Abstract
The electronic structure of ground state oxygen, which is essential for the life of all aerobic organisms, makes it potentially dangerous for those organisms. Atmospheric oxygen contains two unpaired electrons with parallel spin states, which predisposes it to reduction by a univalent pathway. As a consequence, normal aerobic metabolism generates dangerous reactive intermediates of the reduction of O2. These include superoxide radical (O2–), hydrogen peroxide (H2O2), and hydroxyl radical (HO). These reactive oxygen species and others that they can engender can damage all cellular macromolecules and unless opposed by cellular defenses, would make aerobic life impossible. Such defenses include superoxide dismutases, catalases, and peroxidases, enzymes that decrease the concentration of the reactive oxygen species that are their substrates, and others that repair or recycle oxidatively damaged macromolecules. Any factor that stimulates reactive oxygen species production or suppresses the antioxidant systems would inevitably cause cell damage. The role of such oxidative damage in various diseases is well documented. In vivo detection of O2– and other reactive oxygen species is however hampered by the lack of easy, specific, and sensitive analytical methods. Potential artifacts and limitations of the most common detection methods currently in use are briefly discussed.
Key Words: Reactive oxygen species, Superoxide, Singlet oxygen, Hydrogen peroxide, Nitric oxide, Free radical, Oxidative stress, Superoxide dismutase, Superoxide assay
The Basis of Oxygen's Peculiarity
The molecular oxygen that we refer to as the ‘breath of life’ is actually a very peculiar gas. It is paramagnetic in the same way that iron is paramagnetic. If oxygen was a solid rather than a gas, we could observe it being attracted to a magnet. Yet it does respond to a magnetic field and that property is routinely exploited in oximeters that measure the oxygen content of a gas mixture.
Paramagnetism denotes unpaired electrons. Thus an electron has an associated magnetic field whose orientation depends on its spin. Two electrons that occupy the same orbital have opposite spins and so their magnetic fields are oppositely oriented and thus cancel each other. Most stable molecules have all of their electrons as such spin opposed pairs and therefore are not paramagnetic.
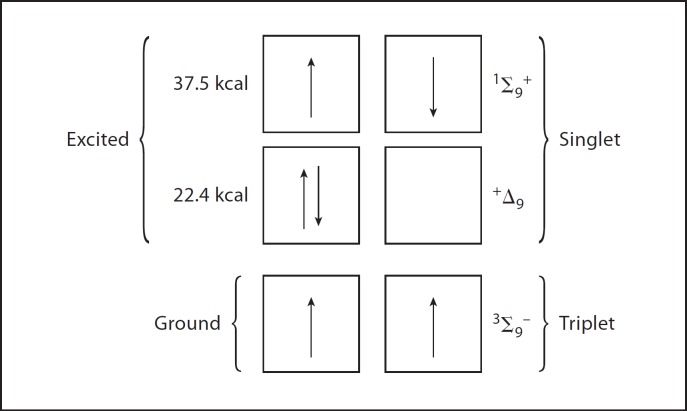
Now let us return to oxygen whose paramagnetism denotes two unpaired electrons. The Pauli exclusion principle teaches that two electrons cannot occupy the same orbital unless their spins are opposed and, in keeping with this principle, each of the two unpaired electrons in oxygen occupies a separate orbital. It further needs to be stated that these two unpaired electrons have the same spin direction or spin state (fig. 1).
Fig. 1.
Electronic configurations energy levels of ground state and singlet molecular oxygen.
To illustrate the importance of these two parallel spin states, try to consider adding a spin opposed pair of electrons from some molecule to oxygen. One of the electrons in the pair that happens to have a spin state opposite to that of the unpaired electrons in oxygen could happily join with one of them, thus creating a stable spin opposed pair. But the other of the electrons seeking to associate with the oxygen would necessarily have a spin that is parallel to that of the remaining unpaired electron in oxygen and so could not pair with it.
