Abstract
Early detection of acute kidney injury is difficult due to lack of known biomarkers; previous studies have tried to identify new biomarkers for detecting acute kidney injury at an early stage. MicroRNA, a 21-23 nucleotide noncoding RNA molecule, has emerged as a desirable marker in the diagnosis and prognosis of various diseases. This study aims to identify the expression profile of microRNA in ischemia–reperfusion-induced kidney injury and determine the possibility of using the candidate microRNA as biomarker for the detection of I/R-induced kidney injury. Based on the established rat model of I/R-induced kidney injury, a microarray analysis of rat urine was performed at the beginning of operation (0 h) as well as 72 h post operation. To validate the results, urine samples from 71 patients who underwent cardiac surgery were collected, after which urinalysis was conducted to determine the microRNA concentration. An alternative expression profile of microRNAs was detected in rat urine. The quantitative validation of microRNA showed that the expression of miR-30c-5p, miR-192-5p, and miR-378a-3p was elevated significantly in urine post operation, which was consistent with those of the microarray analysis and earlier than kidney injury molecule-1 (KIM-1). In patients with acute kidney injury, increased levels of miR-30c-5p and miR-192-5p were also detected 2 h post operation, and miR-30c-5p showed preferable diagnostic value compared with protein-based biomarkers. In conclusion, an aberrant expression profile of microRNA was detected in rat urine based on the established ischemia–reperfusion animal model. Both miR-30c-5p and miR-192-5p served as important potential diagnostic markers for I/R-induced kidney injury.
Impact statement
Firstly, one differentiating factor in our study is that the candidate miRNAs were screened in a controlled animal model rather than in patients with acute kidney injury (AKI) to ensure the purity of the cause of disease and to avoid possible effects of comorbidities on the spectrum of urine miRNA. This ensured the presence of only the relevant candidate miRNA (that associated with I/R injury); and what’s more, the alterative expression of miR-192-5p and miR-30c-5p in animal model, patients with AKI, and cell model was confirmed simultaneously, which is likely to be more convincing. Secondly, the candidate miRNAs were screened sequentially at regular time points, which covered the initiation, progression, and partial repair stages, thus ensuring that no significant miRNAs were omitted in the screening process, and miR-biomarkers in 2 h post operation showed preferable diagnostic performance.
Keywords: Ischemia/reperfusion, kidney injury, microRNAs, biomarkers
Introduction
Acute kidney injury (AKI) is a clinical syndrome with high morbidity and mortality rates. The incidence rate of AKI has steadily been increasing in recent years. Its incidence varies under different clinical settings such as cardiac surgery, intensive care unit, community, etc.1 Several studies have examined the short-term and long-term outcomes of patients with an episode of AKI. The results indicate that a sharp decline in renal function often leads to negative outcomes. Patients requiring renal replacement therapy have higher mortality rates and their condition may progress to chronic kidney disease (CKD).2,3 Nevertheless, AKI is a common clinical condition associated with a number of adverse outcomes. Early diagnosis of this condition could increase the possibility of earlier intervention and, thus, could improve prognosis of AKI patients.4–7
Serum creatinine (SCr) is a marker of renal function; however, kidney injury usually cannot be detected at an early stage because there may not be significant changes in the SCr levels. Much work has been done to search for earlier biomarkers for diagnosis of AKI. Several biomarkers such as neutrophil gelatinase-associated lipocalin (NGAL) and kidney injury molecule-1 (KIM-1) have been thought to provide earlier diagnosis or prognosis of AKI.8–10 Urinary tissue inhibitor of metalloproteinases-2 (TIMP-2) and insulin-like growth factor-binding protein 7 (IGFBP7) were identified as indicators for AKI occurrence or renal recovery.11–13 Unfortunately, increasing evidence indicates that reliable biomarkers sensitive to AKI and specific to etiology are limited and need to be further established.
miRNA was discovered in Caenorhabditis elegans in 1993. It is a 21-23-nucleotide noncoding RNA molecule that plays a negative role in regulating gene expression.4,14 Recently, accumulative evidence supports that miRNA is a favorable biomarker for detecting various diseases at an early stage.15,16 However, the expression profiles of these biomarkers varied in different cohorts.4,17–22 It is a well-known fact that circulating miRNA is very sensitive as diagnostic marker, because it could be affected by the etiology and the underlying disease. Therefore, the aim of this study is to investigate whether miRNA can be used as an early indicator to detect I/R-induced kidney injury on the control of the confounding factors.
Methods and materials
Ethics
The Ethics Committee for Animal Care and Use of Research Center for Experimental Medicine of Ruijin Hospital, Shanghai, China, approved the study techniques involving rats. The experimental procedures involving the samples of patients with AKI were approved by the Ruijin Hospital Ethics Committee, Shanghai Jiao Tong University School of Medicine. The written informed consent letter included the consent to publish from each participant in this study.
I/R rat model and sample collection
Eight-week-old male Sprague-Dawley rats weighing 180 to 200 g were purchased from the Animal Center of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China. A 45-min surgery involving bilateral I/R of renal pedicle vessel was performed according to a previously described procedure.23,24 The control animals underwent a sham surgery, wherein the other procedures were the same as in the I/R group.
The microarray analysis involved experiments using 42 rats, which were divided into two groups, the I/R group (21 rats) and the sham group (21 rats). Using metabolic cages, the urine of the rats was collected prior to the surgery at 0 h, and also following the surgery at 2, 6, 12, 24, 48, and 72 h. The urine did not pooled in microarray analysis. Because rats produce small volumes of urine, which would have limited urine RNA for quantitation in the I/R group, urine was also collected from another set of 15 rats for subsequent real-time polymerase chain reaction (PCR) validation. The urine was then pooled from a set of five rats, so that statistical analysis can be performed on three sets at every time point in real-time PCR experiments. The schematic diagram of urine collection (for microarray analysis and real-time PCR) has been shown in Figure S1.
After urine collection, blood samples of 0.3 to 0.5 mL were collected from the hearts at each time point for the detection of SCr. Blood samples were also collected from the abdominal aorta of the rats under the effect of the isoflurane inhale anesthesia, following which the rats were sacrificed and the kidney cortex harvested. Anesthesia was administered prior to the surgery at 0 h and at 2, 6, 12, 24, 48, and 72 h following the surgery. Both urine and blood samples were centrifuged at 1500 × g for 10 min at 4℃; then the supernatant was stored at –80℃. The kidney cortex was fixed in 10% neutral buffered formalin solution for histopathological staining.
Blood and urine samples of 71 patients who underwent cardiovascular surgery in the intensive care unit were collected to validate results from animal experiments. Urine samples were obtained prior to the surgery at 0 h and at 2, 6, 12, 24, 48, and 72 h following the surgery. Blood samples were obtained prior to the surgery at 0 h and at 6, 24, 48, and 72 h following surgery. The urine samples were obtained using a urinary catheter. The urine and blood samples were centrifuged at 1500 × g for 10 min at 4℃ and the supernatant was stored at –80℃.
