Abstract
Chromosomal translocations that generate oncogenic fusion proteins are causative for most pediatric leukemias and frequently affect the MLL/KMT2A gene. In vivo modeling of bona fide chromosomal translocations in human hematopoietic stem and progenitor cells is challenging but essential to determine their actual leukemogenic potential. We therefore developed an advanced lentiviral CRISPR-Cas9 vector that efficiently transduced human CD34+ hematopoietic stem and progenitor cells and induced the t(11;19)/MLL-ENL translocation. Leveraging this system, we could demonstrate that hematopoietic stem and progenitor cells harboring the translocation showed only a transient clonal growth advantage in vitro. In contrast, t(11;19)/MLL-ENL-harboring CD34+ hematopoietic stem and progenitor cells not only showed long-term engraftment in primary immunodeficient recipients, but t(11;19)/MLL-ENL also served as a first hit to initiate a monocytic leukemia-like disease. Interestingly, secondary recipients developed acute lymphoblastic leukemia with incomplete penetrance. These findings indicate that environmental cues not only contribute to the disease phenotype, but also to t(11;19)/MLL-ENL-mediated oncogenic transformation itself. Thus, by investigating the true chromosomal t(11;19) rearrangement in its natural genomic context, our study emphasizes the importance of environmental cues for the pathogenesis of pediatric leukemias, opening an avenue for novel treatment options.
Introduction
Reciprocal chromosomal translocations are the causative genetic aberration in almost 60% of cases of pediatric acute myeloid leukemia (AML).1 Among these, rearrangements of the MLL1/KMT2A gene located on chromosome 11q23 are most frequent, accounting for almost 25% of pediatric and 50% of infant AML cases.1
The oncogenicity of MLL fusions has been investigated in various mouse models, including human CD34+ xenografts, which provided strong evidence that MLL fusion oncogenes are sufficient to transform human hematopoietic stem and progenitor cells (HSPC).2–4 These studies also highlighted the environmental influence on disease phenotype and the role of the MLL fusion partner on the overall oncogenicity. However, the results were mainly obtained using retroviral expression systems, which invariably express one fusion protein at non-physiological levels, and neglect the loss of the wild-type alleles of both involved genes and a potential contribution of the reciprocal product of the translocation.5,6 Investigation of MLL rearrangements via knock-in in human HSPC supported their oncogenicity at endogenous expression levels, however, the loss of one wild-type allele of each fusion partner, and potential involvement of a reciprocal fusion product, still remain elusive with this approach.4 It is therefore desirable to investigate the oncogenicity of bona fide MLL rearrangements in primary human HSPC both in vitro and in vivo, which can be facilitated with genome editing technologies.7–10
The CRISPR-Cas9 system has only recently been successfully established to modify functional human HSPC.11,12 It was shown to allow the generation of chromosomal inversions in vitro9,10,13 and in murine in vivo models,14 and may thus overcome efficacy hurdles that may have caused the failure of models based on transcription activator-like effector nucleases (TALEN).15
While attempts to recapitulate transformation of primary human HSPC by chromosomal rearrangements in vitro failed,15 and thereby raised the question of whether the efficacy of the technology or an inherent protection of HSPC against chromosomal rearrangements caused the failure,15,16 in vivo experiments with CRISPR-Cas9-induced chromosomal translocations in human HSPC may have the power to answer these open questions, but have not yet been accomplished.
Here we successfully generated chromosomal rearrangements (t[11;19]/MLL-ENL) in CD34+ HSPC, resulting in clonal outgrowth in vitro and monocytic and B-lineage leukemia in vivo, which we accomplished with an improved lentiviral CRISPR-Cas9 system to generate chromosomal rearrangements based on our former work.17 Furthermore, our study highlights the impact of non-cell-autonomous signals influencing not only the phenotype but the overall transformation of HSPC by MLL rearrangements. Thus, our study presents the first human de novo leukemia model with CRISPR-Cas9-engineered chromosomal translocations and highlights the power of this advanced approach.
Methods
Plasmids and viral particle production
A detailed description of the vector construction is provided in the Online Supplementary Methods. CRISPR-Cas9 target sites were selected using the CCTop selection tool.19 Lentiviral vectors were provided via Addgene (# 69146-69148, 69212, 69215, 89392-89395). Lentiviral particles were produced as described before.17
Reporter assay-based testing of the spacers
Reporter-based single-guide RNA (sgRNA) efficacy testing was performed as described elsewhere.17
Cell culture
Cell lines were maintained as described in the Online Supplementary Methods. Human CD34+ cells were isolated from cord blood using the CD34 MicroBead Kit (Miltenyi Biotec) according to the manufacturer’s instructions. The cord blood was provided with the parents’ consent by the Department of Gynecology and Obstetrics, Hannover Medical School, and experiments were approved by the local ethics committee. Details on CD34+ cell maintenance and transduction are described in the Online Supplementary Methods section.
Colony-forming unit assays were performed according to manufacturer’s instructions. Upon initial plating, 1×104 cells were seeded. For replating a total of 3×104 cells per plate were seeded.
Xenotransplantation
All animal experiments were approved by the local authorities. For transplantation, 4×105 CD34+ cells were pre-stimulated and transduced as described above. Twenty-four hours after transduction, the HSPC were transplanted intravenously into sublethally irradiated (2.5 Gy) male or female NSGS mice20 (age: 8–12 weeks).