The Spin Restriction
Spin restriction is the barrier to the reaction of oxygen with all nonparamagnetic molecules. Now it is possible to put enough energy into oxygen to elevate one of its parallel spinning electrons to a higher orbital and in the process to invert its spin. Such an excited state of oxygen is referred to as singlet oxygen, and for singlet oxygen the spin restriction has been eliminated and it is much more reactive than is ground state oxygen (fig. 1). The energy inherent in visible light is enough to convert ground state oxygen into singlet oxygen, but oxygen does not absorb visible light. Dyes such as methylene blue or rose bengal do absorb visible light and then, upon collision with oxygen, can transfer the energy from that light to oxygen. That is one basis of photosensitized oxidations. An illustration of the power of photosensitized oxidation is provided by the observation that NADH is not rapidly oxidized by ground state oxygen, but in the presence of methylene blue plus light it is. That is also an illustration of the spin restriction in limiting the reactivity of ground state oxygen.
There is a way that the spin restriction can be circumvented and that is by adding the electrons to oxygen one at a time. This works because electronic spin can be inverted by interacting with nuclear spin, but it is a relatively slow process operating on a time scale of 10 ns. Since the lifetime of collisional complexes is only in the range of 0.00001 ns, spin inversion is not likely to occur during the lifetime of collisional complexes. But there is lots of time between collisions of potential reactants for spin inversion to happen. That is the basis of the univalent pathway of oxygen reduction. Thus the electrons are added to oxygen one at a time at a rate that allows electronic spin inversions to occur between collisional events.
The Univalent Pathway
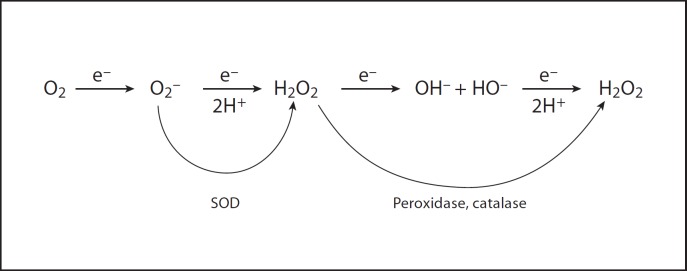
The univalent pathway of oxygen reduction requires that intermediates of oxygen reduction be generated (fig. 2). Thus the complete reduction of oxygen to water requires that four electrons and four protons be added to the oxygen molecule, yielding two molecules of water. Adding the first electron produces O2·–; adding the second electron plus two protons yields H2O2; adding the third electron gives HO· plus OH–; and finally adding the fourth electron plus two more protons produces two water molecules. These intermediates of oxygen reduction are reactive and can damage biological molecules. Indeed, were there not defenses against them, these intermediates would make aerobic life unsustainable.
Fig. 2.
Sequential one-electron reduction of molecular oxygen to water.
The Intermediates of Oxygen Reduction
The first intermediate encountered on the univalent pathway is superoxide (O2·–). It is the conjugate base of the weak acid, the hydroperoxyl radical (HO·2). Its pKa is 4.8, so at neutrality it is 99% ionized. O2·–/HO·2 is not stable in protic solvents and dismutes spontaneously into H2O2 plus O2 with rate constants that depend on pH [1]. Thus at low pH, where it exists primarily as HO·2, the rate constant is close to 105 M−1 s−1. At pH 4.8, where it exists as half HO·2 and half O2·–, the rate constant is 108 M−1 s−1. At high pH it is mostly O2·– and then the rate constant is essentially zero. The reason for this is that the electrostatic repulsion between two O2·– anions prevents close approach and hence prevents the electron transfer between them, which is the basis of the dismutation.
The second intermediate, H2O2, is not a free radical and is relatively stable. However, it can be univalently reduced by metal cations such as ferrous and cuprous to yield the hydroxyl radical (HO·) plus hydroxide ion (OH–). This is known as the Fenton reaction and the HO· it generates is an extraordinarily powerful oxidant. It is also the third intermediate encountered on the univalent pathway of O2 reduction. All of these intermediates are able to damage the components of cells in ways that we will explore next.
