Abstract
Gliomas belong to a group of central nervous system tumors, and consist of various sub-regions. Gold standard labeling of these sub-regions in radiographic imaging is essential for both clinical and computational studies, including radiomic and radiogenomic analyses. Towards this end, we release segmentation labels and radiomic features for all pre-operative multimodal magnetic resonance imaging (MRI) (n=243) of the multi-institutional glioma collections of The Cancer Genome Atlas (TCGA), publicly available in The Cancer Imaging Archive (TCIA). Pre-operative scans were identified in both glioblastoma (TCGA-GBM, n=135) and low-grade-glioma (TCGA-LGG, n=108) collections via radiological assessment. The glioma sub-region labels were produced by an automated state-of-the-art method and manually revised by an expert board-certified neuroradiologist. An extensive panel of radiomic features was extracted based on the manually-revised labels. This set of labels and features should enable i) direct utilization of the TCGA/TCIA glioma collections towards repeatable, reproducible and comparative quantitative studies leading to new predictive, prognostic, and diagnostic assessments, as well as ii) performance evaluation of computer-aided segmentation methods, and comparison to our state-of-the-art method.
Subject terms: CNS cancer, Cancer imaging, Image processing, Translational research, Computational models
Background & Summary
Gliomas are the most common primary central nervous system malignancies. These tumors, which exhibit highly variable clinical prognosis, usually contain various heterogeneous sub-regions (i.e., edema, enhancing and non-enhancing core), with variable histologic and genomic phenotypes. This intrinsic heterogeneity of gliomas is also portrayed in their radiographic phenotypes, as their sub-regions are depicted by different intensity profiles disseminated across multimodal MRI (mMRI) scans, reflecting differences in tumor biology. There is increasing evidence that quantitative analysis of imaging features1–3 extracted from mMRI (i.e., radiomic features), beyond traditionally used clinical measurements (e.g., the largest anterior-posterior, transverse, and inferior-superior tumor dimensions, measured on a subjectively-/arbitrarily-chosen slice), through advanced computational algorithms, leads to advanced image-based tumor phenotyping4. Such phenotyping may enable assessment of reflected biological processes and assist in surgical and treatment planning. Furthermore, its correlation with molecular characteristics established radiogenomic research5–12, leading to improved predictive, prognostic and diagnostic imaging biomarkers9,12–32, hence yielding the potential benefit towards non-invasive precision medicine33. However, it is clear from current literature26,34–38 that such advanced image-based phenotyping requires accurate annotations of the various tumor sub-regions.
Both clinical and computational studies focusing on such research require the availability of ample data to yield significant associations. Considering the value of big data and the potential of publicly available datasets for increased reproducibility of scientific findings, the National Cancer Institute (NCI) of the National Institutes of Health (NIH) created TCGA (cancergenome.nih.gov) and TCIA39 (www.cancerimagingarchive.net). TCGA is a multi-institutional comprehensive collection of various molecularly characterized tumor types, and its data are available in NCI’s Genomic Data Commons portal (gdc-portal.nci.nih.gov). Building upon NIH’s investment in TCGA, the NCI’s Cancer Imaging Program approached sites that contributed tissue samples, to obtain corresponding de-identified routine clinically-acquired radiological data and store them in TCIA. These repositories make available multi-institutional, high-dimensional, multi-parametric data of cancer patients, allowing for radiogenomic analysis. However, the data available in TCIA lack accompanying annotations allowing to fully exploit their potential in clinical and computational studies.
Towards addressing this limitation, this study provides segmentation labels and a panel of radiomic features for the glioma datasets included in the TCGA/TCIA repositories. The main goal is to enable imaging and non-imaging researchers to conduct their analyses and extract measurements in a reproducible and repeatable manner, while eventually allowing for comparison across studies. Specifically, the resources of this study provide i) imaging experts with benchmarks to debate their algorithms, and ii) non-imaging experts (e.g., bioinformaticians, clinicians), who do not have the background to interpret and/or appropriately process the raw images, with data helpful to conduct correlative genomic/clinical studies. Following radiological assessment of both the Glioblastoma Multiforme (TCGA-GBM39, n=262 [Data Citation 1]) and the Low-Grade-Glioma (TCGA-LGG39, n=199 [Data Citation 2]) collections, we identified 135 and 108 pre-operative mMRI scans, respectively. These scans include at least pre- and post-contrast T1-weighted, T2-weighted, and T2 Fluid-Attenuated Inversion Recovery (FLAIR) volumes. The segmentation labels provided for these scans are divided into two categories: a) computer-aided segmentation labels that could be mainly used for computational comparative studies, and b) manually corrected segmentation labels (approved by an expert board-certified neuroradiologist—M.B.) for use in clinically-oriented analyses, as well as for performance evaluation and training of computational models. The method employed to produce the computer-aided labels is named GLISTRboost36,38, which was awarded the 1st prize during the International Multimodal Brain Tumor Image Segmentation challenge 2015 (BraTS’15)36,38,40–56.
The generated data describe two independent datasets [Data Citation 3 and Data Citation 4], one for each glioma collection, and include the computer-aided and manually-revised segmentation labels, coupled with the corresponding co-registered and skull-stripped TCIA scans, in the Neuroimaging Informatics Technology Initiative (NIfTI57) format, allowing for direct analysis. Furthermore, a panel of radiomic features is included entailing intensity, volumetric, morphologic, histogram-based, and textural parameters, as well as spatial information and parameters extracted from glioma growth models58–60. In consistency with the FAIR (Findable, Accessible, Interoperable, Re-usable) principle, these data are made available through TCIA and should enable both clinical and computational quantitative analyses, as well as serve as a resource for i) educational training of neuroradiology and neurosurgery residents, and ii) performance evaluation of segmentation methods. Furthermore, it could potentially lead to predictive, prognostic, and diagnostic imaging markers suitable for enabling oncological treatment models customized on an individual patient basis (precision medicine), through non-invasive quantification of disease processes.
Methods
Data collection
The complete radiological data of the TCGA-GBM and TCGA-LGG collections consist of 262 [Data Citation 1] and 199 [Data Citation 2] mMRI scans provided from 8 and 5 institutions, respectively (Table 1). The data included in this study describe the subset of the pre-operative baseline scans of these collections, with available MRI modalities of at least T1-weighted pre-contrast (T1), T1-weighted post-contrast (T1-Gd), T2, and T2-FLAIR (Fig. 1a). Specifically, we considered 135 and 108 pre-operative baseline scans from the TCIA-GBM and TCIA-LGG collections, respectively. Further detailed information on the diversity of the imaging sequences used for this study is included in Table 2 (available online only). This table covers the TCIA institutional identifier, patient information (i.e., age, sex, weight), scanner information (i.e., manufacturer, model, magnetic field strength, station name), as well as specific imaging volume information extracted from the dicom headers (i.e., modality name, series number, accession number, acquisition/study/series date, scan sequence, type, slice thickness, slice spacing, repetition time, echo time, inversion time, imaging frequency, flip angle, specific absorption rate, numbers of slices, pixel dimensions, acquisition matrix rows/columns).
Table 1. Source of radiographic data for patients (n) provided in TCIA.
| Collection | n | Institutions contributed data—(n) | TCGA ID | Scanner (strength in T) |
|---|---|---|---|---|
| TCGA-GBM | 262 | Henry Ford Hospital, Detroit, MI—(74) | TCGA-06 | GE (1.5, 3): Genesis Signa, Signa Excite |
| CWRU School of Medicine, Cleveland, OH—(38) | TCGA-19 | Siemens (1.5, 3): Avanto, Symphony, Verio | ||
| University of California, San Francisco, CA—(32) | TCGA-08 | GE (1.5, 3): Genesis Signa, Signa Excite | ||
| Emory University, Atlanta, GA—(31) | TCGA-14 | Philips (1.5): InteraSiemens (1.5, 3): Avanto, Trio | ||
| MD Anderson Cancer Center, Houston, TX—(25) | TCGA-02 | GE: Genesis Signa, Signa Excite | ||
| Duke University School of Medicine, Durham, NC—(24) | TCGA-12 | GE (1.5): Genesis Signa, Signa HDx,Signa ExciteSiemens (1.5, 3): Avanto, Trio, Symphony | ||
| Thomas Jefferson University, Philadelphia, PA—(22) | TCGA-76 | Philips (1.5, 3): AchievaSiemens (1.5): Magnetom Vision | ||
| Fondazione IRCCS Instituto Neuroligico C. Besta, Milan, Italy—(16) | TCGA-27 | Philips (0.5): Intera Siemens (1.5): Avanto | ||
| TCGA-LGG | 199 | St Joseph Hospital/Medical Center, Phoenix, AZ—(98) | TCGA-HT | GE (1.5, 3): Signa Excite, Signa HDx, Signa HDxt |
| Henry Ford Hospital, Detroit, MI—(57) | TCGA-DU | Hitachi (1.16): OasisGE (1.5, 3): Genesis, Signa Excite, Signa HDxt,Philips (1.5, 3): Intera, Ingenia | ||
| Case Western Reserve University, Cleveland, OH—(22) | TCGA-FG | Siemens (1.5, 3): Avanto, Symphony, Skyra, Verio | ||
| Thomas Jefferson University, Philadelphia, PA—(20) | TCGA-CS | GE (1.5): Genesis Signa, Signa HDxtPhilips (1.5, 3): AchievaSiemens (1.5): Magnetom Vision | ||
| University of North Carolina, Chapel Hill, NC—(2) | TCGA-EZ | Siemens (3): TrioTim |
Figure 1. Single slice multimodal (T1, T1-Gd, T2, T2-FLAIR) MRI scans of example subjects.
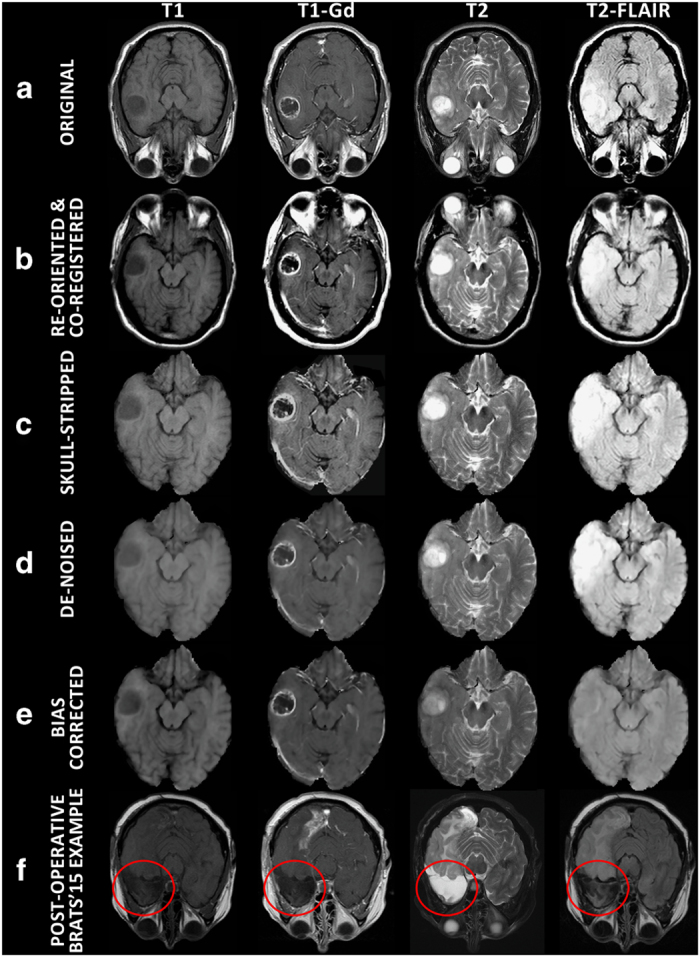
Examples are shown (a) in the original TCIA volume; (b–e) after application of various pre-processing steps; (f) for post-operative volumes in the BraTS ’15 data. Note that the step shown in (e), which is usually used to correct for intensity non-uniformities caused by the inhomogeneity of the scanner’s magnetic field during image acquisition, was not applied in the current data as it obliterated the T2-FLAIR signal.
Table 2. Detailed information capturing the diversity of the used images.
| Collection | TCGA ID | Included in BraTS'15 | BraTS'15 suggested a baseline pre-op scan | We Identified Baseline Pre-operative scans |
|---|---|---|---|---|
| TCGA-GBM | TCGA-02-0003 | Yes (Testing data) | Yes | Yes |
| TCGA-GBM | TCGA-02-0006 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-02-0009 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-02-0011 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-02-0027 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-02-0033 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-02-0034 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-02-0037 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-02-0046 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-02-0047 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-02-0048 | No | N/A | Yes |
| TCGA-GBM | TCGA-02-0054 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-02-0059 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-02-0060 | No | N/A | Yes |
| TCGA-GBM | TCGA-02-0064 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-02-0068 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-02-0069 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-02-0070 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-02-0075 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-02-0085 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-02-0086 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-02-0087 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-02-0102 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-02-0106 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-02-0116 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0119 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0121 | No | N/A | No |
| TCGA-GBM | TCGA-06-0122 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0125 | No | N/A | No |
| TCGA-GBM | TCGA-06-0127 | Yes (Testing data) | Yes | No |
| TCGA-GBM | TCGA-06-0128 | No | N/A | Yes |
| TCGA-GBM | TCGA-06-0129 | No | N/A | No |
| TCGA-GBM | TCGA-06-0130 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0132 | Yes (Training data) | No | Yes |
| TCGA-GBM | TCGA-06-0133 | Yes (Testing data) | No | Yes |
| TCGA-GBM | TCGA-06-0137 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0138 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0139 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0141 | Yes (Training data) | Yes | No |
| TCGA-GBM | TCGA-06-0142 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0143 | Yes (Training data) | No | No |
| TCGA-GBM | TCGA-06-0145 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0147 | Yes (Training data) | Yes | No |
| TCGA-GBM | TCGA-06-0148 | No | N/A | No |
| TCGA-GBM | TCGA-06-0149 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0154 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0156 | Yes (Testing data) | No | No |
| TCGA-GBM | TCGA-06-0157 | Yes (Training data) | Yes | No |
| TCGA-GBM | TCGA-06-0158 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0160 | No | N/A | No |
| TCGA-GBM | TCGA-06-0162 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0164 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0165 | Yes (Training data) | No | No |
| TCGA-GBM | TCGA-06-0166 | Yes (Testing data) | No | Yes |
| TCGA-GBM | TCGA-06-0168 | Yes (Testing data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0171 | Yes (Training data) | No | No |
| TCGA-GBM | TCGA-06-0173 | No | N/A | No |
| TCGA-GBM | TCGA-06-0174 | No | N/A | No |
| TCGA-GBM | TCGA-06-0175 | No | N/A | Yes |
| TCGA-GBM | TCGA-06-0176 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0177 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0178 | Yes (Testing data) | Yes | No |
| TCGA-GBM | TCGA-06-0179 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0182 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0184 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0185 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0187 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0188 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0189 | No | N/A | Yes |
| TCGA-GBM | TCGA-06-0190 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0192 | No | N/A | Yes |
| TCGA-GBM | TCGA-06-0210 | No | N/A | No |
| TCGA-GBM | TCGA-06-0213 | Yes (Testing data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0214 | Yes (Training data) | Yes | No |
| TCGA-GBM | TCGA-06-0216 | No | N/A | No |
| TCGA-GBM | TCGA-06-0221 | No | N/A | No |
| TCGA-GBM | TCGA-06-0237 | Yes (Training data) | Yes | No |
| TCGA-GBM | TCGA-06-0238 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0240 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0241 | Yes (Testing data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0644 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0645 | Yes (Testing data) | Yes | No |
| TCGA-GBM | TCGA-06-0646 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0648 | No | N/A | Yes |
| TCGA-GBM | TCGA-06-0649 | Yes (Testing data) | Yes | Yes |
| TCGA-GBM | TCGA-06-0878 | Yes (Testing data) | No | No |
| TCGA-GBM | TCGA-06-0881 | No | N/A | No |
| TCGA-GBM | TCGA-06-1084 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-1801 | Yes (Training data) | No | No |
| TCGA-GBM | TCGA-06-1802 | Yes (Training data) | No | Yes |
| TCGA-GBM | TCGA-06-1806 | No | N/A | No |
| TCGA-GBM | TCGA-06-2570 | No | N/A | Yes |
| TCGA-GBM | TCGA-06-5408 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-5410 | No | N/A | No |
| TCGA-GBM | TCGA-06-5412 | Yes (Testing data) | Yes | Yes |
| TCGA-GBM | TCGA-06-5413 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-06-5417 | No | N/A | Yes |
| TCGA-GBM | TCGA-06-6389 | No | N/A | Yes |
| TCGA-GBM | TCGA-06-6701 | No | N/A | No |
| TCGA-GBM | TCGA-08-0244 | No | N/A | No |
| TCGA-GBM | TCGA-08-0246 | No | N/A | No |
| TCGA-GBM | TCGA-08-0348 | No | N/A | No |
| TCGA-GBM | TCGA-08-0349 | No | N/A | No |
| TCGA-GBM | TCGA-08-0350 | No | N/A | Yes |
| TCGA-GBM | TCGA-08-0351 | No | N/A | No |
| TCGA-GBM | TCGA-08-0352 | No | N/A | Yes |
| TCGA-GBM | TCGA-08-0353 | No | N/A | Yes |
| TCGA-GBM | TCGA-08-0354 | No | N/A | Yes |
| TCGA-GBM | TCGA-08-0355 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-08-0356 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-08-0357 | No | N/A | Yes |
| TCGA-GBM | TCGA-08-0358 | No | N/A | Yes |
| TCGA-GBM | TCGA-08-0359 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-08-0360 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-08-0380 | No | N/A | No |
| TCGA-GBM | TCGA-08-0385 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-08-0389 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-08-0390 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-08-0392 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-08-0509 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-08-0510 | No | N/A | Yes |
| TCGA-GBM | TCGA-08-0511 | No | N/A | No |
| TCGA-GBM | TCGA-08-0512 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-08-0514 | No | N/A | No |
| TCGA-GBM | TCGA-08-0516 | No | N/A | No |
| TCGA-GBM | TCGA-08-0518 | No | N/A | No |
| TCGA-GBM | TCGA-08-0520 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-08-0521 | No | N/A | Yes |
| TCGA-GBM | TCGA-08-0522 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-08-0524 | No | N/A | Yes |
| TCGA-GBM | TCGA-08-0529 | Yes (Testing data) | Yes | Yes |
| TCGA-GBM | TCGA-12-0616 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-12-0620 | No | N/A | No |
| TCGA-GBM | TCGA-12-0769 | No | N/A | No |
| TCGA-GBM | TCGA-12-0772 | No | N/A | No |
| TCGA-GBM | TCGA-12-0773 | No | N/A | No |
| TCGA-GBM | TCGA-12-0775 | No | N/A | No |
| TCGA-GBM | TCGA-12-0776 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-12-0829 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-12-1092 | No | N/A | No |
| TCGA-GBM | TCGA-12-1093 | No | N/A | Yes |
| TCGA-GBM | TCGA-12-1094 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-12-1096 | Yes (Training data) | Yes | No |
| TCGA-GBM | TCGA-12-1097 | No | N/A | No |
| TCGA-GBM | TCGA-12-1098 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-12-1099 | No | N/A | No |
| TCGA-GBM | TCGA-12-1598 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-12-1599 | No | N/A | No |
| TCGA-GBM | TCGA-12-1600 | No | N/A | No |
| TCGA-GBM | TCGA-12-1601 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-12-1602 | No | N/A | Yes |
| TCGA-GBM | TCGA-12-3646 | No | N/A | No |
| TCGA-GBM | TCGA-12-3648 | No | N/A | No |
| TCGA-GBM | TCGA-12-3649 | No | N/A | No |
| TCGA-GBM | TCGA-12-3650 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-14-0736 | No | N/A | No |
| TCGA-GBM | TCGA-14-0783 | Yes (Training data) | Yes | No |
| TCGA-GBM | TCGA-14-0789 | Yes (Testing data) | No | Yes |
| TCGA-GBM | TCGA-14-0790 | Yes (Training data) | Yes | No |
| TCGA-GBM | TCGA-14-0812 | Yes (Training data) | Yes | No |
| TCGA-GBM | TCGA-14-0813 | Yes (Training data) | Yes | No |
| TCGA-GBM | TCGA-14-0817 | Yes (Testing data) | No | No |
| TCGA-GBM | TCGA-14-0865 | Yes (Training data) | Yes | No |
| TCGA-GBM | TCGA-14-0866 | No | N/A | No |
| TCGA-GBM | TCGA-14-0871 | Yes (Testing data) | Yes | No |
| TCGA-GBM | TCGA-14-1034 | No | N/A | No |
| TCGA-GBM | TCGA-14-1037 | Yes (Testing data) | No | No |
| TCGA-GBM | TCGA-14-1043 | Yes (Training data) | Yes | No |
| TCGA-GBM | TCGA-14-1395 | No | N/A | No |
| TCGA-GBM | TCGA-14-1396 | No | N/A | No |
| TCGA-GBM | TCGA-14-1401 | Yes (Training data) | No | No |
| TCGA-GBM | TCGA-14-1402 | Yes (Testing data) | No | No |
| TCGA-GBM | TCGA-14-1452 | Yes (Testing data) | No | No |
| TCGA-GBM | TCGA-14-1453 | Yes (Training data) | No | No |
| TCGA-GBM | TCGA-14-1454 | Yes (Training data) | No | No |
| TCGA-GBM | TCGA-14-1456 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-14-1458 | Yes (Training data) | No | No |
| TCGA-GBM | TCGA-14-1459 | Yes (Training data) | No | No |
| TCGA-GBM | TCGA-14-1794 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-14-1795 | Yes (Testing data) | No | No |
| TCGA-GBM | TCGA-14-1821 | Yes (Testing data) | No | No |
| TCGA-GBM | TCGA-14-1823 | Yes (Training data) | Yes | No |
| TCGA-GBM | TCGA-14-1825 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-14-1829 | Yes (Testing data) | No | Yes |
| TCGA-GBM | TCGA-14-2555 | Yes (Training data) | No | No |
| TCGA-GBM | TCGA-14-3477 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-19-0955 | Yes (Training data) | Yes | No |
| TCGA-GBM | TCGA-19-0957 | No | N/A | No |
| TCGA-GBM | TCGA-19-0960 | No | N/A | No |
| TCGA-GBM | TCGA-19-0962 | No | N/A | No |
| TCGA-GBM | TCGA-19-0963 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-19-0964 | No | N/A | No |
| TCGA-GBM | TCGA-19-1385 | No | N/A | No |
| TCGA-GBM | TCGA-19-1386 | No | N/A | No |
| TCGA-GBM | TCGA-19-1387 | No | N/A | No |
| TCGA-GBM | TCGA-19-1388 | Yes (Training data) | Yes | No |
| TCGA-GBM | TCGA-19-1389 | No | N/A | No |
| TCGA-GBM | TCGA-19-1390 | No | N/A | Yes |
| TCGA-GBM | TCGA-19-1392 | No | N/A | No |
| TCGA-GBM | TCGA-19-1787 | No | N/A | No |
| TCGA-GBM | TCGA-19-1788 | No | N/A | No |
| TCGA-GBM | TCGA-19-1789 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-19-1791 | No | N/A | No |
| TCGA-GBM | TCGA-19-2619 | No | N/A | No |
| TCGA-GBM | TCGA-19-2620 | No | N/A | No |
| TCGA-GBM | TCGA-19-2621 | No | N/A | No |
| TCGA-GBM | TCGA-19-2623 | No | N/A | No |
| TCGA-GBM | TCGA-19-2624 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-19-2625 | No | N/A | No |
| TCGA-GBM | TCGA-19-2629 | No | N/A | No |
| TCGA-GBM | TCGA-19-2631 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-19-4065 | No | N/A | No |
| TCGA-GBM | TCGA-19-4068 | No | N/A | No |
| TCGA-GBM | TCGA-19-5947 | No | N/A | No |
| TCGA-GBM | TCGA-19-5950 | Yes (Training data) | Yes | No |
| TCGA-GBM | TCGA-19-5951 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-19-5952 | No | N/A | No |
| TCGA-GBM | TCGA-19-5953 | No | N/A | No |
| TCGA-GBM | TCGA-19-5954 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-19-5955 | No | N/A | No |
| TCGA-GBM | TCGA-19-5956 | No | N/A | No |
| TCGA-GBM | TCGA-19-5958 | No | N/A | Yes |
| TCGA-GBM | TCGA-19-5959 | No | N/A | No |
| TCGA-GBM | TCGA-19-5960 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-27-1830 | No | N/A | No |
| TCGA-GBM | TCGA-27-1831 | No | N/A | No |
| TCGA-GBM | TCGA-27-1833 | No | N/A | No |
| TCGA-GBM | TCGA-27-1834 | No | N/A | Yes |
| TCGA-GBM | TCGA-27-1835 | No | N/A | No |
| TCGA-GBM | TCGA-27-1836 | No | N/A | No |
| TCGA-GBM | TCGA-27-1837 | No | N/A | No |
| TCGA-GBM | TCGA-27-1838 | No | N/A | Yes |
| TCGA-GBM | TCGA-27-2518 | No | N/A | No |
| TCGA-GBM | TCGA-27-2519 | No | N/A | No |
| TCGA-GBM | TCGA-27-2521 | No | N/A | No |
| TCGA-GBM | TCGA-27-2523 | No | N/A | No |
| TCGA-GBM | TCGA-27-2524 | No | N/A | No |
| TCGA-GBM | TCGA-27-2526 | No | N/A | Yes |
| TCGA-GBM | TCGA-27-2527 | No | N/A | No |
| TCGA-GBM | TCGA-27-2528 | No | N/A | No |
| TCGA-GBM | TCGA-76-4925 | No | N/A | No |
| TCGA-GBM | TCGA-76-4926 | No | N/A | No |
| TCGA-GBM | TCGA-76-4927 | No | N/A | No |
| TCGA-GBM | TCGA-76-4928 | No | N/A | No |
| TCGA-GBM | TCGA-76-4929 | No | N/A | No |
| TCGA-GBM | TCGA-76-4931 | No | N/A | No |
| TCGA-GBM | TCGA-76-4932 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-76-4934 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-76-4935 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-76-6191 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-76-6192 | No | N/A | No |
| TCGA-GBM | TCGA-76-6193 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-76-6280 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-76-6282 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-76-6285 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-76-6286 | No | N/A | No |
| TCGA-GBM | TCGA-76-6656 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-76-6657 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-76-6661 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-76-6662 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-76-6663 | Yes (Training data) | Yes | Yes |
| TCGA-GBM | TCGA-76-6664 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-CS-4938 | No | N/A | No |
| TCGA-LGG | TCGA-CS-4941 | Yes (Testing data) | Yes | Yes |
| TCGA-LGG | TCGA-CS-4942 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-CS-4943 | No | N/A | Yes |
| TCGA-LGG | TCGA-CS-4944 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-CS-5390 | No | N/A | No |
| TCGA-LGG | TCGA-CS-5393 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-CS-5394 | No | N/A | No |
| TCGA-LGG | TCGA-CS-5395 | Yes (Testing data) | Yes | Yes |
| TCGA-LGG | TCGA-CS-5396 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-CS-5397 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-CS-6186 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-CS-6188 | No | N/A | Yes |
| TCGA-LGG | TCGA-CS-6290 | Yes (Testing data) | Yes | Yes |
| TCGA-LGG | TCGA-CS-6665 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-CS-6666 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-CS-6667 | Yes (Testing data) | Yes | Yes |
| TCGA-LGG | TCGA-CS-6668 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-CS-6669 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-CS-6670 | Yes (Training data) | Yes | No |
| TCGA-LGG | TCGA-DU-5849 | Yes (Testing data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-5851 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-5852 | Yes (Testing data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-5853 | Yes (Testing data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-5854 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-5855 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-5871 | Yes (Testing data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-5872 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-5874 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-6395 | No | N/A | No |
| TCGA-LGG | TCGA-DU-6397 | Yes (Testing data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-6399 | Yes (Testing data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-6400 | Yes (Testing data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-6401 | Yes (Testing data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-6402 | No | N/A | No |
| TCGA-LGG | TCGA-DU-6404 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-6405 | No | N/A | Yes |
| TCGA-LGG | TCGA-DU-6407 | No | N/A | Yes |
| TCGA-LGG | TCGA-DU-6408 | No | N/A | Yes |
| TCGA-LGG | TCGA-DU-6410 | No | N/A | Yes |
| TCGA-LGG | TCGA-DU-6542 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-7008 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-7010 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-7013 | No | N/A | No |
| TCGA-LGG | TCGA-DU-7014 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-7015 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-7018 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-7019 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-7294 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-7298 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-7299 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-7300 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-7301 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-7302 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-7304 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-7306 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-7309 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-8158 | No | N/A | No |
| TCGA-LGG | TCGA-DU-8162 | No | N/A | Yes |
| TCGA-LGG | TCGA-DU-8163 | Yes (Training data) | Yes | No |
| TCGA-LGG | TCGA-DU-8164 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-8165 | No | N/A | Yes |
| TCGA-LGG | TCGA-DU-8166 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-8167 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-8168 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-DU-A5TP | No | N/A | Yes |
| TCGA-LGG | TCGA-DU-A5TR | No | N/A | Yes |
| TCGA-LGG | TCGA-DU-A5TS | No | N/A | Yes |
| TCGA-LGG | TCGA-DU-A5TT | No | N/A | Yes |
| TCGA-LGG | TCGA-DU-A5TU | No | N/A | Yes |
| TCGA-LGG | TCGA-DU-A5TW | No | N/A | Yes |
| TCGA-LGG | TCGA-DU-A5TY | No | N/A | Yes |
| TCGA-LGG | TCGA-DU-A6S2 | No | N/A | Yes |
| TCGA-LGG | TCGA-DU-A6S3 | No | N/A | Yes |
| TCGA-LGG | TCGA-DU-A6S6 | No | N/A | Yes |
| TCGA-LGG | TCGA-DU-A6S7 | No | N/A | Yes |
| TCGA-LGG | TCGA-DU-A6S8 | No | N/A | Yes |
| TCGA-LGG | TCGA-EZ-7264A | No | N/A | No |
| TCGA-LGG | TCGA-EZ-7265A | No | N/A | Yes |
| TCGA-LGG | TCGA-FG-5962 | No | N/A | No |
| TCGA-LGG | TCGA-FG-5963 | No | N/A | No |
| TCGA-LGG | TCGA-FG-5964 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-FG-5965 | No | N/A | No |
| TCGA-LGG | TCGA-FG-6688 | No | N/A | Yes |
| TCGA-LGG | TCGA-FG-6689 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-FG-6690 | Yes (Testing data) | Yes | No |
| TCGA-LGG | TCGA-FG-6691 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-FG-6692 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-FG-7634 | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-FG-7637 | Yes (Testing data) | Yes | No |
| TCGA-LGG | TCGA-FG-7641 | No | N/A | No |
| TCGA-LGG | TCGA-FG-7643 | Yes (Testing data) | Yes | Yes |
| TCGA-LGG | TCGA-FG-8186 | No | N/A | No |
| TCGA-LGG | TCGA-FG-8189 | No | N/A | No |
| TCGA-LGG | TCGA-FG-A4MT | Yes (Training data) | Yes | Yes |
| TCGA-LGG | TCGA-FG-A4MU | No | N/A | No |
| TCGA-LGG | TCGA-FG-A60K | No | N/A | No |
| TCGA-LGG | TCGA-FG-A6IZ | No | N/A | Yes |
| TCGA-LGG | TCGA-FG-A6J1 | No | N/A | No |
| TCGA-LGG | TCGA-FG-A713 | No | N/A | Yes |
| TCGA-LGG | TCGA-FG-A87N | No | N/A | No |
| TCGA-LGG | TCGA-HT-7467 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7468 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7469 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7470 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7471 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7472 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7473 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-7474 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7475 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-7476 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7477 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7478 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7479 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7480 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7481 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7482 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7483 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7485 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7601 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7602 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-7603 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7604 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7605 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7606 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7607 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7608 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7609 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7610 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7611 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7616 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-7620 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7676 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7677 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7680 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-7681 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7684 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-7686 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-7687 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7688 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7689 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7690 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-7691 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7692 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-7693 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-7694 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-7695 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7854 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7855 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-7856 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-7857 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7858 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7860 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-7873 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7874 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-7875 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7877 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7879 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-7880 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7881 | No | N/A | No |
| TCGA-LGG | TCGA-HT-7882 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-7884 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-7902 | No | N/A | No |
| TCGA-LGG | TCGA-HT-8010 | No | N/A | No |
| TCGA-LGG | TCGA-HT-8011 | No | N/A | No |
| TCGA-LGG | TCGA-HT-8012 | No | N/A | No |
| TCGA-LGG | TCGA-HT-8013 | No | N/A | No |
| TCGA-LGG | TCGA-HT-8015 | No | N/A | No |
| TCGA-LGG | TCGA-HT-8018 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-8019 | No | N/A | No |
| TCGA-LGG | TCGA-HT-8104 | No | N/A | No |
| TCGA-LGG | TCGA-HT-8105 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-8106 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-8107 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-8108 | No | N/A | No |
| TCGA-LGG | TCGA-HT-8109 | No | N/A | No |
| TCGA-LGG | TCGA-HT-8110 | No | N/A | No |
| TCGA-LGG | TCGA-HT-8111 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-8113 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-8114 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-8558 | No | N/A | No |
| TCGA-LGG | TCGA-HT-8563 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-8564 | No | N/A | No |
| TCGA-LGG | TCGA-HT-A4DS | No | N/A | No |
| TCGA-LGG | TCGA-HT-A4DV | No | N/A | No |
| TCGA-LGG | TCGA-HT-A5R5 | No | N/A | No |
| TCGA-LGG | TCGA-HT-A5R7 | No | N/A | No |
| TCGA-LGG | TCGA-HT-A5RA | No | N/A | No |
| TCGA-LGG | TCGA-HT-A5RB | No | N/A | No |
| TCGA-LGG | TCGA-HT-A5RC | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-A614 | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-A615 | No | N/A | No |
| TCGA-LGG | TCGA-HT-A616 | No | N/A | No |
| TCGA-LGG | TCGA-HT-A617 | No | N/A | No |
| TCGA-LGG | TCGA-HT-A618 | No | N/A | No |
| TCGA-LGG | TCGA-HT-A619 | No | N/A | No |
| TCGA-LGG | TCGA-HT-A61A | No | N/A | Yes |
| TCGA-LGG | TCGA-HT-A61B | No | N/A | No |
| TCGA-LGG | TCGA-HT-A61C | No | N/A | No |
It should be noted that the diversity of the available scans in NCI/NIH/TCIA is driven by the fact that TCIA collected all available scans for subjects whose tissue specimens had passed the quality evaluation of the NCI/NIH/TCGA program. Due to this collection being retrospective all the MRI scans are considered ‘standard-of-care’, without following any uniform imaging protocol.
Pre-processing
All pre-operative mMRI volumes were re-oriented to the LPS (left-posterior-superior) coordinate system (which is a requirement for GLISTRboost), co-registered to the same T1 anatomic template61 using affine registration through the Oxford center for Functional MRI of the Brain (FMRIB) Linear Image Registration Tool (FLIRT)62,63 of FMRIB Software Library (FSL)64–66, and resampled to 1 mm3 voxel resolution (Fig. 1b). The volumes of all the modalities for each patient were then skull-stripped using the Brain Extraction Tool (BET)67,68 from the FSL64–66 (Fig. 1c). Subsequent skull-stripping, on cases that BET produced insufficient results, was performed using a novel automated method based on a multi atlas registration and label fusion framework69. The template library for this task consisted of 216 MRI scans and their brain masks. This library was then used for target specific template selection and subsequent registrations using an existing strategy of MUlti-atlas Segmentation utilizing Ensembles (MUSE)70. A final region-growing based processing step, guided by T2, was applied to obtain a brain mask that includes the intra-cranial CSF. The resulted volumes are the ones provided in [Data Citation 3 and Data Citation 4].
For producing the computer-aided segmentation labels, further preprocessing steps included the smoothing of all volumes using a low-level image processing method, namely Smallest Univalue Segment Assimilating Nucleus (SUSAN)71, in order to reduce high frequency intensity variations (i.e., noise) in regions of uniform intensity profile while preserving the underlying structure (Fig. 1d). The intensity histograms of all modalities of all patients were then matched72 to the corresponding modality of a single reference patient, using the implemented version in ITK (HistogramMatchingImageFilter).
It should be noted that we did not use any non-parametric, non-uniform intensity normalization algorithm73–75 to correct for intensity non-uniformities caused by the inhomogeneity of the scanner’s magnetic field during image acquisition, as we observed that application of such algorithm obliterated the T2-FLAIR signal (Fig. 1e).
Segmentation labels of glioma sub-regions
Consistent with the BraTS challenge56 the segmentation labels that we consider in the present study, and make available through TCIA [Data Citation 3 and Data Citation 4], delineate the enhancing part of the tumor core (ET), the non-enhancing part of the tumor core (NET), and the peritumoral edema (ED) (Fig. 2). The ET is described by areas that show hyper-intensity in T1-Gd when compared to T1, but also when compared to normal/healthy white matter (WM) in T1-Gd. Biologically, ET is felt to represent regions where there is leakage of contrast through a disrupted blood-brain barrier that is commonly seen in high grade gliomas. The NET represents non-enhancing tumor regions, as well as transitional/pre-necrotic and necrotic regions that belong to the non-enhancing part of the tumor core (TC), and are typically resected in addition to the ET. The appearance of the NET is typically hypo-intense in T1-Gd when compared to T1, but also when compared to normal/healthy WM in T1-Gd. Finally, the ED is described by hyper-intense signal on the T2-FLAIR volumes.
Figure 2. Single slice multimodal MRI scans of example subjects, illustrating all modalities used in GLISTRboost36,38 and example segmentation labels.
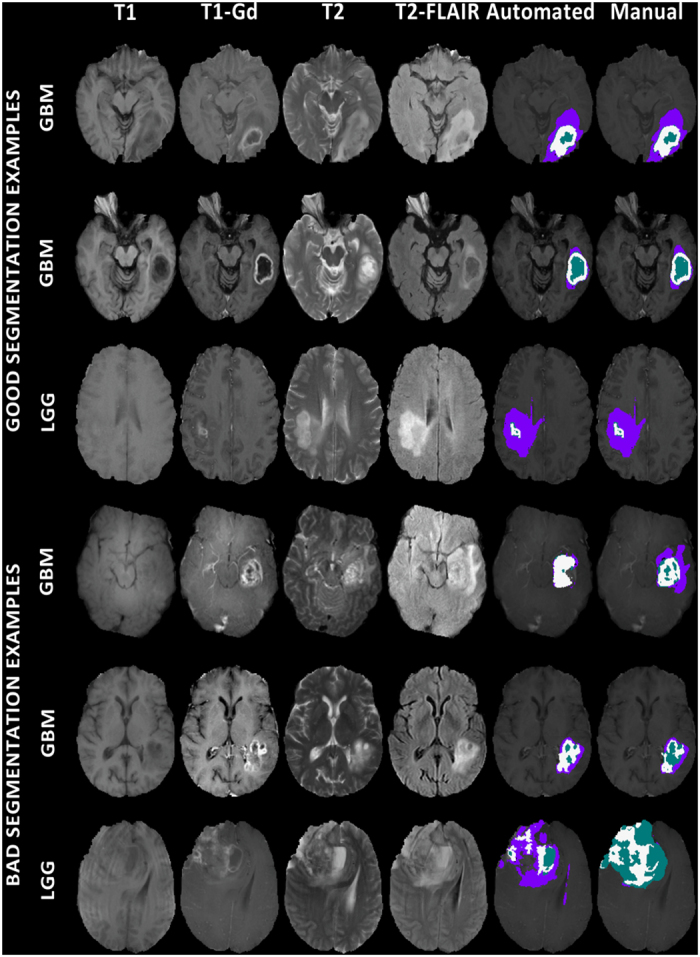
The first three rows depict good segmentation examples, whereas the following three depict bad segmentation examples, produced by GLISTRboost36,38.
Computer-aided segmentation approach
The method used in this study to produce the computer-aided segmentation labels for all pre-operative scans of both TCGA-GBM and TCGA-LGG collections is named GLISTRboost36,38 and it is based on a hybrid generative-discriminative model. The generative part incorporates a glioma growth model58–60, and is based on an Expectation-Maximization (EM) framework to segment the brain scans into tumor (i.e., ET, NET and ED), as well as healthy tissue labels (i.e., WM, gray matter, cerebrospinal fluid, vessels and cerebellum). The discriminative part is based on a gradient boosting76,77 multi-class classification scheme, which was trained on BraTS’15 data (www.virtualskeleton.ch/BRATS/Start2015), to refine tumor labels based on information from multiple patients. Lastly, a Bayesian strategy78 is employed to further refine and finalize the tumor segmentation based on patient-specific intensity statistics from the multiple modalities available. Example segmentation labels are illustrated in Fig. 2.
GLISTRboost36,38 is based on a modified version of the GLioma Image SegmenTation and Registration (GLISTR)79 software. GLISTR jointly performs a) the registration of a healthy population probabilistic atlas to brain scans of patients with gliomas using a tumor growth model to account for mass effects, and b) the segmentation of such scans into healthy and tumor tissues. The whole framework of GLISTR is based on a probabilistic generative model that relies on EM, to recursively refine the estimates of the posteriors for all tissue labels, the deformable mapping to the atlas, and the parameters of the incorporated brain tumor growth model58–60. GLISTR was originally designed to tackle cases with solitary GBMs79–81, and subsequently extended to handle multifocal masses and tumors of complex shapes with heterogeneous texture82. Furthermore, the original version of GLISTR79–82 was based on a single seed-point for each brain tissue label to represent its mean intensity value, while the variance was described by a fixed value for all labels. On the contrary, GLISTRboost incorporates multiple tissue seed-points for each label, to model more accurately the intensity distribution, i.e., mean and variance, for each tissue class. Note that both GLISTR and GLISTRboost take into account only the intensity value of the initialization tissue seed-points on each modality, while they discard spatial information regarding the coordinate position of the respective points. As a consequence, even if the initialized tissue seed-points during two independent segmentation attempts have different coordinates, the output sets of segmentation labels should be identical, given that the modeled intensity distributions during these attempts are the same. In addition to the tissue seed-points, GLISTR and on that account GLISTRboost, requires the definition of a single seed-point and a radius for approximating the center and the bulk volume of each apparent tumor by a sphere. All these seed-points are initialized using the ‘Cancer Imaging Phenomics Toolkit’ (CaPTk)83 (www.med.upenn.edu/sbia/captk.html), which has been primarily developed for this purpose, by the Center for Biomedical Image Computing and Analytics (CBICA) of the University of Pennsylvania. Given the tumor seed-point and radius for a tumor, a growth model is initiated by the parametric model of a sphere. This growth model is used to deform a healthy atlas into one with tumor and edema tissues matching the input scans, while approximating the deformations occurred to all brain tissues due to the mass effect of the tumors. A tumor shape prior is estimated by a random-walk-based generative model, which uses the tumor seed-points as initialization cues. This shape prior is systemically incorporated into the EM framework via an empirical Bayes model82. Furthermore, a minimum of three initialization seed-points is needed for each brain tissue label, in order to capture the intensity variation and model the intensity distribution across all modalities. Use of multiple seed-points improves the initialization of the EM framework, leading to more accurate segmentation labels, when compared to the single seed-point approach82. The output of GLISTR is a posterior probability map for each tissue label, as well as an integrative label map, which describes a very good ‘initial’ segmentation of all different tissues within a patient's brain.
This ‘initial’ segmentation is then refined by taking into account information from multiple patients via a discriminative machine-learning algorithm. Specifically, we used the gradient boosting algorithm76 to perform voxel-level multi-label classification. Gradient boosting produces a prediction model by combining weak learners in an ensemble. We used decision trees of maximum depth 3 as ‘weak learners’, which were trained in a sub-sample of the training set, in order to introduce randomness77. The sampling rate was set equal to 0.6, while additional randomness was introduced by sampling stochastically a subset of imaging (i.e., radiomic) features at each node. The number of sampled features was set equal to the square root of the total number of features. The algorithm was terminated after 100 iterations.
The set of features used for training our model was extracted volumetrically and consists of i) intensity information, ii) image derivative, iii) geodesic information, iv) texture features, and v) the GLISTR posterior probability maps. The intensity information is summarized by the raw intensity value, I, of each image voxel, vi, at each modality, m, (i.e., I(vim)), as well as by the respective differences among all four modalities, i.e., I(viT1)- I(viT1Gd), I(viT1)- I(viT2), I(viT1)- I(viT2FLAIR), I(viT1Gd)- I(viT1), I(viT1Gd)- I(viT2), I(viT1Gd)- I(viT2FLAIR), I(viT2)- I(viT1), I(viT2)- I(viT1Gd), I(viT2)- I(viT2FLAIR), I(viT2FLAIR)- I(viT1), I(viT2FLAIR)- I(viT1Gd), I(viT2FLAIR)- I(viT2). The image derivative component consists of the Laplacian of Gaussians and the image gradient magnitude. Note that in order to ensure that the intensity-based features are comparable, intensity normalization was performed across subjects based on the median intensity value of the cerebrospinal fluid label, as provided by GLISTR. Geodesic information was used to introduce spatial context information. At any voxel vi we calculated the geodesic distance from the seed-point at voxel vs, which was used in GLISTR as the tumor center. The geodesic distance between vi and vs was estimated using the fast marching method84,85 and by taking into account local image gradient magnitude86. Furthermore, we used texture features computed from a gray-level co-occurrence matrix (GLCM)87. Specifically, these texture features describe first-order statistics (i.e., mean and variance of each modality’s intensities within a radius of 2 voxels for each voxel), as well as second-order statistics. To obtain the latter, the image volumes were firstly normalized to 64 different gray levels, and then a bounding box of 5-by-5-by-5 voxels was used for all the voxels of each image as a sliding window. Then, a GLCM was populated by taking into account the intensity values within a radius of 2 pixels and for the 26 main 3D directions to extract the energy, entropy, dissimilarity, homogeneity (i.e., inverse difference moment of order 2), and inverse difference moment of order 1. These features were computed for each direction and their average was used. To avoid overfitting, the gradient boosting machine was trained using simultaneously both LGG and GBM training data of BraTS’15, in a 54-fold cross-validation setting (allowing for using a one out of the 54 available LGGs of the BraTS’15 training data, within each fold).
Finally, the segmentation results were further refined for each patient separately, by assessing the local intensity distribution of the segmentation labels and updating their spatial configuration based on a probabilistic model78. The intensity distributions of the WM, ED, NET and ET, were populated separately using the corresponding voxels of posterior probability equal to 1, as given by GLISTR. Histogram normalization was then performed for the 3 pair-wise distributions considered; ED versus WM in T2-FLAIR, ET versus ED in T1-Gd, and ET versus NET in T1-Gd. Maximum likelihood estimation was used to model the class-conditional probability densities (Pr(I(vi)|Class) by a distinct Gaussian model for each class. In all pair-wise comparisons described before, the former tissue is expected to be brighter than the latter. Voxels of each class with spatial proximity smaller than 4 voxels to the voxels of the paired class, were evaluated by assessing their intensity I(vi) and comparing the (‘Pr(I(vi)|Class1) with Pr(I(vi)|Class2). The voxel vi was then classified into the tissue class with the larger conditional probability. This is equivalent to a classification based on Bayes' Theorem with equal priors for the two classes, i.e., Pr(Class1)=Pr(Class2)=0.5.
Manual revision
The output of GLISTRboost segmentation is expected to yield labels for ET, NET, and ED. However, some gliomas, especially LGG, do not exhibit much contrast enhancement, or ED. Biologically, LGGs may have less blood-brain barrier disruption (leading to less leak of contrast during the scan), and may grow at a rate slow enough to avoid significant edema formation, which results from rapid disruption, irritation, and infiltration of normal brain parenchyma by tumor cells. As such, manual revision of the segmentation labels was performed, particularly for LGG cases lacking ET or ED regions. Specifically, after taking all the above into consideration, in scans of LGGs without an apparent ET area we consider only the NET and ED labels (Fig. 3a,d), whereas in LGG scans without ET and without obvious texture differences across modalities we consider only the NET label, allowing for distinguishing between normal and abnormal brain tissue (Fig. 3e). The difficulty in calculating the accurate boundaries between tumor and healthy tissue in the operating room is reflected in the segmentation labels as well; there is high uncertainty among neurosurgeons, neuroradiologists, and imaging scientists in delineating these boundaries. Therefore, small regions within the segmented labels that were ambiguous of their exact classification, were left as segmented by GLISTRboost.
Figure 3. Single slice multimodal MRI scans of example subjects, illustrating all modalities used in GLISTRboost36,38 and examples of the computer-aided (automated) and the manually-revised (manual) segmentation labels.
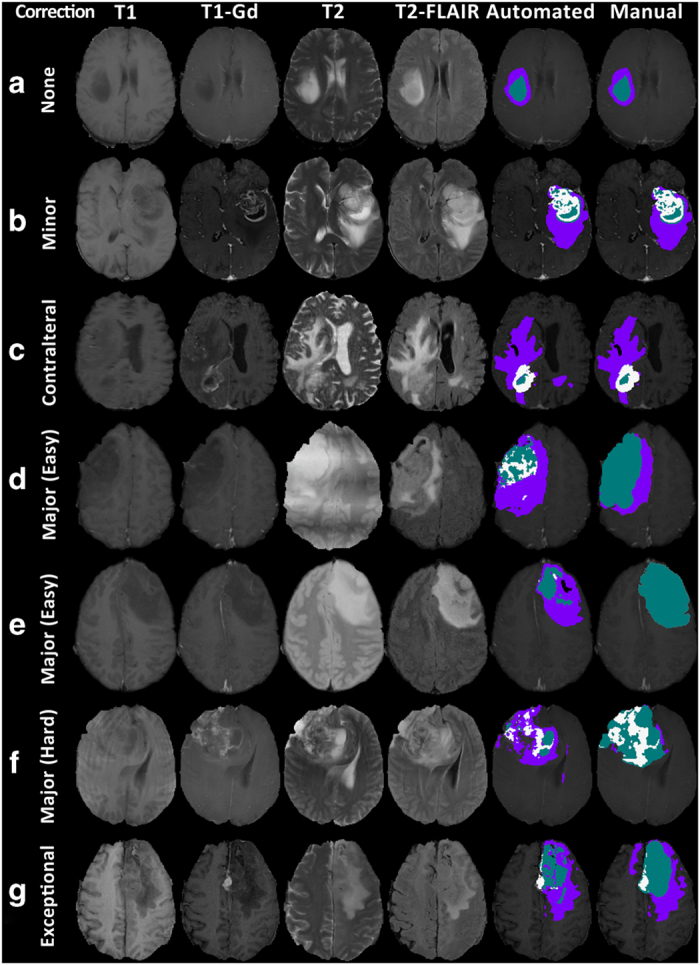
The type of corrections applied during the manual-revision of the segmentation labels is also shown in the left side of each row; (a) no correction, (b) minor corrections, (c) corrections in the contralateral edema, (d) major (easy) correction of LGG without ET, (e) major (easy) correction of LGG without ET or ED, (f) major (hard) corrections, (g) exceptional subject (TCGA-DU-7304) that could have a meningioma in the midline as the apparent lesion seems to raise from the dura.
Manual revisions/corrections applied in the computer-aided segmentation labels include: i) obvious under- or over-segmented ED/ET/NET regions (Fig. 3d–g), ii) voxels classified as ED within the tumor core (Fig. 3b,c,g), iii) unclassified voxels within the tumor core (Fig. 3c–g), iv) voxels classified as NET outside the tumor core. Contralateral and periventricular regions of T2-FLAIR hyper-intensity were excluded from the ED region (Fig. 3c,f), unless they were contiguous with peritumoral ED (Fig. 3g—addition of apparent contralateral ED), as these areas are generally considered to represent chronic microvascular changes, or age-associated demyelination, rather than tumor infiltration88.
Radiomic features panel
An extensive panel of more than 700 radiomic features is extracted volumetrically (in 3D), based on the manually-revised labels of each tumor sub-region that comprised i) intensity, ii) volumetric89, iii) morphologic90–93, iv) histogram-based31, and v) textural parameters, including features based on wavelets94, GLCM87, Gray-Level Run-Length Matrix (GLRLM)93,95–98, Gray-Level Size Zone Matrix (GLSZM)95–97,99, and Neighborhood Gray-Tone Difference Matrix (NGTDM)100, as well as vi) spatial information101, and vii) glioma diffusion properties extracted from glioma growth models58–60, that are already evaluated as having predictive and prognostic value30–32,102,103. The specific features provided are all shown in Table 3 (available online only).
Table 3. Generated radiomic features and their description.
| Collection | Site | Patient ID | GivenModalityForThisStudy | Sex | Age | Weight | ModalityName | SeriesNum | AccessionNum | Acq/Study/SeriesDate | Manufacturer | StationName | ManufacturerModelName | ScanSequence | AcquisitionType | SliceThickness | RepetitionTime | EchoTime | InversionTime | ImagingFreq | MagneticFieldStrength | SliceSpacing | FlipAngle | SpecificAbsorptionRate | Slices | AcqMatRows | AcqMatCols | PixDim | Series/Modalities_ID |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TCGA-GBM | TCGA-02 | TCGA-02-0003 | flair | M | 50 | 99.79 | AX_FLAIR | 11 | 2.8195E+15 | 6/8/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 147 | 2200 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.932588 | 25 | 256 | 160 | 0.781250\0.781250 | TCGA-02-0003/1.3.6.1.4.1.14519.5.2.1.1706.4001.145725991542758792340793681239/1.3.6.1.4.1.14519.5.2.1.1706.4001.166909646801864710106680106577 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0003 | t1Gd | M | 50 | 99.79 | AX_T1_POST | 13 | 2.8195E+15 | 6/8/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 650 | 9 | 6.39E+08 | 1.5 | 6.5 | 90 | 1.972841 | 25 | 256 | 192 | 0.781250\0.781250 | TCGA-02-0003/1.3.6.1.4.1.14519.5.2.1.1706.4001.145725991542758792340793681239/1.3.6.1.4.1.14519.5.2.1.1706.4001.204054528317611873722753919694 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0003 | t1 | M | 50 | 99.79 | AX_T1 | 12 | 2.8195E+15 | 6/8/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 533.332 | 8 | 6.39E+08 | 1.5 | 6.5 | 90 | 1.959656 | 25 | 256 | 128 | 0.781250\0.781250 | TCGA-02-0003/1.3.6.1.4.1.14519.5.2.1.1706.4001.145725991542758792340793681239/1.3.6.1.4.1.14519.5.2.1.1706.4001.769579767788431533139923490444 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0003 | t2 | M | 50 | 99.79 | AX_T2_FSE | 10 | 2.8195E+15 | 6/8/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 3500 | 105 | 6.39E+08 | 1.5 | 6.5 | 90 | 1.697364 | 25 | 256 | 224 | 0.781250\0.781250 | TCGA-02-0003/1.3.6.1.4.1.14519.5.2.1.1706.4001.145725991542758792340793681239/1.3.6.1.4.1.14519.5.2.1.1706.4001.643840351201993277528058523822 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0006 | flair | F | 56 | 70.307 | AX_FLAIR | 4 | 2.8195E+15 | 8/23/1996 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 147 | 2200 | 6.38E+08 | 1.5 | 6.5 | 90 | 0.023404 | 23 | 256 | 160 | 0.781250\0.781250 | TCGA-02-0006/1.3.6.1.4.1.14519.5.2.1.1706.4001.149500105036523046215258942545/1.3.6.1.4.1.14519.5.2.1.1706.4001.315980779833795710131738723922 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0006 | t1Gd | F | 56 | 70.307 | AX_T1_POST | 6 | 2.8195E+15 | 8/23/1996 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 583.332 | 8 | 6.38E+08 | 1.5 | 6.5 | 90 | 0.059235 | 23 | 256 | 192 | 0.781250\0.781250 | TCGA-02-0006/1.3.6.1.4.1.14519.5.2.1.1706.4001.149500105036523046215258942545/1.3.6.1.4.1.14519.5.2.1.1706.4001.170251499565901046332998726504 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0006 | t1 | F | 56 | 70.307 | AX_T1 | 5 | 2.8195E+15 | 8/23/1996 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 416.664 | 8 | 6.38E+08 | 1.5 | 6.5 | 90 | 0.067638 | 23 | 256 | 128 | 0.781250\0.781250 | TCGA-02-0006/1.3.6.1.4.1.14519.5.2.1.1706.4001.149500105036523046215258942545/1.3.6.1.4.1.14519.5.2.1.1706.4001.283912726091613148226144162419 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0006 | t2 | F | 56 | 70.307 | AX_T2_FSE | 3 | 2.8195E+15 | 8/23/1996 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 3500 | 98 | 6.38E+08 | 1.5 | 6.5 | 90 | 0.04572 | 23 | 256 | 224 | 0.781250\0.781250 | TCGA-02-0006/1.3.6.1.4.1.14519.5.2.1.1706.4001.149500105036523046215258942545/1.3.6.1.4.1.14519.5.2.1.1706.4001.929166576410729967676059227597 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0009 | flair | F | 61 | 63.503 | AX_FLAIR | 4 | 2.8195E+15 | 6/14/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 147 | 2200 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.023788 | 23 | 256 | 160 | 0.781250\0.781250 | TCGA-02-0009/1.3.6.1.4.1.14519.5.2.1.1706.4001.743358002952086773602945013452/1.3.6.1.4.1.14519.5.2.1.1706.4001.779013211760175351323192631290 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0009 | t1Gd | F | 61 | 63.503 | AX_T1_POST | 6 | 2.8195E+15 | 6/14/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 583.332 | 8 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.060207 | 23 | 256 | 192 | 0.781250\0.781250 | TCGA-02-0009/1.3.6.1.4.1.14519.5.2.1.1706.4001.743358002952086773602945013452/1.3.6.1.4.1.14519.5.2.1.1706.4001.150346522548097750188958570502 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0009 | t1 | F | 61 | 63.503 | AX_T1 | 5 | 2.8195E+15 | 6/14/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 416.664 | 8 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.068749 | 23 | 256 | 128 | 0.781250\0.781250 | TCGA-02-0009/1.3.6.1.4.1.14519.5.2.1.1706.4001.743358002952086773602945013452/1.3.6.1.4.1.14519.5.2.1.1706.4001.507509430627489084746877138458 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0009 | t2 | F | 61 | 63.503 | AX_T2_FSE | 3 | 2.8195E+15 | 6/14/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 3500 | 98 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.04647 | 23 | 256 | 224 | 0.781250\0.781250 | TCGA-02-0009/1.3.6.1.4.1.14519.5.2.1.1706.4001.743358002952086773602945013452/1.3.6.1.4.1.14519.5.2.1.1706.4001.466873465067673546718804101423 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0011 | flair | F | 18 | 51.256 | AX_FLAIR | 4 | 2.8195E+15 | 2/1/1998 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 147 | 2200 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.024617 | 23 | 256 | 160 | 0.781250\0.781250 | TCGA-02-0011/1.3.6.1.4.1.14519.5.2.1.1706.4001.338073323505507625300877831709/1.3.6.1.4.1.14519.5.2.1.1706.4001.245399232039099875265266953905 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0011 | t1Gd | F | 18 | 51.256 | AX_T1_POST | 6 | 2.8195E+15 | 2/1/1998 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 583.332 | 8 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.062307 | 23 | 256 | 192 | 0.781250\0.781250 | TCGA-02-0011/1.3.6.1.4.1.14519.5.2.1.1706.4001.338073323505507625300877831709/1.3.6.1.4.1.14519.5.2.1.1706.4001.209353288390362628068712523284 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0011 | t1 | F | 18 | 51.256 | AX_T1 | 5 | 2.8195E+15 | 2/1/1998 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 416.664 | 8 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.071146 | 23 | 256 | 128 | 0.781250\0.781250 | TCGA-02-0011/1.3.6.1.4.1.14519.5.2.1.1706.4001.338073323505507625300877831709/1.3.6.1.4.1.14519.5.2.1.1706.4001.101129210060381531266477375594 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0011 | t2 | F | 18 | 51.256 | AX_T2_FSE | 3 | 2.8195E+15 | 2/1/1998 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 3500 | 98 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.048091 | 23 | 256 | 224 | 0.781250\0.781250 | TCGA-02-0011/1.3.6.1.4.1.14519.5.2.1.1706.4001.338073323505507625300877831709/1.3.6.1.4.1.14519.5.2.1.1706.4001.222837342553572147643639077488 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0027 | flair | F | 33 | 61.689 | T2_FLAIR | 4 | 2.8195E+15 | 3/28/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | IR | 2D | 5 | 10002 | 140.244 | 2250 | 127.7415 | 3 | 6.5 | 90 | 0.5753 | 23 | 320 | 224 | 0.46875\0.46875 | TCGA-02-0027/1.3.6.1.4.1.14519.5.2.1.1706.4001.190188151913002985587952372782/1.3.6.1.4.1.14519.5.2.1.1706.4001.218172129878062618773517653321 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0027 | t1Gd | F | 33 | 61.689 | FSPGR_3D | 10 | 2.8195E+15 | 3/28/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 1.5 | 5.872 | 2.1 | 400 | 127.7415 | 3 | 1.5 | 20 | 0.709214 | 124 | 320 | 224 | 0.488281\0.488281 | TCGA-02-0027/1.3.6.1.4.1.14519.5.2.1.1706.4001.190188151913002985587952372782/1.3.6.1.4.1.14519.5.2.1.1706.4001.227653997506797488566901323094 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0027 | t1 | F | 33 | 61.689 | SE_T1 | 5 | 2.8195E+15 | 3/28/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 816.664 | 12 | 0 | 127.7415 | 3 | 6.5 | 90 | 1.755603 | 23 | 320 | 224 | 0.46875\0.46875 | TCGA-02-0027/1.3.6.1.4.1.14519.5.2.1.1706.4001.190188151913002985587952372782/1.3.6.1.4.1.14519.5.2.1.1706.4001.206413929564700806222369874836 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0027 | t2 | F | 33 | 61.689 | FSE_T2 | 3 | 2.8195E+15 | 3/28/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 5200 | 99 | 0 | 127.7415 | 3 | 6.5 | 90 | 1.2148 | 23 | 320 | 256 | 0.46875\0.46875 | TCGA-02-0027/1.3.6.1.4.1.14519.5.2.1.1706.4001.190188151913002985587952372782/1.3.6.1.4.1.14519.5.2.1.1706.4001.198656437193522367812015571836 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0033 | flair | M | 54 | 88.904 | AX_FLAIR | 4 | 2.8195E+15 | 5/26/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 147 | 2200 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.022541 | 23 | 256 | 160 | 0.781250\0.781250 | TCGA-02-0033/1.3.6.1.4.1.14519.5.2.1.1706.4001.211135800355624542661804589744/1.3.6.1.4.1.14519.5.2.1.1706.4001.116289242878713280880004720679 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0033 | t1Gd | M | 54 | 88.904 | AX_T1_POST | 6 | 2.8195E+15 | 5/26/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 583.332 | 8 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.057052 | 23 | 256 | 192 | 0.781250\0.781250 | TCGA-02-0033/1.3.6.1.4.1.14519.5.2.1.1706.4001.211135800355624542661804589744/1.3.6.1.4.1.14519.5.2.1.1706.4001.287753826073472752590065451465 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0033 | t1 | M | 54 | 88.904 | AX_T1 | 5 | 2.8195E+15 | 5/26/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 416.664 | 8 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.065145 | 23 | 256 | 128 | 0.781250\0.781250 | TCGA-02-0033/1.3.6.1.4.1.14519.5.2.1.1706.4001.211135800355624542661804589744/1.3.6.1.4.1.14519.5.2.1.1706.4001.103174687731052142735983046836 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0033 | t2 | M | 54 | 88.904 | AX_T2_FSE | 3 | 2.8195E+15 | 5/26/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 3500 | 98 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.044035 | 23 | 256 | 224 | 0.781250\0.781250 | TCGA-02-0033/1.3.6.1.4.1.14519.5.2.1.1706.4001.211135800355624542661804589744/1.3.6.1.4.1.14519.5.2.1.1706.4001.261091685370401963952512175444 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0034 | flair | M | 60 | 91.626 | AX_FLAIR | 11 | 2.8195E+15 | 7/27/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 147 | 2200 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.769412 | 24 | 256 | 160 | 0.781250\0.781250 | TCGA-02-0034/1.3.6.1.4.1.14519.5.2.1.1706.4001.278960043528371206673900239956/1.3.6.1.4.1.14519.5.2.1.1706.4001.287132853761458303765658181489 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0034 | t1Gd | M | 60 | 91.626 | AX_T1_POST | 13 | 2.8195E+15 | 7/27/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 616.664 | 9 | 6.39E+08 | 1.5 | 6.5 | 90 | 1.921836 | 24 | 256 | 192 | 0.781250\0.781250 | TCGA-02-0034/1.3.6.1.4.1.14519.5.2.1.1706.4001.278960043528371206673900239956/1.3.6.1.4.1.14519.5.2.1.1706.4001.497428918591782710551794403821 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0034 | t1 | M | 60 | 91.626 | AX_T1 | 12 | 2.8195E+15 | 7/27/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 