Abstract
Metabolomics has been used as a powerful tool for the analysis and quality assessment of the natural product (NP)-derived medicines. It is increasingly being used in the quality control and standardization of NP-derived medicines because they are composed of hundreds of natural compounds. The most common techniques that are used in metabolomics consist of NMR, GC-MS, and LC-MS in combination with multivariate statistical analyses including principal components analysis (PCA) and partial least squares-discriminant analysis (PLS-DA). Currently, the quality control of the NP-derived medicines is usually conducted using HPLC and is specified by one or two indicators. To create a superior quality control framework and avoid adulterated drugs, it is necessary to be able to determine and establish standards based on multiple ingredients using metabolic profiling and fingerprinting. Therefore, the application of various analytical tools in the quality control of NP-derived medicines forms the major part of this review. Veregen® (Medigene AG, Planegg/Martinsried, Germany), which is the first botanical prescription drug approved by US Food and Drug Administration, is reviewed as an example that will hopefully provide future directions and perspectives on metabolomics technologies available for the quality control of NP-derived medicines.
Keywords: Metabolomics, Natural product-derived medicines, Quality control, Veregen®
QUALITY CONTROL OF NATURAL PRODUCT DERIVED MEDICINES
A natural product (NP)-derived medicine is a medicinal product that consists of herbal substances or natural remedies such as plants, algae, or macroscopic fungi. The natural remedies such as Camellia sinensis leaves have been used as the important plant sources for the preparation of NP-derived medicines because they are known to have several therapeutic effects such as antioxidant (Yen and Chen, 1995; Babu et al., 2006), anticancer (Cooper et al., 2005; Landis-Piwowar et al., 2007), and hypolipidemic (Lin et al., 1998; Lin and Lin-Shiau, 2006). The use of NP-derived drugs is rapidly growing, and the minority of the population is no longer the main subscriber, but the attention of a considerable number of people has become focused on the eco- and bio-friendly products. Moreover, the belief, which NP-derived drugs are relatively free from the side effects that are frequently associated with synthetic drugs, is an added attraction. Recently, both the Eastern as well as the Western population have sought natural remedies, which are considered safe and effective. Furthermore, there is a continuing effort to discover new NP-derived medicines and develop frameworks for their quality control.
The methods that are currently used for the quality control of NP-derived medicines include morphological assessment, as well as sensory, physicochemical, and biological evaluations (Kunle et al., 2012). Among these evaluations, the determination of the quantitative indicators of the ingredients in NP-derived drug is the most important physicochemical quality evaluation. Currently, the quality control of natural remedies is usually performed using high-performance liquid chromatography (HPLC) and is specified by evaluating one or two indicators. Table 1 shows the existing markers of representative natural ingredients that are recorded in the Korean Pharmaceutical Codex and previous studies. For instance, the apricot kernel (Armeniacae semen), which is a NP-derived drug with antitussive, expectorant, and laxative functions, was analyzed using HPLC, and amygdalin was identified as its major effective ingredient (Lv et al., 2005). Turmeric (Curcuma longa rhizoma), which has the pharmacological properties such as anti-parasitic, anti-mutagenic, and antimicrobial, was also analyzed using HPLC and reported curcumin, desmethyoxycurcuin, and bisdesmethoxycurcuin as the main bioactive substances (Wichitnithad et al., 2009). In addition, berberine is known as the major active ingredient of the coptis rhizome (Coptidis rhizoma), which is used for the treatment of conditions such as diarrhea, jaundice, and sore throat (Selvam, 2008; Hasada et al., 2011). Ginsenoside Rg3 and baicalin were analyzed using HPLC and reported as the marker compounds of red ginseng (Ginseng radix rubra) and scutellaria root (Scutellariae radix), respectively (Court, 2000; Bochořáková et al., 2003).
Table 1.
Marker compounds of natural ingredients that are recorded in the Korean Pharmaceutical Codex and previous studies
| Botanical name | Common name | Marker compounds | Detection method | Reference |
|---|---|---|---|---|
| Areca arecae semen | Areca nut | Arecoline | HPLC | Jantarat et al., 2013 |
| Armeniacae semen | Apricot kernel | Amygdalin | HPLC | Lv et al., 2005 |
| Caulophyllum robustum | Blue cohosh | Aporphinoid N-methylcytisine, Lupanine |
HPLC GC-MS |
Li et al., 2007 |
| Citri unshii pericarpium | Citrus unshiu peel | Hesperidin | HPLC | Nugroho et al., 2009 |
| Coptidis rhizoma | Coptis rhizome | Berberine | 1H-NMR | Hasada et al., 2011 |
| Curcuma longa rhizoma | Turmeric | Curcumine, Desmethoxycurcumin, Bisdesmethoxycurcumin | HPLC | Wichitnithad et al., 2009 |
| Ephedra. sinica | Ephedra | Pseudoephedrine, Norephedrine, Norpseudoephedrine, Methylephedrine | HPLC | Gurley et al., 1998 |
| Gardeniae fructus | Gardenia fruit | Geniposide | HPLC | Tsai et al., 2002 |
| Ginseng radix alba | White ginseng | Ginsenoside Rb1 Ginsenoside Rg1 Panaxadiol |
HPLC HPLC GC |
Samukawa et al., 1995; Court, 2000 |
| Ginseng radix rubra | Red ginseng | Ginsenoside Rg3 | HPLC | Court, 2000 |
| Glycyrrhizae radix et rhizoma | Licorice root | Glycyrrhizinic acid | HPLC, LC-MS | Zhang and Ye, 2009 |
| Lycium Chinese miller | Chinese matrimony vine | Betaine | HPLC | Shin et al., 1999 |
| Moutan cortex | Moutan root bark | Paeonol, paeoniflorin | HPLC | Ding et al., 2009 |
| Paeoniae radix | Peony root | Paeoniflorin | HPLC | Suzuki, 1984 |
| Persicae semen | Peach kernel | Amygdalin | HPLC | Ghiulai et al., 2006 |
| Phellodendri cortex | Phellodendron bark | Berberine | HPLC | Kataoka et al., 2008 |
| Ponciri fructus | Poncirus fruit | Poncirin | HPLC | Avula et al., 2005 |
| Puerariae radix | Pueraria root | Puerarin | HPLC | Oh et al., 1990 |
| Schisandrae fructus | Schisandra fruit | Schizandrin | HPLC | Xu et al., 2007 |
| Scutellariae baicalensis | Scutellaria root | Baicalin | HPLC | Bochořáková et al., 2003 |
| Swertiae herba | Swertia | Swertiamarin | HPLC | Kim et al., 2014 |
HPLC, high-performance liquid chromatography; GC, gas chromatography; GC-MS, gas chromatography-mass spectrometry; NMR, nuclear magnetic resonance; LC-MS, liquid chromatography-mass spectrometry.
Until now, the quality of NP-derived medicines has been determined by the evaluation of one or two kinds of indicators. However, NP-derived drugs usually contain multiple compounds, which indicate that one or two markers would not adequately represent a suitable framework of good quality control. Therefore, quality control involving the evaluation of multiple ingredients using metabolic profiling has increasingly been used to avoid the production of substandard and adulterated NP-derived drugs. The metabolic profiling information can subsequently be applied to the quality assessment of various kinds of medicinal plants and the prediction of various bioactivities of those such as antioxidative, anticancer, and anti-inflammatory, using advanced analytical techniques (such as HPLC, GC, and NMR) and multivariate statistical analyses.