How O2–, H2O2, and HO· Cause Damage
The superoxide radical can act either as a reductant or an oxidant. Thus its ability to reduce cytochrome c was important in its discovery as a product of the aerobic xanthine oxidase reaction and is currently the basis of an assay used to measure the activity of superoxide dismutases. In the reducing environment of cell cytosol, O2·– is more likely to act as an oxidant, and one important target for its action as an oxidant is the iron-sulfur clusters (4Fe4S) of dehydratases, such as that found in aconitase [2]. Superoxide can oxidize these clusters and so inactivate this family of enzymes. This blocks the citric acid cycle that is so essential for the aerobic metabolism of cells, and it has an additional far-reaching consequence. Thus, a (4Fe4S) cluster that has been univalently oxidized by O2·– is unstable and decomposes, releasing free iron that can then participate in the Fenton reaction that generates the powerful oxidant, HO· [3], which can oxidize virtually any biological molecule. Hydroxyl radical is so reactive that it will react with whatever is close by, which would lead one to suppose that it would be consumed by the plethora of relatively unimportant metabolic intermediates. However, when we consider what a cationic metal such as Fe2+ would bind to, polyanionic species such as DNA, RNA, and cell membranes come immediately to mind. Hence, they would be the preferential targets for the HO· generated by the Fenton reaction. Another point to be considered is that HO· is a univalent oxidant. Hence it would produce free radicals derived from its targets, and that opens the door to the propagation of chain reactions that would amplify the damage caused by one HO·. This is specially the case with biological membranes, DNA, RNA and macromolecules in general (for details see [4]).
Defensive Strategies
The first obvious defense is avoidance. Thus, there are enzymes that reduce oxygen divalently to H2O2 and even tetravalently to water without releasing intermediates such as O2·–. D-amino oxidase is in the first category and cytochrome c oxidase is in the second. Yet, because of the spin restriction described above, virtually all auto-oxidations and even some enzyme-catalyzed reactions do generate O2·–. In respiring cells only about 0.1% of the oxygen consumed is released as O2·–[5]. Yet given the large amounts of oxygen utilized in respiration, even so small a fraction as 1/1,000 creates O2·– at intolerable rates. Hence defenses are essential to limit and to repair the damage that O2·– and its progeny H2O2 and HO· would otherwise do. A detailed description of the best studied defense mechanisms and their regulation can be found in a recent review [6].
Superoxide Dismutases
The Cu,ZnSODs
The primary defense is provided by superoxide dismutases (SODs) that catalyze the conversion of O2·– into H2O2 plus oxygen. A dismutation is a reaction that converts two identical substrate molecules into two different products; hence the name. There are several different SODs. One member of this family is the Cu,ZnSODs [7]. These enzymes are found in the eukaryotic cytosol, nuclei, the intermembrane space of mitochondria, in the periplasm of Gram-negative bacteria and in the extracellular spaces of multicellular eukaryotes. This wide distribution indicates that O2·– is encountered in all of these places and presents a threat in all of them.
Mechanism
Cu and Zn are present in an imidazole bridged bimetallic structure at the active sites of this homodimeric protein. Copper is the catalytic metal and is reduced from Cu2+ to Cu1+ during the first interaction with O2·–, and then is reoxidized by the next O2·– encountered [8]. Zn plays a structural role and also holds the bridging imidazolate in place during the catalytic cycle. This is an extraordinarily active enzyme whose interactions with O2·– occur with a rate constant of 2 × 109 M−1 s−1 25°C [9]. At such a diffusion-limited rate, proton conduction would be rate-limiting in the absence of some special facilitating mechanism. The bridging imidazole provides that facilitated proton conduction. Thus, when the Cu2+ is reduced in the first step of the catalytic cycle, it releases the bond to imidazolate, which then binds a proton from water. In the second step of the catalytic cycle, during which the bond to Cu2+ is re-established, that proton is released to form the leaving HO2– and it gets the second proton from the bulk water to form H2O2. There is much homology among all the Cu,ZnSODs, particularly with regard to the active site structure. Nevertheless, there are differences. Thus, some of the bacterial periplasmic Cu,ZnSODs are monomeric [10,11], while the extracellular SOD of mammals is tetrameric and is also glycosylated [12].
The MnSODS and the FeSODs
These SODs exhibit a great deal of homology, but are usually specific for the metal that constitutes the active site [13,14,15]. Thus the FeSOD found in Escherichia coli can be stripped of its metal and the resultant apoenzyme can be reconstituted with either Fe or Mn. If reconstituted with Fe, all of the activity is restored, but if reconstituted with Mn, it remains inactive [16]. In fact, the two metals can be shown to compete for binding to the active site. Some facultative anaerobes do contain a single SOD that can be active with either Fe or Mn at the active site [17]. These are called cambialistic SODs and when the organism is grown anaerobically, Fe is the metal inserted, but when grown aerobically, Mn is inserted. MnSOD is not only found in bacteria, but also in the matrix of mitochondria. Indeed, the parallel between Gram-negative bacteria and mitochondria is striking and can be taken as support for the endosymbiotic origin of these organelles [18]. Thus both E. coli and mitochondria have MnSOD in the cytosol and matrix respectively, and both have Cu,ZnSOD in the periplasm and intermembrane space, respectively (for details see [19]).