483.332 | 8 | 6.39E+08 | 1.5 | 6.5 | 90 | 1.99891 | 24 | 256 | 128 | 0.781250\0.781250 | TCGA-02-0034/1.3.6.1.4.1.14519.5.2.1.1706.4001.278960043528371206673900239956/1.3.6.1.4.1.14519.5.2.1.1706.4001.135129750377370879091413746175 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0034 | t2 | M | 60 | 91.626 | AX_T2_FSE | 10 | 2.8195E+15 | 7/27/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 3500 | 105 | 6.39E+08 | 1.5 | 6.5 | 90 | 1.568419 | 24 | 256 | 224 | 0.781250\0.781250 | TCGA-02-0034/1.3.6.1.4.1.14519.5.2.1.1706.4001.278960043528371206673900239956/1.3.6.1.4.1.14519.5.2.1.1706.4001.185929810886679504848373001030 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0037 | flair | F | 74 | 109.769 | AX_FLAIR | 4 | 2.8195E+15 | 1/13/1998 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 147 | 2200 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.021793 | 23 | 256 | 160 | 0.781250\0.781250 | TCGA-02-0037/1.3.6.1.4.1.14519.5.2.1.1706.4001.170054238455666525856624861567/1.3.6.1.4.1.14519.5.2.1.1706.4001.178667019197289195525849495660 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0037 | t1Gd | F | 74 | 109.769 | AX_T1_POST | 6 | 2.8195E+15 | 1/13/1998 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 583.332 | 8 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.055159 | 23 | 256 | 192 | 0.781250\0.781250 | TCGA-02-0037/1.3.6.1.4.1.14519.5.2.1.1706.4001.170054238455666525856624861567/1.3.6.1.4.1.14519.5.2.1.1706.4001.417797301325658887813420764479 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0037 | t1 | F | 74 | 109.769 | AX_T1 | 5 | 2.8195E+15 | 1/13/1998 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 416.664 | 8 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.062985 | 23 | 256 | 128 | 0.781250\0.781250 | TCGA-02-0037/1.3.6.1.4.1.14519.5.2.1.1706.4001.170054238455666525856624861567/1.3.6.1.4.1.14519.5.2.1.1706.4001.134108057579177395146913192259 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0037 | t2 | F | 74 | 109.769 | AX_T2_FSE | 3 | 2.8195E+15 | 1/13/1998 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 3500 | 98 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.042574 | 23 | 256 | 224 | 0.781250\0.781250 | TCGA-02-0037/1.3.6.1.4.1.14519.5.2.1.1706.4001.170054238455666525856624861567/1.3.6.1.4.1.14519.5.2.1.1706.4001.242653383084895972538145369544 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0046 | flair | M | 61 | 86.183 | AX_FLAIR | 4 | 2.8195E+15 | 11/28/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | IR | 2D | 5 | 10002 | 141.54 | 2200 | 63.88013 | 1.5 | 6.5 | 90 | 0.0226 | 23 | 256 | 160 | 0.78125\0.78125 | TCGA-02-0046/1.3.6.1.4.1.14519.5.2.1.1706.4001.667069087302553300405296434177/1.3.6.1.4.1.14519.5.2.1.1706.4001.509588674123932722082406945181 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0046 | t1Gd | M | 61 | 86.183 | AX_T1_POST | 6 | 2.8195E+15 | 11/28/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 583.332 | 8 | 0 | 63.88013 | 1.5 | 6.5 | 90 | 0.057336 | 23 | 256 | 192 | 0.78125\0.78125 | TCGA-02-0046/1.3.6.1.4.1.14519.5.2.1.1706.4001.667069087302553300405296434177/1.3.6.1.4.1.14519.5.2.1.1706.4001.229600217589805337233000276642 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0046 | t1 | M | 61 | 86.183 | AX_T1 | 5 | 2.8195E+15 | 11/28/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 416.664 | 8 | 0 | 63.88013 | 1.5 | 6.5 | 90 | 0.06547 | 23 | 256 | 128 | 0.78125\0.78125 | TCGA-02-0046/1.3.6.1.4.1.14519.5.2.1.1706.4001.667069087302553300405296434177/1.3.6.1.4.1.14519.5.2.1.1706.4001.266305079892504466765664273320 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0046 | t2 | M | 61 | 86.183 | AX_T2_FSE | 3 | 2.8195E+15 | 11/28/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 3000 | 96.512 | 0 | 63.88013 | 1.5 | 6.5 | 90 | 0.086 | 23 | 256 | 224 | 0.78125\0.78125 | TCGA-02-0046/1.3.6.1.4.1.14519.5.2.1.1706.4001.667069087302553300405296434177/1.3.6.1.4.1.14519.5.2.1.1706.4001.321860854252522153610965271371 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0047 | flair | M | 78 | 71.668 | AX_FLAIR | 4 | 2.8195E+15 | 12/15/1998 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 147 | 2200 | 6.38E+08 | 1.5 | 6.5 | 90 | 0.023332 | 23 | 256 | 160 | 0.78125\0.78125 | TCGA-02-0047/1.3.6.1.4.1.14519.5.2.1.1706.4001.246202953409591552042046870492/1.3.6.1.4.1.14519.5.2.1.1706.4001.123447841249545695967914613161 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0047 | t1Gd | M | 78 | 71.668 | AX_T1_POST | 6 | 2.8195E+15 | 12/15/1998 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 583.332 | 8 | 6.38E+08 | 1.5 | 6.5 | 90 | 0.059053 | 23 | 256 | 192 | 0.78125\0.78125 | TCGA-02-0047/1.3.6.1.4.1.14519.5.2.1.1706.4001.246202953409591552042046870492/1.3.6.1.4.1.14519.5.2.1.1706.4001.245568253298410645742658166184 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0047 | t1 | M | 78 | 71.668 | AX_T1 | 5 | 2.8195E+15 | 12/15/1998 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 416.664 | 8 | 6.38E+08 | 1.5 | 6.5 | 90 | 0.067431 | 23 | 256 | 128 | 0.78125\0.78125 | TCGA-02-0047/1.3.6.1.4.1.14519.5.2.1.1706.4001.246202953409591552042046870492/1.3.6.1.4.1.14519.5.2.1.1706.4001.238166248172920183561871538756 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0047 | t2 | M | 78 | 71.668 | AX_T2_FSE | 3 | 2.8195E+15 | 12/15/1998 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 3500 | 98 | 6.38E+08 | 1.5 | 6.5 | 90 | 0.04558 | 23 | 256 | 224 | 0.78125\0.78125 | TCGA-02-0047/1.3.6.1.4.1.14519.5.2.1.1706.4001.246202953409591552042046870492/1.3.6.1.4.1.14519.5.2.1.1706.4001.489474944906343317155982879920 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0048 | flair | M | 80 | 90.718 | AX_FLAIR | 4 | 2.8195E+15 | 1/29/1999 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 147 | 2200 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.022468 | 23 | 256 | 160 | 0.78125\0.78125 | TCGA-02-0048/1.3.6.1.4.1.14519.5.2.1.1706.4001.271759313632030441078606502900/1.3.6.1.4.1.14519.5.2.1.1706.4001.226227114463442396869459184631 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0048 | t1Gd | M | 80 | 90.718 | AX_T1_POST | 6 | 2.8195E+15 | 1/29/1999 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 583.332 | 8 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.056867 | 23 | 256 | 192 | 0.78125\0.78125 | TCGA-02-0048/1.3.6.1.4.1.14519.5.2.1.1706.4001.271759313632030441078606502900/1.3.6.1.4.1.14519.5.2.1.1706.4001.208892423578919751306703554190 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0048 | t1 | M | 80 | 90.718 | AX_T1 | 5 | 2.8195E+15 | 1/29/1999 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 416.664 | 8 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.064935 | 23 | 256 | 128 | 0.78125\0.78125 | TCGA-02-0048/1.3.6.1.4.1.14519.5.2.1.1706.4001.271759313632030441078606502900/1.3.6.1.4.1.14519.5.2.1.1706.4001.232765711441628100940557835018 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0048 | t2 | M | 80 | 90.718 | AX_T2_FSE | 3 | 2.8195E+15 | 1/29/1999 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 3500 | 98 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.043893 | 23 | 256 | 224 | 0.78125\0.78125 | TCGA-02-0048/1.3.6.1.4.1.14519.5.2.1.1706.4001.271759313632030441078606502900/1.3.6.1.4.1.14519.5.2.1.1706.4001.249496727867125014749061516941 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0054 | flair | F | 44 | 57.153 | AX_FLAIR | 5 | 2.8195E+15 | 5/16/1999 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 147 | 2200 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.024192 | 23 | 256 | 160 | 0.78125\0.78125 | TCGA-02-0054/1.3.6.1.4.1.14519.5.2.1.1706.4001.151634660376289017402983968633/1.3.6.1.4.1.14519.5.2.1.1706.4001.285561797133963558047987589982 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0054 | t1Gd | F | 44 | 57.153 | AX_T1_POST | 9 | 2.8195E+15 | 5/16/1999 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 583.332 | 8 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.061231 | 23 | 256 | 192 | 0.78125\0.78125 | TCGA-02-0054/1.3.6.1.4.1.14519.5.2.1.1706.4001.151634660376289017402983968633/1.3.6.1.4.1.14519.5.2.1.1706.4001.276414996547218243402040733611 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0054 | t1 | F | 44 | 57.153 | AX_T1 | 6 | 2.8195E+15 | 5/16/1999 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 416.664 | 8 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.069917 | 23 | 256 | 128 | 0.78125\0.78125 | TCGA-02-0054/1.3.6.1.4.1.14519.5.2.1.1706.4001.151634660376289017402983968633/1.3.6.1.4.1.14519.5.2.1.1706.4001.281967755247405389888355967057 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0054 | t2 | F | 44 | 57.153 | AX_T2_FSE | 4 | 2.8195E+15 | 5/16/1999 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 3500 | 98 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.04726 | 23 | 256 | 224 | 0.78125\0.78125 | TCGA-02-0054/1.3.6.1.4.1.14519.5.2.1.1706.4001.151634660376289017402983968633/1.3.6.1.4.1.14519.5.2.1.1706.4001.324132997720244415771309925529 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0059 | flair | M | 68 | 66.224 | T2_FLAIR | 12 | 2.8195E+15 | 8/29/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | IR | 2D | 5 | 10002 | 140.244 | 2250 | 127.7374 | 3 | 6.5 | 90 | 0.5816 | 23 | 320 | 224 | 0.46875\0.46875 | TCGA-02-0059/1.3.6.1.4.1.14519.5.2.1.1706.4001.231755935407944517150217116610/1.3.6.1.4.1.14519.5.2.1.1706.4001.152913535526794168392822098633 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0059 | t1Gd | M | 68 | 66.224 | Ax_SE_T1_Post | 14 | 2.8195E+15 | 8/29/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 850 | 12 | 0 | 127.7374 | 3 | 6.5 | 90 | 1.705132 | 23 | 320 | 224 | 0.46875\0.46875 | TCGA-02-0059/1.3.6.1.4.1.14519.5.2.1.1706.4001.231755935407944517150217116610/1.3.6.1.4.1.14519.5.2.1.1706.4001.389966548228828975637733047437 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0059 | t1 | M | 68 | 66.224 | FSPGR_3D | 7 | 2.8195E+15 | 8/29/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 1.2 | 7.02 | 2.1 | 400 | 127.7374 | 3 | 1.2 | 20 | 0.369349 | 124 | 256 | 224 | 0.976562\0.976562 | TCGA-02-0059/1.3.6.1.4.1.14519.5.2.1.1706.4001.231755935407944517150217116610/1.3.6.1.4.1.14519.5.2.1.1706.4001.104793971301967645892117754002 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0059 | t2 | M | 68 | 66.224 | FSE_T2 | 11 | 2.8195E+15 | 8/29/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 4000 | 99 | 0 | 127.7374 | 3 | 6.5 | 90 | 0.8331 | 23 | 320 | 256 | 0.46875\0.46875 | TCGA-02-0059/1.3.6.1.4.1.14519.5.2.1.1706.4001.231755935407944517150217116610/1.3.6.1.4.1.14519.5.2.1.1706.4001.126408749477332517415027496534 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0060 | flair | F | 65 | 48.534 | T2_FLAIR2MM25FOV | 6 | 2.8195E+15 | 2/28/2000 | GE MEDICAL SYSTEMS | SIGNA EXCITE | IR | 2D | 2 | 10002 | 140.778 | 2250 | 127.7412 | 3 | 2 | 90 | 0.6233 | 75 | 256 | 224 | 0.976562\0.976562 | TCGA-02-0060/1.3.6.1.4.1.14519.5.2.1.1706.4001.247522006211308616726493960307/1.3.6.1.4.1.14519.5.2.1.1706.4001.875691179108572562890034722734 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0060 | t1Gd | F | 65 | 48.534 | 3D_AX_FSPGR_POST | 14 | 2.8195E+15 | 2/28/2000 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 1.4 | 4.944 | 2.1 | 400 | 127.7412 | 3 | 0.699999 | 20 | 0.880176 | 216 | 256 | 224 | 0.859375\0.859375 | TCGA-02-0060/1.3.6.1.4.1.14519.5.2.1.1706.4001.247522006211308616726493960307/1.3.6.1.4.1.14519.5.2.1.1706.4001.203293857391455403037115181939 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0060 | t1 | F | 65 | 48.534 | FSPGR_3D_25FOV | 5 | 2.8195E+15 | 2/28/2000 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 1.2 | 6.644 | 2.1 | 400 | 127.7412 | 3 | 1.2 | 20 | 0.426227 | 124 | 256 | 224 | 0.976562\0.976562 | TCGA-02-0060/1.3.6.1.4.1.14519.5.2.1.1706.4001.247522006211308616726493960307/1.3.6.1.4.1.14519.5.2.1.1706.4001.219609462264761522732587573237 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0060 | t2 | F | 65 | 48.534 | FSE_T2 | 8 | 2.8195E+15 | 2/28/2000 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 5500 | 99.268 | 0 | 127.7412 | 3 | 6.5 | 90 | 1.155 | 24 | 320 | 224 | 0.429688\0.429688 | TCGA-02-0060/1.3.6.1.4.1.14519.5.2.1.1706.4001.247522006211308616726493960307/1.3.6.1.4.1.14519.5.2.1.1706.4001.275488470620879346473604871467 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0064 | flair | M | 49 | 88.904 | T2_FLAIR | 12 | 2.8195E+15 | 8/8/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | IR | 2D | 5 | 10002 | 148.118 | 2250 | 127.7414 | 3 | 6.5 | 90 | 0.4916 | 23 | 320 | 224 | 0.46875\0.46875 | TCGA-02-0064/1.3.6.1.4.1.14519.5.2.1.1706.4001.329011203940755039589411572116/1.3.6.1.4.1.14519.5.2.1.1706.4001.289547124248265878668833441740 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0064 | t1Gd | M | 49 | 88.904 | FSPGR_3D | 17 | 2.8195E+15 | 8/8/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 1.5 | 6.24 | 2.1 | 400 | 127.7413 | 3 | 1.5 | 20 | 0.697716 | 124 | 320 | 224 | 0.488281\0.488281 | TCGA-02-0064/1.3.6.1.4.1.14519.5.2.1.1706.4001.329011203940755039589411572116/1.3.6.1.4.1.14519.5.2.1.1706.4001.120605412029452199210260324433 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0064 | t1 | M | 49 | 88.904 | FSPGR_3D | 6 | 2.8195E+15 | 8/8/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 1.2 | 6.636 | 2.1 | 400 | 127.7414 | 3 | 1.2 | 20 | 0.468168 | 124 | 256 | 224 | 0.976562\0.976562 | TCGA-02-0064/1.3.6.1.4.1.14519.5.2.1.1706.4001.329011203940755039589411572116/1.3.6.1.4.1.14519.5.2.1.1706.4001.908751550085828850580923426669 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0064 | t2 | M | 49 | 88.904 | FSE_T2 | 11 | 2.8195E+15 | 8/8/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 4000 | 98.076 | 0 | 127.7414 | 3 | 6.5 | 90 | 0.8718 | 23 | 320 | 256 | 0.46875\0.46875 | TCGA-02-0064/1.3.6.1.4.1.14519.5.2.1.1706.4001.329011203940755039589411572116/1.3.6.1.4.1.14519.5.2.1.1706.4001.305032039524820143164998539810 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0068 | flair | M | 56 | 79.379 | AX_FLAIR | 4 | 2.8195E+15 | 5/16/2000 | GE MEDICAL SYSTEMS | SIGNA EXCITE | IR | 2D | 5 | 10002 | 147.112 | 2200 | 63.8772 | 1.5 | 6.5 | 90 | 0.578 | 23 | 256 | 192 | 0.859375\0.859383 | TCGA-02-0068/1.3.6.1.4.1.14519.5.2.1.1706.4001.239251012204750851180008114061/1.3.6.1.4.1.14519.5.2.1.1706.4001.169273732875851383844768405016 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0068 | t1Gd | M | 56 | 79.379 | AX_T1_POST | 9 | 2.8195E+15 | 5/16/2000 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 550 | 9 | 0 | 63.8772 | 1.5 | 6.5 | 90 | 1.34066 | 23 | 256 | 192 | 0.859375\0.859383 | TCGA-02-0068/1.3.6.1.4.1.14519.5.2.1.1706.4001.239251012204750851180008114061/1.3.6.1.4.1.14519.5.2.1.1706.4001.202828227420706130840978676909 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0068 | t1 | M | 56 | 79.379 | AX_T1 | 6 | 2.8195E+15 | 5/16/2000 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 550 | 9 | 0 | 63.8772 | 1.5 | 6.5 | 90 | 1.34066 | 23 | 256 | 192 | 0.859375\0.859383 | TCGA-02-0068/1.3.6.1.4.1.14519.5.2.1.1706.4001.239251012204750851180008114061/1.3.6.1.4.1.14519.5.2.1.1706.4001.217327932936754121521503964949 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0068 | t2 | M | 56 | 79.379 | AX_T2_FSE | 3 | 2.8195E+15 | 5/16/2000 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 3500 | 93.76 | 0 | 63.8772 | 1.5 | 6.5 | 90 | 0.6985 | 23 | 320 | 224 | 0.429688\0.429692 | TCGA-02-0068/1.3.6.1.4.1.14519.5.2.1.1706.4001.239251012204750851180008114061/1.3.6.1.4.1.14519.5.2.1.1706.4001.164698594795106801737202685813 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0069 | flair | F | 31 | 65.771 | T2_FLAIR | 4 | 2.8195E+15 | 5/28/2000 | GE MEDICAL SYSTEMS | SIGNA EXCITE | IR | 2D | 5 | 10002 | 140.778 | 2250 | 127.7411 | 3 | 6.5 | 90 | 0.4898 | 24 | 256 | 192 | 0.859375\0.859375 | TCGA-02-0069/1.3.6.1.4.1.14519.5.2.1.1706.4001.380770062553415313882886843557/1.3.6.1.4.1.14519.5.2.1.1706.4001.476121618290067004679503813189 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0069 | t1Gd | F | 31 | 65.771 | FSPGR_3D | 12 | 2.8195E+15 | 5/28/2000 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 1.5 | 6.116 | 2.1 | 400 | 127.7411 | 3 | 1.5 | 20 | 0.771899 | 124 | 320 | 224 | 0.488281\0.488281 | TCGA-02-0069/1.3.6.1.4.1.14519.5.2.1.1706.4001.380770062553415313882886843557/1.3.6.1.4.1.14519.5.2.1.1706.4001.787868618042894085065532704205 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0069 | t1 | F | 31 | 65.771 | SE_T1 | 5 | 2.8195E+15 | 5/28/2000 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 700 | 10 | 0 | 127.7411 | 3 | 6.5 | 90 | 2.15934 | 24 | 256 | 192 | 0.859375\0.859375 | TCGA-02-0069/1.3.6.1.4.1.14519.5.2.1.1706.4001.380770062553415313882886843557/1.3.6.1.4.1.14519.5.2.1.1706.4001.249010498119891546309777700223 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0069 | t2 | F | 31 | 65.771 | FSE_T2 | 3 | 2.8195E+15 | 5/28/2000 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 5500 | 99.268 | 0 | 127.7411 | 3 | 6.5 | 90 | 1.2104 | 24 | 320 | 224 | 0.429688\0.429688 | TCGA-02-0069/1.3.6.1.4.1.14519.5.2.1.1706.4001.380770062553415313882886843557/1.3.6.1.4.1.14519.5.2.1.1706.4001.202260155195797715412373698690 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0070 | flair | M | 70 | 99.79 | T2_FLAIR | 4 | 2.8195E+15 | 7/10/2000 | GE MEDICAL SYSTEMS | SIGNA EXCITE | IR | 2D | 5 | 10002 | 146.002 | 2250 | 127.7411 | 3 | 6.5 | 90 | 0.6703 | 24 | 256 | 192 | 0.859371\0.859375 | TCGA-02-0070/1.3.6.1.4.1.14519.5.2.1.1706.4001.288983046313649223240718848290/1.3.6.1.4.1.14519.5.2.1.1706.4001.327171371276055674791400905248 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0070 | t1Gd | M | 70 | 99.79 | AXIAL_T1_+C | 10 | 2.8195E+15 | 7/10/2000 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 700 | 10 | 0 | 127.7411 | 3 | 6.5 | 90 | 1.98231 | 24 | 256 | 192 | 0.859371\0.859375 | TCGA-02-0070/1.3.6.1.4.1.14519.5.2.1.1706.4001.288983046313649223240718848290/1.3.6.1.4.1.14519.5.2.1.1706.4001.333477048103776358657330624985 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0070 | t1 | M | 70 | 99.79 | SE_T1 | 5 | 2.8195E+15 | 7/10/2000 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 700 | 10 | 0 | 127.7411 | 3 | 6.5 | 90 | 1.7422 | 24 | 256 | 192 | 0.859371\0.859375 | TCGA-02-0070/1.3.6.1.4.1.14519.5.2.1.1706.4001.288983046313649223240718848290/1.3.6.1.4.1.14519.5.2.1.1706.4001.223969978304636924462725206105 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0070 | t2 | M | 70 | 99.79 | FSE_T2 | 3 | 2.8195E+15 | 7/10/2000 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 5500 | 99.268 | 0 | 127.7411 | 3 | 6.5 | 90 | 1.2906 | 24 | 320 | 224 | 0.429686\0.429688 | TCGA-02-0070/1.3.6.1.4.1.14519.5.2.1.1706.4001.288983046313649223240718848290/1.3.6.1.4.1.14519.5.2.1.1706.4001.231105368610379750268505671981 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0075 | flair | M | 63 | 73.482 | T2_FLAIR | 11 | 2.8195E+15 | 9/24/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | IR | 2D | 5 | 10002 | 140.778 | 2250 | 127.7412 | 3 | 6.5 | 90 | 0.4983 | 23 | 256 | 192 | 0.859375\0.859375 | TCGA-02-0075/1.3.6.1.4.1.14519.5.2.1.1706.4001.199028178384634677629049332875/1.3.6.1.4.1.14519.5.2.1.1706.4001.327311884902359009396424776096 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0075 | t1Gd | M | 63 | 73.482 | Ax_SE_T1_Post | 17 | 2.8195E+15 | 9/24/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 700 | 10 | 0 | 127.7412 | 3 | 6.5 | 90 | 2.09587 | 23 | 256 | 192 | 0.859375\0.859375 | TCGA-02-0075/1.3.6.1.4.1.14519.5.2.1.1706.4001.199028178384634677629049332875/1.3.6.1.4.1.14519.5.2.1.1706.4001.269081075850064364799501008723 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0075 | t1 | M | 63 | 73.482 | FSPGR_3D | 14 | 2.8195E+15 | 9/24/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 1.5 | 4.984 | 2.1 | 400 | 127.7412 | 3 | 1.5 | 20 | 0.786819 | 124 | 256 | 224 | 0.859375\0.859375 | TCGA-02-0075/1.3.6.1.4.1.14519.5.2.1.1706.4001.199028178384634677629049332875/1.3.6.1.4.1.14519.5.2.1.1706.4001.149622591468669601675018790164 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0075 | t2 | M | 63 | 73.482 | FSE_T2 | 10 | 2.8195E+15 | 9/24/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 5500 | 102.596 | 0 | 127.7412 | 3 | 6.5 | 90 | 1.1799 | 23 | 256 | 192 | 0.859375\0.859375 | TCGA-02-0075/1.3.6.1.4.1.14519.5.2.1.1706.4001.199028178384634677629049332875/1.3.6.1.4.1.14519.5.2.1.1706.4001.317391215776965396207339135736 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0085 | flair | F | 65 | 90.265 | AX_FLAIR | 4 | 2.8195E+15 | 1/30/1999 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 147 | 2200 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.022486 | 22 | 256 | 160 | 0.78125\0.78125 | TCGA-02-0085/1.3.6.1.4.1.14519.5.2.1.1706.4001.289458647018511940499719529853/1.3.6.1.4.1.14519.5.2.1.1706.4001.108905473702454736524787314351 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0085 | t1Gd | F | 65 | 90.265 | AX_T1_POST | 6 | 2.8195E+15 | 1/30/1999 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 583.332 | 8 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.054438 | 22 | 256 | 192 | 0.78125\0.78125 | TCGA-02-0085/1.3.6.1.4.1.14519.5.2.1.1706.4001.289458647018511940499719529853/1.3.6.1.4.1.14519.5.2.1.1706.4001.179485994007439679254001323335 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0085 | t1 | F | 65 | 90.265 | AX_T1 | 5 | 2.8195E+15 | 1/30/1999 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 416.664 | 8 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.06216 | 22 | 256 | 128 | 0.78125\0.78125 | TCGA-02-0085/1.3.6.1.4.1.14519.5.2.1.1706.4001.289458647018511940499719529853/1.3.6.1.4.1.14519.5.2.1.1706.4001.288895680883982235784348384008 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0085 | t2 | F | 65 | 90.265 | AX_T2_FSE | 3 | 2.8195E+15 | 1/30/1999 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 3500 | 98 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.042018 | 22 | 256 | 224 | 0.78125\0.78125 | TCGA-02-0085/1.3.6.1.4.1.14519.5.2.1.1706.4001.289458647018511940499719529853/1.3.6.1.4.1.14519.5.2.1.1706.4001.278352104503675092135094187118 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0086 | flair | F | 45 | 96.162 | AX_FLAIR | 4 | 2.8195E+15 | 7/25/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | IR | 2D | 5 | 10002 | 141.54 | 2200 | 63.87812 | 1.5 | 6.5 | 90 | 0.0222 | 23 | 256 | 160 | 0.78125\0.78125 | TCGA-02-0086/1.3.6.1.4.1.14519.5.2.1.1706.4001.189302178457617024065570049848/1.3.6.1.4.1.14519.5.2.1.1706.4001.223838339019946872254464308895 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0086 | t1Gd | F | 45 | 96.162 | AX_T1_POST | 11 | 2.8195E+15 | 7/25/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 416.664 | 8 | 0 | 63.87817 | 1.5 | 6.5 | 90 | 0.064333 | 23 | 256 | 192 | 0.78125\0.78125 | TCGA-02-0086/1.3.6.1.4.1.14519.5.2.1.1706.4001.189302178457617024065570049848/1.3.6.1.4.1.14519.5.2.1.1706.4001.327450908011286130960738686265 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0086 | t1 | F | 45 | 96.162 | AX_T1 | 6 | 2.8195E+15 | 7/25/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 416.664 | 8 | 0 | 63.87812 | 1.5 | 6.5 | 90 | 0.064333 | 23 | 256 | 128 | 0.78125\0.78125 | TCGA-02-0086/1.3.6.1.4.1.14519.5.2.1.1706.4001.189302178457617024065570049848/1.3.6.1.4.1.14519.5.2.1.1706.4001.300182168244910270765145894367 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0086 | t2 | F | 45 | 96.162 | AX_T2_FSE | 3 | 2.8195E+15 | 7/25/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 3500 | 96.512 | 0 | 63.87812 | 1.5 | 6.5 | 90 | 0.0724 | 23 | 256 | 224 | 0.78125\0.78125 | TCGA-02-0086/1.3.6.1.4.1.14519.5.2.1.1706.4001.189302178457617024065570049848/1.3.6.1.4.1.14519.5.2.1.1706.4001.130793598392145152904761943139 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0087 | flair | F | 27 | 81.647 | T2_FLAIR | 10 | 2.8195E+15 | 12/13/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | IR | 2D | 5 | 10002 | 140.778 | 2250 | 127.7412 | 3 | 6.5 | 90 | 0.5064 | 23 | 256 | 192 | 0.859375\0.859375 | TCGA-02-0087/1.3.6.1.4.1.14519.5.2.1.1706.4001.197977742428665924369847662702/1.3.6.1.4.1.14519.5.2.1.1706.4001.188891971026185310471464105645 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0087 | t1Gd | F | 27 | 81.647 | FSPGR_3D | 18 | 2.8195E+15 | 12/13/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 1.5 | 6.176 | 2.1 | 400 | 127.7411 | 3 | 1.5 | 20 | 0.741433 | 124 | 320 | 224 | 0.488281\0.488281 | TCGA-02-0087/1.3.6.1.4.1.14519.5.2.1.1706.4001.197977742428665924369847662702/1.3.6.1.4.1.14519.5.2.1.1706.4001.130023584613816858008662430540 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0087 | t1 | F | 27 | 81.647 | FSPGR_3D_25FOV | 5 | 2.8195E+15 | 12/13/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 1.2 | 6.644 | 2.1 | 400 | 127.7412 | 3 | 1.2 | 20 | 0.461773 | 124 | 256 | 224 | 0.976562\0.976562 | TCGA-02-0087/1.3.6.1.4.1.14519.5.2.1.1706.4001.197977742428665924369847662702/1.3.6.1.4.1.14519.5.2.1.1706.4001.153352528048877621623084394547 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0087 | t2 | F | 27 | 81.647 | FSE_T2 | 9 | 2.8195E+15 | 12/13/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 5500 | 99.268 | 0 | 127.7412 | 3 | 6.5 | 90 | 1.1992 | 23 | 320 | 224 | 0.429688\0.429688 | TCGA-02-0087/1.3.6.1.4.1.14519.5.2.1.1706.4001.197977742428665924369847662702/1.3.6.1.4.1.14519.5.2.1.1706.4001.297280847905859204769837917360 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0102 | flair | M | 42 | 74.843 | AX_FLAIR | 4 | 2.8195E+15 | 12/15/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 147 | 2200 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.023171 | 23 | 256 | 160 | 0.78125\0.78125 | TCGA-02-0102/1.3.6.1.4.1.14519.5.2.1.1706.4001.240465490805420927209667546432/1.3.6.1.4.1.14519.5.2.1.1706.4001.580630653491671454692169960256 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0102 | t1Gd | M | 42 | 74.843 | AX_T1_POST | 6 | 2.8195E+15 | 12/15/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 583.332 | 9 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.058645 | 23 | 256 | 192 | 0.78125\0.78125 | TCGA-02-0102/1.3.6.1.4.1.14519.5.2.1.1706.4001.240465490805420927209667546432/1.3.6.1.4.1.14519.5.2.1.1706.4001.958674936724987421213786894928 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0102 | t1 | M | 42 | 74.843 | AX_T1 | 5 | 2.8195E+15 | 12/15/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 416.664 | 8 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.066965 | 23 | 256 | 128 | 0.78125\0.78125 | TCGA-02-0102/1.3.6.1.4.1.14519.5.2.1.1706.4001.240465490805420927209667546432/1.3.6.1.4.1.14519.5.2.1.1706.4001.321931575003689210541138128522 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0102 | t2 | M | 42 | 74.843 | AX_T2_FSE | 3 | 2.8195E+15 | 12/15/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 3500 | 105 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.045265 | 23 | 256 | 224 | 0.78125\0.78125 | TCGA-02-0102/1.3.6.1.4.1.14519.5.2.1.1706.4001.240465490805420927209667546432/1.3.6.1.4.1.14519.5.2.1.1706.4001.846686946556146675536736249954 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0106 | flair | M | 54 | 70.307 | AX_FLAIR | 4 | 2.8195E+15 | 10/30/1998 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 147 | 2200 | 6.38E+08 | 1.5 | 6.5 | 90 | 0.023404 | 23 | 256 | 160 | 0.78125\0.78125 | TCGA-02-0106/1.3.6.1.4.1.14519.5.2.1.1706.4001.204859028463062009490599344146/1.3.6.1.4.1.14519.5.2.1.1706.4001.142741494614831385536214775922 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0106 | t1Gd | M | 54 | 70.307 | AX_T1_POST | 6 | 2.8195E+15 | 10/30/1998 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 583.332 | 8 | 6.38E+08 | 1.5 | 6.5 | 90 | 0.059235 | 23 | 256 | 192 | 0.78125\0.78125 | TCGA-02-0106/1.3.6.1.4.1.14519.5.2.1.1706.4001.204859028463062009490599344146/1.3.6.1.4.1.14519.5.2.1.1706.4001.407761314964431500733064308177 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0106 | t1 | M | 54 | 70.307 | AX_T1 | 5 | 2.8195E+15 | 10/30/1998 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 416.664 | 8 | 6.38E+08 | 1.5 | 6.5 | 90 | 0.067638 | 23 | 256 | 128 | 0.78125\0.78125 | TCGA-02-0106/1.3.6.1.4.1.14519.5.2.1.1706.4001.204859028463062009490599344146/1.3.6.1.4.1.14519.5.2.1.1706.4001.257637810777885113989358890547 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0106 | t2 | M | 54 | 70.307 | AX_T2_FSE | 3 | 2.8195E+15 | 10/30/1998 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 3500 | 98 | 6.38E+08 | 1.5 | 6.5 | 90 | 0.04572 | 23 | 256 | 224 | 0.78125\0.78125 | TCGA-02-0106/1.3.6.1.4.1.14519.5.2.1.1706.4001.204859028463062009490599344146/1.3.6.1.4.1.14519.5.2.1.1706.4001.189172039998441866861864803373 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0116 | flair | M | 51 | 99.79 | AX_FLAIR | 4 | 2.8195E+15 | 3/22/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 147 | 2200 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.022128 | 24 | 256 | 160 | 0.781250\0.781250 | TCGA-02-0116/1.3.6.1.4.1.14519.5.2.1.1706.4001.333801404495028917226888170515/1.3.6.1.4.1.14519.5.2.1.1706.4001.511107585415671993882365147439 | |
| TCGA-GBM | TCGA-02 | TCGA-02-0116 | t1Gd | M | 51 | 99.79 | AX_T1_POST | 6 | 2.8195E+15 | 3/22/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 633.332 | 8 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.053814 | 24 | 256 | 192 | 0.781250\0.781250 | TCGA-02-0116/1.3.6.1.4.1.14519.5.2.1.1706.4001.333801404495028917226888170515/1.3.6.1.4.1.14519.5.2.1.1706.4001.324271664399244452369200352648 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0116 | t1 | M | 51 | 99.79 | AX_T1 | 5 | 2.8195E+15 | 3/22/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 433.332 | 8 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.064152 | 24 | 256 | 128 | 0.781250\0.781250 | TCGA-02-0116/1.3.6.1.4.1.14519.5.2.1.1706.4001.333801404495028917226888170515/1.3.6.1.4.1.14519.5.2.1.1706.4001.119332675140408493379026119335 | ||
| TCGA-GBM | TCGA-02 | TCGA-02-0116 | t2 | M | 51 | 99.79 | AX_T2_FSE | 3 | 2.8195E+15 | 3/22/1997 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 3500 | 98 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.045108 | 24 | 256 | 224 | 0.781250\0.781250 | TCGA-02-0116/1.3.6.1.4.1.14519.5.2.1.1706.4001.333801404495028917226888170515/1.3.6.1.4.1.14519.5.2.1.1706.4001.181707635044752935161529942524 | ||
| TCGA-GBM | TCGA-06 | TCGA-06-0119 | flair | F | AxFLAIR-thin_for_surgery | 7 | 2.27963E+15 | 12/26/2003 | BAY2OC0 | IR | 2D | 2.5 | 10004 | 155 | 2200 | 6.38E+08 | 1.5 | 2.5 | 90 | 0.035398 | 71 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0119/1.3.6.1.4.1.14519.5.2.1.4591.4001.283171458436570880779969690543/1.3.6.1.4.1.14519.5.2.1.4591.4001.190846624143899319203062806923 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0119 | t1Gd | F | =+COR_T1 | 12 | 2.27963E+15 | 12/26/2003 | BAY2OC0 | SE | 2D | 5 | 500 | 14 | 6.38E+08 | 1.5 | 5 | 90 | 0.040717 | 37 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0119/1.3.6.1.4.1.14519.5.2.1.4591.4001.283171458436570880779969690543/1.3.6.1.4.1.14519.5.2.1.4591.4001.130997813040669753794699573293 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0119 | t1 | F | AXIAL_T1 | 6 | 2.27963E+15 | 12/26/2003 | BAY2OC0 | SE | 2D | 5 | 500 | 14 | 6.38E+08 | 1.5 | 5 | 90 | 0.037584 | 36 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0119/1.3.6.1.4.1.14519.5.2.1.4591.4001.283171458436570880779969690543/1.3.6.1.4.1.14519.5.2.1.4591.4001.175712352031583034594446384562 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0119 | t2 | F | AXIAL_FSE | 5 | 2.27963E+15 | 12/26/2003 | BAY2OC0 | SE | 2D | 5 | 3500 | 20 | 6.38E+08 | 1.5 | 5 | 90 | 0.039045 | 72 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0119/1.3.6.1.4.1.14519.5.2.1.4591.4001.283171458436570880779969690543/1.3.6.1.4.1.14519.5.2.1.4591.4001.970644432299498858379456667200 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0122 | flair | F | AXIAL_FLAIR | 5 | 1.89289E+15 | 9/14/2004 | MR3T | IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7323 | 3 | 2.5 | 90 | 0.6054 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0122/1.3.6.1.4.1.14519.5.2.1.4591.4001.545613483524570609928056355494/1.3.6.1.4.1.14519.5.2.1.4591.4001.128296557660865063844963462837 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0122 | t1Gd | F | AX_T1_POST_GD_FLAIR | 10 | 1.89289E+15 | 9/14/2004 | MR3T | IR | 2D | 2.5 | 3181.81 | 6.356 | 1238 | 127.7322 | 3 | 2.5 | 90 | 1.4511 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0122/1.3.6.1.4.1.14519.5.2.1.4591.4001.545613483524570609928056355494/1.3.6.1.4.1.14519.5.2.1.4591.4001.151947201227980965067378635297 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0122 | t1 | F | AX_T1_pre_gd | 8 | 1.89289E+15 | 9/14/2004 | MR3T | IR | 2D | 5 | 3000.01 | 6.356 | 1238 | 127.7323 | 3 | 5 | 90 | 1.1543 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0122/1.3.6.1.4.1.14519.5.2.1.4591.4001.545613483524570609928056355494/1.3.6.1.4.1.14519.5.2.1.4591.4001.976676789917195046100044055071 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0122 | t2 | F | AX_T2_FR-FSE_RF2_150 | 6 | 1.89289E+15 | 9/14/2004 | MR3T | SE | 2D | 5 | 3000 | 103.464 | 0 | 127.7323 | 3 | 5 | 90 | 1.3382 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0122/1.3.6.1.4.1.14519.5.2.1.4591.4001.545613483524570609928056355494/1.3.6.1.4.1.14519.5.2.1.4591.4001.284098509532271800980320718400 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0128 | flair | M | 77 | AXIAL_FLAIR | 3 | 2.38081E+15 | 2/18/1999 | General Electric | BAY | GENESIS | flair | 3 | 10004 | 155 | 2200 | 3 | 90 | 53 | 0.938\0.938 | TCGA-06-0128/1.3.6.1.4.1.14519.5.2.1.4591.4001.197146947506099366832803736438/1.3.6.1.4.1.14519.5.2.1.4591.4001.651359394442711367070939266432 | |||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0128 | t1Gd | M | 77 | AXIAL_T1_POST_GD | 9 | 2.38081E+15 | 2/18/1999 | General Electric | BAY | GENESIS | memp | 3 | 500 | 14 | 0 | 3 | 90 | 53 | 0.938\0.938 | TCGA-06-0128/1.3.6.1.4.1.14519.5.2.1.4591.4001.197146947506099366832803736438/1.3.6.1.4.1.14519.5.2.1.4591.4001.155987932372490417929540843368 | |||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0128 | t1 | M | 77 | AXIAL_T1_PRE_GD | 7 | 2.38081E+15 | 2/18/1999 | General Electric | BAY | GENESIS | memp | 3 | 500 | 14 | 0 | 3 | 90 | 53 | 0.938\0.938 | TCGA-06-0128/1.3.6.1.4.1.14519.5.2.1.4591.4001.197146947506099366832803736438/1.3.6.1.4.1.14519.5.2.1.4591.4001.619151632501848799260529175220 | |||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0128 | t2 | M | 77 | AXIAL__FSE | 2 | 2.38081E+15 | 2/18/1999 | General Electric | BAY | GENESIS | fse | 3 | 3500 | 22 | 0 | 3 | 90 | 106 | 0.938\0.938 | TCGA-06-0128/1.3.6.1.4.1.14519.5.2.1.4591.4001.197146947506099366832803736438/1.3.6.1.4.1.14519.5.2.1.4591.4001.120673894695124200101185494671 | |||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0130 | flair | M | AXIAL_FLAIR_RF2__150 | 7 | 2.70782E+15 | 9/11/2001 | MR3T | IR | 2D | 2.5 | 10002 | 123.5 | 2250 | 1.28E+09 | 3 | 2.5 | 90 | 0.090741 | 78 | 256 | 192 | 0.937500\0.937500 | TCGA-06-0130/1.3.6.1.4.1.14519.5.2.1.4591.4001.123247425694007311600825493062/1.3.6.1.4.1.14519.5.2.1.4591.4001.296118904200952959225417317679 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0130 | t1Gd | M | AX_T1_POST_GD_FLAIR | 11 | 2.70782E+15 | 9/11/2001 | MR3T | RM | 2D | 2.5 | 3252.816 | 8.104 | 860 | 1.28E+09 | 3 | 2.5 | 90 | 0.100896 | 80 | 288 | 192 | 0.468750\0.468750 | TCGA-06-0130/1.3.6.1.4.1.14519.5.2.1.4591.4001.123247425694007311600825493062/1.3.6.1.4.1.14519.5.2.1.4591.4001.128853861285611731507706018829 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0130 | t1 | M | AX_T1_pre_gd | 8 | 2.70782E+15 | 9/11/2001 | MR3T | RM | 2D | 5 | 2366.264 | 8.104 | 860 | 1.28E+09 | 3 | 5 | 90 | 0.107373 | 42 | 288 | 192 | 0.468750\0.468750 | TCGA-06-0130/1.3.6.1.4.1.14519.5.2.1.4591.4001.123247425694007311600825493062/1.3.6.1.4.1.14519.5.2.1.4591.4001.838991966145071683848655126039 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0130 | t2 | M | AX_T2_FR-FSE_RF2_150 | 6 | 2.70782E+15 | 9/11/2001 | MR3T | RM | 2D | 5 | 3766.664 | 104.972 | 1.28E+09 | 3 | 5 | 90 | 0.0939 | 40 | 352 | 192 | 0.468750\0.468750 | TCGA-06-0130/1.3.6.1.4.1.14519.5.2.1.4591.4001.123247425694007311600825493062/1.3.6.1.4.1.14519.5.2.1.4591.4001.305701459061918935894190837157 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0132 | flair | M | AXIAL_FLAIR | 5 | 2.64669E+15 | 5/17/2005 | BAY2OC0 | IR | 2D | 2.5 | 10004 | 155 | 2200 | 6.39E+08 | 1.5 | 2.5 | 90 | 0.031318 | 71 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0132/1.3.6.1.4.1.14519.5.2.1.4591.4001.156068272915163991923280385588/1.3.6.1.4.1.14519.5.2.1.4591.4001.288861537619222119588401224045 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0132 | t1Gd | M | AXIAL_T1_GD | 15 | 2.64669E+15 | 5/17/2005 | BAY2OC0 | SE | 2D | 2.5 | 500 | 13 | 6.39E+08 | 1.5 | 2.5 | 90 | 0.044335 | 76 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0132/1.3.6.1.4.1.14519.5.2.1.4591.4001.156068272915163991923280385588/1.3.6.1.4.1.14519.5.2.1.4591.4001.662318710003756037574079830102 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0132 | t1 | M | AXIAL_T1 | 7 | 2.64669E+15 | 5/17/2005 | BAY2OC0 | SE | 2D | 5 | 500 | 14 | 6.39E+08 | 1.5 | 5 | 90 | 0.036023 | 37 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0132/1.3.6.1.4.1.14519.5.2.1.4591.4001.156068272915163991923280385588/1.3.6.1.4.1.14519.5.2.1.4591.4001.249844598065449004919997881569 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0132 | t2 | M | AXIAL_FSE | 4 | 2.64669E+15 | 5/17/2005 | BAY2OC0 | SE | 2D | 5 | 3500 | 20 | 6.39E+08 | 1.5 | 5 | 90 | 0.036463 | 74 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0132/1.3.6.1.4.1.14519.5.2.1.4591.4001.156068272915163991923280385588/1.3.6.1.4.1.14519.5.2.1.4591.4001.148730121880670249624667054880 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0133 | flair | M | AXIAL_FLAIR | 5 | 1.48959E+15 | 5/10/2005 | MR3T | IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7322 | 3 | 2.5 | 90 | 1.0485 | 71 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0133/1.3.6.1.4.1.14519.5.2.1.4591.4001.200432657521696433544702327867/1.3.6.1.4.1.14519.5.2.1.4591.4001.915421561452489655072317720028 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0133 | t1Gd | M | AX_T1_POST_GD_FLAIR | 11 | 1.48959E+15 | 5/10/2005 | MR3T | IR | 2D | 2.5 | 3072.51 | 6.356 | 1238 | 127.7322 | 3 | 2.5 | 90 | 1.5929 | 71 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0133/1.3.6.1.4.1.14519.5.2.1.4591.4001.200432657521696433544702327867/1.3.6.1.4.1.14519.5.2.1.4591.4001.312563117951495282981965119560 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0133 | t1 | M | AX_T1_pre_gd | 8 | 1.48959E+15 | 5/10/2005 | MR3T | IR | 2D | 5 | 3072.58 | 6.356 | 1238 | 127.7323 | 3 | 5 | 90 | 1.5929 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0133/1.3.6.1.4.1.14519.5.2.1.4591.4001.200432657521696433544702327867/1.3.6.1.4.1.14519.5.2.1.4591.4001.188613876158978626356924469119 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0133 | t2 | M | AX_T2_FR-FSE_RF2_150 | 6 | 1.48959E+15 | 5/10/2005 | MR3T | SE | 2D | 5 | 3000 | 103.464 | 0 | 127.7322 | 3 | 5 | 90 | 1.4532 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0133/1.3.6.1.4.1.14519.5.2.1.4591.4001.200432657521696433544702327867/1.3.6.1.4.1.14519.5.2.1.4591.4001.165459028026483730012969806104 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0137 | flair | F | AXIAL_FLAIR | 4 | 1.55442E+15 | 12/24/2001 | BAY2OC0 | IR | 2D | 3 | 10004 | 155 | 2200 | 6.38E+08 | 1.5 | 3 | 90 | 0.028464 | 50 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0137/1.3.6.1.4.1.14519.5.2.1.4591.4001.326662079066250663678557696078/1.3.6.1.4.1.14519.5.2.1.4591.4001.109697696341289807648805483606 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0137 | t1Gd | F | AXIAL_T1_GD | 9 | 1.55442E+15 | 12/24/2001 | BAY2OC0 | SE | 2D | 3 | 500 | 14 | 6.38E+08 | 1.5 | 3 | 90 | 0.039289 | 50 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0137/1.3.6.1.4.1.14519.5.2.1.4591.4001.326662079066250663678557696078/1.3.6.1.4.1.14519.5.2.1.4591.4001.202223923499982894907965771887 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0137 | t1 | F | AXIAL_T1 | 6 | 1.55442E+15 | 12/24/2001 | BAY2OC0 | SE | 2D | 3 | 500 | 14 | 6.38E+08 | 1.5 | 3 | 90 | 0.039289 | 50 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0137/1.3.6.1.4.1.14519.5.2.1.4591.4001.326662079066250663678557696078/1.3.6.1.4.1.14519.5.2.1.4591.4001.305227854284090555965781589764 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0137 | t2 | F | AXIAL_FSE | 3 | 1.55442E+15 | 12/24/2001 | BAY2OC0 | SE | 2D | 3 | 3500 | 20 | 6.38E+08 | 1.5 | 3 | 90 | 0.052326 | 100 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0137/1.3.6.1.4.1.14519.5.2.1.4591.4001.326662079066250663678557696078/1.3.6.1.4.1.14519.5.2.1.4591.4001.157809556490045867232259259088 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0138 | flair | M | AXIAL_T2_FLAIR | 6 | 5.979E+15 | 11/25/2002 | MR3T | IR | 2D | 2.5 | 10002 | 124.9 | 2250 | 127.7324 | 3 | 2.5 | 90 | 1.0139 | 72 | 320 | 224 | 0.46875\0.46875 | TCGA-06-0138/1.3.6.1.4.1.14519.5.2.1.4591.4001.296601565633699364447385491840/1.3.6.1.4.1.14519.5.2.1.4591.4001.312547018558825125677081232576 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0138 | t1Gd | M | AX_T1_POST_GD_FLAIR | 10 | 5.979E+15 | 11/25/2002 | MR3T | IR | 2D | 2.5 | 3285.62 | 6.568 | 1238 | 127.7324 | 3 | 2.5 | 90 | 1.5327 | 69 | 320 | 224 | 0.46875\0.46875 | TCGA-06-0138/1.3.6.1.4.1.14519.5.2.1.4591.4001.296601565633699364447385491840/1.3.6.1.4.1.14519.5.2.1.4591.4001.973243150706573836060823787338 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0138 | t1 | M | AX_T1_pre_gd | 8 | 5.979E+15 | 11/25/2002 | MR3T | IR | 2D | 5 | 3379.57 | 6.584 | 1238 | 127.7324 | 3 | 5 | 90 | 1.2772 | 36 | 320 | 224 | 0.46875\0.46875 | TCGA-06-0138/1.3.6.1.4.1.14519.5.2.1.4591.4001.296601565633699364447385491840/1.3.6.1.4.1.14519.5.2.1.4591.4001.208761379736355493761543330080 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0138 | t2 | M | AX_T2_FR-FSE_RF2_150 | 5 | 5.979E+15 | 11/25/2002 | MR3T | SE | 2D | 5 | 3000 | 104.436 | 0 | 127.7324 | 3 | 5 | 90 | 1.3877 | 36 | 320 | 224 | 0.46875\0.46875 | TCGA-06-0138/1.3.6.1.4.1.14519.5.2.1.4591.4001.296601565633699364447385491840/1.3.6.1.4.1.14519.5.2.1.4591.4001.251176902091719343107685682166 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0139 | flair | M | AXIAL_FLAIR | 5 | 3.34682E+15 | 6/10/2004 | MR3T | IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7323 | 3 | 2.5 | 90 | 0.8274 | 71 | 320 | 224 | 0.46875\0.46875 | TCGA-06-0139/1.3.6.1.4.1.14519.5.2.1.4591.4001.751927611731877561558954106150/1.3.6.1.4.1.14519.5.2.1.4591.4001.191924528389804940209560191592 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0139 | t1Gd | M | AX_T1_POST_GD_FLAIR | 10 | 3.34682E+15 | 6/10/2004 | MR3T | IR | 2D | 2.5 | 3062.5 | 6.356 | 1238 | 127.7323 | 3 | 2.5 | 90 | 1.5454 | 71 | 320 | 224 | 0.46875\0.46875 | TCGA-06-0139/1.3.6.1.4.1.14519.5.2.1.4591.4001.751927611731877561558954106150/1.3.6.1.4.1.14519.5.2.1.4591.4001.229090327759241505815737823916 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0139 | t1 | M | AX_T1_pre_gd | 8 | 3.34682E+15 | 6/10/2004 | MR3T | IR | 2D | 5 | 3062.58 | 6.356 | 1238 | 127.7323 | 3 | 5 | 90 | 1.5453 | 36 | 320 | 224 | 0.46875\0.46875 | TCGA-06-0139/1.3.6.1.4.1.14519.5.2.1.4591.4001.751927611731877561558954106150/1.3.6.1.4.1.14519.5.2.1.4591.4001.332362461346917707181060064705 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0139 | t2 | M | AX_T2_FR-FSE_RF2_150 | 6 | 3.34682E+15 | 6/10/2004 | MR3T | SE | 2D | 5 | 3000 | 103.464 | 0 | 127.7323 | 3 | 5 | 90 | 1.8289 | 36 | 320 | 224 | 0.46875\0.46875 | TCGA-06-0139/1.3.6.1.4.1.14519.5.2.1.4591.4001.751927611731877561558954106150/1.3.6.1.4.1.14519.5.2.1.4591.4001.209789345577718778695745727517 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0142 | flair | M | AXIAL_FLAIR | 7 | 2.8195E+15 | 3/11/2000 | BAY2BAY2 | IR | 2D | 3 | 10004 | 155 | 2200 | 6.38E+08 | 1.5 | 3 | 90 | 0.991425 | 56 | 256 | 192 | 0.937500\0.937500 | TCGA-06-0142/1.3.6.1.4.1.14519.5.2.1.4591.4001.140537559994289172851244415813/1.3.6.1.4.1.14519.5.2.1.4591.4001.265250330319551296448339505489 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0142 | t1Gd | M | AXIAL_T1_GD | 11 | 2.8195E+15 | 3/11/2000 | BAY2BAY2 | SE | 2D | 3 | 500 | 14 | 6.38E+08 | 1.5 | 3 | 90 | 1.228103 | 56 | 256 | 192 | 0.937500\0.937500 | TCGA-06-0142/1.3.6.1.4.1.14519.5.2.1.4591.4001.140537559994289172851244415813/1.3.6.1.4.1.14519.5.2.1.4591.4001.278231207410707039093733128546 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0142 | t1 | M | AXIAL_T1 | 9 | 2.8195E+15 | 3/11/2000 | BAY2BAY2 | SE | 2D | 3 | 500 | 14 | 6.38E+08 | 1.5 | 3 | 90 | 1.228103 | 56 | 256 | 192 | 0.937500\0.937500 | TCGA-06-0142/1.3.6.1.4.1.14519.5.2.1.4591.4001.140537559994289172851244415813/1.3.6.1.4.1.14519.5.2.1.4591.4001.160473331117007298735566672719 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0142 | t2 | M | AXIAL_FSE | 3 | 2.8195E+15 | 3/11/2000 | BAY2BAY2 | SE | 2D | 3 | 3500 | 20 | 6.38E+08 | 1.5 | 3 | 90 | 1.701089 | 112 | 256 | 192 | 0.937500\0.937500 | TCGA-06-0142/1.3.6.1.4.1.14519.5.2.1.4591.4001.140537559994289172851244415813/1.3.6.1.4.1.14519.5.2.1.4591.4001.246556599071909674613941779916 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0145 | flair | F | 52 | 73.482 | AXIAL_FLAIR | 4 | 6.78517E+15 | 11/6/2000 | GE MEDICAL SYSTEMS | BAY2BAY2 | GENESIS_SIGNA | IR | 2D | 3 | 10004 | 155 | 2200 | 6.38E+08 | 1.5 | 3 | 90 | 0.033739 | 54 | 256 | 192 | 0.937500\0.937500 | TCGA-06-0145/1.3.6.1.4.1.14519.5.2.1.4591.4001.312057250385941099348613546751/1.3.6.1.4.1.14519.5.2.1.4591.4001.100984269677348148072147090714 |
| TCGA-GBM | TCGA-06 | TCGA-06-0145 | t1Gd | F | 52 | 73.482 | AXIAL_T1_GD | 10 | 6.78517E+15 | 11/6/2000 | GE MEDICAL SYSTEMS | BAY2BAY2 | GENESIS_SIGNA | SE | 2D | 3 | 500 | 14 | 6.38E+08 | 1.5 | 3 | 90 | 0.041793 | 54 | 256 | 192 | 0.937500\0.937500 | TCGA-06-0145/1.3.6.1.4.1.14519.5.2.1.4591.4001.312057250385941099348613546751/1.3.6.1.4.1.14519.5.2.1.4591.4001.332902950048400716837387278761 | |
| TCGA-GBM | TCGA-06 | TCGA-06-0145 | t1 | F | 52 | 73.482 | AXIAL_T1 | 8 | 6.78517E+15 | 11/6/2000 | GE MEDICAL SYSTEMS | BAY2BAY2 | GENESIS_SIGNA | SE | 2D | 3 | 500 | 14 | 6.38E+08 | 1.5 | 3 | 90 | 0.041793 | 54 | 256 | 192 | 0.937500\0.937500 | TCGA-06-0145/1.3.6.1.4.1.14519.5.2.1.4591.4001.312057250385941099348613546751/1.3.6.1.4.1.14519.5.2.1.4591.4001.315639171460581681490878614022 | |
| TCGA-GBM | TCGA-06 | TCGA-06-0145 | t2 | F | 52 | 73.482 | AXIAL_FSE | 3 | 6.78517E+15 | 11/6/2000 | GE MEDICAL SYSTEMS | BAY2BAY2 | GENESIS_SIGNA | SE | 2D | 3 | 3500 | 20 | 6.38E+08 | 1.5 | 3 | 90 | 0.055821 | 108 | 256 | 192 | 0.937500\0.937500 | TCGA-06-0145/1.3.6.1.4.1.14519.5.2.1.4591.4001.312057250385941099348613546751/1.3.6.1.4.1.14519.5.2.1.4591.4001.124843654110488034987706152331 | |
| TCGA-GBM | TCGA-06 | TCGA-06-0149 | flair | F | AXIAL_T2_FLAIR | 5 | 1.82811E+15 | 3/25/2003 | MR3T | IR | 2D | 2.5 | 10002 | 124.9 | 2250 | 127.7325 | 3 | 2.5 | 90 | 0.5979 | 70 | 320 | 224 | 0.46875\0.46875 | TCGA-06-0149/1.3.6.1.4.1.14519.5.2.1.4591.4001.118072519901339114398931787536/1.3.6.1.4.1.14519.5.2.1.4591.4001.112272323130683081580304494682 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0149 | t1Gd | F | AX_T1_POST_GD_FLAIR | 9 | 1.82811E+15 | 3/25/2003 | MR3T | IR | 2D | 2.5 | 3222.99 | 6.568 | 1238 | 127.7325 | 3 | 2.5 | 90 | 1.6875 | 70 | 320 | 224 | 0.46875\0.46875 | TCGA-06-0149/1.3.6.1.4.1.14519.5.2.1.4591.4001.118072519901339114398931787536/1.3.6.1.4.1.14519.5.2.1.4591.4001.136467249644763011967454408425 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0149 | t1 | F | AX_T1_pre_gd | 7 | 1.82811E+15 | 3/25/2003 | MR3T | IR | 2D | 5 | 3042.13 | 6.584 | 1238 | 127.7325 | 3 | 5 | 90 | 1.3409 | 36 | 320 | 224 | 0.46875\0.46875 | TCGA-06-0149/1.3.6.1.4.1.14519.5.2.1.4591.4001.118072519901339114398931787536/1.3.6.1.4.1.14519.5.2.1.4591.4001.105210083102263310494367161428 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0149 | t2 | F | AX_T2_FR-FSE_RF2_150 | 4 | 1.82811E+15 | 3/25/2003 | MR3T | SE | 2D | 5 | 3000 | 104.436 | 0 | 127.7325 | 3 | 5 | 90 | 1.0799 | 36 | 320 | 224 | 0.46875\0.46875 | TCGA-06-0149/1.3.6.1.4.1.14519.5.2.1.4591.4001.118072519901339114398931787536/1.3.6.1.4.1.14519.5.2.1.4591.4001.250187065726873055656948737218 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0154 | flair | M | 3 | 3.23894E+15 | 4/5/1996 | BAY | flair | 3 | 1000 | 155 | 2200 | 56 | 0.94\0.94 | TCGA-06-0154/1.3.6.1.4.1.14519.5.2.1.4591.4001.163246911709429038918826429573/1.3.6.1.4.1.14519.5.2.1.4591.4001.105686167309383153415308758602 | |||||||||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0154 | t1Gd | M | AXIAL_T1_PRE/POST_GD | 8 | 3.23894E+15 | 4/5/1996 | BAY | memp | 3 | 500 | 8 | 0 | 3 | 90 | 56 | 0.938\0.938 | TCGA-06-0154/1.3.6.1.4.1.14519.5.2.1.4591.4001.163246911709429038918826429573/1.3.6.1.4.1.14519.5.2.1.4591.4001.145285442414651713189537273383 | ||||||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0154 | t1 | M | AXIAL_T1_PRE/POST_GD | 6 | 3.23894E+15 | 4/5/1996 | BAY | memp | 3 | 500 | 8 | 0 | 3 | 90 | 56 | 0.938\0.938 | TCGA-06-0154/1.3.6.1.4.1.14519.5.2.1.4591.4001.163246911709429038918826429573/1.3.6.1.4.1.14519.5.2.1.4591.4001.352404364946080402874319891442 | ||||||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0154 | t2 | M | AXIAL_FSE | 2 | 3.23894E+15 | 4/5/1996 | BAY | fse | 3 | 3500 | 22 | 0 | 3 | 90 | 112 | 0.938\0.938 | TCGA-06-0154/1.3.6.1.4.1.14519.5.2.1.4591.4001.163246911709429038918826429573/1.3.6.1.4.1.14519.5.2.1.4591.4001.693437937502992071234421184617 | ||||||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0158 | flair | M | AXIAL_FLAIR | 3 | 7.69267E+15 | 9/5/1996 | BAY | flair | 3 | 10004 | 155 | 2200 | 3 | 90 | 53 | 0.938\0.938 | TCGA-06-0158/1.3.6.1.4.1.14519.5.2.1.4591.4001.222356408466975831011403030968/1.3.6.1.4.1.14519.5.2.1.4591.4001.274441692198883693078259097052 | ||||||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0158 | t1Gd | M | AXIAL_T1_POST_GD | 5 | 7.69267E+15 | 9/5/1996 | BAY | memp | 3 | 500 | 14 | 0 | 3 | 90 | 53 | 0.938\0.938 | TCGA-06-0158/1.3.6.1.4.1.14519.5.2.1.4591.4001.222356408466975831011403030968/1.3.6.1.4.1.14519.5.2.1.4591.4001.173289171872890553883773995785 | ||||||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0158 | t1 | M | AXIAL_T1_PRE_GD | 2 | 7.69267E+15 | 9/5/1996 | BAY | memp | 3 | 500 | 14 | 0 | 3 | 90 | 53 | 0.938\0.938 | TCGA-06-0158/1.3.6.1.4.1.14519.5.2.1.4591.4001.222356408466975831011403030968/1.3.6.1.4.1.14519.5.2.1.4591.4001.518026839168479419278007929122 | ||||||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0158 | t2 | M | AXIAL_FSE | 2 | 7.69267E+15 | 9/5/1996 | BAY | fse | 3 | 3500 | 22 | 0 | 3 | 90 | 102 | 0.938\0.938 | TCGA-06-0158/1.3.6.1.4.1.14519.5.2.1.4591.4001.222356408466975831011403030968/1.3.6.1.4.1.14519.5.2.1.4591.4001.139278893498918379783151781430 | ||||||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0162 | flair | F | AXIAL_FLAIR | 3 | 2.58526E+15 | 12/7/1998 | BAY | flair | 3 | 10004 | 155 | 2200 | 3 | 90 | 51 | 0.938\0.938 | TCGA-06-0162/1.3.6.1.4.1.14519.5.2.1.4591.4001.680171398593107658050490549032/1.3.6.1.4.1.14519.5.2.1.4591.4001.125093007120701827268088161895 | ||||||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0162 | t1Gd | F | AXIAL_T1_POST_GD | 7 | 2.58526E+15 | 12/7/1998 | BAY | memp | 3 | 500 | 14 | 0 | 3 | 90 | 51 | 0.938\0.938 | TCGA-06-0162/1.3.6.1.4.1.14519.5.2.1.4591.4001.680171398593107658050490549032/1.3.6.1.4.1.14519.5.2.1.4591.4001.235424499795225610529331169998 | ||||||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0162 | t1 | F | AXIAL_T1_PRE_GD | 5 | 2.58526E+15 | 12/7/1998 | BAY | memp | 3 | 500 | 14 | 0 | 3 | 90 | 51 | 0.938\0.938 | TCGA-06-0162/1.3.6.1.4.1.14519.5.2.1.4591.4001.680171398593107658050490549032/1.3.6.1.4.1.14519.5.2.1.4591.4001.198868156954238484147941810221 | ||||||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0162 | t2 | F | AXIAL__FSE | 2 | 2.58526E+15 | 12/7/1998 | BAY | fse | 3 | 3500 | 22 | 0 | 3 | 90 | 102 | 0.938\0.938 | TCGA-06-0162/1.3.6.1.4.1.14519.5.2.1.4591.4001.680171398593107658050490549032/1.3.6.1.4.1.14519.5.2.1.4591.4001.211482278670119430307242095853 | ||||||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0164 | flair | M | AXIAL_FLAIR | 3 | 1.74001E+15 | 2/17/1999 | BAY1OC0 | RM | 2D | 3 | 10004 | 155 | 63.78557 | 1.5 | 3 | 90 | 0.039235 | 55 | 256 | 192 | 0.9375000000\0.9375000000 | TCGA-06-0164/1.3.6.1.4.1.14519.5.2.1.4591.4001.278844795194298910084884570122/1.3.6.1.4.1.14519.5.2.1.4591.4001.598245930322114156315750780139 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0164 | t1Gd | M | AXIAL_T1_POST_GD | 11 | 1.74001E+15 | 2/17/1999 | BAY1OC0 | SE | 2D | 3 | 500 | 14 | 63.78555 | 1.5 | 3 | 90 | 0.041659 | 55 | 256 | 192 | 0.9375000000\0.9375000000 | TCGA-06-0164/1.3.6.1.4.1.14519.5.2.1.4591.4001.278844795194298910084884570122/1.3.6.1.4.1.14519.5.2.1.4591.4001.325638225787739653579401254779 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0164 | t1 | M | AXIAL_T1_PRE | 5 | 1.74001E+15 | 2/17/1999 | BAY1OC0 | SE | 2D | 3 | 500 | 14 | 63.78557 | 1.5 | 3 | 90 | 0.041659 | 55 | 256 | 192 | 0.9375000000\0.9375000000 | TCGA-06-0164/1.3.6.1.4.1.14519.5.2.1.4591.4001.278844795194298910084884570122/1.3.6.1.4.1.14519.5.2.1.4591.4001.248569781958585320389571424266 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0164 | t2 | M | AXIAL__FSE | 2 | 1.74001E+15 | 2/17/1999 | BAY1OC0 | SE | 2D | 3 | 3500 | 22 | 63.78558 | 1.5 | 3 | 90 | 0.05729 | 110 | 256 | 192 | 0.9375000000\0.9375000000 | TCGA-06-0164/1.3.6.1.4.1.14519.5.2.1.4591.4001.278844795194298910084884570122/1.3.6.1.4.1.14519.5.2.1.4591.4001.187337638416414033994910403506 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0166 | flair | M | AXIAL_FLAIR | 3 | 1.46824E+15 | 6/1/1999 | BAY | flair | 3 | 10004 | 155 | 2200 | 3 | 90 | 57 | 0.938\0.938 | TCGA-06-0166/1.3.6.1.4.1.14519.5.2.1.4591.4001.312918919617224104082765445485/1.3.6.1.4.1.14519.5.2.1.4591.4001.721833977766553318114128596643 | ||||||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0166 | t1Gd | M | AXIAL_T1_POST_GD | 7 | 1.46824E+15 | 6/1/1999 | BAY | memp | 3 | 500 | 14 | 0 | 3 | 90 | 57 | 0.938\0.938 | TCGA-06-0166/1.3.6.1.4.1.14519.5.2.1.4591.4001.312918919617224104082765445485/1.3.6.1.4.1.14519.5.2.1.4591.4001.337430353322187787108565325391 | ||||||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0166 | t1 | M | AXIAL_T1_PRE_GD | 5 | 1.46824E+15 | 6/1/1999 | BAY | memp | 3 | 500 | 14 | 0 | 3 | 90 | 57 | 0.938\0.938 | TCGA-06-0166/1.3.6.1.4.1.14519.5.2.1.4591.4001.312918919617224104082765445485/1.3.6.1.4.1.14519.5.2.1.4591.4001.261991934956286396033599671586 | ||||||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0166 | t2 | M | AXIAL__FSE | 2 | 1.46824E+15 | 6/1/1999 | BAY | fse | 3 | 3500 | 22 | 0 | 3 | 90 | 114 | 0.938\0.938 | TCGA-06-0166/1.3.6.1.4.1.14519.5.2.1.4591.4001.312918919617224104082765445485/1.3.6.1.4.1.14519.5.2.1.4591.4001.236385881105540723406863292408 | ||||||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0168 | flair | F | AXIAL_FLAIR | 5 | 2.8195E+15 | 8/12/2000 | BAY2BAY2 | IR | 2D | 3 | 10004 | 155 | 2200 | 6.38E+08 | 1.5 | 3 | 90 | 0.031899 | 56 | 256 | 192 | 0.937500\0.937500 | TCGA-06-0168/1.3.6.1.4.1.14519.5.2.1.4591.4001.183683152249300569710859088871/1.3.6.1.4.1.14519.5.2.1.4591.4001.247852152024838103066111644027 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0168 | t1Gd | F | AXIAL_T1_GD | 10 | 2.8195E+15 | 8/12/2000 | BAY2BAY2 | SE | 2D | 3 | 500 | 14 | 6.38E+08 | 1.5 | 3 | 90 | 0.039514 | 56 | 256 | 192 | 0.937500\0.937500 | TCGA-06-0168/1.3.6.1.4.1.14519.5.2.1.4591.4001.183683152249300569710859088871/1.3.6.1.4.1.14519.5.2.1.4591.4001.839741962266350805891879065337 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0168 | t1 | F | AXIAL_T1 | 8 | 2.8195E+15 | 8/12/2000 | BAY2BAY2 | SE | 2D | 3 | 500 | 14 | 6.38E+08 | 1.5 | 3 | 90 | 0.039514 | 56 | 256 | 192 | 0.937500\0.937500 | TCGA-06-0168/1.3.6.1.4.1.14519.5.2.1.4591.4001.183683152249300569710859088871/1.3.6.1.4.1.14519.5.2.1.4591.4001.231945653959668895422748933210 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0168 | t2 | F | AXIAL_FSE | 4 | 2.8195E+15 | 8/12/2000 | BAY2BAY2 | SE | 2D | 3 | 3500 | 20 | 6.38E+08 | 1.5 | 3 | 90 | 0.054732 | 112 | 256 | 192 | 0.937500\0.937500 | TCGA-06-0168/1.3.6.1.4.1.14519.5.2.1.4591.4001.183683152249300569710859088871/1.3.6.1.4.1.14519.5.2.1.4591.4001.285985999678514345339762054607 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0175 | flair | M | AXIAL_FLAIR | 5 | 2.68236E+15 | 11/20/2001 | MR01MROW | IR | 2D | 3 | 10004 | 149.048 | 2200 | 6.39E+08 | 1.5 | 3 | 90 | 0.0326 | 62 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0175/1.3.6.1.4.1.14519.5.2.1.4591.4001.845891727594474018825776844085/1.3.6.1.4.1.14519.5.2.1.4591.4001.332198392917983887866042069159 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0175 | t1Gd | M | AXIAL_T1_GD | 10 | 2.68236E+15 | 11/20/2001 | MR01MROW | SE | 2D | 3 | 500 | 13 | 6.39E+08 | 1.5 | 3 | 90 | 0.046179 | 62 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0175/1.3.6.1.4.1.14519.5.2.1.4591.4001.845891727594474018825776844085/1.3.6.1.4.1.14519.5.2.1.4591.4001.457501165871046087756658548895 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0175 | t1 | M | AXIAL_T1 | 8 | 2.68236E+15 | 11/20/2001 | MR01MROW | SE | 2D | 3 | 500 | 13 | 6.39E+08 | 1.5 | 3 | 90 | 0.046179 | 62 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0175/1.3.6.1.4.1.14519.5.2.1.4591.4001.845891727594474018825776844085/1.3.6.1.4.1.14519.5.2.1.4591.4001.298011777083077277088460913040 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0175 | t2 | M | AXIAL_FSE | 4 | 2.68236E+15 | 11/20/2001 | MR01MROW | SE | 2D | 3 | 3500 | 65.216 | 6.39E+08 | 1.5 | 3 | 90 | 0.1018 | 124 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0175/1.3.6.1.4.1.14519.5.2.1.4591.4001.845891727594474018825776844085/1.3.6.1.4.1.14519.5.2.1.4591.4001.248832015830985831479055831470 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0176 | flair | M | AXIAL_FLAIR | 4 | 2.61862E+15 | 4/9/2002 | BAY2OC0 | IR | 2D | 3 | 10004 | 155 | 2200 | 6.38E+08 | 1.5 | 3 | 90 | 0.033175 | 60 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0176/1.3.6.1.4.1.14519.5.2.1.4591.4001.198999303901323755527019586055/1.3.6.1.4.1.14519.5.2.1.4591.4001.107610771943798529727458745540 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0176 | t1Gd | M | AXIAL_T1_GD | 9 | 2.61862E+15 | 4/9/2002 | BAY2OC0 | SE | 2D | 3 | 600 | 14 | 6.38E+08 | 1.5 | 3 | 90 | 0.048888 | 60 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0176/1.3.6.1.4.1.14519.5.2.1.4591.4001.198999303901323755527019586055/1.3.6.1.4.1.14519.5.2.1.4591.4001.304427305594571200001961934814 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0176 | t1 | M | AXIAL_T1 | 7 | 2.61862E+15 | 4/9/2002 | BAY2OC0 | SE | 2D | 3 | 600 | 14 | 6.38E+08 | 1.5 | 3 | 90 | 0.048888 | 60 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0176/1.3.6.1.4.1.14519.5.2.1.4591.4001.198999303901323755527019586055/1.3.6.1.4.1.14519.5.2.1.4591.4001.159192685899774190444844541113 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0176 | t2 | M | AXIAL_FSE | 3 | 2.61862E+15 | 4/9/2002 | BAY2OC0 | SE | 2D | 3 | 3266.66 | 20 | 6.38E+08 | 1.5 | 3 | 90 | 0.065345 | 120 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0176/1.3.6.1.4.1.14519.5.2.1.4591.4001.198999303901323755527019586055/1.3.6.1.4.1.14519.5.2.1.4591.4001.137484989470516487260129921893 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0177 | flair | M | Ax_FLAIR_2.5mm_for_surgery | 4 | 1.44338E+15 | 5/25/2002 | MR01MROW | IR | 2D | 2.5 | 10004 | 149.544 | 2200 | 63.88592 | 1.5 | 2.5 | 90 | 0.0331 | 68 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0177/1.3.6.1.4.1.14519.5.2.1.4591.4001.145527987060041086616654852708/1.3.6.1.4.1.14519.5.2.1.4591.4001.176834268304831977485617496866 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0177 | t1Gd | M | Ax_T1_2.5mm_for_surgery | 8 | 1.44338E+15 | 5/25/2002 | MR01MROW | SE | 2D | 2.5 | 500 | 13 | 0 | 63.88592 | 1.5 | 2.5 | 90 | 0.0499 | 68 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0177/1.3.6.1.4.1.14519.5.2.1.4591.4001.145527987060041086616654852708/1.3.6.1.4.1.14519.5.2.1.4591.4001.125672926847564208162176608941 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0177 | t1 | M | AXIAL_T1 | 6 | 1.44338E+15 | 5/25/2002 | MR01MROW | SE | 2D | 5 | 500 | 14 | 0 | 63.88592 | 1.5 | 5 | 90 | 0.0499 | 34 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0177/1.3.6.1.4.1.14519.5.2.1.4591.4001.145527987060041086616654852708/1.3.6.1.4.1.14519.5.2.1.4591.4001.884924697454363427147927460748 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0177 | t2 | M | AXIAL_FSE | 3 | 1.44338E+15 | 5/25/2002 | MR01MROW | SE | 2D | 5 | 3500 | 63.168 | 0 | 63.88592 | 1.5 | 5 | 90 | 0.1135 | 68 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0177/1.3.6.1.4.1.14519.5.2.1.4591.4001.145527987060041086616654852708/1.3.6.1.4.1.14519.5.2.1.4591.4001.227594382691920181249054186980 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0179 | flair | M | AXIAL_T2_FLAIR | 7 | 2.74487E+15 | 7/27/2002 | MR3T | IR | 2D | 2.5 | 10002 | 124.9 | 2250 | 127.7324 | 3 | 2.5 | 90 | 0.6658 | 71 | 320 | 224 | 0.46875\0.46875 | TCGA-06-0179/1.3.6.1.4.1.14519.5.2.1.4591.4001.803812054574937880227208773217/1.3.6.1.4.1.14519.5.2.1.4591.4001.229925037084238292303114803384 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0179 | t1Gd | M | AX_T1_POST_GD_FLAIR | 11 | 2.74487E+15 | 7/27/2002 | MR3T | IR | 2D | 2.5 | 3047.38 | 6.956 | 1238 | 127.7324 | 3 | 2.5 | 90 | 1.484 | 71 | 320 | 224 | 0.46875\0.46875 | TCGA-06-0179/1.3.6.1.4.1.14519.5.2.1.4591.4001.803812054574937880227208773217/1.3.6.1.4.1.14519.5.2.1.4591.4001.186621475598111141744713394004 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0179 | t1 | M | AX_T1_pre_gd | 9 | 2.74487E+15 | 7/27/2002 | MR3T | IR | 2D | 5 | 3047.46 | 6.964 | 1238 | 127.7324 | 3 | 5 | 90 | 1.484 | 36 | 320 | 224 | 0.46875\0.46875 | TCGA-06-0179/1.3.6.1.4.1.14519.5.2.1.4591.4001.803812054574937880227208773217/1.3.6.1.4.1.14519.5.2.1.4591.4001.265101231179155598255544655629 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0179 | t2 | M | AX_T2_FR-FSE_RF2_150 | 5 | 2.74487E+15 | 7/27/2002 | MR3T | SE | 2D | 5 | 3000 | 104.436 | 0 | 127.7324 | 3 | 5 | 90 | 1.1837 | 36 | 320 | 224 | 0.46875\0.46875 | TCGA-06-0179/1.3.6.1.4.1.14519.5.2.1.4591.4001.803812054574937880227208773217/1.3.6.1.4.1.14519.5.2.1.4591.4001.162280835080528945073223289120 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0182 | flair | M | 76 | 83.915 | AxFLAIR-thin_for_surgery | 7 | 1.15217E+15 | 9/1/2002 | GE MEDICAL SYSTEMS | BAY2OC0 | GENESIS_SIGNA | IR | 2D | 2.5 | 10004 | 155 | 2200 | 6.38E+08 | 1.5 | 2.5 | 90 | 0.033029 | 79 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0182/1.3.6.1.4.1.14519.5.2.1.4591.4001.257293214662038104885045596990/1.3.6.1.4.1.14519.5.2.1.4591.4001.310335155097062460898875231783 |
| TCGA-GBM | TCGA-06 | TCGA-06-0182 | t1Gd | M | 76 | 83.915 | =+AXS1-thin_for_surgery | 13 | 1.15217E+15 | 9/1/2002 | GE MEDICAL SYSTEMS | BAY2OC0 | GENESIS_SIGNA | SE | 2D | 2.5 | 500 | 13 | 6.38E+08 | 1.5 | 2.5 | 90 | 0.046759 | 79 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0182/1.3.6.1.4.1.14519.5.2.1.4591.4001.257293214662038104885045596990/1.3.6.1.4.1.14519.5.2.1.4591.4001.328210509441925795754155912089 | |