Flow cytometry
Cells were stained with antibodies specific for human CD45 (APC, V500), CD33 (PE), CD14 (APC-Cy7), CD11b (APC), CD64 (PE), CD117 (PE-Cy7), CD19 (PE-Cy7), CD3 (APC-Cy7), CD10 (BV605), CD20 (V450), CD22 (APC), and CD34 (PerCP-Cy5.5) (all from BD Biosciences). Dead cells were excluded using 4′,6-diamidin-2-phenylindol (DAPI) counterstaining, where applicable. Cells were analyzed on a FACS Canto flow cytometer (BD Biosciences), and data were analyzed with FlowJo V10 software.
RNA extraction and polymerase chain reaction
Total RNA was purified from cells with the RNeasy Micro or Mini Kit (Qiagen), complementary DNA (cDNA) was synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems), according to the manufacturer’s instructions. The primers are summarized in Online Supplementary Table S3.
Genomic DNA extraction and polymerase chain reaction
Genomic DNA was extracted using the QIAmp DNA Mini or Micro Kit (Qiagen) according to the manufacturer’s protocols. Detection of the genomic breakpoint polymerase chain reaction was performed with Extensor 2x Master Mix (Thermo Scientific). Primers are summarized in Online Supplementary Table S3.
T7 endonuclease I based surveyor assay
Polymerase chain reaction primers containing the on-target and off-target of MLL and ENL are listed in Online Supplementary Table S3. The T7 endonuclease I (T7E-I) assay was performed according to the manufacturer’s protocols (New England Biolabs). Digested fragments were separated by DNA gel electrophoresis and imaged with a BioRad GelDoc™ XR+ imaging system. Absolute quantification of DNA fragments was done with the Image Lab 3.0 software (BioRad). The band intensity of the DNA marker (Thermo Scientific) was correlated to its specific amount of input, and subsequently, the absolute quantities of all DNA fragments were defined accordingly and the ratios of cut and uncut fragments were calculated.
Statistical analysis
Statistical analysis was performed with GraphPad Prism software (Graphpad Software).
Results
Advanced lentiviral dual single-guide RNA CRISPR-Cas9 vectors for the generation of chromosomal rearrangements
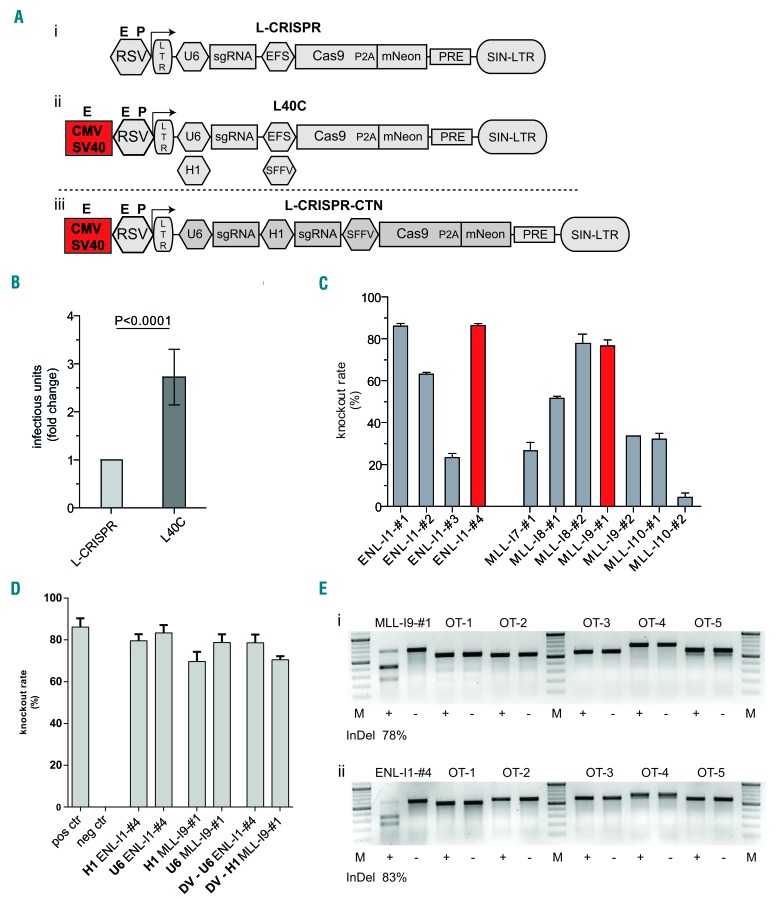
With the aim of elucidating the transformative nature of endogenous chromosomal rearrangements in primary human HSPC, we developed an advanced, all-in-one lentiviral CRISPR-Cas9 system with two sgRNA expression cassettes (L-CRISPR-CTN=CRISPR-Translocations-mNeon) (Figure 1A). We introduced an enhanced sgRNA backbone18 and enhancer elements to boost genomic RNA production,21 resulting in significantly higher titers as tested on multiple hematopoietic cell lines (Figure 1B; Online Supplementary Figure S1).
Figure 1.