METABOLOMICS IN PHARMACETUCIAL INDUSTRY
Metabolomics is an emerging “omics” technology that is primarily concerned with the high-throughput quantification and identification of small-molecule (MW 100–1,000) metabolites in cells, tissues, or fluids. This technology is known to be one of the 10 leading breakthrough technologies according to the Massachusetts Institute of Technology (MIT) Technology Review 2005 (MIT Technology Review, 2005). Recently, the process of drug discovery and development has become very expensive while simultaneously, the number of new drug approvals is steadily declining (Scannell et al., 2012). There is an increasing knowledge that the diseases that are of current interest (including cancer, obesity, Parkinson’s disease, and Alzheimer’s disease) are complex disorders that do not present with simple or single drug targets. These insights have contributed to major transformations that have led to new drug discovery strategies, including the shift from single to multi-compound drugs. NP-derived medicines have been recognized as representative multi-compound drugs and can be investigated by metabolomics technology using various chromatography and spectrometry methods for multi-drug target screening and quality control. In addition, recent improvements in metabolomics technologies have incorporated the valuable tools of gene-function analysis, system biology, and diagnostic platforms. New therapeutic challenges and trends continually create the increasing need for more rapid scientific responses, and metabolomics is currently positioned to be an important tool for providing biomarkers for better diagnosis and prognosis. This paper aims to highlight the superior quality control of NP-derived medicines that can be achieved using metabolomics techniques.
ANALYTICAL PLATFORMS OF METABOLOMICS
A number of analytical platforms, such as nuclear magnetic resonance (NMR), Fourier transform-infrared spectroscopy (FT-IR), and mass spectrometry (MS) coupled to separation techniques including gas chromatography (GC)-MS, liquid chromatography (LC)-MS, and ultra-performance liquid chromatography-mass spectrometry (UPLC-MS), have been used in metabolic fingerprinting and metabolomics. Recently, the technologies that are commonly used for global metabolome studies have been increasingly combined with multi-hyphenated techniques such as GC × GC-time-of-flight (TOF), GC-TOF-MS, and UPLC-quadrupole (Q)-TOF-MS to enable compound analysis using a wider range of metabolome perspectives (Jensen et al., 2002; Okada et al., 2009; Yang et al., 2011; Farag and Wessjohann, 2012; Lee et al., 2014).
NMR is a commonly used analytical technique, which identifies and quantifies a wide range of organic compounds. It has a simple sample preparation step while biofluids such as urine and serum can be directly analyzed without preparation step. In addition, it assesses numerous groups of metabolites, so it is suitable for analyzing the components of unknown medicinal plants for further determination of efficacy. Because NMR is non-selective, all the low molecular weight compounds can be detected concurrently with the structural information, which also helps characterizing components of any complex mixtures (Zhang et al., 2012). The advantages of NMR have made it a useful technique with a long history of use in metabolomics analysis. Therefore, the associated protocols and databases are well developed, providing a rich source of operational information (Weckwerth and Morgenthal, 2005; Kim et al, 2010). The major limitation of NMR is that it has a relatively low sensitivity (micromolar range) compared to MS (picomolar range) (Gromski et al., 2015). Recently, high-throughput NMR techniques have remedied these shortcomings, thereby enabling the detection of disease biomarkers and substitute markers for drug delivery and efficacy (Zhang et al., 2012). Several studies have reported metabolic profiling of natural resources using NMR and multivariate analysis for the quality control. Yang et al. (2006) identified potential markers for the quality control of ginseng (Panax ginseng) using a proton (1H) and two-dimensional NMR metabolomics approaches. They also classified four kinds of ginseng roots for the efficient screening method using soft independent modeling by class analogy and principal components analysis (PCA). In that study, the NMR spectral patterns revealed the presence of alanine, arginine, choline, fumaric acid, inositol, sucrose, and ginsenosides as the important metabolites to differentiate four kinds of ginseng roots, which indicates the potential markers that could be used to characterize these ginseng categories. Kang et al. (2008b) also has provided similar information on differentiation of Panax ginseng from six different origins and ages from China and Korea by using NMR and PCA. From the research, they described that NMR-based metabolomics approach could be applied to detect the adulterated ginseng roots as well as other herbal products from different origins. Furthermore, Kim et al. (2005) used an NMR-based metabolic fingerprinting to distinguish three different Ephedra species (Ephedra sinica, Ephedra intermedia, and Ephedra equisetina) for their quality control. Ephedrine alkaloids and benzoic acid analogs were found to be the important distinguishing metabolites. Other studies using NMR-based methods have been performed for the analysis of metabolites including natural resources like sweet warmwood (Artemisia annua), gingko (Gingko biloba) leaves, and scutellaria root (Scutellaria baicalensis) (Choi et al., 2003; Kang et al., 2008a; Van der Kooy et al., 2008).
FT-IR spectrometry is another valuable metabolic fingerprinting tool, which analyzes a diverse range of sample types and metabolites such as carbohydrates, amino acids, lipids, fatty acids, proteins, and polysaccharides simultaneously (Dunn and Ellis, 2005). It also requires minimum sample preparation and relatively little background training and, therefore, it can be easily used as a highly versatile technique. It works by correlating the absorption and vibration of light at specific wavelengths to the functional groups of molecules for the identification of unknown metabolites. The major limitation of FT-IR spectrometry is the relatively low sensitivity and selectivity. In addition, wet samples are difficult to be analyzed because water can be the issue in mid IR (Gromski et al., 2015). Recently, FT-IR has been used for quality control of natural resources. For instance, Lu et al. (2008) applied FT-near infrared (NIR) in discriminating goji berry (Fructus lycii) of four different geographic regions. Through metabolic fingerprinting, the spectra showed the differences in the range of 4,950–5,700 cm−1 between the samples of different geographic regions. In addition, Kwon et al. (2014) analyzed whole-cell extracts of ginseng leaves using FT-IR and multivariate analysis to distinguish the cultivation age and cultivars of ginseng (Panax ginseng) leaves. The analysis revealed that the most significant spectral variation among four ginseng cultivars was observed in the polysaccharide and amide regions (1,050 to 1,150 and 1,550 to 1,650 cm−1, respectively), which were subsequently identified as potential markers for distinguishing different ginseng cultivars and cultivation ages. FT-IR can also be applied to the discrimination of other NP-derived medicines including areca nuts (Areca catechu) and citrus (Citrus unshiu) of different cultivars and geographic regions for their quality control (Fu et al., 2013; Song et al., 2016).
MS is a widely used technology, which can identify metabolites by providing rapid and selective qualitative and quantitative data with high sensitivity and resolution. It operates by ion formation and separation, and detection of separated ions (Dunn and Ellis, 2005). GC-MS is a combined system where volatile mixtures of compounds are separated by GC, and the eluted compounds are subsequently detected using MS. GC-MS involves derivatization to induce volatility and thermal stability before analyzing volatile metabolites. After derivatization, it is possible to profile hundreds of metabolites simultaneously, including organic acids, amino acids, sugars, sugar alcohols, aromatic amines, and fatty acids, by direct separation and quantification (Zhang et al., 2012). GC-MS handles a large volume of samples and precise peak identification via standard retention times and mass spectra. Due to its high sensitivity, throughput, and comprehensiveness, GC-MS has a history of long-standing use in metabolomics studies; therefore, it is relatively easy to identify compounds based on numerous available databases and protocols. The major limitation of GC-MS is that it is restricted to volatile compounds, and derivatization process is necessary to detect various (2002) metabolites. Derivatization may make difficulties in sample preparation and identification due to any additives and multiple derivative products. In addition, heat sensitive compounds are difficult to be analyzed. GC-MS and multivariate analysis were used in several studies for metabolic profiling and fingerprinting of NP-derived medicine for quality assessment (Jacquemond-Collet et al., 2001; Li et al., 2007; Farag et al., 2012; Lee et al., 2014). Li et al. (2007) used GC-MS to fingerprint Caulophyllum robustum for its quality control, and detected aporphinoid and quinolizidine as the major components of the total alkaloid content, and potential quality control indicators. Furthermore, Farag et al. (2012) used GC-MS for primary metabolite profiling of different species of licorice roots (Glycyrrhiza glabra, Glycyrrhiza uralensis, Glycyrrhiza inflata, and Glycyrrhiza echinata) to distinguish between them and set a framework for their quality control. A total of 33 metabolites including saccharides, as well as amino, fatty, and phenolic acids were detected in the licorice roots. Cadaverine was found only in G. inflata, while the highest myo-inositol content was detected in G. echinata. In addition, sucrose was the major component of all the samples, which suggests that these metabolites could be used as markers to distinguish Glycyrrhiza species. Other studies using GC-MS-based methods have been used in metabolomics including the comparative study of turmeric (Curcuma) and the discrimination of two different cultivars, C. aromatica and C. longa, as well as the identification of alkaloids of rutaceae (Galipea officinalis), by using a GC-TOF-MS, multivariate statistical analysis, and GC-MS, respectively (Jacquemond-Collet et al., 2001; Lee et al., 2014).