One might wonder why E. coli should be able to produce both a FeSOD and a MnSOD. The FeSOD is made at all times, whether the environment is aerobic or anaerobic, while the MnSOD is made under aerobic growth. Facultative microorganisms, such as E. coli, must face the possibility of sudden transfer from anaerobic to aerobic conditions. Hence the FeSOD can be viewed as a standby defense available at all times, while the induction of MnSOD provides for fine tuning of the level of defense to the level of threat. Fe2+ is stable and soluble anaerobically, but is prone to oxidation to Fe3+ aerobically, and Fe3+ forms insoluble hydroxide and phosphate salts. Mn salts, in contrast, remain soluble and thus available under both conditions. Hence it makes good sense that E. coli should make FeSOD at all times even anaerobically and induce the production of MnSOD aerobically. The same reasoning applies to the cambialistic SOD made by Propionibacterium shermanii.
Why So Many Types of SOD?
In addition to the Cu,ZnSODs, MnSODs, and FeSODs already mentioned there is a NiSOD found in Streptomyces[20]. All of these enzymes are comparably active. It is helpful to consider that there must have been a variety of organisms living in a variety of habitats that evolved during the anaerobic phase of this planet's history. Then, with the advent of true photosynthesis in the cyanobacteria, oxygen accumulated in the biosphere and that variety of anaerobes had to evolve SODs or perish. Different SODs evolved, depending on which metal was most available in the environment, and those different SODs are still here [21].
Assays for SOD Activity
Superoxide is not stable in protic solvents such as water, so it is not feasible, except by pulse radiolysis, to assay SOD by following its effect on the rate of consumption of its substrate. One way around this impasse is to have a reaction that produces a constant flux of O2·– and to allow that O2·– to react with some chromogenic substrate. In such a reaction system, SOD will compete with the chromogenic substrate for the flux of O2·– and thus inhibit the rate of color change. The activity of the SOD can then be derived from its inhibition of the color change. In the classical assay, the xanthine oxidase reaction provides the flux of O2·– and cytochrome c acts as the chromogen [22]. The sensitivity of this assay system depends upon the rate of production of O2·– and on the concentration of cytochrome. Obviously, anything that inhibits xanthine oxidase will decrease the rate of reduction of the cytochrome and thus appear to have SOD activity. Likewise, any compound that competes with the cytochrome c for O2·– will result in an apparent decrease in [SOD] or look like an inhibitor of SOD [23]. A control is needed to establish that the material being tested does not directly inhibit xanthine oxidase.
In other assays, the source of the flux of O2·– and the chromogen can be one and the same. Thus a compound that oxidizes spontaneously and so generates O2·– that propagates that oxidation to a final chromogen provides the basis of such an assay. Pyrogallol and epinephrine auto-oxidations are such O2·– propagated chain reactions that are inhibited by SOD and have been used to assay SOD [24,25]. In these assays anything that serves to initiate these chain oxidations will interfere with the measurement as will anything that can serve as a chain-breaking inhibitor. So these assays, although convenient, must be used carefully. There is no substitute for understanding the chemistry involved in the assay being used. Trace metal impurities in the reagents or in the buffer can certainly interfere, and that explains the usual inclusion of EDTA to chelate such trace metals.
Assay Applicable to Polyacrylamide Gels
Photochemically generated fluxes of O2·– combined with a chromogenic detector of O2·– can provide the basis of an activity stain for SOD. An additional requirement is that the chromogen be oxidized to an insoluble colored product. One compound that fulfills these requirements is nitroblue tetrazolium (NBT). In this assay the gel is soaked in a solution containing riboflavin as the photosensitizer, methionine as the photo-oxidizable substrate, and NBT as the chromogen [26]. This soaking is done in the dark and then the gel is rinsed and illuminated. The entire gel becomes blue due to the formazan product of NBT reduction, except at sites containing SOD, which remain colorless. There is a problem with NBT in that it can generate O2·– as well as detect it. For example, the enzyme glucose oxidase reduces oxygen to hydrogen peroxide. It does not generate O2·– but it does reduce NBT to a tetraazoinyl radical that can dismute to yield formazan or can reduce oxygen to O2·– while at the same time yielding NBT. So the reduction of NBT to its formazan by glucose oxidase plus glucose is inhibited by SOD, since the removal of O2·– favors the oxidation and so inhibits the dismutation of the tetrazoinyl radical. This led some authors to the mistaken conclusion that glucose oxidase could make O2·–. This error was subsequently corrected [27]. A tetrazolium that is not reduced to an auto-oxidizable intermediate provides a way to circumvent this artifact. The tetrazolium XTT (2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2-tetrazolium 5-carboxanilide) has these properties and has been used to good advantage [28,29]. Direct XTT reduction by NADPH:XTT reductases, however, makes the XTT assay unreliable in vivo [30].
