Abstract
The calcium ion (Ca2+) is a key intracellular signaling molecule with far-reaching effects on many cellular processes. One of the most important such Ca2+ regulated processes is transcription. A body of literature describes the effect of Ca2+ signaling on transcription initiation as occurring mainly through activation of gene-specific transcription factors by Ca2+-induced signaling cascades. However, the reach of Ca2+ extends far beyond the first step of transcription. In fact, Ca2+ can regulate all phases of transcription, with additional effects on transcription-associated events such as alternative splicing. Importantly, Ca2+ signaling mediates reduced transcription termination in response to certain stress conditions. This reduction allows readthrough transcription, generating a highly inducible and diverse class of downstream of gene containing transcripts (DoGs) that we have recently described.
Keywords: Ca2+ signaling, transcription regulation, readthrough transcription, DoGs
Introduction
In this review, we first provide a brief overview of Ca2+ signaling. Next, we describe how Ca2+ signaling affects the different aspects of transcription downstream of transcription initiation, as well as the generation of enhancer RNAs (eRNAs).
Ca2+ signaling – upstream of Ca2+ release
Ca2+-signaling has previously been reviewed extensively (for example, see [1–5]). See Figure 1 for an overview. Effective Ca2+ signaling requires low cytoplasmic Ca2+ concentrations in resting cells, achieved by active Ca2+ removal. To this end, Ca2+ is pumped into the endoplasmic reticulum (ER) via sarcoendoplasmic reticular Ca2+ ATPase (SERCA) pumps, or pumped out of the cell via plasma membrane Ca2+ ATPase (PMCA) pumps. The resulting low cytoplasmic Ca2+ concentrations of ~100 nM enable rapid cytoplasmic entry once Ca2+ channels are activated by signaling [1.2].
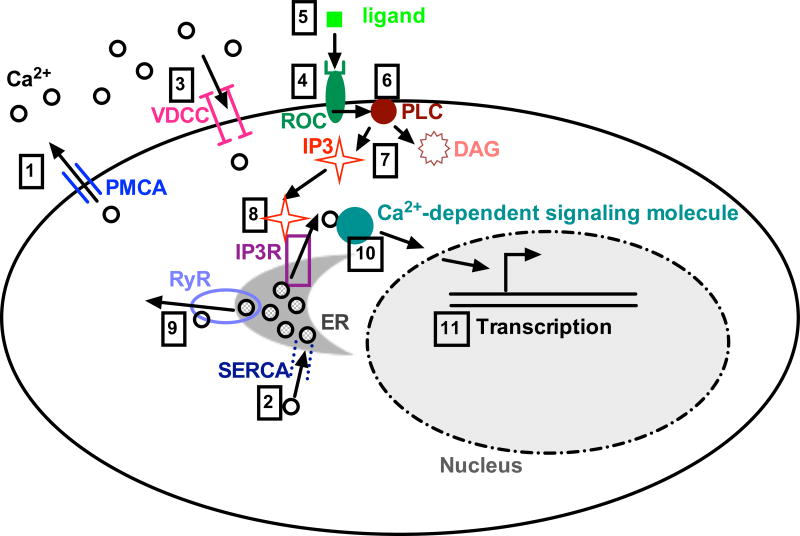
Figure 1. Overview of Ca2+ signaling.
Ca2+ is removed from the cytoplasm by export to the cell exterior by PMCA pumps (1) and by import to the ER by SERCA pumps (2). Signaling induces Ca2+ influx through VDCCs (3) and/or activation of ROCs (4). Ligand binding to ROCs (5) causes activation of PLCs (6), which generate the signaling molecules DAG and IP3 (7). IP3 then activates the IP3R at the ER, leading to Ca2+ release from the ER (8). The ER also contains RyRs that release Ca2+, with the main RyR ligand being Ca2+. Therefore RyRs effectively amplify Ca2+ signaling (9). Once released, Ca2+ binds to and activates downstream signaling molecules (10), ultimately affecting transcription (11). See text for details, abbreviations, and references.
Ca2+ can enter the cytoplasm from either the extracellular environment or intracellular stores. Ca2+ entry from the extracellular environment can occur through two types of plasma membrane Ca2+ channels: voltage-dependent Ca2+ channels (VDCCs) and receptor-operated (ligand-gated) channels (ROCs). The most important intracellular Ca2+ store is the ER; in the ER lumen, Ca2+ buffers bind Ca2+ with high capacity but low affinity, enabling efficient storage of Ca2+ that can be rapidly released upon signaling. The most prominent pathways for Ca2+ release from the ER rely on 1,4,5-inositol trisphosphate (IP3) receptors (IP3Rs) and ryanodine receptors (RyRs) [1–3].
IP3Rs are activated by IP3, which is generated at the plasma membrane by the action of Phospholipase C (PLC) on phosphatidylinositol 4,5 bisphosphate (PIP2). PLC transforms PIP2 into IP3 and diacylglycerol (DAG), another signaling molecule that activates a different set of downstream signaling pathways. Different PLC isoforms are activated by different membrane receptors, such as G-protein-coupled receptors and protein tyrosine kinase-linked receptors [1.2]. The IP3R functions as a homo- or heterotetramer. There are three different IP3R genes (IP3R1–3), which encode IP3R subtypes that have varying binding affinity for IP3; IP3R2 is the most sensitive, whereas both IP3R1 and 2 are significantly more sensitive than IP3R3. However, cell-specific variables such as IP3R isoform abundance also affect the relative importance of each receptor in an individual cell type [3].