| TCGA-GBM | TCGA-06 | TCGA-06-0182 | t1 | M | 76 | 83.915 | AXIAL_T1 | 9 | 1.15217E+15 | 9/1/2002 | GE MEDICAL SYSTEMS | BAY2OC0 | GENESIS_SIGNA | SE | 2D | 5 | 500 | 14 | 6.38E+08 | 1.5 | 5 | 90 | 0.040914 | 40 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0182/1.3.6.1.4.1.14519.5.2.1.4591.4001.257293214662038104885045596990/1.3.6.1.4.1.14519.5.2.1.4591.4001.149790107241376368982697513082 | |
| TCGA-GBM | TCGA-06 | TCGA-06-0182 | t2 | M | 76 | 83.915 | AXIAL_FSE | 6 | 1.15217E+15 | 9/1/2002 | GE MEDICAL SYSTEMS | BAY2OC0 | GENESIS_SIGNA | SE | 2D | 5 | 3500 | 20 | 6.38E+08 | 1.5 | 5 | 90 | 0.04048 | 80 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0182/1.3.6.1.4.1.14519.5.2.1.4591.4001.257293214662038104885045596990/1.3.6.1.4.1.14519.5.2.1.4591.4001.285828690405234989005344319198 | |
| TCGA-GBM | TCGA-06 | TCGA-06-0184 | flair | M | Ax_FLAIR_2.5mm_for_surgery | 6 | 1.89312E+15 | 7/13/2003 | MR01MROW | IR | 2D | 2.5 | 10004 | 149.544 | 2200 | 63.88571 | 1.5 | 2.5 | 90 | 0.033 | 72 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0184/1.3.6.1.4.1.14519.5.2.1.4591.4001.305033536698386255017041052545/1.3.6.1.4.1.14519.5.2.1.4591.4001.298488025067598105207946452228 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0184 | t1Gd | M | Ax_T1_2.5mm_for_surgery | 10 | 1.89312E+15 | 7/13/2003 | MR01MROW | SE | 2D | 2.5 | 516.664 | 13 | 0 | 63.88574 | 1.5 | 2.5 | 90 | 0.050901 | 72 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0184/1.3.6.1.4.1.14519.5.2.1.4591.4001.305033536698386255017041052545/1.3.6.1.4.1.14519.5.2.1.4591.4001.902471635951021018846680182923 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0184 | t1 | M | AXIAL_T1 | 8 | 1.89312E+15 | 7/13/2003 | MR01MROW | SE | 2D | 5 | 550 | 14 | 0 | 63.88574 | 1.5 | 5 | 90 | 0.047805 | 36 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0184/1.3.6.1.4.1.14519.5.2.1.4591.4001.305033536698386255017041052545/1.3.6.1.4.1.14519.5.2.1.4591.4001.421295141066363874489270570138 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0184 | t2 | M | AXIAL_FSE | 5 | 1.89312E+15 | 7/13/2003 | MR01MROW | SE | 2D | 5 | 3500 | 23.688 | 0 | 63.88571 | 1.5 | 5 | 90 | 0.1197 | 72 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0184/1.3.6.1.4.1.14519.5.2.1.4591.4001.305033536698386255017041052545/1.3.6.1.4.1.14519.5.2.1.4591.4001.784491244328130916312511268061 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0185 | flair | M | AxFLAIR-thin_for_surgery | 4 | 2.61836E+15 | 11/7/2003 | BAY2OC0 | IR | 2D | 2.5 | 10004 | 155 | 2200 | 6.38E+08 | 1.5 | 2.5 | 90 | 0.03384 | 73 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0185/1.3.6.1.4.1.14519.5.2.1.4591.4001.232938998404177432213094426945/1.3.6.1.4.1.14519.5.2.1.4591.4001.131233116352308759897551017004 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0185 | t1Gd | M | GD_AxT1-thin_for_surgery | 11 | 2.61836E+15 | 11/7/2003 | BAY2OC0 | SE | 2D | 2.5 | 500 | 13 | 6.38E+08 | 1.5 | 2.5 | 90 | 0.044912 | 71 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0185/1.3.6.1.4.1.14519.5.2.1.4591.4001.232938998404177432213094426945/1.3.6.1.4.1.14519.5.2.1.4591.4001.266324093776988037240038212588 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0185 | t1 | M | AXIAL_T1 | 7 | 2.61836E+15 | 11/7/2003 | BAY2OC0 | SE | 2D | 5 | 500 | 14 | 6.38E+08 | 1.5 | 5 | 90 | 0.038924 | 37 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0185/1.3.6.1.4.1.14519.5.2.1.4591.4001.232938998404177432213094426945/1.3.6.1.4.1.14519.5.2.1.4591.4001.222960602322356165635258148379 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0185 | t2 | M | AXIAL_FSE | 3 | 2.61836E+15 | 11/7/2003 | BAY2OC0 | SE | 2D | 5 | 3500 | 20 | 6.38E+08 | 1.5 | 5 | 90 | 0.039399 | 74 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0185/1.3.6.1.4.1.14519.5.2.1.4591.4001.232938998404177432213094426945/1.3.6.1.4.1.14519.5.2.1.4591.4001.513303216697905784166950817543 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0187 | flair | M | AxFLAIR-thin_for_surgery | 7 | 1.24186E+15 | 7/7/2004 | BAY2OC0 | IR | 2D | 2.5 | 10004 | 155 | 2200 | 6.38E+08 | 1.5 | 2.5 | 90 | 0.031943 | 71 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0187/1.3.6.1.4.1.14519.5.2.1.4591.4001.216767337030769268485462841002/1.3.6.1.4.1.14519.5.2.1.4591.4001.539190347422929234900620483857 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0187 | t1Gd | M | =+AXS1-thin_for_surgery | 12 | 1.24186E+15 | 7/7/2004 | BAY2OC0 | SE | 2D | 2.5 | 500 | 13 | 6.38E+08 | 1.5 | 2.5 | 90 | 0.048048 | 68 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0187/1.3.6.1.4.1.14519.5.2.1.4591.4001.216767337030769268485462841002/1.3.6.1.4.1.14519.5.2.1.4591.4001.222791162703643401365433542560 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0187 | t1 | M | AXIAL_T1 | 9 | 1.24186E+15 | 7/7/2004 | BAY2OC0 | SE | 2D | 5 | 500 | 14 | 6.38E+08 | 1.5 | 5 | 90 | 0.033916 | 36 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0187/1.3.6.1.4.1.14519.5.2.1.4591.4001.216767337030769268485462841002/1.3.6.1.4.1.14519.5.2.1.4591.4001.186471094046575208336353039993 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0187 | t2 | M | AXIAL_FSE | 6 | 1.24186E+15 | 7/7/2004 | BAY2OC0 | SE | 2D | 5 | 3500 | 20 | 6.38E+08 | 1.5 | 5 | 90 | 0.035234 | 72 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0187/1.3.6.1.4.1.14519.5.2.1.4591.4001.216767337030769268485462841002/1.3.6.1.4.1.14519.5.2.1.4591.4001.318345880835704470736633622516 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0188 | flair | M | AxFLAIR-thin_for_surgery | 5 | 2.79037E+15 | 7/17/2004 | BAY2OC0 | IR | 2D | 2.5 | 10004 | 155 | 2200 | 6.38E+08 | 1.5 | 2.5 | 90 | 0.03364 | 68 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0188/1.3.6.1.4.1.14519.5.2.1.4591.4001.236824294948093279948709063675/1.3.6.1.4.1.14519.5.2.1.4591.4001.239261393324265132190998071373 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0188 | t1Gd | M | =+AXS1-thin_for_surgery | 9 | 2.79037E+15 | 7/17/2004 | BAY2OC0 | SE | 2D | 2.5 | 500 | 13 | 6.38E+08 | 1.5 | 2.5 | 90 | 0.050599 | 68 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0188/1.3.6.1.4.1.14519.5.2.1.4591.4001.236824294948093279948709063675/1.3.6.1.4.1.14519.5.2.1.4591.4001.115884482625152677354342118388 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0188 | t1 | M | AXIAL_T1 | 7 | 2.79037E+15 | 7/17/2004 | BAY2OC0 | SE | 2D | 5 | 516.664 | 14 | 6.38E+08 | 1.5 | 5 | 90 | 0.048961 | 34 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0188/1.3.6.1.4.1.14519.5.2.1.4591.4001.236824294948093279948709063675/1.3.6.1.4.1.14519.5.2.1.4591.4001.245392760904119688427626639535 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0188 | t2 | M | AXIAL_FSE | 4 | 2.79037E+15 | 7/17/2004 | BAY2OC0 | SE | 2D | 5 | 3716.664 | 80 | 6.38E+08 | 1.5 | 5 | 90 | 0.065999 | 68 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0188/1.3.6.1.4.1.14519.5.2.1.4591.4001.236824294948093279948709063675/1.3.6.1.4.1.14519.5.2.1.4591.4001.285294068503087038157446001862 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0189 | flair | M | 55 | 97.522 | AXIAL_FLAIR | 5 | 2.17405E+15 | 10/2/2004 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7322 | 3 | 2.5 | 90 | 0.7975 | 74 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0189/1.3.6.1.4.1.14519.5.2.1.4591.4001.271958506783844757321502711697/1.3.6.1.4.1.14519.5.2.1.4591.4001.204121947709766236097014303992 |
| TCGA-GBM | TCGA-06 | TCGA-06-0189 | t1Gd | M | 55 | 97.522 | AX_T1_POST_GD_FLAIR | 14 | 2.17405E+15 | 10/2/2004 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 2.5 | 2720.75 | 6.356 | 1154 | 127.7322 | 3 | 2.5 | 90 | 1.9775 | 76 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0189/1.3.6.1.4.1.14519.5.2.1.4591.4001.271958506783844757321502711697/1.3.6.1.4.1.14519.5.2.1.4591.4001.617019599027525799589736647295 |
| TCGA-GBM | TCGA-06 | TCGA-06-0189 | t1 | M | 55 | 97.522 | AX_T1_pre_gd | 8 | 2.17405E+15 | 10/2/2004 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 5 | 3256.32 | 6.356 | 1238 | 127.7322 | 3 | 5 | 90 | 1.652 | 38 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0189/1.3.6.1.4.1.14519.5.2.1.4591.4001.271958506783844757321502711697/1.3.6.1.4.1.14519.5.2.1.4591.4001.322517941325558672246365758146 |
| TCGA-GBM | TCGA-06 | TCGA-06-0189 | t2 | M | 55 | 97.522 | AX_T2_FR-FSE_RF2_150 | 6 | 2.17405E+15 | 10/2/2004 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 3000 | 103.464 | 0 | 127.7322 | 3 | 5 | 90 | 1.4452 | 39 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0189/1.3.6.1.4.1.14519.5.2.1.4591.4001.271958506783844757321502711697/1.3.6.1.4.1.14519.5.2.1.4591.4001.287445780789038071301584012772 |
| TCGA-GBM | TCGA-06 | TCGA-06-0190 | flair | M | AxFLAIR-thin_for_surgery | 4 | 9.56561E+15 | 12/10/2004 | BAY2OC0 | IR | 2D | 2.5 | 10004 | 155 | 2200 | 6.39E+08 | 1.5 | 2.5 | 90 | 0.033029 | 71 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0190/1.3.6.1.4.1.14519.5.2.1.4591.4001.176500029848577499575043841856/1.3.6.1.4.1.14519.5.2.1.4591.4001.491441365887397929225136700818 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0190 | t1Gd | M | =+AXS1-thin_for_surgery | 8 | 9.56561E+15 | 12/10/2004 | BAY2OC0 | SE | 2D | 2.5 | 516.664 | 13 | 6.39E+08 | 1.5 | 2.5 | 90 | 0.050901 | 71 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0190/1.3.6.1.4.1.14519.5.2.1.4591.4001.176500029848577499575043841856/1.3.6.1.4.1.14519.5.2.1.4591.4001.671165767421867654226548857596 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0190 | t1 | M | AXIAL_T1 | 6 | 9.56561E+15 | 12/10/2004 | BAY2OC0 | SE | 2D | 5 | 550 | 14 | 6.39E+08 | 1.5 | 5 | 90 | 0.047805 | 36 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0190/1.3.6.1.4.1.14519.5.2.1.4591.4001.176500029848577499575043841856/1.3.6.1.4.1.14519.5.2.1.4591.4001.716572214843115361797157088313 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0190 | t2 | M | AXIAL_FSE | 3 | 9.56561E+15 | 12/10/2004 | BAY2OC0 | SE | 2D | 5 | 3800 | 20 | 6.39E+08 | 1.5 | 5 | 90 | 0.067108 | 72 | 256 | 192 | 0.9375\0.9375 | TCGA-06-0190/1.3.6.1.4.1.14519.5.2.1.4591.4001.176500029848577499575043841856/1.3.6.1.4.1.14519.5.2.1.4591.4001.119615138695506529759357598587 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0192 | flair | M | AXIAL_FLAIR | 6 | 2.9077E+15 | 4/8/2005 | MR3T | IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7323 | 3 | 2.5 | 90 | 0.8357 | 71 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0192/1.3.6.1.4.1.14519.5.2.1.4591.4001.678770301318444628129910220947/1.3.6.1.4.1.14519.5.2.1.4591.4001.157734237416927367774289543725 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0192 | t1Gd | M | AX_T1_POST_GD_FLAIR | 10 | 2.9077E+15 | 4/8/2005 | MR3T | IR | 2D | 2.5 | 3189.01 | 6.356 | 1238 | 127.7323 | 3 | 2.5 | 90 | 1.4927 | 71 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0192/1.3.6.1.4.1.14519.5.2.1.4591.4001.297257903497184376204809816721/1.3.6.1.4.1.14519.5.2.1.4591.4001.587950467567034197943180807635 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0192 | t1 | M | AX_T1_pre_gd | 8 | 2.9077E+15 | 4/8/2005 | MR3T | IR | 2D | 5 | 3000.01 | 6.356 | 1238 | 127.7323 | 3 | 5 | 90 | 1.1901 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0192/1.3.6.1.4.1.14519.5.2.1.4591.4001.678770301318444628129910220947/1.3.6.1.4.1.14519.5.2.1.4591.4001.242390157644773510605680848291 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0192 | t2 | M | AX_T2_FR-FSE_RF2_150 | 5 | 2.9077E+15 | 4/8/2005 | MR3T | SE | 2D | 5 | 3000 | 103.464 | 0 | 127.7323 | 3 | 5 | 90 | 1.5901 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0192/1.3.6.1.4.1.14519.5.2.1.4591.4001.678770301318444628129910220947/1.3.6.1.4.1.14519.5.2.1.4591.4001.265674194081211616900213689317 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0213 | flair | F | AXIAL_FLAIR | 3 | 2.24926E+15 | 10/23/1996 | BAY | flair | 3 | 10002 | 155 | 2200 | 3 | 90 | 55 | 0.938\0.938 | TCGA-06-0213/1.3.6.1.4.1.14519.5.2.1.4591.4001.126830789018616922158734856914/1.3.6.1.4.1.14519.5.2.1.4591.4001.868309828837037489455165428643 | ||||||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0213 | t1Gd | F | AXIAL_T1_GD | 9 | 2.24926E+15 | 10/23/1996 | BAY | memp | 3 | 500 | 14 | 0 | 3 | 90 | 56 | 0.938\0.938 | TCGA-06-0213/1.3.6.1.4.1.14519.5.2.1.4591.4001.126830789018616922158734856914/1.3.6.1.4.1.14519.5.2.1.4591.4001.712441540279598808730794710099 | ||||||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0213 | t1 | F | AXIAL_T1_PRE | 7 | 2.24926E+15 | 10/23/1996 | BAY | memp | 3 | 500 | 14 | 0 | 3 | 90 | 56 | 0.938\0.938 | TCGA-06-0213/1.3.6.1.4.1.14519.5.2.1.4591.4001.126830789018616922158734856914/1.3.6.1.4.1.14519.5.2.1.4591.4001.132760894917845514939967527728 | ||||||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0213 | t2 | F | AXIAL_FSE | 2 | 2.24926E+15 | 10/23/1996 | BAY | fse | 3 | 3500 | 88 | 0 | 3 | 90 | 112 | 0.938\0.938 | TCGA-06-0213/1.3.6.1.4.1.14519.5.2.1.4591.4001.126830789018616922158734856914/1.3.6.1.4.1.14519.5.2.1.4591.4001.201340664225508569277190185868 | ||||||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0238 | flair | M | 46 | 68.039 | AXIAL_FLAIR | 5 | 1.73589E+15 | 4/12/2005 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7323 | 3 | 2.5 | 90 | 0.5979 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0238/1.3.6.1.4.1.14519.5.2.1.4591.4001.234150403961428051973172824302/1.3.6.1.4.1.14519.5.2.1.4591.4001.157984242172762905506145896839 |
| TCGA-GBM | TCGA-06 | TCGA-06-0238 | t1Gd | M | 46 | 68.039 | AX_T1_POST_GD_FLAIR | 13 | 1.73589E+15 | 4/12/2005 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 2.5 | 3026.5 | 6.356 | 1238 | 127.7323 | 3 | 2.5 | 90 | 1.8694 | 84 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0238/1.3.6.1.4.1.14519.5.2.1.4591.4001.234150403961428051973172824302/1.3.6.1.4.1.14519.5.2.1.4591.4001.202504047648555593992532497928 |
| TCGA-GBM | TCGA-06 | TCGA-06-0238 | t1 | M | 46 | 68.039 | AX_T1_pre_gd | 11 | 1.73589E+15 | 4/12/2005 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 5 | 3026.5 | 6.356 | 1238 | 127.7323 | 3 | 5 | 90 | 1.8694 | 42 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0238/1.3.6.1.4.1.14519.5.2.1.4591.4001.234150403961428051973172824302/1.3.6.1.4.1.14519.5.2.1.4591.4001.294172607033594086316981791804 |
| TCGA-GBM | TCGA-06 | TCGA-06-0238 | t2 | M | 46 | 68.039 | AX_T2_FR-FSE_RF2_150 | 9 | 1.73589E+15 | 4/12/2005 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 3000 | 103.464 | 0 | 127.7323 | 3 | 5 | 90 | 1.0799 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0238/1.3.6.1.4.1.14519.5.2.1.4591.4001.234150403961428051973172824302/1.3.6.1.4.1.14519.5.2.1.4591.4001.910762318489313615740449757293 |
| TCGA-GBM | TCGA-06 | TCGA-06-0240 | flair | M | AxFLAIR-thin_for_surgery | 4 | 2.74124E+15 | 7/2/2005 | BAY2OC0 | IR | 2D | 2.5 | 10004 | 155 | 2200 | 6.39E+08 | 1.5 | 2.5 | 90 | 0.03262 | 77 | 256 | 192 | 1.015625\1.015625 | TCGA-06-0240/1.3.6.1.4.1.14519.5.2.1.4591.4001.104811886009138088457642404728/1.3.6.1.4.1.14519.5.2.1.4591.4001.329775737865220938751020847036 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0240 | t1Gd | M | =+AXS1-thin_for_surgery | 9 | 2.74124E+15 | 7/2/2005 | BAY2OC0 | SE | 2D | 2.5 | 500 | 13 | 6.39E+08 | 1.5 | 2.5 | 90 | 0.046179 | 77 | 256 | 192 | 1.015625\1.015625 | TCGA-06-0240/1.3.6.1.4.1.14519.5.2.1.4591.4001.104811886009138088457642404728/1.3.6.1.4.1.14519.5.2.1.4591.4001.198392408592904340848985827598 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0240 | t1 | M | AXIAL_T1 | 6 | 2.74124E+15 | 7/2/2005 | BAY2OC0 | SE | 2D | 5 | 500 | 14 | 6.39E+08 | 1.5 | 5 | 90 | 0.037521 | 39 | 256 | 192 | 1.015625\1.015625 | TCGA-06-0240/1.3.6.1.4.1.14519.5.2.1.4591.4001.104811886009138088457642404728/1.3.6.1.4.1.14519.5.2.1.4591.4001.222834836258287210760464320532 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0240 | t2 | M | AXIAL_FSE | 3 | 2.74124E+15 | 7/2/2005 | BAY2OC0 | SE | 2D | 5 | 3500 | 80 | 6.39E+08 | 1.5 | 5 | 90 | 0.039978 | 78 | 256 | 192 | 1.015625\1.015625 | TCGA-06-0240/1.3.6.1.4.1.14519.5.2.1.4591.4001.104811886009138088457642404728/1.3.6.1.4.1.14519.5.2.1.4591.4001.249974415144435604635369691012 | |||||
| TCGA-GBM | TCGA-06 | TCGA-06-0241 | flair | F | AXIAL_FLAIR | 5 | 2.14612E+15 | 8/30/2005 | MR3T | IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7322 | 3 | 2.5 | 90 | 0.6258 | 75 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0241/1.3.6.1.4.1.14519.5.2.1.4591.4001.278928118420898534392751993299/1.3.6.1.4.1.14519.5.2.1.4591.4001.129865339243072579534823799930 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0241 | t1Gd | F | AX_T1_POST_GD_FLAIR | 10 | 2.14612E+15 | 8/30/2005 | MR3T | IR | 2D | 2.5 | 3000 | 6.356 | 1238 | 127.7322 | 3 | 2.5 | 90 | 1.3217 | 75 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0241/1.3.6.1.4.1.14519.5.2.1.4591.4001.278928118420898534392751993299/1.3.6.1.4.1.14519.5.2.1.4591.4001.170539642438067923558826595256 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0241 | t1 | F | AX_T1_pre_gd | 8 | 2.14612E+15 | 8/30/2005 | MR3T | IR | 2D | 5 | 3108.73 | 6.356 | 1238 | 127.7322 | 3 | 5 | 90 | 0.9693 | 38 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0241/1.3.6.1.4.1.14519.5.2.1.4591.4001.278928118420898534392751993299/1.3.6.1.4.1.14519.5.2.1.4591.4001.316434438257717295392918423241 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0241 | t2 | F | AX_T2_FR-FSE_RF2_150 | 6 | 2.14612E+15 | 8/30/2005 | MR3T | SE | 2D | 5 | 3000 | 103.464 | 0 | 127.7322 | 3 | 5 | 90 | 0.9183 | 38 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0241/1.3.6.1.4.1.14519.5.2.1.4591.4001.278928118420898534392751993299/1.3.6.1.4.1.14519.5.2.1.4591.4001.196151036460577742361087055213 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0644 | flair | M | AXIAL_FLAIR | 5 | 3.92717E+15 | 11/28/2005 | MR3T | IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7322 | 3 | 2.5 | 90 | 0.9012 | 75 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0644/1.3.6.1.4.1.14519.5.2.1.4591.4001.146995087154371962686852678030/1.3.6.1.4.1.14519.5.2.1.4591.4001.263636323693757141556368836667 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0644 | t1Gd | M | AX_T1_POST_GD_FLAIR | 10 | 3.92717E+15 | 11/28/2005 | MR3T | IR | 2D | 2.5 | 3228.58 | 6.356 | 1238 | 127.7322 | 3 | 2.5 | 90 | 1.528 | 75 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0644/1.3.6.1.4.1.14519.5.2.1.4591.4001.146995087154371962686852678030/1.3.6.1.4.1.14519.5.2.1.4591.4001.922420967850060003702279065253 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0644 | t1 | M | AX_T1_pre_gd | 8 | 3.92717E+15 | 11/28/2005 | MR3T | IR | 2D | 5 | 3228.66 | 6.356 | 1238 | 127.7322 | 3 | 5 | 90 | 1.5279 | 38 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0644/1.3.6.1.4.1.14519.5.2.1.4591.4001.146995087154371962686852678030/1.3.6.1.4.1.14519.5.2.1.4591.4001.138727768548758840916384891966 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0644 | t2 | M | AX_T2_FR-FSE_RF2_150 | 6 | 3.92717E+15 | 11/28/2005 | MR3T | SE | 2D | 5 | 3000 | 103.464 | 0 | 127.7322 | 3 | 5 | 90 | 1.5033 | 38 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0644/1.3.6.1.4.1.14519.5.2.1.4591.4001.146995087154371962686852678030/1.3.6.1.4.1.14519.5.2.1.4591.4001.621008573828456994425541022047 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0646 | flair | M | AXIAL_FLAIR | 5 | 2.5659E+15 | 12/9/2005 | MR3T | IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7322 | 3 | 2.5 | 90 | 1.0022 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0646/1.3.6.1.4.1.14519.5.2.1.4591.4001.384328429938630524147521856216/1.3.6.1.4.1.14519.5.2.1.4591.4001.140001491379116667324415668587 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0646 | t1Gd | M | AX_T1_POST_GD_FLAIR | 10 | 2.5659E+15 | 12/9/2005 | MR3T | IR | 2D | 2.5 | 3054.22 | 6.356 | 1238 | 127.7322 | 3 | 2.5 | 90 | 1.5065 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0646/1.3.6.1.4.1.14519.5.2.1.4591.4001.384328429938630524147521856216/1.3.6.1.4.1.14519.5.2.1.4591.4001.133122288144320695822686611824 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0646 | t1 | M | AX_T1_pre_gd | 8 | 2.5659E+15 | 12/9/2005 | MR3T | IR | 2D | 5 | 3054.3 | 6.356 | 1238 | 127.7322 | 3 | 5 | 90 | 1.5064 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0646/1.3.6.1.4.1.14519.5.2.1.4591.4001.384328429938630524147521856216/1.3.6.1.4.1.14519.5.2.1.4591.4001.176490286279929598870187285927 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0646 | t2 | M | AX_T2_FR-FSE_RF2_150 | 6 | 2.5659E+15 | 12/9/2005 | MR3T | SE | 2D | 5 | 3000 | 103.464 | 0 | 127.7322 | 3 | 5 | 90 | 1.3661 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0646/1.3.6.1.4.1.14519.5.2.1.4591.4001.384328429938630524147521856216/1.3.6.1.4.1.14519.5.2.1.4591.4001.104763531222810995600952616748 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0648 | flair | M | AXIAL_FLAIR | 6 | 3.12165E+15 | 1/20/2006 | MR3T | IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7322 | 3 | 2.5 | 90 | 0.7669 | 71 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0648/1.3.6.1.4.1.14519.5.2.1.4591.4001.177101472748785866117292551419/1.3.6.1.4.1.14519.5.2.1.4591.4001.956802103164319379282258280030 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0648 | t1Gd | M | AX_T1_POST_GD_FLAIR | 11 | 3.12165E+15 | 1/20/2006 | MR3T | IR | 2D | 2.5 | 3161.55 | 6.356 | 1238 | 127.7322 | 3 | 2.5 | 90 | 1.3317 | 71 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0648/1.3.6.1.4.1.14519.5.2.1.4591.4001.177101472748785866117292551419/1.3.6.1.4.1.14519.5.2.1.4591.4001.461813783107058700607975602197 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0648 | t1 | M | AX_T1_pre_gd | 9 | 3.12165E+15 | 1/20/2006 | MR3T | IR | 2D | 5 | 3000.01 | 6.356 | 1238 | 127.7322 | 3 | 5 | 90 | 1.0526 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0648/1.3.6.1.4.1.14519.5.2.1.4591.4001.177101472748785866117292551419/1.3.6.1.4.1.14519.5.2.1.4591.4001.310583318436210971810347049929 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0648 | t2 | M | AX_T2_FR-FSE_RF2_150 | 7 | 3.12165E+15 | 1/20/2006 | MR3T | SE | 2D | 5 | 3000 | 103.464 | 0 | 127.7322 | 3 | 5 | 90 | 1.4064 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-06-0648/1.3.6.1.4.1.14519.5.2.1.4591.4001.177101472748785866117292551419/1.3.6.1.4.1.14519.5.2.1.4591.4001.560732524941534483904820998846 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-0649 | flair | F | AX._FLAIR | 501 | 2.28347E+15 | 1/26/2006 | PMSN-AA8Y7H70NX | IR | 5 | 11000 | 140 | 2800 | 42.58868 | 1 | 6 | 90 | 25 | 256 | 147 | 0.8984375\0.8984375 | TCGA-06-0649/1.3.6.1.4.1.14519.5.2.1.4591.4001.111226195519163301116460123619/1.3.6.1.4.1.14519.5.2.1.4591.4001.118521252167755472165852492837 | ||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0649 | t1Gd | F | T1_AX_POST | 801 | 2.28347E+15 | 1/26/2006 | PMSN-AA8Y7H70NX | SE | 5 | 696.6778 | 15 | 42.58868 | 1 | 6 | 62 | 26 | 256 | 205 | 0.8984375\0.8984375 | TCGA-06-0649/1.3.6.1.4.1.14519.5.2.1.4591.4001.111226195519163301116460123619/1.3.6.1.4.1.14519.5.2.1.4591.4001.111395755305019629134143006170 | |||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0649 | t1 | F | T1_AX_PRE | 701 | 2.28347E+15 | 1/26/2006 | PMSN-AA8Y7H70NX | SE | 5 | 696.6778 | 15 | 42.58868 | 1 | 6 | 62 | 26 | 256 | 205 | 0.8984375\0.8984375 | TCGA-06-0649/1.3.6.1.4.1.14519.5.2.1.4591.4001.111226195519163301116460123619/1.3.6.1.4.1.14519.5.2.1.4591.4001.253942008984398191974160718322 | |||||||
| TCGA-GBM | TCGA-06 | TCGA-06-0649 | t2 | F | T2W_TSE | 401 | 2.28347E+15 | 1/26/2006 | PMSN-AA8Y7H70NX | SE | 5 | 5780.362 | 110 | 42.58868 | 1 | 6 | 90 | 25 | 288 | 227 | 0.71875\0.71875 | TCGA-06-0649/1.3.6.1.4.1.14519.5.2.1.4591.4001.111226195519163301116460123619/1.3.6.1.4.1.14519.5.2.1.4591.4001.310081522664139753162843331456 | |||||||
| TCGA-GBM | TCGA-06 | TCGA-06-1084 | flair | M | AXIAL_FLAIR | 4 | 3.62419E+15 | 1/15/2007 | MR01MROW | IR | 2D | 5 | 8002 | 152.768 | 2000 | 63.88585 | 1.5 | 5 | 90 | 0.0378 | 34 | 256 | 192 | 0.9375\0.9375 | TCGA-06-1084/1.3.6.1.4.1.14519.5.2.1.4591.4001.205933064051641141719264624760/1.3.6.1.4.1.14519.5.2.1.4591.4001.997008093475972500685701576370 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-1084 | t1Gd | M | AXIAL_T1_GD | 8 | 3.62419E+15 | 1/15/2007 | MR01MROW | SE | 2D | 5 | 516.664 | 14 | 0 | 63.88585 | 1.5 | 5 | 90 | 0.0440378 | 34 | 256 | 192 | 0.9375\0.9375 | TCGA-06-1084/1.3.6.1.4.1.14519.5.2.1.4591.4001.205933064051641141719264624760/1.3.6.1.4.1.14519.5.2.1.4591.4001.184518206794525227519819333834 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-1084 | t1 | M | AXIAL_T1 | 6 | 3.62419E+15 | 1/15/2007 | MR01MROW | SE | 2D | 5 | 516.664 | 14 | 0 | 63.88585 | 1.5 | 5 | 90 | 0.0440378 | 34 | 256 | 192 | 0.9375\0.9375 | TCGA-06-1084/1.3.6.1.4.1.14519.5.2.1.4591.4001.205933064051641141719264624760/1.3.6.1.4.1.14519.5.2.1.4591.4001.253975845398997713297147176708 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-1084 | t2 | M | AXIAL_FSE | 3 | 3.62419E+15 | 1/15/2007 | MR01MROW | SE | 2D | 5 | 3750 | 23.688 | 0 | 63.88585 | 1.5 | 5 | 90 | 0.0966 | 68 | 256 | 192 | 0.9375\0.9375 | TCGA-06-1084/1.3.6.1.4.1.14519.5.2.1.4591.4001.205933064051641141719264624760/1.3.6.1.4.1.14519.5.2.1.4591.4001.225846560104323611908626019440 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-1802 | flair | M | Ax_T2_FLAIR | 6 | 1.85764E+15 | 4/3/2007 | MR3T | SE\IR | 2D | 5 | 9002 | 121.284 | 2250 | 127.7321 | 3 | 6 | 90 | 0.8068 | 25 | 320 | 224 | 0.4297\0.4297 | TCGA-06-1802/1.3.6.1.4.1.14519.5.2.1.4591.4001.319548581969656999665412328642/1.3.6.1.4.1.14519.5.2.1.4591.4001.303785431491112534531563759163 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-1802 | t1Gd | M | AX_T1_POST_GD_FLAIR | 8 | 1.85764E+15 | 4/3/2007 | MR3T | SE\IR | 2D | 4 | 3050.6 | 6.772 | 1238 | 127.7321 | 3 | 5 | 90 | 1.8105 | 30 | 320 | 224 | 0.4297\0.4297 | TCGA-06-1802/1.3.6.1.4.1.14519.5.2.1.4591.4001.319548581969656999665412328642/1.3.6.1.4.1.14519.5.2.1.4591.4001.215549370013921736472676782979 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-1802 | t1 | M | AX_T1_pre_GD_FLAIR | 7 | 1.85764E+15 | 4/3/2007 | MR3T | SE\IR | 2D | 4 | 3050.6 | 6.4 | 1238 | 127.7321 | 3 | 5 | 90 | 1.8105 | 30 | 320 | 224 | 0.4297\0.4297 | TCGA-06-1802/1.3.6.1.4.1.14519.5.2.1.4591.4001.319548581969656999665412328642/1.3.6.1.4.1.14519.5.2.1.4591.4001.165267478442345010741661718562 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-1802 | t2 | M | AX_T2_FR-FSE | 5 | 1.85764E+15 | 4/3/2007 | MR3T | SE | 2D | 5 | 3400 | 110.032 | 0 | 127.7321 | 3 | 7.5 | 90 | 1.2567 | 20 | 352 | 224 | 0.4297\0.4297 | TCGA-06-1802/1.3.6.1.4.1.14519.5.2.1.4591.4001.319548581969656999665412328642/1.3.6.1.4.1.14519.5.2.1.4591.4001.431304703040316559486127609898 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-2570 | flair | F | 21 | 68.492 | AXIAL_FLAIR | 7 | 1.4621E+15 | 7/26/2007 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7322 | 3 | 2.5 | 90 | 0.6048 | 67 | 320 | 224 | 0.4688\0.4688 | TCGA-06-2570/1.3.6.1.4.1.14519.5.2.1.4591.4001.763554173270318063812534542847/1.3.6.1.4.1.14519.5.2.1.4591.4001.875590044906522068491547240162 | |
| TCGA-GBM | TCGA-06 | TCGA-06-2570 | t1Gd | F | 21 | 68.492 | AX_T1_POST_GD_FLAIR | 11 | 1.4621E+15 | 7/26/2007 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 2.5 | 2970.47 | 6.356 | 1238 | 127.7322 | 3 | 2.5 | 90 | 1.6819 | 67 | 320 | 224 | 0.4688\0.4688 | TCGA-06-2570/1.3.6.1.4.1.14519.5.2.1.4591.4001.763554173270318063812534542847/1.3.6.1.4.1.14519.5.2.1.4591.4001.851364973348227212233680415818 | |
| TCGA-GBM | TCGA-06 | TCGA-06-2570 | t1 | F | 21 | 68.492 | AX_T1_pre_gd | 8 | 1.4621E+15 | 7/26/2007 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 2970.61 | 6.356 | 1238 | 127.7322 | 3 | 5 | 90 | 1.5595 | 34 | 320 | 224 | 0.4688\0.4688 | TCGA-06-2570/1.3.6.1.4.1.14519.5.2.1.4591.4001.763554173270318063812534542847/1.3.6.1.4.1.14519.5.2.1.4591.4001.334156106497189532473121151584 | |
| TCGA-GBM | TCGA-06 | TCGA-06-2570 | t2 | F | 21 | 68.492 | AX_T2_FR-FSE_RF2_150 | 6 | 1.4621E+15 | 7/26/2007 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 3000 | 103.464 | 0 | 127.7322 | 3 | 5 | 90 | 1.0316 | 34 | 320 | 224 | 0.4688\0.4688 | TCGA-06-2570/1.3.6.1.4.1.14519.5.2.1.4591.4001.763554173270318063812534542847/1.3.6.1.4.1.14519.5.2.1.4591.4001.181186298778100327141079566062 | |
| TCGA-GBM | TCGA-06 | TCGA-06-5408 | flair | F | Ax_FLAIR_2.5mm_for_surgery | 4 | 1.18161E+15 | 1/11/2008 | MR01MROW | IR | 2D | 2.5 | 10004 | 149.544 | 2200 | 63.88579 | 1.5 | 2.5 | 90 | 0.034 | 67 | 256 | 160 | 0.9375\0.9375 | TCGA-06-5408/1.3.6.1.4.1.14519.5.2.1.4591.4001.694644982891228681410637134135/1.3.6.1.4.1.14519.5.2.1.4591.4001.178021249403048866403790750450 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-5408 | t1Gd | F | Ax_T1_2.5mm_for_surgery | 10 | 1.18161E+15 | 1/11/2008 | MR01MROW | SE | 2D | 2.5 | 500 | 13 | 0 | 63.88584 | 1.5 | 2.5 | 90 | 0.0512679 | 68 | 256 | 160 | 0.9375\0.9375 | TCGA-06-5408/1.3.6.1.4.1.14519.5.2.1.4591.4001.694644982891228681410637134135/1.3.6.1.4.1.14519.5.2.1.4591.4001.255056887939741615994527174538 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-5408 | t1 | F | AXIAL_T1 | 7 | 1.18161E+15 | 1/11/2008 | MR01MROW | SE | 2D | 5 | 500 | 14 | 0 | 63.8858 | 1.5 | 5 | 90 | 0.036189 | 36 | 256 | 192 | 0.9375\0.9375 | TCGA-06-5408/1.3.6.1.4.1.14519.5.2.1.4591.4001.694644982891228681410637134135/1.3.6.1.4.1.14519.5.2.1.4591.4001.366203051096691083339875526418 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-5408 | t2 | F | AXIAL_FSE | 3 | 1.18161E+15 | 1/11/2008 | MR01MROW | SE | 2D | 5 | 3500 | 23.688 | 0 | 63.88579 | 1.5 | 5 | 90 | 0.1235 | 72 | 256 | 192 | 0.9375\0.9375 | TCGA-06-5408/1.3.6.1.4.1.14519.5.2.1.4591.4001.694644982891228681410637134135/1.3.6.1.4.1.14519.5.2.1.4591.4001.190151680889708453331941210343 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-5412 | flair | F | AXIAL_FLAIR | 10 | 2.70336E+15 | 6/3/2008 | MR3T | SE\IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7323 | 3 | 2.5 | 90 | 0.4689 | 73 | 320 | 224 | 0.4688\0.4688 | TCGA-06-5412/1.3.6.1.4.1.14519.5.2.1.4591.4001.221248717333016631625848271272/1.3.6.1.4.1.14519.5.2.1.4591.4001.232178263135282100751854200786 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-5412 | t1Gd | F | AX_T1_POST_GD_FLAIR | 14 | 2.70336E+15 | 6/3/2008 | MR3T | SE\IR | 2D | 2.5 | 2969.99 | 6.356 | 1238 | 127.7323 | 3 | 2.5 | 90 | 1.5969 | 73 | 320 | 224 | 0.4688\0.4688 | TCGA-06-5412/1.3.6.1.4.1.14519.5.2.1.4591.4001.221248717333016631625848271272/1.3.6.1.4.1.14519.5.2.1.4591.4001.164214036339812685287628785362 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-5412 | t1 | F | AX_T1_pre_gd | 11 | 2.70336E+15 | 6/3/2008 | MR3T | SE\IR | 2D | 5 | 2039.94 | 6.356 | 883 | 127.7323 | 3 | 5 | 90 | 1.1943 | 38 | 320 | 224 | 0.4688\0.4688 | TCGA-06-5412/1.3.6.1.4.1.14519.5.2.1.4591.4001.221248717333016631625848271272/1.3.6.1.4.1.14519.5.2.1.4591.4001.120669219115152282285142798139 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-5412 | t2 | F | AX_T2_FR-FSE_RF2_150 | 7 | 2.70336E+15 | 6/3/2008 | MR3T | SE | 2D | 5 | 4000 | 103.464 | 0 | 127.7323 | 3 | 5 | 90 | 0.7542 | 38 | 320 | 224 | 0.4688\0.4688 | TCGA-06-5412/1.3.6.1.4.1.14519.5.2.1.4591.4001.221248717333016631625848271272/1.3.6.1.4.1.14519.5.2.1.4591.4001.142440088173859427055176239317 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-5413 | flair | M | AXIAL_FLAIR | 6 | 7.1913E+15 | 6/17/2008 | MR3T | SE\IR | 2D | 2.5 | 8002 | 125.4 | 2250 | 127.7322 | 3 | 2.5 | 90 | 1.1589 | 75 | 320 | 224 | 0.4688\0.4688 | TCGA-06-5413/1.3.6.1.4.1.14519.5.2.1.4591.4001.397375653956580386291577018002/1.3.6.1.4.1.14519.5.2.1.4591.4001.300669427817208545563445520070 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-5413 | t1Gd | M | AX_T1_POST_GD_FLAIR | 13 | 7.1913E+15 | 6/17/2008 | MR3T | SE\IR | 2D | 2.5 | 3042.02 | 6.484 | 1238 | 127.7322 | 3 | 2.5 | 90 | 1.6205 | 75 | 320 | 224 | 0.4688\0.4688 | TCGA-06-5413/1.3.6.1.4.1.14519.5.2.1.4591.4001.397375653956580386291577018002/1.3.6.1.4.1.14519.5.2.1.4591.4001.234074548712879476847342581821 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-5413 | t1 | M | AX_T1_pre_gd | 9 | 7.1913E+15 | 6/17/2008 | MR3T | SE\IR | 2D | 5 | 2551.56 | 6.484 | 1022 | 127.7322 | 3 | 5 | 90 | 1.6746 | 38 | 320 | 224 | 0.4688\0.4688 | TCGA-06-5413/1.3.6.1.4.1.14519.5.2.1.4591.4001.397375653956580386291577018002/1.3.6.1.4.1.14519.5.2.1.4591.4001.593318939393691383906509606881 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-5413 | t2 | M | AX_T2_FR-FSE_RF2_150 | 8 | 7.1913E+15 | 6/17/2008 | MR3T | SE | 2D | 5 | 4000 | 105.192 | 0 | 127.7322 | 3 | 5 | 90 | 1.9607 | 38 | 320 | 224 | 0.4688\0.4688 | TCGA-06-5413/1.3.6.1.4.1.14519.5.2.1.4591.4001.397375653956580386291577018002/1.3.6.1.4.1.14519.5.2.1.4591.4001.248855779352869249994890192373 | ||||
| TCGA-GBM | TCGA-06 | TCGA-06-5417 | flair | F | 45 | 47.627 | AXIAL_FLAIR | 5 | 3.24633E+15 | 9/3/2008 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 2.5 | 10002 | 128.7 | 2250 | 127.7323 | 3 | 2.5 | 90 | 0.5087 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-06-5417/1.3.6.1.4.1.14519.5.2.1.4591.4001.304604545029494418165835320551/1.3.6.1.4.1.14519.5.2.1.4591.4001.290772413693145619532853352893 | |
| TCGA-GBM | TCGA-06 | TCGA-06-5417 | t1Gd | F | 45 | 47.627 | AX_T1_POST_GD_FLAIR | 13 | 3.24633E+15 | 9/3/2008 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 2.5 | 3066.62 | 6.356 | 1238 | 127.7323 | 3 | 2.5 | 90 | 1.5316 | 76 | 320 | 224 | 0.4688\0.4688 | TCGA-06-5417/1.3.6.1.4.1.14519.5.2.1.4591.4001.304604545029494418165835320551/1.3.6.1.4.1.14519.5.2.1.4591.4001.309906575449439613084040317395 | |
| TCGA-GBM | TCGA-06 | TCGA-06-5417 | t1 | F | 45 | 47.627 | AX_T1_pre_gd | 7 | 3.24633E+15 | 9/3/2008 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 2040.55 | 6.36 | 883 | 127.7323 | 3 | 5 | 90 | 1.1512 | 38 | 320 | 224 | 0.4688\0.4688 | TCGA-06-5417/1.3.6.1.4.1.14519.5.2.1.4591.4001.304604545029494418165835320551/1.3.6.1.4.1.14519.5.2.1.4591.4001.742441362139497895209446188614 | |
| TCGA-GBM | TCGA-06 | TCGA-06-5417 | t2 | F | 45 | 47.627 | AX_T2_FR-FSE_RF2_150 | 6 | 3.24633E+15 | 9/3/2008 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 4000 | 105.732 | 0 | 127.7323 | 3 | 5 | 90 | 0.7272 | 38 | 320 | 224 | 0.4688\0.4688 | TCGA-06-5417/1.3.6.1.4.1.14519.5.2.1.4591.4001.304604545029494418165835320551/1.3.6.1.4.1.14519.5.2.1.4591.4001.165675604340541610850133926501 | |
| TCGA-GBM | TCGA-06 | TCGA-06-6389 | flair | F | 49 | 68.039 | AXIAL_FLAIR | 6 | 2.44088E+15 | 4/4/2009 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7323 | 3 | 2.5 | 90 | 0.5315 | 78 | 320 | 224 | 0.4688\0.4688 | TCGA-06-6389/1.3.6.1.4.1.14519.5.2.1.4591.4001.296868957331552138094328434431/1.3.6.1.4.1.14519.5.2.1.4591.4001.161665405486670689151197029575 | |
| TCGA-GBM | TCGA-06 | TCGA-06-6389 | t1Gd | F | 49 | 68.039 | COR__T1_POST_GD_FLAIR | 12 | 2.44088E+15 | 4/4/2009 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 2907.36 | 6.356 | 1205 | 127.7323 | 3 | 5 | 90 | 1.0657 | 45 | 320 | 224 | 0.4688\0.4688 | TCGA-06-6389/1.3.6.1.4.1.14519.5.2.1.4591.4001.296868957331552138094328434431/1.3.6.1.4.1.14519.5.2.1.4591.4001.272603426520903457320862363374 | |
| TCGA-GBM | TCGA-06 | TCGA-06-6389 | t1 | F | 49 | 68.039 | AX_T1_pre_gd | 8 | 2.44088E+15 | 4/4/2009 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 2105.28 | 6.356 | 883 | 127.7323 | 3 | 5 | 90 | 1.3805 | 39 | 320 | 224 | 0.4688\0.4688 | TCGA-06-6389/1.3.6.1.4.1.14519.5.2.1.4591.4001.296868957331552138094328434431/1.3.6.1.4.1.14519.5.2.1.4591.4001.154154311262940377270912110183 | |
| TCGA-GBM | TCGA-06 | TCGA-06-6389 | t2 | F | 49 | 68.039 | AX_T2_FR-FSE_RF2_150 | 7 | 2.44088E+15 | 4/4/2009 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 4000 | 103.464 | 0 | 127.7323 | 3 | 5 | 90 | 0.8998 | 39 | 320 | 224 | 0.4688\0.4688 | TCGA-06-6389/1.3.6.1.4.1.14519.5.2.1.4591.4001.296868957331552138094328434431/1.3.6.1.4.1.14519.5.2.1.4591.4001.141696374723260511363949850190 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0350 | flair | M | 32 | 95.254 | FLAIR_AXIAL | 6 | 1.74615E+15 | 12/15/1998 | GE MEDICAL SYSTEMS | LPOCOC0 | GENESIS_SIGNA | RM | 2D | 5 | 10002 | 142.5 | 63.87656 | 1.5 | 5 | 90 | 0.027114 | 29 | 256 | 192 | 0.8593750000\0.8593750000 | TCGA-08-0350/1.3.6.1.4.1.14519.5.2.1.7695.4001.569320506186381800578120391569/1.3.6.1.4.1.14519.5.2.1.7695.4001.223790420624969264518080364078 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0350 | t1Gd | M | 32 | 95.254 | 3DSPGR_AXIAL | 14 | 1.74615E+15 | 12/15/1998 | GE MEDICAL SYSTEMS | LPOCOC0 | GENESIS_SIGNA | GR | 3D | 1.5 | 34 | 3 | 63.87656 | 1.5 | 1.5 | 35 | 0.018941 | 124 | 256 | 192 | 0.9375000000\0.9375000000 | TCGA-08-0350/1.3.6.1.4.1.14519.5.2.1.7695.4001.569320506186381800578120391569/1.3.6.1.4.1.14519.5.2.1.7695.4001.149026159959841415765042402601 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0350 | t1 | M | 32 | 95.254 | TI_SAG_!MR_BRAIN!_WHOLE_HEAD | 1 | 1.74615E+15 | 12/15/1998 | GE MEDICAL SYSTEMS | LPOCOC0 | GENESIS_SIGNA | SE | 2D | 4 | 600 | 16 | 63.87656 | 1.5 | 6 | 90 | 0.043908 | 24 | 256 | 192 | 1.1718750000\1.1718750000 | TCGA-08-0350/1.3.6.1.4.1.14519.5.2.1.7695.4001.569320506186381800578120391569/1.3.6.1.4.1.14519.5.2.1.7695.4001.187092894138919886916310352088 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0350 | t2 | M | 32 | 95.254 | 3DFSE_AXIAL | 5 | 1.74615E+15 | 12/15/1998 | GE MEDICAL SYSTEMS | LPOCOC0 | GENESIS_SIGNA | SE | 1.5 | 3000 | 104.576 | 63.87656 | 1.5 | 1.5 | 90 | 0.072917 | 114 | 256 | 192 | 1.0156250000\1.0156250000 | TCGA-08-0350/1.3.6.1.4.1.14519.5.2.1.7695.4001.569320506186381800578120391569/1.3.6.1.4.1.14519.5.2.1.7695.4001.173215282973968589273253996333 | ||
| TCGA-GBM | TCGA-08 | TCGA-08-0352 | flair | M | 79 | 90.718 | AX_FLAIR | 2 | 1.74615E+15 | 12/25/2000 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 127.5 | 2200 | 6.39E+08 | 1.5 | 5 | 90 | 0.024711 | 29 | 256 | 192 | 0.859375\0.859375 | TCGA-08-0352/1.3.6.1.4.1.14519.5.2.1.7695.4001.163028456110647174493384721797/1.3.6.1.4.1.14519.5.2.1.7695.4001.976433586898626163749090098621 |
| TCGA-GBM | TCGA-08 | TCGA-08-0352 | t1Gd | M | 79 | 90.718 | 3D_SPGR_AX | 5 | 1.74615E+15 | 12/25/2000 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 3D | 1.5 | 34 | 3 | 6.39E+08 | 1.5 | 1.5 | 35 | 0.019095 | 124 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0352/1.3.6.1.4.1.14519.5.2.1.7695.4001.163028456110647174493384721797/1.3.6.1.4.1.14519.5.2.1.7695.4001.114566536453601371704250309307 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0352 | t1 | M | 79 | 90.718 | FSPGR_SAG | 1 | 1.74615E+15 | 12/25/2000 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 2D | 4 | 100 | 2.8 | 6.39E+08 | 1.5 | 4 | 70 | 0.00068 | 44 | 256 | 192 | 0.937500\0.937500 | TCGA-08-0352/1.3.6.1.4.1.14519.5.2.1.7695.4001.163028456110647174493384721797/1.3.6.1.4.1.14519.5.2.1.7695.4001.273255727733399439043422333634 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0352 | t2 | M | 79 | 90.718 | 3D_FSE_AX | 3 | 1.74615E+15 | 12/25/2000 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | SE | 3D | 1.5 | 4000 | 104.576 | 6.39E+08 | 1.5 | 1.5 | 90 | 0.055123 | 120 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0352/1.3.6.1.4.1.14519.5.2.1.7695.4001.163028456110647174493384721797/1.3.6.1.4.1.14519.5.2.1.7695.4001.208292216616720295976805439852 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0353 | flair | M | 58 | 71.668 | AX_FLAIR | 2 | 1.74615E+15 | 3/14/2001 | GE MEDICAL SYSTEMS | GEMROW | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 133 | 2200 | 6.39E+08 | 1.5 | 5 | 90 | 0.024837 | 28 | 256 | 192 | 0.937500\0.937500 | TCGA-08-0353/1.3.6.1.4.1.14519.5.2.1.7695.4001.337574128368739942371163019020/1.3.6.1.4.1.14519.5.2.1.7695.4001.140753700428222158006731649445 |
| TCGA-GBM | TCGA-08 | TCGA-08-0353 | t1Gd | M | 58 | 71.668 | 3D_SPGR_AX | 5 | 1.74615E+15 | 3/14/2001 | GE MEDICAL SYSTEMS | GEMROW | GENESIS_SIGNA | GR | 3D | 1.5 | 34 | 3 | 6.39E+08 | 1.5 | 1.5 | 35 | 0.019829 | 124 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0353/1.3.6.1.4.1.14519.5.2.1.7695.4001.337574128368739942371163019020/1.3.6.1.4.1.14519.5.2.1.7695.4001.328234903870025738854180702672 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0353 | t1 | M | 58 | 71.668 | FSPGR_SAG | 1 | 1.74615E+15 | 3/14/2001 | GE MEDICAL SYSTEMS | GEMROW | GENESIS_SIGNA | GR | 2D | 4 | 100 | 2.1 | 6.39E+08 | 1.5 | 4 | 70 | 0.002756 | 44 | 256 | 192 | 0.937500\0.937500 | TCGA-08-0353/1.3.6.1.4.1.14519.5.2.1.7695.4001.337574128368739942371163019020/1.3.6.1.4.1.14519.5.2.1.7695.4001.307993577811064417415017105143 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0353 | t2 | M | 58 | 71.668 | 3D_FSE_AX | 4 | 1.74615E+15 | 3/14/2001 | GE MEDICAL SYSTEMS | GEMROW | GENESIS_SIGNA | SE | 3D | 1.5 | 2500 | 104.64 | 6.39E+08 | 1.5 | 1.5 | 90 | 0.091613 | 114 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0353/1.3.6.1.4.1.14519.5.2.1.7695.4001.337574128368739942371163019020/1.3.6.1.4.1.14519.5.2.1.7695.4001.196310385969371473051006799315 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0354 | flair | F | 52 | 84.822 | AX_FLAIR | 3 | 1.74615E+15 | 5/8/2001 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 142.5 | 2200 | 6.39E+08 | 1.5 | 5 | 90 | 0.024176 | 28 | 256 | 192 | 0.859375\0.859375 | TCGA-08-0354/1.3.6.1.4.1.14519.5.2.1.7695.4001.193272565505328856496292965242/1.3.6.1.4.1.14519.5.2.1.7695.4001.323527541860368517646892071129 |
| TCGA-GBM | TCGA-08 | TCGA-08-0354 | t1 | F | 52 | 84.822 | FSPGR_SAG | 2 | 1.74615E+15 | 5/8/2001 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 2D | 4 | 100 | 2.8 | 6.39E+08 | 1.5 | 4 | 70 | 0.000687 | 44 | 256 | 192 | 0.937500\0.937500 | TCGA-08-0354/1.3.6.1.4.1.14519.5.2.1.7695.4001.193272565505328856496292965242/1.3.6.1.4.1.14519.5.2.1.7695.4001.118503307861787885716018452534 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0354 | t2 | F | 52 | 84.822 | 3D_FSE_AX | 7 | 1.74615E+15 | 5/8/2001 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | SE | 3D | 1.5 | 4000 | 104.016 | 6.39E+08 | 1.5 | 1.5 | 90 | 0.06129 | 126 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0354/1.3.6.1.4.1.14519.5.2.1.7695.4001.193272565505328856496292965242/1.3.6.1.4.1.14519.5.2.1.7695.4001.230224561061739836820209141244 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0355 | flair | F | 30 | 61.235 | AX_FLAIR | 4 | 1.74615E+15 | 6/6/2001 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | IR | 2D | 5 | 8802 | 133 | 2200 | 6.39E+08 | 1.5 | 5 | 90 | 0.033078 | 31 | 256 | 192 | 0.859375\0.859375 | TCGA-08-0355/1.3.6.1.4.1.14519.5.2.1.7695.4001.218361089000532962354851713589/1.3.6.1.4.1.14519.5.2.1.7695.4001.134415699017245527869615131670 |
| TCGA-GBM | TCGA-08 | TCGA-08-0355 | t1Gd | F | 30 | 61.235 | 3D_SPGR_AX | 6 | 1.74615E+15 | 6/6/2001 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 3D | 1.5 | 34 | 3 | 6.39E+08 | 1.5 | 1.5 | 35 | 0.020334 | 108 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0355/1.3.6.1.4.1.14519.5.2.1.7695.4001.218361089000532962354851713589/1.3.6.1.4.1.14519.5.2.1.7695.4001.337847097697226473402589529720 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0355 | t1 | F | 30 | 61.235 | FSPGR_SAG | 2 | 1.74615E+15 | 6/6/2001 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 2D | 4 | 100 | 2.8 | 6.39E+08 | 1.5 | 4 | 70 | 0.000724 | 44 | 256 | 192 | 0.937500\0.937500 | TCGA-08-0355/1.3.6.1.4.1.14519.5.2.1.7695.4001.218361089000532962354851713589/1.3.6.1.4.1.14519.5.2.1.7695.4001.332032456864712781947929643599 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0355 | t2 | F | 30 | 61.235 | 3D_FSE_AX | 7 | 1.74615E+15 | 6/6/2001 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | SE | 3D | 1.5 | 2600 | 104.576 | 6.39E+08 | 1.5 | 1.5 | 90 | 0.081299 | 108 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0355/1.3.6.1.4.1.14519.5.2.1.7695.4001.218361089000532962354851713589/1.3.6.1.4.1.14519.5.2.1.7695.4001.339057153668167454576759289870 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0356 | flair | F | 59 | 104.326 | AX_FLAIR | 3 | 1.74615E+15 | 3/26/2002 | GE MEDICAL SYSTEMS | GEMROW | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 133 | 2200 | 6.39E+08 | 1.5 | 5 | 90 | 0.02673 | 32 | 256 | 160 | 0.859375\0.859375 | TCGA-08-0356/1.3.6.1.4.1.14519.5.2.1.7695.4001.232438853782668841142871631923/1.3.6.1.4.1.14519.5.2.1.7695.4001.145267262126068158967843466003 |
| TCGA-GBM | TCGA-08 | TCGA-08-0356 | t1Gd | F | 59 | 104.326 | AX_3D_SPGR_GAD | 7 | 1.74615E+15 | 3/26/2002 | GE MEDICAL SYSTEMS | GEMROW | GENESIS_SIGNA | GR | 3D | 1.5 | 34 | 3 | 6.39E+08 | 1.5 | 1.5 | 35 | 0.018673 | 124 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0356/1.3.6.1.4.1.14519.5.2.1.7695.4001.232438853782668841142871631923/1.3.6.1.4.1.14519.5.2.1.7695.4001.242840321773074493339785317095 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0356 | t1 | F | 59 | 104.326 | SAG_FSPGR | 2 | 1.74615E+15 | 3/26/2002 | GE MEDICAL SYSTEMS | GEMROW | GENESIS_SIGNA | GR | 2D | 4 | 100 | 2.1 | 6.39E+08 | 1.5 | 4 | 70 | 0.002596 | 44 | 256 | 192 | 0.937500\0.937500 | TCGA-08-0356/1.3.6.1.4.1.14519.5.2.1.7695.4001.232438853782668841142871631923/1.3.6.1.4.1.14519.5.2.1.7695.4001.185236563426956780421070538793 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0356 | t2 | F | 59 | 104.326 | AX_3D_FSE | 4 | 1.74615E+15 | 3/26/2002 | GE MEDICAL SYSTEMS | GEMROW | GENESIS_SIGNA | SE | 3D | 1.5 | 3000 | 104.64 | 6.39E+08 | 1.5 | 1.5 | 90 | 0.071883 | 120 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0356/1.3.6.1.4.1.14519.5.2.1.7695.4001.232438853782668841142871631923/1.3.6.1.4.1.14519.5.2.1.7695.4001.338312619633234820114774625995 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0357 | flair | M | 49 | 81.647 | AX_FLAIR | 2 | 1.74615E+15 | 5/28/2002 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 127.5 | 2200 | 6.39E+08 | 1.5 | 5 | 90 | 0.025131 | 32 | 256 | 160 | 0.937500\0.937500 | TCGA-08-0357/1.3.6.1.4.1.14519.5.2.1.7695.4001.119967927917425608159550386921/1.3.6.1.4.1.14519.5.2.1.7695.4001.209481123456866732766059421408 |
| TCGA-GBM | TCGA-08 | TCGA-08-0357 | t1Gd | M | 49 | 81.647 | 3D_SPGR_AX | 6 | 1.74615E+15 | 5/28/2002 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 3D | 1.5 | 34 | 3 | 6.39E+08 | 1.5 | 1.5 | 35 | 0.01942 | 124 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0357/1.3.6.1.4.1.14519.5.2.1.7695.4001.119967927917425608159550386921/1.3.6.1.4.1.14519.5.2.1.7695.4001.262710894696531450868984668624 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0357 | t1 | M | 49 | 81.647 | FSPGR_SAG | 1 | 1.74615E+15 | 5/28/2002 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 2D | 4 | 100 | 2.8 | 6.39E+08 | 1.5 | 4 | 70 | 0.000691 | 44 | 256 | 160 | 0.937500\0.937500 | TCGA-08-0357/1.3.6.1.4.1.14519.5.2.1.7695.4001.119967927917425608159550386921/1.3.6.1.4.1.14519.5.2.1.7695.4001.659866133278407752982263311318 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0357 | t2 | M | 49 | 81.647 | 3D_FSE_AX | 3 | 1.74615E+15 | 5/28/2002 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | SE | 3D | 1.5 | 3000 | 104.576 | 6.39E+08 | 1.5 | 1.5 | 90 | 0.074759 | 120 | 256 | 160 | 1.015625\1.015625 | TCGA-08-0357/1.3.6.1.4.1.14519.5.2.1.7695.4001.119967927917425608159550386921/1.3.6.1.4.1.14519.5.2.1.7695.4001.205237217403372024720024068239 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0358 | flair | M | 50 | 83.915 | COR_FLAIR | 5 | 1.74615E+15 | 11/26/2002 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 127.5 | 2200 | 6.39E+08 | 1.5 | 5 | 90 | 0.025453 | 32 | 256 | 192 | 0.859375\0.859375 | TCGA-08-0358/1.3.6.1.4.1.14519.5.2.1.7695.4001.178258647502369298089351214604/1.3.6.1.4.1.14519.5.2.1.7695.4001.182758059237808096357467156444 |
| TCGA-GBM | TCGA-08 | TCGA-08-0358 | t1Gd | M | 50 | 83.915 | 3D_SPGR_AX | 7 | 1.74615E+15 | 11/26/2002 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 3D | 3 | 23 | 8 | 6.39E+08 | 1.5 | 3 | 35 | 0.005102 | 60 | 256 | 160 | 0.937500\0.937500 | TCGA-08-0358/1.3.6.1.4.1.14519.5.2.1.7695.4001.178258647502369298089351214604/1.3.6.1.4.1.14519.5.2.1.7695.4001.216730440392063319195987347730 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0358 | t1 | M | 50 | 83.915 | AX_T1 | 2 | 1.74615E+15 | 11/26/2002 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | SE | 2D | 5 | 600 | 9 | 6.39E+08 | 1.5 | 6 | 90 | 0.058411 | 24 | 256 | 192 | 0.859384\0.859375 | TCGA-08-0358/1.3.6.1.4.1.14519.5.2.1.7695.4001.178258647502369298089351214604/1.3.6.1.4.1.14519.5.2.1.7695.4001.179655607006515166822372048602 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0358 | t2 | M | 50 | 83.915 | 3D_FSE_AX | 3 | 1.74615E+15 | 11/26/2002 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | SE | 3D | 1.5 | 4000 | 104.576 | 6.39E+08 | 1.5 | 1.5 | 90 | 0.055123 | 114 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0358/1.3.6.1.4.1.14519.5.2.1.7695.4001.211723469240412331121762942910/1.3.6.1.4.1.14519.5.2.1.7695.4001.302868369039586931692020679777 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0359 | flair | F | 59 | 83.915 | AX_FLAIR_gad | 7 | 1.74615E+15 | 1/19/2003 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 142.5 | 2200 | 6.39E+08 | 1.5 | 5 | 90 | 0.024218 | 27 | 256 | 192 | 0.859375\0.859375 | TCGA-08-0359/1.3.6.1.4.1.14519.5.2.1.7695.4001.291982097074076414376657462951/1.3.6.1.4.1.14519.5.2.1.7695.4001.720373715113146700092274252984 |
| TCGA-GBM | TCGA-08 | TCGA-08-0359 | t1Gd | F | 59 | 83.915 | 3D_SPGR_AX | 6 | 1.74615E+15 | 1/19/2003 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 3D | 1.5 | 34 | 3 | 6.39E+08 | 1.5 | 1.5 | 35 | 0.011393 | 102 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0359/1.3.6.1.4.1.14519.5.2.1.7695.4001.291982097074076414376657462951/1.3.6.1.4.1.14519.5.2.1.7695.4001.441692279775841498698035053980 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0359 | t1 | F | 59 | 83.915 | FSPGR_SAG | 1 | 1.74615E+15 | 1/19/2003 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 2D | 4 | 100 | 2.8 | 6.39E+08 | 1.5 | 4 | 70 | 0.000688 | 44 | 256 | 192 | 0.937500\0.937500 | TCGA-08-0359/1.3.6.1.4.1.14519.5.2.1.7695.4001.291982097074076414376657462951/1.3.6.1.4.1.14519.5.2.1.7695.4001.303658910963991106533940013361 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0359 | t2 | F | 59 | 83.915 | 3D_FSE_AX | 2 | 1.74615E+15 | 1/19/2003 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | SE | 3D | 1.5 | 4000 | 104.576 | 6.39E+08 | 1.5 | 1.5 | 90 | 0.050233 | 102 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0359/1.3.6.1.4.1.14519.5.2.1.7695.4001.291982097074076414376657462951/1.3.6.1.4.1.14519.5.2.1.7695.4001.147430811790810817730832339214 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0360 | flair | M | 76 | 72.575 | AX_FLAIR_gad | 7 | 1.74615E+15 | 2/23/2003 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 127.5 | 2200 | 6.39E+08 | 1.5 | 5 | 90 | 0.025609 | 32 | 256 | 192 | 0.859375\0.859375 | TCGA-08-0360/1.3.6.1.4.1.14519.5.2.1.7695.4001.775866763600199916309126131064/1.3.6.1.4.1.14519.5.2.1.7695.4001.116744760614299221617250951944 |
| TCGA-GBM | TCGA-08 | TCGA-08-0360 | t1Gd | M | 76 | 72.575 | 3D_SPGR_AX | 6 | 1.74615E+15 | 2/23/2003 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 3D | 1.5 | 34 | 3 | 6.39E+08 | 1.5 | 1.5 | 35 | 0.011661 | 124 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0360/1.3.6.1.4.1.14519.5.2.1.7695.4001.775866763600199916309126131064/1.3.6.1.4.1.14519.5.2.1.7695.4001.318817306516501633455833057389 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0360 | t1 | M | 76 | 72.575 | FSPGR_SAG | 1 | 1.74615E+15 | 2/23/2003 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 2D | 4 | 100 | 2.8 | 6.39E+08 | 1.5 | 4 | 70 | 0.000704 | 44 | 256 | 192 | 0.937500\0.937500 | TCGA-08-0360/1.3.6.1.4.1.14519.5.2.1.7695.4001.775866763600199916309126131064/1.3.6.1.4.1.14519.5.2.1.7695.4001.103650870268111201936685511415 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0360 | t2 | M | 76 | 72.575 | 3D_FSE_AX | 2 | 1.74615E+15 | 2/23/2003 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | SE | 3D | 1.5 | 4000 | 104.576 | 6.39E+08 | 1.5 | 1.5 | 90 | 0.062839 | 126 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0360/1.3.6.1.4.1.14519.5.2.1.7695.4001.775866763600199916309126131064/1.3.6.1.4.1.14519.5.2.1.7695.4001.245474303741731760955757807517 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0385 | flair | M | 71 | 70.307 | AX_FLAIR | 2 | 1.74615E+15 | 8/27/2001 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 142.5 | 2200 | 6.39E+08 | 1.5 | 5 | 90 | 0.024913 | 28 | 256 | 192 | 0.859375\0.859375 | TCGA-08-0385/1.3.6.1.4.1.14519.5.2.1.7695.4001.222434989025722161720779872375/1.3.6.1.4.1.14519.5.2.1.7695.4001.470684108406413400813519745909 |
| TCGA-GBM | TCGA-08 | TCGA-08-0385 | t1Gd | M | 71 | 70.307 | 3D_SPGR_AX | 7 | 1.74615E+15 | 8/27/2001 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 3D | 1.5 | 34 | 6 | 6.39E+08 | 1.5 | 1.5 | 35 | 0.01989 | 124 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0385/1.3.6.1.4.1.14519.5.2.1.7695.4001.222434989025722161720779872375/1.3.6.1.4.1.14519.5.2.1.7695.4001.721759255675381060893415325949 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0385 | t1 | M | 71 | 70.307 | FSPGR_SAG | 1 | 1.74615E+15 | 8/27/2001 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 2D | 4 | 100 | 2.8 | 6.39E+08 | 1.5 | 4 | 70 | 0.000708 | 44 | 256 | 192 | 0.937500\0.937500 | TCGA-08-0385/1.3.6.1.4.1.14519.5.2.1.7695.4001.222434989025722161720779872375/1.3.6.1.4.1.14519.5.2.1.7695.4001.204026741851778126255510768761 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0385 | t2 | M | 71 | 70.307 | 3D_FSE_AX | 3 | 1.74615E+15 | 8/27/2001 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | SE | 3D | 1.5 | 2500 | 104.576 | 6.39E+08 | 1.5 | 1.5 | 90 | 0.091895 | 120 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0385/1.3.6.1.4.1.14519.5.2.1.7695.4001.222434989025722161720779872375/1.3.6.1.4.1.14519.5.2.1.7695.4001.280478740522485950254533656856 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0389 | flair | M | 59 | 70.307 | Ax_T2_FLAIR | 5 | 1.74615E+15 | 7/25/2003 | GE MEDICAL SYSTEMS | CBMR1 | SIGNA EXCITE | IR | 2D | 5 | 10002 | 120.3 | 2200 | 63.82921 | 1.5 | 5 | 90 | 0.4477 | 32 | 256 | 224 | 0.429688\0.429688 | TCGA-08-0389/1.3.6.1.4.1.14519.5.2.1.7695.4001.155576204150333185525274253431/1.3.6.1.4.1.14519.5.2.1.7695.4001.231537080539550629623423633749 |
| TCGA-GBM | TCGA-08 | TCGA-08-0389 | t1Gd | M | 59 | 70.307 | 3d_T1_FSPGR__CONT | 10 | 1.74615E+15 | 7/25/2003 | GE MEDICAL SYSTEMS | CBMR1 | SIGNA EXCITE | GR | 3D | 1.5 | 34 | 3 | 0 | 63.82923 | 1.5 | 1.5 | 70 | 0.350961 | 124 | 256 | 192 | 1.01562\1.01562 | TCGA-08-0389/1.3.6.1.4.1.14519.5.2.1.7695.4001.155576204150333185525274253431/1.3.6.1.4.1.14519.5.2.1.7695.4001.332152476495056790815972732567 |
| TCGA-GBM | TCGA-08 | TCGA-08-0389 | t1 | M | 59 | 70.307 | FSPGR_SAG | 2 | 1.74615E+15 | 7/25/2003 | GE MEDICAL SYSTEMS | CBMR1 | SIGNA EXCITE | GR | 2D | 4 | 100 | 2.36 | 0 | 63.8292 | 1.5 | 4 | 70 | 0.192506 | 44 | 256 | 192 | 1.01562\1.01562 | TCGA-08-0389/1.3.6.1.4.1.14519.5.2.1.7695.4001.155576204150333185525274253431/1.3.6.1.4.1.14519.5.2.1.7695.4001.265467282136232588736256233486 |
| TCGA-GBM | TCGA-08 | TCGA-08-0389 | t2 | M | 59 | 70.307 | 3D_FSE_AXIAL | 8 | 1.74615E+15 | 7/25/2003 | GE MEDICAL SYSTEMS | CBMR1 | SIGNA EXCITE | SE | 3D | 1.5 | 3000 | 104.256 | 0 | 63.82923 | 1.5 | 1.5 | 90 | 1.012569 | 114 | 256 | 160 | 1.01562\1.01562 | TCGA-08-0389/1.3.6.1.4.1.14519.5.2.1.7695.4001.155576204150333185525274253431/1.3.6.1.4.1.14519.5.2.1.7695.4001.898120371300749852808668825214 |
| TCGA-GBM | TCGA-08 | TCGA-08-0390 | flair | M | 69 | 74.843 | AX_FLAIR | 5 | 1.74615E+15 | 8/5/2003 | GE MEDICAL SYSTEMS | mrc2 | SIGNA EXCITE | IR | 2D | 5 | 9302 | 140.116 | 2100 | 127.7146 | 3 | 5 | 90 | 0.8505 | 29 | 320 | 192 | 0.429688\0.429685 | TCGA-08-0390/1.3.6.1.4.1.14519.5.2.1.7695.4001.162486595336728686386525070636/1.3.6.1.4.1.14519.5.2.1.7695.4001.215135936895486917436141586502 |
| TCGA-GBM | TCGA-08 | TCGA-08-0390 | t1Gd | M | 69 | 74.843 | AX_SPGR_3D | 8 | 1.74615E+15 | 8/5/2003 | GE MEDICAL SYSTEMS | mrc2 | SIGNA EXCITE | GR | 3D | 1.5 | 26 | 7 | 0 | 127.7147 | 3 | 1.5 | 35 | 0.805193 | 128 | 256 | 192 | 1.01562\1.01562 | TCGA-08-0390/1.3.6.1.4.1.14519.5.2.1.7695.4001.162486595336728686386525070636/1.3.6.1.4.1.14519.5.2.1.7695.4001.339923650761117556933165423898 |
| TCGA-GBM | TCGA-08 | TCGA-08-0390 | t1 | M | 69 | 74.843 | FSPGR_SAG | 2 | 1.74615E+15 | 8/5/2003 | GE MEDICAL SYSTEMS | mrc2 | SIGNA EXCITE | GR | 2D | 4 | 425 | 3.292 | 0 | 127.7147 | 3 | 4 | 90 | 2.345111 | 44 | 256 | 192 | 0.9375\0.9375 | TCGA-08-0390/1.3.6.1.4.1.14519.5.2.1.7695.4001.162486595336728686386525070636/1.3.6.1.4.1.14519.5.2.1.7695.4001.128426618960163084409336878674 |
| TCGA-GBM | TCGA-08 | TCGA-08-0390 | t2 | M | 69 | 74.843 | 3D_FSE_AX_20_slabs | 6 | 1.74615E+15 | 8/5/2003 | GE MEDICAL SYSTEMS | mrc2 | SIGNA EXCITE | SE | 3D | 1.5 | 4300 | 99.288 | 0 | 127.7147 | 3 | 1.5 | 90 | 2.917361 | 120 | 256 | 256 | 1.01562\1.01562 | TCGA-08-0390/1.3.6.1.4.1.14519.5.2.1.7695.4001.162486595336728686386525070636/1.3.6.1.4.1.14519.5.2.1.7695.4001.581616982218423289196570600960 |
| TCGA-GBM | TCGA-08 | TCGA-08-0392 | flair | M | 60 | 81.647 | AX_FLAIR | 4 | 1.74615E+15 | 11/3/2003 | GE MEDICAL SYSTEMS | EXTERNAL_MR | GENESIS_SIGNA | IR | 2D | 5 | 8802 | 127.5 | 2200 | 6.39E+08 | 1.5 | 5 | 90 | 0.028558 | 30 | 256 | 160 | 0.859387\0.859375 | TCGA-08-0392/1.3.6.1.4.1.14519.5.2.1.7695.4001.227489248220590980899973679112/1.3.6.1.4.1.14519.5.2.1.7695.4001.305330809372782012100149087676 |
| TCGA-GBM | TCGA-08 | TCGA-08-0392 | t1Gd | M | 60 | 81.647 | 3D_SPGR_AX | 7 | 1.74615E+15 | 11/3/2003 | GE MEDICAL SYSTEMS | EXTERNAL_MR | GENESIS_SIGNA | GR | 3D | 1.5 | 34 | 3 | 6.39E+08 | 1.5 | 1.5 | 35 | 0.01942 | 124 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0392/1.3.6.1.4.1.14519.5.2.1.7695.4001.227489248220590980899973679112/1.3.6.1.4.1.14519.5.2.1.7695.4001.325303380020247035105126562668 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0392 | t1 | M | 60 | 81.647 | FSPGR_SAG | 2 | 1.74615E+15 | 11/3/2003 | GE MEDICAL SYSTEMS | EXTERNAL_MR | GENESIS_SIGNA | GR | 2D | 4 | 100 | 2.8 | 6.39E+08 | 1.5 | 4 | 70 | 0.000691 | 44 | 256 | 192 | 0.937500\0.937500 | TCGA-08-0392/1.3.6.1.4.1.14519.5.2.1.7695.4001.227489248220590980899973679112/1.3.6.1.4.1.14519.5.2.1.7695.4001.159334401813324113568299278427 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0392 | t2 | M | 60 | 81.647 | 3D_FSE_AX | 5 | 1.74615E+15 | 11/3/2003 | GE MEDICAL SYSTEMS | EXTERNAL_MR | GENESIS_SIGNA | SE | 3D | 1.5 | 4000 | 104.576 | 6.39E+08 | 1.5 | 1.5 | 90 | 0.05606 | 120 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0392/1.3.6.1.4.1.14519.5.2.1.7695.4001.227489248220590980899973679112/1.3.6.1.4.1.14519.5.2.1.7695.4001.285943743580803495108812000858 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0509 | flair | M | 63 | 77.111 | FLAIR_AXIAL | 4 | 1.74615E+15 | 11/13/1997 | GE MEDICAL SYSTEMS | LPOCOC0 | GENESIS_SIGNA | RM | 2D | 5 | 10002 | 152 | 63.87615 | 1.5 | 5 | 90 | 0.028047 | 29 | 256 | 192 | 0.8593851328\0.8593750000 | TCGA-08-0509/1.3.6.1.4.1.14519.5.2.1.7695.4001.292568156105181015740272479103/1.3.6.1.4.1.14519.5.2.1.7695.4001.408596602542998666261259876805 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0509 | t1Gd | M | 63 | 77.111 | 3DSPGR_AXIAL | 2 | 1.74615E+15 | 11/13/1997 | GE MEDICAL SYSTEMS | LPOCOC0 | GENESIS_SIGNA | GR | 3D | 1.5 | 34 | 6 | 63.87616 | 1.5 | 1.5 | 35 | 0.019593 | 124 | 256 | 192 | 1.0156250000\1.0156250000 | TCGA-08-0509/1.3.6.1.4.1.14519.5.2.1.7695.4001.292568156105181015740272479103/1.3.6.1.4.1.14519.5.2.1.7695.4001.258664400961230551340675293298 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0509 | t1 | M | 63 | 77.111 | FMPSPGR_SAG | 1 | 1.74615E+15 | 11/13/1997 | GE MEDICAL SYSTEMS | LPOCOC0 | GENESIS_SIGNA | GR | 2D | 4 | 100 | 2.1 | 63.87618 | 1.5 | 4 | 70 | 0.017739 | 46 | 256 | 192 | 0.9375000000\0.9375000000 | TCGA-08-0509/1.3.6.1.4.1.14519.5.2.1.7695.4001.292568156105181015740272479103/1.3.6.1.4.1.14519.5.2.1.7695.4001.224301481773938865793646319386 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0509 | t2 | M | 63 | 77.111 | 3DFSE_AXIAL | 3 | 1.74615E+15 | 11/13/1997 | GE MEDICAL SYSTEMS | LPOCOC0 | GENESIS_SIGNA | SE | 1.5 | 2500 | 104.576 | 63.87616 | 1.5 | 1.5 | 90 | 0.090521 | 120 | 256 | 192 | 1.0156250000\1.0156250000 | TCGA-08-0509/1.3.6.1.4.1.14519.5.2.1.7695.4001.292568156105181015740272479103/1.3.6.1.4.1.14519.5.2.1.7695.4001.178602050598278251326371799672 | ||
| TCGA-GBM | TCGA-08 | TCGA-08-0510 | flair | M | 75 | 92.986 | AX_FLAIR | 4 | 1.74615E+15 | 1/26/1998 | GE MEDICAL SYSTEMS | GEMROW | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 142.5 | 2200 | 6.39E+08 | 1.5 | 5 | 90 | 0.023893 | 28 | 256 | 192 | 0.859375\0.859375 | TCGA-08-0510/1.3.6.1.4.1.14519.5.2.1.7695.4001.908705038093871340852313648625/1.3.6.1.4.1.14519.5.2.1.7695.4001.680572427201908205055469939074 |
| TCGA-GBM | TCGA-08 | TCGA-08-0510 | t1Gd | M | 75 | 92.986 | 3D_SPGR_AX | 2 | 1.74615E+15 | 1/26/1998 | GE MEDICAL SYSTEMS | GEMROW | GENESIS_SIGNA | GR | 3D | 1.5 | 34 | 3 | 6.39E+08 | 1.5 | 1.5 | 35 | 0.01902 | 124 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0510/1.3.6.1.4.1.14519.5.2.1.7695.4001.908705038093871340852313648625/1.3.6.1.4.1.14519.5.2.1.7695.4001.180669314170322596932458983139 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0510 | t1 | M | 75 | 92.986 | FSPGR_SAG | 1 | 1.74615E+15 | 1/26/1998 | GE MEDICAL SYSTEMS | GEMROW | GENESIS_SIGNA | GR | 2D | 4 | 100 | 2.1 | 6.39E+08 | 1.5 | 4 | 70 | 0.001722 | 44 | 256 | 192 | 0.937500\0.937500 | TCGA-08-0510/1.3.6.1.4.1.14519.5.2.1.7695.4001.908705038093871340852313648625/1.3.6.1.4.1.14519.5.2.1.7695.4001.289052473786821559464871427227 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0510 | t2 | M | 75 | 92.986 | 3D_FSE_AX | 3 | 1.74615E+15 | 1/26/1998 | GE MEDICAL SYSTEMS | GEMROW | GENESIS_SIGNA | SE | 3D | 1.5 | 4000 | 104.016 | 6.39E+08 | 1.5 | 1.5 | 90 | 0.054905 | 120 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0510/1.3.6.1.4.1.14519.5.2.1.7695.4001.908705038093871340852313648625/1.3.6.1.4.1.14519.5.2.1.7695.4001.208630687586140812333680115393 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0512 | flair | M | 48 | 72.575 | FLAIR_AXIAL | 3 | 1.74615E+15 | 5/25/1998 | GE MEDICAL SYSTEMS | LPOCOC0 | GENESIS_SIGNA | RM | 2D | 5 | 10002 | 142.5 | 63.87613 | 1.5 | 5 | 90 | 0.02832 | 29 | 256 | 192 | 0.8593750000\0.8593750000 | TCGA-08-0512/1.3.6.1.4.1.14519.5.2.1.7695.4001.280248392879135411898507571335/1.3.6.1.4.1.14519.5.2.1.7695.4001.740352126423395607820113197890 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0512 | t1Gd | M | 48 | 72.575 | 3DSPGR_AXIAL | 5 | 1.74615E+15 | 5/25/1998 | GE MEDICAL SYSTEMS | LPOCOC0 | GENESIS_SIGNA | GR | 3D | 1.5 | 34 | 6 | 63.87612 | 1.5 | 1.5 | 35 | 0.019784 | 124 | 256 | 192 | 1.0156250000\1.0156250000 | TCGA-08-0512/1.3.6.1.4.1.14519.5.2.1.7695.4001.280248392879135411898507571335/1.3.6.1.4.1.14519.5.2.1.7695.4001.188649667958862219325502314719 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0512 | t1 | M | 48 | 72.575 | FMPSPGR_SAG | 1 | 1.74615E+15 | 5/25/1998 | GE MEDICAL SYSTEMS | LPOCOC0 | GENESIS_SIGNA | GR | 2D | 4 | 100 | 2.1 | 63.87613 | 1.5 | 4 | 70 | 0.017912 | 46 | 256 | 192 | 0.9375000000\0.9375000000 | TCGA-08-0512/1.3.6.1.4.1.14519.5.2.1.7695.4001.280248392879135411898507571335/1.3.6.1.4.1.14519.5.2.1.7695.4001.312239011718497662985901571869 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0512 | t2 | M | 48 | 72.575 | 3DFSE_AXIAL | 2 | 1.74615E+15 | 5/25/1998 | GE MEDICAL SYSTEMS | LPOCOC0 | GENESIS_SIGNA | SE | 1.5 | 2700 | 104.576 | 63.87612 | 1.5 | 1.5 | 90 | 0.093091 | 126 | 256 | 192 | 1.0156250000\1.0156250000 | TCGA-08-0512/1.3.6.1.4.1.14519.5.2.1.7695.4001.280248392879135411898507571335/1.3.6.1.4.1.14519.5.2.1.7695.4001.351239022505812855531771936817 | ||