Examination of renal function and classification of AKI stages in patients with AKI
The SCr levels of rats were detected using liquid chromatography-mass spectrometry (LC-MS/MS) (Agilent 1200 Liquid Chromatography System, Santa Clara, California, USA).23 Firstly, 50 µL of rat serum sample and 5 µL of internal standard were prepared in each tube. Then, 200 µL of acetonitrile was added to each tube and vortex mixing and centrifugation of the suspension were performed. The supernatants were then transferred into sample vials and 5 µL of which was injected and introduced into the LC-MS/MS system. Subsequently, the LC-MS/MS system was used for detecting the SCr levels as per the manufacturer’s instruction.
The SCr levels of patients were detected in a picric acid-based colorimetric autoanalyzer (DxC 800 Autoanalyzer; Beckman Instruments, Germany) at 0, 6, 24, 48, and 72 h. The AKI stages were then defined and classified based on the SCr levels according to the 2012 guidelines of Kidney Disease: Improving Global Outcomes (KDIGO).25 Glomerular filtration rate (GFR) was estimated using the equation of EPI (Epidemiology Collaboration-CKD), as reported in an earlier study.26 The information about demographics, types of surgery, laboratory examination, and comorbidities was obtained from the electronic medical system. Urine concentration of NGAL (NGAL) and KIM-1 was measured by the enzyme-linked immune sorbent assay (ELISA) using the commercially available ELISA test kit (Westang, Shanghai, China) according to the manufacturer’s instructions.
miRNA profiling
Rattus norvegicus GeneChip® miRNA 3.0 Array (Affymetrix, complete coverage of miRBase v17 containing 680 mature miRNA probe sets) was used for detecting the expression profile of miRNA in rat urine.27 Total RNA was extracted from urine using miRNeasy Serum/Plasma Kit (Qiagen, Germany) according to the manufacturer’s instructions. Subsequently, 350 ng of total RNA was used as input in labeling reaction, following which the labeled miRNA was hybridized onto the array for 16 h at 48℃ and 60 r/min as described in the manufacturer’s protocol. This was followed by washing, staining, and scanning of the genechip using the Hewlett-Packard GeneArray Scanner G3000-7. The expression data were generated using the Affymetrix Expression Console software and normalized by the MAS5 method. The candidate miRNA was chosen for subsequent validation according to the P value and fold change.
RNA extraction, reverse transcription, and real-time quantitative PCR
Total RNA was extracted from urine using the miRNeasy Serum/Plasma Kit (Qiagen) according to the manufacturer’s instructions.28 In all, 400 µL of urine was used to extract total RNA containing miRNAs. Then, 7 µL of C. elegans miR-39 miRNA mimic (1.6 × 108 copies, Qiagen) was used for normalizing the extraction procedures right at the beginning. In addition, 30 µL of RNAase-free water was used for dissolving the precipitate.
To verify the expression profile of miRNA in rat urine, Taqman-based quantitative PCR was performed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, CA, USA) and TaqMan MicroRNA Assay (Assay ID for miR-30c-5p 000419/4427975, miR-192-5p 000491/4427975, and miR-378a-3p 002243/4427975, Applied Biosystems).29 A fixed volume of 2 µL RNA sample (scaled-down reaction system) was used to perform the procedure of reverse transcription (RT) followed by continued PCR amplification at a three-time dilution of the RT production. In the following procedure, real-time PCR was performed using the Taqman PCR Kit (Applied Biosystems) and Applied Biosystems® ViiA™ 7 Real-Time PCR System (Applied Biosystems, CA, USA) as per the manufacturer’s protocol. C. elegans miR-39 miRNA mimic, which was spiked at the beginning of the extraction, was used as a normalizer to determine the relative expression change in the 2−ΔCt method.
Histopathology
Hematoxylin-eosin staining was used to determine the degree of tissue damage. The kidney tissues of rats were fixed in 10% neutral buffered formalin solution for 24 h, which was then dehydrated in graded alcohol and finally embedded in paraffin. Subsequently, 4 µm sections were cut from these kidney tissues. Finally, they were stained with hematoxylin-eosin (Baso, Zhuhai, China). The tissue sections were then viewed and images captured with a spot-cam digital camera (Carl Zeiss, Germany). The pathological damage was then assessed for three aspects: (1) degeneration, necrosis, and vacuolation (2) infiltration of inflammation, and (3) tubular atrophy; these aspects were scored according to the following criteria: no lesion, 0; lesions up to 25%, 1; more than 25% and less than 50%, 2; and more than 50%, three points. Two pathologists blinded to the experimental procedures assessed the slides independently. The sum of the scores given by the two pathologists was used as the final evaluation result for each slide.
Cell culture and hypoxia-reoxygenation cell model
HK-2 (human kidney tubular epithelial cell, cortex/proximal tubule) was purchased from ATCC (CRL-2190) and cultured in Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F-12; Gibco), with 10% fetal bovine serum (FBS), Gibco, in an incubator (thermo electron) with 5% CO2 at 37℃. In the process of hypoxia, the cells suffered from a 24-h serum starvation (DMEM only, no FBS), following which it was cultured in a hypoxia incubator (1% O2, 5% CO2, and 94% N2, Thermo Electron) in a 60% to 80% confluent monolayer for 24 h. For reoxygenation, the medium was changed with fresh DMEM/F-12 containing 10% FBS, and the culture flask was moved to normoxia incubator.
Bioinformatics
The miRNA expression profile was screened as follows: a differential expression of miRNAs was obtained at 2, 6, 12, 24, 48, and 72 h using 0 h as the baseline; the expression of miRNA was detected in urine using Affymetrix TAC v1.0 software (CA, USA). One-way ANOVA (unpaired) between subjects was used to input the algorithm. A table containing miRNA as the first row name and the sample label as the column name was drawn; a fold change ratio and P value as value per miRNA per sample were used. Then, a value was set accordingly and recorded as 1 when the sample met the following criteria: upregulation or downregulation of miRNA > 2-fold change (linear) and ANOVA P < .05, otherwise recorded as 0. Furthermore, the total score of the value at each miRNA was counted. The miRNA was considered as a candidate when its total score is ≥3. In this study, a candidate miRNA means that it meets the criterion of upregulation or downregulation of miRNA > 2-fold change (linear) and ANOVA P < .05 for three or more time points.
Statistics
Continuous variables were described as mean ± standard error (SE) for normally distributed variables or median; the interquartile ranges were used for non-normally distributed variables. The normal distribution of the data was evaluated using the Kolmogorov–Smirnov test. Then, the categorical variables were summarized as frequencies with percentages, which were then compared by Pearson’s χ2 analysis or Fisher’s exact test as appropriate. All tests were two-tailed, and the α-level was set as 0.05. SPSS Statistical Software (Version 13.0, SPSS, Inc.) was used to conduct the analysis. Finally, receiver operating characteristic (ROC) curve was generated using SPSS, and other graphs were generated using the Graphpad Prism Software (Graphpad Software Version 6.0c, CA, USA).