RyRs are activated by an increase in cytosolic Ca2+ concentration into the low µM range [1]. Because Ca2+ is the primary RyR agonist, RyRs are mainly involved in propagating already initiated Ca2+ signaling [5]. RyRs exist in three different isoforms encoded by three different genes; they function as homotetramers [3].
Ca2+ signaling – downstream of Ca2+ release
Once Ca2+ is released into the cytosol, it quickly binds to various buffering and signaling molecules. Cytoplasmic Ca2+ buffering proteins bind released Ca2+ to prevent it from spreading far, thus keeping signaling local. Importantly for Ca2+ signaling, Ca2+ in the cytosol also binds a number of signaling molecules including calmodulin and protein kinase C (PKC), as described below.
Ca2+-bound calmodulin binds and activates a large number of downstream effectors, such as Ca2+/calmodulin-dependent protein kinases (CaMKs). CaMKs, most importantly CaMKIV, in turn phosphorylates and activates the transcription factor cyclic AMP (cAMP) response element-binding protein (CREB), which binds to cAMP response elements (CREs) in DNA to activate transcription [1. 2. 4]. Further, Ca2+-bound calmodulin binds and activates the phosphatase calcineurin. Once activated, calcineurin dephosphorylates cytosolic nuclear factor of activated T cells (NFAT), allowing hypophosphorylated NFAT to display a previously buried nuclear localization signal; NFAT enters the nucleus and acts as a transcription factor [1].
Additionally, Ca2+, once released into the cytoplasm, binds PKC. Association with positively charged Ca2+ allows PKC to relocalize to the negatively charged inside of the plasma membrane, where it becomes further activated by DAG and functions as a protein kinase. Once fully activated, PKC phosphorylates and regulates many substrates, including CREB [1.2].
Multiple other pathways are activated by Ca2+ signaling due to complex cross-talk mechanisms. Ca2+ activates calcium-sensitive adenylyl cyclases, which generate the signaling molecule cAMP. Ca2+ signaling can activate mitogen-activated protein kinase (MAPK) and phosphoinositide 3-kinase (PI3K) pathways [5]. For example, Ca2+, once released, stimulates PLC activity to generate DAG. DAG generation can then activate PI3K and protein kinase D (PKD), thus initiating a separate set of signaling cascades [5.6].
Effect of Ca2+ signaling on transcription and co-transcriptional pre-mRNA processing
Transcription and co-transcriptional pre-mRNA processing have been reviewed extensively (for example, see [7–16]).
In short, transcription can be divided into transcription initiation, elongation, and termination, with promoter-proximal pausing and pause release as intermediate steps between initiation and productive elongation. Additionally, pre-mRNA splicing occurs largely co-transcriptionally and is therefore affected by transcriptional regulation. See Figure 2 for an overview of transcription and Figure 3 for an overview of how Ca2+ signaling regulates transcription.
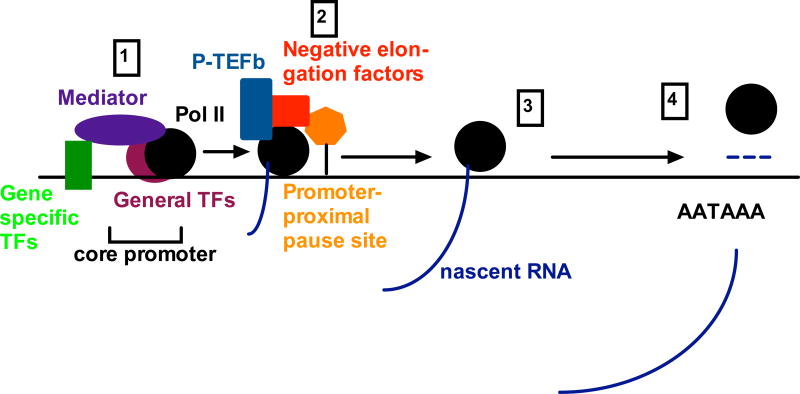
Figure 2. Overview of transcription.
Transcription is initiated when the PIC, consisting of Pol II and general transcription factors (TFs), is recruited to the core promoter by gene-specific transcription factors through interaction with the mediator complex (1). After initiating transcription of many genes, Pol II pauses within 100 nts of the core promoter – promoter proximal pausing – and is released through P-TEFb-mediated phosphorylation of the CTD of Pol II and of negative elongation factors (2). Once released, Pol II continues with productive elongation (3) until RNA synthesis undergoes the process of cleavage and adenylation and transcription termination. This process generates the 3′ end of the mRNA, and Pol II dissociates from the template DNA and from the RNA transcribed downstream of the cleavage and polyadenylation site in a mechanism that either precedes or is preceded by degradation of the downstream RNA (the allosteric or torpedo model, respectively) (4). See text for details, abbreviations, and references.
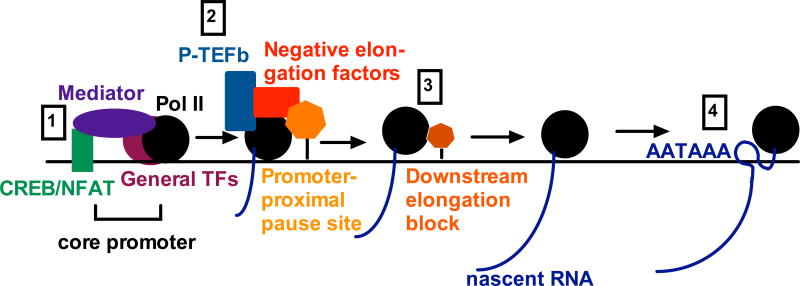
Figure 3. Aspects of transcription affected by Ca2+ signaling.