Superoxide Assays Applied to Cells and Tissues
Investigators have devoutly wished for assays that would measure O2·– within cells and tissues. This is intrinsically difficult. Thus, it would require a cell-permeable compound that was modified specifically by O2·– to a stable product that could be detected spectrophotometrically or fluorimetrically. Furthermore, this compound would have to compete favorably with cellular SODs for the flux of O2·–. Unfortunately, the literature is littered with papers purporting to measure intracellular O2·– and they must all be viewed skeptically. However the finding that O2·– specifically oxidized hydroethidine to 2-hydroxy ethidinium, rather than to ethidinium, does provide a workable method [31].
Oxidative Stress and Medicine
The realization that the reactive and damaging intermediates of oxygen reduction are routinely made in cells led to the awareness that they may be involved in a variety of disease processes. This idea gained considerable traction from the finding that activated leukocytes undergo a respiratory burst that is accompanied by the formation of a large quantity of O2·–[32], and that genetic defects in the ability of the leukocytes to exhibit this respiratory burst are the basis of chronic granulomatous disease associated with a life-threatening susceptibility to infection [33]. At present, the literature is replete with accounts of the involvement of oxidative stress in many pathologies ranging from cancer to inflammations to reperfusion injuries to aging [34,35,36,37,38].
Orchestrated Defenses
The ubiquity of oxidative stress has called forth the evolution of coordinately regulated cohorts of enzymes dedicated to defending cells against these stresses and to repairing the damage due to oxidation. In E. coli, two such orchestrated defenses against oxidative damage have been described. One is called the soxRS regulon and the other is known as the oxyR regulon [19]. The soxRS regulon depends on the oxidation state of an iron-sulfur cluster in the SoxR protein [39]. When this cluster is oxidized, SoxR binds to and activates the operator of SoxS causing SoxS to be made; in turn, SoxS activates the genes for all the members of the regulon [40]. Over tens of genes are members of the soxRS regulon, and that indicates how important it is to defend against oxidative stress. One member of this regulon is the MnSOD that scavenges O2·; another is endonuclease 4 that helps repair oxidative damage to DNA [41]. Dehydratases that contain 4Fe4S clusters, such as fumarases A and B and aconitase B, are particularly susceptible to oxidation by O2·– [42,43], and the regulon includes fumarase C and aconitase A that are less susceptible to such oxidative inactivation [44]. The environment abounds in compounds that can mediate the oxidation of NAD(P)H with the concomitant formation of O2–. One member of the soxRS regulon serves to diminish the permeability of the cell envelope to such compounds [45]. This small sampling of the functions of regulon serves to illustrate the variety of things that contribute to the defense against oxidative stress in E. coli.
OxyR is regulated by the oxidation of a thiol group on the OxyR protein [46,47]. This thiol can be oxidized by H2O2 to a sulfenic acid that then reacts with a nearby thiol to generate a disulfide bond [48]. This changes the conformation of OxyR so that it binds to and activates the genes coding for the members of this regulon that include a catalase, glutathione peroxidase, and an alkylhydroperoxidase, among others [19].
Other Radicals
Nitric oxide (NO·) is produced in many organisms and serves as a signaling molecule. NO·, like O2·–, is a free radical and radical-radical reactions are fast. So it is no surprise that O2·– and NO· react with a diffusion limited rate constant to yield peroxynitrite, and that is a strong oxidant in its own right and can also homolyse to give two reactive radicals, i.e. NO2 and HO· [49]. This leads to the nitration of tyrosine residues in proteins and polyunsaturated fatty acids in phospholipids, and such modified tyrosines and fatty acids can be detected in inflamed tissues [50,51]. Another strongly oxidizing radical likely to be encountered in living things is the carbonate radical [52].