| TCGA-GBM | TCGA-08 | TCGA-08-0520 | flair | M | 70 | 70.307 | AX_FLAIR | 5 | 1.74615E+15 | 10/13/1999 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 142.5 | 2200 | 6.39E+08 | 1.5 | 5 | 90 | 0.024913 | 28 | 256 | 192 | 0.859386\0.859375 | TCGA-08-0520/1.3.6.1.4.1.14519.5.2.1.7695.4001.273090318153644004169216810536/1.3.6.1.4.1.14519.5.2.1.7695.4001.213779719506834152892047845101 |
| TCGA-GBM | TCGA-08 | TCGA-08-0520 | t1Gd | M | 70 | 70.307 | 3D_SPGR_AX | 7 | 1.74615E+15 | 10/13/1999 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 3D | 1.5 | 34 | 3 | 6.39E+08 | 1.5 | 1.5 | 35 | 0.01172 | 124 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0520/1.3.6.1.4.1.14519.5.2.1.7695.4001.273090318153644004169216810536/1.3.6.1.4.1.14519.5.2.1.7695.4001.231723835730198665428126824467 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0520 | t1 | M | 70 | 70.307 | FSPGR_SAG | 1 | 1.74615E+15 | 10/13/1999 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 2D | 4 | 100 | 2.9 | 6.39E+08 | 1.5 | 4 | 70 | 0.000708 | 44 | 256 | 192 | 0.937500\0.937500 | TCGA-08-0520/1.3.6.1.4.1.14519.5.2.1.7695.4001.273090318153644004169216810536/1.3.6.1.4.1.14519.5.2.1.7695.4001.327168483589011701407590503204 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0520 | t2 | M | 70 | 70.307 | 3D_FSE_AX | 4 | 1.74615E+15 | 10/13/1999 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | SE | 3D | 1.5 | 4000 | 104.016 | 6.39E+08 | 1.5 | 1.5 | 90 | 0.063159 | 126 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0520/1.3.6.1.4.1.14519.5.2.1.7695.4001.273090318153644004169216810536/1.3.6.1.4.1.14519.5.2.1.7695.4001.268232372267011104866408504758 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0521 | flair | M | 17 | 68.039 | AX_FLAIR | 2 | 1.74615E+15 | 3/7/2000 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 135 | 2200 | 6.39E+08 | 1.5 | 5 | 90 | 0.034659 | 28 | 256 | 192 | 0.859375\0.859375 | TCGA-08-0521/1.3.6.1.4.1.14519.5.2.1.7695.4001.238470932121913921057408160948/1.3.6.1.4.1.14519.5.2.1.7695.4001.845104538012589376178146557772 |
| TCGA-GBM | TCGA-08 | TCGA-08-0521 | t1Gd | M | 17 | 68.039 | 3D_SPGR_AX | 6 | 1.74615E+15 | 3/7/2000 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 3D | 1.5 | 34 | 3 | 6.39E+08 | 1.5 | 1.5 | 35 | 0.011782 | 124 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0521/1.3.6.1.4.1.14519.5.2.1.7695.4001.238470932121913921057408160948/1.3.6.1.4.1.14519.5.2.1.7695.4001.110099365603373672025357679240 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0521 | t1 | M | 17 | 68.039 | FSPGR_SAG | 1 | 1.74615E+15 | 3/7/2000 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 2D | 4 | 100 | 2.9 | 6.39E+08 | 1.5 | 4 | 70 | 0.000712 | 44 | 256 | 192 | 0.937500\0.937500 | TCGA-08-0521/1.3.6.1.4.1.14519.5.2.1.7695.4001.238470932121913921057408160948/1.3.6.1.4.1.14519.5.2.1.7695.4001.596008525691300018297895420363 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0521 | t2 | M | 17 | 68.039 | 3D_FSE_AX | 3 | 1.74615E+15 | 3/7/2000 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | SE | 3D | 1.5 | 4000 | 104.016 | 6.39E+08 | 1.5 | 1.5 | 90 | 0.057719 | 114 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0521/1.3.6.1.4.1.14519.5.2.1.7695.4001.238470932121913921057408160948/1.3.6.1.4.1.14519.5.2.1.7695.4001.217015902186698075264229258207 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0522 | flair | M | 61 | 72.575 | AX_FLAIR | 9 | 1.74615E+15 | 4/24/2000 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 127.5 | 2200 | 6.39E+08 | 1.5 | 5 | 90 | 0.025609 | 30 | 256 | 192 | 0.859375\0.859375 | TCGA-08-0522/1.3.6.1.4.1.14519.5.2.1.7695.4001.247226492578736624112472654446/1.3.6.1.4.1.14519.5.2.1.7695.4001.158696227902295886606720382429 |
| TCGA-GBM | TCGA-08 | TCGA-08-0522 | t1Gd | M | 61 | 72.575 | 3D_SPGR_AX | 7 | 1.74615E+15 | 4/24/2000 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 3D | 1.5 | 34 | 3 | 6.39E+08 | 1.5 | 1.5 | 35 | 0.019789 | 124 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0522/1.3.6.1.4.1.14519.5.2.1.7695.4001.247226492578736624112472654446/1.3.6.1.4.1.14519.5.2.1.7695.4001.168762703209156995755737783606 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0522 | t1 | M | 61 | 72.575 | FSPGR_SAG | 3 | 1.74615E+15 | 4/24/2000 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 2D | 4 | 100 | 2.9 | 6.39E+08 | 1.5 | 4 | 70 | 0.000704 | 44 | 256 | 192 | 0.937500\0.937500 | TCGA-08-0522/1.3.6.1.4.1.14519.5.2.1.7695.4001.247226492578736624112472654446/1.3.6.1.4.1.14519.5.2.1.7695.4001.243308281114053949145480905289 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0522 | t2 | M | 61 | 72.575 | 3D_FSE_AX | 6 | 1.74615E+15 | 4/24/2000 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | SE | 3D | 1.5 | 3000 | 104.016 | 6.39E+08 | 1.5 | 1.5 | 90 | 0.083799 | 126 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0522/1.3.6.1.4.1.14519.5.2.1.7695.4001.247226492578736624112472654446/1.3.6.1.4.1.14519.5.2.1.7695.4001.160182616952637474240510659084 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0524 | flair | F | 17 | 52.163 | AX_FLAIR | 2 | 1.74615E+15 | 11/2/2000 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | IR | 2D | 3 | 10002 | 133 | 2200 | 6.39E+08 | 1.5 | 3 | 90 | 0.029865 | 32 | 256 | 192 | 0.859375\0.859375 | TCGA-08-0524/1.3.6.1.4.1.14519.5.2.1.7695.4001.195614997200019607573614338362/1.3.6.1.4.1.14519.5.2.1.7695.4001.232360725935275321651480467041 |
| TCGA-GBM | TCGA-08 | TCGA-08-0524 | t1Gd | F | 17 | 52.163 | 3D_SPGR_AX | 4 | 1.74615E+15 | 11/2/2000 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 3D | 1.5 | 34 | 3 | 6.39E+08 | 1.5 | 1.5 | 35 | 0.020863 | 124 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0524/1.3.6.1.4.1.14519.5.2.1.7695.4001.195614997200019607573614338362/1.3.6.1.4.1.14519.5.2.1.7695.4001.267971913863007998166877158233 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0524 | t1 | F | 17 | 52.163 | FSPGR_SAG | 1 | 1.74615E+15 | 11/2/2000 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 2D | 4 | 100 | 2.8 | 6.39E+08 | 1.5 | 4 | 70 | 0.000743 | 44 | 256 | 192 | 0.937500\0.937500 | TCGA-08-0524/1.3.6.1.4.1.14519.5.2.1.7695.4001.195614997200019607573614338362/1.3.6.1.4.1.14519.5.2.1.7695.4001.863645671849260537371170935073 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0524 | t2 | F | 17 | 52.163 | 3D_FSE_AX | 5 | 1.74615E+15 | 11/2/2000 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | SE | 3D | 1.5 | 2600 | 104.576 | 6.39E+08 | 1.5 | 1.5 | 90 | 0.083412 | 108 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0524/1.3.6.1.4.1.14519.5.2.1.7695.4001.195614997200019607573614338362/1.3.6.1.4.1.14519.5.2.1.7695.4001.269722421607288207652501119281 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0529 | flair | F | 56 | 72.575 | AX_FLAIR | 3 | 1.74615E+15 | 10/29/2002 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 127.5 | 2200 | 6.39E+08 | 1.5 | 5 | 90 | 0.025609 | 32 | 256 | 192 | 0.859375\0.859375 | TCGA-08-0529/1.3.6.1.4.1.14519.5.2.1.7695.4001.912696403696942091184547452317/1.3.6.1.4.1.14519.5.2.1.7695.4001.320850822961595333060405852108 |
| TCGA-GBM | TCGA-08 | TCGA-08-0529 | t1Gd | F | 56 | 72.575 | 3D_SPGR_AX | 7 | 1.74615E+15 | 10/29/2002 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 3D | 1.5 | 34 | 3 | 6.39E+08 | 1.5 | 1.5 | 35 | 0.019789 | 124 | 256 | 192 | 1.015625\1.015625 | TCGA-08-0529/1.3.6.1.4.1.14519.5.2.1.7695.4001.912696403696942091184547452317/1.3.6.1.4.1.14519.5.2.1.7695.4001.220439982391729657710258636833 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0529 | t1 | F | 56 | 72.575 | FSPGR_SAG | 2 | 1.74615E+15 | 10/29/2002 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | GR | 2D | 4 | 100 | 2.8 | 6.39E+08 | 1.5 | 4 | 70 | 0.000704 | 42 | 256 | 192 | 0.937500\0.937500 | TCGA-08-0529/1.3.6.1.4.1.14519.5.2.1.7695.4001.912696403696942091184547452317/1.3.6.1.4.1.14519.5.2.1.7695.4001.324296635305794383120984682544 | |
| TCGA-GBM | TCGA-08 | TCGA-08-0529 | t2 | F | 56 | 72.575 | 3D_FSE_AX | 5 | 1.74615E+15 | 10/29/2002 | GE MEDICAL SYSTEMS | GEMSLPMR | GENESIS_SIGNA | SE | 3D | 1.5 | 3000 | 104.576 | 6.39E+08 | 1.5 | 1.5 | 90 | 0.076181 | 120 | 256 | 160 | 1.015625\1.015625 | TCGA-08-0529/1.3.6.1.4.1.14519.5.2.1.7695.4001.912696403696942091184547452317/1.3.6.1.4.1.14519.5.2.1.7695.4001.229810524792550999481504742060 | |
| TCGA-GBM | TCGA-12 | TCGA-12-0616 | flair | F | 36 | 68.1 | ax_flair | 5 | 3.29916E+15 | 4/12/1999 | SIEMENS | Trio | IR\SE | 2D | 5 | 9000 | 121 | 2200 | 123.1898 | 2.893620014 | 7.5 | 150 | 0.301020682 | 22 | 320 | 208 | 0.6875\0.6875 | TCGA-12-0616/1.3.6.1.4.1.14519.5.2.1.8862.4001.266192358731001325275209303138/1.3.6.1.4.1.14519.5.2.1.8862.4001.169997737294570942795903209401 | |
| TCGA-GBM | TCGA-12 | TCGA-12-0616 | t1Gd | F | 36 | 68.1 | cor_mprage_+c | 8 | 3.29916E+15 | 4/12/1999 | SIEMENS | Trio | IR\GR | 3D | 1.25 | 2100 | 5.26 | 1100 | 123.1898 | 2.893620014 | 8 | 0.046533085 | 176 | 256 | 192 | 0.859375\0.859375 | TCGA-12-0616/1.3.6.1.4.1.14519.5.2.1.8862.4001.266192358731001325275209303138/1.3.6.1.4.1.14519.5.2.1.8862.4001.238427576098066516919422281607 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-0616 | t1 | F | 36 | 68.1 | ax_t1 | 2 | 3.29916E+15 | 4/12/1999 | SIEMENS | Trio | SE | 2D | 5 | 800 | 10 | 123.1899 | 2.893620014 | 7.5 | 90 | 0.308275163 | 22 | 384 | 240 | 0.5729166665625\0.5729166665625 | TCGA-12-0616/1.3.6.1.4.1.14519.5.2.1.8862.4001.266192358731001325275209303138/1.3.6.1.4.1.14519.5.2.1.8862.4001.724789870643862849694451333256 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-0616 | t2 | F | 36 | 68.1 | ax_t2 | 3 | 3.29916E+15 | 4/12/1999 | SIEMENS | Trio | SE | 2D | 5 | 3500 | 116 | 123.1899 | 2.893620014 | 7.5 | 180 | 0.255155891 | 22 | 512 | 312 | 0.4296875\0.4296875 | TCGA-12-0616/1.3.6.1.4.1.14519.5.2.1.8862.4001.266192358731001325275209303138/1.3.6.1.4.1.14519.5.2.1.8862.4001.133973750207915213340858829260 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-0776 | flair | M | 52 | 96.162 | Ax_Flair_irFSE_H | 3 | 2.8195E+15 | 5/12/1998 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | IR | 2D | 5 | 9002 | 147 | 2200 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.025085 | 23 | 256 | 160 | 0.4296875\0.4296875 | TCGA-12-0776/1.3.6.1.4.1.14519.5.2.1.8862.4001.252675253473152183963522222391/1.3.6.1.4.1.14519.5.2.1.8862.4001.546462055894409848329223976518 | |
| TCGA-GBM | TCGA-12 | TCGA-12-0776 | t1Gd | M | 52 | 96.162 | AX_T1_FSE_H+C | 5 | 2.8195E+15 | 5/12/1998 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | RM | 2D | 5 | 500 | 11.12 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.0451 | 23 | 320 | 160 | 0.4296875\0.4296875 | TCGA-12-0776/1.3.6.1.4.1.14519.5.2.1.8862.4001.252675253473152183963522222391/1.3.6.1.4.1.14519.5.2.1.8862.4001.242852138496700175683090714079 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-0776 | t1 | M | 52 | 96.162 | Sag_T1_FSE_S | 1 | 2.8195E+15 | 5/12/1998 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 4 | 450 | 14 | 6.39E+08 | 1.5 | 5 | 90 | 0.036675 | 23 | 256 | 160 | 0.46875\0.46875 | TCGA-12-0776/1.3.6.1.4.1.14519.5.2.1.8862.4001.252675253473152183963522222391/1.3.6.1.4.1.14519.5.2.1.8862.4001.269331403973802247544189874015 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-0776 | t2 | M | 52 | 96.162 | Ax_T2_FSE_H | 2 | 2.8195E+15 | 5/12/1998 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | RM | 2D | 5 | 4666.664 | 81.032 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.1077 | 23 | 320 | 192 | 0.4296875\0.4296875 | TCGA-12-0776/1.3.6.1.4.1.14519.5.2.1.8862.4001.252675253473152183963522222391/1.3.6.1.4.1.14519.5.2.1.8862.4001.204050589111893183636316823335 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-0829 | flair | M | 75 | 90 | FLAIR | 4 | 2.8195E+15 | 6/2/1999 | SIEMENS | Symphony | IR\SE | 2D | 5 | 8000 | 108 | 2500 | 63.66644 | 1.494 | 6.5 | 180 | 0.30549785 | 23 | 192 | 133 | 0.59895833\0.59895833 | TCGA-12-0829/1.3.6.1.4.1.14519.5.2.1.8862.4001.161067531590129181313302509460/1.3.6.1.4.1.14519.5.2.1.8862.4001.256774824254590199629447782367 | |
| TCGA-GBM | TCGA-12 | TCGA-12-0829 | t1Gd | M | 75 | 90 | T1_AX_POST | 18 | 2.8195E+15 | 6/2/1999 | SIEMENS | Symphony | SE | 2D | 5 | 635 | 17 | 63.66644 | 1.494 | 6.5 | 75 | 0.73542476 | 23 | 256 | 144 | 0.44921875\0.44921875 | TCGA-12-0829/1.3.6.1.4.1.14519.5.2.1.8862.4001.161067531590129181313302509460/1.3.6.1.4.1.14519.5.2.1.8862.4001.848905768294232154546172773832 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-0829 | t1 | M | 75 | 90 | T1_AX_PRE | 5 | 2.8195E+15 | 6/2/1999 | SIEMENS | Symphony | SE | 2D | 5 | 571 | 17 | 63.66644 | 1.494 | 6.5 | 75 | 0.12039723 | 23 | 256 | 154 | 0.44921875\0.44921875 | TCGA-12-0829/1.3.6.1.4.1.14519.5.2.1.8862.4001.161067531590129181313302509460/1.3.6.1.4.1.14519.5.2.1.8862.4001.229514096688570743417897123096 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-0829 | t2 | M | 75 | 90 | T2_TSE_AX | 3 | 2.8195E+15 | 6/2/1999 | SIEMENS | Symphony | SE | 2D | 5 | 4670 | 104 | 63.6664 | 1.494 | 6.5 | 180 | 0.006011039 | 23 | 512 | 204 | 0.44921875\0.44921875 | TCGA-12-0829/1.3.6.1.4.1.14519.5.2.1.8862.4001.161067531590129181313302509460/1.3.6.1.4.1.14519.5.2.1.8862.4001.126416306775413055857394841475 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-1093 | flair | F | 66 | 54.431 | AX_T2_FLAIR | 4 | 2.8195E+15 | 9/20/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | IR | 2D | 5 | 9002 | 146.816 | 2250 | 63.86599 | 1.5 | 5 | 90 | 0.4465 | 30 | 256 | 192 | 0.859371\0.859374 | TCGA-12-1093/1.3.6.1.4.1.14519.5.2.1.8862.4001.101131717694922416453780234026/1.3.6.1.4.1.14519.5.2.1.8862.4001.114806709082901981977289923516 | |
| TCGA-GBM | TCGA-12 | TCGA-12-1093 | t1Gd | F | 66 | 54.431 | AX_T1+C | 9 | 2.8195E+15 | 9/20/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 466.664 | 20 | 0 | 63.86599 | 1.5 | 5 | 90 | 0.860357 | 30 | 288 | 192 | 0.429685\0.429687 | TCGA-12-1093/1.3.6.1.4.1.14519.5.2.1.8862.4001.101131717694922416453780234026/1.3.6.1.4.1.14519.5.2.1.8862.4001.208421097810800556291170968762 | |
| TCGA-GBM | TCGA-12 | TCGA-12-1093 | t1 | F | 66 | 54.431 | AX_T1 | 8 | 2.8195E+15 | 9/20/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 366.664 | 15 | 0 | 63.86599 | 1.5 | 5 | 90 | 1.09631 | 30 | 288 | 192 | 0.429685\0.429687 | TCGA-12-1093/1.3.6.1.4.1.14519.5.2.1.8862.4001.101131717694922416453780234026/1.3.6.1.4.1.14519.5.2.1.8862.4001.850115408111720965337317146830 | |
| TCGA-GBM | TCGA-12 | TCGA-12-1093 | t2 | F | 66 | 54.431 | AX_T2 | 7 | 2.8195E+15 | 9/20/1999 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 4100 | 85.92 | 0 | 63.86599 | 1.5 | 5 | 90 | 1.3717 | 30 | 256 | 256 | 0.429685\0.429687 | TCGA-12-1093/1.3.6.1.4.1.14519.5.2.1.8862.4001.101131717694922416453780234026/1.3.6.1.4.1.14519.5.2.1.8862.4001.201607716070807113655311579763 | |
| TCGA-GBM | TCGA-12 | TCGA-12-1094 | flair | M | 56 | 136.078 | COR_FLAIR | 3 | 2.8195E+15 | 11/15/1999 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | IR | 2D | 5 | 8002 | 127.5 | 2000 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.025767 | 27 | 256 | 192 | 0.937500\0.937500 | TCGA-12-1094/1.3.6.1.4.1.14519.5.2.1.8862.4001.172549962021947577169833248421/1.3.6.1.4.1.14519.5.2.1.8862.4001.135805150196719046218951097371 | |
| TCGA-GBM | TCGA-12 | TCGA-12-1094 | t1Gd | M | 56 | 136.078 | POST_AX_T1 | 7 | 2.8195E+15 | 11/15/1999 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 550 | 9 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.054078 | 22 | 256 | 224 | 0.937500\0.937500 | TCGA-12-1094/1.3.6.1.4.1.14519.5.2.1.8862.4001.172549962021947577169833248421/1.3.6.1.4.1.14519.5.2.1.8862.4001.195674900023171610747750919749 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-1094 | t1 | M | 56 | 136.078 | AX_T1 | 6 | 2.8195E+15 | 11/15/1999 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 5 | 550 | 9 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.054078 | 22 | 256 | 224 | 0.937500\0.937500 | TCGA-12-1094/1.3.6.1.4.1.14519.5.2.1.8862.4001.172549962021947577169833248421/1.3.6.1.4.1.14519.5.2.1.8862.4001.190799192727661447752457405978 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-1094 | t2 | M | 56 | 136.078 | AX_T2_FS | 5 | 2.8195E+15 | 11/15/1999 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | RM | 2D | 5 | 6650 | 81.928 | 6.39E+08 | 1.5 | 6.5 | 90 | 0.0395 | 22 | 512 | 224 | 0.468750\0.468750 | TCGA-12-1094/1.3.6.1.4.1.14519.5.2.1.8862.4001.172549962021947577169833248421/1.3.6.1.4.1.14519.5.2.1.8862.4001.204812843794603036437575477292 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-1098 | flair | F | 75 | 77.18 | FLAIR_AXIAL | 8 | 2.8195E+15 | 7/23/2000 | SIEMENS | Avanto | IR\SE | 2D | 5 | 8630 | 110 | 2500 | 63.67312 | 1.493999958 | 6.25 | 150 | 0.111189924 | 23 | 256 | 179 | 0.44921875\0.44921875 | TCGA-12-1098/1.3.6.1.4.1.14519.5.2.1.8862.4001.119873216352344754496678099313/1.3.6.1.4.1.14519.5.2.1.8862.4001.444903703803452795088981514764 | |
| TCGA-GBM | TCGA-12 | TCGA-12-1098 | t1Gd | F | 75 | 77.18 | T1_AXIAL__POST | 10 | 2.8195E+15 | 7/23/2000 | SIEMENS | Avanto | IR\GR | 3D | 2 | 1830 | 4.38 | 1100 | 63.67312 | 1.493999958 | 15 | 0.020332757 | 72 | 256 | 157 | 0.48828125\0.48828125 | TCGA-12-1098/1.3.6.1.4.1.14519.5.2.1.8862.4001.119873216352344754496678099313/1.3.6.1.4.1.14519.5.2.1.8862.4001.278563943372409161004573397398 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-1098 | t1 | F | 75 | 77.18 | T1_AXIAL | 9 | 2.8195E+15 | 7/23/2000 | SIEMENS | Avanto | IR\GR | 3D | 2 | 1830 | 4.38 | 1100 | 63.67312 | 1.493999958 | 15 | 0.020332757 | 72 | 256 | 157 | 0.48828125\0.48828125 | TCGA-12-1098/1.3.6.1.4.1.14519.5.2.1.8862.4001.119873216352344754496678099313/1.3.6.1.4.1.14519.5.2.1.8862.4001.222247651601980577893548825337 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-1098 | t2 | F | 75 | 77.18 | T2_TSE_AXIAL | 6 | 2.8195E+15 | 7/23/2000 | SIEMENS | Avanto | SE | 2D | 5 | 4390 | 109 | 63.67309 | 1.493999958 | 6.25 | 150 | 0.265220344 | 23 | 512 | 304 | 0.21502535927022\0.21502535927022 | TCGA-12-1098/1.3.6.1.4.1.14519.5.2.1.8862.4001.119873216352344754496678099313/1.3.6.1.4.1.14519.5.2.1.8862.4001.266821148594345502971893058552 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-1598 | flair | F | 75 | 81.72 | FLAIR_AX | 4 | 2.8195E+15 | 8/23/1999 | SIEMENS | Symphony | IR\SE | 2D | 5 | 10000 | 106 | 2300 | 63.68462 | 1.494 | 6 | 180 | 0.00539074 | 20 | 256 | 179 | 0.8984375\0.8984375 | TCGA-12-1598/1.3.6.1.4.1.14519.5.2.1.8862.4001.146632443531148261788047561414/1.3.6.1.4.1.14519.5.2.1.8862.4001.336849870613672922999907742707 | |
| TCGA-GBM | TCGA-12 | TCGA-12-1598 | t1Gd | F | 75 | 81.72 | T1_AX_gado | 9 | 2.8195E+15 | 8/23/1999 | SIEMENS | Symphony | SE | 2D | 5 | 552 | 17 | 63.68464 | 1.494 | 6.5 | 90 | 0.11230121 | 20 | 256 | 192 | 0.8984375\0.8984375 | TCGA-12-1598/1.3.6.1.4.1.14519.5.2.1.8862.4001.146632443531148261788047561414/1.3.6.1.4.1.14519.5.2.1.8862.4001.342060257312097189842582111091 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-1598 | t1 | F | 75 | 81.72 | T1_AX | 5 | 2.8195E+15 | 8/23/1999 | SIEMENS | Symphony | SE | 2D | 5 | 506 | 17 | 63.68464 | 1.494 | 6.5 | 90 | 0.18678181 | 20 | 256 | 192 | 0.8984375\0.8984375 | TCGA-12-1598/1.3.6.1.4.1.14519.5.2.1.8862.4001.146632443531148261788047561414/1.3.6.1.4.1.14519.5.2.1.8862.4001.102361236469082315415067903736 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-1598 | t2 | F | 75 | 81.72 | T2_AX | 3 | 2.8195E+15 | 8/23/1999 | SIEMENS | Symphony | SE | 2D | 5 | 5760 | 110 | 63.68462 | 1.494 | 6.5 | 180 | 0.005438163 | 20 | 512 | 288 | 0.44921875\0.44921875 | TCGA-12-1598/1.3.6.1.4.1.14519.5.2.1.8862.4001.146632443531148261788047561414/1.3.6.1.4.1.14519.5.2.1.8862.4001.735255270119748511424115051454 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-1601 | flair | M | 71 | 90.71849 | ax_flair | 5 | 1.76067E+15 | 6/19/2000 | SIEMENS | TrioTim | SE\IR | 2D | 5 | 9000 | 134 | 2500 | 123.2367 | 3 | 7.5 | 125 | 0.695210424 | 24 | 320 | 320 | 0.71875\0.71875 | TCGA-12-1601/1.3.6.1.4.1.14519.5.2.1.8862.4001.201486216522847049056957283859/1.3.6.1.4.1.14519.5.2.1.8862.4001.248879959554046988740819326552 | |
| TCGA-GBM | TCGA-12 | TCGA-12-1601 | t1Gd | M | 71 | 90.71849 | ax_t1_+c | 9 | 1.76067E+15 | 6/19/2000 | SIEMENS | TrioTim | SE | 2D | 5 | 720 | 20 | 123.2367 | 3 | 7.5 | 75 | 0.674986112 | 24 | 384 | 240 | 0.59895837306976\0.59895837306976 | TCGA-12-1601/1.3.6.1.4.1.14519.5.2.1.8862.4001.201486216522847049056957283859/1.3.6.1.4.1.14519.5.2.1.8862.4001.146592301314373210626155028600 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-1601 | t1 | M | 71 | 90.71849 | ax_t1 | 3 | 1.76067E+15 | 6/19/2000 | SIEMENS | TrioTim | SE | 2D | 5 | 796 | 10 | 123.2367 | 3 | 7.5 | 75 | 0.694832124 | 24 | 320 | 192 | 0.71875\0.71875 | TCGA-12-1601/1.3.6.1.4.1.14519.5.2.1.8862.4001.201486216522847049056957283859/1.3.6.1.4.1.14519.5.2.1.8862.4001.609705420088117660227141140219 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-1601 | t2 | M | 71 | 90.71849 | ax_t2 | 4 | 1.76067E+15 | 6/19/2000 | SIEMENS | TrioTim | SE | 2D | 5 | 4830 | 113 | 123.2367 | 3 | 7.5 | 140 | 0.695210424 | 24 | 320 | 320 | 0.71875\0.71875 | TCGA-12-1601/1.3.6.1.4.1.14519.5.2.1.8862.4001.201486216522847049056957283859/1.3.6.1.4.1.14519.5.2.1.8862.4001.111322175120195590616905223490 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-1602 | flair | M | 58 | 0 | AX_FLAIR | 7 | 2.8195E+15 | 3/4/2001 | SIEMENS | Trio | IR\SE | 2D | 5 | 10000 | 115 | 2500 | 123.2288 | 3 | 6 | 180 | 0.156334221 | 24 | 256 | 208 | 0.41015625\0.41015625 | TCGA-12-1602/1.3.6.1.4.1.14519.5.2.1.8862.4001.322746683938440731450945038341/1.3.6.1.4.1.14519.5.2.1.8862.4001.239852680542995747210886629062 | |
| TCGA-GBM | TCGA-12 | TCGA-12-1602 | t1Gd | M | 58 | 0 | T1_POST_GD | 19 | 2.8195E+15 | 3/4/2001 | SIEMENS | Trio | SE | 2D | 5 | 720 | 20 | 123.2289 | 3 | 6 | 60 | 0.161792412 | 24 | 256 | 179 | 0.8203125\0.8203125 | TCGA-12-1602/1.3.6.1.4.1.14519.5.2.1.8862.4001.322746683938440731450945038341/1.3.6.1.4.1.14519.5.2.1.8862.4001.325175595450208505943785543644 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-1602 | t1 | M | 58 | 0 | AX_SE_T1 | 6 | 2.8195E+15 | 3/4/2001 | SIEMENS | Trio | SE | 2D | 5 | 678 | 17 | 123.2289 | 3 | 6 | 60 | 0.159116566 | 24 | 256 | 224 | 0.8203125\0.8203125 | TCGA-12-1602/1.3.6.1.4.1.14519.5.2.1.8862.4001.322746683938440731450945038341/1.3.6.1.4.1.14519.5.2.1.8862.4001.504967401532712438614608840804 | ||
| TCGA-GBM | TCGA-12 | TCGA-12-1602 | t2 | M | 58 | 0 | SAG_TSE3D_T2 | 10 | 2.8195E+15 | 3/4/2001 | SIEMENS | Trio | SE | 3D | 1.2 | 3000 | 355 | 123.2289 | 3 | 180 | 0.127274916 | 144 | 256 | 192 | 1\1 | TCGA-12-1602/1.3.6.1.4.1.14519.5.2.1.8862.4001.322746683938440731450945038341/1.3.6.1.4.1.14519.5.2.1.8862.4001.955499093208862961641223883317 | |||
| TCGA-GBM | TCGA-12 | TCGA-12-3650 | flair | M | 46 | 102.058 | Ax_T2Flair | 4 | 2.8195E+15 | 7/29/2001 | GE MEDICAL SYSTEMS | SIGNA HDx | SE\\\\ | 2D | 5 | 8000 | 135.648 | 2000 | 63.82825 | 1.5 | 6 | 90 | 0.846887 | 25 | 288 | 288 | 0.468750\0.468750 | TCGA-12-3650/1.3.6.1.4.1.14519.5.2.1.8862.4001.853973730099261399255070895330/1.3.6.1.4.1.14519.5.2.1.8862.4001.239036983327341632160933559587 | |
| TCGA-GBM | TCGA-12 | TCGA-12-3650 | t1Gd | M | 46 | 102.058 | AX_T1_POST | 9 | 2.8195E+15 | 7/29/2001 | GE MEDICAL SYSTEMS | SIGNA HDx | SE\\\\ | 2D | 5 | 750 | 10.344 | 0 | 63.82825 | 1.5 | 6 | 90 | 1.5028 | 25 | 320 | 224 | 0.4688\0.4688 | TCGA-12-3650/1.3.6.1.4.1.14519.5.2.1.8862.4001.853973730099261399255070895330/1.3.6.1.4.1.14519.5.2.1.8862.4001.259092977773592001212080458593 | |
| TCGA-GBM | TCGA-12 | TCGA-12-3650 | t1 | M | 46 | 102.058 | AX_T1 | 6 | 2.8195E+15 | 7/29/2001 | GE MEDICAL SYSTEMS | SIGNA HDx | SE\\\\ | 2D | 5 | 750 | 10.344 | 0 | 63.82825 | 1.5 | 6 | 90 | 1.5028 | 25 | 320 | 224 | 0.4688\0.4688 | TCGA-12-3650/1.3.6.1.4.1.14519.5.2.1.8862.4001.853973730099261399255070895330/1.3.6.1.4.1.14519.5.2.1.8862.4001.298865676266232133850135994761 | |
| TCGA-GBM | TCGA-12 | TCGA-12-3650 | t2 | M | 46 | 102.058 | Ax_T2 | 5 | 2.8195E+15 | 7/29/2001 | GE MEDICAL SYSTEMS | SIGNA HDx | SE\\\\ | 2D | 5 | 6000 | 115.36 | 63.82825 | 1.5 | 6 | 90 | 1.553567 | 25 | 416 | 416 | 0.468750\0.468750 | TCGA-12-3650/1.3.6.1.4.1.14519.5.2.1.8862.4001.853973730099261399255070895330/1.3.6.1.4.1.14519.5.2.1.8862.4001.250106226507995703345955579876 | ||
| TCGA-GBM | TCGA-14 | TCGA-14-0789 | flair | M | 80 | BRAIN______FLAIR | 301 | 1.3568E+15 | 11/19/1997 | Philips Medical Systems | Gyro01 | Intera | IR | 2D | 5 | 11000 | 140 | 2600 | 63.895 | 1.5 | 6 | 90 | 25 | 256 | 151 | 004.492188e-01\004.492188e-01 | TCGA-14-0789/1.3.6.1.4.1.14519.5.2.1.2783.4001.184309347042506581266884481594/1.3.6.1.4.1.14519.5.2.1.2783.4001.801520796772002669204007145386 | ||
| TCGA-GBM | TCGA-14 | TCGA-14-0789 | t1Gd | M | 80 | BRAIN______T1_AX_POST | 801 | 1.3568E+15 | 11/19/1997 | Philips Medical Systems | Gyro01 | Intera | SE | 2D | 5 | 565 | 12 | 63.895 | 1.5 | 6 | 90 | 25 | 256 | 205 | 004.492188e-01\004.492188e-01 | TCGA-14-0789/1.3.6.1.4.1.14519.5.2.1.2783.4001.184309347042506581266884481594/1.3.6.1.4.1.14519.5.2.1.2783.4001.890592197338541936330714143214 | |||
| TCGA-GBM | TCGA-14 | TCGA-14-0789 | t1 | M | 80 | BRAIN______T1_AX_PRE | 701 | 1.3568E+15 | 11/19/1997 | Philips Medical Systems | Gyro01 | Intera | SE | 2D | 5 | 565 | 12 | 63.895 | 1.5 | 6 | 90 | 25 | 256 | 205 | 004.492188e-01\004.492188e-01 | TCGA-14-0789/1.3.6.1.4.1.14519.5.2.1.2783.4001.184309347042506581266884481594/1.3.6.1.4.1.14519.5.2.1.2783.4001.233041259508543870852056565003 | |||
| TCGA-GBM | TCGA-14 | TCGA-14-0789 | t2 | M | 80 | BRAIN______T2_GRASE | 401 | 1.3568E+15 | 11/19/1997 | Philips Medical Systems | Gyro01 | Intera | SE | 2D | 5 | 5000 | 100 | 63.895 | 1.5 | 6 | 90 | 25 | 256 | 241 | 004.492188e-01\004.492188e-01 | TCGA-14-0789/1.3.6.1.4.1.14519.5.2.1.2783.4001.184309347042506581266884481594/1.3.6.1.4.1.14519.5.2.1.2783.4001.141146456081128930177162025757 | |||
| TCGA-GBM | TCGA-14 | TCGA-14-1456 | flair | M | 80 | STROKE_____FLAIR | 401 | 1.34201E+15 | 4/22/1999 | Philips Medical Systems | Gyro01 | Intera | IR | 2D | 5 | 11000 | 140 | 2600 | 63.90691 | 1.5 | 5 | 90 | 25 | 256 | 159 | 009.375000e-01\009.375000e-01 | TCGA-14-1456/1.3.6.1.4.1.14519.5.2.1.2783.4001.250594672846486970701098971076/1.3.6.1.4.1.14519.5.2.1.2783.4001.321134067484147578724672344840 | ||
| TCGA-GBM | TCGA-14 | TCGA-14-1456 | t1Gd | M | 80 | BRAIN______T1_AX_POST | 1901 | 1.34201E+15 | 4/22/1999 | Philips Medical Systems | Gyro01 | Intera | SE | 2D | 5 | 565 | 12 | 63.90692 | 1.5 | 6 | 90 | 25 | 256 | 205 | 004.492188e-01\004.492188e-01 | TCGA-14-1456/1.3.6.1.4.1.14519.5.2.1.2783.4001.250594672846486970701098971076/1.3.6.1.4.1.14519.5.2.1.2783.4001.784458997482067360267090300516 | |||
| TCGA-GBM | TCGA-14 | TCGA-14-1456 | t1 | M | 80 | Brain______T1W_SE | 201 | 1.34201E+15 | 4/22/1999 | Philips Medical Systems | Gyro01 | Intera | SE | 2D | 5 | 352.1115 | 15 | 63.90692 | 1.5 | 5 | 69 | 25 | 256 | 141 | 009.375000e-01\009.375000e-01 | TCGA-14-1456/1.3.6.1.4.1.14519.5.2.1.2783.4001.250594672846486970701098971076/1.3.6.1.4.1.14519.5.2.1.2783.4001.925674708608577414119113282332 | |||
| TCGA-GBM | TCGA-14 | TCGA-14-1456 | t2 | M | 80 | SEIZURE____T2_GRASE | 301 | 1.34201E+15 | 4/22/1999 | Philips Medical Systems | Gyro01 | Intera | SE | 2D | 5 | 5000 | 100 | 63.90692 | 1.5 | 5 | 90 | 25 | 256 | 216 | 009.375000e-01\009.375000e-01 | TCGA-14-1456/1.3.6.1.4.1.14519.5.2.1.2783.4001.250594672846486970701098971076/1.3.6.1.4.1.14519.5.2.1.2783.4001.139583367552275817017048428824 | |||
| TCGA-GBM | TCGA-14 | TCGA-14-1794 | flair | M | 90 | BRAIN______FLAIR | 301 | 2.13909E+15 | 4/11/1998 | Philips Medical Systems | Gyro01 | Intera | IR | 2D | 5 | 11000 | 140 | 2600 | 63.89496 | 1.5 | 6 | 90 | 25 | 256 | 151 | 004.492188e-01\004.492188e-01 | TCGA-14-1794/1.3.6.1.4.1.14519.5.2.1.2783.4001.867649433395377496837912142545/1.3.6.1.4.1.14519.5.2.1.2783.4001.655280789746543642623011826556 | ||
| TCGA-GBM | TCGA-14 | TCGA-14-1794 | t1Gd | M | 90 | BRAIN______T1_AX_POST | 1101 | 2.13909E+15 | 4/11/1998 | Philips Medical Systems | Gyro01 | Intera | SE | 2D | 5 | 565 | 12 | 63.89496 | 1.5 | 6 | 90 | 25 | 256 | 205 | 004.492188e-01\004.492188e-01 | TCGA-14-1794/1.3.6.1.4.1.14519.5.2.1.2783.4001.867649433395377496837912142545/1.3.6.1.4.1.14519.5.2.1.2783.4001.121223650477077769385830996743 | |||
| TCGA-GBM | TCGA-14 | TCGA-14-1794 | t1 | M | 90 | BRAIN______T1_AX_PRE | 701 | 2.13909E+15 | 4/11/1998 | Philips Medical Systems | Gyro01 | Intera | SE | 2D | 5 | 565 | 12 | 63.89496 | 1.5 | 6 | 90 | 25 | 256 | 205 | 004.492188e-01\004.492188e-01 | TCGA-14-1794/1.3.6.1.4.1.14519.5.2.1.2783.4001.867649433395377496837912142545/1.3.6.1.4.1.14519.5.2.1.2783.4001.101882499143559305814061553019 | |||
| TCGA-GBM | TCGA-14 | TCGA-14-1794 | t2 | M | 90 | BRAIN______T2_GRASE | 401 | 2.13909E+15 | 4/11/1998 | Philips Medical Systems | Gyro01 | Intera | SE | 2D | 5 | 5000 | 100 | 63.89496 | 1.5 | 6 | 90 | 25 | 256 | 241 | 004.492188e-01\004.492188e-01 | TCGA-14-1794/1.3.6.1.4.1.14519.5.2.1.2783.4001.867649433395377496837912142545/1.3.6.1.4.1.14519.5.2.1.2783.4001.466914301691885355538840307808 | |||
| TCGA-GBM | TCGA-14 | TCGA-14-1825 | flair | M | 70 | 76.65712 | t2_flair_ax | 3 | 1.4078E+15 | 2/10/2000 | SIEMENS | MEDPC | Avanto | SE\IR | 2D | 5 | 8000 | 74 | 2500 | 63.67925 | 1.5 | 6 | 180 | 0.278502221 | 27 | 256 | 256 | 0.44921875\0.44921875 | TCGA-14-1825/1.3.6.1.4.1.14519.5.2.1.2783.4001.303234879376974091026102189271/1.3.6.1.4.1.14519.5.2.1.2783.4001.277699748513063132657345876103 |
| TCGA-GBM | TCGA-14 | TCGA-14-1825 | t1Gd | M | 70 | 76.65712 | t1_tra_post | 10 | 1.4078E+15 | 2/10/2000 | SIEMENS | MEDPC | Avanto | SE | 2D | 5 | 430 | 17 | 63.67925 | 1.5 | 6 | 90 | 0.299730615 | 27 | 320 | 144 | 0.71875\0.71875 | TCGA-14-1825/1.3.6.1.4.1.14519.5.2.1.2783.4001.303234879376974091026102189271/1.3.6.1.4.1.14519.5.2.1.2783.4001.258939865753180397841218680382 | |
| TCGA-GBM | TCGA-14 | TCGA-14-1825 | t1 | M | 70 | 76.65712 | t1_tra_pre | 7 | 1.4078E+15 | 2/10/2000 | SIEMENS | MEDPC | Avanto | SE | 2D | 5 | 430 | 17 | 63.67925 | 1.5 | 6 | 90 | 0.299730615 | 27 | 320 | 144 | 0.71875\0.71875 | TCGA-14-1825/1.3.6.1.4.1.14519.5.2.1.2783.4001.303234879376974091026102189271/1.3.6.1.4.1.14519.5.2.1.2783.4001.208384222600208194575599573994 | |
| TCGA-GBM | TCGA-14 | TCGA-14-1825 | t2 | M | 70 | 76.65712 | T2_AX_FS_post | 8 | 1.4078E+15 | 2/10/2000 | SIEMENS | MEDPC | Avanto | SE | 2D | 5 | 6070 | 110 | 63.67922 | 1.5 | 6 | 180 | 0.255179287 | 27 | 512 | 318 | 0.44921875\0.44921875 | TCGA-14-1825/1.3.6.1.4.1.14519.5.2.1.2783.4001.303234879376974091026102189271/1.3.6.1.4.1.14519.5.2.1.2783.4001.642315295692187367949394853394 | |
| TCGA-GBM | TCGA-14 | TCGA-14-1829 | flair | M | 57 | 86.18256 | t2_flair_ax | 3 | 1.63624E+15 | 6/14/2001 | SIEMENS | EUHMR2 | Avanto | SE\IR | 2D | 5 | 10000 | 130 | 2700 | 63.67914 | 1.5 | 6 | 180 | 0.317840978 | 27 | 256 | 172 | 0.44921875\0.44921875 | TCGA-14-1829/1.3.6.1.4.1.14519.5.2.1.2783.4001.103854061130261737009165000972/1.3.6.1.4.1.14519.5.2.1.2783.4001.501460447631638541537723941093 |
| TCGA-GBM | TCGA-14 | TCGA-14-1829 | t1Gd | M | 57 | 86.18256 | T1_3D_FFE | 9 | 1.63624E+15 | 6/14/2001 | SIEMENS | EUHMR2 | Avanto | GR | 3D | 1.5 | 25 | 9 | 63.67914 | 1.5 | 35 | 0.100076854 | 120 | 256 | 256 | 0.9375\0.9375 | TCGA-14-1829/1.3.6.1.4.1.14519.5.2.1.2783.4001.103854061130261737009165000972/1.3.6.1.4.1.14519.5.2.1.2783.4001.216813657680753995205839488022 | ||
| TCGA-GBM | TCGA-14 | TCGA-14-1829 | t1 | M | 57 | 86.18256 | t1_tra_pre_FIL | 7 | 1.63624E+15 | 6/14/2001 | SIEMENS | EUHMR2 | Avanto | SE | 2D | 5 | 454 | 17 | 63.67914 | 1.5 | 6 | 76 | 0.728576006 | 27 | 320 | 144 | 0.71875\0.71875 | TCGA-14-1829/1.3.6.1.4.1.14519.5.2.1.2783.4001.103854061130261737009165000972/1.3.6.1.4.1.14519.5.2.1.2783.4001.281678003339795573732020134118 | |
| TCGA-GBM | TCGA-14 | TCGA-14-1829 | t2 | M | 57 | 86.18256 | T2_AX_FS | 8 | 1.63624E+15 | 6/14/2001 | SIEMENS | EUHMR2 | Avanto | SE | 2D | 5 | 4450 | 110 | 63.6791 | 1.5 | 6 | 180 | 0.608993381 | 27 | 512 | 318 | 0.44921875\0.44921875 | TCGA-14-1829/1.3.6.1.4.1.14519.5.2.1.2783.4001.103854061130261737009165000972/1.3.6.1.4.1.14519.5.2.1.2783.4001.375367856236772946053988995526 | |
| TCGA-GBM | TCGA-14 | TCGA-14-3477 | flair | F | 38 | 58.96702 | t2_flair_ax | 7 | 2.67067E+15 | 5/1/2002 | SIEMENS | MEDPC | TrioTim | SE\IR | 2D | 5 | 8000 | 91 | 2371.5 | 123.2522 | 3 | 6 | 130 | 0.353191561 | 25 | 256 | 205 | 0.44921875\0.44921875 | TCGA-14-3477/1.3.6.1.4.1.14519.5.2.1.2783.4001.123124691157725520037267500618/1.3.6.1.4.1.14519.5.2.1.2783.4001.477389024768558126793730754601 |
| TCGA-GBM | TCGA-14 | TCGA-14-3477 | t1Gd | F | 38 | 58.96702 | t1_ax_post | 15 | 2.67067E+15 | 5/1/2002 | SIEMENS | MEDPC | TrioTim | SE | 2D | 5 | 500 | 8.5 | 123.2523 | 3 | 6 | 70 | 0.225981221 | 25 | 256 | 180 | 0.44921875\0.44921875 | TCGA-14-3477/1.3.6.1.4.1.14519.5.2.1.2783.4001.123124691157725520037267500618/1.3.6.1.4.1.14519.5.2.1.2783.4001.323059567715228362108477826515 | |
| TCGA-GBM | TCGA-14 | TCGA-14-3477 | t1 | F | 38 | 58.96702 | t1_ax_pre | 9 | 2.67067E+15 | 5/1/2002 | SIEMENS | MEDPC | TrioTim | SE | 2D | 5 | 600 | 8.5 | 123.2523 | 3 | 6 | 70 | 0.326445016 | 25 | 256 | 218 | 0.44921875\0.44921875 | TCGA-14-3477/1.3.6.1.4.1.14519.5.2.1.2783.4001.123124691157725520037267500618/1.3.6.1.4.1.14519.5.2.1.2783.4001.260352956461789136764709992439 | |
| TCGA-GBM | TCGA-14 | TCGA-14-3477 | t2 | F | 38 | 58.96702 | t2_fs_ax | 10 | 2.67067E+15 | 5/1/2002 | SIEMENS | MEDPC | TrioTim | SE | 2D | 5 | 6000 | 93 | 123.2522 | 3 | 6 | 120 | 0.479688506 | 25 | 320 | 320 | 0.359375\0.359375 | TCGA-14-3477/1.3.6.1.4.1.14519.5.2.1.2783.4001.123124691157725520037267500618/1.3.6.1.4.1.14519.5.2.1.2783.4001.126087670895542739221764627538 | |
| TCGA-GBM | TCGA-19 | TCGA-19-0963 | flair | M | 61 | 77.18 | MRHG_FLAIR_AX | 3 | 1.06342E+15 | 10/12/2001 | SIEMENS | Symphony | IR\SE | 2D | 5 | 9000 | 129 | 2500 | 63.65864 | 1.5 | 7.5 | 150 | 0.062243182 | 20 | 256 | 224 | 0.8984375\0.8984375 | TCGA-19-0963/1.3.6.1.4.1.14519.5.2.1.5826.4001.182689849979752622358485655607/1.3.6.1.4.1.14519.5.2.1.5826.4001.414254955526029720850488987168 | |
| TCGA-GBM | TCGA-19 | TCGA-19-0963 | t1Gd | M | 61 | 77.18 | MRHG_T1_AX_POST_GAD | 7 | 1.06342E+15 | 10/12/2001 | SIEMENS | Symphony | SE | 2D | 5 | 745 | 17 | 63.65864 | 1.5 | 7.5 | 90 | 0.293716699 | 20 | 256 | 224 | 0.8984375\0.8984375 | TCGA-19-0963/1.3.6.1.4.1.14519.5.2.1.5826.4001.182689849979752622358485655607/1.3.6.1.4.1.14519.5.2.1.5826.4001.203638647034312656830437984768 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-0963 | t1 | M | 61 | 77.18 | MRHG_T1_AX | 6 | 1.06342E+15 | 10/12/2001 | SIEMENS | Symphony | SE | 2D | 5 | 587 | 15 | 63.65864 | 1.5 | 7.5 | 90 | 0.132274136 | 20 | 256 | 224 | 0.8984375\0.8984375 | TCGA-19-0963/1.3.6.1.4.1.14519.5.2.1.5826.4001.182689849979752622358485655607/1.3.6.1.4.1.14519.5.2.1.5826.4001.524833529463103213283998865851 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-0963 | t2 | M | 61 | 77.18 | MRHG_T2_AXIALS | 2 | 1.06342E+15 | 10/12/2001 | SIEMENS | Symphony | SE | 2D | 5 | 4000 | 100 | 63.65863 | 1.5 | 7.5 | 150 | 0.185955927 | 20 | 256 | 224 | 0.8984375\0.8984375 | TCGA-19-0963/1.3.6.1.4.1.14519.5.2.1.5826.4001.182689849979752622358485655607/1.3.6.1.4.1.14519.5.2.1.5826.4001.122111680760213786740031783338 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-1390 | flair | F | 63 | 113.5 | MRHG_FLAIR_AX | 3 | 2.89638E+15 | 3/20/2001 | SIEMENS | Symphony | IR\SE | 2D | 5 | 9000 | 129 | 2500 | 63.50445 | 1.5 | 7 | 150 | 0.110135838 | 20 | 256 | 224 | 0.8984375\0.8984375 | TCGA-19-1390/1.3.6.1.4.1.14519.5.2.1.5826.4001.231567380210698672838678790326/1.3.6.1.4.1.14519.5.2.1.5826.4001.249851327270284089776000264301 | |
| TCGA-GBM | TCGA-19 | TCGA-19-1390 | t1Gd | F | 63 | 113.5 | MP_RAGE_AXIAL | 8 | 2.89638E+15 | 3/20/2001 | SIEMENS | Symphony | IR\GR | 3D | 1 | 2160 | 3.45 | 1100 | 63.50445 | 1.5 | 15 | 0.051605709 | 192 | 256 | 256 | 1\1 | TCGA-19-1390/1.3.6.1.4.1.14519.5.2.1.5826.4001.231567380210698672838678790326/1.3.6.1.4.1.14519.5.2.1.5826.4001.303700331855058891941526823748 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-1390 | t1 | F | 63 | 113.5 | MRHG_T1_AX | 4 | 2.89638E+15 | 3/20/2001 | SIEMENS | Symphony | SE | 2D | 5 | 587 | 15 | 63.50445 | 1.5 | 7 | 90 | 0.234051749 | 20 | 256 | 224 | 0.8984375\0.8984375 | TCGA-19-1390/1.3.6.1.4.1.14519.5.2.1.5826.4001.231567380210698672838678790326/1.3.6.1.4.1.14519.5.2.1.5826.4001.330679245867626368751388377114 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-1390 | t2 | F | 63 | 113.5 | MRHG_T2_AXIALS | 2 | 2.89638E+15 | 3/20/2001 | SIEMENS | Symphony | SE | 2D | 5 | 4000 | 100 | 63.5044 | 1.5 | 7 | 150 | 0.32903868 | 20 | 256 | 224 | 0.8984375\0.8984375 | TCGA-19-1390/1.3.6.1.4.1.14519.5.2.1.5826.4001.231567380210698672838678790326/1.3.6.1.4.1.14519.5.2.1.5826.4001.250425106424807032072862969016 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-1789 | flair | F | 69 | 79.83227 | FLAIR_AX | 8 | 3.12367E+15 | 8/10/2002 | SIEMENS | Avanto | SE\IR | 2D | 3 | 9430 | 109 | 2500 | 63.68429 | 1.5 | 3 | 150 | 0.239955982 | 60 | 256 | 232 | 1\1 | TCGA-19-1789/1.3.6.1.4.1.14519.5.2.1.5826.4001.231928631261949063719336921407/1.3.6.1.4.1.14519.5.2.1.5826.4001.235249827946511179930616208703 | |
| TCGA-GBM | TCGA-19 | TCGA-19-1789 | t1Gd | F | 69 | 79.83227 | VOLUMAXGAD | 12 | 3.12367E+15 | 8/10/2002 | SIEMENS | Avanto | GR\IR | 3D | 1 | 2160 | 2.81 | 1100 | 63.68429 | 1.5 | 15 | 0.059041861 | 192 | 256 | 256 | 1\1 | TCGA-19-1789/1.3.6.1.4.1.14519.5.2.1.5826.4001.231928631261949063719336921407/1.3.6.1.4.1.14519.5.2.1.5826.4001.110732246637966706387432991466 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-1789 | t1 | F | 69 | 79.83227 | MP_RAGE_AXIAL_VOLUMETRIC | 2 | 3.12367E+15 | 8/10/2002 | SIEMENS | Avanto | GR\IR | 3D | 1 | 2160 | 2.81 | 1100 | 63.68429 | 1.5 | 15 | 0.059041861 | 192 | 256 | 256 | 1\1 | TCGA-19-1789/1.3.6.1.4.1.14519.5.2.1.5826.4001.231928631261949063719336921407/1.3.6.1.4.1.14519.5.2.1.5826.4001.248150557952959587380829005187 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-1789 | t2 | F | 69 | 79.83227 | TSE_T2_ax | 9 | 3.12367E+15 | 8/10/2002 | SIEMENS | Avanto | SE | 2D | 3 | 3580 | 91 | 63.68434 | 1.5 | 3 | 180 | 0.703742258 | 60 | 256 | 232 | 0.5\0.5 | TCGA-19-1789/1.3.6.1.4.1.14519.5.2.1.5826.4001.231928631261949063719336921407/1.3.6.1.4.1.14519.5.2.1.5826.4001.296790073644899137258609675950 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-2624 | flair | M | 51 | 90.71849 | MRHR_FLAIR_AX | 3 | 1.73254E+15 | 12/10/2002 | SIEMENS | Avanto | SE\IR | 2D | 5 | 9000 | 109 | 2500 | 63.68411 | 1.5 | 7 | 150 | 0.13693494 | 25 | 256 | 192 | 0.95703125\0.95703125 | TCGA-19-2624/1.3.6.1.4.1.14519.5.2.1.5826.4001.209294630890894137037336921815/1.3.6.1.4.1.14519.5.2.1.5826.4001.237957332039616981825055417941 | |
| TCGA-GBM | TCGA-19 | TCGA-19-2624 | t1Gd | M | 51 | 90.71849 | MP_RAGE_AXIAL_VOLUMETRIC | 12 | 1.73254E+15 | 12/10/2002 | SIEMENS | Avanto | GR\IR | 3D | 1 | 2160 | 2.81 | 1100 | 63.68411 | 1.5 | 15 | 0.05179356 | 192 | 256 | 256 | 1\1 | TCGA-19-2624/1.3.6.1.4.1.14519.5.2.1.5826.4001.209294630890894137037336921815/1.3.6.1.4.1.14519.5.2.1.5826.4001.315244591826595784983776021079 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-2624 | t1 | M | 51 | 90.71849 | MRHR_T1_AX | 11 | 1.73254E+15 | 12/10/2002 | SIEMENS | Avanto | SE | 2D | 5 | 586 | 14 | 63.68411 | 1.5 | 7 | 70 | 0.419917791 | 25 | 256 | 192 | 0.95703125\0.95703125 | TCGA-19-2624/1.3.6.1.4.1.14519.5.2.1.5826.4001.209294630890894137037336921815/1.3.6.1.4.1.14519.5.2.1.5826.4001.288482729261153427645680784384 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-2624 | t2 | M | 51 | 90.71849 | MRHR_T2_AX | 2 | 1.73254E+15 | 12/10/2002 | SIEMENS | Avanto | SE | 2D | 5 | 4650 | 96 | 63.68408 | 1.5 | 7 | 150 | 0.236890556 | 25 | 256 | 192 | 0.95703125\0.95703125 | TCGA-19-2624/1.3.6.1.4.1.14519.5.2.1.5826.4001.209294630890894137037336921815/1.3.6.1.4.1.14519.5.2.1.5826.4001.648135048726787046712784361279 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-2631 | flair | F | 74 | 102.0583 | MRHR_FLAIR_AX | 4 | 2.58878E+15 | 3/2/2003 | SIEMENS | Avanto | SE\IR | 2D | 5 | 9000 | 109 | 2500 | 63.68498 | 1.5 | 5.5 | 150 | 0.125045094 | 28 | 256 | 192 | 0.8984375\0.8984375 | TCGA-19-2631/1.3.6.1.4.1.14519.5.2.1.5826.4001.113291808789528092767840707216/1.3.6.1.4.1.14519.5.2.1.5826.4001.152727517643491829458173260916 | |
| TCGA-GBM | TCGA-19 | TCGA-19-2631 | t1Gd | F | 74 | 102.0583 | MRHR_T1_AX_POST_GAD | 8 | 2.58878E+15 | 3/2/2003 | SIEMENS | Avanto | SE | 2D | 5 | 718 | 17 | 63.68498 | 1.5 | 5.5 | 90 | 0.170915377 | 28 | 256 | 192 | 0.8984375\0.8984375 | TCGA-19-2631/1.3.6.1.4.1.14519.5.2.1.5826.4001.113291808789528092767840707216/1.3.6.1.4.1.14519.5.2.1.5826.4001.328461846490310318014149271099 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-2631 | t1 | F | 74 | 102.0583 | MRHR_T1_AX | 7 | 2.58878E+15 | 3/2/2003 | SIEMENS | Avanto | SE | 2D | 5 | 532 | 10 | 63.68498 | 1.5 | 5.5 | 90 | 0.230387199 | 28 | 256 | 192 | 0.8984375\0.8984375 | TCGA-19-2631/1.3.6.1.4.1.14519.5.2.1.5826.4001.113291808789528092767840707216/1.3.6.1.4.1.14519.5.2.1.5826.4001.171072817207494283871306430547 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-2631 | t2 | F | 74 | 102.0583 | MRHR_T2_AX | 3 | 2.58878E+15 | 3/2/2003 | SIEMENS | Avanto | SE | 2D | 5 | 3800 | 95 | 63.68497 | 1.5 | 5.5 | 150 | 0.327169674 | 28 | 256 | 192 | 0.8984375\0.8984375 | TCGA-19-2631/1.3.6.1.4.1.14519.5.2.1.5826.4001.113291808789528092767840707216/1.3.6.1.4.1.14519.5.2.1.5826.4001.114530788702118112150801713711 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-5951 | flair | M | 76 | 93.89363 | FLAIR_AX | 3 | 3.39069E+15 | 9/14/2003 | SIEMENS | Verio | SE\IR | 2D | 4 | 9000 | 94 | 2500 | 123.2487 | 3 | 4.52 | 150 | 0.132235472 | 31 | 256 | 169 | 0.8984375\0.8984375 | TCGA-19-5951/1.3.6.1.4.1.14519.5.2.1.5826.4001.194554107490457210178645733278/1.3.6.1.4.1.14519.5.2.1.5826.4001.321497813789636909159148294851 | |
| TCGA-GBM | TCGA-19 | TCGA-19-5951 | t1Gd | M | 76 | 93.89363 | T1_AXIAL_Gd | 7 | 3.39069E+15 | 9/14/2003 | SIEMENS | Verio | GR | 2D | 4 | 250 | 2.48 | 123.2487 | 3 | 4.52 | 70 | 0.258163656 | 31 | 320 | 211 | 0.71875\0.71875 | TCGA-19-5951/1.3.6.1.4.1.14519.5.2.1.5826.4001.194554107490457210178645733278/1.3.6.1.4.1.14519.5.2.1.5826.4001.330225254008219946706138731112 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-5951 | t1 | M | 76 | 93.89363 | TIR_T1_AX | 6 | 3.39069E+15 | 9/14/2003 | SIEMENS | Verio | SE\IR | 2D | 4 | 2800 | 19 | 701.1 | 123.2487 | 3 | 4.52 | 150 | 0.309723401 | 31 | 256 | 169 | 0.8984375\0.8984375 | TCGA-19-5951/1.3.6.1.4.1.14519.5.2.1.5826.4001.194554107490457210178645733278/1.3.6.1.4.1.14519.5.2.1.5826.4001.555701715534477096875939011679 | |
| TCGA-GBM | TCGA-19 | TCGA-19-5951 | t2 | M | 76 | 93.89363 | T2_AX | 2 | 3.39069E+15 | 9/14/2003 | SIEMENS | Verio | SE | 2D | 4 | 5500 | 96 | 123.2487 | 3 | 4.52 | 150 | 0.265902937 | 31 | 320 | 211 | 0.71875\0.71875 | TCGA-19-5951/1.3.6.1.4.1.14519.5.2.1.5826.4001.194554107490457210178645733278/1.3.6.1.4.1.14519.5.2.1.5826.4001.932737691207898989363089745632 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-5954 | flair | F | 72 | 77.11071 | FLAIR_AX | 3 | 2.97506E+15 | 12/30/2003 | SIEMENS | Verio | SE\IR | 2D | 3 | 9000 | 94 | 2500 | 123.2487 | 3 | 3 | 150 | 0.330161553 | 54 | 256 | 256 | 1\1 | TCGA-19-5954/1.3.6.1.4.1.14519.5.2.1.5826.4001.321057407286032186200454220971/1.3.6.1.4.1.14519.5.2.1.5826.4001.253159030516730311576836577490 | |
| TCGA-GBM | TCGA-19 | TCGA-19-5954 | t1Gd | F | 72 | 77.11071 | VOLUMETRIC_AXIAL_Gd | 11 | 2.97506E+15 | 12/30/2003 | SIEMENS | Verio | GR\IR | 3D | 1 | 1900 | 2.75 | 900 | 123.2487 | 3 | 9 | 0.054662186 | 192 | 256 | 251 | 1\1 | TCGA-19-5954/1.3.6.1.4.1.14519.5.2.1.5826.4001.321057407286032186200454220971/1.3.6.1.4.1.14519.5.2.1.5826.4001.560018405631653221233558769839 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-5954 | t1 | F | 72 | 77.11071 | VOLUMETRIC_AXIAL | 2 | 2.97506E+15 | 12/30/2003 | SIEMENS | Verio | GR\IR | 3D | 1 | 1900 | 2.75 | 900 | 123.2487 | 3 | 9 | 0.054662186 | 192 | 256 | 251 | 1\1 | TCGA-19-5954/1.3.6.1.4.1.14519.5.2.1.5826.4001.321057407286032186200454220971/1.3.6.1.4.1.14519.5.2.1.5826.4001.295910608118826040218967173851 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-5954 | t2 | F | 72 | 77.11071 | T2_AX | 4 | 2.97506E+15 | 12/30/2003 | SIEMENS | Verio | SE | 2D | 3 | 5500 | 93 | 123.2487 | 3 | 3 | 150 | 0.334439301 | 54 | 256 | 256 | 1\1 | TCGA-19-5954/1.3.6.1.4.1.14519.5.2.1.5826.4001.321057407286032186200454220971/1.3.6.1.4.1.14519.5.2.1.5826.4001.301348699602097269733909521821 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-5958 | flair | M | 56 | 77.11071 | FLAIR_AX | 3 | 2.64703E+15 | 3/9/2004 | SIEMENS | Verio | SE\IR | 2D | 3 | 9000 | 94 | 2500 | 123.2486 | 3 | 3 | 150 | 0.253902656 | 54 | 256 | 256 | 1\1 | TCGA-19-5958/1.3.6.1.4.1.14519.5.2.1.5826.4001.192041630414624709736670800727/1.3.6.1.4.1.14519.5.2.1.5826.4001.151132655513156536888042112741 | |
| TCGA-GBM | TCGA-19 | TCGA-19-5958 | t1Gd | M | 56 | 77.11071 | VOLUMETRIC_AXIAL_Gd | 11 | 2.64703E+15 | 3/9/2004 | SIEMENS | Verio | GR\IR | 3D | 1 | 1900 | 2.75 | 900 | 123.2487 | 3 | 9 | 0.042036613 | 192 | 256 | 251 | 1\1 | TCGA-19-5958/1.3.6.1.4.1.14519.5.2.1.5826.4001.192041630414624709736670800727/1.3.6.1.4.1.14519.5.2.1.5826.4001.157222381431362159457522834384 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-5958 | t1 | M | 56 | 77.11071 | VOLUMETRIC_AXIAL | 2 | 2.64703E+15 | 3/9/2004 | SIEMENS | Verio | GR\IR | 3D | 1 | 1900 | 2.75 | 900 | 123.2487 | 3 | 9 | 0.042036613 | 192 | 256 | 251 | 1\1 | TCGA-19-5958/1.3.6.1.4.1.14519.5.2.1.5826.4001.192041630414624709736670800727/1.3.6.1.4.1.14519.5.2.1.5826.4001.179990125364012900028568210197 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-5958 | t2 | M | 56 | 77.11071 | T2_AX | 4 | 2.64703E+15 | 3/9/2004 | SIEMENS | Verio | SE | 2D | 3 | 5500 | 93 | 123.2487 | 3 | 3 | 150 | 0.257192359 | 54 | 256 | 256 | 1\1 | TCGA-19-5958/1.3.6.1.4.1.14519.5.2.1.5826.4001.192041630414624709736670800727/1.3.6.1.4.1.14519.5.2.1.5826.4001.307938529330684497954612875613 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-5960 | flair | M | 56 | 72.57479 | FLAIR_AX | 7 | 3.03248E+15 | 3/15/2004 | SIEMENS | Verio | SE\IR | 2D | 4 | 9000 | 94 | 2500 | 123.2486 | 3 | 4.52 | 150 | 0.12577795 | 34 | 256 | 183 | 0.8984375\0.8984375 | TCGA-19-5960/1.3.6.1.4.1.14519.5.2.1.5826.4001.135874542095979859687918030388/1.3.6.1.4.1.14519.5.2.1.5826.4001.186796014901942467105889390386 | |
| TCGA-GBM | TCGA-19 | TCGA-19-5960 | t1Gd | M | 56 | 72.57479 | T1_AXIAL_Gd | 9 | 3.03248E+15 | 3/15/2004 | SIEMENS | Verio | GR | 2D | 4 | 250 | 2.48 | 123.2487 | 3 | 4.52 | 70 | 0.246707612 | 34 | 256 | 183 | 0.8984375\0.8984375 | TCGA-19-5960/1.3.6.1.4.1.14519.5.2.1.5826.4001.135874542095979859687918030388/1.3.6.1.4.1.14519.5.2.1.5826.4001.330951756415447864524009831768 | ||
| TCGA-GBM | TCGA-19 | TCGA-19-5960 | t1 | M | 56 | 72.57479 | TIR_T1_AX | 8 | 3.03248E+15 | 3/15/2004 | SIEMENS | Verio | SE\IR | 2D | 4 | 2990 | 19 | 715.6 | 123.2487 | 3 | 4.52 | 150 | 0.24781075 | 34 | 256 | 183 | 0.8984375\0.8984375 | TCGA-19-5960/1.3.6.1.4.1.14519.5.2.1.5826.4001.135874542095979859687918030388/1.3.6.1.4.1.14519.5.2.1.5826.4001.566265127405363595842987702076 | |
| TCGA-GBM | TCGA-19 | TCGA-19-5960 | t2 | M | 56 | 72.57479 | T2_AX | 3 | 3.03248E+15 | 3/15/2004 | SIEMENS | Verio | SE | 2D | 4 | 5500 | 93 | 123.2487 | 3 | 4.52 | 150 | 0.243332034 | 34 | 256 | 183 | 0.8984375\0.8984375 | TCGA-19-5960/1.3.6.1.4.1.14519.5.2.1.5826.4001.135874542095979859687918030388/1.3.6.1.4.1.14519.5.2.1.5826.4001.139985413894688673535204262164 | ||
| TCGA-GBM | TCGA-27 | TCGA-27-1834 | flair | M | 57 | 65 | T2_Flair_cor_5mm_5_9_07 | 4 | 3.05234E+15 | 11/3/1987 | SIEMENS | Avanto | SE\IR | 2D | 5 | 8200 | 91 | 2500 | 63.63845 | 1.5 | 6.5 | 150 | 0.077359584 | 25 | 320 | 230 | 0.78125\0.78125 | TCGA-27-1834/1.3.6.1.4.1.14519.5.2.1.3775.4001.257288270999874248292312942337/1.3.6.1.4.1.14519.5.2.1.3775.4001.220362068065489352287173639632 | |
| TCGA-GBM | TCGA-27 | TCGA-27-1834 | t1Gd | M | 57 | 65 | t1_se_tra_5_9_07 | 7 | 3.05234E+15 | 11/3/1987 | SIEMENS | Avanto | SE | 2D | 6 | 477 | 10 | 63.63845 | 1.5 | 7.8 | 70 | 0.270370927 | 20 | 256 | 224 | 0.44921875\0.44921875 | TCGA-27-1834/1.3.6.1.4.1.14519.5.2.1.3775.4001.257288270999874248292312942337/1.3.6.1.4.1.14519.5.2.1.3775.4001.578556393784694438698235299037 | ||
| TCGA-GBM | TCGA-27 | TCGA-27-1834 | t1 | M | 57 | 65 | t1_se_tra_5_9_07 | 5 | 3.05234E+15 | 11/3/1987 | SIEMENS | Avanto | SE | 2D | 6 | 477 | 10 | 63.63845 | 1.5 | 7.8 | 70 | 0.270370927 | 20 | 256 | 224 | 0.44921875\0.44921875 | TCGA-27-1834/1.3.6.1.4.1.14519.5.2.1.3775.4001.257288270999874248292312942337/1.3.6.1.4.1.14519.5.2.1.3775.4001.187906847315134178012322222812 | ||
| TCGA-GBM | TCGA-27 | TCGA-27-1834 | t2 | M | 57 | 65 | pd+t2_tse_tra_5_9_07 | 3 | 3.05234E+15 | 11/3/1987 | SIEMENS | Avanto | SE | 2D | 5 | 3621.9 | 13 | 63.63845 | 1.5 | 6.5 | 150 | 0.30795125 | 50 | 320 | 198 | 0.71875\0.71875 | TCGA-27-1834/1.3.6.1.4.1.14519.5.2.1.3775.4001.257288270999874248292312942337/1.3.6.1.4.1.14519.5.2.1.3775.4001.364247940891429788631023542309 | ||
| TCGA-GBM | TCGA-27 | TCGA-27-1838 | flair | M | 75 | ET2W/FLAIR_ | 302 | 2.86841E+15 | 8/29/1988 | Philips Medical Systems | Intera | IR | 6 | 6000 | 120 | 1900 | 21.29607 | 0.5 | 7.2 | 90 | 20 | 256 | 176 | 0.8984375\0.8984375 | TCGA-27-1838/1.3.6.1.4.1.14519.5.2.1.3775.4001.111712322912660120243025601400/1.3.6.1.4.1.14519.5.2.1.3775.4001.119287179819363124512383566810 | ||||
| TCGA-GBM | TCGA-27 | TCGA-27-1838 | t1Gd | M | 75 | Egad | 502 | 2.86841E+15 | 8/29/1988 | Philips Medical Systems | Intera | SE | 6 | 542.022 | 15 | 21.29607 | 0.5 | 6.6 | 60 | 20 | 224 | 179 | 0.9375\0.9375 | TCGA-27-1838/1.3.6.1.4.1.14519.5.2.1.3775.4001.111712322912660120243025601400/1.3.6.1.4.1.14519.5.2.1.3775.4001.994851682380835865056380711802 | |||||
| TCGA-GBM | TCGA-27 | TCGA-27-1838 | t1 | M | 75 | Et1_se_tra | 402 | 2.86841E+15 | 8/29/1988 | Philips Medical Systems | Intera | SE | 6 | 542.022 | 15 | 21.29607 | 0.5 | 6.6 | 60 | 20 | 224 | 179 | 0.9375\0.9375 | TCGA-27-1838/1.3.6.1.4.1.14519.5.2.1.3775.4001.111712322912660120243025601400/1.3.6.1.4.1.14519.5.2.1.3775.4001.788024501395553679565117126823 | |||||
| TCGA-GBM | TCGA-27 | TCGA-27-1838 | t2 | M | 75 | Edp+t2_se_t | 202 | 2.86841E+15 | 8/29/1988 | Philips Medical Systems | Intera | SE | 6 | 2219.564 | 20 | 21.29607 | 0.5 | 6.6 | 90 | 40 | 256 | 192 | 0.9375\0.9375 | TCGA-27-1838/1.3.6.1.4.1.14519.5.2.1.3775.4001.111712322912660120243025601400/1.3.6.1.4.1.14519.5.2.1.3775.4001.957862673647515802653015684121 | |||||
| TCGA-GBM | TCGA-27 | TCGA-27-2526 | flair | M | 77 | EFLAIR/long | 402 | 2.29145E+15 | 3/7/1988 | Philips Medical Systems | Intera | IR | 6 | 11000 | 140 | 2800 | 21.29607 | 0.5 | 7 | 90 | 20 | 256 | 161 | 0.9375\0.9375 | TCGA-27-2526/1.3.6.1.4.1.14519.5.2.1.3775.4001.338505699209290103023897865510/1.3.6.1.4.1.14519.5.2.1.3775.4001.242527138671194700564129592420 | ||||
| TCGA-GBM | TCGA-27 | TCGA-27-2526 | t1Gd | M | 77 | neuronavigaT1W/3D/GAD | 501 | 2.29145E+15 | 3/7/1988 | Philips Medical Systems | Intera | GR | 2 | 30.3 | 13.81153 | 21.29607 | 0.5 | 2 | 30 | 80 | 256 | 256 | 1.0\1.0 | TCGA-27-2526/1.3.6.1.4.1.14519.5.2.1.3775.4001.338505699209290103023897865510/1.3.6.1.4.1.14519.5.2.1.3775.4001.199545238646153862339050303771 | |||||
| TCGA-GBM | TCGA-27 | TCGA-27-2526 | t1 | M | 77 | Et1_se_tra | 302 | 2.29145E+15 | 3/7/1988 | Philips Medical Systems | Intera | SE | 6 | 541.926 | 15 | 21.29607 | 0.5 | 6.6 | 60 | 20 | 256 | 179 | 0.9375\0.9375 | TCGA-27-2526/1.3.6.1.4.1.14519.5.2.1.3775.4001.338505699209290103023897865510/1.3.6.1.4.1.14519.5.2.1.3775.4001.290126392868809844278609708560 | |||||
| TCGA-GBM | TCGA-27 | TCGA-27-2526 | t2 | M | 77 | Edp+t2_se_t | 202 | 2.29145E+15 | 3/7/1988 | Philips Medical Systems | Intera | SE | 6 | 2219.564 | 20 | 21.29607 | 0.5 | 6.6 | 90 | 40 | 256 | 179 | 0.9375\0.9375 | TCGA-27-2526/1.3.6.1.4.1.14519.5.2.1.3775.4001.338505699209290103023897865510/1.3.6.1.4.1.14519.5.2.1.3775.4001.337166470300809460748886608281 | |||||
| TCGA-GBM | TCGA-76 | TCGA-76-4932 | flair | F | 50 | 77 | FAST_BRAIN/FLAIR_AXIAL | 6 | 2.08702E+15 | 3/16/1997 | SIEMENS | mrscan | MAGNETOM VISION | IR | 2D | 5 | 7000 | 105 | 2250 | 63.61525 | 1.493806 | 2.5 | 180 | 0 | 178 | 512 | 140 | 008.984375E-01\08.984375E-01 | TCGA-76-4932/1.3.6.1.4.1.14519.5.2.1.1188.4001.696081929184000436121119244803/1.3.6.1.4.1.14519.5.2.1.1188.4001.325184889034836529092840230248 |
| TCGA-GBM | TCGA-76 | TCGA-76-4932 | t1Gd | F | 50 | 77 | BRAIN_STRYKER/t1_AX | 8 | 2.08702E+15 | 3/16/1997 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 880 | 12 | 63.61548 | 1.493806 | 2.5 | 70 | 0 | 258 | 512 | 144 | 008.593750E-01\08.593750E-01 | TCGA-76-4932/1.3.6.1.4.1.14519.5.2.1.1188.4001.696081929184000436121119244803/1.3.6.1.4.1.14519.5.2.1.1188.4001.127105277778356529905172144511 | |
| TCGA-GBM | TCGA-76 | TCGA-76-4932 | t1 | F | 50 | 77 | BRAIN_STRYKER/T1_AXIAL | 3 | 2.08702E+15 | 3/16/1997 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 540 | 12 | 63.61548 | 1.493806 | 2.5 | 70 | 0 | 39 | 512 | 144 | 008.593750E-01\08.593750E-01 | TCGA-76-4932/1.3.6.1.4.1.14519.5.2.1.1188.4001.696081929184000436121119244803/1.3.6.1.4.1.14519.5.2.1.1188.4001.336336678889681314725089183401 | |
| TCGA-GBM | TCGA-76 | TCGA-76-4932 | t2 | F | 50 | 77 | BRAIN_STRYKER/T2_AX_DOUBLE | 4 | 2.08702E+15 | 3/16/1997 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 2020 | 20 | 63.61548 | 1.493806 | 2.5 | 90 | 0 | 79 | 512 | 221 | 004.296875E-01\04.296875E-01 | TCGA-76-4932/1.3.6.1.4.1.14519.5.2.1.1188.4001.696081929184000436121119244803/1.3.6.1.4.1.14519.5.2.1.1188.4001.210657790793239037253399861860 | |
| TCGA-GBM | TCGA-76 | TCGA-76-4934 | flair | F | 100 | T2_AX_FLAIR | 601 | 7.26288E+15 | 10/8/2000 | Philips Medical Systems | PHILIPS-13EEFF6 | Achieva | IR | 2D | 5 | 11000 | 125 | 2850 | 127.796 | 3 | 6 | 90 | 0.90577203 | 25 | 336 | 255 | 0.68452382087707\0.68452382087707 | TCGA-76-4934/1.3.6.1.4.1.14519.5.2.1.1188.4001.280345518346345066284488429203/1.3.6.1.4.1.14519.5.2.1.1188.4001.286635506916337591478062137823 | |
| TCGA-GBM | TCGA-76 | TCGA-76-4934 | t1Gd | F | 100 | T1_AX_SE_FS_POST | 1401 | 7.26288E+15 | 10/8/2000 | Philips Medical Systems | PHILIPS-13EEFF6 | Achieva | SE | 2D | 4 | 699.84 | 10 | 127.7961 | 3 | 5 | 90 | 3.200000048 | 32 | 284 | 228 | 0.71875\0.71875 | TCGA-76-4934/1.3.6.1.4.1.14519.5.2.1.1188.4001.280345518346345066284488429203/1.3.6.1.4.1.14519.5.2.1.1188.4001.301507599146459685277894885768 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-4934 | t1 | F | 100 | T1_AX__SE | 901 | 7.26288E+15 | 10/8/2000 | Philips Medical Systems | PHILIPS-13EEFF6 | Achieva | SE | 2D | 4 | 565.0768 | 10.3 | 127.7961 | 3 | 5 | 90 | 3.199997187 | 32 | 316 | 255 | 0.68452382087707\0.68452382087707 | TCGA-76-4934/1.3.6.1.4.1.14519.5.2.1.1188.4001.280345518346345066284488429203/1.3.6.1.4.1.14519.5.2.1.1188.4001.338412102935503631330631798910 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-4934 | t2 | F | 100 | AXIAL__T2 | 701 | 7.26288E+15 | 10/8/2000 | Philips Medical Systems | PHILIPS-13EEFF6 | Achieva | SE | 2D | 4 | 3361.032 | 100 | 127.796 | 3 | 5 | 90 | 3.160874844 | 32 | 404 | 340 | 0.44921875\0.44921875 | TCGA-76-4934/1.3.6.1.4.1.14519.5.2.1.1188.4001.280345518346345066284488429203/1.3.6.1.4.1.14519.5.2.1.1188.4001.134660201518103802336514714033 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-4935 | flair | F | 65 | T2_AX_FLAIR | 801 | 2.56097E+15 | 1/22/2001 | Philips Medical Systems | PHILIPS-13EEFF6 | Achieva | IR | 2D | 5 | 11000 | 125 | 2850 | 127.7953 | 3 | 6 | 90 | 1.033632755 | 28 | 348 | 272 | 0.68181818723678\0.68181818723678 | TCGA-76-4935/1.3.6.1.4.1.14519.5.2.1.1188.4001.279290817710613414079827510861/1.3.6.1.4.1.14519.5.2.1.1188.4001.198703612394476952929652973732 | |
| TCGA-GBM | TCGA-76 | TCGA-76-4935 | t1Gd | F | 65 | T1_AX_SE_FS_POST | 1301 | 2.56097E+15 | 1/22/2001 | Philips Medical Systems | PHILIPS-13EEFF6 | Achieva | SE | 2D | 4 | 533.63 | 10 | 127.7954 | 3 | 5 | 90 | 3.199999571 | 30 | 296 | 266 | 0.75\0.75 | TCGA-76-4935/1.3.6.1.4.1.14519.5.2.1.1188.4001.279290817710613414079827510861/1.3.6.1.4.1.14519.5.2.1.1188.4001.120015554026401136166447740917 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-4935 | t1 | F | 65 | T1_AX__SE | 1101 | 2.56097E+15 | 1/22/2001 | Philips Medical Systems | PHILIPS-13EEFF6 | Achieva | SE | 2D | 4 | 719.0448 | 10.3 | 127.7954 | 3 | 5 | 90 | 3.199997187 | 32 | 332 | 266 | 0.68181818723678\0.68181818723678 | TCGA-76-4935/1.3.6.1.4.1.14519.5.2.1.1188.4001.279290817710613414079827510861/1.3.6.1.4.1.14519.5.2.1.1188.4001.210457495932820162711563788216 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-4935 | t2 | F | 65 | AXIAL__T2 | 901 | 2.56097E+15 | 1/22/2001 | Philips Medical Systems | PHILIPS-13EEFF6 | Achieva | SE | 2D | 4 | 3367.982 | 100 | 127.7953 | 3 | 5 | 90 | 3.15435195 | 32 | 420 | 337 | 0.42857143282890\0.42857143282890 | TCGA-76-4935/1.3.6.1.4.1.14519.5.2.1.1188.4001.279290817710613414079827510861/1.3.6.1.4.1.14519.5.2.1.1188.4001.165274213290821040312747820547 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-6191 | flair | M | 57 | 136 | BRAIN_CONTRAST/FLAIR | 3 | 9.71383E+15 | 10/20/2000 | SIEMENS | mrscan | MAGNETOM VISION | IR | 2D | 5 | 9000 | 105 | 2340 | 63.62077 | 1.493806 | 3.25 | 180 | 0 | 43 | 512 | 112 | 009.765625E-01\09.765625E-01 | TCGA-76-6191/1.3.6.1.4.1.14519.5.2.1.1188.4001.237134987835312583841357983114/1.3.6.1.4.1.14519.5.2.1.1188.4001.269462493891859236294268767034 |
| TCGA-GBM | TCGA-76 | TCGA-76-6191 | t1Gd | M | 57 | 136 | BRAIN_CONTRAST/T1_AXIAL | 4 | 9.71383E+15 | 10/20/2000 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 600 | 14 | 63.62077 | 1.493806 | 3.25 | 70 | 0 | 63 | 512 | 115 | 009.765625E-01\09.765625E-01 | TCGA-76-6191/1.3.6.1.4.1.14519.5.2.1.1188.4001.237134987835312583841357983114/1.3.6.1.4.1.14519.5.2.1.1188.4001.193088623684162190916524071714 | |
| TCGA-GBM | TCGA-76 | TCGA-76-6191 | t1 | M | 57 | 136 | BRAIN_STRYKER/t1_AX | 9 | 9.71383E+15 | 10/20/2000 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 440 | 12 | 63.62068 | 1.493806 | 3.25 | 70 | 0 | 343 | 512 | 134 | 009.765625E-01\09.765625E-01 | TCGA-76-6191/1.3.6.1.4.1.14519.5.2.1.1188.4001.237134987835312583841357983114/1.3.6.1.4.1.14519.5.2.1.1188.4001.139468407540359589585598023466 | |
| TCGA-GBM | TCGA-76 | TCGA-76-6191 | t2 | M | 57 | 136 | BRAIN_CONTRAST/DOUBLE_ECHO | 5 | 9.71383E+15 | 10/20/2000 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 2020 | 20 | 63.62077 | 1.493806 | 3.25 | 90 | 0 | 103 | 512 | 115 | 004.882813E-01\04.882813E-01 | TCGA-76-6191/1.3.6.1.4.1.14519.5.2.1.1188.4001.237134987835312583841357983114/1.3.6.1.4.1.14519.5.2.1.1188.4001.714380615105580420167556298073 | |
| TCGA-GBM | TCGA-76 | TCGA-76-6193 | flair | M | 78 | 86 | BRAIN_NON_CONTRAST/FLAIR_A | 4 | 2.29039E+15 | 6/2/2001 | SIEMENS | mrscan | MAGNETOM VISION | IR | 2D | 5 | 9000 | 105 | 2340 | 63.62074 | 1.493806 | 2.5 | 180 | 0 | 59 | 512 | 126 | 008.593750E-01\08.593750E-01 | TCGA-76-6193/1.3.6.1.4.1.14519.5.2.1.1188.4001.532365074075118365845331160175/1.3.6.1.4.1.14519.5.2.1.1188.4001.331171186011270999664762183981 |
| TCGA-GBM | TCGA-76 | TCGA-76-6193 | t1Gd | M | 78 | 86 | BRAIN_STRYKER/AX-RAGE-STRY | 9 | 2.29039E+15 | 6/2/2001 | SIEMENS | mrscan | MAGNETOM VISION | IR | 3D | 1.5 | 15 | 7 | 300 | 63.62069 | 1.493806 | 15 | 0 | 339 | 512 | 256 | 005.078125E-01\05.078125E-01 | TCGA-76-6193/1.3.6.1.4.1.14519.5.2.1.1188.4001.532365074075118365845331160175/1.3.6.1.4.1.14519.5.2.1.1188.4001.140712239861920670139072389080 | |
| TCGA-GBM | TCGA-76 | TCGA-76-6193 | t1 | M | 78 | 86 | BRAIN_STRYKER/t1_AX | 10 | 2.29039E+15 | 6/2/2001 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 540 | 12 | 63.62069 | 1.493806 | 2.500004 | 70 | 0 | 359 | 512 | 134 | 008.593750E-01\08.593750E-01 | TCGA-76-6193/1.3.6.1.4.1.14519.5.2.1.1188.4001.532365074075118365845331160175/1.3.6.1.4.1.14519.5.2.1.1188.4001.164007274133639277695926891010 | |
| TCGA-GBM | TCGA-76 | TCGA-76-6193 | t2 | M | 78 | 86 | BRAIN_CONTRAST/DOUBLE_ECHO | 7 | 2.29039E+15 | 6/2/2001 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 2020 | 20 | 63.62074 | 1.493806 | 2.499951 | 90 | 0 | 139 | 512 | 115 | 004.296875E-01\04.296875E-01 | TCGA-76-6193/1.3.6.1.4.1.14519.5.2.1.1188.4001.532365074075118365845331160175/1.3.6.1.4.1.14519.5.2.1.1188.4001.520134226758299999468011999899 | |
| TCGA-GBM | TCGA-76 | TCGA-76-6280 | flair | M | 79 | T2_AX_FLAIR | 401 | 1.39014E+15 | 7/21/1998 | Philips Medical Systems | PMSN-1DL0VL9SN4 | Achieva | IR | 5 | 6000 | 120 | 2000 | 63.90027 | 1.5 | 6 | 90 | 28 | 256 | 161 | 0.859375\0.859375 | TCGA-76-6280/1.3.6.1.4.1.14519.5.2.1.1188.4001.259303451742499057480927151400/1.3.6.1.4.1.14519.5.2.1.1188.4001.274502557202127285948259115378 | |||
| TCGA-GBM | TCGA-76 | TCGA-76-6280 | t1Gd | M | 79 | T1_SE_POST | 701 | 1.39014E+15 | 7/21/1998 | Philips Medical Systems | PMSN-1DL0VL9SN4 | Achieva | SE | 5 | 589.224 | 12 | 63.90028 | 1.5 | 7 | 69 | 25 | 256 | 204 | 0.859375\0.859375 | TCGA-76-6280/1.3.6.1.4.1.14519.5.2.1.1188.4001.259303451742499057480927151400/1.3.6.1.4.1.14519.5.2.1.1188.4001.259307244299991614762344622590 | ||||
| TCGA-GBM | TCGA-76 | TCGA-76-6280 | t1 | M | 79 | T1_SE_PRE | 601 | 1.39014E+15 | 7/21/1998 | Philips Medical Systems | PMSN-1DL0VL9SN4 | Achieva | SE | 5 | 413.72 | 12 | 63.90027 | 1.5 | 7 | 69 | 25 | 256 | 204 | 0.859375\0.859375 | TCGA-76-6280/1.3.6.1.4.1.14519.5.2.1.1188.4001.259303451742499057480927151400/1.3.6.1.4.1.14519.5.2.1.1188.4001.261189460146477809142077908700 | ||||
| TCGA-GBM | TCGA-76 | TCGA-76-6280 | t2 | M | 79 | DUAL_AX_T2 | 501 | 1.39014E+15 | 7/21/1998 | Philips Medical Systems | PMSN-1DL0VL9SN4 | Achieva | SE | 5 | 2991.893 | 30 | 63.90027 | 1.5 | 7.5 | 90 | 48 | 256 | 153 | 0.859375\0.859375 | TCGA-76-6280/1.3.6.1.4.1.14519.5.2.1.1188.4001.259303451742499057480927151400/1.3.6.1.4.1.14519.5.2.1.1188.4001.113253988688109445728345358337 | ||||
| TCGA-GBM | TCGA-76 | TCGA-76-6282 | flair | M | 63 | 91 | BRAIN_STRYKER/FLAIR | 6 | 5.47453E+15 | 8/24/1998 | SIEMENS | mrscan | MAGNETOM VISION | IR | 2D | 5 | 9000 | 105 | 2340 | 63.61581 | 1.493806 | 2.5 | 180 | 0 | 104 | 512 | 140 | 008.593750E-01\08.593750E-01 | TCGA-76-6282/1.3.6.1.4.1.14519.5.2.1.1188.4001.623989292006918600441736922866/1.3.6.1.4.1.14519.5.2.1.1188.4001.337742353862340790005797114133 |
| TCGA-GBM | TCGA-76 | TCGA-76-6282 | t1Gd | M | 63 | 91 | BRAIN_STRYKER/AX-RAGE-STRY | 8 | 5.47453E+15 | 8/24/1998 | SIEMENS | mrscan | MAGNETOM VISION | IR | 3D | 1.5 | 15 | 7 | 300 | 63.61575 | 1.493806 | 15 | 0 | 304 | 512 | 179 | 004.687500E-01\04.687500E-01 | TCGA-76-6282/1.3.6.1.4.1.14519.5.2.1.1188.4001.623989292006918600441736922866/1.3.6.1.4.1.14519.5.2.1.1188.4001.120631863329002959798073074121 | |
| TCGA-GBM | TCGA-76 | TCGA-76-6282 | t1 | M | 63 | 91 | BRAIN_STRYKER/T1_AXIAL | 4 | 5.47453E+15 | 8/24/1998 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 540 | 12 | 63.61574 | 1.493806 | 2.5 | 70 | 0 | 44 | 512 | 134 | 008.593750E-01\08.593750E-01 | TCGA-76-6282/1.3.6.1.4.1.14519.5.2.1.1188.4001.623989292006918600441736922866/1.3.6.1.4.1.14519.5.2.1.1188.4001.288195363293492828132191428304 | |
| TCGA-GBM | TCGA-76 | TCGA-76-6282 | t2 | M | 63 | 91 | BRAIN_STRYKER/T2_AX_DOUBLE | 5 | 5.47453E+15 | 8/24/1998 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 2020 | 20 | 63.61574 | 1.493806 | 2.5 | 90 | 0 | 84 | 512 | 134 | 004.296875E-01\04.296875E-01 | TCGA-76-6282/1.3.6.1.4.1.14519.5.2.1.1188.4001.623989292006918600441736922866/1.3.6.1.4.1.14519.5.2.1.1188.4001.404945593685724778366507370745 | |