Ca2+ signaling regulates gene-specific transcription factors (TFs) such as CREB and NFAT, which activate transcriptional initiation (1). Further, Ca2+ signaling regulates phosphorylation of the PTEF-b subunit CDK9) on Thr-186, affecting PTEF-b activity in pause release (2). Ca2+ can also regulate release from downstream elongation blocks (3) and inhibit transcription termination (4), thus allowing DoG transcription. See text for details, abbreviations, and references.
Calcium signaling and transcript pause release
Transcription initiation is divided into two steps. First, a pre-initiation complex (PIC) forms on the core promoter, defined as the shortest DNA sequence (~50–100 nt) sufficient to initiate transcription [8]. The PIC contains RNA polymerase II (Pol II) and the general transcription factors. The PIC assembles on the core promoter through interaction with the mediator complex, which in turn contacts gene-specific transcription factors. Once the PIC is assembled, Pol II is released from the mediator complex and the core promoter sequence to start transcribing, concurrent with phosphorylation of the Pol II C-terminal domain (CTD) on serine-5 [7].
Recent studies show that on about a third of human genes, Pol II pauses after escaping the promoter. This pausing occurs within 100 nt of the transcription start site (TSS) in a process known as promoter-proximal pausing, which is induced by the association of Pol II with the factors DRB Sensitivity-Inducing Factor (DSIF) and Negative ELongation Factor (NELF). Promoter-proximal pausing is rate-limiting for transcription and transcriptional output can be regulated by varying the length of the pause [9.10].
The transition of Pol II from paused to productive elongation is mediated by Positive Transcription Elongation Factor-b (P-TEFb). P-TEFb phosphorylates the Pol II CTD at Serine-2 residues, as well as DSIF and NELF, which reduces suppression of elongation mediated by negative transcription factors and further causes NELF release from the elongation complex, allowing Pol II to enter into productive elongation.
P-TEFb can be directly recruited to the elongation complex by positive transcription factors. Further, P-TEFb activity is inhibited by its binding to 7SK RNA and the HEXIM1 protein [9.10]. Phosphorylation of Threonine 186 (Thr-186) of the CDK9 subunit of P-TEFb is required both for P-TEFb activity as a positive elongation factor and for P-TEFb interaction with 7SK and HEXIM1 [14]. Phosphorylation of CDK9 at Thr-186 is affected by Ca2+ signaling in several ways. First, Chen and colleagues reported that the Ca2+/calmodulin-activated phosphatase calcineurin is necessary but not sufficient to cause dephosphorylation of CDK9-Thr186 in HeLa cells [17]. The authors found that when calcineurin or calmodulin was inhibited by cyclosporine A or W-7, P-TEFb release from HEXIM1 and 7SK in response to UV- or hexamethylene bisacetamide (HMBA) treatment was prevented. They further observed that calcineurin affects Thr-186 phosphorylation by inducing a conformational change in the 7SK/HEXIM1/P-TEFb RNP, which in turn allows the phosphatase PP1α to access and dephosphorylate Thr-186, causing disassociation of the 7SK/HEXIM1/PTEFb complex. P-TEFb is then presumably re-phosphorylated, allowing it to function in pause release. In support for this model, the authors demonstrated that after calcineurin/PP1α-mediated dephosphorylation and release of P-TEFb from 7SK/HEXIM1, the released P-TEFb could act to enhance elongation in a luciferase reporter system [17].
Additionally, Ca2+ signaling may play a direct role in the phosphorylation of CDK9 at Thr-186. Resting CD4+ T-lymphocytes exhibit low levels of Thr-186 phosphorylation, whereas, after T-cell activation, Thr-186 is rapidly phosphorylated. Ramakrishnan and Rice found that inhibiting either CamK or calmodulin using the inhibitors KN-93 or W-7, respectively, reduced CDK9 Thr-186 phosphorylation in HeLa cells [18]. They further demonstrated the importance of Ca2+ signaling for Thr-186 phosphorylation in activated CD4+ T-cells by treating cells with Thapsigargin prior to activation. Thapsigargin prevents SERCA-mediated Ca2+ reuptake into the ER and therefore depletes ER Ca2+ stores, reducing the ER’s ability to release Ca2+ in response to signaling. Thapsigargin pretreatment led to reduced CDK9 Thr-186 phosphorylation in response to T-cell activation. Further, calmodulin inhibition led to decreased PTEFb activity in promoting elongation, measured by a luciferase reporter assay. In a screen for kinases involved in CDK9 Thr-186 phosphorylation, Ramakrishnan and Rice identified CaMK1D, although the effect may be indirect [18].
Ca2+ and release of transcription elongation blocks
Once Pol II has been released from pausing and becomes committed to productive elongation, the Pol II complex is generally stable, as demonstrated by the fact that it can continue transcribing for hundreds of kilobases [9]. However, several studies have reported Ca2+ regulated blocks to elongation after of pause-release. The best-studied instance occurs in the fos gene close to the exon1/intron1 border, as mapped by nuclear run-on assays and in vitro transcription assays in nuclear extracts [19.20]. The role of Ca2+ signaling in overcoming this block has been demonstrated in several systems. Elongation of fos transcripts is induced by treatment with a Ca2+ ionophore, which enables extracellular Ca2+ to cross the plasma membrane and thus activate Ca2+ signaling. Conversely, fos induction is prevented either by using cell culture media lacking Ca2+ or by pretreating cells with the Ca2+ chelator EGTA [20–24].