Epilogue
We now realize that the oxygen that is now so abundant in the earth's atmosphere, and is so essential for the life of aerobes such as we, is at the same time the progenitor of threatening reactive species. The study of the manifold effects of these reactive oxygen species and the defenses that allow aerobes to harvest the benefits of oxygen while surviving its onslaughts is still in its infancy. Pursuit of such studies will expand our understanding and provide solutions to currently intractable problems. As the Count de La Rochefoucauld once said, ‘Knowledge is the only way out of the cages of life’.
References
- 1.Bielski BHJ, Arudi RL. Preparation and stabilization of aqueous/ethanolic superoxide solutions. Anal Biochem. 1983;133:170–178. doi: 10.1016/0003-2697(83)90239-7. [DOI] [PubMed] [Google Scholar]
- 2.Gardner PR. Superoxide-driven aconitase FE-S center cycling. Biosci Rep. 1997;17:33–42. doi: 10.1023/a:1027383100936. [DOI] [PubMed] [Google Scholar]
- 3.Liochev SI. The role of iron-sulfur clusters in in vivo hydroxyl radical production. Free Radic Res. 1996;25:369–384. doi: 10.3109/10715769609149059. [DOI] [PubMed] [Google Scholar]
- 4.Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 5.Imlay JA, Fridovich I. Assay of metabolic superoxide production in Escherichia coli. J Biol Chem. 1991;266:6957–6965. [PubMed] [Google Scholar]
- 6.Chiang SM, Schellhorn HE. Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch Biochem Biophys. doi: 10.1016/j.abb.2012.02.007. DOI: 10.1016/j.abb.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 7.McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 8.Hodgson EK, Fridovich I. The interaction of bovine erythrocyte superoxide dismutase with hydrogen peroxide: inactivation of the enzyme. Biochemistry. 1975;14:5294–5299. doi: 10.1021/bi00695a010. [DOI] [PubMed] [Google Scholar]
- 9.Klug D, Rabani J, Fridovich I. A direct demonstration of the catalytic action of superoxide dismutase through the use of pulse radiolysis. J Biol Chem. 1972;247:4839–4842. [PubMed] [Google Scholar]
- 10.Benov L, Sage H, Fridovich I. The copper- and zinc-containing superoxide dismutase from Escherichia coli: Molecular weight and stability. Arch Biochem Biophys. 1997;340:305–310. doi: 10.1006/abbi.1997.9940. [DOI] [PubMed] [Google Scholar]
- 11.Battistoni A, Rotilio G. Isolation of an active and heat-stable monomeric form of Cu,Zn superoxide dismutase from the periplasmic space of Escherichia coli. FEBS Lett. 1995;374:199–202. doi: 10.1016/0014-5793(95)01106-o. [DOI] [PubMed] [Google Scholar]
- 12.Marklund SL. Extracellular superoxide dismutase. Methods Enzymol. 2002;349:74–80. doi: 10.1016/s0076-6879(02)49322-6. [DOI] [PubMed] [Google Scholar]
- 13.Gregory EM, Yost FJ, Jr, Fridovich I. Superoxide dismutases of Escherichia coli: intracellular localization and functions. J Bacteriol. 1973;115:987–991. doi: 10.1128/jb.115.3.987-991.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keele BB, Jr, McCord JM, Fridovich I. Superoxide dismutase from Escherichia coli B. A new manganese-containing enzyme. J Biol Chem. 1970;245:6176–6181. [PubMed] [Google Scholar]
- 15.Yost FJ, Jr, Fridovich I. An iron containing superoxide dismutase from Escherichia coli. J Biol Chem. 1973;248:4905–4908. [PubMed] [Google Scholar]
- 16.Privalle CT, Fridovich I. Transcriptional and maturational effects of manganese and iron on the biosynthesis of manganese-superoxide dismutase in Escherichia coli. J Biol Chem. 1992;267:9140–9145. [PubMed] [Google Scholar]
- 17.Gabbianelli R, Battistoni A, Polizio F, Carrì MT, De Martino A, Meier B, Desideri A, Rotilio G. Metal uptake of recombinant cambialistic superoxide dismutase from Propionibacterium shermanii is affected by growth conditions of host Escherichia coli cells. Biochem Biophys Res Commun. 1995;216:841–847. doi: 10.1006/bbrc.1995.2698. [DOI] [PubMed] [Google Scholar]