| TCGA-GBM | TCGA-76 | TCGA-76-6285 | flair | F | 64 | 136 | BRAIN_STRYKER/FLAIR | 4 | 8.29785E+15 | 12/19/1998 | SIEMENS | mrscan | MAGNETOM VISION | IR | 2D | 5 | 9000 | 105 | 2340 | 63.6157 | 1.493806 | 2 | 180 | 0 | 61 | 512 | 98 | 009.375000E-01\09.375000E-01 | TCGA-76-6285/1.3.6.1.4.1.14519.5.2.1.1188.4001.440169796153998480809455332999/1.3.6.1.4.1.14519.5.2.1.1188.4001.284225597658034426109919290461 |
| TCGA-GBM | TCGA-76 | TCGA-76-6285 | t1Gd | F | 64 | 136 | BRAIN_STRYKER/t1_AX | 7 | 8.29785E+15 | 12/19/1998 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 440 | 12 | 63.61561 | 1.493806 | 2 | 70 | 0 | 181 | 512 | 134 | 009.375000E-01\09.375000E-01 | TCGA-76-6285/1.3.6.1.4.1.14519.5.2.1.1188.4001.440169796153998480809455332999/1.3.6.1.4.1.14519.5.2.1.1188.4001.239825616284727719466680894711 | |
| TCGA-GBM | TCGA-76 | TCGA-76-6285 | t1 | F | 64 | 136 | BRAIN_STRYKER/T1_AXIAL | 3 | 8.29785E+15 | 12/19/1998 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 440 | 12 | 63.61568 | 1.493806 | 2 | 70 | 0 | 41 | 512 | 134 | 009.375000E-01\09.375000E-01 | TCGA-76-6285/1.3.6.1.4.1.14519.5.2.1.1188.4001.440169796153998480809455332999/1.3.6.1.4.1.14519.5.2.1.1188.4001.175007799198843363270738924370 | |
| TCGA-GBM | TCGA-76 | TCGA-76-6285 | t2 | F | 64 | 136 | RETRO_ORBIT/T2_LATE_ECHO_B | 5 | 8.29785E+15 | 12/19/1998 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 4700 | 120 | 63.6157 | 1.493806 | 2 | 180 | 0 | 81 | 512 | 150 | 009.375000E-01\09.375000E-01 | TCGA-76-6285/1.3.6.1.4.1.14519.5.2.1.1188.4001.440169796153998480809455332999/1.3.6.1.4.1.14519.5.2.1.1188.4001.140445044250167916734736842790 | |
| TCGA-GBM | TCGA-76 | TCGA-76-6656 | flair | M | 68 | T2_AX_FLAIR | 601 | 2.06192E+15 | 6/2/2001 | Philips Medical Systems | PHILIPS-13EEFF6 | Achieva | IR | 2D | 5 | 11000 | 125 | 2850 | 127.7946 | 3 | 6 | 90 | 0.984625876 | 28 | 336 | 255 | 0.68452382087707\0.68452382087707 | TCGA-76-6656/1.3.6.1.4.1.14519.5.2.1.1188.4001.313245752188924211777085602901/1.3.6.1.4.1.14519.5.2.1.1188.4001.313956114391186647662214330900 | |
| TCGA-GBM | TCGA-76 | TCGA-76-6656 | t1Gd | M | 68 | T1_AX_SE_FS_POST | 1101 | 2.06192E+15 | 6/2/2001 | Philips Medical Systems | PHILIPS-13EEFF6 | Achieva | SE | 2D | 4 | 656.1 | 10 | 127.7947 | 3 | 5 | 90 | 3.200000048 | 30 | 284 | 228 | 0.71875\0.71875 | TCGA-76-6656/1.3.6.1.4.1.14519.5.2.1.1188.4001.313245752188924211777085602901/1.3.6.1.4.1.14519.5.2.1.1188.4001.307641110251975013714713829108 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-6656 | t1 | M | 68 | T1_AX__SE | 901 | 2.06192E+15 | 6/2/2001 | Philips Medical Systems | PHILIPS-13EEFF6 | Achieva | SE | 2D | 4 | 529.7595 | 10.3 | 127.7946 | 3 | 5 | 90 | 3.199996948 | 30 | 316 | 255 | 0.68452382087707\0.68452382087707 | TCGA-76-6656/1.3.6.1.4.1.14519.5.2.1.1188.4001.313245752188924211777085602901/1.3.6.1.4.1.14519.5.2.1.1188.4001.234057418002688075397382125444 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-6656 | t2 | M | 68 | AXIAL__T2 | 701 | 2.06192E+15 | 6/2/2001 | Philips Medical Systems | PHILIPS-13EEFF6 | Achieva | SE | 2D | 4 | 3143.824 | 100 | 127.7946 | 3 | 5 | 90 | 3.168056727 | 30 | 404 | 340 | 0.44921875\0.44921875 | TCGA-76-6656/1.3.6.1.4.1.14519.5.2.1.1188.4001.313245752188924211777085602901/1.3.6.1.4.1.14519.5.2.1.1188.4001.175702182191848905428876073118 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-6657 | flair | M | 90 | T2_AX_FLAIR | 501 | 6.29841E+15 | 6/11/2001 | Philips Medical Systems | MR1 | Achieva | IR | 2D | 5 | 11000 | 125 | 2850 | 127.7817 | 3 | 6 | 90 | 0.985684812 | 28 | 336 | 249 | 0.68452382087707\0.68452382087707 | TCGA-76-6657/1.3.6.1.4.1.14519.5.2.1.1188.4001.114621594146207945121756272697/1.3.6.1.4.1.14519.5.2.1.1188.4001.281208541851434233735133229295 | |
| TCGA-GBM | TCGA-76 | TCGA-76-6657 | t1Gd | M | 90 | T1_AX_SE | 1301 | 6.29841E+15 | 6/11/2001 | Philips Medical Systems | MR1 | Achieva | SE | 2D | 5 | 481.14 | 10 | 127.7817 | 3 | 7.5 | 90 | 3.200000048 | 22 | 284 | 227 | 0.71875\0.71875 | TCGA-76-6657/1.3.6.1.4.1.14519.5.2.1.1188.4001.114621594146207945121756272697/1.3.6.1.4.1.14519.5.2.1.1188.4001.139759446384249479928559633803 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-6657 | t1 | M | 90 | T1_AX__SE | 901 | 6.29841E+15 | 6/11/2001 | Philips Medical Systems | MR1 | Achieva | SE | 2D | 5 | 423.8076 | 10 | 127.7817 | 3 | 7.5 | 90 | 3.199997187 | 24 | 316 | 254 | 0.68452382087707\0.68452382087707 | TCGA-76-6657/1.3.6.1.4.1.14519.5.2.1.1188.4001.114621594146207945121756272697/1.3.6.1.4.1.14519.5.2.1.1188.4001.133273809379709831292592700914 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-6657 | t2 | M | 90 | T2W_TSE | 701 | 6.29841E+15 | 6/11/2001 | Philips Medical Systems | MR1 | Achieva | SE | 2D | 5 | 3000 | 105 | 127.7817 | 3 | 6 | 90 | 2.275380373 | 26 | 420 | 380 | 0.41071429848670\0.41071429848670 | TCGA-76-6657/1.3.6.1.4.1.14519.5.2.1.1188.4001.114621594146207945121756272697/1.3.6.1.4.1.14519.5.2.1.1188.4001.231168282285866175377263571095 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-6661 | flair | M | 70 | T2_AX_FLAIR | 701 | 9.0359E+15 | 12/8/2001 | Philips Medical Systems | MR1 | Achieva | IR | 2D | 5 | 11000 | 125 | 2850 | 127.7744 | 3 | 6 | 90 | 0.984946549 | 28 | 320 | 218 | 0.6875\0.6875 | TCGA-76-6661/1.3.6.1.4.1.14519.5.2.1.1188.4001.102058737511198476066014834840/1.3.6.1.4.1.14519.5.2.1.1188.4001.281066854198433689922490047868 | |
| TCGA-GBM | TCGA-76 | TCGA-76-6661 | t1Gd | M | 70 | T1_SAG_SE | 1501 | 9.0359E+15 | 12/8/2001 | Philips Medical Systems | MR1 | Achieva | SE | 2D | 5 | 400 | 10 | 127.7744 | 3 | 7.5 | 90 | 1.644433618 | 16 | 256 | 219 | 0.9375\0.9375 | TCGA-76-6661/1.3.6.1.4.1.14519.5.2.1.1188.4001.102058737511198476066014834840/1.3.6.1.4.1.14519.5.2.1.1188.4001.474873019581159388054539531313 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-6661 | t1 | M | 70 | T1_AX__SE | 1101 | 9.0359E+15 | 12/8/2001 | Philips Medical Systems | MR1 | Achieva | SE | 2D | 5 | 423.8076 | 10.2 | 127.7744 | 3 | 7.5 | 90 | 3.199997187 | 24 | 304 | 242 | 0.6875\0.6875 | TCGA-76-6661/1.3.6.1.4.1.14519.5.2.1.1188.4001.102058737511198476066014834840/1.3.6.1.4.1.14519.5.2.1.1188.4001.279952068981139943403600145423 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-6661 | t2 | M | 70 | T2W_TSE | 901 | 9.0359E+15 | 12/8/2001 | Philips Medical Systems | MR1 | Achieva | SE | 2D | 5 | 3000 | 105 | 127.7744 | 3 | 6 | 90 | 3.033840179 | 24 | 400 | 355 | 0.4296875\0.4296875 | TCGA-76-6661/1.3.6.1.4.1.14519.5.2.1.1188.4001.102058737511198476066014834840/1.3.6.1.4.1.14519.5.2.1.1188.4001.270839976577326958300328675313 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-6662 | flair | M | 95 | T2_AX_FLAIR | 501 | 1.52941E+15 | 12/13/2001 | Philips Medical Systems | MR1 | Achieva | IR | 2D | 5 | 11000 | 125 | 2850 | 127.7742 | 3 | 6 | 90 | 0.906080961 | 27 | 348 | 262 | 0.68181818723678\0.68181818723678 | TCGA-76-6662/1.3.6.1.4.1.14519.5.2.1.1188.4001.313762558732585076631143086043/1.3.6.1.4.1.14519.5.2.1.1188.4001.248075422254562638743658647594 | |
| TCGA-GBM | TCGA-76 | TCGA-76-6662 | t1Gd | M | 95 | T1_AX_SE | 1001 | 1.52941E+15 | 12/13/2001 | Philips Medical Systems | MR1 | Achieva | SE | 2D | 5 | 409.404 | 10 | 127.7742 | 3 | 7 | 90 | 3.200001001 | 24 | 296 | 239 | 0.75\0.75 | TCGA-76-6662/1.3.6.1.4.1.14519.5.2.1.1188.4001.313762558732585076631143086043/1.3.6.1.4.1.14519.5.2.1.1188.4001.187826247221510809421836351871 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-6662 | t1 | M | 95 | T1_AX__SE | 801 | 1.52941E+15 | 12/13/2001 | Philips Medical Systems | MR1 | Achieva | SE | 2D | 5 | 375 | 10 | 127.7742 | 3 | 7 | 90 | 2.631093741 | 24 | 332 | 266 | 0.68181818723678\0.68181818723678 | TCGA-76-6662/1.3.6.1.4.1.14519.5.2.1.1188.4001.313762558732585076631143086043/1.3.6.1.4.1.14519.5.2.1.1188.4001.998300758126981185243547729186 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-6662 | t2 | M | 95 | T2W_TSE | 701 | 1.52941E+15 | 12/13/2001 | Philips Medical Systems | MR1 | Achieva | SE | 2D | 5 | 3318.263 | 105 | 127.7742 | 3 | 6 | 90 | 3.199999571 | 27 | 436 | 401 | 0.42857143282890\0.42857143282890 | TCGA-76-6662/1.3.6.1.4.1.14519.5.2.1.1188.4001.313762558732585076631143086043/1.3.6.1.4.1.14519.5.2.1.1188.4001.152280069325527575285353872757 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-6663 | flair | F | 74 | T2_AX_FLAIR | 501 | 2.30791E+15 | 12/27/2001 | Philips Medical Systems | MR1 | Achieva | IR | 2D | 5 | 11000 | 125 | 2850 | 127.7735 | 3 | 6 | 90 | 0.984931111 | 28 | 320 | 218 | 0.6875\0.6875 | TCGA-76-6663/1.3.6.1.4.1.14519.5.2.1.1188.4001.461523921338830081291431565499/1.3.6.1.4.1.14519.5.2.1.1188.4001.178083867370599982819220236278 | |
| TCGA-GBM | TCGA-76 | TCGA-76-6663 | t1Gd | F | 74 | T1_AX_SE | 1001 | 2.30791E+15 | 12/27/2001 | Philips Medical Systems | MR1 | Achieva | SE | 2D | 5 | 586.993 | 10 | 127.7735 | 3 | 7.5 | 90 | 3.199999809 | 22 | 272 | 217 | 0.76388889551162\0.76388889551162 | TCGA-76-6663/1.3.6.1.4.1.14519.5.2.1.1188.4001.461523921338830081291431565499/1.3.6.1.4.1.14519.5.2.1.1188.4001.166315135512753068373556679698 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-6663 | t1 | F | 74 | T1_AX__SE | 801 | 2.30791E+15 | 12/27/2001 | Philips Medical Systems | MR1 | Achieva | SE | 2D | 5 | 388.4903 | 10 | 127.7735 | 3 | 7.5 | 90 | 3.199997187 | 22 | 304 | 242 | 0.6875\0.6875 | TCGA-76-6663/1.3.6.1.4.1.14519.5.2.1.1188.4001.461523921338830081291431565499/1.3.6.1.4.1.14519.5.2.1.1188.4001.106189376158731728515656074602 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-6663 | t2 | F | 74 | T2W_TSE | 701 | 2.30791E+15 | 12/27/2001 | Philips Medical Systems | MR1 | Achieva | SE | 2D | 5 | 2555.412 | 105 | 127.7735 | 3 | 6 | 90 | 3.200000048 | 24 | 400 | 374 | 0.4296875\0.4296875 | TCGA-76-6663/1.3.6.1.4.1.14519.5.2.1.1188.4001.461523921338830081291431565499/1.3.6.1.4.1.14519.5.2.1.1188.4001.185350532654453205286429576409 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-6664 | t1 | F | 47 | T1W_3D_stryker | 301 | 2.68045E+15 | 1/10/2002 | Philips Medical Systems | PHILIPS-DE02D8D | Achieva | GR | 3D | 1 | 8.0406 | 3.746 | 127.793 | 3 | 1 | 8 | 0.016735241 | 167 | 300 | 300 | 0.75\0.75 | TCGA-76-6664/1.3.6.1.4.1.14519.5.2.1.1188.4001.280508857811965887839758381790/1.3.6.1.4.1.14519.5.2.1.1188.4001.182497160692414636856926373398 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-6664 | flair | F | 47 | T2_AX_FLAIR | 601 | 2.68045E+15 | 1/10/2002 | Philips Medical Systems | PHILIPS-DC8D0AA | Achieva | IR | 2D | 5 | 6000 | 120 | 2000 | 63.90021 | 1.5 | 6 | 90 | 1.003544927 | 28 | 256 | 161 | 0.859375\0.859375 | TCGA-76-6664/1.3.6.1.4.1.14519.5.2.1.1188.4001.280508857811965887839758381790/1.3.6.1.4.1.14519.5.2.1.1188.4001.172605629671945162895985475280 | |
| TCGA-GBM | TCGA-76 | TCGA-76-6664 | t1Gd | F | 47 | T1_SE_POST | 1201 | 2.68045E+15 | 1/10/2002 | Philips Medical Systems | PHILIPS-DC8D0AA | Achieva | SE | 2D | 5 | 589.512 | 12 | 63.90021 | 1.5 | 7 | 69 | 1.466690779 | 22 | 256 | 204 | 0.859375\0.859375 | TCGA-76-6664/1.3.6.1.4.1.14519.5.2.1.1188.4001.280508857811965887839758381790/1.3.6.1.4.1.14519.5.2.1.1188.4001.302763847255950328481343215267 | ||
| TCGA-GBM | TCGA-76 | TCGA-76-6664 | t2 | F | 47 | T2-WHOLE_BRAIN | 801 | 2.68045E+15 | 1/10/2002 | Philips Medical Systems | PHILIPS-DC8D0AA | Achieva | SE | 2D | 5 | 4530.271 | 100 | 63.90021 | 1.5 | 7.5 | 90 | 1.586941838 | 22 | 256 | 225 | 0.41015625\0.41015625 | TCGA-76-6664/1.3.6.1.4.1.14519.5.2.1.1188.4001.280508857811965887839758381790/1.3.6.1.4.1.14519.5.2.1.1188.4001.101392134704233686294499449068 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-4941 | flair | M | 67 | 86.183 | AXIAL_FLAIR | 5 | 2.6496E+15 | 9/9/1996 | GE MEDICAL SYSTEMS | mr1OW | GENESIS_SIGNA | IR | 2D | 5 | 10002 | 157.5 | 2500 | 6.39E+08 | 1.5 | 6 | 90 | 0.027182 | 28 | 256 | 192 | 0.8593798876\0.859375 | TCGA-CS-4941/1.3.6.1.4.1.14519.5.2.1.1188.4001.128551799994885838421577683863/1.3.6.1.4.1.14519.5.2.1.1188.4001.311513693456544543531535133857 |
| TCGA-LGG | TCGA-CS | TCGA-CS-4941 | t1Gd | M | 67 | 86.183 | *AX_3D_FOR_STRYKER_PROTOCOL | 9 | 2.6496E+15 | 9/9/1996 | GE MEDICAL SYSTEMS | mr1OW | GENESIS_SIGNA | GR | 3D | 1.5 | 7.68 | 1.692 | 6.39E+08 | 1.5 | 1.5 | 30 | 0.032676 | 124 | 256 | 192 | 0.9375\0.9375 | TCGA-CS-4941/1.3.6.1.4.1.14519.5.2.1.1188.4001.128551799994885838421577683863/1.3.6.1.4.1.14519.5.2.1.1188.4001.211133122498306244529495926656 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-4941 | t1 | M | 67 | 86.183 | AXIAL_T1 | 2 | 2.6496E+15 | 9/9/1996 | GE MEDICAL SYSTEMS | mr1OW | GENESIS_SIGNA | SE | 2D | 5 | 600 | 9 | 6.39E+08 | 1.5 | 7.5 | 90 | 0.066078 | 23 | 256 | 192 | 0.7812440991\0.78125 | TCGA-CS-4941/1.3.6.1.4.1.14519.5.2.1.1188.4001.128551799994885838421577683863/1.3.6.1.4.1.14519.5.2.1.1188.4001.132691411436475486878420509309 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-4941 | t2 | M | 67 | 86.183 | AXIAL_T2 | 3 | 2.6496E+15 | 9/9/1996 | GE MEDICAL SYSTEMS | mr1OW | GENESIS_SIGNA | SE | 2D | 5 | 3000 | 30 | 6.39E+08 | 1.5 | 7.5 | 90 | 0.022943 | 50 | 256 | 192 | 0.8593685627\0.859375 | TCGA-CS-4941/1.3.6.1.4.1.14519.5.2.1.1188.4001.128551799994885838421577683863/1.3.6.1.4.1.14519.5.2.1.1188.4001.724227449850990988403354297278 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-4942 | flair | F | 15 | T2_AX_FLAIR | 501 | 3.05011E+15 | 2/22/1997 | Philips Medical Systems | MR3T | Intera Achieva | IR | 5 | 11000 | 125 | 2800 | 127.7969 | 3 | 6 | 90 | 24 | 272 | 189 | 0.69444441795349\0.69444441795349 | TCGA-CS-4942/1.3.6.1.4.1.14519.5.2.1.1188.4001.205805171270283233290247304689/1.3.6.1.4.1.14519.5.2.1.1188.4001.203315309793102340695695217917 | |||
| TCGA-LGG | TCGA-CS | TCGA-CS-4942 | t1Gd | F | 15 | TI_AX_SE | 801 | 3.05011E+15 | 2/22/1997 | Philips Medical Systems | MR3T | Intera Achieva | SE | 5 | 500.0002 | 10 | 127.7968 | 3 | 7.5 | 90 | 20 | 256 | 179 | 0.8984375\0.8984375 | TCGA-CS-4942/1.3.6.1.4.1.14519.5.2.1.1188.4001.205805171270283233290247304689/1.3.6.1.4.1.14519.5.2.1.1188.4001.147319190030218663096857357676 | ||||
| TCGA-LGG | TCGA-CS | TCGA-CS-4942 | t1 | F | 15 | T1_AX__SE | 301 | 3.05011E+15 | 2/22/1997 | Philips Medical Systems | MR3T | Intera Achieva | SE | 5 | 500.0002 | 10 | 127.7969 | 3 | 7.5 | 90 | 20 | 256 | 205 | 0.8984375\0.8984375 | TCGA-CS-4942/1.3.6.1.4.1.14519.5.2.1.1188.4001.205805171270283233290247304689/1.3.6.1.4.1.14519.5.2.1.1188.4001.122619603792498033279228114936 | ||||
| TCGA-LGG | TCGA-CS | TCGA-CS-4942 | t2 | F | 15 | DUAL_AX_SE | 401 | 3.05011E+15 | 2/22/1997 | Philips Medical Systems | MR3T | Intera Achieva | SE | 5 | 3187.174 | 20 | 127.7969 | 3 | 7.5 | 90 | 48 | 256 | 192 | 0.50925928354263\0.50925928354263 | TCGA-CS-4942/1.3.6.1.4.1.14519.5.2.1.1188.4001.205805171270283233290247304689/1.3.6.1.4.1.14519.5.2.1.1188.4001.309224177898458617251995007653 | ||||
| TCGA-LGG | TCGA-CS | TCGA-CS-4943 | flair | M | 37 | 91 | BRAIN_STRYKER/FLAIR | 5 | 7.24147E+15 | 9/2/2000 | SIEMENS | mrscan | MAGNETOM VISION | IR | 2D | 5 | 9000 | 105 | 2340 | 63.62082 | 1.493806 | 2.499996 | 180 | 0 | 101 | 512 | 140 | 008.593750E-01\08.593750E-01 | TCGA-CS-4943/1.3.6.1.4.1.14519.5.2.1.1188.4001.340234692744555909933537052921/1.3.6.1.4.1.14519.5.2.1.1188.4001.147804566525382153225080630011 |
| TCGA-LGG | TCGA-CS | TCGA-CS-4943 | t1Gd | M | 37 | 91 | BRAIN_STRYKER/AX-RAGE-STRY | 7 | 7.24147E+15 | 9/2/2000 | SIEMENS | mrscan | MAGNETOM VISION | IR | 3D | 1.5 | 15 | 7 | 300 | 63.62078 | 1.493806 | 15 | 0 | 301 | 512 | 134 | 005.078125E-01\05.078125E-01 | TCGA-CS-4943/1.3.6.1.4.1.14519.5.2.1.1188.4001.340234692744555909933537052921/1.3.6.1.4.1.14519.5.2.1.1188.4001.123016478892109436983134203477 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-4943 | t1 | M | 37 | 91 | BRAIN_STRYKER/T1_SAG | 2 | 7.24147E+15 | 9/2/2000 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 400 | 12 | 63.62078 | 1.493806 | 2 | 70 | 0 | 21 | 512 | 179 | 009.375000E-01\09.375000E-01 | TCGA-CS-4943/1.3.6.1.4.1.14519.5.2.1.1188.4001.340234692744555909933537052921/1.3.6.1.4.1.14519.5.2.1.1188.4001.290529229678888668035398419158 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-4943 | t2 | M | 37 | 91 | BRAIN_STRYKER/T2_AX_DOUBLE | 4 | 7.24147E+15 | 9/2/2000 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 2020 | 20 | 63.62078 | 1.493806 | 2.499996 | 90 | 0 | 81 | 512 | 115 | 004.296875E-01\04.296875E-01 | TCGA-CS-4943/1.3.6.1.4.1.14519.5.2.1.1188.4001.340234692744555909933537052921/1.3.6.1.4.1.14519.5.2.1.1188.4001.287263919814542538289235456512 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-4944 | flair | M | 50 | 77 | BRAIN_STRYKER/FLAIR | 5 | 2.19794E+15 | 2/8/2001 | SIEMENS | mrscan | MAGNETOM VISION | IR | 2D | 5 | 9000 | 105 | 2340 | 63.62075 | 1.493806 | 2.5 | 180 | 0 | 103 | 512 | 140 | 008.593750E-01\08.593750E-01 | TCGA-CS-4944/1.3.6.1.4.1.14519.5.2.1.1188.4001.129145242695024660859453065472/1.3.6.1.4.1.14519.5.2.1.1188.4001.203804375049412380660273335819 |
| TCGA-LGG | TCGA-CS | TCGA-CS-4944 | t1Gd | M | 50 | 77 | BRAIN_STRYKER/AX-RAGE-STRY | 8 | 2.19794E+15 | 2/8/2001 | SIEMENS | mrscan | MAGNETOM VISION | IR | 3D | 1.5 | 15 | 7 | 300 | 63.62066 | 1.493806 | 15 | 0 | 323 | 512 | 256 | 005.078125E-01\05.078125E-01 | TCGA-CS-4944/1.3.6.1.4.1.14519.5.2.1.1188.4001.129145242695024660859453065472/1.3.6.1.4.1.14519.5.2.1.1188.4001.339210546101357716851441307311 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-4944 | t1 | M | 50 | 77 | BRAIN_STRYKER/T1_AXIAL | 3 | 2.19794E+15 | 2/8/2001 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 540 | 12 | 63.62067 | 1.493806 | 2.5 | 70 | 0 | 43 | 512 | 134 | 008.593750E-01\08.593750E-01 | TCGA-CS-4944/1.3.6.1.4.1.14519.5.2.1.1188.4001.129145242695024660859453065472/1.3.6.1.4.1.14519.5.2.1.1188.4001.207297865468602642550067713550 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-4944 | t2 | M | 50 | 77 | BRAIN_NON_CONTRAST/FL_2D_A | 6 | 2.19794E+15 | 2/8/2001 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 800 | 26 | 63.62075 | 1.493806 | 2.499996 | 15 | 0 | 123 | 512 | 96 | 008.593750E-01\08.593750E-01 | TCGA-CS-4944/1.3.6.1.4.1.14519.5.2.1.1188.4001.129145242695024660859453065472/1.3.6.1.4.1.14519.5.2.1.1188.4001.162789104395443584625385186701 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-5393 | flair | M | 39 | 100 | BRAIN_CONTRAST/FLAIR | 4 | 8.62427E+15 | 6/6/1999 | SIEMENS | mrscan | MAGNETOM VISION | IR | 2D | 5 | 9000 | 105 | 2340 | 63.61585 | 1.493806 | 2.5 | 180 | 0 | 61 | 512 | 112 | 008.593750E-01\08.593750E-01 | TCGA-CS-5393/1.3.6.1.4.1.14519.5.2.1.1188.4001.270866039761941345997853128540/1.3.6.1.4.1.14519.5.2.1.1188.4001.132954609680080850667151992756 |
| TCGA-LGG | TCGA-CS | TCGA-CS-5393 | t1Gd | M | 39 | 100 | BRAIN_STRYKER/AX-RAGE-STRY | 8 | 8.62427E+15 | 6/6/1999 | SIEMENS | mrscan | MAGNETOM VISION | IR | 3D | 1.5 | 15 | 7 | 300 | 63.61583 | 1.493806 | 15 | 0 | 321 | 512 | 134 | 005.078125E-01\05.078125E-01 | TCGA-CS-5393/1.3.6.1.4.1.14519.5.2.1.1188.4001.270866039761941345997853128540/1.3.6.1.4.1.14519.5.2.1.1188.4001.229538992522004896365823528826 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-5393 | t1 | M | 39 | 100 | BRAIN_CONTRAST/T1_AXIAL | 3 | 8.62427E+15 | 6/6/1999 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 600 | 14 | 63.61582 | 1.493806 | 2.500004 | 70 | 0 | 41 | 512 | 115 | 008.593750E-01\08.593750E-01 | TCGA-CS-5393/1.3.6.1.4.1.14519.5.2.1.1188.4001.270866039761941345997853128540/1.3.6.1.4.1.14519.5.2.1.1188.4001.290034506973629211391468552664 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-5393 | t2 | M | 39 | 100 | BRAIN_CONTRAST/hemosiderin | 6 | 8.62427E+15 | 6/6/1999 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 800 | 26 | 63.61585 | 1.493806 | 2.5 | 15 | 0 | 121 | 512 | 134 | 008.593750E-01\08.593750E-01 | TCGA-CS-5393/1.3.6.1.4.1.14519.5.2.1.1188.4001.270866039761941345997853128540/1.3.6.1.4.1.14519.5.2.1.1188.4001.335851554045791851017805707229 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-5395 | flair | M | 43 | 91 | BRAIN_CONTRAST/FLAIR | 3 | 1.65611E+15 | 10/4/1998 | SIEMENS | mrscan | MAGNETOM VISION | IR | 2D | 5 | 9000 | 105 | 2340 | 63.61564 | 1.493806 | 2.5 | 180 | 0 | 41 | 512 | 140 | 008.593750E-01\08.593750E-01 | TCGA-CS-5395/1.3.6.1.4.1.14519.5.2.1.1188.4001.202878570115416620116241845955/1.3.6.1.4.1.14519.5.2.1.1188.4001.187917860252441163272978739749 |
| TCGA-LGG | TCGA-CS | TCGA-CS-5395 | t1Gd | M | 43 | 91 | BRAIN_STRYKER/AX-RAGE-STRY | 13 | 1.65611E+15 | 10/4/1998 | SIEMENS | mrscan | MAGNETOM VISION | IR | 3D | 1.5 | 15 | 7 | 300 | 63.6156 | 1.493806 | 15 | 0 | 332 | 512 | 179 | 004.687500E-01\04.687500E-01 | TCGA-CS-5395/1.3.6.1.4.1.14519.5.2.1.1188.4001.202878570115416620116241845955/1.3.6.1.4.1.14519.5.2.1.1188.4001.174104235571112005602820353561 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-5395 | t1 | M | 43 | 91 | BRAIN_STRYKER/T1_AXIAL | 10 | 1.65611E+15 | 10/4/1998 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 540 | 12 | 63.61564 | 1.493806 | 2.5 | 70 | 0 | 92 | 512 | 134 | 008.593750E-01\08.593750E-01 | TCGA-CS-5395/1.3.6.1.4.1.14519.5.2.1.1188.4001.202878570115416620116241845955/1.3.6.1.4.1.14519.5.2.1.1188.4001.977609850409468267249989944139 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-5395 | t2 | M | 43 | 91 | BRAIN_PITUITARY/PD_T2_AXIA | 11 | 1.65611E+15 | 10/4/1998 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 3200 | 15 | 63.61564 | 1.493806 | 2.5 | 180 | 0 | 132 | 1024 | 196 | 004.296875E-01\04.296875E-01 | TCGA-CS-5395/1.3.6.1.4.1.14519.5.2.1.1188.4001.202878570115416620116241845955/1.3.6.1.4.1.14519.5.2.1.1188.4001.154523005230294777772339327074 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-5396 | flair | F | 81 | T2_AX_FLAIR | 601 | 7.91565E+15 | 3/2/2001 | Philips Medical Systems | MR1 | Achieva | IR | 2D | 5 | 11000 | 125 | 2850 | 127.7858 | 3 | 6 | 90 | 0.984979868 | 28 | 320 | 250 | 0.6875\0.6875 | TCGA-CS-5396/1.3.6.1.4.1.14519.5.2.1.1188.4001.866856253970500879015300047605/1.3.6.1.4.1.14519.5.2.1.1188.4001.170311900627152897231718498890 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-5396 | t1Gd | F | 81 | T1_AX_SE | 1101 | 7.91565E+15 | 3/2/2001 | Philips Medical Systems | MR1 | Achieva | SE | 2D | 5 | 640.356 | 10 | 127.7858 | 3 | 7.5 | 90 | 3.199999571 | 24 | 272 | 217 | 0.76388889551162\0.76388889551162 | TCGA-CS-5396/1.3.6.1.4.1.14519.5.2.1.1188.4001.866856253970500879015300047605/1.3.6.1.4.1.14519.5.2.1.1188.4001.250829490408448799557825354608 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-5396 | t1 | F | 81 | T1_AX__SE | 1001 | 7.91565E+15 | 3/2/2001 | Philips Medical Systems | MR1 | Achieva | SE | 2D | 5 | 423.8076 | 10 | 127.7858 | 3 | 7.5 | 90 | 3.199997187 | 24 | 304 | 243 | 0.6875\0.6875 | TCGA-CS-5396/1.3.6.1.4.1.14519.5.2.1.1188.4001.866856253970500879015300047605/1.3.6.1.4.1.14519.5.2.1.1188.4001.121538176450529977959006375237 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-5396 | t2 | F | 81 | T2W_TSE | 701 | 7.91565E+15 | 3/2/2001 | Philips Medical Systems | MR1 | Achieva | SE | 2D | 5 | 3000 | 105 | 127.7858 | 3 | 6 | 90 | 3.033840179 | 24 | 400 | 380 | 0.4296875\0.4296875 | TCGA-CS-5396/1.3.6.1.4.1.14519.5.2.1.1188.4001.866856253970500879015300047605/1.3.6.1.4.1.14519.5.2.1.1188.4001.186406456919519776851284170754 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-5397 | flair | F | 49 | T2_AX_FLAIR | 501 | 8.75582E+15 | 3/15/2001 | Philips Medical Systems | MR1 | Achieva | IR | 2D | 5 | 11000 | 125 | 2850 | 127.7853 | 3 | 6 | 90 | 0.985324383 | 28 | 336 | 255 | 0.68452382087707\0.68452382087707 | TCGA-CS-5397/1.3.6.1.4.1.14519.5.2.1.1188.4001.157283642653722420546464271991/1.3.6.1.4.1.14519.5.2.1.1188.4001.186076363067152633547598779834 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-5397 | t1Gd | F | 49 | T1W_3D_stryker | 1101 | 8.75582E+15 | 3/15/2001 | Philips Medical Systems | MR1 | Achieva | GR | 3D | 3 | 8.3691 | 4.1335 | 127.7853 | 3 | 1.5 | 8 | 0.276564628 | 120 | 272 | 240 | 0.83333331346511\0.83333331346511 | TCGA-CS-5397/1.3.6.1.4.1.14519.5.2.1.1188.4001.157283642653722420546464271991/1.3.6.1.4.1.14519.5.2.1.1188.4001.309416896241519723753279836949 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-5397 | t1 | F | 49 | T1_AX__SE | 901 | 8.75582E+15 | 3/15/2001 | Philips Medical Systems | MR1 | Achieva | SE | 2D | 5 | 399.9996 | 10 | 127.7853 | 3 | 7.5 | 90 | 3.107922792 | 22 | 316 | 255 | 0.68452382087707\0.68452382087707 | TCGA-CS-5397/1.3.6.1.4.1.14519.5.2.1.1188.4001.157283642653722420546464271991/1.3.6.1.4.1.14519.5.2.1.1188.4001.148929021964876442187111599680 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-5397 | t2 | F | 49 | T2W_TSE | 601 | 8.75582E+15 | 3/15/2001 | Philips Medical Systems | MR1 | Achieva | SE | 2D | 5 | 3318.263 | 105 | 127.7853 | 3 | 6 | 90 | 3.199999571 | 28 | 420 | 380 | 0.41071429848670\0.41071429848670 | TCGA-CS-5397/1.3.6.1.4.1.14519.5.2.1.1188.4001.157283642653722420546464271991/1.3.6.1.4.1.14519.5.2.1.1188.4001.993332431135153833493944928693 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-6186 | flair | M | 85 | T2_AX_FLAIR | 401 | 4.696E+15 | 6/1/2000 | Philips Medical Systems | PHILIPS-8D9B7F3 | Achieva | IR | 2D | 5 | 6000 | 120 | 2000 | 63.90019 | 1.5 | 6 | 90 | 1.003544927 | 30 | 256 | 161 | 0.859375\0.859375 | TCGA-CS-6186/1.3.6.1.4.1.14519.5.2.1.1188.4001.242366823329087276501028197608/1.3.6.1.4.1.14519.5.2.1.1188.4001.206296150819865714212366072445 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-6186 | t1Gd | M | 85 | STRYKER | 1101 | 4.696E+15 | 6/1/2000 | Philips Medical Systems | PHILIPS-8D9B7F3 | Achieva | GR | 3D | 3 | 25 | 4.5994 | 63.90019 | 1.5 | 1.5 | 30 | 0.319924802 | 118 | 256 | 244 | 0.9375\0.9375 | TCGA-CS-6186/1.3.6.1.4.1.14519.5.2.1.1188.4001.242366823329087276501028197608/1.3.6.1.4.1.14519.5.2.1.1188.4001.171955373280735242099311227064 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-6186 | t1 | M | 85 | T1_SE_PRE | 901 | 4.696E+15 | 6/1/2000 | Philips Medical Systems | PHILIPS-8D9B7F3 | Achieva | SE | 2D | 5 | 413.944 | 12 | 63.90019 | 1.5 | 7 | 69 | 1.721659899 | 25 | 256 | 204 | 0.859375\0.859375 | TCGA-CS-6186/1.3.6.1.4.1.14519.5.2.1.1188.4001.242366823329087276501028197608/1.3.6.1.4.1.14519.5.2.1.1188.4001.269684106183325571291783916483 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-6186 | t2 | M | 85 | T2-WHOLE_BRAIN | 501 | 4.696E+15 | 6/1/2000 | Philips Medical Systems | PHILIPS-8D9B7F3 | Achieva | SE | 2D | 5 | 4938.149 | 100 | 63.90019 | 1.5 | 7.5 | 90 | 1.588215947 | 24 | 256 | 225 | 0.41015625\0.41015625 | TCGA-CS-6186/1.3.6.1.4.1.14519.5.2.1.1188.4001.242366823329087276501028197608/1.3.6.1.4.1.14519.5.2.1.1188.4001.664999071910196574827570132557 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-6188 | flair | M | 113 | T2_AX_FLAIR | 501 | 2.21267E+15 | 8/12/2001 | Philips Medical Systems | MR1 | Achieva | IR | 2D | 5 | 11000 | 125 | 2850 | 127.7793 | 3 | 6 | 90 | 0.984957218 | 28 | 320 | 250 | 0.6875\0.6875 | TCGA-CS-6188/1.3.6.1.4.1.14519.5.2.1.1188.4001.203713524649110849540001495530/1.3.6.1.4.1.14519.5.2.1.1188.4001.102732308226606493016714027780 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-6188 | t1Gd | M | 113 | COR_MPR | 1002 | 2.21267E+15 | 8/12/2001 | Philips Medical Systems | Achieva | GR | 3D | 1 | 7.6244 | 3.5515 | 127.7793 | 3 | 1 | 8 | 0.246233359 | 200 | 300 | 300 | 0.75\0.75 | TCGA-CS-6188/1.3.6.1.4.1.14519.5.2.1.1188.4001.203713524649110849540001495530/1.3.6.1.4.1.14519.5.2.1.1188.4001.255282194301702091567144865881 | |||
| TCGA-LGG | TCGA-CS | TCGA-CS-6188 | t1 | M | 113 | T1W___SE | 401 | 2.21267E+15 | 8/12/2001 | Philips Medical Systems | MR1 | Achieva | SE | 2D | 5 | 400 | 10 | 127.7792 | 3 | 7.5 | 90 | 1.644433618 | 16 | 256 | 205 | 0.9375\0.9375 | TCGA-CS-6188/1.3.6.1.4.1.14519.5.2.1.1188.4001.203713524649110849540001495530/1.3.6.1.4.1.14519.5.2.1.1188.4001.663981468162513582180008232641 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-6188 | t2 | M | 113 | T2W_TSE | 701 | 2.21267E+15 | 8/12/2001 | Philips Medical Systems | MR1 | Achieva | SE | 2D | 5 | 3000 | 105 | 127.7793 | 3 | 6 | 90 | 3.033840179 | 24 | 400 | 380 | 0.4296875\0.4296875 | TCGA-CS-6188/1.3.6.1.4.1.14519.5.2.1.1188.4001.203713524649110849540001495530/1.3.6.1.4.1.14519.5.2.1.1188.4001.237410497056644642573784487223 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-6290 | flair | M | 31 | 113 | BRAIN_CONTRAST/FLAIR | 11 | 3.38024E+15 | 9/17/2000 | SIEMENS | mrscan | MAGNETOM VISION | IR | 2D | 5 | 9000 | 105 | 2340 | 63.62079 | 1.493806 | 2.5 | 180 | 0 | 351 | 512 | 112 | 008.593750E-01\08.593750E-01 | TCGA-CS-6290/1.3.6.1.4.1.14519.5.2.1.1188.4001.103822155980636301332134926774/1.3.6.1.4.1.14519.5.2.1.1188.4001.752531645793998918722907278370 |
| TCGA-LGG | TCGA-CS | TCGA-CS-6290 | t1Gd | M | 31 | 113 | BRAIN_CONTRAST/T1_CORONAL_ | 15 | 3.38024E+15 | 9/17/2000 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 440 | 12 | 63.62079 | 1.493806 | 3 | 70 | 0 | 451 | 512 | 134 | 008.593750E-01\08.593750E-01 | TCGA-CS-6290/1.3.6.1.4.1.14519.5.2.1.1188.4001.103822155980636301332134926774/1.3.6.1.4.1.14519.5.2.1.1188.4001.331156113378797656070442234457 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-6290 | t1 | M | 31 | 113 | BRAIN_CONTRAST/T1_AXIAL | 7 | 3.38024E+15 | 9/17/2000 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 600 | 14 | 63.62079 | 1.493806 | 2.500004 | 70 | 0 | 309 | 512 | 115 | 008.593750E-01\08.593750E-01 | TCGA-CS-6290/1.3.6.1.4.1.14519.5.2.1.1188.4001.103822155980636301332134926774/1.3.6.1.4.1.14519.5.2.1.1188.4001.184518783244690052738829109319 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-6290 | t2 | M | 31 | 113 | BRAIN_CONTRAST/hemosiderin | 13 | 3.38024E+15 | 9/17/2000 | SIEMENS | mrscan | MAGNETOM VISION | RM | 2D | 5 | 800 | 26 | 63.62079 | 1.493806 | 2.5 | 15 | 0 | 411 | 512 | 134 | 008.593750E-01\08.593750E-01 | TCGA-CS-6290/1.3.6.1.4.1.14519.5.2.1.1188.4001.103822155980636301332134926774/1.3.6.1.4.1.14519.5.2.1.1188.4001.296212542270973986107566466837 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-6665 | flair | F | 68 | T2_AX_FLAIR | 501 | 1.29453E+15 | 8/17/2001 | Philips Medical Systems | MR1 | Achieva | IR | 2D | 5 | 11000 | 125 | 2850 | 127.7791 | 3 | 6 | 90 | 0.827241838 | 24 | 320 | 247 | 0.65476191043853\0.65476191043853 | TCGA-CS-6665/1.3.6.1.4.1.14519.5.2.1.1188.4001.159768644950365298898163950114/1.3.6.1.4.1.14519.5.2.1.1188.4001.173192188712863545223866126615 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-6665 | t1Gd | F | 68 | T1_AX_SE | 1101 | 1.29453E+15 | 8/17/2001 | Philips Medical Systems | MR1 | Achieva | SE | 2D | 5 | 600 | 10 | 127.7791 | 3 | 6 | 90 | 2.799360037 | 24 | 272 | 217 | 0.76388889551162\0.76388889551162 | TCGA-CS-6665/1.3.6.1.4.1.14519.5.2.1.1188.4001.159768644950365298898163950114/1.3.6.1.4.1.14519.5.2.1.1188.4001.304734519877038894320972032214 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-6665 | t1 | F | 68 | T1_AX__SE | 901 | 1.29453E+15 | 8/17/2001 | Philips Medical Systems | MR1 | Achieva | SE | 2D | 5 | 423.8076 | 10 | 127.7791 | 3 | 6 | 90 | 3.199997187 | 24 | 304 | 243 | 0.6875\0.6875 | TCGA-CS-6665/1.3.6.1.4.1.14519.5.2.1.1188.4001.159768644950365298898163950114/1.3.6.1.4.1.14519.5.2.1.1188.4001.291878956429584404970815668083 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-6665 | t2 | F | 68 | T2W_TSE | 701 | 1.29453E+15 | 8/17/2001 | Philips Medical Systems | MR1 | Achieva | SE | 2D | 5 | 3000 | 105 | 127.7791 | 3 | 6 | 90 | 3.033840179 | 24 | 400 | 365 | 0.4296875\0.4296875 | TCGA-CS-6665/1.3.6.1.4.1.14519.5.2.1.1188.4001.159768644950365298898163950114/1.3.6.1.4.1.14519.5.2.1.1188.4001.120098837774084096221182816485 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-6666 | flair | M | 83 | T2_AX_FLAIR | 701 | 5.29894E+15 | 11/9/2001 | Philips Medical Systems | PHILIPS-DE02D8D | Achieva | IR | 2D | 5 | 11000 | 125 | 2850 | 127.7934 | 3 | 6 | 90 | 0.067167066 | 28 | 320 | 247 | 0.6875\0.6875 | TCGA-CS-6666/1.3.6.1.4.1.14519.5.2.1.1188.4001.562796873094346571498215071602/1.3.6.1.4.1.14519.5.2.1.1188.4001.128846142536225521942208108756 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-6666 | t1Gd | M | 83 | T1W_3D_STRYKER | 1001 | 5.29894E+15 | 11/9/2001 | Philips Medical Systems | PHILIPS-DE02D8D | Achieva | GR | 3D | 1 | 7.6774 | 3.583 | 127.7934 | 3 | 1 | 8 | 0.017320534 | 200 | 300 | 300 | 0.75\0.75 | TCGA-CS-6666/1.3.6.1.4.1.14519.5.2.1.1188.4001.562796873094346571498215071602/1.3.6.1.4.1.14519.5.2.1.1188.4001.160531909116163750684978784137 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-6666 | t1 | M | 83 | T1_AX__SE | 901 | 5.29894E+15 | 11/9/2001 | Philips Medical Systems | PHILIPS-DE02D8D | Achieva | SE | 2D | 5 | 461.4519 | 10.3 | 127.7934 | 3 | 6 | 90 | 0.217081264 | 26 | 304 | 242 | 0.6875\0.6875 | TCGA-CS-6666/1.3.6.1.4.1.14519.5.2.1.1188.4001.562796873094346571498215071602/1.3.6.1.4.1.14519.5.2.1.1188.4001.143563584178390497626882678879 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-6666 | t2 | M | 83 | AXIAL__T2 | 801 | 5.29894E+15 | 11/9/2001 | Philips Medical Systems | PHILIPS-DE02D8D | Achieva | SE | 2D | 5 | 3000 | 100 | 127.7934 | 3 | 6 | 90 | 0.196178049 | 26 | 384 | 324 | 0.4296875\0.4296875 | TCGA-CS-6666/1.3.6.1.4.1.14519.5.2.1.1188.4001.562796873094346571498215071602/1.3.6.1.4.1.14519.5.2.1.1188.4001.309761309865874206034533043433 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-6667 | flair | F | 39 | 58.967 | AX_FLAIR | 7 | 4.45891E+15 | 11/5/2001 | GE MEDICAL SYSTEMS | GEMSOW | Signa HDxt | SE\IR | 2D | 5 | 10002 | 144.884 | 2500 | 63.84815 | 1.5 | 6 | 90 | 0.5505 | 26 | 288 | 192 | 0.4297\0.4297 | TCGA-CS-6667/1.3.6.1.4.1.14519.5.2.1.1188.4001.190019357332393685119456096919/1.3.6.1.4.1.14519.5.2.1.1188.4001.144509347774443554281904614614 |
| TCGA-LGG | TCGA-CS | TCGA-CS-6667 | t1Gd | F | 39 | 58.967 | Ax_T1_FS_BRAIN_POST | 9 | 4.45891E+15 | 11/5/2001 | GE MEDICAL SYSTEMS | GEMSOW | Signa HDxt | SE | 2D | 5 | 483.336 | 12 | 0 | 63.84816 | 1.5 | 7.5 | 90 | 0.916982 | 20 | 256 | 160 | 0.4297\0.4297 | TCGA-CS-6667/1.3.6.1.4.1.14519.5.2.1.1188.4001.190019357332393685119456096919/1.3.6.1.4.1.14519.5.2.1.1188.4001.270532183341952426047363273660 |
| TCGA-LGG | TCGA-CS | TCGA-CS-6667 | t1 | F | 39 | 58.967 | Ax_T1 | 5 | 4.45891E+15 | 11/5/2001 | GE MEDICAL SYSTEMS | GEMSOW | Signa HDxt | SE | 2D | 5 | 533.336 | 12 | 0 | 63.84815 | 1.5 | 7.5 | 90 | 1.51747 | 20 | 256 | 192 | 0.4297\0.4297 | TCGA-CS-6667/1.3.6.1.4.1.14519.5.2.1.1188.4001.190019357332393685119456096919/1.3.6.1.4.1.14519.5.2.1.1188.4001.342053548701630640583211031183 |
| TCGA-LGG | TCGA-CS | TCGA-CS-6667 | t2 | F | 39 | 58.967 | AX_FSE_T2 | 6 | 4.45891E+15 | 11/5/2001 | GE MEDICAL SYSTEMS | GEMSOW | Signa HDxt | SE | 2D | 5 | 3000 | 91.52 | 0 | 63.84815 | 1.5 | 7.5 | 90 | 0.7956 | 20 | 320 | 224 | 0.4297\0.4297 | TCGA-CS-6667/1.3.6.1.4.1.14519.5.2.1.1188.4001.190019357332393685119456096919/1.3.6.1.4.1.14519.5.2.1.1188.4001.118303589976037481805625826661 |
| TCGA-LGG | TCGA-CS | TCGA-CS-6668 | flair | F | 83 | T2_AX_FLAIR | 501 | 2.03626E+15 | 10/25/2001 | Philips Medical Systems | PHILIPS-DE02D8D | Achieva | IR | 2D | 5 | 11000 | 125 | 2850 | 127.7935 | 3 | 6 | 90 | 0.067193471 | 28 | 320 | 247 | 0.6875\0.6875 | TCGA-CS-6668/1.3.6.1.4.1.14519.5.2.1.1188.4001.980162626547355442928625529896/1.3.6.1.4.1.14519.5.2.1.1188.4001.169889219500152307085456851345 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-6668 | t1Gd | F | 83 | T1W_3D_STRYKER | 1101 | 2.03626E+15 | 10/25/2001 | Philips Medical Systems | PHILIPS-DE02D8D | Achieva | GR | 3D | 1 | 8.1329 | 3.824 | 127.7934 | 3 | 1 | 8 | 0.017320534 | 170 | 300 | 300 | 0.75\0.75 | TCGA-CS-6668/1.3.6.1.4.1.14519.5.2.1.1188.4001.980162626547355442928625529896/1.3.6.1.4.1.14519.5.2.1.1188.4001.114078538471828186024243211217 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-6668 | t1 | F | 83 | T1W_3D_STRYKER | 701 | 2.03626E+15 | 10/25/2001 | Philips Medical Systems | PHILIPS-DE02D8D | Achieva | GR | 3D | 1 | 8.0072 | 3.736 | 127.7935 | 3 | 1 | 8 | 0.017320534 | 170 | 300 | 300 | 0.75\0.75 | TCGA-CS-6668/1.3.6.1.4.1.14519.5.2.1.1188.4001.980162626547355442928625529896/1.3.6.1.4.1.14519.5.2.1.1188.4001.142459179818453226386988487183 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-6668 | t2 | F | 83 | AXIAL__T2 | 601 | 2.03626E+15 | 10/25/2001 | Philips Medical Systems | PHILIPS-DE02D8D | Achieva | SE | 2D | 4 | 3584.453 | 100 | 127.7936 | 3 | 5 | 90 | 0.214710966 | 33 | 384 | 324 | 0.4296875\0.4296875 | TCGA-CS-6668/1.3.6.1.4.1.14519.5.2.1.1188.4001.980162626547355442928625529896/1.3.6.1.4.1.14519.5.2.1.1188.4001.323550053999956700006878511084 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-6669 | flair | F | 68 | T2_AX_FLAIR | 501 | 3.05642E+15 | 1/2/2002 | Philips Medical Systems | PHILIPS-DE02D8D | Achieva | IR | 2D | 5 | 11000 | 125 | 2850 | 127.7932 | 3 | 6 | 90 | 0.067156747 | 28 | 320 | 250 | 0.6875\0.6875 | TCGA-CS-6669/1.3.6.1.4.1.14519.5.2.1.1188.4001.158950330763169507420209045633/1.3.6.1.4.1.14519.5.2.1.1188.4001.331668859120900125509759340193 | |
| TCGA-LGG | TCGA-CS | TCGA-CS-6669 | t1Gd | F | 68 | T1_AX_SE | 901 | 3.05642E+15 | 1/2/2002 | Philips Medical Systems | PHILIPS-DE02D8D | Achieva | SE | 2D | 5 | 600.0005 | 10 | 127.7932 | 3 | 7.5 | 90 | 0.174959749 | 22 | 272 | 217 | 0.76388889551162\0.76388889551162 | TCGA-CS-6669/1.3.6.1.4.1.14519.5.2.1.1188.4001.158950330763169507420209045633/1.3.6.1.4.1.14519.5.2.1.1188.4001.975564333883987761901816698053 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-6669 | t1 | F | 68 | T1_AX__SE | 801 | 3.05642E+15 | 1/2/2002 | Philips Medical Systems | PHILIPS-DE02D8D | Achieva | SE | 2D | 5 | 388.4903 | 10.344 | 127.7932 | 3 | 7.5 | 90 | 0.218181506 | 22 | 304 | 243 | 0.6875\0.6875 | TCGA-CS-6669/1.3.6.1.4.1.14519.5.2.1.1188.4001.158950330763169507420209045633/1.3.6.1.4.1.14519.5.2.1.1188.4001.270701452492080803456322167850 | ||
| TCGA-LGG | TCGA-CS | TCGA-CS-6669 | t2 | F | 68 | T2W_TSE | 701 | 3.05642E+15 | 1/2/2002 | Philips Medical Systems | PHILIPS-DE02D8D | Achieva | SE | 2D | 5 | 3081.244 | 105 | 127.7932 | 3 | 6 | 90 | 0.218181595 | 26 | 400 | 380 | 0.4296875\0.4296875 | TCGA-CS-6669/1.3.6.1.4.1.14519.5.2.1.1188.4001.158950330763169507420209045633/1.3.6.1.4.1.14519.5.2.1.1188.4001.478644020758691119117361200831 | ||
| TCGA-LGG | TCGA-DU | TCGA-DU-5849 | flair | U | 48 | 90.718 | AXIAL_FLAIR | 8 | 1.61956E+15 | 4/5/1995 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7323 | 3 | 2.5 | 90 | 0.9008 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-5849/1.3.6.1.4.1.14519.5.2.1.4591.4003.109364397081642293014271520866/1.3.6.1.4.1.14519.5.2.1.4591.4003.216135639247172328000547905561 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5849 | t1Gd | U | 48 | 90.718 | AX_T1_POST_GD_FLAIR | 12 | 1.61956E+15 | 4/5/1995 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 3236.34 | 6.356 | 1238 | 127.7323 | 3 | 2.5 | 90 | 1.7636 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-5849/1.3.6.1.4.1.14519.5.2.1.4591.4003.109364397081642293014271520866/1.3.6.1.4.1.14519.5.2.1.4591.4003.141732357113095863404589053586 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5849 | t1 | U | 48 | 90.718 | AX_T1_pre_gd | 9 | 1.61956E+15 | 4/5/1995 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 5 | 2152.82 | 6.356 | 883 | 127.7323 | 3 | 5 | 90 | 1.4365 | 38 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-5849/1.3.6.1.4.1.14519.5.2.1.4591.4003.109364397081642293014271520866/1.3.6.1.4.1.14519.5.2.1.4591.4003.326882926621335107573626721019 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5849 | t2 | U | 48 | 90.718 | AX_T2_FR-FSE_RF2_150 | 7 | 1.61956E+15 | 4/5/1995 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 4000 | 103.464 | 0 | 127.7323 | 3 | 5 | 90 | 1.3993 | 38 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-5849/1.3.6.1.4.1.14519.5.2.1.4591.4003.109364397081642293014271520866/1.3.6.1.4.1.14519.5.2.1.4591.4003.280912757798474012433274591281 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5851 | flair | F | 40 | 72.575 | Ax_FLAIR_2.5mm_for_surgery | 4 | 1.32058E+15 | 4/28/1995 | GE MEDICAL SYSTEMS | MR01MROW | Signa HDxt | IR | 2D | 2.5 | 10002 | 149.544 | 2200 | 63.88583 | 1.5 | 2.5 | 90 | 0.039 | 79 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-5851/1.3.6.1.4.1.14519.5.2.1.4591.4003.129680596450908220610617702930/1.3.6.1.4.1.14519.5.2.1.4591.4003.121163298773025279216831005877 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5851 | t1Gd | F | 40 | 72.575 | COR_T1_POST_GD | 9 | 1.32058E+15 | 4/28/1995 | GE MEDICAL SYSTEMS | MR01MROW | Signa HDxt | SE | 2D | 5 | 483.336 | 14 | 0 | 63.88583 | 1.5 | 5 | 90 | 0.0495152 | 48 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-5851/1.3.6.1.4.1.14519.5.2.1.4591.4003.129680596450908220610617702930/1.3.6.1.4.1.14519.5.2.1.4591.4003.304925718859314734932464300032 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5851 | t1 | F | 40 | 72.575 | Ax_T1_2.5mm_for_surgery | 8 | 1.32058E+15 | 4/28/1995 | GE MEDICAL SYSTEMS | MR01MROW | Signa HDxt | SE | 2D | 2.5 | 600 | 13 | 0 | 63.88583 | 1.5 | 2.5 | 90 | 0.054777 | 79 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-5851/1.3.6.1.4.1.14519.5.2.1.4591.4003.129680596450908220610617702930/1.3.6.1.4.1.14519.5.2.1.4591.4003.269931978354736840234304467989 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5851 | t2 | F | 40 | 72.575 | AXIAL_FSE | 3 | 1.32058E+15 | 4/28/1995 | GE MEDICAL SYSTEMS | MR01MROW | Signa HDxt | SE | 2D | 5 | 3516.67 | 23.688 | 0 | 63.88583 | 1.5 | 5 | 90 | 0.1355 | 80 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-5851/1.3.6.1.4.1.14519.5.2.1.4591.4003.129680596450908220610617702930/1.3.6.1.4.1.14519.5.2.1.4591.4003.198926562240285905428979227040 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5852 | flair | F | 61 | 85.275 | AXIAL_FLAIR | 7 | 8.68519E+15 | 7/9/1995 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7322 | 3 | 2.5 | 90 | 0.8357 | 70 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-5852/1.3.6.1.4.1.14519.5.2.1.4591.4003.149176124210739275471526262702/1.3.6.1.4.1.14519.5.2.1.4591.4003.750066108904337108875868387535 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5852 | t1Gd | F | 61 | 85.275 | AX_T1_POST_GD_FLAIR | 11 | 8.68519E+15 | 7/9/1995 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 3207.73 | 6.356 | 1238 | 127.7322 | 3 | 2.5 | 90 | 1.6004 | 70 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-5852/1.3.6.1.4.1.14519.5.2.1.4591.4003.149176124210739275471526262702/1.3.6.1.4.1.14519.5.2.1.4591.4003.369433803261020392440363841253 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5852 | t1 | F | 61 | 85.275 | AX_T1_pre_gd | 8 | 8.68519E+15 | 7/9/1995 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 5 | 2208.66 | 6.356 | 883 | 127.7322 | 3 | 5 | 90 | 1.7437 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-5852/1.3.6.1.4.1.14519.5.2.1.4591.4003.149176124210739275471526262702/1.3.6.1.4.1.14519.5.2.1.4591.4003.421016340465864461716578061202 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5852 | t2 | F | 61 | 85.275 | AX_T2_FR-FSE_RF2_150 | 5 | 8.68519E+15 | 7/9/1995 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 4000 | 103.464 | 0 | 127.7322 | 3 | 5 | 90 | 1.1923 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-5852/1.3.6.1.4.1.14519.5.2.1.4591.4003.149176124210739275471526262702/1.3.6.1.4.1.14519.5.2.1.4591.4003.258127417527584037720287408573 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5853 | flair | M | 29 | 90.265 | AXIAL_FLAIR | 6 | 1.49065E+15 | 8/23/1995 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7323 | 3 | 2.5 | 90 | 0.8954 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-5853/1.3.6.1.4.1.14519.5.2.1.4591.4003.488910637254672352689793378436/1.3.6.1.4.1.14519.5.2.1.4591.4003.179001448691349455487193384159 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5853 | t1Gd | M | 29 | 90.265 | AX_T1_POST_GD_FLAIR | 11 | 1.49065E+15 | 8/23/1995 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 3233.94 | 6.356 | 1238 | 127.7323 | 3 | 2.5 | 90 | 1.7498 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-5853/1.3.6.1.4.1.14519.5.2.1.4591.4003.488910637254672352689793378436/1.3.6.1.4.1.14519.5.2.1.4591.4003.864077196769485883942711381300 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5853 | t1 | M | 29 | 90.265 | AX_T1_pre_gd | 8 | 1.49065E+15 | 8/23/1995 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 5 | 2040.27 | 6.356 | 883 | 127.7323 | 3 | 5 | 90 | 1.3873 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-5853/1.3.6.1.4.1.14519.5.2.1.4591.4003.488910637254672352689793378436/1.3.6.1.4.1.14519.5.2.1.4591.4003.185137173531988376064097714019 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5853 | t2 | M | 29 | 90.265 | AX_T2_FR-FSE_RF2_150 | 7 | 1.49065E+15 | 8/23/1995 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 4000 | 103.464 | 0 | 127.7323 | 3 | 5 | 90 | 1.3144 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-5853/1.3.6.1.4.1.14519.5.2.1.4591.4003.488910637254672352689793378436/1.3.6.1.4.1.14519.5.2.1.4591.4003.118506229786576611944108655295 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5854 | flair | F | 57 | 86 | Ax_FLAIR_2.5mm_for_surgery | 5 | 2.27863E+15 | 11/4/1995 | GE MEDICAL SYSTEMS | MR01MROW | Signa HDxt | IR | 2D | 2.5 | 10004 | 149.544 | 2200 | 63.8858 | 1.5 | 2.5 | 90 | 0.0328 | 71 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-5854/1.3.6.1.4.1.14519.5.2.1.4591.4003.765125757509176996485268928000/1.3.6.1.4.1.14519.5.2.1.4591.4003.295508881770666613087332596147 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5854 | t1Gd | F | 57 | 86 | COR_T1_POST_GD | 9 | 2.27863E+15 | 11/4/1995 | GE MEDICAL SYSTEMS | MR01MROW | Signa HDxt | SE | 2D | 5 | 500 | 14 | 0 | 63.88579 | 1.5 | 5 | 90 | 0.0436644 | 43 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-5854/1.3.6.1.4.1.14519.5.2.1.4591.4003.765125757509176996485268928000/1.3.6.1.4.1.14519.5.2.1.4591.4003.496112103419126320360113889954 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5854 | t1 | F | 57 | 86 | AXIAL_T1 | 6 | 2.27863E+15 | 11/4/1995 | GE MEDICAL SYSTEMS | MR01MROW | Signa HDxt | SE | 2D | 5 | 500 | 14 | 0 | 63.88579 | 1.5 | 5 | 90 | 0.0349315 | 36 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-5854/1.3.6.1.4.1.14519.5.2.1.4591.4003.765125757509176996485268928000/1.3.6.1.4.1.14519.5.2.1.4591.4003.106838409056090834066289704722 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5854 | t2 | F | 57 | 86 | AXIAL_FSE | 3 | 2.27863E+15 | 11/4/1995 | GE MEDICAL SYSTEMS | MR01MROW | Signa HDxt | SE | 2D | 5 | 3500 | 23.16 | 0 | 63.88577 | 1.5 | 5 | 90 | 0.1192 | 72 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-5854/1.3.6.1.4.1.14519.5.2.1.4591.4003.765125757509176996485268928000/1.3.6.1.4.1.14519.5.2.1.4591.4003.249770102183222124726802201732 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5855 | flair | F | 80 | RA_ROUTINE_FLAIR_AX | 501 | 2.90902E+15 | 12/17/1995 | Philips Medical Systems | PMSN-9KQRJRSGE8 | Intera | IR | 5 | 11000 | 140 | 2800 | 63.89301 | 1.5 | 6 | 90 | 26 | 256 | 176 | 0.9765625\0.9765625 | TCGA-DU-5855/1.3.6.1.4.1.14519.5.2.1.4591.4003.852581990127740690043374426777/1.3.6.1.4.1.14519.5.2.1.4591.4003.290438072272990333107581688659 | |||
| TCGA-LGG | TCGA-DU | TCGA-DU-5855 | t1Gd | F | 80 | BrainLab_onBL_3DT1axC+ | 1201 | 2.90902E+15 | 12/17/1995 | Philips Medical Systems | PMSN-9KQRJRSGE8 | Intera | GR | 2 | 25 | 4.5874 | 63.89301 | 1.5 | 1 | 30 | 180 | 256 | 256 | 0.48828125\0.48828125 | TCGA-DU-5855/1.3.6.1.4.1.14519.5.2.1.4591.4003.852581990127740690043374426777/1.3.6.1.4.1.14519.5.2.1.4591.4003.112592252232666023998953315710 | ||||
| TCGA-LGG | TCGA-DU | TCGA-DU-5855 | t1 | F | 80 | RA_ROUTINE_T1_AX | 701 | 2.90902E+15 | 12/17/1995 | Philips Medical Systems | PMSN-9KQRJRSGE8 | Intera | SE | 5 | 450.0002 | 14 | 63.89301 | 1.5 | 6.074518 | 90 | 26 | 256 | 203 | 0.9375\0.9375 | TCGA-DU-5855/1.3.6.1.4.1.14519.5.2.1.4591.4003.852581990127740690043374426777/1.3.6.1.4.1.14519.5.2.1.4591.4003.304501649602313541252309917008 | ||||
| TCGA-LGG | TCGA-DU | TCGA-DU-5855 | t2 | F | 80 | RA_ROUTINE_T2_AX | 601 | 2.90902E+15 | 12/17/1995 | Philips Medical Systems | PMSN-9KQRJRSGE8 | Intera | SE | 5 | 5729.641 | 110 | 63.89301 | 1.5 | 6 | 90 | 26 | 352 | 264 | 0.46875\0.46875 | TCGA-DU-5855/1.3.6.1.4.1.14519.5.2.1.4591.4003.852581990127740690043374426777/1.3.6.1.4.1.14519.5.2.1.4591.4003.478972965056203795981794302830 | ||||
| TCGA-LGG | TCGA-DU | TCGA-DU-5871 | flair | F | 37 | 86.183 | AXIAL_FLAIR | 6 | 1.41281E+15 | 12/6/1994 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7322 | 3 | 2.5 | 90 | 0.8466 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-5871/1.3.6.1.4.1.14519.5.2.1.4591.4003.247530214976989618914834177531/1.3.6.1.4.1.14519.5.2.1.4591.4003.275038399338610640375595922910 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5871 | t1Gd | F | 37 | 86.183 | AX_T1_POST_GD_FLAIR | 11 | 1.41281E+15 | 12/6/1994 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 3212.43 | 6.356 | 1238 | 127.7322 | 3 | 2.5 | 90 | 1.6273 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-5871/1.3.6.1.4.1.14519.5.2.1.4591.4003.247530214976989618914834177531/1.3.6.1.4.1.14519.5.2.1.4591.4003.109993263227573949563103763849 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5871 | t1 | F | 37 | 86.183 | AX_T1_pre_gd | 8 | 1.41281E+15 | 12/6/1994 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 5 | 2040.27 | 6.356 | 883 | 127.7322 | 3 | 5 | 90 | 1.2815 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-5871/1.3.6.1.4.1.14519.5.2.1.4591.4003.247530214976989618914834177531/1.3.6.1.4.1.14519.5.2.1.4591.4003.264195765064795376195430575838 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5871 | t2 | F | 37 | 86.183 | AX_T2_FR-FSE_RF2_150 | 7 | 1.41281E+15 | 12/6/1994 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 4000 | 103.464 | 0 | 127.7322 | 3 | 5 | 90 | 1.2142 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-5871/1.3.6.1.4.1.14519.5.2.1.4591.4003.247530214976989618914834177531/1.3.6.1.4.1.14519.5.2.1.4591.4003.313460819031107627612831965483 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5872 | flair | F | 43 | 102.965 | Ax_FLAIR_2.5mm_for_surgery | 5 | 6.32252E+15 | 2/23/1995 | GE MEDICAL SYSTEMS | Signa HDxt | IR | 2D | 2.5 | 10004 | 149.544 | 2200 | 63.8858 | 1.5 | 2.5 | 90 | 0.0316 | 71 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-5872/1.3.6.1.4.1.14519.5.2.1.4591.4003.120185163731564589832419288714/1.3.6.1.4.1.14519.5.2.1.4591.4003.261961539641753291673667731127 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-5872 | t1Gd | F | 43 | 102.965 | Ax_T1_2.5mm_for_surgery | 13 | 6.32252E+15 | 2/23/1995 | GE MEDICAL SYSTEMS | Signa HDxt | SE | 2D | 2.5 | 433.336 | 13 | 0 | 63.88584 | 1.5 | 2.5 | 90 | 0.0489826 | 71 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-5872/1.3.6.1.4.1.14519.5.2.1.4591.4003.120185163731564589832419288714/1.3.6.1.4.1.14519.5.2.1.4591.4003.312067442126966343281611085579 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-5872 | t1 | F | 43 | 102.965 | AXIAL_T1 | 11 | 6.32252E+15 | 2/23/1995 | GE MEDICAL SYSTEMS | Signa HDxt | SE | 2D | 2.5 | 433.336 | 13 | 0 | 63.88584 | 1.5 | 2.5 | 90 | 0.0489826 | 71 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-5872/1.3.6.1.4.1.14519.5.2.1.4591.4003.120185163731564589832419288714/1.3.6.1.4.1.14519.5.2.1.4591.4003.292214029866915931071160355226 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-5872 | t2 | F | 43 | 102.965 | AXIAL_FSE | 8 | 6.32252E+15 | 2/23/1995 | GE MEDICAL SYSTEMS | Signa HDxt | SE | 2D | 5 | 3500 | 23.688 | 0 | 63.88584 | 1.5 | 5 | 90 | 0.1158 | 72 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-5872/1.3.6.1.4.1.14519.5.2.1.4591.4003.120185163731564589832419288714/1.3.6.1.4.1.14519.5.2.1.4591.4003.560328669711494922031263596257 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-5874 | flair | F | 62 | 72.575 | AXIAL_FLAIR | 8 | 1.72824E+15 | 5/10/1995 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7323 | 3 | 2.5 | 90 | 0.5936 | 75 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-5874/1.3.6.1.4.1.14519.5.2.1.4591.4003.146738498094443311044095973527/1.3.6.1.4.1.14519.5.2.1.4591.4003.313704071781391494489476699641 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5874 | t1Gd | F | 62 | 72.575 | AX_T1_POST_GD_FLAIR | 12 | 1.72824E+15 | 5/10/1995 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 3042 | 6.356 | 1238 | 127.7323 | 3 | 2.5 | 90 | 1.3335 | 75 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-5874/1.3.6.1.4.1.14519.5.2.1.4591.4003.146738498094443311044095973527/1.3.6.1.4.1.14519.5.2.1.4591.4003.911828461315012354254276528074 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5874 | t1 | F | 62 | 72.575 | AX_T1_pre_gd | 9 | 1.72824E+15 | 5/10/1995 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 5 | 2039.94 | 6.356 | 883 | 127.7323 | 3 | 5 | 90 | 1.5118 | 38 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-5874/1.3.6.1.4.1.14519.5.2.1.4591.4003.146738498094443311044095973527/1.3.6.1.4.1.14519.5.2.1.4591.4003.244192045261781363315969668886 |
| TCGA-LGG | TCGA-DU | TCGA-DU-5874 | t2 | F | 62 | 72.575 | AX_T2_FR-FSE_RF2_150 | 6 | 1.72824E+15 | 5/10/1995 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 4000 | 103.464 | 0 | 127.7323 | 3 | 5 | 90 | 0.9547 | 38 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-5874/1.3.6.1.4.1.14519.5.2.1.4591.4003.146738498094443311044095973527/1.3.6.1.4.1.14519.5.2.1.4591.4003.282335375849209407307272273321 |
| TCGA-LGG | TCGA-DU | TCGA-DU-6397 | flair | M | 92 | AXIAL_FLAIR | 3 | 3.39228E+15 | 1/30/1985 | General Electric | GENESIS | flair | 3 | 10004 | 155 | 2200 | 3 | 90 | 60 | 0.938\0.938 | TCGA-DU-6397/1.3.6.1.4.1.14519.5.2.1.4591.4003.258077058743333147571656830833/1.3.6.1.4.1.14519.5.2.1.4591.4003.188438482998491912137083743459 | ||||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6397 | t1Gd | M | 92 | AXIAL_T1_POST_GD | 9 | 3.39228E+15 | 1/30/1985 | General Electric | GENESIS | memp | 3 | 500 | 14 | 0 | 3 | 90 | 60 | 0.938\0.938 | TCGA-DU-6397/1.3.6.1.4.1.14519.5.2.1.4591.4003.284484161156328443723397198607/1.3.6.1.4.1.14519.5.2.1.4591.4003.270565032379072141870325726078 | ||||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6397 | t1 | M | 92 | AXIAL_T1_PRE_GD | 7 | 3.39228E+15 | 1/30/1985 | General Electric | GENESIS | memp | 3 | 500 | 14 | 0 | 3 | 90 | 60 | 0.938\0.938 | TCGA-DU-6397/1.3.6.1.4.1.14519.5.2.1.4591.4003.258077058743333147571656830833/1.3.6.1.4.1.14519.5.2.1.4591.4003.274264134427812998608655897757 | ||||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6397 | t2 | M | 92 | AXIAL__FSE | 2 | 3.39228E+15 | 1/30/1985 | General Electric | GENESIS | fse | 3 | 3500 | 22 | 0 | 3 | 90 | 120 | 0.938\0.938 | TCGA-DU-6397/1.3.6.1.4.1.14519.5.2.1.4591.4003.284484161156328443723397198607/1.3.6.1.4.1.14519.5.2.1.4591.4003.899617347532250132137213482681 | ||||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6399 | flair | M | 81 | AXIAL_FLAIR | 3 | 1.11121E+15 | 4/16/1983 | General Electric | BAY | GENESIS | flair | 3 | 10004 | 155 | 2200 | 3 | 90 | 53 | 0.938\0.938 | TCGA-DU-6399/1.3.6.1.4.1.14519.5.2.1.4591.4003.299248386745518212707766902195/1.3.6.1.4.1.14519.5.2.1.4591.4003.130842300032756201956702941632 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6399 | t1Gd | M | 81 | AXIAL_T1_PRE/POST_GD | 7 | 1.11121E+15 | 4/16/1983 | General Electric | BAY | GENESIS | memp | 3 | 500 | 8 | 0 | 3 | 90 | 53 | 0.938\0.938 | TCGA-DU-6399/1.3.6.1.4.1.14519.5.2.1.4591.4003.299248386745518212707766902195/1.3.6.1.4.1.14519.5.2.1.4591.4003.278779321592049493927377920886 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6399 | t1 | M | 81 | AXIAL_T1_PRE/POST_GD | 9 | 1.11121E+15 | 4/16/1983 | General Electric | BAY | GENESIS | memp | 3 | 500 | 8 | 0 | 3 | 90 | 53 | 0.938\0.938 | TCGA-DU-6399/1.3.6.1.4.1.14519.5.2.1.4591.4003.299248386745518212707766902195/1.3.6.1.4.1.14519.5.2.1.4591.4003.272344052386358898114067225953 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6399 | t2 | M | 81 | AXIAL_FSE | 2 | 1.11121E+15 | 4/16/1983 | General Electric | BAY | GENESIS | fse | 3 | 3500 | 22 | 0 | 3 | 90 | 106 | 0.938\0.938 | TCGA-DU-6399/1.3.6.1.4.1.14519.5.2.1.4591.4003.299248386745518212707766902195/1.3.6.1.4.1.14519.5.2.1.4591.4003.217995210743248085634186942839 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6400 | flair | F | 90 | AXIAL_FLAIR | 3 | 2.10335E+15 | 5/18/1983 | General Electric | BAY | GENESIS | flair | 3 | 10004 | 155 | 2200 | 3 | 90 | 57 | 0.938\0.938 | TCGA-DU-6400/1.3.6.1.4.1.14519.5.2.1.4591.4003.908985028885715482708348438359/1.3.6.1.4.1.14519.5.2.1.4591.4003.566925305517233881271079424180 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6400 | t1Gd | F | 90 | axial_t1/gd | 9 | 2.10335E+15 | 5/18/1983 | General Electric | BAY | GENESIS | memp | 3 | 500 | 8 | 0 | 3 | 90 | 57 | 0.938\0.938 | TCGA-DU-6400/1.3.6.1.4.1.14519.5.2.1.4591.4003.908985028885715482708348438359/1.3.6.1.4.1.14519.5.2.1.4591.4003.192875840084482262754628153598 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6400 | t1 | F | 90 | axial_t1_pre_gd | 7 | 2.10335E+15 | 5/18/1983 | General Electric | BAY | GENESIS | memp | 3 | 500 | 8 | 0 | 3 | 90 | 57 | 0.938\0.938 | TCGA-DU-6400/1.3.6.1.4.1.14519.5.2.1.4591.4003.908985028885715482708348438359/1.3.6.1.4.1.14519.5.2.1.4591.4003.189029122549488052602811406393 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6400 | t2 | F | 90 | AXIAL_FSE | 2 | 2.10335E+15 | 5/18/1983 | General Electric | BAY | GENESIS | fse | 3 | 3500 | 22 | 0 | 3 | 90 | 114 | 0.938\0.938 | TCGA-DU-6400/1.3.6.1.4.1.14519.5.2.1.4591.4003.908985028885715482708348438359/1.3.6.1.4.1.14519.5.2.1.4591.4003.706430347214749666182639541093 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6401 | flair | F | 105 | AXIAL_FLAIR | 3 | 1.81358E+15 | 10/1/1983 | General Electric | GENESIS | flair | 3 | 10004 | 155 | 2200 | 3 | 90 | 51 | 0.938\0.938 | TCGA-DU-6401/1.3.6.1.4.1.14519.5.2.1.4591.4003.313654124851567890245405685812/1.3.6.1.4.1.14519.5.2.1.4591.4003.277524703900606944630060015579 | ||||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6401 | t1Gd | F | 105 | AXIAL_T1_POST_GD | 7 | 1.81358E+15 | 10/1/1983 | General Electric | GENESIS | memp | 3 | 500 | 14 | 0 | 3 | 90 | 58 | 0.938\0.938 | TCGA-DU-6401/1.3.6.1.4.1.14519.5.2.1.4591.4003.313654124851567890245405685812/1.3.6.1.4.1.14519.5.2.1.4591.4003.119828237865293463375263609301 | ||||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6401 | t1 | F | 105 | AXIAL_T1_PRE_GD | 5 | 1.81358E+15 | 10/1/1983 | General Electric | GENESIS | memp | 3 | 500 | 14 | 0 | 3 | 90 | 58 | 0.938\0.938 | TCGA-DU-6401/1.3.6.1.4.1.14519.5.2.1.4591.4003.313654124851567890245405685812/1.3.6.1.4.1.14519.5.2.1.4591.4003.193992828285897381024209543565 | ||||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6401 | t2 | F | 105 | AXIAL__FSE | 2 | 1.81358E+15 | 10/1/1983 | General Electric | GENESIS | fse | 3 | 3500 | 22 | 0 | 3 | 90 | 102 | 0.938\0.938 | TCGA-DU-6401/1.3.6.1.4.1.14519.5.2.1.4591.4003.313654124851567890245405685812/1.3.6.1.4.1.14519.5.2.1.4591.4003.747042361635308948495529305328 | ||||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6404 | flair | F | 72 | AXIAL_FLAIR | 3 | 3.3067E+15 | 6/29/1985 | General Electric | BAY | GENESIS | flair | 3 | 10004 | 155 | 2200 | 3 | 90 | 53 | 0.938\0.938 | TCGA-DU-6404/1.3.6.1.4.1.14519.5.2.1.4591.4003.261147184642760980049477062392/1.3.6.1.4.1.14519.5.2.1.4591.4003.123957563256151384189601599356 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6404 | t1Gd | F | 72 | AXIAL_T1_POST_GD | 10 | 3.3067E+15 | 6/29/1985 | General Electric | BAY | GENESIS | memp | 3 | 500 | 14 | 0 | 3 | 90 | 55 | 0.938\0.938 | TCGA-DU-6404/1.3.6.1.4.1.14519.5.2.1.4591.4003.261147184642760980049477062392/1.3.6.1.4.1.14519.5.2.1.4591.4003.416743692444202760044121743862 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6404 | t1 | F | 72 | AXIAL_T1_PRE_GD | 7 | 3.3067E+15 | 6/29/1985 | General Electric | BAY | GENESIS | memp | 3 | 500 | 14 | 0 | 3 | 90 | 55 | 0.938\0.938 | TCGA-DU-6404/1.3.6.1.4.1.14519.5.2.1.4591.4003.261147184642760980049477062392/1.3.6.1.4.1.14519.5.2.1.4591.4003.318797057168031444167840441025 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6404 | t2 | F | 72 | AXIAL__FSE | 2 | 3.3067E+15 | 6/29/1985 | General Electric | BAY | GENESIS | fse | 3 | 3500 | 22 | 0 | 3 | 90 | 110 | 0.938\0.938 | TCGA-DU-6404/1.3.6.1.4.1.14519.5.2.1.4591.4003.261147184642760980049477062392/1.3.6.1.4.1.14519.5.2.1.4591.4003.187814519293639539595282245500 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6405 | flair | F | 74 | AXIAL_FLAIR | 3 | 1.72784E+15 | 10/5/1985 | General Electric | BAY | GENESIS | flair | 3 | 10004 | 155 | 2200 | 3 | 90 | 60 | 0.938\0.938 | TCGA-DU-6405/1.3.6.1.4.1.14519.5.2.1.4591.4003.185886426431608624150226113206/1.3.6.1.4.1.14519.5.2.1.4591.4003.165112875200401239584250174943 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6405 | t1Gd | F | 74 | AXIAL_T1_POST_GD | 10 | 1.72784E+15 | 10/5/1985 | General Electric | BAY | GENESIS | memp | 3 | 500 | 14 | 0 | 3 | 90 | 60 | 0.938\0.938 | TCGA-DU-6405/1.3.6.1.4.1.14519.5.2.1.4591.4003.185886426431608624150226113206/1.3.6.1.4.1.14519.5.2.1.4591.4003.652945086744147125437089852534 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6405 | t1 | F | 74 | AXIAL_T1_PRE_GD | 7 | 1.72784E+15 | 10/5/1985 | General Electric | BAY | GENESIS | memp | 3 | 500 | 14 | 0 | 3 | 90 | 60 | 0.938\0.938 | TCGA-DU-6405/1.3.6.1.4.1.14519.5.2.1.4591.4003.185886426431608624150226113206/1.3.6.1.4.1.14519.5.2.1.4591.4003.322175775658634614307924880128 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6405 | t2 | F | 74 | AXIAL__FSE | 2 | 1.72784E+15 | 10/5/1985 | General Electric | BAY | GENESIS | fse | 3 | 3500 | 22 | 0 | 3 | 90 | 120 | 0.938\0.938 | TCGA-DU-6405/1.3.6.1.4.1.14519.5.2.1.4591.4003.185886426431608624150226113206/1.3.6.1.4.1.14519.5.2.1.4591.4003.233437082494886273190709309681 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6407 | flair | F | 35 | 75.296 | AXIAL_FLAIR | 3 | 4.68473E+15 | 5/14/1986 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | RM | 2D | 3 | 10004 | 155 | 63.85846 | 1.5 | 3 | 90 | 0.033598 | 58 | 256 | 192 | 0.9375000000\0.9375000000 | TCGA-DU-6407/1.3.6.1.4.1.14519.5.2.1.4591.4003.115454172245963376362112828234/1.3.6.1.4.1.14519.5.2.1.4591.4003.118594670002293851625094179535 | ||
| TCGA-LGG | TCGA-DU | TCGA-DU-6407 | t1Gd | F | 35 | 75.296 | AXIAL_T1_POST_GD | 1 | 2.58298E+15 | 5/14/1986 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 3 | 500 | 14 | 63.85847 | 1.5 | 3 | 90 | 0.044591 | 58 | 256 | 192 | 0.9375000000\0.9375000000 | TCGA-DU-6407/1.3.6.1.4.1.14519.5.2.1.4591.4003.189340939734965094898503523740/1.3.6.1.4.1.14519.5.2.1.4591.4003.279601843629640300146176277810 | ||
| TCGA-LGG | TCGA-DU | TCGA-DU-6407 | t1 | F | 35 | 75.296 | AXIAL_T1_PRE-GAD/SAME_LOCS | 7 | 4.68473E+15 | 5/14/1986 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 3 | 500 | 14 | 63.85846 | 1.5 | 3 | 90 | 0.044591 | 58 | 256 | 192 | 0.9375000000\0.9375000000 | TCGA-DU-6407/1.3.6.1.4.1.14519.5.2.1.4591.4003.115454172245963376362112828234/1.3.6.1.4.1.14519.5.2.1.4591.4003.231046420302507784804996585380 | ||
| TCGA-LGG | TCGA-DU | TCGA-DU-6407 | t2 | F | 35 | 75.296 | AXIAL__FSE | 2 | 4.68473E+15 | 5/14/1986 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 3 | 3500 | 22 | 63.85846 | 1.5 | 3 | 90 | 0.059279 | 116 | 256 | 192 | 0.9375000000\0.9375000000 | TCGA-DU-6407/1.3.6.1.4.1.14519.5.2.1.4591.4003.115454172245963376362112828234/1.3.6.1.4.1.14519.5.2.1.4591.4003.651809519364398826987125576597 | ||