Another Ca2+-regulated transcription elongation block has been identified in exon 1 of rat MAP Kinase Phosphatase-1 (MKP-1), again mapped by nuclear run-on assays [25]. In rat pituitary GH4C1 cells treated with thyrotropin-releasing hormone (TRH), MKP-1 is rapidly induced upon Ca2+ -mediated release of this block to elongation. Ca2+-dependency was shown by pretreatment with EGTA prior to TRH stimulation, which caused reduced MKP-1 induction [25]. The distance between the TSSs and the sites of the fos or MKP-1 transcription elongation block – estimated at 300–400 nt downstream of the 5′ end of both transcripts – is not compatible with promoter-proximal pausing, which occurs within 100 nt of the TSS. Therefore, these situations might represent a different kind of transcription elongation control, but the nature of the block and the Ca2+-regulated pathways involved remain to be investigated.
Ca2+ transcription elongation rate, and alternative splicing
The nascent RNA generated through elongation is further processed by splicing: introns are removed and exons are joined together. Nearly all human genes are subject to alternative splicing, where alternative exons are included or excluded, or alternative splice sites generate longer or shorter exon variants. Recently, it has become evident that a large proportion of splicing occurs co-transcriptionally. Interestingly, Pol II elongation rate can affect splice site choice. A slower Pol II elongation rate promotes recognition of weaker splice sites [10.13.15.16]. In chromatin, DNA is wrapped around histone proteins to form nucleosomes, and nucleosomes need to be partially disassembled to allow ongoing transcription. The Pol II elongation rate is affected by nucleosome density and chromatin structure, which in turn correlate with chromatin and histone modifications. Histone acetylation is known to relax chromatin structure, increasing the Pol II elongation rate [26.27].
Ca2+ signaling has repeatedly been implicated in regulating histone modification. For example, Ca2+ signaling induced by depolarization of rat hippocampal cells has been coupled to histone 2B acetylation through a mechanism requiring CamK activity [28]. Further, ROCs activated by NMDA (NMDA receptors) cause Ca2+ influx, which ultimately leads to histone H3 acetylation through activation of the MAPK ERK [29.30].
Although the effects of these Ca2+-induced modifications on alternative splicing have not been explored in rat hippocampus, similar studies in mouse cells have connected Ca2+-induced chromatin modifications and alternative splicing. Depolarization of the mouse neuroblastoma cell line N2 causes Ca2+ influx through VDCCs and induces skipping of NCAM exon 18 [31]. This exon skipping event is dependent on increased histone H3 and H4 acetylation. Specifically, H3K9ac was increased in a region of the NCAM gene encompassing the skipped exon and was accompanied by increased chromatin accessibility, suggesting less rigid chromatin structure in response to Ca2+ signaling [31]. An additional study found that depolarization of mouse cardiomyocytes induced histone H3 and H4 hyperacetylation, resulting in splicing changes in a number of genes [32]. Histone H3 and H4 acetylation was increased in the two alternatively spliced genes selected by the authors for detailed study, and this hyperacetylation was associated with increased Pol II elongation rate. Further, the authors observed decreased association of type II histone deacetylases (HDACs) – which act to remove histone acetyl modifications – and these genes after depolarization. Because phosphorylation of class II HDACs leads to their nuclear exclusion (reviewed in [33]), the authors hypothesized that a Ca2+-dependent kinase was involved in regulating class II HDAC localization in response to Ca2+-signaling. Indeed, they observed reduced histone acetylation in response to the combined inhibition of both CamKIIδ and PKD1, either by using inhibitors or through siRNA-mediated knockdown [32].
Ca2+ and transcription termination versus DoG production
The process of transcription ends with transcription termination. Transcription termination requires elongation through a cleavage and polyadenylation (polyA) signal (most commonly AAUAAA). Shortly after the polyA sequence, the nascent RNA is cleaved by the process of cleavage and polyadenylation. Non-templated A residues are added to the 3′ end of what will become the mRNA, while the 5′ end of the downstream RNA that is still attached to, and being extended by, Pol II remains unprotected. Actual transcription termination – the dissociation of Pol II from the template DNA and from the RNA – occurs within nucleotides to kilobases downstream of the polyA signal. Two models for the mechanism of transcription termination have been proposed: the allosteric model and the torpedo model. In the allosteric model, transcription through the polyA signal causes a conformation change in Pol II, which later leads to dissociation from the template DNA. In the alternative torpedo model, the unprotected 5′ end of the downstream transcript generated by the cleavage of the nascent RNA serves as an entry point for an exonuclease, which degrades the nascent transcript and causes Pol II dissociation from the template DNA [11.12]. Two recent conflicting studies have provided new support for each model [34.35]; however, as these models are not mutually exclusive, the actual mechanism for termination is likely a combination of the two. In addition to the transcription of the polyA signal and the process of cleavage and polyadenylation, Pol II pausing near the end of genes may also favor transcription termination [11.12].
We have recently discovered a link between Ca2+ signaling and transcription termination that affects more than 10% of human genes [36]. Our interest in Ca2+-mediated regulation of transcription termination originated from our studies of a putative ncRNA associated with bad-prognosis neuroblastoma. We found that this ncRNA was induced ~30-fold in response to osmotic stress in SK-N-BE(2)C neuroblastoma cells as well as in several other human cell lines. Using RNA-Seq and bioinformatic analyses, we determined that the ncRNA was part of a transcript that appeared to be generated by transcriptional readthrough from the upstream protein-coding gene. By extending our analyses to encompass the whole transcriptome, we found that osmotic stress widely impacts transcription downstream of genes. We coined the term DoGs for these downstream of gene containing transcripts. We found that DoGs are generated downstream of more than 10% of human protein-coding genes. Their prevalence, in combination with the length of each individual DoG (often >45 kb), suggested that DoGs could explain a significant portion of intergenic transcripts. Further study confirmed this hypothesis: DoGs can account for as much as 20% of intergenic transcription in human cells.