Results
Rats showed an evident kidney injury after the I/R surgery
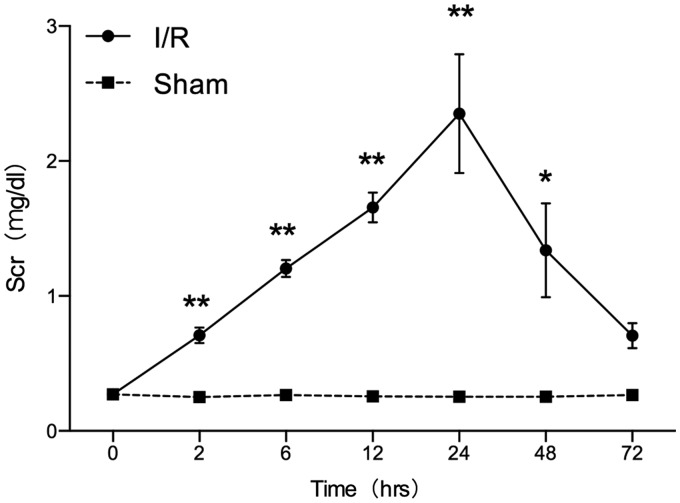
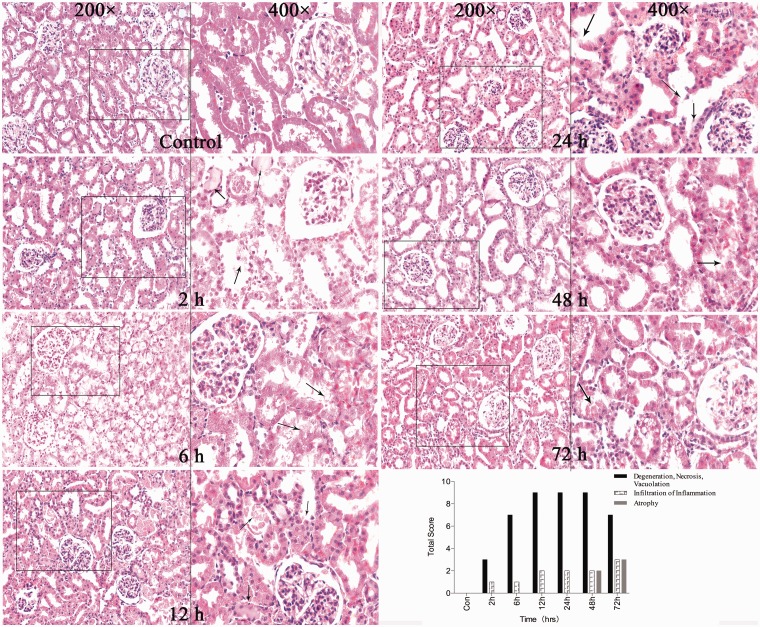
To investigate whether the I/R-induced AKI is associated with miRNA, an I/R-induced AKI animal model was established. The surgery induced an evident increase in the SCr levels. The results of this experiment indicate that the SCr levels increased in animals at 2 h after surgery; it peaked at 24 h and returned to baseline at 72 h as shown in Figure 1. The SCr levels in the experimental group showed a significant increase at 2, 6, 12, 24, and 48 h compared with that in the sham group. This indicates that the animal model can successfully be established. The renal tubular-interstitial injury was confirmed by performing histopathological staining as shown in Figure 2. The histopathological change was assessed according to the range and degree of the damage. Swelling of renal tubule, cellular degeneration, and necrosis were observed significantly at 12, 24, and 48 h. At 72 h, a slight tubular atrophy could be detected in the rodent model. During the post-operative period, a few inflammatory cells were infiltrated in the interstitial tubule as the extension of time.
Figure 1.
Comparison of serum creatinine in the rat model (I/R vs Sham). *P < .05, **P < .01, compared with the sham group at the same time points. The data represent the level of SCr of rats in both the I/R group and sham group. SCr of I/R group was elevated in 2 h (0.71 ± 0.06 vs. 0.25 ± 0.01 mg/dL, P < .01) post-surgery, and it peaked at 24 h (2.35 ± 0.44 vs. 0.25 ± 0.02 mg/dL, P < .01). Results indicate that compared with the sham group, SCr showed significant statistical difference from 2 h to 48 h, and it returned to baseline at 72 h
Figure 2.
Histopathological staining of the kidney cortex in rat model. Damage in the graph was marked using arrow. In the I/R-induced AKI model, degeneration, necrosis, vacuolation were present at 2 h and 6 h after surgery, and peaked at 12 and 24 h post-surgery. As the time after surgery extended, inflammation was infiltrated and atrophy was formed gradually. (A color version of this figure is available in the online journal.)
Aberrant expression profiles of miRNA was detected in rat urine
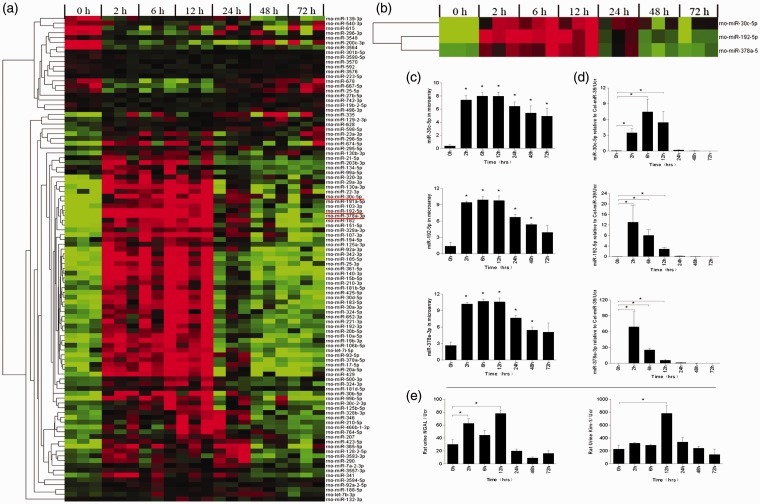
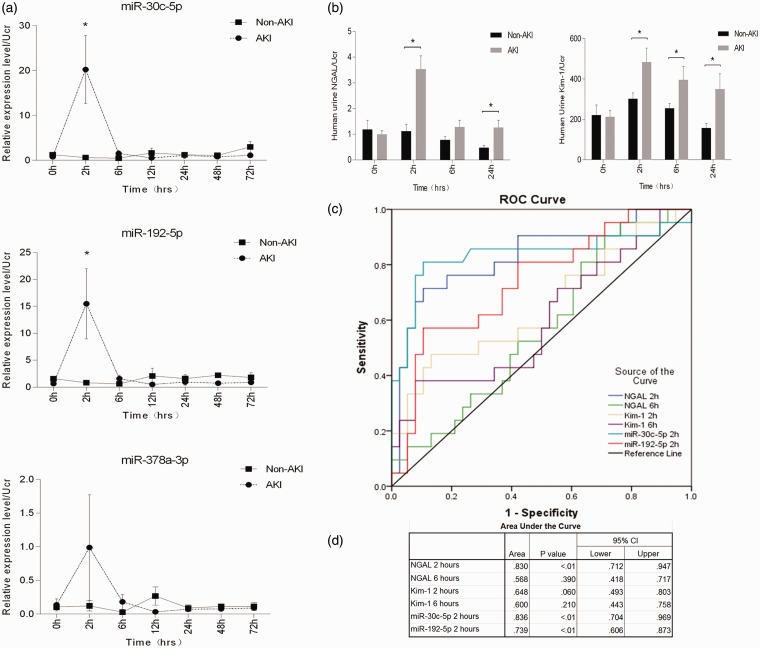
To explore miRNA expression profile in urine, urinalysis of miRNA was performed using the microarray method. For the primary candidates, ANOVA test was used for screening. In all, 101 miRNAs met the criterion of P < 0.05 in three or more time points as shown in Figure 3(a). Further screening was done using the criterion of miRNAs > 2-fold change upregulation or downregulation in three or more time points for which 125 miRNAs met the above criterion as shown in Figure S2. In addition, 71 miRNAs that satisfied the aforementioned 2 requirements simultaneously have been presented in Table S1. Most of the candidate miRNAs have a tendency to rise or fall after undergoing surgery and then gradually returning to the baseline. The top three (miR-192-5p, miR-378a-3p, and miR-30c-5p) miRNAs were selected by the sum of fold change of all time points for subsequent verification as shown in Figure 3(b) and (c).