An improved lentiviral vector system for generation of CRISPR-Cas9-induced chromosomal rearrangements. (A) Schematic presentation of lentiviral vector architecture including genomic RNA-generating promoter assembly: published architecture (L-CRISPR) (i), improved architecture with cytomegalovirus enhancer (CMV) and simian virus 40 enhancer (SV40) and exchangeability of the hU6 promoter for a H1 promoter (L40C) (ii), and lentiviral vector for dual sgRNA delivery (L-CRISPR-CTN (iii). (B) Analysis of viral titers in three independent cell lines with two different sgRNAs and three replicates each. (C) Knock-out efficacies (fluorescence reporter assay) of sgRNAs targeting intronic sequences of ENL and MLL. Selected sgRNAs are marked. (D) Knock-out efficacies of selected sgRNAs expressed from a human U6 or H1 promoter, as indicated. Knock-out efficacies of selected sgRNAs in L-CRISPR-CTN dual sgRNA configuration (DV) (neg ctrl= anti-luciferase (Luc) sgRNA, pos ctrl= Tet2 sgRNA). (E) T7-endonuclease-I assay for on-target sites and the top five predicted off-target sites in HEL cells (OT-1-OT-5). Indel frequencies at endogenous loci are indicated below. Analysis in cells transduced with targeting (+) and Luc (-) sgRNAs: MLL-I9-#1(i) and ENL-I1-#4 (ii).
Utilizing fluorescence reporter-based spacer testing (Online Supplementary Figure S2), we established highly efficient sgRNAs (>80% cleavage activity) targeting MLL/KMT2A and ENL intronic sequences to generate the t(11;19)/MLL-ENL translocation (Figure 1C; Online Supplementary Table S1). To prospectively achieve CRISPR-Cas9-induced chromosomal rearrangement from a single vector (Figure 1A), the knock-out efficacy of the various selected sgRNAs expressed from a H1 promoter was tested. Only minor, non-significant differences in the cleavage efficacy were detectable with sgRNA expression either from a H1 or U6 promoter (Figure 1D). Importantly, the activity of the sgRNA was retained in the dual sgRNA vector configuration (Figure 1D). No recombination of the integrated provirus was observed (Online Supplementary Figure S3), warranting high confidence delivery of both sgRNAs.
Off-target cleavage activity is a major concern with the application of genome editing, but is mostly abrogated by three or more mismatches between the spacer and protospacer.22,23 T7E-I assays for the top five off-target sites and the on-target sites of our pre-selected sgRNAs verified high on-target and no detectable off-target activity at the endogenous loci (Figure 1E, Online Supplementary Table S2).
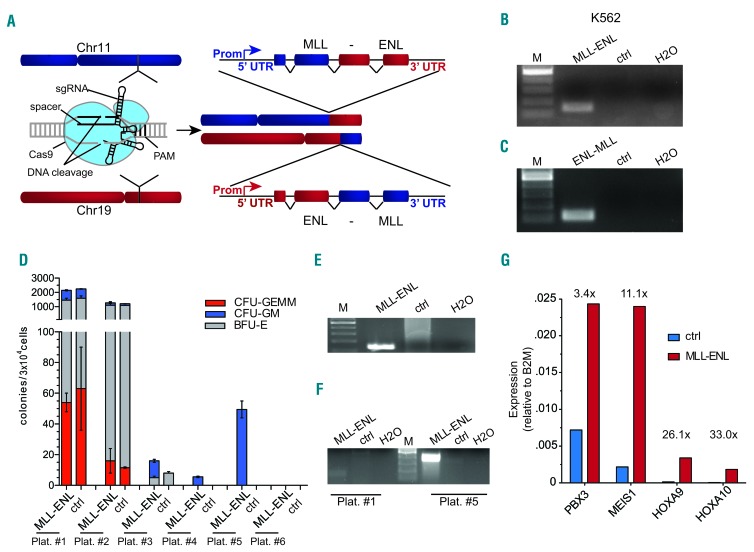
Based on these results, we tested the generation of chromosomal rearrangements [t[11;19]; L-CRISPR-CTN(11;19)] in hematopoietic cells (Figure 2A). In L-CRISPR-CTN(11;19)-transduced K562 cells, we could readily detect expression of the MLL-ENL and reciprocal ENL-MLL transcript in the bulk population (Figure 2B,C). Amplification of the genomic breakpoint and sequence analysis further verified polyclonal t(11;19) induction at the targeted genomic sequences (Online Supplementary Figure S4). We could thereby establish an all-in-one lentiviral CRISPR-Cas9 system for the efficient induction of chromosomal translocations (L-CRISPR-CTN).
Figure 2.
CRISPR-Cas9-induced MLL-ENL rearrangements cause clonal expansion of human CD34+ hematopoietic stem and progenitor cells. (A) Schematic depiction of CRISPR-Cas9-induced chromosomal rearrangements at the MLL and ENL loci. (B) Reverse transcriptase polymerase chain reaction-based detection of MLL-ENL transcript in K562 cells. Ctrl = MLL-I9-#1 + Luc sgRNAs. (C) RT-PCR-based detection of reciprocal ENL-MLL transcript in K562 cells. Ctrl = MLL-I9-#1 + Luc sgRNA. (D) Serial plating of CD34+ HSPC after transduction with L-CRISPR-CTN. (E) Detection of MLL-ENL expression in CD34+ HSPCs at fifth plating. Control (MLL-I9-#1 + Luc sgRNAs) template from third plating. (F) Detection of the genomic MLL-ENL breakpoint in CD34+ HSPCs at fifth plating (control (MLL-I9-#1 + Luc sgRNAs) template from second plating) compared to the first plating. (G) Analysis of MLL target genes in MLL-ENL-expressing cells (fifth plating) compared to controls (third plating). Differential regulation marked above. Ctrl = MLL-I9-#1 + Luc sgRNAs.