DoGs appeared to be generated by transcriptional readthrough. However, alternatively, DoG transcription could be the result of initiation at TSSs overlapping or just downstream of the upstream DoG-associated protein-coding transcript. To distinguish between these two possibilities, we applied a high-throughput method of sequencing and mapping 5′-capped RNAs (Cap Seq) that we had recently developed [37]. Using this approach, we demonstrated that DoGs do not initiate at downstream, stress-inducible TSSs, but indeed result from transcriptional readthrough. This point was further confirmed by 1) using biotinylated probes targeting upstream transcripts combined with pull-down on streptavidin beads and detection of both the upstream transcript and DoG by qRT-PCR, and 2) by inhibiting transcription through the upstream gene using catalytically inactive CRISPR/Cas9 [38], which also prevented DoG generation. Together, these data demonstrated that DoG production depends on transcription of the upstream gene, and that the upstream transcript and the transcribed DoG region reside in the same molecule [36].
We found that DoGs are induced at the level of transcription after stress, as demonstrated by an increase in DoGs using the uridine analogue 5-ethynyl uridine to label newly synthesized RNA. After incorporation and cell lysis, 5-ethynyl uridine can be coupled to biotin, allowing the labeled RNA to be isolated on streptavidin beads. We observed full induction of selected DoGs already after 25 minutes of labeling, while transcript levels from the upstream genes that generate DoGs were unchanged. DoG half-lives of ~1 hour were unaffected by KCl treatment, as demonstrated by an actinomycin D chase. The fact that DoG upstream transcripts are not increased after stress indicates that DoG induction is due to reduced transcription termination of the upstream transcripts, not increased initiation of the upstream gene [36].
Further, we found Ca2+ signaling to be necessary for DoG induction; specifically, the release of intracellular Ca2+. This conclusion rests on pretreating cells with BAPTAAM, a membrane-permeable Ca2+ chelator that chelates intracellular as well as extracellular Ca2+, or with EGTA, a Ca2+ chelator that does not penetrate cell membranes. We found that BAPTA-AM prevented DoG induction by osmotic stress, whereas pretreating cells with EGTA did not affect the increase in DoG levels. We thus excluded extracellular Ca2+ as the agent for DoG-inducing Ca2+ signaling, indicating that the effect is due instead to release of intracellular Ca2+ [36].
As mentioned above, intracellular Ca2+ is mainly released from the ER through IP3Rs or RyRs. We investigated the effect of IP3R signaling on DoG induction by pretreating cells with the IP3R inhibitor 2-ABP. We found that in the presence of 2-ABP, DoG induction by KCl was abolished, indicating that DoG induction is mediated by Ca2+ release from the ER by the IP3R. To confirm this conclusion, we knocked down the three IP3R isoforms using siRNA and found that knocking down IP3R1 prevented DoG induction in response to osmotic stress. Next, we investigated downstream Ca2+-dependent pathways required for DoG induction and found that the combined inhibition of both CamKII and PKC/PKD using the inhibitors KN-93 and Gö6976 prevented DoG induction in response to stress [36]. See Figure 4 for an overview of the role of Ca2+ signaling in DoG induction.
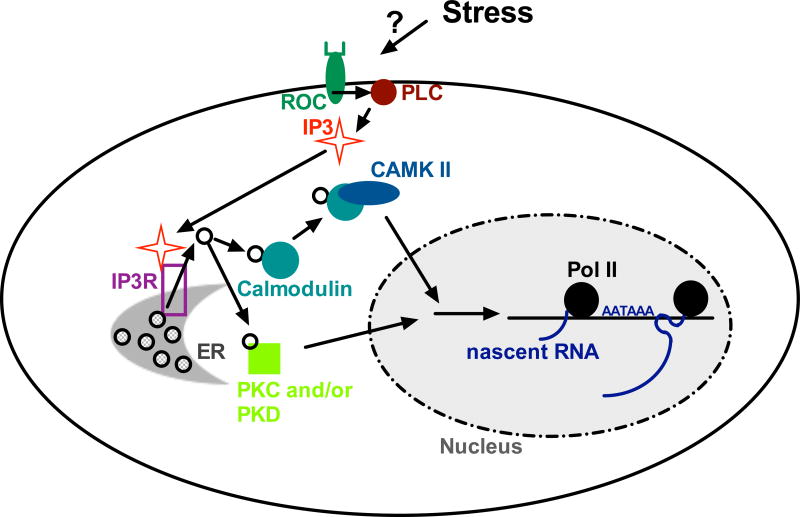
Figure 4. Ca2+ signaling results in DoG transcription.
IP3/IP3R signaling causes Ca2+ release from the ER, activating downstream CAMKII and PKC/PKD pathways, ultimately decreasing transcription termination. The mechanism remains to be elucidated. See text for details, abbreviations, and references.
Outstanding questions with regard to Ca2+-regulated DoG induction concern the mechanism through which osmotic stress induces IP3R-mediated Ca2+-release, as well as the nature of the actual perturbation of transcription termination. Several studies report increased cytosolic Ca2+ and IP3 release after osmotic stress [39] or heat shock [40]. Surprisingly, pretreatment of human neuroblastoma SK-N-BE(2)C cells with U73122, which is generally considered to be an inhibitor of receptor-mediated PLC activation, did not prevent DoG induction in response to osmotic stress (our unpublished results). This result suggests that receptor-activated IP3 generation does not increase in response to osmotic stress in SK-N-BE(2)C cells. However, U73122 effects on PLC activation may be complex [41], raising the possibility that U73122 may not prevent all PLC activation. For example, osmotic stress might activate PLCs by a mechanism that is not dependent on agonist binding to a membrane receptor. It is unclear how U73122 would affect such atypical PLC activation.