Figure 3.
miRNA expression, NGAL. and KIM-1 level in rat urine. (a) The heat map shows the expression level of 101 miRNAs at seven time points after I/R injury (n = 3). Each column represents a sample and each row represents a miRNA in the graph. Red and blue represent the relative up and down expression, respectively. The preliminary candidates were screened out according to the criterion that ANOVA P < .05 in three or more time points. (b) and (c) Relative expression levels of miR-30c-5p, miR-192-5p, and miR-378a-3p in microarray. miR-192-5p peaked at 12 h (fold change 325.74, P < .001) and slipped down at the following 24, 48, and 72-h time points; miR-378a-3p peaked at 6 h (fold change 257.51, P < .001) and slipped down at the following 12, 24, 48, and 72-h time points; miR-30c-5p peaked at 12 h (fold change 190.35, P < .001) and slipped down at the following 24, 48, and 72-h time points. (d) Dynamic change trend of candidate miRNA detected by real-time PCR. Based on the average fold change in microarray, miR-30c-5p, miR-192-5p, and miR-378a-3p were screened for further validation. *P < .05, compared with the control group. Statistics in the graph represent the relative expression level/urine creatinine. The temporal tendency of real-time PCR agreed with the genechip in general. (e) Dynamic change trend of NGAL and KIM-1 detected by Elisa in rat model. *P < .05, compared with the control group. NGAL in rat urine elevated at 2 h post-surgery, and peaked at 12 h (78.01 ± 5.37 vs. 30.17 ± 7.45, P < .01). KIM-1 peaked at 12 h (782.74 ± 118.54 vs. 227.72 ± 61.70, P < .05) and slipped down at following time points. (A color version of this figure is available in the online journal.)
Candidate miRNAs were elevated in rat urine and earlier than KIM-1
In order to verify the plausibility of the performance of miR-192-5p, miR-378a-3p, and miR-30c-5p in the microarray data, TaqMan-based qPCR was performed that confirmed their expression levels in urine. Using the cel-miR-39 mimic as the normalizer and the sham group as the control, it was found that miR-192-5p was elevated at 2, 6, and 12 h significantly, and peaked at 2 h, after which it slipped to baseline at the following 24, 48, and 72-h time points. miR-378a-3p had an analogical shifting tendency. Compared with the first two, the expression of miR-30c-5p varied slightly; although it elevated at 2, 6, and 12 h, it peaked at 6 h and returned to baseline at 24 h (Figure 3(d)). In addition, we examined the urine NGAL and KIM-1 in rat model. Compared with the control group, NGAL in rat urine elevated at 2 h post-surgery, and peaked at 12 h. KIM-1 peaked at 12 h and slipped down at following time points (Figure 3(e)).
Candidate miRNAs were elevated in human urine and miR-30c-5p showed presentable diagnostic value
To explore the possibility of using miR-192-5p, miR-378a-3p, or miR-30c-5p as a sensitive and specific biomarker in I/R-induced AKI, urine samples were collected from patients who underwent cardiac operation. The clinical parameters are presented in Table 1 and Figure 4. Different types of surgeries were performed on patients in two groups. The results indicated that compared with others, patients with cardiac valve surgery and on-pump tend to suffer more from AKI. Of 71 post-cardiac patients, 27 developed AKI according to the criterion proposed in KDIGO 2012. Of 27 patients with AKI, 21 were classified under stage 1; 4 under stage 2, and 2 under stage 3. Of 71 post-cardiac patients, 30 underwent extracorporeal circulation (16 patients developed AKI and 14 patients with non-AKI). No patients received renal replacement therapy and no patients died during the in-hospital period. Results showed that eGFR of patients with AKI began to decrease at 6 h post operation; it reached its minimum level in 24 h and sprung back at 48 h after surgery.
Table 1.
Comparison of clinical characteristics of patients who underwent cardiac surgery (AKI vs non-AKI)
| Non-AKI | AKI | P | |
|---|---|---|---|
| N = 44 | N = 27 | ||
| Sex,male(%) | 26 (59.1) | 21 (77.8) | 0.13 |
| Age | 59.23 (2.21) | 57.59 (2.48) | 0.48 |
| BMI | 23.71 (0.43) | 23.72 (0.65) | 0.99 |
| Length of stay | 17.82 (0.76) | 25.59 (2.37) | 0.004 |
| Comorbidities, n(%) | |||
| Hypertension | 28 (63.6) | 16 (59.3) | 0.71 |
| Diabetes | 13 (29.5) | 8 (29.6) | 0.99 |
| CVD | 3 (6.8) | 1 (3.7) | 0.58 |
| Stroke | 2 (4.5) | 5 (18.5) | 0.10 |
| Prior operation | |||
| Baseline eGFR (ml/min/1.73 m2) | 81.22 (2.66) | 71.26 (4.75) | 0.052 |
| Serum creatinine, mg/dl | 0.94 (0.02) | 1.16 (0.07) | 0.050 |
| Hemoglobin, g/L | 127.09 (2.30) | 128.59 (4.50) | 0.77 |
| Albumin, g/L | 39.16 (0.98) | 40.74 (1.21) | 0.32 |
| Types of surgery, n(%) | |||
| CABG | 26 (59.1) | 9 (33.3) | |
| Valve | 10 (22.7) | 16 (59.3) | |
| CABG + Valve | 1 (2.3) | 0 | |
| Others | 7 (15.9) | 2 (7.4) | 0.012 |
| Extracorporeal circulation, n(%)a | 14 (31.8) | 16 (59.3) | 0.028 |
| Post operation | |||
| eGFR (ml/min/1.73 m2) | |||
| 6 h | 81.21 (3.05) | 49.05 (4.04) | <0.01 |
| 24 h | 88.36 (2.84) | 48.86 (3.97) | <0.01 |
| 48 h | 86.23 (3.96) | 52.68 (6.16) | <0.01 |
| 72 h | 87.61 (3.81) | 56.44 (6.59) | <0.01 |
| 7 day | 87.77 (3.14) | 61.89 (6.57) | 0.001 |
| Serum creatinine | |||
| 6 h | 0.93 (0.03) | 1.69 (0.64) | 0.08 |
| 24 h | 0.84 (0.03) | 1.69 (0.12) | <0.01 |
| 48 h | 0.85 (0.03) | 1.80 (0.18) | <0.01 |
| 72 h | 0.83 (0.03) | 1.79 (0.22) | <0.01 |
| 7 day | 0.86 (0.03) | 1.61 (0.23) | <0.01 |
| Hemoglobin-7 day, g/L | 106.45 (2.73) | 104.11 (3.27) | 0.58 |
| Albumin-7 day, g/L | 32.18 (0.69) | 32.59 (0.69) | 0.69 |
| Pump time, minb | 87.71 (10.84) | 99.25 (7.04) | 0.37 |
| Cross clamp time, minc | 60.73 (8.82) | 59.67 (7.13) | 0.92 |
In 71 post cardiac patients, 30 patients underwent extracorporeal circulation.