- 18.Fridovich I. Superoxide dismutases. An adaptation to a paramagnetic gas. J Biol Chem. 1989;264:7761–7764. [PubMed] [Google Scholar]
- 19.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim EJ, Kim HP, Hah YC, Roe JH. Differential expression of superoxide dismutases containing Ni and Fe/Zn in Streptomyces coelicolor. Eur J Biochem. 1996;241:178–185. doi: 10.1111/j.1432-1033.1996.0178t.x. [DOI] [PubMed] [Google Scholar]
- 21.Fridovich I. Superoxide dismutases. Adv Enzymol Relat Areas Mol Biol. 1986;58:61–97. doi: 10.1002/9780470123041.ch2. [DOI] [PubMed] [Google Scholar]
- 22.Okado-Matsumoto A, Fridovich I. Assay of superoxide dismutase: cautions relevant to the use of cytochrome c, a sulfonated tetrazolium, and cyanide. Anal Biochem. 2001;298:337–342. doi: 10.1006/abio.2001.5385. [DOI] [PubMed] [Google Scholar]
- 23.Hassan HM, Dougherty H, Fridovich I. Inhibitors of superoxide dismutases: a cautionary tale. Arch Biochem Biophys. 1980;199:349–354. doi: 10.1016/0003-9861(80)90290-8. [DOI] [PubMed] [Google Scholar]
- 24.Misra HP, Fridovich I. The role of superoxide anion in the autoxidation of epinephrine and a simple assay for superoxide dismutase. J Biol Chem. 1972;247:3170–3175. [PubMed] [Google Scholar]
- 25.Marklund S, Marklund G. Involvement of the superoxide anion radical in the autoxidation of pyrogallol and a convenient assay for superoxide dismutase. Eur J Biochem. 1974;47:469–474. doi: 10.1111/j.1432-1033.1974.tb03714.x. [DOI] [PubMed] [Google Scholar]
- 26.Beauchamp C, Fridovich I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem. 1971;44:276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- 27.Liochev SI, Fridovich I. Superoxide from glucose oxidase or from nitroblue tetrazolium. Arch Biochem Biophys. 1995;318:408–410. doi: 10.1006/abbi.1995.1247. [DOI] [PubMed] [Google Scholar]
- 28.Sutherland MW, Learmonth BA. The tetrazolium dyes MTS and XTT provide new quantitative assays for superoxide and superoxide dismutase. Free Radic Res. 1997;27:283–289. doi: 10.3109/10715769709065766. [DOI] [PubMed] [Google Scholar]
- 29.Ukeda H, Maeda S, Ishii T, Sawamura M. Spectrophotometric assay for superoxide dismutase based on tetrazolium salt 3′-{1-[(phenylamino)-carbonyl]-3,4-tetrazolium}-bis(4-methoxy-6-nitro)benzenesulfonic acid hydrate reduction by xanthine-xanthine oxidase. Anal Biochem. 1997;251:206–209. doi: 10.1006/abio.1997.2273. [DOI] [PubMed] [Google Scholar]
- 30.Benov L, Fridovich I. Is reduction of the sulfonated tetrazolium 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2-tetrazolium 5-carboxanilide a reliable measure of intracellular superoxide production? Anal Biochem. 2002;310:186–190. doi: 10.1016/s0003-2697(02)00310-x. [DOI] [PubMed] [Google Scholar]
- 31.Zielonka J, Vasquez-Vivar J, Kalyanaraman B. Detection of 2-hydroxyethidium in cellular systems: a unique marker product of superoxide and hydroethidine. Nat Protoc. 2008;3:8–21. doi: 10.1038/nprot.2007.473. [DOI] [PubMed] [Google Scholar]
- 32.Babior BM, Kipnes RS, Curnutte JT. Biological defense mechanisms. The production by leukocytes of superoxide, a potential bactericidal agent. J Clin Invest. 1973;52:741–744. doi: 10.1172/JCI107236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roos D, Van Bruggen R, Meischl C. Oxidative killing of microbes by neutrophils. Microbes Infect. 2003;5:1307–1315. doi: 10.1016/j.micinf.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 34.Elmarakby AA, Sullivan JC. Relationship between oxidative stress and inflammatory cytokines in diabetic nephropathy. Cardiovasc Ther. 2012;30:49–59. doi: 10.1111/j.1755-5922.2010.00218.x. [DOI] [PubMed] [Google Scholar]