L-CRISPR-CTN-induced t(11;19) translocations increase the re-plating efficiency of primary human hematopoietic stem and progenitor cells in vitro
To determine the impact of endogenous t(11;19) on HSPC, we transduced cord blood-derived CD34+ HSPC (16.6±5.2%, n=10). FACS-sorted cells grown in methylcellulose were tested for MLL-ENL expression at the first re-plating and transcript identity was validated via sequencing (Online Supplementary Figure S5). Of note, vector expression was not monitored over time since genome editing does not rely on continuous expression of the Cas9 or the sgRNA. In three independent experiments, MLL-ENL messenger RNA (mRNA) was detectable, resulting in a rearrangement efficacy of at least 1.6×10−3 (±0.26×10−3). Upon serial re-plating we detected the extended plating capacity of t(11;19)-containing cells (Figure 2D), accompanied by robust expression of the MLL-ENL transcript (Figure 2E, Online Supplementary Figure S6) and detection of the genomic breakpoint (Figure 2F, Online Supplementary Figure S6). The latter was not detectable at the first re-plating, likely due to lower sensitivity at the genomic level compared to mRNA expression. Our experiments thus provide evidence that CRISPR-Cas9-induced t(11;19) translocations can provide self-renewal capacity to human HSPC. However, the cord blood CD34+ cells with t(11;19) formed normal hematopoietic colonies and eventually ceased proliferating. Results in methylcellulose were further supported by the transient clonal outgrowth of MLL-ENL-expressing cells in one out of four experiments performed in liquid culture (Online Supplementary Figure S7). Long-term tracking of MLL-ENL-expressing cells in other experiments without growth advantage indicates the need for additional stimuli to induce transformation (Online Supplementary Figure S8A,B).
When clonal outgrowth was observed it was accompanied by a robust up-regulation of known downstream effectors of MLL-ENL, such as HOXA9, HOXA10, MEIS1, and PBX3 (Figure 2G, Online Supplementary Figure S7D),24 which was absent in samples without clonal outgrowth (Online Supplementary Figure S8C). In contrast to former studies, genes associated with a leukemic stem cell phenotype in mice (CBX5, HMGB3, MYBL2)24 were not upregulated in samples with clonal outgrowth (Online Supplementary Figure S7D).
Overall, these experiments underline both the potential and insufficiencies of endogenous t(11;19) to modulate self-renewal and growth of human HSPC in vitro.
This is in line with a recent report on TALEN-induced MLL rearrangements in CD34+ HSPC in vitro.15 This cumulative evidence points towards a non-cell-autonomous component in cellular transformation by MLL rearrangements beyond phenotypic manifestation.
In vivo environment affects the oncogenic transformation of primary human hematopoietic stem and progenitor cells by t(11;19)
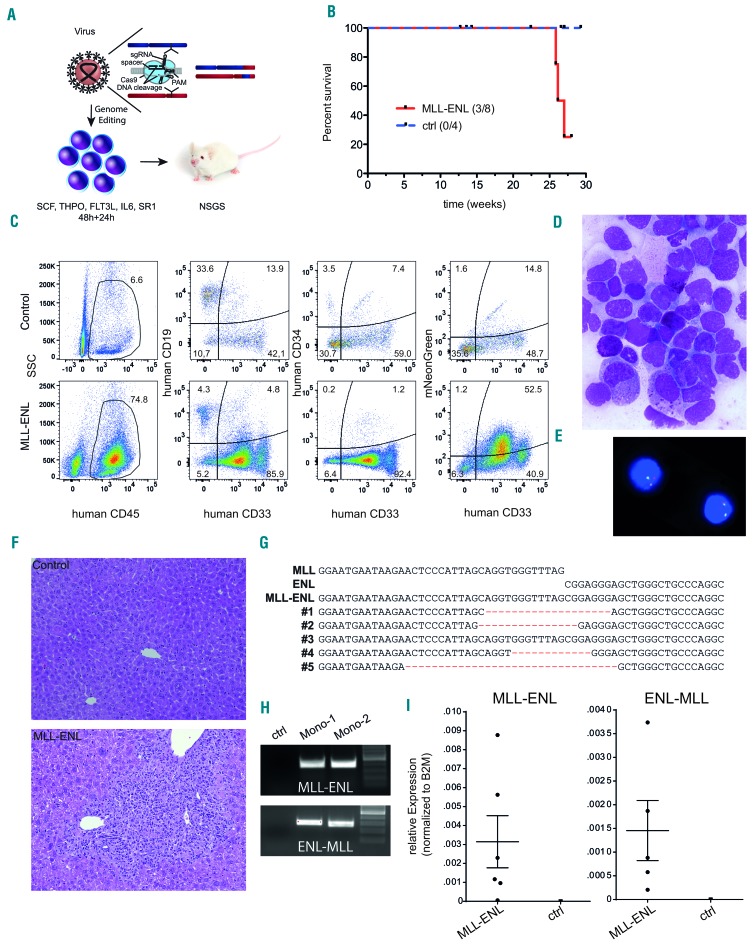
To further test this hypothesis, we performed transplantation of freshly-transduced, non-sorted L-CRISPR-CTN(11;19) and L-CRISPR-CTN(ctrl) CD34+ cells into immunodeficient mice (Figure 3A, Online Supplementary Figure S9). At weeks 25, 26, and 27 after transplantation, three moribund mice were analyzed (Figure 3B). One of the mice presented with intermediate human engraftment, mixed lineage reconstitution and only moderate expression of the vector backbone, excluding leukemia as the cause of sickness (Online Supplementary Figure S10AC). Two of the mice presented with a hCD45+CD33+ monocytic phenotype (Figure 3C, Online Supplementary Figure S10B), robust vector expression (Figure 3C, Online Supplementary Figure S10B), and the MLL rearrangement was confirmed by fluorescence in situ hybridization in cultured cells from one mouse (Figure 3E, MLL-split probe 7% positivity). An excess of monocytic cells with immature features in the bone marrow was verified by nuclear staining (>80%, Figure 3D). Severe infiltration of myeloid cells in the liver further supported hematologic disease (Figure 3F). Sequence analysis of the genomic MLL-ENL breakpoints detected in all analyzed mice (diseased and healthy, Sanger sequencing) indicated a clonal origin and outgrowth of t(11;19)-containing cells (Figure 3G). MLL-ENL and ENL-MLL breakpoints (Figure 3H), expression and identity of MLL-ENL and ENL-MLL transcripts (Figure 3I, Online Supplementary Figure S11), and presence of human cells expressing the introduced vector were confirmed (Figure 3C, Online Supplementary Figures S10 and S12A). Notably, while no correlation between MLL-ENL expression and human cell content was observed, MLL-ENL expression significantly correlated with the percentage of vector-expressing cells, further supporting our approach (Online Supplementary Figure S12B,C).