Another analytical strategy is the LC-MS, where LC separates the metabolites. LC-MS requires lower analysis temperatures and simpler sample preparation than GC-MS does. It does not require sample volatility, so sample derivatization is generally not required. Metabolites are normally detected in positive or negative ion modes and, therefore, a wide range of metabolites including polar, semi-polar, and non-polar compounds and secondary metabolites can be detected by analysis in both modes. The high throughput and comprehensiveness of LC-MS have contributed to its versatility, particularly in targeted metabolite identification and quantification in complex mixtures (Dunn and Ellis, 2005). However, LC-MS has a restricted applicability to compound identification in the non-targeted analysis because of some undifferentiated isomers and formation of multiple adducts. Recently, hydrophilic interaction LC (HILIC)-MS, LC-atmospheric pressure chemical ionization (APCI)-MS, and UPLC-TOF-MS have been successfully used for comprehensive metabolic profiling of natural products (Jensen et al., 2002; Okada et al., 2009; Montoro et al., 2011; Yang et al., 2011; Farag and Wessjohann, 2012; Farag et al., 2012).
Previous studies have shown the metabolic profiling and the discrimination of natural products, such as liquorice roots (Glycyrrhiza glabra) of different cultivars and geographical areas using LC-ESI-MS in both positive and negative ionization modes (Montoro et al., 2011; Farag et al., 2012). Glycyrrhizin, 4-hydroxyphenyl acetic acid, and glycosidic conjugates of liquiritigenin showed significantly different content between the four kinds of glycyrrhiza genus (G. glabra, G. uralensis, inflata, echinata) (Farag et al., 2012). Furthormore, phenolic constituents and saponins related to glycyrrhizic acid were identified as the comparative metabolites for the determination of geographical areas of liquorice roots (Montoro et al., 2011). Jensen et al. (2002) also used LC-APCI-MS to evaluate the composition of the active terpene constituents of Ginkgo biloba (ginkgolides and bilobalide) in the negative ion mode. Ginkgolides and bilobalide were detected as the main active terpene constituents and suggested as suitable quality assessment markers of NP-derived medicine (Jensen et al., 2002). Other studies using LC-MS-based methods have been applied to metabolic profiling and fingerprinting processes such as the identification, quantitation, and principal component analysis of Senecio plants, Ephedra plants, and St. John’s Wort (Hypericum perforatum) using the UPLC-diode array detector (DAD)-ESI-MS and UPLC-Q-TOF-MS (Okada et al., 2009; Yang et al., 2011; Farag and Wessjohann, 2012).
MULTIVARIATE STATISTICAL ANALYSIS
Quality control of herbal extracts is a challenging endeavor because they usually contain numerous phytochemicals (Kim et al., 2016). Furthermore, multivariate statistical analyses are needed to reduce the complexity of the data from metabolic profiling and facilitate the detection of the pattern of changes related to the environmental or genetic factors in metabolite compositions.
PCA, partial least squares-discriminant analysis (PLS-DA), and PLS-regression (PLS-R) are widely used multivariate data analysis methods. The PCA is an unsupervised multivariate analysis method, which is widely used in metabolic fingerprinting and profiling. It shows the overview of the obtained data by representing the original multivariate data as an unbiased, lower dimensional output data. This overview provides information not only on the groups of observations, trends, and outliers but also on the relationships between the observations (Eriksson et al., 2006). PCA is determined using the score plot that shows the statistical differences between the groups and the loading plot that displays the compounds that are responsible for the differences between the groups. PCA, however, has a limit to validate statistical models because it cannot assign the class membership of unknown test samples (Kang et al., 2008a, 2008b). Thus, additional multivariate data analysis methods, such as PLS-DA and PLS-R should be performed in order to investigate class differentiation of unknown samples or determination of origins of the natural products.
The PLS analysis is a regression extension of the PCA, which is used to relate the information on two matrices (independent and dependent X and Y variables, respectively) using regression analysis (Eriksson et al., 2006). It is usually used to observe the spaces and maximized correlation between the X and Y groups and to estimate the specific activities of the data sets. PLS-DA is often used and involves suggesting the class membership of each observation to maximize the separation direction among the classes of observations. The main advantage of the PLS-DA is its ability to improve the separation between the groups of observations and analyze which variables convey the class separation (Perez-Enciso and Tenenhaus, 2003). Therefore, it enables the provision of more information about the observation. On the other hand, it can also cause the model validation to be overlooked or overfitting. It may also present overoptimistic view of the class separation on the score plot (Gromski et al., 2015). In contributing to the separation process of the PLS-DA model, the variable importance in projection (VIP) is performed to reflect the influence of the variables from the metabolic profiling data results. The optimal PLS-DA model is determined by the goodness-fit parameter (R2Y) and predictive ability parameter (Q2Y) with values close to 1. Since the PLS-DA model is supervised, it needs to be validated by performing a random permutation test.
PLS-R represents the relationship between two X and Y groups of a PLS model and interprets the model to predict Y by analyzing X (Abdi, 2003; Eriksson et al., 2006). To validate the PLSR model, a cross-validation is first performed to determine the suitability of the model (Eriksson et al., 2006; Kim et al., 2016). R2Y is a parameter that represents the goodness of fit and indicates how close the values from the PLSR are to the actual values. In other words, the R2Y determines the margin of error, and it ranges from 0 to 1, where 1 is considered the perfect fit. The Q2Y is another parameter that represents the goodness of prediction and indicates how accurate the Y-variable prediction is. The values that are greater than 0.5 and 0.9 are considered as good and excellent, respectively (Eriksson et al., 2006). If the model is considered suitable, a permutation test is performed as the next step. The permutation test is necessary to evaluate the statistical significance of the estimated predictive power of the Q2Y values from the cross-validation. When the permutation test is performed, the PLSR models including the samples with the randomly selected Y-variable values inevitably emerge with low cross-validation values, which are the R2Y and Q2Y values. Therefore, the permutation test determines how the cross-validation value of the PLS model using the normal samples values is statistically significant compared to the other random models. As determining criteria, R2Y and Q2Y intercepts <0.3-0.4 and <0.05, respectively indicate the validity of the model (Eriksson et al., 2006). If the model is not statistically significant, the number of components set by the autofit should be checked to ensure their suitability. Otherwise, it should be reduced manually until it passes the permutation test. In this case, the autofit cannot be performed because the R2Y intercept would approach 1 if the autofit is performed, and then there would be almost no difference between the actual and random models. This suggests that the model can be overfitted when an autofit is performed.