- 35.Jaeschke H, McGill MR, Ramachandran A. Oxidant stress, mitochondria, and cell death mechanisms in drug-induced liver injury: lessons learned from acetaminophen hepatotoxicity. Drug Metab Rev. 2012;44:88–106. doi: 10.3109/03602532.2011.602688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sohal RS, Orr WC. The redox stress hypothesis of aging. Free Radic Biol Med. 2012;52:539–555. doi: 10.1016/j.freeradbiomed.2011.10.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhimaraj A, Tang WHW. Role of oxidative stress in disease progression in stage B, a pre-cursor of heart failure. Heart Fail Clin. 2012;8:101–111. doi: 10.1016/j.hfc.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kryston TB, Georgiev AB, Pissis P, Georgakilas AG. Role of oxidative stress and DNA damage in human carcinogenesis. Mutat Res. 2011;711:193–201. doi: 10.1016/j.mrfmmm.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 39.Zheng M, Storz G. Redox sensing by prokaryotic transcription factors. Biochem Pharmacol. 2000;59:1–6. doi: 10.1016/s0006-2952(99)00289-0. [DOI] [PubMed] [Google Scholar]
- 40.Nunoshiba T, Hidalgo E, Cuevas CFA, Demple B. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J Bacteriol. 1992;174:6054–6060. doi: 10.1128/jb.174.19.6054-6060.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nunoshiba T. Two-stage gene regulation of the superoxide stress response soxRS system in Escherichia coli. Crit Rev Eukaryot Gene Expr. 1996;6:377–389. doi: 10.1615/critreveukargeneexpr.v6.i4.30. [DOI] [PubMed] [Google Scholar]
- 42.Gardner PR, Fridovich I. Superoxide sensitivity of the Escherichia coli aconitase. J Biol Chem. 1991;266:19328–19333. [PubMed] [Google Scholar]
- 43.Liochev SI, Fridovich I. Modulation of the fumarases of Escherichia coli in response to oxidative stress. Arch Biochem Biophys. 1993;301:379–384. doi: 10.1006/abbi.1993.1159. [DOI] [PubMed] [Google Scholar]
- 44.Liochev SI, Fridovich I. Fumarase C, the stable fumarase of Escherichia coli, is controlled by the soxRS regulon. Proc Natl Acad Sci USA. 1992;89:5892–5896. doi: 10.1073/pnas.89.13.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chou JH, Greenberg JT, Demple B. Posttranscriptional repression of Escherichia coli OmpF protein in response to redox stress: positive control of the micF antisense RNA by the soxRS locus. J Bacteriol. 1993;175:1026–1031. doi: 10.1128/jb.175.4.1026-1031.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antelmann H, Helmann JD. Thiol-based redox switches and gene regulation. Antioxid Redox Signal. 2011;14:1049–1063. doi: 10.1089/ars.2010.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pomposiello PJ, Demple B. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 2001;19:109–114. doi: 10.1016/s0167-7799(00)01542-0. [DOI] [PubMed] [Google Scholar]
- 48.Åslund F, Zheng M, Beckwith J, Storz G. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc Natl Acad Sci USA. 1999;96:6161–6165. doi: 10.1073/pnas.96.11.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merényi G, Lind J. Free radical formation in the peroxynitrous acid (ONOOH)/peroxynitrite (ONOO-) system. Chem Res Toxicol. 1998;11:243–246. doi: 10.1021/tx980026s. [DOI] [PubMed] [Google Scholar]
- 50.Radi R. Nitric oxide, oxidants, and protein tyrosine nitration. Proc Natl Acad Sci USA. 2004;101:4003–4008. doi: 10.1073/pnas.0307446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schopfer FJ, Baker PRS, Freeman BA. NO-dependent protein nitration: a cell signaling event or an oxidative inflammatory response? Trends Biochem Sci. 2003;28:646–654. doi: 10.1016/j.tibs.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 52.Goss SPA, Singh RJ, Kalyanaraman B. Bicarbonate enhances the peroxidase activity of Cu,Zn-superoxide dismutase. Role of carbonate anion radical. J Biol Chem. 1999;274:28233–28239. doi: 10.1074/jbc.274.40.28233. [DOI] [PubMed] [Google Scholar]