadden D, Rappaport L, Kunkel L, et al. The co-morbidity burden of children and young adults with autism spectrum disorders. PLoS One. 2012;7(4):e33224. doi: 10.1371/journal.pone.0033224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajmalnik-Brown R, Lozupone C, Kang DW, Adams JB. Gut bacteria in children with autism spectrum disorders: challenges and promise of studying how a complex community influences a complex disease. Microb Ecol Health Dis. 2015;26:26914. doi: 10.3402/mehd.v26.26914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krey JF, Pasca SP, Shcheglovitov A, Yazawa M, Schwemberger R, Rasmusson R, et al. Timothy syndrome is associated with activity-dependent dendritic retraction in rodent and human neurons. Nat Neurosci. 2013;16(2):201–209. doi: 10.1038/nn.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Cook EH, Leventhal BL, Amaral DG. Autism spectrum disorders. Neuron. 2000a;28(2):355–363. doi: 10.1016/s0896-6273(00)00115-x. [DOI] [PubMed] [Google Scholar]
- Lord C, Risi S, Lambrecht L, Cook EH, Jr, Leventhal BL, DiLavore PC, et al. The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J Autism Dev Disord. 2000b;30(3):205–223. [PubMed] [Google Scholar]
- Masi A, DeMayo MM, Glozier N, Guastella AJ. An Overview of Autism Spectrum Disorder, Heterogeneity and Treatment Options. Neurosci Bull. 2017;33(2):183–193. doi: 10.1007/s12264-017-0100-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermet JM. Limit of quantitation in atomic spectrometry: An unambiguous concept? Spectrochimica Acta Part B-Atomic Spectroscopy. 2008;63(2):166–182. doi: 10.1016/j.sab.2007.11.029. [DOI] [Google Scholar]
- Ming X, Stein TP, Barnes V, Rhodes N, Guo L. Metabolic perturbance in autism spectrum disorders: a metabolomics study. J Proteome Res. 2012;11(12):5856–5862. doi: 10.1021/pr300910n. [DOI] [PubMed] [Google Scholar]
- Ming X, Stein TP, Brimacombe M, Johnson WG, Lambert GH, Wagner GC. Increased excretion of a lipid peroxidation biomarker in autism. Prostaglandins Leukot Essent Fatty Acids. 2005;73(5):379–384. doi: 10.1016/j.plefa.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Mulle JG, Sharp WG, Cubells JF. The gut microbiome: a new frontier in autism research. Curr Psychiatry Rep. 2013;15(2):337. doi: 10.1007/s11920-012-0337-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngounou Wetie AG, Wormwood KL, Charette L, Ryan JP, Woods AG, Darie CC. Comparative two-dimensional polyacrylamide gel electrophoresis of the salivary proteome of children with autism spectrum disorder. J Cell Mol Med. 2015;19(11):2664–2678. doi: 10.1111/jcmm.12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ospina MB, Krebs Seida J, Clark B, Karkhaneh M, Hartling L, Tjosvold L, et al. Behavioural and developmental interventions for autism spectrum disorder: a clinical systematic review. PLoS One. 2008;3(11):e3755. doi: 10.1371/journal.pone.0003755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozonoff S, Goodlin-Jones BL, Solomon M. Evidence-based assessment of autism spectrum disorders in children and adolescents. J Clin Child Adolesc Psychol. 2005;34(3):523–540. doi: 10.1207/s15374424jccp3403_8. [DOI] [PubMed] [Google Scholar]
- Parracho HM, Bingham MO, Gibson GR, McCartney AL. Differences between the gut microflora of children with autistic spectrum disorders and that of healthy children. J Med Microbiol. 2005;54(Pt 10):987–991. doi: 10.1099/jmm.0.46101-0. [DOI] [PubMed] [Google Scholar]