| TCGA-LGG | TCGA-DU | TCGA-DU-6408 | flair | F | 68 | AXIAL_FLAIR | 3 | 2.17053E+15 | 5/21/1986 | General Electric | BAY | GENESIS | flair | 3 | 10004 | 155 | 2200 | 3 | 90 | 56 | 0.938\0.938 | TCGA-DU-6408/1.3.6.1.4.1.14519.5.2.1.4591.4003.112304111943954735219267840424/1.3.6.1.4.1.14519.5.2.1.4591.4003.206509980716702405576239322793 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6408 | t1Gd | F | 68 | AXIAL_T1_POST_GD | 8 | 2.17053E+15 | 5/21/1986 | General Electric | BAY | GENESIS | memp | 3 | 500 | 14 | 0 | 3 | 90 | 56 | 0.938\0.938 | TCGA-DU-6408/1.3.6.1.4.1.14519.5.2.1.4591.4003.112304111943954735219267840424/1.3.6.1.4.1.14519.5.2.1.4591.4003.192752058381051614023466272787 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6408 | t1 | F | 68 | AXIAL_T1_PRE-GAD. | 7 | 2.17053E+15 | 5/21/1986 | General Electric | BAY | GENESIS | memp | 3 | 500 | 14 | 0 | 3 | 90 | 56 | 0.938\0.938 | TCGA-DU-6408/1.3.6.1.4.1.14519.5.2.1.4591.4003.112304111943954735219267840424/1.3.6.1.4.1.14519.5.2.1.4591.4003.232902849705284049546633334954 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6408 | t2 | F | 68 | AXIAL__FSE | 2 | 2.17053E+15 | 5/21/1986 | General Electric | BAY | GENESIS | fse | 3 | 3500 | 22 | 0 | 3 | 90 | 112 | 0.938\0.938 | TCGA-DU-6408/1.3.6.1.4.1.14519.5.2.1.4591.4003.112304111943954735219267840424/1.3.6.1.4.1.14519.5.2.1.4591.4003.244913018716376399862060897029 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-6410 | flair | M | 56 | 81.647 | Ax_FLAIR_2.5mm_for_surgery | 4 | 3.17478E+15 | 12/28/1995 | GE MEDICAL SYSTEMS | MR01MROW | Signa HDxt | IR | 2D | 2.5 | 10004 | 151.8 | 2200 | 63.88581 | 1.5 | 2.5 | 90 | 0.0351 | 74 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-6410/1.3.6.1.4.1.14519.5.2.1.4591.4003.265163022076336806171223776637/1.3.6.1.4.1.14519.5.2.1.4591.4003.617078350735972730657422815388 |
| TCGA-LGG | TCGA-DU | TCGA-DU-6410 | t1Gd | M | 56 | 81.647 | Ax_T1_2.5mm_for_surgery | 11 | 3.17478E+15 | 12/28/1995 | GE MEDICAL SYSTEMS | MR01MROW | Signa HDxt | SE | 2D | 2.5 | 500 | 13 | 0 | 63.88583 | 1.5 | 2.5 | 90 | 0.0440288 | 74 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-6410/1.3.6.1.4.1.14519.5.2.1.4591.4003.265163022076336806171223776637/1.3.6.1.4.1.14519.5.2.1.4591.4003.704707103419055538089427941121 |
| TCGA-LGG | TCGA-DU | TCGA-DU-6410 | t1 | M | 56 | 81.647 | AXIAL_T1 | 8 | 3.17478E+15 | 12/28/1995 | GE MEDICAL SYSTEMS | MR01MROW | Signa HDxt | SE | 2D | 5 | 500 | 14 | 0 | 63.88581 | 1.5 | 5 | 90 | 0.038159 | 38 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-6410/1.3.6.1.4.1.14519.5.2.1.4591.4003.265163022076336806171223776637/1.3.6.1.4.1.14519.5.2.1.4591.4003.506942221024780688931689457526 |
| TCGA-LGG | TCGA-DU | TCGA-DU-6410 | t2 | M | 56 | 81.647 | AXIAL_FSE | 3 | 3.17478E+15 | 12/28/1995 | GE MEDICAL SYSTEMS | MR01MROW | Signa HDxt | SE | 2D | 5 | 3500 | 23.16 | 0 | 63.88581 | 1.5 | 5 | 90 | 0.1269 | 76 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-6410/1.3.6.1.4.1.14519.5.2.1.4591.4003.265163022076336806171223776637/1.3.6.1.4.1.14519.5.2.1.4591.4003.334542090086353921649284997370 |
| TCGA-LGG | TCGA-DU | TCGA-DU-6542 | flair | U | 25 | 72.575 | AXIAL_FLAIR | 4 | 1.33377E+15 | 5/8/1996 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7323 | 3 | 2.5 | 90 | 0.6678 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-6542/1.3.6.1.4.1.14519.5.2.1.4591.4003.179855774303237007643875027308/1.3.6.1.4.1.14519.5.2.1.4591.4003.341644629709955365244089935548 |
| TCGA-LGG | TCGA-DU | TCGA-DU-6542 | t1Gd | U | 25 | 72.575 | AX_T1_POST_GD_FLAIR | 11 | 1.33377E+15 | 5/8/1996 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 3145.81 | 6.356 | 1238 | 127.7323 | 3 | 2.5 | 90 | 1.2379 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-6542/1.3.6.1.4.1.14519.5.2.1.4591.4003.179855774303237007643875027308/1.3.6.1.4.1.14519.5.2.1.4591.4003.666216402911624214152103699692 |
| TCGA-LGG | TCGA-DU | TCGA-DU-6542 | t1 | U | 25 | 72.575 | AX_T1_pre_gd | 8 | 1.33377E+15 | 5/8/1996 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 5 | 2039.94 | 6.356 | 883 | 127.7323 | 3 | 5 | 90 | 1.5118 | 37 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-6542/1.3.6.1.4.1.14519.5.2.1.4591.4003.179855774303237007643875027308/1.3.6.1.4.1.14519.5.2.1.4591.4003.767410396026066548941611317760 |
| TCGA-LGG | TCGA-DU | TCGA-DU-6542 | t2 | U | 25 | 72.575 | AX_T2_FR-FSE_RF2_150 | 7 | 1.33377E+15 | 5/8/1996 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 4000 | 103.464 | 0 | 127.7323 | 3 | 5 | 90 | 0.9547 | 37 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-6542/1.3.6.1.4.1.14519.5.2.1.4591.4003.179855774303237007643875027308/1.3.6.1.4.1.14519.5.2.1.4591.4003.827513524462400327174945110201 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7008 | flair | F | 67 | AXIAL_FLAIR | 3 | 2.46393E+15 | 7/23/1983 | General Electric | BAY | GENESIS | flair | 3 | 10004 | 155 | 2200 | 3 | 90 | 52 | 0.938\0.938 | TCGA-DU-7008/1.3.6.1.4.1.14519.5.2.1.4591.4003.122287415106636818029092960413/1.3.6.1.4.1.14519.5.2.1.4591.4003.238576995972747484602053820193 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-7008 | t1Gd | F | 67 | AXIAL_T1_POST_GD | 9 | 2.46393E+15 | 7/23/1983 | General Electric | BAY | GENESIS | memp | 3 | 500 | 14 | 0 | 3 | 90 | 52 | 0.938\0.938 | TCGA-DU-7008/1.3.6.1.4.1.14519.5.2.1.4591.4003.122287415106636818029092960413/1.3.6.1.4.1.14519.5.2.1.4591.4003.326331027531602715966185898176 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-7008 | t1 | F | 67 | AXIAL_T1_PRE_GD | 7 | 2.46393E+15 | 7/23/1983 | General Electric | BAY | GENESIS | memp | 3 | 500 | 14 | 0 | 3 | 90 | 52 | 0.938\0.938 | TCGA-DU-7008/1.3.6.1.4.1.14519.5.2.1.4591.4003.122287415106636818029092960413/1.3.6.1.4.1.14519.5.2.1.4591.4003.512451012844469919337165237750 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-7008 | t2 | F | 67 | AXIAL_FSE | 2 | 2.46393E+15 | 7/23/1983 | General Electric | BAY | GENESIS | fse | 3 | 3500 | 22 | 0 | 3 | 90 | 104 | 0.781\0.781 | TCGA-DU-7008/1.3.6.1.4.1.14519.5.2.1.4591.4003.122287415106636818029092960413/1.3.6.1.4.1.14519.5.2.1.4591.4003.287636355705793503766039754021 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-7010 | flair | F | 58 | 131.542 | AXIAL_FLAIR | 3 | 1.02385E+15 | 3/7/1986 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | RM | 2D | 3 | 10004 | 155 | 63.78261 | 1.5 | 3 | 90 | 0.030729 | 58 | 256 | 192 | 0.9375000000\0.9375000000 | TCGA-DU-7010/1.3.6.1.4.1.14519.5.2.1.4591.4003.143081416024357624170479022330/1.3.6.1.4.1.14519.5.2.1.4591.4003.264069163339825265971003353649 | ||
| TCGA-LGG | TCGA-DU | TCGA-DU-7010 | t1Gd | F | 58 | 131.542 | AXIAL_T1_POST_GD | 10 | 1.02385E+15 | 3/7/1986 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 3 | 600 | 14 | 63.7826 | 1.5 | 3 | 90 | 0.045284 | 58 | 256 | 192 | 0.9375000000\0.9375000000 | TCGA-DU-7010/1.3.6.1.4.1.14519.5.2.1.4591.4003.143081416024357624170479022330/1.3.6.1.4.1.14519.5.2.1.4591.4003.586339173501030344454818020544 | ||
| TCGA-LGG | TCGA-DU | TCGA-DU-7010 | t1 | F | 58 | 131.542 | AXIAL_T1_PRE-GAD. | 6 | 1.02385E+15 | 3/7/1986 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 3 | 500 | 14 | 63.7826 | 1.5 | 3 | 90 | 0.040783 | 58 | 256 | 192 | 0.9375000000\0.9375000000 | TCGA-DU-7010/1.3.6.1.4.1.14519.5.2.1.4591.4003.143081416024357624170479022330/1.3.6.1.4.1.14519.5.2.1.4591.4003.323984999799359680736512212053 | ||
| TCGA-LGG | TCGA-DU | TCGA-DU-7010 | t2 | F | 58 | 131.542 | AXIAL__FSE | 2 | 1.02385E+15 | 3/7/1986 | GE MEDICAL SYSTEMS | GENESIS_SIGNA | SE | 2D | 3 | 3500 | 22 | 63.78261 | 1.5 | 3 | 90 | 0.054217 | 116 | 256 | 192 | 0.9375000000\0.9375000000 | TCGA-DU-7010/1.3.6.1.4.1.14519.5.2.1.4591.4003.143081416024357624170479022330/1.3.6.1.4.1.14519.5.2.1.4591.4003.530247852057132627671867267641 | ||
| TCGA-LGG | TCGA-DU | TCGA-DU-7014 | flair | M | 135 | AXIAL_FLAIR | 5 | 4.3765E+15 | 6/18/1986 | General Electric | BAY | GENESIS | flair | 3 | 10004 | 155 | 2200 | 3 | 90 | 60 | 0.938\0.938 | TCGA-DU-7014/1.3.6.1.4.1.14519.5.2.1.4591.4003.241850919885310339695393380617/1.3.6.1.4.1.14519.5.2.1.4591.4003.153756522270512057920568006644 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-7014 | t1Gd | M | 135 | AXIAL_T1_POST_GD | 11 | 4.3765E+15 | 6/18/1986 | General Electric | BAY | GENESIS | memp | 3 | 500 | 14 | 0 | 3 | 90 | 60 | 0.938\0.938 | TCGA-DU-7014/1.3.6.1.4.1.14519.5.2.1.4591.4003.241850919885310339695393380617/1.3.6.1.4.1.14519.5.2.1.4591.4003.190391970345142170156174344067 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-7014 | t1 | M | 135 | AXIAL_T1_PRE-GAD. | 9 | 4.3765E+15 | 6/18/1986 | General Electric | BAY | GENESIS | memp | 3 | 500 | 14 | 0 | 3 | 90 | 60 | 0.938\0.938 | TCGA-DU-7014/1.3.6.1.4.1.14519.5.2.1.4591.4003.241850919885310339695393380617/1.3.6.1.4.1.14519.5.2.1.4591.4003.182726989500629952477447903736 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-7014 | t2 | M | 135 | AXIAL__FSE | 3 | 4.3765E+15 | 6/18/1986 | General Electric | BAY | GENESIS | fse | 3 | 3500 | 22 | 0 | 3 | 90 | 118 | 0.938\0.938 | TCGA-DU-7014/1.3.6.1.4.1.14519.5.2.1.4591.4003.241850919885310339695393380617/1.3.6.1.4.1.14519.5.2.1.4591.4003.196810229656728396616579184712 | |||||||
| TCGA-LGG | TCGA-DU | TCGA-DU-7015 | flair | F | 41 | 90.718 | AXIAL_FLAIR | 8 | 2.22697E+15 | 6/18/1989 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 2.5 | 10002 | 124.9 | 2250 | 127.7324 | 3 | 2.5 | 90 | 0.8978 | 72 | 320 | 224 | 0.46875\0.46875 | TCGA-DU-7015/1.3.6.1.4.1.14519.5.2.1.4591.4003.213617385466211233370402716160/1.3.6.1.4.1.14519.5.2.1.4591.4003.933475500875157813970798042112 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7015 | t1Gd | F | 41 | 90.718 | AX_T1_POST_GD_FLAIR | 10 | 2.22697E+15 | 6/18/1989 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 2.5 | 2965.64 | 6.956 | 1238 | 127.7324 | 3 | 2.5 | 90 | 1.8622 | 85 | 320 | 224 | 0.46875\0.46875 | TCGA-DU-7015/1.3.6.1.4.1.14519.5.2.1.4591.4003.213617385466211233370402716160/1.3.6.1.4.1.14519.5.2.1.4591.4003.286591706137405018110350719430 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7015 | t1 | F | 41 | 90.718 | AX_T1_pre_gd | 7 | 2.22697E+15 | 6/18/1989 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 5 | 2965.71 | 6.964 | 1238 | 127.7325 | 3 | 5 | 90 | 1.8622 | 51 | 320 | 224 | 0.46875\0.46875 | TCGA-DU-7015/1.3.6.1.4.1.14519.5.2.1.4591.4003.213617385466211233370402716160/1.3.6.1.4.1.14519.5.2.1.4591.4003.108609639777979340339125393841 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7015 | t2 | F | 41 | 90.718 | AX_T2_FR-FSE_RF2_150 | 5 | 2.22697E+15 | 6/18/1989 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 3000 | 104.436 | 0 | 127.7324 | 3 | 5 | 90 | 1.3494 | 42 | 320 | 224 | 0.46875\0.46875 | TCGA-DU-7015/1.3.6.1.4.1.14519.5.2.1.4591.4003.213617385466211233370402716160/1.3.6.1.4.1.14519.5.2.1.4591.4003.716639613302745992208497298258 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7018 | flair | F | 57 | 59.874 | AXIAL_FLAIR | 8 | 1.77649E+15 | 12/20/1991 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7323 | 3 | 2.5 | 90 | 0.4305 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7018/1.3.6.1.4.1.14519.5.2.1.4591.4003.244030423007851267821957098852/1.3.6.1.4.1.14519.5.2.1.4591.4003.330490545332144580881833428777 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7018 | t1Gd | F | 57 | 59.874 | AX_T1_POST_GD_FLAIR | 13 | 1.77649E+15 | 12/20/1991 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 2.5 | 3115.28 | 6.356 | 1238 | 127.7323 | 3 | 2.5 | 90 | 1.5811 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7018/1.3.6.1.4.1.14519.5.2.1.4591.4003.244030423007851267821957098852/1.3.6.1.4.1.14519.5.2.1.4591.4003.185659203907953115031317947091 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7018 | t1 | F | 57 | 59.874 | AX_T1_pre_gd | 10 | 1.77649E+15 | 12/20/1991 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 5 | 3115.42 | 6.356 | 1238 | 127.7323 | 3 | 5 | 90 | 1.581 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7018/1.3.6.1.4.1.14519.5.2.1.4591.4003.244030423007851267821957098852/1.3.6.1.4.1.14519.5.2.1.4591.4003.171723091698922856226026800693 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7018 | t2 | F | 57 | 59.874 | AX_T2_FR-FSE_RF2_150 | 5 | 1.77649E+15 | 12/20/1991 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 3000 | 103.464 | 0 | 127.7323 | 3 | 5 | 90 | 0.9517 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7018/1.3.6.1.4.1.14519.5.2.1.4591.4003.244030423007851267821957098852/1.3.6.1.4.1.14519.5.2.1.4591.4003.292747299771299775245149391572 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7019 | flair | M | 39 | 83.915 | AXIAL_FLAIR | 6 | 3.3163E+15 | 9/8/1994 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7322 | 3 | 2.5 | 90 | 0.7285 | 76 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7019/1.3.6.1.4.1.14519.5.2.1.4591.4003.307537448187255340523133336813/1.3.6.1.4.1.14519.5.2.1.4591.4003.151973570528562002920929536919 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7019 | t1Gd | M | 39 | 83.915 | AX_T1_POST_GD_FLAIR | 11 | 3.3163E+15 | 9/8/1994 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 3152.26 | 6.356 | 1238 | 127.7322 | 3 | 2.5 | 90 | 1.7163 | 76 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7019/1.3.6.1.4.1.14519.5.2.1.4591.4003.307537448187255340523133336813/1.3.6.1.4.1.14519.5.2.1.4591.4003.261334161791196607349981234098 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7019 | t1 | M | 39 | 83.915 | AX_T1_pre_gd | 8 | 3.3163E+15 | 9/8/1994 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 5 | 3149.54 | 6.356 | 1238 | 127.7322 | 3 | 5 | 90 | 1.164 | 38 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7019/1.3.6.1.4.1.14519.5.2.1.4591.4003.307537448187255340523133336813/1.3.6.1.4.1.14519.5.2.1.4591.4003.240955732907901933945193246981 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7019 | t2 | M | 39 | 83.915 | AX_T2_FR-FSE_RF2_150 | 5 | 3.3163E+15 | 9/8/1994 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 3000 | 103.464 | 0 | 127.7322 | 3 | 5 | 90 | 1.1172 | 38 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7019/1.3.6.1.4.1.14519.5.2.1.4591.4003.307537448187255340523133336813/1.3.6.1.4.1.14519.5.2.1.4591.4003.288258067481390732955397456476 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7294 | flair | F | 53 | 77.111 | AXIAL_FLAIR_RF2__150 | 7 | 3.10478E+15 | 1/4/1989 | GE MEDICAL SYSTEMS | MR3T | GENESIS_SIGNA | IR | 2D | 2.5 | 10002 | 123.5 | 2250 | 1.28E+09 | 3 | 2.5 | 90 | 0.085563 | 68 | 256 | 192 | 0.937500\0.937500 | TCGA-DU-7294/1.3.6.1.4.1.14519.5.2.1.4591.4003.559420023378630100798761419917/1.3.6.1.4.1.14519.5.2.1.4591.4003.184784670334461777798195407121 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7294 | t1Gd | F | 53 | 77.111 | AX_T1_POST_GD_FLAIR | 13 | 3.10478E+15 | 1/4/1989 | GE MEDICAL SYSTEMS | MR3T | GENESIS_SIGNA | RM | 2D | 2.5 | 2382.272 | 8.104 | 860 | 1.28E+09 | 3 | 2.5 | 90 | 0.120246 | 68 | 288 | 192 | 0.468750\0.468750 | TCGA-DU-7294/1.3.6.1.4.1.14519.5.2.1.4591.4003.559420023378630100798761419917/1.3.6.1.4.1.14519.5.2.1.4591.4003.359311293105706905697963367377 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7294 | t1 | F | 53 | 77.111 | AX_T1_pre_gd | 8 | 3.10478E+15 | 1/4/1989 | GE MEDICAL SYSTEMS | MR3T | GENESIS_SIGNA | RM | 2D | 5 | 2382.476 | 8.104 | 860 | 1.28E+09 | 3 | 5 | 90 | 0.125032 | 34 | 288 | 192 | 0.468750\0.468750 | TCGA-DU-7294/1.3.6.1.4.1.14519.5.2.1.4591.4003.559420023378630100798761419917/1.3.6.1.4.1.14519.5.2.1.4591.4003.365826625949084795369137973491 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7294 | t2 | F | 53 | 77.111 | AX_T2_FR-FSE_RF2_150 | 4 | 3.10478E+15 | 1/4/1989 | GE MEDICAL SYSTEMS | MR3T | GENESIS_SIGNA | RM | 2D | 5 | 3500 | 104.972 | 1.28E+09 | 3 | 5 | 90 | 0.09 | 34 | 352 | 192 | 0.468750\0.468750 | TCGA-DU-7294/1.3.6.1.4.1.14519.5.2.1.4591.4003.559420023378630100798761419917/1.3.6.1.4.1.14519.5.2.1.4591.4003.152901242239851224383447108103 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-7298 | flair | F | 38 | 95.254 | Ax_FLAIR_2.5mm_for_surgery | 4 | 3.94591E+15 | 3/24/1991 | GE MEDICAL SYSTEMS | MR01MROW | SIGNA EXCITE | IR | 2D | 2.5 | 10004 | 149.544 | 2200 | 63.88599 | 1.5 | 2.5 | 90 | 0.0323 | 65 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-7298/1.3.6.1.4.1.14519.5.2.1.4591.4003.850439737313047023965195610889/1.3.6.1.4.1.14519.5.2.1.4591.4003.617708870507397603144839116562 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7298 | t1Gd | F | 38 | 95.254 | Ax_T1_2.5mm_for_surgery | 8 | 3.94591E+15 | 3/24/1991 | GE MEDICAL SYSTEMS | MR01MROW | SIGNA EXCITE | SE | 2D | 2.5 | 500 | 13 | 0 | 63.88599 | 1.5 | 2.5 | 90 | 0.0513142 | 65 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-7298/1.3.6.1.4.1.14519.5.2.1.4591.4003.850439737313047023965195610889/1.3.6.1.4.1.14519.5.2.1.4591.4003.522835700137350932664864037553 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7298 | t1 | F | 38 | 95.254 | AXIAL_T1 | 6 | 3.94591E+15 | 3/24/1991 | GE MEDICAL SYSTEMS | MR01MROW | SIGNA EXCITE | SE | 2D | 5 | 500 | 14 | 0 | 63.886 | 1.5 | 5 | 90 | 0.04582 | 32 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-7298/1.3.6.1.4.1.14519.5.2.1.4591.4003.850439737313047023965195610889/1.3.6.1.4.1.14519.5.2.1.4591.4003.227924142993589728677636698859 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7298 | t2 | F | 38 | 95.254 | AXIAL_FSE | 3 | 3.94591E+15 | 3/24/1991 | GE MEDICAL SYSTEMS | MR01MROW | SIGNA EXCITE | SE | 2D | 5 | 3500 | 23.688 | 0 | 63.88599 | 1.5 | 5 | 90 | 0.1075 | 66 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-7298/1.3.6.1.4.1.14519.5.2.1.4591.4003.850439737313047023965195610889/1.3.6.1.4.1.14519.5.2.1.4591.4003.328014632509126852764708350605 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7299 | flair | M | 33 | 95.708 | AXIAL_FLAIR | 5 | 6.19481E+15 | 4/17/1991 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7323 | 3 | 2.5 | 90 | 0.8754 | 72 | 320 | 224 | 0.46875\0.46875 | TCGA-DU-7299/1.3.6.1.4.1.14519.5.2.1.4591.4003.100550055416544983972223484114/1.3.6.1.4.1.14519.5.2.1.4591.4003.280133920197978134668503903289 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7299 | t1Gd | M | 33 | 95.708 | AX_T1_POST_GD_FLAIR | 10 | 6.19481E+15 | 4/17/1991 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 2.5 | 3079.21 | 6.356 | 1238 | 127.7323 | 3 | 2.5 | 90 | 1.6242 | 72 | 320 | 224 | 0.46875\0.46875 | TCGA-DU-7299/1.3.6.1.4.1.14519.5.2.1.4591.4003.100550055416544983972223484114/1.3.6.1.4.1.14519.5.2.1.4591.4003.208743704368662518972303449564 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7299 | t1 | M | 33 | 95.708 | AX_T1_pre_gd | 8 | 6.19481E+15 | 4/17/1991 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 5 | 3079.28 | 6.356 | 1238 | 127.7323 | 3 | 5 | 90 | 1.6242 | 36 | 320 | 224 | 0.46875\0.46875 | TCGA-DU-7299/1.3.6.1.4.1.14519.5.2.1.4591.4003.100550055416544983972223484114/1.3.6.1.4.1.14519.5.2.1.4591.4003.213644167210595395557913686144 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7299 | t2 | M | 33 | 95.708 | AX_T2_FR-FSE_RF2_150 | 6 | 6.19481E+15 | 4/17/1991 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 3000 | 103.464 | 0 | 127.7323 | 3 | 5 | 90 | 1.9378 | 36 | 320 | 224 | 0.46875\0.46875 | TCGA-DU-7299/1.3.6.1.4.1.14519.5.2.1.4591.4003.100550055416544983972223484114/1.3.6.1.4.1.14519.5.2.1.4591.4003.243528672587003301299349211418 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7300 | flair | F | 53 | 61.235 | AXIAL_FLAIR | 5 | 1.32855E+15 | 8/14/1991 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7323 | 3 | 2.5 | 90 | 0.432 | 71 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7300/1.3.6.1.4.1.14519.5.2.1.4591.4003.182337016313469829681151533968/1.3.6.1.4.1.14519.5.2.1.4591.4003.610344381534068539694672916329 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7300 | t1Gd | F | 53 | 61.235 | AX_T1_POST_GD_FLAIR | 10 | 1.32855E+15 | 8/14/1991 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 2.5 | 3126.94 | 6.356 | 1238 | 127.7323 | 3 | 2.5 | 90 | 1.6882 | 71 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7300/1.3.6.1.4.1.14519.5.2.1.4591.4003.182337016313469829681151533968/1.3.6.1.4.1.14519.5.2.1.4591.4003.107063738082189385510017517011 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7300 | t1 | F | 53 | 61.235 | AX_T1_pre_gd | 8 | 1.32855E+15 | 8/14/1991 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 5 | 3116 | 6.356 | 1238 | 127.7323 | 3 | 5 | 90 | 1.5862 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7300/1.3.6.1.4.1.14519.5.2.1.4591.4003.182337016313469829681151533968/1.3.6.1.4.1.14519.5.2.1.4591.4003.145651249822981603023440375477 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7300 | t2 | F | 53 | 61.235 | AX_T2_FR-FSE_RF2_150 | 6 | 1.32855E+15 | 8/14/1991 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 3000 | 103.464 | 0 | 127.7323 | 3 | 5 | 90 | 0.955 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7300/1.3.6.1.4.1.14519.5.2.1.4591.4003.182337016313469829681151533968/1.3.6.1.4.1.14519.5.2.1.4591.4003.240459259364124440138958818382 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7301 | flair | M | 53 | 102.058 | AXIAL_FLAIR | 6 | 2.67272E+15 | 11/12/1991 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7323 | 3 | 2.5 | 90 | 0.9521 | 69 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7301/1.3.6.1.4.1.14519.5.2.1.4591.4003.186750792022738934663242822050/1.3.6.1.4.1.14519.5.2.1.4591.4003.161378909706211331451432266613 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7301 | t1Gd | M | 53 | 102.058 | AX_T1_POST_GD_FLAIR | 10 | 2.67272E+15 | 11/12/1991 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 2.5 | 3099.22 | 6.356 | 1238 | 127.7323 | 3 | 2.5 | 90 | 1.7204 | 69 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7301/1.3.6.1.4.1.14519.5.2.1.4591.4003.186750792022738934663242822050/1.3.6.1.4.1.14519.5.2.1.4591.4003.159697020502195515104591556964 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7301 | t1 | M | 53 | 102.058 | AX_T1_pre_gd | 8 | 2.67272E+15 | 11/12/1991 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 5 | 3099.3 | 6.356 | 1238 | 127.7323 | 3 | 5 | 90 | 1.7204 | 35 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7301/1.3.6.1.4.1.14519.5.2.1.4591.4003.186750792022738934663242822050/1.3.6.1.4.1.14519.5.2.1.4591.4003.168130876918360040497499340072 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7301 | t2 | M | 53 | 102.058 | AX_T2_FR-FSE_RF2_150 | 5 | 2.67272E+15 | 11/12/1991 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 3000 | 103.464 | 0 | 127.7323 | 3 | 5 | 90 | 1.4421 | 35 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7301/1.3.6.1.4.1.14519.5.2.1.4591.4003.186750792022738934663242822050/1.3.6.1.4.1.14519.5.2.1.4591.4003.242378501402238333467282668026 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7302 | flair | F | 48 | 86.183 | AXIAL_FLAIR | 5 | 2.53184E+15 | 12/3/1991 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7323 | 3 | 2.5 | 90 | 0.7325 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7302/1.3.6.1.4.1.14519.5.2.1.4591.4003.103908736272347635807818341440/1.3.6.1.4.1.14519.5.2.1.4591.4003.339742571964695966242385646378 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7302 | t1Gd | F | 48 | 86.183 | AX_T1_POST_GD_FLAIR | 11 | 2.53184E+15 | 12/3/1991 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 2.5 | 3230.38 | 6.356 | 1238 | 127.7323 | 3 | 2.5 | 90 | 1.7293 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7302/1.3.6.1.4.1.14519.5.2.1.4591.4003.103908736272347635807818341440/1.3.6.1.4.1.14519.5.2.1.4591.4003.102325590172330913084588545183 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7302 | t1 | F | 48 | 86.183 | AX_T1_pre_gd | 8 | 2.53184E+15 | 12/3/1991 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 5 | 3028.59 | 6.356 | 1238 | 127.7323 | 3 | 5 | 90 | 1.3835 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7302/1.3.6.1.4.1.14519.5.2.1.4591.4003.103908736272347635807818341440/1.3.6.1.4.1.14519.5.2.1.4591.4003.319430134100597162181769253768 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7302 | t2 | F | 48 | 86.183 | AX_T2_FR-FSE_RF2_150 | 6 | 2.53184E+15 | 12/3/1991 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 3000 | 103.464 | 0 | 127.7323 | 3 | 5 | 90 | 1.6192 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7302/1.3.6.1.4.1.14519.5.2.1.4591.4003.103908736272347635807818341440/1.3.6.1.4.1.14519.5.2.1.4591.4003.214930116826612964540175085470 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7304 | flair | M | 43 | 77.111 | AXIAL_FLAIR | 5 | 9.06509E+15 | 3/25/1993 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7322 | 3 | 2.5 | 90 | 0.7409 | 71 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7304/1.3.6.1.4.1.14519.5.2.1.4591.4003.233329049923110225958892646100/1.3.6.1.4.1.14519.5.2.1.4591.4003.963112459262733039117409779801 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7304 | t1Gd | M | 43 | 77.111 | AX_T1_POST_GD_FLAIR | 10 | 9.06509E+15 | 3/25/1993 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 2.5 | 3151.38 | 6.356 | 1238 | 127.7322 | 3 | 2.5 | 90 | 1.2713 | 71 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7304/1.3.6.1.4.1.14519.5.2.1.4591.4003.233329049923110225958892646100/1.3.6.1.4.1.14519.5.2.1.4591.4003.278231939792427678289216579006 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7304 | t1 | M | 43 | 77.111 | AX_T1_pre_gd | 8 | 9.06509E+15 | 3/25/1993 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 5 | 3000.01 | 6.356 | 1238 | 127.7322 | 3 | 5 | 90 | 1.0016 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7304/1.3.6.1.4.1.14519.5.2.1.4591.4003.233329049923110225958892646100/1.3.6.1.4.1.14519.5.2.1.4591.4003.338852786223329525878990954697 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7304 | t2 | M | 43 | 77.111 | AX_T2_FR-FSE_RF2_150 | 6 | 9.06509E+15 | 3/25/1993 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 3000 | 103.464 | 0 | 127.7322 | 3 | 5 | 90 | 1.3382 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7304/1.3.6.1.4.1.14519.5.2.1.4591.4003.233329049923110225958892646100/1.3.6.1.4.1.14519.5.2.1.4591.4003.157034137886992535107544502554 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7306 | flair | U | 67 | 88.451 | AXIAL_FLAIR | 6 | 1.75684E+15 | 5/12/1993 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7322 | 3 | 2.5 | 90 | 0.8735 | 84 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7306/1.3.6.1.4.1.14519.5.2.1.4591.4003.127855909272597883231465711323/1.3.6.1.4.1.14519.5.2.1.4591.4003.130974444495303333550284072811 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7306 | t1Gd | U | 67 | 88.451 | AX_T1_POST_GD_FLAIR | 11 | 1.75684E+15 | 5/12/1993 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 2.5 | 3115.46 | 6.356 | 1238 | 127.7322 | 3 | 2.5 | 90 | 1.4234 | 84 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7306/1.3.6.1.4.1.14519.5.2.1.4591.4003.127855909272597883231465711323/1.3.6.1.4.1.14519.5.2.1.4591.4003.259382038351091849536381817891 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7306 | t1 | U | 67 | 88.451 | AX_T1_pre_gd | 9 | 1.75684E+15 | 5/12/1993 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | IR | 2D | 5 | 3115.54 | 6.356 | 1238 | 127.7322 | 3 | 5 | 90 | 1.4233 | 42 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7306/1.3.6.1.4.1.14519.5.2.1.4591.4003.127855909272597883231465711323/1.3.6.1.4.1.14519.5.2.1.4591.4003.155963458783583162763789728619 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7306 | t2 | U | 67 | 88.451 | AX_T2_FR-FSE_RF2_150 | 7 | 1.75684E+15 | 5/12/1993 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 3000 | 103.464 | 0 | 127.7322 | 3 | 5 | 90 | 1.3167 | 42 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7306/1.3.6.1.4.1.14519.5.2.1.4591.4003.127855909272597883231465711323/1.3.6.1.4.1.14519.5.2.1.4591.4003.103388284169265511747126541748 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7309 | flair | F | 41 | 76.657 | AXIAL_FLAIR | 6 | 2.57831E+15 | 8/31/1996 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7323 | 3 | 2.5 | 90 | 0.652 | 80 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7309/1.3.6.1.4.1.14519.5.2.1.4591.4003.168648868285787837108458883234/1.3.6.1.4.1.14519.5.2.1.4591.4003.492234721143934975701574344187 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7309 | t1Gd | F | 41 | 76.657 | AX_T1_POST_GD_FLAIR | 12 | 2.57831E+15 | 8/31/1996 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 2970.27 | 6.356 | 1238 | 127.7323 | 3 | 2.5 | 90 | 1.62 | 80 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7309/1.3.6.1.4.1.14519.5.2.1.4591.4003.168648868285787837108458883234/1.3.6.1.4.1.14519.5.2.1.4591.4003.179698976018344713629324575778 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7309 | t1 | F | 41 | 76.657 | AX_T1_pre_gd | 10 | 2.57831E+15 | 8/31/1996 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 5 | 2141.76 | 6.356 | 883 | 127.7323 | 3 | 5 | 90 | 1.6646 | 40 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7309/1.3.6.1.4.1.14519.5.2.1.4591.4003.168648868285787837108458883234/1.3.6.1.4.1.14519.5.2.1.4591.4003.992012705357304347023368682760 |
| TCGA-LGG | TCGA-DU | TCGA-DU-7309 | t2 | F | 41 | 76.657 | AX_T2_FR-FSE_RF2_150 | 4 | 2.57831E+15 | 8/31/1996 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 4000 | 103.464 | 0 | 127.7322 | 3 | 5 | 90 | 1.1038 | 40 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-7309/1.3.6.1.4.1.14519.5.2.1.4591.4003.168648868285787837108458883234/1.3.6.1.4.1.14519.5.2.1.4591.4003.156894594838434855648349079703 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8162 | flair | F | 61 | 56.699 | AXIAL_FLAIR | 4 | 2.43944E+15 | 10/29/1996 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7323 | 3 | 2.5 | 90 | 0.5225 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8162/1.3.6.1.4.1.14519.5.2.1.4591.4003.239858237185580206387331844912/1.3.6.1.4.1.14519.5.2.1.4591.4003.232666278628494820601580634659 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8162 | t1Gd | F | 61 | 56.699 | AX_T1_POST_GD_FLAIR | 11 | 2.43944E+15 | 10/29/1996 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 3103.47 | 6.356 | 1238 | 127.7322 | 3 | 2.5 | 90 | 1.4727 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8162/1.3.6.1.4.1.14519.5.2.1.4591.4003.239858237185580206387331844912/1.3.6.1.4.1.14519.5.2.1.4591.4003.120802060567647850144606478295 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8162 | t1 | F | 61 | 56.699 | AX_T1_pre_gd | 8 | 2.43944E+15 | 10/29/1996 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 5 | 2039.94 | 6.356 | 883 | 127.7323 | 3 | 5 | 90 | 1.1829 | 37 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8162/1.3.6.1.4.1.14519.5.2.1.4591.4003.239858237185580206387331844912/1.3.6.1.4.1.14519.5.2.1.4591.4003.199135420531148912087697063463 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8162 | t2 | F | 61 | 56.699 | AX_T2_FR-FSE_RF2_150 | 7 | 2.43944E+15 | 10/29/1996 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 4000 | 103.464 | 0 | 127.7323 | 3 | 5 | 90 | 0.747 | 37 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8162/1.3.6.1.4.1.14519.5.2.1.4591.4003.239858237185580206387331844912/1.3.6.1.4.1.14519.5.2.1.4591.4003.233930895945610254694242682140 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8164 | flair | M | 51 | 99.79 | AXIAL_FLAIR | 4 | 2.13629E+15 | 1/11/1997 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7322 | 3 | 2.5 | 90 | 1.0139 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8164/1.3.6.1.4.1.14519.5.2.1.4591.4003.286153250156424635720891098366/1.3.6.1.4.1.14519.5.2.1.4591.4003.263357156153587255427662103012 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8164 | t1Gd | M | 51 | 99.79 | AX_T1_POST_GD_FLAIR | 10 | 2.13629E+15 | 1/11/1997 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 3081.66 | 6.356 | 1238 | 127.7322 | 3 | 2.5 | 90 | 1.6356 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8164/1.3.6.1.4.1.14519.5.2.1.4591.4003.286153250156424635720891098366/1.3.6.1.4.1.14519.5.2.1.4591.4003.505191286789266005167052602819 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8164 | t1 | M | 51 | 99.79 | AX_T1_pre_gd | 8 | 2.13629E+15 | 1/11/1997 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 5 | 2198.48 | 6.356 | 883 | 127.7322 | 3 | 5 | 90 | 1.6562 | 37 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8164/1.3.6.1.4.1.14519.5.2.1.4591.4003.286153250156424635720891098366/1.3.6.1.4.1.14519.5.2.1.4591.4003.304093308417890058498224439637 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8164 | t2 | M | 51 | 99.79 | AX_T2_FR-FSE_RF2_150 | 7 | 2.13629E+15 | 1/11/1997 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 4000 | 103.464 | 0 | 127.7322 | 3 | 5 | 90 | 1.6476 | 37 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8164/1.3.6.1.4.1.14519.5.2.1.4591.4003.286153250156424635720891098366/1.3.6.1.4.1.14519.5.2.1.4591.4003.227082676099967810370197194767 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8165 | flair | F | 60 | 58.967 | AXIAL_FLAIR | 6 | 8.26478E+15 | 2/5/1997 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7322 | 3 | 2.5 | 90 | 0.5424 | 69 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8165/1.3.6.1.4.1.14519.5.2.1.4591.4003.140730945904429213927161894305/1.3.6.1.4.1.14519.5.2.1.4591.4003.958512138684118440272310116811 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8165 | t1Gd | F | 60 | 58.967 | AX_T1_POST_GD_FLAIR | 12 | 8.26478E+15 | 2/5/1997 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 3104.34 | 6.356 | 1238 | 127.7322 | 3 | 2.5 | 90 | 1.4813 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8165/1.3.6.1.4.1.14519.5.2.1.4591.4003.140730945904429213927161894305/1.3.6.1.4.1.14519.5.2.1.4591.4003.317099727468822952520944196133 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8165 | t1 | F | 60 | 58.967 | AX_T1_pre_gd | 9 | 8.26478E+15 | 2/5/1997 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 5 | 2101.38 | 6.356 | 883 | 127.7322 | 3 | 5 | 90 | 1.0148 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8165/1.3.6.1.4.1.14519.5.2.1.4591.4003.140730945904429213927161894305/1.3.6.1.4.1.14519.5.2.1.4591.4003.305131916722305493965002769017 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8165 | t2 | F | 60 | 58.967 | AX_T2_FR-FSE_RF2_150 | 8 | 8.26478E+15 | 2/5/1997 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 4000 | 103.464 | 0 | 127.7322 | 3 | 5 | 90 | 0.712 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8165/1.3.6.1.4.1.14519.5.2.1.4591.4003.140730945904429213927161894305/1.3.6.1.4.1.14519.5.2.1.4591.4003.486167804750109627510369135709 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8166 | flair | U | 29 | 61.235 | AXIAL_FLAIR | 7 | 2.32218E+15 | 3/22/1997 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7323 | 3 | 2.5 | 90 | 0.5456 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8166/1.3.6.1.4.1.14519.5.2.1.4591.4003.336568303900495873094738894974/1.3.6.1.4.1.14519.5.2.1.4591.4003.172004762305844447401871420431 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8166 | t1Gd | U | 29 | 61.235 | AX_T1_POST_GD_FLAIR | 12 | 2.32218E+15 | 3/22/1997 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 2556.99 | 6.356 | 1076 | 127.7323 | 3 | 2.5 | 90 | 1.8091 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8166/1.3.6.1.4.1.14519.5.2.1.4591.4003.336568303900495873094738894974/1.3.6.1.4.1.14519.5.2.1.4591.4003.141004776239267801712876247718 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8166 | t1 | U | 29 | 61.235 | AX_T1_pre_gd | 9 | 2.32218E+15 | 3/22/1997 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 5 | 2540.29 | 6.356 | 1067 | 127.7323 | 3 | 5 | 90 | 1.821 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8166/1.3.6.1.4.1.14519.5.2.1.4591.4003.336568303900495873094738894974/1.3.6.1.4.1.14519.5.2.1.4591.4003.270588047395699670537024032082 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8166 | t2 | U | 29 | 61.235 | AX_T2_FR-FSE_RF2_150 | 8 | 2.32218E+15 | 3/22/1997 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 4000 | 103.464 | 0 | 127.7323 | 3 | 5 | 90 | 0.7582 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8166/1.3.6.1.4.1.14519.5.2.1.4591.4003.336568303900495873094738894974/1.3.6.1.4.1.14519.5.2.1.4591.4003.180440215937530195248658513948 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8167 | flair | F | 69 | 74.843 | AXIAL_FLAIR | 6 | 3.35479E+15 | 4/2/1997 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7322 | 3 | 2.5 | 90 | 0.6258 | 73 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8167/1.3.6.1.4.1.14519.5.2.1.4591.4003.168817713276868679404712029125/1.3.6.1.4.1.14519.5.2.1.4591.4003.973607962673153304494674925902 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8167 | t1Gd | F | 69 | 74.843 | AX_T1_POST_GD_FLAIR | 10 | 3.35479E+15 | 4/2/1997 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 3042 | 6.356 | 1238 | 127.7322 | 3 | 2.5 | 90 | 1.4057 | 73 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8167/1.3.6.1.4.1.14519.5.2.1.4591.4003.168817713276868679404712029125/1.3.6.1.4.1.14519.5.2.1.4591.4003.158150700999497317857027374207 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8167 | t1 | F | 69 | 74.843 | AX_T1_pre_gd | 8 | 3.35479E+15 | 4/2/1997 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 5 | 2039.94 | 6.356 | 883 | 127.7322 | 3 | 5 | 90 | 1.5937 | 37 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8167/1.3.6.1.4.1.14519.5.2.1.4591.4003.168817713276868679404712029125/1.3.6.1.4.1.14519.5.2.1.4591.4003.200746342658024033787157836450 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8167 | t2 | F | 69 | 74.843 | AX_T2_FR-FSE_RF2_150 | 7 | 3.35479E+15 | 4/2/1997 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 4000 | 103.464 | 0 | 127.7322 | 3 | 5 | 90 | 0.9534 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8167/1.3.6.1.4.1.14519.5.2.1.4591.4003.168817713276868679404712029125/1.3.6.1.4.1.14519.5.2.1.4591.4003.130537543563011500866320702988 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8168 | flair | F | 55 | 47.627 | AXIAL_FLAIR | 7 | 4.84646E+15 | 5/3/1997 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7323 | 3 | 2.5 | 90 | 0.5249 | 71 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8168/1.3.6.1.4.1.14519.5.2.1.4591.4003.246416124390990436653999855138/1.3.6.1.4.1.14519.5.2.1.4591.4003.211295928669757772419648940636 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8168 | t1Gd | F | 55 | 47.627 | AX_T1_POST_GD_FLAIR | 12 | 4.84646E+15 | 5/3/1997 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 2.5 | 3099.44 | 6.356 | 1238 | 127.7323 | 3 | 2.5 | 90 | 1.4356 | 71 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8168/1.3.6.1.4.1.14519.5.2.1.4591.4003.246416124390990436653999855138/1.3.6.1.4.1.14519.5.2.1.4591.4003.207704217831644522002629479320 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8168 | t1 | F | 55 | 47.627 | AX_T1_pre_gd | 8 | 4.84646E+15 | 5/3/1997 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE\IR | 2D | 5 | 2107.14 | 6.356 | 883 | 127.7323 | 3 | 5 | 90 | 1.0561 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8168/1.3.6.1.4.1.14519.5.2.1.4591.4003.246416124390990436653999855138/1.3.6.1.4.1.14519.5.2.1.4591.4003.227113158557470572673750614297 |
| TCGA-LGG | TCGA-DU | TCGA-DU-8168 | t2 | F | 55 | 47.627 | AX_T2_FR-FSE_RF2_150 | 4 | 4.84646E+15 | 5/3/1997 | GE MEDICAL SYSTEMS | MR3T | SIGNA EXCITE | SE | 2D | 5 | 4000 | 103.464 | 0 | 127.7323 | 3 | 5 | 90 | 0.6889 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-8168/1.3.6.1.4.1.14519.5.2.1.4591.4003.246416124390990436653999855138/1.3.6.1.4.1.14519.5.2.1.4591.4003.197335199152369341066172598020 |
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TP | flair | M | 33 | 106.594 | AXIAL_FLAIR | 6 | 8.32022E+15 | 6/14/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7322 | 3 | 2.5 | 90 | 1.1015 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-A5TP/1.3.6.1.4.1.14519.5.2.1.4591.4003.291142487825047263671923414542/1.3.6.1.4.1.14519.5.2.1.4591.4003.974831159092121399697175807910 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TP | t1Gd | M | 33 | 106.594 | AX_T1_POST_GD_FLAIR | 10 | 8.32022E+15 | 6/14/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 2.5 | 3042.02 | 6.356 | 1238 | 127.7322 | 3 | 2.5 | 90 | 1.5357 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-A5TP/1.3.6.1.4.1.14519.5.2.1.4591.4003.291142487825047263671923414542/1.3.6.1.4.1.14519.5.2.1.4591.4003.240056898333026509898056790264 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TP | t1 | M | 33 | 106.594 | AX_T1_pre_gd | 7 | 8.32022E+15 | 6/14/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 2060.84 | 6.356 | 883 | 127.7322 | 3 | 5 | 90 | 1.5117 | 38 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-A5TP/1.3.6.1.4.1.14519.5.2.1.4591.4003.291142487825047263671923414542/1.3.6.1.4.1.14519.5.2.1.4591.4003.414793790467374440698643449092 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TP | t2 | M | 33 | 106.594 | AX_T2_FR-FSE_RF2_150 | 5 | 8.32022E+15 | 6/14/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 4000 | 103.464 | 0 | 127.7322 | 3 | 5 | 90 | 1.8447 | 38 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-A5TP/1.3.6.1.4.1.14519.5.2.1.4591.4003.291142487825047263671923414542/1.3.6.1.4.1.14519.5.2.1.4591.4003.125375269087118703971208415488 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TR | flair | M | 51 | 90.718 | AXIAL_FLAIR | 5 | 1.80456E+15 | 7/26/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7322 | 3 | 2.5 | 90 | 0.9008 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-A5TR/1.3.6.1.4.1.14519.5.2.1.4591.4003.425990241084610539458158617402/1.3.6.1.4.1.14519.5.2.1.4591.4003.279028600034741547478221979506 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TR | t1Gd | M | 51 | 90.718 | AX_T1_POST_GD_FLAIR | 10 | 1.80456E+15 | 7/26/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 2.5 | 3236.34 | 6.356 | 1238 | 127.7322 | 3 | 2.5 | 90 | 1.7636 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-A5TR/1.3.6.1.4.1.14519.5.2.1.4591.4003.425990241084610539458158617402/1.3.6.1.4.1.14519.5.2.1.4591.4003.149558942269897008551830323755 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TR | t1 | M | 51 | 90.718 | AX_T1_pre_gd | 7 | 1.80456E+15 | 7/26/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 2152.82 | 6.356 | 883 | 127.7323 | 3 | 5 | 90 | 1.4365 | 38 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-A5TR/1.3.6.1.4.1.14519.5.2.1.4591.4003.425990241084610539458158617402/1.3.6.1.4.1.14519.5.2.1.4591.4003.421746639107968090165101173601 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TR | t2 | M | 51 | 90.718 | AX_T2_FR-FSE_RF2_150 | 4 | 1.80456E+15 | 7/26/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 4000 | 103.464 | 0 | 127.7322 | 3 | 5 | 90 | 1.3993 | 38 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-A5TR/1.3.6.1.4.1.14519.5.2.1.4591.4003.425990241084610539458158617402/1.3.6.1.4.1.14519.5.2.1.4591.4003.796723886140993479035572234223 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TS | flair | M | 42 | 90.718 | Ax_FLAIR_2.5mm_for_surgery | 4 | 2.8282E+15 | 7/26/1997 | GE MEDICAL SYSTEMS | Signa HDxt | IR | 2D | 2.5 | 10004 | 149.544 | 2200 | 63.88574 | 1.5 | 2.5 | 90 | 0.9489 | 69 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-A5TS/1.3.6.1.4.1.14519.5.2.1.4591.4003.506469428889289487430866900379/1.3.6.1.4.1.14519.5.2.1.4591.4003.133906818467918965336047375341 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TS | t1Gd | M | 42 | 90.718 | Ax_T1_2.5mm_for_surgery | 11 | 2.8282E+15 | 7/26/1997 | GE MEDICAL SYSTEMS | Signa HDxt | SE | 2D | 2.5 | 516.668 | 13 | 0 | 63.88574 | 1.5 | 2.5 | 90 | 1.46247 | 69 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-A5TS/1.3.6.1.4.1.14519.5.2.1.4591.4003.506469428889289487430866900379/1.3.6.1.4.1.14519.5.2.1.4591.4003.108198143538126559661143849338 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TS | t1 | M | 42 | 90.718 | AXIAL_T1 | 5 | 2.8282E+15 | 7/26/1997 | GE MEDICAL SYSTEMS | Signa HDxt | SE | 2D | 5 | 550 | 14 | 0 | 63.88574 | 1.5 | 5 | 90 | 1.37351 | 35 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-A5TS/1.3.6.1.4.1.14519.5.2.1.4591.4003.506469428889289487430866900379/1.3.6.1.4.1.14519.5.2.1.4591.4003.318078055811576334314843090781 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TS | t2 | M | 42 | 90.718 | Ax_T2_FSE | 7 | 2.8282E+15 | 7/26/1997 | GE MEDICAL SYSTEMS | Signa HDxt | SE | 2D | 5 | 3500 | 59.456 | 0 | 63.88575 | 1.5 | 5 | 90 | 1.2952 | 35 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-A5TS/1.3.6.1.4.1.14519.5.2.1.4591.4003.506469428889289487430866900379/1.3.6.1.4.1.14519.5.2.1.4591.4003.221097235994186959722655915953 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TT | flair | M | 85 | FLAIR_LongTR | 601 | 1.13728E+15 | 3/18/1998 | Philips Healthcare | Ingenia | IR | 2D | 2.5 | 11000 | 125 | 2800 | 127.7473 | 3 | 2.5 | 90 | 0.234652877 | 79 | 320 | 232 | 0.625\0.625 | TCGA-DU-A5TT/1.3.6.1.4.1.14519.5.2.1.4591.4003.166583820652373876958420163650/1.3.6.1.4.1.14519.5.2.1.4591.4003.644099169794791264778319370500 | ||
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TT | t1Gd | M | 85 | =+ax_t1_1mm | 1205 | 1.13728E+15 | 3/18/1998 | Philips Healthcare | Ingenia | GR | 3D | 1 | 8.0445 | 3.685 | 127.7472 | 3 | 1 | 8 | 0.024283046 | 215 | 240 | 222 | 0.9375\0.9375 | TCGA-DU-A5TT/1.3.6.1.4.1.14519.5.2.1.4591.4003.166583820652373876958420163650/1.3.6.1.4.1.14519.5.2.1.4591.4003.293175010350662286369185988684 | |||
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TT | t1 | M | 85 | Pre_AX_T1W_IR_TSE | 801 | 1.13728E+15 | 3/18/1998 | Philips Healthcare | Ingenia | IR | 2D | 2.5 | 2000 | 20 | 900 | 127.7473 | 3 | 2.5 | 90 | 0.178819239 | 65 | 308 | 233 | 0.46875\0.46875 | TCGA-DU-A5TT/1.3.6.1.4.1.14519.5.2.1.4591.4003.166583820652373876958420163650/1.3.6.1.4.1.14519.5.2.1.4591.4003.591742088771199762537683503915 | ||
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TT | t2 | M | 85 | COR_T2_1MM | 1303 | 1.13728E+15 | 3/18/1998 | Philips Healthcare | Ingenia | SE | 3D | 0.977 | 2500 | 252.046 | 127.7472 | 3 | 0.977 | 90 | 0.032736853 | 235 | 252 | 252 | 0.9765625\0.9765625 | TCGA-DU-A5TT/1.3.6.1.4.1.14519.5.2.1.4591.4003.166583820652373876958420163650/1.3.6.1.4.1.14519.5.2.1.4591.4003.319562880877655611034521122625 | |||
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TU | flair | F | 62 | 158.76 | Axial_FLAIR | 10 | 1.46469E+15 | 3/12/1998 | Hitachi Medical Corporation | OASIS | IR | 2D | 5 | 6600 | 100 | 2000 | 49.40144 | 1.16 | 6 | 90 | 0.96 | 24 | 256 | 230 | 0.390625\0.390625 | TCGA-DU-A5TU/1.3.6.1.4.1.14519.5.2.1.4591.4003.141072676039453911508495577342/1.3.6.1.4.1.14519.5.2.1.4591.4003.312699074942345945201654113367 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TU | t1Gd | F | 62 | 158.76 | Axial_T1_FSE_Post_Gad | 15 | 1.46469E+15 | 3/12/1998 | Hitachi Medical Corporation | OASIS | SE | 2D | 5 | 479 | 12 | 0 | 49.40143 | 1.16 | 7 | 90 | 1.48 | 23 | 256 | 230 | 0.390625\0.390625 | TCGA-DU-A5TU/1.3.6.1.4.1.14519.5.2.1.4591.4003.141072676039453911508495577342/1.3.6.1.4.1.14519.5.2.1.4591.4003.171470113945364268606527917816 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TU | t1 | F | 62 | 158.76 | Axial_T1_FSE | 9 | 1.46469E+15 | 3/12/1998 | Hitachi Medical Corporation | OASIS | SE | 2D | 5 | 479 | 12 | 0 | 49.40144 | 1.16 | 7 | 90 | 1.48 | 23 | 256 | 230 | 0.390625\0.390625 | TCGA-DU-A5TU/1.3.6.1.4.1.14519.5.2.1.4591.4003.141072676039453911508495577342/1.3.6.1.4.1.14519.5.2.1.4591.4003.203628204006459721121981606321 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TU | t2 | F | 62 | 158.76 | Axial_T2_FSE_Hi-Res | 7 | 1.46469E+15 | 3/12/1998 | Hitachi Medical Corporation | OASIS | SE | 2D | 5 | 4829 | 96 | 0 | 49.40144 | 1.16 | 7 | 90 | 1.52 | 23 | 512 | 288 | 0.390625\0.390625 | TCGA-DU-A5TU/1.3.6.1.4.1.14519.5.2.1.4591.4003.141072676039453911508495577342/1.3.6.1.4.1.14519.5.2.1.4591.4003.254982119512511066350514016668 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TW | flair | F | 73 | AX_T2_FLAIR | 501 | 2.42768E+15 | 2/28/1998 | Philips Healthcare | Ingenia | IR | 2D | 5 | 11000 | 140 | 2800 | 63.8746 | 1.5 | 6 | 90 | 0.056122024 | 31 | 256 | 182 | 0.65340906381607\0.65340906381607 | TCGA-DU-A5TW/1.3.6.1.4.1.14519.5.2.1.4591.4003.151177506773916107803967580988/1.3.6.1.4.1.14519.5.2.1.4591.4003.999479300108182485740869637956 | ||
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TW | t1Gd | F | 73 | POST_COR_T1 | 1001 | 2.42768E+15 | 2/28/1998 | Philips Healthcare | Ingenia | SE | 2D | 5 | 475.1262 | 15 | 63.8746 | 1.5 | 6 | 69 | 0.191281557 | 35 | 308 | 232 | 0.53240740299224\0.53240740299224 | TCGA-DU-A5TW/1.3.6.1.4.1.14519.5.2.1.4591.4003.151177506773916107803967580988/1.3.6.1.4.1.14519.5.2.1.4591.4003.232216335634337768854876330351 | |||
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TW | t1 | F | 73 | AX_T1 | 601 | 2.42768E+15 | 2/28/1998 | Philips Healthcare | Ingenia | SE | 2D | 5 | 422.3344 | 15 | 63.8746 | 1.5 | 6 | 69 | 0.191281542 | 31 | 308 | 232 | 0.51339286565780\0.51339286565780 | TCGA-DU-A5TW/1.3.6.1.4.1.14519.5.2.1.4591.4003.151177506773916107803967580988/1.3.6.1.4.1.14519.5.2.1.4591.4003.774128579215354606663851701946 | |||
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TW | t2 | F | 73 | AX_T2 | 401 | 2.42768E+15 | 2/28/1998 | Philips Healthcare | Ingenia | SE | 2D | 5 | 6251.354 | 100 | 63.8746 | 1.5 | 6 | 90 | 0.142390534 | 31 | 384 | 289 | 0.41071429848670\0.41071429848670 | TCGA-DU-A5TW/1.3.6.1.4.1.14519.5.2.1.4591.4003.151177506773916107803967580988/1.3.6.1.4.1.14519.5.2.1.4591.4003.184593776366081692952346590558 | |||
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TY | flair | F | 46 | 58.967 | AXIAL_FLAIR | 5 | 1.23443E+15 | 7/9/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 2.5 | 10002 | 125.4 | 2250 | 127.7322 | 3 | 2.5 | 90 | 0.5257 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-A5TY/1.3.6.1.4.1.14519.5.2.1.4591.4003.100575669289135379476050880103/1.3.6.1.4.1.14519.5.2.1.4591.4003.337101806553456931837410645628 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TY | t1Gd | F | 46 | 58.967 | AX_T1_POST_GD_FLAIR | 10 | 1.23443E+15 | 7/9/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 2.5 | 3104.34 | 6.356 | 1238 | 127.7323 | 3 | 2.5 | 90 | 1.4813 | 72 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-A5TY/1.3.6.1.4.1.14519.5.2.1.4591.4003.100575669289135379476050880103/1.3.6.1.4.1.14519.5.2.1.4591.4003.245172832153486701331950115937 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TY | t1 | F | 46 | 58.967 | AX_T1_pre_gd | 7 | 1.23443E+15 | 7/9/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 2111.74 | 6.356 | 883 | 127.7323 | 3 | 5 | 90 | 1.0891 | 36 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-A5TY/1.3.6.1.4.1.14519.5.2.1.4591.4003.100575669289135379476050880103/1.3.6.1.4.1.14519.5.2.1.4591.4003.240559113302056176406494179303 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A5TY | t2 | F | 46 | 58.967 | AX_T2_FR-FSE_RF2_150 | 4 | 1.23443E+15 | 7/9/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 4000 | 103.464 | 0 | 127.7322 | 3 | 5 | 90 | 0.7515 | 37 | 320 | 224 | 0.4688\0.4688 | TCGA-DU-A5TY/1.3.6.1.4.1.14519.5.2.1.4591.4003.100575669289135379476050880103/1.3.6.1.4.1.14519.5.2.1.4591.4003.275135018804563456372957805186 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A6S2 | flair | F | 136 | FLAIR_LongTR | 501 | 1.09867E+15 | 4/4/1998 | Philips Healthcare | Ingenia | IR | 2D | 2.5 | 11000 | 125 | 2800 | 127.7472 | 3 | 2.5 | 90 | 0.221153647 | 75 | 320 | 232 | 0.625\0.625 | TCGA-DU-A6S2/1.3.6.1.4.1.14519.5.2.1.4591.4003.141199374739420890948176927768/1.3.6.1.4.1.14519.5.2.1.4591.4003.293109290224113648309168335981 | ||
| TCGA-LGG | TCGA-DU | TCGA-DU-A6S2 | t1Gd | F | 136 | POST_AX_T1_BRAIN_LAB_1MM | 1102 | 1.09867E+15 | 4/4/1998 | Philips Medical Systems | Ingenia | GR | 3D | 1 | 8.0456 | 3.685 | 127.7473 | 3 | 1 | 8 | 0.024283046 | 194 | 240 | 222 | 0.9375\0.9375 | TCGA-DU-A6S2/1.3.6.1.4.1.14519.5.2.1.4591.4003.141199374739420890948176927768/1.3.6.1.4.1.14519.5.2.1.4591.4003.135589548347858256981695184436 | |||
| TCGA-LGG | TCGA-DU | TCGA-DU-A6S2 | t1 | F | 136 | AX_T1W_IR_TSE | 601 | 1.09867E+15 | 4/4/1998 | Philips Healthcare | Ingenia | IR | 2D | 5 | 2000 | 20 | 800 | 127.7472 | 3 | 5 | 90 | 0.211403593 | 38 | 352 | 262 | 0.42857143282890\0.42857143282890 | TCGA-DU-A6S2/1.3.6.1.4.1.14519.5.2.1.4591.4003.141199374739420890948176927768/1.3.6.1.4.1.14519.5.2.1.4591.4003.179292058159580586521876864087 | ||
| TCGA-LGG | TCGA-DU | TCGA-DU-A6S2 | t2 | F | 136 | AX_T2W_DRIVE | 401 | 1.09867E+15 | 4/4/1998 | Philips Healthcare | Ingenia | SE | 2D | 2.5 | 3000 | 80 | 127.7472 | 3 | 2.5 | 90 | 0.325780809 | 75 | 420 | 333 | 0.42857143282890\0.42857143282890 | TCGA-DU-A6S2/1.3.6.1.4.1.14519.5.2.1.4591.4003.141199374739420890948176927768/1.3.6.1.4.1.14519.5.2.1.4591.4003.616725239162147941590320402703 | |||
| TCGA-LGG | TCGA-DU | TCGA-DU-A6S3 | flair | M | 170 | FLAIR_LongTR | 701 | 1.80811E+15 | 7/11/1998 | Philips Healthcare | Ingenia | IR | 2D | 2.5 | 11000 | 125 | 2800 | 127.7472 | 3 | 2.5 | 90 | 0.207654387 | 70 | 332 | 234 | 0.625\0.625 | TCGA-DU-A6S3/1.3.6.1.4.1.14519.5.2.1.4591.4003.248355137484740846341982351715/1.3.6.1.4.1.14519.5.2.1.4591.4003.292224188967208352325450551998 | ||
| TCGA-LGG | TCGA-DU | TCGA-DU-A6S3 | t1Gd | M | 170 | POST_AX_T1__3MM | 1303 | 1.80811E+15 | 7/11/1998 | Philips Medical Systems | Ingenia | GR | 3D | 3 | 8.0451 | 3.685 | 127.7473 | 3 | 3 | 8 | 0.025058357 | 64 | 248 | 222 | 0.85714286565780\0.85714286565780 | TCGA-DU-A6S3/1.3.6.1.4.1.14519.5.2.1.4591.4003.248355137484740846341982351715/1.3.6.1.4.1.14519.5.2.1.4591.4003.118759847392627168669666819967 | |||
| TCGA-LGG | TCGA-DU | TCGA-DU-A6S3 | t1 | M | 170 | AX_T1W_IR_TSE | 801 | 1.80811E+15 | 7/11/1998 | Philips Healthcare | Ingenia | IR | 2D | 5 | 2000 | 20 | 800 | 127.7472 | 3 | 5 | 90 | 0.211403593 | 36 | 368 | 293 | 0.390625\0.390625 | TCGA-DU-A6S3/1.3.6.1.4.1.14519.5.2.1.4591.4003.248355137484740846341982351715/1.3.6.1.4.1.14519.5.2.1.4591.4003.295602931149011312134948745170 | ||
| TCGA-LGG | TCGA-DU | TCGA-DU-A6S3 | t2 | M | 170 | AX_T2W_DRIVE | 601 | 1.80811E+15 | 7/11/1998 | Philips Healthcare | Ingenia | SE | 2D | 2.5 | 3000 | 80 | 127.7472 | 3 | 2.5 | 90 | 0.350840807 | 70 | 440 | 337 | 0.390625\0.390625 | TCGA-DU-A6S3/1.3.6.1.4.1.14519.5.2.1.4591.4003.248355137484740846341982351715/1.3.6.1.4.1.14519.5.2.1.4591.4003.176031418834958083041704052058 | |||
| TCGA-LGG | TCGA-DU | TCGA-DU-A6S6 | flair | F | 35 | 56.699 | Ax_FLAIR_2.5mm_for_surgery | 4 | 2.44522E+15 | 5/21/1992 | GE MEDICAL SYSTEMS | SIGNA HDx | IR | 2D | 2.5 | 10004 | 149.544 | 2200 | 63.88597 | 1.5 | 2.5 | 90 | 0.0351 | 70 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-A6S6/1.3.6.1.4.1.14519.5.2.1.4591.4003.157755470529359814735610642529/1.3.6.1.4.1.14519.5.2.1.4591.4003.247515038873804783086202408555 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A6S6 | t1Gd | F | 35 | 56.699 | Ax_T1_2.5mm_for_surgery | 8 | 2.44522E+15 | 5/21/1992 | GE MEDICAL SYSTEMS | SIGNA HDx | SE | 2D | 2.5 | 516.664 | 13 | 0 | 63.886 | 1.5 | 2.5 | 90 | 0.0541962 | 70 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-A6S6/1.3.6.1.4.1.14519.5.2.1.4591.4003.157755470529359814735610642529/1.3.6.1.4.1.14519.5.2.1.4591.4003.147732384686361520756078194258 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A6S6 | t1 | F | 35 | 56.699 | AXIAL_T1 | 6 | 2.44522E+15 | 5/21/1992 | GE MEDICAL SYSTEMS | SIGNA HDx | SE | 2D | 5 | 550 | 14 | 0 | 63.88598 | 1.5 | 5 | 90 | 0.0508993 | 35 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-A6S6/1.3.6.1.4.1.14519.5.2.1.4591.4003.157755470529359814735610642529/1.3.6.1.4.1.14519.5.2.1.4591.4003.238129359139198844765626935458 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A6S6 | t2 | F | 35 | 56.699 | AXIAL_FSE | 3 | 2.44522E+15 | 5/21/1992 | GE MEDICAL SYSTEMS | SIGNA HDx | SE | 2D | 5 | 3500 | 23.688 | 0 | 63.88597 | 1.5 | 5 | 90 | 0.1239 | 70 | 256 | 192 | 0.9375\0.9375 | TCGA-DU-A6S6/1.3.6.1.4.1.14519.5.2.1.4591.4003.157755470529359814735610642529/1.3.6.1.4.1.14519.5.2.1.4591.4003.315196201662757798532330052767 | |
| TCGA-LGG | TCGA-DU | TCGA-DU-A6S7 | flair | F | 57 | FLAIR_LongTR | 601 | 3.08723E+15 | 5/13/1998 | Philips Healthcare | Ingenia | IR | 2D | 2.5 | 11000 | 125 | 2800 | 127.7472 | 3 | 2.5 | 90 | 0.234652787 | 76 | 320 | 232 | 0.625\0.625 | TCGA-DU-A6S7/1.3.6.1.4.1.14519.5.2.1.4591.4003.222463746204839267807160781139/1.3.6.1.4.1.14519.5.2.1.4591.4003.153209951824448019360838381161 | ||
| TCGA-LGG | TCGA-DU | TCGA-DU-A6S7 | t1Gd | F | 57 | POST_AX_T1_BRAIN_LAB_1MM | 1102 | 3.08723E+15 | 5/13/1998 | Philips Medical Systems | Ingenia | GR | 3D | 1 | 8.0445 | 3.685 | 127.7472 | 3 | 1 | 8 | 0.024283046 | 194 | 240 | 222 | 0.9375\0.9375 | TCGA-DU-A6S7/1.3.6.1.4.1.14519.5.2.1.4591.4003.222463746204839267807160781139/1.3.6.1.4.1.14519.5.2.1.4591.4003.174567062740096698672017092596 | |||
| TCGA-LGG | TCGA-DU | TCGA-DU-A6S7 | t1 | F | 57 | AX_T1W_IR_TSE | 701 | 3.08723E+15 | 5/13/1998 | Philips Healthcare | Ingenia | IR | 2D | 5 | 2000 | 20 | 800 | 127.7472 | 3 | 5 | 90 | 0.211403564 | 41 | 352 | 262 | 0.42857143282890\0.42857143282890 | TCGA-DU-A6S7/1.3.6.1.4.1.14519.5.2.1.4591.4003.222463746204839267807160781139/1.3.6.1.4.1.14519.5.2.1.4591.4003.318036549395257841823120919980 | ||
| TCGA-LGG | TCGA-DU | TCGA-DU-A6S7 | t2 | F | 57 | AX_T2W_DRIVE | 501 | 3.08723E+15 | 5/13/1998 | Philips Healthcare | Ingenia | SE | 2D | 2.5 | 3000 | 80 | 127.7472 | 3 | 2.5 | 90 | 0.325780809 | 76 | 420 | 333 | 0.42857143282890\0.42857143282890 | TCGA-DU-A6S7/1.3.6.1.4.1.14519.5.2.1.4591.4003.222463746204839267807160781139/1.3.6.1.4.1.14519.5.2.1.4591.4003.240158011787928914076090296018 | |||
| TCGA-LGG | TCGA-DU | TCGA-DU-A6S8 | flair | F | 82 | FLAIR_LongTR | 701 | 3.23035E+15 | 6/20/1998 | Philips Healthcare | Ingenia | IR | 2D | 2.5 | 11000 | 125 | 2800 | 127.7473 | 3 | 2.5 | 90 | 0.221153691 | 73 | 320 | 232 | 0.625\0.625 | TCGA-DU-A6S8/1.3.6.1.4.1.14519.5.2.1.4591.4003.217867204129018050748395707083/1.3.6.1.4.1.14519.5.2.1.4591.4003.204516936842937081704572758933 | ||
| TCGA-LGG | TCGA-DU | TCGA-DU-A6S8 | t1Gd | F | 82 | POST_AX_T1_BRAIN_LAB_1MM | 1302 | 3.23035E+15 | 6/20/1998 | Philips Medical Systems | Ingenia | GR | 3D | 1 | 8.0445 | 3.685 | 127.7473 | 3 | 1 | 8 | 0.024283046 | 194 | 240 | 222 | 0.9375\0.9375 | TCGA-DU-A6S8/1.3.6.1.4.1.14519.5.2.1.4591.4003.217867204129018050748395707083/1.3.6.1.4.1.14519.5.2.1.4591.4003.132452867851880683397728197269 | |||
| TCGA-LGG | TCGA-DU | TCGA-DU-A6S8 | t1 | F | 82 | AX_T1W_IR_TSE | 801 | 3.23035E+15 | 6/20/1998 | Philips Healthcare | Ingenia | IR | 2D | 5 | 2000 | 20 | 800 | 127.7473 | 3 | 5 | 90 | 0.211403742 | 37 | 352 | 262 | 0.42857143282890\0.42857143282890 | TCGA-DU-A6S8/1.3.6.1.4.1.14519.5.2.1.4591.4003.217867204129018050748395707083/1.3.6.1.4.1.14519.5.2.1.4591.4003.889250232340214087576015361713 | ||
| TCGA-LGG | TCGA-DU | TCGA-DU-A6S8 | t2 | F | 82 | AX_T2W_DRIVE | 601 | 3.23035E+15 | 6/20/1998 | Philips Healthcare | Ingenia | SE | 2D | 2.5 | 3000 | 80 | 127.7473 | 3 | 2.5 | 90 | 0.325780809 | 73 | 420 | 333 | 0.42857143282890\0.42857143282890 | TCGA-DU-A6S8/1.3.6.1.4.1.14519.5.2.1.4591.4003.217867204129018050748395707083/1.3.6.1.4.1.14519.5.2.1.4591.4003.261214969156460346846015628974 | |||
| TCGA-LGG | TCGA-EZ | TCGA-EZ-7265A | flair | M | 26 | 97.06878 | FLAIR_AXIAL | 5 | 2.5083E+15 | 9/15/2001 | SIEMENS | TrioTim | SE\IR | 2D | 4 | 8500 | 112 | 2500 | 123.2411 | 3 | 4.8 | 160 | 0.594111835 | 34 | 256 | 208 | 0.859375\0.859375 | TCGA-EZ-7265A/1.3.6.1.4.1.14519.5.2.1.8421.4003.118065813881052850602362195650/1.3.6.1.4.1.14519.5.2.1.8421.4003.306492368085941976344452042305 | |
| TCGA-LGG | TCGA-EZ | TCGA-EZ-7265A | t1Gd | M | 26 | 97.06878 | T1_MPRAGE_AX | 7 | 2.5083E+15 | 9/15/2001 | SIEMENS | TrioTim | GR\IR | 3D | 1 | 1900 | 2.63 | 900 | 123.2411 | 3 | 9 | 0.094795003 | 176 | 256 | 256 | 1\1 | TCGA-EZ-7265A/1.3.6.1.4.1.14519.5.2.1.8421.4003.118065813881052850602362195650/1.3.6.1.4.1.14519.5.2.1.8421.4003.471672703542665098957555277296 | ||
| TCGA-LGG | TCGA-EZ | TCGA-EZ-7265A | t1 | M | 26 | 97.06878 | T1_FL_COR | 14 | 2.5083E+15 | 9/15/2001 | SIEMENS | TrioTim | GR | 2D | 5 | 267 | 2.46 | 123.2412 | 3 | 6 | 80 | 0.707966423 | 32 | 320 | 224 | 0.71875\0.71875 | TCGA-EZ-7265A/1.3.6.1.4.1.14519.5.2.1.8421.4003.118065813881052850602362195650/1.3.6.1.4.1.14519.5.2.1.8421.4003.556090517143317008209723958322 | ||
| TCGA-LGG | TCGA-EZ | TCGA-EZ-7265A | t2 | M | 26 | 97.06878 | T2_AX_TSE | 6 | 2.5083E+15 | 9/15/2001 | SIEMENS | TrioTim | SE | 2D | 3 | 4780 | 87 | 123.2411 | 3 | 3 | 150 | 0.455488092 | 55 | 512 | 256 | 0.4296875\0.4296875 | TCGA-EZ-7265A/1.3.6.1.4.1.14519.5.2.1.8421.4003.118065813881052850602362195650/1.3.6.1.4.1.14519.5.2.1.8421.4003.105472801741628196191958426767 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-5964 | flair | M | 62 | 90.71849 | MRHG_FLAIR_AX | 5 | 6.20494E+15 | 5/11/2001 | Siemens | Symphony | IR\SE | 2D | 5 | 9590 | 129 | 2500 | 63.65037 | 1.5 | 6.5 | 150 | 0.104002565 | 26 | 256 | 224 | 0.8984375\0.8984375 | TCGA-FG-5964/1.3.6.1.4.1.14519.5.2.1.5826.4003.238471712497372270512892955981/1.3.6.1.4.1.14519.5.2.1.5826.4003.404178152704844737198656421158 | |
| TCGA-LGG | TCGA-FG | TCGA-FG-5964 | t1Gd | M | 62 | 90.71849 | MP_RAGE_AXIAL | 7 | 6.20494E+15 | 5/11/2001 | Siemens | Symphony | IR\GR | 3D | 1 | 2160 | 3.45 | 1100 | 63.65038 | 1.5 | 15 | 0.040618185 | 192 | 256 | 256 | 1\1 | TCGA-FG-5964/1.3.6.1.4.1.14519.5.2.1.5826.4003.238471712497372270512892955981/1.3.6.1.4.1.14519.5.2.1.5826.4003.213387747040169487656654950911 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-5964 | t1 | M | 62 | 90.71849 | MP_RAGE_AXIAL | 2 | 6.20494E+15 | 5/11/2001 | Siemens | Symphony | IR\GR | 3D | 1 | 2160 | 3.45 | 1100 | 63.65037 | 1.5 | 15 | 0.039926529 | 192 | 256 | 256 | 1\1 | TCGA-FG-5964/1.3.6.1.4.1.14519.5.2.1.5826.4003.238471712497372270512892955981/1.3.6.1.4.1.14519.5.2.1.5826.4003.247020117690444362792912756464 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-5964 | t2 | M | 62 | 90.71849 | TSE_T2_AXIALS | 3 | 6.20494E+15 | 5/11/2001 | Siemens | Symphony | SE | 2D | 2 | 5700 | 93 | 63.65036 | 1.5 | 2 | 150 | 0.185237393 | 92 | 256 | 256 | 1\1 | TCGA-FG-5964/1.3.6.1.4.1.14519.5.2.1.5826.4003.238471712497372270512892955981/1.3.6.1.4.1.14519.5.2.1.5826.4003.272421027569753270309595372912 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-6688 | flair | F | 59 | 99.79033 | FLAIR_AXIALS | 3 | 2.23104E+15 | 2/15/2002 | Siemens | Symphony | IR\SE | 2D | 4 | 8790 | 114 | 2500 | 63.49749 | 1.5 | 4 | 150 | 0.153146952 | 36 | 256 | 208 | 1\1 | TCGA-FG-6688/1.3.6.1.4.1.14519.5.2.1.5826.4003.323004807957544310211707018551/1.3.6.1.4.1.14519.5.2.1.5826.4003.224031693818178176078411166389 | |
| TCGA-LGG | TCGA-FG | TCGA-FG-6688 | t1Gd | F | 59 | 99.79033 | MP_RAGE_AXIAL_PG | 7 | 2.23104E+15 | 2/15/2002 | Siemens | Symphony | IR\GR | 3D | 1 | 2160 | 3.45 | 1100 | 63.49749 | 1.5 | 15 | 0.058381364 | 192 | 256 | 256 | 1\1 | TCGA-FG-6688/1.3.6.1.4.1.14519.5.2.1.5826.4003.323004807957544310211707018551/1.3.6.1.4.1.14519.5.2.1.5826.4003.305496105341737447254851785503 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-6688 | t1 | F | 59 | 99.79033 | MP_RAGE_AXIAL | 2 | 2.23104E+15 | 2/15/2002 | Siemens | Symphony | IR\GR | 3D | 1 | 2160 | 3.45 | 1100 | 63.49749 | 1.5 | 15 | 0.058381364 | 192 | 256 | 256 | 1\1 | TCGA-FG-6688/1.3.6.1.4.1.14519.5.2.1.5826.4003.323004807957544310211707018551/1.3.6.1.4.1.14519.5.2.1.5826.4003.315562198227737027776970803095 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-6688 | t2 | F | 59 | 99.79033 | MRHG_T2_AXIALS | 5 | 2.23104E+15 | 2/15/2002 | Siemens | Symphony | SE | 2D | 5 | 4190 | 94 | 63.49745 | 1.5 | 5.5 | 150 | 0.462320268 | 26 | 256 | 224 | 0.8984375\0.8984375 | TCGA-FG-6688/1.3.6.1.4.1.14519.5.2.1.5826.4003.323004807957544310211707018551/1.3.6.1.4.1.14519.5.2.1.5826.4003.222396944059496713742105306085 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-6689 | flair | M | 30 | 83.9146 | FLAIR_AXIALS | 3 | 7.53987E+15 | 3/26/2002 | Siemens | Symphony | IR\SE | 2D | 4 | 9980 | 114 | 2500 | 63.49709 | 1.5 | 4 | 150 | 0.127931744 | 48 | 256 | 256 | 1\1 | TCGA-FG-6689/1.3.6.1.4.1.14519.5.2.1.5826.4003.844510743144337710585638347983/1.3.6.1.4.1.14519.5.2.1.5826.4003.194766601320226262498310199471 | |