Usually, the model is obtained by setting the component in the highest Q2Y value, and the R2Y has the tendency to become close to 1 as the number of components increases. Since the autofit is the process used to obtain the highest Q2Y value, the model naturally passes through the cross-validation test. Autofit, however, can have a limitation in passing the permutation test. The external validation is performed as the first step in determining the accuracy of the model by importing an independent test data set, which was not used in building the model, into the corresponding PLSR model (Kim et al., 2016). The root-mean-square error of estimation and root-mean-square error of prediction represent the precision of the PLS projection to the PLSR with the training and test sets, respectively, and values close to zero are considered to depict a high-precision model. This established model through all the verification processes can be reliable as a validated model in terms of accuracy and predictability.
CASE STUDY: VEREGEN® (Medigene AG, Planegg/Martinsried, Germany)
Numerous natural resources have been used as medicinal drugs for many centuries worldwide. Various benefits to human health exhibited by natural remedies or herb-derived products have led to these substances being investigated by numerous pharmaceutical companies. For instance, artemisinin extracted from the sweet wormwood (Artemisia annua) plant was discovered and developed as an anti-malarial agent with specific activity against Plasmodium falciparum, by Chinese scientists (Cui and Su, 2009). Legalon® SIL was developed by Rottapharm/Madaus (Cologne, Germany) from silibinin from milk thistle (Silybum marianum) fruit seeds and is used for the treatment of acute hepatotoxicity (Mengs et al., 2012).
Veregen® is the first botanical ointment produced by MediGene AG and was originally approved as a prescription drug by the US Food and Drug Administration (FDA) on October 31, 2006 (US Food and Drug Administration, 2006b) (http://www.accessdata.fda.gov/). It is indicated for the topical treatment of external and perianal warts (condylomata acuminata) (Chen et al., 2008). Vergen is an extract of the Chinese green tea (C. sinensis) leaf, containing a mixture of active compounds and its active ingredient is sinecatechin, which is the major chemical constituent of Chinese green tea. Although it is a relatively simple NP-derived drug compared to other botanicals, the quality control is still not as easy as that of other pure drugs. Quality control measures should be made and implemented by the FDA on the commercial production of this native plant. Therefore, Veregen needs to be carefully evaluated in setting the quality specification by analyzing each individual major and minor catechin as well as the unknown metabolites using various techniques, rather than just controlling the total catechin.
The chemistry and manufacturing controls of Veregen 15% ointment have been reviewed and approved by the US FDA and reported in the Chemistry Review application for the Center for Drug Evaluation and Research (Application number: NDA 21-902) (US Food and Drug Administration, 2006a)(http://www.accessdata.fda.gov/). In the chemistry assessment, Veregen was analyzed using HPLC to quantify and identify all the catechin components to assess the overall quality of the drug product. It was found to contained 15% kunecatechins drug substance as well as excipients consisting of isopropyl myristate, white petrolatum, beeswax (cera alba), propylene glycol palmitostearate (propylene glycol monopalmitostearate), and oleyl alcohol. Kunecatechins is a mixture of catechins, and a total of eight were identified and quantitated, including epigallocatechin gallate (EGCG), epicatechin (EC), epicatechin gallate (ECG), epigallocatechin (EGC), gallocatechin gallate (GCG), gallocatechin (GC), catechin gallate (CG), and catechin (C) (http://www.accessdata.fda.gov/). Among them, the major component was EGCG, which constitutes approximately 55% of the total catechin content. Kunecatechins also contain gallic acid, caffeine, and theobromine, which comprise approximately 2.5% of the drug substances. Although these measures contribute to ensuring the quality control of Veregen, future changes in specification and improvement must be implemented to maintain the consistency of batches for the safety and efficacy of Veregen.
Furthermore, C. sinensis green tea leaves can have different chemical composition based on several factors including the processing techniques, climatic conditions, soil, and genetic strain. Therefore, it is important to evaluate the quality and characteristics of Veregen batches by analyzing the entire metabolite composition of the tea leaves using various instruments and prediction models. Recently, numerous studies have conducted metabolic profiling and fingerprinting of various kinds of green teas using NMR-, GC-MS-, and LC-MS-based techniques (Table 2).
Table 2.
List of marker compounds and detection methods of various kinds of tea
| Common name | Detection methoda | Number of identified compounds | Research group location | Marker compoundsb | Reference |
|---|---|---|---|---|---|
| Japanese green tea | PY-GC-MS | 7 | Japan | Caffeine, phenol, toluene, hexadecanoic acid, indole, hydroquinone, 4-methylphenol | Pongsuwan et al., 2008b |
| GC-TOFMS | 7 | Japan | Sucrose, glucose, quinic acid, fructose, caffeine, malic acid, theanine | Pongsuwan et al., 2007 | |
| UPLC-TOFMS | 3 | Japan | EGCG, ECG, EGC | Pongsuwan et al., 2008a | |
| GC-TOFMS | 10 | Japan | Quinic acid, theanine, sucrose, EGC, caffeine, polyphenol, fructose, phosphoric acid, glucose, disaccharide | Jumtee et al., 2009 | |
| GC-FID | 10 | Japan | Quinic acid, polyphenol, EGCg, sucrose, disaccharide, EC, ECG, phosphoric acid, fructose, EGC | Jumtee et al., 2009 | |
| 1H-NMR | 11 | Japan | Theanine, quinic acid, caffeine, arginine, myo-inositol, chlorogenic acid, 2-O-β-L-arabinopyranosyl-myo-inositol, p-coumaryl quinic acid, cinnamic acid, EGCG, ECG | Tarachiwin et al., 2007 | |
| GC-MS | 71 | Japan | Alanine, oxalic acid, malonic acid, urea, serine, methyl 5-oxo-2-pyrrolidinecarboxylate, ethanolamine, leucine, phosphoric acid, isoleucine, etc. | Sakamoto et al., 2010 | |
| Chinese green tea | 1H-NMR | 8 | United Kingdom | Theanine, gallic acid, caffeine, EGCG, ECG, theogallin, theobromine, 2-O-(β-L-arabinopyranosyl)myo-inositol | Le Gall et al., 2004 |
| HILIC-MS | 5 | New Zealand | Arginine, histidine, aspartic acid, glutamine, and glutamic acid | Fraser et al., 2012 | |
| UPLC-QTOF-MS | 5 | China, United States | ECG, EGCG, theaflavine, leucine, asparagine | Xie et al., 2009 | |
| UPLC-DAD-MS | 3 | US, China | Epigallocatechin, quercetin 3-O-dirhamnosylglucoside, kaempferol 3-O-p-coumaroyldirhamnosylhexoside | Zhao et al., 2011 |
1H-NMR, proton nuclear magnetic resonance; GC-TOF-MS, gas chromatography-time-of-flight-mass spectrometry; FID, flame ionization detector; PY, pyrolysis-gas; UPLC-TOF-MS, ultra-performance liquid chromatography-time-of-flight-mass spectrometry; DAD, diode array detection; Q, quadrupole; HIILC, hydrophilic interaction-liquid chromatography.
ECG: epicatechin gallate; EGCG: epigallocatechin gallate; EC: epicatechin; EGC: epigallocatechin.