As to the exact mechanism by which the IP3R signaling affects the process of transcription termination, there are a number of possibilities. We found that knockdown of CPSF73, the catalytic subunit of the cleavage and polyadenylation complex, leads to modest DoG induction (about 4-fold) [36], suggesting that the cleavage and polyadenylation complex may be targeted by Ca2+ signaling to reduce transcription termination. Further, joint signaling of the PKD and CamKII pathways have been found to enhance Pol II elongation through reducing the nuclear localization of class II HDACs (see the discussion on Ca2+ and alternative splicing above) [32], which may reduce the amount of Pol II pausing near the end of genes, thus preventing termination. However, it is unclear whether an indirect effect on chromatin modification would occur quickly enough to explain rapid DoG induction after stress. Additionally, our preliminary data do not suggest any changes in HDAC localization after osmotic stress in our system. Finally, it is possible that other, perhaps as-of-yet unidentified, factors involved in transcription termination are affected by Ca2+ signaling. Further studies are required to elucidate IP3R activation and effect on termination in response to osmotic stress.
DoG Function
To pursue the putative function(s) of DoGs, we used the Ca2+ dependency of DoG transcription as a tool to prevent global DoG induction in response to stress. DoG localization on chromatin – demonstrated by cellular fractionation assays and RNA FISH – suggests that DoGs may function in a chromatin stress response. Additionally, the diversity among DoGs suggests that their putative function(s) is/are sequence independent. Because hyperosmotic stress – the type of stress used in [36] – forces water to leave the cell, nuclear shrinkage and chromatin condensation result. Recently, ncRNAs have been implicated in reinforcing the nuclear scaffold and maintaining euchromatin [42]. Thus, we hypothesized that DoGs may similarly reinforce the nuclear scaffold after stress. After exposure of SK-N-BE(2)C neuroblastoma cells to osmotic stress in the presence of the IP3R inhibitor 2-ABP, which prevents DoG induction (see above), nuclear phenotypes associated with osmotic stress were aggravated. These preliminary indications that DoGs are involved in reinforcing the nuclear scaffold after stress suggest the exciting possibility of sequence-independent, structural roles for this new large class of ncRNAs [36].
Interestingly, we found induction of selected DoGs also in response to heat shock [36]. Similar transcriptional readthrough has been observed in response to viral stress induced by HSV-1 infection [43] and in renal carcinoma [44]. Together these observations suggest that DoG induction is not limited to osmotic stress but may be a feature of a more general nuclear stress response. Although cytoplasmic responses to proteotoxic stresses such as heat shock and osmotic stress have been studied extensively, less is known about how the nucleus copes with such stress. Evidence from yeast suggests that proteotoxic stress responses are connected to pathways protecting DNA and RNA integrity (reviewed in [45]). Perhaps DoGs represent such a mechanism through which the nucleus protects its DNA content and RNA output from the deleterious effects of stress.
Ca2+ signaling and other types of non-coding transcripts
In recent years, a wealth of non-coding RNAs (ncRNAs) has been discovered (reviewed in [46.47]). Considering the abundance of types of ncRNA species, it is perhaps not surprising that other Ca2+-regulated ncRNAs apart from DoGs have been identified. An important class of ncRNAs, namely eRNAs was identified through Ca2+-mediated enhancer activation. Kim and co-authors [48] induced Ca2+ signaling by KCl-mediated depolarization of mouse primary neurons in culture, which leads to influx of Ca2+ through VDCCs. They observed that a large fraction (~40%) of activity-induced enhancers were transcribed bi-directionally, generating short (<2 kb) transcripts, referred to as eRNAs. Moreover, eRNA induction by Ca2+ signaling correlated with induction of neighboring genes [48]. Further studies have confirmed that at least a fraction of eRNAs contribute to enhancer function, potentially by facilitating chromatin looping to enable contacts between enhancers and promoters (reviewed in [49]).
Conclusions and outlook
As our understanding of transcriptional regulation deepens, it is becoming increasingly clear that all steps of the transcription cycle are subject to tight regulation. Such regulation affects the transcriptional output of a given gene, but may also lead to the generation of alternative, often non-coding, transcripts. Clearly, regulation of any and all stages of transcription has the potential to significantly affect cellular processes. Because Ca2+ signaling is such a conserved and important regulatory circuit, it is not surprising that it influences so many aspects of transcription. Additionally, dysregulated Ca2+ signaling is associated with a number of human diseases, for instance heart disease and neurological pathologies [2]. Further research will identify the molecular targets of Ca2+ signaling that execute the various effects of Ca2+ on transcription and will aid our understanding of how these effects can be manipulated to improve treatment of Ca2+ signaling-related disease.
Acknowledgments
We thank Kazimierz Tycowski and Mingyi Xie for critical discussion and Angela Miccinello for editorial assistance. This work was supported by grant GM026154 from the National Institutes of Health. A.V. was supported by the Wenner-Gren Foundations, Swedish Society for Medical Research and Sweden-America Foundation. J.A.S is an Investigator at the Howard Hughes Medical Institute.