Figure 3.
CRISPR-Cas9-induced MLL-ENL rearrangements are leukemogenic in a CD34+ hematopoietic stem and progenitor cell xenotransplantation model. (A) Schematic depiction of the xenotransplantation model. (B) Survival of mice transplanted with L-CRISPR-CTN-transduced CD34+ HSPC. Mice that succumbed to non-hematopoietic disease were censored and are indicated (ticked). (C) Flow cytometric analysis of one mouse with hematopoietic disease compared to a control, with markers as indicated (ctrl = MLL-I9-#1 + Luc sgRNAs). (D) Bone marrow (BM) cytospin analysis of a diseased mouse (MGG, 1000X). (E) Detection of an MLL translocation with fluorescence in situ hybridization on interphase nuclei (Vysis LSI MLL probe; Abbott Laboratories) in BM cells from one diseased mouse. (F) Histopathological analysis of liver tissue from a healthy control mouse (ctrl = MLL-I9-#1 + Luc sgRNA) (top) and a mouse with monocytic leukemia-like disease transplanted with L-CRISPR-CTN(11;19)-containing CD34+ HSPC (bottom) (HE, 100x). (G) Alignment of Sanger sequencing-derived genomic t(11;19)/MLL-ENL breakpoints of mice with a detectable MLL-ENL breakpoint. (H) MLL-ENL and ENL-MLL genomic breakpoints detected in the BM of two mice with a monocytic leukemia-like disease. (I) Expression of the MLL-ENL (left) and reciprocal ENL-MLL (right) fusion genes, measured by quantitative polymerase chian reaction from the BM of mice with a detectable MLL-ENL breakpoint compared to control mice (MLL-I9-#1 + Luc sgRNAs).
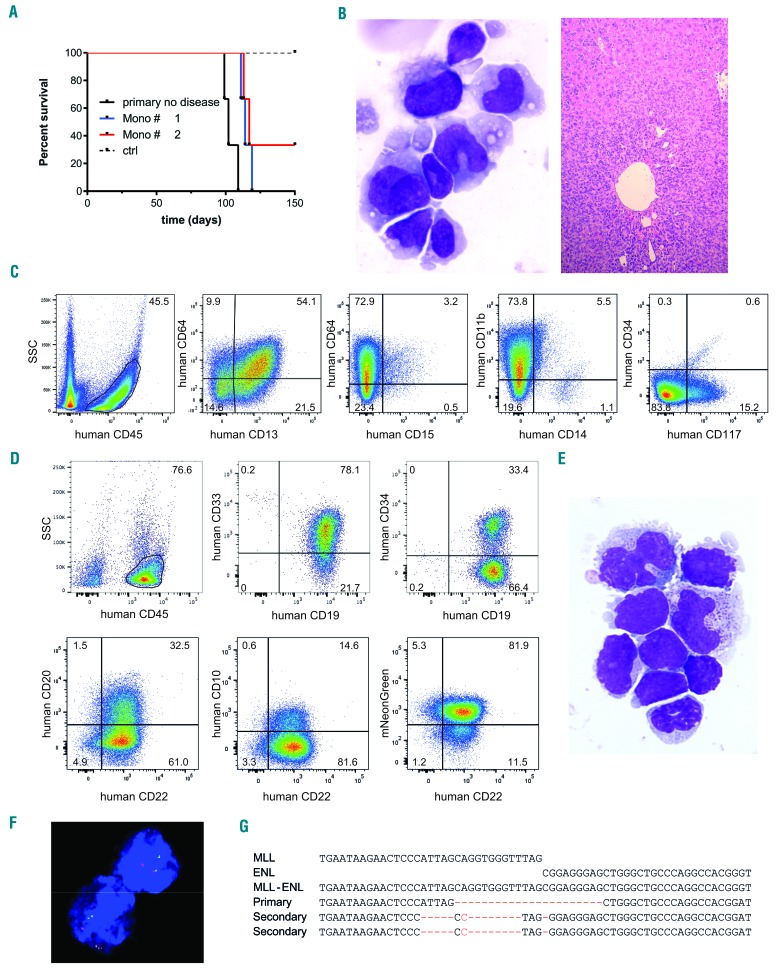
To validate a malignant phenotype, secondary transplantation of the monocytic samples was performed, assuming that healthy committed myelo-monocytic progenitors would fail to engraft, which was indeed the case for two control mice transplanted with L-CRISPR-CTN(ctrl)-transduced CD34+ cells (Figure 4A). Surprisingly, five out of six recipients from both donors with a monocytic phenotype presented with disease symptoms 15–16 weeks after transplantation (Figure 4A). The immature monocytic phenotype was confirmed, with CD64 expression, absence of CD14 and CD15 expression, and partial CD117 expression by flow cytometric analysis, as well as by morphological analyses of hepatic infiltration (Figure 4B,C, Online Supplementary Figure S13). Of note, while disease latency was slightly shortened (25–26 weeks versus 16–17 weeks), the absence of blast morphology and CD34 expression indicated no progression of the monocytic leukemia-like disease to a more immature AML phenotype, despite the transplantation-induced stress.