Marker compounds of Japanese green tea have been identified using 1H-NMR, GC-MS, GC-TOF-MS, GC-flame ionization detector (FID), pyrolysis-gas (PY)-GC-MS, and UPLC-TOFMS by Japanese research groups. Tarachiwin et al. (2007) applied the 1H-NMR metabolomics approach and PCA to evaluate Japanese green tea quality. Theanine and quinic acid were identified in the low-frequency region at δ 0.5–3.0 ppm; caffeine, arginine, myo-inositol, chlorogenic acid, and quinic acid were observed in the middle-frequency region at δ 3.0–4.5 ppm; and 2-O-β-L-arabinopyranosyl-myo-inositol, p-coumaryl quinic acid, cinnamic acid, EGCG, and ECG were detected in the high-frequency region at δ 5.0–8.0 ppm. When the PCA was performed, it clearly distinguished between the high- and the low-quality Japanese green tea products (Tarachiwin et al., 2007). Therefore, the detected compounds could be considered as suitable markers, which can be associated with the quality evaluation of Japanese green tea with a simple preparation and short analysis time.
Pongsuwan et al. (2007, 2008a, 2008b) used GC-TOFMS, PY-GC-MS, and UPLC-TOFMS to perform metabolite fingerprinting and comprehensive metabolic analysis and predicted Japanese green tea quality by constructing quality prediction models. When the PY-GC-MS was performed, the seven detected major compounds were identified as caffeine, phenol, toluene, hexadecanoic acid, indole, hydroquinone, 4-methylphenol (Pongsuwan et al., 2008b). When GC-TOFMS was used, seven compounds, which were sucrose, glucose, quinic acid, fructose, caffeine, malic acid, and theanine, were found to be significant in creating a quality prediction model (Pongsuwan et al., 2007). Furthermore, UPLC-TOFMS was used to identify a group of epicatechins (EGCG, ECG, and EGC), which were found to be the most influential variables for quality prediction (Pongsuwan et al., 2008a). In addition, Jumtee et al. (2009) compared the metabolic fingerprinting of Japanese green tea using GC-TOFMS and GC-FID for its quality ranking prediction. Quinic acid, sucrose, disaccharide, phosphoric acid, fructose, and EGC were commonly found to be the important compounds that contributed to the quality predictive model. In addition to the same compounds contributing to both models, theanine, caffeine, polyphenol, and glucose were discovered as key compounds that influenced the GC-TOFMS predictive model construction, while polyphenol, EGCG, EC, and ECG contributed to the GC-FID predictive model construction (Jumtee et al., 2009).
In addition to Japanese green tea and Chinese green tea were also analyzed using metabolite profiling and multivariate statistics for quality assessment using 1H-NMR, UPLC-QTOFMS, and UPLC-DAD-MS, HILIC, and GC × GC-TOF (Le Gall et al., 2004; Xie et al., 2009; Zhao et al., 2011; Fraser et al., 2012). When high quality Chinese green tea (Longing tea) was investigated using 1H-NMR, eight compounds, which were theanine, gallic acid, caffeine, EGCG, ECG, theogallin, theobromine, and 2-O-(β-L-arabinopyranosyl)-myo-inositol, showed high levels and were identified as suitable markers for the authentication of the quality of the tea (Le Gall et al., 2004). The non-targeted analysis of Chinese green using HILIC-MS in both the positive and negative ionization modes was developed and optimized (Fraser et al., 2012). Its metabolic fingerprinting showed significant differences between the different tea types. The Chinese green tea contained five compounds; arginine, histidine, aspartic acid, glutamine, and glutamic acid, with a significantly higher content than was found in other types of tea.
Fraser et al. (2012) also performed a PCA to distinguish the different types of tea and determine the compounds that contributed most to the differentiation of the tea samples. There was a clear distinction between the green and other types of tea when the PCA analysis of the negative ESI data was performed. In addition, when the PC1 loadings from the PCA analysis were examined, sucrose was identified as an important compound that facilitated the differentiation of the tea types. Xie et al. (2009) used UPLC-QTOFMS to analyze and compare Chinese green tea to other types of tea (pu-erh and black teas) using an orthogonal PLS-DA (OPLS-DA). The characteristic constituents of the teas were identified as ECG, EGCG, theaflavine, leucine, and asparagine in the Chinese green tea. When the OPLS-DA was performed, the Chinese green tea and those different tea types showed a clear distinction, which indicates that the differences in tea processing resulted in differences in the characteristic markers. Furthermore, a UPLC-DAD-MS was performed to identify and quantitate Chinese green tea and other kinds of tea (pu-erh and white teas) (Zhao et al., 2011). The identified components were ECG, quercetin 3-O-dirhamnosylglucoside, kaempferol, and 3-O-p-coumaroyldirhamnosylhexoside in Chinese green tea; 3-caffeoylquinic acid, 5-caffeoylquinic acid, 4-caffeoylquinic acid, catechin, quercetin 3-O-rhamnosylgalactoside, rutin, quercetin 3-O-glucoside, kaempferol-3-O-rutinoside, and quercetin in pu-erh tea; and gallic acid methyl ester, 1,6-digalloylglucose, 1,2,6-trigalloylglucose, caffeine, digallocatechin-catechin, quercetin 3-O-glucosylrutinoside, qu-ercetin 3-O-rhamnoside, myricetin, kaempferol, kaempferol 3-O-p-coumaroylglucoside, kaempferol 3-O-6″-p-coumaroylglucoside, kaempferol 3-O-di-p-coumaroylhexoside, and kaempferol 3-O-2,6″-di-p-coumaroylhexoside in white tea. The PCA score plot was used, and it clearly separated between the tea types.
The results of the metabolic fingerprinting and profiling of various kinds of tea provide evidence to support the need for improved quality control of raw materials for the production of Veregen. Furthermore, the standardization of the marker compounds, which can be achieved by regulating several factors including the processing techniques, environmental conditions, and cultivars, will be required to ensure better quality control of Veregen and other NP-derived medicines.
CONCLUSIONS
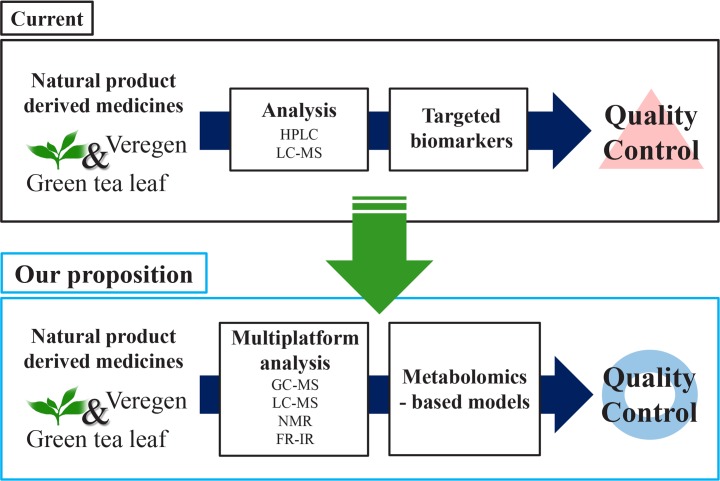
Metabolomics has been developing in response to the need for standardization and quality control of NP-derived medicines as well as the characterization and identification of their underlying molecular mechanism. Moreover, the development of metabolomics techniques including multivariate statistical analyses for evaluating profiling data have has provided further promoted the value of metabolomics. NP-derived medicines usually contain multiple compounds that need to be identified and quantified using various metabolomics techniques. Currently, the quality control of Veregen is performed using HPLC with identification and quantification of eight catechins. However, the active ingredient in this case, which is green tea, contains multiple compounds depending on the origin and analytical techniques. Therefore, it needs to be analyzed using diverse developed metabolomics techniques with multivariate statistical analyses to ensure its batch-to-batch consistency for quality control. We summarized our proposition as Fig. 1. The metabolomics technique will provide a major framework for the quality control of NP-derived medicines.