| TCGA-LGG | TCGA-FG | TCGA-FG-6689 | t1Gd | M | 30 | 83.9146 | MP_RAGE_AXIAL | 6 | 7.53987E+15 | 3/26/2002 | Siemens | Symphony | IR\GR | 3D | 1 | 2160 | 3.45 | 1100 | 63.49709 | 1.5 | 15 | 0.041316584 | 192 | 256 | 256 | 1\1 | TCGA-FG-6689/1.3.6.1.4.1.14519.5.2.1.5826.4003.844510743144337710585638347983/1.3.6.1.4.1.14519.5.2.1.5826.4003.791850473131565744847058876558 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-6689 | t1 | M | 30 | 83.9146 | MP_RAGE_AXIAL | 5 | 7.53987E+15 | 3/26/2002 | Siemens | Symphony | IR\GR | 3D | 1 | 2160 | 3.45 | 1100 | 63.49709 | 1.5 | 15 | 0.041316584 | 192 | 256 | 256 | 1\1 | TCGA-FG-6689/1.3.6.1.4.1.14519.5.2.1.5826.4003.844510743144337710585638347983/1.3.6.1.4.1.14519.5.2.1.5826.4003.321356076384546982705007068721 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-6689 | t2 | M | 30 | 83.9146 | T2_AXIALS | 2 | 7.53987E+15 | 3/26/2002 | Siemens | Symphony | SE | 2D | 4 | 4300 | 91 | 63.49709 | 1.5 | 4 | 150 | 0.297007412 | 48 | 256 | 256 | 1\1 | TCGA-FG-6689/1.3.6.1.4.1.14519.5.2.1.5826.4003.844510743144337710585638347983/1.3.6.1.4.1.14519.5.2.1.5826.4003.502089644490762861281022782793 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-6691 | flair | F | 23 | 81.64664 | FLAIR_AXIALS | 4 | 7.02986E+15 | 4/5/2002 | Siemens | Symphony | IR\SE | 2D | 4 | 9980 | 114 | 2500 | 63.49715 | 1.5 | 4 | 150 | 0.146310553 | 48 | 256 | 256 | 1\1 | TCGA-FG-6691/1.3.6.1.4.1.14519.5.2.1.5826.4003.329102386198066869568471530457/1.3.6.1.4.1.14519.5.2.1.5826.4003.256505097264461536420482208985 | |
| TCGA-LGG | TCGA-FG | TCGA-FG-6691 | t1Gd | F | 23 | 81.64664 | MP_RAGE_AXIAL | 6 | 7.02986E+15 | 4/5/2002 | Siemens | Symphony | IR\GR | 3D | 1 | 2160 | 3.45 | 1100 | 63.49715 | 1.5 | 15 | 0.047252156 | 192 | 256 | 256 | 1\1 | TCGA-FG-6691/1.3.6.1.4.1.14519.5.2.1.5826.4003.329102386198066869568471530457/1.3.6.1.4.1.14519.5.2.1.5826.4003.656879804613220409889901206605 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-6691 | t1 | F | 23 | 81.64664 | MP_RAGE_AXIAL | 2 | 7.02986E+15 | 4/5/2002 | Siemens | Symphony | IR\GR | 3D | 1 | 2160 | 3.45 | 1100 | 63.49715 | 1.5 | 15 | 0.047252156 | 192 | 256 | 256 | 1\1 | TCGA-FG-6691/1.3.6.1.4.1.14519.5.2.1.5826.4003.329102386198066869568471530457/1.3.6.1.4.1.14519.5.2.1.5826.4003.529788387304099813337988254846 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-6691 | t2 | F | 23 | 81.64664 | T2_AXIALS | 3 | 7.02986E+15 | 4/5/2002 | Siemens | Symphony | SE | 2D | 4 | 4300 | 91 | 63.49715 | 1.5 | 4 | 150 | 0.339675784 | 48 | 256 | 256 | 1\1 | TCGA-FG-6691/1.3.6.1.4.1.14519.5.2.1.5826.4003.329102386198066869568471530457/1.3.6.1.4.1.14519.5.2.1.5826.4003.943440743719674101532157437216 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-6692 | flair | M | 63 | 65.31731 | MRHR_FLAIR_AX | 5 | 7.0351E+15 | 6/6/2002 | Siemens | Avanto | SE\IR | 2D | 5 | 9000 | 109 | 2500 | 63.62652 | 1.5 | 5.5 | 150 | 0.124829227 | 26 | 256 | 208 | 0.8984375\0.8984375 | TCGA-FG-6692/1.3.6.1.4.1.14519.5.2.1.5826.4003.319380682030030732733259154931/1.3.6.1.4.1.14519.5.2.1.5826.4003.192830341660611753009089717349 | |
| TCGA-LGG | TCGA-FG | TCGA-FG-6692 | t1Gd | M | 63 | 65.31731 | MRHR_T1_SAG_POST_GAD | 14 | 7.0351E+15 | 6/6/2002 | Siemens | Avanto | SE | 2D | 5 | 690 | 17 | 63.62568 | 1.5 | 6.15 | 90 | 0.430428356 | 25 | 256 | 256 | 0.8984375\0.8984375 | TCGA-FG-6692/1.3.6.1.4.1.14519.5.2.1.5826.4003.319380682030030732733259154931/1.3.6.1.4.1.14519.5.2.1.5826.4003.186672506079531283321680579916 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-6692 | t1 | M | 63 | 65.31731 | T1_AX | 10 | 7.0351E+15 | 6/6/2002 | Siemens | Avanto | SE | 2D | 5 | 529 | 12 | 63.62568 | 1.5 | 5.5 | 70 | 0.531351037 | 26 | 256 | 224 | 0.8984375\0.8984375 | TCGA-FG-6692/1.3.6.1.4.1.14519.5.2.1.5826.4003.319380682030030732733259154931/1.3.6.1.4.1.14519.5.2.1.5826.4003.186281903647307431209277892227 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-6692 | t2 | M | 63 | 65.31731 | MRHR_T2_AX | 2 | 7.0351E+15 | 6/6/2002 | Siemens | Avanto | SE | 2D | 5 | 5000 | 91 | 63.6265 | 1.5 | 5.5 | 150 | 0.301251205 | 26 | 256 | 224 | 0.8984375\0.8984375 | TCGA-FG-6692/1.3.6.1.4.1.14519.5.2.1.5826.4003.319380682030030732733259154931/1.3.6.1.4.1.14519.5.2.1.5826.4003.234091914086236121186030369038 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-7634 | flair | M | 28 | 83.9146 | FLAIR_AX | 3 | 2.63434E+15 | 1/28/2000 | Siemens | Verio | SE\IR | 2D | 4 | 9000 | 94 | 2500 | 123.2494 | 3 | 5.2 | 150 | 0.144740377 | 27 | 256 | 169 | 0.8984375\0.8984375 | TCGA-FG-7634/1.3.6.1.4.1.14519.5.2.1.5826.4003.112089609616478176328783523271/1.3.6.1.4.1.14519.5.2.1.5826.4003.181265280864825404459789253505 | |
| TCGA-LGG | TCGA-FG | TCGA-FG-7634 | t1Gd | M | 28 | 83.9146 | VOLUMETRIC_AXIAL | 9 | 2.63434E+15 | 1/28/2000 | Siemens | Verio | GR\IR | 3D | 1 | 5500 | 2.75 | 900 | 123.2494 | 3 | 9 | 0.016694271 | 192 | 256 | 251 | 1\1 | TCGA-FG-7634/1.3.6.1.4.1.14519.5.2.1.5826.4003.112089609616478176328783523271/1.3.6.1.4.1.14519.5.2.1.5826.4003.120522077294140993470551701861 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-7634 | t1 | M | 28 | 83.9146 | VOLUMETRIC_AXIAL | 8 | 2.63434E+15 | 1/28/2000 | Siemens | Verio | GR\IR | 3D | 1 | 1900 | 2.75 | 900 | 123.2494 | 3 | 9 | 0.048193246 | 192 | 256 | 251 | 1\1 | TCGA-FG-7634/1.3.6.1.4.1.14519.5.2.1.5826.4003.112089609616478176328783523271/1.3.6.1.4.1.14519.5.2.1.5826.4003.162682992466409363718204747302 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-7634 | t2 | M | 28 | 83.9146 | T2_AX | 2 | 2.63434E+15 | 1/28/2000 | Siemens | Verio | SE | 2D | 4 | 5000 | 96 | 123.2494 | 3 | 5.2 | 150 | 0.399312478 | 27 | 320 | 211 | 0.71875\0.71875 | TCGA-FG-7634/1.3.6.1.4.1.14519.5.2.1.5826.4003.112089609616478176328783523271/1.3.6.1.4.1.14519.5.2.1.5826.4003.290432067045012638044885643345 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-7643 | flair | // | TCGA-FG-7643/1.3.6.1.4.1.14519.5.2.1.5826.4003.543441953908033691486687933312/1.3.6.1.4.1.14519.5.2.1.5826.4003.566940196246926075314106414146 | ||||||||||||||||||||||||
| TCGA-LGG | TCGA-FG | TCGA-FG-7643 | t1 | // | TCGA-FG-7643/1.3.6.1.4.1.14519.5.2.1.5826.4003.543441953908033691486687933312/1.3.6.1.4.1.14519.5.2.1.5826.4003.229152749348243416870891129832 | ||||||||||||||||||||||||
| TCGA-LGG | TCGA-FG | TCGA-FG-7643 | t1Gd | // | TCGA-FG-7643/1.3.6.1.4.1.14519.5.2.1.5826.4003.543441953908033691486687933312/1.3.6.1.4.1.14519.5.2.1.5826.4003.216434171023938431138472406051 | ||||||||||||||||||||||||
| TCGA-LGG | TCGA-FG | TCGA-FG-7643 | t2 | // | TCGA-FG-7643/1.3.6.1.4.1.14519.5.2.1.5826.4003.543441953908033691486687933312/1.3.6.1.4.1.14519.5.2.1.5826.4003.179485783112349332939448183328 | ||||||||||||||||||||||||
| TCGA-LGG | TCGA-FG | TCGA-FG-A4MT | flair | F | 27 | 61.68857 | FLAIR_AXIALS | 3 | 2.45097E+15 | 2/12/2002 | Siemens | Symphony | IR\SE | 2D | 3 | 9050 | 114 | 2500 | 63.64688 | 1.5 | 3 | 150 | 0.10171707 | 50 | 256 | 192 | 1\1 | TCGA-FG-A4MT/1.3.6.1.4.1.14519.5.2.1.5826.4003.329714707736603564770655892800/1.3.6.1.4.1.14519.5.2.1.5826.4003.108791662326014870116459702285 | |
| TCGA-LGG | TCGA-FG | TCGA-FG-A4MT | t1Gd | F | 27 | 61.68857 | MP_RAGE_AXIAL | 6 | 2.45097E+15 | 2/12/2002 | Siemens | Symphony | IR\GR | 3D | 1 | 2160 | 3.45 | 1100 | 63.64688 | 1.5 | 15 | 0.03827221 | 192 | 256 | 256 | 1\1 | TCGA-FG-A4MT/1.3.6.1.4.1.14519.5.2.1.5826.4003.329714707736603564770655892800/1.3.6.1.4.1.14519.5.2.1.5826.4003.810239147090844490746341825104 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-A4MT | t1 | F | 27 | 61.68857 | MP_RAGE_AXIAL | 2 | 2.45097E+15 | 2/12/2002 | Siemens | Symphony | IR\GR | 3D | 1 | 2160 | 3.45 | 1100 | 63.64688 | 1.5 | 15 | 0.03827221 | 192 | 256 | 256 | 1\1 | TCGA-FG-A4MT/1.3.6.1.4.1.14519.5.2.1.5826.4003.329714707736603564770655892800/1.3.6.1.4.1.14519.5.2.1.5826.4003.208492381534690436622701341883 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-A4MT | t2 | F | 27 | 61.68857 | TSE_T2_AXIALS | 4 | 2.45097E+15 | 2/12/2002 | Siemens | Symphony | SE | 2D | 3 | 4230 | 90 | 63.64687 | 1.5 | 3 | 150 | 0.12996757 | 50 | 256 | 192 | 1\1 | TCGA-FG-A4MT/1.3.6.1.4.1.14519.5.2.1.5826.4003.329714707736603564770655892800/1.3.6.1.4.1.14519.5.2.1.5826.4003.323518390920997145878509984446 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-A6IZ | flair | M | 60 | 79.37867 | FLAIR_AX | 11 | 6.76664E+15 | 2/20/2004 | SIEMENS | Avanto | SE\IR | 2D | 3 | 9430 | 109 | 2500 | 63.62654 | 1.5 | 3 | 150 | 0.195461899 | 60 | 256 | 232 | 1\1 | TCGA-FG-A6IZ/1.3.6.1.4.1.14519.5.2.1.5826.4003.294186107439947951642908363316/1.3.6.1.4.1.14519.5.2.1.5826.4003.702011097062990231893856395088 | |
| TCGA-LGG | TCGA-FG | TCGA-FG-A6IZ | t1Gd | M | 60 | 79.37867 | MP_RAGE_AXIAL | 13 | 6.76664E+15 | 2/20/2004 | SIEMENS | Avanto | GR\IR | 3D | 1 | 2160 | 2.81 | 1100 | 63.62654 | 1.5 | 15 | 0.048093966 | 192 | 256 | 256 | 1\1 | TCGA-FG-A6IZ/1.3.6.1.4.1.14519.5.2.1.5826.4003.294186107439947951642908363316/1.3.6.1.4.1.14519.5.2.1.5826.4003.331410843391598749669849855225 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-A6IZ | t1 | M | 60 | 79.37867 | MP_RAGE_AXIAL_VOLUMETRIC | 12 | 6.76664E+15 | 2/20/2004 | SIEMENS | Avanto | GR\IR | 3D | 1 | 2160 | 2.81 | 1100 | 63.62654 | 1.5 | 15 | 0.048093966 | 192 | 256 | 256 | 1\1 | TCGA-FG-A6IZ/1.3.6.1.4.1.14519.5.2.1.5826.4003.294186107439947951642908363316/1.3.6.1.4.1.14519.5.2.1.5826.4003.277684855659716713037896125841 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-A6IZ | t2 | M | 60 | 79.37867 | MRHR_T2_AX | 10 | 6.76664E+15 | 2/20/2004 | SIEMENS | Avanto | SE | 2D | 3 | 5000 | 91 | 63.62649 | 1.5 | 3 | 150 | 0.374650202 | 60 | 256 | 232 | 1\1 | TCGA-FG-A6IZ/1.3.6.1.4.1.14519.5.2.1.5826.4003.294186107439947951642908363316/1.3.6.1.4.1.14519.5.2.1.5826.4003.178027193899941193791707458901 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-A713 | flair | F | 74 | 79.37867 | FLAIR_axial | 4 | 8.51776E+15 | 7/9/2004 | SIEMENS | Skyra | SE\IR | 2D | 3 | 9000 | 94 | 2498.5 | 123.2567 | 3 | 3 | 150 | 0.657520147 | 64 | 256 | 208 | 1\1 | TCGA-FG-A713/1.3.6.1.4.1.14519.5.2.1.5826.4003.239451224442205099709649006810/1.3.6.1.4.1.14519.5.2.1.5826.4003.103848826726158954491300808517 | |
| TCGA-LGG | TCGA-FG | TCGA-FG-A713 | t1Gd | F | 74 | 79.37867 | t1_mprage_tra_Gd | 11 | 8.51776E+15 | 7/9/2004 | SIEMENS | Skyra | GR\IR | 3D | 1 | 2200 | 2.45 | 900 | 123.2567 | 3 | 8 | 0.070773934 | 192 | 256 | 256 | 1\1 | TCGA-FG-A713/1.3.6.1.4.1.14519.5.2.1.5826.4003.239451224442205099709649006810/1.3.6.1.4.1.14519.5.2.1.5826.4003.780276319244830396853648645030 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-A713 | t1 | F | 74 | 79.37867 | t1_mprage_tra | 2 | 8.51776E+15 | 7/9/2004 | SIEMENS | Skyra | GR\IR | 3D | 1 | 2200 | 2.45 | 900 | 123.2567 | 3 | 8 | 0.070773934 | 192 | 256 | 256 | 1\1 | TCGA-FG-A713/1.3.6.1.4.1.14519.5.2.1.5826.4003.239451224442205099709649006810/1.3.6.1.4.1.14519.5.2.1.5826.4003.291884242340469108197706129148 | ||
| TCGA-LGG | TCGA-FG | TCGA-FG-A713 | t2 | F | 74 | 79.37867 | T2_axial | 3 | 8.51776E+15 | 7/9/2004 | SIEMENS | Skyra | SE | 2D | 3 | 6000 | 104 | 123.2567 | 3 | 3 | 150 | 0.734946399 | 64 | 256 | 208 | 1\1 | TCGA-FG-A713/1.3.6.1.4.1.14519.5.2.1.5826.4003.239451224442205099709649006810/1.3.6.1.4.1.14519.5.2.1.5826.4003.127828297999920935064228260226 | ||
| TCGA-LGG | TCGA-HT | TCGA-HT-7473 | flair | M | 28 | 68.039 | Ax_Flair_irFSE_H | 8 | 6.82232E+15 | 8/26/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 9002 | 121.752 | 2200 | 63.86294 | 1.5 | 5 | 90 | 0.9491 | 32 | 256 | 224 | 0.9375\0.9375 | TCGA-HT-7473/1.3.6.1.4.1.14519.5.2.1.2531.4003.339148669016426179130214847712/1.3.6.1.4.1.14519.5.2.1.2531.4003.235597736552717483361225257005 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7473 | t1Gd | M | 28 | 68.039 | AX_3D_SPGR+C | 12 | 6.82232E+15 | 8/26/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 9.428 | 4.54 | 450 | 63.86294 | 1.5 | 1 | 20 | 0.293884 | 212 | 320 | 192 | 0.4688\0.4688 | TCGA-HT-7473/1.3.6.1.4.1.14519.5.2.1.2531.4003.339148669016426179130214847712/1.3.6.1.4.1.14519.5.2.1.2531.4003.333980853070789124033221676794 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7473 | t1 | M | 28 | 68.039 | AX_3D_SPGR | 5 | 6.82232E+15 | 8/26/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 9.428 | 4.54 | 450 | 63.86294 | 1.5 | 1 | 20 | 0.293884 | 212 | 320 | 192 | 0.4688\0.4688 | TCGA-HT-7473/1.3.6.1.4.1.14519.5.2.1.2531.4003.339148669016426179130214847712/1.3.6.1.4.1.14519.5.2.1.2531.4003.233544485840464225800386044386 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7473 | t2 | M | 28 | 68.039 | 3-pl_T2*_FGRE_S | 1 | 6.82232E+15 | 8/26/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | RM | 2D | 5 | 5.104 | 1.46 | 0 | 63.86291 | 1.5 | 10 | 30 | 0.242037 | 9 | 256 | 128 | 0.9375\0.9375 | TCGA-HT-7473/1.3.6.1.4.1.14519.5.2.1.2531.4003.339148669016426179130214847712/1.3.6.1.4.1.14519.5.2.1.2531.4003.479342730015683457990678269518 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7475 | flair | M | 67 | 106 | Ax_Flair_irFSE_H | 6 | 2.30316E+15 | 9/18/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 9002 | 121.524 | 2200 | 63.86295 | 1.5 | 5 | 90 | 1.1742 | 32 | 256 | 224 | 0.9375\0.9375 | TCGA-HT-7475/1.3.6.1.4.1.14519.5.2.1.2531.4003.306463775653742362436970431460/1.3.6.1.4.1.14519.5.2.1.2531.4003.174114720577685561285251606527 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7475 | t1Gd | M | 67 | 106 | AX_3D_SPGR+C | 11 | 2.30316E+15 | 9/18/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 9.388 | 4.488 | 450 | 63.86295 | 1.5 | 1 | 20 | 0.325404 | 212 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-7475/1.3.6.1.4.1.14519.5.2.1.2531.4003.306463775653742362436970431460/1.3.6.1.4.1.14519.5.2.1.2531.4003.241979134763350176634365733028 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7475 | t1 | M | 67 | 106 | AX_3D_SPGR | 5 | 2.30316E+15 | 9/18/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 9.388 | 4.488 | 450 | 63.86295 | 1.5 | 1 | 20 | 0.325404 | 212 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-7475/1.3.6.1.4.1.14519.5.2.1.2531.4003.306463775653742362436970431460/1.3.6.1.4.1.14519.5.2.1.2531.4003.221279757470705906693181410807 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7475 | t2 | M | 67 | 106 | 3-pl_T2*_FGRE_S | 1 | 2.30316E+15 | 9/18/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | RM | 2D | 5 | 5.104 | 1.46 | 0 | 63.86294 | 1.5 | 10 | 30 | 0.299437 | 9 | 256 | 128 | 0.9375\0.9375 | TCGA-HT-7475/1.3.6.1.4.1.14519.5.2.1.2531.4003.306463775653742362436970431460/1.3.6.1.4.1.14519.5.2.1.2531.4003.187157052835080951633840760211 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7602 | flair | M | 21 | 68.039 | Ax_Flair_irFSE_H | 4 | 5.214E+15 | 11/3/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 9002 | 121.98 | 2200 | 63.86308 | 1.5 | 5 | 90 | 0.8304 | 28 | 256 | 224 | 0.9375\0.9375 | TCGA-HT-7602/1.3.6.1.4.1.14519.5.2.1.2531.4003.204317324583381684838188896833/1.3.6.1.4.1.14519.5.2.1.2531.4003.935616829107968413051604663620 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7602 | t1Gd | M | 21 | 68.039 | AX_3D_SPGR | 10 | 5.214E+15 | 11/3/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 9.344 | 4.476 | 450 | 63.86308 | 1.5 | 2 | 20 | 0.295292 | 106 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-7602/1.3.6.1.4.1.14519.5.2.1.2531.4003.204317324583381684838188896833/1.3.6.1.4.1.14519.5.2.1.2531.4003.171677757382127546017310356128 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7602 | t1 | M | 21 | 68.039 | AX_T1 | 7 | 5.214E+15 | 11/3/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 666.664 | 17 | 0 | 63.86308 | 1.5 | 7.5 | 90 | 1.09596 | 20 | 256 | 192 | 0.9375\0.9375 | TCGA-HT-7602/1.3.6.1.4.1.14519.5.2.1.2531.4003.204317324583381684838188896833/1.3.6.1.4.1.14519.5.2.1.2531.4003.242761211097040357467172205806 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7602 | t2 | M | 21 | 68.039 | 3-pl_T2*_FGRE_S | 1 | 5.214E+15 | 11/3/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | RM | 2D | 5 | 5.104 | 1.46 | 0 | 63.86306 | 1.5 | 10 | 30 | 0.242037 | 9 | 256 | 128 | 0.9375\0.9375 | TCGA-HT-7602/1.3.6.1.4.1.14519.5.2.1.2531.4003.204317324583381684838188896833/1.3.6.1.4.1.14519.5.2.1.2531.4003.235986455034628242616079902294 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7616 | flair | M | 75 | 106.594 | Ax_Flair_irFSE_H | 8 | 3.14339E+15 | 8/13/1994 | GE MEDICAL SYSTEMS | SIGNA EXCITE | IR | 2D | 5 | 9002 | 121.676 | 2200 | 63.86317 | 1.5 | 5 | 90 | 1.1773 | 30 | 256 | 224 | 0.9375\0.9375 | TCGA-HT-7616/1.3.6.1.4.1.14519.5.2.1.2531.4003.282021490454584883892357674533/1.3.6.1.4.1.14519.5.2.1.2531.4003.246158931363136936871049530437 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7616 | t1Gd | M | 75 | 106.594 | AX_3D_SPGR+C | 10 | 3.14339E+15 | 8/13/1994 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 23 | 4.6 | 0 | 63.86318 | 1.5 | 2 | 26 | 0.135368 | 92 | 256 | 160 | 1.0156\1.0156 | TCGA-HT-7616/1.3.6.1.4.1.14519.5.2.1.2531.4003.282021490454584883892357674533/1.3.6.1.4.1.14519.5.2.1.2531.4003.187307241768156692958489188568 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7616 | t1 | M | 75 | 106.594 | AX_3D_SPGR | 4 | 3.14339E+15 | 8/13/1994 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 23 | 4.6 | 0 | 63.86317 | 1.5 | 2 | 26 | 0.499826 | 92 | 256 | 160 | 1.0156\1.0156 | TCGA-HT-7616/1.3.6.1.4.1.14519.5.2.1.2531.4003.282021490454584883892357674533/1.3.6.1.4.1.14519.5.2.1.2531.4003.283487006692286623042316208228 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7616 | t2 | M | 75 | 106.594 | 3-pl_T2*_FGRE_S | 1 | 3.14339E+15 | 8/13/1994 | GE MEDICAL SYSTEMS | SIGNA EXCITE | RM | 2D | 5 | 5.104 | 1.46 | 0 | 63.86315 | 1.5 | 10 | 30 | 0.300241 | 9 | 256 | 128 | 0.9375\0.9375 | TCGA-HT-7616/1.3.6.1.4.1.14519.5.2.1.2531.4003.282021490454584883892357674533/1.3.6.1.4.1.14519.5.2.1.2531.4003.318187228786416199345869204369 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7680 | flair | F | 32 | 61.235 | Ax_Flair_irFSE_H | 6 | 2.71385E+15 | 2/2/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 9002 | 121.524 | 2200 | 63.86295 | 1.5 | 5 | 90 | 0.9023 | 32 | 256 | 224 | 0.9375\0.9375 | TCGA-HT-7680/1.3.6.1.4.1.14519.5.2.1.2531.4003.231158914302938549165293612810/1.3.6.1.4.1.14519.5.2.1.2531.4003.135075318828198110810035695027 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7680 | t1Gd | F | 32 | 61.235 | C+AX_3D_SPGR | 12 | 2.71385E+15 | 2/2/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 9.336 | 4.472 | 450 | 63.86295 | 1.5 | 2 | 20 | 0.286776 | 106 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-7680/1.3.6.1.4.1.14519.5.2.1.2531.4003.231158914302938549165293612810/1.3.6.1.4.1.14519.5.2.1.2531.4003.234480128631300125982061092286 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7680 | t1 | F | 32 | 61.235 | AX_T1 | 5 | 2.71385E+15 | 2/2/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 683.332 | 17 | 0 | 63.86295 | 1.5 | 7.5 | 90 | 1.06725 | 21 | 256 | 192 | 0.9375\0.9375 | TCGA-HT-7680/1.3.6.1.4.1.14519.5.2.1.2531.4003.231158914302938549165293612810/1.3.6.1.4.1.14519.5.2.1.2531.4003.201090454229010635061782438562 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7680 | t2 | F | 32 | 61.235 | COR_GRE_T2 | 7 | 2.71385E+15 | 2/2/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 2D | 5 | 550 | 35 | 0 | 63.86295 | 1.5 | 7.5 | 20 | 0.00509736 | 48 | 256 | 192 | 0.9375\0.9375 | TCGA-HT-7680/1.3.6.1.4.1.14519.5.2.1.2531.4003.231158914302938549165293612810/1.3.6.1.4.1.14519.5.2.1.2531.4003.109629960664151193660949097585 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7684 | flair | M | 58 | 90.718 | Ax_Flair_irFSE_H | 7 | 7.00231E+15 | 8/16/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 9002 | 121.828 | 2200 | 63.86312 | 1.5 | 5 | 90 | 1.0896 | 32 | 256 | 224 | 0.9375\0.9375 | TCGA-HT-7684/1.3.6.1.4.1.14519.5.2.1.2531.4003.186054156253219473364551929908/1.3.6.1.4.1.14519.5.2.1.2531.4003.326875482336787381981274326694 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7684 | t1Gd | M | 58 | 90.718 | POST_AX_3D_SPGR | 12 | 7.00231E+15 | 8/16/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 9.364 | 4.48 | 450 | 63.86311 | 1.5 | 2 | 20 | 0.319206 | 106 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-7684/1.3.6.1.4.1.14519.5.2.1.2531.4003.186054156253219473364551929908/1.3.6.1.4.1.14519.5.2.1.2531.4003.324057765300350182002585590682 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7684 | t1 | M | 58 | 90.718 | AX_T1 | 4 | 7.00231E+15 | 8/16/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 450 | 17 | 0 | 63.86312 | 1.5 | 7.5 | 90 | 1.2134 | 25 | 256 | 192 | 0.9375\0.9375 | TCGA-HT-7684/1.3.6.1.4.1.14519.5.2.1.2531.4003.186054156253219473364551929908/1.3.6.1.4.1.14519.5.2.1.2531.4003.316825701703669893555766169473 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7684 | t2 | M | 58 | 90.718 | 3-pl_T2*_FGRE_S | 1 | 7.00231E+15 | 8/16/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | RM | 2D | 5 | 5.104 | 1.46 | 0 | 63.86308 | 1.5 | 10 | 30 | 0.277876 | 9 | 256 | 128 | 0.9375\0.9375 | TCGA-HT-7684/1.3.6.1.4.1.14519.5.2.1.2531.4003.186054156253219473364551929908/1.3.6.1.4.1.14519.5.2.1.2531.4003.248152573378052966975211701886 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7686 | flair | F | 29 | 64.41 | Ax_Flair_irFSE_H | 7 | 2.71269E+15 | 6/29/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | IR | 2D | 5 | 9002 | 121.6 | 2200 | 63.86313 | 1.5 | 5 | 90 | 0.8089 | 28 | 256 | 224 | 0.9375\0.9375 | TCGA-HT-7686/1.3.6.1.4.1.14519.5.2.1.2531.4003.244405970705727872326343639199/1.3.6.1.4.1.14519.5.2.1.2531.4003.164288781276096644376995835931 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7686 | t1Gd | F | 29 | 64.41 | AX_3D_SPGR | 10 | 2.71269E+15 | 6/29/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 9.336 | 4.472 | 450 | 63.86313 | 1.5 | 2 | 20 | 0.29382 | 106 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-7686/1.3.6.1.4.1.14519.5.2.1.2531.4003.244405970705727872326343639199/1.3.6.1.4.1.14519.5.2.1.2531.4003.617892831080967474932515439020 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7686 | t1 | F | 29 | 64.41 | AX_T1 | 5 | 2.71269E+15 | 6/29/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 666.664 | 17 | 0 | 63.86313 | 1.5 | 7.5 | 90 | 1.0675 | 20 | 256 | 192 | 0.9375\0.9375 | TCGA-HT-7686/1.3.6.1.4.1.14519.5.2.1.2531.4003.244405970705727872326343639199/1.3.6.1.4.1.14519.5.2.1.2531.4003.244921667827712799297721626042 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7686 | t2 | F | 29 | 64.41 | COR_GRE_T2 | 9 | 2.71269E+15 | 6/29/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 2D | 5 | 467 | 35 | 0 | 63.86313 | 1.5 | 7.5 | 20 | 0.0051273 | 40 | 256 | 192 | 0.9375\0.9375 | TCGA-HT-7686/1.3.6.1.4.1.14519.5.2.1.2531.4003.244405970705727872326343639199/1.3.6.1.4.1.14519.5.2.1.2531.4003.198314247552871385304859040626 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7690 | flair | M | 29 | 113.398 | Ax_Flair_irFSE_H | 7 | 9.81991E+15 | 3/12/1996 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 9002 | 121.524 | 2200 | 63.86304 | 1.5 | 5 | 90 | 1.2128 | 32 | 256 | 224 | 0.9375\0.9375 | TCGA-HT-7690/1.3.6.1.4.1.14519.5.2.1.2531.4003.244290172471315468690716464118/1.3.6.1.4.1.14519.5.2.1.2531.4003.813549114384769615209313988757 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7690 | t1Gd | M | 29 | 113.398 | C+AX_3D_SPGR | 12 | 9.81991E+15 | 3/12/1996 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 9.396 | 4.488 | 450 | 63.86304 | 1.5 | 2 | 20 | 0.330267 | 106 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-7690/1.3.6.1.4.1.14519.5.2.1.2531.4003.244290172471315468690716464118/1.3.6.1.4.1.14519.5.2.1.2531.4003.166257326692653730359194508511 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7690 | t1 | M | 29 | 113.398 | AX_T1 | 5 | 9.81991E+15 | 3/12/1996 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 783.332 | 17 | 0 | 63.86304 | 1.5 | 7.5 | 90 | 1.22579 | 24 | 256 | 192 | 0.9375\0.9375 | TCGA-HT-7690/1.3.6.1.4.1.14519.5.2.1.2531.4003.244290172471315468690716464118/1.3.6.1.4.1.14519.5.2.1.2531.4003.250335993934930482214933635923 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7690 | t2 | M | 29 | 113.398 | COR_GRE_T2 | 8 | 9.81991E+15 | 3/12/1996 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 2D | 5 | 567 | 35 | 0 | 63.86304 | 1.5 | 7.5 | 20 | 0.00719977 | 52 | 256 | 192 | 0.9375\0.9375 | TCGA-HT-7690/1.3.6.1.4.1.14519.5.2.1.2531.4003.244290172471315468690716464118/1.3.6.1.4.1.14519.5.2.1.2531.4003.114844545936671776285659054250 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7692 | flair | M | 43 | 70.307 | Ax_Flair_irFSE_H | 6 | 1.49904E+15 | 7/24/1996 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 9002 | 121.828 | 2200 | 63.86302 | 1.5 | 5 | 90 | 0.9641 | 30 | 256 | 224 | 0.9375\0.9375 | TCGA-HT-7692/1.3.6.1.4.1.14519.5.2.1.2531.4003.211398814274191438581429613406/1.3.6.1.4.1.14519.5.2.1.2531.4003.222350740835087785989639436055 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7692 | t1 | M | 43 | 70.307 | AX_T1 | 5 | 1.49904E+15 | 7/24/1996 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 666.664 | 17 | 0 | 63.86302 | 1.5 | 7.5 | 90 | 1.11334 | 20 | 256 | 192 | 0.9375\0.9375 | TCGA-HT-7692/1.3.6.1.4.1.14519.5.2.1.2531.4003.211398814274191438581429613406/1.3.6.1.4.1.14519.5.2.1.2531.4003.957195987351897640611427265940 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7692 | t2 | M | 43 | 70.307 | AX_FSE_T2_inter | 8 | 1.49904E+15 | 7/24/1996 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 2 | 4766.66 | 93.248 | 0 | 63.86302 | 1.5 | 2 | 90 | 1.8934 | 84 | 256 | 160 | 1.0156\1.0156 | TCGA-HT-7692/1.3.6.1.4.1.14519.5.2.1.2531.4003.211398814274191438581429613406/1.3.6.1.4.1.14519.5.2.1.2531.4003.326142621012177190108169467120 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7693 | flair | F | 51 | 83.915 | Ax_Flair_irFSE_H | 7 | 2.56082E+15 | 5/20/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | IR | 2D | 5 | 9002 | 121.6 | 2200 | 63.86308 | 1.5 | 5 | 90 | 1.0496 | 32 | 256 | 224 | 0.9375\0.9375 | TCGA-HT-7693/1.3.6.1.4.1.14519.5.2.1.2531.4003.628801887079472592322253871036/1.3.6.1.4.1.14519.5.2.1.2531.4003.216268246131326829920251118767 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7693 | t1Gd | F | 51 | 83.915 | C+AX_3D_SPGR | 11 | 2.56082E+15 | 5/20/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 9.336 | 4.472 | 450 | 63.86308 | 1.5 | 2 | 20 | 0.3336 | 106 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-7693/1.3.6.1.4.1.14519.5.2.1.2531.4003.628801887079472592322253871036/1.3.6.1.4.1.14519.5.2.1.2531.4003.748980782452317231270307024306 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7693 | t1 | F | 51 | 83.915 | AX_T1 | 5 | 2.56082E+15 | 5/20/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 683.332 | 17 | 0 | 63.86308 | 1.5 | 7.5 | 90 | 1.2415 | 21 | 256 | 192 | 0.9375\0.9375 | TCGA-HT-7693/1.3.6.1.4.1.14519.5.2.1.2531.4003.628801887079472592322253871036/1.3.6.1.4.1.14519.5.2.1.2531.4003.322960220935957352201340032031 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7693 | t2 | F | 51 | 83.915 | 3-pl_T2*_FGRE_S | 1 | 2.56082E+15 | 5/20/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | RM | 2D | 5 | 5.104 | 1.46 | 0 | 63.8631 | 1.5 | 10 | 30 | 0.267671 | 9 | 256 | 128 | 0.9375\0.9375 | TCGA-HT-7693/1.3.6.1.4.1.14519.5.2.1.2531.4003.628801887079472592322253871036/1.3.6.1.4.1.14519.5.2.1.2531.4003.289315312435476678143013547067 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7694 | flair | M | 60 | 107 | Ax_Flair_irFSE_H | 7 | 2.73244E+15 | 4/4/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | IR | 2D | 5 | 9002 | 121.6 | 2200 | 63.86312 | 1.5 | 5 | 90 | 1.1795 | 32 | 256 | 224 | 0.9375\0.9375 | TCGA-HT-7694/1.3.6.1.4.1.14519.5.2.1.2531.4003.287964934328207007202059293153/1.3.6.1.4.1.14519.5.2.1.2531.4003.131371499353948533109942982301 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7694 | t1Gd | M | 60 | 107 | C+AX_3D_SPGR | 13 | 2.73244E+15 | 4/4/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 9.128 | 4.468 | 450 | 63.86312 | 1.5 | 2 | 20 | 0.356997 | 106 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-7694/1.3.6.1.4.1.14519.5.2.1.2531.4003.287964934328207007202059293153/1.3.6.1.4.1.14519.5.2.1.2531.4003.125678417374893795437733747898 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7694 | t1 | M | 60 | 107 | AX_T1 | 5 | 2.73244E+15 | 4/4/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 683.332 | 17 | 0 | 63.86312 | 1.5 | 7.5 | 90 | 1.19617 | 21 | 256 | 192 | 1.0156\1.0156 | TCGA-HT-7694/1.3.6.1.4.1.14519.5.2.1.2531.4003.287964934328207007202059293153/1.3.6.1.4.1.14519.5.2.1.2531.4003.326296038155567068613921506545 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7694 | t2 | M | 60 | 107 | COR_GRE_T2 | 8 | 2.73244E+15 | 4/4/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 2D | 5 | 550 | 35 | 0 | 63.86312 | 1.5 | 7.5 | 20 | 0.00666331 | 48 | 256 | 192 | 1.0156\1.0156 | TCGA-HT-7694/1.3.6.1.4.1.14519.5.2.1.2531.4003.287964934328207007202059293153/1.3.6.1.4.1.14519.5.2.1.2531.4003.866906654890810756461307550000 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7855 | flair | M | 39 | 94.347 | Ax_T2_Flair | 5 | 2.90984E+15 | 10/20/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 9002 | 152.964 | 2250 | 127.7268 | 3 | 5.5 | 90 | 0.6902 | 28 | 512 | 192 | 0.4688\0.4688 | TCGA-HT-7855/1.3.6.1.4.1.14519.5.2.1.2531.4003.198324244153534584390623424581/1.3.6.1.4.1.14519.5.2.1.2531.4003.326558553266212493966722036356 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7855 | t1Gd | M | 39 | 94.347 | Ax_T1_MP_SPGR+C | 10 | 2.90984E+15 | 10/20/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 7.76 | 3.08 | 300 | 127.7268 | 3 | 2 | 13 | 0.643089 | 106 | 320 | 224 | 0.5078\0.5078 | TCGA-HT-7855/1.3.6.1.4.1.14519.5.2.1.2531.4003.198324244153534584390623424581/1.3.6.1.4.1.14519.5.2.1.2531.4003.181614742987205173574912986643 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7855 | t1 | M | 39 | 94.347 | Ax_T1_SE | 7 | 2.90984E+15 | 10/20/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 550 | 17 | 0 | 127.7268 | 3 | 7.5 | 90 | 2.02031 | 21 | 512 | 256 | 0.4688\0.4688 | TCGA-HT-7855/1.3.6.1.4.1.14519.5.2.1.2531.4003.198324244153534584390623424581/1.3.6.1.4.1.14519.5.2.1.2531.4003.199095925660425709421413760410 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7855 | t2 | M | 39 | 94.347 | Cor_2D_T2*GRE | 9 | 2.90984E+15 | 10/20/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 2D | 5 | 600 | 10 | 0 | 127.7268 | 3 | 7.5 | 25 | 0.0315047 | 44 | 512 | 192 | 0.4688\0.4688 | TCGA-HT-7855/1.3.6.1.4.1.14519.5.2.1.2531.4003.198324244153534584390623424581/1.3.6.1.4.1.14519.5.2.1.2531.4003.350021473582796468994279160988 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7856 | flair | M | 35 | 92.079 | Ax_Flair_irFSE_H | 6 | 2.8409E+15 | 8/31/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 9002 | 121.752 | 2200 | 63.86313 | 1.5 | 5 | 90 | 1.0974 | 32 | 256 | 224 | 0.9375\0.9375 | TCGA-HT-7856/1.3.6.1.4.1.14519.5.2.1.2531.4003.100877690152591433926158971037/1.3.6.1.4.1.14519.5.2.1.2531.4003.335140292213978326526348108103 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7856 | t1Gd | M | 35 | 92.079 | COR_T1+C | 11 | 2.8409E+15 | 8/31/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 416.664 | 17 | 0 | 63.86313 | 1.5 | 7.5 | 90 | 1.21878 | 24 | 256 | 192 | 0.9375\0.9375 | TCGA-HT-7856/1.3.6.1.4.1.14519.5.2.1.2531.4003.100877690152591433926158971037/1.3.6.1.4.1.14519.5.2.1.2531.4003.881801191410038591436680403595 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7856 | t1 | M | 35 | 92.079 | SAG_T1 | 4 | 2.8409E+15 | 8/31/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 7 | 566.664 | 17 | 0 | 63.86313 | 1.5 | 9 | 90 | 1.26795 | 17 | 256 | 192 | 0.9375\0.9375 | TCGA-HT-7856/1.3.6.1.4.1.14519.5.2.1.2531.4003.100877690152591433926158971037/1.3.6.1.4.1.14519.5.2.1.2531.4003.882113785187560242328167389560 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7856 | t2 | M | 35 | 92.079 | COR_GRE_T2 | 7 | 2.8409E+15 | 8/31/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 2D | 5 | 500 | 35 | 0 | 63.86313 | 1.5 | 7.5 | 20 | 0.0045474 | 48 | 256 | 192 | 0.9375\0.9375 | TCGA-HT-7856/1.3.6.1.4.1.14519.5.2.1.2531.4003.100877690152591433926158971037/1.3.6.1.4.1.14519.5.2.1.2531.4003.298451687529064117671386672478 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7860 | flair | F | 60 | 68 | Ax_Flair_irFSE_H | 7 | 1.97569E+15 | 5/13/1996 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 9002 | 121.524 | 2200 | 63.86304 | 1.5 | 5 | 90 | 0.9488 | 32 | 256 | 224 | 0.9375\0.9375 | TCGA-HT-7860/1.3.6.1.4.1.14519.5.2.1.2531.4003.290722495921577895242204914721/1.3.6.1.4.1.14519.5.2.1.2531.4003.301059638139812997612342918747 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7860 | t1Gd | F | 60 | 68 | C+AX_3D_SPGR | 12 | 1.97569E+15 | 5/13/1996 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 9.344 | 4.476 | 450 | 63.86304 | 1.5 | 2 | 20 | 0.29521 | 106 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-7860/1.3.6.1.4.1.14519.5.2.1.2531.4003.290722495921577895242204914721/1.3.6.1.4.1.14519.5.2.1.2531.4003.100799255779837914036141600878 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7860 | t1 | F | 60 | 68 | AX_T1 | 5 | 1.97569E+15 | 5/13/1996 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 716.664 | 17 | 0 | 63.86304 | 1.5 | 7.5 | 90 | 1.12092 | 22 | 256 | 192 | 0.9375\0.9375 | TCGA-HT-7860/1.3.6.1.4.1.14519.5.2.1.2531.4003.290722495921577895242204914721/1.3.6.1.4.1.14519.5.2.1.2531.4003.255671469725080757792140904566 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7860 | t2 | F | 60 | 68 | Prop_T2_TRF | 6 | 1.97569E+15 | 5/13/1996 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 5200 | 113.256 | 63.86303 | 1.5 | 7.5 | 90 | 1.2053772 | 22 | 384 | 384 | 0.468750\0.468750 | TCGA-HT-7860/1.3.6.1.4.1.14519.5.2.1.2531.4003.290722495921577895242204914721/1.3.6.1.4.1.14519.5.2.1.2531.4003.228588646024368852629824081803 | ||
| TCGA-LGG | TCGA-HT | TCGA-HT-7874 | flair | F | 47 | 62 | Ax_Flair_irFSE_H | 6 | 2.64153E+15 | 9/2/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 9002 | 121.6 | 2200 | 63.86313 | 1.5 | 5 | 90 | 0.9077 | 30 | 256 | 224 | 0.9375\0.9375 | TCGA-HT-7874/1.3.6.1.4.1.14519.5.2.1.2531.4003.253968930679883438764511461816/1.3.6.1.4.1.14519.5.2.1.2531.4003.114059255086495939980670300373 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7874 | t1Gd | F | 47 | 62 | AX_3D_SPGR | 11 | 2.64153E+15 | 9/2/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 9.42 | 4.536 | 450 | 63.86313 | 1.5 | 2 | 20 | 0.287115 | 106 | 320 | 192 | 0.4688\0.4688 | TCGA-HT-7874/1.3.6.1.4.1.14519.5.2.1.2531.4003.253968930679883438764511461816/1.3.6.1.4.1.14519.5.2.1.2531.4003.271081801017486100580794738668 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7874 | t1 | F | 47 | 62 | AX_T1 | 5 | 2.64153E+15 | 9/2/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 666.664 | 17 | 0 | 63.86313 | 1.5 | 7.5 | 90 | 1.04814 | 20 | 256 | 192 | 0.9375\0.9375 | TCGA-HT-7874/1.3.6.1.4.1.14519.5.2.1.2531.4003.253968930679883438764511461816/1.3.6.1.4.1.14519.5.2.1.2531.4003.172208561596137136210902304486 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7874 | t2 | F | 47 | 62 | 3-pl_T2*_FGRE_S | 1 | 2.64153E+15 | 9/2/1995 | GE MEDICAL SYSTEMS | SIGNA EXCITE | RM | 2D | 5 | 5.104 | 1.46 | 0 | 63.86308 | 1.5 | 10 | 30 | 0.231476 | 9 | 256 | 128 | 0.9375\0.9375 | TCGA-HT-7874/1.3.6.1.4.1.14519.5.2.1.2531.4003.253968930679883438764511461816/1.3.6.1.4.1.14519.5.2.1.2531.4003.108904271616553624903958841695 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7879 | flair | M | 31 | 74.843 | Ax_T2_Flair | 5 | 1.40238E+15 | 10/9/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 9002 | 150.528 | 2250 | 127.7265 | 3 | 5 | 90 | 0.7351 | 28 | 512 | 192 | 0.4688\0.4688 | TCGA-HT-7879/1.3.6.1.4.1.14519.5.2.1.2531.4003.208179366605438767398250387116/1.3.6.1.4.1.14519.5.2.1.2531.4003.390124297613614320003204742124 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7879 | t1Gd | M | 31 | 74.843 | POST_Ax_T1_MP_SPGR | 12 | 1.40238E+15 | 10/9/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 6.773 | 2.772 | 0 | 127.7265 | 3 | 1 | 13 | 0.473884 | 212 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-7879/1.3.6.1.4.1.14519.5.2.1.2531.4003.208179366605438767398250387116/1.3.6.1.4.1.14519.5.2.1.2531.4003.287469028595190429124363364353 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7879 | t1 | M | 31 | 74.843 | Ax_T1_MP_SPGR | 6 | 1.40238E+15 | 10/9/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 6.773 | 2.772 | 0 | 127.7265 | 3 | 1 | 13 | 0.473884 | 212 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-7879/1.3.6.1.4.1.14519.5.2.1.2531.4003.208179366605438767398250387116/1.3.6.1.4.1.14519.5.2.1.2531.4003.218683428988186850780269340361 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7879 | t2 | M | 31 | 74.843 | (OPTIONAL)Ax_T2_FSE_INTER | 7 | 1.40238E+15 | 10/9/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 2 | 4200 | 82.752 | 0 | 127.7265 | 3 | 2 | 90 | 1.6163 | 78 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-7879/1.3.6.1.4.1.14519.5.2.1.2531.4003.208179366605438767398250387116/1.3.6.1.4.1.14519.5.2.1.2531.4003.723937203870259397937020184430 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7882 | flair | M | 66 | 67.132 | Ax_Flair_irFSE_H | 5 | 2.48389E+15 | 1/25/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 9002 | 121.752 | 2200 | 63.863 | 1.5 | 5 | 90 | 0.943 | 32 | 256 | 224 | 0.9375\0.9375 | TCGA-HT-7882/1.3.6.1.4.1.14519.5.2.1.2531.4003.553374666251335749299587381378/1.3.6.1.4.1.14519.5.2.1.2531.4003.111610015608806980339206664631 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7882 | t1Gd | M | 66 | 67.132 | POSTCOR_T1_SE | 14 | 2.48389E+15 | 1/25/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 500 | 16 | 0 | 63.863 | 1.5 | 5 | 90 | 1.12453 | 37 | 256 | 160 | 0.9375\0.9375 | TCGA-HT-7882/1.3.6.1.4.1.14519.5.2.1.2531.4003.553374666251335749299587381378/1.3.6.1.4.1.14519.5.2.1.2531.4003.143978018567514063128225625202 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7882 | t1 | M | 66 | 67.132 | REPEAT_AX_3D_SPGR | 9 | 2.48389E+15 | 1/25/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 9.344 | 4.476 | 450 | 63.863 | 1.5 | 2 | 20 | 0.293396 | 106 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-7882/1.3.6.1.4.1.14519.5.2.1.2531.4003.553374666251335749299587381378/1.3.6.1.4.1.14519.5.2.1.2531.4003.131861920354865041057455029355 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7882 | t2 | M | 66 | 67.132 | COR_GRE_T2 | 6 | 2.48389E+15 | 1/25/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 2D | 5 | 567 | 35 | 0 | 63.863 | 1.5 | 7.5 | 20 | 0.00559802 | 50 | 256 | 192 | 0.9375\0.9375 | TCGA-HT-7882/1.3.6.1.4.1.14519.5.2.1.2531.4003.553374666251335749299587381378/1.3.6.1.4.1.14519.5.2.1.2531.4003.110745866062969069187735984363 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7884 | flair | F | 44 | 68.039 | Ax_Flair_irFSE_H | 7 | 3.32409E+15 | 9/13/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 9002 | 131.328 | 2200 | 63.8609 | 1.5 | 5 | 90 | 0.8304 | 28 | 288 | 224 | 0.4688\0.4688 | TCGA-HT-7884/1.3.6.1.4.1.14519.5.2.1.2531.4003.213534942777481053514809821174/1.3.6.1.4.1.14519.5.2.1.2531.4003.321207014837740778483316363283 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7884 | t1Gd | F | 44 | 68.039 | AX_3D_SPGR+C | 11 | 3.32409E+15 | 9/13/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 9.344 | 4.476 | 450 | 63.8609 | 1.5 | 1 | 20 | 0.295292 | 212 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-7884/1.3.6.1.4.1.14519.5.2.1.2531.4003.213534942777481053514809821174/1.3.6.1.4.1.14519.5.2.1.2531.4003.318439416511462348230053052662 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7884 | t1 | F | 44 | 68.039 | AX_T1 | 5 | 3.32409E+15 | 9/13/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 666.664 | 17 | 0 | 63.8609 | 1.5 | 7 | 90 | 1.09596 | 20 | 288 | 192 | 0.4688\0.4688 | TCGA-HT-7884/1.3.6.1.4.1.14519.5.2.1.2531.4003.213534942777481053514809821174/1.3.6.1.4.1.14519.5.2.1.2531.4003.207800235363459841669208898660 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-7884 | t2 | F | 44 | 68.039 | (OPT)_AX_T2_FSE_inter | 12 | 3.32409E+15 | 9/13/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 2 | 4550 | 93.248 | 0 | 63.8609 | 1.5 | 2 | 90 | 1.8596 | 80 | 256 | 160 | 1.0156\1.0156 | TCGA-HT-7884/1.3.6.1.4.1.14519.5.2.1.2531.4003.213534942777481053514809821174/1.3.6.1.4.1.14519.5.2.1.2531.4003.333883177788958445968782847214 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8018 | flair | F | 40 | 92.986 | Ax_Flair_irFSE_H | 6 | 1.76778E+15 | 4/11/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 9002 | 126.588 | 2200 | 63.86295 | 1.5 | 5 | 90 | 0.0531 | 32 | 256 | 224 | 0.9375\0.9375 | TCGA-HT-8018/1.3.6.1.4.1.14519.5.2.1.2531.4003.112684169989572671934693953563/1.3.6.1.4.1.14519.5.2.1.2531.4003.750153295669091838671413843337 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8018 | t1Gd | F | 40 | 92.986 | AX_3D_SPGR_+_C | 9 | 1.76778E+15 | 4/11/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 9.336 | 4.472 | 450 | 63.86295 | 1.5 | 2 | 20 | 0.0118572 | 106 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-8018/1.3.6.1.4.1.14519.5.2.1.2531.4003.112684169989572671934693953563/1.3.6.1.4.1.14519.5.2.1.2531.4003.314673445079965605329585246765 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8018 | t1 | F | 40 | 92.986 | AX_T1 | 4 | 1.76778E+15 | 4/11/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 666.664 | 17 | 0 | 63.86295 | 1.5 | 7.5 | 90 | 0.0430794 | 20 | 256 | 192 | 0.9375\0.9375 | TCGA-HT-8018/1.3.6.1.4.1.14519.5.2.1.2531.4003.112684169989572671934693953563/1.3.6.1.4.1.14519.5.2.1.2531.4003.626802034424465313475732328979 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8018 | t2 | F | 40 | 92.986 | (OPTIONAL)_AX_FSE_T2_inter | 5 | 1.76778E+15 | 4/11/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 2 | 4100 | 87.424 | 0 | 63.86295 | 1.5 | 2 | 90 | 0.1135 | 84 | 256 | 160 | 1.0156\1.0156 | TCGA-HT-8018/1.3.6.1.4.1.14519.5.2.1.2531.4003.112684169989572671934693953563/1.3.6.1.4.1.14519.5.2.1.2531.4003.757368868058675693748842299676 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8105 | flair | M | 54 | 149.685 | Ax_Flair_irFSE_H | 6 | 1.40311E+15 | 8/26/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 9002 | 120.156 | 2200 | 63.86083 | 1.5 | 5 | 90 | 1.1596 | 32 | 288 | 224 | 0.4688\0.4688 | TCGA-HT-8105/1.3.6.1.4.1.14519.5.2.1.2531.4003.666089736017948642080519950202/1.3.6.1.4.1.14519.5.2.1.2531.4003.521767595065028541607661249239 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8105 | t1Gd | M | 54 | 149.685 | =+C_AX_3D_SPGR | 10 | 1.40311E+15 | 8/26/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 9.456 | 4.504 | 450 | 63.86083 | 1.5 | 1 | 20 | 0.332902 | 212 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-8105/1.3.6.1.4.1.14519.5.2.1.2531.4003.666089736017948642080519950202/1.3.6.1.4.1.14519.5.2.1.2531.4003.261424643169894361999089010789 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8105 | t1 | M | 54 | 149.685 | SAG_T1 | 4 | 1.40311E+15 | 8/26/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 4 | 800 | 17 | 0 | 63.86083 | 1.5 | 6 | 90 | 1.37125 | 24 | 256 | 192 | 0.9375\0.9375 | TCGA-HT-8105/1.3.6.1.4.1.14519.5.2.1.2531.4003.666089736017948642080519950202/1.3.6.1.4.1.14519.5.2.1.2531.4003.359499344977834153037880376188 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8105 | t2 | M | 54 | 149.685 | COR_GRE_T2 | 7 | 1.40311E+15 | 8/26/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 2D | 5 | 517 | 15 | 0 | 63.86083 | 1.5 | 7.5 | 20 | 0.00763495 | 44 | 288 | 192 | 0.4688\0.4688 | TCGA-HT-8105/1.3.6.1.4.1.14519.5.2.1.2531.4003.666089736017948642080519950202/1.3.6.1.4.1.14519.5.2.1.2531.4003.290098899120495776459456484100 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8106 | flair | M | 53 | 79.379 | Ax_Flair_irFSE_H | 6 | 3.00856E+15 | 7/27/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 9002 | 121.6 | 2200 | 63.86295 | 1.5 | 5 | 90 | 1.022 | 32 | 256 | 224 | 0.9375\0.9375 | TCGA-HT-8106/1.3.6.1.4.1.14519.5.2.1.2531.4003.193992540899316552400927467522/1.3.6.1.4.1.14519.5.2.1.2531.4003.223047379880676549838224939528 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8106 | t1Gd | M | 53 | 79.379 | C+AX_3D_SPGR | 12 | 3.00856E+15 | 7/27/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 9.36 | 4.48 | 450 | 63.86295 | 1.5 | 1 | 20 | 0.305234 | 212 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-8106/1.3.6.1.4.1.14519.5.2.1.2531.4003.193992540899316552400927467522/1.3.6.1.4.1.14519.5.2.1.2531.4003.208453668054401410366698146928 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8106 | t1 | M | 53 | 79.379 | AX_T1 | 5 | 3.00856E+15 | 7/27/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 716.664 | 17 | 0 | 63.86295 | 1.5 | 7.5 | 90 | 1.20734 | 22 | 256 | 192 | 0.9375\0.9375 | TCGA-HT-8106/1.3.6.1.4.1.14519.5.2.1.2531.4003.193992540899316552400927467522/1.3.6.1.4.1.14519.5.2.1.2531.4003.227475575209664270665791110082 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8106 | t2 | M | 53 | 79.379 | Prop_T2_TRF | 8 | 3.00856E+15 | 7/27/1997 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 5300 | 112.736 | 63.86295 | 1.5 | 7.5 | 90 | 1.273803 | 22 | 384 | 384 | 0.468750\0.468750 | TCGA-HT-8106/1.3.6.1.4.1.14519.5.2.1.2531.4003.193992540899316552400927467522/1.3.6.1.4.1.14519.5.2.1.2531.4003.268245415182347752143774434147 | ||
| TCGA-LGG | TCGA-HT | TCGA-HT-8107 | flair | M | 62 | 86 | Ax_T2_Flair | 7 | 2.31066E+15 | 7/8/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 9002 | 150.864 | 2250 | 127.7266 | 3 | 5 | 90 | 0.7109 | 28 | 512 | 192 | 0.4688\0.4688 | TCGA-HT-8107/1.3.6.1.4.1.14519.5.2.1.2531.4003.126549621125671515633918730177/1.3.6.1.4.1.14519.5.2.1.2531.4003.742126079765319223078547725684 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8107 | t1Gd | M | 62 | 86 | Ax_T1_MP_SPGR_+C | 13 | 2.31066E+15 | 7/8/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 6.773 | 2.772 | 0 | 127.7266 | 3 | 1 | 13 | 0.484134 | 212 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-8107/1.3.6.1.4.1.14519.5.2.1.2531.4003.126549621125671515633918730177/1.3.6.1.4.1.14519.5.2.1.2531.4003.140725228801837632444136864481 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8107 | t1 | M | 62 | 86 | Ax_T1_SE | 9 | 2.31066E+15 | 7/8/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 550 | 18 | 0 | 127.7266 | 3 | 7.5 | 90 | 2.06216 | 20 | 320 | 224 | 0.4688\0.4688 | TCGA-HT-8107/1.3.6.1.4.1.14519.5.2.1.2531.4003.126549621125671515633918730177/1.3.6.1.4.1.14519.5.2.1.2531.4003.339572109113075228569870448997 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8107 | t2 | M | 62 | 86 | (OPTIONAL)Ax_T2_FSE_INTER | 6 | 2.31066E+15 | 7/8/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 2 | 4500 | 82.752 | 0 | 127.7266 | 3 | 2 | 90 | 1.6202 | 82 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-8107/1.3.6.1.4.1.14519.5.2.1.2531.4003.126549621125671515633918730177/1.3.6.1.4.1.14519.5.2.1.2531.4003.175934860067825647876040050083 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8111 | flair | M | 32 | 89.811 | Ax_Flair_irFSE_H | 6 | 1.48464E+15 | 3/30/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 9002 | 131.48 | 2200 | 63.8609 | 1.5 | 5 | 90 | 0.9488 | 28 | 288 | 224 | 0.4688\0.4688 | TCGA-HT-8111/1.3.6.1.4.1.14519.5.2.1.2531.4003.242435921414571703883231168651/1.3.6.1.4.1.14519.5.2.1.2531.4003.207964003557221916122969204644 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8111 | t1Gd | M | 32 | 89.811 | AX_3D_SPGR+C | 11 | 1.48464E+15 | 3/30/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 9.364 | 4.48 | 450 | 63.8609 | 1.5 | 1 | 20 | 0.31767 | 212 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-8111/1.3.6.1.4.1.14519.5.2.1.2531.4003.242435921414571703883231168651/1.3.6.1.4.1.14519.5.2.1.2531.4003.320743579510829717547301347886 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8111 | t1 | M | 32 | 89.811 | AX_T1 | 5 | 1.48464E+15 | 3/30/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 750 | 17 | 0 | 63.8609 | 1.5 | 7 | 90 | 1.22394 | 22 | 288 | 192 | 0.4688\0.4688 | TCGA-HT-8111/1.3.6.1.4.1.14519.5.2.1.2531.4003.242435921414571703883231168651/1.3.6.1.4.1.14519.5.2.1.2531.4003.908347256312619710522332501164 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8111 | t2 | M | 32 | 89.811 | 3-pl_T2*_FGRE_S | 1 | 1.48464E+15 | 3/30/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | RM | 2D | 5 | 5.104 | 1.46 | 0 | 63.86089 | 1.5 | 10 | 30 | 0.276539 | 15 | 256 | 128 | 0.9375\0.9375 | TCGA-HT-8111/1.3.6.1.4.1.14519.5.2.1.2531.4003.242435921414571703883231168651/1.3.6.1.4.1.14519.5.2.1.2531.4003.269592914235352345887545312963 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8113 | flair | F | 49 | 79.379 | Ax_Flair_irFSE_H | 5 | 1.58118E+15 | 8/9/1993 | GE MEDICAL SYSTEMS | SIGNA EXCITE | IR | 2D | 5 | 9002 | 126.252 | 2200 | 63.86342 | 1.5 | 5 | 90 | 0.0544 | 32 | 256 | 224 | 0.937513\0.937488 | TCGA-HT-8113/1.3.6.1.4.1.14519.5.2.1.2531.4003.119195295177370323580160059884/1.3.6.1.4.1.14519.5.2.1.2531.4003.875941858876296194253422981944 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8113 | t1Gd | F | 49 | 79.379 | AX_3D_SPGR | 9 | 1.58118E+15 | 8/9/1993 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 23 | 4.6 | 0 | 63.86342 | 1.5 | 2 | 26 | 0.0162443 | 86 | 256 | 160 | 1.01562\1.01562 | TCGA-HT-8113/1.3.6.1.4.1.14519.5.2.1.2531.4003.119195295177370323580160059884/1.3.6.1.4.1.14519.5.2.1.2531.4003.107596026517862198471059681510 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8113 | t1 | F | 49 | 79.379 | AX_T1 | 3 | 1.58118E+15 | 8/9/1993 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 683.332 | 17 | 0 | 63.86342 | 1.5 | 7.5 | 90 | 0.0452583 | 21 | 256 | 192 | 0.937488\0.937495 | TCGA-HT-8113/1.3.6.1.4.1.14519.5.2.1.2531.4003.119195295177370323580160059884/1.3.6.1.4.1.14519.5.2.1.2531.4003.255471461929192148244064707039 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8113 | t2 | F | 49 | 79.379 | COR_T2__FSE | 7 | 1.58118E+15 | 8/9/1993 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 3 | 3850 | 82.56 | 0 | 63.86341 | 1.5 | 3.5 | 90 | 0.1288 | 22 | 320 | 256 | 0.351561\0.351562 | TCGA-HT-8113/1.3.6.1.4.1.14519.5.2.1.2531.4003.119195295177370323580160059884/1.3.6.1.4.1.14519.5.2.1.2531.4003.213902046445134007901271843900 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8114 | flair | M | 36 | 77.111 | Ax_Flair_irFSE_H | 7 | 3.2038E+15 | 10/30/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 9002 | 127.176 | 2200 | 63.86084 | 1.5 | 5 | 90 | 1.1059 | 32 | 288 | 224 | 0.4688\0.4688 | TCGA-HT-8114/1.3.6.1.4.1.14519.5.2.1.2531.4003.558276508192500356115090222947/1.3.6.1.4.1.14519.5.2.1.2531.4003.108490552093857556325409386866 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8114 | t1Gd | M | 36 | 77.111 | AX_3D_SPGR | 11 | 3.2038E+15 | 10/30/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 9.352 | 4.476 | 450 | 63.86084 | 1.5 | 1 | 20 | 0.30714 | 212 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-8114/1.3.6.1.4.1.14519.5.2.1.2531.4003.558276508192500356115090222947/1.3.6.1.4.1.14519.5.2.1.2531.4003.342300001999257561480858599436 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8114 | t2 | M | 36 | 77.111 | 3-pl_T2*_FGRE_S | 6 | 3.2038E+15 | 10/30/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | RM | 2D | 5 | 5.104 | 1.46 | 0 | 63.86081 | 1.5 | 10 | 30 | 0.257024 | 15 | 256 | 128 | 0.9375\0.9375 | TCGA-HT-8114/1.3.6.1.4.1.14519.5.2.1.2531.4003.558276508192500356115090222947/1.3.6.1.4.1.14519.5.2.1.2531.4003.262039158313498745895460022053 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8563 | flair | F | 30 | 77.111 | Ax_Flair_irFSE_H | 6 | 4.02868E+15 | 12/9/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE\IR | 2D | 5 | 9002 | 126.924 | 2200 | 63.86088 | 1.5 | 5 | 90 | 1.1059 | 32 | 288 | 224 | 0.4688\0.4688 | TCGA-HT-8563/1.3.6.1.4.1.14519.5.2.1.2531.4003.104336479544128599145573158636/1.3.6.1.4.1.14519.5.2.1.2531.4003.147276557455303984775492047930 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8563 | t1Gd | F | 30 | 77.111 | AX_3D_SPGR | 10 | 4.02868E+15 | 12/9/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | GR | 3D | 2 | 9.352 | 4.476 | 450 | 63.86088 | 1.5 | 1 | 20 | 0.30714 | 212 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-8563/1.3.6.1.4.1.14519.5.2.1.2531.4003.104336479544128599145573158636/1.3.6.1.4.1.14519.5.2.1.2531.4003.315705360410181524285206535180 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8563 | t1 | F | 30 | 77.111 | AX_T1 | 5 | 4.02868E+15 | 12/9/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | SE | 2D | 5 | 766.664 | 17 | 0 | 63.86088 | 1.5 | 7 | 90 | 1.16336 | 23 | 288 | 192 | 0.4688\0.4688 | TCGA-HT-8563/1.3.6.1.4.1.14519.5.2.1.2531.4003.104336479544128599145573158636/1.3.6.1.4.1.14519.5.2.1.2531.4003.146200342358276156961672468450 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-8563 | t2 | F | 30 | 77.111 | 3-pl_T2*_FGRE_S | 1 | 4.02868E+15 | 12/9/1998 | GE MEDICAL SYSTEMS | SIGNA EXCITE | RM | 2D | 5 | 5.104 | 1.46 | 0 | 63.86082 | 1.5 | 10 | 30 | 0.257024 | 15 | 256 | 128 | 0.9375\0.9375 | TCGA-HT-8563/1.3.6.1.4.1.14519.5.2.1.2531.4003.104336479544128599145573158636/1.3.6.1.4.1.14519.5.2.1.2531.4003.491822277939696706830446210807 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-A5RC | flair | F | 70 | 74.843 | Ax_Flair_irFSE_H | 5 | 1.89125E+15 | 8/31/1999 | GE MEDICAL SYSTEMS | Signa HDxt | SE\IR | 2D | 5 | 9002 | 127.176 | 2200 | 63.86087 | 1.5 | 5 | 90 | 1.0902 | 32 | 288 | 224 | 0.4688\0.4688 | TCGA-HT-A5RC/1.3.6.1.4.1.14519.5.2.1.2531.4003.231610681115360230926274787101/1.3.6.1.4.1.14519.5.2.1.2531.4003.106462291730893974080562229741 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-A5RC | t1Gd | F | 70 | 74.843 | AX_3D_SPGR | 3 | 1.89125E+15 | 8/31/1999 | GE MEDICAL SYSTEMS | Signa HDxt | GR | 3D | 2 | 9.344 | 4.476 | 450 | 63.86086 | 1.5 | 1 | 20 | 0.304583 | 212 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-A5RC/1.3.6.1.4.1.14519.5.2.1.2531.4003.231610681115360230926274787101/1.3.6.1.4.1.14519.5.2.1.2531.4003.324452218716694155037396009138 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-A5RC | t1 | F | 70 | 74.843 | SAG_T1 | 4 | 1.89125E+15 | 8/31/1999 | GE MEDICAL SYSTEMS | Signa HDxt | SE | 2D | 4 | 766.668 | 17 | 0 | 63.86087 | 1.5 | 6 | 90 | 1.14681 | 23 | 288 | 192 | 0.4688\0.4688 | TCGA-HT-A5RC/1.3.6.1.4.1.14519.5.2.1.2531.4003.231610681115360230926274787101/1.3.6.1.4.1.14519.5.2.1.2531.4003.109664598871282097695886250609 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-A5RC | t2 | F | 70 | 74.843 | 3-pl_T2*_FGRE_S | 1 | 1.89125E+15 | 8/31/1999 | GE MEDICAL SYSTEMS | Signa HDxt | GR | 2D | 5 | 5.512 | 1.692 | 0 | 63.86085 | 1.5 | 10 | 30 | 0.231173 | 15 | 256 | 128 | 1.0156\1.0156 | TCGA-HT-A5RC/1.3.6.1.4.1.14519.5.2.1.2531.4003.231610681115360230926274787101/1.3.6.1.4.1.14519.5.2.1.2531.4003.121974323729426673427319472220 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-A614 | flair | M | 47 | 120.202 | Ax_T2_Flair | 5 | 6.74674E+15 | 12/24/1999 | GE MEDICAL SYSTEMS | Signa HDxt | SE\IR | 2D | 5 | 9002 | 145.008 | 2250 | 127.7263 | 3 | 5 | 90 | 0.5385 | 28 | 512 | 192 | 0.4688\0.4688 | TCGA-HT-A614/1.3.6.1.4.1.14519.5.2.1.2531.4003.176367534254999887804695059404/1.3.6.1.4.1.14519.5.2.1.2531.4003.918114974441949509180480975934 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-A614 | t1Gd | M | 47 | 120.202 | =+C_Ax_T1_MP_SPGR | 11 | 6.74674E+15 | 12/24/1999 | GE MEDICAL SYSTEMS | Signa HDxt | GR | 3D | 2 | 6.792 | 2.772 | 0 | 127.7263 | 3 | 1 | 13 | 0.488775 | 212 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-A614/1.3.6.1.4.1.14519.5.2.1.2531.4003.176367534254999887804695059404/1.3.6.1.4.1.14519.5.2.1.2531.4003.657538458835042249023564628808 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-A614 | t1 | M | 47 | 120.202 | Ax_T1_MP_SPGR | 9 | 6.74674E+15 | 12/24/1999 | GE MEDICAL SYSTEMS | Signa HDxt | GR | 3D | 2 | 6.792 | 2.772 | 0 | 127.7263 | 3 | 1 | 13 | 0.488775 | 212 | 320 | 192 | 0.5078\0.5078 | TCGA-HT-A614/1.3.6.1.4.1.14519.5.2.1.2531.4003.176367534254999887804695059404/1.3.6.1.4.1.14519.5.2.1.2531.4003.925945533711458317260480099964 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-A614 | t2 | M | 47 | 120.202 | Sag_T2_FSE | 4 | 6.74674E+15 | 12/24/1999 | GE MEDICAL SYSTEMS | Signa HDxt | SE | 2D | 4 | 3583.34 | 80.208 | 0 | 127.7263 | 3 | 6 | 90 | 1.0185 | 24 | 512 | 224 | 0.4688\0.4688 | TCGA-HT-A614/1.3.6.1.4.1.14519.5.2.1.2531.4003.176367534254999887804695059404/1.3.6.1.4.1.14519.5.2.1.2531.4003.556634184158803243157182204153 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-A61A | flair | F | 20 | 73.936 | (OPT)_FLAIR_AX | 14 | 2.14144E+15 | 1/27/2000 | GE MEDICAL SYSTEMS | SIGNA HDx | SE\IR | 2D | 2 | 9002 | 127.7 | 2200 | 127.7243 | 3 | 2 | 90 | 1.2709 | 88 | 256 | 224 | 1.0156\1.0156 | TCGA-HT-A61A/1.3.6.1.4.1.14519.5.2.1.2531.4003.176077465885263584703190609361/1.3.6.1.4.1.14519.5.2.1.2531.4003.548888566112287264452476674687 | |
| TCGA-LGG | TCGA-HT | TCGA-HT-A61A | t1Gd | F | 20 | 73.936 | =+C_3D_AXIAL IRSPGR Fast | 18 | 2.14144E+15 | 1/27/2000 | GE MEDICAL SYSTEMS | SIGNA HDx | GR | 3D | 1.2 | 6.392 | 2.524 | 800 | 127.7243 | 3 | 1.2 | 148 | 224 | 192 | 1.0156\1.0156 | TCGA-HT-A61A/1.3.6.1.4.1.14519.5.2.1.2531.4003.176077465885263584703190609361/1.3.6.1.4.1.14519.5.2.1.2531.4003.102441147751659163225639430837 | |||
| TCGA-LGG | TCGA-HT | TCGA-HT-A61A | t1 | F | 20 | 73.936 | 3D_AXIAL IRSPGR Fast | 16 | 2.14144E+15 | 1/27/2000 | GE MEDICAL SYSTEMS | SIGNA HDx | GR | 3D | 1.2 | 6.392 | 2.524 | 800 | 127.7243 | 3 | 1.2 | 148 | 224 | 192 | 1.0156\1.0156 | TCGA-HT-A61A/1.3.6.1.4.1.14519.5.2.1.2531.4003.176077465885263584703190609361/1.3.6.1.4.1.14519.5.2.1.2531.4003.325665435231731333959547617273 | |||
| TCGA-LGG | TCGA-HT | TCGA-HT-A61A | t2 | F | 20 | 73.936 | (OPTIONAL)Ax_T2_FSE_INTER | 15 | 2.14144E+15 | 1/27/2000 | GE MEDICAL SYSTEMS | SIGNA HDx | SE | 2D | 2 | 4500 | 81.072 | 0 | 127.7244 | 3 | 2 | 90 | 1.3554 | 83 | 256 | 192 | 1.0156\1.0156 | TCGA-HT-A61A/1.3.6.1.4.1.14519.5.2.1.2531.4003.176077465885263584703190609361/1.3.6.1.4.1.14519.5.2.1.2531.4003.225099482324673279939207452331 |
These radiomic features are provided on an ‘as-is’ basis, and are distinct from the panel of features used in GLISTRboost. The biological significance of these individual radiomic features remains unknown, but we include them here to facilitate research on their association with molecular markers, clinical outcomes, treatment responses, and other endpoints, by researchers without sufficient computational background to extract such features. Although researchers can derive their own radiomic features from our segmentation labels, and the corresponding images we included a collection of features that have been shown in various studies to relate to clinical outcome31 and underlying tumor molecular characteristics30,32. Note that the radiomic features we provide are extracted from the denoised images, and the users might also want to consider extracting features from the unsmoothed images provided in [Data Citation 3 and Data Citation 4].
Code availability
All software tools used for pre-processing, initialization, and generation of the hereby described segmentation labels are based on publicly available tools. Specifically, the tools used for the pre-processing steps of skull-stripping (BET)67,68 and co-registration (FLIRT)62,63 are publicly available from the FMRIB Software Library (FSL)64–66, in: fsl.fmrib.ox.ac.uk. The software used for the further skull-stripping approaches, i.e., Multi-Atlas Skull-Stripping (MASS)69 and MUSE70, are publicly available in www.med.upenn.edu/sbia/mass.html and www.med.upenn.edu/sbia/muse.html, respectively.
We developed CaPTk83 as a toolkit to facilitate translation of complex research algorithms into clinical practice, by enabling operators to conduct quantitative analyses without requiring substantial computational background. Towards this end CaPTk is a dynamically growing software platform, with various integrated applications, allowing 1) interactive definition of coordinates and regions, 2) generic image analysis (e.g., registration, feature extraction), and 3) specialized analysis algorithms (e.g., identification of genetic mutation imaging markers12). Specifically for this study, CaPTk was used to 1) manually initialize seed-points required for the initialization of GLISTRboost36,38, 2) apply the de-noising approach (SUSAN)71 used for smoothing images before their input to GLISTRboost, as well as 3) to extract the radiomic features released in TCIA [Data Citation 3 and Data Citation 4]. The exact version used for initializing the required seed-points in this study was released on the 14th of October 2016 and the code source, as well as executable installers, are available in: www.med.upenn.edu/sbia/captk.html.
Finally, our segmentation approach, GLISTRboost36,38, has been made available for public use through the Online Image Processing Portal (IPP—ipp.cbica.upenn.edu) of the CBICA. CBICA's IPP allows users to perform their data analysis using integrated algorithms, without any software installation, whilst also using CBICA's High Performance Computing resources. It should be noted that we used the Python package scikit-learn104 for the implementation of the gradient boosting algorithm.
Data Records
We selected only the pre-operative multimodal scans of the TCGA-GBM [Data Citation 1] and TCGA-LGG [Data Citation 2] glioma collections, from the publicly available TCIA repository. The generated data, which is made publicly available through TCIA’s Analysis Results Directory (wiki.cancerimagingarchive.net/x/sgH1) [Data Citation 3 and Data Citation 4], comprise pre-operative baseline re-oriented, co-registered and skull-stripped mMRI scans together with their corresponding computer-aided and manually-revised segmentation labels in NIfTI57 format. We have further enriched the file containers to include an extensive panel of radiomic features, which we hope may facilitate radiogenomic research using the TCGA portal, as well as comparison of segmentation methods, even among those scientists without image analysis resources.
A subset of the pre-operative scans included in the generated data [Data Citation 3 and Data Citation 4] was also part of the BraTS’15 dataset (Table 4 (available online only)), which were skull-stripped, co-registered to the same anatomical template and resampled to 1 mm3 voxel resolution by the challenge organizers. For this subset, we provide the identical MRI volumes as provided by the BraTS’15 challenge, allowing other researchers to compare their segmentation labels to the leaderboard of the BraTS’15 challenge. Furthermore, the manually-revised segmentation labels provided in [Data Citation 3 and Data Citation 4] are included in the datasets of the BraTS’17 challenge, for benchmarking computational segmentation algorithms against tumor delineation validated by expert neuroradiologists, allowing for repeatable research.
Table 4. List of TCG-GBM and TCGA-LGG identifiers included in BraTS’15 and this study.