Figure 4.
In vivo environment affects the oncogenic transformation of primary human hematopoietic stem and progenitor cells by t(11;19) (A) Survival of serially transplanted mice with human cell engraftment. Donor: primary recipients with [L-CRISPR-CTN(11;19)] a monocytic leukemia-like disease (Mono #1/#2), a healthy mouse with a detectable MLL-ENL breakpoint but no disease in the primary recipient, and two control donors (MLL-I9-#1 + Luc sgRNAs) (n = 3 per donor). (B) Analysis of bone marrow (BM) cell morphology (left: MGG, 1000x) and liver histopathology (right: HE, 100X) showing severe infiltration of a secondary recipient transplanted with monocytic leukemia-like disease cells. (C) Flow cytometry analysis of monocytic leukemia cells for monocytic and progenitor cell surface marker expression. (D) Flow cytometry analysis of B-ALL cells. (E) Analysis of BM cell morphology (MGG, 1000x) of a secondary recipient with B-ALL. (F) Detection of an MLL translocation with fluorescence in situ hybridization on interphase nuclei of B-ALL cells (Vysis LSI MLL probe; Abbott Laboratories). (G) Alignment of Sanger sequencing-derived genomic t(11;19)/MLL-ENL breakpoints of mice with B-ALL (Secondary) compared to the healthy primary (Primary) mouse.
Next, we asked whether EVI1 expression – an indicator of stem cell origin in MLL-rearranged leukemia25–27 – and known downstream MLL targets were deregulated in the mice. While EVI1 was not detectable in our samples or MLL-rearranged control cell lines, we found upregulation of PBX3 in the majority of mice. Some samples also showed upregulation of HOXA10, MEIS1, and MYB (Online Supplementary Figure S14).
Our experiments thereby provide compelling evidence that endogenous t(11;19) can initiate a monocytic leukemia-like phenotype in our xenotransplantation setting, but lacks the capacity to initiate an immature AML. The detection of t(11;19) in non-diseased mice also strongly indicates that additional events are either required for the development of frank leukemia or influence the latency of disease appearance.
To test this hypothesis of inherent stress and environmental cues as contributing factors in disease manifestation caused by t(11;19), we harvested bone marrow from primary recipients freshly-transplanted with L-CRISPR-CTN(11;19)- and L-CRISPR-CTN(ctrl)-transduced CD34+ cells 8–10 weeks after transplantation – before the onset of leukemia – and transplanted the bone marrow into secondary recipients. Supposedly-clonal genomic MLL-ENL breakpoints were detectable in the bone marrow of two out of three mice at the time of transplantation (Online Supplementary Figure S15).
Indeed, we monitored human cell engraftment in the secondary animals and observed rapid expansion indicating leukemic transformation (Figure 4A). Detailed analysis of the cells revealed the development of B-cell acute lymphoblastic leukemia (B-ALL) with expression of CD19, CD22, CD33 and partial expression of CD20 and CD34 (Figure 4D). Of note, the expression of CD33 is a common feature of B-ALL cells.28,29
The occurrence of B-ALL was further confirmed by morphological analysis of bone marrow cells showing an excess of blasts (Figure 4E). The presence of the MLL rearrangement was additionally verified by fluorescence in situ hybridization (Figure 4F, MLL-split probe 67% positivity). Interestingly, the genomic MLL-ENL breakpoint revealed that the leukemic clone developed independently from the dominant clone in the primary recipients, further supporting our hypothesis that environmental signals may alter the transformation of MLL-ENL-harboring cells (Figure 4G). Utilizing serial transplantation and tracking of NHEJ-scarring at genomic breakpoints, as with our system, potentially provides a tool to investigate the clonal evolution of leukemia with potentially variable stress conditions, such as drug treatments, in vivo.
Discussion
By leveraging CRISPR-Cas9 genome editing, we induced bona fide chromosomal rearrangements in primary human HSPC in vivo, resulting in a de novo leukemia-like disease reflecting all the oncogenic properties of MLL-rearrangements: namely the combined, endogenous expression of both resulting fusion mRNA and concurrent loss of one wild-type allele of each fusion partner. To achieve efficient genome editing in CD34+ HSPC we improved our formerly constructed lentiviral CRISPR-Cas9 delivery system17 for the generation of chromosomal translocations and transduction of cord blood-derived CD34+ HSPC. Successful transduction of human HSPC with our system reproducibly resulted in both MLL-ENL and ENL-MLL mRNA expression, providing strong evidence for the formation of true t(11;19)/MLL-ENL translocations. Intriguingly, functional studies revealed only a transient and variable growth advantage affecting the minority of cultures. No full transformation was observed in liquid cultures or methylcellulose-based in vitro assays using CD34+ HSPC, which is in line with a recent study utilizing TALEN to induce MLL translocations or knock-in of AF9/ENL cDNA into the MLL locus in CD34+ HSPC.4,15 With MLL-ENL causing cellular senescence32 and an intrinsic protection of early HSPC against MLL-ENL transformation,16 MLL rearrangements may only serve as a first hit insufficient to cause full transformation without additional oncogenic events. Isolation of clonal lines and testing of additional cytokine combinations may help to understand the lack of in vitro transformation in consecutive studies.