Fig. 1.
Proposition of quality control method of natural product derived medicines by metabolomics.
Acknowledgments
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIP) (NRF-2015R1A5A1008958), and by the Chung-Ang University Graduate Research Scholarship in 2015.
REFERENCES
- Abdi H. Partial least square regression (PLS regression) In: Salkind N, editor. Encyclopedia for Research Methods for the Social Sciences. Sage Publications, Inc; USA: 2003. pp. 792–795. [Google Scholar]
- Avula B, Joshi VC, Weerasooriya A, Khan IA. Liquid chromatography for separation and quantitative determination of adrenergic amines and flavonoids from Poncirus trifoliatus Raf. fruits at different stages of growth. Chromatographia. 2005;62:379–383. doi: 10.1365/s10337-005-0639-z. [DOI] [Google Scholar]
- Babu PV, Sabitha KE, Shyamaladevi CS. Therapeutic effect of green tea extract on oxidative stress in aorta and heart of streptozotocin diabetic rats. Chem Biol Interact. 2006;162:114–120. doi: 10.1016/j.cbi.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Bochořáková H, Paulova H, Slanina J, Musil P, Táborská E. Main flavonoids in the root of Scutellaria baicalensis cultivated in Europe and their comparative antiradical properties. Phytother Res. 2003;17:640–644. doi: 10.1002/ptr.1216. [DOI] [PubMed] [Google Scholar]
- Chen ST, Dou J, Temple R, Agarwal R, Wu KM, Walker S. New therapies from old medicines. Nat Biotechnol. 2008;26:1077–1083. doi: 10.1038/nbt1008-1077. [DOI] [PubMed] [Google Scholar]
- Choi YH, Choi HK, Hazekamp A, Bermejo P, Schilder Y, Erkelens C, Verpoorte R. Quantitative analysis of bilobalide and ginkgolides from Ginkgo biloba leaves and Ginkgo products using 1H-NMR. Chem Pharm Bull. 2003;51:158–161. doi: 10.1248/cpb.51.158. [DOI] [PubMed] [Google Scholar]
- Cooper R, Morré DJ, Morré DM. Medicinal benefits of green tea: part II. Review of anticancer properties. J Altern Complement Med. 2005;11:639–652. doi: 10.1089/acm.2005.11.639. [DOI] [PubMed] [Google Scholar]
- Court WE. In: Ginseng: The Genus Panax. Hardman R, editor. Taylor & Francise-Library; The Netherlands: 2000. pp. 205–220. [Google Scholar]
- Cui L, Su XZ. Discovery, mechanisms of action and combination therapy of artemisinin. Expert Rev Anti Infect Ther. 2009;7:999–1013. doi: 10.1586/eri.09.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Wu E, Chen J, Nguyen HT, Do TH, Park KL, Ba KH, Kim HY, Kang JS. Quality evaluation of moutan cortex radicis using multiple component analysis by high performance liquid chromatography. Bull Korean Chem Soc. 2009;30:2240–2244. doi: 10.5012/bkcs.2009.30.10.2240. [DOI] [Google Scholar]
- Dunn WB, Ellis DI. Metabolomics: current analytical platforms and methodologies. Trends Analyt Chem. 2005;24:285–294. doi: 10.1016/j.trac.2004.11.021. [DOI] [Google Scholar]
- Eriksson L, Johansson E, Kettaneh-Wold N, Trygg J, Wikstrom C, Wold S. Multi and Megavariate Data Analysis: Part I: Basic Principles and Applications. Umetrics AB; Sweden: 2006. pp. 39–101. [Google Scholar]
- Farag MA, Porzel A, Wessjohann LA. Comparative metabolite profiling and fingerprinting of medicinal licorice roots using a multiplex approach of GC-MS, LC-MS and 1D NMR techniques. Phytochemistry. 2012;76:60–72. doi: 10.1016/j.phytochem.2011.12.010. [DOI] [PubMed] [Google Scholar]
- Farag MA, Wessjohann LA. Metabolome classification of commercial Hypericum perforatum (St. John’s Wort) preparations via UPLC-qTOF-MS and chemometrics. Planta Med. 2012;78:488–496. doi: 10.1055/s-0031-1298170. [DOI] [PubMed] [Google Scholar]
- Fraser K, Harrison SJ, Lane GA, Otter DE, Hemar Y, Quek SY, Rasmussen S. Non-targeted analysis of tea by hydrophilic interaction liquid chromatography and high resolution mass spectrometry. Food Chem. 2012;134:1616–1623. doi: 10.1016/j.foodchem.2012.03.045. [DOI] [PubMed] [Google Scholar]
- Fu HY, Huang DC, Yang TM, She YB, Zhang H. Rapid recognition of Chinese herbal pieces of Areca catechu by different concocted processes using Fourier transform mid-infrared and near-infrared spectroscopy combined with partial least-squares discriminant analysis. Chinese Chem Lett. 2013;24:639–642. doi: 10.1016/j.cclet.2013.04.019. [DOI] [Google Scholar]
- Ghiulai VM, Socaciu C, Jianu I, Ranga F, Fetea F. Identification and quantitative evaluation of amygdalin from apricot, plum and peach oils and kernels. Bulletin UASVM Agriculture. 2006;62:246–253. [Google Scholar]
- Gromski PS, Muhamadali H, Ellis DI, Xu Y, Correa E, Turner ML, Goodacre R. A tutorial review: Metabolomics and partial least squares-discriminant analysis--a marriage of convenience or a shotgun wedding. Anal. Chim. Acta. 2015;879:10–23. doi: 10.1016/j.aca.2015.02.012. [DOI] [PubMed] [Google Scholar]
- Gurley BJ, Wang P, Gardner SF. Ephedrine-type alkaloid content of nutritional supplements containing Ephedra sinica (Ma-huang) as determined by high performance liquid chromatography. J Pharm Sci. 1998;87:1547–1553. doi: 10.1021/js9801844. [DOI] [PubMed] [Google Scholar]
- Hasada K, Yoshida T, Yamazaki T, Sugimoto N, Nishimura T, Nagatsu A, Mizukami H. Application of 1H-NMR spectroscopy to validation of berberine alkaloid reagents and to chemical evaluation of Coptidis rhizoma. J Nat Med. 2011;65:262–267. doi: 10.1007/s11418-010-0490-x. [DOI] [PubMed] [Google Scholar]
- Jacquemond-Collet I, Bessière JM, Hannedouche S, Bertrand C, Fourasté I, Moulis C. Identification of the alkaloids of Galipea officinalis by gas chromatography-mass spectrometry. Phytochem Anal. 2001;12:312–329. doi: 10.1002/pca.594. [DOI] [PubMed] [Google Scholar]