In the non-AKI group, 14 patients have pump time value. And in the AKI group, 16 patients have the pump time value (patients with extracorporeal circulation).
In the non-AKI group, 11 patients have cross clamp time value. And in the AKI group, 15 patients have the cross clamp time value (patients with extracorporeal circulation). Four values missed.
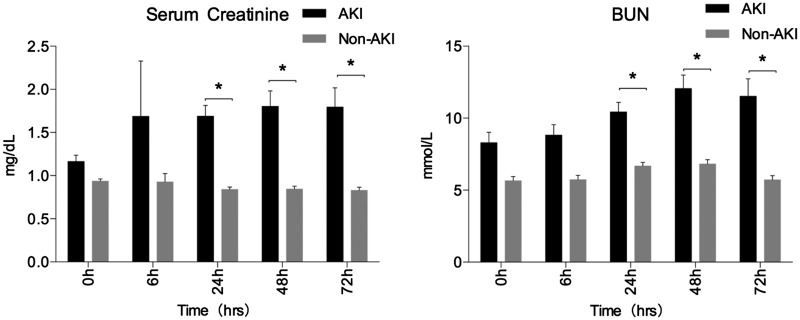
Figure 4.
Comparison of SCr and BUN in patients who underwent cardiac surgery (AKI vs non-AKI). *P < .05, compared with the non-AKI group at each time points. In patients who underwent cardiac surgery, SCr of AKI patients were elevated at 6 h (1.69 ± 0.64 vs. 0.93 ± 0.09 mg/dL) and made a difference at 24 h (1.69 ± 0.12 vs. 0.84 ± 0.03 mg/dL, P < .05). Serum BUN had similar changes; they were elevated at 24 h (10.44 ± 0.65 vs. 6.67 ± 0.26 mmol/L), and they peaked at 48 h (12.06 ± 0.93 vs. 6.82 ± 0.29 mmol/L, P < .05)
Then, the expression levels of miR-192-5p, miR-378a-3p, and miR-30c-5p were examined in human urine. Using the cel-miR-39 mimic as the normalizer and the non-AKI group as the control, we found that miR-192-5p peaked at 2 h (P < .05); however, it slipped to baseline at the following 6, 12, 24, 48, and 72-h time points. miR-30c-5p had a similar changing tendency. However, the expression of miR-378a-3p varied slightly; it peaked at 2 h and returned to baseline at 6 h, and there was no statistically significant difference at any time points as shown in Figure 5(a). We also analyzed the candidate miRNAs’ expression at 2 h post operation in patients with different AKI stages (Figure S3). We found that the miRNAs levels were higher in patients with AKI stage 1 than in non-AKI patients. Similar trend also existed in patients with AKI stage 2 and 3, although the difference was not significant due to limited sample size.
Figure 5.
Candidate miRNAs were elevated in human urine and miR-30c-5p showed presentable diagnostic value compared with protein-based markers in patients who underwent cardiac surgery (AKI vs. non-AKI). Statistics in the graph represent the relative expression level/urine creatinine. *P < .05, compared with the non-AKI group at each time point. (a) miR-192-5p was elevated at 2 h (P = 0.035) and returned to baseline at 6 to72 h. miR-30c-5p was elevated at 2 h (P = 0.01) and returned to baseline at 6 to 72 h. miR-378a-3p did not show any statistically significant difference at any time points. (b) NGAL was elevated at 2 h (P < .001) and slipped down at following time points (left). KIM-1 has the similar shifting tendency with NGAL (Right). (c–d) ROC curve of miR-30c-5p (2 h), miR-192-5p (2 h), NGAL (2 h), NGAL (6 h), KIM-1 (2 h), KIM-1 (6 h) was generated. Area under curve (AUC) of miR-30c-5p (2 h) was 0.836 (P < .01), which is higher than NGAL at 2 h (0.830, P < .01). (A color version of this figure is available in the online journal.)
To investigate the diagnostic value of miRNA as biomarker, we examined the urine concentration of NGAL and KIM-1 in patients and compared the sensitivity and specificity between miRNA and protein-based biomarker (NGAL, KIM-1). The results suggested that NGAL and KIM-1 elevated at 2 h post-surgery (P < .05), and slipped down at 24 h (Figure 5(b)). Then the ROC curve of miR-30c-5p (2 h), miR-192-5p (2 h), NGAL (2 h), NGAL (6 h), KIM-1 (2 h), KIM-1 (6 h) was generated. Area under curve (AUC) of miR-30c-5p (2 hours) was 0.836 (P < .01), which is higher than NGAL at 2 h (0.830, P < .01). The detailed data are presented in Figure 5(c) and (d).
Source of miR-30c-5p and miR-192-5p might be different
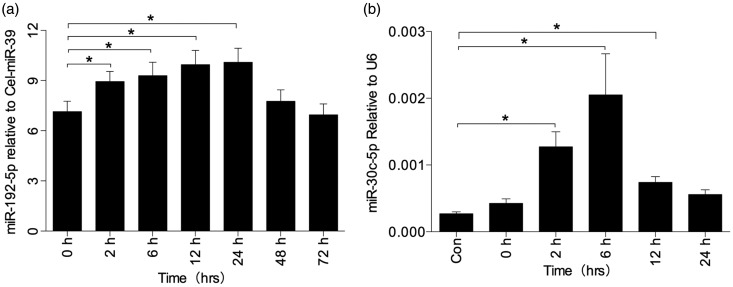
With a belief that miRNA in urine might be associated with plasma filtration or injured tubular epithelial cells, the serum concentration levels of miR-30c-5p, miR-192-5p, and miR-378a-3p in rat model were evaluated in this study. It was found that miR-192-5p was elevated in the serum at 2 h; it returned to baseline at 48 h as shown in Figure 6(a). However, there was no visible difference in the levels of miR-30c-5p and miR-378a-3p (data not shown). To further clarify the sources of urinary miRNA, HK-2 cell line was used to build the hypoxia/reoxygenation (H/R) model. Elevated levels of miR-30c were evaluated after treating the cell with 1% O2 and incubating for 24 h (Figure 6(b)). miR-378a-3p did not show any significant differences in the H/R model (data not shown). Therefore, it was inferred that urine contains elevated miR-30c-5p owing to injured renal tubules. Furthermore, elevated urinary miR-192-5p levels were thought to be due to the rise in serum concentration levels.