Leukemic transformation in patients lacking additional known driver mutations34 prompted us to hypothesize that environmental cues may not only affect the phenotype of MLL rearrangement-induced leukemia,3 but may also determine the overall transformation capacity. Of note, MLL rearrangements are particularly frequent in pediatric and infant AML,1 in which both the environment and the cell of origin differ from those in adults.35,36 The latter was already shown to affect the transformation capacity of retrovirally expressed MLL-AF9.36 Supporting the idea that yet unknown external stimuli of the in vivo environment alter transformation permissiveness, we noted successful engraftment and persistence of t(11;19)-harboring cells after long-term observation, exceeding the time of the in vitro experiments. This engraftment culminated in the development of a monocytic leukemia-like disease in primary recipients, contrasting with the lack of transformation in vitro, despite similar input cell numbers. Taking into account that leukemic transformation exceeded culture time about 3-fold, the lack of supportive signals for maintaining a pre-leukemic cell in vitro, or a prolonged time-frame for the acquisition of additional genetic lesions in vivo could explain the observed phenomenon, although similar conditions have been used to transform human HSPC with retroviral overexpression of MLL fusion oncogenes.2,3 The development of B-ALL in secondary recipients of a healthy primary donor may either indicate a disturbance of cellular homeostasis contributing to leukemic outgrowth, or result from B-ALL having a higher proportion of leukemia-initiating cells.39 The detection of MLL rearrangements in mice without leukemia-like disease in the same time-frame strongly indicates a progressive acquisition of cooperating mutations needed for full leukemic transformation. Sequencing of NRAS and KRAS hotspots, which are frequently mutated in MLL-rearranged leukemia,34 did not reveal any mutations (data not shown). More comprehensive sequencing to detect additional oncogenic hits, tracking of clonal t(11;19) isolates in serial transplantations, and a larger cohort are required to identify cooperating events in the future.
Former studies have made major contributions to the understanding of leukemogenesis induced by MLL fusion oncogenes.2,3,24 However, several aspects may have limited the grasp on MLL rearrangement-guided leukemogenicity and could hamper the development of novel treatment options. First, retroviral expression of MLL fusion oncogenes poses the risk of overexpression-induced phenotypes, which may be amplified by the use of murine models over human xenotransplantation models.2,3 To overcome these limitations of retroviral studies, another recent study has utilized TALEN-induced DNA-DSB-supported knock-in of AF9 and ENL cDNA into the MLL locus in HSPC, thereby recapitulating endogenous expression of the fusion oncogene.4 Yet, lack of expression of the reciprocal fusion gene, in some cases shown to be essential for the phenotype,5 loss of one wild-type allele of the fusion partner, potential effects of the translocation of enhancer elements,40 and microRNA binding sites in the untranslated region of the fusion partner can only be covered by the induction of true chromosomal translocations, as shown in our study. Furthermore, an excess of input cells insufficiently reflecting the clonal disease development and homeostatic mechanisms regulating clonal outgrowth in patients could have influenced the outcomes of former studies, which may be indicated by our detection of chromosomal translocations in mice not showing disease. Notably, CRISPR-Cas9-induced NHEJ-scaring of the chromosomal breakpoints allowed tracking of clonal fluctuation and revealed a clonal shift during leukemic outgrowth in secondary recipients.
Our study reporting a humanized, CRISPR-Cas9-induced cancer model with in vivo transformation uncovers new aspects of the oncogenic potency and limitations of endogenous MLL rearrangements in human HSPC, and advances disease modeling closer to patient-specific disease progression than before. Leveraging our approach to generate precise cancer models with the CRISPR-Cas9 system will allow more detailed analysis of potential homeostatic mechanisms and oncogenic cooperations during leukemic transformation, as well as the development of targeted therapies and investigation of drug resistance mechanisms.