- Jantarat C, Sirathanarun P, Songsrm W, Srinornate W, Daengprom S. A simple and rapid HPLC technique for determination of arecoline in areca nut (Areca catechu L.) extract. Walailak J Sci & Tech. 2013;10:57–66. [Google Scholar]
- Jensen AG, Ndjoko K, Wolfender JL, Hostettmann K, Camponovo F, Soldati F. Liquid chromatography-atmospheric pressure chemical ionisation/mass spectrometry: a rapid and selective method for the quantitative determination of ginkgolides and bilobalide in ginkgo leaf extracts and phytopharmaceuticals. Phytochem Anal. 2002;13:31–38. doi: 10.1002/pca.614. [DOI] [PubMed] [Google Scholar]
- Jumtee K, Bamba T, Fukusaki E. Fast GC-FID based metabolic fingerprinting of Japanese green tea leaf for its quality ranking prediction. J Sep Sci. 2009;32:2296–2304. doi: 10.1002/jssc.200900096. [DOI] [PubMed] [Google Scholar]
- Kang J, Choi MY, Kang S, Kwon HN, Wen H, Lee CH, Park S, Wiklund S, Kim HJ, Kwon SW, Park S. Application of a 1H nuclear magnetic resonance (NMR) metabolomics approach combined with orthogonal projections to latent structure-discriminant analysis as an efficient tool for discriminating between Korean and Chinese herbal medicines. J Agric Food Chem. 2008a;56:11589–11595. doi: 10.1021/jf802088a. [DOI] [PubMed] [Google Scholar]
- Kang J, Lee S, Kang S, Kwon HN, Park JH, Kwon SW, Park S. NMR-based metabolomics approach for the differentiation of ginseng (Panax ginseng) roots from different origins. Arch Pharm Res. 2008b;31:330–336. doi: 10.1007/s12272-001-1160-2. [DOI] [PubMed] [Google Scholar]
- Kataoka M, Tokuyama E, Miyanaga Y, Uchida T. The taste sensory evaluation of medicinal plants and Chinese medicines. Int J Pharm. 2008;351:36–44. doi: 10.1016/j.ijpharm.2007.09.017. [DOI] [PubMed] [Google Scholar]
- Kim HK, Choi YH, Erkelens C, Lefeber AW, Verpoorte R. Metabolic fingerprinting of Ephedra species using 1H-NMR spectroscopy and principal component analysis. Chem Pharm Bull. 2005;53:105–109. doi: 10.1248/cpb.53.105. [DOI] [PubMed] [Google Scholar]
- Kim HK, Choi YH, Verpoorte R. NMR-based metabolomic analysis of plants. Nat Protoc. 2010;5:536–549. doi: 10.1038/nprot.2009.237. [DOI] [PubMed] [Google Scholar]
- Kim SH, Shin YS, Choi HK. NanoESI-MS-based lipidomics to discriminate between cultivars, cultivation ages, and parts of Panax ginseng. Anal Bioanal Chem. 2016;408:2109–2121. doi: 10.1007/s00216-016-9314-5. [DOI] [PubMed] [Google Scholar]
- Kim TH, Jang S, Lee AR, Lee AY, Choi G, Kim HK. Optimization of extraction conditions for swertiamarin in Swertia japonica makino. Korea J Herbol. 2014;29:13–18. [Google Scholar]
- Kunle OF, Egharevba HO, Ahmadu PO. Standardization of herbal medicines - A review. Int J Biodivers Conserv. 2012;4:101–112. doi: 10.5897/IJBC11.163. [DOI] [Google Scholar]
- Kwon YK, Ahn MS, Park JS, Liu JR, In DS, Min BW, Kim SW. Discrimination of cultivation ages and cultivars of ginseng leaves using Fourier transform infrared spectroscopy combined with multivariate analysis. J Ginseng Res. 2014;38:52–58. doi: 10.1016/j.jgr.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis-Piwowar KR, Huo C, Chen DI, Milacic V, Shi G, Chan TH, Dou QP. A novel prodrug of the green tea polyphenol (−)-epigallocatechin-3-gallate as a potential anticancer agent. Cancer Res. 2007;67:4303–4310. doi: 10.1158/0008-5472.CAN-06-4699. [DOI] [PubMed] [Google Scholar]
- Le Gall G, Colquhoun IJ, Defernez M. Metabolite profiling using (1)H NMR spectroscopy for quality assessment of green tea, Camellia sinensis (L.). J Agric Food Chem. 2004;52:692–700. doi: 10.1021/jf034828r. [DOI] [PubMed] [Google Scholar]
- Lee J, Jung Y, Shin JH, Kim HK, Moon BC, Ryu DH, Hwang GS. Secondary metabolite profiling of Curcuma species grown at different locations using GC/TOF and UPLC/Q-TOF MS. Molecules. 2014;19:9535–9551. doi: 10.3390/molecules19079535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hu Z, He L. An approach to develop binary chromatographic fingerprints of the total alkaloids from Caulophyllum robustum by high performance liquid chromatography/diode array detector and gas chromatography/mass spectrometry. J Pharm Biomed Anal. 2007;43:1667–1672. doi: 10.1016/j.jpba.2006.12.028. [DOI] [PubMed] [Google Scholar]
- Lin JK, Lin-Shiau SY. Mechanisms of hypolipidemic and anti-obesity effects of tea and tea polyphenols. Mol Nutr Food Res. 2006;50:211–217. doi: 10.1002/mnfr.200500138. [DOI] [PubMed] [Google Scholar]
- Lin YL, Cheng CY, Lin YP, Lau YW, Juan IM, Lin JK. Hypolipidemic effect of green tea leaves through induction of antioxidant and phase II enzymes including superoxide dismutase, catalase, and glutathione S-transferase in rats. J Agric Food Chem. 1998;46:1893–1899. doi: 10.1021/jf970963q. [DOI] [Google Scholar]
- Lu J, Xiang B, Liu H, Xiang S, Xie S, Deng H. Application of two-dimensional near-infrared correlation spectroscopy to the discrimination of Chinese herbal medicine of different geographic regions. Spectrochim Acta A Mol Biomol Spectrosc. 2008;69:580–586. doi: 10.1016/j.saa.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Lv WF, Ding MY, Zheng R. Isolation and quantitation of amygdalin in Apricot-kernel and Prunus Tomentosa Thunb. by HPLC with solid-phase extraction. J Chromatogr Sci. 2005;43:383–387. doi: 10.1093/chromsci/43.7.383. [DOI] [PubMed] [Google Scholar]
- Mengs U, Pohl T, Mitchell T. Legalon® SIL: the antidote of choice in patients with acute hepatotoxicity from amatoxin poisoning. Curr Pharm Biotechnol. 2012;13:1964–1970. doi: 10.2174/138920112802273353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIT Technology Review (2005) 10 Breakthrough Technology. 2015 Available from: http://www2.technologyreview.com/news/404001/10-emerging-technologies.