- Pastural E, Ritchie S, Lu Y, Jin W, Kavianpour A, Khine Su-Myat K, et al. Novel plasma phospholipid biomarkers of autism: mitochondrial dysfunction as a putative causative mechanism. Prostaglandins Leukot Essent Fatty Acids. 2009;81(4):253–264. doi: 10.1016/j.plefa.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Quinn KD, Nguyen NQ, Wach MM, Wood TD. Tandem mass spectrometry of bilin tetrapyrroles by electrospray ionization and collision-induced dissociation. Rapid Commun Mass Spectrom. 2012;26(16):1767–1775. doi: 10.1002/rcm.6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey JM, Guest PC, Broek JA, Glennon JC, Rommelse N, Franke B, et al. Identification of an age-dependent biomarker signature in children and adolescents with autism spectrum disorders. Mol Autism. 2013;4(1):27. doi: 10.1186/2040-2392-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SH, Pothoulakis C, Mayer EA. Principles and clinical implications of the brain-gut-enteric microbiota axis. Nat Rev Gastroenterol Hepatol. 2009;6(5):306–314. doi: 10.1038/nrgastro.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinehart NJ, Bradshaw JL, Brereton AV, Tonge BJ. A clinical and neurobehavioural review of high-functioning autism and Asperger’s disorder. Aust N Z J Psychiatry. 2002;36(6):762–770. doi: 10.1046/j.1440-1614.2002.01097.x. [DOI] [PubMed] [Google Scholar]
- Rogers GB, Keating DJ, Young RL, Wong ML, Licinio J, Wesselingh S. From gut dysbiosis to altered brain function and mental illness: mechanisms and pathways. Molecular Psychiatry. 2016;21(6):738–748. doi: 10.1038/mp.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph HL, Friesen WL, Wood TD. The Hunt for Biomarkers of Autism Spectrum Disorders. J Metabol Sys Biol. 2013;1(1):1–11. [Google Scholar]
- Rudolph HL, Sekera ER, Wood TD. Stable (18) O-labeling method for stercobilin and other bilins for metabolomics. Rapid Commun Mass Spectrom. 2016;30(13):1469–1474. doi: 10.1002/rcm.7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwin E, Sandhu KV, Dinan TG, Cryan JF. May the Force Be With You: The Light and Dark Sides of the Microbiota-Gut-Brain Axis in Neuropsychiatry. CNS Drugs. 2016;30(11):1019–1041. doi: 10.1007/s40263-016-0370-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Liu C, Finegold SM. Real-time PCR quantitation of clostridia in feces of autistic children. Appl Environ Microbiol. 2004;70(11):6459–6465. doi: 10.1128/AEM.70.11.6459-6465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splawski I, Timothy KW, Decher N, Kumar P, Sachse FB, Beggs AH, et al. Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations. Proc Natl Acad Sci U S A. 2005;102(23):8089–8096. doi: 10.1073/pnas.0502506102. discussion 8086–8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitek L, Majer F, Muchova L, Zelenka J, Jiraskova A, Branny P, et al. Identification of bilirubin reduction products formed by Clostridium perfringens isolated from human neonatal fecal flora. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;833(2):149–157. doi: 10.1016/j.jchromb.2006.01.032. [DOI] [PubMed] [Google Scholar]
- Wang L, Christophersen CT, Sorich MJ, Gerber JP, Angley MT, Conlon MA. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol Autism. 2013;4(1):42. doi: 10.1186/2040-2392-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CJ. The Origin of Natural Crystalline Urobilin (Stercobilin) J Biol Chem. 1936;114(1):47–57. [Google Scholar]
- Watson CJ, Lowry PT, Sborov VE, Hollinshead WH, Kohan S, Matte HO. A simple method of isolation of crystalline stercobilin or urobilin from feces. J Biol Chem. 1953;200(2):697–701. [PubMed] [Google Scholar]
- Wood TD, Pennington CL, Choi YS. Stercobilin: A Possible Biomarker for Autism?. Paper presented at the Proc. 55th ASMS Conference on Mass Spectrometry & Allied Topics; Indianapolis, IN. June 3–7, 2007.2007. [Google Scholar]
- Woods AG, Wormwood KL, Wetie AG, Aslebagh R, Crimmins BS, Holsen TM, et al. Autism spectrum disorder: an omics perspective. Proteomics Clin Appl. 2015;9(1–2):159–168. doi: 10.1002/prca.201400116. [DOI] [PubMed] [Google Scholar]