Supplementary Material
Acknowledgments
We thank Dr. Dr. A. Schambach and Michelle Ng for discussions and D. Trono of EPFL, Lausanne, Switzerland, for kindly providing both pMD2.G (Addgene plasmid 12259) and psPAX2 (Addgene plasmid 12260). DH is supported by the German Cancer Aid (111743). JHK is a fellow of the Emmy Noether-Programme from the DFG (KL-2374/2-1). This work was supported by grants to DH and JHK from the DFG (HE-7482/1-1, KL-2374/2-1; KL-2374/1-3) and to GG and BS for the Cluster of Excellence REBIRTH (DFG, EXC 62/3). JR and ML were supported by the Hannover Biomedical Research School.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/102/9/1558
References
- 1.Creutzig U, van den Heuvel-Eibrink MM, Gibson B, et al. Diagnosis and management of acute myeloid leukemia in children and adolescents: recommendations from an international expert panel. Blood. 2012;120(16):3187–3205. [DOI] [PubMed] [Google Scholar]
- 2.Barabe F, Kennedy JA, Hope KJ, Dick JE. Modeling the initiation and progression of human acute leukemia in mice. Science. 2007;316(5824):600–604. [DOI] [PubMed] [Google Scholar]
- 3.Wei J, Wunderlich M, Fox C, et al. Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia. Cancer Cell. 2008;13(6):483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buechele C, Breese EH, Schneidawind D, et al. MLL leukemia induction by genome editing of human CD34+ hematopoietic cells. Blood. 2015;126(14):1683–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bursen A, Schwabe K, Ruster B, et al. The AF4.MLL fusion protein is capable of inducing ALL in mice without requirement of MLL.AF4. Blood. 2010;115(17):3570–3579. [DOI] [PubMed] [Google Scholar]
- 6.Chen W, Kumar AR, Hudson WA, et al. Malignant transformation initiated by Mll-AF9: gene dosage and critical target cells. Cancer Cell. 2008;13(5):432–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piganeau M, Ghezraoui H, De Cian A, et al. Cancer translocations in human cells induced by zinc finger and TALE nucleases. Genome Res. 2013;23(7):1182–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torres R, Martin MC, Garcia A, Cigudosa JC, Ramirez JC, Rodriguez-Perales S. Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR-Cas9 system. Nat Commun. 2014;5:3964. [DOI] [PubMed] [Google Scholar]
- 10.Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun. 2014;5:3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandal PK, Ferreira LM, Collins R, et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell. 2014;15(5): 643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gundry MC, Brunetti L, Lin A, et al. Highly Efficient genome editing of murine and human hematopoietic progenitor cells by CRISPR/Cas9. Cell Rep. 2016;17(5):1453–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanden Bempt M, Demeyer S, Mentens N, et al. Generation of the Fip1l1-Pdgfra fusion gene using CRISPR/Cas genome editing. Leukemia. 2016;30(9):1913–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blasco RB, Karaca E, Ambrogio C, et al. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep. 2014;9(4):1219–1227. [DOI] [PubMed] [Google Scholar]
- 15.Breese EH, Buechele C, Dawson C, Cleary ML, Porteus MH. Use of genome engineering to create patient specific MLL translocations in primary human hematopoietic stem and progenitor cells. PLoS One. 2015;10(9): e0136644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ugale A, Norddahl GL, Wahlestedt M, et al. Hematopoietic stem cells are intrinsically protected against MLL-ENL-mediated transformation. Cell Rep. 2014;9(4):1246–1255. [DOI] [PubMed] [Google Scholar]
- 17.Heckl D, Kowalczyk MS, Yudovich D, et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol. 2014;32(9):941–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen B, Gilbert LA, Cimini BA, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155(7):1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stemmer M, Thumberger T, Del Sol Keyer M, Wittbrodt J, Mateo JL. CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One. 2015;10(4): e0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wunderlich M, Chou FS, Link KA, et al. AML xenograft efficiency is significantly improved in NOD/SCID-IL2RG mice constitutively expressing human SCF, GM-CSF and IL-3. Leukemia. 2010;24(10):1785–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schambach A, Mueller D, Galla M, et al. Overcoming promoter competition in packaging cells improves production of self-inactivating retroviral vectors. Gene Ther. 2006;13(21):1524–1533. [DOI] [PubMed] [Google Scholar]
- 22.Fu Y, Foden JA, Khayter C, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu PD, Scott DA, Weinstein JA, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Somervaille TC, Matheny CJ, Spencer GJ, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4(2):129–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arai S, Yoshimi A, Shimabe M, et al. Evi-1 is a transcriptional target of mixed-lineage leukemia oncoproteins in hematopoietic stem cells. Blood. 2011;117(23):6304–6314. [DOI] [PubMed] [Google Scholar]
- 26.Bindels EM, Havermans M, Lugthart S, et al. EVI1 is critical for the pathogenesis of a subset of MLL-AF9-rearranged AMLs. Blood. 2012;119(24):5838–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stavropoulou V, Kaspar S, Brault L, et al. MLL-AF9 expression in hematopoietic stem cells drives a highly invasive AML expressing EMT-related genes linked to poor outcome. Cancer Cell. 2016;30(1):43–58. [DOI] [PubMed] [Google Scholar]
- 28.Mejstrikova E, Kalina T, Trka J, Stary J, Hrusak O. Correlation of CD33 with poorer prognosis in childhood ALL implicates a potential of anti-CD33 frontline therapy. Leukemia. 2005;19(6):1092–1094. [DOI] [PubMed] [Google Scholar]
- 29.Suggs JL, Cruse JM, Lewis RE. Aberrant myeloid marker expression in precursor B-cell and T-cell leukemias. Exp Mol Pathol. 2007;83(3):471–473. [DOI] [PubMed] [Google Scholar]
- 30.Takacova S, Slany R, Bartkova J, et al. DNA damage response and inflammatory signaling limit the MLL-ENL-induced leukemogenesis in vivo. Cancer Cell. 2012;21(4):517–531. [DOI] [PubMed] [Google Scholar]
- 31.Andersson AK, Ma J, Wang J, et al. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet. 2015;47(4):330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132(4):631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horton SJ, Jaques J, Woolthuis C, et al. MLL-AF9-mediated immortalization of human hematopoietic cells along different lineages changes during ontogeny. Leukemia. 2013;27(5):1116–1126. [DOI] [PubMed] [Google Scholar]
- 34.Rehe K, Wilson K, Bomken S, et al. Acute B lymphoblastic leukaemia-propagating cells are present at high frequency in diverse lymphoblast populations. EMBO Mol Med. 2013;5(1):38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Groschel S, Sanders MA, Hoogenboezem R, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157(2): 369–381. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.