Figure 6.
miRNA expression in serum of rat model (a) and H/R cell model (b). (a) Expression of miR-192-5p was elevated in rat serum at 2, 6, 12, and 24. Statistics in the graph represent the relative expression level/cel-miR-39. *P < .05, compared with the 0-h group. (b) Expression of miR-30c-5p was elevated in H/R cell model. Statistics in the graph represent the relative expression level/U6. *P < .05, compared with the control group. After incubation in the hypoxia chamber for 24 h, miR-30c-5p did not show any statistical significance. Therefore, miR-30c-5p showed an elevated trend after incubation in the hypoxia chamber for 24 h as well as reoxygenation with fresh medium and normoxia chamber for 2, 6, 12, and 24 h
Discussion
In this screening study, the miRNA expression levels were determined within 72 h following surgery to observe the trend of time-dependent variation. The results of this study showed that the aberrant expression of miRNA in I/R animal model could be detected as early as 2 h post operation. This finding was further validated in patients with AKI to verify the possibility of clinical application, and results indicated that miR-30c-5p showed better performance than protein-based markers. In addition, the expression levels of miR-30c-5p and miR-192-5p in rat serum and cell model were also investigated, and it was found that the sources of miR-30c-5p and miR-192-5p are likely to be different.
Therefore, there are evidence suggesting that miRNA showed an abnormal expression profile in patients with a coexisting underlying disease.30–32 Hence, one differentiating factor of this study is that the candidate miRNAs were screened in a controlled animal model rather than in patients with AKI to ensure the purity of the cause of disease and to avoid possible effects of comorbidities on the spectrum of urine miRNA. This ensured the presence of only the relevant candidate miRNA (that associated with I/R injury). Subsequently, the elevated level of miR-30c-5p and miR-192-5p 2 h after the operation was verified in clinical settings and be founded in agreement with the findings in the animal model, which could correct misguidance that caused by specie difference. Moreover, the expression levels of miR-192-5p and miR-30c-5p were also detected in the serum of rat model and cell model (H/R). Hence, the alterative expression of miR-192-5p and miR-30c-5p in animal model, patients with AKI, and cell model was confirmed simultaneously, which is likely to be more convincing.
SCr is widely accepted as overall index for kidney function but often failed for early detection of AKI. In current study, a statistically significant increase of miRNA (2 h post operation) is earlier than SCr (24 h post operation) in AKI patients (Figures 4 and 5), indicating an evident benefit over SCr. Noninvasive urine-based markers have been proposed as early indicators for identifying AKI, including NGAL, Kim-1, L-FABP, TIMP2/IGFBP7 etc.8,12,13 Of late, emerging evidence indicated that urine miRNA could act as novel and sensitive biomarker for detecting kidney injury, mainly because it is stable in body fluids and extremely sensitive to alteration of organism. Therefore, researchers carried out a lot of experiments in this field. Wang et al.33 proved that miR-10a and miR-30d were elevated in patients with focal segmental glomerular sclerosis compared to normal volunteers. Saikumar et al.22 found that miR-21 and miR-155 might serve as translational biomarkers potentially for AKI detection. In a pilot study34 that involved post-cardiac surgery patients, urine miR-21 could predict AKI outcome and they were associated with higher risk of AKI progression. Another study21 identified miR-21, miR-200c, miR-423, and miR-4640 might be sensitive to predict for kidney injury. Our study found that miRNA biomarkers showed preferable performance in diagnosing IR-AKI compared with NGAL and Kim-1. However, our study did not confirm any association between urine miRNAs and clinical outcomes. It might be due to the short follow-up period of our cohort.
Act as a negative regulator of gene expression, modulating the translational efficiency and/or the stability of target messenger RNAs, miRNAs is involved in a wide range of biological processes including normal development of organism, pathogenesis, and progression of disease.35–37 As reported previously,38 miR-30c directs target ADAM19 to inhibit colon cancer, which may serve as a promising therapeutic strategy in the treatment of colon cancer. Zhou et al.39 also indicated that miR-30c acts as a tumor negative suppressor by regulating endometrial cancer cells targeting MTA1. In addition, there is evidence suggesting that miR-30-5p function as a novel therapeutic tool for targeting the oncogenic Wnt/β-catenin/BCL9 pathway.40 From these studies, it might be presumed that miR-30c is an inhibitor of cell proliferation or an accelerator of cell death. Hence, it may play a key role during the pathophysiological process of kidney injury or repair. Taken into account the elevation of miRNAs in the current study is remarkable, the literature from Jeon et al.41 seems to be able to explain the phenomenon partially. They found that miR-30c activated by NF-κB pathway could reduce the tumor growth by modulating tumor suppressor genes. Therefore, the important signaling pathways were involved in pathophysiological process of AKI frequently,42 so it is reasonable about the fact that expression level of miRNA make a difference in the AKI model.
The definition and classification of AKI remain controversial, especially for AKI stage 1, because the slight fluctuation of SCr could be caused by some other factor such as state of hydration. Thus, a transient SCr increase (returning to no AKI within 72 h) was observed and analyzed separately in Vanmassenhove’s study43; and in another study only stage 2 and 3 (based on AKI) was classified to AKI12, mainly due to the fact that the diagnosis of AKI needs a strong evidence of kidney injury. Our data (Figure 4) showed that SCr concentration at 72 h after surgery in the AKI group persistently elevated significantly compared to non-AKI patients, which means the diagnosis of patients with AKI in current study is credible. In addition, all patients received sufficient fluid infusion during operation (more than 1000 ml within 4 h) and most of them had normal urine output (>2000 ml/24 h) after surgery. Thus, we believe hydration status is unlikely occurred in patients included in this study.
Previous studies indicate that miR-192 is involved in liver damage such as ischemia/reperfusion-induced and drug-induced damage.44–49 Then we reviewed liver damage index of subjects in our cohort. There is no damage of baseline liver function, and no statistical significance of hepatic enzymatic indices after they underwent cardiac surgery between the groups of AKI and non-AKI patients (data not shown). So we thought rise of miRNA in the current population is independent of the effect of liver damage. Given these evidence, we could presume that miR-192-5p is associated with AKI credibly. However, the source of urinary miR-192 remains unclear. As the next step, the intent of this study is to further elucidate the effects of intervention of miR-30c-5p and miR-192-5p on the I/R and H/R model to confirm their sources and functional roles.
This study also has several limitations to be considered. Firstly, the evaluations performed in this study are of fairly short-time points post-surgery. Hence, a relationship between the elevated miRNA and a long-term renal recovery is not established. Secondly, the applicability of miRNAs as a diagnostic marker in other types of kidney injury such as contrast-induced kidney injury and nephrotoxic kidney injury should be validated.
In summary, this study provides a dynamic profiling of urinary miRNA within 72 h post-surgery in I/R-induced AKI in both a rodent model as well as in humans with AKI. The levels of miR-30c-5p, miR-378a-3p, and miR-192-5p in urine of rats and patients with AKI were quantitatively evaluated. Both miR-30c-5p and miR-192-5p show potential to serve as diagnostic markers for I/R-induced AKI; however, more studies are necessary to further validate its diagnostic value for the detection of kidney injury.