| Feature Label | Type of Feature | Description |
|---|---|---|
| ID | Subject identifier | The TCGA subject's identifier. |
| Date | Date of scan | The date of the scan considered. |
| VOLUME_ET | Volumetric | The volume of the enhancing (ET) part of the tumor core (TC). |
| VOLUME_NET | Volumetric | The volume of the non-enhancing (NET) parts of the tumor core (TC). |
| VOLUME_ED | Volumetric | The volume of the peritumoral edematous region (ED). |
| VOLUME_TC | Volumetric | The volume of the tumor core (TC), comprising the ET and NET. |
| VOLUME_WT | Volumetric | The volume of the whole tumor (WT), comprising ET, NET, and ED. |
| VOLUME_BRAIN | Volumetric | The volume of the complete brain, excluding skull structures. |
| VOLUME_ET_OVER_NET | Volumetric | The volume of the ET relative to the volume of the NET. |
| VOLUME_ET_OVER_ED | Volumetric | The volume of the ET relative to the volume of the ED. |
| VOLUME_NET_OVER_ED | Volumetric | The volume of the NET relative to the volume of the ED. |
| VOLUME_ET_over_TC | Volumetric | The volume of the ET relative to the volume of the TC. |
| VOLUME_NET_over_TC | Volumetric | The volume of the NET relative to the volume of the TC. |
| VOLUME_ED_over_TC | Volumetric | The volume of the ED relative to the volume of the TC. |
| VOLUME_ET_OVER_WT | Volumetric | The volume of the ET relative to the volume of the WT. |
| VOLUME_NET_OVER_WT | Volumetric | The volume of the NET relative to the volume of the WT. |
| VOLUME_ED_OVER_WT | Volumetric | The volume of the ED relative to the volume of the WT. |
| VOLUME_TC_OVER_WT | Volumetric | The volume of the TC relative to the volume of the WT. |
| VOLUME_ET_OVER_BRAIN | Volumetric | The volume of the ET relative to the volume of the complete brain. |
| VOLUME_NET_OVER_BRAIN | Volumetric | The volume of the NET relative to the volume of the complete brain. |
| VOLUME_ED_over_BRAIN | Volumetric | The volume of the ED relative to the volume of the complete brain. |
| VOLUME_TC_over_BRAIN | Volumetric | The volume of the TC relative to the volume of the complete brain. |
| VOLUME_WT_OVER_BRAIN | Volumetric | The volume of the WT relative to the volume of the complete brain. |
| DIST_Vent_TC | Spatial | The closest distance of the TC from the ventricles. |
| DIST_Vent_ED | Spatial | The closest distance of the ED from the ventricles. |
| INTENSITY_Mean_ET_T1Gd | Intensity | The raw average intensity of the ET in the post-contrast T1 (T1-Gd) volume. |
| INTENSITY_STD_ET_T1Gd | Intensity | The standard deviation of the raw intensities of the ET in the T1-Gd volume. |
| INTENSITY_Mean_ET_T1 | Intensity | The raw average intensity of the ET in the pre-contrast T1 (T1) volume. |
| INTENSITY_STD_ET_T1 | Intensity | The standard deviation of the raw intensities of the ET in the T1 volume. |
| INTENSITY_Mean_ET_T2 | Intensity | The raw average intensity of the ET in the T2-weighted (T2) volume. |
| INTENSITY_STD_ET_T2 | Intensity | The standard deviation of the raw intensities of the ET in the T2 volume. |
| INTENSITY_Mean_ET_FLAIR | Intensity | The raw average intensity of the ET in the T2-weighted Fluid Attenuated Inversion Recovery (FLAIR) volume. |
| INTENSITY_STD_ET_FLAIR | Intensity | The standard deviation of the raw intensities of the ET in the FLAIR volume. |
| INTENSITY_Mean_NET_T1Gd | Intensity | The raw average intensity of the NET in the T1-Gd volume. |
| INTENSITY_STD_NET_T1Gd | Intensity | The standard deviation of the raw intensities of the NET in the T1-Gd volume. |
| INTENSITY_Mean_NET_T1 | Intensity | The raw average intensity of the NET in the T1 volume. |
| INTENSITY_STD_NET_T1 | Intensity | The standard deviation of the raw intensities of the NET in the T1 volume. |
| INTENSITY_Mean_NET_T2 | Intensity | The raw average intensity of the NET in the T2 volume. |
| INTENSITY_STD_NET_T2 | Intensity | The standard deviation of the raw intensities of the NET in the T2 volume. |
| INTENSITY_Mean_NET_FLAIR | Intensity | The raw average intensity of the NET in the FLAIR volume. |
| INTENSITY_STD_NET_FLAIR | Intensity | The standard deviation of the raw intensities of the NET in the FLAIR volume. |
| INTENSITY_Mean_ED_T1Gd | Intensity | The raw average intensity of the ED in the T1-Gd volume. |
| INTENSITY_STD_ED_T1Gd | Intensity | The standard deviation of the raw intensities of the ED in the T1-Gd volume. |
| INTENSITY_Mean_ED_T1 | Intensity | The raw average intensity of the ED in the T1 volume. |
| INTENSITY_STD_ED_T1 | Intensity | The standard deviation of the raw intensities of the ED in the T1 volume. |
| INTENSITY_Mean_ED_T2 | Intensity | The raw average intensity of the ED in the T2 volume. |
| INTENSITY_STD_ED_T2 | Intensity | The standard deviation of the raw intensities of the ED in the T2 volume. |
| INTENSITY_Mean_ED_FLAIR | Intensity | The raw average intensity of the ED in the FLAIR volume. |
| INTENSITY_STD_ED_FLAIR | Intensity | The standard deviation of the raw intensities of the ED in the FLAIR volume. |
| HISTO_ET_T1Gd_Bin1 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T1-Gd volume (Bin 1). |
| HISTO_ET_T1Gd_Bin2 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T1-Gd volume (Bin 2). |
| HISTO_ET_T1Gd_Bin3 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T1-Gd volume (Bin 3). |
| HISTO_ET_T1Gd_Bin4 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T1-Gd volume (Bin 4). |
| HISTO_ET_T1Gd_Bin5 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T1-Gd volume (Bin 5). |
| HISTO_ET_T1Gd_Bin6 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T1-Gd volume (Bin 6). |
| HISTO_ET_T1Gd_Bin7 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T1-Gd volume (Bin 7). |
| HISTO_ET_T1Gd_Bin8 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T1-Gd volume (Bin 8). |
| HISTO_ET_T1Gd_Bin9 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T1-Gd volume (Bin 9). |
| HISTO_ET_T1Gd_Bin10 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T1-Gd volume (Bin 10). |
| HISTO_ED_T1Gd_Bin1 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T1-Gd volume (Bin 1). |
| HISTO_ED_T1Gd_Bin2 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T1-Gd volume (Bin 2). |
| HISTO_ED_T1Gd_Bin3 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T1-Gd volume (Bin 3). |
| HISTO_ED_T1Gd_Bin4 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T1-Gd volume (Bin 4). |
| HISTO_ED_T1Gd_Bin5 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T1-Gd volume (Bin 5). |
| HISTO_ED_T1Gd_Bin6 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T1-Gd volume (Bin 6). |
| HISTO_ED_T1Gd_Bin7 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T1-Gd volume (Bin 7). |
| HISTO_ED_T1Gd_Bin8 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T1-Gd volume (Bin 8). |
| HISTO_ED_T1Gd_Bin9 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T1-Gd volume (Bin 9). |
| HISTO_ED_T1Gd_Bin10 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T1-Gd volume (Bin 10). |
| HISTO_NET_T1Gd_Bin1 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T1-Gd volume (Bin 1). |
| HISTO_NET_T1Gd_Bin2 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T1-Gd volume (Bin 2). |
| HISTO_NET_T1Gd_Bin3 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T1-Gd volume (Bin 3). |
| HISTO_NET_T1Gd_Bin4 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T1-Gd volume (Bin 4). |
| HISTO_NET_T1Gd_Bin5 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T1-Gd volume (Bin 5). |
| HISTO_NET_T1Gd_Bin6 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T1-Gd volume (Bin 6). |
| HISTO_NET_T1Gd_Bin7 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T1-Gd volume (Bin 7). |
| HISTO_NET_T1Gd_Bin8 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T1-Gd volume (Bin 8). |
| HISTO_NET_T1Gd_Bin9 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T1-Gd volume (Bin 9). |
| HISTO_NET_T1Gd_Bin10 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T1-Gd volume (Bin 10). |
| HISTO_ET_T1_Bin1 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T1 volume (Bin 1). |
| HISTO_ET_T1_Bin2 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T1 volume (Bin 2). |
| HISTO_ET_T1_Bin3 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T1 volume (Bin 3). |
| HISTO_ET_T1_Bin4 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T1 volume (Bin 4). |
| HISTO_ET_T1_Bin5 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T1 volume (Bin 5). |
| HISTO_ET_T1_Bin6 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T1 volume (Bin 6). |
| HISTO_ET_T1_Bin7 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T1 volume (Bin 7). |
| HISTO_ET_T1_Bin8 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T1 volume (Bin 8). |
| HISTO_ET_T1_Bin9 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T1 volume (Bin 9). |
| HISTO_ET_T1_Bin10 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T1 volume (Bin 10). |
| HISTO_ED_T1_Bin1 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T1 volume (Bin 1). |
| HISTO_ED_T1_Bin2 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T1 volume (Bin 2). |
| HISTO_ED_T1_Bin3 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T1 volume (Bin 3). |
| HISTO_ED_T1_Bin4 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T1 volume (Bin 4). |
| HISTO_ED_T1_Bin5 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T1 volume (Bin 5). |
| HISTO_ED_T1_Bin6 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T1 volume (Bin 6). |
| HISTO_ED_T1_Bin7 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T1 volume (Bin 7). |
| HISTO_ED_T1_Bin8 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T1 volume (Bin 8). |
| HISTO_ED_T1_Bin9 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T1 volume (Bin 9). |
| HISTO_ED_T1_Bin10 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T1 volume (Bin 10). |
| HISTO_NET_T1_Bin1 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T1 volume (Bin 1). |
| HISTO_NET_T1_Bin2 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T1 volume (Bin 2). |
| HISTO_NET_T1_Bin3 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T1 volume (Bin 3). |
| HISTO_NET_T1_Bin4 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T1 volume (Bin 4). |
| HISTO_NET_T1_Bin5 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T1 volume (Bin 5). |
| HISTO_NET_T1_Bin6 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T1 volume (Bin 6). |
| HISTO_NET_T1_Bin7 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T1 volume (Bin 7). |
| HISTO_NET_T1_Bin8 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T1 volume (Bin 8). |
| HISTO_NET_T1_Bin9 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T1 volume (Bin 9). |
| HISTO_NET_T1_Bin10 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T1 volume (Bin 10). |
| HISTO_ET_T2_Bin1 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T2 volume (Bin 1). |
| HISTO_ET_T2_Bin2 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T2 volume (Bin 2). |
| HISTO_ET_T2_Bin3 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T2 volume (Bin 3). |
| HISTO_ET_T2_Bin4 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T2 volume (Bin 4). |
| HISTO_ET_T2_Bin5 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T2 volume (Bin 5). |
| HISTO_ET_T2_Bin6 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T2 volume (Bin 6). |
| HISTO_ET_T2_Bin7 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T2 volume (Bin 7). |
| HISTO_ET_T2_Bin8 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T2 volume (Bin 8). |
| HISTO_ET_T2_Bin9 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T2 volume (Bin 9). |
| HISTO_ET_T2_Bin10 | Histogram-based | Binned histogram of the raw intensity values of the ET in the T2 volume (Bin 10). |
| HISTO_ED_T2_Bin1 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T2 volume (Bin 1). |
| HISTO_ED_T2_Bin2 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T2 volume (Bin 2). |
| HISTO_ED_T2_Bin3 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T2 volume (Bin 3). |
| HISTO_ED_T2_Bin4 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T2 volume (Bin 4). |
| HISTO_ED_T2_Bin5 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T2 volume (Bin 5). |
| HISTO_ED_T2_Bin6 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T2 volume (Bin 6). |
| HISTO_ED_T2_Bin7 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T2 volume (Bin 7). |
| HISTO_ED_T2_Bin8 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T2 volume (Bin 8). |
| HISTO_ED_T2_Bin9 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T2 volume (Bin 9). |
| HISTO_ED_T2_Bin10 | Histogram-based | Binned histogram of the raw intensity values of the ED in the T2 volume (Bin 10). |
| HISTO_NET_T2_Bin1 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T2 volume (Bin 1). |
| HISTO_NET_T2_Bin2 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T2 volume (Bin 2). |
| HISTO_NET_T2_Bin3 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T2 volume (Bin 3). |
| HISTO_NET_T2_Bin4 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T2 volume (Bin 4). |
| HISTO_NET_T2_Bin5 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T2 volume (Bin 5). |
| HISTO_NET_T2_Bin6 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T2 volume (Bin 6). |
| HISTO_NET_T2_Bin7 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T2 volume (Bin 7). |
| HISTO_NET_T2_Bin8 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T2 volume (Bin 8). |
| HISTO_NET_T2_Bin9 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T2 volume (Bin 9). |
| HISTO_NET_T2_Bin10 | Histogram-based | Binned histogram of the raw intensity values of the NET in the T2 volume (Bin 10). |
| HISTO_ET_FLAIR_Bin1 | Histogram-based | Binned histogram of the raw intensity values of the ET in the FLAIR volume (Bin 1). |
| HISTO_ET_FLAIR_Bin2 | Histogram-based | Binned histogram of the raw intensity values of the ET in the FLAIR volume (Bin 2). |
| HISTO_ET_FLAIR_Bin3 | Histogram-based | Binned histogram of the raw intensity values of the ET in the FLAIR volume (Bin 3). |
| HISTO_ET_FLAIR_Bin4 | Histogram-based | Binned histogram of the raw intensity values of the ET in the FLAIR volume (Bin 4). |
| HISTO_ET_FLAIR_Bin5 | Histogram-based | Binned histogram of the raw intensity values of the ET in the FLAIR volume (Bin 5). |
| HISTO_ET_FLAIR_Bin6 | Histogram-based | Binned histogram of the raw intensity values of the ET in the FLAIR volume (Bin 6). |
| HISTO_ET_FLAIR_Bin7 | Histogram-based | Binned histogram of the raw intensity values of the ET in the FLAIR volume (Bin 7). |
| HISTO_ET_FLAIR_Bin8 | Histogram-based | Binned histogram of the raw intensity values of the ET in the FLAIR volume (Bin 8). |
| HISTO_ET_FLAIR_Bin9 | Histogram-based | Binned histogram of the raw intensity values of the ET in the FLAIR volume (Bin 9). |
| HISTO_ET_FLAIR_Bin10 | Histogram-based | Binned histogram of the raw intensity values of the ET in the FLAIR volume (Bin 10). |
| HISTO_ED_FLAIR_Bin1 | Histogram-based | Binned histogram of the raw intensity values of the ED in the FLAIR volume (Bin 1). |
| HISTO_ED_FLAIR_Bin2 | Histogram-based | Binned histogram of the raw intensity values of the ED in the FLAIR volume (Bin 2). |
| HISTO_ED_FLAIR_Bin3 | Histogram-based | Binned histogram of the raw intensity values of the ED in the FLAIR volume (Bin 3). |
| HISTO_ED_FLAIR_Bin4 | Histogram-based | Binned histogram of the raw intensity values of the ED in the FLAIR volume (Bin 4). |
| HISTO_ED_FLAIR_Bin5 | Histogram-based | Binned histogram of the raw intensity values of the ED in the FLAIR volume (Bin 5). |
| HISTO_ED_FLAIR_Bin6 | Histogram-based | Binned histogram of the raw intensity values of the ED in the FLAIR volume (Bin 6). |
| HISTO_ED_FLAIR_Bin7 | Histogram-based | Binned histogram of the raw intensity values of the ED in the FLAIR volume (Bin 7). |
| HISTO_ED_FLAIR_Bin8 | Histogram-based | Binned histogram of the raw intensity values of the ED in the FLAIR volume (Bin 8). |
| HISTO_ED_FLAIR_Bin9 | Histogram-based | Binned histogram of the raw intensity values of the ED in the FLAIR volume (Bin 9). |
| HISTO_ED_FLAIR_Bin10 | Histogram-based | Binned histogram of the raw intensity values of the ED in the FLAIR volume (Bin 10). |
| HISTO_NET_FLAIR_Bin1 | Histogram-based | Binned histogram of the raw intensity values of the NET in the FLAIR volume (Bin 1). |
| HISTO_NET_FLAIR_Bin2 | Histogram-based | Binned histogram of the raw intensity values of the NET in the FLAIR volume (Bin 2). |
| HISTO_NET_FLAIR_Bin3 | Histogram-based | Binned histogram of the raw intensity values of the NET in the FLAIR volume (Bin 3). |
| HISTO_NET_FLAIR_Bin4 | Histogram-based | Binned histogram of the raw intensity values of the NET in the FLAIR volume (Bin 4). |
| HISTO_NET_FLAIR_Bin5 | Histogram-based | Binned histogram of the raw intensity values of the NET in the FLAIR volume (Bin 5). |
| HISTO_NET_FLAIR_Bin6 | Histogram-based | Binned histogram of the raw intensity values of the NET in the FLAIR volume (Bin 6). |
| HISTO_NET_FLAIR_Bin7 | Histogram-based | Binned histogram of the raw intensity values of the NET in the FLAIR volume (Bin 7). |
| HISTO_NET_FLAIR_Bin8 | Histogram-based | Binned histogram of the raw intensity values of the NET in the FLAIR volume (Bin 8). |
| HISTO_NET_FLAIR_Bin9 | Histogram-based | Binned histogram of the raw intensity values of the NET in the FLAIR volume (Bin 9). |
| HISTO_NET_FLAIR_Bin10 | Histogram-based | Binned histogram of the raw intensity values of the NET in the FLAIR volume (Bin 10). |
| SPATIAL_Frontal | Spatial | Percentage of TC volume in the frontal lobe. |
| SPATIAL_Temporal | Spatial | Percentage of TC volume in the temporal lobe. |
| SPATIAL_Parietal | Spatial | Percentage of TC volume in the parietal lobe. |
| SPATIAL_Basal_G | Spatial | Percentage of TC volume in the regions of putamen, caudate nucleus, globus pallidus, subthalamic nucleus, nucleus accumbens, internal capsule, and thalamus. |
| SPATIAL_Insula | Spatial | Percentage of TC volume in the region of the insula. |
| SPATIAL_CC_Fornix | Spatial | Percentage of TC volume in the regions of corpus callosum, fornix, and cingulate. |
| SPATIAL_Occipital | Spatial | Percentage of TC volume in the occipital lobe. |
| SPATIAL_Cere | Spatial | Percentage of TC volume in the region of the cerebellum. |
| SPATIAL_Brain_stem | Spatial | Percentage of TC volume in the region of brain stem. |
| ECCENTRICITY_ET | Volumetric | Eccentricity metric for ET: sqrt[1 - a*b/c^2], where c is the longest semi-principal axes of an ellipsoid fitted on the ET, and a and b are the 2nd and 3rd longest semi-principal axes of the ellipsoid. |
| ECCENTRICITY_ED | Volumetric | Eccentricity metric for ED: sqrt[1 - a*b/c^2], where c is the longest semi-principal axes of an ellipsoid fitted on the ED, and a and b are the 2nd and 3rd longest semi-principal axes of the ellipsoid. |
| ECCENTRICITY_NET | Volumetric | Eccentricity metric for NET: sqrt[1 - a*b/c^2], where c is the longest semi-principal axes of an ellipsoid fitted on the NET, and a and b are the 2nd and 3rd longest semi-principal axes of the ellipsoid. |
| SOLIDITY_ET | Volumetric | Ratio of the number of voxels in the ET to the number of voxels in the 3D convex hull of the ET. |
| SOLIDITY_ED | Volumetric | Ratio of the number of voxels in the ED to the number of voxels in the 3D convex hull of the ED. |
| SOLIDITY_NET | Volumetric | Ratio of the number of voxels in the NET to the number of voxels in the 3D convex hull of the NET. |
| TEXTURE_GLOBAL_ET_T1Gd_Variance | Textural (Global) | Variance of the histogram of the ET in the T1-Gd volume. |
| TEXTURE_GLOBAL_ET_T1Gd_Skewness | Textural (Global) | Skewness of the histogram of the ET in the T1-Gd volume. |
| TEXTURE_GLOBAL_ET_T1Gd_Kurtosis | Textural (Global) | Kurtosis of the histogram of the ET in the T1-Gd volume. |
| TEXTURE_GLOBAL_ET_T1_Variance | Textural (Global) | Variance of the histogram of the ET in the T1 volume. |
| TEXTURE_GLOBAL_ET_T1_Skewness | Textural (Global) | Skewness of the histogram of the ET in the T1 volume. |
| TEXTURE_GLOBAL_ET_T1_Kurtosis | Textural (Global) | Kurtosis of the histogram of the ET in the T1 volume. |
| TEXTURE_GLOBAL_ET_T2_Variance | Textural (Global) | Variance of the histogram of the ET in the T2 volume. |
| TEXTURE_GLOBAL_ET_T2_Skewness | Textural (Global) | Skewness of the histogram of the ET in the T2 volume. |
| TEXTURE_GLOBAL_ET_T2_Kurtosis | Textural (Global) | Kurtosis of the histogram of the ET in the T2 volume. |
| TEXTURE_GLOBAL_ET_FLAIR_Variance | Textural (Global) | Variance of the histogram of the ET in the FLAIR volume. |
| TEXTURE_GLOBAL_ET_FLAIR_Skewness | Textural (Global) | Skewness of the histogram of the ET in the FLAIR volume. |
| TEXTURE_GLOBAL_ET_FLAIR_Kurtosis | Textural (Global) | Kurtosis of the histogram of the ET in the FLAIR volume. |
| TEXTURE_GLOBAL_ED_T1Gd_Variance | Textural (Global) | Variance of the histogram of the ED in the T1-Gd volume. |
| TEXTURE_GLOBAL_ED_T1Gd_Skewness | Textural (Global) | Skewness of the histogram of the ED in the T1-Gd volume. |
| TEXTURE_GLOBAL_ED_T1Gd_Kurtosis | Textural (Global) | Kurtosis of the histogram of the ED in the T1-Gd volume. |
| TEXTURE_GLOBAL_ED_T1_Variance | Textural (Global) | Variance of the histogram of the ED in the T1 volume. |
| TEXTURE_GLOBAL_ED_T1_Skewness | Textural (Global) | Skewness of the histogram of the ED in the T1 volume. |
| TEXTURE_GLOBAL_ED_T1_Kurtosis | Textural (Global) | Kurtosis of the histogram of the ED in the T1 volume. |
| TEXTURE_GLOBAL_ED_T2_Variance | Textural (Global) | Variance of the histogram of the ED in the T2 volume. |
| TEXTURE_GLOBAL_ED_T2_Skewness | Textural (Global) | Skewness of the histogram of the ED in the T2 volume. |
| TEXTURE_GLOBAL_ED_T2_Kurtosis | Textural (Global) | Kurtosis of the histogram of the ED in the T2 volume. |
| TEXTURE_GLOBAL_ED_FLAIR_Variance | Textural (Global) | Variance of the histogram of the ED in the FLAIR volume. |
| TEXTURE_GLOBAL_ED_FLAIR_Skewness | Textural (Global) | Skewness of the histogram of the ED in the FLAIR volume. |
| TEXTURE_GLOBAL_ED_FLAIR_Kurtosis | Textural (Global) | Kurtosis of the histogram of the ED in the FLAIR volume. |
| TEXTURE_GLOBAL_NET_T1Gd_Variance | Textural (Global) | Variance of the histogram of the NET in the T1-Gd volume. |
| TEXTURE_GLOBAL_NET_T1Gd_Skewness | Textural (Global) | Skewness of the histogram of the NET in the T1-Gd volume. |
| TEXTURE_GLOBAL_NET_T1Gd_Kurtosis | Textural (Global) | Kurtosis of the histogram of the NET in the T1-Gd volume. |
| TEXTURE_GLOBAL_NET_T1_Variance | Textural (Global) | Variance of the histogram of the NET in the T1 volume. |
| TEXTURE_GLOBAL_NET_T1_Skewness | Textural (Global) | Skewness of the histogram of the NET in the T1 volume. |
| TEXTURE_GLOBAL_NET_T1_Kurtosis | Textural (Global) | Kurtosis of the histogram of the NET in the T1 volume. |
| TEXTURE_GLOBAL_NET_T2_Variance | Textural (Global) | Variance of the histogram of the NET in the T2 volume. |
| TEXTURE_GLOBAL_NET_T2_Skewness | Textural (Global) | Skewness of the histogram of the NET in the T2 volume. |
| TEXTURE_GLOBAL_NET_T2_Kurtosis | Textural (Global) | Kurtosis of the histogram of the NET in the T2 volume. |
| TEXTURE_GLOBAL_NET_FLAIR_Variance | Textural (Global) | Variance of the histogram of the NET in the FLAIR volume. |
| TEXTURE_GLOBAL_NET_FLAIR_Skewness | Textural (Global) | Skewness of the histogram of the NET in the FLAIR volume. |
| TEXTURE_GLOBAL_NET_FLAIR_Kurtosis | Textural (Global) | Kurtosis of the histogram of the NET in the FLAIR volume. |
| TEXTURE_GLCM_ET_T1Gd_Energy | Textural (Grey-Level Co-occurrence Matrix) | The Energy within the ET in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T1Gd_Contrast | Textural (Grey-Level Co-occurrence Matrix) | The Contrast within the ET in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T1Gd_Entropy | Textural (Grey-Level Co-occurrence Matrix) | The Entropy within the ET in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T1Gd_Homogeneity | Textural (Grey-Level Co-occurrence Matrix) | The Homogeneity within the ET in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T1Gd_Correlation | Textural (Grey-Level Co-occurrence Matrix) | The Correlation within the ET in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T1Gd_SumAverage | Textural (Grey-Level Co-occurrence Matrix) | The SumAverage within the ET in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T1Gd_Variance | Textural (Grey-Level Co-occurrence Matrix) | The Variance within the ET in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T1Gd_Dissimilarity | Textural (Grey-Level Co-occurrence Matrix) | The Dissimilarity within the ET in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T1Gd_AutoCorrelation | Textural (Grey-Level Co-occurrence Matrix) | The Autocorrelation within the ET in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T1_Energy | Textural (Grey-Level Co-occurrence Matrix) | The Energy within the ET in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T1_Contrast | Textural (Grey-Level Co-occurrence Matrix) | The Contrast within the ET in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T1_Entropy | Textural (Grey-Level Co-occurrence Matrix) | The Entropy within the ET in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T1_Homogeneity | Textural (Grey-Level Co-occurrence Matrix) | The Homogeneity within the ET in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T1_Correlation | Textural (Grey-Level Co-occurrence Matrix) | The Correlation within the ET in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T1_SumAverage | Textural (Grey-Level Co-occurrence Matrix) | The SumAverage within the ET in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T1_Variance | Textural (Grey-Level Co-occurrence Matrix) | The Variance within the ET in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T1_Dissimilarity | Textural (Grey-Level Co-occurrence Matrix) | The Dissimilarity within the ET in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T1_AutoCorrelation | Textural (Grey-Level Co-occurrence Matrix) | The Autocorrelation within the ET in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T2_Energy | Textural (Grey-Level Co-occurrence Matrix) | The Energy within the ET in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T2_Contrast | Textural (Grey-Level Co-occurrence Matrix) | The Contrast within the ET in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T2_Entropy | Textural (Grey-Level Co-occurrence Matrix) | The Entropy within the ET in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T2_Homogeneity | Textural (Grey-Level Co-occurrence Matrix) | The Homogeneity within the ET in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T2_Correlation | Textural (Grey-Level Co-occurrence Matrix) | The Correlation within the ET in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T2_SumAverage | Textural (Grey-Level Co-occurrence Matrix) | The SumAverage within the ET in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T2_Variance | Textural (Grey-Level Co-occurrence Matrix) | The Variance within the ET in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T2_Dissimilarity | Textural (Grey-Level Co-occurrence Matrix) | The Dissimilarity within the ET in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_T2_AutoCorrelation | Textural (Grey-Level Co-occurrence Matrix) | The Autocorrelation within the ET in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_FLAIR_Energy | Textural (Grey-Level Co-occurrence Matrix) | The Energy within the ET in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_FLAIR_Contrast | Textural (Grey-Level Co-occurrence Matrix) | The Contrast within the ET in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_FLAIR_Entropy | Textural (Grey-Level Co-occurrence Matrix) | The Entropy within the ET in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_FLAIR_Homogeneity | Textural (Grey-Level Co-occurrence Matrix) | The Homogeneity within the ET in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_FLAIR_Correlation | Textural (Grey-Level Co-occurrence Matrix) | The Correlation within the ET in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_FLAIR_SumAverage | Textural (Grey-Level Co-occurrence Matrix) | The SumAverage within the ET in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_FLAIR_Variance | Textural (Grey-Level Co-occurrence Matrix) | The Variance within the ET in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_FLAIR_Dissimilarity | Textural (Grey-Level Co-occurrence Matrix) | The Dissimilarity within the ET in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ET_FLAIR_AutoCorrelation | Textural (Grey-Level Co-occurrence Matrix) | The Autocorrelation within the ET in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T1Gd_Energy | Textural (Grey-Level Co-occurrence Matrix) | The Energy within the ED in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T1Gd_Contrast | Textural (Grey-Level Co-occurrence Matrix) | The Contrast within the ED in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T1Gd_Entropy | Textural (Grey-Level Co-occurrence Matrix) | The Entropy within the ED in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T1Gd_Homogeneity | Textural (Grey-Level Co-occurrence Matrix) | The Homogeneity within the ED in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T1Gd_Correlation | Textural (Grey-Level Co-occurrence Matrix) | The Correlation within the ED in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T1Gd_SumAverage | Textural (Grey-Level Co-occurrence Matrix) | The SumAverage within the ED in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T1Gd_Variance | Textural (Grey-Level Co-occurrence Matrix) | The Variance within the ED in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T1Gd_Dissimilarity | Textural (Grey-Level Co-occurrence Matrix) | The Dissimilarity within the ED in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T1Gd_AutoCorrelation | Textural (Grey-Level Co-occurrence Matrix) | The Autocorrelation within the ED in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T1_Energy | Textural (Grey-Level Co-occurrence Matrix) | The Energy within the ED in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T1_Contrast | Textural (Grey-Level Co-occurrence Matrix) | The Contrast within the ED in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T1_Entropy | Textural (Grey-Level Co-occurrence Matrix) | The Entropy within the ED in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T1_Homogeneity | Textural (Grey-Level Co-occurrence Matrix) | The Homogeneity within the ED in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T1_Correlation | Textural (Grey-Level Co-occurrence Matrix) | The Correlation within the ED in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T1_SumAverage | Textural (Grey-Level Co-occurrence Matrix) | The SumAverage within the ED in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T1_Variance | Textural (Grey-Level Co-occurrence Matrix) | The Variance within the ED in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T1_Dissimilarity | Textural (Grey-Level Co-occurrence Matrix) | The Dissimilarity within the ED in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T1_AutoCorrelation | Textural (Grey-Level Co-occurrence Matrix) | The Autocorrelation within the ED in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T2_Energy | Textural (Grey-Level Co-occurrence Matrix) | The Energy within the ED in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T2_Contrast | Textural (Grey-Level Co-occurrence Matrix) | The Contrast within the ED in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T2_Entropy | Textural (Grey-Level Co-occurrence Matrix) | The Entropy within the ED in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T2_Homogeneity | Textural (Grey-Level Co-occurrence Matrix) | The Homogeneity within the ED in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T2_Correlation | Textural (Grey-Level Co-occurrence Matrix) | The Correlation within the ED in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T2_SumAverage | Textural (Grey-Level Co-occurrence Matrix) | The SumAverage within the ED in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T2_Variance | Textural (Grey-Level Co-occurrence Matrix) | The Variance within the ED in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T2_Dissimilarity | Textural (Grey-Level Co-occurrence Matrix) | The Dissimilarity within the ED in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_T2_AutoCorrelation | Textural (Grey-Level Co-occurrence Matrix) | The Autocorrelation within the ED in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_FLAIR_Energy | Textural (Grey-Level Co-occurrence Matrix) | The Energy within the ED in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_FLAIR_Contrast | Textural (Grey-Level Co-occurrence Matrix) | The Contrast within the ED in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_FLAIR_Entropy | Textural (Grey-Level Co-occurrence Matrix) | The Entropy within the ED in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_FLAIR_Homogeneity | Textural (Grey-Level Co-occurrence Matrix) | The Homogeneity within the ED in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_FLAIR_Correlation | Textural (Grey-Level Co-occurrence Matrix) | The Correlation within the ED in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_FLAIR_SumAverage | Textural (Grey-Level Co-occurrence Matrix) | The SumAverage within the ED in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_FLAIR_Variance | Textural (Grey-Level Co-occurrence Matrix) | The Variance within the ED in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_FLAIR_Dissimilarity | Textural (Grey-Level Co-occurrence Matrix) | The Dissimilarity within the ED in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_ED_FLAIR_AutoCorrelation | Textural (Grey-Level Co-occurrence Matrix) | The Autocorrelation within the ED in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T1Gd_Energy | Textural (Grey-Level Co-occurrence Matrix) | The Energy within the NET in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T1Gd_Contrast | Textural (Grey-Level Co-occurrence Matrix) | The Contrast within the NET in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T1Gd_Entropy | Textural (Grey-Level Co-occurrence Matrix) | The Entropy within the NET in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T1Gd_Homogeneity | Textural (Grey-Level Co-occurrence Matrix) | The Homogeneity within the NET in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T1Gd_Correlation | Textural (Grey-Level Co-occurrence Matrix) | The Correlation within the NET in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T1Gd_SumAverage | Textural (Grey-Level Co-occurrence Matrix) | The SumAverage within the NET in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T1Gd_Variance | Textural (Grey-Level Co-occurrence Matrix) | The Variance within the NET in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T1Gd_Dissimilarity | Textural (Grey-Level Co-occurrence Matrix) | The Dissimilarity within the NET in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T1Gd_AutoCorrelation | Textural (Grey-Level Co-occurrence Matrix) | The Autocorrelation within the NET in the T1-Gd volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T1_Energy | Textural (Grey-Level Co-occurrence Matrix) | The Energy within the NET in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T1_Contrast | Textural (Grey-Level Co-occurrence Matrix) | The Contrast within the NET in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T1_Entropy | Textural (Grey-Level Co-occurrence Matrix) | The Entropy within the NET in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T1_Homogeneity | Textural (Grey-Level Co-occurrence Matrix) | The Homogeneity within the NET in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T1_Correlation | Textural (Grey-Level Co-occurrence Matrix) | The Correlation within the NET in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T1_SumAverage | Textural (Grey-Level Co-occurrence Matrix) | The SumAverage within the NET in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T1_Variance | Textural (Grey-Level Co-occurrence Matrix) | The Variance within the NET in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T1_Dissimilarity | Textural (Grey-Level Co-occurrence Matrix) | The Dissimilarity within the NET in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T1_AutoCorrelation | Textural (Grey-Level Co-occurrence Matrix) | The Autocorrelation within the NET in the T1 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T2_Energy | Textural (Grey-Level Co-occurrence Matrix) | The Energy within the NET in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T2_Contrast | Textural (Grey-Level Co-occurrence Matrix) | The Contrast within the NET in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T2_Entropy | Textural (Grey-Level Co-occurrence Matrix) | The Entropy within the NET in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T2_Homogeneity | Textural (Grey-Level Co-occurrence Matrix) | The Homogeneity within the NET in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T2_Correlation | Textural (Grey-Level Co-occurrence Matrix) | The Correlation within the NET in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T2_SumAverage | Textural (Grey-Level Co-occurrence Matrix) | The SumAverage within the NET in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T2_Variance | Textural (Grey-Level Co-occurrence Matrix) | The Variance within the NET in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T2_Dissimilarity | Textural (Grey-Level Co-occurrence Matrix) | The Dissimilarity within the NET in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_T2_AutoCorrelation | Textural (Grey-Level Co-occurrence Matrix) | The Autocorrelation within the NET in the T2 volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_FLAIR_Energy | Textural (Grey-Level Co-occurrence Matrix) | The Energy within the NET in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_FLAIR_Contrast | Textural (Grey-Level Co-occurrence Matrix) | The Contrast within the NET in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_FLAIR_Entropy | Textural (Grey-Level Co-occurrence Matrix) | The Entropy within the NET in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_FLAIR_Homogeneity | Textural (Grey-Level Co-occurrence Matrix) | The Homogeneity within the NET in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_FLAIR_Correlation | Textural (Grey-Level Co-occurrence Matrix) | The Correlation within the NET in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_FLAIR_SumAverage | Textural (Grey-Level Co-occurrence Matrix) | The SumAverage within the NET in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_FLAIR_Variance | Textural (Grey-Level Co-occurrence Matrix) | The Variance within the NET in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_FLAIR_Dissimilarity | Textural (Grey-Level Co-occurrence Matrix) | The Dissimilarity within the NET in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLCM_NET_FLAIR_AutoCorrelation | Textural (Grey-Level Co-occurrence Matrix) | The Autocorrelation within the NET in the FLAIR volume, considering 26-connected neighboring voxels in the 3D volume. |
| TEXTURE_GLRLM_ET_T1Gd_SRE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Emphasis" within the ET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1Gd_LRE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Emphasis" within the ET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1Gd_GLN | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Nonuniformity" within the ET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1Gd_RLN | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Nonuniformity" within the ET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1Gd_RP | Textural (Gray-Level Run-Length Matrix) | The "Run Percentage" within the ET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1Gd_LGRE | Textural (Gray-Level Run-Length Matrix) | The "Low Gray-Level Run Emphasis" within the ET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1Gd_HGRE | Textural (Gray-Level Run-Length Matrix) | The "High Gray-Level Run Emphasis" within the ET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1Gd_SRLGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Low Gray-Level Emphasis" within the ET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1Gd_SRHGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run High Gray-Level Emphasis" within the ET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1Gd_LRLGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Low Gray-Level Emphasis" within the ET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1Gd_LRHGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run High Gray-Level Emphasis" within the ET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1Gd_GLV | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Variance" within the ET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1Gd_RLV | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Variance" within the ET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1_SRE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Emphasis" within the ET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1_LRE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Emphasis" within the ET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1_GLN | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Nonuniformity" within the ET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1_RLN | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Nonuniformity" within the ET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1_RP | Textural (Gray-Level Run-Length Matrix) | The "Run Percentage" within the ET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1_LGRE | Textural (Gray-Level Run-Length Matrix) | The "Low Gray-Level Run Emphasis" within the ET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1_HGRE | Textural (Gray-Level Run-Length Matrix) | The "High Gray-Level Run Emphasis" within the ET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1_SRLGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Low Gray-Level Emphasis" within the ET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1_SRHGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run High Gray-Level Emphasis" within the ET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1_LRLGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Low Gray-Level Emphasis" within the ET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1_LRHGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run High Gray-Level Emphasis" within the ET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1_GLV | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Variance" within the ET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T1_RLV | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Variance" within the ET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T2_SRE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Emphasis" within the ET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T2_LRE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Emphasis" within the ET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T2_GLN | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Nonuniformity" within the ET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T2_RLN | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Nonuniformity" within the ET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T2_RP | Textural (Gray-Level Run-Length Matrix) | The "Run Percentage" within the ET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T2_LGRE | Textural (Gray-Level Run-Length Matrix) | The "Low Gray-Level Run Emphasis" within the ET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T2_HGRE | Textural (Gray-Level Run-Length Matrix) | The "High Gray-Level Run Emphasis" within the ET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T2_SRLGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Low Gray-Level Emphasis" within the ET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T2_SRHGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run High Gray-Level Emphasis" within the ET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T2_LRLGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Low Gray-Level Emphasis" within the ET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T2_LRHGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run High Gray-Level Emphasis" within the ET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T2_GLV | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Variance" within the ET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_T2_RLV | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Variance" within the ET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_FLAIR_SRE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Emphasis" within the ET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_FLAIR_LRE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Emphasis" within the ET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_FLAIR_GLN | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Nonuniformity" within the ET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_FLAIR_RLN | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Nonuniformity" within the ET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_FLAIR_RP | Textural (Gray-Level Run-Length Matrix) | The "Run Percentage" within the ET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_FLAIR_LGRE | Textural (Gray-Level Run-Length Matrix) | The "Low Gray-Level Run Emphasis" within the ET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_FLAIR_HGRE | Textural (Gray-Level Run-Length Matrix) | The "High Gray-Level Run Emphasis" within the ET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_FLAIR_SRLGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Low Gray-Level Emphasis" within the ET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_FLAIR_SRHGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run High Gray-Level Emphasis" within the ET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_FLAIR_LRLGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Low Gray-Level Emphasis" within the ET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_FLAIR_LRHGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run High Gray-Level Emphasis" within the ET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_FLAIR_GLV | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Variance" within the ET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ET_FLAIR_RLV | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Variance" within the ET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1Gd_SRE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Emphasis" within the ED in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1Gd_LRE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Emphasis" within the ED in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1Gd_GLN | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Nonuniformity" within the ED in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1Gd_RLN | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Nonuniformity" within the ED in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1Gd_RP | Textural (Gray-Level Run-Length Matrix) | The "Run Percentage" within the ED in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1Gd_LGRE | Textural (Gray-Level Run-Length Matrix) | The "Low Gray-Level Run Emphasis" within the ED in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1Gd_HGRE | Textural (Gray-Level Run-Length Matrix) | The "High Gray-Level Run Emphasis" within the ED in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1Gd_SRLGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Low Gray-Level Emphasis" within the ED in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1Gd_SRHGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run High Gray-Level Emphasis" within the ED in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1Gd_LRLGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Low Gray-Level Emphasis" within the ED in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1Gd_LRHGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run High Gray-Level Emphasis" within the ED in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1Gd_GLV | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Variance" within the ED in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1Gd_RLV | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Variance" within the ED in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1_SRE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Emphasis" within the ED in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1_LRE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Emphasis" within the ED in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1_GLN | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Nonuniformity" within the ED in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1_RLN | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Nonuniformity" within the ED in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1_RP | Textural (Gray-Level Run-Length Matrix) | The "Run Percentage" within the ED in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1_LGRE | Textural (Gray-Level Run-Length Matrix) | The "Low Gray-Level Run Emphasis" within the ED in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1_HGRE | Textural (Gray-Level Run-Length Matrix) | The "High Gray-Level Run Emphasis" within the ED in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1_SRLGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Low Gray-Level Emphasis" within the ED in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1_SRHGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run High Gray-Level Emphasis" within the ED in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1_LRLGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Low Gray-Level Emphasis" within the ED in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1_LRHGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run High Gray-Level Emphasis" within the ED in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1_GLV | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Variance" within the ED in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T1_RLV | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Variance" within the ED in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T2_SRE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Emphasis" within the ED in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T2_LRE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Emphasis" within the ED in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T2_GLN | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Nonuniformity" within the ED in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T2_RLN | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Nonuniformity" within the ED in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T2_RP | Textural (Gray-Level Run-Length Matrix) | The "Run Percentage" within the ED in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T2_LGRE | Textural (Gray-Level Run-Length Matrix) | The "Low Gray-Level Run Emphasis" within the ED in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T2_HGRE | Textural (Gray-Level Run-Length Matrix) | The "High Gray-Level Run Emphasis" within the ED in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T2_SRLGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Low Gray-Level Emphasis" within the ED in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T2_SRHGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run High Gray-Level Emphasis" within the ED in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T2_LRLGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Low Gray-Level Emphasis" within the ED in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T2_LRHGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run High Gray-Level Emphasis" within the ED in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T2_GLV | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Variance" within the ED in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_T2_RLV | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Variance" within the ED in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_FLAIR_SRE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Emphasis" within the ED in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_FLAIR_LRE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Emphasis" within the ED in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_FLAIR_GLN | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Nonuniformity" within the ED in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_FLAIR_RLN | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Nonuniformity" within the ED in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_FLAIR_RP | Textural (Gray-Level Run-Length Matrix) | The "Run Percentage" within the ED in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_FLAIR_LGRE | Textural (Gray-Level Run-Length Matrix) | The "Low Gray-Level Run Emphasis" within the ED in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_FLAIR_HGRE | Textural (Gray-Level Run-Length Matrix) | The "High Gray-Level Run Emphasis" within the ED in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_FLAIR_SRLGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Low Gray-Level Emphasis" within the ED in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_FLAIR_SRHGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run High Gray-Level Emphasis" within the ED in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_FLAIR_LRLGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Low Gray-Level Emphasis" within the ED in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_FLAIR_LRHGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run High Gray-Level Emphasis" within the ED in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_FLAIR_GLV | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Variance" within the ED in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_ED_FLAIR_RLV | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Variance" within the ED in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1Gd_SRE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Emphasis" within the NET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1Gd_LRE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Emphasis" within the NET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1Gd_GLN | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Nonuniformity" within the NET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1Gd_RLN | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Nonuniformity" within the NET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1Gd_RP | Textural (Gray-Level Run-Length Matrix) | The "Run Percentage" within the NET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1Gd_LGRE | Textural (Gray-Level Run-Length Matrix) | The "Low Gray-Level Run Emphasis" within the NET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1Gd_HGRE | Textural (Gray-Level Run-Length Matrix) | The "High Gray-Level Run Emphasis" within the NET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1Gd_SRLGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Low Gray-Level Emphasis" within the NET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1Gd_SRHGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run High Gray-Level Emphasis" within the NET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1Gd_LRLGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Low Gray-Level Emphasis" within the NET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1Gd_LRHGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run High Gray-Level Emphasis" within the NET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1Gd_GLV | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Variance" within the NET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1Gd_RLV | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Variance" within the NET in the T1-Gd volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1_SRE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Emphasis" within the NET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1_LRE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Emphasis" within the NET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1_GLN | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Nonuniformity" within the NET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1_RLN | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Nonuniformity" within the NET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1_RP | Textural (Gray-Level Run-Length Matrix) | The "Run Percentage" within the NET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1_LGRE | Textural (Gray-Level Run-Length Matrix) | The "Low Gray-Level Run Emphasis" within the NET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1_HGRE | Textural (Gray-Level Run-Length Matrix) | The "High Gray-Level Run Emphasis" within the NET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1_SRLGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Low Gray-Level Emphasis" within the NET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1_SRHGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run High Gray-Level Emphasis" within the NET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1_LRLGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Low Gray-Level Emphasis" within the NET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1_LRHGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run High Gray-Level Emphasis" within the NET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1_GLV | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Variance" within the NET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T1_RLV | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Variance" within the NET in the T1 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T2_SRE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Emphasis" within the NET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T2_LRE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Emphasis" within the NET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T2_GLN | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Nonuniformity" within the NET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T2_RLN | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Nonuniformity" within the NET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T2_RP | Textural (Gray-Level Run-Length Matrix) | The "Run Percentage" within the NET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T2_LGRE | Textural (Gray-Level Run-Length Matrix) | The "Low Gray-Level Run Emphasis" within the NET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T2_HGRE | Textural (Gray-Level Run-Length Matrix) | The "High Gray-Level Run Emphasis" within the NET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T2_SRLGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Low Gray-Level Emphasis" within the NET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T2_SRHGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run High Gray-Level Emphasis" within the NET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T2_LRLGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Low Gray-Level Emphasis" within the NET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T2_LRHGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run High Gray-Level Emphasis" within the NET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T2_GLV | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Variance" within the NET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_T2_RLV | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Variance" within the NET in the T2 volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_FLAIR_SRE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Emphasis" within the NET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_FLAIR_LRE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Emphasis" within the NET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_FLAIR_GLN | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Nonuniformity" within the NET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_FLAIR_RLN | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Nonuniformity" within the NET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_FLAIR_RP | Textural (Gray-Level Run-Length Matrix) | The "Run Percentage" within the NET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_FLAIR_LGRE | Textural (Gray-Level Run-Length Matrix) | The "Low Gray-Level Run Emphasis" within the NET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_FLAIR_HGRE | Textural (Gray-Level Run-Length Matrix) | The "High Gray-Level Run Emphasis" within the NET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_FLAIR_SRLGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run Low Gray-Level Emphasis" within the NET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_FLAIR_SRHGE | Textural (Gray-Level Run-Length Matrix) | The "Short Run High Gray-Level Emphasis" within the NET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_FLAIR_LRLGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run Low Gray-Level Emphasis" within the NET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_FLAIR_LRHGE | Textural (Gray-Level Run-Length Matrix) | The "Long Run High Gray-Level Emphasis" within the NET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_FLAIR_GLV | Textural (Gray-Level Run-Length Matrix) | The "Gray-Level Variance" within the NET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLRLM_NET_FLAIR_RLV | Textural (Gray-Level Run-Length Matrix) | The "Run-Length Variance" within the NET in the FLAIR volume, considering the 13 main directions in the 3D volume. |
| TEXTURE_GLSZM_ET_T1Gd_SZE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Emphasis" within the ET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1Gd_LZE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Emphasis" within the ET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1Gd_GLN | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Nonuniformity" within the ET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1Gd_ZSN | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Nonuniformity" within the ET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1Gd_ZP | Textural (Gray-Level Size Zone Matrix) | The "Zone Percentage" within the ET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1Gd_LGZE | Textural (Gray-Level Size Zone Matrix) | The "Low Gray-Level Zone Emphasis" within the ET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1Gd_HGZE | Textural (Gray-Level Size Zone Matrix) | The "High Gray-Level Zone Emphasis" within the ET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1Gd_SZLGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Low Gray-Level Emphasis" within the ET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1Gd_SZHGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone High Gray-Level Emphasis" within the ET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1Gd_LZLGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Low Gray-Level Emphasis" within the ET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1Gd_LZHGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone High Gray-Level Emphasis" within the ET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1Gd_GLV | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Variance" within the ET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1Gd_ZSV | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Variance" within the ET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1_SZE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Emphasis" within the ET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1_LZE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Emphasis" within the ET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1_GLN | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Nonuniformity" within the ET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1_ZSN | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Nonuniformity" within the ET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1_ZP | Textural (Gray-Level Size Zone Matrix) | The "Zone Percentage" within the ET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1_LGZE | Textural (Gray-Level Size Zone Matrix) | The "Low Gray-Level Zone Emphasis" within the ET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1_HGZE | Textural (Gray-Level Size Zone Matrix) | The "High Gray-Level Zone Emphasis" within the ET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1_SZLGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Low Gray-Level Emphasis" within the ET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1_SZHGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone High Gray-Level Emphasis" within the ET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1_LZLGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Low Gray-Level Emphasis" within the ET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1_LZHGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone High Gray-Level Emphasis" within the ET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1_GLV | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Variance" within the ET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T1_ZSV | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Variance" within the ET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T2_SZE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Emphasis" within the ET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T2_LZE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Emphasis" within the ET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T2_GLN | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Nonuniformity" within the ET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T2_ZSN | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Nonuniformity" within the ET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T2_ZP | Textural (Gray-Level Size Zone Matrix) | The "Zone Percentage" within the ET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T2_LGZE | Textural (Gray-Level Size Zone Matrix) | The "Low Gray-Level Zone Emphasis" within the ET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T2_HGZE | Textural (Gray-Level Size Zone Matrix) | The "High Gray-Level Zone Emphasis" within the ET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T2_SZLGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Low Gray-Level Emphasis" within the ET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T2_SZHGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone High Gray-Level Emphasis" within the ET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T2_LZLGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Low Gray-Level Emphasis" within the ET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T2_LZHGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone High Gray-Level Emphasis" within the ET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T2_GLV | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Variance" within the ET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_T2_ZSV | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Variance" within the ET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_FLAIR_SZE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Emphasis" within the ET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_FLAIR_LZE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Emphasis" within the ET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_FLAIR_GLN | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Nonuniformity" within the ET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_FLAIR_ZSN | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Nonuniformity" within the ET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_FLAIR_ZP | Textural (Gray-Level Size Zone Matrix) | The "Zone Percentage" within the ET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_FLAIR_LGZE | Textural (Gray-Level Size Zone Matrix) | The "Low Gray-Level Zone Emphasis" within the ET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_FLAIR_HGZE | Textural (Gray-Level Size Zone Matrix) | The "High Gray-Level Zone Emphasis" within the ET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_FLAIR_SZLGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Low Gray-Level Emphasis" within the ET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_FLAIR_SZHGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone High Gray-Level Emphasis" within the ET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_FLAIR_LZLGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Low Gray-Level Emphasis" within the ET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_FLAIR_LZHGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone High Gray-Level Emphasis" within the ET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_FLAIR_GLV | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Variance" within the ET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ET_FLAIR_ZSV | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Variance" within the ET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1Gd_SZE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Emphasis" within the ED in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1Gd_LZE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Emphasis" within the ED in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1Gd_GLN | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Nonuniformity" within the ED in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1Gd_ZSN | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Nonuniformity" within the ED in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1Gd_ZP | Textural (Gray-Level Size Zone Matrix) | The "Zone Percentage" within the ED in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1Gd_LGZE | Textural (Gray-Level Size Zone Matrix) | The "Low Gray-Level Zone Emphasis" within the ED in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1Gd_HGZE | Textural (Gray-Level Size Zone Matrix) | The "High Gray-Level Zone Emphasis" within the ED in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1Gd_SZLGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Low Gray-Level Emphasis" within the ED in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1Gd_SZHGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone High Gray-Level Emphasis" within the ED in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1Gd_LZLGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Low Gray-Level Emphasis" within the ED in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1Gd_LZHGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone High Gray-Level Emphasis" within the ED in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1Gd_GLV | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Variance" within the ED in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1Gd_ZSV | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Variance" within the ED in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1_SZE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Emphasis" within the ED in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1_LZE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Emphasis" within the ED in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1_GLN | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Nonuniformity" within the ED in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1_ZSN | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Nonuniformity" within the ED in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1_ZP | Textural (Gray-Level Size Zone Matrix) | The "Zone Percentage" within the ED in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1_LGZE | Textural (Gray-Level Size Zone Matrix) | The "Low Gray-Level Zone Emphasis" within the ED in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1_HGZE | Textural (Gray-Level Size Zone Matrix) | The "High Gray-Level Zone Emphasis" within the ED in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1_SZLGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Low Gray-Level Emphasis" within the ED in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1_SZHGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone High Gray-Level Emphasis" within the ED in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1_LZLGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Low Gray-Level Emphasis" within the ED in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1_LZHGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone High Gray-Level Emphasis" within the ED in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1_GLV | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Variance" within the ED in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T1_ZSV | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Variance" within the ED in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T2_SZE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Emphasis" within the ED in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T2_LZE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Emphasis" within the ED in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T2_GLN | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Nonuniformity" within the ED in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T2_ZSN | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Nonuniformity" within the ED in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T2_ZP | Textural (Gray-Level Size Zone Matrix) | The "Zone Percentage" within the ED in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T2_LGZE | Textural (Gray-Level Size Zone Matrix) | The "Low Gray-Level Zone Emphasis" within the ED in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T2_HGZE | Textural (Gray-Level Size Zone Matrix) | The "High Gray-Level Zone Emphasis" within the ED in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T2_SZLGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Low Gray-Level Emphasis" within the ED in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T2_SZHGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone High Gray-Level Emphasis" within the ED in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T2_LZLGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Low Gray-Level Emphasis" within the ED in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T2_LZHGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone High Gray-Level Emphasis" within the ED in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T2_GLV | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Variance" within the ED in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_T2_ZSV | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Variance" within the ED in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_FLAIR_SZE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Emphasis" within the ED in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_FLAIR_LZE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Emphasis" within the ED in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_FLAIR_GLN | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Nonuniformity" within the ED in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_FLAIR_ZSN | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Nonuniformity" within the ED in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_FLAIR_ZP | Textural (Gray-Level Size Zone Matrix) | The "Zone Percentage" within the ED in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_FLAIR_LGZE | Textural (Gray-Level Size Zone Matrix) | The "Low Gray-Level Zone Emphasis" within the ED in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_FLAIR_HGZE | Textural (Gray-Level Size Zone Matrix) | The "High Gray-Level Zone Emphasis" within the ED in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_FLAIR_SZLGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Low Gray-Level Emphasis" within the ED in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_FLAIR_SZHGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone High Gray-Level Emphasis" within the ED in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_FLAIR_LZLGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Low Gray-Level Emphasis" within the ED in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_FLAIR_LZHGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone High Gray-Level Emphasis" within the ED in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_FLAIR_GLV | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Variance" within the ED in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_ED_FLAIR_ZSV | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Variance" within the ED in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1Gd_SZE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Emphasis" within the NET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1Gd_LZE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Emphasis" within the NET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1Gd_GLN | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Nonuniformity" within the NET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1Gd_ZSN | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Nonuniformity" within the NET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1Gd_ZP | Textural (Gray-Level Size Zone Matrix) | The "Zone Percentage" within the NET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1Gd_LGZE | Textural (Gray-Level Size Zone Matrix) | The "Low Gray-Level Zone Emphasis" within the NET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1Gd_HGZE | Textural (Gray-Level Size Zone Matrix) | The "High Gray-Level Zone Emphasis" within the NET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1Gd_SZLGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Low Gray-Level Emphasis" within the NET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1Gd_SZHGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone High Gray-Level Emphasis" within the NET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1Gd_LZLGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Low Gray-Level Emphasis" within the NET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1Gd_LZHGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone High Gray-Level Emphasis" within the NET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1Gd_GLV | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Variance" within the NET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1Gd_ZSV | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Variance" within the NET in the T1-Gd volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1_SZE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Emphasis" within the NET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1_LZE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Emphasis" within the NET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1_GLN | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Nonuniformity" within the NET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1_ZSN | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Nonuniformity" within the NET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1_ZP | Textural (Gray-Level Size Zone Matrix) | The "Zone Percentage" within the NET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1_LGZE | Textural (Gray-Level Size Zone Matrix) | The "Low Gray-Level Zone Emphasis" within the NET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1_HGZE | Textural (Gray-Level Size Zone Matrix) | The "High Gray-Level Zone Emphasis" within the NET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1_SZLGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Low Gray-Level Emphasis" within the NET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1_SZHGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone High Gray-Level Emphasis" within the NET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1_LZLGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Low Gray-Level Emphasis" within the NET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1_LZHGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone High Gray-Level Emphasis" within the NET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1_GLV | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Variance" within the NET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T1_ZSV | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Variance" within the NET in the T1 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T2_SZE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Emphasis" within the NET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T2_LZE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Emphasis" within the NET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T2_GLN | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Nonuniformity" within the NET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T2_ZSN | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Nonuniformity" within the NET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T2_ZP | Textural (Gray-Level Size Zone Matrix) | The "Zone Percentage" within the NET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T2_LGZE | Textural (Gray-Level Size Zone Matrix) | The "Low Gray-Level Zone Emphasis" within the NET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T2_HGZE | Textural (Gray-Level Size Zone Matrix) | The "High Gray-Level Zone Emphasis" within the NET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T2_SZLGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Low Gray-Level Emphasis" within the NET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T2_SZHGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone High Gray-Level Emphasis" within the NET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T2_LZLGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Low Gray-Level Emphasis" within the NET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T2_LZHGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone High Gray-Level Emphasis" within the NET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T2_GLV | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Variance" within the NET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_T2_ZSV | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Variance" within the NET in the T2 volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_FLAIR_SZE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Emphasis" within the NET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_FLAIR_LZE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Emphasis" within the NET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_FLAIR_GLN | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Nonuniformity" within the NET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_FLAIR_ZSN | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Nonuniformity" within the NET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_FLAIR_ZP | Textural (Gray-Level Size Zone Matrix) | The "Zone Percentage" within the NET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_FLAIR_LGZE | Textural (Gray-Level Size Zone Matrix) | The "Low Gray-Level Zone Emphasis" within the NET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_FLAIR_HGZE | Textural (Gray-Level Size Zone Matrix) | The "High Gray-Level Zone Emphasis" within the NET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_FLAIR_SZLGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone Low Gray-Level Emphasis" within the NET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_FLAIR_SZHGE | Textural (Gray-Level Size Zone Matrix) | The "Small Zone High Gray-Level Emphasis" within the NET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_FLAIR_LZLGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone Low Gray-Level Emphasis" within the NET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_FLAIR_LZHGE | Textural (Gray-Level Size Zone Matrix) | The "Large Zone High Gray-Level Emphasis" within the NET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_FLAIR_GLV | Textural (Gray-Level Size Zone Matrix) | The "Gray-Level Variance" within the NET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_GLSZM_NET_FLAIR_ZSV | Textural (Gray-Level Size Zone Matrix) | The "Zone-Size Variance" within the NET in the FLAIR volume, whilst zones of different sizes are computed using a 26-voxel connectivity. |
| TEXTURE_NGTDM_ET_T1Gd_Coarseness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Coarseness within the ET in the T1-Gd volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ET_T1Gd_Contrast | Textural (Neighborhood Gray-Tone Difference Matrix) | The Contrast within the ET in the T1-Gd volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ET_T1Gd_Busyness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Busyness within the ET in the T1-Gd volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ET_T1Gd_Complexity | Textural (Neighborhood Gray-Tone Difference Matrix) | The Complexity within the ET in the T1-Gd volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ET_T1Gd_Strength | Textural (Neighborhood Gray-Tone Difference Matrix) | The Strength within the ET in the T1-Gd volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ET_T1_Coarseness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Coarseness within the ET in the T1 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ET_T1_Contrast | Textural (Neighborhood Gray-Tone Difference Matrix) | The Contrast within the ET in the T1 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ET_T1_Busyness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Busyness within the ET in the T1 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ET_T1_Complexity | Textural (Neighborhood Gray-Tone Difference Matrix) | The Complexity within the ET in the T1 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ET_T1_Strength | Textural (Neighborhood Gray-Tone Difference Matrix) | The Strength within the ET in the T1 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ET_T2_Coarseness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Coarseness within the ET in the T2 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ET_T2_Contrast | Textural (Neighborhood Gray-Tone Difference Matrix) | The Contrast within the ET in the T2 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ET_T2_Busyness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Busyness within the ET in the T2 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ET_T2_Complexity | Textural (Neighborhood Gray-Tone Difference Matrix) | The Complexity within the ET in the T2 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ET_T2_Strength | Textural (Neighborhood Gray-Tone Difference Matrix) | The Strength within the ET in the T2 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ET_FLAIR_Coarseness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Coarseness within the ET in the FLAIR volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ET_FLAIR_Contrast | Textural (Neighborhood Gray-Tone Difference Matrix) | The Contrast within the ET in the FLAIR volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ET_FLAIR_Busyness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Busyness within the ET in the FLAIR volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ET_FLAIR_Complexity | Textural (Neighborhood Gray-Tone Difference Matrix) | The Complexity within the ET in the FLAIR volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ET_FLAIR_Strength | Textural (Neighborhood Gray-Tone Difference Matrix) | The Strength within the ET in the FLAIR volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ED_T1Gd_Coarseness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Coarseness within the ED in the T1-Gd volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ED_T1Gd_Contrast | Textural (Neighborhood Gray-Tone Difference Matrix) | The Contrast within the ED in the T1-Gd volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ED_T1Gd_Busyness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Busyness within the ED in the T1-Gd volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ED_T1Gd_Complexity | Textural (Neighborhood Gray-Tone Difference Matrix) | The Complexity within the ED in the T1-Gd volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ED_T1Gd_Strength | Textural (Neighborhood Gray-Tone Difference Matrix) | The Strength within the ED in the T1-Gd volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ED_T1_Coarseness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Coarseness within the ED in the T1 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ED_T1_Contrast | Textural (Neighborhood Gray-Tone Difference Matrix) | The Contrast within the ED in the T1 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ED_T1_Busyness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Busyness within the ED in the T1 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ED_T1_Complexity | Textural (Neighborhood Gray-Tone Difference Matrix) | The Complexity within the ED in the T1 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ED_T1_Strength | Textural (Neighborhood Gray-Tone Difference Matrix) | The Strength within the ED in the T1 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ED_T2_Coarseness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Coarseness within the ED in the T2 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ED_T2_Contrast | Textural (Neighborhood Gray-Tone Difference Matrix) | The Contrast within the ED in the T2 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ED_T2_Busyness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Busyness within the ED in the T2 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ED_T2_Complexity | Textural (Neighborhood Gray-Tone Difference Matrix) | The Complexity within the ED in the T2 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ED_T2_Strength | Textural (Neighborhood Gray-Tone Difference Matrix) | The Strength within the ED in the T2 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ED_FLAIR_Coarseness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Coarseness within the ED in the FLAIR volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ED_FLAIR_Contrast | Textural (Neighborhood Gray-Tone Difference Matrix) | The Contrast within the ED in the FLAIR volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ED_FLAIR_Busyness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Busyness within the ED in the FLAIR volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ED_FLAIR_Complexity | Textural (Neighborhood Gray-Tone Difference Matrix) | The Complexity within the ED in the FLAIR volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_ED_FLAIR_Strength | Textural (Neighborhood Gray-Tone Difference Matrix) | The Strength within the ED in the FLAIR volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_NET_T1Gd_Coarseness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Coarseness within the NET in the T1-Gd volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_NET_T1Gd_Contrast | Textural (Neighborhood Gray-Tone Difference Matrix) | The Contrast within the NET in the T1-Gd volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_NET_T1Gd_Busyness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Busyness within the NET in the T1-Gd volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_NET_T1Gd_Complexity | Textural (Neighborhood Gray-Tone Difference Matrix) | The Complexity within the NET in the T1-Gd volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_NET_T1Gd_Strength | Textural (Neighborhood Gray-Tone Difference Matrix) | The Strength within the NET in the T1-Gd volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_NET_T1_Coarseness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Coarseness within the NET in the T1 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_NET_T1_Contrast | Textural (Neighborhood Gray-Tone Difference Matrix) | The Contrast within the NET in the T1 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_NET_T1_Busyness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Busyness within the NET in the T1 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_NET_T1_Complexity | Textural (Neighborhood Gray-Tone Difference Matrix) | The Complexity within the NET in the T1 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_NET_T1_Strength | Textural (Neighborhood Gray-Tone Difference Matrix) | The Strength within the NET in the T1 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_NET_T2_Coarseness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Coarseness within the NET in the T2 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_NET_T2_Contrast | Textural (Neighborhood Gray-Tone Difference Matrix) | The Contrast within the NET in the T2 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_NET_T2_Busyness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Busyness within the NET in the T2 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_NET_T2_Complexity | Textural (Neighborhood Gray-Tone Difference Matrix) | The Complexity within the NET in the T2 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_NET_T2_Strength | Textural (Neighborhood Gray-Tone Difference Matrix) | The Strength within the NET in the T2 volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_NET_FLAIR_Coarseness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Coarseness within the NET in the FLAIR volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_NET_FLAIR_Contrast | Textural (Neighborhood Gray-Tone Difference Matrix) | The Contrast within the NET in the FLAIR volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_NET_FLAIR_Busyness | Textural (Neighborhood Gray-Tone Difference Matrix) | The Busyness within the NET in the FLAIR volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_NET_FLAIR_Complexity | Textural (Neighborhood Gray-Tone Difference Matrix) | The Complexity within the NET in the FLAIR volume, considering a 26-voxel connectivity. |
| TEXTURE_NGTDM_NET_FLAIR_Strength | Textural (Neighborhood Gray-Tone Difference Matrix) | The Strength within the NET in the FLAIR volume, considering a 26-voxel connectivity. |
| TGM_p1 | Tumor Growth Model Parameter | The mass-effect parameters considering all apparent tumors. |
| TGM_dw | Tumor Growth Model Parameter | The estimated diffusion coefficient of white matter, considering all apparent tumors. |
| TGM_Cog_X_1 | Tumor Growth Model Parameter | The 3D coordinates (in Sagital plane) of the apparent tumor's center of gravity. |
| TGM_Cog_Y_1 | Tumor Growth Model Parameter | The 3D coordinates (in Coronal plane) of the apparent tumor's center of gravity. |
| TGM_Cog_Z_1 | Tumor Growth Model Parameter | The 3D coordinates (in Axial plane) of the apparent tumor's center of gravity. |
| TGM_T_1 | Tumor Growth Model Parameter | The estimated growth time of the apparent tumor in this location. |
| TGM_Cog_X_2 | Tumor Growth Model Parameter | The 3D coordinates (in Sagital plane) of the apparent tumor's center of gravity. |
| TGM_Cog_Y_2 | Tumor Growth Model Parameter | The 3D coordinates (in Coronal plane) of the apparent tumor's center of gravity. |
| TGM_Cog_Z_2 | Tumor Growth Model Parameter | The 3D coordinates (in Axial plane) of the apparent tumor's center of gravity. |
| TGM_T_2 | Tumor Growth Model Parameter | The estimated growth time of the apparent tumor in this location. |
| TGM_Cog_X_3 | Tumor Growth Model Parameter | The 3D coordinates (in Sagital plane) of the apparent tumor's center of gravity. |
| TGM_Cog_Y_3 | Tumor Growth Model Parameter | The 3D coordinates (in Coronal plane) of the apparent tumor's center of gravity. |
| TGM_Cog_Z_3 | Tumor Growth Model Parameter | The 3D coordinates (in Axial plane) of the apparent tumor's center of gravity. |
| TGM_T_3 | Tumor Growth Model Parameter | The estimated growth time of the apparent tumor in this location. |
| TGM_Cog_X_4 | Tumor Growth Model Parameter | The 3D coordinates (in Sagital plane) of the apparent tumor's center of gravity. |
| TGM_Cog_Y_4 | Tumor Growth Model Parameter | The 3D coordinates (in Coronal plane) of the apparent tumor's center of gravity. |
| TGM_Cog_Z_4 | Tumor Growth Model Parameter | The 3D coordinates (in Axial plane) of the apparent tumor's center of gravity. |
| TGM_T_4 | Tumor Growth Model Parameter | The estimated growth time of the apparent tumor in this location. |
| TGM_Cog_X_5 | Tumor Growth Model Parameter | The 3D coordinates (in Sagital plane) of the apparent tumor's center of gravity. |
| TGM_Cog_Y_5 | Tumor Growth Model Parameter | The 3D coordinates (in Coronal plane) of the apparent tumor's center of gravity. |
| TGM_Cog_Z_5 | Tumor Growth Model Parameter | The 3D coordinates (in Axial plane) of the apparent tumor's center of gravity. |
| TGM_T_5 | Tumor Growth Model Parameter | The estimated growth time of the apparent tumor in this location. |
| TGM_Cog_X_6 | Tumor Growth Model Parameter | The 3D coordinates (in Sagital plane) of the apparent tumor's center of gravity. |
| TGM_Cog_Y_6 | Tumor Growth Model Parameter | The 3D coordinates (in Coronal plane) of the apparent tumor's center of gravity. |
| TGM_Cog_Z_6 | Tumor Growth Model Parameter | The 3D coordinates (in Axial plane) of the apparent tumor's center of gravity. |
| TGM_T_6 | Tumor Growth Model Parameter | The estimated growth time of the apparent tumor in this location. |
Technical Validation
Data collection
Our expert board-certified neuroradiologist (M.B.) identified 135 and 108 pre-operative baseline scans of the TCGA-GBM and the TCGA-LGG glioma collections, via radiological assessment and while blinded to the glioma grade. Since it is not always easy to determine if a scan is pre-operative or post-operative only by visually assessing MRI volumes, and the radiological reports were not available through the TCGA/TCIA repositories, whenever we mention ‘pre-operative scans’ in this study, we refer to those that radiographically do not have clear evidence of prior instrumentation. Specifically, the main evaluation criterion for classifying scans as pre-operative, was absence of obvious skull defect and of operative cavity through either biopsy or resection.
We note that a mixed (pre- and post-operative) subset of 223 and 59 scans from the TCIA-GBM and TCIA-LGG datasets, respectively, were included in the BraTS’15 challenge, as part of their training (nGBM=200, nLGG=44) and testing (nGBM=23, nLGG=15) datasets, via the Virtual Skeleton Database (VSD) platform56,105 (www.virtualskeleton.ch). Since an explicit distinction as pre- or post-operative was not provided for the BraTS’15 dataset, we conducted the radiological assessment of the complete TCIA collections, blind to whether a scan was part of the BraTS challenge, and only included the BraTS’15 volumes identified as pre-operative (Fig. 1f) (Table 4 (available online only)).
Segmentation labels
The segmentation method we developed to produce the segmentation labels, GLISTRboost36,38, was ranked as the best performing method and awarded the 1st prize during the International Multimodal Brain Tumor Image Segmentation challenge 2015 (BraTS’15)36,38,40–56. Specifically, the performance of the computer-aided segmentation labels was assessed during the challenge for the test data, through the VSD platform, by comparing the voxel-level overlap between the segmentation labels produced by GLISTRboost and the ground truth labels provided by the BraTS organizers in three regions, i.e., the whole tumor (WT), the tumor core (TC) and the ET. The WT describes the union of the ET, NET and ED, whereas the TC describes the union of the ET and NET. The performance was quantitatively validated by the per-voxel overlap between respective regions, using the DICE coefficient and the robust Hausdorff distance (95% quantile), as suggested by the BraTS’15 organizers56. The former metric takes values between 0 and 1, with higher values corresponding to increased overlap, whereas lower values in the latter correspond to segmentation labels closer to the gold standard labels. Note that the quantitative results for the test data were not provided to the participants, until a manuscript summarizing the results of BraTS’14 and BraTS’15 is published. However, for reporting the performance of our method, we report here the cross-validated results of the same metrics used in BraTS’15 for the subset of GBM subjects included in the training set of BraTS’15 and identified as pre-operative in this study (Table 4 (available online only)). The median DICE values with their corresponding inter-quartile ranges (IQR) for the three evaluated regions, i.e., WT, TC, ET, were equal to 0.92 (IQR: 0.88–0.94), 0.88 (IQR: 0.81–0.93) and 0.88 (IQR: 0.81–0.91), respectively. Equivalently, the 95th percentile of the Hausdorff distance for WT, TC and ET were equal to 3.61 (IQR: 2.39–8.15), 4.06 (IQR: 2.39–7.29), and 2 (IQR: 1.41–2.83), respectively.
Furthermore, we used the Jaccard coefficient, in order to quantify the difference between the computer-aided segmentation labels produced for all the scans identified as pre-operative and all the manually-corrected labels that we provide in [Data Citation 3 and Data Citation 4]. The median (mean±std.dev) Jaccard values for the three regions of interest i.e., WT, TC, ET, were equal to 0.96 (0.93±0.1), 0.87 (0.78±0.23), and 0.86 (0.73±0.29), respectively.
Manual correction
The classification scheme of segmentation labels considered for the manual corrections of the GBM and LGG cases describe all three segmentation labels (i.e., ET, NET, and ED) for both GBMs and LGGs with an apparent ET area. However, whenever we note LGG scans without an apparent ET area and not obvious texture differences, we considered only the NET label, allowing for distinguishing between normal and abnormal brain tissue, as slowly growing tumors are not expected to induce ED. Furthermore, due to high uncertainty (reported by neurosurgeons, neuroradiologists, and imaging scientists) on the exact boundaries between the various tumor labels, particularly between NET and ED, small regions that visual assessment was ambiguous of their exact classification, were left as segmented by GLISTRboost.
Manual revisions/corrections applied in the computer-aided segmentation labels comprise: i) obvious under- or over-segmented ED/ET/NET regions (Fig. 3d–g), ii) voxels classified as ED within the tumor core (Fig. 3b,c,g), iii) unclassified voxels within the tumor core (Fig. 3c–g), iv) voxels classified as NET outside the tumor core. Note that during the manual corrections only peritumoral ED was considered, and both contralateral, and periventricular ED was deleted (Fig. 3c,f), unless it was a clear continuation of the peritumoral ED, in which cases was added (Fig. 3g). The rationale for this is that contralateral and periventricular white matter hyper-intensities regions might be considered pre-existing conditions, related to small vessel ischemic disease, especially in older patients.
The scheme followed for the manual correction included two computational imaging scientists (S.B., A.S.) and a medical doctor (H.A.) working in medical image computing and analysis for 10, 12 and 8 years, respectively. These operators corrected mislabeled voxels following the rules set by our expert board-certified neuroradiologist (M.B.) with 14 years of experience. The corrected labels were then iteratively re-evaluated by the latter and re-iterated until they were satisfactory segmented.
Additional Information
How to cite this article: Bakas, S. et al. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 4:170117 doi: 10.1038/sdata.2017.117 (2017).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
Martin Rozycki (who did the final QC of the submitted data), Michel Bilello (who validated all manually-revised segmentation labels), Spyridon Bakas, and Christos Davatzikos had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. The authors would like to thank Dr Gaurav Shukla (Department of Radiation Oncology, Sidney Kimmel Cancer Center, Thomas Jefferson University, Philadelphia, PA, USA) for assisting in the extraction of the radiomic features. Furthermore, the authors would like to acknowledge the effort of Ke Zeng, Saima Rathore, Bilwaj Gaonkar, and Sarthak Pati, who all contributed in successfully developing GLISTRboost. This work was supported in part by the National Institutes of Health (NIH) R01 grant on ‘Predicting brain tumor progression via multiparametric image analysis and modeling’ (R01-NS042645), and in part by the NIH U24 grant of ‘Cancer imaging phenomics software suite: application to brain and breast cancer’ (U24-CA189523).
Footnotes
The authors declare no competing financial interests.
Data Citations
- Scarpace L. 2016. The Cancer Imaging Archive. http://doi.org/10.7937/K9/TCIA.2016.RNYFUYE9
- Pedano N. 2016. The Cancer Imaging Archive. http://doi.org/10.7937/K9/TCIA.2016.L4LTD3TK
- Bakas S. 2017. The Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2017.KLXWJJ1Q
- Bakas S. 2017. The Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2017.GJQ7R0EF
References
- Kumar V. et al. Radiomics: the process and the challenges. Magnetic Resonance Imaging 30, 1234–1248 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambin P. et al. Radiomics: Extracting more information from medical images using advanced feature analysis. European Journal of Cancer 48, 441–446 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies R. J., Kinahan P. E. & Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 278, 563–577 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L. S. et al. Multi-Parametric MRI and Texture Analysis to Visualize Spatial Histologic Heterogeneity and Tumor Extent in Glioblastoma. PLOS ONE 10, e0141506 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutman A. M. & Kuo M. D. Radiogenomics: Creating a link between molecular diagnostics and diagnostic imaging. European Journal of Radiology 70, 232–241 (2009). [DOI] [PubMed] [Google Scholar]
- Jaffe C. C. Imaging and Genomics: Is There a Synergy? Radiology 264, 329–331 (2012). [DOI] [PubMed] [Google Scholar]
- Proud C. Radiogenomics: The Promise of Personalized Treatment in Radiation Oncology? Clinical Journal of Oncology Nursing 18, 185 (2014). [DOI] [PubMed] [Google Scholar]
- Rosenstein B. S. et al. Radiogenomics: Radiobiology Enters the Era of Big Data and Team Science. International Journal of Radiation Oncology • Biology • Physics 89, 709–713 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellingson B. M. Radiogenomics and imaging phenotypes in glioblastoma: novel observations and correlation with molecular characteristics. Curr Neurol Neurosci Rep 15, 506 (2015). [DOI] [PubMed] [Google Scholar]
- Mahajan A., Moiyadi A. V., Jalali R. & Sridhar E. Radiogenomics of glioblastoma: a window into its imaging and molecular variability. Cancer Imaging 15, P14 (2015). [Google Scholar]
- Mazurowski M. A. Radiogenomics: What It Is and Why It Is Important. Journal of the American College of Radiology 12, 862–866 (2015). [DOI] [PubMed] [Google Scholar]
- Bakas S. et al. In vivo detection of EGFRvIII in glioblastoma via perfusion magnetic resonance imaging signature consistent with deep peritumoral infiltration: the φ-index. Clinical Cancer Research, doi:10.1158/1078-0432.ccr-16-1871 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tykocinski E. S. et al. Use of magnetic perfusion-weighted imaging to determine epidermal growth factor receptor variant III expression in glioblastoma. Neuro-oncology 14, 613–623 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert O. et al. in Proceedings of the 103rd Annual Meeting of the American Association for Cancer Research.
- Gutman D. A. et al. MR Imaging Predictors of Molecular Profile and Survival: Multi-institutional Study of the TCGA Glioblastoma Data Set. Radiology 267, 560–569 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurowski M. A., Desjardins A. & Malof J. M. Imaging descriptors improve the predictive power of survival models for glioblastoma patients. Neuro-Oncology 15, 1389–1394 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari H. et al. Pattern Analysis of Dynamic Susceptibility Contrast MRI Reveals Peritumoral Tissue Heterogeneity. Radiology 273, 502–510 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert O. et al. Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology 273, 168–174 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill B. J. et al. MRI-localized biopsies reveal subtype-specific differences in molecular and cellular composition at the margins of glioblastoma. Proc Natl Acad Sci USA 111, 12550–12555 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R. et al. Outcome Prediction in Patients with Glioblastoma by Using Imaging, Clinical, and Genomic Biomarkers: Focus on the Nonenhancing Component of the Tumor. Radiology 272, 484–493 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arevalo-Perez J. et al. T1-Weighted Dynamic Contrast-Enhanced MRI as a Noninvasive Biomarker of Epidermal Growth Factor Receptor vIII Status. AJNR Am J Neuroradiol 36, 2256–2261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakas S. et al. Identification of Imaging Signatures of the Epidermal Growth Factor Receptor Variant III (EGFRvIII) in Glioblastoma. Neuro-oncology 17, 154–154 (2015). [Google Scholar]
- Bonekamp D. et al. Association of overall survival in patients with newly diagnosed glioblastoma with contrast-enhanced perfusion MRI: Comparison of intraindividually matched T1- and T2*-based bolus techniques. Journal of Magnetic Resonance Imaging 42, 87–96 (2015). [DOI] [PubMed] [Google Scholar]
- Itakura H. et al. Magnetic resonance image features identify glioblastoma phenotypic subtypes with distinct molecular pathway activities. Sci Transl Med 7, 303ra138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolasjilwan M. et al. Addition of MR imaging features and genetic biomarkers strengthens glioblastoma survival prediction in TCGA patients. Journal of Neuroradiology 42, 212–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios Velazquez E. et al. Fully automatic GBM segmentation in the TCGA-GBM dataset: Prognosis and correlation with VASARI features. Scientific Reports 5, 16822 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari H. et al. Imaging Surrogates of Infiltration Obtained Via Multiparametric Imaging Pattern Analysis Predict Subsequent Location of Recurrence of Glioblastoma. Neurosurgery 78, 572–580 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakas S. et al. Highly-expressed wild-type EGFR and EGFRvIII mutant glioblastomas have similar MRI signature, consistent with deep peritumoral infiltration. Neuro-Oncology 18, vi125 (2016). [Google Scholar]
- Batmanghelich N., Dalca A., Quon G., Sabuncu M. & Golland P. Probabilistic Modeling of Imaging, Genetics and Diagnosis. IEEE Trans Med Imaging 35, 1765–1779 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder Z. A. et al. Extracellular EGFR289 activating mutations confer poorer survival and exhibit radiographic signature of enhanced motility in primary glioblastoma. Neuro-Oncology 18, vi105–vi106 (2016). [Google Scholar]
- Macyszyn L. et al. Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro-oncology 18, 417–425 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathore S., Akbari H., Rozycki M., Bakas S. & Davatzikos C. Imaging pattern analysis reveals three distinct phenotypic subtypes of GBM with different survival rates. Neuro-Oncology 18, vi128 (2016). [Google Scholar]
- Aerts H. J. The Potential of Radiomic-Based Phenotyping in Precision Medicine: A Review. JAMA oncology 2, 1636–1642 (2016). [DOI] [PubMed] [Google Scholar]
- Kaus M. R. et al. Medical Image Computing and Computer-Assisted Intervention—MICCAI’99: Second International Conference, Cambridge, UK, September 19-22, 1999. Proceedings (eds Taylor C. & Colchester A. 1–10 (Springer Berlin Heidelberg, 1999). [Google Scholar]
- Akkus Z. et al. Semi-automated segmentation of pre-operative low grade gliomas in magnetic resonance imaging. Cancer Imaging 15, 12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakas S. et al. Proceedings of the Multimodal Brain Tumor Image Segmentation Challenge held in conjunction with MICCAI 2015 (MICCAI-BRATS 2015) (eds Menze B. H. et al.) 5–12 (Technische Universität München (T.U.M.), 2015). [Google Scholar]
- Simi V. R. & Joseph J. Segmentation of Glioblastoma Multiforme from MR Images—A comprehensive review. The Egyptian Journal of Radiology and Nuclear Medicine 46, 1105–1110 (2015). [Google Scholar]
- Bakas S. et al. GLISTRboost: Combining Multimodal MRI Segmentation, Registration, and Biophysical Tumor Growth Modeling with Gradient Boosting Machines for Glioma Segmentation. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries 9556, 144–155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K. et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. Journal of Digital Imaging 26, 1045–1057 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agn M., Puonti O., Law I., Rosenschold P. M. A. & Leemput K. V. In Proceedings of the Multimodal Brain Tumor Image Segmentation Challenge held in conjunction with MICCAI 2015 (MICCAI-BRATS 2015) (eds Menze B. H. et al.1–4 (Technische Universität München (T.U.M.), 2015). [Google Scholar]
- Dvorak P. & Menze B. H. In Proceedings of the Multimodal Brain Tumor Image Segmentation Challenge held in conjunction with MICCAI 2015 (MICCAI-BRATS 2015) (eds Menze B. H. et al. 13–24 (Technische Universität München (T.U.M.), 2015). [Google Scholar]
- Haeck T., Maes F. & Suetens P. In Proceedings of the Multimodal Brain Tumor Image Segmentation Challenge held in conjunction with MICCAI 2015 (MICCAI-BRATS 2015) (eds Menze B. H. et al.25–28 (Technische Universität München (T.U.M.), 2015). [Google Scholar]
- Havaei M., Dutil F., Pal C., Larochelle H. & Jodoin P.-M. In Proceedings of the Multimodal Brain Tumor Image Segmentation Challenge held in conjunction with MICCAI 2015 (MICCAI-BRATS 2015) (eds Menze B. H. et al. 29–33 (Technische Universität München (T.U.M.), 2015). [Google Scholar]
- Hoogi A., Lee A., Bharadwaj V. & Rubin D. L. In Proceedings of the Multimodal Brain Tumor Image Segmentation Challenge held in conjunction with MICCAI 2015 (MICCAI-BRATS 2015) (eds Menze B. H. et al.34–37 (Technische Universität München (T.U.M.), 2015). [Google Scholar]
- Maier O., Wilms M. & Handels H. In Proceedings of the Multimodal Brain Tumor Image Segmentation Challenge held in conjunction with MICCAI 2015 (MICCAI-BRATS 2015), (eds Menze B. H. et al. 38–41 (Technische Universität München (T.U.M.), 2015). [Google Scholar]
- Malmi E., Parambath S., Peyrat J.-M., Abinahed J. & Chawla S. In Proceedings of the Multimodal Brain Tumor Image Segmentation Challenge held in conjunction with MICCAI 2015 (MICCAI-BRATS 2015) (eds Menze B. H. et al. 42–47 (Technische Universität München (T.U.M.), 2015). [Google Scholar]
- Meier R., Karamitsou V., Habegger S., Wiest R. & Reyes M. In Proceedings of the Multimodal Brain Tumor Image Segmentation Challenge held in conjunction with MICCAI 2015 (MICCAI-BRATS 2015) (eds Menze B. H. et al. 48–51 (Technische Universität München (T.U.M.), 2015). [Google Scholar]
- Pereira S., Pinto A., Alves V. & Silva C. A. In Proceedings of the Multimodal Brain Tumor Image Segmentation Challenge held in conjunction with MICCAI 2015 (MICCAI-BRATS 2015) (eds Menze B. H. et al. 52–55 (Technische Universität München (T.U.M.), 2015). [Google Scholar]
- Rao V., Sarabi M. S. & Jaiswal A. In Proceedings of the Multimodal Brain Tumor Image Segmentation Challenge held in conjunction with MICCAI 2015 (MICCAI-BRATS 2015) (eds Menze B. H. et al. 56–59 (Technische Universität München (T.U.M.), 2015). [Google Scholar]
- Vaidhya K., Santhosh R., Thirunavukkarasu S., Alex V. & Krishnamurthi G. In Proceedings of the Multimodal Brain Tumor Image Segmentation Challenge held in conjunction with MICCAI 2015 (MICCAI-BRATS 2015) (eds Menze B. H. et al. 60–64 (Technische Universität München (T.U.M.), 2015). [Google Scholar]
- Agn M., Puonti O., Rosenschöld P. M. A., Law I. & Leemput K. V. Brain Tumor Segmentation Using a Generative Model with an RBM Prior on Tumor Shape. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries 9556, 168–180 (2016). [Google Scholar]
- Havaei M., Dutil F., Pal C., Larochelle H. & Jodoin P.-M. A Convolutional Neural Network Approach to Brain Tumor Segmentation. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries 9556, 195–208 (2016). [Google Scholar]
- Maier O., Wilms M. & Handels H. Image Features for Brain Lesion Segmentation Using Random Forests. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries 9556, 119–130 (2016). [Google Scholar]
- Pereira S., Pinto A., Alves V. & Silva C. A. Deep Convolutional Neural Networks for the Segmentation of Gliomas in Multi-sequence MRI. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries 9556, 131–143 (2016). [Google Scholar]
- Pereira S., Pinto A., Alves V. & Silva C. A. Brain Tumor Segmentation Using Convolutional Neural Networks in MRI Images. IEEE Transactions on Medical Imaging 35, 1240–1251 (2016). [DOI] [PubMed] [Google Scholar]
- Menze B. H. et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Transactions on Medical Imaging 34, 1993–2024 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R. W. et al. in Tenth Annual Meeting of the Organization for Human Brain Mapping.
- Hogea C., Biros G., Abraham F. & Davatzikos C. A robust framework for soft tissue simulations with application to modeling brain tumor mass effect in 3D MR images. Physics in medicine and biology 52, 6893–6908 (2007). [DOI] [PubMed] [Google Scholar]
- Hogea C., Davatzikos C. & Biros G. Brain-tumor interaction biophysical models for medical image registration. SIAM Journal on Scientific Computing 30, 3050–3072 (2008). [Google Scholar]
- Hogea C. S., Biros G. & Davatzikos C. An image-driven parameter estimation problem for a reaction-diffusion glioma growth model with mass effects. Journal of Mathematical Biology 56, 793–825 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlfing T., Zahr N. M., Sullivan E. V. & Pfefferbaum A. The SRI24 Multi-Channel Atlas of Normal Adult Human Brain Structure. Human brain mapping 31, 798–819 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M. & Smith S. A global optimisation method for robust affine registration of brain images. Medical image analysis 5, 143–156 (2001). [DOI] [PubMed] [Google Scholar]
- Jenkinson M., Bannister P. R., Brady J. M. & Smith S. M. Improved optimisation for the robust and accurate linear registration and motion correction of brain images. NeuroImage 17, 825–841 (2002). [DOI] [PubMed] [Google Scholar]
- Smith S. M. et al. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage 23, 208–219 (2004). [DOI] [PubMed] [Google Scholar]
- Woolrich M. W. et al. Bayesian analysis of neuroimaging data in FSL. NeuroImage 45, S173–S186 (2009). [DOI] [PubMed] [Google Scholar]
- Jenkinson M., Beckmann C. F., Behrens T. E. & Woolrich M. W. FSL. NeuroImage 62, 782–790 (2012). [DOI] [PubMed] [Google Scholar]
- Smith S. M. Fast robust automated brain extraction. Hum Brain Mapp 17, 143–155 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M., Pechaud M. & Smith S. in Eleventh Annual Meeting of the Organization for Human Brain Mapping.
- Doshi J., Erus G., Ou Y., Gaonkar B. & Davatzikos C. Multi-Atlas Skull-Stripping. Acad Radiol 20, 1566–1576 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doshi J. et al. MUSE: MUlti-atlas region Segmentation utilizing Ensembles of registration algorithms and parameters, and locally optimal atlas selection. NeuroImage 127, 186–195 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. M. & Brady J. M. SUSAN—a new approach to low level image processing. International Journal of Computer Vision 23, 45–78 (1997). [Google Scholar]
- Nyul L. G., Udupa J. K. & Zhang X. New Variants of a Method of MRI Scale Standardization. IEEE transactions on medical imaging 19, 143–150 (2000). [DOI] [PubMed] [Google Scholar]
- Sled J. G., Zijdenbos A. P. & Evans A. C. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE transactions on medical imaging 17, 87–97 (1998). [DOI] [PubMed] [Google Scholar]
- Tustison N. J. et al. N4ITK: Improved N3 Bias Correction. IEEE Transactions on Medical Imaging 29, 1310–1320 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen C. T., Iglesias J. E. & Van Leemput K. Bayesian and grAphical Models for Biomedical Imaging: First International Workshop, BAMBI 2014, Cambridge, MA, USA, September 18, 2014, Revised Selected Papers (eds Jorge Cardoso M. et al.1–12 (Springer International Publishing, 2014). [Google Scholar]
- Friedman J. H. Greedy function approximation: a gradient boosting machine. Annals of statistics 1189–1232 (2001). [Google Scholar]
- Friedman J. H. Stochastic gradient boosting. Computational Statistics & Data Analysis 38, 367–378 (2002). [Google Scholar]
- Bakas S. et al. Fast Semi-Automatic Segmentation of Focal Liver Lesions in Contrast-Enhanced Ultrasound, Based on a Probabilistic Model. TCIV Computer Methods in Biomechanics and Biomedical Engineering: Imaging & Visualization 5, 329–338 (2017). [Google Scholar]
- Gooya A. et al. GLISTR: Glioma Image Segmentation and Registration. IEEE transactions on medical imaging 31, 1941–1954 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooya A., Pohl K., Billelo M., Biros G. & Davatzikos C. in International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) Vol. 14 532-540 (Canada, 2011). [DOI] [PMC free article] [PubMed]
- Gooya A., Biros G. & Davatzikos C. Deformable Registration of Glioma Images Using EM Algorithm and Diffusion Reaction Modeling. IEEE transactions on medical imaging 30, 375–390 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon D., Shinohara R. T., Akbari H. & Davatzikos C. in Medical Image Computing and Computer-Assisted Intervention—MICCAI 2014. 763-770 (Lecture Notes in Computer Science). [DOI] [PMC free article] [PubMed]
- Pati S. et al. in Radiological Society of North America Scientific Assembly and Annual Meeting (RSNA) (Chicago, IL, 2016).
- Sethian J. A. A fast marching level set method for monotonically advancing fronts. Proceedings of the National Academy of Sciences of the United States of America 93, 1591–1595 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps T. & Cohen L. D. Fast extraction of minimal paths in 3D images and applications to virtual endoscopy1. Medical Image Analysis 5, 281–299 (2001). [DOI] [PubMed] [Google Scholar]
- Gaonkar B. et al. Automated tumor volumetry using computer-aided image segmentation. Acad Radiol 22, 653–661 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralick R. M., Shanmugam K. & Dinstein I. H. Textural features for image classification. IEEE Transactions on Systems, Man, and Cybernetics 3, 610–621 (1973). [Google Scholar]
- Haller S. et al. Do brain T2/FLAIR white matter hyperintensities correspond to myelin loss in normal aging? A radiologic-neuropathologic correlation study. Acta Neuropathologica Communications 1, 14–14 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallières M., Freeman C., Skamene S. & El Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Physics in medicine and biology 60, 5471 (2015). [DOI] [PubMed] [Google Scholar]
- Max J. Quantizing for minimum distortion. IRE Transactions on Information Theory 6, 7–12 (1960). [Google Scholar]
- Lloyd S. Least squares quantization in PCM. IEEE transactions on information theory 28, 129–137 (1982). [Google Scholar]
- Li Q. & Griffiths J. G. in Geometric modeling and processing, proceedings 335–340 (IEEE, 2004). [Google Scholar]
- Thibault G. et al. Shape and texture indexes application to cell nuclei classification. International Journal of Pattern Recognition and Artificial Intelligence 27, 1357002 (2013). [Google Scholar]
- Collewet G., Strzelecki M. & Mariette F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magnetic Resonance Imaging 22, 81–91 (2004). [DOI] [PubMed] [Google Scholar]
- Galloway M. M. Texture analysis using grey level run lengths. Computer Graphics and Image Processing 4, 172–179 (1975). [Google Scholar]
- Chu A., Sehgal C. M. & Greenleaf J. F. Use of gray value distribution of run lengths for texture analysis. Pattern Recognition Letters 11, 415–419 (1990). [Google Scholar]
- Dasarathy B. V. & Holder E. B. Image characterizations based on joint gray level—run length distributions. Pattern Recognition Letters 12, 497–502 (1991). [Google Scholar]
- Tang X. Texture information in run-length matrices. IEEE transactions on image processing 7, 1602–1609 (1998). [DOI] [PubMed] [Google Scholar]
- Thibault G. Indices de forme et de texture: de la 2D vers la 3D: application au classement de noyaux de cellules. Aix Marseille2 (2009). [Google Scholar]
- Amadasun M. & King R. Textural features corresponding to textural properties. IEEE Transactions on systems, man, and Cybernetics 19, 1264–1274 (1989). [Google Scholar]
- Bilello M. et al. Population-based MRI atlases of spatial distribution are specific to patient and tumor characteristics in glioblastoma. NeuroImage. Clinical 12, 34–40 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assefa D. et al. Robust texture features for response monitoring of glioblastoma multiforme on T1‐weighted and T2‐FLAIR MR images: A preliminary investigation in terms of identification and segmentation. Medical physics 37, 1722–1736 (2010). [DOI] [PubMed] [Google Scholar]
- Aerts H. J. et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nature communications 5, 4006 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F. et al. Scikit-learn: Machine Learning in {P}ython. Journal of Machine Learning Research 12, 2825–2830 (2011). [Google Scholar]
- Kistler M., Bonaretti S., Pfahrer M., Niklaus R. & Büchler P. The Virtual Skeleton Database: An Open Access Repository for Biomedical Research and Collaboration. Journal of Medical Internet Research 15, e245 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Scarpace L. 2016. The Cancer Imaging Archive. http://doi.org/10.7937/K9/TCIA.2016.RNYFUYE9
- Pedano N. 2016. The Cancer Imaging Archive. http://doi.org/10.7937/K9/TCIA.2016.L4LTD3TK
- Bakas S. 2017. The Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2017.KLXWJJ1Q
- Bakas S. 2017. The Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2017.GJQ7R0EF