Abbreviations
- cAMP
cyclic AMP
- CaMKs
Ca2+/calmodulin-dependent protein kinases
- CREs
cAMP response elements
- CREB
CRE-binding protein
- CTD
C-terminal domain
- DAG
diacylglycerol
- DoG
downstream of gene containing transcript
- DSIF
DRB Sensitivity-Inducing Factor
- ER
endoplasmic reticulum
- eRNAs
enhancer RNAs
- HDACs
histone deacetylases
- HMBA
hexamethylene bisacetamide
- IP3
1,4,5-inositol trisphosphate
- IP3R
IP3 receptors
- MAPK
mitogen-activated protein kinase
- MKP-1
MAP Kinase Phosphatase-1
- ncRNAs
non-coding RNAs
- NELF
Negative ELongation Factor
- NFAT
nuclear factor of activated T cells
- PI3K
phosphoinositide 3-kinase
- PIP2
phosphatidylinositol 4,5 bisphosphate
- PLC
Phospholipase C
- PKC
protein kinase C
- PMCA
plasma membrane Ca2+ ATPase pumps
- Pol II
RNA polymerase II
- polyA signal
polyadenylation signal
- P-TEFb
Positive Transcription Elongation Factor-b
- ROC
receptor-operated channel
- RyRs
ryanodine receptors
- SERCA
sarcoendoplasmic reticular Ca2+ ATPase pumps
- Thr-186
Threonine 186
- TSS
transcription start site
- VDCC
voltage-dependent Ca2+ channel
Footnotes
Conflicting interests
The authors have declared that no competing interests exist.
References
- 1.Clapham DE. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 2.Berridge MJ. Calcium signalling remodelling and disease. Biochem Soc Trans. 2012;40:297–309. doi: 10.1042/BST20110766. [DOI] [PubMed] [Google Scholar]
- 3.Kawamoto EM, Vivar C, Camandola S. Physiology and pathology of calcium signaling in the brain. Front Pharmacol. 2012;3:61. doi: 10.3389/fphar.2012.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbado M, Fablet K, Ronjat M, De Waard M. Gene regulation by voltage-dependent calcium channels. Biochim Biophys Acta. 2009;1793:1096–1104. doi: 10.1016/j.bbamcr.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Bading H. Nuclear calcium signalling in the regulation of brain function. Nat Rev Neurosci. 2013;14:593–608. doi: 10.1038/nrn3531. [DOI] [PubMed] [Google Scholar]
- 6.Kunkel MT, Toker A, Tsien RY, Newton AC. Calcium-dependent regulation of protein kinase D revealed by a genetically encoded kinase activity reporter. J Biol Chem. 2007;282:6733–6742. doi: 10.1074/jbc.M608086200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allen BL, Taatjes DJ. The Mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol. 2015;16:155–166. doi: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danino YM, Even D, Ideses D, Juven-Gershon T. The core promoter: At the heart of gene expression. Biochim Biophys Acta. 2015;1849:1116–1131. doi: 10.1016/j.bbagrm.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta. 2011;1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porrua O, Libri D. Transcription termination and the control of the transcriptome: why where and how to stop. Nat Rev Mol Cell Biol. 2015;16:190–202. doi: 10.1038/nrm3943. [DOI] [PubMed] [Google Scholar]
- 13.Carrillo Oesterreich F, Bieberstein N, Neugebauer KM. Pause locally, splice globally. Trends Cell Biol. 2011;21:328–335. doi: 10.1016/j.tcb.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Diribarne G, Bensaude O. 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor. RNA Biol. 2009;6:122–128. doi: 10.4161/rna.6.2.8115. [DOI] [PubMed] [Google Scholar]
- 15.Sharma A, Lou H. Depolarization-mediated regulation of alternative splicing. Front Neurosci. 2011;5:141. doi: 10.3389/fnins.2011.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dujardin G, Lafaille C, Petrillo E, Buggiano V, Gomez Acuna LI, Fiszbein A, et al. Transcriptional elongation and alternative splicing. Biochim Biophys Acta. 2013;1829:134–140. doi: 10.1016/j.bbagrm.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 17.Chen R, Liu M, Li H, Xue Y, Ramey WN, He N, et al. PP2B and PP1alpha cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev. 2008;22:1356–1368. doi: 10.1101/gad.1636008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramakrishnan R, Rice AP. Cdk9 T-loop phosphorylation is regulated by the calcium signaling pathway. J Cell Physiol. 2012;227:609–617. doi: 10.1002/jcp.22760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mechti N, Piechaczyk M, Blanchard JM, Jeanteur P, Lebleu B. Sequence requirements for premature transcription arrest within the first intron of the mouse c-fos gene. Mol Cell Biol. 1991;11:2832–2841. doi: 10.1128/mcb.11.5.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collart MA, Tourkine N, Belin D, Vassalli P, Jeanteur P, Blanchard JM. c-fos gene transcription in murine macrophages is modulated by a calcium-dependent block to elongation in intron 1. Mol Cell Biol. 1991;11:2826–2831. doi: 10.1128/mcb.11.5.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coulon V, Veyrune JL, Tourkine N, Vie A, Hipskind RA, Blanchard JM. A novel calcium signaling pathway targets the c-fos intragenic transcriptional pausing site. J Biol Chem. 1999;274:30439–30446. doi: 10.1074/jbc.274.43.30439. [DOI] [PubMed] [Google Scholar]
- 22.Werlen G, Belin D, Conne B, Roche E, Lew DP, Prentki M. Intracellular Ca2+ and the regulation of early response gene expression in HL-60 myeloid leukemia cells. J Biol Chem. 1993;268:16596–16601. [PubMed] [Google Scholar]