Supplementary Material
Authors’ contributions
YFZ carried out the experiments on animal model, molecular studies, data collection on cardiac patients, Elisa Assay and drafted the manuscript; DW carried out the data analyses of cardiac patients, performed pathological staining, and participated in the molecular studies; QZ provided technological guidance; PYS conducted the screening of cardiac patients and provided pathological consultation; HS performed the statistical analyses; QZ contributed to the screening of cardiac patients and processing of the sample; YXC participated in the study design and helped draft the manuscript; and WZ conceived the study, participated in its design, and helped draft the manuscript. All authors have read and approved the final manuscript.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
This study was supported by the grants from the National Natural Science Foundation of China (No 81470967, No 81470041) and the Science and Technology Commission of Shanghai Municipality (12DJ1400303, 11JC1407901, 15411963800).
References
- 1.Bellomo R, Kellum JA, Ronco C. Acute kidney injury. Lancet 2012; 380: 756–66. [DOI] [PubMed] [Google Scholar]
- 2.Chertow GM, Burdick E, Honour M, Bonventre JV, Bates DW. Acute kidney injury, mortality, length of stay, and costs in hospitalized patients. J Am Soc Nephrol 2005; 16: 3365–70. [DOI] [PubMed] [Google Scholar]
- 3.Lafrance JP, Miller DR. Acute kidney injury associates with increased long-term mortality. J Am Soc Nephrol 2010; 21: 345–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ranganathan P, Jayakumar C, Tang Y, Park KM, Teoh JP, Su H, Li J, Kim IM, Ramesh G. MicroRNA-150 deletion in mice protects kidney from myocardial infarction-induced acute kidney injury. Am J Physiol Renal Physiol 2015; 309: F551–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balasubramanian G, Al-Aly Z, Moiz A, Rauchman M, Zhang Z, Gopalakrishnan R, Balasubramanian S, El-Achkar TM. Early nephrologist involvement in hospital-acquired acute kidney injury: A pilot study. Am J Kidney Dis 2011; 57: 228–34. [DOI] [PubMed] [Google Scholar]
- 6.Porter CJ, Juurlink I, Bisset LH, Bavakunji R, Mehta RL, Devonald MA. A real-time electronic alert to improve detection of acute kidney injury in a large teaching hospital. Nephrol Dial Transplant 2014; 29: 1888–93. [DOI] [PubMed] [Google Scholar]
- 7.Alge JL, Arthur JM. Biomarkers of AKI: A review of mechanistic relevance and potential therapeutic implications. Clin J Am Soc Nephrol 2015; 10: 147–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martensson J, Martling CR, Bell M. Novel biomarkers of acute kidney injury and failure: Clinical applicability. Br J Anaesth 2012; 109: 843–50. [DOI] [PubMed] [Google Scholar]
- 9.Parikh CR, Thiessen-Philbrook H, Garg AX, Kadiyala D, Shlipak MG, Koyner JL, Edelstein CL, Devarajan P, Patel UD, Zappitelli M, Krawczeski CD, Passik CS, Coca SG. Performance of kidney injury molecule-1 and liver fatty acid-binding protein and combined biomarkers of AKI after cardiac surgery. Clin J Am Soc Nephrol 2013; 8: 1079–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huen SC, Parikh CR. Molecular phenotyping of clinical AKI with novel urinary biomarkers. Am J Physiol Renal Physiol 2015; 309: F406–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meersch M, Schmidt C, Van Aken H, Martens S, Rossaint J, Singbartl K, Gorlich D, Kellum JA, Zarbock A. Urinary TIMP-2 and IGFBP7 as early biomarkers of acute kidney injury and renal recovery following cardiac surgery. PLoS One 2014; 9: e93460–e93460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunnerson KJ, Shaw AD, Chawla LS, Bihorac A, Al-Khafaji A, Kashani K, Lissauer M, Shi J, Walker MG, Kellum JA. TIMP2*IGFBP7 biomarker panel accurately predicts acute kidney injury in high-risk surgical patients. J Trauma Acute Care Surg 2016; 80: 243–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pajenda S, Ilhan-Mutlu A, Preusser M, Roka S, Druml W, Wagner L. NephroCheck data compared to serum creatinine in various clinical settings. BMC Nephrol 2015; 16: 206–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graves P, Zeng Y. Biogenesis of mammalian microRNAs: A global view. Genom Proteom Bioinform 2012; 10: 239–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khalyfa A, Gozal D. Exosomal miRNAs as potential biomarkers of cardiovascular risk in children. J Transl Med 2014; 12: 162–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mirzaei H, Gholamin S, Shahidsales S, Sahebkar A, Jaafari MR, Mirzaei HR, Hassanian SM, Avan A. MicroRNAs as potential diagnostic and prognostic biomarkers in melanoma. Eur J Cancer 2016; 53: 25–32. [DOI] [PubMed] [Google Scholar]
- 17.Weiland M, Gao XH, Zhou L, Mi QS. Small RNAs have a large impact: Circulating microRNAs as biomarkers for human diseases. RNA Biol 2012; 9: 850–9. [DOI] [PubMed] [Google Scholar]
- 18.Lan YF, Chen HH, Lai PF, Cheng CF, Huang YT, Lee YC, Chen TW, Lin H. MicroRNA-494 reduces ATF3 expression and promotes AKI. J Am Soc Nephrol 2012; 23: 2012–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorenzen JM, Kielstein JT, Hafer C, Gupta SK, Kumpers P, Faulhaber-Walter R, Haller H, Fliser D, Thum T. Circulating miR-210 predicts survival in critically ill patients with acute kidney injury. Clin J Am Soc Nephrol 2011; 6: 1540–6. [DOI] [PubMed] [Google Scholar]
- 20.Molitoris JK, Molitoris BA. Circulating micro-RNAs in acute kidney injury: Early observations. Clin J Am Soc Nephrol 2011; 6: 1517–9. [DOI] [PubMed] [Google Scholar]
- 21.Ramachandran K, Saikumar J, Bijol V, Koyner JL, Qian J, Betensky RA, Waikar SS, Vaidya VS. Human miRNome profiling identifies microRNAs differentially present in the urine after kidney injury. Clin Chem 2013; 59: 1742–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saikumar J, Hoffmann D, Kim TM, Gonzalez VR, Zhang Q, Goering PL, Brown RP, Bijol V, Park PJ, Waikar SS, Vaidya VS. Expression, circulation, and excretion profile of microRNA-21, -155, and -18a following acute kidney injury. Toxicol Sci 2012; 129: 256–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, Zhang L, Chen Y-X, Xie Y-Y, Zou Y-F, Zhang M-J, Gao Y-H, Liu Y, Zhao Q, Huang Q-H. Identification of nestin as a urinary biomarker for acute kidney injury. Am J Nephrol 2014; 39: 110–21. [DOI] [PubMed] [Google Scholar]
- 24.Reeves WB, Kwon O, Ramesh G. Netrin-1 and kidney injury. II. Netrin-1 is an early biomarker of acute kidney injury. Am J Physiol Renal Physiol 2008; 294: F731–8. [DOI] [PubMed] [Google Scholar]