- Montoro P, Maldini M, Russo M, Postorino S, Piacente S, Pizza C. Metabolic profiling of roots of liquorice (Glycyrrhiza glabra) from different geographical areas by ESI/MS/MS and determination of major metabolites by LC-ESI/MS and LC-ESI/MS/MS. J Pharm Biomed Anal. 2011;54:535–544. doi: 10.1016/j.jpba.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Nugroho A, Park MG, Jin SE, Choi JS, Park HJ. Quantitative analsysis of flavanone glycosides and peroxynitrite scavenging effect of the five oriental medicinal drugs (Aurantii nobilis Pericarpium, Citrii unshiu Pericarpium, Citrii unshiu Semen, Aurantii Fructus, Poncirii Fructus). Korean J Pharmacogn. 2009;40:370–375. [Google Scholar]
- Oh MJ, Lee KS, Son HY, Kim SY. Antioxidative components of Pueraria root. Korean J Food Sci Technol. 1990;22:793–798. [Google Scholar]
- Okada T, Nakamura Y, Kanaya S, Takano A, Malla KJ, Nakane T, Kitayama M, Sekita S. Metabolome analysis of Ephedra plants with different contents of ephedrine alkaloids by using UPLC-Q-TOF-MS. Planta Med. 2009;75:1356–1362. doi: 10.1055/s-0029-1185577. [DOI] [PubMed] [Google Scholar]
- Perez-Enciso M, Tenenhaus M. Prediction of clinical outcome with microarray data: a partial least squares discriminant analysis (PLS-DA) approach. Hum Genet. 2003;112:581–592. doi: 10.1007/s00439-003-0921-9. [DOI] [PubMed] [Google Scholar]
- Pongsuwan W, Bamba T, Harada K, Yonetani T, Kobayashi A, Fukusaki E. High-throughput technique for comprehensive analysis of Japanese green tea quality assessment using ultra-performance liquid chromatography with time-of-flight mass spectrometry (UPLC/TOF MS) J Agric Food Chem. 2008a;56:10705–10708. doi: 10.1021/jf8018003. [DOI] [PubMed] [Google Scholar]
- Pongsuwan W, Bamba T, Yonetani T, Kobayashi A, Fukusaki E. Quality prediction of Japanese green tea using pyrolyzer coupled GC/MS based metabolic fingerprinting. J Agric Food Chem. 2008b;56:744–750. doi: 10.1021/jf072791v. [DOI] [PubMed] [Google Scholar]
- Pongsuwan W, Fukusaki E, Bamba T, Yonetani T, Yamahara T, Kobayashi A. Prediction of Japanese green tea ranking by gas chromatography/mass spectrometry-based hydrophilic metabolite fingerprinting. J Agric Food Chem. 2007;55:231–236. doi: 10.1021/jf062330u. [DOI] [PubMed] [Google Scholar]
- Sakamoto Y, Nakagawa K, Kawana S, Lingga N, Lai Chin HL, Miyagawa H. GC/MS Technical Report (No. 1) 2010 Available from: https://www.shimadzu.it/sites/default/files/Profiling_of_Japanese_Green_Tea_Metabolites_by_GC-MS.pdf/.
- Samukawa K, Yamashita H, Matsuda H, Kubo M. Simultaneous analysis of ginsenosides of various ginseng radix by HPLC. Yakugaku Zasshi. 1995;43:137–141. doi: 10.1248/yakushi1947.115.3_241. [DOI] [PubMed] [Google Scholar]
- Scannell JW, Blanckley A, Boldon H, Warrington B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat Rev Drug Discov. 2012;11:191–200. doi: 10.1038/nrd3681. [DOI] [PubMed] [Google Scholar]
- Selvam ABD. Inventory of vegetable crude drug samples housed in botanical survey of India, Howrah. Pharmacogn Rev. 2008;2:61–94. [Google Scholar]
- Shin YG, Cho KH, Kim JM, Park MK, Park JH. Determination of betaine in Lycium chinense fruits by liquid chromatography-electrospray ionization mass spectrometry. J. Chromatogr. A. 1999;857:331–335. doi: 10.1016/S0021-9673(99)00720-7. [DOI] [PubMed] [Google Scholar]
- Song SY, Lee YK, Kim IJ. Sugar and acid content of Citrus prediction modeling using FT-IR fingerprinting in combination with multivariate statistical analysis. Food Chem. 2016;190:1027–1032. doi: 10.1016/j.foodchem.2015.06.068. [DOI] [PubMed] [Google Scholar]
- Suzuki H. Standard compounds for quantitative determination of principles of crude drugs-1-Paeoniflorin, a major principle of peony root. Jap J Pharmacogn. 1984;38:144–148. [Google Scholar]
- Tarachiwin L, Ute K, Kobayashi A, Fukusaki E. 1H NMR based metabolic profiling in the evaluation of Japanese green tea quality. J Agric Food Chem. 2007;55:9330–9336. doi: 10.1021/jf071956x. [DOI] [PubMed] [Google Scholar]
- Tilton R, Paiva AA, Guan JQ, Marathe R, Jiang Z, van Eyndhoven W, Bjoraker J, Prusoff Z, Wang H, Liu SH, Cheng YC. A comprehensive platform for quality control of botanical drugs (PhytomicsQC): a case study of Huangqin Tang (HQT) and PHY906. Chin Med. 2010;5:30. doi: 10.1186/1749-8546-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai TR, Tseng TY, Chen CF, Tsai TH. Identification and determination of geniposide contained in Gardenia jasminoides and in two preparations of mixed traditional Chinese medicines. J. Chromatogr. A. 2002;961:83–88. doi: 10.1016/S0021-9673(02)00365-5. [DOI] [PubMed] [Google Scholar]
- U.S. Food Drug Administration Chemistry Review Data Sheet. 2006a Available from: http://www.accessdata.fda.gov/drugsatfda_docs/nda/2006/021902s000_chemr.pdf/.
- U.S. Food and Drug Administration Drugs@FDA. 2006b Available from: http://www.accessdata.fda.gov/scripts/cder/drugsatfda/index.cfm?fuseaction=Search.DrugDetails/.
- Van der Kooy F, Verpoorte R, Meyer JM. Metabolomic quality control of claimed anti-malarial Artemisia afra herbal remedy and A. afra and A. annua plant extracts. S Afr J Bot. 2008;74:186–189. doi: 10.1016/j.sajb.2007.10.004. [DOI] [Google Scholar]
- Weckwerth W, Morgenthal K. Metabolomics: from pattern recognition to biological interpretation. Drug Discov. Today. 2005;10:1551–1558. doi: 10.1016/S1359-6446(05)03609-3. [DOI] [PubMed] [Google Scholar]
- Wichitnithad W, Jongaroonngamsang N, Pummangura S, Rojsitthisak P. A simple isocratic HPLC method for the simultaneous determination of curcuminoids in commercial turmeric extracts. Phytochem Anal. 2009;20:314–319. doi: 10.1002/pca.1129. [DOI] [PubMed] [Google Scholar]
- Xie G, Ye M, Wang Y, Ni Y, Su M, Huang H, Qiu M, Zhao A, Zheng X, Chen T, Jia W. Characterization of pu-erh tea using chemical and metabolic profiling approaches. J Agric Food Chem. 2009;57:3046–3054. doi: 10.1021/jf804000y. [DOI] [PubMed] [Google Scholar]
- Xu L, Li J, Yan C, Shan A. Study on HPLC method to determine contents of Schisandrin A andSchisandrin B in Schisandra chinensis extraction. J Northeast Agric Univ. 2007;14:323–326. [Google Scholar]
- Yang SY, Kim HK, Lefeber AW, Erkelens C, Angelova N, Choi YH, Verpoorte R. Application of two-dimensional nuclear magnetic resonance spectroscopy to quality control of ginseng commercial products. Planta Med. 2006;72:364–369. doi: 10.1055/s-2005-916240. [DOI] [PubMed] [Google Scholar]
- Yang X, Yang L, Xiong A, Li D, Wang Z. Authentication of Senecio scandens and S. vulgaris based on the comprehensive secondary metabolic patterns gained by UPLC-DAD/ESI-MS. J Pharm Biomed Anal. 2011;56:165–172. doi: 10.1016/j.jpba.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Yen GC, Chen HY. Antioxidant activity of various tea extracts in relation to their antimutagenicity. J Agric Food Chem. 1995;43:27–32. doi: 10.1021/jf00049a007. [DOI] [Google Scholar]
- Zhang A, Sun H, Wang P, Han Y, Wang X. Modern analytical techniques in metabolomics analysis. Analyst. 2012;137:293–300. doi: 10.1039/C1AN15605E. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Ye M. Chemical analysis of the Chinese herbal medicine Gan-Cao (licorice) J. Chromatogr. A. 2009;1216:1954–1969. doi: 10.1016/j.chroma.2008.07.072. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Chen P, Lin L, Harnly JM, Yu LL, Li Z. Tentative identification, quantitation, and principal component analysis of green pu-erh, green, and white teas using UPLC/DAD/MS. Food Chem. 2011;126:1269–1277. doi: 10.1016/j.foodchem.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]