- 23.Lee G, Gilman M. Dual modes of control of c-fos mRNA induction by intracellular calcium in T cells. Mol Cell Biol. 1994;14:4579–4587. doi: 10.1128/mcb.14.7.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Haasteren G, Li S, Ryser S, Schlegel W. Essential contribution of intron sequences to Ca(2+)-dependent activation of c-fos transcription in pituitary cells. Neuroendocrinology. 2000;72:368–378. doi: 10.1159/000054606. [DOI] [PubMed] [Google Scholar]
- 25.Ryser S, Tortola S, van Haasteren G, Muda M, Li S, Schlegel W. MAP kinase phosphatase-1 gene transcription in rat neuroendocrine cells is modulated by a calcium-sensitive block to elongation in the first exon. J Biol Chem. 2001;276:33319–33327. doi: 10.1074/jbc.M102326200. [DOI] [PubMed] [Google Scholar]
- 26.Carey M, Li B, Workman JL. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol Cell. 2006;24:481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 28.Maharana C, Sharma KP, Sharma SK. Depolarization induces acetylation of histone H2B in the hippocampus. Neuroscience. 2010;167:354–360. doi: 10.1016/j.neuroscience.2010.02.023. [DOI] [PubMed] [Google Scholar]
- 29.Levenson JM, O'Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- 30.Chandramohan Y, Droste SK, Arthur JS, Reul JM. The forced swimming-induced behavioural immobility response involves histone H3 phospho-acetylation and c-Fos induction in dentate gyrus granule neurons via activation of the N-methyl-D-aspartate/extracellular signal-regulated kinase/mitogen- and stress-activated kinase signalling pathway. Eur J Neurosci. 2008;27:2701–2713. doi: 10.1111/j.1460-9568.2008.06230.x. [DOI] [PubMed] [Google Scholar]
- 31.Schor IE, Rascovan N, Pelisch F, Allo M, Kornblihtt AR. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc Natl Acad Sci U S A. 2009;106:4325–4330. doi: 10.1073/pnas.0810666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma A, Nguyen H, Geng C, Hinman MN, Luo G, Lou H. Calcium-mediated histone modifications regulate alternative splicing in cardiomyocytes. Proc Natl Acad Sci U S A. 2014;111:E4920–4928. doi: 10.1073/pnas.1408964111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKinsey TA. Derepression of pathological cardiac genes by members of the CaM kinase superfamily. Cardiovasc Res. 2007;73:667–677. doi: 10.1016/j.cardiores.2006.11.036. [DOI] [PubMed] [Google Scholar]
- 34.Fong N, Brannan K, Erickson B, Kim H, Cortazar MA, Sheridan RM, et al. Effects of Transcription Elongation Rate and Xrn2 Exonuclease Activity on RNA Polymerase II Termination Suggest Widespread Kinetic Competition. Mol Cell. 2015;60:256–267. doi: 10.1016/j.molcel.2015.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, Rigo F, Martinson HG. Poly(A) Signal-Dependent Transcription Termination Occurs through a Conformational Change Mechanism that Does Not Require Cleavage at the Poly(A) Site. Mol Cell. 2015;59:437–448. doi: 10.1016/j.molcel.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 36.Vilborg A, Passarelli MC, Yario TA, Tycowski KT, Steitz JA. Widespread Inducible Transcription Downstream of Human Genes. Mol Cell. 2015;59:449–461. doi: 10.1016/j.molcel.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie M, Li M, Vilborg A, Lee N, Shu MD, Yartseva V, et al. Mammalian 5'-capped microRNA precursors that generate a single microRNA. Cell. 2013;155:1568–1580. doi: 10.1016/j.cell.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Erickson GR, Alexopoulos LG, Guilak F. Hyper-osmotic stress induces volume change and calcium transients in chondrocytes by transmembrane, phospholipid, and G-protein pathways. J Biomech. 2001;34:1527–1535. doi: 10.1016/s0021-9290(01)00156-7. [DOI] [PubMed] [Google Scholar]
- 40.Calderwood SK, Stevenson MA, Hahn GM. Heat stress stimulates inositol trisphosphate release and phosphorylation of phosphoinositides in CHO and Balb C 3T3 cells. J Cell Physiol. 1987;130:369–376. doi: 10.1002/jcp.1041300309. [DOI] [PubMed] [Google Scholar]
- 41.Klein RR, Bourdon DM, Costales CL, Wagner CD, White WL, Williams JD, et al. Direct activation of human phospholipase C by its well known inhibitor u73122. J Biol Chem. 2011;286:12407–12416. doi: 10.1074/jbc.M110.191783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hall LL, Carone DM, Gomez AV, Kolpa HJ, Byron M, Mehta N, et al. Stable C0T-1 repeat RNA is abundant and is associated with euchromatic interphase chromosomes. Cell. 2014;156:907–919. doi: 10.1016/j.cell.2014.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rutkowski AJ, Erhard F, L'Hernault A, Bonfert T, Schilhabel M, Crump C, et al. Widespread disruption of host transcription termination in HSV-1 infection. Nat Commun. 2015;6:7126. doi: 10.1038/ncomms8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grosso AR, Leite AP, Carvalho S, Matos MR, Martins FB, Vitor AC, et al. Pervasive transcription read-through promotes aberrant expression of oncogenes and RNA chimeras in renal carcinoma. Elife. 2015;4 doi: 10.7554/eLife.09214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shibata Y, Morimoto RI. How the nucleus copes with proteotoxic stress. Curr Biol. 2014;24:R463–474. doi: 10.1016/j.cub.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 47.Bonasio R, Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu Rev Genet. 2014;48:433–455. doi: 10.1146/annurev-genet-120213-092323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lam MT, Li W, Rosenfeld MG, Glass CK. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci. 2014;39:170–182. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]