- 25.Khwaja A. KDIGO clinical practice guidelines for acute kidney injury. Nephron Clin Pract 2012; 120: c179–84. [DOI] [PubMed] [Google Scholar]
- 26.Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF, 3rd, Feldman HI, Kusek JW, Eggers P, Van Lente F, Greene T, Coresh J. A new equation to estimate glomerular filtration rate. Ann Intern Med 2009; 150: 604–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McClintick JN, Edenberg HJ. Effects of filtering by Present call on analysis of microarray experiments. BMC Bioinform 2006; 7: 49–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spornraft M, Kirchner B, Haase B, Benes V, Pfaffl MW, Riedmaier I. Optimization of extraction of circulating RNAs from plasma – enabling small RNA sequencing. PLoS One 2014; 9: e107259–e107259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luna-Aguirre CM, de la Luz Martinez-Fierro M, Mar-Aguilar F, Garza-Veloz I, Trevino-Alvarado V, Rojas-Martinez A, Jaime-Perez JC, Malagon-Santiago GI, Gutierrez-Aguirre CH, Gonzalez-Llano O, Salazar-Riojas R, Hidalgo-Miranda A, Martinez-Rodriguez HG, Gomez-Almaguer D, Ortiz-Lopez R. Circulating microRNA expression profile in B-cell acute lymphoblastic leukemia. Cancer Biomark 2015; 15: 299–310. [DOI] [PubMed] [Google Scholar]
- 30.Al-Kafaji G, Al-Mahroos G, Alsayed NA, Hasan ZA, Nawaz S, Bakhiet M. Peripheral blood microRNA-15a is a potential biomarker for type 2 diabetes mellitus and pre-diabetes. Mol Med Rep 2015; 12: 7485–90. [DOI] [PubMed] [Google Scholar]
- 31.Romaine SP, Tomaszewski M, Condorelli G, Samani NJ. MicroRNAs in cardiovascular disease: An introduction for clinicians. Heart 2015; 101: 921–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonventre JV, Yang L. Cellular pathophysiology of ischemic acute kidney injury. J Clin Invest 2011; 121: 4210–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang N, Zhou Y, Jiang L, Li D, Yang J, Zhang CY, Zen K. Urinary MicroRNA-10a and MicroRNA-30d serve as novel, sensitive and specific biomarkers for kidney injury. PLoS One 2012; 7: e51140–e51140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du J, Cao X, Zou L, Chen Y, Guo J, Chen Z, Hu S, Zheng Z. MicroRNA-21 and risk of severe acute kidney injury and poor outcomes after adult cardiac surgery. PLoS One 2013; 8: e63390–e63390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato M, Arce L, Natarajan R. MicroRNAs and their role in progressive kidney diseases. Clin J Am Soc Nephrol 2009; 4: 1255–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du T, Zamore PD. microPrimer: The biogenesis and function of microRNA. Development 2005; 132: 4645–52. [DOI] [PubMed] [Google Scholar]
- 37.Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and function. Thromb Haemost 2012; 107: 605–605. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Q, Yu L, Qin D, Huang R, Jiang X, Zou C, Tang Q, Chen Y, Wang G, Wang X, Gao X. Role of microRNA-30c targeting ADAM19 in colorectal cancer. PLoS One 2015; 10: e0120698–e0120698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou H, Xu X, Xun Q, Yu D, Ling J, Guo F, Yan Y, Shi J, Hu Y. microRNA-30c negatively regulates endometrial cancer cells by targeting metastasis-associated gene-1. Oncol Rep 2012; 27: 807–12. [DOI] [PubMed] [Google Scholar]
- 40.Zhao JJ, Lin J, Zhu D, Wang X, Brooks D, Chen M, Chu ZB, Takada K, Ciccarelli B, Admin S, Tao J, Tai YT, Treon S, Pinkus G, Kuo WP, Hideshima T, Bouxsein M, Munshi N, Anderson K, Carrasco R. miR-30-5p functions as a tumor suppressor and novel therapeutic tool by targeting the oncogenic Wnt/beta-catenin/BCL9 pathway. Cancer Res 2014; 74: 1801–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeon YJ, Middleton J, Kim T, Lagana A, Piovan C, Secchiero P, Nuovo GJ, Cui R, Joshi P, Romano G, Di Leva G, Lee BK, Sun HL, Kim Y, Fadda P, Alder H, Garofalo M, Croce CM. A set of NF-kappaB-regulated microRNAs induces acquired TRAIL resistance in lung cancer. Proc Natl Acad Sci U S A 2015; 112: E3355–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meldrum KK, Hile K, Meldrum DR, Crone JA, Gearhart JP, Burnett AL. Simulated ischemia induces renal tubular cell apoptosis through a nuclear factor-kappaB dependent mechanism. J Urol 2002; 168: 248–52. [PubMed] [Google Scholar]
- 43.Vanmassenhove J, Glorieux G, Lameire N, Hoste E, Dhondt A, Vanholder R, Van Biesen W. Influence of severity of illness on neutrophil gelatinase-associated lipocalin performance as a marker of acute kidney injury: A prospective cohort study of patients with sepsis. BMC Nephrol 2015; 16: 18–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fang L, Ellims AH, Moore XL, White DA, Taylor AJ, Chin-Dusting J, Dart AM. Circulating microRNAs as biomarkers for diffuse myocardial fibrosis in patients with hypertrophic cardiomyopathy. J Transl Med 2015; 13: 314–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pek SL, Tavintharan S, Woon K, Lin L, Ong CN, Lim SC, Sum CF. MicroRNAs as biomarkers of hepatotoxicity in a randomized placebo-controlled study of simvastatin and ubiquinol supplementation. Exp Biol Med 2016; 241: 317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Motawi TK, Shaker OG, El-Maraghy SA, Senousy MA. Serum MicroRNAs as potential biomarkers for early diagnosis of hepatitis C virus-related hepatocellular carcinoma in egyptian patients. PLoS One 2015; 10: e0137706–e0137706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silakit R, Loilome W, Yongvanit P, Thongchot S, Sithithaworn P, Boonmars T, Koonmee S, Titapun A, Khuntikeo N, Chamadol N, Techasen A, Namwat N. Urinary microRNA-192 and microRNA-21 as potential indicators for liver fluke-associated cholangiocarcinoma risk group. Parasitol Int 2015. Epub ahead of print 9 October. DOI: 10.1016/j.parint.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 48.Chen Q, Ge X, Zhang Y, Xia H, Yuan D, Tang Q, Chen L, Pang X, Leng W, Bi F. Plasma miR-122 and miR-192 as potential novel biomarkers for the early detection of distant metastasis of gastric cancer. Oncol Rep 2014; 31: 1863–70. [DOI] [PubMed] [Google Scholar]
- 49.Hu J, Wang Z, Tan CJ, Liao BY, Zhang X, Xu M, Dai Z, Qiu SJ, Huang XW, Sun J, Sun QM, He YF, Song K, Pan Q, Wu Y, Fan J, Zhou J. Plasma microRNA, a potential biomarker for acute rejection after liver transplantation. Transplantation 2013; 95: 991–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.